
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 April 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1126940
This article is part of the Research TopicInsights Into the Role of Microorganisms on Food Quality and Food SafetyView all 17 articles
Fresh produce contaminated with Listeria monocytogenes has caused major listeriosis outbreaks in the last decades. Our knowledge about components of the listerial biofilms formed on fresh produce and their roles in causing foodborne illness remains incomplete. Here, we investigated, for the first time, the role of the listerial Pss exopolysaccharide (EPS) in plant surface colonization and stress tolerance. Pss is the main component of L. monocytogenes biofilms synthesized at elevated levels of the second messenger c-di-GMP. We developed a new biofilm model, whereby L. monocytogenes EGD-e and its derivatives are grown in the liquid minimal medium in the presence of pieces of wood or fresh produce. After 48-h incubation, the numbers of colony forming units of the Pss-synthesizing strain on pieces of wood, cantaloupe, celery and mixed salads were 2−12-fold higher, compared to the wild-type strain. Colonization of manmade materials, metals and plastics, was largely unaffected by the presence of Pss. The biofilms formed by the EPS-synthesizing strain on cantaloupe rind were 6−16-fold more tolerant of desiccation, which resembles conditions of whole cantaloupe storage and transportation. Further, listeria in the EPS-biofilms survived exposure to low pH, a condition encountered by bacteria on the contaminated produce during passage through the stomach, by 11−116-fold better than the wild-type strain. We surmise that L. monocytogenes strains synthesizing Pss EPS have an enormous, 102−104-fold, advantage over the non-synthesizing strains in colonizing fresh produce, surviving during storage and reaching small intestines of consumers where they may cause disease. The magnitude of the EPS effect calls for better understanding of factors inducing Pss synthesis and suggests that prevention of listerial EPS-biofilms may significantly enhance fresh produce safety.
Listeria monocytogenes is one of the most notorious foodborne pathogens. Listeriosis, a severe systemic illness caused by this bacterium, affects primarily the elderly, pregnant women, newborns, and the immune compromised individuals. While the number of listeriosis cases is relatively modest, e.g., approximately 1,600 cases per annum in the USA, mortality rates are extremely high, 15−20% (Centers for Disease Control and Prevention, 2022). This justifies the “zero L. monocytogenes tolerance” policy of the US Department of Agriculture (Archer, 2018), which means that large shipments of ready-to-eat food products that are contaminated or suspected to be contaminated with L. monocytogenes are often recalled. The high socioeconomic costs associated with contaminated food make L. monocytogenes the third “most expensive” pathogen in the USA among all bacterial, viral, and eukaryotic foodborne pathogens (Hoffmann and Ahn, 2021).
While traditional sources of listerial contamination have included ready-to-eat meat, poultry, seafood, and dairy products, the number of listeriosis outbreaks associated with contaminated fresh produce has been growing (Zhu et al., 2017). Major outbreaks in the recent years have been caused by contaminated cantaloupes (also known as rock melons), celery, packaged salads, bean sprouts, caramelized apples, and frozen vegetables (Centers for Disease Control and Prevention, 2022; NSW Government). Among these outbreaks, the 2011 cantaloupe-associated outbreak stands out. It affected people in 28 states resulting in 33 fatalities and one miscarriage, making it one of the highest mortality food-poisoning incidents in the modern US history. That outbreak involved whole cantaloupes contaminated with L. monocytogenes at a Colorado cantaloupe processing facility (McCollum et al., 2013). Currently, we poorly understand what factors may have contributed to L. monocytogenes colonization of the whole cantaloupes, and helped bacteria survive on the inhospitable surface of cantaloupe rind during extended periods of storage and transportation throughout the USA. This situation applies to other kinds of fresh produce, which impedes our ability to prevent fresh produce-associated listeriosis outbreaks (Marik et al., 2020).
On the surfaces of various materials bacteria grow in biofilms, where aggregated cells are surrounded by the secreted extracellular matrix that enhances their survival (Costerton et al., 1999; Flemming and Wingender, 2010; Flemming et al., 2016). In the environment, listeria, which is frequently associated with the decaying plant matter (Welshimer and Donker-Voet, 1971; Fenlon, 1986; Ivanek et al., 2007; Freitag, 2009), likely grows in biofilms as well, yet we do not know composition of listerial biofilms in the environment. The studies of plant-associated biofilms in various other bacteria revealed that the extracellular biofilm matrix comprises exopolysaccharide (EPS), extracellular DNA (eDNA), fimbriae/pili and amyloid fibers (Flemming and Wingender, 2010; Limoli et al., 2015; Karygianni et al., 2020). Fimbriae/pili and amyloid fibers were never observed in L. monocytogenes. eDNA is an abundant component of listerial biofilms on various materials (Harmsen et al., 2010; Zetzmann et al., 2015). The role of EPS as component of plant-associated listerial biofilms has not been studied, until this work. The presence of EPS in biofilms formed by various listerial strains was inferred via indirect methods (Borucki et al., 2003; Hefford et al., 2005; Combrouse et al., 2013; Tiensuu et al., 2013), yet until recently, the ability of L. monocytogenes to synthesize EPS has remained controversial (Renier et al., 2011).
Our earlier work (Chen et al., 2014) uncovered the ability of L. monocytogenes to synthesize EPS, designated Pss, when cellular levels of the second messenger, cyclic dimeric GMP (c-di-GMP), are elevated. The Pss EPS is a unique polymer composed of the repeating trisaccharide unit, {4)-β-ManpNAc-(1−4)-[α-Galp-(1−6)]-β-ManpNAc-(1-}, and is the only cell-attached, insoluble, EPS synthesized by listeria (Köseoğlu et al., 2015). In liquid media, Pss causes cell aggregation, clumping but does not promote attachment to glass or polystyrene materials (Chen et al., 2014). The second messenger c-di-GMP, that activates Pss synthesis, controls EPS synthesis and production of other biofilm components not only in listeria, but in a variety of bacteria (Römling et al., 2013). At present, environmental factors that turn on c-di-GMP synthesis in L. monocytogenes are unknown, therefore, Pss synthesis is induced by genetic means, e.g., by expressing a heterologous c-di-GMP synthase, diguanylate cyclase (DGC), or by inactivating c-di-GMP phosphodiesterases (PDEs) that hydrolyze c-di-GMP (Chen et al., 2014). The rise in cellular c-di-GMP levels is conveyed to the Pss biosynthetic apparatus by the c-di-GMP-effector protein, PssE, whose gene is encoded within the pssA-E biosynthesis operon (Chen et al., 2014).
To investigate the role of Pss in L. monocytogenes colonization of plant surfaces, we developed a listeria-plant biofilm model that involves incubation of the autoclaved wood coupons (disks), as substitutes for partially degraded wood present in the environment, in the minimal medium inoculated with listeria. We also used pieces of fresh produce in place of wood coupons. The plant material was incubated in liquid medium to allow not only bacterial attachment but also biofilm formation. We used the EGD-e wild-type strain and its derivatives that express varying EPS levels. Strain ΔpdeB/C/D constitutively expresses EPS because it lacks c-di-GMP-specific PDEs, PdeB, PdeC and PdeD (Chen et al., 2014). Strain ΔpdeB/C/DΔpssC contains elevated c-di-GMP levels but carries a mutation in the gene encoding the key Pss biosynthetic enzyme, glycosyltransferase PssC, which impairs Pss synthesis (Köseoğlu et al., 2015). Yet another strain, engineered in this work, lacks all DGCs and PDEs, ΔpdeB/C/D ΔdgcA/B/C, is devoid of c-di-GMP and therefore, impaired in Pss synthesis.
We found that the Pss EPS promotes L. monocytogenes colonization of wood, cantaloupe rind and pulp, celery, and lettuce, but does not affect colonization of abiotic, manmade materials. We also found that listeria growing in the EPS-biofilms on plant surfaces are much more tolerant of desiccation and acid stress. The enhanced colonization of plant matter, increased tolerance of desiccation and acid stress (conditions that mimic, respectively, fresh produce storage and exposure to stomach acid following produce consumption), may give L. monocytogenes EPS-biofilms enormous advantage in causing foodborne illness. Results of this work imply that L. monocytogenes EPS-biofilms may present a serious problem for fresh produce industry.
To study listerial biofilms on plant surfaces, we departed from the commonly used models. First, instead of growing L. monocytogenes in rich media that contains peptides, amino acids, lipids, and nucleotides or nucleic acids, we switched to minimal medium containing glucose as carbon source, because such medium better approximates conditions under which listeria interact with plant matter in nature and in fresh produce processing facilities. Second, we incubated autoclaved wood coupons (as substrates mimicking partially degraded wood in the environment) and sterilized pieces of fresh produce in the liquid medium inoculated with L. monocytogenes for extended period, 48 h, to allow biofilm formation.
Following incubation, a significant fraction of biomass of the Pss EPS-synthesizing ΔpdeB/C/D strain was attached to the wood coupons (Figures 1A, B). This contrasted poor colonization of surfaces of the manmade materials, i.e., stainless steel, aluminum or acrylic (Figures 1A, B). This finding is consistent with our earlier observations that listerial EPS-aggregates formed in liquid medium poorly attach to polystyrene and glass surfaces (Chen et al., 2014). To test if surface roughness, as opposed to the nature of the material, played a major role in attachment, we scratched the surfaces of manmade materials thus bringing surfaces roughness closer to the roughness of the wood coupons. This did not improve bacterial attachment (not shown), suggesting that EPS-synthesizing listeria strongly prefer wood over manmade materials.
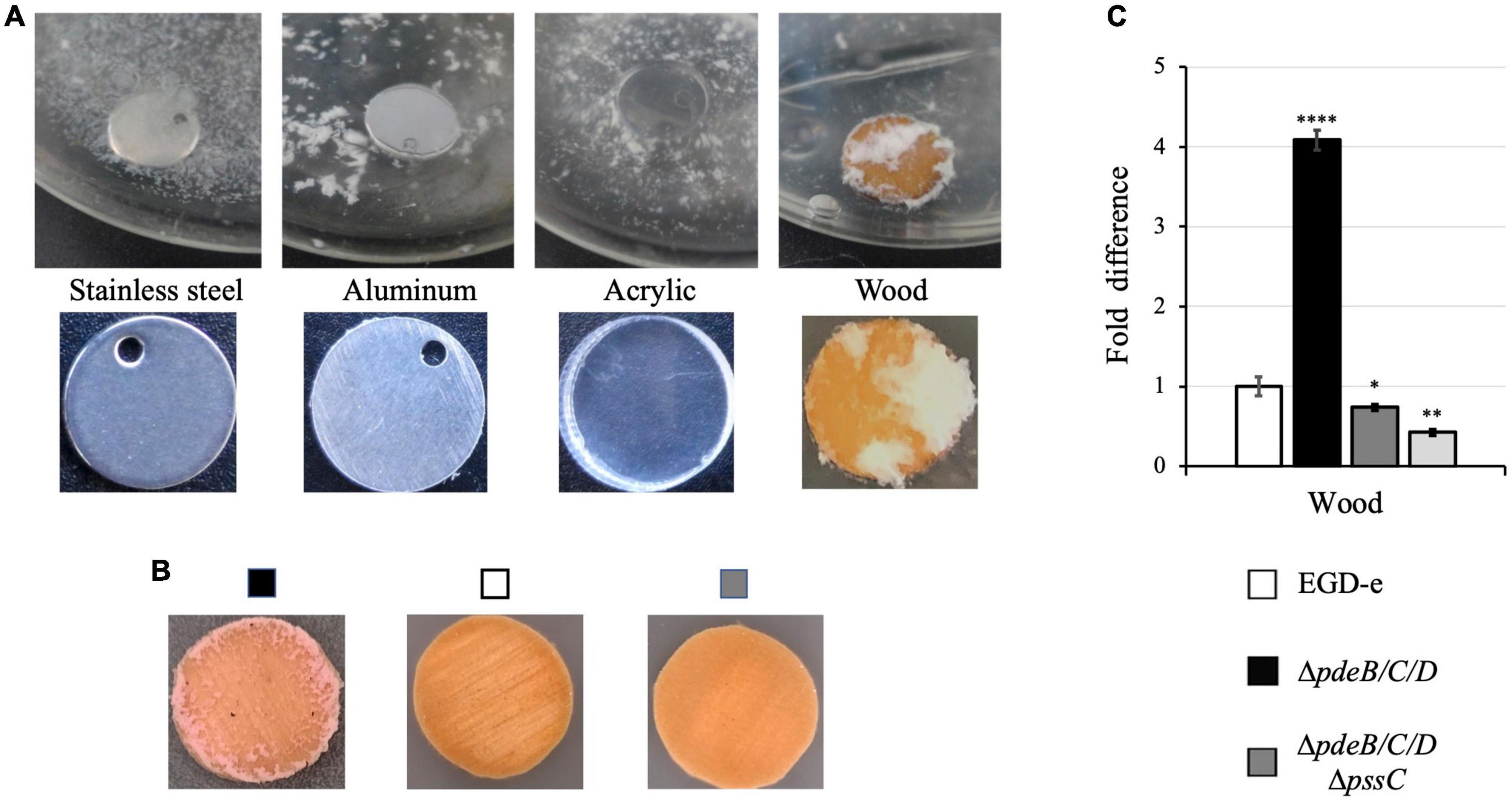
Figure 1. The listerial Pss EPS enhances colonization of wood but not manmade materials. L. monocytogenes strains were grown in liquid HTM/GY medium at 30°C for 48 h in the presence of coupons (disks) made from wood and manmade materials. (A) The EPS-synthesizing strain (ΔpdeB/C/D) grown in the presence of coupons made from stainless steel, aluminum and acrylic, and (unknown tree) wood forms voluminous biofilms only on wood coupons. (B) The EPS-synthesizing strain, but not wild type or the EPS-impaired strain (ΔpdeB/C/DΔpssC), form biofilms on wood coupons. All strains express red fluorescent protein mScarlet, which turns biomass pink (Table 1; Bindels et al., 2017). (C) Comparison of the CFU numbers in biofilms formed by various strains on the wood coupons following removal of the loosely attached biomass by rinsing. Shown are fold-differences compared to the wild type (EGD-e). The average CFU numbers of the wild type were 8.0 × 108 CFU per coupon. Data are from three independent experiments with at least two replicates in each experiment. Error bars represent standard deviations. Significant differences, compared to EGD-e, are indicated as follows: *P < 0.05, **P < 0.01, and ****P < 0.0001.
In contrast to the voluminous biofilms formed on wood coupons by the Pss-synthesizing strain, 48-h old biofilms of the wild-type strain, EGD-e, or the ΔpdeB/C/DΔpssC mutant impaired in EPS synthesis were not visible to the naked eye (Figures 1A, B). To quantify attached cells, the biofilm biomass was scraped off from the surface of wood coupons, vigorously vortexed in liquid medium, and serial dilutions were plated for enumerating colony forming units (CFUs). Figure 1C shows that the Pss EPS strongly, by at least fourfold, promotes colonization of wood coupons. Because prior to biomass scraping, loosely associated aggregates were removed by rinsing in sterile medium, the reported fold-difference is an underestimate.
To assess whether preference for wood is specific to the (unknown) tree species from which coupons were made, we tested coupons made from three known tree types. The EPS-synthesizing strain showed improved attachment to all tested wood coupons, albeit to varying extent (Figure 2). Therefore, listerial Pss increases attachment to various kinds of wood.
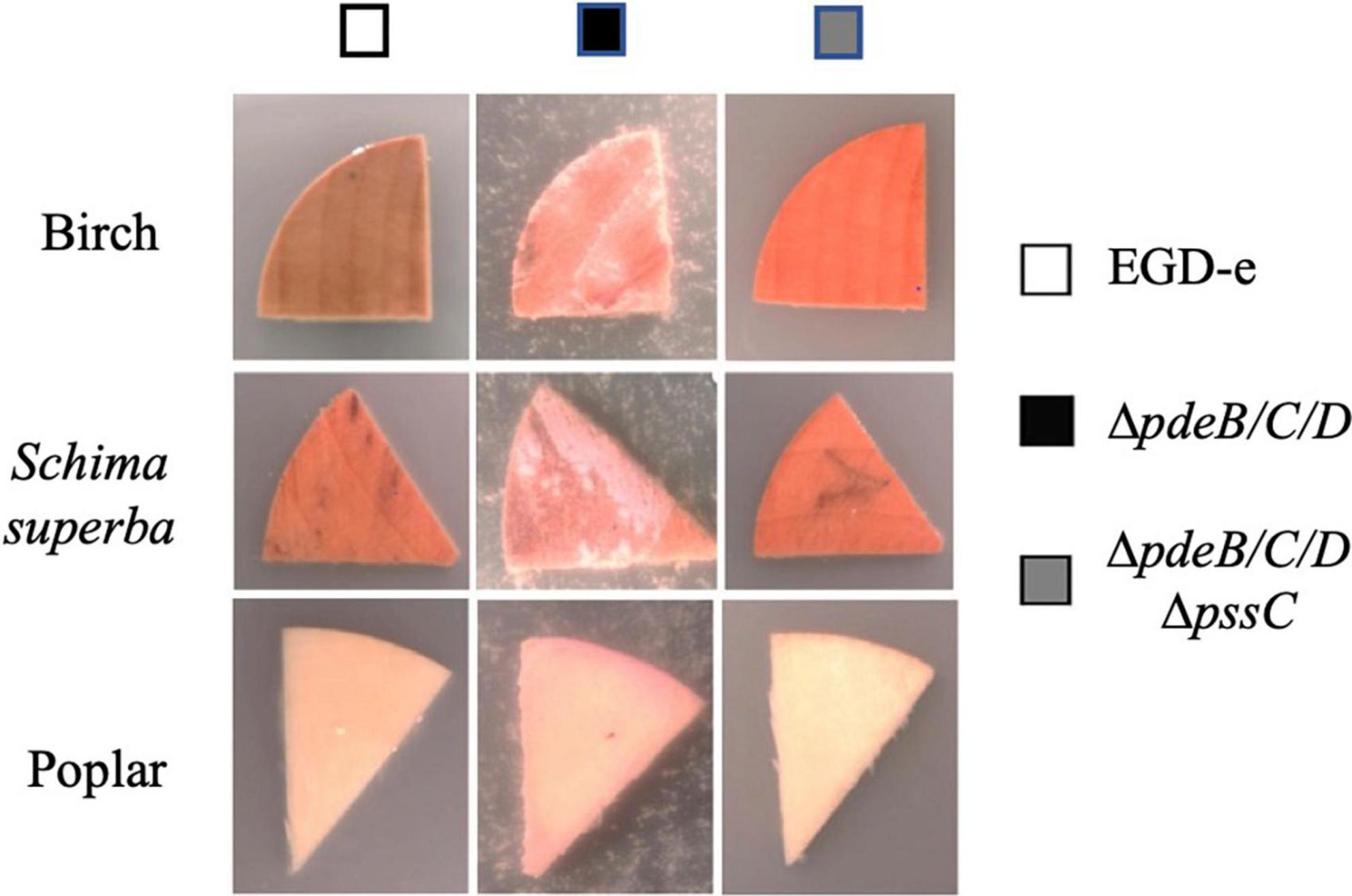
Figure 2. The listerial EPS-enhanced colonization of wood from various tree species. Coupon pieces from three tree species (birch, Schima superba and poplar) show higher colonization by the L. monocytogenes EPS-synthesizing strain (ΔpdeB/C/D), compared to the wild type (EGD-e) and EPS-impaired strain (ΔpdeB/C/DΔpssC). The pink color of the biomass is due to the expressed mScarlet protein. Wood pieces were incubated in the HTM/GY minimal medium in the presence of listeria for 48 h at 30°C.
Next, we investigated the role of the Pss EPS in listerial colonization of fresh produce using produce types that were associated with the past listeriosis outbreaks. We tested the role of Pss in biofilm formation on cantaloupe rind, the cause of major listeriosis outbreaks in the US and Australia (McCollum et al., 2013; NSW Government, 2018), as well as on cut pieces of fresh produce, which caused the majority of listeriosis outbreaks (Marik et al., 2020; Centers for Disease Control and Prevention, 2022). Sterilized pieces of approximately the same sizes were inoculated with L. monocytogenes in flasks containing minimal medium at 30°C.
After 48-h incubation, the EPS-synthesizing strain formed visible biofilms on cantaloupe rind (Figure 3A). Such biofilms were also visible on the surfaces of celery and lettuce (Figure 3A), most prominently on the cutting edges. No biofilms were visible on the pieces of produce grown in the presence of the wild-type and EPS-impaired strains (Figure 3A). Biofilms of the EPS-synthesizing strain contained ∼2−12-fold higher CFUs, compared to the wild type, and ∼4−22-fold higher, compared to the EPS-impaired strain (Figure 3B), suggesting that EPS strongly promotes colonization of various plant surfaces. The effect of EPS on colonization of cantaloupe rind versus pulp was stronger (12-fold versus 2-fold) (Figure 3B), suggesting that plant surface composition and roughness play important roles in EPS-dependent L. monocytogenes colonization.
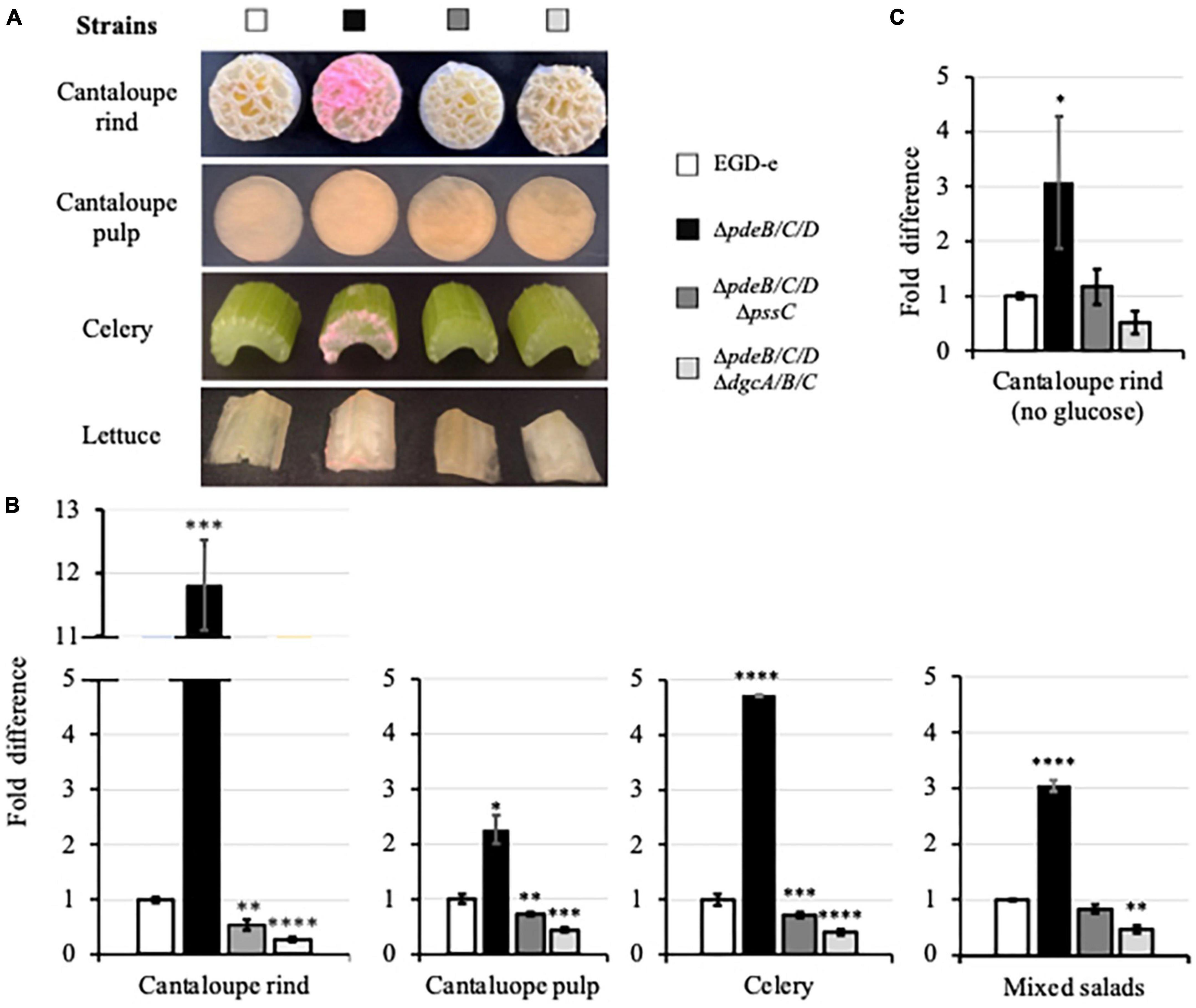
Figure 3. The EPS-enhanced colonization of the pieces of fresh produce. (A) Pieces of cantaloupe (containing or lacking rind), celery and Iceberg lettuce (a component of mixed salad) following incubation in the HTM/GY medium with L. monocytogenes for 48 h at 30°C. The biomass appears pink due to the expressed mScarlet protein. (B) Comparison of the CFUs of L. monocytogenes in biofilms formed on the fresh produce pieces incubated in the HTM/GY medium. Shown are fold-differences compared to the wild type (EGD-e). The average CFU numbers for the wild type were 7.0 × 109 CFU per cantaloupe rind, 1.2 × 1010 CFU per cantaloupe pulp, 1.9 × 108 CFU per piece of celery, and 8.0 × 108 CFU per three pieces (lettuce, carrot, cabbage) of mixed salad. Error bars represent standard deviations. (C) Comparison of the L. monocytogenes CFUs in biofilms formed on cantaloupe rind in the HTM medium lacking glucose and yeast extract. The average CFUs of the wild type was 7.4 × 106 CFU per cantaloupe rind. In all figures, data are from three independent experiments with at least two replicates in each experiment. Standard deviations are shown on the graph as error bars. Significant differences, compared to the wild type, are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001, and ****P < 0.0001.
In the experiments described above, minimal media contained glucose as a carbon source. To mimic conditions at cantaloupe storage and processing facilities, where water/moisture may be present, but carbon source is likely absent or limiting, we repeated the experiment using minimal medium lacking glucose. Residual bacterial growth under those conditions was possible due to the juice leaking from cantaloupe pieces. The lack of glucose in the medium significantly, by almost 103-fold, decreased colonization of cantaloupe rind (7.4 × 106 versus 7.0 × 109 CFU, Figures 3B, C). However, the trend in surface colonization was preserved, i.e., the EPS-synthesizing strain colonized cantaloupe rind several-fold more efficiently than the wild-type or the EPS-impaired strains (Figure 3C).
For a closer look at the biofilms on cantaloupe rind, we employed scanning electron microscopy (SEM). All strains formed extracellular appendages on cantaloupe rind, which may represent a combination of eDNA, nanotubes and flagella (Figure 4) and will not be discussed further. The biofilms formed by the EPS-synthesizing strain showed abundant EPS matrix in which listeria were embedded (Figure 4). We were unable to unambiguously determine if the wild type formed any Pss. It showed somewhat higher attachment to plant matter, compared to the EPS-impaired strain, ΔpdeB/C/DΔpssC (Figures 1C, 3B), however, whether this was due to low levels of Pss is unclear.
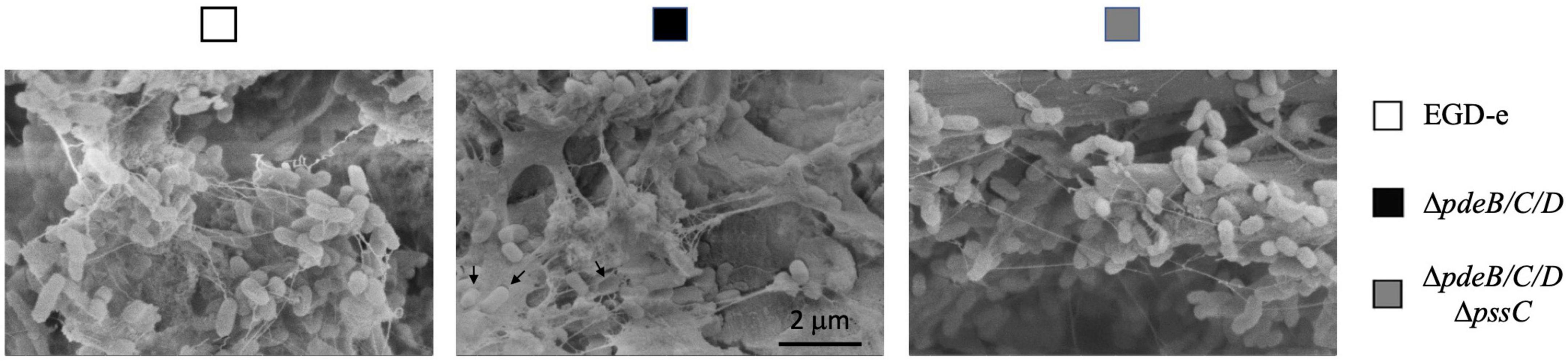
Figure 4. The SEM images of L. monocytogenes biofilms formed on cantaloupe rind. Bacterial cells embedded in the EPS matrix and presumed empty shells around bacteria removed during sample preparation (black arrows) are seen in the EPS-synthesizing strain but not in the wild type and EPS-impaired strains. Magnification, 40,000×.
Earlier we showed that the Pss EPS enhances L. monocytogenes tolerance to desiccation, when cell aggregates grown in liquid medium were compared to planktonically grown cells (Chen et al., 2014). Desiccation is the condition experienced by listeria during storage of fresh produce, such as whole cantaloupes, at the processing facilities and transportation to supermarkets and consumer homes and restaurants (Esbelin et al., 2018). To assess the role of the Pss EPS-biofilms on fresh produce surfaces in desiccation tolerance, we subjected the 48-h old biofilms formed on cantaloupe rind to drying on air at room temperature for up to 3 weeks. For ease of comparisons, we adjusted CFUs of surface-attached bacteria of all strains to approximately the same initial values by mechanically removing ∼90% biofilm of the EPS-synthesizing strain (Figure 3B) prior to the desiccation experiment. We realize that much thinner EPS-biofilms likely resulted in our underestimating the magnitude of desiccation tolerance benefits rendered by the Pss EPS.
After 7 days, CFUs decreased in the wild type from 100 to ∼10%, in the EPS-impaired strain to ∼7.4%, and in the EPS-synthesizing strain to 74%, i.e., by ∼10, ∼13, and 1.35-fold, respectively, (Figure 5). During prolonged desiccation, the EPS-synthesizing strain continued to maintain significant survival advantage over other strains (Figure 5). Desiccation tolerance in this strain was 6−16-fold higher, compared to the wild type, over the whole 3 week time course. This implies that the listerial EPS-biofilms formed on fresh produce could survive storage and transportation of fresh produce much better than biofilms formed by the strains not synthesizing Pss.
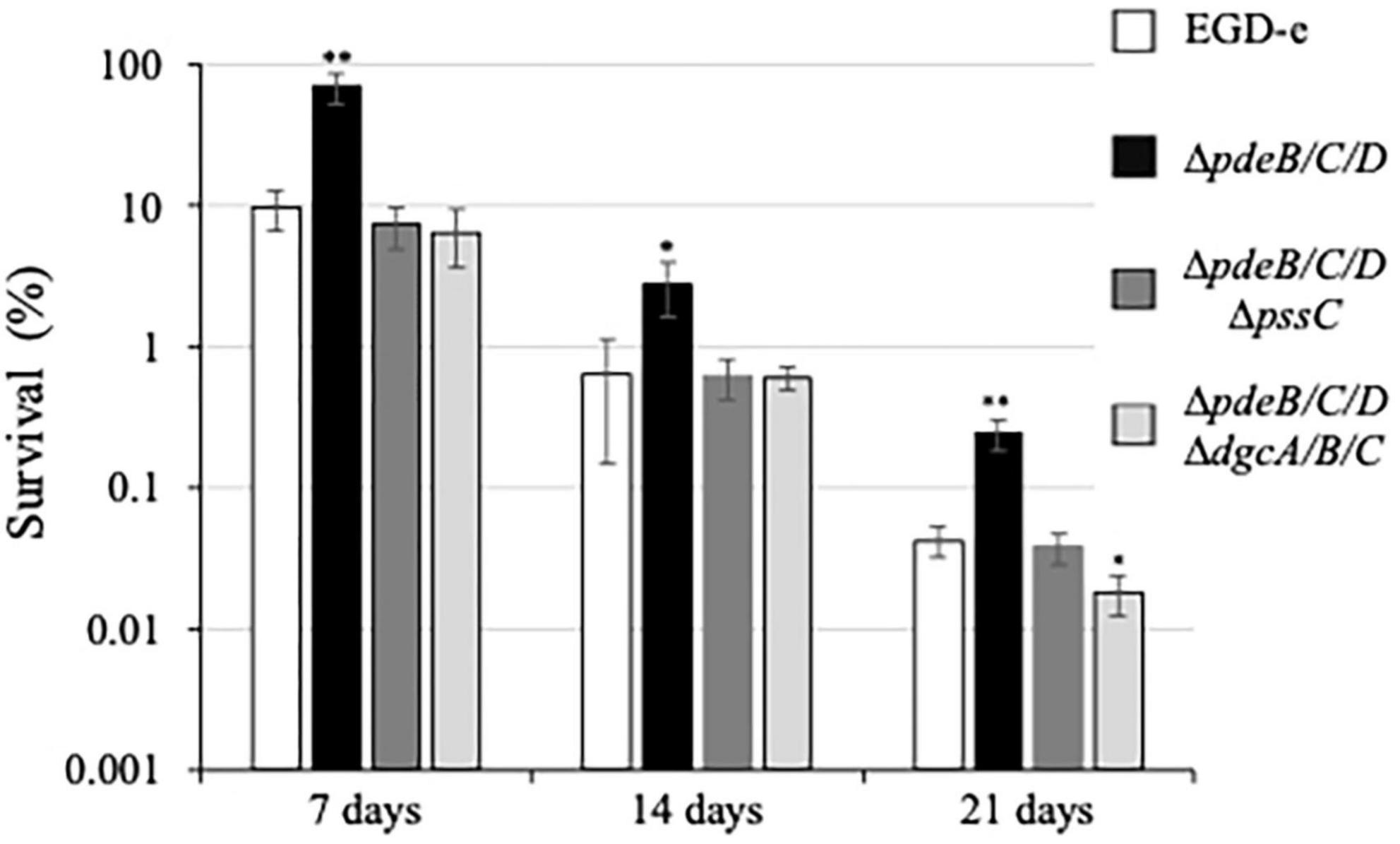
Figure 5. Survival under desiccation of the L. monocytogenes biofilms. The 48-h old biofilms formed on cantaloupe coupons containing rind and pulp (diameter, 20 mm; depth 4 mm) were placed in Petri dishes rind up and allowed to dry on open air. A coupon was cut into four quarters. One quarter was used for enumeration of initial CFUs, other quarters were subjected to desiccation and withdrawn on days 7, 14, and 21 days, respectively. Following withdrawal, the coupon quarters were rehydrated, homogenized, and subjected to CFU enumeration. For each strain, survival was calculated based on the initial CFU number of that strain on day 0 (assigned to 100%). Prior to the experiment, ∼90% of the biofilm biomass of the EPS-synthesizing strain, ΔpdeB/C/D, was removed from rind to bring the initial CFU numbers close to those of other strains. The average initial CFUs per coupon quarter for all strains were within the range of (1.1–3.4) × 108. Significant differences, compared to EGD-e, are indicated as follows: *P < 0.05 and **P < 0.01.
Prior to reaching hospitable small intestine, the site of the gastrointestinal tract where L. monocytogenes causes disease, it faces the challenge of stomach (gastric) acid. To mimic the process of consumption of the contaminated produce, we tested acid stress survival of the listerial biofilms formed on the cut fresh produce. For this experiment, we switched from cantaloupe rind as a model to cantaloupe pulp and celery, because rind is not consumed. The pieces of pulp and celery containing 48-h old listerial biofilms were exposed to acid stress, approximating pH of the stomach acid, i.e., pH 1.5−3.5 (Marieb and Hoehn, 2018). One stress regimen involved a 15-min exposure of the produce pieces to pH 2.5. Another regimen involved a 15-min exposure to pH 5.0, followed by 15 min at pH 2.5 (Guerreiro et al., 2022).
As shown in Figures 6A, B, exposure to pH 5 alone resulted in only moderate decreases in bacterial survival in all strains. However, exposure to pH 2.5 uncovered a large difference in survival, i.e., ∼20% for the EPS-synthesizing strain versus ∼1.8% for the wild type and ∼1.4% for the EPS-impaired strain (Figure 6A). Thus gives the EPS-synthesizing strain an ∼11-fold survival advantage over the wild type. Similarly, after a two-step regimen (pH 5.0, then pH 2.5), ∼1% of the EPS-synthesizing strain survived but only ∼0.085% of the wild type and ∼0.081% of the EPS-impaired strain, thus revealing an ∼12-fold advantage of the EPS-synthesizing strain over the wild type. Similar results were obtained when listerial survival in biofilms on the celery pieces was measured (Figure 6B), i.e., ∼25% survival of the EPS-synthesizing strain versus ∼2% for the wild type and ∼1.7% for the EPS-impaired strain following exposure to pH 2.5, and ∼4.7% versus 0.04% and 0.03% survival following a two-step regimen. Thus, the EPS-synthesizing strain has a 12−116-fold survival advantage on celery pieces, compared to the wild-type. These results suggest that from ∼11- to ∼116-fold higher number of L. monocytogenes in the EPS-biofilms on cut fresh produce could survive exposure to stomach acid, compared to the biofilms lacking Pss EPS.
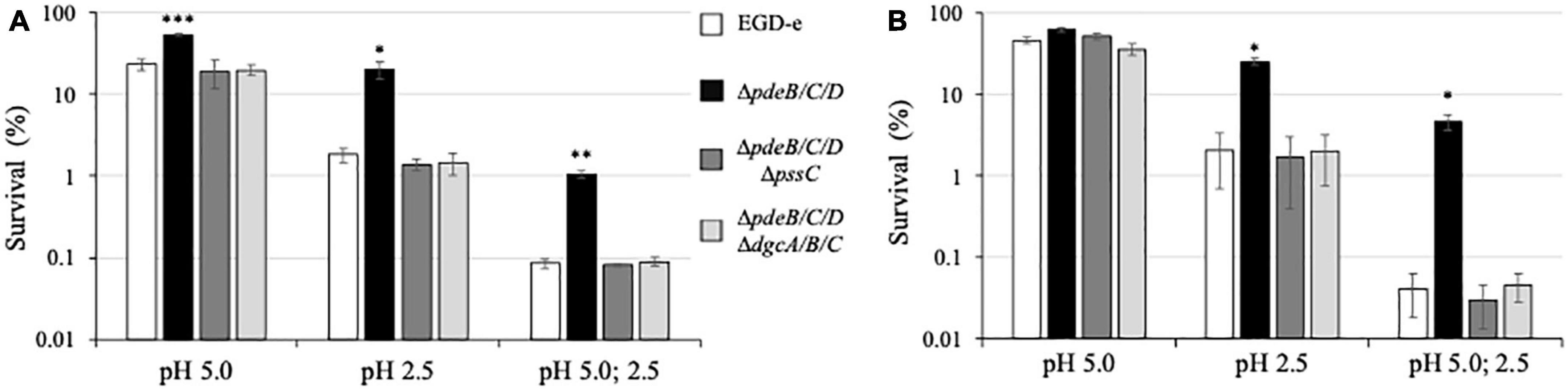
Figure 6. Survival under acid stress of the L. monocytogenes biofilms. The 48-h old biofilms formed on pieces of cantaloupe pulp (A) and celery (B) were exposed for 15 min to pH 5.0 or pH 2.5, or sequentially to pH 5.0 (15 min) and pH 2.5 (15 min). Following medium neutralization, cantaloupe and celery pieces were collected, homogenized, and CFU were enumerated. For each strain, survival percentages were calculated based on the initial CFU prior to exposure to acid (assigned to 100%). The average initial CFUs for all strains were within the range of (5.3–7.3) × 109 per cantaloupe pulp piece and (1.1–1.4) × 109 per celery piece. Significant differences, compared to EGD-e, are indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
In L. monocytogenes, EPS synthesis is the dominant, albeit not the only phenotype associated with the elevated c-di-GMP levels (Chen et al., 2014; Elbakush et al., 2018). In other bacteria, elevated c-di-GMP levels, in addition to upregulating EPS synthesis, affect such biofilm components as protein adhesins, pili, and fimbria (Römling et al., 2013). To access the impact of c-di-GMP-dependent factors other than Pss EPS that contribute to listerial colonization and survival on plant matter, we constructed the L. monocytogenes c-di-GMP null strain, ΔpdeB/C/DΔdgcA/B/C. In addition to lacking c-di-GMP-specific PDEs, this mutant also lacks all DGCs involved in c-di-GMP synthesis (Chen et al., 2014). We found that the c-di-GMP null mutant behaved similarly to the high c-di-GMP strain impaired in Pss synthesis, ΔpdeB/C/D ΔpssC, in survival assays (Figures 5, 6), thus suggesting that Pss is the key factor affecting stress tolerance. In plant surface colonization assays the c-di-GMP null strain appeared somewhat, ∼2-fold, inferior, compared to the ΔpdeB/C/D ΔpssC strain (Figures 1C, 3B, C). From these experiments, it is evident that the Pss EPS is the dominant c-di-GMP-dependent factor affecting listerial colonization of plant matter and stress tolerance.
In this study, we investigated the role of the Pss EPS in L. monocytogenes colonization of plant surfaces. We discovered that Pss significantly, from 2 to 12-fold, improves colonization of diverse plant matter, from different kinds of wood to cantaloupe rind, cantaloupe pulp, celery, and lettuce. Pss has little or no effect on colonization of non-plant, manmade materials (Figures 1–3). While the ability of several kinds of EPS to facilitate attachment to plant surfaces has been reported in various bacteria (Danhorn and Fuqua, 2007; Yaron and Römling, 2014; Castiblanco and Sundin, 2016), this is the first report describing such function of the L. monocytogenes Pss EPS.
Colonization efficiency depended on the properties of plant surface. The Pss EPS had the largest effect on rough, fiber-rich surfaces of wood and netted cantaloupe rind, and the smallest – on smooth surfaces of cantaloupe pulp (Figure 3B). More EPS-biofilms were formed on the edges than on the smooth surfaces of the same produce (Figure 3A). Because strains used in this study grow at the same rate, differences in colonization reflect differences in initial surface attachment, therefore Pss is involved in attachment to plant matter. While we did not directly interrogate the mechanism of Pss-mediated plant surface binding, the ability of Pss to bind to diverse plant matter, but not to manmade materials, and bind better to fiber-rich surfaces of wood and cantaloupe rind suggests that Pss interacts with the insoluble plant (ligno)cellulose fibers. Only few additional factors promoting attachment to plant matter are known in L. monocytogenes. One of these is functional flagella. They enhance initial attachment to plant surfaces in some L. monocytogenes strains (Lemon et al., 2007; Gorski et al., 2009). Another factor is a cellulose binding protein, Lcp (LMOf2365_0859). Strain L. monocytogenes F2365 attached to cantaloupe rind, lettuce, and spinach better than the lcp mutant by several-fold (Bae et al., 2013). Lmo0842, an Lcp homolog, is encoded in the EGD-e genome and may have the same function. Note, however, that voluminous Pss EPS synthesized by the ΔpdeB/C/D strain likely masked surface proteins, including Lmo0842. Because the EPS-synthesizing strain is impaired in motility (Chen et al., 2014), Pss also likely negated the effects of flagella.
In addition to enhancing colonization, the Pss EPS strongly protected L. monocytogenes grown in the EPS-biofilms on the surfaces of fruit and vegetables against desiccation and acid stresses. These stresses are arguably most relevant from the standpoint of fresh produce safety because they mimic, respectively, conditions of produce storage and transportation, and passage of the contaminated produce pieces through the stomach following consumption. In our experimental setup, EPS-biofilms showed a 6−16-fold increase in desiccation tolerance (Figure 5), and a 11−116-fold increase in acid stress survival (Figure 6). The findings that listerial EPS-biofilms are much more tolerant to stresses are in line with the role of EPS-biofilms in other bacteria (Mao et al., 2006; Deka et al., 2019). These findings are also consistent with our earlier observations that clumps of the ΔpdeB/C/D strain formed in liquid media are much more tolerant of desiccation and exposure to harmful chemicals, compared to planktonically grown cells (Chen et al., 2014).
To estimate the overall effect that the L. monocytogenes EPS on safety of such produce as cantaloupes, we combined the effects of Pss on produce colonization (Figures 1C, 3B, C; 2−12-fold increase in CFUs), ability to survive during storage and transportation (Figure 5, 6−16-fold increase in desiccation tolerance) and ability to survive stomach acid exposure (Figure 6, 11−116-fold increase in acid stress survival). By multiplying the low- and high-end values, we estimate that the EPS-synthesizing strain has an enormous, 102- to 104-fold, advantage over EPS-impaired strains in reaching the small intestine of consumers where they may cause disease. If desiccation is not part of processing, the advantage would remain very large. Note that these numbers may be underestimates, because the bulk (∼90%) of the EPS-biofilms was removed prior to the desiccation experiment, in order to bring pre-desiccation CFUs of all strains to the same range. Further, the 48-h biofilms tested in this study may not have fully developed protective potential, compared to older biofilms (Hingston et al., 2013; Chen et al., 2020). We surmise that the enormous advantage imparted by EPS to listeria on fresh produce makes EPS an extremely problematic factor from the standpoint of fresh produce safety.
The key question not addressed in this work is how often listerial EPS-biofilms are formed on plant matter, including fresh produce. On the one hand, Listeria are frequently associated with the decaying plant matter in nature (Welshimer and Donker-Voet, 1971; Fenlon, 1986; Ivanek et al., 2007; Freitag, 2009). The benefits rendered by Pss regarding enhanced colonization and stress tolerance point to the high likelihood that EPS is synthesized on plant matter. Further, the pssA-E operon encoding the Pss EPS biosynthetic machinery is part of the core L. monocytogenes genome, deciphered from 1,696 genomes of strains isolated from diverse sources and geographic locations (Moura et al., 2016). Strong conservation of the pssA-E operon in the genomes of pathogenic and non-pathogenic listeria supports the notion that Pss is important for environmental survival (Chen et al., 2014). On the other hand, the Pss EPS has been unambiguously detected only in the L. monocytogenes strains with artificially elevated c-di-GMP levels, either via mutations (e.g., ΔpdeB/C/D) or via expression of a foreign DGC (Chen et al., 2014). Under the conditions used in this study, the wild type, EGD-e, showed no solid evidence of Pss. If it was synthesized, the amounts were not readily detectable by SEM (Figure 4) and were low to make a major difference in colonization and stress survival, compared to the EPS-impaired strains (Figures 1C, 3, 5, 6). It is of course possible that EGD-e is not an optimal strain for investigating EPS synthesis, because it has been propagated under the laboratory conditions for several decades (Murray et al., 1926). Such maintenance is known to select against biofilm phenotypes resulting in domesticated strains (McLoon et al., 2011). L. monocytogenes strains isolated from the recent fresh produce outbreaks may therefore offer more relevant insights into EPS synthesis.
Additional factors that may have obscured EPS presence in L. monocytogenes biofilms concern experimental design of most studies of listerial attachment to plant matter. L. monocytogenes in these studies was usually grown in rich liquid media, spotted on the surfaces of fresh produce, and allowed to dry. Bacterial biofilms and survival were then investigated over time at different environmental conditions, such as temperature and humidity (Marik et al., 2020). This setup undervalues time needed for planktonic, single-cell bacteria from liquid medium to switch to the surface-attached, biofilm lifestyle, when EPS synthesis is expected to take place (Römling et al., 2013). Further, rich media used in those studies may not adequately mimic conditions under which listerial biofilms are formed on plant matter, whether in the environment or in fresh produce processing facilities. It is also important to note that the majority of listerial biofilm studies involved manmade materials used in food processing, such as stainless steel, aluminum, rubber, glass, and plastics (Blackman and Frank, 1996; Bonaventura et al., 2008; Lee et al., 2017). The extracellular biofilm matrices on these materials contain mostly eDNA and secreted proteins (Blackman and Frank, 1996; Rieu et al., 2008; Harmsen et al., 2010; Ferreira et al., 2014; Guilbaud et al., 2015; Zetzmann et al., 2015; Oloketuyi and Khan, 2017; Rodríguez-López et al., 2018), and biofilm forming capacity is highly dependent on specific strains and experimental conditions (Borucki et al., 2003; Cherifi et al., 2017; Lee et al., 2017; Gorski et al., 2021; Gray et al., 2021). The lack of EPS under these conditions is consistent with the results of this work that showed that Pss does not affect colonization of manmade materials (Chen et al., 2014; Figure 1).
To better understand the role of the L. monocytogenes Pss EPS in nature and in fresh produce industry, we need to uncover environmental factors that increase c-di-GMP levels and thus turn on Pss synthesis. Having a Pss-specific probe would also be helpful for detecting Pss EPS on plant matter. Work in these areas is ongoing, as is the search for compounds that inhibit formation of EPS-biofilms. Even if such biofilms are formed on fresh produce only by some strains and only under specific conditions, from the standpoint of fresh produce safety, the 102−104-fold advantage that EPS may render L. monocytogenes in causing foodborne illness seems too large to ignore.
Strains and plasmids used in this study are listed in Table 1. The c-di-GMP null strain, ΔpdeB/C/D ΔdgcA/B/C, was constructed from strain ΔpdeB/C/D using sequential deletion of dgcA/B (with the help of pKSV7-ΔdgcAB) and dgcC (with the help of pKSV7-ΔdgcC), as described earlier (Chen et al., 2014). Plasmid pAM-mScarlet expressing the red fluorescent protein mScarlet (Bindels et al., 2017) was constructed to enhance visualization of L. monocytogenes on the surfaces of wood and fresh produce. The mScarlet gene was synthesized with L. monocytogenes-optimized codons (Twist Bioscience) and cloned downstream of the strong Bacillus subtilis Pveg promoter (Guiziou et al., 2016) in the pAM101-derived vector (Fujimoto and Ike, 2001). The listerial strains were grown in the liquid minimum HTM medium (Tsai and Hodgson, 2003) containing 3% glucose and 0.2% yeast extract, HTM/GY, at 30°C under shaking (220 rpm). When strains containing pAM-mScarlet were used, the media was supplemented with 10 μg/mL chloramphenicol (Cm). For enumerating CFUs, cultures were routinely plated onto Brain Heart Infusion (BHI) agar (Millipore Sigma, Burlington, MA, United States) and incubated at 37°C for 36 h.
All coupons (disks for do-it-yourself arts and crafts projects) were purchased on from various vendors, including coupons made from unspecified wood (32 mm diameter × 1.2 mm thickness, Woodpile), birch (50 mm × 8 mm, Woodpeckers), poplar (50 mm × 2.5 mm, Juvale), Schima superba (38 mm × 4 mm, Axe Sickle), stainless steel (38 × 1.55 mm, PH Pandahall, CA, USA), aluminum (38 × 1.55 mm, RPM Stamping Blanks, Rose Metal Products, Springfield, MO, USA), and acrylic (25.4 × 1.8 mm, Tupalizy, PTC-Office). Prior to use, all non-wood coupons were immersed in 70% ethanol for 20 min for disinfection, rinsed twice in sterile deionized water and air dried. The wood coupons were autoclaved (121°C, 30 min). In some experiments, prior to sterilization, surfaces of steel, aluminum and acrylic coupons were roughened up by a scrapper to imitate surface roughness of wood coupons.
The overnight L. monocytogenes cultures were diluted 1:100 into 10 mL HTM/GY medium in 125-mL flasks and grown at 30°C until optical density of ∼ 0.4 at 600 nm. Sterile wood coupons or pieces of fresh produce were added at this point and cultivation was continued for 48 h. The coupons were subsequently withdrawn and rinsed twice in sterile HTM to remove loosely attached biofilms. The biomass attached to the coupons and produce pieces was thoroughly scrapped off into HTM medium using a sterile scalpel, and the suspensions were vortexed. Serial dilutions were plated onto BHI agar plates and grown at 37°C for 36 h followed by CFU enumeration.
Whole cantaloupes, celery, and mixed salads (Iceberg lettuce, red cabbage, carrots) used in this study were purchased from local (Laramie, WY) retail stores. Cantaloupe coupons (20 mm × 4 mm) were obtained by using a cork borer. Some pieces were cut out to contain rind, others were cut out to contain cantaloupe pulp only. Celery was cut in pieces of varying lengths from similar sized stocks. Prior to processing, fresh produce was thoroughly washed. Cutout pieces were sterilized by complete submersion into 8.25% sodium hypochlorite (CloroxPro, Clorox Professional Products Company) in the 50-mL centrifuge tubes placed on a rocker platform for 20 min. Following sterilization, fresh produce pieces were submerged in 50-mL sterile distilled water overnight and subsequently rinsed in fresh sterile distilled water. Sterile pieces were added to listerial cultures, as described above for coupons.
Following 48-h incubation, produce pieces were aseptically removed from the cultures and rinsed by dipping in three sequential beakers with sterile HTM media. To measure listerial colonization, cantaloupe rind was carefully separated from pulp and processed separately, while the remaining pulp was discarded. To measure colonization of cantaloupe pulp, pulp-only pieces lacking rind were used. The produce pieces were mechanically macerated in the Stomacher bags in 5 mL HTM medium and homogenized for 10 min at high speed (Stomacher® 80 Biomaster, Seward, UK). The serial dilutions of the homogenates were plated onto BHI agar plates for CFU enumeration.
Small (5 mm diameter x 1 mm thickness) pieces of cantaloupe rind containing L. monocytogenes biofilms were used for SEM (Scanning Electron Microscope FEI Quanta 250). Briefly, rind pieces were fixed for 2 h in 2% glutaraldehyde-PBS buffer at room temperature, followed by dehydration steps in a series of ethanol baths (10 min each) containing 25, 50, 75, 95, and 100% (3 times) ethanol, and postfixed by incubation with 1% osmium tetroxide in PBS for 1 h in the dark. The samples were then dried using Balzers Critical Point Dryer (CPD 020) before coating with gold-palladium, as described earlier (Muravnik et al., 2016).
Rind-containing cantaloupe pieces were removed from the liquid L. monocytogenes cultures after 48-h incubation. To adjust biomass levels to approximately the same initial cell numbers, the voluminous EPS-biofilms of the ΔpdeB/C/D strain were gently brushed off cantaloupe rind, resulting in the removal of ∼ 90% biomass. Coupons were then aseptically cut into four equal-sized quarters. One quarter was used to determine initial CFU (day 0) following homogenization in Stomacher. The remaining quarters were placed on sterile blotting paper in open Petri dishes and kept at room temperature inside the biological safety cabinet with the switched off blower. After 7, 14, and 21 days of storage the quarters were placed in Stomacher bags with 5 mL HTM medium, soaked for 1 h, homogenized and serial dilutions were plated for CFU enumeration.
Biofilms formed on the pieces of celery and cantaloupe pulp were used in acid stress experiments. The sizes of celery pieces were adjusted to have similar initial number of listeria in biofilms, as follows: 20 mm (EGD-e), 5 mm (ΔpdeB/C/D), 22 mm (ΔpdeB/C/DΔpssC), and 24 mm (ΔpdeB/C/DΔdgcA/B/C). The 4-mm thick cantaloupe pulp pieces had the following diameters: 18 mm (EGD-e), 15 mm (ΔpdeB/C/D), 19 mm (ΔpdeB/C/DΔpssC), and 20 mm (ΔpdeB/C/DΔdgcA/B/C). Following 48-h incubation in liquid L. monocytogenes cultures, celery or cantaloupe pulp pieces were removed and placed in HTM/GY acidified to pH 5.0 or 2.5 with hydrochloric acid for 15 min at 37°C. A preconditioning protocol involved exposing celery pieces to pH 5.0 for 15 min followed by exposure to pH 2.5 for additional 15 min, as described (Guerreiro et al., 2022). The acid was neutralized to pH 7.0 by addition of the sodium hydroxide solution, celery pieces were homogenized, and serial dilutions were plated for CFU enumeration.
Microsoft Excel was used for data processing analysis and presentation. The bar charts display a mean ± standard deviation (SD) from three independent experiments, each of which had at least two replicates. Unpaired Student’s t-tests were performed using Prism 9 for Mac (GraphPad). Significant differences, compared to the wild type, EGD-e, are indicated in all figures as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001, and ****, P < 0.0001.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AF performed the experiments, analyzed the data, and wrote the manuscript. AE performed the experiments and analyzed the data. L-HC constructed the L. monocytogenes c-di-GMP null strain. MG conceived, designed, coordinated the research, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by USDA-NIFA-AFRI-1946224 and in part by the NIFA HATCH Program via the University of Wyoming Agriculture Experimental Station grant WYO-583-17 and Integrated Microscopy Core is supported by NIH P20 GM121310.
We acknowledge Dr. Zhaojie Zhang (Integrated Microscopy Core) and Dr. Tyler C. Brown (Materials Characterization Laboratory) at the University of Wyoming for technical support in SEM studies, and Kiet Tran for the pAM-mScarlet plasmid.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Archer, D. L. (2018). The evolution of FDA’s policy on Listeria monocytogenes in ready-to-eat foods in the United States. Curr. Opin. Food Sci. 20, 64–68. doi: 10.1016/j.cofs.2018.03.007
Bae, D., Seo, K. S., Zhang, T., and Wang, C. (2013). Characterization of a potential Listeria monocytogenes virulence factor associated with attachment to fresh produce. Appl. Environ. Microbiol. 79, 6855–6861. doi: 10.1128/AEM.01006-13
Bindels, D., Haarbosch, L., Weeren, L., Postma, M., Wiese, K., Mastop, M., et al. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56. doi: 10.1038/nmeth.4074
Blackman, I. C., and Frank, J. F. (1996). Growth of Listeria monocytogenes as a biofilm on various food-processing surfaces. J. Food Prot. 59, 827–831. doi: 10.4315/0362-028X-59.8.827
Bonaventura, G. D., Piccolomini, R., Paludi, D., D'Orio, V., Vergara, A., Conter, M., et al. (2008). Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 104, 1552–1561. doi: 10.1111/j.1365-2672.2007.03688.x
Borucki, M. K., Peppin, J. D., White, D., Loge, F., and Call, D. R. (2003). Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69, 7336–7342. doi: 10.1128/aem.69.12.7336-7342.2003
Castiblanco, L. F., and Sundin, G. W. (2016). New insights on molecular regulation of biofilm formation in plant-associated bacteria. J. Integr. Plant Biol. 58, 362–372. doi: 10.1111/jipb.12428
Centers for Disease Control and Prevention (2022). Available online at: https://www.cdc.gov/listeria/. (accessed February 6, 2022).
Chen, L., Köseoğlu, V., Güvener, Z., Myers-Morales, T., Reed, J., Orazio, S., et al. (2014). Cyclic di-GMP-dependent signaling pathways in the pathogenic firmicute Listeria monocytogenes. PLoS Pathog. 10:e1004301. doi: 10.1371/journal.ppat.1004301
Chen, X., Thomsen, T. R., Winkler, H., and Xu, Y. (2020). Influence of biofilm growth age, media, antibiotic concentration and exposure time on Staphylococcus aureus and Pseudomonas aeruginosa biofilm removal in vitro. BMC Microbiol. 20:264. doi: 10.1186/s12866-020-01947-9
Cherifi, T., Jacques, M., Quessy, S., and Fravalo, P. (2017). Impact of nutrient restriction on the structure of Listeria monocytogenes biofilm grown in a microfluidic system. Front. Microbiol. 8:864. doi: 10.3389/fmicb.2017.00864
Combrouse, T., Sadovskaya, I., Faille, C., Kol, O., Guérardel, Y., and Midelet-Bourdin, G. (2013). Quantification of the extracellular matrix of the Listeria monocytogenes biofilms of different phylogenic lineages with optimization of culture conditions. J. Appl. Microbiol. 114, 1120–1131. doi: 10.1111/jam.12127
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Deka, P., Goswami, G., Das, P., Gautom, T., Chowdhury, N., Boro, R., et al. (2019). Bacterial exopolysaccharide promotes acid tolerance in Bacillus amyloliquefaciens and improves soil aggregation. Mol. Biol. Rep. 46, 1079–1091. doi: 10.1007/s11033-018-4566-0
Elbakush, A. M., Miller, K. W., and Gomelsky, M. (2018). CodY-mediated c-di-GMP-dependent inhibition of mammalian cell invasion in Listeria monocytogenes. J. Bacteriol. 200:e457. doi: 10.1128/JB.00457-17
Esbelin, J., Santos, T., and Hébraud, M. (2018). Desiccation: an environmental and food industry stress that bacteria commonly face. Food Microbiol. 69, 82–88. doi: 10.1016/j.fm.2017.07.017
Fenlon, D. R. (1986). Growth of naturally occurring Listeria spp. in silage: a comparative study of laboratory and farm ensiled grass. Grass Forage Sci. 41, 375–378. doi: 10.1111/j.1365-2494.1986.tb01828.x
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Freitag, N. E. (2009). Complete transcriptional profile of an environmental pathogen. Future Microbiol. 4, 779–782. doi: 10.2217/fmb.09.56
Fujimoto, S., and Ike, Y. (2001). pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 67, 1262–1267. doi: 10.1128/AEM.67.3.1262-1267.2001
Gorski, L., Duhé, J. M., and Flaherty, D. (2009). The use of flagella and motility for plant colonization and fitness by different strains of the foodborne pathogen Listeria monocytogenes. PLoS One 4:e5142. doi: 10.1371/journal.pone.0005142
Gorski, L., Walker, S., Romanolo, K. F., and Kathariou, S. (2021). Growth and survival of attached Listeria on lettuce and stainless steel varies by strain and surface type. J. Food Prot. 84, 903–911. doi: 10.4315/JFP-20-434
Gray, J., Chandry, P. S., Kaur, M., Kocharunchitt, C., Fanning, S., Bowman, J., et al. (2021). Colonisation dynamics of Listeria monocytogenes strains isolated from food production environments. Sci. Rep. 11:12195. doi: 10.1038/s41598-021-91503-w
Guerreiro, D. N., Pucciarelli, M. G., Tiensuu, T., Gudynaite, D., Boyd, A., Johansson, J., et al. (2022). Acid stress signals are integrated into the σB - dependent general stress response pathway via the stressosome in the food-borne pathogen Listeria monocytogenes. PLoS Pathog. 18:e1010213. doi: 10.1371/journal.ppat.1010213
Guilbaud, M., Piveteau, P., Desvaux, M., Brisse, S., and Briandet, R. (2015). Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl. Environ. Microbiol. 81, 1813–1819. doi: 10.1128/AEM.03173-14
Guiziou, S., Sauveplane, V., Chang, H., Clert, C., Declerck, N., Jules, M., et al. (2016). A part toolbox to tune genetic expression in Bacillus subtilis. Nucl. Acids Res. 44, 7495–7508. doi: 10.1093/nar/gkw624
Harmsen, M., Lappann, M., Knøchel, S., and Molin, S. (2010). Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 76, 2271–2279. doi: 10.1128/AEM.02361-09
Hefford, M. A., Aoust, S., Cyr, T. D., Austin, J. W., Sanders, G., Kheradpir, E., et al. (2005). Proteomic and microscopic analysis of biofilms formed by Listeria monocytogenes 568. Can. J. Microbiol. 51, 197–208. doi: 10.1139/w04-129
Hingston, P. A., Stea, E. C., Knøchel, S., and Hansen, T. (2013). Role of initial contamination levels, biofilm maturity and presence of salt and fat on desiccation survival of Listeria monocytogenes on stainless steel surfaces. Food Microbiol. 36, 46–56. doi: 10.1016/j.fm.2013.04.011
Hoffmann, S., and Ahn, J. W. (2021). Updating economic burden of foodborne diseases estimates for inflation and income growth, ERR-297. Department of Agriculture, Economic Research Service. Available online at: https://www.ers.usda. gov/webdocs/publications/102640/err-297.pdf?v=1403.6 (accessed March 18, 2023).
Ivanek, R., Gröhn, Y. T., and Wiedmann, M. (2007). Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Food. Pathog. Dis. 3, 319–336. doi: 10.1089/fpd.2006.3.319
Karygianni, L., Ren, Z., Koo, H., and Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Köseoğlu, V., Heiss, C., Azadi, P., Topchiy, E., Güvener, Z., Lehmann, T., et al. (2015). Listeria monocytogenes exopolysaccharide: origin, composition, biosynthetic machinery, and c-di-GMP-dependent regulation. Mol. Microbiol. 96, 728–743. doi: 10.1111/mmi.12966
Lee, B. H., Hébraud, M., and Bernardi, T. (2017). Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol. 8:2221. doi: 10.3389/fmicb.2017.02221
Lemon, K. P., Higgins, D. E., and Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/JB.01967-06
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3:11. doi: 10.1128/microbiolspec.MB-0011
Mao, Y., Doyle, M. P., and Chen, J. (2006). Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett. Appl. Microbiol. 42, 642–647. doi: 10.1111/j.1472-765X.2006.01875.x
Marik, C. M., Zuchel, J., Schaffner, D. W., and Strawn, L. K. (2020). Growth and survival of Listeria monocytogenes on intact fruit and vegetable surfaces during postharvest handling: a systematic literature review. J. Food. Prot. 83, 108–128. doi: 10.4315/0362-028X.JFP-19-283
McCollum, J. T., Cronquist, A. B., Silk, B. J., Jackson, K. A., Connor, K. A., Cosgrove, S., et al. (2013). Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 369, 944–953. doi: 10.1056/NEJMoa121583
McLoon, A. L., Guttenplan, S. B., Kearns, D. B., Kolter, R., and Losick, R. (2011). Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 193, 2027–2034. doi: 10.1128/JB.00722-08
Moura, A., Criscuolo, A., Pouseele, H., Maury, M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Muravnik, L. E., Kostina, O. V., and Shavarda, A. L. (2016). Glandular trichomes of tussilago farfara (senecioneae, asteraceae). Planta 244, 737–752. doi: 10.1007/s00425-016-2539-x
Murray, E. G. D., Webb, R. A., and Swann, M. B. R. (1926). A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes. J. Pathol. 29, 407–439. doi: 10.1002/path.1700290409
NSW Government, (2018). Department of primary industries. listeria outbreak investigation. summary report for the melon industry. Available online at: https://www.foodauthority.nsw.gov.au/sites/default/files/_Documents/foodsafetyandyou/listeria_outbreak_investigation.pdf (accessed March 18, 2023).
Oloketuyi, S. F., and Khan, F. (2017). Inhibition strategies of Listeria monocytogenes biofilms-current knowledge and future outlooks. J. Basic Microbiol. 57, 728–743. doi: 10.1002/jobm.201700071
Renier, S., Hébraud, M., and Desvaux, M. (2011). Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic gram-positive foodborne pathogen. Environ. Microbiol. 13, 835–850. doi: 10.1111/j.1462-2920.2010.02378.x
Rieu, A., Briandet, R., Habimana, O., Garmyn, D., Guzzo, J., and Piveteau, P. (2008). Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl. Environ. Microbiol. 74, 4491–4497. doi: 10.1128/AEM.00255-08
Rodríguez-López, P., Rodríguez-Herrera, J. J., Vázquez-Sánchez, D., and López, C. M. (2018). Current knowledge on Listeria monocytogenes biofilms in food-related environments: incidence, resistance to biocides, ecology and biocontrol. Foods 7:E85. doi: 10.3390/foods7060085
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Tiensuu, T., Andersson, C., Rydén, P., and Johansson, J. (2013). Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 87, 909–924. doi: 10.1111/mmi.12140
Tsai, H. N., and Hodgson, D. A. (2003). Development of a synthetic minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 69, 6943–6945. doi: 10.1128/AEM.69.11.6943-6945.2003
Welshimer, H. J., and Donker-Voet, J. (1971). Listeria monocytogenes in nature. Appl. Microbiol. 21, 516–519. doi: 10.1128/am.21.3.516-519.1971
Yaron, S., and Römling, U. (2014). Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 7, 496–516. doi: 10.1111/1751-7915.12186
Zetzmann, M., Okshevsky, M., Endres, J., Sedlag, A., Caccia, N., Auchter, M., et al. (2015). DNase-sensitive and -resistant modes of biofilm formation by Listeria monocytogenes. Front. Microbiol. 6:1428. doi: 10.3389/fmicb.2015.01428
Keywords: foodborne pathogen, listeriosis, food safety, c-di-GMP, desiccation, acid stress, biofilm, vegetable and fruit
Citation: Fulano AM, Elbakush AM, Chen L-H and Gomelsky M (2023) The Listeria monocytogenes exopolysaccharide significantly enhances colonization and survival on fresh produce. Front. Microbiol. 14:1126940. doi: 10.3389/fmicb.2023.1126940
Received: 18 December 2022; Accepted: 11 April 2023;
Published: 27 April 2023.
Edited by:
Hongshun Yang, Jiangnan University (Shaoxing) Industrial Technology Research Institute, ChinaReviewed by:
Soraya Chaturongakul, Mahidol University, ThailandCopyright © 2023 Fulano, Elbakush, Chen and Gomelsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Gomelsky, Z29tZWxza3lAdXd5by5lZHU=
†Present address: Li-Hong Chen, College of Horticulture and Plant Protection, Inner Mongolia Agricultural University, Hohhot, China
‡These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.