- 1Department of Biotechnology and Life Sciences, Faculty of Biotechnology and Life Sciences, Sojo University, Kumamoto, Japan
- 2Division of Infection and Host Defense, Tohoku Medical and Pharmaceutical University, Sendai, Japan
- 3Department of Biotechnology, The University of Tokyo, Tokyo, Japan
- 4Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Tokyo, Japan
- 5Department of Applied Biochemistry and Food Science, Faculty of Agriculture, Saga University, Saga, Japan
Filamentous fungi belonging to the genus Aspergillus are known to possess galactomannan in their cell walls. Galactomannan is highly antigenic to humans and has been reported to be involved in the pathogenicity of pathogenic filamentous fungi, such as A. fumigatus, and in immune responses. In this study, we aimed to confirm the presence of D-galactofuranose-containing glycans and to clarify the biosynthesis of D-galactofuranose-containing glycans in Aspergillus oryzae, a yellow koji fungus. We found that the galactofuranose antigen is also present in A. oryzae. Deletion of ugmA, which encodes UDP-galactopyranose mutase in A. oryzae, suppressed mycelial elongation, suggesting that D-galactofuranose-containing glycans play an important role in cell wall integrity in A. oryzae. Proton nuclear magnetic resonance spectrometry revealed that the galactofuranose-containing sugar chain was deficient and that core mannan backbone structures were present in ΔugmA A. oryzae, indicating the presence of fungal-type galactomannan in the cell wall fraction of A. oryzae. The findings of this study provide new insights into the cell wall structure of A. oryzae, which is essential for the production of fermented foods in Japan.
1. Introduction
Galactomannan (GM) is a polysaccharide composed of D-mannose (Man) and D-galactofuranose (Galf). GM functions as a component of the cell wall in filamentous fungi (Tefsen et al., 2012; Gow et al., 2017; Oka, 2018). The detailed structure of GM has been elucidated in Aspergillus fumigatus, a major pathogenic fungus that causes invasive pulmonary aspergillosis (Latgé et al., 1994; Kudoh et al., 2015). A. fumigatus possesses two types of GM: O-mannose–type GM (OMGM) and fungal-type GM (FTGM) (Kudoh et al., 2015; Katafuchi et al., 2017; Oka, 2018). OMGM is a galactomannoprotein consisting of an α-(1,2)-mannosyl chain attached to the hydroxyl group of a serine and/or threonine residue in the protein and a galactofuran side chain comprising a β-(1,5)−/β-(1,6)-galactofuranosyl chain attached to a Man residue (Oka et al., 2004; Goto et al., 2009; Kudoh et al., 2015). FTGM has a linear α-mannan backbone consisting of 9–10 α-(1,2)-mannotetraose units linked by α-(1,6) bonds (Latgé et al., 1994; Kudoh et al., 2015; Kadooka et al., 2022a), and its galactofuran side chains are β-(1,2)-, β-(1,3)-, and/or β-(1,6)-linked to this α-core-mannan (Latgé et al., 1994; Kudoh et al., 2015). FTGM is biosynthesized in the Golgi apparatus and is presumed to be transported to the cell surface via the glycosylphosphatidylinositol (GPI) anchor as a carrier molecule (Costachel et al., 2005; Fontaine and Latgé, 2020). By consolidating the findings from recent studies, it can be speculated that FTGMs that are transported to the cell surface can covalently bind to β-glucan, further solidifying the cell wall structure, and some may be released into extracellular compartments, such as the culture supernatant (Muszkieta et al., 2019; Fontaine and Latgé, 2020; Vogt et al., 2020).
Galf is highly antigenic to humans and is commonly detected in the blood of patients with invasive aspergillosis (Stynen et al., 1992). Therefore, Galf is considered a virulence factor for the pathogenicity of A. fumigatus. Single-gene disruption of glfA, which encodes UDP-galactopyranose mutase, the primary enzyme involved in the biosynthesis of Galf-containing polysaccharides, was reported to induce temperature sensitivity and to significantly reduce the pathogenicity of A. fumigatus to mice (Bakker et al., 2005; Schmalhorst et al., 2008). Subsequently, glfA (renamed Afugm1) was analyzed in different strains of A. fumigatus, and the loss of Galf antigens was found to have little effect on pathogenicity (Lamarre et al., 2009). Although the relevance of Galf antigens to pathogenicity is unclear, their antigenic properties suggest that they are involved in certain immune responses in host cells.
The yellow koji fungus, Aspergillus oryzae, has been listed as “generally recognized as safe” by the US Food and Drug Administration (Machida, 2002). A. oryzae is a filamentous fungus used in the production of sake, miso, and soy sauce in Japan and in many fermentation industries owing to its safety (Kitamoto, 2015; Ichishima, 2016; Gomi, 2019; Kitagaki, 2021). Because A. oryzae, like A. fumigatus, is a filamentous fungus belonging to the subphylum Pezizomycotina, it may contain Galf-containing polysaccharides as cell wall components. In a previous study, Galf-containing glycan structures were detected in the cell wall alkali-soluble fraction of A. oryzae from which O-linked glycans were removed and β-(1,2)-Galf was added to the N-glycan outer chain structures (Nakajima and Ichishima, 1994). Furthermore, the genome of A. oryzae has been reported to contain the genes AougmA and AougmB, which are presumed to encode UDP-galactopyranose mutase (Damveld et al., 2008). Because A. oryzae is used in food production, it is important to analyze the Galf-containing glycan structure in detail to ensure its safety.
Thus, this study aimed to clarify the structure of Galf-containing polysaccharides in A. oryzae. Our immunoblot analysis using an anti-Galf antibody revealed the presence of a small amount of galactomannoprotein, a Galf antigen, in A. oryzae. We also found that A. oryzae ugmA encodes UDP-galactopyranose mutase and that Galf-containing polysaccharides are important for normal mycelial elongation in A. oryzae. In addition, proton nuclear magnetic resonance (1H-NMR) spectrometry revealed the presence of an FTGM-like structure in A. oryzae.
2. Materials and methods
2.1. Strains and growth conditions
The Aspergillus strains used in this study are listed in Supplementary Table S1. The strains were grown on minimal medium (MM) containing 1% w/v glucose, 10 mM sodium glutamate, 0.052% w/v KCl, 0.052% w/v MgSO4·7H2O, and 0.152% w/v KH2PO4, plus Hunter’s trace elements (pH 6.5). To cultivate A. oryzae NSPlD1 (Maruyama and Kitamoto, 2008), 1.5 g/l methionine, 1.22 g/l uracil, and 1.21 g/l uridine were added to MM. To cultivate the Aspergillus nidulans strains, 1 mg/l biotin was added to MM.
2.2. Construction of the ugmA-disrupted strain
ugmA was disrupted in A. oryzae NSPlD1 by inserting AnpyrG. A gene replacement cassette encompassing the homology arms at the 5′ and 3′ ends of ugmA was amplified by recombinant polymerase chain reaction (PCR) using A. oryzae RIB40 genomic DNA as the template and the primer pairs ugmA-1/ugmA-2 and ugmA-3/ugmA-4, respectively (Supplementary Table S2). The A. nidulans pyrG (AnpyrG) marker was amplified by recombinant PCR using pHSG396-AnpyrG (Kadooka et al., 2022b) as the template and the primer pair pHSG396-F/pHSG396-R. The resultant DNA fragment, amplified using the primers ugmA-1 and ugmA-4, was used to transform A. oryzae NSPlD1, yielding the ΔugmA strain. MM agar plates without uracil and uridine were used to select the transformants. The introduction of AnpyrG into each gene locus was confirmed by PCR using the primer pair ugmA-F/ugmA-R (Supplementary Figure S1).
2.3. Construction of pPTR-II-ugmA
ugmA that included 1.5 kbp upstream of ugmA was amplified by PCR using A. oryzae RIB40 genomic DNA as the template and the primer pair pPTR-II-ugmA-IF-F/pPTR-II-ugmA-IF-R. The amplified fragment was inserted into the SmaI site of pPTR-II using the In-Fusion HD Cloning Kit (Takara, Kusatsu, Shiga, Japan) to yield pPTR-II-ugmA.
2.4. Preparation of the GM fraction
Total GM (FTGM and galactomannoproteins) from A. oryzae was prepared as previously described (Katafuchi et al., 2017). Briefly, the hot-water-soluble extract from cells was fractionated using cetyl trimethyl ammonium bromide. The resultant fraction was precipitated at pH 9.0 with NaOH in the presence of borate and resolved in distilled water as the total GM fraction. A β-elimination reaction was performed to remove O-glycans from the galactomannoproteins under reducing alkali conditions (500 mM NaBH4 /100 mM NaOH, 10 ml, at 25°C for 24 h). After neutralization with 50% acetic acid, the samples were dialyzed overnight against distilled water. The purified samples were then lyophilized, resuspended in distilled water, and clarified using 0.45-μm-pore filters. The resultant samples were prepared as GM fractions.
2.5. 1H-NMR spectroscopy
Samples for 1H-NMR were exchanged twice in D2O with intervening lyophilization and then dissolved in D2O (99.97% atom 2H). The 1H-NMR spectra were recorded using a JNM-LA600 spectrometer (JEOL, Akishima, Tokyo, Japan) at 45°C. The proton chemical shifts were referenced relative to internal acetone at δ 2.225.
2.6. Immunoblotting
Immunoblotting was performed as previously described (Komachi et al., 2013). The EB-A2 antibody of the Platelia Aspergillus enzyme immunoassay (Bio-Rad Laboratories, Hercules, CA, United States) was used at a dilution of 1:10 to detect β-Galf.
2.7. Analysis of surface adhesion
Hyphal surface adhesion assay was performed as previously described with slight modifications (Lamarre et al., 2009; Alam et al., 2014). Briefly, 0.5-μm-diameter polystyrene beads (Sigma) were diluted to 1:100 in sterile phosphate-buffered saline (PBS). Mycelia were grown for 18 h at 30°C with shaking at 127 rpm in liquid potato dextrose medium, harvested into PBS-containing polystyrene beads for 1 h, and then washed five times with PBS. Mycelium images were acquired using a microscope equipped with a digital camera.
3. Results
3.1. Detection of Galf-containing glycoprotein in koji fungi
To investigate the presence of Galf-containing sugar chains in A. oryzae, Aspergillus luchuensis (Aspergillus awamori var. kawachi) and A. luchuensis mut. Kawachii (A. kawachii), galactomannoproteins were extracted from mycelia and subjected to immunoblotting to detect Galf-containing glycoproteins using the anti-Galf antibody EB-A2 (Oka et al., 2005). Smeared bands indicated the presence of Galf-containing glycoproteins (Figure 1), which were thought to be mainly attributable to the O-glycans among the glycoproteins (Komachi et al., 2013). Densitometric quantification of the immunoblot bands was performed using ImageJ software (Schneider et al., 2012). The signal intensity ratios for EB-A2 were then calculated, and the ratio for A. fumigatus was normalized to 1.0. The ratio of EB-A2 intensity to Galf-containing glycoprotein in A. oryzae was 0.6-fold less than that in A. fumigatus (Figure 1), suggesting the presence of few Galf-containing glycoproteins in A. oryzae (Figure 1). Interestingly, the intensities of A. luchuensis and A. kawachii were more than four times higher than that of A. fumigatus (Figure 1). These data indicate that the quantities of Galf-containing glycoproteins differ among different species of koji fungi.
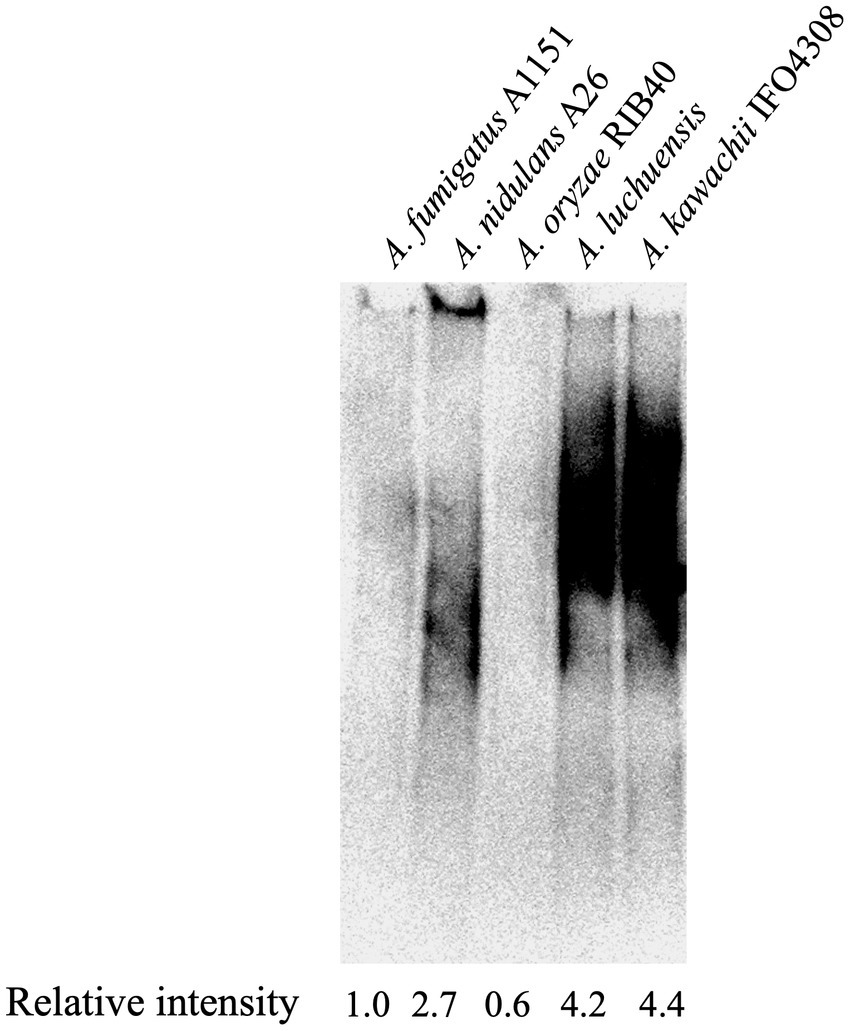
Figure 1. Immunoblot analysis of galactomannoproteins from Aspergillus spp. The presence of Galf-containing glycoproteins was detected using EB-A2. Lanes 1–5: 20 μg of galactomannoproteins from Aspergillus fumigatus A1151 (Lane 1), Aspergillus nidulans A26 (Lane 2), Aspergillus oryzae RIB40 (Lane 3), Aspergillus luchuensis (Lane 4), and Aspergillus kawachii IFO4308 (Lane 5) were loaded. Densitometric quantification of immunoblotting bands was performed using ImageJ software (Schneider et al., 2012). The intensity ratios of EB-A2 for galactomannoproteins were calculated, and the ratio for A. fumigatus A1151 was normalized to 1.0.
3.2. Putative UDP-galactopyranose mutase in koji fungi
To investigate whether the genes encoding UDP-galactopyranose mutase are present in the genome of koji fungi, we performed an NCBI protein BLAST1 search using the amino acid sequence of A. nidulans UgmA (AN3112) as a query sequence. AO090012000855 of A. oryzae RIB40, RIB2604_01707610 of A. luchuensis RIB2604, and AKAW2_10730A of A. kawachii IFO4308 exhibited strong homology to A. nidulans UgmA (91.75, 91.18, and 91.35% identities, respectively).
To clarify the diversity of GlfA/UgmA in detail, we performed a phylogenetic analysis of UDP-galactopyranose mutase conserved in fungi (Figure 2). The dataset for analysis was obtained by an NCBI protein BLAST search using the amino acid sequence of A. nidulans UgmA as a query sequence. The BLAST search identified the UDP-galactopyranose mutase of A. oryzae, which was named AoUgmA by Damveld et al. (2008). In the present study, AoUgmA is referred to as UgmA. Damveld et al. (2008) also revealed the presence of a putative UDP-galactopyranose mutase named AoUgmB in A. oryzae. However, as AoUgmB encodes only 168 amino acids, it probably does not function as a UDP-galactopyranose mutase because its protein size is too small. Therefore, AoUgmB was excluded from the present analysis. Phylogenetic analysis revealed that UgmA is widely distributed in the subphylum Pezizomycotina and the phylum Basidiomycota. In the phylum Ascomycota, Saccharomycotina and Taphrinomycotina do not carry UgmA. These results are consistent with previously reported findings and with the fact that galactofuranosyl chains are not present in Saccharomycotina and Taphrinomycotina (Tefsen et al., 2012). UgmA is present in Agaricomycotina, Ustilaginomycotina, and Pucciniomycotina in the phylum Basidiomycota, whereas it is absent in most Agaricomycotina in the phylum Basidiomycota. Functional analysis of UgmA in basidiomycetes is currently limited, and further functional analysis is warranted in future studies.
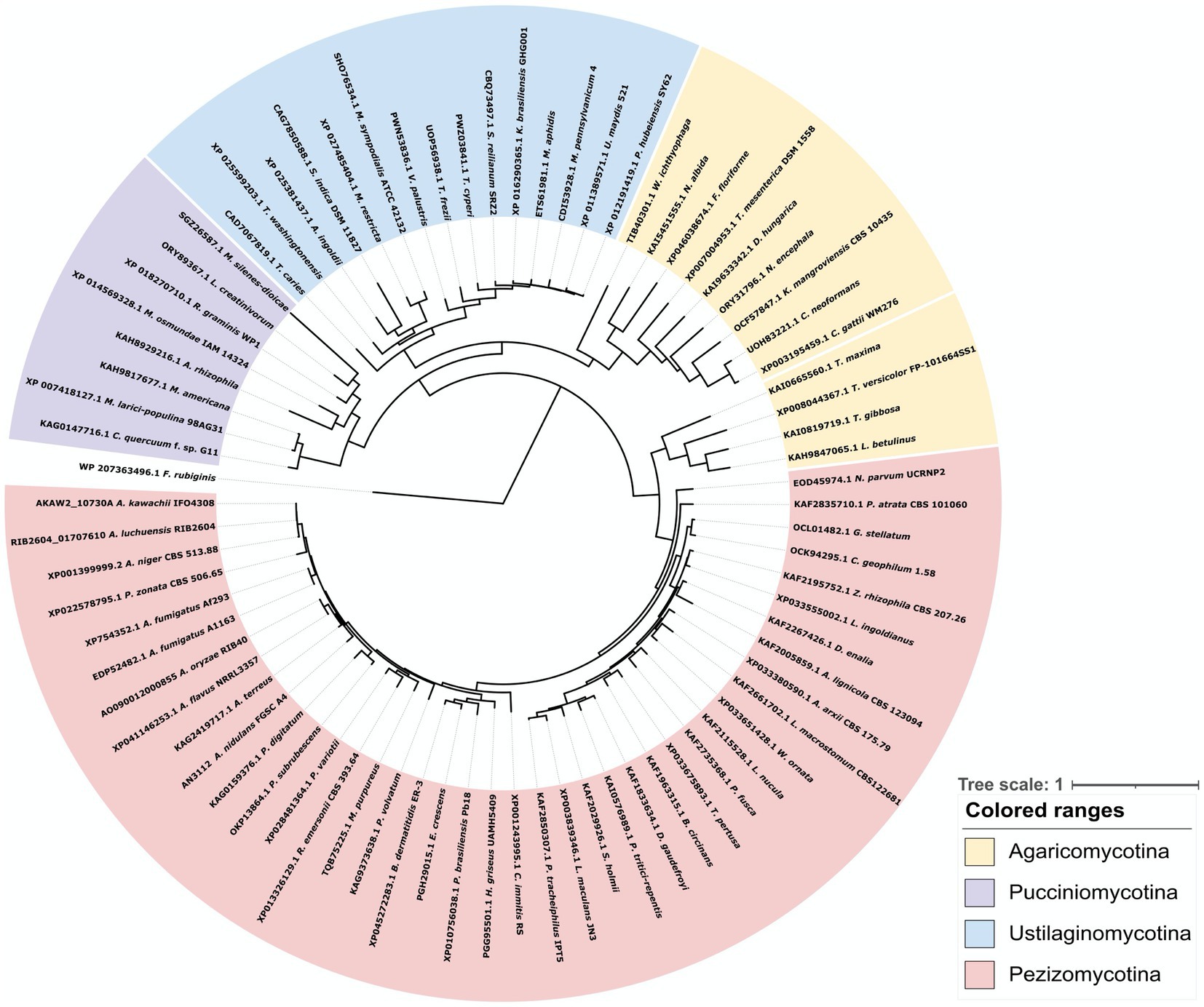
Figure 2. Phylogenetic analysis of UDP-galactopyranose mutase family proteins from bacteria, Basidiomycota, and Pezizomycotina species. Protein sequences were downloaded from NCBI. The phylogenetic tree was drawn using iTOL, and the alignment and phylogenetic tree inference were performed using MAFFT and RAxML, included in ETE v3. The UgmA homologous protein from the bacteria Fibrella rubiginis, presumably with a common ancestor, was used as the outgroup.
3.3. Phenotypic analysis of the Aspergillus oryzae ugmA disruptant
To investigate the physiological roles of ugmA in A. oryzae, we constructed a ugmA gene disruptant and observed the colonial morphology following culture at 30°C, 37°C, and 42°C for 5 days (Figure 3A). ΔugmA had a smaller colony size than the parental strain, indicating the important role of UgmA in normal mycelial formation in A. oryzae. The growth of the parental and ∆ugmA strains was delayed at 42°C compared with that at 30°C on MM. The colony diameters of the parental strain were 34% smaller after culture for 4 days at 42°C compared with that at 30°C, while those of the ∆ugmA strain were 23% smaller after culture for 4 days at 42°C compared with that at 30°C. These results indicate that the ∆ugmA strain exhibits a temperature-sensitive phenotype (Figure 3A; Supplementary Figure S2). This temperature-sensitive phenotype tended to improve under high osmotic support conditions (Figure 3A), suggesting that the ΔugmA strain exhibits temperature sensitivity attributable to the absence of some cell wall components. To confirm that the ΔugmA phenotype was caused by the disruption of ugmA (Figure 3B), we constructed a ugmA complementation strain (ΔugmA + A. oryzae ugmA) and observed the colonies (Figure 3B). The ΔugmA + A. oryzae ugmA strain had a similar colony size and morphology as the parental strain on MM (Figure 3B). This result indicated that the abnormal phenotypes of ΔugmA were truly attributable to ugmA disruption.
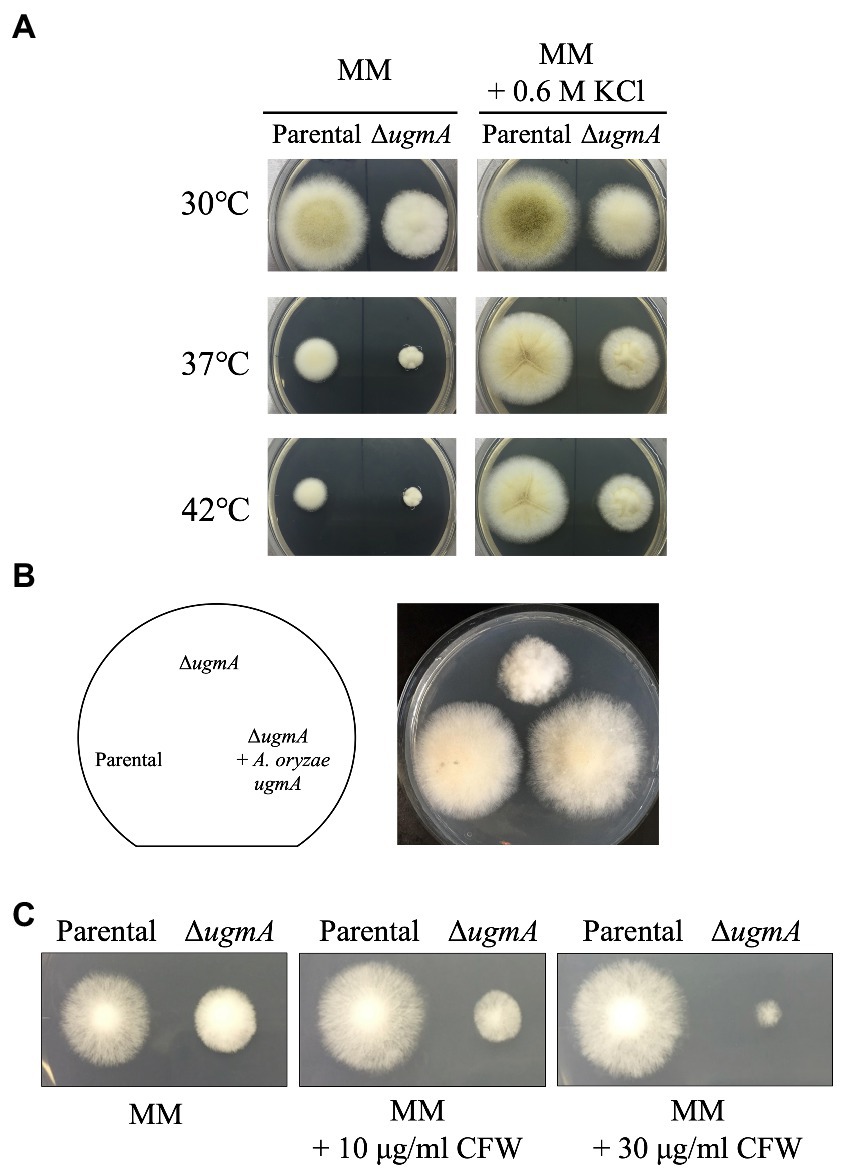
Figure 3. Phenotypic analysis of the ΔugmA strain. (A) Colonial morphology of parental (NSPlD1) and ΔugmA strains on minimal medium (MM) agar and MM agar supplemented with 0.6 M KCl after culture at 30°C, 37°C, or 42°C for 4 days. Agar medium was inoculated with 1.0 × 104 conidiospores. (B) Colonial morphology of the parental, ΔugmA, and ΔugmA + A. oryzae ugmA strains after culture on MM agar for 4 days. Agar medium was inoculated with 1.0 × 104 conidiospores. (C) Sensitivity to the cell wall stress inducer calcofluor white (CFW). Parental and ΔugmA strains were grown on MM agar supplemented with 10 or 30 μg/ml CFW at 30°C for 3 days.
In addition, we investigated the drug sensitivity of ΔugmA. In A. nidulans and A. niger, ugmA-disrupted strains were shown to be sensitive to higher concentrations of calcofluor white (CFW), a chitin-binding reagent (Damveld et al., 2008; El-Ganiny et al., 2008). Therefore, we investigated whether ugmA disruption also affected CFW resistance in A. oryzae. The growth of ΔugmA was delayed on MM supplemented with 30 μg/ml CFW compared with that of the parental strain (Figure 3C), suggesting that ugmA disruption changes the balance of cell wall components, such as chitin and glucan, in A. oryzae.
It was previously reported that ΔglfA and ∆gfsABC strains of A. fumigatus exhibited increased hyphae branching (Lamarre et al., 2009; Chihara et al., 2020). Therefore, we examined whether abnormal mycelial branching also occurred in the ΔugmA strain of A. oryzae. Branching structures at the hyphal tips were observed at a high frequency in the ΔugmA strain (Figure 4A), indicating that the loss of Galf-containing oligosaccharides increases abnormal mycelial branching. It has also been reported that the deletion of Galf-containing oligosaccharides from cells increases cell surface hydrophobicity (Lamarre et al., 2009; Chihara et al., 2020). To confirm the increase in cell surface hydrophobicity of the ΔugmA strain, we examined the level of attachment of latex beads to the mycelium in the ΔugmA strain and found that the level of attachment was clearly increased (Figure 4B), suggesting that Galf-containing oligosaccharides are involved in the cell surface hydrophobicity of A. oryzae.
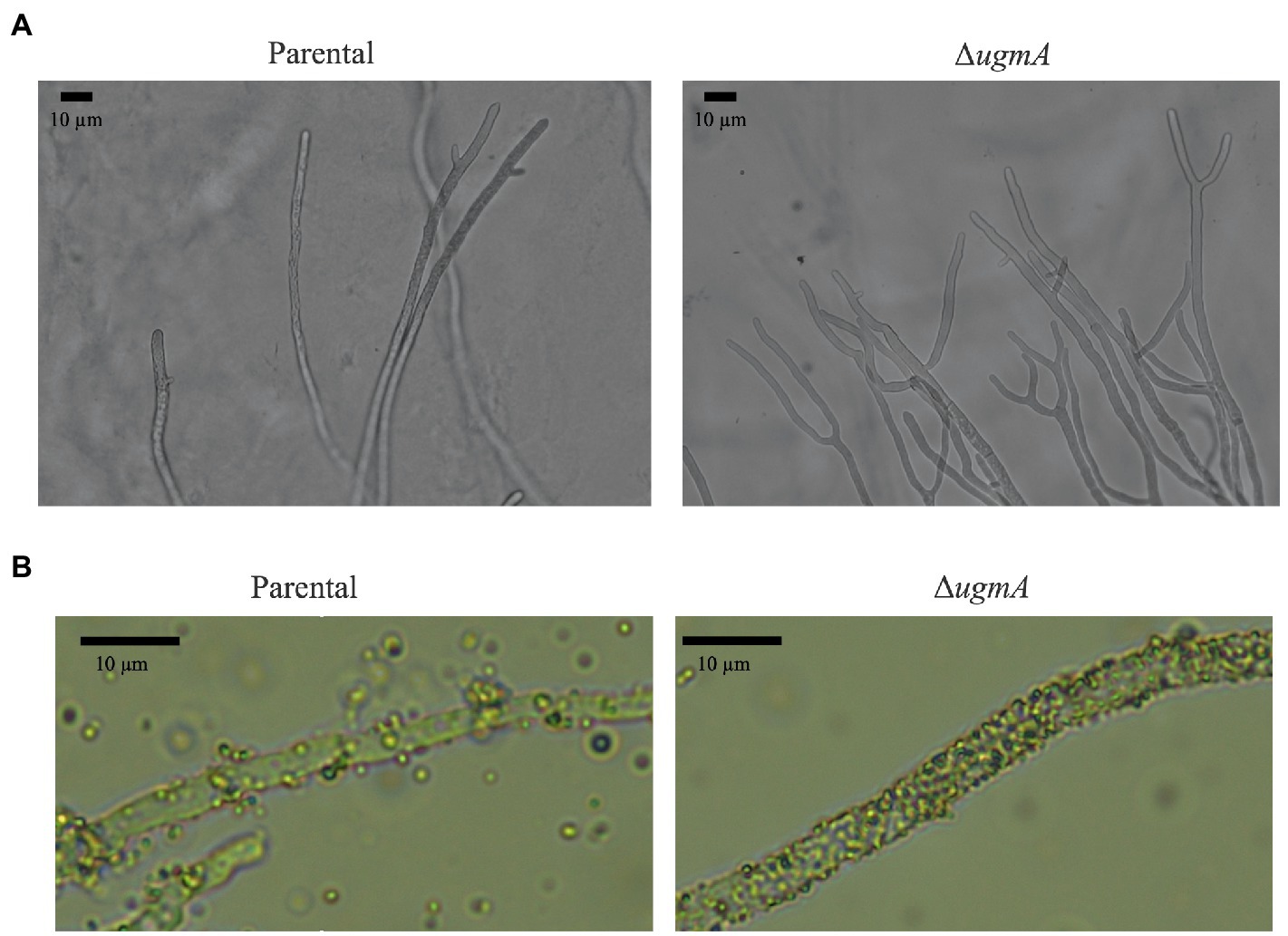
Figure 4. Morphology of the parental and ΔugmA strains. (A) Morphology of the hyphae of the parental and ΔugmA strains. (B) Hydrophobicity of the hyphae of the parental and ΔugmA strains. Hydrophobicity was indicated by the adherence of latex beads to the hyphae.
3.4. Complementation test of ugmA expression in Aspergillus nidulans ΔAnugmA
To determine whether A. oryzae ugmA can complement the growth defect and Galf antigen-lacking phenotype in A. nidulans ΔAnugmA, ugmA was expressed in A. nidulans ΔAnugmA. First, we evaluated whether the expression of A. oryzae ugmA could reverse the growth defects and aberrant conidial formation of A. nidulans ΔugmA. Each conidium was inoculated on MM agar and cultivated at 37°C for 3 days. Expression of A. oryzae ugmA recovered the phenotype of A. nidulans ΔAnugmA, suggesting that A. oryzae ugmA complements the function of A. nidulans ΔAnugmA (Figure 5A). Next, we analyzed the presence of the Galf antigen on the glycoprotein using EB-A2 antibody. Disruption of AnugmA results in the loss of Galf antigens in glycoproteins in A. nidulans (Komachi et al., 2013), but expression of A. oryzae ugmA restored the Galf antigen (Figure 5B). These results indicated that A. oryzae ugmA encodes a UDP-galactopyranose mutase.
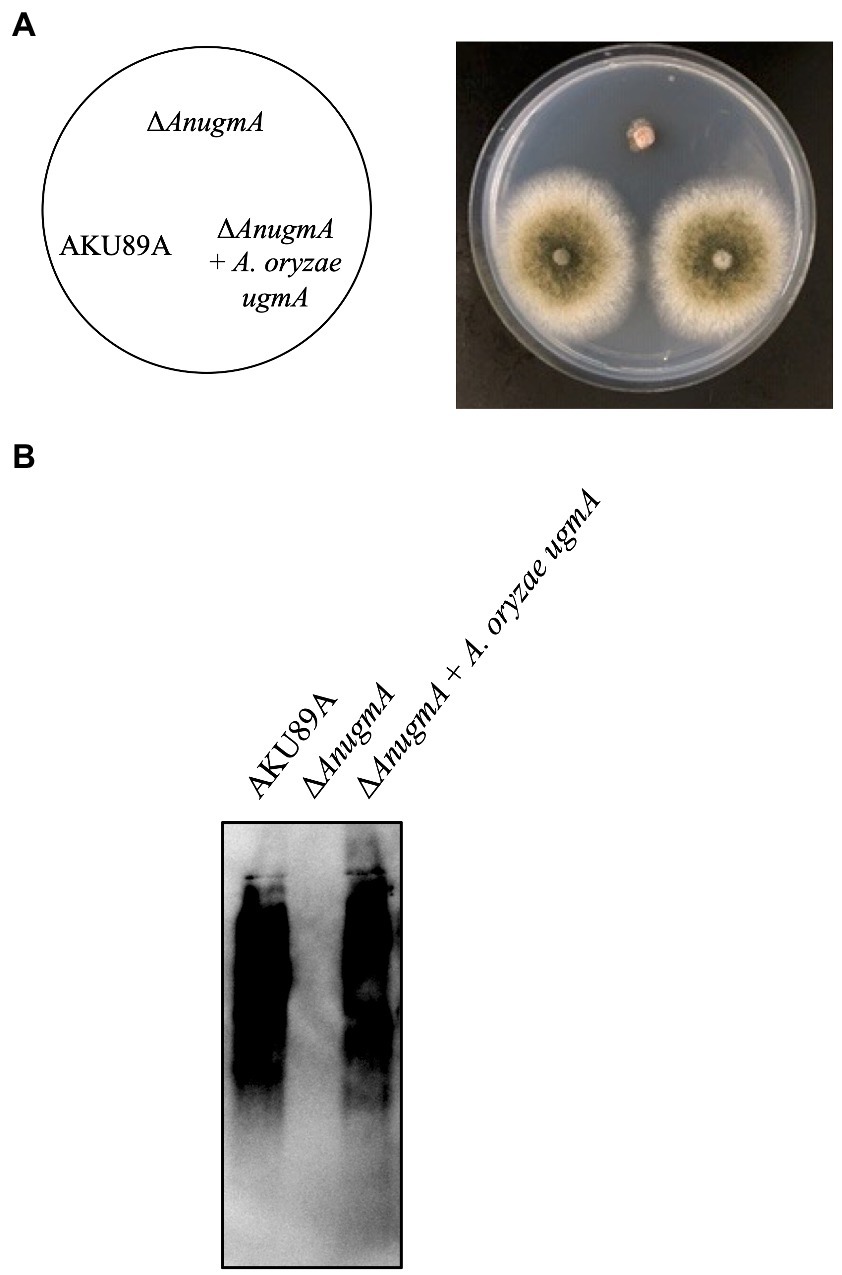
Figure 5. Expression of Aspergillus oryzae ugmA complements the A. nidulans ΔAnugmA phenotype. (A) Colonial morphology of AKU89A (control), ΔAnugmA, and ΔAnugmA + A. oryzae ugmA strains after culture on MM agar at 30°C for 4 days. Agar medium was inoculated with 1.0 × 104 conidiospores. (B) Immunoblot analysis of galactomannoproteins using EB-A2. Lanes 1–3: 20 μg of galactomannoproteins extracted from AKU89A (Lane 1), ΔAnugmA (Lane 2), and ΔAnugmA + A. oryzae ugmA (Lane 3) were loaded.
3.5. Deficiency of galactofuranose residues on FTGM in ΔugmA in the A. oryzae cell wall
To determine whether the galactofuranose-containing sugar chain is deficient in the ∆ugmA strain of A. oryzae, the galactofuranosyl residues were detected by 1H-NMR (Figure 6). The chemical shift signals at 5.195 and 5.05 ppm of the 1H-NMR spectra represented the H-1 signal of the underlined Galf residue in the β-Galf-(1,5)-β-Galf-(1,5)-β-Gal and β-Galf-(1,5)-β-Galf-(1,6)-β-Galf structures, respectively, according to previous studies on A. fumigatus (Shibata et al., 2009; Kudoh et al., 2015). Chemical shift signals at 5.195 and 5.05 ppm were detected in the GM fraction of the parental strain, consistent with the chemical shift signals of the β-(1,5)−/β-(1,6)-galactofuran side chain of FTGM in A. fumigatus (Figure 6). This result indicated the presence of the β-(1,5)−/β-(1,6)-galactofuran side chain structure in A. oryzae. In contrast, these signals were absent in the GM fraction of the ∆ugmA strain, indicating that the galactofuranose-containing sugar chain is deficient in the ∆ugmA strain of A. oryzae (Figure 6). In addition, four unique chemical shifts at 5.0–5.2 ppm (signals A–D), indicating the presence of core mannan backbone structures, emerged in the GM fraction of ΔugmA, indicating the presence of a core mannan chain in FTGM (Kudoh et al., 2015; Onoue et al., 2018; Kadooka et al., 2022b). In the ΔugmA + ugmA strain, the chemical shift indicating the presence of the α-core-mannan backbone was masked, and the chemical shifts indicating the galactofuran chain reappeared (Figure 6). These results clearly indicate that a structure consistent with the FTGM of A. fumigatus is also present in A. oryzae.
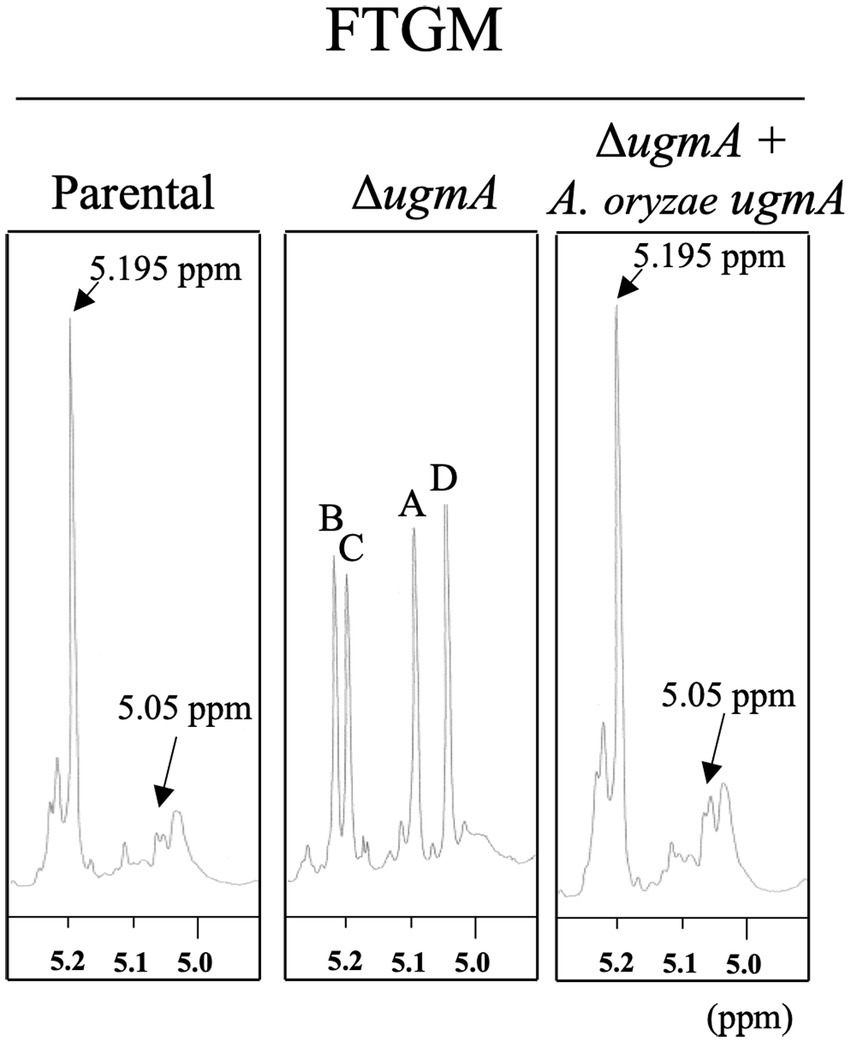
Figure 6. Proton nuclear magnetic resonance (1H-NMR) spectrometry of fungal-type galactomannan (FTGM) fraction from Aspergillus oryzae. The signal at 5.195 and 5.05 ppm of the 1H-NMR spectra is the H-1 signal of the C-1 position of the underlined Galf residue in the β-Galf-(1,5)-β-Galf-(1,5)-β-Galf and β-Galf-(1,5)-β-Galf-(1,6)-β-Galf structures (Kudoh et al., 2015). Signals A (5.104 ppm), B (5.233 ppm), C (5.216 ppm), and D (5.054 ppm) from the 1 H-NMR spectra were derived from H-1 at the C-1 position of the underlined D-Mannose (Man) residues in the structures -(1,6)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,6)- (A), −(1,6)-α-Man-(1,2)- α-Man-(1,2)- α-Man-(1,2)-α-Man-(1,6)- (B), −(1,6)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,6)- (C), and -(1,6)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,2)-α-Man-(1,6)- (D). The proton chemical shifts were referenced relative to internal acetone at δ 2.225 ppm.
4. Discussion
It is important to analyze the substances produced by the microorganisms used in the production of fermented foods to ensure the foods’ safety. In this study, we focused on glycan structures and found that A. oryzae produces Galf-containing glycans, such as galactomannoprotein and FTGM. Galf-containing glycans are found in animal and plant pathogenic fungi such as A. fumigatus and Fusarium spp. (Takegawa et al., 1997; Tefsen et al., 2012; Oka, 2018; de Oliveira et al., 2019; Lee et al., 2019), and their relevance to the demonstration of virulence has attracted attention. Therefore, the Galf-containing glycan of A. oryzae is likely a remnant of its phytopathogenic ancestor before its domestication. A. oryzae is commonly used in the production of Japanese fermented foods, such as miso, soy sauce, and sake, and Japanese people are likely to habitually consume large quantities of Galf antigen. Although the positive and/or negative effects of ingesting Galf antigens on the human body are unknown, these Japanese foods are considered healthy and may have immunostimulatory effects and/or improve intestinal flora (Kitagaki, 2021). Indeed, Nomura et al. (2021) reported that heat-killed A. oryzae spores and cell wall extracts from A. oryzae spores had a soothing effect on dextran sodium sulfate-induced colitis. It is hoped that the health benefits of habitual Galf antigen ingestion will be reported in future studies.
Immunoblot analysis using EB-A2 revealed the presence of Galf-containing glycoproteins in koji fungi. A. oryzae contained lower amounts of Galf-containing glycoproteins than A. nidulans and A. fumigatus, whereas A. luchuensis and A. kawachii produced large amounts of Galf-containing glycoproteins. Because it is known that ugmA deletion results in severe growth defects in A. niger, it is likely that Galf-containing glycans, including glycoproteins, function as important cell wall components in Aspergillus section Nigri (Damveld et al., 2008; Park et al., 2016). Conversely, Galf-containing glycoproteins tend to be less abundant in A. oryzae than in other filamentous fungi. Such differences of phenotype among Aspergillus species could indicate that the contributions of Galf-containing glycoproteins to cell wall composition and normal mycelial formation differ among different Aspergillus species.
This study demonstrated that disruption of ugmA causes growth defects in A. oryzae. This inhibition of growth in ∆ugmA strains has been observed in other Aspergillus species and appears to be a common phenomenon (Damveld et al., 2008; El-Ganiny et al., 2008; Schmalhorst et al., 2008), thus indicating that Galf-containing glycan structures play an important role in cell wall integrity in A. oryzae. In fact, ΔugmA exhibited greater sensitivity to CFW, suggesting that the abundance of cell wall components, such as chitin and glucan, was altered in this strain. In A. niger, it has been reported that ugmA disruption increases the expression of various genes involved in the biosynthesis of α-glucan, β-glucan, and chitin (Park et al., 2016; Arentshorst et al., 2020). Although Galf-containing glycans are not major components of the cell wall, they might be important in maintaining the balance between glucans and chitin in filamentous fungi.
1H-NMR spectrometry suggested that a structure similar to the FTGM of A. fumigatus is present in the cell wall of A. oryzae (Figure 6). The signal indicating the presence of Galf residues was completely lost in the A. oryzae ∆ugmA strain, indicating that UgmA is the only UDP-galactopyranose mutase in A. oryzae and that AoUgmB does not have the same enzymatic function as UDP-galactopyranose mutase (Figure 6). The level of Galf-containing glycoproteins is lower in A. oryzae than in other filamentous fungi, suggesting that the growth defect of ΔugmA is largely attributable to the loss of the galactofuran side chain of FTGM. The predicted biosynthesis map of FTGM in A. oryzae was illustrated based on previous studies (Figure 7). UDP-Galf is synthesized from UDP-glucose via UDP-galactose by UgeA (AO090010000463) and UgmA in the cytosol (Damveld et al., 2008; El-Ganiny et al., 2008; Schmalhorst et al., 2008; El-Ganiny et al., 2010; Lee et al., 2014; Park et al., 2014). The glfB/ugtA homolog gene (AO090012000853), encoding a Golgi-localized UDP-Galf transporter, is conserved adjacent to ugmA in the A. oryzae genome, as reported for A. fumigatus (Engel et al., 2009; Afroz et al., 2011; Park et al., 2015). Unlike other Aspergillus spp., in A. oryzae, only gfsA (AO090120000096) is present in the genome as a gfsA/B/C homolog encoding β-(1,5)-galactofuranosyltransferase (Komachi et al., 2013; Katafuchi et al., 2017; Chihara et al., 2020). Therefore, it is likely that only gfsA is involved in the biosynthesis of the β-(1,5)-galactofuran side chain of FTGM in A. oryzae. AO090009000688 is conserved as a homolog of the Golgi-localized GDP-Man transporter gene, gmtA (Jackson-Hayes et al., 2008; Engel et al., 2012). In addition, cmsA (AO090010000463) and cmsB (AO090120000333), encoding α-(1,2)-mannosyltransferase, and anpA (AO090023000901), encoding α-(1,6)-mannosyltransferase, which are involved in the biosynthesis of the FTGM core mannan backbone, are conserved in the A. oryzae genome (Onoue et al., 2018; Kadooka et al., 2022b). Although the enzymes that add Galf to the core mannan backbone (α-mannoside β-galactofuranosyltransferase: Mgf) have not yet been identified in filamentous fungi, it can be inferred that the aforementioned genes are essential for the biosynthesis of FTGM in A. oryzae. Recent studies have postulated that FTGM is transported to the cell surface via the GPI-anchor and then cross-linked to β-glucan by Dfg proteins, which are mannosidases belonging to the GH76 family (Muszkieta et al., 2019; Fontaine and Latgé, 2020; Vogt et al., 2020). These FTGM-conjugated β-glucan structures are thought to play an important role in normal mycelial elongation in A. oryzae.
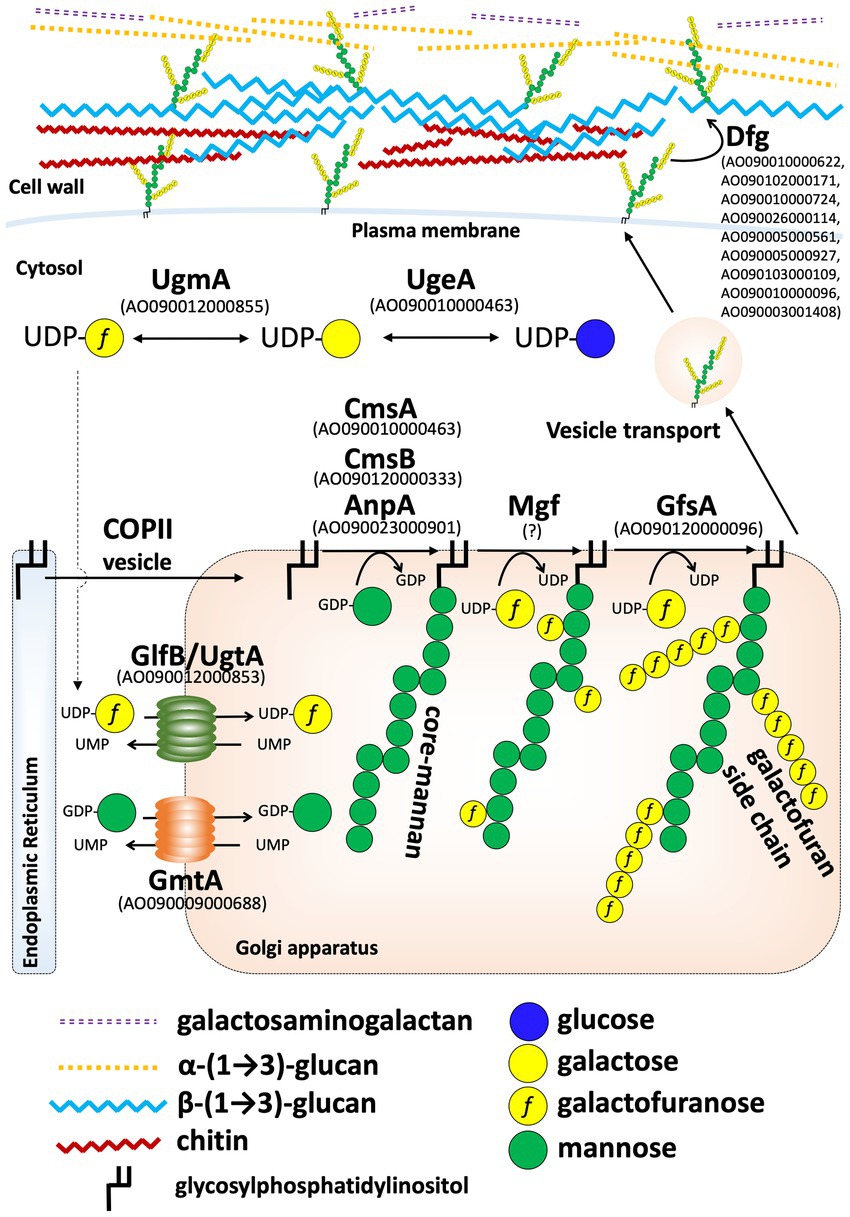
Figure 7. Predicted model of the fungal-type galactomannan (FTGM) biosynthetic pathway in Aspergillus oryzae. Homologs of each FTGM biosynthesis-related protein in A. oryzae were identified by BLASTP at NCBI using Uge5 (Afu5g10780/AFUB_058380), GlfB (Afu3g12700/AFUB_036470), GfsA (Afu6g02120/AFUB_096220), GmtA (Afu5g05740/AFUB_053290), CmsA (Afu5g02740/AFUB_051270), CmsB (Afu5g12160/AFUB_059750), and AnpA (Afu4g06870/AFUB_063940) of A. fumigatus as queries.
Previous studies have shown that the polysaccharides that compose the A. oryzae cell wall include β-(1,3)-glucan, α-(1,3)-glucan, chitin, and galactosaminogalactan (Müller et al., 2003; Zhang et al., 2017; Miyazawa et al., 2019, Figure 7). Through an analysis of ugmA, we revealed that A. oryzae cell walls also contain FTGM structures and galactomannoproteins. Our findings provide novel insights into the structure of the A. oryzae cell wall and the health-promoting effects of fermented foods made from the yellow koji fungus in Japan.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
CK and TO designed and performed the experiments. YT performed the nuclear magnetic response analysis. DH performed the evolutionary phylogenetic analysis. CK, YT, and TO analyzed and interpreted the data. JM contributed materials. MG and JM participated in discussion of the study. TO planned and designed the project. CK and TO wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS KAKENHI; 18K05418 to TO, 21K05373 to TO, 22K06600 to YT, and 19K05835 to DH) and by Grant-in-Aid for Early-Career Scientists from Japan Society for the Promotion of Science (JSPS KAKENHI; 22K14817 to CK and 20K15997 to YT).
Acknowledgments
We thank Ms. Maria Igawa and Ms. Chihiro Kuroda (Sojo University) for assisting with some experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1110996/full#supplementary-material
Footnotes
References
Afroz, S., El-Ganiny, A. M., Sanders, D. A., and Kaminskyj, S. G. (2011). Roles of the Aspergillus nidulans UDP-galactofuranose transporter, UgtA in hyphal morphogenesis, cell wall architecture, conidiation, and drug sensitivity. Fungal Genet. Biol. 48, 896–903. doi: 10.1016/j.fgb.2011.06.001
Alam, M. K., van Straaten, K. E., Sanders, D. A., and Kaminskyj, S. G. (2014). Aspergillus nidulans cell wall composition and function change in response to hosting several Aspergillus fumigatus UDP-galactopyranose mutase activity mutants. PLoS One 9:e85735. doi: 10.1371/journal.pone.0085735
Arentshorst, M., de Lange, D., Park, J., Lagendijk, E. L., Alazi, E., van den Hondel, C. A. M. J. J., et al. (2020). Functional analysis of three putative galactofuranosyltransferases with redundant functions in galactofuranosylation in Aspergillus Niger. Arch. Microbiol. 202, 197–203. doi: 10.1007/s00203-019-01709-w
Bakker, H., Kleczka, B., Gerardy-Schahn, R., and Routier, F. H. (2005). Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol. Chem. 386, 657–661. doi: 10.1515/BC.2005.076
Chihara, Y., Tanaka, Y., Izumi, M., Hagiwara, D., Watanabe, A., Takegawa, K., et al. (2020). Biosynthesis of β-(1→5)-galactofuranosyl chains of fungal-type and O-mannose-type galactomannans within the invasive pathogen Aspergillus fumigatus. mSphere 5, e00770–e00719. doi: 10.1128/mSphere.00770-19
Costachel, C., Coddeville, B., Latgé, J. P., and Fontaine, T. (2005). Glycosylphosphatidylinositol-anchored fungal polysaccharide in Aspergillus fumigatus. J. Biol. Chem. 280, 39835–39842. doi: 10.1074/jbc.M510163200
Damveld, R. A., Franken, A., Arentshorst, M., Punt, P. J., Klis, F. M., van den Hondel, C. A., et al. (2008). A novel screening method for cell wall mutants in Aspergillus Niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178, 873–881. doi: 10.1534/genetics.107.073148
de Oliveira, N. F., Santos, G. R. C., Xisto, M. I. D. S., Pires Dos Santos, G. M., Nucci, M., Haido, R. M. T., et al. (2019). β-1,6-linked galactofuranose- rich peptidogalactomannan of Fusarium oxysporum is important in the activation of macrophage mechanisms and as a potential diagnostic antigen. Med. Mycol. 57, 234–245. doi: 10.1093/mmy/myx167
El-Ganiny, A. M., Sanders, D. A., and Kaminskyj, S. G. (2008). Aspergillus nidulans UDP-galactopyranose mutase, encoded by ugmA plays key roles in colony growth, hyphal morphogenesis, and conidiation. Fungal Genet. Biol. 45, 1533–1542. doi: 10.1016/j.fgb.2008.09.008
El-Ganiny, A. M., Sheoran, I., Sanders, D. A., and Kaminskyj, S. G. (2010). Aspergillus nidulans UDP-glucose-4-epimerase UgeA has multiple roles in wall architecture, hyphal morphogenesis, and asexual development. Fungal Genet. Biol. 47, 629–635. doi: 10.1016/j.fgb.2010.03.002
Engel, J., Schmalhorst, P. S., Dörk-Bousset, T., Ferrières, V., and Routier, F. H. (2009). A single UDP-galactofuranose transporter is required for galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 284, 33859–33868. doi: 10.1074/jbc.M109.070219
Engel, J., Schmalhorst, P. S., and Routier, F. H. (2012). Biosynthesis of the fungal cell wall polysaccharide galactomannan requires intraluminal GDP-mannose. J. Biol. Chem. 287, 44418–44424. doi: 10.1074/jbc.M112.398321
Fontaine, T., and Latgé, J. P. (2020). Galactomannan produced by Aspergillus fumigatus: an update on the structure, biosynthesis and biological functions of an emblematic fungal biomarker. J. Fungi (Basel) 6:283. doi: 10.3390/jof6040283
Gomi, K. (2019). Regulatory mechanisms for amylolytic gene expression in the Koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 83, 1385–1401. doi: 10.1080/09168451.2019.1625265
Goto, M., Harada, Y., Oka, T., Matsumoto, S., Takegawa, K., and Furukawa, K. (2009). Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot. Cell 8, 1465–1474. doi: 10.1128/EC.00371-08
Gow, N. A. R., Latge, J. P., and Munro, C. A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.FUNK-0035-2016
Ichishima, E. (2016). Development of enzyme technology for Aspergillus oryzae, A. sojae, and A. luchuensis, the national microorganisms of Japan. Biosci. Biotechnol. Biochem. 80, 1681–1692. doi: 10.1080/09168451.2016.1177445
Jackson-Hayes, L., Hill, T. W., Loprete, D. M., Fay, L. M., Gordon, B. S., Nkashama, S. A., et al. (2008). Two GDP-mannose transporters contribute to hyphal form and cell wall integrity in Aspergillus nidulans. Microbiology 154, 2037–2047. doi: 10.1099/mic.0.2008/017483-0
Kadooka, C., Hira, D., Tanaka, Y., Chihara, Y., Goto, M., and Oka, T. (2022a). Mnt1, an α-(1 → 2)-mannosyltransferase responsible for the elongation of N-glycans and O-glycans in Aspergillus fumigatus. Glycobiology 32, 1137–1152. doi: 10.1093/glycob/cwac049
Kadooka, C., Hira, D., Tanaka, Y., Miyazawa, K., Bise, M., Takatsuka, S., et al. (2022b). Identification of the α-(1→6)-mannosyltransferase contributing to biosynthesis of the fungal-type galactomannan α-core-mannan structure in Aspergillus fumigatus. mSphere 7:e0048422. doi: 10.1128/msphere.00484-22
Katafuchi, Y., Li, Q., Tanaka, Y., Shinozuka, S., Kawamitsu, Y., Izumi, M., et al. (2017). GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 27, 568–581. doi: 10.1093/glycob/cwx028
Kitagaki, H. (2021). Medical application of substances derived from non-pathogenic fungi Aspergillus oryzae and A. luchuensis-containing Koji. J. Fungi (Basel) 7:243. doi: 10.3390/jof7040243
Kitamoto, K. (2015). Cell biology of the Koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 79, 863–869. doi: 10.1080/09168451.2015.1023249
Komachi, Y., Hatakeyama, S., Motomatsu, H., Futagami, T., Kizjakina, K., Sobrado, P., et al. (2013). GfsA encodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen of O-glycan in Aspergillus nidulans and Aspergillus fumigatus. Mol. Microbiol. 90, 1054–1073. doi: 10.1111/mmi.12416
Kudoh, A., Okawa, Y., and Shibata, N. (2015). Significant structural change in both O- and N-linked carbohydrate moieties of the antigenic galactomannan from Aspergillus fumigatus grown under different culture conditions. Glycobiology 25, 74–87. doi: 10.1093/glycob/cwu091
Lamarre, C., Beau, R., Balloy, V., Fontaine, T., Hoi, W. S., Guadagnini, S., et al. (2009). Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 11, 1612–1623. doi: 10.1111/j.1462-5822.2009.01352.x
Latgé, J. P., Kobayashi, H., Debeaupuis, J. P., Diaquin, M., Sarfati, J., Wieruszeski, J. M., et al. (1994). Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62, 5424–5433. doi: 10.1128/iai.62.12.5424-5433.1994
Lee, M. J., Gravelat, F. N., Cerone, R. P., Baptista, S. D., Campoli, P. V., Choe, S. I., et al. (2014). Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides. J. Biol. Chem. 289, 1243–1256. doi: 10.1074/jbc.M113.522516
Lee, D. J., O’Donnell, H., Routier, F. H., Tiralongo, J., and Haselhorst, T. (2019). Glycobiology of human fungal pathogens: new avenues for drug development. Cells 8:1348. doi: 10.3390/cells8111348
Machida, M. (2002). Progress of Aspergillus oryzae genomics. Adv. Appl. Microbiol. 51, 81–106. doi: 10.1016/S0065-2164(02)51002-9
Maruyama, J., and Kitamoto, K. (2008). Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol. Lett. 30, 1811–1817. doi: 10.1007/s10529-008-9763-9
Miyazawa, K., Yoshimi, A., Sano, M., Tabata, F., Sugahara, A., Kasahara, S., et al. (2019). Both galactosaminogalactan and α-1,3-glucan contribute to aggregation of Aspergillus oryzae hyphae in liquid culture. Front. Microbiol. 10:2090. doi: 10.3389/fmicb.2019.02090
Müller, C., Hansen, K., Szabo, P., and Nielsen, J. (2003). Effect of deletion of chitin synthase genes on mycelial morphology and culture viscosity in Aspergillus oryzae. Biotechnol. Bioeng. 81, 525–534. doi: 10.1002/bit.10491
Muszkieta, L., Fontaine, T., Beau, R., Mouyna, I., Vogt, M. S., Trow, J., et al. (2019). The glycosylphosphatidylinositol-anchored DFG family is essential for the insertion of galactomannan into the β-(1,3)-glucan-chitin core of the cell wall of Aspergillus fumigatus. mSphere 4:e00397–19. doi: 10.1128/mSphere.00397-19
Nakajima, T., and Ichishima, E. (1994). Chemical structure of the galactomannan moiety in the cell wall glycoproteins of Aspergillus oryzae. J. Ferment. Bioeng. 78, 472–475. doi: 10.1016/0922-338X(94)90050-7
Nomura, R., Tsuzuki, S., Kojima, T., Nagasawa, M., Sato, Y., Uefune, M., et al. (2021). Administration of Aspergillus oryzae suppresses DSS-induced colitis. Food Chem 4:100063. doi: 10.1016/j.fochms.2021.100063
Oka, T. (2018). Biosynthesis of galactomannans found in filamentous fungi belonging to Pezizomycotina. Biosci. Biotechnol. Biochem. 82, 183–191. doi: 10.1080/09168451.2017.1422383
Oka, T., Hamaguchi, T., Sameshima, Y., Goto, M., and Furukawa, K. (2004). Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 150, 1973–1982. doi: 10.1099/mic.0.27005-0
Oka, T., Sameshima, Y., Koga, T., Kim, H., Goto, M., and Furukawa, K. (2005). Protein O-mannosyltransferase a of Aspergillus awamori is involved in O-mannosylation of glucoamylase I. Microbiology 151, 3657–3667. doi: 10.1099/mic.0.28088-0
Onoue, T., Tanaka, Y., Hagiwara, D., Ekino, K., Watanabe, A., Ohta, K., et al. (2018). Identification of two mannosyltransferases contributing to biosynthesis of the fungal-type galactomannan α-core-mannan structure in Aspergillus fumigatus. Sci. Rep. 8:16918. doi: 10.1038/s41598-018-35059-2
Park, J., Hulsman, M., Arentshorst, M., Breeman, M., Alazi, E., Lagendijk, E. L., et al. (2016). Transcriptomic and molecular genetic analysis of the cell wall salvage response of Aspergillus Niger to the absence of galactofuranose synthesis. Cell. Microbiol. 18, 1268–1284. doi: 10.1111/cmi.12624
Park, J., Tefsen, B., Arentshorst, M., Lagendijk, E., van den Hondel, C. A., van Die, I., et al. (2014). Identification of the UDP-glucose-4-epimerase required for galactofuranose biosynthesis and galactose metabolism in A. niger. Fungal biol. Biotechnology 1:6. doi: 10.1186/s40694-014-0006-7
Park, J., Tefsen, B., Heemskerk, M. J., Lagendijk, E. L., van den Hondel, C. A., van Die, I., et al. (2015). Identification and functional analysis of two Golgi-localized UDP-galactofuranose transporters with overlapping functions in Aspergillus Niger. BMC Microbiol. 15:253. doi: 10.1186/s12866-015-0541-2
Schmalhorst, P. S., Krappmann, S., Vervecken, W., Rohde, M., Müller, M., Braus, G. H., et al. (2008). Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 7, 1268–1277. doi: 10.1128/EC.00109-08
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Shibata, N., Saitoh, T., Tadokoro, Y., and Okawa, Y. (2009). The cell wall galactomannan antigen from Malassezia furfur and Malassezia pachydermatis contains beta-1,6-linked linear galactofuranosyl residues and its detection has diagnostic potential. Microbiology 155, 3420–3429. doi: 10.1099/mic.0.029967-0
Stynen, D., Sarfati, J., Goris, A., Prévost, M. C., Lesourd, M., Kamphuis, H., et al. (1992). Rat monoclonal antibodies against Aspergillus galactomannan. Infect. Immun. 60, 2237–2245. doi: 10.1128/iai.60.6.2237-2245.1992
Takegawa, K., Satoh, K., Ramli, N., Jikibara, T., and Iwahara, S. (1997). Production and characterization of extracellular uronic acid-containing glycoproteins from Fusarium oxysporum. J. Ferment. Bioeng. 83, 197–200. doi: 10.1016/S0922-338X(97)83583-0
Tefsen, B., Ram, A. F., van Die, I., and Routier, F. H. (2012). Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact. Glycobiology 22, 456–469. doi: 10.1093/glycob/cwr144
Vogt, M. S., Schmitz, G. F., Varón Silva, D., Mösch, H. U., and Essen, L. O. (2020). Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls. Proc. Natl. Acad. Sci. U. S. A. 117, 22061–22067. doi: 10.1073/pnas.2010661117
Keywords: UDP-galactopyranose mutase (UGM), fungal-type galactomannan, galactofuranose, cell wall, Aspergillus oryzae
Citation: Kadooka C, Tanaka Y, Hira D, Maruyama J, Goto M and Oka T (2023) Identification of galactofuranose antigens such as galactomannoproteins and fungal-type galactomannan from the yellow koji fungus (Aspergillus oryzae). Front. Microbiol. 14:1110996. doi: 10.3389/fmicb.2023.1110996
Edited by:
Giancarlo Perrone, Institute of Sciences of Food Production (CNR), ItalyReviewed by:
Shuji Tani, Osaka Metropolitan University, JapanMotoyuki Shimizu, Meijo University, Japan
Copyright © 2023 Kadooka, Tanaka, Hira, Maruyama, Goto and Oka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takuji Oka, b2thQGJpby5zb2pvLXUuYWMuanA=
 Chihiro Kadooka
Chihiro Kadooka Yutaka Tanaka
Yutaka Tanaka Daisuke Hira1
Daisuke Hira1 Jun-ichi Maruyama
Jun-ichi Maruyama Masatoshi Goto
Masatoshi Goto Takuji Oka
Takuji Oka