
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 March 2023
Sec. Microbial Symbioses
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1119887
This article is part of the Research Topic Rumen Microbiome: Interacting with Host Genetics, Dietary Nutrients Metabolism, Animal Production, and Environment View all 15 articles
Background: This study examined the effects of substituting cottonseed meal (CSM) or rapeseed meal (RSM) for soybean meal (SBM) on Hu sheep performance, rumen fermentation, and bacterial composition. 51 four-month-old indigenous male Hu sheep with starting body weights of 22.51 ± 2.84 kg and similar origins were randomly assigned to three treatments; (1) non-fermented total mixed ration (TMR) with SBM (CK), (2) fermented TMR containing CSM (F-CSM group), and (3) fermented TMR containing RSM (F-RSM group).
Results: The three groups’ intake of dry matter differed significantly (p < 0.05). In terms of average daily gain, the F-RSM group outperformed the CK and F-CSM groups (p < 0.05). The pH of the rumen was substantially lower in the CK group than in the F-CSM and F-RSM groups (p < 0.05), and the F-CSM group had greater amounts of volatile fatty acids (VFA) than the F-RSM and CK groups. In comparison to the CK group, the microbial crude protein yield was significantly higher in the F-CSM and F-RSM groups (p < 0.05). The F-CSM group significantly outperformed the F-RSM group of pepsin and cellulose enzyme activity (p < 0.05). The relative abundance of Bacteroidetes was greater in the CK and F-RSM groups compared to the F-CSM group (p < 0.05). In comparison to the other groups, Firmicutes were less abundant in the CK group (p < 0.05). Prevotella was present in a higher relative abundance in the F-CSM and F-RSM groups than in the CK group (p < 0.05). Prevotella was greater in relative abundance in the F-CSM and F-RSM groups than in the CK group (p < 0.05). The relative abundances of Veillonellaceae_UCG-001 and Lachnospiraceae_XPB1014 correlated with rumen butyric acid content and NH3-N content (p < 0.05). Gene function prediction revealed that replacing SBM with F-CSM or F-RSM in the diet of Hu sheep can promote glycan biosynthesis and metabolism.
Conclusion: The replacement of F-CSM and F-RSM for SBM has an influence on the richness and diversity of rumen bacteria at the phylum and genus levels. Replacement of SBM with F-CSM increased VFA yield and further promoted the performance of Hu sheep.
The worldwide population is forecast to spread 9.7 billion (Olimid and Olimid, 2019), while universal need for meat and milk is expected to grow by 57 and 48%, respectively (Alexandratos and Bruinsma, 2012). As an outcome, it is estimated that livestock production will rise by 21% between 2010 and 2025 (Mottet et al., 2017). The universal need for livestock protein for human nourishment is increasing exponentially, increasing the price of animal feed concentrates (Tufarelli et al., 2018). To overcome the problem, it is vital to adopt different sources of feed and strategies using of alternative feeding methods. By-products are the sole viable option for making low-cost feed ingredients; they are generated during food processing and production and thus are not appropriate for human consumption (Xu et al., 2007). In ruminant diets, CSM and RSM may be appropriate alternatives to SBM as they are less expensive and relatively more widely accessible regionally (Zeng, 2020).
China has a large supply of CSM and has long been measured as a reasonable alternative to SBM (Wang et al., 2017). As a by-product of cottonseed oil manufacturing, CSM exhibits a crude protein content of 34–40%, crude fibre content of 11%, vitamin B, and organic phosphorus, among other nutrients (Zhang et al., 2018). However, due of the presence of Free gossypol (FG), a poisonous that may significantly influence animal development and reproductive and digestive development, as well as cause anomalies in internal organs, the use of CSM in animal diets is restricted (Francis et al., 2001; Robinson et al., 2001). Cotton-rich feed is a common source of protein for ruminants, particularly in cotton-growing regions such as India, China, and the United States, where it is used to replace soybean meal (SBM) (Heuze et al., 2015). However, CSM is now a cost-effective source of high protein (Nie et al., 2015). RSM, a by-product of the rapeseed oil industry, has a high protein concentration and a well-balanced amino acid composition (Wanasundara et al., 2017; Ashayerizadeh et al., 2018), making RSM appropriate for use in livestock feed. Nonetheless, because of the presence of glucosinolates (Huisman and Tolman, 1992; Tripathi and Mishra, 2007) and other anti-nutritional factors, RSM can poison animals and impair their development (Nagalakshmi et al., 2003). As a result, there is a limited usage of CSM and RSM in livestock diets.
Several approaches had been used to decrease the anti-nutritiona factors of CSM, such as calcium hydroxide (Nagalakshmi et al., 2003), chemical treatment with ferrous sulfate (Barrett et al., 1997), and microbial fermentation (Weng and Sun, 2006). To reduce the anti-nutritional factors of RSM, methods such as inactivation of myrosinase, solvent extraction, steam removal, and liquidation have been applied. Still, such methods also have some disadvantages including loss of protein, high expense, commercial relative unimportance, and environmental pollution (Pal and Walia, 2001). Among the different methods, microbial fermentation can help in increasing crude protein and other nutritional content, reduce toxin content, and maintain stomach microbial stability of the plants (McSweeney et al., 2001; Xu et al., 2020). Furthermore, our preliminary study showed that the nutritious value of CSM and RSM can be increased by microbial fermentation, while the anti-nutritional factors (FG and glucosinolate) in them can be reduced.
The diverse and adaptable rumen microbiota, which includes bacteria, archaea, fungus, protozoa, and viruses, allows for the utilization of a variety of food materials, including recalcitrant plant wall elements, and the production of assimilable energy and nutrients needed by ruminants (Kolver et al., 2001). Undoubtedly, the rumen microbiome also allows for nutritional manipulations. In fact, functions of the rumen microbiota and a number of the production phenotypes of ruminants are linked (Zhang et al., 2017; Li et al., 2019; Zhang Y. et al., 2020). VFAs, which supply 70% of the required energy, can be produced by ruminants from solid diet. The microbes in the rumen enable this (Anantasook et al., 2013; Khan et al., 2016). The variations in feed efficiency among individual cattle can be primarily attributed to rumen bacteria and VFAs (Liang et al., 2017; Zhou et al., 2018; Zhang Y. X. et al., 2020). In a study by Zhang et al. (2018), the amount of FG and total gossypol in fermented CSM fell by 78 and 49%, respectively, under the best fermentation conditions in comparison to the control. These results indicate that rumen bacteria have considerable capacity to digest gossypol, which helps to explain why ruminants have a high tolerance for gossypol (Tang et al., 2018). However, the microbes involved in such rumen fermentation are not known. Moreover, the impact of CSM and RSM on rumen fermentation and rumen microorganisms has only been studied by a few researchers. Therefore, this study’s objective was to determine whether replacement of SBM with F-CSM or F-RSM affected growth performance, rumen fermentation and bacterial composition in Hu sheep.
The experiment was conducted in compliance with the Guidelines for Experimental Animals (2006) established by the Ministry of Science and Technology (Beijing, China), and it was given the go-ahead by the Chinese Academy of Agricultural Sciences’ Animal Ethics Committee (AEC-CAAS-20190517).
51 four-month-old native male Hu sheep with ancestries that were identical and an initial weight of 22.51 ± 2.84 kg were acquired from China’s Inner Mongolia autonomous region. The Hu sheep were divided into three groups (17 Hu sheep each) using a single-factor randomized block design: CK group, F-CSM group, and F-RSM group. Subsequently, they were fed the total mixed ration (TMR). A stall (3 m × 1.5 m) equipped with individual feed and water buckets was utilized to maintain the Hu sheep. Unrestricted access to TMR and clean water was given to the Hu sheep.
Along with SBM, CSM, or RSM meals, wheat bran, maize, and corn stalks were used in the TMR formulation. Additionally, urea, premixed vitamins, and fat powder were added. The dietary feed was formulated according to the guidelines for meeting the nutrient requirements of sheep and allowing them to gain 300 g per day (NRC, 2007). Table 1 is a list of the TMRs’ components and chemical composition. Based on our previous studies (Yusuf et al., 2021), F-CSM was prepared with a 50% moisture content and a 1:5 mixture of microbial strains (1.0 × 109 CFU/kg DM (dry matter) of Bacillus clausii and 5.0 × 109 CFU/kg DM of Saccharomyces cariocanus). F-RSM was also prepared with a 50% moisture content and a mixture of microbial strains (1.0 × 1010 CFU/kg DM of B. clausii and 5.0 × 1010 CFU/kg DM of S. cariocanus). In a 500 kg fermenter machine, the mixture was fermented (Model SSJX-WH-3.0; Shengshun Machinery Manufacturing Co., Ltd., Shenyang, China) for 60 h at 32°C (F-CSM group) or 28°C (F-RSM group). Following fermentation, the mixture was uniformly mixed with whole corn silage before being bagged in plastic. Every 3 days, TMR (CK group) without fermentation was mixed.
During July to October of 2021, the experiment was conducted at the Chinese Academy of Agricultural Sciences’ Nankou Experimental Pilot Test Base (China’s Beijing). The study lasted for 97 days, including 10 days for adaptation, 80 days for the feeding trial, and 7 days for the apparent digestion trial. At 07:00 and 17:00 each day, ad libitum feedings were given, with the delivered amount adjusted considering a nearly 10% refusal rate.
During all feeding periods, the diets and ort samples of individual Hu sheep were harvested daily to calculate the nutrient intake. The initial and final body weights were measured before the morning feed on the start and finish days. Afterwards, the dry matter intake (DMI), average daily gain (ADG) and feed conversion ratio (FCR) were calculated (Hassan et al., 2022a).
At the end of the experiment, 2 h after the morning feeding, using an oral stomach tube, rumen fluid was taken (Anscitech Co., Wuhan, Hubei, China). A sample of rumen fluid (1.0 ml) and 0.3 ml of 25% metaphosphoric acid were mixed and stored at-20°C.till the VFA, microbial crude protein (MCP) and ammonia nitrogen (NH3-N) concentrations were investigated. The pH of rumen fluid was quickly determined using a portable pH detector (Testo-206-pH, Testo Co., Germany). The VFA concentration was determined using an Agilent Technology-78-90A gas chromatograph (Agilent Technologies, Waldbronn, Germany) coupled to an attached silica vessel column (30 mm × 0.25 mm × 0.25 μm film thickness; SP-3420A; Beifenrili Analyzer Associates, Beijing, China). A modified colorimetric technique was used to measure the NH3-N content (Chaney and Marbach, 1962). The microbial protein component of the rumen fluid was identified using an updated colourimetric technique at 595 nm (Verdouw et al., 1978). Utilizing materials acquired from the Chinese company Nanjing Jiancheng Institute of Biological Engineering Co., we determined the activities of amylase (CNPG3 method), pepsin (A080-1), and cellulase (A) in rumen fluid. The main instruments used were an HWS-12 water bath (Shanghai Blue Pard Instruments Co., Ltd.), a 721 spectrophotometer, a thermostat, and a PL-6906 microplate meter (Shanghai Youke Instrument Co., Ltd).
The V3-V4 region of the 16S rDNA was amplified using specific primers with a barcode; the primer sequences were 338F: ACTCCTACGGGAGGCAGCAG; and 806R: GGACTACHVGGGTWTCTAAT. DNA was extracted using the kit (Omega Biotek, Norcross, GA, United States). By using 1% agarose gel electrophoresis to identify PCR products, they were then purified using the Agencourt AMPure XP nucleic acid purification kit. Equal quantities of the purified amplified products were mixed, and the “Y” shaped connection was connected. Magnetic bead screening was used to remove the connector’s self-connecting parts. The library template was enriched by PCR amplification to generate single-stranded DNA fragments, and MiSeq was used to construct and sequence the library.
As stated previously, sequencing data was analyzed using the Quantitative Insights into Microbial Ecology (QIIME, v1.8.0) pipeline (Caporaso et al., 2010). In brief, raw sequencing reads that exactly matched the barcodes on the samples tested were assigned to them and identified as valid sequences. Low-quality sequences were detected by the following parameters: length of 150 bp, average Phred scores of 20, ambiguous bases, and > 8 bp of mononucleotide repetitions. FLASH was used to assemble paired-end reads (Magoc and Salzberg, 2011). Following chimera identification, UCLUST was used to group the remaining high-quality sequences into operational taxonomic units (OTUs) with 97% sequence identity. Using the default parameters, we selected a representative sample sequence from each OTU. By comparing the representative sequence set to the Greengenes Database and selecting the best hit, BLAST was used to classify OTUs. Each OTU’s abundance and taxonomy were then recorded in an OTU table for each sample. OTUs with less than 0.001% sequence content across all samples were discarded. A typical, rounded rarefied OTU table to even out sample-to-sample sequencing depth differences. We calculated richness estimates and diversity indices using the QIIME v1.8, including Shannon, Simpson, Chao 1, Observed species, Goods coverage, and PD entire tree. We obtained the species classification information corresponding to each OTU by annotating specific information for the communities at various levels and comparing OTU representative sequences using BLAST (2.6.0+). We used linear discriminant analysis (LDA) effect size analysis to identify species with significant differences in abundance among groups (Langille et al., 2013). TThe first method for identifying species with statistically significant differences in abundance among groups was the analysis of variance (ANOVA) test. The Wilcoxon rank-sum method was then used to investigate group differences. For the purpose of data reduction and an investigation of the significance of significant phenotypic differences between species, the algebraic linear discriminant analysis score was set to 3.0. To compare all samples, we used Principal Coordinates Analysis (PCoA) based on weighted UniFrac distances, and a distance-based matrices analysis was performed to identify sample differences.
To estimate the functions of the rumen microbial community, we used the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt v2.0.01). The abundance of each functional category was determined using data on the functional orthologues, pathways, and enzyme commission numbers collected in accordance with the analysis module of the Kyoto Encyclopedia of Genes and Genomes (Langille et al., 2013).
All data were represented using the mean standard deviation. A randomized complete block design with repeated measurements was utilized to analyze the data on rumen fermentation parameters. The Kruskal-Wallis technique in the R program was used to analyze the differences in alpha diversity, relative abundance at the phylum, family, and genus levels, and microbiota function among the three groups (v 4.0.3). The “ape” and “ggplot2” packages in R (v 4.0.3) were used to display PCoA, Venn, column, and linear discriminant analysis effect size (LEfSe) data, respectively. Using R (v 4.0.3), package “corrplot,” we investigated the relationship between the top 20 genera in all data (based on Spearman’s coefficient). The “igraph” package of the R (v 4.0.3) language was used to show the network of the top 20 genera. At P 0.05, all results related to intergroup differences were statistically significant.
The Hu sheep in the CK, F-CSM, and F-RSM groups exhibited ADG of 192.10 g/d, 210.12 g/d, and 226.03 g/d, respectively; Compared to the CK group, Hu sheep ADG in F-RSM group significantly higher than CK and F-CSM group (p < 0.05). The F-CSM group had significantly higher DMI than the F-RSM and CK groups (p < 0.05). Additionally, the difference between both the FCR in the CK and F-CSM groups and the F-RSM group was statistically significant (p < 0.05; Table 2).
In comparison to the CK group, the pH of the rumen fluid was considerably lower in the F-CSM and F-RSM groups (p < 0.05; Table 3). Total VFA, acetate, propionate, and butyrate concentrations differed (p < 0.05) among the groups, with the F-CSM group having higher concentrations than the F-RSM and CK groups. However, the F-CSM group exhibited a higher proportion of propionate than that exhibited by the CK and F-RSM groups (p < 0.05). When compared to the CK group, the F-CSM and F-RSM groups had substantially greater NH3-N contents and MCP concentrations, respectively (p < 0.05).
The enzyme activities in each group are shown in Table 4. Adding different proteins to the TMR did not affect rumen α-amylase activity (p > 0.05). Pepsin and cellulose activities in the F-CSM group were substantially greater than those in the F-RSM group (p < 0.05). There was no significant difference between the CK and F-CSM groups (p > 0.05).
The OUT rank curve graphic gradually stabilized, indicating that the test samples’ high coverage (Figure 1). A total of 1,831,937 high-quality reads were obtained, for a sample size of 76,330 reads, composed the rumen microbiota. Nucleotide sequence identification (97%) resulted in a total of 2,285 OTUs (Figure 1A). Among these, 1,448 OTUs (63.37%) were shared among samples from different groups. There were no dietary effects observed, and PCoA analysis based on Weighted UniFrac or Bray–Curtis dissimilarity revealed that the rumen liquid fraction was not completely differentiated from other liquids (Figure 1B).
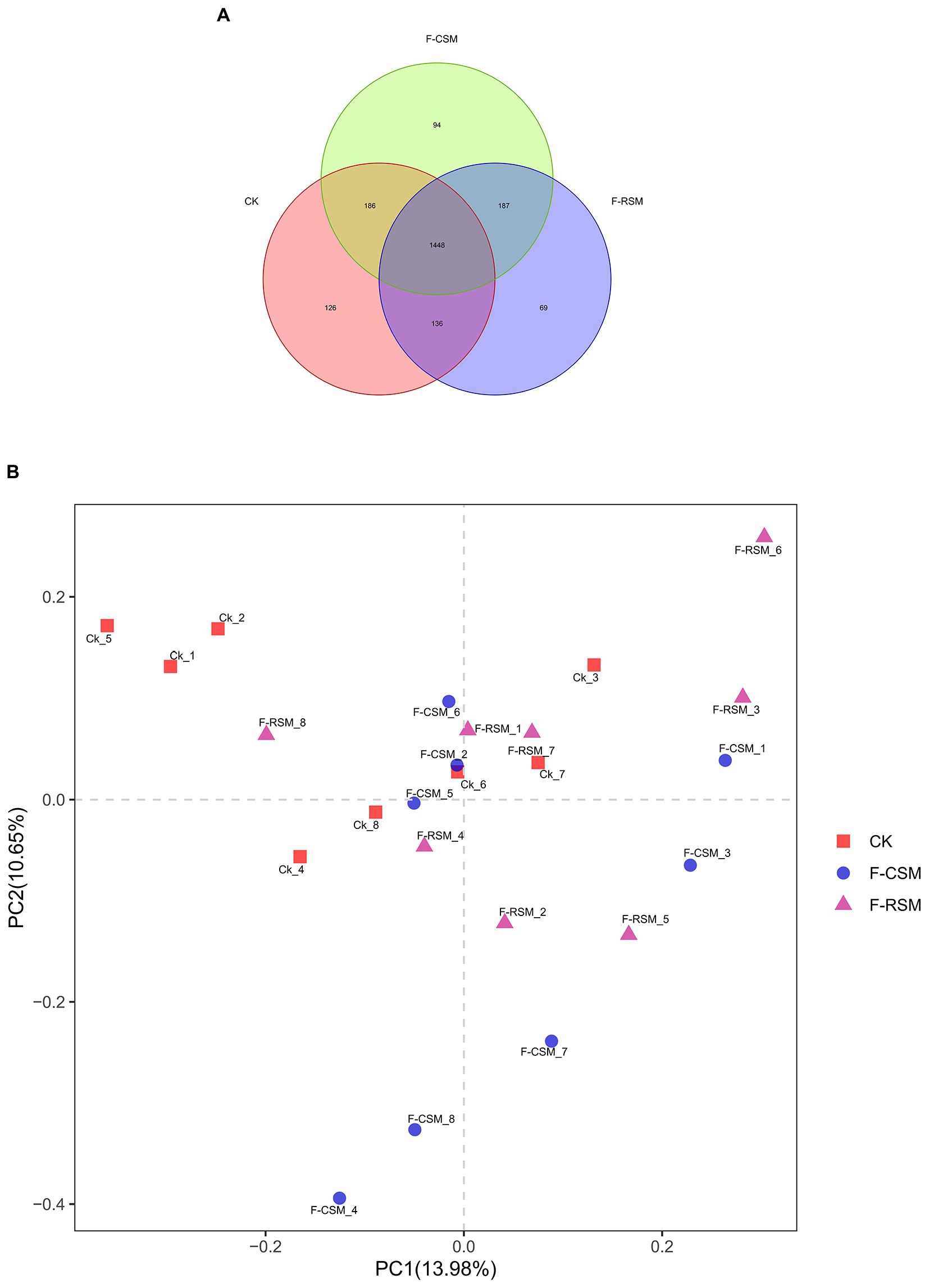
Figure 1. Effects of the diets on rumen microbiota of Hu sheep (A) Composition of rumen microbiota from Hu sheep fed three diets (OTU-level analysis). (B) PCoA analysis of rumen microbiota from Hu sheep fed different diets (%); CK, non-fermented group; F-CSM, fermented cotton seed meal TMR; F-RSM, fermented rapeseed meal TMR.
Analysis of alpha diversity (Figure 2) showed that the F-CSM group had numerically higher Shannon and community richness (Chao) indices than those in the CK and F-RSM groups; however, the differences were not significantly different (p > 0.05). The detected species were not substantially different among groups (p > 0.05). The Simpson index showed no discernible differences between the three groups (p > 0.05).
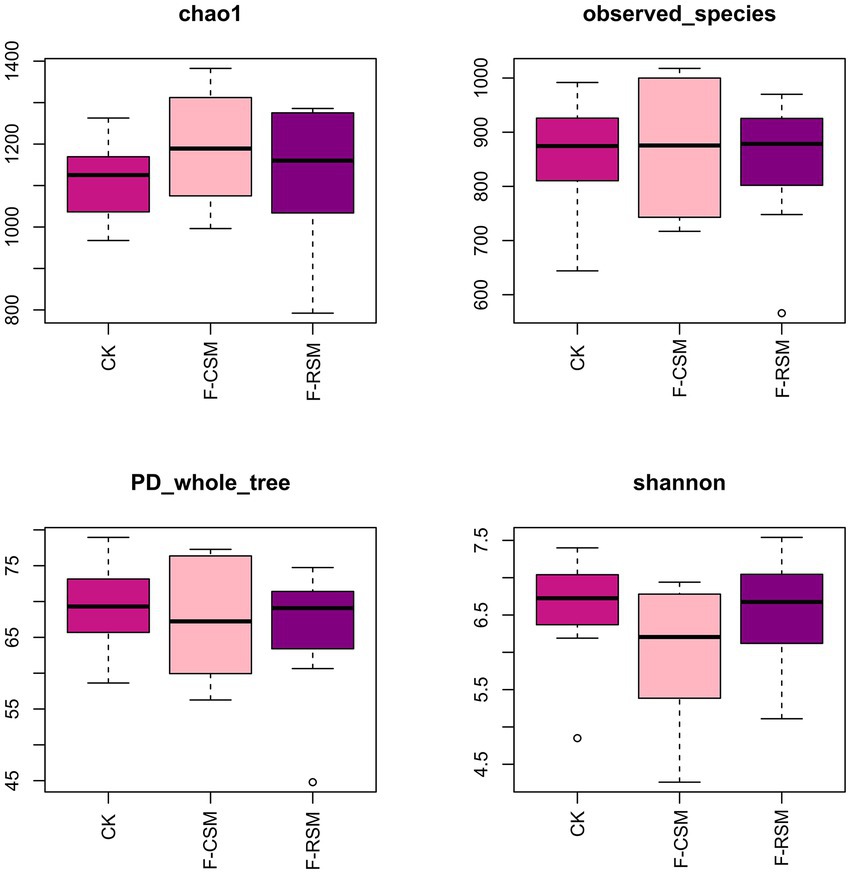
Figure 2. Effects of the diets on rumen alpha diversity of Hu sheep. CK, non-fermented group; F-CSM, fermented cotton seed meal TMR; F-RSM, fermented rapeseed meal TMR.
Seventeen bacterial phyla were found among all treatments, with Bacteroidota, Firmicutes, and Proteobacteria being the three most predominant. In comparison to that in the F-CSM group (58.72%), in the CK and F-RSM groups, the relative abundance of Bacteroidetes was greater (71.41 and 79.89%, respectively; p < 0.05). Firmicutes showed lower abundance in the CK group (18.30%) than in the F-CSM and F-RSM groups (p < 0.05). There were 195 diverse microbial genera found among all groups, but only 14 of them were determined to be the “major genera” since they each accounted for more than 0.5% of the total sequences through at least one treatment (Figure 3). Except for the genera Prevotella and Christensenellaceae_R-7 (p < 0.05), no interaction (p > 0.05) was found for any bacterial abundance at the genus level. When compared to the CK group, Prevotella was more abundant in the F-CSM and F-RSM groups (p < 0.05), but the TMR type had no influence on their relative abundance (Figures 4, 5).
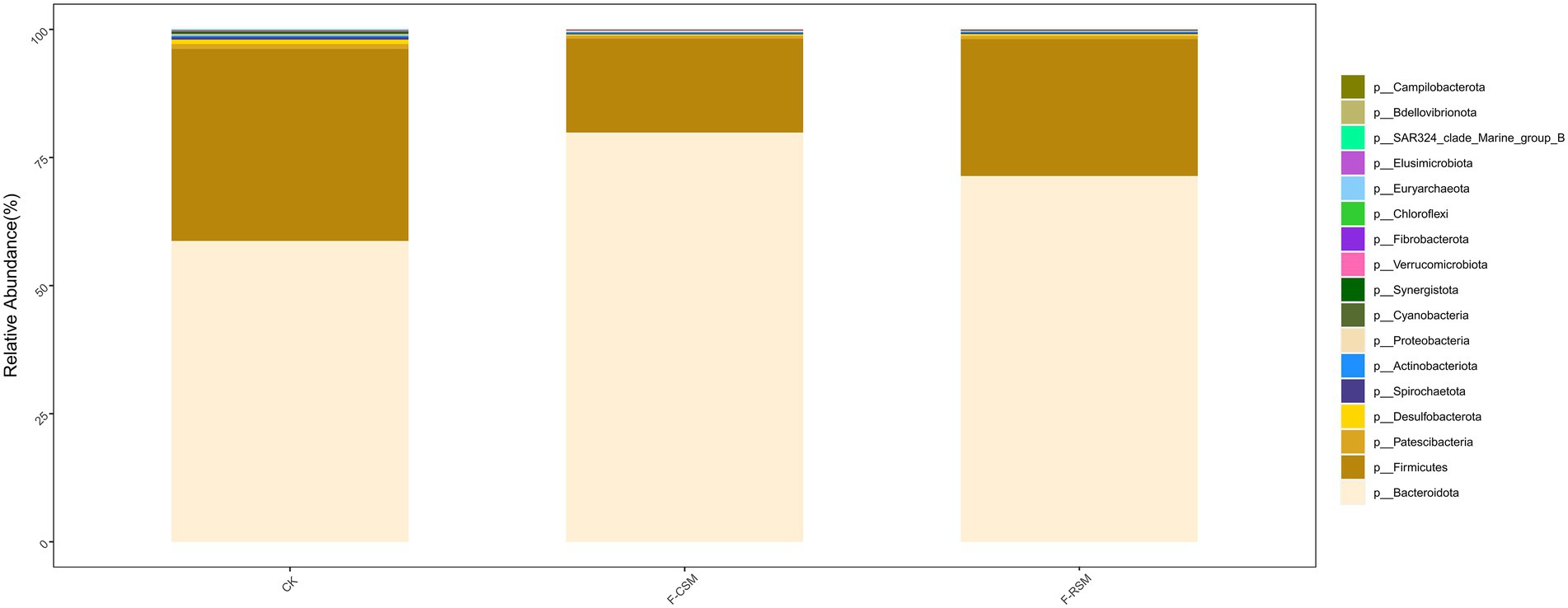
Figure 3. Effects of the diets on compositional profifiles of rumen microbiota of Hu sheep (Phylum level; %). CK, non-fermented group; F-CSM, fermented cotton seed meal TMR; F-RSM, fermented rapeseed meal TMR.
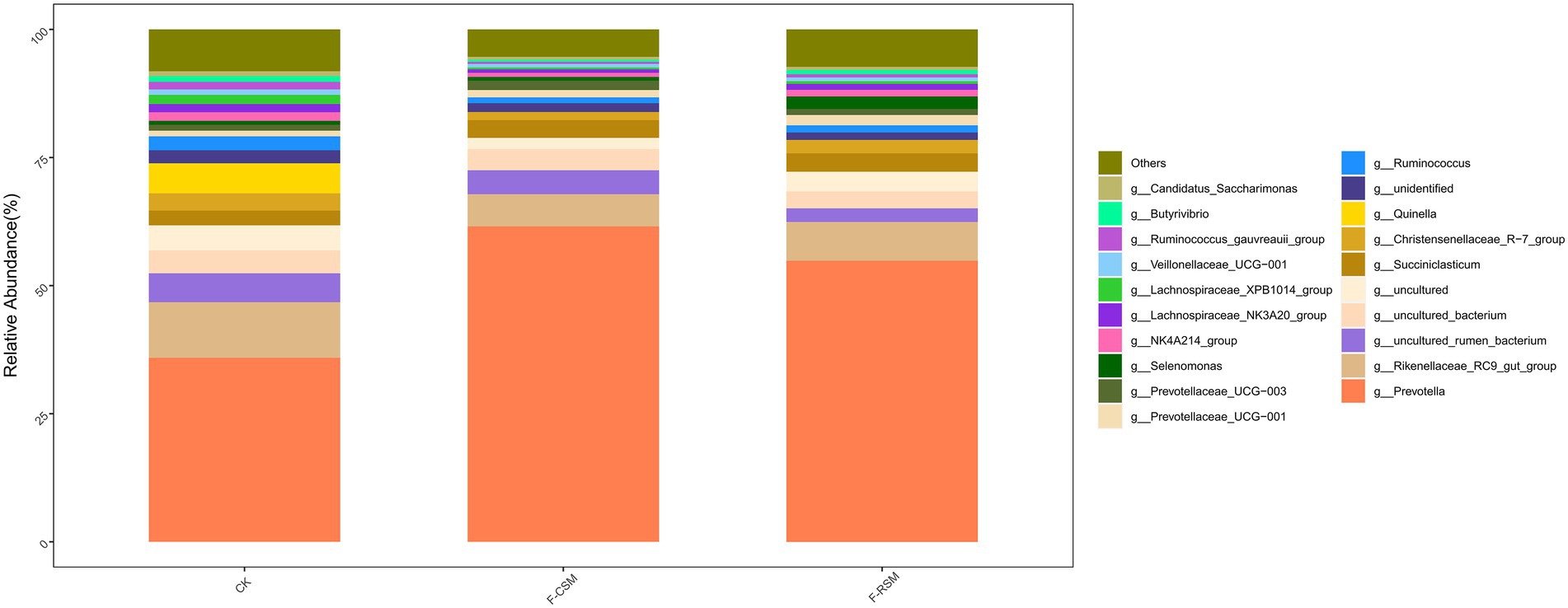
Figure 4. Effects of the diets on compositional profifiles of rumen microbiota of Hu sheep (Genus level; %); CK, non-fermented group; F-CSM, fermented Cotton seed meal TMR; F-RSM, fermented Rapeseed meal TMR.
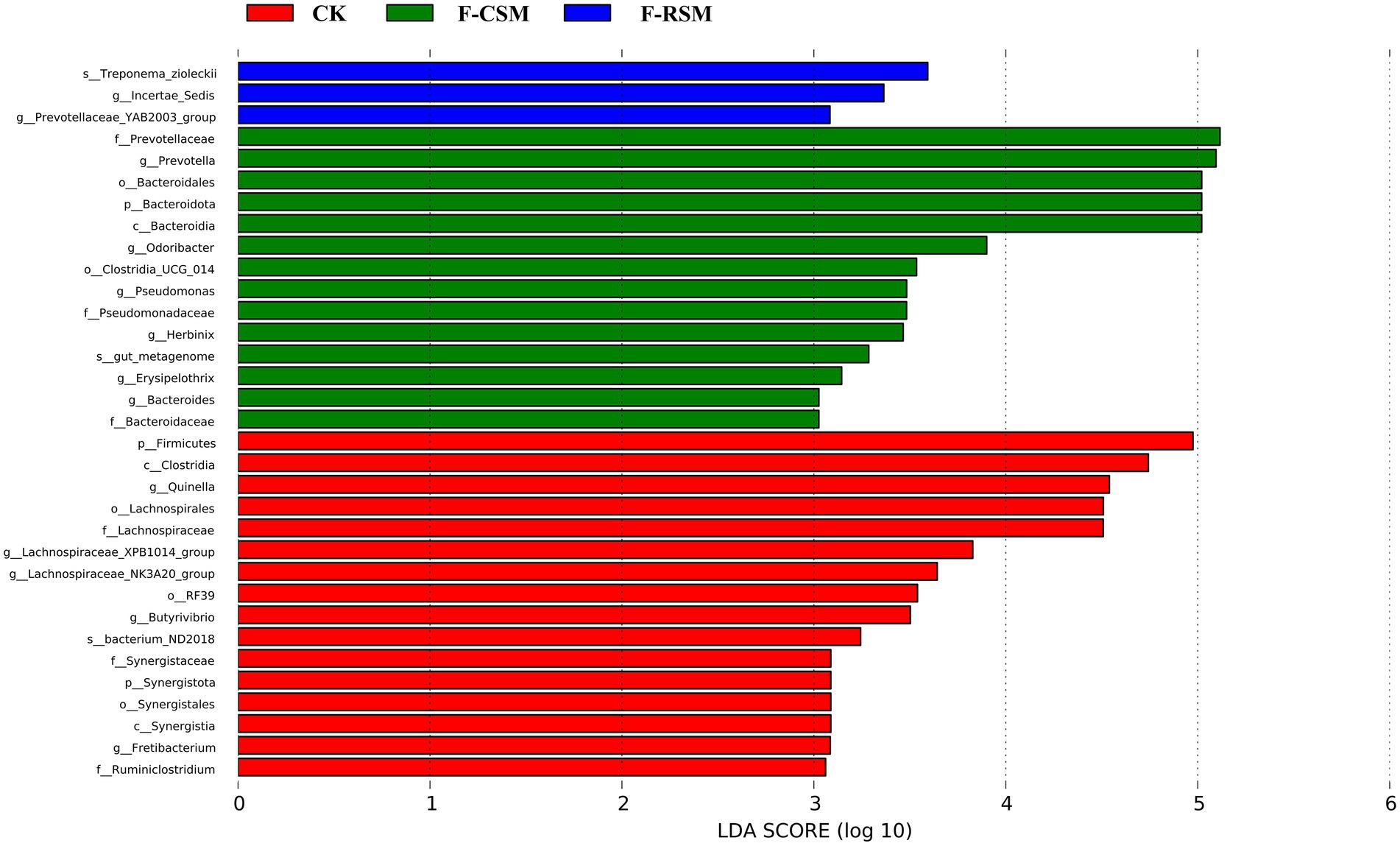
Figure 5. Effect of potential biomarkers was defined by LEfSe at Hu sheep rumen liquite (log10); CK, non-fermented group; F-CSM, fermented Cotton seed meal TMR; F-RSM, fermented Rapeseed meal TMR.
In Table 5, the top 20 metabolic pathways are listed. The TMR type had a significant effect on 8 out of the 20 pathways. F-CSM and F-RSM exhibited higher (p < 0.05) relative abundance of the functions related to terpenoid, polyketide, lipid, and folding and transcription-associated metabolisms, despite the fact that there was no discernible difference between the two groups (p > 0.05).
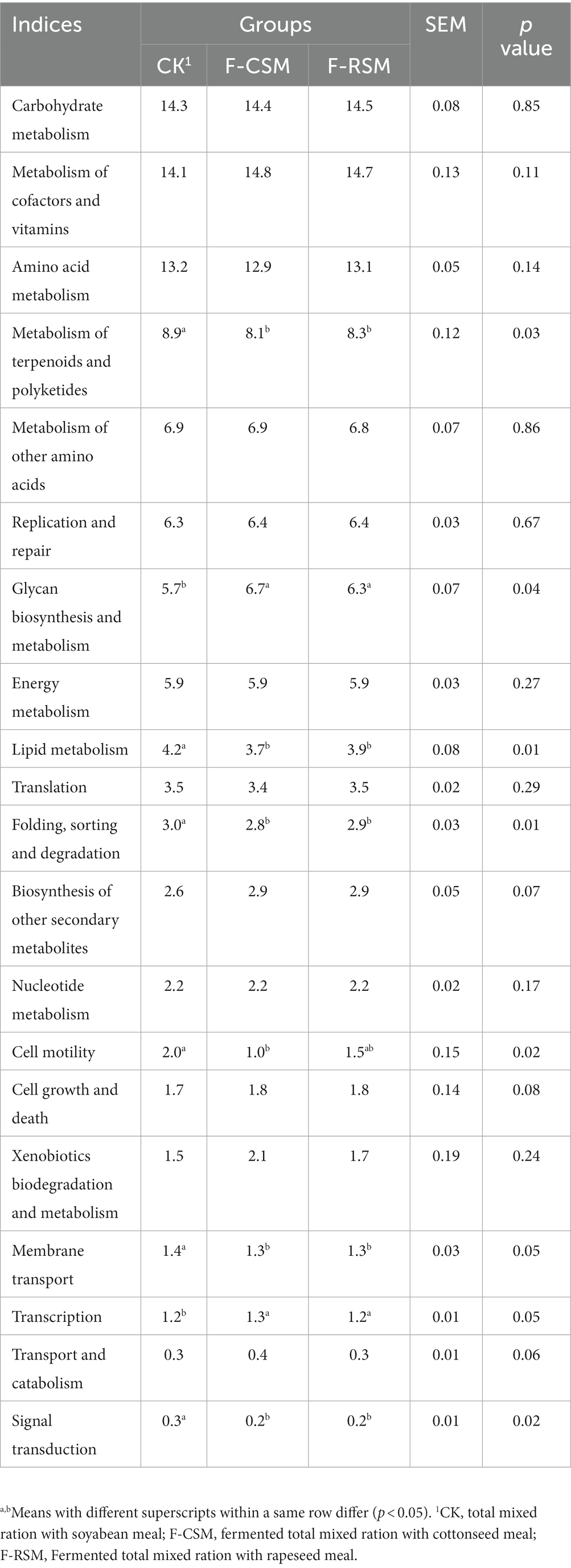
Table 5. Effect of feeding different diets on predominant predicted gene pathways in the different ruminal ecological niches of growing Hu sheep.
The relationship between the fermentation-related parameters and the relative abundance of the top 20 bacterial genera is shown in Figure 6. The relative abundances of Veillonellaceae_UCG-001 and Lachnospiraceae_XPB1014 correlated with rumen fermentation and NH3-N content (p < 0.05). While the relative abundance of Lachnospiraceae_XPB1014 negatively correlated with NH3-N concentration (r = 0.53), the relative abundance of Veillonellaceae_UCG-001 positively correlated with butyric acid content (r = 0.72).
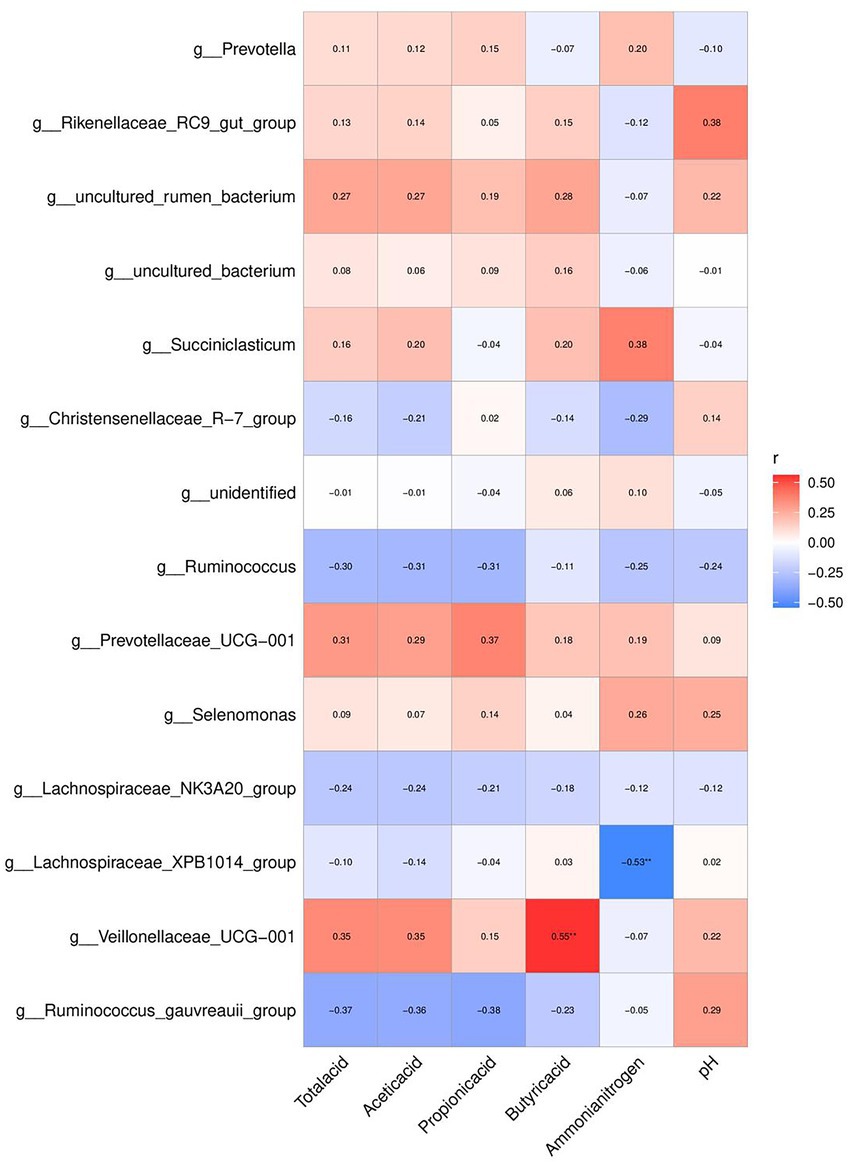
Figure 6. Effects of the diets on correlation between the top 20 relative abundances of genera, rumen VFA and enzyme activity mpositional profifiles of Hu sheep. *p < 0.05; **p < 0.01.
Due to the presence of anti-nutritional factors and the characteristics of the available substrates, diet composition has a significant impact on nutrient utilization and gastrointestinal physiology (Kiaie et al., 2014; Kim et al., 2017). Although it is generally recognized that TMR fermentation with microorganisms improves feed quality, there is an energy cost and a dry matter loss (Liang et al., 2017). Microbial fermentation can significantly reduce anti-nutritional factor content while improving palatability and nutritional value. CSM contains FG, while RSM contains glucosinolates, which affect the palatability and nutritional value of feed, reduce the production performance of animals, and also harm physical organs such as the liver and thyroid, greatly limiting its popularization and application in production. Hu et al. (2010) found that replacing SBM with 10% fermented CSM significantly increased the weight gain of sheep and reduced meat production costs. In a study by Xu et al. (2020), fermented feed partly breaks down the protein into the minor peptides & free amino acids, that imparts a pleasant acidic aroma to the feeds, enhances palatability as well as increases diet digestibility, which finally changes growth performance (Xu et al., 2020). However, our study showed that the F-CSM or F-RSM diet increased the overall body weight and daily weight gain in Hu sheep.
Rumen pH is the result of the interaction between VFAs in rumen digesta and buffer salts in saliva, absorption of VFAs by the rumen epithelium, and excretion of rumen digesta (Segata et al., 2011). It has an important impact on the rumen microbiota, rumen fermentation products, and rumen function, and is the most direct manifestation of rumen physiological conditions (Feng, 2004). Rumen pH is closely related to dietary composition and nutrient composition, and can comprehensively reflect the production, absorption, discharge, and neutralization of rumen microorganisms, metabolites, and organic acids (Nagaraja and Titgemeyer, 2007). The ruminal fluid pH varied from 5.93 to 6.39, which was within the normal range (Gilani et al., 2005), and among the groups, there was no significant different. These results indicate that various beneficial components contained in the fermented mixed meals were conducive to maintaining the stability of rumen pH, which could partially or completely replace SBM in the diet.
The VFAs produced during fermentation in the rumen are used as a source of energy in ruminants. VFAs produced by rumen fermentation, can provide up to 80% of the body’s energy and serve as the primary source of carbon for the proliferation of rumen microbes (Zhou et al., 2018). Acetic acid is the main precursor for milk fat synthesis, whereas propionic acid synthesizes the glucose required by the body through gluconeogenesis. As a result, propionic acid fermentation can provide the body more energy, which is beneficial for improving animal productivity and feed utilization (Spears et al., 2004). Studies have shown that microbial fermented feed can regulate rumen fermentation mode (Guo, 2016). Zhang Z. (2017) found that microbial fermented feed could significantly increase the contents of T-VFA, acetic acid and propionic acid and the ratio of acetate to propionate in the rumen of dairy cows through in vitro experiments, Guo (2016) also obtained similar results, and Chen et al. (2015) reported that fermented feed could increase the contents of rumen VFA, acetic acid and propionic acid. However, it had no significant effect on the ratio of acetate to propionate. Meanwhile, Li Z. et al. (2015) found that fermented feed could increase the content of rumen VFA of sheep and reduce the ratio of acetate to propionate. The present results show that rumen VFA concentration was the highest and the acetate/propionate ratio was the lowest in the animals fed with F-CSM. The F-CSM group showed lower VFA concentration than the F-RSM and CK groups. These results indicate that the substitution of fermented TMR contain CSM for SBM in Hu sheep diet is beneficial to the transformation of rumen fermentation type into propionic acid fermentation type and improve energy utilization efficiency.
The NH3-N concentration reflects the rate of decomposition of nitrogen-containing substances to produce NH3 and the uptake and utilization of NH3 by rumen microorganisms. A maximum concentration over 5 mg/dl can satisfy the needs of rumen microbial development. The NH3-N concentration in the rumen is essential for protein degradation and microbial protein synthesis (Li M. S. et al., 2015). In this experiment the replacement of SBM with F-CSM or F-RSM had a significant effect on rumen NH3-N content; however, the rumen NH3-N content was 7.09 mmol/l and 7.11 mmol/l, respectively, in the F-CSM and F-RSM groups, which is within the normal physiological range of rumen NH3-N (6–30 mg/d L; Yang et al., 2001). In our study, the diversity and richness of rumen bacteria did not significantly change when SBM was replaced with F-CSM or F-RSM in the diet of Hu sheep.
Rumen microorganisms decompose and utilize various secreted enzymes. Enzyme activity directly reflects the decomposition efficiency of feed nutrients in the rumen and determines the utilization efficiency of feed nutrients by animals. In this study, the activities of pepsin and cellulose in rumen fluid increased after replacing SBM with F-CSM. This may be because fermented TMR changes the pH of rumen fluid and provides favourable conditions for the reaction of digestive enzymes, thus improving the activity of digestive enzymes (Bowman and Asplund, 1988). In the F-RSM group, enzyme activity decreased. This could be the result of the anti-nutritional factors in RSM (Hassan et al., 2022b).
Researchers (Diao et al., 2013; Zhang et al., 2016; Zhou et al., 2017; Bi et al., 2018; Guo et al., 2021; Liu et al., 2021; Lu et al., 2021) have shown that the two dominant phyla in ruminants are Bacteroidetes and Firmicutes. The two most frequent taxa discovered in this study were Bacteroidetes and Firmicutes; the findings are consistent with other studies. In this study, we found that Prevotella and Christensenellaceae R-7 were successively dominant at the genus level. Studies have shown that Bacteroides plays an important role in the degradation of non-fibrous substances, and Firmicutes mainly participate in the decomposition of fibrous substances (Evans et al., 2011), while non-fibrous substances produce propionic acid after rumen fermentation, while cellulose substances mainly produce acetic acid after degradation (Li et al., 2017). Our study the relative abundances of Bacteroidetes and Firmicutes in the F-CSM and F-RSM groups were greater than those in the CK group. These results indicate that fermented TMR can promote the growth and proliferation of Bacteroidetes in rumen, accelerate the degradation of non-fiber substances in diet, produce more propionic acid, provide more energy for the body, and promote the growth of animals.
The most abundant genus of Bacteroidetes is Prevotella (Zou et al., 2019). It participates in the metabolism of various microorganisms, has a high hemicellulose degradation ability, and can adapt to different dietary structures (Wolff et al., 2017). Prevotella is important in the degradation of crude protein, starch, xylan, and pectin (Li Z. et al., 2015; Jami et al., 2013). Prevotella relative abundance in the rumen was significantly greater in the F-CSM and F-RSM groups in this study. This suggests that F-CSM and F-RSM can replace SBM and help Hu sheep in improved nutrient absorption and digestion, thereby improving feed conversion rate. Veillonellaceae belongs to the phylum Firmicutes and can degrade and utilize cellulose (Long et al., 2022). We also found that the proportion of butyric acid was significantly positively correlated with the relative abundance of Veillonellaceae UCG-001. This indicated that Veillonellaceae_UCG-001 was related to rumen VFA metabolism, which is consistent with the results of Thoetkiattikul et al. (2013). These results indicate that F-CSM and F-RSM can promote the growth of microorganisms, maintain intestinal structure and function, and regulate immunity in Hu sheep. Lachnospiraceae_XPB1014’s relative abundance and NH3-N content showed a negative correlation. These findings suggest a relationship between NH3-N concentrations and rumen bacteria in Hu sheep. Therefore, replacing SBM with F-CSM or F-RSM likely influenced the VFA and NH3-N contents, and ultimately impacted the rumen microbiota. Microbiota functional properties affect host-microbiome interaction (Lin et al., 2019; Zeng, 2020). The gene function prediction results showed that F-CSM and F-RSM can replace SBM in the diet of Hu sheep and have an important influence on their metabolism. Particularly, it promoted glycan biosynthesis and metabolism, indicating that F-CSM and F-RSM contributed to the maturation and stability of Hu sheep rumen microflora.
Substituting SBM with F-CSM or F-RSM in the diet of Hu sheep is beneficial for the conversion of microbiota from rumen fermentation type to propionic acid fermentation type and improving energy utilization efficiency. No negative effects were observed on rumen microbiota, and substituting SBM with F-CSM resulted in the best effect.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Committee of IFRCAAS (AEC-IFR-CAAS, Beijing, China). Written informed consent was obtained from the owners for the participation of their animals in this study.
HR and HY: animal trial, data collection and evaluation, laboratory and statistical analysis, and writing. TM: critical manuscript review. QD: data evaluation and manuscript review. LxK and LyK: animal trial, laboratory analysis, and data collection. YT: study design, feed formulation, data evaluation, and critical manuscript review. All authors contributed to the article and approved the submitted version.
This study was supported by the Innovation Project of Inner Mongolia (grant number: NMKJXM202110) and China Agriculture Research System of MOF and MARA (CARS-38).
We acknowledge everyone at the Beijing Feed Research Institute and the Nankou Base of the Institute of Feed Research for their help with animal experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alexandratos, N., and Bruinsma, J. (2012). World agriculture towards 2030/2050: The 2012 Revision. ESA Working Paper No. 12-03. Rome: Food and Agriculture Organization.
Anantasook, N., Wanapat, M., Cherdthong, A., and Gunun, P. (2013). Changes of microbial population in the rumen of dairy steers as influenced by plant containing tannins and saponins and roughage to concentrate ratio. Asian Australas. J. Anim. Sci. 26, 1583–1591. doi: 10.5713/ajas.2013.13182
Ashayerizadeh, A., Dastar, B., Shargh, M. S., Mahoonak, A. S., and Zerehdaran, S. (2018). Effects of feeding fermented rapeseed meal on growth performance, gastrointestinal microflora population, blood metabolites, meat quality, and lipid metabolism in broiler chickens. Livest. Sci. 216, 183–190. doi: 10.1016/j.livsci.2018.08.012
Barrett, J. E., Klopfenstein, C. F., and Leipold, H. W. (1997). Detoxification of rapeseed meal by extrusion with an added basic salt. Cereal Chem. 74, 168–170. doi: 10.1094/CCHEM.1997.74.2.168
Bi, Y. L., Zeng, S. Q., Zhang, R., Diao, Q. Y., and Tu, Y. (2018). Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 18:69. doi: 10.1186/s12866-018-1213-9
Bowman, J. G. P., and Asplund, J. M. (1988). Nitrogen utilization, ruminal fermentation and abomasal nitrogen flow in sheep fed Caucasian bluestem hay supplemented with lucerne or urea. Anim. Feed Sci. Technol. 20, 33–44. doi: 10.1016/0377-8401(88)90125-3
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chaney, A. L., and Marbach, E. P. (1962). Modified Reagents for Determination of Urea and Ammonia. Clin. Chem. 8, 130–132.
Chen, G. J., Peng, Z. L., Song, S. D., Wang, B. X., Guo, C. H., Zhang, Z. F., et al. (2015). Effects of the fermented distillers’ on performance, apparent nutrient digestibility, ruminal fermentation and serum biochemical indices in house-fed yaks. Chin. J. Anim. Nutr. 27, 2920–2927. doi: 10.3969/j.issn.1006-267x.2015.09.032
Diao, H., Zheng, P., Yu, B., He, J., Mao, X. B., Yu, J., et al. (2013). Effects of benzoic acid on growth performance, serum biochemical parameters, nutrient digestibility and digestive enzyme activities of jejunal digesta in weaner piglets. Chin. J. Anim. Nutr. 25, 768–777. doi: 10.3969/j.issn.1006-267x.2013.04.014
Evans, N. J., Brown, J. M., Murray, R. D., Getty, B., Birtles, R. J., Hart, C. A., et al. (2011). Characterization of novel bovine gastrointestinal tract treponema isolates and comparison with bovine digital dermatitis treponemes. Appl. Environ. Microbiol. 77, 138–147. doi: 10.1128/AEM.00993-10
Francis, G., Makkar, H. P. S., and Becker, K. (2001). Anti nutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture 199, 197–227. doi: 10.1016/S0044-8486(01)00526-9
Gilani, G. S., Cockell, K. A., and Sepehr, E. (2005). Effects of anti nutritional factors on protein digestibility and amino acid availability in foods. J. AOAC Int. 88, 967–987. doi: 10.1093/jaoac/88.3.967
Guo, P., (2016). Study on Microbial Fermentation Feed Formula of Dairy Cows and the Matched Application Technique. Inner Mongolia Agricultural University, Hohhot, China.
Guo, N., Wu, Q., Shi, F., Niu, J., Zhang, T., Degen, A. A., et al. (2021). Seasonal dynamics of diet-gut microbiota interaction in adaptation of yaks to life at high altitude. NPJ Biofifilms Microbiomes 7:38. doi: 10.1038/s41522-021-00207-6
Hassan, A. Y., Halidai, R., Tao, M., Min, Y. P., Rui, Y. H., and Yan, T. (2022a). Fermented Total mixed ration with cottonseed meal or rapeseed meal improved growth performance and meat quality of Hu lamb compared to Total mixed ration with soybean meal. Fermentation 8:576. doi: 10.3390/fermentation8110576
Hassan, A. Y., Halidai, R., Tao, M., Min, Y. P., Rui, Y. H., and Yan, T. (2022b). Effect of lactic acid bacteria and yeast supplementation on anti-nutritional factors and chemical composition of fermented total mixed ration containing cottonseed meal or rapeseed meal. Anim. Biosci. 35, 556–566. doi: 10.5713/ab.21.0270
Heuze, V., Tran, G., Noziere, P., Lessire, M., and Lebas, F. (2015). Linseed Meal. Feedipedia, a Programme by INRA, CIRAD, AFZ and FAO. Rome, Italy: FAO.
Hu, P. F., Wu, Z. H., S, T., and Y, G. Q. (2010). Effect of microbial treatment of cottonseed meal on fattening effect of lambs. Chin. Heilongjiang Anim. Sci. Vet. 24, 105–106.
Huisman, J., and Tolman, G. (1992). “Anti nutritional factors in the plant proteins of diets for non-ruminants” in Recent Developments in Pig Nutrition. eds. P. C. Garnsworthy and J. Wiseman, vol. 68 (Nottingham: Nottingham University Press), 101–110.
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Khan, M. A., Bach, A., Weary, D. M., and Keyserlingk, M. A. G. (2016). Invited review: transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 99, 885–902. doi: 10.3168/jds.2015-9975
Kiaie, E., Romero, L. F., and Ravindran, V. (2014). Growth performance, nutrient utilization, and digesta characteristics in broiler chickens fed corn or wheat diets without or with supplemental xylanase. Poult. Sci. 93, 1186–1196. doi: 10.3382/ps.2013-03715
Kim, E., Leung, H., Akhtar, N., Li, J., Barta, J. R., and Wang, Y. (2017). Growth performance and gastrointestinal responses of broiler chickens fed corn-soybean meal diet without or with exogenous epidermal growth factor upon challenge with Eimeria. Poult. Sci. 96, 3676–3686. doi: 10.3382/ps/pex192
Kolver, E. S., Roche, J. R., DMiller, D., and Densley, R. (2001). Maize Silage for Dairy Cows. In: Proceedings of the Conference-New Zealand Grassland Association. pp. 195–201.
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., and Reyes, J. A. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Li, L. J., Cheng, S. R., Diao, Q. Y., Fu, T., Bi, Y. L., Wang, A. S., et al. (2017). Effects of diets ith different NFC/NDF levels on the rumen fermentation parameters and bacterial community in male calves. Chin. J. Acta Veterinaria et Zootechnica Sinica. 48. 2347–2357. doi: 10.11843/j.issn.0.66-6964.2017.12.014
Li, F. H., Ding, Z. T., Ke, W. C., Xu, D. M., Zhang, P., and Bai, J. (2019). Ferulic acid esterase producing lactic acid bacteria and cellulase pretreatments of corn stalk silage at two different temperatures: ensiling characteristics, carbohydrates composition and enzymatic saccharification. Bioresour. Technol. 282, 211–221. doi: 10.1016/j.biortech.2019.03.022
Li, Z., Wright, A. D., Liu, H., Fan, Z., Yang, F., Zhang, Z., et al. (2015). Response of the rumen microbiota of sika deer (Cervus nippon) fed different concentrations of tannin rich plants. PLoS One 10:e0123481. doi: 10.1371/journal.pone.0123481
Li, M. S., Xue, S. Y., Wang, C., Wang, L. M., Guo, T. L., and Jin, H. (2015). Effects of Salix mixed fermented feed on ruminal environmental parameters by in vitro gas production method. Chin. J. Anim. Nutr. 27, 1943–1953. doi: 10.3969/j.issn.1006-267x.2015.02.039
Liang, Y. S., Li, G. Z., Li, X. Y., Lv, J. Y., Li, F. D., and Tang, D. F. (2017). Growth performance, rumen fermentation, bacteria composition, and gene expressions involved in intracellular pH regulation of rumen epithelium in finishing hu lambs differing in residual feed intake phenotype. J. Anim. Sci. 95, 1727–1738. doi: 10.2527/jas.2016.1134
Lin, L., Xie, F., Sun, D. M., Liu, J. H., Zhu, W. Y., and Mao, S. Y. (2019). Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 7:83. doi: 10.1186/s40168-019-0701-y
Liu, W. W., Wang, Q., Song, J. J., Xin, J. W., Zhang, S. S., Lei, Y. H., et al. (2021). Comparison of gut microbiota of yaks from different geographical regions. Front. Microbiol. 12:666940. doi: 10.3389/fmicb.2021.666940
Long, P. L., Lei, Q., and Tuo, L. (2022). Supplemental dietary selenohomolanthionine affects growth and rumen bacterial population of Shaanbei white cashmere wether goats. Front. Microbiol. 13:942848. doi: 10.3389/fmicb.2022.942848
Lu, L. H., Wan, C. X., Han, W. C., Xiao, Y. C., Fa, S. Q., Tao, M., et al. (2021). Comparison of rumen fermentation parameters and microbiota of yaks from different altitude regions in Tibet, China. Front. Virol. 12, 1–12. doi: 10.3389/fmicb.2021.807512
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
McSweeney, C. S., Palmer, B., McNeill, D. M., and Krause, D. O. (2001). Microbial interactions with tannins: nutritional consequences for ruminants. Anim. Feed Sci. Technol. 91, 83–93. doi: 10.1016/S0377-8401(01)00232-2
Mottet, A., Dehaan, C., Falcucci, A., Tempio, G., Opio, C., and Gerber, P. (2017). Livestock: on our plates or eating at our table? A new analysis of the feed/food debate. Glob. Food Sec. 14, 1–8. doi: 10.1016/j.gfs.2017.01.001
Nagalakshmi, D., Sastry, V. R. B., and Pawde, A. (2003). Rumen fermentation patterns and nutrient digestion in lambs fed cottonseed meal supplemental diets. Anim. Feed Sci. Technol. 103, 1–14. doi: 10.1016/S0377-8401(02)00140-2
Nagaraja, T. G., and Titgemeyer, E. C. (2007). Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90, E17–E38. doi: 10.3168/jds.2006-478
Nie, C., Zhang, W., Ge, W., Liu, Y., Wang, Y., and Liu, J. (2015). Effect of cottonseed meal fermented with yeast on the lipid-related gene expression in broiler chickens. Braz. J. Poult. Sci. 17, 57–64. doi: 10.1590/1516-635XSPECIALISSUENutrition-PoultryFeedingAdditives057-064
Olimid, A. P., and Olimid, D. A. (2019). Societal challenges, population trends and human security: evidence from the public governance within the United Nations publications (2015-2019). Rev. Stiint. Polit. 64, 53–64.
Pal, V. A., and Walia, A. (2001). Beneficial effects of Rhizopus oligosporus fermentation on reduction of glucosinolates, fibre and phytic acid in rapeseed (Brassica napus) meal. Bioresour. Technol. 78, 309–312. doi: 10.1016/S0960-8524(01)00030-X
Robinson, P. H., Getachew, G., De, P. E. J., and Calhoun, M. C. (2001). Influence of variety and storage for up to 22 days on nutrient composition and gossypol level of Pima cottonseed (Gossypium spp.). Anim. Feed Sci. Technol. 91, 149–156. doi: 10.1016/S0377-8401(01)00202-4
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., and Garrett, W. S. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Spears, J. W., Schlegel, P., Seal, M. C., and Lloyda, K. E. (2004). Bioavailability of zinc from zinc sulfate and different organic zinc sources and their effects on ruminal volatile fatty acid proportions. Livest. Prod. Sci. 90, 211–217. doi: 10.1016/j.livprodsci.2004.05.001
Tang, C. H., Liu, J., Zhao, Q. Y., and Zhang, J. M. (2018). Pharmacokinetic comparison of gossypol isomers in cattle: transfer from diet to plasma and degradation by rumen microbes. J. Zhejiang Univ. Sci. B 19, 471–480. doi: 10.1631/jzus.B1700289
Thoetkiattikul, H., Mhuantong, W., Laothanachareon, T., Tangphatsornruang, S., Pattarajinda, V., Eurwilaichitr, L., et al. (2013). Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S r RNA gene pyrosequencing. Curr. Microbiol. 67, 130–137. doi: 10.1007/s00284-013-0336-3
Tripathi, M. K., and Mishra, A. S. (2007). Glucosinolates in animal nutrition: a review. Anim. Feed Sci. Technol. 132, 1–27. doi: 10.1016/j.anifeedsci.2006.03.003
Tufarelli, V., Ragni, M., and Laudadio, V. (2018). Feeding forage in poultry: a promising alternative for the future of production systems. Agriculture 8:81. doi: 10.3390/agriculture8060081
Verdouw, H., Van Echteld, C. J. A., and Dekkers, E. M. J. (1978). Ammonia determination based on indophenol formation with sodium salicylate. Water Res. 12, 399–402. doi: 10.1016/0043-1354(78)90107-0
Wanasundara, J. P. D., Tan, S., Alashi, A. M., Pudel, F., and Blanchard, C. (2017). “Proteins from canola/rapeseed” in Sustainable Protein Sources. eds. S. R. Nadathur, J. P. D. Wanasundara, and L. Scanlin (San Diego, CA, USA: Elsevier), 285–304.
Wang, D. Q., Song, D., Wang, W., Zhou, H., Wang, L., and Li, A. (2017). Effects of fermented cottonseed meal on growth performance, serum biochemical parameters, immune functions, antioxidative abilities, and cecal microflora in broilers. Food Agric. Immunol. 28, 725–738. doi: 10.1080/09540105.2017.1311308
Weng, X. Y., and Sun, J. Y. (2006). Biodegradation of free gossypol by a new strain of Candida tropicalis under solid state fermentation: effects of fermentation parameters. Process Biochem. 41, 1663–1668. doi: 10.1016/j.procbio.2006.03.015
Wolff, S. M., Ellison, M. J., Hao, Y., Cockrum, R. R., Austin, K. J., and Baraboo, M. (2017). Diet shifts provoke complex and variable changes in the metabolic networks of the ruminal microbiome. Microbiome 5:60. doi: 10.1186/s40168-017-0274-6
Xu, X., Ma, L., Fan, S., Ma, W., and Zhang, X. (2020). Effects of fermented Caragana korshinskii feed on meat quality characteristics in different muscles of Tan sheep. Ital. J. Anim. Sci. 19, 1036–1045. doi: 10.1080/1828051X.2020.1816505
Xu, C. Y., Moriya, N., and Ogawa, M. (2007). Nutritive value for ruminants of green tea grounds as a replacement of brewers’ grains in totally mixed ration silage. Anim. Feed Sci. Technol. 138, 228–238. doi: 10.1016/j.anifeedsci.2006.11.014
Yang, W. Z., Beauchemin, K. A., and Rode, L. M. (2001). Effects of grain processing, forage to concentrate ratio, and forage particle size on rumen pH and digestion by dairy cows. J. Dairy Sci. 84, 2203–2216. doi: 10.3168/jds.s0022-0302(01)74667-x
Yusuf, H. A., Piao, M. Y., Ma, T., Huo, R. Y., and Tu, Y. (2021). Enhancing the quality of Total mixed ration containing cottonseed or rapeseed meal by optimization of fermentation conditions. Fermentation 7:234. doi: 10.3390/fermentation7040234
Zeng, Y. (2020). Effects of Fermented Mixed Meals to Replace Soybean Meal and Small Peptide Products Supplementation in Diets on Growth Performance, Nutrient Digestion, Rumen Fermentation and Bacterial Composition of Beef Cattle. Southwest Minzu University, Chengdu, China.
Zhang, Z. (2017). Effects of Active Yeeast and its Fermented Feeds on the Rumen Fermentation and Nutrient Digestibility. Inner Mongolia Agricultural University, Hohhot, China.
Zhang, Y. X., Ke, W. C., Bai, J., Li, F. H., Xu, D. M., and Ding, Z. T. (2020). The effect of Pediococcus acidilactici J17 with high-antioxidant activity on antioxidant, α-tocopherol, β-carotene, fatty acids, and fermentation profiles of alfalfa silage ensiled at two different dry matter contents. Anim. Feed Sci. Technol. 268:114614. doi: 10.1016/j.anifeedsci.2020.114614
Zhang, Y., Li, F., Chen, Y., Wu, H., Meng, Q., and Guan, L. L. (2020). Metatranscriptomic profifiling reveals the effect of breed on active rumen eukaryotic composition in beef cattle with varied feed efficiency. Front. Microbiol. 11:367. doi: 10.3389/fmicb.2020.00367
Zhang, L. Y., Ma, Q. G., Ma, S. S., Zhang, J. Y., Jia, R., and Ji, C. (2017). Ameliorating effects of Bacillus subtilis ANSB060 on growth performance, antioxidant functions, and aflatoxin residues in ducks fed diets contaminate with aflatoxins. Toxins 9:1. doi: 10.3390/toxins9010001
Zhang, Z. G., Xu, D. M., Wang, L., Hao, J. J., Wang, J. F., and Zhou, X. (2016). Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 26, 1873–1879. doi: 10.1016/j.cub.2016.05.012
Zhang, Y., Zhang, Z., Dai, L., Liu, Y., Cheng, M., and Chen, L. (2018). Isolation and characterization of a novel gossypol-degrading bacteria Bacillus subtilis strain rumen Bacillus Subtilis. Asian Australas. J. Anim. Sci. 31, 63–70. doi: 10.5713/ajas.17.0018
Zhou, Z. M., Fang, L., Meng, Q. X., Li, S. L., Chai, S. T., Liu, S. J., et al. (2017). Assessment of ruminal bacterial and archaeal community structure in yak (Bos grunniens). Front. Microbiol. 8:179. doi: 10.3389/fmicb.2017.00179
Zhou, M., Peng, Y. J., Chen, Y. H., Klinger, C. M., Oba, M., and Liu, J. X. (2018). Assessment of microbiome changes after rumen transfaunation: implications on improving feed efficiency in beef cattle. Microbiome 6:62. doi: 10.1186/s40168-018-0447-y
Keywords: fermented total mixed ration, cottonseed meal, rapeseed meal, Hu sheep, rumen microbiota
Citation: Rehemujiang H, Yusuf HA, Ma T, Diao Q, Kong L, Kang L and Tu Y (2023) Fermented cottonseed and rapeseed meals outperform soybean meal in improving performance, rumen fermentation, and bacterial composition in Hu sheep. Front. Microbiol. 14:1119887. doi: 10.3389/fmicb.2023.1119887
Received: 09 January 2023; Accepted: 02 February 2023;
Published: 16 March 2023.
Edited by:
Qingbiao Xu, Huazhong Agricultural University, ChinaReviewed by:
Rongzhen Zhong, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Rehemujiang, Yusuf, Ma, Diao, Kong, Kang and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Tu, dHV5YW5AY2Fzcy5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.