
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 January 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1124770
This article is part of the Research TopicInsights Into the Role of Microorganisms on Food Quality and Food SafetyView all 17 articles
The physicochemical property, volatile flavor compounds, and microbial community structure of Jinhua fatty ham (FH) and lean ham (LH) were investigated and compared by high-throughput sequencing and HS-GC-IMS. Results showed that FH had higher pH and slightly lighter and yellower color than LH. Meanwhile, 33 volatile flavor compounds were identified from FH and LH, among which LH showed higher abundance of total alcohols and acids, but FH had generally richer aldehydes, ketones, esters, heterocyclic, and sulfur-containing compounds. Moreover, FH and LH did not have significant difference in α-diversity of bacterial community, but LH presented a much lower α-diversity of fungal community than FH. Besides, the dominant microorganisms (relative abundance >2%) in FH were Ruminococcaceae UCG-005, Staphylococcus, Ruminococcaceae UCG-014, Meyerozyma, and Aspergillus at the genus level, while in LH were Staphylococcus, Psychrobacter, Halomonas, Propionicicella, Ruminococcaceae UCG-005, Meyerozyma, Yamadazyma, and Aspergillus. Furthermore, the analysis of Pearson’s correlation and metabolic network confirmed that the discriminative flavor compounds of FH were mainly β-oxidation and degradation products of fatty acids, while those of LH were mostly derived from the Strecker reaction or microbial metabolism of amino acids. The present study could help understand the potential pathway of characteristic microorganisms affecting flavor formation of fat-deficient dry-cured hams and provide theoretical supports for developing healthier fermented meat products.
Chinese dry-cure ham is a traditional fermented meat product with a long history of over 1,000 years (Li et al., 2022). Jinhua ham, considered one of the most representative Chinese dry-cured hams, is widely prevalent around the world due to its unique flavor and abundant nutrients (Li W. et al., 2021; Wang et al., 2021). Jinhua hams are produced from fresh hind legs of pigs through a long processing procedures, mainly including raw material selection, salting, dry-ripening, and post-ripening, which usually spend several months (Zhou et al., 2021a). The fats, proteins, and glycogens in raw hams can be degraded into large amounts of amino acids, fatty acids, and pyruvates during the dry-ripening, which further produce the characteristic flavor compounds of dry-cured hams through diverse chemical reactions, such as β-oxidation, deamination, Strecker degradation, and Millard reaction (Martínez-Onandi et al., 2017; Bosse et al., 2018; Shi et al., 2019). Hence, fat plays a crucial role in the formation of unique flavor of Jinhua hams. However, high-level fats or polyunsaturated fatty acids in meats are sensitive to oxidative deterioration, which are related with the rancid smell and may have a side effect on consumers’ acceptance (Benet et al., 2015; Zhou et al., 2021b). In addition, the consumption of foods with high-level saturated fats could be associated with an increased risk of cholesterol-related cardiovascular pathologies (Trevisan et al., 1990; Benet et al., 2016). Therefore, the lean ham (LH), manufactured from the pork hind leg removed skin and fat tissues, are more and more popular among consumers, especially populations with health concerns. Nevertheless, the physicochemical property and flavor quality were rarely studied for LH, even though sufficiently reported for normal fatty hams (FH). Moreover, the effect of fat deficiency on the flavor characteristics of dry-cured hams was also not investigated yet.
On the other hand, the aforementioned complex reactions producing unique flavor compounds are dependent on the enzymatic actions of not only endogenous enzymes but also microbial enzymes (Petrova et al., 2015; Wang et al., 2021). Recent studies reported that various microorganisms could promote the formation of characteristic flavor and quality property of dry-cured hams by performing proteolysis, lipolysis, and oxidation activities (Toledano et al., 2019; Chen et al., 2021; Li et al., 2022). Meanwhile, several reports focused on the composition of microbial communities and relative content of volatiles in Jinhua FH (Ge et al., 2017; Wang et al., 2021; Deng et al., 2022). Moreover, the core microorganisms in Jinhua FH, such as Staphylococcus, Lactobacillus, Debaryomyces, and Apiotrichum, have been identified positively associated with the flavor compounds derived from the catabolism of amino acids, such as some branched aldehydes and heterocyclic compounds (Zhou et al., 2022). However, there is still a lack of comprehensive knowledge on microbial community of Jinhua LH. Furthermore, the potential metabolic pathway of core bacteria/fungi affecting flavor formation in LH was also not understood.
Therefore, the objective of this study was firstly, to compare the physicochemical property, volatile flavor quality, and microbial community composition of Jinhua FH and LH, and secondly, to explore the correlations and connections between core microbes and differential flavor compounds. The color, pH, water activity, chemical composition, and nitrite residue of both FH and LH were determined. The headspace-gas chromatography-ion mobility spectrometry (HS-GC-IMS) and high-throughput sequencing of 16S rRNA/ITS genes were also utilized to analyze volatile flavor profiles and bacterial/fungal communities, respectively. Furthermore, the analysis of correlation and metabolic network was applied for the exploration of relationship between core microorganisms and volatiles. The information obtained from this study could help understand the potential pathway of characteristic microorganisms influencing flavor formation of dry-cured hams under the condition of fat deficiency.
The Jinhua hams used in the present study were prepared and sampled in Jinhua Jinnian Ham Co., Ltd (Zhejiang, China). Six fresh hind legs (14.5 ± 0.5 kg, pH = 5.9 ± 0.2) of domestic pigs (Large White × Landrace) were used to prepare Jinhua FH and LH following the procedures of Zhou et al. (2021a) with some modifications. For Jinhua FH, three whole hind legs were salted for 75 days with 0.014% NaNO2 and 10% NaCl per leg, followed by soak cleaning for 1 day and sun-drying for 1 day. Subsequently, hind legs were dehydrated for 7 days in a dehydration plant, sun-dried for another day, and then ripened for 180 days in a dry-ripening room. During the ripening, the ambient temperature progressively increased from 5°C to 35°C, while the relative humidity gradually decreased from 85 to 65%. Afterward, legs were further post-ripened for about 30 days at room temperature (25°C), and the hams were finally obtained when the weight loss reached approximately 40% of the initial weight. For Jinhua LH, the skins and fat tissues were firstly trimmed off from the other three hind legs. The final hams were also obtained following the gradual procedures of salting, soak cleaning, sun-drying, dehydrating, secondary sun-drying, dry-ripening, and post-ripening, which were all performed under the same conditions with Jinhua FH. Furthermore, as described by Chen et al. (2021) with minor modifications, approximately 5 cm × 2 cm × 0.2 cm pieces were cut from the surface of the biceps femoris muscle of each ham, which were used for the fungal community determination. Meanwhile, about 20 g interior samples were taken from the central fraction (about 3–4 cm depth) of the biceps femoris muscle of each ham, which were used for the evaluation of physicochemical parameters, volatile flavor compounds, and bacterial community. All samples were vacuum-packaged and frozen at-80°C until further analysis.
The color, pH and water aw were measured following the methods of our previous studies (Li H. et al., 2021; Zhang et al., 2022) with minor modifications. The color of samples was directly detected using a colorimeter (NH310, 3NH Technology Co., Ltd., China), where the L, a*, and b* values represent lightness, redness/greenness, and yellowness/blueness, respectively. A white standard plate with the L, a*, and b* values of 99.46, 0.19, and −1.98, respectively, was used for calibration before color detection. The pH value was determined using a portable pH meter (Testo 205, Testo Instruments Co., Ltd., Shenzhen, China) equipped with a piercing pH probe and a temperature-compensated temperature probe. The aw value was determined with an intelligent water activity meter (HD-4, Huake Instrument and Meter Co., Ltd., Wuxi, China).
The chemical composition of ham samples was detected as described by our previous study (Zhang et al., 2017) with minor modifications. Briefly, the total contents of moisture, protein, fat, and minerals were analyzed by the method of direct drying (GB/T 5009.3–2016), Kjeldahl (ISO 5983-1997), Soxhlet extraction (ISO 1444-1996), and dry-ashing (ISO 5984-2002), respectively.
According to the method of Liu et al. (2019) with minor modifications, the content of residual nitrite was detected with a nitrite assay kit following the manufacturer’s instruction (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Briefly, ham samples were dispersed into distilled water and filtrated, followed by the reaction with sulphanilamide solution and N-naphthyl-ethylenediamine dihydrochloride. The nitrite content was calculated based on the absorbance of supernatant at 550 nm, which was assayed using an ultraviolet–visible spectrophotometer (UV-1750, Shimadzu, Kyoto, Japan).
As described by Liu et al. (2020) with minor modifications, the volatile flavor compounds of ham samples were identified by a headspace-gas chromatography-ion mobility spectrometer (HS-GC-IMS; Flavorspec, G.A.S. Instrument, Germany) equipped with a SE-54 capillary column (15 m × 0.53 mm × 1 μM). Samples were firstly cut into approximately 1 cm × 1 cm × 1 cm cubes and minced by an analytical grinder (Aika Instrument Equipment Co., Ltd., Guangzhou, China). Then 2 g samples were put into a 20 ml headspace sampling vial and incubated at 60°C for 20 min. Afterward, 500 μL headspace was injected into the injector automatically with a heated syringe at 85°C. Subsequently, samples were transferred into the capillary column by high-purity nitrogen (>99.99%) at the following programmed flow rates: initially 2 ml/min for first 2 min, then 10 ml/min for 8 min, next 100 ml/min for 10 min, and eventually 150 ml/min for 5 min. Meanwhile, the temperature of column and drift tube was kept as 60°C and 45°C, respectively, and the flow rate of drift gas (nitrogen gas, >99.99% purity) was maintained as 150 mL/min.
The IMS data were analyzed using the instrumental laboratory analysis view (LAV) software with the Reporter, Gallery Plot, and Dynamic PCA plug-in applications. The volatile flavor compound was identified by comparing the retention index (RI) and drift time (DT) with the NIST library and IMS database retrieval software obtained from G.A.S. The intensities of these compounds were calculated based on the height of selected signal peaks.
The analysis of microbial communities, including the compositions of both fungi and bacteria, were performed by high-throughput sequencing following the procedures of Chen et al. (2021) and Wang et al. (2021) with some modifications. The total genome DNA of bacteria and fungi was extracted using a Cetyltrimethylammonium Bromide (CTAB) method following the manufacturer’s instructions of the genomic DNA extraction kit. The V3-V4 hypervariable regions of bacterial 16S rRNA genes were amplified with the forward primer 5’-CCTAYGGGRBGCASCAG-3′ and reverse primer 5′-GGACTACNNGGGTATCTAAT-3′. The ITS1-1\u00B0F regions of fungal ITS genes were amplified with the forward primer 5′-CTTGGTCATTTAGAGGAAGTAA-3′ and reverse primer 5′-GCTGCGTTCTTCATCGATGC-3′. All PCR reactions were conducted in 30 μL reactions with 15 μL Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM forward and reverse primers, and approximately 10 ng template DNA. PCR products were then mixed in equidensity ratios and purified through the Axy Prep DNA Gel Extraction Kit (AXYGEN; for bacteria) or Qiagen Gel Extraction Kit (Qiagen, Germany; for fungi).
The data sequencing was carried out according to the procedures of Wang et al. (2021) and Li et al. (2022) with some modifications. Sequencing libraries were generated using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, United States; for bacteria) or TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, United States; for fungi) following the manufacturer’s recommendations and index codes were added. The library quality was evaluated on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyzer 2,100 system. Then the library was sequenced on the Illumina Miseq/HiSeq2500 (for bacteria) or NovaSeq (for fungi) platform. Afterward, the QIIME (V1.9.1) quality controlled process was performed to filter raw reads to obtain high-quality clean reads. The reads were compared with the reference database (Silva database) using the UCHIME algorithm to detect chimera sequences, and the chimera sequences were removed to acquire the effective clean reads. The sequence analysis was performed using the UPARSE algorithms. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs), and representative sequences for each OTU were screened for further annotation. The significance of differences in microbial communities was statistically analyzed based on the relative abundance of bacteria or fungi at various levels, mainly including phylum, genus, and species. The α-diversity was presented via the Shannon index, Simpson index, ACE index, Chao 1 index, and observed species, while the β-diversity was assayed by the principal coordinate analysis (PCoA).
All experiments were performed in triplicate with the results shown as average ± standard deviation. Tables were made by the Microsoft Excel 2016 software, while figures were drawn with the Origin V2021 (Origin-Lab, Northampton, United States) and Microsoft PowerPoint 2016. Analysis of variance (ANOVA) was performed using the SAS V8 software (SAS Institute Inc., Carry, United States), while analysis of Pearson’s correlation and hierarchical cluster (HCA) was conducted by the plug-in applications in Origin V2021. Differences among mean values were established with the Duncan multiple range test. The significant difference was confirmed when p < 0.05.
The physicochemical property of dry-cure ham was closely associated with the flavor quality and microbial diversity (Domínguez et al., 2022). The physicochemical parameters of internal samples from Jinhua FH and LH, including color, water activity (aw), pH, chemical composition, and nitrite residue, are shown in Table 1. It is clear that there is no significant distinction between the a* values (redness) of FH and LH (p > 0.05). However, FH showed slightly higher L and b* values than LH (p < 0.05), suggesting that the color of FH was slightly lighter and yellower than that of FH. This result may be attributed to their difference in the redox degree of proteins, especially the myoglobin and hemoglobin (Parolari et al., 2016). Specifically, myoglobin and hemoglobin are the main pigments responsible for the color of dry-cured hams (Parolari et al., 2016; Zhou et al., 2021b), and the lack of protection by fat and skin tissues might allow more myoglobins/hemoglobins in LH to be oxidized during dry-ripening, possibly resulting in the darker color of LH.
Besides, the aw values of FH and LH were not obviously different (p > 0.05) and both far under 0.85, which was a low aw that protects hams from most harmful microbes and meanwhile allows the growth of salt-consuming and flavor-generating bacteria/fungi (Chen et al., 2021). However, LH exhibited a remarkable lower pH than FH (p < 0.05). Bosse et al. (2018) reported that a pH of 6.0–6.2 would cause a higher microbial risk and reduced water diffusion ability in hams, while hams with pH of 5.6–6.0 have more desirable saltiness, color, and texture. The pH values of FH and LH were 6.08 ± 0.03 and 5.75 ± 0.04, respectively, suggesting that LH might has higher sensory quality and edible safety than FH. The distinct pH values of FH and LH was probably attributed to their distinction in the intensity of acids generated during ripening (Table 2; Figure 1).
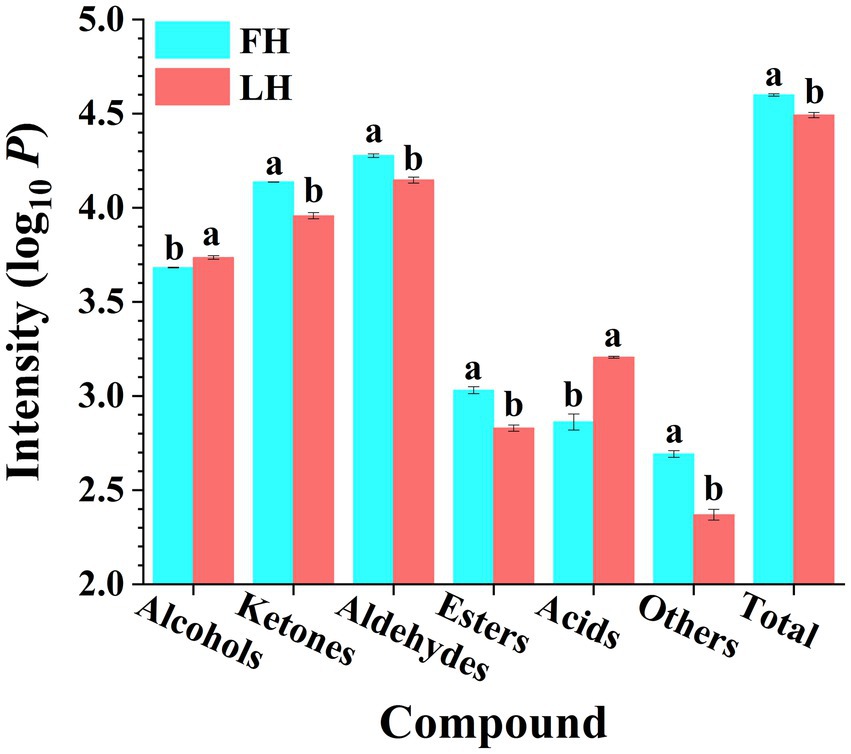
Figure 1. Comparison of volatile flavor compound intensities of Jinhua FH and LH in different chemical families. Different lowercases above the error bar denote significant differences between Jinhua FH and LH (p < 0.05). FH, fatty ham; LH, lean ham. Log10 P is the logarithmic value of total intensity for compounds of each chemical family in HS-GC-IMS spectra.
Furthermore, these two hams also exhibited no significant difference in chemical compositions (p > 0.05). The moisture content of both FH and LH (around 31.5%) was similar with that of Mianning hams reported by Chen et al. (2021), which is low enough to prevent spoilage and improve quality of hams. This result was also consistent with the aw data as mentioned above, since the water content is usually considered positively correlated with the water activity. Meanwhile, the minerals in hams can be mainly salts (Domínguez et al., 2022), hence FH and LH showed similar mineral contents (about 8.50%; p > 0.05) because of the same adding amount of NaCl during salting. Additionally, the nitrite residues were 1.13 ± 0.21 and 1.57 ± 0.12 for FH and LH, respectively, without marked distinction (p > 0.05) and far under the secure nitrite residue limits (30 mg/kg) for meat products in China (Chen et al., 2021). On the other hand, the pigments contributing to the rose-red color of dry-cured hams are mainly nitromyoglobins and nitrohemoglobins, which are the product of reactions between nitrites/nitrates and myoglobins/hemoglobins (Parolari et al., 2016). Therefore, the similar nitrite residue of FH and LH suggested a same nitrite consuming amount for the production of red pigments, which can be the main reason for their similar redness (a* value) as mentioned above. Overall, the data shown in Table 1 indicate that LH had an obviously lower lightness, yellowness, and pH than FH, probably resulting from their discrimination in oxidation and fermentation products.
The volatile flavor profiles of internal samples from Jinhua FH and LH were analyzed by HS-GC-IMS and results are illustrated as a two-dimension spectra plot (Figure 2). The y-axis and x-axis in Figure 2 showed the retention time (RT) of GC and the relative drift time (DT) of ions, respectively (Liu et al., 2020). Meanwhile, the red line representing x = 1.0 was the reactive ion peak (RIP). Besides, each data point corresponded to a volatile flavor compound, and its color indicated the intensity of volatile (Li et al., 2019). Specifically, the blue and white colors represented a low intensity of volatiles, whereas the yellow and red colors showed a high intensity of compounds. It is exhibited that the two hams showed similar number of ion peaks within the DT range of 1.0–1.7 ms, suggesting that the numbers of main identified volatiles were not obviously different in FH and LH. However, the data points (ion peaks) of FH mostly showed a redder color or bigger size than those of LH, indicating that FH had an overall higher intensity of volatile flavor than LH.
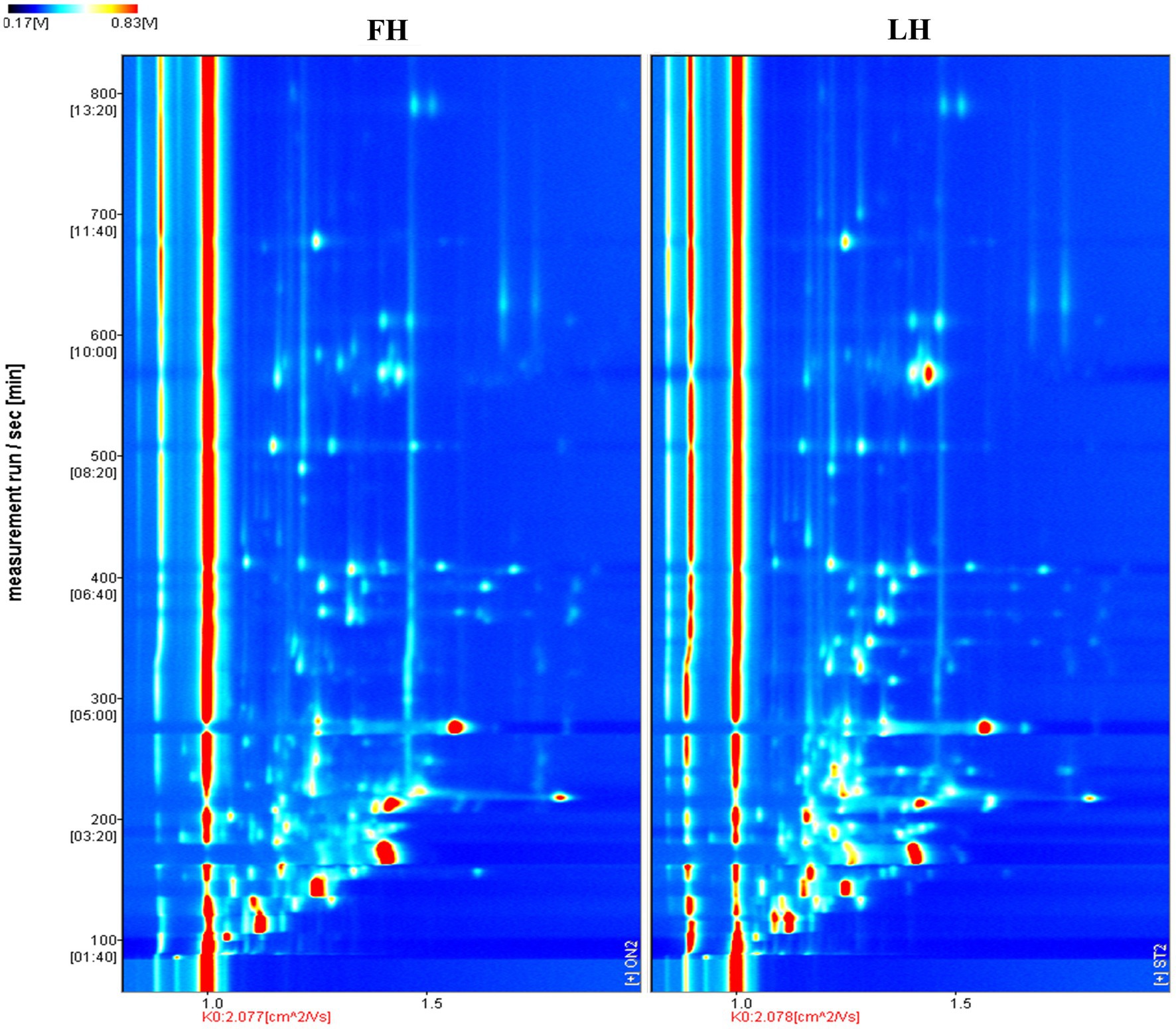
Figure 2. Comparison of volatile flavor profiles of Jinhua FH and LH with two-dimension HS-GC-IMS spectra. FH, fatty ham; LH, lean ham.
Furthermore, to compare the finger-print of characteristic volatiles in FH and LH, the Gallery plot plug-in was utilized with results presented in Figure 3. Each row in Figure 3 represented a single volatile flavor compound, which could correspond to a single signal (monomer) or a pot (dimer) in the IMS spectra plot (Figure 2) depending on its concentration (Arroyo-Manzanares et al., 2018). As shown in Figure 3, a total of 33 volatile compounds were identified, which can be classified into 7 chemical families. In details, there were 7 alcohols, 8 ketones, 10 aldehydes, 4 esters, 2 acids, 1 heterocyclic compound, and 1 sulfur-containing compound. These findings were in accordance with the reports of Liu et al. (2020), Wang et al. (2021), and Li et al. (2022) on the volatile profiles of Jinhua hams.
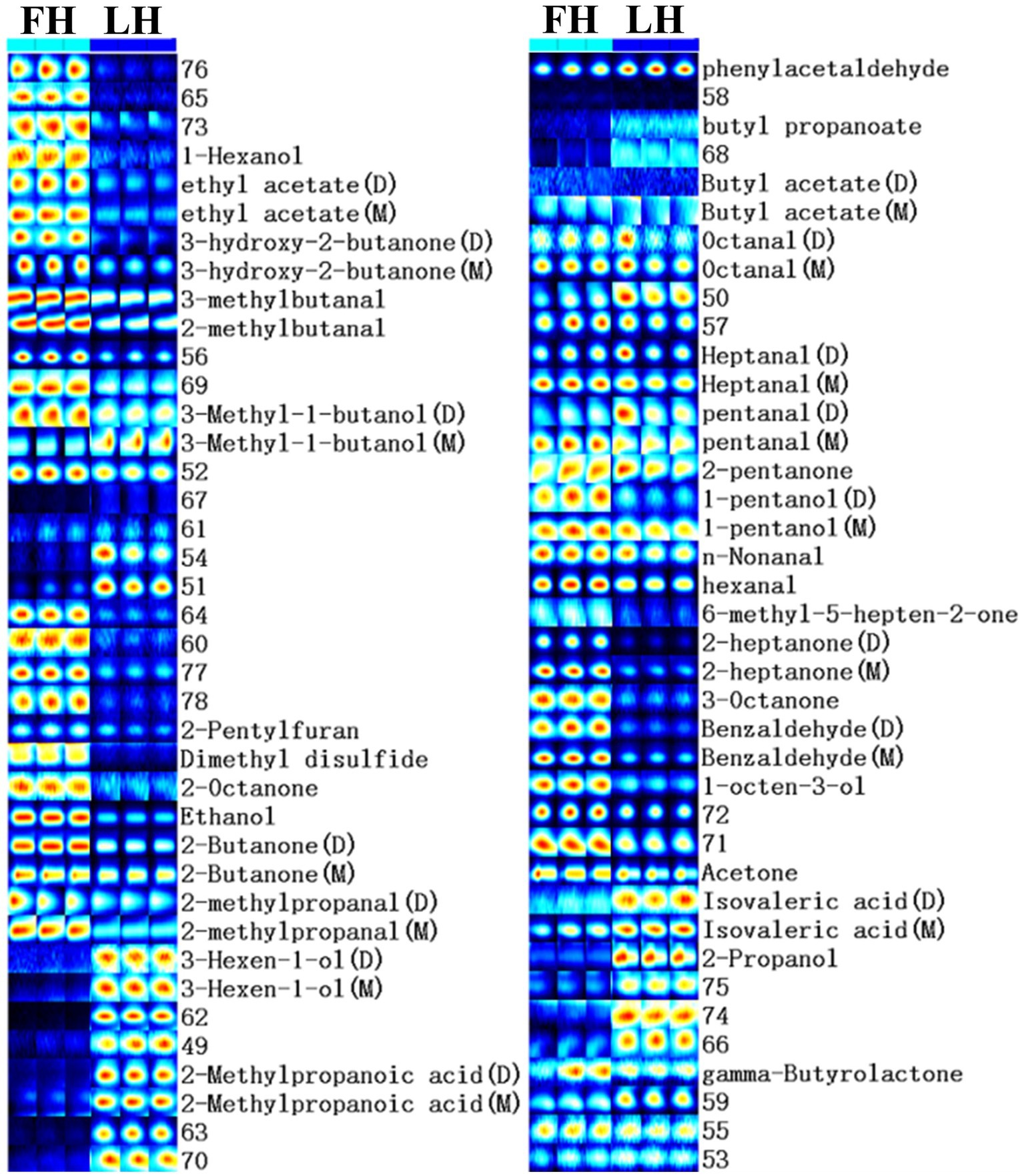
Figure 3. Gallery plot of characteristic flavor finger-print of Jinhua FH and LH determined by HS-GC-IMS. FH, fatty ham; LH, lean ham.
The profiles of individual volatile flavor compounds were further analyzed with data shown in Table 2. Aldehydes are known as the major contributors to the unique flavor of fermented meats due to their high concentrations and low aroma thresholds (Liu et al., 2020; Wang et al., 2021). As exhibited in Table 2, hexanal, 2-methylbutanal, and 3-methylbutanal were the richest aldehydes in both FH and LH, followed by 2-methylpropanal (for FH) or phenylacetaldehyde (for LH), in accordance with the findings of Wang et al. (2021) on the volatiles of Jinhua hams. Hexanal is regarded as the major lipid oxidation product in dry-cured hams, and a low-content hexanal generally provides a pleasant grassy, fruity, and green odor (Benet et al., 2015; Domínguez et al., 2022). 2-methylbutanal, 3-methylbutanal, and 2-methylpropanal are all branched aldehydes, deriving from oxidative deamination and decarboxylation of valine, leucine, and isoleucine through Strecker degradation (Narváez-Rivas et al., 2014). Meanwhile, they are also medium-chain (C4-C9) aliphatic aldehydes, which were considered responsible for grassy, fatty, and/or nutty flavors of meat products (Liu et al., 2020; Wang et al., 2021). Furthermore, the abundances of most identified aldehydes were significantly lower (p < 0.05) or similar (p > 0.05) in LH compared with those in FH. Noteworthily, only the phenylacetaldehyde showed a relatively higher richness in LH (p < 0.05). Phenylacetaldehyde is originated from the Strecker degradation of phenylalanine and might contribute to the spicy sensation of hams (García-González et al., 2008).
Ketones are another important flavor component in dry-cured meat products, and the high-intensity ketones are usually associated with the creamy, fruity, cooked, and spicy flavor characteristics (Zhu et al., 2018). The most abundant ketones in the two hams were 2-butanone and acetone (p < 0.05; Table 2). Acetone could be mainly transferred from acetyl-CoA, one of the major degradation products of glycogens in the muscle, and imparts a buttery taste and fruity aroma to meat products (Shi et al., 2019; Li W. et al., 2021). 2-butanone and other methyl ketones are produced via the decarboxylation of β-keto acids or the β-oxidation of saturated fatty acids, and act as precursors in the formation of fatty flavor during the ripening of meats (Pham et al., 2008; Shi et al., 2019). Besides, FH had significantly higher intensities in almost all identified ketones than LH (p < 0.05) excluding 2-pentanone. The 2-pentanone did not show noteworthy distinction between the two hams (p > 0.05; Table 2).
Alcohols are considered contributing less to the aroma of hams because of their relatively higher odor thresholds than aldehydes and ketones, but they can also impart to an herbal, woody, and oily flavor at high concentrations (Liu et al., 2020). The linear-chain aliphatic alcohols are the oxidation products of lipids, whereas the branched alcohols are mostly reported as the microbial degradation products of corresponding branched aldehydes (Shi et al., 2019). It is clear in Table 2 that the most abundant alcohol was obviously distinct for the two hams, which was 2-propanol in LH but ethanol in FH. Meanwhile, most identified linear-chain alcohols showed a higher richness in FH (p < 0.05), while 2-propanol, 3-methyl-1-butanol (monomer), and 3-hexen-1-ol presented higher abundances in LH (p < 0.05).
Two short-chain fatty acids (SCFAs; C1-C6), isovaleric acid and 2-methylpropanoic acid, were also identified in both FH and LH, which probably originated from the deamination of amino acids or the secondary metabolism by microbes (Liu et al., 2020; Li et al., 2022). Interestingly, both the two identified acids were more abundant in LH (p < 0.05; Table 2), which can be a key reason for its relatively lower pH (p < 0.05; Table 1). These acids could contribute to a sour taste and cheese flavor, and neutralize some deleterious alkaline compounds in fermented meats such as amine and pyrazine (García-González et al., 2008; Liu et al., 2020). Esters were found with relatively high aroma thresholds, generated via the esterification between carboxylic acids and alcohols, and partially provided the sweet, fruit, and/or fatty flavors in meat products (Carrapiso et al., 2015). The predominant esters were ethyl acetate and γ-butyrolactone in both FH and LH. The intensity of γ-butyrolactone was similar in FH and LH (p > 0.05), but the richness of ethyl acetate was relatively higher in FH than that in LH (p < 0.05).
Moreover, only one heterocyclic compound, 2-pentylfuran, was found in the two hams, which was also determined as the most abundant furans in Chinese bone-less hams by Li et al. (2022). 2-pentylfuran is regarded as an odor-active compound with a green and fruity flavor in dry-cured hams (García-González et al., 2008). In addition, dimethyl disulfide was the only identified sulfur-containing volatile in the two hams, which was in line with the report of Liu et al. (2020) on Jinhua hams. This chemical family is generally products of sulfur-containing amino acid catabolism or microbial metabolism, and usually provides an unpleasant flavor with low odor threshold (Pérez-Santaescolástica et al., 2018). However, dimethyl disulfide has a vegetable aroma and important contribution to the characteristic flavors of cured meats (Liu et al., 2020). Besides, both 2-pentylfuran and dimethyl disulfide showed higher concentrations in FH than those in LH (p < 0.05).
The total abundance of volatiles belonged to each chemical family was further summarized with results illustrated in Figure 1. It is shown that FH had significantly higher total intensity of ion peaks for volatiles than LH (p < 0.05). On one hand, aldehydes were the most abundant volatiles among all chemical families for both FH and LH, followed by ketones and alcohols, which were consistent with the findings of Liu et al. (2020) and Li W. et al. (2021) on volatiles from various Chinese dry-cured hams. On the other hand, LH showed remarkably higher total intensities of acids and alcohols (p < 0.05), whereas other types of volatiles were obviously less abundant for LH (p < 0.05).
The high-throughput sequencing of 16S rRNA genes was performed to investigate the bacterial community structures of Jinhua FH and LH. The α-diversity of bacteria from FH and LH, including Shannon, Simpson, ACE, and Chao 1 indexes, are shown in Table 3. Shannon and Simpson indexes represent the community diversity, and the ACE and Chao 1 indexes are associated with the community richness (Mu et al., 2020). It is clear that the two hams did not have significantly different α-diversities in terms of all indexes (p > 0.05). Besides, the numbers of observed bacterial species in FH and LH were 1,058 ± 307 and 1,262 ± 71, respectively, without marked difference (p > 0.05). These data indicate that FH and LH had an overall similar diversity and richness in bacterial communities.
Figure 4 compares the bacterial community structure of two hams at various levels, including phylum, genus, and species. As illustrated in Figure 4A, Firmicutes was the most abundant bacteria at the phylum level in FH with a relative abundance of 87.23% (p < 0.05), followed by Bacteroidetes (6.06%) and Proteobacteria (3.46%; p < 0.05). This result was in consistence with the reports of Wang et al. (2021) and Li et al. (2022) on the bacterial composition of Jinhua hams. However, the dominant bacterial phyla (>5%) in LH showed a greater variety, which included Proteobacteria (41.39%), Firmicutes (20.33%), Cyanobacteria (16.26%), Actinobacteria (8.92%), and Bacteroidetes (5.57%; p < 0.05). Compared with the bacteria in FH, the richness of Firmicutes was sharply decreased in LH (p < 0.05), but Proteobacteria, Cyanobacteria, Actinobacteria, Chloroflexi, and ε-acteraeota exhibited a significant elevation in relative abundance (p < 0.05).
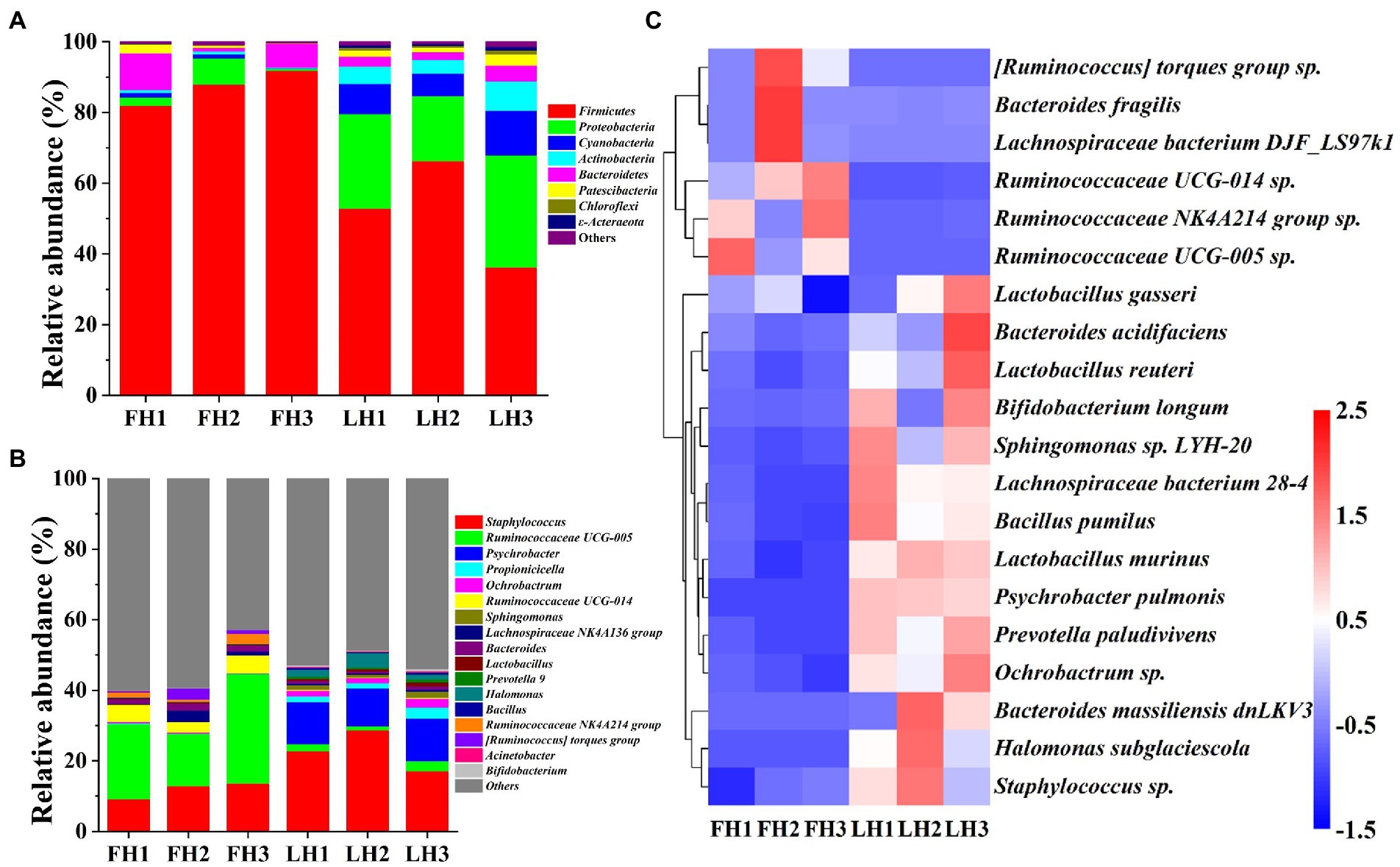
Figure 4. Bacterial community structure of Jinhua FH and LH at the phylum, genus, and species levels. (A) Relative abundance at the phylum level. (B) Relative abundance at the genus level. (C) Heatmap of HCA for the relative abundance of main species (top 20). The color gradation in panel C represents the Z-scores of corresponding relative abundances. FH, fatty ham; LH, lean ham.
At the genus level (Figure 4B), Ruminococcaceae UCG-005 (22.39%), Staphylococcus (11.91%), and Ruminococcaceae UCG-014 (4.27%) were the dominant bacteria (>2%) in FH (p < 0.05), while Staphylococcus (22.88%) and Psychrobacter (11.58%) were the richest bacterial genera in LH (p < 0.05), followed by Halomonas (2.50%), Propionicicella (2.07%), and Ruminococcaceae UCG-005 (2.02%; p < 0.05). Wang et al. (2021) reported that Staphylococcus, Psychrobacter, Halomonas, and unidentified Ruminococcaceae were among dominant bacterial genera of Jinhua hams ripened for different months, which were corresponded to our above findings. Furthermore, among the main bacterial genera with a relative abundance >1%, FH had richer Ruminococcaceae UCG-005, Ruminococcaceae UCG-014, Bacteroides, Ruminococcaceae NK4A214 group, and [Ruminococcus] torques group (p < 0.05), whereas LH showed higher richness of Staphylococcus, Psychrobacter, Propionicicella, Ochrobactrum, Sphingomonas, Lactobacillus, and Halomonas (p < 0.05).
At the species level (Figure 4C), the dominant bacteria (>1%) were Staphylococcus sp., Ruminococcaceae UCG-005 sp., and Ruminococcaceae NK4A214 group sp. for FH (p < 0.05), whereas Staphylococcus sp., Ochrobactrum sp., Psychrobacter pulmonis, and Halomonas subglaciescola for LH (p < 0.05). On the other hand, among the core bacterial species with a richness >0.5%, the richness of Ruminococcaceae UCG-005 sp. and Ruminococcaceae NK4A214 group sp. were more abundant in FH (p < 0.05), while Staphylococcus sp., Ochrobactrum sp., Psychrobacter pulmonis, Prevotella paludivivens, and Halomonas subglaciescola presented a relatively higher richness in LH (p < 0.05).
The high-throughput sequencing of ITS genes was applied to analyze the fungal community structures of Jinhua FH and LH. The α-diversity of fungi from the two hams are exhibited in Table 4. Generally, FH presented much higher Shannon, ACE, and Chao 1 indexes than LH (p < 0.05). Moreover, the number of fungal species observed from FH was 697 ± 147, approximately 2.8 times of that observed from LH (246 ± 5; p < 0.05). These results reveal that both diversity and richness of fungal community in LH was dramatically reduced in comparison to FH.
Figure 5 illustrates the fungal community structure of FH and LH at the phylum, genus, and species level. As illustrated in Figure 5A, Ascomycota was the richest fungus phylum in FH and accounted for a relative abundance of 82.79%, followed by Basidiomycota (6.06%) and Mortierellomycota (2.66%; p < 0.05). Meanwhile, Ascomycota was the only dominant fungal phylum (>5%) in LH (p < 0.05), accounting for a much higher relative abundance (97.69%) than that in FH (p < 0.05). In accordance with this finding, Lin et al. (2020) and Chen et al. (2021) also found that Ascomycota was the absolutely predominant fungus at the phylum level in Laowo dry-cured hams and Mianning hams, respectively. Besides, the relative abundances of many fungal phyla, mainly including Basidiomycota, Rozellomycota, Chytridiomycota, and Glomeromycota were significantly lower in LH than those in FH (p < 0.05).
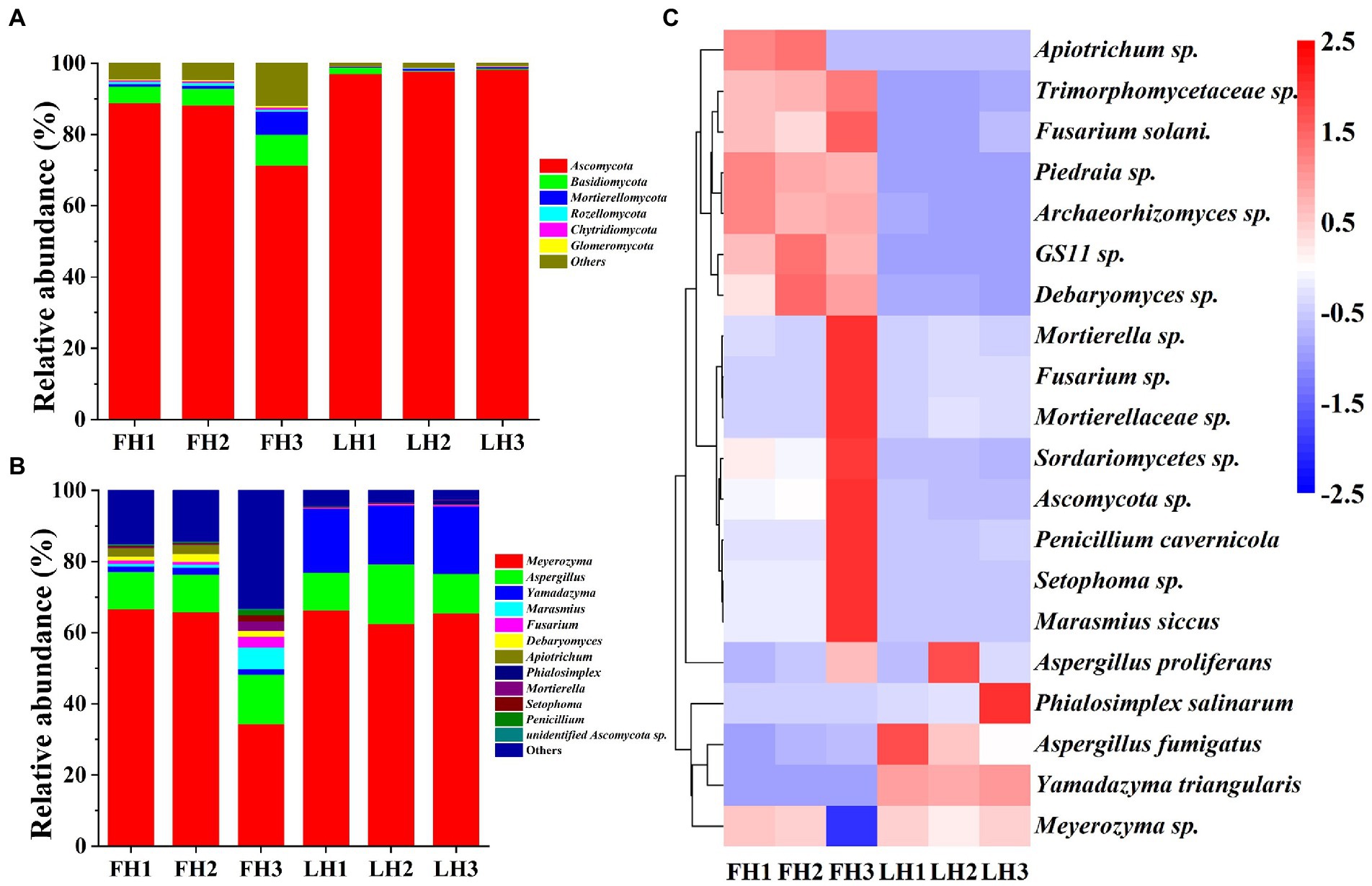
Figure 5. Fungal community structure of Jinhua FH and LH at the phylum, genus, and species levels. (A) Relative abundance at the phylum level. (B) Relative abundance at the genus level. (C) Heatmap of HCA for the relative abundance of main species (top 20). The color gradation in panel C represents the Z-scores of relative abundances. FH, fatty ham; LH, lean ham.
At the genus level (Figure 5B), Meyerozyma (55.61%) was the most abundant fungus in FH (p < 0.05), followed by Aspergillus (11.67%; p < 0.05). While Meyerozyma (64.79%), Yamadazyma (17.76%), and Aspergillus (12.82%) were the dominant fungal genera (>2%) in LH (p < 0.05). Consistent with these findings, Lin et al. (2020) also found that Yamadazyma, Meyerozyma, and Aspergillus was the most abundant fungal genus in Laowo dry-cured hams ripened for 1–3 years, respectively. On the other hand, among the core fungal genera with a relative abundance >1%, the richness of Fusarium, Debaryomyces, Marasmius, and Apiotrichum were relatively higher in FH (p < 0.05), and Yamadazyma was the only more abundant fungus in LH (p < 0.05).
At the species level (Figure 5C), Meyerozyma sp., Aspergillus proliferans, and Yamadazyma triangularis were all among the core fungi (>1%) of both FH and LH (p < 0.05). Besides, Debaryomyces sp. and Apiotrichum sp. were also the dominant fungi in FH (p < 0.05). Moreover, among the core fungal species with a relative abundance >0.5%, FH showed more abundant Marasmius siccus, Debaryomyces sp., Apiotrichum sp., Archaeorhizomyces sp., and Fusarium solani (p < 0.05), whereas LH presented a higher relative abundance in Yamadazyma triangularis (p < 0.05).
Figure 6A shows the Pearson’s correlations between 17 differential core microbial genera (p < 0.05 and relative abundance >1%; 12 bacterial genera, 2 yeast genera, and 3 mold genera) and 29 discriminative volatile flavor compounds (p < 0.05) in Jinhua FH and LH. Surprisingly, 12 of the 17 differential core microorganisms, including 10 bacterial genera and 2 yeast genera, showed significant correlations with the distinct volatiles (p < 0.05 and |r| > 0.82). Among these microbial genera, Staphylococcus, Lactobacillus, Yamadazyma, Ochrobactrum, and Sphingomonas were one type of microbes mainly positively correlating with acids, branched alcohols, and a few linear-chain alcohols (2-propanol and 3-hexen-1-ol) in the two hams (p < 0.05 and r > 0.82). Staphylococcus has been considered the most important factor in the sensory characteristics development of dry-cured hams, owning to its strong abilities of nitrate reductase, catalase, protease, and lipase (Landeta et al., 2013; Xiao et al., 2020). Lactobacillus could extend the shelf life of hams by inhibiting the growth of spoilage bacteria via its product lactic acid, which could be further utilized by other microorganisms for SCFA synthesis (Figure 6A; Table 1; Xiao et al., 2020; Hu et al., 2022). Yamadazyma has been reported positively related to the organic acid level and negatively associated with the amino acid content in hams (Lin et al., 2020; Mu et al., 2020), suggesting its potential capability of producing SCFAs from amino acids (Figure 6A). Furthermore, Lactobacillus and Yamadazyma were both core microorganisms with higher relative abundances in LH (p < 0.05; Figures 4B, 5B), which could be an important contributor to the relatively higher acid volatile level (Figure 1; Table 2) and lower pH (Table 1) of LH (p < 0.05), owning to their potential association with SCFA generation.
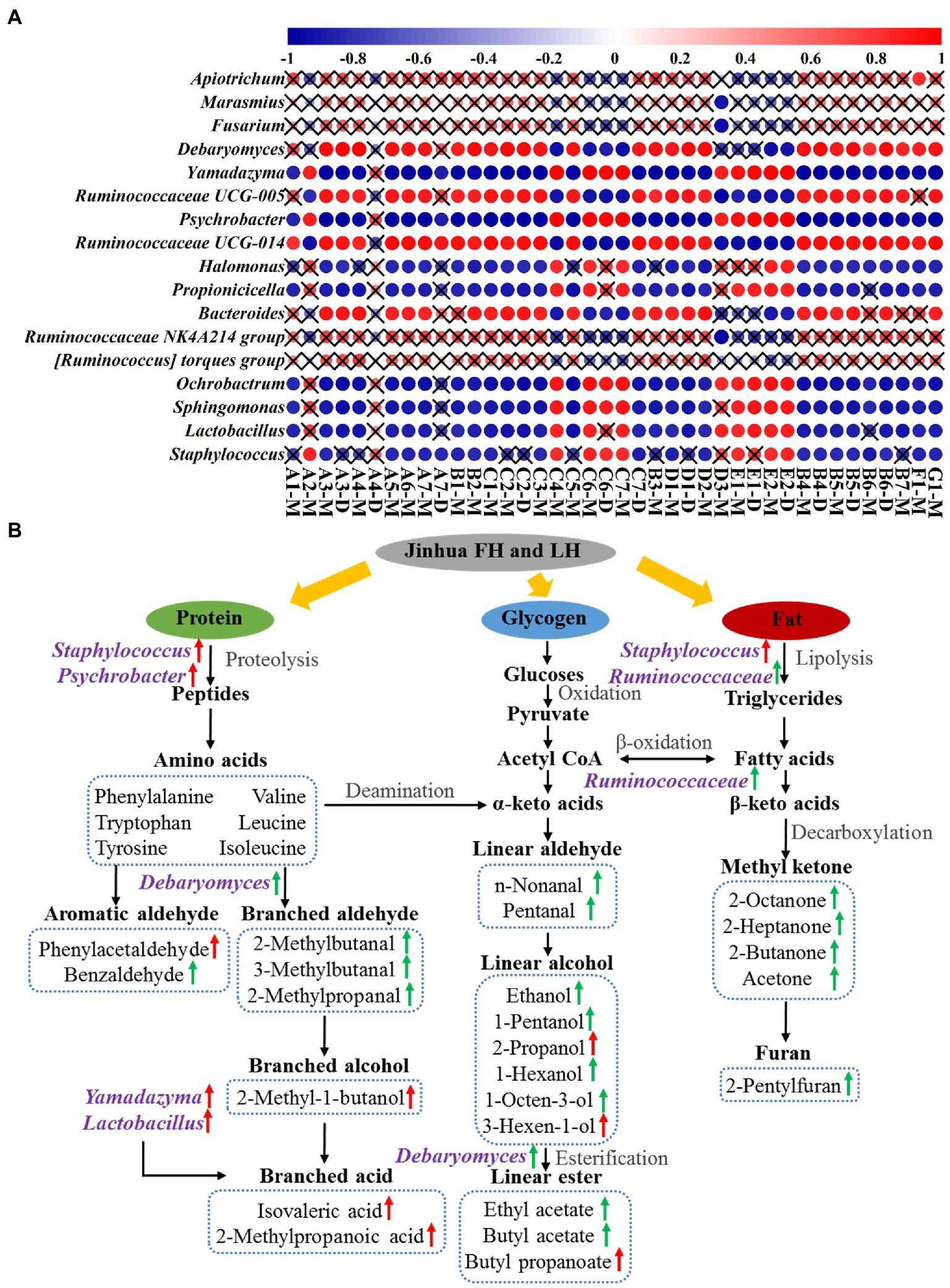
Figure 6. Pearson’s correlations (A) and potential pathways (B) between differential core microorganisms and distinct volatile flavor compounds. The color gradation and circle size in panel A represent the correlation coefficient (r value), while the pattern without a “×” symbol indicates a significant correlation (p < 0.05). The green and red arrows in panel B indicate an up-regulation in FH and LH, respectively. A1, n-nonanal; A2, phenylacetaldehyde; A3, benzaldehyde; A4, pentanal; A5, 2-methylbutanal; A6, 3-methylbutanal; A7, 2-methylpropanal; B1, 3-octanone; B2, 6-methyl-5-hepten-2-one; B3, 2-octanone; B4, 2-heptanone; B5, 3-hydroxy-2-butanone; B6, 2-butanone; B7, acetone; C1, 1-octen-3-ol; C2, 1-pentanol; C3, ethanol; C4, 2-propanol; C5, 1-hexanol; C6, 3-hexen-1-ol; C7, 3-methyl-1-butanol; D1, ethyl acetate; D2, butyl acetate; D3, butyl propanoate; E1, isovaleric acid; E2, 2-methylpropanoic acid; F1, 2-pentylfuran; G1, dimethyl disulfide; M, monomer; D, dimer.
On the other hand, Ruminococcaceae UCG-005, Psychrobacter, Ruminococcaceae UCG-014, and Debaryomyces were another type of microorganisms that positively correlated with aldehydes, ketones, most linear-chain alcohols, esters, and the other two compounds (2-pentylfuran and dimethyl disulfide) in the two hams (p < 0.05 and r > 0.82). Psychrobacter is a common bacterium in low-temperature fermented foods with substantial proteolysis ability (Broekaert et al., 2013). Zhang et al. (2021) has reported that Debaryomyces could produce branched aldehydes, such as 2-methylbutanal and 3-methylbutanal, by utilizing amino acids. Furthermore, Flores et al. (2004) reported that Debaryomyces could promote the generation of ethyl esters in dry-fermented meats. The intensities of 2-methylbutanal, 3-methylbutanal, and ethyl acetate were all positively correlated with the richness of Debaryomyces (Figure 6A), which were consistent with their findings. Moreover, the role of Ruminococcaceae family was rarely reported in fermented meat products, but it is known with capacities of fat degradation and fatty acid β-oxidation (Baars et al., 2018), which might contribute to the high-level volatiles originated from fats in FH, such as methyl ketones, furans, and some liner-chain aldehydes/alcohols (Figure 6A).
According to the present data and associated previous reports (Chen et al., 2017; Shi et al., 2019; Mu et al., 2020; Li et al., 2022), the potential metabolic network of discriminative volatiles’ formation under the involvement of core microorganisms in Jinhua FH and LH is further illustrated in Figure 6B. Briefly, the absence of fats significantly affected the microbial community structure and the flavor formation process in LH. By lipases from Staphylococcus and Ruminococcaceae in FH, fats can be degraded into fatty acids, which further generated methyl ketones and furans through decarboxylation and other reactions. Besides, the unsaturated fatty acids can be broken down through the β-oxidation by Ruminococcaceae to produce linear-chain aldehydes, which can be reduced to linear-chain alcohols and further esterified under the promotion of Debaryomyces. Whereas in LH, the protein metabolism was relatively enhanced due to the lack of fats and the action of characteristic core microbes. Under the proteolysis of Staphylococcus and Psychrobacter, more proteins were degraded into amino acids, which were then subjected to Strecker degradation and deamination for further generation of some aromatic aldehydes or linear-chained alcohols. Besides, the branched acids could be generated by Yamadazyma and some other microbes through utilizing amino acids or Lactobacillus-produced lactic acids. In summary, this metabolic network confirmed that the discriminative flavor compounds of FH were mainly β-oxidation and degradation products of fatty acids, whereas those of LH were mostly derived from the Strecker reaction or microbial metabolism of amino acids.
In conclusion, the lack of fats obviously influenced the microbial composition and flavor formation of LH, which further affected some physiochemical parameters. FH and LH did not show significant differences in redness, water activity, chemical composition, and nitrite residue, but FH had higher pH and a slightly lighter and yellower color. Besides, a total of 33 volatile flavor compounds were identified. FH and LH exhibited significant differences in 29 identified volatiles, among which LH showed higher total abundance of alcohols and acids, whereas FH had generally richer aldehydes, ketones, esters, heterocyclic compounds, and sulfur-containing compounds. Meanwhile, FH and LH also showed no significant difference in α-diversity of bacterial community, but LH had a both lower richness and diversity of fungal community than FH. The dominant microorganisms (>2%) were Ruminococcaceae UCG-005, Staphylococcus, Ruminococcaceae UCG-014, Meyerozyma, and Aspergillus in FH at the genus level, whereas Staphylococcus, Psychrobacter, Halomonas, Propionicicella, Ruminococcaceae UCG-005, Meyerozyma, Yamadazyma, and Aspergillus for LH. The analysis of Pearson’s correlation and metabolic network confirmed that the discriminative flavor compounds of FH were mainly β-oxidation and degradation products of fatty acids, while those of LH were mostly derived from the Strecker reaction or microbial metabolism of amino acids. Furthermore, Staphylococcus, Yamadazyma, Ruminococcaceae, Debaryomyces, Lactobacillus, and Psychrobacter might be the core microorganisms contributing to their differences in flavor characteristics and pH value. This work could help understand the potential pathway of characteristic microorganisms influencing flavor formation of fat-deficient dry-cured hams and provide theoretical supports for developing healthier fermented meat products.
The data presented in the study are deposited in the Sequence Read Archive of National Center for Biotechnology Information repository (https://www.ncbi.nlm.nih.gov/sra), accession number PRJNA917481.
JZ: conceptualization, methodology, writing-original draft, writing-review and editing, visualization, and funding acquisition. KZ: data curation. HL: software. SL: validation. WX: resources. LC: funding acquisition. JX: formal analysis and resources. HT: conceptualization, resources, and funding acquisition. All authors contributed to the article and approved the submitted version.
This research was financially supported by the Zhejiang Key Research and Development Program (Nos. 2021C04024 and 2022C02060), Zhejiang Natural Science Foundation (No. LY21C200003), International Cooperation Fund of Zhejiang Academy of Agricultural Science (No. 10411040122GJ0101F), Young Professionals Promotion Funds of Zhejiang Academy of Agricultural Sciences (No. 2020R15R08E01), and Scientific Research Fund for Advanced Scholar of Zhejiang Academy of Agricultural Science (No. 3300002172744).
We thank Shanghai Applied Protein Technology Co., Ltd. Co., Ltd., Shanghai, China and GAS Instrument Co., Ltd., Germany for their assistance in the analysis of microbial communities and volatile flavor compounds, respectively.
WX is employed by Jinhua Jinnian Ham Co., Ltd (Jinhua, China).
The remaining authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arroyo-Manzanares, N., Martín-Gómez, A., Jurado-Campos, N., Garrido-Delgado, R., Arce, C., and Arce, L. (2018). Target vs. spectral fingerprint data analysis of Iberian ham samples for avoiding labelling fraud using headspace-gas chromatography-ion mobility spectrometry. Food Chem. 246, 65–73. doi: 10.1016/j.foodchem.2017.11.008
Baars, A., Oosting, A., Lohuis, M., Koehorst, M., El Aidy, S., Hugenholtz, F., et al. (2018). Sex differences in lipid metabolism are affected by presence of the gut microbiota. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-31695-w
Benet, I., Guàrdia, M. D., Ibañez, C., Solà, J., Arnau, J., and Roura, E. (2015). Analysis of SPME or SBSE extracted volatile compounds from cooked cured pork ham differing in intramuscular fat profiles. LWT Food Sci. Technol. 60, 393–399. doi: 10.1016/j.lwt.2014.08.016
Benet, I., Guàrdia, M. D., Ibañez, C., Solà, J., Arnau, J., and Roura, E. (2016). Low intramuscular fat (but high in PUFA) content in cooked cured pork ham decreased Maillard reaction volatiles and pleasing aroma attributes. Food Chem. 196, 76–82. doi: 10.1016/j.foodchem.2015.09.026
Bosse, R., Müller, A., Gibis, M., Weiss, A., Schmidt, H., and Weiss, J. (2018). Recent advances in cured raw ham manufacture. Crit. Rev. Food Sci. Nutr. 58, 610–630. doi: 10.1080/10408398.2016.1208634
Broekaert, K., Noseda, B., Heyndrickx, M., Vlaemynck, G., and Devlieghere, F. (2013). Volatile compounds associated with Psychrobacter spp. and Pseudoalteromonas spp., the dominant microbiota of brown shrimp (Crangon crangon) during aerobic storage. Int. J. Food Microbiol. 166, 487–493. doi: 10.1016/j.ijfoodmicro.2013.08.013
Carrapiso, A. I., Noseda, B., García, C., Reina, R., Del Pulgar, J. S., and Devlieghere, F. (2015). SIFT-MS analysis of Iberian hams from pigs reared under different conditions. Meat Sci. 104, 8–13. doi: 10.1016/j.meatsci.2015.01.012
Chen, Q., Kong, B., Han, Q., Xia, X., and Xu, L. (2017). The role of bacterial fermentation in lipolysis and lipid oxidation in Harbin dry sausages and its flavor development. LWT Food Sci. Technol. 77, 389–396. doi: 10.1016/j.lwt.2016.11.075
Chen, L., Wang, Z., Ji, L., Zhang, J., Zhao, Z., Zhang, R., et al. (2021). Flavor composition and microbial community structure of Mianning ham. Front. Microbiol. 11:623775. doi: 10.3389/fmicb.2020.623775
Deng, J., Xu, H., Li, X., Wu, Y., and Xu, B. (2022). Correlation of characteristic flavor and microbial community in Jinhua ham during the post-ripening stage. LWT Food Sci. Technol. 171:114067. doi: 10.1016/j.lwt.2022.114067
Domínguez, R., Pateiro, M., Teixeira, A., Perez-Alvarez, J. A., Santos, E. M., Trindade, M. A., et al. (2022). “Dry-cured ham” in Production of traditional Mediterranean meat products. ed. J. M. Lorenzo (New York, NY: Humana), 57–65. doi: 10.1007/978-1-0716-2103-5_7
Flores, M., Durá, M. A., Marco, A., and Toldrá, F. (2004). Effect of Debaryomyces spp. on aroma formation and sensory quality of dry-fermented sausages. Meat Sci. 68, 439–446. doi: 10.1016/j.meatsci.2003.04.001
García-González, D. L., Tena, N., Aparicio-Ruiz, R., and Morales, M. T. (2008). Relationship between sensory attributes and volatile compounds qualifying dry-cured hams. Meat Sci. 80, 315–325. doi: 10.1016/j.meatsci.2007.12.015
Ge, Q., Gu, Y., Zhang, W., Yin, Y., Yu, H., Wu, M., et al. (2017). Comparison of microbial communities from different Jinhua ham factories. AMB Express 7, 1–8. doi: 10.1186/s13568-017-0334-0
Hu, Y., Zhang, L., Wen, R., Chen, Q., and Kong, B. (2022). Role of lactic acid bacteria in flavor development in traditional Chinese fermented foods: a review. Crit. Rev. Food Sci. Nutr. 62, 2741–2755. doi: 10.1080/10408398.2020.1858269
Landeta, G., Curiel, J. A., Carrascosa, A. V., Muñoz, R., and De Las Rivas, B. (2013). Characterization of coagulase-negative staphylococci isolated from Spanish dry cured meat products. Meat Sci. 93, 387–396. doi: 10.1016/j.meatsci.2012.09.019
Li, W., Chen, Y. P., Blank, I., Li, F., Li, C., and Liu, Y. (2021). GC×GC-ToF-MS and GC-IMS based volatile profile characterization of the Chinese dry-cured hams from different regions. Food Res. Int. 142:110222. doi: 10.1016/j.foodres.2021.110222
Li, H., Chen, Y., Tang, H., Zhang, J., Zhang, L., Yang, X., et al. (2021). Effect of lysozyme and Chinese liquor on Staphylococcus aureus growth, microbiome, flavor profile, and the quality of dry fermented sausage. LWT Food Sci. Technol. 150:112059. doi: 10.1016/j.lwt.2021.112059
Li, Z., Wang, Y., Pan, D., Geng, F., Zhou, C., and Cao, J. (2022). Insight into the relationship between microorganism communities and flavor quality of Chinese dry-cured boneless ham with different quality grades. Food Biosci. 50:102174. doi: 10.1016/j.fbio.2022.102174
Li, M., Yang, R., Zhang, H., Wang, S., Chen, D., and Lin, S. (2019). Development of a flavor fingerprint by HS-GC–IMS with PCA for volatile compounds of Tricholoma matsutake singer. Food Chem. 290, 32–39. doi: 10.1016/j.foodchem.2019.03.124
Lin, F., Cai, F., Luo, B., Gu, R., Ahmed, S., and Long, C. (2020). Variation of microbiological and biochemical profiles of laowo dry-cured ham, an indigenous fermented food, during ripening by GC-TOF-MS and UPLC-QTOF-MS. J. Agric. Food Chem. 68, 8925–8935. doi: 10.1021/acs.jafc.0c03254
Liu, D., Bai, L., Feng, X., Chen, Y. P., Zhang, D., Yao, W., et al. (2020). Characterization of Jinhua ham aroma profiles in specific to aging time by gas chromatography-ion mobility spectrometry (GC-IMS). Meat Sci. 168:108178. doi: 10.1016/j.meatsci.2020.108178
Liu, P., Wang, S., Zhang, H., Wang, H., and Kong, B. (2019). Influence of glycated nitrosohaemoglobin prepared from porcine blood cell on physicochemical properties, microbial growth and flavor formation of Harbin dry sausages. Meat Sci. 148, 96–104. doi: 10.1016/j.meatsci.2018.10.008
Martínez-Onandi, N., Rivas-Cañedo, A., Ávila, M., Garde, S., Nuñez, M., and Picon, A. (2017). Influence of physicochemical characteristics and high pressure processing on the volatile fraction of Iberian dry-cured ham. Meat Sci. 131, 40–47. doi: 10.1016/j.meatsci.2017.04.233
Mu, Y., Su, W., Mu, Y., and Jiang, L. (2020). Combined application of high-throughput sequencing and metabolomics reveals metabolically active microorganisms during Panxian ham processing. Front. Microbiol. 10:3012. doi: 10.3389/fmicb.2019.03012
Narváez-Rivas, M., Gallardo, E., and León-Camacho, M. (2014). Chemical changes in volatile aldehydes and ketones from subcutaneous fat during ripening of Iberian dry-cured ham: Prediction of the curing time. Food Res. Int. 55, 381–390. doi: 10.1016/j.foodres.2013.11.029
Parolari, G., Aguzzoni, A., and Toscani, T. (2016). Effects of processing temperature on color properties of dry-cured hams made without nitrite. Foods, 5:33 33, doi: 10.3390/foods5020033
Pérez-Santaescolástica, C., Carballo, J., Fulladosa, E., Garcia-Perez, J. V., Benedito, J., and Lorenzo, J. M. (2018). Effect of proteolysis index level on instrumental adhesiveness, free amino acids content and volatile compounds profile of dry-cured ham. Food Res. Int. 107, 559–566. doi: 10.1016/j.foodres.2018.03.001
Petrova, I., Aasen, I. M., Rustad, T., and Eikevik, T. M. (2015). Manufacture of dry-cured ham: a review. Part 1. Biochemical changes during the technological process. Eur. Food Res. Technol. 241, 587–599. doi: 10.1007/s00217-015-2490-2
Pham, A. J., Schilling, M. W., Mikel, W. B., Williams, J. B., Martin, J. M., and Coggins, P. C. (2008). Relationships between sensory descriptors, consumer acceptability and volatile flavor compounds of American dry-cured ham. Meat Sci. 80, 728–737. doi: 10.1016/j.meatsci.2008.03.015
Shi, Y., Li, X., and Huang, A. (2019). A metabolomics-based approach investigates volatile flavor formation and characteristic compounds of the Dahe black pig dry-cured ham. Meat Sci. 158:107904. doi: 10.1016/j.meatsci.2019.107904
Toledano, A. M., Jordano, R., Medina, L. M., and López-Mendoza, M. C. (2019). Behavior and effect of combined starter cultures on microbiological and physicochemical characteristics of dry-cured ham. J. Food Sci. Technol. 56, 122–131. doi: 10.1007/s13197-018-3465-7
Trevisan, M., Krogh, V., Freudenheim, J. L., Blake, A., Muti, P., Panico, S., et al. (1990). Diet and coronary heart disease risk factors in a population with varied intake. Prev. Med. 19, 231–241. doi: 10.1016/0091-7435(90)90024-E
Wang, Y., Li, F., Chen, J., Sun, Z., Wang, F., Wang, C., et al. (2021). High-throughput sequencing-based characterization of the predominant microbial community associated with characteristic flavor formation in Jinhua ham. Food Microbiol. 94:103643. doi: 10.1016/j.fm.2020.103643
Xiao, Y., Liu, Y., Chen, C., Xie, T., and Li, P. (2020). Effect of lactobacillus plantarum and staphylococcus xylosus on flavor development and bacterial communities in Chinese dry fermented sausages. Food Res. Int. 135:109247. doi: 10.1016/j.foodres.2020.109247
Zhang, J., He, S., Kong, F., Huang, S., Xiong, S., Yin, T., et al. (2017). Size reduction and calcium release of fish bone particles during nanomilling as affected by bone structure. Food Bioprocess Technol. 10, 2176–2187. doi: 10.1007/s11947-017-1987-z
Zhang, L., Huang, C., Johansen, P. G., Petersen, M. A., Poojary, M. M., Lund, M. N., et al. (2021). The utilization of amino acids by Debaryomyces hansenii and Yamadazyma triangularis associated with cheese. Int. Dairy J. 121:105135. doi: 10.1016/j.idairyj.2021.105135
Zhang, J., Zhu, L., Li, H., Tang, H., Yang, H., Zhao, K., et al. (2022). Effects of micro−/nano-scaled chicken bones on heat-induced gel properties of low-salt pork batter: physicochemical characteristics, water distribution, texture, and microstructure. Food Chem. 373:131574. doi: 10.1016/j.foodchem.2021.131574
Zhou, C. Y., Bai, Y., Wang, C., Li, C. B., Xu, X. L., Pan, D. D., et al. (2021a). 1H NMR-based metabolomics and sensory evaluation characterize taste substances of Jinhua ham with traditional and modern processing procedures. Food Control 126:107873. doi: 10.1016/j.foodcont.2021.107873
Zhou, C. Y., Pan, D. D., Cao, J. X., and Zhou, G. H. (2021b). A comprehensive review on molecular mechanism of defective dry-cured ham with excessive pastiness, adhesiveness, and bitterness by proteomics insights. Compr. Rev. Food Sci. Food Saf. 20, 3838–3857. doi: 10.1111/1541-4337.12779
Zhou, C., Xia, Q., Du, L., He, J., Sun, Y., Dang, Y., et al. (2022). Recent developments in off-odor formation mechanism and the potential regulation by starter cultures in dry-cured ham. Crit. Rev. Food Sci. Nutr. 4, 1–15. doi: 10.1080/10408398.2022.2057418
Keywords: Jinhua ham, fatty ham, lean ham, volatile flavor compound, bacterial community, fungal community, GC-IMS, high-throughput sequencing
Citation: Zhang J, Zhao K, Li H, Li S, Xu W, Chen L, Xie J and Tang H (2023) Physicochemical property, volatile flavor quality, and microbial community composition of Jinhua fatty ham and lean ham: A comparative study. Front. Microbiol. 14:1124770. doi: 10.3389/fmicb.2023.1124770
Received: 15 December 2022; Accepted: 13 January 2023;
Published: 27 January 2023.
Edited by:
Changyu Zhou, Ningbo University, ChinaReviewed by:
Ruiming Luo, Ningxia University, ChinaCopyright © 2023 Zhang, Zhao, Li, Li, Xu, Chen, Xie and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honggang Tang, ✉ emFhc3RhbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.