- 1Anhui Provincial Laboratory of Microbiology and Parasitology, Anhui Key Laboratory of Zoonoses, Department of Microbiology and Parasitology, School of Basic Medical Sciences, Anhui Medical University, Hefei, China
- 2The Second Clinical Medical College, Anhui Medical University, Hefei, China
- 3The First Clinical Medical College, Anhui Medical University, Hefei, China
Malaria caused by Plasmodium is still a serious public health problem. Genomic editing is essential to understand parasite biology, elucidate mechanical pathways, uncover gene functions, identify novel therapeutic targets, and develop clinical diagnostic tools. Recent advances have seen the development of genomic diagnostic technologies and the emergence of genetic manipulation toolbox comprising a host of several systems for editing the genome of Plasmodium at the DNA, RNA, and protein level. Genomic manipulation at the RNA level is critical as it allows for the functional characterization of several transcripts. Of notice, some developed artificial RNA genome editing tools hinge on the endogenous RNA interference system of Plasmodium. However, Plasmodium lacks a robust RNAi machinery, hampering the progress of these editing tools. CRISPR-Cas13, which belongs to the VI type of the CRISPR system, can specifically bind and cut RNA under the guidance of crRNA, with no or minimal permanent genetic scar on genes. This review summarizes CRISPR-Cas13 system from its discovery, classification, principle of action, and diagnostic platforms. Further, it discusses the application prospects of Cas13-based systems in Plasmodium and highlights its advantages and drawbacks.
1. Introduction
Malaria has been a major global health problem for humans throughout their history and is a leading cause of mortality across many tropical and subtropical countries (Hemingway and Bates, 2003; Cowman et al., 2016; Monroe et al., 2022). Malaria control efforts have been undermined by the decline in the effectiveness of the primary malaria-figurehting tools, increasing resistance to treated-insecticide nets and anti-malarial drugs, and no effective vaccine (Olotu et al., 2013; Haldar et al., 2018; Memvanga and Nkanga, 2021). Spotlights from the 2022 world malaria report by WHO showed that cases of malaria increased from 245 million in 2020 to 247 million in 2021, although malaria-related deaths slightly decreased from 625,000 to 619,000 (World Health Organization, 2022). The etiological agent of malaria, Plasmodium, mainly thrives in human and mosquito hosts. Of all Plasmodium parasites, five species are responsible for global malaria infections in humans: Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi (Lalremruata et al., 2017; Memvanga and Nkanga, 2021; Sato, 2021).
Microscopic examination of Giemsa-stained peripheral blood smears has over the years been the “gold standard” for the diagnosis of malaria (Oboh et al., 2021; Fitri et al., 2022). However, its usefulness has been confronted with a host of challenges as results generated from microscopy are dependent on the technical know-how of the microscopist, the quality of blood sample, the parasite density, and the subjectivity of results interpretation (Barber et al., 2013; Berzosa et al., 2018; Ospina-Villa et al., 2018), indicating a void in diagnosis and the need to develop new effective tools. The Plasmodium falciparum genome encompasses 22.8 mega-bases with an average gene density of 1 gene per 4,338 base pairs—distributed among a total of 14 chromosomes (Gardner et al., 2002). Almost two-thirds of the proteins coded by these genes are unique to Plasmodium (Gardner et al., 2002) providing peculiar targets for molecular detection. In an attempt to fill the gaps in malaria diagnosis, several molecular diagnostic tools have been established to target various unique genes in Plasmodium. Molecular diagnosis notwithstanding remains a big huddle as the performance of these tools varies depending on the epidemiological setting (Oboh et al., 2018).
Interestingly, 74% of the total genes in Plasmodium remain functionally uncharacterized despite the availability of the parasite’s genome (Tan and Mutwil, 2020). To address this paucity, forward and reverse functional genomic tools have been developed to manipulate the parasites’ genomes to functionally characterize these genes (Zhang et al., 2014; Walker and Lindner, 2019; Cárdenas et al., 2022; Thiam et al., 2022). In particular, RNA-targeting tools hold great potential to understand the biological functions of genes and identify novel anti-malarial targets and effective vaccines (Nisbet et al., 2016; Liu et al., 2019). RNA level manipulation/editing affects the timing and/or level of transcription and alters the stability of gene expression, thereby offering scientists the platform to study the basic cellular functions of mRNAs, and non-coding RNAs (ncRNA) such as small nuclear RNA (snRNA), ribosomal RNA (rRNA), microRNA (miRNA), and piwi interaction RNA (piRNA; Briquet et al., 2022).
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR-associated protein) system is found in archaea and bacteria—responsible for coordinating defense against foreign genetic elements (Mojica et al., 2000; Marraffini, 2015; Moon et al., 2019). The CRISPR-Cas system is a powerful biological tool whose usefulness scientists have been unlocking in the last few decades. It is a double-edged tool that scientists have applied for both diagnosis and gene editing to unravel the function of many genes at the DNA and RNA levels in several organisms (Graham and Root, 2015; Hartenian and Doench, 2015; Zhang D. et al., 2021). Currently, the CRISPR-Cas system is structurally and functionally diverse and divided into 2 classes, 6 types, and more than 30 subtypes. The first class includes type I, type III, and type IV, and the second class includes type II, type V, and type VI (O'Connell, 2019). The small size, atypical hypercompact architecture, and high efficiency of class 2 multi-domain Cas proteins have made them a prime focus of many researchers compared to class I proteins (Cao et al., 2022; Liu and Pei, 2022). Of these, the majority of research in the past decade has focused on CRISPR-Cas9 belonging to the type II category (Atkins et al., 2021), and its properties have been harnessed for gene knockdown, gene editing, epitranscriptomics modification, and sensing of RNA targets (Zhang H. et al., 2021). CRISPR-Cas13 systems—which constitute the type VI group—have been demonstrated to generally lack DNase (Deoxyribonuclease) activity, but are fixated on RNA cleaving, and could offer distinct targeting advantages to RNA. Thus they represent a clinically promising platform capable of efficiently characterizing genes at the RNA level (Gaj, 2021).
In this review, we have summarized the CRISPR-Cas13 system from its discovery, classification, principle of action, and its application in malaria diagnosis. Given that malaria parasites lack relevant interference systems (Baum et al., 2009) thereby limiting the use of endogenous interference-dependent RNA editing systems, we will further discuss the application prospects of Cas13 in Plasmodium at the RNA level.
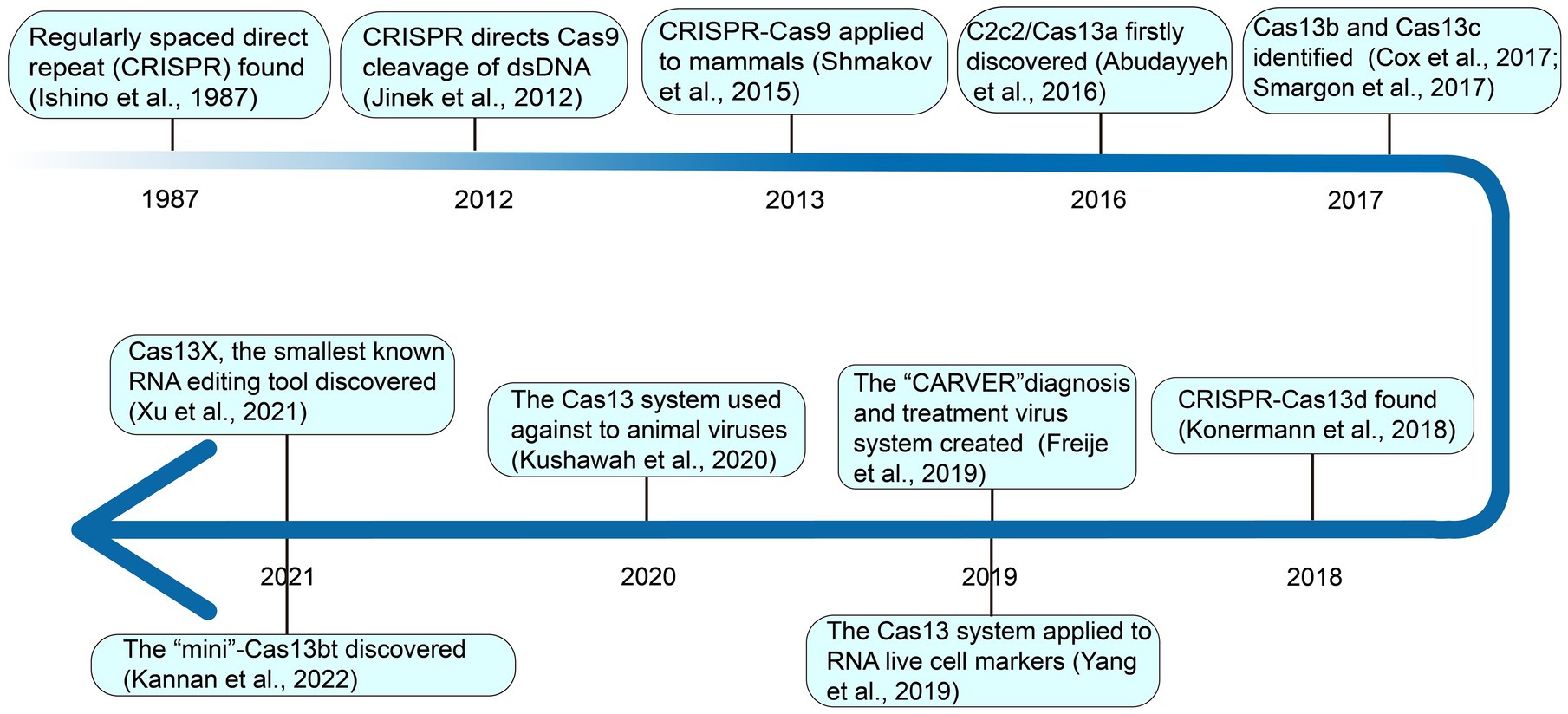
2. Discovery of CRISPR-Cas systems, especially for Casl3
In 1987, Yoshizumi Ishino of Osaka University in Japan detected regularly spaced direct palindromic repeats downstream of the stop codon of the iap (isoenzymes of alkaline phosphate) genes of E. coli K-12 cells (Ishino et al., 1987). This event provided the blueprint for the discovery of new CRISPR-Cas systems, ushering scientists into a new era of gene editing and genomic engineering. At the time, the relevance of these repeat sequences remained obscure. Mojica and his team detected a similar sequence in phylogenetically distinct organisms: Haloferax mediterranei and Haloferax volcanii of the Archaea domain (Mojica et al., 1993). They later postulated that these repeats are involved in replicon partitioning (Mojica et al., 1995). In 2002, substantial progress was made by Jansen and collaborators resulting in the official naming of this mysterious repeat as CRISPR which is now commonly accepted by the scientific community (Jansen et al., 2002). Soon after, spurred on by his previous findings, a major leap toward the understanding of the function of CRISPR was made by Mojica and his team (Mojica et al., 2005). They pointed out that the spacer sequence in CRISPR had homology with foreign phages or plasmids, and that the virus could not infect cells carrying homologous spacer sequences, but was easy to invade cells without spacer sequences. Against this background, it was posited that CRISPR might participate in the immune function of bacteria and it requires a precise sequence match between the spacer and the target sequence of the virus (Mojica et al., 2005; Lander, 2016). The functionality of the second component of the CRISPR-Cas complex, CRISPR-associated protein (Cas), was computationally demonstrated later by Marakova and collaborators as a composite of the DNA repair machinery (Makarova et al., 2006). In a later study by Horvath, he noticed a palindromic repeat that coded for an endonuclease protein, Cas9, and intimated that the protein cleaves viral DNA and thus grants immunity to the host bacteria (Horvath and Barrangou, 2010). A few years later, in 2012, Doudna and Charpentier’s team discovered the third component of the CRISPR system—the trans-acting crRNA (tracrRNA) which oversees the maturation of precursor crRNA into crRNA and also directs Cas9 cleavage of dsDNA (Jinek et al., 2012). Subsequent development saw the exploitation of the CRISPR-Cas system as a genomic editing tool. In 2013, the gene-editing tool CRISPR-Cas9 was applied in mammalian cells and became popular all over the world, because of its novelty, efficiency, and versatility (Shmakov et al., 2015).
Different from CRISPR-Cas9, which binds and cuts at the DNA level, as shown in Figure 1, in 2016, deeply searching for microbial genome data, it was found that a new CRISPR-Cas system effector protein, named c2c2 (now referred to as Casl3a), could specifically bind and cut RNA under the guidance of crRNA, and regulate the activity of genes at the RNA level (Abudayyeh et al., 2016). This opened up a new avenue for editing targeted RNA. In 2017, through rigorous biochemical experiments and genetic computations, Zhang Feng’s team identified two new Cas13 subtypes, Cas13b and Cas13c (Cox et al., 2017; Smargon et al., 2017). Subsequent investigation revealed that CRISPR-Cas13 systems uniquely lack the third component, tracrRNA (Yan et al., 2019). Going further, they demonstrated that Cas13b could be applied to precisely edit the full length of transcript harboring deleterious mutations in mammalian cells (Cox et al., 2017). In 2018, Konermarm et al. analyzed a prokaryote genome and discovered the CRISPR-Casl3d system, which has the smallest molecular weight of all CRISPR-Casl3 effectors, and was suitable for virus vectors, and had great application potential in vivo (Konermann et al., 2018). Through the exploration of bulk metagenomic data, Hu et al. (2022) identified novel hypercompact Cas13 systems and therein named them Cas13e, Cas13f, Cas13g, Cas13h, and Cas13i. However, their functional relevance remains to be described in future studies.
Since its advent, Cas13 has been widely used in various fields. For example, in 2019, Zhang Feng’s team created a new CRISPR-Cas13 system called CARVER (Cas13-assisted Restriction of Viral Expression and Readout) antiviral, which combines the antiviral activity of Cas13 and its diagnostic capabilities, as a promising system for the diagnosis and treatment of viral infections (Freije et al., 2019). In the same year, Chen Lingling’s group used the CRISPR-Casl3d system for RNA live cell labeling (Yang et al., 2019). In 2020, Gopal Kushawah and others used CRISPR-Cas13d to induce high-efficiency nucleoprotein degradation in animal embryos, providing new conditions for animal antiviral research (Kushawah et al., 2020). More recently, in 2021, Yang Hui’s team identified Cas13X and Cas13Y from high-salinity samples and designed an RNA interference experiment for Cas13X.1 in a mammalian cell line (Xu et al., 2021). During the same period, Zhang Feng’s team discovered “Mini”-Cas13bt, functionalizing Cas13bt through the use of adenosine and cytosine deaminase (Kannan et al., 2022).
Collectively, the catalog of CRISPR-Cas13 systems keeps expanding as novel types and orthologs are being described in current studies. At present, based on phylogeny, this important class of effector proteins is categorized into VI-A (Cas13a), VI-B (Cas13b), VI-C (Cas13c), VI-D (Cas13d; Zhang H. et al., 2021), VI-E to I (Cas13e-Cas13i; Hu et al., 2022), and the latest CRISPR-Cas13 system like Cas13X, Cas13Y (Xu et al., 2021) and Cas13bt (Kannan et al., 2022). Here, the classification and average size of Cas13 proteins, which have been widely used in various fields, were summarized (Figure 2).
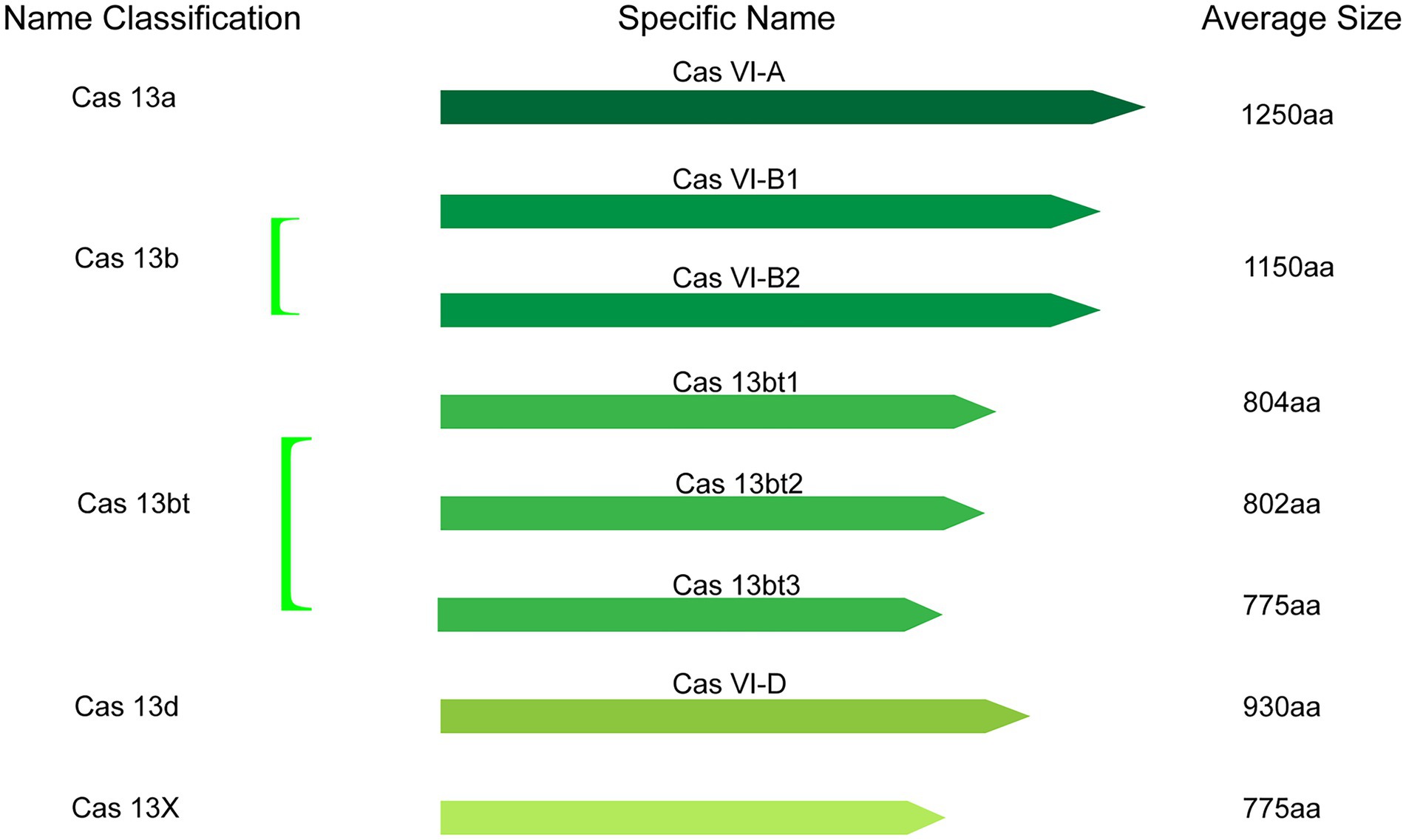
Figure 2. The classification and average size of Cas13 proteins with having been widely used in various fields.
3. The components of the CRISPR-Cas13 system
As shown in Figure 3, using Cas13a as an example, the CRISPR-Cas13 system mainly consists of crRNA (CRISPR RNA) and CRISPR-related nucleases (Cas1, Cas2, and Cas13). The biology of these components is briefly discussed below.
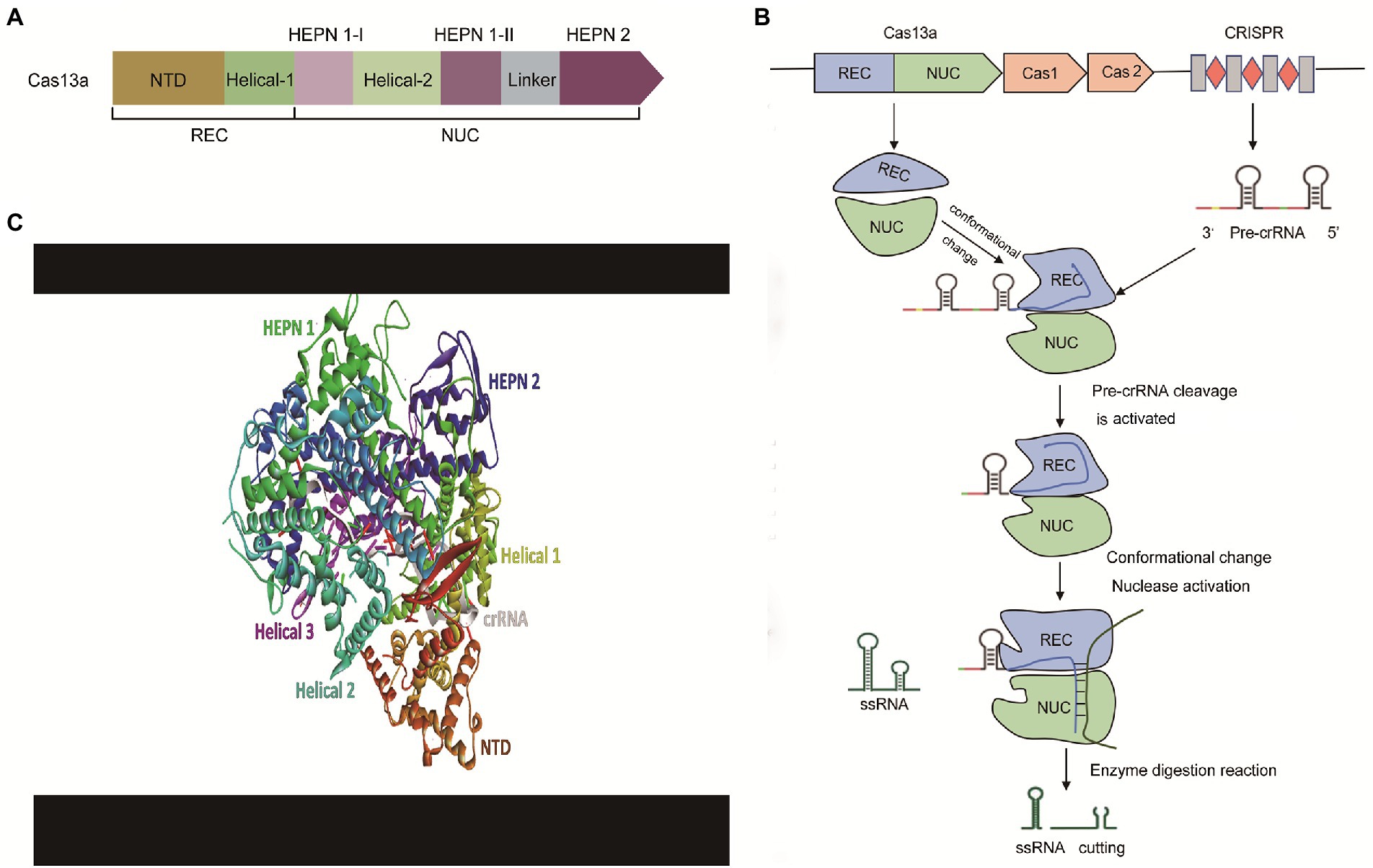
Figure 3. The effector modules and mechanism of action of the CRISPR-Cas13a. (A) The architecture of effector modules of Cas13a. Cas13a has a bi-lobed structure comprising a ‘Nuclease’ (NUC) lobe and a ‘Recognition’ (REC) lobe. NUC Lobe contains two distinct but functionally linked endonuclease sites—Higher Eukaryotes and Prokaryotes Nucleotide-binding Domain 1 and 2 (HEPN-1 and HEPN-2) separated by a basic linker/Helical-3 domain, and a Helical-2 domain. The REC lobe encompasses the N-terminal domain (NTD) and the Helical-1 domain, (B) The mechanism of action of the CRISPR-Cas13a. The pre-crRNA recognizes and binds to the REC leaves of Cas13a. Next, Cas13a processes its pre-crRNA to form a stable crRNA-Cas13a complex. Subsequently, the target ssRNA is recruited to the crRNA-Cas13a complex to undergo base-complementary pairing with the matured crRNA resulting in the activation of the Cas13a endonuclease enzyme. Finally, the activated catalytic domain of Cas13cleaves the target sequence, and (C) Crystal structure of Cas13a complex with crRNA. REC, Recognition lobe; NUC, Nuclease lobe; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; Pre-crRNA, Precursor CRISPR RNA; ssRNA, Single-stranded RNA.
3.1. CRISPR-related nuclease (Cas13)
Cas13a has a bi-lobed structure comprising a ‘Nuclease’ (NUC) lobe and a ‘Recognition’ (REC) lobe. These two domains form a cavity that accommodates the crRNA. NUC Lobe contains two distinct but functionally linked endonuclease sites—Higher Eukaryotes and Prokaryotes Nucleotide-binding Domain 1 and 2 (HEPN-1 and HEPN-2) separated by a basic linker/Helical-3 domain, and a Helical-2 domain (Zhu and Huang, 2018; O'Connell, 2019; Figure 3A). The REC lobe encompasses the N-terminal domain (NTD) and the Helical-1 domain (Liu and Pei, 2022). The HEPN-1 acts as a scaffold that links NUC and REC lobes (Zhang et al., 2018). The formation of the spacer-protospacer binary complex is driving the recognition of the stem-loop (5’handle) of crRNA by NTD. In Cas13d effectors, the α1 component of the HEPN1 and the C-terminal of HEPN-2 domains form a catalytic site that primes the spacer-protospacer binary complex for both targeted and collateral cleavage (Zhang et al., 2018). The 5′ end repeat region of crRNA binds to the REC leaf so that the guide region sequence of crRNA is guided into the cavity, formed in the NUC area (nuclease lobe). The NUC leaf has two different domains, NUC1 and NUC2. The function of these two domains is to “sandwich” the guide region of crRNA, thereby forming a plane, allowing the complementary sequence of the target ssRNA (single-stranded RNA) to bind for cleavage.
3.2. CRISPR RNA
A typical crRNA landscape includes a promoter within an AT-rich leader sequence adjacent to the first CRISPR repeat (Jiang and Doudna, 2015). CRISPR RNA (crRNA) is synthesized from the transcriptions of the CRISPR array into a long precursor crRNA (pre-crRNA) and subsequently processed enzymatically by removing the repeat region and part of the spacer sequence into a smaller matured crRNA (Kenchappa et al., 2013; Jiang and Doudna, 2015). The crRNA is composed of a single variable spacer flanked by short direct CRISPR repeat sequences at the 5′ and 3′ ends. The 5′ end direct repeat (5′ handle) morphs into a short hairpin loop (handle) with a guide segment which when in complex with Cas13 proteins is sandwiched by the HEPN2 of the Nuclease domain and NTD of the Recognition domain (Zhu and Huang, 2018). Downstream, the hairpin loop is a 3–5 nucleotide sequence which promotes firm binding and Cas13 catalytic activities (O'Connell, 2019). The spacer region of crRNA is clamped between the Helical-1 and Helical-2 domains (Zhang et al., 2018). Embedded at the center of the 3′ end of the guide sequence is a “seed” region that directs the scanning-for-target process and ensures firm hybridization with the protospacer of target RNA (Zhu and Huang, 2018; Bandaru et al., 2020). Mismatch at the middle of the seed region of Cas13a maximally reduces the binding affinity, while mismatches elsewhere result in a subtle reduction in binding affinity (Tambe et al., 2018). Thus precise complementarity, particularly in the middle of the seed region, is key for target binding and subsequent cleavage. Of importance, a single base-pair mismatch is sufficient to attenuate Cas13a HEPN activation despite the firm binding affinity between crRNA and target RNA (Tambe et al., 2018).
4. The mechanism of action of the CRISPR-Cas13 system
During bacteria immune surveillance and cleavage of exogenous RNA, CRISPR-Cas13 requires a guide from a matured crRNA (Bayoumi and Munir, 2021; Krohannon et al., 2022). At a glance, this seems a challenge since CRISPR type V1 lacks Cas6 or Cas5d endonuclease activity for processing pre-crRNA into matured crRNA (crRNA biogenesis). Now, it has been shown that Cas13 performs both crRNA biogenesis and RNA cleavage employing two chemically and mechanistically distinct mechanisms for both tasks (East-Seletsky et al., 2016). Using three purified recombinant Cas13 proteins, it was revealed that Cas13 cut at two to five nucleotides upstream of the target pre-crRNA to produce a 60–66 nucleotide-long matured crRNA (Huynh et al., 2020). The resulting matured crRNA contains a single spacer sequence (20–30 nucleotide long) that is complementary to the target RNA sequence and a direct repeat region (Ai et al., 2022).
A generalized molecular action of CRISPR-Cas13 is briefly described in Figures 3B,C. CRISPR-Cas13-mediated cleavage is initiated as pre-crRNA recognizes and binds to the REC leaves of Cas13 to form an intermediate transition state complex. This event induces a conformational change in the conserved residues between Helical-1 and HEPN2 in the NUC region (Zhang et al., 2019). HEPN-2 provides an acid–base catalytic site that catalyzes the processing of pre-crRNA into a matured crRNA (Zhang et al., 2019). Upon complementary pairing between the matured crRNA and target ssRNA, a conformational rearrangement is triggered, as the catalytic HEPN domain of HEPN-2 moves close to the catalytic HEPN domain of HEPN-1 (Zhang et al., 2019). The two HEPN domains combine to form a single catalytically competent active site—which subsequently cleaves the RNA target sequence (Makarova et al., 2017; O'Connell, 2019).
4.1. The collateral damage pitfall of Cas13 system
The utility of CRISPR-Cas13 has been hampered by its promiscuous degradation of unintended RNA targets by the active HEPN domain resulting in collateral damage (Abudayyeh et al., 2016; Smargon et al., 2017; Yan et al., 2018; Ai et al., 2022). This setback seems not typical of all Cas13 systems, but it has been observed in Cas13a of Leptotrichia shahii (Shmakov et al., 2015) and Leptotrichia buccalis (East-Seletsky et al., 2016). It is expected that any efficient Cas13-based editing platform should invariably eliminate the Cas13 off-target activity to increase specificity. To avert this undesirable property, different engineering strategies have been adopted. In one recent study, the truncation of the crRNA spacer length led to the loss of off-target catalytic activities but maintained target binding specificity, and this property was harnessed to effectively knock down multiple transcripts with reduced collateral damage (Abudayyeh et al., 2017). In a different study, transgenic constructs of cas13 (a-d) variants with altered codons were shown to be specific with very low tolerance to off-RNA targets but high fidelity in Drosophila SG4_CD cells (Huynh et al., 2020). Interestingly, Lin et al. have discovered and described novel anti-CRISPR-Cas13a inhibitors (arcVI-A) capable of attenuating RNA targeting and editing in human cells. The application of these molecules could modulate precise RNA editing and also inhibit unintended cleavage (Lin et al., 2020). Altogether, it appears that the off-target property of Cas13 effector proteins is amendable and thus could be efficiently used to edit RNA targets.
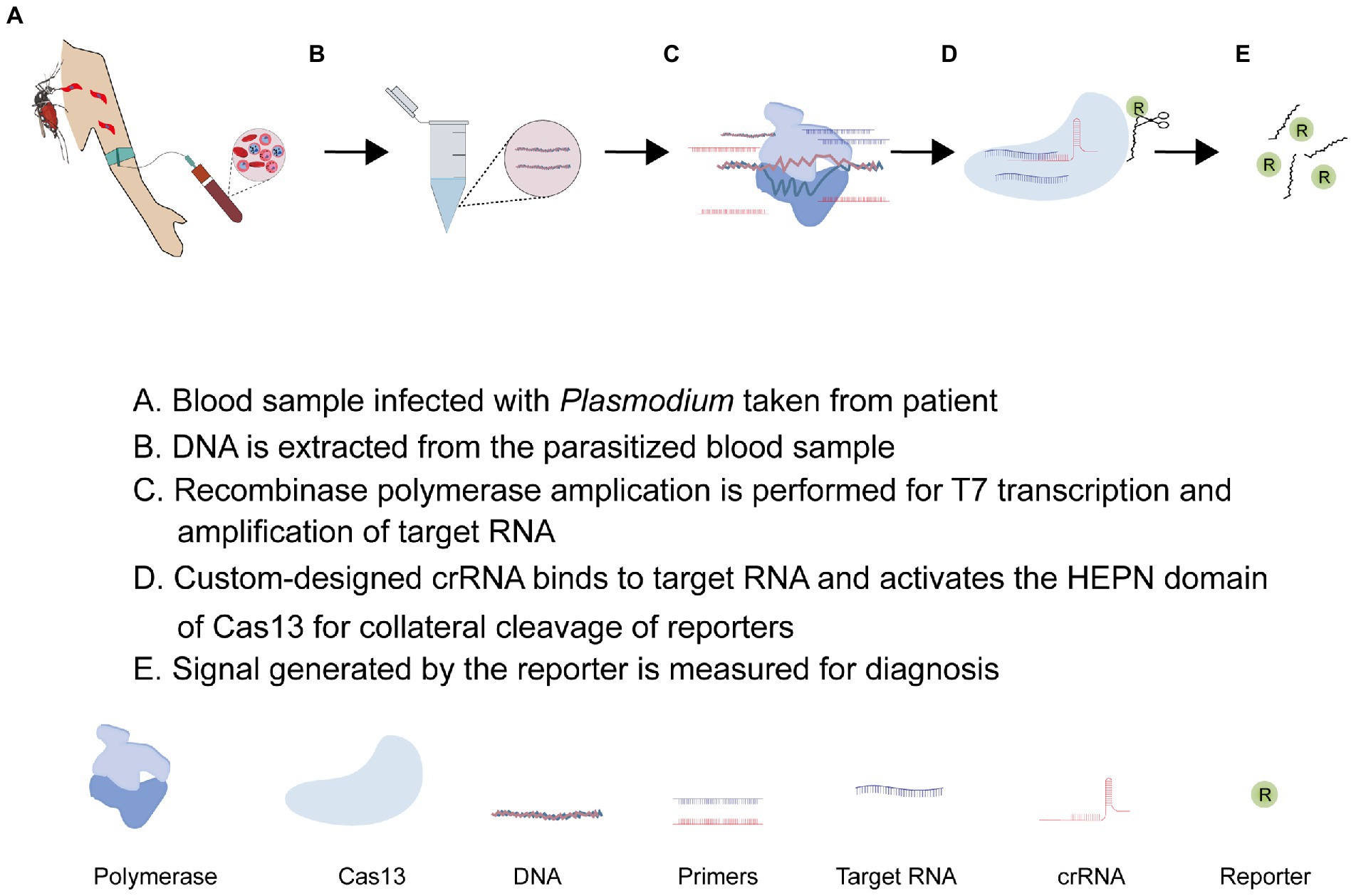
5. CRISPR-Cas13 system as a diagnostic tool
As described earlier, the promiscuous nuclease activity of some Cas13 systems possess a central challenge in their application. This challenge is not essentially negative but opens up new ways for nucleic acid detection if it is carefully harnessed and appropriately applied. This is what the recently developed SHERLOCK platform does. As shown in Figure 4, the SHERLOCK CRISPR tool combines an isothermal nucleic acid amplification technique, and recombinant polymerase amplification technique (RPA), and leverages the indiscriminate endonuclease activity of Cas13 effector protein for clinical diagnosis of diseases (Kellner et al., 2019; Mahas et al., 2021; Mustafa and Makhawi, 2021). While many Cas13 orthologs have been identified and functionally characterized in different bacteria, LwCas13a is the most widely used type in the SHERLOCK applications (Patchsung et al., 2020). Recent advances have seen LwCas13a-based SHERLOCK adopted in different ways (Patchsung et al., 2020). Regardless of the approach, the Cas13-based SHERLOCK platform essentially makes use of: (1) target sequence found in the test sample; (2) T7 RNA polymerase promoter and T7 RNA polymerase for RPA; (3) RNA reporters or sensors sensitive to Cas13 collateral activities; and (4) custom-designed crRNA linked to a programmed Cas13 effector protein for guided detection of target RNA and collateral cleavage of reporters, respectively. The SHERLOCK protocol usually starts with the pre-amplification of the DNA/RNA using RPA primers and the conversion of target dsDNA into ssRNA using the T7 transcriptase enzyme. This is followed by the detection of RNA targets with custom Cas13-crRNA. The custom-designed crRNA is generally composed of a variable spacer region complementary to the target RNA, a constant region that recruits Cas13a, and a T7 promoter sequence that enhances in vitro transcription (Gootenberg et al., 2017; Kellner et al., 2019). Once the spacer of the crRNA aligns with the target RNA, the constant region binds to the Cas13 effector protein resulting in the activation of the off-target HEPN endonuclease activity of Cas13 which consequently cleaves an RNA-tagged reporter to generate a signal for detection (Rajan et al., 2022). To improve signal generation, Yang et al. (2022) engineered a LwCas13a with improved collateral activity by inserting different RNA-binding domains into a unique site within the HEPN domain. The SHERLOCK application has been developed into a cheap and convenient paper-based CRISPR-Cas13-base diagnosing assay capable of detecting and differentiating RNAs of viruses (Zika virus and Dengue virus) and genotyping DNA of bacteria (Escherichia coli and Pseudomonas aeruginosa; Gootenberg et al., 2017; Myhrvold et al., 2018; Manning et al., 2021).
5.1. CRISPR-Cas13-based diagnosis of Plasmodium infection
Besides Cas13a, Class II type V CRISPR-Cas12a (cpf1) has been described to possess a weak collateral activity making it useful in detecting target ssDNA or dsDNA (Nguyen et al., 2020; Kim et al., 2021; Lv et al., 2021; Wei et al., 2021). A Cas12-based SHERLOCK platform has been employed in detecting and differentiating Plasmodium species. In 2020, Lee and his team adopted this application to detect and distinguish P. falciparum (Pfr364 gene)-, P. vivax (18s rRNA)-, P. ovale (18s rRNA)-, and P. malaria (18s rRNA)-specific dsDNA using programmed Cas12a (cpf1) effector proteins (Lee et al., 2020). Noteworthy, the T7 transcription step is skipped, allowing for the direct detection of amplified targeted DNA instead of RNA. This assay could detect less than two parasites per microliter of blood and showed high sensitivity and specificity in differentiating clinical samples of P. falciparum and P. vivax with 100% accuracy. Based on these data, a field-applied diagnostic method for asymptomatic carriers was established for a rapid clinical diagnosis of non-falciparum malaria and low-density P. falciparum infections (Lee et al., 2020). Of notice, their fluorescence read-out assay demonstrated a relatively low fluorescence signal at 50 attomolar (30 parasites per microliter) possibly due to the weak collateral activity of Cas12a. Against this backdrop, the strong collateral activity of Cas13a provides an appropriate alternative to efficiently detect Plasmodium infections using the Cas13-based SHERLOCK platform. An additional advantage of Cas13 is its auto-catalytic ability to process its pre-cRNA without the involvement of tracrRNA and thus allows for the use of multiple guide crRNA in a single streamlined multiplex assay. Accordingly, these properties have been leveraged for a triple-purpose assay involving Plasmodium detection, species differentiation, and drug resistance genotyping using custom-designed crRNA with spacers specific to P. falciparum, P. vivax, and dhps (dihydropteroate synthetase) variants in a single multiplex assay (Cunningham et al., 2021). In briefly, 30–35 nucleotide-long RPA primers tagged with T7 promoter sequences were used to amplify a dhps sequence and/or Plasmodium 18S ribosomal RNA sequence conserved across human-infecting Plasmodium species to generate a short dsDNA. The assay utilized a custom-designed 67 nucleotide-long crRNA composed of a variable spacer region complementary to the target RNA, a constant region that recruits LwCas13a, and a T7 promoter sequence to enhance in vitro transcription. Using T7 polymerase, the resulting short dsDNA of the target sequence is transcribed into ssRNAs in vitro. Once the spacer of the crRNA aligns with the target RNA, the constant region binds to LwCas13a resulting in the activation of the off-target endonuclease activity of LwCas13a which consequently cleaves a fluorescent or colorimetric reporter RNA to generate a signal. Predefined nucleotide variations in dhps are known to confer resistance to sulfadoxine or sulfadoxine-pyrimethamine (SP; Okell et al., 2017), which are the main anti-malarial drug for intermittent preventive treatment in pregnant women. Thus, when the Cas13a-based SHERLOCK platform is improved to a point-of-care diagnostic tool, it will inevitably benefit the quest to track the continuous evolution of anti-malarial drug resistance in endemic areas.
6. Gene manipulating tools at the RNA level in Plasmodium
Most eukaryotic organisms possess an evolutionarily conserved mechanism—RNAi, which regulates genes post-transcriptionally and guards against intrinsic and extrinsic threats by degrading the coding regions of target genes. Typical RNAi-related genes such as Dicer and Argonaute have been identified in Plasmodium, but they lack endogenous RNAi machinery, which hinders gene annotation (Mueller et al., 2014). In this regard, non-canonical RNAi systems have been designed to target RNAs in Plasmodium. In 2020, Franziska Hentzschel’s team designed the rodent Plasmodium berghei to express a minimal, unconventional RNAi mechanism that only requires Ago2 (argonaute 2) and a modified short loop RNA, AgoshRNA. The non-canonical AgoshRNA for target genes structurally encompasses a 5 bp hairpin loop (CTTCA) with a sense sequence (having G-U and G-C mismatches initial and terminal codons, respectively) and an antisense sequence attached to either end of the construct. By integrating an AgoshRNA episomally maintained in plasmid into an RNAi-competent Plasmodium, they achieved an efficient gradient gene-knockout of several genes although its application is currently limited to the erythrocytic-stage parasites (Hentzschel et al., 2020).
To circumvent the occurrence of cell death when essential genes are detected for editing/manipulation, conditional manipulating tools such as glms ribozyme system and the Tet operating system have been designed to manipulate the RNA of Plasmodium (Prommana et al., 2013; Briquet et al., 2022). In Tet systems, the promoter of the target gene is replaced with unstable multiple tetracycline operating sites that bind to a transactivator domain for transcription. This system can be turned off or on depending on the presence or absence of anhydrotetracycline (de Koning-Ward et al., 2015). The glms/riboswitch system is designed to integrate an auto-cleaving ribozyme gene into the target gene sequence which upon expression cleaves the UTR region of the target mRNA to induce degradation (Prommana et al., 2013; de Koning-Ward et al., 2015).
6.1. RNA editing advantages of Cas13
In the last decade, steady progress has been made in the use of CRISPR-Cas13 systems to systematically interrogate the genome of several different organisms. For instance, Cas13 (Cas13a, Cas13d, cas13X, and cas13Y) RNAi systems have been applied for functional characterization of various genes in zebrafish embryos, medaka embryos, mouse embryos, and other mammalian cells demonstrating their practicability in varied cell lines (Cox et al., 2017; Konermann et al., 2018; Kushawah et al., 2020; Xu et al., 2021). Although the CRISPR-Cas13-based system has not been widely applied to RNAs of Plasmodium it would be of great importance if adapted to Plasmodium owing to the advantages discussed below.
The CRISPR-Cas9 system is by far the most established CRISPR-Cas system for interrogating Plasmodium’s genome at the DNA level (Crawford et al., 2017; Nishi et al., 2021). Based on the observation that the HNH catalytic domain of Cas9 is homologous to RNA-cleaving HNH, Cas9 has been repurposed to cleave RNA targets (O’Connell et al., 2014) adding to its already known DNA cleaving property. It has however been shown that the binding affinity and programmable cleaving of target RNA by Cas9 require the presence of sequence-specific PAM-presenting oligonucleotide as a separate DNA, an additional component that is not needed when using Cas13 (O’Connell et al., 2014). In the absence of PAM-presenting oligonucleotides, Cas9 only exhibits steric inhibition of protein translation with no obvious effect on mRNA architecture or levels indicating a limitation to its transcript editing prospects (Liu et al., 2016). Like Cas13, Cas9 is also limited by its unexpected off-target effects (Fu et al., 2013, 2014; Yang et al., 2014) although there have been attempts to experimentally avert this undesirable catalytic property (Fu et al., 2014; Kleinstiver et al., 2016; Slaymaker et al., 2016). Due to the affinity of HNH and RuvC domains of Cas9 to DNA, off-target mutagenesis by Cas9 could imprint unwanted permanent genomic scars on genomic DNA sites having corresponding RNA PAMs or any proximal endogenous locus (Haeussler et al., 2016; Ayabe et al., 2019), which may hinder attempts to associate intended genetic edits to observed experimental results. Cas13 is not constrained by PAM sequence at the target locus (Cox et al., 2017; Tang et al., 2021), making it a flexible tool to use compared to Cas9. Another impediment to the use of Cas9 is that, for DNA targets, edits cannot be made distal (50–100 bp away from) to the point of cut (Elison and Acar, 2018). It is however unknown if this limitation also exists during RNA editing with Cas9. Another peculiar RNA editing advantage of Cas13 is its auto-catalytic ability of Cas13 to process its pre-crRNA, which allows for streamlined delivery of targeting crRNAs for large-scale mRNA transcript editing without loss of specificity and efficiency (Gootenberg et al., 2018; Ackerman et al., 2020; Li et al., 2022; Thakku et al., 2022). This property has already been applied to manipulate a host of genes in live cells (East-Seletsky et al., 2016).
Although the already discussed RNAi systems (Dicer, Argonaute, Tet, and glms ribozyme systems) have great potential, Cas13-based RNA systems have advantages: (1) it does not depend on the cellular RNAi machinery; (2) it is not prone to toxicity from exogenous additives (as in the case of the use of glucosamine-6-phosphate in the glms system); and (3) it can be used in strains the lacks inducible expression systems.
Cas13-based systems however have drawbacks. Firstly, many investigators have shown that the effectiveness of Cas13a, Cas13b, and Cas13d is dependent on the length of crRNA (Abudayyeh et al., 2017; Bandaru et al., 2020; Huynh et al., 2020). It has been demonstrated that the ideal crRNA length is 23–30 nucleotides and any guide shorter or longer may have compromised effectiveness (Abudayyeh et al., 2017; Bandaru et al., 2020; Huynh et al., 2020). Secondly, it has been revealed that crRNA designed to target intronic splice sites or 3’ UTR regions activates lower cleavage activity relative to crRNAs targeting coding regions (Wessels et al., 2020). Thus, an improvement on the combined HEPN catalytic activity of HEPN-1 and HEPN-2 may be required when targeting splice sites or 3’ UTR regions. Thirdly, effective cleavage of mRNAs of essential genes may be toxic to the host cell (Abudayyeh et al., 2016). Lastly, the delivery of less compact Cas13 effectors is derailed by the size of the RNA editors as they exceed the loading capacity of the adeno-associated virus vectors (Hu et al., 2022). Thus, the size of Cas13 effectors is a critical factor to consider in future applications of the CRISPR-Cas13 genomic tool in Plasmodium.
6.2. Prospects of Cas13 for manipulating RNAs in Plasmodium
As earlier mentioned, >70% of Plasmodium’s genome remains functionally uncharacterized. It has indeed been posited that the sparse application of functionally characterizing genetic tools in Plasmodium forecasts that this proportion is unlikely to change if no new methods are developed (Le Roch et al., 2003). In light of this, the addition of the promising CRISPR-Cas13-based tools to the existing tool set for investigating the parasites’ genome will be immensely useful (Figure 5). Below we discuss the prospects of emerging Cas13 systems in alternative RNA editing and identify “hotspots” in Plasmodium research where they could be adapted.
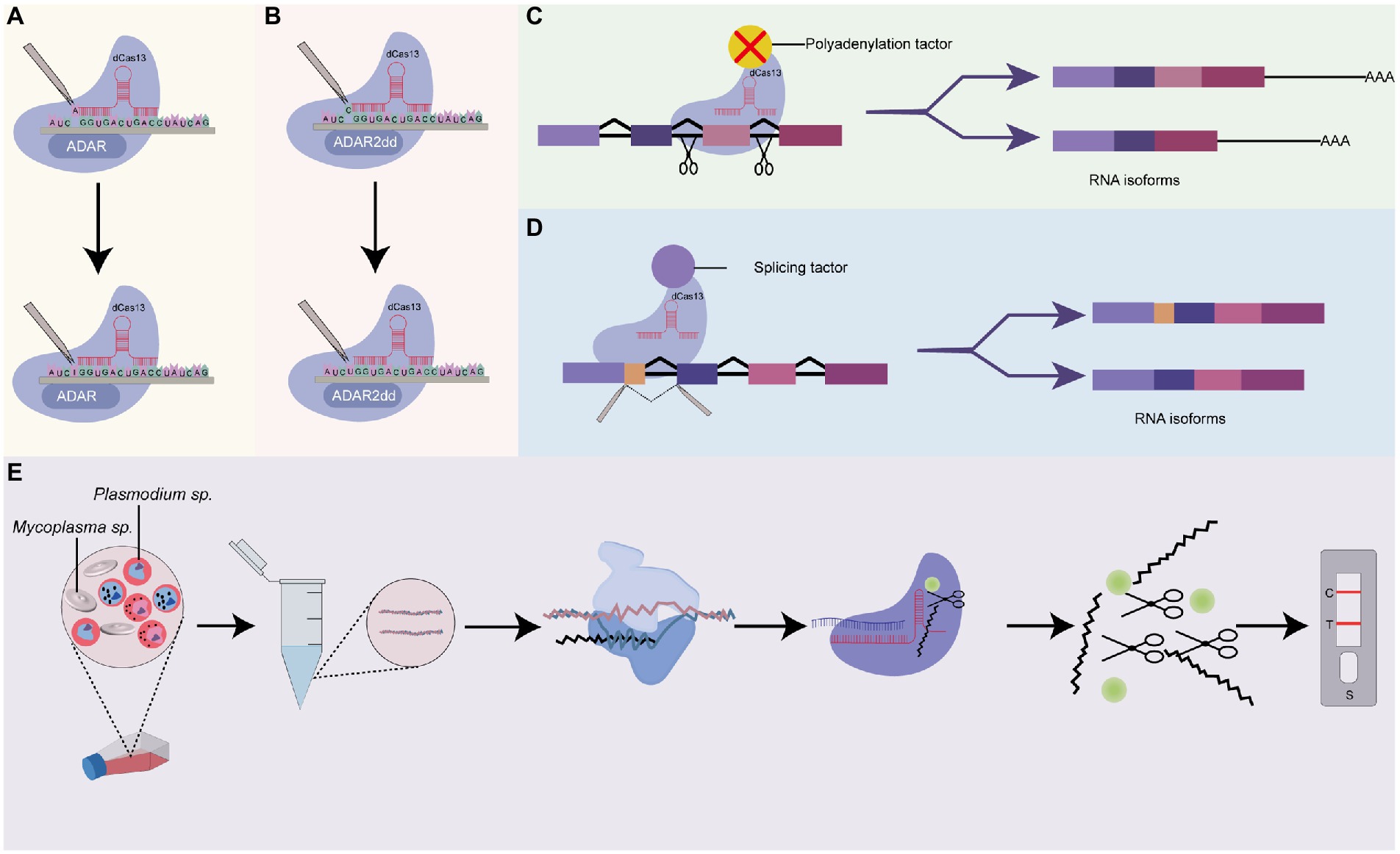
Figure 5. Prospects of Cas13-base systems for RNA editing in Plasmodium. (A) dCas13 tethered to ADARs for a single-nucleotide base editing in RNA. This system uses dCas13 effector-deaminase acting on RNAs (ADARs) domains fusion, for crRNA-guided programmable adenosine to inosine RNA editing, (B) dCas13 tethered to ADAR2dds for a single-nucleotide base editing in RNA. The dCas13 effector is fused with deaminase acting on RNA2 (ADAR2dd) for programmable Cytosine to Uracil RNA editing, (C) dCas13 tethered to a polyadenylation site or a polyadenylation factor. This system uses a dCas13 effector fused with an alternative splicing factor or AAUAAA polyadenylation sites to block access by the polyadenylation machinery. Depending on the positions of the polyadenylation site, it could be used to include or exclude specific exons during pre-mRNA processing, (D) dCas13 tethered to a splicing factor. This system could be used to characterize the functions of RNA isoforms of a specific gene, and (E) Cas13 system for rapid detection of Mycoplasma sp. as a contaminant in Plasmodium culture. CRISPR-Cas13 can be used to target Mycoplasma sp. AfterT7 transcription and amplification of target Mycoplasma sp. RNA, the transcript is subjected to CRISPR-Cas13 detection. The binding of crRNA to the target RNA activates the solvent-exposed HEPN site for non-specific cleavage of nearby RNA-linked reporters to emit a signal. The resulting signal could be measured using lateral flow detection with antigen-labeled reporters.
6.2.1. Hybrid Cas13-based systems as prospective tools for single-base editing in Plasmodium
Recent advances have seen the design of hybrid Cas13-based editing platforms capable of performing single-nucleotide editing in target locus. Generally, as shown in Figures 5A,B, these platforms make use of genetically engineered (mutated) Cas13 effectors (Cas13b or Cas13x.1) fused or tethered to other catalytic domains. One such application is RNA Editing for Programmable A-to-I Replacement version 1/2 (REPAIRv1 and REPAIRv2). The REPAIR systems introduce dead Cas13 effectors (Cas13 with a mutation in HEPN lacking RNase activity) fused with either endogenous exogenous adenosine deaminase acting on RNAs (ADARs) catalytic domain, which substitute adenosine for inosine into target transcripts and thereby stalls translation (Cox et al., 2017). Following the advent of the REPAIR systems, a new platform, RNA Editing for Specific C-to-U Exchange (RESCUE) system and its variants which performs both A-to-I and C-to-U RNA editing have been designed and applied to several genes (Abudayyeh et al., 2019; Li et al., 2021). These systems utilize the fusion of Cas13b with ADR2dd to create synergistic effectors with precise single-base editing capabilities (Abudayyeh et al., 2019; Li et al., 2021).
Although the development of these platforms is still at the embryonic stage (Li et al., 2021; Tang et al., 2021), they may be useful for the study of single-nucleotide polymorphism (SNP) variants and point mutations in Plasmodium if expanded and their efficiencies are improved. In Plasmodium parasites, there is a wealth of SNPs that could be used as proxies for genomic surveillance of drug resistance, and tracking the movement of Plasmodium strains on a global scale (Campino et al., 2011). For instance, a genome-wide integrated analysis of just three parasite strains from the Netherlands, Honduras, and Indochina unraveled 27,000 SNP of which many were uncharacterized (Miles et al., 2016). While few SNPs have been used to genotypically distinguish between parasites originating from different geographical locations, their phenotypic relevance is unknown (Campino et al., 2011; Bankole et al., 2018), suggesting the need to apply novel tools like REPAIR and RESCUE for characterization.
6.2.2. Cas13, a prospective tool for characterizing alternative polyadenylation in Plasmodium
Alternative polyadenylation (APA) is conserved post-transcriptional machinery that promotes transcriptome and proteome diversity through the cleaving of alternative polyadenylation sites, followed by the adding of poly (A) tail to generate isoforms differing at the 3′ ends (Arora et al., 2022). APA shields mRNA from enzymatic degradation and regulates nuclear exports and translation (Zhang Y. et al., 2021). Depending on the site of cleavage, APA events could result in the inclusion or exclusion of exon (exon skipping) and intron retention (Yang et al., 2021) which consequently contributes to a diverse protein repertoire. Employing long-read sequencing techniques, Yang et al. (2021) identified 1,555 alternative polyadenylation sites in transcripts of asexual blood stages of Plasmodium parasites. Further results from this team indicated that 369 (23.7%) of the total transcripts harbored more than five (5) poly (A) sites. A different study showed the presence of AT-rich hexamer, AAUAUU, which putatively serves as a positive signal for polyadenylation in Plasmodium falciparum (Stevens et al., 2018). Nonetheless, the relevance of these APA sites and their corresponding isoforms remains largely unexplored in Plasmodium perhaps due to the unavailability of suitable tools for APA perturbation. Further, the types of APA (tandem 3’ UTR APA, alternative terminal exon APA, intronic APA, internal exon APA) events in Plasmodium remains largely unclassified. Recently, as shown in Figure 5C, Tian et al. (2022) developed a CRISPR-dCas13 system (CRISPR iPAS) that can be used to functionally characterize APA sites. They achieved this by combining EGFP-tagged dCasf13b from Porphyromonas gulae with crRNA-targeting upstream UGUA element or AAUAAA polyadenylation sites core regulation region to block access by the polyadenylation machinery. They demonstrated that, in HEK293T cells, CRISPR iPAS could regulate tandem 3’UTR, alternative terminal exon, and intronic polyadenylation site APA types with high specificity and efficiency. This system could be harnessed to study isoforms of genes that modulate drug resistance, transcriptional and post-transcriptional modification, and cellular differentiation in Plasmodium parasites.
6.2.3. Cas13, a prospective tool for characterizing alternative splicing in Plasmodium
As established, alternative splicing plays an essential role in stage-specific cellular differentiation and the transition of the Plasmodium parasite from the sexual stage to the asexual stage and vice versa (Yeoh et al., 2019; Yang et al., 2021). It has been demonstrated to be a common phenomenon that usually controls sexual dimorphisms in Plasmodium species. Alternative splicing results in the generation of many different matured mRNA transcripts from a single pre-mRNA transcript; creating an important layer of regulation at the RNA level (Iriko et al., 2009). Indeed, a myriad of investigators had sought to quantify and explore the role of alternative splicing in essential genes of Plasmodium using RNA sequencing techniques (Sorber et al., 2011; Gabriel et al., 2015; Turnbull et al., 2017; Yeoh et al., 2019). Excitingly, recent advances in Cas13-based systems have seen the development of programmable mRNA splicing tools that can be used to efficiently characterize splicing events in Plasmodium. Generally, these Cas13-based splicing systems are designed to fuse dCas13 effectors with distinct splice elements for isoform perturbation (Figure 5D). Via this approach, Konermann and co-workers achieved >80% exon exclusion efficiency in human cells using multiple crRNAs targeting intronic branch point, splice acceptor site, exonic splice enhancer, and splice donor (Konermann et al., 2018). Interestingly, this system was adopted differently by Du et al. (2020). In their study, they successfully engineered a CRISPR artificial splicing factor by fusing dCas13 orthologs with splicing regulatory domains. They designed two crRNA that simultaneously target different positions on the sequence element which efficiently excludes or includes exons of interest during the processing of pre-mRNA. These programmable systems could be adapted to interrogate the role of alternative splicing in ApiAP2 transcriptional factors and how they contribute to the stage-dependent functions of ApiAP2 members in Plasmodium species (Quansah et al., 2022). Conceptually, these systems could be adapted for a bulk interrogation of alternative splicing targeting many isoforms of ApiAP2 genes in a single assay.
6.2.4. Cas13, a prospective tool for detecting contaminants in cell cultures
Contaminants such as Mycoplasma usually remain inconspicuous in continuous Plasmodium cultures but substantially alter the property and behavior of infected cells making them potential significant confounders in experimental studies (Malave-Ramos et al., 2022). There is the prospect of repurposing the off-target property of Cas13 for detecting contaminants in long-term in vitro Plasmodium cell cultures (Figure 5E). Cas12a system has already been applied in Mycoplasma as a diagnostic tool proving the applicability of the CRISPR-Cas system in Mycoplasma. The relatively robust collateral activity of Cas13 over Cas12 makes it a more robust tool for the detection of Mycoplasma contaminants in in vitro Plasmodium cultures.
7. Conclusion
This review has summarized the discovery, classification, principle of action, and diagnostic platforms of CRISPR-Cas13 system. Further, it has shown that Cas13-based systems could be prospectively used for single-base editing, and characterizing alternative polyadenylation or alternative splicing in Plasmodium and detecting contaminants in in vitro Plasmodium cultures. The promising CRISPR-Cas13-based tools will be immensely useful for investigating the parasites’ genome.
Author contributions
CZ: conceptualization. YC, EQ, CZ, and LY: writing–original draft. CZ, LY, DS, YC, EQ, SY, JW, YZ, and MC: writing– review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82072304 and 81871671 to LY), grants from Scientific Research of BSKY from Anhui Medical University (XJ201807 to CZ), and the Foundation of Education Department of Anhui province (KJ2021A0213 to CZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR–Cas13. Nature 550, 280–284. doi: 10.1038/nature24049
Abudayyeh, O. O., Gootenberg, J. S., Franklin, B., Koob, J., Kellner, M. J., Ladha, A., et al. (2019). A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386. doi: 10.1126/science.aax7063
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Ackerman, C. M., Myhrvold, C., Thakku, S. G., Freije, C. A., Metsky, H. C., Yang, D. K., et al. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282. doi: 10.1038/s41586-020-2279-8
Ai, Y., Liang, D., and Wilusz, J. E. (2022). CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 50:e65. doi: 10.1093/nar/gkac159
Arora, A., Goering, R., Lo, H. Y. G., Lo, J., Moffatt, C., and Taliaferro, J. M. (2022). The role of alternative polyadenylation in the regulation of subcellular RNA localization. Front. Genet. 12:818668. doi: 10.3389/fgene.2021.818668
Atkins, A., Chung, C. H., Allen, A. G., Dampier, W., Gurrola, T. E., Sariyer, I. K., et al. (2021). Off-target analysis in gene editing and applications for clinical translation of CRISPR/Cas9 in HIV-1 therapy. Front. Genome Ed. 3:673022. doi: 10.3389/fgeed.2021.673022
Ayabe, S., Nakashima, K., and Yoshiki, A. (2019). Off- and on-target effects of genome editing in mouse embryos. J. Reprod. Dev. 65, 1–5. doi: 10.1262/jrd.2018-128
Bandaru, S., Tsuji, M. H., Shimizu, Y., Usami, K., Lee, S., Takei, N. K., et al. (2020). Structure-based design of gRNA for Cas13. Sci. Rep. 10:11610. doi: 10.1038/s41598-020-68459-4
Bankole, B. E., Kayode, A. T., Nosamiefan, I. O., Eromon, P., Baniecki, M. L., Daniels, R. F., et al. (2018). Characterization of plasmodium falciparum structure in Nigeria with malaria SNPs barcode. Malar. J. 17:472. doi: 10.1186/s12936-018-2623-8
Barber, B. E., William, T., Grigg, M. J., Yeo, T. W., and Anstey, N. M. (2013). Limitations of microscopy to differentiate plasmodium species in a region co-endemic for plasmodium falciparum, plasmodium vivax and plasmodium knowlesi. Malar. J. 12:8. doi: 10.1186/1475-2875-12-8
Baum, J., Papenfuss, A. T., Mair, G. R., Janse, C. J., Vlachou, D., Waters, A. P., et al. (2009). Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37, 3788–3798. doi: 10.1093/nar/gkp239
Bayoumi, M., and Munir, M. (2021). Potential use of CRISPR/Cas13 machinery in understanding virus–host interaction. Front. Microbiol. 12:743580. doi: 10.3389/fmicb.2021.743580
Berzosa, P., de Lucio, A., Romay-Barja, M., Herrador, Z., González, V., García, L., et al. (2018). Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar. J. 17:333. doi: 10.1186/s12936-018-2481-4
Briquet, S., Gissot, M., and Silvie, O. J. M. M. (2022). A toolbox for conditional control of gene expression in apicomplexan parasites. Mol. Microbiol. 117, 618–631. doi: 10.1111/mmi.14821
Campino, S., Auburn, S., Kivinen, K., Zongo, I., Ouedraogo, J. B., Mangano, V., et al. (2011). Population genetic analysis of plasmodium falciparum parasites using a customized Illumina GoldenGate genotyping assay. PLoS One 6:e20251. doi: 10.1371/journal.pone.0020251
Cao, H., Wang, Y., Zhang, N., Xia, S., Tian, P., Lu, L., et al. (2022). Progress of CRISPR-Cas13 mediated live-cell RNA imaging and detection of RNA-protein interactions. Front. Cell Dev. Biol. 10:866820. doi: 10.3389/fcell.2022.866820
Cárdenas, P., Esherick, L. Y., Chambonnier, G., Dey, S., Turlo, C. V., Nasamu, A. S., et al. (2022). GeneTargeter: automated in silico Design for Genome Editing in the malaria parasite, Plasmodium falciparum. CRISPR J. 5, 155–164. doi: 10.1089/crispr.2021.0069
Cowman, A. F., Healer, J., Marapana, D., and Marsh, K. (2016). Malaria: biology and disease. Cells 167, 610–624. doi: 10.1016/j.cell.2016.07.055
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. doi: 10.1126/science.aaq0180
Crawford, E. D., Quan, J., Horst, J. A., Ebert, D., Wu, W., and DeRisi, J. L. (2017). Plasmid-free CRISPR/Cas9 genome editing in plasmodium falciparum confirms mutations conferring resistance to the dihydroisoquinolone clinical candidate SJ733. PLoS One 12:e0178163. doi: 10.1371/journal.pone.0178163
Cunningham, C. H., Hennelly, C. M., Lin, J. T., Ubalee, R., Boyce, R. M., Mulogo, E. M., et al. (2021). A novel CRISPR-based malaria diagnostic capable of plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine 68:103415. doi: 10.1016/j.ebiom.2021.103415
de Koning-Ward, T. F., Gilson, P. R., and Crabb, B. S. (2015). Advances in molecular genetic systems in malaria. Nat. Rev. Microbiol. 13, 373–387. doi: 10.1038/nrmicro3450
Du, M., Jillette, N., Zhu, J. J., Li, S., and Cheng, A. W. (2020). CRISPR artificial splicing factors. Nat. Commun. 11:2973. doi: 10.1038/s41467-020-16806-4
East-Seletsky, A., O’ Connell, M. R., Knight, S. C., Burstein, D., Cate, J. H. D., Tjian, R., et al. (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. doi: 10.1038/nature19802
Elison, G. L., and Acar, M. (2018). Scarless genome editing: progress towards understanding genotype-phenotype relationships. Curr. Genet. 64, 1229–1238. doi: 10.1007/s00294-018-0850-8
Fitri, L. E., Widaningrum, T., Endharti, A. T., Prabowo, M. H., Winaris, N., and Nugraha, R. Y. B. (2022). Malaria diagnostic update: from conventional to advanced method. J. Clin. Lab. Anal. 36:e24314. doi: 10.1002/jcla.24314
Freije, C. A., Myhrvold, C., Boehm, C. K., Lin, A. E., Welch, N. L., Carter, A., et al. (2019). Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 76, 826–837.e11. doi: 10.1016/j.molcel.2019.09.013
Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D., Joung, J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. doi: 10.1038/nbt.2623
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808
Gabriel, H. B., De Azevedo, M. F., Palmisano, G., Wunderlich, G., Kimura, E. A., Katzin, A. M., et al. (2015). Single-target high-throughput transcription analyses reveal high levels of alternative splicing present in the FPPS/GGPPS from plasmodium falciparum. Sci. Rep. 5:18429. doi: 10.1038/srep18429
Gaj, T. (2021). Next-generation CRISPR technologies and their applications in gene and cell therapy. Trends Biotechnol. 39, 692–705. doi: 10.1016/j.tibtech.2020.10.010
Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., et al. (2002). Genome sequence of the human malaria parasite plasmodium falciparum. Nature 419, 498–511. doi: 10.1038/nature01097
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444. doi: 10.1126/science.aaq0179
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Graham, D. B., and Root, D. E. (2015). Resources for the design of CRISPR gene editing experiments. Genome Biol. 16:260. doi: 10.1186/s13059-015-0823-x
Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J. B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17:148. doi: 10.1186/s13059-016-1012-2
Haldar, K., Bhattacharjee, S., and Safeukui, I. (2018). Drug resistance in plasmodium. Nat. Rev. Microbiol. 16, 156–170. doi: 10.1038/nrmicro.2017.161
Hartenian, E., and Doench, J. G. (2015). Genetic screens and functional genomics using CRISPR/Cas9 technology. FEBS J. 282, 1383–1393. doi: 10.1111/febs.13248
Hemingway, J., and Bates, I. (2003). Malaria: past problems and future prospects. After more than a decade of neglect, malaria is finally black on the agenda for both biomedical research and public health politics. EMBO Rep. 4, S29–S31. doi: 10.1038/sj.embor.embor841
Hentzschel, F., Mitesser, V., Fraschka, S. A., Krzikalla, D., Carrillo, E. H., Berkhout, B., et al. (2020). Gene knockdown in malaria parasites via non-canonical RNAi. Nucleic Acids Res. 48:e2. doi: 10.1093/nar/gkz927
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Hu, Y., Chen, Y., Xu, J., Wang, X., Luo, S., Mao, B., et al. (2022). Metagenomic discovery of novel CRISPR-Cas13 systems. Cell Discov. 8:107. doi: 10.1038/s41421-022-00464-5
Huynh, N., Depner, N., Larson, R., and King-Jones, K. (2020). A versatile toolkit for CRISPR-Cas13-based RNA manipulation in drosophila. Genome Biol. 21:279. doi: 10.1186/s13059-020-02193-y
Iriko, H., Jin, L., Kaneko, O., Takeo, S., Han, E. T., Tachibana, M., et al. (2009). A small-scale systematic analysis of alternative splicing in plasmodium falciparum. Parasitol. Int. 58, 196–199. doi: 10.1016/j.parint.2009.02.002
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Jansen, R., Embden, J. D. A., Gaastra, W., and Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43, 1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x
Jiang, F., and Doudna, J. A. (2015). The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 30, 100–111. doi: 10.1016/j.sbi.2015.02.002
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kannan, S., Altae-Tran, H., Jin, X., Madigan, V. J., Oshiro, R., Makarova, K. S., et al. (2022). Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 40, 194–197. doi: 10.1038/s41587-021-01030-2
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., and Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Kenchappa, C. S., Heidarsson, P. O., Kragelund, B. B., Garrett, R. A., and Poulsen, F. M. (2013). Solution properties of the archaeal CRISPR DNA repeat-binding homeodomain protein Cbp2. Nucleic Acids Res. 41, 3424–3435. doi: 10.1093/nar/gks1465
Kim, H., Lee, S., Yoon, J., Song, J., and Park, H. G. (2021). CRISPR/Cas12a collateral cleavage activity for simple and rapid detection of protein/small molecule interaction. Biosens. Bioelectron. 194:113587. doi: 10.1016/j.bios.2021.113587
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526
Konermann, S., Lotfy, P., Brideau, N. J., Oki, J., Shokhirev, M. N., and Hsu, P. D. (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cells 173, 665–676.e14. doi: 10.1016/j.cell.2018.02.033
Krohannon, A., Srivastava, M., Rauch, S., Srivastava, R., Dickinson, B. C., and Janga, S. C. (2022). CASowary: CRISPR-Cas13 guide RNA predictor for transcript depletion. BMC Genomics 23:172. doi: 10.1186/s12864-022-08366-2
Kushawah, G., Hernandez-Huertas, L., Del Prado, J. A. N., Martinez-Morales, J. R., DeVore, M. L., Hassan, H., et al. (2020). CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell 54, 805–817. doi: 10.1016/j.devcel.2020.07.013
Lalremruata, A., Jeyaraj, S., Engleitner, T., Joanny, F., Lang, A., Bélard, S., et al. (2017). Species and genotype diversity of plasmodium in malaria patients from Gabon analysed by next generation sequencing. Malar. J. 16:398. doi: 10.1186/s12936-017-2044-0
Le Roch, K. G., Zhou, Y., Blair, P. L., Grainger, M., Moch, J. K., Haynes, J. D., et al. (2003). Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508. doi: 10.1126/science.1087025
Lee, R. A., Puig, H. D., Nguyen, P. Q., Angenent-Mari, N. M., Donghia, N. M., McGee, J. P., et al. (2020). Ultrasensitive CRISPR-based diagnostic for field-applicable detection of plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. U. S. A. 117, 25722–25731. doi: 10.1073/pnas.2010196117
Li, L., Duan, C., Weng, J., Qi, X., Liu, C., Li, X., et al. (2022). A field-deployable method for single and multiplex detection of DNA or RNA from pathogens using Cas12 and Cas13. Sci. China Life Sci. 65, 1456–1465. doi: 10.1007/s11427-021-2028-x
Li, G., Wang, Y., Li, X., Wang, Y., Huang, X., Gao, J., et al. (2021). Developing PspCas13b-based enhanced RESCUE system, eRESCUE, with efficient RNA base editing. Cell Commun. Signal. 19:84. doi: 10.1186/s12964-021-00716-z
Lin, P., Qin, S., Pu, Q., Wang, Z., Wu, Q., Gao, P., et al. (2020). CRISPR-Cas13 inhibitors block RNA editing in bacteria and mammalian cells. Mol. Cell 78, 850–861.e5. doi: 10.1016/j.molcel.2020.03.033
Liu, Y., Chen, Z., He, A., Zhan, Y., Li, J., Liu, L., et al. (2016). Targeting cellular mRNAs translation by CRISPR-Cas9. Sci. Rep. 6:29652. doi: 10.1038/srep29652
Liu, M., Lu, B., Fan, Y., He, X., Shen, S., Jiang, C., et al. (2019). TRIBE uncovers the role of Dis3 in shaping the dynamic transcriptome in malaria parasites. Front. Cell Dev. Biol. 7:264. doi: 10.3389/fcell.2019.00264
Liu, L., and Pei, D. S. (2022). Insights gained from RNA editing targeted by the CRISPR-Cas13 family. Int. J. Mol. Sci. 23:11400. doi: 10.3390/ijms231911400
Lv, H., Wang, J., Zhang, J., Chen, Y., Yin, L., Jin, D., et al. (2021). Definition of CRISPR Cas12a trans-cleavage units to facilitate CRISPR diagnostics. Front. Microbiol. 12:766464. doi: 10.3389/fmicb.2021.766464
Mahas, A., Wang, Q., Marsic, T., and Mahfouz, M. M. (2021). A novel miniature CRISPR-Cas13 system for SARS-CoV-2 diagnostics. ACS Synth. Biol. 10, 2541–2551. doi: 10.1021/acssynbio.1c00181
Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I., and Koonin, E. V. (2006). A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 1:7. doi: 10.1186/1745-6150-1-7
Makarova, K. S., Zhang, F., and Koonin, E. V. J. C. (2017). SnapShot: class 2 CRISPR-Cas systems. Cells 168, 328–328.e1. doi: 10.1016/j.cell.2016.12.038
Malave-Ramos, D. R., Kennedy, K., Key, M. N., Dou, Z., and Kafsack, B. F. (2022). Safe, effective, and inexpensive clearance of mycoplasma contamination from cultures of apicomplexan parasites with Sparfloxacin. Microbiol. Spectr. 10:e03497-22. doi: 10.1128/spectrum.03497-22
Manning, B. J., Khan, W. A., Peña, J. M., Fiore, E. S., Boisvert, H., Tudino, M. C., et al. (2021). High-throughput CRISPR-Cas13 SARS-CoV-2 test. Clin. Chem. 68, 172–180. doi: 10.1093/clinchem/hvab238
Marraffini, L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. doi: 10.1038/nature15386
Memvanga, P. B., and Nkanga, C. I. J. M. J. (2021). Liposomes for malaria management: the evolution from 1980 to 2020. Malar. J. 20:327. doi: 10.1186/s12936-021-03858-0
Miles, A., Iqbal, Z., Vauterin, P., Pearson, R., Campino, S., Theron, M., et al. (2016). Indels, structural variation, and recombination drive genomic diversity in plasmodium falciparum. Genome Res. 26, 1288–1299. doi: 10.1101/gr.203711.115
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Mojica, F. J., Díez-Villaseñor, C., Soria, E., and Juez, G. (2000). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36, 244–246. doi: 10.1046/j.1365-2958.2000.01838.x
Mojica, F. J. M., Ferrer, C., Juez, G., and Rodríguez‐Valera, F. (1995). Long stretches of short tandem repeats are present in the largest replicons of the archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17, 85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x
Mojica, F. J., Juez, G., and Rodríguez-Valera, F. (1993). Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 9, 613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x
Monroe, A., Williams, N. A., Ogoma, S., Karema, C., and Okumu, F. (2022). Reflections on the 2021 world malaria report and the future of malaria control. Malar. J. 21:154. doi: 10.1186/s12936-022-04178-7
Moon, S. B., Kim, D. Y., Ko, J. H., and Kim, Y. S. (2019). Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 51, 1–11. doi: 10.1038/s12276-019-0339-7
Mueller, A. K., Hammerschmidt-Kamper, C., and Kaiser, A. (2014). RNAi in Plasmodium. Curr. Pharm. Des. 20, 278–283. doi: 10.2174/13816128113199990027
Mustafa, M. I., and Makhawi, A. M. (2021). SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases. J. Clin. Microbiol. 59:e00745-20. doi: 10.1128/JCM.00745-20
Myhrvold, C., Freije, C. A., Gootenberg, J. S., Abudayyeh, O. O., Metsky, H. C., Durbin, A. F., et al. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448. doi: 10.1126/science.aas8836
Nguyen, L. T., Smith, B. M., and Jain, P. K. (2020). Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 11:4906. doi: 10.1038/s41467-020-18615-1
Nisbet, R. E. R., Kurniawan, D. P., Bowers, H. D., and Howe, C. J. (2016). Transcripts in the plasmodium Apicoplast undergo cleavage at tRNAs and editing, and include antisense sequences. Protist 167, 377–388. doi: 10.1016/j.protis.2016.06.003
Nishi, T., Shinzawa, N., Yuda, M., and Iwanaga, S. (2021). Highly efficient CRISPR/Cas9 system in plasmodium falciparum using Cas9-expressing parasites and a linear donor template. Sci. Rep. 11:18501. doi: 10.1038/s41598-021-97984-z
O’Connell, M. R., Oakes, B. L., Sternberg, S. H., East-Seletsky, A., Kaplan, M., and Doudna, J. A. (2014). Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266. doi: 10.1038/nature13769
Oboh, M. A., Badiane, A. S., Ntadom, G., Ndiaye, Y. D., Diongue, K., Diallo, M. A., et al. (2018). Molecular identification of plasmodium species responsible for malaria reveals plasmodium vivax isolates in Duffy negative individuals from southwestern Nigeria. Malar. J. 17:439. doi: 10.1186/s12936-018-2588-7
Oboh, M. A., Oriero, E. C., Ndiaye, T., Badiane, A. S., Ndiaye, D., and Amambua-Ngwa, A. (2021). Comparative analysis of four malaria diagnostic tools and implications for malaria treatment in southwestern Nigeria. Int. J. Infect. Dis. 108, 377–381. doi: 10.1016/j.ijid.2021.05.049
O'Connell, M. R. J. J. O. M. B. (2019). Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 431, 66–87. doi: 10.1016/j.jmb.2018.06.029
Okell, L. C., Griffin, J. T., and Roper, C. (2017). Mapping sulphadoxine-pyrimethamine-resistant plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci. Rep. 7:7389. doi: 10.1038/s41598-017-06708-9
Olotu, A., Fegan, G., Wambua, J., Nyangweso, G., Awuondo, K. O., Leach, A., et al. (2013). Four-year efficacy of RTS, S/AS01E and its interaction with malaria exposure. N. Engl. J. Med. 368, 1111–1120. doi: 10.1056/NEJMoa1207564
Ospina-Villa, J. D., López-Camarillo, C., Castañón-Sánchez, C. A., Soto-Sánchez, J., Ramírez-Moreno, E., and Marchat, L. A. (2018). Advances on aptamers against protozoan parasites. Genes (Basel) 9:584. doi: 10.3390/genes9120584
Patchsung, M., Jantarug, K., Pattama, A., Aphicho, K., Suraritdechachai, S., Meesawat, P., et al. (2020). Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 4, 1140–1149. doi: 10.1038/s41551-020-00603-x
Prommana, P., Uthaipibull, C., Wongsombat, C., Kamchonwongpaisan, S., Yuthavong, Y., Knuepfer, E., et al. (2013). Inducible knockdown of plasmodium gene expression using the glmS ribozyme. PLoS One 8:e73783. doi: 10.1371/journal.pone.0073783
Quansah, E., Pappoe, F., Shen, J., Liu, M., Yang, S., Yu, L., et al. (2022). ApiAP2 gene-network regulates Gametocytogenesis in plasmodium parasites. Cell. Microbiol. 2022:5796578. doi: 10.1155/2022/5796578
Rajan, A., Shrivastava, S., Kumar, A., Singh, A. K., and Arora, P. K. (2022). CRISPR-Cas system: from diagnostic tool to potential antiviral treatment. Appl. Microbiol. Biotechnol. 106, 5863–5877. doi: 10.1007/s00253-022-12135-2
Sato, S. (2021). Plasmodium—a brief introduction to the parasites causing human malaria and their basic biology. J. Physiol. Anthropol. 40:1. doi: 10.1186/s40101-020-00251-9
Shmakov, S., Abudayyeh, O. O., Makarova, K. S., Wolf, Y. I., Gootenberg, J. S., Semenova, E., et al. (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397. doi: 10.1016/j.molcel.2015.10.008
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. doi: 10.1126/science.aad5227
Smargon, A. A., Cox, D. B. T., Pyzocha, N. K., Zheng, K., Slaymaker, I. M., Gootenberg, J. S., et al. (2017). Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7. e7. doi: 10.1016/j.molcel.2016.12.023
Sorber, K., Dimon, M. T., and DeRisi, J. L. (2011). RNA-Seq analysis of splicing in plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res. 39, 3820–3835. doi: 10.1093/nar/gkq1223
Stevens, A. T., Howe, D. K., and Hunt, A. G. (2018). Characterization of mRNA polyadenylation in the apicomplexa. PLoS One 13:e0203317. doi: 10.1371/journal.pone.0203317
Tambe, A., East-Seletsky, A., Knott, G. J., Doudna, J. A., and O’Connell, M. R. (2018). RNA binding and HEPN-nuclease activation are decoupled in CRISPR-Cas13a. Cell Rep. 24, 1025–1036. doi: 10.1016/j.celrep.2018.06.105
Tan, Q. W., and Mutwil, M. (2020). Malaria.Tools-comparative genomic and transcriptomic database for plasmodium species. Nucleic Acids Res. 48, D768–d775. doi: 10.1093/nar/gkz662
Tang, T., Han, Y., Wang, Y., Huang, H., and Qian, P. (2021). Programmable system of Cas13-mediated RNA modification and its biological and biomedical applications. Front. Cell Dev. Biol. 9:677587. doi: 10.3389/fcell.2021.677587
Thakku, S. G., Ackerman, C. M., Myhrvold, C., Bhattacharyya, R. P., Livny, J., Ma, P., et al. (2022). Multiplexed detection of bacterial nucleic acids using Cas13 in droplet microarrays. PNAS Nexus. 1:pgac021. doi: 10.1093/pnasnexus/pgac021
Thiam, L. G., Mangou, K., Ba, A., Mbengue, A., and Bei, A. K. (2022). Leveraging genome editing to functionally evaluate plasmodium diversity. Trends Parasitol. 38, 558–571. doi: 10.1016/j.pt.2022.03.005
Tian, S., Zhang, B., He, Y., Sun, Z., Li, J., Li, Y., et al. (2022). CRISPR-iPAS: a novel dCAS13-based method for alternative polyadenylation interference. Nucleic Acids Res. 50:e26. doi: 10.1093/nar/gkac108
Turnbull, L. B., Siwo, G. H., Button-Simons, K. A., Tan, A., Checkley, L. A., Painter, H. J., et al. (2017). Simultaneous genome-wide gene expression and transcript isoform profiling in the human malaria parasite. PLoS One 12:e0187595. doi: 10.1371/journal.pone.0187595
Walker, M. P., and Lindner, S. E. (2019). Ribozyme-mediated, multiplex CRISPR gene editing and CRISPR interference (CRISPRi) in rodent-infectious plasmodium yoelii. J. Biol. Chem. 294, 9555–9566. doi: 10.1074/jbc.RA118.007121
Wei, Y., Zhou, Y., Liu, Y., Ying, W., Lv, R., Zhao, Q., et al. (2021). Indiscriminate ssDNA cleavage activity of CRISPR-Cas12a induces no detectable off-target effects in mouse embryos. Protein Cell 12, 741–745. doi: 10.1007/s13238-021-00824-z
Wessels, H.-H., Méndez-Mancilla, A., Guo, X., Legut, M., Daniloski, Z., and Sanjana, N. E. (2020). Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727. doi: 10.1038/s41587-020-0456-9
World Health Organization (2022). Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022#:~:text=Despite%20continued%20impact%20of%20COVID,further%20setbacks%20to%20malaria%20control
Xu, C., Zhou, Y., Xiao, Q., He, B., Geng, G., Wang, Z., et al. (2021). Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods 18, 499–506. doi: 10.1038/s41592-021-01124-4
Yan, W. X., Chong, S., Zhang, H., Makarova, K. S., Koonin, E. V., Cheng, D. R., et al. (2018). Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5. doi: 10.1016/j.molcel.2018.02.028
Yan, F., Wang, W., and Zhang, J. (2019). CRISPR-Cas12 and Cas13: the lesser known siblings of CRISPR-Cas9. Cell Biol. Toxicol. 35, 489–492. doi: 10.1007/s10565-019-09489-1
Yang, L., Grishin, D., Wang, G., Aach, J., Zhang, C. Z., Chari, R., et al. (2014). Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 5:5507. doi: 10.1038/ncomms6507
Yang, M., Shang, X., Zhou, Y., Wang, C., Wei, G., Tang, J., et al. (2021). Full-length transcriptome analysis of plasmodium falciparum by single-molecule long-read sequencing. Front. Cell. Infect. Microbiol. 11:631545. doi: 10.3389/fcimb.2021.631545
Yang, J., Song, Y., Deng, X., Vanegas, J. A., You, Z., Zhang, Y., et al. (2022). (Online ahead of print)). Engineered LwaCas13a with enhanced collateral activity for nucleic acid detection. Nat. Chem. Biol. 19, 45–54. doi: 10.1038/s41589-022-01135-y
Yang, L. Z., Wang, Y., Li, S. Q., Yao, R. W., Luan, P. F., Wu, H., et al. (2019). Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell 76, 981–997. doi: 10.1016/j.molcel.2019.10.024
Yeoh, L. M., Goodman, C. D., Mollard, V., McHugh, E., Lee, V. V., Sturm, A., et al. (2019). Alternative splicing is required for stage differentiation in malaria parasites. Genome Biol. 20:151. doi: 10.1186/s13059-019-1756-6
Zhang, C., Konermann, S., Brideau, N. J., Lotfy, P., Wu, X., Novick, S. J., et al. (2018). Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cells 175, 212–223.e17. doi: 10.1016/j.cell.2018.09.001
Zhang, Y., Liu, L., Qiu, Q., Zhou, Q., Ding, J., Lu, Y., et al. (2021). Alternative polyadenylation: methods, mechanism, function, and role in cancer. J. Exp. Clin. Cancer Res. 40:51. doi: 10.1186/s13046-021-01852-7
Zhang, H., Qin, C., An, C., Zheng, X., Wen, S., Chen, W., et al. (2021). Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol. Cancer 20:126. doi: 10.1186/s12943-021-01431-6
Zhang, C., Xiao, B., Jiang, Y., Zhao, Y., Li, Z., Gao, H., et al. (2014). Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5:e01414. doi: 10.1128/mBio.01414-14
Zhang, B., Ye, Y., Ye, W., Perčulija, V., Jiang, H., Chen, Y., et al. (2019). Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat. Commun. 10:2544. doi: 10.1038/s41467-019-10507-3
Zhang, D., Zhang, Z., Unver, T., and Zhang, B. (2021). CRISPR/Cas: a powerful tool for gene function study and crop improvement. J. Adv. Res. 29, 207–221. doi: 10.1016/j.jare.2020.10.003
Keywords: malaria, Plasmodium , CRISPR-Cas system, CRISPR-Cas13 diagnosis, CRISPR-Cas13 RNA editing
Citation: Quansah E, Chen Y, Yang S, Wang J, Sun D, Zhao Y, Chen M, Yu L and Zhang C (2023) CRISPR-Cas13 in malaria parasite: Diagnosis and prospective gene function identification. Front. Microbiol. 14:1076947. doi: 10.3389/fmicb.2023.1076947
Edited by:
Jian Li, Hubei University of Medicine, ChinaReviewed by:
Paul R. Gilson, Burnet Institute, AustraliaKarine Gaelle Le Roch, University of California, Riverside, United States
Copyright © 2023 Quansah, Chen, Yang, Wang, Sun, Zhao, Chen, Yu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Yu, ✉ bGlseXl1MzNAMTI2LmNvbQ==; Chao Zhang, ✉ aGFwcHkyMDA4Y29uQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
 Elvis Quansah
Elvis Quansah Yihuan Chen
Yihuan Chen Shijie Yang
Shijie Yang Junyan Wang2
Junyan Wang2 Yangxi Zhao
Yangxi Zhao Chao Zhang
Chao Zhang