
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 18 October 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.996031
This article is part of the Research TopicCommunity series in Insights of Gut Microbiota: Probiotics and Bioactive Compounds, Volume IIView all 9 articles
 Yeboah Kwaku Opoku1*
Yeboah Kwaku Opoku1* Kwame Kumi Asare2
Kwame Kumi Asare2 George Ghartey-Quansah2
George Ghartey-Quansah2 Justice Afrifa3
Justice Afrifa3 Felicity Bentsi-Enchill1
Felicity Bentsi-Enchill1 Eric Gyamerah Ofori1
Eric Gyamerah Ofori1 Charles Kwesi Koomson4
Charles Kwesi Koomson4 Rosemary Kumi-Manu1
Rosemary Kumi-Manu1Rheumatoid arthritis (RA) is a common systemic autoimmune disease with a global health importance. It is characterized by long-term complications, progressive disability and high mortality tied to increased social-economic pressures. RA has an inflammatory microenvironment as one of the major underlying factors together with other complex processes. Although mechanisms underlying the triggering of RA remain partially elusive, microbiota interactions have been implicated. Again, significant alterations in the gut microbiome of RA patients compared to healthy individuals have intimated a chronic inflammatory response due to gut dysbiosis. Against this backdrop, myriads of studies have hinted at the prospective therapeutic role of probiotics as an adjuvant for the management of RA in the quest to correct this dysbiosis. In this article, the major gut microbiome alterations associated with RA are discussed. Subsequently, the role of the gut microbiome dysbiosis in the initiation and progression of RA is highlighted. Lastly, the effect and mechanism of action of probiotics in the amelioration of symptoms and severity of RA are also espoused. Although strain-specific, probiotic supplementation as adjuvant therapy for the management of RA is very promising and warrants more research.
Rheumatoid arthritis (RA) is a common systemic autoimmune disease with a global prevalence of about 0.5–1%. It is characterized by long-term complications, progressive disability and high mortality tied to increased social-economic pressures (Firestein, 2003; Mikuls, 2003). Irrespective of the devastating effect of RA on populations, its etiology remains elusive. However, environmental and genetic effects have been intimated as the major culprits. These culminate in the pathophysiological destruction of joints due to the inability of the immune system to distinguish self-antigens of the joint and its associated tissues from non-self, this initiating a cascade of inflammatory reactions (Opoku et al., 2020). The observed inflammatory cascade results in articular damage mainly driven by lymphocytes, macrophages and fibroblast-like synoviocytes (FLS; McInnes et al., 2011). These underlying reactions lead to pathological changes which result in the destruction of cartilage and subchondral bone, infiltration of inflammatory cells and synovitis with associated hyperplasia (Gu et al., 2014).
Like many other chronic diseases and their sequelae, RA has an inflammatory microenvironment as one of the major underlying factors (Opoku et al., 2020). Although inflammation is part of the innate immune system and plays a critical role in the removal of pathogens, damaged cells and irritants, it is only advantageous when short-termed and under control (acute inflammation). However, when prolonged than necessary, known as chronic inflammation, it becomes disadvantageous and contributes to concurrent obliteration and curing of tissues (Aggarwal et al., 2009).
Although mechanisms underlying the triggering of RA remain partially elusive, microbiota interactions have been implicated (Ruff and Kriegel, 2015; Corrêa et al., 2019). These microbiome alterations have been reported especially in the gut and oral microbiome. Again, these alterations or changes in the microbiome have been recently reported as key environmental risk factors in the establishment of RA (Scher et al., 2016). It is highly believed that the neonatal period is critical for the maturation of T and B cells and the development of lymphoid structures tied to the acquisition of immune tolerance to gut commensals (Zeissig and Blumberg, 2014; Zhao and Elson, 2018). Research has intimated the possibility of dysregulation and subsequent immune-mediated diseases such as autoimmunity and allergies when microbial communities are altered during this time of development (Dehner et al., 2019). Bacteroidetes and Firmicutes are the two major phyla which are known to dominate the microbiota of humans even though their abundance in fecal samples remains relatively constant in healthy individuals.
Humans from inception have constantly coexisted and coevolved with trillions of microbes colonizing most body surfaces and cavities. This is normally referred to as the human microbiome. The gastrointestinal tract harbors most of the bacteria present in the body with most of these organisms from the phyla Bacteroidetes and Firmicutes (Li and Wang, 2021). Together with other organisms from different phyla such as Cyanobacteria, Actinobacteria, Proteobacteria, Fusobacteria, Verrucomicrobia etc., they play key roles in the establishment and maintenance of homeostasis in the immune system (De Santis et al., 2015).
Although this microbiome can remain resilient through the life of an individual, its composition is liable to alterations by factors such as age, sex, genes, drugs, diet, diseases, etc. (Clemente et al., 2018). Previously difficult to study, the advent of modern technologies such as shotgun sequencing has made it possible to study the possible constitution and alteration of the microbiome.
Several animal model experiments gave earlier intimation of the role of gut microbiota in the initiation and progression of RA (Taurog et al., 1994; Abdollahi-Roodsaz et al., 2008; Wu et al., 2010). In these studies, it was observed that most of these susceptible animal models of RA under germ-free conditions remained healthy only to develop RA after exposure to certain microbes. Yamamoto et al. (2000), hinted at the role of the gut microbiome in arthritis when they reported a chronic animal model of polyarthritis by hyper-immunization with attenuated Enterococcus faecalis. In a related study by Dorożyńska et al. (2014), escalated arthritic symptoms were reported in patients with partial depletion of their gut microbiome induced by antibiotics. Other studies have reported a significant decrease in Haemophilus spp. tied to an upsurge in the levels of Lactobacillus salivarius in saliva, gut and dental microbiome of patients with RA compared to their healthy counterparts. Interestingly, this alteration or dysregulation was partly restored after treatment with Disease-modifying antirheumatic drugs (DMARDs; Zhang et al., 2015). Recently, a study has reported a significant decrease in Lactobacillus, Enterobacter, Alloprevotella and Odoribacter coupled with an upsurge in the number of genera Escherichia–Shigella and Bacteroides in some Chinese patients with RA. In a similar Japanese cohort of RA patients, however, there was an increase in the abundance of the genus Prevotella (Sun et al., 2019). Again, anticitrullinated peptide antibody (ACPA)-positive RA patients were associated with an increased amount of Clostridiales, Blautia and Akkermansia compared to ACPA-negative RA individuals (Chiang et al., 2019). Also, other specific oral bacteria responsible for periodontal diseases such as Prevotella intermedia, Aggregatibacter actinomycetemcomitans and P. gingivalis have all been associated with the initiation of RA (Mei et al., 2021). In a current related study, the genus Bifidobacterium from the phylum Actinobacteria and genus Dialister reported significant increases in patients with RA compared to healthy controls (Wang et al., 2022).
To further buttress the alteration in the gut microbiome, Mei et al. (2021), associated a rich composition of Neisseria spp., Streptococcus spp. and Haemophilus spp. with RA patients. However, in the same patients, significant depletion of other species such as Fusobacterium varium, Clostridium celatum and Enterococcus faecalis were recorded. A careful analysis of these species clearly shows that species enriched under RA conditions belonged to Proteobacteria and Firmicutes while healthy controls are enriched with species from the Firmicutes genera. Again, Enterococcus faecalis have been observed in both synovial tissues and blood cultures of patients with RA (Luo et al., 2017; Chandradevan et al., 2020).
Again, evidence from both clinical and experimental studies has suggested alterations in the gut microbiome during the treatment of patients with RA (Mei et al., 2021). In one study, etanercept, a tumor necrosis factor-alpha (TNF-α) inhibitor, made significant alterations and partly improved the microbiota in patients with RA (Picchianti-Diamanti et al., 2018). Similarly, in animal studies involving collagen-induced arthritic mice, etanercept significantly reduced Escherichia/Shigella while increasing the composition of Tannerella, Lactobacillus and Clostridium XIVa (Wang et al., 2020). Furthermore, other natural compounds which have demonstrated enormous prospects for the alleviation of arthritis-associated dysbiosis in gut microbiome composition such as Clematis triterpenoid saponins have also proved effective in the improvement of arthritis symptoms (Guo et al., 2019). Recently, the emergence of data on microbiomes associated with different treatments has provided new insights regarding the associations among RA treatments, the gut microbiota, and clinical outcomes.
The gut microbiota play a critical role in the complexity and dynamics of host immune homeostasis (D’Amelio et al., 2018). According to Wu et al. (2016), the immune system of the mucosa is involved in the development and maintenance of a healthy microbiome in the gut. The gut microbiome alters colonic regulatory T cells (Tregs) to maintain the immune homeostasis and functions (Tanoue et al., 2016). Tregs play a vital role in the immune system by secreting anti-inflammatory cytokines to aid in the suppression of unwarranted activation of effector T cells (Kayama et al., 2020). The interdependence and relationship of the gut microbiome and dysbiosis influence individual susceptibility to immune-mediated diseases such as type 1 diabetes, RA etc. (de Oliveira et al., 2017). This is possible as disruption of immune response due to dysbiosis of gut microbiome is often not localized but rather can affect other unrelated anatomical sites some distance away (Cho and Blaser, 2012).
In the development of RA, different species of microbes in the gut microbiota may either play a protective or a pathogenic role (Mizuno et al., 2017). Species of Prevotella such as P. copri are suggested to influence the initiation and progression of RA. This is made evident as microbes such as P. copri highly stimulates the upregulation of Th17-related cytokines including IL-6 and IL-23 tied to the elevated responsiveness of Treg cells to dendritic cells (Maeda et al., 2016). This leads to an increase in the recruitment of dendritic cells to synovial fluid and tissues in the joint in RA. These dendritic cells are responsible for the secretion of chemokines such as CCL3, CCL17, CXCL19 and CXCL10 which leads to the influx of proinflammatory immune cells and recruitment of T cells to the RA synovium (Moret et al., 2013; Yu and Langridge, 2017). Again, in human RA patients, CD4 + CD25 + Foxp3+ Treg cells with high plasticity for the production of IL-17 has been documented to accumulate in the synovium (Möttönen et al., 2005; Komatsu et al., 2014). Other filamentous bacteria have demonstrated a robust ability to incite the gastrointestinal and systemic follicular helper T cell responses leading to massive production of autoantibodies in K/BxN mice (Teng et al., 2016).
The innate immune system relies mainly on receptors such as pattern recognition receptors (PRRs) which include Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs). The interaction of TLRs and pathogen-associated molecular patterns (PAMPs) on microbes leads to a cascade of reactions leading to the initiation of inflammatory responses and the production of proinflammatory cytokines such as interleukin-6, TNF-α, or interleukin-1β to eliminate the pathogen (Bennike et al., 2017). Individual species of microbes in the microbiome of the gut stimulate the innate immune system differently which ultimately affects the release of either pro-inflammatory or anti-inflammatory cytokines (Horta-Baas et al., 2017). The upregulation of Th17 differentiation tied to the suppression of Treg cells leads to a shift in homeostasis towards inflammation (Horta-Baas et al., 2017).
Gut microbiome alterations may potentiate an increase in the permeability of the epithelia in the gut. These changes allow microbes and their products to cross into the lamina propria and sub-epithelial spaces. RA, like many other autoimmune diseases is preceded by autoimmunity marked by the presence autoantibodies. However, not all patients with these autoantibodies develop into RA and even in some patients there is seroreversion from positive to negative. This implies that the development of autoantibodies is reversible and requires additional triggers to transition into a chronic autoimmune state that eventually leads to RA. This transition is believed to be mediated by HLA class II with T cells playing a crucial role. Again, before the onset of arthritis in experimental animals, Th17 cells aggregate in germinal centers thereby suppressing the expression of sialyltransferases in plasmablasts. This causes the expression of IgG molecules with Fc-glycans that lack sialic acid thereby affecting the activity of the antibody (Nimmerjahn et al., 2007; Raju, 2008; Hayes et al., 2014). These interactions although poorly understood are believed to the associated the production of autoantibodies by pro-inflammatory T cells such as Th1 and Th17 cells (Zhong et al., 2018). The release of these autoantibodies together with other cytokines into circulation carries them to tissues and organs leading to the activation of macrophages culminating in the release of pro-inflammatory cytokines such as IL-6, IL-1, TNF-α, and IL-17. On the other hand, some microbes within the gut microbiome play a protective role in RA. Thus, enhance the polarization of regulatory T cells (Tregs) leading to the activation of an anti-inflammatory pathways by the production of cytokines such as transforming growth factor-β (TGF-β), IL-4 and IL-10 (Zhong et al., 2018).
Again, protein molecules from the gut microbiome such as filamin A (FLNA) and N-Acetyl-glucosamine-6-sulfatase (GNS) can potentiate and induce autoimmunity in RA through molecular mimicry. This stems from the high degree of epitope homology between HLA-DR-presented GNS peptide and sulfatase proteins of Parabacteroides sp. and Prevotella sp. In a similar manner, the HLA-DR-presented FLNA peptide is also homologous with epitopes from proteins of the Butyricimonas sp. and Prevotella sp. (Pianta et al., 2017). This observed homology presents some form of cross-reactivity of T cell epitopes of some bacteria with self-epitopes. Other mechanisms implicating gut microbiota with RA is the increased permeability of the gut. This is buttressed by the research work of Chen et al. (2016) who demonstrated an increase in the permeability of the gut through the downregulation of the expression of tight junction proteins due to bacteria such as Collinsella sp. These observations can partly be attributed to the changes in the redox environment, transport and metabolism of molecules such as zinc, iron and sulphur (X. Zhang et al., 2015).
Aside these mechanisms, current studies have suggested enzymatic initiation of peptide citrullination, antigenic mimicry and stimulation of APCs by influencing TLRs or NOD-like receptors (NLRs) as other means by which the gut microbiome lead to the initiation of RA (Li and Wang, 2021; Figure 1).
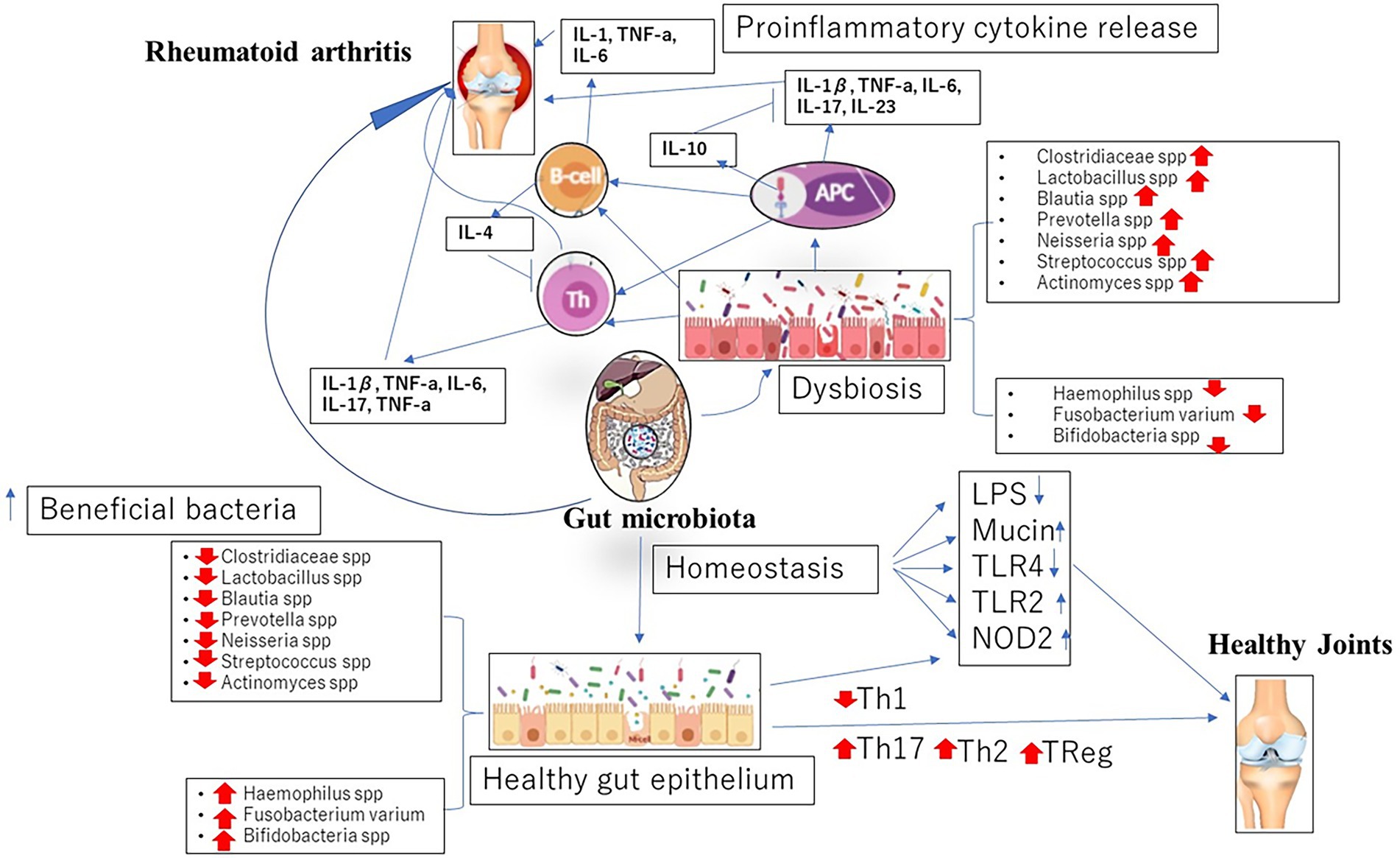
Figure 1. Dysbiosis in gut microbiome associated with the initiation and progression of rheumatoid arthritis (RA).
Probiotics refer to a range of live microbes that are administered in acceptable amounts to provide essential health benefits to the host (Hill et al., 2014). Moreover, myriads of studies have opined on the use of probiotics as a prospective adjuvant for the therapeutic management of RA (Ciccia et al., 2016; Wang et al., 2016; Reyes-Castillo et al., 2021). Although the mechanisms behind these observations remain partly unclear, it is believed that exogenous bacteria exert a temporary effect on the gut microbiome causing alterations to correct and improve dysbiosis-related diseases of which RA is one (Zhang et al., 2016). This effect is tied to the correction of imbalance in the cytokine levels in RA patients (Mohammed et al., 2017). The upregulation of pro-inflammatory cytokines coupled with a decrease in anti-inflammatory cytokines is a determining factor for the initiation and progression of RA. However, studies have documented in RA patients a reduction in the levels of IL-6, a key cytokine associated with the progression and severity of RA after being on probiotics (Brzustewicz and Bryl, 2015).
Probiotics-rich diet has been reported to ameliorate some of the symptoms of RA and this observation has been linked to the restoration of barrier mechanisms in the gut mucosal due to dysregulation (Mohammed et al., 2017). These barriers are restored by maintaining a healthy balance between gut microflora whiles resisting the colonization and translocation of harmful microbes. Again, barrier mechanisms are further enhanced by probiotics through the stimulation of mucus secretion from intestinal epithelial cells (Liu et al., 2016).
Again, probiotics have proven to directly modulate the immune system (La Fata et al., 2018; Kalinkovich and Livshits, 2019). Here, they have been reported to reduce inflammation by significantly downregulating the expression of TLRs (Gómez-Llorente et al., 2010) which are key components in the activation of signaling pathways leading to the production of cytokines for immune modulation (Cristofori et al., 2021). Furthermore, not only TLRs have been implicated to be stimulated by probiotics to produce cytokines, antigen presenting cells (APCs) have also been reported (Azad et al., 2018). In a related study, Kwon et al. (2010) also have reported on the induction of Treg immune response in experimental models of RA by probiotics. This was evident in the conversion of T cells into Tregs bearing FOXP3 which can regulate and suppress inflammatory cascade (Kwon et al., 2010). To our dismay, latest studies have collaborated with these earlier findings as probiotics have been implicated in the modulation of tryptophan metabolism and adenosine signaling in some experimental models. Here, specific probiotic strains have been noted for the activation of adenosine receptors which suppress the effect of Tregs on effectors T cells (He et al., 2017; Gao et al., 2018).
Interestingly, metabolites such as short-chain fatty acids of the gut microbiome established due to the effect of probiotics reportedly have both antimicrobial and anti-inflammatory effects contributing to the alleviation of some symptoms of RA as observed in myriads of studies (Pirozzi et al., 2018; Lee et al., 2019; Figure 2).
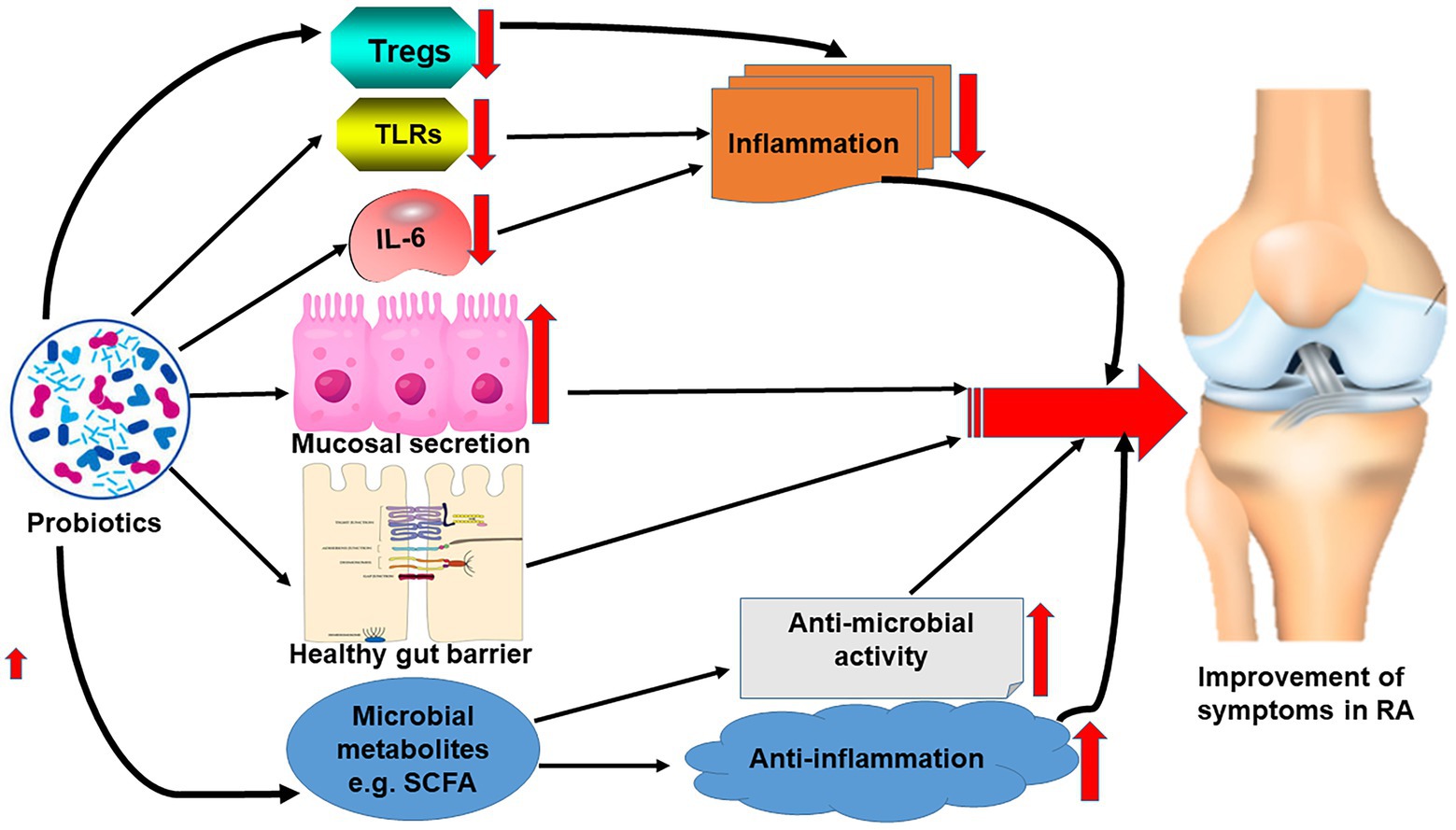
Figure 2. Effect of probiotics on the correction of gut microbiome dysbiosis leading to the reduction of the symptoms of RA.
The alarming incidence of RA coupled with the difficulties associated with its management is a worrying challenge to stakeholders. With the implication of gut microbiome dysbiosis at the core of RA, supplements and adjuvants that can correct these alterations will be more than welcome. With a plethora of studies suggestive of the protective and corrective role of probiotics in the dysbiosis associated with RA, more research will be needed for its prospective therapeutic application. Again, probiotic strains with this beneficial role will have to be studied extensively to ensure their lack of pathogenicity.
YO developed the idea. YO, KA, GG-Q, JA, FB-E, EO, CK, and RK-M wrote and reviewed the manuscript. YO and KA designed the figures. All authors read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdollahi-Roodsaz, S., Joosten, L. A., Koenders, M. I., Devesa, I., Roelofs, M. F., Radstake, T. R., et al. (2008). Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 118, 205–216. doi: 10.1172/JCI32639
Aggarwal, B. B., Kunnumakkara, A. B., Harikumar, K. B., Gupta, S. R., Tharakan, S. T., Koca, C., et al. (2009). Signal transducer and activator of transcription-3, inflammation, and cancer: How intimate is the relationship? Ann. N. Y. Acad. Sci. 1171, 59–76. doi: 10.1111/j.1749-6632.2009.04911.x
Azad, M., Kalam, A., Sarker, M., and Wan, D. J. B. r. i. (2018). Immunomodulatory effects of probiotics on cytokine profiles. BioMed. Res. Int. 2018, 1–10. doi: 10.1155/2018/8063647
Bennike, T. B., Carlsen, T. G., Ellingsen, T., Bonderup, O. K., Glerup, H., Bøgsted, M., et al. (2017). Proteomics dataset: the colon mucosa from inflammatory bowel disease patients, gastrointestinal asymptomic rheumatoid arthritis patients, and controls. Data Brief 15, 511–516. doi: 10.1016/j.dib.2017.09.059
Brzustewicz, E., and Bryl, E. J. C. (2015). The role of cytokines in the pathogenesis of rheumatoid arthritis–practical and potential application of cytokines as biomarkers and targets of personalized therapy. Cytokine 76, 527–536. doi: 10.1016/j.cyto.2015.08.260
Chandradevan, R., Takeda, H., Lim, T., and Patel, N. J. I. (2020). Mycobacterium tuberculosis concealed by Enterococcal sacroiliitis. IDCases 21:e00858. doi: 10.1016/j.idcr.2020.e00858
Chen, J., Wright, K., Davis, J. M., Jeraldo, P., Marietta, E. V., Murray, J., et al. (2016). An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome. Med. 8, 1–14. doi: 10.1186/s13073-016-0299-7
Chiang, H.-I., Li, J.-R., Liu, C.-C., Liu, P.-Y., Chen, H.-H., Chen, Y.-M., et al. (2019). An association of gut microbiota with different phenotypes in Chinese patients with rheumatoid arthritis. J. Clin. Med. 8:1770. doi: 10.3390/jcm8111770
Cho, I., and Blaser, J. M. (2012). The human microbiome: at the interface of health disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Ciccia, F., Ferrante, A., Guggino, G., Triolo, G. J. B. P., and Rheumatology, R. C. (2016). The role of the gastrointestinal tract in the pathogenesis of rheumatic diseases. Best Pract. Res. Clin. Rheumatol. 30, 889–900. doi: 10.1016/j.berh.2016.10.003
Clemente, J. C., Manasson, J., and Scher, J. U. J. B. (2018). The role of the gut microbiome in systemic inflammatory disease. BMJ 360. doi: 10.1136/bmj.j5145
Corrêa, J. D., Fernandes, G. R., Calderaro, D. C., Mendonça, S. M. S., Silva, J. M., Albiero, M. L., et al. (2019). Oral microbial dysbiosis linked to worsened periodontal condition in rheumatoid arthritis patients. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-44674-6
Cristofori, F., Dargenio, V. N., Dargenio, C., Miniello, V. L., Barone, M., Francavilla, R. J. F., et al. (2021). Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body. Front Immunol. 12:178. doi: 10.3389/fimmu.2021.578386
D’Amelio, P., and Sassi, F. J. C. (2018). Gut microbiota, immune system. Bone 102, 415–425. doi: 10.1007/s00223-017-0331-y
de Oliveira, G. L. V., Leite, A. Z., Higuchi, B. S., Gonzaga, M. I., and Mariano, V. S. J. I. (2017). Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 152, 1–12. doi: 10.1111/imm.12765
De Santis, S., Cavalcanti, E., Mastronardi, M., Jirillo, E., Chieppa, M. J. F., et al. (2015). Nutritional keys for intestinal barrier modulation. Front. Immunol. 6:612. doi: 10.3389/fimmu.2015.00612
Dehner, C., Fine, R., and Kriegel, M. A. (2019). The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol. 31, 201–207. doi: 10.1097/BOR.0000000000000574
Dorożyńska, I., Majewska-Szczepanik, M., Marcińska, K., and Szczepanik, M. J. P. R. (2014). Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice. Phrmacol. Rep. 66, 250–255. doi: 10.1016/j.pharep.2013.09.007
Firestein, G. S. J. N. (2003). Evolving concepts of rheumatoid arthritis. Nature 423, 356–361. doi: 10.1038/nature01661
Gao, J., Xu, K., Liu, H., Liu, G., Bai, M., Peng, C., et al. (2018). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell Infect. Microbviol. 8:13. doi: 10.3389/fcimb.2018.00013
Gómez-Llorente, C., Munoz, S., Gil, A. J. P., et al. (2010). Role of toll-like receptors in the development of immunotolerance mediated by probiotics. Proc. Nutr. Soc. 69, 381–389. doi: 10.1017/S0029665110001527
Gu, Y., Fan, W., and Yin, G. J. M. O. I. (2014). The study of mechanisms of protective effect of rg1 against arthritis by inhibiting osteoclast differentiation and maturation in cia mice. Mediators Inflamm. 2014:305071. doi: 10.1155/2014/305071
Guo, L.-X., Wang, H.-Y., Liu, X.-D., Zheng, J.-Y., Tang, Q., Wang, X.-N., et al. (2019). Saponins from Clematis mandshurica Rupr. Regulates gut microbiota and its metabolites during alleviation of collagen-induced arthritis in rats. Pharmacol. Res. 149:104459. doi: 10.1016/j.phrs.2019.104459
Hayes, J. M., Cosgrave, E. F., Struwe, W. B., Wormald, M., Davey, G. P., Jefferis, R., et al. (2014). Glycosylation and fc receptors. Curr. Top. Microbiol. Immunol. 165–199. doi: 10.1007/978-3-319-07911-0_8
He, B., Hoang, T. K., Tran, D. Q., Rhoads, J. M., and Liu, Y. J. F. I. I. (2017). Adenosine A2A receptor deletion blocks the beneficial effects of lactobacillus reuteri in regulatory T-deficient scurfy mice. Front. Immunol. 8:1680. doi: 10.3389/fimmu.2017.01680
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The, International scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gatroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Horta-Baas, G., Romero-Figueroa, M. D. S., Montiel-Jarquín, A. J., Pizano-Zárate, M. L., García-Mena, J., and Ramírez-Durán, N. J. J. O. I. R. (2017). Intestinal dysbiosis and rheumatoid arthritis: a link between gut microbiota and the pathogenesis of rheumatoid arthritis. Immunol. Res. 2017:4835189. doi: 10.1155/2017/4835189
Kalinkovich, A., and Livshits, G. (2019). A cross talk between dysbiosis and gut-associated immune system governs the development of inflammatory arthropathies. Semin. Arthritis Rheum. 49, 474–484. doi: 10.1016/j.semarthrit.2019.05.007
Kayama, H., Okumura, R., and Takeda, K. J. A. (2020). Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu. Rev. Immunol. 38, 23–48. doi: 10.1146/annurev-immunol-070119-115104
Komatsu, N., Okamoto, K., Sawa, S., Nakashima, T., Oh-Hora, M., Kodama, T., et al. (2014). Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20, 62–68. doi: 10.1038/nm.3432
Kwon, H.-K., Lee, C.-G., So, J.-S., Chae, C.-S., Hwang, J.-S., Sahoo, A., et al. (2010). Generation of regulatory dendritic cells and CD4+ Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. U. S. A. 107, 2159–2164. doi: 10.1073/pnas.0904055107
La Fata, G., Weber, P., and Mohajeri, M. H. J. P. (2018). Probiotics and the gut immune system indirect regulation. Probiotics Antimicrob. Proteins 10, 11–21. doi: 10.1007/s12602-017-9322-6
Lee, S., Koh, J., Chang, Y., Kim, H. Y., and Chung, D. H. J. T. J. (2019). Invariant NKT cells functionally link microbiota-induced butyrate production and joint inflammation. J. Immunol. 203, 3199–3208. doi: 10.4049/jimmunol.1801314
Li, M., and Wang, F. J. J. o. I. R. (2021). Role of intestinal microbiota on gut homeostasis and rheumatoid arthritis. J. Immunol. Res. 2021. doi: 10.1155/2021/8167283
Liu, D., Jiang, X.-Y., Zhou, L.-S., Song, J.-H., and Zhang, X. J. M. (2016). Effects of probiotics on intestinal mucosa barrier in patients with colorectal cancer after operation: meta-analysis of randomized controlled trials. Oxidative Medicine And Cellular Longevity 95. doi: 10.1097/MD.0000000000003342
Luo, J.-M., Guo, L., Chen, H., Yang, P.-F., Xiong, R., Peng, Y., et al. (2017). A study of pre-operative presence of micro-organisms in affected knee joints of rheumatoid arthritis patients who need total knee arthroplasty. Knee 24, 409–418. doi: 10.1016/j.knee.2016.11.001
Maeda, Y., Kurakawa, T., Umemoto, E., Motooka, D., Ito, Y., Gotoh, K., et al. (2016). Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661. doi: 10.1002/art.39783
McInnes, I. B., and Schett, G. J. N. E. J. (2011). The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 365, 365, 2205–2219. doi: 10.1056/NEJMra1004965
Mei, L., Yang, Z., Zhang, X., Liu, Z., Wang, M., Wu, X., et al. (2021). Sustained drug treatment alters the gut microbiota in rheumatoid arthritis. Front. Immunol. 12:704089. doi: 10.3389/fimmu.2021.704089
Mikuls, T. R. J. B. P. (2003). Co-morbidity in rheumatoid arthritis. Rheumatology 17, 729–752. doi: 10.1016/s1521-6942(03)00041-x
Mizuno, M., Noto, D., Kaga, N., Chiba, A., and Miyake, S. J. P. O. (2017). The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One 12:e0173032. doi: 10.1371/journal.pone.0173032
Mohammed, A. T., Khattab, M., Ahmed, A. M., Turk, T., Sakr, N., MKhalil, A., et al. (2017). The therapeutic effect of probiotics on rheumatoid arthritis: a systematic review and meta-analysis of randomized control trials. Clin. Rheumatol. 36, 2697–2707. doi: 10.1007/s10067-017-3814-3
Moret, F. M., Hack, C. E., van der Wurff-Jacobs, K. M., de Jager, W., Radstake, T. R., Lafeber, F. P., et al. (2013). Intra-articular CD1c-expressing myeloid dendritic cells from rheumatoid arthritis patients express a unique set of T cell-attracting chemokines and spontaneously induce Th1, Th17 and Th2 cell activity. Arthritis Res.Ther. 15, 1–10. doi: 10.1186/ar4338
Möttönen, M., Heikkinen, J., Mustonen, L., Isomäki, P., Luukkainen, R., Lassila, O. J. C., et al. (2005). CD4+ CD25+ T cells with the phenotypic and functional characteristics of regulatory T cells are enriched in the synovial fluid of patients with rheumatoid arthritis. Clin. Exp. Immunol. 140, 360–367. doi: 10.1111/j.1365-2249.2005.02754.x
Nimmerjahn, F., Anthony, R. M., and Ravetch, J. V. J. P. O. T. N. A. O. S. (2007). Agalactosylated IgG antibodies depend on cellular fc receptors for in vivo activity. Curr. Opin. Organ. Transplant. 104, 8433–8437. doi: 10.1073/pnas.0702936104
Opoku, Y. K., Liu, Z., Liu, H., Afrifa, J., Koranteng, H., Ren, G., et al. (2020). Fibroblast growth factor–21 ameliorates rheumatoid arthritis by maintaining articular integrity. Int. J. Pept. Res. Ther. 26, 651–659. doi: 10.1007/s10989-019-09872-w
Pianta, A., Arvikar, S. L., Strle, K., Drouin, E. E., Wang, Q., Costello, C. E., et al. (2017). Two rheumatoid arthritis–specific autoantigens correlate microbial immunity with autoimmune responses in joints. J. Clin. Investig. 127, 2946–2956. doi: 10.1172/JCI93450
Picchianti-Diamanti, A., Panebianco, C., Salemi, S., Sorgi, M. L., Di Rosa, R., Tropea, A., et al. (2018). Analysis of gut microbiota in rheumatoid arthritis patients: disease-related dysbiosis and modifications induced by etanercept. Int. J. Mol. Sci. 19:2938. doi: 10.3390/ijms19102938
Pirozzi, C., Francisco, V., Di Guida, F., Gómez, R., Lago, F., Pino, J., et al. (2018). Butyrate modulates inflammation in chondrocytes via GPR43 receptor. Cell. Physiol. Biochem. 51, 228–243. doi: 10.1159/000495203
Raju, T. S. J. C. O. I. I. (2008). Terminal sugars of fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 20, 471–478. doi: 10.1016/j.coi.2008.06.007
Reyes-Castillo, Z., Valdés-Miramontes, E., Llamas-Covarrubias, M., Muñoz-Valle, J. J. C., and Medicine, E. (2021). Troublesome friends within us: the role of gut microbiota on rheumatoid arthritis etiopathogenesis and its clinical and therapeutic relevance. Clin. Exp. Med. 21, 1–13. doi: 10.1007/s10238-020-00647-y
Ruff, W. E., and Kriegel, M. A. (2015). Autoimmune host–microbiota interactions at barrier sites and beyond. Trends Mol. Med. 21, 233–244. doi: 10.1016/j.molmed.2015.02.006
Scher, J. U., Littman, D. R., and Abramson, S. B. (2016). Microbiome in inflammatory arthritis and human rheumatic diseases. Arthritis Rheumatol. 68, 35–45. doi: 10.1002/art.39259
Sun, Y., Chen, Q., Lin, P., Xu, R., He, D., Ji, W., et al. (2019). Characteristics of gut microbiota in patients with rheumatoid arthritis in Shanghai. China 369. doi: 10.3389/fcimb.2019.00369
Tanoue, T., Atarashi, K., and Honda, K. J. N. R. I. (2016). Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16, 295–309. doi: 10.1038/nri.2016.36
Taurog, J. D., Richardson, J. A., Croft, J., Simmons, W. A., Zhou, M., Fernández-Sueiro, J. L., et al. (1994). The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 180, 2359–2364. doi: 10.1084/jem.180.6.2359
Teng, F., Klinger, C. N., Felix, K. M., Bradley, C. P., Wu, E., Tran, N. L., et al. (2016). Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 44, 875–888. doi: 10.1016/j.immuni.2016.03.013
Wang, B., He, Y., Tang, J., Ou, Q., and Lin, J. J. I. J. (2020). Alteration of the gut microbiota in tumor necrosis factor-α antagonist-treated collagen-induced arthritis mice. Int. J. Rheum. Dis. 23, 472–479. doi: 10.1111/1756-185X.13802
Wang, P., Tao, J.-H., and Pan, H.-F. J. I. (2016). Probiotic bacteria: a viable adjuvant therapy for relieving symptoms of rheumatoid arthritis. Inflammopharmacology 24, 189–196. doi: 10.1007/s10787-016-0277-0
Wang, Q., Zhang, S.-X., Chang, M.-J., Qiao, J., Wang, C.-H., Li, X.-F., et al. (2022). Characteristics of the gut microbiome and its relationship with peripheral CD4+ T cell subpopulations and cytokines in rheumatoid arthritis. Front. Microbiol. 64. doi: 10.3389/fmicb.2022.799602
Wu, X., He, B., Liu, J., Feng, H., Ma, Y., Li, D., et al. (2016). Molecular insight into gut microbiota and rheumatoid arthritis. Int. J. Mol. Sci. 17:431. doi: 10.3390/ijms17030431
Wu, H.-J., Ivanov, I. I., Darce, J., Hattori, K., Shima, T., Umesaki, Y., et al. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. doi: 10.1016/j.immuni.2010.06.001
Yamamoto, H., Konishi, Y., Mineo, S., Sekiya, M., Kohno, T., Kohno, M. J. E., et al. (2000). Characteristics of polyarthritis in rabbits by hyperimmunization with attenuated Enterococcus faecalis. J. Toxicol. Pathol. 52, 247–255. doi: 10.1016/S0940-2993(00)80041-X
Yu, M. B., and Langridge, W. H. J. R. I. (2017). The function of myeloid dendritic cells in rheumatoid arthritis. Rheumatol. Int. 37, 1043–1051. doi: 10.1007/s00296-017-3671-z
Zeissig, S., and Blumberg, R. S. (2014). Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 15, 307–310. doi: 10.1038/ni.2847
Zhang, C., Derrien, M., Levenez, F., Brazeilles, R., Ballal, S. A., Kim, J., et al. (2016). Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 10, 2235–2245. doi: 10.1038/ismej.2016.13
Zhang, X., Zhang, D., Jia, H., Feng, Q., Wang, D., Liang, D., et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Food Sci. Nutr. 21, 895–905. doi: 10.1038/nm.3914
Zhao, Q., and Elson, C. O. (2018). Adaptive immune education by gut microbiota antigens. Immunology 154, 28–37. doi: 10.1111/imm.12896
Keywords: rheumatoid arthritis, gut microbiome, probiotics, dysbiosis, therapy, inflammation
Citation: Opoku YK, Asare KK, Ghartey-Quansah G, Afrifa J, Bentsi-Enchill F, Ofori EG, Koomson CK and Kumi-Manu R (2022) Intestinal microbiome–rheumatoid arthritis crosstalk: The therapeutic role of probiotics. Front. Microbiol. 13:996031. doi: 10.3389/fmicb.2022.996031
Received: 16 July 2022; Accepted: 21 September 2022;
Published: 18 October 2022.
Edited by:
Smith Etareri Evivie, University of Benin, NigeriaReviewed by:
Mariana Carmen Chifiriuc, University of Bucharest, RomaniaCopyright © 2022 Opoku, Asare, Ghartey-Quansah, Afrifa, Bentsi-Enchill, Ofori, Koomson and Kumi-Manu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yeboah Kwaku Opoku, eWtvcG9rdUB1ZXcuZWR1Lmdo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.