- State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
Vibrio cholerae serogroup O1 (V. cholerae O1) is closely associated with cholera epidemics and has two main immunologically distinguishable serotypes, Ogawa and Inaba. Isolates serotype as Ogawa if the O-antigen polysaccharide (O-PS) is methylated or as Inaba if the O-PS is not methylated. This methylation is mediated by a methyltransferase encoded by the rfbT gene, and the mutation and low expression of rfbT results in serotype switch from Ogawa to Inaba. Previously, we have shown that cAMP receptor protein (CRP) activates rfbT. In this study, we demonstrated that histone-like nucleoid structuring protein (H-NS) is directly involved in the transcriptional repression of rfbT. This finding is supported by the analyses of rfbT mRNA level, rfbT-lux reporter fusions, electrophoretic mobility shift assay (EMSA), and DNase I footprinting assay. The rfbT mRNA abundances were significantly increased by deleting hns rather than fis which also preferentially associates with AT-rich sequences. A single-copy chromosomal complement of hns partly restored the down-regulation of rfbT. Analysis of rfbT-lux reporter fusions validated the transcriptional repression of hns. Subsequent EMSA and DNase I footprinting assay confirmed the direct binding of H-NS to rfbT promoter and mapped the exact binding site which was further verified by site-directed mutagenesis and promoter functional analysis. Furthermore, we found that in hns deletion mutant, CRP is no longer required for transcriptionally activating rfbT, suggesting that CRP functions as a dedicated transcription factor to relieve H-NS repression at rfbT. Together, this study expanded our understanding of the genetic regulatory mechanism of serotype conversion by global regulators in V. cholerae O1.
1. Introduction
Cholera is an acute watery diarrheal disease caused by Vibrio cholerae, which is naturally present in the environment and autochthonous to coastal and estuarine ecosystems. People are usually infected by ingesting V. cholerae-contaminated water or food. Upon colonization of the host, V. cholerae produces cholera toxin (CT), which acts on intestinal epithelial cells, resulting in secretory diarrhea and even death within a few days if without treatment (Chiang and Mekalanos, 2000). Based on the heat-stable somatic O antigen, the species V. cholerae is divided into more than 200 serogroups. Among them, only two serogroups, toxigenic O1 and O139, have been demonstrated to cause epidemic and pandemic cholera. O1 serogroups have caused seven pandemics, and O139 emerged in the seventh pandemic (Longini et al., 2002). Serogroup O1 has two biotypes, El Tor and classical. The first six pandemics are believed to be caused by the classical biotype, whereas the seventh pandemic that started in the early 1960s is caused by the El Tor biotype.
Vibrio cholerae O1 antigen consists of at least three types of antigenic factors: A, B, and C. According to the differences of the antigenic factors, each of the two biotypes can be further classified into two major cross-reacting serotypes, Ogawa and Inaba. Ogawa serotype strain expresses A and B antigens as well as a small amount of C antigens, while Inaba only expresses A and C antigens. A third serotype, Hikojima expressing both the B and C antigens, is also reported but is rare and unstable (Chatterjee and Chaudhuri, 2003). Ogawa and Inaba serotypes differ only by a single 2-O-methyl group that is present in the upstream (nonreducing) terminal perosamine unit of the Ogawa O-antigen polysaccharide (O-PS) but is absent in Inaba (Chatterjee and Chaudhuri, 2003). An isolate is serotyped as Ogawa if its O1 serogroup O-PS is methylated and as Inaba, if its O-PS is not methylated (Ito et al., 1993). This methylation is catalyzed by a methyltransferase encoded by the rfbT gene (also known as webT or tsfB; Rijpkema et al., 2004). Therefore, genetic alterations of rfbT resulted from various mutational events, such as specific point mutation, single nucleotide or short fragment insertion/deletion, or transposase insertion can all lead to the serotype shift from Ogawa to Inaba (Sharifnia et al., 2012; Liang et al., 2013; Karlsson et al., 2016). Once the complete rfbT is replenished, the Inaba type can be reverted to the Ogawa.
The serotype shifts can occur during subculture in vitro, passage in vivo, or even during pandemics (Ito et al., 1993; Liang et al., 2013; Karlsson et al., 2016). Recently, a major serotype switch (ranging from 7 to 100%) from Ogawa to Inaba was discovered after 5 years of the onset of cholera in Haiti in October, 2010 (Alam et al., 2016). Such serotransitions are nonrandom processes and thought to be related to selective pressures of serotype-specific immunity within the host population or environmental stress, the specific intrinsic drivers and regulation mechanism remain to be investigated (Longini et al., 2002; Karlsson et al., 2016). Previously, we demonstrated that a global regulator, cAMP receptor protein (CRP), positively regulates rfbT transcription through directly binding to a non-canonical CRP binding site (CBS) in its promoter region (Li et al., 2019).
The histone-like nucleoid structuring protein (H-NS) is a global regulator of environmentally controlled gene expression. It belongs to a small family of nucleoid-associated proteins (NAPs; Winardhi et al., 2015). This family comprises a group of basic, low molecular weight DNA binding proteins that participate in chromatin organization, restraining of DNA supercoiling, and transcription regulation. The factor for inversion stimulation (Fis), leucine-responsive protein (Lrp), heat-labile protein (HU) and integration host factor (IHF) are all members of this family. H-NS consists of a coiled-coil N-terminal domain that mediates the protein oligomerization and a C-terminal DNA-binding domain, which binds to promoters exhibiting AT-rich and highly curved regions as transcriptional inhibitors, affecting a broad spectrum of physiological processes including virulence-related genes at multiple phases of the V. cholerae life cycle (Winardhi et al., 2015; Ayala et al., 2017). Totally 701 genes have been identified to be regulated by H-NS in V. cholerae (Wang et al., 2015).
In this study, we show that H-NS negatively regulates the transcription of the serotype-switching gene rfbT by directly binding to its promoter region, whereas fis does not affect its expression. CRP likely activates transcription of rfbT through derepression of H-NS.
2. Materials and methods
2.1. Bacterial strains, culture conditions, and plasmids
Bacterial strains and plasmids used in this study are shown in Table 1. The V. cholerae O1 El Tor biotype, Ogawa serotype strain C7258 was used as wild-type (WT) precursor (Peru isolate, 1991). The mutant Δcrp (WL7258) was generated previously (Li et al., 2019). E. coli DH5αλpir and S17-1λpir were, respectively, used for cloning and conjugation purposes, ER2566 is used as host for the expression and purification of hns cloned into the pTXB1 vector. All strains were grown in Luria–Bertani (LB) broth (Oxoid, Basingstoke, United Kingdom) containing 1% NaCl (170 mM) at 37°C. When necessary, culture media were supplemented with ampicillin (Amp, 100 μg/ml), chloramphenicol (Cm, 10 μg/ml for E. coli, 2.5 μg/ml for V. cholerae) or polymyxin B (100 units/ml). Isopropyl-β-D-thiogalactopyranoside (IPTG) was used at a concentration of 0.5 mM for induction purposes.
2.2. Construction of mutants and complementation plasmids
Mutants Δhns, Δfis were constructed by homologous recombination mediated by suicide plasmid using C7258 as a precursor, while ΔcrpΔhns and ΔcrpΔfis used Δcrp (WL7258) as a precursor. Upstream and downstream chromosomal DNA fragments flanking the hns or fis were amplified from C7258 genomic DNA using the corresponding primers listed in Table 2. The amplicons were stitched together by overlapping PCR. Δhns or Δfis fragments were cloned into pWM91 and constructed in DH5αλpir. The resulting pWM91-Δhns or pWM91-Δfis was introduced into C7258 or Δcrp (WL7258) by conjugation from S17-1λpir. Exconjugants and mutants were selected as described previously (Wu et al., 2015; Li et al., 2019).
Chromosomal complementation strain Δhns::hns was constructed using a temperature-sensitive transposable plasmid PGRG25. pGRG25 contains a Tn7 transposon that can carry the target fragment for specific recombination with the chromosome of the host bacterium, directionally inserted into the downstream of the glms in the chromosome (McKenzie and Craig, 2006). For this purpose, hns promotor region and open reading frame (ORF) was amplified and cloned into pGRG25 to generate pGRG25-hns which was mobilized into Δhns by conjugation. The chromosomal insertion of the transposon was induced by 0.1% arabinose in LB broth at 30°C for 16 h, then screened on LB agar by a temperature at 42°C. Grown colonies were tested for Amp sensitivity, and proper insertion of hns downstream of glms was verified by PCR with primers targeting the glms and hns sequences. Primer sequences used here are shown in Table 2.
2.3. RNA extraction and quantitative reverse transcription PCR
Vibrio cholerae strains were cultured to OD600 1.0. Total RNA extraction, removal of chromosomal DNA contamination and cDNA synthesis were performed as previously described (Wu et al., 2015). Equation R = 2− (ΔCq rfbT-ΔCq thyA) was used to calculate the relative expression values (R) of rfbT, where Cq is the threshold cycle fraction and thyA was used as an internal reference. A control reaction with total RNA as a template was performed for each sample to exclude contamination from chromosomal DNA. Primers used were listed in Table 2.
2.4. Transcriptional reporter fusion construction and bioluminescence assay
Two different length fragments of rfbT promoter region were, respectively, amplified and cloned into pBBRlux upstream of the promoterless luxCDABE operon. The resultant fusion plasmids pBBRlux-rfbT1 and pBBRlux-rfbT2 were constructed in DH5αλpir and then mobilized into V. cholerae strains C7258 and ∆hns by conjugation from S17-1λpir. pBBRlux-rfbT1-M1, pBBRlux-rfbT1-M3-1, and pBBRlux-rfbT1-M3-2 reporter fusions containing the specific mutations in the predicted H-NS binding sites were generated by PCR-based site-directed mutagenesis with pBBRlux-rfbT1 as a template. Overnight cultures of V. cholerae strains containing lux reporter fusion plasmid were diluted at 1:100 in fresh LB and incubated at 37°C with shaking to grow to exponential phase. 200 μl of the broth was transferred into 96 well microtiter plates (Costar 3,917) every 1 h, and luminescence and OD600 were measured using a microplate reader (Infinite M200 Pro, Tecan, Austria). Luminescence activity was calculated as light unit/OD600 as previously described (Pan et al., 2018).
2.5. Expression and purification of H-NS protein
E. coli strain ER2566 containing the recombinant expression plasmid pXTB1-HNS (Wang H. et al., 2012) was cultured to OD600 of 0.5 with shaking at 37°C and then protein expression was induced with 0.4 mM IPTG for 4 h at 28°C. The cells were collected by centrifugation, resuspended in ice-cold Column buffer (20 mM Tris–HCl, pH 8.0, 0.5 M NaCl, and 1 mM EDTA), and lysed by sonication. The cell debris was removed by centrifugation, and H-NS-intein fusion protein with chitin binding domain (CBD) was purified using IMPACT™ Kit (New England Biolabs, United Kingdom) according to the manufacturer’s instructions. The clarified lysate was slowly loaded onto the equilibrated chitin column, and then the chitin column was washed with 20 bed volumes of Column Buffer. Subsequently, the column was quickly washed with 5 bed volumes of the Cleavage Buffer (Column Buffer containing 80 mM DTT), and then incubated at 4°C overnight for full cleavage reaction on-column. Finally, the H-NS was eluted with Column Buffer. H-NS-containing fractions were combined and dialyzed against Column Buffer at 4°C to remove DTT. The purity of the recombinant H-NS was analyzed by SDS-PAGE (Figure 1A), and the protein concentration was determined by a Pierce BCA protein assay kit (Thermo Fisher Scientific, United States). The protein was stored in 20% glycerol at - 80°C.
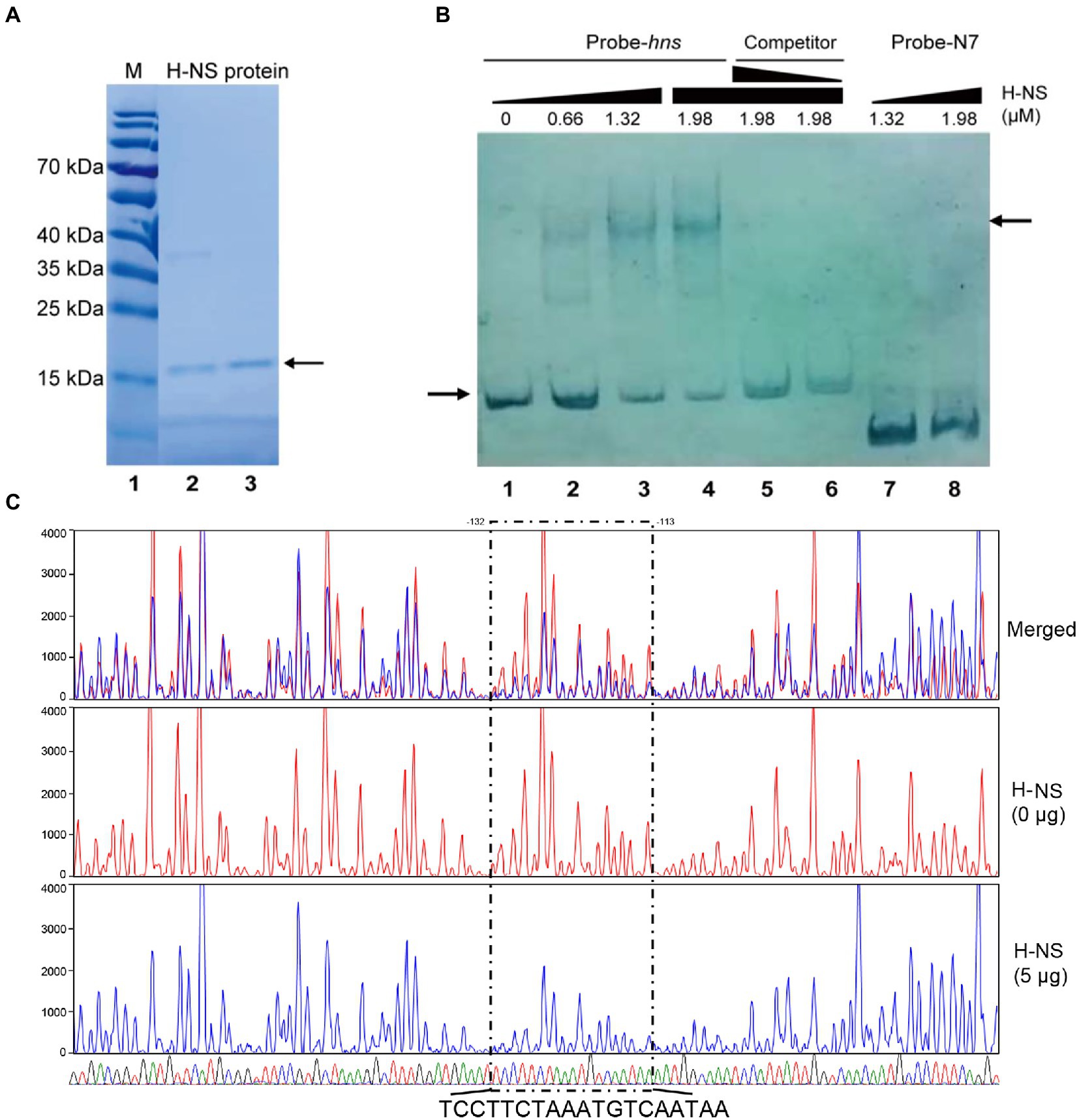
Figure 1. H-NS physically binds to the promoter region of rfbT. (A) H-NS protein purification. Lane 1 is protein marker, and lanes 2–3 are purified H-NS proteins. The right arrow indicates purified bands. (B) EMSA of H-NS bound to rfbT promoter regions. EMSA was described in the “Materials and Methods.” Biotin-labeled 256-bp DNA probe (20 ng) was incubated with increasing amounts of purified H-NS protein. For competitive analysis, the identical but unlabelled probe was added at 10 or 50-fold concentration relative to the labeled one (5–6 channels). Lanes 7 and 8 are Probe-N7 with H-NS. The left arrow indicated the free probe, whereas the right arrow referred to H-NS-rfbT bound one. (C) DNase I footprinting assay of H-NS binding to the promoter region of rfbT. As described in the “Materials and Methods,” purified H-NS protein were incubated with FAM-labelled fragments of the rfbT promoter region, and then the fragments were digested with optimized DNase I. Finally, the digested fragments were analyzed, and the protected regions were boxed and marked. The colored traces representing the different concentrations of H-NS used (red, 0 μg and blue, 5 μg) are indicated separately and then merged, together with the DNA sequencing results (G, T, A, and C) displayed by four different colors. The region where the blue traces drop is the binding region of H-NS to the probe.
2.6. Electrophoretic mobility shift assays
Probe-hns was a 256 bp fragment of rfbT promoter region containing all of the predicted H-NS binding sequences, which was amplified with 5′ biotin-labeled primers. Competing cold probe was amplified using the same primers without a biotin label. Probe-N7 (Li et al., 2019) was a 140 bp fragment of rfbT promoter region containing the previously determined CRP binding site and used as a nonspecific control probe for H-NS binding (Li et al., 2019). The reaction mixture of 15 ng biotin-labeled probe with increasing amounts of purified H-NS protein in reaction buffer (10 mM Hepes, 150 mM KCl, 1 mM EDTA, 1 mM DTT, 10 mM (NH4)2SO4, 0.2% Tween 20) together with 100 ng BSA and 100 ng CF-DNA in each reaction (20 μl) was incubated at 28°C for 30 min and then separated on a 6% native polyacrylamide gel. The free and H-NS-bound probes were visualized with the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, United States) according to the manufacturer’s instruction after transferring them onto nylon membranes.
2.7. DNase I footprinting assay
For preparation of fluorescent FAM-labeled probes, the promoter region was PCR amplified with 2 × HIFI DNA polymerase premix from the plasmid pBBRlux-rfbT1 using primers of rfbT-up (FAM)and rfbT-dn. The FAM-labeled probes were purified by the Wizard® SV Gel and PCR Clean-Up System (Promega, United States) and were quantified with NanoDrop 2000C (Thermo, United States).
DNase I footprinting assay was performed as previously described (Wang Y. et al., 2012). For each assay, 250 ng probes were incubated with different amounts of protein in a total volume of 40 μl. After incubation for 30 min at 25°C, 10 μl solution containing about 0.015 unit DNase I (Promega, United States) and 100 nmol freshly prepared CaCl2 was added and further incubation was performed at 37°C for 1 min. The reaction was stopped by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). Samples were first extracted with phenol/chloroform, and then precipitated with ethanol. Pellets were dissolved in 30 μl MiniQ water. The preparation of the DNA ladder, electrophoresis, and data analysis were the same as described before (Wang Y. et al., 2012), except that the GeneScan-LIZ600 size standard (Applied Biosystems) was used.
2.8. Statistical analysis
GraphPad Prism 9 software was used for statistical analysis and graphical representation of data. Statistical significance was determined by an unpaired two-tailed Student’s t-test.
3. Results
3.1. Characterization of the promoter region of rfbT
Though the gene rfbT has been recognized as the genetic determinant of Ogawa serotype of V. cholerae O1 serogroup for more than 30 years (Stroeher et al., 1992) and various kinds of mutations were revealed in the rfbT coding sequence of isolates from different space–time sources (Sharifnia et al., 2012; Liang et al., 2013; Karlsson et al., 2016), its regulation and the molecular structural features except the transcriptional start site, putative −35 and − 10 elements of the promoter-regulatory region remain unclear. Previously, we identified a cis-regulatory element, i.e., a non-canonical CBS in the promoter region, through which global regulator CRP exerts an activational effect (Li et al., 2019). Further sequence analysis revealed that the G + C content of promoter-intergenic region of rfbT (41.2%) is quite low compared with the V. cholerae genome in general (47.7% for chromosome I and 46.9% for chromosome II). In other words, the promoter-intergenic region of rfbT is AT rich and probably prone to be regulated by small nucleoid associated proteins such as H-NS and Fis which tend to bind AT rich sequences. Indeed, subsequent Virtual Footprint and PRODORIC analysis using the 10-bp H-NS consensus (Bouffartigues et al., 2007; Figure 2A) revealed 3 potential H-NS binding elements, site 1 (5’-CCTATTAAAG-3′), site 2 (5’-TATCAAACGT-3′), and site 3 (5’-TCAATAAAAT-3′) in the rfbT promoter-intergenic region (Figure 2B). The three binding sites are, respectively, located at nucleotides −283 to −273, −250 to −240, and − 120 to −110 relative to the rfbT start codon, and are all downstream of nonclassical CBS (Figure 2B). Of these, 8 of the 10 bp at site 3 is consistent with the consensus, followed by site 1 with 5 bp, and finally site 2 with only 4 bp (Figure 2A). It’s worth noting that the site 3 overlaps the predicted −10 promoter element (Figure 2B). These findings strongly indicated the possibility that H-NS regulates serotype-shifting gene rfbT expression.

Figure 2. Nucleotide sequence analysis of the rfbT promoter region. (A) Conservation analysis of H-NS binding sites. Virtual Footprint was used to predict the binding sites of H-NS at the rfbT promoter (https://www.prodoric.de/vfp/). (B) Characteristics of the rfbT promoter region. Underlined sequences show the H-NS binding site predicted by Virtual Footprint. Dashed underlined sequences show the CRP binding sites characterized previously (Li et al., 2019). The black box marks the Shine-Dalgarno sequence (SD) (Ito et al., 1991). The triangle and the bold T represent the proved TSS of the rfbT, and the gray shaded sequences represent its −10 and −35 regions (Stroeher et al., 1992). The negative numbers at the bottom of sequences indicate the nucleotide positions relative to the start codon of rfbT.
3.2. H-NS negatively regulates rfbT expression
To determine whether H-NS is involved in the regulation of rfbT, we constructed an hns deletion mutant using C7258 as a precursor and detected the rfbT mRNA level in WT C7258, and Δhns mutant. As shown in Figure 3A, compared to the C7258, Δhns mutant statistically produced more rfbT mRNA. To further confirm the result, we constructed an hns complementation strain C7258Δhns::hns where a single copy of hns gene with its native promoter region was integrated downstream of chromosomal glms. As displayed, the rfbT mRNA abundance was reduced in C7258Δhns::hns compared to Δhns mutant, though the expression level was not restored to the WT level (Figure 3A). Together, these results showed that H-NS negatively regulates rfbT expression.
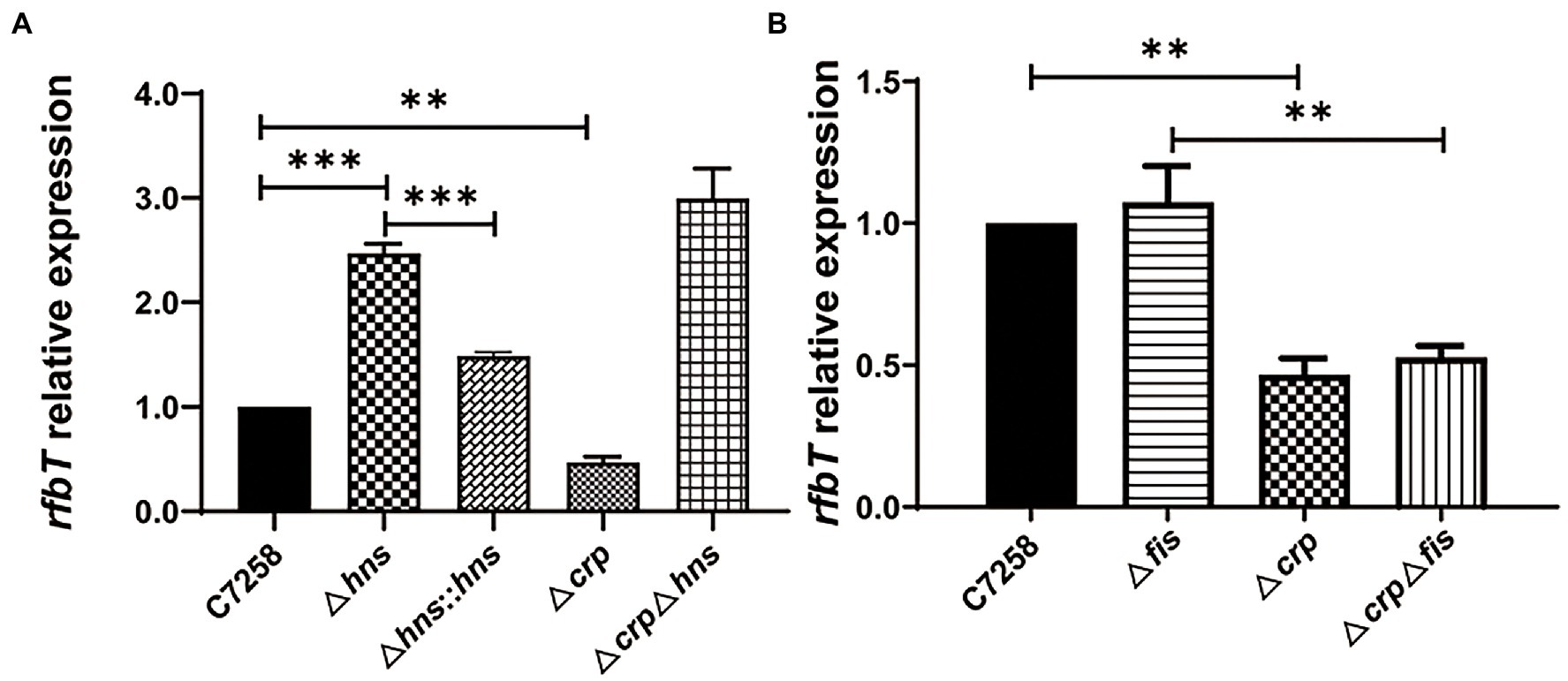
Figure 3. Effects of H-NS, CRP and Fis on rfbT expression. The mRNA abundances around OD600 of 1.0 were determined using qPCR. (A) Comparison of mRNA level of rfbT in V. cholerae strains C7258, Δhns, Δhns::hns, Δcrp, and ΔcrpΔhns. The equation R = 2−(ΔCq rfbT - ΔCq thyA) is used to calculate the mRNA levels. **p < 0.01, ***p < 0.001. (B) The relative mRNA levels of rfbT in C7258, Δfis, Δcrp, and ΔcrpΔfis. The calculation method is the same as (A). **p < 0.01.
In our previous study, we demonstrated that CRP could positively regulate rfbT transcription, and then we wondered whether H-NS regulates rfbT expression in a CRP-dependent manner (Li et al., 2019). For this purpose, we constructed a ΔcrpΔhns double mutant and compared its rfbT mRNA expression with those of Δhns and Δcrp mutants. We found that, contrary to the Δcrp mutant where rfbT expression was obviously lower than its WT and Δhns, the rfbT mRNA level was significantly increased in ΔcrpΔhns (Figure 3A), and additionally, the Δhns and ΔcrpΔhns mutants displayed roughly similar rfbT mRNA level. These results proved that H-NS negatively regulates the expression of rfbT in a CRP-independent manner. From another perspective, these results also indicated that CRP is not required for rfbT activation in H-NS negative background.
3.3. Fis does not affect rfbT expression
Like H-NS, Fis belongs to the small family of nucleoid-associated proteins and is widely implicated in the control of gene expression through binding to the A−/AT-tracts-constituted binding site (Cho et al., 2008). To find out whether Fis takes part in the regulation of rfbT, we constructed Δfis and ΔcrpΔfis deletion mutants and measured the rfbT mRNA levels. As displayed in Figure 3B, the rfbT mRNA level in Δfis is similar to the WT, and deletion of crp significantly reduced rfbT expression regardless of the presence or absence of fis, implying that Fis is not involved in the regulation of rfbT.
3.4. H-NS represses the promoter activities of rfbT
To determine whether the H-NS-mediated repression of rfbT occurs at transcription level, we constructed two transcriptional reporter plasmids by fusing the different length fragments of promoter region of rfbT to the promoterless bioluminescence reporter genes luxCDABE. The 552-bp promoter fragment in pBBRlux-rfbT1 contains both the previously identified CBS and the predicted H-NS binding sites, while the 403-bp promoter region in pBBRlux-rfbT2 lacks the CBS. Consistent with the rfbT mRNA expression, bioluminescence activities of both pBBRlux-rfbT1 and pBBRlux-rfbT2 in Δhns were significantly higher than in its WT (Figure 4). These results demonstrated that H-NS negatively regulates rfbT at the promoter level. H-NS represses the transcription of rfbT probably through binding to the predicted binding sites and therefore the direct interaction still needs to be clarified.
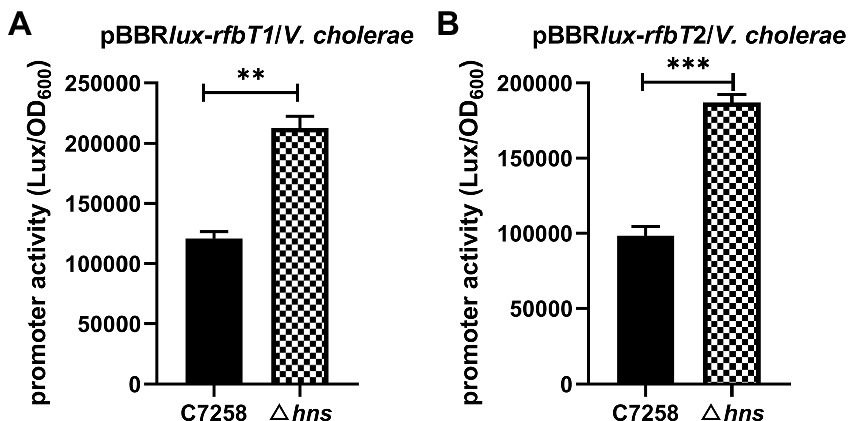
Figure 4. Luminescence activities of lux reporter fusion plasmids in V. cholerae C7258 and Δhns. (A) Luminescence activities of pBBRlux-rfbT1 in C7258 and Δhns. rfbT1 is a 554 bp fragment of rfbT promoter region. (B) Luminescence activity of pBBRlux-rfbT2 in C7258 and Δhns. rfbT2 is a 403 bp fragment of the rfbT promoter region without CRP binding site. The luminescent activities were reported as luminescence/OD600 at the designated time points. ***p < 0.001.
3.5. H-NS directly binds to the promoter of rfbT
To verify the direct binding of H-NS to the rfbT promoter region, we performed EMSA with purified H-NS protein. A 256-bp DNA fragment of rfbT promoter region encompassing the three predicted H-NS binding sites were labeled with biotin at 5′ end and used as specific probe. The same fragment without biotin label was employed as competing cold probe. The 135 amino acids V. cholerae H-NS protein was induced and purified from pTXB1-HNS recombinant plasmid (Wang H. et al., 2012) in E. coli host ER2566 and reached a purity of higher than 90% (Figure 1A). As displayed in Figure 1B, inclusion of H-NS (0.66 μM) generated two shifted bands with slower mobility. With the increment of H-NS (>1.32 μM), the intensity of the higher-shifted band was increased, concomitantly with the decrease of the amount of free probe. Adding the same, but unlabeled 256-bp DNA fragment greatly competed with the labeled probe in a dose-dependent manner. Both of the 10-fold and 50-fold addition of the competing cold probe completely abolished the shifted-band, and concurrently the labeled probe was released as the free one. As expected, the amount of the released free probe is more in 50-fold cold probe reaction mixture than the 10-fold one. To further validate the specificity of H-NS binding, we adopted Probe-N7 which is a 140-bp DNA fragment of rfbT promoter region containing the intact CBS but without the predicted H-NS binding element (Li et al., 2019). No apparent shifted band was observed even when Probe-N7 was incubated with higher amount of H-NS (1.32 μM or 1.98 μM) under the same reaction condition, indicating that H-NS could not bind to the rfbT promoter fragment lack of specific binding site.
To further clarify the real H-NS binding site in the rfbT promoter and dissect the binding sequences, we conducted DNase I footprinting analysis. As shown in Figure 1C, the assay revealed one clearly protected region against DNase I digestion which is composed of TCCTTCTAAATGTCAATAA extending from −132 to −113 relative to the start codon of rfbT, i.e., this region completely overlaps the predicted binding site 3 extending from −120 to −110. Though EMSA displayed two H-NS retarded bands that seems somehow in accordance with the predicted existence of two more conserved binding sites 1 and 3, unexpectedly, the less conserved binding site 1 was not confirmed in the DNase I footprinting assay, which suggested that the more conserved site 3 is intrinsically the real binding site with high-affinity. Of course, we cannot exclude the possibility that the current assay condition did not favor binding to the low-conservation site 1.
To further validate that site 3 is the real functional H-NS binding site and the less conserved site 1 has no function, we set out to introduce mutations at sites 1 and 3 through PCR-based site-directed mutagenesis with pBBRlux-rfbT1 as the template (Figure 5A). We introduced 4-bp changes in the site 1 by replacing the ATT and the antepenultimate A with CAC and C, respectively, to generate a new construct pBBRlux-rfbT1-M1. Considering that site 3 overlaps the predicted −10 promoter element, we introduced two sets of mutations to construct pBBRlux-rfbT1-M3-1 and pBBRlux-rfbT1-M3-2. In pBBRlux-rfbT1-M3-1, the conserved ATA and the antepenultimate A were changed to CAC and C, where TA and A are involved in the −10 motif of rfbT promoter. In pBBRlux-rfbT1-M3-2, the TC and second A were changed to GA and C. These new constructs were mobilized into V. cholerae WT and Δhns to measure the corresponding bioluminescence activities. As depicted in Figure 5B, pBBRlux-rfbT1-M1 and pBBRlux-rfbT1 had similar bioluminescence activities in the WT background which indicated that the mutation of sites 1 has no effect on the rfbT promoter, i.e., site 1 is indeed not a H-NS binding site. However, compared to the pBBRlux-rfbT1, pBBRlux-rfbT1-M3-2 displayed significantly high bioluminescence activity in WT as same as in the Δhns, implying that the introduced mutation affects H-NS binding and thus relieves its repression. Not surprisingly, pBBRlux-rfbT1-M3-1 almost completely lost the bioluminescence signal due to the mutations that destroyed the −10 motif of rfbT promoter by changing TAAAAT to ACACAT. Altogether, these results further experimentally validated that site 3 partially overlaps the −10 motif of rfbT promoter and is the intrinsically functional H-NS binding site.
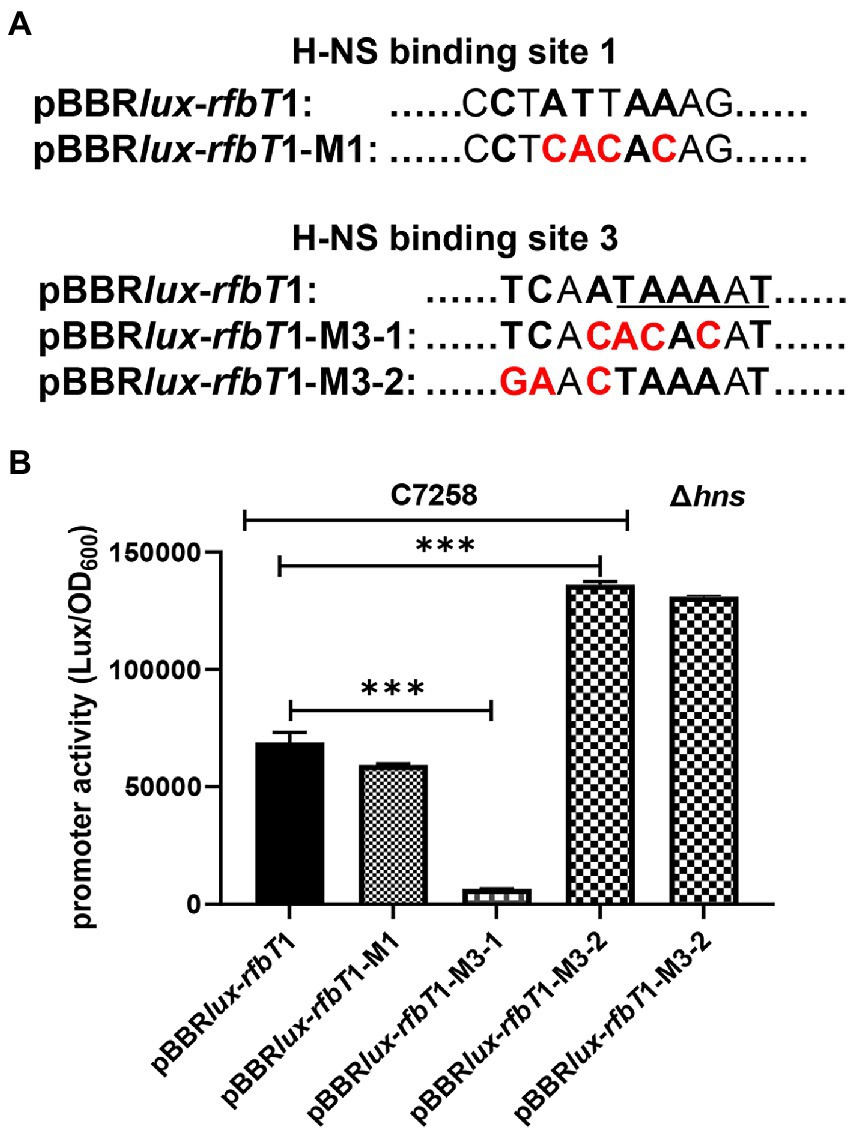
Figure 5. Schematics of the site-directed mutagenesis of the H-NS binding sites at rfbT and luminescence activities of lux reporter fusion plasmids containing mutations in V. cholerae. (A) The pBBRlux-rfbT1-M1, pBBRlux-rfbT1-M3-1, and pBBRlux-rfbT1-M3-2 were constructed by introducing specific mutations in the relevant H-NS sites at the rfbT promoter. The bases identical to the 10-bp H-NS binding consensus were indicated in boldface letters and the changed ones in red color. The underlined sequence is the predicted −10 motif of rfbT promoter. (B) Luminescence activities of pBBRlux-rfbT1, pBBRlux-rfbT1-M1, pBBRlux-rfbT1-M3-1, and pBBRlux-rfbT1-M3-2 in C7258 and pBBRlux-rfbT1-M3-2 in Δhns. The luminescence activities were reported as luminescence/OD600 as described in the “Materials and Methods.” ***p < 0.001.
4. Discussion
rfbT (also named webT or tsfB) is a genetic determinant of Ogawa serotype of V. cholerae O1 by encoding an enzyme that methylates the O-PSs-terminal peraminoglycans of surface LPS. Genetic alteration of rfbT results in impaired function of the enzyme, causing serotype shift from Ogawa to Inaba. Various mutational events of rfbT leading to serotype switching have been reported under different circumstance around the world (Longini et al., 2002; Liang et al., 2013; Karlsson et al., 2016). The epidemiological significance of the two serotype variants shift, the influencing factors and regulation mechanism remain largely uninvestigated. Driven by the serotype-specific immunity acquired within the infected host population (Longini et al., 2002) is a commonly accepted speculation.
In our previous study, we reported the global regulator CRP activates the expression of rfbT and further revealed the underlying genetic mechanism (Li et al., 2019). In this study, we reported another global regulator H-NS which is involved in the transcriptional repression of rfbT by directly binding to its AT-rich promoter region (Figures 1, 3–5). Though more than one potential binding sites were predicted, binding site 3 was finally validated to be the functional H-NS binding site by DNase I footprinting assay and site-directed mutagenesis (Figures 1C, 5). This site overlaps the −10 promoter element (Figures 1C, 2B, 5), thus strongly implying that H-NS functions to block rfbT transcription by interfering with RNA polymerase binding and activity. In addition, we showed that though CRP activates the transcription of rfbT (Li et al., 2019), it loses this function in the H-NS minus background (Figures 3A, 4), indicating that CRP may act as an antisilencer by antagonizing H-NS repression in the rfbT promoter. A number of virulence regulators, such as ToxT, ToxR, and IHF have been reported to act as antirepressors by displacing H-NS at specific promoters (Stonehouse et al., 2008, 2011; Kazi et al., 2016). In these cases, the H-NS binding sites generally overlap or are adjacent to the binding sites of specific activators, such as at V. cholerae tcpA promoter, the binding site of IHF is located between the two sites of H-NS with 6 bp intervals with the up one and overlapping the down one (Stonehouse et al., 2008). At cholera toxin ctx promoter, competitive binding to the overlapping H-NS/ToxT binding sites was proved in EMSA where ToxT could displace H-NS from the ctx promoter (Stonehouse et al., 2008). However, in our case, the detailed molecular mechanism of CRP antagonizing H-NS repression on rfbT remains investigated considering the CRP-specific CBS and H-NS binding site 3 are separated by 220 bp. We reasoned the alleviation of H-NS repression by CRP at rfbT is mechanistically distinct from tcpA and ctx and will be investigated in the future.
Selective silencing of horizontally acquired genes is a common theme in H-NS transcription regulation (Navarre et al., 2006). Horizontally acquired foreign DNA generally has a lower GC-content than its progenitor genome (Kazi et al., 2016). The G + C content of rfbT (31.7%) is quite low compared with the rest of the rfb region (39.1%) and with V. cholerae genome (47% in average). These observations suggest that rfbT is acquired as a foreign DNA and its expression is normally silenced by H-NS. This repression effect could be alleviated by other regulators such as CRP under appropriate environmental conditions. The preference for binding low GC-content DNA is also shared by IHF and Fis. Fis has been proved to be unable to affect the expression of rfbT, while the regulation effect of IHF remains to be determined.
The silencing function of H-NS is dependent on its oligomerization properties. It is believed that an H-NS dimer is the minimal functional binding unit (Badaut et al., 2002). Environmental stimuli such as temperature and osmolality can alter the oligomerization states of H-NS in vivo and hence affect its gene-silencing properties (Amit et al., 2003; Stella et al., 2006; Bouffartigues et al., 2007). At a certain osmolality, the ability of H-NS to bind DNA decreased significantly with increasing temperature (Amit et al., 2003; Bouffartigues et al., 2007). In a word, a variety of factors can affect the function of the H-NS. Whether these environmental factors affect the phenotype of Ogawa serotype strain through H-NS remains to be clarified.
In summary, we demonstrated that V. cholerae Ogawa serotype specific gene rfbT is transcriptionally repressed by the global regulator H-NS through directly binding to a specific cis regulatory element in the promoter region. This work expanded our knowledge of understanding the genetic determinants and complicated regulatory mechanism of V. cholerae O1 serotype shift.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
WL and BK conceived and designed this study. YH, JL, HG, XL, and RD contributed to the experiment. YH and WL contributed to writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the National Key R&D Program of China under grant 2021YFC2300302.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alam, M. T., Ray, S. S., Chun, C. N., Chowdhury, Z. G., Rashid, M. H., Madsen Beau De Rochars, V. E., et al. (2016). Major shift of toxigenic V. cholerae O1 from Ogawa to Inaba serotype isolated from clinical and environmental samples in Haiti. PLoS Negl. Trop. Dis. 10:e0005045. doi: 10.1371/journal.pntd.0005045
Amit, R., Oppenheim, A. B., and Stavans, J. (2003). Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 84, 2467–2473. doi: 10.1016/S0006-3495(03)75051-6
Ayala, J. C., Silva, A. J., and Benitez, J. A. (2017). H-NS: an overarching regulator of the vibrio cholerae life cycle. Res. Microbiol. 168, 16–25. doi: 10.1016/j.resmic.2016.07.007
Badaut, C., Williams, R., Arluison, V., Bouffartigues, E., Robert, B., Buc, H., et al. (2002). The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 277, 41657–41666. doi: 10.1074/jbc.M206037200
Bouffartigues, E., Buckle, M., Badaut, C., Travers, A., and Rimsky, S. (2007). H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14, 441–448. doi: 10.1038/nsmb1233
Chatterjee, S. N., and Chaudhuri, K. (2003). Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639, 65–79. doi: 10.1016/j.bbadis.2003.08.004
Chiang, S. L., and Mekalanos, J. J. (2000). Construction of a Vibrio cholerae vaccine candidate using transposon delivery and FLP recombinase-mediated excision. Infect. Immun. 68, 6391–6397. doi: 10.1128/IAI.68.11.6391-6397.2000
Cho, B. K., Knight, E. M., Barrett, C. L., and Palsson, B. O. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A−/AT-tracts. Genome Res. 18, 900–910. doi: 10.1101/gr.070276.107
Ito, T., Hiramatsu, K., Ohshita, Y., and Yokota, T. (1993). Mutations in the rfbT gene are responsible for the Ogawa to Inaba serotype conversion in Vibrio cholerae O1. Microbiol. Immunol. 37, 281–288. doi: 10.1111/j.1348-0421.1993.tb03211.x
Ito, T., Ohshita, Y., Hiramatsu, K., and Yokota, T. (1991). Identification and nucleotide sequence determination of the gene responsible for Ogawa serotype specificity of V. cholerae O1. FEBS Lett. 286, 159–162. doi: 10.1016/0014-5793(91)80964-5
Karlsson, S. L., Thomson, N., Mutreja, A., Connor, T., Sur, D., Ali, M., et al. (2016). Retrospective analysis of serotype switching of vibrio cholerae O1 in a cholera endemic region shows it is a non-random process. PLoS Negl. Trop. Dis. 10:e0005044. doi: 10.1371/journal.pntd.0005044
Kazi, M. I., Conrado, A. R., Mey, A. R., Payne, S. M., and Davies, B. W. (2016). ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog. 12:e1005570. doi: 10.1371/journal.ppat.1005570
Li, J., Lu, S., Kan, B., and Liang, W. (2019). Serotype-shifting gene rfbT is a direct transcriptional target of cAMP receptor protein (CRP) in V. cholerae O1. Biochem. Biophys. Res. Commun. 519, 874–879. doi: 10.1016/j.bbrc.2019.09.080
Liang, W., Pascual-Montano, A., Silva, A. J., and Benitez, J. A. (2007). The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology (Reading) 153, 2964–2975. doi: 10.1099/mic.0.2007/006668-0
Liang, W., Wang, L., Liang, P., Zheng, X., Zhou, H., Zhang, J., et al. (2013). Sequence polymorphisms of rfbT among the Vibrio cholerae O1 strains in the Ogawa and Inaba serotype shifts. BMC Microbiol. 13:173. doi: 10.1186/1471-2180-13-173
Longini, I. M., Yunus, M., Zaman, K., Siddique, A. K., Sack, R. B., and Nizam, A. (2002). Epidemic and Endemic Cholera Trends over a 33-year period in Bangladesh. J. Infect. Dis. 186, 246–251. doi: 10.1086/341206
McKenzie, G. J., and Craig, N. L. (2006). Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 6:39. doi: 10.1186/1471-2180-6-39
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Pan, J., Zhao, M., Huang, Y., Li, J., Liu, X., Ren, Z., et al. (2018). Integration host factor modulates the expression and function of T6SS2 in Vibrio fluvialis. Front. Microbiol. 9:962. doi: 10.3389/fmicb.2018.00962
Rijpkema, S. G., Durrani, Z., Ramamurthy, T., and Balakrish Nair, G. (2004). Assessing clonality of Vibrio cholerae Inaba isolates by characterization of nonsense mutations in wbeT. J. Med. Microbiol. 53, 1105–1107. doi: 10.1099/jmm.0.45744-0
Sharifnia, A., Bakhshi, B., and Pourshafie, M. R. (2012). wbeT sequence typing and IS1004 profiling of Vibrio cholerae isolates. Lett. Appl. Microbiol. 54, 267–271. doi: 10.1111/j.1472-765X.2012.03204.x
Stella, S., Falconi, M., Lammi, M., Gualerzi, C. O., and Pon, C. L. (2006). Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 355, 169–174. doi: 10.1016/j.jmb.2005.10.034
Stonehouse, E. A., Hulbert, R. R., Nye, M. B., Skorupski, K., and Taylor, R. K. (2011). H-NS binding and repression of the ctx promoter in Vibrio cholerae. J. Bacteriol. 193, 979–988. doi: 10.1128/JB.01343-09
Stonehouse, E., Kovacikova, G., Taylor, R. K., and Skorupski, K. (2008). Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J. Bacteriol. 190, 4736–4748. doi: 10.1128/JB.00089-08
Stroeher, U. H., Karageorgos, L. E., Morona, R., and Manning, P. A. (1992). Serotype conversion in Vibrio cholerae O1. Proc. Natl. Acad. Sci. U. S. A. 89, 2566–2570. doi: 10.1073/pnas.89.7.2566
Wang, H., Ayala, J. C., Benitez, J. A., and Silva, A. J. (2012). Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J. Bacteriol. 194, 1205–1215. doi: 10.1128/JB.05900-11
Wang, H., Ayala, J. C., Benitez, J. A., and Silva, A. J. (2015). RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One 10:e0118295. doi: 10.1371/journal.pone.0118295
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Winardhi, R. S., Yan, J., and Kenney, L. J. (2015). H-NS regulates gene expression and compacts the nucleoid: insights from single-molecule experiments. Biophys. J. 109, 1321–1329. doi: 10.1016/j.bpj.2015.08.016
Keywords: rfbT, H-NS, V. cholerae, transcriptional regulation, serotype shift
Citation: Han Y, Li J, Gao H, Li X, Duan R, Cheng Q, Kan B and Liang W (2023) Serotype conversion gene rfbT is directly regulated by histone-like nucleoid structuring protein (H-NS) in V. cholerae O1. Front. Microbiol. 14:1111895. doi: 10.3389/fmicb.2023.1111895
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Masatoshi Miyakoshi, University of Tsukuba, JapanSaswat S. Mohapatra, Berhampur University, India
Pengfei Ding, University of Maryland, United States
Copyright © 2023 Han, Li, Gao, Li, Duan, Cheng, Kan and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weili Liang,  bGlhbmd3ZWlsaUBpY2RjLmNu; Biao Kan,
bGlhbmd3ZWlsaUBpY2RjLmNu; Biao Kan,  a2FuYmlhb0BpY2RjLmNu
a2FuYmlhb0BpY2RjLmNu
 Yu Han
Yu Han Jing Li
Jing Li He Gao
He Gao Ran Duan
Ran Duan Biao Kan
Biao Kan Weili Liang
Weili Liang