
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 15 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.994941
This article is part of the Research TopicInteractions Between Filamentous Plant Pathogens with their Host Plants and Biocontrol AgentsView all 16 articles
The aversive behavior of Caenorhabditis elegans is an important strategy that increases their survival under pathogen infection, and the molecular mechanisms underlying this behavior have been described. However, whether this defensive response occurs in plant-parasitic nematodes (PPNs), which have quite different life cycles and genomic sequences from the model nematode, against biocontrol microbes and affects interspecific interactions in ecological environments remains unclear. Here, we showed that Meloidogyne incognita, one of the most common PPNs, engaged in lawn-leaving behavior in response to biocontrol bacteria such as Bacillus nematocida B16 and B. thuringiensis Bt79. Genomic analysis revealed that the key genes responsible for the aversive behavior of C. elegans, such as serotonin-and TGF-β-related genes in canonical signaling pathways, were homologous to those of M. incognita, and the similarity between these sequences ranged from 30% to 67%. Knockdown of the homologous genes impaired avoidance of M. incognita to varying degrees. Calcium ion imaging showed that the repulsive response requires the involvement of the multiple amphid neurons of M. incognita. In situ hybridization specifically localized Mi-tph-1 of the serotonin pathway to ADF/NSM neurons and Mi-dbl-1 of the TGF-β pathway to AVA neurons. Our data suggested that the repulsive response induced by different biocontrol bacteria strongly suppresses the invasion of tomato host plants by M. incognita. Overall, our study is the first to clarify the pathogen-induced repulsive response of M. incognita and elucidate its underlying molecular mechanisms. Our findings provide new insights into interspecific interactions among biocontrol bacteria, PPNs, and host plants.
Plant parasitic nematodes (PPNs) are some of the most devastating agricultural pests. Root-knot nematodes (Meloidogyne spp.) are the most omnivorous PPNs, and they can parasitize more than 5,500 host plants and cause over $100 billion agriculture losses worldwide annually (Wang et al., 2021b). Meloidogyne preferentially invades plant roots and forms root galls, which impede the normal uptake of water and nutrients, and further facilitates the infection of some other soil-borne phytopathogens (Makhubu et al., 2021).
PPNs accomplish their life cycles through a robust chemosensory system, which allows them to sense a variety of chemical signals from soil and plants, and their responses to these signals are mediated via the synergistic functions of chemoreceptors, neuropeptides, and molecular signaling pathways. For example, root exudates secreted by plants strongly attract the second-stage juveniles (J2s) of Meloidogyne, which play an indispensable role when nematodes migrate in soil and search for suitable feeding sites on plants (Oota et al., 2020). During this process, chemical gradients of volatile chemicals such as pinene, limonene, and tridecane, and organic acids, such as malic acid, oxalic acid, and lactic acid have been reported to be tracked by Meloidogyne (Wang et al., 2021a). Meloidogyne also shows a chemotactic response toward fats and their derivatives, including low concentrations of lauric acid from crown daisy (Dong et al., 2014). A few phytohormones, such as zeatin, ethylene, jasmonic acid, salicylic acid, and abscisic acid, contribute to the development of feeding cells and alter the attractiveness of plant roots (Wubben et al., 2004; Swiecicka et al., 2009; Fudali et al., 2013; Cabrera et al., 2014; Kirwa et al., 2018). However, repellents, such as dibutyl phthalate, palmitic acid and linoleic acid derived from plant roots, induce repulsive response in M. incognita J2 and thus provide protection against parasitism by PPNs (Yang et al., 2016; Dong et al., 2018).
In C. elegans, pathogen-induced aversive behavior is an important defense strategy that confers resistance to virulent bacteria, in addition to innate immunity. Pathogen-induced aversive behavior also keeps the nematodes away from predators or tainted food. Several canonical molecular signaling pathways, such as the serotonin or TGF-β pathways, are responsible for the pathogen-induced aversive behavior of C. elegans (Shivers et al., 2009; Melo and Ruvkun, 2012; Zhang and Zhang, 2012). However, whether PPNs use their chemosensory system to induce the similar repulsive response in the presence of biocontrol bacteria, or whether this response affects their ability to infect host plants and subsequently alter the efficiency of biocontrol agents remains unclear (Zhou et al., 2019).
In 2008, the draft genomes of M. incognita and M. hapla were published, and bioinformatic analyses have suggested that the size of the genomes of these two root-knot nematodes is significantly reduced compared with the genome of C. elegans; this reduction is especially pronounced for families of genes involved in stress responses and innate immunity, such as glutathione transferase, cytochrome P450, immune effector elements, G protein-coupled receptors, and chitin-degrading enzymes, which might be attributed to the parasitic lifestyle of Meloidogyne within host plants (Abad et al., 2008; Opperman et al., 2008). The significant differences in the genome sequences of Meloidogyne and C. elegans suggest that the repulsive response of M. incognita might differ from that of C. elegans, or even be stronger than that of C. elegans to compensate for the loss of genes involved in stress resistance and innate immunity. Previous studies had shown that Bacillus and Pseudomonas spp. were the most common populations that colonized the rhizosphere and could effectively antagonize root-knot nematodes for their nematocidal activity (Zhu et al., 2021; Antil et al., 2022; Gowda et al., 2022). Here, the repulsive response of M. incognita against two species of biocontrol bacteria, including B. nematocida B16 and B. thuringiensis Bt79, was investigated and the signal pathways underlying this behavior were elucidated. Our findings provide new insights into interspecific interactions among biocontrol bacteria, PPNs, and host plants and have implications for the development of new biocontrol strategies.
A pure isolate of M. incognita was maintained on tomatoes (Solanum lycopersicum cv. Jingfen Champion) in the greenhouse at Yunnan University, Kunming, China. Egg masses were collected from infected plants and hatched in distilled water. The freshly hatched M. incognita J2s were used in subsequent experiments.
Chemotactic assays were used to test the repulsive response of M. incognita on pluronic gel medium of PF-127 (Sigma-Aldrich, St. Louis, United States; Figure 1A). In the avoidance assay, 4 ml of 23% (w/v) PF-127 (liquid at 4°C) was added to a 6 cm diameter Petri dish at room temperature for the gel to solidify. Five μl of bacterial culture and LB fluid medium without bacteria, which were used as the sample and control groups, respectively, were added to opposite sides at a distance of 2 cm from the center of the plate. Approximately 100 M. incognita J2s were added to the center of the plate, and the lid was closed. After 6, 12, and 24 h, the numbers on both sides were counted under stereomicroscope (Nikon Co, Tokyo, Japan). The avoidance index (AI) was calculated using the following formula: (number of nematodes on the control side – number of nematodes on the sample side) /total number of nematodes; the index ranged from −1.0 to 1.0 (Shivakumara et al., 2018; Kuang et al., 2020). Locomotion, including the crawling pattern and movement tracks, was also assessed on PF-127 medium under the stereomicroscope.
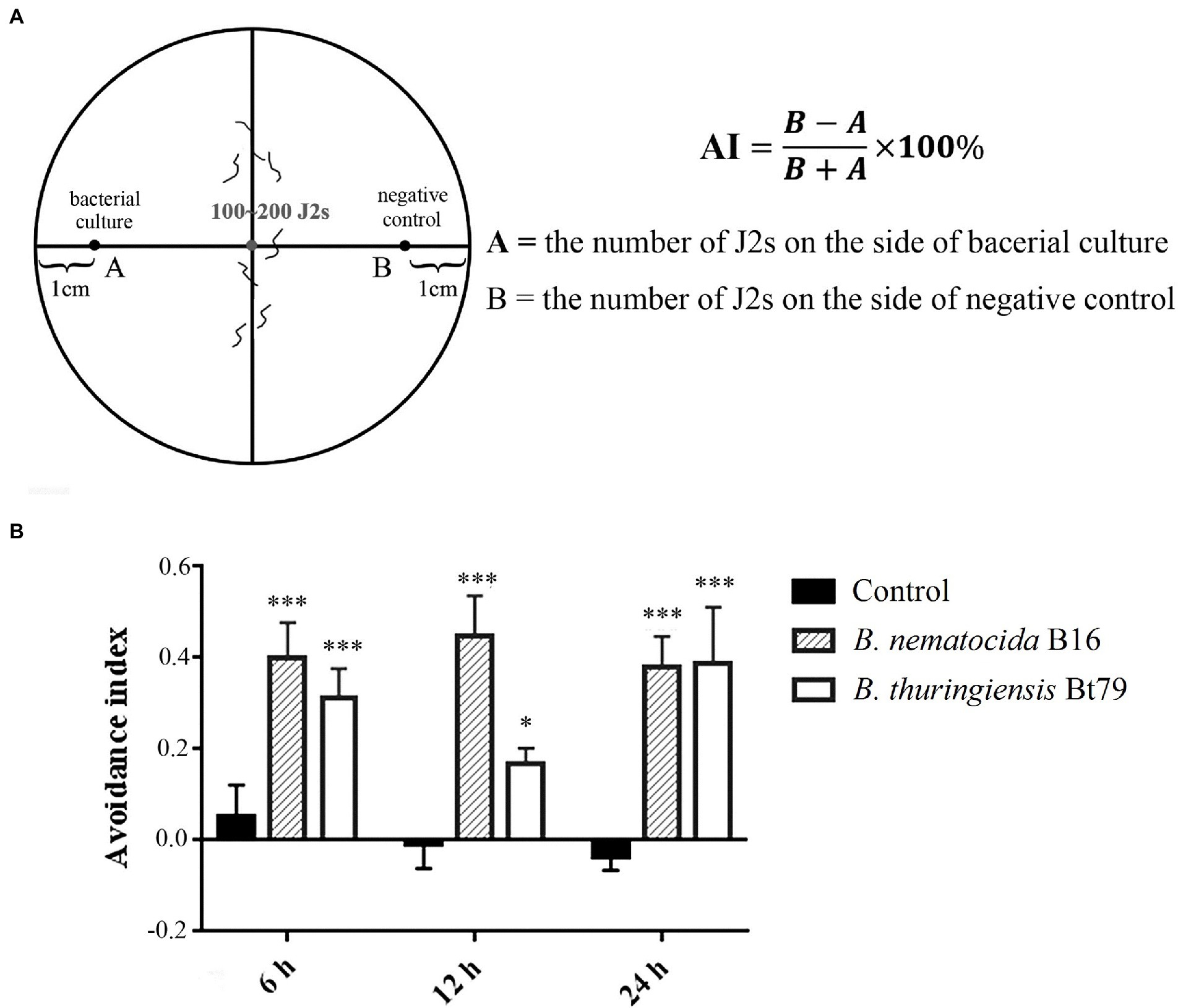
Figure 1. Repulsive response of Meloidogyne incognita second-stage juveniles (J2s) induced by different biocontrol bacteria. (A) Schematic diagram of the chemotaxis assay of M. incognita J2s on PF-127 medium. (B) Avoidance index of M. incognita J2s against different biocontrol bacteria. *p < 0.05 and ***p < 0.001.
Fura-2-AM (Beyotime, Shanghai, China), a fluorescent indicator with high affinity for Ca2+, was used to detect the intracellular Ca2+ concentration (Shuang et al., 2010). Approximately 300 freshly hatched J2s were soaked in 500 μl of M9 buffer containing 2 μl of 20 mM Fura-2-AM and 10 μl of 0.5% resorcinol, and they were incubated in the dark at room temperature on a rotary mixer for 6 h and then washed several times with sterile M9 buffer. A fluorescent microscope of Nikon E800 (Nikon Co, Tokyo, Japan) was used to record the fluorescence intensity in the head neurons under 330–350 nm excitation wavelengths before and after the bacterial culture of B. nematocida B16 and B. thuringiensis Bt79 was added near J2. The software ImageJ was used to quantify the mean fluorescence intensity.
Total RNA was extracted from 10,000 J2s by Trizol reagent (Tiangen Co., Tianjin, China). Synthesis of cDNA was performed using random primers with the Prime Script RT Reagent Kit (Takara, Dalian, China). The target cDNA of Mi-tph-1, Mi-mod-1, Mi-dbl-1, and Mi-sma-6 was amplified using EX Taq DNA polymerase (Takara, Dalian, China). The components of the PCR reaction were 0.5 μl of each primer, 0.5 μl of cDNA, 2.5 μl of 10 × Ex Taq buffer, 2 μl of dNTP mix, 0.2 μl of Ex Taq DNA Polymerase, and 18.8 μl of double distilled water. The primers are shown in Table 1. The amplified fragment was then cloned into pGEM-T Easy Vector (Promega, Madison, United States), and the inserted fragment was confirmed by sequencing.
Purified PCR products were used as the template to synthesize the dsRNA of each target gene using the MEGAscript™ SP6 Transcription Kit following the method described by Shivakumara et al. (2018) (Thermo Fisher, Waltham, MA, United States). RNA interference (RNAi) treatment of nematodes was performed following a previously described method (Bakhetia et al., 2005) with minor modifications. Briefly, the freshly hatched J2s were immersed in soaking buffer with 1 mg/ml dsRNA supplemented with 0.1 mg/ml FITC, 0.1% resorcinol, 20 mM octopamine, 30 mM spermidine, and 1% gelatin on a rotary mixer in the dark at room temperature for 24 h. Fluorescein isothiocyanate (Beyotime, Shanghai, China) was used as a tracer to assess the uptake efficiency of dsRNA by observing the fluorescence intensity. Meloidogyne incognita J2s incubated in the soaking buffer without dsRNA probes were used as the negative control. After that, the soaked nematodes were recovered in the nuclease-free water for another 12 h. Real-time qPCR was used to evaluate the efficiency of the transcriptional knockdown of target genes.
Probe preparation and hybridization were performed using the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Basel, Switzerland). The probes of Mi-tph-1 and Mi-dbl-1 were generated by PCR using previously designed primers (see Table 1) to amplify a 200–250 base pair region from cDNA cloned in pGEM-TEasy. PCR products were then visualized on a 1.2% (w/v) agarose gel, and sequences were verified as described above. One microgram of purified PCR product was added to an RNAase-free PCR tube and mixed with ddH2O to a volume of 16 μl. dsDNA was denatured in a boiling water bath for 10 min. The PCR tube was then quickly inserted into an ice-water mixture for 5 min to prevent the renaturation of DNA. Next, 4 μl of DIG-High Prime was added to the above PCR tube. The probe was then labeled with DIG-11-dUTP after incubation at 37°C for 20 h and at 65°C for 10 min to end the reaction. The labeled probe could be immediately used or stored at −20°C for standby application.
Fixation, permeabilization, probe hybridization, and detection were conducted following the procedures of Kimber et al. (2002) with slight modifications. In short, freshly hatched M. incognita J2s were thoroughly washed in sterile water and fixed in 4% paraformaldehyde overnight at 4°C. After fixation, the nematodes were washed twice in M9 buffer and permeabilized using 5 mg/ml proteinase K for 25 min on a rotator at room temperature, followed by incubation in an ice bath for 20 min, incubation in methanol for 1 min (−20°C), and incubation in acetone for 1 min (−20°C).
The permeabilized nematodes were thoroughly washed with M9 buffer and resuspended in 1,000 μl of hybridization buffer for prehybridization at 42°C for 2 h. Five μl of digoxigenin-labeled dsDNA probe was diluted with 50 μl of double distilled water and heat-denatured at 95°C for 10 min, followed by an ice bath for 2 min to maintain the melted state of the probe. The denatured probe was then added to the prehybridization system for hybridization at 42°C overnight, and DIC-labeled DNA was added to the control group. The nematodes were washed two times with 2× saline sodium citrate (SSC) with 0.1% SDS at room temperature and then twice with 0.2 × SSC/0.1% SDS at 42°C, followed by one wash with maleic acid buffer for 3 min at room temperature. Hybridization was conducted for 30 min at room temperature in 1 × blocking solution diluted by maleic acid buffer. The antibody of alkaline phosphatase-conjugated anti-digoxigenin was diluted 1:5,000 by 1 × blocking solution and incubated at room temperature for 2 h, followed by washing with maleic acid buffer for 2 min and 2–5 min by detection buffer. After overnight incubation in a color substrate solution of nitroblue tetrazolium salt and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) diluted 1:50 by detection buffer, the nematodes were washed twice with 0.1 × M9 buffer for 1 min each wash. The nematode specimens were then mounted on a glass slide covered with a cover slip and observed under a microscope.
The infection assay was performed on PF-127 medium to evaluate the effect of the repulsive response induced by different biocontrol bacteria on infection of the plant roots (Dash et al., 2017). Using a pipette, ~300 J2s were added to 1.5 cm posterior to the root tips of tomato seedlings on 23% PF-127 medium in a Petri dish with a diameter of 3 cm. Approximately 5 μl bacterial culture was added near the root tips. Roots were stained by acid fuchsin at 48 h after inoculation, and the number of M. incognita J2s that invaded the host plant roots was counted.
At least three technical and biological replicates were performed for all assays. All data were expressed as mean ± standard deviation. Statistical analyses were performed using one-or two-way ANOVA and t-tests.
To determine whether the repulsive response of PPNs could be induced by biocontrol bacteria, a PF-127 chemotactic assay was conducted, and the AI was calculated for M. incognita J2s after sensing the volatile molecules secreted by the bacterial strains (Figure 1A). Compared with the negative control (blank media), the lawn-leaving behavior of M. incognita was observed after 6 h of exposure to the two biocontrol bacteria, and the AI was 0.40 ± 0.08 against B. nematocida B16 and 0.3 ± 0.06 against B. thuringiensis Bt79. As the time course was extended, the AI was increased to 0.45 ± 0.09 at 12 h and slightly decreased to 0.38 ± 0.09 at 24 h against B. nematocida B16. To B. thuringiensis Bt79, the AI was firstly decreased to 0.17 ± 0.03 at 12 h and increased to 0.39 ± 0.12 again at 24 h (Figure 1B). Thus, the PF-127 chemotactic assay demonstrated that M. incognita J2s can escape efficiently from the different biocontrol bacteria, which is similar with the defensive response of C. elegans against its bacterial pathogens (Zhang et al., 2005).
The repulsive response of M. incognita J2 was observed after 6 h of exposure to biocontol bacteria, suggesting that it might involve learning-associated avoidance. We analyzed the homology of key genes of C. elegans in canonical signaling pathways, such as tph-1 encoding serotonin synthase, mod-1 encoding the GPCR receptor in the serotonin pathway, and dbl-1 and sma-6 encoding the ligand and receptor of the TGF-β pathway, respectively, to the genomic sequence of M. incognita. Alignment to these four genes revealed low similarities (30%–67%) in amino acid sequences between M. incognita and C. elegans (Table 2), which urged us to further validate their roles in repulsive response of M. incognita.
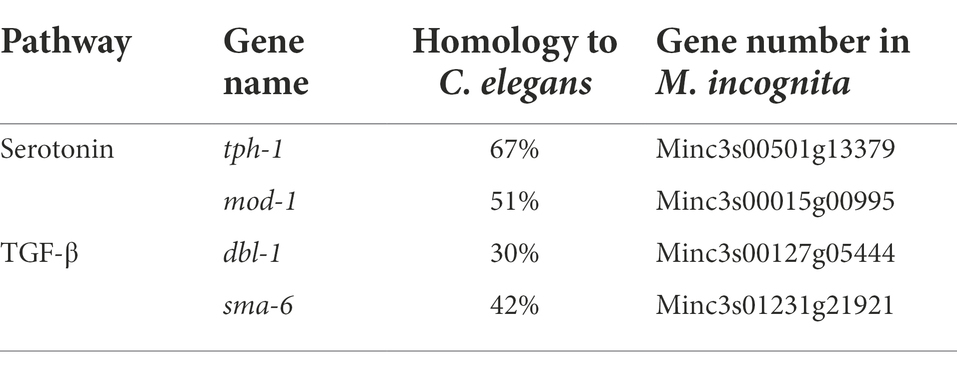
Table 2. Homology of key genes in canonical pathways related to learning-associated avoidance between Meloidogyne incognita and Caenorhabditis elegans.
To validate whether the homologous genes of M. incognita play the similar roles in repulsive response against biocontol bacteria, RNAi targeting of Mi-tph-1 and Mi-mod-1 in the serotonin pathway, as well as Mi-dbl-1 and Mi-sma-6 in the TGF-β pathway, was conducted by soaking M. incognita J2s in buffer containing dsRNA probes and FITC. In this reaction system, the green fluorescence of FITC was detected in esophagus and intestinal tract of M. incognita J2, suggesting that dsRNA of the target gene was successfully absorbed by M. incognita (Figure 2A). To determine the efficiency of gene knockdown, we used qRT-PCR to analyze transcription levels and found that the expression of each gene, including Mi-tph-1, Mi-mod-1, Mi-dbl-1, and Mi-sma-6, was significantly down-regulated as expected (p < 0.001; Figure 2B).
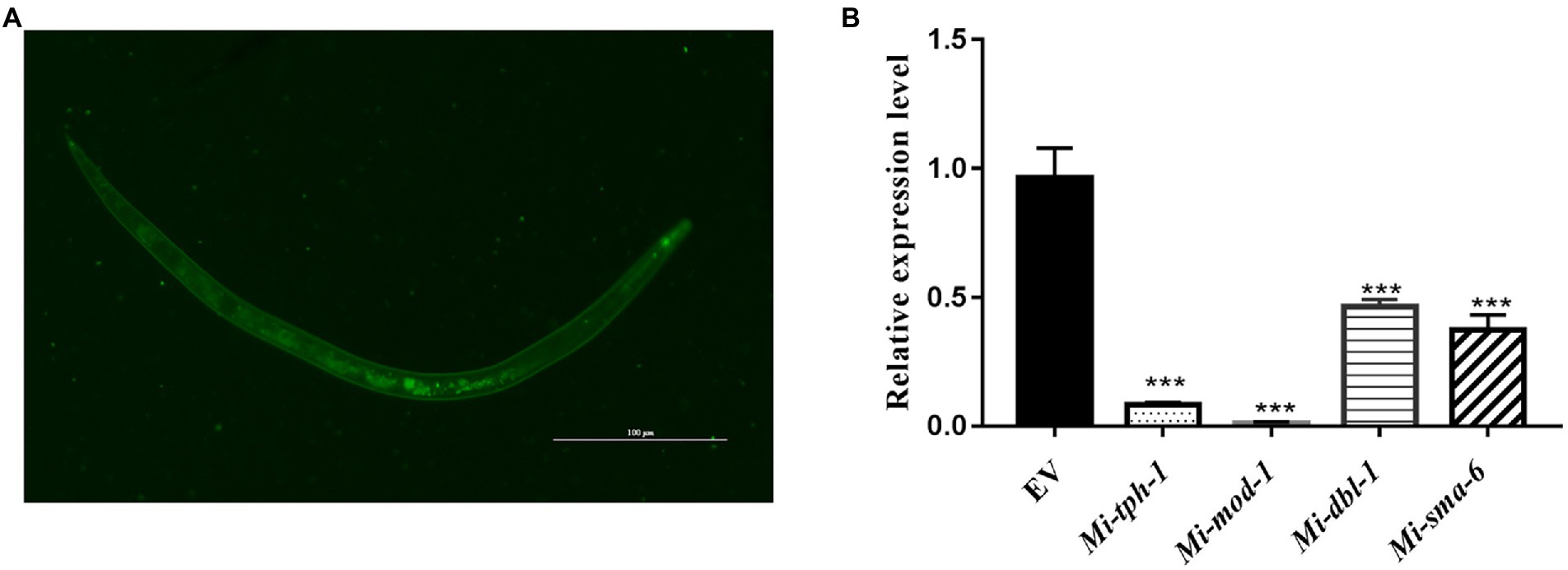
Figure 2. Expression levels of target genes in the RNAi treatment of Meloidogyne incognita second-stage juveniles (J2s). (A) dsRNA uptake of M. incognita J2s determined by the green fluorescence of FITC in the stylet, esophageal gland, and intestine. (B) Relative expressional levels of target genes, including Mi-tph-1, Mi-mod-1, Mi-dbl-1 and Mi-sma-6, quantified by qRT-PCR. EV refers to the control group with empty vector. ***p < 0.001.
Next, the PF-127 chemotactic assay was performed again for the roles of the candidate genes in repulsive response. Knockdown of genes in either the serotonin or TGF-β pathway weakened the lawn-leaving behavior of M. incognita to varying degrees: the AIs of Mi-tph-1 RNAi decreased from 0.59 ± 0.16 to-0.01 ± 0.15 and from 0.40 ± 0.18 to 0.12 ± 0.24 when M. incognita J2s was exposed to B. nematocida B16 and B. thuringiensis Bt79, respectively (Figure 3A); knockdown of the receptor gene Mi-mod-1 also attenuated aversive response, and the AIs decreased to-0.03 ± 0.12 and 0.08 ± 0.03 against B. nematocida B16 and B. thuringiensis Bt79, respectively (Figure 3B). Similarly, RNAi of the other two genes in the TGF-β signaling pathway, Mi-dbl-1 and Mi-sma-6, caused deficiencies in the lawn-leaving behavior against B. nematocida and B. thuringiensis (Figures 3C,D). Overall, our data indicate that both the serotonin and TGF-β pathways of M. incognita play the same roles in the repulsive response to biocontrol bacteria as in C. elegans despite their relatively low homology of gene sequences.
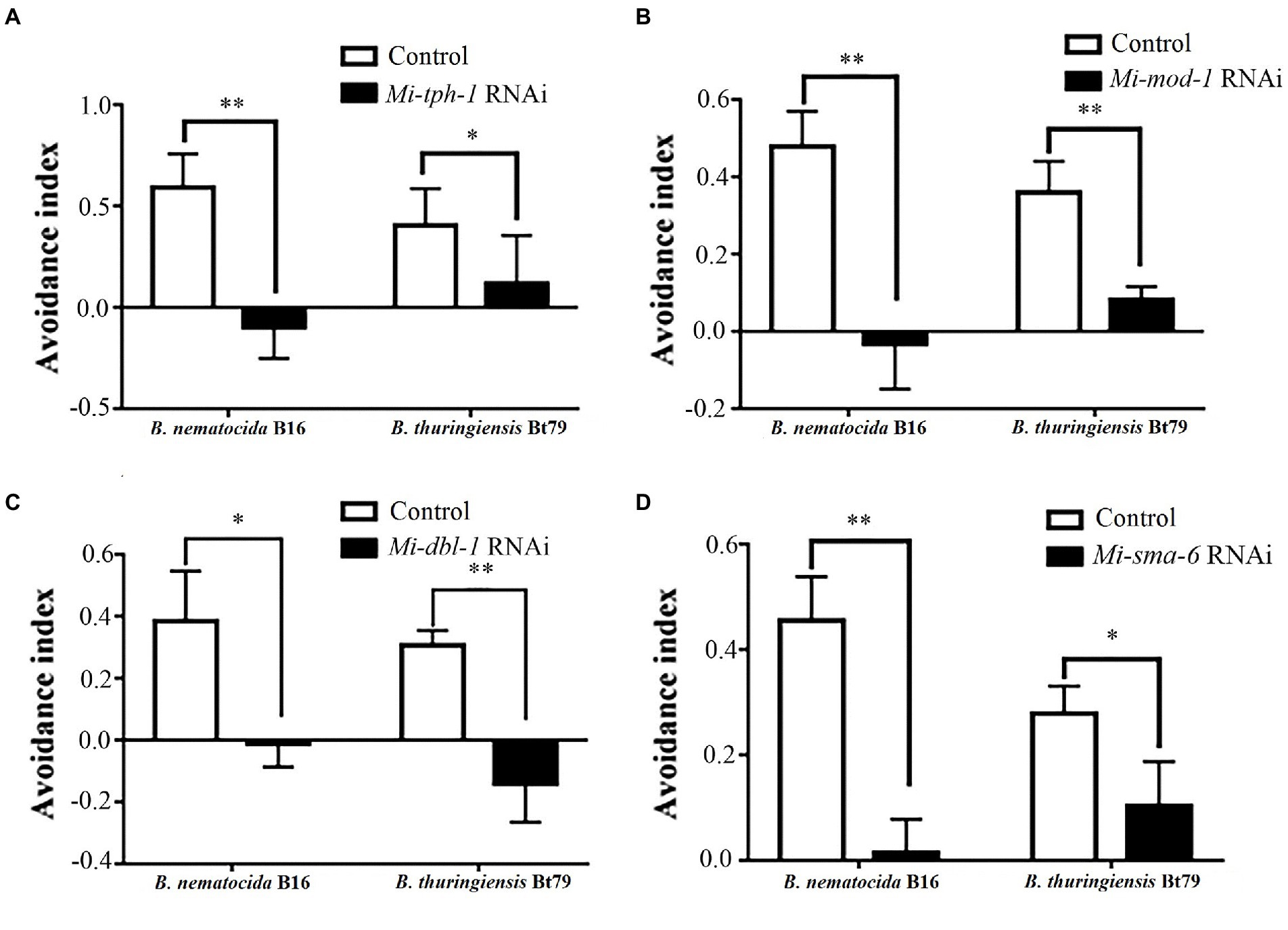
Figure 3. Avoidance index of Meloidogyne incognita against different biocontrol bacteria after RNAi treatment of target genes. RNAi of Mi-tph-1 (A), Mi-mod-1 (B), Mi-dbl-1 (C), and Mi-sma-6 (D). *p < 0.05 and **p < 0.01.
RNAi of Mi-tph-1 and Mi-mod-1 also altered the movements of M. incognita, and no similar effect has been described for alterations to the serotonin pathway of C. elegans. To confirm the potentially new function of the serotonin pathway in M. incognita, we tracked the movements of nematodes with Mi-tph-1 and Mi-mod-1 knocked down by RNAi. Compared with the control group of M. incognita with only empty vector, which displayed crooked movement trajectories on PF-127 medium, the movement patterns of M. incognita with either Mi-tph-1 or Mi-mod-1 knocked down were altered; specifically, the swing amplitude of the body was reduced, and the traces of the nematodes were more flattened (Figure 4A). No differences in body rigidity and movement patterns were observed in N2, tph-1(mg280), and mod-1(ok103) of C. elegans (Figure 4B).
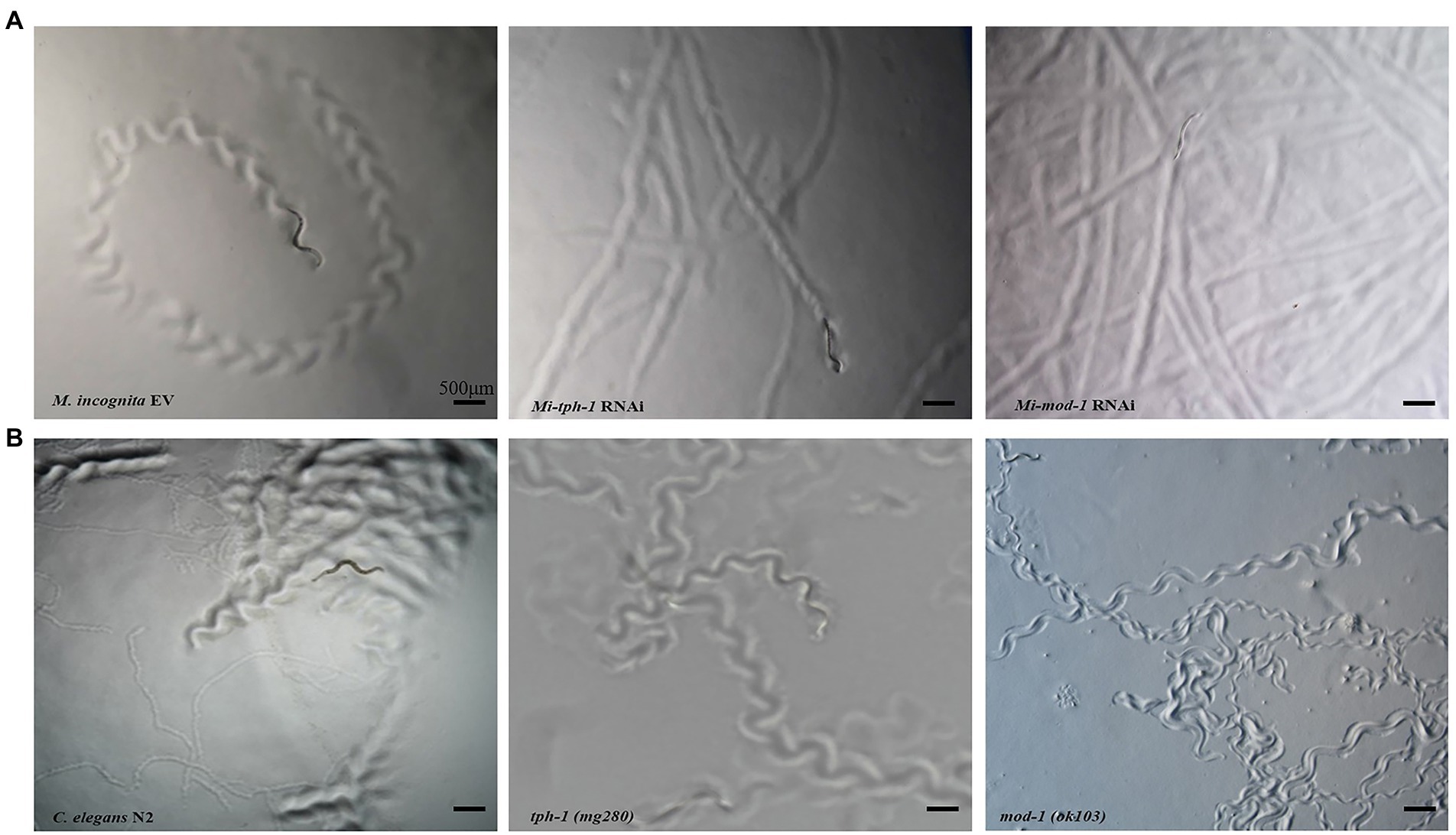
Figure 4. Effects of inactivating the serotonin signaling pathway on the movement trajectory of Meloidogyne incognita. (A) Movement traces of the control with empty vector, Mi-tph-1 RNAi, and Mi-mod-1 RNAi of M. incognita. (B) Movement traces of the wild type nematodes (N2), tph-1(mg280), and mod-1(ok103) of Caenorhabditis elegans.
To identify the neuron(s) involved in the repulsive response of M. incognita against biocontrol bacteria, we measured intracellular changes in Ca2+ using Fura-2-AM imaging. Once the bacterial culture of either B. nematocidal B16 or B. thuringiensis Bt79 was sensed by M. incognita J2 and the repulsive response was stimulated, the fluorescence enhancement of Ca2+ was detected in multiple amphid neurons of M. incognita J2 (Figure 5A), which was further supported by a statistical significance of the average fluorescence intensities within the amphid neurons at 0 min and 5 min (Figure 5B).
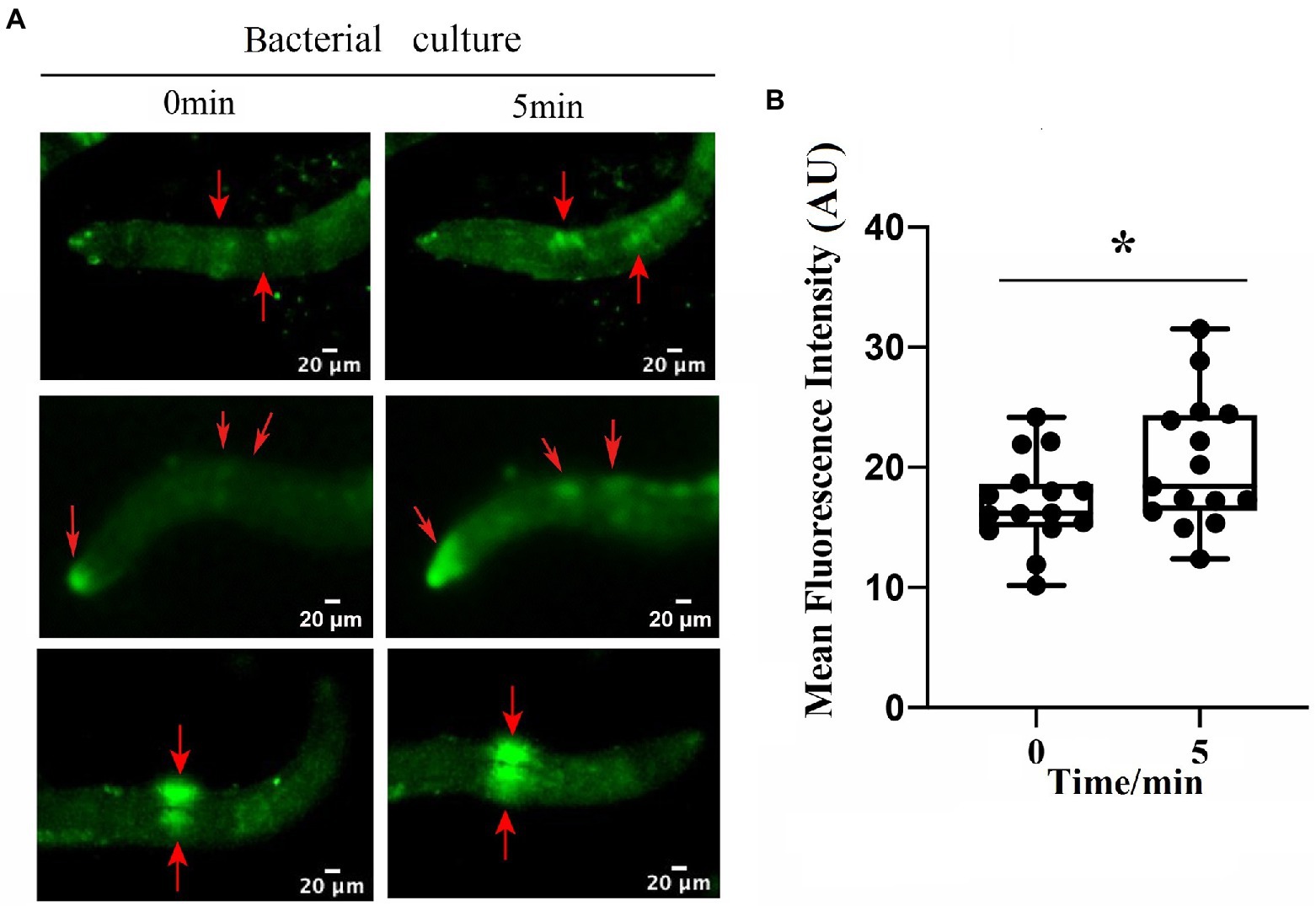
Figure 5. Involvement of multiple amphid neurons in the repulsive response of Meloidogyne incognita. (A) Images of intracellular Ca2+ measurements on the amphid neurons using Fura-2-AM. (B) Quantitative analysis to the mean fluorescence intensity in the amphid neurons before and after adding the bacterial culture (n = 15). *p < 0.05.
As previous studies have reported that the aversive behavior of C. elegans depends on the functioning of tph-1 in ADF neurons and dbl-1 in AVA neurons (Zhang et al., 2005; Zhang and Zhang, 2012), we characterized their neuronal localization in M. incognita. ISH revealed that these two genes exhibited similar expressional patterns in the amphid neurons, with Mi-tph-1 mainly localized in ADF/NSM neurons (Figures 6A,B) and Mi-dbl-1 in AVA neurons (Figures 6C,D).
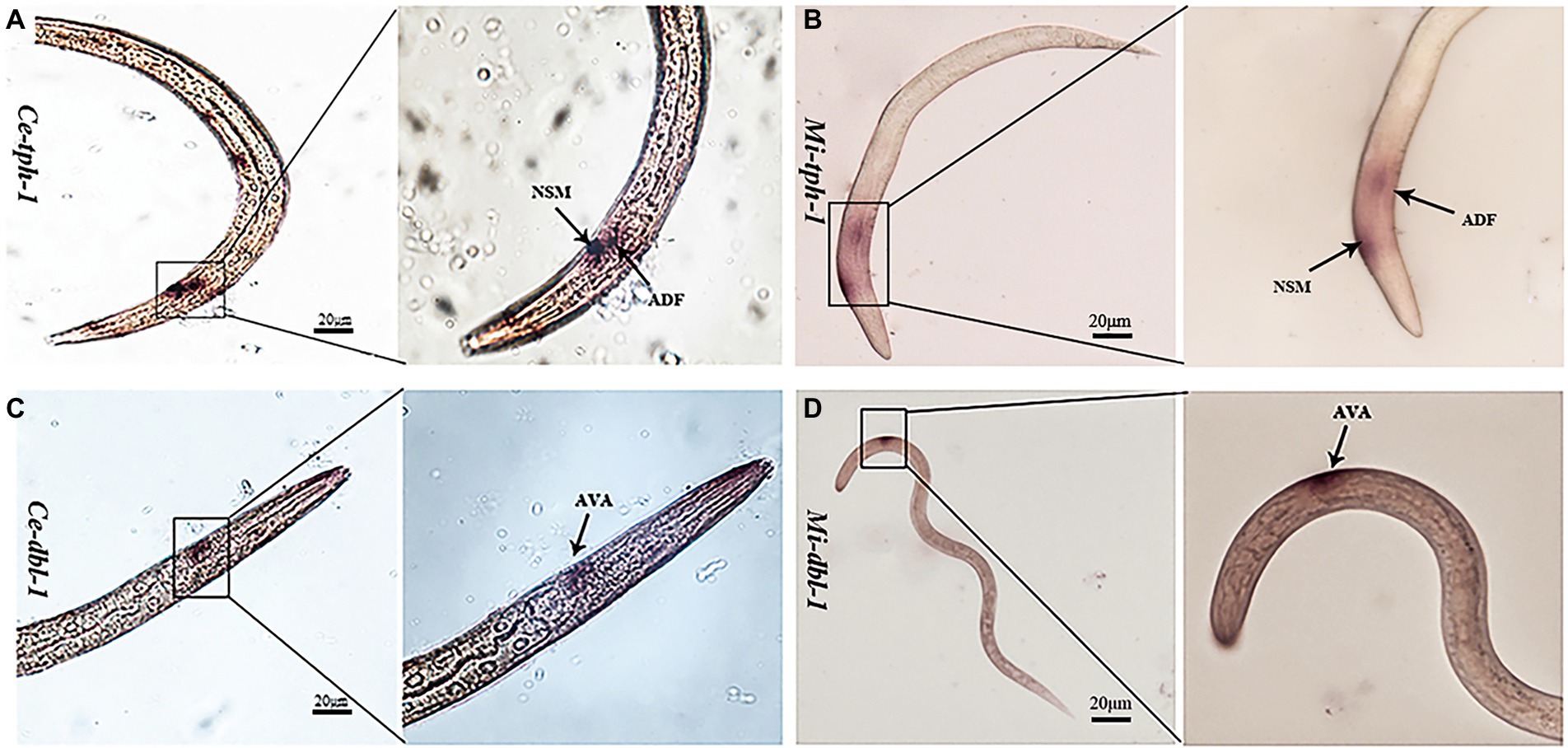
Figure 6. Comparisons of the neuronal localization. In situ hybridization with the DIG-labeled probes of Ce-tph-1 in Caenorhabditis elegans (A) and Mi-tph-1 in Meloidogyne incognita (B) on NSM and ADF neurons, Ce-dbl-1 in C. elegans (C) and Mi-dbl-1 in M. incognita (D) on AVA neurons.
Infection experiments was designed to investigate whether the repulsive response affects the colonization of PPNs within host plants or alters the efficiency of biocontrol bacteria (Figure 7A). Treatment of either B. nematocida B16 or B. thuringiensis Bt79 significantly decreased the number of nematodes inside tomato roots 48 h (Figure 7B). Statistical analysis to 10 samples in each treatment suggested that the average numbers of nematodes in the root tips were 1.30 ± 1.34 and 2.07 ± 1.62 in the groups treated with B. nematocida B16 and B. thuringiensis Bt79, which were 4.42 and 2.78 times lower than that of the control (5.75 ± 2.44), respectively (Figure 7C).
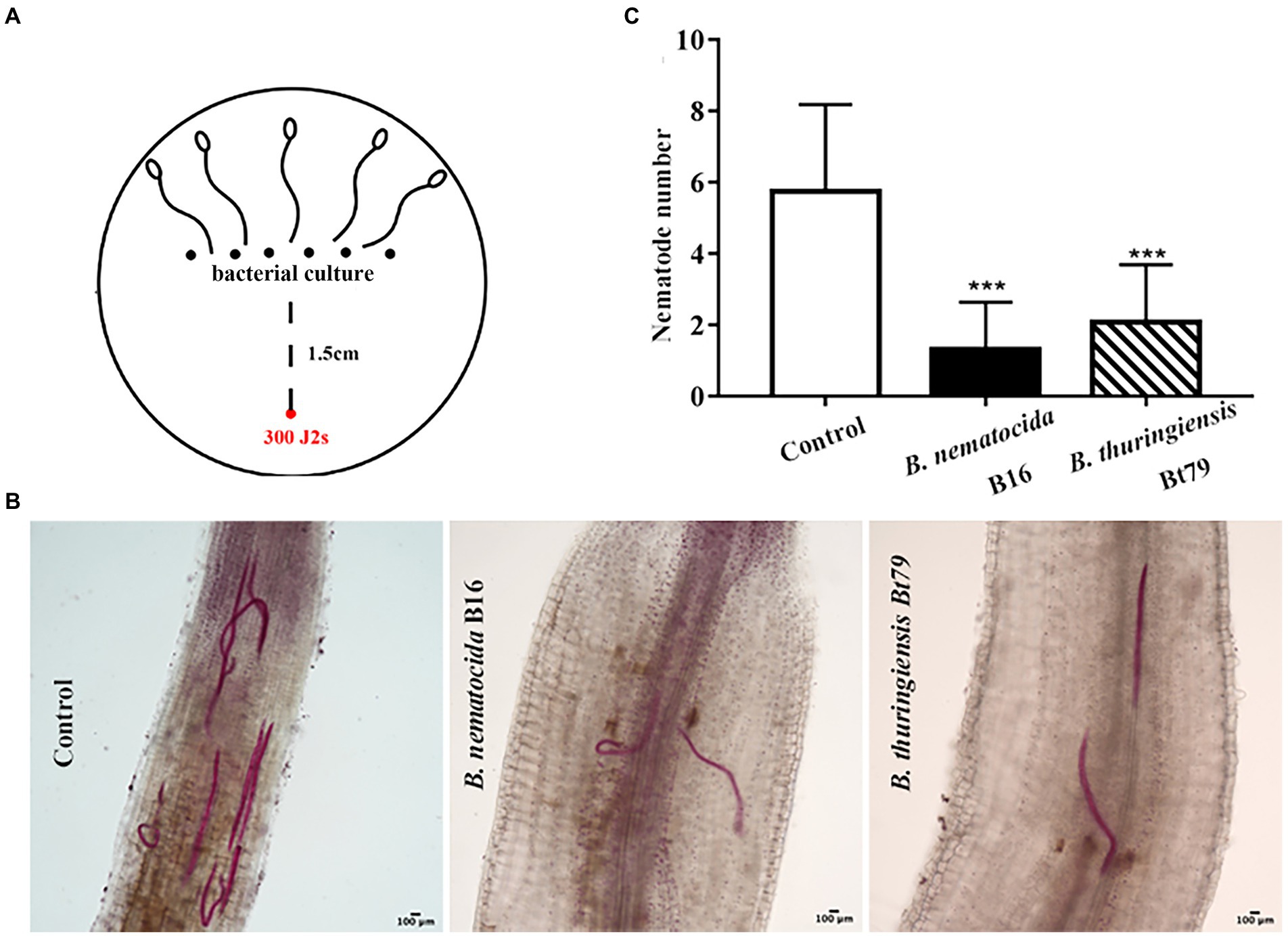
Figure 7. Induction of the repulsive response of Meloidogyne incognita by biocontrol bacteria suppresses the invasion of M. incognita into host plants. (A) Schematic diagram of the assay for evaluating the effects of repulsive response on the invasion of M. incognita second-stage juveniles (J2s) into tomato roots. (B) Stained nematodes that have invaded tomato roots after 48 h. (C) The numbers of nematodes inside tomato roots (n = 10). ***p < 0.001.
In natural ecosystems, rhizosphere microbiota participates in interactions between nematodes and plants. The symbiotic bacteria of plants, such as Bacterillus and Pseudomonas, synthesize hundreds of compounds to either stimulate plant growth or enhance the resistance to nematodes (Santoyo, 2021). Additionally, a few non-virulence bacteria, acting as the food of bacterivorous nematodes, have developed a strategy to mobilize their ally of nematode-trapping fungi to help them kill nematodes (Wang et al., 2014).
The biocontrol bacteria, B. nematocidal and B. thuringiensis, can induce the repulsive response of M. incognita J2s after 6 h of exposure to inhibit their invasion of host plants. But what’s interesting is that the AI index for B. thuringiensis Bt79 was once decreased at 12 h, and then continued to increase at 24 h. This change trend seems different from that of B. nematocidal B16, which retains a constant increase at all time points. It may be due to the differential volatile compounds between B. nematocidal and B. thuringiensis.
The pathogenic bacterium P. aeruginosa PA14 can stimulate learning-associated avoidance in C. elegans after 4 h of exposure, and this might increase their survival rate during infection (Zhang et al., 2005). A few molecular mechanisms have been suggested to underlie these behaviors. First, serotonin from ADF chemosensory neurons is increased by transcriptional and post-transcriptional mechanisms. After binding to its receptor MOD-1, a serotonin-gated chloride channel localized to the sensory interneurons AIY and AIZ, serotonin promotes the aversive learning of C. elegans against P. aeruginosa PA14 (Zhang et al., 2005; Melo and Ruvkun, 2012). The module DBL-1/SMA-6 in the TGF-β signaling pathway is also thought to be required for the lawn-escaping behavior of C. elegans. After aversive training with P. aeruginosa PA14, the repressed activity of AVA interneurons triggers an increase in the expression of the ligand DBL-1, which binds to the type I TGF-β receptor SMA-6 on ASI neurons, promotes olfactory plasticity, and induces a strong aversive response (Zhang and Zhang, 2012). In light of differences in the genomic sequences between M. incognita and C. elegans and limitations in current research techniques, the genes, molecular pathways, and neural circuits involved in the repulsive response of M. incognita require further study.
Sequence alignment of the genes involved in the above two classical pathways between C. elegans and M. incognita revealed a similarity >50% for genes in the serotonin pathway and 30%–42% for genes in the TGF-β signaling pathway. Silencing of the key genes in both pathways decreased the AIs of M. incognita J2 against biocontrol bacteria, which confirmed that they played similar roles in the repulsive response of M. incognita. Ca2+ imaging with Fura-2-AM and ISH showed that both sets of genes functioned in the same neural circuits in M. incognita as in C. elegans. However, the contributions of these signaling pathways vary to the repulsive response induced by the different biocontrol bacteria. Additionally, a novel role of serotonin pathway has also been suggested to modulate the muscle tone of M. incognita, as decreased body swing amplitude and more flattened traces were observed following RNAi of Mi-tph-1 and Mi-mod-1, and such changes are absent in C. elegans. We speculate that this might be explained by the fact that the reduced genome of M. incognita possesses genes that are responsible for more biological activities.
The repulsive response of M. incognita induced by biocontrol bacteria likely affects interspecific interactions among bacteria, PPNs, and host plants. In agriculture, the control of PPNs is fairly difficult because the nematodes generally inhabit soil and attack the underground parts of plants. Chemical nematicides have long been thought to be the most effective approach for controlling PPNs. Due to their high toxicity, as well as the ease with which they form residues and can be abused, the use of chemical nematicides can have deleterious effects on the environment and cause the serious pollution for agricultural products. Therefore, biological control, which is a more environmentally friendly alternative that exploits the interactions between nematode-antagonistic microorganisms and their hosts, provides a more robust complementary approach. Nematode-antagonistic microorganisms employ different strategies to alleviate PPN infections in plants. Theoretical studies of their interactions are of great value to improve the efficacy of biocontrol by enhancing the virulence factors of biocontrol microorganisms and weakening the defensive response of PPNs.
Since semiochemicals have been successfully used to control pests (Park et al., 2003; Lee et al., 2004), and the infection efficiencies of M. incognita can be greatly reduced via induction of the repulsive response by biocontrol bacteria, we believe the results of our study provide new insights into the biocontrol control of PPNs.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
XH conceived and designed the study. YZ and QZ conducted experiments, searched the literature and wrote the manuscript. XH, CZ, and KZ revised the manuscript. XH and CZ supplied funding. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant number 32170184, 32060632, and U1802233) and the Department of Science and Technology of Yunnan Province (grant number 2019FA046).
We would like to acknowledge the State Key Lab for Conservation and Utilization of Bio-Resources for supplying the experimental platform.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abad, P., Gouzy, J., Aury, J. M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Antil, S., Kumar, R., Pathak, D. V., Kumar, A., Panwar, A., and Kumari, A. (2022). Plant growth-promoting rhizobacteria – Bacillus cereus KMT-5 and B. megaterium KMT-8 effectively suppressed Meloidogyne javanica infection. Appl. Soil Ecol. 174:104419. doi: 10.1016/j.apsoil.2022.104419
Bakhetia, M., Charlton, W., Atkinson, H. J., and Mc Pherson, M. J. (2005). RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol. Plant Microbe. 18, 1099–1106. doi: 10.1094/MPMI-18-1099
Cabrera, J., Barcala, M., Fenoll, C., and Escobar, C. (2014). Transcriptomic signatures of transfer cells in early developing nematode feeding cells of Arabidopsis focused on auxin and ethylene signaling. Front. Plant Sci. 5:107. doi: 10.3389/fpls.2014.00107
Dash, M., Dutta, T. K., Phani, V., Papolu, P. K., Shivakumara, T. N., and Rao, U. (2017). RNAi-mediated disruption of neuropeptide genes, nlp-3 and nlp-12, cause multiple behavioral defects in Meloidogyne incognita. Biochem. Biophys. Res. Commun. 490, 933–940. doi: 10.1016/j.bbrc.2017.06.143
Dong, L. L., Li, X. L., Huang, L., Gao, Y., Zhong, L. N., Zheng, Y. Y., et al. (2014). Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts mi-flp-18 expression to block infection. J. Exp. Bot. 65, 131–141. doi: 10.1093/jxb/ert356
Dong, L., Li, X., Huang, C., Lu, Q., Li, B., Yao, Y., et al. (2018). Reduced Meloidogyne incognita infection of tomato in the presence of castor and the involvement of fatty acids. Sci. Hortic. 237, 169–175. doi: 10.1016/j.scienta.2018.03.066
Fudali, S. L., Wang, C. L., and Williamson, V. M. (2013). Ethylene signaling pathway modulates attractiveness of host roots to the root-knot nematode Meloidogyne hapla. Mol. Plant Microbe In. 26, 75–86. doi: 10.1094/mpmi-05-12-0107-r
Gowda, M. T., Meena, B. R., Krishnan, N., Manjunath, M., Sellaperumal, C., Rai, A. B., et al. (2022). Antimicrobial peptides producing native bacillus spp. for the management of root-knot nematode Meloidogyne incognita infecting okra (Abelmoschus esculentus L. Moench). Biol. Control 171:104951. doi: 10.1016/j.biocontrol.2022.104951
Kimber, M. J., Fleming, C. C., Prior, A., Jones, J. T., Halton, D. W., and Maule, A. G. (2002). Localisation of Globodera pallid FMRFamide-related peptide encoding genes using in situ hybridisation. Int. J. Parasitol. 32, 1095–1105. doi: 10.1016/s0020-7519(02)00084-x
Kirwa, H. K., Murungi, L. K., Beck, J. J., and Torto, B. (2018). Elicitation of differential responses in the root-knot nematode Meloidogyne incognita to tomato root exudate cytokinin, flavonoids, and alkaloids. J. Agric. Food Chem. 66, 11291–11300. doi: 10.1021/acs.jafc.8b05101
Kuang, M. S., Liu, T. T., Wu, H. B., Lan, H. P., Wen, Y. X., Wu, H. B., et al. (2020). Constituents leached by tomato seeds regulate the behavior of root-knot nematodes and their antifungal effects against seed-borne fungi. J. Agric. Food Chem. 68, 9061–9069. doi: 10.1021/acs.jafc.0c01797
Lee, B. H., Annis, P. C., Tumaalii, F., and Choi, W. S. (2004). Fumigant toxicity of essential oils from the Myrtaceae family and 1, 8-cineole against 3 major stored-grain insects. J. Sto. Pro. Res. 40, 553–564. doi: 10.1016/j.jspr.2003.09.001
Makhubu, F. N., Khosa, M. C., and McGaw, L. J. (2021). South African plants with nematicidal activity against root-knot nematodes: a review. S. Afr. J. Bot. 139, 183–191. doi: 10.1016/j.sajb.2021.02.010
Melo, J., and Ruvkun, G. (2012). Inactivation of conserved C. elegans genes engages pathogen-and xenobiotic-associated defenses. Cell 149, 452–466. doi: 10.1016/j.cell.2012.02.050
Oota, M., Tsai, A. Y., Aoki, D., Matsushita, Y., Toyoda, S., Fukushima, K., et al. (2020). Identification of naturally occurring polyamines as root-knot nematode attractants. Mol. Plant 13, 658–665. doi: 10.1016/j.molp.2019.12.010
Opperman, C. H., Bird, D. M., Williamson, V. M., Rokhsar, D. S., Burke, M., Cohn, J., et al. (2008). Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. U. S. A. 105, 14802–14807. doi: 10.1073/pnas.0805946105
Park, I. K., Lee, S. G., Choi, D. H., Park, J. D., and Ahn, Y. J. (2003). Insecticidal activities of constituents identified in the from leaves of Chamaecyparis obtusa against Callosobruchus chinensis (L.) and Sitophilus oryzae (L.). J. Sto. Pro. Res. 39, 375–384. doi: 10.1016/S0022-474X(02)00030-9
Santoyo, G. (2021). How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. doi: 10.1016/j.jare.2021.11.020 (in press).
Shivakumara, T. N., Dutta, T. K., and Rao, U. (2018). A novel in vitro chemotaxis bioassay to assess the response of Meloidogyne incognita towards various test compounds. J. Nematol. 50, 487–494. doi: 10.21307/jofnem-2018-047
Shivers, R. P., Kooistra, T., Chu, S. W., Pagano, D. J., and Kim, D. H. (2009). Tissue-specific activities of an immune signaling module regulate physical response to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6, 321–330. doi: 10.1016/j.chom.2009.09.001
Shuang, S. M., Qiao, X. N., Wang, D. D., Fan, L., Lu, D. T., Mu, L. X., et al. (2010). Analytical behavior of Fura-2 and its determination of [Ca2+]i in lymphocytes treated with cefotaxime. J. Meas. Sci. Instru. 1, 395–400. doi: 10.3969/j.issn.1674-8042.2010.04.21
Swiecicka, M., Filipecki, M., Lont, D., Vliet, J. V., Qin, L., Goverse, A., et al. (2009). Dynamics in the tomato root transcriptome on infection with the potato cyst nematode Globodera rostochiensis. Mol. Plant Pathol. 10, 487–500. doi: 10.1111/j.1364-3703.2009.00550.x
Wang, J. X., Ding, Z. J., Bian, J., Bo, T. T., and Liu, Y. J. (2021a). Chemotaxis response of Meloidogyne incognita to volatiles and organic acids from root exudates. Rhizosphere 17:100320. doi: 10.1016/j.rhisph.2021.100320
Wang, S., Fan, H. Y., Zhao, D., Zhu, X. F., Wang, Y. Y., Liu, X. Y., et al. (2021b). Multifunctional efficacy of the nodule endophyte Pseudomonas fragi in stimulating tomato immune response against Meloidogyne incognita. Biol. Control 164:104773. doi: 10.1016/j.biocontrol.2021.104773
Wang, X., Li, G. H., Zou, C. G., Ji, X. L., Liu, T., Zhao, P. J., et al. (2014). Bacteria can mobilize nematode-trapping fungi to kill nematodes. Nat. Commun. 5:5776. doi: 10.1038/ncomms6776
Wubben, M. J. E., Rodermel, S. R., and Baum, T. J. (2004). Mutation of a UDP-glucose-4-epimerase alters nematode susceptibility and ethylene responses in Arabidopsis roots. Plant J. 40, 712–724. doi: 10.1111/j.1365-313x.2004.02257.x
Yang, G. D., Zhou, B. L., Zhang, X. Y., Zhang, Z. J., Wu, Y. Y., Zhang, Y. M., et al. (2016). Effects of tomato root exudates on Meloidogyne incognita. PLoS One 11:e0154675. doi: 10.1371/journal.pone.0154675
Zhang, Y., Hang, L., and Bargmann, C. I. (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184. doi: 10.1038/nature04216
Zhang, X., and Zhang, Y. (2012). DBL-1, a TGF-β, is essential for Caenorhabditis elegans aversive olfactory learning. Proc. Natl. Acad. Sci. U. S. A. 109, 17081–17086. doi: 10.1073/pnas.1205982109
Zhou, Q. Y., Zhu, M., Huang, H., and Huang, X. W. (2019). Chemosensory system of plant parasitic nematodes. Sci. Sin. Vitae 49, 828–838. doi: 10.1360/SSV-2019-0092
Keywords: plant parasitic nematodes, Meloidogyne incognita, biocontrol bacteria, repulsive response, host plant, interspecific interactions
Citation: Zhao Y, Zhou Q, Zou C, Zhang K and Huang X (2022) Repulsive response of Meloidogyne incognita induced by biocontrol bacteria and its effect on interspecific interactions. Front. Microbiol. 13:994941. doi: 10.3389/fmicb.2022.994941
Received: 15 July 2022; Accepted: 22 August 2022;
Published: 15 September 2022.
Edited by:
Ryan Kessens, Corteva Agriscience™, United StatesReviewed by:
Hari S. Gaur, Sharda University, IndiaCopyright © 2022 Zhao, Zhou, Zou, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowei Huang, eHdodWFuZ0B5bnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.