
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 March 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1056790
Background: Studies on Citrobacter spp. are limited, hindering our understanding of its species evolution and medical relevance.
Methods: A total of 164 clinical Citrobacter spp. isolates were collected from 2017 to 2020 and identified by VITEK MALDI-TOF MS or VITEK-2 Gram-Negative Identification Card. All isolates were further analyzed by whole-genome sequencing using a HiSeq sequencer. All sequences were processed using different modules of the PGCGAP integrated package: Prokka and fastANI were used for annotation and average nucleotide identification (ANI), respectively. Antibiotic resistance and virulence genes were identified by searching CARD, ResFinder, and VFDB databases, respectively. Strains were identified using Ribosomal Multi-locus Sequence Typing (rMLST) classification based on 53 ribosome protein subunits (rps). The evolutionary relationship was analyzed using kSNP3 and visualized by iTOL editor v1_1. Genetic environments were compared by BLAST and visualized by Easyfig 2.2.5. The pathogenicity of some Citrobacter freundii isolates was confirmed by Galleria mellonella larvae infection test.
Results: A total of 14 species of Citrobacter spp. were identified from 164 isolates. However, 27 and 11 isolates were incorrectly identified as C. freundii and Citrobacter braakii by MALDI-TOF MS, respectively. In addition, MS also failed to identify Citrobacter portucalensis. The virulence genes mainly encoded proteins related to flagella and iron uptake systems. Citrobacter koseri isolates (n = 28) contained two iron uptake systems, coding yersiniabactin and aerobactin, respectively. C. braakii isolates (n = 32), like Salmonella, carried Vi capsule polysaccharide synthesis genes. The yersiniabactin gene clusters identified in five C. freundii isolates are located on various ICEkp elements and have not been reported previously. Moreover, ICEkp-carrying C. freundii showed diverse pathogenic features.
Conclusion: Conventional methods have significant defects in identifying Citrobacter spp. ICEkp-like elements-mediated acquirement of the Yersinia high-pathogenicity island was identified for the first time in C. freundii.
Citrobacter spp. are facultatively anaerobic, Gram-negative Enterobacterales bacteria. They usually colonized soils, waters, and animal or human intestines (Yuan et al., 2019b). Although Citrobacter spp. have yet to be considered important hospital-associated pathogens, they are related to hospitals and community-acquired infections of the respiratory tract, urinary tract, bloodstream, and central nervous system, especially in immunocompromised populations (Liu L. H. et al., 2018; Qian et al., 2018).
To date, 18 species have been named in the genus Citrobacter (Wang et al., 2021). Citrobacter spp. are often misidentified by traditional identification methods (Hasan et al., 2019; Oberhettinger et al., 2020), hindering our understanding of their evolution and medical relevance. With the development of whole-genome sequencing (WGS), Citrobacter spp. species can be distinguished more precisely, which advances research on their species identification, evolutionary relationships, and genome structures (Yap et al., 2020; Dziri et al., 2021). However, most studies on Citrobacter spp. have focused on the common species Citrobacter koseri and Citrobacter freundii and their phenotypic multi-drug resistance, pathogenicity, and related mechanisms (Liu L. et al., 2018; Yuan et al., 2019a; Liu et al., 2021).
Multiple siderophore systems, including enterobactin, yersiniabactin, salmochelin, and aerobactin in the family Enterobacteriaceae, have been associated with greater pathogenic potentials (Kramer et al., 2020). The Yersinia high-pathogenicity island (HPI) is widely distributed among different members of the family Enterobacteriaceae (Bach et al., 2000). The role of HPI is to synthesize iron carriers through gene clusters in an iron-deficient environment, compete for iron binding in their environment, form a complex that is recognized by recognition sites on the cell surface, and enter cell for bacterial use (Kramer et al., 2020).
Mobile genetic elements (MGEs) play a role in resistance genes’ and virulence factors’ capture, accumulation and transmission. They mainly include insertion sequences, transposons, gene cassettes/integrons, plasmids, and integrative conjugative elements (ICEs) (Partridge et al., 2018). The ICEs are self-transmissible MGEs that can be transmitted and expressed in different bacteria, allowing them to acquire different genotypes and phenotypes, facilitating their adaptation to environmental changes, and increasing their pathogenicity (Johnson and Grossman, 2015).
There is an HPI-associated ICE in Enterobacteriaceae, mainly in Klebsiella pneumoniae and Escherichia coli, named ICEkp (Lin et al., 2008). The ICEkp consists of a 5′ end, which contains a P4-like int and an HPI, a type IV secretion system (T4SS), and a 3′ end which is associated with DNA conjugative transfer. ICEkp is mainly distributed in Klebsiella spp., while ICEkp-like elements are also present in E. coli, C. koseri, Yersinia pestis, Yersinia pseudotuberculosis, and Enterobacter hormaechei. There are many subfamilies in the ICEkp family (Lam et al., 2018). Among them, ICEkp1 family contains fourteen members (ICEkp1.1–1.14), and yersiniabactin-encoding genes (ybt) contain 17 members (ybt1–ybt17) (Lin et al., 2008; Lam et al., 2018). Recent studies have identified ICEkp2 family, which is completely different from the ICEkp1 family but also possesses modules mediating the movement of elements and other accessory genes (Farzand et al., 2019). Although ICEkp2 family cannot mediate its own transmission between bacteriophages, it enhances the transmission of ICEkp1 family when coexisting (Farzand et al., 2019). Similar phenomenon also exists between ICETh1 and ICETh2 in Thermus thermophilus (Baquedano et al., 2020).
Citrobacter koseri is the only Citrobacter spp. that has been reported to contain an HPI (Yuan et al., 2019b). Its HPI is mainly related to the HPI among Y. pestis and is located on the chromosomes for conserved clonal transmission and pathogenicity enhancement (Yuan et al., 2019b). In addition, C. koseri also possesses HPI-related ICEkp1 family with unverified pathogenicity (Lam et al., 2018). In this study, for the first time, we identified ICE-mediated HPI transmission in C. freundii. We also found that ICE might spread across strains and even genera to allow the same genotype or phenotype to cross. Furthermore, we also investigated the evolutionary relationships of Citrobacter spp. through genome-wide sequence comparisons for species identification.
A total of 164 clinical Citrobacter spp. isolates were collected from our hospital from 2017 to 2020. The species of the isolates was first identified by using VITEK MALDI-TOF MS. The isolates were spotted on the target plate of VITEK MALDI-TOF MS, and then 1 μl VITEK MS-CHCA matrix was applied over the sample until the matrix and sample were dried and co-crystallized. The target plate with all prepared samples then was loaded into the VITEK MS system to acquire the mass spectra of whole bacterial cell protein. Based on the characteristics of ribosomal protein, the species of the isolates were determined by the comparison of the known mass spectra contained in the database. Moreover, we confirmed the species and tested the antimicrobial susceptibility of the isolates by biochemical assays with the VITEK 2 Compact system. The isolates were adjusted to the concentration of McFarland standard 0.5∼0.63. In 3.0 ml sodium chloride solution (0.45%), and then 145 μl bacterial solution was taken into 3.0 ml phosphate buffer solution with the insertion of VITEK-2 Gram-Negative Identification Card and AST-GN13 card for the bacterial identification and antimicrobial susceptibility, respectively. AST-GN13 card contains cephalosporins, carbapenems, aminoglycosides, quinolones, and other antibiotics. The results were interpreted with reference to CLSI M100. E. coli 8739 and 25922 were used as reference strains.
All isolates were subjected to WGS using a paired-end library with an average insert size of 350 bp (ranging from 150 to 600 bp) on a HiSeq sequencer (Illumina, CA, USA). All sequences were processed using different modules of the PGCGAP integrated package. Data were assembled using SPAdes, annotated using Prokka and subjected to fastANI for whole-genome ANI. ANI-related heatmaps were generated using self-developed scripts “triangle2list.pl,” “get_ANImatrix.pl” and “Plot_ANIheatmap.R.” Resistance genes and virulence genes were identified using CARD, ResFinder, and VFDB databases, respectively. Strains were identified using the Ribosomal Multi-locus Sequence Typing (rMLST) classification based on 53 ribosome protein subunits (rps) available on the PubMLST website. The evolutionary relationship was analyzed using kSNP3 and visualized by iTOL editor v5. Genetic environments were compared by BLAST and visualized by Easyfig 2.2.5. The bacterial genome data has been uploaded to NCBI under BioProject accession PRJNA885261.
Fresh greater wax moth larvae were used for virulence testing as previously described (Gu et al., 2018). Overnight C. freundii cultures were suspended in phosphate-buffered solution at a concentration of 1.5 × 108 CFU/ml. After injecting about 10 μl of the suspension into the ventral cavity above the anterior left hind leg, the larvae were placed in a 37°C incubator to observe their survival. Ten injections of each strain were made, and the test was repeated three times.
All 164 Citrobacter spp. isolates were collected from clinical specimens, including urine (n = 92, 56.10%), bile (n = 49, 29.88%), and blood (n = 23, 14.02%). Analysis of sequencing data (rMLST and FastANI), revealed that 27 and 11 isolates were incorrectly identified as C. freundii and Citrobacter braakii by MALDI-TOF MS, respectively. In addition, MS failed to identify Citrobacter portucalensis (Table 1). The majority of Citrobacter spp. isolates exhibited higher resistance to the third-generation cephalosporins ceftazidime and ceftriaxone and were susceptible to other clinically commonly used antibiotics (Table 1). Several C. freundii and C. portucalensis isolates were resistant to carbapenems, and most of them carried blaNDM–1 or blaNDM–5 (Supplementary Table 1). C. koseri was highly susceptible to all types of antibiotics, possibly because it contained fewer resistance genes (Supplementary Table 1).
The k-mer based analysis of whole-genome SNPs revealed that strains belonging to the same species showed higher genetic relationships (Figure 1). The isolate Cpo90 was distant from other C. portucalensis and more closely related to C. freundii clade. Individual evolutionary trees of each species showed that C. koseri and C. freundii had small outbreaks, while the other strains were more dispersed (Figure 2).
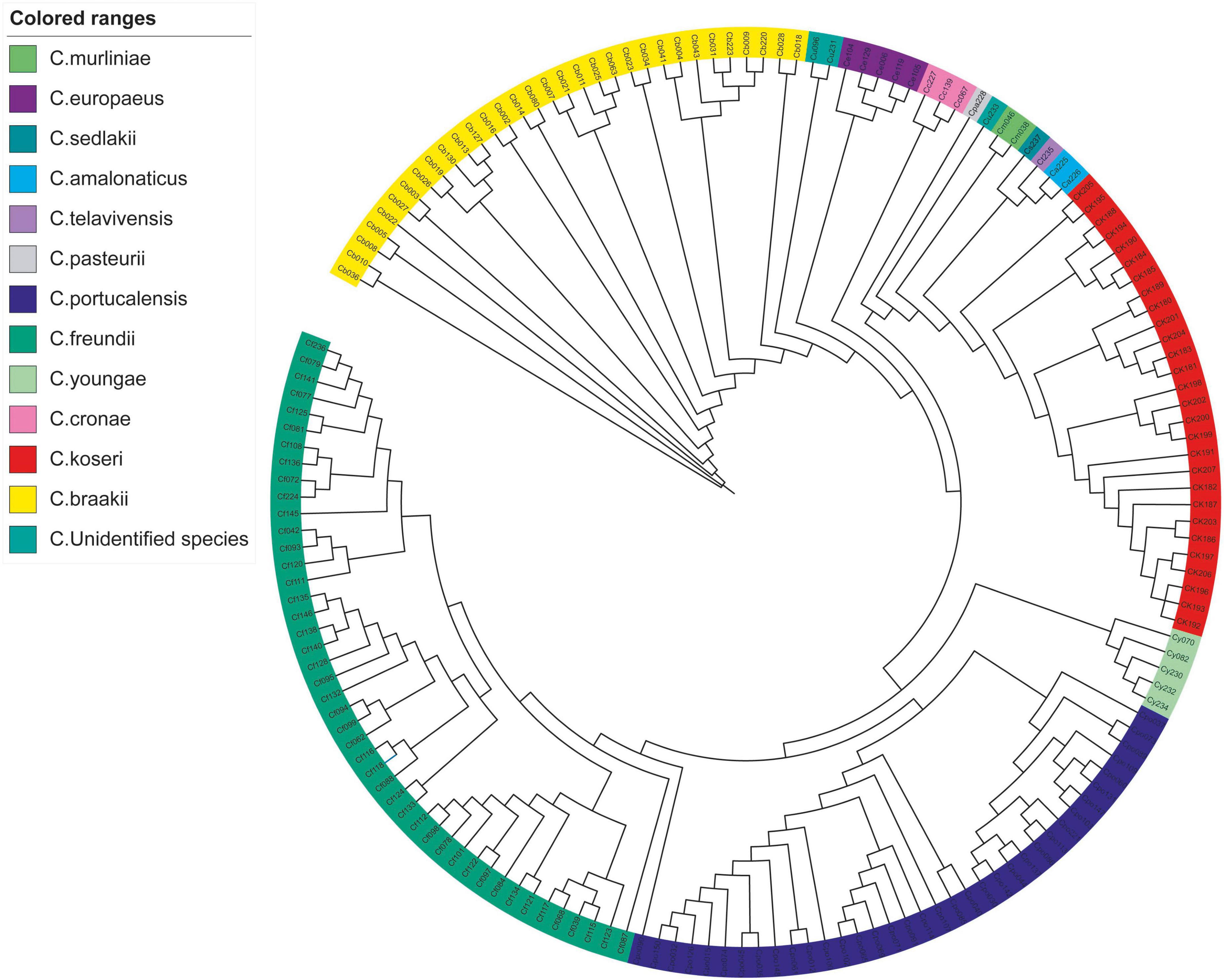
Figure 1. Whole-genome SNP phylogeny of Citrobacter spp. A phylogenetic tree of 164 Citrobacter spp. isolates was constructed based on whole-genome SNP using kSNP3.
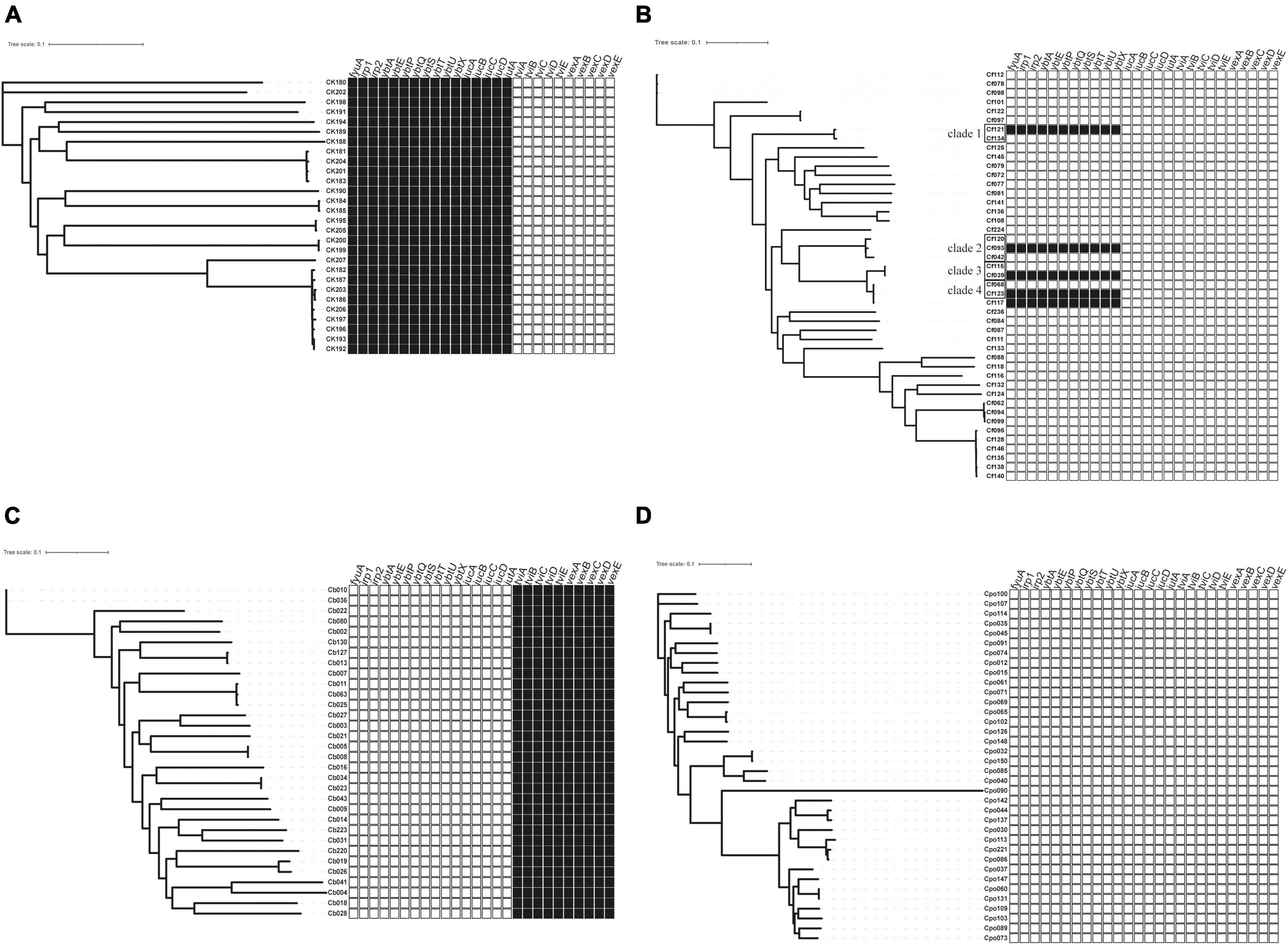
Figure 2. The distribution of virulence factors across Citrobacter spp. Boxes represent iron uptake and Vi capsule virulence factors derived from whole-genome data comparison with VFDB. (A) C. koseri, (B) C. freundii, (C) C. braakii, and (D) C. portucalensis.  , exist;
, exist;  , absence. For a full outline of sources, see Supplementary Table 1.
, absence. For a full outline of sources, see Supplementary Table 1.
The virulence genes mainly encoded flagella and iron uptake-related systems. C. koseri contained two iron uptake systems, coding yersiniabactin and aerobactin (Figure 2A). The complete genomes and GenBank annotations of 11 C. koseri strains were downloaded from NCBI. These genomes possessed multiple copies of structure-uncorrelated genes encoding yersiniabactin and aerobactin. A comparison of our sequencing data with the above complete genomes using BLAST showed similar molecular characteristics (data not shown). C. braakii, like Salmonella, carried Vi capsule polysaccharide synthesis genes (Figure 2C), while C. portucalensis contained the lowest number of virulence genes (Figure 2D). Surprisingly, we, for the first time, identified the yersiniabactin gene cluster in five C. freundii isolates (Figure 2B).
BLAST and annotation analysis revealed that all C. koseri isolates carried a cluster of yersiniabactin genes as conserved elements. However, the Yersinia HPI in four C. freundii isolates was located on various ICEkp (Figure 3). The HPI structure of Cf039 and Cf123 was highly homologous to ICEkp1.3, which has an insertion of IS630 family transposase ISEc33 in the putative functional region between HPI and T4SS secretion system (Figure 3A). Genetic structure of Cf093 was similar to that of ICEkp1.2, which has an insertion of IS5 family transposase IS903 before mobB (Figure 3B). The HPI structure of Cf121 had an ISEc33 inserted between ybt and T4SS of ICEkp1.5 (Figure 3C). Interestingly, Cf117 carried only partial ICEkp1.3 sequence, which might result in functional defects (Figure 3A).
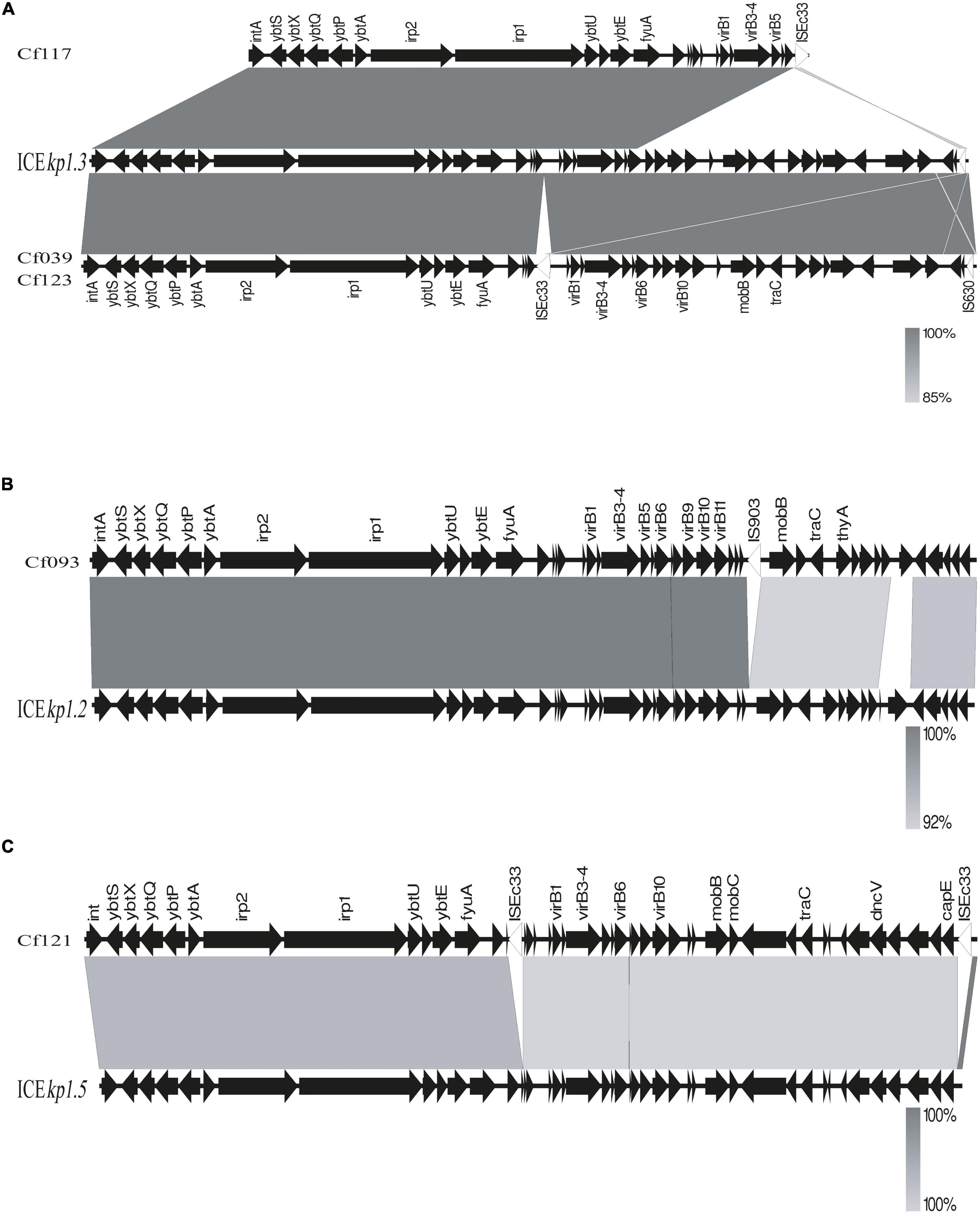
Figure 3. Linear comparison of ICEkp sequences in five C. freundii. Sequences were aligned to one another using Easyfig. Genes are denoted by arrows with a gradient from gray (low identity) to black (high identity). (A) Genomic comparison of Cf117, Cf039, Cf123 with ICEkp1.3, (B) Cf093 with ICEkp1.2 and (C) Cf121 with ICEkp1.5.
Several C. freundii isolates with HPI were used to infect Galleria mellonella larvae. Meanwhile, HPI-negative C. freundii isolates of the same clade (Figure 2B) were used as controls. The survival rates of HPI-carrying Cf039 and Cf093 were 20 and 40%, respectively. The survival rates of Cf123 and Cf121 were 100% at 48 h after infection (Figure 4). The survival rates of five HPI-negative strains Cf134, Cf042, Cf115, Cf068, and Cf120 were 50, 50, 90, 20, and 90%, respectively (Figure 4).
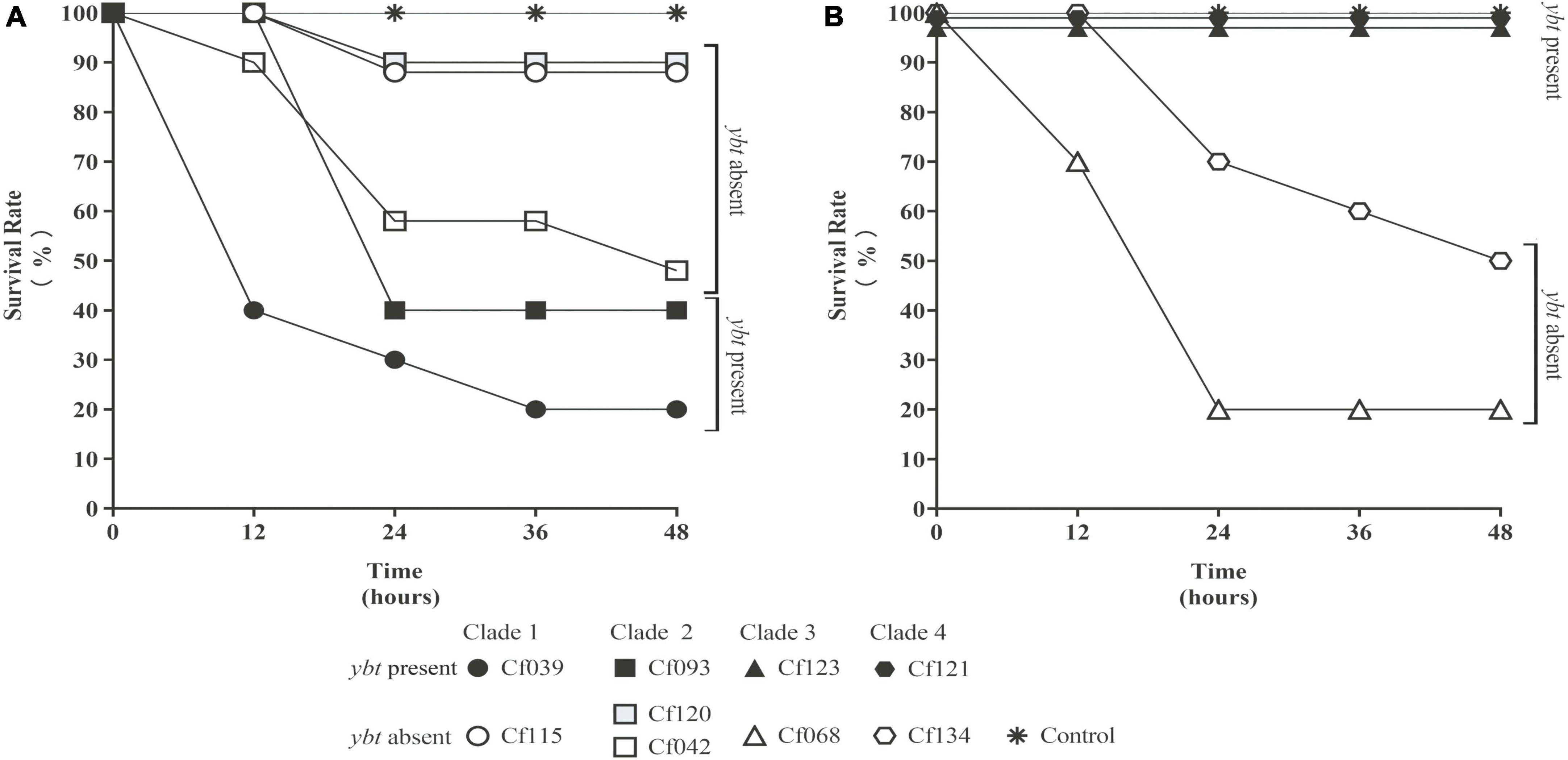
Figure 4. The survival rate of Galleria mellonella larvae. The solid black symbol represents C. freundii carrying ybt. The hollow white and gray symbol represent ybt-negative strains. The same shape represents the strains within the same clade. The values were fine-tuned to differentiate the folds with the same survival rate. All results were rounded to the nearest integer values. Strains with ybt had a lower (A) or higher (B) survival rate.
In this study, 164 clinical Citrobacter spp. isolates were identified by MALDI-TOF MS, rMLST and ANI. rMLST and ANI can identify most Citrobacter species more accurately, while C. portucalensis were incorrectly identified as C. freundii by MALDI-TOF MS (Table 1). Previous studies have also shown that MALDI-TOF MS has a lower identification accuracy and can misidentify intraspecies such as C. portucalensis and C. freundii, which are generally considered the most prevalent Citrobacter species in the clinics (Rödel et al., 2019). Since MS is the main identification method routinely used in most clinical laboratories, and rMLST and ANI are difficult to carry out, the low identification accuracy of MS may lead to large errors in the epidemiological studies of Citrobacter spp. and may interfere with drug selection in empirical anti-infection therapies. However, Citrobacter farmeri and Citrobacter werkmanii are exceptions because they can be accurately identified by MALDI-TOF MS, but not by rMLST and ANI (Table 1), indicating that different identification methods have different accuracy for different species. Therefore, supplementary methods, such as digital DNA-DNA hybridization computation and biochemical characterization are needed for identifying some species (Oberhettinger et al., 2020).
Phylogenetic analysis has shown that clinical Citrobacter spp. exhibits high level of clonal diversity. The majority of strains exhibit interspecies differences (Figure 1). The isolates of the main species also show intraspecific differences (Figure 2), while only C. koseri (Figure 2A) and C. freundii (Figure 2B) caused small-scale outbreaks. Several C. freundii and C. portucalensis isolates exhibit resistance to carbapenems due to the acquisition of various types of blaNDM. This suggests we should pay attention to the risk of transmission caused by some species, such as C. portucalensis, which can acquire foreign resistance genes (Cao et al., 2021).
Most Citrobacter spp. carry various virulence factors related to flagella apparatus biosynthesis and iron uptake system (Figure 2). C. koseri contains the most virulence factors and uniquely possesses two groups of iron uptake systems (yersiniabactin and aerobactin). HPI is an important factor closely related to the pathogenicity of C. koseri (Yuan et al., 2019b). We have revealed that ybt carried by C. koseri is rarely present in ICEkp, which is mainly carried by various ICEkp in other Enterobacteriaceae (Lam et al., 2018). In addition, most C. koseri isolates also contain aerobactin. However, analysis of their complete sequence revealed that the genes coding aerobactin and yersiniabactin are not genetically linked, and the biological effects of carrying multiple siderophores need to be further confirmed. Interestingly, C. koseri isolates carry fewer drug resistance genes (Supplementary Table 1) and exhibited lower drug resistance than other species (Table 1). Conversely, C. portucalensis contains fewer virulence factors (Figure 2D) and relatively more drug resistance genes (Supplementary Table 1). It has been suggested that high virulence and high resistance populations do not overlap. However, the possibility of dual expression of high virulence and high resistance is increasing through genomic changes (Bialek-Davenet et al., 2014). In addition, all C. braakii isolates contain clusters of genes associated with Vi capsular polysaccharide, (Hu et al., 2017; Qian et al., 2018; Yuan et al., 2019b).
Citrobacter freundii can produce large amounts of Vi and attach to the cell surface, and the viaB locus can affect changes in Vi antigen expression (Snellings et al., 1981). However, our results showed that C. freundii does not contain any Vi capsular synthesis gene cluster (Figure 2B), suggesting that the Vi cluster may be externally obtained but not transmitted by a conserved clone.
Through BLAST analysis, we found that five strains of C. freundii in this study contained yersiniabactin gene cluster (HPI) (Figure 2), and seven of all C. freundii sequences downloaded on NCBI also contained HPI. The genomic comparative analysis found that all but one HPIs are present in various ICEkp of ICEkp1 family with high similarity (Figure 3; Lam et al., 2018). This is the first report on ICEkp-mediated HPI among C. freundii. The ICEkp differs from the hosts in genomic GC content, suggesting that these HPIs may be transmitted through ICE level. C. freundii is the most prevalent Citrobacter spp. with the most carbapenem-resistant genes (Arana et al., 2017). Once multi-drug resistant C. freundii acquires HPI and spreads widely, it will make treatment more difficult. However, our results showed that clinical C. freundii isolates with HPI do not exhibit higher pathogenicity than HPI-negative isolates (Figure 4). The possible relevant mechanisms include (1) ICE may not exhibit its functions when bound to the host’s chromosome and only functions after it has been sheared down to form a loop (Johnson and Grossman, 2015) and (2) HPI-containing strains activate host’s autophagy, making the HPI-carrying host at a disadvantage stage at certain point (Dalmasso et al., 2021). From the perspective of virulence factors contained in the whole, through comparison of VFDB database, it was found that clade3 and clade4 had no difference in virulence factor except HPI gene cluster. Except that clade1 and clade2 contain different HPI gene clusters, Cf039 in clade1 contains gene clusters encoding fimbriae while Cf115 does not. The results in clade2 are opposite (Figure 4A). Fimbriae are important factors for bacteria to adhere to host cells and play a crucial role in their colonization on the host. However, according to the results of this study, it may be considered that the pathogenicity of fimbriae on bacteria needs further study.
Our study has several limitations. First, Citrobacter spp. were collected from single hospital. Second, the transmission ability of ICEkp in C. freundii, and whether the insertion of IS affects the propagation of ICEkp were not analyzed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA885261.
JY designed the study. GZ, QZ, KY, LY, and YM did phenotypic and genotypic analysis. GZ and JY drafted the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1056790/full#supplementary-material
Arana, D. M., Ortega, A., González-Barberá, E., Lara, N., Bautista, V., and Gómez-Ruíz, D. (2017). Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 72, 3283–3287. doi: 10.1093/jac/dkx325
Bach, S., de Almeida, A., and Carniel, E. (2000). The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183, 289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x
Baquedano, I., Mencía, M., Blesa, A., Burrus, V., and Berenguerfi, J. (2020). ICETh1 and ICETh2, two interdependent mobile genetic elements in thermus thermophilus transjugation. Environ. Microbiol. 22, 158–169. doi: 10.1111/1462-2920.14833
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., and Delannoy-Vieillard, A.-S. (2014). Nicolas-Chanoine, D. Decré, and Brisse, S., genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 20, 1812–1820. doi: 10.3201/eid2011.140206
Cao, X., Xie, H., Huang, D., Zhou, W., Liu, Y., Shen, H., et al. (2021). Detection of a clinical carbapenem-resistant Citrobacter portucalensis strain and the dissemination of C. portucalensis in clinical settings. J. Global Antimicrob. Resist. 27, 79–81. doi: 10.1016/j.jgar.2021.04.027
Dalmasso, G., Nguyen, H. T. T., Faïs, T., Massier, S., Chevarin, C., and Vazeille, E. (2021). Yersiniabactin siderophore of Crohn’s disease-associated adherent-invasive is involved in autophagy activation in host cells. Int. J. Mol. Sci. 22:3152. doi: 10.3390/ijms22073512
Dziri, R., Kuskucu, M. A., Arfaoui, A., Fethi, M., Ifaoui, S., and Bellaaj, R. (2021). Whole genome sequencing of a Citrobacter freundii strain isolated from the hospital environment: an extremely multiresistant NDM-1 and VIM-48 coproducing isolate. Microb. Drug Resist. 28, 18–22. doi: 10.1089/mdr.2020.0417
Farzand, R., Rajakumar, K., Zamudio, R., Oggioni, M. R., Barer, M. R., and O’Hare, H. M. (2019). ICEKp2: description of an integrative and conjugative element in Klebsiella pneumoniae, co-occurring and interacting with ICEKp1. Sci. Rep. 9:13892. doi: 10.1038/s41598-019-50456-x
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., and Wang, L. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Hasan, M. S., Sultana, M., and Hossain, M. A. (2019). Complete genome arrangement revealed the emergence of a poultry origin superbug Citrobacter portucalensis strain NR-12. J. Glob Antimicrob. Resist. 18, 126–129. doi: 10.1016/j.jgar.2019.05.031
Hu, X., Chen, Z., Xiong, K., Wang, J., Rao, X., and Cong, Y. (2017). Vi capsular polysaccharide: synthesis, virulence, and application. Crit. Rev. Microbiol. 43, 440–452.
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Kramer, J., Özkaya, Ö, and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163.
Lam, M. M. C., Wick, R. R., Wyres, K. L., Gorrie, C. L., Judd, L. M., Jenney, A. W. J., et al. (2018). Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom 4:e000196. doi: 10.1099/mgen.0.000196
Lin, T. L., Lee, C. Z., Hsieh, P. F., Tsai, S. F., and Wang, J. T. (2008). Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190, 515–526. doi: 10.1128/JB.01219-07
Liu, L., Chen, D., Liu, L., Lan, R., Hao, S., and Jin, W. (2018). Genetic diversity, multidrug resistance, and virulence of citrobacter freundii from diarrheal patients and healthy individuals. Front. Cell Infect. Microbiol. 8:233. doi: 10.3389/fcimb.2018.00233
Liu, L., Zhang, L., Zhou, H., Yuan, M., Hu, D., Wang, Y., et al. (2021). Antimicrobial resistance and molecular characterization of spp. causing extraintestinal infections. Front. Cell. Infect. Microbiol. 11:737636.
Liu, L. H., Wang, N. Y., Wu, A. Y., Lin, C. C., Lee, C. M., and Liu, C. P. (2018). Citrobacter freundii bacteremia: risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 51, 565–572. doi: 10.1016/j.jmii.2016.08.016
Oberhettinger, P., Schule, L., Marschal, M., Bezdan, D., Ossowski, S., and Dorfel, D. (2020). Description of Citrobacter cronae sp. nov., isolated from human rectal swabs and stool samples. Int. J. Syst. Evol. Microbiol. 70, 2998–3003. doi: 10.1099/ijsem.0.004100
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088–17. doi: 10.1128/CMR.00088-17
Qian, C., Du, Y., Li, H., Wu, P., Wang, L., Wei, Y., et al. (2018). Development of rapid and simple experimental and in silico serotyping systems for Citrobacter. Future Microbiol. 13, 1511–1522. doi: 10.2217/fmb-2018-0187
Rödel, J., Mellmann, A., Stein, C., Alexi, M., Kipp, F., and Edel, B. (2019). Use of MALDI-TOF mass spectrometry to detect nosocomial outbreaks of Serratia marcescens and Citrobacter freundii. Eur. J. Clin. Microbiol. Infect. Dis. 38, 581–591. doi: 10.1007/s10096-018-03462-2
Snellings, N. J., Johnson, E. M., Kopecko, D. J., Collins, H. H., and Baron, L. S. (1981). Genetic regulation of variable Vi antigen expression in a strain of Citrobacter freundii. J. Bacteriol. 145, 1010–1017. doi: 10.1128/jb.145.2.1010-1017.1981
Wang, H., Hou, H., and Huang, J. (2021). Citrobacter arsenatis sp. nov., an arsenate-reducing bacterium isolated from freshwater sediment. Antonie Van Leeuwenhoek 114, 1285–1292. doi: 10.1007/s10482-021-01601-y
Yap, P. S. X., Ahmad Kamar, A., Chong, C. W., Yap, I. K. S., and Teh, C. S. J. (2020). Whole genome analysis of multidrug-resistant Citrobacter freundii B9-C2 isolated from preterm neonate’s stool in the first week. J. Glob Antimicrob. Resist. 21, 246–251. doi: 10.1016/j.jgar.2020.03.024
Yuan, C., Yang, P., Wang, J., and Jiang, L. (2019a). Myo-inositol utilization by Citrobacter koseri promotes brain infection. Biochem. Biophys. Res. Commun. 517, 427–432. doi: 10.1016/j.bbrc.2019.07.112
Keywords: Citrobacter, integrative conjugative element, yersiniabactin, high-pathogenicity island, Citrobacter freundii
Citation: Zhang G, Zhao Q, Ye K, Ye L, Ma Y and Yang J (2023) Molecular analysis of clinical Citrobacter spp. isolates: Acquisition of the Yersinia high-pathogenicity island mediated by ICEkp in C. freundii. Front. Microbiol. 14:1056790. doi: 10.3389/fmicb.2023.1056790
Received: 29 September 2022; Accepted: 27 February 2023;
Published: 16 March 2023.
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Li Han, Chinese Center for Disease Control and Prevention, ChinaCopyright © 2023 Zhang, Zhao, Ye, Ye, Ma and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiyong Yang, eWFuZ2p5MzAxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.