
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 23 March 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1099098
 Shujian Xiao1†
Shujian Xiao1† Zhen Wang2†
Zhen Wang2† Bangxu Wang1
Bangxu Wang1 Bo Hou1
Bo Hou1 Jie Cheng1*
Jie Cheng1* Ting Bai1
Ting Bai1 Yin Zhang1
Yin Zhang1 Wei Wang1
Wei Wang1 Lixiu Yan3*
Lixiu Yan3* Jiamin Zhang1*
Jiamin Zhang1*Tryptophan derivatives are various aromatic compounds produced in the tryptophan metabolic pathway, such as 5-hydroxytryptophan, 5-hydroxytryptamine, melatonin, 7-chloro-tryptophan, 7-bromo-tryptophan, indigo, indirubin, indole-3-acetic acid, violamycin, and dexoyviolacein. They have high added value, widely used in chemical, food, polymer and pharmaceutical industry and play an important role in treating diseases and improving life. At present, most tryptophan derivatives are synthesized by biosynthesis. The biosynthesis method is to combine metabolic engineering with synthetic biology and system biology, and use the tryptophan biosynthesis pathway of Escherichia coli, Corynebacterium glutamicum and other related microorganisms to reconstruct the artificial biosynthesis pathway, and then produce various tryptophan derivatives. In this paper, the characteristics, applications and specific biosynthetic pathways and methods of these derivatives were reviewed, and some strategies to increase the yield of derivatives and reduce the production cost on the basis of biosynthesis were introduced in order to make some contributions to the development of tryptophan derivatives biosynthesis industry.
The basic metabolic pathways of aromatic compound biosynthesis involve glycolytic pathway (EMP), pentose phosphate pathway (PPP), and shikimate pathway. In shikimate pathway, phosphoenolpyruvate (PEP) produced by glycolytic pathway, and D-erythrose 4-phosphate (E4P) produced by pentose phosphate pathway are used as precursors to condense to form 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP). DAHP then undergoes a six step catalytic reaction via shikimate pathway to generate chorismate (Wu et al., 2021). With chorismate as precursor, chorismate is transformed into three aromatic amino acids through two ways. One way is first transformed into prephenylalanine, and then L-phenylalanine or L-tyrosine were synthesized, respectively. The other way is to generate L-tryptophan (L-Trp) from o-aminobenzoic acid (Barik, 2020; Wu et al., 2021).
Tryptophan belongs to one of the three aromatic amino acids and is the only amino acid containing an indole ring. Tryptophan not only participates in the biosynthesis and turnover of proteins and peptides, but also is absorbed into the body and transformed into a series of bioactive small multi effect compounds (Barik, 2020). It is mainly degraded through two parallel pathways, which are 5-hydroxytryptamine (5-HT) pathway and kynurenine pathway. These two pathways will produce a series of secondary metabolites. The metabolites of serotonin pathway include 5-hydroxytryptophan (5-HTP), serotonin and melatonin. The metabolites of kynurenine pathway include kynurenine and niacin. Melatonin and niacin are the final products of the above two parallel pathways, while 5-HTP, 5-HT, and inulin are intermediate metabolites (Barik, 2020; Cas et al., 2021). In addition, in plants and microorganisms, tryptophan derivatives also include chlorotryptophan, bromotryptophan (Lee and Lee, 2020), and indole alkaloids such as indole-3-acetic acid (IAA), indirubin, indigo (Xu et al., 2014).
Tryptophan derivatives are widely needed because of their various functions, such as serotonin and melatonin, which can treat insomnia (Arnao and Hernandez-Ruiz, 2018). Halogenated tryptophan is an important intermediate or component of active substances related to the pharmaceutical, chemical and pesticide industries (Lee and Lee, 2020). Auxin (IAA) affects the root growth of plants and plays an important role in the interaction between plants and microorganisms (Leontovycova et al., 2020; Figure 1). However, due to the problems of cost, pollution and complex steps in the chemical synthesis of tryptophan derivatives, the development of modern biotechnology and synthetic biology has opened up another way for us to synthesize tryptophan derivatives (Choi et al., 2003; Lee and Lee, 2020). This paper mainly reviews the biosynthesis of common tryptophan derivatives, such as 5-HTP, serotonin, melatonin, IAA, halotryptophan, violacein, indirubin, indigo, etc. (Figure 2; Table 1).
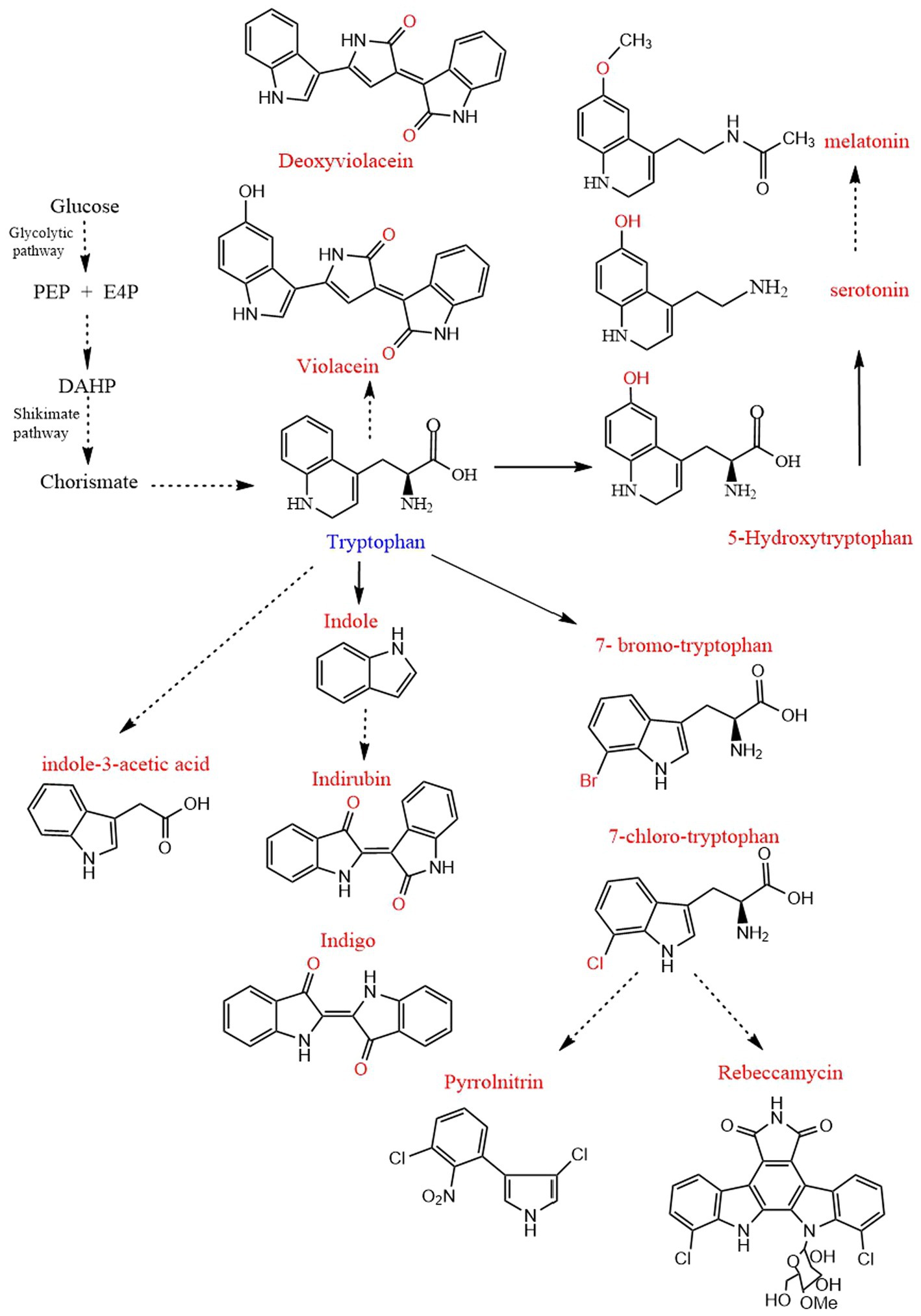
Figure 2. Biosynthesis pathway of tryptophan derivatives. PEP, Phosphoenolpyruvate; E4P, D-erythrose 4-phosphate; DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate. The straight line and dotted line represent one-step and multi-step, respectively.
5-HTP, serotonin and melatonin are products of the same tryptophan metabolic pathway (Cas et al., 2021). Tryptophan is converted to 5-HTP through tryptophan hydroxylase (TPH), and 5-HTP is converted to serotonin through aromatic acid decarboxylase. The serotonin is converted to N-acetylserotonin through arylalkylamine N-acetyltransferase, which is converted to melatonin through hydroxyindole-O-methyltransferase (Zheng et al., 2021; Figure 3).
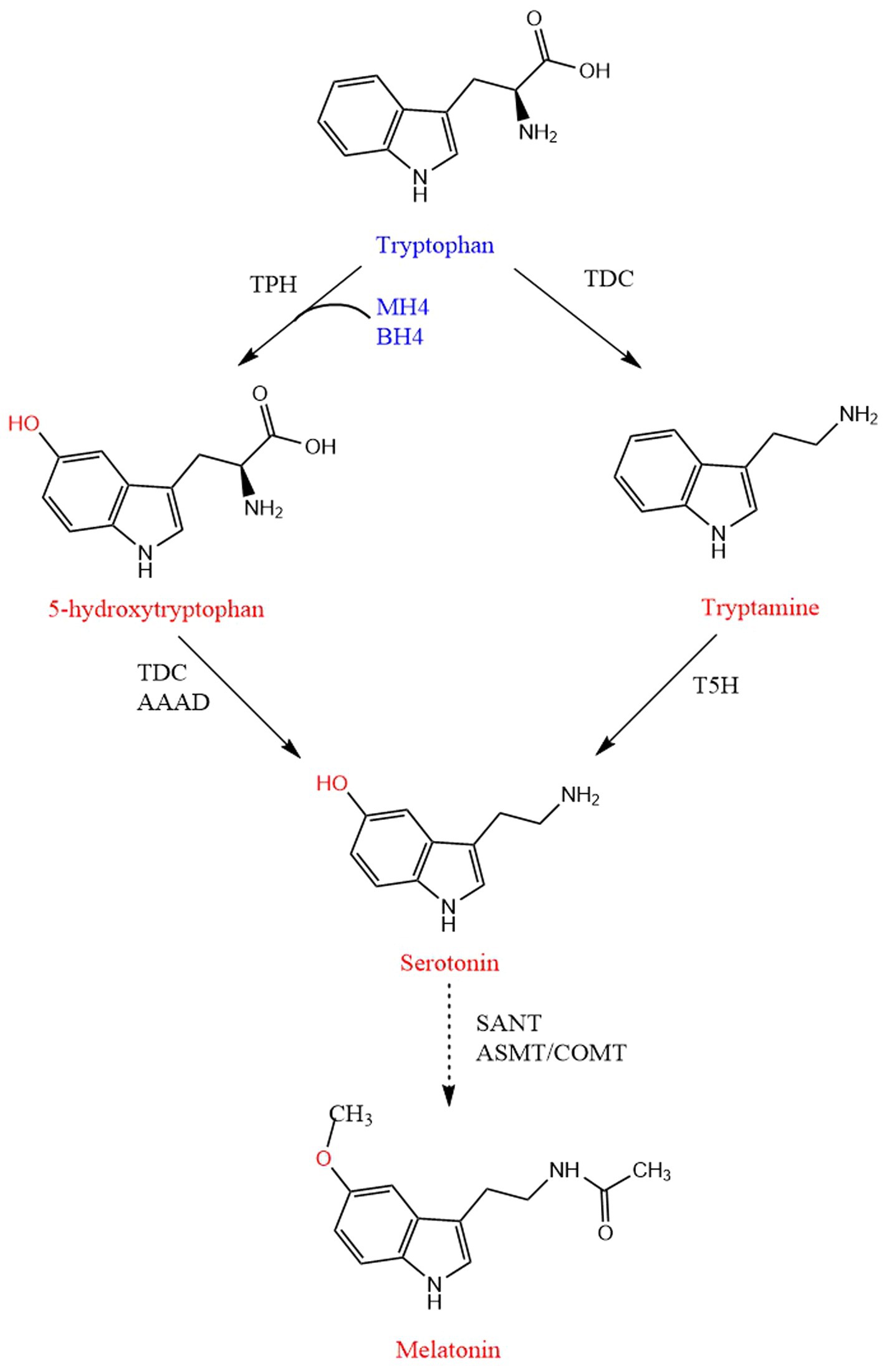
Figure 3. Biosynthetic pathway of 5-HTP, serotonin, and melatonin. Linear and dashed lines mean a single-step and multi-steps, respectively. TPH, Tryptophan 5-hydroxylase; TDC, Tryptophan decarboxylase; AAAD, Aromatic amino acid decarboxylase; T5H, Tryptamine 5-hydroxylase; SNAT, Serotonin N-acetyltransferase; ASMT, N-acetylserotonin O-methyltransferase; COMT, Caffeic acid O-methyltransferase; BH4, Tetrahydrobiopterin; MH4, Mtetrahydromonapterin.
5-HTP is a natural amino acid (AA) that does not participate in protein synthesis. It is derived from tryptophan, and the hydrogen atoms at the 5′-position on the benzene ring of tryptophan are replaced by hydroxyl groups (Liu et al., 2021). 5-HTP is the precursor of serotonin and melatonin, which can be used to treat depression, insomnia, migraine and other diseases (Wang et al., 2018). 5-HTP was originally extracted from Griffonia simplicifolia and other plants, but this method is expensive and raw materials are rare. Besides, the use of chemical synthesis method is cumbersome and harsh. With the progress of biotechnology, the use of microorganisms, especially E. coli, to synthesize 5-HTP has become the mainstream (Liu et al., 2021).
In human and mammalian cells, 5-HTP is synthesized by L-Trp hydroxylation with Fe2+ and BH4 as cofactors and O2 as cosubstrate catalyzed by TPH. BH4 is oxidized to pterin-4α-carbinolamine (BH3OH) during L-Trp hydroxylation and regenerated through the function of pterin-4α-carbinolamine dehydratase and dihydropteridine reductase (DHPR; Wang et al., 2018). Knight et al. found that the co-expression of the animal BH4 biosynthesis pathway and the truncated tryptamine 5-hydroxylase (T5H) from Oryctolagus cuniculus in E. coli produced 198 mg/L 5-HTP (Knight et al., 2013). However, most bacteria such as E. coli cannot naturally produce BH4, and they can only synthesize BH4 analogue tetrahydromonapterin (MH4; Lin et al., 2014). It is generally necessary to add exogenous BH4, or realize the biosynthesis and regeneration of bacterial BH4 through gene recombination (Germann et al., 2016).
An artificial MH4 recycling system was established by the expression of phhB from P. aeruginosa and folM [encoding dihydromonasin reductase (DHMR)] from E. coli. With this circulating system, E. coli cells could use tryptophan to produce 1114.8 mg/L 5-HTP in shake flasks (Lin et al., 2014; Figure 3). The tryptophan synthesis pathway was successfully introduced into E. coli to realize the de novo production of 5-HTP. After a series of optimization such as improving the hydroxylation activity of TPH through enzyme modification, the titer of 5-HTP was significantly increased to 1.29 g/L (Wang et al., 2018; Liu et al., 2021). Furthermore, by designing the strength of the 3-deoxy-7-phosphate synthase promoter and adjusting the copy number of the L-Trp hydroxylation plasmid, the output of 5-HTP in shake flask was increased to 1.61 g/L (Xu et al., 2020).
Serotonin, also known as 5-HT, is an amino acid derivative with high added value. It can participate in emotional regulation, behavior management, and sleep cycle maintenance. It also can promote plant seed germination and growth and other physiological processes (Shen et al., 2020). Serotonin is synthesized in different ways in animals and plants. In animals, tryptophan is hydroxylated to 5-HTP through tryptophan 5-hydroxylase, and then tryptophan decarboxylase (TDC) converts 5-HTP to 5-HT, namely serotonin (Gaddum and Giarman, 1956; Cao et al., 2020). In plants, tryptophan is first converted to tryptamine by TDC, and then serotonin is produced by T5H (Goncalves et al., 2022; Figure 3).
TDC from rice was overexpressed in transgenic rice, recombinant E. coli (pET28b TDC), and recombinant yeast (pYES-TDC), and serotonin accumulation was detected, which confirmed that serotonin was produced under the condition of 5-HTP as substrate (Park et al., 2008). A functional T5H enzyme (GSTΔ37T5H) was constructed by a series of n-terminal deletion or labeling proteins, and then, 24 mg/L serotonin was produced by GSTΔ37T5H and TDC (Park et al., 2011). The semi rational engineering recombinant strain of aromatic amino acid hydroxylase was used to produce 5-HTP, and then the recombinant strain containing tryptophan decarboxylase was used for biotransformation of 5-HTP to produce about 154 mg/L serotonin, which was the first time to realize the production of serotonin from a simple carbon source (Mora-Villalobos and Zeng, 2018). In the first step, about 962 mg/L 5-HTP was produced by using a recombinant strain with a semi-rationally engineered aromatic amino acid hydroxylase. In the second step, biotransformation of 5HTP using recombinant strains containing TDC, about 154 mg/L of serotonin was produced. A method of producing 5-HT from tryptophan through two enzyme cascades in one pot has also been proposed (Wang et al., 2022). The tryptophan hydroxylase from Schistosoma mansoni, the artificial endogenous BH4 module and the dopa decarboxylase from Harminia axyridis, are heterologously expressed in E. coli. The recombinant E. coli can produce about 414 mg/L of 5-HT from 2 g/L of tryptophan.
Melatonin, a natural product derived from tryptophan, is a major biomolecule synthesized in almost all biological organisms, including animals and plants (Back et al., 2016). Melatonin can affect circadian rhythm, mood, sleep, etc. it can also be used as a plant biological stimulant to resist biological and abiotic stress and regulate the ability of plant growth (Arnao and Hernandez-Ruiz, 2018).
Its synthesis goes through four steps. In plants, tryptophan is converted to tryptamine by TDC, and then tryptamine is converted to serotonin by T5H. Serotonin is catalyzed by serotonin N-acetyltransferase (SNAT) to complete N-acetylation, and then N-acetylserotonin is methylated by acetylserotonin methyl transferase (ASMT, a hydroxyindole-O-methyltransferase) to produce melatonin (Arnao and Hernandez-Ruiz, 2018). In animals, tryptophan produces 5-HTP under the combined action of TPH, cofactor BH4 and oxygen. Next, 5-HTP is converted to serotonin by tryptophan carboxylase. Subsequently, aralkylamine N-acetyltransferase produces N-acetyl 5-hydroxytryptamine at the expense of acetyl-CoA. Finally, N-acetyl 5-hydroxytryptamine methyltransferase is accompanied by the conversion of cofactor SAM to SAH to produce the final product melatonin (Xie et al., 2022).
In addition to animals and plants, many microorganisms can also synthesize melatonin. As seen in Figure 3, a strain of Saccharomyces cerevisiae has been cultivated, which used glucose as the only carbon source to ferment in the culture medium and produce 14.5 mg/L melatonin (Germann et al., 2016). E. coli can also be used for melatonin production. In several double expression box combinations, the recombinant E. coli expressing sheep SNAT with rice O-methyltransferase (COMT) produced a large amount of melatonin, which is the first report using E. coli to heterologously produce melatonin (Byeon and Back, 2016). Moreover, the biosynthetic pathway of melatonin was introduced into E. coli, and then the engineered strain produced about 2.0 g/L of melatonin through protein engineering of rate-limiting tryptophan hydroxylase, chromosomal integration of aromatic amino acid decarboxylase, and deletion of tryptophan export protein YddG (Luo et al., 2020).
Indole and indole alkaloids belong to the same pathway. Tryptophan is degraded into indole by tryptophanase. Indole can be converted into a variety of indole alkaloids by different enzymes through different pathways, among which IAA, indigo and indirubin are common (Chen et al., 2016).
Indole, also known as 2,3-benzopyrrole, is widely used in chemical, pharmaceutical, dye and other industries. It is an important precursor in industry, but it is also a typical nitrogen heterocyclic pollutant released into the environment (Li et al., 2020). It is a signal molecule that regulates a variety of physiological processes, including movement, biofilm formation, antibiotic resistance, plasmid stability, sustained cell formation (Han T. H. et al., 2011), indole and its derivatives strongly affect the physiological functions of bacteria and animals (Ferrer et al., 2022b). Tryptophan has been proved to be completely degraded by tryptophanase to produce indole (Watanabe and Snell, 1972; Figure 4). Li and Young (2013) reported that the final yield of indole in E. coli depends on the amount of exogenous tryptophan, and the transformation process was mainly dependent on the tryptophanase TnaA. On the other hand, excessive indole may also inhibit the activity of TnaA and the transport process of tryptophan.
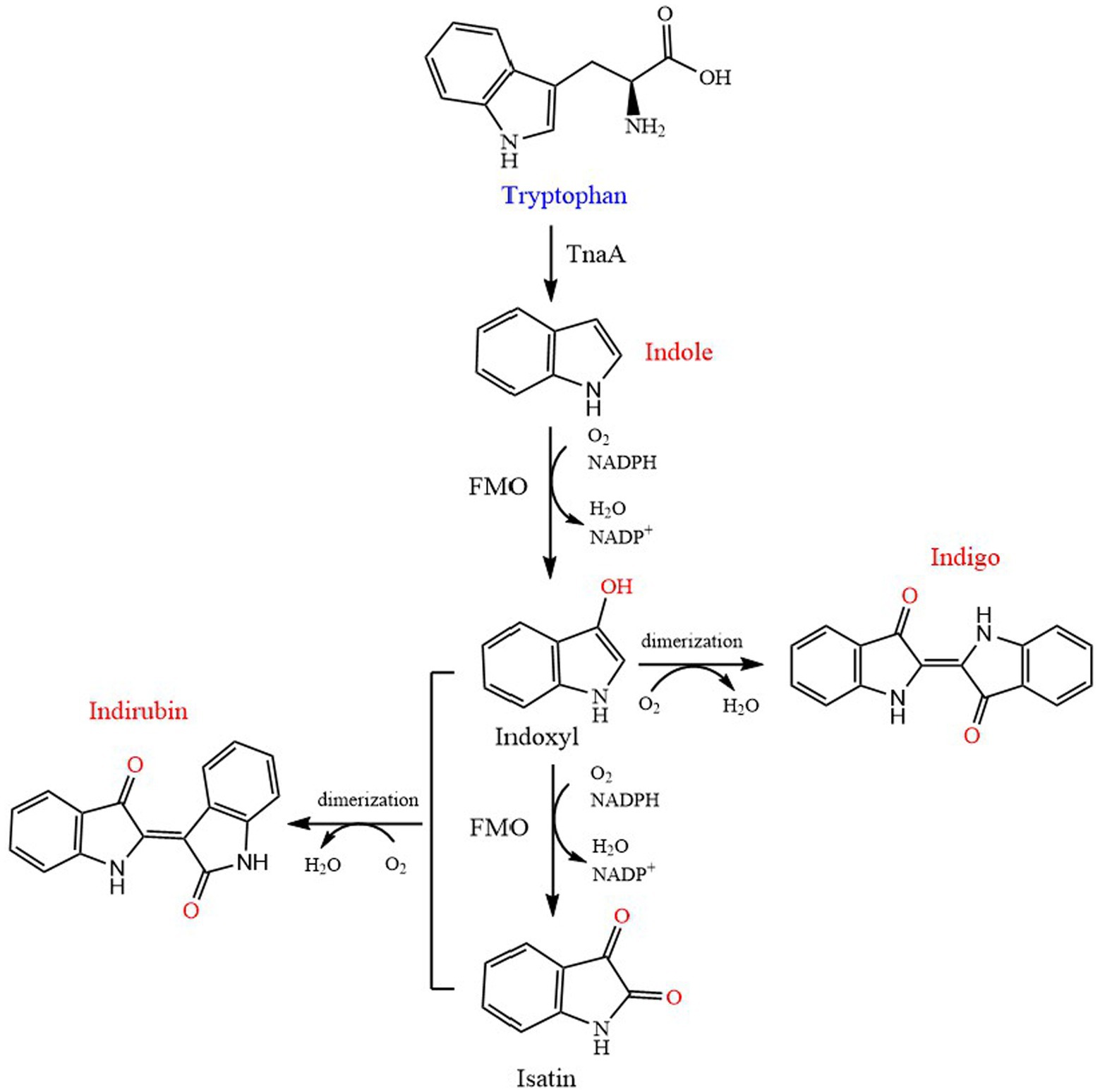
Figure 4. Biosynthetic pathway of indole, indigo and indirubin. TnaA, tryptophanase; FMO, flavin-containing monooxygenase.
In addition to E. coli, some heterologous strains can also be used to produce indole, for example, indole synthesis-related gene was introduced into C. glutamicum. With the expression of endogenous TSA gene or IGL gene of wheat, about 0.7 g/L indole were produced (Ferrer et al., 2022b). 5.7 g/L of indole can be produced by co-expressing the natural aromatic amino acid permease gene aroP and the tryptophanase from Providencia rettgeri in C. glutamicum (Mindt et al., 2022).
Indirubin and indigo belong to Indole alkaloids, which are secondary metabolites derived from plants. Many of them have important medicinal properties and have been used as drugs and dyes since ancient times (Cao et al., 2020). In addition, IAA, as plant auxin, belongs to indole alkaloids too, and IAA, indigo and indirubin belong to simple indole alkaloids (Chen et al., 2016).
IAA is the most abundant auxin-active natural hormone in plants, which controls many physiological processes, such as cell proliferation and division, tissue differentiation, phototropism and geotropism reactions (Leontovycova et al., 2020). Some plant-related bacteria, fungi and yeasts, including Agrobacterium tumefaciens, Azospirillum brasilense, Bradyrhizobium spp. and Enterobacter cloacae, are known to synthesize IAA in the presence of tryptophan (Romasi and Lee, 2013).
According to the main intermediates in the IAA synthesis process, the Trp-dependent biosynthesis process in plants is usually divided into four branches: indole-3-acetaldoxime (IAOx) pathway, tryptamine pathway, indole-3-acetamide (IAM) pathway and indole-3-pyruvic acid (IPA) pathway (Jiali et al., 2012). IPA pathway is the main and generally conserved biosynthetic pathway in plants, while other redundant pathways run in parallel (Casanova-Saez et al., 2021). ① IAOx pathway (also known as CYP79B pathway): Firstly, tryptophan is catalyzed by Cytochrome P450 Mono-oxygenase CYP79B2 and CYP79B3 to generate indole 3-acetaldoxime, which is then converted into indole-3-acetonitrile and indole-3-acetaldehyde (IAAld), and then IAA is generated under the catalysis of nitrilase and aldehyde oxidase, respectively. ② IPA pathway: Indole-3-pyruvate, an intermediate product, is decarboxylated to form IAAld under the action of indolepyruvate decarboxylase (IpdC), and then oxidized to IAA. ③ Tryptamine pathway: the tryptamine pathway starts with tryptophan passing through TDC catalyzes the formation of tryptamine, and then generates IAA through the intermediate product IAAld. ④ IAM pathway: The pathway consists of two distinct steps. In the first step tryptophan monooxygenase (encoded by iaaM gene, the gene has not been found in plants) converts tryptophan to IAM; in the second step IAM is hydrolyzed to IAA and ammonia by an IAM hydrolase (encoded by iaaH gene; Spaepen and Vanderleyden, 2011; Jiali et al., 2012; Romasi and Lee, 2013). The synthetic pathway of IAA in bacteria is highly similar to that in plants, except for the addition of a tryptophan side-chain oxidase pathway and it has only been demonstrated in Pseudomonas fluorescens CHA0 (Spaepen and Vanderleyden, 2011). In this pathway tryptophan is directly converted to IAAld bypassing IPyA, which can be oxidized to IAA (Spaepen et al., 2007). In addition, the tryptamine pathway in bacteria is opposite to that in plants. Tryptophan is first decarboxylated to tryptamine by a TDC, which is directly converted to IAAld by amine oxidase (Spaepen and Vanderleyden, 2011; Figure 5).
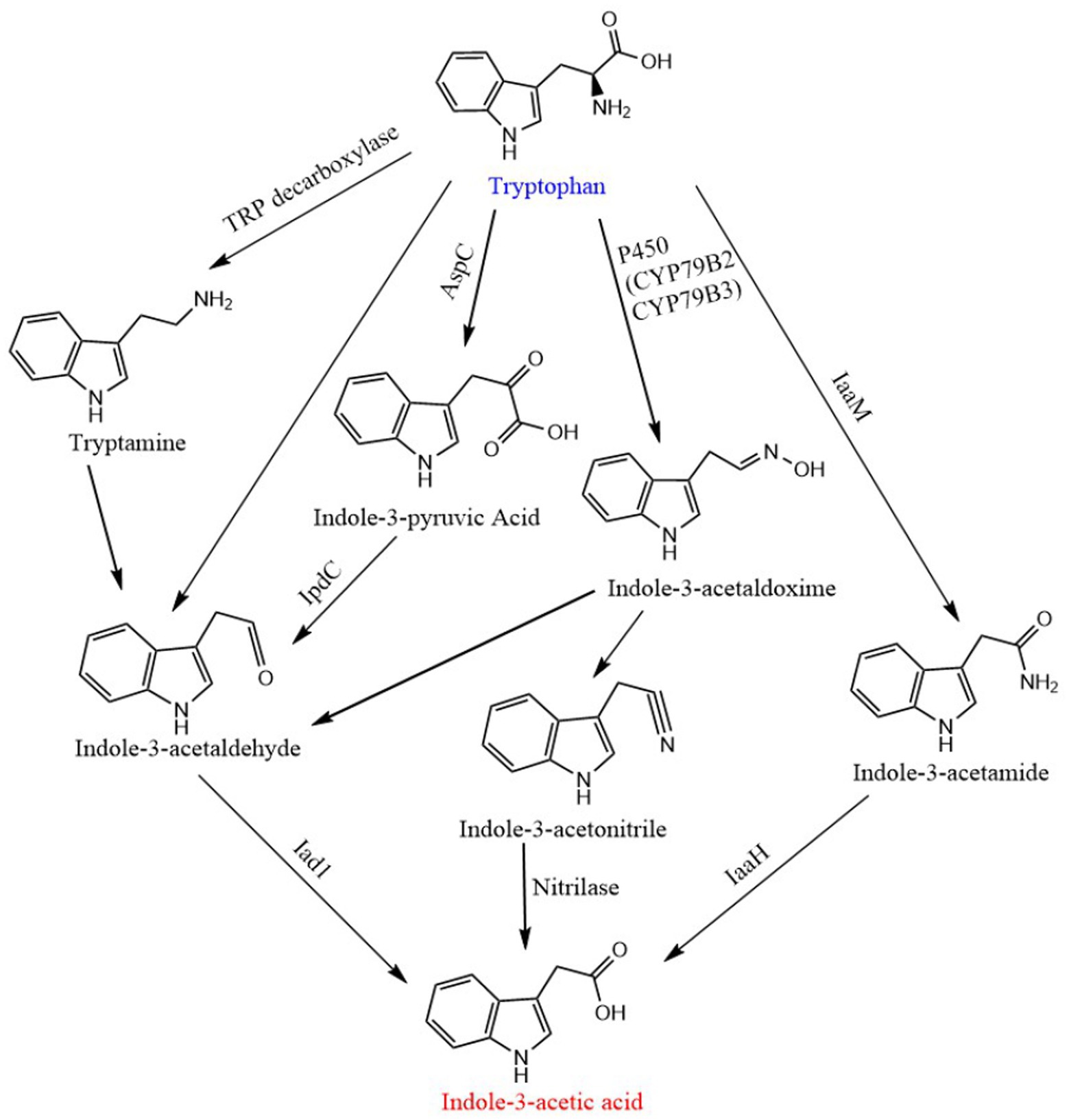
Figure 5. Biosynthetic pathway of in-dole-3-acetic acid. CYP79B2 and CYP79B3, cytochrome P450 monooxygenase; AapC, aminotransferase; Ipdc, indole-3-pyruvic acid decarboxylase; Iad1, indole-3-acetic acid dehydrogenase; IaaM, tryptophan 2-monooxygenase; IAAH, IAM hydrolase.
E. coli also can be used for IAA production. The ipdC (encoding indole-3-pyruvic acid decarboxylase) from Enterobacter cloacae ATCC 13047, aspC (encoding aminotransferase) from E. coli and iad1 (encoding indole-3-acetic acid dehydrogenase) from Ustilago maydis were cloned and expressed in E. coli using tac and sod promoters, and deleted a tnaA gene that mediates indole formation from tryptophan, recombinant E. coli produced 3.0 g/L IAA (Romasi and Lee, 2013). Similarly, The above method could also be used for C. glutamicum, and the recombinant strain produced 2.3 and 7.3 g/L IAA from 10 g/L L-Trp in flask culture and 5-L bioreactor, respectively (Kim et al., 2019).
Indigo is a kind of blue dye which has been used for thousands of years, it is mainly used for the production of blue jeans denim (Lolita Ameria et al., 2015). It also has medicinal value, as well as hemostatic, antipyretic, anti-inflammatory and sedative properties, and can be used for anti-tumor or anti-leukemia activity (Heine et al., 2019). Indigo undergoes three stages of biosynthesis in L-Trp. First, L-Trp is decomposed into indole by tryptophanse, and then indole is oxidized to indoxyl by various oxygenase catalytic reactions, finally, indoxyl spontaneous reaction of 2 molecules generates indigo (Zhang et al., 2014; Lee and Lee, 2020; Figure 4). The representative enzymes involved in this indole oxidation reaction were naphthalene dioxygenase (NDO) and toluene dioxygenase from Pseudomonas putida, phenol hydroxylase from Acinetobacter sp. and cytochrome P450 monooxygenase from Bacillus megaterium,toluene monooxygenase from Burkholderia cepacia, flavin-containing monooxygenase (cFMO) from C. glutamicum and flavin-containing monooxygenase (mFMO) from Methylophaga aminisulfidivorans et al. (Kang and Lee, 2009; Lee and Lee, 2020).
Recombinant E. coli DH5α containing flavone monooxygenase (FMO) gene has been successfully cultivated, the indigo titer was 911 mg/L by batch fermentation in a 3,000 L fermenter, and the continuous fermentation in a 5 L fermenter for 110 h accumulated 23 g indigo (Han G. H. et al., 2011). In addition, a self-sufficient cytochrome P450 monooxygenase CYP102A (CYP102A_scat) cloned from Streptomyces cattleya was also successfully recombined in E. coli strain BL21(DE3), and the strain could synthesize about 1.0 g/L indigo in LB medium. This is the first self-sufficient CYP exhibiting indole hydroxylation activity to produce indigo without mutating the wild-type enzyme (Kim et al., 2017). The indole oxygenase indAB genes in Cupriavidus sp. SHE were also successfully cloned and heterologously expressed in E. coli BL21(DE3),and the recombinant bacteria could produce 307 mg/L indigo in 1.0 g/L tryptophan medium (Dai et al., 2019).
Indirubin, a 3,2-bisindole isomer of indigo, is one of the main active ingredients of Danggui longhui Wan, which is traditionally used in China to treat chronic myeloid leukemia (Cao et al., 2020). Furthermore, indirubin and its derivatives have considerable therapeutic effects on a variety of cancers, Alzheimer ‘s disease and delayed hypersensitivity (Lee and Lee, 2020). The production of indirubin in tryptophan is the same as that in indigo. Firstly, tryptophan is oxidized to indole by tryptophanase, and then indole can be converted to 3-hydroxyindoxyl, isatin and/or 2-oxindole by heterologous oxygenases, such as NDO. Two molecules of indoxyl are spontaneously dimerized in the presence of oxygen to form indigo, whereas indoxyl and 2-oxindole/isatin are condensed to generate indirubin (Hu et al., 2010; Han et al., 2012; Zhang et al., 2014; Figure 4).
In 5 l fermentation broth containing tryptophan medium, recombinant E. coli DH5α cells containing FMO gene were fermented in batches to produce 5.0 mg/L indirubin. Moreover, it was found that adding 0.36 g/L cysteine to tryptophan medium could significantly increase the yield of indirubin (Han et al., 2012). There is a possible way to increase the output of indirubin. Recombinant E. coli expressing naphthalene dioxygenase (NDO) gene from Comamonas sp. MQ, induced by 2-oxindole, produced about 58 mg/L indirubin (Zhang et al., 2014). There are also some methods, such as introducing cFMO gene into E. coli, 103 mg/L indirubin was produced after 48 h fermentation in LB medium containing 2.5 g/L tryptophan (Lolita Ameria et al., 2015). Or introducing flavin-reducing enzyme Fre, tryptophan-lysing and -importing enzymes TnaA, TnaB and H2O2-degrading enzyme KatE, after adding 5 mmol/L tryptophan and 10 mmol/L 2-hydroxyindole, 250.7 mg/L indirubin was obtained after 48 h fermentation (Yin et al., 2021).
However, all the above methods involve the addition of tryptophan to produce indirubin, which has a high cost. Producing indirubin directly from glucose can be considered as a way to reduce production costs (Cao et al., 2020). For instance, the introduction of Methylophaga aminothioxanthans FMO and E.coli tryptophanase TnaA into E. coli could directly produce indirubin 0.056 g/L from glucose through fed batch fermentation (Dua et al., 2018).
Violacein and deoxyviolacein are biindole pigments with application value of anti-bacterial, anti-virus, anti-oxidation and anti-cancer (Zhou et al., 2018). They are secondary metabolites of bacteria such as Alteromonas luteoviolacea, Chromobacterium violaceum, Janthinobacterium lividum, and Pseudoalteromonas luteoviolacea (Yang et al., 2011; Lee and Lee, 2020).
Violacein is a purple natural indole derivative, was first isolated from C. violaceum. It is synthesized by condensation of two tryptophan molecules in several bacterial genera to respond to quorum sensing signals (Ahmed et al., 2021). At first, through gene cluster separation, sequencing and heterologous expression, the production of violacein was considered to rely mainly on four adjacent genes VioA-D. Then, the fifth gene VioE was supplemented, which played an important role in the formation of violacein (Balibar and Walsh, 2006). The complete synthesis pathway of violacein was formed: VioA (flavin-dependent tryptophan-2 monooxygenase enzyme) catalyzes the oxidation of tryptophan to indole 3-pyruvic acid (IPA) imine, and reduces FAD cofactors. VioB further converts IPA into short-lived imine dimer through dimerization reaction. The imine dimer is either spontaneously converted to chromopyrrolic acid (CPA), or VioE converts the imine dimer into protodeoxyviolaceinic acid (PDVA) through the 1,2-displacement of the indole ring. PDVA is converted to protoviolaceinic acid (PVA) by adding a hydroxyl at the C5 position of an indole ring through nadp-dependent oxygenase VioD. PVA is converted into violaceinic acid (VA) by adding a hydroxyl group to the C2 position of another indole ring via another nadp-dependent oxygenase VioC, and then the final product violacein is generated by spontaneous oxidative decarboxylation. In addition, VioC can also use PDVA as the substrate to produce the main by-product deoxyviolacein (Ahmed et al., 2021; Park et al., 2021; Figure 6). The above five enzymes involve five coding genes vio ABCDE, and the successful expression of the operon composed of these genes requires CviI synthetase to catalyze the conversion of fatty acids or S-adenosyl methionine into AHL, which is triggered by the complex formed by AHL and CviR (a receptor; Kothari et al., 2017).
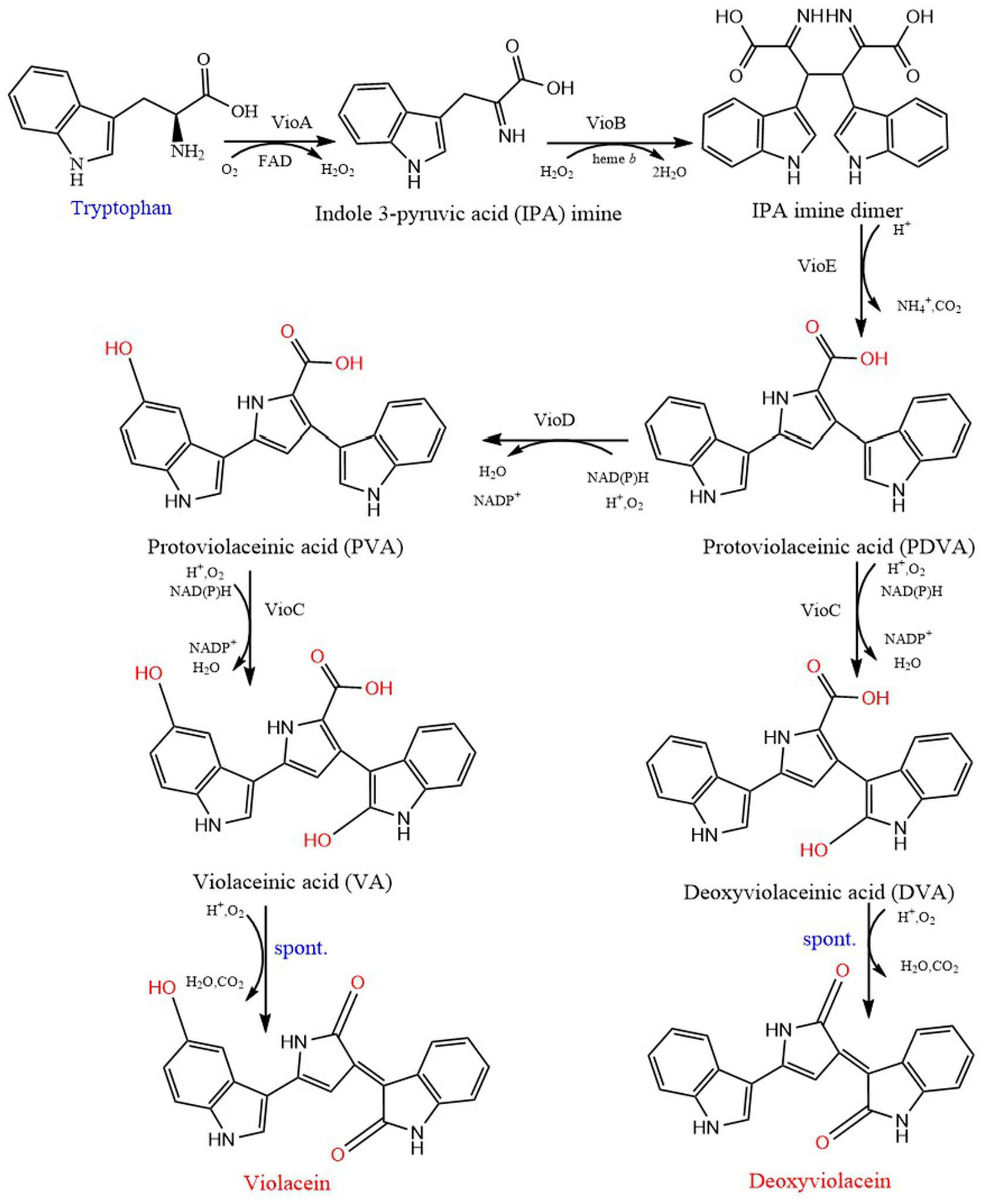
Figure 6. Biosynthetic pathway of violacein and deoxyviolacein. VioA, tryptophan oxidase; VioB, iminophenyl-pyruvate dimer synthase; VioE, violacein biosynthesis enzyme; VioD, protodeoxyviolaceinate monooxygenase; VioC, violacein synthase.
Violacein can be produced by natural production bacteria such as Chromobacterium violaceum (Rodrigues et al., 2012) and Janthinobacterium lividum (Sun et al., 2016). But the relatively low productivity of natural wild strains greatly limits the functional analysis and industrial application of violacein (Yang et al., 2011), and the violacein producing strains of Chromo-bacterium violaceum and Janthinobacterium lividum can cause rare but highly lethal infections in humans (Rodrigues et al., 2013). Therefore, the technology for heterologous expression of violacein gene cluster and production of violacein by genetic engineering has been developed and gradually matured. Pemberton et al. (1991) reported that the violacein gene cluster from C. violaceum was cloned and successfully expressed in E. coli for the first time. Later, it was reported that violacein-synthesizing gene cluster can also be heterologously expressed in Citrobacter freundii, the final concentration of violacein reached 4.13 g/L. This is the first report on the efficient production of violacein by genetic engineering strains in fermentation tanks (Yang et al., 2011).
In order to control the cost, increase the supply of tryptophan and improve the yield of violacein, most people began to choose the combination of the upstream pathway of tryptophan production and the downstream pathway of purple mold production (Fang et al., 2015; Park et al., 2021). By combining knockout of trpR/tnaA/pheA gene and overexpression of trpEfbr/trpD, then, the gene cluster of violacein biosynthetic pathway was introduced into the downstream of tryptophan production pathway. Recombinant E. coli B2/PED+pVio produced 1.75 g/L of purpomycin with glucose as the carbon source (Fang et al., 2015). After the discovery that VioE is the rate limiting step of Aspergillus purpureus synthesis, a strain of E. coli B8/PTRPH1-PVIo-Vioe was obtained by overexpressing VioE, 4.45 g/L violacein was obtained after fed-batch fermentation (Zhou et al., 2018). Fuethermore, the production of recombinant E. coli violacein could be pushed to a new level of 6.19 g/L through integrated system metabolic engineering, cell morphology engineering, inner- and outer-membrane vesicle formation, and fermentation optimization (Yang et al., 2021).
Deoxyviolacein is a structural analogue of violacein and a microbial metabolite. It lacks one oxygen atom at the 6th position of indole ring (Wanga et al., 2012). It has attracted much attention due to its biological activities against tumor, Gram-positive bacteria and plant pathogenic fungi. However, the production of deoxyviolacein in wild Vio bacteria is very low, which is difficult to meet the practical needs (Andre Luis Rodrigues et al., 2014). The vioABCDE pathway was successfully expressed in E.coli, creating a new way for heterologous synthesis of violacein and deoxyviolacein (Pemberton et al., 1991). In addition, the production pathway of deoxyviolacein is mostly coincident with that of purplemycin. Only after PDVA is produced, VioC can directly use PDVA as the substrate to produce deoxyviolacein (Ahmed et al., 2021). Therefore, under conventional conditions, violacein produced by various bacteria is crude violacein, that is, the mixture of violacein and deoxyviolacein (Sun et al., 2016). In order to obtain pure deoxyviolacein, further purification is needed, such as silica gel (SiO2) column chromatography (Bilsland et al., 2018). Pathway summary shows that the expression of VioABCE without VioD would lead to a single end-product of deoxyviolacein (Park et al., 2021). Although E. coli with pLvioABCE (vioDdeleted Vio gene cluster) completely eliminate the production of violacein, however, the presence of a small amount of intermediate PDV associated with deoxyviolacein may regulate the violacein pathway, leading to inefficient production of deoxyviolacein (Sánchez et al., 2006).
A stable and efficient biosynthesis system for the synthesis of pure deoxyviolacein was first attempted and developed. The vioABCE gene cluster from Duganella sp.B2 was spliced and introduced into C. freundii, the recombinant strain produced 1.9 g/L pure deoxyviolacein in the shake flask (Jiang et al., 2012).
The synthesis of deoxyviolacein can also start directly from the synthesis of tryptophan. The araBAD promoter, which controls the expression of deoxyviolomycin cluster vioABCE, was deleted, deoxyviolacein biosynthesis was induced by pentose. Then, 1.6 g/L deoxyviolacein was obtained from E. coli dvio-8 with glycerol as the carbon source (Andre Luis Rodrigues et al., 2014). On the other hand, by integrating system metabolic engineering, cell morphology engineering, internal and external membrane vesicle formation and fermentation optimization, the yield of deoxyviolacein was further increased to 11.26 g/L (Yang et al., 2021).
Halogenated amino acids are widely used in pharmaceutical, chemical and agrochemical industries. They exist in a variety of natural products, including antibiotics chloramphenicol and pyrrolidomycin, plant growth regulating thienodolin and anti Eubacterium pyrrolnitrin, and rebeccamycin, which inhibits DNA topoisomerase I (Veldmann et al., 2019b). Among them, halides derived from tryptophan include 7-chloro-tryptophan and 7-bromo-tryptophan. 7-chloro-tryptophan (Karabencheva-Christova et al., 2017) is the precursor of pyrrolnitrin and rebeccamycin, 7-bromo-tryptophan (Ferrer et al., 2022a) is a precursor of the bioactive protease inhibitor TMC-95A.
In traditional chemical synthesis, the halogenation reaction is usually not an environmentally friendly reaction, so a green method is used by many scholars, that is, halogenase to catalyze the halogenation reaction (Phintha et al., 2021). The enzymatic catalysis of 7-halotryptophan can be completed by FADH-dependent halogenase RebH, NADH-dependent flavin reductase RebF from the biosynthesis of rebeccamycin, or tryptophan 7-halogenase PrnA and its partner flavin reductase Fre from the biosynthesis of pyrrolnitrin (Dong et al., 2005; Yeh et al., 2005; van Pee and Patallo, 2006; Figure 7). They can catalyze the regioselective chlorination/bromination of tryptophan at the 7-position of indole ring (Glenn et al., 2011; van Pee, 2012). The halogenation mechanism is that FAD, O2, and halogen ions are used as substrates, FAD is reduced to FADH2 by flavin reductase, combined with halogenase, and reacts with O2, halogen ions (Cl−, Br−) and substrates at the active site of halogenase to generate halogenated products (Dong et al., 2005; Yeh et al., 2006). For the detailed reaction principle, some scholars have proposed nucleophilic and electrophilic mechanisms respectively, but they have been proved incorrect (Chen and van Pee, 2008).
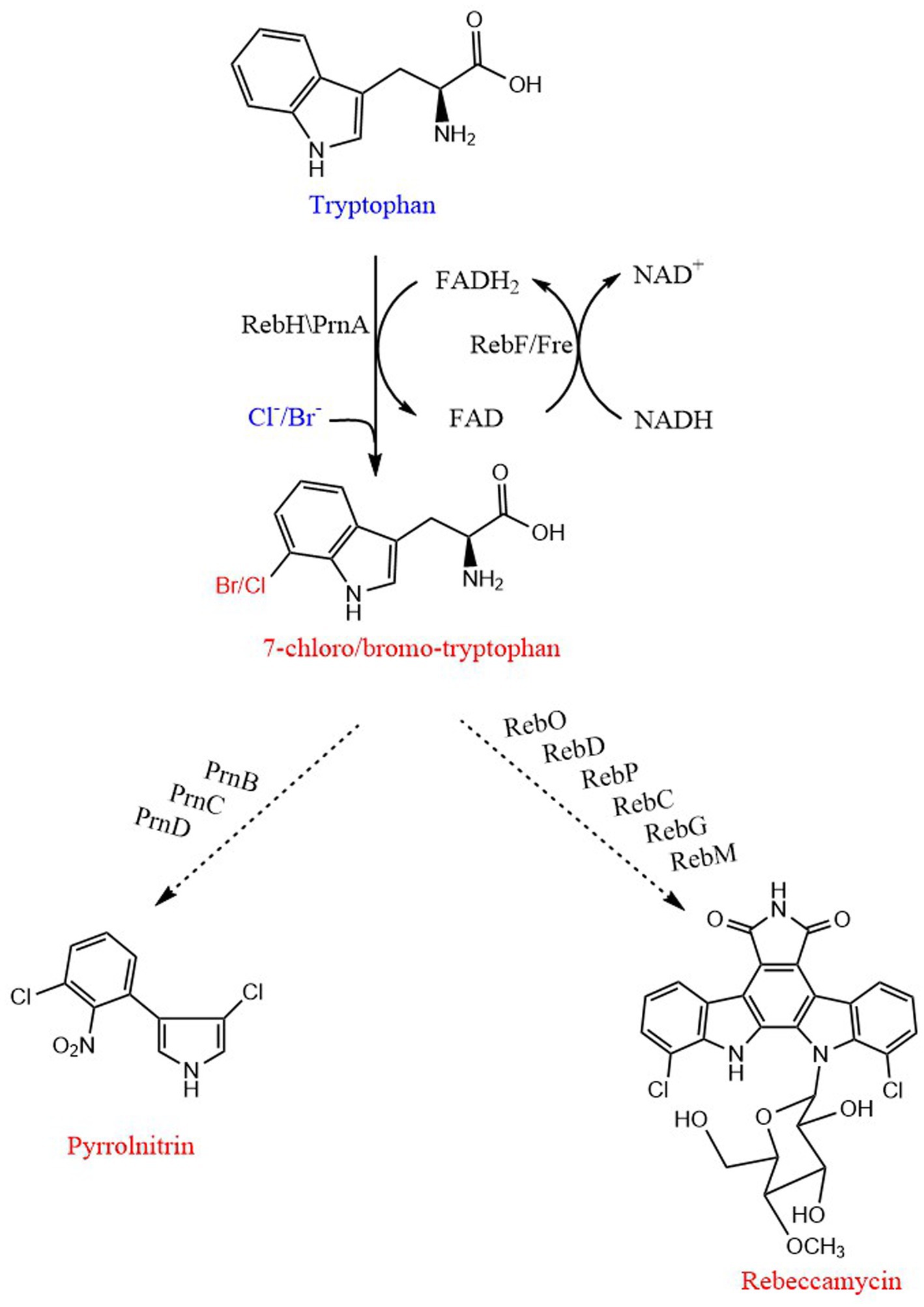
Figure 7. Biosynthetic pathway of 7-Halo-tryptophan, pyrrolnitrin and rebeccamycin. PrnA and RebH are FAD-dependent halogenases. RebF and Fre are NADH-dependent flavin reductases. PrnB, monodechloroaminopyrrolnitrin synthase; PrnC, monodechloroaminopyrrolnitrin halogenase; PrnD, aminopyrrolnitrin oxygenase; RebD acts as both a catalase and a CPA synthase; RebO, FAD-dependent L-tryptophan oxidase; RebP, cytochrome P450 enzyme; RebC, monooxygenase; RebG, N-glycosyltransferase; RebM, methyltransferase. The order of action of enzymes is from top to bottom. The straight line and dotted line represent one-step and multi-step, respectively.
Based on the information obtained from the structural study, the reaction mechanism of tryptophan enzymatic chlorination was proposed (Karabencheva-Christova et al., 2017). In the catalytic cycle of tryptophan 7-halogenase, the reduced form of its cofactor flavin adenine dinucleotide (FAD)-FADH2 first reacts with molecular oxygen O2 to generate C4a-peroxyflavin, and then reacts with chlorine to generate HOCl. The HOCl formed at the FAD binding site moves through the tunnel within the enzyme and is activated to chlorinate the substrate at the tryptophan binding site. The hydrogen bond between HOCl and lysine 79 activates HOCl by increasing its electrophilicity, thereby promoting the chlorination reaction (Dong et al., 2005; Karabencheva-Christova et al., 2017).
The gram-level synthesis of halogenated tryptophan by RebH reaction has been reported, but the efficiency is too low, and it takes 8 days to complete the transformation (Frese and Sewald, 2015). Veldmann et al. (2019b) reported that a trpE gene variant encoding feedback resistant anthranilate synthase component 1, trpD encoding E. coli anthranilate phosphoribosyltransferase and the genes encoding RebH and RebF were overexpressed in C. glutamicum, could obtain 108 mg/L 7-Cl-Trp. According to the above method, with NaBr as the bromine source, the recombinant C. glutamicum could be used to culture in 2 L working volume to obtain 1.2 g/L 7-Br-Trp (Veldmann et al., 2019a).
In the use of enzymes, it was found that when the ratio of RebF and RebH was 3:1, the activity was the best (Yeh et al., 2005). Perhaps we can increase the production of halogenated tryptophan through this idea. In addition, tryptophan can not only be halogenated by enzyme PrnA and RebH at position 7, but also can be halogenated by tryptophan 5-halogenase (PyrH; Zhu et al., 2009) and tryptophan 6-halogenase (Thal; Moritzer et al., 2019), (SttH; Shepherd et al., 2016; Lee et al., 2021) at positions 5 and 6.
Pyrrolnitrin (3-chloro-4-(2′-nitro-3′-chlorophenyl)-pyrrole) is a tryptophan-derived secondary metabolite (Hamill et al., 1970), it was first isolated from Burkholderia pyrrocinia (Pseudomonas pyrrocinia) by Arima et al. (1964) and this compound and its derivatives can also be isolated from rhizospheric fluorescent or non-fluorescent pseudomonads, Serratia and Burkholderia (Pawar et al., 2019). Pyrrolnitrin has been used to treat skin fungal infections due to its strong antifungal activity, and has also been developed as an agricultural fungicide to inhibit soil borne fungal pathogens that affect crop yield (Kwak and Shin, 2015).
The gene cluster necessary for the synthesis of pyrrolnitrin was isolated from Pseudomonas fluorescens BL915, which can produce pyrrolidinitroprotein. It was composed of four genes (ORF1234), named as prnABCD, respectively (Hammer et al., 1997). Combined with the synthesis pathway of pyrrolnitrin speculated by van Pée et al. (1980), the complete gene coding catalytic synthesis pathway of pyrrolnitrin was proposed: the prnA gene product catalyzes the chlorination reaction of L-Trp to produce 7-chloro-tryptophan, and the prnB gene product catalyzes the ring rearrangement and decarboxylation to convert 7-chloro-tryptophan to monochloroaminopyrrolitrin, the prnC gene product chlorinates monodechloroaminopyrrolnitrin at the 3 position to form aminopyrrolnitrin, and the prnD gene product catalyzes the oxidation of the amino group of aminopyrrolnitrin to a nitro group to form pyrrolnitrin (Kirner et al., 1998; Figure 7).
The prnABCD operon was cloned from plymuthica G3 and expressed in E. coli DH5α, the mutant was able to overproduce pyrrolnitrin with isopropyl β-D-thiogalactoside (IPTG) induction by overexpressing prnABCD (Liu et al., 2018). In the wild-type strain, the amount of pyrrolnitrin secreted is small (Pawar et al., 2019). For example, the pyrrolnitrin production of P. aureofaciens ATCC 15926 strain was less than 0.3 μg/ml when grown in minimal medium, but it could be induced by N-methyl-N′-nitro-N nitrosoguanidine to increase its yield (Salcher and Lingens, 1980). In addition, the yield of pyrrolnitrin was also affected by pH, the shake flask fermentation of P. cepacia LT4-12-W showed that the final yield of pyrrolnitrin (168 h) almost doubled at pH 5.8 (Pawar et al., 2019). Therefore, it is necessary to fully consider the influence of various factors in the production process of pyrrolnitrin to improve the output of pyrrolnitrin as much as possible.
Rebeccamycin is a yellow crystalline hydrophobic substance, which was isolated from lechevalieria aerogenes 92 in 1985 (Nettleton et al., 1985; Bush et al., 1987). It is a halogenated natural product of indolcarbazole family, with antibiotic and anti-tumor effects. It has antibacterial activity against several Gram-positive bacteria, such as Staphylococcus aureus and Streptococcus faecalis and can also cause DNA double strand breaks and inhibit topoisomerase I, and inhibit the growth of some tumor cell lines (Sanchez et al., 2002; Waliskoa et al., 2017).
Rebeccamycin is derived from one unit of glucose, one of methionine, and two of tryptophan (Lain et al., 1990). The gene cluster of its biosynthesis was determined by Sanchez et al. (2002). Based on sequence analysis and database searches, they proposed four indole carbazole biosynthetic genes (rebO, rebD, rebC and rebP), two halogenation genes (rebH and rebF), glycosylation gene (rebG, renamed by ngt) and sugar methylation gene (rebM), as well as one regulatory gene (rebR) and two resistance and secretion genes (rebU and rebT), a total of 11 genes involved in rebeccamycin biosynthesis. Subsequently, mutants of the above genes were constructed, after the products of each gene were studied, through the identification of several key biosynthetic intermediates, the biosynthetic pathway of rebekamycin was found, but the RebD catalytic product was not clear (Onaka et al., 2003a). After studying the heterologous expression of RebO and RebD, a mechanism of converting IPA imines through RebO/RebD and a mechanism of generating CPA from L-Trp through two enzyme RebO/RebD system were proposed (Jones and Walsh, 2005). Furthermore, these mechanisms have been proved through experiments (Spolitak and Ballou, 2015). Therefore, the biosynthetic pathway of rebeccamycin is more complete and credible.
The biosynthesis of rebekamycin begins with L-Trp, and forms bisindole pyrrole CPA in a three-step process. The first step is the halogenation reaction of tryptophan, which is catalyzed by RebF/RebH to generate 7-chloro-tryptophan. And then, RebO, a fad-dependent L-Trp oxidase, converts 7-chloro-tryptophan to 7-chloroindole-3-pyruvic acid imine by releasing hydrogen peroxide. The oxidase RebD converts two molecules of 7-chloroindole-3-pyruvate imine to 11,11′-dichlorochromopyrrolic acid. The monooxygenase RebC and cytochrome P450 enzyme RebP perform decarboxylative ring closure. Rebeccamycin aglycon is glycosylated by RebG to 4′-o-dimethyl-rebeccamycin, and then RebM partially methylates glucose to rebeccamycin (Sanchez et al., 2002, 2006; Pommerehne et al., 2019; Figure 7).
As early as 1987, the production of rebeccamycin was attempted by using a strain with aeromycelium c-38,383-RK-2, the strain produced 663 mg/L rebeccamycin after fermentation (Bush et al., 1987). Of course, in addition to the use of wild-type bacteria producing rebekamycin, other types of bacteria can also be used to heterologously express the rebekamycin gene. For example, S. lividans pTOYAMAcos was used to express the whole gene cluster from Lechevalieria aerogenes that synthesized rebeccamycin, and the production of rebeccamycin was detected in transformed S. lividans (Onaka et al., 2003b). Jana et al. have reported that the enhanced culture of micro- and macroparticle has a positive effect on the production of rebeccamycin in the pellet-like morphology of bacteria particles (Waliskoa et al., 2017). In addition, Hiroyasu et al. (Onaka et al., 2015) reported that the yield of biosynthetic gene clusters of goadsporin, staurosporine and rebekamycin was significantly higher in co-culture than in pure culture. Therefore, we may improve the production of rebeccamycin by co-culturing gene clusters expressing different natural products or adding particles of different sizes in the process of culture.
It can be seen from the above that tryptophan derivatives have been applied in various fields such as medicine, agriculture and life. For example, they are used to make cosmetics, textile dyeing products, drugs, pesticides, etc. And they have become an indispensable part of various fields. With the progress of metabolic engineering and synthetic biology, the chemical synthesis methods of tryptophan derivatives with pollution problems have been gradually replaced by green and healthy biosynthesis, such as the research and use of more serotonin, melatonin, indigo, indirubin and so on, they are produced by heterologous expression of genes, and their titer and efficiency are also improved. At present, the biosynthesis methods of tryptophan derivatives can be roughly divided into three categories: one is the production of tryptophan derivatives by native bacteria or plants; the second is the cultivation of mutants on the basis of native bacteria; the third is the heterologous expression of related genes. In substrate selection, tryptophan can be directly added as a substrate to generate various derivatives, and glucose, glycerol, etc. can also be used as carbon sources to integrate the tryptophan synthesis pathway and the tryptophan derivative pathway in the same strain to achieve de novo synthesis.
In the process of biosynthesis of tryptophan derivatives, high yield has always been the pursuit of everyone. For how to improve the yield, the methods of various derivatives described above are different. Here, the author summarizes these methods: the first is to control the proportion of various enzymes. The appropriate proportion of enzymes can maximize the synthesis of catalytic products. Second, the substrate concentration should be controlled. High substrate concentration would inhibit the biological activity and reduce the productivity of the final product. The third is to inhibit the consumption of the final product, when the bacteria produce tryptophan derivatives, the bacteria will also use the final product to meet their own life activities. The fourth is the combined culture of bacteria, there is a mutual promotion between the enzymes and their catalytic products in several bacteria, the combined culture of multiple colonies is expected to increase the yield, but further purification may be needed when various products are harvested. In addition, there is also a complex relationship between cell morphology and yield. Culture parameters including inoculum size and age, genetic factors, culture medium composition, pH, mechanical stress, mass transfer, viscosity, osmotic pressure, solid particles, addition of polymers, surfactants or chelates, temperature, and the geometry of reactors and agitators can significantly affect their morphology, which in turn affects yield.
The biosynthesis of tryptophan derivatives is indeed a green and safe way than chemical synthesis. However, the cost of using tryptophan as the substrate is higher. Therefore, it is a major trend to synthesize tryptophan derivatives from the source using glucose, glycerol and other carbon sources. However, the yield of synthesize tryptophan derivatives from glucose is low. It is also because of this problem that many tryptophan derivatives, such as pyrrolnitrin, have not yet met the requirements of industrial biological manufacturing. According to the above description, we can try to improve the enzyme activity and optimize the culture conditions to improve the yield. On this basis, we can also try to cultivate more suitable strains or use genetic engineering to optimize the synthesis pathway to achieve efficient synthesis of tryptophan derivatives from the source. Finally, because of the wide variety of secondary metabolites of tryptophan, there are still blank in the function, synthesis pathway, and relationship between many secondary metabolites. The tryptophan derivative system needs to be further expanded and improved, and they are also worth exploring for more efficacy and more functions in various fields.
SX, ZW, BW, BH, TB, YZ, and WW drafted the manuscript. JC, LY, and JZ finalized the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (22108017), the Natural Science Foundation of Sichuan Province (2022NSFSC1614), the Starting Grant from Hebei Agricultural University, China (YJ201950) to ZW, the Key Research and Development Program Projects of Hebei Province (22322905D), and the Open Funding Project of Meat Processing Key Laboratory of Sichuan Province (22-R-11 and 22-R-24).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmed, A., Li, R., AL-Ansi, W., Ansi, W. A., Fatima, M., Mushtaq, B. S., et al. (2021). Recent advances in synthetic, industrial and biological applications of violacein and its heterologous production. J. Microbiol. Biotechnol. 31, 1465–1480. doi: 10.4014/jmb.2107.07045
Andre Luis Rodrigues, A. L., Judith Becker, J., Lima, A. O. D. S., Porto, L. M., and Wittman, C. (2014). Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol. Biotechnol. Bioeng. 111, 2280–2289. doi: 10.1002/bit.25297
Arima, K., Imanaka, H., and Kousaka, M. (1964). Pyrrolnitrin, a new antibiotic substance produced by pseudomonas. Agric. Biol. Chem. 28, 575–576. doi: 10.1080/00021369.1964.10858275
Arnao, M. B., and Hernandez-Ruiz, J. (2018). Melatonin and its relationship to plant hormones. Ann. Bot. 121, 195–207. doi: 10.1093/aob/mcx114
Back, K., Tan, D. X., and Reiter, R. J. (2016). Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437. doi: 10.1111/jpi.12364
Balibar, C. J., and Walsh, C. T. (2006). In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium Violaceum. Biochemist 45, 15444–15457. doi: 10.1021/bi061998z
Barik, S. (2020). The uniqueness of tryptophan in biology: properties, metabolism, interactions and localization in proteins. Int. J. Mol. Sci. 21:8776. doi: 10.3390/ijms21228776
Bilsland, E., Tavella, T. A., Krogh, R., Stokes, J. E., Roberts, A., Ajioka, J., et al. (2018). Antiplasmodial and trypanocidal activity of violacein and deoxyviolacein produced from synthetic operons. BMC Biotechnol. 18:22. doi: 10.1186/s12896-018-0428-z
Bush, J. A., Long, B. H., Catino, J. J., and Bradner, W. T. (1987). Production and biological activity of rebeccamycin, a novel antitumor agent. J. Antibiot. 40, 668–678. doi: 10.7164/antibiotics.40.668
Byeon, Y., and Back, K. (2016). Melatonin production in Escherichia coli by dual expression of serotonin N-acetyltransferase and caffeic acid O-methyltransferase. Appl. Microbiol. Biotechnol. 100, 6683–6691. doi: 10.1007/s00253-016-7458-z
Cao, M. F., Gao, M. R., Suástegui, M., Meie, Y. Z., and Shao, Z. Y. (2020). Building microbial factories for the production of aromatic amino acid pathway derivatives: from commodity chemicals to plant-sourced natural products. Metab. Eng. 58, 94–132. doi: 10.1016/j.ymben.2019.08.008
Cas, D. M., Vigentini, I., Vitalini, S., Laganaro, A., Iriti, M., Paroni, R., et al. (2021). Tryptophan derivatives by Saccharomyces cerevisiae EC1118: evaluation, optimization, and production in a soybean-based medium. Int. J. Mol. Sci. 22:472. doi: 10.3390/ijms22010472
Casanova-Saez, R., Mateo-Bonmati, E., and Ljung, K. (2021). Auxin metabolism in plants. CSH Perspect. Biol. 13:a039867
Chen, X., and van Pee, K. H. (2008). Catalytic mechanisms, basic roles, and biotechnological and environmental significance of halogenating enzymes. Acta Biochim. Biophys. Sin. Shanghai 40, 183–193. doi: 10.1111/j.1745-7270.2008.00390.x
Chen, Y., Zhang, Q. L., and Huang, Y. X. (2016). Research progress on biosynthesis of indole alkaloids. Mod. Tradit. Chin. Med. Mat. Medi.-World Sci. Tech. 18, 1914–1920.
Choi, H. S., Kim, J. K., Cho, E. H., Kim, Y. C., Kim, J. I., and Kim, S. W. (2003). A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 306, 930–936. doi: 10.1016/S0006-291X(03)01087-8
Dai, C., Ma, Q., Li, Y., Zhou, D., Yang, B., and Qu, Y. (2019). Application of an efficient indole oxygenase system from Cupriavidus sp. SHE for indigo production. Bioprocess Biosyst. Eng. 42, 1963–1971. doi: 10.1007/s00449-019-02189-4
Dong, C., Flecks, S., Unversucht, S., Haupt, C., van Pee, K. H., and Naismith, J. H. (2005). Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science 309, 2216–2219. doi: 10.1126/science.1116510
Dua, J. K., Yang, D. S., Luo, Z. W., and Lee, S. Y. (2018). Metabolic engineering of Escherichia coli for the production of indirubin from glucose. J. Biotechnol. 267, 19–28. doi: 10.1016/j.jbiotec.2017.12.026
Fang, M. Y., Zhang, C., Yang, S., Cui, J. Y., Jiang, P. X., Lou, K., et al. (2015). High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microb. Cell Factories 14:8. doi: 10.1186/s12934-015-0192-x
Ferrer, L., Elsaraf, M., Mindt, M., and Wendisch, V. F. (2022a). L-serine biosensor-controlled fermentative production of L-tryptophan derivatives by Corynebacterium glutamicum. Biology 11:744.
Ferrer, L., Mindt, M., Suarez-Diez, M., Jilg, T., Zagorscak, M., Lee, J. H., et al. (2022b). Fermentative indole production via bacterial tryptophan synthase alpha subunit and plant indole-3-glycerol phosphate lyase enzymes. J. Agric. Food Chem. 70, 5634–5645. doi: 10.1021/acs.jafc.2c01042
Frese, M., and Sewald, N. (2015). Enzymatic halogenation of tryptophan on a gram scale. Angew. Chem. Int. Ed. Eng. 54, 298–301. doi: 10.1002/anie.201408561
Gaddum, J. H., and Giarman, N. J. (1956). Preliminary studies on the biosynthesis of 5-hydroxytryptamine. Br. J. Pharmacol. 11, 88–92.
Germann, S. M., Baallal Jacobsen, S. A., Schneider, K., Harrison, S. J., Jensen, N. B., Chen, X., et al. (2016). Glucose-based microbial production of the hormone melatonin in yeast Saccharomyces cerevisiae. Biotechnol. J. 11, 717–724. doi: 10.1002/biot.201500143
Glenn, W. S., Nims, E., and O'Connor, S. E. (2011). Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J. Am. Chem. Soc. 133, 19346–19349. doi: 10.1021/ja2089348
Goncalves, S., Nunes-Costa, D., Cardoso, S. M., Empadinhas, N., and Marugg, J. D. (2022). Enzyme promiscuity in serotonin biosynthesis, from bacteria to plants and humans. Front. Microbiol. 13:873555. doi: 10.3389/fmicb.2022.873555
Hamill, R. H., Elander, R. P., and Mabe, J. A. (1970). Metabolism of tryptophan by Pseudomonas aureofaciens: III. Production of substituted pyrrolnitrins from tryptophan analogues. J. Appl. Microbiol. 19, 721–725. doi: 10.1128/am.19.5.721-725.1970
Hammer, P. E., Hill, D. S., Lam, S. T., Van Pee, K. H., and Ligon, J. M. (1997). Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63, 2147–2154. doi: 10.1128/aem.63.6.2147-2154.1997
Han, G. H., Bang, S. E., Babu, B. K., Chang, M., Shin, H.-J., and Kim, S. W. (2011). Bio-indigo production in two different fermentation systems using recombinant Escherichia coli cells harboring a flavin-containing monooxygenase gene (fmo). Process Biochem. 46, 788–791. doi: 10.1016/j.procbio.2010.10.015
Han, G. H., Gim, G. H., Kim, W., Seo, S., and Kim, S. W. (2012). Enhanced indirubin production in recombinant Escherichia coli harboring a flavin-containing monooxygenase gene by cysteine supplementation. J. Biotechnol. 164, 179–187. doi: 10.1016/j.jbiotec.2012.08.015
Han, T. H., Lee, J. H., Cho, M. H., Wood, T. K., and Lee, J. (2011). Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 162, 108–116. doi: 10.1016/j.resmic.2010.11.005
Heine, T., Grossmann, C., Hofmann, S., and Tischler, D. (2019). Indigoid dyes by group E monooxygenases: mechanism and biocatalysis. Biol. Chem. 400, 939–950. doi: 10.1515/hsz-2019-0109
Hu, S., Huang, J., Mei, L. H., Yu, Q., Yao, S. J., and Jin, Z. H. (2010). Altering the regioselectivity of cytochrome P450 BM-3 by saturation mutagenesis for the biosynthesis of indirubin. J. Mol. Catal. B-Enzym. 67, 29–35. doi: 10.1016/j.molcatb.2010.07.001
Jiali, W., Dongcheng, L., and Xiaoli, G. (2012). Research progress of auxin synthesis pathway. Chin. Bull. Bot. 47, 292–301.
Jiang, P. X., Wang, H. S., Xiao, S., Fang, M. Y., Zhang, R. P., He, S. Y., et al. (2012). Pathway redesign for deoxyviolacein biosynthesis in Citrobacter freundii and characterization of this pigment. Appl. Microbiol. Biotechnol. 94, 1521–1532. doi: 10.1007/s00253-012-3960-0
Jones, A. R. H., and Walsh, C. T. (2005). Enzymatic generation of the chromopyrrolicacid scaffold of rebeccamycin by the tandem action of RebO and RebD. Biochemistry 44, 15652–15663. doi: 10.1021/bi051706e
Kang, M. S., and Lee, J. (2009). Cloning and expression of indole oxygenase gene derived from Rhodococcus sp. RHA1. Kor. J. Microbiol. Biotechnol. 37, 197–203.
Karabencheva-Christova, T. G., Torras, J., Mulholland, A. J., Lodola, A., and Christov, C. Z. (2017). Mechanistic insights into the reaction of chlorination of tryptophan catalyzed by tryptophan 7-halogenase. Sci. Rep. 7:17395. doi: 10.1038/s41598-017-17789-x
Kim, H.-J., Jang, S., Kim, J., Yang, Y.-H., Kim, Y.-G., Kim, B.-G., et al. (2017). Biosynthesis of indigo in Escherichia coli expressing self-sufficient CYP102A from Streptomyces cattleya. Dyes Pigments 140, 29–35. doi: 10.1016/j.dyepig.2017.01.029
Kim, Y.-M., Kwak, M.-H., Kim, H.-S., and Lee, J.-H. (2019). Production of Indole-3-acetate in Corynebacterium glutamicum by heterologous expression of the Indole-3-pyruvate pathway genes. Microbiol. Biotechnol. Lett. 47, 242–249. doi: 10.4014/mbl.1901.01013
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., et al. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180, 1939–1943. doi: 10.1128/JB.180.7.1939-1943.1998
Knight, E.M., Zhu, J.F., Förster, J., and Luo, H. (2013). Microorganisms for the production of 5-hydroxytryptophan, Vol. WO2013127914.
Kothari, V., Sharma, S., and Padia, D. (2017). Recent research advances on Chromobacterium violaceum. Asian Pac J Trop Med 10, 744–752. doi: 10.1016/j.apjtm.2017.07.022
Kwak, Y. Y., and Shin, J. H. (2015). Complete genome sequence of Burkholderia pyrrocinia 2327(T), the first industrial bacterium which produced antifungal antibiotic pyrrolnitrin. J. Biotechnol. 211, 3–4. doi: 10.1016/j.jbiotec.2015.06.420
Lain, K. S., Forenza, S., Doyle, T. W., and Pearce, C. J. (1990). Identification of indolepyruvic acid as an intermediate of rebeccamycin biosynthesis. J. Ind. Microbiol. Biotechnol. 6, 291–294.
Lee, J., Kim, J., Song, J. E., Song, W. S., Kim, E. J., Kim, Y. G., et al. (2021). Production of Tyrian purple indigoid dye from tryptophan in Escherichia coli. Nat. Chem. Biol. 17, 104–112. doi: 10.1038/s41589-020-00684-4
Lee, J. Y., and Lee, J. H. (2020). Recent research progress in the microbial production of aromatic compounds derived from L-tryptophan. J. Life Sci. 30, 919–929.
Leontovycova, H., Trda, L., Dobrev, P. I., Sasek, V., Gay, E., Balesdent, M. H., et al. (2020). Auxin biosynthesis in the phytopathogenic fungus Leptosphaeria maculans is associated with enhanced transcription of indole-3-pyruvate decarboxylase LmIPDC2 and tryptophan aminotransferase LmTAM1. Res. Microbiol. 171, 174–184. doi: 10.1016/j.resmic.2020.05.001
Li, Y., Dai, C. X., and Yang, J. (2020). Microbial metabolism of indole and its research progress as a new signal molecule. Microbiol. Chin. 47, 3622–3633.
Li, G., and Young, K. D. (2013). Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159, 402–410. doi: 10.1099/mic.0.064139-0
Lin, Y. H., Sun, X. X., and Yuan, Q. P. (2014). Engineering bacterial phenylalanine 4-hydroxylase for microbial synthesis of human neurotransmitter precursor 5-hydroxytryptophan. ACS Synth. Biol. 3, 497–505. doi: 10.1021/sb5002505
Liu, X. G., Gao, K. X., Yu, X. L., Heeb, S., Gao, S., Chan, K. G., et al. (2018). Functional identification of the prnABCD operon and its regulation in Serratia plymuthica. Appl. Microbiol. Biotechnol. 102, 3711–3721. doi: 10.1007/s00253-018-8857-0
Liu, X. X., Zhang, B., and Ai, L. Z. (2021). Advances in the microbial synthesis of 5-hydroxytryptophan. Front. Bioeng. Biotechnol. 9:624503. doi: 10.3389/fbioe.2021.624503
Lolita Ameria, S. P., Jung, H. S., Kim, H. S., Han, S. S., Kim, H. S., and Lee, J. H. (2015). Characterization of a flavin-containing monooxygenase from Corynebacterium glutamicum and its application to production of indigo and indirubin. Biotechnol. Lett. 37, 1637–1644. doi: 10.1007/s10529-015-1824-2
Luo, H., Schneider, K., Christensen, U., Lei, Y., Herrgard, M., and Palsson, B. O. (2020). Microbial synthesis of human-hormone melatonin at gram scales. ACS Synth. Biol. 9, 1240–1245. doi: 10.1021/acssynbio.0c00065
Mindt, M., Beyraghdar Kashkooli, A., Suarez-Diez, M., Ferrer, L., Jilg, T., Bosch, D., et al. (2022). Production of indole by Corynebacterium glutamicum microbial cell factories for flavor and fragrance applications. Microb. Cell Factories 21:45. doi: 10.1186/s12934-022-01771-y
Mora-Villalobos, J. A., and Zeng, A. P. (2018). Synthetic pathways and processes for effective production of 5-hydroxytryptophan and serotonin from glucose in Escherichia coli. J. Biol. Eng. 12:3. doi: 10.1186/s13036-018-0094-7
Moritzer, A. C., Minges, H., Prior, T., Frese, M., Sewald, N., and Niemann, H. H. (2019). Structure-based switch of regioselectivity in the flavin-dependent tryptophan 6-halogenase Thal. J. Biol. Chem. 294, 2529–2542. doi: 10.1074/jbc.RA118.005393
Nettleton, D. E., Doyle, T. W., and Krishnan, B. (1985). Isolation and structure of rebeccamycin - a new antitumor antibiotic from nocardia aerocoligenes. Tetrahedron Lett. 26, 4011–4014. doi: 10.1016/S0040-4039(00)89280-1
Onaka, H., Ozaki, T., Mori, Y., Izawa, M., Hayashi, S., and Asamizu, S. (2015). Mycolic acid-containing bacteria activate heterologous secondary metabolite expression in Streptomyces lividans. J. Antibiot. 68, 594–597. doi: 10.1038/ja.2015.31
Onaka, H., Taniguchi, S., Igarashi, Y., and Furumai, T. (2003a). Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci. Biotechnol. Biochem. 67, 127–138. doi: 10.1271/bbb.67.127
Onaka, H., Taniguchi, S., and Ikeda, H. (2003b). pTOYAMAcos, pTYM18, and pTYM19, actinomycete-Escherichia coli integrating vectors for heterologous gene expression. J. Antibiot. 56, 950–956. doi: 10.7164/antibiotics.56.950
Park, S., Kang, K., Lee, S. W., Ahn, M. J., Bae, J. M., and Back, K. (2011). Production of serotonin by dual expression of tryptophan decarboxylase and tryptamine 5-hydroxylase in Escherichia coli. Appl. Microbiol. Biotechnol. 89, 1387–1394. doi: 10.1007/s00253-010-2994-4
Park, M., Kang, K., Park, S., and Back, K. (2008). Conversion of 5-hydroxytryptophan into serotonin by tryptophan decarboxylase in plants, Escherichia coli, and yeast. Biosci. Biotechnol. Biochem. 72, 2456–2458. doi: 10.1271/bbb.80220
Park, H., Park, S., Yang, Y. H., and Choi, K. Y. (2021). Microbial synthesis of violacein pigment and its potential applications. Crit. Rev. Biotechnol. 41, 879–901. doi: 10.1080/07388551.2021.1892579
Pawar, S., Chaudhari, A., Prabha, R., Shukla, R., and Singh, D. P. (2019). Microbial pyrrolnitrin: natural metabolite with immense practical utility. Biomol. Ther. 9:443. doi: 10.3390/biom9090443
Pemberton, J. M., Vincent, K. M., and Penfold, R. J. (1991). Cloning and heterologous expression of the violacein biosynthesis gene cluster from Chromobacterium violaceum. Curr. Microbiol. 22, 355–358. doi: 10.1007/BF02092154
Phintha, A., Prakinee, K., Jaruwat, A., Lawan, N., Visitsatthawong, S., Kantiwiriyawanitch, C., et al. (2021). Dissecting the low catalytic capability of flavin-dependent halogenases. J. Biol. Chem. 296:100068. doi: 10.1074/jbc.RA120.016004
Pommerehne, K., Walisko, J., Ebersbach, A., and Krull, R. (2019). The antitumor antibiotic rebeccamycin-challenges and advanced approaches in production processes. Appl. Microbiol. Biotechnol. 103, 3627–3636. doi: 10.1007/s00253-019-09741-y
Rodrigues, A. L., Gocke, Y., Bolten, C., Brock, N. L., Dickschat, J. S., and Wittmann, C. (2012). Microbial production of the drugs violacein and deoxyviolacein: analytical development and strain comparison. Biotechnol. Lett. 34, 717–720. doi: 10.1007/s10529-011-0827-x
Rodrigues, A. L., Trachtmann, N., Becker, J., Lohanatha, A. F., Blotenberg, J., Bolten, C. J., et al. (2013). Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 20, 29–41. doi: 10.1016/j.ymben.2013.08.004
Romasi, E. F., and Lee, J. (2013). Development of indole-3-acetic acid-producing Escherichia coli by functional expression of IpdC, AspC, and Iad1. J. Microbiol. Biotechnol. 23, 1726–1736. doi: 10.4014/jmb.1308.08082
Salcher, O., and Lingens, F. (1980). Isolation and characterization of a mutant of Pseudomonas aureofaciens ATCC 15926 with an increased capacity for synthesis of pyrrolnitrin. J. Gen. Microbiol. 118, 509–513.
Sánchez, C., Braña, A. F., and Méndez, C. (2006). Reevaluation of the violacein biosynthetic pathway and its relationship to indolocarbazole biosynthesis. Chembiochem 7, 1231–1240. doi: 10.1002/cbic.200600029
Sanchez, C., Butovich, I. A., Brana, A. F., Rohr, J., Mendez, C., and Salas, J. A. (2002). The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem. Biol. 9, 519–531.
Sanchez, C., Mendez, C., and Salas, J. A. (2006). Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. Nat. Prod. Rep. 23, 1007–1045. doi: 10.1039/B601930G
Shen, P. J., Qi, F., and Huang, J. Z. (2020). Progress in serotonin biosynthesis. Pharm. Biotechnol. 27, 68–72.
Shepherd, S. A., Menon, B. R., Fisk, H., Struck, A. W., Levy, C., Leys, D., et al. (2016). A structure-guided switch in the regioselectivity of a tryptophan halogenase. Chembiochem 17, 821–824. doi: 10.1002/cbic.201600051
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a004138. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Spolitak, T., and Ballou, D. P. (2015). Evidence for catalytic intermediates involved in generating the chromopyrrolic acid scaffold of rebeccamycin by RebO and RebD. Arch. Biochem. Biophys. 573, 111–119. doi: 10.1016/j.abb.2015.03.020
Sun, H. N., Zhao, D. D., Xiong, B., Zhang, C. Z., and Bi, C. H. (2016). Engineering Corynebacterium glutamicum for violacein hyper production. Microb. Cell Factories 15:148. doi: 10.1186/s12934-016-0545-0
van Pee, K. H. (2012). Enzymatic chlorination and bromination. Methods Enzymol. 516, 237–257. doi: 10.1016/B978-0-12-394291-3.00004-6
van Pee, K. H., and Patallo, E. P. (2006). Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl. Microbiol. Biotechnol. 70, 631–641.
van Pée, K. H., Salcher, O., and Lingens, F. (1980). Formation of pyrrolnitrin and 3-(2-amino-3-chlorophenyl)pyrrole from 7-chlorotryptophan. Angew. Chem. Int. Ed. Eng. 19, 828–829.
Veldmann, K. H., Dachwitz, S., Risse, J. M., Lee, J. H., Sewald, N., and Wendisch, V. F. (2019a). Bromination of L-tryptophan in a fermentative process with Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 7:219. doi: 10.3389/fbioe.2019.00219
Veldmann, K. H., Minges, H., Sewald, N., Lee, J. H., and Wendisch, V. F. (2019b). Metabolic engineering of Corynebacterium glutamicum for the fermentative production of halogenated tryptophan. J. Biotechnol. 291, 7–16. doi: 10.1016/j.jbiotec.2018.12.008
Waliskoa, J., Vernenb, F., and Pommerehne, K. (2017). Particle-based production of antibiotic rebeccamycin with Lechevalieria aerocolonigenes. Process Biochem. 53, 1–9. doi: 10.1016/j.procbio.2016.11.017
Wang, Y., Chen, X., Chen, Q., Zhou, N., Wang, X., Zhang, A., et al. (2022). Construction of cell factory capable of efficiently converting L-tryptophan into 5-hydroxytryptamine. Microb. Cell Factories 21:47. doi: 10.1186/s12934-022-01745-0
Wang, H. J., Liu, W. Q., and Shi, F. (2018). Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli. Metab. Eng. 48, 279–287. doi: 10.1016/j.ymben.2018.06.007
Wanga, A. S., Wang, F. Z., Hu, X. F., Yan, Y. C., Yu, X. H., Jiang, P. X., et al. (2012). Biosynthesis and characterization of violacein, deoxyviolacein and oxyviolacein in heterologous host, and their antimicrobial activities. Biochem. Eng. J. 67, 148–155. doi: 10.1016/j.bej.2012.06.005
Watanabe, T., and Snell, E. E. (1972). Reversibility of tryptophanase reaction-synthesis of tryptophan from indole, pyruvate, and ammonia. Proc. Nat. Acad. Sci. U. S. A. 69, 1086–1090. doi: 10.1073/pnas.69.5.1086
Wu, F. L., Wang, X. S., Song, F. Q., Peng, Y. F., and Wang, Q. H. (2021). Research progress in microbial metabolic engineering of aromatic compounds. Chin. J. Biotechnol. 37, 1771–1793.
Xie, X., Ding, D., Bai, D., Zhu, Y., Sun, W., Sun, Y., et al. (2022). Melatonin biosynthesis pathways in nature and its production in engineered microorganisms. Synth. Syst. Biotechnol. 7, 544–553. doi: 10.1016/j.synbio.2021.12.011
Xu, D., Fang, M. J., and Wang, H. J. (2020). Enhanced production of 5-hydroxytryptophan through the regulation of L-tryptophan biosynthetic pathway. Appl. Microbiol. Biotechnol. 104, 2481–2488. doi: 10.1007/s00253-020-10371-y
Xu, W., Gavia, D. J., and Tang, Y. (2014). Biosynthesis of fungal indole alkaloids. Nat. Prod. Rep. 31, 1474–1487. doi: 10.1039/C4NP00073K
Yang, C., Jiang, P. X., Xiao, S., Zhang, C., Kai, L., and Xing, X. H. (2011). Fed-batch fermentation of recombinant Citrobacter freundii with expression of a violacein-synthesizing gene cluster for efficient violacein production from glycerol. Biochem. Eng. J. 57, 55–62. doi: 10.1016/j.bej.2011.08.008
Yang, D. S., Park, S. Y., and Lee, S. Y. (2021). Production of rainbow colorants by metabolically engineered Escherichia coli. Adv. Sci. 8:e2100743. doi: 10.1002/advs.202100743
Yeh, E., Cole, L. J., Barr, E. W., Bollinger, J. J. M., Ballou, D. P., and Walsh, C. T. (2006). Flavin redox chemistry precedes substrate chlorination during the reaction of the flavin-dependent halogenase RebH. Biochemistry 45, 7904–7912. doi: 10.1021/bi060607d
Yeh, E., Garneau, S., and Walsh, C. T. (2005). Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 102, 3960–3965. doi: 10.1073/pnas.0500755102
Yin, H. F., Chen, H. P., Yan, M., Li, Z. K., Yang, R. D., Li, Y. J., et al. (2021). Efficient bioproduction of indigo and indirubin by optimizing a novel terpenoid cyclase XiaI in Escherichia coli. ACS Omega 6, 20569–20576. doi: 10.1021/acsomega.1c02679
Zhang, X. W., Qu, Y. Y., Ma, Q., Kong, C. L., Zhou, H., Cao, X. Y., et al. (2014). Production of indirubin from tryptophan by recombinant Escherichia coli containing naphthalene dioxygenase genes from Comamonas sp. MQ. Appl. Biochem. Biotechnol. 172, 3194–3206. doi: 10.1007/s12010-014-0743-3
Zheng, S., Zhu, Y., Liu, C., Fan, W., Xiang, Z., and Zhao, A. (2021). Genome-wide identification and characterization of genes involved in melatonin biosynthesis in Morus notabilis (wild mulberry). Phytochemistry 189:112819. doi: 10.1016/j.phytochem.2021.112819
Zhou, Y., Fang, M. Y., Li, G., Zhang, C., and Xing, X. H. (2018). Enhanced production of crude violacein from glucose in Escherichia coli by overexpression of rate-limiting key enzyme(S) involved in violacein biosynthesis. Appl. Biochem. Biotechnol. 186, 909–916. doi: 10.1007/s12010-018-2787-2
Keywords: tryptophan derivatives, 5-hydroxytryptophan, indigo, indoleacetic acid, halotryptophan
Citation: Xiao S, Wang Z, Wang B, Hou B, Cheng J, Bai T, Zhang Y, Wang W, Yan L and Zhang J (2023) Expanding the application of tryptophan: Industrial biomanufacturing of tryptophan derivatives. Front. Microbiol. 14:1099098. doi: 10.3389/fmicb.2023.1099098
Received: 15 November 2022; Accepted: 08 March 2023;
Published: 23 March 2023.
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Zhi-Gang Jeff Qian, Shanghai Jiao Tong University, ChinaCopyright © 2023 Xiao, Wang, Wang, Hou, Cheng, Bai, Zhang, Wang, Yan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Cheng, Y2hlbmdqaWVAY2R1LmVkdS5jbg==; Lixiu Yan, OTMyMTc1NTc0QHFxLmNvbQ==; Jiamin Zhang, NDkyMzQ2ODg0QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.