
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 February 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1064838
Fusarium root rot (FRR) caused by Fusarium graminearum poses a threat to global food security. Biological control is a promising control strategy for FRR. In this study, antagonistic bacteria were obtained using an in-vitro dual culture bioassay with F. graminearum. Molecular identification of the bacteria based on the 16S rDNA gene and whole genome revealed that the species belonged to the genus Bacillus. We evaluated the strain BS45 for its mechanism against phytopathogenic fungi and its biocontrol potential against FRR caused by F. graminearum. A methanol extract of BS45 caused swelling of the hyphal cells and the inhibition of conidial germination. The cell membrane was damaged and the macromolecular material leaked out of cells. In addition, the mycelial reactive oxygen species level increased, mitochondrial membrane potential decreased, oxidative stress-related gene expression level increased and oxygen-scavenging enzyme activity changed. In conclusion, the methanol extract of BS45 induced hyphal cell death through oxidative damage. A transcriptome analysis showed that differentially expressed genes were significantly enriched in ribosome function and various amino acid transport pathways, and the protein contents in cells were affected by the methanol extract of BS45, indicating that it interfered with mycelial protein synthesis. In terms of biocontrol capacity, the biomass of wheat seedlings treated with the bacteria increased, and the BS45 strain significantly inhibited the incidence of FRR disease in greenhouse tests. Therefore, strain BS45 and its metabolites are promising candidates for the biological control of F. graminearum and its related root rot diseases.
Fusarium graminearum can cause disease on many economically important crops, including maize and wheat (Goswami and Kistler, 2004; Tillmann et al., 2017). Wheat (Triticum aestivum L.) is the most widely cultivated crop globally and is one of the three major grains in the world (Jabran et al., 2017). Its plants may be infected by F. graminearum at all stages, and the infections cause two epidemic diseases, wheat root rot and wheat head blight, which seriously affecting the yield and quality of grain worldwide (Stephen et al., 2015; Wang and Gottwald, 2017). To effectively kill pathogenic microorganisms in crops, the commonly used control method is to spray chemical fungicides into the fields (Cruppe et al., 2021; Edwards, 2022), but these synthetic fungicides may remain on grain, resulting in food safety issues (Chen and Ying, 2015; Rjiba-Touati et al., 2022). In addition, chemical agents may affect the growth of other microorganisms in the field, which can be detrimental to the structure of soil species diversity (Bacmaga et al., 2018). Furthermore, the effectiveness of fungicides is reduced when exposed to rainy weather conditions, whereas a warm environment is conducive to the growth of Fusarium (Kriss et al., 2010). Therefore, it is very important to find effective biological control agents to replace chemical synthetic agents in the control F. graminearum. At present, biocontrol agent development is a new strategy to replace chemical fungicides. Microbial active products can be used as ideal alternatives to chemically synthesized antifungal agents to improve the quality of agricultural products (Abdalla, 2016). Several major bacteria, including Bacillus (Chen et al., 2020), Pseudomonas (Dugassa et al., 2021), Trichoderma (Hasan et al., 2021), Lactobacillus (Muhialdin et al., 2020) and Streptomyces (Passari et al., 2016, 2020), have been demonstrated to be effective in controlling pathogenic fungi and decreasing the incidence and severity of diseases. Bacillus has received extensive attention as an agricultural biological control agent because of its ability to simultaneously fight plant pathogens and promote plant growth (Shafi et al., 2017). Recently, Bacillus amyloliquefaciens GB03 has been used as a representative commercial strain of Bacillus for the biological control of broad-spectrum plant pathogens and as a biofertilizer to promote the growth and yield of field crops (Choi et al., 2014).
Members of the Bacillus genus are widely distributed and abundant in nature, resulting from their good environmental adaptability. Bacillus subtilis itself can achieve biocontrol effects by competing with pathogenic fungi for growth sites, and it produces a variety of antibacterial substances (Hamdache et al., 2011), including ribosomal synthetic peptides (lanosulfur antibiotics) and non-ribosomal synthetic peptides (lipopeptides), as well as non-peptide compounds, such as polyketones (Rabbee and Baek, 2020). Lipopeptide compounds, such as surfactants, iturins, and fengycins, which are amphiphilic low molecular weight compounds, have been intensively studied. These lipopeptide antibiotics have high inhibitory activity levels against insects, mites, nematodes, and pathogens that are harmful to plants. In addition, they can also induce the pathogen-associated molecular pattern-triggered immunity of plants (Sarowar et al., 2019), which has a biological control effect on crops. Thus, they have good development and application values.
Metabolites produce by B. subtilis have high antibacterial activities against a variety of human and plant pathogenic fungi (Sarowar et al., 2019). A biological control agent produces antagonistic metabolites mainly focusing on inhibiting fungal conidial germination, damaging membranes, and cell walls (Wu et al., 2020), interacting with a variety of intracellular targets, leaking macromolecular substances, and inducing accumulation of endogenous active substances and mitochondrial membrane potential (MMP) collapse in mycelial cells to damage pathogenic fungi (Han et al., 2015; Zhang and Sun, 2018). Low-concentration reactive oxygen species (ROS) act as intracellular messengers of many molecular events in cells and also play an important role in secondary metabolic synthesis (Cadenas and Davies, 2000; Zhu et al., 2020). However, the accumulation of excessive ROS in vivo attributed to high production or insufficient removal of ROS will cause oxidative stress (OS) and then damage cells (Zhang and Sun, 2018). The ROS can be eliminated by antioxidases to protect the body from damage. Superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) are common antioxidases. Studies have used transcriptome analysis to explore the inhibitory effect of Bacillus on Aspergillus carbonarius, and indicated that Bacillus alters the fungal cell structure, and perturbs energy, transport and osmotic pressure metabolism (Jiang et al., 2020). Other studies have used transcriptome analysis to investigate the control mechanism of metabolites from Streptomyces or plant extracts, such as thymol and glabridin, against Fusarium (Han S. et al., 2021; Yang et al., 2021; Wang et al., 2022). However, few studies have used transcriptome analysis to explore the signaling pathway changes and intracellular responses to Bacillus on F. graminearum. Pathogens treated with Bacillus elicit varying stress responses. Therefore, further study of the corresponding metabolic pathway changes in F. graminearum in vivo is necessary. Herein, the antifungal mechanism of the newly isolated Bacillus BS45 against F. graminearum was revealed through physiological and biochemical experiments and comparative transcriptomic analysis.
Root infections are considered important components of the F. graminearum life cycle (Beccari et al., 2011; Wang et al., 2015). Wheat seeds infected with FRR at an early stage die before or after seedling emergence with browning of the sheath, whereas late infection results in brown spots between the first two or three nodes. Damage due to FRR can result in production losses as high as 50% in some areas (Balmas et al., 2014; Williamson and Dhingra, 2021). A variety of laboratory or greenhouse experiments have confirmed that the use of biocontrol bacteria can effectively control a pathogen. Pseudomonas fluorescens and its fermentation broth had good inhibitory effects on Penicillium digitorum in the control of green mold decay of citrus fruits. Studies have also confirmed that using microorganisms or metabolites to produce biological agents can effectively reduce fungal infections of fruits, and the infection rates of cucumbers and tomatoes treated with biological agents were greatly reduced (Li and Chen, 2019).
The aims of this study were to develop effective inhibitory sites against F. graminearum, investigate the interactions between Bacillus and F. graminearum, explore the action targets and antibacterial mechanisms of antimicrobial substances of isolated strain BS45 against F. graminearum, and provide new ideas for biological control application.
Bacillus subtilis strain BS45 were isolated from soil (36.94°N, 100.86°E) in Xining City, Qinghai Province, PR China. Bacteria were maintained on Luria-Bertani (LB) medium at 37°C. Pathogen F. graminearum NRRL 31084 (Preservation No. CCTCC AF2015031) was obtained from China Center for Type Culture Collection (CCTCC). potato dextrose agar (PDA) was inoculated with F. graminearum at 28°C for 7 days. Conidial suspensions of F. graminearum were obtained using carboxymethylcellulose (CMC) liquid medium as previously described (Bi et al., 2020).
The collected soil was suspended in sterile distilled water, and the re-suspension was diluted in a gradient of 10−4, 10−5, and 10−6. The diluted liquid was spread on LB solid medium and cultured at 37°C. Single colonies were picked and transferred to LB solid medium and incubated at 37°C. After colony growth, the previous steps were repeated until a pure culture was obtained. The antagonistic activity of the isolated strain against F. graminearum was tested in vitro. A 5-mm diameter agar plug of F. graminearum was placed in the middle of a PDA (90-mm diameter), and bacterial solutions of 10 μl were independently added 30-mm away from the cake and incubated at 28°C for 5 days. The strains with antifungal activity were observed and recorded.
The DNA of the antifungal strain was extracted using a Bacterial Genomic DNA Extraction Kit (Solarbio, Beijing, China) in accordance with the manufacturer’s instructions. The 16S rRNA gene fragment of strain BS45 was amplified using the universal primer pair of 27F (5′-AGAGTTTGATCCTGGCTCA-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′; Lopez and Alippi, 2019). All PCR reactions were performed in a total volume of 50 μl including 25 μl of Taq Mix (Dream TaqTM Green PCR Master Mix (2×), Thermo Fisher, United States), a total of 2 μl of 10 mM primers, 1 μl of DNA template, and 22 μl of nuclease-free water. PCR was performed with a T100™ Thermal Cycler (Bio-Rad, America) with pre-deformation at 95°C for 3 min, 30 cycles of denaturation for 15 s at 95°C, annealing for 30 s at 55°C, and extension for 1 min at 72°C, followed by a final step of 72°C for 5 min. The amplified products were sequenced (Sangon Biotech Co., Ltd., Shanghai, China). The 16S rRNA gene sequence of the antifungal strain was compared with the sequences deposited in the GenBank database.1 Phylogenetic trees were constructed with the neighbor-joining method using MEGA X (Kumar et al., 2018). Genomic DNA of strain BS45 was single-molecule sequenced using PromethION, an Oxford Nanopore Technology sequencer, to obtain original sequencing data. The genome sequence has been deposited at the China National GeneBank DataBase (CNGBdb) with accession number CNP0001678. The homologous average nucleotide identity (orthoANI) was determined using the JSpeciesWS website2 online, and heat maps were constructed using the omicstudio web service.3 The antiSMASH web service4 was used to predict the secondary metabolites of strain BS45 (Blin et al., 2019; Singh et al., 2021).
The gram-reaction test was performed using a Gram stain kit (Hopebio Co., Ltd., Shandong), and the results were observed by light microscopy. Cell morphology was observed using scanning electron microscopy (SEM, Hitachi S-3400 Tokyo, Japan). The SEM samples were prepared as follows: strain BS45 was cultured in LB medium at 37°C for 16 h. Then, 1 ml of liquid culture was absorbed and added into centrifuge tube, centrifuged at 3,000 × g for 2 min, and bacterial cells were collected after the supernatant was discarded. Bacterial cells were suspended in a 2% glutaraldehyde solution at 4°C for 4 h. Glutaraldehyde was discarded after centrifugation at 4,000 × g for 5 min. The fixed bacteria were dehydrated step-by-step with 30, 50, 60, 70 and 80% concentrations of ethylalcohol for 10 min per gradient step. The final bacterial solution was suspended in 100% anhydrous ethanol. The dehydrated samples were mounted onto a SEM scaffold and subsequently sputter-coated with Au. SEM was used to observe the samples, and images were taken.
Strain BS45 was cultured on LB medium at 37°C for 4 days. Sterile fermentation broth was collected by centrifugation at 13,000 × g for 10 min. The liquid was then filtered through a 0.22-μm microporous filtration membrane and stored in a medical freezer at 4°C for subsequent tests.
Samples of the sterile fermentation broth of BS45 were placed independently at 40°C, 60°C, 80°C and 100°C and then sampled at 0.5, 1, 1.5, and 2 h to measure the thermal stability of the strain BS45 bacteriostatic substance. The untreated sterile fermentation broth was used as the control, and F. graminearum acted as the indicator strain. The inhibitory activity of the sterile fermentation broth under different treatment conditions against pathogenic bacteria was determined.
The pH of the sterile fermentation broth of BS45 was adjusted independently to 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12 using 1 mol/L NaOH and HCl solutions, and placed at room temperature for 1 h. A total of 12 subsamples with different pH values were obtained. The untreated sterile fermentation broth was used as the control, and F. graminearum acted as the indicator strain. The inhibitory activity of the sterile fermentation broth under different treatment conditions against pathogenic bacteria was determined.
The trypsin solution was prepared with phosphate buffer and then added to the sterile fermentation broth to adjust the final concentration to 1 mg/ml trypsin. The sterile fermentation broth was treated at 37°C for 1 h and then treated at 65°C for 20 min. Aseptic fermentation broth with phosphate buffer was used as the control to test the inhibitory activity of aseptic fermentation broth under different treatment conditions against pathogenic fungi.
The sterile fermentation broth was poured into six disposable sterile petri dishes without covers, and placed in a sterile environment 30 cm away from UV irradiation for 2, 4, 6, 8, 10, or 12 h. The sterile fermentation broth without UV irradiation was used as a control, and F. graminearum served as the indicator strain. The inhibitory activity of the sterile fermentation broth under different treatment conditions against pathogenic bacteria was determined.
Strain BS45 was cultured on LB medium at 37°C for 4 days. Sterile fermentation liquid was collected by centrifugation at 13,000 × g for 10 min. The extraction method was acid-precipitation extraction. 6 mol/L HCl was gradually added into the fermentation supernatant to adjust the pH of the supernatant to 2 and placed at 4°C overnight. Then, the supernatant was centrifuged at 4°C and 12,000 × g for 10 min to collect the precipitant. Methanols (50 ml × 3) were used for extraction precipitations. A rotary evaporator was used to evaporate the methanol extract of BS45 at 37°C to obtain a yellow solid, which was dissolved in methanol to obtain the crude methanol extract (Liu et al., 2022). The crude methanol extract of BS45 was sterilized using a 0.22-μm filter membrane.
Inhibition of fungi was performed with the PDA-amendment method as described by Lyu et al. (2017). Mycelia of F. graminearum agar plugs with a 5-mm diameter were then transferred to PDA containing a series of concentrations (0, 25, 50, 100, 200, and 400 μg/ml) of the extract dissolved independently in methanol. PDA amended with methanol (1%, v/v) was used as a control. Growth was observed after incubation at 25°C for 5 days. Each concentration was repeated three times. The colonies with the maximum and minimum diameters were measured, and the average maximum and minimum diameters were calculated. The formula for the inhibitory rate is as follows (Zhao et al., 2017):
The toxicity regression equation, correlation coefficient and concentration for the 50% of maximal effect were calculated using IBM SPSS statistical software (International Business Machines Corp, United States; https://www.ibm.com/cn-zh/analytics/spss-statistics-software).
Fusarium graminearum tip mycelia were selected, inoculated onto PDA solid plates and cultured at 28°C for 7 days. CMC liquid medium was selected as the sporulation medium, and a 5-mm cake of F. graminearum was taken from the F. graminearum culture plate using a hole punch. The cake was added into the CMC liquid medium and cultured at 25°C for 4 days. Conidial suspensions were collected using eight layers of sterile cheese cloth. The conidial concentration was calculated using a hemocytometer, and the conidial concentration was adjusted to 1 × 106 spores/ml.
The conidial suspension and methanol extract of BS45 were added to the PDB medium at final concentrations of 1 × 105 spores/ml and 100 μg/ml, respectively. PDB amended with methanol was used as a control. The spores were observed under a light microscope (BM2000, Nanjing Jiangnan Novel Optics Co., Ltd., Nanjing) at 3 and 24 h of incubation.
The mycelial growth of F. graminearum was measured using the dry cell weight method. Crude methanol extract with a final concentration of 100 μg/ml was added to PDB with 1 × 105 spores/ml spores and then cultured at 28°C with shaking at 180 r/min. The same volume fraction of methanol was added as a control. Measurements were collected every 24 h, on three parallel samples each time. The fermentation broth was filtered with a vacuum pump, washed with sterilized ultra-pure water and then dried at 65°C to constant weight. A growth curve was constructed with incubation time as the abscissa and mycelial weight as the ordinate.
The micromorphology of F. graminearum mycelia was observed using an SEM (Hitachi S-3400 Tokyo, Japan) after the mycelia were cultured for 3 and 24 h with or without treatment with the crude methanol extract of BS45. Mycelia were selected, treated with 2% glutaraldehyde for 4 h and then placed on SEM scaffolds. After drying, the mycelia were coated by Au sputtering. SEM was used to observe the samples, and images were taken (Jiang et al., 2018).
Mycelia of F. graminearum cultured in PDB medium at 28°C with or without 100 μg/ml of crude methanol extract of BS45 for 48 h at 28°C. The cell suspension was mixed with 0.4% Trypan blue staining solution (Sangon Biotech Co., Ltd., Shanghai, China) in an appropriate proportion (0.04% final concentration of Trypan blue), stained for 3 min and observed under a light microscope (BM2000, Nanjing Jiangnan Novel Optics Co., Ltd., Nanjing).
Mycelia of F. graminearum were cultured in PDB medium at 28°C with or without 100 μg/ml of crude methanol extract of BS45 for 48 h at 28°C. The supernatant was obtained by centrifugation at 5,000 × g for 5 min. Then the OD260 and OD280 values were detected to evaluate the leakage of nucleic acids and proteins from F. graminearum. Mycelia of F. graminearum were cultured in PDB medium at 28°C with or without 100 μg/ml of crude methanol extract of BS45 at 28°C. After being treated for 48 and 72 h, the mycelia were obtained by centrifugation at 5,000 × g for 5 min. The soluble protein content in mycelia was determined with the Coomassie brilliant blue G-250 method according to previous research (Grintzalis et al., 2015).
Mycelia of F. graminearum cultured in PDB medium at 28°C with or without 100 μg/ml of crude methanol extract of BS45 were collected at 12-h intervals. The mycelia were washed with isotonic, ice-cold NaCl (0.9%, w/v) solution, blotted to dryness, and weighed to 0.1 g. Then, 1-ml NaCl (0.9%, w/v) solution was mixed with mycelia and homogenized using a homogenizer under cold conditions. The supernatant was obtained using a refrigerated centrifuge at 8,000 × g for 10 min. The supernatants were used for protein content estimation, determination of SOD, CAT and POD activity levels, and for malondialdehyde (MDA) content determination.
Reactive oxygen species were detected using the fluorescent probe 2′,7′,-dichlorofluorescein diacetate (DCFH-DA; Parent et al., 2017). Normal cultured mycelia and mycelia treated with crude methanol extract (100 μg/ml) were selected and cover mycelia with 0.01 M DCFH-DA. The mycelia were incubated in the dark at 28°C for 20 min. The mycelia were cleaned with ultrapure water and observed with a fluorescence microscope (Olympus IX53, China) after being dried.
The MMP was detected using fluorescent probe JC-1. Normal cultured mycelia and mycelia treated with crude methanol extract of BS45 were used with JC-1 for 15 min at 28°C in the dark. The residual dye was washed with ultrapure water and observed using a fluorescence microscope (Olympus IX53, China) with 515-nm and 529-nm filters.
The MDA content was determined using the thiobarbituric acid (TBA) and can be measured by producing 3,5,5′-trimethyloxazol2, 4-dione with an absorption peak at 532 nm using MDA and TBA under acidic and high-temperature conditions. A 2-ml preparation of supernatant was mixed with 2-ml 0.67% TBA solution for 30 min at 100°C. The absorbance values of the supernatant at 450, 532 and 600 nm were determined, and each treatment was repeated three times (Khoubnasabjafari et al., 2016).
POD, SOD, and CAT activity levels of the prepared supernatant were determined using the respective activity detection kits (Solarbio, Beijing, China) in accordance with the manufacturer’s instructions.
Mycelia of F. graminearum cultured in PDB medium together with (ET group) or without (control group, CK) 100 μg/ml of crude methanol extract of BS45 for 48 h at 28°C were collected. The mycelia were quickly frozen with liquid nitrogen and then total RNA was extracted from the tissue with TRIzol® Reagent (Invitrogen, CA). The mRNA from the total RNA was isolated with magnetic beads (Invitrogen) with oligo (dT). The cDNA obtained from the extracted mRNA with a SuperScript double-stranded cDNA synthesis kit (Illumina, CA) with random hexamer primers (Illumina). The cDNA library was constructed with a Truseq™ RNA sample preparation Kit (Illumina, San Diego, CA). Transcriptome sequencing using an Illumina NovaSeq 6,000 platform (Majorbio, Shanghai, China). Each group had three biological replicates. Quality control of raw data using fastp software (version 0.19.5, https://github.com/OpenGene/fastp) to obtain high-quality clean data. After the Read Counts of genes were obtained, DESeq2 software was used to analyze the differentially expressed genes (DEGs) among multiple (≥2) samples and identify DEGs between samples to study the DEG functions. Absolute fold change ≥2 and p-adjust <0.05 were used as thresholds to identify the DEGs. All the DEGs obtained by transcriptome assembly were compared with six databases—Non-redundant (NR), Swiss-Prot, Pfam, Clusters of Orthologous Groups (COG), Gene Ontology Consortium (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG)—to comprehensively obtain functional information. Goatools software (Version 0.6.5, https://files.pythonhosted.org/packages) were used for GO enrichment analyses of DEGs. Fisher tests were used to determine the accuracy, and the FDR with Benjamini/Hochberg correction was determined. At a corrected p-value (Padjust) < 0.05, the GO function was considered significantly enriched. A KEGG pathway enrichment analysis was performed on DEGs using R script, and the calculation principle that used for the GO functional enrichment analysis. At a corrected p-value (Padjust) < 0.05, the KEGG pathway was considered significantly enriched. The raw data for both experiments have been deposited at the National Genomics Data Center (NGDC) with accession number CRA007209.
Total RNA was extracted using RNAiso Plus (TaKaRa) in accordance with the manufacturer’s instructions. RNA was transcribed into cDNA using the reverse transcription kit ToloScript RT EasyMix for qPCR (ToloBio, Shanghai, China). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Premix ExTaq™ (TaKaRa) and the ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The PCR conditions were as follows: pre-denaturation at 95°C for 10 min, denaturation at 95°C for 10 s, and 40 cycles of amplification (95°C for 15 s, 55°C for 30 s). β-Tubulin was used as an endogenous reference gene (Scala et al., 2017), and the relative expression levels of target genes were calculated using the 2−△△Ct method. Primer sequences used for qRT-PCR are listed in Supplementary Table S1.
The steps of wheat disinfection are as follows: (1) place in 70% ethanol for 1 min; (2) place in 3% sodium hypochlorite for 6 min; and (3) wash six times with sterile water. A final application of sterile water to the LB medium determines whether disinfection is complete. Then, 200 identically sized, plump, healthy wheat seeds were picked. In total, 100 seeds were soaked in 10 ml bacterial suspension (1 × 108 CFU/ml) for 4 h, and the experimental group was named T. The control group, which consisted of 100 seeds soaked in 10 ml sterile water for 4 h, was named CK. Wheat seeds were placed in a 150-mm sterile petri dishes. The petri dishes were then exposed to light for 16 h and darkness for 8 h under aseptic conditions. The germination times of seeds were observed and recorded. After wheat seeds were cultured for 9 days, their biomass was determined. The root and stem lengths of wheat seedlings were measured using Vernier calipers. An electronic balance was used to measure dry weights.
Wheat seeds were incubated in a sterile 150 mm chamber covered with sterilized filter paper and wetted with 10 ml of sterile water in the dish. The petri dishes were placed under sterile conditions of 16 h light and 8 h dark, and the growth of wheat was observed regularly. When the primary root roots of seedlings reached approximately 50 mm, the wheat roots were immersed in a suspension of F. graminearum at a concentration of 1 × 105 conidia. Three groups of experiments were performed: in the first group, suspension of strain BS45 was added; in the second group, sterile water was added as a control; in the third group, carbendazim solution was added. Each group contained 15 wheat seedlings. After 4 and 8 days, the extent of root necrosis was measured. The formulae to determine the inhibitory ability of Bacillus BS45 against root rot incorporating a reference rating index (0 = asymptomatic; 1 = mild necrosis; 2 = moderate necrosis; 3 = severe necrosis; 4 = complete necrosis) were as follows (Wang and Gottwald, 2017):
IBM SPSS statistical software (Armonk, New York, United States) was used for the statistical analyses. The results of all determinations are expressed as mean ± standard deviation (SD). Statistical significance was evaluated with one-way ANOVA, followed by t tests. p < 0.05 was considered to indicate a statistically significant difference. Origin5 and R software [R: The R Project for Statistical Computing (r-project.org)] were using for constructing graphics.
In total, 24 strains with inhibitory activity against F. graminearum were isolated and screened (Supplementary Table S2). Colony morphology is shown in Supplementary Figure S1A. Strain BS45 was opalescent, opaque and wrinkled on the surface with an irregular contour on LB solid medium. Strain BS45 was also Gram-stain positive Supplementary Figure S1B. The bacteria were rod-shaped with no pods or flagella observed by SEM, and they were (2.15 ± 0.23) μm × (0.70 ± 0.03) μm in size, as shown in Supplementary Figure S1C.
The 16S rRNA gene of strain BS45 was amplified using 27F and 1492R primers, and the PCR products were sequenced. A phylogenetic tree was constructed using the 16S rRNA results, and it showed that the strain BS45 was closely related to the Bacillus genus (Figure 1A).
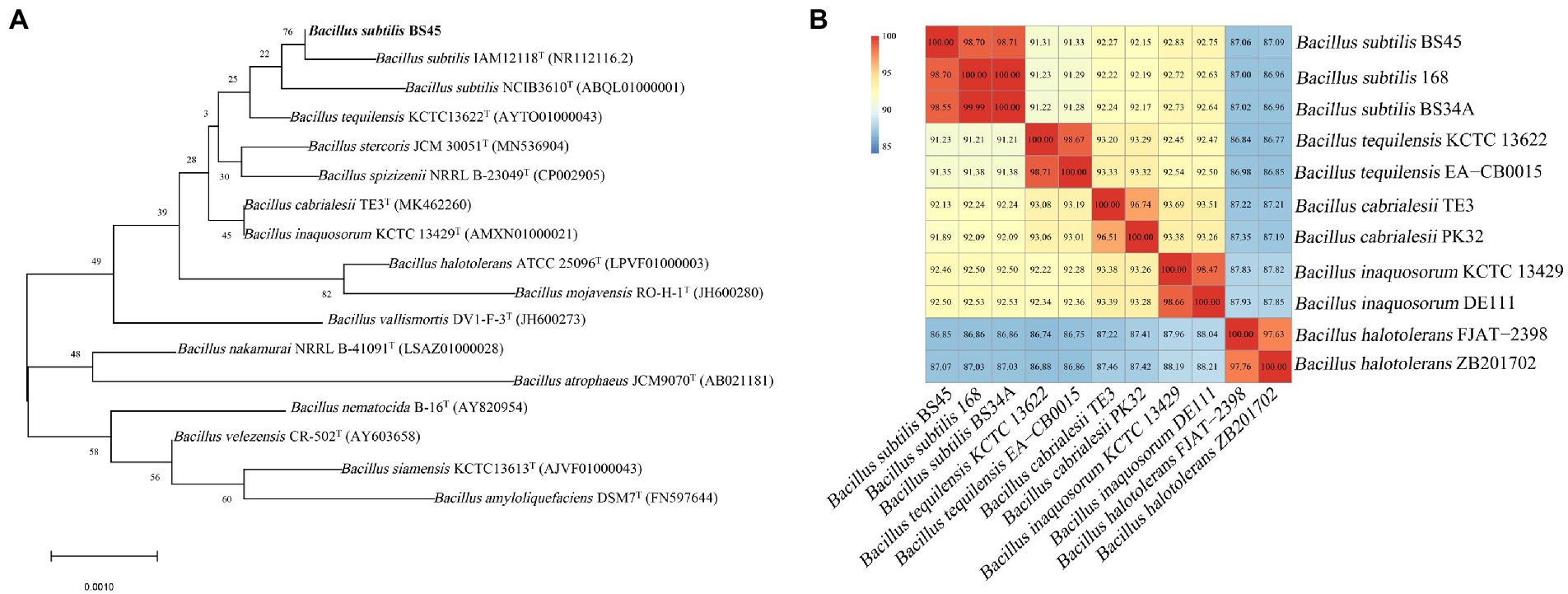
Figure 1. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences, showing the phylogenetic relationships of strain BS45 and related strains (A). Heat map of the average nucleotide identity based on the complete genome sequence of strain 45 and related strains (B).
The genome size of strain BS45 is 4,048,190 bp. Comparisons of the whole genome of strain BS45 with those of other strains of the same genus revealed that the average nucleotide identity (ANI) value between strain BS45 and B. subtilis was the highest, exceeding 98% (Figure 1B). The ANI values between the strain and the whole genome of B. subtilis 168 and B. subtilis BS34A were 98.7 and 98.55%, respectively, higher than the threshold of species delineation (95%). These results were consistent with the 16S rDNA-based phylogenetic tree. Therefore, strain BS45 was classified as B. subtilis.
The whole genome of strain BS45 was sequenced using PromethION, a third-generation high-throughput sequencing platform. After quality control, a whole genome composed of one contig was obtained, with a size of 4,048,190 bp. The average GC content was 43.85%, including 30 rRNAs and 86 tRNAs (Table 1). Microorganisms are potential sources of unique low-molecular weight bioactive secondary metabolites, which can play important roles in many fields, such as medicine and agriculture. In accordance with the comparative genomic analysis, the antiSMASH web service was used to predict the secondary metabolites of strain BS45. In total, 11 gene clusters related to the synthesis of secondary metabolites were predicted, including 2 terpene gene clusters and 2 T3PK gene clusters (Supplementary Table S3). Additionally, one CDPS, one RRE-containing and one sactipeptide gene cluster, as well as four non-ribosomal peptide synthase gene clusters, were predicted. The latter were responsible independently for the biosynthesis of surfactin, bacillaene, fengycin, and bacillibactin (Supplementary Figure S2).
The regression equation of inhibitory toxicity was y = 5.02 + 2.57x, the correlation coefficient was 0.99, and the concentration for the 50% of maximal effect was 90.974 μg/ml (Table 2). These values indicated that the active inhibitory components of metabolites were enriched in the methanol solvent. The metabolites of strain BS45 had a good inhibitory effect on F. graminearum, and strain BS45 had potential to be developed into an agricultural biological control agent.
The antibacterial activity of strain BS45’s fermentation broth was stable at pH 2–10, and the statistical analysis showed that there were no significant differences in the inhibitory rates. The inhibitory rates of the bacterial fermentation broth at pH 11 and 12 decreased compared with at other pH levels (Supplementary Figure S3A). Thus, the tolerance of the active components in strain BS45’s fermentation broth to strong alkalinity was lower than that to acidity, and it had good stability under acidic to weak-base conditions.
The antibacterial activity of BS45’s fermentation broth was affected by high temperature and treatment time. During a short time period, the temperature increase did not significantly affect the antibacterial activity of strain BS45’s fermentation broth Within 1 h of treatment time, there were no significant differences in the bacteriostatic rates of the fermentation broth at 40–100°C, and the bacteriostatic rates were the same as that of the untreated original fermentation broth (Supplementary Figure S3B).
The fermentation broth of strain BS45 was resistant to trypsin. After 1.5 h of trypsin treatment, the fermentation broth still had good antibacterial activity, and there was no significant difference compared with the control group (Supplementary Figure S3C).
Additionally, UV light had little effect on the active components of the fermentation broth (Supplementary Figure S3D).
The crude methanol extract of strain BS45 inhibited the conidial germination rate of F. graminearum. After 3 h of culturing, conidia began to germinate in the control group (Figure 2A), whereas conidia treated with the extract did not germinate (Figure 2B). After 24 h of culturing, all the conidia in the control group grew into normal hyphae (Figure 2C). Although the length of the conidia bud tube after the extract treatment increased, obvious swelling was observed (Figure 2D), which indicated that morphogenesis was significantly abnormal compared with the control group. Thus, the metabolites of strain BS45 inhibited the conidial germination process and induced changes in conidial morphology, thereby inhibiting the normal growth of F. graminearum.
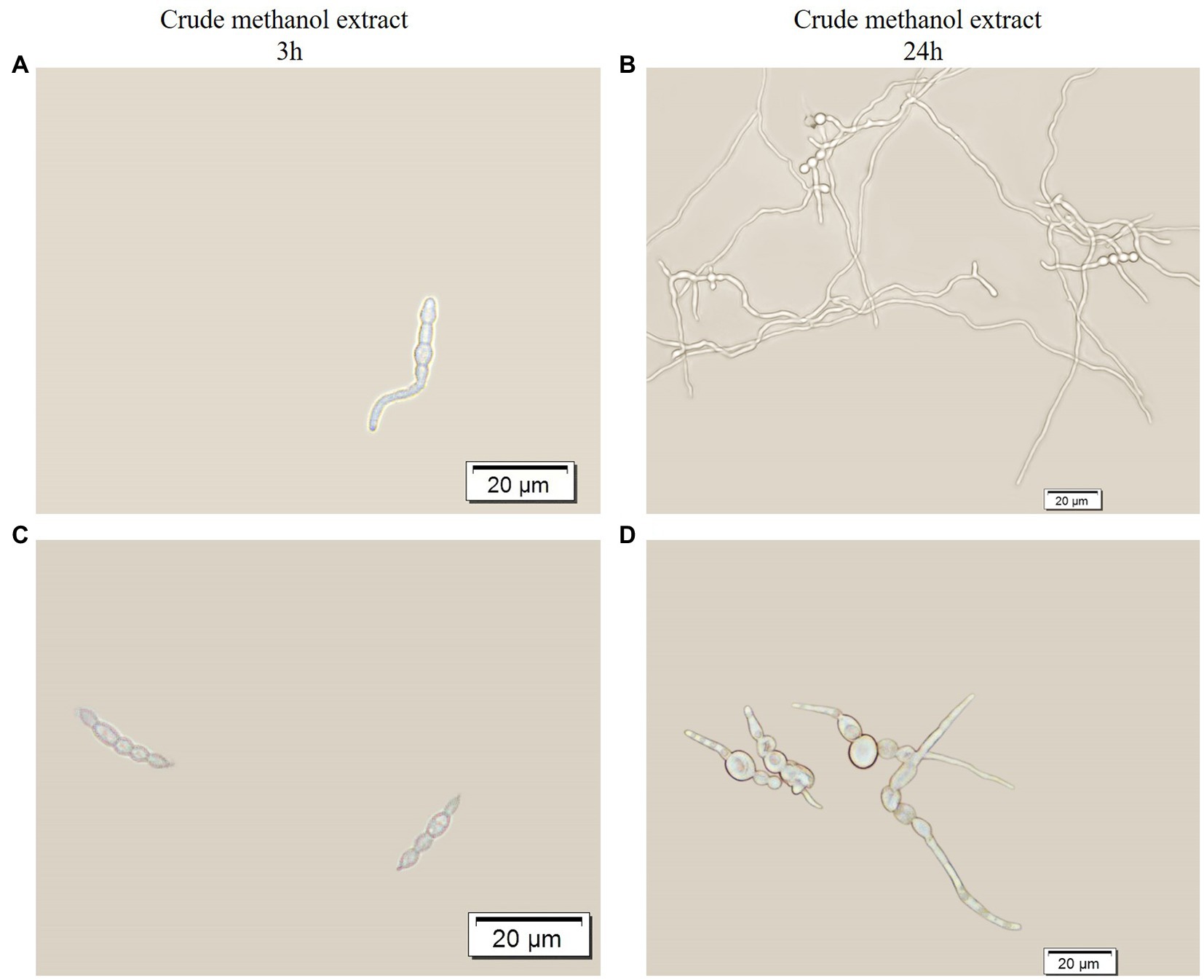
Figure 2. Optical micrograph showing the germination of spores with or without treatment with crude methanol extract (100 μg/ml) for 3 h (A,C) and 24 h (B,D).
The experimental results of the influence of the crude methanol extract of BS45 on the growth of F. graminearum are shown in Figure 3. The dry weight of mycelia treated with methanol extract was significantly different from that of the control group from the 3rd day of culturing. The dry weights of treated mycelia increased slowly, by only 0.17 g from 2 to 8 days. Thus, the methanol extract of BS45 had a significant effect on the growth of F. graminearum.
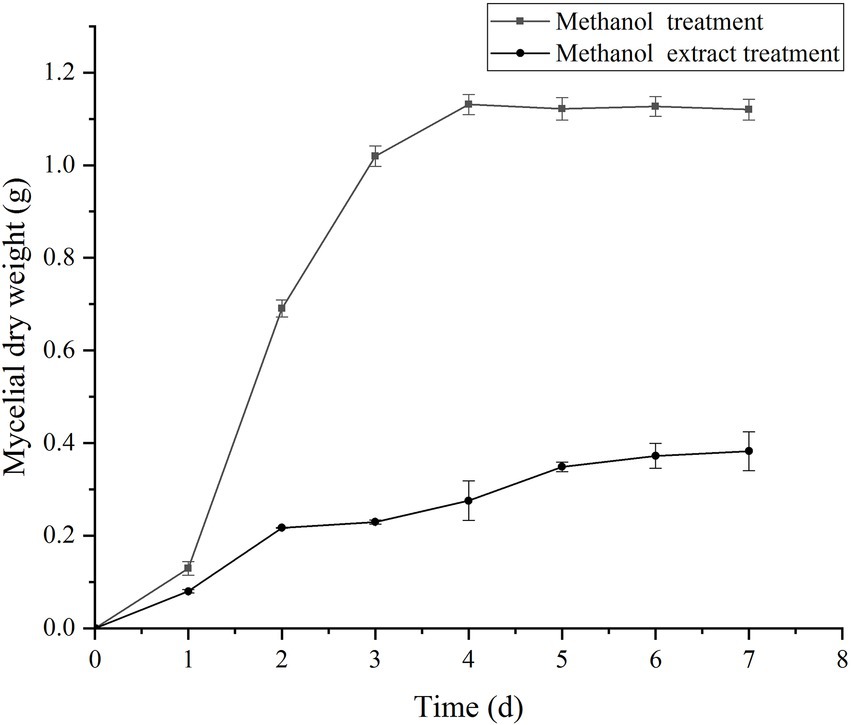
Figure 3. Effect of crude methanol extract (100 μg/ml) on mycelial growth of F. graminearum. The error bars in the figures indicate the standard deviations (±SD) from three independent samples.
The micromorphological changes of mycelia treatment with or without crude methanol extract were observed using SEM. Mycelia incubated without crude methanol extract showed equal widths, even surfaces and healthy growing branches (Figures 4A,B). The mycelia in the treatment group underwent tip and node swelling, followed by collapse and shrinkage after expansion (Figures 4C,D).
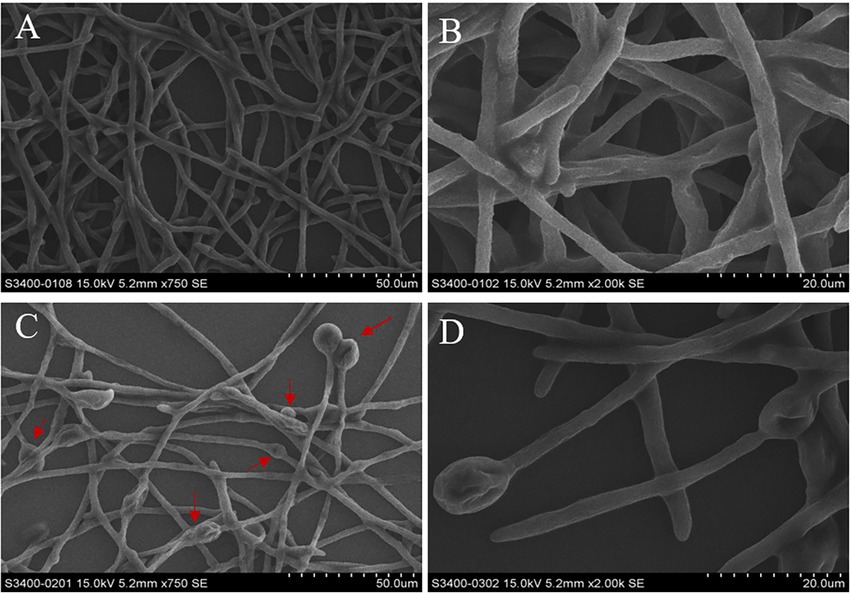
Figure 4. Micromorphology of mycelia from 48 h old cultures of F. graminearum under scanning electron microscopy at 750× and 2,000× magnification. Mycelial structure of F. graminearum. (A,B) In the absence of methanol extract treatment. (C,D) In the presence of treatment with crude methanol extract (100 μg/ml). The red arrows indicate areas of swelling.
Trypan blue dye can penetrate through a damaged cell membrane into the cell and bind to DNA, resulting in a blue color. After methanol extraction, the membranes of the swollen mycelia were damaged, thus allowing Trypan blue to enter and bind the DNA, and making the cells appear blue, whereas the DNA of the control cells was not stained (Figures 5A,B).
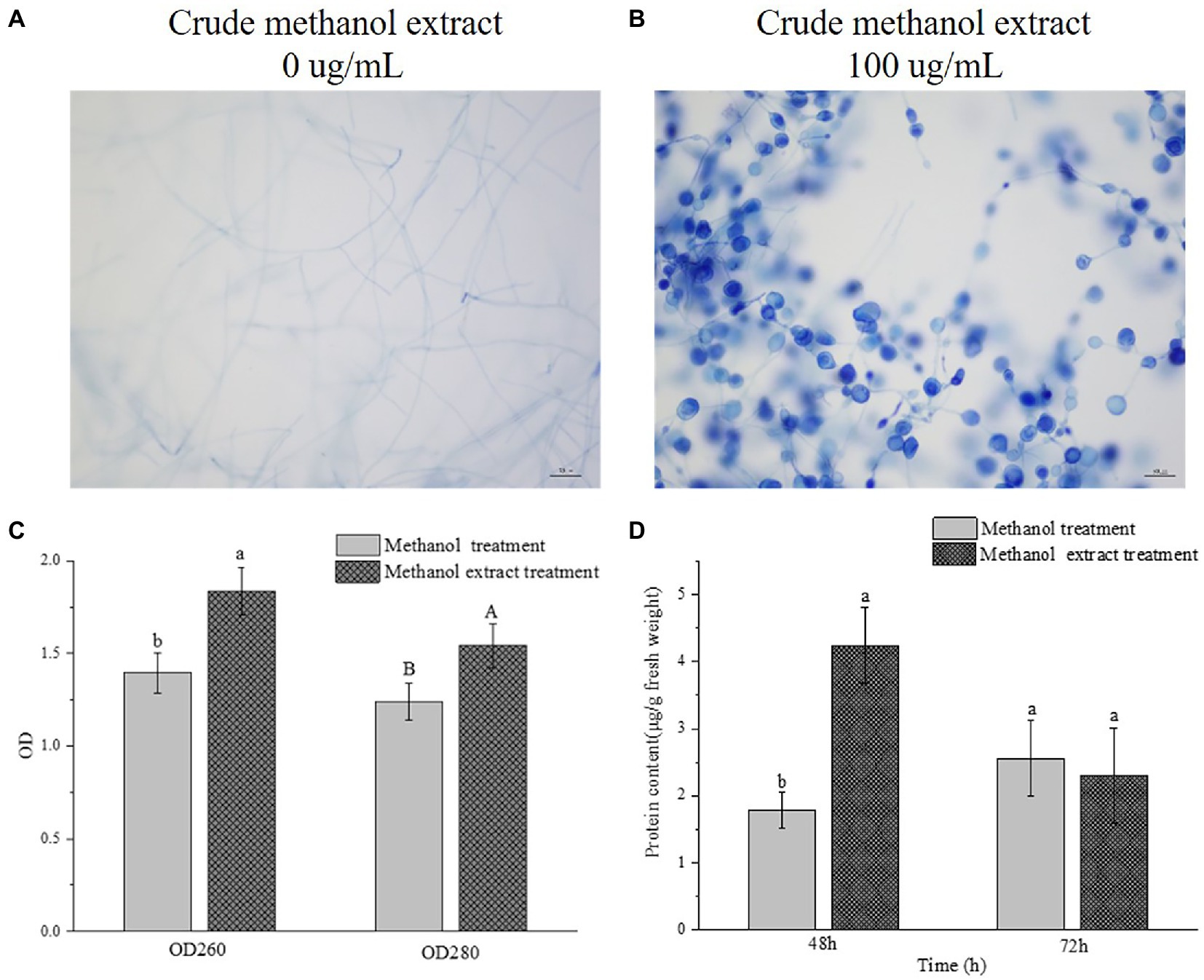
Figure 5. Effects of crude methanol extract on the membrane permeability and protein content of F. graminearum hyphal cells. (A,B) 40× optical micrograph of Trypan blue stained mycelia treated with crude methanol extract for 48 h. (C) Leakage levels of nucleic acid (OD260) and proteins (OD280) from F. graminearum suspensions, with or without treatment with crude methanol extract (100 μg/ml). (D) In vivo protein content of F. graminearum. Different letters indicate significant differences between the lines (p < 0.05).
The leakage of macromolecular substances from the cells after methanol extraction was measured. Because nucleic acids and proteins have strong absorbency values at OD260 and OD280, the changes at OD260 and OD280 can reflect the leakage of nucleic acids and proteins from cells, allowing the integrity of cell membranes to be investigated. The amounts of nucleic acid and protein leakage of F. graminearum after crude methanol extraction are shown in Figure 5C. Both OD values of the extracellular fluid in the treatment group were significantly increased compared with those of the control group. Thus, the methanol extraction affected the integrity of bacterial somatic membranes. One way in which the metabolites of strain BS45 are inhibited is by inducing cell content leakage. The protein content in a thallic unit at different periods was determined and calculated, as shown in Figure 5D. The protein content in the treated group was greater than that in the control group after 48 h, and they were the same at 72 h.
Low-concentration ROS act as intracellular messengers of many molecular events in cells (Cadenas and Davies, 2000), and they also play important roles in secondary metabolite synthesis in fungi (Zhu et al., 2020). However, the accumulation of excessive ROS damages cells (Ongena and Jacques, 2008). To investigate whether ROS is involved in the process of F. graminearum cell death induced by metabolites of strain BS45, the fluorescence probe DCFH-DA was used to detect the ROS content in vivo. The detection principle of DCFH-DA has two parts: first, the probe is hydrolyzed by lactase to form DCFH without fluorescence. Then, DCFH is converted into fluorescent DCF123 by ROS. The fluorescence intensity of the control group was weak, whereas mycelia treated with the methanol extract produced strong green fluorescence, indicating that there was an obvious accumulation of ROS in the cells, and the fluorescence was strongest in the swollen areas (Figure 6). Thus, the normal metabolic activities of mycelia appear to be destroyed by high ROS levels, resulting in cell damage and apoptosis.
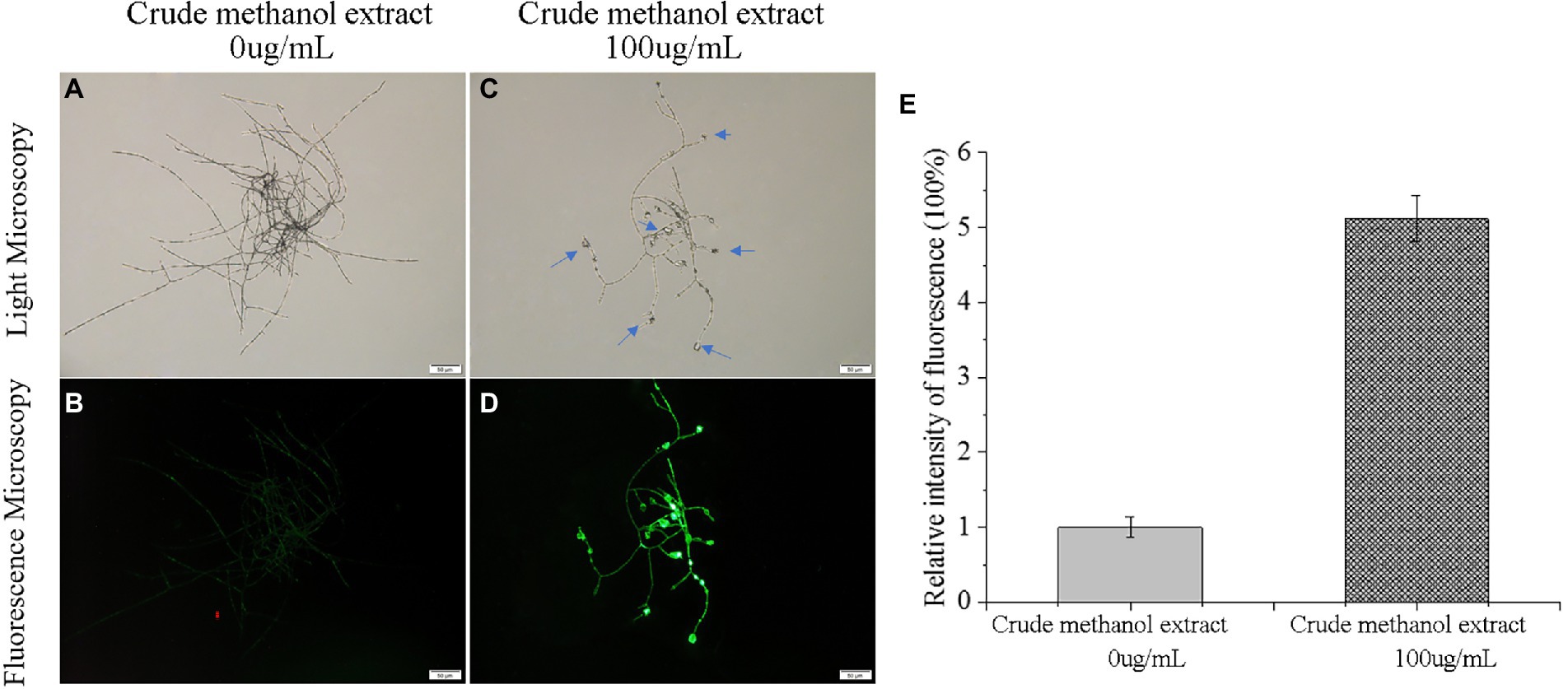
Figure 6. Effects of crude methanol extract (100 μg/ml) on reactive oxygen species and mitochondrial membrane potential in F. graminearum hyphal cells, observed by light microscopy (A,C) and fluorescence microscopy (B,D). Reactive oxygen species level fluorescence intensity (E).
The fluorescence probe JC-1 can be used to detect mitochondrial membrane potential. JC-1 forms a polymer emitting red fluorescence (Ex = 585 nm, Em = 590 nm) in normal mitochondria, and it produces green fluorescence as a monomer (Ex = 514 nm, Em = 529 nm) in mitochondria with reduced or lost membrane potential. Therefore, changes in mitochondrial membrane potential can be measured by changes in red/green fluorescence intensity. The ratio of red/green fluorescence intensity in the control group was greater than that in the treatment group (Figure 7), indicating that the membrane potential decreased. Thus, mitochondria were affected by the methanol extract, and the intracellular potential showed depolarization.
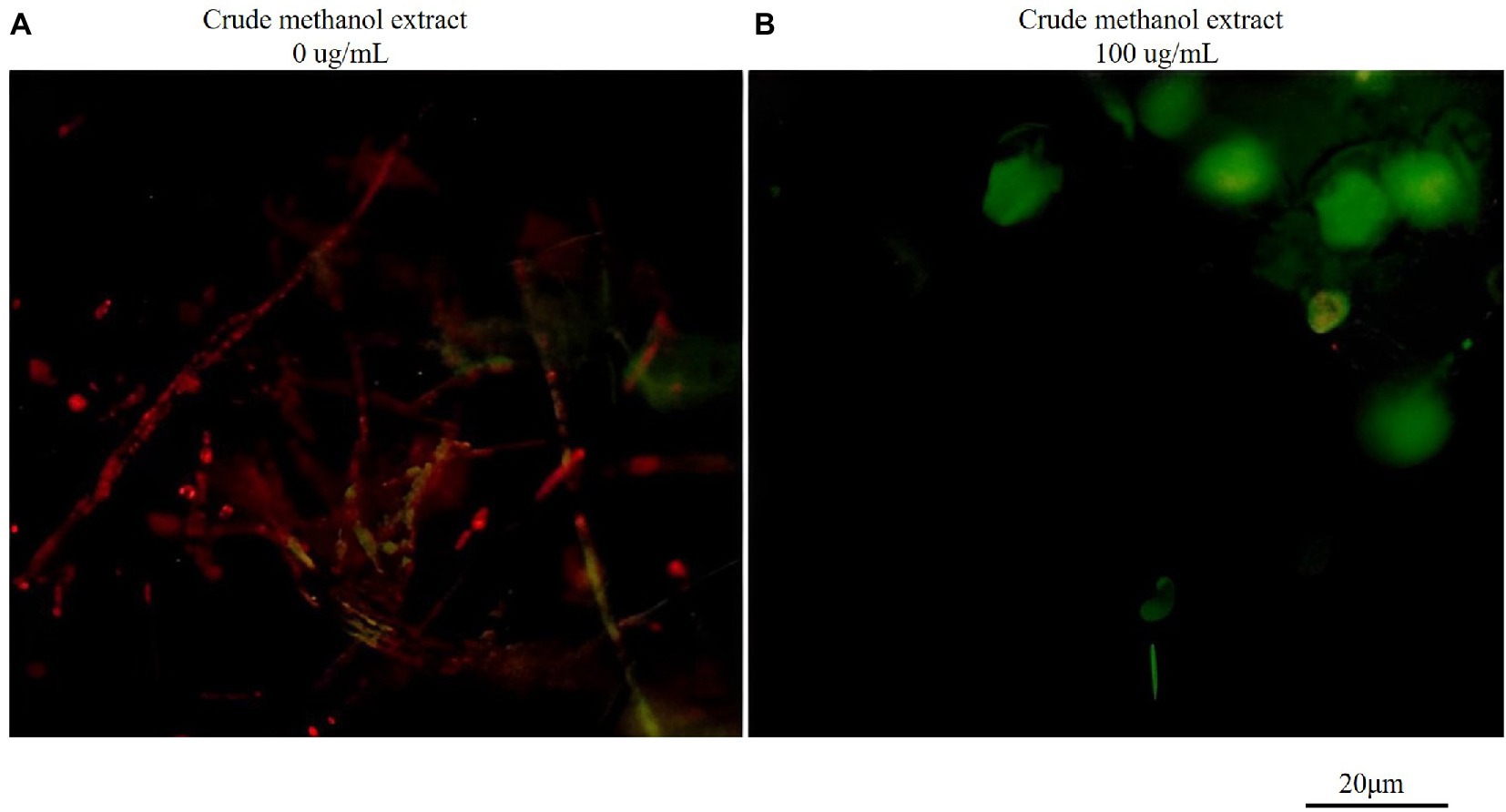
Figure 7. Effects of crude methanol extract (100 μg/ml) on mitochondrial membrane potential in hyphal cells of F. graminearum, observed by fluorescence microscopy (A,B). The scale bar applies to all panels.
The SOD enzyme converts superoxide anion into hydrogen peroxide, and CAT catalyzes hydrogen peroxide into water. The activity levels of SOD, POD and CAT in mycelia increased and then decreased (Figures 8A–C). SOD was maintained at a low level in the later stage (Figure 8A), and catalase and peroxidase activity decreased to levels similar to those in the control group (Figures 8B,C). These results indicated that mycelia were subjected to oxygen damage induced by the methanol extract, which triggered the oxidative stress response of the mycelia, and a series of ROS-related genes and proteases in the body were regulated and changed[24]. After a 24-h methanol extract treatment, the MDA concentration in mycelia was significantly higher than in the control group (Figure 8D). The MDA levels represented membranous peroxidation products. The determination of MDA contents in mycelia treated with methanol extracts compared with the control group also indirectly indicated the fungal cell oxidation levels. After exposure to methanol extract, the active oxygen levels in mycelia increased, resulting in cell membrane peroxidation, thus inhibiting the growth of F. graminearum.
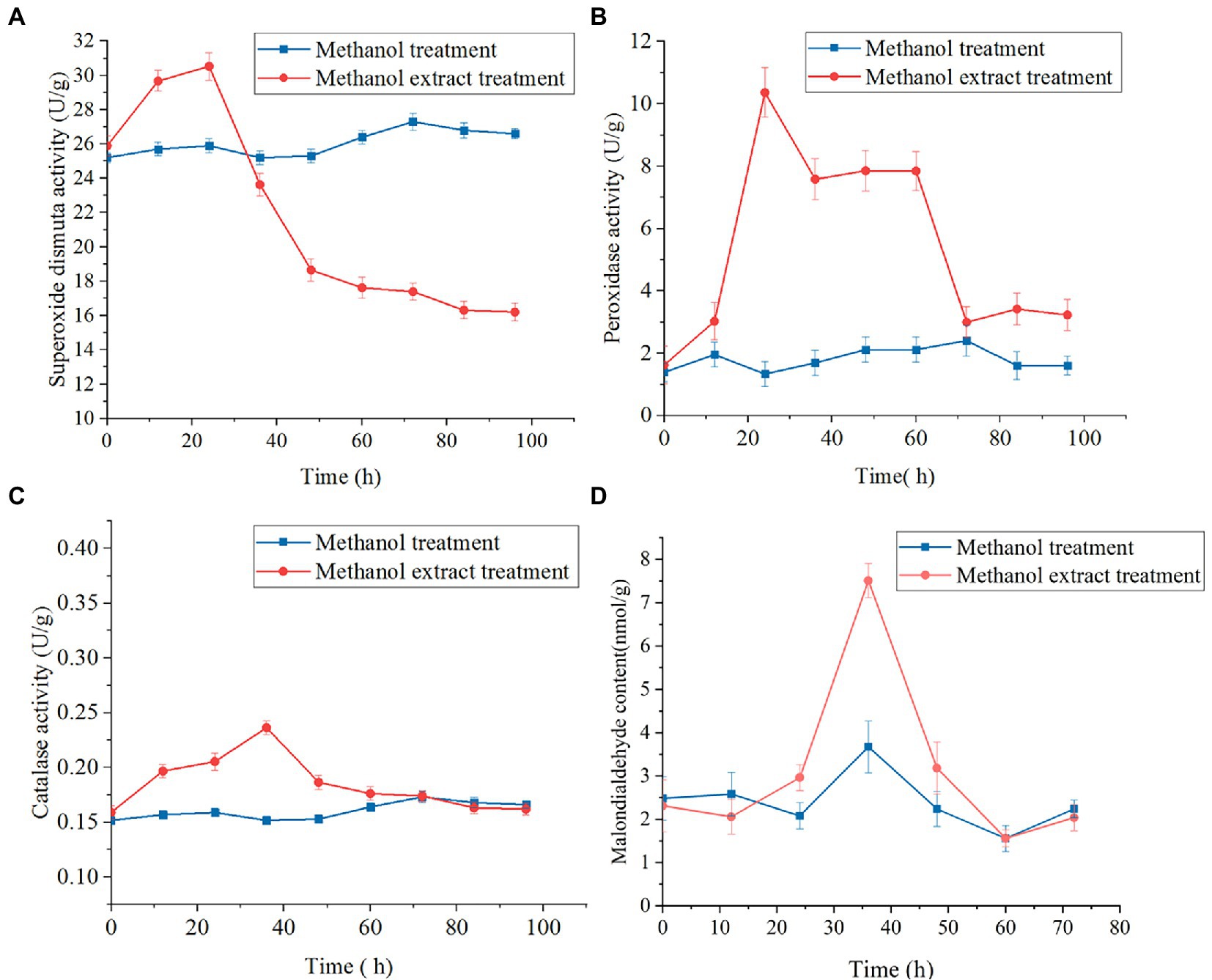
Figure 8. Effects of methanol extract (100 μg/ml) on SOD (A), POD (B) and CAT (C) activity and MDA content (D) in Fusarium graminearum. The hyphal cells of F. graminearum with or without treatment with methanol extract (100 μg/ml) are shown. Methanol (0.04% [vol/vol]) served as the control treatment. The error bars in the figures indicate the standard deviations (±SD) from three independent samples.
A transcriptome sequencing analysis was conducted on F. graminearum cultured in a PDB medium with or without crude methanol extract of BS45. It was conducted to study the expression patterns of related genes in F. graminearum and to explore the F. graminearum genes that responded to stimulation by metabolites of strain BS45. Furthermore, their antifungal mechanisms were analyzed.
Clean read data were filtered from raw reads having connectors and low-quality data to obtain high-quality data (Clean reads) for statistical analyses, and 51,423,806, 52,016,132, 47,952,218, 55,065,336, 54,654,234 and 46,375,852 clean reads were obtained after the removal of connectors and filtration. Clean read accounted for more than 99% of the sequences (Supplementary Table S4), indicating that the quality of the sequencing data was good and met the requirements for subsequent analyses. RSEM soft6 was used to calculate the expression levels of all the genes in each sample, and then, the differential expression level of the CK vs. ET group was calculated in accordance with the expression levels. A total of 3,709 differentially expressed genes (DEGs) were obtained with p ≤ 0.05, and a differential multiple higher than two was used as the screening threshold. Among the DEGs, 2,082 were down-regulated and 1,708 were up-regulated. In addition, genes with higher differential expression levels were down-regulated (Supplementary Figure S4).
To investigate the detailed effects of crude methanol extract of BS45 on F. graminearum, the GO terms and KEGG pathways were analyzed. All the DEGs were subjected to GO and KEGG pathway enrichment analyses. A total of 20 GO terms (Table 3) and 18 KEGG pathways (Table 4) were significantly enriched on the basis of p value <0.05 from 365 GO terms and 115 KEGG pathways. Some of the top 20 GO terms and all 18 of the KEGG pathways were associated with the ribosome structure, amino acid metabolism and transcript pathways. The terms of translation preinitiation complex, integral component of the membrane, transporter activity, transmembrane transporter activity, nutrient reservoir activity, oxidoreductase activity, transferase activity, hydrolase activity, and catalytic activity were highly enriched in the GO analysis. The GO enrichment analysis also showed that ribosomal RNA genes, which are associated with ribosomal biosynthesis, were significantly enriched (Table 3). In addition, the KEGG pathway enrichment analysis revealed that a large number of significantly expressed genes were annotated into the ribosome pathway and various amino acid metabolic pathways, which are associated with protein biosynthesis (Table 4).
In this study, multiple genes involved in oxidative phosphorylation and the electron transport chain were significantly upregulated (Table 5). Cytochrome C enzyme oxidase and NADH dehydrogenase metabolic pathways in the oxidative phosphorylation pathway were increased. All the peroxisome-related DEGs were significantly decreased, except for FGSG_07069 (Table 5). The expression levels of oxygen removal enzymes, such as superoxide oxidase and CAT, decreased significantly. This indicated that the mycelia of pathogenic fungi were subjected to oxygen stress after the methanol extract of BS45 treatment.
Trichothecene mycotoxins are the main toxin species produced by F. graminearum. Therefore, the synthesis pathway of trichothecene mycotoxins was studied. Consistent biosynthetic pathways for trichothecene mycotoxins were obtained by comparative genomic analysis and gene function validation. We analyzed the DEGs in the trichothecene mycotoxin synthesis pathway and found that the expression of several genes in this pathway was down-regulated after methanol extract treatment (Figure 9). Notably, the expression of TRI5, a key gene in the synthesis pathway, was down-regulated by 2.749 fold, thus indicating that strain BS45 might affect the final toxin synthesis of F. graminearum.
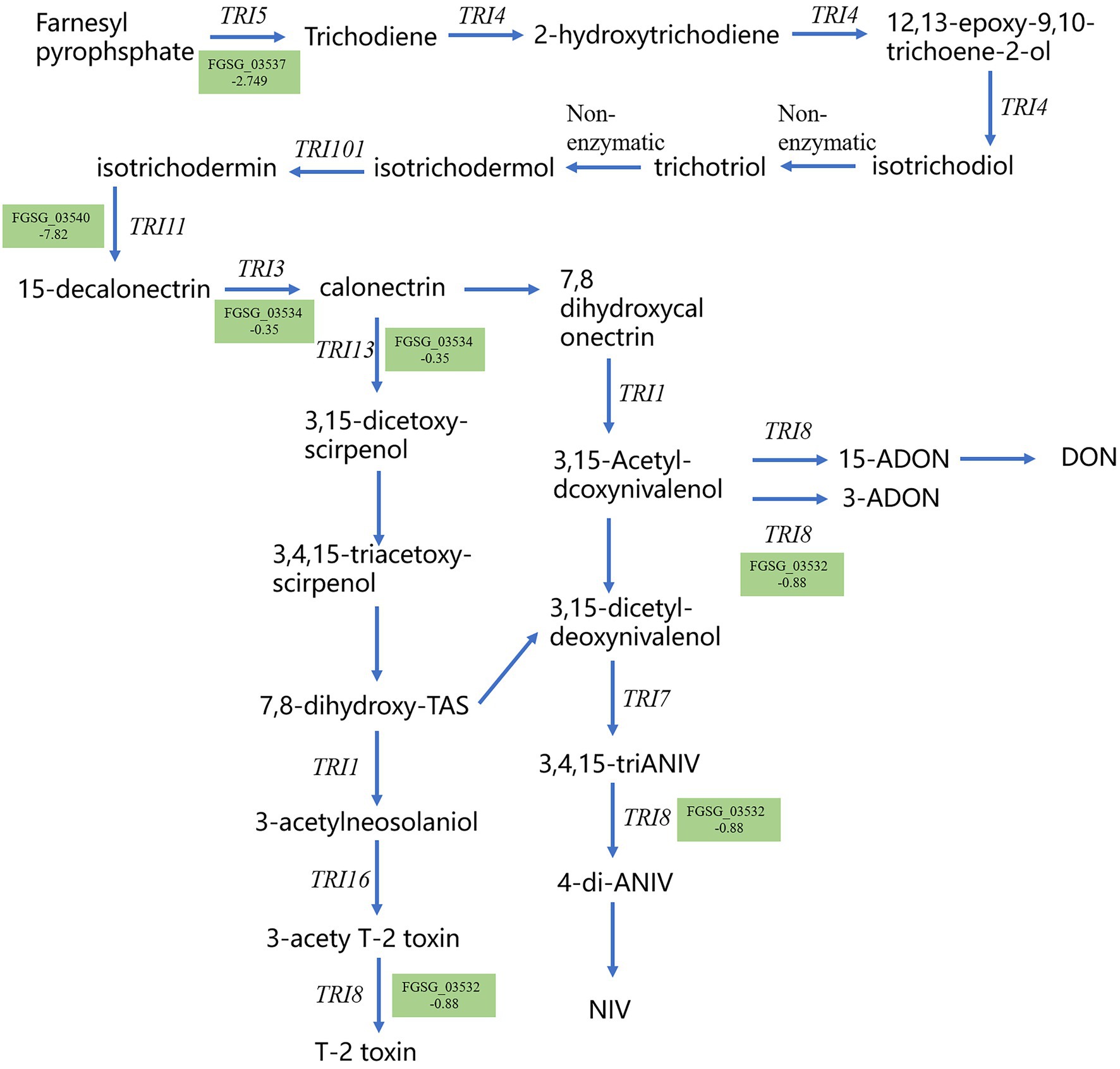
Figure 9. Gene expression for the trichothecene mycotoxin metabolism pathway. Gene names in green indicate downregulated gene expression; number represents the fold decrease.
To validate the transcriptome sequencing data, a subset of five genes was selected and expression assessed by real-time qRT-PCR. The selected genes are mainly involved in the oxidation–reduction process, ribosome regulating pathway. The upregulated and downregulated changes determined by qRT-PCR were consistent with the transcriptome data (Figure 10), indicating that the transcriptome sequencing data were reliable.
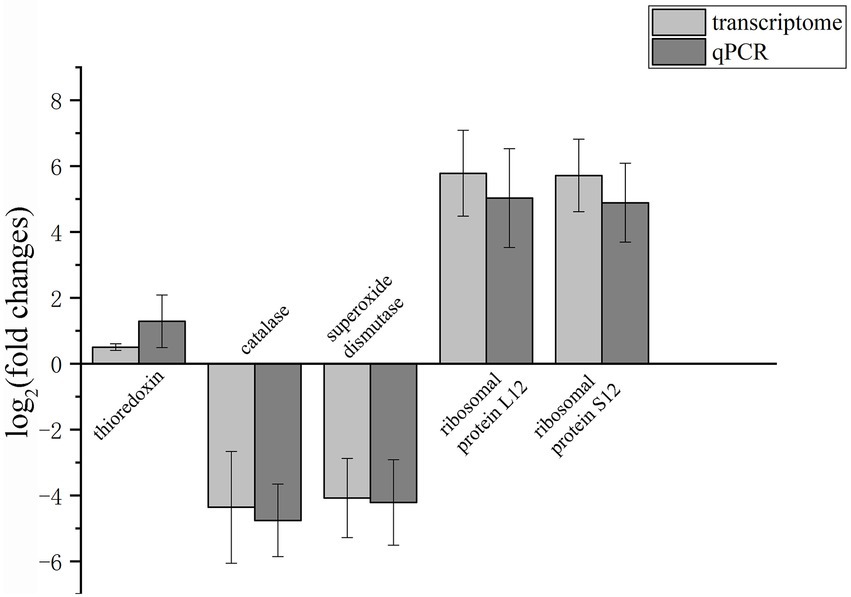
Figure 10. Validation of transcriptomic data by qRT-PCR of five selected genes. β-Tubulin was selected as the reference gene. The upregulation and downregulation of gene expression are shown above and below the X-axis, respectively.
At 9 days after inoculating wheat seeds with Bacillus BS45, wheat seedlings were observed. There was a significant difference between the experimental and control groups. Inoculation with Bacillus BS45 significantly increased the root (Figure 11A) and stem (Figure 11B) lengths of wheat seedlings. The fresh and dry weights of seedlings revealed that the plant biomass of the experimental group was greater than that of the control group, indicating that Bacillus BS45 had a growth promoting effect on wheat (Figure 11C).
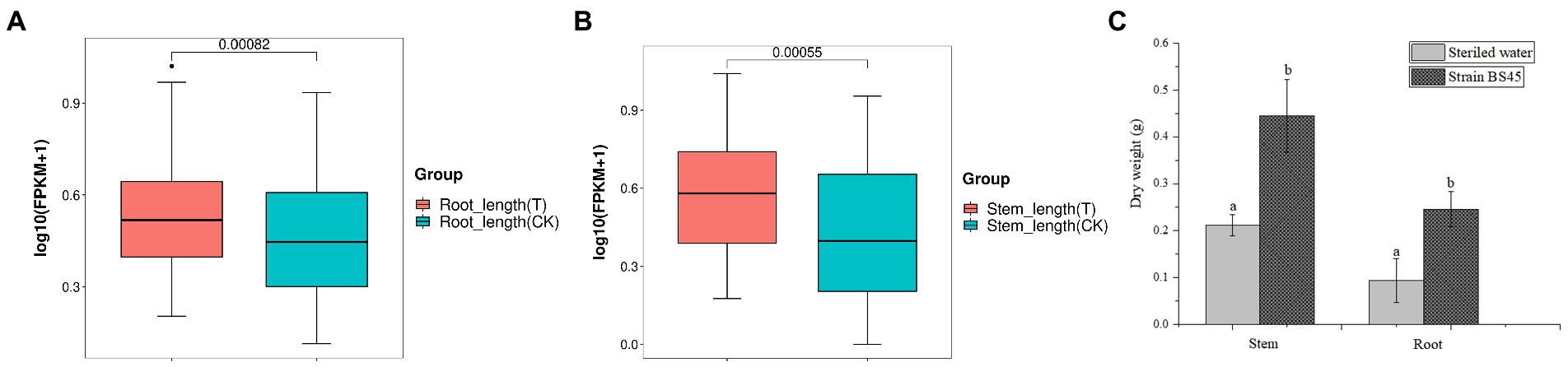
Figure 11. Effects of B. subtilis BS45 suspension on root length (A), stem length (B), and root and stem dry weights (C) of wheat. T: wheat seedlings inoculated with suspension of B. subtilis BS45; CK: wheat seedlings inoculated with sterile water. Three repeated experiments were performed, and the values are the means ± SD of the results from three independent experiments. Different letters indicate significant differences between the lines (p < 0.05).
The severity of Fusarium root rot (FRR) disease may be assessed using the rate of visible root necrosis. The biological control effect of F. graminearum and Bacillus BS45 on FRR was determined by co-culturing F. graminearum and B. subtilis BS45 during wheat growth. As shown in Table 6, the FRR disease index of wheat seedlings inoculated with strain BS45 was lower than that of wheat seedlings inoculated with water after 4 and 8 days. The biological control effect of strain BS45 was consistent with that of carbendazim after 4 days. The control effect of inoculated strain BS45 after 8 days was 40%, indicating that strain BS45 had a good biological control effect on FRR.
Fusarium graminearum is known for causing devastating spike infections and grain yield damage. Plant diseases caused by pathogenic fungi severely threaten global food production (Goswami and Kistler, 2004; Siamak and Zheng, 2018). Currently, biological control is a new strategy to control crop diseases using microorganisms and microorganism-derived metabolites. In this study, several strains of bacteria inhibiting F. graminearum were isolated. Here, we obtained a newly identified Bacillus subtilis BS45 through ANI analysis, which efficiently inhibited the growth of F. graminearum (Zheng et al., 2020). The metabolites of strain BS45 showed physical stability. The physical stability of metabolites contributes to the selection of conditions used for mass fermentation culturing (Wang et al., 2018). The biosynthetic gene clusters responsible for metabolite production in B. subtilis BS45, as predicted by genomics and bioinformatics, include lipopeptides and siderophores. These metabolites play important roles in defending against pathogens. This finding may explain the relationships between genes involved in biocontrol mechanisms and biocontrol goals and efficacy. Therefore, B. subtilis BS45 has potential for biocontrol applications. Inhibition of spore germination—a key step for the development of fungal disease during the early stage (Karlsson et al., 2021)—had a favorable effects in protecting plants from fungal infection. In this study, a crude methanol extract of B. subtilis BS45 perturbed the conidial germination of F. graminearum. Meanwhile, healthy mycelia became swollen at the ends and nodes when exposed directly to the crude extract of B. subtilis BS45. Similar results have indicated that the fungal spores of Aspergillus carbonarius form hyphae, but the hyphae are swollen after treatment with metabolites of Bacillus subtilis (Jiang et al., 2020). Bacillus metabolites have been confirmed to interact with the cytoplasmic membranes of conidia, and to interfere with conidia germination and bud tube elongation (Jin et al., 2020). Furthermore, SEM revealed that the swollen sites had collapsed. Further experiments indicated that the OD260 and OD280 values of the fermentation liquid increased, thus indicating leakage of cell contents, such as intracellular nucleic acid and proteins, which may be the reason for the observed shriveling of F. graminearum. The shrinking phenomenon has also been observed in a study of F. graminearum treated with glabridin (Yang et al., 2021) Meanwhile, in accordance with the elevated protein content in this early stage, we speculated that this self-protection mechanism might be initiated by microorganisms to cope with external environmental stress. In the later stage, lipid peroxidation of the cell membrane led to the destruction of the membrane, thereby resulting in the release of the cell contents and a decrease in protein content. Similar results have been found in studies on the protein content of other pathogenic fungi (Zhou et al., 2017).
Bacillus has demonstrated complex inhibition mechanisms against a variety of pathogenic fungi. In this experiment, we demonstrated that ROS levels were induced by the crude extract, and the observed fluorescence intensity was five times higher in the treated group than in the control group of F. graminearum. Recent studies have indicated high levels of ROS in pathogenic fungi such as Fusarium, Magnaporthe grisea, Phytophthora infestans, and Verticillium dahliae, thus suggesting that the death of pathogenic fungi through the induction of ROS accumulation by Bacillus may be a common mechanism (Han et al., 2015; Zhang and Sun, 2018; Liu et al., 2019; Wang et al., 2020). In eukaryotes, mitochondria are the sites of ROS production. Heat-induced ROS production and cell death in Saccharomyces cerevisiae cells are accompanied by hyperpolarization of the mitochondrial inner membrane (Pyatrikas et al., 2015). However, in response to the strain Magnaporthe grisea under fengycin stress, ROS levels increase while MMP decrease; however, the correlation between them remains to be determined (Zhang and Sun, 2018). The relationship between MMP and ROS is complex. Changes in mitochondrial inner membrane potential and ROS production under different stresses do not appear to show fixed patterns, and the association between them depends on the mode of action. Mitochondrial ROS production is generally assumed to increase with an excessive reduction of the respiratory chain; however, ROS production is sometimes accompanied by a decrease in MMP (Kowaltowski et al., 2009; Pyatrikas et al., 2015). Gottwald has suggested that damaged mitochondria produce excessive ROS, thereby leading to oxidative stress and activation of cell death (Gottwald et al., 2018). Although we have not determined the relationship between MMP and cell death, the change in ROS levels was clearly related to the change in MMP potential. ROS might potentially interfere with the change in MMP through a series of activities, and more ROS may be generated by the affected MMP. However, this process requires further study. Organisms attempt to maintain relatively low endogenous ROS levels to achieve cellular homeostasis. Under normal conditions, ROS are produced by peroxisome and electron transport chain pathways. The mitochondrial membrane potential was changed by the crude extract; in addition, the expression of cytochrome C enzyme oxidase and NADH dehydrogenase metabolism pathways in the oxidative phosphorylation pathway increased. Both pathways are involved in electron leakage during electron transfer during mitochondrial oxidation (Wu et al., 2022). Superoxide radicals can be formed by the combination of a small amount of “leaking” electrons with oxygen and further produce hydrogen peroxide and singlet oxygen. Thus, the oxidative phosphorylation pathway of mycelia was affected by treatment with methanol extract of BS45, thereby increasing the production of intracellular reactive oxygen species. The death of pathogenic fungi due to oxidative damage was further verified through transcriptome analysis and antioxidant enzyme activity determination. The activity of three typical antioxidant enzymes, SOD, POD and CAT, markedly decreased after treatment with crude methanol extract. This finding is similar to those reported by Zhang and Sun (2018). However, our results differ in that the antioxidant enzyme activity of pathogenic fungi was significantly elevated in the early stage, with respect to that in the control group (0 μg/ml). These findings indicated that the organism was actively coping with this OS to maintain its level. However, under the continuous action, F. graminearum cells could not cope with this OS, resulting in oxidative damage. The increase in the MDA content further confirmed the occurrence of oxidative damage. Transcriptomic results also confirmed the oxidative damage of pathogenic fungi. The peroxisome is a complex organelle that is closely related to mitochondria and chloroplast. It is an important source of ROS and active nitrogen for organisms and contains a complex array of antioxidant defenses that regulate the accumulation of ROS and active nitrogen. Organisms can protect themselves against toxicity through the activities of organelles such as peroxisomes and their proteomes (Piacentini et al., 2021). A transcriptome analysis showed that all the genes in the peroxisome pathway were down-regulated, except for one up-regulated SOD gene FGSG_07069. The main CAT enzyme genes were significantly down-regulated, indicating that peroxisome was not able to control the regulation of high oxygen levels.
The KEGG enrichment analysis showed significant enrichment in the ribosome pathway and various amino acid metabolic pathways. Amino acids are the raw materials for protein synthesis and are widely and closely related to the growth and development of organisms. The ribosome is the site of protein translation. Here, we speculated that F. graminearum interfered with the normal synthesis and regulation of ribosomes and amino acids in the early stage of BS45 metabolite treatments, causing abnormal changes in the bacterial protein contents. Meanwhile, most DEGs (log2FC < −10) were related to cell permeability. This included FGSG_00219, which is a Major Facilitator Superfamily gene, that is involved in the expulsion of toxic molecules into cells. Transporters are important factors associated with pathogenicity. Limited transporter function leads to the accumulation of toxic substances in the cell, thus inhibiting the growth and reproduction of mycelia, and affecting pathogenicity (Han S. et al., 2021). However, the significant downregulation of this gene seems to indicate that the defense system in the pathogenic fungi was affected. The gene with the greatest down-regulation was FGSG_07836, a mitogen active protein kinase gene, that has important effects on several developmental processes related to sexual reproduction, plant infection and cell wall integrity in F. graminearum (Hou et al., 2002). This finding suggests that crude extracts from strain BS45 interfere with the metabolic regulation of pathogenic fungi in many ways and cause cell death.
Fusarium graminearum is a major root-infecting pathogen (Wang et al., 2015). Root diseases are difficult to diagnose, because they occur consistently in the subsurface parts of the soil and are difficult to observe until they reach the above-ground organs, thus causing necrosis of the lower stem tissue (Looseley and Newton, 2014). Biological control is an effective measure to address root rot. Dugassa has indicated that Trichoderma and Pseudomonas strains can be used against bean black root rot disease caused by Fusarium solani (Dugassa et al., 2021). Treating soybeans with a strains of B. subtilis HSY21, screened from rhizosphere soil, have been found to inhibit soybean root rot caused by F. oxysporum by 63.83 and 57.07% in greenhouse and field conditions, respectively (Han C. Y. et al., 2021). The BS45-induced inhibition of Fusarium root rot in a greenhouse setting reached 40% after 8 days. Our research confirmed the effects of Bacillus BS45 on growth promotion and root rot control in wheat seedlings. Moreover, the biomass of wheat seedings significantly increased. Our findings may provide a basis for the control of wheat diseases caused by F. graminearum.
Altogether, the comparative transcriptome and biochemical analyses in this study suggested that crude methanol extract of B. subtilis BS45 induces high ROS accumulation and a collapse of the MMP, thus resulting in oxidative damage and perturbing related protein synthesis, and leading to F. graminearum cell death. Our study also highlights the antagonistic potential of strains of B. subtilis BS45, which may be explored for wheat protection against Fusarium root rot disease, and might find future dual applications as biocontrol agents and growth-promoting bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
HY: conceptualization, project administration, resources, funding acquisition, and supervision. ZL: writing—original draft preparation. ZL: writing—review and editing. DZ: supervision. MC: visualization. ZL and XL: investigation, methodology. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China, grant number 32260016; Natural Science Foundation of Jiangxi Province, grant number 20181BAB214003; and Project of the Graduate Innovation Foundation from Education Department of Jiangxi Province, grant number YC2020-S184.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1064838/full#supplementary-material
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi/
2. ^http://jspecies.ribohost.com/jspeciesws/
3. ^https://www.omicstudio.cn/tool
4. ^https://antismash.secondarymetabolites.org/#!/start
Abdalla, M. A. (2016). Three new cyclotetrapeptides isolated from Streptomyces sp. 447. Nat. Prod. Res. 31, 1014–1021. doi: 10.1080/14786419.2016.1263849
Bacmaga, M., Wyszkowska, J., Kucharski, J., and Kaczyński, P. (2018). Changes in microbiological properties of soil during fungicide degradation. Soil Sci. Annu. 69, 169–176. doi: 10.2478/ssa-2018-0017
Balmas, V., Scherm, B., Marcello, A., Beyer, M., Hoffmann, L., Migheli, Q., et al. (2014). Fusarium species and chemotypes associated with fusarium head blight and fusarium root rot on wheat in Sardinia. Plant Pathol. 64, 972–979. doi: 10.1111/ppa.12337
Beccari, G., Covarelli, L., and Nicholson, P. (2011). Infection processes and soft wheat response to root rot and crown rot caused by Fusarium culmorum. Plant Pathol. 60, 671–684. doi: 10.1111/j.1365-3059.2011.02425.x
Bi, Y. Q., Wang, Y. H., Luo, S. C., Guo, R. J., and Wang, J. (2020). Effects of MB and CMC growth media on the sporulation of Fusarium graminearum. Plant Prot. 46, 155–158.
Blin, K., Kim, H. U., Medema, M. H., and Weber, T. (2019). Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 20, 1103–1113. doi: 10.1093/bib/bbx146
Cadenas, E., and Davies, K. J. (2000). Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230. doi: 10.1016/s0891-5849(00)00317-8
Chen, M. C., Wang, J. P., Liu, B., Zhu, Y. J., Xiao, R. F., Yang, W. J., et al. (2020). Biocontrol of tomato bacterial wilt by the new strain Bacillus velezensis FJAT-46737 and its lipopeptides. BMC Microbiol. 20:160. doi: 10.1186/s12866-020-01851-2
Chen, Z. F., and Ying, G. G. (2015). Occurrence, fate and ecological risk of five typical azole fungicides as therapeutic and personal care products in the environment: a review. Environ. Int. 84, 142–153. doi: 10.1016/j.envint.2015.07.022
Choi, S. K., Jeong, H., Kloepper, J. W., and Ryu, C. M. (2014). Genome sequence of Bacillus amyloliquefaciens GB03, an active ingredient of the first commercial biological control product. Genome Announc. 2:5. doi: 10.1128/genomeA.01092-14
Cruppe, G., Dewolf, E., Jaenisch, B. R., Onofre, K. A., Valent, B., Fritzb, A. K., et al. (2021). Experimental and producer-reported data quantify the value of foliar fungicide to winter wheat and its dependency on genotype and environment in the U.S. central great plains. Field Crop Res. 273:108300. doi: 10.1016/j.fcr.2021.108300
Dugassa, A., Alemu, T., and Woldehawariat, Y. (2021). In-vitro compatibility assay of indigenous Trichoderma and Pseudomonas species and their antagonistic activities against black root rot disease (Fusarium solani) of faba bean (Vicia faba L.). BMC Microbiol. 21:115. doi: 10.1186/s12866-021-02181-7
Edwards, S. G. (2022). Pydiflumetofen co-formulated with prothioconazole: a novel fungicide for Fusarium head blight and deoxynivalenol control. Toxins 14:34. doi: 10.3390/toxins14010034
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/j.1364-3703.2004.00252.x
Gottwald, E. M., Duss, M., Bugarski, M., Haenni, D., Schuh, C. D., Landau, E. M., et al. (2018). The targeted anti-oxidant MitoQ causes mitochondrial swelling and depolarization in kidney tissue. Phys. Rep. 6:e13667. doi: 10.14814/phy2.13667
Grintzalis, K., Georgiou, C. D., and Schneider, Y. J. (2015). An accurate and sensitive coomassie brilliant blue G-250-based assay for protein determination. Anal. Biochem. 480, 28–30. doi: 10.1016/j.ab.2015.03.024
Hamdache, A., Lamarti, A., Aleu, J., and Collado, I. J. (2011). Non-peptide metabolites from the genus Bacillus. J. Nat. Prod. 74, 893–899. doi: 10.1021/np100853e
Han, S., Chen, J., Zhao, Y., and Cai, H. (2021). Bacillus subtilis HSY21 can reduce soybean root rot and inhibit the expression of genes related to the pathogenicity of Fusarium oxysporum. Pestic. Biochem. Physiol. 178:104916. doi: 10.1016/j.pestbp.2021.104916
Han, Q., Wu, F., Wang, X., Qi, H., Shi, L., and Ren, A. (2015). The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ. Microbiol. 17, 1166–1188. doi: 10.1111/1462-2920.12538
Han, C. Y., Yu, Z. Y., Zhang, Y. T., Wang, Z. Y., Zhao, J. W., Huang, S. X., et al. (2021). Discovery of Frenolicin B as potential agrochemical fungicide for controlling Fusarium head blight on wheat. J. Agric. Food Chem. 68, 2108–2117. doi: 10.1021/acs.jafc.0c04277
Hasan, Z., Zainudin, N. A. I. M., Aris, A., Ibrahim, M. H., and Yusof, M. T. (2021). Evaluation of Trichoderma asperellum for inhibiting growth of Fusarium oxysporum f. sp. lycopersici and enhancing growth of tomato and fruit quality. Arch. Phytopathol. Plant Protect. 54, 1360–1371. doi: 10.1080/03235408.2021.1910416
Hou, Z., Xue, C., Peng, Y., Katan, T., Kistler, H. C., and Xu, J. R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 15:1119. doi: 10.1094/MPMI.2002.15.11.1119
Jabran, K., Mahmood, K., Melander, B., Bajwa, A. A., and Kudak, P. (2017). Chapter three - weed dynamics and management in wheat. Adv. Agron. 145, 97–166. doi: 10.1016/bs.agron.2017.05.002
Jiang, C. M., Li, Z. Z., Shi, Y. H., Guo, D., Pang, B., Chen, X. Q., et al. (2020). Bacillus subtilis inhibits Aspergillus carbonarius by producing iturin A, which disturbs the transport, energy metabolism, and osmotic pressure of fungal. Int. J. Food Microbiol. 330:108783. doi: 10.1016/j.ijfoodmicro.2020.108783
Jiang, C. H., Liao, M. J., Wang, H. K., Zheng, M. Z., Xu, J. J., and Guo, J. H. (2018). Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 126, 147–157. doi: 10.1016/j.biocontrol.2018.07.017
Jin, P., Wang, H., Tan, Z., Dahar, G.,. Y., Li, Q. X., Xuan, Z., et al. (2020). Antifungal mechanism of bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides Penz. Pestic. Biochem. Physiol. 163, 102–107. doi: 10.1016/j.pestbp.2019.11.004
Karlsson, I., Persson, P., and Friberg, H. (2021). Fusarium head blight from a microbiome perspective. Front. Microbiol. 12:628373. doi: 10.3389/fmicb.2021.628373
Khoubnasabjafari, M., Ansarin, K., and Jouyban, A. (2016). Critical review of malondialdehyde analysis in biological samples. Curr. Pharm. Anal. 12, 4–17. doi: 10.2174/1573412911666150505185343
Kowaltowski, A. J., Souza-Pinto, N., Castilho, R. F., and Vercesi, A. E. (2009). Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 47, 333–343. doi: 10.1016/j.freeradbiomed.2009.05.004
Kriss, A. B., Paul, P. A., and Madden, L. V. (2010). Relationship between yearly fluctuations in fusarium head blight intensity and environmental variables: a window-pane analysis. Phytopathology 100, 784–797. doi: 10.1094/PHYTO-100-8-0784
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 6, 1547–1549. doi: 10.1093/molbev/msy096
Li, Y., and Chen, S. (2019). Fusaricidin produced by Paenibacillus polymyxa WLY78 induces systemic resistance against Fusarium wilt of cucumber. Int. J. Mol. Sci. 20:5240. doi: 10.3390/ijms20205240
Liu, Y., Lu, J., Sun, J., Zhu, X., Zhou, L., Lu, Z., et al. (2019). C16-Fengycin A affect the growth of Candida albicans by destroying its cell wall and accumulating reactive oxygen species. Appl. Microbiol. Biotechnol. 103, 8963–8975. doi: 10.1007/s00253-019-10117-5
Liu, S., Tang, M., and Cheng, J. (2022). Fermentation optimization of surfactin production of Bacillus amyloliquefaciens HM618. Biotechnol. Appl. Biochem. 2, 1–13. doi: 10.1002/bab.2327
Looseley, M. E., and Newton, A. C. (2014). Assessing the consequences of microbial infection in field trials: seen, unseen, beneficial, parasitic and pathogenic. Agronomy 4, 302–321. doi: 10.3390/agronomy4020302
Lopez, A. C., and Alippi, A. M. (2019). Feasibility of using RFLP of PCR-amplified 16S rRNA gene(s) for rapid differentiation of isolates of aerobic spore-forming bacteria from honey. J. Microbiol. Methods 165:105690. doi: 10.1016/j.mimet.2019.105690
Lyu, A., Liu, H., and Che, H. J. (2017). Reveromycins A and B from Streptomyces sp. 3–10: antifungal activity against plant pathogenic fungi in vitro and in a strawberry food model system. Front. Microbiol. 8:550. doi: 10.3389/fmicb.2017.00550
Muhialdin, B. J., Algboory, H. L., Kadum, H., Mohammed, N. K., Saari, N., Hassan, Z., et al. (2020). Antifungal activity determination for the peptides generated by Lactobacillus plantarum TE10 against Aspergillus flavus in maize seeds. Food Control 109:106898. doi: 10.1016/j.foodcont.2019.106898
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Parent, Z. K., Anthony, A. A., Laurent, F., Steels, S., and Ongena, M. (2017). Stimulation of fengycin-type antifungal lipopeptides in Bacillus amyloliquefaciens in the presence of the maize fungal pathogen Rhizomucor variabilis. Front. Microbiol. 8:850. doi: 10.3389/fmicb.2017.00850
Passari, A. K., Leo, V. V., Singh, G., Samanta, L., Ram, H., Siddaiah, C. N., et al. (2020). In vivo studies of inoculated plants and in vitro studies utilizing methanolic extracts of endophytic Streptomyces sp. strain DBT34 obtained from Mirabilis jalapa L. exhibit ROS-scavenging and other bioactive properties. Int. J. Mol. Sci. 21:7364. doi: 10.3390/ijms21197364
Passari, A. K., Mishra, V. K., Gupta, V. K., Saikia, R., and Singh, B. P. (2016). Distribution and identification of endophytic Streptomyces species from Schima wallichii as potential biocontrol agents against fungal plant pathogens. Pol. J. Microbiol. 65, 319–329. doi: 10.5604/17331331.1215611
Piacentini, D., Della, R. F., Bertoldi, I., Massimi, L., Sofo, A., Altamura, M. M., et al. (2021). Peroxisomal PEX7 receptor affects cadmium-induced ROS and auxin homeostasis in Arabidopsis root system. Antioxidants 10:1494. doi: 10.3390/antiox10091494
Pyatrikas, D. V., Fedoseeva, I. V., Varakina, N. N., Rusaleva, T. M., Stepanov, A. V., Fedyaeva, A. V., et al. (2015). Relation between cell death progression, reactive oxygen species production and mitochondrial membrane potential in fermenting Saccharomyces cerevisiae cells under heat-shock conditions. FEMS Microbiol. Lett. 362:fnv082. doi: 10.1093/femsle/fnv082
Rabbee, M. F., and Baek, K. H. (2020). Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 25:4973. doi: 10.3390/molecules25214973
Rjiba-Touati, K., Hamdi, H., M’nassri, A., Guedri, Y., Mokni, M., and Abid, S. (2022). Bromuconazole caused genotoxicity and hepatic and renal damage via oxidative stress process in wistar rats. Environ. Sci. Pollut. Res. 29, 14111–14120. doi: 10.1007/s11356-021-16091-8
Sarowar, S., Alam, S. T., Makandar, R., Lee, H., Trick, H. N., and Dong, Y. (2019). Targeting the pattern-triggered immunity pathway to enhance resistance to Fusarium graminearum. Mol. Plant Pathol. 20, 626–640. doi: 10.1111/mpp.12781
Scala, V., Visentin, I., and Cardinale, F. (2017). Evaluating fumonisin gene expression in Fusarium verticillioides. Methods Mol. Biol. 1542, 249–257. doi: 10.1007/978-1-4939-6707-0_16
Shafi, J., Tian, H., and Ji, M. S. (2017). Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip. 31, 446–459. doi: 10.1080/13102818.2017.1286950
Siamak, S. B. S., and Zheng, S. (2018). Banana Fusarium wilt (Fusarium oxysporum f. sp. cubense) control and resistance, in the context of developing wilt-resistant bananas within sustainable production systems. Hortic. Plant J. 4, 208–281. doi: 10.1016/j.hpj.2018.08.001
Singh, T. A., Passari, A. K., Jajoo, A., Bhasin, S., Gupta, V. K., Hashem, A., et al. (2021). Tapping into actinobacterial genomes for natural product discovery. Front. Microbiol. 12:655620. doi: 10.3389/fmicb.2021.655620
Stephen, N. W., Baenziger, P. S., John, H. N., William, W. B., and Heather, H. A. (2015). Management of Fusarium head blight of wheat and barley. Crop Prot. 73, 100–107. doi: 10.1016/j.cropro.2015.02.025
Tillmann, M., von Tiedemann, A., and Winter, M. (2017). Crop rotation effects on incidence and diversity of Fusarium species colonizing stem bases and grains of winter wheat. J. Plant Dis. Prot. 124, 121–130. doi: 10.1007/s41348-016-0064-6
Wang, Q., Buxa, S. V., Furch, A., Friedt, W., and Gottwald, S. (2015). Insights into Triticum aestivum seedling root rot caused by Fusarium graminearum. Mol. Plant-Microbe Interact. 28, 1288–1303. doi: 10.1094/MPMI-07-15-0144-R
Wang, Q., and Gottwald, S. (2017). Wheat root-dip inoculation with Fusarium graminearum and assessment of root rot disease severity. Bio-protocol 7:e2189. doi: 10.21769/BioProtoc.2189
Wang, K., Tian, Y., Zhou, N., Liu, D., and Zhang, D. (2018). Studies on fermentation optimization, stability and application of prolyl aminopeptidase from Bacillus subtilis. Process Biochem. 74, 10–20. doi: 10.1016/j.procbio.2018.08.035
Wang, L. Q., Wu, K. T., Yang, P., Hou, F., Rajput, S. A., Qi, D. S., et al. (2022). Transcriptomics reveals the effect of thymol on the growth and toxin production of Fusarium graminearum. Toxins 14:142. doi: 10.3390/toxins14020142
Wang, Y., Zhang, C., Liang, J., Wang, L., Gao, W. B., Jiang, J., et al. (2020). Surfactin and fengycin B extracted from Bacillus pumilus W-7 provide protection against potato late blight via distinct and synergistic mechanisms. Appl. Microbiol. Biotechnol. 104, 7467–7481. doi: 10.1007/s00253-020-10773-y
Williamson, B. B., and Dhingra, A. (2021). Understanding root rot disease in agricultural crops. Horticulturae 7:33. doi: 10.3390/horticulturae7020033
Wu, T., Chen, M., Zhou, L., Lu, F., Bie, X., and Lu, Z. (2020). Bacillomycin D effectively controls growth of Malassezia globosa by disrupting the cell membrane. Appl. Microbiol. Biotechnol. 104, 3529–3540. doi: 10.1007/s00253-020-10462-w
Wu, R., Li, S., Hudlikar, R., Wang, L., Shannar, A., Peter, R., et al. (2022). Redox signaling, mitochondrial metabolism, epigenetics and redox active phytochemicals. Free Radic. Biol. Med. 179, 328–336. doi: 10.1016/j.freeradbiomed.2020.12.007
Yang, C., Xie, L., Ma, Y., Cai, X., Yue, G., Qin, G., et al. (2021). Study on the fungicidal mechanism of glabridin against Fusarium graminearum. Pestic. Biochem. Physiol. 179:104963. doi: 10.1016/j.pestbp.2021.104963
Zhang, L., and Sun, C. (2018). Fengycins, cyclic Lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 84, e00445–e00418. doi: 10.1128/AEM.00445-18
Zhao, S. S., Zhang, Y., Yan, W., and Cao, L. L. (2017). Chaetomium globosum CDW7, a potential biological control strain and its antifungal metabolites. FEMS Microbiol. Lett. 364:w287. doi: 10.1093/femsle/fnw287
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Zhou, J. C., Zhou, G. Y., Liu, J. A., He, Y. H., and Dong, W. T. (2017). Antifungal mechanisms of the ethyl acetate extracts from Phytolacca americana and Ageratum conyzoides against Colletotrichum gloeosporioides of Dalbergia odorifera. Plant Prot. 43:60.
Keywords: Bacillus subtilis, biological control, Fusarium graminearum, reactive oxygen species, Fusarium root rot
Citation: Lu Z, Chen M, Long X, Yang H and Zhu D (2023) Biological potential of Bacillus subtilis BS45 to inhibit the growth of Fusarium graminearum through oxidative damage and perturbing related protein synthesis. Front. Microbiol. 14:1064838. doi: 10.3389/fmicb.2023.1064838
Received: 08 October 2022; Accepted: 31 January 2023;
Published: 20 February 2023.
Edited by:
Reeta Goel, GLA University, IndiaReviewed by:
Lipi Parikh, Montana State University, United StatesCopyright © 2023 Lu, Chen, Long, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huilin Yang, ✉ eWFuZ2hsQGp4bnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.