- Forschungszentrum Jülich GmbH, Institute for Bio- and Geosciences 1, Jülich, Germany
Heme is an essential cofactor for almost all living cells by acting as prosthetic group for various proteins or serving as alternative iron source. However, elevated levels are highly toxic for cells. Several corynebacterial species employ two paralogous, heme-responsive two-component systems (TCS), ChrSA and HrrSA, to cope with heme stress and to maintain intracellular heme homeostasis. Significant cross-talk at the level of phosphorylation between these systems was previously demonstrated. In this study, we have performed a laboratory evolution experiment to adapt Corynebacterium glutamicum to increasing heme levels. Isolated strains showed a highly increased tolerance to heme growing at concentrations of up to 100 μM. The strain featuring the highest heme tolerance harbored a frameshift mutation in the catalytical and ATPase-domain (CA-domain) of the chrS gene, converting it into a catalytically-inactive pseudokinase (ChrS_CA-fs). Reintroduction of the respective mutation in the parental C. glutamicum strain confirmed high heme tolerance and showed a drastic upregulation of hrtBA encoding a heme export system, conserved in Firmicutes and Actinobacteria. The strain encoding the ChrS pseudokinase variant showed significantly higher heme tolerance than a strain lacking chrS. Mutational analysis revealed that induction of hrtBA in the evolved strain is solely mediated via the cross-phosphorylation of the response regulator (RR) ChrA by the kinase HrrS and BACTH assays revealed the formation of heterodimers between HrrS and ChrS. Overall, our results emphasize an important role of the ChrS pseudokinase in high heme tolerance of the evolved C. glutamicum and demonstrate the promiscuity in heme-dependent signaling of the paralogous two-component systems facilitating fast adaptation to changing environmental conditions.
Introduction
Heme constitutes 95% of functional iron in the human body and is a key molecule for almost all living cells (Ponka, 1999; Andrews et al., 2003) acting as cofactor for many important proteins, including cytochromes, hydroxylases, catalases, peroxidases (Ajioka et al., 2006; Layer et al., 2010), and serving as alternative iron source (Anzaldi and Skaar, 2010). Nevertheless, elevated levels of this iron-bound protoporphyrin are highly cytotoxic. While this toxicity partially originates from the redox-active iron, causing the formation of reactive oxygen species (Kumar and Bandyopadhyay, 2005), a not yet unraveled porphyrin-related toxicity is furthermore suggested (Stojiljkovic et al., 1999). Consequently, organisms have evolved sophisticated mechanisms ensuring heme homeostasis (Padmanaban et al., 1989; Anzaldi and Skaar, 2010). Among those, systems enhancing heme tolerance play an important role in both pathogenic and non-pathogenic prokaryotes. Known strategies include mechanisms of (i) heme sequestration (e.g., HemS of Yersinia enteroliticia Stojiljkovic and Hantke, 1994), (ii) heme degradation (e.g., IsdG of Bacillus anthracis Skaar et al., 2006), and (iii) heme export by the HrtBA system (Stauff and Skaar, 2009b; Heyer et al., 2012; Nakamura et al., 2022). The heme-dedicated ATP-binding cassette efflux pump HrtBA is a highly conserved system and predominantly found in Firmicutes and Actinobacteria (Krüger et al., 2022). Recent structural studies shed light on the mechanism HrtBA employs to sequester and extrude heme from the cytoplasmic membrane (Nakamura et al., 2022).
In Gram-positive bacteria, two-component systems (TCS) play a predominant role in the regulation of heme homeostasis (Bibb et al., 2007; Stauff and Skaar, 2009a; Hentschel et al., 2014; Keppel et al., 2019; Krüger et al., 2022). The prototypical TCS consists of a membrane-bound histidine kinase (HK), which undergoes autophosphorlyation at a conserved histidine residue upon stimulus perception. Subsequently, the phosphoryl group is transferred to a conserved aspartate residue of a cytoplasmic response regulator (RR) resulting in an appropriate output, e.g., altering gene expression (Stock et al., 2000; Mascher et al., 2006; Laub and Goulian, 2007). HKs may be composed of multiple domains with a significant architectural diversity, but typically consist of an N-terminal transmembrane domain and a C-terminal transmitter domain. The transmitter domain can be split up in the dimerization and histidine phosphotransfer (DHp) domain and the catalytical and ATPase (CA) domain (Dutta et al., 1999; Gao and Stock, 2009). The CA-domain comprises four sequence motifs, including N, G1, F, and G2 boxes, which bind ATP in a pocket using an ATP lid and are consequently necessary for the autophosphorylation reaction (Kim and Forst, 2001; Wolanin et al., 2002). The DHp-domain possesses the H box motif harboring the conserved histidine residue which is phosphorylated upon stimulus perception (Dutta et al., 1999), as well as the X box, which is required for dimerization. The DHp- and CA-domain are connected via a flexible linker, which probably also supports keeping the RR in place during the phosphotransfer reaction (Casino et al., 2009). Many HKs are bifunctional possessing also a phosphatase motif and subsequently acting both as kinase and phosphatase for the RR (Perego and Hoch, 1996; Laub and Goulian, 2007; Hentschel et al., 2014). Furthermore, also catalytically inactive variants of kinases have been identified, referred to as pseudokinases, that can act as important signaling modulators by various mechanisms (Raju and Shaw, 2015; Kung and Jura, 2019; Kwon et al., 2019; Mace and Murphy, 2021).
Gene duplication events facilitate the evolution of TCS signaling enabling the integration of new input signals and diversification of the gene regulatory network. Members of the Corynebacteriaceae family, including the Gram-positive soil bacterium Corynebacterium glutamicum, represent an interesting example of a recent gene duplication event, encoding two paralogous two-component systems that both respond to the multifaceted molecule heme (Figure 1). After sensing heme availability via intramembrane interaction (Ito et al., 2009; Keppel et al., 2018), the TCS ChrSA acts as an activator of the hrtBA operon encoding the heme export system (Heyer et al., 2012). In contrast, the paralogous TCS HrrSA is a global regulator of heme homeostasis controlling more than 200 genomic targets including inter alia genes involved in heme biosynthesis, respiration as well as hmuO, encoding heme oxygenase (Keppel et al., 2020). Strikingly, a high level of cross-phosphorylation between the systems was observed (Keppel et al., 2019), while phosphatase activity of these HKs remains specific to their cognate RR (Hentschel et al., 2014).
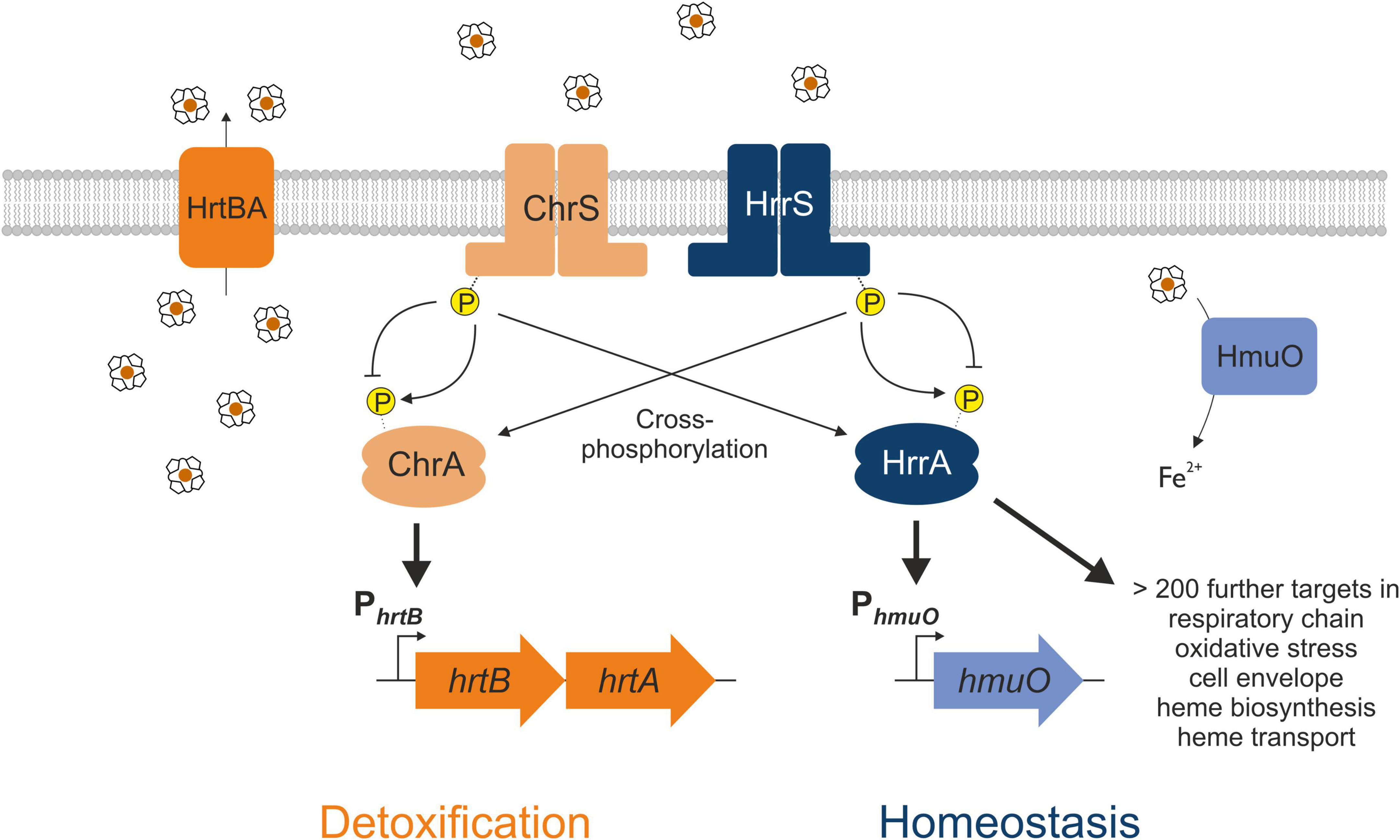
Figure 1. Interaction between the two heme-responsive two-component systems (TCS) ChrSA and HrrSA in Corynebacterium glutamicum. The simplified schematic representation shows the interaction of the two TCS ChrSA (orange) and HrrSA (blue) and each one representative target gene. ChrSA is solely responsible for the detoxification from heme via activating the hrtBA operon and autoregulates its own expression (Heyer et al., 2012). The HrrSA system was recently shown to act as regulator for heme homeostasis, controlling hmuO as well as more than 200 further genes involved in heme biosynthesis, respiratory chain, oxidative stress, or cell envelope remodeling (Keppel et al., 2020). The histidine kinases ChrS as well as HrrS undergo autophosphorylation in response to heme, activating the respective response regulator ChrA and HrrA. Cross-phosphorylation of the non-cognate response regulator has been demonstrated, while the phosphatase activity remains specific for the respective cognate response regulator (Hentschel et al., 2014).
In this study, we addressed the question how this underlying signaling cascade consisting of two paralogous TCSs facilitates fast adaptation to high heme levels, such as encountered by pathogenic species in the mammalian host, using the non-pathogenic C. glutamicum as a model. Understanding the mechanisms underlying microbial heme tolerance are not only important for the control of bacterial infections but also of biotechnological relevance for the engineering of a microbial production host demanding high product tolerance (Ko et al., 2021). Adaptive laboratory evolution (ALE) of C. glutamicum to increased heme levels resulted in the isolation of highly heme-tolerant clones bearing mutations in the CA-domain of ChrS. The catalytically inactive ChrS pseudokinase was shown to be required for the efficient activation of ChrA via the paralogous HK HrrS. Overall, this study demonstrated the potential of this ALE approach to provide new mechanistic insights in heme-dependent signaling and highlights the flexibility of paralogous TCS signaling facilitating the fast adaptation to enhanced heme levels.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Supplementary Table 1. For standard cultivation, C. glutamicum cells ATCC 13032 (wild type) and derivatives were streaked on agar plates (17 g/l) containing brain heart infusion (BHI) (Difco, BD, Heidelberg, Germany) (37 g/l) and inoculated at 30°C overnight. One single colony was picked and incubated for approximately 8 h at 30°C in 5 ml BHI in reaction tubes (for cultivation in shake flasks) or in 1 ml BHI in deep-well plates (VWR International, PA, United States) (for microtiter cultivation). This first pre-culture was used to inoculate the second pre-culture 1:10 in CGXII minimal medium (Keilhauer et al., 1993) supplemented with 2% (w/v) glucose but without any iron source to starve the cells from iron allowing the usage of heme as alternative iron source in the main culture. CGXII medium without FeSO4 is in the following referred to as “iron-free CGXII.” If appropriate, 25 μg/ml kanamycin was added to the medium. Incubation followed shaking at 120 rpm over night at 30°C. For the main experiment, cultures were inoculated to an OD600 of 1 in iron-free CGXII with 2% (w/v) glucose, and the respective amount of hemin (Sigma-Aldrich, St. Louis, United States). For simplicity, hemin is further referred to as heme throughout this study.
For the ALE experiment, the main culture was grown in deep-well plates for 1–3 days and then freshly transferred at an OD600 of 1 for the next batch. After the 13th inoculation, glycerol stocks of each population were frozen at −80°C. This allowed a restreaking of each potentially heterogeneous population on BHI-agar plates and picking of single evolved clones. Online monitoring of bacterial growth was performed using the BioLector® microtiter cultivation system of Beckman Coulter GmbH (Baesweiler) (Kensy et al., 2009). Backscatter (a.u.) was measured in 30 min intervals as scattered light with a wavelength of 620 nm (gain: 20); YFP-fluorescence was measured at an excitation wavelength of 508 nm and an emission wavelength of 532 nm (gain: 80). Specific fluorescence (a.u.) was calculated by dividing the YFP-signal by the backscatter signal for each measurement.
Escherichia coli strains including DH5α and BTH101 were cultivated in Lysogeny Broth (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl) medium at 37°C in a rotary shaker and if needed for selection, 50 μg/ml kanamycin or 100 μg/ml ampicillin was added to the medium.
Recombinant DNA work
Standard molecular methods were performed according to Sambrook and Russell (2001). For polymerase chain reactions (PCR) amplification of DNA fragments, chromosomal DNA of C. glutamicum ATCC 13032 was used as template and prepared as described previously (Eikmanns et al., 1994). Synthesis of oligonucleotides as well as sequencing was performed by Eurofins Genomics (Ebersberg, Germany).
Plasmids were constructed by amplifying DNA fragments using the respective oligonucleotides (Supplementary Tables 2, 3) and enzymatically ligated into a pre-cut vector backbone using Gibson assembly (Gibson et al., 2009).
For the deletion of genes in the genome of C. glutamicum, the suicide vector pK19-mobsacB was used (Schäfer et al., 1994). Electrocompetent C. glutamicum cells were transformed with the respective isolated plasmids by electroporation (van der Rest et al., 1999). Subsequently, the first and second recombination events were performed and verified as described in previous studies (Niebisch and Bott, 2001). The respective deletion was confirmed by amplification and sequencing.
Whole genome sequencing
Whole genome resequencing of C. glutamicum strains isolated during the ALE experiment was performed using next generation sequencing (NGS). Genomic DNA was prepared using the NucleoSpin microbial DNA kit (Macherey-Nagel, Düren, Germany) according to manufacturer’s instructions. Concentrations of the purified genomic DNA were measured using Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, United States) according to manufacturer’s instructions. The purified genomic DNA was used for the preparation for genome sequencing using NEBNext Ultra II DNA Library Kit for Illumina (New England BioLabs, Frankfurt am Main) and MiSeq Reagent Kit v2 (300-cycles) (Illumina, San Diego, CA, United States), according to manufacturer’s instructions. A MiSeq system (Illumina, San Diego, CA, United States) was used for sequencing. Data analysis and base calling were accomplished with the Illumina instrument software. FASTQ output files were analyzed for single nucleotide polymorphisms using PathoSystems Resource Integration Center (PATRIC) 3.6.12 (Davis et al., 2020).
Gradient plates
For heme gradient plates, the different mutant strains were cultivated in triplicates as described above using deep-well plates for the first and second pre-culture. Subsequently, cultures were harvested and resuspended to an OD600 of 1 in 0.9% NaCl. For each spot, 2 μl of the respective samples were spotted on the gradient plates. The gradient plates were always prepared freshly. Therefore, 30 ml of iron-free CGXII with 2% (w/v) glucose, and 17 g/l Bacto Agar (Difco, BD, Heidelberg, Germany) was poured into a squared agar plate, which was in inclined position. Then, after drying of the first layer, the incline was removed and further 30 ml of iron-free CGXII containing 2% glucose, and 15 μM heme were poured in the plates so that a heme gradient results.
Bacterial two-hybrid assays
Bacterial two-hybrid plate assays for the qualitative assessment of protein-protein interactions
Bacterial two-hybrid assays were performed based on the BACTH kit according to manufacturer’s instructions (Euromedex, Souffelweyersheim, France). This method is based on the two fragments T25 and T18 of the catalytical domain of the adenylate cyclase from Bordetella pertussis, which is only active when these two fragments are physically in close contact. Therefore, T25 and T18 were each fused once to ChrS, ChrS-Ala245fs, and HrrS. If the HKs interact with each other, this allows a functional complementation of T25 and T18, leading to cAMP synthesis, which binds to the catabolic activator protein (CAP). cAMP/CAP complexes are pleiotropic regulators of gene transcription in E. coli and therefore turn on the expression of e.g., the lac operon.
Therefore, E. coli BTH101, which lack adenylate cyclase activity, were transformed with two plasmids of heterologous proteins fused once to T25 and once to T18. This approach was directly diluted and spotted as 100, 10–1, and 10–2 dilutions on LB plates containing 40 μg/ml X-Gal, 50 μg/ml kanamycin, 100 μg/ml ampicillin, and 0.5 mM IPTG and incubated approximately 24 h at 30°C. Bacteria producing interacting proteins will form blue colonies. Otherwise, the colonies remain white. This allowed to check also for heterogeneity in expression. Additionally, the approach was also plated on LB plates only with antibiotics, to allow picking for further biological replicates for the β-galactosidase assay.
Bacterial two-hybrid β-galactosidase measurements
Transformed E. coli strains (compare 1.5.1) were re-cultivated in deep-well plates as triplicates in LB media with 50 μg/ml kanamycin, 100 μg/ml ampicillin and 0.5 mM IPTG. OD600 of the overnight cultures was measured in a Tecan Reader (Thermo Fisher Scientific, Massachusetts, United States). The β-galactosidase assay was adapted according to a previous study for 96-well plates (Griffith and Wolf, 2002). Per sample, 1 ml Z-buffer (60 mM Na2HPO4 × 12H2O, 40 mM NaH2PO4 × H2O, 10 mM KCl, 1 mM MgSO4 × 7H2O, 50 mM β-mercaptoethanol) was mixed with 20 μl 0.1% SDS, 40 μl chloroform and 200 μl of cell culture. The solution was resuspended 15 times for permeabilization. 100 μl of the permeabilized cells were put into a microtiter plate, where each 20 μl of 4 mg/ml o-nitrophenol-ß-galactosidase (ONPG) was added. When a yellowish color was observable (10 min), 50 μl of 1 M Na2CO3 was added for reaction termination. Using the Tecan Reader (Thermo Fisher Scientific, Massachusetts, United States), A420 and A550 was measured. The Miller Unit, representing the standardized amount of β-Gal activity, was calculated using the following formula:
With A420 being the absorbance of the yellow o-nitrophenol, A550 the scatter from the cell debris, 1.75 is the factor which needs to be multiplied with A550 to approximate scatter observed at 420 nm, t is time in minutes, v is the volume of the culture employed in the plate and A600 for the cell density. The value of A600 was calibrated to proper OD600.
DNA microarrays
For the analysis of the transcriptome, C. glutamicum wild type and C. glutamicum ChrS-Ala245fs were cultivated in triplicates as described above in 50 ml CGXII medium containing 2% glucose, no FeSO4 and 4 μM heme in shake flasks. Cells were harvested after 6 h, when the wild type reached an OD600 around 2.5 and the mutated strain around 5. The cell suspension was centrifuged at 4,250 × g, 10 min, 4°C in falcons filled with ice. The resulting pellets were frozen in liquid nitrogen and stored at −80°C. The following RNA preparation, cDNA synthesis, microchip hybridization, scanning, and overall evaluation was performed as described in previous studies (Baumgart et al., 2013). The microarray data described in this study are available at NCBI’s Gene Expression Omnibus under the GEO accession number GSE206796.
Results
Adaptive laboratory evolution of Corynebacterium glutamicum toward tolerance of high heme levels depends on the heme exporter HrtBA
Previous studies reported a crucial role of the two-component system ChrSA for heme tolerance of C. glutamicum (Heyer et al., 2012). Figures 2A,B show characteristic growth curves of the C. glutamicum wild type (Figure 2A) and the deletion mutant ΔhrtBA (Figure 2B) on increasing concentrations of heme. Growing on standard conditions with 36 μM FeSO4 as iron source, the wild type displayed a growth rate of 0.42 ± 0.01 h–1. Lower heme concentrations between 2.5 and 5 μM showed strongly reduced growth rates of 0.16 ± 0.003 and 0.20 ± 0.003 h–1, respectively, and reduced backscatter levels compared to the cultivation under non-limiting conditions. Higher concentrations between 10 and 15 μM heme exhibited restored backscatter levels and growth rates of 0.22 ± 0.005 and 0.27 ± 0.012 h–1, but were accompanied by an elongated lag phase as a result of heme toxicity. This effect of toxicity was even more evident for the growth of a strain lacking the operon hrtBA. While growth on standard conditions and low concentrations of heme were comparable to the WT, higher concentrations led to a significantly elongated lag phase of ∼35 and 50 h, respectively.
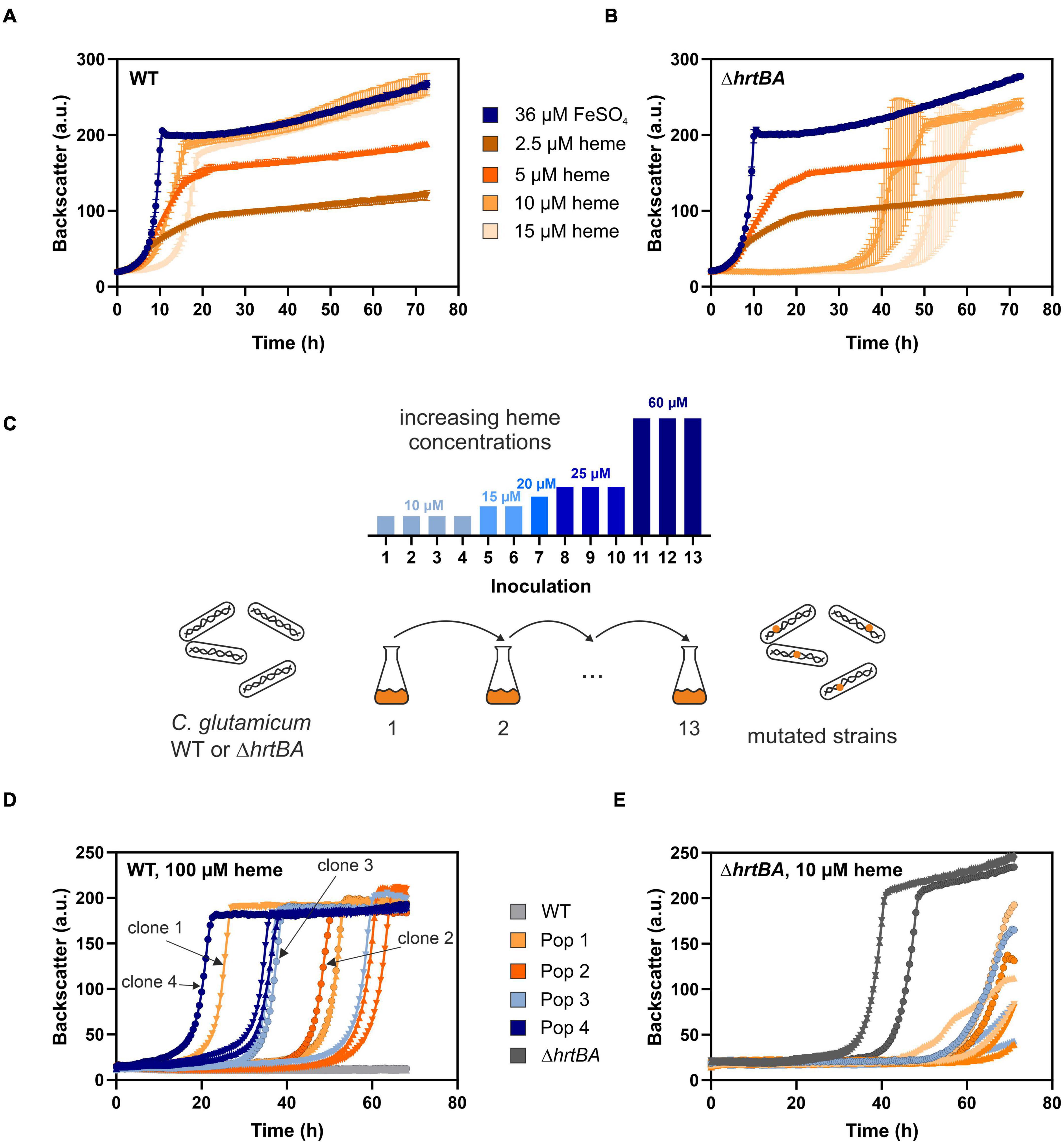
Figure 2. Adaptive laboratory evolution of C. glutamicum to high heme levels. The C. glutamicum wild type (WT) (A) as well as the ΔhrtBA strain (B) were inoculated at a starting-OD600 of 1 in CGXII medium containing 2% glucose and either 36 μM iron (blue) or increasing concentrations of heme (2.5–15 μM, shades of orange). Data represent the average of three biological replicates including standard deviations depicted as error bars. (C) Schematic representation of the adaptive laboratory evolution (ALE) experiment. The heme concentration was increased from 10 to 60 μM heme for the WT in overall 13 repetitive batch cultures (depicted in the bar graph). For the ΔhrtBA strain, concentrations > 15 μM remained toxic throughout the experiment and therefore were not further increased. (D) Growth of each three single clones derived from four different evolved C. glutamicum wild type populations on 100 μM heme. (E) Growth of each three single clones of the three evolved ΔhrtBA populations on 10 μM heme.
To elucidate mechanisms promoting high heme tolerance, we performed an ALE experiment applying increasing concentrations of heme to C. glutamicum wild type and the ΔhrtBA strain (Figure 2C). The ALE was accomplished in CGXII minimal medium, with 13 repetitive batch cultivations and started from each four independent single colonies, yielding four evolving populations. Batch cultures were started on 10 μM heme and were stepwise increased to finally reach 60 μM heme in the case of C. glutamicum wild type. From C. glutamicum wild type populations, four single clones were isolated from agar plates after the 13th batch cultivation and then further characterized in liquid culture (Figure 2D). Further analysis on earlier inoculation steps was also performed (Supplementary Figure 1). Remarkably, all isolates from the 13th inoculation were able to grow in the re-cultivation on medium containing up to 100 μM heme where growth of the parental strain was completely inhibited. By contrast, heme levels above 15 μM heme remained toxic to the ΔhrtBA strain and could not be increased throughout the ALE experiment without killing the cells. Only three of the four starting populations survived 13 inoculations and for these, no significant adaptation was observed throughout the ALE experiment (Figure 2E). These results already underlined that HrtBA represents a key factor for the adaptation of C. glutamicum to high heme levels.
Mutations in the catalytic- and ATPase domain of ChrS lead to significantly improved growth on heme
Whole genome sequencing of the four isolated clones from the ALE experiment revealed that all of them possess a mutation in the gene encoding the HK ChrS (Table 1). Remarkably, in all four cases, the catalytic- and ATPase (CA) domain was affected. For clone 2, a stop codon was inserted directly at the beginning of the CA-domain, while clone 3 possesses an amino acid exchange inside and close to the start of this domain. Strikingly, clone 1 and 4 (evolved in two independent cell lines) showed the exact same frameshift mutation for alanine at position 245, immediately after the dimerization and histidine phosphotransfer (DHp) domain of ChrS. This mutation also affects the linker between the CA- and DHp-domain. Since the strains carrying this mutation showed the highest heme tolerance, evolved clone number 1, in the following referred to as 1.fs, was further analyzed within this study.
Reintegration of this mutation into the wild type parental strain confirmed that this frameshift mutation in chrS led to highly improved growth on heme (Figure 3A and Supplementary Figure 2). Protein structure prediction via AlphaFold2 shows the truncated CA-domain of ChrS-Ala245fs with additional 60 amino acids caused by the frameshift resulting in a presumably catalytically-inactive pseudokinase variant (Figure 3B; Jumper et al., 2021; Varadi et al., 2021).
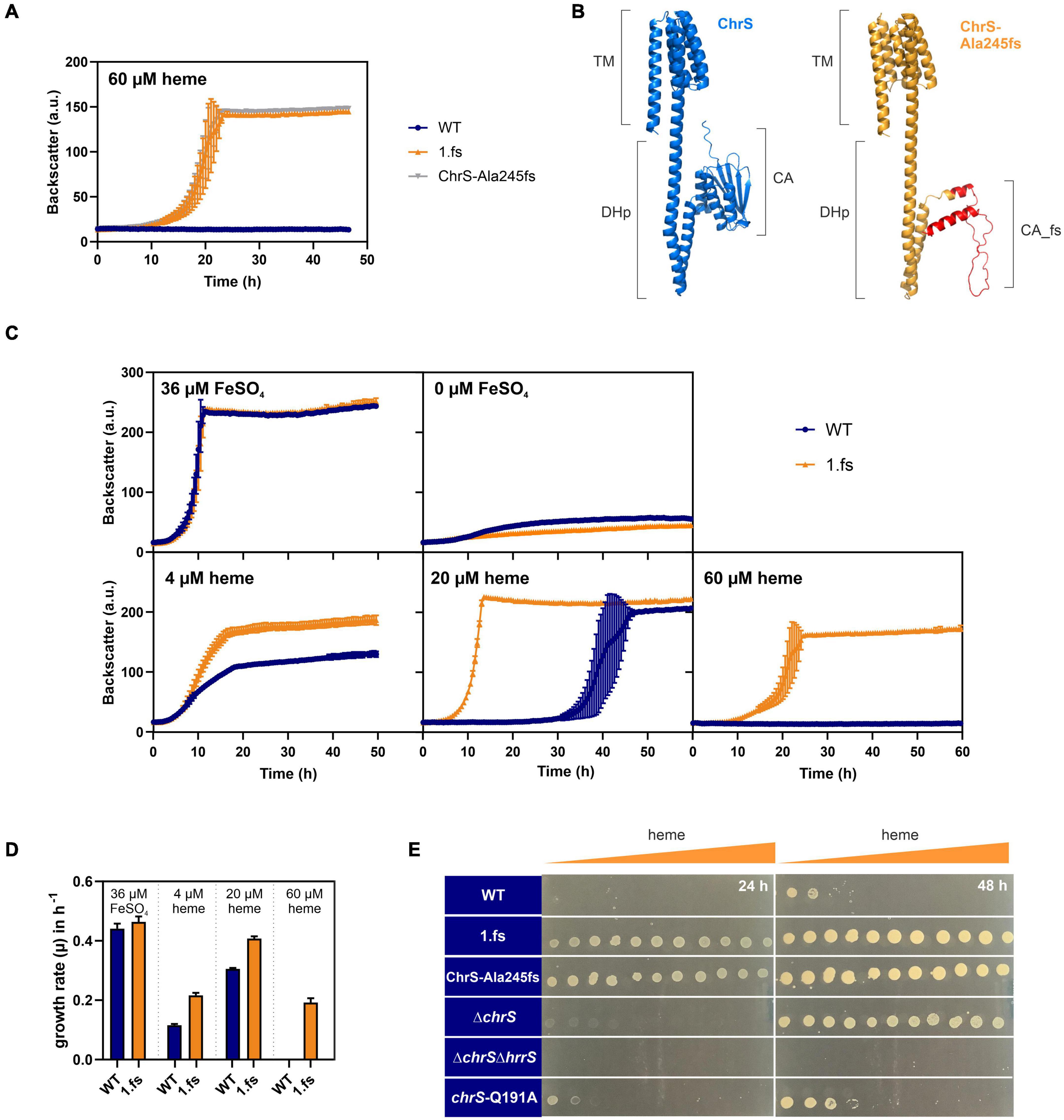
Figure 3. The ChrS-Ala245fs pseudokinase promotes heme tolerance. Data represent the average of three biological replicates including standard deviations depicted as error bars. Cells were inoculated at a starting-OD600 of 1 in CGXII medium containing 2% glucose and the indicated amount of heme or iron. (A) Growth of C. glutamicum carrying the reintegrated ChrS-Ala245fs allele (gray) compared to the evolved clone 1.fs (orange) and the wild type (WT) (blue). (B) Predicted protein structures of ChrS (blue) and the truncated ChrS-Ala245fs variant (orange, with 60 additional amino acids caused by the frameshift, shown in red). Prediction was performed using AlphaFold2 (Jumper et al., 2021; Varadi et al., 2021). Domain arrangements are shown next to the protein structures. TM, transmembrane domain; DHp, dimerization- and histidine-phosphotransfer-domain; CA, catalytic- and ATPase-domain. (C) Growth of the WT (blue) and the evolved clone 1.fs (orange) on different heme and iron concentrations; further conditions shown in Supplementary Figure 2. (D) Comparison of growth rates μ in h– 1 of WT (blue) and 1.fs (orange) at different heme and iron conditions. (E) Different strains were spotted on heme gradient plates. The WT was compared to the evolved clone (1.fs), the reintegration for the evolved clone (ChrS-Ala245fs), the chrS deletion strain (ΔchrS), the chrS and hrrS deletion strain (ΔchrS ΔhrrS) and the phosphatase mutant (chrS-Q191A). Photos of plates were taken after 24 and 48 h. A representative experiment out of three is shown.
The 1.fs strain showed normal growth under standard conditions. However, it outcompeted the wild type in the presence of heme at all concentrations tested, but not under iron starved conditions (Figures 3C,D and Supplementary Figure 2). A heme gradient plate experiment further confirmed the improved heme tolerance of the evolved clone 1.fs, and showed that this is also the case for the reintegrated pseudokinase variant ChrS-Ala245fs (Figure 3E).
The ChrS pseudokinase promotes increased hrtBA expression and is crucial for the improved growth on heme
The main known target of ChrA is the hrtBA operon encoding a heme export system. To unravel the impact of the frame-shift mutation in chrS on its activation kinetics, hrtBA reporter assays were performed, using the reporter plasmid pJC1-PhrtBA-eyfp (Heyer et al., 2012). In fact, reporter assays (Figure 4A) showed a > 10-fold elevated expression of hrtB in the evolved clone 1.fs compared to the WT. This also applies for strain carrying the reintegrated gene variant (Supplementary Figure 3A) and was further confirmed by qPCR (Supplementary Figure 4). Strikingly, hrtBA expression was constitutively high also during cultivation under standard conditions, i.e., without external addition of heme (Supplementary Figure 3).
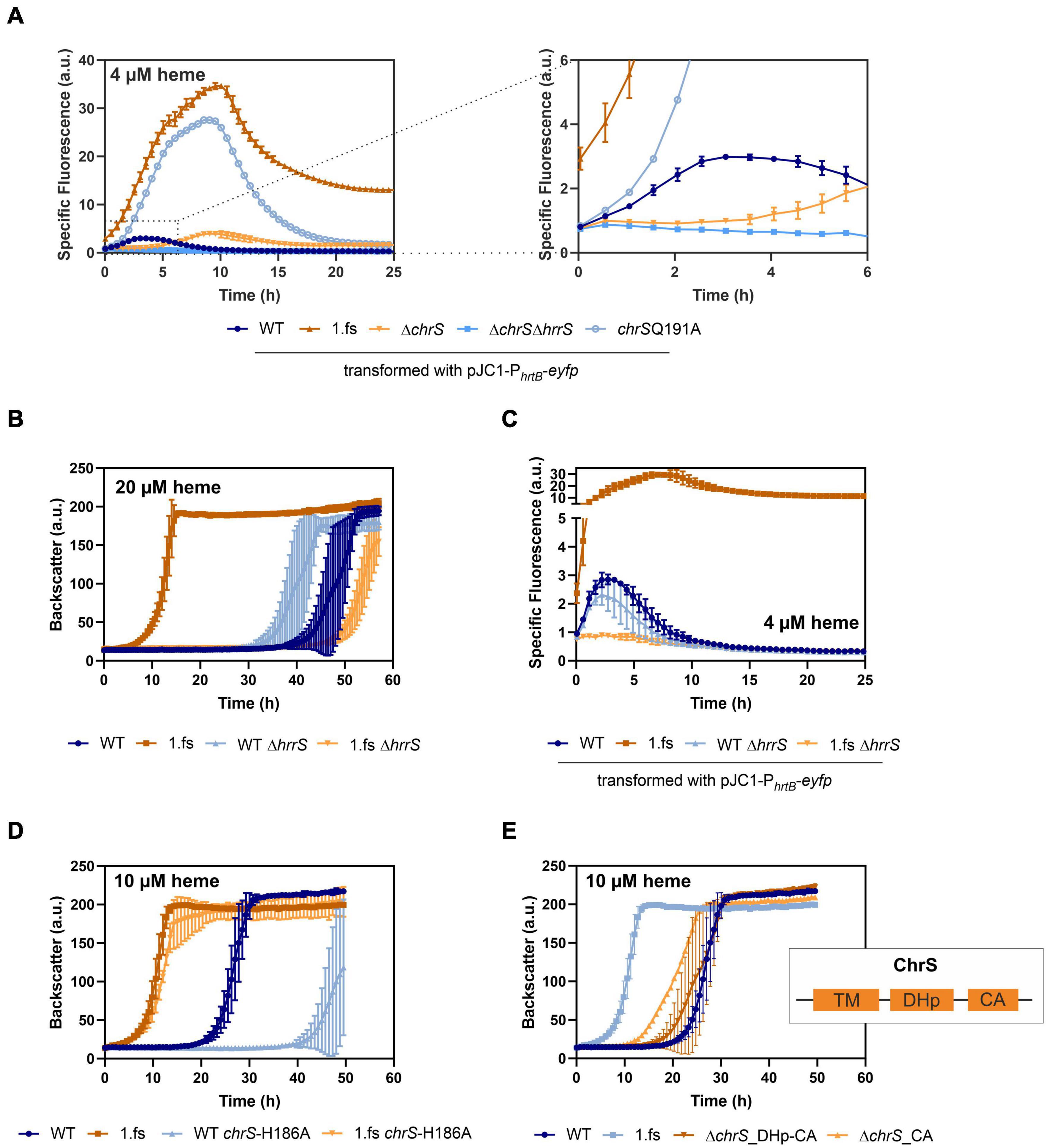
Figure 4. Mutational analysis provides insights in the hrtBA activating cascade of the evolved C. glutamicum 1.fs clone. Data represent the average of three biological replicates including standard deviations depicted as error bars. Cells were inoculated at a starting-OD600 of 1 in CGXII medium containing 2% glucose and the indicated amount of heme or iron. (A) Reporter assays visualizing hrtBA-expression using the plasmid pJC1-PhrtB-eyfp for transformation of the wild type (WT) (dark blue), the evolved clone 1.fs (dark orange), ΔchrS (light orange), ΔchrSΔhrrS (mid-blue), and chrS-Q191A (light blue). (B) Growth of the evolved clone (shades of orange) and the WT (shades of blue) natively or with each a deletion of hrrS. (C) Reporter assays for hrtB-expression using pJC1-PhrtB-eyfp upon deletion of hrrS in 1.fs (shades of orange) and the WT (shades of blue). (D) Growth of the wild type strain (shades of blue) and the evolved 1.fs strain (shades of orange) natively or possessing an amino acid exchange of the autophosphorylation histidine (H186) of ChrS. (E) Impact of ChrS truncation on growth; ChrS variants lacking only the CA-domain (light orange) or CA-domain and DHp-domain (dark orange) compared to the WT and 1.fs strain (shades of blue). TM, transmembrane domain; DHp, dimerization- and histidine-phosphotransfer-domain; CA, catalytic- and ATPase-domain.
In the following, we further analyzed the growth and hrtBA expression of the evolved clone in comparison to different mutant strains, including the deletion mutants ΔchrS and ΔchrSΔhrrS as well as the phosphatase mutant chrS-Q191A (Figures 3E, 4A; Hentschel et al., 2014). Heme gradient plates showed that the complete deletion of the chrS gene also led to an improved growth compared to the WT. This is caused by the abolished dephosphorylation of ChrA by its cognate kinase/phosphatase ChrS, allowing constant hrtBA expression due to cross talk with HrrS. The delay in hrtB expression is explained by the less sensitive response of HrrS to heme (Keppel et al., 2019). Interestingly, the ΔchrS deletion strain grew worse than the evolved strain 1.fs. This is in line with the lower hrtBA expression level in comparison to 1.fs. These results indicated that the remaining part of ChrS, which is present in the evolved clone, must play a crucial role for this high activation. Strikingly, 1.fs also significantly outperformed the phosphatase mutant chrS-Q191A in terms of growth and hrtBA expression.
Considering that the frame-shift mutation in chrS likely abolished the catalytic activity of the ChrS kinase, we wondered whether activation of ChrA is solely dependent on HrrS. Cross-talk between the kinases was previously described (Hentschel et al., 2014) and might explain the activation of the hrtBA operon in the evolved clone. To test this hypothesis, the hrrS gene was deleted in evolved strain 1.fs and compared to a ΔhrrS mutant in the parental background. Figure 4B shows that upon deletion of hrrS, 1.fs grew with an elongated lag phase of ∼50 h, which is even longer than for the WT. This result was in agreement with reporter assays showing that hrrS deletion also led to an abolishment of the hrtBA expression in 1.fs (Figure 4C). These results confirmed our hypothesis that HrrS is essential for activating hrtBA expression in the evolved clone encoding the catalytically-inactive pseudokinase of ChrS. Deletion of hrrA in the evolved variant did not influence its improved heme tolerance, therefore indicating that this is mainly an effect of HrrS activating ChrA (Supplementary Figure 5A,B). Overexpression of chrA also improved the heme tolerance to some extent (Supplementary Figure 5C). Interestingly, a plasmid-based overexpression of hrtBA in the WT background led to growth defects, probably caused by severe iron/heme starvation due to the excessive heme export (Supplementary Figure 6). This is further supported by comparative transcriptome analyses showing a higher upregulation in the strain overexpressing hrtBA compared to the evolved clone. This demonstrates the necessity of a tight balance between export and intracellular iron availability to maintain homeostasis while achieving optimal heme tolerance.
To investigate if the truncated version of ChrS still plays a role in the phosphotransfer to ChrA, we exchanged the conserved histidine residue at position 186 to an alanine (Figure 4D). This amino acid exchange did, however, not significantly influence the heme tolerance of strain 1.fs. Therefore, it can be assumed that the truncated version of ChrS does not participate in the phosphotransfer via autophosphorylation at the histidine 186.
Based on these results, we postulated that a lack of the catalytic activity of ChrS is beneficial for C. glutamicum heme tolerance. In line with this hypothesis, an in-frame deletion of the CA-domain of chrS led to improved growth on heme compared to the wild type. However, this strain did not reach comparably high heme tolerance like the 1.fs strain (Figure 4E). Upon additional deletion of the DHp-domain, this growth advantage was abolished suggesting the necessity of homo- or heterodimerization and/or interaction with ChrA.
ChrS and HrrS form heterodimers in vivo
In general, HKs act as homodimers. However, based on the fact that the autophosphorylation histidine residue of ChrS in the evolved mutant 1.fs was not relevant for its growth benefit on heme (Figure 4D), we aimed to investigate the homo- and heterodimerization properties of the C. glutamicum HKs ChrS and HrrS and the pseudokinase ChrS (ChrS_CA-fs).
To assess these protein-protein interactions between the respective monomers ChrS, HrrS and the truncated HK variant ChrS_CA-fs, we performed bacterial two-hybrid (BACTH) assays (Euromedex, Souffelweyersheim, France). The plate assays in Figure 5A as well as the quantitative β-galactosidase assay in Figure 5B show that homodimerization for both the native and the evolved HKs was observed when these fusion proteins were produced as C-terminal fusions in E. coli. The assays also revealed heterodimerization of the native HKs, while there was no significant evidence for heterodimerization of the truncated ChrS_CA-fs version. Similar results were observed when the proteins were produced as N-terminal fusions (Supplementary Figure 7).
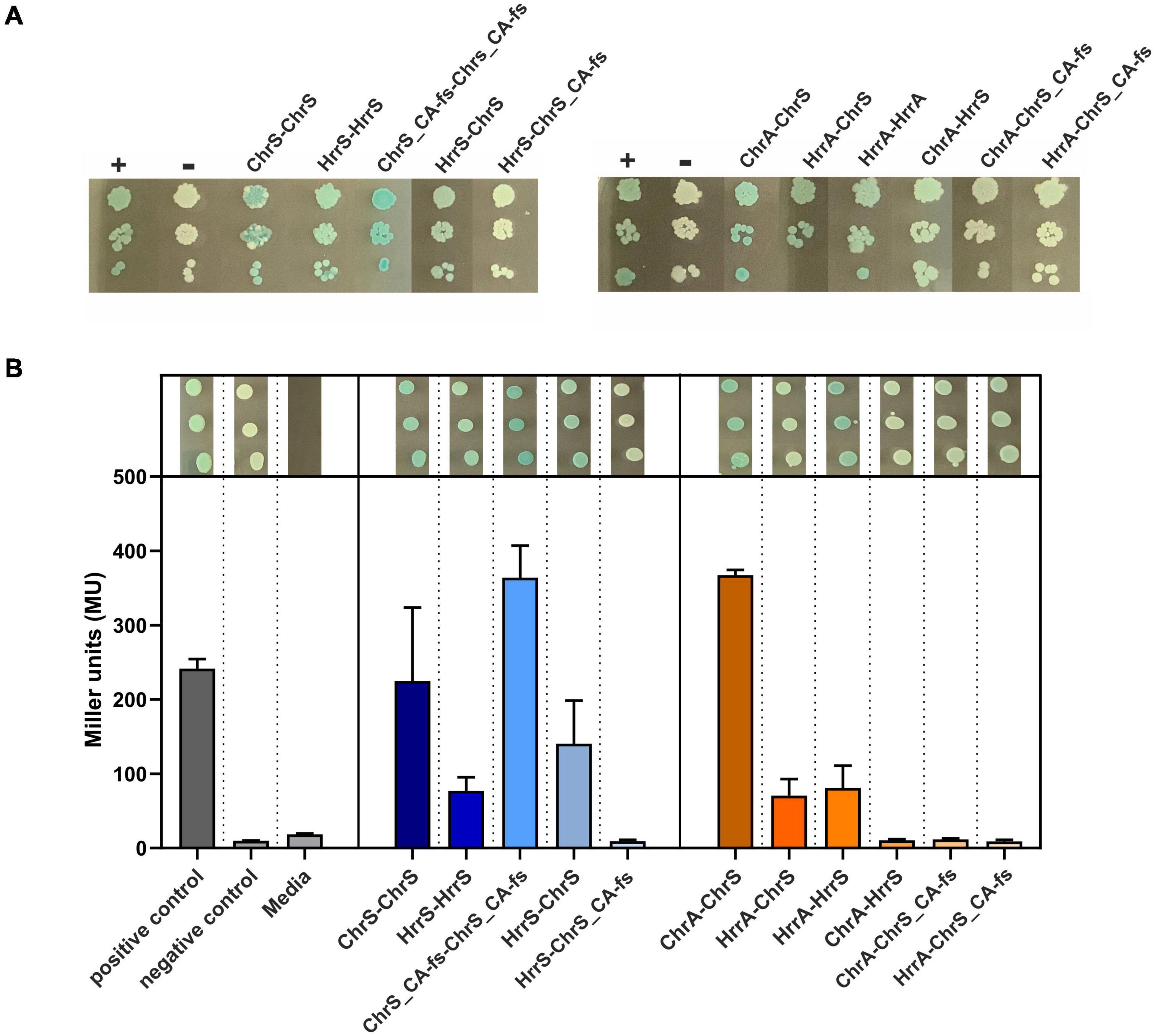
Figure 5. Bacterial two-hybrid assays of interactions between ChrSA and HrrSA. (A) BACTH interactions between the histidine kinases ChrS, HrrS and the evolved ChrS variant (here ChrS_CA-fs) were analyzed as C-terminal fusions; results for N-terminal variants are shown in Supplementary Figure 7. Blueish color of the colonies indicate interaction, while white colonies indicate no interaction (Euromedex, Souffelweyersheim, France). First histidine kinase represents the T25-fusion, the second the T18-fusion. + = pKTN25-zip with pUT18-zip (leucine zipper, positive control), – = pKTN25 with pUT18 (negative control). (B) Quantitative analysis using a β-galactosidase assay. Triplicates were cultivated and treated according to Griffith and Wolf (2002) to measure colorimetric β-galactosidase activity (given in Miller units). Gray bars represent the controls, blue bars show the interaction between the histidine kinases, and orange bars show the interaction of histidine kinase with the response regulators. Drop assays on top of the bar plots represent the triplicates picked after re-cultivation.
In a next set of experiments, we investigated the interaction between the sensor kinases and the RRs. Here, BACTH assays confirmed the interaction of ChrS with ChrA and HrrS with HrrA, as well as the cross-talk between ChrS and HrrA. Interaction was not observed for ChrS_CA-fs and neither ChrA nor HrrA within the β-galactosidase assay, although a slight signal appeared to be visible on plates (Figure 5). However, the assays did also not reveal the already reported cross-talk between HrrS and ChrA (Hentschel et al., 2014) demonstrating also the limitations of the in vivo approach based on the E. coli system. Therefore, a direct interaction between ChrA and the evolved ChrS_CA-fs in the C. glutamicum in vivo background should not be excluded.
Heme-binding proteins contribute to heme tolerance
Within this study, we showed the crucial role of the HrtBA export system for C. glutamicum heme tolerance. To identify further potentially relevant factors, we performed a comparative transcriptome analysis of the ChrS-Ala245fs strain and C. glutamicum wild type (Table 2). As expected, the hrtBA operon showed significantly increased mRNA levels in the evolved clone (∼150-fold increase). Besides hrtBA, several other heme-related targets also showed increased expression levels, including the TCS chrSA itself, hmuO encoding heme oxygenase and the heme transport system hmuU. Quantitative PCR confirmed unaltered expression levels of hrrS (Supplementary Figure 4). Strikingly, all genes encoding known heme-binding proteins were significantly upregulated. Furthermore, many targets of the DtxR regulon were upregulated (Wennerhold and Bott, 2006), while targets of the RipA regulon were downregulated (Wennerhold et al., 2005). This indicates that the evolved clone encounters a strong iron depletion, most likely caused by the extreme heme export. This is in line with a growth defect of this strain under iron starvation conditions (Figure 3C).
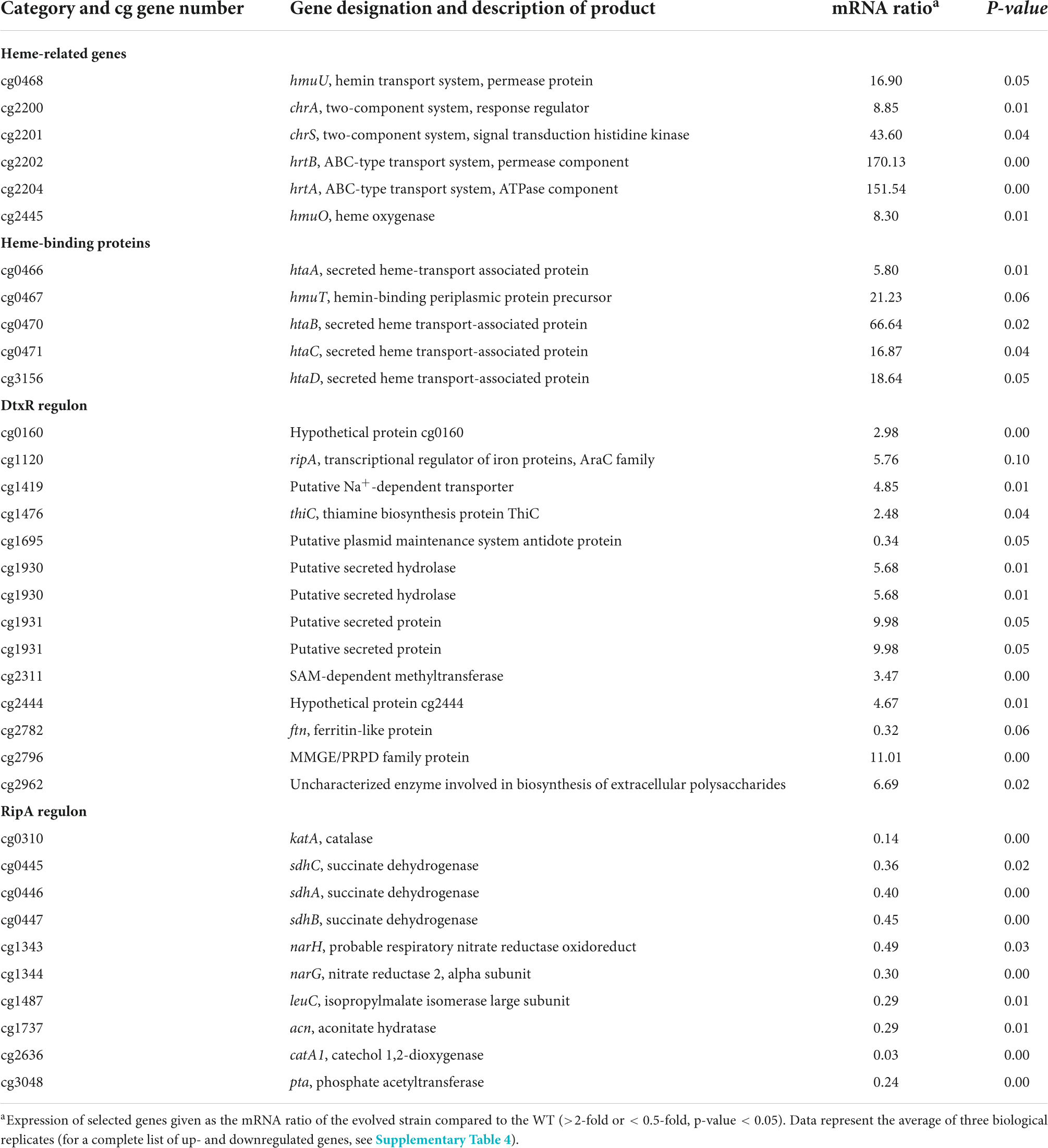
Table 2. Comparative transcriptome analysis of C. glutamicum wild type and C. glutamicum chrS-Ala245fs growing on 4 μM heme.
To test whether the upregulation of heme-binding proteins could also contribute to heme tolerance mediated by heme sequestration, we further analyzed the impact of heme binding proteins by the construction of serial deletions (Figure 6). A strain lacking the heme binding proteins hmuT, htaA, htaB, htaC, and htaD showed wild typic growth at low (4 μM) and high (20 μM) heme levels. However, at moderate (10 μM) heme concentrations, the mutant showed a significant growth defect. These results suggested that heme sequestration via heme-binding proteins could promote heme tolerance at moderate levels.
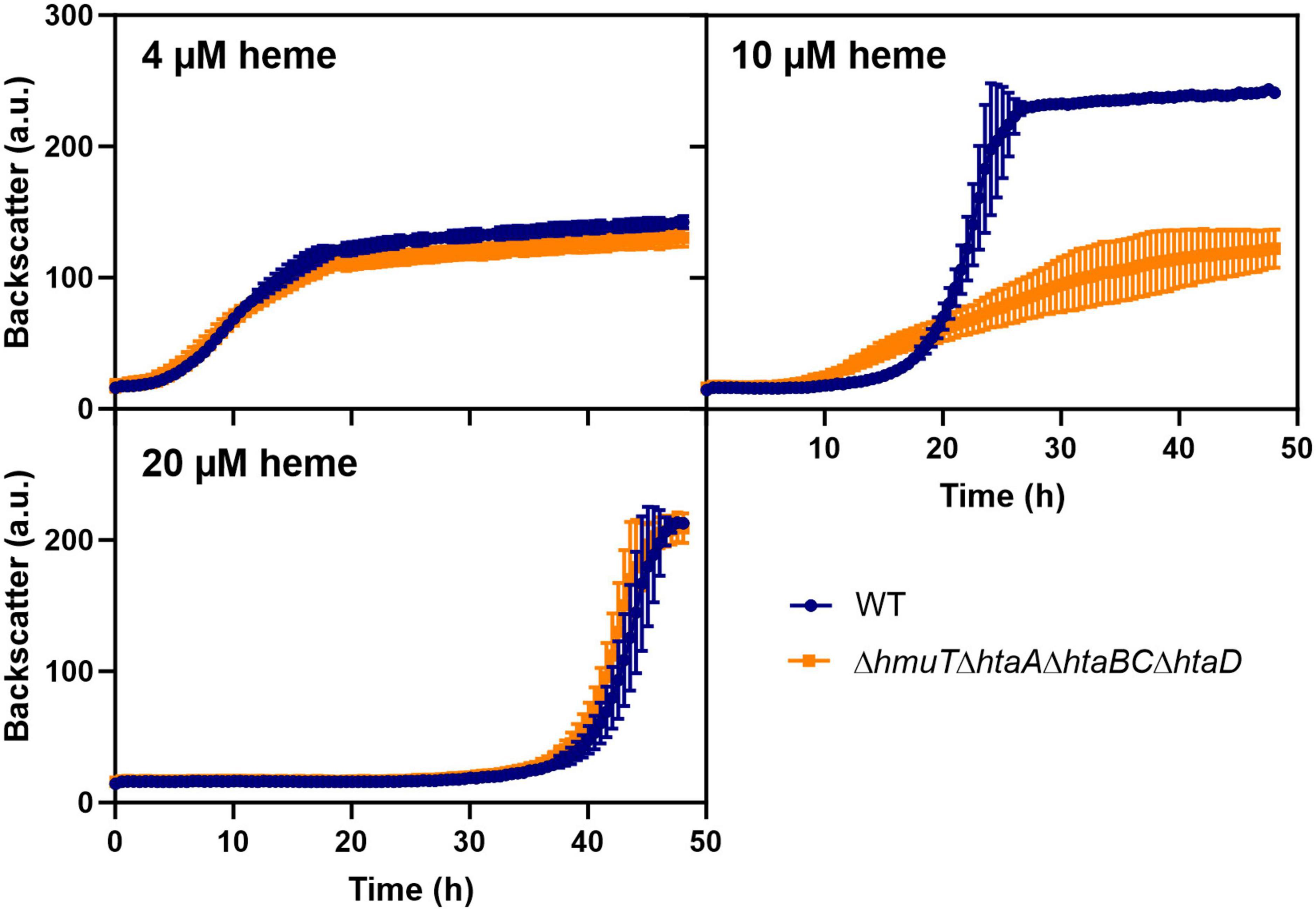
Figure 6. Heme binding proteins contribute to heme tolerance. The C. glutamicum wild type (WT) (blue) and the heme binding deficient mutant ΔhmuTΔhtaAΔhtaBCΔhtaD strain (orange) were inoculated at a starting-OD600 of 1 in CGXII medium containing 2% glucose and either 4, 10, or 20 μM heme. Data represent the average of three biological replicates including standard deviations depicted as error bars.
Discussion
In this study, we pursued a laboratory evolution approach to adapt C. glutamicum ATCC 13032 to high heme levels. This ALE approach resulted in the isolation of strains harboring a frameshift mutation in the chrS HK gene yielding a catalytically inactive pseudokinase, which was shown to promote high heme tolerance of up to 100 μM. This effect could mainly be attributed to the strong upregulation of the heme exporter HrtBA and was strictly dependent on phosphotransfer via the non-cognate HK HrrS to the RR ChrA. Further mutational analysis confirmed that the conserved histidine residue of the ChrS pseudokinase (ChrS_CA-fs) was not involved in this phosphotransfer reaction.
Continuously high hrtBA expression levels observed in our evolved 1.fs strain are in agreement with a defect in ChrS phosphatase function of this strain. Remarkably, the evolved strains encoding the truncated pseudokinase variant ChrS showed significantly higher heme tolerance and higher hrtBA expression levels compared to a phosphatase deficient strain chrS-Q191A or a strain lacking chrS completely (ΔchrS) (Hentschel et al., 2014). Consequently, dephosphorylation of ChrA seems to be abolished in the mutant clones, but the presence of the ChrS pseudokinase is apparently further beneficial for enhancing heme tolerance.
Our results indicate that the catalytically inactive version of ChrS promotes—directly or indirectly—the efficient phosphotransfer reaction from the paralogous HrrS to ChrA leading to the constitutive activation of hrtBA (due to the absence of ChrS phosphatase activity). Gene duplication is a powerful evolutionary driving force and the presence of paralogs has previously been shown to be beneficial for adaptations to new environmental conditions (Gevers et al., 2004; Bratlie et al., 2010). In our study, the interaction between the two paralogous TCSs HrrSA and ChrSA enabled fast adaptation and the evolution of novel functionality of the signaling cascade based on the pseudokinase version of ChrS.
Pseudokinases are described as kinases lacking catalytic functions, but can contribute to signaling via functioning as allosteric modulators, dynamic scaffolds, or competitors of protein-protein interactions (Reiterer et al., 2014; Kwon et al., 2019; Tomoni et al., 2019). An example for a bacterial pseudokinase is DivL from Caulobacter crescentus. DivL controls the autophosphorylation of another HK CckA mediated by the phosphorylation status of the RR DivK. The direct interaction with DivL is required for maximal kinase activity of CckA, but this is achieved by a yet unknown mechanism (Iniesta et al., 2010; Tsokos et al., 2011; Francis and Porter, 2019). It was shown that neither the ATPase domain nor the autophosphorylation residue of DivL is necessary for its function (Reisinger et al., 2007)—which is similar to the scenario observed for ChrS in this study. It can therefore be hypothesized that the catalytically inactive ChrS pseudokinase has a stimulating effect on HrrS activity, e.g., by influencing it’s “on” or “off” states. An alternative or additional reason for enhanced phosphotransfer could also be the recruitment of ChrA via the ChrS pseudokinase fostering phosphotransfer from HrrS by the formation of heterodimers. In fact, heterodimerization between the native versions of ChrS and HrrS could be demonstrated using BACTH assays speaking for a direct signaling between ChrS and HrrS in vivo. However, interaction could not be observed with the truncated version of ChrS, but it has to be kept in mind that the E. coli based BACTH does not perfectly reflect the C. glutamicum in vivo situation. Here, we also observed high upregulation of the chrSA operon itself offering also enhanced levels of RR acceptor protein.
Although only a few reports on heterodimerization of HKs exist up to date (Capra and Laub, 2012; Willett and Crosson, 2017), studies in e.g., Pseudomonas aeruginosa showed that the HK RetS directly controls the HK GacS on three different levels by heterooligomerization (Goodman et al., 2009; Jing et al., 2010; Francis et al., 2018). Such pivotal (multiple) regulatory roles of heterodimers or –oligomers could also play a role in the native C. glutamicum system. In previous studies, HrrS was shown to act as “kickstarter” of the ChrSA-mediated response and the absence of HrrS led to a delayed promoter activation of hrtBA (Keppel et al., 2019). Consistently, heterooligomerization of HrrS with ChrS could support the fast activation of ChrA when heme becomes available, e.g., by cross-phosphorylation on the HK-level. An alternative option could be an indirect communication between ChrS and HrrS via another—yet unknown—component involved in cross-signaling (Buelow and Raivio, 2010; Salvado et al., 2012).
Apart from the conserved heme export system HrtBA, transcriptome analysis gave further hints for additional players contributing to heme tolerance in C. glutamicum. Here, we observed a high upregulation of all genes encoding heme-binding proteins and a deletion mutant showed reduced tolerance to intermediate heme levels. In fact, heme-binding proteins could serve a detoxifying role via heme sequestration, as described throughout the literature for other organisms, including HbpC of Bartonella henselae (Roden et al., 2012), HemS of Yersinia enterocolitica (Stojiljkovic and Hantke, 1994) or HupZ of Group A Streptococcus, which has been recently hypothesized to function as heme chaperone (Lyles et al., 2022).
Moreover, several transport systems are differently expressed in the evolved strains. Export of further toxic heme-related products, or even import of neutralizing compounds coping with H2O2, like e.g., described for the ribulose-5-phosphate 3-epimerase in Escherichia coli or the Mn(II) uptake system of Neisseria gonorrhoeae importing manganese (Horsburgh et al., 2002; Seib et al., 2004; Sobota and Imlay, 2011), could also aid at tolerating heme. Of special interest is e.g., the operon cg2675-cg2678, which encodes an ABC-type transport system and was found also to be regulated by the heme-responsive RR HrrA (Keppel et al., 2020). Future studies on these systems, are however, required to elucidate their role in heme tolerance or homeostatic processes.
The appearance of a ChrS pseudokinase was not yet described to occur naturally in corynebacterial strains (Bott and Brocker, 2012). However, our experimental approach demonstrated their ability to quickly adapt to high heme levels, which is for example relevant for virulence conditions in the mammalian host (Stauff and Skaar, 2009a). In the soil environment, a constant expression of the hrtBA operon is likely too costly as reflected by the strong upregulation of the iron starvation response in the evolved clones, including the regulon of DtxR (Wennerhold and Bott, 2006) and the downregulation of RipA targets (Wennerhold et al., 2005). The native cascade consequently rather facilitates a fast but transient activation of hrtBA in response to heme levels (Keppel et al., 2019).
Strains featuring an elevated heme tolerance are also highly interesting for the biotechnological production of heme, which is commercially produced for medical uses or the food sector for artificial meat products. Recent metabolic engineering efforts resulted in E. coli (Kwon et al., 2003; Zhao et al., 2018) and C. glutamicum heme-producing strains (Ko et al., 2021) achieving heme yields of about 0.14–0.61 mmol mol–1. In this context, the pseudokinase variant of ChrS described in this study might aid future metabolic engineering approaches to promote efficient heme export and high product tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JF and AK conceived and designed the analysis and wrote the manuscript. AK performed the experiments and collected the data, contributed to data and analysis tools, and performed the analysis. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Helmholtz Association (grant no. W2/W3-096).
Acknowledgments
We thank Ulrike Viets for whole genome sequencing, Tino Polen for help with the microarrays and upload to databases and Elvira Mukinovic for assisting with the qPCR experiments.
Conflict of interest
AK and JF were employed by the Forschungszentrum Jülich GmbH.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.997448/full#supplementary-material
References
Ajioka, R. S., Phillips, J. D., and Kushner, J. P. (2006). Biosynthesis of heme in mammals. Biochim. Biophys. Acta 1763, 723–736. doi: 10.1016/j.bbamcr.2006.05.005
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Anzaldi, L. L., and Skaar, E. P. (2010). Overcoming the heme paradox: Heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78, 4977–4989. doi: 10.1128/IAI.00613-10
Baumgart, M., Luder, K., Grover, S., Gätgens, C., Besra, G. S., and Frunzke, J. (2013). IpsA, a novel LacI-type regulator, is required for inositol-derived lipid formation in Corynebacteria and Mycobacteria. BMC Biol. 11:122. doi: 10.1186/1741-7007-11-122
Bibb, L. A., Kunkle, C. A., and Schmitt, M. P. (2007). The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75, 2421–2431. doi: 10.1128/IAI.01821-06
Bott, M., and Brocker, M. (2012). Two-component signal transduction in Corynebacterium glutamicum and other corynebacteria: On the way towards stimuli and targets. Appl. Microbiol. Biotechnol. 94, 1131–1150. doi: 10.1007/s00253-012-4060-x
Bratlie, M. S., Johansen, J., Sherman, B. T., Huang, D. W., Lempicki, R. A., and Drabløs, F. (2010). Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11:588. doi: 10.1186/1471-2164-11-588
Buelow, D. R., and Raivio, T. L. (2010). Three (and more) component regulatory systems – auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75, 547–566. doi: 10.1111/j.1365-2958.2009.06982.x
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Casino, P., Rubio, V., and Marina, A. (2009). Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336. doi: 10.1016/j.cell.2009.08.032
Davis, J. J., Wattam, A. R., Aziz, R. K., Brettin, T., Butler, R., Butler, R. M., et al. (2020). The PATRIC bioinformatics resource center: Expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612. doi: 10.1093/nar/gkz943
Dutta, R., Qin, L., and Inouye, M. (1999). Histidine kinases: Diversity of domain organization. Mol. Microbiol. 34, 633–640. doi: 10.1046/j.1365-2958.1999.01646.x
Eikmanns, B. J., Thum-Schmitz, N., Eggeling, L., Lüdtke, K.-U., and Sahm, H. (1994). Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140, 1817–1828. doi: 10.1099/13500872-140-8-1817
Francis, V. I., and Porter, S. L. (2019). Multikinase networks: Two-component signaling networks integrating multiple stimuli. Annu. Rev. Microbiol. 73, 199–223. doi: 10.1146/annurev-micro-020518-115846
Francis, V. I., Waters, E. M., Finton-James, S. E., Gori, A., Kadioglu, A., Brown, A. R., et al. (2018). Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 9:2219. doi: 10.1038/s41467-018-04640-8
Gao, R., and Stock, A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154. doi: 10.1146/annurev.micro.091208.073214
Gevers, D., Vandepoele, K., Simillion, C., and Van de Peer, Y. (2004). Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12, 148–154. doi: 10.1016/j.tim.2004.02.007
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Goodman, A. L., Merighi, M., Hyodo, M., Ventre, I., Filloux, A., and Lory, S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259. doi: 10.1101/gad.1739009
Griffith, K. L., and Wolf, R. E. Jr. (2002). Measuring beta-galactosidase activity in bacteria: Cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290, 397–402. doi: 10.1006/bbrc.2001.6152
Hentschel, E., Mack, C., Gatgens, C., Bott, M., Brocker, M., and Frunzke, J. (2014). Phosphatase activity of the histidine kinases ensures pathway specificity of the ChrSA and HrrSA two-component systems in Corynebacterium glutamicum. Mol. Microbiol. 92, 1326–1342. doi: 10.1111/mmi.12633
Heyer, A., Gatgens, C., Hentschel, E., Kalinowski, J., Bott, M., and Frunzke, J. (2012). The two-component system ChrSA is crucial for haem tolerance and interferes with HrrSA in haem-dependent gene regulation in Corynebacterium glutamicum. Microbiology 158, 3020–3031. doi: 10.1099/mic.0.062638-0
Horsburgh, M. J., Wharton, S. J., Karavolos, M., and Foster, S. J. (2002). Manganese: Elemental defence for a life with oxygen. Trends Microbiol. 10, 496–501. doi: 10.1016/S0966-842X(02)02462-9
Iniesta, A. A., Hillson, N. J., and Shapiro, L. (2010). Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. U.S.A. 107, 7012–7017. doi: 10.1073/pnas.1001767107
Ito, Y., Nakagawa, S., Komagata, A., Ikeda-Saito, M., Shiro, Y., and Nakamura, H. (2009). Heme-dependent autophosphorylation of a heme sensor kinase, ChrS, from Corynebacterium diphtheriae reconstituted in proteoliposomes. FEBS Lett. 583, 2244–2248. doi: 10.1016/j.febslet.2009.06.001
Jing, X., Jaw, J., Robinson, H. H., and Schubot, F. D. (2010). Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins 78, 1631–1640. doi: 10.1002/prot.22679
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Keilhauer, C., Eggeling, L., and Sahm, H. (1993). Isoleucine synthesis in Corynebacterium glutamicum: Molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175, 5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993
Kensy, F., Zang, E., Faulhammer, C., Tan, R.-K., and Büchs, J. (2009). Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb. Cell Fact. 8:31. doi: 10.1186/1475-2859-8-31
Keppel, M., Davoudi, E., Gätgens, C., and Frunzke, J. (2018). Membrane topology and heme binding of the histidine kinases HrrS and ChrS in Corynebacterium glutamicum. Front. Microbiol. 9:183. doi: 10.3389/fmicb.2018.00183
Keppel, M., Hünnefeld, M., Filipchyk, A., Viets, U., Davoudi, C. F., Krüger, A., et al. (2020). HrrSA orchestrates a systemic response to heme and determines prioritization of terminal cytochrome oxidase expression. Nucleic Acids Res. 48, 6547–6562. doi: 10.1093/nar/gkaa415
Keppel, M., Piepenbreier, H., Gatgens, C., Fritz, G., and Frunzke, J. (2019). Toxic but tasty - temporal dynamics and network architecture of heme-responsive two-component signaling in Corynebacterium glutamicum. Mol. Microbiol. 111, 1367–1381. doi: 10.1111/mmi.14226
Kim, D. J., and Forst, S. (2001). Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology (Reading) 147, 1197–1212. doi: 10.1099/00221287-147-5-1197
Ko, Y. J., Kim, M., You, S. K., Shin, S. K., Chang, J., Choi, H. J., et al. (2021). Animal-free heme production for artificial meat in Corynebacterium glutamicum via systems metabolic and membrane engineering. Metabolic Eng. 66, 217–228. doi: 10.1016/j.ymben.2021.04.013
Krüger, A., Keppel, M., Sharma, V., and Frunzke, J. (2022). The diversity of heme sensor systems - heme-responsive transcriptional regulation mediated by transient heme protein interactions. FEMS Microbiol. Rev. 46:fuac002. doi: 10.1093/femsre/fuac002
Kumar, S., and Bandyopadhyay, U. (2005). Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157, 175–188. doi: 10.1016/j.toxlet.2005.03.004
Kung, J. E., and Jura, N. (2019). Prospects for pharmacological targeting of pseudokinases. Nat. Rev. Drug Discov. 18, 501–526. doi: 10.1038/s41573-019-0018-3
Kwon, A., Scott, S., Taujale, R., Yeung, W., Kochut, K. J., Eyers, P. A., et al. (2019). Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal. 12:eaav3810. doi: 10.1126/scisignal.aav3810
Kwon, S. J., Boer, A. L. D., Petri, R., and Schmidt-Dannert, C. (2003). High-Level production of porphyrins in metabolically engineered Escherichia coli: Systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 69, 4875–4883. doi: 10.1128/AEM.69.8.4875-4883.2003
Laub, M. T., and Goulian, M. (2007). Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41, 121–145. doi: 10.1146/annurev.genet.41.042007.170548
Layer, G., Reichelt, J., Jahn, D., and Heinz, D. W. (2010). Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161. doi: 10.1002/pro.405
Lyles, K. V., Thomas, L. S., Ouellette, C., Cook, L. C. C., and Eichenbaum, Z. (2022). HupZ, a unique heme-binding protein, enhances group A Streptococcus fitness during mucosal colonization. Front. Cell. Infect. Microbiol. 12:867963. doi: 10.3389/fcimb.2022.867963
Mace, P. D., and Murphy, J. M. (2021). There’s more to death than life: Noncatalytic functions in kinase and pseudokinase signaling. J. Biol. Chem. 296:100705. doi: 10.1016/j.jbc.2021.100705
Mascher, T., Helmann, J. D., and Unden, G. (2006). Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938. doi: 10.1128/MMBR.00020-06
Nakamura, H., Hisano, T., Rahman, M. M., Tosha, T., Shirouzu, M., and Shiro, Y. (2022). Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 119:e2123385119. doi: 10.1073/pnas.2123385119
Niebisch, A., and Bott, M. (2001). Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175, 282–294. doi: 10.1007/s002030100262
Padmanaban, G., Venkateswar, V., and Rangarajan, P. N. (1989). Haem as a multifunctional regulator. Trends Biochem. Sci. 14, 492–496. doi: 10.1016/0968-0004(89)90182-5
Perego, M., and Hoch, J. A. (1996). Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet. 12, 97–101. doi: 10.1016/0168-9525(96)81420-X
Ponka, P. (1999). Cell biology of heme. Am. J. Med. Sci. 318, 241–256. doi: 10.1016/S0002-9629(15)40628-7
Raju, S., and Shaw, A. S. (2015). What is the point of pseudokinases? Elife 4:e07771. doi: 10.7554/eLife.07771
Reisinger, S. J., Huntwork, S., Viollier, P. H., and Ryan, K. R. (2007). DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J. Bacteriol. 189, 8308–8320. doi: 10.1128/JB.00868-07
Reiterer, V., Eyers, P. A., and Farhan, H. (2014). Day of the dead: Pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505. doi: 10.1016/j.tcb.2014.03.008
Roden, J. A., Wells, D. H., Chomel, B. B., Kasten, R. W., and Koehler, J. E. (2012). Hemin binding protein C is found in outer membrane vesicles and protects Bartonella henselae against toxic concentrations of hemin. Infect. Immun. 80, 929–942. doi: 10.1128/IAI.05769-11
Salvado, B., Vilaprinyo, E., Karathia, H., Sorribas, A., and Alves, R. (2012). Two component systems: Physiological effect of a third component. PLoS One 7:e31095. doi: 10.1371/journal.pone.0031095
Sambrook, J. F., and Russell, D. (2001). Molecular cloning: A laboratory manual (3-Volume Set), Vo. 1. New York, NY: Cold Spring Harbor Laboratory Press.
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Seib, K. L., Tseng, H. J., McEwan, A. G., Apicella, M. A., and Jennings, M. P. (2004). Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: Distinctive systems for different lifestyles. J. Infect. Dis. 190, 136–147. doi: 10.1086/421299
Skaar, E. P., Gaspar, A. H., and Schneewind, O. (2006). Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188, 1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006
Sobota, J. M., and Imlay, J. A. (2011). Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U.S.A. 108, 5402–5407. doi: 10.1073/pnas.1100410108
Stauff, D. L., and Skaar, E. P. (2009b). The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 16, 120–135. doi: 10.1159/000219376
Stauff, D. L., and Skaar, E. P. (2009a). Bacillus anthracis HssRS signaling to HrtAB regulates heme resistance during infection. Mol. Microbiol. 72, 763–778. doi: 10.1111/j.1365-2958.2009.06684.x
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Stojiljkovic, I., and Hantke, K. (1994). Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13, 719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x
Stojiljkovic, I., Kumar, V., and Srinivasan, N. (1999). Non-iron metalloporphyrins: Potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31, 429–442. doi: 10.1046/j.1365-2958.1999.01175.x
Tomoni, A., Lees, J., Santana, A. G., Bolanos-Garcia, V. M., and Bastida, A. (2019). Pseudokinases: From allosteric regulation of catalytic domains and the formation of macromolecular assemblies to emerging drug targets. Catalysts 9:778. doi: 10.3390/catal9090778
Tsokos, C. G., Perchuk, B. S., and Laub, M. T. (2011). A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev. Cell 20, 329–341. doi: 10.1016/j.devcel.2011.01.007
van der Rest, M. E., Lange, C., and Molenaar, D. (1999). A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545. doi: 10.1007/s002530051557
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2021). AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Wennerhold, J., and Bott, M. (2006). The DtxR regulon of Corynebacterium glutamicum. J. Bacteriol. 188, 2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006
Wennerhold, J., Krug, A., and Bott, M. (2005). The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR*. J. Biol. Chem. 280, 40500–40508. doi: 10.1074/jbc.M508693200
Willett, J. W., and Crosson, S. (2017). Atypical modes of bacterial histidine kinase signaling. Mol. Microbiol. 103, 197–202. doi: 10.1111/mmi.13525
Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002). Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013
Keywords: adaptive laboratory evolution (ALE), heme, pseudokinase, two-component system (TCS), histidine kinase, CA-domain
Citation: Krüger A and Frunzke J (2022) A pseudokinase version of the histidine kinase ChrS promotes high heme tolerance of Corynebacterium glutamicum. Front. Microbiol. 13:997448. doi: 10.3389/fmicb.2022.997448
Received: 18 July 2022; Accepted: 18 August 2022;
Published: 07 September 2022.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Miaomiao Shi, The University of Chicago, United StatesHaibi Wang, Cornell University, United States
Copyright © 2022 Krüger and Frunzke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Frunzke, ai5mcnVuemtlQGZ6LWp1ZWxpY2guZGU=
 Aileen Krüger
Aileen Krüger Julia Frunzke
Julia Frunzke