
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 March 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1106739
This article is part of the Research Topic Biological Soil Crusts: Spatio-temporal Development and Ecological Functions of Soil Surface Microbial Communities across Different Scales View all 25 articles
Introduction: Biological soil crusts (BSCs) constitute a substantial portion of primary production in dryland ecosystems. They successionally mature to deliver a series of ecosystem services. Bacteria, as an important community in BSCs, play critical roles in maintaining the structure and functions of BSCs. However, the process by which bacterial diversity and community are altered with BSC development is not fully understood.
Methods: In this study, amplicons sequencing was used to investigate bacterial diversity and community compositions across five developmental stages of BSCs (bare sand, microbial crusts, algae crusts, lichen crusts, and moss crusts) and their relationship with environmental variables in the Gonghe basin sandy land in Qinghai-Tibet Plateau, northwestern China.
Results: The results showed that Proteobacteria, Actinobacteria, Cyanobacteria, Acidobacteria, Bacteroidetes, and Firmicutes were predominant in different developmental stages of BSCs, accounting for more than 77% of the total relative abundance. The phyla of Acidobacteria and Bacteroidetes were abundant in this region. With BSC development, bacterial diversity significantly increased, and the taxonomic community composition significantly altered. The relative abundance of copiotrophic bacteria, such as Actinobacteria, Acidobacteria, Bacteroidetes, Verrucomicrobia, Planctomycetes, and Gemmatimonadetes significantly increased, whereas the relative abundance of oligotrophic bacteria, such as Proteobacteria and Firmicutes significantly decreased. The relative abundance of Cyanobacteria in the algae crusts was significantly higher than that in the other developmental stages (p < 0.05).
Conclusion: Variations in bacterial composition suggested that the potential ecological functions of the bacterial community were altered with BSC development. The functions varied from enhancing soil surface stability by promoting soil particle cementation in the early stages to promoting material circulation of the ecosystem by fixing carbon and nitrogen and decomposing litter in the later stages of BSC development. Bacterial community is a sensitive index of water and nutrient alterations during BSC development. SWC, pH value, TC, TOC, TN, NO3−, TP and soil texture were the primary environmental variables that promoted changes in the bacterial community composition of BSCs.
Drylands are extremely important for achieving global sustainability as they cover more than 40% of the land surface on Earth and host more than a third of the total human population (Reynolds et al., 2007). Approximately half of arid and semiarid drylands are devoid of plants and are instead occupied by biological soil crusts (BSCs), an assemblage of soil fine particles, cyanobacteria, green algae, lichen, mosses and microbes in various proportions (Belnap, 2001). BSCs support a wide range of ecosystem functions, such as enhancing soil stability, reducing soil and wind erosion, improving the nutrient cycling of ecosystems, regulating water availability and redistribution, and influencing the emergence and survival of vascular plants (Belnap and Lange, 2003; Maestre et al., 2011; Wang et al., 2021; Barrera et al., 2022). Given the global distribution of BSCs and their key functional roles in the ecosystems where they are prevalent, understanding the developmental mechanisms and ecological functions of BSCs is critical in formulating sustainable natural resource management and conservation policies in drylands.
Biological soil crusts colonize bare grounds and thus constitute the first successional stage in the development of arid and semiarid area ecosystems (Garcia-Pichel et al., 2003). BSCs are categorized into main successional stages according to the predominant organisms present, including microbial, algae, lichen and moss crusts (Zhang et al., 2015; Liu et al., 2017). Microbial and algae crusts represent the earliest developmental stages of biocrusts, whereas lichen and mosses appear during the later stages. Bacteria play an important role in the formation and development of BSCs (Belnap, 2001). In arid and semiarid areas, many drought-tolerant and high temperature-resistant bacteria (up to 70°C) can colonize the sand surface, alter the physical and chemical properties of the soil through their physiological metabolic mechanisms, and lay a strong foundation for the formation and development of BSCs (Jorge-Villar and Edwards, 2013). Additionally, some bacterial phyla, such as Bacteroides and Cyanobacteria, can secrete metabolites or form centimeter-long filament bundles to maintain the physical structure of BSCs (Garcia-Pichel and Wojciechowski, 2009). Moreover, as primary producers, bacteria (primarily cyanobacteria) increase soil fertility by fixing carbon and nitrogen in the atmosphere, providing a strong foundation for the development of BSCs (Wang et al., 2021).
Bacterial communities present in BSCs have been investigated in many studies worldwide. Among them, owing to the significant colonization status, cyanobacteria in BSCs were studied most frequently (Steven et al., 2014; Zhang et al., 2018). However, the trends in the variation of bacterial diversity with BSC development remain inconclusive. Evidence from some studies suggested that bacterial diversity gradually increased with the development of BSCs (Gundlapally and Garcia-Pichel, 2006; Zhang et al., 2016), whereas that from other studies suggested that bacterial diversity peaked in the middle stages of BSC development and gradually declined thereafter (Redfield et al., 2006; Liu et al., 2017). In addition, significant changes occurred in the composition of the microbial community with the development of BSCs. Specifically, Cyanobacteria was the most abundant phylum in the algae and lichen crusts (Nagy et al., 2005; Abed et al., 2010; Zhang et al., 2016), whereas the abundances of Proteobacteria and Bacteroidetes were higher in the moss crusts (Maier et al., 2014; Zhang et al., 2016). Nevertheless, changes in the bacterial community composition with the development of BSCs are not fully understood. Findings from previous studies also highlighted that the bacterial community in BSCs is primarily affected by cyanobacterial abundance, soil water content, soil particle composition, and electrical conductivity (Zhang et al., 2018; Zhou et al., 2020). However, the results of the study widely varied based on the study area. In recent years, owing to the development of high-throughput sequencing technology, the accuracy of microbial component surveys has considerably improved (Atrache, 2017; Li et al., 2020). This approach facilitates a more precise analysis of the shift in microbial populations at different developmental stages of BSCs, and also makes the analyses of key ecological driving factors feasible.
Alpine sandy land regions, such as the Qinghai-Tibet Plateau, are recognized as regions very sensitive to climate change (Wang et al., 2010). Gonghe Basin, located in the northeastern part of the Qinghai-Tibet Plateau, forms an important ecological barrier in northwest China. Caragana microphylla is one of the most typical sandy shrubs found in the sandy land of Gonghe Basin and plays an important role in desertification control and biodiversity conservation (Cheng et al., 2022). BSCs are widely distributed in the C. microphylla community. Until now, only a few studies conducted in these regions have discussed the BSC carbon flux, its response to climate change, and the effects of BSCs on moisture absorption in soil, among other parameters (Jia et al., 2016; Cheng et al., 2019). However, our understanding of bacterial community varies with BSC development in the regions remains limited. Studies of this knowledge gap could improve our understanding of BSC developmental mechanism and functions in the process of ecological restoration of sandy land, and are critical to predicting alpine desert ecosystem responses under climate change scenarios.
In this study, 16S rRNA gene amplicon sequencing was used to systematically explore the bacterial diversity and community composition at the different developmental stages of BSCs in the Gonghe basin sandy land in northwestern China. We aimed to answer the following questions: (i) What is the uniqueness of the BSC bacterial communities in this study area (alpine sandy land)? (ii) Do microbial taxa change with the development of BSCs, and how? and, (iii) What are the key environmental variables affecting bacterial communities of BSCs?
The study site (100°25′ E, 36°24′ N) is located in the northwest of the Gonghe Basin sandy land, in Gonghe County, Qinghai Province. It is managed by the Qinghai Republican Desert Ecosystem Research Station, National Forestry and Grassland Administration (Figure 1). The Gonghe Basin (~13,800 km2) is the area of the Qinghai-Tibet Plateau that is most severely affected and threatened by land desertification; 33.5% of the basin area has undergone desertification. The average altitude of the Gonghe Basin is more than 2,900 m. This area is characterized by a plateau mountain climate, with a mean annual temperature of 1.0–5.2°C and mean annual precipitation of 311.1–402.1 mm. At our study site, the annual mean temperature is 2.4°C and the annual average precipitation is 246.3 mm, with approximately 70% of precipitation occurring between May and September. The mean annual potential evaporation is 1,716.7 mm. Soil type is aeolian sand and soil salinity is 28.7 g/kg. The Gonghe Basin sandy land comprises fixed, semi-fixed, and shifting dunes, with vegetation cover rates of 30–50, 10–30, and <10%, respectively. The vegetation type is temperate desert shrubland. Shrubs primarily consist of cultivated vegetation, and Caragana korshinskii, Hippophae rhamnoides, and Tamarix chinensis are the predominant species, with Lycium chinense being spatially scattered. Additionally, different types of BSCs, including microbial, algae, lichen, and moss crusts, are widely distributed on the fixed and semi-fixed dunes (with a cover of 40–80%; Figure 1).
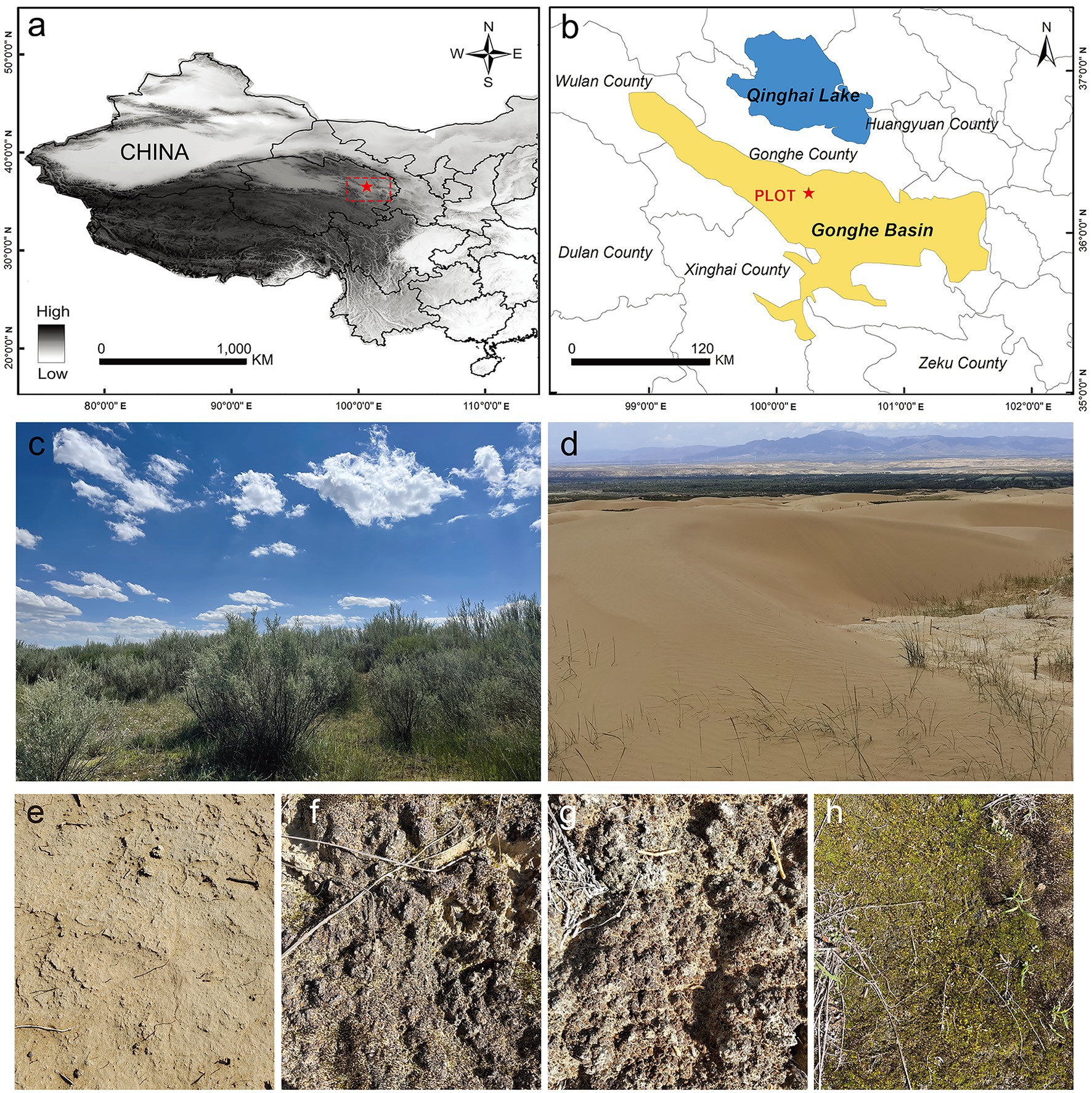
Figure 1. Study site. (A,B) Geographical location of the sampling plot in the Gonghe Basin, western China; (C) revegetation using desert shrubs for stabilizing mobile dunes; (D–H) bare sand, microbial crusts, algae crusts, lichen crusts, and moss crusts, respectively.
Sampling was conducted in August 2021. The experimental region had not received any rainfall in 72 h before sampling. Intact samples included a series of different developmental and successional BSCs. A relatively flat area (approximately 150 m × 200 m) was selected to ensure greater homogeneity in site conditions and reduce the variability associated with small-scale differences. Each developmental stage was considered from three individual plots (at least 20 m between two adjacent plots). Within each subplot, five subsamples were randomly collected and combined to form one composite sample for each BSC developmental stage. Samples were collected from the interspaces between shrubs (0.5 m away from the shrubs) using a sterile cutting ring (diameter of 9.0 cm) according to a protocol reported by Zhou et al. (2020). Bare sand samples were collected from shifting dunes with a thickness of 2 cm. Fifteen types of different developmental BSCs, including bare sand, were collected (five developmental stages × three individual subplots), and the samples were preserved in an ice box. These samples were sieved (2.0 mm mesh) to remove visible roots and stones and categorized in three parts for further analysis. The first part was air-dried, the second part was stored at 4°C to analyze soil properties, and the third part was freeze-dried at −80°C for DNA extraction.
Soil pH was determined using a potentiometer with a pH electrode. A 1:5 soil/water (w/w) suspension was prepared previously. The soil water content (SWC) was determined by oven-drying the soil to a constant mass at 105°C. The total carbon (TC) and total nitrogen (TN) contents were measured by dry combustion using an elemental analyzer (2400II CHN elemental analyzer; Perkin-Elmer, Waltham, MA, United States). Total phosphorus (TP) was digested in 1 mol L−1 H2SO4 after ignition at 550°C in a muffle furnace, and then measured using the molybdate colorimetric method at 880 nm on a spectrophotometer (UV-2550; Shimadzu, Japan; Pang, 2003). Total organic carbon (TOC) was determined using a wet combustion method with a mixture of potassium dichromate (K2Cr2O7) and sulfuric acid (H2SO4) under external heating (Lemonnier et al., 2010). Nitrate (NO3−) and ammonium (NH4+) were extracted using 50 ml of 2 M KCl. After shaking for 30 min, the solution was filtered and analyzed using a continuous flow analyzer (Skalar, Breda, Netherlands). A standard soil hydrometer method (Klute, 1986) was used to determine the sand, silt, and clay composition in the crust samples. Microbial biomass carbon (MBC) and nitrogen (MBN) content in soil were measured using the chloroform fumigation direct extraction method, as described by Witt et al. (2000).
DNA extraction was performed using the PowerSoil®DNA Isolation Kit (MO BIO, Carlsbad, CA, United States) per the manufacturer’s instructions. Total DNA was then eluted and dissolved in 100 μl of elution buffer. DNA samples were purified using a 0.8% (w/v) agarose gel. DNA bands were excised from the gel, and DNA was extracted using a gel extraction kit (Bioer, China) and quantified with a NanoDrop™8,000 spectrophotometer (Thermo Scientific, Waltham, MA, United States). Bacterial 16S rRNA gene fragments were PCR-amplified using conserved domain-specific primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTA CHVGGGTWTCTAAT-3′), as described by Caporaso et al. (2012). PCR amplification and tag-encoded high-throughput sequencing of 16S genes were conducted at Novogene Company in Beijing, China, using the Illumina HiSeq platform (PE 2500).
Sequence processing, clustering, taxonomic assignments, and biodiversity calculations were performed using the QIIME pipeline.1 Both forward and reverse primers were trimmed. Sequences with high quality (length > 200 bp, without ambiguous base “N,” and average base quality score > 30) were used for downstream analyses. Operational taxonomic units (OTUs) were clustered with 97% similarity cut-off using UPARSE (Edgar, 2013), and chimeric sequences were identified and removed using UCHIME (Edgar et al., 2011). Taxonomic assignments were performed using the SILVA database as a reference (Pruesse et al., 2007). The non-bacterial sequences were discarded. Lastly, to correct the bias caused by different sequencing depths, the sequence data were normalized to 13,509 sequences per sample.
Bacterial alpha-diversity was assessed based on the observed species (species richness), the Shannon-Wiener and Chao1 indexes. Differences in bacterial community composition (Bray–Curtis dissimilarity) in the different developmental stages of BSCs were assessed using permutational multivariate ANOVA (PerMANOVA) and visualized using principal coordinate analysis (PCoA). After performing normality and equality of variance tests, repeated-measures analysis of variance (ANOVA) was conducted to compare the effects of soil physicochemical characteristics and microbial community composition in the different developmental stages of BSCs. Post-hoc analyses were performed using Fisher’s least significant difference (LSD) test. Differences were considered to be statistically significant at p < 0.05. Mantel tests based on the Bray–Curtis distances were performed to correlate the microbial communities with environmental variables.
Statistical analyses and figure preparation were performed using R 3.6.0 (R.Team, 2011). We used the ggplot2 package (Wickham, 2011) for generating figures.
The soil water content did not significantly differ between bare sand and microbial crusts but significantly increased from 2.04% (bare sand) to 3.40% (algae crusts), 8.48% (lichen crusts), and 13.65% (moss crusts) with the development of BSCs (Table 1). Similarly, the TOC, TC, TN, TP, and ammonium (NH4+) concentrations in the samples also increased in a stepwise manner with BSC development. From bare sand to moss crusts, the TOC, TC, TN, and TP increased 4.61, 4.76, 2.65, and 0.39 times, respectively. In contrast, the concentration of nitrate (NO3−) and the pH value in moss crusts decreased from that in bare sand (p < 0.05). The soil texture clearly differed between bare sand and BSCs. The sand content was the highest in the bare sand sample (95.36%) and decreased in microbial crusts (88.97%), algae crusts (86.64%), lichen crusts (75.54%), and moss crusts (75.33%). On the contrary, the silt and clay contents significantly increased with the development of BSCs (p < 0.05).
Microbial biomass carbon content was lower in the bare sand (36.98 mg/kg) and microbial crusts (34.94 mg/kg), and significantly increased (141.80 mg/kg) in the lichen crusts, and then decreased in the moss crusts (120.67 mg/kg). MBN increased with the development of BSCs and peaked in the moss crusts (from 4.04 mg/kg in bare sand to 21.03 mg/kg in moss crusts), being 5.21 times higher than that in the bare sand (Supplementary Figure S1).
To identify the OTUs that contributed to the divergence in the bacterial community composition in the five developmental stages, we observed overlaps and distinctions in the abundant OTUs in the five developmental stages (Figure 2). As shown in Figure 2, 13,590 bacterial OTUs were detected in all developmental stages of BSCs. There were 2,476 bacterial OTUs common with those in the bare sand and in the different developmental stages of BSCs. Moss crusts were the most abundant in unique OTUs (1,251), followed by lichen crusts (950), bare sand (679), and microbial crusts (555). Algae crusts had the lowest number of unique OTUs (419). Bacterial diversity in different developmental stages of BSCs showed significant differences (Figure 3). The number of observed species significantly increased from 3,200 in bare sand to 3,611 in microbial crusts, 3,810 in algae crusts, 4,124 in lichen crusts, and 4,507 in moss crusts (p < 0.05), with increment rates of 12.8, 19.1, 28.9, and 40.8%, respectively. Shannon–Wiener and Chao1 indexes also gradually increased with the development, with the highest values observed in the moss crusts (Figure 3).
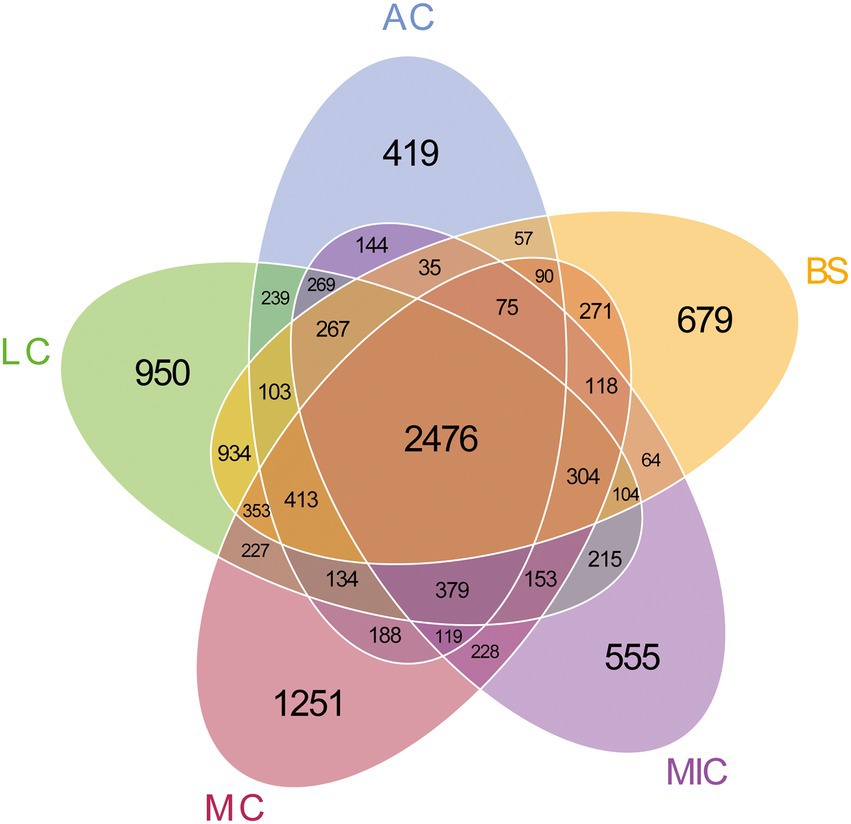
Figure 2. Number of operational taxonomic units (OTUs) of bacteria in each developmental stage of BSCs. BS, bare sand; MIC, microbial crusts; AC, algae crusts; LC, lichen crusts; MC, moss crusts.
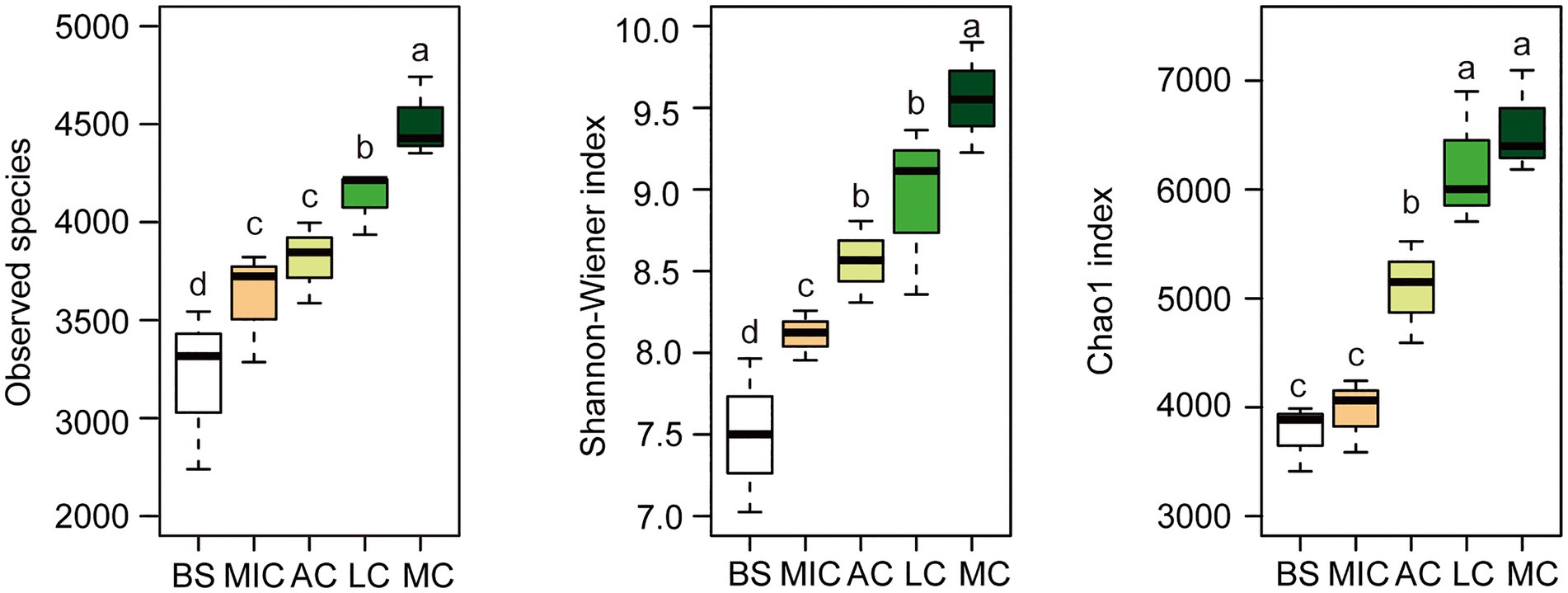
Figure 3. Alpha diversity of the bacterial taxonomic community. The box was drawn to represent values from the lower 1/4 quantile to the upper 1/4 quantile. Whiskers above and below the box represent the upper 95% CI and lower 95% CI, respectively. Medians are indicated using black horizontal bars within boxes. We used one-way ANOVA to observe significant changes between treatments. Letters depict significant differences across compartments. BS, bare sand; MIC, microbial crusts; AC, algae crusts; LC, lichen crusts; MC, moss crusts.
In total, 58 bacteria phyla were detected in our study. Bacterial communities in all five developmental stages were dominated by Actinobacteria (15.1% ± 4.6%), Proteobacteria (24.6% ± 7.0%), Cyanobacteria (6.5% ± 6.2%), Acidobacteria (10.6% ± 6.0%), Bacteroidetes (15.2% ± 3.0%), and Firmicutes (5.3% ± 2.3%; Figure 4A). The relative abundances of these six phyla accounted for more than 77% of the total relative abundance.
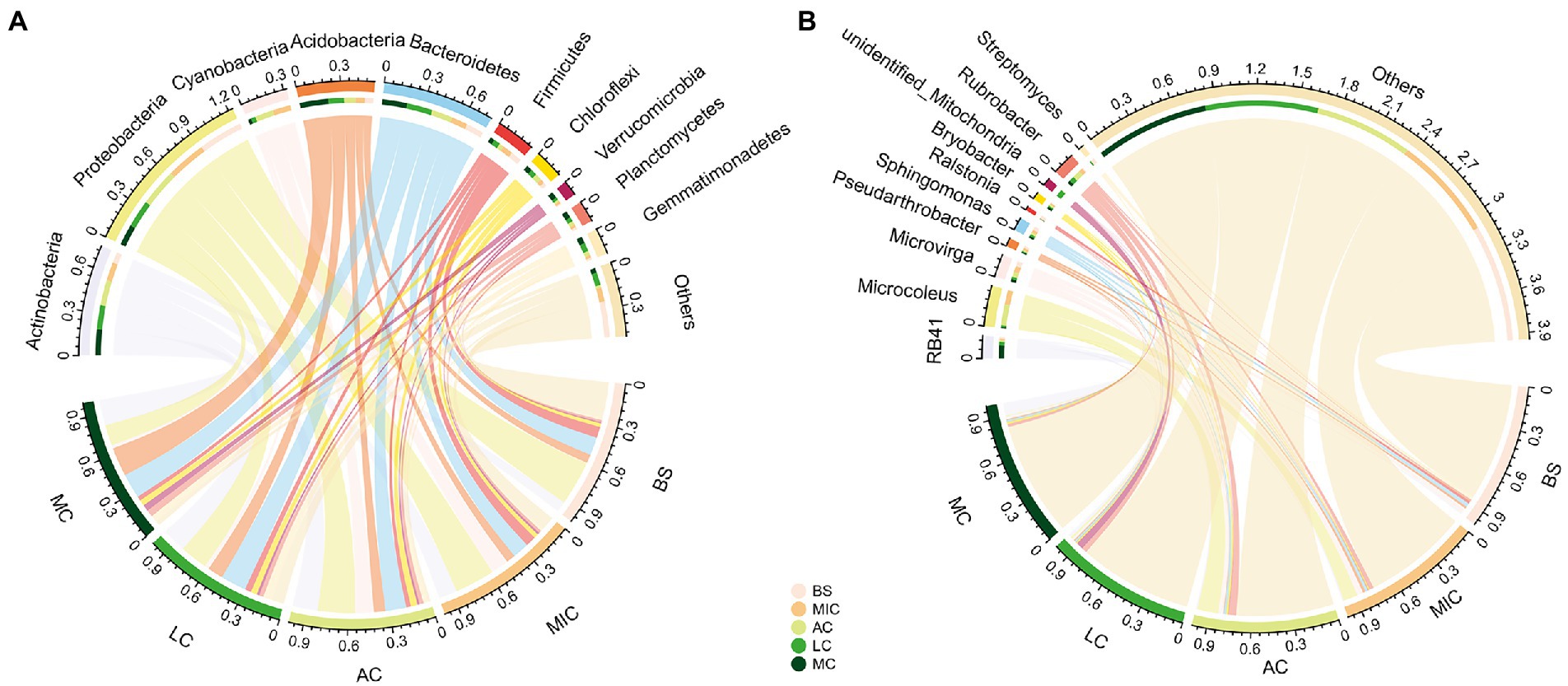
Figure 4. Relative abundance of bacterial communities at the phylum (A) and genus (B) levels in different developmental stages of BSCs. BS, bare sand; MIC, microbial crusts; AC, algae crusts; LC, lichen crusts; MC, moss crusts.
The compositions of bacterial communities distinctly differed in the five developmental stages (Figure 3). Specifically, the relative abundance of Actinobacteria, Acidobacteria, Bacteroidetes, Verrucomicrobia, Planctomycetes, and Gemmatimonadetes significantly increased with the development of BSCs (p < 0.05; Figure 4A; Supplementary Table S2). On the contrary, the relative abundance of Proteobacteria and Firmicutes significantly decreased with the development of BSCs. From bare sand to moss crusts, the relative abundance of Proteobacteria decreased from 32.1 to 15.0% and that of Firmicutes decreased from 8.0 to 3.2%. The relative abundance of Cyanobacteria was the lowest in bare sand (0.4% ± 0.1%), significantly increased and peaked (13.3% ± 2.0%) in the algae crusts, and significantly decreased in lichen crusts (3.2% ± 1.0%) and moss crusts (2.3% ± 0.4%). The relative abundance of Chloroflexi showed no significant differences with the development of BSCs (p > 0.05; Figure 4A; Supplementary Table S2).
To further investigate taxonomic compositions of different BSC bacterial communities, we compared the dominant bacterial taxa in the different developmental stages of BSCs at the finer levels of classification. At the genus level, 825 genera were identified. We also observed notable differences among the four developmental stages (Figure 4B; Supplementary Table S2). The abundance of two genera significantly increased with the development of BSCs (p < 0.05; Supplementary Table S2), including the acidobacterial genera RB41 and Bryobacter. On the contrary, the abundance of two proteobacterial genera Ralstonia and Sphingomonas significantly decreased with BSC development (p < 0.05). Algae crusts had a significantly greater proportion of the cyanobacterial genera Microcoleus than other stages (p < 0.05).
Unconstrained principal coordinate analyses (PCoAs) based on the Bray–Curtis distance matrix of OTU relative abundances were performed to investigate the patterns of bacterial communities (Figure 5). PCoA plots showed that bacterial communities among bare sand and the four developmental stages of BSCs were well separated from each other. Bacterial axes 1 and 2 accounted for 30.37 and 15.51% of the variance, respectively (Figure 5). Results of permutational multivariate analysis of variance (PerMANOVA), Adonis function, confirmed that the bacterial taxonomic communities were significantly different among bare sand and different developmental stages of BSCs (R2 = 0.422, p = 0.001, Supplementary Table S1).

Figure 5. Principal coordinate analysis (PCoA) plots of bacterial communities based on the Bray–Curtis dissimilarities. Circles represent samples from each developmental stage of BSCs. BS, bare sand; MIC, microbial crusts; AC, algae crusts; LC, lichen crusts; MC, moss crusts.
Mantel tests were applied to evaluate the correlations between each environmental factor and bacterial communities in BSCs (Table 2). Among the environmental variables examined, the SWC (p = 0.002), pH (p = 0.006), TC (p = 0.001), TOC (p = 0.049), TN (p = 0.001), NO3− (p = 0.008), TP (p = 0.001), sand content (p = 0.012), silt content (p = 0.044), and clay content (p = 0.003) were significantly correlated with the bacterial community composition of BSCs. Among these environmental variables, SWC exerted the strongest impact on bacterial community composition (R = 0.683). However, the NH4− content (p = 0.243) showed no significant correlation with the bacterial community composition.
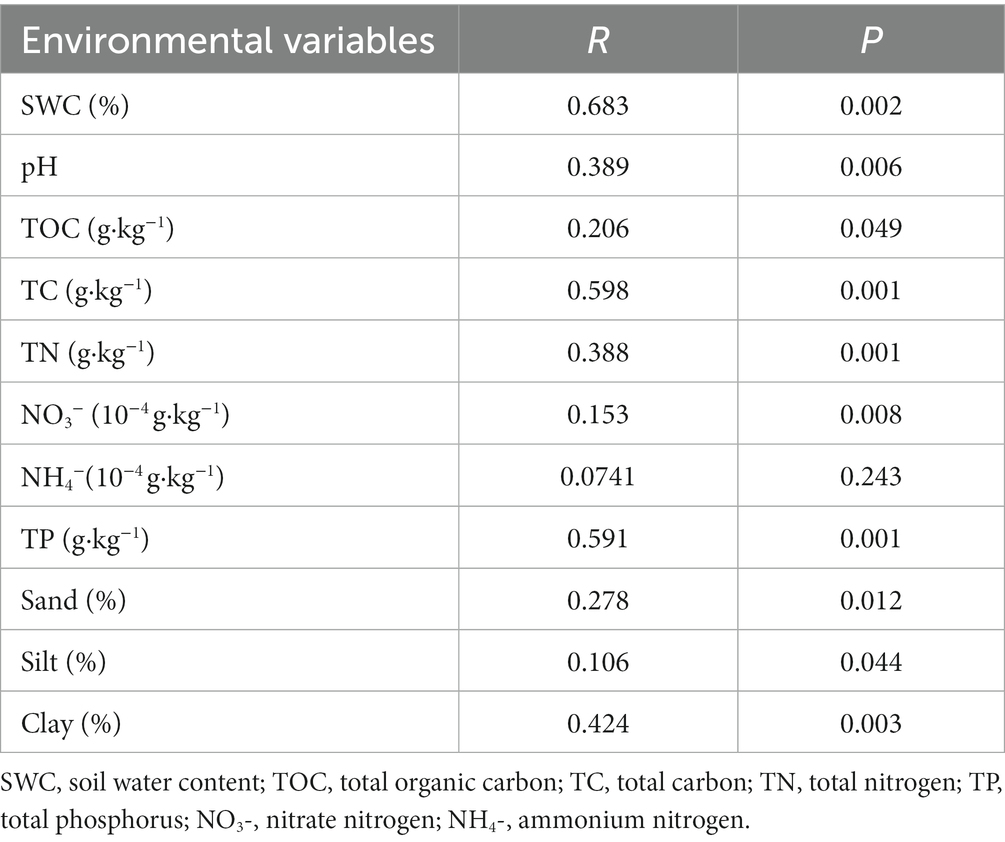
Table 2. Mantel tests for determining the bacterial community composition and environmental variables.
Mantel tests were also performed to examine the potential correlations between individual bacterial phyla and environmental variables (Figure 6). For example, the TC, TN, TOC, and clay contents were positively correlated with the compositions of Actinobacteria, Cyanobacteria, Acidobacteria, Bacteroidetes, Planctomycetes, and Gemmatimonadetes but negatively correlated with that of Proteobacteria (p < 0.05). Sand and NO3− contents in the BSCs were positively correlated with the composition of Proteobacteria (p = 0.004 and p = 0.017) and negatively correlated with that of other phyla, except Firmicutes (p < 0.05). Additionally, the SWC showed significant correlations with the bacterial phyla composition, except Cyanobacteria (Figure 6). TP showed significant positive correlations with the compositions of Actinobacteria, Cyanobacteria, Planctomycetes, and Gemmatimonadetes (p < 0.05) but a significant negative correlation with that of Proteobacteria (p = 0.013).
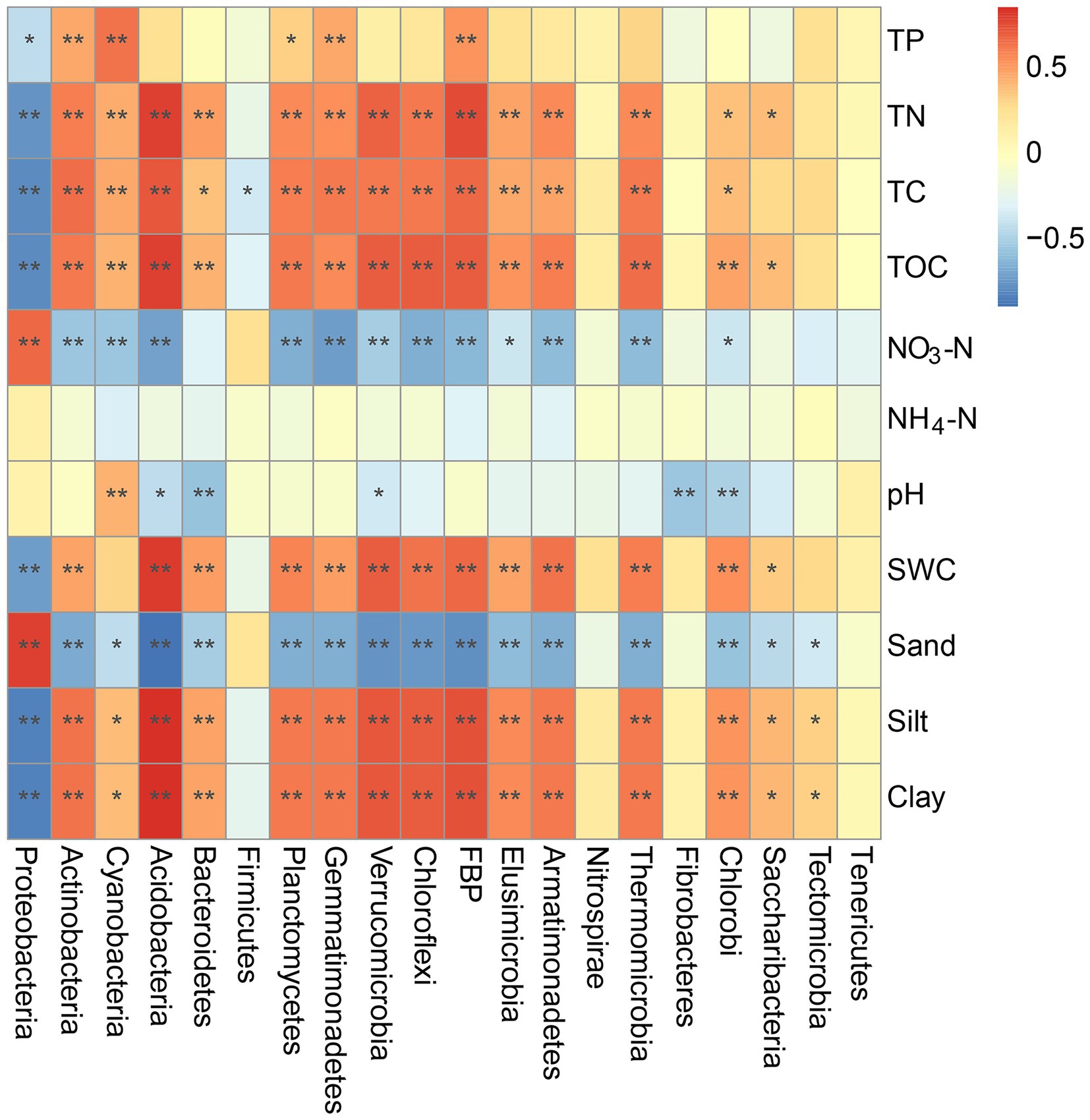
Figure 6. Mantel tests of environmental variables and different bacterial phyla in BSCs. *p < 0.05, **p < 0.01. SWC, soil water content; TOC, total organic carbon; TC, total carbon; TN, total nitrogen; TP, total phosphorus; NO3-N, nitrate nitrogen; NH4-N, ammonium nitrogen.
Given the high phylogenetic diversity, abundance, ubiquity, and biogeochemical importance of microbes (Harris, 2009), studies on microbial communities help us understand how microbes play essential roles in biogeochemical cycling and ecosystem functioning of BSCs. Our data collected from the alpine sandy land of the Qinghai-Tibet Plateau revealed that BSC bacterial communities were diverse and abundant in this region. Higher bacterial diversity in BSCs has been reported in many regions of desert and plateau ecosystems. For example, in the Shapotou area of Tengger Desert (China; average altitude is 1,377 m), the number of OTUs in four different developmental stages of BSCs reached about 4,000, and the Shannon-Wiener indexes were more than 7.2 (Zhang et al., 2018). Here, the total amount of OTUs in BSCs we observed in alpine sandy land was up to 3,600, and their Shannon-Wiener indexes were more than 8.0 (Figure 3), thus much higher than that in the Shapotou area. Ren et al. (2018) observed that soil bacterial alpha diversity was significantly higher at medium altitude (2,060 ~ 3,300 m) than that in low altitudes (less than 2,000 m), which was consistent with our results.
Proteobacteria, Actinobacteria, Cyanobacteria, Acidobacteria, Bacteroidetes, and Firmicutes were predominant in the bacterial communities in BSCs (Figure 4A), similar to observations in the Tengger Desert and Gurbantunggut Desert (Zhang et al., 2016, 2018). However, the relative abundances of Acidobacteria and Bacteroidetes (6.3–20.8% and 11.8–18.6%) in this study were significantly higher than that in Tengger Desert (2.0–9.1% and 2.5–7.9%) and Gurbantunggut Desert (2.0–8.3% and 1.7–5.8%). According to findings from previous studies, the relative abundance of Acidobacteria was significantly unimodal with altitude, with a declining pattern in the middle and low altitudes (1,000 ~ 2,000 m) and an increasing pattern at high altitudes (above 2,000 m; Liu et al., 2016). The altitude of Gonghe Basin (over 2,900 m) was significantly higher than that of the Tengger Desert (1,377 m) and Gurbantunggut Desert (550 m). This could explain the relatively high abundance of Acidobacteria in the BSCs in our study. The phylum Bacteroidetes had a strong ability to adapt to cold environments (Zhang et al., 2009). In addition, Junge et al. (2004) observed that with a decrease in temperature, the relative abundance of Bacteroides increased, which was consistent with our results. An increase in altitude usually corresponds to a change in the hydrothermal conditions, with a higher altitude associated with a colder climate. The Gonghe Basin sandy land is located in the Qinghai-Tibet Plateau, and its annual mean temperature (2.4°C) is lower than that of the Tengger Desert (7.8°C) and Gurbantunggut Desert (5–5.7°C). Thus, the high relative abundance of Bacteroidetes in our study could be explained by the relatively colder environmental conditions of the Gonghe Basin. All these environmental variables may underpin differences in BSC bacterial communities between the alpine sandy land and other regions.
MBC and MBN significantly increased with BSC development (Supplementary Figure S1), which was similar to the findings of a study conducted in the Tengger Desert (Yang et al., 2017) and Gurbantunggut Desert (Zhang et al., 2015). These results could be attributed to two potential reasons. First, with the development of BSCs, the thickness of the crusts increased, and the influence of wind erosion on the microbial community gradually weakened. This provided a relatively stable environment for the survival and reproduction of microbes. Second, the organic carbon and water content exerted the strongest influence on microbial biomass, including MBC and MBN (Xu et al., 2013). As the results showed, both the total organic carbon and water content significantly increased with the development of BSCs (p < 0.05, Table 1), resulting in higher bacterial biomass in this study.
Through the comparison of diversity based on OTUs, we found that the abundance and diversity of the bacterial community significantly increased with the development of BSCs and peaked in the moss crusts (Figure 3). Although similar holistic surveys of microbial diversity in different developmental stages of BSCs have been conducted in a few studies, there is evidence that the bacterial diversity index significantly increased with BSC development. For example, Mogul et al. (2017) recorded the bacterial diversity of early algae crusts in different land cover regions and showed that it increased with the development of BSCs in the central Mojave Desert. Li and Zhang (2017) observed that the diversity of the bacterial community was higher in lichen crusts than in algae crusts. Evidence from previous study suggested that microbial diversity positively influences multiple ecosystem functions related to nutrient cycling, and greater microbial community diversity support multiple functions simultaneously (Cameron et al., 2019). Thus, with the development of BSCs, richer and more diverse bacterial communities in our study may provide more functional attributes.
The bacterial community composition significantly changed with BSC development (Figure 4). The relative abundance of eutrophic groups, such as Actinobacteria, Acidobacteria, Bacteroidetes, and Planctomycetes, significantly increased with the development of BSCs (Supplementary Table S2). The increase in the abundance of these phyla could be explained by the improvement in nutritional conditions. Notably, most Actinobacteria are mycelial and may play a major role in maintaining the structure of BSCs in the later developmental stages (Maier et al., 2016). Meanwhile, we suggested that the high abundance of Actinobacteria in lichen and moss crusts may imply that some members contribute to primary productivity. Based on other findings from the Antarctic desert surface soil communities, the primary producers were proposed to be Actinobacteria (Mukan et al., 2017). Additionally, findings by Větrovsky et al. (2014) indicated that, Actinobacteria members can degrade complex compounds, such as polysaccharides and phenolic compounds, and improve the nutritional status of BSCs. Acidobacteria are heterotrophic bacteria with highly diverse functions (Lauber et al., 2009). As acidophilic bacteria, Acidobacteria had the greatest relative abundance in moss crusts with the lowest pH value in our study (Table 1). Bacteroidetes can secrete several exopolysaccharides, and the increase in their relative abundance may be related to the increase in fine particulate matter and plant growth in the later stages of BSC development (Zhang et al., 2018). In contrast, highly stress-resistant oligotrophic groups (Barnard et al., 2013), such as Proteobacteria and Firmicutes, were abundant in bare sand and microbial crusts, and their relative abundances decreased with further development (Supplementary Table S2). Gundlapally and Garcia-Pichel (2006) recorded that bacteria from phylum Proteobacteria can secrete extracellular polysaccharides to bind sand grains. These Proteobacteria were also shown to be the major contributors to N-fixation under nitrogen restriction conditions for BSC formation (Gundlapally and Garcia-Pichel, 2006; Zhou et al., 2020). The dominance of Firmicutes in the early stages of BSCs can be explained by its endospore-forming ability and the high G + C content, which enables them to survive in challenging dry conditions in bare sand and early stages of BSC development (Schimel et al., 2007). In addition, both Proteobacteria and Firmicutes showed a negative correlation with the soil water content, total carbon, and total organic carbon in our study (Figure 6), indicating that these bacteria can survive in an environment with low nutrition and water deficiency. This explains why their relative abundances were found to be higher in bare sand and microbial crusts. As the important oxygen-producing photosynthetic organism in BSCs, Cyanobacteria play the role of primary producers (Zhang et al., 2016). We found that the relative abundance of Cyanobacteria peaked in the algae crusts and decreased in the moss crusts (Supplementary Table S2). This may be related to the fact that the density and thickness of the pseudo roots of bryophytes are higher in the moss crusts, which affect the living space of Cyanobacteria. Additionally, members of the Cyanobacteria genus Microcoleus were also more abundant in algae crusts than in other stages (Figure 4B and Supplementary Table S2), and previous studies have shown that it can bind soil particles using filament bundles (Weber et al., 2016).
In general, microbial communities can respond more rapidly than plant communities to change conditions during succession, and they emit early signals of the recovery trajectory. Findings from our study showed that the bacterial community composition of BSCs was significantly affected by the SWC, pH, TC, TOC, TN, NO3−, TP, and soil texture, among which SWC exerted the strongest effect on bacterial composition (p < 0.05; Table 2). This finding was consistent with the results reported by Zhang et al. (2018). According to previous studies, in arid and semi-arid regions, water availability and soil nutrient content are the key limiting factors affecting microbial communities (Yahdjian et al., 2011; Cheng et al., 2022). In the early stage of BSC formation, the bacterial community was restricted by water and nutrients. As the water and nutrient conditions gradually improved with the development of BSCs, the composition of the bacterial community gradually changed. In addition, in late developmental stages of BSCs, the increase in water content makes nutrients (Table 1), such as nitrate nitrogen, more easily dissoluble and utilizable by bacteria. This has a significant impact on bacterial community composition.
We also suggest that the bacterial community composition is a sensitive indicator of the water and nutrient conditions of BSCs at different developmental stages. Changes in water and nutrient conditions during BSC development are the primary reason for differences in bacterial communities in different developmental stages of BSC. In addition, different bacterial groups exhibit different responses to environmental processes in BSCs (Figure 6). This phenomenon may be attributed to the unique ecological niche of each group. Findings from previous studies have suggested that niches play a crucial role in the construction of bacterial community structures (Dumbrell et al., 2010; Yao et al., 2014). Therefore, changes in the bacterial community composition with BSC development may be attributed to the fact that BSCs at different developmental stages select species that are better adapted to a particular ecological niche, thus outperforming less adapted species in competition.
In this study, we identified the changes in bacterial communities with the development of BSCs in the Gonghe basin. Our findings provide novel insights that can help understand the development mechanism and ecological function of BSCs in the alpine sandy areas. We found that Acidobacteria and Bacteroidetes were abundant in different developmental stages of BSCs in this region. With the development of BSCs, the diversity of the bacterial community significantly increased, which is essential for maintaining various ecological functions during BSC development. As the water and nutrient conditions improved with BSC development, the bacterial community composition significantly changed, with an increase in the abundance of copiotrophic taxa and a decrease in the abundance of oligotrophic taxa. As different ecological functions are performed by each group of microbes, our results indicate potential changes in the ecological functions of bacterial communities with BSC development. These results further suggest that the soil water and nutrient contents are key factors driving the shift in bacterial communities with BSC development. Given the predominant role of microbes in BSC formation and development, our findings improve our understanding of the BSC developmental mechanisms and ecological functions, helping us assess the impact of climate change on Alpine desert ecosystems. We propose that the BSC development status can be used as an indicator to evaluate the degradation or health of alpine sandy land.
Publicly available datasets were analyzed in this study. This data can be found here: Sequence reads generated in this study were archived in the sequence read archive database of the National Center for Biotechnology Information under accession number PRJNA902024.
HZ designed the experiments. HZ and YL conducted the sample collection and sample site survey. HZ, YL, and LL performed the experiments and analyzed the data. HZ wrote the manuscript along with YL and LL. All authors reviewed the article.
This work was supported by the Natural Science Foundation of Qinghai Province (2022-ZJ-959Q) and the Open Project of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University (2021-ZZ-04).
We sincerely thank all research colleagues who collected soil samples on our behalf. We are grateful to the referees for valuable comments and suggestions that helped to improve the quality of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1106739/full#supplementary-material
Abed, R. M. M., Samiha, A. K., Angela, S., and Robinson, M. D. (2010). Bacterial diversity, pigments and nitrogen fixation of biological desert crusts from the Sultanate of Oman. FEMS Microbiol. Ecol. 72, 418–428. doi: 10.1111/j.1574-6941.2010.00854.x
Atrache, R. (2017). Comparative Metagenomics of freshwater cyanobacteria bloom and non-bloom sites in Ontario and the investigation into the utilization of conserved signature proteins for identification of cyanobacteria. Doctoral thesis. Canada: McMaster University.
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Barrera, A., Acuña-Rodríguez, I. S., Ballesteros, G. I., Atala, C., and Molina-Montenegro, M. A. (2022). Biological soil crusts as ecosystem engineers in Antarctic ecosystem. Front. Microbiol. 13:755014. doi: 10.3389/fmicb.2022.755014
Belnap, J. (2001). “Comparative structure of physical and biological soil crusts” in Biological soil crusts, structure, function, and management. eds. J. Belnap and O. L. Lange (Berlin: Springer) doi: 10.1007/978-3-642-56475-8_15
Belnap, J., and Lange, O.L. (2003). Biological Soil Crusts: Structure, Function, and Management. Berlin: Springer.
Cameron, W., Klaus, S., Samiran, B., Eiko, E. K., and Marcel, G. A. V. D. H. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10, 4841–4810. doi: 10.1038/s41467-019-12798-y
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cheng, L., Jia, X., Wu, B., Li, Y., and Zhao, X. (2019). Composition analysis of condensation water in biological soil crusts covering area in alpine sandy lands. Plateau Meteorol. 38, 439–447. doi: 10.7522/j.issn.1000-0534.2018.00089
Cheng, L., Yue, Y., Zhou, H., Fei, B., Xiu, X., Sun, Y., et al. (2022). Biological soil crusts enhance the role of non-rainfall water in the water input in alpine sandy land ecosystems. J. Hydrol. 610:127966. doi: 10.1016/j.jhydrol.2022.127966
Dumbrell, A. J., Nelson, M., Helgason, T., Dytham, C., and Fitter, A. H. (2010). Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4, 337–345. doi: 10.1038/ismej.2009.122
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Christopher, Q., and Rob, K. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Garcia-Pichel, F., Belnap, J., Neuer, S., and Schanz, F. (2003). Estimates of global cyanobacterial biomass and its distribution. Algol. Stud. 109, 213–227. doi: 10.1127/1864-1318/2003/0109-0213
Garcia-Pichel, F., and Wojciechowski, M. F. (2009). The evolution of a capacity to build supracellular ropes enabled filamentous cyanobacteria to colonize highly erodible substrate. PLoS One 4:e7801. doi: 10.1371/journal.pone.0007801
Gundlapally, S. R., and Garcia-Pichel, F. (2006). The community and phylogenetic diversity of biological soil crusts in the Colorado plateau studied by molecular fingerprinting and intensive cultivation. Microb. Ecol. 52, 345–357. doi: 10.2307/25153385
Harris, J. (2009). Soil microbial communities and restoration ecology: facilitators or followers? Science 325, 573–574. doi: 10.1126/science.1172975
Jia, X., Gu, C., Wu, B., Li, Y., Cheng, L., and Li, X. (2016). Responses of carbon dioxide fluxes from biological soil crusted soils to pulse rain in arid desert ecosystem. J. Desert Res. 36, 423–432. doi: 10.7522/j.issn.1000-694X.2015.00199
Jorge-Villar, S., and Edwards, H. (2013). Microorganism response to stressed terrestrial environments: a Raman spectroscopic perspective of extremophilic life strategies. Life Open Access J. 3, 276–294. doi: 10.3390/life3010276
Junge, K., Eicken, H., and Deming, J. W. (2004). Bacterial activity at −2 to −20°C in arctic wintertime sea ice. Appl. Environ. Microbiol. 70, 550–557. doi: 10.1128/AEM.70.1.550-557.2004
Klute, A. (1986). Methods of soil analysis: Part 1: Physical and mineralogical methods, 2nd. Maddison: Soil Science Society of America
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/aem.00335-09
Lemonnier, H., Courties, C., Mugnier, C., Torréton, J.-P., and Herbland, A. (2010). Nutrient and microbial dynamics in eutrophying shrimp ponds affected or unaffected by vibriosis. Mar. Pollut. Bull. 60, 402–411. doi: 10.1016/j.marpolbul.2009.10.012
Li, J. Y., Jin, X. Y., Zhang, X. C., Chen, L., Liu, J. L., Zhang, H. M., et al. (2020). Comparative metagenomics of two distinct biological soil crusts in the Tengger Desert. China. Soil Biol. Biochem. 140:107637. doi: 10.1016/j.soilbio.2019.107637
Li, J., and Zhang, X. (2017). Microbial diversity analysis of different biological soil crusts in Tengger desert. Ecol. Sci. 36, 36–42. doi: 10.14108/j.cnki.1008-8873.2017.03.006
Liu, C., Dong, Y., and Jiao, R. (2016). Research Progress in Acidobacteria diversity in Forest soil. World For. Res. 29, 17–22. doi: 10.13348/j.cnki.sjlyyj.2016.0041.y
Liu, L. C., Liu, Y. B., Peng, Z., Song, G., Rong, H., Wang, Z. R., et al. (2017). Development of bacterial communities in biological soil crusts along a revegetation chronosequence in the Tengger Desert, Northwest China. Biogeosciences 14, 3801–3814. doi: 10.5194/bg-14-3801-2017
Maestre, F. T., Bowker, M. A., Cantón, Y., Castillo-Monroy, A. P., Cortina, J., Escolar, C., et al. (2011). Ecology and functional roles of biological soil crusts in semi-arid ecosystems of Spain. J. Arid Environ. 75, 1282–1291. doi: 10.1016/j.jaridenv.2010.12.008
Maier, S., Muggia, L., Kuske, C.R., and Grube, M. (2016). Bacteria and Non-lichenized Fungi within Biological Soil Crusts, Biological Soil Crusts: An Organizing Principle in Drylands. Cham: Springer.
Maier, S., Schmidt, T. S. B., Zheng, L., Peer, T., Wagner, V., and Grube, M. (2014). Analyses of dryland biological soil crusts highlight lichens as an important regulator of microbial communities. Biodivers. Conserv. 23, 1735–1755. doi: 10.1007/s10531-014-0719-1
Mogul, R., Vaishampayan, P., Bashir, M., McKay, C. P., and Schubert, K. (2017). Microbial community and biochemical dynamics of biological soil crusts across a gradient of surface coverage in the Central Mojave Desert. Front. Microbiol. 8:1974. doi: 10.3389/fmicb.2017.01974
Mukan, G., Chris, V., Inka, C., Carlo, R., Sean, K., Steen, J., et al. (2017). Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552, 400–403. doi: 10.1038/nature25014
Nagy, M. L., Pérez, A., and Garcia-Pichel, F. (2005). The prokaryotic diversity of biological soil crusts in the Sonoran Desert (organ pipe cactus National Monument, AZ). FEMS Microbiol. Ecol. 54, 233–245. doi: 10.1016/j.femsec.2005.03.011
Pang, P. (2003). Determination of Total phosphorus in soil by ammonium Molybdate spectrophotometry. Chinese Journal of Spectroscopy Laboratory 20, 697–699. doi: 10.3969/j.issn.1004-8138.2003.05.014
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
R.Team (2011). R: a language and environment for statistical computing. Computing 1, 12–21. doi: 10.1890/0012-9658(2002)083[3097:CFHIWS]2.0.CO;2
Redfield, E., Barns, S. M., Belnap, J., Daane, L. L., and Kuske, C. R. (2006). Comparative diversity and composition of cyanobacteria in three predominant soil crusts of the Colorado plateau. FEMS Microbiol. Ecol. 40, 55–63. doi: 10.1111/j.1574-6941.2002.tb00936.x
Ren, C., Zhang, W., Zhong, Z., Han, X., Yang, G., Feng, Y., et al. (2018). Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 610-611, 750–758. doi: 10.1016/j.scitotenv.2017.08.110. Epub 2017 Aug 17
Reynolds, J. F., Smith, D., Lambin, E. F., Turner, B. L., Mortimore, M., Batterbury, S., et al. (2007). Global desertification: building a science for Dryland development. Science 316, 847–851. doi: 10.1126/science.1131634
Schimel, J., Balser Teri, C., and Wallenstein, M. (2007). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.2307/27651246
Steven, B., Gallegos-Graves, L. V., Yeager, C., Belnap, J., and Kuske, C. R. (2014). Common and distinguishing features of the bacterial and fungal communities in biological soil crusts and shrub root zone soils. Soil Biol. Biochem. 69, 302–312. doi: 10.1016/j.soilbio.2013.11.008
Větrovsky, T., Steffen, K. T., and Baldrian, P. (2014). Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS One 9:e89108. doi: 10.1371/journal.pone.0089108
Wang, Q., Han, Y., Lan, S., and Hu, C. (2021). Metagenomic insight into patterns and mechanism of nitrogen cycle during biocrust succession. Front. Microbiol. 12:633428. doi: 10.3389/fmicb.2021.633428
Wang, S. P., Luo, C. Y., Xu, G. P., Chao, Z. G., Lin, X. W., Hu, Y. G., et al. (2010). Effect of warming and grazing on litter mass loss and temperature sensitivity of litter and dung mass loss on the Tibetan plateau. Glob. Chang. Biol. 16, 1606–1617. doi: 10.1111/j.1365-2486.2009.02026.x
Weber, B., Büdel, B., and Belnap, J. (2016). Biological Soil Crusts: An Organizing Principle in Drylands. Cham: Springer
Wickham, H. (2011). ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185. doi: 10.1002/wics.147
Witt, C., Gaunt, J. L., Galicia, C. C., Ottow, J. C. G., and Neue, H. U. (2000). A rapid chloroform-fumigation extraction method for measuring soil microbial biomass carbon and nitrogen in flooded rice soils. Biol. Fertil. Soils 30, 510–519. doi: 10.1007/s003740050030
Xu, X. F., Thornton, P. E., and Post, W. M. (2013). A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 22, 737–749. doi: 10.1111/geb.12029
Yahdjian, L., Gherardi, L., and Sala, O. E. (2011). Nitrogen limitation in arid-subhumid ecosystems: A meta-analysis of fertilization studies. J. Arid Environ. 75, 675–680. doi: 10.1016/j.jaridenv.2011.03.003
Yang, H., Liu, Y., Wang, Y., and Hui, R. (2017). Effects of biological soil crusts on the amount and activities of soil microbes in desert areas. J. Desert Res. 37, 950–960. doi: 10.7522/j.issn.1000-694X.2016.00071
Yao, M., Rui, J., Li, J., Dai, Y., Bai, Y., Heděnec, P., et al. (2014). Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol. Biochem. 79, 81–90. doi: 10.1016/j.soilbio.2014.09.009
Zhang, B. C., Kong, W. D., Wu, N., and Zhang, Y. M. (2016). Bacterial diversity and community along the succession of biological soil crusts in the Gurbantunggut Desert, northern China. J. Basic Microbiol. 56, 670–679. doi: 10.1002/jobm.201500751
Zhang, X., Ma, X., Wang, N., and Yao, T. (2009). New subgroup of Bacteroidetes and diverse microorganisms in Tibetan plateau glacial ice provide a biological record of environmental conditions. FEMS Microbiol. Ecol. 67, 21–29. doi: 10.1111/j.1574-6941.2008.00604.x
Zhang, Q. Y., Wang, Q., Ouyang, H. L., Lan, S. B., and Hu, C. X. (2018). Pyrosequencing reveals significant changes in microbial communities along the ecological successions of biological soil crusts in Tengger Desert of China. Pedosphere 28, 350–362. doi: 10.1016/s1002-0160(17)60477-6
Zhang, B., Zhou, X., and Zhang, Y. (2015). Responses of microbial activities and soil physical-chemical properties to the successional process of biological soil crusts in the Gurbantunggut Desert, Xinjiang. J. Arid. Land 7, 101–109. doi: 10.1007/s40333-014-0035-3
Keywords: biological soil crusts, developmental process, bacterial community, Illumina sequencing, Gonghe basin
Citation: Zhou H, Li L and Liu Y (2023) Biological soil crust development affects bacterial communities in the Caragana microphylla community in alpine sandy areas. Front. Microbiol. 14:1106739. doi: 10.3389/fmicb.2023.1106739
Received: 24 November 2022; Accepted: 28 February 2023;
Published: 15 March 2023.
Edited by:
David R. Elliott, University of Derby, United KingdomReviewed by:
Artur Banach, The John Paul II Catholic University of Lublin, PolandCopyright © 2023 Zhou, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunxiang Liu, MTc3OTEzOTQ0NTJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.