- 1Laboratory of Veterinary Mycoplasmology, College of Veterinary Medicine, Southwest University, Chongqing, China
- 2Institute of Veterinary Sciences and Pharmaceuticals, Chongqing Academy of Animal Sciences, Chongqing, China
- 3Agricultural Service Center, Sub-District of Rongchang, Chongqing, China
Salmonella is one of the most important foodborne zoonotic pathogens, causing global morbidity and mortality in both humans and animals. Due to the extensive use of antimicrobials in food-producing animals, the antimicrobial resistance of Salmonella has attracted increasing attention globally. There have been many reports concerning the antimicrobial resistance of Salmonella from food-producing animals, meats and the environment. However, few studies on Salmonella from food-producing animals have been reported in Chongqing municipality, China. The aim of the present study was to determine the prevalence, serovar diversity, sequence types, and antimicrobial resistance of Salmonella isolated from livestock and poultry in Chongqing. Meanwhile, we also want to know the presence of β-lactamase genes, plasmid-mediated quinolone resistance (PMQR) genes and quinolone resistance-determining region (QRDR) mutations of Salmonella isolates. A total of 129 Salmonella strains were recovered from 2,500 fecal samples at 41 farms from pigs, goats, beef cattle, rabbits, chickens, and ducks. Fourteen serovars were identified, with S. Agona and S. Derby being the dominant serovars. The 129 isolates had high resistance to doxycycline (87.6%), ampicillin (80.6%), tetracycline (79.8%), trimethoprim (77.5%), florfenicol (76.7%) chloramphenicol (72.9%), and trimethoprim-sulfamethoxazole (71.3%), but were susceptible to cefepime. A total of 114 (88.4%) isolates showed multidrug resistant phenotypes. The prevalence of β-lactamase genes in Salmonella isolates was 89.9% (116/129), and among these isolates, 107 (82.9%) harbored blaTEM, followed by blaOXA (26, 20.2%), blaCTX-M (8, 6.2%), and blaCMY (3, 2.3%). In addition, qnrB, qnrD, qnrS, oqxA, oqxB, and aac(6′)-Ib-cr were detected in 11, 2, 34, 34, 43, and 72 PMQR-producing isolates, respectively. Moreover, QRDR mutations were very common in PMQR-positive Salmonella isolates (97.2%, 70/72) with mutation(s) in parC or combinative mutations in gyrA and parC. More significantly, 32 extended spectrum beta-lactamase (ESBL)-producing isolates were identified, and 62.5% of them were found to harbor one to four PMQR genes. Furthermore, 11 sequence types were identified from the isolates, and most of ESBL-producing isolates were attributed to ST34 (15.6%) and ST40 (62.5%). The coexistence of PMQR genes with β-lactamase genes and the extensive mutations in QRDR present in Salmonella isolates from food-producing animals suggest a potential threat to public health. Reasonable utilization and strict control strategies for antimicrobials in animal husbandry and animal treatment are necessary to reduce the emergence and dissemination of drug-resistant Salmonella isolates.
1. Introduction
Salmonella is considered one of the most important foodborne pathogens causing global morbidity and mortality in both humans and animals (Nichols et al., 2021; Harrison et al., 2022; Li et al., 2022). According to data from the World Health Organization, nontyphoidal Salmonella infection causes 155,000 deaths worldwide, and approximately 93.8 million gastroenteritis cases are closely related to nontyphoidal Salmonella infection per year (Majowicz et al., 2010). According to unpublished data from Chinese Center for Disease Control and Prevention surveillance system, 549 out of 100,000 people carried Salmonella in 2013. As one of the most important carriers, food-producing animals can transfer Salmonella to humans through the food chain (Foley and Lynne, 2008; Marus et al., 2019).
Adding antimicrobials to feed and drinking water has long been an important means for farms to prevent and treat bacterial diseases, including salmonellosis. However, under long-term exposure to antimicrobials, bacteria have developed resistance to antimicrobials and even multidrug resistance (MDR), which has become a serious challenge to public health (Nichols et al., 2022). Extended-spectrum cephalosporins and quinolones are two classes of antimicrobials often used at farms, where resistance to Salmonella is emerging due to the misuse and overuse of antimicrobials. Resistance to cephalosporins is primarily due to the acquisition of β-lactamases that are mainly carried by transferable plasmids and transposons (Ghafourian et al., 2015). The major mechanisms of quinolone resistance have been elucidated to be chromosomal mutations in the quinolone resistance-determining regions (QRDRs) and the presence of plasmid-mediated quinolone resistance (PMQR) genes, which might aid the selection and facilitate the mutation of fluoroquinolone resistance genes (Kuang et al., 2018a). There have been numerous reports of antimicrobial resistance (AMR), a major global public health concern that has the potential to destroy the effectiveness of antimicrobials, and resistance genes about Salmonella around the world (Hetman et al., 2022; Canning et al., 2023; Igbinosa et al., 2023). In China, there are also many reports of AMR in Salmonella from food-producing animals and their products, such as pigs, poultry and cattle, involving many provinces (Tang et al., 2022; Wang et al., 2023). Moreover, many of these Salmonella isolates were resistant to β-lactamases and quinolone, and harbor β-lactam resistance genes and PMQR genes, as well as the chromosomal mutations in the QRDRs. However, there are few reports of AMR and resistance genes of Salmonella from food-producing animals in Chongqing, a province-level municipality.
In this study, we investigated Salmonella strains isolated from food-producing animal feces and analyzed their serovar diversity, sequence types, and AMR. Furthermore, we compared the relationship between phenotypes and genotypes regarding the resistance towards β-lactams and quinolones.
2. Materials and methods
2.1. Isolation and identification of Salmonella
A total of 2,500 fecal samples were collected from 1,600 pigs, 400 goats, 100 beef cattle, 50 rabbits, 300 chickens, and 50 ducks between September 2016 and May 2019 (Supplementary Table S1). Since numbers of rabbits and ducks were far lower than those of pigs and chickens in Chongqing, to avoid repetitive clones, we collected 50 fecal samples each from rabbits and ducks. Approximately 1 g of each sample was pre-enriched in 10 mL of sterile buffered peptone water (BPW) at 37°C for 8 h. Then, 0.1 mL of the suspension was added to 5 mL of tetrathionate broth (TTB) and incubated at 42°C for 18–24 h. After selective enrichment, the suspensions were streaked onto xylose lysine tergitol 4 (XLT-4) plates and incubated at 37°C for 18–48 h. Five suspected colonies were picked from each plate and confirmed by amplifying the invA gene, which is specific for Salmonella. Primers designed by our group were invA-F (5’-GAAATTATCGCCACGTTCGGGCA-3′) and invA-R (5’-TCATCGCACCGTCAAAGGA-3′). Finally, one confirmed clone was selected randomly, even if more than one clone was identified as Salmonella at the same plate. All identified isolates were aliquoted and stored at −80°C in Luria-Bertani (LB) broth containing 50% glycerol (Cody et al., 2008).
2.2. Salmonella serotyping
Salmonella isolates were serotyped using slide agglutination with hyperimmune sera (Tianrun, Ningbo, China), and the results were interpreted according to the Kauffmann-White scheme.
2.3. Multilocus sequence typing
Seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were selected to carry out MLST. Primers (Supplementary Table S2) were synthetized according to the published data (General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China, 2017) and the sequence type (ST) was assigned according to the MLST database.1
2.4. Antimicrobial susceptibility testing
The antimicrobial susceptibility of Salmonella isolates was determined using the disk diffusion method on Mueller-Hinton agar plates and broth dilution method with cation-adjusted Mueller-Hinton broth according to the guidelines of Clinical and Laboratory Standards Institute standards CLSI M100-S32 (CLSI, 2022) and VET08Ed4E (CLSI, 2019). Twenty-seven antimicrobials were tested by the disk diffusion method: ampicillin (AMP), cephalexin (LEX), cefazolin (CFZ), cefoxitin (FOX), cefotaxime (CTX), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), imipenem (IPM), aztreonam (ATM), streptomycin (STR), kanamycin (KAN), gentamicin (GEN), amikacin (AMK), tetracycline (TET), doxycycline (DOX), chloramphenicol (CHL), florfenicol (FFC), nalidixic acid (NAL), norfloxacin (NOR), ciprofloxacin (CIP), enrofloxacin (ENO), ofloxacin (OFX), enoxacin (ENX), gatifloxacin (GAT), trimethoprim-sulfamethoxazole (SXT), and trimethoprim (TMP). However, the broth dilution method tested 26 antimicrobials, except for enrofloxacin, for there was no interpretive criterion of this drug by using broth dilution method. The results were interpreted according to the standards described by CLSI M100-S32 (CLSI, 2022) and VET08Ed4E (CLSI, 2019). Escherichia coli ATCC®* 25922 was used as the quality control.
2.5. Detection of β-lactamase genes, PMQR genes and mutations within the QRDR
To analyze the resistance mechanisms, we detected nine β-lactamase genes, 10 PMQR genes and identified mutations in the QRDRs of DNA gyrase and topoisomerase IV by means of PCR and sequencing. The β-lactamase genes were blaTEM, blaCTX-M, blaSHV (Chen et al., 2004), blaOXA, blaCMY, blaPSE (Qiao et al., 2017), blaPER (Qiao et al., 2017), blaVEB (Dallenne et al., 2010), and blaGES (Dallenne et al., 2010), while the PMQR genes included qnrA (Robicsek et al., 2006), qnrB, qnrC (Cattoir et al., 2007), qnrD, qnrVC, qnrS, aac(6′)-Ib-cr, oqxA, oqxB, and qepA. Two DNA gyrase genes were gyrA (Kim et al., 2016) and gyrB (Hansen and Heisig, 2003), and two topoisomerase IV genes comprised parC (Kim et al., 2016) and parE (Kim et al., 2016). The primers and related parameters were listed in Supplementary Table S2. DNA was extracted using a DNA Extraction Kit (Tiangen Biotech, Beijing, China). Sequences of PCR products were aligned and analyzed using BLAST.2 The resulting DNA sequences of all PCR products by amplifying QRDR genes were compared with the S. typhimurium LT2 genome as a reference.
2.6. Statistical analysis
Statistical analysis was performed using GraphPad Prism (Version 8.0). Differences between proportions were calculated using the Chi-square test. p ≤ 0.05 were considered to be statistically significant.
3. Results
3.1. Salmonella prevalence and serovars
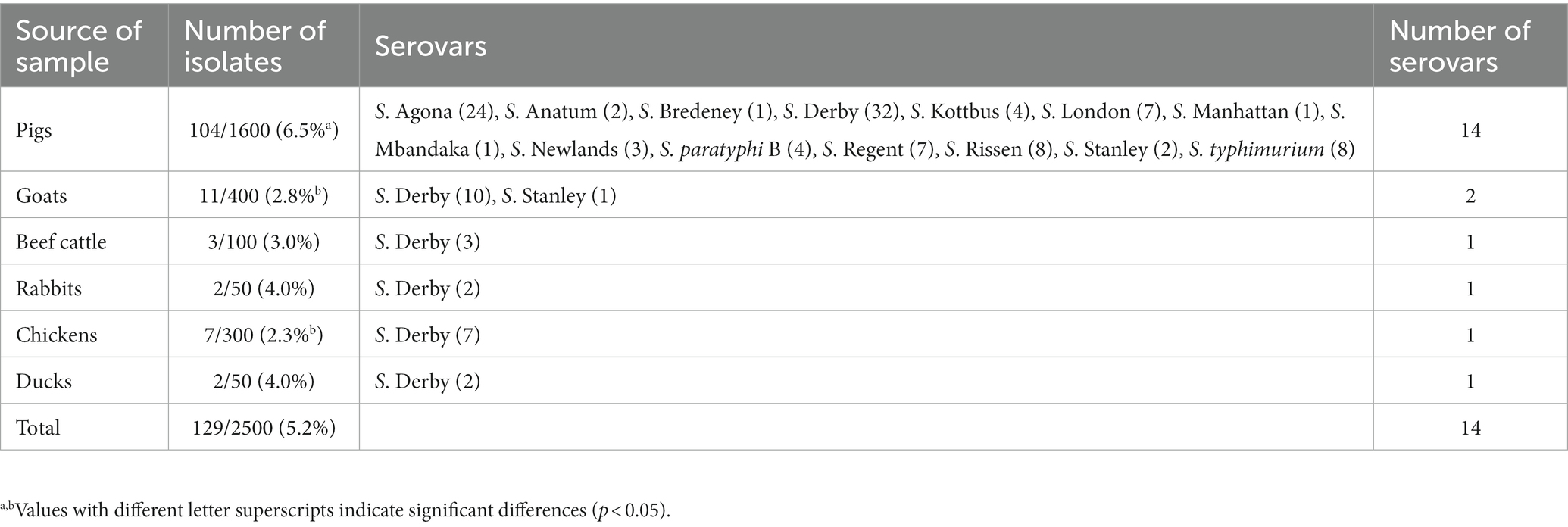
In this study, 129 (5.2%) isolates of Salmonella were obtained from 2,500 fecal samples, among which 104 (6.5%) were recovered from pigs, 11 (2.8%) from goats, three (3.0%) from beef cattle, two (4.0%) from rabbits, seven (2.3%) from chickens, and two (4.0%) from ducks (Table 1; Supplementary Table S1). Overall, the isolation rate among pigs was higher than that for other sources (p < 0.05). Moreover, a significantly higher prevalence of Salmonella was detected in pigs than in goats and chickens (p < 0.05). However, there was no difference in the isolation rate of strains in different years, even from different sources. Fourteen different serovars were identified from the isolates, and S. Derby (56, 43.4%) and S. Agona (24, 18.6%) were the two most common serovars (Table 1). The distribution of serovars varied among different sources. S. Derby (32, 30.8%) and S. Agona (24, 23.1%) were more prevalent than other serovars in pigs. However, most isolates recovered from goats (90.9%, 10/11) and all strains acquired from beef cattle, rabbits, and poultry were S. Derby, although the number of these isolates from these animals was much less than that of isolates from pigs.
3.2. Antimicrobial susceptibility testing of 129 Salmonella isolates
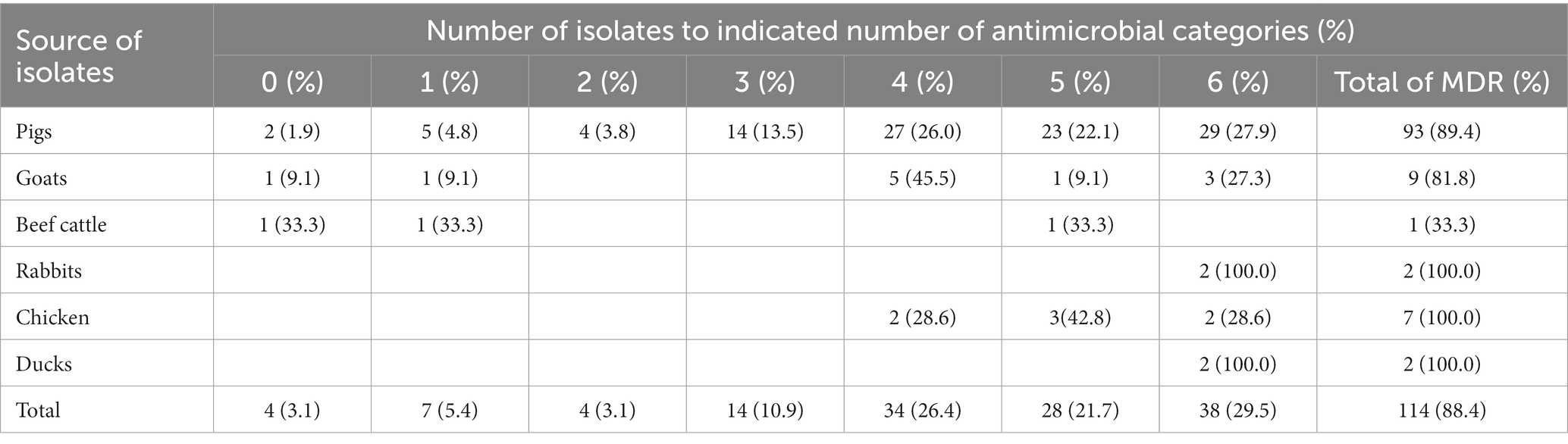
The AMR phenotypes of 129 Salmonella isolates exhibited high rates of resistance to doxycycline (87.6%, 113/129), ampicillin (80.6%, 104/129), tetracycline (79.8%, 103/129), trimethoprim (77.5%, 100/129), florfenicol (76.7%, 99/129), chloramphenicol (72.9%, 94/129), and trimethoprim-sulfamethoxazole (71.3%, 92/129). However, all isolates were susceptible to cefepime and less resistant to amikacin (0.8%, 1/129), ceftazidime (1.6%, 2/129), cefoxitin (2.3%, 3/129), gatifloxacin (3.1%, 4/129), ceftriaxone (5.4%, 7/129), ofloxacin (5.4%, 7/129), aztreonam (6.2%, 8/129), and enoxacin (6.2%, 8/129; Figure 1; Table 2; Supplementary Tables S2, S3). Moreover, three (2.3%) isolates were susceptible to all antimicrobial agents, while 126 (97.7%) isolates showed resistance to at least one drug.
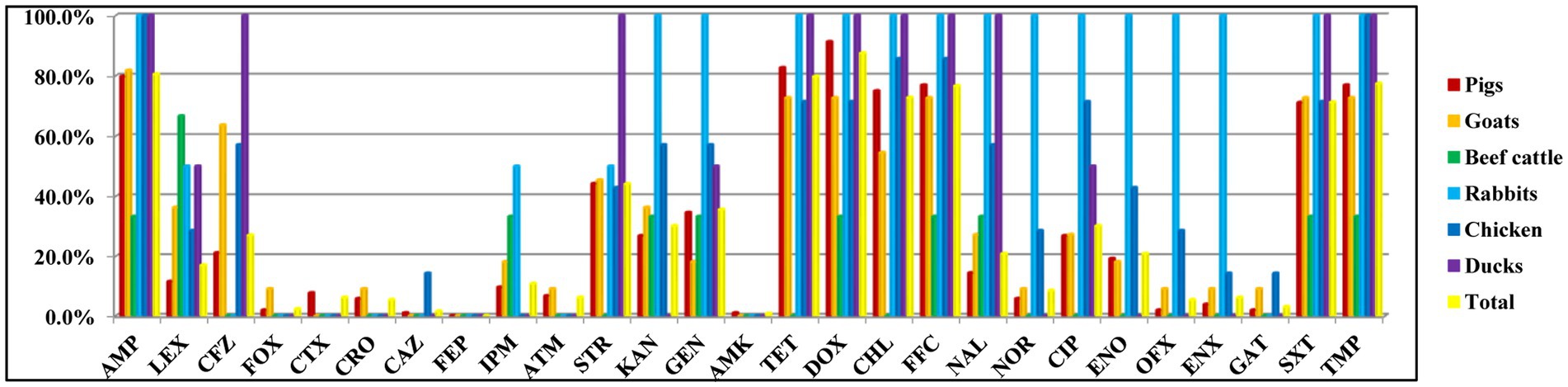
Figure 1. Antimicrobial resistance of Salmonella isolated from food-producing animals was determined by disk diffusion method. AMP (ampicillin), LEX (cephalexin), CFZ (cefazolin), FOX (cefoxitin), CTX (cefotaxime), CRO (ceftriaxone), CAZ (ceftazidime), FEP (cefepime), IPM (imipenem), ATM (aztreonam), STR (streptomycin), KAN (kanamycin), GEN (gentamicin), AMK (amikacin), TET (tetracycline), DOX (doxycycline), CHL (chloramphenicol), FFC (florfenicol), NAL (nalidixic acid), NOR (norfloxacin), CIP (ciprofloxacin), ENO (enrofloxacin), OFX (ofloxacin), ENX (enoxacin), GAT (gatifloxacin), SXT (trimethoprim-sulfamethoxazole), and TMP (trimethoprim).
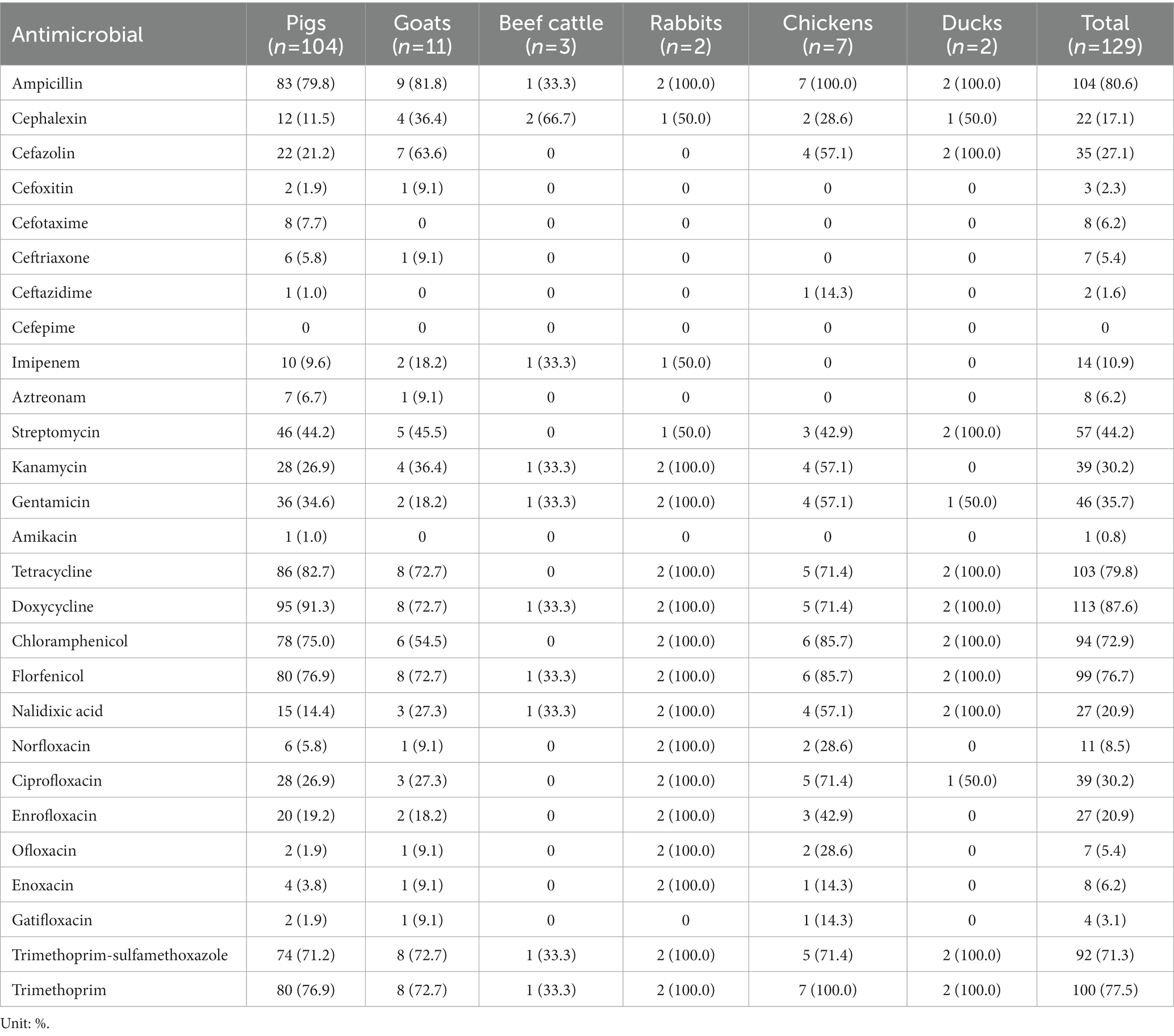
Table 2. Antimicrobial resistance profiles of Salmonella isolates recovered from pigs, goats, beef cattle, rabbits, chickens, and ducks as determined by disk diffusion method.
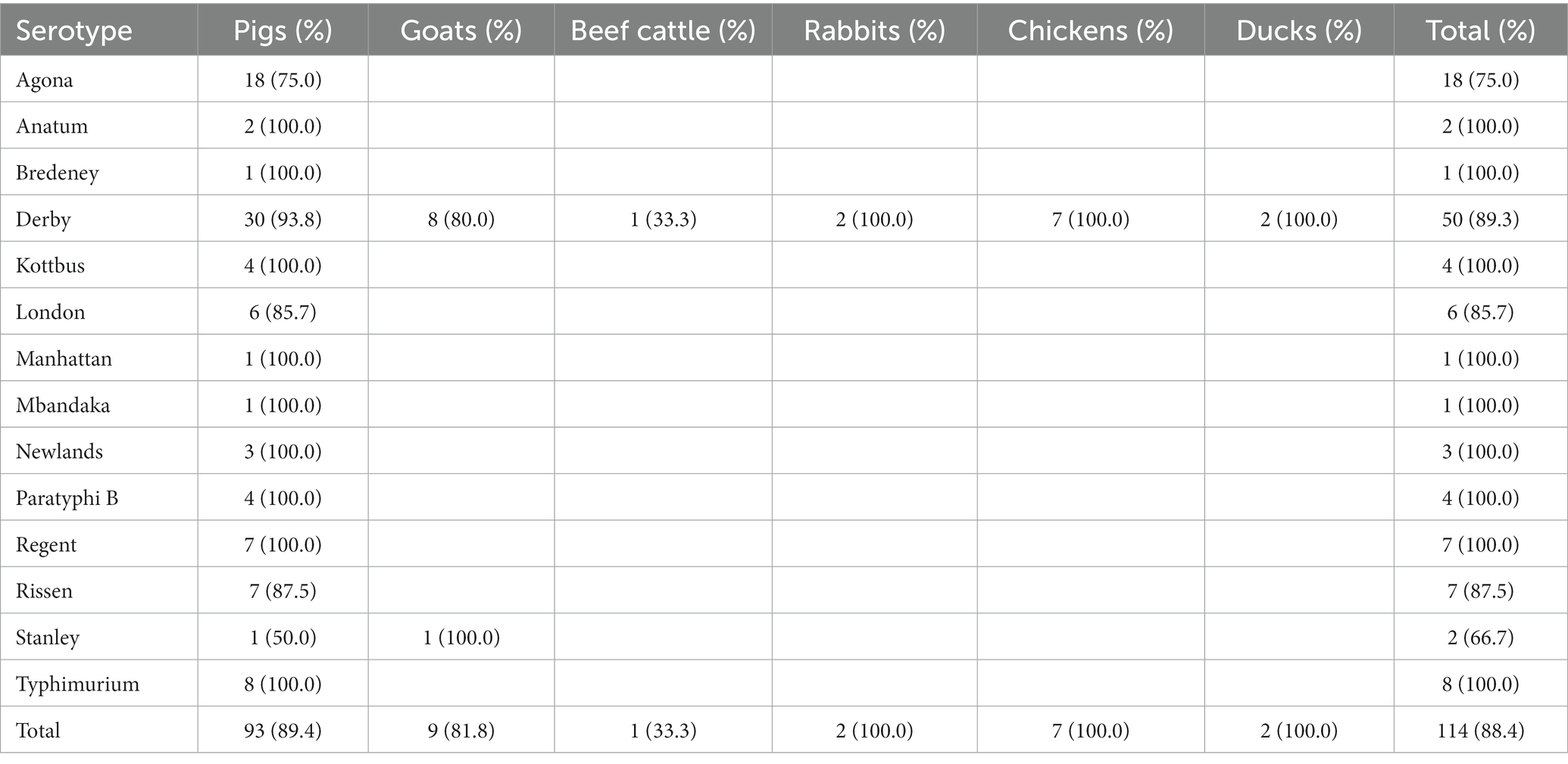
A total of 114 (88.4%) isolates exhibited MDR, which was defined as resistant to three or more classes of antimicrobials (Table 3). All strains belonging to S. Derby obtained from rabbits and poultry showed MDR profiles. A higher prevalence of MDR isolates from pigs was found among S. Derby (93.8%, 30/32), compared with S. Agona (75.0%, 18/24), although there was no significant difference. Serovars S. Anatum (2), S. Bredeney (1), S. Kottbus (4), S. London (6), S. Manhattan (1), S. Mbandaka (1), S. Newlands (3), S. paratyphi B (4), S. Regent (7), S. Rissen (7), S. Stanley (1), and S. typhimurium (8) from pigs were MDR (Table 4).
3.3. Prevalence of β-lactamase genes in Salmonella isolates
The prevalence of β-lactamase genes in Salmonella isolates was 89.9% (116/129), and of these, all isolates collected from rabbits and poultry, and 90.4% (94/104) of pig-associated strains harbored at least one β-lactamase gene. Among the isolates, 107 (82.9%) harbored blaTEM, followed by blaOXA (26, 20.2%), blaCTX-M (8, 6.2%), and blaCMY (3, 2.3%; Table 5). Strains from goats, beef cattle, rabbits and poultry carried blaTEM genes and were resistant to at least one β-lactam drug except for 1 strain isolated from beef cattle. The allele of 104 blaTEM was TEM-1 (44.2%, 57/129), TEM-1a (12.4%, 16/129), TEM-1b (7.8%, 10/129), TEM-116 (0.8%, 1/129), and TEM-171 (17.8%, 23/129). blaOXA was found in pigs, goats, rabbits and chickens, and its genetic genotypes were blaOXA-1 (76.9%, 20/26) and blaOXA-10 (23.1%, 6/26). Three blaCMY-116-harboring strains were detected, and two were isolated from pigs and one from duck. blaCTX-M was amplified from eight pig-associated strains, and the genotypes were blaCTX-M-65. As the predominant serovars, 79.2% (19/24) S. Agona and 87.5% (49/56) S. Derby harbored blaTEM, and the proportions of blaTEM-positive isolates in both serovars were lower than the proportion of blaTEM-positive isolates in the total number of Salmonella isolates. Beta-lactamase genes in isolates carried one to three bla genes (Supplementary Table S4). The most prevalent genotype form was TEM-1 (31.0%, 40/129), followed by TEM-1a (11.6%, 15/129), TEM-171 (10.9%, 14/129), TEM-1b (7.8%, 10/129), and TEM-171 + OXA-1 (7.0%, 9.129). Extended spectrum β-lactamases (ESBLs) are defined as enzymes produced by certain bacteria that are able to hydrolyze β-lactam ring of broad-spectrum β-lactams such as oxyimino-cephalosporins including cefotaxime, ceftriaxone, and ceftazidime (Kawamura et al., 2017). The detection result showed that six isolates carrying blaCTX-M-65 gene were resistant to the third-generation cephalosporins cefotaxime and ceftriaxone.
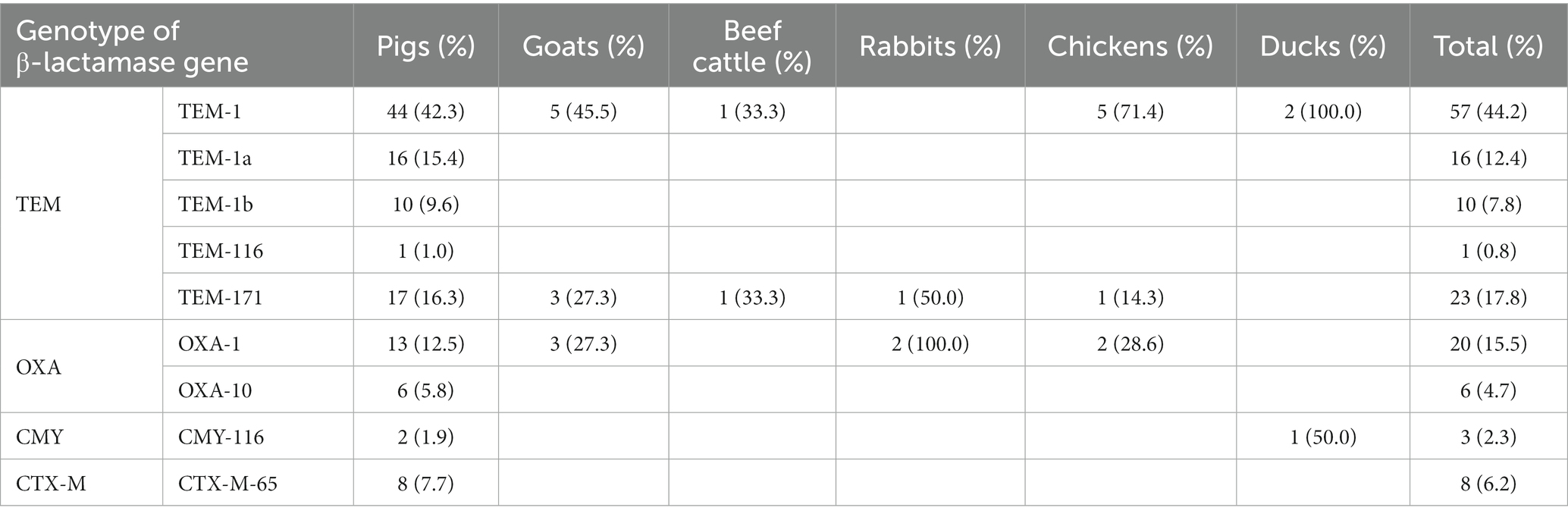
Table 5. The prevalence of β-lactamase genes in Salmonella isolates originating from different animals.
3.4. Distribution of PMQR and QRDR mutations among strains
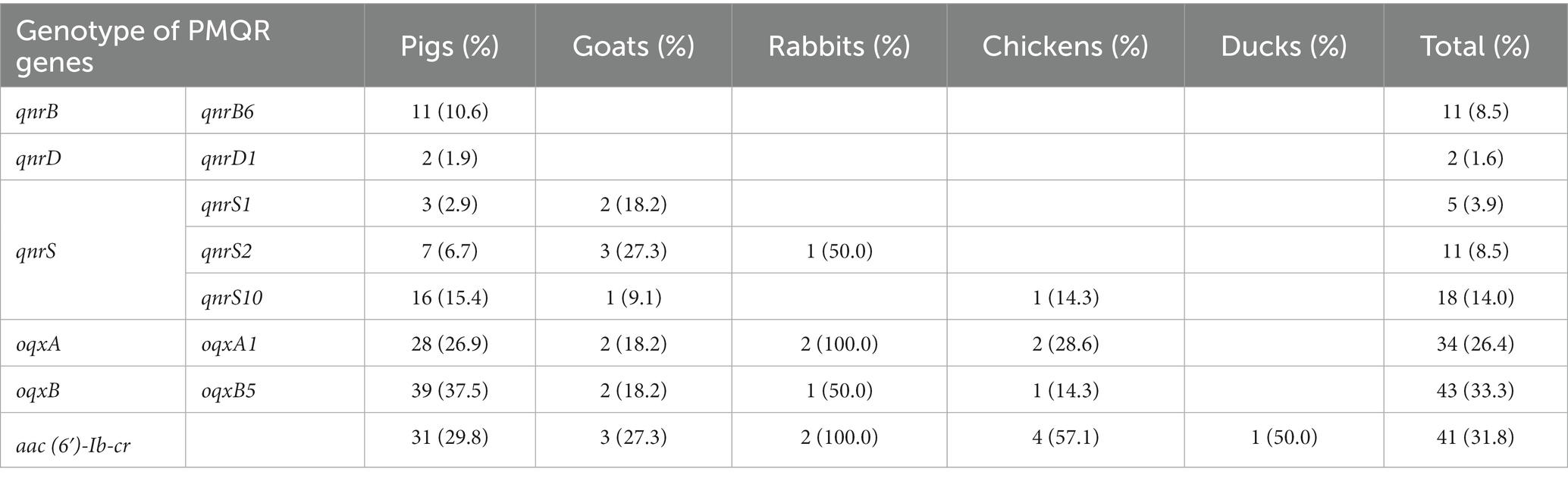
Fifty-seven strains were resistant to at least one tested quinolone, and most of these strains harbored PMQR gene(s) (82.5%, 47/57) and/or exhibited mutation(s) in QRDR gene(s) (98.2%, 56/57). However, PMQR gene(s) and QRDR point mutation(s) were detected in 82 (63.6%) and 121 (93.8%) Salmonella isolates, regardless of whether they were resistant to the quinolones tested. Only qnrB, qnrD, qnrS, oqxA, oqxB, and aac(6′)-Ib-cr were detected in 13.4% (11), 2.4% (2), 41.5% (34), 41.5% (34), 52.4% (43), and 50.0% (41) of the PMQR-producing isolates, respectively (Table 6). qnrA, qnrC, qnrVC, and qepA were not present in any isolate. The genotypes of qnrB, qnrD, oqxA, and oqxB were qnrB6, qnrD1, oqxA1, and oqxB5. Nevertheless, qnrS represented qnrS1 (6.1%, 5/82), qnrS2 (13.4%, 11/82) and qnrS10 (22.0%, 18/82). Similar to β-lactamase gene-containing isolates, there were 20 genotype forms of PMQR genes in 82 PMQR-positive strains (Supplementary Table S5). There were 14 and six isolates carrying a single PMQR gene, qnrS10 and oqxB5, respectively. Moreover, 10 isolates carried the oqxA1 and oqxB5 genes, and eight isolates carried qnrS2, oqxA1, oqxB5, and aac(6′)-Ib-cr simultaneously. Interestingly, most qnrS2-positive strains (90.9%, 10/11) were positive for aac(6′)-Ib-cr.
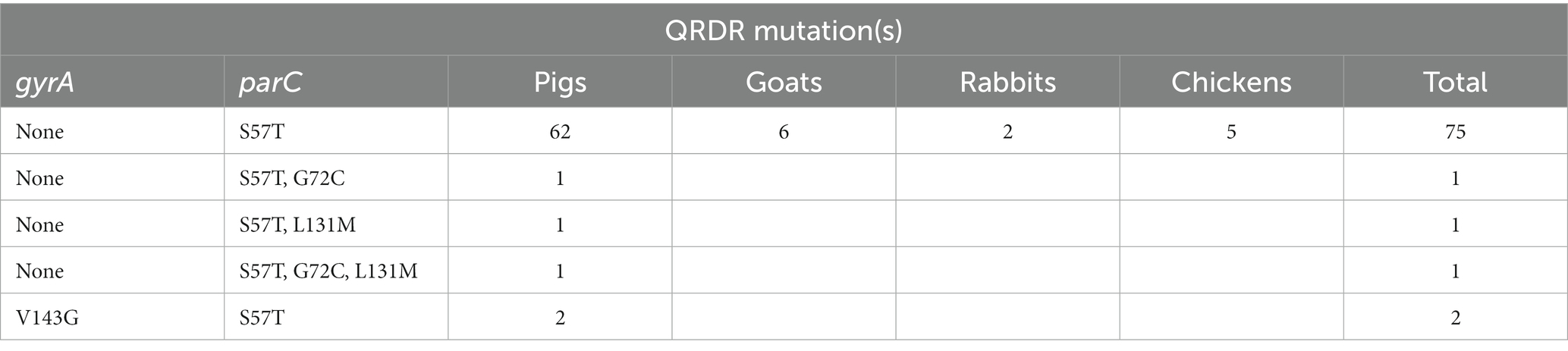
QRDR mutations were very common in PMQR-positive Salmonella isolates (97.5%, 80/82; Table 7). A single mutation in parC (T57S) was detected in isolates obtained from goats (6), rabbits (2) and chickens (5), and 91.2% (62) pig-associated PMQR-positive isolates. Double mutations and triple mutations in parC were found in two (S57T/G72C and S57T/L131M) and one (S57T/G72C/L131M) isolates, respectively. A combination of mutations in gyrA (V143G) and in parC (S57T) was found in two isolates. However, both mutations in gyrA (S83L) and parC (S57T) were detected in one isolate.
3.5. Mechanisms of ciprofloxacin resistance among the isolates
To detect the main mechanism for the presence of 39 CIP-resistant isolates, we found that the most frequent mutation in parC was S57T (89.7%, 35/39; Table 8). Moreover, double mutations in gyrA and parC of three isolates mentioned above were also resistant to ciprofloxacin. Most of CIP-resistant isolates harbored at least one PMQR gene, except for three strains. Surprisingly, one isolate, resistant to ciprofloxacin, only contained aac(6′)-Ib-cr and no mutation was identified at any QRDR genes.
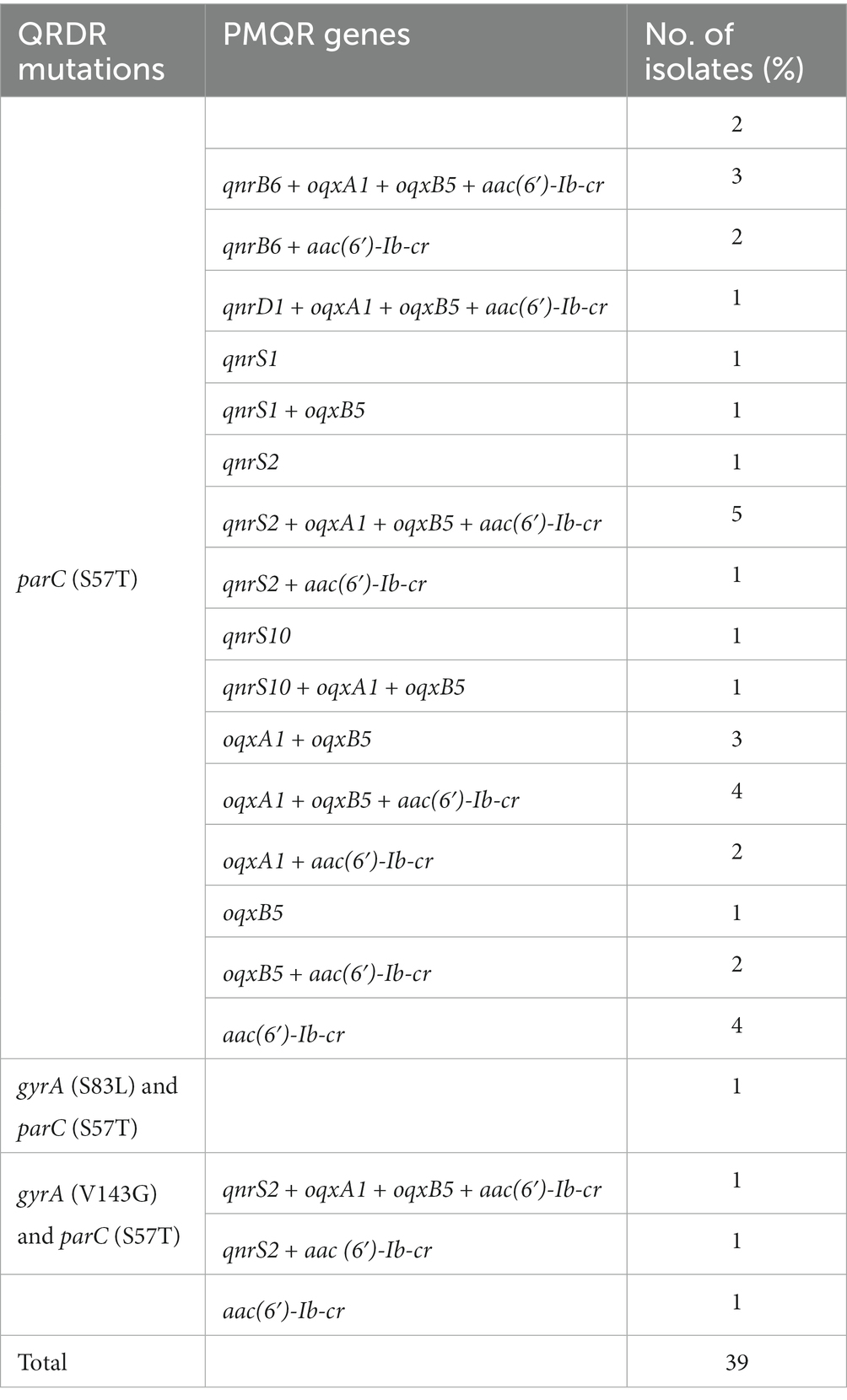
Table 8. Distribution of QRDR mutations and PMQR genes among the 39 ciprofloxacin-resistant Salmonella isolates.
3.6. Characterization of PMQR genes in ESBL-producing isolates
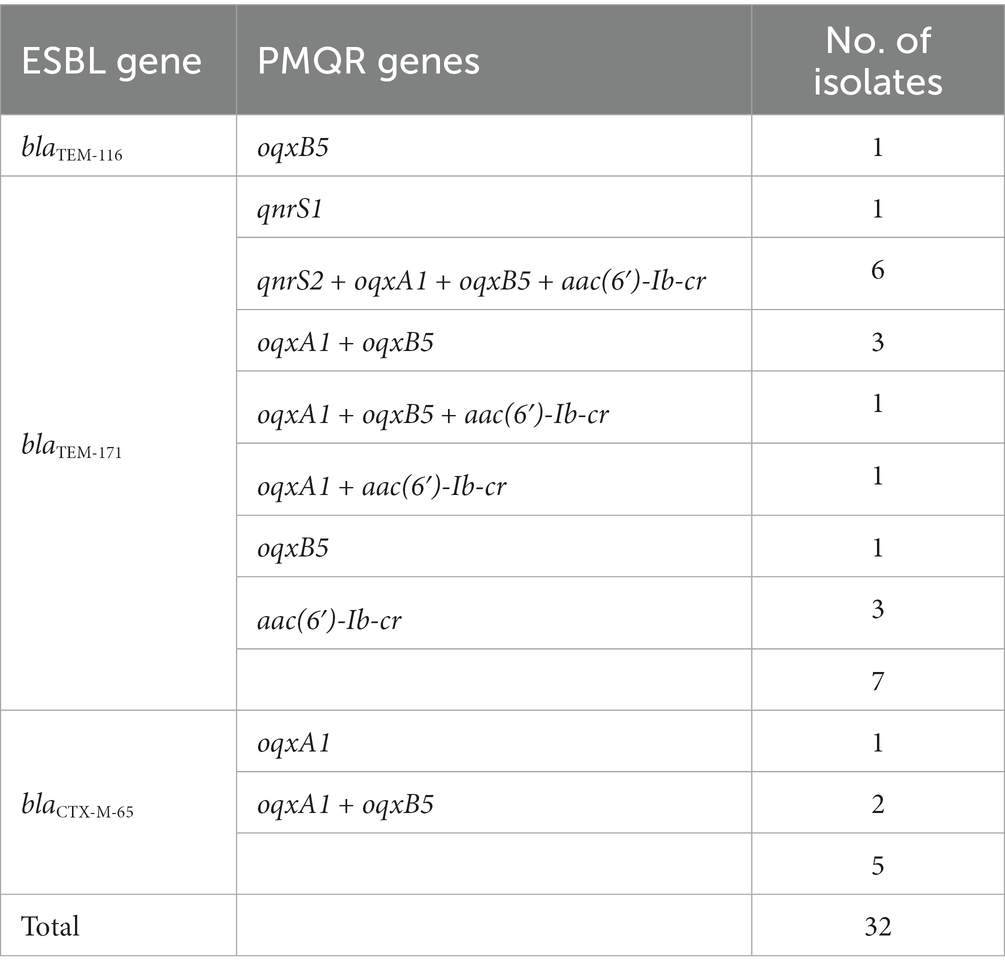
Thirty-two ESBL-producing strains were identified from the isolates, and each strain carried one of ESBL genes from three genotypes, namely blaTEM-116, blaTEM-171, and blaCTX-M-65. The distribution of PMQR-encoding genes among 32 ESBL-producing Salmonella isolates was shown in Table 9. Twenty (62.5%) ESBL-producing Salmonella isolates were found to harbor one to four PMQR genes, and combinations of qnrS2 + oqxA1 + oqxB5 + aac(6′)-Ib-cr (30%, 6/20) was the most common combination type, followed by oqxA1 + oqxB5 (25.0%, 5/20).
3.7. Sequence types and the relatedness with ESBL-producing isolates
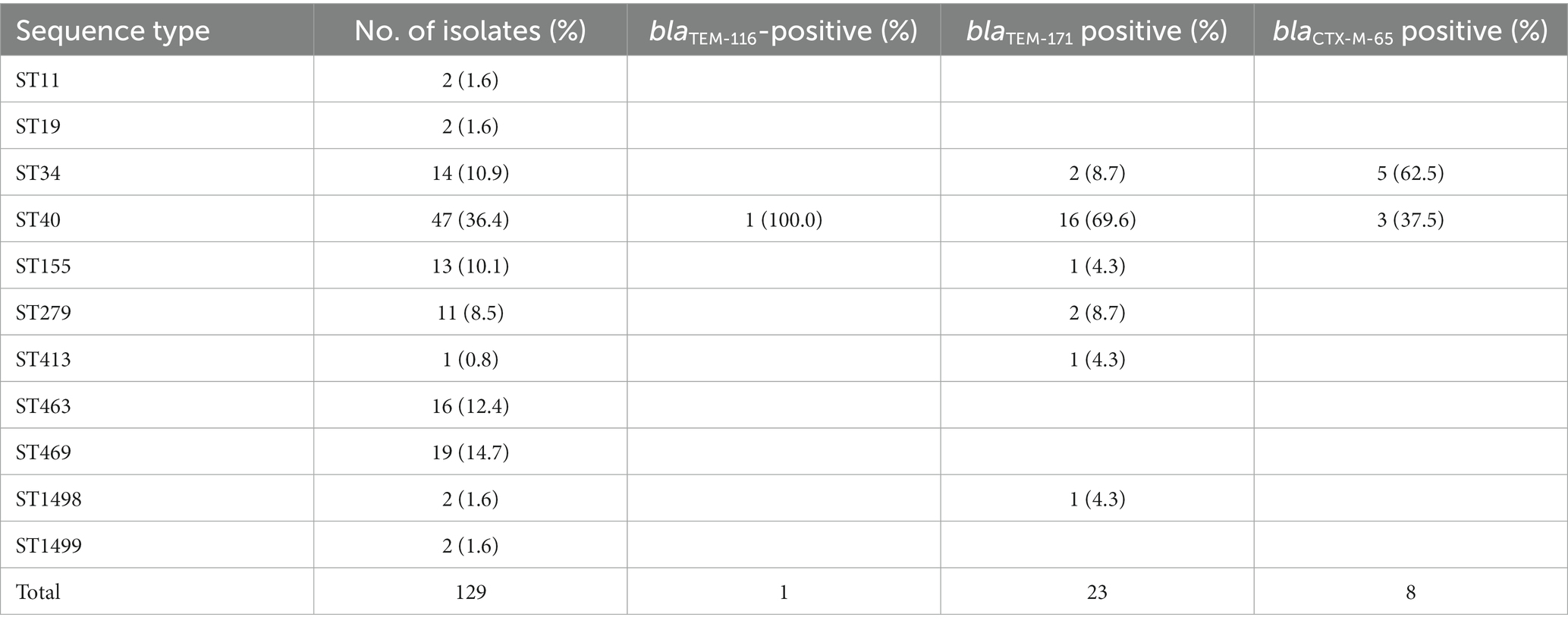
In total, 11 sequence types were identified from the Salmonella isolates, including ST11, ST19, ST34, ST40, ST155, ST279, ST413, ST463, ST469, ST1498, and ST1499 (Table 10). ST34 (10.9%, 14/129), ST40 (36.4%, 47/129), ST463 (12.4%, 16/129), and ST469 (14.7%, 19/129) were the more popular sequence types. Most of isolates containing blaTEM-171 (69.6%, 16/23) and the blaTEM-116-positive isolate were identified as ST40. However, isolates harboring blaCTX-M-65 were attributed to ST34 (62.5%, 5/8) and ST40 (37.5%, 3/8).
4. Discussion
As major reservoirs of bacteria and an important part of food chain, food-producing animals play an irreplaceable role in the transmission of Salmonella. It is of great significance for the prevention and control of salmonellosis in humans and animals to monitor the prevalence and dynamics of antimicrobial resistance and resistant genes of Salmonella originating from food-producing animals in countries or regions with extensive livestock and/or poultry breeding. In this study, we investigated the prevalence, serovar distribution, sequence types, antimicrobial susceptibility phenotypes, the emergence of β-lactamase and PMQR genes, and the occurrence of mutations in the QRDR of Salmonella isolates acquired from pigs, goats, beef cattle, rabbits, and poultry in Chongqing, China.
The number of samples collected from different animals was roughly consistent with the breeding density of different animals in Chongqing, and the number of samples collected from pigs (1600) and chickens (300) was much higher than those from beef cattle (100), rabbits (50) and ducks (50). Overall, 104 Salmonella isolates were recovered from 1,600 fecal samples of pigs, showing a prevalence of 6.5%, higher than the prevalence in Italy (3.4%; Siddi et al., 2021) but much lower than the prevalence in England (19.5%; Wales et al., 2013) and Thailand (37.54%; Phongaran et al., 2019). Moreover, the prevalence of Salmonella in pigs was lower than the prevalence in Shanghai (26.3%; Tian et al., 2021), Shandong (11.1%; Zhao et al., 2017) and other Chinese provinces (Li et al., 2013; Zhang et al., 2016; Kuang et al., 2018b; Zhang et al., 2019). Similarly to the prevalence of Salmonella from pigs, the prevalence of Salmonella isolated from chickens in Chongqing is lower than that of Salmonella from chickens in other countries and regions (Zhao et al., 2017; Caffrey et al., 2021; Sarker et al., 2021), even with a few exceptions (Li et al., 2013).
Previous studies have reported that ducks are important reservoirs of Salmonella (Li et al., 2013; Zhao et al., 2017; Zhang et al., 2019; Kim et al., 2021; Kang et al., 2022), and Chinese duck production exceeds 90% of all ducks globally; therefore, we investigated Salmonella in duck samples. The results indicate that the isolation rate of Salmonella in ducks is lower than in other regions of China (Li et al., 2013; Zhao et al., 2017; Zhang et al., 2019; Kim et al., 2021; Kang et al., 2022). Moreover, we isolated Salmonella from herbivores, including goats, beef cattle, and rabbits, with isolation rates of 2.8, 3.0 and 4.0%, respectively. Compared with other countries, the isolation rate of Salmonella in beef cattle and goats is low (Bosilevac et al., 2015; Thomas et al., 2020; Gutema et al., 2021). Such differences in isolation rates among different animals in our study and other reports can be interpreted based on differences in region, animal species, sample types, collection seasons, culture methods, isolation methodologies, culture media, and local environmental conditions (Kuang et al., 2015).
Among the strains isolated from pigs, 12 serovars were identified in addition to the dominant serovars Agona and Derby. These, previously reported to having been recovered in pigs, included S. Anatum (Li et al., 2013; Siddi et al., 2021), S. Bredeney (Grafanakis et al., 2001; Siddi et al., 2021), S. Kottbus (Toboldt et al., 2014), S. London (Grafanakis et al., 2001; Bonardi et al., 2016), S. Manhattan (Wales et al., 2013; Bonardi et al., 2016), S. Mbandaka (Tian et al., 2021), S. Newlands (Kuang et al., 2015; Li et al., 2019), S. paratyphi B (Phongaran et al., 2019), S. Rissen (Bonardi et al., 2016; Tian et al., 2021), S. Stanley (Kuang et al., 2015; Bonardi et al., 2016) and S. typhimurium (Li et al., 2013; Zhao et al., 2017; Tian et al., 2021). However, to the best of our knowledge, this is the first study reporting S. Regent recovered from pigs, although this serovar was first reported to be isolated from ducks in Korea (Yoon et al., 2014). This demonstrates a serovar of Salmonella that is not common at pig farms in other places, with clearly reported pathogenicity (Duan, 2006). Moreover, there were few reports about S. paratyphi B isolated from pigs (Phongaran et al., 2019), and no report about the isolation of S. paratyphi B from pigs in China prior to our research. Therefore, monitoring of S. Regent and S. paratyphi B in pig farms and pork products should be strengthened to prevent potential transmissions to humans through the food chain. In addition, S. Derby and S. Stanley isolated from goats have not been reported before. The main reason may be that, compared with studies on Salmonella obtained from pigs and poultry, there have been fewer studies on goat-associated Salmonella.
The resistance of isolates to different antimicrobials was positively correlated with the frequency of antimicrobial use (private communication with farmers or workers). For example, isolates have a high resistance rate to ampicillin and doxycycline, which are often used in the breeding process. However, some antimicrobials are rarely used in farms, but the isolates have high resistance to them. For instance, the use of tetracycline is far lower than the use of doxycycline at farms in China. However, our results show that Salmonella isolates had almost identical resistant phenotypes to tetracycline and doxycycline. The reason may be that strains resistant to doxycycline were also resistant to tetracycline because they have the same resistance genes, such as tet genes (Pavelquesi et al., 2021). Moreover, the resistant phenotype of isolates to chloramphenicol, which has been banned for more than 20 years in China, was similar to that of florfenicol, which should also be related to the same resistance genes of these two antimicrobials. In general, pig, chicken and rabbit producers use antimicrobials much more frequently than producers of ruminants, reflected in the resistance to antimicrobials of isolates obtained from these two kinds of animals. The resistance rates of Salmonella isolated from pigs, chickens and rabbits to most tested antimicrobials were higher than those of Salmonella isolated from ruminants. The proportion of multidrug-resistant strains of Salmonella isolated from pigs, chickens and rabbits was also higher than that of Salmonella strains isolated from ruminants.
In this study, β-lactamase genes were amplified in 89.9% of the strains, of which 82.9% carried the blaTEM gene, whose alleles were TEM-1, TEM-1a, TEM-1b, TEM-116, and TEM-171. However, we found that only eight isolates carried blaCTX-M gene (6.2%). Most reports on Salmonella isolated from food-producing animals, including those in China, and even studies in Sichuan Province adjacent to Chongqing, have shown that blaCTX-M was the most widespread ESBL gene. Therefore, the prevalence of blaTEM was generally lower than blaCTX-M (Zhang et al., 2016; Luk-In et al., 2018; Kuang et al., 2018b; Zhang et al., 2019), although there are a few reports that the prevalence of blaTEM was higher than that of blaCTX-M (Zhao et al., 2017; Al-Gallas et al., 2022). The prevalence of the β-lactamase gene, especially blaTEM, is very worrisome. China’s transportation network is very developed, and every village has cement concrete pavements, which makes it very convenient for people to travel but also provides convenience for pathogens to use the same developed transportation network to spread through carriers, such as animals, meat products, contaminated vehicles, and even people. Salmonella from food-producing animals is very easy to spread out of Chongqing through the carriers mentioned above. The blaTEM gene in the Salmonella strains can be transferred into other blaTEM-negative strains through plasmid conjugation, transformation, transduction, etc., (data not shown), resulting in the widespread prevalence of blaTEM-positive Salmonella outside Chongqing.
The coexistence of PMQR genes in CTX-M-producing isolates has been widely reported worldwide (Li et al., 2014; Zhang et al., 2016). However, there has been no demonstrated linkage between the emergence of blaCTX-M and resistance to quinolones, and the emergence of blaCTX-M and coexistence of PMQR genes in Salmonella isolates. The reason was that all CTX-M-positive isolates were susceptible to seven tested quinolones and 62.5% (5/8) of isolates did not carry any PMQR gene. This indicated that blaCTX-M and PMQR are not necessarily cotransferred in some regions or that blaCTX-M may be located on chromosomes, such as CTX-M-14 (Zhang et al., 2019; Hamamoto et al., 2020).
The ESBLs phenotype mediates the resistance to third or fourth-generation cephalosporins. In the ESBL-producing isolates, 75% (6/8) of strains carrying blaCTX-M-65 showed resistance to cefotaxime and ceftriaxone, while only one strain (4.3%, 1/23) harboring blaTEM-171 exhibited resistance to ceftriaxone. This indicates that blaCTX-M-65 plays a more important role in resistance to broad-spectrum β-lactams. A research from United States also supported this point (Tate et al., 2017). Therefore, although the proportion of Salmonella carrying the blaCTX-M-65 gene in food-producing animals in Chongqing is low, its resistance to broad-spectrum β-lactams could not be ignored.
The mutations of QRDR genes often occurred in gyrA, such as S83Y, S83F, D87G, D87N, and D87Y (Zhang et al., 2016; Wang et al., 2020). The mutations of parC were less than the mutations found in gyrA, and the amino acid substitution of ParC was usually at the position of 80th (S80R; Zhang et al., 2016; Wang et al., 2020). However, in our study, mutations mainly occurred at the 57th amino acid of the ParC. Similar study has also been reported in Salmonella isolated from Guangdong in recent years (Chen et al., 2021), indicating an increasing trend of mutation in the 57th amino acid of the ParC.
The Ministry of Agriculture of China requested that the usage of lomefloxacin, pefloxacin, ofloxacin and norfloxacin in food-producing animals be forbidden beginning December 31, 2016. On October 21, 2021, the Ministry of Agriculture and Rural Affairs of China issued the “national action plan for reducing the use of veterinary antimicrobials (2021–2025).” These actions provided powerful measures for reducing drug-resistant bacteria from food-producing animals. Further longitudinal monitoring of antimicrobial susceptibility, serotypes and resistance genes of Salmonella isolates from food-producing animals in the same geographic region should be carried out to compare and evaluate the trends of above parameters.
5. Conclusion
In summary, we first analyzed the prevalence, serovar diversity, sequence types, and antimicrobial resistance and examined the β-lactamase, QRDR, and PMQR genes of Salmonella strains isolated from livestock and poultry at farms in Chongqing, China. Our findings demonstrated the diversity of serovars of Salmonella isolates. Notably, various MDR serovars of Salmonella are widespread, which highlights the potential risk of antimicrobial-resistant Salmonella foodborne infections. This study emphasizes the significant roles of blaTEM genes in β-lactam-resistant isolates and parC mutations in quinolone-resistant isolates, especially those that carry PMQR genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by IACUC-20190305-01.
Author contributions
HD, HM, and YH conceived and designed the experiments. JL, HM, BZ, JH, XC, YG, MZ, MX, YZ, and YH performed the experiments. HD and HW analyzed the data. JL, JW, and YY contributed reagents/materials/analysis tools. HD and HM wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Chongqing’s Modern Agricultural Industry Technology System Program for Herbivore (2022[12]).
Acknowledgments
The authors would like to acknowledge all the study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1011719/full#supplementary-material
Footnotes
References
Al-Gallas, N., Belghouthi, K., Barratt, N. A., Ghedira, K., Hotzel, H., Tomaso, H., et al. (2022). Identification and characterization of multidrug-resistant ESBL-producing Salmonella enterica serovars Kentucky and typhimurium isolated in Tunisia CTX-M-61/TEM-34, a novel cefotaxime-hydrolysing β-lactamase of Salmonella. J. Appl. Microbiol. 132, 279–289. doi: 10.1111/jam.15211
Bonardi, S., Alpigiani, I., Bruini, I., Barilli, E., Brindani, F., Morganti, M., et al. (2016). Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Int. J. Food Microbiol. 218, 44–50. doi: 10.1016/j.ijfoodmicro.2015.11.005
Bosilevac, J. M., Gassem, M. A., al Sheddy, I. A., Almaiman, S. A., al-Mohizea, I. S., Alowaimer, A., et al. (2015). Prevalence of Escherichia coli O157:H7 and Salmonella in camels, cattle, goats, and sheep harvested for meat in Riyadh. J. Food Prot. 78, 89–96. doi: 10.4315/0362-028X.JFP-14-176
Caffrey, N., Agunos, A., Gow, S., Liljebjelke, K., Mainali, C., and Checkley, S. L. (2021). Salmonella spp. prevalence and antimicrobial resistance in broiler chicken and Turkey flocks in Canada from 2013 to 2018. Zoonoses Public Health 68, 719–736. doi: 10.1111/zph.12769
Canning, M., Birhane, M. G., Dewey-Mattia, D., Lawinger, H., Cote, A., Gieraltowski, L., et al. (2023). Salmonella outbreaks linked to beef, United States, 2012-2019. J. Food Prot. 86:100071. doi: 10.1016/j.jfp.2023.100071
Cattoir, V., Poirel, L., Rotimi, V., Soussy, C. J., and Nordmann, P. (2007). Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60, 394–397. doi: 10.1093/jac/dkm204
Chen, Z., Bai, J., Zhang, X., Wang, S., Chen, K., Lin, Q., et al. (2021). Highly prevalent multidrug resistance and QRDR mutations in Salmonella isolated from chicken, pork and duck meat in southern China, 2018-2019. Int. J. Food Microbiol. 340:109055. doi: 10.1016/j.ijfoodmicro.2021.109055
Chen, S., Zhao, S., White, D. G., Schroeder, C. M., Lu, R., Yang, H., et al. (2004). Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70, 1–7. doi: 10.1128/AEM.70.1.1-7.2004
CLSI. (2019). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, Approved Standard-Fourth Edition (VET08-A4). Wayne, PA: CLSI.
CLSI. (2022). Performance Standards for Antimicrobial Susceptibility Testing: Thirty-Second Informational Supplement M100-S32. Wayne, PA: CLSI.
Cody, W. L., Wilson, J. W., Hendrixson, D. R., McIver, K. S., Hagman, K. E., Ott, C. M., et al. (2008). Skim milk enhances the preservation of thawed −80 degrees C bacterial stocks. J. Microbiol. Methods 75, 135–138. doi: 10.1016/j.mimet.2008.05.006
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Duan, Z. (2006). Detection and analysis of repeated food poisoning caused by Salmonella regent. Chin. J. Health. Lab. Tech. 16, 471–472. doi: 10.3969/j.issn.1004-8685.2006.04.042
Foley, S. L., and Lynne, A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86, E173–E187. doi: 10.2527/jas.2007-0447
General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China. (2017). Multilocus Sequence Typing Detection Method for Pathogens in Export Food – Part 1: Salmonella. Industry Standards for Entry-Exit Inspection and Quarantine of the People’s Republic of China: No. SN/T 4525.1-2016.
Ghafourian, S., Sadeghifard, N., Soheili, S., and Sekawi, Z. (2015). Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr. Issues Mol. Biol. 17, 11–21.
Grafanakis, E., Leontides, L., and Genigeorgis, C. (2001). Seroprevalence and antibiotic sensitivity of serotypes of Salmonella enterica in Greek pig herds. Vet. Rec. 148, 407–411. doi: 10.1136/vr.148.13.407
Gutema, F. D., Rasschaert, G., Agga, G. E., Merera, O., Duguma, A. B., Abdi, R. D., et al. (2021). Prevalence, antimicrobial resistance, and molecular characterization of Salmonella in cattle, beef, and diarrheic patients in Bishoftu, Ethiopia. Foodborne. Pathog. Dis. 18, 283–289. doi: 10.1089/fpd.2020.2869
Hamamoto, K., Tokunaga, T., Yagi, N., and Hirai, I. (2020). Characterization of blaCTX-M-14 transposition from plasmid to chromosome in Escherichia coli experimental strain. Int. J. Med. Microbiol. 310:151395. doi: 10.1016/j.ijmm.2020.151395
Hansen, H., and Heisig, P. (2003). Topoisomerase IV mutations in quinolone-resistant salmonellae selected in vitro. Microb. Drug Resist. 9, 25–32. doi: 10.1089/107662903764736319
Harrison, O., Rensing, S., Jones, C. K., and Trinetta, V. (2022). Salmonella enterica 4,[5],12:i:- an emerging threat for the swine feed and pork production industry. J. Food Prot. 85, 660–663. doi: 10.4315/JFP-21-400
Hetman, B. M., Pearl, D. L., Barker, D. O. R., Robertson, J., Nash, J. H. E., Reid-Smith, R., et al. (2022). Combining analytical epidemiology and genomic surveillance to identify risk factors associated with the spread of antimicrobial resistance in Salmonella enterica subsp. enterica serovar Heidelberg. Microb. Genom. 8:mgen000891. doi: 10.1099/mgen.0.000891
Igbinosa, I. H., Amolo, C. N., Beshiru, A., Akinnibosun, O., Ogofure, A. G., el-Ashker, M., et al. (2023). Identification and characterization of MDR virulent Salmonella spp isolated from smallholder poultry production environment in Edo and Delta states, Nigeria. PLoS. One. 18:e0281329. doi: 10.1371/journal.pone.0281329
Kang, X., Wang, M., Meng, C., Li, A., Jiao, X., and Pan, Z. (2022). Prevalence and whole-genome sequencing analysis of Salmonella reveal its spread along the duck production chain. Poult. Sci. 101:101993. doi: 10.1016/j.psj.2022.101993
Kawamura, K., Nagano, N., Suzuki, M., Wachino, J. I., Kimura, K., and Arakawa, Y. (2017). ESBL-producing Escherichia coli and its rapid rise among healthy people. Food. Saf (Tokyo). 5, 122–150. doi: 10.14252/foodsafetyfscj.2017011
Kim, T. S., Kim, G. S., Son, J. S., Lai, V. D., Mo, I. P., and Jang, H. (2021). Prevalence, biosecurity factor, and antimicrobial susceptibility analysis of Salmonella species isolated from commercial duck farms in Korea. Poult. Sci. 100:100893. doi: 10.1016/j.psj.2020.12.006
Kim, S. Y., Lee, S. K., Park, M. S., and Na, H. T. (2016). Analysis of the fluoroquinolone antibiotic resistance mechanism of Salmonella enterica isolates. J. Microbiol. Biotechnol. 26, 1605–1612. doi: 10.4014/jmb.1602.02063
Kuang, X., Hao, H., Dai, M., Wang, Y., Ahmad, I., Liu, Z., et al. (2015). Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front. Microbiol. 6:602. doi: 10.3389/fmicb.2015.00602
Kuang, D., Zhang, J., Xu, X., Shi, W., Chen, S., Yang, X., et al. (2018a). Emerging high-level ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int. J. Food Microbiol. 280, 1–9. doi: 10.1016/j.ijfoodmicro.2018.05.001
Kuang, D., Zhang, J., Xu, X., Shi, W., Yang, X., Su, X., et al. (2018b). Increase in ceftriaxone resistance and widespread extended-spectrum β-lactamases genes among Salmonella enterica from human and nonhuman sources. Foodborne Pathog. Dis. 15, 770–775. doi: 10.1089/fpd.2018.2468
Li, R., Lai, J., Wang, Y., Liu, S., Li, Y., Liu, K., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan province, China. Int. J. Food. Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Li, L., Liao, X. P., Liu, Z. Z., Huang, T., Li, X., Sun, J., et al. (2014). Co-spread of oqxAB and blaCTX-M-9G in non-Typhi Salmonella enterica isolates mediated by ST2-IncHI2 plasmids. Int. J. Antimicrob. Agents 44, 263–268. doi: 10.1016/j.ijantimicag.2014.05.014
Li, Q., Yin, J., Li, Z., Li, Z., du, Y., Guo, W., et al. (2019). Serotype distribution, antimicrobial susceptibility, antimicrobial resistance genes and virulence genes of Salmonella isolated from a pig slaughterhouse in Yangzhou, China. AMB. Express. 9:210. doi: 10.1186/s13568-019-0936-9
Li, P., Zhang, M., Hao, G., and Sun, S. (2022). Research note: hypervirulent arthritis-causing Salmonella Pullorum isolated from Chinese native chicken breeds significantly decreased growth performance of chicks. Poult. Sci. 101:101575. doi: 10.1016/j.psj.2021.101575
Luk-In, S., Chatsuwan, T., Pulsrikarn, C., Bangtrakulnonth, A., Rirerm, U., and Kulwichit, W. (2018). High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype Choleraesuis isolates in Thailand: the emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. choleraesuis Isolates. Int. J. Med. Microbiol. 308, 447–453. doi: 10.1016/j.ijmm.2018.03.008
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Marus, J. R., Magee, M. J., Manikonda, K., and Nichols, M. C. (2019). Outbreaks of Salmonella enterica infections linked to animal contact: demographic and outbreak characteristics and comparison to foodborne outbreaks-United States, 2009-2014. Zoonoses Public Health 66, 370–376. doi: 10.1111/zph.12569
Nichols, M., Gollarza, L., Palacios, A., Stapleton, G. S., Basler, C., Hoff, C., et al. (2021). Salmonella illness outbreaks linked to backyard poultry purchasing during the COVID-19 pandemic: United States, 2020. Epidemiol. Infect. 149:e234. doi: 10.1017/S0950268821002132
Nichols, M., Gollarza, L., Sockett, D., Aulik, N., Patton, E., Francois Watkins, L. K., et al. (2022). Outbreak of multidrug-resistant Salmonella Heidelberg infections linked to dairy calf exposure, United States, 2015-2018. Foodborne Pathog. Dis. 19, 199–208. doi: 10.1089/fpd.2021.0077
Pavelquesi, S. L. S., de Oliveira Ferreira, A. C. A., Rodrigues, A. R. M., de Souza Silva, C. M., Orsi, D. C., and da Silva, I. C. R. (2021). Presence of tetracycline and sulfonamide resistance genes in Salmonella spp.: literature review. Antibiotics (Basel). 10:1314. doi: 10.3390/antibiotics10111314
Phongaran, D., Khang-Air, S., and Angkititrakul, S. (2019). Molecular epidemiology and antimicrobial resistance of Salmonella isolates from broilers and pigs in Thailand. Vet. World. 12, 1311–1318. doi: 10.14202/vetworld.2019.1311-1318
Qiao, J., Zhang, Q., Alali, W. Q., Wang, J., Meng, L., Xiao, Y., et al. (2017). Characterization of extended-spectrum β-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 248, 72–81. doi: 10.1016/j.ijfoodmicro.2017.02.016
Robicsek, A., Strahilevitz, J., Sahm, D. F., Jacoby, G. A., and Hooper, D. C. (2006). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50, 2872–2874. doi: 10.1128/AAC.01647-05
Sarker, B. R., Ghosh, S., Chowdhury, S., Dutta, A., Chandra Deb, L., Krishna Sarker, B., et al. (2021). Prevalence and antimicrobial susceptibility profiles of non-typhoidal Salmonella isolated from chickens in Rajshahi, Bangladesh. Vet. Med. Sci. 7, 820–830. doi: 10.1002/vms3.440
Siddi, G., Piras, F., Spanu, V., Demontis, M., Meloni, M. P., Sanna, R., et al. (2021). Trend of Salmonella enterica occurrence and serotypes in Sardinian pig slaughterhouses. Ital. J. Food. Saf. 10:9362. doi: 10.4081/ijfs.2021.9362
Tang, B., Elbediwi, M., Nambiar, R. B., Yang, H., Lin, J., and Yue, M. (2022). Genomic characterization of antimicrobial-resistant Salmonella enterica in duck, chicken, and pig farms and retail markets in eastern China. Microbiol. Spectr. 10:e0125722. doi: 10.1128/spectrum.01257-22
Tate, H., Folster, J. P., Hsu, C. H., Chen, J., Hoffmann, M., Li, C., et al. (2017). Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 61, e00488–e00417. doi: 10.1128/AAC.00488-17
Thomas, K. M., de Glanville, W. A., Barker, G. C., Benschop, J., Buza, J. J., Cleaveland, S., et al. (2020). Prevalence of Campylobacter and Salmonella in African food animals and meat: a systematic review and meta-analysis. Int. J. Food Microbiol. 315:108382. doi: 10.1016/j.ijfoodmicro.2019.108382
Tian, Y., Gu, D., Wang, F., Liu, B., Li, J., Kang, X., et al. (2021). Prevalence and characteristics of Salmonella spp. from a pig farm in Shanghai, China. Foodborne. Pathog. Dis. 18, 477–488. doi: 10.1089/fpd.2021.0018
Toboldt, A., Tietze, E., Helmuth, R., Junker, E., Fruth, A., and Malorny, B. (2014). Molecular epidemiology of Salmonella enterica serovar Kottbus isolated in Germany from humans, food and animals. Vet. Microbiol. 170, 97–108. doi: 10.1016/j.vetmic.2014.01.020
Wales, A., Weaver, J., McLaren, I. M., Smith, R. P., Mueller-Doblies, D., and Davies, R. H. (2013). Investigation of the distribution of Salmonella within an integrated pig breeding and production organisation in the United Kingdom. ISRN. Vet. Sci. 2013:943126. doi: 10.1155/2013/943126
Wang, W., Zhao, L., Hu, Y., Dottorini, T., Fanning, S., Xu, J., et al. (2020). Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob. Agents Chemother. 64, e00092–e00020. doi: 10.1128/AAC.00092-20
Wang, J., Zhu, X., Wang, Z., Chen, Y., Robertson, I. D., Guo, A., et al. (2023). Prevalence and antimicrobial resistance of Salmonella and the enumeration of ESBL E. coli in dairy farms in Hubei Province, China. Prev. Vet. Med. 212:105822. doi: 10.1016/j.prevetmed.2022.105822
Yoon, R. H., Cha, S. Y., Wei, B., Roh, J. H., Seo, H. S., Oh, J. Y., et al. (2014). Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea. J. Food Prot. 77, 1579–1582. doi: 10.4315/0362-028X.JFP-14-018
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located blaCTX-M-55 in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Zhang, W. H., Lin, X. Y., Xu, L., Gu, X. X., Yang, L., Li, W., et al. (2016). CTX-M-27 producing Salmonella enterica serotypes typhimurium and Indiana are prevalent among food-producing animals in China. Front. Microbiol. 7:436. doi: 10.1016/10.3389/fmicb.2016.00436
Keywords: serovar, multidrug resistance, ESBL, PMQR, blaTEM , parC
Citation: Lai J, Mu H, Zhou B, He J, Cheng X, Gan Y, Zhao M, Xie M, Zhang Y, He Y, Yang Y, Wang J, Wang H and Ding H (2023) BlaTEM-positive Salmonella enterica serovars Agona and Derby are prevalent among food-producing animals in Chongqing, China. Front. Microbiol. 14:1011719. doi: 10.3389/fmicb.2023.1011719
Edited by:
Jianmin Zhang, South China Agricultural University, ChinaReviewed by:
Jiyun Li, Hunan Agricultural University, ChinaMaojun Liu, Jiangsu Academy of Agricultural Sciences, China
Bao-Tao Liu, Qingdao Agricultural University, China
Copyright © 2023 Lai, Mu, Zhou, He, Cheng, Gan, Zhao, Xie, Zhang, He, Yang, Wang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honglei Ding, aG9uZ2xlaWRpbmdAc3d1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Jiacui Lai1†
Jiacui Lai1† Hao Mu
Hao Mu Haoju Wang
Haoju Wang Honglei Ding
Honglei Ding