- Department of Biology, College of Arts and Sciences, Emory University, Atlanta, GA, United States
Streptomyces are soil dwelling bacteria that are notable for their ability to sporulate and to produce antibiotics and other secondary metabolites. Antibiotic biosynthesis is controlled by a variety of complex regulatory networks, involving activators, repressors, signaling molecules and other regulatory elements. One group of enzymes that affects antibiotic synthesis in Streptomyces is the ribonucleases. In this review, the function of five ribonucleases, RNase E, RNase J, polynucleotide phosphorylase, RNase III and oligoribonuclease, and their impact on antibiotic production will be discussed. Mechanisms for the effects of RNase action on antibiotic synthesis are proposed.
Introduction
Members of the genus Streptomyces are Gram-positive, soil dwelling bacteria notable for their ability to undergo morphological differentiation (sporulation) and for their production of antibiotics (Chater, 2016; Hopwood, 2019). Both sporulation and antibiotic biosynthesis are tightly and elegantly regulated. A recent review has posited four levels of the regulatory cascade that controls Streptomyces antibiotic synthesis: (1) signals that initiate antibiotic synthesis; (2) global regulators of antibiotic synthesis; (2) cluster-situated regulators CSRs, formerly referred to as pathway-specific regulators [PSRs] or SARPs [Streptomyces antibiotic regulatory proteins]; (4) feedback inhibition (Xia et al., 2020). These various levels of regulation involve the participation of signaling molecules such as the γ-butyrolactones and (p)ppGpp, activator and repressor proteins, two-component regulatory systems, RNA polymerase sigma factors and anti-sigma factors, regulatory RNAs and other regulatory molecules (Bibb, 1996; Liu et al., 2013; Niu et al., 2016).
One group of enzymes that affects antibiotic biosynthesis in Streptomyces is the RNases, enzymes that degrade RNAs either exo- or endonucleolytically (Jones, 2010). This review will focus on the roles of five specific RNases, their function in Streptomyces and their roles in the control of antibiotic production, viz. RNase E, RNase J, polynucleotide phosphorylase, RNase III and oligoribonuclease. Mechanisms to explain the effects of these enzymes on antibiotic production in Streptomyces are proposed. This subject was last reviewed in 2010 (Jones, 2010) and the current review will focus on experimental results obtained since that date and on their interpretation. In some cases, the analysis of more recent experiments will require referencing studies published prior to 2010.
RNase E
RNase E is a single-strand specific endoribonuclease that is widely distributed in bacteria (Cohen and Mcdowall, 1997). RNase E from Escherichia coli is a 1,061 amino acid protein (Casaregola et al., 1992) that is organized as two domains, an N-terminal catalytic domain (NTH) and a C-terminal scaffold domain (CTH) (Mcdowall and Cohen, 1996; Mackie, 2013). The crystal structures of the RNase E catalytic domain (Callaghan et al., 2005) and of the RNase E apoprotein (Koslover et al., 2008) have been determined. These structural studies showed that the catalytic domain is comprised of several subdomains that are involved in catalysis, RNA binding and sensing the 5′-end of target RNA molecules (Koslover et al., 2008). The C-terminal scaffold domain of RNase E binds polynucleotide phosphorylase (PNPase), the RNA helicase, RhlB, and enolase (Py et al., 1996), to form the RNA-degrading complex known as the degradosome (Vanzo et al., 1998). RNase E is tethered to the cell inner membrane in E. coli (Khemici et al., 2008).
Hagege and Cohen demonstrated the presence of an RNase E-like activity in the model species for the study of streptomycetes, Streptomyces coelicolor (Hagege and Cohen, 1997). Lee and Cohen identified the gene encoding the enzyme responsible for that activity, sco2599 (rns) and overexpressed and characterized the gene product, designating it RNase ES (Lee and Cohen, 2003). S. coelicolor RNase ES is even larger that RNase E with 1,340 amino acids (Lee and Cohen, 2003). The RNase E and RNase ES catalytic domains are 33% identical and 50% similar in amino acid sequence. Key residues involved in catalysis at the active site, in sensing the 5′-end of target RNA molecules and in Zn ion binding (Callaghan et al., 2004; Koslover et al., 2008) are strongly conserved between the two enzymes (Table 1).
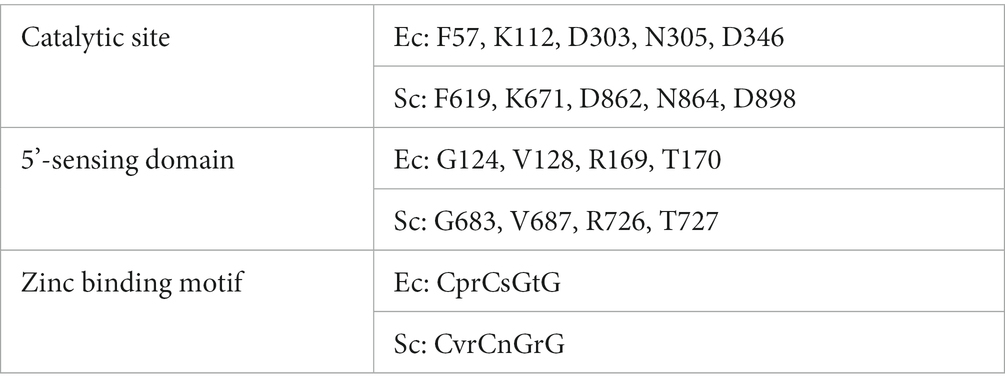
Table 1. Conserved amino acids in the domains of E. coli (Ec) RNase E and S. coelicolor (Sc) RNase ES.
Unlike RNase E, the catalytic domain of RNase ES is located near the center rather than at the N-terminus of the enzyme (Lee and Cohen, 2003). Lee and Cohen provided evidence indicating that regions near the N- and C-termini of RNase ES interact with polynucleotide phosphorylase. Western blots of immunoprecipitates obtained by treating mycelial extracts from S. coelicolor with antibody to RNase ES revealed a co-precipitated protein that reacted with antibody to PNPase from Streptomyces antibioticus (Jones, 1994). Western blotting experiments utilizing truncated forms of RNase ES, lacking ca. 800 residues from the N-terminal end or ca. 400 residues from the C-terminal end of full length RNase ES, indicated that sequences interacting with PNPase were present in both the N- and C-terminal regions of RNase ES. Lee and Cohen argued that the N- and C-terminal regions of RNase ES are functionally equivalent to the C-terminal scaffold domain of RNase E. The authors further speculated that S. coelicolor might contain a macromolecular complex similar to the degradosome (Lee and Cohen, 2003), but no additional evidence supporting this suggestion has been adduced. It should also be noted that a subsequent study, Yeom et al. were unable to demonstrate the interaction of E. coli PNPase with RNase ES (Yeom et al., 2008). The structural basis for the discrimination by RNase ES between E. coli and S. antibioticus PNPases remains to be determined.
RNase ES activity increased as S. coelicolor mycelium progressed from exponential growth to stationary phase in liquid cultures and from mycelial growth to spore formation on solid media (Hagege and Cohen, 1997). RNase E is essential in E. coli (Apirion and Lassar, 1978) but the corresponding gene (sco2599, rns) could be disrupted in S. coelicolor with only minimal impact on the physiology of the null mutant (Lee and Cohen, 2003). Lee and Cohen noted only a 10% decrease in the growth rate of the rns null mutant as compared with the parental strain.
The effects of the rns null mutation on the transcriptome of S. coelicolor have not been examined, thus not much is known about the role of RNase ES in RNA processing and degradation. In in vitro studies, Hagege and Cohen and Lee and Cohen demonstrated RNase ES cleavage of RNA I, an antisense repressor of the replication of ColE1-type plasmids in E. coli (Hagege and Cohen, 1997; Lee and Cohen, 2003). RNase ES cleaved that RNA at the same site as did RNase E. RNase E and ES also cleaved the E. coli 9S RNA precursor of 5S ribosomal RNA. However, the cleavage sites recognized by the two enzymes were not identical. RNase ES correctly processed the E. coli pM1 RNA, the precursor of the RNA component of RNase P (Inagawa et al., 2003).
Perhaps the most significant observation regarding RNase ES function is that the enzyme can substitute for RNase E in an E. coli rne (RNase E) null mutant. Lee and Cohen demonstrated that the rne null mutant when complemented by rns grew at rates that were comparable to the wild type strain. They showed further that the truncated RNase ES derivatives described above complemented the rne null mutation, since those derivatives contained the RNase ES catalytic domain (Lee and Cohen, 2003). Inagawa et al. confirmed the observation that rns complemented the rne null mutation and demonstrated further that both the 9 s rRNA precursor and the pM1 RNA were processed normally in the complemented mutant. These authors also showed that rns complemented a null mutation in the gene for RNase G, a paralog of RNase E (Inagawa et al., 2003). Taken together these observations suggest that RNase E and RNase ES play similar roles in their respective hosts. It is likely, therefore, that as with RNase E (Mackie, 2013), RNase ES is involved in mRNA degradation, ribosomal RNA processing, tRNA processing and the processing of small regulatory RNAs, among other likely functions. S. coelicolor contains RNase P (Kim et al., 2000) so it is possible that RNase ES is involved in processing the RNA component of that ribozyme.
The effects of the rns null mutation on antibiotic production by S. coelicolor were not examined in the studies of the Cohen group. Comparative studies in E. coli and S. coelicolor may bear on that issue. Lee et al. and Gao et al. characterized two proteins that modulate the activity of RNase E in E. coli (Lee et al., 2003; Gao et al., 2006). These proteins were designated RraA and RraB (Rra = Regulator of ribonuclease activity). RraA and RraB inhibit the activity of RNase E in vivo and in vitro, albeit by different mechanisms. RraA inhibits the enzyme by interacting with the NTH catalytic domain. Although inhibition of the catalytic activity of the isolated NTH by RraA could be demonstrated, that inhibition was enhanced by the presence of the scaffold domain (Lee et al., 2003; Gao et al., 2006). RraB binds to the RNase E scaffold domain and Gao et al. identified a specific region of the CTH that is essential for RraB binding (Gao et al., 2006). Both RraA and RraB inhibited the processing of the RNase P pM1 RNA in vivo and in vitro, although to different extents (Gao et al., 2006). An interesting property of both RraA and RraB is their ability to remodel the E. coli degradosome, changing the ratios of RNase E, PNPase, RhlB and enolase in that complex (Gao et al., 2006).
Yeom et al. (2008) obtained the interesting result that RraA and RraB from E. coli can inhibit the activity of RNase ES. This observation suggested that S. coelicolor might also contain proteins capable of regulating the activity of RNase ES in trans. Two such proteins have been found to date and were designated RraAS1 and RraAS2 (Ahn et al., 2008). RraAS1 is encoded by sco5940 and RraAS2 by sco7163. The two proteins are ca. 25 and 17% identical to RraA, respectively, and are ca. 35 and 27% similar to RraA. They are less identical and similar to RraB. RraAS1 and S2 are also ca. 60 amino acids longer than RraA and B.
Heo et al. demonstrated that an E. coli rne null mutant expressing rns instead grew more slowly when RraAS2 was overexpressed in the mutant strain than in the absence of RraAS2 overexpression (Heo et al., 2016). Moreover, RraAS2 inhibited RNase ES cleavage of a truncated form of the pM1 RNA in vitro (Heo et al., 2016). Thus, as with RraA and B, RraAS2 is an inhibitor of RNase ES activity. Immunoprecipitation experiments, using extracts of E. coli cells expressing RraAS2 showed that, unlike RraA and B, RraAS2 did not affect the ratios of the enzymes contained in the E. coli degradosome (Yeom et al., 2008). Instead, RraAS2 decreased the affinity of RNase ES for its RNA substrates. Immunoprecipitation experiments demonstrated that RraAS2 binds directly to RNase ES and that this binding requires the N- and C- terminal scaffold domains of RNase ES. Truncated RNase ES derivatives lacking those domains did not bind RraAS2 (Heo et al., 2016).
Seo et al. (2017) studied the function of RraAS1 and its role in the physiology of S. coelicolor. RraAS1 overexpression increased the copy number of a ColE1-type plasmid in the E. coli rne null mutant expressing rns, presumably reflecting the inhibition of RNase ES by RraS1 thereby decreasing the levels of RNA I. Moreover, RraAS1 inhibited the RNase ES-catalyzed in vitro cleavage of a model substrate, a synthetic RNA containing the E. coli RNA I cleavage site recognized by RNase E. As with RraAS2, RraAS1 co-precipitated with RNase ES when extracts of E. coli cells expressing both proteins were treated with antibody to Myc-tagged RraAS1. Unlike the situation with RraAS2, truncated derivatives of RraAS1 lacking the N- and/or C-terminal scaffold domains were as functional as the full-length protein in inhibiting RNase ES activity in vivo and in vitro. Moreover, truncated RNase ES derivatives, lacking the N- and/or C-terminal scaffold domains, co-immunoprecipitated with RraAS1. These observations indicate that while RraAS2 interacts with the N- and C-terminal scaffold domains of RNase ES, RraAS1 interacts with the catalytic domain (Seo et al., 2017).
What then is the connection between these analyses and antibiotic production by S. coelicolor? On laboratory growth media, wild type S. coelicolor produces four antibiotics, two pigmented ones, actinorhodin (act) (Cole et al., 1987) and undecylprodigiosin (red) (Feitelson et al., 1985) and two unpigmented ones, calcium dependent antibiotic (cda) (Hopwood and Wright, 1983) and methylenomycin (mmy) (Wright and Hopwood, 1976). Seo et al. (2017) constructed an rraAS1 null mutant of S. coelicolor and examined the effects of the null mutation on the physiology of the organism. They observed a slightly increased growth rate of the mutant as compared with the parental strain. They also observed that both act and red were overproduced in the rraAS1 null mutant. Overproduction began during vegetative growth of the mutant strain and continued for several days post-inoculation (Figure 1). At their maxima, red was produced at a level ca. twice that of the parental strain and act at almost four times the level observed for the parental strain (Figure 1). The authors examined the transcripts for two of the CSRs involved in act and red production, viz. actII-orf4 and redZ. The levels of those transcripts were essentially identical in the rraAS1 null mutant and the parental strain (Seo et al., 2017). It should be noted that the authors only examined CSR transcript levels at a single time point, mid-log phase. It is possible that levels for these transcripts did vary at other times during the growth of the organisms. Taken at face value, though, the results suggest that RraAS1 exerts its effect on act and red production at a level other than the CSRs. That level might be RNase ES.
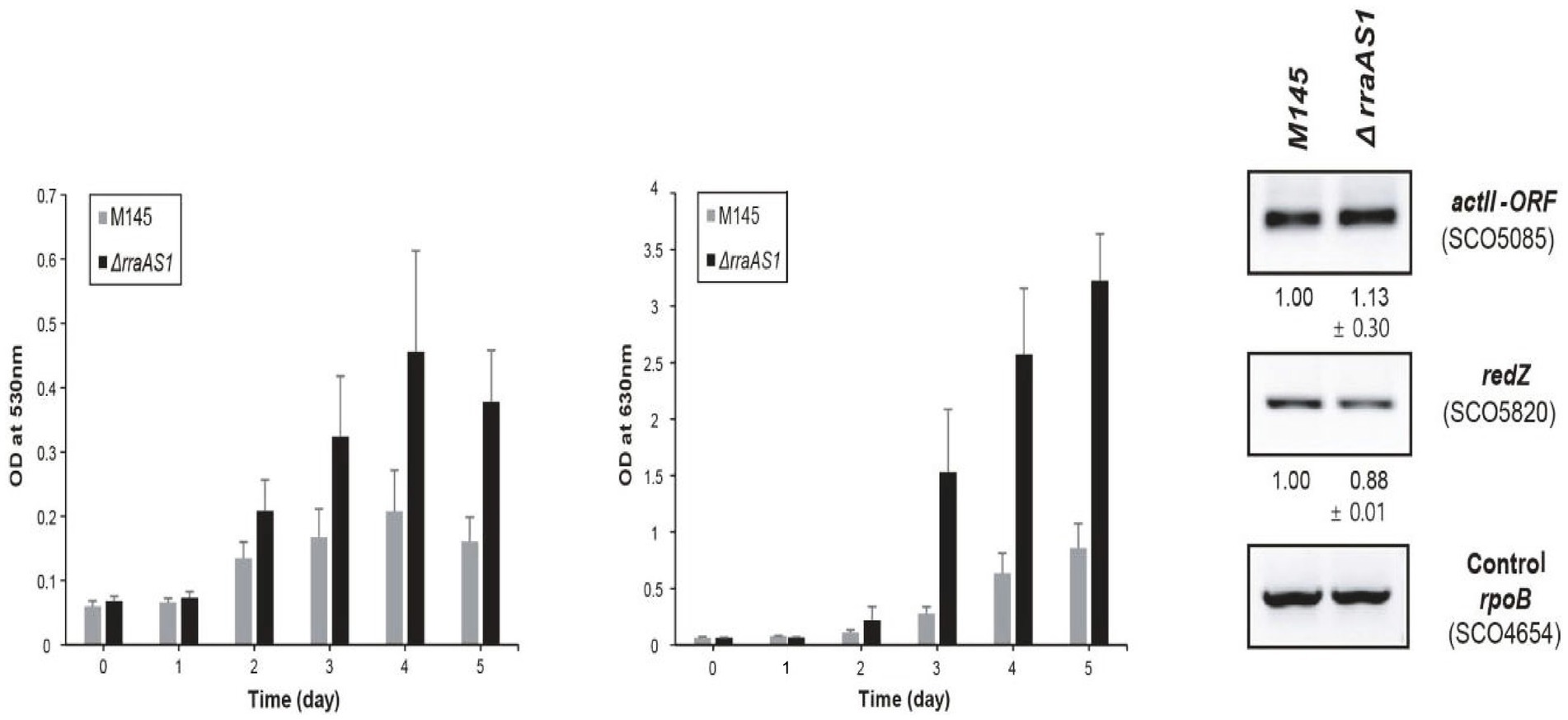
Figure 1. Overproduction of red (right bar graph) and act (left bar graph) by an rraAS1 null mutant of S. coelicolor. The antibiotics were measured using spectroscopic methods from mycelium harvested at the times indicated on the x-axis of the figures. The antibiotic concentrations were normalized relative to mycelial dry weights. The rightmost part of the figure shows the results of an RT-PCR analysis of the levels of the CSRs, actII-orf4 and redZ in the parental and rraAS1 null mutant strains of S. coelicolor. Reprinted with permission from Seo et al. (2017).
RNase J
“The lack of a bacterial 5′-to-3′-exonuclease was the accepted dogma, even for years after bacterial genomes sequences began to appear in the mid-1990s (Bechhofer and Deutscher, 2019).” That dogma was shown to be erroneous by the discovery in 2007 of two RNases (RNase J1 and J2) with 5′-3′-exonuclease activity in Bacillus subtilis (Mathy et al., 2007). These two enzymes were first identified in B. subtilis in 2005 (Even et al., 2005), but their exonuclease activities were not demonstrated until 2007 (Mathy et al., 2007). RNase J1 and J2 are paralogs, and they not only possess 5′-3′-exonuclease activity, they are bifunctional enzymes possessing endonuclease activities as well (Even et al., 2005; Mathy et al., 2007). RNase J1 has been shown to function in mRNA turnover, rRNA processing and in the processing of small RNAs in B. subtilis (Condon, 2010; Bechhofer and Deutscher, 2019). The role of RNase J2 remains somewhat controversial, at least in B. subtilis, since an rnjB (RNase J2) null mutant grew normally under laboratory conditions (Mader et al., 2008), while an rnjA (RNase J1) null mutant grew poorly (Figaro et al., 2013). RNase J1 and J2 can form heteromeric complexes in B. subtilis but the biological significance of this association remains unclear (Newman et al., 2011).
Since the discovery of RNases J1 and J2 in B. subtilis, orthologs have been identified in a number of other organisms, including Gram-negative bacteria, Archaea, and plant chloroplasts (Even et al., 2005). RNases J1 and J2 have been shown to be involved in a variety of biological processes in addition to RNA degradation and processing, including the conferral of multidrug resistance (Martini et al., 2022), controlling plasmid copy number (Guimaraes et al., 2021) and the modulation of cell morphology, primary metabolism and virulence (Luong et al., 2021).
RNase J was identified in S. coelicolor by Bralley et al. (2014a). Unlike B. subtilis (but like several other systems), S. coelicolor contains only one RNase J ortholog. S. coelicolor RNase J is ca. 38% identical and 60% similar to RNases J1 and J2 from B. subtilis. S. coelicolor RNase J was overexpressed and purified and its activities were examined using a model substrate, the thrS RNA, derived from the leader region of the threonyl-tRNA synthetase gene of B. subtilis (Condon et al., 1996). The thrS RNA was chosen to allow comparison of the results obtained with S. coelicolor RNase J with those of other studies using thrS RNA (Even et al., 2005). Bralley et al. (2014a) demonstrated that S. coelicolor RNase J possessed both 5′-3′-exonuclease and endonuclease activities. As was observed in studies with other RNases J, the S. coelicolor enzyme preferred a 5′-monophosphorylated substrate to a 5′-triphosphorylated substrate. Thus, the S. coelicolor enzyme did not produce GTP when challenged with 5′-triphosphorylated thrS RNA with G as the 5’terminal base and instead generated a series of oligonucleotides 2–10 residues in length. In studies with RNase J from Mycobacterium smegmatis, Taverniti et al. (2011) showed that the enzyme did produce GTP from the 5′-triphosphorylated thrS substrate. These differences relate to the mechanisms proposed for the exo- and endoribonuclease activities of RNase J and will be considered in detail below.
The crystal structure of S. coelicolor RNase J was determined by Pei et al. (2015) and consideration of that structure suggests a mechanism for exonuclease cleavage and for the preference for monophosphorylated substrates. In the model proposed by Pei et al. to describe substrate interactions for exo- and endonuclease cleavages by S. coelicolor RNase J, an RNA substrate is bound at the catalytic site adjacent to which is a binding pocket which can accommodate a 5′-monophosphorylated end. Binding of that 5′-end then leads to exonuclease cleavage of the substrate at the active site, producing a mononucleotide and a new 5′-monophosphorylated end that can be further processed by the enzyme. A triphosphorylated 5′-end cannot be accommodated by the binding pocket, the RNA substrate slides through the active site in consequence, and oligonucleotides are produced by endonuclease cleavage (Pei et al., 2015). This model may also explain the results described by Taverniti et al. (2011), who suggested that the substrate binding pocket of M. smegmatis RNase J was unable to accommodate a 5′-triphosphorylated substrate, that the substrate RNA slid past the active site and that GTP was released by an endonucleolytic cleavage. If this model is correct, it is interesting that M. smegmatis RNase J could release the 5’-GTP, but S. coelicolor RNase J could not.
RNase J is not essential in S. coelicolor and Bralley et al. (2014a). were able to construct an rnj null mutant. The authors reported no obvious differences in the growth of the null mutant on solid or liquid media as compared with the parental strain nor in its ability to sporulate. Bralley et al. observed that the rnj null mutant showed significantly altered capacities to produce antibiotics as compared with the parent. In particular, act production was delayed compared to the parental strain (Figure 2). Act was observed after 3 days of growth on production medium in the parent but no act was observed at that time point in the null mutant. Production of cda was significantly diminished in the null mutant as compared with the parental strain (Figure 2). The rnj null mutation led to substantial overproduction of the red antibiotic (Bralley et al., 2014a). The authors did not examine the effects of the mutation on the CSRs that are involved in regulating production of act, red and cda.
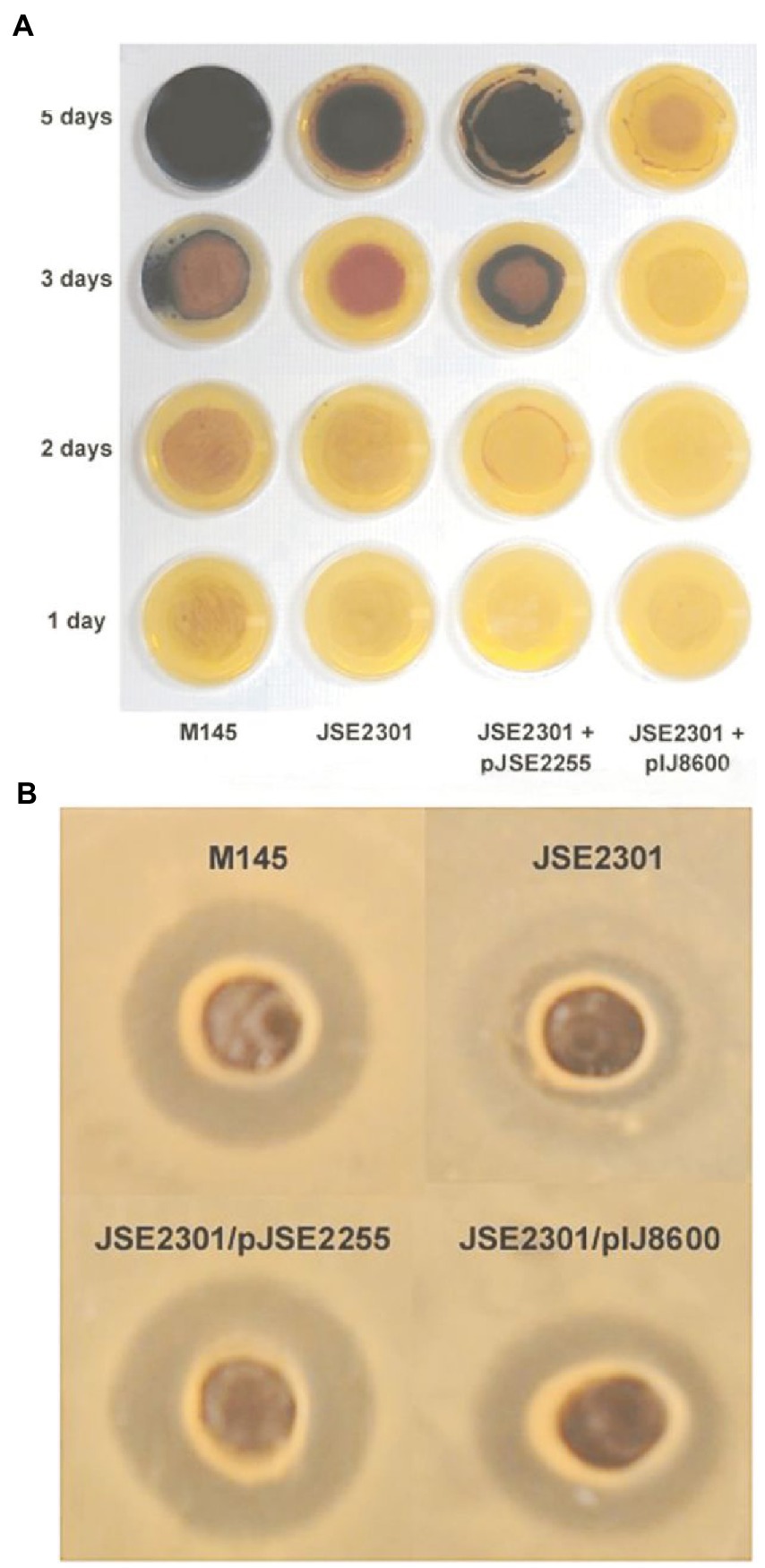
Figure 2. Effects of an rnj null mutation on antibiotic production in S. coelicolor. The top panel (A) shows the temporal progress of act and red production in the parental (M145) and mutant (JSE2301) strains cultured on solid medium that supports antibiotic biosynthesis. pJSE2255 is a plasmid containing the wild type RNase J gene cloned into the streptomycete expression vector, pIJ8600. In the rnj null mutant, act production is suppressed while red is overproduced. The bottom panel (B) shows the levels of cda production in the wild type and parental S. coelicolor strains. The figure shows zones of inhibition produced by cda action against a cda-sensitive bacterial strain, B. subtilis BG267. Reprinted with permission from Pei et al. (2015).
Jones et al. (2014) examined that role of RNase J in another streptomycete, Streptomyces venezuelae. As with S. coelicolor, rnj is not essential in S. venezuelae but unlike S. coelicolor, the rnj null mutation affected strain morphology. Jones et al. observed several sporulation defects in the rnj null mutant. They reported that mutant spores were unpigmented while wild types spores were associated with a green pigment. In liquid medium, Jones et al. found that the onset of sporulation was significantly delayed as compared with the wild type, that fewer spore chains were formed by sporulating cultures of the rnj mutant as compared with the wild type strain and that the mutant spores were shorter and more sensitive to heat than wild type. Jones et al. examined ribosome profiles in the rnj null mutant and reported that the mutant ribosome population contained a significant fraction of ribosome dimers, sedimenting at ca. 100S in sucrose gradients. The authors did not examine the ability of the rnj mutant to synthesize proteins in vivo or in vitro, either in terms of overall amino acid incorporation or by looking at the synthesis of specific gene products.
Jones et al. found that the rnj mutation led to changes in the antibiotic sensitivity of mutant strains, leading to increased sensitivity to some antibiotics and decreased sensitivity to others (Jones et al., 2014). They examined the effects of the rnj mutation on the production of the antibiotic jadomycin by S. venezuelae. They observed a 20–30% decrease in jadomycin production as compared to the wild type strain but no difference in the transcript levels of a CSR, jadJ, or two biosynthetic genes, jadA an jadM, involved in jadomycin production in S. venezuelae.
RNase J function has also been studied recently in the lincomycin producing species, Streptomyces lincolnensis. Wang et al. observed that deletion of rnj led to a 22.4-fold increase in lincomycin production as compared with the wild type strain and that this increase was 85% of that previously observed in a commercial overproducing strain of S. lincolnensis obtained by traditional mutation and screening methods (Wang et al., 2020).
It seems clear from the foregoing analyses that RNase J plays a significant role in cellular economy in Streptomyces and in the production of streptomycete antibiotics. Further studies are required to illuminate the roles of RNase J in these processes.
Polynucleotide phosphorylase
Polynucleotide phosphorylase (PNPase) is a 3′-5′-exoribonuclease that is found in bacteria, protists and in plant and animal organelles (Wright et al., 1977; Leszczyniecka et al., 2004). The PNPase reaction is as follows:
where N is any of the four bases found in RNA. The forward reaction describes the phosphorolysis of RNA chains, in which the enzyme processively removes one residue at a time from the 3′-end, concomitantly generating nucleoside diphosphates (Grunberg-Manago and Ochoa, 1955; Singer, 1958; Godefroy-Colburn and Grunberg-Manago, 1972; Littauer and Soreq, 1982; Littauer, 2005). The reaction is fully reversible, however, and PNPase will synthesize polyribonucleotide chains de novo (polymerization) using nucleoside diphosphates as substrates, generating inorganic phosphate as a by-product (Grunberg-Manago and Ochoa, 1955; Grunberg-Manago et al., 1956; Godefroy-Colburn and Grunberg-Manago, 1972; Littauer and Soreq, 1982; Littauer, 2005).
Streptomyces PNPase has been characterized in terms of biochemical function and structure. Indeed, the first published PNPase crystal structure was that of the enzyme from S. antibioticus (Symmons et al., 2000). The enzyme was described as a trimer of dimers. The trimer designation derives from the observation that the native enzyme is composed of three PNPase monomers. Each monomer consists of two RNase PH domains which presumably arose by gene duplication and fusion, thus a trimer of dimers (Symmons et al., 2000). The enzyme also contains KH and S1 domains at its C-terminal end (Bycroft et al., 1997; Lewis et al., 2000). These domains have been shown to be involved in RNA binding and in the autoregulation of PNPase expression (see further below). In addition to the catalysis of phosphorolysis and polymerization, Streptomyces PNPases have several novel features including function as RNA 3′-polyribonucleotide (poly(A)) polymerases; stimulation of phosphorolysis by nucleoside diphosphates; and, inhibition of phosphorolysis and polymerization by ppGpp. These features have been recently reviewed (Jones, 2018) and will not be discussed in detail here (but see further below regarding the possible connection between ppGpp inhibition and antibiotic synthesis).
In E. coli and other bacteria, the PNPase gene, pnp, is transcribed as a part of an operon that also includes rpsO, the gene for ribosomal protein S15 (Régnier and Portier, 1986; Clarke and Dowds, 1994; Luttinger et al., 1996). In many of these organisms, the rpsO-pnp operon is transcribed from two promoters, one upstream of rpsO and one in the intergenic region between rpsO and pnp (Portier and Regnier, 1984). Thus, transcription of the operon produces an rpsO transcript and a readthrough transcript from PrpsO, and a pnp transcript from Ppnp. The intergenic region also contains a stem-loop structure or hairpin which is involved in the regulation of expression of the operon (Régnier and Portier, 1986).
The organization of the rpsO-pnp operon in Streptomyces resembles that of E. coli and other bacteria but there is at least one important difference. While the operon is transcribed from two promoters in E. coli, rpsO-pnp is transcribed from four promoters in S. coelicolor, two upstream of rpsO, PrpsO A and B, and two in the intergenic region, Ppnp A and B (Bralley et al., 2014b). As in E. coli, an rpsO transcript, a readthrough transcript and a pnp transcript are produced from the operon (Gatewood et al., 2011). These four promoters are temporally regulated and they respond differentially to stress in the form of cold shock. The precise reasons for the presence of four rpsO-pnp promoters in S. coelicolor are unknown but may reflect the need for Streptomyces to respond to changing conditions in the soil environments in which they normally grow. The S. coelicolor intergenic region also contains a hairpin that is involved in operon regulation (Gatewood et al., 2011). The details of that regulation will be described below in the section on RNase III.
PNPase is essential in Streptomyces (Bralley et al., 2006). Thus, it has not been possible to examine antibiotic synthesis in a pnp null mutant. Using a different approach, pnp was overexpressed in S. antibioticus leading to a 2-3-fold higher level of PNPase in the overexpression strains than in the parental strain (Bralley and Jones, 2003). Actinomycin levels, however, were lower in strains overexpressing PNPase and the levels in those strains were comparable to those observed in control strains containing only the overexpression vector. Thus, it was not possible in these studies to discern a specific effect of pnp overexpression on actinomycin biosynthesis. There are other studies, involving ppGpp, that bear on the role of PNPase in antibiotic production.
ppGpp is an alarmone that mediates the stringent response to amino acid starvation in bacteria (Cashel et al., 1996). The stringent response has been shown to occur in Streptomyces (Strauch et al., 1991). Moreover, ppGpp plays an important role in antibiotic biosynthesis. In Streptomyces, ppGpp production begins during vegetative growth of mycelium, increases during the transition phase prior to the onset of antibiotic production, reaches a maximum during stationary phase, when antibiotics are actively produced and declines during the later stages of stationary phase (Ochi, 1987; Kelly et al., 1991; Takano et al., 1992; Hoyt and Jones, 1999). Despite the decline during the late stages of stationary phase, ppGpp levels do not fall to zero.
In E. coli and other bacteria, including Streptomyces, ppGpp is synthesized by the RelA protein from GTP and ATP (Cashel et al., 1996). RelA interacts with ribosomes during the process of ppGpp synthesis in vivo and that interaction involves ribosomal protein L11 (Yang and Ishiguro, 2001). Thus, relA null mutations and mutations in L11 (relC mutations) decrease or abolish ppGpp synthesis (Cashel et al., 1996). It has been observed in several Streptomyces species, producing different antibiotics, that relA and relC mutations decrease or abolish antibiotic production. Thus, a relA mutation decreased act and red production in S. coelicolor (Chakraburtty and Bibb, 1997), relA and relC mutations abolished actinomycin synthesis in S. antibioticus (Kelly et al., 1991; Hoyt and Jones, 1999) and a relC mutation decreased the production of streptomycin by Streptomyces griseus (Ochi, 1987).
ppGpp has also been shown to affect the half-life of bulk mRNA in S. coelicolor. These experiments involved an S. coelicolor parental strain and a strain containing an inducible relA gene (Gatewood and Jones, 2010). The half-life of bulk mRNA was almost twice as long in the S. coelicolor parental strain during stationary phase as in exponential phase (5.7 min vs. 3.2 min). In the strain containing an inducible relA gene, producing increased levels of (p)ppGpp, induction occasioned a ca. two-fold increase in the half-life of bulk mRNA in stationary phase, from 6.6 to 11.8 min. No such changes in mRNA half-life were observed in a parental E. coli strain or its corresponding relA null mutant (Gatewood and Jones, 2010). Taken together, these observations suggest that (p)ppGpp may stabilize mRNAs in stationary phase S. coelicolor cells, as compared with cells growing exponentially.
How do these observations relate to the impact of PNPase on antibiotic production? ppGpp has been shown to inhibit both phosphorolysis and polymerization by S. coelicolor and S. antibioticus PNPases in vitro (Gatewood and Jones, 2010). No such inhibition was observed with PNPase from E. coli. Moreover, ppGpp inhibits both phosphorolysis and polymerization by PNPase from the rare actinomycete, Nonomuraea sp. ATCC 39727, a producer of A40926, a glycopeptide antibiotic (Siculella et al., 2010). Siculella et al. measured the half-lives of the mRNA encoding a CSR, dpgA, in the A40926 gene cluster in the parental Nonomuraea strain and in a relC mutant strain, producing decreased levels of ppGpp and of the glycopeptide antibiotic, A40926. Siculella et al. observed a 1.5-2-fold longer half-life (13–18 min) for dpgA in stationary phase in the parental strain, containing ppGpp, as compared with the relC mutant, with lower levels of ppGpp (9 min). These observations and those obtained in the studies of S. antibioticus and S. coelicolor suggest that the effects of ppGpp on the activity of PNPase may be involved in the stabilization of mRNAs during stationary phase and thus, in the control of antibiotic production in Streptomyces and other actinomycetes.
It is well established that although levels of RNA and protein synthesis decrease dramatically as Streptomyces cultures move from the exponential to the stationary phase of growth, a basal level of synthesis is maintained throughout stationary phase (Jones, 1976; Liras et al., 1977). This basal level of macromolecular synthesis is presumably required to produce enzymes and other proteins involved in the synthesis of the secondary metabolites, such as antibiotics, that these organisms produce in stationary phase. Stabilization of the transcripts for these proteins would represent one strategy the organisms could employ to ensure the persistence of macromolecular synthesis to support secondary metabolite production. It is known that (p)ppGpp is present in significant amounts, even in stationary phase streptomycete cultures (Takano et al., 1992; Hoyt and Jones, 1999). Thus, the inhibition of PNPase by ppGpp might represent a strategy used by actinomycetes to stabilize essential mRNAs during stationary phase, ensuring the continued production of necessary antibiotics. It would be interesting to determine whether (p)ppGpp inhibits the activity of other Streptomyces exo- and endonucleases.
RNase III
Ribosomal, messenger and transfer RNAs all contain regions of secondary structure. It is not surprising, then, that ribonucleases exist that recognize and cleave in these regions. One such enzyme is RNase III, a double-strand specific endoribonuclease that is found in bacteria and eukaryotes (Nicholson, 1999; Drider and Condon, 2004). S. coelicolor RNase III was characterized by Chang et al. and the enzyme was shown to cleave synthetic double stranded polyribonucleotides with a preference for poly(G)-poly(C) over poly(A)-poly(U) (Chang et al., 2005). This preference is not surprising given the relatively high GC content of streptomycete genomes (Frontali et al., 1965). S. coelicolor RNase III also cleaves the readthrough transcript from the rpsO-pnp operon (Chang et al., 2005). However, the cleavage sites for the S. coelicolor enzyme differ from those of E. coli RNase III (Régnier and Portier, 1986). S. coelicolor cleaves on the two sides of a [4/4] internal loop in the rpsO-pnp intergenic hairpin. The primary cleavage site is on the 5′-side of that loop and the secondary site is on the 3′-side. In contrast, E. coli RNase III cleaves the intergenic hairpin within a base-paired stem situated just below an internal loop in the hairpin (Régnier and Portier, 1986). The internal loop in the E. coli intergenic hairpin is larger than that from the S. coelicolor hairpin and Calin-Jagerman and Nicholson have suggested that the features of internal loops determine the specificity of RNase III cleavage (Calin-Jageman and Nicholson, 2003).
As is also true for E. coli (Régnier and Portier, 1986), RNase III cleavage of the intergenic hairpin plays an important role in the regulation of expression of the rpsO-pnp operon in S. coelicolor. RNase III is not essential in S. coelicolor (or in E. coli) and it was possible to construct an RNase III (rnc) null mutant in that organism (Gravenbeek and Jones, 2008). In subsequent experiments, PNPase activity was measured and 2-4-fold higher activity levels were observed in the mutant strain during all phases of growth as compared with the parental strain (Gatewood et al., 2011). These increases correlated with increased stabilities of the transcripts derived from the rpsO-pnp operon. Thus, the half-life of the rpsO-pnp readthrough transcript increased from <<4 min (at the 4 min sampling time point, the readthrough transcript was virtually undetectable) in the parental strain to ca. 7 min in the null mutant, the half-life of the pnp transcript increased from <<4 min to 3.3 min and the rpsO transcript increased in half-life from ca. 4 min to 7 min (Gatewood et al., 2011). These results indicate that the increased PNPase activity observed in the S. coelicolor rnc null mutant was due to the stabilization of the mRNAs that are transcribed to produce the enzyme, viz. the readthrough and pnp transcripts. Unlike the situation in E. coli, the S. coelicolor pnp open reading frame also contained a cleavage site for RNase III (Gatewood et al., 2011). Thus, in the parental S. coelicolor strain, RNase III cleavage produces fragments of the transcripts from the rpsO-pnp operon with 3′-ends that can be subsequently digested by PNPase itself. RNase III thus contributes to the autoregulation of pnp expression in S. coelicolor, as it does in E. coli (Régnier and Portier, 1986).
RNase III also autoregulates its own expression in S. coelicolor. Xu et al. (2008) demonstrated increased levels of a polycistronic transcript containing rnc in a strain with a point mutation in rnc (the C120 mutation, see further below) as compared with the parental strain. They also showed that the half-life of the polycistronic transcript increased in the point mutant as compared with the parental strain. The authors synthesized the polycistronic rnc transcript in vitro and treated that transcript with isolated RNase III or with the protein containing the point mutation. The wild type protein cleaved the synthetic transcript at two positions while the mutant protein was only weakly active (but not completely inactive) against this substrate. Xu et al. used primer extension to identify the cleavage sites for RNase III in the polycistronic transcript. One of the cleavage sites was situated in the rnc cistron whereas the second was situated upstream of rnc. Both cleavages occurred in the loop regions of stem-loop structures (Xu et al., 2008).
In other studies, using an RNA Seq approach, coupled with RNA immunoprecipitation, Gatewood et al. (2012) identified ca. 800 transcripts from S. coelicolor that were bound by RNase III and therefore, were potential targets for RNase III cleavage. That number represents ca. 10% of the genes in the S. coelicolor genome (Bentley et al., 2002). By way of comparison, and using a different approach and a much smaller experimental sample size, Gitelman and Apirion identified 21 of 80 E. coli proteins whose abundance was affected by a point mutation in rnc (Gitelman and Apirion, 1980). It is noteworthy that Setinova et al. (2017) demonstrated the association of cognate antisense RNAs with 17 of the mRNAs identified in the RNA Seq experiments described by Gatewood et al.
The genetic locus encoding RNase III in S. coelicolor was first identified by virtue of the effect of a mutation in that locus on antibiotic production. Using N-methyl-N-nitro-N-nitrosoguanidine mutagenesis, Adamidis and Champness isolated a mutant, designated C120, that was defective in the production of all four antibiotics normally produced by S. coelicolor under laboratory conditions (Adamidis and Champness, 1992; Aceti and Champness, 1998). They designated this mutant absB (for antibiotic synthesis deficient). Act production by the mutant was reduced to only 2% of that observed in the parental strain and red production was reduced to 15% of normal levels. Cda and mmy production were also reduced (Adamidis and Champness, 1992). They subsequently showed that actII-orf4 and redD levels and the level of the transcript for an act biosynthetic enzyme, actVI-orf1, were substantially reduced in the mutant. Huang et al. demonstrated reduced levels of the CSRs for act (actII-orf4), red (redD and redZ) and cda (cdaR) in C120 in a microarray analysis of the S. coelicolor transcriptome (Huang et al., 2005).
In a later study, Price et al. (1999) complemented the absB mutation with cloned DNA from S. coelicolor and showed that the absB gene was that for S. coelicolor RNase III. The C120 mutation in the RNase III gene (rnc) was identified as a point mutation, T188P. Price et al. constructed an rnc disruptant and demonstrated even lower levels of antibiotic production in that mutant as compared with C120. C120 was shown to be defective in ribosomal RNA processing, as demonstrated by the accumulation of the 30S rRNA precursor of 16S and 23S rRNAs in the mutant (Price et al., 1999).
Another level of rnc regulation in Streptomyces involves the regulatory protein, AdpA, which responds to the bacterial hormone, A-factor, a C13-γ-butyrolactone (Horinouchi and Beppu, 2007). AdpA was identified in S. coelicolor by Takano et al. (2003). Xu et al. (2010) demonstrated that the adpA transcript was cleaved by RNase III in vitro and in vivo, implicating the enzyme in the regulation of AdpA levels in S. coelicolor. Moreover, Xu et al. demonstrated that despite the presence of elevated levels of the mutant rnc transcript in the C120 rnc point mutant (Adamidis and Champness, 1992), the levels of the mutant RNase III protein actually decreased at later times during the C120 growth cycle. This decrease was partially reversed by the presence of an adpA null mutation in the C120 strain. The authors also observed a decreased expression of genes for various S. coelicolor proteases in the adpA null mutant, and argued that the lower level of mutant RNase III protein in C120 was due to the action of proteases on the mutant RNase III protein, proteases whose expression was at least partially controlled by adpA. Thus, the authors argue that rnc and adpA are components of a feedback loop, in which each gene product regulates the abundance of the other (Xu et al., 2010).
Xu et al. also noted precocious production of act and red in the adpA null mutant. The authors argued that this observation indicates that the normal function of RNase III in the control of antibiotic production in S. coelicolor is not dependent on AdpA. To support that point, Xu et al. (2010) demonstrated that act and red production were not restored by an adpA/rnc double mutant.
Several hypotheses have been advanced to explain the effects of an endoribonuclease, like RNase III, on antibiotic production in S. coelicolor. The most straightforward hypothesis would posit that RNase III is required for the processing or degradation of the transcript for an activator or repressor of antibiotic synthesis. In this regard, it is not known whether the transcripts for any of the CSRs required for antibiotic production in S. coelicolor are cleaved by RNase III. Price et al. suggested the possibility that RNase III itself might function as a global regulator of antibiotic production by binding to regulatory transcripts without RNA processing or degradation, thus affecting the expression of genes that are critical for antibiotic production (Price et al., 1999). There is some evidence for the regulation of gene expression in E. coli via the binding of RNase III to dsRNA targets without cleaving them (Oppenheim et al., 1993; Dasgupta et al., 1998).
To examine this hypothesis, Gravenbeek and Jones (2008) constructed an rnc null mutant in S. coelicolor and demonstrated that this mutant was unable to produce act and red. These authors also constructed a point mutant of S. coelicolor rnc that was unable to cleave a transcript that was readily cleaved by the wild type enzyme, viz. the rpsO-pnp readthrough transcript. This mutant, D70A, nevertheless retained the ability to bind the rpsO-pnp transcript. Gravenbeek and Jones observed that D70A could not restore act and red production to the S. coelicolor rnc null mutant, whereas wild type rnc restored full levels of antibiotic production. The authors concluded that dsRNA binding alone was insufficient to control antibiotic production in S. coelicolor and that the endoribonuclease activity of RNase III was required for this function.
Based on the observation that rnc null mutations affected ribosomal RNA processing, Sello and Buttner suggested that defective ribosomes might be formed in the null mutants and that those ribosomes could be unable to translate the long polycistronic transcripts that are characteristic of S. coelicolor antibiotic gene clusters (Sello and Buttner, 2008a). To examine this possibility, Gatewood and Jones (2011) cloned an operon derived from the act cluster of S. coelicolor, producing a transcript of ca. 5,700 bases in length. A kanamycin resistance gene was placed at the 3′-end of the cloned operon, replacing the normal 3′-gene, and that construct was then placed in a reporter plasmid which would allow transcription and translation of the cloned operon. The authors observed the expression of kanamycin resistance when the plasmid construct was introduced into S. coelicolor, indicating that the act cluster operon was being transcribed and translated in the cloning host. In addition to kanamycin resistance, Gatewood and Jones (2011) demonstrated the presence of aminoglycoside phosphotransferase, the enzyme that confers kanamycin resistance. The data support the conclusion that ribosomes in the rnc null mutant could translate a long polycistronic S. coelicolor mRNA.
Thus, two proposed mechanisms for the effects of RNase III on antibiotic synthesis in S. coelicolor have been eliminated and it has been demonstrated that the endoribonuclease activity of the enzyme is required for its function in antibiotic synthesis. Other possible mechanisms will be considered below. It should be noted that RNase III is required for actinomycin production in S. antibioticus (Lee et al., 2013) and for jadomycin production in S. venezuelae (Jones et al., 2014).
Oligoribonuclease
While exoribonucleases release mononucleotides from digested substrates, endoribonucleases release oligonucleotides and larger fragments. Thus, an additional activity is required to convert oligoribonucleotides to mononucleotides which can be utilized in RNA synthesis. Oligoribonuclease possesses such an activity. The enzyme was identified in E. coli some years ago (Datta and Niyogi, 1975) and has subsequently been found in bacteria and eukaryotes (Zhang et al., 1998). The enzyme releases mononucleotides from the 3′-end of oligoribonucleotides 2–8 residues in length (Datta and Niyogi, 1975).
Ohnishi et al. characterized oligoribonuclease in Streptomyces (Ohnishi et al., 2000). These workers identified the enzyme initially in the streptomycin producer, S. griseus, and designated the enzyme OrnA. OrnA was shown to possess 3′-5′- exoribonuclease activity against a synthetic substrate, and its gene, ornA, was shown to lie just downstream of adpA in S. griseus. This suggested the possibility that adpA and ornA might be transcriptionally coupled and Ohnishi et al. demonstrated that adpA and ornA were, indeed, cotranscribed from a promoter upstream of adpA (Ohnishi et al., 2000).
An ornA null mutant grew slowly and produced sparse amounts of aerial mycelium and spores. The null mutant also produced reduced amounts of streptomycin as compared with the parental strain but the authors attributed this effect to the lower growth rate, concluding that ornA has no direct impact on streptomycin production in S. griseus (Ohnishi et al., 2000). Ohnishi et al. also demonstrated the presence of ornA in S. coelicolor and constructed a null mutant in that organism. As with S. griseus, the S. coelicolor null mutant grew slowly and formed sparse aerial mycelium, but there was essentially no effect of the null mutation on the production of act and red (Ohnishi et al., 2000).
Sello and Buttner also constructed an ornA null mutation in S. coelicolor (Sello and Buttner, 2008b). In contrast to the observations of Ohnishi et al., these workers reported that the null mutant underwent morphological differentiation and sporulation, at least on minimal medium, although more poorly than in the parental strain. The null mutant also overproduced actinorhodin. As in S. griseus, ornA is situated just downstream of adpA in S. coelicolor. Sello and Buttner performed a transcriptional analysis of ornA and concluded that, unlike the situation in S. griseus, ornA in S. coelicolor was not cotranscribed with adpA. This conclusion has been challenged by Xu et al. (2010) who performed RT-PCR experiments which revealed an apparent readthrough transcript in S. coelicolor, originating upstream of adpA and which contained both adpA and ornA sequences. The reason for the discrepancy between the results of Sello and Buttner and those of Xu et al. is not known at this time.
Possible mechanisms for the function of RNases in antibiotic synthesis
Any mechanisms to explain the effects of RNases on antibiotic production in Streptomyces must account for at least three critical observations: (1) a null mutation in a gene encoding an RNase can affect the production of multiple antibiotics simultaneously; (2) null mutations can lead to an increase or decrease in antibiotic production; (3) the effect of an RNase on antibiotic production can apparently be achieved without affecting the CSRs in the relevant pathways.
Given that the nuclease activities of the Streptomyces RNases are likely to be required for their effect on antibiotic production, a straightforward but comprehensive mechanism for those effects is presented in Figure 3. Figure 3 posits global regulators as one class of targets for Streptomyces RNases. In the case of RNase III for example, there may a single global regulator of act, red, cda and mmy production, whose transcript requires RNase III processing or degradation. That regulator, if it exists, remains elusive. Alternatively, it is possible that RNase III is involved in the processing of four different transcripts, each of which is required to produce one of the four S. coelicolor antibiotics. In similar fashion, any of the RNases discussed above might cleave the transcripts from any of the classes depicted in Figure 3, leading to inhibition or overproduction of antibiotics. It seems highly likely, although it is yet unproven, that RNase action on transcripts other than CSRs and other known regulators affects Streptomyces antibiotic production.

Figure 3. Proposed mechanism for the action of RNases on the transcripts of the various regulatory factors that have been shown to be involved in the regulation and production of antibiotics in Streptomyces. The model proposes that any of the transcript classes shown might be targets for RNase processing or degradation and that the effects of the RNases would then impact antibiotic production.
The field awaits the development of strategies to globally identify those transcripts.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aceti, D. J., and Champness, W. (1998). Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180, 3100–3106. doi: 10.1128/JB.180.12.3100-3106.1998
Adamidis, T., and Champness, W. (1992). Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J. Bacteriol. 174, 4622–4628. doi: 10.1128/jb.174.14.4622-4628.1992
Ahn, S., Shin, E. K., Yeom, J. H., and Lee, K. (2008). Modulation of Escherichia coli RNase E activity by RraAS2, a Streptomyces coelicolor ortholog of RraA. Korean J. Microbiol. 44, 93–97.
Apirion, D., and Lassar, A. B. (1978). A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 253, 1738–1742. doi: 10.1016/S0021-9258(17)34927-X
Bechhofer, D. H., and Deutscher, M. P. (2019). Bacterial ribonucleases and their roles in RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 54, 242–300. doi: 10.1080/10409238.2019.1651816
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bibb, M. J. (1996). The regulation of antibiotic production in Streptomyces coelicolor. Microbiology 142, 1335–1344. doi: 10.1099/13500872-142-6-1335
Bralley, P., Aseem, M., and Jones, G. H. (2014a). SCO5745, a bifunctional RNase J ortholog, affects antibiotic production in Streptomyces coelicolor. J. Bacteriol. 196, 1197–1205. doi: 10.1128/JB.01422-13
Bralley, P., Gatewood, M. L., and Jones, G. H. (2014b). Transcription of the rpsO-pnp operon of Streptomyces coelicolor involves four temporally regulated, stress responsive promoters. Gene 536, 177–185. doi: 10.1016/j.gene.2013.10.055
Bralley, P., Gust, B., Chang, S. A., Chater, K. F., and Jones, G. H. (2006). RNA 3′-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and PNPase. Microbiology-SGM 152, 627–636. doi: 10.1099/mic.0.28363-0
Bralley, P., and Jones, G. H. (2003). Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149, 2173–2182. doi: 10.1099/mic.0.26334-0
Bycroft, M., Hubbard, T. J., Proctor, M., Freund, S. M., and Murzin, A. G. (1997). The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cells 88, 235–242. doi: 10.1016/S0092-8674(00)81844-9
Calin-Jageman, I., and Nicholson, A. W. (2003). RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucl. Acids. Res. 31, 2381–2392. doi: 10.1093/nar/gkg329
Callaghan, A. J., Aurikko, J. P., Ilag, L. L., Gunter Grossmann, J., Chandran, V., Kuhnel, K., et al. (2004). Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340, 965–979. doi: 10.1016/j.jmb.2004.05.046
Callaghan, A. J., Marcaida, M. J., Stead, J. A., Mcdowall, K. J., Scott, W. G., and Luisi, B. F. (2005). Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191. doi: 10.1038/nature04084
Casaregola, S., Jacq, A., Laoudj, D., Mcgurk, G., Margarson, S., Tempete, M., et al. (1992). Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 228, 30–40. doi: 10.1016/0022-2836(92)90489-7
Cashel, M., Gentry, D. R., Hernandez, V. J., and Vinella, D. (1996). “The stringent response” in Escherichia coli and Salmonella: cellular and molecular biology. eds. F. C. Neidhardt, R. Curtiss Iii, J. L. Ingraham, E. C. C. Lin, and K. B. Low, et al. (Washington, DC: ASM Press)
Chakraburtty, R., and Bibb, M. J. (1997). The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179, 5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997
Chang, S. A., Bralley, P., and Jones, G. H. (2005). The absB gene encodes a double-strand specific endoribonuclease that cleaves the readthrough transcript of the rspO-pnp operon in Streptomyces coelicolor. J. Biol. Chem. 280, 33213–33219. doi: 10.1074/jbc.M503440200
Chater, K. F. (2016). Recent advances in understanding Streptomyces. F1000Res 5, 2795–2810. doi: 10.12688/f1000research.9534.1
Clarke, D. J., and Dowds, B. C. (1994). The gene coding for polynucleotide phosphorylase in Photorhabdus sp. strain K122 is induced at low temperatures. J. Bacteriol. 176, 3775–3784. doi: 10.1128/jb.176.12.3775-3784.1994
Cohen, S. N., and Mcdowall, K. J. (1997). RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 23, 1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x
Cole, S. P., Rudd, B. A., Hopwood, D. A., Chang, C. J., and Floss, H. G. (1987). Biosynthesis of the antibiotic actinorhodin. Analysis of blocked mutants of Streptomyces coelicolor. J. Antibiot. 40, 340–347. doi: 10.7164/antibiotics.40.340
Condon, C. (2010). What is the role of RNase J in mRNA turnover? RNA Biol. 7, 316–321. doi: 10.4161/rna.7.3.11913
Condon, C., Putzer, H., and Grunberg-Manago, M. (1996). Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 93, 6992–6997. doi: 10.1073/pnas.93.14.6992
Dasgupta, S., Fernandez, L., Kameyama, L., Inada, T., Nakamura, Y., Pappas, A., et al. (1998). Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III--the effect of dsRNA binding on gene expression. Mol. Microbiol. 28, 629–640. doi: 10.1046/j.1365-2958.1998.00828.x
Datta, A. K., and Niyogi, K. (1975). A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J. Biol. Chem. 250, 7313–7319. doi: 10.1016/S0021-9258(19)40946-0
Drider, D., and Condon, C. (2004). The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8, 195–200. doi: 10.1159/000086700
Even, S., Pellegrini, O., Zig, L., Labas, V., Vinh, J., Brechemmier-Baey, D., et al. (2005). Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33, 2141–2152. doi: 10.1093/nar/gki505
Feitelson, J. S., Malpartida, F., and Hopwood, D. A. (1985). Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 131, 2431–2441. doi: 10.1099/00221287-131-9-2431
Figaro, S., Durand, S., Gilet, L., Cayet, N., Sachse, M., and Condon, C. (2013). Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 195, 2340–2348. doi: 10.1128/JB.00164-13
Frontali, C., Hill, L. R., and Silvestri, L. G. (1965). The base composition of deoxyribonucleic acids of Streptomyces. J. Gen. Microbiol. 38, 243–250. doi: 10.1099/00221287-38-2-243
Gao, J., Lee, K., Zhao, M., Qiu, J., Zhan, X., Saxena, A., et al. (2006). Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol. Microbiol. 61, 394–406. doi: 10.1111/j.1365-2958.2006.05246.x
Gatewood, M. L., Bralley, P., and Jones, G. H. (2011). RNase III-dependent expression of the rpsO-pnp operon of Streptomyces coelicolor. J. Bacteriol. 193, 4371–4379. doi: 10.1128/JB.00452-11
Gatewood, M. L., Bralley, P., Weil, M. R., and Jones, G. H. (2012). RNA-Seq and RNA immunoprecipation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J. Bacteriol. 194, 2228–2237. doi: 10.1128/JB.06541-11
Gatewood, M. L., and Jones, G. H. (2010). (p)ppGpp inhibits polynucleotide phosphorylase from Streptomyces but not from Escherichia coli and increases the stability of bulk mRNA in Streptomyces coelicolor. J. Bacteriol. 192, 4275–4280. doi: 10.1128/JB.00367-10
Gatewood, M. L., and Jones, G. H. (2011). Expression of a polycistronic messenger RNA involved in antibiotic production in an rnc mutant of Streptomyces coelicolor. Arch. Microbiol. 194, 147–155. doi: 10.1007/s00203-011-0740-7
Gitelman, D. R., and Apirion, D. (1980). The synthesis of some proteins is affected in RNA processing mutants of Escherichia coli. Biochem. Biophys. Res. Commun. 96, 1063–1070. doi: 10.1016/0006-291X(80)90060-1
Godefroy-Colburn, T., and Grunberg-Manago, M. (1972). Polynucleotide phosphorylase. Enzymes 7, 533–574. doi: 10.1016/S1874-6047(08)60462-X
Gravenbeek, M. L., and Jones, G. H. (2008). The endonuclease activity of RNase III is required for the regulation of antibiotic production by Streptomyces coelicolor. Microbiology 154, 3547–3555. doi: 10.1099/mic.0.2008/022095-0
Grunberg-Manago, M., and Ochoa, S. (1955). Enzymatic synthesis and breakdown of polynucleotides: polynucleotide phosphorylase. J. Am. Chem. Soc. 77, 3165–3166. doi: 10.1021/ja01616a093
Grunberg-Manago, M., Ortiz, P. J., and Ochoa, S. (1956). Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of Azotobacter vinelandii. Biochim. Biophys. Acta 20, 269–285. doi: 10.1016/0006-3002(56)90286-4
Guimaraes, V. A., Le Scornet, A., Khemici, V., Hausmann, S., Armitano, J., Prados, J., et al. (2021). RNase J1 and J2 are host-encoded factors for plasmid replication. Front. Microbiol. 12:586886. doi: 10.3389/fmicb.2021.586886
Hagege, J. M., and Cohen, S. N. (1997). A developmentally regulated Streptomyces endoribonuclease resembles ribonuclease E of Escherichia coli. Mol. Microbiol. 25, 1077–1090. doi: 10.1046/j.1365-2958.1997.5311904.x
Heo, J., Kim, D., Joo, M., Lee, B., Seo, S., Lee, J., et al. (2016). RraAS2 requires both scaffold domains of RNase ES for high-affinity binding and inhibitory action on the ribonucleolytic activity. J. Microbiol. 54, 660–666. doi: 10.1007/s12275-016-6417-9
Hopwood, D. A. (2019). Highlights of Streptomyces genetics. Heredity 123, 23–32. doi: 10.1038/s41437-019-0196-0
Hopwood, D. A., and Wright, H. M. (1983). CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2). J. Gen. Microbiol. 129, 3575–3579. doi: 10.1099/00221287-129-12-3575
Horinouchi, S., and Beppu, T. (2007). Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83, 277–295. doi: 10.2183/pjab.83.277
Hoyt, S., and Jones, G. H. (1999). relA is required for actinomycin production in Streptomyces antibioticus. J. Bacteriol. 181, 3824–3829. doi: 10.1128/JB.181.12.3824-3829.1999
Huang, J., Shi, J., Molle, V., Sohlberg, B., Weaver, D., Bibb, M. J., et al. (2005). Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58, 1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x
Inagawa, T., Okamoto, S., Wachi, M., and Ochi, K. (2003). RNase ES of Streptomyces coelicolor A3(2) can complement the rne and rng mutations in Escherichia coli. Biosci. Biotechnol. Biochem. 67, 1767–1771. doi: 10.1271/bbb.67.1767
Jones, G. H. (1976). RNA synthesis in Streptomyces antibioticus: in vitro effects of actinomycin and transcriptional inhibitors. Biochemistry 15, 3331–3341. doi: 10.1021/bi00660a025
Jones, G. H. (1994). Purification and properties of ATP:GTP 3′-pyrophosphotransferase (guanosine pentaphosphate synthetase) from Streptomyces antibioticus. J. Bacteriol. 176, 1475–1481. doi: 10.1128/jb.176.5.1475-1481.1994
Jones, G. H. (2010). RNA degradation and the regulation of antibiotic synthesis in Streptomyces. Future Microbiol. 5, 419–429. doi: 10.2217/fmb.10.14
Jones, G. H. (2018). Novel aspects of polynucleotide phosphorylase function in Streptomyces. Antibiotics 7, 25–37. doi: 10.3390/antibiotics7010025
Jones, S. E., Leong, V., Ortega, J., and Elliot, M. A. (2014). Development, antibiotic production, and ribosome assembly in Streptomyces venezuelae are impacted by RNase J and RNase III deletion. J. Bacteriol. 196, 4253–4267. doi: 10.1128/JB.02205-14
Kelly, K. S., Ochi, K., and Jones, G. H. (1991). Pleiotropic effects of a relC mutation in Streptomyces antibioticus. J. Bacteriol. 173, 2297–2300. doi: 10.1128/jb.173.7.2297-2300.1991
Khemici, V., Poljak, L., Luisi, B. F., and Carpousis, A. J. (2008). The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70, 799–813. doi: 10.1111/j.1365-2958.2008.06454.x
Kim, H. J., Calcutt, M. J., Schmidt, F. J., and Chater, K. F. (2000). Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182, 1313–1320. doi: 10.1128/JB.182.5.1313-1320.2000
Koslover, D. J., Callaghan, A. J., Marcaida, M. J., Garman, E. F., Martick, M., Scott, W. G., et al. (2008). The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure 16, 1238–1244. doi: 10.1016/j.str.2008.04.017
Lee, J. H., Gatewood, M. L., and Jones, G. H. (2013). RNase III is required for actinomycin production in Streptomyces antibioticus. Appl. Environ. Microbiol. 79, 6447–6451. doi: 10.1128/AEM.02272-13
Lee, K., and Cohen, S. N. (2003). A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 48, 349–360. doi: 10.1046/j.1365-2958.2003.03435.x
Lee, K., Zhan, X., Gao, J., Qiu, J., Feng, Y., Meganathan, R., et al. (2003). RraA. A protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cells 114, 623–634. doi: 10.1016/j.cell.2003.08.003
Leszczyniecka, M., Desalle, R., Kang, D. C., and Fisher, P. B. (2004). The origin of polynucleotide phosphorylase domains. Mol. Phylogenet. Evol. 31, 123–130. doi: 10.1016/j.ympev.2003.07.012
Lewis, H. A., Musunuru, K., Jensen, K. B., Edo, C., Chen, H., Darnell, R. B., et al. (2000). Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cells 100, 323–332. doi: 10.1016/S0092-8674(00)80668-6
Liras, P., Villanueva, J. R., and Martin, J. F. (1977). Sequential expression of macromolecule biosynthesis and candicidin formation in Streptomyces griseus. J. Gen. Microbiol. 102, 269–277. doi: 10.1099/00221287-102-2-269
Littauer, U. Z. (2005). From polynucleotide phosphorylase to neurobiology. J. Biol. Chem. 280, 38889–38897. doi: 10.1074/JBC.X500007200
Littauer, U. Z., and Soreq, H. (1982). Polynucleotide phosphorylase. Enzymes 15, 517–553. doi: 10.1016/S1874-6047(08)60289-9
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
Luong, T. T., Nguyen, M. T., Chen, Y. W., Chang, C., Lee, J. H., Wittchen, M., et al. (2021). Ribonuclease J-mediated mRNA turnover modulates cell shape, metabolism and virulence in Corynebacterium diphtheriae. Microorganisms 9, 389–400. doi: 10.3390/microorganisms9020389
Luttinger, A., Hahn, J., and Dubnau, D. (1996). Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19, 343–356. doi: 10.1046/j.1365-2958.1996.380907.x
Mackie, G. A. (2013). RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11, 45–57. doi: 10.1038/nrmicro2930
Mader, U., Zig, L., Kretschmer, J., Homuth, G., and Putzer, H. (2008). mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 70, 183–196. doi: 10.1111/j.1365-2958.2008.06400.x
Martini, M. C., Hicks, N. D., Xiao, J., Alonso, M. N., Barbier, T., Sixsmith, J., et al. (2022). Loss of RNase J leads to multi-drug tolerance and accumulation of highly structured mRNA fragments in Mycobacterium tuberculosis. PLoS Pathog. 18:e1010705. doi: 10.1371/journal.ppat.1010705
Mathy, N., Benard, L., Pellegrini, O., Daou, R., Wen, T., and Condon, C. (2007). 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cells 129, 681–692. doi: 10.1016/j.cell.2007.02.051
Mcdowall, K. J., and Cohen, S. N. (1996). The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255, 349–355. doi: 10.1006/jmbi.1996.0027
Newman, J. A., Hewitt, L., Rodrigues, C., Solovyova, A., Harwood, C. R., and Lewis, R. J. (2011). Unusual, dual endo- and exonuclease activity in the degradosome explained by crystal structure analysis of RNase J1. Structure 19, 1241–1251. doi: 10.1016/j.str.2011.06.017
Nicholson, A. W. (1999). Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23, 371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Ochi, K. (1987). Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 169, 3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987
Ohnishi, Y., Nishiyama, Y., Sato, R., Kameyama, S., and Horinouchi, S. (2000). An oligoribonuclease gene in Streptomyces griseus. J. Bacteriol. 182, 4647–4653. doi: 10.1128/JB.182.16.4647-4653.2000
Oppenheim, A. B., Kornitzer, D., Altuvia, S., and Court, D. L. (1993). Posttranscriptional control of the lysogenic pathway in bacteriophage lambda. Prog. Nucleic Acid Res. Mol. Biol. 46, 37–49. doi: 10.1016/S0079-6603(08)61017-X
Pei, X. Y., Bralley, P., Jones, G. H., and Luisi, B. F. (2015). Linkage of catalysis and 5′ end recognition in ribonuclease RNase. J. Nucleic Acids Res 43, 8066–8076. doi: 10.1093/nar/gkv732
Portier, C., and Regnier, P. (1984). Expression of the rpsO and pnp genes: structural analysis of a DNA fragment carrying their control regions. Nucl. Acids Res. 12, 6091–6099. doi: 10.1093/nar/12.15.6091
Price, B., Adamidis, T., Kong, R., and Champness, W. (1999). A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J. Bacteriol. 181, 6142–6151. doi: 10.1128/JB.181.19.6142-6151.1999
Py, B., Higgins, C. F., Krisch, H. M., and Carpousis, A. J. (1996). A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169–172. doi: 10.1038/381169a0
Régnier, P., and Portier, C. (1986). Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 187, 23–32. doi: 10.1016/0022-2836(86)90403-1
Sello, J. K., and Buttner, M. J. (2008a). The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J. Bacteriol. 190, 4079–4083. doi: 10.1128/JB.01889-07
Sello, J. K., and Buttner, M. J. (2008b). The oligoribonuclease gene in Streptomyces coelicolor is not transcriptionally or translationally coupled to adpA, a key bldA target. FEMS Microbiol. Lett. 286, 60–65. doi: 10.1111/j.1574-6968.2008.01260.x
Seo, S., Kim, D., Song, W., Heo, J., Joo, M., Lim, Y., et al. (2017). RraAS1 inhibits the ribonucleolytic activity of RNase ES by interacting with its catalytic domain in Streptomyces coelicolor. J. Microbiol. 55, 37–43. doi: 10.1007/s12275-017-6518-0
Setinova, D., Smidova, K., Pohl, P., Music, I., and Bobek, J. (2017). RNase III-binding-mRNAs revealed novel complementary transcripts in Streptomyces. Front. Microbiol. 8, 2693–2703. doi: 10.3389/fmicb.2017.02693
Siculella, L., Damiano, F., Di Summa, R., Tredici, S. M., Alduina, R., Gnoni, G. V., et al. (2010). Guanosine 5′-diphosphate 3′-diphosphate (ppGpp) as a negative modulator of polynucleotide phosphorylase activity in a ‘rare’ actinomycete. Mol. Microbiol. 77, 716–729. doi: 10.1111/j.1365-2958.2010.07240.x
Singer, M. F. (1958). Phosphorolysis of oligoribonucleotides by polynucleotide phosphorylase. J. Biol. Chem. 232, 211–228. doi: 10.1016/S0021-9258(18)70389-X
Strauch, E., Takano, E., Bayliss, H. A., and Bibb, M. J. (1991). The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5, 289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x
Symmons, M., Jones, G. H., and Luisi, B. (2000). A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity and regulation. Structure 8, 1215–1226. doi: 10.1016/S0969-2126(00)00521-9
Takano, E., Gramajo, H. C., Strauch, E., Andres, N., White, J., and Bibb, M. J. (1992). Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6, 2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x
Takano, E., Tao, M., Long, F., Bibb, M. J., Wang, L., Li, W., et al. (2003). A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50, 475–486. doi: 10.1046/j.1365-2958.2003.03728.x
Taverniti, V., Forti, F., Ghisotti, D., and Putzer, H. (2011). Mycobacterium smegmatis RNase J is a 5′-3′ exo−/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol. Microbiol. 82, 1260–1276. doi: 10.1111/j.1365-2958.2011.07888.x
Vanzo, N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., et al. (1998). Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12, 2770–2781. doi: 10.1101/gad.12.17.2770
Wang, R., Kong, F., Wu, H., Hou, B., Kang, Y., Cao, Y., et al. (2020). Complete genome sequence of high-yield strain S. lincolnensis B48 and identification of crucial mutations contributing to lincomycin overproduction. Synth. Syst. Biotechnol. 5, 37–48. doi: 10.1016/j.synbio.2020.03.001
Wright, B. E., Killick, K. A., and Tai, A. (1977). Fourth expansion and glucose perturbation of the Dictyostelium kinetic model. Eur. J. Biochem. 74, 217–225. doi: 10.1111/j.1432-1033.1977.tb11384.x
Wright, L. F., and Hopwood, D. A. (1976). Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 95, 96–106. doi: 10.1099/00221287-95-1-96
Xia, H., Li, X., Li, Z., Zhan, X., Mao, X., and Li, Y. (2020). The application of regulatory cascades in Streptomyces: yield enhancement and metabolite mining. Front. Microbiol. 11:406. doi: 10.3389/fmicb.2020.00406
Xu, W., Huang, J., and Cohen, S. N. (2008). Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 190, 5526–5530. doi: 10.1128/JB.00558-08
Xu, W., Huang, J., Lin, R., Shi, J., and Cohen, S. N. (2010). Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75, 781–791. doi: 10.1111/j.1365-2958.2009.07023.x
Yang, X., and Ishiguro, E. E. (2001). Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J. Bacteriol. 183, 6532–6537. doi: 10.1128/JB.183.22.6532-6537.2001
Yeom, J. H., Go, H., Shin, E., Kim, H. L., Han, S. H., Moore, C. J., et al. (2008). Inhibitory effects of RraA and RraB on RNAse E-related enzymes imply conserved functions in the regulated enzymatic cleavage of RNA. FEMS Microbiol. Lett. 285, 10–15. doi: 10.1111/j.1574-6968.2008.01205.x
Keywords: Streptomyces, antibiotic, regulation, ribonuclease, RNA decay, RNA processing
Citation: Jones GH (2023) Streptomyces RNases – Function and impact on antibiotic synthesis. Front. Microbiol. 14:1096228. doi: 10.3389/fmicb.2023.1096228
Edited by:
Huangen Ding, Louisiana State University, United StatesReviewed by:
Rute G. Matos, Universidade Nova de Lisboa, PortugalYvonne Mast, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), Germany
Copyright © 2023 Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George H. Jones, Z2VvcmdlLmguam9uZXNAZW1vcnkuZWR1
 George H. Jones
George H. Jones