
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 October 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.997587
This article is part of the Research Topic Genetically Modified Organisms: Tricks or Threats, Volume II View all 4 articles
Advances in synthetic biology and the clinical application of bacteriotherapy enable the use of genetically engineered bacteria (GEB) to combat various diseases. GEB act as a small ‘machine factory’ in the intestine or other tissues to continuously produce heterologous proteins or molecular compounds and, thus, diagnose or cure disease or work as an adjuvant reagent for disease treatment by regulating the immune system. Although the achievements of GEBs in the treatment or adjuvant therapy of diseases are promising, the practical implementation of this new therapeutic modality remains a grand challenge, especially at the initial stage. In this review, we introduce the development of GEBs and their advantages in disease management, summarize the latest research advances in microbial genetic techniques, and discuss their administration routes, performance indicators and the limitations of GEBs used as platforms for disease management. We also present several examples of GEB applications in the treatment of cancers and metabolic diseases and further highlight their great potential for clinical application in the near future.
Despite the success of modern medical technologies in the prevention and treatment of most human diseases, the rapid increase in antibiotic-resistant microorganisms (Cassini et al., 2019) and chronic patient populations (Bernell and Howard, 2016; Bray et al., 2018; Blüher, 2019; Khan et al., 2020; Powell-Wiley et al., 2021), adverse effects caused by chemical drugs, exorbitant medical costs (Gheorghe et al., 2018; Lentz et al., 2019), cancers and other incurable diseases urgently require highly economic, convenient and efficient methods for addressing these issues. Increasing evidence demonstrates the links between the dysfunction of the human microbial community and the onset and development of many human diseases (Gilbert et al., 2016; Dang and Marsland, 2019), signifying the potential use of microorganisms as an alternative strategy to conquer these issues (Mazhar et al., 2020). Remarkably, bacteria account for over 90% of intestinal microbes, and advances in synthetic biology enable the precise manipulation of bacteria for diverse purposes (Lawson et al., 2019). For instance, technical improvement in Clustered regularly interspaced short palindromic repeats-cas9 (CRISPR-cas9; Li et al., 2020; Wang et al., 2020) and molecular biological methods (e.g., gene synthesis, DNA/RNA sequence, DNA transfection, clone techniques of large DNA fragment, etc.; Tavanti, 2022) greatly contribute to generating various engineered bacteria for producing many proteins and molecular compounds that originate from uncultivated microorganisms, fungi, archaea, microeukaryotes and eukaryotes (Çelik and Çalık, 2012; Gupta and Shukla, 2016; Charbonneau et al., 2020). Mounting evidence demonstrates that genetically engineered bacteria (GEB) can be orally or intravenously administered in clinical trials to cure different diseases (Liu et al., 2016; Zhang et al., 2018). Due to their special colonization ability in solid tumors, intestinal tracts, respiratory tracts, genital tracts, and the oral cavity, GEBs perform their activities by supplying active molecules, interfering with immune cells, restraining pathogenic bacteria, or killing tumor cells by expressing foreign genes or enhancing endogenous gene expression, thus preventing, diagnosing or curing diseases (Fan et al., 2019). Here, we systemically summarize the recent advances in bacteriotherapy, which uses GEBs as the main body, including the introduction of genetically engineered bacteria, engineering techniques, administration strategies, performance indicators, and biological safety. Finally, we describe the preclinical and clinical applications of GEBs and several probiotics in the treatment of cancers and metabolic diseases and discuss their limitations and prospects.
A GEB is defined as a bacterium with the ability to efficiently express heterologous proteins or molecular compounds for a specific purpose after genetic engineering. As early as the 19th century, Coley revealed the therapeutic effects of inactivated bacterial mixtures in sarcoma therapy (Coley, 1893). By the end of the 20th century, genetic engineering techniques have been widely used to modify bacteria to obtain the expected compounds. After decades of effort, numerous GEBs have been established for various applications in the food industry, disease treatment, chemical synthesis, environmental protection, etc. Notably, E. coli (Michael Schultz, 2008; He et al., 2017; Christofi et al., 2019; Terol et al., 2021), Lactobacillus (Yang et al., 2015; Lin et al., 2016; Chen et al., 2017; Oh et al., 2020), Salmonella (Chirullo et al., 2015; Zheng and Min, 2016; Li et al., 2017; Lim et al., 2017; Kawaguchi et al., 2018) are the most popular bacteria used as chassis tools for constructing different GEBs. In particular, more than 50 bacterial species, such as Bifidobacterium (Liu et al., 2018), Bacillus subtilis (Westbrook et al., 2016), Listeria monocytogenes (Selvanesan et al., 2022), and Lactobacillus brevis CD2 (He et al., 2017; Alfano et al., 2020), are being used in health care and scientific research (Figure 1).
To date, GEBs have made great achievements in the management of various diseases, such as infectious diseases, antibiotic-related diarrhea, allergies, and metabolic syndromes, in health care for daily life (Ma et al., 2017; Mazhar et al., 2020). Importantly, GEBs have more systematic and comprehensive therapeutic effects than traditional methods in the prevention and treatment of some diseases (Duan et al., 2015; Isabella et al., 2018; Kurtz et al., 2019) partly because the colonized microorganisms in the body are almost equal to tiny living factories that can autonomously reduplicate, detect abnormal homeostasis, produce therapeutic agents, and initiate self-destruction at a defined time (Pedrolli et al., 2019). Accordingly, GEBs have promising potential to be used at different stages of disease for different purposes. Genetically engineered bacterial vaccines, such as Salmonella, Vibrio cholera, Listeria, and Neisseria meningitidis, are capable of activating in vivo immune protective responses by introducing some effective antigens into low- or nonvirulence bacteria (Le et al., 2015; Herzog, 2016; Sharma et al., 2020). Using cholera as an example, the oral administration of attenuated Haiti V to rabbits efficiently restrained the colonization of wild vibrio cholera in the small intestine, thus reducing the incidence of Vibrio cholera infection (Hubbard et al., 2018; Satchell, 2018). Additionally, genetically engineered Lactobacillus succeeded in detecting Vibrio cholerae in stool samples via the specific phenomenon of “quorum sensing” in pathogenic microorganisms (Carignan et al., 2016; Mao et al., 2018). In cancer therapy, some bacteria, such as Salmonella, Clostridium novyi-NT, and E. coli, have a special ability to colonize solid tumors, enabling them to be excellent candidates for drug delivery and drug production (Wei et al., 2016). For example, transforming growth factor alpha-pseudomonas exotoxin-expressing S. typhimurium displayed significant inhibitory effects on the growth of CT26, MC38, and 4 T1 solid tumors (Lim et al., 2017). In melanoma therapy, recombinant S. typhimurium transfected with the interferon-gamma gene plasmid integrated into the N-terminal region (residues 1–160) of a surface immunogenic protein demonstrated obvious toxicity to cancer cells (Yoon et al., 2017). In addition, S. typhimurium loaded with CpG ODN and PD-1-siRNA induced innate immunity and inhibited PD-1 expression, thus killing cancer cells (Jia et al., 2021).
To date, the generally used GEBs are mainly confined to some intestinal diseases, such as inflammatory bowel disease (IBD; Saez-Lara et al., 2015) and cholera (Duan and March, 2010; Satchell, 2018). For instance, Lactobacillus transfected with interleukin 1 receptor antagonist was capable of reducing CD4+ IL-17A+ cells in mesenteric lymph nodes and blocking IL-1-cell signaling, thus alleviating the symptoms of acute colitis (Namai et al., 2020); E. coli Nissle 1917 (EcN) with the expression of cholera autoinducer 1 could restrain the virulence gene expression of Vibrio cholera and, thus, reduce its colonization in the gut (Duan and March, 2010). Essentially, the advantages of GEBs could be briefly summarized as follows: reduced cost of health care because of manufacturing scale-up and long-term effects of GEBs in colonization sites; decreased adverse effects, especially when they are orally administered; and for structurally unstable or environmentally sensitive compounds, free of drug purification and low-temperature storage. Notably, GEBs can produce multiple foreign proteins or compounds in one strain instead of requiring several drugs to achieve synergistic treatments (Peters et al., 2019).
Microbial genetic engineering uses genetic operation tools to shear, splice, and integrate the target genes and then introduce them into chassis cells. Thus, the recombinant genes are transferred into the desired products or endow the bacteria with new phenotypes. Due to the great progress in sequencing technologies and bioinformatics, a growing body of functional genes and gene clusters from nonculturable microorganisms are being excavated. How to express functional genes or gene clusters in a chassis cell is becoming a new research hotspot in the study of GEBs.
Indeed, GEB construction mainly includes the following two stages: upstream (functional gene acquisition) and downstream (heterogeneous expression). The acquisition of target genes or large gene clusters is an important part of the upstream stage of constructing an expected GEB (Cobb and Zhao, 2012). Due to the national microbial genome projects launched in multiple countries, numerous potential genes are available to be used for fabricating various GEBs. In fact, the size of the targeted genes determines the choice of MGE for GEB construction. The acquirement and modification of small gene fragments (<10 kb) can be performed using the general or long PCR method or direct DNA synthesis and restriction enzyme digestion (Fahnøe and Bukh, 2019). However, when the gene size exceeds 50 kb, some recombination methods, such as CRISPR–Cas9 and the Red/ET recombination system, are the optimal methods to alter, replace, delete or add bases or gene fragments of plasmids or genomes (Li and Elledge, 2012; Luo et al., 2016; Alberti and Corre, 2019; Strain-Damerell et al., 2021). Specifically, CRISPR–Cas9 can edit bacterial DNA fragments up to 100 kb in a single step in which RNA-guided Cas9 nuclease targets and cleaves DNA fragments, and the final large gene fragments are assembled via Gibson assembly (Jiang et al., 2015).
Cloning target genes or gene clusters into bacterial chassis includes gene transfer and genetic recombination using the techniques of transfection, transduction, conjugative transfer, lysogenic conversion, and protoplast fusion. The DNA size and property of the bacterial chassis determine the transfer methods. Heat-shock and electroporation transfection are widely used to transfer plasmids in E. coli, Salmonella, Bacillus thuringiensis, Pseudomonas aeruginosa, etc. Conjugative transfer and protoplast fusion often involve the transfer of plasmids from donor bacteria to recipient bacteria (Mukai et al., 2020). For instance, the conjugative type IV secretion system is synergistic with DNA-processing machinery termed the “relaxosome,” and a large extracellular tube termed the “pilus” is capable of orchestrating directional conjugated plasmid transfer (Waksman, 2019). Additionally, homologous recombination technologies, mainly including homologous recombination, site-specific recombination, transposable recombination and the CRISPR–Cas9 technique, enable the direct integration of target genes into the host chromosome in an expected strain. The typical homologous recombination methods require more than 1 kb homologous sequences to realize the recombination of target genes into the chromosomal genome (Beumer et al., 2013). Due to CRISPR–Cas9 technology, DNA operation has become much more efficient and accurate, which greatly benefits the extraction and heterologous expression of gene clusters in chassis cells, especially of those with a size of over 50 kb. Transposition (Martínez-García and de Lorenzo, 2012) and homologous recombination (Wang et al., 2012; Davy et al., 2017) are also available to integrate exogenous DNA fragments into the bacterial genome. Remarkably, when selection marker or CRISPR editing alleles are not applicable, transposition is an optimal choice for integrating target genes into the bacterial genome (Jiang et al., 2013; Vo et al., 2021). To date, the generally used transposases mainly include sleeping beauty, piggyBac (Tschorn et al., 2020), Tol2 (Ni et al., 2016), Tn5 (Balasubramanian et al., 2021; Xu et al., 2021), Tn7 (Kaczmarska et al., 2022), and ICEBs1 (Ni et al., 2016; Peters et al., 2019; Strecker et al., 2019; Wu et al., 2021), all of which possess the ability to integrate small or large DNA fragments into bacterial genomes (Figure 2). However, the transposition efficiency slightly decreases as the size of the inserted DNA fragments increases (Kowalczykowski, 2015). Interestingly, several studies have shown that the synthetic Himar1 transposase-dead Cas9 fusion protein, which is characterized by DNA integration ability by Himar1 transposase (a Tn7-like transposon) and targeted localization by programmable dead Cas9, is capable of achieving targeted transposition under cell-free condition, thus avoiding the random insertion of transposons (Bhatt and Chalmers, 2019). This recombination method succeeded in accomplishing the transfer of transposons larger than 7 kb, the accuracy of which was as high as 80% (Chen and Wang, 2019). Similarly, another integrated system originating from Tn6677 transposase utilized a Tn7-like transposon and CRISPR protein to guide RNA-assisted targeted insertion of transposable factors, enabling the accurate insertion of over 10 kb DNA sequences into bacterial genomes (Nelson et al., 2021).
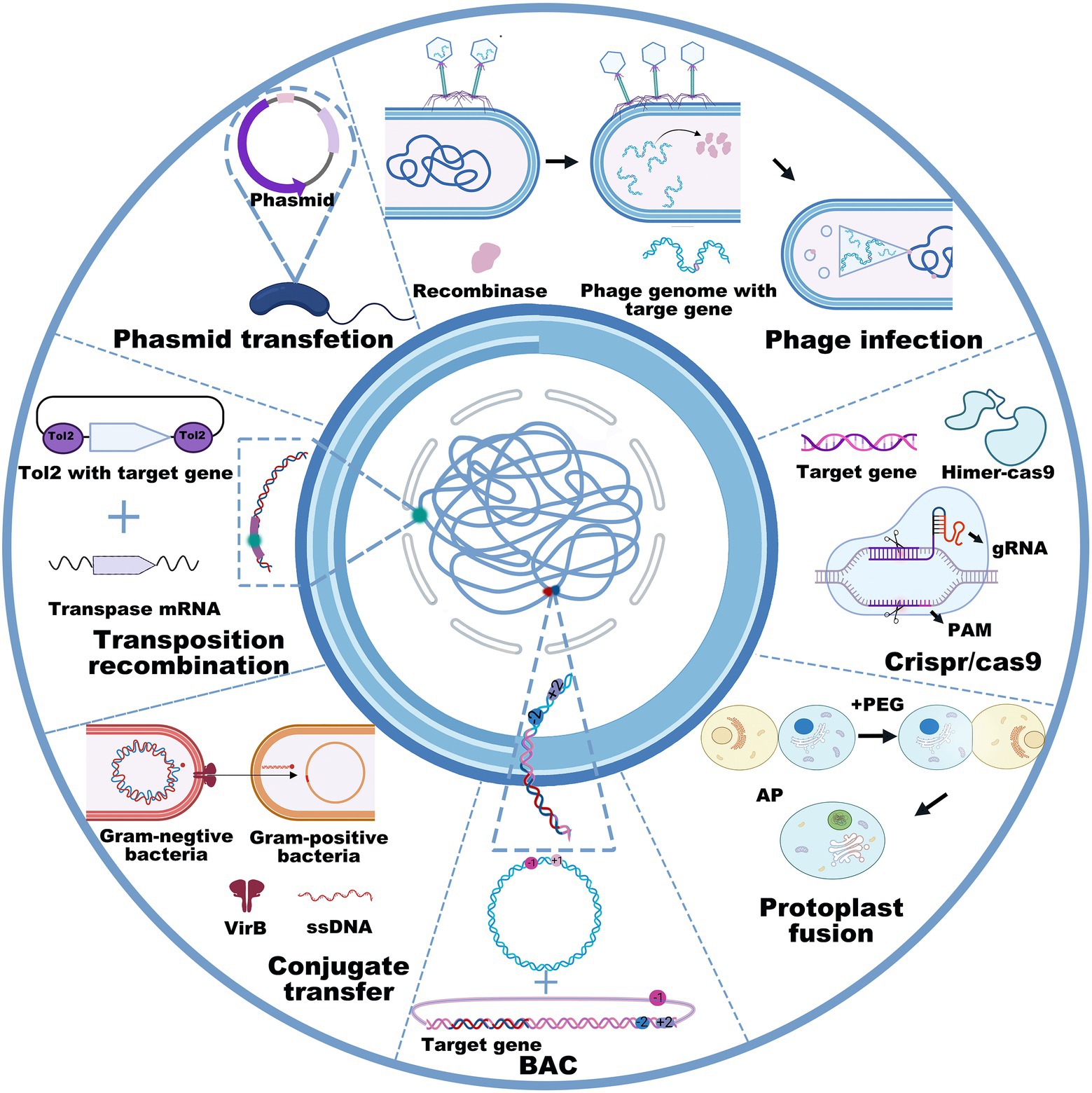
Figure 2. General used recombination technologies, including phage infection, phasmid transfection, transposition recombination, conjugate transfer, BAC, protoplast fusion, and CRISPR-Cas9 in counter-clockwise order.
However, the plasmid carrying capacity and transposition method do not allow for operating large DNA fragments, especially when their sizes are larger than 100 kb (Hashimoto et al., 2015). To achieve the heterologous expression of large gene clusters, researchers are more inclined to use transformation-associated recombination (Bilyk et al., 2016; Zhang et al., 2019), bacterial artificial chromosomes (BAC; Huo et al., 2019; Hashimoto et al., 2020b), phage recombination systems (Oßwald et al., 2014; Nah et al., 2017), or integrase-mediated recombination systems (Du et al., 2015; Ke et al., 2021; Figure 2). For example, the BAC technique enabled the integration of over 181 kb DNA fragments into the genome of Streptomyces lividans TK23 (Hashimoto et al., 2020a). Concomitantly, the protoplast fusion method is also an option to acquire new functions for GEBs by integrating two bacterial genomes (Zhang et al., 2002). Although the insertion of foreign genes into the genome is more complex than the plasmid-based expression system, it possesses incomparable stability (Miyazaki and van der Meer, 2013; Pedrolli et al., 2019). Collectively, these methods, such as DNA extraction, site-specific mutation, gene insertion, gene deletion, and DNA transfection, have the potential to cater to most genetic manipulations in GEB construction, even when their size exceeds 100 kb. However, it is still a considerable challenge to establish GEBs for reliably and efficiently expressing foreign proteins or compounds mainly because interspecific differences disable the microbial chassis from expressing most foreign genes or gene clusters, even after codon optimization. Therefore, the principal contradiction of establishing GEBs is that gene sequencing reveals an increasing number of functional genes or gene clusters from microorganisms characterized by nonculture, a long growth cycle, or harsh culture, but not enough microbial chassis are available to express or deliver them for disease diagnosis and treatment.
The route of GEB administration depends on multiple factors, including the target tissues, disease types, properties of heterogeneous proteins or compounds, and chassis cells (Hosseinidoust et al., 2016). However, considering the potential pathogenicity of microorganisms, especially when administered via intravenous administration, the delivery route should be seriously considered at the initial stage of GEB construction in health care and disease management. Intensive clinical trials demonstrate that therapeutic and adverse effects are closely associated with drug delivery models (Ott et al., 2004). To date, the routes of GEB administration mainly included oral administration, intravenous injection, intratumoral injection, nasal administration, and subcutaneous injection.
Oral administration is the most widely used method in bacteriotherapy because of its simple operation, noninvasiveness, and wide applicability. However, the gastrointestinal tract is known to be a complex environment, including drastic pH changes among different organs [pH 1.0–2.5 in the stomach (Lund et al., 2014), pH 7–7.4 in the small intestine, and pH 6–6.7 in the colon (Kamada et al., 2013; Koziolek et al., 2015)] and differences in oxygen percentages (Zheng et al., 2015), nutrient enrichment, and flora diversity, which greatly affect GEB activity when orally administered (Sender et al., 2016). Notably, the survival rates of GEBs have a great influence on their therapeutic efficacies, but they do not exceed 50% when they are orally taken without extra protection, highlighting the key role of shielding bacteria from a complex gastrointestinal environment (Heavey et al., 2021). Indeed, the emergence of new technologies, such as polysaccharides (alginate, κ-carrageenan, locust bean gum, gellan gum, and xanthan gum; Ta et al., 2021), cationic liposomes (Chowdhuri et al., 2016), and mammalian cell membranes (Cao et al., 2019a), enable the isolation of bacteria from hostile environments. However, different packaging methods have different effects on the survival rates of GEBs after oral administration (Chowdhuri et al., 2016; Han et al., 2016; Sánchez et al., 2017; Cao et al., 2019b; Gharibzahedi and Smith, 2021). For instance, liposomal emulsified bacterial vaccines exhibited higher immune-boosting capacity and therapeutic efficiency than the uncoated vaccines in animal experiments (Naciute et al., 2021).
Comparatively, the intravenous administration of live bacteria only occurs in preclinical and clinical cancer therapy due to the high pathogenicity of the systematic administration of GEBs (Forbes et al., 2018). Additionally, nasal drip is another administration route of GEBs. Due to the specific safeguard function of nasal mucosa, intranasal vaccination becomes capable of activating local humoral and cellular immune responses at the entrance of the respiratory mucosa, distal mucosal sites, and their associated lymphoid tissues, thereby exerting prevention or therapeutic effects (Riese et al., 2014; Xu et al., 2021). Notably, bacterial administration via blood, nasal or intestinal routes is very beneficial for them to quickly reach disease sites, and thus, in most cases, their therapeutic effects are superior to those of oral administration (Wells and Mercenier, 2008; Pandey et al., 2022). However, some studies signified that the oral administration of GEBs was able to produce a higher immune response than intranasal immunization (Wan and Ping, 2021). The controversial issue is probably attributed to the use of distinct disease molds in these studies. In contrast to the treatment of enteric disease through oral administration, intranasal immunization is more suitable for the prevention of systemic allergy and airway inflammation (Sarate et al., 2019). In fact, various recombinant bacteria, such as Streptococcus gordonii, Staphylococci, and Lactobacillus, have shown great potential as active carriers of nasal vaccines (Mojgani et al., 2020; Dadar et al., 2021).
Furthermore, intratumoral injection is another key administration route for live GEBs to treat solid tumors. This method is greatly beneficial for reducing the potential systemic toxicity of live GEBs (Taniguchi et al., 2010). The intratumoral injected bacteria actively or passively colonize necrosis because of their homing instincts, chemotactic effects (Mirkhani et al., 2021), and cumulative effects (Ganai et al., 2011). Then, they activate immunogenicity and release toxic molecules to induce cell apoptosis and restrain tumor growth (Yaghoubi et al., 2020; Lin et al., 2021). However, this treatment may lead to highly malignant adverse effects because the biological toxicity caused by rapid and massive cell death in highly colonized bacterial tumors is extremely harmful to other normal organs and even induces a cytokine storm, thus leading to patient death (Karbach et al., 2012; Pandey et al., 2022). Therefore, the administration route is a key factor in GEB construction for different purposes.
To date, there are still no official standards for evaluating the efficacy of GEBs in disease diagnosis, prevention, or treatment. Increasing evidence has demonstrated that the intrinsic biological property, colonization ability, dose tolerance, and potential pathogenicity of GEBs all influence their activities. Establishing standard performance indicators plays a key role in advancing GEB clinical transition.
GEB colonization ability refers to their survival and biological inheritance at the expected sites of disease after they enter the body and is an important indicator for evaluating GEB efficacy in disease management. Various techniques, mainly including tissue sectioning, fluorescent labeling, 16S rRNA sequencing, quantitative polymerase chain reaction, etc., have been developed to evaluate GEB conization ability (Cronin et al., 2012; Shen et al., 2015). Fluorescently labeled GEBs can be traced and recorded from the initial administration to fecal samples under a fluorescence microscope; sequencing technology could use 16S rRNA gene tags to obtain the current bacterial lineage in fecal samples, thus evaluating the colonization ability of GEBs by comparison with the initial fecal flora (Shen et al., 2015).
Recent studies have identified some bacteria with high intestinal colonization (Dosoky et al., 2020). For instance, E. coli NGF-1 has been found to colonize in vivo for up to 6 months in a stable and persistent state (Riglar et al., 2017). However, various external factors may influence the in vivo colonization ability of GEBs. The key influencing factor is the location of GEBs in the body because each bacterium in our body has a corresponding colonization area in which the microenvironment formed by long-period interactions with other bacteria and mammalian cells provides a safe place for them to proliferate with high genetic stability (O’Toole and Claesson, 2010; Zou et al., 2019; Tochitani, 2021). Therefore, the disease location in the body determines the species of chassis bacterium, thus enhancing their colonization and reducing the off-target effects of their secreted substances (Dosoky et al., 2020).
Indeed, some studies have taken advantage of organ-specific GEBs to exert their best therapeutic effects (Tarahomjoo, 2012), such as using colon-colonizing strains to treat ulcerative colitis (Conrad et al., 2014) and employing Lactobacillus bacteria, which colonize in the small intestine and colon, to treat Crohn’s disease (Donaldson et al., 2016). More importantly, the administration method also influences GEB colonization ability. The intravenous injected Salmonella typhi exhibited higher tumor colonization ability than the intraperitoneal administered ones. In addition, the tumor size and bacterial number affect GEB colonization in target sites (Mei et al., 2002). Currently, the studies investigating GEB colonization ability, genetic stability, and the mechanisms underlying their activities are still in their infancy, thereby requiring more basic and clinical studies to advance the clinical transition of GEBs.
Intensive studies have emphasized the key role of dose in the therapeutic effects of GEBs against diseases (Ott et al., 2004). Bifidobacterium infantis 35,624, which is generally used to treat irritable bowel syndrome, exhibited the best performance at a dose of 108 cfu per day, while 106 cfu and 1010 cfu per day did not result in significant differences from the placebo group (Whorwell et al., 2006), thus indicating that the use of bacteria against disease is closely related to the bacterial number. Although a high amount of bacteria generally exhibits enhanced therapeutic effects, potential side effects also increase (Leventhal et al., 2020). The oral administration of SYNB1020 at a concentration of 1012 cfu per person induced adverse reactions, such as nausea and vomiting, in healthy volunteers and hyperammonemia patients (Kurtz et al., 2019). Some studies further demonstrated that an increasing number of bacteria generally leads to the development of dose-dependent problems and may even restrain immune system memories (Sivick et al., 2018). Therefore, how to achieve better therapeutic effects with low doses of GEBs remains a hot topic in the development of therapeutically engineered bacteria. Increasing their targeting ability is a selective strategy to decrease the administered dose. For example, the expression of tumor-targeting adhesins on the membrane surface of E. coli significantly decreased the intravenous injected bacterial number required for the minimum effective dose (Piñero-Lambea et al., 2015).
Simultaneously, different administration methods also influence the effective doses of GEBs in disease treatment. Lactobacilli were administered via intraperitoneal injection, intestinal administration, and oral administration, but the doses required to reach the same therapeutic efficacy differed by nearly 10,000 times among them (Steidler et al., 2000). Notably, each bacterium has an optimal dose for producing the best therapeutic efficacy. The recommended dose for Bifidobacterium infantis 35,264 is 108 CFU per day, while the optimal dose for probiotic preparation VSL#3 (VSL Pharmaceuticals) is 1.8 × 1012 CFU per day when taken by oral administration (Gionchetti et al., 2003). Although daily probiotic use has a long history, the use of bacteria for the prevention and treatment of disease is still at its initial stage.
Using bacteria to improve intestinal function and treat diseases has been proven to be a safe and effective modality (Sharif et al., 2017). However, GEBs prepared by genetic modification, chemical capsulation or other methods need to be fully evaluated for metabolic pathways and toxicological effects in vivo before clinical application. The pathogenicity of GEBs is mostly derived from the bacteria, but this defect could be partly removed or mitigated by the gene knockout or mutation of virulence genes. Typical examples include lipopolysaccharide deletion in gram-negative bacteria, virulence elimination in Listeria monocytogenes, and exotoxin gene knockout in Clostridium novyi (Zhou et al., 2018).
Additionally, the regulation of bacterial population provides another strategy to further enhance the safety of bacteriotherapy. A synchronous lysis circuit consisting of positive and negative feedback genes, and an inducible promoter can precisely control the proliferation and lysis of bacteria, thus restraining the bacterial population within a specified scope (Din et al., 2016). Moreover, population competition between the intrinsic bacterial flora and orally administered GEBs is always inevitable. Once foreign bacteria dominate the original flora, they probably alter the physiology of the host, thus leading to dysfunction of the gastrointestinal tract, such as inflammation (Spees et al., 2013) and pathogenic infection (Kamada et al., 2013). Therefore, purging foreign bacteria from the host after treatment is another key issue in GEB application. Biocontainment strategies, alternative selection markers and the use of homologous DNA have been performed to inhibit potential transmission in the environment and purge residual foreign bacteria (Plavec and Berlec, 2020). For instance, SYNB1618 must rely on exogenous diaminopimelate for cell wall synthesis in the case of dapA gene deletion, thus ensuring the complete purge of the bacteria after treatment (Isabella et al., 2018).
As early as 100 years ago, bacteria were used to treat tumors and clostridial enteritis (Coley, 1893; Hoffman, 2016). Engineered bacteria at their initial stages mainly focused on the treatment of gastrointestinal inflammation and tumors (Steidler et al., 2000; Geirnaert et al., 2017; Zhou et al., 2018). Advances in genetic technology endowed GEBs with more functions and broad application prospects. An alternative method is to deliver foreign therapeutic drugs, thus alleviating the shortcomings of natural drugs in the low production level, short action time, and nonoral administration property. In this paragraph, we conclude and describe the use of GEBs for treating multiple diseases, such as IBD, obesity, diabetes and cancer, in detail (Table 1).
The dysregulation of intestinal microbes induces gastrointestinal diseases and, in some cases, damages normal organs via the tissue-gut axis (Fassarella et al., 2021; Lee and Chang, 2021). Routine supplementation with probiotics successfully improved or cured types of diseases, such as acute diarrhea (Mu and Cong, 2019), IBD (Saez-Lara et al., 2015; Jakubczyk et al., 2020), and diabetes (Kobyliak et al., 2016; Razmpoosh et al., 2016; Kocsis et al., 2020; van de Wijgert and Verwijs, 2020; Davidson et al., 2021). Specifically, IBD, as an autoimmune disease, is characterized by chronic inflammation of the gastrointestinal tract and the loss of epithelial barrier integrity in the intestine (Fakhoury et al., 2014). Ulcerative colitis and Crohn’s disease are both classified as IBD. IBD has a high incidence worldwide, but there is still no efficient treatment method. Considering the key role of microorganisms in the intestinal microecological balance and IBD inflammatory characteristics, an anti-inflammatory cytokine, interleukin-10, was expressed in Lactobacillus to treat chronic disease, which finally led to a 50% therapeutic efficacy in a mouse IBD model (Steidler et al., 2000). Similarly, interleukin-4 (IL-4)-expressing Lactobacillus also significantly alleviated inflammatory responses caused by increased Th1 cells (Souza et al., 2016). Additionally, the oral administration of interleukin-35 (IL-35) expressing E. coli obviously attenuated inflammatory damage in mouse colon tissue, thereby improving the symptoms of IBD (Zhang et al., 2018). Furthermore, EcN has a long history in treating intestinal tract diseases in infants and toddlers. Using EcN to colonize and secrete intestinal trefoil factor could significantly improve the integrity of the intestinal epithelium and reduce the dextran sodium sulfate-induced intestinal inflammatory response in a mouse model (Pedrolli et al., 2019).
Moreover, another potential way to treat colitis is to increase superoxide dismutase or catalase in the intestine, thereby reducing reactive oxygen species, which is a key factor in inflammation (Hwang et al., 2020; Wan et al., 2022). Utilizing GEBs to produce foreign proteins to enhance the delivery of superoxide dismutase or catalase to the intestine successfully reduced the inflammatory reaction in a trinitrobenzene sulfonic acid-induced colitis model (Del Carmen et al., 2014). Additionally, the expression of manganese superoxide dismutase in Bifidobacterium greatly decreased dextran sodium sulfate-induced IBD (Liu et al., 2018). Furthermore, elastin is generally used to treat IBD because of its inhibitory effects on serine protease activity and anti-inflammatory effects (Bermúdez-Humarán et al., 2015). The oral administration of elastin-expressing Lactobacillus strains exhibited a strong inhibitory effect on dextran sodium sulfate-induced IBD (Bermúdez-Humarán et al., 2015). Similarly, genetically modified Lactobacillus strains could use mucosa to deliver an anti-inflammatory molecule, recombinant mouse heme oxygenase-1, and thus reduce the incidence of acute colitis (Shigemori et al., 2015). In addition to IBD, GEBs have been used to express foreign proteins, such as antibacterial pancreatitis-related protein, to alleviate or treat chemotherapy-induced intestinal diseases and mucositis in mice (Carvalho et al., 2017).
The specific tumor microenvironment enables the colonization of facultative anaerobic or anaerobic bacteria in the hypoxic regions of solid tumors. The use of tumor-targeting bacteria, such as Bifidobacterium (Chen et al., 2021), Salmonella (Chirullo et al., 2015; Kim et al., 2015; Li et al., 2017), Clostridium novyi-NT (Roberts et al., 2014; Staedtke et al., 2016; Janku et al., 2021), and E. coli (Afkhami-Poostchi et al., 2020; Chiang and Hong, 2021), to deliver various proteins, chemical molecules, preenzymes, etc. for cancer therapy is collectively referred to as bacteriolytic therapy. Detailing all anticancer GEBs is beyond the scope of this review, and this topic has been recently reviewed by others (Badie et al., 2021; Huang et al., 2021; Kalia et al., 2021).
The increasing prevalence of obesity, diabetes and other metabolic diseases in modern society places a heavy burden on medical care. These metabolic diseases are generally characterized by hyperglycemia, hyperlipidemia and high-density lipoprotein, and simultaneously, they could trigger each other (Guo, 2014). Notably, most patients with metabolic diseases must take drugs for the rest of their lives to alleviate or slow disease progression. In contrast, the oral administration of GEBs has exhibited few side effects and better acceptance. Indeed, many GEBs have already been constructed and evaluated for curing inborn or acquired metabolic diseases.
Diabetes is one of the greatest public health problems worldwide and causes a substantial burden on the socioeconomic development of the world (Lin et al., 2020). Diabetes is clinically divided into type 1 diabetes (T1D) and type 2 diabetes (T2D; Zimmet et al., 2014). The treatment for T1D relies on the regular injection of insulin to slow the progression of the diabetic process (Miller et al., 2015), whereas T2D patients take hypoglycemic drugs, such as metformin, to restrain disease progression. However, the long-term use of glucose-lowering drugs usually results in serious side effects and economic burdens (Khunti et al., 2019). Numerous studies have found that disease progression in T2D is closely associated with an imbalance in gut microflora (Forslund et al., 2015). Therefore, the regulation of the intestine microenvironment using probiotics or GEBs could be an alternative strategy to cure or slow diabetes, especially T2D. Given the underlying mechanisms of diabetes, treatment could be achieved by using an engineered bacterium to produce proteins with hypoglycemic effects. For example, glucagon-like peptide 1 (GLP-1) is an efficient drug for diabetes treatment, but its short half-life time and high cost greatly prevent its extensive clinical use. However, GEBs characterized by gut colonization have the potential to address the above problems. Leveraging GLP-1 (1–37) with the ability to transfer intestinal epithelial cells into insulin-secreting cells (Duan et al., 2015), GLP-1-expressing Lactobacillus successfully normalized blood glucose levels in diabetic Goto-Kakizaki rats (Lin et al., 2016). Other proteins or compounds against diabetes can also achieve similar therapeutic effects by using probiotics as delivery vectors. For example, recombinant Lactobacillus expressing heat shock protein 65 and IA2P2 (a 23 amino acid peptide) effectively alleviated the symptoms of pancreatitis and improved diabetes by inhibiting the antigen-specific proliferation of T cells in T1D and regulating the balance between Th17/Tregs and Th1/Th2 cells (Liu et al., 2016).
Obesity is a complex multifactorial disease and a key factor in other chronic diseases, such as cardiovascular disease, cancer, and diabetes (Pan et al., 2021). Generally, obesity occurs in middle-aged and elderly people. Increasing reports demonstrate that the prevalence of obesity in children and adolescents aged 2 to 19 years has been gradually increasing worldwide, especially in developed countries (Fryar et al., 2018, 2020). Most traditional anti-obesity drugs, such as sibutramine and orlistat, act by suppressing appetite or blocking the absorption of body fat, but they produce obvious adverse effects on normal physiological function (Huang et al., 2019). Fortunately, several studies have developed GEBs to express anti-obesity factors for the purpose of alleviating obesity. For instance, GLP-1-expressing Lactobacillus could reduce the incidence of high-fat diet-induced obesity in mice by restraining lipid accumulation, enhancing GLP-1 resistance to glucose intolerance, and increasing the expression of genes involved in the triglyceride degradation pathway (Wang et al., 2021). Additionally, Bacillus subtilis SCK6 utilized Coenzyme A transferase acetate to increase butyric acid production and stimulate the butyric acid kinase pathway, thus attenuating hepatic steatosis and fat accumulation in high-fat diet mice (Bai et al., 2020). Similarly, interleukin-22 (IL-22)-expressing Lactobacillus reuteri obviously decreased the incidence of nonalcoholic fatty liver disease in high-fat diet mice (Oh et al., 2020).
Hyperammonemia, as a metabolic disease, is used to describe patients with high levels of plasma ammonia levels (>50 μmol/l in adults and >100 μmol/l in the neonatal period). Under normal circumstances, ammonia is mainly produced in the intestine and excreted by the liver, but severe metabolic disorders probably induce the massive accumulation of ammonia in the body and, thus, affect the urea cycle and may even trigger hepatic encephalopathy (Auron and Brophy, 2012). The current treatment strategies for patients with hyperammonemia include hemodialysis, peritoneal dialysis, and antibiotic therapy. However, these treatments greatly increase infection probabilities and lead to drug resistance (Matoori and Leroux, 2015). However, oral probiotic use could significantly decrease ammonia levels in mice with hepatic encephalopathy (Lunia et al., 2014), demonstrating the feasibility of establishing GEBs to treat Hyperammonemia. In addition, by deleting the negative regulator of I-arginine synthesis and adding a feedback-resistant I-arginine biosynthetic enzyme in EcN, GEBs successfully achieved the conversion of NH3 to I-arginine in bacteria, thereby blocking ammonia accumulation in vivo and slowing Hyperammonemia progression (Kurtz et al., 2019).
Phenylketonuria is an autosomal recessive genetic disease characterized by mental and growth retardation. Phenylketonuria results from the gene deficiency of phenylalanine hydroxyls, thus preventing the conversion of phenylalanine to tyrosine. The Phenylketonuria therapeutic methods include dietary restriction, gene therapy, and enzyme replacement. However, even a well-controlled diet cannot completely prevent the occurrence of psychiatric problems in Phenylketonuria patients, while gene therapy is extremely costly, and enzyme replacement therapy is obviously affected by the drug dose and administration schedule (Camp et al., 2014). However, probiotics have the potential to be modified to produce deficient phenylalanine hydroxyl enzymes to supplement physiological requirements. The insertion of the genes of phenylalanine-ammonia-lyase and L-amino acid deaminase into the EcN genome enables probiotics to convert phenylalanine into trans-cinnamic acid salt in the gastrointestinal tract, thus leading to a 38% reduction in orally administered phenylalanine in blood (Isabella et al., 2018; Puurunen et al., 2021).
Furthermore, hypertension is a common chronic disease that is not only a major cause of cardiovascular disease but also damages the brain, kidney, and other organs (Fuchs and Whelton, 2020). Many therapeutic agents, including β-blockers, calcium channel blockers, diuretics, and renin-angiotensin converting enzyme inhibitors, have been developed to treat hypertension. However, the long-term use of these antihypertensive drugs often leads to potential side effects, such as arterial damage, angioedema, arrhythmias, impotence, hyperkalemia, and cough. More importantly, some drugs, such as angiotensin-converting enzyme inhibitory peptides, require large doses to exert therapeutic effects because their activity is inclined to be inhibited by other enzymes (Laurent, 2017). However, probiotics engineered to express the enzyme have the potential to address this issue. For example, the introduction of the genes encoding tuna frame protein and yellowfin sole frame protein into Lactobacillus plantarum NC8 enabled the probiotics to synthesize angiotensin-converting enzyme inhibitory peptides, and the oral administration of angiotensin-converting enzyme inhibitory peptides significantly reduced systolic blood pressure and triglyceride levels in a spontaneously hypertensive mouse model, displaying good antihypertensive ability (Yang et al., 2015).
Microbes have been implicated in almost all fundamental activities of physiological homeostasis. One example of the increasingly important role of bacteriotherapy is the regulation of the intestinal flora for curing intestinal diseases and adjuvant therapy of other diseases, such as cancer and metabolic diseases (Andrade-Oliveira et al., 2015; Alexander et al., 2017; Yuan et al., 2018; Molina et al., 2021; Si et al., 2021). Notably, advances in gene sequencing technologies have partly revealed the mysteries of human microbes and the interaction between microbes and various mammalian cells, greatly advancing GEB preclinical studies. To date, most existing therapeutic GEBs have been modified from human intestinal probiotics, demonstrating their potential safety compared to traditional chemical drugs, especially for long-term use. GEBs are not only effective therapeutic agents against chronic diseases due to their intestinal colonization ability but also expand the scope of the application of tumor-targeting treatment.
The use of engineered bacteria in disease treatment is still in the infancy stage and has limitations for incurable diseases. In addition, the clinical translation of GEBs is still hindered by potential pathogenicity and local laws and regulations, which only allow clinical trials to use bacteria without any genetic modification. A future challenge will be to determine whether the genetically inserted genes of GEBs could spill over into the genomes of other bacteria or mammalian cells; how GEBs could stably colonize and produce the expected substrates in the targeted sites; how they interact with the intestinal flora, thus normalizing them into the health status; and how to lock them into the expected tissues and clear them once they accomplish their mission. Such questions underscore the importance of investigating the genetic stability of the inserted genes in GEBs under normal physiological conditions. However, as biological technologies continue to evolve, newly available bacterial tools and the upgrading of therapeutic strategies of GEBs will further alleviate potential safety concerns and enhance their depth and breadth in disease prevention and treatment.
Collectively, given the importance of the microbiota in normal physiological function, the long historical use of probiotics and bacterial-derived products for health care, and our increasingly better understanding of the mechanisms underlying the various gut-brain similar axes, we expect that GEBs will make an important contribution to the prevention and treatment of various diseases that current drugs cannot address or cause serious side effects.
YL, JF, and HP searched references and drafted the manuscript. XZ and YZ conceived, supervised, and improved the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (grant no. 81971726) and the Nanjing Healthcare Science and Technology Development Special Funded Project (YKK20191).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afkhami-Poostchi, A., Mashreghi, M., Iranshahi, M., and Matin, M. M. (2020). Use of a genetically engineered E. coli overexpressing β-glucuronidase accompanied by glycyrrhizic acid, a natural and anti-inflammatory agent, for directed treatment of colon carcinoma in a mouse model. Int. J. Pharm. 579:119159. doi: 10.1016/j.ijpharm.2020.119159
Alberti, F., and Corre, C. (2019). Editing streptomycete genomes in the CRISPR/Cas9 age. Nat. Prod. Rep. 36, 1237–1248. doi: 10.1039/c8np00081f
Alexander, J. L., Wilson, I. D., Teare, J., Marchesi, J. R., Nicholson, J. K., and Kinross, J. M. (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365. doi: 10.1038/nrgastro.2017.20
Alfano, A., Perillo, F., Fusco, A., Savio, V., Corsaro, M. M., Donnarumma, G., et al. (2020). Lactobacillus brevis CD2: fermentation strategies and extracellular metabolites characterization. Probiotics and Antimicrobial Proteins 12, 1542–1554. doi: 10.1007/s12602-020-09651-w
Andrade-Oliveira, V., Amano, M. T., Correa-Costa, M., Castoldi, A., Felizardo, R. J., de Almeida, D. C., et al. (2015). Gut bacteria products prevent AKI induced by ischemia-reperfusion. J. Am. Soc. Nephrol. 26, 1877–1888. doi: 10.1681/asn.2014030288
Auron, A., and Brophy, P. D. (2012). Hyperammonemia in review: pathophysiology, diagnosis, and treatment. Pediatr. Nephrol. 27, 207–222. doi: 10.1007/s00467-011-1838-5
Badie, F., Ghandali, M., Tabatabaei, S. A., Safari, M., Khorshidi, A., Shayestehpour, M., et al. (2021). Use of salmonella bacteria in cancer therapy: direct, drug delivery and combination approaches. Front. Oncol. 11:142. doi: 10.3389/fonc.2021.624759
Bai, L., Gao, M., Cheng, X., Kang, G., Cao, X., and Huang, H. (2020). Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice. Microb. Cell Factories 19, 94–13. doi: 10.1186/s12934-020-01350-z
Balasubramanian, D., Murcia, S., Ogbunugafor, C. B., Gavilan, R., and Almagro-Moreno, S. (2021). Cholera dynamics: lessons from an epidemic. J. Med. Microbiol. 70:298. doi: 10.1099/jmm.0.001298
Bermúdez-Humarán, L. G., Motta, J.-P., Aubry, C., Kharrat, P., Rous-Martin, L., Sallenave, J.-M., et al. (2015). Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb. Cell Factories 14, 26–11. doi: 10.1186/s12934-015-0198-4
Bernell, S., and Howard, S. W. (2016). Use your words carefully: what is a chronic disease? Front. Public Health 4:159. doi: 10.3389/fpubh.2016.00159
Beumer, K. J., Trautman, J. K., Mukherjee, K., and Carroll, D. (2013). Donor DNA utilization during gene targeting with zinc-finger nucleases. G3: Genes, Genomes, Genetics 3, 657–664. doi: 10.1534/g3.112.005439
Bhatt, S., and Chalmers, R. (2019). Targeted DNA transposition in vitro using a dCas9-transposase fusion protein. Nucleic Acids Res. 47, 8126–8135. doi: 10.1093/nar/gkz552
Bilyk, O., Sekurova, O. N., Zotchev, S. B., and Luzhetskyy, A. (2016). Cloning and heterologous expression of the grecocycline biosynthetic gene cluster. PLoS One 11:e0158682. doi: 10.1371/journal.pone.0158682
Blüher, M. (2019). Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298. doi: 10.1038/s41574-019-0176-8
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J. Clinicians 68, 394–424. doi: 10.3322/caac.21492
Camp, K. M., Parisi, M. A., Acosta, P. B., Berry, G. T., Bilder, D. A., Blau, N., et al. (2014). Phenylketonuria scientific review conference: state of the science and future research needs. Mol. Genet. Metab. 112, 87–122. doi: 10.1016/j.ymgme.2014.02.013
Cao, Z., Cheng, S., Wang, X., Pang, Y., and Liu, J. (2019a). Camouflaging bacteria by wrapping with cell membranes. Nat. Commun. 10, 3452–3410. doi: 10.1038/s41467-019-11390-8
Cao, Z., Wang, X., Pang, Y., Cheng, S., and Liu, J. (2019b). Biointerfacial self-assembly generates lipid membrane coated bacteria for enhanced oral delivery and treatment. Nat. Commun. 10:5783. doi: 10.1038/s41467-019-13727-9
Carignan, B. M., Brumfield, K. D., and Son, M. S. (2016). Single nucleotide polymorphisms in regulator-encoding genes have an additive effect on virulence gene expression in a vibrio cholerae clinical isolate. Msphere 1, e00253–e00216. doi: 10.1128/msphere.00253-16
Carvalho, R. D., Breyner, N., Menezes-Garcia, Z., Rodrigues, N. M., Lemos, L., Maioli, T. U., et al. (2017). Secretion of biologically active pancreatitis-associated protein I (PAP) by genetically modified dairy Lactococcus lactis NZ9000 in the prevention of intestinal mucositis. Microb. Cell Factories 16, 27–11. doi: 10.1186/s12934-017-0624-x
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
Çelik, E., and Çalık, P. (2012). Production of recombinant proteins by yeast cells. Biotechnol. Adv. 30, 1108–1118. doi: 10.1016/j.biotechadv.2011.09.011
Charbonneau, M. R., Isabella, V. M., Li, N., and Kurtz, C. B. (2020). Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 11:11. doi: 10.1038/s41467-020-15508-1
Chen, J., Chen, X., and Loong, H. C. (2021). Recent development of probiotic Bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 9:248. doi: 10.3389/fbioe.2021.770248
Chen, S. P., and Wang, H. H. (2019). An engineered Cas-transposon system for programmable and site-directed DNA transpositions. CRISPR J. 2, 376–394. doi: 10.1089/crispr.2019.0030
Chen, H. Q., Xia, Y., Zhu, S. B., Yang, J., Yao, J., Di, J. Z., et al. (2017). Lactobacillus plantarum LP-Onlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin-10 knockout mice. Mol. Med. Rep. 16, 5979–5985. doi: 10.3892/mmr.2017.7351
Chiang, C.-J., and Hong, Y.-H. (2021). In situ delivery of biobutyrate by probiotic Escherichia coli for cancer therapy. Sci. Rep. 11, 18172–18117. doi: 10.1038/s41598-021-97457-3
Chirullo, B., Ammendola, S., Leonardi, L., Falcini, R., Petrucci, P., Pistoia, C., et al. (2015). Attenuated mutant strain of salmonella typhimurium lacking the ZnuABC transporter contrasts tumor growth promoting anti-cancer immune response. Oncotarget 6, 17648–17660. doi: 10.18632/oncotarget.3893
Chowdhuri, S., Cole, C. M., and Devaraj, N. K. (2016). Encapsulation of living cells within Giant phospholipid liposomes formed by the inverse-emulsion technique. Chembiochem 17, 886–889. doi: 10.1002/cbic.201500643
Christofi, T., Panayidou, S., Dieronitou, I., Michael, C., and Apidianakis, Y. (2019). Metabolic output defines Escherichia coli as a health-promoting microbe against intestinal Pseudomonas aeruginosa. Sci. Rep. 9, 14463–14413. doi: 10.1038/s41598-019-51058-3
Cobb, R. E., and Zhao, H. (2012). Direct cloning of large genomic sequences. Nat. Biotechnol. 30, 405–406. doi: 10.1038/nbt.2207
Coley, W. B. (1893). The treatment of malignant tumors by repeated inoculations of erysipelas: with a report of ten original cases. 1. American J. Medical Sci. (1827–1924) 105, 487–510. doi: 10.1097/00000441-189305000-00001
Conrad, K., Roggenbuck, D., and Laass, M. W. (2014). Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 13, 463–466. doi: 10.1016/j.autrev.2014.01.028
Cronin, M., Akin, A. R., Collins, S. A., Meganck, J., Kim, J.-B., Baban, C. K., et al. (2012). High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One 7:e30940. doi: 10.1371/journal.pone.0030940
Dadar, M., Shahali, Y., and Mojgani, N. (2021). “Probiotic bacteria as a functional delivery vehicle for the development of live Oral vaccines” in Probiotic bacteria and Postbiotic metabolites: Role in animal and human health (Germany: Springer), 319–335. doi: 10.1007/978-981-16-0223-8_13
Dang, A. T., and Marsland, B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
Davidson, S. J., Barrett, H. L., Price, S. A., Callaway, L. K., and Nitert, M. D. (2021). Probiotics for preventing gestational diabetes. Cochrane Database Syst. Rev. 4:CD009951. doi: 10.1002/14651858.cd009951.pub2
Davy, A. M., Kildegaard, H. F., and Andersen, M. R. (2017). Cell factory engineering. Cell systems 4, 262–275. doi: 10.1016/j.cels.2017.02.010
Del Carmen, S., De Moreno de LeBlanc, A., Martin, R., Chain, F., Langella, P., Bermúdez-Humarán, L. G., et al. (2014). Genetically engineered immunomodulatory Streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced anti-inflammatory activities. Appl. Environ. Microbiol. 80, 869–877. doi: 10.1128/aem.03296-13
Din, M. O., Danino, T., Prindle, A., Skalak, M., Selimkhanov, J., Allen, K., et al. (2016). Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536:81. doi: 10.1038/nature18930
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Dosoky, N. S., May-Zhang, L. S., and Davies, S. S. (2020). Engineering the gut microbiota to treat chronic diseases. Appl. Microbiol. Biotechnol. 104, 7657–7671. doi: 10.1007/s00253-020-10771-0
Du, D., Wang, L., Tian, Y., Liu, H., Tan, H., and Niu, G. (2015). Genome engineering and direct cloning of antibiotic gene clusters via phage ϕBT1 integrase-mediated site-specific recombination in Streptomyces. Sci. Rep. 5, 1–8. doi: 10.1038/srep08740
Duan, F. F., Liu, J. H., and March, J. C. (2015). Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 64, 1794–1803. doi: 10.2337/db14-0635
Duan, F., and March, J. C. (2010). Engineered bacterial communication prevents vibrio cholerae virulence in an infant mouse model. Proc. Natl. Acad. Sci. 107, 11260–11264. doi: 10.1073/pnas.1001294107
Fahnøe, U., and Bukh, J. (2019). Full-length open reading frame amplification of hepatitis C Virus. Methods Mol. Biol,1911, 85–91. doi: 10.1007/978-1-4939-8976-8_5.
Fakhoury, M., Negrulj, R., Mooranian, A., and Al-Salami, H. (2014). Inflammatory bowel disease: clinical aspects and treatments. J. Inflamm. Res. 7, 113–120. doi: 10.2147/JIR.S65979
Fan, J. X., Peng, M. Y., Wang, H., Zheng, H. R., Liu, Z. L., Li, C. X., et al. (2019). Engineered bacterial bioreactor for tumor therapy via Fenton-like reaction with localized H2O2 generation. Adv. Mater. 31:1808278. doi: 10.1002/adma.201808278
Fassarella, M., Blaak, E. E., Penders, J., Nauta, A., Smidt, H., and Zoetendal, E. G. (2021). Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut 70, 595–605. doi: 10.1136/gutjnl-2020-321747
Forbes, N. S., Coffin, R. S., Deng, L., Evgin, L., Fiering, S., Giacalone,, et al. (2018). White paper on microbial anti-cancer therapy and prevention. J. Immunother. Cancer 6, 78–24. doi: 10.1186/s40425-018-0381-3
Forslund, K., Hildebrand, F., Nielsen, T., Falony, G., Le Chatelier, E., Sunagawa, S., et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266. doi: 10.1038/nature15766
Fryar, C. D., Carroll, M. D., and Afful, J. (2020). Prevalence of overweight, obesity, and severe obesity among children and adolescents aged 2–19 years: United States, 1963–1965 through 2017–2018. National Center for Health Statistics.
Fryar, C. D., Carroll, M. D., and Ogden, C. L. (2018). Prevalence of overweight, obesity, and severe obesity among children and adolescents aged 2–19 years: United States, 1963–1965 through 2015–2016.
Fuchs, F. D., and Whelton, P. K. (2020). High blood pressure and cardiovascular disease. Hypertension 75, 285–292. doi: 10.1161/hypertensionaha.119.14240
Ganai, S., Arenas, R. B., Sauer, J. P., Bentley, B., and Forbes, N. S. (2011). In tumors salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 18, 457–466. doi: 10.1038/cgt.2011.10
Geirnaert, A., Calatayud, M., Grootaert, C., Laukens, D., Devriese, S., Smagghe, G., et al. (2017). Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 7, 11450–11414. doi: 10.1038/s41598-017-11734-8
Gharibzahedi, S. M. T., and Smith, B. (2021). Legume proteins are smart carriers to encapsulate hydrophilic and hydrophobic bioactive compounds and probiotic bacteria: a review. Compr. Rev. Food Sci. Food Saf. 20, 1250–1279. doi: 10.1111/1541-4337.12699
Gheorghe, A., Griffiths, U., Murphy, A., Legido-Quigley, H., Lamptey, P., and Perel, P. (2018). The economic burden of cardiovascular disease and hypertension in low- and middle-income countries: a systematic review. BMC Public Health 18:975. doi: 10.1186/s12889-018-5806-x
Gilbert, J. A., Quinn, R. A., Debelius, J., Xu, Z. J. Z., Morton, J., Garg, N., et al. (2016). Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103. doi: 10.1038/nature18850
Gionchetti, P., Rizzello, F., Helwig, U., Venturi, A., Lammers, K. M., Brigidi, P., et al. (2003). Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124, 1202–1209. doi: 10.1016/S0016-5085(03)00171-9
Guo, S. (2014). Insulin signaling, resistance, and the metabolic syndrome: insights from mouse models to disease mechanisms. J. Endocrinol. 220, T1–T23. doi: 10.1530/joe-13-0327
Gupta, S. K., and Shukla, P. (2016). Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit. Rev. Biotechnol. 36, 1089–1098. doi: 10.3109/07388551.2015.1084264
Han, J.-W., Choi, Y. J., Cho, S., Zheng, S., Ko, S. Y., Park, J.-O., et al. (2016). Active tumor-therapeutic liposomal bacteriobot combining a drug (paclitaxel)-encapsulated liposome with targeting bacteria (salmonella typhimurium). Sensors Actuators B Chem. 224, 217–224. doi: 10.1016/j.snb.2015.09.034
Hashimoto, T., Hashimoto, J., Teruya, K., Hirano, T., Shin-ya, K., Ikeda,, et al. (2015). Biosynthesis of versipelostatin: identification of an enzyme-catalyzed [4+ 2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J. Am. Chem. Soc. 137, 572–575. doi: 10.1021/ja510711x
Hashimoto, T., Kozone, I., Hashimoto, J., Suenaga, H., Fujie, M., Satoh, N., et al. (2020a). Identification, cloning and heterologous expression of biosynthetic gene cluster for desertomycin. J. Antibiot. 73, 650–654. doi: 10.1038/s41429-020-0319-0
Hashimoto, T., Kozone, I., Hashimoto, J., Ueoka, R., Kagaya, N., Fujie, M., et al. (2020b). Novel macrolactam compound produced by the heterologous expression of a large cryptic biosynthetic gene cluster of Streptomyces rochei IFO12908. J. Antibiot. 73, 171–174. doi: 10.1038/s41429-019-0265-x
He, L., Yang, H., Liu, F., Chen, Y., Tang, S., Ji, W., et al. (2017). Escherichia coli Nissle 1917 engineered to express Tum-5 can restrain murine melanoma growth. Oncotarget 8, 85772–85782. doi: 10.18632/oncotarget.20486
Heavey, M. K., Durmusoglu, D., Crook, N., and Anselmo, A. C. (2021). Discovery and delivery strategies for engineered live biotherapeutic products. Trends Biotechnol. 40, 354–369. doi: 10.1016/j.tibtech.2021.08.002
Herzog, C. (2016). Successful comeback of the single-dose live oral cholera vaccine CVD 103-HgR. Travel Med. Infect. Dis. 14, 373–377. doi: 10.1016/j.tmaid.2016.07.003
Hoffman, R. M. (2016). Future of bacterial therapy of cancer. Methods Mol. Biol. 1409, 177–184. doi: 10.1007/978-1-4939-3515-4_15
Hosseinidoust, Z., Mostaghaci, B., Yasa, O., Park, B.-W., Singh, A. V., and Sitti, M. (2016). Bioengineered and biohybrid bacteria-based systems for drug delivery. Adv. Drug Deliv. Rev. 106, 27–44. doi: 10.1016/j.addr.2016.09.007
Huang, D., Deng, M., and Kuang, S. (2019). Polymeric carriers for controlled drug delivery in obesity treatment. Trends Endocrinol. Metab. 30, 974–989. doi: 10.1016/j.tem.2019.09.004
Huang, X., Pan, J., Xu, F., Shao, B., Wang, Y., Guo, X., et al. (2021). Bacteria-based cancer immunotherapy. Advanced Science 8:2003572. doi: 10.1002/advs.202003572
Hubbard, T. P., Billings, G., Dörr, T., Sit, B., Warr, A. R., Kuehl, C. J., et al. (2018). A live vaccine rapidly protects against cholera in an infant rabbit model. Sci. Transl. Med. 10:423. doi: 10.1126/scitranslmed.aap8423
Huo, L., Hug, J. J., Fu, C., Bian, X., Zhang, Y., and Müller, R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 36, 1412–1436. doi: 10.1039/c8np00091c
Hwang, J., Jin, J., Jeon, S., Moon, S. H., Park, M. Y., Yum, D.-Y., et al. (2020). SOD1 suppresses pro-inflammatory immune responses by protecting against oxidative stress in colitis. Redox Biol. 37:101760. doi: 10.1016/j.redox.2020.101760
Isabella, V. M., Ha, B. N., Castillo, M. J., Lubkowicz, D. J., Rowe, S. E., Millet, Y. A., et al. (2018). Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864. doi: 10.1038/nbt.4222
Jakubczyk, D., Leszczyńska, K., and Górska, S. (2020). The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)—a critical review. Nutrients 12:1973. doi: 10.3390/nu12071973
Janku, F., Zhang, H. H., Pezeshki, A., Goel, S., Murthy, R., Wang-Gillam, A., et al. (2021). Intratumoral injection of clostridium novyi-NT spores in patients with treatment-refractory advanced solid TumorsPhase I study of Intratumoral clostridium novyi-NT. Clin. Cancer Res. 27, 96–106. doi: 10.1158/1078-0432.ccr-20-2065
Jia, X., Guo, J., Guo, S., Zhao, T., Liu, X., Cheng, C., et al. (2021). Antitumor effects and mechanisms of CpG ODN combined with attenuated salmonella-delivered siRNAs against PD-1. Int. Immunopharmacol. 90:107052. doi: 10.1016/j.intimp.2020.107052
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi: 10.1038/nbt.2508
Jiang, W., Zhao, X., Gabrieli, T., Lou, C., Ebenstein, Y., and Zhu, T. F. (2015). Cas9-assisted targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 6, 1–8. doi: 10.1038/ncomms9101
Kaczmarska, Z., Czarnocki-Cieciura, M., Górecka-Minakowska, K. M., Wingo, R. J., Jackiewicz, J., Zajko, W., et al. (2022). Structural basis of transposon end recognition explains central features of Tn7 transposition systems. Mol. Cell 82, 2618–2632.e7. doi: 10.1016/j.molcel.2022.05.005
Kalia, V. C., Patel, S. K., Cho, B.-K., Wood, T. K., and Lee, J.-K. (2021). Emerging applications of bacteria as antitumor agents. Semin. Cancer Biol. doi: 10.1016/j.semcancer.2021.05.012
Kamada, N., Chen, G. Y., Inohara, N., and Nunez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Karbach, J., Neumann, A., Brand, K., Wahle, C., Siegel, E., Maeurer, M., et al. (2012). Phase I clinical trial of mixed bacterial vaccine (Coley's toxins) in patients with NY-ESO-1 expressing cancers: immunological effects and clinical activity. Clin. Cancer Res. 18, 5449–5459. doi: 10.1158/1078-0432.ccr-12-1116
Kawaguchi, K., Miyake, K., Zhao, M., Kiyuna, T., Igarashi, K., Miyake, M., et al. (2018). Tumor targeting salmonella typhimurium A1-R in combination with gemcitabine (GEM) regresses partially GEM-resistant pancreatic cancer patient-derived orthotopic xenograft (PDOX) nude mouse models. Cell Cycle 17, 2019–2026. doi: 10.1080/15384101.2018.1480223
Ke, J., Wang, B., and Yoshikuni, Y. (2021). Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 39, 244–261. doi: 10.1016/j.tibtech.2020.07.008
Khan, M. A. B., Hashim, M. J., King, J. K., Govender, R. D., Mustafa, H., and Al Kaabi, J. (2020). Epidemiology of type 2 diabetes–global burden of disease and forecasted trends. J. epidemiology and global health 10, 107–111. doi: 10.2991/jegh.k.191028.001
Khunti, S., Khunti, K., and Seidu, S. (2019). Therapeutic inertia in type 2 diabetes: prevalence, causes, consequences and methods to overcome inertia. Ther. Adv. Endocrinol. Metab. 10:2042018819844694. doi: 10.1177/2042018819844694
Kim, J.-E., Phan, T. X., Nguyen, V. H., Dinh-Vu, H.-V., Zheng, J. H., Yun, M., et al. (2015). Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1β. Theranostics 5, 1328–1342. doi: 10.7150/thno.11432
Kobyliak, N., Conte, C., Cammarota, G., Haley, A. P., Styriak, I., Gaspar, L., et al. (2016). Probiotics in prevention and treatment of obesity: a critical view. Nutrition Metabol. 13:14. doi: 10.1186/s12986-016-0067-0
Kocsis, T., Molnár, B., Németh, D., Hegyi, P., Szakács, Z., Bálint, A., et al. (2020). Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: a meta-analysis of randomized clinical trials. Sci. Rep. 10, 11787–11714. doi: 10.1038/s41598-020-68440-1
Kowalczykowski, S. C. (2015). An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7:a016410. doi: 10.1101/cshperspect.a016410
Koziolek, M., Grimm, M., Becker, D., Iordanov, V., Zou, H., Shimizu, J., et al. (2015). Investigation of pH and temperature profiles in the GI tract of fasted human subjects using the Intellicap® system. J. Pharm. Sci. 104, 2855–2863. doi: 10.1002/jps.24274
Kurtz, C. B., Millet, Y. A., Puurunen, M. K., Perreault, M., Charbonneau, M. R., Isabella, V. M., et al. (2019). An engineered E-coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 11:14. doi: 10.1126/scitranslmed.aau7975
Laurent, S. (2017). Antihypertensive drugs. Pharmacol. Res. 124, 116–125. doi: 10.1016/j.phrs.2017.07.026
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Loffler, F. E., O'Malley, M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi: 10.1038/s41579-019-0255-9
Le, D. T., Wang-Gillam, A., Picozzi, V., Greten, T. F., Crocenzi, T., Springett, G., et al. (2015). Safety and survival with GVAX pancreas prime and listeria monocytogenes-expressing Mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 33, 1325–1333. doi: 10.1200/jco.2014.57.4244
Lee, M., and Chang, E. B. (2021). Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology 160, 524–537. doi: 10.1053/j.gastro.2020.09.056
Lentz, R., Benson, A. B., and Kircher, S. (2019). Financial toxicity in cancer care: prevalence, causes, consequences, and reduction strategies. J. Surg. Oncol. 120, 85–92. doi: 10.1002/jso.25374
Leventhal, D. S., Sokolovska, A., Li, N., Plescia, C., Kolodziej, S. A., Gallant, C. W., et al. (2020). Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 11, 2739–2715. doi: 10.1038/s41467-020-16602-0
Li, M., and Elledge, S. J. (2012). SLIC: a method for sequence- and ligation-independent cloning. In Methods Mol Biol, 852, 51–9. doi: 10.1007/978-1-61779-564-0_5
Li, Y., Sun, Y.-h., Ison, C., Levine, M. M., and Tang, C. M. (2004). Vaccination with attenuated Neisseria meningitidis strains protects against challenge with live meningococci. Infect. Immun., 72, 345–351. doi: 10.1128/iai.72.1.345-351.2004
Li, H. Y., Yang, Y., Hong, W. Q., Huang, M. Y., Wu, M., and Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 5:1. doi: 10.1038/s41392-019-0089-y
Li, C. X., Yu, B., Shi, L., Geng, W., Lin, Q. B., Ling, C. C., et al. (2017). ‘Obligate’anaerobic salmonella strain YB1 suppresses liver tumor growth and metastasis in nude mice. Oncol. Lett. 13, 177–183. doi: 10.3892/ol.2016.5453
Lim, D., Kim, K. S., Kim, H., Ko, K.-C., Song, J. J., Choi, J. H., et al. (2017). Anti-tumor activity of an immunotoxin (TGFα-PE38) delivered by attenuated salmonella typhimurium. Oncotarget 8, 37550–37560. doi: 10.18632/oncotarget.17197
Lin, D., Feng, X., Mai, B., Li, X., Wang, F., Liu, J., et al. (2021). Bacterial-based cancer therapy: an emerging toolbox for targeted drug/gene delivery. Biomaterials 277:121124. doi: 10.1016/j.biomaterials.2021.121124
Lin, Y., Krogh-Andersen, K., Pelletier, J., Marcotte, H., Östenson, C.-G., and Hammarström, L. (2016). Oral delivery of pentameric glucagon-like peptide-1 by recombinant lactobacillus in diabetic rats. PLoS One 11:e0162733. doi: 10.1371/journal.pone.0162733
Lin, X., Xu, Y., Pan, X., Xu, J., Ding, Y., Sun, X., et al. (2020). Global, regional, and national burden and trend of diabetes in 195 countries and territories: an analysis from 1990 to 2025. Sci. Rep. 10, 14790–14711. doi: 10.1038/s41598-020-71908-9
Liu, M., Li, S., Zhang, Q., Xu, Z., Wang, J., and Sun, H. (2018). Oral engineered Bifidobacterium longum expressing rhMnSOD to suppress experimental colitis. Int. Immunopharmacol. 57, 25–32. doi: 10.1016/j.intimp.2018.02.004
Liu, K.-F., Liu, X.-R., Li, G.-L., Lu, S.-P., Jin, L., and Wu, J. (2016). Oral administration of Lactococcus lactis-expressing heat shock protein 65 and tandemly repeated IA2P2 prevents type 1 diabetes in NOD mice. Immunol. Lett. 174, 28–36. doi: 10.1016/j.imlet.2016.04.008
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38, 1091–1125. doi: 10.1111/1574-6976.12076
Lunia, M. K., Sharma, B. C., Sharma, P., Sachdeva, S., and Srivastava, S. (2014). Probiotics prevent hepatic encephalopathy in patients with cirrhosis: a randomized controlled trial. Clin. Gastroenterol. Hepatol. 12:e1001. doi: 10.1016/j.cgh.2013.11.006
Luo, Y., Enghiad, B., and Zhao, H. (2016). New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat. Prod. Rep. 33, 174–182. doi: 10.1039/c5np00085h
Ma, B., Pan, Q., and Peppelenbosch, M. P. (2017). Genetically engineered bacteria for treating human disease. Trends Pharmacol. Sci. 38, 763–764. doi: 10.1016/j.tips.2017.07.001
Mao, N., Cubillos-Ruiz, A., Cameron, D. E., and Collins, J. J. (2018). Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 10:586. doi: 10.1126/scitranslmed.aao2586
Martínez-García, E., and de Lorenzo, V. (2012). Transposon-based and plasmid-based genetic tools for editing genomes of gram-negative bacteria. Methods Mol., 813, 267–83. doi: 10.1007/978-1-61779-412-4_16
Matoori, S., and Leroux, J.-C. (2015). Recent advances in the treatment of hyperammonemia. Adv. Drug Deliv. Rev. 90, 55–68. doi: 10.1016/j.addr.2015.04.009
Mazhar, S. F., Afzal, M., Almatroudi, A., Munir, S., Ashfaq, U. A., Rasool, M., et al. (2020). The prospects for the therapeutic implications of genetically engineered probiotics. J. Food Qual. 2020, 1–11. doi: 10.1155/2020/9676452
Mei, S., Theys, J., Landuyt, W., Anne, J., and Lambin, P. (2002). Optimization of tumor-targeted gene delivery by engineered attenuated salmonella typhimurium. Anticancer Res. 22, 3261–3266.
Miller, K. M., Foster, N. C., Beck, R. W., Bergenstal, R. M., DuBose, S. N., DiMeglio, L. A., et al. (2015). Current state of type 1 diabetes treatment in the US: updated data from the T1D exchange clinic registry. Diabetes Care 38, 971–978. doi: 10.2337/dc15-0078
Mirkhani, N., Gwisai, T., and Schuerle, S. (2021). Engineering cell-based Systems for Smart Cancer Therapy. Advanced Intelligent Systems 4:2100134. doi: 10.1002/aisy.202100134
Miyazaki, R., and van der Meer, J. R. (2013). A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl. Environ. Microbiol. 79, 4440–4447. doi: 10.1128/aem.00711-13
Mojgani, N., Shahali, Y., and Dadar, M. (2020). Immune modulatory capacity of probiotic lactic acid bacteria and applications in vaccine development. Benefic. Microbes 11, 213–226. doi: 10.3920/bm2019.0121
Molina, B., Mastroianni, J., Suarez, E., Soni, B., Forsberg, E., and Finley, K. (2021). Treatment with Bacterial Biologics Promotes Healthy Aging and Traumatic Brain Injury Responses in Adult Drosophila, Modeling the Gut–Brain Axis and Inflammation Responses. Cells 10:900. doi: 10.3390/cells10040900
Mu, Y., and Cong, Y. (2029). Bacillus coagulans and its applications in medicine. Beneficial microbes, 10:679–688. doi: 10.3920/bm2019.0016
Mukai, T., Yoneji, T., Yamada, K., Fujita, H., Nara, S., and Su’etsugu, M. (2020). Overcoming the challenges of megabase-sized plasmid construction in Escherichia coli. ACS Synth. Biol. 9, 1315–1327. doi: 10.1021/acssynbio.0c00008
Naciute, M., Kiwitt, T., Kemp, R. A., and Hook, S. (2021). Bacteria biohybrid oral vaccines for colorectal cancer treatment reduce tumor growth and increase immune infiltration. Vaccine 39, 5589–5599. doi: 10.1016/j.vaccine.2021.08.028
Nah, H.-J., Pyeon, H.-R., Kang, S.-H., Choi, S.-S., and Kim, E.-S. (2017). Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front. Microbiol. 8:394. doi: 10.3389/fmicb.2017.00394
Namai, F., Shigemori, S., Ogita, T., Sato, T., and Shimosato, T. (2020). Microbial therapeutics for acute colitis based on genetically modified Lactococcus lactis hypersecreting IL-1Ra in mice. Exp. Mol. Med. 52, 1627–1636. doi: 10.1038/s12276-020-00507-5
Nelson, M. T., Charbonneau, M. R., Coia, H. G., Castillo, M. J., Holt, C., Greenwood, E. S., et al. (2021). Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nat. Commun. 12:2805. doi: 10.1038/s41467-021-23072-5
Ni, J., Wangensteen, K. J., Nelsen, D., Balciunas, D., Skuster, K. J., Urban, M. D., et al. (2016). Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mob. DNA 7, 6–11. doi: 10.1186/s13100-016-0062-z
Oh, J.-H., Schueler, K. L., Stapleton, D. S., Alexander, L. M., Yen, C.-L. E., Keller, M. P., et al. (2020). Secretion of recombinant interleukin-22 by engineered lactobacillus reuteri reduces fatty liver disease in a mouse model of diet-induced obesity. Msphere 5, e00183–e00120. doi: 10.1128/msphere.00183-20
Oßwald, C., Zipf, G., Schmidt, G., Maier, J., Bernauer, H. S., Müller, R., et al. (2014). Modular construction of a functional artificial epothilone polyketide pathway. ACS Synth. Biol. 3, 759–772. doi: 10.1021/sb300080t
O'Toole, P. W., and Claesson, M. J. (2010). Gut microbiota: changes throughout the lifespan from infancy to elderly. Int. Dairy J. 20, 281–291. doi: 10.1016/j.idairyj.2009.11.010
Ott, S. J., Musfeldt, M., Wenderoth, D. F., Hampe, J., Brant, O., Folsch, U. R., et al. (2004). Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53, 685–693. doi: 10.1136/gut.2003.025403
Pan, X.-F., Wang, L., and Pan, A. (2021). Epidemiology and determinants of obesity in China. The lancet Diabetes & endocrinology 9, 373–392. doi: 10.1016/s2213-8587(21)00045-0
Pandey, M., Choudhury, H., Vijayagomaran, P. A. P., Lian, P. N. P., Ning, T. J., Wai, N. Z., et al. (2022). Recent update on bacteria as a delivery carrier in cancer therapy: from evil to allies. Pharm. Res. 39, 1115–1134. doi: 10.1007/s11095-022-03240-y
Pedrolli, D. B., Ribeiro, N. V., Squizato, P. N., de Jesus, V. N., Cozetto, D. A., Tuma, R. B., et al. (2019). Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 37, 100–115. doi: 10.1016/j.tibtech.2018.09.005
Peters, J. M., Koo, B.-M., Patino, R., Heussler, G. E., Hearne, C. C., Qu, J., et al. (2019). Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat. Microbiol. 4, 244–250. doi: 10.1038/s41564-018-0327-z
Piñero-Lambea, C., Bodelón, G., Fernández-Periáñez, R., Cuesta, A. M., Álvarez-Vallina, L., Fernández, L. A., et al. (2015). Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth. Biol. 4, 463–473. doi: 10.1021/sb500252a
Plavec, T. V., and Berlec, A. (2020). Safety aspects of genetically modified lactic acid bacteria. Microorganisms 8:297. doi: 10.3390/microorganisms8020297
Powell-Wiley, T. M., Poirier, P., Burke, L. E., Despres, J. P., Gordon-Larsen, P., Lavie, C. J., et al. (2021). Obesity and cardiovascular disease: a scientific statement from the American Heart Association. Circulation 143, E984–E1010. doi: 10.1161/cir.0000000000000973
Puurunen, M. K., Vockley, J., Searle, S. L., Sacharow, S. J., Phillips, J. A., Denney, W. S., et al. (2021). Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nat. Metab. 3, 1125–1132. doi: 10.1038/s42255-021-00430-7
Razmpoosh, E., Javadi, M., Ejtahed, H. S., and Mirmiran, P. (2016). Probiotics as beneficial agents in the management of diabetes mellitus: a systematic review. Diabetes Metab. Res. Rev. 32, 143–168. doi: 10.1002/dmrr.2665
Riese, P., Sakthivel, P., Trittel, S., and Guzmán, C. A.. (2014). Intranasal formulations: promising strategy to deliver vaccines. Expert Opinion on Drug Delivery, 11, 1619–1634. doi: 10.1517/17425247.2014.931936
Riglar, D. T., Giessen, T. W., Baym, M., Kerns, S. J., Niederhuber, M. J., Bronson, R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35, 653–658. doi: 10.1038/nbt.3879
Roberts, N. J., Zhang, L., Janku, F., Collins, A., Bai, R.-Y., Staedtke, V., et al. (2014). Intratumoral injection of clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 35, 653–658. doi: 10.1038/nbt.3879
Saez-Lara, M. J., Gomez-Llorente, C., Plaza-Diaz, J., and Gil, A. (2015). The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. Biomed. Res. Int. 2015, 1–15. doi: 10.1155/2015/505878
Sánchez, M. T., Ruiz, M. A., Lasserrot, A., Hormigo, M., and Morales, M. E. (2017). An improved ionic gelation method to encapsulate lactobacillus spp. bacteria: protection, survival and stability study. Food Hydrocoll. 69, 67–75. doi: 10.1016/j.foodhyd.2017.01.019
Sarate, P., Heinl, S., Poiret, S., Drinić, M., Zwicker, C., Schabussova, I., et al. (2019). E. coli Nissle 1917 is a safe mucosal delivery vector for a birch-grass pollen chimera to prevent allergic poly-sensitization. Mucosal Immunol. 12, 132–144. doi: 10.1038/s41385-018-0084-6
Satchell, K. J. (2018). Engineered bacteria for cholera prophylaxis. Cell Host Microbe 24, 192–194. doi: 10.1016/j.chom.2018.07.013
Schultz, M. (2008). Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 14, 1012–1018. doi: 10.1002/ibd.20377
Selvanesan, B. C., Chandra, D., Quispe-Tintaya, W., Jahangir, A., Patel, A., Meena, K., et al. (2022). Listeria delivers tetanus toxoid protein to pancreatic tumors and induces cancer cell death in mice. Sci. Transl. Med. 14:eabc1600. doi: 10.1126/scitranslmed.abc1600
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Sharif, A., Kashani, H. H., Nasri, E., Soleimani, Z., and Sharif, M. R. (2017). The role of probiotics in the treatment of dysentery: a randomized double-blind clinical trial. Probiotics and Antimicrobial Proteins 9, 380–385. doi: 10.1007/s12602-017-9271-0
Sharma, T., Joshi, N., Mandyal, A. K., Nordqvist, S. L., Lebens, M., Kanchan, V., et al. (2020). Development of Hillchol®, a low-cost inactivated single strain Hikojima oral cholera vaccine. Vaccine 38, 7998–8009. doi: 10.1016/j.vaccine.2020.10.043
Shen, T.-C. D., Albenberg, L., Bittinger, K., Chehoud, C., Chen, Y.-Y., Judge, C. A., et al. (2015). Engineering the gut microbiota to treat hyperammonemia. J. Clin. Invest. 125, 2841–2850. doi: 10.1172/jci79214
Shigemori, S., Watanabe, T., Kudoh, K., Ihara, M., Nigar, S., Yamamoto, Y., et al. (2015). Oral delivery of Lactococcus lactis that secretes bioactive heme oxygenase-1 alleviates development of acute colitis in mice. Microb. Cell Factories 14:189. doi: 10.1186/s12934-015-0378-2
Si, H., Yang, Q., Hu, H., Ding, C., Wang, H., and Lin, X. (2021). Colorectal cancer occurrence and treatment based on changes in intestinal flora. Semin. Cancer Biol. 70, 3–10. doi: 10.1016/j.semcancer.2020.05.004
Sivick, K. E., Desbien, A. L., Glickman, L. H., Reiner, G. L., Corrales, L., Surh, N. H., et al. (2018). Magnitude of therapeutic STING activation determines CD8+ T cell-mediated anti-tumor immunity. Cell Rep. 25:e3075. doi: 10.1016/j.celrep.2018.11.047
Souza, B. M., Preisser, T. M., Pereira, V. B., Zurita-Turk, M., de Castro, C. P., da Cunha, V. P., et al. (2016). Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb. Cell Factories 15, 150–118. doi: 10.1186/s12934-016-0548-x
Spees, A. M., Wangdi, T., Lopez, C. A., Kingsbury, D. D., Xavier, M. N., Winter, S. E., et al. (2013). Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio 4, e00430–e00413. doi: 10.1128/mbio.00430-13
Staedtke, V., Roberts, N. J., Bai, R.-Y., and Zhou, S. (2016). Clostridium novyi-NT in cancer therapy. Genes & diseases 3, 144–152. doi: 10.1016/j.gendis.2016.01.003
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. doi: 10.1126/science.289.5483.1352
Strain-Damerell, C., Mahajan, P., Fernandez-Cid, A., Gileadi, O., and Burgess-Brown, N. A. (2021). Screening and production of recombinant human proteins: ligation-independent cloning, Structural Genomics, 2199, 23–43. doi: 10.1007/978-1-0716-0892-0_3
Strecker, J., Ladha, A., Gardner, Z., Schmid-Burgk, J. L., Makarova, K. S., Koonin, E. V., et al. (2019). RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53. doi: 10.1126/science.aax9181
Ta, L. P., Bujna, E., Antal, O., Ladanyi, M., Juhasz, R., Szecsi,, et al. (2021). Effects of various polysaccharides (alginate, carrageenan, gums, chitosan) and their combination with prebiotic saccharides (resistant starch, lactosucrose, lactulose) on the encapsulation of probiotic bacteria lactobacillus casei 01 strain. Int. J. Biol. Macromol. 183, 1136–1144. doi: 10.1016/j.ijbiomac.2021.04.170
Taniguchi, S. I., Fujimori, M., Sasaki, T., Tsutsui, H., Shimatani, Y., Seki, K., et al. (2010). Targeting solid tumors with non-pathogenic obligate anaerobic bacteria. Cancer Sci. 101, 1925–1932. doi: 10.1111/j.1349-7006.2010.01628.x
Tarahomjoo, S. (2012). Development of vaccine delivery vehicles based on lactic acid bacteria. Mol. Biotechnol. 51, 183–199. doi: 10.1007/s12033-011-9450-2
Tavanti, M. (2022). Synthetic DNA libraries for protein engineering toward process improvement in drug synthesis. In F. Magnani, C. Marabelli, and F. Paradisi (Eds.), Enzyme engineering: Methods and protocols (pp. 33–45). New York, NY: Springer US.
Terol, G. L., Gallego-Jara, J., Martínez, R. A. S., Vivancos, A. M., Díaz, M. C., and de Diego Puente, T. (2021). Impact of the expression system on recombinant protein production in Escherichia coli BL21. Front. Microbiol. 12:001. doi: 10.3389/fmicb.2021.682001
Tochitani, S. (2021). Vertical transmission of gut microbiota: points of action of environmental factors influencing brain development. Neurosci. Res. 168, 83–94. doi: 10.1016/j.neures.2020.11.006
Tschorn, N., Berg, K., and Stitz, J. (2020). Transposon vector-mediated stable gene transfer for the accelerated establishment of recombinant mammalian cell pools allowing for high-yield production of biologics. Biotechnol. Lett. 42, 1103–1112. doi: 10.1007/s10529-020-02889-y
van de Wijgert, J. H., and Verwijs, M. C. (2020). Lactobacilli-containing vaginal probiotics to cure or prevent bacterial or fungal vaginal dysbiosis: a systematic review and recommendations for future trial designs. BJOG Int. J. Obstet. Gynaecol. 127, 287–299. doi: 10.1111/1471-0528.15870
Vo, P. L. H., Ronda, C., Klompe, S. E., Chen, E. E., Acree, C., Wang, H. H., et al. (2021). CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39, 480–489. doi: 10.1038/s41587-020-00745-y
Waksman, G. (2019). From conjugation to T4S systems in gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20:e47012. doi: 10.15252/embr.201847012
Wan, T., and Ping, Y. (2021). Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv. Drug Deliv. Rev. 168, 196–216. doi: 10.1016/j.addr.2020.05.002
Wan, Y., Yang, L., Jiang, S., Qian, D., and Duan, J. (2022). Excessive apoptosis in ulcerative colitis: crosstalk between apoptosis, ROS, ER stress, and intestinal homeostasis. Inflamm. Bowel Dis. 28, 639–648. doi: 10.1093/ibd/izab277
Wang, L., Chen, T., Wang, H., Wu, X., Cao, Q., Wen,, et al. (2021). Engineered bacteria of MG1363-pMG36e-GLP-1 attenuated obesity-induced by high fat diet in mice. Front. Cell. Infect. Microbiol. 11:62. doi: 10.3389/fcimb.2021.595575
Wang, H. H., Kim, H., Cong, L., Jeong, J., Bang, D., and Church, G. M. (2012). Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593. doi: 10.1038/NMETH.1971
Wang, D., Zhang, F., and Gao, G. P. (2020). CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cells 181, 136–150. doi: 10.1016/j.cell.2020.03.023
Wei, T., Cheng, B.-Y., and Liu, J.-Z. (2016). Genome engineering Escherichia coli for L-DOPA overproduction from glucose. Sci. Rep. 6, 1–9. doi: 10.1038/srep30080
Wells, J. M., and Mercenier, A. (2008). Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nature Reviews Microbiology 6, 349–362. doi: 10.1038/nrmicro1840
Westbrook, A. W., Moo-Young, M., and Chou, C. P. (2016). Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl. Environ. Microbiol. 82, 4876–4895. doi: 10.1128/aem.01159-16
Whorwell, P. J., Altringer, L., Morel, J., Bond, Y., Charbonneau, D., Omahony, L., et al. (2006). Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Official J. American College of Gastroenterology| ACG 101, 1581–1590. doi: 10.1111/j.1572-0241.2006.00734.x
Wu, J., Xin, Y., Kong, J., and Guo, T. (2021). Genetic tools for the development of recombinant lactic acid bacteria. Microb. Cell Factories 20, 118–110. doi: 10.1186/s12934-021-01607-1
Xu, T., Xiao, M., and Yu, L. (2021). Method for efficient soluble expression and purification of recombinant hyperactive Tn5 transposase. Protein Expr. Purif. 183:105866. doi: 10.1016/j.pep.2021.105866
Yaghoubi, A., Khazaei, M., Jalili, S., Hasanian, S. M., Avan, A., Soleimanpour, S., et al. (2020). Bacteria as a double-action sword in cancer. Biochimica et Biophysica Acta (BBA) - reviews on. Cancer 1874:188388. doi: 10.1016/j.bbcan.2020.188388
Yang, G., Jiang, Y., Yang, W., Du, F., Yao, Y., Shi, C., et al. (2015). Effective treatment of hypertension by recombinant lactobacillus plantarum expressing angiotensin converting enzyme inhibitory peptide. Microb. Cell Factories 14, 1–9. doi: 10.1186/s12934-015-0394-2
Yoon, W., Park, Y. C., Kim, J., Chae, Y. S., Byeon, J. H., Min, S.-H., et al. (2017). Application of genetically engineered salmonella typhimurium for interferon-gamma–induced therapy against melanoma. Eur. J. Cancer 70, 48–61. doi: 10.1016/j.ejca.2016.10.010
Yuan, X., Ni, H., Chen, X., Feng, X., Wu, Q., and Chen, J. (2018). Identification of therapeutic effect of glucagon-like peptide 1 in the treatment of STZ-induced diabetes mellitus in rats by restoring the balance of intestinal flora. J. Cell. Biochem. 119, 10067–10074. doi: 10.1002/jcb.27343
Zhang, B., Liu, Y., Lan, X., Xu, X., Zhang, X., Li, X., et al. (2018). Oral Escherichia coli expressing IL-35 meliorates experimental colitis in mice. J. Transl. Med. 16, 71–10. doi: 10.1186/s12967-018-1441-7
Zhang, Y.-X., Perry, K., Vinci, V. A., Powell, K., Stemmer, W. P., and del Cardayré, S. B. (2002). Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415, 644–646. doi: 10.1038/415644a
Zhang, J. J., Yamanaka, K., Tang, X., and Moore, B. S. (2019). Direct cloning and heterologous expression of natural product biosynthetic gene clusters by transformation-associated recombination. Methods Enzymol., 621, 87–110. doi: 10.1016/bs.mie.2019.02.026
Zhao, C., He, J., Cheng, H., Zhu, Z., and Xu, H. (2016). Enhanced therapeutic effect of an antiangiogenesis peptide on lung cancer in vivo combined with salmonella VNP20009 carrying a Sox2 shRNA construct. J. Exp. Clin. Cancer Res. 35, 107–111. doi: 10.1186/s13046-016-0381-4
Zheng, L., Kelly, C. J., and Colgan, S. P. (2015). Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am. J. Phys. Cell Phys. 309, C350–C360. doi: 10.1152/ajpcell.00191.2015
Zheng, J. H., and Min, J.-J. (2016). Targeted cancer therapy using engineered salmonella typhimurium. Chonnam Med. J. 52, 173–184. doi: 10.4068/cmj.2016.52.3.173
Zhou, S., Gravekamp, C., Bermudes, D., and Liu, K. (2018). Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743. doi: 10.1038/s41568-018-0070-z
Zimmet, P. Z., Magliano, D. J., Herman, W. H., and Shaw, J. E. (2014). Diabetes: a 21st century challenge. The lancet Diabetes & endocrinology 2, 56–64. doi: 10.1016/S2213-8587(13)70112-8
Keywords: bacteriotherapy, genetically engineered bacterium, clinical application, synthetic biotechnology, heterologous expression
Citation: Liu Y, Feng J, Pan H, Zhang X and Zhang Y (2022) Genetically engineered bacterium: Principles, practices, and prospects. Front. Microbiol. 13:997587. doi: 10.3389/fmicb.2022.997587
Received: 19 July 2022; Accepted: 23 September 2022;
Published: 13 October 2022.
Edited by:
Elisa Michelini, University of Bologna, ItalyReviewed by:
Hirokazu Suzuki, Tottori University, JapanCopyright © 2022 Liu, Feng, Pan, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuwei Zhang, emhhbmd4aXV3ZWl5d3lAMTI2LmNvbQ==; Yunlei Zhang, eXVubGVpemhhbmdAbmptdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.