
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 February 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1035669
This article is part of the Research TopicUsing Virus Specific-Signatures During Infection to Characterize Host-Pathogen InteractionsView all 5 articles
Polydnaviruses (PDVs) exhibit species-specific mutualistic relationships with endoparasitoid wasps. PDVs can be categorized into bracoviruses and ichnoviruses, which have independent evolutionary origins. In our previous study, we identified an ichnovirus of the endoparasitoid Diadegma fenestrale and named it DfIV. Here, DfIV virions from the ovarian calyx of gravid female wasps were characterized. DfIV virion particles were ellipsoidal (246.5 nm × 109.0 nm) with a double-layered envelope. Next-generation sequencing of the DfIV genome revealed 62 non-overlapping circular DNA segments (A1–A5, B1–B9, C1–C15, D1–D23, E1–E7, and F1–F3); the aggregate genome size was approximately 240 kb, and the GC content (43%) was similar to that of other IVs (41%–43%). A total of 123 open reading frames were predicted and included typical IV gene families such as repeat element protein (41 members), cysteine motif (10 members), vankyrin (9 members), polar residue-rich protein (7 members), vinnexin (6 members), and N gene (3 members). Neuromodulin N (2 members) was found to be unique to DfIV, along with 45 hypothetical genes. Among the 62 segments, 54 showed high (76%–98%) sequence similarities to the genome of Diadegma semiclausum ichnovirus (DsIV). Three segments, namely, D22, E3, and F2, contained lepidopteran host genome integration motifs with homologous regions of about 36–46 bp between them (Diadegma fenestrale ichnovirus, DfIV and lepidopteran host, Plutella xylostella). Most of the DfIV genes were expressed in the hymenopteran host and some in the lepidopteran host (P. xylostella), parasitized by D. fenestrale. Five segments (A4, C3, C15, D5, and E4) were differentially expressed at different developmental stages of the parasitized P. xylostella, and two segments (C15 and D14) were highly expressed in the ovaries of D. fenestrale. Comparative analysis between DfIV and DsIV revealed that the genomes differed in the number of segments, composition of sequences, and internal sequence homologies.
Parasitoid wasps of the family Ichneumonidae represent a diverse, species-rich group of insects (Gómez et al., 2018) that can regulate their hosts to ensure successful parasitism (Stoltz and Vinson, 1979). The genus Diadegma represents a large group of parasitoid wasps with 409 known species worldwide (Sharkey et al., 2013; Klopfstein et al., 2019), including 55 species within 13 genera in the Korean peninsula (Choi and Lee, 2018). In Korea, two koinobiont endoparasitoids of the diamondback moth, Plutella xylostella, exist: D. semiclausum (Hellen) and D. fenestrale (Holmgren; Hymenoptera: Ichneumonidae; Nam et al., 2022). Diadegma semiclausum is thought to have originated in Eurasia (Fitton, 1992) and is a specific endoparasitoid of P. xylostella larvae. The generalist D. fenestrale is polyphagous in nature and attacks over 90 herbivore species (Thompson, 1951; Azidah et al., 2000), including P. xylostella and the potato tuber moth, Phthorimaea operculella.
During oviposition, parasitoid wasps lay eggs containing a parasitoid-associated polydnavirus (PDV; Tanaka et al., 2007; Dicke et al., 2020). PDVs belong to the family Polydnaviridae, which comprises two genera, namely, Bracovirus and Ichnovirus (Stoltz and Vinson, 1979; Federici and Bigot, 2003; Lorenzi et al., 2022). The PDV genome comprises encapsidated double-stranded DNA and is 187–567 kb in length (Dupuy et al., 2006; Webb et al., 2006; Tanaka et al., 2007; Wang et al., 2021). PDV particles exist in the parasitoid wasp host ovaries, particularly in the calyx tissues located at the base of the oviducts, where they assemble with the eggs (Stoltz and Vinson, 1979). PDV virions are then injected into hosts during oviposition and can replicate alone or in conjunction with other factors (i.e., virulence factors and integration of genes into the wasp genome) that actively suppress host immunity, promoting the survival of the parasitoids (Edson et al., 1981; Luckhart and Webb, 1996; Moreau et al., 2009; Kashkouli et al., 2021). The host specificity of PDVs depends on their evolutionary lineage (Stoltz and Vinson, 1979). Several PDV virulence genes have seemed to be transcribed in hosts, possibly providing functions on PDV role in parasitism (Strand and Burke, 2013). PDV parasitism rates vary depending on the host, with Diadegma fenestrale ichnovirus (DfIV) showing a different parasitism rate in P. xylostella and P. operculella (Kim et al., 2015).
Campoletis sonorensis ichnovirus (CsIV) was the first identified PDV (Norton et al., 1975). To date, eight encapsidated IV genomes, including the Campoplegine and Banchine IV groups, have been fully sequenced and appear to share the canonical ichnovirus gene families (Webb et al., 2006; Lapointe et al., 2007; Tanaka et al., 2007; Djoumad et al., 2013; Doremus et al., 2014; Wang et al., 2021). These two IV groups differ morphologically, with campoplegine IVs possessing singly enveloped nucleocapsids and banchine IVs possessing several smaller nucleocapsids. Banchine IV genome, including Glypta fumiferanae ichnovirus (GfIV) and Apophua simplicipes ichnovirus (AsIV) genome, shares an approximately 260-bp region sequence not found in campoplegine IV circles (Djoumad et al., 2013). The function of this region is currently unknown, but it is thought to be involved in the integration of GfIV genome segments into the genome of the lepidopteran host, similar to how host integration motifs in D. semiclausum Ichnovirus genome segments (Wang et al., 2021). In addition, eight encapsidated genomes have similar genome sizes (~250 kb) except Diadegma semiclausum ichnovirus (DsIV, 208 kb), with the number of DNA segments ranging from 20 (Tranosema rostrale ichnovirus, TrIV) to 62 (DfIV). DfIV and DsIV exhibited all the known major gene groups representative of ichnoviruses, and their overall gene counts were similar to those of HfIV and CsIV (Kim et al., 2015; Nam et al., 2022). Nevertheless, the individual Tranosema rostrale (TrIV) and Hyposoter fugitivus (HfIV) ichnoviruses have special features, including similar genome lengths (∼250 kb), nested genome segments, and members of six conserved gene families (Tanaka et al., 2007). In addition, TrIV has a distinct gene family and more open reading frames (ORFs) than HfIV. The interspecific variations could be related to differences in parasitoid host biology (Tanaka et al., 2007).
DsIV was identified in D. semiclausum, and its viral gene contents were partially assessed from the parasitized host, P. xylostella (Wang et al., 2021), but the expression of PDV genes in parasitized hosts remains poorly explored (Etebari et al., 2011). The gene expression pattern of DfIV varies across hosts, with higher expression in P. operculella than in P. xylostella (Kim et al., 2015). However, the factors determining host range (i.e., lepidopteran host) and preference in IVs remain to be understood. Previous studies have suggested that the virus uses virulence factors to manipulate host immune systems to enable the survival of parasitoid eggs and larvae (Herniou et al., 2013; Kashkouli et al., 2021). PDVs contain functional genes that assist in manipulating host physiological functions, for example through transcription inhibition (Shelby et al., 1998; Barandoc and Kim, 2009). Several research groups have attempted to investigate the genomic composition of PDVs to isolate functional genes, and genomic characterization has been attempted to gain insight into the mechanisms of action of these viruses (Barat-Houari et al., 2006; Lapointe et al., 2007; Tanaka et al., 2007; Choi et al., 2009). However, few genes have been identified to characterize those that may be involved in physiological functions.
Although DfIV was identified before DsIV and DfIV gene expression has been studied (Kim et al., 2015), its genome has not yet been identified. In this study, we attempted to elucidate the relationship between parasitoid organisms and PDV from an evolutionary point of view by sequencing the DfIV genome and comparing DfIV to those of symbiotic DsIV genome, DsIV from D. semiclausum, which have very closely related evolutionary lineages. Furthermore, we compared PDV gene expression in two major PDV hosts, the lepidopteran moth P. xylostella, and the parasitoid wasp D. fenestrale. To confirm the presence of DfIV in the ovary, microscopic analysis was performed by using transmission electron microscopy (TEM). We identified three integration motifs in lepidopteran hosts, as well as a novel gene family, hypothetical genes, their putative functions, and gene expression patterns in respective hosts. Our findings highlight new genes and elucidate their interactions, integration, and expression, improving our understanding of the coevolution of hosts and viruses.
Diadegma fenestrale was obtained by infesting potato fields with the parasitized larval stage of Phthorimaea operculella (Zeller), in Jeju, Korea, in May 2009. Specimens were collected and maintained in the laboratory in Pyeongchang, Korea (Choi et al., 2013). Diadegma semiclausum was donated by the World Vegetable Center (former Asian Vegetable Research and Development Center, AVRDC), Taiwan, in 2001 (Kwon et al., 2003; Nam et al., 2022). Diadegma fenestrale was reared on P. operculella or P. xylostella as hosts in transparent plastic cages (30 × 30 × 30 cm) under the conditions of 25°C ± 2°C, 16:8-h light:dark photoperiod, and 50%–70% relative humidity. Third instar P. operculella or P. xylostella larvae (5 days after hatch) were parasitized by D. fenestrale in an open-type cylindrical plastic cage (15 cm diameter, 30 cm height) for 24 h, and parasitized hosts were reared in conditions similar to those used for unparasitized hosts. The emerged D. fenestrale adults were collected every day and allowed to mate for 24 h before use for parasitization. Adult wasps were fed with a 10% sucrose solution. Rearing methods and conditions for D. semiclausum were similar to those used for D. fenestrale, but P. xylostella was used as a moth host.
Diadegma fenestrale ovaries from 1-day-old female wasps were dissected, and tissues from more than 100 female parasitoids were collected in phosphate-buffered saline (PBS). The dissected ovary tissues were homogenized using a glass–glass micro-tissue grinder (Radnoti, Monrovia, CA, United States), and the homogenate was passed through a 0.45 μm syringe filter (Advantec MFS Inc., Dublin, CA, United States) and centrifuged for 30 min at 15,000 ×g and 4°C. The pellet was resuspended in DNAzol (Molecular Research Center Inc., Cincinnati, Ohio, United States) and homogenized using a disposable tissue grinder. The resulting homogenate was centrifuged for 15 min at 12,000 ×g and 4°C, and then, the supernatant was transferred to a new tube. The DfIV genomic DNA (gDNA) was precipitated by adding the same volume of ethanol and centrifuging for 10 min at 10,000 ×g and 4°C. The pellet was washed with 75% ethanol, dried, and then resuspended in nuclease-free water. DfIV gDNA was quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). To visualize the viral segment DNAs, 2 μg of DfIV gDNA was separated on 0.5% agarose gel at 30 V for 9 h. The gDNA of DsIV was isolated following the methods used for DfIV.
To characterize the genome composition of DfIV viral particles, extracted gDNA was subjected to 454 pyrosequencing (Science, 2008). Whole-genome shotgun sequencing of DfIV was performed at Macrogen Inc. (Seoul, Korea) using the Roche 454 GS-FLX sequencer (Roche, Basel, Switzerland) with FLX-plus chemistry sets according to the GS-FLX manual. After shotgun sequencing, the adapter and primer sequences were removed, and the DfIV genome was assembled using the Newbler ver. 2.6 GS de novo assembler (Roche). To improve the genome sequences, resequencing of the DfIV genome was performed through Illumina paired-end library construction and MiSeq platform sequencing (Illumina, San Diego, CA, United States). The sequencing data were trimmed to remove the adapter and low-quality reads (Q20, 1-bp error read in each 100-bp sequencing; call accuracy of 99% and Q30% indicates virtual reads will be perfect with no ambiguities) and then assembled using SOAPdenovo ver. 2.0 (Xie et al., 2014) and DNASTAR Lasergene ver. 14.1.0 (DNASTAR, Inc., Madison, WI, United States) with default parameters. Contigs with similarity to the DsIV genome were selected based on BLAST (Zhang et al., 2000) searches against the non-redundant nucleotide sequence in GenBank. The DfIV genome was annotated using BLAST against Diadegma semiclausum ichnovirus sequences,1 ab initio gene prediction in GeneMark (Besemer and Borodovsky, 2005) and FGENESV0 (Solovyev et al., 2006), and final manual curation. The DsIV genome was sequenced through the same methods using the Illumina platform. Sequence similarity between DfIV and DsIV genome segments was analyzed using reciprocal BLAST searches and visualized using Circos (http://circos.ca/; Krzywinski et al., 2009). Open reading frames (ORFs; Stothard, 2000) were predicted using the NCBI ORF finder program.2 Functional gene prediction was performed using domain-enhanced lookup time accelerated BLAST (Boratyn et al., 2012), and a cluster analysis was conducted through the cluster W method using Lasergene v14 (DNASTAR) with the Pfam database (Sonnhammer et al., 1997; El-Gebali et al., 2019). Genome assembly and annotation were conducted with the cooperation of Phyzen (Phyzen Genomics Institute, Seongnam, Korea), a bioinformatics company in Korea.
The proteins in DfIV, including repeat elements (Reps), cysteine motif (Cys), vankyrins, polar residue-rich protein (PRRP), vinnexins, and neuromodulin N sequences, were used for alignment via Lasergene v14 (DNASTAR). Neuromodulin N was found to represent a distinct gene family in the DfIV genome. Unrooted phylogenetic tree construction with the protein sequences of Reps was performed by using MEGA 11.2 (Tamura et al., 2021) for comparison between the DfIV and DsIV genomes (Supplementary Figure 4A). The Jones–Taylor–Thornton (JTT) + Gamma-distributed (G) model was selected as the best-fit model and was used for maximum likelihood analysis with 1,000 bootstrap replicates. DfIV was distinguished from DsIV by the presence of colored (red color DfIV and green color DsIV proteins; cys-motifs, n = 10; vankyrins, n = 9; PRRP, n = 7; vinnexins, n = 6 proteins; n = copy number of each gene; Supplementary Figures 4B–E). Three duplicated copies of the vinnexins protein sequence were also used for the analysis, including vinnexin 3 (C12 and C13), vinnexin 4 (D6 and D11), and vinnexin 5 (12 and 13). In this study, 41 DfIV Rep protein sequences were identified, but one short protein sequence was excluded (rep 1–1, ULM71580.1) from the phylogenetic analysis.
Gene expression analyses in different hosts were performed to understand the molecular mechanisms of complex interaction (i.e., internal sequence homologies, number of segments, composition of sequences, integration motif and sites, and expression levels of genes). Gene expression patterns were analyzed in D. fenestrale and P. xylostella 1 and 5 days after inoculation. We used inoculated third and fifth instar P. xylostella on days 1 and 5, respectively, after inoculation. Five P. xylostella larvae were used as a biological replicate (pool of larvae). After 1 and 5 days of parasitization with D. fenestrale, total RNA was extracted from P. xylostella samples and the ovarian tissue of 30 parasitoids in each replicate (replicate consisting of ovaries from a pool of ovaries from several individuals) using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The 2100 Bioanalyzer RNA 6000 NANO chip (Agilent) was used to confirm the quality of the RNA samples. RNAseq library preparation and main sequencing were performed following the procedures described in section 2.3. RNA-seq data were trimmed using Trimmomatic v0.36 (Bolger et al., 2014). Parameters of p-value < 0.05 and|log2 (fold change)| > 1 were used to determine significantly differentially expressed transcripts (DNASTAR; EdgeR). Statistical analysis was performed based on read counts or fragments per kilobase million values. Trimmed and assembled RNA-seq data were mapped (BWA tools, version 0.7.10) to DfIV genome (Supplementary Table 1), and the P. xylostella genome (GCA_000330985.1) was also used for comparative analysis.
After 1 day of parasitization with D. fenestrale, gDNAs were extracted from P. xylostella samples hosting parasitoid eggs. A single larva was used as a biological replicate, and three larvae genomes were re-sequenced. Library preparation and genome sequencing were performed following the procedures for virus genome sequencing described in section 2.3. Resequencing data were trimmed to remove the adapter and low-quality reads using Trimmomatic v0.36 (Bolger et al., 2014) and the trimmed high-quality data were mapped to the genome using the Burrows–Wheeler Aligner (BWA) software (version 0.7.17, Li and Durbin, 2009) with default parameters. SAMtools software (version 1.15, Li et al., 2009) was used to convert mapping results into the BAM format and to remove the unmapped and non-unique reads. The Picard package (version 2.27)3 was used to filter duplicated reads. Trimmed re-sequence data were mapped to the DfIV reference genome, and the P. xylostella genome (GCA_000330985.1) was used for comparison. The investigated integration motifs in the host genome revealed microhomologies (≥15 read count/number of repeat sequences, and 10-, 20-, and 50-motif length) between the host genome and the DfIV circles at integration breakpoints. If an integration motif matched a P. xylostella genome read count of 15 bp, it was not counted. Microhomologies (threshold level ≥ 15 read count/ number of repeat sequences, and 10-, 20-, and 50-motif length, see Supplementary Table 4) were revealed between the host genome and the DfIV circles at integration breakpoints.
The DfIV genome data (62 segments) were deposited in the NCBI database under accession numbers MZ129220–MZ129281. The DsIV genome sequences (47 segments) were previously deposited by Wang et al. (2021) under the GenBank accession numbers KF156214–KF156260, and the new segment identified in this study (segment no. 48) was deposited under GenBank accession number MZ129282. Three segments of Saesbyeol virus genome data were deposited: large (MH698456), medium (MH823676), and small (MH698457).
DfIV morphology was observed within the nuclei of calyx epithelial cells in female reproductive organs (Figure 1). To confirm the presence of DfIV in the ovary, ultrathin cross-sections of the distal ovary, proximal ovary, and calyx regions were prepared and examined using transmission electron microscopy (TEM; Figures 1A–C, and E). Each ovary comprised more than 10 ovarioles, each containing developing oocytes (Figure 1A1). Nuclei were observed inside the oocytes (Figures 1B1,C1). More than five developed oocytes were observed (Figure 1A2). Each oocyte had a relatively large nucleus surrounded by follicular epithelium (Figures 1B2,C2). DfIV particles were identified within the calyx, alongside immature oocytes (Figures 1A3,B3). Each ovarian epithelial cell contained a large nucleus with highly stained materials. The viral particles were ellipsoidal, with an average size of 246.45 nm × 1.20 nm (long axis) and 108.95 nm × 1.64 nm (short axis), and possessed double membranes in the form of bilayer envelopes that assembled to form an intranuclear virogenic stroma (Figure 1C3). Each pair of ovaries was filled with fully grown oocytes at the ovarian calyx; lateral oviducts are shown in Figure 1D. Blue-colored materials were also detected in the lumen, representing fully grown oocytes. Stem cells and early immature oocytes were observed in the distal ovariole region.
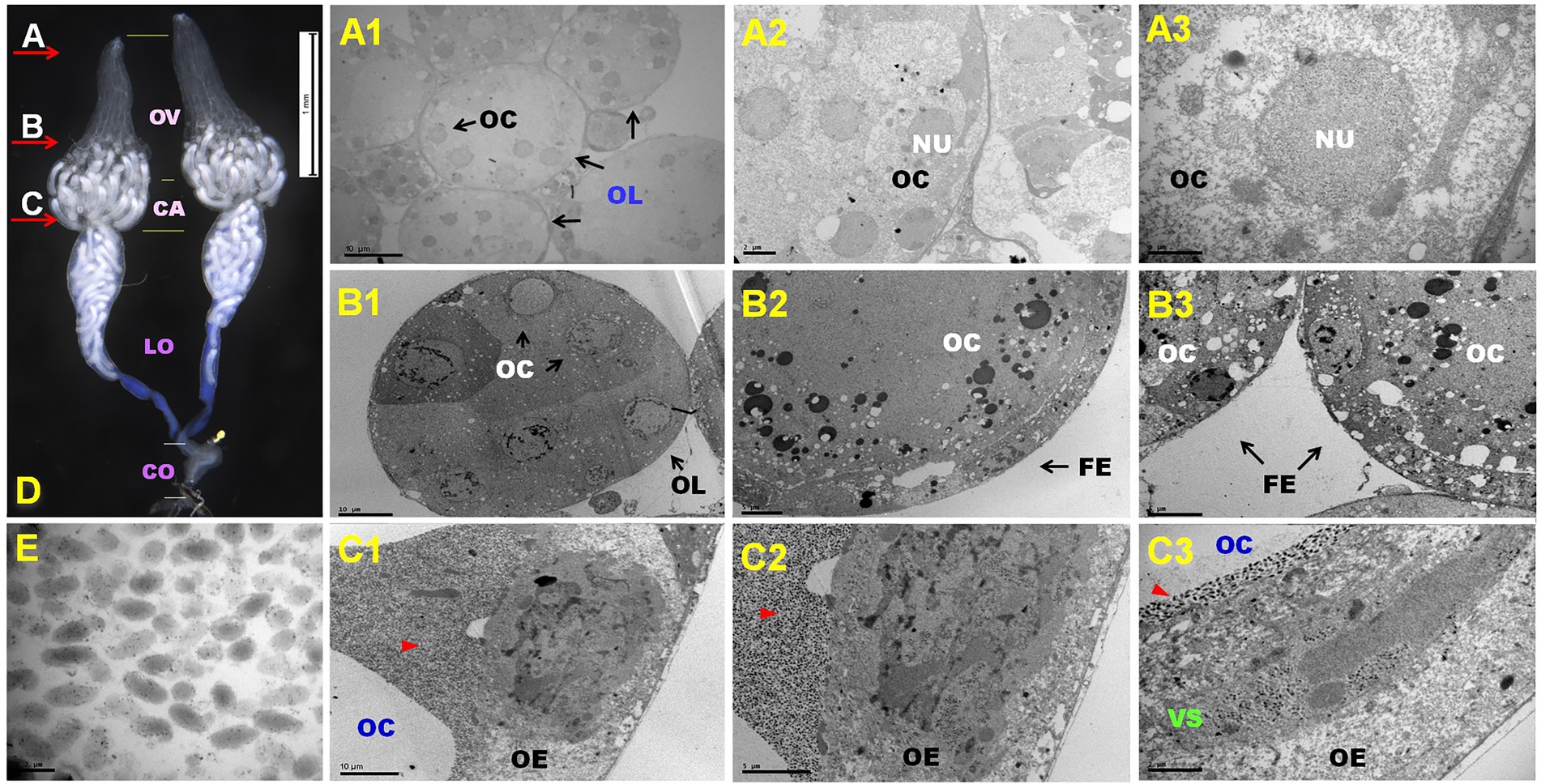
Figure 1. (A–E) Representative ovary structure of Diadegma fenestrale (D); red arrow indicates the dissection position and direction of the cutting sites (A–C). The reproductive organ comprised four major parts: ovary (OV), calyx (CA), lateral oviduct (LO), and common oviduct (CO). Cutting site (A): oogenesis at the germarium of the D. fenestrale ovary (A1–A3). A single ovary contained about 10 ovarioles. More than five immature oocytes (OC) were located in each ovariole (OL) (A1). A detailed view of the ovariole is shown in (A2). Five oocytes and their nuclei (NU) were observed in an ovariole. A detailed view of the oocyte is shown in (A3). Cutting site (B): OCs in vitellogenesis are surrounded by the follicular epithelium (FE). Six OCs were present in an OL (B1). A detailed view of the OCs is shown in (B2,B3). Cutting site (C): OCs surrounded by virion particles at the calyx area after vitellogenesis. The ovarian epithelium (OE) contains virogenic stroma (VS) and releases the virion particle to the calyx chamber. Red triangles indicate the presence of DfIV particles (C1,C2). DfIVs were located within the OE with OCs (C3). Newly growing oocytes were abundant at the ovarian calyx and lateral oviducts (D). DfIV showed a double membrane envelope, but the inner membrane could not be distinguished clearly (E).
The analyzed genome size determined by gel electrophoresis may be skewed due to the poor quality of separation of supercoiled genome segments and differences in the abundance of genome segments. Therefore, electrophoresis confirmed only that the DfIV genome comprised segments of various sizes, but the exact number and size of segments could not be determined (Supplementary Figure 1). The Illumina-based genome sequence confirmed the precise number (n = 62, number of segments) and size of segments (1.4–6.6 kb; Supplementary Table 2).
A total of 20,810,524 bp was sequenced via Roche 454 from 51,684 reads and assembled to approximately 120 contigs. Following the Illumina-based genome sequence, 62 circular DNA segments were assembled with an approximate total size of 247 kb (Supplementary Table 2). A total of 126 ORFs were predicted, but three ORF gene sequences were located in different loci of gene segments and had identical or slightly modified sequences from the original. Therefore, we trimmed the overlapping sequences and predicted a total of 123 ORFs (Table 1). The complete genome size, GC content, number of segments, and number of genes were compared between DfIV and other PDVs. Genome size and GC content were highly similar among DfIV and ichnoviruses of other Campopleginae family parasitoid hosts, H. fugitivus ichnovirus (HfIV) and C. sonorensis ichnovirus (CsIV). Glypta fumiferanae ichnovirus (GfIV) and Apophua simplicipes ichnovirus (AsIV; Banchinae) showed slight differences in genome size and number of segments when compared with DfIV. The total genome size of AsIV was estimated to be 300 kb, similar to the 292 kb reported for GfIV (Lapointe et al., 2007) and with comparable gene families and variation in a number of genome segments (AsIV > 132 and GfIV 105). However, the numbers of segments (15–132) and genes (27–186) were highly variable (Table 1). The DfIV genome segment size ranged from 1.5 to 8.2 kb with a median of 3.77 kb. DfIV genome characteristics were different from those of bracoviruses, including CcBV, MdBV, and CvBV (Table 1).
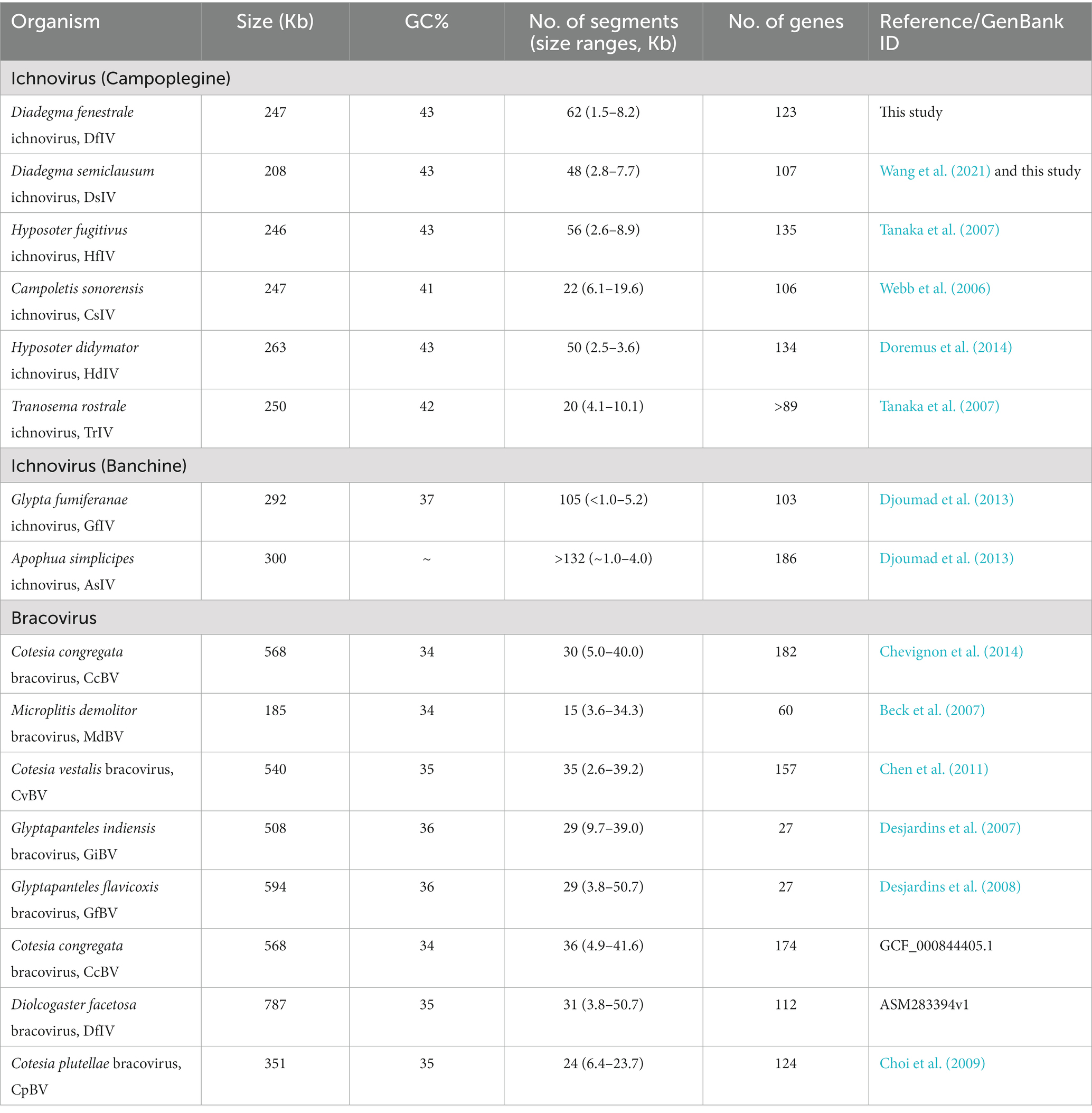
Table 1. List of comparable data from diverse ichnoviruses and bracoviruses regarding genome sequence length, gene segment abundance, and number of genes.
In the DfIV genome, we identified 123 ORFs/genes, including rep (n = 41), cys (n = 10), vankyrin (n = 9), vinnexin (n = 6), PRRP (n = 7), neuromodulin N1 (n = 2), and unassigned ORFs (n = 45), which we numbered based on the genome segments of DfIV (N = 62) and their segment-specific order. Forty-one of these putative ORFs overlapped in at least one direction (Figure 2). Approximately one-third (21/62; A1–5; B1, 3, 4, 6, 8; C1, 7, 8, 11, 14, 15; D4, 10, 12, 22; and E4) of the DfIV genome segments contained only one putative ORF (Figure 2). In the DsIV genome, 103 ORFs/genes were annotated, including rep (n = 36), cys (n = 8), vankyrin (n = 6), vinnexin (n = 7), PRRP (n = 7), N-protein (n = 3), and unassigned ORFs (n = 36). The genome of DsIV had already been sequenced (N = 47 segments; Wang et al., 2021), but our analysis identified an extra segment (segment number 48, GenBank accession no: MZ129282.1) containing three hypothetical proteins (ULM71713, 171; ULM71712, 109; and ULM71711, 79 amino acids). No neuromodulin N genes were identified in DsIV. We aligned the two gene sequences of neuromodulin N from DfIV with those of other closely related three sequences (DfIV_Neuromodulin_N1_C12, DfIV_Neuromodulin_N2_C13, HdIV_U2 [AIK25642], HfIV_b7.1 [YP001031227], and DsIV_vinnexin1 [AHY21952]), based on BLAST sequence similarity searches and observed indels on the aligned gene sequences (Supplementary Figure 2).

Figure 2. Graphical representation of the DfIV genome and its annotated genes, with 62 non-redundant (NR) circular genome segments shown as linear molecules. DfIV genome segments were 1.5–8.2 kp in length. Colored boxes show the sizes (length) and arbitrary locations of gene families, with directions indicated by the arrowheads on each box. Red boxes indicate integration motifs of the lepidopteran host genome. Gray regions represent non-coding DNA. Hp, hypothetical protein; PRRP, polar residue-rich protein 1; Rep, repeat element protein; Neuro-1, neuromodulin N1.
Whole-genome sequence similarity was analyzed and visualized of corresponding genomic areas between 62 genomic segments of DfIV and 48 genomic segments of DsIV (Figure 3). The sequence similarity between DfIV and DsIV was high, ranging from 85% to 96% with an average of 90% similarity. Nine DfIV-specific segments (A1, A2, B1, B2, B6, C10, D23, E2, and E4) and two DsIV-specific segments (4 and 5) were non-homologous. Phylogenetic analysis of commonly observed proteins (Rep, Cys, PRRP, Vankyrin, and Vinnexin) in DfIV and DsIV confirmed a high degree of similarity (Supplementary Figure 3). The diversity of rep genes (41 members) was higher than that of cys, vankyrin, PRRP, and vinnexin genes (10, 9, 7, and 6 members, respectively). Therefore, we developed an unrooted phylogenetic tree of rep genes (Supplementary Figure 4A). Maximum likelihood analysis was conducted based on the deduced amino acid sequences of DfIV and DsIV rep proteins (>150 aa). In addition, reps were found in all ichnoviruses. The 40 protein sequences used for phylogenetic analyses were well cladded and clustered. Four commonly found ichnovirus genes, including cys, PRRP, vankyrin, and vinnexin, were downloaded from NCBI to construct phylogenetic trees (Supplementary Figures 4B–E) and clarify the evolutionary origins of ichnovirus and bracovirus lineages. The most abundant (41 members) rep gene family was encoded in 20 different segments. However, three members (7, 25, and 27) existed as duplications in different segments, including rep 7 in C5 and D18, rep 25 in F2 and D20, and rep 27 in F2 and D20 segments. Furthermore, one segment (A3) contained two rep genes (rep1-1 and rep1-2; see accession no MZ129222.1, segment A3). Two copies of DfIV vinnexins were observed in the DfIV genome, vinnexin 3 (segments C12 and C13), vinnexin 4 (segments, D1 and D6), and vinnexin 5 (segments 12 and 13; Figure 3). No duplicated genes were observed in DsIV.
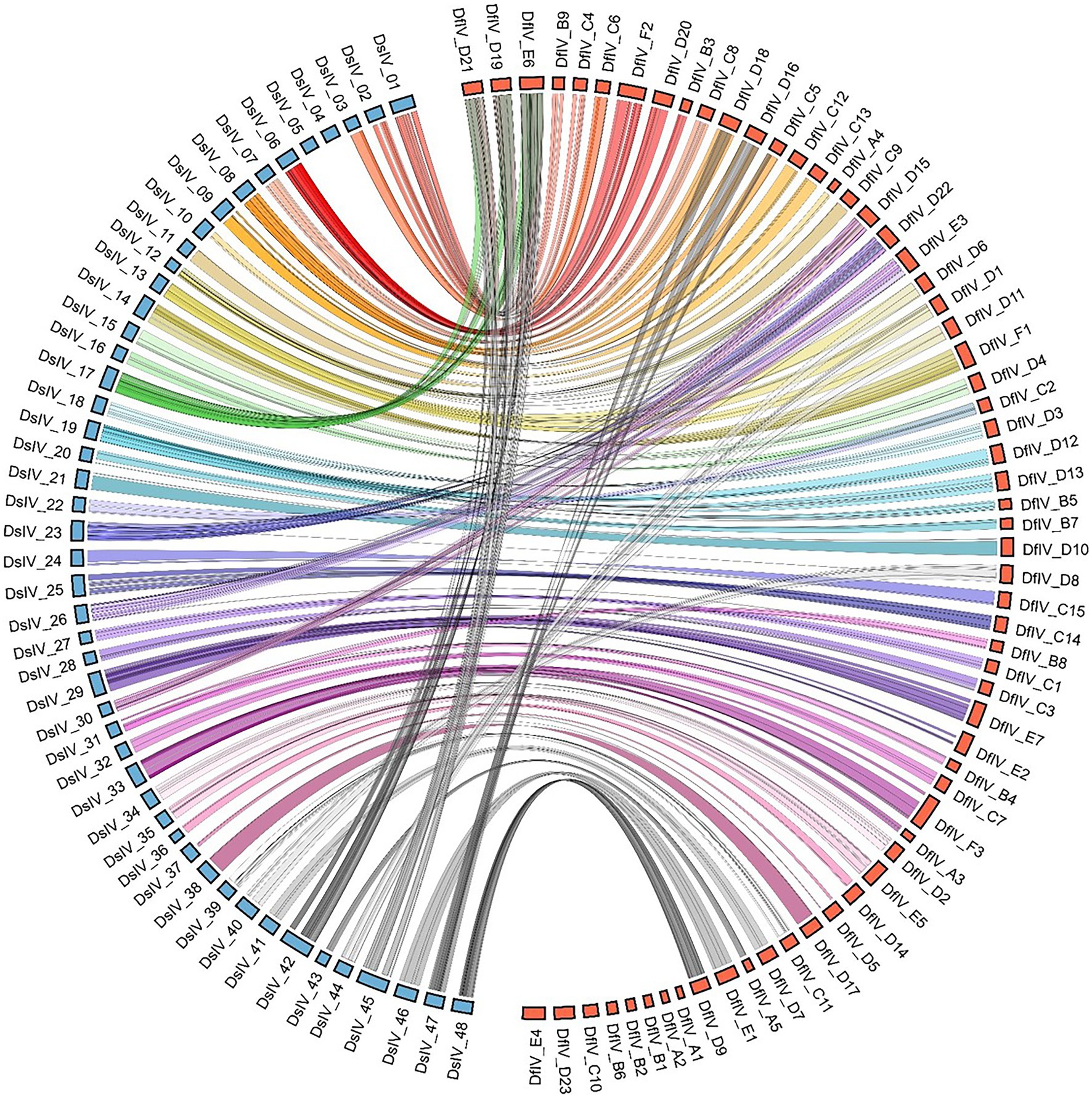
Figure 3. Comparison of sequence similarity between 62 genomic segments of DfIV (red) and 48 genomic segments of DsIV (blue) based on BLAST results. Eight specific segments of DfIV and two of DsIV were identified. The DsIV genome was re-sequenced using the AVRDC strain as a template (Taiwan strain, Kim et al., 2015). Comparative analyses revealed a high degree of synteny between loci coding for DfIV and DsIV proviral segments, as well as flanking DNA.
Gene expression patterns were examined in different viral hosts and life stages (hymenopteran host, D. fenestrale; lepidopteran host, P. xylostella). A total of 62 genome segments were expressed in D. fenestrale ovaries, and segments C15 and D14 were particularly highly expressed (Figure 4). Five segments (E4, D5, C15, C3, and A4) were differentially expressed in P. xylostella of different developmental stages (third and fifth instar). The most highly expressed gene in the third instar P. xylostella was hypothetical gene Hp 30 in segment D5 (P. xylostella _Df-1, 1 day after parasitization by D. fenestrale) and that in fifth instar larvae was Rep 33 in segment E4 (P. xylostella Df-5, 5 days after parasitization by D. fenestrale). The highest expressed gene in D. fenestrale was N1 in segment C15, followed by Rep15 in segment D14.
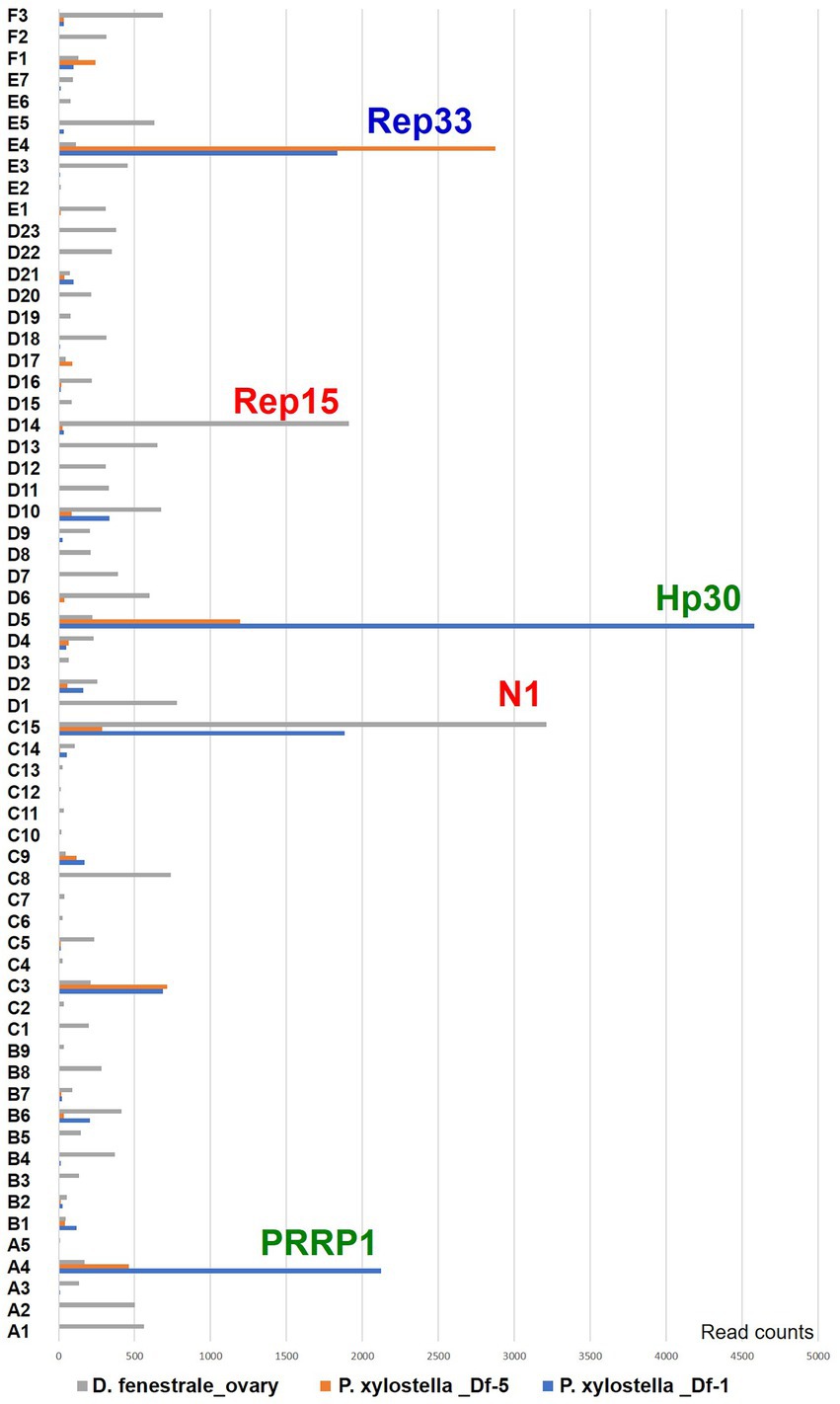
Figure 4. Gene expression pattern in two different hosts (hymenopteran host, Diadegma fenestrale, and lepidopteran host, Plutella xylostella) and developmental stages of P. xylostella based on RNAseq. The segments are expressed in the ovaries of hymenopteran host, D. fenestrale; only two segments (C15 and D14) were highly expressed in D. fenestrale ovaries. Five segments (E4, D5, C15, C3, and A4) were differentially expressed according to the developmental stage of P. xylostella (1 and 5 days after parasitization by D. fenestrale; third and fourth larval instar).
A P. xylostella genome was re-sequenced 1 day after parasitization by D. fenestrale, and the resequencing results of the host genome integration motif are shown in Supplementary Table 3. PDV (DfIV) cannot replicate within its lepidopteran host, P. xylostella, but genes are integrated and expressed throughout the parasitism. Genome integration is also dependent on host immunity and physiology under high selection pressure because DNA integration requires the parasitoid’s success. Only virus encapsidation and expression sequences in the genome of the lepidopteran host (P. xylostella) are required to be present on the DNA circles (Drezen et al., 2003). The homologous recombination was evidenced between left and right direct repeat junctions (DRJs) to combine one DRJ sequence. The presence of multiple repeats that differ in sequence and position suggests that homologous recombination may result in a mix of overlapping and/or nested segments (Legeai et al., 2020). However, IV replication and integration mechanisms in the host genome are largely unknown. In this study, host genome integration motifs were compared between the DfIV and P. xylostella genomes, and two complementary conserved motifs were surveyed at their breaking sites. The general mechanism of motif integration was as follows: (1) The homologous region between host and viral genomes ranged from 36 to 46 bp, and the integration motifs existed on both sides of DfIV and P. xylostella; (2) each homologous site on the host and viral genome was integrated into the host genome; (3) using the process of homologous recombination (Legeai et al., 2020), the homologous region and DfIV genome with integrated motifs were combined into the host genome (Supplementary Figure 5). Across three biological replicates of P. xylostella genome (DBM-Df-1, DBM-Df-3, and DBM-Df-7), resequencing results are shown in Supplementary Table 3, and only three genomic segments (DfIV-D22, -E3 and -F2) were predicted to be integrated. Two genomic segments (D22 and E3) were integrated into one P. xylostella chromosome, but F2 integration motifs were found at five P. xylostella chromosome sites (F2–1 to F2–5; Supplementary Table 4). Of the investigated integration motifs in the host genome, the D20 segment was not matched to the P. xylostella genome within the threshold parameters, so it was not counted (data not shown). DfIV genomic segment replication was not detected in the lepidopteran host (data not shown).
In this study, one new virus was found in D. fenestrale and Plutella xylostella through transcriptome analysis; this was denoted as “Saesbyeol virus.” The aggregate genome sequence length of Saesbyeol virus was 14, 447 bp, and the genome contained three independent segments, large, L (MH698456, 6,987 bp), small, S (MH698457, 1,912 bp), and medium, M (MH823676, 5,548 bp), similar to Bunyaviridae family, which is divided into three segments called L, M, and S based on their segment size (Amroun et al., 2017). The sequence MH698456 contains a conserved protein domain, based on NCBI’s conserved domain database (CDD) search, and it encodes a bunyavirus RdRP (RNA-dependent RNA polymerase). MH823676 BLAST matches are viral sequences only, of which most are glycoproteins of bunyaviruses. In addition, MH698457 matches virus sequences only, all bunyavirus nucleocapsid proteins. It seems very likely that the three segments are derived from the same virus as they match sequences of bunyaviruses which typically have a genome consisting of several segments. The genome assembly (ASM412811v1) was deposited to the NCBI database.4 The function of Saesbyeol virus and its interactions with DfIV and its hosts (D. fenestrale and P. xylostella) should be investigated in future studies.
PDVs are obligatory symbionts of thousands of endoparasitoid wasp species and are essential for their successful parasitism by modulating host immunity (Ignesti et al., 2018). Here, TEM analysis showed that the viral replication was likely to occur in the ovarian calyx, with replicated viral particles released into the oviduct lumen. In PDV-harboring wasps, the ovary is well developed in the calyx region where the PDVs are replicated (Volkoff et al., 1995). DfIV morphogenesis was observed within the nuclei of calyx epithelial cells in female reproductive organs (Figure 1), which is likely to be present in other closely related PDV (Lorenzi et al., 2022). Furthermore, ichnoviruses and bracoviruses share a common life cycle but are morphologically different. Unique morphological characteristics include a layer of cell membrane that discriminates ichnoviruses from bracoviruses (Lapointe et al., 2007). Bracoviruses have cylindrical nucleocapsids surrounded by a single envelope, while ichnoviruses have lenticular nucleocapsids with two membranous envelopes (Krell, 1987). The morphology was detected in this study, though the inner membrane of DfIV was not distinguished.
The DfIV genome sequence was too complicated to be fully sequenced because of segmentation and internal sequence homologies between segments (Webb, 1998; Federici and Bigot, 2003). Unequal replication of different segments is a characteristic of PDVs (Beck et al., 2007), though little is understood about the exact mechanism explaining this unique kind of viral replication. The resulting 65 genomic segments were re-sequenced using the Illumina platform with a high depth of genome sequence coverage, narrowing the results to 62 genomic segments. However, in ichnoviruses, an unequal number of segments can be partially explained by a segment nesting process (Cui and Webb, 1997; Kadash, 2000). Ichnovirus genome segments have been observed to range in size from 2 to 28 kb (Webb and Cui, 1998), with estimated genome sizes from 75 to 250 kb. In accordance with this, the DfIV genome segments analyzed here ranged from 1.5 to 8.2 kb with a total genome size of approximately 247 kb.
For most parasitoids, successful parasitism depends on ecological, biological, and anthropological factors, and molecular mechanisms are not well understood (Vinson and Iwantsch, 1980). However, it has been suggested that some parasitoids can extend their host survival as a consequence of coevolution between parasitoids and their hosts. For ichnovirus-harboring parasitoids, successful parasitism is dependent on the expression of certain ichnovirus genes exceeding a threshold value (Doremus et al., 2014). In one study, the differential expression of two cysteine proteins, CsIV VHv1.1 and VHv1.4, influenced lepidopteran hosts to ensure successful parasitism by C. sonorensis (Cui et al., 2000; Kim, 2005). Therefore, we focused on the expression patterns of common (cysteine motif protein, cys; viral ankyrin, vankyrin; viral innexin, vinnexin; and unassigned genes) and novel (PRRP, N gene, and neuromodulin N) PDV gene families that may contribute to host immune response. The diversity of gene expression was dependent on the nature of the host species (permissive hosts, where the host cellular immune response cannot eliminate parasitoid eggs, and non-permissive hosts, where the host cellular immune response can eliminate parasitoid eggs).
In our previous study (Kim et al., 2015), D. fenestrale was observed to prefer P. operculella to P. xylostella hosts, according to ovipositional and survival rate analyses. DfIV gene expression in P. operculella was higher than that in P. xylostella after parasitization by D. fenestrale, suggesting that higher DfIV gene expression promoted the survival of parasitoids. However, the exact mechanisms of host preferences have not yet been explored. Moreover, qRT-PCR analysis results of gene expression were observed after parasitization by D. fenestrale, but many DfIV genes were not expressed, including PRRP, neuromodulin N1, hypothetical genes (Hp), and several rep genes (identified rep genes in the present research, n = 41; rep genes identified in previous research, n = 28; Kim et al., 2015). Interestingly, Rep33 gene was more highly expressed in segment E4 of DfIV at 5 day post-parasitization than at 1 day post-parasitization of P. xylostella (Figure 4). This result indicated that Illumina-based analysis yielded better resolution than the Roche 454 with the qRT-PCR strategy used previously (Kim et al., 2015). In an earlier study, the segments of PDV that encode virulence genes were demonstrated to be transcribed in the parasitized hosts but not in the wasps themselves (Strand and Burke, 2013). This raises a question as to why a large amount of N gene transcript, which is known to be involved in host immune suppression, was expressed in the ovary of the parasitoid wasp. One possible hypothesis is that expression of a single gene may vary depending on the interaction with the host, but further research is still needed.
Five gene families (rep, cys, PRRP, vankyrins, and vinnexins) were identified in both DsIV and DfIV genomes, as previously reported in many studies (Webb et al., 2006; Tanaka et al., 2007; Doremus et al., 2014; Wang et al., 2021). Among these families, rep was the most abundant in DfIV, with 41 predicted members. Reps have been previously observed in several ichnoviruses, including HfIV, CsIV, and TrIV (Galibert et al., 2006; Rasoolizadeh et al., 2009). Although the function of rep genes has not yet been fully elucidated, their conservation and abundance in viral genomes indicate that they play an important role in viral maintenance (Galibert et al., 2006). The second most common family, cys-motif, was represented by 10 members in DfIV. The cys-motif family is found in other ichnoviruses (eight members in DsIV) but not in bracoviruses (Zhang and Turnbull, 2021). Two cys-motif genes in CsIV (VHv1.1 and VHv1.4) affect the hemocytes so that the cellular immune system would be suppressed, to the benefit of the hymenopteran insect (Kim, 2005; Ye et al., 2018). Therefore, cys-motif genes in DfIV may have similar molecular functions (Kim, 2005). Genes of the third most represented family, PRRP, contain approximately 63–77% polar amino acids (Tanaka et al., 2005), but their function is unknown (Tanaka et al., 2007). The fourth most abundant family, vankyrins, included nine members in DfIV and six in DsIV. Unlike other ichnovirus genes, vankyrins are also encoded in bracoviruses (Webb, 1998). The nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is an important transcription factor that is expressed in almost all cells. The CsIV vankyrin proteins are thought to be involved in the suppression of NF-κB activity during immune responses and/or parasitized host development. The fifth gene family, vinnexins, exhibits homology with the insect gap junction innexin genes and was predicted in DfIV with six members and in DsIV with seven. Vinnexin is a highly conserved protein in ichnoviruses and may interfere with host cell–cell communication (Phelan et al., 1998), alteration of cytoskeletal networks (Hasegawa et al., 2020), host cell encapsulation, molting, and larval maturation (Zhang and Turnbull, 2021). Here, the phylogenetic relationships of these five DfIV and DsIV gene families (rep, cys, PRRP, vankyrin, and vinnexin) were compared and found to be closely related. Phylogenetic analyses can reveal important information related to the evolutionary origin of viruses (Federici and Bigot, 2003). In addition, other canonical ichnovirus genes, such as neuromodulin N, and 45 unassigned genes were predicted in DfIV. Neuromodulin is also referred to as growth-associated protein 43 (Gap-43) and is considered a “growth” or “plasticity” protein in humans because it is expressed at high levels in neuronal growth cones during development (Apel et al., 1990; Benskin et al., 2009) and interacts with many proteins. The neuromodulin gene is known as a gap junction gene and may be related to immune suppression (Glatz et al., 2004). Until now, no functional information has been elucidated regarding viral neuromodulin, but we observed moderate sequence similarity with both viral innexin and vinnexin. Moreover, neuromodulin genes identified in this study showed similarity to HdIV_U2 and HfIV_b7.1, genes from generalist ichnoviruses in Hyposoter didymator and H. fugitivus (Doremus et al., 2014). Some indels in protein-coding regions (i.e., neuromodulin genes, unknown function based on BLAST) of DfIV may be beneficial for understanding evolutionary processes (Sung et al., 2016).
Host integration motifs of DsIV circles have been observed in the parasitized P. xylostella genome (Wang et al., 2021), and similar integration motifs of DfIV were observed here. The precise mechanism of genome integration remains unknown, and the exact functions of the integration motif, integration sites, and homologous sequences in the DfIV genome are still poorly understood. Previous research demonstrated that PDVs can exist in host cell episomes with full capability to express virulence genes or immunosuppressive agents (Beck et al., 2011; Burke and Strand, 2012), promoting virus survival. The integration process into the host genome appears to be site-specific and is mediated by the wasp genome; it is also dependent on the integration machinery of specific hosts (Burke and Strand, 2012). The ability of segments to integrate into host cells explains how PDV gene products are expressed in hosts for long periods (Beck et al., 2011; Burke and Strand, 2012). In a past study, Wang et al. (2021) showed that DsIV genome segments or circles existed in P. xylostella host hemocytes and were integrated into the host genome either conservatively or randomly. However, they were unable to determine the precise location of the repeat motif and homology sequences, nor could they identify the position of the virus and host genomes. Here, we compared host integration motifs and homology sequences between DfIV and P. xylostella genomes and proposed a mechanism for their integration based on previous reports on HdIV and CsIV (Legeai et al., 2020). Further research should be conducted to enhance the understanding of integration motifs and their patterns in DfIV and its host. According to our present findings, some DfIV genome segments can integrate into the P. xylostella genome, as suggested in previous research (Kim et al., 1996; Volkoff et al., 2002). Because many lepidopteran hosts can be parasitized by generalists, DfIV is thought to have a high probability of integration in various species (Doucet et al., 2007) while accelerating the evolution of the viral genome (Kim et al., 1996; Volkoff et al., 2002). Furthermore, we observed microhomologies (≥15 read count, see Supplementary Table 4) between the P. xylostella host genome and the DfIV circles at integration breakpoints, suggesting a potential mechanism similar to that of human papillomavirus integration (Hu et al., 2015). Further research is needed to determine how host genome segments can become complete circles.
This study did have some limitations. Both DfIV and DsIV have been maintained under lab conditions for over a decade with their hymenopteran hosts. It is, therefore, possible that these PDVs have begun to develop unique evolutionary lineages. Our findings would be reinforced by employing purer or more recent lines of DfIV and DsIV. Alternatively, we can compare our findings against results from Chinese and Taiwanese DsIV strains to understand strain-specific variations. Further investigations should focus on long-term evolutionary consequences of coevolution (natural selection-driven reciprocal evolutionary changes) between a virus and its hosts, analysis of known functional genes, putative functional analysis of unknown genes, and gene expression analysis in viral hosts. Our present findings provide insight for further investigations regarding viral evolution and virus–host coevolution.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JK and YK: conceptualization and experimental design. M-MR and JK: software and writing the original draft. JK, M-MR, A-YK, SR, MK, and YK: writing, reviewing, and editing. JK, A-YK, SR, MK, and YK: formal analysis and validation. JK, SR, MK, and YK: resources and editing. JK: supervision and project administration. All authors contributed to the article and approved the submitted version.
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2021R1A6A1A03044242), Republic of Korea.
We are grateful to Sangchun Lee in PHYZEN and Phyzen Genomics Institute staff for technical support in NGS and bioinformatics and to Sunghee Lee, Eunjoo Hong, and Hongju Moon for supporting lepidopteran hosts and parasitoid collection and rearing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1035669/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/nuccore/?term=txid993492[Organism:noexp]
2. ^https://www.ncbi.nlm.nih.gov/orffinder/
3. ^http://broadinstitute.github.io/picard/
4. ^www.ncbi.nlm.nih.gov/search/all/?term=Saesbyeol%20virus%20, accessed date 02 August 2022.
Amroun, A., Priet, S., de Lamballerie, X., and Quérat, G. (2017). Bunyaviridae RdRps: structure, motifs, and RNA synthesis machinery. Crit. Rev. Microbiol. 43, 753–778. doi: 10.1080/1040841X.2017.1307805
Apel, E. D., Byford, M. F., Au, D., Walsh, K. A., and Storm, D. R. (1990). Identification of the protein kinase C phosphorylation site in neuromodulin. Biochemistry 29, 2330–2335. doi: 10.1021/bi00461a017
Azidah, A. A., Fitton, M. G., and Quicke, D. L. J. (2000). Identification of the Diadegma species (hymenoptera: Ichneumonidae, Campopleginae) attacking the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Bull. Entomol. Res. 90, 375–389. doi: 10.1017/S0007485300000511
Barandoc, K. P., and Kim, Y. (2009). Identification of three host translation inhibitory factors encoded in Cotesia glomerata bracovirus. Comp. Biochem. Physiol. Part D Genomics Proteomics 4, 218–226. doi: 10.1016/j.cbd.2009.03.003
Barat-Houari, M., Hilliou, F., Jousset, F. X., Sofer, L., Deleury, E., Rocher, J., et al. (2006). Gene expression profiling of Spodoptera frugiperda hemocytes and fat body using cDNA microarray reveals polydnavirus-associated variations in lepidopteran host genes transcript levels. BMC Genomics 7, 1–20. doi: 10.1186/1471-2164-7-160
Beck, M. H., Inman, R. B., and Strand, M. R. (2007). Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359, 179–189. doi: 10.1016/j.virol.2006.09.002
Beck, M. H., Zhang, S., Bitra, K., Burke, G. R., and Strand, M. R. (2011). The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens. J. Virol. 85, 11685–11696. doi: 10.1128/JVI.05726-11
Benskin, C. M. H., Wilson, K., Jones, K., and Hartley, I. R. (2009). Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biol. Rev. 84, 349–373. doi: 10.1111/j.1469-185X.2008.00076.x
Besemer, J., and Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boratyn, G. M., Schäffer, A. A., Agarwala, R., Altschul, S. F., Lipman, D. J., and Madden, T. L. (2012). Domain enhanced lookup time accelerated BLAST. Biol. Direct 7, 12–14. doi: 10.1186/1745-6150-7-12
Burke, G. R., and Strand, M. R. (2012). Polydnaviruses of parasitic wasps: domestication of viruses to act as gene delivery vectors. Insects 3, 91–119. doi: 10.3390/insects3010091
Chen, Y., Gao, F., Ye, X., Wei, S., Shi, M., Zheng, H., et al. (2011). Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology 414, 42–50. doi: 10.1016/j.virol.2011.03.009
Chevignon, G., Thézé, J., Cambier, S., Poulain, J., Da Silva, C., Bézier, A., et al. (2014). Functional annotation of Cotesia congregata bracovirus: identification of viral genes expressed in parasitized host immune tissues. J. Virol. 88, 8795–8812. doi: 10.1128/JVI.00209-14
Choi, J.-K., Kim, J. I., Kwon, M., and Lee, J.-W. (2013). Description of the Diadegma fenestrale (hymenoptera: Ichneumonidae: Campopleginae) attacking the potato tuber moth, Phthorimaea operculella (Lep.: Gelechiidae) new to Korea. Anim. Syst. Evol. Divers. 29, 70–73. doi: 10.5635/ASED.2013.29.1.70
Choi, J. Y., Kwon, S.-J., Roh, J. Y., Yang, T. J., Yoon, S. H., Kim, H., et al. (2009). Sequence and gene organization of 24 circles from the Cotesia plutellae bracovirus genome. Arch. Virol. 154, 1313–1327. doi: 10.1007/s00705-009-0441-6
Choi, J.-K., and Lee, J.-W. (2018). Five new records of the subfamily Campopleginae (hymenoptera: Ichneumonidae) from South Korea. Anim. Syst. Evol. Diversity 34, 27–36. doi: 10.5635/ASED.2018.34.1.047
Cui, L., Soldevila, A. I., and Webb, B. A. (2000). Relationships between polydnavirus gene expression and host range of the parasitoid wasp Campoletis sonorensis. J. Insect Physiol. 46, 1397–1407. doi: 10.1016/S0022-1910(00)00059-7
Cui, L., and Webb, B. A. (1997). Homologous sequences in the Campoletis sonorensis polydnavirus genome are implicated in replication and nesting of the W segment family. J. Virol. 71, 8504–8513. doi: 10.1128/jvi.71.11.8504-8513.1997
Desjardins, C. A., Gundersen-Rindal, D. E., Hostetler, J. B., Tallon, L. J., Fadrosh, D. W., Fuester, R. W., et al. (2008). Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9, R183–R117. doi: 10.1186/gb-2008-9-12-r183
Desjardins, C. A., Gundersen-Rindal, D. E., Hostetler, J. B., Tallon, L. J., Fuester, R. W., Schatz, M. C., et al. (2007). Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol. 7, 61–17. doi: 10.1186/1471-2180-7-61
Dicke, M., Cusumano, A., and Poelman, E. H. (2020). Microbial symbionts of parasitoids. Annu. Rev. Entomol. 65, 171–190. doi: 10.1146/annurev-ento-011019-024939
Djoumad, A., Stoltz, D., Beliveau, C., Boyle, B., Kuhn, L., and Cusson, M. (2013). Ultrastructural and genomic characterization of a second banchine polydnavirus confirms the existence of shared features within this ichnovirus lineage. J. Gen. Virol. 94, 1888–1895. doi: 10.1099/vir.0.052506-0
Doremus, T., Cousserans, F., Gyapay, G., Jouan, V., Milano, P., Wajnberg, E., et al. (2014). Extensive transcription analysis of the Hyposoter didymator ichnovirus genome in permissive and non-permissive lepidopteran host species. PLoS One 9:e104072. doi: 10.1371/journal.pone.0104072
Doucet, D., Levasseur, A., Béliveau, C., Lapointe, R., Stoltz, D., and Cusson, M. (2007). In vitro integration of an ichnovirus genome segment into the genomic DNA of lepidopteran cells. J. Gen. Virol. 88, 105–113. doi: 10.1099/vir.0.82314-0
Drezen, J.-M., Provost, B., Espagne, E., Cattolico, L., Dupuy, C., Poirie, M., et al. (2003). Polydnavirus genome: integrated vs. free virus. J. Insect Physiol. 49, 407–417. doi: 10.1016/S0022-1910(03)00058-1
Dupuy, C., Huguet, E., and Drezen, J.-M. (2006). Unfolding the evolutionary story of polydnaviruses. Virus Res. 117, 81–89. doi: 10.1016/j.virusres.2006.01.001
Edson, K. M., Vinson, S. B., Stoltz, D. B., and Summers, M. D. (1981). Virus in a parasitoid wasp: suppression of the cellular immune response in the parasitoid’s host. Science 211, 582–583. doi: 10.1126/science.7455695
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Etebari, K., Palfreyman, R. W., Schlipalius, D., Nielsen, L. K., Glatz, R. V., and Asgari, S. (2011). Deep sequencing-based transcriptome analysis of Plutella xylostella larvae parasitized by Diadegma semiclausum. BMC Genomics 12, 1–18. doi: 10.1186/1471-2164-12-446
Federici, B. A., and Bigot, Y. (2003). Origin and evolution of polydnaviruses by symbiogenesis of insect DNA viruses in endoparasitic wasps. J. Insect Physiol. 49, 419–432. doi: 10.1016/S0022-1910(03)00059-3
Fitton, M. (1992). Hymenopterous parasitoids associated with diamondback moth: the taxonomic dilemma. in Diamondback Moth and Other Crucifer Pests: Proceedings the Second International Workshop (Asian Vegetable Research and Development Center), 225–232.
Galibert, L., Devauchelle, G., Cousserans, F., Rocher, J., Cérutti, P., Barat-Houari, M., et al. (2006). Members of the Hyposoter didymator Ichnovirus repeat element gene family are differentially expressed in Spodoptera frugiperda. Virol. J. 3, 1–11. doi: 10.1186/1743-422X-3-48
Glatz, R. V., Asgari, S., and Schmidt, O. (2004). Evolution of polydnaviruses as insect immune suppressors. Trends Microbiol. 12, 545–554. doi: 10.1016/j.tim.2004.10.004
Gómez, I. C., Sääksjärvi, I. E., Mayhew, P. J., Pollet, M., Rey del Castillo, C., Nieves-Aldrey, J., et al. (2018). Variation in the species richness of parasitoid wasps (Ichneumonidae: Pimplinae and Rhyssinae) across sites on different continents. Insect Conserv. Divers. 11, 305–316. doi: 10.1111/icad.12281
Hasegawa, D. K., Zhang, P., and Turnbull, M. W. (2020). Intracellular dynamics of polydnavirus innexin homologues. Insect Mol. Biol. 29, 477–489. doi: 10.1111/imb.12657
Herniou, E. A., Huguet, E., Thézé, J., Bézier, A., Periquet, G., and Drezen, J.-M. (2013). When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. B Biol. Sci. 368:20130051. doi: 10.1098/rstb.2013.0051
Hu, Z., Zhu, D., Wang, W., Li, W., Jia, W., Zeng, X., et al. (2015). Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 47, 158–163. doi: 10.1038/ng.3178
Ignesti, M., Ferrara, R., Romani, P., Valzania, L., Serafini, G., Pennacchio, F., et al. (2018). A polydnavirus-encoded ANK protein has a negative impact on steroidogenesis and development. Insect Biochem. Mol. Biol. 95, 26–32. doi: 10.1016/j.ibmb.2018.03.003
Kadash, K. (2000). Mutualism between parasitoids and Polydnaviruses [dissertation]. Madison, WI: The University of Wisconsin-Madison.
Kashkouli, M., Mehrabadi, M., and Fathipour, Y. (2021). “The symbionts” in Microbial approaches for insect Pest management. ed. Omkar (Singapore: Springer), 217–269.
Kim, Y. (2005). Identification of host translation inhibitory factor of Campoletis sonorensis ichnovirus on the tobacco budworm, Heliothis virescens. Arch. Insect Biochem. Physiol. Publ. Collab. Entomol. Soc. Am. 59, 230–244. doi: 10.1002/arch.20074
Kim, J. I., Kwon, M., Lee, S. H., and Kim, Y. (2015). Parasitism and survival rate of Diadegma fenestrale (hymenoptera: Ichneumonidae) and DfIV gene expression patterns in two lepidopteran hosts. Biochem. Biophys. Res. Commun. 459, 579–584. doi: 10.1016/j.bbrc.2015.02.150
Kim, M.-K., Sisson, G., and Stoltz, D. (1996). Ichnovirus infection of an established gypsy moth cell line. J. Gen. Virol. 77, 2321–2328. doi: 10.1099/0022-1317-77-9-2321
Klopfstein, S., Riedel, M., and Schwarz, M. (2019). Checklist of ichneumonid parasitoid wasps in Switzerland (hymenoptera, Ichneumonidae): 470 species new for the country and an appraisal of the alpine diversity. Alp. Entomol. 3, 51–81. doi: 10.3897/alpento.3.31613
Krell, P. J. (1987). Replication of long virus-like particles in the reproductive tract of the ichneumonid wasp Diadegma terebrans. J. Gen. Virol. 68, 1477–1483. doi: 10.1099/0022-1317-68-5-1477
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kwon, M., Park, K.-R., and Kwon, H.-J. (2003). Developmental characteristics of Diadegma semiclausum Hellen (hymenoptera: Ichneumonidae), a larval parasitoid of Plutella xylostella L.(Lepidoptera: Yponomeutidae). J. Asia Pac. Entomol. 6, 105–110. doi: 10.1016/S1226-8615(08)60175-2
Lapointe, R., Tanaka, K., Barney, W. E., Whitfield, J. B., Banks, J. C., Béliveau, C., et al. (2007). Genomic and morphological features of a banchine polydnavirus: comparison with bracoviruses and ichnoviruses. J. Virol. 81, 6491–6501. doi: 10.1128/JVI.02702-06
Legeai, F., Santos, B. F., Robin, S., Bretaudeau, A., Dikow, R. B., Lemaitre, C., et al. (2020). Genomic architecture of endogenous ichnoviruses reveals distinct evolutionary pathways leading to virus domestication in parasitic wasps. BMC Biol. 18, 1–23. doi: 10.1186/s12915-020-00822-3
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lorenzi, A., Strand, M. R., Burke, G. R., and Volkoff, A.-N. (2022). Identifying bracovirus and ichnovirus genes involved in virion morphogenesis. Curr. Opin. insect Sci. 49, 63–70. doi: 10.1016/j.cois.2021.11.006
Luckhart, S., and Webb, B. A. (1996). Interaction of a wasp ovarian protein and polydnavirus in host immune suppression. Dev. Comp. Immunol. 20, 1–21. doi: 10.1016/0145-305X(95)00040-Z
Moreau, S., Huguet, E., and Drezen, J.-M. (2009). “Polydnaviruses as tools to deliver wasp virulence factors to impair lepidopteran host immunity” in Insect Infection and Immunity: Evolution, Ecology, and Mechanisms. eds. J. Rolff and S. Reynolds (New York, NY: Oxford Oxford University Press Inc), 137–158.
Nam, H., Kwon, M., Ramasamy, S., and Kim, J. (2022). Identification of two diamondback moth parasitoids, Diadegma fenestrale and Diadegma semiclausum, using LAMP for application in biological control. Horticulturae 8:366. doi: 10.3390/horticulturae8050366
Norton, W. N., Vinson, S. B., and Stoltz, D. B. (1975). Nuclear secretory particles associated with the calyx cells of the ichneumonid parasitoid Campoletis sonorensis (Cameron). Cell Tissue Res. 162, 195–208. doi: 10.1007/BF00209207
Phelan, P., Bacon, J. P., Davies, J. A., Stebbings, L. A., Todman, M. G., Avery, L., et al. (1998). Innexins: a family of invertebrate gap-junction proteins. Trends Genet. 14, 348–349. doi: 10.1016/S0168-9525(98)01547-9
Rasoolizadeh, A., Beliveau, C., Stewart, D., Cloutier, C., and Cusson, M. (2009). Tranosema rostrale ichnovirus repeat element genes display distinct transcriptional patterns in caterpillar and wasp hosts. J. Gen. Virol. 90, 1505–1514. doi: 10.1099/vir.0.008664-0
Roche Applied, Science. (2008). Genome sequencer data analysis software manual. Software Version 2.0.00. Berlin, Germany: Roche Diagnostics GmbH.
Sharkey, M. J., Janzen, D. H., Hallwachs, W., Chapman, E. G., Smith, M. A., Dapkey, T., et al. (2013). Minimalist revision and description of 403 new species in 11 subfamilies of costa Rican braconid parasitoid wasps, including host records for 219 species. Zookeys 1013, 1–665. doi: 10.3897/zookeys.1013.55600
Shelby, K. S., Cui, L., and Webb, B. A. (1998). Polydnavirus-mediated inhibition of lysozyme gene expression and the antibacterial response. Insect Mol. Biol. 7, 265–272. doi: 10.1111/j.1365-2583.1998.00071.x
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, S10–S12. doi: 10.1186/gb-2006-7-s1-s10
Sonnhammer, E. L. L., Eddy, S. R., and Durbin, R. (1997). Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins Struct. Funct. Bioinforma. 28, 405–420. doi: 10.1002/(SICI)1097-0134(199707)28:3<405::AID-PROT10>3.0.CO;2-L
Stoltz, D. B., and Vinson, S. B. (1979). Viruses and parasitism in insects. Adv. Virus Res. 24, 125–171. doi: 10.1016/S0065-3527(08)60393-0
Stothard, P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102–1104. doi: 10.2144/00286ir01
Strand, M. R., and Burke, G. R. (2013). Polydnavirus-wasp associations: evolution, genome organization, and function. Curr. Opin. Virol. 3, 587–594. doi: 10.1016/j.coviro.2013.06.004
Sung, W., Ackerman, M. S., Dillon, M. M., Platt, T. G., Fuqua, C., Cooper, V. S., et al. (2016). Evolution of the insertion-deletion mutation rate across the tree of life. G3 6, 2583–2591. doi: 10.1534/g3.116.030890
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tanaka, K., Barney, W. E., Makkay, A. M., Stoltz, D. B., and Webb, B. A. (2005). Annotation of the Hyposoter fugitivus ichnovirus (HfIV) genome. in The 2005 ESA annual meeting and exhibition.
Tanaka, K., Lapointe, R., Barney, W. E., Makkay, A. M., Stoltz, D., Cusson, M., et al. (2007). Shared and species-specific features among ichnovirus genomes. Virology 363, 26–35. doi: 10.1016/j.virol.2006.11.034
Thompson, W. R. (1951). A catalogue of the parasites and predators of insect pests. Section 2. Host parasite catalogue. Parts 1-5. A cat. Parasites predators insect pests. Ottawa: Commonwealth Inst. Bio.Control.
Vinson, S. B., and Iwantsch, G. F. (1980). Host suitability for insect parasitoids. Annu. Rev. Entomol. 25, 397–419. doi: 10.1146/annurev.en.25.010180.002145
Volkoff, A.-N., Béliveau, C., Rocher, J., Hilgarth, R., Levasseur, A., Duonor-Cerutti, M., et al. (2002). Evidence for a conserved polydnavirus gene family: ichnovirus homologs of the CsIV repeat element genes. Virology 300, 316–331. doi: 10.1006/viro.2002.1535
Volkoff, A., Ravallec, M., Bossy, J., Cerutti, P., Rocher, J., Cerutti, M., et al. (1995). The replication of Hyposoter didymator polydnavirus: cytopathology of the calyx cells in the parasitoid. Biol. Cell 83, 1–13. doi: 10.1016/0248-4900(96)89926-6
Wang, Z., Zhou, Y., Yang, J., Ye, X., Shi, M., Huang, J., et al. (2021). Genome-wide profiling of Diadegma semiclausum ichnovirus integration in parasitized Plutella xylostella hemocytes identifies host integration motifs and insertion sites. Front. Microbiol. 11:608346. doi: 10.3389/fmicb.2020.608346
Webb, B. A. (1998). “Polydnavirus biology, genome structure, and evolution” in The insect viruses. eds. L. K. Miller and L. Andrew Ball (Berlin: Springer), 105–139.
Webb, B. A., and Cui, L. (1998). Relationships between polydnavirus genomes and viral gene expression. J. Insect Physiol. 44, 785–793. doi: 10.1016/S0022-1910(98)00011-0
Webb, B. A., Strand, M. R., Dickey, S. E., Beck, M. H., Hilgarth, R. S., Barney, W. E., et al. (2006). Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347, 160–174. doi: 10.1016/j.virol.2005.11.010
Xie, Y., Wu, G., Tang, J., Luo, R., Patterson, J., Liu, S., et al. (2014). SOAPdenovo-trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. doi: 10.1093/bioinformatics/btu077
Ye, X., Shi, M., Huang, J., and Chen, X. (2018). Parasitoid polydnaviruses and immune interaction with secondary hosts. Dev. Comp. Immunol. 83, 124–129. doi: 10.1016/j.dci.2018.01.007
Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. doi: 10.1089/10665270050081478
Keywords: polydnavirus, koinobiont, Campopleginae, coevolution, host adaptation, DfIV, Saesbyeol virus
Citation: Kim J, Rahman M-M, Kim A-Y, Ramasamy S, Kwon M and Kim Y (2023) Genome, host genome integration, and gene expression in Diadegma fenestrale ichnovirus from the perspective of coevolutionary hosts. Front. Microbiol. 14:1035669. doi: 10.3389/fmicb.2023.1035669
Received: 03 September 2022; Accepted: 25 January 2023;
Published: 17 February 2023.
Edited by:
Rajarshi Kumar Gaur, Deen Dayal Upadhyay Gorakhpur University, IndiaReviewed by:
Lumi Viljakainen, University of Oulu, FinlandCopyright © 2023 Kim, Rahman, Kim, Ramasamy, Kwon and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juil Kim, ✉ Zm9yd2Vla0BrYW5nd29uLmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.