
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 March 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1121564
This article is part of the Research Topic Prevalence, Transmission and Control of Clinically Important Antimicrobial-Resistant Bacteria/Genes within One Health Framework View all 22 articles
 Idris Nasir Abdullahi1
Idris Nasir Abdullahi1 Carmen Lozano1
Carmen Lozano1 Myriam Zarazaga1
Myriam Zarazaga1 Andre Becker Simoes Saidenberg2,3
Andre Becker Simoes Saidenberg2,3 Marc Stegger2
Marc Stegger2 Carmen Torres1*
Carmen Torres1*Introduction: Nasal carriage of coagulase-positive staphylococci (CoPS) in healthy dogs could indicate increased risks of colonization for in-contact people or vice versa. This study determined the nasal carriage rate of CoPS among healthy dogs and in-contact people, their genotypic characteristics and phylogenetic relatedness.
Methods: Nasal samples were collected from 27 households (34 dogs and 41 humans) in Spain. Staphylococci were identified by MALDI-TOF-MS, their antimicrobial resistance (AMR) genes and spa-types were tested by PCR/sequencing. The relatedness of CoPS from the same households was assessed by core genome single nucleotide polymorphisms (SNPs) analyses.
Results: Staphylococcus aureus carriage was found in 34.1% of humans (including one methicillin-resistant S. aureus MRSA-CC5-t2220-SCCmec type-IV2B) and 5.9% of dogs; Staphylococcus pseudintermedius in 2.4% of humans and 32.4% of dogs; while Staphylococcus coagulans was only detected in dogs (5.4%). Remarkably, one human co-carried S. aureus/S. pseudintermedius, while a dog co-carried the three CoPS species. Household density was significantly associated with S. pseudintermedius carriage in households with > than 1 dog and >than 1 human (OR = 18.10, 95% CI: 1.24–260.93, p = 0.034). Closely related (<15 SNPs) S. aureus or S. pseudintermedius were found in humans or dogs in three households. About 56.3% S. aureus carriers (dog or human) harboured diverse within-host spa-types or AMR genotypes. Ten clonal complexes (CCs) were detected among the S. aureus, of which methicillin-susceptible S. aureus-CC398-IEC-type C (t1451 and t571) was the most frequent, but exclusive to humans. S. aureus and S. pseudintermedius isolates harboured resistance genes or mutations associated to 9 classes of antimicrobials including linezolid (G2261A & T1584A point mutations in 23S rDNA). The S. coagulans isolates were susceptible to all antimicrobials. Most of the S. pseudintermedius carried lukS/F-I, siet, and sient genes, and all S. aureus were negative for lukS/F-PV, tst-1, eta and etb genes.
Discussion: Clonally related human-to-human MSSA and dog-to-human MSSP were found. The detection of the MSSA-CC398 clade highlights the need for its continuous surveillance from One Health perspective.
Resident bacteria of the mucosal surfaces (such as nasal or oral) and skin of dogs could easily be transmitted to in-contact persons or owners by direct contact or from the household environment (Trinh et al., 2018). Moreover, transmission from humans to pets could also be possible (Orsini et al., 2022). These events (zoonosis and anthroponosis) represent a growing public health problem (Rahman et al., 2020; Orsini et al., 2022).
Two main groups of staphylococci are defined based on rabbit’s plasma coagulase activity, viz, coagulase-negative staphylococci (CoNS) and coagulase-positive staphylococci (CoPS) (Carroll et al., 2021). Of the two groups, CoPS are more pathogenic (Carroll et al., 2021). Staphylococcus aureus is the archetype of the CoPS and relevant animal and human pathogen (Tong et al., 2015); however, other CoPS related to animals have been described. One of the emerging CoPS of clinical importance is S. pseudintermedius, a member of the Staphylococcus intermedius group (SIG) that comprises four other species, viz.; Staphylococcus intermedius, Staphylococcus delphini, the recently described Staphylococcus cornubiensis, and S. ursi (Murray et al., 2018; Perreten et al., 2020; Carroll et al., 2021). In addition to these CoPS, Staphylococcus coagulans is another species that was originally described in 1990 as Staphylococcus schleiferi subsp. coagulans before being classified, based on genomic studies, into a separate species in 2020 (Madhaiyan et al., 2020).
Staphylococcus aureus is an opportunist pathogen frequently found in humans, which colonizes the anterior nose and nasal vestibule of 25–50% of healthy people (Liu et al., 2015; Sakr et al., 2018). S. aureus nasal colonization in humans and animals could place individuals at risk of opportunistic infections, and the carriage of specific virulence genes by the isolates can increase their pathogenic potential. In this respect, the following virulence genes are especially relevent in S. aureus: tst (encodes the toxic shock syndrome toxin, associated with sepsis), eta and etb (encode the exfoliatin A and B, associated with the staphylococcal scalded skin syndrome), and the luk-S/F-PV (encodes Panton Valentine Leucocidin, associated with abscesses and community-acquired pneumonia) (Tam and Torres, 2019; Hu et al., 2021).
The population genetics of S. aureus have shown the presence of several clonal complexes (CCs). Remarkably, some of these CCs have somewhat animal host specificity, while others are less host-specific (Chaguza et al., 2022). Methicillin-Resistant S. aureus (MRSA) is a globally epidemic bacterium in nosocomial settings and is referred to as healthcare-associated MRSA (HA-MRSA) (Cuny et al., 2010; Cuny et al., 2022). MRSA has also emerged in the community without any relation to the healthcare facilities [community associated (CA)-MRSA] and in the last decade, some CCs (mainly CC398) have been associated with livestock animals (LA-MRSA) (Matuszewska et al., 2022).
Staphylococcus aureus frequently colonizes the skin and nasal cavity of healthy humans, while S. pseudintermedius appears to predominantly colonize pets (especially dogs) but could occasionally be found in humans (Cuny et al., 2022). Although infrequent, S. pseudintermedius has been also isolated from human skin lesions (Lozano et al., 2017), due en ocassions to dog bites (Börjesson et al., 2015), as well as causing septicemia (Somayaji et al., 2016). Previous reports on nasal colonization in humans by S. pseudintermedius in dog-owning households have reported a prevalence of 25–65.9% in dogs and 3–4.5% in humans (Gómez-Sanz et al., 2013c; Han et al., 2016; Rodrigues et al., 2018). Certain isolates of S. pseudintermedius have become a major veterinary pathogen of concern due to their frequent multidrug-resistance phenotypes (Priyantha, 2022). Also, severe human infections with methicillin-resistant S. pseudintermedius (MRSP) have recently been reported (Priyantha, 2022).
Data about the co-colonization of dogs and in-contact persons with CoPS at the community level are still scarce. Hanselman et al. (2009) reported the presence of S. pseudintermedius, S. aureus, and S. schleiferi subsp. coagulans (including methicillin-resistant isolates) in dogs at the household level. In addition, concurrent animal and human colonization by indistinguishable S. pseudintermedius and S. aureus isolates have been observed (Hanselman et al., 2009). In a previous study performed a decade ago, our research group investigated the household nasal carriage of CoPS in dogs and humans, determining the prevalence of genetic lineages and their recovery rate over a year in animals and owners (Gómez-Sanz et al., 2013c). Nowadays, the abundance of dog ownership in Spain has significantly changed and the number of households with pets has increased. According to available data, there were 20 million registered pets in Spain in 2019 and about 26% of all households own dogs (Asociación Madrileña de Veterinarios de Animales de Compañía, 2019). By implication, sharing and transmission of nasal microorganisms (such as CoPS) between dogs and dog-owning households could be more frequent (Van Balen et al., 2017).
Nasal carriage of CoPS in healthy dogs could be an indicator of increased risk of colonization/infection for people in-contact with these animals, especially if S. aureus carries key virulence determinants of special relevance such as lukS/F-PV, tst-1, eta and etb (González-Martín et al., 2020). However, the influence of pet ownership on the diversity of CoPS isolates circulating among dog owners still needs to be fully elucidated (Van Balen et al., 2017). Consequently, this study sought to understand the current epidemiological situation of CoPS nasal carriage in dog-owning households by characterization of the genetic lineages, relatedness between isolates from the same households, antimicrobial resistance (AMR) determinants, virulence factors, and immune evasion cluster (IEC) types of S. aureus isolates.
A total of 41 humans and 34 dogs from 27 households were prospectively studied to determine the nasal carriage of CoPS; the sampling was performed in La Rioja region (Northern Spain) between January to March 2022. Household density was classified into four, viz: (a) household with a dog and a human (b) household with >1 dog and a human, (c) household with 1 dog and >than 1 human, and (d) household with > than 1 dog and >than 1 human. The sampled humans and dogs did not have recent hospital stays prior to the study or received antibiotics (at least 3 months before sampling) and the humans had no professional contact with health institutions and did not work in microbiology laboratories. None of the participants had consultations or visits to hospitals in the last 3 months before sample collection. Nasal samples were obtained using sterile swabs with conservation media (Amies BD Life sciences®, New Jersey, USA). All procedures were approved by the ethical committee of the University of La Rioja (Spain) and were carried out following all applicable international, national, and/or institutional guidelines for human samples experiments (as described in the revised Helsinki declaration) and for ethical use of animals (directive 2010/63/EU, Spanish laws 9/2003 and 32/2007, and RD 178/2004 and RD 1201/2005).
Samples were enriched in Brain Heart Infusion broth (BHI; Condalab, Madrid, Spain) supplemented with 6.5% (w/v) NaCl and incubated for 24 h at 37°C. After 24 h of incubation, the broth samples were diluted and carefully dispensed onto four different bacteriological media: blood agar, mannitol salt agar (MSA; Condalab, Madrid, Spain), oxacillin resistance screening agar base (ORSAB with 2 mg/L oxacillin; Oxoid, Hampshire, UK) and ChromAgar LIN (Paris, France) and incubated for 24 h at 37°C. After overnight growth, 3 to 8 different colonies (presenting staphylococci morphology) were randomly selected per sample and identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF, Bruker Daltonics, Bremen, Germany) using the standard extraction protocol. Those isolates identified as CoPS were included in this study.
Susceptibility testing for 13 antimicrobial agents was performed by the disk diffusion method following the recommendations and breakpoints of the European Committee on Antimicrobial Susceptibility Testing (European Committee on Antimicrobial Susceptibility Testing (EUCAST), 2022). The antimicrobial agents tested were as follows (μg/disk): penicillin (PEN) (1 or 10 μg, depending on the CoPS species), cefoxitin (FOX) (30 μg to detect methicillin-resistant S. aureus (MRSA) isolates), oxacillin (OXA) (1 μg to detect methicillin-resistant S. pseudintermedius [MRSP] or S. coagulans isolates), erythromycin (ERY) (15 μg), clindamycin (CLI) (2 μg), gentamicin (GEN) (10 μg), tobramycin (TOB) (10 μg), tetracycline (TET) (30 μg), ciprofloxacin (CIP) (5 μg), chloramphenicol (CHL) (30 μg), linezolid (LZD) (10 μg), mupirocin (MUP) (200 μg), and trimethoprim-sulfamethoxazole (SXT) (1.25 μg + 23.75 μg).
Once the antimicrobial resistance phenotype of all CoPS was determined, distinct isolates were selected for further studies (defined as those of different samples or those from the same sample but of different species, and/or different antimicrobial resistance phenotypes).
For DNA extraction, isolates were seeded on BHI agar and incubated for 24 h at 37°C. An isolated colony was suspended in 45 μL of sterile MiliQ water and added 5 μL of lysostaphin (1 mg/mL) (Sigma). The mixture was vortexed and incubated for 10 min at 37°C. Forty-five μL of sterile MiliQ water, 150 μL of Tris–HCl (0.1 M, pH 8) and 5 μL of proteinase K (2 mg/mL) (Sigma) were added. The final mixture was vortexed and incubated for 10 min at 60°C, then boiled for 5 min at 100°C. To separate and obtain the DNA (supernatant) from debris, the final mixture was centrifuged at 12,000 revolutions per minute for 3 min, and stored at −20°C.
The presence of the following resistance genes was tested by single PCRs, selected according to the antimicrobial resistance phenotype of isolates: beta-lactams (blaZ, mecA, and mecC), erythromycin and clindamycin (ermA, ermB, ermC, ermT, mphC, msrA, lnuA, and lnuB), aminoglycosides (aac6′-aph2″, and ant4′), tetracycline [tet(L), tet(M), and tet(K)], trimethoprim (dfrA, dfrD, dfrG and dfrK), and chloramphenicol (catpC221, catpC223, catpC194, catA, fexA, and fexB). Linezolid or chloramphenicol-resistant isolates were screened for the presence of the linezolid transferable resistance genes (cfr, cfrB, cfrD, poxtA, and optrA). Mutations in 23S rDNA were also investigated by PCR and amplicon sequencing. The obtained sequences were compared with those of linezolid-susceptible S. aureus NCTC 8325 (GenBank accession number CP000253) using the EMBOSS Needle software for nucleotide and amino acid (BLOSUM62 cost matrix) alignments. Primers and conditions of PCRs performed in this study are included in Supplementary Table S1. Isolates were considered multi-drug resistant (MDR) when they were resistant to ≥3 classes of the antimicrobial agents tested (Magiorakos et al., 2012).
The presence of the genes tst (toxin of shock toxic syndrome), lukS/F-PV (Panton-Valentine leucocidin), and eta and etb (exfoliative toxins A and B), was tested by PCR. Immune evasion cluster (IEC) genes (scn, chp, sak, sea, and sep) were analysed and classified into seven different IEC types (A–G), based on the combination of the positive genes. The scn gene (encoding the staphylococcal complement inhibitor) was used as a marker of the IEC system (van Wamel et al., 2006). The presence of the lukS/F-I, siet, and sient genes was analysed for the S. pseudintermedius isolates. Primers and conditions of PCRs performed in this study are included in Supplementary Table S1.
All recovered S. aureus isolates were characterized by spa typing by the PCR/Sanger sequencing. New repeat combinations were submitted to the Ridom spa Server.1 CC398 clone was determined by a specific PCR protocol for the sau1-hsdS1 variant (Stegger et al., 2011). The clonal complex of the isolates was assigned, when possible, according to the spa-types. Moreover, multilocus sequence typing (MLST) was performed in selected S. aureus isolates (isolates with spa-types repeatedly found and those with new spa-types). The seven housekeeping genes of S. pseudintermedius (pta, cpn60, tuf, ack, purA, sar and fdh) were amplified, and the sequence type (ST) was assigned according to the MLST database.2 The selection of S. pseudintermedius isolates for MLST was only on household with dog and human carriers. Staphylococcal Cassette Chromosome mec (SCCmec) types were determined by multiplex PCRs. Primers and conditions of PCRs performed in this study are included in Supplementary Table S1.
Positive controls (confirmed by sequencing) from the collection of the Universidad de La Rioja were included in all the PCR assays in this study.
Fourteen selected isolates (11 S. aureus and two S. pseudintermedius) were whole genome sequenced on the Illumina NextSeq platform. The selection was based on: (a) all MSSA-CC398 isolates, (b) all isolates from households with two or more humans and/or dogs’ carriers of either S. aureus or S. pseudintermedius, (c) the MRSA isolate.
The MagNA Pure 96 DNA Multi-Sample Kit (Life Technologies, Carlsbad, CA, USA, 4413021) was used to extract genomic DNA according to instructions provided by the manufacturers. The Qubit 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, Scoresby, VIC, Australia) was used for DNA quantification, while Sequencing libraries were prepared using the Illumina Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA, FC-131-1096) and sequenced on the NextSeq 500 platform (Illumina, San Diego, CA, USA) using a 300-cycle kit to obtained paired-end 150 bp reads, as previously described (Saidenberg et al., 2022).
All the genomes analysed in this study were de novo assembled using SPAdes (v.3.15.5), performing the in silico typing with the settings of a minimum of 90% coverage and 80% identity. Core-genome single nucleotide polymorphisms (SNPs) were detected with the NASP pipeline v.1.0.0 (Sahl et al., 2016), mapping against the chromosome reference sequences CA-347 (GenBank accession ID CP006044, S. aureus), and SP_11304-3A (GenBank accession ID CP065921.1, S. pseudintermedius) for each separate phylogenetic analysis. GATK (v.4.2.2) was used to call SNPs and excluded positions featuring <90% unambiguous variant calls and <10 depth. IQ-TREE (v.2.1.2), was used to construct the phylogenetic trees using ModelFinder with 100 bootstraps. The graphical data was added to the phylogenies with iTOL v.6.6 (Letunic and Bork, 2021).3
The STs were determined with MLST v.2.16 (Jolley et al., 2018),4 and undefined STs were submitted to the PubMLST database for ST assignment (Larsen et al., 2012). Virulence factors, plasmid replicons, and antimicrobial resistance genes were identified using ABRicate v.0.9.0 and the respective databases VFDB, Plasmidfinder, and Resfinder databases from the Center for Genomic Epidemiology.5,6 The spaTyper v1.0 was used to confirm the spa types (Bartels et al., 2014) and mutations associated with AMR were identified using ResFinder v4.1 (Bortolaia et al., 2020) and PointFinder (Zankari et al., 2017).
All the raw genome reads generated from this study have been deposited at European Nucleotide Archive under Study Accession no. PRJEB57210.7
Data collected were verified and processed and the Statistical Package for Social Sciences (SPSS) Version 26 (IBM, California, U.S.A) was used for analysis. Data were reported as numbers and percentages (for categorical variables) and presented on tables and charts. Data were subjected to univariate logistic regression to compute odd ratio (OR) at a 95% confidence interval (95%CI) between the carriage rate of S. aureus/S. pseudintermedius and the household densities with significant association (p < 0.05).
A total of 73 S. aureus, 31 S. pseudintermedius and two S. coagulans isolates were recovered from 75 nasal samples of humans and dogs. After AMR phenotype determination, 52 isolates were selected for further characterization (31 S. aureus, 19 S. pseudintermedius and 2 S. coagulans), corresponding to one isolate per sample or more than one if they presented different species and/or different AMR phenotype (Supplementary Table S2).
Staphylococcus aureus was found in 14 humans (34.1%) (including one individual with MRSA) and two dogs (5.9%) (Figure 1). S. pseudintermedius was identified in one human (2.4%) and 11 dogs (32.4%). However, S. coagulans was solely identified in two dogs (5.9%) of the same household. Apart from these three species, no other CoPS species were detected in the cultures. Remarkably, one human presented S. aureus/S. pseudintermedius co-carriage (2.4%) while a dog had co-carriage of all three CoPS species (2.9%) (Figure 1). In total, 14 humans and 12 dogs carried CoPS. Household density was significantly associated with S. pseudintermedius carriage in households with > than 1 dog and >than 1 human (OR = 18.10, 95% CI: 1.24–260.93, p = 0.034) (Table 1).
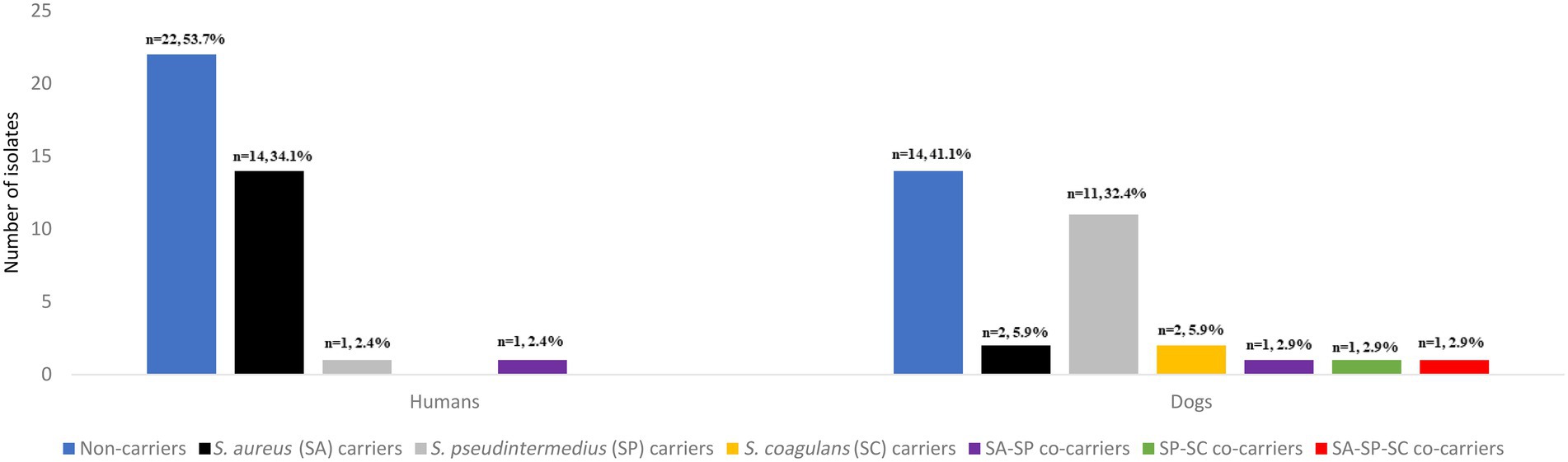
Figure 1. Number and prevalence of Staphylococcus aureus, Staphylococcus pseudintermedius, Staphylococcus coagulans isolates and co-carriage detected in the nasal cavities of healthy humans and dogs.
The 31 distinct S. aureus isolates harboured AMR as follows [percentage of resistant isolates/resistance genotype]: penicillin [77.4/blaZ], cefoxitin [9.7/mecA], erythromycin-clindamycin-inducible [19.4/ermT], erythromycin [9.7/msrA, mphC], clindamycin [3.2/lnuA], gentamicin-tobramycin [22.6/aac6′-aph2″], tetracycline [3.2/tet(K)], sulfonamide [3.2/dfrA], fluoroquinolones [22.5/amino acid changes in GrlA: S80F, GyrA: S84L], mupirocin (3.2/mupA) and linezolid [3.2/G2261A & T1584A point mutations in 23S rDNA] (Figure 2; Table 2). Moreover, the 19 distinct S. pseudintermedius isolates harboured AMR as follows [percentage of resistant isolates/resistance genotype]: penicillin [57.9/blaZ], erythromycin-clindamycin-constitutive [26.3/ermB], tobramycin [15.8/ant4′], tetracycline [26.3/tet(M)], trimethoprim-sulfamethoxazole [63.2/dfrA, dfrD, dfrG, dfrK], and chloramphenicol [5.3/catA] (Figure 2; Table 3). No resistance markers were detected in the S. coagulans isolates, that were susceptible to all antimicrobial agents tested (Figure 2).
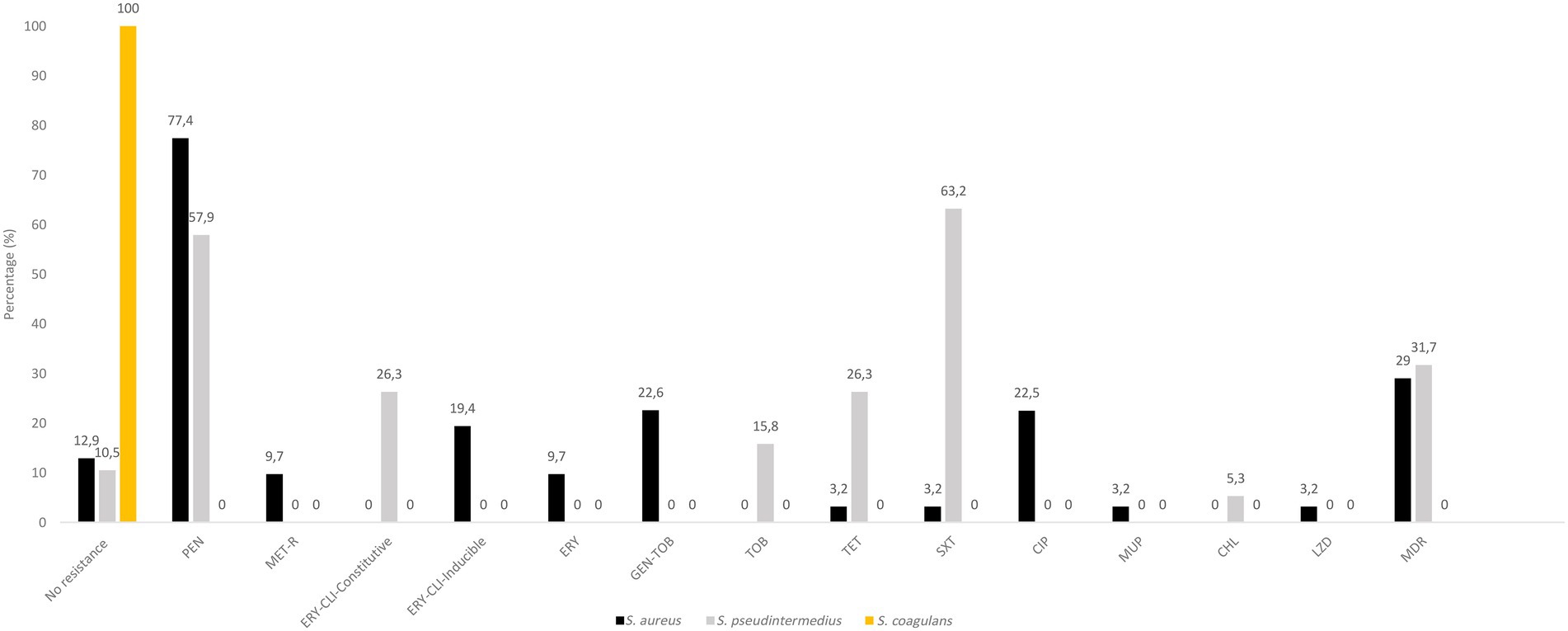
Figure 2. Antimicrobial resistance rates in S. aureus, S. pseudintermedius and S. coagulans isolates. Percentages were based on the collection of CoPS (31 S. aureus, 19 S. pseudintermedius and 2 S. coagulans) obtained of different samples or those of the same sample but with different species and/or AMR phenotype. CHL, chloramphenicol; CLI, clindamycin; CIP, ciprofloxacin; ERY, erythromycin; GEN, gentamicin; MET-R, methicillin-resistant; MDR, multidrug resistance (resistance to three or more classes of antibiotics); PEN, penicillin; SXT, sulfamethoxazole/trimethoprim; TET, tetracycline, TOB, tobramycin.
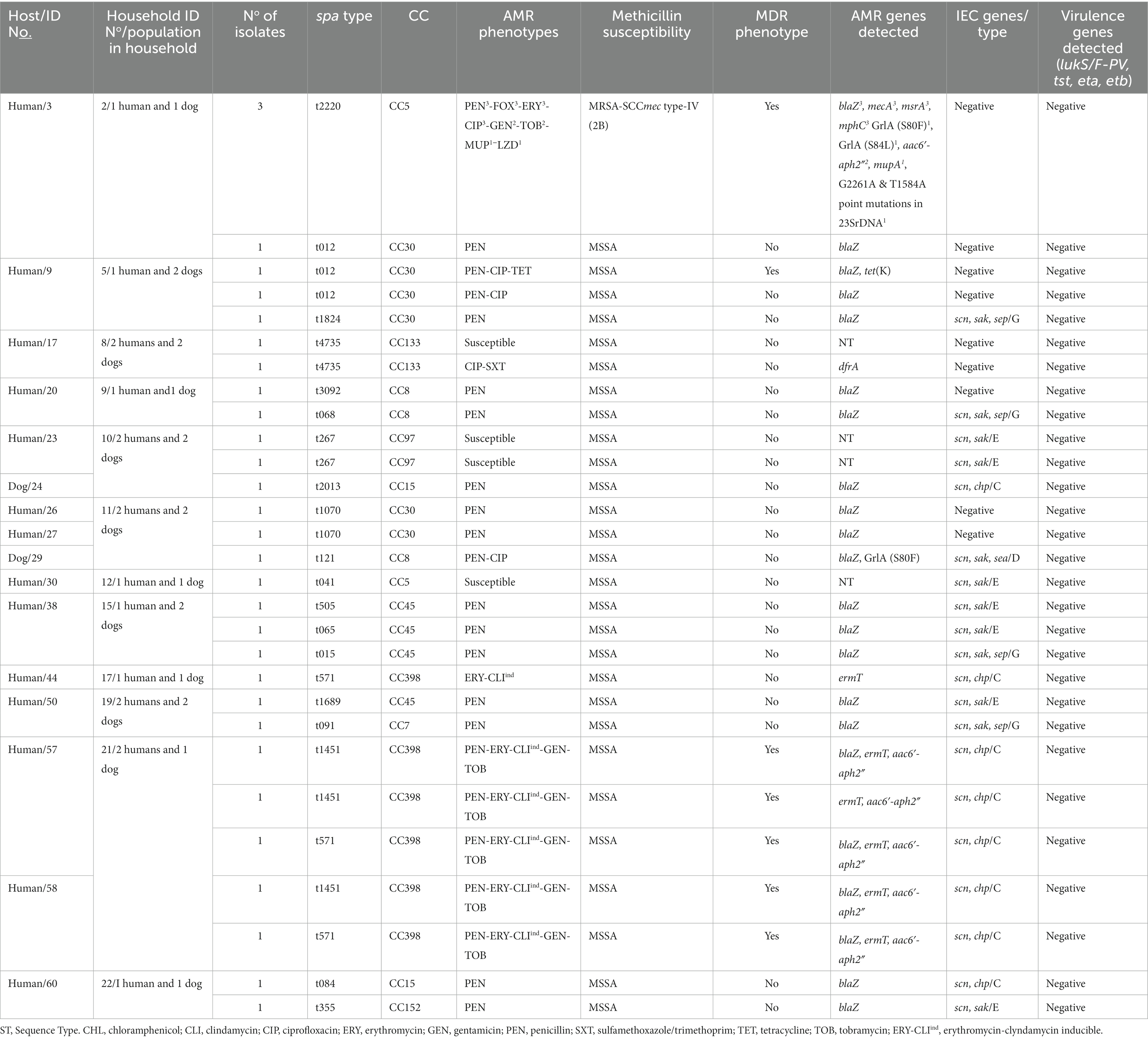
Table 2. Molecular characterization, antimicrobial resistance and virulence profile of the S. aureus isolates from humans and dogs.
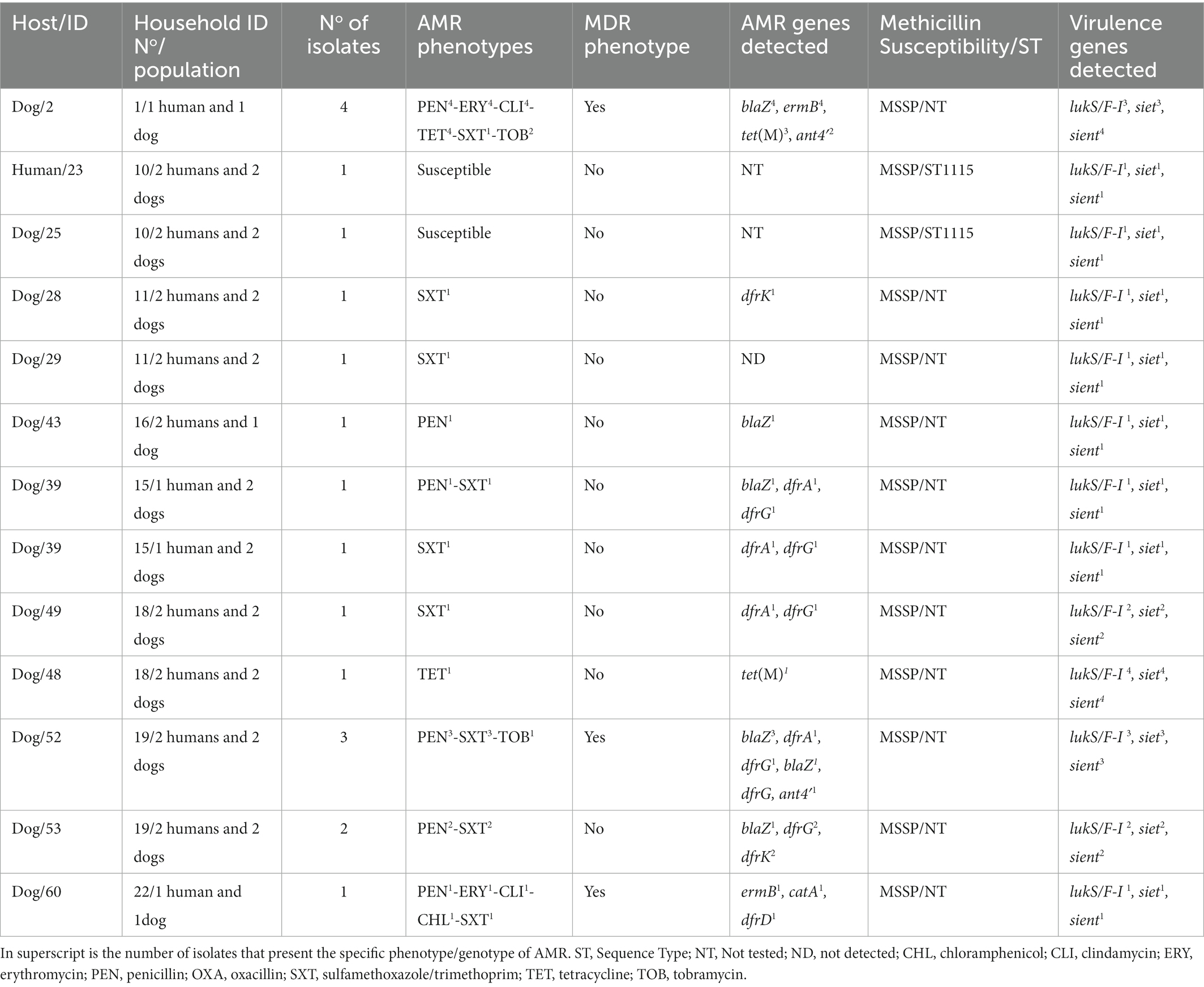
Table 3. Intra-host variation of AMR determinant and virulence factors of S. pseudintermedius isolates.
Regarding the genetic lineages of S. aureus isolates, the three MRSA isolates from humans (same individual but different AMR phenotypes/genotypes) belonged to the spa type t222, associated with CC5. All other isolates were methicillin-susceptible S. aureus (MSSA) with 19 different spa-types assigned to 10 different CCs. The MSSA-CC398 clone (t1451 and t571) was the most frequently identified (18.8% of S. aureus carriers); these isolates were all IEC-type C. Other CCs (spa-types) detected were as follows: CC5 (t041), CC7 (t091), CC8 (t121, t126, t1070, t3092), CC15 (t084, t2013), CC30 (t012, t1824), CC45 (t015, t065, t505, t1689), CC97 (t267), CC133 (t4735) and CC152 (t355) (Table 2). For S. pseudintermedius isolates, all isolates were methicillin susceptible (MSSP) (including two ST1115) (Table 3).
Clonally related S. aureus or S. pseudintermedius isolates were found in humans or dogs among 11.1% of households (n = 3). Two of the 16 households (household No11 and 21) positive for nasal S. aureus had human carriers with similar clonal complexes (CCs), spa-types and IEC types (Table 2). In one of these households (No 11), MSSA-CC30-spa-type t1070 isolates (scn-negative) were identified in two humans, however, a different lineage, MSSA-CC8 of the spa-type t121 (IEC type-D), was identified in their dog (Table 4). In the second household (N° 21), two humans carried MSSA-CC398 isolates of different spa-types (t1451 and t571), although the dog was not S. aureus carrier. Moreover, in another household (No 10), a dog and a human were carriers of the same genetic lineage of S. pseudintermedius (MSSP-ST1115); in this household, the human also carried MSSA-CC97-t267 and a dog MSSA-t2013-CC15 (Table 4). All the S. aureus isolates were negative for lukS/F-PV, tst-1, eta and etb genes (Table 2). However, all the S. pseudintermedius isolates were positive for lukS/F-I, siet, and sient virulence genes, but one was only sient-positive (Tables 2, 3).
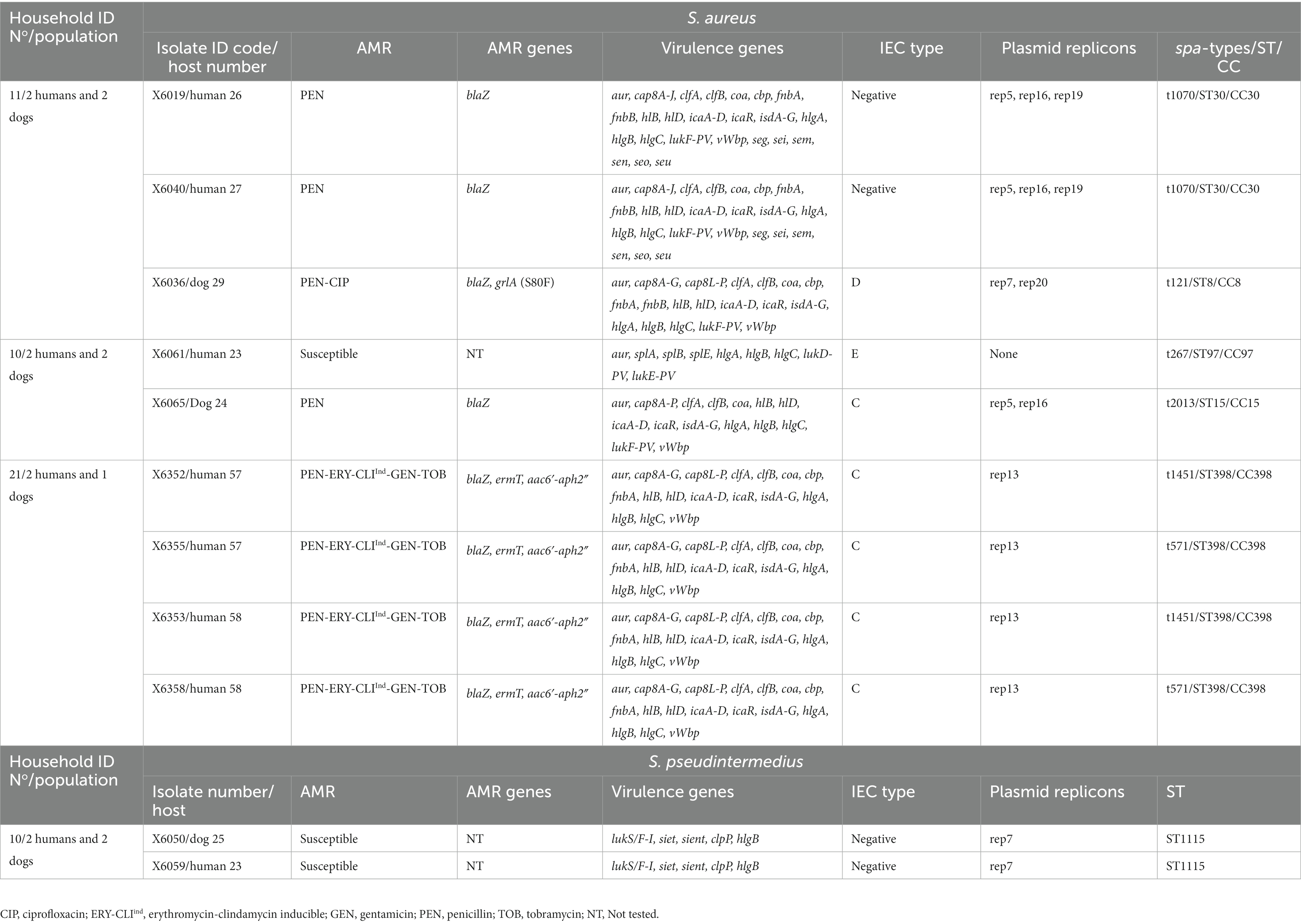
Table 4. Genetic lineages, virulence and AMR genes of CoPS isolates among households with both dog and human carriers.
Nine of the 16 S. aureus (56.3%) carriers harboured diverse spa-types or AMR genotypes in the same individual (dog or human). Of these, two to four genetically distinct S. aureus isolates were detected in these hosts (Table 2). In one human (ID number 3) with both MSSA and MRSA-SCCmec type-IV (2B) nasal carriage, three different MRSA-CC5-t2220 isolates with different AMR phenotypes/genes were detected: PEN-FOX-ERY-CLI-CIP-TOB-MUP-LZD/blaZ, mecA, lnuA, msrA, mphC, mupA, G2261A point mutation in 23S rDNA; PEN-FOX-ERY-CIP-GEN-TOB/blaZ, mecA, aac6′-aph2″, msrA, mphC; and PEN-FOX-ERY-CIP/blaZ, mecA, msrA, mphC, respectively; moreover, the MSSA isolate was typed as CC30-t012 and showed resistance only to PEN (blaZ positive) (Table 2). Two other humans (ID numbers 57 and 58) from the same household (No 21) carried S. aureus isolates both with similar genetic lineage (CC398) but different spa types (t571 and t1451) and similar AMR phenotypes (PEN-ERY-CLIind-GEN-TOB) (Table 2). In another human S. aureus carrier (ID number 60) from a different household (No 22), isolates with different genetic lineages (CC15 and CC152) were detected (Table 2).
In the S. pseudintermedius isolates, 6 of the 12 carriers showed differences and intra-host variations in the AMR phenotypes or AMR genotypes. For instance, one of the dogs (ID number 52) harboured two different MSSP isolates (PEN-SXT/blaZ, dfrA, dfrG and PEN-SXT-TOB/blaZ, dfrG, ant4′) (Table 3). About 31.7% of the S. pseudintermedius had a MDR phenotype (Figure 2). All the three MRSA isolates and some MSSA isolates (20.0%) presented a MDR profile (Tables 2).
Upon WGS, very few SNPs difference (<15) were detected among S. aureus isolates from human carriers within the same household (No 10 and 21) (Figure 3), and these all shared the same repertoire of AMR genes, IEC types and virulence genes and plasmid replicons (Table 4; Figure 3). Concerning the MSSA-CC398 isolates, one isolate from a single person without a co-carrier in their household (No 17) had >250 SNP differences with those from another household (No 21) with two human MSSA-CC398 carriers (Supplementary Table S3). The major difference between these MSSA-CC398 isolates was the absence of the blaZ, aac6′-aph2″, and fnbA genes in the isolate from household number 17 (Figure 3).
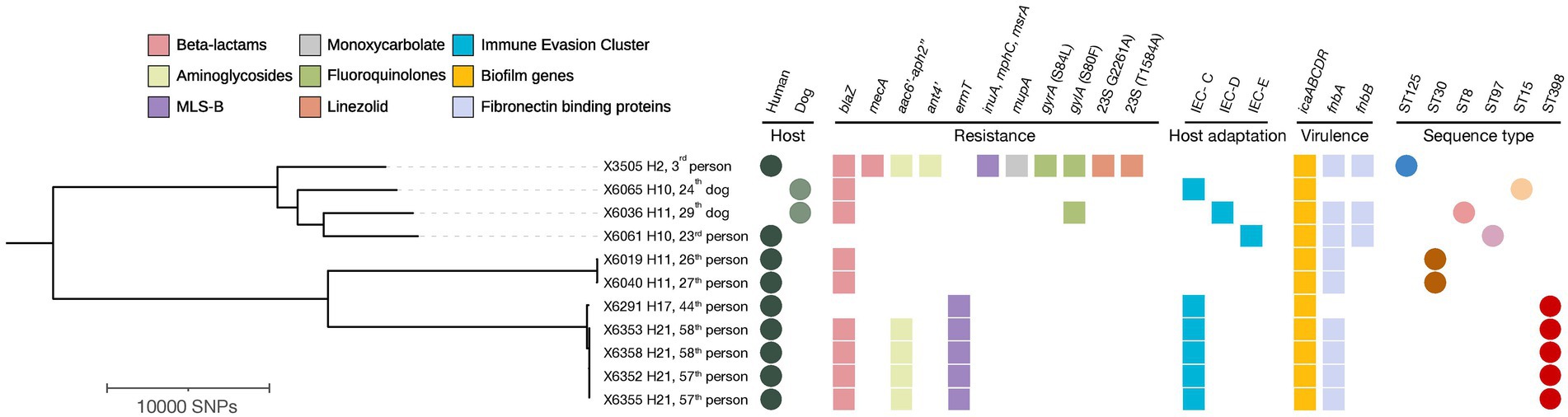
Figure 3. Phylogenetic tree based on core genome SNP analysis of 11 MRSA, MSSA-CC398, and other MSSA isolates from households with ≥2 humans and/or dogs’ carriers. The presence of antimicrobial resistance genes, immune evasion cluster (IEC), icaABCDR operon, fibronectin binding proteins, host type and sequence types (ST) are indicated. The filled circles indicate hosts and sequence types while the filled and squares indicate the confirmed antimicrobial resistance, host adaptation markers and virulence determinants extracted from MLST 2.0, ResFinder, PointFinder and VirulenceFinder. Visuals were obtained using iTOL v6.6. MLS-B, Macrolide-Lincosamide-Streptogramin-B. H, Household.
Concerning the two S. pseudintermedius, each from a dog and human from the same household (No 10), they were both ST1115 NS identical (zero SNP differences) and shared identical virulence genes (Table 4).
Several studies have reported the transmission of CoPS between pets and their owners (Gómez-Sanz et al., 2013c; Han et al., 2016; Sahin-Tóth et al., 2021; Cuny et al., 2022). However, the present study is among the few that studied intra-species and within-host genetic diversities of CoPS in these hosts. Such information can better illustrate the complexity of challenges in the control of AMR in healthy dog-owning households.
The potential influence of dog-ownership on the nasal staphylococcal community (especially S. aureus and S. pseudintermedius colonization) of dogs and humans needs continuous surveillance. The present S. aureus household carriage rate (at least one dog and/or one human) in our study (44.4%) is lower than the 51.2% detected in a previous study performed a decade ago in Spain (Gómez-Sanz et al., 2013c), and the S. aureus carriage rate in humans in our study was also lower (34.1% vs 41.8%) (Gómez-Sanz et al., 2013c). Concerning studies from other European countries, our S. aureus prevalence in humans was higher than those reported in Germany (22%) and Hungary (23.8%) (Holtfreter et al., 2016; Sahin-Tóth et al., 2021).
Data on S. aureus nasal carriage rate among healthy dogs in community settings are sparsely available, varying between 2–8% according to different sources (Fazakerley et al., 2010; Rubin and Chirino-Trejo, 2010; Walther et al., 2012; Sahin-Tóth et al., 2021). Our results further support these numbers: we found a 5.9% carriage rate in dogs. However, in some African and Asian countries, high S. aureus nasal carriage is reported in healthy dogs, such as in Nigeria (36.9%), Indonesia (48.0%), Bangladesh (25.0%) and India (35.0%) (Mustapha et al., 2016; Sekhar et al., 2017; Rahman et al., 2018; Decline et al., 2020). The wide variation in nasal S. aureus carriage in dogs across the continents could be influenced by the local epidemiology of the S. aureus, differences in methodologies, dogs’ hygiene, environmental sanitation, antibiotic use in animals, and/or the health status of owners (Collignon and Voss, 2015; Fletcher, 2015; Valiakos et al., 2020).
As it was observed in this study, human S. aureus carriers were much more prevalent than dog carriers and indicating that transmission between humans and dogs occurs less frequently when compared to possible intrahousehold human-to-human transmission. Taking into consideration the molecular typing results, the genotypic profiles of S. aureus between our studied dog and human isolates were not similar, but those originating from humans in the same households were closely related in several cases, strongly suggesting human-to-human transmission. Moreover, there was a significant association between the household densities and nasal carriage of S. pseudintermedius in households with > than 1 dog and >than 1 human, pointing out for the possibility that a relatively higher household population is related to a higher detection rate of S. pseudintermedius.
The diverse pattern of the CCs of the MSSA isolates from dogs and in-contact humans corroborated the findings from a previous study in Germany and Spain (Gómez-Sanz et al., 2013c; Holtfreter et al., 2016). Here, a few human S. aureus isolates were scn-negative which indicates animal-adapted (perhaps dog-adapted) CCs of S. aureus. Nevertheless, dogs were carriers of MSSA-scn-positive isolates (CC8-IEC-type G and CC15-IEC-type C) suggesting an anthroponotic route of transmission. This is consistent with findings from a previous longitudinal study on S. aureus colonization dynamics in humans and dogs in Spain (Gómez-Sanz et al., 2013b). When compared to the current study, this suggests that humans are the major reservoir of S. aureus contributing to transmission to dogs. However, it is important to mention that zoonotic transmission from dogs to humans is also possible (Abdullahi et al., 2022).
Concerning S. pseudintermedius, 2.4 and 32.4% nasal carriage rates were detected in our studied humans and dogs, respectively. The S. pseudintermedius carriage rate among dogs in this study was higher than previously reported for dogs from Spain (22.7%) (Gómez-Sanz et al., 2013c), but lower than those reported in Canada (46%), Germany (37.5%), Australia (85.4%), Korea (65.9%), or Hungary (34.3%) (Hanselman et al., 2009; Bean and Wigmore, 2016; Han et al., 2016; Sahin-Tóth et al., 2021; Cuny et al., 2022). This suggests that the transfer between nasal S. pseudintermedius among healthy dogs could depend on the number of dogs in the household. This corroborated the previous reports of diverse MSSP lineages from Germany and France (Haenni et al., 2020; Cuny et al., 2022). Moreover, we did find nasal carriage of S. pseudintermedius among a person living with dogs in one of the investigated households. However, this is lower than previously reported in Korea, Spain, and Canada (Hanselman et al., 2009; Gómez-Sanz et al., 2013c; Han et al., 2016). It appears that humans are not natural hosts for S. pseudintermedius, but adaptation to humans could occur. A recent large study has shown diversity between the S. pseudintermedius isolates of human and dog infections with similar pathogenicity islands and virulence gene-containing prophages (Phumthanakorn et al., 2021). The household in our study with both human and dog S. pseudintermedius carriers strongly suggested intrahousehold transmission, as the isolates had no SNP (zero) difference and were confirmed as clones by our genomic analyses. To our knowledge, this lineage (ST1115) has not been reported so far for MSSP from dogs.
In our study, no MRSP was detected among healthy dogs and similar results were obtained in an study in Sweden (Börjesson et al., 2015). However, other studies refer low rates of MRSP nasal carriage in dogs, as 0.9% in Germany, 4.5% in Canada and 2.6% in Norway (Hanselman et al., 2009; Kjellman et al., 2015; Cuny et al., 2022). Moreover, a pooled 4.6% MRSP was reported among healthy dogs in Spain (Gómez-Sanz et al., 2011); of the nine MRSP nasal carriers, one was from a household dog, while the remaining eight were from stray dogs (Gómez-Sanz et al., 2011). The absence of MRSP in our study (healthy animals and household members) and the previously low MRSP in healthy dog studies are remarkably different to the high prevalence in dogs receiving treatment in veterinary clinics in France (16.9%) (Haenni et al., 2014). MRSP seems to be associated with animal-hospital-lineages (Ruiz-Ripa et al., 2021b), whereas isolates that are susceptible or have low AMR levels may represent natural colonizers of dogs. However, among our 19 isolates, three were found to harbour dfrK, a gene that is rarely detected in S. pseudintermedius, which could be linked to the mobile genetic element Tn559 (Ruzauskas et al., 2016; Ruiz-Ripa et al., 2021b). Further characterization of our isolate employing long-read sequencing could help to elucidate this possibility.
The MSSA-CC398 was a predominant lineage in our study, although detected only in humans. Evolutionarily, there are two clades of the CC398 based on the acquisition of SCCmec mobile element (carrying mecA), Tn916 (carrying tet(M)), or prophage φsa3 (carrying IEC), viz: (a) MRSA-CC398, often considered the predominant LA-MRSA clade (mostly IEC-negative) in Europe, (b) livestock-independent (human-adapted) clade of the MSSA-CC398 (often IEC-positive) (Price et al., 2012; Matuszewska et al., 2022). However, on very rare occasions, MRSA-CC398 carrying the φsa3 (scn-positive) and MSSA-CC398-scn-negative have been reported among certain spa types within the CC398 lineage (Price et al., 2012). Hence, these phenomena make it difficult to categorically classify the CC398 lineages into animal-adapted or human-adapted clades. However, it has been established that the CC398 lineage originated in MSSA from humans, acquired the tetracycline (tet(M)) and methicillin (mecA) genes and spread to livestock (Price et al., 2012). Thus, the absence of tet(M) in all the S. aureus and the detection of MSSA-CC398 clade could be inferred as humans-adapted strains. In many cases, MSSA-CC398 clade is associated with the predominant spa type t571 and the macrolide resistance gene ermT (Mama et al., 2021b). Worryingly, this MSSA-CC398 human clade has been recently considered an emergent lineage in invasive human infections in Spain and other countries (Laumay et al., 2021; Mama et al., 2021b). Concerning MSSA-CC398 in dog-owning households, a previous study by Gómez-Sanz et al. (2013c) also reported MSSA-CC398 of the spa type t571. An important similarity between the previous study and the current is that here, MSSA-CC398 isolates were only detected in humans and all were IEC type C of the spa type t571 and t1451. Recently, increased detection of penicillin susceptibility phenotype has been detected among invasive MSSA human isolates that have been causing clinical infections (Mama et al., 2021a). This phenotype has been frequently found among CC398 isolates (Mama et al., 2021a), however, most of our isolates are phenotypically resistant to penicillin. In another study by Gómez-Sanz et al. (2013a), about 7.1% of 98 kennel dogs also carried MSSA-CC398-scn-negative isolates but of different spa types (t034, t5883 and t108), and all were pan-susceptible. Our MSSA CC398 isolates were resistant at least to one antibiotic and worryingly 50% of the isolates were MDR. A major difference between the MSSA-CC398 isolates reported by Gómez-Sanz et al. (2013a) and ours was that here, human isolates were scn-positive (IEC type C). Recently in France, the MSSA-CC398 lineage (t571, t1451 and t18379) was also reported in 14.6 and 27.3% of dogs and cats, respectively (Tegegne et al., 2022). Worthy mentioning is the detection of MSSA-CC398-t571-scn-negative in a cat (Tegegne et al., 2022). The reason for this variation is not fully understood, however, it could be attributed to the spa type associated with the MSSA CC398 isolates or due to the carriage status of the Sa3 prophage (Gómez et al., 2020). The loss of Sa3int prophages in the scn-negative isolates is a major determinant for the human-to-animal transmission of MSSA CC398 (Price et al., 2012; Matuszewska et al., 2022). The findings of Gómez-Sanz et al. (2013a,c) and our study suggest the persistence of MSSA CC398 in humans and dogs.
Another finding of special epidemiological relevance is the dual MRSA/MSSA carriage detected in our study as both MSSA-CC30 and MRSA-CC5 were identified in a human household member. In a previous Spanish study, simultaneous carriage of both MRSA and MSSA of the CC398 lineage was reported in a farm worker with occupational exposure (Gómez et al., 2020). In another study among healthcare students in Portugal, concurrent detection of MRSA and MSSA in a single person was also reported (Coelho et al., 2021).
Though all the MRSA isolates had an MDR phenotype, more than 20% of the MSSA and MSSP isolates were also MDR. Generally, the AMR rate was moderate, but the most common AMR in S. aureus isolates were to penicillin, aminoglycosides, and erythromycin-clindamycin. Conversely, similarly to previous studies in S. pseudintermedius, the predominant AMR phenotype was to sulfamethoxazole-trimethoprim, erythromycin, and tetracycline (Rynhoud et al., 2021). Novel mutations (G2261A & T1584A) in the domain V region of the 23SrDNA of one MRSA isolate was observed, and although the predicted in silico resistance did not reveal a currently known AMR phenotype attributable to this mutation, the strain was phenotypically linezolid resistant. The inability to detect the linezolid phenotype from the genome database could be due to that this mutation has not been fully characterized (not previously reported and deposited in the genome database), as opposed to the most frequently detected 23S rDNA point mutation in staphylococci (G2576T) (Gostev et al., 2021; Ruiz-Ripa et al., 2021a). However, recently, novel point mutations in 23S rRNA associated with linezolid resistance in staphylococci have been reported in S. epidermidis in Austria (Huber et al., 2021) and in S. capitis in China (Han et al., 2022). It could be those novel mutations in domain V of 23S rRNA are silently emerging and mandates the need for close surveillance.
Regarding S. aureus virulence factors, all were negative for TSST-1, PVL, ETA and ETB encoding genes. All except one of the nasal S. pseudintermedius isolates of dogs and human origins carried the lukS/F-I, siet, and expA genes. These leucocidins and exfoliatins are responsible for host-specific clinical infections in dogs (Gharsa et al., 2013; Gómez-Sanz et al., 2013c).
Contrary to the findings of Penna et al. (2013), which reported S. schleiferi subsp. schleiferi carriage among ~32% of healthy dogs in Brazil, only 2 dogs from our study had S. schleiferi nasal carriage (5.4%). This finding is similar to the S. coagulans-positive dogs (4.9%) from the study of Lee et al. (2019) in Korea, but relatively higher than the 1.0% previously reported in Spain (Gómez-Sanz et al., 2013c). Since its first identification in humans in 1988 (Freney et al., 1988), several S. coagulans infections have been reported in humans and pets (May et al., 2005; Abraham et al., 2007; Yarbrough et al., 2017). Furthermore, the recent emergence of S. coagulans among healthy and pyodermic dogs has been a relevant global health issue in veterinary medicine due to its high AMR and diverse virulence factors (May et al., 2012; Abraham et al., 2007; Lee et al., 2019). Contrary to this assertion, all S. coagulans identified in our study were susceptible to antibiotics tested, likely because the isolates were cultured from healthy dogs.
It is worth mentioning a limitation of this study. As this was a one-point prospective study on the potential transmission of CoPS between dogs and dog-owner, randomly selected households were used. The sample size was relatively small and this could affect the detection of rare AMR phenotypes and virulence genes of CoPS in healthy individuals such as MRSP, eta and tst.
The nasal carriage of S. aureus and S. pseudintermedius in healthy dogs’ households were moderate. Low rates of MRSA and S. coagulans, and no MRSP carriage were detected. Human-to-human MSSA and dog-to-human MSSP transmissions were identified in this study. S. pseudintermedius isolates had a homogeneous profile of virulence determinants. The detection of MSSA CC398, an emergent clade that has been implicated in invasive human infections is a relevant health concern and suggests the need for its continuous surveillance of humans, different species of animals and their shared environment from the One Health perspective.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving human and animal participants were reviewed and approved by Ethic Committe of the University of La Rioja, Spain. The patients/participants provided their written informed consent to participate in this study.
IA and CT: conceptualization, methodology, and writing — original draft preparation. IA: laboratory experiments and software analysis. CT, IA, MZ, and CL: validation. IA, CT, MS, and AS: formal analysis and data curation. CT, IA, MS, AS, MZ, and CL: writing — review and editing. CT, CL, and MS: supervision. CT: project administration. CT, MZ, and IA: funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the project PID2019-106158RB-I00 of the MCIN/AEI/10.13039/501100011033 of Spain. Also, it received funding from the European Union’s H2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement N° 801586.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1121564/full#supplementary-material
1. ^https://spa.ridom.de/submission.shtml
4. ^https://github.com/tseemann/mlst
5. ^https://github.com/tseemann/abricate
Abdullahi, I. N., Zarazaga, M., Campaña-Burguet, A., Eguizábal, P., Lozano, C., and Torres, C. (2022). Nasal Staphylococcus aureus and S. pseudintermedius carriage in healthy dogs and cats: a systematic review of their antibiotic resistance, virulence and genetic lineages of zoonotic relevance. J. Appl. Microbiol. 133, 3368–3390. doi: 10.1111/jam.15803
Abraham, J. L., Morris, D. O., Griffeth, G. C., Shofer, F. S., and Rankin, S. C. (2007). Surveillance of healthy cats and cats with inflammatory skin disease for colonization of the skin by methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi ssp. schleiferi. Vet. Dermatol. 18, 252–259. doi: 10.1111/j.1365-3164.2007.00604.x
Asociación Madrileña de Veterinarios de Animales de Compañía (2019). Available at: https://www.aedpac.com/
Bartels, M. D., Petersen, A., Worning, P., Nielsen, J. B., Larner-Svensson, H., Johansen, H. K., et al. (2014). Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 52, 4305–4308. doi: 10.1128/JCM.01979-14
Bean, D. C., and Wigmore, S. M. (2016). Carriage rate and antibiotic susceptibility of coagulase-positive staphylococci isolated from healthy dogs in Victoria, Australia. Austr. Vet. J. 94, 456–460. doi: 10.1111/avj.12528
Börjesson, S., Gómez-Sanz, E., Ekström, K., Torres, C., and Grönlund, U. (2015). Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur. J. Clin. Microbiol. Infect. Dis. 34, 839–844. doi: 10.1007/s10096-014-2300-y
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carroll, K. C., Burnham, C. D., and Westblade, L. F. (2021). From canines to humans: clinical importance of Staphylococcus pseudintermedius. PLoS Pathog. 17:e1009961. doi: 10.1371/journal.ppat.1009961
Chaguza, C., Smith, J. T., Bruce, S. A., Gibson, R., Martin, I. W., and Andam, C. P. (2022). Prophage-encoded immune evasion factors are critical for Staphylococcus aureus host infection, switching, and adaptation. Cell Genomics 2:100194. doi: 10.1016/j.xgen.2022.100194
Coelho, M. J., Magalhães, R., Lopes Cardoso, I., Pina, C., and Pia Ferraz, M. (2021). Identification of nasal carriage of Staphylococcus aureus among nursing students during curricular clinical internships: an observational study. Integrat. J. Med. Sci. 8, 1–16. doi: 10.15342/ijms.2021.440
Collignon, P., and Voss, A. (2015). China, what antibiotics and what volumes are used in food production animals? Antimicrob. Resist. Infect. Control 4:16. doi: 10.1186/s13756-015-0056-5
Cuny, C., Friedrich, A., Kozytska, S., Layer, F., Nübel, U., Ohlsen, K., et al. (2010). Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300, 109–117.
Cuny, C., Layer-Nicolaou, F., Weber, R., Köck, R., and Witte, W. (2022). Colonization of dogs and their owners with Staphylococcus aureus and Staphylococcus pseudintermedius in households, veterinary practices, and healthcare facilities. Microorganisms 10:677. doi: 10.3390/microorganisms10040677
Decline, V., Effendi, M. H., Rahmaniar, R. P., Yanestria, S. M., and Harijani, N. (2020). Profile of antibiotic-resistant and presence of methicillin-resistant Staphylococcus aureus from nasal swab of dogs from several animal clinics in Surabaya, Indonesia. Int. J. One Health 6, 90–94. doi: 10.14202/IJOH.2020.90-94
European Committee on Antimicrobial Susceptibility Testing (EUCAST). (2022). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. Available at: http://www.eucast.org (Accessed May 6, 2022).
Fazakerley, J., Williams, N., Carter, S., McEwan, N., and Nuttall, T. (2010). Heterogeneity of Staphylococcus pseudintermedius isolates from atopic and healthy dogs. Vet. Dermatol. 21, 578–585. doi: 10.1111/j.1365-3164.2010.00894.x
Fletcher, S. (2015). Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ. Health Prev. Med. 20, 243–252. doi: 10.1007/s12199-015-0468-0
Freney, J., Brun, Y., Bes, M., Meugnier, H., Grimont, F., Grimont, P. A., et al. (1988). Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. System. Bacteriol. 38, 168–172. doi: 10.1099/00207713-38-2-168
Gharsa, H., Ben Slama, K., Gómez-Sanz, E., Lozano, C., Klibi, N., Jouini, A., et al. (2013). Antimicrobial resistance, virulence genes, and genetic lineages of Staphylococcus pseudintermedius in healthy dogs in Tunisia. Microb. Ecol. 66, 363–368. doi: 10.1007/s00248-013-0243-y
Gómez, P., Aspiroz, C., Hadjirin, N. F., Benito, D., Zarazaga, M., Torres, C., et al. (2020). Simultaneous nasal carriage by methicillin-resistant and methicillin susceptible Staphylococcus aureus of lineage ST398 in a live pig transporter. Pathogens 9:401. doi: 10.3390/pathogens9050401
Gómez-Sanz, E., Torres, C., Benito, D., Lozano, C., and Zarazaga, M. (2013a). Animal and human Staphylococcus aureus associated clonal lineages and high rate of Staphylococcus pseudintermedius novel lineages in Spanish kennel dogs: predominance of S. aureus ST398. Vet. Microbiol. 166, 580–589. doi: 10.1016/j.vetmic.2013.07.014
Gómez-Sanz, E., Torres, C., Ceballos, S., Lozano, C., and Zarazaga, M. (2013b). Clonal dynamics of nasal Staphylococcus aureus and Staphylococcus pseudintermedius in dog-owning household members. Detection of MSSA ST(398). PLoS One 8:e69337. doi: 10.1371/journal.pone.0069337
Gómez-Sanz, E., Torres, C., Lozano, C., Sáenz, Y., and Zarazaga, M. (2011). Detection and characterization of methicillin-resistant Staphylococcus pseudintermedius in healthy dogs in La Rioja, Spain. Comp. Immunol. Microbiol. Infect. Dis. 34, 447–453. doi: 10.1016/j.cimid.2011.08.002
Gómez-Sanz, E., Torres, C., Lozano, C., and Zarazaga, M. (2013c). High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp. Immunol. Microbiol. Infect. Dis. 36, 83–94. doi: 10.1016/j.cimid.2012.10.001
González-Martín, M., Corbera, J. A., Suárez-Bonnet, A., and Tejedor-Junco, M. T. (2020). Virulence factors in coagulase-positive staphylococci of veterinary interest other than Staphylococcus aureus. Vet. Q. 40, 118–131. doi: 10.1080/01652176.2020.1748253
Gostev, V., Leyn, S., Kruglov, A., Likholetova, D., Kalinogorskaya, O., Baykina, M., et al. (2021). Global expansion of linezolid-resistant coagulase-negative staphylococci. Front. Microbiol. 12:661798. doi: 10.3389/fmicb.2021.661798
Haenni, M., de Moraes, N. A., Châtre, P., Médaille, C., Moodley, A., and Madec, J. Y. (2014). Characterisation of clinical canine meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius in France. J. Global Antimicrob. Resist. 2, 119–123. doi: 10.1016/j.jgar.2014.02.002
Haenni, M., El Garch, F., Miossec, C., Madec, J. Y., Hocquet, D., and Valot, B. (2020). High genetic diversity among methicillin-susceptible Staphylococcus pseudintermedius in dogs in Europe. J. Glob. Antimicrob. Resist. 21, 57–59. doi: 10.1016/j.jgar.2020.02.016
Han, J. I., Yang, C. H., and Park, H. M. (2016). Prevalence and risk factors of Staphylococcus spp. carriage among dogs and their owners: a cross-sectional study. Vet. J., 212, 15–221. doi: 10.1016/j.tvjl.2015.10.059
Han, X., Zou, G., Liu, J., Yang, C., Du, X., Chen, G., et al. (2022). Mechanisms of linezolid resistance in Staphylococcus capitis with the novel mutation C2128T in the 23S rRNA gene in China. BMC Microbiol. 22:203. doi: 10.1186/s12866-022-02616-9
Hanselman, B. A., Kruth, S. A., Rousseau, J., and Weese, J. S. (2009). Coagulase positive staphylococcal colonization of humans and their household pets. Can. Vet. J. 50, 954–958.
Holtfreter, S., Grumann, D., Balau, V., Barwich, A., Kolata, J., Goehler, A., et al. (2016). Molecular epidemiology of Staphylococcus aureus in the general population in Northeast Germany: results of the study of health in Pomerania (SHIP-TREND-0). J. Clin. Microbiol. 54, 2774–2785. doi: 10.1128/JCM.00312-16
Hu, D., Li, S., Fang, R., and Ono, H. K. (2021). Update on molecular diversity and multipathogenicity of staphylococcal superantigen toxins. Anim. Dis. 1:7. doi: 10.1186/s44149-021-00007-7
Huber, S., Knoll, M. A., Berktold, M., Würzner, R., Brindlmayer, A., Weber, V., et al. (2021). Genomic and phenotypic analysis of linezolid-resistant Staphylococcus epidermidis in a tertiary hospital in Innsbruck, Austria. Microorganisms 9:1023. doi: 10.3390/microorganisms9051023
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kjellman, E. E., Slettemeås, J. S., Small, H., and Sunde, M. (2015). Methicillin-resistant Staphylococcus pseudintermedius (MRSP) from healthy dogs in Norway – occurrence, genotypes and comparison to clinical MRSP. Microbiol. Open 4, 857–866. doi: 10.1002/mbo3.258
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Laumay, F., Benchetrit, H., Corvaglia, A. R., van der Mee-Marquet, N., and François, P. (2021). The Staphylococcus aureus CC398 lineage: an evolution driven by the acquisition of prophages and other mobile genetic elements. Genes 12:1752. doi: 10.3390/genes12111752
Lee, G. Y., Lee, H. H., Hwang, S. Y., Hong, J., Lyoo, K. S., and Yang, S. J. (2019). Carriage of Staphylococcus schleiferi from canine otitis externa: antimicrobial resistance profiles and virulence factors associated with skin infection. J. Vet. Sci. 20:e6. doi: 10.4142/jvs.2019.20.e6
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liu, C. M., Price, L. B., Hungate, B. A., Abraham, A. G., Larsen, L. A., Christensen, K., et al. (2015). Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 1:e1400216. doi: 10.1126/sciadv.1400216
Lozano, C., Rezusta, A., Ferrer, I., Pérez-Laguna, V., Zarazaga, M., Ruiz-Ripa, L., et al. (2017). Staphylococcus pseudintermedius human infection cases in Spain: Dog-to-human transmission. Vector. Borne Zoonotic Dis. 17(4, 268–270. doi: 10.1089/vbz.2016.2048
Madhaiyan, M., Wirth, J. S., and Saravanan, V. S. (2020). Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 70, 5926–5936. doi: 10.1099/ijsem.0.004498
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mama, O. M., Aspiroz, C., Lozano, C., Ruiz-Ripa, L., Azcona, J. M., Seral, C., et al. (2021a). Penicillin susceptibility among invasive MSSA infections: a multicentre study in 16 Spanish hospitals. J. Antimicrob. Chemother. 76, 2519–2527. doi: 10.1093/jac/dkab208
Mama, O. M., Aspiroz, C., Ruiz-Ripa, L., Ceballos, S., Iñiguez-Barrio, M., Cercenado, E., et al. (2021b). Prevalence and genetic characteristics of Staphylococcus aureus CC398 isolates from invasive infections in Spanish hospitals, focusing on the livestock-independent CC398-MSSA clade. Front. Microbiol. 12:623108. doi: 10.3389/fmicb.2021.623108
Matuszewska, M., Murray, G. G. R., Ba, X., Wood, R., Holmes, M. A., and Weinert, L. A. (2022). Stable antibiotic resistance and rapid human adaptation in livestock-associated MRSA. elife 11:e74819. doi: 10.7554/eLife.74819
May, E. R., Hnilica, K. A., Frank, L. A., Jones, R. D., and Bemis, D. A. (2005). Isolation of Staphylococcus schleiferi from healthy dogs and dogs with otitis, pyoderma, or both. J. Am. Vet. Med. Assoc. 227, 928–931. doi: 10.2460/javma.2005.227.928
May, E. R., Kinyon, J. M., and Noxon, J. O. (2012). Nasal carriage of Staphylococcus schleiferi from healthy dogs and dogs with otitis, pyoderma or both. Vet. Microbiol. 160, 443–448. doi: 10.1016/j.vetmic.2012.06.020
Murray, A. K., Lee, J., Bendall, R., Zhang, L., Sunde, M., Schau Slettemeås, J., et al. (2018). Staphylococcus cornubiensis sp. nov., a member of the Staphylococcus intermedius Group (SIG). Int. J. Syst. Evol. Microbiol. 68, 3404–3408. doi: 10.1099/ijsem.0.002992
Mustapha, M., Bukar-Kolo, Y. M., Geidam, Y. A., and Gulani, I. A. (2016). Phenotypic and genotypic detection of methicillin-resistant Staphylococcus aureus in hunting dogs in Maiduguri metropolitan, Borno State, Nigeria. Vet. World 9, 501–506. doi: 10.14202/vetworld.2016.501-506
Orsini, M., Petrin, S., Corrò, M., Baggio, G., Spagnolo, E., and Losasso, C. (2022). Anthroponotic-based transfer of Staphylococcus to dog: a case study. Pathogens 11:802. doi: 10.3390/pathogens11070802
Penna, B., Mendes, W., Rabello, R., and Lilenbaum, W. (2013). Carriage of methicillin susceptible and resistant Staphylococcus schleiferi among dog with or without topic infections. Vet. Microbiol. 162, 298–299. doi: 10.1016/j.vetmic.2012.08.022
Perreten, V., Kania, S. A., and Bemis, D. (2020). Staphylococcus ursi sp. nov., a new member of the ‘Staphylococcus intermedius group’ isolated from healthy black bears. Int. J. Syst. Evol. Microbiol. 70, 4637–4645. doi: 10.1099/ijsem.0.004324
Phumthanakorn, N., Schwendener, S., Donà, V., Chanchaithong, P., Perreten, V., and Prapasarakul, N. (2021). Genomic insights into methicillin-resistant Staphylococcus pseudintermedius isolates from dogs and humans of the same sequence types reveals diversity in prophages and pathogenicity islands. PLoS One 16:e0254382. doi: 10.1371/journal.pone.0254382
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3, e00305–e00311. doi: 10.1128/mBio.00305-11
Priyantha, M. (2022). An overview of human infections caused by Staphylococcus pseudintermedius: a zoonotic risk of the oldest friend. Sri Lankan J. Infect Dis. 12:1. doi: 10.4038/sljid.v12i1.8415
Rahman, M. M., Amin, K. B., Rahman, S. M. M., Khair, A., Rahman, M., Hossain, A., et al. (2018). Investigation of methicillin-resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR. BMC Vet. Res. 14:300. doi: 10.1186/s12917-018-1611-0
Rahman, M. T., Sobur, M. A., Islam, M. S., Ievy, S., Hossain, M. J., El Zowalaty, M. E., et al. (2020). Zoonotic diseases: etiology, impact, and control. Microorganisms 8:1405. doi: 10.3390/microorganisms8091405
Rodrigues, A. C., Belas, A., Marques, C., Cruz, L., Gama, L. T., and Pomba, C. (2018). Risk factors for nasal colonization by methicillin-resistant staphylococci in healthy humans in professional daily contact with companion animals in Portugal. Microb. Drug Resist. 24, 434–446. doi: 10.1089/mdr.2017.0063
Rubin, J. E., and Chirino-Trejo, M. (2010). Pharyngeal, rectal and nasal colonization of clinically healthy dogs with Staphylococcus aureus. Vet. Microbiol. 143, 440–441. doi: 10.1016/j.vetmic.2009.11.012
Ruiz-Ripa, L., Feßler, A. T., Hanke, D., Eichhorn, I., Azcona-Gutiérrez, J. M., Alonso, C. A., et al. (2021a). Mechanisms of linezolid resistance among clinical Staphylococcus spp. in Spain: spread of methicillin-and linezolid-resistant S. epidermidis ST2. Microb. Drug Resist. 27(2, 145–153. doi: 10.1089/mdr.2020.0122
Ruiz-Ripa, L., Simón, C., Ceballos, S., Ortega, C., Zarazaga, M., Torres, C., et al. (2021b). S. pseudintermedius and S. aureus lineages with transmission ability circulate as causative agents of infections in pets for years. BMC Vet. Res. 17:42. doi: 10.1186/s12917-020-02726-4
Ruzauskas, M., Couto, N., Pavilonis, A., Klimiene, I., Siugzdiniene, R., Virgailis, M., et al. (2016). Characterization of Staphylococcus pseudintermedius isolated from diseased dogs in Lithuania. Pol. J. Vet. Sci. 19, 7–14. doi: 10.1515/pjvs-2016-0002
Rynhoud, H., Forde, B. M., Beatson, S. A., Abraham, S., Meler, E., Soares Magalhães, R. J., et al. (2021). Molecular epidemiology of clinical and colonizing methicillin-resistant staphylococcus isolates in companion animals. Front. Vet. Sci. 8:620491. doi: 10.3389/fvets.2021.620491
Sahin-Tóth, J., Kovács, E., Tóthpál, A., Juhász, J., Forró, B., Bányai, K., et al. (2021). Whole genome sequencing of coagulase positive staphylococci from a dog-and-owner screening survey. PLoS One 16:e0245351. doi: 10.1371/journal.pone.0245351
Sahl, J. W., Lemmer, D., Travis, J., Schupp, J. M., Gillece, J. D., Aziz, M., et al. (2016). NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2:e000074. doi: 10.1099/mgen.0.000074
Saidenberg, A. B. S., Van Vliet, A. H. M., Stegger, M., Johannesen, T. B., Semmler, T., Cunha, M., et al. (2022). Genomic analysis of the zoonotic ST73 lineage containing avian and human extraintestinal pathogenic Escherichia coli (ExPEC). Vet. Microbiol. 267:109372. doi: 10.1016/j.vetmic.2022.109372
Sakr, A., Brégeon, F., Mège, J. L., Rolain, J. M., and Blin, O. (2018). Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 9:2419. doi: 10.3389/fmicb.2018.02419
Sekhar, M., Rao, T., and Sharif, N. (2017). Nasal colonization of methicillin resistant Staphylococcus aureus (MRSA) among dogs and dog handlers in Andhra Pradesh, India. Pharm. Innov. 6, 86–90.
Somayaji, R., Priyantha, M. A., Rubin, J. E., and Church, D. (2016). Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn. Microbiol. Infect. Dis. 85, 471–476. doi: 10.1016/j.diagmicrobio.2016.05.008
Stegger, M., Lindsay, J. A., Moodley, A., Skov, R., Broens, E. M., and Guardabassi, L. (2011). Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J. Clin. Microbiol. 49, 732–734. doi: 10.1128/JCM.01970-10
Tam, K., and Torres, V. J. (2019). Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectrum 7, 1–34. doi: 10.1128/microbiolspec.GPP3-0039-2018
Tegegne, H. A., Madec, J. Y., and Haenni, M. (2022). Is methicillin-susceptible Staphylococcus aureus (MSSA) CC398 a true animal-independent pathogen? J. Glob. Antimicrob. Resist. 29, 120–123. doi: 10.1016/j.jgar.2022.02.017
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Trinh, P., Zaneveld, J. R., Safranek, S., and Rabinowitz, P. M. (2018). One health relationships between human, animal, and environmental microbiomes: a mini-review. Front. Public Health 6:235. doi: 10.3389/fpubh.2018.00235
Valiakos, G., Pavlidou, E., Zafeiridis, C., Tsokana, C. N., and Del Rio Vilas, V. J. (2020). Antimicrobial practices among small animal veterinarians in Greece: a survey. One Health Outlook 2:7. doi: 10.1186/s42522-020-00013-8
Van Balen, J. C., Landers, T., Nutt, E., Dent, A., and Hoet, A. E. (2017). Molecular epidemiological analysis to assess the influence of pet-ownership in the biodiversity of Staphylococcus aureus and MRSA in dog-and non-dog-owning healthy households. Epidemiol. Infect. 145, 1135–1147. doi: 10.1017/S0950268816003228
van Wamel, W. J., Rooijakkers, S. H., Ruyken, M., van Kessel, K. P., and van Strijp, J. A. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188, 1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006
Walther, B., Hermes, J., Cuny, C., Wieler, L. H., Vincze, S., Abou Elnaga, Y., et al. (2012). Sharing more than friendship–nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS One 7:e35197. doi: 10.1371/journal.pone.0035197
Yarbrough, M. L., Hamad, Y., Burnham, C. A., and George, I. A. (2017). The brief case: bacteremia and vertebral osteomyelitis due to Staphylococcus schleiferi. J. Clin. Microbiol. 55, 3157–3161. doi: 10.1128/JCM.00500-17
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Keywords: Staphylococcus, zoonosis, pets, MSSA-CC398, Staphylococcus pseudintermedius carriage, Staphylococcus aureus carriage, linezolid resistance
Citation: Abdullahi IN, Lozano C, Zarazaga M, Saidenberg ABS, Stegger M and Torres C (2023) Clonal relatedness of coagulase-positive staphylococci among healthy dogs and dog-owners in Spain. Detection of multidrug-resistant-MSSA-CC398 and novel linezolid-resistant-MRSA-CC5. Front. Microbiol. 14:1121564. doi: 10.3389/fmicb.2023.1121564
Received: 11 December 2022; Accepted: 31 January 2023;
Published: 02 March 2023.
Edited by:
Qixia Luo, Zhejiang University, ChinaReviewed by:
Amjad Islam Aqib, Cholistan University of Veterinary and Animal Sciences, PakistanCopyright © 2023 Abdullahi, Lozano, Zarazaga, Saidenberg, Stegger and Torres. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Torres, Y2FybWVuLnRvcnJlc0B1bmlyaW9qYS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.