- 1Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, China
- 2College of Food Science, Southwest University, Chongqing, China
- 3Engineering Research Center for Replacement Technology of Feed Antibiotics of Yunnan College, Kunming, Yunnan, China
Lactic acid bacteria are generally regarded as alternatives to antibiotics in livestock and poultry farming, especially Lactobacillus strains, which are safe and have probiotic potential. Although Lactobacillus salivarius has long been proposed to be a probiotic, the understanding of the roles of this species is still in its infancy. Here, a strain of L. salivarius CGMCC20700 isolated from the intestinal mucosa of Yunnan black-bone chicken broilers was investigated in the context of its safety and probiotic characteristics by whole-genome sequencing in parallel with phenotypic analysis. Whole-genome sequencing results showed that L. salivarius CGMCC20700 has a single scaffold of 1,737,577 bp with an average guanine-to-cytosine (GC) ratio of 33.51% and 1,757 protein-coding genes. The annotation of Clusters of Orthologous Groups (COG) classified the predicted proteins from the assembled genome as possessing cellular, metabolic, and information-related functions. Sequences related to risk assessment, such as antibiotic resistance and virulence genes, were identified, and the strain was further confirmed as safe according to the results of antibiotic resistance, hemolytic, and acute oral toxicology tests. Two gene clusters of antibacterial compounds and broad-spectrum antimicrobial activity were identified using genome mining tools and antibacterial spectrum tests. Stress resistance genes, active stressor removal genes, and adhesion related genes that were identified and examined with various phenotypic assays (such as stress tolerance tests in acids and bile salts and auto aggregation and hydrophobicity assays). The strain showed a high survival rate in the presence of bile salts and under acidic conditions and exhibited significant auto aggregation capacity and hydrophobicity. Overall, L. salivarius CGMCC20700 demonstrated excellent safety and probiotic potential at both the genomic and physiological levels and can be considered an appropriate candidate probiotic for livestock and poultry farming.
Introduction
For decades, antibiotics have been widely used in livestock and poultry farming-related fields and are commonly used to prevent or treat bacterial infections and as antimicrobial growth promoters (Al-Khalaifa et al., 2019). The United Nations Food and Agriculture Organization (FAO) reported that the total amount of antibiotics used in farming processes such as poultry, animal husbandry, and aquaculture is alarming (Pierce et al., 2020). The global usage of antibiotics in food animals reached 93,309 ton in 2017 and was projected to increase by 11.5% to 104,079 tons by 2030 (Tiseo et al., 2020). However, the widespread use of antibiotics has created problems, including animal gut microbiome disorders, the development of antibiotic resistance, and environmental issues (Al-Khalaifa et al., 2019; Ramirez et al., 2020; Tiseo et al., 2020). For instance, previous studies have shown that vancomycin treatment in mice can promote the proliferation of pathogenic gram-negative bacteria, leading to an imbalance in the gut microbiome (Yamaguchi et al., 2020). In a study of Enterococcus in fecal samples from broilers fed antibiotics, VanA transposons were found to be transported from animals to humans (Van et al., 2002). Many countries have now promulgated laws to ban the nontherapeutic use of antibiotics in poultry and livestock farming processes, including the European Union, United States of America, and China (FDA, 2009; Ricke et al., 2020). Therefore, there is an urgent need to find effective alternatives to antibiotics for application in the livestock and poultry industries.
Probiotics are defined by the WHO/FAO as “live microorganisms,” which can provide health benefits to the host when given in sufficient amounts (García-Hernández et al., 2016; Kai et al., 2020). For example, dietary supplementation with Lactobacillus fermentum in a mouse model of colitis was proven to initiate signaling pathways involved in epithelial barrier protection (Paveljšek et al., 2018). Similarly, supplementation with Lactobacillus rhamnosus GG and Lactobacillus casei IMAU60214 triggered innate immune responses and improved the phagocytic and bactericidal activities of human macrophages (Rocha-Ramírez et al., 2017). Moreover, many probiotics have been shown to have positive effects in livestock and poultry farming processes (Cerezuela et al., 2012; Hiremath and Pragasam, 2022). Bacillus subtilis feeding for 2 weeks enhanced the serum IgM level and leukocyte phagocytosis activity in gilthead seabream (Cerezuela et al., 2012). Lacticaseibacillus paracasei NSMJ56 feeding for 10 days increased the abundance of CD4+ T cells in the small intestinal lamina propria and gut microbial diversity in early-age broiler chickens (Joo et al., 2022). Therefore, this evidence indicates that probiotics can serve as functional microbiological resources for use as alternatives to antibiotics.
Lactobacillus salivarius, an important member of lactic acid bacteria (LAB), is widely distributed in traditional fermented food and animal gastrointestinal tracts, particularly in the avian intestine (Chiu et al., 2017; Li H. W. et al., 2021). Previous studies have shown that L. salivarius strains, as potential probiotic strains, possess inhibitory activity against intestinal pathogens and regulate the balance of the intestinal microbiome due to the production of many selectively stimulating metabolites, as well as antimicrobial compounds, antioxidants, and organic acids (Chen et al., 2022; Xu et al., 2022). For instance, in broilers, L. salivarius Erya conferred resistance to Salmonella pullorum infection and alleviated the negative effects of aflatoxin B1, while adding L. salivarius Erya also improved growth performance, liver function, and meat quality (Chen et al., 2022); in laying hens, L. salivarius CML352 was considered a suitable probiotic with positive effects on intestinal health and performance (Xu et al., 2022). These scattered examples indicate that L. salivarius is possibly a probiotic species. However, in recent years, a growing body of research has revealed that the functions of probiotics are highly strain specific and that their biological effects should be individually evaluated (Tanizawa et al., 2015; Li et al., 2020). Particularly, for newly isolated probiotics, it is necessary to analyze and evaluate the related characteristics of their probiotic function at the gene level to explore more potential biological functions and information (Tanizawa et al., 2015; Saroj and Gupta, 2020).
Although many whole genome sequences of LAB probiotics have been reported, their whole genome participation and in vivo probiotic effects are still poorly understood (Goel et al., 2020; Qureshi et al., 2020; Saroj and Gupta, 2020). In our previous study, a strain of L. salivarius CGMCC20700 with significant antibacterial ability was isolated from the intestinal mucosa of Yunnan black-bone chicken broilers and confirmed to produce active antimicrobial substances as a novel bacteriocin (Li H. W. et al., 2021). However, the safety level, probiotic capabilities, and practical potential for this strain to be used as an alternative to antibiotics remain unknown. Therefore, the aim of this study was to evaluate the safety and potential probiotic characteristics of L. salivarius CGMCC20700 using a series of in vitro tests. Additionally, the whole genome sequence was analyzed to provide a deeper understanding and insight into the full breadth of its biological capabilities for an assessment of safety and probiotic-associated capacity.
Materials and methods
Bacterial strains and growth conditions
Lactobacillus salivarius CGMCC20700 was isolated from the intestinal mucosa of Yunnan black-bone chickens (Gallus gallus) and is deposited at the China General Microbiological Culture Collection Center (CGMCC). The strain was cultured in de Man Rogosa Sharpe (MRS) medium (Solarbio, Beijing, China) at 37°C for 24 h in anaerobic jars for routine use, as previously reported (Li H. W. et al., 2021).
Identification of Lactobacillus salivarius CGMCC20700
The morphology and phylogenetics of L. salivarius CGMCC20700 were assessed as previously reported (Fu et al., 2022; Jiang et al., 2022a). Briefly, L. salivarius CGMCC20700 was cultured on MRS solid medium plates at 37°C for 24 h, the colonies on the plates were observed, Gram staining was conducted, and the bacterial cell morphology was observed by a scanning electron microscope (S-3000 N, Hitachi) (Jiang et al., 2022a). Finally, the genotypic identification of L. salivarius CGMCC20700 was conducted by comparison of its 16S rRNA sequence analysis with the sequences deposited in the National Center Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST). The phylogenetic tree was reconstructed using MEGA6 software with the neighbor-joining method (Tamura et al., 2011).
Whole genome sequencing, assembly, and annotation
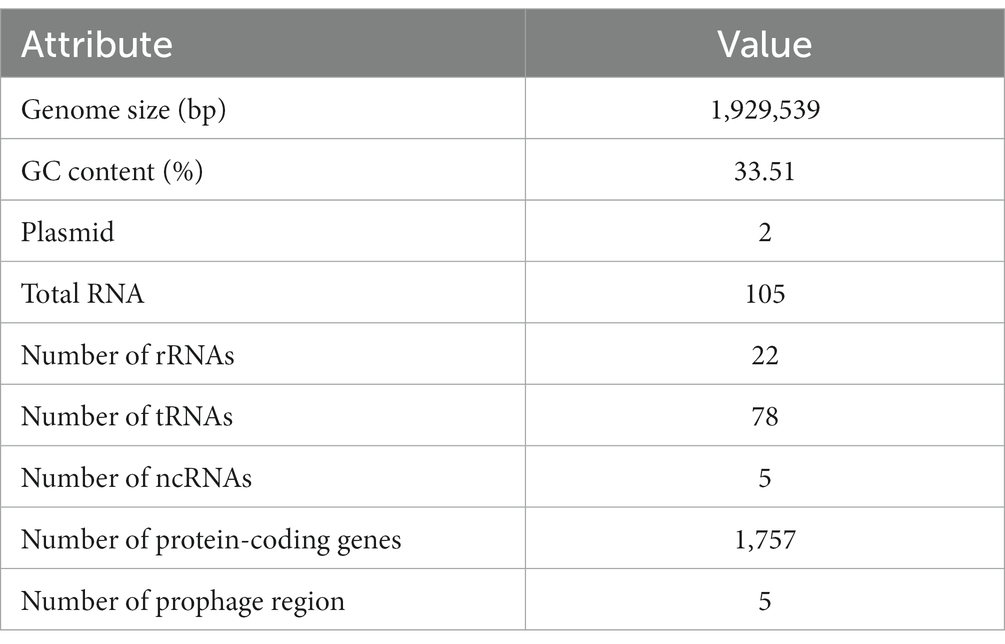
Whole-genome sequencing, assembly and annotation of L. salivarius CGMCC20700 were conducted by Beijing Genomics Institute (Shenzhen, China). Briefly, L. salivarius CGMCC20700 was cultured to exponential phase and collected by centrifugation at 8000 × g for 5 min, and the total DNA of bacterial cells was extracted using a DNA Purification kit (Solarbio, Beijing, China). Subsequently, whole-genome sequencing was carried out using a combination of the second-generation BGISEQ platform and the third-generation PacBio platform sequencing technology (Huada Gene Co., Ltd., Shenzhen, China) (Dai et al., 2021; Gao et al., 2021). The assembly of the completed sequence was performed by using GATK v. v1.6–13 and SMRT Analysis v. v2.2.0 software to assemble the main complete and continuous contigs. After single-base correction, loop judgment and other analyzes based on the obtained contigs, we generated credible complete map sequences. The genome annotation of L. salivarius CGMCC20700 was performed using the Prokaryotic Genome Annotation Pipeline (PGAP) algorithm of the National Center for Biotechnological Information (NCBI) (Kai et al., 2020; Vyacheslav et al., 2020). Then, GeneMark software (V4.17)1 was adopted for the prediction of protein-coding RNA in the whole genome. The prediction of sRNA, rRNA, and tRNA was determined using the cmsearch program V1.1rc4, RNAmmer 1.2 and tRNAscan-SE V1.3.1. The CRISPR regions were identified using CRISPR digger V1.0, and plasmid information was obtained using an online tool with Plasmid Finder. The Cluster of Orthologous Groups of Proteins (COG) database was used for general function annotation. The genome sequences of L. salivarius CGMCC20700 have been submitted to GenBank under accession number CP101685.
Safety assessment of Lactobacillus salivarius CGMCC20700
Identification of safety-related genes
Safety-related genes were identified to evaluate the potential safety at the genomic level of L. salivarius CGMCC20700, as previously reported (Fu et al., 2022; Wu et al., 2022). Putative virulence genes of L. salivarius CGMCC20700 were analyzed by comparison with the VFDB. Antibiotic resistance genes of L. salivarius CGMCC20700 were identified using the ARDB.
Antibiotic resistance analysis
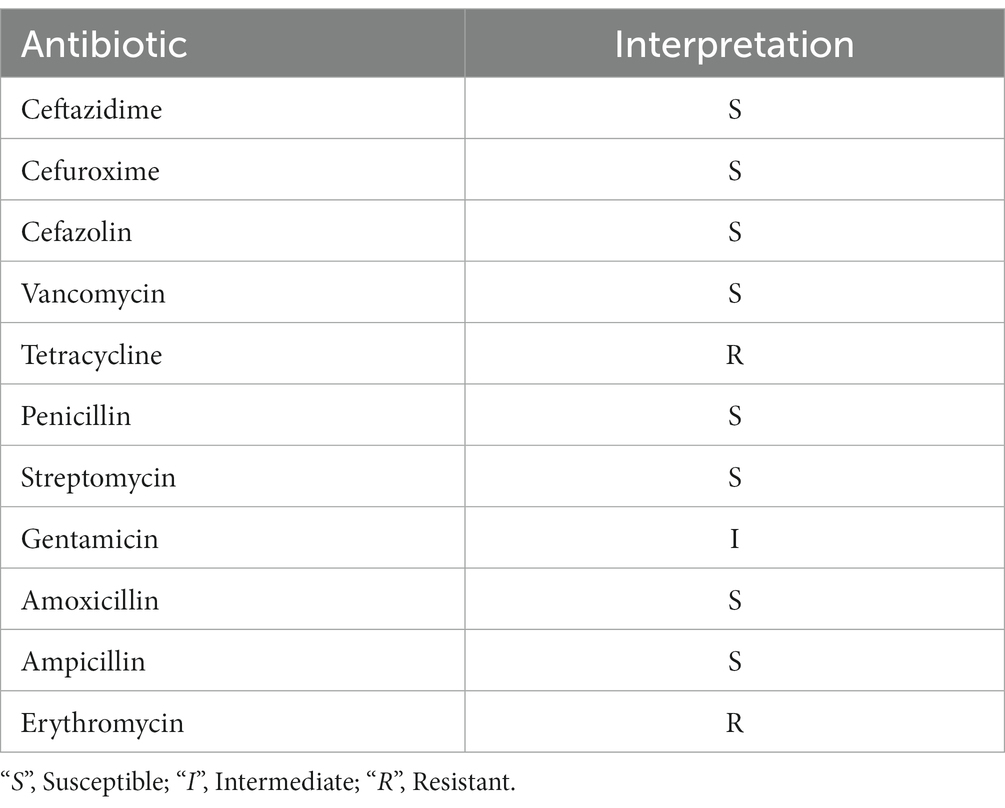
Antibiotic resistance was evaluated by adopting the method from a previous study (Zheng et al., 2021). Briefly, susceptibility to the following 13 antibiotics was assessed using filter paper disks infused with the following: 30 μg each of ceftazidime, cefuroxime, cefazolin, vancomycin, and tetracycline; 10 μg each of penicillin, streptomycin, gentamicin, and amoxicillin; 100 μg of ampicillin; and 15 μg of erythromycin (Shanghai Yibaiju Economic and Trade Co., Ltd., Shanghai, China). A volume of 100 μl of L. salivarius CGMCC20700 cultures (107 CFU/mL) was spread on MRS solid medium plates, and the antibiotic-infused paper disks were adhered to the surface of MRS medium and cultured at 37°C for 24 h. The inhibition zones (mm) were measured using a Vernier caliper. According to the guidelines of the Institute of Clinical and Laboratory Standards Institute (CLSI), the drug resistance susceptibility was determined as follows: S = sensitive (zone diameter ≥ 17 mm); I = intermediate (zone diameter 12 to 17 mm); R = resistant (zone diameter ≤ 1.2 cm).
Hemolytic activity analysis
The hemolytic activity assays were performed by adopting the method from Zheng et al. (2021). Briefly, L. salivarius CGMCC20700 and Escherichia coli CMCC(B)44102 cultures were crossed, streaked on Columbia blood agar containing fresh sheep blood (Shanghai Yibaiju Economic and Trade Co., Ltd., Shanghai, China) and incubated at 37°C for 24 h. Hemolytic activity was determined according to the following rules: if the colony (strain) showed a grass-green ring on the plate, the strain was identified as α-hemolytic; if the colony showed a completely clear hemolytic ring on the plate, the strain was identified as β-hemolytic; and if no changes were observed, the colony was identified as nonhemolytic.
Broiler acute toxicity assay
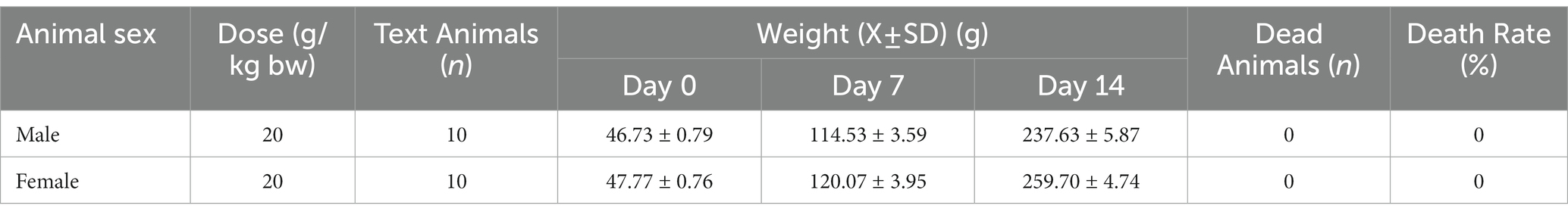
An acute toxicity assay was conducted to assess the safety of L. salivarius CGMCC20700 in broilers. Briefly, L. salivarius CGMCC20700 cultures (107 CFU/mL) were harvested by centrifugation (8,000 × g, 5 min), washed several times with sterile water, mixed well with freeze-dried protection solutions (10% trehalose and 10% skim milk) and then freeze-dried to the desired concentration (1× 1010 CFU/g). Twenty healthy broilers (half male, 3 days old) were provided by Kunming Yuankang Food Agriculture and Animal Husbandry Co., Ltd. The freeze-dried L. salivarius CGMCC20700 powders were prepared with sterile water to a concentration of 2 g/mL, and a 20 mL/kg body weight dose was gavaged two times a day at 4-h intervals after a 6-h fast. The diet composition and housing conditions for broiler feeding were followed as presented in our previous study (Jiang et al., 2022b). The experiment lasted for 14 days, and internal tissues and organs were immediately observed and evaluated by the naked eye after exposure.
Assessment of probiotic properties
Analysis of antimicrobial compounds in the genome
Antimicrobial genes were identified to confirm the antimicrobial ability of L. salivarius CGMCC20700, as previously reported (Goel et al., 2020; Zheng et al., 2021). The presence of gene clusters of nonribosomally synthesized secondary metabolites (NRPS) was evaluated using AntiSMASH 5.2 The potential bacteriocin synthesis gene clusters were identified using the BAGEL4 webserver.3
Assessment of antimicrobial spectrum
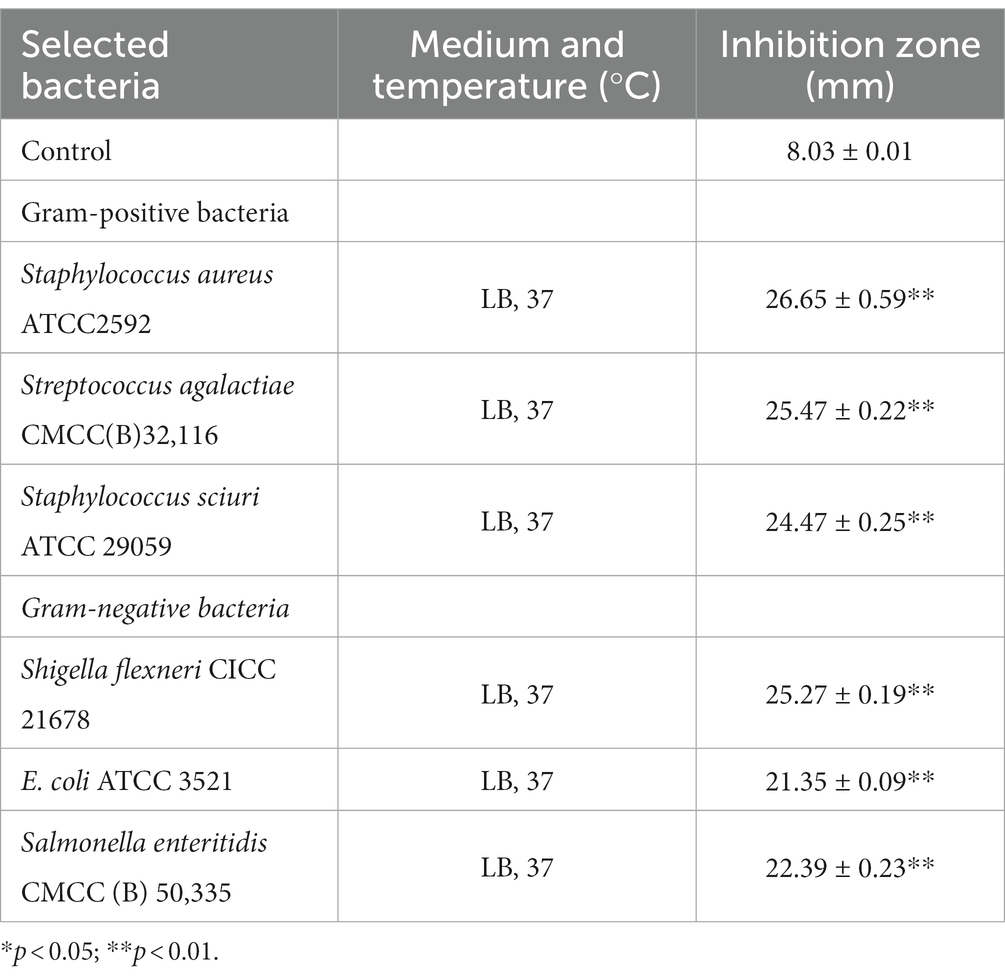
The antimicrobial activity of L. salivarius CGMCC20700 against six common pathogen indicator strains was determined as previously reported (Jiang et al., 2022a). Briefly, all indicator strains (as shown in Table 1) were precultured in Luria-Bertani (LB) liquid medium (Solarbio, Beijing, China) at 37°C for 12 h. Later, the antimicrobial activity of the L. salivarius CGMCC20700 cell-free supernatant (200 μL) against each indicator strain (107 CFU/mL) was determined by the Oxford cup double-plate method. The inhibition zone was measured using a Vernier caliper. MRS broth medium was used as a control.
Identification of probiotic-related genes in the genome
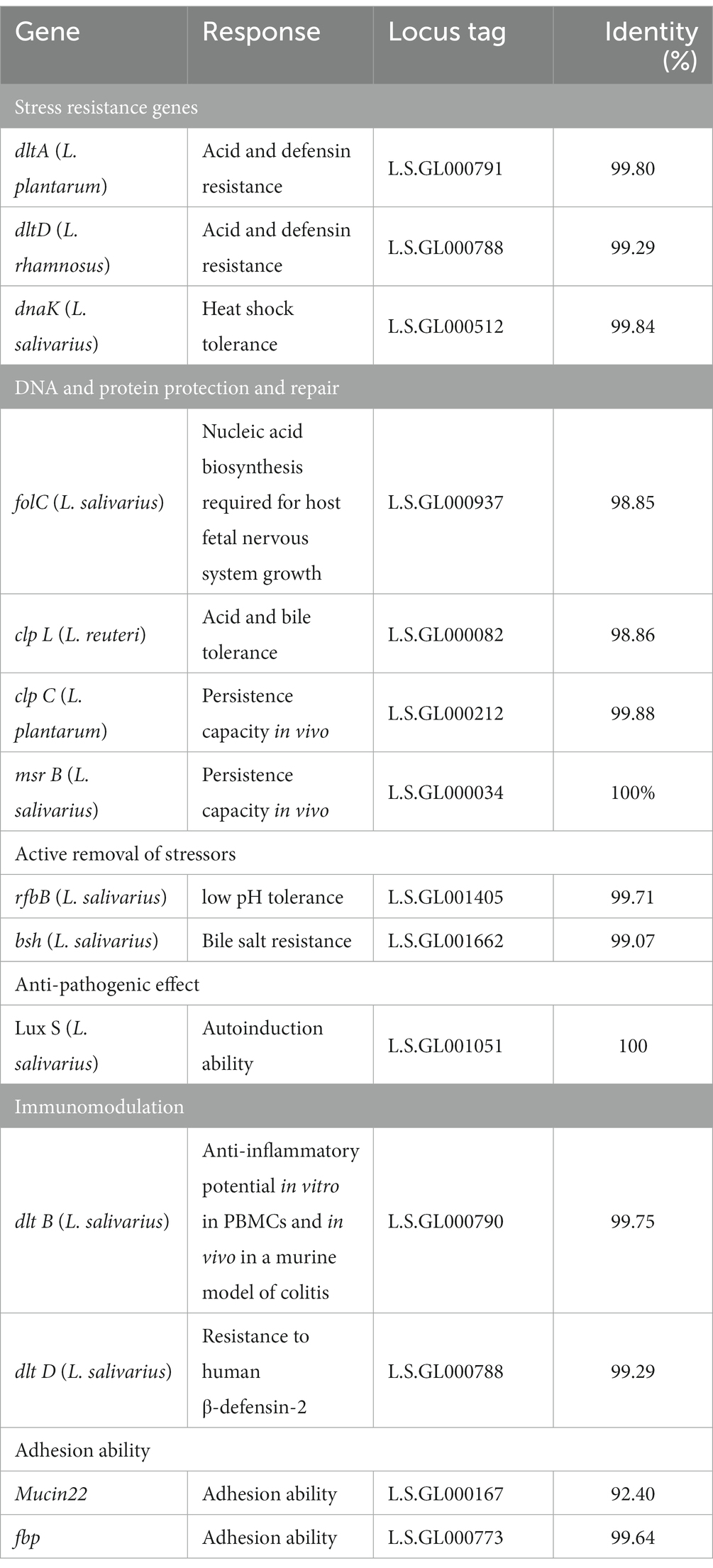
Probiotic genes were identified to evaluate the potential probiotic functions of L. salivarius CGMCC20700, as previously reported (Goel et al., 2020; Zheng et al., 2021). Briefly, different probiotic genes were obtained using BLASTP in the NCBI database and compared with known probiotic genes. Subsequently, the genes of L. salivarius CGMCC20700 were classified into functions related to stress resistance, DNA and protein protection and repair, active removal of stressors, antipathogenic effects, immunomodulation, and adhesion ability.
Bile salt and acid tolerance assay
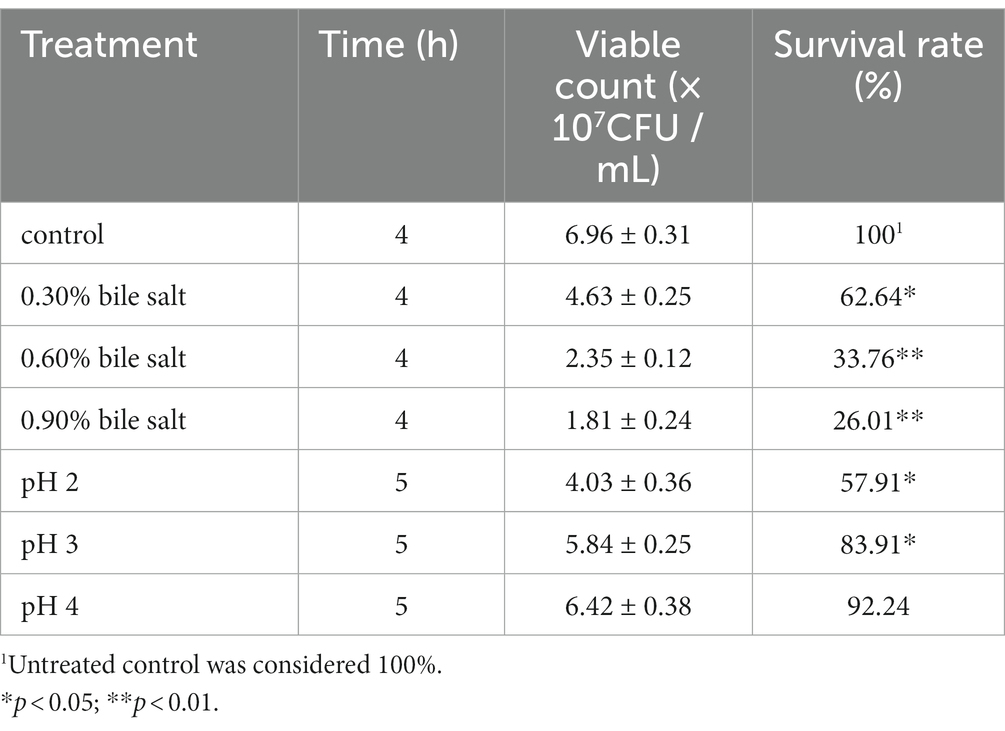
The bile salt and acid tolerance assay were performed by adopting the method described by Li X. Y. et al. (2021). Briefly, for bile salt tolerance tests, different concentrations (0.3, 0.6, and 0.9% (w/v)) of bile salts (Sangon Biotech, Shanghai, China) were added to MRS broth medium; for acid resistance tests, MRS broth medium was adjusted to different pH values (pH 2.0, 3.0 and 4.0) using 2 mol/mL HCl. Subsequently, L. salivarius CGMCC20700 cultures (107 CFU/mL) were added to MRS broth medium and cultured at 37°C for 4 h and 5 h, respectively. After incubation, 100 μL of each bacterial suspension was separately coated on MRS solid plates by the serial dilution method and inverted incubation at 37°C for 24 h. The viable counts of 30 ~ 300 colonies were counted, and bile salt tolerance was determined by calculating the ratio (%) of viable cells compared to the control without bile salt survival rates (%).
Auto aggregation capacity assay
The auto aggregate capability was determined according to the method described by Qureshi et al. (2020). Briefly, L. salivarius CGMCC20700 was added to MRS broth medium, and after culturing at 37°C for 12 h, the cells were collected by centrifugation at 8000 × g for 10 min, washed twice with PBS (pH 7.0), and resuspended to OD600 = 0.6. The initial absorbance value (Ab0) was measured. Then, the absorbance of 1 mL of bacterial suspension from each Eppendorf (EP) tube was measured as the OD600 value (Abt) of the supernatant after allowing it to stand at 37°C for 2–3 h. The percentage of auto aggregation was as follows: .
Cell surface hydrophobicity
The hydrophobicity of bacteria was determined according to the method described by Qureshi et al. (2020), with slight modifications. Briefly, L. salivarius CGMCC20700 was cultured in MRS broth at 37°C for 12 h and then centrifuged at 8000 × g for 10 min to collect the cells. The pellet was washed twice with PBS buffer, and the cells were resuspended in PBS to a cell OD600 = 0.7. The initial absorbance (Abi) was recorded. Afterward, the bacterial suspension was mixed with xylene (3:1) and incubated at 37°C for 10 min. The mixture was left standing at 37°C for 1 h, the aqueous phase was separated, and its absorbance (Abf) was measured at 600 nm. Surface hydrophobicity was calculated according to the following formula:
Statistical analysis
All experiments were conducted in triplicate, and each sample was evaluated in triplicate. The results are presented as the mean ± standard deviation (SD). Statistical significance was determined by one-way analysis of variance (ANOVA) in SPSS 22.0 statistical software (IBM Software Inc., NY, United States). p values <0.05 were considered indicative of a significant difference.
Results
Identification of Lactobacillus salivarius CGMCC20700
The results of morphological identification showed that the L. salivarius CGMCC20700 strain had colonies with a round, medium-sized, raised, whitish, moist, entire edge (Figure 1A). The L. salivarius CGMCC20700 strain is gram-positive (Figure 1B) and arranged in short rods without spores or flagella under SEM observation (Figures 1C,D). Based on 16S rRNA analysis, the L. salivarius CGMCC20700 strain showed ≥99% similarity with the L. salivarius 3,158 strain. The phylogenetic tree was constructed using the neighbor-joining method in MEGA 6 software (Figure 1E).
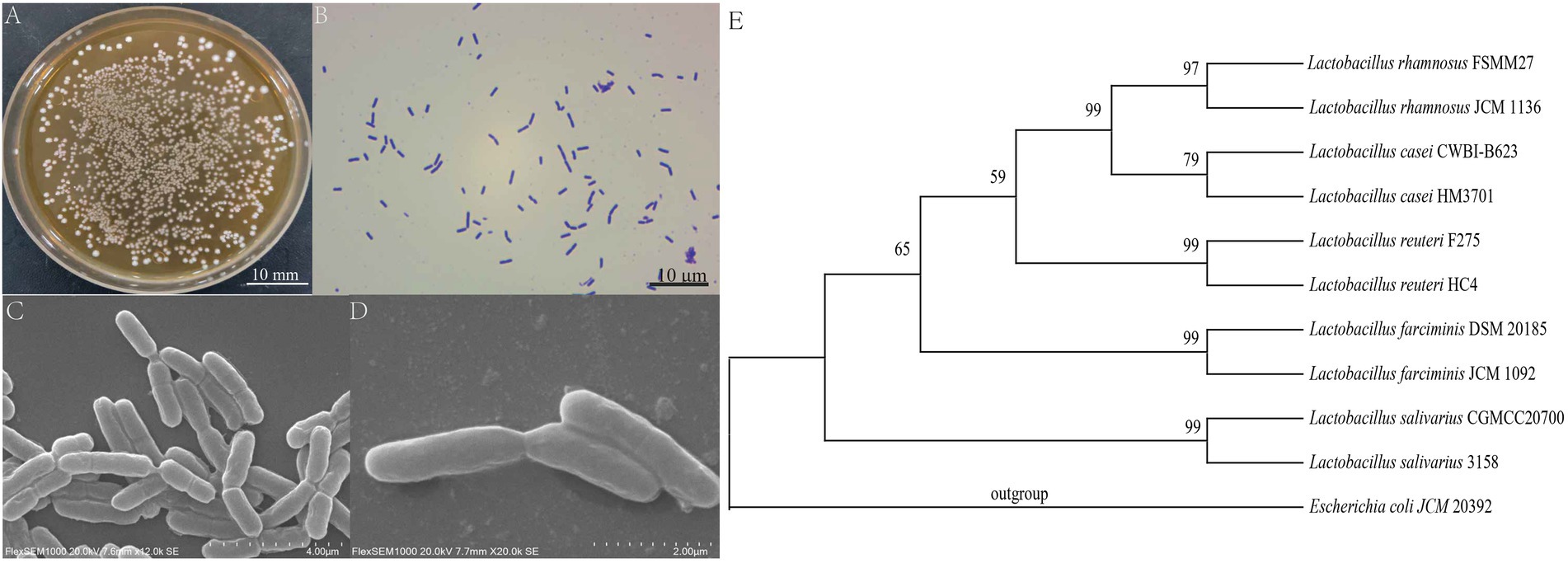
Figure 1. Morphological identification and phylogenetic tree of Lactobacillus salivarius CGMCC20700. (A) Colony observation, (B) Observation of Gram staining, (C,D) SEM observation, and (E) phylogenetic tree. All sequences originated from Lactobacillus strains, and other Lactobacillus species were used as outgroups. The numbers at the nodes indicate the bootstrap values of neighbor joining analyzes with 1,000 replicates.
Genome properties of Lactobacillus salivarius CGMCC20700
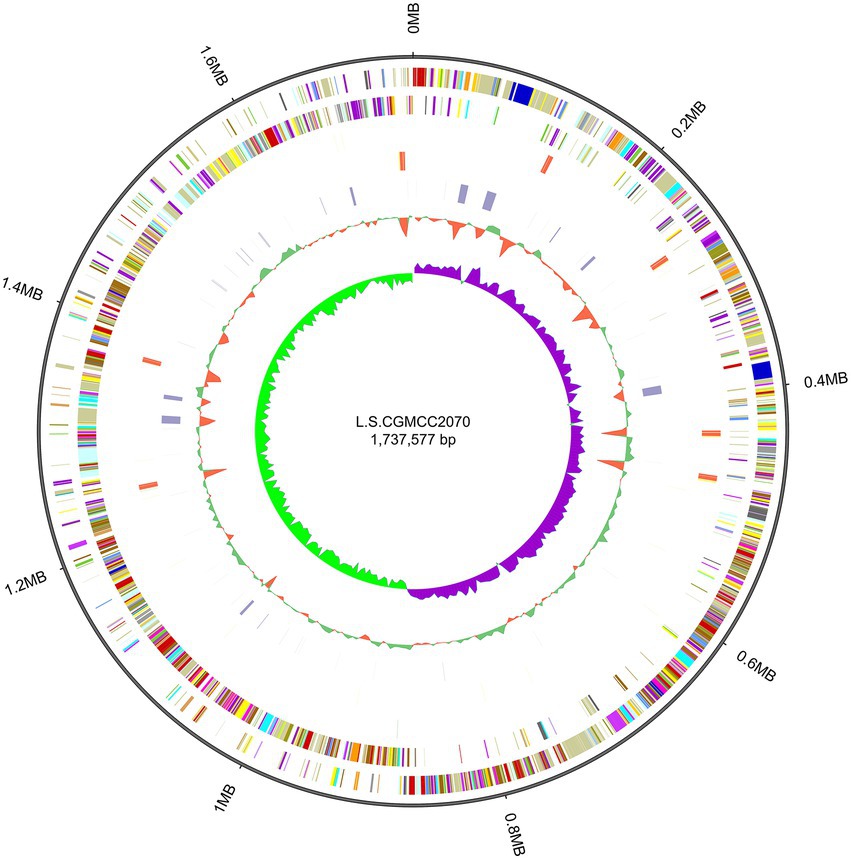
Whole-genome sequencing of L. salivarius CGMCC20700 showed that its genome size was 1.92 Mb with a single, circular chromosome with a GC content of 33.51% and two circular plasmids named plasmid1 (169,139 bp) and plasmid2 (22,823 bp) (Figure 2), which matched the results reported by Chiu et al. (2017). A total of 1,757 protein-coding sequences, 78 tRNA genes, 22 rRNA genes and 5 sRNA genes were identified, as shown in Table 1. Based on the COG database, 1,450 protein-coding genes were assigned to families comprising 22 functional categories into four types, including cellular, metabolism, information, and assembled genome. COG classification showed that L. salivarius CGMCC20700 was involved in the following aspects: (1) translation/ribosomal structure and biogenesis, (2) amino acid transport and metabolism, (3) carbohydrate transport and metabolism, (4) energy production and conversion, (5) coenzyme transport and metabolism, and (6) secondary metabolite biosynthesis, transport, and catabolism (Figure 3).
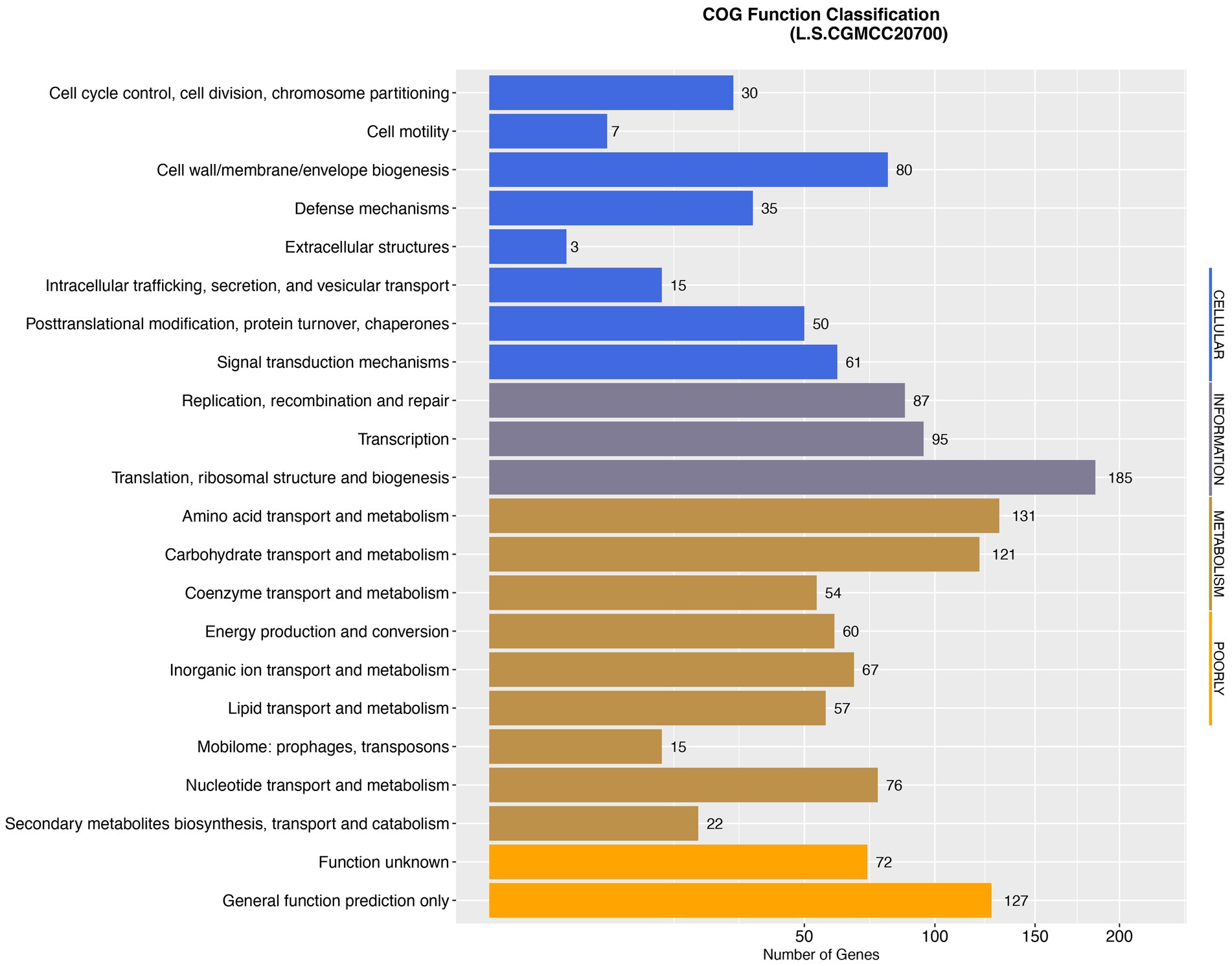
Figure 3. Distribution of genes across COG functional categories in the genome of L. salivarius CGMCC20700.
Safety analysis of Lactobacillus salivarius CGMCC20700
Identification of antibiotic resistance and toxicological factors
The genes related to antibiotic resistance and toxin production in the L. salivarius CGMCC20700 genome were identified according to the VFDB and ARDB databases, respectively. Based on the ARDB, 10 genes associated with antibiotic resistance were identified, and only three resistance genes with more than 90% similarity were covered, including tetracycline (tetm, tetl) and macrolide (ermc) resistance-related genes (Supplementary Table S1). Meanwhile, a total of 83 putative virulence factor genes were identified based on the VFDB database. The similarity of most putative virulence factor genes with VFDB was less than 80% (Supplementary Table S2). Furthermore, L. salivarius CGMCC20700 was sensitive to the antibiotics tested, as shown in Table 2, and was found to be sensitive only to vancomycin and erythromycin. The positive control bacteria (E. coli CMCC(B)44102) showed significant inhibition zones, which were identified as β hemolysis, while the L. salivarius CGMCC20700 strain did not show any hemolytic activity (Figure 4).
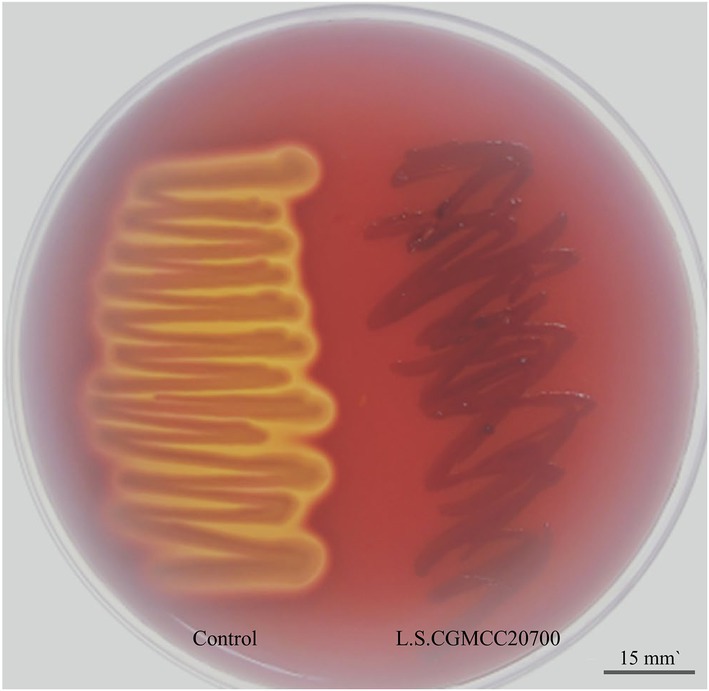
Figure 4. Hemolysis ability of L. salivarius CGMCC20700. The positive control Escherichia coli CMCC(B)44102 produced an obvious zone of β-hemolysis (left); CGMCC20700 showed γ-hemolysis (right).
Broiler acute toxicity test
The broilers were fed 20 g/kg body weight L. salivarius CGMCC20700 solution every day. The weight of broilers gradually increased during 14 days of observation, and no poisoning or mortality was found, as shown in Table 3. After the experiment, the broilers were dissected, and no internal tissue and organ lesions were observed by the naked eye.
Assessment of probiotic properties
Antimicrobial compound genes
The AntiSMASH 5.0 and BAGEL 4.0 databases were used to identify putative genes in the L. salivarius CGMCC20700 genome involved in antimicrobial compound production. In the two databases, two genes associated with T3PKS and enterococcin A were identified by antiSMASH and BAGEL4, respectively (Figure 5). Meanwhile, the cell-free supernatant of L. salivarius CGMCC20700 showed significant antibacterial activity against selected common pathogenic bacteria, both Gram-positive and Gram-negative, compared with the control (p < 0.01), particularly against Staphylococcus aureus ATCC2592, Staphylococcus sciuri ATCC 29059 and Salmonella enteritidis CMCC (B) 50,335, with an inhibitory zone reaching up to 24 mm (Table 4).
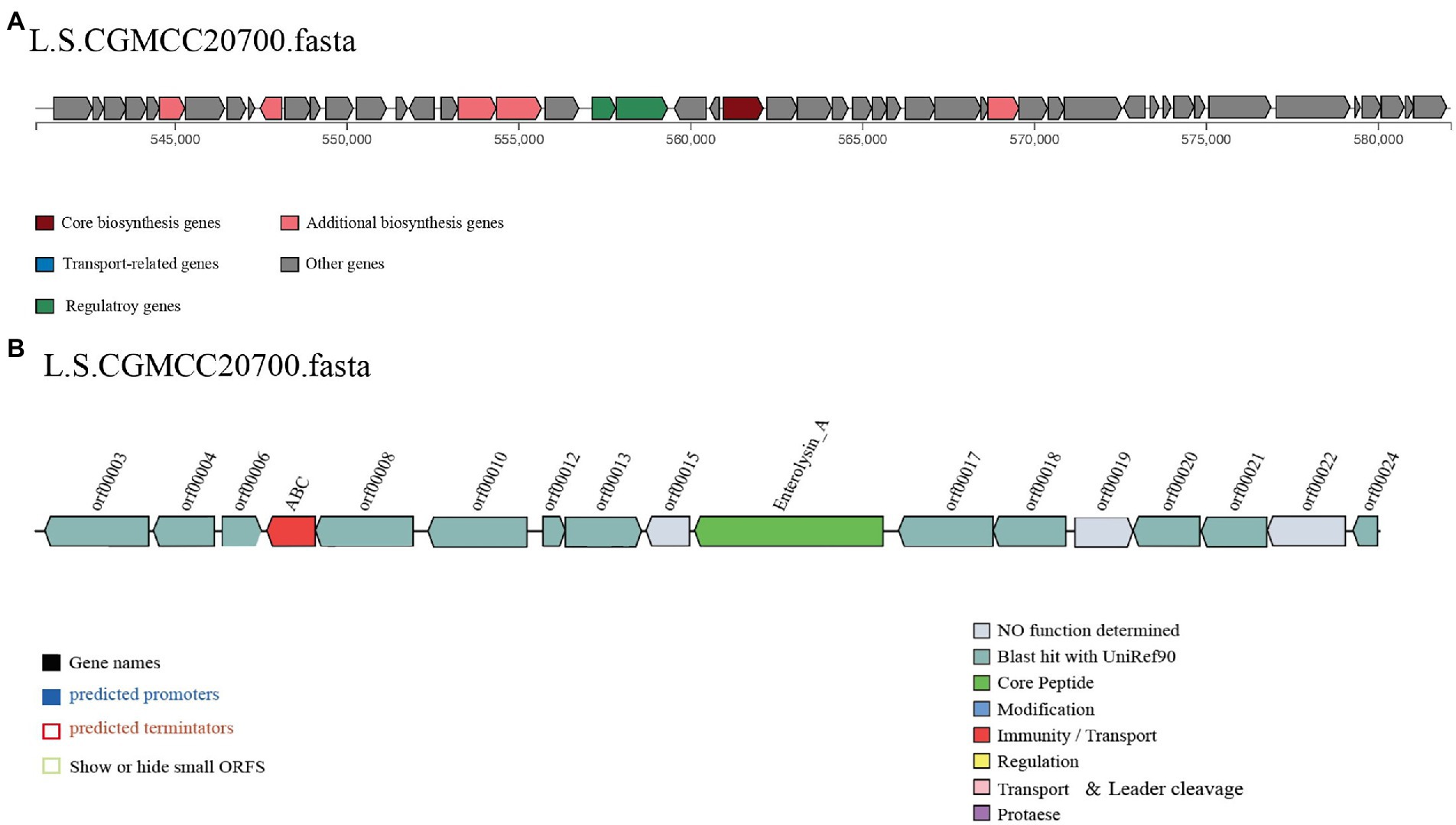
Figure 5. Predicted biosynthetic gene clusters encoding antibacterial compounds in the L. salivarius CGMCC20700 genome. The gene clusters encoding T3PKS (A) and enteroccin A (B) are represented by arrows with different colors corresponding to the operons of different functions.
Identification of probiotic genes
The genes related to probiotic properties were identified by annotation in the whole genome of L. salivarius CGMCC20700, as shown in Table 5. Of these, genes responsible for stress resistance included dltA, dltD and dnaK; genes responsible for DNA and protein protection and repair included folC, aclp L and msr B; genes responsible for the active removal of stressors included rfbB and bsh; genes responsible for immunomodulation included dlt B and dlt D; and genes responsible for anti-pathogenic effects included Lux S. Genes responsible for adhesion ability included Mucin22 and fbp. In addition to the adhesive ability-related gene Mucin22, the similarity of all other genes related to probiotic properties was over 98%.
Bile salt and acid tolerance
The treatment results of CGMCC20700 at different concentrations of acid-resistant and bile salts are shown in Table 6. When treated with bile salt concentrations of 0.3, 0.6 and 0.9% for 4 h, the survival rate of L. salivarius CGMCC20700 significantly decreased to 62.64, 33.76, and 26.01%, respectively, compared with the control (p < 0.05). At pH values of 2, 3 and 4 for 5 h, the survival rate of L. salivarius CGMCC20700 decreased to 57.91% (p < 0.05), 83.91% (p < 0.05) and 92.24%, respectively, compared with the control.
Auto aggregation and hydrophobic capability
According to the results, the aggregation rate of the L. salivarius CGMCC20700 strain was 57.12 ± 1.23%. The hydrophobicity of the L. salivarius CGMCC20700 strain was determined, and the hydrophobicity index was 61.16 ± 1.19%.
Discussion
Probiotics are considered biotherapeutic agents owing to their potential to bestow various health benefits (Al-Khalaifa et al., 2019). Numerous studies have shown that probiotics may adhere and survive in the gastrointestinal tract of humans and animals and contribute to maintaining a microecological balance of the gut microbiome, promoting digestive and metabolic processes, and modulating the immune response, thereby enhancing host immunity and improving human and animal health (Kai et al., 2020; Zheng et al., 2021). However, since the effectiveness of probiotics is species or strain dependent, they should meet a series of specific characteristics, such as safety, functional and beneficial characteristics (Li et al., 2020; Wu et al., 2022). Thus, in this study, we focused on the safety and potential probiotic properties of L. salivarius CGMCC20700 using a series of in vitro tests combined with whole-genome sequencing to reveal their potential biological functions.
The development of new strain resources and evaluation of the safety of strains is necessary to obtain the most effective probiotics (Kai et al., 2020; Wu et al., 2022). It has been reported that candidate probiotics should not transport antibiotic resistance genes for hosts (García-Hernández et al., 2016; Goel et al., 2020). However, previous studies have found that LAB strains may develop resistance to tetracycline, 4-quinolones, rifampicin, and macrolides due to ribosome protection, antibiotic efflux and associated efflux pump formation (Ma et al., 2021; Fu et al., 2022). In this study, the ARDB and a variety of antibiotic susceptibility tests found that the strain contained macrolide and tetracycline antibiotic genes and was sensitive to tetracycline and erythromycin, showing antibiotic resistance similar to or lower than that of other known probiotic strains (Zheng et al., 2021; Fu et al., 2022). For instance, E. lactis JDM1 was resistant to erythromycin, quinupristin-dalfopristin 1R and furantoin and contained six highly similar resistance genes, efmA, aac and msrC (Fu et al., 2022); Lactobacillus paracasei CY2 is resistant to four types of antibiotics, kanamycin, gentamicin, and vancomycin (Zheng et al., 2021). Furthermore, the presence of virulence factors and hemolytic activity are important indicators of potentially beneficial strains (Goel et al., 2020; Kai et al., 2020). The ARDB database and hemolytic tests revealed that L. salivarius CGMCC20700 lacked highly similar virulence factor genes and showed nonhemolytic activity, which implied that the strains were not toxic. In particular, Lactobacillus virulence determinants that were confirmed by previous studies include cytohemolysin (cyl), aggregates (AS), and gelatinases (Kai et al., 2020; Vyacheslav et al., 2020); however, none of these were identified in L. salivarius CGMCC20700. Additionally, no harmful effects were found on the growth performance and overall health of broilers after feeding a L. salivarius CGMCC20700 supplementary diet. Thus, these results comprehensively indicated that L. salivarius CGMCC20700 has good safety for use in livestock and poultry farming.
Antimicrobial activity is one of the most important criteria for selecting new probiotic strains because probiotics can maintain intestinal homeostasis by inhibiting the growth of intestinal pathogenic bacteria (Grosu-Tudor et al., 2014). Due to the convenience of whole-genome sequencing and the diversity of genome mining tools, it is possible to predict a strain’s capability for producing antimicrobial compounds (Fu et al., 2022; Wu et al., 2022). In this study, AntiSMASH 5.0 and BAGEL 4.0 prediction results showed that two putative genes associated with antimicrobial compounds, T3PKS and enteroccin A, were identified in the L. salivarius CGMCC20700 genome. Previous studies have shown that LAB are known to have a well-developed secretion system that can produce a variety of metabolites, including antimicrobial peptides synthesized by ribosomes (RiPPs), nonribosomal synthetic peptides (NRPs) and polyketides (PKs) (Weber et al., 2015). This evidence showed that L. salivarius CGMCC20700 was capable of producing a variety of antibacterial compounds to help livestock and poultry against pathogenic infections. Additionally, the cell-free supernatant of L. salivarius CGMCC20700 showed high antibacterial activity against uncommon types of pathogens, and the maximum inhibition circle size was up to 26 mm, which further demonstrated the antibacterial compounds produced by L. salivarius CGMCC20700 with excellent broad-spectrum antibacterial activity. Thus, these results demonstrated that L. salivarius CGMCC20700 could effectively influence the balance of the intestinal flora and occupy a good competitive position.
Moreover, tolerance to bile salts and acidic conditions are two key characteristics when assessing beneficial traits, as the presence of bile salts and highly acidic conditions constitute the greatest barriers to the survival of Lactobacillus in the animal host gastrointestinal tract (Kai et al., 2020; Vyacheslav et al., 2020). Previous studies have shown that the dltA, dltD and rfbB genes mainly contribute to acid tolerance and the survival of bacteria in acidic environments; the rfbB gene encodes dTDP-glucose 4,6-dehydratase activity and plays an important role in the response of bacteria to low-pH conditions (Behera et al., 2018; Goel et al., 2020). In this study, L. salivarius CGMCC20700 genomic analysis showed that all the above bile salt and acid tolerance-related genes were obtained and that the similarity of these genes was over 90%, implying that L. salivarius CGMCC20700 had good bile salt and acid tolerance. Similarly, after treatment with 0.90% bile salts for 4 h and pH = 2 for 5 h, the survival rate of L. salivarius CGMCC20700 was maintained at 26.01 and 57.91%, respectively, demonstrating higher tolerance efficacy compared to the other partial probiotic LAB strains. For instance, Lactobacillus plantarum CY2 and Lactobacillus paracasei CY3 isolated from yak milk were only maintained at 20.10 and 12.38%, respectively, after treatment with 0.5% bile salts for 4 h, and L. paracasei CY3 was only maintained at 36.09% after treatment at pH 2 for 3 h (Zheng et al., 2021). Thus, these findings confirmed that L. salivarius CGMCC20700 can survive typical animal gastrointestinal tract conditions.
Additionally, self-agglomeration and hydrophobicity are also important characteristics for the efficient colonization of probiotics in the animal gut (Goel et al., 2020; Qureshi et al., 2020). In this study, the self-agglomeration and hydrophobicity of L. salivarius CGMCC20700 were 57.12 and 61.16%, respectively. Furthermore, an anti-pathogenic effect and adhesion ability with related genes were also identified in the L. salivarius CGMCC20700 genome, which associated functional genes involved Lux S, Mucin22 and fbp. Generally, Mucin22 and fbp genes are responsible for adhesion ability to the intestinal epithelial layer, likely excluding the adhesion of pathogenic species (Garcia-Gonzalez et al., 2018). Additionally, the genome of L. salivarius CGMCC20700 also contains dltB and dltD genes, and these genes are involved in human immunity and anti-inflammatory processes (Elbanna et al., 2018; Galdeano et al., 2019; Goel et al., 2020). Collectively, the combined in vitro probiotic characterization and genetic analysis showed that L. salivarius CGMCC20700 has good potential as a probiotic with resistance to intestinal and gastric fluids, adherence to intestinal epithelial tissues and robust immunomodulatory and anti-inflammatory effects. Notably, the probiotic potential of L. salivarius CGMCC20700 needs to be systematically investigated in further experiments, both at the cellular level and in vivo in animal experiments.
Conclusion
In the present study, we identified a L. salivarius CGMCC20700 strain and investigated its safety and probiotic properties. The genome screenings indicated the absence of active antibiotic resistance genes and virulence factor genes. Hemolytic assays, acute oral toxicology, and antibiotic resistance tests further confirmed its safety. The detection of antimicrobial gene clusters, adhesion-related genes and stressor-reducing genes, such as extreme acids and bile salts, and the simulation of gastric and intestinal fluid stresses revealed potential probiotic properties. Additionally, L. salivarius CGMCC20700 is highly self-agglomerative and hydrophobic, and in silico analysis demonstrated the genes responsible for adhesion, immunity, and anti-inflammation. Collectively, this study provides experimental evidence that L. salivarius CGMCC20700 can serve as an effective probiotic candidate to replace antibiotic applications in livestock and poultry farming.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Y-HJ: investigation, methodology, data curation, software, and writing review and editing. R-SY: methodology, investigation, and writing review and editing. Y-CL: methodology and writing review and editing. W-GX: methodology, investigation, data curation, and software. H-YZ, FW, and Q-LZ: methodology and investigation. L-BL: conceptualization, methodology, resource, data curation, writing original draft, and writing review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Yunnan Major Scientific and Technological Projects (grant no. 202202AG050008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1120263/full#supplementary-material
Footnotes
References
Al-Khalaifa, H., Al-Nasser, A., Al-Surayee, T., Al-Kandari, S., Al-Enzi, N., Al-Sharrah, T., et al. (2019). Effect of dietary probiotics and prebiotics on the performance of broiler chickens. Poult.Sci. 98, 4465–4479. doi: 10.3382/ps/pez282
Behera, S. S., Ray, R. C., and Nevijo, Z. (2018). Lactobacillus plantarum with functional properties: An approach to increase safety and shelf-life of fermented foods. Biomed. Res. Int. 2018, 1–18. doi: 10.1155/2018/9361614
Cerezuela, R., Guardiola, F. A., Meseguer, J., and Esteban, M. Á. (2012). Increases in immune parameters by inulin and Bacillus subtilis dietary administration to gilthead seabream (Sparus aurata L.) did not correlate with disease resistance to Photobacterium damselae. Fish. Shellfish. Immu. 32, 1032–1040. doi: 10.1016/j.fsi.2012.02.025
Chen, X., Ishfaq, M., and Wang, J. (2022). Effects of Lactobacillus salivarius supplementation on the growth performance, liver function, meat quality, immune responses and salmonella Pullorum infection resistance of broilers challenged with Aflatoxin B1. Poultry. Sci. 101:101651. doi: 10.1016/j.psj.2021.101651
Chiu, S. H., Chen, C. C., Wang, L. T., and Huang, L. (2017). Whole-genome sequencing of Lactobacillus salivarius strains BCRC 14759 and BCRC 12574. Genome Announc. 5:e01336-17-e01336-17. doi: 10.1128/genomeA.01336-17
Dai, L. M., Li, L. L., Liu, Y. X., Shi, Y. P., and Cai, Z. Y. (2021). Whole genome sequencing and genomics analysis of bacillus amyloliquefaciens BS-3 with biocontrol activity. Microbiol. China. 48, 2073–2088. doi: 10.13344/j.microbiol.china.220589
Elbanna, K., Hadad, S. E., Assaeedi, A., Aldahlawi, A., Khider, M., and Alhebshi, A. (2018). In vitro and in vivo evidences for innate immune stimulators lactic acid bacterial starters isolated from fermented camel dairy products. Sci. Pep. 8, 12553–12515. doi: 10.1038/s41598-018-31006-3
FDA. (2009). Summary report on antimicrobials sold or distributed for use in food producing animals. Food and Drug Administration, Available at: http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM231851.pdf (Accessed August 31, 2011).
Fu, X., Yu, L. L., Wang, Y., Zhang, Y., Guo, X., Chen, Q., et al. (2022). Safety assessment and probiotic characteristics of enterococcus lactis JDM1. Microb. Pathogenesis 163:105380. doi: 10.1016/j.micpath.2021.105380
Galdeano, C. M., Cazorla, S. I., Dumit, J. M. L., Vélez, E., and Perdigón, G. (2019). Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 74, 115–124. doi: 10.1159/000496426
Gao, S. F., Xu, B. S., Lu, D. Q., Liu, A. Q., Gou, Y. F., Sun, S. W., et al. (2021). Whole genome sequencing and analysis of the bio-control strain bacillus velezensis Z. Chi. J. Trop. Crops. 42, 1216–1222.
Garcia-Gonzalez, N., Prete, R., Battista, N., and Corsetti, A. (2018). Adhesion properties of food-associated lactobacillus plantarum strains on human intestinal epithelial cells and modulation of IL-8 release. Front. Microbiol. 9:2392. doi: 10.3389/fmicb.2018.02392
García-Hernández, Y., Pérez-Sánchez, T., Boucourt, R., Balcázar, J. L., Nicoli, J. R., Moreira-Silva, J., et al. (2016). Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 108, 125–132. doi: 10.1016/j.rvsc.2016.08.009
Goel, A., Halami, P., and Tamang, J. P. (2020). Genome analysis of lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front. Microbiol. 11:40. doi: 10.3389/fmicb.2020.00040
Grosu-Tudor, S. S., Stancu, M. M., Pelinescu, D., and Zamfir, M. (2014). Characterization of some bacteriocins produced by lactic acid bacteria isolated from fermented foods. World. J. Microb. Bio. 30, 2459–2469. doi: 10.1007/s11274-014-1671-7
Hiremath, S., and Pragasam, V. (2022). Oxalobacter formigenes: a new hope as a live biotherapeutic agent in the management of calcium oxalate renal stones. Anaerobe 75:102572. doi: 10.1016/j.anaerobe.2022.102572
Jiang, Y. H., Xin, W. G., Yang, L. Y., Ying, J. P., Zhao, Z. S., Lin, L. B., et al. (2022a). A novel bacteriocin against Staphylococcus aureus from lactobacillus paracasei isolated from Yunnan traditional fermented yogurt: purification, antibacterial characterization, and antibiofilm activity. J. Dairy Sci. 105, 2094–2107. doi: 10.3168/jds.2021-21126
Jiang, Y. H., Xin, W. G., Zhang, G. L., Wang, F., Wang, C., Zhang, Q. L., et al. (2022b). Effects of phagelyase on growth performance, organ index and major antioxidant enzyme activities in white-feather broilers. Animal. Husb. Vet. Med. 54, 46–51.
Joo, S. S., Yoon, J. H., Jung, J. Y., Joo, S. Y., An, S. H., Ban, B. C., et al. (2022). The modulatory effects of Lacticaseibacillus paracasei strain NSMJ56 on gut immunity and microbiome in early-age broiler chickens. Animals 12:3413. doi: 10.3390/ANI12233413
Kai, Y., Ping, L., and Qing, G. (2020). Complete genome sequence analysis of a strain lactobacillus pentosus ZFM94 and its probiotic characteristics. Genomics 112, 3142–3149. doi: 10.1016/j.ygeno.2020.05.015
Li, X. Y., Li, L. X., Li, Y., Zhou, R. C., Li, B., Gu, X., et al. (2021). Complete genome sequencing of Peyer's patches-derived lactobacillus taiwanensis CLG01, a potential probiotic with antibacterial and immunomodulatory activity. BMC Microbiol. 21:68. doi: 10.1186/s12866-021-02127-z
Li, M., Wang, Y., Cui, H., Li, Y., Sun, Y., and Qiu, H. J. (2020). Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. Vet. Sci. 7:49. doi: 10.3389/fvets.2020.00049
Li, H. W., Xiang, Y. Z., Zhang, M., Jiang, Y. H., Zhang, Y., Liu, Y. Y., et al. (2021). A novel bacteriocin from Lactobacillus salivarius against Staphylococcus aureus: isolation, purification, identification, antibacterial and antibiofilm activity. LWT-Food. Sci. Technol. 140:110826. doi: 10.1016/J.LWT.2020.110826
Ma, Q. Q., Pei, Z. M., Fang, Z. F., Wang, H. C., Zhu, J. L., Lee, Y. K., et al. (2021). Evaluation of tetracycline resistance and determination of the tentative microbiological cutoff values in lactic acid bacterial species. Microorganisms 9:2128. doi: 10.3390/MICROORGANISMS9102128
Paveljšek, D., Juvan, P., Košir, R., Rozman, D., Hacin, B., Ivičak-Kocjan, K., et al. (2018). Lactobacillus fermentum L930BB and Bifidobacterium animalis subsp. animalis IM386 initiate signalling pathways involved in intestinal epithelial barrier protection. Benef. Microbes. 9, 515–525. doi: 10.3920/BM2017.0107
Pierce, J., Apisarnthanarak, A., Schellack, N., Cornistein, W., and Stevens, M. P. (2020). Global antimicrobial stewardship with a focus on low-and middle-income countries. Iint. J. Infect. Dis. 96, 621–629. doi: 10.1016/j.ijid.2020.05.126
Qureshi, N., Gu, Q., and Li, P. (2020). Whole genome sequence analysis and in vitro probiotic characteristics of a lactobacillus strain lactobacillus paracasei ZFM54. J. Appl. Microbiol. 129, 422–433. doi: 10.1111/jam.14627
Ramirez, J., Guarner, F., Fernandez, L. B., Maruy, A., Sdepanian, V. L., and Cohen, H. (2020). Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 10:572912. doi: 10.3389/FCIMB.2020.572912
Ricke, S. C., Sang, I. L., Sun, A. K., Si, H. P., and Shi, Z. (2020). Prebiotics and the poultry gastrointestinal tract microbiome. Poultry Sci. 99, 670–677. doi: 10.1016/j.psj.2019.12.018
Rocha-Ramírez, L., Pérez-Solano, R., Castañón-Alonso, S., Moreno, G. S., Ramírez, P. A., Garibay, M. G., et al. (2017). Probiotic lactobacillus strains stimulate the inflammatory response and activate human macrophages. J Immunol Res 2017:4607491. doi: 10.1155/2017/4607491
Saroj, D. B., and Gupta, A. K. (2020). Genome based safety assessment for Bacillus coagulans strain LBSC (DSM 17654) for probiotic application. Int. J. Food Microbiol. 318:108523. doi: 10.1016/j.ijfoodmicro.2020.108523
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanizawa, Y., Tohno, M., Kaminuma, E., Nakamura, Y., and Arita, M. (2015). Complete genome sequence and analysis of lactobacillus hokkaidonensis LOOC260T, a psychrotrophic lactic acid bacterium isolated from silage. BMC.Genomics 16, 1–11. doi: 10.1186/s12864-015-1435-2
Tiseo, K., Huber, L., Gilbert, M., Robinson, T. P., and Boeckel, T. (2020). Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 9:918. doi: 10.3390/ANTIBIOTICS9120918
Van, D. B., Willems, A., London, R., Top, N. J., and Stobberingh, E. (2002). Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J. Antumicrob. Chemoth. 49, 497–505. doi: 10.1093/jac/49.3.497
Vyacheslav, T., Elena, D., Olga, S., Stanislav, S., and Timur, V. (2020). Whole-genome sequencing of lactobacillus helveticus D75 and D76 confirms safety and probiotic potential. Microorganisms 8:329. doi: 10.3390/microorganisms8030329
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H., Bruccoleri, U. R., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Wu, Y. P., Liu, D. M., Zhao, S., Huang, Y. Y., Yu, J. J., and Zhou, Q. Y. (2022). Assessing the safety and probiotic characteristics of Bacillus coagulans 13002 based on complete genome and phenotype analysis. LWT-Food. Sci. Technol. 155:112847. doi: 10.1016/J.LWT.2021.112847
Xu, C., Wei, F. X., Yang, X. Y., Feng, Y. Q., Liu, D., and Hu, Y. F. (2022). Lactobacillus salivarius CML352 isolated from Chinese local breed chicken modulates the gut microbiota and improves intestinal health and egg quality in late-phase laying hens. Microorganisms. 10:726. doi: 10.3390/MICROORGANISMS10040726
Yamaguchi, T., Konishi, H., Aoki, K., Ishii, Y., Chono, K., and Tateda, K. (2020). The gut microbiome diversity of Clostridioides difficile-inoculated mice treated with vancomycin and fidaxomicin. J. Infect. Chemother. 26, 483–491. doi: 10.1016/j.jiac.2019.12.020
Keywords: Lactobacillus salivarius, whole-genome sequence, phenotypic analysis, safety, probiotic characteristics
Citation: Jiang Y-H, Yang R-S, Lin Y-C, Xin W-G, Zhou H-Y, Wang F, Zhang Q-L and Lin L-B (2023) Assessment of the safety and probiotic characteristics of Lactobacillus salivarius CGMCC20700 based on whole-genome sequencing and phenotypic analysis. Front. Microbiol. 14:1120263. doi: 10.3389/fmicb.2023.1120263
Edited by:
Renpeng Du, Heilongjiang University, ChinaReviewed by:
Naheed Mojgani, Razi Vaccine and Serum Research Institute, IranJing Chen, Sichuan University, China
Hanghui Ye, University of Texas MD Anderson Cancer Center, United States
Copyright © 2023 Jiang, Yang, Lin, Xin, Zhou, Wang, Zhang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lian-Bing Lin, bGlubGJAa211c3QuZWR1LmNu
†These authors have contributed equally to this work
 Yu-Hang Jiang1,2†
Yu-Hang Jiang1,2† Wei-Gang Xin
Wei-Gang Xin