- 1Department of Minimally Invasive Gynecology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing Maternal and Child Health Care Hospital, Beijing, China
- 2Department of Gynecology, Shengli Clinical Medical College of Fujian Medical University, Fujian Provincial Hospital, Fuzhou, China
Introduction: The endometrial microbiota plays an essential role in the health of the female reproductive system. However, the interactions between the microbes in the endometrium and their effects on adenomyosis remain obscure.
Materials and methods: We profile endometrial samples from 38 women with (n=21) or without (n=17) adenomyosis to characterize the composition of the microbial community and its potential function in adenomyosis using 5R 16S rRNA gene sequencing.
Results: The microbiota profiles of patients with adenomyosis were different from the control group without adenomyosis. Furthermore, analysis identified Lactobacillus zeae, Burkholderia cepacia, Weissella confusa, Prevotella copri, and Citrobacter freundii as potential biomarkers for adenomyosis. In addition, Citrobacter freundii, Prevotella copri, and Burkholderia cepacia had the most significant diagnostic value for adenomyosis. PICRUSt results identified 30 differentially regulated pathways between the two groups of patients. In particular, we found that protein export, glycolysis/gluconeogenesis, alanine, aspartate, and glutamate metabolism were upregulated in adenomyosis. Our results clarify the relationship between the endometrial microbiota and adenomyosis.
Discussion: The endometrial microbiota of adenomyosis exhibits a unique structure and Citrobacter freundii, Prevotella copri, and Burkholderia cepacia were identified as potential pathogenic microorganisms associated with adenomyosis. Our findings suggest that changes in the endometrial microbiota of patients with adenomyosis are of potential value for determining the occurrence, progression, early of diagnosis, and treatment oadenomyosis.
1. Introduction
Adenomyosis (ADS) is one of the most challenging problems in benign uterine disease. This is because the mechanisms associated with this condition remain unclear and there are no unified and effective treatment options at present. The Endometriosis Committee of the Chinese Medical Doctor Association of Obstetricians and Gynecologists defines ADS as an eutopic endometrium that invades and migrates into the uterine myometrium, a condition that affects 7–23% of women of reproductive age (García-Solares et al., 2018; Leng et al., 2020). The recognized mechanisms of ADS include the invasion of the endometrial basalis into the myometrium, microtrauma of the endometrial-myometrial interface, genetics, immune disorders, and inflammatory mediators (An et al., 2017; Vannuccini et al., 2017; Guo, 2020; Zhai et al., 2020; Guo et al., 2022). However, the clinical presentations of patients with ADS are heterogeneous, thus indicating that unknown pathological mechanisms may exist (Bourdon et al., 2021).
Several studies have proposed that ADS is associated with an imbalance in the vagina microbiota (Chen et al., 2020; Chao et al., 2021; Kunaseth et al., 2022). Lactobacillus is known to dominate the bacterial composition of a healthy vagina and deters the growth of opportunistic pathogens; however, dysbiosis can potentially lead to the occurrence of disease. Chronic pelvic pain (CPP) has been associated with both endometriosis (EM) and ADS. The vaginal microbiota of patients with EM/ADS-associated CPP is known to feature a higher abundance of Clostridium butyricum, Clostridium disporicum, Alloscardovia omnicolens, and Veillonella montpellierensis (Chao et al., 2021). In a previous study, Kunaseth et al. (2022) reported that Alloscardovia, Oscillospirales, Ruminoccoccaceae, Oscillospiraceae, Enhydrobacter, Megamonas, Selenomonadaceae, and Faecalibacterium all showed an increased abundance in the vaginal microbiota of patients with ADS. Atopobium is a known biomarker for patients with EM combined with ADS of the vaginal microbiota (Chen et al., 2020).
The uterus has long considered as a sterile environment. However, several studies have shown that the endometrium is colonized by bacterial strains that differ from the vaginal microbiota (Moreno et al., 2016; Chen et al., 2017). Dysbiosis of the endometrial microbiota is likely to lead to endometrial cancer (Walther-António et al., 2016; Sobstyl et al., 2022), polyps (Fang et al., 2016), endometriosis (Akiyama et al., 2019; Wessels et al., 2021), endometritis (Moreno et al., 2018; Hernandes et al., 2020), and recurrent pregnancy loss (Łaniewski et al., 2020). A recent study confirmed that Porphyromonas somerae represents a suitable biomarker for endometrial cancer (Walsh et al., 2019). Fang et al. (2016) found that Lactobacillus, Bifidobacterium, Gardnerella, and Streptococcus were increased in women with endometrial polyps. A previous study found that the relative abundances of the Streptococcaceae, Moraxellaceae, Staphylococcaceae, and Enterobacteriaceae families were significantly increased among females with endometriosis (Akiyama et al., 2019). However, research on the endometrial microbiota in patients with ADS is rare. In this study, we aimed to investigate the composition and biomarkers of the microbiota in endometrial samples from females with and without ADS using 5R 16S rRNA gene sequencing (Nejman et al., 2020; Poore et al., 2020). We also investigated whether biomarkers of the endometrial microbiota could be used to diagnose ADS and then predict the potential interactions between microbiota and ADS.
2. Materials and methods
The study was performed according to the guidelines of the Declaration of Helsinki and was approved by the Ethics Committee of the Beijing Obstetrics and Gynecology Hospital, Capital Medical University No. 2016-KY-012-02.
2.1. Patients
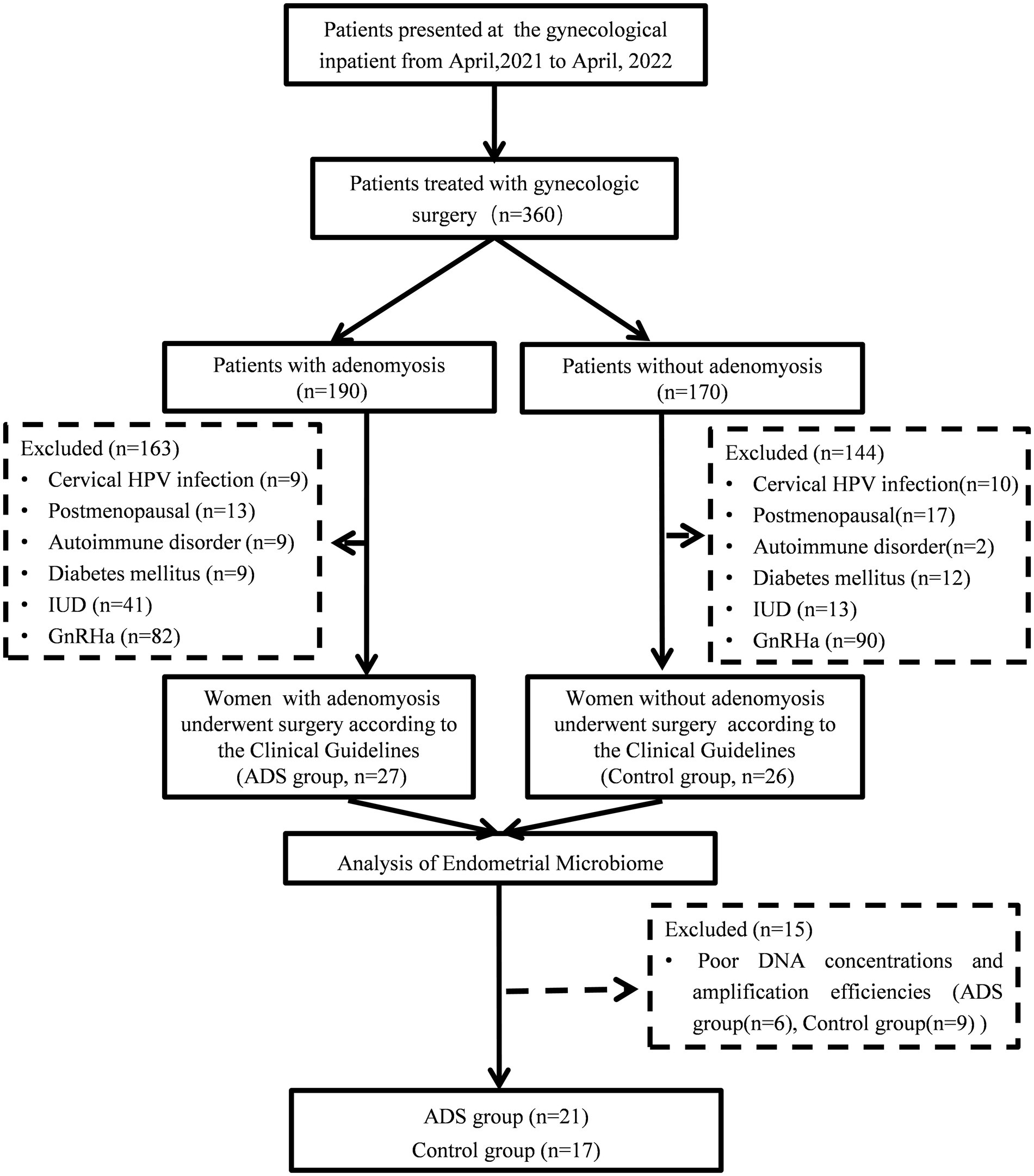
In total, we recruited 360 females who received laparoscopic hysterectomy and were pathologically diagnosed with and without adenomyosis between April 2021 and April 2022 at the Institute of Minimally Invasive Gynecology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University. All participants were fully informed and provided signed informed consent in accordance with the Declaration of Helsinki. The diagnosis of adenomyosis was confirmed by histological examination by two pathologists as the presence of ectopic endometrial tissue (endometrial stroma and glands) within the myometrium. The exclusion criteria were as follows: (1) postmenopausal women; (2) women who received preoperative hormone or gonadotropin-releasing hormone agonist (GnRHa) therapy for at least 3 months prior to sampling; (3) women who were diagnosed with endometrial disease, such as endometrial carcinoma and atypical endometrial hyperplasia; (4) women who had been confirmed with acute genital tract inflammation or with diseases of the lower urinary tract; (5) women with chronic diseases, including autoimmune disorders, diabetes mellitus, coronary heart disease, and hypertension; (6) women with cervical HPV infection and fever; (7) women with intrauterine devices, and (8) patients who had taken antibiotics over the last 3 months before sample collection (except mandatory prophylactic antibiotic treatment before surgery). Three hundred and seven patients (163 from the ADS group and 144 from the control group) were excluded from this study. Besides, 15 endometrial samples (6 from the ADS group and 9 from the control group) did not present PCR products for sequencing and were considered negative. Thirty-eight participants (twenty-one from the ADS group and seventeen from the control group) were ultimately included in the final analysis. The overall procedure is outlined in Figure 1.
2.2. Endometrial samples
Samples were collected from participants during surgery. Following hysterectomy, the ‘Y’ shape uterus was immediately opened with a sterile knife by incising along the central line from the fundus to the external uterine ostium and then exposing each uterine horn. Specimens for subsequent research were obtained from the fundus uteri with a sterile cotton swab, including the posterior and anterior wall of the endometrium. Each specimen was placed in a sterile tube and immediately transported to the laboratory in an icebox. Samples were subsequently stored in a −80°C refrigerator to await DNA extraction.
2.3. DNA extraction, 5R 16S rRNA gene amplification, and sequencing
Genomic DNA was extracted from samples by CTAB methods and 1% agarose gels were used to determine the concentration and quality of DNA. The five (V2, V3, V5, V6, and V8) regions on the 16S rDNA were amplified with specific primers (F1, 5′-TGGCGAACGGGTGAGTAA-3′; F2, 5′-ACTCCTACGGGAGGCAGC-3′; F3, 5′-GTGTAGCGGTGRAATGCG-3′; F4, 5′-GGAGCATGTGGWTTAATTCGA-3′; F5, 5′-GGAGGAAGGTGGGGATGAC-3′; R1, 5′-AGACGTGTGCTCTTCCGAAGACGTGTGCTCTTCCGA-3′; R2, 5′-AGACGTGTGCTCTTCCGATCTGTATTACCGCGGCTGCTG-3′; R3, 5′-AGACGTGTGCTCTTCCGATCTCCCGTCAATTCMTTTGAGTT-3′; R4, 5′-AGACGTGTGCTCTTCCGATCTCGTTGCGGGACTTAACCC-3′; R5,5′-AGACGTGTGCTCTTCCGATCTAAGGCCCGGGAACGTATT-3′; FF1,5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGGCGAACGGGTGAGTAA-3′; FF2, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTACTCCTACGGGAGGCAGC-3′; FF3, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTGTAGCGGTGRAATGCG-3′; FF4, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGGAGCATGTGGWTTAATTCGA-3′; FF5, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGGAGGAAGGTGGGGATGAC-3′; RR5, 5′-CAAGCAGAAGACGGCATACGAGAT-NNNNNNNN-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′). Polymerase chain reaction (PCR) was carried out with Phusion Hot Start Flex 2X Master Mix (New England Biolabs, M0536L). The cycling conditions for PCR were as follows: 98°C for 30 s, followed by 30 cycles of 98°C for 10 s, 62°C for 10 s, 62°C for 15 s, and another 5 min at 72°C. Subsequently, PCR products were purified by applying the E.Z.N.A. Gel Extraction Kit and libraries were sequenced on an Illumina NovaSeq 6,000 system (LC-Bio, Hangzhou, China). Sampling controls (blank control), DNA extraction controls (negative control), and no-template PCR amplification controls (negative control) were also utilized. The Short Multiple Regions Framework (SMURF) method was used to read counts from the five regions into coherent profiling results, classify bacterial taxonomy, and analyze the relative abundance of bacteria (Fuks et al., 2018; Nejman et al., 2020). Sequence reads were normalized to reduce the impact of low abundance noise when the raw reads were less than 1,000 and the relative abundance of bacteria was less than 10−4. Moreover, we used the prevalence of bacteria in the negative control samples to determine the abundance of contaminating bacteria from sampling and experimental processes. The threshold for identifying contaminated bacteria was set at a prevalence of 50%. The GreenGenes database (version: 20131) was used as a reference database for the SMURF method.
2.4. Statistical analysis
In this study, we used a blinded approach that the analyst unaware of the patients group allocation in all statistical analyses. Clinical statistical analysis was performed in SPSS software (version 25.0). Continuous variables were analyzed by t-tests while categorical variables were analyzed by Fisher’s exact test; p < 0.05 was considered statistically significant. The raw sequence reads were analyzed by the vsearch and usearch tools in Rstudio. A bacterial abundance table was generated to analyze the differences in bacterial profiles between the ADS and control group. R and STAMP v.2.1.3. were used to visualize the taxonomic composition of microbial communities. Alpha diversity was computed using the Mann–Whitney test and p < 0.05 was considered statistically. Rarefaction curves were used to estimate whether the sample size could comprehensively reflect diversity. Community clustering was measured by principal component analysis (PCA) and analyzed by the Adonis test; p < 0.05 was considered statistically significant. The Linear Discriminative Analysis Effect Size (LEfSe) method was used to identify distinct endometrial microbiota between different groups and explore potential biomarkers (Segata et al., 2011; Parks et al., 2014).2 Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, version 2.03) (Douglas et al., 2019) was performed to identify the different functions of the endometrial bacteria and to profile Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology functions.4 Statistically significant results (p < 0.05) were presented in the figures by STAMP v2.1.3 (Parks et al., 2014).
3. Results
3.1. Sociodemographic and samples
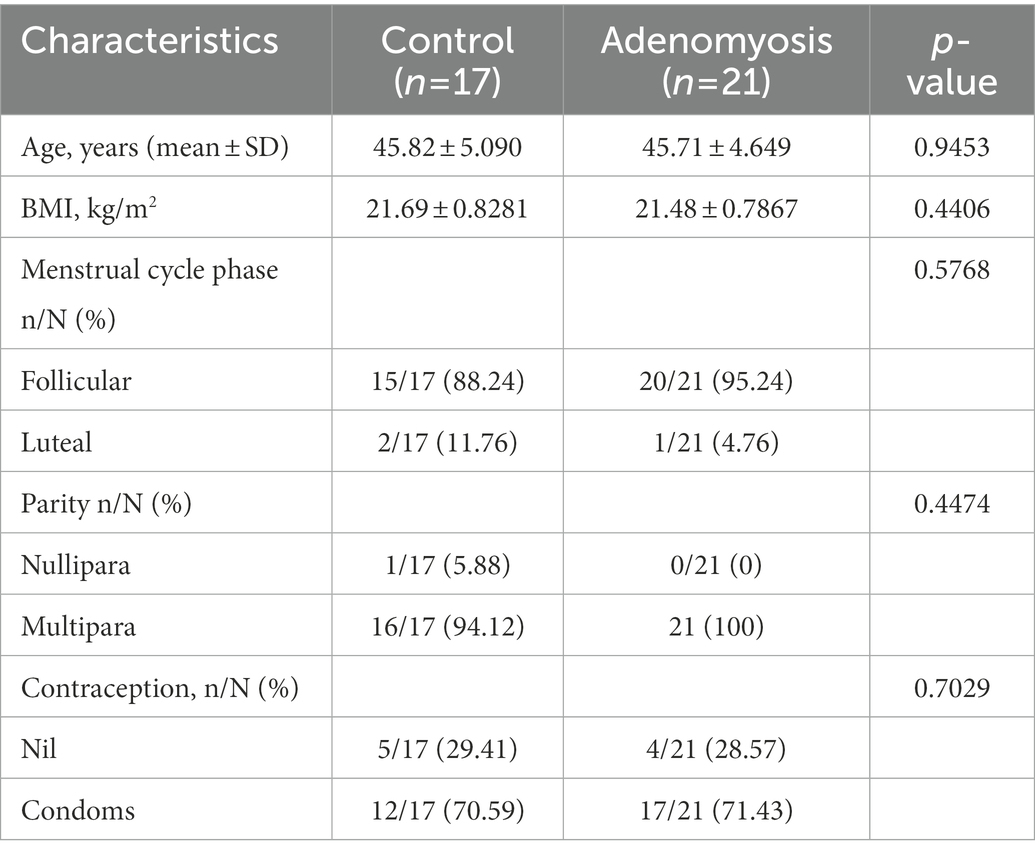
A total of 53 participants were included in this study and each participant provided endometrial samples; 21 samples were obtained from ADS patients and 17 samples obtained from patients without adenomyosis (the control group). These samples were used for PCR and produced high qualify DNA. All the recruited participants received hysterectomy and their surgical and pathological reports were available for analysis. The mean age was 45.82 ± 5.090 years in the control group and 45.71 ± 4.649 years in the ADS group; there was no significant difference between the two groups in terms of age (p = 0.9453). Furthermore, no significant differences were found in terms of body mass index (BMI), menstrual cycle phase, parity, or contraceptive method when compared between the two groups (p = 0.4406, p = 0.5768, p = 0.4474, and p = 0.7029, respectively). Table 1 shows the clinical features of the patients at baseline.
3.2. Sequencing coverage
5R 16S rRNA gene analysis was performed on the remaining 21 patients from the ADS group and 17 from the control group. A total of 3,703,825 raw reads (97,469.07895 reads per sample) and 3,277,880 clean reads (86,260 reads per sample) were obtained from 38 samples with an average Q30 percentage of 91.12% (Supplementary Table S1). The alpha rarefaction curve indicated a reasonable sequencing depth and Good’s coverage was almost 0.99 in the two groups (Supplementary Figure S1).
3.3. Identification of endometrial microbiota
The distribution of endometrial microbiota differed between the two groups at the phylum, genus, and species levels. A total of 24 phyla, 651 genera, and 1,931 species were detected (Figures 2A–C). The distribution of the top 10 taxa of the endometrial microbiota at the phylum, genus, and species levels is depicted in Figures 2D–F. At the phylum level (Figure 2D), the bacterial distribution in the ADS group was dominated by Firmicutes (43.61%), followed by Proteobacteria (31%), Actinobacteria (17.11%), and Bacteroidetes (6.68%). However, most species in the control group were Proteobacteria (62.94%), followed by Firmicutes (24.68%), Actinobacteria (6.09%), Bacteroidetes (3.86%), and Cyanobacteria (1.66%). At the genus level (Figure 2E), Lactobacillus (22.58%), Corynebacterium (5.32%), Streptococcus (4.17%), Prevotella (3.94%), Hyphomicrobium (3.90%), Geobacillus (3.78%), and Vibrio (3.34%) were the predominant abundant bacterial in the ADS group. In contrast, Vibrio (29.71%) were predominant in the control group. Lactobacillus iners (L. iners), belonging to the Firmicutes phylum, was the most dominant bacteria in the ADS group at the species level, while the main proportion in the control group was Vibrio metschnikovii (V. metschnikovii; phylum: Proteobacteria) (Figure 2F).
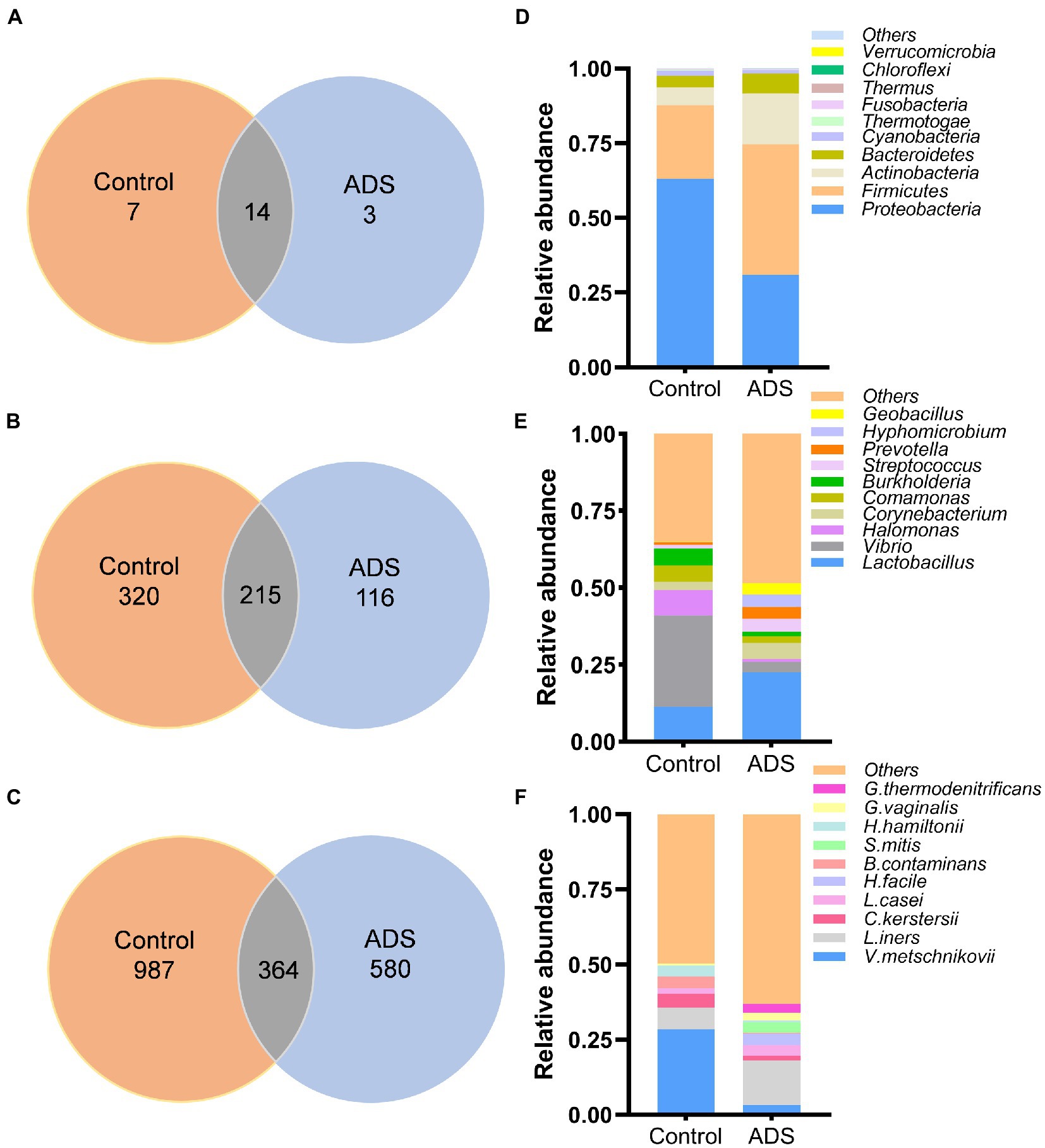
Figure 2. Disparities in bacterial distribution between the adenomyosis (ADS) group and control group. Venn diagram representing the total number of bacterial at the phylum (A), genus (B), and species (C) levels; Colum chart showing the top 10 taxa of endometrial microbiota between the two groups at the phylum (D), genus (E), and species (F) levels. V. metschnikovii, Vibrio metschnikovii; L. iners, Lactobacillus iners; C. kerstersii, Comamonas kerstersii; B. contaminans, Burkholderia contaminans; H. hamiltonii, Halomonas hamiltonii; L. casei, Lactobacillus casei; G. vaginalis, Gardnerella vaginalis; G. thermodenitrificans, Geobacillus thermodenitrificans; H. facile, Hyphomicrobium facile; S. mitis, Streptococcus mitis.
3.4. Diversity analysis of the endometrial microbiota between the two groups: Resource identification initiative
To estimate and determine the endometrial microbiota richness and diversity, we performed alpha and beta diversity analysis. Alpha diversity, including Chao1, Shannon index, Simpson index, and Observed species, was used to generate four column diagrams. As shown in Figure 3, there was no difference in the Shannon and Simpson indices when compared between the two groups (p > 0.05). In contrast, the Chao1 (p = 0.0303) and Observed species (p = 0.0340) in the control group were significantly higher than in the ADS group (p < 0.05). These results demonstrated a significant reduction in the endometrial microbial richness in patients with adenomyosis.
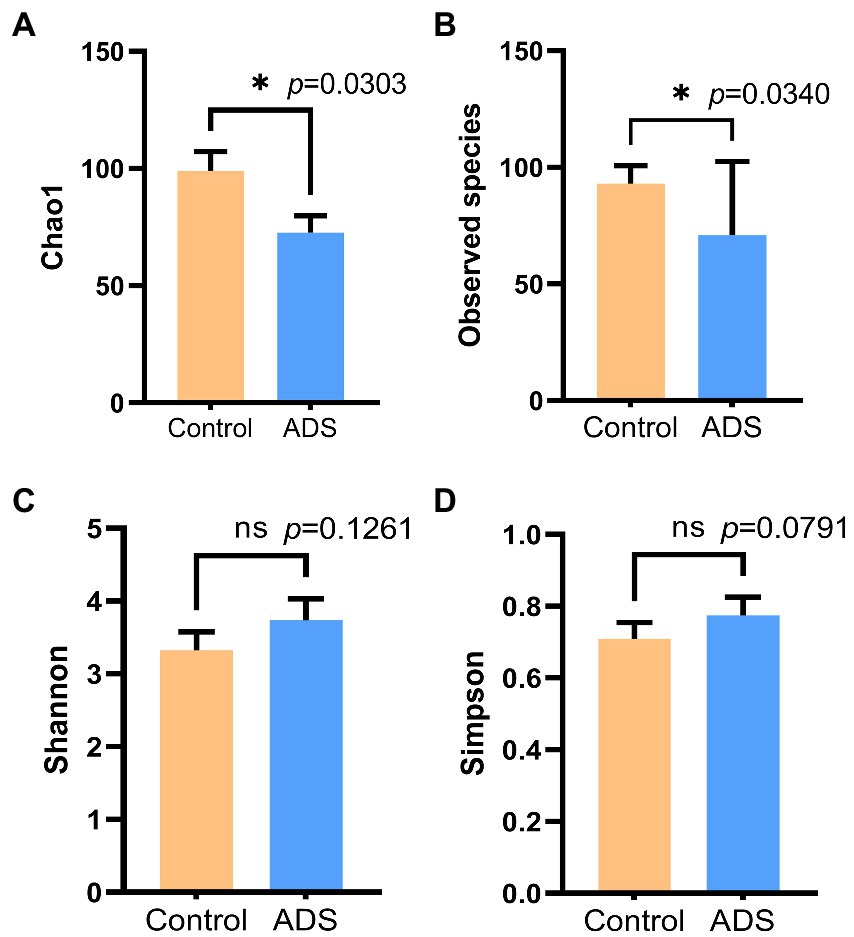
Figure 3. Alpha diversity analysis of the endometrial microbiota of women with or without adenomyosis. Chao1 (A), Observed species (B), Shannon index (C), and Simpson index (D) were analyzed by the Mann–Whitney test. *p < 0.05 was statistically significant.
In addition, we also performed beta diversity analysis to reveal differences in the endometrial microbiota structure between the two groups. As previously shown in Figure 2, for patients with adenomyosis, Firmicutes and Actinobacteria were the most significant bacteria at the phylum level when compared with the control group. Lactobacillus was more dominant than the control group at the genus level. At the species level, Lactobacillus iners (L. iners), Halomonas hamiltonii (H. hamiltonii), and Streptococcus mitis (S. mitis) were more abundant than in the control group. The relative abundance of the five top-ranked bacteria in the endometrial microbiota was selected for further analysis among the two groups. In total, five differential bacterial phyla, genera, and species were depicted in a circular diagram (Figures 4A–C). These results are consistent with the previous conclusion that Lactobacillus and Corynebacterium dominated in adenomyosis. In contrast, Vibrio, Halomonas, and Comamonas were dominant in the control group. Principal component analysis (PCA) was used to analyze the relative abundance of bacteria and beta diversity index was determined by Bray–Curtis analysis at the species level. Figure 4D shows that the distribution and structure of the endometrial flora in the ADS group were significantly separate and different from the control group (p = 0.005).
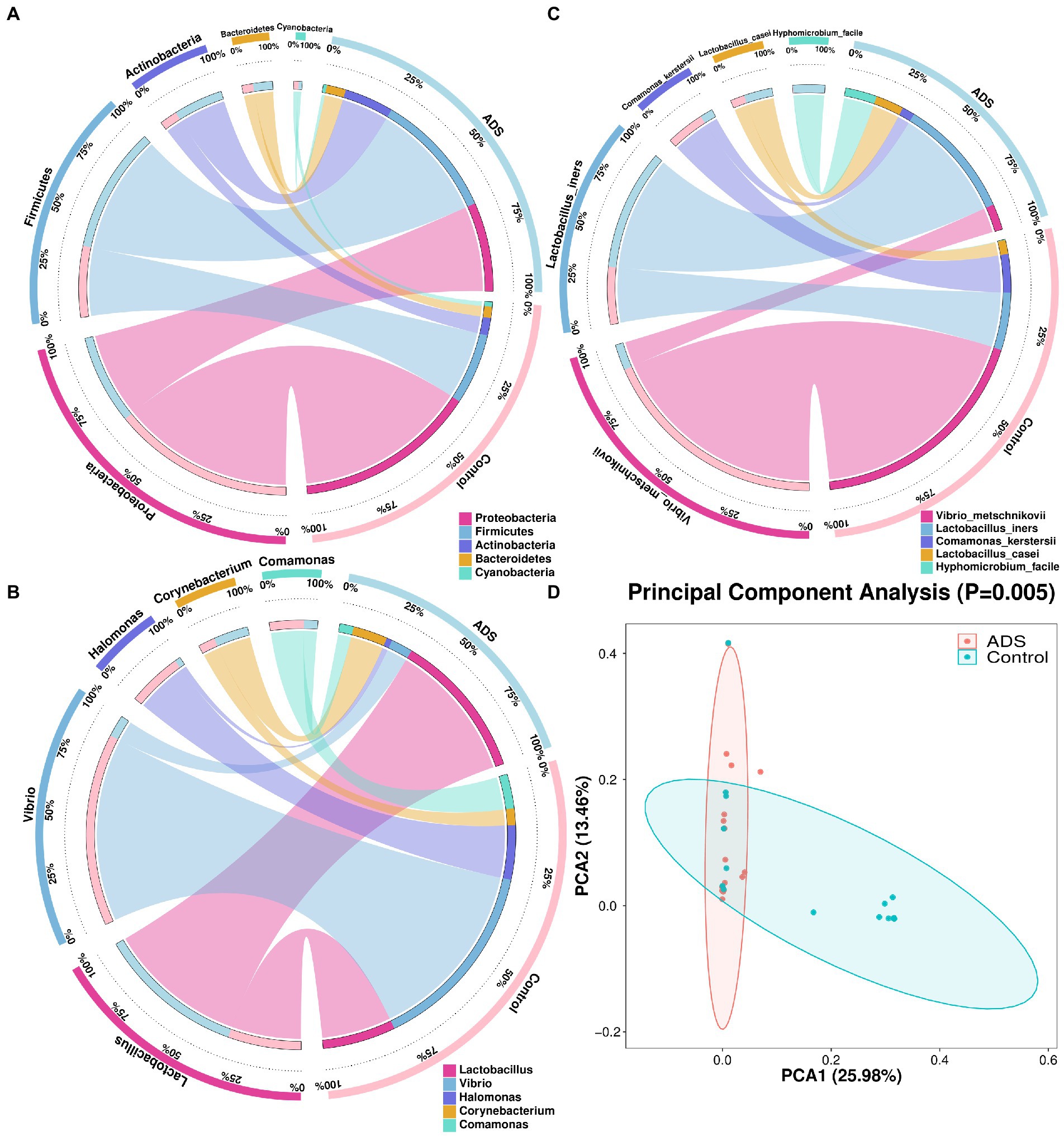
Figure 4. Endometrial microbiota were clustered according to adenomyosis or control grouping. Five different bacteria are presented in this figure with the relative abundance of the top five (TOP 5) at the phylum (A), genus (B), and species (C) levels. The two groups and the relative proportions of each bacterium are shown in the right half of the circle. The five bacteria and the proportions occupied by each group are shown in the left half part of the circle. Principal Components Analysis (PCA) plots were also conducted (D). Red dots indicate the adenomyosis (ADS) group while blue dots indicate the control group. Ovals represent the 95% CI for each group. The percentage on the X (PCA1) and Y (PCA2) axis explains the degree of each variation. Adonis tests were performed to calculate the statistical significance.
3.5. Analysis of biomarkers
Next, we attempted to identify differences in the composition of microbiota between groups by using the linear discriminant analysis (LDA) effect size (LEfSe) algorithm to discover potential biomarkers between the two groups (Figure 5). Using an LDA > 2.0 as a threshold for discriminative features, our analysis identified significant differences in the abundance of Firmicutes and Actinobacteria at the phylum level; these were more abundant in the ADS group than in the control group (Figure 5A). In addition, we found that Corynebacterium, Enterococcus, Staphylococcus, Caulobacter, and Citrobacter were significantly more abundant in the ADS group when compared with the control group at the genus level (Figure 5B). Furthermore, Lactobacillus zeae (L. zeae), Burkholderia cepacia (B. cepacia), Weissella confusa (W. confusa), Citrobacter freundii (C. freundii), and Prevotella copri (P. copri) were significantly more abundant in the ADS group at the species level (Figure 5C). Identical results were generated by cladogram analysis (Figure 6).
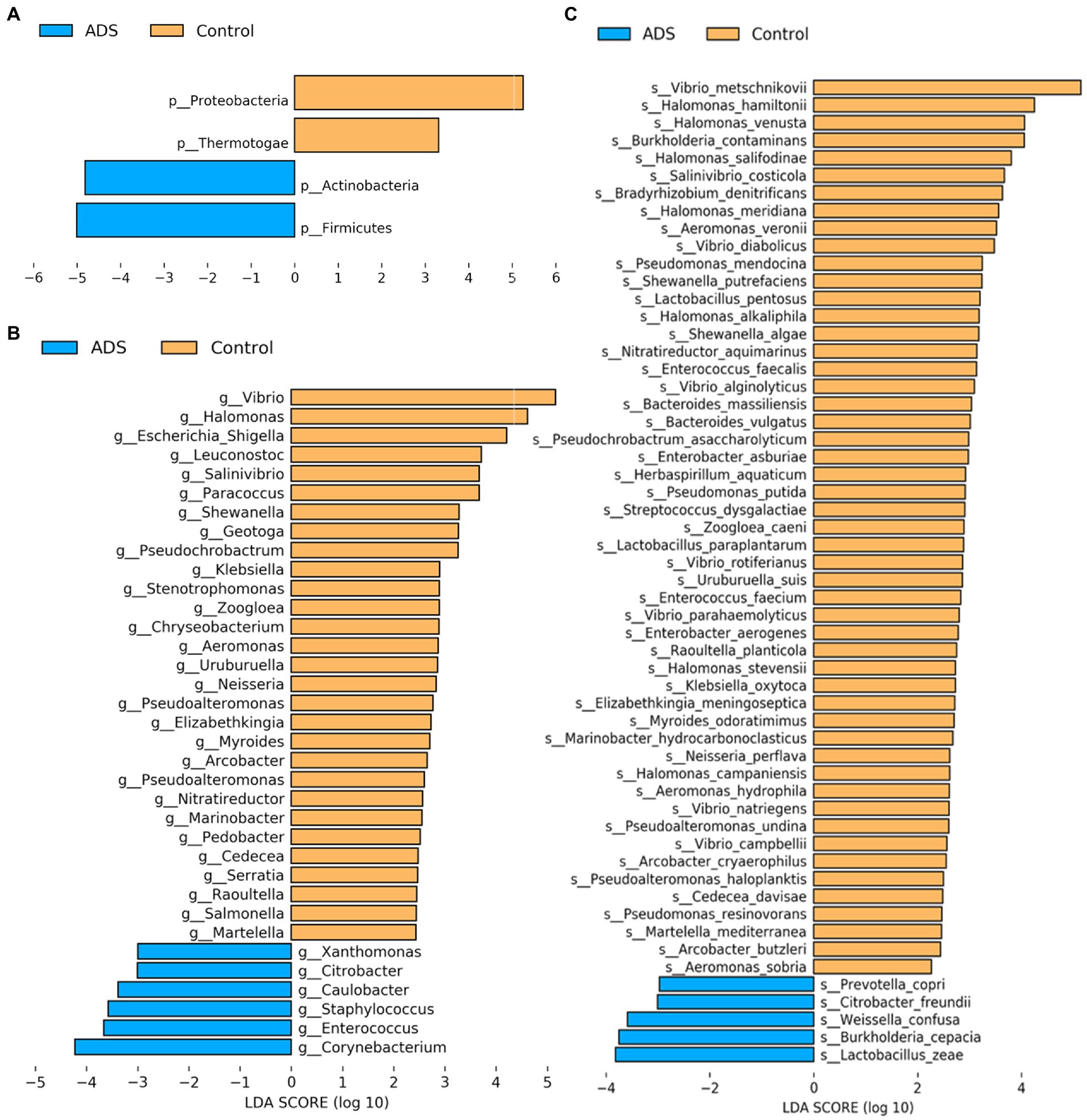
Figure 5. Analysis of putative biomarkers for adenomyosis. LEfSe analysis revealed the enrichment of taxa, including bacteria in the Firmicutes and Actinobacteria phylum, Corynebacterium, Enterococcus, Staphylococcus, Caulobacter, and Citrobacter genus, and Lactobacillus zeae, Burkholderia cepacia, Weissella confusa, Citrobacter freundii, and Prevotella copri species in patients with adenomyosis. The control group showed enrichment of the Proteobacteria and Thermotogae phylum, Vibrio, Halomonas, Escherichia, Shigella, Leuconostoc, Salinivibrio, Paracoccus, Shewanella, Geotoga, and Pseudochrobactrum genus. Histogram of LDA scores (log10) showing bacteria that presented with higher relative abundance in the ADS group (blue) when compared to the control group (orange) at the phylum (A), genus (B), and species (C) levels. Only statistically significant differences (p < 0.05) are shown.
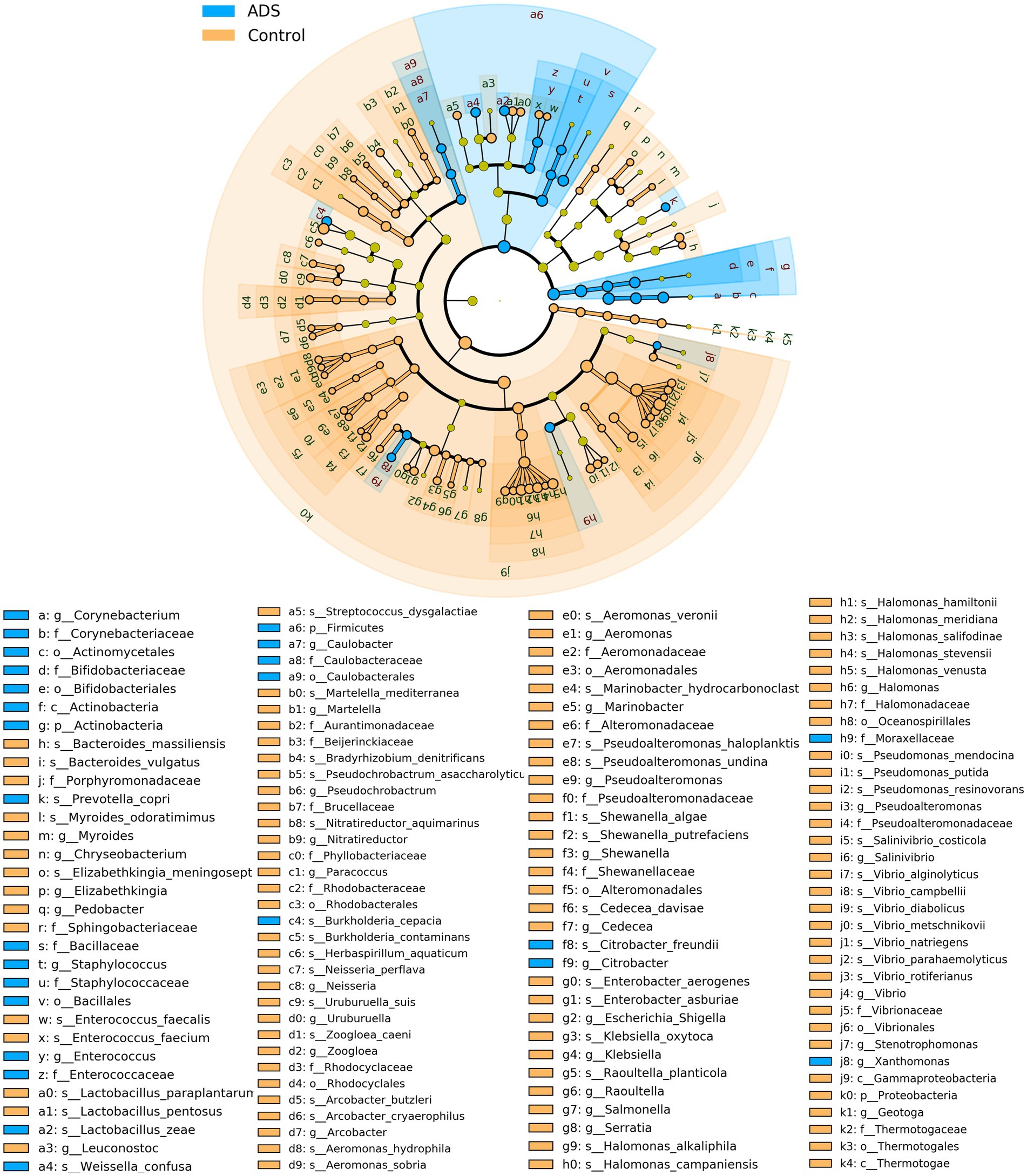
Figure 6. Cladogram report. The circle radiating from the inside to outside represents the classification level (from phylum to genus or species). Species without significant differences are colored yellow. Biomarkers were shown by specific colors in each group; the blue nodes indicate the bacteria that play an important role in the adenomyosis (ADS) group while the orange nodes indicate the microbial groups that play an important role in the control group. Prefixes represent abbreviations for each taxon: phylum (p_), class (c_), order (o_), family (f_), genus (g_), species(s_). Only statistically significant differences (p < 0.05) are shown.
3.6. Analysis of biomarkers for the prediction and diagnosis of adenomyosis
Next, receiver operating characteristic (ROC) curves were created to identify potential microbiome biomarkers at the phylum, genus, and species level for distinguishing patients with adenomyosis from those without adenomyosis (Figure 7). Area under the curve (AUC) analysis was then used to quantify the accuracy of adenomyosis diagnosis; an AUC > 0.7 was acceptable. At the phylum level, the AUC values for Firmicutes and Actinobacteria for the diagnosis of adenomyosis were 0.726 and 0.765, respectively (Figure 7A). At the genus level, Figure 7C depicts five curves representing Caulobacter, Corynebacterium, Staphylococcus, Enterococcus, and Citrobacter, respectively; the AUC values were 0.755, 0.753, 0.750, 0.699, and 0.691, respectively. At the species level, ROC curve analyses indicated the following diagnostic accuracies: Citrobacter freundii (AUC = 0.756), Prevotella copri (AUC = 0.753), and Burkholderia cepacia (AUC = 0.701) (Figure 7E). Notably, the integration of significantly different bacteria into logistic regression models indicated significantly better AUC values at the phylum (AUC = 0.839, sensitivity = 90.5%, specificity = 68.80%), genus (AUC = 0.878, sensitivity = 71.4%, specificity = 100%), and species levels (AUC = 0.970, sensitivity = 81%, specificity = 100%) (Figures 7B,D,F). These results indicate that Citrobacter freundii, Burkholderia cepacia, and Prevotella copri might serve as predictive markers for adenomyosis. Combined with biomarkers at the genus level, the diagnostic sensitivity and specificity for adenomyosis were 71.4 and 100.0%, respectively. However, the diagnostic sensitivity of adenomyosis at the species level increased to 81%.
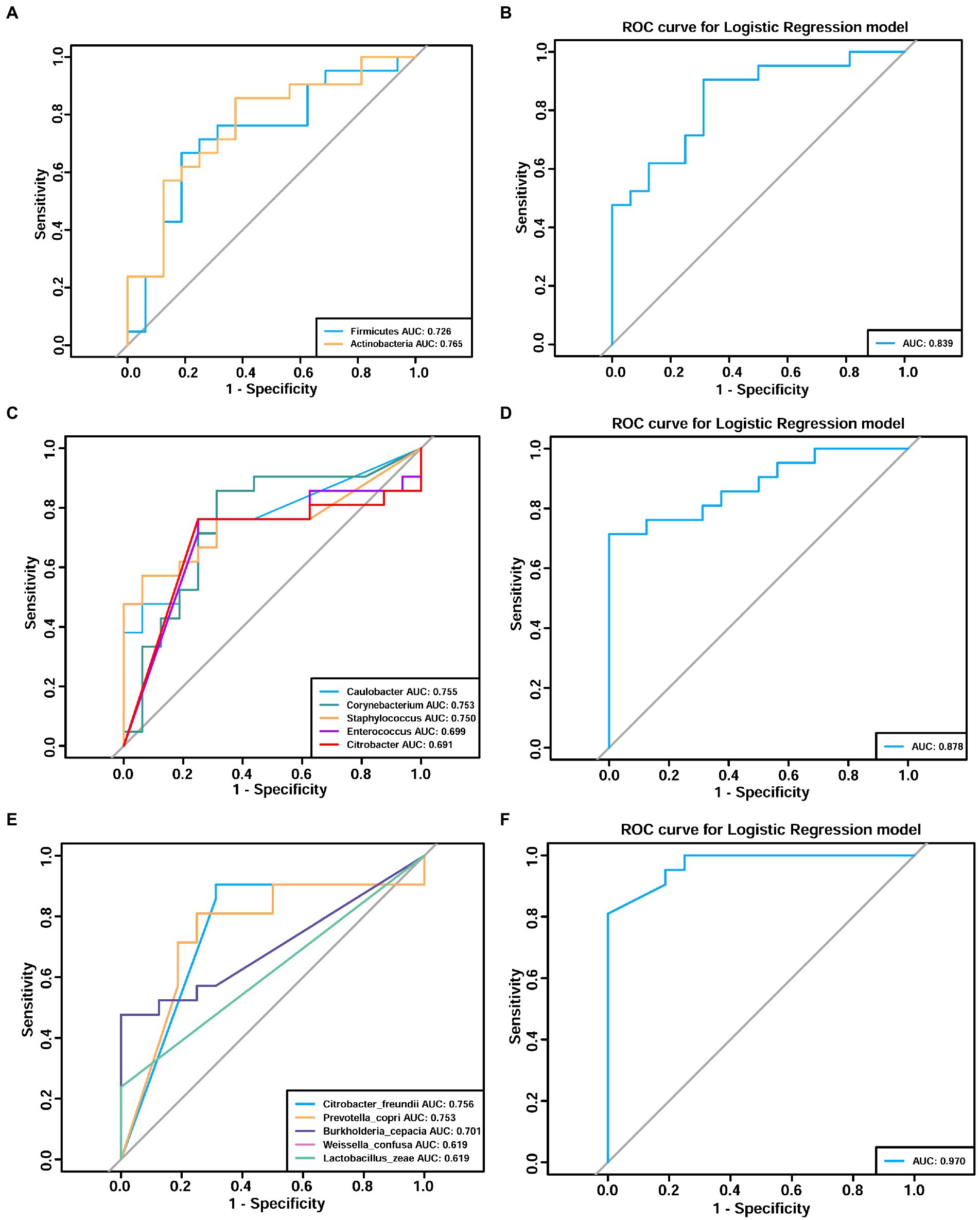
Figure 7. Receiver operating characteristic (ROC) analyses demonstrating the adenomyosis diagnosis efficacy of the endometrial microbiome at the phylum (A,B), genus (C,D), and species (E,F) levels. Different colors correspond to different taxa. Area under the curve (AUC) analysis was used to quantify the accuracy of adenomyosis diagnosis (an AUC value >0.7 was acceptable).
3.7. Predictive functional profiling of endometrial microbiota
PICRUSt was performed to explore the potential functional profiling of endometrial microbiota from the two groups. Supplementary Figure S2 illustrates several dysfunctional pathways among the two groups. On level 1, five functions identified as being statistically significant (p < 0.05): metabolism and genetics and genetic information processing were upregulated in the ADS group when compared with the control group (p = 0.0074 to p = 0.02446, mean proportion > 1%), while environmental information processing was upregulated in the control group when compared with the ADS group (p = 0.00168, mean proportion > 1%) (Supplementary Figure S3). On level 2, 18 functions were shown to be statistically significant (p < 0.05); the ADS patients exhibited upregulation in several pathways, including nucleotide metabolism, replication and repair, translation, enzyme families, and carbohydrate metabolism (p = 0.00267 to p = 0.021, mean proportion > 1%). In contrast, the control group exhibited upregulation in other pathways, including metabolism, signal transduction, cellular processes and signaling, and cell motility (p < 0.00001 to p = 0.02658, mean proportion > 1%) (Supplementary Figure S4). On level 3, 30 functions were identified as being statistically significant (p < 0.05). Compared with the control group, the ADS group exhibited upregulation in several pathways, including protein export, glycolysis/gluconeogenesis, alanine, aspartate, and glutamate metabolism (p < 0.001 to p = 0.00176, mean proportion > 1%). Compared with the ADS group, the control group exhibited upregulation in certain other pathways, including the two-component system, transcription factors, the secretion system, ABC transporters, other ion-coupled transporters, and bacterial motility proteins (p = 0.00011 to p = 0.0016, mean proportion > 1%) (Figure 8).
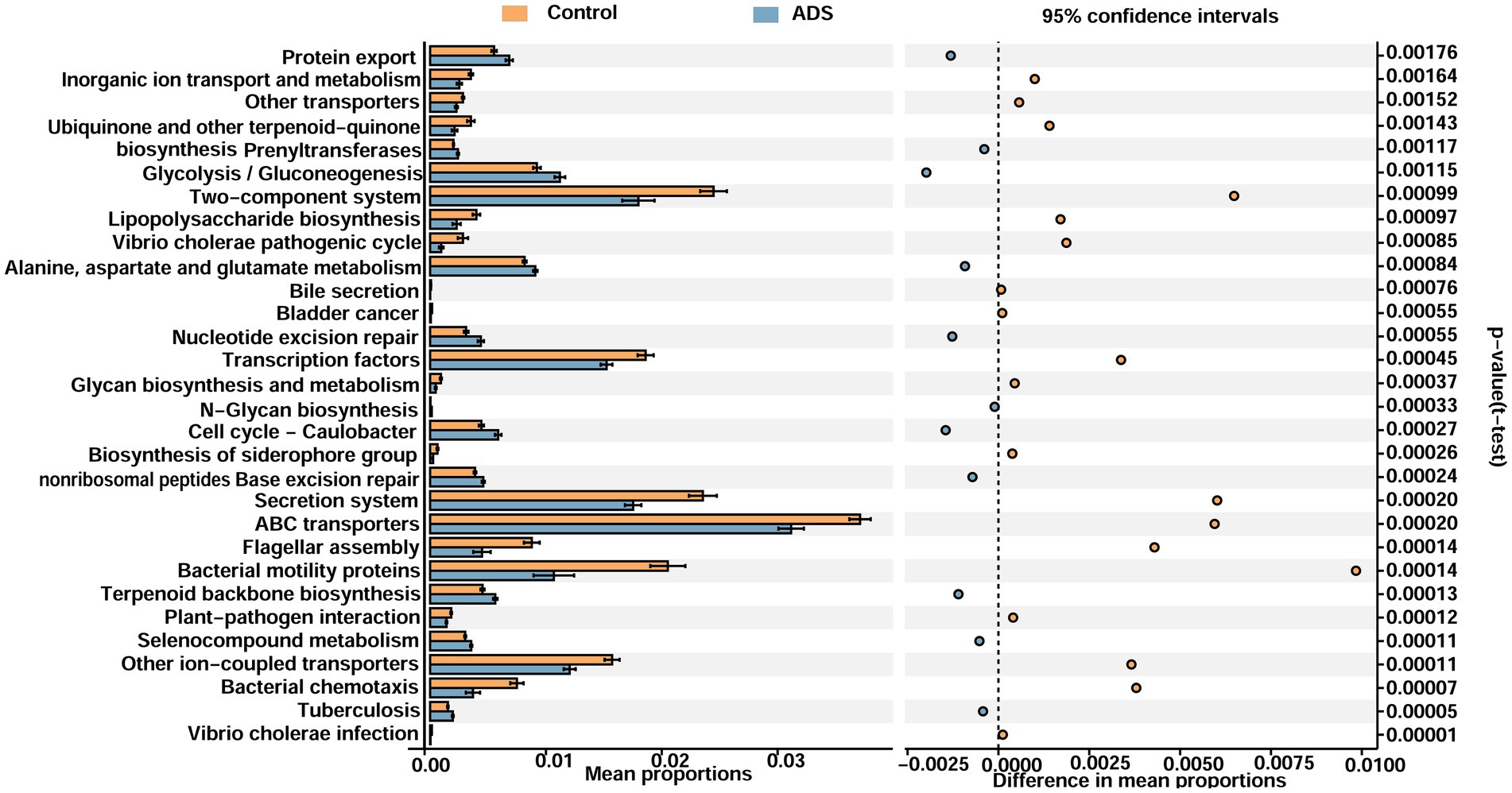
Figure 8. Functional analyses of the endometrial microbiota of participants in the two groups (level 3). Microbiota differences between groups as shown by differentially functional pathways. Downregulated pathways imply a lower ratio of the mean proportion of expression. Upregulated pathways suggest a higher ratio of the mean proportion of expression. White’s non-parametric t-test was used to calculate p-values.
4. Discussion
In this study, we compared the endometrial microbiota from patients with and without adenomyosis, as confirmed by pathological analysis. We demonstrated the presence of an endometrial microbiota in the uteri of patients with adenomyosis that had a significantly lower richness than the control group. Until now, we know little about the precise etiology involved in the reduced richness of the endometrial microbiota in patients with adenomyosis. There were no significant differences between the two groups of patients with regard to their age, BMI, menstrual cycle phase, parity, and methods of contraception. In addition, we excluded patients with autoimmune disorders and genital infections. In contrast to our findings (mean age > 45 years old), one previous study found that the richness of the endometrial flora in patients with endometriosis (mean age < 36 years old) was increased (Wessels et al., 2021); this finding might be due to differences in participant age. Wang et al. (2021) reported that the richness of the uterine microbiome in the 40–50-year age group was lower than that in younger women. To a certain extent, such changes in the diversity of the endometrial microbiota may be the result of changes in the endometrial immune microenvironment, including endometrial immune response and inflammation (abnormal activation and differentiation of dendritic cells, macrophages, and Natural killer cells) which ultimately reduce endometrial receptivity, disrupt embryo implantation, and promote the progression of disease (Wira et al., 2005; Chen et al., 2021).
Furthermore, we found that the majority of samples could be clustered according to disease status by performing beta diversity analysis; this indicated that the structure and composition of the endometrial microbiota of patients with or without adenomyosis were different. LEfSe analysis also identified some biomarkers that were strictly associated with adenomyosis that might be useful for the diagnosis of adenomyosis. A high abundance of Firmicutes and Actinomycetes was detected in endometrial samples from patients with adenomyosis, whereas Proteobacteria and Thermotogae were present in high abundance in patients from the control group. These results are consistent with previous studies of endometrial cancer, intrauterine adhesions, endometrial polyps, and patients with infertility (Fang et al., 2016; Benner et al., 2018; Walsh et al., 2019; Qiu et al., 2021). In a previous study, Khan et al. studied patients with endometriotic cysts and observed a higher proportion of Corynebacterium (Actinomycetes, Gram-positive), Enterococcus (Firmicutes, Gram-positive), Staphylococcus (Firmicutes, Gram-positive), Caulobacter (Proteobacteria, Gram-negative), and Citrobacter (Proteobacteria, Gram-negative) in patients with adenomyosis (Akiyama et al., 2019). However, Chen et al. (2017) reported that Methylophilaceae (Proteobacteria, Gram-negative) was a useful biomarker for the endometrial flora in patients with adenomyosis. In the present study, blank and negative controls were used to eliminate potential bacterial contamination from sampling collection, DNA processing, library preparation, and from the local environment. Differences between our results and those published previously may be due to analytical differences, along with different collection methods and study populations. Moreover, our results align with the bacterial contamination hypothesis of endometriosis that Gram-negative bacteria show enrichment in the endometrium of patients with endometriosis. Lipopolysaccharide (LPS) is a component of the outer membrane of Gram-negative bacteria and regulates the uncontrolled activation of inflammatory and immune responses in the pelvis and the growth of endometriosis via the LPS/TLR4 cascade (Khan et al., 2018). The major taxa of relative richness (TOP 10) represented in the population of patients with adenomyosis were non-Lactobacillus dominant (Lactobacillus species <90%, NLD), including Lactobacillus, Corynebacterium, Streptococcus, Prevotella, Hyphomicrobium, Geobacillus (3.78%), and Vibrio. These results are consistent with those reported previously by Chen et al. (2017) but differed from several other studies which reported that the most dominant endometrial microbiota are Lactobacillus (Lactobacillus species >90%, LD) (Moreno et al., 2016; Tao et al., 2017; Kyono et al., 2018).
Although the precise mechanisms underlying the endometrial microbiota with regard to human health and disease remain unclear, an imbalance of the microbiota is closely related to the pathophysiological mechanisms of endometriosis, endometrial cancer, and uterine fibroids. Currently, most of our knowledge relating to the interaction between bacteria and their host is derived from the highly diverse intestinal microbiota (Belkaid and Hand, 2014). The diversity of the endometrial microbiota is lower than that of the intestinal microbiota and higher than that of the vaginal microbiota (Chen et al., 2017). At the species level, our results indicated that Citrobacter freundii (Gram-negative), Burkholderia cepacia (Gram-negative), and Prevotella copri (Gram-negative) could be biomarkers for adenomyosis. Citrobacter freundii is a symbiotic bacterium found in the intestines of humans and animals (Liu et al., 2015). However, another study reported that Citrobacter freundii activated inflammation via the nucleotide oligomeric Domain-Like receptor family, a pyridine domain containing 3 (NLRP3) inflammatory body, in a T6SS-dependent manner (Liu et al., 2021). Citrobacter freundii can also invade, reproduce, and transfect human microvascular endothelial cells in vitro (Badger et al., 1999); this was associated with neonatal meningitis and brain abscesses (Joaquin et al., 1991). Burkholderia cepacia was previously shown to secrete BceF Tyrosine Kinase which participates in the formation biofilm and contributes to respiratory tract infection. BceF tyrosine kinase does not show any homology with human kinase and is therefore useful for the development of target antibacterial drugs (Mayer et al., 2021). Another study revealed that ‘quorum sensing’ (QS) enabled Burkholderia cepacia to produce virulent factors and biofilms, thus destroying the host’s innate immunity, especially epithelial cells, neutrophils, and megaphagocytes in patients with cystic fibrosis (Ganesh et al., 2020). Similar results were observed in patients with keratitis (Ho et al., 2021). Wessels et al. also reported that Burkholderia enriched the endometrium of symptomatic patients with pelvic pain but without endometriosis, as confirmed by surgery (Wessels et al., 2021).
In addition, Prevotella copri, a potential pro-inflammatory bacterium, has been associated with inflammation and autoimmunity. In a mouse model of glucan sulfate-induced colitis, elevated levels of Prevotella copri richness were associated with more severe intestinal inflammation than mice without Prevotella copri (Cao et al., 2020). Prevotella copri was also positively correlated with the severity of rheumatoid arthritis (Scher et al., 2013). Recent studies have revealed that Prevotella copri play an important role in anti-tumor therapy; patients with a high abundance of Prevotella copri could benefit from treatment (Jin et al., 2019). In addition, Yu et al. (2019) proved that Prevotella copri caused damage to the mucosal barrier by expressing reduced levels of tight junction proteins (ZO-1, Occludin, and Claudin-1) and increased expression levels of inflammatory factors (IFN-γ, IL-1 β, IL-6, and MCP-1). The eutopic endometrium is thought to play a vital role in the pathogenesis of adenomyosis. Studies have predicted that the IL-6 and ERK/MAPK pathways are abnormally activated in the eutopic endometrium of patients with adenomyosis (Xiang et al., 2019). Furthermore, the numbers of natural killer cells and macrophages have been found to be elevated in the eutopic endometrium of patients with adenomyosis along with increased levels of inflammatory markers (IL-1b and IL-6) (Zhihong et al., 2016; Carrarelli et al., 2017).
Although the role of the microbiota has been revealed in some diseases, we explored the potential mechanisms that may act between the host and microbes by applying the PICRUSt algorithm. Differentially regulated pathways between the two groups that may be of relevance included genetic information processing and metabolism; these were both enriched in the ADS group when compared to the control group. Furthermore, nucleotide metabolism, replication and repair, translation, enzyme families, and carbohydrate metabolism pathways were all upregulated in the ADS group when compared to the control group. Furthermore, protein export, glycolysis/gluconeogenesis, alanine aspartate, and glutamate metabolism were all enriched in the ADS group when compared to the control group. Only limited research has targeted the endometrial microbiota of women with adenomyosis. Citrobacter freundii, Burkholderia cepacia, and Prevotella copri are pathogenic microbiota that may be associated with adenomyosis; these are Gram-negative bacteria that secrete LPS via protein export, which causes mucosal inflammation and damages the mucosal barrier integrity (Mobley et al., 2001). Microorganisms and their metabolites can also trigger DNA damage and mutation of the epithelial cells and directly regulate the biological function of host cells via increased proliferation or reduced apoptosis (Lim et al., 2017; Chattopadhyay et al., 2019; Helmink et al., 2019).
Our study has several strengths that should be considered. We used a blinded approach to ensure the accuracy in all statistical analyses. We also took multiple methods to minimize the contamination. Furthermore, all patients in this study received pathological examination to guarantee the precision and accuracy of diagnosis. However, this study has several limitations that should be considered. First, the patients in this study were ethnically and genetically homogenous. Second, the endometrial microbiota was susceptible to contamination as a specimen with low microbial abundance. Researchers tried to avoid contamination at all times during sampling; however, environmental contamination cannot be ruled out. Third, all participants received hysterectomy in this study; therefore, we were unable to recruit healthy samples for the control group; this would have provided a better understanding of the differences in endometrial microbiota between the patients with adenomyosis and healthy women. Finally, another limitation of this study is its small sample size; however, other studies on similar topics also involved small sample sizes. In the future, studies should aim to recruit a larger population size for investigation.
5. Conclusion
In conclusion, females possess an endometrial microbiota that contains various microbes. Adenomyosis is highly associated with endometrial bacterial dysbiosis. A potential variation in the microbial environment of the endometrium might contribute to adenomyosis. Pathogenic bacteria, such as Citrobacter freundii, Burkholderia cepacia, and Prevotella copri, could be considered as potential emerging causes of adenomyosis. Further studies should investigate the cross-talk between potential pathogenic microbiota in patients with adenomyosis, evaluate whether the early diagnosis of adenomyosis could be assessed according to the endometrial microbiota profile, and investigate whether the endometrial microbiota could be modified to increase treatment outcomes.
Data availability statement
The data presented in the study are deposited in the Sequence Read Archive repository (https://www.ncbi.nlm.nih.gov/sra/), accession number PRJNA891223.
Ethics statement
The studies involving human participants were reviewed and approved by Medical Ethics Committee of Beijing Obstetrics and Gynecology Hospital, Capital Medical University No. 2016-KY-012-02. The patients/participants provided their written informed consent to participate in this study.
Author contributions
QL, HD, ShW, ZG, and YC conceived the study and reviewed and edited the manuscript. QL, SiW, CC, MS, HS, and CZ participated in sample collection. QL and ZG conducted the data analysis and prepared the figures and tables. QL helped to write the original draft. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (grant number 81571412) and supported by the Beijing Obstetrics and Gynecology Hospital, Capital Medical University (grant number FCYY201920).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1075900/full#supplementary-material
Footnotes
2. ^http://www.ehbio.com/ImageGP/index.php/Home/Index/LEFSe.html
References
Akiyama, K., Nishioka, K., Khan, K. N., Tanaka, Y., Mori, T., Nakaya, T., et al. (2019). Molecular detection of microbial colonization in cervical mucus of women with and without endometriosis. Am. J. Reprod. Immunol. 82:e13147. doi: 10.1111/aji.13147
An, M., Li, D., Yuan, M., Li, Q., Zhang, L., and Wang, G. (2017). Interaction of macrophages and endometrial cells induces epithelial-mesenchymal transition-like processes in adenomyosis. Biol. Reprod. 96, 46–57. doi: 10.1095/biolreprod.116.144071
Badger, J. L., Stins, M. F., and Kim, K. S. (1999). Citrobacter freundii invades and replicates in human brain microvascular endothelial cells. Infect. Immun. 67, 4208–4215. doi: 10.1128/IAI.67.8.4208-4215.1999
Belkaid, Y., and Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cells 157, 121–141. doi: 10.1016/j.cell.2014.03.011
Benner, M., Ferwerda, G., Joosten, I., and van der Molen, R. G. (2018). How uterine microbiota might be responsible for a receptive, fertile endometrium. Hum. Reprod. Update 24, 393–415. doi: 10.1093/humupd/dmy012
Bourdon, M., Santulli, P., Marcellin, L., Maignien, C., Maitrot-Mantelet, L., Bordonne, C., et al. (2021). Adenomyosis: an update regarding its diagnosis and clinical features. J. Gynecol. Obstet. Hum. Reprod. 50:102228. doi: 10.1016/j.jogoh.2021.102228
Cao, Y., Wang, X., Yang, Q., Deng, H., Liu, Y., Zhou, P., et al. (2020). Critical role of intestinal microbiota in ATF3-mediated gut immune homeostasis. J. Immunol. 205, 842–852. doi: 10.4049/jimmunol.1901000
Carrarelli, P., Yen, C. F., Funghi, L., Arcuri, F., Tosti, C., Bifulco, G., et al. (2017). Expression of inflammatory and neurogenic mediators in adenomyosis. Reprod. Sci. 24, 369–375. doi: 10.1177/1933719116657192
Chao, X., Liu, Y., Fan, Q., Shi, H., Wang, S., and Lang, J. (2021). The role of the vaginal microbiome in distinguishing female chronic pelvic pain caused by endometriosis/adenomyosis. Ann. Transl. Med. 9:771. doi: 10.21037/atm-20-4586
Chattopadhyay, I., Verma, M., and Panda, M. (2019). Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol. Cancer Res. Treat. 18. doi: 10.1177/1533033819867354
Chen, P., Chen, P., Guo, Y., Fang, C., and Li, T. (2021). Interaction between chronic endometritis caused endometrial microbiota disorder and endometrial immune environment change in recurrent implantation failure. Front. Immunol. 12:748447. doi: 10.3389/fimmu.2021.748447
Chen, S., Gu, Z., Zhang, W., Jia, S., Wu, Y., Zheng, P., et al. (2020). Microbiome of the lower genital tract in Chinese women with endometriosis by 16s-rRNA sequencing technique: a pilot study. Ann. Transl. Med. 8:1440. doi: 10.21037/atm-20-1309
Chen, C., Song, X., Wei, W., Zhong, H., Dai, J., Lan, Z., et al. (2017). The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 8:875. doi: 10.1038/s41467-017-00901-0
Douglas, G. M., Maffei, V. J., Zaneveld, J., Yurge, S. N., Brown, J. R., Taylor, C. M., et al. (2019). PICRUSt2: An improved and extensible approach for metagenome inference. BioRxiv :672295. doi: 10.1101/672295
Fang, R. L., Chen, L. X., Shu, W. S., Yao, S. Z., Wang, S. W., and Chen, Y. Q. (2016). Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am. J. Transl. Res. 8, 1581–1592.27186283.
Fuks, G., Elgart, M., Amir, A., Zeisel, A., Turnbaugh, P. J., Soen, Y., et al. (2018). Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome. 6:17. doi: 10.1186/s40168-017-0396-x
Ganesh, P. S., Vishnupriya, S., Vadivelu, J., Mariappan, V., Vellasamy, K. M., and Shankar, E. M. (2020). Intracellular survival and innate immune evasion of Burkholderia cepacia: improved understanding of quorum sensing-controlled virulence factors, biofilm, and inhibitors. Microbiol. Immunol. 64, 87–98. doi: 10.1111/1348-0421.12762
García-Solares, J., Donnez, J., Donnez, O., and Dolmans, M. M. (2018). Pathogenesis of uterine adenomyosis: invagination or metaplasia? Fertil. Steril. 109, 371–379. doi: 10.1016/j.fertnstert.2017.12.030
Guo, S. W. (2020). The pathogenesis of adenomyosis vis-à-vis endometriosis. J. Clin. Med. 9:485. doi: 10.3390/jcm9020485
Guo, Z., Duan, H., Wang, S., Wang, S., Lin, Q., and Li, Y. (2022). RNA-seq reveals co-dysregulated circular RNAs in the adenomyosis eutopic endometrium and endometrial-myometrial interface. BMC Womens Health 22:293. doi: 10.1186/s12905-022-01871-2
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V., and Wargo, J. A. (2019). The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388. doi: 10.1038/s41591-019-0377-7
Hernandes, C., Silveira, P., Rodrigues Sereia, A. F., Christoff, A. P., Mendes, H., Valter de Oliveira, L. F., et al. (2020). Microbiome profile of deep endometriosis patients: comparison of vaginal fluid, endometrium and lesion. Diagnostics 10:163. doi: 10.3390/diagnostics10030163
Ho, M. C., Kang, E. Y., Yeh, L. K., Ma, D., Lin, H. C., Tan, H. Y., et al. (2021). Clinico-microbiological profile of Burkholderia cepacia keratitis: a case series. Ann. Clin. Microbiol. Antimicrob. 20:6. doi: 10.1186/s12941-020-00407-6
Jin, Y., Dong, H., Xia, L., Yang, Y., Zhu, Y., Shen, Y., et al. (2019). The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol. 14, 1378–1389. doi: 10.1016/j.jtho.2019.04.007
Joaquin, A., Khan, S., Russel, N., and Al Fayez, N. (1991). Neonatal meningitis and bilateral cerebellar abscesses due to Citrobacter freundii. Pediatr. Neurosurg. 17, 23–24. doi: 10.1159/000120561
Khan, K. N., Fujishita, A., Hiraki, K., Kitajima, M., Nakashima, M., Fushiki, S., et al. (2018). Bacterial contamination hypothesis: a new concept in endometriosis. Reprod. Med. Biol. 17, 125–133. doi: 10.1002/rmb2.12083
Kunaseth, J., Waiyaput, W., Chanchaem, P., Sawaswong, V., Permpech, R., Payungporn, S., et al. (2022). Vaginal microbiome of women with adenomyosis: a case-control study. PLoS One 17:e0263283. doi: 10.1371/journal.pone.0263283
Kyono, K., Hashimoto, T., Nagai, Y., and Sakuraba, Y. (2018). Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: a single-center pilot study. Reprod. Med. Biol. 17, 297–306. doi: 10.1002/rmb2.12105
Łaniewski, P., Ilhan, Z. E., and Herbst-Kralovetz, M. M. (2020). The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 17, 232–250. doi: 10.1038/s41585-020-0286-z0286-z
Leng, J. H., Chen, Z. J., Dai, J., Dai, Y., Deng, C. Y., Di, W., et al. (2020). Chinese experts’ consensus on diagnosis and treatment of adenomyosis. Zhonghua Fu Chan Ke Za Zhi 55, 376–383. doi: 10.3760/cma.j.cn112141-20200228-00150
Lim, Y., Totsika, M., Morrison, M., and Punyadeera, C. (2017). Oral microbiome: a new biomarker reservoir for Oral and oropharyngeal cancers. Theranostics 7, 4313–4321. doi: 10.7150/thno.21804
Liu, L., Hao, S., Lan, R., Wang, G., Xiao, D., Sun, H., et al. (2015). The type VI secretion system modulates flagellar gene expression and secretion in Citrobacter freundii and contributes to adhesion and cytotoxicity to host cells. Infect. Immun. 83, 2596–2604. doi: 10.1128/IAI.03071-14
Liu, L., Song, L., Deng, R., Lan, R., Jin, W., Van Nhieu, T., et al. (2021). Citrobacter freundii activation of NLRP3 inflammasome via the type VI secretion system. J. Infect. Dis. 223, 2174–2185. doi: 10.1093/infdis/jiaa692
Mayer, M., Matiuhin, Y., Nawatha, M., Tabachnikov, O., Fish, I., Schutz, N., et al. (2021). Structural and functional insights into the biofilm-associated Bce F tyrosine kinase domain from Burkholderia cepacia. Biomol. Ther. 11:1196. doi: 10.3390/biom11081196
Mobley, H. L. T.., Mendz, G. L., and Hazell, S. L.. (2001). Helicobacter pylori: Physiology and Genetics. Washington, DC: ASM Press.
Moreno, I., Cicinelli, E., Garcia-Grau, I., Gonzalez-Monfort, M., Bau, D., Vilella, F., et al. (2018). The diagnosis of chronic endometritis in infertile asymptomatic women: a comparative study of histology, microbial cultures, hysteroscopy, and molecular microbiology. Am. J. Obstet. Gynecol. 218, 602.e1–602.e16. doi: 10.1016/j.ajog.2018.02.012
Moreno, I., Codoñer, F. M., Vilella, F., Valbuena, D., Martinez-Blanch, J. F., Jimenez-Almazán, J., et al. (2016). Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 215, 684–703. doi: 10.1016/j.ajog.2016.09.075
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Poore, G. D., Kopylova, E., Zhu, Q., Carpenter, C., Fraraccio, S., Wandro, S., et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574. doi: 10.1038/s41586-020-2095-1
Qiu, T., Liu, L., Zhou, H., Sheng, H., He, Y., Liu, M., et al. (2021). Analysis of endometrial microbiota in intrauterine adhesion by high-throughput sequencing. Ann. Transl. Med. 9:195. doi: 10.21037/atm-20-2813
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2:e01202. doi: 10.7554/eLife.01202
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sobstyl, M., Brecht, P., Sobstyl, A., Mertowska, P., and Grywalska, E. (2022). The role of microbiota in the immunopathogenesis of endometrial cancer. Int. J. Mol. Sci. 23:5756. doi: 10.3390/ijms23105756
Tao, X., Franasiak, J. M., Zhan, Y. P., Scott, R. T., Rajchel, J., Bedard, J., et al. (2017). Characterizing the endometrial microbiome by analyzing the ultra-low bacteria from embryo transfer catheter tips in IVF cycles: next generation sequencing (NGS) analysis of the 16S ribosomal gene. Hum. Micro J. 3, 15–21. doi: 10.1016/j.humic.2017.01.004
Vannuccini, S., Tosti, C., Carmona, F., Huang, S. J., Chapron, C., Guo, S. W., et al. (2017). Pathogenesis of adenomyosis: an update on molecular mechanisms. Reprod. Biomed. Online 35, 592–601. doi: 10.1016/j.rbmo.2017.06.016
Walther-António, M. R., Chen, J., Multinu, F., Hokenstad, A., Distad, T. J., Cheek, E. H., et al. (2016). Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 8:122. doi: 10.1186/s13073-016-0368-y
Walsh, D. M., Hokenstad, A. N., Chen, J., Sung, J., Jenkins, G. D., Chia, N., et al. (2019). Postmenopause as a key factor in the composition of the endometrial cancer microbiome (ECbiome). Sci. Rep. 9:19213. doi: 10.1038/s41598-019-55720-8
Wang, J., Li, Z., Ma, X., Du, L., Jia, Z., Cui, X., et al. (2021). Translocation of vaginal microbiota is involved in impairment and protection of uterine health. Nat. Commun. 12:4191. doi: 10.1038/s41467-021-24516-8
Wessels, J. M., Domínguez, M. A., Leyland, N. A., Agarwal, S. K., and Foster, W. G. (2021). Endometrial microbiota is more diverse in people with endometriosis than symptomatic controls. Sci. Rep. 11:18877. doi: 10.1038/s41598-021-98380-3
Wira, C. R., Fahey, J. V., Sentman, C. L., Pioli, P. A., and Shen, L. (2005). Innate and adaptive immunity in female genital tract: cellular responses and interactions. Immunol. Rev. 206, 306–335. doi: 10.1111/j.0105-2896.2005.00287.x
Xiang, Y., Sun, Y., Yang, B., Yang, Y., Zhang, Y., Yu, T., et al. (2019). Transcriptome sequencing of adenomyosis eutopic endometrium: a new insight into its pathophysiology. J. Cell. Mol. Med. 23, 8381–8391. doi: 10.1111/jcmm.14718
Yu, C., Zhou, B., Xia, X., Chen, S., Deng, Y., Wang, Y., et al. (2019). Prevotella copri is associated with carboplatin-induced gut toxicity. Cell Death Dis. 10:714. doi: 10.1038/s41419-019-1963-9
Zhai, J., Vannuccini, S., Petraglia, F., and Giudice, L. C. (2020). Adenomyosis: mechanisms and pathogenesis. Semin. Reprod. Med. 38, 129–143. doi: 10.1055/s-0040-1716687
Keywords: adenomyosis, endometrium, endometrial microbiota, microbiota diversity, microbiota function
Citation: Lin Q, Duan H, Wang S, Guo Z, Wang S, Chang Y, Chen C, Shen M, Shou H and Zhou C (2023) Endometrial microbiota in women with and without adenomyosis: A pilot study. Front. Microbiol. 14:1075900. doi: 10.3389/fmicb.2023.1075900
Edited by:
Carlos Simon, University of Valencia, SpainReviewed by:
Nicoletta Di Simone, Humanitas University, ItalyPaola Villa, Università Cattolica del Sacro Cuore, Italy
Copyright © 2023 Lin, Duan, Wang, Guo, Wang, Chang, Chen, Shen, Shou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Duan, ✉ ZHVhbmh1YUBjY211LmVkdS5jbg==
 Qi Lin
Qi Lin Hua Duan
Hua Duan Sha Wang1
Sha Wang1 Yanan Chang
Yanan Chang Chang Zhou
Chang Zhou