- Biological Resource Center, Korean Collection for Type Cultures (KCTC), Korea Research Institute of Bioscience and Biotechnology (KRIBB), Jeongeup, Republic of Korea
A novel, nostoxanthin-producing, endophytic bacterium, designated as AK-PDB1-5T, was isolated from the needle-like leaves of the Korean fir (Abies koreana Wilson) collected from Mt. Halla in Jeju, South Korea. A 16S rRNA sequence comparison indicated that the closest phylogenetic neighbors were Sphingomonas crusticola MIMD3T (95.6%) and Sphingomonas jatrophae S5-249T (95.3%) of the family Sphingomonadaceae. Strain AK-PDB1-5T had a genome size of 4,298,284 bp with a 67.8% G + C content, and digital DNA–DNA hybridization and OrthoANI values with the most closely related species of only 19.5–21% and 75.1–76.8%, respectively. Cells of the strain AK-PDB1-5T were Gram-negative, short rods, oxidase- and catalase-positive. Growth occurred at pH 5.0–9.0 (optimum pH 8.0) in the absence of NaCl at 4–37°C (optimum 25–30°C). Strain AK-PDB1-5T contained C14:0 2OH, C16:0 and summed feature 8 as the major cellular fatty acids (> 10%), while sphingoglycolipid, phosphatidylethanolamine, phosphatidylglycerol, phospholipids and lipids were found to be the major polar lipids. The strain produces a yellow carotenoid pigment; natural products prediction via AntiSMASH tool found zeaxanthin biosynthesis clusters in the entire genome. Biophysical characterization by ultraviolet–visible absorption spectroscopy and ESI-MS studies confirmed the yellow pigment was nostoxanthin. In addition, strain AK-PDB1-5T was found significantly promote Arabidopsis seedling growth under salt conditions by reducing reactive oxygen species (ROS). Based on the polyphasic taxonomic analysis results, strain AK-PDB1-5T was determined to be a novel species in the genus Sphingomonas with the proposed name Sphingomonas nostoxanthinifaciens sp. nov. The type strain is AK-PDB1-5T (= KCTC 82822T = CCTCC AB 2021150T).
1. Introduction
The genus Sphingomonas belonging to the family Sphingomonadaceae of the class Alphaproteobacteria was first proposed by Yabuuchi et al. (1990). The type strain is Sphingomonadaceae paucimobilis VKM B-2241T, and four other members were proposed at the same time (Holmes et al., 1977). The genus Sphingomonas has been divided into four clusters based on phylogenetic and chemotaxonomic analysis in 2001, and three new genera Sphingobium, Novosphingobium, and Sphingopyxis were proposed in addition to the genus Sphingomonas sensu stricto (Takeuchi et al., 2001). As of August 2022, this genus contains 146 published species.1 The common cellular characteristics of the genus Sphingomonas are Gram-negative, a yellow or white-color, rod shape, strictly aerobic, motile or non-motile, and non-fermentative. Q-10 is the predominant isoprenoid quinone in this genus, while the major fatty acids are C16:0, C14:0 2OH, as well as summed feature 8 (comprising C18:1ω7c and/or C18:1ω6c). Species in this genus have been isolated from various sources, including the soil (Kim et al., 2014), water, air (Kim et al., 2014; Xue et al., 2018), plant endophytes, rice paddy and noni, and a medical clinic (Busse et al., 2005). Some Sphingomonas species such as Sphingomonas laterariae and Sphingomonas wittichii have been found in contaminated environments, probably due to their high survivability and superior facility for degradation (Kaur et al., 2012; Moreno-Forero and van der Meer, 2015). Many Sphingomonas species have been determined to play important roles in plant abiotic stress tolerance, bioremediation, and biodegradation (Chen et al., 2008; Yu et al., 2013; Khan et al., 2014), as well as promoting plant growth (Yang et al., 2014; Luo et al., 2019, 2020).
Whole-genome sequencing is currently an important methodology for the discovery of useful secondary metabolites. Many secondary metabolites are natural products encoded by gene groups known as biosynthetic gene clusters (BGCs), which can be identified via whole-genome annotation. Various secondary metabolites including non-ribosomal peptide synthases, type I and type II polyketide synthases, lanthipeptides, lasso peptides, sactipeptides, and thiopeptides can be detected using the antiSMASH tool for genome mining (Blin et al., 2021). To survive in diverse environments, some members of the genus Sphingomonas are known to produce carotenoids such as lycopene, β-carotene, zeaxanthin, caloxanthin, astaxanthin, and nostoxanthin (Jenkins et al., 1979; Kawahara and Kuraishi, 1999; Zhu et al., 2012; Kim et al., 2014), whose antioxidant and anticancer properties reduce stress arising from the environments. Carotenoids are members of the isoprenoids group with diverse structures and functions, which are all synthesized via the common precursor isopentenyl diphosphate (IPP) or its isomer dimethylallyl diphosphate (DMAPP). The carotenoid biosynthesis pathway begins with the condensation of two molecules of geranylgeranyl pyrophosphate to produce phytoene, then converted into lycopene via phytofluene, ζ-carotene, and neurosporene in four desaturation steps, and finally to lycopene, then cyclization produces the various carotenes, such as β-carotene, xanthophylls such as zeaxanthin are oxygenation products of carotenes. Among these carotenoids, nostoxanthin ((2R,3R,2′R,3′R)-β or β-Carotene-2,3,2′,3′-tetrol) is a yellow pigment in the xanthophyll group of the carotenoid family having the molecular formula C40H56O4. It is biosynthesized from the precursor zeaxanthin. Two hydroxylations produce caloxanthin followed by nostoxanthin (Zhu et al., 2012). Nostoxanthin is a natural pigment and is found in many strains of the genus Pseudomonas (Jenkins et al., 1979), some Sphingomonas strains (Kikukawa et al., 2021), Sphingobium sp. (Liu et al., 2021), and cyanobacteria (Silkina et al., 2019). The marine bacteria Erythrobacter flavus (Setiyono et al., 2019) was also reported to produce nostoxanthin. Although one excellent review analysis of the genetic diversity of carotenogenesis in bacteria of the order Sphingmonasdales, is presumptive that some Sphingomonas members produce nostoxanthin based on the color of strain and presence of crt ORF, still a need to identify the pigment and well-describe the biosynthesis pathway and other genomic details in genus Sphingomonas (Siddaramappa et al., 2018). Besides, many research suggests that carotenoids such as zeaxanthin, and astaxanthin play the antioxidant role that scavenges ROS and free radicals and/or free radicals in the lipid phase of the plant cell membrane, helping the plant alleviate salt stress (Zhang et al., 2012, 2016; Ren et al., 2021; Song et al., 2022). In the present study, a new strain designated as AK-PDB1-5T was isolated from the needle-like leaves of the Korean fir. The Korean fir is an endemic, rare, and valuable tree species that suffered large dieback and a severe decline for unknown reasons. As a traditional medicine plant (Yeşilada et al., 1995), the Korean fir can produce many important secondary metabolites such as essential oils (Yoon et al., 2009), lanostane terpenoids (Kim et al., 2004), and secocycloartenoids (Kim et al., 2001) that can be used to treat hypertension, uterine bleeding, human skin and lung cells (Ahn et al., 2020), and tuberculosis (Chinthanom et al., 2021). More and more researchers support that some biological properties of plant-based compounds could be due to the interaction of endophytes and plants (Zhao et al., 2011; Aly et al., 2013; Singh et al., 2021; Urumbil and Anilkumar, 2021). Endophytes provide bioactive secondary metabolites to the host. The anti-inflammatory properties of some plants are due to compounds with anti-inflammatory activity that are secreted by endophytes (Fadiji and Babalola, 2020; Urumbil and Anilkumar, 2021). In our study, we investigated endophyte-derived natural compounds from a native medicinal plant. The bacterial community of the Korean fir was studied. Eighty-one endophytic strains were isolated from surface-sterilized needle leaves of Abies koreana. Among all the isolates, 10 strains belonged to the genus Sphingomonas. Genus Sphingomonas with the most diverse and the second most popular that attracted us to study. Strain AK-PDB1-5T was isolated and characterized as a novel species of the genus Sphingomonas, based on the results obtained from a polyphasic taxonomic study, and genome mining detected a zeaxanthin gene cluster tightly linked to the zeaxanthin biosynthesis pathway. Finally, based on its chemical structure and whole-genome annotation, a yellow pigment produced by this bacterium has been characterized as nostoxanthin. The nostoxanthin-producing strain AK-PDB1-5T alleviates the salt stress of Arabidopsis seedlings by scavenging the reactive oxygen species (ROS).
2. Materials and methods
2.1. Bacterial strains
Strain AK-PDB1-5T is an endophytic bacterium isolated from the needle-like leaves of the Korean fir (Abies koreana Wilson) which grows on the upper part of the Mt. Halla on Jeju island in Korea (33°21′42″N 126°31′45″E). Samples were collected and placed into sterile plastic bags. Five grams of sample were surface-sterilized with 1.05% sodium hypochlorite for 10 min, followed by rinsing 5 times in sterile distilled water. After grinding with 10 ml of PBS (phosphate buffered saline) buffer, then serially diluted from 10−1 to 10−4 with PBS buffer. An aliquot of 100 μl of diluted sample was spread on potato dextrose agar (PDA, Difco) and incubated at 25°C for 7 days. Single colonies were obtained and subsequently streaked on fresh PDA. A circular, smooth, opaque strain containing a light yellow pigment was designated as AK-PDB1-5T and selected for further study. The strain was preserved in sterile skimmed milk (10%, w/v) at −80°C, and deposited in the Korean Collection for Type Cultures (KCTC) and the China Center for Type Culture Collection (CCTCC) with the respective accession numbers KCTC 82822T and CCTCC AB 2021150T. Unless otherwise stated, the cells were grown on PDA for 4 days for subsequent tests.
2.2. Phenotypic and biochemical characterization
The morphological characteristics of strain AK-PDB1-5T were observed using scanning electron microscopy (Quanta 250 FEG) at the KRIBB Microscopy Core Facility after the strain was grown on PDA medium for 3 days. Cellular motility was tested by growth on semi-solid PDA medium (0.4% agar). Gram reactions were determined with a Gram staining kit (Difco, Korea) following the manufacturer’s instructions. Bubble production indicated positive catalase activity after 3% (v/v) hydrogen peroxide solution was added to fresh cells (Lee and Jeon, 2017), and an oxidase reagent kit (bioMérieux) was used to determine oxidase activity. Various media were used to optimize the growth of strain AK-PDB1-5T, including nutrient agar (NA, beef extract 3 g/l, peptone 5 g/l, and agar 15 g/l), PDA, Luria-Bertani agar (LB), Trypticase Soy Agar (TSA), marine agar 2216 (MA), reasoner’s 2A agar (R2A), and YEP medium. The optimal temperature was determined by growing the strain at various temperatures including 4, 10, 15, 20, 25, 30, 37, 40, 45, 50, and 60°C for 7 days. NaCl tolerance and the pH range for growth were measured in PD broth (PDB), and pH values ranging from 3.0 to 12.0 were adjusted with 1 N HCl and NaOH, while tested salt concentrations encompassed 0–15% (w/v; 1% concentration increments; Lee et al., 2019). Other biochemical characteristics, such as the substrate utilization, acid production from carbohydrates, and enzyme activities were tested using API 20NE (bioMérieux) or API ZYM with NaCl 0.85% medium (bioMérieux) and API 50CH according to the manufacturer’s protocol. All closely related type strains were tested under the same conditions.
2.3. 16S rRNA gene sequence analysis
Amplification of the 16S rRNA gene was done using PCR with bacterial DNA as the template and the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′; Lane, 1991). Sequencing of the PCR products was carried out by Macrogen, Inc. (Republic of Korea) using the 27F, 1492R, 518F (5′-CCAGCAGCCGCGGTAATACG-3′), and 800R (5′-TACCAGGGTATCTAATCC-3′) primers. The Vector NTI software (1.6.1) was used for assembling the sequence results and finally, nearly a full-length 16S rRNA sequence was obtained. The GenBank2 and the EzBioCloud databases3 (Yoon et al., 2017) were used to compare 16S rRNA sequence similarity. The 16S rRNA genes of closely related strains were downloaded from EZbiocloud. After multiple alignments by clustalW, gaps at the 5′ and 3′ ends were deleted by BioEdit. A 16S rRNA gene-based phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA 10.0) software (Kumar et al., 2018) employing the neighbor-joining (NJ), minimum-evolution (ME), and maximum-likelihood (ML) algorithms with 1,000 bootstrap iterations and Kimura two-parameter model. Usitatibacter rugosus 0125-3T was used as an outgroup.
2.4. Chemotaxonomic characterization
Chemotaxonomic features of strain AK-PDB1-5T, including cellular fatty acids, polar lipids, and quinones, were investigated. To determine cellular fatty acids, fresh cells were harvested from PDA medium, and the whole-cell fatty acid methyl esters (FAME) were extracted according to the instructions for the standard MIDI (Sherlock Microbial Identification System version 6.0), and then analyzed by gas chromatography (Model 6,890 N; Agilent) based on the Microbial Identification software package (Sasser, 2006). Isoprenoid quinones were extracted from 100 mg of freeze-dried cells by shaking them in a chloroform/methanol mixture (2:1, v/v), and purified by thin-layer chromatography as described by Collins et al. (1980). Finally, reverse-phase high-performance liquid chromatography with ultraviolet (UV) absorbance detection at 270 nm was carried out. Polar lipids were extracted from 100 mg of freeze-dried cells with a chloroform/methanol mixture (1:2, v/v), followed by identification by two-dimensional thin-layer chromatography on Kieselgel 60 F254 plates (silica gel, 10 × 10 cm; Merck). In addition, 0.2% ninhydrin (Sigma-Aldrich), molybdenum blue (Sigma-Aldrich), 4% phosphomolybdic acid reagent, and Dragendorff’s solution were sprayed onto the plates to detect amino group-containing lipids, sugar-containing lipids, phosphorus-containing lipids, total lipids, and quaternary nitrogen-containing lipids, respectively. Polyamines were extracted from freeze-dried cells and analyzed as described by Busse and Auling (1988).
2.5. Genomic sequencing and annotation
A genomic DNA purification kit (MGmed, Republic of Korea) was used to isolate the genomic DNA, and the genomic DNA quantity and quality were evaluated with the PicoGreen and Nanodrop (radio A260/A280). The whole genome was sequenced by the Macrogen facility (Macrogen, Korea) on the PacBio RSII (Pacific Biosciences, Inc.) and the Illumina sequencing platforms, followed by assembly with the SMRT Portal (version 2.3) de novo assembler. The assembled genome was checked for potential contamination by testing the 16S rRNA in the genome using the ContEst16S algorithm (Lee et al., 2017). Genome annotation was performed via the National Center for Biotechnology Information Prokaryotic Genome Annotation Pipeline (PGAP) and Rast SEED (Tatusova et al., 2016). Metabolic pathways were reconstructed using BlastKOALA, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al., 2016). Genome mining for the presence of secondary metabolite gene clusters was performed via the antiSMASH tool4 (Blin et al., 2021). The dDDH, ANI values, and orthoANI values between AK-PDB1-5T and closely related strains were determined using the Genome-to-Genome Distance Calculation (GGDC) webserver5 (Meier-Kolthoff et al., 2013), the ANI calculator,6 and the standalone Orthologous Average Nucleotide Identity (OAT) software (Lee et al., 2016), respectively. The closely related strains were obtained from NCBI GenBank websites that were publicly available, and genome quality was evaluated by the Microbial Genomes Atlas (MiGA) webserver (Rodriguez et al., 2018). A whole-genome-based phylogenomic tree was constructed using the up-to-date bacterial core gene set and pipeline (UBCG) as described by Na et al. (2018), Usitatibacter rugosus 0125-3T was used as an outgroup.
2.6. Nostoxanthin structure determination
Freeze-dried cells were used for carotenoid identification. An aliquot of 100 mg of freeze-dried sample was mixed with 10 ml of methanol, then shaken at 200 rpm at 25°C for 12 h. The resultant methanol extract was centrifuged at 8,000 rpm for 10 min. The UV–Vis absorption spectra were recorded from 200 to 800 nm using a microplate spectrophotometer (Multiskan Skyhigh, Thermo Fisher Scientific). The methanol extract was filtered through a 0.2 μm membrane filter and analyzed using an HPLC system (Shimadzu Corporation, Kyoto, Japan), with a YMC carotenoid (250 × 4.6 mm. D, S-5 μm, YMC) column. The separation column temperature was 35°C, with solvent A consisting of methanol-MTBE-water (85:10:5, v/v/v), and solvent B of 100% MTBE, as the mobile phase. The flow rate was 1 ml/min, the wavelength was set at 450 nm, and a 20 μl sample was injected. For identification of the structure, the sample was five-fold dilution and stored at −20°C until a subsequent analysis by an ultra-performance liquid chromatography-electrospray ionization-mass spectrometry (UPLC-ESI-MS) method. HR-ESI-MS spectra were obtained using a Waters ACQUITY UPLC system (Waters Corp., Milford, MA, United States) connected to a quadrupole time of flight mass spectrometer (Xevo G2-XS QTOF, Waters Corp.) from the Korea Basic Science Institute (KBSI, Chuncheon Center).
2.7. Assessment of plant growth and salt stress condition
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized with 70% ethanol for 5 min, 2.5% sodium hypochlorite for 15 min, and rinsed three times with sterile distilled water for 5 min. All sterilized seeds were sown on half-strength Murashige & Skooog (MS) medium (MS basal 2.2 g/l, sucrose 10 g/l, 2-MES 0.5 g/l, bacto-agar 15 g/l) and plants were grown in a growth chamber at 22°C and 55–60% relative humidity (RH) with a 16-h light/8-h dark cycle. Seven-day-old seedlings were co-cultured with strain AK-PDB1-5T and untreated control for 10 days.
The root fresh weight, leaf fresh weight as well as chlorophyll contents (equation: y = 8.02 × A663 + 20.2 × A645) were recorded to estimate the plant growth condition. Differences between untreated controls and AK-PDB1-5T co-cultivation under the normal and salt condition were analyzed using a two-way ANOVA followed by Sidak’s multiple comparison test in GraphPad Prism (v.8.4.3) program.
2.8. Reactive oxygen species (ROS) detection
To detect the reactive oxygen species, plant roots grown on 1/2MS medium containing 0 mM and 100 mM NaCl were collected from the plates, washed with 1×PBS buffer, stained root cells by adding 5 mM DCFH-DA in 5 mM sodium phosphate buffer (pH 7.0) and incubated for 30 min. The stained root tips were observed under fluorescent microscopy (ECLIPSE 80i) with FITC filter (515 nm). The fluorescent intensities were measured by Image J. Differences were analyzed by multiple t-tests in GraphPad Prism (v.8.4.3) program.
3. Results and discussion
3.1. Phenotypic and biochemical characterization
Strain AK-PDB1-5T was isolated from the needle-like leaves of Korean fir. It was found to grow well on PDA, R2A, and grow poorly on LB, NA, TSA, and YEP (optimum PDA and R2A), but not on MA. Strain AK-PDB1-5T was observed to be Gram-negative, non-motile, and rod-shaped, without flagella, 0.3–0.5 μm in width and 0.9–1.2 μm in length (Supplementary Figures S1AB). The colonies were 2–6 mm in diameter, and creamy white to yellowish. The organism grew at 4–37°C (optimal temperature 25–30°C) in the 0–0.5% salt (optimal 0%), and at pH 6.0–9.0 (optimum pH 8.0). The strain tested oxidase- and catalase-positive. Other characteristics that distinguished strain AK-PDB1-5T from other closely related strains are shown in Table 1.
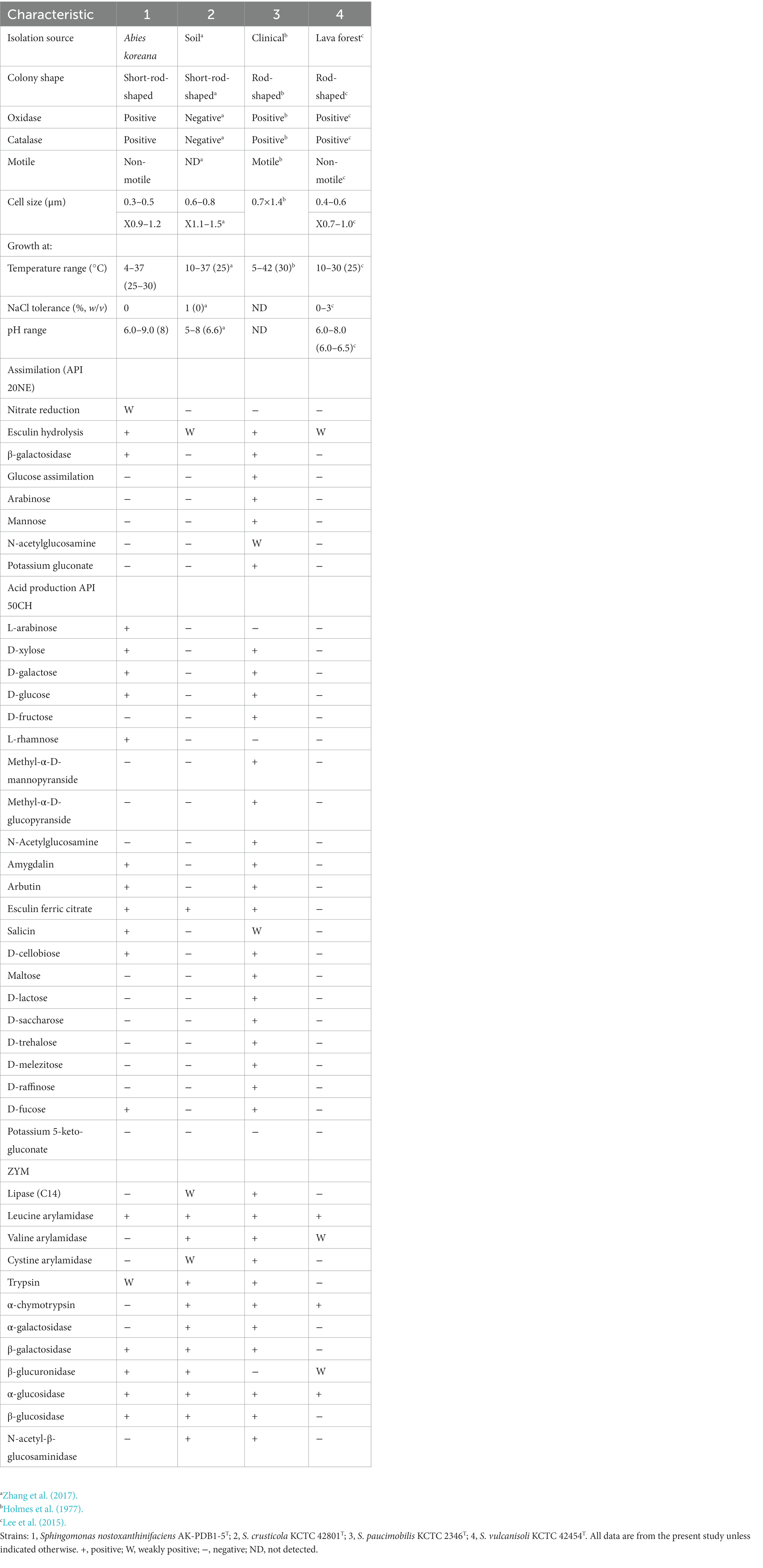
Table 1. Differential physiological and biochemical characteristics of strain AK-PDB1-5T from closely related type strains.
3.2. Phylogenetic analyzes
The nearly full-length 16S rRNA gene amplicon of strain AK-PDB1-5T was 1,419 nucleotides long (MZ956805). Based on the 16S rRNA gene sequence analysis, strain AK-PDB1-5T appeared to belong to the genus Sphingomonas. A comparative analysis of the 16S rRNA sequence showed the highest similarity to S. crusticola MIMD3T (95.6%) and S. jatrophae S5-249T (95.3%) with values between 93.9 and 95.2% to the top 25 type strains in the genus Sphingomonas. To provide an ecological insight, the 16S rRNA gene sequence of strain AK-PDB1-5T was compared to all the publicly available sequences on NCBI including environmental sequences and sequences from non-type strains. Results showed strain AK-PDB1-5T shared a high similarity (16S rRNA gene sequence more than 97% identity) with two uncultured Sphingomonas isolates from environmental samples (strain Plot29-2F10, accession number: EU202874), and one uncultured bacterium from pig deep litter system (clone A702, accession number: KM456152). Based on the novel species recognition threshold of 98.6% (Kim et al., 2014), strain AK-PDB1-5T was designated as a novel species in the genus Sphingomonas. The 16S rRNA gene sequence-based phylogenetic trees using NJ showed that strain AK-PDB1-5T formed clusters with S. crusticola MIMD3T, S. jatrophae S5-249T, and S. vulcanisoli SN6-13T. The ML and ME algorithms phylogenetic tree also supported this result (Figure 1). Therefore, S. crusticola MIMD3T (KCTC 42801T) and S. vulcanisoli SN6-13T (KCTC 42454T) as well as the type species S. paucimobilis KCTC 2346T were selected as reference species for further comparative testing.
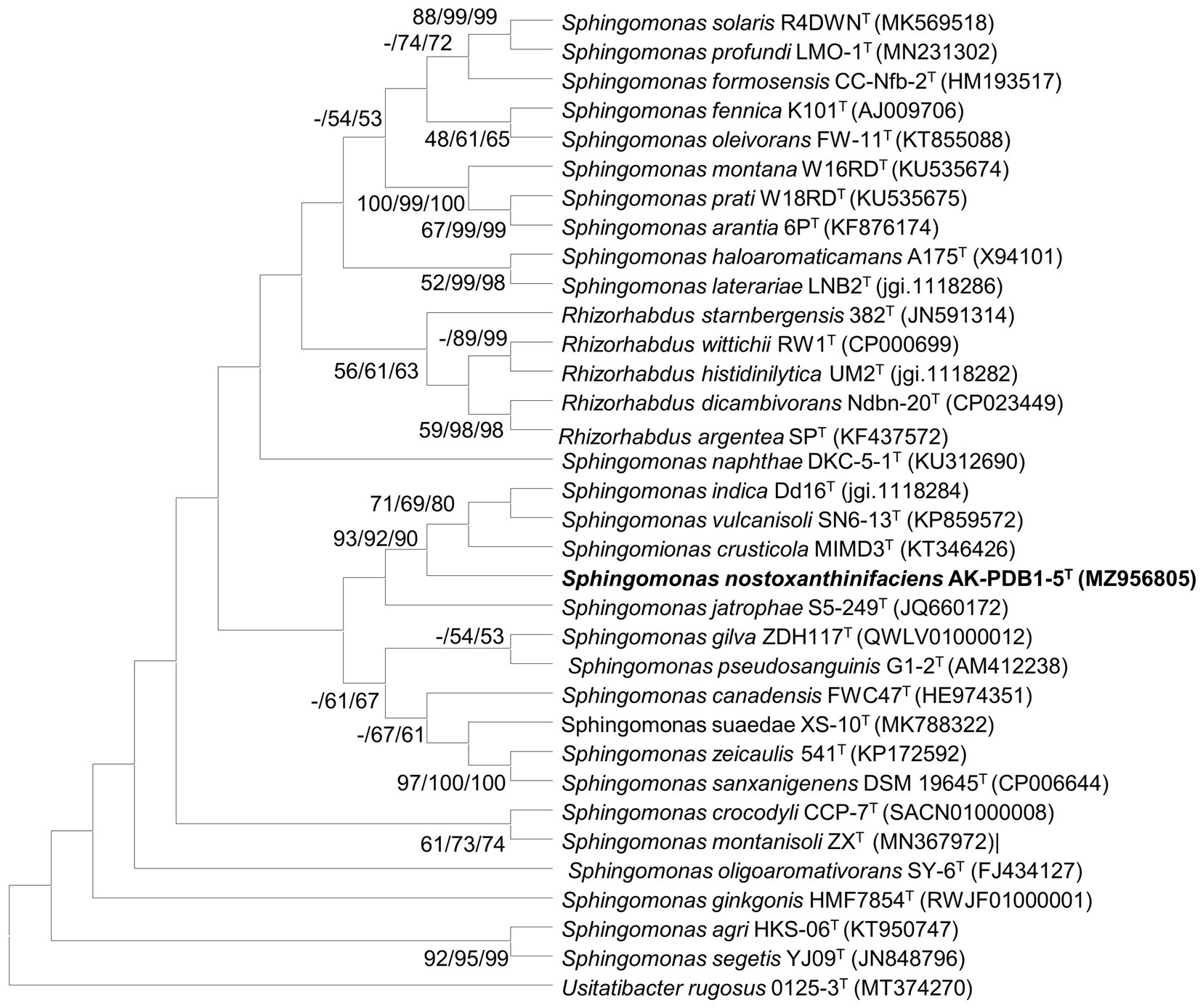
Figure 1. Maximum-likelihood (ML) algorithm phylogenetic tree based on 16S rRNA gene of strain AK-PDB1-5T. Bootstrap values (> 70%) calculated using the NJ, ML, and ME algorithms are shown.
3.3. Chemotaxonomic characterization
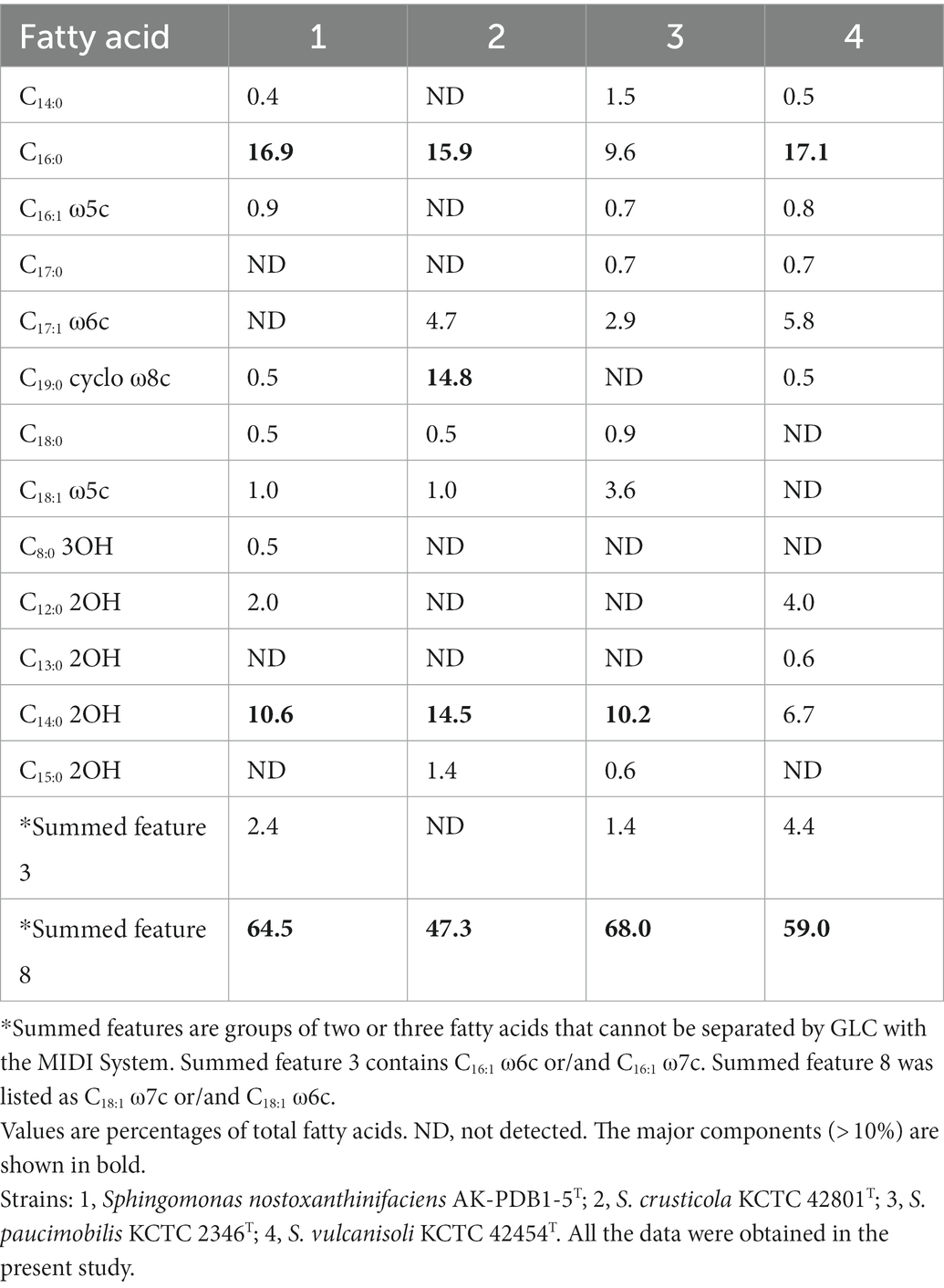
The major cellular fatty acids (> 10%) of strain AK-PDB1-5T and closely related strains are listed in Table 2. The most abundant fatty acids of strain AK-PDB1-5T were C14:0 2OH (10.6%), C16:0 (16.9%) and summed featured 8 (64.5%). Similar cellular fatty acid contents are consistent with closely related strains. However, the results for S. crusticola MIMD3T (KCTC 42801T) also include C19:0 cyclo ω8c, while the cellular fatty acid profile of S. vulcanisoli SN6-13T (KCTC 42454T) does not contain C16:0, and S. paucimobilis KCTC 2346T does not contain C14:0 2OH, which can differentiate strain AK-PDB1-5T from the most closely related strains.
Consistent with the genus Sphingomonas, Q-10 was found as the predominant isoprenoid of strain AK-PDB1-5T, and the polar lipid profile consisted of a mixture of phosphatidylethanolamine (PE), sphingoglycolipid (SGL), phosphatidylglycerol (PG), phospholipids (PL), and lipids (L; Supplementary Figure S2). The polar lipid profile of strain AK-PDB1-5T was similar to those of profiles identified from the closely related strains (Supplementary Figure S2). Although the enzyme for PC biosynthesis was annotated from the whole genome, PC was not detected in the polar lipids test. The polyamine patterns detected in strain AK-PDB1-5T were for homospermidine (53.4%), putrescine (41.0%), and spermidine (15.6%; Supplementary Figure S3). The major polyamines were consistent with those reported for members of the genus Sphingomonas (Zhang et al., 2010; Margesin et al., 2012; Sheu et al., 2015).
3.4. Genome properties and genetic relatedness
The whole genome of strain AK-PDB1-5T comprised a single circular chromosome of 4,298,284 bp in size after assembly by the SMRT Portal de novo assembler (version 2.3). A comparison of two copies of 16S rRNA gene fragments from the whole-genome sequence indicated that no contaminating DNA sequences occurred during genome assembly. The G + C content calculated from the genome of AK-PDB1-5T was 67.0%, similar to other Sphingomonas species, which had high G + C contents as indicated in a previous report for the genus Sphingomonas (57.4–70.5 mol%).7 The whole genome was deposited in NCBI under the accession number CP082839.1. A total of 3,994 protein-coding genes and 60 RNA genes, including two 5S rRNA genes, two 16S rRNA genes, two 23S rRNA genes, four ncRNAs, and 50 tRNA genes were annotated by PGAP (Supplementary Table S1). According to the cluster orthologous group (COG) annotation results, most CDSs were classified as unknown (29.5% of total assigned COGs), and general function prediction only (8.2% of the total assigned COGs), amino acid transport, and metabolism (6.0% of the total assigned COGs; Supplementary Figures S4, S5). The genome comparison between strain AK-PDB1-5T and the closest species showed ANI values in a range of 75.1–76.8%. The dDDH values ranged from 19.5 to 21%, and the OrthoANI values ranged from 73.8 to 76.5 (Supplementary Figure S6), which also supported that the strain was a novel species in the genus Sphingomonas, considering that the values obtained were significantly lower than the proposed dDDH (< 70%) and ANI cutoff (95–96%) values for bacterial species delineation (Chun et al., 2018). A circular map of the stain AK-PDB1-5T is shown in Supplementary Figure S2. The whole genome of closely related strains obtained from NCBI GenBank showed a relatively high quality (more than 83.2) by employing MiGA webserver to assess the quality/completeness/contamination of the genome assembly (Supplementary Table S1). The whole-genome phylogenomic tree based on 92 core genes also supported that strain AK-PDB1-5T formed a phylogenetic lineage within the genus Sphingomonas, consistent with the 16S rRNA-based phylogenomic tree (Figure 2).

Figure 2. Maximum-likelihood (ML) algorithm phylogenomic tree based on up-to-date bacterial core genes (UBCGs; concatenated alignment of 92 core genes) showing the relationship between strain AK-PDB1-5T and other members of the genus Sphingomonas. Gene support index (GSI, left) and bootstrap values (right) are indicated at the nodes. Scale bar, 0.050 substitutions per position. The 92 bacterial core genes were alaS, argS, aspS, cgtA, coaE, cysS, dnaA, dnaG, dnaX, engA, ffh, fmt, frr, ftsY, gmk, hisS, ileS, infB, infC, ksgA, lepA, leuS, ligA, nusA, nusG, pgk, pheS, pheT, prfA, pyrG, recA, rbfA, rnc, rplA, rplB, rplC, rplD, rplE, rplF, rplI, rplJ, rplK, rplL, rplM, rplN, rplO, rplP, rplQ, rplR, rplS, rplT, rplU, rplV, rplW, rplX, rpmA, rpmC, rpmI, rpoA, rpoB, rpoC, rpsB, rpsC, rpsD, rpsE, rpsF, rpsG, rpsH, rpsI, rpsJ, rpsK, rpsL, rpsM, rpsO, rpsP, rpsQ, rpsR, rpsS, rpsT, secA, secG, secY, serS, smpB, tig, tilS, truB, tsaD, tsf, uvrB, ybeY, and ychF.
Based on data obtained by the phenotypic, physiological, phylogenomic, and biochemical results support that strain AK-PDB1-5T is a member of the genus Sphingomonas. However, a low 16S rRNA similarity (below 95.6%), ANI values (less than 75.1%), as well as low dDDH values (less than 19.5%), leads to the conclusion that strain AK-PDB1-5T differs from the most closely related strains. This conclusion was further supported by the phylogenetic trees based on the 16S rRNA and core gene sets from the whole-genome sequence. Taken together, this data suggests that AK-PDB1-5T is a new member of the genus Sphingomonas.
3.5. Genome-derived features of strain AK-PDB1-5T
Genome annotation was carried out using the RAST server and the BlastKOALA pipeline to reconstruct the metabolic pathways. The following sections are predicted from the genome sequences (Supplementary Figure S7).
3.5.1. Phenotypic and biochemical features
3.5.1.1. Motility
Genes related to flagellar motility in prokaryota were detected in the whole genome, such as the flagellar motor switch protein FliM, the basal-body rod protein FlgC, the flagellin protein FlaA, the flagellar L-ring protein FlgH, the flagellar motor switch protein FliN, the flagellar biosynthesis proteins FlhB, FlhA, FliR, the flagellar motor rotation proteins MotA, MotB, the flagellum specific ATP synthase FliI, and the flagella basal-body rod modification protein FlgD. More than 11,000 amino acids in the genus Lactobacillus (Cousin et al., 2015), and 27 proteins in the genus Flavobacteriaceae were related to flagella motility (Gavriilidou et al., 2020). The gene cluster of flagellar motility for Sphingomonas is poorly characterized. Although many flagellar assembly genes were annotated in the genome of strain AK-PDB1-5T, it may absence of some genetic elements since it lacks flagella and non-motile.
3.5.1.2. Respiration
Genome annotation indicated the presence of all the subcategory genes related to terminal electron-accepting reactions and electron-donating reactions. Some oxidase subunits such as terminal cytochrome O ubiquinol oxidase, terminal cytochrome C ubiquinol oxidase, terminal cytochrome oxidase, and anaerobic respiratory reductases such as vanillate O-demethylase oxidoreductase (EC 1.14.13), flavodoxin reductases (ferredoxin-NADPH reductases) family 1, and ferredoxin reductase were annotated from the whole genome. The electron-donating reactions included respiratory complex I, NADH ubiquinone oxidoreductase (chain A-N), and others.
Stress response: Genes related to stress response including oxidative stress, detoxification, and periplasmic stress were detected. Osmotic stress-related genes coding for the outer membrane protein A precursor, the synthesis of osmoregulated periplasmic glucans, protection from reactive oxygen species, oxidative stress, biosynthesis, and gamma-glutamyl cycle as well as glutaredoxins were seen. Detoxification-related genes included a glutathione-dependent pathway for formaldehyde detoxification. Periplasmic stress-related genes containing the periplasmic stress response were also noted.
Polar lipid metabolism: phosphatidylethanolamine (PE), Phosphatidylcholine (PC) which is biosynthesized from PE, and sphingolipid (SGL) biosynthesis pathway were reconstructed from the whole genome via the BlastKOALA pipeline. The PC biosynthesis enzymes phosphatidylethanolamine/phosphatidyl-N-methylethanolamine N-methyltransferase [EC:2.1.1.17 2.1.1.71], and the PE biosynthesis enzymes phosphatidate cytidylyltransferase [EC:2.7.7.41]; CDP-diacylglycerol-serine O-phosphatidyltransferase [EC:2.7.8.8]; phosphatidylserine decarboxylase [EC:4.1.1.65] were annotated in the genome. However, a polar lipid pattern consisting of phosphatidylethanolamine (PE), sphingoglycolipid (SGL), phosphatidylglycerol (PG), phospholipids (PL), and lipid (L) was detected for strain AK-PDB1-5T in two-dimensional TLC.
3.5.2. Metabolite biosynthesis gene clusters analysis
Although the genus Sphingomonas contains 146 published species, only limited secondary metabolites were found (Mnich et al., 2017), and it remains untapped for potential applications for those natural products. AntiSMASH is an important tool for the mining of gene clusters of natural products. BGCs encode enzymes related to the secondary metabolite biosynthesis pathway. The whole genome of strain AK-PDB1-5T was used to predict the secondary metabolite gene clusters by the online tool antiSMASH (Blin et al., 2021). Six BGCs regions were identified from the whole genome of AK-PDB1-5T (Supplementary Table S2 and Supplementary Figures S8–S13), which contained sphingan polysaccharide, malleobactin A/malleobactin B/malleobactin C/malleobactin D and zeaxanthin. Sphingan polysaccharide is a very common compound in this genus, exemplified by compounds such as WL gum, gallen gum, rhamsan gum, and welan gum (Zheng et al., 2018; Baez et al., 2019; Li et al., 2019; Zhu et al., 2019 2020, 2021; Huang et al., 2020; Liu et al., 2020; Xu et al., 2020; Ke et al., 2021; Zhao et al., 2021). The sphingans group has applications in a variety of industries such as food, cement, personal care, abiotic stress tolerance, bioremediation, and biodegradation, etc. Malleobactin is a member of the siderophore family and has an important role in plant resistance to pathogens (Xiao et al., 2020; Saboe et al., 2021). Zeaxanthin has significant commercial value for its antioxidant potential and is widely used in the food, cosmetic and pharmaceutical industries. Some species in the genus Sphingomonas have been reported to produce zeaxanthin (Asker et al., 2007; Thawornwiriyanun et al., 2012); however, the biosynthesis pathway and related gene clusters are not clear. We provide the BGCs information and valuable insights into those natural products.
3.5.3. Identification of the carotenoid pigment
Strain AK-PDB1-5T grown on PDA medium was white-yellow in color and showed carotenoid accumulation. The genome mining tool antiSMASH was used to predict the secondary metabolite gene clusters of carotenoids, and the results suggested that the yellow color produced by strain AK-PDB1-5T could be zeaxanthin, caloxanthin (zeaxanthin with one hydroxyl group added), or nostoxanthin (zeaxanthin with two hydroxyl groups added).
We analyzed a methanol extract using HPLC and some carotenoid standards including zeaxanthin, lutein, cryptoxanthin, β-carotene, α-carotene, violaxanthin, and neoxanthin were tested by HPLC at the same time. The results showed the pigment from strain AK-PDB1-5T had a retention time of 4.5 min and a minor peak with a retention time of 5.5 min (Supplementary Figure S14). However, these peaks did not correspond to the peaks produced by authentic standards of zeaxanthin, lutein, cryptoxanthin, β-carotene, α-carotene, violaxanthin, or neoxanthin (Supplementary Figure S14). Compared to zeaxanthin, the main peak of AK-PDB1-5T had a shorter retention time, because the carotenoid from strain AK-PDB1-5T has higher hydrophilicity than zeaxanthin.
UV–Vis absorption spectroscopy was performed to determine the absorption spectra of a methanol extract from strain AK-PDB1-5T, and showed highly absorbent peaks at 452 and 480 nm (Figure 3A). A previous study showed that β-carotene produced a strong band with subpeaks at 481, 454, and near 425 nm. On the other hand, zeaxanthin and caloxanthin, a hydroxyl-substituted derivative of β-carotene, has three bands with a shape almost identical to those of β-carotene (λmax values at 422, 450, and 476 nm; Zhu et al., 2012; Takaya et al., 2018), canthaxanthin (473 nm), nostoxanthin (λmax at 427, 452, and 480 nm; Kikukawa et al., 2021), and astaxanthin (477 nm) all carbonyl-substituted derivatives of β-carotene show absorption bands much less structured than those of β-carotene (Takaya et al., 2018). The strain AK-PDB1-5T spectra were highly matched with nostoxanthin (λmax values at 452 and 480 nm; Takaya et al., 2018) and close to that of zeaxanthin (λmax values at 422, 450, and 476 nm; Pavelková et al., 2020).
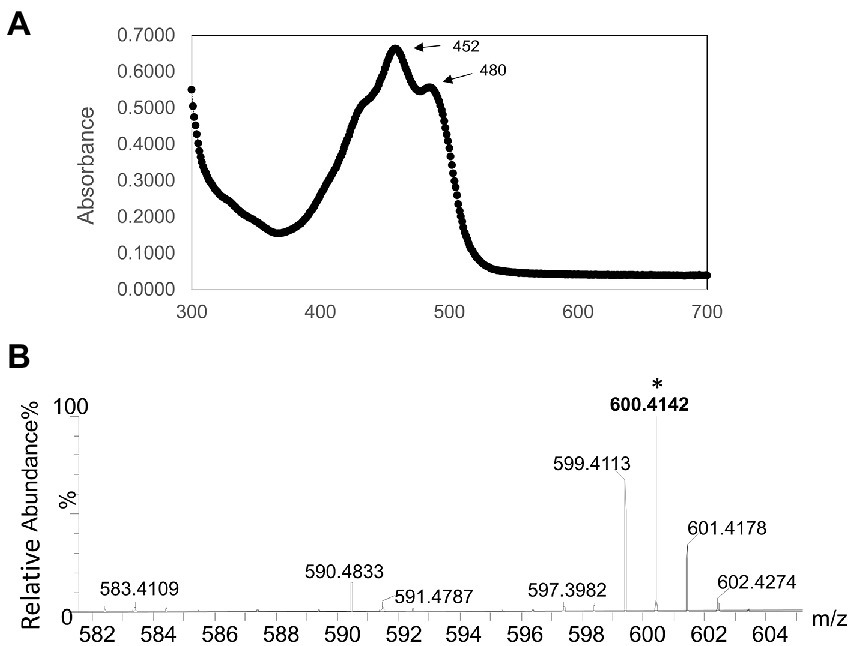
Figure 3. Identification of the carotenoid produced by strain AK-PDB1-5T (A) Ultraviolet-visible absorption spectra (UV–Vis) of the major carotenoid from AK-PDB1-5T (B) ESI-MS ion spectra of the carotenoids from AK-PDB1-5T.
To identify the carotenoid produced by strain AK-PDB1-5T, an analysis was conducted by UPLC equipped with an electro-spray ionization (ESI) time-of-flight (TOF) mass spectrometer (MS), and MS/MS systems. The ESI-MS profile of all the extracts was found to be comparable with the standard, the zeaxanthin MS/MS spectra data of [M + H]+: 569, caloxanthin MS/MS spectra data of [M + H]+: 569, β-carotene MS/MS spectra data of [M + H]+: 537, while nostoxanthin spectra data of m/z 600.4243 [M + H]+ (Zhu et al., 2012). The molecular formula of the carotenoid from strain AK-PDB1-5T was determined to be C40H56O4, with ESI MS spectra data of m/z 600.4243 [M + H]+ and m/z 583.4109 [M + H-18]+ (Figure 3B). The ion patterns produced were same as the MassBank Record: CA000155 that is available online Nostoxanthin Mass Spectrum (massbank.jp) and as those from previous studies (Zhu et al., 2012). Based on the results above, the carotenoid produced by strain AK-PDB1-5T was identified as nostoxanthin.
3.5.4. Nostoxanthin biosynthetic pathway
Nostoxanthin ((2R,3R,2′R,3′R)-β, β-Carotene-2,3,2′,3′-tetrol) is biosynthesized from zeaxanthin. Zeaxanthin, the precursor of nostoxanthin, is hydroxylzed twice, first to caloxanthin and finally to nostoxanthin (Zhu et al., 2012). Although many different carotenoid (crt) synthetic enzymes have been cloned from bacteria, plants and algae, much further study is still necessary to fully elucidate the biosynthesis of carotenoids as well as modifications of the final structures. To better understand the biosynthesis pathway in strain AK-PDB1-5T, the whole genome was used to predict it. Two separate pathways exist for the biosynthesis of IPP, including the mevalonate (MVA) pathway and the 2-C-Methyl-D-erythritol 4-phosphate (MEP) pathway, both of which are carotenoid precursors. Genome annotation analysis via PGAP, RAST SEED, and AntiSMASH annotations revealed that IPP from strain AK-PDB1-5T is only generated via the MEP pathway. Some genes for some enzymes in the MEP pathway were predicted; one copy of 1-deoxy-D-xylose 5-phosphate synthase (dxs), one copy of 1-deoxy-D-xylose 5-phosphate reductoisomerase (dxr), putative intracellular septation protein A (ispZ), two copies of 4-hydroxy-3-methylbut-2-enyl diphosphate (ispH), one copy of flavodoxin-dependent(E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ispG), one copy of phosphoenolpyruvate carboxykinase (pck), one copy of phosphoenolpyruvate carboxylase (ppc), and one copy of pyruvate kinase (pykFA), all for the MEP pathway. No genes for enzymes in the MVA pathway were predicted. The absence of genes for the MVA pathway reinforces the conclusion that in strain AK-PDB1-5T, IPP is biosynthesized via the MEP pathway. These results were consistent with a previous study that indicated that the MEP pathway is typically found in most bacteria and on plant plastids, while the MVA pathway is mostly present in plants and archaea (Li et al., 2020).
After IPP has been synthesized, carotenoid biosynthesis continues by growing the polyprenyl pyrophosphate chain. IPP is isomerized to geranyl PP (GPP, C10), farnesyl PP (FPP, C15), or geranylgeranyl PP (GGPP, C20) by ispA and crtE or gps, which are precursors of mono-, di-, and tri-terpenes and carotenoids. Next, two GGPP molecules are condensed head-to-head by phytoene synthase, resulting in the formation of the first carotene, phytoene (C40) by crtB (Figure 4A). Four double bonds are introduced into phytoene to form lycopene, which then becomes a precursor to a variety of carotenoids, e.g., lycopene (crtI), β-carotene (crtL), zeaxanthin (crtZ), and astaxanthin (crtW; Kim et al., 2014; Lee et al., 2018). Zeaxanthin is converted to the yellow pigment nostoxanthin by the gene product of crtG. The crtG gene encodes 2,2-β-hydroxylase which is responsible for hydroxylation of β-carotene (Zhu et al., 2012). In comparison with the carotenoid biosynthesis gene cluster of “Sphingomonas elodea” ATCC 31461 (JN224892.1), which has been reported to produce nostoxanthin, strain AK-PDB1-5T showed 71.1% gene similarity to S. elodea ATCC 31461. The carotenoid biosynthesis gene cluster as well as nostoxanthin biosynthesis pathway of stain AK-PDB1-5T has been proposed (Figure 4). PGAP, RAST SEED and AntiSMASH annotations suggest that the strain AK-PDB1-5T analyzed in this study possesses the gene cluster crtB, crtG, crtI, crtY, crtZ to produce the carotenoid nostoxanthin from the precursor GGPP (Figure 4B). Although we did not directly find crtG from the whole genome, by blasting the crtG gene sequence of “Sphingomonas elodea” ATCC 31461 (JN224892.1; Zhu et al., 2012), a 76.3% gene similarity to crtG of “Sphingomonas elodea” ATCC 31461 was found in the whole genome of AK-PDB1-5T. Based on the annotation results from the whole genome, strain AK-PDB1-5T was confirmed to produce nostoxanthin.
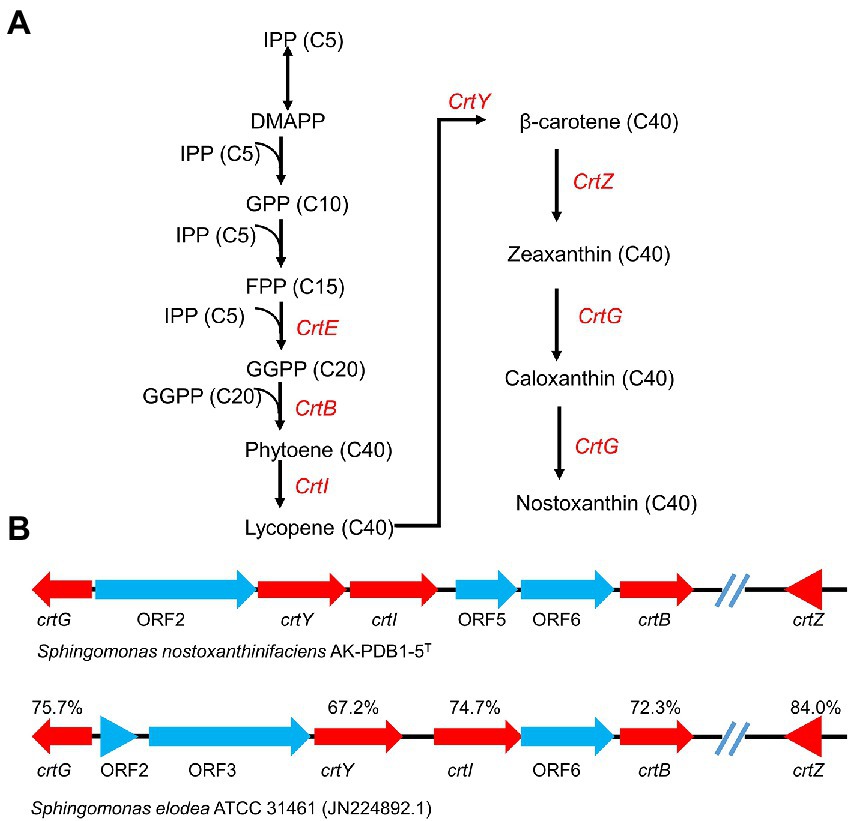
Figure 4. Carotenogenic gene clusters and the proposed biosynthetic pathway of nostoxanthin in strain AK-PDB1-5T. (A) comparison of the gene cluster in strain AK-PDB1-5T (up) and “Sphingomonas elodea” ATCC 31461 (JN224892.1; bottom). The genes are presented as arrows pointing in the direction of transcription; the value below each gene designates the similarity of nucleotides between the AK-PDB1-5T nostoxanthin biosynthetic enzyme and the corresponding gene from the S. elodea ATCC 31461. (B) nostoxanthin biosynthetic pathway in strain AK-PDB1-5T. IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate. crtE, GGPP synthase; crtB, phytoene synthase; crtG, carotenoid 2,2-β-hydroxylase; crtI, phytoene desaturase/dehydrogenase; crtY, lycopene cyclase; crtZ, and β-carotene hydroxylase.
3.6. Effect of strain AK-PDB1-5T on plant growth under salt condition
To evaluate the effect of strain AK-PDB1-5T on plant growth under salt conditions, 7-day-old Arabidopsis seedlings were co-cultivated with strain AK-PDB1-5T under 0 mM and 100 mM of NaCl. The leaf fresh weight and root fresh weight were examined after 10 days of co-cultivation (Figure 5). The results showed strain AK-PDB1-5T significantly improved the Arabidopsis seedling biomass under 0 mM and 100 mM of NaCl. Shoot fresh weight was increased 1.69-fold under salt condition (100 mM NaCl), which was increased higher compared to normal conditions (1.34-fold), while root fresh weight was increased 1.02-fold under salt condition. A similar result was found in chlorophyll content. These results showed that strain AK-PDB1-5T has not only plant growth-promoting activity under normal condition, but also enhances salt tolerance when plants are exposed to salt stress.
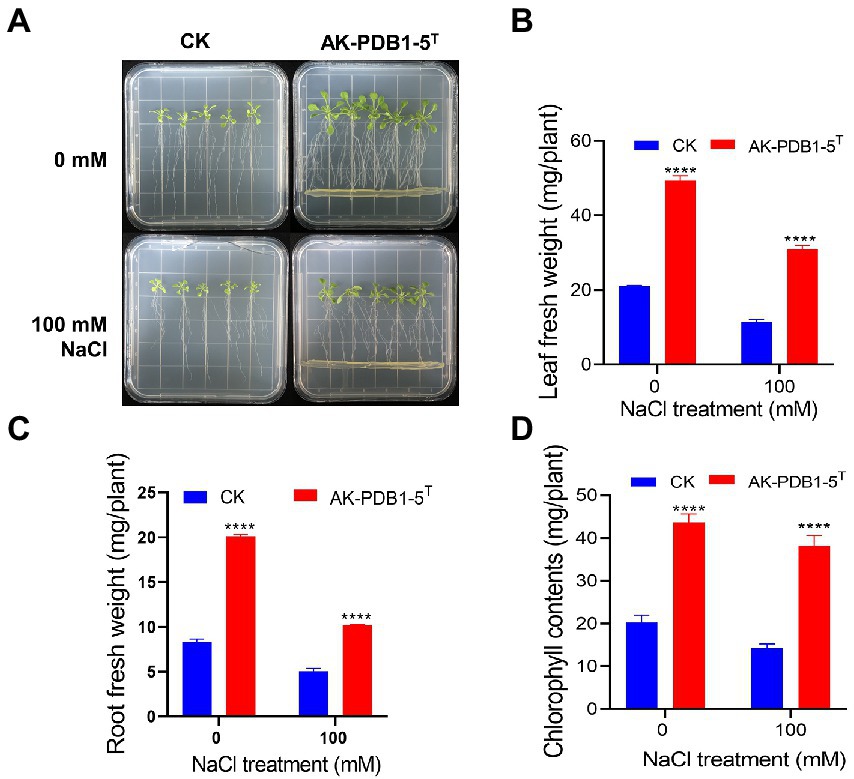
Figure 5. Plant growth-promoting effects of Arabidopsis seedling by strain AK-PDB1-5T under normal and salt stress conditions. (A) Comparison of Arabidopsis seedling growth by control (no treatment, CK) and AK-PDB1-5T co-cultivated at 22°C for 10 days under normal and salt stress conditions (100 mM NaCl). (B–D) Quantification of leaf fresh weight (B), root fresh weight (C), and chlorophyll contents (D) Error bars represent the standard error. Asterisks indicate significant differences between the control and AK-PDB1-5T treatment under salt stress (100 mM NaCl; ****p < 0.0001).
Furthermore, we examined ROS production in Arabidopsis root tips under salt stress condition (Figure 6). We observed that ROS levels in roots were significantly increased under salt stress condition (100 mM NaCl; Figure 6), but the fluorescence intensity of ROS in root tips was greatly decreased by strain AK-PDB1-5T (Figure 6). These results suggest that strain AK-PDB1-5T alleviates salt stress in Arabidopsis roots by scavenging the ROS. In the previous study, it has well studied that carotenoid is known lipid protectants and active oxygen scavengers, and play an important role in antioxidant caused by ROS, these carotenoid can scavenge ROS and free radicals and/or free radicals in the lipid phase of the plant cell membrane, help the plant alleviate salt stress (Zhang et al., 2012, 2016; Ren et al., 2021; Song et al., 2022). For instance, pretreated with carotenoids (β-carotene) improved the Lepidium sativum growth under salt stress, as β-carotene increased the antioxidant activity of the plant and mitigated salt stress (Babaei et al., 2022); exogenous zeaxanthin alleviates pepper plant grown under low temperature and low light by reduced ROS accumulation in pepper seedlings. In the present study, nostoxanthin, as one type of yellow carotenoid, biosynthesized from zeaxanthin by the addition of two hydroxyl groups, was produced by strain AK-PDB1-5T, was found to reduce ROS accumulation caused by salt stress, promoted plant growth under salt stress. Except for nostoxanthin, we try to find some specific metabolic pathways that may reduce the abundance of ROS, such as a complete trehalose biosynthesis pathway, including glgB gene (1,4-alpha-glucan branching enzyme [EC:2.4.1.18]), glgA gene (starch synthase [EC:2.4.1.21]), glgC (glucose-1-phosphate adenylyltransferase [EC:2.7.7.27]), treX (isoamylase [EC:3.2.1.68]), glgZ (maltooligosyltrehalose trehalohydrolase [EC:3.2.1.141]), and glgY ((1- > 4)-alpha-D-glucan 1-alpha-D-glucosylmutase [EC:5.4.99.15]). Trehalose as a key organic osmolyte was proven to be effectively involved in plant abiotic stress. The plant enhanced tolerance to cold, salinity, and drought tolerance by exogenous trehalose. However, more studies need to be performed to understand the mechanism.
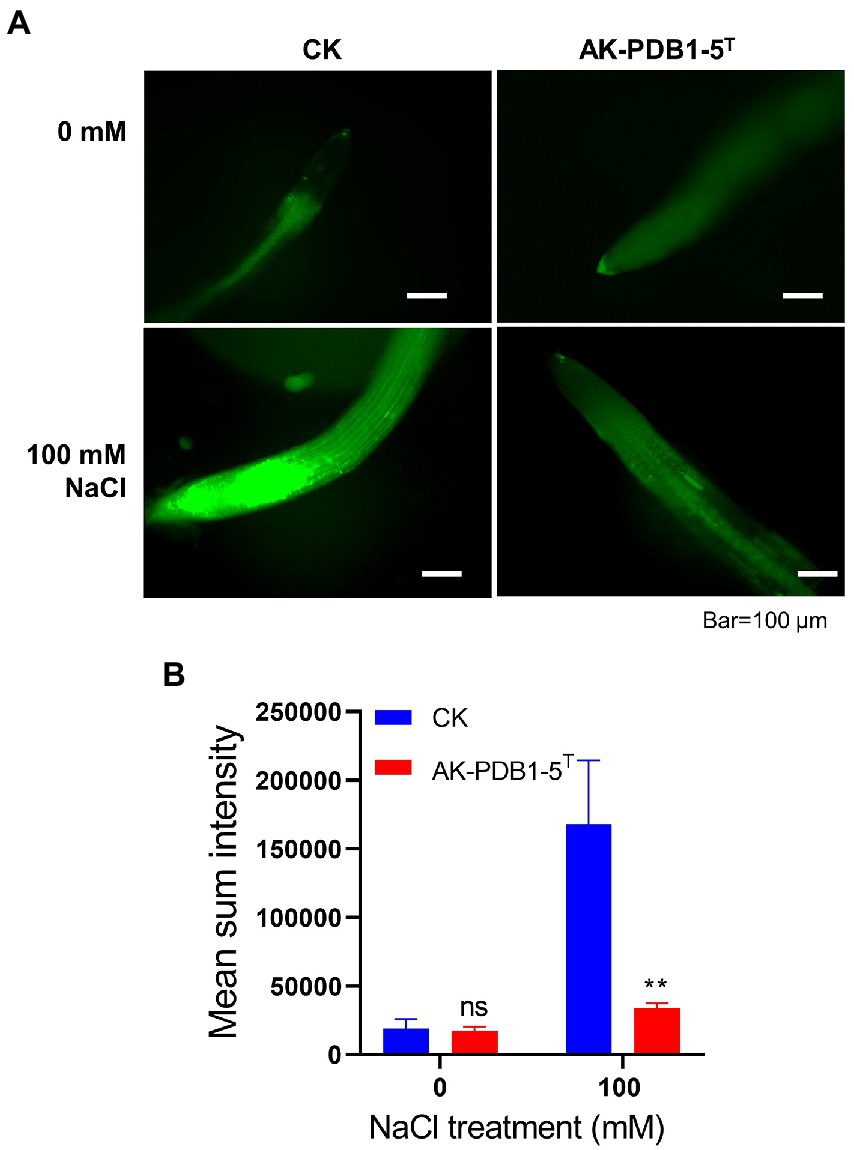
Figure 6. DCFH-DA staining of ROS in roots. (A) DCFH-DA straining of root tips after co-inoculation with strain AK-PDB1-5T upon different salt conditions (0 and 100 mM NaCl). Scale bar: 100 μm. (B) The fluorescence intensities of roots were measured by ImageJ. Error bars represent the standard error. Asterisks indicate significant differences between the control and AK-PDB1-5T treatment under salt stress condition (100 mM NaCl, **p < 0.01).
3.6.1. Description of Sphingomonas nostoxanthinifaciens sp. nov
Sphingomonas nostoxanthinifaciens (nos.to.xan.thi.ni.fa’ci.ens. N.L. neut. n. nostoxanthinum, nostoxanthin; L. pres. part. faciens, making/producing; N.L. part. Adj. nostoxanthinifaciens, nostoxanthin-producing).
Colonies are white-yellow pigmented, circular, smooth, and opaque (2–6 mm in diameter) after growth on PDA medium at 25°C for 3 days. Strain AK-PDB1-5T is found to grow well on PDA, R2A, can grow poorly on LB, NA, TSA, and YEP (optimum PDA and R2A), but not on MA. Cells are Gram-negative, catalase-, oxidase-positive, non-motile, and rod-shaped (0.3–0.5 μm in width and 0.9–1.2 μm in length), without flagella. Optimal growth occurs at 25–30°C, pH 8.0, and in 0% (w/v) NaCl.
In the API ZYM test, positive reactions for alkaline phosphate, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphate, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucuronidase, α-glucosidase, and β-glucosidase, while the other substrates were negative. On API 20NE strips, esculin hydrolysis, nitrate reduction, and β-galactosidase showed positive reactions, while the other reactions are negative. On API 50CH strips, L-arabinose, D-galactose, D-glucose, L-rhamnose, amygdalin, arbutin, esculin ferric citrate, salicin, D-cellobiose, and D-fucose show positive reactions, and other substrates are negative. The major fatty acids are C14:0 2OH, C16:0, and summed featured 8. The respiratory quinone detected in strain AK-PDB1-5T was Q-10, while sphingoglycolipid and phosphatidylethanolamine are found as the major polar lipids.
The type strain is Sphingomonas nostoxanthinifaciens AK-PDB1-5T (= KCTC 82822T = CCTCC AB 2021150T), isolated from needle-like leaves of the Korean fir taken from Mt. Halla, Jeju island, South Korea. The accession numbers for 16S rRNA gene and the whole-genome sequences of strain AK-PDB1-5T are MZ956805.1 and CP082839.1.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JL designed the research and supervised the project. LJ and JS carried out the experiment. LJ analyzed the results and wrote the manuscript. YP, DJ, JHL, and CK edited the manuscript. JL and LJ modified the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM5282223) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R111A2072308).
Acknowledgments
We thank Seon Ju Park at the Korea Basic Science Institute (KBSI, Chuncheon Center) for high-resolution mass data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1101150/full#supplementary-material
Footnotes
1. ^https://lpsn.dsmz.de/genus/sphingomonas
2. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^http://antismash.secondarymetabolites.org
5. ^http://ggdc.dsmz.de/distcalc2.php
References
Ahn, C., Lee, J. H., Park, M. J., Kim, J. W., Yang, J., Yoo, Y. M., et al. (2020). Cytostatic effects of plant essential oils on human skin and lung cells. Exp. Ther. Med. 19, 2008–2018. doi: 10.3892/etm.2020.8460
Aly, A. H., Debbab, A., and Proksch, P. (2013). Fungal endophytes-secret producers of bioactive plant metabolites. Pharmazie 68, 499–505. doi: 10.1691/ph.2013.6517
Asker, D., Beppu, T., and Ueda, K. (2007). Sphingomonas jaspsi sp. nov., a novel carotenoid-producing bacterium isolated from Misasa, Tottori, Japan. Int. J. Syst. Evol. Microbiol. 57, 1435–1441. doi: 10.1099/ijs.0.64828-0
Babaei, M., Shabani, L., and Hashemi-Shahraki, S. (2022). Improving the effects of salt stress by beta-carotene and gallic acid using increasing antioxidant activity and regulating ion uptake in Lepidium sativum L. Bot. Stud. 63:22. doi: 10.1186/s40529-022-00352-x
Baez, L. A., Santos, J., Ramirez, P., Trujillo-Cayado, L. A., and Munoz, J. (2019). Development of emulgels formulated with sweet fennel oil and rhamsan gum, a biological macromolecule produced by Sphingomonas. Int. J. Biol. Macromol. 129, 326–332. doi: 10.1016/j.ijbiomac.2019.01.114
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Busse, J., and Auling, G. (1988). Polyamine pattern as a chemotaxonomic marker within the proteobacteria. Syst. App. Microbiol. 11, 1–8. doi: 10.1016/s0723-2020(88)80040-7
Busse, H. J., Hauser, E., and Kämpfer, P. (2005). Description of two novel species, Sphingomonas abaci sp. nov. and Sphingomonas panni sp. nov. Int. J. Syst. Evol. Microbiol. 55, 2565–2569. doi: 10.1099/ijs.0.63872-0
Chen, J., Wong, M. H., Wong, Y. S., and Tam, N. F. (2008). Multi-factors on biodegradation kinetics of polycyclic aromatic hydrocarbons (PAHs) by Sphingomonas sp. a bacterial strain isolated from mangrove sediment. Mar. Pollut. Bull. 57, 695–702. doi: 10.1016/j.marpolbul.2008.03.013
Chinthanom, P., Vichai, V., Dokladda, K., Sappan, M., Thongpanchang, C., and Isaka, M. (2021). Semisynthetic modifications of antitubercular lanostane triterpenoids from Ganoderma. J. Antibiot. (Tokyo) 74, 435–442. doi: 10.1038/s41429-021-00422-5
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Collins, M., Shah, H., and Minnikin, D. (1980). A note on the separation of natural mixtures of bacterial menaquinones using reverse phase thin-layer chromatography. J. Appl. Bacteriol. 48, 277–282. doi: 10.1111/j.1365-2672.1980.tb01227.x
Cousin, F. J., Lynch, S. M., Harris, H. M., McCann, A., Lynch, D. B., Neville, B. A., et al. (2015). Detection and genomic characterization of motility in lactobacillus curvatus: confirmation of motility in a species outside the lactobacillus salivarius clade. Appl. Environ. Microbiol. 81, 1297–1308. doi: 10.1128/AEM.03594-14
Fadiji, A. E., and Babalola, O. O. (2020). Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 8:467. doi: 10.3389/fbioe.2020.00467
Gavriilidou, A., Gutleben, J., Versluis, D., Forgiarini, F., Passel, M. W. J., Ingham, C. J., et al. (2020). Comparative genomic analysis of Flavobacteriaceae: insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genomics. 21:569. doi: 10.1186/s12864-020-06971-7
Holmes, B., Owen, R. J., Evand, A., Malnick, H., and Willcox, W. R. (1977). Pseudomonas paucimobilis, a new species isolated from human clinical specimens, the hospital environment, and other sources. Int. J. Syst. Evol. Bacteriol. 27, 133–146. doi: 10.1099/00207713-27-2-133
Huang, J., Zhu, S., Li, C., Zhang, C., and Ji, Y. (2020). Cost-effective optimization of gellan gum production by Sphingomonas paucimobilis using corn steep liquor. Prep. Biochem. Biotechnol. 50, 191–197. doi: 10.1080/10826068.2019.1692215
Jenkins, C. L., Andrewes, A. G., McQuade, T. J., and Start, M. P. (1979). The pigment of Pseudomonas paucimobilis is a carotenoid (nostoxanthin), rather than a brominated aryl-polyene (xanthomonadin). Curr. Microbiol. 3, 1–4. doi: 10.1007/BF02603124
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kaur, J., Kaur, J., Niharika, N., and Lal, R. (2012). Sphingomonas laterariae sp. nov., isolated from a hexachlorocyclohexane-contaminated dump site. Int. J. Syst. Evol. Microbiol. 62, 2891–2896. doi: 10.1099/ijs.0.034686-0
Kawahara, K., and Kuraishi, H. (1999). Chemical structure and function of glycosphingolipids of Sphingomonas spp. and their distribution among members of the alpha-4 subclass of Proteobacteria. J. Ind. Microbiol. Biotechnol. 23, 408–413. doi: 10.1038/sj.jim.2900708
Ke, C., Wei, L., Wang, M., Li, Q., Liu, X., Guo, Y., et al. (2021). Effect of NaCl addition on the production of welan gum with the UV mutant of Sphingomonas sp. Carbohydr. Polym. 265:118110. doi: 10.1016/j.carbpol.2021.118110
Khan, A. L., Waqas, M., Kang, S. M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52, 689–695. doi: 10.1007/s12275-014-4002-7
Kikukawa, H., Okaya, T., Maoka, T., Miyazaki, M., Murofushi, K., Kato, T., et al. (2021). Carotenoid nostoxanthin production by Sphingomonas sp. SG73 isolated from deep sea sediment. Mar. Drugs 19:274. doi: 10.3390/md19050274
Kim, H. J., Choi, E. H., and Lee, I. S. (2004). Two lanostane triterpenoids from Abies koreana. Phytochemistry 65, 2545–2549. doi: 10.1016/j.phytochem.2004.07.007
Kim, S. H., Kim, J. H., Lee, B. Y., and Lee, P. C. (2014). The astaxanthin dideoxyglycoside biosynthesis pathway in Sphingomonas sp. PB304. Appl. Microbiol. Biotechnol. 98, 9993–10003. doi: 10.1007/s00253-014-6050-7
Kim, H. J., Le, Q. K., Lee, M. H., Kim, T. S., Lee, H. K., Kim, Y. H., et al. (2001). A cytotoxic secocycloartenoid from Abies koreana. Arch. Pharm. Res. 24, 527–531. doi: 10.1007/BF02975159
Kim, S. J., Moon, J. Y., Lim, J. M., Ahn, J. H., Weon, H. Y., Ahn, T. Y., et al. (2014). Sphingomonas aerophila sp. nov. and Sphingomonas naasensis sp. nov., isolated from air and soil, respectively. Int. J. Syst. Evol. Microbiol. 64, 926–932. doi: 10.1099/ijs.0.055269-0
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platform. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lane, D. (1991). “16S/23S rRNA sequencing” in Nucleic Acid Techniques in Bacterial Systematics. eds. E. Stackebrandt and M. Goodfellow (New York, NY: Wiley).
Lee, I., Chalita, M., Ha, S. M., Na, S. I., Yoon, S. H., and Chun, J. (2017). ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 67, 2053–2057. doi: 10.1099/ijsem.0.001872
Lee, K. C., Kim, K. K., Kim, J. S., Kim, D. S., Ko, S. H., Yang, S. H., et al. (2015). Sphingomonas vulcanisoli sp. nov., isolated from soil of a lava forest. Int. J. Syst. Evol. Microbiol. 65, 3320–3325. doi: 10.1099/ijsem.0.000414
Lee, Y., and Jeon, C. O. (2017). Cohnella algarum sp. nov., isolated from a freshwater green alga Paulinella chromatophora. Int. J. Syst. Evol. Microbiol. 67, 4767–4772. doi: 10.1099/ijsem.0.002377
Lee, J. H., Kim, J. W., and Lee, P. C. (2018). Complete genome sequence of Flavobacterium kingsejongi WV39, a type species of the genus Flavobacterium and a microbial C40 carotenoid zeaxanthin producer. J. Biotechnol. 266, 9–13. doi: 10.1016/j.jbiotec.2017.11.012
Lee, S. A., Kim, Y., Sang, M. K., Song, J., Kwon, S. W., and Weon, H. Y. (2019). Chryseolinea soli sp. nov., isolated from soil. J. Microbiol. 57, 122–126. doi: 10.1007/s12275-019-8562-4
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, H., Li, J., Jiao, X., Li, K., Sun, Y., Zhou, W., et al. (2019). Characterization of the biosynthetic pathway of nucleotide sugar precursor UDP-glucose during sphingan WL gum production in Sphingomonas sp. WG. J. Biotechnol. 302, 1–9. doi: 10.1016/j.jbiotec.2019.06.005
Li, C., Swofford, C. A., and Sinskey, A. J. (2020). Modular engineering for microbial production of carotenoids. Metab. Eng. Commun. 10:e00118. doi: 10.1016/j.mec.2019.e00118
Liu, M., Yang, Y., Li, L., Ma, Y., Huang, J., and Ye, J. (2021). Engineering Sphingobium sp. to accumulate various carotenoids using agro-industrial byproducts. Front Bioeng. Front. Bioeng. Biotechnol. 9:784559. doi: 10.3389/fbioe.2021.784559
Liu, X., Zhao, M., Xu, Z., Xu, H., and Li, S. (2020). Construction of a robust Sphingomonas sp. strain for welan gum production via the expression of global transcriptional regulator IrrE. Front Bioeng. Front. Bioeng. Biotechnol. 8:674. doi: 10.3389/fbioe.2020.00674
Luo, Y., Wang, F., Huang, Y., Zhou, M., Gao, J., Yan, T., et al. (2019). Sphingomonas sp. Cra20 increases plant growth rate and alters rhizosphere microbial community structure of Arabidopsis thaliana under drought stress. Front. Microbiol. 10:1221. doi: 10.3389/fmicb.2019.01221
Luo, Y., Zhou, M., Zhao, Q., Wang, F., Gao, J., Sheng, H., et al. (2020). Complete genome sequence of Sphingomonas sp. Cra20, a drought resistant and plant growth promoting rhizobacteria. Genomics 112, 3648–3657. doi: 10.1016/j.ygeno.2020.04.013
Margesin, R., Zhang, D. C., and Busse, H. J. (2012). Sphingomonas alpina sp. nov., a psychrophilic bacterium isolated from alpine soil. Int. J. Syst. Evol. Microbiol. 62, 1558–1563. doi: 10.1099/ijs.0.035964-0
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14:60. doi: 10.1186/1471-2105-14-60
Mnich, E., Vanholme, R., Oyarce, P., Liu, S., Lu, F., Goeminne, G., et al. (2017). Degradation of lignin beta-aryl ether units in Arabidopsis thaliana expressing LigD, LigF and LigG from Sphingomonas paucimobilis SYK-6. Plant Biotechnol. J. 15, 581–593. doi: 10.1111/pbi.12655
Moreno-Forero, S. K., and van der Meer, J. R. (2015). Genome-wide analysis of Sphingomonas wittichii RW1 behaviour during inoculation and growth in contaminated sand. ISME J. 9, 150–165. doi: 10.1038/ismej.2014.101
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Pavelková, P., Krmela, A., and Schulzová, V. (2020). Determination of carotenoids in flowers and food supplements by HPLC-DAD. Acta Chimica Slovaca 13, 6–12. doi: 10.2478/acs-2020-0002
Ren, Y., Sun, H., Deng, J., Huang, J., and Chen, F. (2021). Carotenoid production from microalgae: biosynthesis, salinity responses and novel biotechnologies. Mar. Drugs 19:713. doi: 10.3390/md19120713
Rodriguez, R. L., Gunturu, S., Harvey, W. T., Rossello-Mora, R., Tiedje, J. M., Cole, J. R., et al. (2018). The microbial genomes atlas (MiGA) webserver: taxonomic and gene diversity analysis of archaea and bacteria at the whole genome level. Nucleic Acids Res. 46, W282–W288. doi: 10.1093/nar/gky467
Saboe, A., Adrian, Y., Widyatmoko, L., Hasan, M., Cool, C. J., Hartantri, Y., et al. (2021). A fatal case of early prosthetic valve endocarditis caused by multidrug-resistant (MDR)-Sphingomonas paucimobilis. IDCases 24:e01152. doi: 10.1016/j.idcr.2021.e01152
Sasser, M. (2006). Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). MIDI Technical MIDI Technical Note #101 MIDI Inc, Newark, DE, USA.
Setiyono, E., Heriyanto, P. D., Shioi, Y., Kanesaki, Y., Awai, K., and Brotosudarmo, T. H. P. (2019). Sulfur-containing carotenoids from a marine coral symbiont erythrobacter flavus strain KJ5. Mar. Drugs 17:349. doi: 10.3390/md17060349
Sheu, S. Y., Chen, Y. L., and Chen, W. M. (2015). Sphingomonas fonticola sp. nov., isolated from spring water. Int. J. Syst. Evol. Microbiol. 65, 4495–4502. doi: 10.1099/ijsem.0.000602
Siddaramappa, S., Viswanathan, V., Thiyagarajan, S., and Narjala, A. (2018). Genomewide characterisation of the genetic diversity of carotenogenesis in bacteria of the order Sphingomonadales. Microb. Genom. 4:e000172. doi: 10.1099/mgen.0.000172
Silkina, A., Kultschar, B., and Llewellyn, C. A. (2019). Far-red light acclimation for improved mass cultivation of cyanobacteria. Metabolites 9:170. doi: 10.3390/metabo9080170
Singh, A., Singh, D. K., Kharwar, R. N., White, J. F., and Gond, S. K. (2021). Fungal endophytes as efficient sources of plant-derived bioactive compounds and their prospective applications in natural product drug discovery: insights, avenues, and challenges. Microorganisms 9:197. doi: 10.3390/microorganisms9010197
Song, Y., Zheng, C., Basnet, R., Li, S., Chen, J., and Jiang, M. (2022). Astaxanthin synthesized gold nanoparticles enhance salt stress tolerance in rice by enhancing tetrapyrrole biosynthesis and scavenging reactive oxygen species in vitro. Plant Stress. 6:100122. doi: 10.1016/j.stress.2022.100122
Takaya, T., Anan, M., and Iwata, K. (2018). Vibrational relaxation dynamics of beta-carotene and its derivatives with substituents on terminal rings in electronically excited states as studied by femtosecond time-resolved stimulated Raman spectroscopy in the near-IR region. Phys. Chem. Chem. Phys. 20, 3320–3327. doi: 10.1039/c7cp06343a
Takeuchi, M., Hamana, K., and Hiraishi, A. (2001). Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 51, 1405–1417. doi: 10.1099/00207713-51-4-1405
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thawornwiriyanun, P., Tanasupawat, S., Dechsakulwatana, C., Techkarnjanaruk, S., and Suntornsuk, W. (2012). Identification of newly zeaxanthin-producing bacteria isolated from sponges in the Gulf of Thailand and their zeaxanthin production. Appl. Biochem. Biotechnol. 167, 2357–2368. doi: 10.1007/s12010-012-9760-2
Urumbil, S. K., and Anilkumar, M. N. (2021). Anti-inflammatory activity of endophytic bacterial isolates from Emilia sonchifolia (Linn.) DC. J. Ethnopharmacol. 281:114517. doi: 10.1016/j.jep.2021.114517
Xiao, B., Zhang, C., Song, X., Wu, M., Mao, J., Yu, R., et al. (2020). Rationally engineered prolyl endopeptidases from Sphingomonas capsulata with improved hydrolytic activity towards pathogenic peptides of celiac diseases. Eur. J. Med. Chem. 202:112499. doi: 10.1016/j.ejmech.2020.112499
Xu, X., Nie, Z., Zheng, Z., Zhu, L., and Zhan, X. (2020). Effect of different nitrogen sources on the viscosity and rheological properties of welan gum produced by Sphingomonas sp. ATCC 31555. J. Texture Stud. 51, 642–649. doi: 10.1111/jtxs.12519
Xue, H., Piao, C. G., Wang, X. Z., Lin, C. L., Guo, M. W., and Li, Y. (2018). Sphingomonas aeria sp. nov., isolated from air. Int. J. Syst. Evol. Microbiol. 68, 2866–2871. doi: 10.1099/ijsem.0.002910
Yabuuchi, E., Yano, I., Oyaizu, H., Hashimoto, Y., Ezaki, T., and Yamamoto, H. (1990). Proposals of Sphingomonas paucimobilis gen. Nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol. Immunol. 34, 99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x
Yang, S., Zhang, X., Cao, Z., Zhao, K., Wang, S., Chen, M., et al. (2014). Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 7, 611–620. doi: 10.1111/1751-7915.12148
Yeşilada, E., Honda, G., Sezik, E., Tabata, M., Fujita, T., Tanaka, T., et al. (1995). Traditional medicine in Turkey. V. Folk medicine in the inner Taurus Mountains. J. Ethnopharmacol. 46, 133–152. doi: 10.1016/0378-8741(95)01241-5
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, W.-J., Kim, S.-S., Oh, T.-H., Lee, N. H., and Hyun, C.-G. (2009). Abies koreana essential oil inhibits drug-resistant skin pathogen growth and LPS-induced inflammatory effects of murine macrophage. Lipids 44, 471–476. doi: 10.1007/s11745-009-3297-3
Yu, F. B., Shan, S. D., Luo, L. P., Guan, L. B., and Qin, H. (2013). Isolation and characterization of a Sphingomonas sp. strain F-7 degrading fenvalerate and its use in bioremediation of contaminated soil. J. Environ. Sci. Health B 48, 198–207. doi: 10.1080/03601234.2013.730299
Zhang, J. Y., Liu, X. Y., and Liu, S. J. (2010). Sphingomonas changbaiensis sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 60, 790–795. doi: 10.1099/ijs.0.011486-0
Zhang, K., Tang, K., Feng, F., Yuan, B., Zhang, X., and Meng, J. (2017). Sphingomonas crusticola sp. nov., isolated from biological soil crusts. Int. J. Syst. Evol. Microbiol. 67, 3033–3037. doi: 10.1099/ijsem.0.002074
Zhang, Z., Wang, Y., Chang, L., Zhang, T., An, J., Liu, Y., et al. (2016). MsZEP, a novel zeaxanthin epoxidase gene from alfalfa (Medicago sativa), confers drought and salt tolerance in transgenic tobacco. Plant Cell Rep. 35, 439–453. doi: 10.1007/s00299-015-1895-5
Zhang, Q. Y., Wang, L. Y., Kong, F. Y., Deng, Y. S., Li, B., and Meng, Q. W. (2012). Constitutive accumulation of zeaxanthin in tomato alleviates salt stress-induced photoinhibition and photooxidation. Physiol. Plant. 146, 363–373. doi: 10.1111/j.1399-3054.2012.01645.x
Zhao, J., Shan, T., Mou, Y., and Zhou, L. (2011). Plant-derived bioactive compounds produced by endophytic fungi. Mini Rev. Med. Chem. 11, 159–168. doi: 10.2174/138955711794519492
Zhao, M., Zhang, H., Xu, X., Li, S., and Xu, H. (2021). A strategy for the synthesis of low-molecular-weight welan gum by eliminating capsule form of Sphingomonas strains. Int. J. Biol. Macromol. 178, 11–18. doi: 10.1016/j.ijbiomac.2021.02.157
Zheng, J., Feng, J. Q., Zhou, L., Mbadinga, S. M., Gu, J. D., and Mu, B. Z. (2018). Characterization of bacterial composition and diversity in a long-term petroleum contaminated soil and isolation of high-efficiency alkane-degrading strains using an improved medium. World J. Microbiol. Biotechnol. 34:34. doi: 10.1007/s11274-018-2417-8
Zhu, G., Cheng, X., Fu, Z., Zhang, Z., and Tong, Q. (2020). The mechanism of improved gellan gum production by two-stage culture of Sphingomonas paucimobilis. 3 Biotech. 10:70. doi: 10.1007/s13205-019-2047-3
Zhu, G., Guo, N., Yong, Y., Xiong, Y., and Tong, Q. (2019). Effect of 2-deoxy-D-glucose on gellan gum biosynthesis by Sphingomonas paucimobilis. Bioprocess Biosyst. Eng. 42, 897–900. doi: 10.1007/s00449-019-02078-w
Zhu, L., Wu, X., Li, O., Qian, C., and Gao, H. (2012). Cloning and characterization of genes involved in nostoxanthin biosynthesis of Sphingomonas elodea ATCC 31461. PLoS One 7:e35099. doi: 10.1371/journal.pone.0035099
Keywords: polyphasic taxonomy, Abies koreana, phylogeny, orthologous average nucleotide identity, whole-genome annotation, antiSMASH
Citation: Jiang L, Seo J, Peng Y, Jeon D, Lee JH, Kim CY and Lee J (2023) A nostoxanthin-producing bacterium, Sphingomonas nostoxanthinifaciens sp. nov., alleviates the salt stress of Arabidopsis seedlings by scavenging of reactive oxygen species. Front. Microbiol. 14:1101150. doi: 10.3389/fmicb.2023.1101150
Edited by:
James T. Tambong, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Heng-Lin Cui, Jiangsu University, ChinaYang Liu, Guangdong Academy of Sciences, China
Izhar Ul-Haq Khan, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2023 Jiang, Seo, Peng, Jeon, Lee, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiyoung Lee, ✉ aml5b3VuZzFAa3JpYmIucmUua3I=
 Lingmin Jiang
Lingmin Jiang Jiyoon Seo
Jiyoon Seo Yuxin Peng
Yuxin Peng Doeun Jeon
Doeun Jeon Ju Huck Lee
Ju Huck Lee Cha Young Kim
Cha Young Kim Jiyoung Lee
Jiyoung Lee