
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 April 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1097173
This article is part of the Research Topic MicroRNA: the swift development in infectious diseases View all 6 articles
 Gokulakannan Venkatesan1
Gokulakannan Venkatesan1 Wan Suriana Wan Ab Rahman1*
Wan Suriana Wan Ab Rahman1* Wan Nazatul Shima Shahidan1
Wan Nazatul Shima Shahidan1 Salfarina Iberahim2
Salfarina Iberahim2 Alwi bin Muhd Besari@Hashim3
Alwi bin Muhd Besari@Hashim3Early disease diagnosis is critical for better management and treatment outcome of patients. Therefore, diagnostic methods should ideally be accurate, consistent, easy to perform at low cost and preferably non-invasive. In recent years, various biomarkers have been studied for the detection of cardiovascular diseases, cerebrovascular diseases, infectious diseases, diabetes mellitus and malignancies. Exosomal microRNA (miRNA) are small non-coding RNA molecules that influence gene expression after transcription. Previous studies have shown that these types of miRNAs can potentially be used as biomarkers for cancers of the breast and colon, as well as diffuse large B-cell lymphoma. It may also be used to indicate viral and bacterial infections, such as the human immunodeficiency virus (HIV), tuberculosis and hepatitis. However, its use in the diagnosis of vector-borne diseases is rather limited. Therefore, this review aims to introduce several miRNAs derived from exosomal plasma that may potentially serve as a disease biomarker due to the body’s immune response, with special focus on the early detection of vector-borne diseases.
Extracellular vesicles (EVs) are a heterogeneous group of membrane vesicles secreted by nearly all cell types in normal physiology, and the secretion of their content is influenced by the cells’ current pathophysiological state (Yáñez-Mó et al., 2015). They are classified based on their generation mechanism, including ectosomes, apoptotic bodies and exosomes (Martelli et al., 2018). Ectosomes, known as microvesicles, typically range from 200 to 2,000 nm in size and is generated through outward buddings from the plasma membrane (Wu et al., 2021). Microvesicles mainly contain cytosolic and plasma membrane-associated proteins, such as tetraspanins. They also carry cytoskeletal proteins, heat-shock proteins, integrins and post-translationally modified proteins that have undergone glycosylation and phosphorylation (Doyle and Wang, 2019). The discovery of new proteins in microvesicles have spurred global interest in fully understanding their contents for diagnostic and therapeutic potential. In malaria infection caused by Plasmodium vivax, a rise in the level of plasma-derived microvesicles has been observed in the blood of patients, which potentially allows disease detection at an earlier stage (Campos et al., 2010). Apoptotic bodies, meanwhile, are vesicles with a size ranging from 500 nm to 2 μm, and are discharged by dying cells into the extracellular space. They contain broken down cellular particles, such as intact organelles, chromatin and small amounts of glycosylated proteins (Borges et al., 2013). A recent study has demonstrated that these vesicles may be used as a non-invasive method to track apoptosis in patients with cerebrovascular and neurodegenerative diseases for prognostic purposes (Serrano-Heras et al., 2020).
Exosomes are nanometer-sized (50–150 nm) intraluminal vesicles (ILV) of multivesicular bodies (MVB) secreted by various cells, including the microglia (Mashouri et al., 2019; Fan et al., 2022). They have double membrane structure (Saadatpour et al., 2016) that originates from endocytic compartments and are matured in the MVB of late endosomes (Statello et al., 2018). MVBs can fuse with the plasma membrane, leading to the release of ILVs into the extracellular space as exosomes (Colombo et al., 2014). Exosomes are surrounded by a lipid bilayer, carrying a variety of biomolecules, such as glycans, proteins, metabolites, lipids, DNA, and RNA, including miRNA (Chung et al., 2020). They can potentially serve as an important medium for cell-to-cell or cell-to-internal environment communication (Jeppesen et al., 2019), and the biomolecules may alter physiological responses by mediating short- and long-distance inter-organ communication (Fatima and Nawaz, 2017). Exosomes are less immunogenic, non-cytotoxic and non-mutagenic to their target cells compared with other gene delivery vehicles (O’Loughlin et al., 2012). Depending on the molecules they carry, exosomes can perform a variety of roles, such as encouraging cell proliferation, migration, lowering oxidative stress and preventing apoptosis, besides mediating immune responses like cytokine secretion and inhibition or stimulation of inflammasomes (Noonin and Thongboonkerd, 2021). Emerging findings have suggested that exosomes may play a key role in mediating infection and inflammatory processes (Xie et al., 2022).
Infection by a vector-borne disease will trigger the effects of a host-parasite interaction, which facilitates the parasite’s long-term survival in the host body (Podolska and Nadolna, 2014). The host-parasite interaction can also trigger the host immune response and enhance pathogen diffusion (Cox et al., 2012). As parasites interact with host cells through excretory-secretory channels, the exosomes within excretory-secretory products will naturally serve as a route for successfully mediating the parasite’s contact for host manipulation. It also makes sense that the host would utilize this pathway as a defense mechanism (Nawaz et al., 2019).
Recently, exosomes have gained much research attention in biomarker development as they contain specific proteins, microRNAs (miRNAs) and signaling molecules that are associated with disease progression (Hoshino et al., 2015). Exosome miRNA is highly accessible and stable, thus raising its potential use in precision medicine (Muinelo-Romay et al., 2018). However, studies on the role of exosomes in vector-borne diseases are nonetheless limited by inefficient separation techniques, a lack of distinctive biomarkers, and challenges in obtaining high-resolution imaging methods. Exosomes contain identical protein and RNA profiles to the cells that produce them. However, very few distinct cell-specific proteins and miRNAs exist (Li et al., 2017). Furthermore, exosomes from pathogen-infected cells are heterogeneous, and their concentration exists in a different range than the standard level (Reyes-Ruiz et al., 2019). In this review, we collated and updated the role of exosomal plasma miRNA in various diseases, and then examined the scope of miRNA-derived from plasma exosomes as a reliable diagnostic tool for vector-borne diseases.
Article search and data collection were performed on PubMed, Google Scholar, and World Health Organization (WHO) databases. It included peer-reviewed articles with original data from 2010 to 2022. Non-peer-reviewed articles, conference abstracts, letters, articles without full-text were excluded. All articles were collected and compiled using the Mendeley application to eliminate duplicates. A PubMed database search using the keywords “extracellular vesicles and vector bore diseases” resulted in a total of 225 articles. After subsequently applying the keywords “exosomes and vector-borne disease,” a total of 115 articles were produced. As the search was narrowed down with the keywords “miRNA biomarker and vector bore diseases,” the articles were further reduced to 79. Finally, 31 articles remained when the second parameter “plasma miRNA and vector-borne diseases” was added. The articles were manually divided into two groups: “original articles” and “others” (not related to topic, reviews, reports, editorials, commentaries, etc.). The original articles addressing plasma exosome miRNA and vector-borne diseases became the focus of the study and were used as material in this review.
Exosomes are critical paracrine mediators that may be detected in bodily fluids, including plasma (Carrozzo et al., 2021). The content of human plasma exosomes have been linked to a variety of physiological and pathological conditions (Yáñez-Mó et al., 2015). Such exosomes may be isolated using ultracentrifugation, polymer-based precipitation kits, size exclusion chromatography, immunoaffinity, and ultrafiltration (Kalra et al., 2013). Ultracentrifugation and polymer-based precipitation kits are the most popular methods, but the former has several limitations, including low exosome concentration and considerable albumin contamination (Baranyai et al., 2015). These may be resolved by changing the isolation parameters, such as viscosity, sedimentation distance and temperature, or by prolonging the ultracentrifugation time to increase yield (Koh et al., 2018). The presence of exosomes may be characterized using complex methods, such as nanoparticle tracking analysis, electrophoretic light scattering, transmission electron microscopy and EXOCET colorimetric assay, or simple protein quantification like Western blotting (Martins et al., 2018). In blood samples, plasma exosomes have a higher concentration of miRNA compared with serum, and this may serve as a better sample for blood-based proteomics research of exosomes (Cao et al., 2019). Plasma cells may be more protected from degradation and this enables more accurate distribution of target cells, providing a distinctive perspective into pertinent diseases, or defining novel diagnostic and prognostic potential for different disorders (Ludwig et al., 2017). Hence, plasma-derived exosomes are a clinically valuable biomarker to assess the immune system of patients because its molecular profiling and protein monitoring may show the degree of immunological suppression before and during treatment (Ludwig et al., 2017).
MiRNA are naturally occurring short non-coding RNA molecules with a length of about 21–25 nucleotides (Ha and Kim, 2014). MiRNA from exosomes may affect the body’s physiological modulation and disease progression (Liu et al., 2020). Depending on disease condition, the level of miRNA in bodily fluids will be up- or down-regulated (Abdellatif, 2012). The miRNA level in blood, for example, is affected by various factors and will differ from sample to sample (e.g., plasma, serum, and other fluids) (Blondal et al., 2013). Hemolysis can influence the expression of miRNA in serum, but plasma-derived miRNA is unaffected (Foye et al., 2017). MiRNA in the exosomal plasma are in more stable form than any other bodily fluids because they are protected from endogenous RNase degradation (Köberle et al., 2013). This makes them useful in the diagnosis of infectious diseases, besides diseases of the central nervous system and respiratory systems, including malignancy (Alipoor et al., 2016).
During the early stage of human immunodeficiency virus-1 (HIV-1) infection, changes in exosomal miR-155 expression play a role in the development of an effective antiviral effector response by CD8 T cells and at the same time increase the number of regulatory T cells (Lind et al., 2013). Thus, exosomal miRNA involved in the regulation of immune and inflammatory responses, and also serve as an excellent companion in the diagnosis of HIV infection (Hubert et al., 2015). Tuberculosis is another infectious disease that is currently screened by microscopic examination and confirmed using genotypic assays (line probe assay, cartridge-based nucleic acid amplification test, or loop-mediated isothermal amplification), besides the gold standard culture methods (liquid culture media) (Chopra and Singh, 2020). Still, there is research on finding a reproducible, efficient, cost-effective tool with minimal infrastructure requirements for this infectious disease. Recently, upregulated miRNA-185-5p expression in exosome plasma has been identified in tuberculosis (Kaushik et al., 2021). In hepatitis C (HCV), immunomodulatory miRNA enrichment exosomes are found to be associated with their inhibitory activity on innate immune cell function. This may provide valuable biomarkers to monitor immune response recovery (Santangelo et al., 2018).
Most recently, reverse-transcription polymerase chain reaction (RT-PCR) has been mainly used to confirm COVID-19 infection. However, false-negatives may occur in a small percentage of tests due to a low viral load in patient samples (Anand et al., 2021). The use of exosomes as a diagnostic tool for COVID-19 has yet to be widely studied. Therefore, there is an immediate need to study and develop an alternative method that is more sensitive, specific and easy to apply. The study of the exosomal cargo may provide key information with respect to differential secretion of miRNA in SARS-CoV 2 infected cells compared with uninfected cells. Table 1 summarizes the exosomal plasma-derived miRNA biomarkers for diagnostic applications in various diseases. The preceding studies support the notion that plasma-derived exosomal miRNAs play important roles in diseases as a promising biomarker and provide knowledge for developing a new diagnostic tool for the diseases.
From DNA sequences, miRNAs are translated into precursor miRNAs (pre-miRNAs), mature miRNAs and primary miRNAs (pri-miRNAs). MiRNAs regulate protein translation by binding to the 3’ untranslated regions of mRNA template (MacFarlane and Murphy, 2010). However, it has been reported that they can also interact with other regions, including the 5’ UTR coding sequence and gene promoters (Broughton et al., 2016). These miRNAs play a crucial role in influencing molecular and cellular processes in cancer and inflammation (O’Connell et al., 2012).
Enterotoxigenic Bacteroides Fragilis (ETBF) is an enterotoxin-producing bacterium that is strongly associated with inflammatory bowel disease (IBD), colitis-associated colorectal cancer and colorectal cancer (CRC) as it has been observed to change the mucosal immune response and induce epithelial cell changes (Zamani et al., 2020). A recent study on ETBF shows that the exosomal down- regulated miR-149-3p derived from ETBF-treated cells facilitated T-helper type 17 cell differentiation and promoted the PHF5A (Plant homeodomain finger protein 5A) mediated RNA alternative splicing of the KAT2A gene in CRC cells. Thus, the ETBF/miR-149-3p pathway showed a potential solution for treatment of patients with intestinal inflammation and CRC with a high amount of ETBF (Cao et al., 2021). Plasma-derived exosomal miR-216a-5p may also inhibit rTp17-induced inflammatory cytokine production and the TLR4-MYD88 signaling pathway in syphilis infection, suggesting a possible therapeutic target for inflammation caused by the sexually transmitted disease (Peng et al., 2019).
Medulloblastoma (MB) is a malignant brain tumor that commonly affects children. Exosomal miR-101-3p and miR-423-5p from plasma of MB patients have been identified to act as tumor suppressors by directly targeting FOXP4 and EZH2 oncogenes (Xue et al., 2022). This shows that plasma-derived exosomal miRNA can control the expression of oncogenes, making them a potential therapeutic agent. An interesting study on long-term exercise-derived plasma exosome found that miR-342-5p may prevent ischemic heart disease by suppressing the level of Caspase 9 and JNK2, thus reducing hypoxia/reoxygenation-induced cardiomyocyte apoptosis; moreover, it increased survival signaling (p-Akt) by targeting the phosphatase gene Ppm1f (Hou et al., 2019). These studies provided the information needed to understand the role plasma-derived exosomal miRNA as a therapeutic agent by controlling the expression of genes that cause various diseases.
Vector-borne diseases are infections caused by pathogens that are usually transmitted by arthropods, such as mosquitoes, triatomine bugs, black flies, tsetse flies, sand flies, lice and ticks (Golding et al., 2015). However, they may also include other carriers such water snails, which serve as a reservoir for Schistosomiasis. In the case of insects, they include dengue, Chagas disease, Japanese encephalitis, leishmaniasis, lymphatic filariasis, malaria, and yellow fever, which all together threaten more than 80% of the world’s population. They are inordinately distributed in the tropics and subtropics (Chala and Hamde, 2021), and the World Health Organization (WHO) estimated that they account for more than 700,000 deaths annually (World Health Organization, 2022a). Figure 1 shows the number of vector-borne cases reported worldwide from 2011 to 2020. Table 2 summarizes the laboratory method for vector-borne disease detection in the host and its vectors.
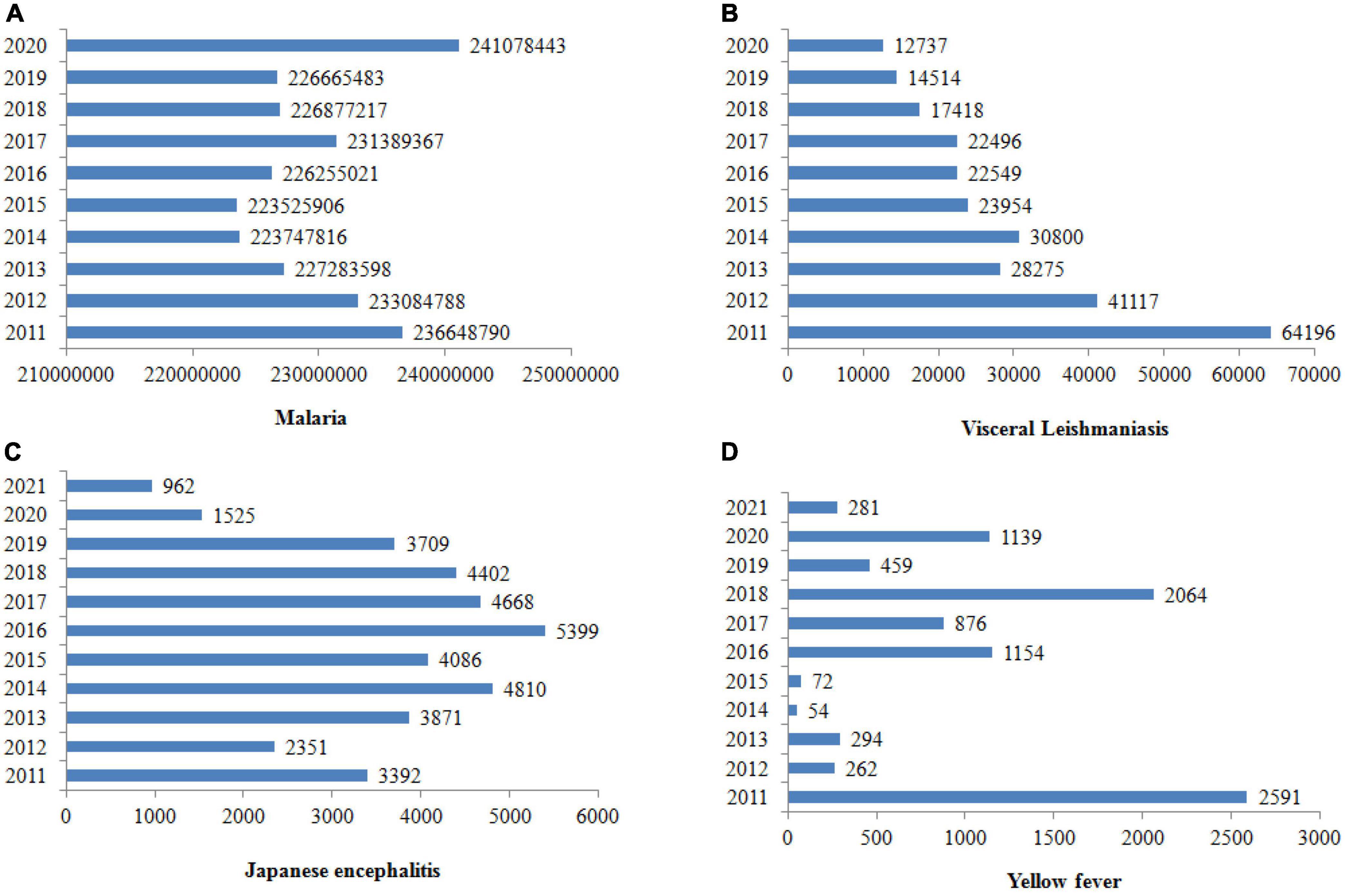
Figure 1. Cases of vector-borne diseases worldwide from 2011 to 2020. The prevalence of malaria is rising, but leishmaniasis, Japanese encephalitis and yellow fever seem to be declining (World Health Organization, 2020a,b, 2021, 2022d).
When feeding on their hosts, arthropods release exosomes into the host blood through their saliva (Figure 2; Oliva Chávez et al., 2021). In response, the host cells will also release their own exosomes to inhibit the transmission of pathogens (Alenquer and Amorim, 2015). This increases exosome concentrations in the plasma and other body fluids (Opadokun and Rohrbach, 2021). As a result, exosomes from the host and pathogen will play distinct roles in the survival and suppression of vector-borne diseases.
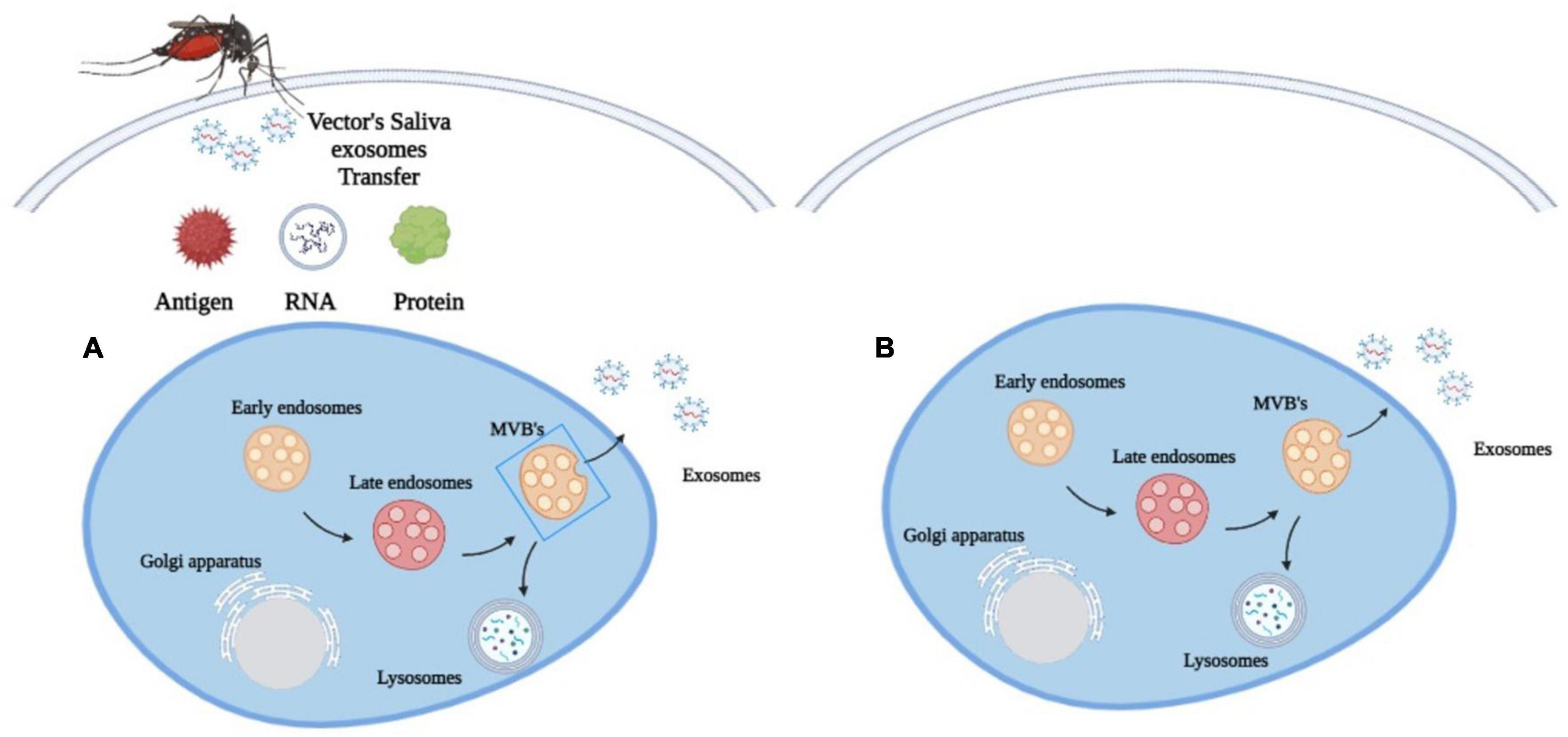
Figure 2. Exosomes biogenesis in (A) vector-borne diseases. Arthropods release exosomes into the host blood circulation through their saliva, which transfer antigens, proteins and RNA to modulate immune responses, gene expression and metabolic processes and (B) in other diseases. Exosomes are released only by the diseased cell.
The exosomes in the arthropod’s saliva can be considered a carrier for various biological processes in vector-borne diseases. They transfer antigens, proteins and RNA into the host cell to influence metabolic processes, gene expression and immunological responses (Figure 2; Yuan et al., 2021). Through this process, these exosomes may facilitate the transmission of arboviruses that cause dengue and encephalitis to mammals (Zhou et al., 2019). The discovery of exosomes produced by arthropods has spurred the development of new methods to prevent the spread of vector-borne diseases through the understanding of molecular determinants in the mode of pathogen transmission. A Malaysian study has found that the EV secreted by Aedes albopictus and Aedes aegypti contains a tetraspanin domain-containing glycoprotein known as Tsp29Fb that could facilitate the transmission of Dengue Virus 2 (DENV2) in host cells (Vora et al., 2018). This study has led to a novel proposal to restrict the transmission of DENV2 and other mosquito-borne flaviviruses to mammalian cells by inhibiting the effects of Tsp29Fb in mosquitoes using the GW4869 inhibitor. A proteomic study on exosomes of Drosophila cells infected by the cricket paralysis virus (CrPV) also revealed that they contained viral RNA to facilitate the spread of the pathogen’s genetic material in the host (Kerr et al., 2018).
Pathogen surveillance in arthropods may provide essential details for disease control. A central American study tried to discover the best method to survey the prevalence of Trypanosoma cruzi that causes Chagas disease in the vector Triatoma dimidiata (blood-sucking bug) (Stevens et al., 2021). The researchers concluded that using genotype-by-sequencing (GBS) was twice superior in detecting the pathogen in its vector compared with PCR assay (based on a test with the significant agreement; 53% vs. 19%) However, the difficulty of pathogen detection in insects may be caused by genetic variations in the parasite itself, and this can affect the disease surveillance and control process. Meanwhile, detection of the EV secreted by pathogens may also assist in the development of new clinical biomarkers and therapeutic agents. The study of exosomes in Brugia malayi has found that miRNA let-7 (lethal 7) could function as a biomarker for lymphatic filariasis disease (Zamanian et al., 2015). Table 3 summarizes the potential miRNA biomarkers for vector-borne diseases. Table 4 summarizes the exosomal miRNA biomarkers for diagnostic applications in vector-borne diseases.
Dengue fever is a febrile illness caused by Flavivirus of the family Flaviviridae. This virus has a common size of between 40 and 65 nm and contains a positive single-stranded RNA genome (Murugesan and Manoharan, 2019). The WHO estimates its prevalence at 50–100 million cases each year in tropical countries (WHO Dengue, 2022). In the 21st century, dengue has emerged as one of the most significant mosquito-borne diseases that pose a severe threat to public health (Gurugama et al., 2010). Therefore, finding predictive indicators of severe infection will greatly aid in patient management and treatment allocations. Conventional diagnostic methods include the structural protein 1 (NS1) test, immunoglobulin G/M test and real-time PCR (Kabir et al., 2021). However, a comprehensive analysis of miRNAs circulating in the plasma of dengue patients has identified miR-122-5p as a potential biomarker for diagnosis and prognosis of the disease (Saini et al., 2020). Integrated bioinformatic analysis predicted that interferon-induced protein 44-like (IFI44L) and interferon-α inducible protein-6 (IFI6) could also be used as potential biomarkers of infection (Xie et al., 2021).
MiRNA expression may change during the different stages of infection, thus providing the potential biomarkers for treatment monitoring (Soe et al., 2018). Dengue virus infection causes platelet cells to release exosomes via the C-type lectin-like receptor 2 (CLEC2) signaling (Sung et al., 2019). Exosomes released from mosquito cells infected with dengue virus were observed to have an effect on native C6/36 cells, which shows that the exosomes played a role in virus dissemination (Reyes-Ruiz et al., 2019). Exosomes secreted from dengue virus infected cells has been observed to carry LC3-phosphatidylethanolamine conjugates (LC3 II), which is an autophagy marker (Wu et al., 2016). However, further knowledge on plasma exosomes and their exact function in the transmission dengue virus is still lacking.
Malaria is a vector-borne disease caused by plasmodium parasites carried by the female Anopheles mosquito (Walter and John, 2022). It is most prevalent in Africa, where 95% of the fatality occurs. In 2020 alone, it was estimated that there were about 241 million cases worldwide, with a death toll of 627,000 (WHO Malaria, 2022). To overcome this disease, an in-depth understanding of the parasite biology and mechanisms underlying the disease is urgently needed (Chen et al., 2021). Conventionally, microscopy or a rapid diagnostic kit is used to confirm the presence of malaria-causing parasites (CDC Malaria, 2022). Lately, studies have shown that plasma exosome also increases in malaria patients compared with healthy individuals (Nantakomol et al., 2011). These biomolecules play many important roles in intercellular communication during the infection process (Schorey et al., 2015). Exosomal plasma-derived hsa-miR-150-5p and hsa-miR-15b-5p have been studied for their potential use as disease biomarkers (Ketprasit et al., 2020). In the case of clinical management, malaria is difficult to treat due to the ability of the plasmodium parasite to develop drug resistance. One study has observed the production of biomolecules with potential function to pass on drug-resistance markers to susceptible parasites in gametocytogenesis stage, which is critical for disease transmission (Regev-Rudzki et al., 2013).
Rickettsia is a species of highly pleomorphic, non-motile, Gram-negative bacteria that may exist in coccus, bacilli and thread forms, and is transmitted via the bites of mites, fleas, lice, and ticks (Moreira et al., 2018). Being an obligate intracellular parasite, it has to infect eukaryotic host cells to survive and replicate, particularly endothelial cells. Even though rickettsial infections have similar clinical presentations, the causative species and epidemiology may differ regionally. Some examples include epidemic typhus (Rickettsia prowazekii, worldwide), the Australian tick typhus (Rickettsia australis, Australia), Rocky Mountain spotted fever (Rickettsia rickettsii, North America), rickettsialpox (Rickettsia akari, United States and Russia), oriental spotted fever (Rickettsia japonica, Japan) and African tick bite fever (Rickettsia africae, South Africa). To accurately diagnose and treat these diseases, it is crucial to understand both the typical symptoms and epidemiology of a specific area (Khamesipour et al., 2018). In severe rickettsiosis, endothelial barrier dysfunction occurs in the brain and lungs that results in edema. A recent study showed that endothelial exosomal miR-23a and miR-30b have potential role in targeting the endothelial barriers and may serve as an early diagnostic tool (Liu et al., 2021).
Chagas disease, also known as American trypanosomiasis, is a tropical parasitic disease caused by the protozoa Trypanosoma cruzi. It is transmitted mainly by bugs of the triatominae subfamily (also known as kissing bugs or vampire bugs) (Rassi et al., 2010). More than seven million people are infected with Chagas disease annually, mainly in Mexico, Central America and South America. It has been estimated to be responsible for 12,500 fatalities per year since 2006 (World Health Organization, 2022b). Chagas disease is diagnosed using microscopy and serological tests to detect trypomastigotes in blood (CDC Chagas, 2022). The significance of miR-208a levels in plasma as a potential biomarker for clinical prognosis of Chagas disease is illustrated in the chronic indeterminate phase, which is a progressive phase implicated in the development of chagas cardiomyopathy (Linhares-Lacerda et al., 2018). Plasma exosomes produced during disease infection may also induce different interferon (IFN) and Interleukin-17 (IL-17) expression, which may be used to track the development of the disease (Madeira et al., 2021).
Schistosomiasis is the second leading parasitic disease after malaria, which is caused by trematodes of the genus Schistosoma (Amoah et al., 2020). Schistosomiasis is prevalent in tropical and subtropical areas, with 236.6 million cases reported worldwide in 2019 (WHO Schistosomiasis, 2022). It can excrete certain proteins into the circulation of a final host (humans) to facilitate their parasitism (Liao et al., 2011). Currently, the diagnostic methods for this disease include stool and urine microscopy, besides serological testing for antischistosomal antibody (CDC Schistosomiasis, 2022). Different schistosome-specific miRNAs, like miR-10, miR-3479, miR-0001, and miR-10, have been discovered to describe the pathogen-specific short RNA community in the plasma of a final host infected with Schistosoma japonicum (Cheng et al., 2013). Currently, serum exosome-derived miR-142-3p and miR-223-3p are being used as diagnostic markers of schistosomiasis at an early stage (Patent number: CN110760589-A and CN110760590-A) (Wu et al., 2020). Some studies have shown that exosomal products can serve as prognostic and stage-predictive biomarkers of the disease (Meningher et al., 2017).
Zika virus (ZIKV) is a mosquito-borne virus transmitted by Aedes mosquitoes. It is closely related to dengue and other diseases related to viruses in the Flaviviridae family. The virus is enveloped and icosahedral, containing a positive sense single-stranded RNA as its genetic material (Marbán-Castro et al., 2021). Conventionally, ZIKV was detected in blood and other bodily fluids using RT-PCR; enzyme-linked immunosorbent assay (ELISA) for NS1 antigen; enzyme immunoassay (EIA) for IgM/IgG; immunofluorescence assay (IFA); and rapid point-of-care testing, such as the plaque-reduction neutralization test (PRNT) (World Health Organization, 2022c).
It is vital to differentiate ZIKV from other closely related flaviviruses that can cross-react when using conventional diagnostic methods like ELISA and EIA. And to this end, RT-PCR seems to be the most accurate tool. The detection of ZIKV in Aedes aegypti and Aedes albopictus mosquitoes using RT-PCR in São Paulo, Brazil, has helped to understand the transmission dynamics of ZIKV and potential risk of future outbreaks in several neighborhoods in the city (Parra et al., 2022). However, in the clinical setting, there is a need for a method that is fast and low cost. The EVs from the ZIKV-infected human saliva express typical EV biomarker tetraspanins CD9, CD63, and CD81 to prevent target cells (Conzelmann et al., 2020). The induction of miR-145 and miR-148a in postmortem brain samples from stillborn with severe congenital Zika syndrome cause neuronal dysfunction and malformation (Castro et al., 2019). These studies showed the role of extracellular vesicles and the miRNA in Zika infection.
Japanese encephalitis virus (JEV) is a mosquito-borne disease transmitted by Culex mosquitoes. Like dengue, West Nile, Yellow Fever and Zika counterparts, it is part of the family Flaviviridae that causes viral encephalitis (Sharma et al., 2021). Conventionally, it may be diagnosed using ELISA, RT-PCR, and the plaque reduction neutralization test. However, there is still a need to develop more efficient, inexpensive, quicker, sensitive and time-saving techniques to detect JEV (Roberts and Gandhi, 2020). Exosome study in acute patients has found upregulated miRNAs like miR-21-5p, miR-150-5p, and miR-342-3p in the cerebrospinal fluid, which can be a potential diagnostic biomarker (Goswami et al., 2017). The host microRNA miR-34c-5p in JEV infection is downregulated and activates a notch pathway to modulate the microglia and mediate inflammation (Kumari et al., 2016). This showed that miRNA can regulate host response to JEV in microglial cells.
Chikungunya virus (CHIKV) is an alphavirus transmitted by Aedes and some Culex mosquitoes. It is a positive-sense, single-stranded RNA virus in the family Togaviridae that causes Chikungunya disease (Schwartz and Albert, 2010). Conventionally, Chikungunya is mainly diagnosed using RT-PCR. The study on extracellular vesicles released by CHIKV-infected cells contained the immunogenic components of virus as well as its genomic RNA. These EVs then helped other epithelial cells produce contagious virions (Le et al., 2022). Since there is a limited study on the role of exosomal miRNA in chikungunya, this study initiated to characterize extracellular vesicles in chikungunya infection.
Lymphatic filariasis (LF) is a mosquito-borne tropical disease caused by parasitic nematodes, such as Wuchereria bancrofti, Brugia malayi, and Brugia timori (Lourens and Ferrell, 2019). These parasites can modulate the host immune response. However, there is still a lack of knowledge to understand this mechanism. In lymphatic filariasis, the first study in parasite B. malayi exosome reported the mechanism of host response on parasite infection. This study also suggested that the further investigation in extracellular vesicles may led to the discovery of novel treatment approaches and diagnostic biomarkers for lymphatic filariasis infection (Zamanian et al., 2015). The parasite-derived EV study on filarial nematode B. malayi has suggested new screening platforms for anti-filarial drug development (Harischandra et al., 2018).
Leishmaniasis is a vector-borne disease transmitted by the protozoan parasites infected adult female phlebotomine sand flies. It is caused by the leishmania parasite that belongs to the family Trypanosomatidae. The conventional methods to diagnose leishmaniasis are parasitological tests (microscopic examination, PCR detection) and serological tests, such as an indirect IFA, ELISA, and Western blot (WHO Leishmaniasis, 2023). The exosomal miRNA study in eukaryotic Leishmania infection cell reported that the exosomal miR122 can interfere with lipoprotein status in the host cell after hepatic dysfunctions induced by the parasite (Di Loria et al., 2020). This study showed that exosomal miRNA from the vector can involve in the host metabolism modulation. The parasite detection of Leishmania donovani in arthropod Phlebotomus argentipes using DNA amplification (PCR) and sequencing methods help to study the disease epidemiology in Kurunegala District, Sri Lanka (Wijerathna et al., 2021). The parasite Leishmania-infected dendritic cells release the exosomes to protect against the parasite infection. It provokes new knowledge to develop a cell-free vaccine for immune prophylaxis against leishmaniasis (Schnitzer et al., 2010).
The currently available diagnostic methods for many vector-borne diseases are insufficient and have many limitations. Most of the detection is dependent on the existence of signs and symptoms, or antibodies of particular pathogen. Therefore, there is an urgent need to support the development of new biomarker techniques in diagnostic platforms. Plasma-derived exosome miRNAs are more stable, reliable and highly concentrated than any other bodily fluids. Recent studies have indicated that exosomal miRNA within excretory-secretory products may serve as a route for successfully mediating parasite-host contacts for host manipulation. Interestingly, previous studies discovered certain differentially expressed host plasma-derived exosome miRNAs that may be potential diagnostic and prognostic biomarker of vector-borne diseases. However, there is still limited research of plasma exosome in vector-borne diseases as compared with other diseases such as malignancy. The lack of consistent definitions and a standard method causes considerable confusion in the laboratory study of exosomes. We believe that further research in this area will solve the problem, and provide the new ideas and approaches for the application of plasma-derived exosome miRNA in clinical practice.
GV conducted the literature search and drafted the manuscript. WS conceived the idea of this review and gathered the data. WNS, SI, and AM gave critical ideas in drafting the manuscript. All authors approved the final submission of this manuscript.
The writing and research to produce this review was funded by the Ministry of Higher Education Malaysia under the Fundamental Research Grant Scheme (Project code: FRGS/1/2020/SKK0/USM/03/16).
We thank the staff members of the Hematology Laboratory Unit of Hospital Universiti Sains Malaysia for their help.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdellatif, M. (2012). Differential expression of microRNAs in different disease states. Circ. Res. 110, 638–650. doi: 10.1161/CIRCRESAHA.111.247437
Alenquer, M., and Amorim, M. J. (2015). Exosome biogenesis, regulation, and function in viral infection. Viruses 7, 5066–5083. doi: 10.3390/V7092862
Ali, E. O. M., Babalghith, A. O., Bahathig, A. O. S., Dafalla, O. M., Al-Maghamsi, I. W., Mustafa, N. E. A. G., et al. (2022). Detection of dengue virus from Aedes aegypti (Diptera, Culicidae) in field-caught samples from Makkah Al-Mokarramah, kingdom of Saudi Arabia, using RT-PCR. Front. Public Heal. 10:850851. doi: 10.3389/FPUBH.2022.850851
Alipoor, S. D., Mortaz, E., Garssen, J., Movassaghi, M., Mirsaeidi, M., and Adcock, I. M. (2016). Exosomes and exosomal miRNA in respiratory diseases. Mediat. Inflamm. 2016:5628404. doi: 10.1155/2016/5628404
Amoah, A. S., Hoekstra, P. T., Casacuberta-Partal, M., Coffeng, L. E., Corstjens, P. L. A. M., Greco, B., et al. (2020). Sensitive diagnostic tools and targeted drug administration strategies are needed to eliminate schistosomiasis. Lancet Infect. Dis. 20, e165–e172. doi: 10.1016/S1473-3099(20)30254-1
Anand, K., Vadivalagan, C., Joseph, J. S., Singh, S. K., Gulati, M., Shahbaaz, M., et al. (2021). A novel nano therapeutic using convalescent plasma derived exosomal (CP Exo) for COVID-19: A combined hyperactive immune modulation and diagnostics. Chem. Biol. Interact. 344:109497. doi: 10.1016/J.CBI.2021.109497
Baranyai, T., Herczeg, K., Onódi, Z., Voszka, I., Módos, K., Marton, N., et al. (2015). Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One 10:e0145686. doi: 10.1371/JOURNAL.PONE.0145686
Bavia, L., Mosimann, A. L. P., Aoki, M. N., and Duarte Dos Santos, C. N. (2016). A glance at subgenomic flavivirus RNAs and microRNAs in flavivirus infections. Virol. J. 13:84. doi: 10.1186/S12985-016-0541-3
Blondal, T., Jensby Nielsen, S., Baker, A., Andreasen, D., Mouritzen, P., Wrang Teilum, M., et al. (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. doi: 10.1016/J.YMETH.2012.09.015
Borges, F. T., Reis, L. A., and Schor, N. (2013). Extracellular vesicles: Structure, function, and potential clinical uses in renal diseases. Braz. J. Med. Biol. Res. 46, 824–830. doi: 10.1590/1414-431X20132964
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W., Pasquinelli, A. E., Broughton, J. P., et al. (2016). Pairing beyond the seed supports MicroRNA targeting specificity article pairing beyond the seed supports MicroRNA targeting specificity. Mol. Cell 64, 320–333. doi: 10.1016/j.molcel.2016.09.004
Campos, F. M., Franklin, B. S., Teixeira-Carvalho, A., Filho, A. L., De Paula, S. C., Fontes, C. J., et al. (2010). Augmented plasma microparticles during acute Plasmodium vivax infection. Malar. J. 9:327. doi: 10.1186/1475-2875-9-327
Caner, V., Cetin, G. O., Hacioglu, S., Baris, I. C., Tepeli, E., Turk, N., et al. (2021). The miRNA content of circulating exosomes in DLBCL patients and in vitro influence of DLBCL-derived exosomes on miRNA expression of healthy B-cells from peripheral blood. Cancer Biomark. 32, 519–529. doi: 10.3233/CBM-210110
Cao, F., Gao, Y., Chu, Q., Wu, Q., Zhao, L., Lan, T., et al. (2019). Proteomics comparison of exosomes from serum and plasma between ultracentrifugation and polymer-based precipitation kit methods. Electrophoresis 40, 3092–3098. doi: 10.1002/ELPS.201900295
Cao, Y., Wang, Z., Yan, Y., Ji, L., He, J., Xuan, B., et al. (2021). Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology 161, 1552–1566.e12. doi: 10.1053/J.GASTRO.2021.08.003
Carrozzo, A., Casieri, V., Di Silvestre, D., Brambilla, F., De Nitto, E., Sardaro, N., et al. (2021). Plasma exosomes characterization reveals a perioperative protein signature in older patients undergoing different types of on-pump cardiac surgery. Geroscience 43, 773–789. doi: 10.1007/S11357-020-00223-Y
Castro, F. L., Geddes, V. E. V., Monteiro, F. L. L., Gonçalves, R. M. D. T., Campanati, L., Pezzuto, P., et al. (2019). MicroRNAs 145 and 148a Are upregulated during congenital Zika virus infection. ASN Neuro 11:1759091419850983. doi: 10.1177/1759091419850983
CDC Malaria (2022). Centers for disease control and prevention, malaria diagnostic tests. Atlanta, GA: CDC.
CDC Schistosomiasis (2022). Centers for disease control and prevention, schistosomiasis. Atlanta, GA: CDC.
Chala, B., and Hamde, F. (2021). Emerging and re-emerging vector-borne infectious diseases and the challenges for control: A review. Front. Public Heal. 9:715759. doi: 10.3389/FPUBH.2021.715759
Chen, J. Y., Zhou, J. K., and Pan, W. (2021). Immunometabolism: Towards a better understanding the mechanism of parasitic infection and immunity. Front. Immunol. 12:661241. doi: 10.3389/FIMMU.2021.661241
Cheng, G., Luo, R., Hu, C., Cao, J., and Jin, Y. (2013). Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology 140, 1751–1761. doi: 10.1017/S0031182013000917
Chopra, K. K., and Singh, S. (2020). Newer diagnostic tests for tuberculosis, their utility, and their limitations. Curr. Med. Res. Pract. 10, 8–11. doi: 10.1016/J.CMRP.2020.01.004
Chung, I. M., Rajakumar, G., Venkidasamy, B., Subramanian, U., and Thiruvengadam, M. (2020). Exosomes: Current use and future applications. Clin. Chim. Acta 500, 226–232. doi: 10.1016/J.CCA.2019.10.022
Colombo, M., Raposo, G., and Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. doi: 10.1146/ANNUREV-CELLBIO-101512-122326
Conzelmann, C., Groß, R., Zou, M., Krüger, F., Görgens, A., Gustafsson, M. O., et al. (2020). Salivary extracellular vesicles inhibit Zika virus but not SARS-CoV-2 infection. J. Extracell. Vesicles 9:1808281. doi: 10.1080/20013078.2020.1808281
Cox, J., Mota, J., Sukupolvi-Petty, S., Diamond, M. S., and Rico-Hesse, R. (2012). Mosquito bite delivery of dengue virus enhances immunogenicity and pathogenesis in humanized mice. J. Virol. 86, 7637–7649. doi: 10.1128/JVI.00534-12
Cruz, A. C. R., Neto, J. P. N., da Silva, S. P., da Silva, E. V. P., Pereira, G. J. G., Santos, M. M., et al. (2020). Chikungunya virus detection in Aedes aegypti and Culex quinquefasciatus during an outbreak in the amazon region. Viruses 12:853. doi: 10.3390/V12080853
Di Loria, A., Dattilo, V., Santoro, D., Guccione, J., De Luca, A., Ciaramella, P., et al. (2020). Expression of serum exosomal miRNA 122 and lipoprotein levels in dogs naturally infected by Leishmania infantum: A preliminary study. Animals 10:100. doi: 10.3390/ANI10010100
Doyle, L. M., and Wang, M. Z. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8:727. doi: 10.3390/CELLS8070727
Entonu, M. E., Muhammad, A., Ndams, I. S., and Franciosa, G. (2020). Evaluation of actin-1 expression in wild caught wuchereria bancrofti-infected mosquito vectors. J. Pathog. 2020, 1–9. doi: 10.1155/2020/7912042
Fan, C., Li, Y., Lan, T., Wang, W., Long, Y., and Yu, S. Y. (2022). Microglia secrete miR-146a-5p-containing exosomes to regulate neurogenesis in depression. Mol. Ther. 30, 1300–1314. doi: 10.1016/J.YMTHE.2021.11.006
Fatima, F., and Nawaz, M. (2017). Long distance metabolic regulation through adipose-derived circulating exosomal miRNAs: A trail for RNA-based therapies? Front. Physiol. 8:545. doi: 10.3389/FPHYS.2017.00545
Foye, C., Yan, I. K., David, W., Shukla, N., Habboush, Y., Chase, L., et al. (2017). Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS One 12:e0189165. doi: 10.1371/JOURNAL.PONE.0189165
Garcia-Contreras, M., Shah, S. H., Tamayo, A., Robbins, P. D., Golberg, R. B., Mendez, A. J., et al. (2017). Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration type 1 diabetes. Sci. Rep. 7:5998. doi: 10.1038/S41598-017-05787-Y
Gaye, P. M., Doucoure, S., Senghor, B., Faye, B., Goumballa, N., Sembène, M., et al. (2021). Bulinus senegalensis and Bulinus umbilicatus snail infestations by the Schistosoma haematobium group in Niakhar, Senegal. Pathogens 10:860. doi: 10.3390/PATHOGENS10070860
Golding, N., Wilson, A. L., Moyes, C. L., Cano, J., Pigott, D. M., Velayudhan, R., et al. (2015). Integrating vector control across diseases. BMC Med. 13:249. doi: 10.1186/S12916-015-0491-4
Goswami, S., Banerjee, A., Kumari, B., Bandopadhyay, B., Bhattacharya, N., Basu, N., et al. (2017). Differential expression and significance of circulating microRNAs in cerebrospinal fluid of acute encephalitis patients infected with Japanese encephalitis virus. Mol. Neurobiol. 54, 1541–1551. doi: 10.1007/s12035-016-9764-y
Gurugama, P., Garg, P., Perera, J., Wijewickrama, A., and Seneviratne, S. (2010). Dengue viral infections. Indian J. Dermatol. 55, 68–78. doi: 10.4103/0019-5154.60357
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524. doi: 10.1038/NRM3838
Hannafon, B. N., Trigoso, Y. D., Calloway, C. L., Zhao, Y. D., Lum, D. H., Welm, A. L., et al. (2016). Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 18:90. doi: 10.1186/S13058-016-0753-X
Harischandra, H., Yuan, W., Loghry, H. J., Zamanian, M., and Kimber, M. J. (2018). Profiling extracellular vesicle release by the filarial nematode Brugia malayi reveals sex-specific differences in cargo and a sensitivity to ivermectin. PLoS Negl. Trop. Dis. 2:e0006438. doi: 10.1371/journal.pntd.0006438
He, X., Sai, X., Chen, C., Zhang, Y., Xu, X., Zhang, D., et al. (2013). Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasit. Vectors 6:272. doi: 10.1186/1756-3305-6-272
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. doi: 10.1038/NATURE15756
Hou, Z., Qin, X., Hu, Y., Zhang, X., Li, G., Wu, J., et al. (2019). Longterm exercise-derived exosomal miR-342-5p: A novel exerkine for cardioprotection. Circ. Res. 124, 1386–1400. doi: 10.1161/CIRCRESAHA.118.314635
Hubert, A., Subra, C., Jenabian, M. A., Labrecque, P. F. T., Tremblay, C., Laffont, B., et al. (2015). Elevated abundance, size, and MicroRNA Content of plasma extracellular vesicles in viremic HIV-1+ patients: Correlations with known markers of disease progression. J. Acquir. Immune Defic. Syndr. 70, 219–227. doi: 10.1097/QAI.0000000000000756
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of exosome composition. Cell 177, 428–445.e18. doi: 10.1016/J.CELL.2019.02.029
Kabir, M. A., Zilouchian, H., Younas, M. A., and Asghar, W. (2021). Dengue detection: Advances in diagnostic tools from conventional technology to point of care. Biosensors 11:206. doi: 10.3390/BIOS11070206
Kalra, H., Adda, C. G., Liem, M., Ang, C. S., Mechler, A., Simpson, R. J., et al. (2013). Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13, 3354–3364. doi: 10.1002/PMIC.201300282
Kaur, H., Sehgal, R., Kumar, A., Sehgal, A., Bansal, D., and Sultan, A. A. (2018). Screening and identification of potential novel biomarker for diagnosis of complicated Plasmodium vivax malaria. J. Transl. Med. 16:272. doi: 10.1186/S12967-018-1646-9
Kaushik, A. C., Wu, Q., Lin, L., Li, H., Zhao, L., Wen, Z., et al. (2021). Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis. Brief. Bioinform. 22:bbab210. doi: 10.1093/BIB/BBAB210
Kerr, C. H., Dalwadi, U., Scott, N. E., Yip, C. K., Foster, L. J., and Jan, E. (2018). Transmission of cricket paralysis virus via exosome-like vesicles during infection of Drosophila cells. Sci. Rep. 8:17353. doi: 10.1038/S41598-018-35717-5
Ketprasit, N., Cheng, I. S., Deutsch, F., Tran, N., Imwong, M., Combes, V., et al. (2020). The characterization of extracellular vesicles-derived microRNAs in Thai malaria patients. Malar. J. 19:285. doi: 10.1186/S12936-020-03360-Z
Khamesipour, F., Dida, G. O., Anyona, D. N., Razavi, M., and Rakhshandehroo, E. (2018). Tick-borne zoonoses in the order Rickettsiales and Legionellales in Iran: A systematic review. PLoS Negl. Trop. Dis. 12:e0006722. doi: 10.1371/JOURNAL.PNTD.0006722
Köberle, V., Pleli, T., Schmithals, C., Augusto Alonso, E., Haupenthal, J., Bönig, H., et al. (2013). Differential stability of cell-free circulating microRNAs: Implications for their utilization as biomarkers. PLoS One 8:e75184. doi: 10.1371/JOURNAL.PONE.0075184
Koh, Y. Q., Almughlliq, F. B., Vaswani, K., Peiris, H. N., and Mitchell, M. D. (2018). Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front. Biosci. 23:865–874. doi: 10.2741/4621
Kumari, B., Jain, P., Das, S., Ghosal, S., Hazra, B., Trivedi, A. C., et al. (2016). Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese encephalitis virus in microglial cells. Sci. Rep. 6:20263. doi: 10.1038/SREP20263
Le, B. C. T., Burassakarn, A., Tongchai, P., Ekalaksananan, T., Aromseree, S., Phanthanawiboon, S., et al. (2022). Characterization and involvement of exosomes originating from Chikungunya virus-infected epithelial cells in the transmission of infectious viral elements. Int. J. Mol. Sci. 23:12117. doi: 10.3390/ijms232012117
Lessard, B. D., Kurucz, N., Rodriguez, J., Carter, J., and Hardy, C. M. (2021). Detection of the Japanese encephalitis vector mosquito Culex tritaeniorhynchus in Australia using molecular diagnostics and morphology. Parasit. Vectors 14:411. doi: 10.1186/S13071-021-04911-2
Li, P., Kaslan, M., Lee, S. H., Yao, J., and Gao, Z. (2017). Progress in exosome isolation techniques. Theranostics 7, 789–804. doi: 10.7150/THNO.18133
Liang, M., Yu, S., Tang, S., Bai, L., Cheng, J., Gu, Y., et al. (2020). A panel of plasma exosomal miRNAs as potential biomarkers for differential diagnosis of thyroid nodules. Front. Genet. 11:449. doi: 10.3389/FGENE.2020.00449
Liao, Q., Yuan, X., Xiao, H., Liu, C., Lv, Z., Zhao, Y., et al. (2011). Identifying Schistosoma japonicum excretory/secretory proteins and their interactions with host immune system. PLoS One 6:e23786. doi: 10.1371/JOURNAL.PONE.0023786
Lind, E. F., Elford, A. R., and Ohashi, P. S. (2013). Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J. Immunol. 190, 1210–1216. doi: 10.4049/JIMMUNOL.1202700
Linhares-Lacerda, L., Granato, A., Gomes-Neto, J. F., Conde, L., Freire-de-Lima, L., de Freitas, E. O., et al. (2018). Circulating plasma MicroRNA-208a as potential biomarker of chronic indeterminate phase of Chagas disease. Front. Microbiol. 9:269. doi: 10.3389/FMICB.2018.00269
Liu, W., Yang, D., Chen, L., Liu, Q., Wang, W., Yang, Z., et al. (2020). Plasma exosomal miRNA-139-3p is a novel biomarker of colorectal cancer. J. Cancer 11, 4899–4906. doi: 10.7150/JCA.45548
Liu, Y., Zhou, C., Su, Z., Chang, Q., Qiu, Y., Bei, J., et al. (2021). endothelial exosome plays a functional role during rickettsial infection. MBio 12, e769–e721. doi: 10.1128/MBIO.00769-21
Lourens, G. B., and Ferrell, D. K. (2019). Lymphatic filariasis. Nurs. Clin. North Am. 54, 181–192. doi: 10.1016/J.CNUR.2019.02.007
Ludwig, S., Floros, T., Theodoraki, M. N., Hong, C. S., Jackson, E. K., Lang, S., et al. (2017). Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin. Cancer Res. 23, 4843–4854. doi: 10.1158/1078-0432.CCR-16-2819
Lugli, G., Cohen, A. M., Bennett, D. A., Shah, R. C., Fields, C. J., Hernandez, A. G., et al. (2015). Plasma exosomal miRNAs in persons with and without Alzheimer disease: Altered expression and prospects for biomarkers. PLoS One 10:e0139233. doi: 10.1371/JOURNAL.PONE.0139233
MacFarlane, L.-A., and Murphy, P. R. (2010). MicroRNA: Biogenesis, function and role in cancer. Curr. Genomics 11:537. doi: 10.2174/138920210793175895
Madeira, R. P., Dal’Mas Romera, L. M., De Cássia Buck, P., Mady, C., Ianni, B. M., and Torrecilhas, A. C. (2021). New biomarker in Chagas disease: Extracellular vesicles isolated from peripheral blood in chronic Chagas disease patients modulate the human immune response. J. Immunol. Res. 2021:6650670. doi: 10.1155/2021/6650670
Marbán-Castro, E., Goncé, A., Fumadó, V., Romero-Acevedo, L., and Bardají, A. (2021). Zika virus infection in pregnant women and their children: A review. Eur. J. Obstet. Gynecol. Reprod. Biol. 265, 162–168. doi: 10.1016/J.EJOGRB.2021.07.012
Martelli, F., Macera, L., Spezia, P. G., Medici, C., Pistello, M., Guasti, D., et al. (2018). Torquetenovirus detection in exosomes enriched vesicles circulating in human plasma samples 11 medical and health sciences 1108 medical microbiology. Virol. J. 15:145. doi: 10.1186/S12985-018-1055-Y
Martins, T. S., Catita, J., Rosa, I. M., A. B Da Cruz E Silva, O., and Henriques, A. G. (2018). Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One 13:e0198820. doi: 10.1371/JOURNAL.PONE.0198820
Mashouri, L., Yousefi, H., Reza Aref, A., Mohammad Ahadi, A., Molaei, F., and Alahari, S. K. (2019). Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 18:75. doi: 10.1186/s12943-019-0991-5
Meningher, T., Lerman, G., Regev-Rudzki, N., Gold, D., Ben-Dov, I. Z., Sidi, Y., et al. (2017). Schistosomal MicroRNAs isolated from extracellular vesicles in sera of infected patients: A new tool for diagnosis and follow-up of human schistosomiasis. J. Infect. Dis. 215, 378–386. doi: 10.1093/INFDIS/JIW539
Moreira, J., Bressan, C. S., Brasil, P., and Siqueira, A. M. (2018). Epidemiology of acute febrile illness in Latin America. Clin. Microbiol. Infect. 24, 827–835. doi: 10.1016/J.CMI.2018.05.001
Muinelo-Romay, L., Casas-Arozamena, C., and Abal, M. (2018). Liquid biopsy in endometrial cancer: New opportunities for personalized oncology. Int. J. Mol. Sci. 19:2311. doi: 10.3390/IJMS19082311
Murugesan, A., and Manoharan, M. (2019). Dengue virus. Emerg. Reemerg. Viral Pathog. 2020, 281–359. doi: 10.1016/B978-0-12-819400-3.00016-8
Nabet, C., Chaline, A., Franetich, J. F., Brossas, J. Y., Shahmirian, N., Silvie, O., et al. (2020). Prediction of malaria transmission drivers in Anopheles mosquitoes using artificial intelligence coupled to MALDI-TOF mass spectrometry. Sci. Rep. 10:11379. doi: 10.1038/S41598-020-68272-Z
Nantakomol, D., Dondorp, A. M., Krudsood, S., Udomsangpetch, R., Pattanapanyasat, K., Combes, V., et al. (2011). Circulating red cell-derived microparticles in human malaria. J. Infect. Dis. 203, 700–706. doi: 10.1093/INFDIS/JIQ104
Nawaz, M., Malik, M. I., Hameed, M., and Zhou, J. (2019). Research progress on the composition and function of parasite-derived exosomes. Acta Trop. 196, 30–36. doi: 10.1016/J.ACTATROPICA.2019.05.004
Nawaz, M., Malik, M. I., Zhang, H., Hassan, I. A., Cao, J., Zhou, Y., et al. (2020). Proteomic analysis of exosome-like vesicles isolated from saliva of the tick Haemaphysalis longicornis. Front. Cell Infect. Microbiol. 10:585. doi: 10.3389/FCIMB.2020.542319/BIBTEX
Noonin, C., and Thongboonkerd, V. (2021). Exosome-inflammasome crosstalk and their roles in inflammatory responses. Theranostics 11, 4436–4451. doi: 10.7150/THNO.54004
O’Connell, R. M., Rao, D. S., and Baltimore, D. (2012). microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312. doi: 10.1146/ANNUREV-IMMUNOL-020711-075013
O’Loughlin, A., Woffindale, C., and Wood, M. (2012). Exosomes and the emerging field of exosome-based gene therapy. Curr. Gene Ther. 12, 262–274. doi: 10.2174/156652312802083594
Oliva Chávez, A. S., Wang, X., Marnin, L., Archer, N. K., Hammond, H. L., Carroll, E. E., et al. (2021). Tick extracellular vesicles enable arthropod feeding and promote distinct outcomes of bacterial infection. Nat. Commun. 12:3696. doi: 10.1038/S41467-021-23900-8
Opadokun, T., and Rohrbach, P. (2021). Extracellular vesicles in malaria: An agglomeration of two decades of research. Malar. J. 20:442. doi: 10.1186/S12936-021-03969-8
Ouyang, X., Jiang, X., Gu, D., Zhang, Y., Kong, S. K., Jiang, C., et al. (2016). Dysregulated serum MiRNA profile and promising biomarkers in dengue-infected patients. Int. J. Med. Sci. 13, 195–205. doi: 10.7150/IJMS.13996
Parashar, D., Paingankar, M. S., More, A., Patil, P., and Amdekar, S. (2018). Altered microRNA expression signature in Chikungunya-infected mammalian fibroblast cells. Virus Genes 54, 502–513. doi: 10.1007/S11262-018-1578-8
Parra, M. C. P., Lorenz, C., de Aguiar Milhim, B. H. G., Dibo, M. R., Guirado, M. M., Chiaravalloti-Neto, F., et al. (2022). Detection of Zika RNA virus in Aedes aegypti and Aedes albopictus mosquitoes, São Paulo, Brazil. Infect. Genet. Evol. 98:105226. doi: 10.1016/J.MEEGID.2022.105226
Peng, R. R., Shang, S. X., Zhao, L. S., and Long, F. Q. (2019). MiR-216a-5p-containing exosomes suppress rTp17-induced inflammatory response by targeting TLR4. Biosci. Rep. 39:BSR20190686. doi: 10.1042/BSR20190686
Podolska, M., and Nadolna, K. (2014). Acetylcholinesterase secreted by Anisakis simplex larvae (Nematoda: Anisakidae) parasitizing herring, Clupea harengus: An inverse relationship of enzyme activity in the host-parasite system. Parasitol. Res. 113, 2231–2238. doi: 10.1007/S00436-014-3878-9
Qi, Y., Ai, L., Jiao, J., Wang, J., Wu, D., Wang, P., et al. (2022). High prevalence of Rickettsia spp. in ticks from wild hedgehogs rather than domestic bovine in Jiangsu province, Eastern China. Front. Cell. Infect. Microbiol. 12:954785. doi: 10.3389/fcimb.2022.954785
Rassi, A., Rassi, A., and Marin-Neto, J. A. (2010). Chagas disease. Lancet 375, 1388–1402. doi: 10.1016/S0140-6736(10)60061-X
Regev-Rudzki, N., Wilson, D. W., Carvalho, T. G., Sisquella, X., Coleman, B. M., Rug, M., et al. (2013). Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 153, 1120–1133. doi: 10.1016/J.CELL.2013.04.029
Reyes-Ruiz, J. M., Osuna-Ramos, J. F., De Jesús-González, L. A., Hurtado-Monzón, A. M., Farfan-Morales, C. N., Cervantes-Salazar, M., et al. (2019). Isolation and characterization of exosomes released from mosquito cells infected with dengue virus. Virus Res. 266, 1–14. doi: 10.1016/J.VIRUSRES.2019.03.015
Roberts, A., and Gandhi, S. (2020). Japanese encephalitis virus: A review on emerging diagnostic techniques. Front. Biosci. 25:1875–1893. doi: 10.2741/4882
Saadatpour, L., Fadaee, E., Fadaei, S., Nassiri Mansour, R., Mohammadi, M., Mousavi, S. M., et al. (2016). Glioblastoma: Exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 23, 415–418. doi: 10.1038/CGT.2016.48
Saini, J., Bandyopadhyay, B., Pandey, A. D., Ramachandran, V. G., Das, S., Sood, V., et al. (2020). High-throughput RNA sequencing analysis of plasma samples reveals circulating microRNA signatures with biomarker potential in dengue disease progression. mSystems 5, e00724–20. doi: 10.1128/MSYSTEMS.00724-20
Santangelo, L., Bordoni, V., Montaldo, C., Cimini, E., Zingoni, A., Battistelli, C., et al. (2018). Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int. 38, 1741–1750. doi: 10.1111/LIV.13700
Schnitzer, J. K., Berzel, S., Fajardo-Moser, M., Remer, K. A., and Moll, H. (2010). Fragments of antigen-loaded dendritic cells (DC) and DC-derived exosomes induce protective immunity against Leishmania major. Vaccine 28, 5785–5793. doi: 10.1016/j.vaccine.2010.06.077
Schorey, J. S., Cheng, Y., Singh, P. P., and Smith, V. L. (2015). Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 16, 24–43. doi: 10.15252/EMBR.201439363
Schwartz, O., and Albert, M. L. (2010). Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8, 491–500. doi: 10.1038/NRMICRO2368
Serrano-Heras, G., Díaz-Maroto, I., Castro-Robles, B., Carrión, B., Perona-Moratalla, A. B., Gracia, J., et al. (2020). Isolation and quantification of blood apoptotic bodies, a non-invasive tool to evaluate apoptosis in patients with ischemic stroke and neurodegenerative diseases. Biol. Proced. Online 22, 1–17. doi: 10.1186/S12575-020-00130-8/FIGURES/6
Sharma, K. B., Vrati, S., and Kalia, M. (2021). Pathobiology of Japanese encephalitis virus infection. Mol. Aspects Med. 81:100994. doi: 10.1016/J.MAM.2021.100994
Soe, H. J., Yong, Y. K., Al-Obaidi, M. M. J., Raju, C. S., Gudimella, R., Manikam, R., et al. (2018). Identifying protein biomarkers in predicting disease severity of dengue virus infection using immune-related protein microarray. Medicine 97:e9713. doi: 10.1097/MD.0000000000009713
Statello, L., Maugeri, M., Garre, E., Nawaz, M., Wahlgren, J., Papadimitriou, A., et al. (2018). Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One 13:e0195969. doi: 10.1371/JOURNAL.PONE.0195969
Stevens, L., Lima-Cordón, R. A., Helms Cahan, S., Dorn, P. L., Monroy, M. C., Axen, H. J., et al. (2021). Catch me if you can: Under-detection of Trypanosoma cruzi (Kinetoplastea: Trypanosomatida) infections in Triatoma dimidiata s.l. (Hemiptera: Reduviidae) from Central America. Acta Trop. 224:106130. doi: 10.1016/J.ACTATROPICA.2021.106130
Sung, P. S., Huang, T. F., and Hsieh, S. L. (2019). Extracellular vesicles from CLEC2-activated platelets enhance dengue virus-induced lethality via CLEC5A/TLR2. Nat. Commun. 10:2402. doi: 10.1038/S41467-019-10360-4
Takahasi, K., Iinuma, H., Wada, K., Minezaki, S., Kawamura, S., Kainuma, M., et al. (2018). Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J. Hepatobiliary Pancreat. Sci. 25, 155–161. doi: 10.1002/JHBP.524
Vora, A., Zhou, W., Londono-renteria, B., Woodson, M., and Sherman, M. B. (2018). Arthropod EVs mediate dengue virus transmission through interaction with a tetraspanin domain containing glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. U.S.A. 115, E6604–E6613. doi: 10.1073/pnas.1720125115
Wang, J., Yan, F., Zhao, Q., Zhan, F., Wang, R., Wang, L., et al. (2017). Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 7:4150. doi: 10.1038/S41598-017-04386-1
Wijerathna, T., Gunathilaka, N., Gunawardena, K., Fujii, Y., and Gunasekara, D. (2021). Detection of Leishmania donovani DNA within field-caught phlebotomine sand flies (Diptera: Psychodidae) in three cutaneous Leishmaniasis endemic foci of Kurunegala district, Sri Lanka. J. Trop. Med. 2021:6650388. doi: 10.1155/2021/6650388
World Health Organization (2020a). WHO visceral-leishmaniasis Report. Geneva: World Health Organization.
World Health Organization (2020b). WHO world Japanese encephalitis report 2020. Geneva: World Health Organization.
World Health Organization (2022c). Laboratory testing for Zika virus and dengue virus infections. Geneva: World Health Organization, 1–14.
Wu, L. F., Zhang, Q., Mo, X. B., Lin, J., Wu, Y. L., Lu, X., et al. (2022). Identification of novel rheumatoid arthritis-associated MiRNA-204-5p from plasma exosomes. Exp. Mol. Med. 54, 334–345. doi: 10.1038/S12276-022-00751-X
Wu, P., Zhang, B., Ocansey, D. K. W., Xu, W., and Qian, H. (2021). Extracellular vesicles: A bright star of nanomedicine. Biomaterials 269:120467. doi: 10.1016/J.BIOMATERIALS.2020.120467
Wu, Y. W., Mettling, C., Wu, S. R., Yu, C. Y., Perng, G. C., Lin, Y. S., et al. (2016). Autophagy-associated dengue vesicles promote viral transmission avoiding antibody neutralization. Sci. Rep. 6:32243. doi: 10.1038/SREP32243
Wu, Z., Sun, X., Wang, L., Gao, J., and Yu, Z. (2020). Inventors; Univ Sun Yat-Sen, Assignee. Use of serum exosome microRNA (miR)-142a-3p as diagnostic marker for Schistosoma japonicum infection patent CN110760589-a (2020), Vol. 147. 2934–2947.
Xie, L. M., Yin, X., Bi, J., Luo, H. M., Cao, X. J., Ma, Y. W., et al. (2021). Identification of potential biomarkers in dengue via integrated bioinformatic analysis. PLoS Negl. Trop. Dis. 15:e0009633. doi: 10.1371/JOURNAL.PNTD.0009633
Xie, Y., Yang, L., Cao, P., Li, S., Zhang, W., Dang, W., et al. (2022). Plasma exosomal proteomic pattern of epstein-barr virus-associated hemophagocytic lymphohistiocytosis. Front. Microbiol. 13:821311. doi: 10.3389/FMICB.2022.821311
Xue, P., Huang, S., Han, X., Zhang, C., Yang, L., Xiao, W., et al. (2022). Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 29, 82–95. doi: 10.1038/S41418-021-00838-4
Yáñez-Mó, M., R-M Siljander, P., Andreu, Z., Bedina Zavec, A., Borràs, F. E., Buzas, E. I., et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4:27066. doi: 10.3402/jev.v4.27066
Yuan, Y., Zhao, J., Chen, M., Liang, H., Long, X., Zhang, B., et al. (2021). Understanding the pathophysiology of exosomes in schistosomiasis: a new direction for disease control and prevention. Front. Immunol. 12:634138. doi: 10.3389/FIMMU.2021.634138
Zamani, S., Taslimi, R., Sarabi, A., Jasemi, S., Sechi, L. A., and Feizabadi, M. M. (2020). Enterotoxigenic Bacteroides fragilis: A possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front. Cell Infect. Microbiol. 9:449. doi: 10.3389/FCIMB.2019.00449
Zamanian, M., Fraser, L. M., Agbedanu, P. N., Harischandra, H., Moorhead, A. R., Day, T. A., et al. (2015). Release of small RNA-containing exosome-like vesicles from the human filarial parasite Brugia malayi. PLoS Negl. Trop. Dis. 9:e0004069. doi: 10.1371/journal.pntd.0004069
Zhao, X., Jia, Y., Chen, H., Yao, H., and Guo, W. (2019). Plasma-derived exosomal miR-183 associates with protein kinase activity and may serve as a novel predictive biomarker of myocardial ischemic injury. Exp. Ther. Med. 18, 179–187. doi: 10.3892/ETM.2019.7555
Keywords: vector-borne diseases, exosomes, plasma, miRNA—microRNA, biomarker
Citation: Venkatesan G, Wan Ab Rahman WS, Shahidan WNS, Iberahim S and Muhd Besari@Hashim Ab (2023) Plasma-derived exosomal miRNA as potential biomarker for diagnosis and prognosis of vector-borne diseases: A review. Front. Microbiol. 14:1097173. doi: 10.3389/fmicb.2023.1097173
Received: 13 November 2022; Accepted: 21 March 2023;
Published: 13 April 2023.
Edited by:
Siddhartha Pati, NatNov Bioscience Pvt. Ltd., IndiaReviewed by:
Marcel I. Ramirez, Carlos Chagas Institute (ICC)/(FIOCRUZ), BrazilCopyright © 2023 Venkatesan, Wan Ab Rahman, Shahidan, Iberahim and Muhd Besari@Hashim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan Suriana Wan Ab Rahman, c3VyaWFuYUB1c20ubXk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.