- 1Department of Microbiology, Maharshi Dayanand University, Rohtak, India
- 2Centre for Bioinformatics, Maharshi Dayanand University, Rohtak, India
- 3Centre for Medical Biotechnology, Maharshi Dayanand University, Rohtak, India
Reduced agricultural production as well as issues like nutrient-depleted soils, eutrophication, and groundwater contamination have drawn attention to the use of endophyte-based bioformulations to restore soil fertility. Pantoea agglomerans CPHN2, a non-rhizobial nodule endophyte isolated from Cicer arietinum, exhibited a variety of plant growth-promoting traits. In this study, we used NextSeq500 technology to analyze whole-genome sequence information of this plant growth-promoting endophytic bacteria. The genome of P. agglomerans CPHN2 has a length of 4,839,532 bp and a G + C content of 55.2%. The whole genome comprises three different genomic fractions, comprising one circular chromosome and two circular plasmids. A comparative analysis between P. agglomerans CPHN2 and 10 genetically similar strains was performed using a bacterial pan-genome pipeline. All the predicted and annotated gene sequences for plant growth promotions (PGPs), such as phosphate solubilization, siderophore synthesis, nitrogen metabolism, and indole-3-acetic acid (IAA) of P. agglomerans CPHN2, were identified. The whole-genome analysis of P. agglomerans CPHN2 provides an insight into the mechanisms underlying PGP by endophytes and its potential applications as a biofertilizer.
Introduction
Pantoea, a highly versatile member of family Enterobacteriaceae, capable of adapting to diverse range of environment conditions, has more than twenty recognized species assigned to it (Walterson and Stavrinides, 2015). It has also been reported that, in addition to inhabiting a variety of hosts as parasites, Pantoea occurs as a mutualistic associate, exhibiting the true versatility of its nature. It has been isolated from different habitats, such as soil, water, plants, humans, and animals (Lee et al., 2010; Walterson and Stavrinides, 2015). Pantoea was previously considered an enemy because of its pathogenicity, but it is now recognized as a friend after the establishment of its beneficial effects such as its potential for biocontrol and plant growth promotion (PGP) (Dutkiewicz et al., 2016; Kukreti et al., 2020; Agri et al., 2021, 2022).
Pantoea agglomerans CPHN2, non-rhizobial plant growth promoting endophytic bacteria (PGPEB) was isolated from Cicer arietinum nodules. This strain possessed the ability to produce ammonia, phytohormone indole-3-acetic acid (IAA), and also able to solubilise phosphate (Maheshwari et al., 2019; Rani et al., 2021). Indole acetic acid (IAA), an important phytohormone, plays a crucial role in plant physiology and development. Under optimized conditions, this strain was also able to produce high amount of IAA (unpublished). Multiple IAA biosynthetic pathways have been reported in both PGPEBs and plants. These pathways can be characterized by metabolic intermediates, enzymes, and mutants or by genomic studies (Duca et al., 2014; Navarro-Torre et al., 2017; Chi et al., 2018; Duca and Glick, 2020; Dudeja et al., 2021).
Whole-genome shotgun (WGS) sequencing provides insights into complete data of chromosomes and helps in the identification of significant target genes in terms of PGP activity in addition to yielding information on their interactions with plants. Recently, a large number of whole-genome sequences of PGPEBs, such as Bacillus, Pseudomonas, Pantoea, and Enterobacter, have been studied (Verheggen et al., 2016). In the present study, whole-genome sequencing of P. agglomerans CPHN2 was carried out with the aim of unraveling the underlying molecular mechanisms, functional potential, and taxonomy. Comparative genomics was carried out by evaluating its proximity to related strains and by comparing its whole genome to 10 closely related strains in terms of the core and pan-genomes.
Materials and methods
Bacterial sample
Pantoea agglomerans CPHN2 for this study was taken from the Plant Microbe Interaction Laboratory, Department of Microbiology, Maharshi Dayanand University, Rohtak. The isolate was initially grown at 30°C ± 2°C on tryptone soya agar (TSA) medium and then purified.
Genome sequencing and annotations
The colonies were sent for whole genome sequencing to Eurofins Genomics India Pvt. Ltd. Bacterial deoxyribonucleic acid (DNA) was isolated from pure culture using the Quick-DNA Miniprep Plus kit (Zymo Research), according to the kit procedure. The quality and quantity of the extracted genomic DNA (gDNA) were tested using the NanoDrop Spectrophotometer.
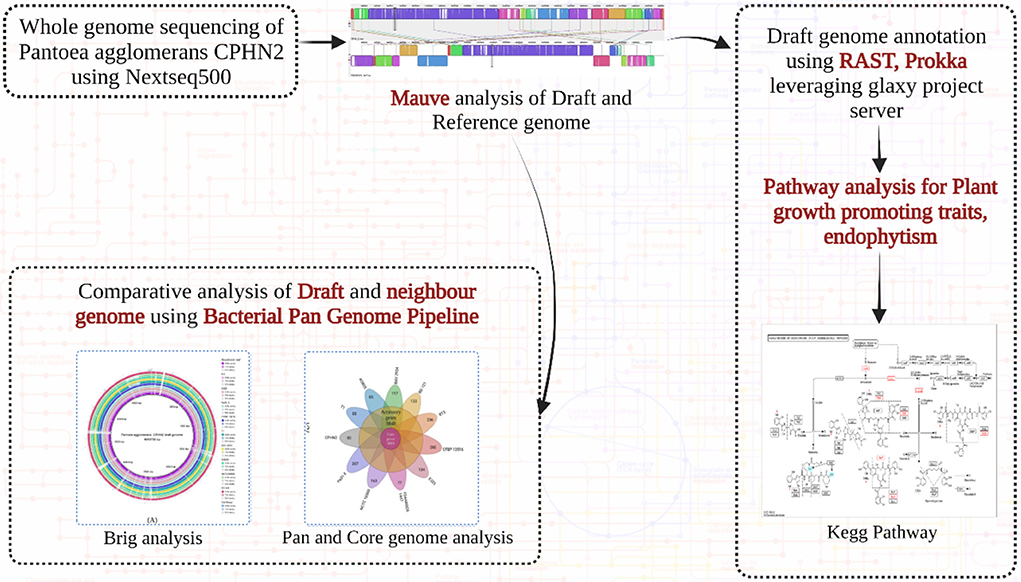
The whole genome was sequenced using the Illumina NextSeq500 platform. The paired-end (PE) sequencing library was constructed from 1 mg of genomic DNA samples using the illuminaTruSeq Nano DNA Library Prep kit as per the manufacturer's protocol. The quality of raw reads was checked with FastQC v0.11.8 (Andrews, 2010) and filtered and trimmed with fastp v0.201 (Chen et al., 2018), and de novo genome assemblies were performed with Shovill v1.0.4 (Seemann, 2017). Genome identification and annotations were performed with Galaxy (https://usegalaxy.org/) as well as with RAST (https://rast.nmpdr.org/). Several general features of genomes such as transfer ribonucleic acid (tRNA) and ribosomal RNA (rRNA) were filtered and reported using the in-house script provided by the annotation tools. The annotated genes were then analyzed to determine their role in IAA production and in other plant growth-promoting pathways.
Whole-genome and pan-genome analysis
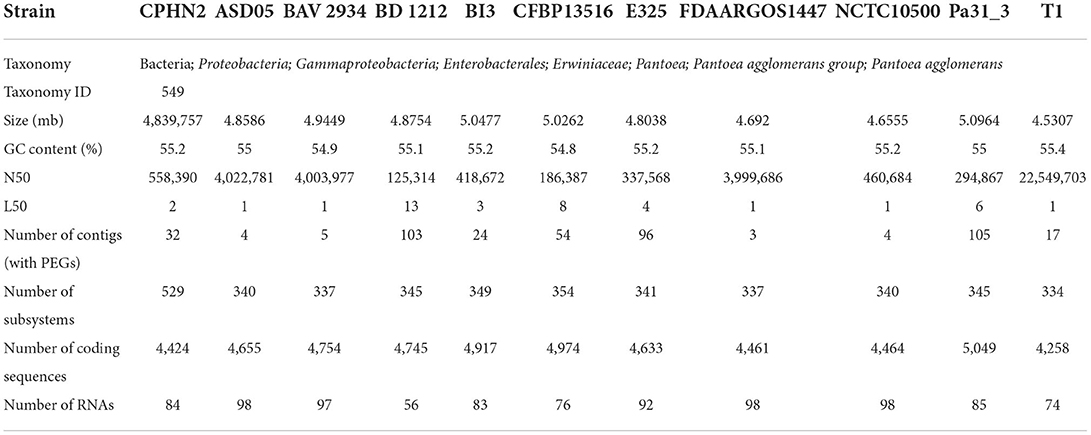
The genome of P. agglomerans CPHN2 was compared to 10 closely related genomes available in NCBI databases using 16s rRNA with WGS contigs. Several genomic features, such as genome size, gene number, GC content, and the number of tRNAs and rRNA, were compared. BRIG 0.95 was used to construct a pairwise genomic alignment of all the 11 strains and an out-group (Alikhan et al., 2011).
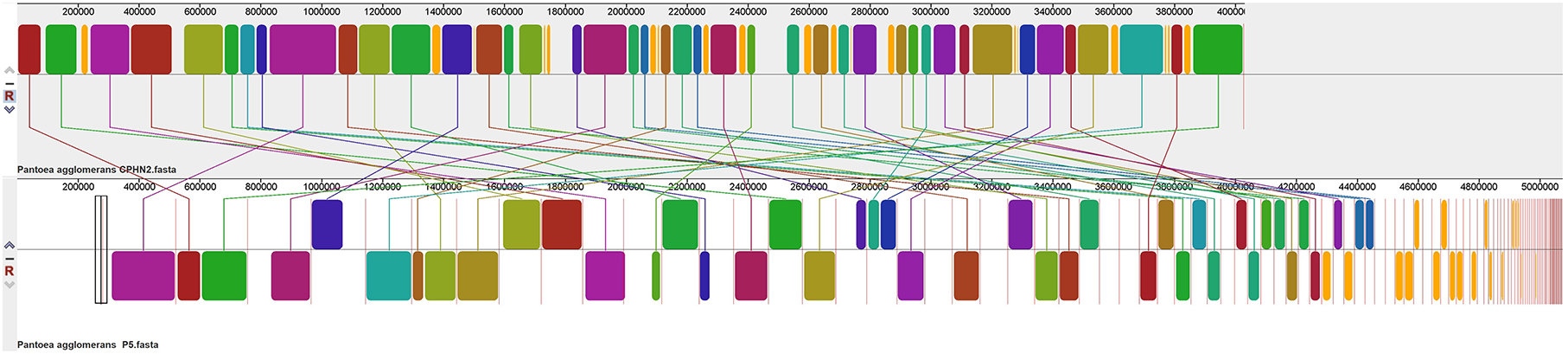
To study the rearrangement and alignment of genomes, P. agglomerans CPHN2 (draft genome) was compared with P. agglomerans FDAARGOS1447 (GenBank accession no. CP077366.1) using the Mauve program in Mauve v 2.3.1 (Darling et al., 2004). By taking the genome of P. agglomerans CPHN2 and its 10 neighbor strains, BPGA was used to perform pan-genome analysis (Chaudhari et al., 2016) to identify strain-specific and core genes (Shariati et al., 2017; Pinski et al., 2020).
Pathway analysis
To determine the presence of specific pathways for plant growth-promoting traits, annotated and predicted gene sequences were evaluated. The ascribed gene functions were manually analyzed from previously reported studies (Glick, 2012; Ahemad and Kibret, 2014; Ke et al., 2016; Liu et al., 2016; Dudeja et al., 2021) and compared with the relevant pathways available in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway databases (https://www.genome.jp/kegg/pathway.html).
Results
P. agglomerans CPHN2 sequencing statistics
Illumina NextSeq500 technology was used to sequence the whole genome of P. agglomerans CPHN2, which resulted in 100-fold coverage. In total, 9,731,611 PE reads with a minimum length of 100 bp were filtered for low-quality sequences (reads with more than 10% quality threshold (QV) 20 Phred score) and ambiguous reads (reads with unknown nucleotides “N” >5%) (Supplementary Table 1). Finally, in the downstream analysis, a total of 2,898,434,450 bp were used. The reads were assembled, and the total length of 4,839,532 bp was generated. In the final assembly, a total of 32 contigs were obtained, each with a total of 4,424 coding sequences (CDS), including N50 of size 558,390 bp (Supplementary Table 2).
Genomic characteristics of P. agglomerans CPHN2 and comparative genomic analysis
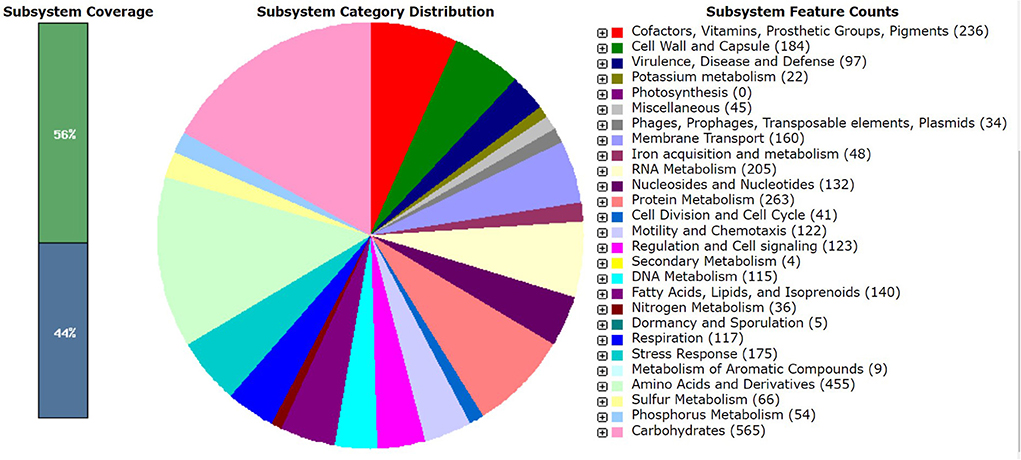
Pantoea agglomerans CPHN2 has a single circular chromosome approximately 4.8 Mbp long, with a GC content of 55.2% and two plasmids. RAST (https://rast.nmpdr.org/) was used to predict a total of 4,468 putative CDS (Figure 1). The chromosomal sequence predicted 71 tRNA-coding genes, one rRNA gene, and one CRISPR. The closely related genome of P. agglomerans FDAARGOS 1447 was used as the reference genome for preliminary comparative analysis based on 16S rRNA sequences. The ordered genome assembly of CPHN2 and strain FDAARGOS 1447 (GenBank accession no. CP077366.1) were compared with progressive Mauve from Mauvev 2.3.1 software. The genomic alignment identified 12 collinear blocks, as well as many inversion and rearrangement sites (Figure 2). By mapping most of the portions of the two genomes, large areas of high similarity were observed, indicating that the chromosome alignments of both strains are approximately identical. However, a region in the chromosome scaffolding between contigs 4–9 and 11 has an inverted orientation, indicating differences in their synteny relationships. In addition, the genomic features of P. agglomerans CPHN2 and 10 closely related genomes were analyzed based on genome size, the number of genes, the predicted CDS, GC content distribution, and the number of tRNA and rRNA genes (Figures 3A,B, Table 1). The genome size of CPHN2 was lower than that of the strains BI3, Pa31 3, and CFBP13516 with a genome size of more than 5 Mb. Of these, Pa31_3 has approximately the genome size of 5.0964 Mb, which is relatively large among all genomes. The draft genome of CPHN2 has 83 unique CDS (Figures 4A,B).
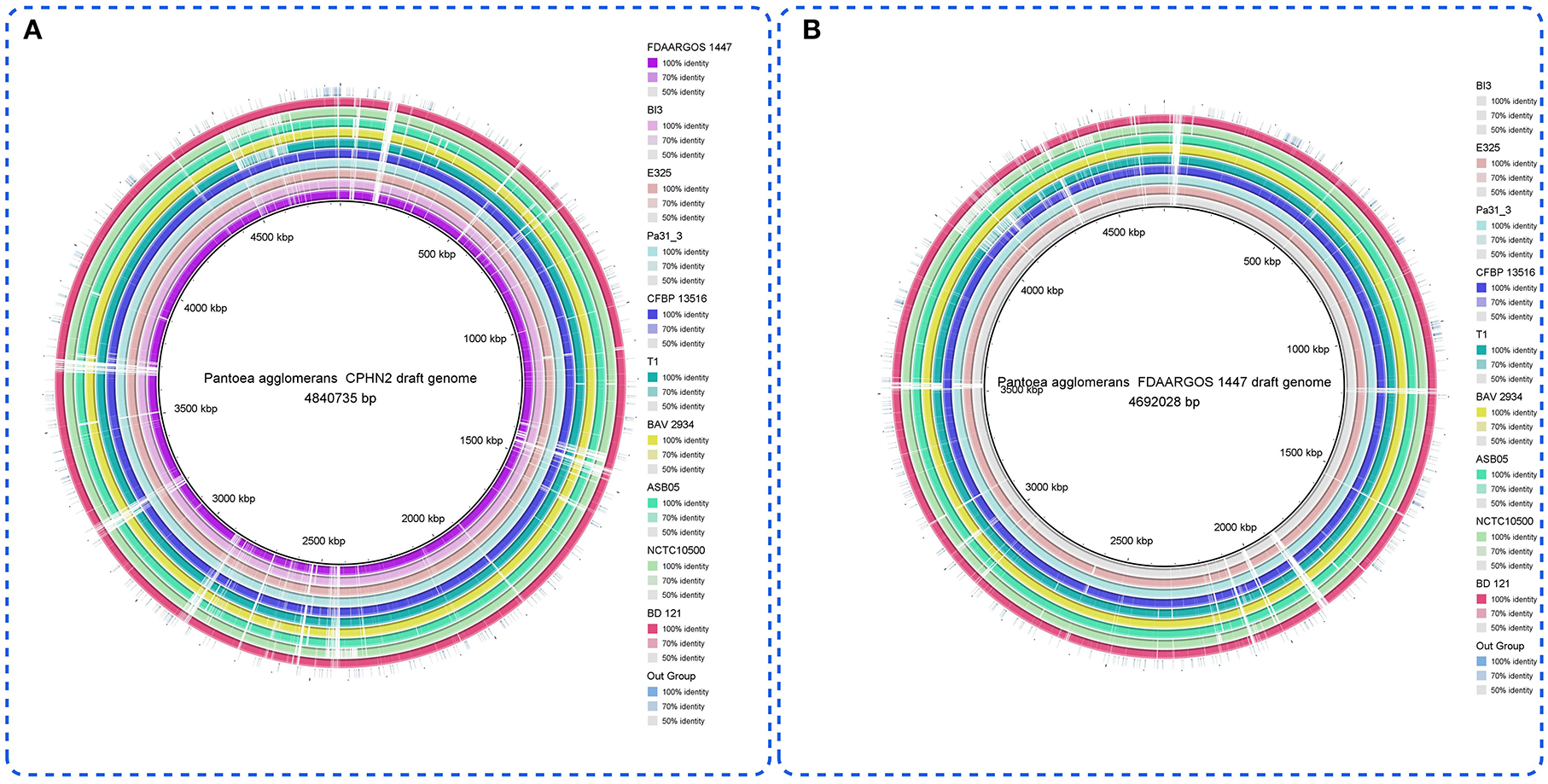
Figure 3. (A) Genome comparisons of different P. agglomerans strains to the drafted CPHN2 genome created by BRIG 0.95. The inner cycle (back) reflects the whole genome of the reference strain CPHN2, and the shade of each color indicates the similarities between all strains with the strain CPHN2. (B) With genome comparisons of other P. agglomerans strains except for CPHN2 vs. the reference genome (FDAARGOS 1447), the circular map depicts the full-genome comparison of the strain FDAARGOS 1447 against the other 10 sequenced P. agglomerans and an out group.
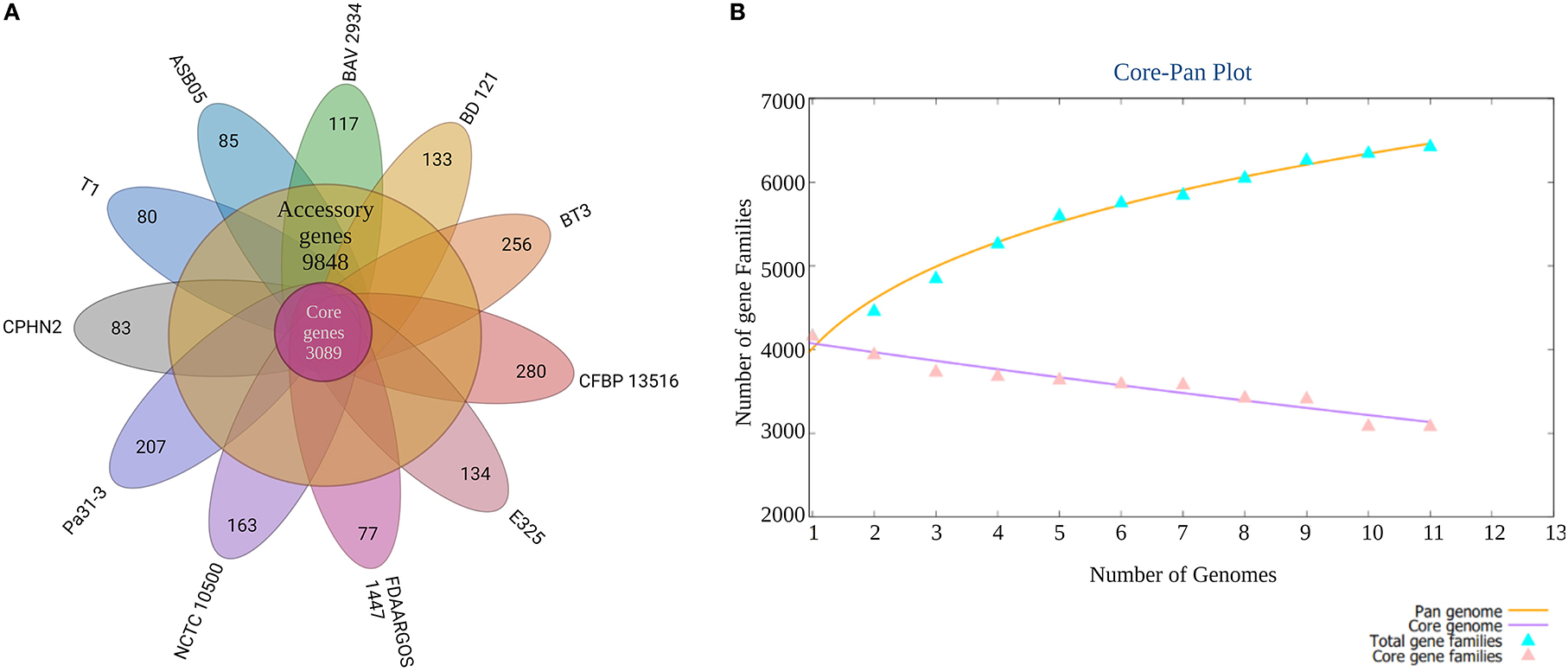
Figure 4. A plot of the core/pan-genome of P. agglomerans strains and a Venn diagram of the core-genome and strain-specific coding sequences (CDS). (A) The number of distinct CDS for each strain of the pan-genome of P. agglomerans. (B) Genome comparisons using n = 11 genomes were performed to identify the core-genome and strain-specific CDS for the 11 P. agglomerans strains sequenced.
Intra-species phylogenetic tree analysis
The phylogenetic relationships of P. agglomerans CPHN2 and 10 other P. agglomerans strains were shown separately based on the pan genome (Figure 5A) and the core genome (Figure 5B). This analysis depicted P. agglomerans strain CPHN2, genetically close to the NCTC10500 and T1 genomes, clustered in the same clade based on the pan-genome. The analysis also revealed a distant phylogenetic relationship between the strain FDAARGOS 1447 and other strains. In a core-genome-based phylogenetic study, CPHN2 did not cluster in any clade, while FDAARGOS 1447 clustered along with BI3 and CFBP13516 (Figure 5).
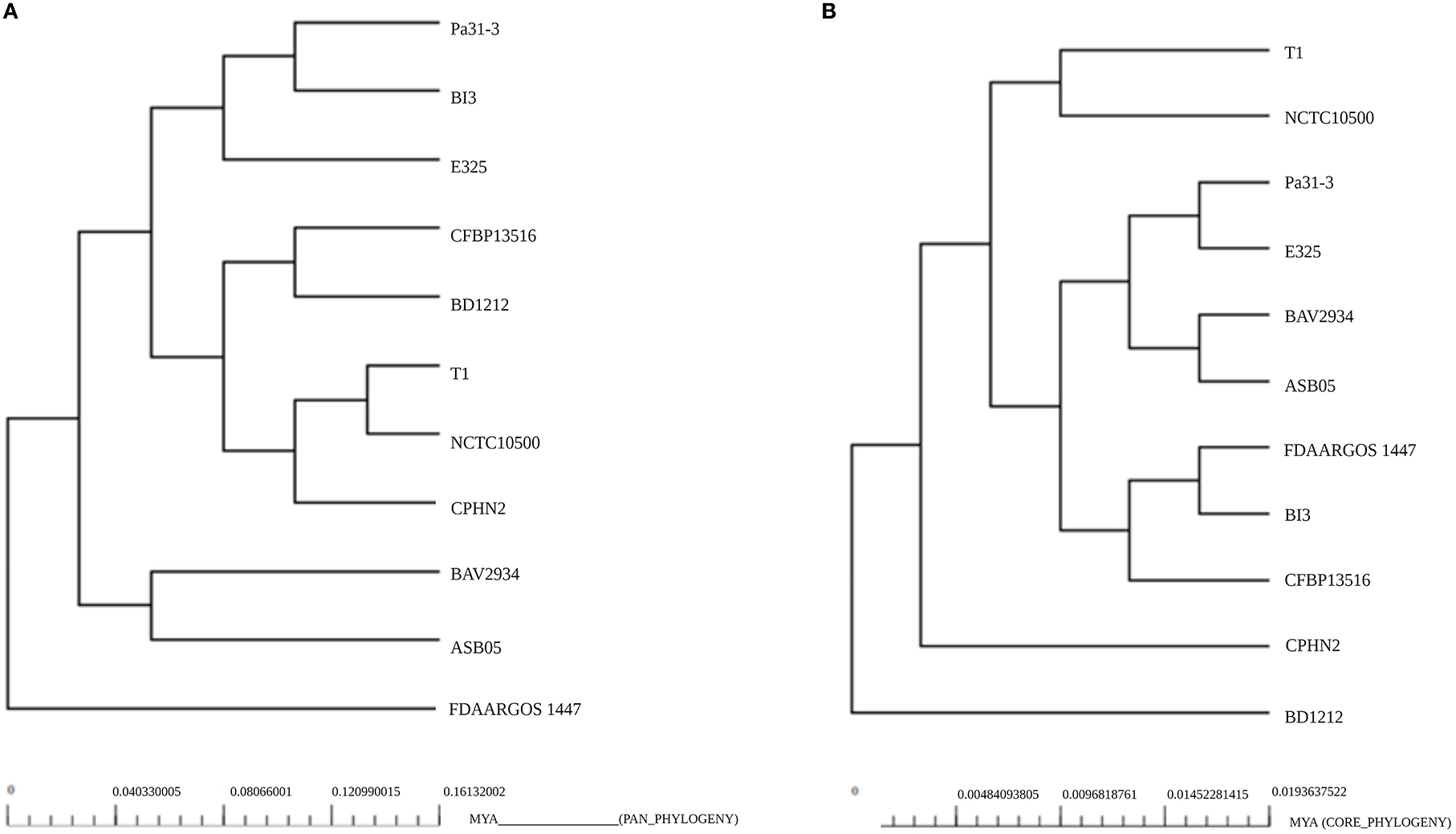
Figure 5. Phylogeny of the P. agglomerans strains based on the analysis of (A) pan-genomes and (B) core genomes.
Carbohydrate metabolism
The CPHN2 genome comprises a number of carbohydrate metabolism subsystems that encompass around 12.7% of the coding area. The genome has a number of metabolic pathways for carbohydrate metabolisms such as glycolysis, gluconeogenesis, Entner–Doudoroff, the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway, and acetyl CoA fermentation. It also has genes that metabolize the amount of other sugars and their derivatives (Supplementary Table 3).
Genes involved in plant growth stimulation
Genes involved in the solubilization of phosphates and in the production of organic acids, siderophores, ammonia, and indole acetic acid (IAA) have been annotated and identified in the P. agglomerans CPHN2 genome (Glick, 2012; Ahemad and Kibret, 2014; Liu et al., 2016).
Phosphate and organic acids
It is well-known that, after being applied as a fertilizer, a significant portion of inorganic phosphates is immobilized, leaving the phosphate unavailable to plants (Xie et al., 2016). As a consequence, it is crucial for some bacterial species to produce acid phosphatases and organic acids, notably gluconic acid (GA), and to solubilize insoluble or poorly soluble mineral phosphates (Rodríguez et al., 2007). GA is a kind of organic acid that aids in converting the immobilized mineral phosphates that contain phosphorus into a biologically available form. Glucose-1-dehydrogenase (gcd) along with its cofactor, pyrrolo-quinolone quinine (pqq), catalyzes the synthesis of GA. Gluconate dehydrogenase is another enzyme that also helps in the production of GA and its conversion to 2-ketogluconate (Ramachandran et al., 2006; Eastman et al., 2014). CPHN2 genomic annotation showed the existence of a number of GA and cofactor genes, including pqqABCDE (Eastman et al., 2014).
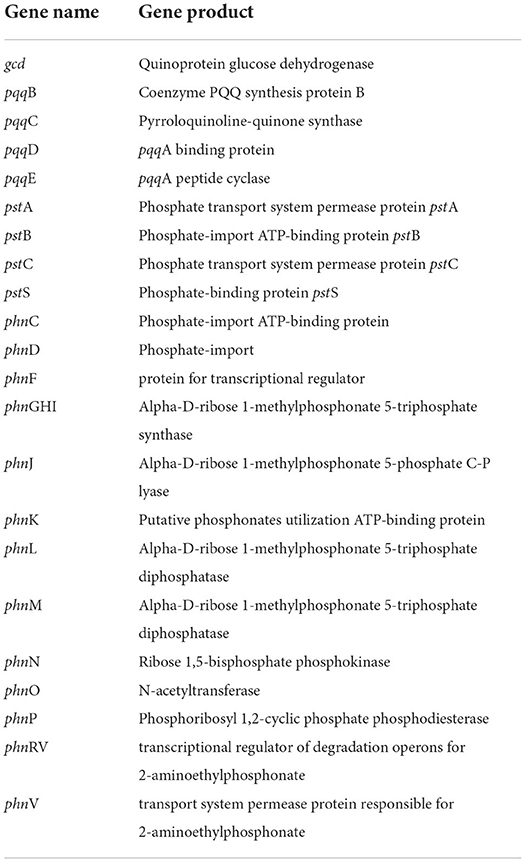
Phosphonates, which are organophosphorus molecules, have a direct carbon–phosphorus (C–P) bond instead of a typical C–O–P linkage that is often observed in phosphate esters (C–O–P) (Parker et al., 1999). Bacterial breakdown of phosphonates results in the release of physiologically available phosphates, which are controlled by the phosphonate-related phn gene cluster. CPHN2 has numerous phn genes, including phnCDFGHIJKLMNOPRV, which catalyze the hydrolysis of phosphonate into phosphate and an alkane, according to genomic analysis. Phosphorus compounds must be transported across the plasma membrane before they may be used. Two high-affinity phosphate transport systems, pstBAC (phosphate transporter) and phnDC (phosphate dehydrogenase), have also been observed in the CPHN2 genome (phosphonate transporter) (Figure 6, Table 2, Supplementary Table 4).
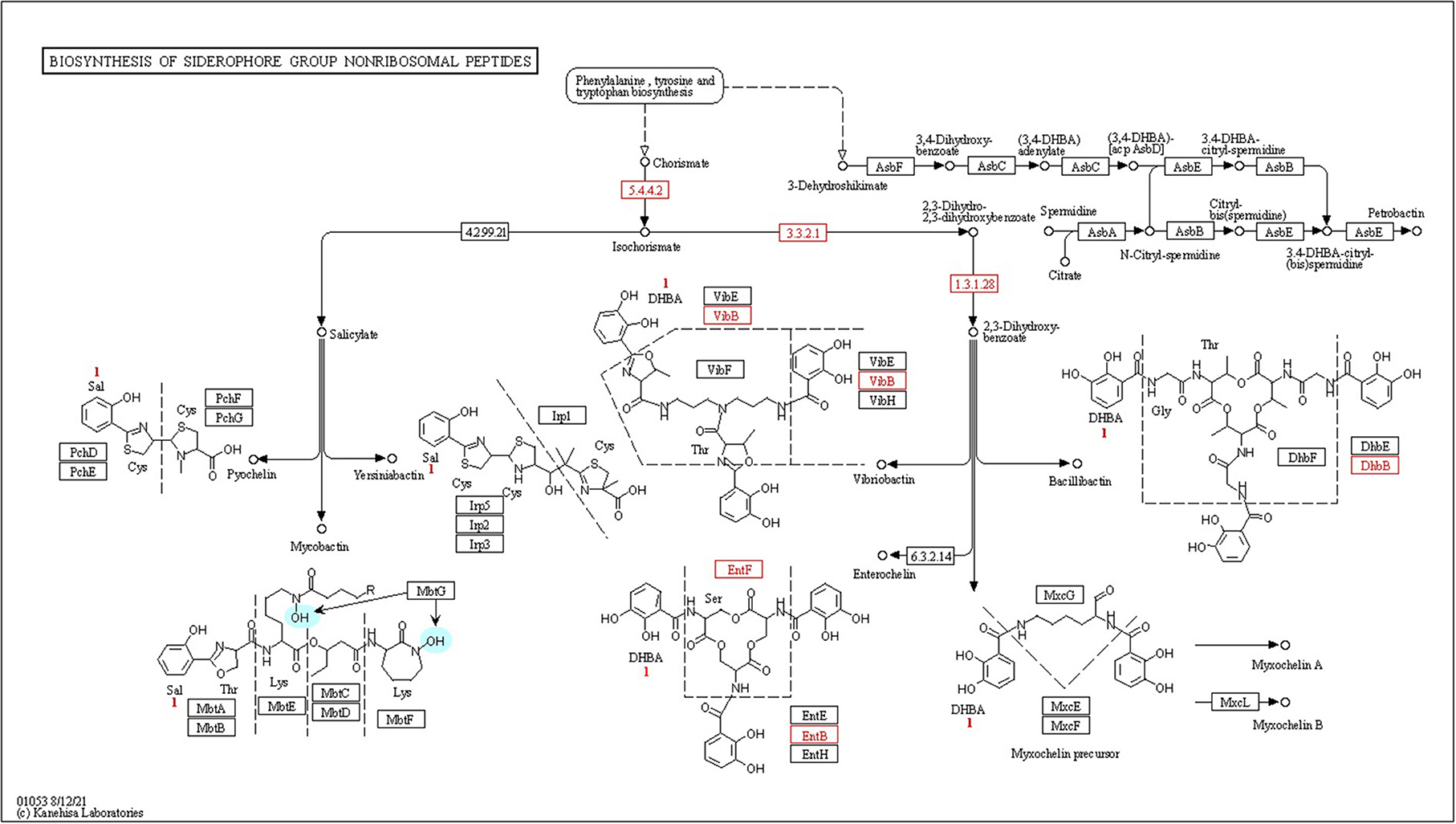
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene mapping of the strain CPHN2 for siderophore synthesis.
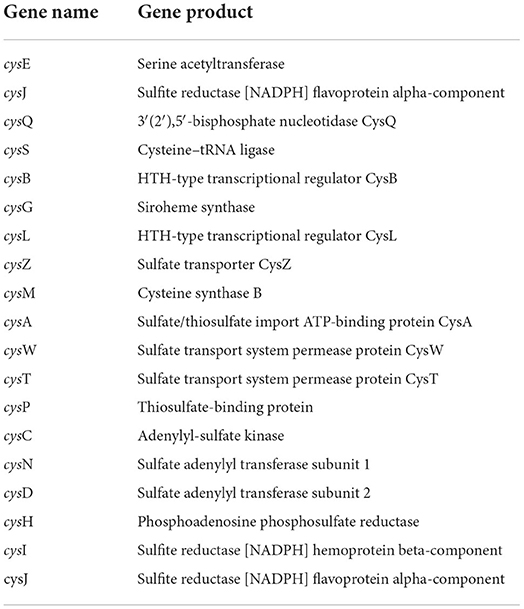
Hydrogen sulfide (H2S) has emerged as an essential chemical for phosphate solubilization. H2S interacts with ferric phosphate to produce ferrous sulfate and release phosphate. The CPHN2 genome encodes a collection of genes involved in H2S production (Table 3), including a cluster of cys genes involved in reduction, transportation, protein binding, and acetyl and adenyl group transfer. The CPHN2 genome also has genes related to sulfur metabolism: mineralization and transport of sulfite, sulfate, and phosphoadenosine phosphosulfate.
Siderophores
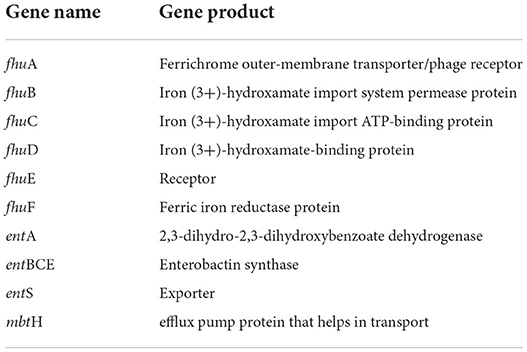
High-affinity iron chelating compounds have been shown to be produced mostly by plant-associated bacterial strains (de Souza et al., 2015) and may assist them by collecting iron from the environment (Niazi et al., 2014). CPHN2 has the ability to synthesize an enterobactin siderophore, and the entABCEF genes for the same have been annotated. This siderophore is responsible for the recovery of iron through complex formation once it is exported from the cell by entS. P. agglomerans. CPHN2 also has many receptors for siderophores, mbtH, an efflux pump protein, which helps in transport, and fhuABCDEF, which helps in transport and binding (Figure 6, Table 4, Supplementary Table 4).
Nitrogen metabolism
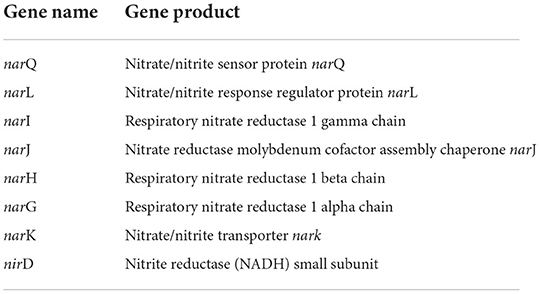
Ammonia production, another important trait of PGPEB, indirectly promotes plant growth and biomass accumulation. Our genomic study has shown that CPHN2 includes narP, narJ, and narL, which converts atmospheric nitrogen to nitrite, as well as nirB and nirD, nitrite to nitrate, and nrtA is responsible for nitrate transportation (Figure 7, Table 5, Supplementary Table 4).
Indole-3-acetic acid
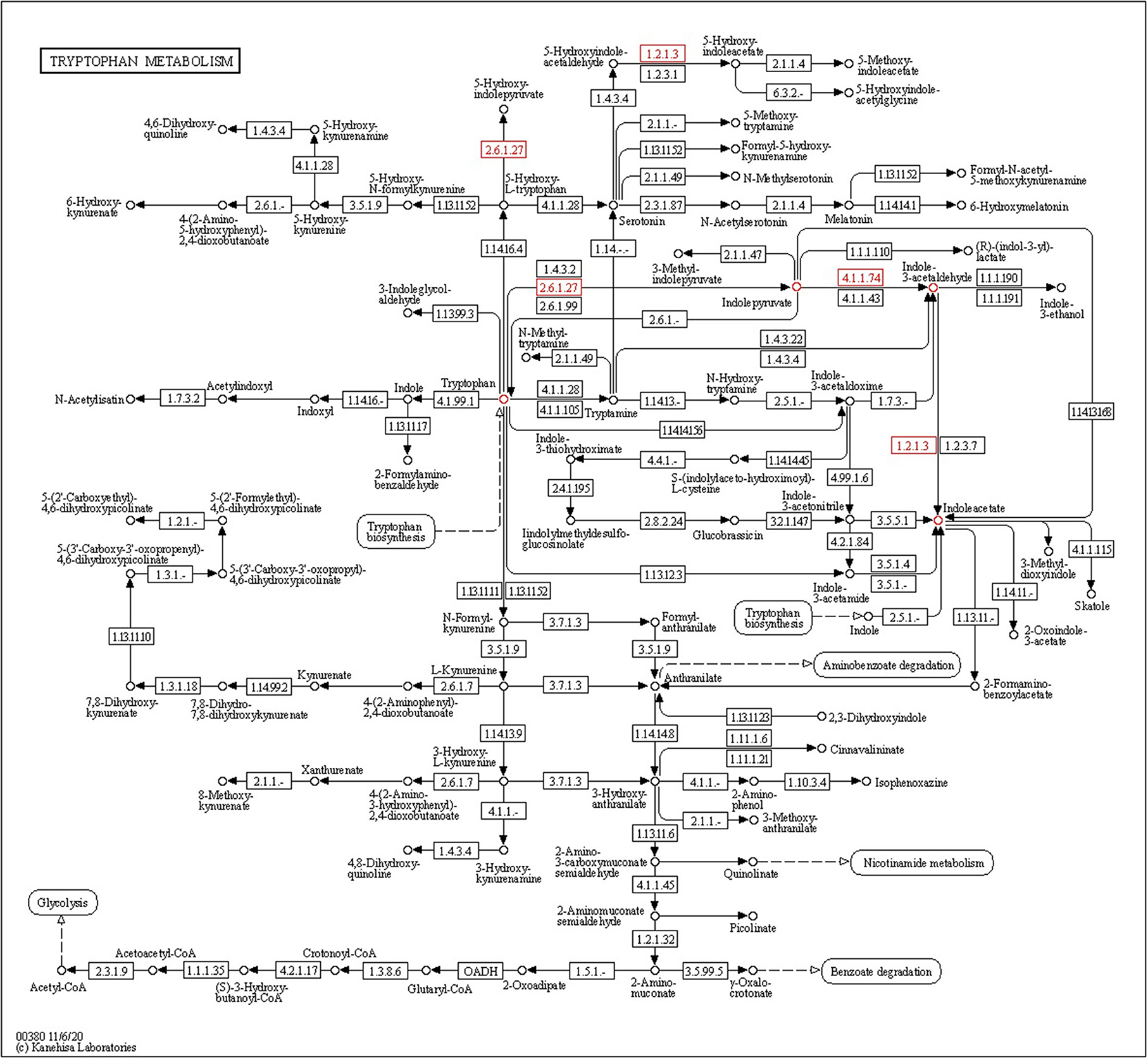
Endophytic bacteria produce indole-3-acetic acid (IAA), which is necessary for plant development processes and play an important role in plant–microbe interactions. Four tryptophan-dependent pathways, namely, indole-3-acetamide (IAM), indole-3-pyruvic acid (IPA), tryptamine (TAM), and indole-3-acetaldoxime (IAOx), are involved in the biosynthesis of IAA present in bacteria. The indole-3-pyruvate decarboxylase (ipdC) gene is present in CPHN2 and is involved in the conversion of indole-3-pyruvate to IAA in the IPA pathway. The iaaM and iaaH genes were involved in the synthesis of IAA using the IAM pathway. Of these, only the iaaH gene is present in the CPHN2 genome. These data show that IPA is the sole route for the synthesis of IAA using this strain (Figure 8, Supplementary Table 4).
Chemotaxis, motility, and attachment of genes
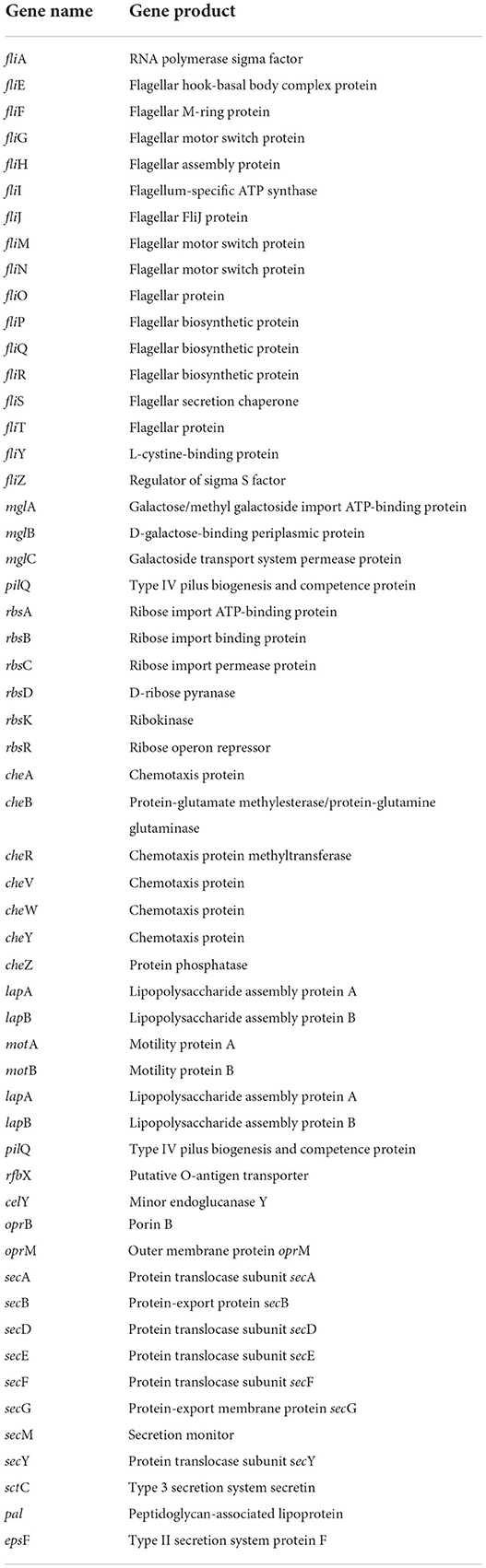
In the whole genome of CPHN2, genes, such as the cluster of fli gene (fliAEFGHIJMNOPQRSTYZ), sec gene (secABDEFGMY), mgl gene (mglABC), rbs gene (rbsABCDKR), che gene (cheABRVWYZ), lapAB, motAB, lapAB, pilQ, rfbX, oprBM, pal, epsF, and sctC, responsible for endophytic behavior, like chemotaxis movement and attachment to the host, have been annotated (Table 6, Supplementary Table 4).
Discussion
Plant-associated endophytic microbes have evolved unique biosynthetic pathways to aid in their interactions with their host. These different metabolites of endophytic origin promote plant growth; therefore, their genomic studies recently gained substantial interest (Patel et al., 2016; Kandel et al., 2022). In our previous study, we isolated non-rhizobial endophytic bacteria P. agglomerans CPHN2, which possessed multiple plant growth-promoting traits such as the production of ammonia, siderophores, and IAA, and phosphate solubilization (Maheshwari et al., 2019) and produced a high amount of IAA under optimized conditions (Unpublished). Genomic mining of their genes might help us develop commercial bioinoculants so as to reduce the use of chemical fertilizers. The presence of these positive traits in P. agglomerans CPHN2 prompted whole-genome sequencing and genomic annotations. The genome of P. agglomerans CPHN2 was also compared with the already reported genomic sequences of 10 strains to unravel the similarities between the two. A phylogenetic tree constructed based on the pan-genome showed that P. agglomerans CPHN2, NCTC10500, and T1 belonged to the same clade (Figure 4). This indicates that these isolates share a large set of similar genes. Core-genome-based phylogenetic tree shows that these two strains NCTC10500 and T1 belong to the same clade but CPHN2 has a separate niche. The closely related FDAARGOS 1447 was present in a separate niche upon the pan-genome analysis and in a clade upon the core-genome analysis.
Genomic mining of CPHN2 supported a previous study on its ability to possess multiple PGP traits and promote plant growth (Maheshwari et al., 2019). Analysis of CPHN2 provided clues to the presence of specific genes for phosphate solubilization, nitrogen metabolism, and the production of siderophores and IAA. PGPR promotes plant growth by solubilizing the phosphate mineral and making it accessible to plants. They employed different solubilization mechanisms, either through the production of extracellular enzymes or by the release of organic acids, protons, and hydroxyl ions. Phosphate-solubilizing bacteria such as Bacillus, Pseudomonas, and Pantoea are reported to release GA most commonly (Rodríguez et al., 2007). It has also been reported that Enterobacter asburiae mutants, which lack glucose dehydrogenase (GDH) activity, were unable to release phosphate (Gyaneshwar et al., 1999). Analysis of the CPHN2 genome revealed the presence of the genes gcd and pqq responsible for the synthesis of GA and the solubilization of inorganic mineral phosphates.
Microorganisms have also evolved mechanisms to use other sources of phosphorous, such as phosphonates, during phosphate starvation. Enzymes involved in phosphonate catabolic pathways are conserved and encoded by ortholog genes in bacteria. The strain CPHN2 has phn genes like phnCDFGHIJKLMNOPRV responsible for phosphate solubilization. phnCD is a gene for phosphate-import protein, phnF is a gene for transcriptional regulation, phnGHILMNOP is a gene for the involvement of enzymes in the phosphonate degradation pathway, and phnRV is a gene for transcriptional regulation of degrading operons, and phnV is a gene for permease protein. The strain CPHN2 also has pstABCS genes, which are responsible for phosphate binding, import, and transportation. Such similar clusters have also been reported for phosphonate binding, transport, and degradation in Escherichia coli and P. agglomerans strain P5 (Metcalf and Wanner, 1993; Shariati et al., 2017).
Siderophores are low molecular weight compounds, which are produced by a large number of bacterial endophytes to scavenge iron under iron-destitute state. Siderophore production is an important plant growth-promoting trait as it helps the microbes to obtain iron, which in turn is directly transferred to plants and indirectly suppresses the growth of phytopathogens. In addition to this, genes associated with the transport and production of siderophores are also present. fhuABCDEF genes are responsible for binding and transport, entA gene is for 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, entBCE gene is for Enterobactin synthase, entS gene is an exporter, and mbtH gene is an efflux pump protein that helps in transport. Along with this, we were also able to find out the cytoplasmic membrane and periplasmic proteins involved in the uptake of siderophores (Stevens et al., 1999) and tonB-dependent outer-membrane receptors are coded for by the fhuDCB operon in Rhizobium (Shariati et al., 2017). In addition to this, CPHN2 is also capable of carrying out dissimilatory nitrate reduction. It is a two-step process in which, during anoxic conditions, nitrate acts as an electron acceptor and is converted to ammonia. The main enzymes involved are nitrate reductase (encoded by narGHIJ) and nitrite reductase (nirD).
Several endophytic bacterial genera are known to produce phytohormones, the most common of which is IAA. Multiple pathways have been used by both plants as well as bacteria for the biosynthesis of IAA (Spaepen et al., 2007). There are several pathways in which the amino acid tryptophan is a direct precursor of IAA and modulates the level of IAA synthesis (Zaidi et al., 2009). Among all these Trp-dependent pathways, IAM and IPA pathways are widespread among bacteria (Mano and Nemoto, 2012). The IAM pathway is common among phytopathogens such as Pseudomonas syringae, Agrobacterium tumefaciens, and Agrobacterium rhizogenes (Szkop and Bielawski, 2013), while most PGPRs such as Pseudomonas putida, Enterobacter cloacae, and Azospirillum brasilense use the IPA pathway for the biosynthesis of IAA. The IPA pathway has been well-characterized in bacteria, and the conversion of indole-3-pyruvate to indole-3-acetaldehyde by ipdC is the rate-limiting step (Zhao, 2010). The expression of the key gene ipdC has been found to increase in P. agglomerans while growing on the surfaces of plants, depicting an interesting case of interaction between plants and microbes (Jasim et al., 2014). This study also aimed to identify the complete IAA biosynthetic pathways in CPHN2. Genomic mining revealed the presence of all the genes, including the involvement of the key gene ipdC in the IPA pathway. A previous study on the genomic study of P. agglomerans strain P5 revealed that IAM was the only pathway for the production of IAA (Shariati et al., 2017). Meanwhile, our CPHN2 has an incomplete IAM pathway in which the iaaH gene is present.
Endophytic colonization is a conglomerate of processes such as entry to the host plant, growth on to the plant, and subsequent multiplication there. Root exudates are various chemicals, ranging from organic acids and amino acids to sugars, which help in the recruitment of bacterial endophytes from a pool of rhizospheric bacteria. According to our annotation, CPHN2 contains the cluster of FLI proteins, which are responsible for the production of the flagellar protein and motor system. It also has a cluster of che, mgl, and rbs genes, responsible for galactose and ribose binding, transport, and chemotaxis, as previously reported (Harayama et al., 1983; Barroga et al., 1996). In addition, the Sec gene cluster responsible for protein translocation, export, and monitoring was also found to be present.
The lip, pil, rfb, cel, opr, sct, and pal genes were also annotated in CPHN2. Previous studies reported the role of these genes in chemotaxis, motility, adhesion, pilli formation, dehydratase synthesis, and porin creation in the outer membrane and in the formation of exopolysaccharides (EPSs). Recent studies demonstrated that endophytic bacteria frequently secrete EPSs for their adherence to the root surface (Dudeja et al., 2021). EPS also decreases the concentration of free radicals in the plant cell wall and protects cells against oxidative stress. CPHN2 also carries the gene for the production of EPS.
As members of the genus Pantoea were previously recognized as phytopathogens causing wilting and necrosis, we also analyzed the presence of the type III secretion system (T3SSs) coded by the hrc/hrp gene cluster responsible for pathogenicity. A large number of Gram-negative bacteria use this secretion system to convey effector molecules directly to the cytosol of their host. Several studies reported the role of the whole hrc/hrp gene cluster in pathogenicity and hypersensitive response in plants (Buonaurio et al., 2015; Shariati et al., 2017). However, CPHN2 does not contain the complete gene cluster and lacks pathogenicity. The study also revealed the occurrence of only one pathway, i.e., IPA pathway, associated with useful bacteria for the production of IAA, out of the five pathways reported in bacteria. Therefore, this study provides in-depth information about genomic pathways and helps in understanding the mechanisms of PGP by endophytic bacteria.
Conclusion
Genomic analysis of P. agglomerans CPHN2 revealed the presence of several genes engaged in PGPs, such as phosphate solubilization and nitrate reduction, and in the production of siderophores and IAA. Moreover, it has genes related to chemotaxis, motility, adhesion, and pilli formation, which are required for effective colonization. It lacks the complete pathogenic gene cluster. This research provides insights into the plant growth-promoting ability of CPHN2 and highlights its potential as a biofertilizer. Therefore, this strain is deemed safe and can be further explored as a bioinoculant. However, more research is needed to provide a detailed interpretation of the functions and the regulation of these genes.
Data availability statement
The whole genome of P. agglomerans CPHN2 has been deposited in GenBank under Accession nos. CP098412, CP098413 for plasmids, and CP098414 for genome and is annotated by a prokaryotic genomic annotation pipeline (PGAP) (Kumar et al., 2022) (Tatusova et al., 2016; Kumar et al., 2022). Bio Project accession number PRJNA811747 (Sequence Read Archive Accession No. SRR18189611) (https://www.ncbi.nlm.nih.gov/sra/SRR18189611).
Author contributions
PK: data curation, investigation, methodology, software, validation, visualization, and writing–original draft. SR and PD: formal analysis, writing, and software analysis. AK, AD, and PS: conceptualization, funding acquisition, supervision, validation, and writing–review and editing. All authors contributed to the article and approved the submitted version.
Acknowledgments
PK, SR, and PD would like to acknowledge the Council of Scientific and Industrial Research, New Delhi, India, for the CSIR fellowship. PS acknowledges support from the Department of Science and Technology (DST), Government of India, New Delhi. The authors would like to acknowledge the Department of Science and Technology, Government of India, for providing a FIST grant (Grant No. 1196 SR/FST/LS-I/2017/4) to the Department of Microbiology, MDU, Rohtak.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.998821/full#supplementary-material
References
Agri, U., Chaudhary, P., and Sharma, A. (2021). In vitro compatibility evaluation of agriusable nanochitosan on beneficial plant growth-promoting rhizobacteria and maize plant. Natl. Acad. Sci. Lett. 44, 555–559. doi: 10.1007/s40009-021-01047-w
Agri, U., Chaudhary, P., Sharma, A., and Kukreti, B. (2022). Physiological response of maize plants and its rhizospheric microbiome under the influence of potential bioinoculants and nanochitosan. Plant Soil 474, 451–468. doi: 10.1007/s11104-022-05351-2
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 1–10. doi: 10.1186/1471-2164-12-402
Andrews, S. (2010). Data from: FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=17.+Andrews+S.+2010.+FastQC%3A+a+quality+control+tool+for+high+throughput+sequence+data.+http%3A%2F%2F (accessed April 4, 2022).
Barroga, C. F., Zhang, H., Wajih, N., Bouyer, J. H., and Hermodson, M. A. (1996). The proteins encoded by the rbs operon of Escherichia coli: I. Overproduction, purification, characterization, and functional analysis of RbsA. Protein Sci. 5, 1093–1099. doi: 10.1002/PRO.5560050611
Buonaurio, R., Moretti, C., Da Silva, D. P., Cortese, C., Ramos, C., and Venturi, V. (2015). The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front. Plant Sci. 6, 434. doi: 10.3389/fpls.2015.00434
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 1–10. doi: 10.1038/srep24373
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chi, H., Liu, C., Yang, H., Zeng, W. F., Wu, L., Zhou, W. J., et al. (2018). Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 36, 1059–1061. doi: 10.1038/nbt.4236
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394. doi: 10.1101/gr.2289704
de Souza, R., Ambrosini, A., and Passaglia, L. M. P. (2015). Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 38, 401–419. doi: 10.1590/S1415-475738420150053
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek. 106, 85–125. doi: 10.1007/s10482-013-0095-y
Duca, D. R., and Glick, B. R. (2020). Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Appl. Microbiol. Biotechnol. 104, 8607–8619. doi: 10.1007/s00253-020-10869-5
Dudeja, S. S., Suneja-Madan, P., Paul, M., Maheswari, R., and Kothe, E. (2021). Bacterial endophytes: molecular interactions with their hosts. J. Basic Microbiol. 61, 475–505. doi: 10.1002/jobm.202000657
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Eastman, A. W., Heinrichs, D. E., and Yuan, Z. C. (2014). Comparative and genetic analysis of the four sequenced Paenibacillus polymyxa genomes reveals a diverse metabolism and conservation of genes relevant to plant-growth promotion and competitiveness. BMC Genomics 15, 1–22. doi: 10.1186/1471-2164-15-851
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012, 1–15. doi: 10.6064/2012/963401
Gyaneshwar, P., Parekh, L. J., Archana, G., Poole, P. S., Collins, M. D., Hutson, R. A., et al. (1999). Involvement of a phosphate starvation inducible glucose dehydrogenase in soil phosphate solubilization by Enterobacter asburiae. FEMS Microbiol. Lett. 171, 223–229. doi: 10.1111/j.1574-6968.1999.tb13436.x
Harayama, S., Bollinger, J., Iino, T., and Hazelbauer, G. L. (1983). Characterization of the mgl operon of Escherichia coli by transposon mutagenesis and molecular cloning. J. Bacteriol. 153, 408–415. doi: 10.1128/jb.153.1.408-415.1983
Jasim, B., Jimtha John, C., Shimil, V., Jyothis, M., and Radhakrishnan, E. K. (2014). Studies on the factors modulating indole-3-acetic acid production in endophytic bacterial isolates from Piper nigrum and molecular analysis of ipdc gene. J. Appl. Microbiol. 117, 786–799. doi: 10.1111/jam.12569
Kandel, P. P., Naumova, M., Fautt, C., Patel, R. R., Triplett, L. R., and Hockett, K. L. (2022). Genome mining shows ubiquitous presence and extensive diversity of toxin-antitoxin systems in pseudomonas syringae. Front. Microbiol. 12, 815911. doi: 10.3389/fmicb.2021.815911
Ke, M., Shen, H., Wang, L., Luo, S., Lin, L., Yang, J., et al. (2016). Identification, quantification, and site localization of protein posttranslational modifications via mass spectrometry-based proteomics. Adv. Exp. Med. Biol. 919, 345–382. doi: 10.1007/978-3-319-41448-5_17
Kukreti, B., Sharma, A., Chaudhary, P., Agri, U., and Maithani, D. (2020). Influence of nanosilicon dioxide along with bioinoculants on Zea mays and its rhizospheric soil. 3 Biotech 10, 345. doi: 10.1007/s13205-020-02329-8
Kumar, P., Chauhan, V., Dang, A. S., Kumar, A., and Suneja, P. (2022). Draft genome sequence of pantoea agglomerans CPHN2, a potential plant-growth-promoting endophyte. Microbiol. Resour. Announc. 11, e0019222. doi: 10.1128/mra.00192-22
Lee, J. E., Lee, B. J., Chung, J. O., Hwang, J. A., Lee, S. J., Lee, C. H., et al. (2010). Geographical and climatic dependencies of green tea (Camellia sinensis) metabolites: A 1H NMR-based metabolomics study. J. Agric. Food Chem. 58, 10582–10589. doi: 10.1021/jf102415m
Liu, W., Wang, Q., Hou, J., Tu, C., Luo, Y., and Christie, P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 26710. doi: 10.1038/srep26710
Maheshwari, R., Bhutani, N., Bhardwaj, A., and Suneja, P. (2019). Functional diversity of cultivable endophytes from Cicer arietinum and Pisum sativum: bioprospecting their plant growth potential. Biocatal. Agric. Biotechnol. 20, 101229. doi: 10.1016/j.bcab.2019.101229
Mano, Y., and Nemoto, K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63, 2853–2872. doi: 10.1093/jxb/ers091
Metcalf, W. W., and Wanner, B. L. (1993). Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129, 27–32. doi: 10.1016/0378-1119(93)90692-V
Navarro-Torre, S., Barcia-Piedras, J. M., Mateos-Naranjo, E., Redondo-Gómez, S., Camacho, M., Caviedes, M. A., et al. (2017). Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant Biol. 19, 249–256. doi: 10.1111/plb.12521
Niazi, A., Manzoor, S., Asari, S., Bejai, S., Meijer, J., and Bongcam-Rudloff, E. (2014). Genome analysis of Bacillus amyloliquefaciens Subsp. plantarum UCMB5113: a rhizobacterium that improves plant growth and stress management. PLoS ONE 9, e104651. doi: 10.1371/journal.pone.0104651
Parker, G. F., Higgins, T. P., Hawkes, T., and Robson, R. L. (1999). Rhizobium (Sinorhizobium) meliloti phn genes: characterization and identification of their protein products. J. Bacteriol. 181, 389–395. doi: 10.1128/JB.181.2.389-395.1999
Patel, R. R., Thakkar, V. R., and Subramanian, R. B. (2016). Simultaneous detection and quantification of phytohormones by a sensitive method of separation in culture of Pseudomonas sp. Curr. Microbiol. 72, 744–751. doi: 10.1007/s00284-016-1012-1
Pinski, A., Zur, J., Hasterok, R., and Hupert-Kocurek, K. (2020). Comparative genomics of Stenotrophomonas maltophilia and Stenotrophomonas rhizophila revealed characteristic features of both species. Int. J. Mol. Sci. 21, 4922. doi: 10.3390/ijms21144922
Ramachandran, S., Fontanille, P., Pandey, A., and Larroche, C. (2006). Gluconic Acid: Properties, Applications and Microbial Production. Food Technol. Biotechnol. 44. Available online at: https://www.researchgate.net/publication/228342155_Gluconic_Acid_Properties_Applications_and_Microbial_Production (accessed April 5, 2022).
Rani, S., Kumar, P., and Suneja, P. (2021). Biotechnological interventions for inducing abiotic stress tolerance in crops. Plant Gene 27, 100315. doi: 10.1016/j.plgene.2021.100315
Rodríguez, H., Fraga, R., Gonzalez, T., and Bashan, Y. (2007). First International Meeting on Microbial Phosphate Solubilization. Available online at: https://link.springer.com/book/10.1007/978-1-4020-5765-6 (accessed April 4, 2022).
Seemann, T. (2017). Shovill: faster SPAdes assembly of Illumina reads (v0. 9.0). Available online at: https://github.com/tseemann/shovill
Shariati, V. J., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owilia, P., and Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7, 15610. doi: 10.1038/s41598-017-15820-9
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Stevens, J. B., Carter, R. A., Hussain, H., Carson, K. C., Dilworth, M. J., and Johnston, A. W. B. (1999). The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: FhuDCB are adjacent to a pseudogene version of fhuA. Microbiology 145, 593–601. doi: 10.1099/13500872-145-3-593
Szkop, M., and Bielawski, W. (2013). TyrB-2 and phhC genes of Pseudomonas putida encode aromatic amino acid aminotransferase isozymes: evidence at the protein level. Amino Acids 45, 351–358. doi: 10.1007/s00726-013-1508-y
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Verheggen, K., Martens, L., Berven, F. S., Barsnes, H., and Vaudel, M. (2016). Database search engines: paradigms, challenges and solutions. Adv. Exp. Med. Biol. 919, 147–156. doi: 10.1007/978-3-319-41448-5_6
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/femsre/fuv027
Xie, J., Shi, H., Du, Z., Wang, T., Liu, X., and Chen, S. (2016). Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 6, 21329. doi: 10.1038/srep21329
Zaidi, A., Khan, M. S., Ahemad, M., and Oves, M. (2009). Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol. Immunol. Hung. 56, 263–284. doi: 10.1556/AMicr.56.2009.3.6
Keywords: KEGG pathway, next-generation sequencing, Pantoea agglomerans, PGPEB, whole genome
Citation: Kumar P, Rani S, Dahiya P, Kumar A, Dang AS and Suneja P (2022) Whole genome analysis for plant growth promotion profiling of Pantoea agglomerans CPHN2, a non-rhizobial nodule endophyte. Front. Microbiol. 13:998821. doi: 10.3389/fmicb.2022.998821
Received: 20 July 2022; Accepted: 13 September 2022;
Published: 07 November 2022.
Edited by:
Vijay K. Sharma, Agricultural Research Organization (ARO), IsraelReviewed by:
U. M. Aruna Kumara, University of Colombo, Sri LankaParul Chaudhary, G. B. Pant University of Agriculture and Technology, India
Salma Mukhtar, Connecticut Agricultural Experiment Station, United States
Copyright © 2022 Kumar, Rani, Dahiya, Kumar, Dang and Suneja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pooja Suneja, cG9vamFwYXZpdEBnbWFpbC5jb20=
 Pradeep Kumar
Pradeep Kumar Simran Rani
Simran Rani Priyanka Dahiya
Priyanka Dahiya Ajit Kumar
Ajit Kumar Amita Suneja Dang
Amita Suneja Dang Pooja Suneja
Pooja Suneja