
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 May 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1105924
Phage therapy was taken as an alternative strategy to antibiotics in shrimp farming for the control of Vibrio species of Vibrio parahaemolyticus and Vibrio alginolyticus, which cause substantial mortality and significant economic losses. In this study, a new Vibrio phage vB_ValM_PVA8 (PVA8), which could efficiently infect pathogenic isolates of V. alginolyticus and V. parahaemolyticus, was isolated from sewage water and characterized by microbiological and in silico genomic analyses. The phage was characterized to be a member of the Straboviridae family with elongated head and contractile tail by transmission electron microscopy. Genome sequencing showed that PVA8 had a 246,348-bp double-stranded DNA genome with a G + C content of 42.6%. It harbored totally 388 putative open reading frames (ORFs), among them 92 (23.71%) assigned to functional genes. Up to 27 transfer RNA (tRNA) genes were found in the genome, and the genes for virulence, antibiotic resistance, and lysogeny were not detected. NCBI genomic blasting results and the phylogenetic analysis based on the sequences of the large terminase subunits and the DNA polymerase indicated that PVA8 shared considerable similarity with Vibrio phage V09 and bacteriophage KVP40. The phage had a latent period of 20 min and a burst size of 309 PFUs/infected cell with the host V. alginolyticus, and it was stable over a broad pH range (4.0–11.0) and a wide temperature span (−80°C to 60°C), respectively, which may benefit its feasibility for phage therapy. In addition, it had the minimum multiplicity of infection (MOI) of 0.0000001, which revealed its strong multiplication capacity. The shrimp cultivation lab trials demonstrated that PVA8 could be applied in treating pathogenic V. parahaemolyticus infection disease of shrimp with a survival rate of 88.89% comparing to that of 34.43% in the infected group, and the pond application trails confirmed that the implementation of PVA8 could rapidly yet effectively reduce the level of the Vibrio. Taken together, PVA8 may be potential to be explored as a promising biological agent for Vibrio control in aquaculture farming industry.
Facilitated with the adoption of high stocking density farming practice, the global aquaculture industry has experienced a rapid development worldwide in the past 30 years. Unfortunately, the high stocking density mode is often accompanied by the incidence of pathogenic bacteria caused diseases which results in severe economic losses. Family Vibrionaceae is one of the major sources of the pathogens, which are normal ubiquitous bacteria of the aquatic system and responsible for most of the bacterial outbreaks recorded across the countries (Seghal Kiran et al., 2016). Among them, a panel of species poses as pathogens causing disease in commercially crustacean, shellfish, and fish (Thompson et al., 2004). Vibrio alginolyticus is one of the most famous disease-causing Vibrio species that can infect a variety of aquatic animals, and it has been implicated in several types of mass mortality cases in major aquaculture species, such as fish, mollusks, and crustaceans (Letchumanan et al., 2016). The bacterium is also pathogenic to humans, causing skin and ear infections as well as acute gastroenteritis (Tan et al., 2015). Vibrio parahaemolyticus is another important disease-causing Vibrio species, which is pathogenic to both humans and marine animals (Su and Liu, 2007). It has been considered as a major concern for human health because it often causes acute gastroenteritis when people consume raw seafoods contaminated with this bad bug (Su and Liu, 2007; Iwamoto et al., 2010). Strikingly, V. parahaemolyticus can cause acute hepatopancreatic necrosis disease (AHPND) or early mortality syndrome (EMS), which is one of the most serious diseases in shrimp aquaculture which has led to devasting economic losses in the shrimp industry worldwide (Kalatzis et al., 2018).
Routinely, disinfectants and antibiotics are widely used in farming practice to prevent Vibrio. But these chemicals are becoming ineffective due to the emergence of the drug resistant by Vibrio. Moreover, the use of these chemicals has brought a series of negative effects, such as chemical pollution, the dysbiosis due to accumulated antibiotic residues, and the emergence of resistant bacteria derived in seawater (Kalatzis et al., 2018; Kosznik-Kwaśnicka et al., 2019; Matamp and Bhat, 2019). Therefore, alternative strategies to combat pathogens in aquaculture are in need (Weber-Dabrowska et al., 2016).
Bacteriophages, or phages for short, are viruses that can infect and kill their specific host bacteria. They are highly abundant in marine environments (Rasmussen et al., 2019). As a natural biocontrol method, phage therapy is characterized by high efficiency, specificity, and environmental friendliness. With the emerging of antibiotic resistance, phage therapy has been revitalized and considered as a potential alternative to antibiotic therapy in coping with bacterial infections (Choudhury et al., 2016; Sieiro et al., 2020). Nowadays, phage therapy has been explored in medicine (Onsea et al., 2019; Ooi et al., 2019), agriculture (Svircev et al., 2018; Kahn et al., 2019), food (Clavijo et al., 2019; Vaz et al., 2020), and environmental fields (Ayyaru et al., 2018; Sun et al., 2019). Similarly, phage therapy has great potential in aquaculture against pathogenic bacteria (Le and Kurtböke, 2019; Le et al., 2018). Recently, many successful cases of phage therapy for preventive and therapeutic purposes in aquaculture have been reported, using phages targeting Vibrio species including V. parahaemolyticus (Wong et al., 2019; Tan et al., 2021), V. alginolyticus (Li et al., 2019; Chen et al., 2020), Vibrio harveyi (Misol et al., 2020; Wu et al., 2021), Vibrio vulnificus (Kim et al., 2021), Vibrio anguillarum (Mauritzen et al., 2020), Vibrio campbellii (Lomeli-Ortega et al., 2021), Vibrio mediterranean (Xu et al., 2021) and other species including Pseudomonas aeruginosa (Cafora et al., 2019), Aeromonas hydrophila (Le et al., 2018; Pan et al., 2022; SumeetRai et al., 2023), and Flavobacterium columnare and other fish pathogens.
For phage to serve as biocontrol agent against Vibrio, a full characterization of their genomes is needed so that the presence of potentially harmful genes related to virulence or antibiotic resistance are ruled out as well as their lytic or temperate nature are revealed (Kokkari et al., 2018). To date, approximately hundreds of phages infecting Vibrio hosts have been isolated, completely sequenced, and deposited in the National Center for Biotechnology Information (NCBI) genome database. Most of them are phages infecting V. parahaemolyticus, V. cholerae, and V. vulnificus. Until recently, the complete genomes of several V. alginolyticus phages had been reported, including those myovirus-like phages [VP01 (Sasikala and Srinivasan, 2016), PVA1 (Cui et al., 2014), ValKK3 (Lal et al., 2016), and PVA23 (Fu et al., 2022)], those podovirus-like phages [IM271 (Li et al., 2019), VEN (Kokkari et al., 2018), ϕA318 (Lin et al., 2012), and vB_ValP_VA-RY-3 (Ren et al., 2022)], and those siphovirus-like phages [ValSw3-3 (Chen et al., 2020) and Artemius (Droubogiannis et al., 2022)]. Adding to this list, some potent V. alginolyticus phages have been documented, however, the complete genomic information of them still remain unavailable (Sasikala and Srinivasan, 2016; Le et al., 2020). To enlarge the arsenals for Vibrio control, more fully characterized phages are in need.
Previously, a potent Vibrio phage vB_ValM_PVA8 (PVA8) was isolated from local sewage samples. It exhibited decent efficiency in lysing V. alginolyticus and V. parahaemolyticus. In current study, we reported the phenotypical determination and the genomic characterization of PVA8, together with the evaluation of its application against pathogens including V. alginolyticus and V. parahaemolyticus.
Vibrio parahaemolyticus ATCC 17802 purchased from Guangdong Microbial Culture Collection Center. Other strains used, including V. alginolyticus (Table 1), V. parahaemolyticus (Table 2) and other species of Vibrio and non-Vibrio strains (Table 3), were from the stocks of our lab. All Vibrio strains were isolated from seawater and hepatopancreas of diseased shrimps collected from different regions of China using the methods as reported (George et al., 2005). The Vibrio strains were stored at −80°C in 2216E liquid medium (Qingdao Hope Bio-Technology Co., China) with 30% glycerol. They were sub-cultured from −80°C stocks onto 2216E agar plates (2216E with 2% agar) and cultivated at 37°C for a minimum of 24 h. Unless otherwise specified, the Vibrio strains were cultured under standard conditions: liquid 2216E, 220 rpm aeration, and at 37°C.
A panel of common virulence genes including alkaline serine protease gene (AspA), ferric uptake system gene (fur), and hemolysin genes (tlh, tdh, and trh) of V. alginolyticus and photorhabdus insect related toxin A gene (PirA), virulence-regulating genes (toxR), and hemolysin genes (tlh, tdh, and trh) of V. parahaemolyticus were detected by PCR assay (Kahla-Nakbi et al., 2006; Fethi et al., 2011; Table 4). The PCR amplification was performed under the following conditions: initial denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1.5 min; and a final extension at 72°C for 8 min. Amplified PCR products were visualized on a 1% agarose gel stained with ethidium bromide, running at 90 V for 25 min. The presence of virulence genes was confirmed by gel electrophoresis that showing the corresponding target band by the gel imaging system after repeating the PCR three times.
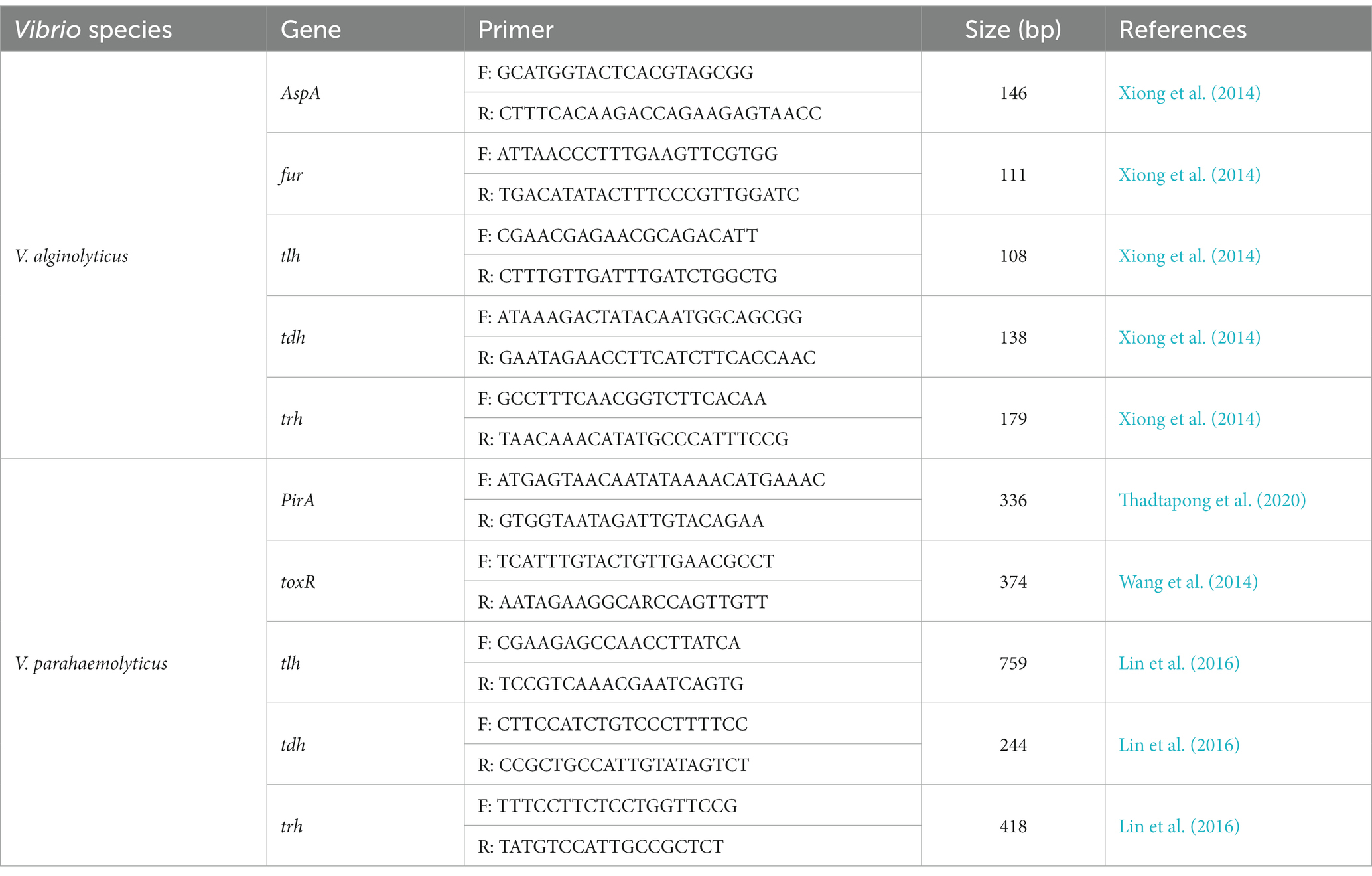
Table 4. Primers used in this study for amplification of virulence genes of the Vibrio alginolyticus and Vibrio parahaemolyticus strains.
Isolation of phages was conducted using the conventional double-layer agar method (Chen et al., 2019). All the strains of V. alginolyticus and V. parahaemolyticus mentioned above were used as hosts to isolate phages. Sewage samples were collected from a large-scale aquatic market in Qingdao, China. First, sewage water (20 mL) was centrifuged at 8,000 × g for 10 min, and filtered through a 0.22 μm membrane to get rid of bacteria. Next, 5 mL of the filtrate and 5 mL of Vibrio culture in logarithmic phase (107 CFU/mL) were added to 50 mL of sterilized 2216E medium. The mixture was incubated at 37°C with orbital shaking for 6 h to enrich possible phages, then it was centrifuged at 8,000 × g for 10 min and filtered through a 0.22 μm membrane to remove the remaining bacteria. 100 μL of the filtrate, mixed with 100 μL of Vibrio culture, was poured into 5 mL of molten soft 2216E upper agar (2216E liquid medium with 0.7% agar) and then poured onto the surface of a prepared and cooled 2216E lower plate (2216E liquid medium with 1.5% agar). Phage plaques appearance was observed after the overnight incubation at 37°C.
A single plaque was picked into 1 mL of sterilized PBS buffer (8 g NaCl, 0.2 g KCL, 1.44 g Na2HPO4 12H2O, 0.24 g KH2PO4, 0.11 g CaCl2, adding water to 1,000 mL and adjusting pH to 7.2 with Na2HPO4 12H2O or KH2PO4) and bathed in water at 40°C for 30 min to harvest phages. Then the phage-containing PBS buffer was centrifuged at 12,000 × g for 5 min and filtered through a 0.22 μm membrane. This filtrate was diluted at a 10-fold gradient with PBS buffer to a suitable multiple which could obtain single plaques on the double-agar plate. The single-plaque isolation process by the double-layer agar method was repeated four more times until plaques of uniform size, shape, and clarity were obtained.
The lysate of the pure single plaque in 100 μL PBS was mixed with equal amount of fresh culture of the host strain, and then was added into 5 mL of 2216E tube and incubated at 37°C with shaking at 220 rpm to getting phage proliferation solution. Cell debris of the solution was removed by centrifugation and filtration. The bacteriophage titer of the filtrate was determined using the double-layer agar method and expressed as plaque-forming units (PFU) per mL filtrate. Finally, the proliferation solution was stored at 4°C for further experiments.
Phage host ranges was determined by the method of standard spot tests assay (Feng et al., 2021). All the strains of V. alginolyticus and V. parahaemolyticus mentioned above were tested. Briefly, 100 μL of each bacterium in logarithmic phase (107 CFU/mL) was added to 5 mL of molten soft 2216E upper agar and then poured onto the surface of a prepared and cooled 2216E lower plate. 2 μL of phage stock (109 PFU/mL) was spotted onto the lawn and incubated at 37°C for 12 h. Finally, the spots observed were classified into four categories based on their clarity, which were clear, slight turbidity, heavy turbidity, and unresponsive, respectively. The test was repeated three times to obtain reliable results.
In addition, a panel of other bacterial species of Vibrio and non-Vibrio, commonly found in pond waters, were also tested to determine the host ranges of the phage using the same method.
The morphology of the phage was characterized by using transmission electron microscopy (Chen et al., 2019). Phage particles were precipitated with 10% polyethylene glycol 8,000 (PEG 8000) at 4°C overnight, centrifuged at 10,000 × g for 15 min, and subsequently suspended in PBS buffer. One drop of the concentrated phage particles was spotted onto the surface of carbon grids for 15 min and then negatively stained with 2% phosphotungstic acid for 10 min. Then the phage was observed using a Hitachi H-7700 biological transmission electron microscope (Hitachi High-Technologies Corporation, Tokyo, Japan) at an accelerating voltage of 80 kV. It was classified and identified on the basis of International Committee on Taxonomy of Viruses (ICTV) guidelines.
The optimum adsorption time of the phage to the host bacteria was determined using methods described previously (Qi et al., 2020). Briefly, 50 μL of phage stock (109 PFU/mL) and 50 ml of log-phase V. alginolyticus VA10 culture (107 CFU/mL) were mixed evenly in the test tube to achieve an MOI at 0.1, then followed by immediate incubation at 37°C with shaking. This moment was defined as t = 0 min, and 1 mL of the mixture was collected at every 1 min before 10 min and every 5 min after 10 min until 30 min (1 tube per time point). The titer of phage particles was conducted using the double-layer agar method. All experiments were performed in triplicate. The percentage of free phage particles was calculated by the ratio of the average number of phage plagues at each time point to the average number of phage plagues at 0 min. The percentage of adsorbed phage particles was obtained by 1 minus the percentage of free phage particles. The time of producing the highest percentage of adsorbed phage particles is the optimum adsorption time of the phage to the host.
The optimal MOI for the phage was determined using methods described previously (Xi et al., 2019). Briefly, phage stock (109 PFU/mL) were added to 5 mL of log-phase V. alginolyticus VA10 culture (107 CFU/mL) to achieve an MOI of 0.0000001, 0.000001, 0.00001, 0.0001, 0.001, 0.01, 0.1, 1, 10, and 100, respectively, and then incubated at 37°C, 220 rpm for 6 h. Culture supernatant was then filtered through a 0.22-μm filter, and the titer of the phage in the supernatant was using the double-layer agar method. The experiment was carried out in triplicate. The MOI resulting in the highest phage titer was considered the optimal MOI of the phage.
In this study, the one-step growth curve experiment was conducted as previously described by Lee et al. (2016) with some modifications (Lee et al., 2016). First, the phage stock (109 PFU/mL) was mixed with the host V. alginolyticus VA10 culture in logarithmic phase (107 CFU/mL) at a MOI of 0.1 and incubated at 37°C for 10 min to allow the phage to adsorb the bacteria. After incubation, the broth was centrifuged at 12,000 × g for 30 s to remove unabsorbed free phage. Next, the pellets were washed with 2216E for twice at 37°C, and the suspension was transferred to 20 ml of 2216E followed by immediate incubation at 37°C with shaking. This moment was defined as t = 0 s, and 200-μL sample was collected every 10 min up to 180 min. The titer of phage particles was conducted using the double-layer agar method. All experiments were performed in triplicate. The burst size of phage was calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells.
The thermal stability assay of the phage was performed as previously described (Shahin and Bouzari, 2017). Phage stock (109 PFU/mL) was incubated at different temperatures of 40°C, 50°C, 60°C, 70°C, and 80°C, respectively. Aliquots of 200-μL were taken at 20, 40, and 60 min during the incubation at each temperature, respectively. In addition, the phage lysate is kept at −80°C for a long time, and aliquots of 200-μL are taken every one month to test the activity. To evaluate the pH stability of the phage, phage stock (109 PFU/mL) was treated with 10 ml PBS buffers with varied pH values (2–13, adjusted with NaOH or HCl) at 37°C for 2 h. Phage titers were determined using the double-layer agar method. The experiments were conducted in triplicate.
To determine the chloroform sensitivity of the phage, PVA8 stock (5 ml, 109 PFU/mL) was mixed with chloroform to get final concentration at 1%, 2%, 3%, 4%, and 5%, respectively, and the mix was incubated at 37°C for 30 min by shaking. After incubation, 200-μL aliquots was taken for phage titer measuring. To determine the impact of Ultraviolet (UV) irradiation, phage stock (109 PFU/mL) was exposed to UV irradiation (20 W, 30 cm) and phage tittering was carried out at irregular intervals before 3 min and every 3 min after 3 min until 30 min. The experiments were conducted in triplicate.
The ability of the phage to lyse the V. alginolyticus cells was determined in in vitro dose-dependent assay. The host V. alginolyticus VA10 culture in logarithmic phase (107 CFU/mL) was mixed with a 10-fold serial dilution of the phage stock (109 PFU/mL) to make MOIs at 0.1, 1, and 10, respectively. The mix was incubated at 37°C for 300 min with orbital shaking. Host culture untouched with the phage was served as the control. Aliquots were collected at 10-min intervals and the optical density (OD600) was measured and recorded. The experiment was conducted in triplicate.
To determine the bacterial phage-insensitive mutation frequency (BIMF), the phage was used to lyse the host V. alginolyticus VA10 culture for resistance observation by following the method previously described (Lu et al., 2003; Gutierrez et al., 2015). The fresh cultures of the host in logarithmic phase (107 CFU/mL, 100 μL) were used to coculture with phage stock (109 PFU/mL, 100 μL) at 37°C for 10 min, the host bacterium without phage was used as the control. After serial dilution, the mixture was coated on TCBS plates and incubated at 37°C for 18 h, and the number of phage-resistant colonies in the plate was recorded (the number of assumed insensitive colonies). Then all the resistant colonies were picked for 2216E broth, and tolerance to the phage was confirmed by the spot assay method mentioned above. The number of colonies without lysis circle was recorded (the number of determined insensitive colonies). BIMF is the ratio of the number of determined insensitive colonies to the total number of colonies. The experiment was conducted in triplicate.
To increase our understanding of the phage at the genomic level, the phage DNA was extracted as previously described (Chen et al., 2019). First, phage particles were treated with DNase I and RNase A (New England BioLab, England) followed by incubating at 37°C for 30 min to remove any exogenous host genomic DNA and RNA. Then phage PVA8 genomic DNA was extracted using the λ Bacteriophage Genomic DNA Rapid Extracted Kit (Shanghai Yuanye Biotechnology Co., Ltd., China) following the manufacturer’s instructions. The purified phage PVA8 DNA was performed on an Illumina NovaSeq sequencing platform (Qingdao Weilai Biotechnology Co., Ltd., China). The filtered reads were assembled using SOAPdenovo by the default parameters. The complete genome of phage PVA8 was finished and then manually inspected.
All open reading frames (ORFs) of the complete phage genome sequence are predicted by the program GeneMarkS (Besemer and Borodovsky, 2005). Annotation and functional analysis of all predicted ORFs were conducted using Subsystem Technology (RAST, https://rast.nmpdr.org/rast.cgi). BLASTp algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi, protein–protein BLAST) against the non-redundant (nr) protein database of the National Center for Biotechnology Information [NCBI; (Song et al., 2009)] was used for further verification of the predicted proteins. The genome circle map was drawn by CGView server (Grant and Stothard, 2008). The genome sequences of 11 other closely related homologous phages were downloaded from NCBI database, and similarities between the genomic sequences were determined using the average nucleotide identity MUMmer (ANIm; Pritchard et al., 2016). Comparative whole-genome maps of phages were also visualized by CGView (Grant and Stothard, 2008). The putative tRNA-encoding genes were searched using tRNAscan-SE (Schattner et al., 2005).1 The genes for virulence and antibiotic resistance were detected by Virulence Factor Database (VFDB)2 and Antibiotic Resistance Database (ARDB),3 respectively (Underwood et al., 2005).
To determine the taxonomy of the phage, ML algorithm in FastTree software4 was used to construct phylogenetic tree based on the nucleotide sequence of the complete genome of phage, and the reliability of the branches was verified (bootstrap, 1,000 replications) after the establishment. Multiple-sequence alignment of the phage with the highest similar reference phages was performed using Easyfig 2.2 (Sullivan et al., 2011).
To validate the therapeutic performance of the phage in controlling the pathogenic bacterial infections in practical applications, 450 healthy 60-day-old shrimp (Lenaeus vannamei; weight = 10 ± 0.5 g), cultured in 20 L glass tanks under appropriate conditions with aquatic water temperature maintaining at 25°C ± 1°C during the study, were equally divided into five groups. Each group contained 90 shrimps and was then divided equally into three parallel subgroups as replicates (30 shrimp per subgroup). The V. parahaemolyticus VP8 culture was poured into the glass tanks to challenge shrimps in the experiment. To prepare the cells of V. parahaemolyticus VP8, the culture in logarithmic phase was spined down and washed with sterile saline for three times and then resuspended to make a stock at a concentration of 2 × 109 CFU/mL by measuring the optical density (OD600) and coating TCBS plates. The phage stock (2 × 1010 PFU/mL) was poured into the glass tanks to treat shrimps. Group 1 was set up without any treatment. Group 2 was supplemented with the phage stock at a final concentration of 2 × 107 PFU/mL in the water but without the challenge of VP8. Group 3 was challenged with VP8 suspension at a final concentration of 2 × 106 CFU/mL in the water for 2 days. Group 4 was treated with 10 mg/L doxycycline after a two-day challenge of VP8 at 2 × 106 CFU/mL. Group 5 received the treatment of the phage stock at a final concentration of 2 × 107 PFU/mL in the water after a 2-day challenge of VP8 at 2 × 106 CFU/mL. The survival rate of the shrimp of each group was subsequently summarized after 7-day cultivation, and the status of shrimps was observed and monitored during the experiment.
The experiment was made in a shrimp farming plant located in Yantai, China, which was farming for shrimps (P. vannamei, average weight = 6 g ± 0.5 g), kept in 30-m3 pond under appropriate conditions (water temperature 26°C ± 1°C, salinity 25‰). The pond water in this plant was changed by 30% every day. To control the propagation of pathogenic Vibrio, the pond was regularly added with agents, such as povidone-iodine, complex iodine, chlorine dioxide and probiotic products, etc., which were stopped at least 72 h before the phage tests.
The experiments contained four groups, and the Vibrio contents of the ponds in each group were detected by coating TCBS plates before the start. Group 1 was the blank control receiving no control agent. Group 2 was supplemented with an iodic disinfectant (found in market) at dose of 5 mg/L. Group 3 was supplemented with a probiotic product (Qingdao Bioantai Biological Technology Co., LTD, Implementation standard Q/370212BAAT001-2019) at dose of 5 mg/L. Group 4 was supplemented with the phage stock (1010 PFU/mL) with the MOI at 0.1. Each group of the tests repeated twice, with pond water sampled every 4 h and the Vibrio content checked.
The phage was preserved in the China General Microbiological Culture Collection Center (Beijing, China) with CGMCC number of No.24098. The complete genome sequence of the phage has been submitted to the NCBI GenBank database under accession number of OQ164647.
Data were analyzed by one-way analysis of variance (ANOVA) using GraphPad Prism 8.0. Significant differences were determined using Tukey’s Multiple Comparison Test. Statistical significance was considered at the p < 0.05 level, and the results were expressed as mean ± standard deviation (SD).
In this work, up to 40 V. alginolyticus strains and 28 V. parahaemolyticus strains were used. In order to investigate the virulent features of these strains, a panel of virulence-associated genes including alkaline serine protease gene (AspA), ferric uptake system gene (fur), and hemolysin genes (tlh, tdh, and trh) of V. alginolyticus and photorhabdus insect related toxin A gene (PirA), virulence-regulating genes (toxR), and hemolysin genes (tlh, tdh, and trh) of V. parahaemolyticus, were checked by PCR assay. For the V. alginolyticus strains (Figure 1A), AspA and fur were the most prevalent genes with the detection frequencies of 100% (40/40), followed by tlh (92.5%, 37/40), trh (37.5%, 15/40), and tdh (27.5%, 11/40), respectively. All the V. alginolyticus strains carried virulent genes of AspA and fur, and the majority also carried the tlh gene. Among them, two strains, V. alginolyticus VA15 and VA17, carried all the five virulence genes. For the V. parahaemolyticus strains (Figure 1B), toxR and tlh were the most prevalent genes with the detection frequencies of 100% (28/28), followed by PirA (14.29%, 4/28) and trh (3.57%, 1/28), respectively, while the tdh gene had the detection frequency of 0% (0/28). Among them, four V. parahaemolyticus strains, VP5, VP7, VP8, and VP15, carried the PirA virulence gene, thus they were selected as the presentative strains for later phage lysis testing.
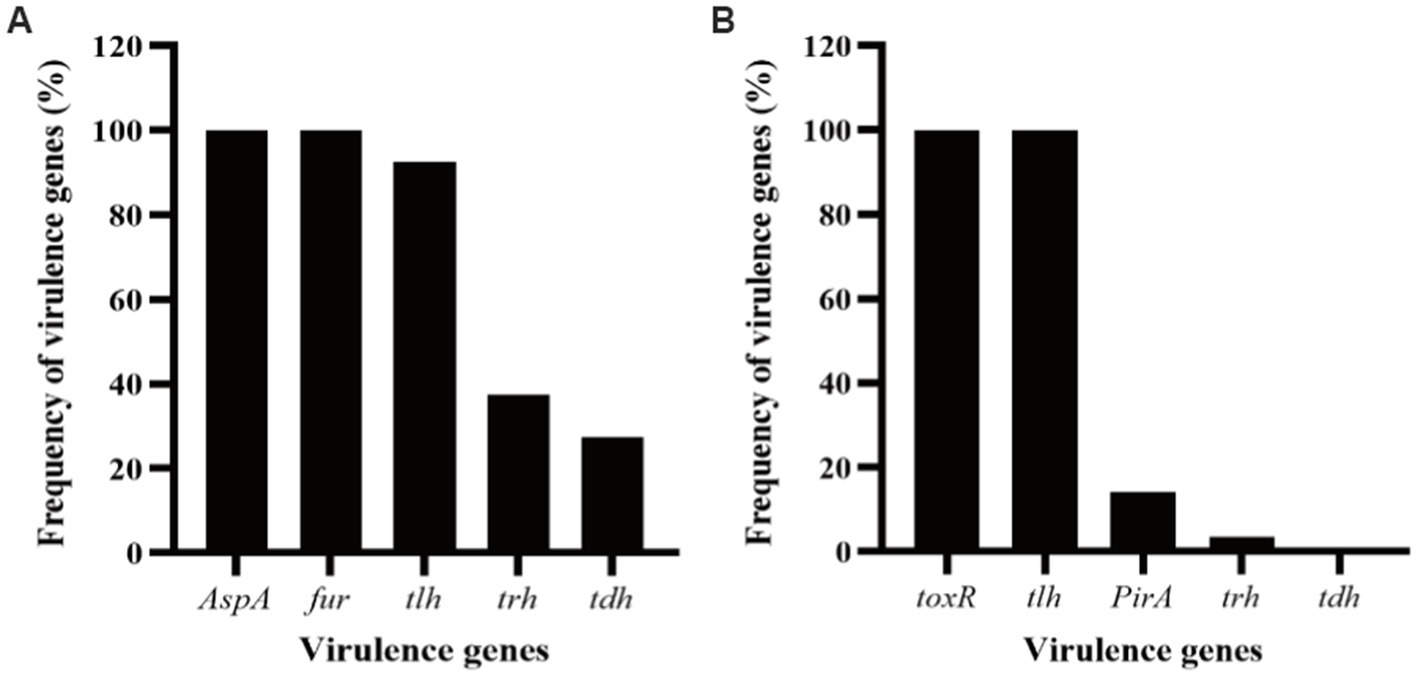
Figure 1. Frequency of virulence genes detected in the strains of Vibrio alginolyticus and Vibrio parahaemolyticus. (A) Five virulence genes, AspA, fur, tlh, tdh, and trh, were selected for PCR detection for the V. alginolyticus strains. (B) Five virulence genes, toxR, PirA, tlh, tdh, and trh, were selected for PCR detection for the V. parahaemolyticus strains.
Sewage samples were collected from a large-scale aquatic market in Qingdao, China, and the V. alginolyticus and V. parahaemolyticus strains mentioned above were used to isolate and screen phages. Using these Vibrio strains as hosts, 124 bacteriophages were successfully isolated and purified from the sewage samples, including 63 V. alginolyticus-infecting bacteriophages (Supplementary Table S1) and 61 V. parahaemolyticus-infecting bacteriophages (Supplementary Table S2). These phages formed clear plaques on the double-layer plates after overnight incubation at 37°C. The host ranges of the isolated phages were then determined using these Vibrio strains by the method of spot tests. According to the results (Supplementary Tables S1, S2), a phage, named PVA8, with the VA10 as the host was screened for its high lysis rate of 82.5% (33/40) to the V. alginolyticus strains (Table 1). Then, we found that it could also effectively infect the V. parahaemolyticus strains with the lysis rates of 71.43% (20/28; Table 2).
In addition, the result of the host ranges of PVA8 using a panel of bacterial species of the other Vibrio and non-Vibrio is shown in Table 3. Except the Vibrio species of V. alginolyticus and V. parahaemolyticus, PVA8 could also infect other Vibrio species, but it did not exhibit any infectivity to bacterial species in genus other than Vibrio. This cross-species lytic ability and specificity for Vibrio species of phage PVA8 can be used to control different pathogenic Vibrio species and is a suitable choice for further study.
Phage PVA8 formed clear plaques on the lawn of the V. alginolyticus VA10, with an averaged diameter about 0.5 mm after 8 h incubation (Figure 2A). Purified phage was subjected to transmission electron microscopy observation. It was found that PVA8 had an elongated head (prolate capsid) with about 139.34 nm long and 71.72 nm wide and a contractile tail with about 112.45 nm long (Figure 2B). Based on the classification standards issued by International Committee on Taxonomy of Viruses (ICTV), PVA8 was identified as one member of the Straboviridae family.
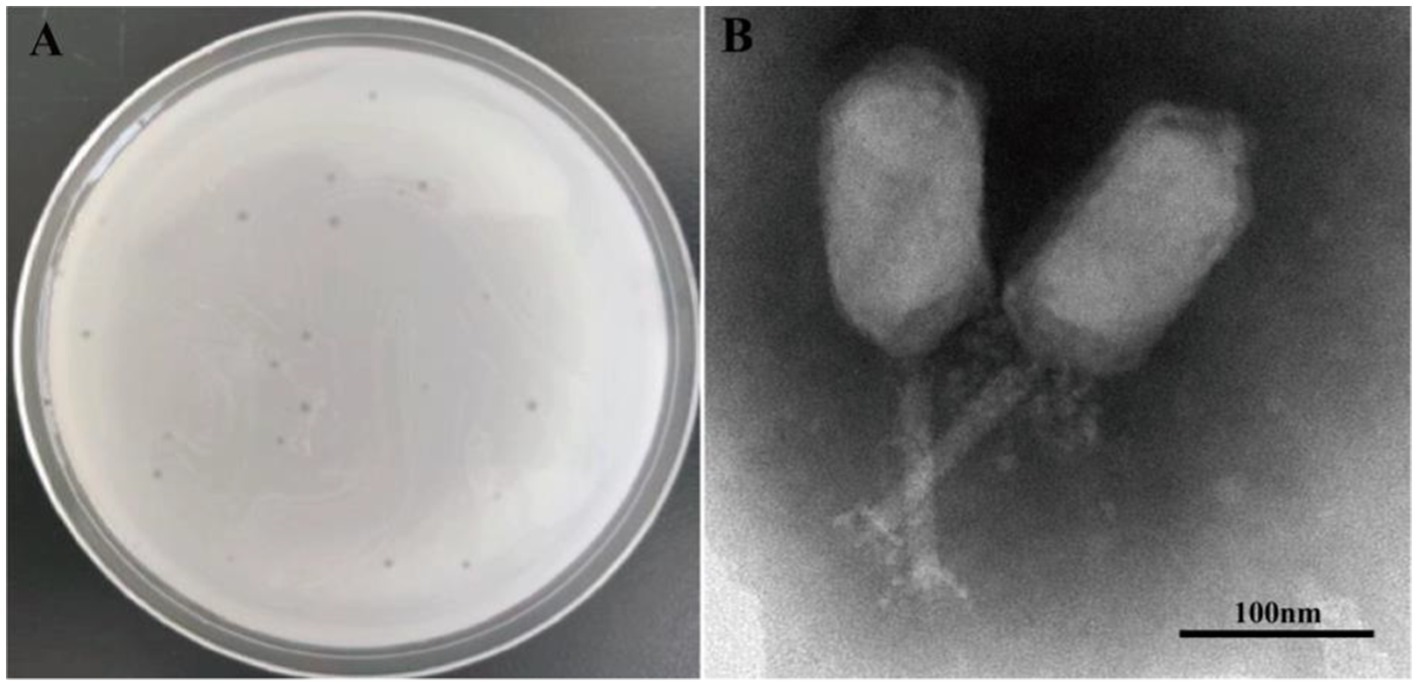
Figure 2. Phage plaques and transmission electron microscopy of phage PVA8. (A) Phage plagues formed on double-layer agar plate of Vibro alginolyticus VA10; (B) Transmission electron microscopy of PVA8, scale bar = 100 nm.
The percentage of adsorbed phages of the mixture reached to (88.5 ± 5.3)% at 8 min (Table 5), and after that the progeny of the phages began to release. Thus, the percentage of adsorbed phages could not be calculated since 9 min. The percentage of adsorbed phages of the mixture is the highest at 8 min, which is the optimum adsorption time of PVA8 to the host V. alginolyticus strain VA10.
With V. alginolyticus strain VA10 as the host, the highest titer of PVA8 amounted to 2.79 × 1012 PFU/mL at MOI of 0.001, and the lowest MOI of PVA8 was as low as 0.0000001, indicating that the phage could achieve high yield yet strong proliferative capacity (Figure 3A).
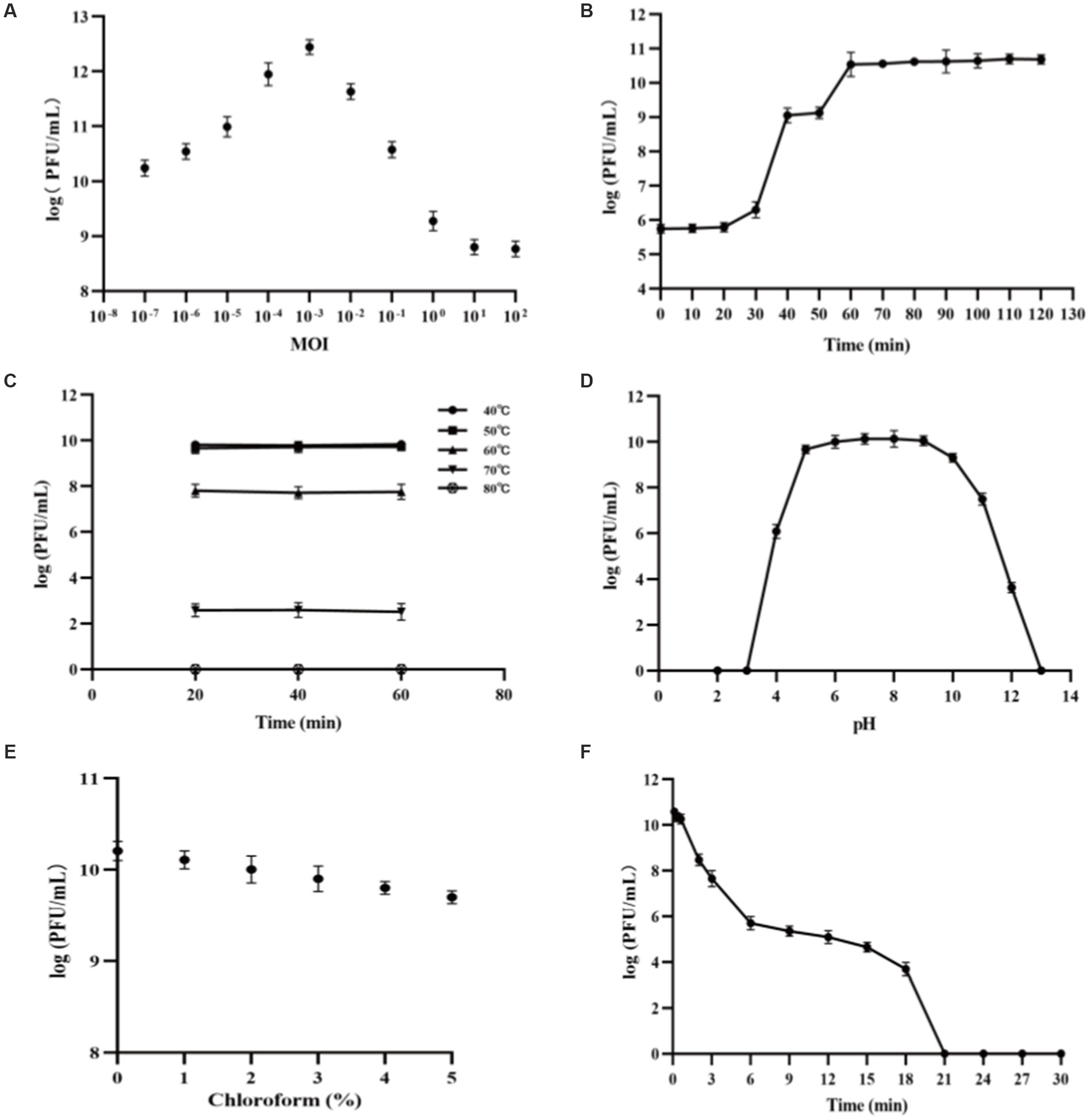
Figure 3. Biological characteristics and stability features of PVA8. (A) The MOI test of PVA8; (B) The one-step growth curve of PVA8; (C) The thermal stability test of PVA8; (D) The pH stability test of PVA8; (E) Chloroform sensitivity test of PVA8, and (F) Ultraviolet (UV) sensitivity test of PVA8.
PVA8 was confirmed to have very strong lysis ability by the one-step growth curve determination, in which a short latent time (20 min) and a big burst size (309 PFUs/infected cell) were demonstrated, respectively (Figure 3B).
The thermal stability assay showed that PVA8 was fairly stable at temperatures below 60°C, but it was completely inactivated as temperature was at 80°C (Figure 3C). In addition, the titer of this phage remains stable for a long time in fridge at −80°C, indicating that it can be stored at this temperature for long time.
The pH stability test showed that PVA8 remained active over a broad pH range, from 4.0 to 12.0, for at least 2 h, but it was completely inactivated under harsh pH conditions (below pH 3.0 or above pH 13.0; Figure 3D).
As shown in Figure 3E, PVA8 was sensitive to the chloroform treatment by a concentration-dependent mode, which indicated the phage capsid protein might contain lipid-like structure.
The survival curve of PVA8 under UV irradiation was depicted in Figure 3F. The results demonstrated that PVA8 was sensitive to the UV irradiation, by following a time based PFU declination trend as observed. It was completely inactivated at 21 min after the treatment.
In vitro tests were conducted to evaluate the lysis potential of phage PVA8, using V. alginolyticus VA10 strain as the host and by applying three MOIs at 0.1, 1, and 10, respectively (Figure 4). In the control, it was observed that the OD600 kept increasing and it amounted to 1.44 at 300 min. In contrast, the OD600 of all the phage treatments reduced significantly after the initial growth in the first few hours, indicating the lysis of the host cells caused by PVA8. It was also noticed that the lysis time was negatively correlated to the MOIs with a rough S-shape OD600 trend observed for each MOI, respectively. The OD600 rebounding might reflect the establishment of host resistance to the phage infection via some undefined escaping ways such as mutation or alteration.
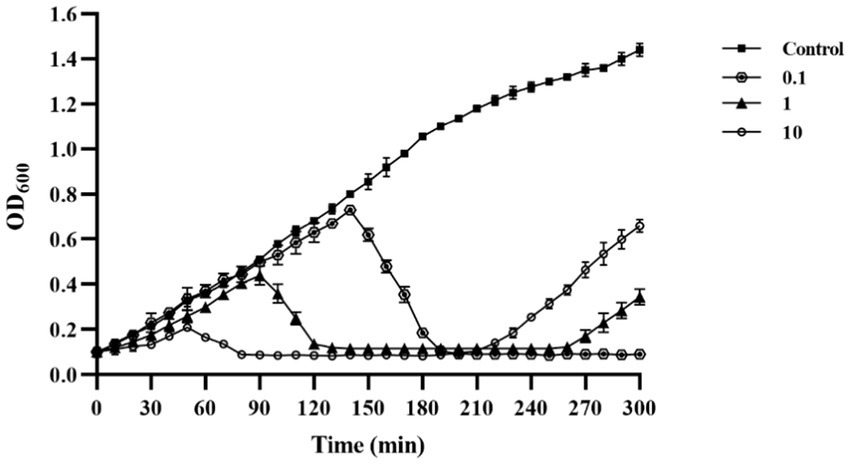
Figure 4. Lysis ability in vitro of phage PVA8 against the host organism VA10 at three different MOIs (10, 1, and 0.1).
In order to investigate the rebounding of the host strain, the bacterial phage-insensitive mutation frequency (BIMF) test was conducted. As shown in Table 6, the BIMF of V. alginolyticus VA10 to PVA8 was (6.39 ± 0.78) × 10−7, which was similar to those seen in previous reports (Gutierrez et al., 2015). This result reasoned why the rebounding of the host bacteria in the phage application experiment as observed, which implied that a certain mutation might occur when the target Vibrio strain was challenged with the phage. Thus, for the control of V. alginolyticus VA10, phage cocktails were suggested in practice to reduce the possible resistance derived due to mutations in the targeted Vibrio.
The complete genome of PVA8 was 246, 348 bp long, with 99.63% of the reads matched to the complete genome (3,909,018 out of 3,923,346).
The genome circle map of phage PVA8 with NR annotations is shown in Figure 5. Sequencing analysis showed that the genome was a linear, double-stranded DNA molecule of 246, 348 bp in length with a GC content of 42.6%. It assigned 388 coding sequences (CDSs) with online RAST annotation. Among the 388 CDSs, 378 CDSs begin with the start codon ATG, only seven begin the start codon TTG, and three begin with the start codon GTG. Blastp analysis against the NCBI nr database showed that 92 ORFs (23.71%) were annotated to have corresponding functions. These ORFs related to the basic functions of phages can be identified and categorized, which covered proteins as DNA replication/modification, structure protein, packing protein, metabolism/regulation, and host lysis, respectively.
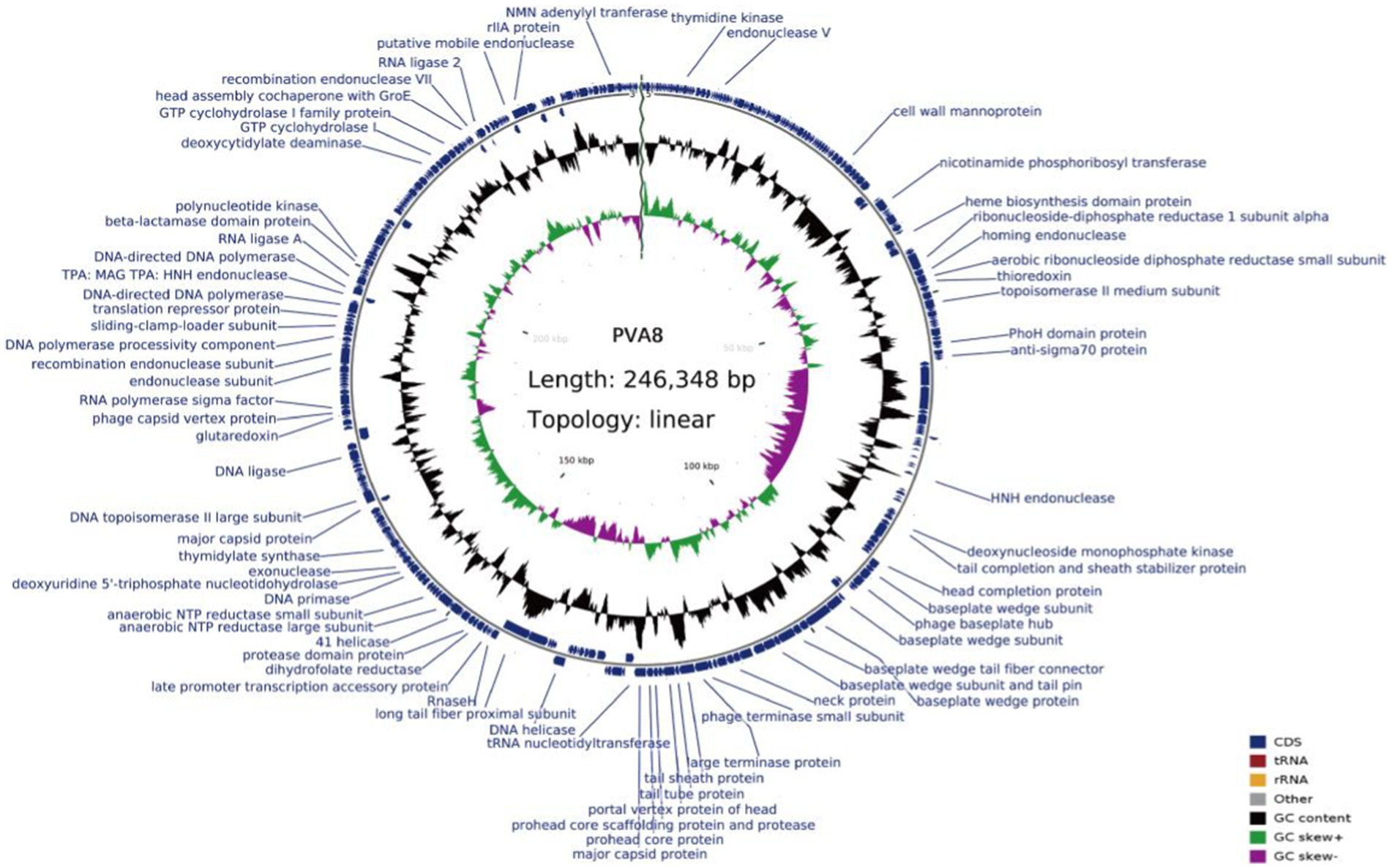
Figure 5. Genome circle map of phage PVA8. From inside to outside, the first circle represents the scale, the second circle represents GC Skew, the third circle represents G + C content, and the outermost circles represent the positions of CDs on the genome. In the second circle, the outward direction indicates that the GC-skew [(G − C)/(G + C)] is larger than zero, and the inward direction indicates that the GC-skew [(G − C)/(G + C)] is lower than zero. In the third circle, the outward direction indicates that the G + C content of this region is larger than the average G + C content of the whole genome, and the inward direction indicates that the G + C content of that region is lower than the average G + C content.
Several typical enzymes and proteins in DNA replication/modification module were predicted in the genome of PVA8, which are similar with those of other Vibrio phages. ORF207 was predicted to encode ribonuclease H, an enzyme that cleaves the RNA strand of an RNA/DNA hybrid (Ohyama et al., 2014), showing 100% identity to that of bacteriophage KVP40. DNA polymerase encoded by ORF290 has 97.17% homology to that of Vibrio phage V09, which can be used to fill DNA gaps generated during DNA repair, recombination, and replication. DNA helicase, an enzyme that utilizes ATP hydrolysis to separate the DNA double helix into individual strands, was encoded by ORF200, showing 99.8% identity to that of Vibrio phage phi-pp2. DNA primase encoded by ORF226 has 100% identity to that of bacteriophage KVP40, which can initiate DNA synthesis. In addition, enzymes involved in nucleotide metabolism including the exonuclease and the endonuclease, were encoded by ORF229 and ORF279, respectively, both showing 100% identity to those of bacteriophage KVP40. These enzymes and proteins were responsible for generating deoxyribonucleotides for phage DNA synthesis by hydrolyzing host genomic DNA and RNA (Gao et al., 2020). In structural protein module, ORF186 was predicted to be a major capsid protein with 100% identity to that of bacteriophage KVP40, and the typical tail structural proteins including tail tube protein, tail sheath protein, and baseplate protein were encoded by ORF180, ORF179, and ORF 163–179, respectively. In the packing protein module, the large terminase subunit was encoded by ORF178 with 100% identity to that of bacteriophage KVP40, and the small terminase subunit was encoded by ORF177 with 100% identity to that of Vibrio phage phi-pp2. ORF181 was predicted to encode a portal protein with 100% identity to that of Vibrio phage phi-pp2, which can infect DNA into the host cell through a pathway formed by portal protein (Yasui et al., 2017). Just as reported (Paul et al., 1998), terminase plays an important role in the DNA packing processes of the phage genome. The large terminase subunit plays a major role in packaging, while the small terminase subunit binds specifically to the genome and directs the packaging process. The proteins for metabolic and regulated functions were found in ORF18 and ORF237, which encoded thymidylate kinase and thymidylate synthase, respectively, involving in the metabolism of nucleic acids and show100% identity to that of bacteriophage KVP40 and Vibrio phage phi-pp2, respectively. In addition, protein rllA, the one affecting or avoiding the lysis of another phage, was encoded by ORF360 with 95.36% identity to that of Vibrio phage phi-pp2. It allows the cell infected by the first invader to avoid or exclude the invasion of another phage virion (Moussa et al., 2014).
In the genome of PVA8, the genes for virulence, antibiotic resistance, and lysogeny were not found, suggesting that this phage could be used to control Vibrio without much safety concern. According to the analysis using tRNAscan-SE, totally up to 27 tRNAs were found in the genome of PVA8, which was on the high end of tRNA numbers as found in other similar phages genome. The high number of tRNA may benefit the phage in the synthesis of its own proteins (Lu et al., 2020).
The BLASTn comparison result showed that the genome of PVA8 had high nucleotide homology with V. parahaemolyticus-infecting bacteriophage KVP40 (98.99% identity, 97% coverage), V. parahaemolyticus-infecting phage V09 (98.72% identity, 96% coverage), V. parahaemolyticus-infecting V07 (98.71% identity, 96% coverage), Vibrio natriegens-infecting phage VH1_2019 (98.48% identity, 95% coverage), V. parahaemolyticus-infecting phage V05 (98.17% identity, 92% coverage), and V. parahaemolyticus-infecting and V. alginolyticus-infecting phage phi-pp2 (98.16 identity, 96% coverage), respectively. By using the ANI heatmap analysis, PVA8 was aligned with a panel of bacteriophages including KVP40, V09, V07, VH1_2019, V05, and phi-pp2, with the ANIm percentage identity each of the phages derived as 98.30%, 98.49%, 97.95%, 97.44%, 97.40%, and 97.70%, respectively (Figure 6). CGView tool was employed to charting comparative map of the whole-genomes of phage PVA8 and the most similar reference phages selected from the NCBI database. As presented in Figures 7, we have used the phage PVA8 as a reference genome and BLAST with the most similar reference phages selected from the NCBI database. The similarities among these phages were plotted by a solid color circle and dissimilarities among the phages were plotted by a white color circle. For the seven phages, high similarities have been observed.
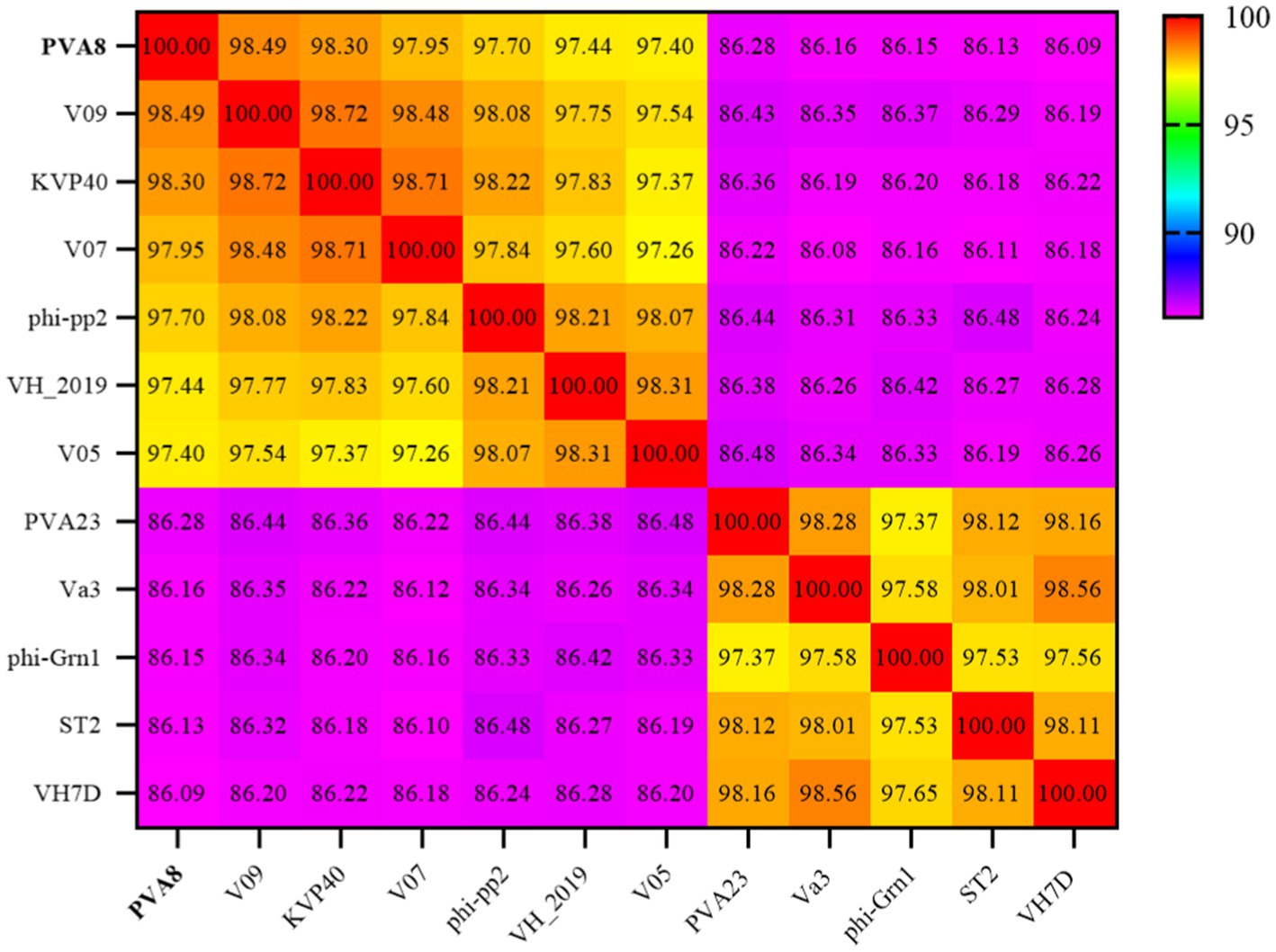
Figure 6. Average nucleotide identity heatmap. The percentage identity values range from 86.09% (purple) to 100% (red).
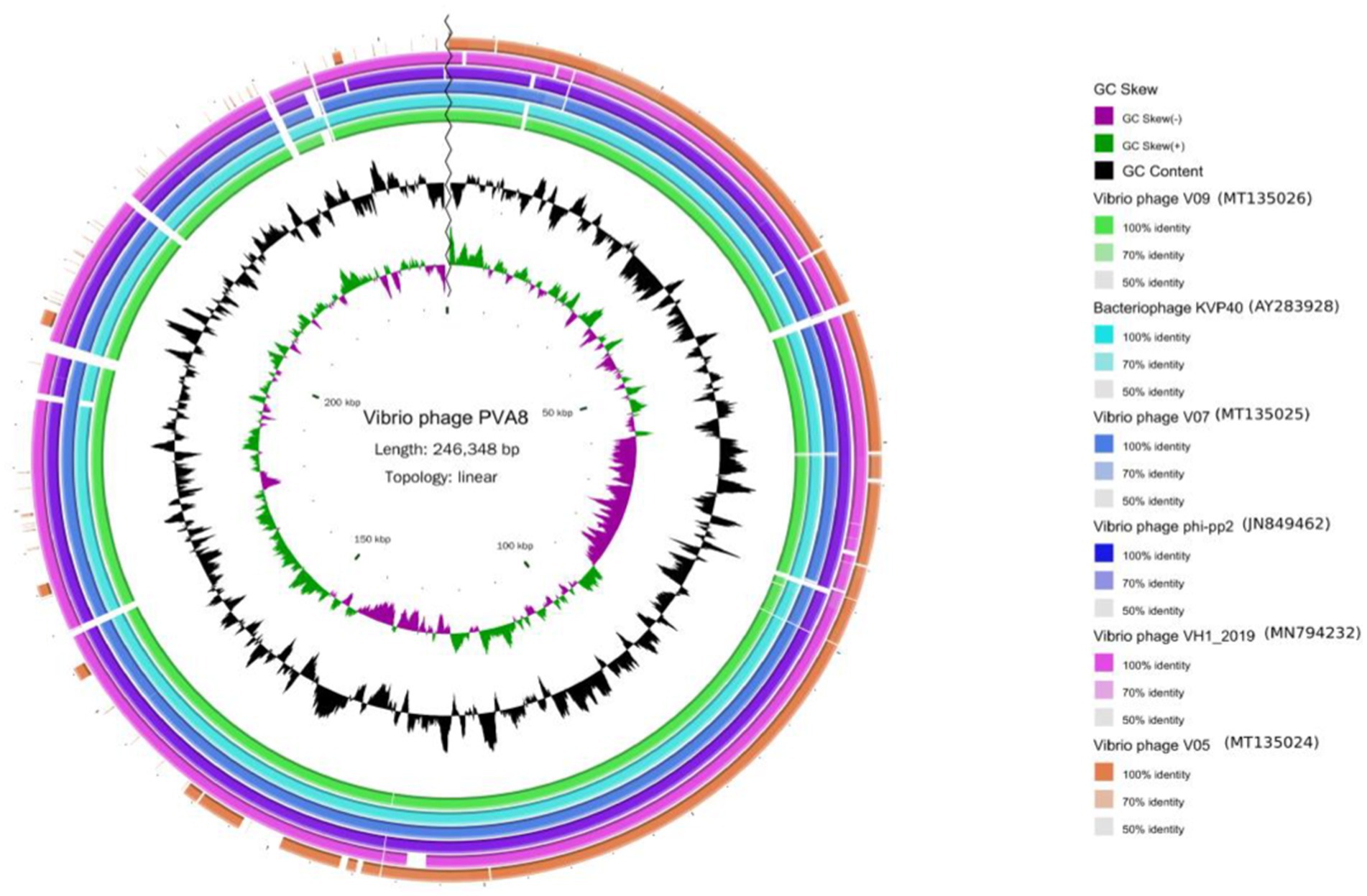
Figure 7. The genomic comparison map between phage PVA8 and six other most similar reference phages. From the outside to the inside: (1) The first circle represents the Vibrio phage V05 sample; the second circle represents the Vibrio phage VH1_2019 sample; the third circle represents the Vibrio phage phi-pp2 sample; the fourth circle represents the Vibrio phage V07 sample; the fifth circle represents the bacteriophage KVP40 sample and the sixth circle represents the Vibrio phage V09 sample. (2) GC content represented by the seventh circle. (3) G + C skew represented by the eighth circle. (4) The ninth circle is the scale.
In order to decipher the evolutionary relationships of PVA8 with the other closely related homologous phages, sequences of the complete genome of PVA8 were aligned with that of the other closely related phages downloaded from NCBI database. The phylogenetic tree of the complete genome (Figure 8) demonstrated that phage PVA8 was more closely related to V07 and V05, forming a clear clade and showing the homologous coverage with 89%, and following V09 and KVP40. The comparison of whole genomic sequence alignments of PVA8, KVP40, and V09 was conducted, which enabled the depiction of the relationships of the phages at the genomic levels. As aforementioned, the ORFs assigned to basic phage-related functions can be identified and categorized. Therefore, we categorize the genes of PVA8 into five groups: DNA replication/modification, structure and packing proteins, metabolism/regulation, host lysis, and hypothetical protein. Similarly, the genome sequences of KVP40 and V09 were down loaded from NCBI and grouped in the same way. As shown in Figure 9, PVA8 was homologous to KVP40 and V09 in a very big portion of the genome, and the nucleotide identities were both not less than 69%.

Figure 8. Phylogenetic analysis based on the nucleotide sequence of the complete genome of phage PVA8. The phylogenetic tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replicates.
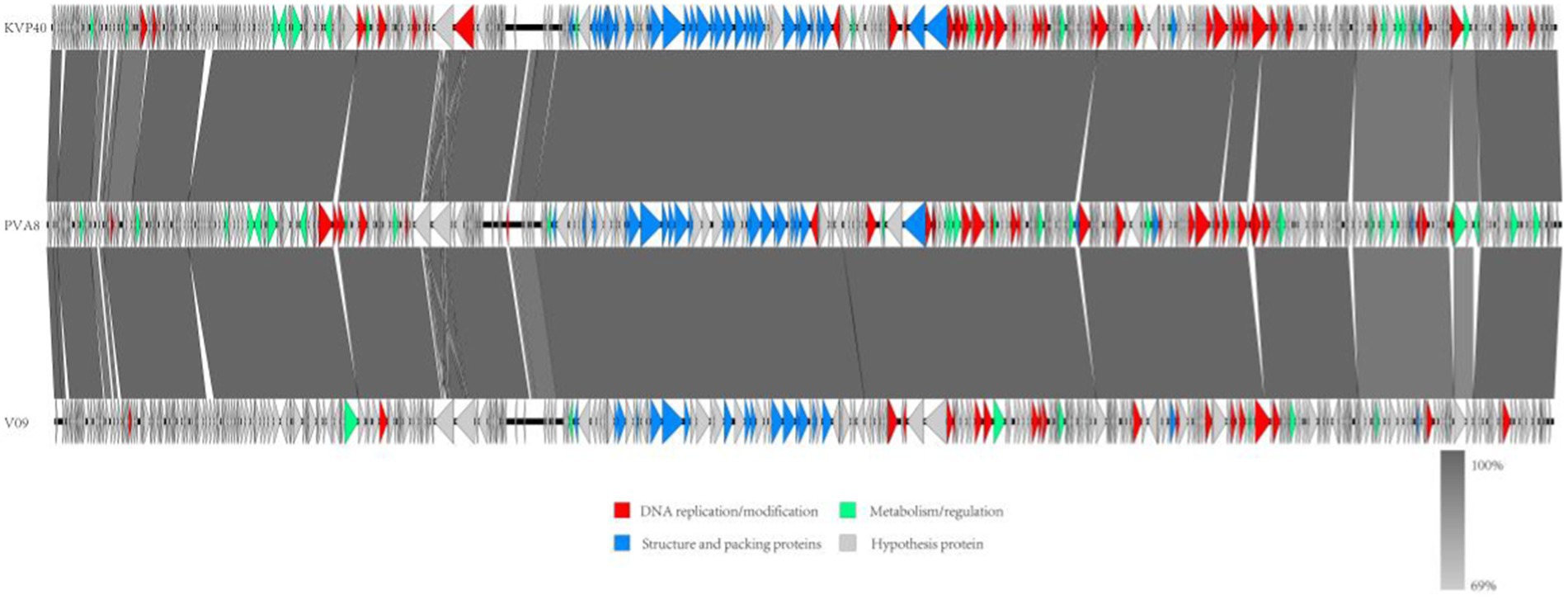
Figure 9. Comparative genome map of a multiple-genome comparison (amino acid level) performed using Easyfig software. Open reading frames (ORFs) are shown as arrows to indicate the direction of transcription and are colored in accordance with their predicted functions: DNA replication/modification (red arrows), structure and packing proteins (blue arrows) and metabolism/regulation (green arrows). Gray arrows represent ORFs of unknown functions (hypothetical protein). Gray shading indicates nucleotide identity between sequences (69%–100%).
The laboratory shrimp cultivation trials were conducted to evaluate the effectiveness of phage PVA8 against the pathogenic infections of the shrimp. In the trials, one blank control group and four experimental groups were arranged. It was observed that in the blank group shrimp bodies were transparent, rigid and cyan-black in color, with clearly-edged and brownish black hepatopancreas shaped like an inverted gourd. The survival rate of the shrimp in this group was 100% (G-1 in Figures 10A,B). When the shrimp challenged and infected with VP8, the skin color of the infected shrimp became white and turbid and the body was soft, meanwhile the hepatopancreas was enlarged and light yellow in color. The shrimp survival rate in this group declined to 34.43% in 7 days (G-3 in Figures 10A,B). In comparison, in the group with phage treatment, the shrimp survival rate amounted to 88.89% (G-5 in Figures 10A,B), which was comparable or superior to that of the antibiotics group (80%; G-4 in Figures 10A,B). To exclude the adverse effect of PVA8 to shrimp, phage alone was used for the injection. The survival rate in G-2 was 98.89% (G-2 in Figures 10A,B), very close to that of group 1, suggesting that phage alone does not bring about adverse effects to shrimp.
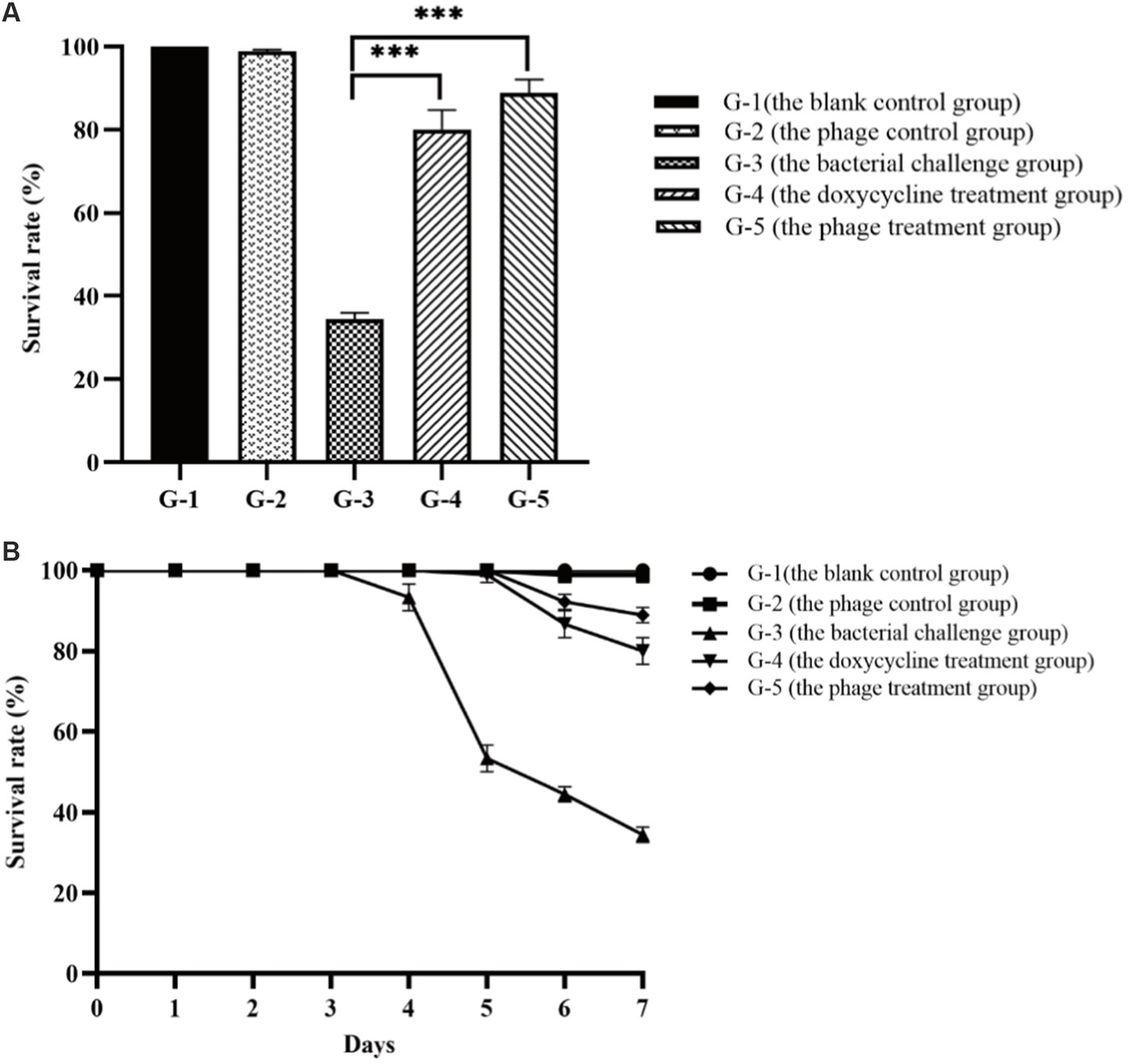
Figure 10. Comparison of the survival rates of the shrimp in different treatment groups. (A) Survival rates of the shrimp of each group after the 7-day treatment; (B) Survival rates of the shrimp of each group in the experiment duration. G-1: Group 1 was the blank control group without any treatment. G-2: Group 2 received of the phage stock at the final concentration of 2 × 107 PFU/mL. G-3: Group 3 was challenged with Vibrio parahaemolyticus strain VP8 at concentration of 2 × 106 CFU/mL. G-4: Group 4 was treated with 10 mg/l doxycycline after two-day challenge of strain VP8 at 2 × 106 CFU/mL. G-5: Group 5 received treatment of the phage stock at the final concentration of 2 × 107 PFU/mL after two-day challenge of strain VP8 at 2 × 106CFU/mL. Asterisks above the columns indicate significant differences at the p < 0.01 level.
To evaluate the effectiveness of phage PVA8 in the commercial shrimp farming practice, application trials were designed. In the trials, one blank control group and three experimental groups were included. It was found that Vibrio content in each blank control kept constant or increasing with time (G-1 in Figure 11), which reflected the challenge presented by Vibrio if no intervention applied. The group 2 that received the treatment of an iodic disinfectant showed valid effect at 4 h, but then the Vibrio kept bouncing and elevated to the same level as in the control at 24 h (G-2 in Figure 11). The application of the probiotic product showed obvious effect at 12 h and kept constant decrease after that (G-3 in Figure 11). In comparison, the Vibrio in the ponds with phages maintained a tendency of decreasing during the whole trial (G-4 in Figure 11). These results of phage PVA8 against the Vibrio, especially V. alginolyticus and V. parahaemolyticus for specifically, in farming plant trials confirmed that the phage could serve as an effective biological agent for the control of Vibrio in farming plant practice.
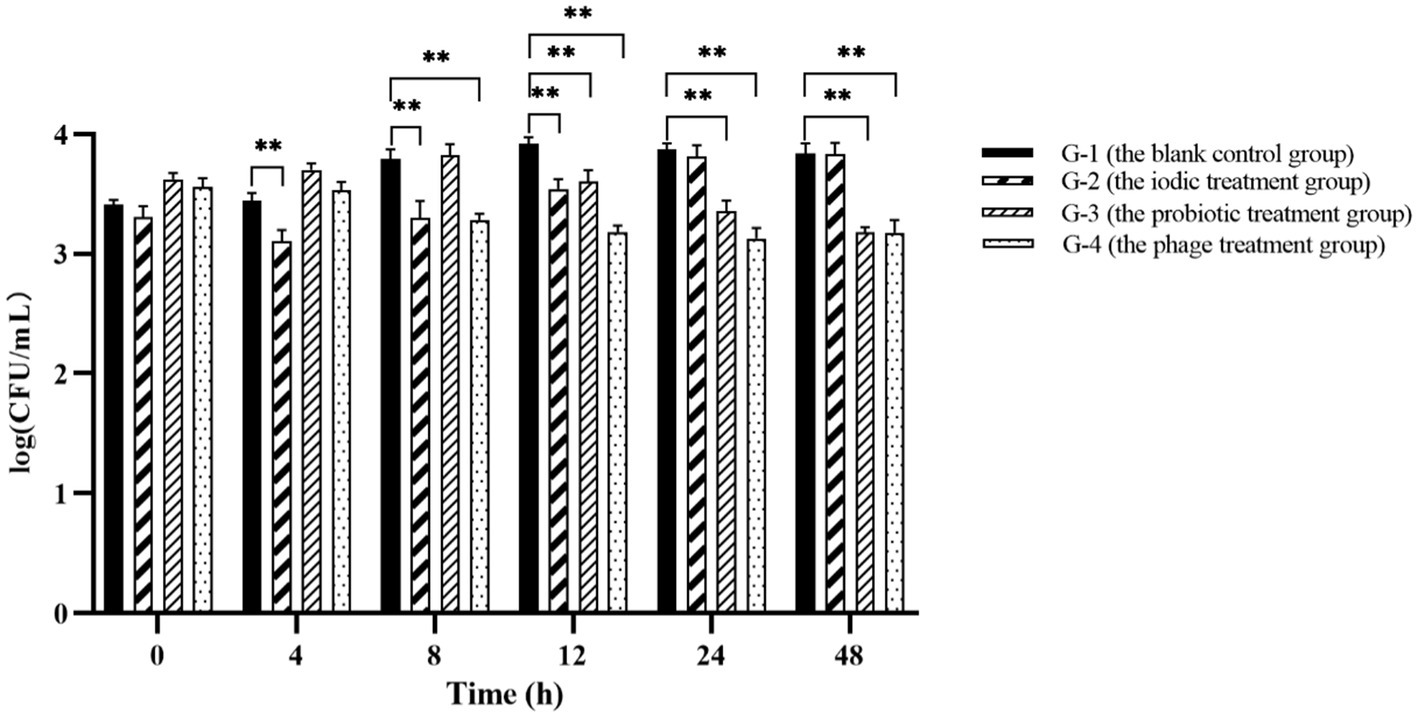
Figure 11. The result of phage PVA8 application in the shrimp farming plant. G-1: Group 1 was the blank control group receiving no control agent. G-2: Group 2 was the positive control group 1 receiving an iodic disinfectant (found in market) at dose of 5 mg/L. G-3: Group 3 was the positive control group 2 receiving a probiotic product (Qingdao Bioantai Biological Technology Co., LTD) at dose of 5 mg/L. G-4: Group 4 was the phage treatment group receiving the phage stock with MOI at 0.1. Asterisks above the columns indicate significant differences at the p < 0.01 level.
The emergence of antibiotic-resistant pathogens and the awareness of the adverse effects of antibiotics to animals have necessitated alternatives to antibiotics used in aquaculture (Wang et al., 2017). Phages have long been regarded as the safe yet effective substitute to antibiotics used in aquaculture. Phages exist in natural aquatic environments, serving as native antibacterial agents that maintain the balance of ecosystem. There are many cases with exemplified the success of phage therapy against Vibrio species (Mateus et al., 2014). Generally, in order to screen a truly useful bacteriophage, a panel of crucial factors need to be taken into considerations, such as its effectiveness against the pathogens that found in real farming practice, its safety for use as well as its feasibility for engineering manipulations.
Vibrio alginolyticus and V. alginolyticus are the two most important opportunistic pathogens that infect a variety of aquatic animals, such as fish, shellfish, and shrimp, causing mass mortality cases and considerable economic losses (Letchumanan et al., 2016; Sasikala and Srinivasan, 2016). They are also among the major foodborne pathogens in seafood that very often bring about severe risks to the health of consumers (Gao et al., 2020). The main virulence factors of V. alginolyticus include extracellular products (alkaline serine protease, collagenase), outer membrane proteins, hemolysins, ferric uptake system and virulence expression regulation gene system (Fethi et al., 2011). Vibrio parahaemolyticus is one of the major cause of disease, mortality, and economical losses in the shrimp aquaculture industry (Lomelí-Ortega and Martínez-Díaz, 2014). It can cause AHPND, one of the most serious diseases in shrimp aquaculture, resulting in a near 100% mortality rate within a week after the first symptoms appearing in cultured shrimp (Lai et al., 2015). The causative agents of the disease have been identified as two Pir-like toxins, PirA and PirB, which are encoded by a plasmid (Lee et al., 2015).
The purpose of this work is to identify the potential phages that could be used for the control of two pathogenic Vibrio, V. alginolyticus and V. alginolyticus. In order to obtain the real virulent Vibrio, a panel of Vibrio was isolated and selected. A group of common virulence genes including alkaline serine protease gene (AspA), ferric uptake system gene (fur), and hemolysin genes (tlh, tdh, and trh) of V. alginolyticus and photorhabdus insect related toxin A gene (PirA), virulence-regulating genes (toxR), and hemolysin genes (tlh, tdh, and trh) of V. parahaemolyticus were used for selecting the pathogenic V. alginolyticus. Among the isolates, two strains, V. alginolyticus VA15 and VA17, were found carrying all the five virulence genes, thus they were used as the host for the followed phage screening. Similarly, the signature genes for V. parahaemolyticus (Han et al., 2015), toxR and tlh, were applied for search V. parahaemolyticus in general, whereas the gene causative to the AHPND disease (Lee et al., 2015), PirA, was used for obtain the virulent V. parahaemolyticus in specific. Among the isolates of V. parahaemolyticus strains, VP5, VP7, VP8, and VP15, were found carrying the PirA virulence gene, thus they were also used as the host for phage screening.
Through screening against the six pathogenic Vibrio host, a potent phage PVA8 was identified. It was isolated from the shrimp farming sewage samples containing V. alginolyticus. The bacteriophage belongs to the Straboviridae family with a double-stranded DNA genome sequence in length of 246,348 bp, a prolate icosahedral capsid, and a long contractile tail with associated baseplate and extended tail fibers. Subsequent genome analysis showed that neither toxic genes nor antibiotic resistant genes identified in the genome of PVA8, which supposed its low safety concern for use.
The results of the BLASTn, genomic comparison and the phylogenetic tree based on the complete genome all showed that PVA8 had highest nucleotide homology with bacteriophage KVP40 and Vibrio phage V09. All the three phages are giant phages with linear dsDNA and large genome sizes, and are all belonged to the Straboviridae family with same morphological characteristics. Both of PVA8 and KVP40 are broad host range phages, different with V09 which is specific for V. parahaemolyticus. PVA8 could infect at least seven Vibrio, comparable to KVP40 that infect at least eight Vibrio plus one Photobacterium species.
The three phages all encoded high numbers of tRNAs with 27 (PVA8), 30 (KVP40), and 27 (V09), respectively. The tRNAs as translation-related genes contributing in the phage infection process and the synthesis of phage capsid and tail proteins are usually found in phage genomes (Mei and Zhu, 2015). In general, virulent phages contain more tRNAs than lysogenic phages, the reason might be that there is a significant association between tRNA distribution and codon usage which leading to an enhanced translational efficiency (Li et al., 2012). The high number of tRNA might benefit the phage in synthesis of its own proteins, compared with other phages that encoded fewer tRNAs, which mainly depend of the host translation machinery. Therefore, PVA8 was considered as a strong virulent phage (Lu et al., 2020).
It is noteworthy that phage PVA8 and bacteriophage KVP40 are alike in a panel of aspects, including phenotypic characteristics, similarities in genomic features, broad host ranges, etc. The tail is an important component of the phage, and its adsorption devices specialize in identifying and binding receptors on the surface of bacteria, initiating virus infection and DNA transferring. The complexity of this devices varies from single tail fiber to complex tail tip combination or complex base plate (Ding et al., 2020). Interestingly, it was found that the annotation information of most tail proteins of phage PVA8 have 100% similarity to those found in bacteriophage KVP40. The resemblance in tail devices may partial reason why the broad host range nature of the two phages.
For double-stranded DNA bacteriophages, the bacterial lysis is achieved by the phage-encoded muralytic enzyme called endolysins that hydrolyze the peptidoglycan (PG) layer of the host bacterial cell wall during the final stage of the phage reproduction events (O'Flaherty et al., 2009). There are three different mechanisms explaining the lysis events by endolysins. The most explicitly demonstrated mechanism evolves a lytic system called holin-endolysin, it involves a second phage-encoded protein called holin to assist endolysin acts on PG layer (Matamp and Bhat, 2019). Holin allows endolysin to diffuse through pores in the membrane and target the PG layer by depolarizing the cytoplasmic membrane, as most of endolysins lack the signal peptide sequence and cannot cross the cell membrane (Matamp and Bhat, 2019). Based on the mechanism of action, endolysins have at least four different hydrolase activities that degrade the cell wall, which are glycoside transferase, lysozyme, amidase and endopeptidase, respectively (Matamp and Bhat, 2019). These four hydrolase activities can all be found in the endolysins of Vibrio phages, especially glycoside transferase (Misol et al., 2020). However, none of endolysins mentioned above was found in the annotation results of the PVA8 genome. Similarly, neither endolysins were identified in bacteriophage KVP40. For bacteriophage KVP40, the presence of several copies of genes encoding proteins linked with phage tail or tail fibers in the genome suggest an increased flexibility in host range adaptation (Miller et al., 2003). Thus, here we suppose that similar host range adaptation might also happen to phage PVA8. Further evidences could be furnished in the future via the comparison of the tail-related genes as used elsewhere (Jacobs-Sera et al., 2012).
In this work, we succeeded in evaluating the preparation of phage PVA8 for the use against Vibrio pathogens in aquatic farming practice, with therapeutic and prophylactic of Vibrio infections trials arranged. After phage treatment, the shrimp survival rate in control was improved to 88.89% (from 34.43% in the control receiving Vibrio only), which is comparable to the best result reported elsewhere (Ding et al., 2020; Wu et al., 2021). It was noteworthy that preparation of phage PVA8 outperformed the chemical disinfectant in Vibrio control, and it even achieve faster inhibition of the pathogenic +than that of the best probiotic product currently used in the market. Therefore, we suggested that PVA8 based phage therapy might be a potential alternative to antibiotics regarding the control of pathogenic Vibrio in aquaculture (Stalin and Srinivasan, 2017).
In this study, a potential Vibrio lytic bacteriophage PVA8 with broad host ranges was screened and characterized. The phage was belonged to the Straboviridae family with a high lytic capability. It demonstrated a panel of features like broad host ranges, low MOI, large burst size, and decent tolerances to adverse conditions, which are promising merits for antibiotics substitution. Genome analysis revealed that there was neither virulence nor antibiotic resistance genes in the genome, further convincing its environmental-friendly nature and safety for application. The anti-Vibrio potency of the phage was indicated by lab shrimp trials and was further proved by the pond application. The fact that PVA8 could rapidly yet effectively reduce the level of Vibrio in pond practice doubtlessly provided evidence that PVA8 might serve as a biological agent for Vibrio control in aquaculture farming prophylactically and therapeutically.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
ZH and JF conceived and designed the study. JF and YL performed the experiments. JF, LZ, and CW analyzed the data. JF and ZH wrote the paper. ZH managed the project. All authors contributed to the article and approved the submitted version.
This research was funded by Special Fund for TaiShan Industrial Experts Program (tscy202006017) and Science and Technology Projects in Key Areas of Guangdong (2020B0202010001).
JF and ZH were employed by Marine Biomedical Research Institute of Qingdao Co., Ltd. JF, CW, and ZH were employed by Qingdao Bioantai Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1105924/full#supplementary-material
1. ^http://trna.ucsc.edu/tRNAscan-SE/
2. ^http://www.mgc.ac.cn/VFs/main.htm
Ayyaru, S., Choi, J., and Ahn, Y.-H. (2018). Biofouling reduction in a MBR by the application of a lytic phage on a modified nanocomposite membrane. Environ Sci Water Res Technol 4, 1624–1638. doi: 10.1039/C8EW00316E
Besemer, J., and Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Cafora, M., Deflorian, G., Forti, F., Ferrari, L., Binelli, G., Briani, F., et al. (2019). Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci. Rep. 9:1527. doi: 10.1038/s41598-018-37636-x
Chen, L., Fan, J., Yan, T., Liu, Q., Yuan, S., Zhang, H., et al. (2019). Isolation and characterization of specific Phages to prepare a cocktail preventing Vibrio sp. Va-F3 infections in shrimp (Litopenaeus vannamei). Front. Microbiol. 10:2337. doi: 10.3389/fmicb.2019.02337
Chen, L., Liu, Q., Fan, J., Yan, T., Zhang, H., Yang, J., et al. (2020). Characterization and genomic analysis of ValSw3-3, a new Siphoviridae bacteriophage infecting Vibrio alginolyticus. J. Virol. 94, e00066–20. doi: 10.1128/JVI.00066-20
Choudhury, G., Tanmoy, V. T., Nagaraju, S. G., Paria, A., and Parhi, J. (2016). Advances in bacteriophage research for bacterial disease control in aquaculture. Rev Fish Sci Aquacult 25, 113–125. doi: 10.1080/23308249.2016.1241977
Clavijo, V., Baquero, D., Hernandez, S., Farfan, J. C., Arias, J., Arevalo, A., et al. (2019). Phage cocktail SalmoFREE(R) reduces Salmonella on a commercial broiler farm. Poult. Sci. 98, 5054–5063. doi: 10.3382/ps/pez251
Cui, H., Cong, C., Wang, L., Li, X., Li, J., Yang, H., et al. (2014). Complete genomic sequence of the Vibrio alginolyticus lytic bacteriophage PVA1. Arch. Virol. 159, 3447–3451. doi: 10.1007/s00705-014-2207-z
Ding, T., Sun, H., Pan, Q., Zhao, F., Zhang, Z., and Ren, H. (2020). Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res. 286:198080. doi: 10.1016/j.virusres.2020.198080
Droubogiannis, S., Pavlidi, L., Tsertou, M. I., Kokkari, C., Skliros, D., Flemetakis, E., et al. (2022). Vibrio phage Artemius, a novel phage infecting Vibrio alginolyticus. Pathogens 11, 848–864. doi: 10.3390/pathogens11080848
Feng, T., Leptihn, S., Dong, K., Loh, B., Zhang, Y., Stefan, M. I., et al. (2021). JD419, a Staphylococcus aureus phage with a unique morphology and broad host range. Front. Microbiol. 12:602902. doi: 10.3389/fmicb.2021.602902
Fethi, B. A., Ali, E., Rihab, L., Heacute, K., and Bakhrouf, A. (2011). Virulence gene expression, proteins secreted and morphological alterations of Vibrio parahaemolyticus and Vibrio alginolyticus in response to long-term starvation in seawater. Afr. J. Microbiol. Res. 5, 792–801. doi: 10.5897/AJMR10.653
Fu, J., Li, Y., Zhao, L., Chunguang, W., and He, Z. (2022). Characterization and genomic analysis of a bacteriophage with potential in lysing Vibrio alginolyticus. Viruses 15, 135–158. doi: 10.3390/v15010135
Gao, M., Qin, Y., Fan, H., Zhang, X., Li, P., Liu, H., et al. (2020). Characteristics and complete genome sequence of the virulent Vibrio alginolyticus phage VAP7, isolated in Hainan China. Arch. Virol. 165, 947–953. doi: 10.1007/s00705-020-04535-4
George, M. R., John, K. R., Iyappan, T., and Jeyaseelan, M. J. (2005). Genetic heterogeneity among Vibrio alginolyticus isolated from shrimp farms by PCR fingerprinting. Lett. Appl. Microbiol. 40, 369–372. doi: 10.1111/j.1472-765X.2005.01675.x
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Gutierrez, D., Vandenheuvel, D., Martinez, B., Rodriguez, A., Lavigne, R., and Garcia, P. (2015). Two Phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species staphylococcal biofilms. Appl. Environ. Microbiol. 81, 3336–3348. doi: 10.1128/AEM.03560-14
Han, H., Li, N., and Guo, Y. (2015). Molecular genetic makers for Vibrio parahaemolyticus-a review. Acta Microbiol. Sin. 55, 12–21. doi: 10.13343/j.cnki.wsxb.20140203
Iwamoto, M., Ayers, T., Mahon, B. E., and Swerdlow, D. L. (2010). Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 23, 399–411. doi: 10.1128/CMR.00059-09
Jacobs-Sera, D., Marinelli, L. J., Bowman, C., Broussard, G. W., Guerrero Bustamante, C., Boyle, M. M., et al. (2012). On the nature of mycobacteriophage diversity and host preference. Virology 434, 187–201. doi: 10.1016/j.virol.2012.09.026
Kahla-Nakbi, A. B., Chaieb, K., Besbes, A., Zmantar, T., and Bakhrouf, A. (2006). Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet. Microbiol. 117, 321–327. doi: 10.1016/j.vetmic.2006.06.012
Kahn, L. H., Bergeron, G., Bourassa, M. W., De Vegt, B., Gill, J., Gomes, F., et al. (2019). From farm management to bacteriophage therapy: strategies to reduce antibiotic use in animal agriculture. Ann. N. Y. Acad. Sci. 1441, 31–39. doi: 10.1111/nyas.14034
Kalatzis, P. G., Castillo, D., Katharios, P., and Middelboe, M. (2018). Bacteriophage interactions with marine pathogenic Vibrios: implications for phage therapy. Antibiotics 7, 15–38. doi: 10.3390/antibiotics7010015
Kim, H. J., Kim, Y. T., Kim, H. B., Choi, S. H., and Lee, J. H. (2021). Characterization of bacteriophage VVP001 and its application for the inhibition of Vibrio vulnificus causing seafood-borne diseases. Food Microbiol. 94:103630. doi: 10.1016/j.fm.2020.103630
Kokkari, C., Sarropoulou, E., Bastias, R., Mandalakis, M., and Katharios, P. (2018). Isolation and characterization of a novel bacteriophage infecting Vibrio alginolyticus. Arch. Microbiol. 200, 707–718. doi: 10.1007/s00203-018-1480-8
Kosznik-Kwaśnicka, K., Topka, G., Dydecka, A., Necel, A., Nejman-Faleńczyk, B., Bloch, S., et al. (2019). “The use of bacteriophages in animal health and food protection” in Phage Therapy: A Practical Approach eds. A. Górski, R. Międzybrodzki, and J. Borysowski Phage Therapy: A Practical Approach. Springer, Cham. doi: 10.1007/978-3-030-26736-0_9
Lai, H. C., HannNg, T., and Masahiro, A. (2015). Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish Immunol. 47, 1006–1014. doi: 10.1016/j.fsi.2015.11.008
Lal, T. M., Sano, M., Hatai, K., and Ransangan, J. (2016). Complete genome sequence of a giant Vibrio phage ValKK3 infecting Vibrio alginolyticus. Genom Data 8, 37–38. doi: 10.1016/j.gdata.2016.03.002
Le, S. T., and Kurtböke, I. (2019). Bacteriophages as biocontrol agents in aquaculture. Focus 40, 37–41. doi: 10.1071/MA19003
Le, T. S., Nguyen, T. H., Vo, H. P., Doan, V. C., Nguyen, H. L., Tran, M. T., et al. (2018). Protective effects of bacteriophages against Aeromonas hydrophila species causing motile Aeromonas septicemia (MAS) in striped catfish. Antibiotics 7:16. doi: 10.3390/antibiotics7010016
Le, T. S., Southgate, P. C., O'Connor, W., Vu, S. V., and Kurtboke, D. I. (2020). Application of bacteriophages to control Vibrio alginolyticus contamination in oyster (Saccostrea glomerata) larvae. Antibiotics 9:415. doi: 10.3390/antibiotics9070415
Lee, C. T., Chen, I. T., Yang, Y. T., Ko, T. P., Huang, Y. T., Huang, J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. U. S. A. 112, 10798–10803. doi: 10.1073/pnas.1503129112
Lee, H., Ku, H. J., Lee, D. H., Kim, Y. T., Shin, H., Ryu, S., et al. (2016). Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157:H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS One 11:e0168985. doi: 10.1371/journal.pone.0168985
Letchumanan, V., Chan, K. G., Pusparajah, P., Saokaew, S., Duangjai, A., Goh, B. H., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. doi: 10.3389/fmicb.2016.01114
Li, P., Chen, B., Song, Z., Song, Y., Yang, Y., Ma, P., et al. (2012). Bioinformatic analysis of the Acinetobacter baumannii phage AB1 genome. Gene 507, 125–134. doi: 10.1016/j.gene.2012.07.029
Li, F., Xing, S., Fu, K., Zhao, S., Liu, J., Tong, Y., et al. (2019). Genomic and biological characterization of the Vibrio alginolyticus-infecting "Podoviridae" bacteriophage, vB_ValP_IME271. Virus Genes 55, 218–226. doi: 10.1007/s11262-018-1622-8
Lin, Y. R., Chiu, C. W., Chang, F. Y., and Lin, C. S. (2012). Characterization of a new phage, termed varphiA318, which is specific for Vibrio alginolyticus. Arch. Virol. 157, 917–926. doi: 10.1007/s00705-012-1244-8
Lin, J., Guocheng, S., and Huang, J. (2016). Estabilishment of a quadruple PCR method for detecting pathogenic genes in Vibrio parahaemolyticus. Microbiol China 43, 2521–2529.
Lomelí-Ortega, C. O., and Martínez-Díaz, S. F. (2014). Parahaemolyticus infection in the whiteleg shrimp (Litopenaeus vannamei) larvae. Aquaculture 46, 81–85.
Lomeli-Ortega, C. O., Martinez-Sandez, A. J., Barajas-Sandoval, D. R., Reyes, A. G., Magallon-Barajas, F., Veyrand-Quiros, B., et al. (2021). Isolation and characterization of vibriophage vB_Vc_SrVc9: an effective agent in preventing Vibrio campbellii infections in brine shrimp nauplii (Artemia franciscana). J. Appl. Microbiol. 131, 36–49. doi: 10.1111/jam.14937
Lu, Z., Breidt, F., Fleming, H. P., Altermann, E., and Klaenhammer, T. R. (2003). Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 84, 225–235. doi: 10.1016/S0168-1605(03)00111-9
Lu, H., Yan, P., Xiong, W., Wang, J., and Liu, X. (2020). Genomic characterization of a novel virulent phage infecting Shigella fiexneri and isolated from sewage. Virus Res. 283:197983. doi: 10.1016/j.virusres.2020.197983
Matamp, N., and Bhat, S. G. (2019). Phage Endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms 7, 84–95. doi: 10.3390/microorganisms7030084
Mateus, L., Costa, L., Silva, Y. J., Pereira, C., Cunha, A., and Almeida, A. (2014). Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 424-425, 167–173. doi: 10.1016/j.aquaculture.2014.01.001
Mauritzen, J. J., Castillo, D., Tan, D., Svenningsen, S. L., and Middelboe, M. (2020). Beyond cholera: characterization of zot-encoding filamentous Phages in the marine fish pathogen Vibrio anguillarum. Viruses 12, 730–737. doi: 10.3390/v12070730
Mei, S., and Zhu, H. (2015). Multi-label multi-instance transfer learning for simultaneous reconstruction and cross-talk modeling of multiple human signaling pathways. BMC Bioinformatics 16:417. doi: 10.1186/s12859-015-0841-4
Miller, E. S., Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Durkin, A. S., Ciecko, A., et al. (2003). ‘Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage’, J Bacteriol 185, 5220–5233.
Misol, G. N., Kokkari, C., and Katharios, P. (2020). Biological and genomic characterization of a novel jumbo bacteriophage, vB_VhaM_pir03 with broad host lytic activity against Vibrio harveyi. Pathogens 9, 1051–1089. doi: 10.3390/pathogens9121051
Moussa, S. H., Lawler, J. L., and Young, R. (2014). Genetic dissection of T4 lysis. J. Bacteriol. 196, 2201–2209. doi: 10.1128/JB.01548-14
O'Flaherty, S., Ross, R. P., and Coffey, A. (2009). Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33, 801–819. doi: 10.1111/j.1574-6976.2009.00176.x
Ohyama, H., Sakai, T., Agari, Y., Fukui, K., Nakagawa, N., Shinkai, A., et al. (2014). 'The role of ribonucleases in regulating global mRNA levels in the model organism Thermus thermophilus HB8. BMC Genomics 15:386. doi: 10.1186/1471-2164-15-386
Onsea, J., Soentjens, P., Djebara, S., Merabishvili, M., Depypere, M., Spriet, I., et al. (2019). Bacteriophage application for difficult-to-treat musculoskeletal infections: development of a standardized multidisciplinary treatment protocol. Viruses 11, 891–903. doi: 10.3390/v11100891
Ooi, M. L., Drilling, A. J., Morales, S., Fong, S., Moraitis, S., Macias-Valle, L., et al. (2019). Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to Staphylococcus aureus. JAMA Otolaryngol. Head Neck Surg. 145, 723–729. doi: 10.1001/jamaoto.2019.1191
Pan, L., Li, D., Lin, W., Liu, W., Qu, C., Qian, M., et al. (2022). Novel Aeromonas phage Ahy-Yong1 and its protective effects against Aeromonas hydrophila in brocade carp (Cyprinus aka koi). Viruses 14, 2498–2520. doi: 10.3390/v14112498
Paul, J., Jardine, M., McCormick, C., Lutze-Wallace, C., and Coombs, D. H. (1998). 'The bacteriophage T4 DNA packaging apparatus targets the unexpanded Prohead. J. Mol. Biol. 284, 647–659.
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G., and Toth, I. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/C5AY02550H
Qi, Z., Yang, S., Dong, S., Zhao, F., Qin, J., and Jun, X. (2020). Biological characteristics and genomic information of a bacteriophage against pan-drug resistant Klebsiella pneumoniae in a burn patient and its effects on bacterial biofilm. Chin J Burns 36, 14–23. doi: 10.3760/cma.j.issn.1009-2587.2020.01.004
Rasmussen, B. B., Kalatzis, P. G., Middelboe, M., and Gram, L. (2019). Combining probiotic Phaeobacter inhibens DSM17395 and broad-host-range vibriophage KVP40 against fish pathogenic vibrios. Aquaculture 513:734415. doi: 10.1016/j.aquaculture.2019.734415
Ren, Y., Wang, L., Chen, R., Li, X., Li, S., Li, J., et al. (2022). Isolation and characterization of a novel phage vB_ValP_VA-RY-3 infecting Vibrio alginolyticus. Virus Res. 322:198945. doi: 10.1016/j.virusres.2022.198945
Sasikala, D., and Srinivasan, P. (2016). Characterization of potential lytic bacteriophage against Vibrio alginolyticus and its therapeutic implications on biofilm dispersal. Microb. Pathog. 101, 24–35. doi: 10.1016/j.micpath.2016.10.017
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi: 10.1093/nar/gki366
Seghal Kiran, G., Priyadharshini, S., Dobson, A. D. W., Gnanamani, E., and Selvin, J. (2016). Degradation intermediates of polyhydroxy butyrate inhibits phenotypic expression of virulence factors and biofilm formation in luminescent Vibrio sp. PUGSK8. NPJ Biofilms Microbiomes 2:16002. doi: 10.1038/npjbiofilms.2016.2
Shahin, K., and Bouzari, M. (2017). Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Technol. 55, 550–559. doi: 10.1007/s13197-017-2964-2
Sieiro, C., Areal-Hermida, L., Pichardo-Gallardo, A., Almuina-Gonzalez, R., de Miguel, T., Sanchez, S., et al. (2020). A hundred years of bacteriophages: can Phages replace antibiotics in agriculture and aquaculture? Antibiotics 9:493. doi: 10.3390/antibiotics9080493
Song, D., Yang, Y., Yu, B., Zheng, B., Deng, Z., Lu, B. L., et al. (2009). Computational prediction of novel non-coding RNAs in Arabidopsis thaliana. BMC Bioinformatics 10:S36. doi: 10.1186/1471-2105-10-S1-S36
Stalin, N., and Srinivasan, P. (2017). Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet. Microbiol. 207, 83–96. doi: 10.1016/j.vetmic.2017.06.006
Su, Y.-C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
SumeetRai, A., Naveen Kumar, B. T., and Reddy, V. K. (2023). Isolation and characterization of Aeromonas hydrophila lytic phage, and evaluation of a phage cocktail against A. hydrophila contamination in fish fillet. Food Control 145:109460. doi: 10.1016/j.foodcont.2022.109460
Sun, M., Ye, M., Zhang, Z., Zhang, S., Zhao, Y., Deng, S., et al. (2019). Biochar combined with polyvalent phage therapy to mitigate antibiotic resistance pathogenic bacteria vertical transfer risk in an undisturbed soil column system. J. Hazard. Mater. 365, 1–8. doi: 10.1016/j.jhazmat.2018.10.093
Svircev, A., Roach, D., and Castle, A. (2018). Framing the future with bacteriophages in agriculture. Viruses 10:218. doi: 10.3390/v10050218
Tan, D., Dahl, A., and Middelboe, M. (2015). Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl. Environ. Microbiol. 81, 4489–4497. doi: 10.1128/AEM.00518-15
Tan, C. W., Rukayadi, Y., Hasan, H., Abdul-Mutalib, N. A., Jambari, N. N., Hara, H., et al. (2021). Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front. Microbiol. 12:616548. doi: 10.3389/fmicb.2021.616548
Thadtapong, N., Salinas, M. B. S., Charoensawan, V., Saksmerprome, V., and Chaturongakul, S. (2020). Genome characterization and comparison of early mortality syndrome causing Vibrio parahaemolyticus pirABvp− mutant from Thailand with V. parahaemolyticus pirABvp+ AHPND isolates. Front. Mar. Sci. 7:00290. doi: 10.3389/fmars.2020.00290
Thompson, F. L., Iida, T., and Swings, J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
Underwood, A. P., Mulder, A., Gharbia, S., and Green, J. (2005). Virulence searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 11, 770–772. doi: 10.1111/j.1469-0691.2005.01210.x
Vaz, C. S. L., Voss-Rech, D., Alves, L., Coldebella, A., Brentano, L., and Trevisol, I. M. (2020). Effect of time of therapy with wild-type lytic bacteriophages on the reduction of Salmonella Enteritidis in broiler chickens. Vet. Microbiol. 240:108527. doi: 10.1016/j.vetmic.2019.108527
Wang, Y., Mary, B., Elliott, L., Li, X. X., Abraham, S., O'Dea, M., et al. (2017). Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473, 251–258. doi: 10.1016/j.aquaculture.2017.01.003
Wang, Q., Teng, Y.-y., and He, S. (2014). Rapid detection of 5 important pathogenic Vibrio species by multiplex PCR. Chin J Health Inspect 24, 3497–3504. doi: 10.1016/j.diagmicrobio.2007.05.016
Weber-Dabrowska, B., Jonczyk-Matysiak, E., Zaczek, M., Lobocka, M., Lusiak-Szelachowska, M., and Gorski, A. (2016). Bacteriophage procurement for therapeutic purposes. Front. Microbiol. 7:1177. doi: 10.3389/fmicb.2016.01177
Wong, H. C., Wang, T. Y., Yang, C. W., Tang, C. T., Ying, C., Wang, C. H., et al. (2019). Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus. Res. Microbiol. 170, 13–23. doi: 10.1016/j.resmic.2018.07.003
Wu, L., Tian, Y., Pang, M., Yang, Z., Bao, H., Zhou, Y., et al. (2021). A novel vibriophage vB_VhaS_PcB-1G capable of inhibiting virulent Vibrio harveyi pathogen. Aquaculture 542:736854. doi: 10.1016/j.aquaculture.2021.736854
Xi, H., Dai, J., Tong, Y., Cheng, M., Zhao, F., Fan, H., et al. (2019). The characteristics and genome analysis of vB_AviM_AVP, the first phage infecting Aerococcus viridans. Viruses 11:104. doi: 10.3390/v11020104
Xiong, P., Peng, X., Wei, S., Chen, Y., Zhao, H., Tang, S., et al. (2014). Virulence-related genes of Vibrio alginolyticus and its virulence in mice. Acta Microbiol. Sin. 54, 80–88.
Xu, L., Li, D., Tong, Y., Fang, J., Yang, R., Qin, W., et al. (2021). A novel singleton Giant phage Yong-XC31 lytic to the Pyropia pathogen Vibrio mediterranei. Appl. Sci. 11
Keywords: phage therapy, Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio phage, Vibrio control, shrimp
Citation: Fu J, Li Y, Zhao L, Wu C and He Z (2023) Characterization of vB_ValM_PVA8, a broad-host-range bacteriophage infecting Vibrio alginolyticus and Vibrio parahaemolyticus. Front. Microbiol. 14:1105924. doi: 10.3389/fmicb.2023.1105924
Received: 23 November 2022; Accepted: 12 April 2023;
Published: 12 May 2023.
Edited by:
Milan Surjit, Translational Health Science and Technology Institute (THSTI), IndiaCopyright © 2023 Fu, Li, Zhao, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengguo He, YmlvYW50YWk4OEB2aXAuMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.