- National Key Laboratory of Green Pesticide, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Center for Research and Development of Fine Chemicals, Guizhou University, Guiyang, China
Camellia spiny whitefly, Aleurocanthus camelliae (Hemiptera: Aleyrodidae), is a major pest in tea, which poses a serious threat to tea production. Similar to many insects, various bacterial symbioses inside A. camelliae may participate in the reproduction, metabolism, and detoxification of the host. However, few reports included research on the microbial composition and influence on A. camelliae growth. We first applied high-throughput sequencing of the V4 region in the 16S rRNA of symbiotic bacteria to study its component and effect on the biological trait of A. camelliae by comparing it with the antibiotic treatment group. The population parameters, survival rate, and fecundity rate of A. camelliae were also analyzed using the age–stage two-sex life table. Our results demonstrated that phylum Proteobacteria (higher than 96.15%) dominated the whole life cycle of A. camelliae. It unveiled the presence of Candidatus Portiera (primary endosymbiont) (67.15–73.33%), Arsenophonus (5.58–22.89%), Wolbachia (4.53–11.58%), Rickettsia (0.75–2.59%), and Pseudomonas (0.99–1.88%) genus. Antibiotic treatment caused a significant decrease in the endosymbiont, which negatively affected the host's biological properties and life process. For example, 1.5% rifampicin treatment caused a longer preadult stage in the offspring generation (55.92 d) compared to the control (49.75d) and a lower survival rate (0.36) than the control (0.60). The decreased intrinsic rate of increase (r), net reproductive rate (R0), and prolonged mean generation time (T) were signs of all disadvantageous effects associated with symbiotic reduction. Our findings confirmed the composition and richness of symbiotic bacteria in larva and adult of A. camelliae by an Illumina NovaSeq 6000 analysis and their influence on the development of the host by demographic research. Together, the results suggested that symbiotic bacteria play an important role in manipulating the biological development of their hosts, which might help us for developing new pest control agents and technologies for better management of A. camelliae.
1. Introduction
The Camellia spiny whitefly, Aleurocanthus camelliae (Hemiptera: Aleyrodidae) (Figure 1), is a major pest to the Theaceae plants that originated in China (Kanmiya et al., 2011; Chen et al., 2018; Andrianto and Kasai, 2022) and is currently considered to be an invasive species over the world tea producing area (Uesugi et al., 2016; Jansen and Porcelli, 2018; Adi and Susanti, 2020; Rizzo et al., 2021). The main harm caused by the A. camelliae whitefly is the sucking of tea sap and the excretion of honeydew that covers the surfaces of tea leaves which subsequently promotes the growth of sooty mold. The combination of all these factors causes a decline in the health and vigor of the tea tree and a reduction in yield (Tian et al., 2020). In fact, citrus whitefly A. spiniferus had been denoted as A. camelliae before 2011 till the Japanese researchers proved the difference between the spiny whitefly on tea trees and the whitefly A. spiniferus on citrus and defined the new scientific name of Aleurocanthus camelliae for Camellia spiny whitefly in tea plant (Kanmiya et al., 2011; Uesugi and Sato, 2011).
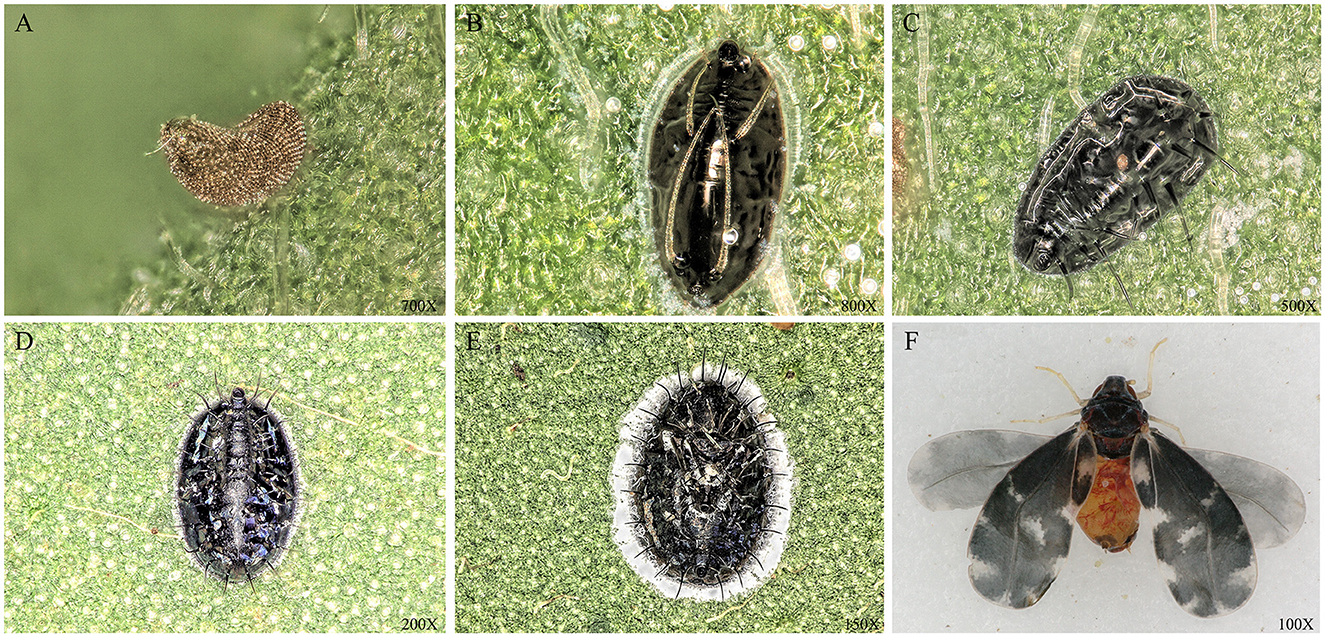
Figure 1. Morphology of A. camelliae at different stages. (A) egg; (B) the first instar nymph; (C) the second instar nymph; (D) the third instar nymph; (E) Pupa; and (F) Adult. The picture was taken under the Ultra-Depth Microscope (Leica DVM5, Germany).
Insects generally carry heritable symbiotic bacteria including primary and secondary symbionts which coevolve with host insects (Wang et al., 2020), and endosymbionts play an important role in insect growth and reproduction (Hillman et al., 2017; Akami et al., 2019; Zhang et al., 2019). Cooperation between the host insect and its microbiota is largely beneficial for all partners, but shifts in microbiota composition can also violate this mutualism (dysbiosis) and result in severe adverse effects on insects (Engl and Kaltenpoth, 2018; Grenier and Leulier, 2020). It is known that Aleyrodidae species harbor endosymbiont Bacteroides playing fundamental roles in their fitness. In addition, it has been demonstrated that endosymbionts can both protect their host against enemies (i.e., parasitoids, other bacteria, and viruses) and influence some biological traits (e.g., fitness, sex ratio, and host range), usually ameliorating the growth rate of the host under certain stress conditions (Bing et al., 2013; Su et al., 2014; Bubici et al., 2020). Limited studies related to bacterial symbionts of other close species have been examined, including A. woglumi (Pandey et al., 2013) and A. spiniferus (Bubici et al., 2020). However, bacterial symbionts of A. camelliae are poorly understood except for a report on the Wolbachia infection in Aleurocanthus cryptic species complex (Andrianto and Kasai, 2022). Therefore, this study aimed to examine the infection status and diversity of symbionts and their influence on A. camelliae.
Many 16S rRNA-based surveys of intestinal microbiota use taxonomic classification as the primary tool for comparative analysis to identify taxa that are indicative of a certain host phenotype. For example, 16S rRNA gene sequencing reveals the diversity and dynamic change of bacterial symbionts of Adelphocoris suturalis (Xue et al., 2021) and a shift in the microbiota of Diaphorina citri during the psyllid life cycle (Meng et al., 2019) and environmental-associated microbiota composition in honey bees (Jones et al., 2018). However, there was a lack of systematic research on the microorganisms hosted in A. camelliae and their function. We, therefore, by using high throughput sequencing of the bacterial 16S rRNA genes, detected the composition and diversity of bacteria in the A. camelliae to grasp the resource status of bacterial microorganisms in this whitefly.
The functions of the symbionts in whiteflies have attracted increasing research efforts in recent years. For example, the Cardinium provides protection of Bemisia tabaci against heat stress and can increase the longevity of both female and male adults and oviposition periods (Yang et al., 2021). The abundance of the Portiera in the whitefly population decreased with the increase in the nitrogen levels of the host plant (Liu et al., 2020). Hamiltonella–Cardinium coinfection induced lower fecundity, egg hatchability, and a number of female offspring, leading to a male-biased sex ratio in B. tabaci (Zhao et al., 2018). In addition, Hamiltonella elimination from the whitefly host by heat treatment and antibiotics influences the sex ratio of an insect host (Shan et al., 2019). Symbiotic bacteria of whitefly together affected host resistance to neonicotinoids (Barman et al., 2022). Reductions of primary and secondary symbionts in whiteflies following an antibiotic treatment negatively affect the host's fitness (Zhang et al., 2015).
Because most of the bacterial symbionts found in whiteflies and other sap-sucking insects are uncultivable, one way to study the functions of these symbionts is to establish host lines that differ in symbiosis treatment and then compare the performance between different host lines. For this purpose, antibiotic treatment of the hosts has frequently been employed to cure symbionts (Zhang et al., 2020; Weiland et al., 2022). This approach has also been tried with whiteflies in B. tabaci (Shan et al., 2016; Lv et al., 2018; Andreason et al., 2020). The antibiotic rifampicin was used in the experiments because all the previous articles, which present successful elimination or suppression of whitefly symbionts (Shan et al., 2016; Zhao et al., 2020), report that rifampicin was the most selective and efficient agent in their experiments between the several antibiotics tested, including tetracycline and ampicillin. In this study, we examined the effects of rifampicin treatments on the demographic development of the Camellia whitefly and its symbionts, including the primary symbiont Portiera and other secondary symbionts.
Life table analysis is the most widely used method to study life history characteristics and population dynamics, which can provide a powerful tool to describe the impact of external factors on the growth, development, reproductive capacity, and population growth of insects (Chi et al., 2020; Yang et al., 2020). However, the demographic evaluation of A. camelliae and its changes caused by symbionts have not been studied so far. This research on symbionts of A. camelliae revealed the endosymbionts and their function on host reproduction, survival, population dynamics, and adaptation to the environment. It will provide new insight into the biological function of endosymbionts and the theoretical basis for the potential biological control of A. camelliae.
2. Materials and methods
2.1. Insect rearing and maintenance
The A. camelliae individuals used in this study were collected from a tea garden in Qingzhen (N 26°55′, E 106°47′), Guizhou, China. The population was maintained on 2-year tea plants at 25 ± 1°C, 70 ± 5% relative humidity, and a photoperiod of 14:10 h (L:D) in climatic chambers, and the insects are raised for more than two generations for research. Tea plants with 6–8 true-leaf stages were selected for consumption in experiments. Molecular identification of the mtDNA COI gene and morphological observation to study the species identification of A. camelliae were referred to in previous studies (Chen et al., 2018).
2.2. Diversity analysis of symbiotic bacteria in A. camelliae
2.2.1. Sampling and DNA extraction
All A. camelliae at the third larva and 1-day-old adult stage were starved for 6 h and then were surface sterilized with 75% ethanol for 2 min and rinsed with distilled sterile water three times before DNA extraction. Insects were classified as larva (Ac. L) and adult (Ac. A) with three duplicates for each group. Total bacterial DNAs of A. camelliae were isolated from 50 individuals using the Insect DNA Isolation kit (Omega, United States), according to the manufacturer's instructions, and stored at −80°C. Before library construction, the concentration and integrality of all DNA samples were analyzed by microspectrophotometer (Implen, Germany) and agarose gel electrophoresis.
2.2.2. PCR amplification and sequence data processing
The V4 region of bacterial 16S rRNA genes was amplified from the DNA extracts using the bacterial-specific primers (V4-515F: 5' -GTGCCAGCMGCCGCGGTAA−3', V4-806R: 5'-GGACTACHVGGGTWTCTAAT-3'). PCR enrichment was performed in a 30 μl reaction containing a 10 μl template, a fusion PCR primer, and a PCR master mix. PCR cycling conditions were as follows: 98°C for 1 min, 30 cycles of 98°C for 10 s, 50°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min. After validation by 2% agarose gel electrophoresis, PCR products were purified using a Gel Extraction kit (Qiagen, Germany) and then pooled in equimolar concentrations. Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation kit (Illumina, United States), following the manufacturer's recommendations and qualified by the Agilent 5400 bioanalyzer (Agilent, United States). The validated libraries were used for sequencing on an Illumina NovaSeq 6000 platform (Illumina, United States), according to the standard Illumina procedures.
2.3. Symbiotic reduction of A. camelliae by antibiotic treatment
2.3.1. Symbiotic reduction
We used plant-mediated antibiotic delivery methods to obtain aposymbiotic whitefly samples by treating the natural adult A. camelliae, according to the previous report (Ayoubi et al., 2020), and minor improvement. Rifampicin was applied as an antibiotic agent and resolved in 2% glycerol to prepare a 1.5% antibiotic solution. Each 15 ml rifampicin (1.5%) solution was sprayed onto a tea plant with 6–8 leaves. After the solution on the surface of the leaves was completely dry (~5 h), the A. camelliae adults were subjected to each plant. In addition, the eggs (F1) produced by these adults in 24 h were kept on the plant for the following observation (the parental adults were cleared). In total, a 2% glycerol solution served as the control, and six repetitions were set for the control and rifampicin treatment (aposymbiotic) groups.
2.3.2. DNA extraction and real-time quantitative PCR (qPCR)
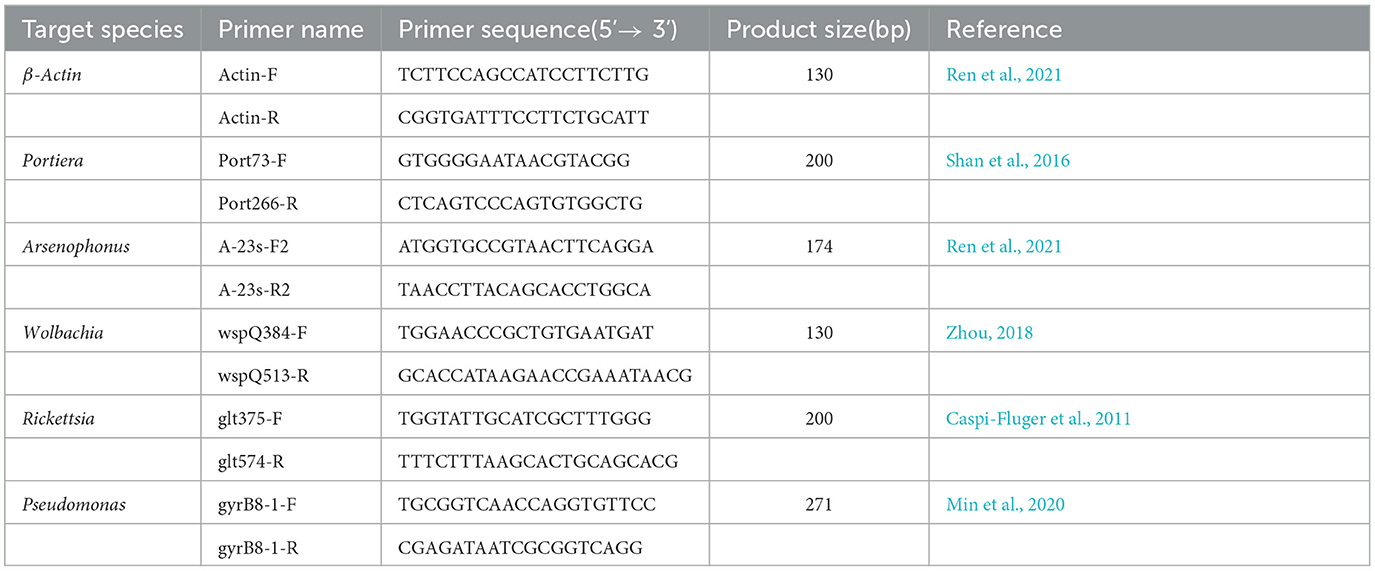
DNA from control and symbionts-suppressed treatments of insects were obtained by the same procedure as described earlier. Extraction samples were subjected to real-time quantitative PCR (qPCR) to check the bacterial content and change by using the specific primers to each endosymbiont as presented in Table 1 with the β-Actin gene serving as the selected reference gene. The PCR reaction mixture was prepared as follows: 10 μl of 2× Real Star Green Fast Mixture (GenStar, China), 0.5 μl of Primer F and Primer R, 1μl of DNA template, and proper ddH2O were added to achieve a 20 μl final reaction solution. The reaction mixture was initially denatured at 95°C for 120 s, followed by 40 cycles of amplification at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s using a Light Cycler®96 (Roche, Switzerland). qPCR data were analyzed using the 2−ΔCt method (Livak and Schmittgen, 2001), and the relative quantity of symbionts was achieved.
2.4. Life table study of A. camelliae compared with aposymbiotic treatment
Aleurocanthus camelliae pairs that had enclosed within 12 h of the collection were placed in a cage (50 cm × 50 cm × 50 cm) with 2-year tea seedlings for both control and aposymbiotic treatments as described earlier. After 24 h egg-laying on the leaves, 100 eggs were kept in each group, and the excess eggs and adult pairs were cleared. The tea seedlings with the eggs of A. camelliae were kept in climatic chambers at 25 ± 1°C, 70 ± 5% RH, and 14:10 h (L:D).
Once hatched, the first instar nymphs will settle on the leaf underside and stay till their pupa stage (the immature stages of A. camelliae begin life as mobile individuals but soon attach to host plants), so the positions of the whole nymph stage could be photographically marked and recorded. Their developmental duration and survival were then daily observed during the whole generation. After the emergence of adult A. camelliae, each pair of them was switched to the rear on a fresh tea shoot in an isolated insect box (20 × 25 × 10 cm). Fresh new tea shoots were replaced daily for adults to count the amount of newly laid eggs until all the adults died.
2.5. Data static analysis
2.5.1. Sequencing analysis of bacterial symbionts
The data obtained from independent sequencing were analyzed separately. Replicate samples of the third instar larva (Ac. L 1/2/3) and 1-day-old adult (Ac. A 1/2/3) were included in this study. Raw reads were filtered to remove adaptors, and low-quality and ambiguous bases and paired-end reads were added to tags using the Fast Length Adjustment of Short reads program (Magoc and Salzberg, 2011) to obtain the tags. The Uparse (Edgar, 2013) algorithm was used to cluster all effective tags of all samples, and a nucleotide similarity threshold of 97% was used to assign sequence reads to operational taxonomic units (OTUs). Taxonomic information was annotated for representative sequences of each OTU by using the Mothur method (Schloss, 2020) and SSU rRNA database in SILVA138.1 (Quast et al., 2013) to obtain the community composition of each sample. The Shannon, Simpson, Chao1 (Kuczynski et al., 2012), and Ace diversity indices were used to assess species diversity among samples. Alpha diversity was estimated by QIIME (Caporaso et al., 2010), and the dilution curves were drawn using R software (v 2.15.3). We calculated the abundance of each functional category based on the information in the KEGG database and the OTU abundance information. In addition, PICRUSt was used to obtain the levels of metabolic pathway information, and the abundance of each level was obtained.
2.5.2. Two-sex life table analysis
The analysis of life table raw data was performed based on the age–stage, two-sex life table theory (Chi and Liu, 1984; Chi, 1988) by using the computer program, TWOSEX-MSChart. The parameters including the age–stage-specific survival rate (sxj), age-specific survival rate (lx), age–stage-specific fecundity (fxj), age-specific fecundity (mx), net maternity (lxmx), age–stage life expectancy (exj), intrinsic rate of increase (r), finite rate of increase (λ), net reproductive rate (R0), and mean generation time (T) were calculated accordingly referring to the report (Khanamani et al., 2017). The age–stage-specific survival rate (sxj) is the probability that a newborn individual survives to age x and stage j. The age–stage-specific fecundity (fxj) represents the daily number of eggs produced per female of age x. The age–stage life expectancy (exj) indicates the total expected survival time of individuals in age x. We also determined the adult and nymphal stage longevities, the durations of preoviposition periods including the adult preoviposition period (APOP) and total preoviposition period (TPOP), as well as the number of egg masses produced during the lifetime of the females at each treatment (fecundity). The mean and standard errors of the population parameters were calculated by using the bootstrap procedure with 100,000 resamplings.
3. Results
3.1. Diversity analysis of symbiotic bacteria in A. camelliae
3.1.1. Pyrosequencing of bacterial 16S rRNA gene
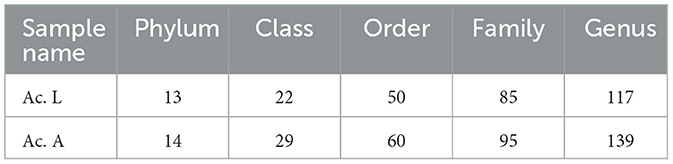
The analysis of symbiotic bacteria composition and abundance in A. camelliae was analyzed by Illumina NovaSeq 6000 via the sequencing of the 16S rRNA gene. We obtained a total of 520,336 raw tags and 491,297 effective tags, with a mean length of 425 bp in six samples (Table 2). Based on 97% species similarity, we clustered the spliced tags into OTU. The number of OTUs at the larva and adult stages is presented in Table 2. We constructed dilution curves for Ace, Chao1, Shannon, Simpson, and Good's coverage and observed species, which demonstrated the quality and credibility of sequencing quantity (Supplementary Figure 1). Good's coverage of all samples was above 99%, indicating that our sequencing results were sufficient to fully estimate the diversity of the A. camelliae bacterial community (Table 2).
3.1.2. Bacterial diversity in the A. camelliae
Among all the larvae, 13 bacterial phyla, 22 classes, 50 orders, 85 families, and 117 genera were detected. However, there were 14 bacterial phyla, 29 classes, 60 orders, 95 families, and 139 genera in the adults (Table 3). The relative abundance of the 10 most abundant bacterial groups at the phylum and genus levels within each library is shown in Figure 2. The 10 most represented phyla were Proteobacteria, Firmicutes, Bacteroidota, Verrucomicrobiota, Actinobacteria, unidentified_Bacteria, Cyanobacteria, Actinobacteriota, Spirochaetota, and Desulfobacterota. Among them, the Proteobacteria were most abundant in all the libraries and comprised 96.82% and 96.15% of the bacterial communities in the larva and adult communities, respectively. Firmicutes were the next most abundant phylum and comprised 1.22% and 2.85% of the corresponding communities (Figure 2A). At the class level, the most abundant bacterial community was Gammaproteobacteria and comprising 91.40% and 81.90% of the larva and adult communities, respectively. Alphaproteobacteria and Clostridia were the next most abundant classes. At the order level, the three most abundant bacterial communities are Pseudomonadales, Enterobacterales, and Rickettsiales and represented 68.18%, 23.12%, and 5.29% of all of the OTUs in the larvae, respectively, and 75.35%, 6.51%, and 14.17% in adults, respectively. Overall, the 10 most abundant families identified were the Halomonadaceae, Morganellaceae, Anaplasmataceae, Rickettsiaceae, Pseudomonadaceae, Enterobacteriaceae, Ruminococcaceae, Lachnospiraceae, Enterococcaceae, and Hafniaceae. Halomonadaceae dominated the whole community and represented 67.15% of all OTUs in the larva community and 73.33% of those in the adult community. The Morganellaceae (22.90%) accounted second in the larva while the Anaplasmataceae (11.58%) accounted second in the adult stage (data for class, order, and family are available in Supplementary Figure 2).
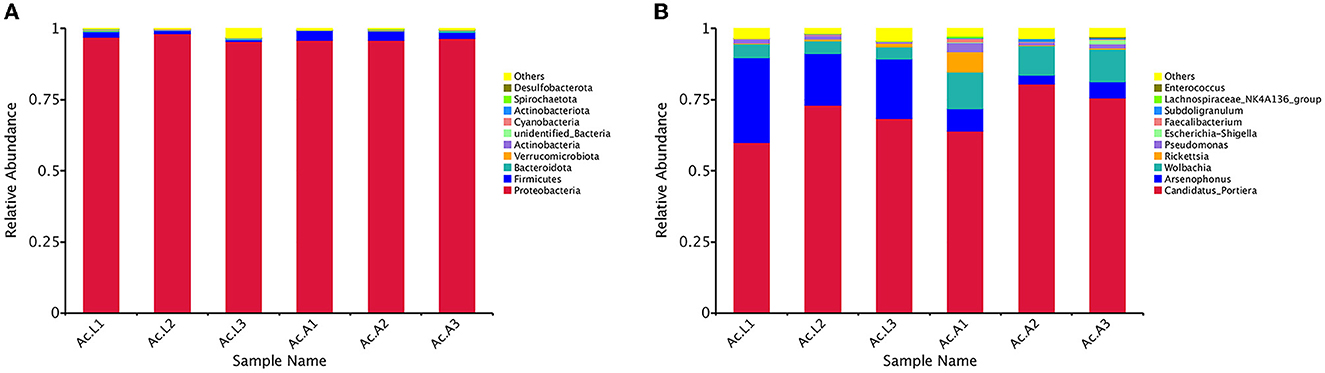
Figure 2. Bacterial community dynamics among larvae and adults in A. camelliae. (A) Relative abundance of bacteria communities at the phylum level in different groups. (B) Relative abundance of bacteria communities at the genus level in different groups.
At the general level, the first 10 compositions were Candidatus_Portiera, Arsenophonus, Wolbachia, Rickettsia, Pseudomonas, Escherichia-Shigella, Faecalibacterium, Subdoligranulum, Lachnospiraceae_NK4A136_group, and Enterococcus (Figure 2B). Candidatus_Portiera ranked first and comprised 67.15% and 73.33% of the bacterial communities in the larva and adult communities, respectively. Arsenophonus was the most abundant secondary symbiont in the larva stage A. camelliae and accounted for 22.89%, while Wolbachia was the most abundant secondary symbiont in adults A. camelliae and accounted for 11.58%. In addition, with the growth of A. camelliae from larvae to adults, the quantity of secondary symbiotic bacteria Arsenophonus decreased and Wolbachia increased (Figure 2B).
The abundance and diversity of symbiotic bacteria are reflected through multiple indicators (Chao1, Ace, Shannon, and Simpson indices). The results showed that the abundance of symbiotic bacteria in adults was higher than that in larvae, and the diversity is not different from that of larvae (Table 2). We selected the bacterial genera with the top 35 abundance ratios and drew heat maps based on their relative abundance in larvae and adults (Figure 3). Different colors in the same row indicate the difference in abundance between larvae and adults in A. camelliae. The left side showed the similarity between bacteria. The analysis shows that the relative abundance of the most symbiotic bacteria in the adult stage is higher than that in the larva stage which is consistent with the species analysis results. The relative abundance of Arsenophonus in the larva population is higher than that in the adult population. The relative abundance of Portiera, Wolbachia, Rickettsia, and Pseudomonas in the adult population is higher than that in the larva population (Figure 3). It further indicates that the characteristics of symbiotic bacteria under different insect states are different at the two principal component levels.
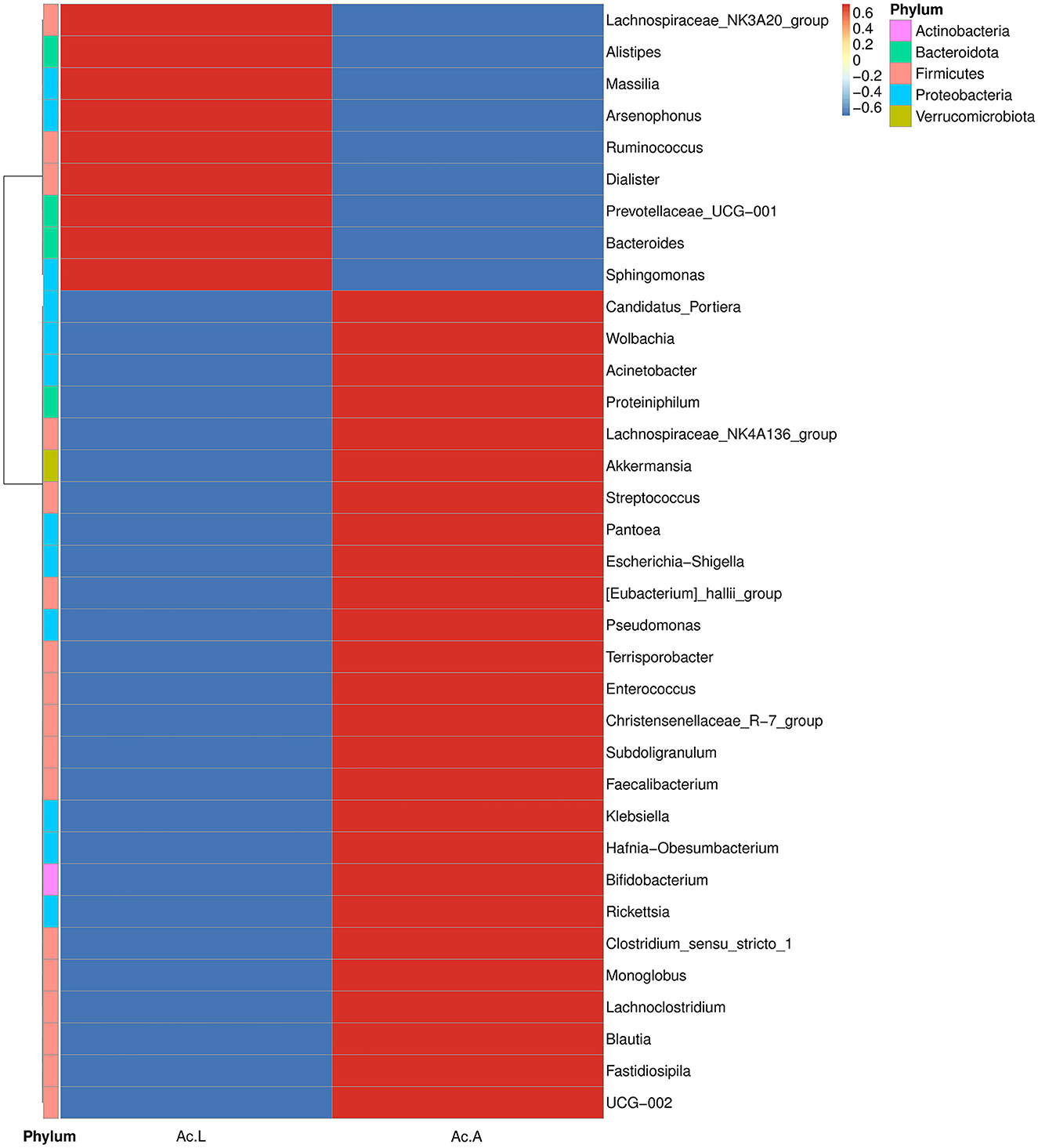
Figure 3. Heat map analysis of the top 35 microbial populations with relative abundance at A. camelliae. Ac. L and Ac. A refers to larvae and adults of A. camelliae, respectively.
3.2. Suppressed symbiont titer under antibiotics treatment
We confirmed the abundance and reduction of the bacterial symbionts in A. camelliae by qPCR in control and plant-mediated antibiotic treatment. This bar chart compared the relative symbionts' abundance in the third nymph and adult stages in A. camelliae considering the aposymbiotic treatment (Figure 4). Consistent with the NovaSeq 6000 sequencing, all five targeted symbionts detected by qPCR represented the same relative density and change trend except Wolbachia and Arsenophonus. Portiera as the primary symbiont accounted overwhelmingly highest in both stages. For the secondary symbionts, the relative density of Wolbachia is lower than Arsenophonus in both stages by qPCR, but Wolbachia is higher than Arsenophonus in adults by 16s rRNA sequencing. The bacterial symbionts including Portiera, Arsenophonus, Rickettsia, Wolbachia, and Pseudomonas in the rif (rifampicin treatment) were all dramatically declined but not completely cleared (Figure 4). For the third nymph A. camelliae, the levels of Portiera, Arsenophonus, Wolbachia, Rickettsia, and Pseudomonas were ~3-, 10-, 14-, 17-, and 60-folds lower than that of the control group, respectively. For the adult stage, the symbionts were approximately 3-, 2-, 26-, 13-, and 1-folds reduced than that of the control group.
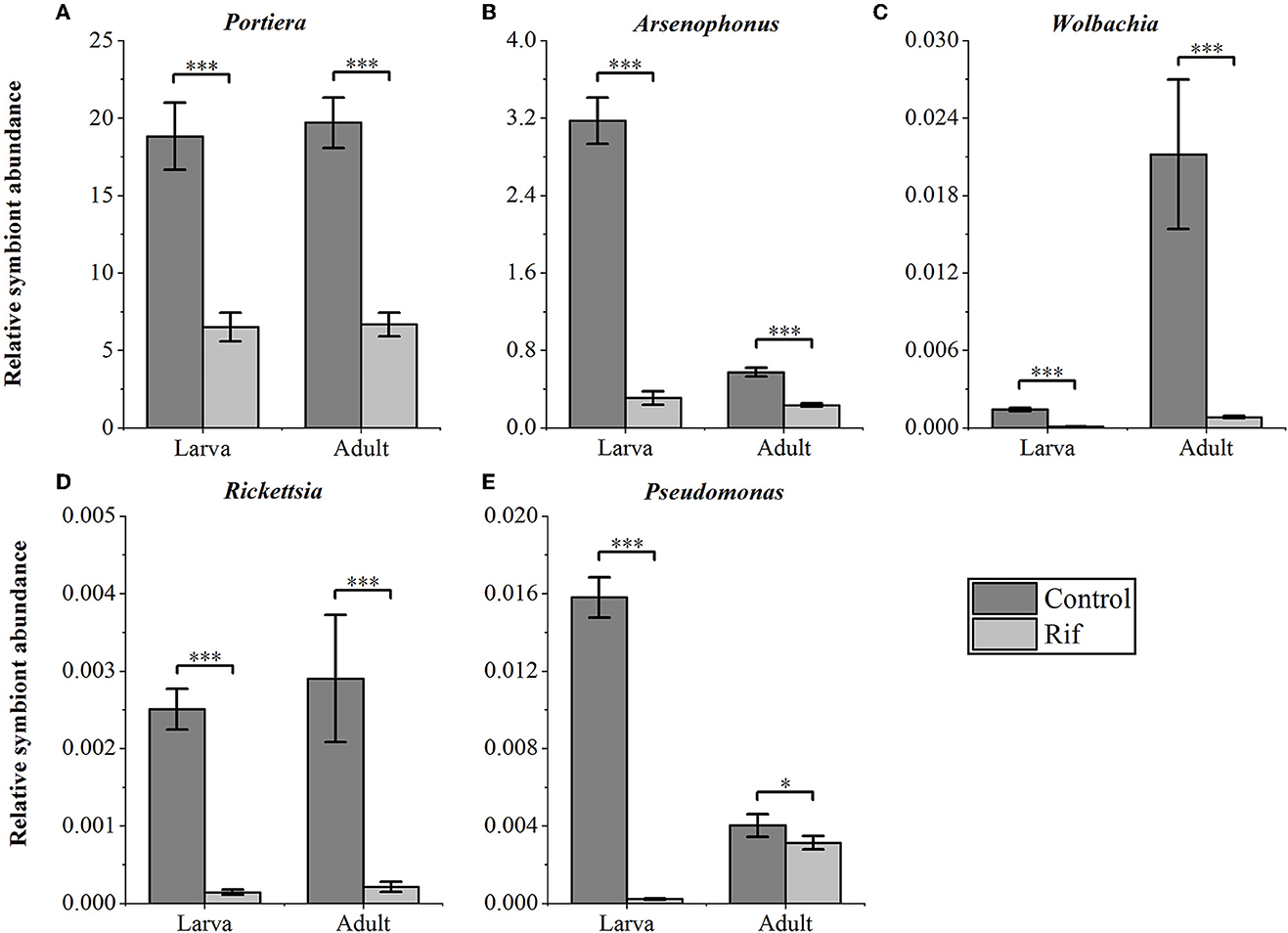
Figure 4. qPCR quantification of bacterial symbionts in A. camelliae accompanied by rifampicin (Rif) treatments in the larva (3rd nymph) and adult stages. (A) Portiera, (B) Arsenophonus, (C) Wolbachia, (D) Rickettsia, and (E) Pseudomonas. The error bars indicate the standard errors (*P < 0.05, ***P < 0.001, t-test. N = 6).
3.3. Life table study of A. camelliae and influence of rifampicin
3.3.1. Development of the A. camelliae in different age stages
Eggs collected from rif (rifampicin treatment) and the control group were involved to check their fitness analysis including population growth parameters, duration of different stages, adult preoviposition period (APOP), total preoviposition period (TPOP), and fecundity to achieve the life table of A. camelliae.
The duration of the stage for egg, first to third instar nymph, and total preadult of F1 A. camelliae in aposymbiotic insect (rifampicin treatment) were significantly longer than those of the symbiotic insect (control), though with no significant difference in the pupal stage (Table 4). However, the TPOP of the aposymbiotic insect was ~7 days extended compared with 50.29 days in the control group (Table 4), while the following APOP had no significant difference for the two groups. The fecundity of aposymbiotic A. camelliae was 21.88%, and ~32.2% decreased from 31.32% offspring in the control group. In addition, the number of female and male offspring was close in the control group with 34 female and 30 male (F: M = 1.13:1) insects and changed to 24 female and 14 male (F: M = 1.71:1) insects when the symbiont was suppressed. The offspring of both sexes dropped significantly, and an obvious disequilibrium toward the dominance of females (1.71:1) happened after the symbiont was suppressed. These observations suggested that the males are more susceptible to antibiotic treatment than the females.
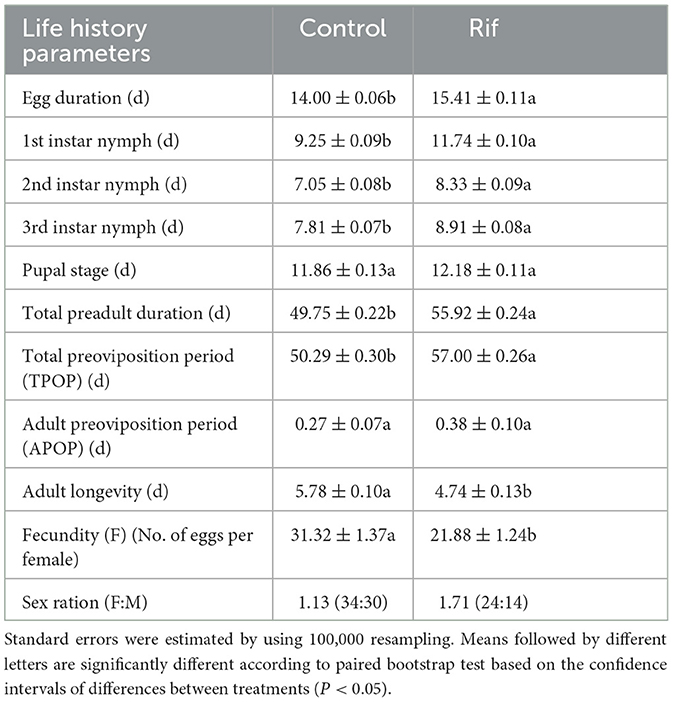
Table 4. Stages and developmental periods (days) (mean ± SE) of A. camelliae from the rifampicin treatment and control group.
3.3.2. Survival and reproduction rate of the A. camelliae
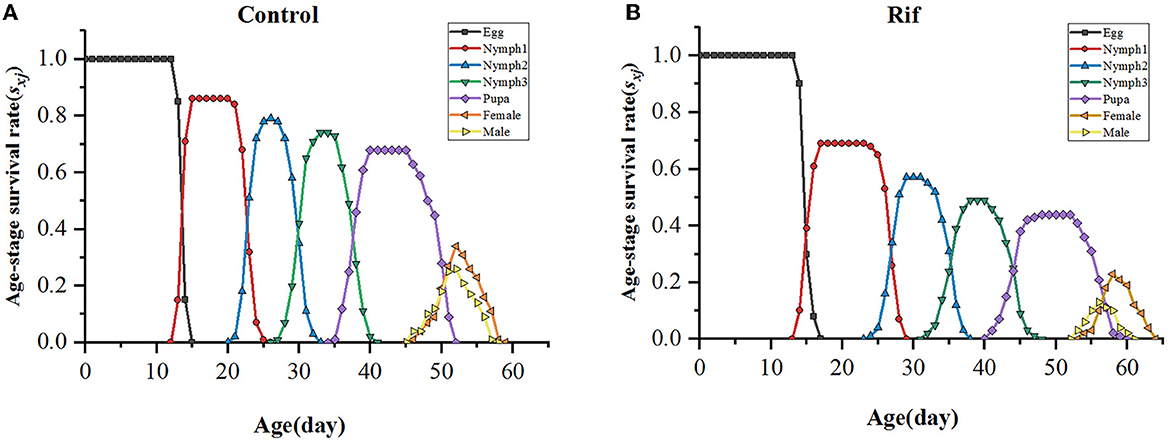
The age–stage-specific survival rate (sxj) of A. camelliae (Figure 5) between aposymbiotic insect and control showed different survivorship and various developmental rates. The survival rate of the offspring of A. camelliae exposed to rifampicin decreased rapidly and was systemically lower than that of the control. The probability of the aposymbiotic insects developing from egg to first instar nymph was 0.69, which is lower than 0.86 in symbiotic insects. The second instar nymph and third instar nymph were 0.57 and 0.49, respectively, which is also lower than the survival probability of the symbiotic insect (0.78 and 0.74). Similarly, the probability for adult survival (0.36) remarkably declined to that of the symbiotic insect (0.60). This indicated that the emergence or survival rate of both females and males in the aposymbiotic whitefly was consistently lower than the natural insects. Especially, male survival was strongly eliminated resulting in sex ratios biased to females.
The probability for age-specific survival rate and fecundity was also evaluated, as shown in Figure 6. The age-specific survival rate (lx) shows the potentiality that a newborn individual will survive to age x and is calculated by pooling all individuals of both sexes. Therefore, the curve of lx is a simplified version of the sxj curve, which is presented in Figure 6. It could be seen that aposymbiotic insects caused a dramatic decline in the lx curve and covered a longer age (65 days) when paralleling to that of symbiotic insects. Considering the data in the sxj curve, the larger slope of the lx curve in aposymbiotic insects was mainly due to the lower egg hatchability and higher mortality of the 1st and 2nd nymph stages. The age–stage-specific fecundity (fxj), age-specific fecundity (mx), and net maternity (lxmx) of female adults increased first and then decreased with the extension of time (Figure 6). With respect to the fxj curve, the oviposition in the symbiotic insect started on the 47th day and lasted for 13 days till death, and in the aposymbiotic insect, the oviposition started on the 55th day and lasted for 10 days. However, the top fecundity in the natural group reached 7.3 and the apo-group was 6.1. It is clear aposymbiotic insects caused the oviposition period to shrink and fecundity to fall. It can be referred that the future population of symbiont removal insects would be eliminated. Similarly, the age-specific fecundity (mx) and net maternity (lxmx) covered the same oviposition duration and lower fecundity in aposymbiotic insects. Though the mx curve showed an exceptionally top fecundity with 4.92 eggs oviposited at day 60, the net maternity (lxmx) was still lower when accompanied by an age-specific survival rate (lx), which was quite low (Figure 6).
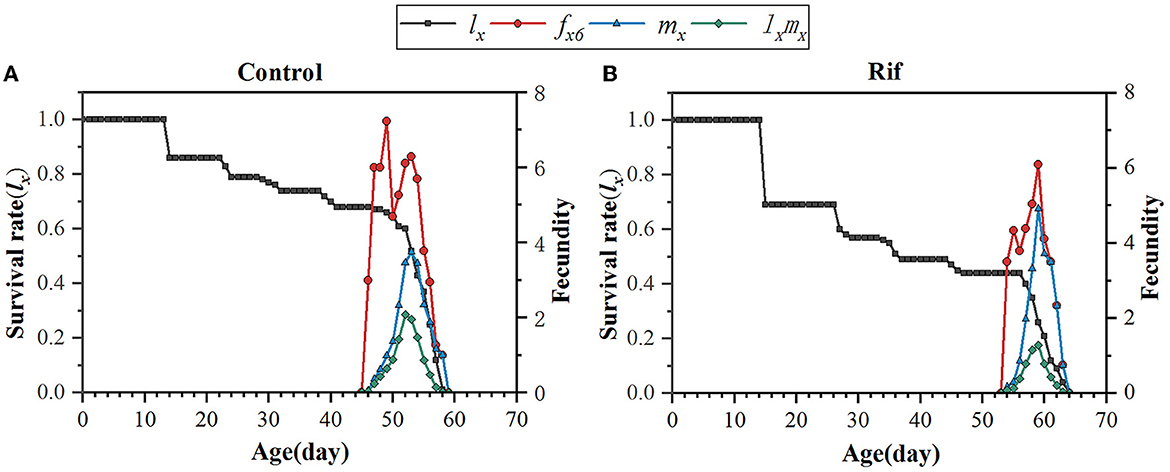
Figure 6. Age-specific survival rate (lx), age–stage-specific fecundity (fxj), age-specific fecundity (mx), and net maternity (lxmx) of A. camelliae in the control (A) and the rif-treatment (B).
In addition, the age–stage-specific reproductive value (vxj) is estimated and illustrated in Figure 7. As data showed, the climbing trend of reproductive value (vxj) with age increasing in the two groups was similar except for the longer stage duration in the aposymbiotic group, and reproductive value reached a maximum around the time of TPOP. For example, the highest reproduction value occurred on the 47th day in the control group, which was close to the day of TPOP (50.29) (Table 4) and vxj in rif treatment arrived at peak value at day 55 where the TPOP was 57.00 (Table 4). The remarkable difference between both appeared in the adult stage where the top reproductive value of female A. camelliae was dropped by 33% in aposymbiotic insects compared with the control group (Figure 7).
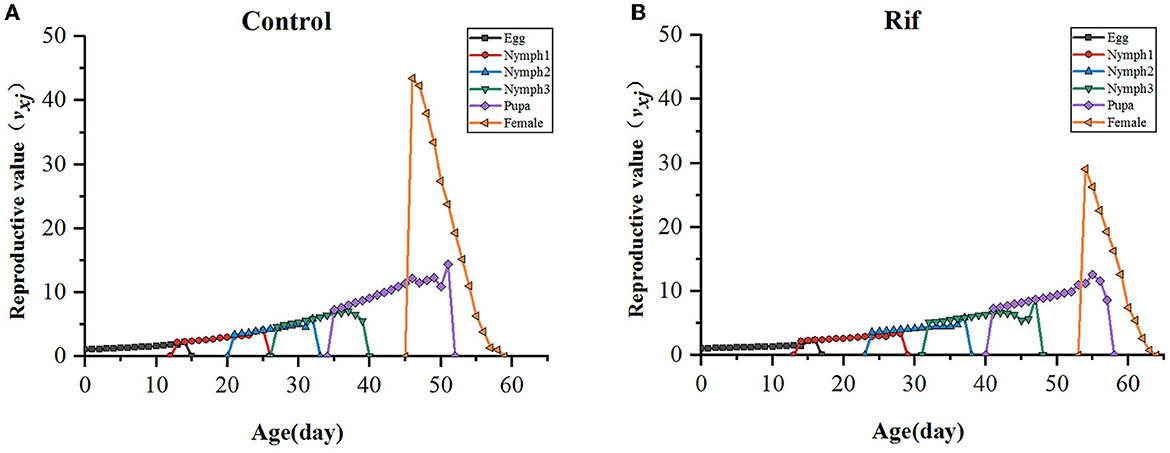
Figure 7. Age–stage-specific reproductive value (vxj) of A. camelliae. (A) Control, (B) Rif-treatment.
3.3.3. Life expectancy of A. camelliae
The life expectancy (exj) indicates the length of time that an individual A. camelliae of age x and stage j is expected to live. The curve in both groups decreased with age and increased with a degraded level in aposymbiotic insects. The life expectancy of newly hatched eggs of the control and the aposymbiotic insects was estimated to be 45.07 and 39.64 days, respectively (Figure 8).
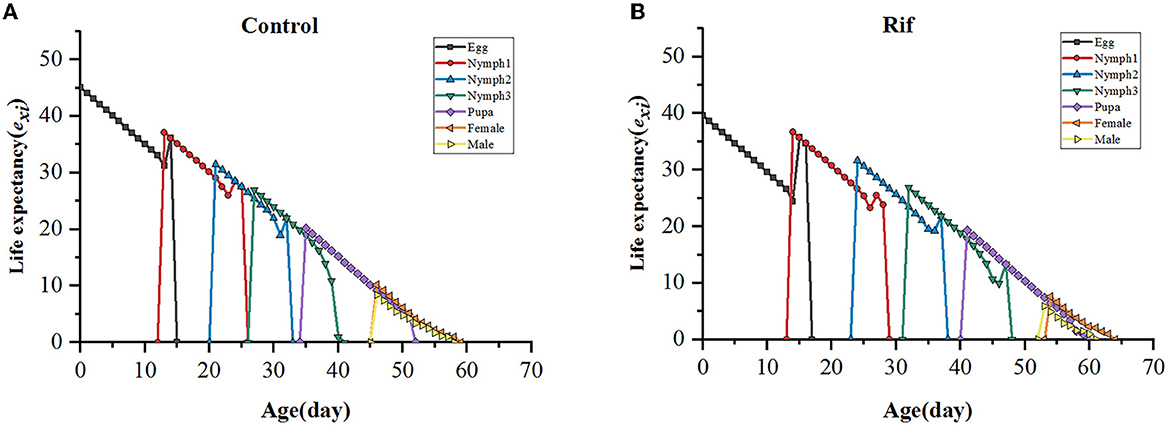
Figure 8. Age–stage life expectancy (exj) of A. camelliae in the control (A) and the rif-treatment (B).
3.3.4. Life table parameters of A. camelliae
The intrinsic rate of increase (r) was 0.0279 in the aposymbiotic group that is significantly lower than 0.0446 in the control population. In addition, the net reproductive rate (R0) and finite rate of increase (λ) of aposymbiotic insects were significantly lower than that of control insects. The mean generation time (T) was significantly prolonged in aposymbiotic insects, whereas there was no significant difference in gross reproductive rates (GRR) in the two groups of A. camelliae (Table 5).
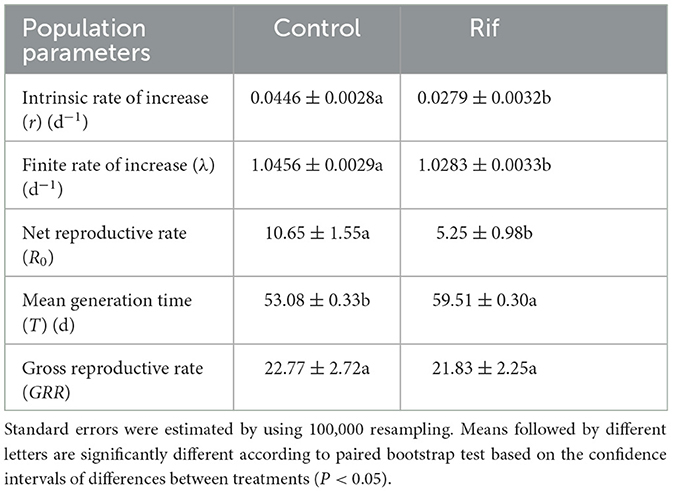
Table 5. Population parameters (means ± SE) of A. camelliae in the control and the rif-treatment calculated by using the age–stage, two-sex life table.
4. Discussion
Bacterial symbionts play prominent roles in their insect hosts. Many reports related to other whitefly insects, such as Bemisia tabaci (Shan et al., 2016; Su et al., 2016; Hashmi et al., 2019; Zhao et al., 2020), proved the effects of symbiotic bacteria on the host growth and development. However, research on symbionts of spiny whitefly and their function was lacking. To make clear this respect, we checked the community composition and diversity of symbiotic bacteria of A. camelliae by 16S rRNA high-throughput sequencing and investigated their biological implications by using the age–stage two-sex life table with comparing the aposymbiotic and natural groups.
Our present results proved that the symbiotic bacteria of A. camelliae were abundant. Interestingly, it can be observed from the aspect of symbiotic bacteria diversity that larva and adult endosymbiont community diversities did not show obvious changes. We inferred that endosymbionts are in a relatively stable state in A. camelliae to mutualism with host insects (Harris et al., 2010). We found that the main five symbiotic bacteria genus were Portiera, Arsenophonus, Wolbachia, Rickettsia, and Pseudomonas, which together composed approximately 94.83–96.24% in A. camelliae. Those type of symbiotic bacteria composition in A. camelliae is similar to that in Bemisia tabaci (Zchori-Fein et al., 2014; Hashmi et al., 2019) though different in relative content. However, there is an exception that no symbiotic Hamiltonella was found in A. camelliae, which was reported to exist in Bemisia tabaci (Bello et al., 2019). Therefore, we deduced that insects of the same genus have similar symbiotic bacteria.
Proteobacteria are a major group (phylum) of gram-negative bacteria and most important endosymbionts and include a wide variety of bacterial genera in different insects (Yun et al., 2014). It was also found to be dominated in A. camelliae in this present result, which was similar to the findings in other Hemipteran insects (Lim and Ab Majid, 2020; Liu et al., 2022). All five main genera, Portiera, Arsenophonus, Wolbachia, Rickettsia, and Pseudomonas, belonged to the Proteobacteria phylum. Among them, Portiera as the primary endosymbiont was the most abundant genera in a stable content level throughout the development cycle of A. camellia, indicating that Portiera plays a lasting symbiotic role in the growth, development, and survival of A. camellia. Portiera can evade autophagy of the whitefly bacteriocytes by activating the TOR (target of rapamycin) pathway by providing essential nutrients (Wang et al., 2022). Interestingly, Portiera can facilitate the whitefly performance by cooperation with the panBC gene for pantothenate production (Sun et al., 2022). Other secondary endosymbionts' content and relative level would differ among the nymph and adult stages. For example, the relative abundance of Arsenophonus (22.89%) was higher than that of Wolbachia (4.52%) in the individual larva but lower in the adult stage where Arsenophonus accounted for 5.57% and Wolbachia increased to 11.58%. This fluctuation suggested a competition between Arsenophonus and Wolbachia in the growth of A. camelliae. This mutual exclusion relationship may be caused by its similar harbor in host cells and competition for limited resources as reported in other insects (Gottlieb et al., 2008; Qu, 2013). Furthermore, the substantial increase in Wolbachia level from the larva to adult stages in A. camelliae agreed with Wolbachia changes in Diaphorina citri (Meng et al., 2019) in our early report where Wolbachia symbiotic level kept increasing with the growth of tea green leafhopper (Zhang et al., 2022). These results from 16S rRNA sequences were confirmed by the qPCR method. The relative content was consistent in both ways, and all five investigated endosymbionts showed the same relative abundance level and changing tendency from the larva to adult stages in two analysis ways (Figures 2, 4). For example, the Portiera content kept at the highest level in both stages (Figure 4A), Arsenophonus dropped in adults (Figure 4B), and Wolbachia increased (Figure 4C). This indicated that this qPCR result was consistent with Illumina NovaSeq 6000, and the comparison was reliable.
Rifampicin was often applied to achieve aposymbiotic insects to check the change and influence of symbionts (Ahmed et al., 2010; Ren et al., 2021). We also employed rifampicin treatment to perform the symbiotic reduction in A. camelliae. We failed to selectively remove secondary symbionts without affecting the primary symbiont in A. camelliae. Some articles reported success in selectively removing the secondary symbiont Hamiltonella or Wolbachia in B. tabaci, using the same antibiotic rifampicin without affecting the primary symbiont Portiera (Xue et al., 2012; Su et al., 2015). Our results indicated that rifampicin could sufficiently reduce all investigated symbiotic bacteria of A. camelliae. Furthermore, Portiera was greatly reduced in rif-treatment but inhibited to less extent compared to secondary symbiotic bacteria changes. This phenomenon was also reported in Bemisia caused by rifampicin treatment (Shan et al., 2016; Zhao et al., 2020).
The age–stage two-sex life table can help researchers systematically understand the impact of removing symbiotic bacteria on the survival rate, development rate, and reproduction rate of insect populations, as well as the comprehensive impact of various factors on population growth (Chi et al., 2020). We first studied the role of symbiotic bacteria in A. camelliae by using the age–stage two-sex life table method. Our result indicated that rif-treatment markedly affected their biological parameters including the net reproductive rate (R0), intrinsic rate of increase (r), and finite rate of increase (λ) (Table 5). Among them, the intrinsic rate of increase (r) is a basic ecological feature helpful to estimate the population growth potential (Roy et al., 2003). In our present result, (r) was decreased about 2-fold in the rifampicin treatment compared to the control. After symbiotic reduction, this indicated that the preoviposition period (including TPOP and APOP) was prolonged, and the adult longevity was shortened. Moreover, the control group had longer life expectancy and age–stage-specific survival rates in comparison to the rifampicin treatment. These changed biological traits negatively affected the development of A. camelliae, ultimately resulting in the reduction of the offspring population. The present results showed that aposymbiotic action had a clear unfavorable influence on A. camelliae, which is highly consistent with early reports (Karamipour et al., 2021). For example, bacterial symbiont elimination had negative effects on nymphal development and adult emergence of Brachynema germari, Acrosternum heegeri, and Acrosternum arabicum (Kashkouli et al., 2019) and adverse effects on life history traits of Graphosoma lineatum (Karamipour et al., 2021).
Symbiotic bacteria can help host insects degrade nutrient macromolecules in food and convert them into small molecules that can be directly used to provide missing nutrients for host insects. Endosymbiotic bacteria play an important role in the nutrition utilization of host insects (Frago et al., 2012). Luan et al. (2015) proved that Portiera, the primary symbiotic bacteria of Bemisia tabaci, was mainly involved in the nutrient metabolism of the host and could provide essential amino acids for the host. Previous studies have shown that Portiera may help whitefly improve their performance, and Portiera has a close nutritional relationship with their host during host plant acclimation (Hu and Tsai, 2020). The removal of Arsenophonus increased the developmental time of the immature stages and reduced the values of different life history parameters in Ommatissus lybicus (Karimi et al., 2019). We simply illustrated the function of symbiotic bacteria of A. camelliae by metagenomic analysis served by PICRUSt (Supplementary Figure 2D), which allows inference of the functional profile of the symbiotic bacterial consortia. It suggested that metabolism is highly responsible for the relative abundance of function. Whether the effect of symbiotic bacteria of A. camelliae is related to the synthesis of amino acids or other respect needs further research. It was needed to mention that rifampicin is broad spectrum and resulted in sufficient changes in symbiont titers of A. camelliae and downstream severe fitness defects, including elevated nymphal mortality and reduced population growth parameters. These changes in the population growth may be caused by the collaboration of all consortia of a specific bacterium. However, each specific bacterium and its function could not be specified in this present way. Though it has been reported that rif could specifically reduce Buchnera without affecting other bacterial compositions of Aphis gossypii (Ayoubi et al., 2020). Classification of the specific bacterium may be realized by the combination of sterilization and infection. On the other hand, this biological change of A. camelliae might not only involve bacterial clearance but also other changes in the microenvironment of the host.
5. Conclusion
Taken together, we sequenced the A. camelliae 16S rRNA gene through the Illumina NovaSeq 6000 platform, which directly revealed the structure of the bacterial community in the larva and adult stages and proved that Candidatus Portiera, Arsenophonus, Wolbachia, Rickettsia, and Pseudomonas genera of Proteobacteria dominated the whole life cycle of this whitefly. In addition, we confirmed that the rifampicin treatment can suppress the endosymbiont of A. camelliae and cause defective host fitness including a longer preadult stage in the offspring generation, a lower survival rate, decreased intrinsic rate of increase (r), and net reproductive rate (R0).
The present study indicated that endosymbionts played an important role in the normal development and growth of A. camelliae. Regarding the applied aspect of Camellia spiny whitefly control, the rapid reduction in the primary and secondary symbionts and consequent death of the whitefly hosts indicate that antibiotics or agents targeted on these bacteria may possess the properties of rational insecticides to be used as part of a whitefly management program.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA910681.
Author contributions
YT and BG conducted the experiments. YT and JW designed and performed the experiments. YT, QZ, and CL analyzed the data. LJ and XZ conceived and supervised the project. XZ and YT wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Transformation Projects of Major Scientific and Technological Achievements of Guizhou Province, [2022]006, the Science and Technology Project of Guizhou Province, [2015]5020, and the Scientific Research Projects of Major Agricultural Industries of Guizhou Province, [2021]1105.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1124386/full#supplementary-material
References
Adi, M. B., and Susanti, D. (2020). Short communication: first record of Aleurocanthus camelliae (Homoptera: Aleyrodidae) in Indonesia, an invasive pest on various medicinal plants. J. Tumbuhan Obat. Indonesia 13, 94–100. doi: 10.22435/jtoi.v13i2.3869
Ahmed, M. Z., Ren, S. X., Xue, X., Li, X. X., Jin, G. H., and Qiu, B. L. (2010). Prevalence of endosymbionts in Bemisia tabaci populations and their in vivo sensitivity to antibiotics. Curr. Microbiol. 61, 322–328. doi: 10.1007/s00284-010-9614-5
Akami, M., Ren, X. M., Qi, X., Mansour, A., Gao, B., Cao, S., et al. (2019). Symbiotic bacteria motivate the foraging decision and promote fecundity and survival of Bactrocera dorsalis (Diptera: Tephritidae). BMC Microbiol. 19, 229. doi: 10.1186/s12866-019-1607-3
Andreason, S. A., Shelby, E. A., Moss, J. B., Moore, P. J., Moore, A. J., and Simmons, A. M. (2020). Whitefly endosymbionts: biology, evolution, and plant virus interactions. Insects 11, 775. doi: 10.3390/insects11110775
Andrianto, E., and Kasai, A. (2022). Wolbachia in black spiny whiteflies and their new parasitoid wasp in Japan: evidence of the distinct infection status on Aleurocanthus camelliae cryptic species complex. Insects 13, 788. doi: 10.3390/insects13090788
Ayoubi, A., Talebi, A. A., Fathipour, Y., and Mehrabadi, M. (2020). Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Sci. 27, 86–98. doi: 10.1111/1744-7917.12603
Barman, M., Samanta, S., Upadhyaya, G., Thakur, H., Chakraborty, S., Samanta, A., et al. (2022). Unraveling the basis of neonicotinoid resistance in whitefly species complex: role of endosymbiotic bacteria and insecticide resistance genes. Front. Microbiol. 13, 1793. doi: 10.3389/fmicb.2022.901793
Bello, V. H., Watanabe, L. F. M., Santos, B. R., Marubayashi, J. M., Yuki, V. A., De Marchi, B. R., et al. (2019). Evidence for increased efficiency of virus transmission by populations of Mediterranean species of Bemisia tabaci with high Hamiltonella prevalence. Phytoparasitica 47, 293–300. doi: 10.1007/s12600-019-00729-y
Bing, X. L., Ruan, Y. M., Rao, Q., Wang, X. W., and Liu, S. S. (2013). Diversity of secondary endosymbionts among different putative species of the whitefly Bemisia tabaci. Insect Sci. 20, 194–206. doi: 10.1111/j.1744-7917.2012.01522.x
Bubici, G., Prigigallo, M. I., Garganese, F., Nugnes, F., Jansen, M., and Porcelli, F. (2020). First report of Aleurocanthus spiniferus on Ailanthus altissima: profiling of the insect microbiome and microRNAs. Insects 11, 161. doi: 10.3390/insects11030161
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Mouton, L., Hunter, M. S., and Zchori-Fein, E. (2011). Rickettsia 'in' and 'out': two different localization patterns of a bacterial symbiont in the same insect species. PLoS ONE 6, 1096. doi: 10.1371/journal.pone.0021096
Chen, Y., Yang, W., Zhou, Y., Liu, H., and Yang, M. (2018). Identification of the citrus spiny whitefly in tea-infesting populations in Guizhou. J. Jiangxi Agricult. Univ. 40, 494–501. doi: 10.13836/j.jjau.2018065
Chi, H. (1988). Life-table analysis incorporating both sexes and variable development rates among individuals. Environ. Entomol. 17, 26–34. doi: 10.1093/ee/17.1.26
Chi, H., and Liu, H. S. I. (1984). Two new methods for study of insect population ecology. Bull. Inst. Zool Acad. Sin. 24, 35.
Chi, H., You, M. S., Atlihan, R., Smith, C. L., Kavousi, A., Ozgokce, M. S., et al. (2020). Age-stage, two-sex life table: an introduction to theory, data analysis, and application. Entomol. Gen. 40, 103–124. doi: 10.1127/entomologia/2020/0936
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Engl, T., and Kaltenpoth, M. (2018). Influence of microbial symbionts on insect pheromones. Nat. Prod. Rep. 35, 386–397. doi: 10.1039/C7NP00068E
Frago, E., Dicke, M., and Godfray, H. C. J. (2012). Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Gottlieb, Y., Ghanim, M., Gueguen, G., Kontsedalov, S., Vavre, F., Fleury, F., et al. (2008). Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. Faseb. J. 22, 2591–2599. doi: 10.1096/fj.07-101162
Grenier, T., and Leulier, F. (2020). How commensal microbes shape the physiology of Drosophila melanogaster. Curr. Opin. Insect. Sci. 41, 92–99. doi: 10.1016/j.cois.2020.08.002
Harris, H. L., Brennan, L. J., Keddie, B. A., and Braig, H. R. (2010). Bacterial symbionts in insects: balancing life and death. Symbiosis 51, 37–53. doi: 10.1007/s13199-010-0065-3
Hashmi, T. R., Devi, S. R., Ahmad, A., Meshram, N. M., and Prasad, R. (2019). Genetic status and endosymbionts diversity of Bemisia tabaci (Gennadius) on hosts belonging to family malvaceae in India. Neotrop. Entomol. 48, 207–218. doi: 10.1007/s13744-018-0639-y
Hillman, E. T., Lu, H., Yao, T., and Nakatsu, C. H. (2017). Microbial ecology along the gastrointestinal tract. Microb. Environ. 32, 300–313. doi: 10.1264/jsme2.ME17017
Hu, F. Y., and Tsai, C. W. (2020). Nutritional relationship between bemisia tabaci and its primary endosymbiont, Portiera Aleyrodidarum, during host plant acclimation. Insects 11, 498. doi: 10.3390/insects11080498
Jansen, M., and Porcelli, F. (2018). Aleurocanthus camelliae (Hemiptera: Aleyrodidae), a species possibly new for the European fauna of a genus in great need of revision. Tijdschrift voor Entomol. 161, 63–78. doi: 10.1163/22119434-00002075
Jones, J. C., Fruciano, C., Hildebrand, F., Al Toufalilia, H., Balfour, N. J., Bork, P., et al. (2018). Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evolut. 8, 441–451. doi: 10.1002/ece3.3597
Kanmiya, K., Ueda, S., Kasai, A., Yamashita, K., Sato, Y., and Yoshiyasu, Y. (2011). Proposal of new specific status for tea-infesting populations of the nominal citrus spiny whitefly Aleurocanthus spiniferus (Homoptera: Aleyrodidae). Zootaxa 25–44. doi: 10.11646/zootaxa.2797.1.3
Karamipour, N., Fathipour, Y., and Mehrabadi, M. (2021). Removal of gut symbiotic bacteria negatively affects life history traits of the shield bug, Graphosoma lineatum. Ecol. Evol. 11, 2515–2523. doi: 10.1002/ece3.7188
Karimi, S., Askari Seyahooei, M., Izadi, H., Bagheri, A., and Khodaygan, P. (2019). Effect of Arsenophonus endosymbiont elimination on fitness of the date palm hopper, Ommatissus lybicus (Hemiptera: Tropiduchidae). Environ. Entomol. 48, 614–622. doi: 10.1093/ee/nvz047
Kashkouli, M., Fathipour, Y., and Mehrabadi, M. (2019). Potential management tactics for pistachio stink bugs, Brachynema germari, Acrosternum heegeri and Acrosternum arabicum (Hemiptera: Pentatomidae): high temperature and chemical surface sterilants leading to symbiont suppression. J. Econ. Entomol. 112, 244–254. doi: 10.1093/jee/toy324
Khanamani, M., Fathipour, Y., Talebi, A. A., and Mehrabadi, M. (2017). Linking pollen quality and performance of Neoseiulus californicus (Acari: Phytoseiidae) in two-spotted spider mite management programmes. Pest. Manag. Sci. 73, 452–461. doi: 10.1002/ps.4305
Kuczynski, J., Stombaugh, J., Walters, W. A., Gonzalez, A., Caporaso, J. G., and Knight, R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. Chapter 1, Unit 1E 5. doi: 10.1002/9780471729259.mc01e05s27
Lim, L., and Ab Majid, A. H. (2020). Metagenomic 16S rDNA amplicon data of microbial diversity of guts of fully fed tropical bed bugs, Cimex hemipterus (F.) (Hemiptera: Cimicidae). Data Brief 30, 105575. doi: 10.1016/j.dib.2020.105575
Liu, Q., Zhang, H., and Huang, X. L. (2022). Strong linkage between symbiotic bacterial community and host age and morph in a hemipteran social insect. Microbial. Ecol. 3, 2114. doi: 10.1007/s00248-022-02114-5
Liu, Y. H., Shah, M. M. R., Song, Y., and Liu, T. X. (2020). Host plant affects symbiont abundance in bemisia tabaci (Hemiptera: Aleyrodidae). Insects 11, 501. doi: 10.3390/insects11080501
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, J. B., Chen, W. B., Hasegawa, D. K., Simmons, A. M., Wintermantel, W. M., Ling, K. S., et al. (2015). Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Gen. Biol. Evolut. 7, 2635–2647. doi: 10.1093/gbe/evv170
Lv, Z. H., Wei, X. Y., Tao, Y. L., and Chu, D. (2018). Differential susceptibility of whitefly-associated bacteria to antibiotic as revealed by metagenomics analysis. Infect. Genet. Evolut. 63, 24–29. doi: 10.1016/j.meegid.2018.04.024
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Meng, L., Li, X., Cheng, X., and Zhang, H. (2019). 16S rRNA gene sequencing reveals a shift in the microbiota of Diaphorina citri during the psyllid life cycle. Front. Microbiol. 10, 1948. doi: 10.3389/fmicb.2019.01948
Min, K., Zhang, Z., Zhang, Y., Hou, W., Wang, H., and Zhou, M. (2020). Establishment of a real-time PCR method for detecting Pseudomonas fluorescens in food samples. Food Sci. 41, 304–309. doi: 10.7506/spkx1002-6630-20191018-185
Pandey, N., Singh, A., Rana, V. S., and Rajagopal, R. (2013). Molecular characterization and analysis of bacterial diversity in Aleurocanthus woglumi (Hemiptera: Aleyrodidae). Environ. Entomol. 42, 1257–1264. doi: 10.1603/EN13110
Qu, L. (2013). Competitive Relationship Analysis of Endosymbiont Wolbachia and Arsenophonus in the Brown Planthopper Nilaparvata Lugens. Zhejiang University. doi: 10.1007/s13199-013-0256-9
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic. Acids. Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ren, F. R., Sun, X., Wang, T. Y., Yan, J. Y., Yao, Y. L., Li, C. Q., et al. (2021). Pantothenate mediates the coordination of whitefly and symbiont fitness. Isme J. 15, 1655–1667. doi: 10.1038/s41396-020-00877-8
Rizzo, D., Suma, P., Rossi, E., Farina, P., Daniele, Da Lio, D., et al. (2021). First record of Aleurocanthus camelliae Kanmiya and Kasai, 2011 (Hemiptera, Aleyrodidae) from Italy, on ornamental Camellia spp. plants. Bullet. OEPP/EPPO Bullet. 51, 1–7. doi: 10.1111/epp.12760
Roy, M., Brodeur, J., and Cloutier, C. (2003). Effect of temperature on intrinsic rates of natural increase (rm) of a coccinellid and its spider mite prey. Biocontrol 48, 57–72. doi: 10.1023/A:1021289832664
Schloss, P. D. (2020). Reintroducing mothur: 10 years later. Appl. Environ. Microbiol. 86, 19. doi: 10.1128/AEM.02343-19
Shan, H. W., Luan, J. B., Liu, Y. Q., Douglas, A. E., and Liu, S. S. (2019). The inherited bacterial symbiont Hamiltonella influences the sex ratio of an insect host. Proceed. Royal Soc. B-Biol. Sci. 286, 1677. doi: 10.1098/rspb.2019.1677
Shan, H. W., Zhang, C. R., Yan, T. T., Tang, H. Q., Wang, X. W., Liu, S. S., et al. (2016). Temporal changes of symbiont density and host fitness after rifampicin treatment in a whitefly of the Bemisia tabaci species complex. Insect Sci. 23, 200–214. doi: 10.1111/1744-7917.12276
Su, M. M., Guo, L., Tao, Y. L., Zhang, Y. J., Wan, F. H., and Chu, D. (2016). Effects of host plant factors on the bacterial communities associated with two whitefly sibling species. PLoS ONE 11, 2183. doi: 10.1371/journal.pone.0152183
Su, Q., Oliver, K. M., Xie, W., Wu, Q. J., Wang, S. L., and Zhang, Y. J. (2015). The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Func. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Su, Q., Xie, W., Wang, S. L., Wu, Q. J., Ghanim, M., and Zhang, Y. J. (2014). Location of symbionts in the whitefly Bemisia tabaci affects their densities during host development and environmental stress. PLoS ONE 9, 1802. doi: 10.1371/journal.pone.0091802
Sun, X., Liu, B. Q., Li, C. Q., Chen, Z. B., Xu, X. R., and Luan, J. B. (2022). A novel microRNA regulates cooperation between symbiont and a laterally acquired gene in the regulation of pantothenate biosynthesis within Bemisia tabaci whiteflies. Mol. Ecol. 31, 2611–2624. doi: 10.1111/mec.16416
Tian, Y., Chen, Z., Huang, X., Zhang, L., and Zhang, Z. (2020). Evaluation of botanicals for management of piercing-sucking pests and the effect on beneficial arthropod populations in tea trees camellia sinensis (L.) O. Kuntze (Theaceae). J. Insect. Sci. 20, 101. doi: 10.1093/jisesa/ieaa101
Uesugi, R., and Sato, Y. (2011). Differentiation of the tea-infesting population of citrus spiny whitefly aleurocanthus spiniferus (Homoptera: Aleyrodidae) from the citrus-infesting population in japan on the basis of differences in the mitochondrial cytochrome c oxidase subunit i gene. Jpn. J. Appl. Entomol. Z 55, 155–161. doi: 10.1303/jjaez.2011.155
Uesugi, R., Sato, Y., Han, B. Y., Huang, Z. D., Yara, K., and Furuhashi, K. (2016). Molecular evidence for multiple phylogenetic groups within two species of invasive spiny whiteflies and their parasitoid wasp. Bull. Entomol. Res. 106, 328–340. doi: 10.1017/S0007485315001030
Wang, Y. B., Li, C., Yan, J. Y., Wang, T. Y., Yao, Y. L., Ren, F. R., et al. (2022). Autophagy regulates whitefly-symbiont metabolic interactions. Appl. Environ. Microb. 88, 21. doi: 10.1128/AEM.02089-21
Wang, Y. B., Ren, F. R., Yao, Y. L., Sun, X., Walling, L. L., Li, N. N., et al. (2020). Intracellular symbionts drive sex ratio in the whitefly by facilitating fertilization and provisioning of B vitamins. ISME J. 14, 2923–2935. doi: 10.1038/s41396-020-0717-0
Weiland, S. O., Detcharoen, M., Schlick-Steiner, B. C., and Steiner, F. M. (2022). Analyses of locomotion, wing morphology, and microbiome in Drosophila nigrosparsa after recovery from antibiotics. Microbiologyopen 11, e1291. doi: 10.1002/mbo3.1291
Xue, H., Zhu, X., Wang, L., Zhang, K., Li, D., Ji, J., et al. (2021). Gut bacterial diversity in different life cycle stages of Adelphocoris suturalis (Hemiptera: Miridae). Front. Microbiol. 12, 670383. doi: 10.3389/fmicb.2021.670383
Xue, X., Li, S. J., Ahmed, M. Z., De Barro, P. J., Ren, S. X., and Qiu, B. L. (2012). Inactivation of Wolbachia reveals its biological roles in whitefly host. PLoS ONE 7, e48148. doi: 10.1371/journal.pone.0048148
Yang, K., Yuan, M. Y., Liu, Y., Guo, C. L., Liu, T. X., Zhang, Y. J., et al. (2021). First evidence for thermal tolerance benefits of the bacterial symbiont Cardinium in an invasive whitefly, Bemisia tabaci. Pest. Manag. Sci. 77, 5021–5031. doi: 10.1002/ps.6543
Yang, X., Sun, L., Chi, H., Kang, G., and Zheng, C. (2020). Demography of thrips palmi (Thysanoptera: Thripidae) reared on brassica oleracea (Brassicales: Brassicaceae) and phaseolus vulgaris (Fabales: Fabaceae) with discussion on the application of the bootstrap technique in life table research. J. Econ. Entomol. 113, 2390–2398. doi: 10.1093/jee/toaa171
Yun, J. H., Roh, S. W., Whon, T. W., Jung, M. J., Kim, M. S., Park, D. S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Zchori-Fein, E., Lahav, T., and Freilich, S. (2014). Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front. Microbiol. 5, 310. doi: 10.3389/fmicb.2014.00310
Zhang, C. R., Shan, H. W., Xiao, N., Zhang, F. D., Wang, X. W., Liu, Y. Q., et al. (2015). Differential temporal changes of primary and secondary bacterial symbionts and whitefly host fitness following antibiotic treatments. Scient. Reports 5, 5898. doi: 10.1038/srep15898
Zhang, J. H., Yu, N., Xu, X. X., and Liu, Z. W. (2019). Community structure, dispersal ability and functional profiling of microbiome existing in fat body and ovary of the brown planthopper, Nilaparvata lugens. Insect. Sci. 26, 683–694. doi: 10.1111/1744-7917.12575
Zhang, Q. Q., Lan, R. M., Ji, D. Z., Tan, Y. N., Zhou, X., Tan, X. F., et al. (2022). The detection of Wolbachia in tea green leafhopper (Empoasca onukii Matsuda) and its influence on the host. Agriculture-Basel 12, 36. doi: 10.3390/agriculture12010036
Zhang, X., Li, T. P., Zhou, C. Y., Zhao, D. S., Zhu, Y. X., Bing, X. L., et al. (2020). Antibiotic exposure perturbs the bacterial community in the small brown planthopper Laodelphax striatellus. Insect Sci 27, 895–907. doi: 10.1111/1744-7917.12675
Zhao, D. X., Hoffmann, A. A., Zhang, Z. C., Niu, H. T., and Guo, H. F. (2018). Interactions between facultative symbionts hamiltonella and cardinium in bemisia tabaci (Hemiptera: Aleyrodoidea): cooperation or conflict? J. Econ. Entomol. 111, 2660–2666. doi: 10.1093/jee/toy261
Zhao, D. X., Zhang, Z. C., Niu, H. T., and Guo, H. F. (2020). Selective and stable elimination of endosymbionts from multiple-infected whiteflyBemisia tabaciby feeding on a cotton plant cultured in antibiotic solutions. Insect Sci. 27, 964–974. doi: 10.1111/1744-7917.12703
Keywords: Aleurocanthus camelliae, 16S rRNA, symbiotic bacteria, rifampicin, age-stage two-sex life table
Citation: Tan Y, Gong B, Zhang Q, Li C, Weng J, Zhou X and Jin L (2023) Diversity of endosymbionts in camellia spiny whitefly, Aleurocanthus camelliae (Hemiptera: Aleyrodidae), estimated by 16S rRNA analysis and their biological implications. Front. Microbiol. 14:1124386. doi: 10.3389/fmicb.2023.1124386
Received: 15 December 2022; Accepted: 16 March 2023;
Published: 17 April 2023.
Edited by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandReviewed by:
Manoj Kumar Karnena, Gandhi Institute of Technology and Management (GITAM), IndiaBalasubramanian Cibichakravarthy, Yale University, United States
Shalini Rai, Society for Higher Education and Practical Applications, India
Copyright © 2023 Tan, Gong, Zhang, Li, Weng, Zhou and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linhong Jin, bGhqaW5AZ3p1LmVkdS5jbg==
 Yanni Tan
Yanni Tan Linhong Jin
Linhong Jin