- 1Guangdong Key Laboratory for New Technology Research of Vegetables, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
Listeria monocytogenes is a major foodborne pathogen that is well-known for its high mortality rate upon infection. In recent years, the edible mushroom has also been found to be an important source of L. monocytogenes, but the contamination sources in Pleurotus eryngii (the king oyster mushroom) were unclear. In this study, a total of 203 edible mushrooms and environmental samples from four P. eryngii production plants were obtained. As a result, 29 samples (14.3%) were positive for L. monocytogenes, including eight mushroom samples (13.3%, 8/60) and 21 associated environmental samples (14.7%, 21/143). The contamination of L. monocytogenes in plants A and B was more severe and was likely to originate from the mycelium stimulation machine. The isolates belonged to serogroups II.1 (4b-4d-4e), I.1 (1/2a-3a), and I.2 (1/2c-3c), and multilocus sequence typing (MLST) revealed that these L. monocytogenes strains belonged to five different sequence types (ST3, ST121, ST9, ST87, and ST224). The ST121 and ST3 isolates were only found in plants A and B, respectively. The isolates were carried by hly (29/29, 100%), inlB (23/29, 79.3%), inlA (29/29, 100%), inlC (29/29, 100%), inlJ (29/29, 100%), actA (19/29, 65.5%), iap (29/29, 100%), plcA (26/29, 100%), plcB (29/29, 100%), prfA (27/29, 93.1%), and mpl (29/29, 100%). Further study of inlA sequencing showed that 65.5% of strains (19/29) contained full-length InlA that was required for host cell invasion, whereas the mutation led to premature stop codons (PMSCs) at position 492 (type 6) on inlA alleles. All isolates in this survey were sensitive to gentamicin, kanamycin, sulbactam/ampicillin, trimethoprim-sulfamethoxazole, tetracycline, and doxycycline. The drug with the highest resistance is rifampicin (37.9%), followed by penicillin (24.1%) and ciprofloxacin (10.3%). Most multiply resistant strains are isolated from raw materials and equipment of the P. eryngii processing lines. Our study reflects the contamination patterns and potential risk of L. monocytogenes infection in P. eryngii production plants. The persistence of specific L. monocytogenes isolates (such as ST121 and ST3) may assist with contamination. In accordance with these results, the control of L. monocytogenes should focus on the environmental materials, especially in the mycelium stimulation stage. However, effective Listeria monitoring programs will allow for the improved development of Listeria control measures to minimize cross-contamination in the processing of P. eryngii.
Introduction
Listeria monocytogenes is a major foodborne pathogen causing human listeriosis, a severe foodborne disease with high hospitalization rates (97.0%) and mortality rates (15.6%) (European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC], 2019). Although L. monocytogenes accounted for 1% of foodborne disease outbreaks, it caused the highest percentage of foodborne disease-related deaths in the United States from 2009 to 2015 (Dewey-Mattia et al., 2018). The infection of L. monocytogenes involves the elderly, immunocompromised people, newborns, and pregnant women, which leads to sepsis, meningitis, encephalitis, abortion, and stillbirth (Lomonaco et al., 2009; Camargo et al., 2019). Its ability to survive and grow under a wide range of environmental conditions, including refrigeration temperatures, high salt concentrations, and low pH, makes it a potential hazard in foods (Ryser and Marth, 2007; Wu et al., 2016). L. monocytogenes causes listeriosis, with most cases caused by consuming contaminated food (Pérez-Rodríguez et al., 2017; El-Hajjaji et al., 2021). Meat, milk, and vegetables represent important sources of contamination, and L. monocytogenes can survive and multiply in raw products or ready-to-eat food made with such raw items, even if refrigerated (Filiousis et al., 2009).
Edible mushrooms, such as Lentinula edodes, Auricularia auricula-judae, Pleurotus ostreatus, Flammulina velutipes, Pleurotus eryngii, Agaricus bisporus, and Auricularia polytricha, are popular worldwide according to their taste and nutritional value. In 2020, the total output was 40.6 million tons in China, with an economic output of RMB ¥346.6 billion, accounting for ∼75% of global mushroom production. However, contaminations detected in food processing plants have led to rigid recalls of products and serious economic losses. In recent years, several recalls have occurred after detectable levels of L. monocytogenes were found on whole or sliced fresh mushrooms (Viswanath et al., 2013). More recently, L. monocytogenes was found contaminated in enoki mushrooms, which are responsible for 36 reported cases of listeriosis, including 31 hospitalizations and four deaths in the United States (Byun et al., 2022). Meanwhile, the occurrence of L. monocytogenes in edible mushroom products has been reported in several countries (Cordano and Jacquet, 2009; Venturini et al., 2011; Viswanath et al., 2013; Chen et al., 2018). However, it is necessary to understand how L. monocytogenes is entering mushroom production facilities through incoming raw materials and/or surviving and growing in farm and packing house environments.
Different molecular typing methods have been employed to assess the source of contamination and the routes of transmission of L. monocytogenes. The multilocus sequence typing (MLST) method is one of the most robust tools for investigating the global epidemiology of microbial populations (Sullivan et al., 2005). In addition, differentiation between virulent and hypo-virulent strains is significant for evaluating the potential implications of the presence of this microorganism for food safety and public health (Jensen et al., 2008). Among the 14 serotypes of L. monocytogenes identified, the results of multiplex polymerase chain reaction (PCR) indicated that these major serogroups could be separated into distinct groups: I.1 (1/2a-3a), I.2 (1/2c-3c), II.1 (4b-4d-4e), II.2 (1/2b-3b-7), and III (4a-4c) (Doumith et al., 2004). Serotypes 4b, 1/2a, and 1/2b are mostly associated with listeriosis in humans (Orsi et al., 2011; Poimenidou et al., 2018; Pérez-Baltar et al., 2021). Moreover, many virulence traits that participate in host invasion and cellular proliferation are mainly distributed in Listeria pathogenicity islands (LIPI), which have been identified depending on the pathogenicity of the strain and severity of the infection (Radoshevich and Cossart, 2018; Al et al., 2022). The prfA-regulated virulence gene cluster (pVGC), which consists of a monocistronic hly in the center of the locus, a lecithinase operon comprising mpl, actA, and plcB genes, and the plcA-prfA operon located upstream from hly and transcribed in the reverse direction are often referred to as Listeria pathogenicity island 1 (LIPI-1) (Vázquez-Boland et al., 2001a; Ward et al., 2004; Poimenidou et al., 2018). In addition, the internalins (InlA, InlB, InlC, and InlJ) play key roles in adhesion and invasion, which are essential for the pathogen to invade intestinal cells (Bonazzi et al., 2009). Furthermore, the determination of the genetic profile and antimicrobial susceptibility profile of L. monocytogenes isolates present in food sold in open markets may also help to trace future listeriosis outbreaks.
In a previous study, it was demonstrated that various retail edible mushroom samples such as F. velutipes, P. eryngii, Volvariella volvacea, Hypsizygus marmoreus, Pleurotus geesteranus, P. ostreatus, and L. edodes could both contaminate L. monocytogenes (Chen et al., 2018). Few other studies explored the contamination sources in F. velutipes, H. marmoreus, or other edible mushroom production facilities (Viswanath et al., 2013; Chen et al., 2014; Murugesan et al., 2015; Sun et al., 2021), but in P. eryngii, the situation is unclear. Considering that the processing of production for P. eryngii was similar to that of F. velutipes or H. marmoreus, this study was designed to investigate the prevalence of L. monocytogenes in four P. eryngii production plants and processing environments to explore the contamination sources and potential risk of the L. monocytogenes isolates in P. eryngii based on their genotypes determined by molecular serotyping and MLST, their antibiotic susceptibility, and their virulence gene profiles.
Materials and methods
Sampling and isolation of L. monocytogenes
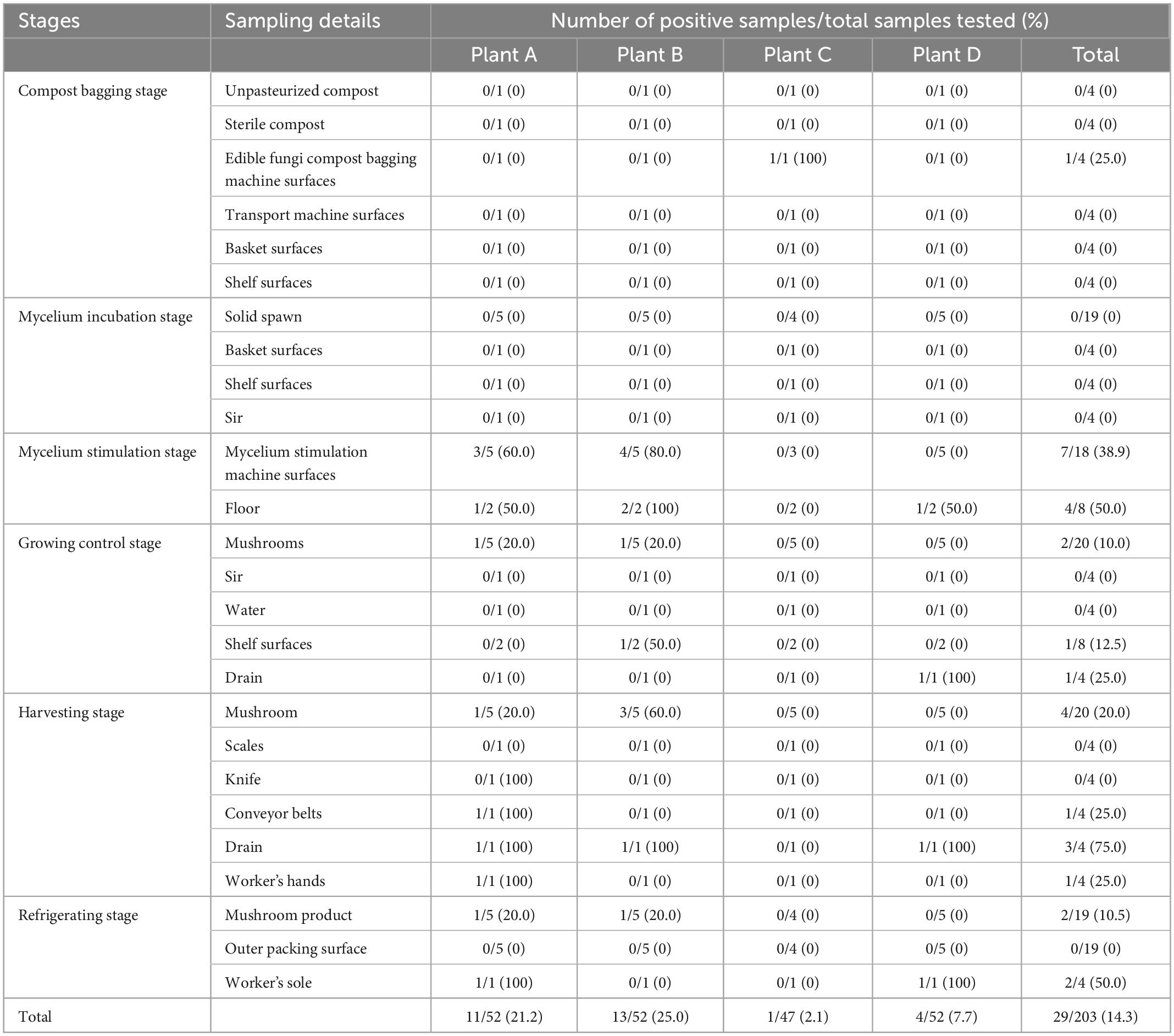
A total of 203 samples were detected from edible mushrooms and environment samples in four king oyster mushroom (P. eryngii) production plants (A, B, C, and D) for contamination analyses. A sampling schematic of this study is shown in Table 1. Overall, each plant collected 47–52 samples, including 52 samples in plant A (Shaoguan, Guangdong), 52 samples in plant B (Yichun, Jiangxi), 47 samples in plant C (Suizhou, Hubei), and 52 samples in plant D (Chengdu, Sichuan). The samples were collected from different phases of production, including composting, bagging, the inoculation room (solid spawn, sterile compost, inoculating machinery surfaces, and inoculation room air), the breeding and mycelium stimulation machinery, the growing room (atomized water, shelf surfaces, mushrooms, and air), and the harvesting and refrigerating room (packaging machinery surfaces, scales, conveyor belts, and packaged mushrooms). The samples were placed in a cold box at a temperature of approximately 4°C, tightly sealed with sterile plastic wrap, transported to an accredited laboratory, and subjected to microbiological analysis within 24 h.
Strains were isolated in accordance with the following protocol based on GB/T4789.30-2010 of food microbiological examination of L. monocytogenes (National Food Safety Standards of China) with slight modification for qualitative detection. Approximately 25 g of sample was added to 225 ml of LB1 enrichment broth culture (Huankai, Guangzhou, China), which was incubated at 30°C for 24 h after homogenization in a stomacher bag. Then, 0.1 ml of LB1 enrichment broth culture was transferred to 10 ml of LB2 enrichment broth culture at 30°C for 24 h. A portion (10 μl) of LB2 enrichment broth culture was streaked onto Listeria-selective plates (CHROM-agar, Paris, France), which were incubated at 37°C for 24 h. Colonies with a blue halo were analyzed by Gram stain, catalase, and oxidase tests, and colonies were identified using a Micro ID Listeria identification system (Microgen, Camberley, UK).
Molecular serotyping and multilocus sequence typing
Polymerase chain reaction assays were employed for molecular serogroups. A multiplex-PCR serogrouping of L. monocytogenes isolates was performed according to the instructions of Doumith et al. (2004) using five primers (lmo0737, lmo1118, ORF2819, ORF2110, and prs). This convenient method can characterize L. monocytogenes into five serogroups as follows: I.1 (1/2a-3a), I.2 (1/2c-3c), II.1 (1/2b-3b-7), II.2 (4b-4d-4e), and III (4a-4c).
The MLST scheme used to characterize L. monocytogenes isolates is based on the sequence analysis of the following seven housekeeping genes: abcZ (ABC transporter), bglA (beta-glucosidase), cat (catalase), dapE (succinyl-diaminopimelate desuccinylase), dat (D-amino acid aminotransferase), ldh (L-lactate dehydrogenase), and lhkA (histidine kinase) (Salcedo et al., 2003). The PCR amplification conditions were as follows: an initial cycle of 94°C for 4 min; 35 cycles of 94°C for 30 s, 52°C for 30 s (45°C for bglA), 72°C for 2 min, and a final extension at 72°C for 10 min. The DNA fragments were purified by using a PCR purification kit (Qiagen, Germany) and were sequenced in each direction with Big Dye fluorescent terminators on an ABI3730XL sequencer (Applied BioSystems).
For each MLST locus, an allele number was given to each distinct sequence variant, and a distinct sequence type (ST) number was attributed to each distinct combination of alleles at the seven genes. STs were determined by using the Listeria monocytogenes MLST database (Jolley et al., 2001). Sequence Type Analysis and Recombinational Tests software (S.T.A.R.T. ver. 2) was used to analyze the data of MLST.
Determination of virulence-associated genes
Eleven virulence genes (hly, inlB, inlA, inlC, inlJ, actA, iap, plcA, mpl, plcB, and prfA) encoding the virulence proteins were chosen based on their importance in L. monocytogenes pathogenesis. PCR was performed to detect the presence of these virulence-related genes in the L. monocytogenes isolates. The relevant information is described in Supplementary Table 1. The amplicons were stained with GoldView, electrophoresed in 1.5% agarose at 120 V for 0.5 h, and visualized under a UV transilluminator gel imaging system (GE Healthcare, WI, USA). The images were saved as TIFF files for analysis.
The 2,400 bp long inlA gene was also sequenced in L. monocytogenes isolates. External primers were used for amplification covering the whole inlA ORF, and internal primers were used for sequencing (Supplementary Table 2). The inlA sequences were assembled using SeqMan (DNASTAR, Lasergene). Mutation types were determined according to the site of the mutation that leads to a premature stop codon (PMSC) in inlA (Nightingale et al., 2005) and by comparing the obtained inlA sequence data to that of the L. monocytogenes EGDe reference strain (Glaser et al., 2001).
Antimicrobial susceptibility test
Antimicrobial susceptibility tests were performed using the standard disk diffusion Kirby–Bauer method on Mueller–Hinton agar, following the guidelines of the Clinical and Laboratory Standards Institute (Bauer et al., 1966; Granier et al., 2011; The Clinical and Laboratory Standards Institute [CLSI], 2018). Seventeen antibiotic disks (Oxoid, UK) were selected for the strains. These antibiotic disks were gentamicin (GEN, 10 μg), streptomycin (STR, 25 μg), kanamycin (KAN, 30 μg), chloramphenicol (CHL, 30 μg), rifampicin (RIF, 5 μg), cephalothin (CEF, 30 μg), levofloxacin (LEV, 5 μg), ciprofloxacin (CIP, 5 μg), vancomycin (VAN, 30 μg), clindamycin (CLI, 2 μg), erythromycin (ERY, 15 μg), ampicillin (AMP, 10 μg), penicillin (PEN, 10 U), sulbactam/ampicillin (SAM, 10/10 μg), trimethoprim-sulfamethoxazole (SXT, 23.75/1.25 μg), tetracycline (TET, 30 μg), and doxycycline (DOX, 30 μg). Staphylococcus aureus ATCC29213 and Escherichia coli ATCC25922 were used as quality control organisms (The Clinical and Laboratory Standards Institute [CLSI], 2018).
Results
Prevalence of L. monocytogenes in P. eryngii production plants
As shown in Table 1, it was found that 29 positive L. monocytogenes samples out of 203 detected samples showed a 14.3% prevalence in P. eryngii production plants in the present study. A total of 11 mushrooms and environment samples (21.2%, 11/52), 13 mushrooms and environment samples (25.0%, 13/52), 1 environment sample (2.1%, 1/47), and 4 environment samples (7.7%, 4/52) detected L. monocytogenes in plant A, plant B, plant C, and plant D, respectively. A total of 8 mushroom samples (13.3%, 8/60) were positive for L. monocytogenes in plants A and B, whereas 21 environmental samples (14.7%, 21/143) were positive in both four plants. The environment samples for detected L. monocytogenes involved compost bagging machine surfaces (25.0%, 1/4), mycelium stimulation machine surfaces (38.9%, 7/18), floor (50.0%, 4/8), drain (50.0%, 4/8), shelf surfaces (25.0%, 1/4), conveyor belts (25.0%, 1/4), worker’s hands (25.0%, 1/4), and worker’s sole (50.0%, 2/4). In different stages, the compost bagging stage and mycelium incubation stage were both free for L. monocytogenes, except one environment sample was positive in plant C. The two major stages of contamination were the mycelium stimulation stage and the harvesting stage. Meanwhile, the L. monocytogenes detected in mushroom samples focused on the growing control stage, the harvesting stage, and the refrigerating stage.
Genotype diversity of L. monocytogenes isolates
A total of 29 L. monocytogenes isolates were collected from each positive sample. The isolates were carried out by molecular serotyping and MLST. All isolates were serotyped into three serogroups. Approximately 10 isolates (34.5%) belonged to serogroup I.1 (1/2a and 3a), four isolates (13.8%) belonged to serogroup I.2 (1/2c and 3c), and 15 isolates (51.7%) belong to serogroup II.1 (4b, 4d, and 4e), respectively. Serogroup II.2 (1/2b, 3b, and 7) and serogroup III (4a and 4c) did not exist in these isolates. Interestingly, all serogroup I.1 (1/2a and 3a) strains were isolated from plant A and most serogroup II.1 (4b, 4d, and 4e) from plant B, respectively.
By the MLST method, there were five different STs for all isolates. The most common allelic profile was ST3 (13/29, 44.8% of isolates), followed by ST121 (10/29, 34.5% of isolates), ST9 (4/29, 13.8% of isolates), ST87 (1/29, 3.5% of isolates), and ST224 (1/29, 3.5% of isolates). STs correlated well with molecular serotypes. Of the STs, ST121 belonged to serogroup I.1 (1/2a-3a), ST3, ST87, and ST224 belonged to serogroup II.1 (4b-4d-4e), and ST9 belonged to serogroup I.2 (1/2c-3c). The ST121 isolates were only found in plant A. Similarly, the ST3 isolates were only found in plant B. Except for one isolate (A2601LM) from plant A belonging to ST9, the other STs (ST9, ST224, and ST87) were respectively obtained from environmental samples in plant C and plant D. A phylogenetic tree based on the seven concatenated MLST sequences (Figure 1) shows the relatedness between the isolates.
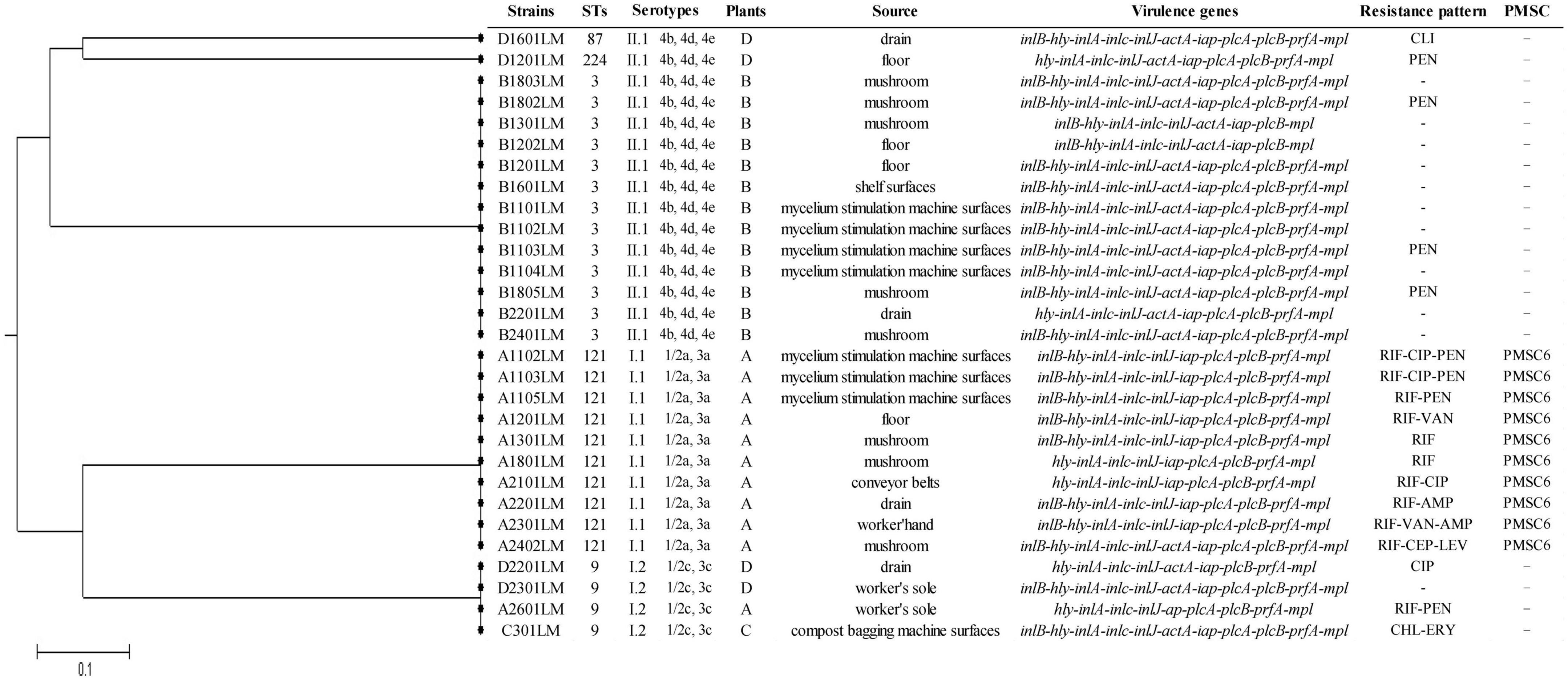
Figure 1. The unweighted pair group method with arithmetic mean (UPGMA) tree of the seven multilocus sequence typing loci of L. monocytogenes isolates from Pleurotus eryngii production plants. This tree was generated using the S.T.A.R.T (version 2).
Virulence characterization of L. monocytogenes isolates
All 29 isolates in this study were examined for the presence of 11 virulence marker genes by PCR. As a result, all strains harbored hly, inlA, inlC, inlJ, iap, mpl, and plcB sequences, whereas 93.1% (27/29) of isolates harbored prfA, 89.7% (26/29) of isolates harbored plcA, 79.3% (23/29) of isolates harbored inlB, and 65.5% of isolates harbored actA. The full-length sequence of inlA was determined in all 29 isolates by DNA sequencing, and 65.5% (19/29) of isolates contained full-length InlA. It was found that 10 isolates harbor the same PMSC at position 492 (PMSC type 6) on inlA alleles. PMSC6 was detected in all tested ST121 strains.
Antimicrobial susceptibility profiles
The antimicrobial susceptibility results of all isolates are shown in Table 2. Overall, the isolates in this survey were sensitive to gentamicin, kanamycin, sulbactam/ampicillin, trimethoprim-sulfamethoxazole, tetracycline, and doxycycline, except for two isolates with intermediate resistance to streptomycin. For 17 antibiotics, the highest resistance of L. monocytogenes isolates was rifampicin (11/29, 37.9%), followed by penicillin (7/29, 24.1%), ciprofloxacin (3/29, 10.3%), erythromycin (2/29, 6.9%), vancomycin (2/29, 6.9%), clindamycin (1/29, 3.5%), chloramphenicol (1/29, 3.5%), cephalothin (1/29, 3.5%), levofloxacin (1/29, 3.5%), and ampicillin (1/29, 3.5%). Ten isolates were susceptible to all 17 tested antibiotics, thirteen isolates were resistant to more than two antibiotics, and most of them were isolated from raw material and equipment of the edible mushroom processing lines.
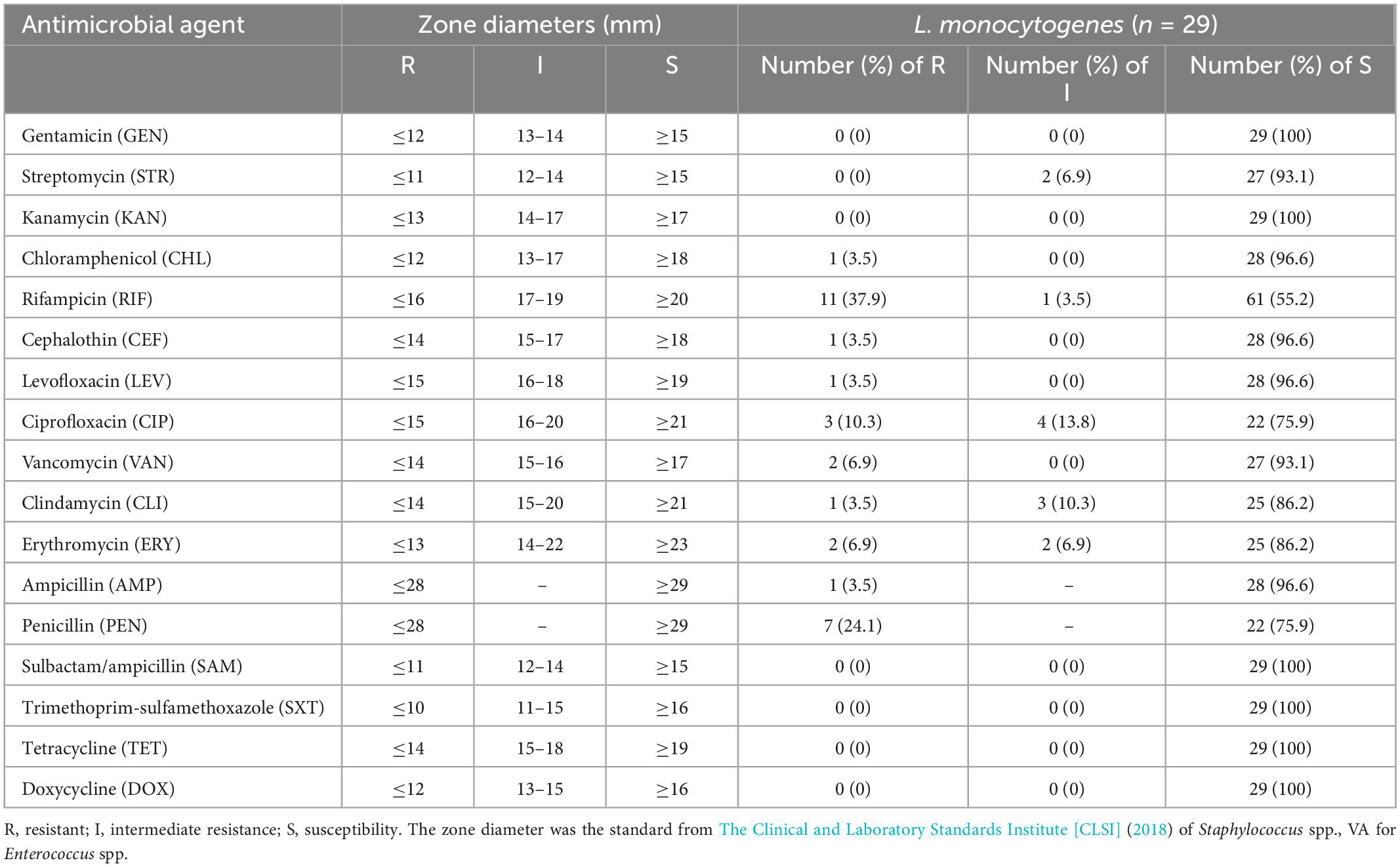
Table 2. Results of antimicrobial susceptibility tests of Listeria monocytogenes isolates obtained from Pleurotus eryngii production plants.
Discussion
In recent years, the contamination of L. monocytogenes has been found in edible mushrooms. Although few listeriosis outbreaks attributed to fresh mushrooms have been reported, recalls of mushroom products are common, caused by contamination with L. monocytogenes (Sun et al., 2021). P. eryngii, also known as the king oyster mushroom or cardoncello, is becoming increasingly popular with consumers in Europe, Asia, and North America. In recent times, it has become one of the most important commercial producers and exporters of edible mushrooms in China. However, the contamination source of L. monocytogenes in P. eryngii remains unclear. In this study, we investigated the prevalence of L. monocytogenes in four P. eryngii production plants from four cities and found that 14.3% of samples were positive for L. monocytogenes, including 13.3% of mushroom samples and 14.7% of environmental samples. The contamination is likely to originate from the mycelium stimulation machine, which coincides with previous research in the F. velutipes and H. marmoreus production facilities (Chen et al., 2014; Sun et al., 2021). In addition, the prevalence rate of L. monocytogenes in the P. eryngii production facility (14.3%) was higher than the A. bisporus production facility (1.6%) (Viswanath et al., 2013) but lower than the H. marmoreus production facility (27.0%) and the F. velutipes production facility (18.6%) (Chen et al., 2014). Compared with the mushroom samples (including the growing stage, harvesting stage, and products), our results were similar to those of H. marmoreus but lower than those of retail F. velutipes products (55.5%) (Chen et al., 2018). To our knowledge, the fruiting temperature of F. velutipes (6–12°C) is lower than that of P. eryngii (10–18°C) and H. marmoreus (12–16°C) in modern industrial cultivation of edible mushrooms. When L. monocytogenes was exposed to or persisted in the associated environment, its ability to proliferate at low temperatures may also play a key role in the persistence of this bacterium in harvesting and refrigerating stage product. Thus, the temperature accompanied by cross-contamination was probably the major reason for L. monocytogenes contamination in edible mushrooms.
As we know, different molecular typing methods have been employed to assess the source of contamination and the routes of transmission of L. monocytogenes. In this study, MLST was used and proved to be particularly suitable for the epidemiology investigations of food processing for L. monocytogenes. Overall, the source of L. monocytogenes contamination in the P. eryngii production plant was different and occurred inside the plant. Except for one isolate (A2601LM) belonging to ST9 from plant A, the isolates of L. monocytogenes focus on ST121 and ST3, which are found in plants A and B and originate from mycelium stimulation machines. The contamination was focused on P. eryngii production plants A and B, which are likely to originate from the mycelium stimulation stage. Meanwhile, in plants C and D, isolates from various STs (ST9, ST224, and ST87) were isolated from surfaces, floors, or drains, respectively. Thus, the L. monocytogenes contamination in the P. eryngii production plant could be caused by a wide variety of environments, but the contamination of the mycelium stimulation machine was the most important factor in this study. This is consistent with previous research in the F. velutipes and H. marmoreus production facility, which showed that the mycelium-scraping machine was the main transmission source in the production process (Chen et al., 2014; Sun et al., 2021).
Furthermore, the contamination of L. monocytogenes may also persist in P. eryngii production plants. Persistence has been defined as “repeated isolation on different dates of L. monocytogenes strains that are identified as identical subtypes (as determined by phenotypic or genotypic methods)” (Ferreira et al., 2014; Maury et al., 2016). According to previous studies, the L. monocytogenes ST121 strains, also found in this study, have previously been isolated from food and food processing facilities over several years in processing plants (Hein et al., 2011; Holch et al., 2013; Stoller et al., 2019). In Hein et al. (2011) found that the ST121 strains were isolated in Austria and Belgium from different ecological niches, including food and human cases, over several years. Similarly, Holch et al. (2013) reported that the ST121 strains isolated from four of eight different Danish fish processing industries were the persistent and dominant type in three plants over a period of 6 years, and Stoller et al. (2019) found that L. monocytogenes ST121 strains persisted in a meat processing facility over a 4-year period. According to the research of Harter et al. (2017), L. monocytogenes ST121 strains have the stress survival islet 2 (SSI-2) that is beneficial for survival under alkaline and oxidative stress conditions and is commonly encountered in food processing environments (Harter et al., 2017). All L. monocytogenes ST121 strains isolated from our study occurred PMSC mutations in inlA, which indicates that the ability of these persistent strains to attach to human host cells may decrease and also virulence attenuation (Van Stelten and Nightingale, 2008). Except for ST121, we also found ST3 isolates persisting in P. eryngii production plant B. These isolates belonged to serogroup II.1 (4b, 4d, and 4e), which was mostly related to food or humans with listeriosis (Sosnowski et al., 2019). According to the population structure analysis of L. monocytogenes isolates in Australia from 1931 to 2015, ST1 and ST3 were the most abundant strains (Jennison et al., 2017). In Poland, ST3 was the second most common type in L. monocytogenes isolates from various types of food of animal origin during 2013–2016 (Sosnowski et al., 2019). CC3 is the fourth most common clonal complex in Europe, is predominant in Australia, and was also detected in South and North America, Japan, and Oceania (Bespalova et al., 2021). Thus, the persistence of ST3 L. monocytogenes in the P. eryngii production plant deserves published attention. However, the reasons why ST3 L. monocytogenes persist in P. eryngii production plants remain to be further explored.
Listeria monocytogenes is classified into at least four evolutionary lineages (I, II, III, and IV) with different serotypes. Isolates of serotypes 1/2b, 3b, 3c, and 4b were assigned to lineage I, whereas isolates of serotypes 1/2a, 3a, and 1/2c were assigned to lineage II, and isolates of serotypes 4a and 4c were assigned to lineage III (Orsi et al., 2011). In this study, the L. monocytogenes isolates were serogroup II.1 (4b, 4d, and 4e), serogroup I.1 (1/2a and 3a), and serogroup I.2 (1/2c and 3c), which both belonged to lineage I and lineage II. As previously studied, most L. monocytogenes isolates belong to lineages I and II, which harbor the serotypes more commonly associated with human clinical cases, including serotype 1/2a (lineage II) and serotypes 1/2b and 4b (lineage I) (Orsi et al., 2011). Lineage I strains are mostly associated with the majority of human listeriosis outbreaks worldwide (Aureli et al., 2000; Jeffers et al., 2001; Gray et al., 2004), whereas lineage II strains are common in foods, seem to be widespread in natural and farm environments, and are also commonly isolated from animal listeriosis cases and sporadic human clinical cases (Lukinmaa et al., 2003; Parihar et al., 2008). Although L. monocytogenes has 14 serotypes that differ in virulence potential, serotypes 3a, 3b, 3c, 4a, 4c, 4e, and seven are very infrequent in food (Kathariou, 2002; Doumith et al., 2004; Yin et al., 2019). Thus, the dominant serotypes of L. monocytogenes isolated from the P. eryngii production facility should be 4b (15/29, 51.4%) and 1/2a (10/29, 34.5%), whereas 1/2c (4/29, 13.8%) is limited. However, the serotypes of L. monocytogenes isolates found in P. eryngii production plants have a close connection with human listeriosis outbreaks and should receive much attention.
In this study, the virulence genes, such as hly, inlB, inlA, inlC, inlJ, actA, iap, plcA, mpl, plcB, and prfA, were determined in L. monocytogenes isolates by PCR. These genes proved to play an essential role in the pathogenicity of L. monocytogenes. As a result, all isolates harbored at least nine of these genes. LIPI-1 (prfA, plcA, hly, mpl, actA, and plcB) genes were found in 55.2% (16/29) of the isolates, as well as the iap gene, which has an indirect role in their pathogenesis by codifying a product that is responsible for entering the host cell (Vázquez-Boland et al., 2001b; Iglesias et al., 2017). In addition, the inlA gene encodes a product that is responsible for the entry of the bacterium into the host cell, and the genes inlB, inlC, and inlJ are directly involved in the subsequent infection stages (Doumith et al., 2004; Iglesias et al., 2017). The L. monocytogenes strain isolated from the P. eryngii production plant contained inlA, inlC, and inlJ, whereas inlB was detected in 79.3% of isolates (23/29). However, the presence of either or both LIPI-1, inlA, inlB, inlC, and inlJ genes involved in the L. monocytogenes strain indicates that they had the ability to enter via the intraperitoneal route or through the use of contaminated food and cause infection. Furthermore, the mutations in inlA leading to a PMSC significantly reduce the invasion of the strain by human epithelial cells (Nightingale et al., 2005). However, only ten isolates from the sequencing of inlA genes had PMSC mutations, which means most of the strains (19/29, 65.5%) can produce full-length InlA, which is required for host cell invasion.
Regardless of the source, antimicrobial resistance within the isolates was significant. In this study, 17 clinically common antibiotics were tested for antimicrobial susceptibility in all L. monocytogenes strains. Compared with previous studies, the results of this study suggest that the overall incidence of antibiotic resistance in L. monocytogenes is relatively low (Pesavento et al., 2010; Rahimi et al., 2010; Camargo et al., 2015; Chen et al., 2018). Most L. monocytogenes isolates were sensitive to selected antibiotics. As the first-choice treatment for invasive listeriosis, only one isolate (3.5%, 1/29) and seven isolates (24.1%, 7/29) were resistant to ampicillin and penicillin. Besides, there was no resistance to the association of trimethoprim-sulfamethoxazole (an alternative treatment to invasive listeriosis). Erythromycin, a molecule recommended for the treatment of listeriosis in pregnant women (Charpentier and Courvalin, 1999), showed a mild level of resistance among the isolates (6.9% resistant and 6.9% intermediate resistance). L. monocytogenes is naturally susceptible to a wide range of antibiotics except for cephalosporin and fosfomycin (Hof et al., 1997). However, single- or multiple-resistant L. monocytogenes strains isolated from food, the environment, and clinics have been reported on a regular basis in recent years (Barbosa et al., 2013; Cufaoglu et al., 2021). Thus, as foodborne L. monocytogenes is the main cause of listeriosis, an antimicrobial susceptibility test for L. monocytogenes in food requires constant monitoring.
Conclusion
In summary, the results of this study suggested that the contamination of L. monocytogenes in P. eryngii (king oyster mushroom) production plants is most likely caused by mycelium stimulation machines, with low temperatures during growth being the most likely cause. In addition, the persistence of specific L. monocytogenes isolates (such as ST121 and ST3) may assist with contamination. Most isolates belonged to molecular serotypes 4b-4d-4e and 1/2a-3a, which are associated with human listeriosis, suggesting that this pathogen may represent a potential danger to public health. Furthermore, the presence of either or both LIPI-1, inlA, inlB, inlC, and inlJ genes involved in the L. monocytogenes strain indicates that they had the ability to enter via the intraperitoneal route or using contaminated food and cause infection. Further study of inlA sequencing showed that most of the selected strains contained PMSC-lacking inlA gene sequences required for the encoding of the InlA factor and bacterial invasion of the host cell. Although the isolates we have found were not highly resistant, antimicrobial susceptibility tests for L. monocytogenes in food require constant monitoring. Our study reflects the potential risk of L. monocytogenes contamination in P. eryngii production plants. However, effective Listeria monitoring programs will allow for the improved development of Listeria control measures to minimize cross-contamination in processing P. eryngii.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SW and JX conceived and designed the experiments and analyzed the data. JX and ML performed the experiments. HH and YP contributed to reagents, materials, and analysis tools. SW, JX, and ZX contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31900013), the National Natural Science Foundation of Guangdong Province (2022A1515010059), and the GDAS’ Special Project of Science and Technology Development (2020GDASYL-20200104015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1064575/full#supplementary-material
References
Al, S., Disli, H. B., Hizlisoy, H., Ertas Onmaz, N., Yildirim, Y., and Gonulalan, Z. (2022). Prevalence and molecular characterization of Listeria monocytogenes isolated from wastewater of cattle slaughterhouses in Turkey. J. Appl. Microbiol. 132, 1518–1525. doi: 10.1111/jam.15261
Aureli, P., Fiorucci, G. C., Caroli, D., Marchiaro, G., Novara, O., Leone, L., et al. (2000). An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med. 342, 1236–1241. doi: 10.1056/NEJM200004273421702
Barbosa, J., Magalhães, R., Santos, I., Ferreira, V., Brandão, T. R. S., Silva, J., et al. (2013). Evaluation of antibiotic resistance patterns of food and clinical Listeria monocytogenes isolates in Portugal. Foodborne Pathog. Dis. 10, 861–866. doi: 10.1089/fpd.2013.1532
Bauer, A., Kirby, W., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493. doi: 10.1093/ajcp/45.4_ts.493
Bespalova, T. Y., Mikhaleva, T. V., Meshcheryakova, N. Y., Kustikova, O. V., Matovic, K., Dmitrić, M., et al. (2021). Novel sequence types of Listeria monocytogenes of different origin obtained in the republic of serbia. Microorganisms 9:1289. doi: 10.3390/microorganisms9061289
Bonazzi, M., Lecuit, M., and Cossart, P. (2009). Listeria monocytogenes internalin and E-cadherin: From structure to pathogenesis. Cell Microbiol. 11, 693–702. doi: 10.1111/j.1462-5822.2009.01293.x
Byun, K.-H., Han, S. H., Choi, M. W., Park, S. H., and Ha, S.-D. (2022). Isolation, characterization, and application of bacteriophages to reduce and inhibit Listeria monocytogenes in celery and enoki mushroom. Food Control 135:108826. doi: 10.1016/j.foodcont.2022.108826
Camargo, A. C., De Castilho, N. P. A., Da Silva, D. A. L., Vallim, D. C., Hofer, E., and Nero, L. A. (2015). Antibiotic resistance of Listeria monocytogenes isolated from meat-processing environments, beef products, and clinical cases in Brazil. Microb. Drug Resist. 21, 458–462. doi: 10.1089/mdr.2014.0270
Camargo, A. C., Moura, A., Avillan, J., Herman, N., Mcfarland, A. P., Sreevatsan, S., et al. (2019). Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ. Microbiol. 21, 4478–4487. doi: 10.1111/1462-2920.14726
Charpentier, E., and Courvalin, P. (1999). Antibiotic Resistance in Listeria spp. Antimicrob. Agents Chemother. 43, 2103–2108. doi: 10.1128/AAC.43.9.2103
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H., et al. (2018). Prevalence, potential virulence, and genetic diversity of listeria monocytogenes isolates from edible mushrooms in chinese markets. Front. Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, M., Wu, Q., Zhang, J., Guo, W., Wu, S., and Yang, X. (2014). Prevalence and contamination patterns of Listeria monocytogenes in Flammulina velutipes plants. Foodborne Pathog. Dis. 11, 620–627. doi: 10.1089/fpd.2013.1727
Cordano, A. M., and Jacquet, C. (2009). Listeria monocytogenes isolated from vegetable salads sold at supermarkets in Santiago, Chile: Prevalence and strain characterization. Int. J. Food Microbiol. 132, 176–179. doi: 10.1016/j.ijfoodmicro.2009.04.008
Cufaoglu, G., Ambarcioglu, P., and Ayaz, N. D. (2021). Meta-analysis of the prevalence of Listeria spp. and antibiotic resistant L. monocytogenes isolates from foods in Turkey. LWT 144:111210. doi: 10.1016/j.lwt.2021.111210
Dewey-Mattia, D., Manikonda, K., Hall, A. J., Wise, M. E., and Crowe, S. J. (2018). Surveillance for foodborne disease outbreaks – United States, 2009-2015. MMWR Surveill. Summ. 67, 1–11. doi: 10.15585/mmwr.ss6710a1
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
El-Hajjaji, S., Gérard, A., De Laubier, J., Lainé, A., Patz, V., and Sindic, M. (2021). Study of the bacterial profile of raw milk butter, made during a challenge test with Listeria monocytogenes, depending on cream maturation temperature. Food Microbiology 98:103778. doi: 10.1016/j.fm.2021.103778
European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC] (2019). The European Union one health 2018 zoonoses report. EFSA J. 17:e05926. doi: 10.2903/j.efsa.2019.5926
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Filiousis, G., Johansson, A., Frey, J., and Perreten, V. (2009). Prevalence, genetic diversity and antimicrobial susceptibility of Listeria monocytogenes isolated from open-air food markets in Greece. Food Control 20, 314–317. doi: 10.1016/j.foodcont.2008.05.018
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Granier, S., Moubareck, C., Colaneri, C., Lemire, A., Roussel, S., Dao, T., et al. (2011). Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl. Environ. Microbiol. 77, 2788–2790. doi: 10.1128/AEM.01381-10
Gray, M. J., Zadoks, R. N., Fortes, E. D., Dogan, B., Cai, S., Chen, Y., et al. (2004). Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70, 5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004
Harter, E., Wagner, E. M., Zaiser, A., Halecker, S., Wagner, M., and Rychli, K. (2017). Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 83:e00827–17. doi: 10.1128/AEM.00827-17
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stessl, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi: 10.1128/AEM.02159-10
Hof, H., Nichterlein, T., and Kretschmar, M. (1997). Management of listeriosis. Clin. Microbiol. Rev. 10, 345–357.
Holch, A., Webb, K., Lukjancenko, O., Ussery, D., Rosenthal, B. M., and Gram, L. (2013). Genome sequencing identifies two nearly unchanged strains of persistent listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 79, 2944–2951. doi: 10.1128/AEM.03715-12
Iglesias, M. A., Kroning, I. S., Decol, L. T., De Melo Franco, B. D. G., and Silva, W. P. D. (2017). Occurrence and phenotypic and molecular characterization of Listeria monocytogenes and Salmonella spp. in slaughterhouses in southern Brazil. Food Res. Int. 100, 96–101.
Jeffers, G. T., Bruce, J. L., Mcdonough, P. L., Scarlett, J., Boor, K. J., and Wiedmann, M. (2001). Comparative genetic characterization of Listeria monocytogenes isolates from human and animal Listeriosis cases. Microbiology (Reading) 147, 1095–1104.
Jennison, A. V., Masson, J. J., Fang, N.-X., Graham, R. M., Bradbury, M. I., Fegan, N., et al. (2017). Analysis of the Listeria monocytogenes population structure among isolates from 1931 to 2015 in Australia. Front. Microbiol. 8:603. doi: 10.3389/fmicb.2017.00603
Jensen, A., Thomsen, L. E., Jørgensen, R. L., Larsen, M. H., Roldgaard, B. B., Christensen, B. B., et al. (2008). Processing plant persistent strains of Listeria monocytogenes appear to have a lower virulence potential than clinical strains in selected virulence models. Int. J. Food Microbiol. 123, 254–261. doi: 10.1016/j.ijfoodmicro.2008.02.016
Jolley, K. A., Feil, E. J., Chan, M.-S., and Maiden, M. C. J. (2001). Sequence type analysis and recombinational tests (START). Bioinformatics 17, 1230–1231. doi: 10.1093/bioinformatics/17.12.1230
Kathariou, S. (2002). Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65, 1811–1829.
Lomonaco, S., Decastelli, L., Nucera, D., Gallina, S., Manila Bianchi, D., and Civera, T. (2009). Listeria monocytogenes in gorgonzola: Subtypes, diversity and persistence over time. Int. J. Food Microbiol. 128, 516–520. doi: 10.1016/j.ijfoodmicro.2008.10.009
Lukinmaa, S., Miettinen, M., Nakari, U. M., Korkeala, H., and Siitonen, A. (2003). Listeria monocytogenes isolates from invasive infections: Variation of sero- and genotypes during an 11-year period in Finland. J. Clin. Microbiol. 41, 1694–1700. doi: 10.1128/JCM.41.4.1694-1700.2003
Maury, M., Tsai, Y., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313.
Murugesan, L., Kucerova, Z., Knabel, S. J., and Laborde, L. F. (2015). Predominance and distribution of a persistent Listeria monocytogenes clone in a commercial fresh mushroom processing environment. J. Food Prot. 78, 1988–1998. doi: 10.4315/0362-028X.JFP-15-195
Nightingale, K., Windham, K., Martin, K., Yeung, M., and Wiedmann, M. (2005). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005
Orsi, R. H., Bakker, H. C. D., and Wiedmann, M. (2011). Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Parihar, V. S., Lopez-Valladares, G., Danielsson-Tham, M. L., Peiris, I., Helmersson, S., Unemo, M., et al. (2008). Characterization of human invasive isolates of Listeria monocytogenes in Sweden 1986-2007. Foodborne Pathog. Dis. 5, 755–761. doi: 10.1089/fpd.2008.0123
Pérez-Baltar, A., Pérez-Boto, D., Medina, M., and Montiel, R. (2021). Genomic diversity and characterization of Listeria monocytogenes from dry-cured ham processing plants. Food Microbiol. 99:103779. doi: 10.1016/j.fm.2021.103779
Pérez-Rodríguez, F., Carrasco, E., Bover-Cid, S., Jofré, A., and Valero, A. (2017). Closing gaps for performing a risk assessment on Listeria monocytogenes in ready-to-eat (RTE) foods: Activity 2, a quantitative risk characterization on L. monocytogenes in RTE foods; starting from the retail stage. EFSA Support. Publ. 14:1252E.
Pesavento, G., Ducci, B., Nieri, D., Comodo, N., and Lo Nostro, A. (2010). Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 21, 708–713.
Poimenidou, S. V., Dalmasso, M., Papadimitriou, K., Fox, E. M., Skandamis, P. N., and Jordan, K. (2018). Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 9:1103. doi: 10.3389/fmicb.2018.01103
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Rahimi, E., Ameri, M., and Momtaz, H. (2010). Prevalence and antimicrobial resistance of Listeria species isolated from milk and dairy products in Iran. Food Control 21, 1448–1452. doi: 10.1016/j.foodcont.2010.03.014
Ryser, E. T., and Marth, E. H. (2007). Listeria, listeriosis, and food safety. Boca Raton, FL: CRC Press. doi: 10.1201/9781420015188
Salcedo, C., Arreaza, L., Alcala, B., De La Fuente, L., and Vazquez, J. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/JCM.41.2.757-762.2003
Sosnowski, M., Lachtara, B., Wieczorek, K., and Osek, J. (2019). Antimicrobial resistance and genotypic characteristics of Listeria monocytogenes isolated from food in Poland. Int. J. Food Microbiol. 289, 1–6. doi: 10.1016/j.ijfoodmicro.2018.08.029
Stoller, A., Stevens, M. J. A., Stephan, R., and Guldimann, C. (2019). Characteristics of Listeria Monocytogenes strains persisting in a meat processing facility over a 4-year period. Pathogens 8:32. doi: 10.3390/pathogens8010032
Sullivan, C. B., Diggle, M. A., and Clarke, S. C. (2005). Multilocus sequence typing. Mol. Biotechnol. 29, 245–254.
Sun, Q., Cai, S., Cheng, J., Zhang, Y., Lin, R., Ye, Q., et al. (2021). Distribution, contamination routes, and seasonal influence of persistent Listeria monocytogenes in a commercial fresh Hypsizigus marmoreus production facility. Food Control 127:108118.
The Clinical and Laboratory Standards Institute [CLSI] (2018). Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Approved standard-M02-A12. Wayne, PA: The Clinical and Laboratory Standards Institute.
Van Stelten, A., and Nightingale, K. (2008). Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74, 7365–7375. doi: 10.1128/AEM.01138-08
Vázquez-Boland, J. A., Domínguez-Bernal, G., González-Zorn, B., Kreft, J., and Goebel, W. (2001a). Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3, 571–584. doi: 10.1016/s1286-4579(01)01413-7
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., DomıìNguez-Bernal, G., Goebel, W., et al. (2001b). Listeria pathogenesis and molecular virulence determinants. Clinical Microbiology Reviews 14, 584–640. doi: 10.1128/CMR.14.3.584-640.2001
Venturini, M. E., Reyes, J. E., Rivera, C. S., Oria, R., and Blanco, D. (2011). Microbiological quality and safety of fresh cultivated and wild mushrooms commercialized in Spain. Food Microbiol. 28, 1492–1498. doi: 10.1016/j.fm.2011.08.007
Viswanath, P., Murugesan, L., Knabel, S. J., Verghese, B., Chikthimmah, N., and Laborde, L. F. (2013). Incidence of Listeria monocytogenes and Listeria spp. in a small-scale mushroom production facility. J. Food Prot. 76, 608–615. doi: 10.4315/0362-028X.JFP-12-292
Ward, T. J., Gorski, L., Borucki, M. K., Mandrell, R. E., Hutchins, J., and Pupedis, K. (2004). Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186, 4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Keywords: Listeria monocytogenes, Pleurotus eryngii, antimicrobial resistance, MLST, virulence genes
Citation: Xu J, Wu S, Liu M, Xiao Z, Peng Y and He H (2023) Prevalence and contamination patterns of Listeria monocytogenes in Pleurotus eryngii (king oyster mushroom) production plants. Front. Microbiol. 14:1064575. doi: 10.3389/fmicb.2023.1064575
Received: 08 October 2022; Accepted: 03 January 2023;
Published: 27 January 2023.
Edited by:
Anne Brisabois, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceReviewed by:
Haiyan Zeng, Guangdong University of Technology, ChinaYanhong Liu, United States Department of Agriculture (USDA), United States
Copyright © 2023 Xu, Wu, Liu, Xiao, Peng and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi Wu,  d3VzaGlsb3ZleW91QDEyNi5jb20=
d3VzaGlsb3ZleW91QDEyNi5jb20=
 Jiang Xu
Jiang Xu Shi Wu
Shi Wu Ming Liu
Ming Liu Zitian Xiao1
Zitian Xiao1