- 1Department of Bacteriology, Capital Institute of Pediatrics, Beijing, China
- 2Department of Life Science and Medicine, University of Science and Technology of China, Hefei, China
- 3Military Supplies and Energy Quality Supervision Station of NV, PLA, Beijing, China
Staphylococcus aureus is an opportunistic pathogen that shows a unique ability to quickly respond to a variety of antibiotics. The Crp/Fnr family transcriptional regulator ArcR controls expression of arginine deiminase pathway genes arcABDC, which enable the utilization of arginine as an energy source for cell growth under anaerobic conditions. However, ArcR shares low overall similarity with other Crp/Fnr family proteins, suggesting that they differ in the response to environmental stress. In this study, MIC and survival assays were performed to determine the role of ArcR in antibiotic resistance and tolerance. The results showed that deletion of arcR reduced tolerance of S.aureus to fluoroquinolone antibiotics, mainly through a defect in the response to oxidative stress. In ΔarcR mutant, the expression of the major catalase gene katA was downregulated, and katA overexpression restored bacterial resistance to oxidative stress and antibiotics. We showed that ArcR directly regulated katA transcription by binding to the promoter region of katA. Therefore, our results revealed the contribution of ArcR in bacterial tolerance to oxidative stress and subsequently to fluoroquinolones antibiotics. This study added our understanding on the role of Crp/Fnr family in bacterial susceptibility to antibiotics.
Introduction
Staphylococcus aureus is one of the most common pathogens worldwide and causes a variety of diseases from mild skin infection to fatal endocarditis and sepsis (Levy and Marshall, 2004; Cheung et al., 2021). Antibacterial therapy is one of the only curative pharmacological treatments. Antibiotics exert a bacteriostatic or bactericidal effect by interacting with their targets, via three main mechanisms: inhibition of DNA replication or RNA transcription; disruption of protein synthesis; and prevention of cell wall renewal (Walsh, 2000). Several studies showed that aminoglycoside, β-lactam, and quinolone antibiotics, regardless of drug-target interactions, could stimulate the formation of highly deleterious hydroxyl radicals that assist the bactericidal process (Imlay and Fridovich, 1991; Dwyer et al., 2007; Kohanski et al., 2007). It was well accepted that majority of superoxide generation in E. coli occurs through oxidation of the respiratory electron transport chain driven by oxygen and the conversion of NADH to NAD+. NADH is generated from NAD+ during the TCA cycle. Aminoglycoside, β-lactam, and quinolone antibiotics could result in catabolic NADH depletion, which facilitated the strengthening of the TCA cycle and the generation of superoxide such as hydrogen peroxide (H2O2). Then, superoxide could stimulate the Fenton reaction that produced hydroxyl radicals (Imlay and Fridovich, 1991; Dwyer et al., 2007). All three of these types of bactericidal antibiotics stimulate the production of highly harmful reactive oxygen species (ROS) in both Gram-negative and Gram-positive bacteria, which ultimately help to kill bacteria (Kohanski et al., 2007; Singh et al., 2021).
In natural or host immune system environment, bacteria respond to various reactive species that could modify the bacterial proteome by post-translational modifications and result in bacterial defense through dedicated signal transduction pathways. The reactive species include ROS that are generated in bacteria as the unavoidable consequence of the aerobic life, by incomplete reduction of molecular oxygen during respiration (Imlay, 2003, 2008). The ROS are composed by superoxide anion O2•−, H2O2, and the highly reactive OH•. Catalases and peroxidases are the major peroxide scavengers that convert H2O2 to H2O and O2 through different mechanisms. Catalases have an extremely high peroxide turnover rate and make function at high H2O2 concentrations. In S. aureus, the major catalase KatA is present that is peroxide-inducible and confers H2O2 resistance. Mutations in the gene encoding catalase increase antibiotic susceptibility in Burkholderia. In Pseudomonas aeruginosa, mutation of katA results in increased susceptibility to ciprofloxacin, aminoglycosides, and β-lactam antibiotics (Yu and Deutscher, 1995).
Members of the Crp/Fnr family are DNA-binding proteins that function as transcriptional regulators which primarily activate the expression of genes, although some also repress the expression (Körner et al., 2003). ArcR belongs to the Crp/Fnr family regulators, whose gene arcR and arginine deiminase pathway genes arcABDC form an operon in S. aureus. There was 19% identity and 45% similarity between the ArcR open reading frame of S. aureus and Bacillus licheniformis, but less similarity compared with Crp of Escherichia coli. Under anaerobic conditions, ArcR binds to the upstream regulatory regions of arcABDC to positively regulate their transcription, and this process can be influenced by glucose and arginine (Makhlin et al., 2007). Apart from this regulatory process, the role of ArcR in the response of S. aureus to environmental stress, such as antibiotics, remains unknown.
In this study, we found that ArcR played an important role in the resistance of S. aureus to fluoroquinolones antibiotics. When arcR was deleted, S. aureus had a reduced response to oxidative stress. We further demonstrated that ArcR positively regulated the transcription level of katA by directly binding to the promoter region of katA, thereby affecting the tolerance of bacteria to oxidative stress and fluoroquinolone antibiotics.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used are listed in Table 2. When necessary, the corresponding antibiotics (100 μg/ml ampicillin and 50 μg/ml kanamycin) were added to the medium. S. aureus was usually cultured in Tryptone Soy Broth medium at 37°C and 220 rpm, to which 15 μg/ml chloramphenicol and 10 μg/ml erythromycin were added when necessary.
Construction of knockout and complementary strains
To construct arcR knockout plasmid, the upstream and downstream homologous arm fragments were amplified from wild-type strain NCTC8325 and then ligated by overlap PCR. The obtained fragment was cloned into the pBTs plasmid digested with EcoR I and Hind III. Ampicillin was used to screen the correct transformants. The pBTs derivative pBTs-arcR was electroporated into RN4220 for modification and transformed into NCTC8325 for allelic replacement as described (Hu et al., 2015). PCR and sequencing were used to verify the mutation. For the construction of complementary plasmids, arcR and katA genes were amplified from NCTC8325, and the obtained fragments were ligated into pCN51 plasmid digested with Pst I. The derivates pCN51-arcR and pCN51-katA were transformed into RN4220 and subsequently electroporated into ΔarcR mutant. Expression of arcR and katA was induced by 10 μM CdCl2 if necessary.
Minimal inhibit concentration and survival assays
The MICs of S. aureus to a variety of antibiotics were determined in Mueller-Hinton Broth medium as described previously (Borrero et al., 2014). For the determination of time-dependent bactericidal curves, overnight culture was diluted into fresh TSB medium at a ratio of 1:100 and incubated at 37°C and 220 rpm until OD600 reached 1.0. The MICs of ciprofloxacin, ofloxacin, norfloxacin, levofloxacin, moxifloxacin, and garenoxacin were 0.32, 0.25, 0.48, 0.19, 0.125, and 0.03 μg/ml, respectively. Thiourea (150 mM) was added when the strains grew to OD600 0.6. Samples were taken out at certain time points, diluted 10-fold, and colonies counted by dropping plate.
H2O2 susceptibility assay
The overnight cultures were diluted with fresh TSB medium to OD600 0.05 and then, H2O2 at a final concentration of 0.4 mM was added to the cultures as described previously (Oogai et al., 2016). They were incubated at 37°C with 150 rpm and 1 ml aliquots were removed at the indicated times. The supernatant was removed by centrifugation and pellet was washed twice with sterile 1 × PBS. The precipitates were diluted 10-fold and then dropped onto TSB plates. The plates were cultured overnight at 37°C, and colony counts were performed on the next day.
Neutrophil extraction and ROS measurement
The whole blood of mice was collected with an anticoagulant tube, and neutrophils were extracted using a mouse peripheral blood neutrophil isolation kit (Solarbio, Beijing, China). The concentration of extracted cells was counted by cell counter. Measurement of ROS levels was determined as previously described with minor modifications (Wu et al., 2009). Neutrophil cells were diluted to 105/ml using warm Hanks’ balanced salt solution (HBSS) containing 100 mM luminol and 1 U/ml horseradish peroxidase. The diluted cell suspension was added to a 96-well plate at 200 μl/well, and the reaction was performed at 37°C for 10 min. The bacterial strains were added at a multiplicity of infection (MOI) of 5. At the same time, 106 cells were removed and infected with an MOI of 10. The reaction was carried out in a cell incubator at 37°C, and 20 μl reactant was removed every hour for dilution and dropping plate counting, to which 100 mM NAC was added if necessary.
RNA extraction, reverse transcription, and quantitative real-time PCR
When the bacteria were cultured in TSB medium to OD600 1.0, they were treated in PBS containing 0.4 mM H2O2 for 10 min and collected by centrifugation. Total RNA was extracted using RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). Then, cDNA was synthesized with 1 μg RNA and PrimeScript™ IV 1st strand cDNA Synthesis Mix (Takara, Dalian, China). For the qRT-PCR, cDNA was mixed with specific primers (Table 2) and chamQ universal SYBR quantitative RT-PCR master mix (Vazyme, Nanjing, China). 16S rRNA gene was used as internal controls.
Catalase activity assay
Bacteria were cultured to OD600 1.0 using TSB medium, and 1 ml of culture was collected by centrifugation and washed twice with PBS. The cultures were incubated in PBS with or without 0.4 mM H2O2 for 30 min. The reaction samples were collected by centrifugation, washed twice with PBS, and broken by Ultrasonic Cell Disruptor (Xinzhi, Ningbo, China). Catalase activity was detected using a catalase assay kit (Beyotime, Shanghai, China).
ArcR protein expression and purification
For the construction of ArcR expression plasmid, the arcR gene fragment was amplified from NCTC8325 genome and the PCR product was ligated with pET28a digested with EcoRI. Kanamycin was used to screen the correct transformants. PCR and sequencing were used for verification to obtain the correct expression of pET28A-his6-ArcR. The correctly constructed pET28a-His6-ArcR plasmid was transferred into E. coli BL21(DE3). The transformers were grown in LB medium containing 50 μg/ml kanamycin at 37°C until OD600 reached 0.6 and were induced with 0.5 mM IPTG at 37°C for 4 h. The cells were harvested and lysed by sonication with lysis buffer (25 mM HEPES, 5 mM β-mercaptoethanol, 500 mM NaCl, and 10% glycerol, pH 7.8). The His-tagged fusion proteins were purified in nickel nitrogen triacetate agarose solution (Qiagen, Beijing, China). The bounding protein was eluted by elution buffer (25 mM HEPES, 5 mM β-mercaptoethanol, 500 mM NaCl, 10% glycerol, and 500 mM imidazole, pH 7.8). Target protein was stored at −80°C until use. The concentration of purified proteins was determined by SDS-PAGE and bicinchoninic acid as the standard protein.
Electrophoretic mobility shift assay
EMSA was performed as previously described with minor modifications (Hellman and Fried, 2007). We used NCTC8325 as the template to amplify katA promoter region DNA fragment. The 30 ng DNA fragment was incubated with 0, 100, 200, and 400 ng purified ArcR in binding buffer (25 mM HEPES, 1 mM dithiothreitol, 200 mM NaCl, and 10% glycerol, pH 7.8) at 37°C for 30 min. The 8% polyacrylamide gel was pre-electrophoresed in 1× Tris-borate-EDTA buffer (0.044 M Tris, 0.044 M boric acid, and 0.001 M EDTA, pH 8.0) for 1 h to remove impurities. After adding the sample, electrophoresis was performed for 1 h 40 min on ice. At the end of the electrophoresis, the glue was stained with 0.5 μg/ml ethidium bromide. Imaging was performed using a gel imager (Bio-Rad, Hercules, CA, USA).
Results
ArcR is involved in tolerance of Staphylococcus aureus to fluoroquinolone antibiotics
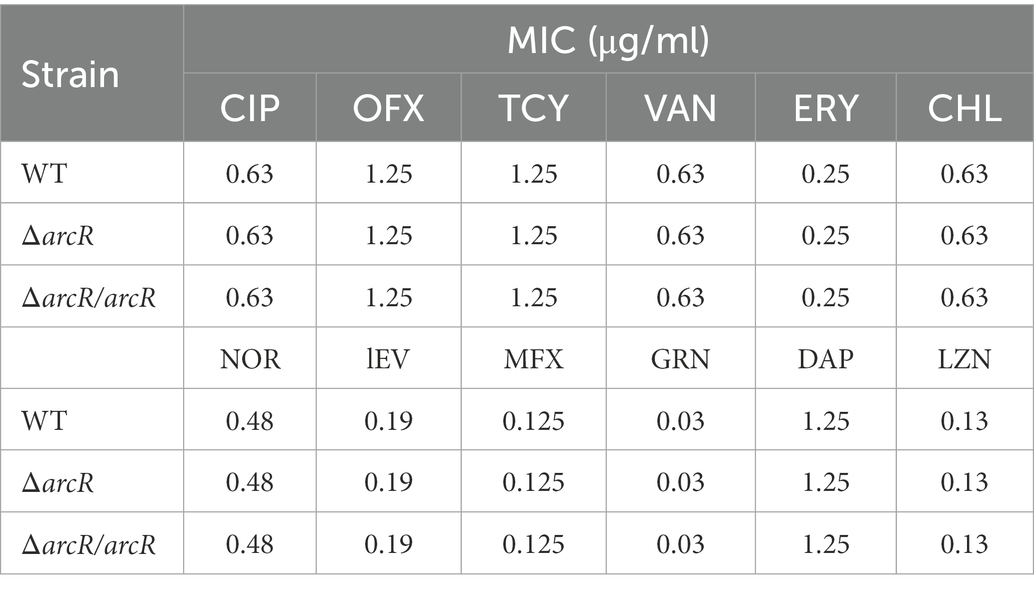
To understand whether ArcR affected antibiotic resistance of S. aureus, the MICs of ΔarcR mutants to ciprofloxacin, ofloxacin, linezolid, tetracycline, vancomycin, erythromycin, chloramphenicol, norfloxacin, levofloxacin, moxifloxacin, garenoxacin, and daptomycin were determined. Wild-type NCTC8325 and ΔarcR mutant displayed similar levels of resistance (MICs) to the above antibiotics (Table 3). However, following treatment with 0.32 μg/ml ciprofloxacin, 0.5 μg/ml ofloxacin, 0.5 μg/ml norfloxacin, 0.5 μg/ml levofloxacin, 0.2 μg/ml moxifloxacin, or 0.06 μg/ml garenoxacin, the survival rate of ΔarcR mutant was approximately 342-, 158-, 74-, 20-, 93-, or 87-fold lower than that of wild-type NCTC8325 strain after 10 h sterilization (Figures 1A–F).
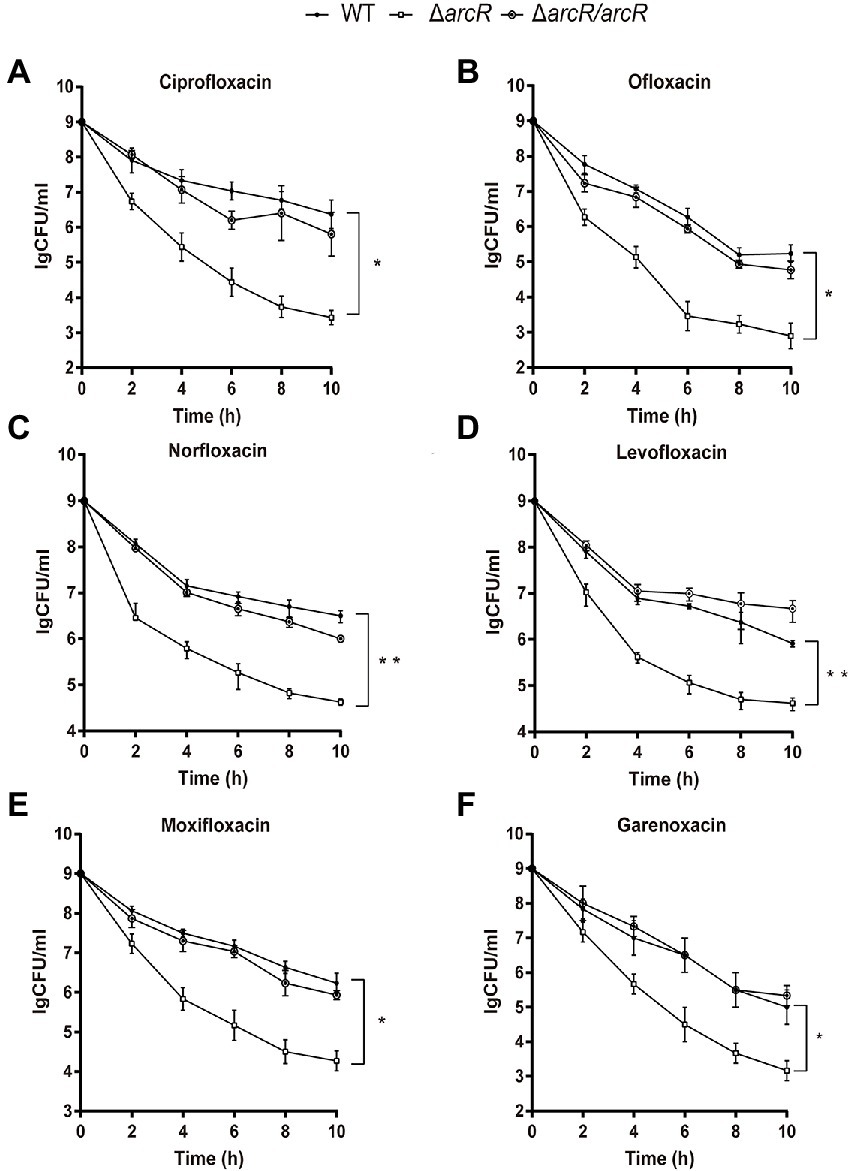
Figure 1. Roles of ArcR in bacterial resistance to fluoroquinolone antibiotics. Bacteria at OD600 1.0 were treated with (A) 0.32 μg/ml ciprofloxacin, (B) 0.5 μg/ml ofloxacin, (C) 0.5 μg/ml norfloxacin, (D) 0.5 μg/ml levofloxacin, (E) 0.2 μg/ml moxifloxacin, or (F) 0.06 μg/ml garenoxacin in TSB at 37°C with agitation. Bacteria were collected at indicated times, and CFUs were determined by serial dilution and plating. ΔarcR/arcR represented the complementary strain, pCN51-arcR in ΔarcR mutant. Error bars represent standard deviations. The data represent results from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, by Student’s t-test.
Defective oxidative stress response leads to increased susceptibility to fluoroquinolone antibiotics in ΔarcR mutant
The tolerance of S. aureus to fluoroquinolones is usually associated with mutations of topoisomerase II genes, including gyrA and gyrB, and high expression of NorA, NorB, NorC, and SdrM belonging to MFS superfamily efflux pump (Lowy, 2003; Ding et al., 2008; Xu et al., 2011). Expression of norA, norB, norC, and sdrM was detected in ΔarcR mutant, and there was no significant difference compared with the wild-type strain (Supplementary Figures 1A–D). These results suggest that ArcR influenced the resistance of S. aureus against fluoroquinolone antibiotics through other mechanisms.
It was reported that ROS was involved in the bactericidal process of aminoglycoside, β-lactam, and fluoroquinolone antibiotics (Kohanski et al., 2007). To determine the role of ROS in the increased sensitivity of ΔarcR mutant to fluoroquinolones, thiourea that neutralize intracellular ROS was added into the medium when killing curves were detected. Thiourea supplementation increased the survival rate of the ΔarcR mutant strain to a level similar to that of the wild type when treated with the same concentration of antibiotics (Figures 2A,B). These results suggest that a defective oxidative stress response leads to increased sensitivity to fluoroquinolone antibiotics in ΔarcR mutant.
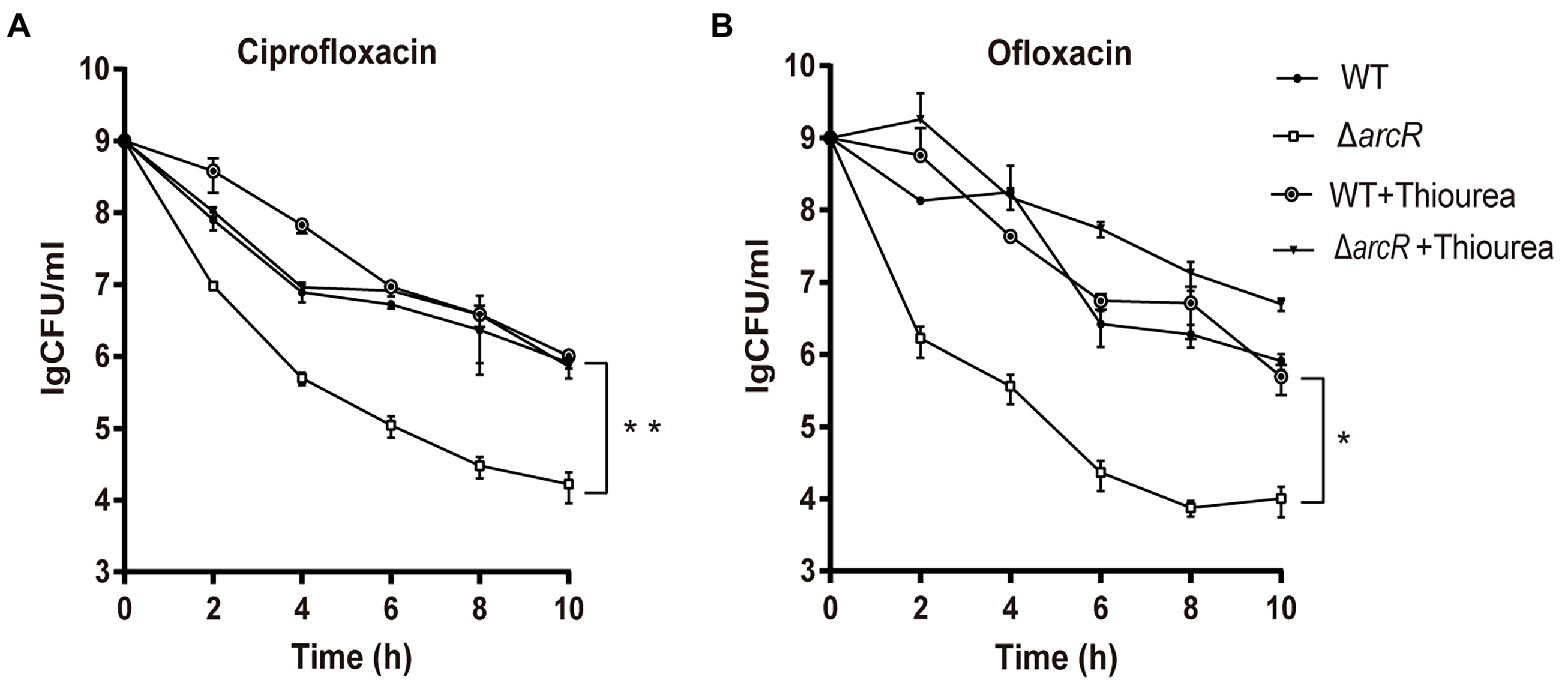
Figure 2. Effects of hydroxyl radical scavenger on bacterial resistance to fluoroquinolone antibiotics. Bacteria were treated with (A) 0.32 μg/ml ciprofloxacin or (B) 0.5 μg/ml ofloxacin in the presence or absence of 150 mM thiourea in TSB at 37°C with agitation. At the indicated times, the live bacterial counts were determined by serial dilution and plating. Error bars represent standard deviations. The data represent results from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 by Student’s t-test.
ArcR is involved in bacterial tolerance to oxidative stress
The bacterial response to oxidative stress is a manifestation of virulence, which can help bacteria to resist ROS produced by phagocytes and enhance their infectivity. Activated neutrophils produce O2•−, H2O2, nitric oxide (NO), and hypochlorite (HOCl), which kill invading pathogenic bacteria through oxidative burst (Forman et al., 2002; van der Veen et al., 2009). To understand the role of ArcR in the bacterial oxidative stress response, bacteria were incubated with neutrophils. The wild type and ΔarcR mutant could introduce the same level ROS produced by neutrophils when they were incubated with neutrophils (Supplementary Figure 2). However, the survival rate of ΔarcR mutant was significantly lower than that of wild-type strain when they were treated with neutrophils (Figure 3A). The presence of ROS-neutralizing N-acetylcysteine (NAC) increased the survival rate of ΔarcR mutant after incubation with neutrophils (Figure 3B). We used H2O2 to verify the role of ArcR in bacterial tolerance to oxidative stress, and the ΔarcR mutant showed higher sensitivity to H2O2 than wild-type strain (Figure 3C). These results suggest that ArcR contributes to bacterial resistance to oxidative stress.
Figure 3. Roles of ArcR in bacterial tolerance to neutrophils and H2O2. (A) Bacteria were incubated with mouse neutrophils at an MOI of 5 in HBSS or (B) HBSS containing 80 mM NAC. Live bacterial counts were determined every hour for 3 h. (C) Bacteria were treated with 0.4 mM H2O2 for 12 h in TSB at 37°C. Live bacterial counts were determined by serial dilution and plating. The survival rate was calculated as the number of CFUs of each strain after individual treatment divided by the number of CFUs in the corresponding sample before treatment. The data represent results from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, by Student’s t-test.
ArcR participates in regulation of oxidative stress resistance genes
The increased sensitivity to ROS suggested that the expression of genes involved in ROS response might be reduced in ΔarcR mutant. PerR is a major global regulator of H2O2 response, which is a repressor of peroxide reaction (Dubbs and Mongkolsuk, 2012). The expression of PerR was not significantly altered in ΔarcR mutant (Figure 4A). We measured the expression of other ROS resistance genes. In the presence or absence of H2O2, mRNA levels of katA, which encodes primary catalase, were approximately fourfold lower in the ΔarcR mutant than in the wild-type strain (Figure 4B). Whereas, the mRNA levels of ahpC, ahpF, dps, and hmp were similar between the wild-type and ΔarcR mutant (Figures 4C–F). Total catalase activity was lower in ΔarcR mutant than that in wild-type strains when treated with or without H2O2, which consistent with the change of katA mRNA levels (Figure 4G). Complement with arcR or katA in ΔarcR mutant restored the catalase to wild-type level (Figure 4G).
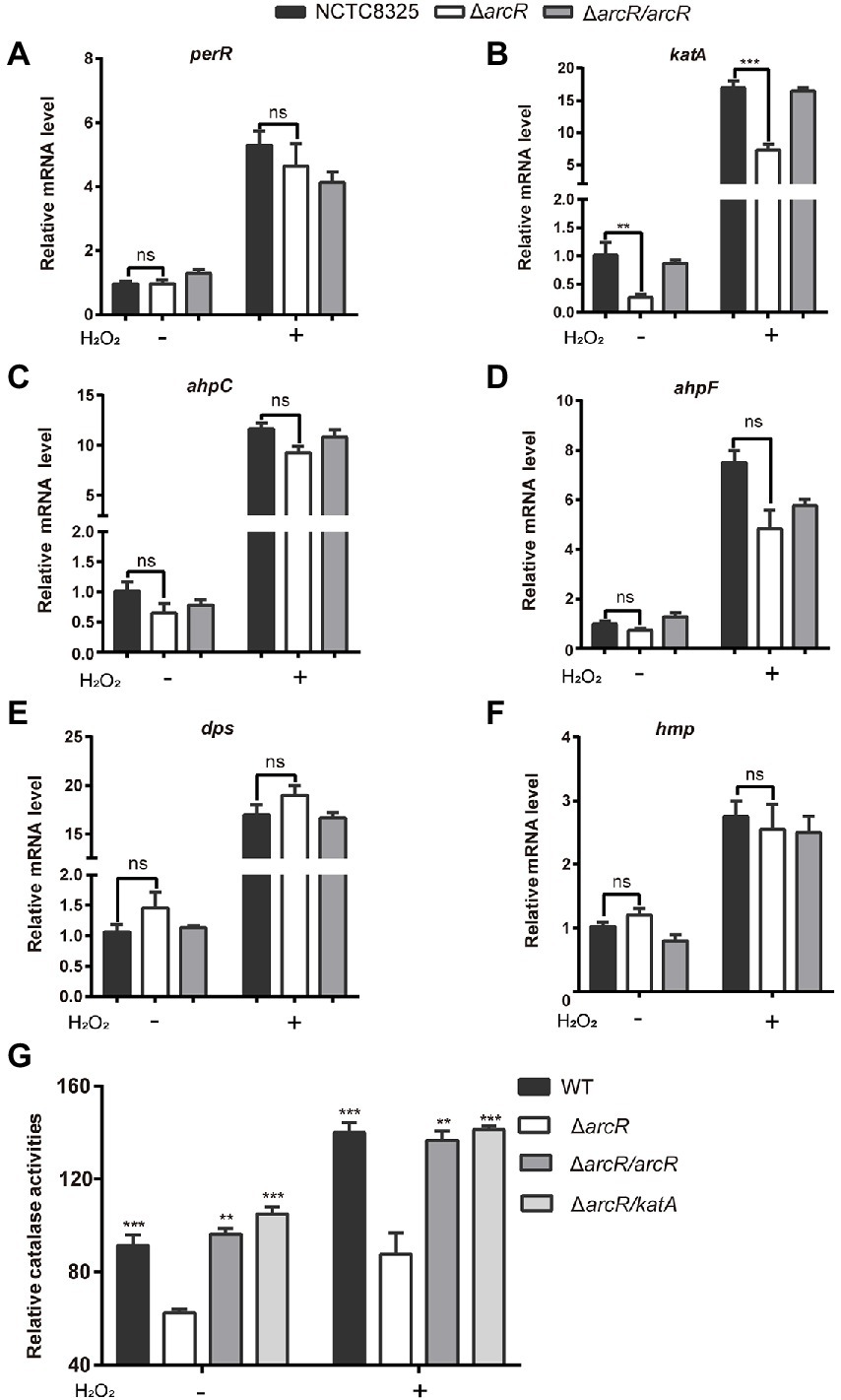
Figure 4. Expression of oxidative stress responsive genes. Bacteria were treated with or without 0.4 mM H2O2 for 10 min in PBS. The mRNA levels of perR (A), katA (B), ahpC (C), ahpF (D), dpS (E), and hmp (F) were determined by real-time PCR. The 16S rRNA gene was used as an internal control. (G) Bacteria were lysed by sonication, and catalase activity was determined with a catalase assay kit. The data represent results from three independent experiments. ns, not significant. *p < 0.05, **p < 0.01, and ***p < 0.001, by Student’s t-test.
Overexpression of katA in ΔarcR mutant strain restored bacterial tolerance to neutrophils, H2O2, ofloxacin, and ciprofloxacin (Figures 5A–D). These results confirmed that downregulation of katA expression was responsible for bacterial susceptibility to H2O2 which increased Fenton reaction and OH• production and finally resulted in decreased tolerance to fluoroquinolone antibiotics.
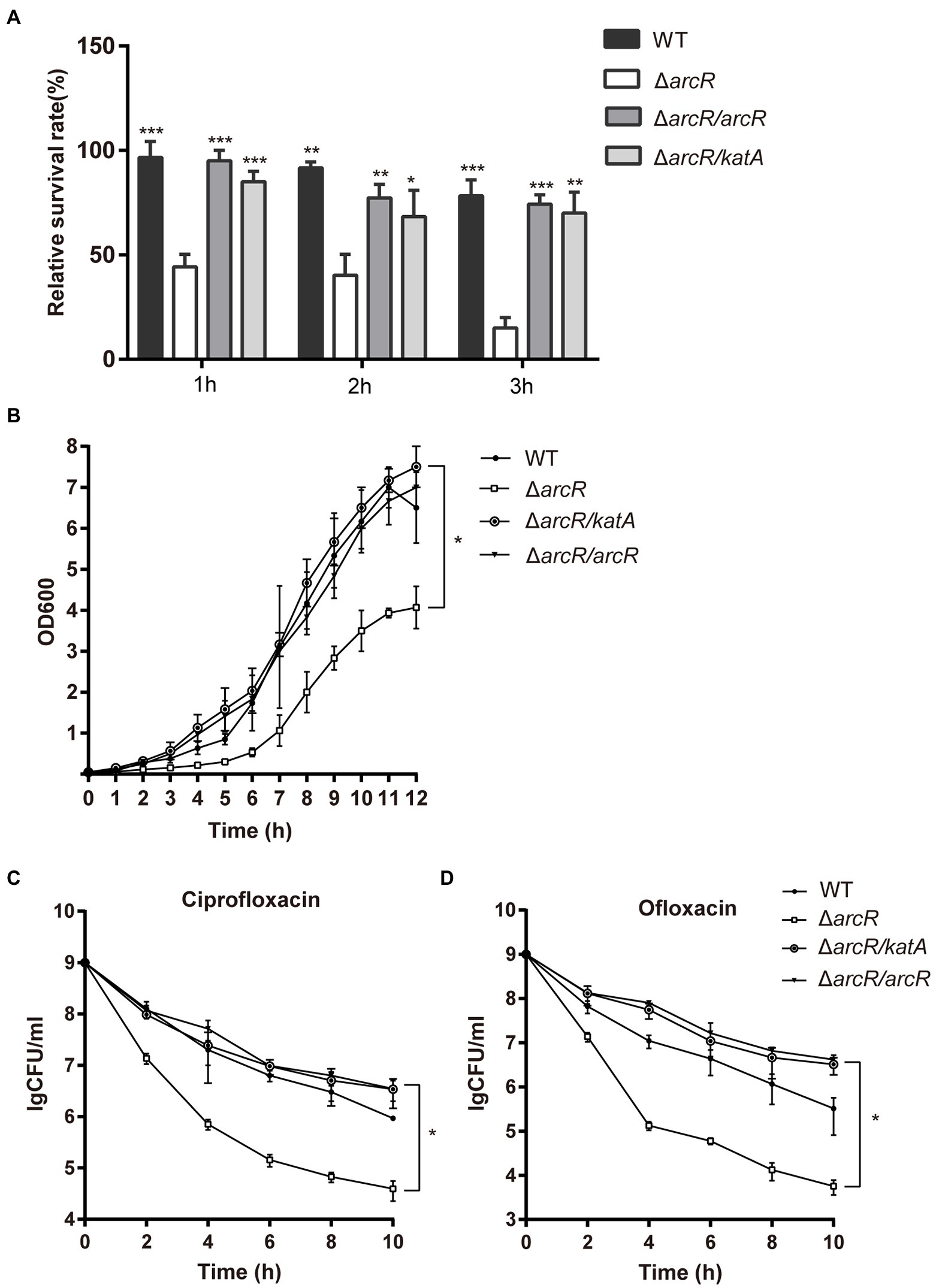
Figure 5. Overexpression of katA restored resistance of ΔarcR mutant against antibiotics and oxidative stress. (A) Bacteria were incubated with mouse neutrophils at MOI 5 in HBSS for 3 h. (B) Bacteria were treated with 0.4 mM H2O2 for 12 h in TSB at 37°C. (C and D) Bacteria were treated with (C) 0.32 μg/ml ciprofloxacin or (D) 0.5 μg/ml ofloxacin for 10 h in TSB with agitation. Live counts were determined by plating. ΔarcR/katA represented the katA overexpression strain, pCN51-katA in ΔarcR mutant. The data represent results from three independent experiments. ns, not significant. *p < 0.05, **p < 0.01, and ***p < 0.001, by Student’s t-test.
ArcR promotes expression of katA at the transcriptional level by directly binding to the katA promoter region
The ArcR regulator of S. aureus contains two domains that are characteristic of members of the Crp family of regulatory proteins. Structural prediction showed that the N terminus of ArcR had a circular nucleotide binding domain, and the C terminus contained a highly conserved helix-turn-helix DNA-binding domain. To explore how ArcR regulated katA transcription, we used pET 28a to express ArcR protein in E. coli BL21. Electrophoretic mobility shift assay (EMSA) was performed to detect the binding of ArcR to 244-bp DNA in the promoter region of katA. The amount of free DNA gradually decreased as the amount of ArcR protein increased (Figure 6A). The specificity of binding was reflected in the fact that DNA mobility was not affected when an unrelated His-tagged TetR protein was used. It also did not bind to 267-bp DNA of tetA promoter region (Figures 6B,C). These results suggested that the binding of ArcR to the katA promoter region was specific.
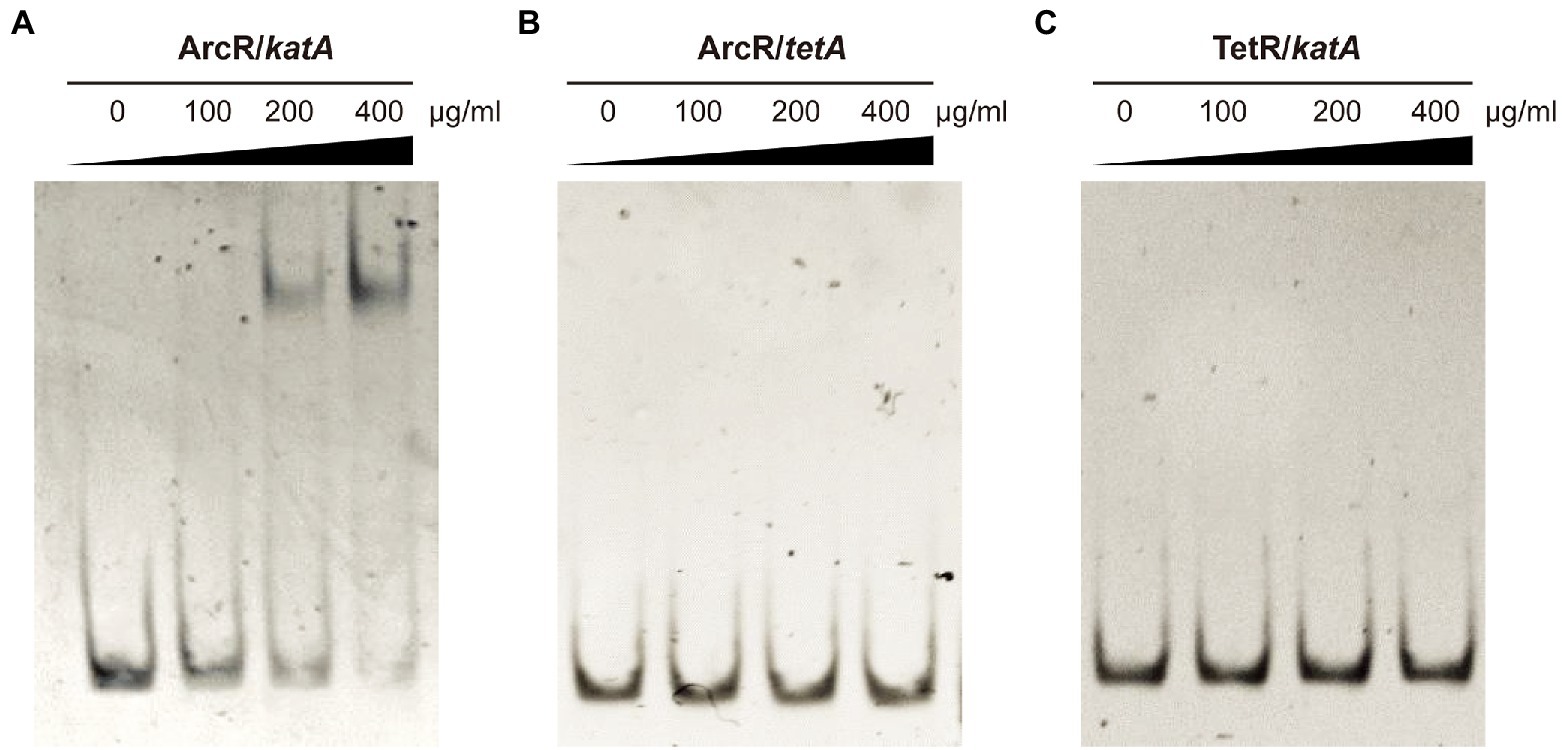
Figure 6. ArcR promotes expression of katA by directly binding to its promoter region. Interaction between ArcR and katA promoter region was examined by EMSA. ArcR was incubated with katA promoter region (A), ArcR was incubated with tetA promoter region (B), and TetR was incubated with katA promoter region (C) at 37°C for 30 min. The mixtures were electrophoresed on agarose gels and the bands were visualized under UV light after ethidium bromide staining. The data represent results from three independent experiments.
Discussion
We found that ArcR controlled the resistance of bacteria to fluoroquinolones by modulating catalase KatA. Enzymes involved in the oxidative stress response have been shown to contribute to antibiotic resistance. KatA and superoxide dismutase are involved in bacterial antibiotic resistance. In Enterococcus faecalis, the absence of superoxide dismutase gene sodA reduced the bacterial tolerance to vancomycin and penicillin (Bizzini et al., 2009). In Acinetobacter baumannii, the sod2343 mutant strains were more susceptible to colistin and tetracycline (Heindorf et al., 2014). In Pseudomonas aeruginosa, katA plays an important role in bacterial tolerance to aminoglycoside and β-lactam antibiotics (Xia et al., 2019). KatA is the major catalase in S. aureus. Here, we demonstrated that KatA was involved in bacterial tolerance to oxidative stress and subsequent tolerance to fluoroquinolone antibiotics. Additionally, KatA affected the survival of S. aureus at low temperature (Masmoudi et al., 2010; Suo et al., 2022).
PerR functions as a major repressed regulator of the response to H2O2 in S. aureus, which replaces OxyR in many Gram-positive bacteria (Mongkolsuk and Helmann, 2002; Dubbs and Mongkolsuk, 2012). In S. aureus, Fur is a regulator interacting with PerR, which participates in the oxidative stress response by positively regulating catalase and iron homeostasis (Horsburgh et al., 2001). Sigma B controls the general stress response, and there are sigma B recognition sites in the katA promoter region, which in some cases regulate katA expression (Horsburgh et al., 2002). ppGpp regulates katA expression in a PerR-independent manner (Horvatek et al., 2020). The srrAB two-component regulatory system can inhibit expression of katA, and the srrA promoter region is predicted by bioinformatics to contain an ArcR binding site (Makhlin et al., 2007; Oogai et al., 2016). To determine whether ArcR also regulated expression of katA by repressing srrAB, we detected expression of srrAB in ΔarcR mutant by quantitative RT-PCR and found that it was not significantly different from that in the wild-type strain. This indicates that ArcR directly regulates expression of katA, and this process is independent of srrAB.
Glucose in combination with aminoglycosides could be used to treat the biofilms of E. coli and S. aureus. Glucose catabolism generated NADH and then NADH was oxidized in the electron transport chain, which, in turn, contributed to PMF. The elevated PMF facilitated the uptake of aminoglycoside antibiotics (Allison et al., 2011; Peng et al., 2015). Cyclic lipopeptide resistance can also be affected by glucose, which induces bacterial resistance to polymyxin B by enhancing glycolytic flux to maintain intracellular ATP levels in PB-treated bacteria. Similar results were observed in S. aureus, where glucose enhanced daptomycin resistance (Yu et al., 2019). In S. aureus, glucose could inhibit the expression of arcR. Meanwhile, arcR affect the response of S. aureus to ROS and then affect the sensitivity to fluoroquinolone antibiotics. Therefore, glucose most likely influences the tolerance of S. aureus to fluoroquinolone antibiotics through ArcR. This study may add to our understanding on the role of carbon metabolism in bacterial susceptibility to antibiotics.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
TF, ZF, and YL designed the experiments, performed the experiments, and wrote the manuscript. JY designed the experiments and revised the manuscript. All authors performed the experiments, analyzed the results, or revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation for Key Programs of China Grants (82130065), National Natural Science Foundation of China (32170201 and 82002191), Beijing Natural Science Foundation (7222014), FENG foundation (FFBR 202103), the Research Foundation of Capital Institute of Pediatrics (CXYJ-2021-04), Public service development and reform pilot project of the Beijing Medical Research Institute (BMR2019-11), and Postdoctoral Research Fund of Chaoyang District, Beijing, China in 2021.
Acknowledgments
We thank Cathel Kerr from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1106340/full#supplementary-material
References
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. doi: 10.1038/nature10069
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Bizzini, A., Zhao, C., Auffray, Y., and Hartke, A. (2009). The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J. Antimicrob. Chemother. 64, 1196–1202. doi: 10.1093/jac/dkp369
Borrero, N. V., Bai, F., Perez, C., Duong, B. Q., Rocca, J. R., Jin, S., et al. (2014). Phenazine antibiotic inspired discovery of potent bromophenazine antibacterial agents against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem. 12, 881–886. doi: 10.1039/c3ob42416b
Charpentier, E., Anton, A. I., Barry, P., Fang, Y., and Novick, R. P. (2004). Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70, 6076–6085. doi: 10.1128/aem.70.10.6076-6085.2004
Cheung, G. Y. C., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569. doi: 10.1080/21505594.2021.1878688
Ding, Y., Onodera, Y., Lee, J. C., and Hooper, D. C. (2008). NorB, an efflux pump in Staphylococcus aureus strain MW2, contributes to bacterial fitness in abscesses. J. Bacteriol. 190, 7123–7129. doi: 10.1128/jb.00655-08
Dubbs, J. M., and Mongkolsuk, S. (2012). Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 194, 5495–5503. doi: 10.1128/jb.00304-12
Dwyer, D. J., Kohanski, M. A., Hayete, B., and Collins, J. J. (2007). Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91. doi: 10.1038/msb4100135
Forman, H. J., Torres, M., and Fukuto, J. (2002). Redox signaling. Mol. Cell. Biochem. 234/235, 49–62. doi: 10.1023/A:1015913229650
Heindorf, M., Kadari, M., Heider, C., Skiebe, E., and Wilharm, G. (2014). Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One 9:e101033. doi: 10.1371/journal.pone.0101033
Hellman, L. M., and Fried, M. G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861. doi: 10.1038/nprot.2007.249
Horsburgh, M. J., Aish, J. L., White, I. J., Shaw, L., Lithgow, J. K., and Foster, S. J. (2002). sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184, 5457–5467. doi: 10.1128/jb.184.19.5457-5467.2002
Horsburgh, M. J., Ingham, E., and Foster, S. J. (2001). In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. doi: 10.1128/jb.183.2.468-475.2001
Horvatek, P., Salzer, A., Hanna, A. M. F., Gratani, F. L., Keinhörster, D., Korn, N., et al. (2020). Inducible expression of (pp)pGpp synthetases in Staphylococcus aureus is associated with activation of stress response genes. PLoS Genet. 16:e1009282. doi: 10.1371/journal.pgen.1009282
Hu, J., Zhang, X., Liu, X., Chen, C., and Sun, B. (2015). Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob. Agents Chemother. 59, 1352–1355. doi: 10.1128/aac.04290-14
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Imlay, J. A., and Fridovich, I. (1991). Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem. 266, 6957–6965. doi: 10.1016/S0021-9258(20)89596-9
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A., and Collins, J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cells 130, 797–810. doi: 10.1016/j.cell.2007.06.049
Körner, H., Sofia, H. J., and Zumft, W. G. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592. doi: 10.1016/s0168-6445(03)00066-4
Kreiswirth, B. N., Löfdahl, S., Betley, M. J., O’Reilly, M., Schlievert, P. M., Bergdoll, M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712. doi: 10.1038/305709a0
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Lowy, F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273. doi: 10.1172/jci18535
Makhlin, J., Kofman, T., Borovok, I., Kohler, C., Engelmann, S., Cohen, G., et al. (2007). Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 189, 5976–5986. doi: 10.1128/jb.00592-07
Masmoudi, S., Denis, M., and Maalej, S. (2010). Inactivation of the gene katA or sodA affects the transient entry into the viable but non-culturable response of Staphylococcus aureus in natural seawater at low temperature. Mar. Pollut. Bull. 60, 2209–2214. doi: 10.1016/j.marpolbul.2010.08.017
Mongkolsuk, S., and Helmann, J. D. (2002). Regulation of inducible peroxide stress responses. Mol. Microbiol. 45, 9–15. doi: 10.1046/j.1365-2958.2002.03015.x
Oogai, Y., Kawada-Matsuo, M., and Komatsuzawa, H. (2016). Staphylococcus aureus SrrAB affects susceptibility to hydrogen peroxide and co-existence with Streptococcus sanguinis. PLoS One 11:e0159768. doi: 10.1371/journal.pone.0159768
Peng, B., Su, Y. B., Li, H., Han, Y., Guo, C., Tian, Y. M., et al. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Singh, V., Pal, A., and Darokar, M. P. (2021). Glabridin synergy with norfloxacin induces ROS in multidrug resistant Staphylococcus aureus. J. Gen. Appl. Microbiol. 67, 269–272. doi: 10.2323/jgam.2021.06.002
Suo, B., Guan, P., Dong, Z., Zeng, Y., Fan, S., Fan, H., et al. (2022). Comparative transcriptomic analysis of Staphylococcus aureus reveals the genes involved in survival at low temperature. Foods 11:996. doi: 10.3390/foods11070996
van der Veen, B. S., de Winther, M. P., and Heeringa, P. (2009). Myeloperoxidase: molecular mechanisms of action and their relevance to human health and disease. Antioxid. Redox Signal. 11, 2899–2937. doi: 10.1089/ars.2009.2538
Walsh, C. (2000). Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781. doi: 10.1038/35021219
Wu, W., Hsu, Y. M., Bi, L., Songyang, Z., and Lin, X. (2009). CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat. Immunol. 10, 1208–1214. doi: 10.1038/ni.1788
Xia, B., Li, M., Tian, Z., Chen, G., Liu, C., Xia, Y., et al. (2019). Oligoribonuclease contributes to tolerance to aminoglycoside and β-lactam antibiotics by regulating KatA in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63:e00212–19. doi: 10.1128/aac.00212-19
Xu, Z., Li, L., Shirtliff, M. E., Peters, B. M., Li, B., Peng, Y., et al. (2011). Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001-2006. Clin. Microbiol. Infect. 17, 714–718. doi: 10.1111/j.1469-0691.2010.03379.x
Yu, D., and Deutscher, M. P. (1995). Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J. Bacteriol. 177, 4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995
Keywords: Staphylococcus aureus, ArcR, fluoroquinolone antibiotics, oxidative stresses, catalase, KatA
Citation: Fu T, Fan Z, Li Y, Li Z, Du B, Liu S, Cui X, Zhang R, Zhao H, Feng Y, Xue G, Cui J, Yan C, Gan L, Feng J, Xu Z, Yu Z, Tian Z, Ding Z, Chen J, Chen Y and Yuan J (2023) ArcR contributes to tolerance to fluoroquinolone antibiotics by regulating katA in Staphylococcus aureus. Front. Microbiol. 14:1106340. doi: 10.3389/fmicb.2023.1106340
Edited by:
Rami M. Elshazli, Horus University, EgyptReviewed by:
Marat R. Sadykov, University of Nebraska Medical Center, United StatesJianhua Yin, Zhejiang University of Technology, China
Copyright © 2023 Fu, Fan, Li, Li, Du, Liu, Cui, Zhang, Zhao, Feng, Xue, Cui, Yan, Gan, Feng, Xu, Yu, Tian, Ding, Chen, Chen and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yuan, ✉ eXVhbmppbmc2MjE2QDE2My5jb20=
†These authors have contributed equally to this work
 Tongtong Fu1†
Tongtong Fu1† Zheng Fan
Zheng Fan Yujie Li
Yujie Li Yanling Feng
Yanling Feng Guanhua Xue
Guanhua Xue Jinghua Cui
Jinghua Cui Chao Yan
Chao Yan Junxia Feng
Junxia Feng Ziying Xu
Ziying Xu Zihui Yu
Zihui Yu Jing Yuan
Jing Yuan