
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 September 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.993721
This article is part of the Research Topic Trends of Microbial Technologies in Rehabilitation of Contaminated Environments View all 6 articles
Diquat is used in agricultural contexts to control the growth of broadleaf and grassy weeds in both terrestrial and aquatic areas. Diquat can be readily absorbed by the soil and can remain therein for extended periods of time, altering the local microenvironment. In this study, the Meyerozyma guilliermondii Wyslmt yeast strain, which has the capacity to degrade Diquat, was isolated from soil exposed to long-term Diquat treatment. Over a 7-day incubation period, this strain was able to remove 42.51% of available Diquat (100 mg/L). RNA-Seq was performed to assess changes in gene expression in this yeast strain over the course of Diquat degradation, revealing 63 and 151 upregulated and downregulated genes, respectively. KEGG pathway enrichment analysis revealed these genes to be most highly enriched in the carbohydrate metabolism pathway. Through functional annotation and gene expression analyses, we identified seven genes were predicted to be involved in Diquat biodegradation. Results of qRT-PCR assays indicated that the relative mRNA expression levels of these seven genes were significantly higher relative to the control group. Together these analyses led to the identification of DN676 as a candidate Diquat-degrading gene. When a pET-DN676 vector was expressed in E. coli BL21, this strain was able to remove 12.49% of provided Diquat (100 mg/L) over the course of a 7-day incubation. These results thus confirmed that the DN676 gene can promote Diquat degradation, with these studies having yielded an engineered BL21-pET-DN676 bacterial strain capable of degrading Diquat.
Diquat (1,1′-ethylene-2,2′-bipyridylium) is a non-selective bipyridylium herbicide (Wang et al., 2017). It is widely used to impair photosynthetic activity and thereby kill exposed plants (Yuan et al., 2020). In agricultural contexts, Diquat is applied to control the growth of broadleaf and grassy weeds in both aquatic and terrestrial non-crop regions (Ducrot et al., 2010). Diquat is immediately inactivated upon reaching the soil via its absorption by clay minerals and organic matter. In this state, it is protected from leaching and can accumulate in the soil system. As it is highly water-soluble, establishing effective and secure procedures for purifying soil and water polluted by Diquat is crucial.
Microbial remediation is a process whereby microbes convert environmental toxicants to yield less toxic byproducts (Chen et al., 2015; Liu et al., 2015; Cycon and Piotrowska-Seget, 2016), offering a promising approach to removing chemical pollutants from contaminated soil and water biomes. Relative to traditional chemical or physiochemical approaches, microbial remediation is a more efficient means of controlling or eliminating environmental pollution (Chen et al., 2014; Arora et al., 2018; Yang et al., 2018). Both Diquat and paraquat are bipyridine herbicides, and the limited number of studies exploring their degradation to date have largely focused on paraquat. It has been established that a number of fungus species are effective paraquat degraders. For instance, T. versicolor ECS-79, T. pavonia ECS-67, and H. dispersum ECS-705 can degrade paraquat at similar 55, 55, and 71% rates, respectively (Camachomorales et al., 2017). Funderburk (1969) found that L. starkeyi was able to degrade both Diquat and paraquat. Furthermore, a bacterial consortium capable of achieving 97% paraquat (100 mg/L) degradation (Li et al., 2017), while Lipomyces yeast species were able to degrade 100% of available paraquat (27 mg/L) over a 3-day culture period in liquid medium (Hata et al., 1986). These prior studies concentrated on isolating and characterizing microbes that metabolized paraquat and Diquat. However, studies regarding the metabolic genes that are involved in the degradation of Diquat are limited. In the present study, we identified the Diquat-degrading Meyerozyma guilliermondii Wyslmt yeast strain and found that 42.51% of provided Diquat (100 mg/L) was degraded by this strain over the course of a 7-day incubation. In order to discover the genes facilitating Diquat degradation, we further sequenced the genome of this Wyslmt strain (deposited in GenBank under the name “Wyslmt” with the accession number MZ520358; Bioproject accession: PRJNA809846). A novel degradation gene, DN676, was discovered to be involved in Diquat degradation, and its expression and degradation phenotype were confirmed. As such, the DN676 gene may represent a promising candidate gene for use in future herbicide degradation efforts.
Soil samples were collected from soil that had been exposed to Diquat for extended periods of time and were sieved using a 5.0 mm mesh to remove debris and loose stones. Samples were transferred into plastic bags and stored at 4°C.
Diquat (99.9%) was purchased from the Putian Genesis Biotechnology co., Ltd. (Beijing, China). Other chemicals used in this study were of analytical grade, and those used for HPLC analysis were of HPLC grade. Liquid enrichment medium (EM) used for this study was composed of a 0.5 g MgSO4⋅7H2O, 1.0 g KH2PO4, and 5.0 g peptone per liter. Potato dextrose broth (PDB) medium was composed of 200.0 g potato and 20.0 g glucose per liter. Luria-Bertani (LB) broth was composed of 10.0 g peptone, 5.0 g yeast powder, and 5.0 g NaCl per liter. Diquat was added to the medium at appropriate concentrations to yield Diquat-supplemented EM, PDB, or LB (DEM, DPDB, and DLB, respectively). Medium was sterilized via autoclaving for 30 min at 121°C.
To enrich for Diquat-degrading microbes, 5 g samples of soil were added to 100 mL of DEM containing 500 mg/L Diquat in a 250 mL Erlenmeyer flask, and samples were then incubated for 5 days with constant shaking (150 rpm) at 30°C under aerobic conditions. Next, 10 mL of the enriched culture was transferred into fresh DEM containing 1,000 mg/L Diquat, and this process was repeated three times to a final Diquat concentration of 2,500 mg/L. After being serially diluted, these enriched cultures were subsequently plated onto PDB agar plates with Diquat (2,500 mg/L) added and cultivated at 28°C. Then, to separate pure cultures, morphologically distinct colonies were selected and streaked onto PDB agar plates. One fungal isolate, known as strain Wyslmt, showed high activity and was thus chosen for more analysis. All strains were kept on DPDB containing 2,500 mg/L of Diquat, and they all were kept frozen in 30% glycerol stocks at −80°C.
A combination of conventional biochemical methods and 26S rDNA sequencing was used to identify the Wyslmt strain. Total genomic DNA was isolated from this fungi with a Yeast Ezup Column Yeast Genomic DNA Extraction Kit (Sangon Biotech Co., Ltd., Shanghai, China), and sequencing was performed with the NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) forward primer and the NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) reverse primer. PCR settings were as follows: 94°C for 4 min; 30 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 60 s; 72°C for 10 min. The resultant amplified products were separated via 1% agarose gel electrophoresis and purified with a SanPrep column DNAJ gel recovery kit (Sangon Biotech), after which sequencing was performed by Sangon Biotech Co., Ltd. (Shanghai, China).
After sequencing, 26S rDNA sequences were aligned and compared to those in the National Center for Biotechnology Information (NCBI) database using the BLAST program. MEGA version 6.0 was used for phylogenetic and molecular evolutionary analysis. The phylogenetic tree was built using the neighbor-joining method, and bootstrapping was done for 1,000 repetitions.
Dichloromethane was used three times to extract Diquat. The mixed organic extracts were subsequently concentrated, dried using an evaporator, and the volume was adjusted using methanol of the High Performance Liquid Chromatography (HPLC) grade. A 0.22 μm nylon filter was used to filter the aqueous samples after extraction in order to prepare them for HPLC analysis. The concentration of Diquat in each sample was quantified via HPLC (Ultimate 3000). For Diquat detection analyses, HPLC was performed using a variable wavelength UV detector set to 308 nm and fitted with a reverse-phase C18 column (4.6 × 250 mm, 5 μm) at a flow rate of 1.0 mL/min (acetonitrile/water = 40/60, v/v) and a column temperature of 30°C. All injection volumes were 10 μL.
The initial and final concentrations of Diquat were measured via HPLC, and the percentage of Diquat removed was calculated. The percentage of Diquat degraded was calculated as follows:
where X is the degradation rate of Diquat, CCK is the original concentration of Diquat (mg/L), and CX is the final concentration of Diquat (mg/L).
Wyslmt strain yeast were cultured to the logarithmic phase of growth (OD600 = 0.6) to prepare a stock solution from which a 1% inoculum was added into 100 mL of DPDB containing Diquat (100 mg/L). The concentration of Diquat and yeast growth were measured on days 1, 3, 5, and 7 of culture at 28°C with constant agitation (180 rpm). Yeast growth was monitored by measuring the absorbance of culture supernatants at 600 nm using a TU-1901 spectrophotometer (Beijing Purkinje General Instrument Co., Ltd., China). The concentration of Diquat was measured via HPLC. Experiments were repeated in triplicate, and control solutions were prepared without any starting inoculum.
For RNA-Seq analyses, a 1% Wyslmt yeast inoculum was added to 100 mL of DPDB (2,500 mg/L), with a sample without any Diquat being prepared as a control. Both treatments were prepared in triplicate, and were incubated for 24 h at 28°C with constant shaking (180 rpm) (Zhang et al., 2019). Samples were then centrifuged for 10 min (4°C) at 8,000 × g, snap-frozen with liquid nitrogen, and sent to Sangon Biological Co., Ltd., for RNA-Seq analysis. Briefly, total RNA was extracted from these yeast, mRNA was enriched from these extracts, and the purified mRNA was fragmented and utilized as a template for cDNA synthesis. After preparation, cDNA libraries were sequenced with an Illumina HiSeq device (Illumina, CA, United States).
Base calling was used to turn the raw image data from Illumina sequencing into sequence data. FastQC (version 0.11.2) was used to evaluate the quality of sequenced data. Raw reads were filtered using Trimmomatic (version 0.36) through the several steps: (1) The removal of sequences with N bases; (2) The removal of adaptor sequences when present in detected reads; (3) The removal of low-quality bases from reads in a 3′ to 5′ direction (Q < 20); (4) The removal of low-quality bases from reads in a 5′ to 3′ direction (Q < 20); (5) The use of a sliding window method to remove base values < 20 from read tails (window size: 5 bp); (6) Removing reads with reads length less than 35 nt and paired reads. The remaining clean data were used for further analysis.
HISAT2 (version 2.1.0) was used to align the clean reads with the reference genome. Blast2GO (Götz et al., 2008) was used to annotate gene functions in the Gene Ontology (GO) database (Harris et al., 2004), and the euKaryotic Orthologous Groups (KOG) database was used to phylogenetically classify the encoded proteins, and the associated metabolic pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000).
StringTie (version 1.3.3b) was used to assemble transcripts and GffCompare (version 0.10.1) was used to align these with known gene modules to detect novel transcript regions. To identified significant DEGs, the following settings were used: q-value < 0.05, Log2| FoldChange| > 1 (Reiner et al., 2003; Trapnell et al., 2011, 2013).
An qRT-PCR approach was employed to confirm the differential expression of key genes of interest identified by RNA-Seq in the Wyslmt strain. These qRT-PCR assays were performed using a 4S Red Plus Nucleic Acid Stain (A606695, Sangon Biotech, Shanghai, China) with the primers being listed in Table 1. Briefly, control Wyslmt cells and cells cultured in the presence of Diquat (2,500 mg/L) were grown for 24 h at 28°C and 180 rpm. Total RNA was extracted from these cells using the same protocols employed prior to RNA-seq analyses. Random primers and Maxima Reverse Transcriptase (EP0743, Thermo Scientific, Shanghai, China) were then used to prepare cDNA, which was used as a template for qRT-PCR. Three biological replicates were analyzed per sample, and the 2–ΔΔCT method was used to assess relative gene expression (Cheng et al., 2018).
Following the cloning and expression testing of the 10 candidate genes indicated above, the gene DN676 c0 g3—also known as DN676—was chosen for further study. The DN676 gene was combined with the expression vector pET-4T-1 and digested with BamHI and SmaI to create the recombinant plasmid pET-DN676. This plasmid was then transformed into capable E. coli BL21 (DE3) cells via heat shock (37°C, 200 rpm). Isopropy-D-thiogalactoside (IPTG) was added at a final concentration of 0.5 mM at 20 and 37°C when the OD600 value reached 0.6. Cells were collected and centrifuged at 4,000 rpm for 10 min after being cultured for 2 h. The protein was then added to the buffer for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) separation after the supernatants had been removed.
To confirm the relationship between this gene and Diquat degradation, the BL21-pET strain served as a control group, while the BL21-pET-DN676 group was the treatment group. These BL21-pET and BL21-pET-DN676 bacteria were cultured until reaching the logarithmic phase of growth (OD600 = 0.6), at which point a 1% inoculum was transferred into 100 mL of DLB containing Diquat (100 mg/L). The concentration of Diquat and bacterial growth were measured on days 1, 3, 5, and 7 of incubation at 37°C with constant agitation (200 rpm). Bacterial growth was monitored by measuring the absorbance of the culture supernatant at 600 nm using a TU-1901 spectrophotometer (Beijing Purkinje General Instrument Co., Ltd., China). The concentration of Diquat was measured via HPLC. Experiments were repeated in triplicate.
DEGseq (version 1.26.0) was used to detect DEGs (q-value < 0.05, Log2| FoldChange| > 1) in RNA-seq studies between the control group and the treatment group. ANOVA (SPSS version 21.0) were used to analyze KEGG enrichment data, with P < 0.05 being considered the threshold of significant pathway enrichment.
The Wyslmt yeast strain, which exhibited robust Diquat degrading activity, was isolated and selected for further investigation. Physiological and morphological characterization indicated that Wyslmt cells were ovoid and without flagella, consistent with their characterization as a yeast strain (Figure 1A).
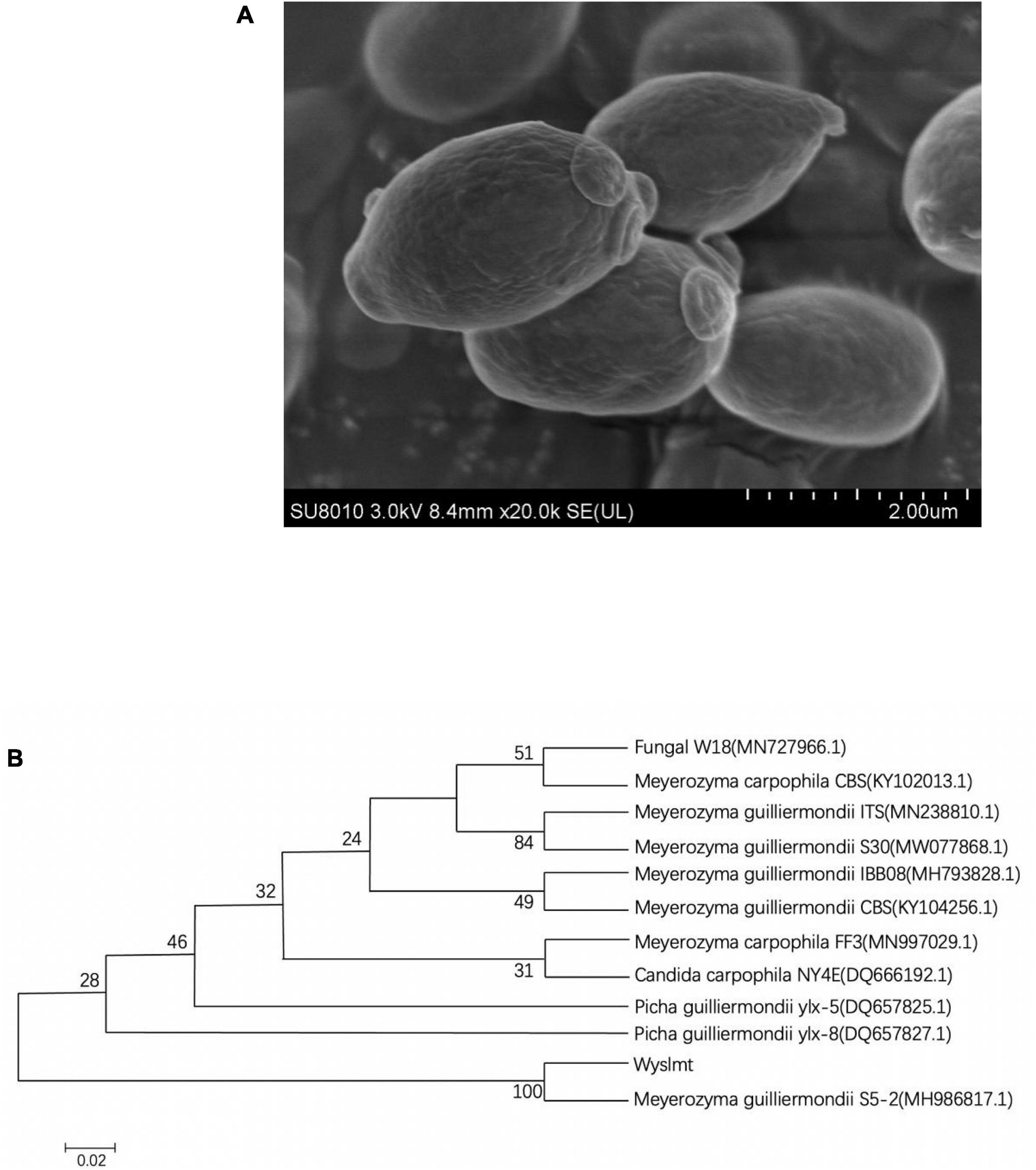
Figure 1. Scanning electron micrograph of the Wyslmt strain (A). Phylogenetic tree based on the 26S rDNA gene sequences from the Wyslmt yeast strain (B). The neighbor-joining method was used to construct a phylogenetic tree (bootstrap number = 1,000), with the scale bar denoting 0.02 substitutions per nucleotide position.
The 26S rDNA gene isolated from Wyslmt yeast was 579 bp in length, and a comparison of this sequence to other sequences available in the GenBank database indicated it to be 100% homologous to sequences form Meyerozyma guilliermondii S5-2. A phylogenetic tree was then constructed via the neighbor-joining approach. In the resultant tree (Figure 1B), this Wyslmt strain clustered with Meyerozyma guilliermondii.
Next, we assessed the ability of this Wyslmt strain to degrade Diquat. Samples collected from the culture medium were analyzed for growth at 600 nm via ultraviolet spectrophotometry and also subjected to HPLC analyses. These yeast cells grew in accordance with a typical growth curve, and the concentration of Diquat in the culture medium decreased throughout the experimental period (Figure 2). In total, 42.51% of the provided Diquat (100 mg/L) was removed by this strain after a 7-day incubation period.
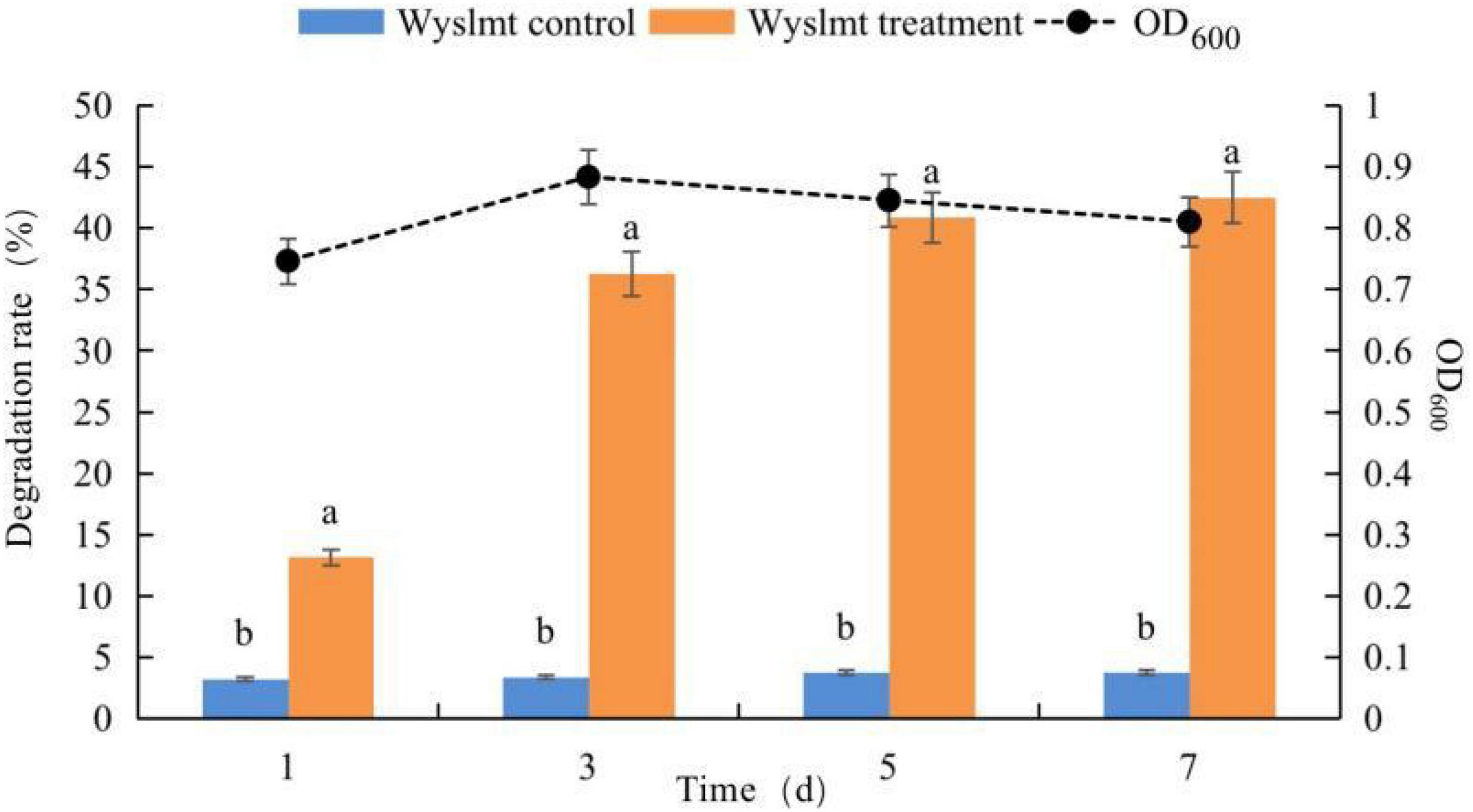
Figure 2. Growth of the Wyslmt strain and associated Diquat degradation. Error bars represent the standard deviation for three replicate samples. Different letters indicate significant differences between treatments (P < 0.05).
In total, 44,132,249 and 47,354,547 raw RNA-Seq reads were obtained from the Diquat-treated and control groups, respectively. After filtering, 42,760,279 and 45,769,503 clean reads of 5,859,601,868 bp (GC content: 46.27%) and 6,320,620,062 bp (GC content: 46.19%) were obtained for the treatment and control groups, respectively. The Q20 and Q30% values for the clean data were 98.53 and 98.48% for the treatment group, respectively, and 96.20 and 96.28% for the control group, respectively. Following the removal of sequences < 20 bp in length, 2,931 non-reductant transcripts were retained. These differentially expressed genes were next subjected to GO and KEGG analyses aimed at elucidating their roles in the context of Diquat biodegradation.
The GO database was used to annotate 2,310 genes of the Wyslmt strain in total (Supplementary Figure 1). In the GO database, a significant number of genes were annotated in 8 out of the 64 groups of GO terms divided in the 3 categories (molecular functions, biological processes, and cellular components) of the gene products, including the binding (1,361 genes assigned, accounting for 58.92% of the 2,310 genes annotated), catalytic activity (1,301, 56.32%), cellular process (1,743, 75.45%), metabolic process (1,527, 66.10%), cell (1,997, 86.45%), cell part (1,995, 86.36%), organelle (1,652, 71.52%), and organelle part (1,124, 48.66%) terms. In contrast, relatively few genes (1–2) were associated with 9 groups of GO terms including the chemoattractant activity, protein tag, channel regulator activity, receptor regulator activity, behavior, biological phase, cell aggregation, cell killing, and extracellular matrix terms.
Using sequence homology as the basis, 1,784 genes in all were mapped onto 24 potential protein categories in the KOG database (Supplementary Figure 2). The greatest numbers of mapped genes were connected with eight gene function classes, including the RNA processing and modification (100 genes assigned, accounting for 5.61% of the 1,784 mapped genes), energy production and conversion (130, 7.29%), amino acid transport and metabolism (134, 7.51%), carbohydrate transport and metabolism (108, 6.05%), translation, ribosomal structure and biogenesis (191, 10.71%), general function prediction only (211, 11.83%), signal transduction mechanisms (105, 5.89%), and intracellular trafficking, secretion, and vesicular transport (113, 6.33%) classes. In contrast, just 0–1 mapped genes were associated with the cell motility class.
In the KEGG database, a total of 895 genes were categorized into 32 metabolic pathways to identify their biological roles (Supplementary Figure 3), with the translation pathway being associated with the greatest number of these genes (154, accounting for 17.21% of the 895 classified genes), followed by 127 genes (14.19%) associated with signal transduction and 124 genes (13.85%) associated with carbohydrate metabolism. Many other metabolic pathways were associated with fewer than 10 genes, including the membrane transport (8), development (9), environmental adaptation (9), and sensory system (5) pathways.
To have a comprehensive understanding of the Wyslmt strain’s transcriptome response to the degradation of Diquat, DEGseq (version 1.26.0) was used to further analyze differences in gene expression profiles between the treatment and control groups. In total, 214 DEGs (q-value < 0.05, Log2| FoldChange| > 1) were identified between the treatment and control groups, of which 63 (29.44%) were upregulated and 151 (70.56%) were downregulated. The upregulated genes may play a positive role in the metabolic processing of Diquat, while the downregulated genes may be associated with the toxicity of Diquat to this Wyslmt strain (Figure 3).
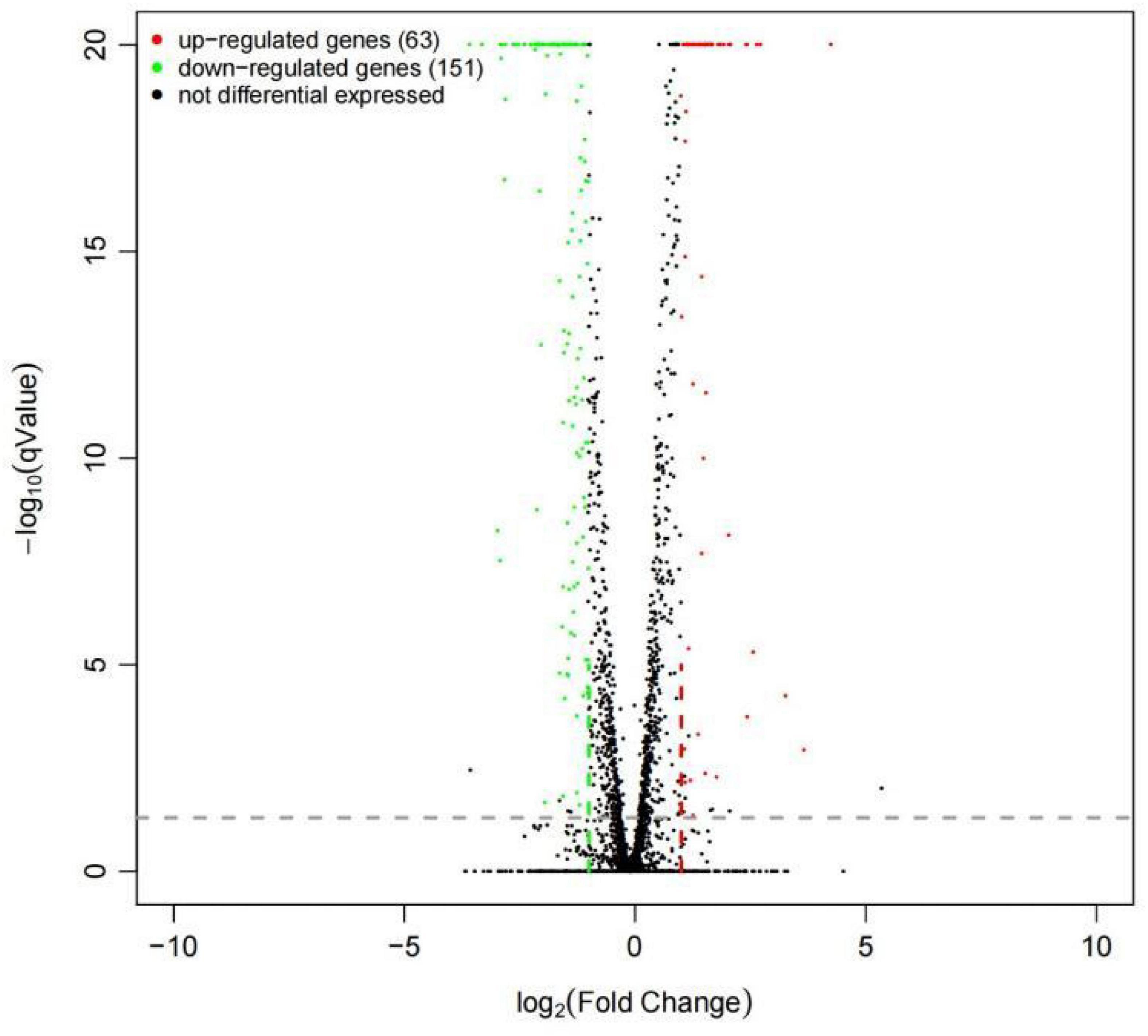
Figure 3. Differential gene expression in Wyslmt cells in the context of Diquat degradation. Individual points correspond to specific genes, with upregulated, downregulated, and non-differentially expressed genes being shown in red, green, and black, respectively.
By mapping the genes onto corresponding GO terms to explore the 214 DEGs’ functions (Figure 4). In total, the mapping of 185 DEGs onto 64 groups of GO terms in three main categories (molecular functions, cell components, and biological processes). Genes differentially expressed in response to Diquat were associated with regulatory and metabolic processes, including the cellular process, metabolic process, cell part, cell, organelle, binding, and catalytic activity GO terms. Most DEGs were associated with the biological process terms metabolic process and cellular process. Specifically, 29 upregulated genes and 90 downregulated genes were related to the metabolic process GO term, while 32 upregulated genes and 105 downregulated genes were associated with the cellular process term. These results demonstrated that the Wyslmt strain’s metabolic functions were impacted by growth in Diquat-containing medium, with the DEGs associated with these GO terms potentially being involved in the process of Diquat degradation. Furthermore, the catalytic activity GO term was associated with 34 upregulated and 58 downregulated genes, while the binding term was associated with 26 upregulated and 82 downregulated genes. These results showed a possible connection between the DEGs linked to these two GO terms and the Diquat-associated catalytic and binding activity in the Wyslmt strain.
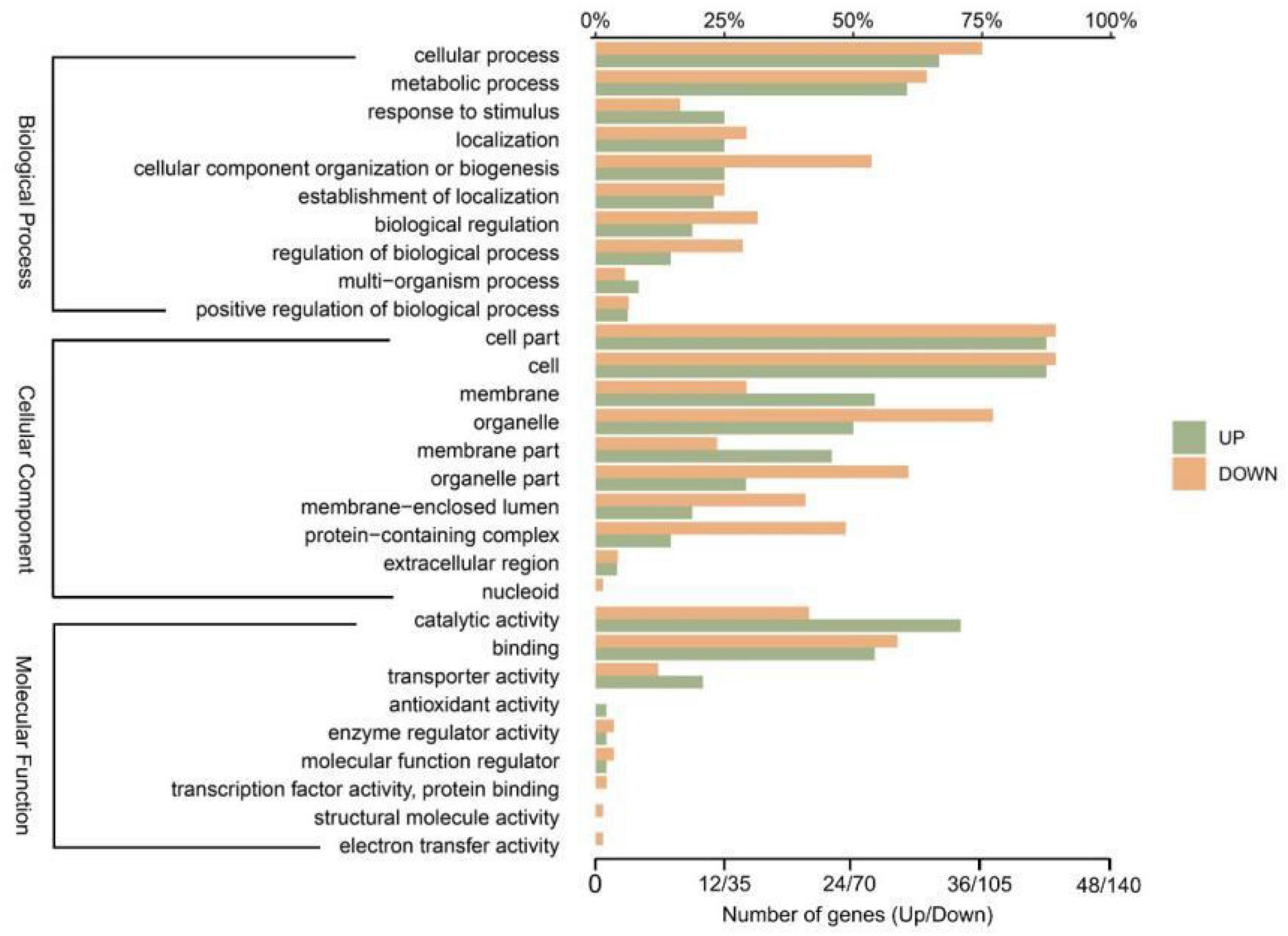
Figure 4. GO annotation histograms for the Wyslmt strain’s DEGs. The precise classification of GO terms is represented by the vertical coordinates. The three brackets show three secondary GO database classifications (biological process, cellular component, and molecular function). The percentage of genes annotated with GO terms in relation to all genes annotated with GO terms is shown by the upper abscissa. The number of genes with GO annotations is shown in the bottom abscissa. Horizontal green and yellow bars, respectively, show the up-regulated and down-regulated genes.
These 214 DEGs were also subjected to a KEGG enrichment analysis aimed at identifying associated metabolic pathways (Figure 5). The results of these analyses revealed that the 32 enriched metabolic pathways (P-value < 0.05) between the treatment and control groups were associated with five categories, including Environmental Information Processing (EIP), Genetic Information Processing (GIP), Cellular Processes (CP), Organismal Systems (OS), and Metabolism (M) (Figure 5). A total of 66 DEGs were associated with 32 KEGG pathways, and most of these DEGs were enriched in the metabolism category, including amino acid metabolism, carbohydrate metabolism, energy metabolism, lipid metabolism, metabolism of cofactors and vitamins, metabolism of other amino acids, and nucleotide metabolism.
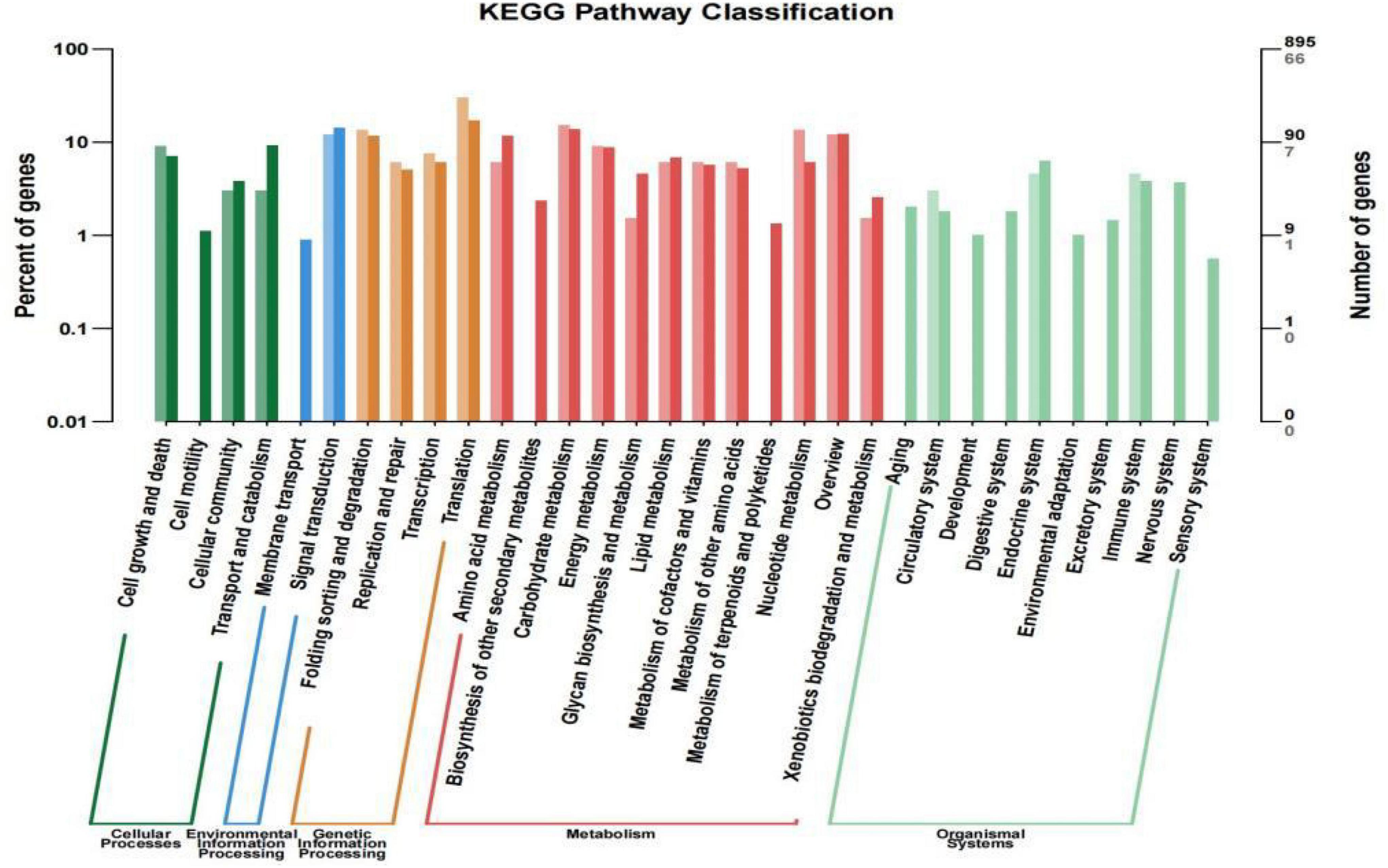
Figure 5. KEGG pathway enrichment analysis results for the Wyslmt strain. Functional categories are shown on the horizontal axis, while the vertical axis corresponds to the number of genes per category (right) and the percentage of the total number of annotated genes (left). Different categories are marked with different colors. Differentially expressed genes and all genes are shown in lighter and darker colors, respectively.
Of these DEGs, 63 upregulated genes may be involved in the process of Diquat degradation. According to annotation results, numerous genes are probably predicted to encode transfer/transport proteins or enzymes that might either directly or indirectly contribute to the metabolism of Diquat before being involved in its degradation. Based on Log2| FoldChange| values for these of these 63 upregulated DEGs, we screened 10 candidate genes that may facilitate Diquat degradation (Table 2). Among these, gene DN573_c0_g4 was identified as allantoicase and associated with purine metabolism, gene DN554_c0_g13 was identified as Choline/ethanolaminephosphotransferase 1 and associated with phosphonate and phosphinate metabolism, glycerophospholipid metabolism, and ether lipid metabolism, and gene DN95_c0_g1 was identified as gamma-glutamyltransferase and associated with cyanoamino acid metabolism, glutathione metabolism, and taurine and hypotaurine metabolism. While the 7 remaining genes were not specifically associated with any metabolic pathways, we cannot rule out the potential that these enzymes are involved in the degradation of Diquat.
Next, the mRNA expression levels of the Diquat degrading genes were measured to validate the transcriptome results via qRT-PCR, as shown in Figure 6. To determine the expression levels of these genes during the biodegradation process, the RNA-Seq results were utilized as a reference. The resultant | Log2FC| values (>1) confirmed the upregulation of the genes in the treatment group identified via RNA-Seq. The Diquat concentration utilized for preparing samples for RNA-Seq and qRT-PCR was 2,500 mg/L, and RNA extraction was performed following a 24 h culture period to further validate RNA-Seq findings. As the Wyslmt strain was capable of degrading Diquat when cultured for 24 h, the genes upregulated during this process probably connected to the Wyslmt strain’s ability to metabolize Diquat or to increase its tolerance to Diquat. Overall, the mRNA expression levels for the seven genes under investigation are considerably greater in the treatment group than in the control group (DN678_c2_g13, DN678_c2_g10, DN95_c0_g1, DN1001_c0_g1, DN573_c0_g4, DN554_c0_g13, and DN676_c0_g3). Accordingly, we hypothesize that these seven genes may be crucial in the Wyslmt strain’s ability to degrade Diquat, in line with our above findings.
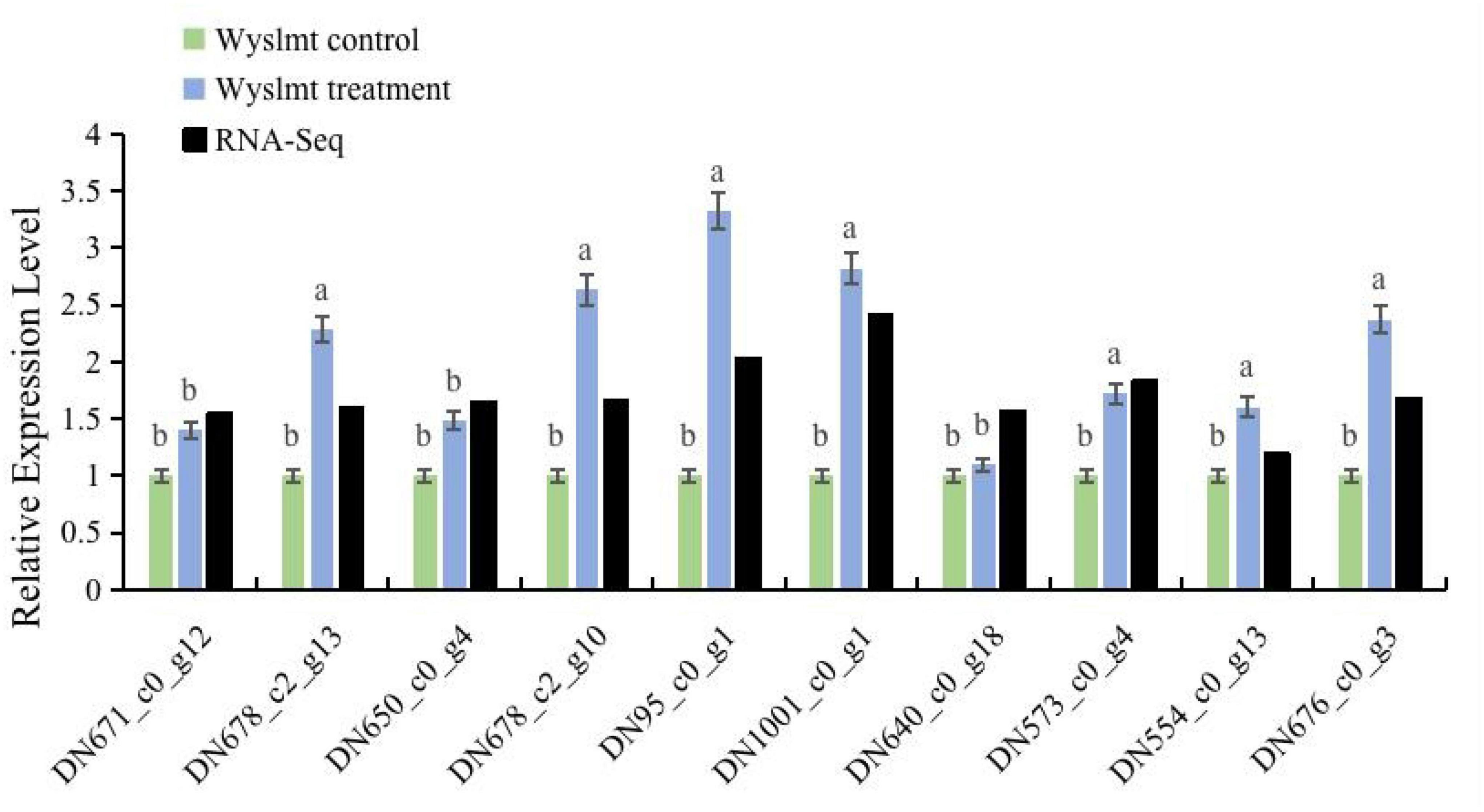
Figure 6. The relative mRNA expression levels of DEGs in both the treatment and control groups. Black bars represent log2| FoldChange| (treatment group/control group) values from RNA-seq analyses. Error bars represent the standard deviation for three replicate samples. Different letters indicate significant differences between treatments (P < 0.05).
Other qualified candidates were cloned and found to not increase Diquat degradation. The functions of this gene were predicted using the https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi website, which identified DN676 as a member of the Pleiotropic Drug Resistance (PDR) family. A clear band was evident at 140 kDa, with its molecular weight being close to the predicted molecular weight (GST fusion protein 26 kDa + 112 kDa = 138 kDa), confirming the successful expression of the this protein in E. coli BL21, although it was primarily present in the precipitate in the form of inclusion bodies (Figure 7).
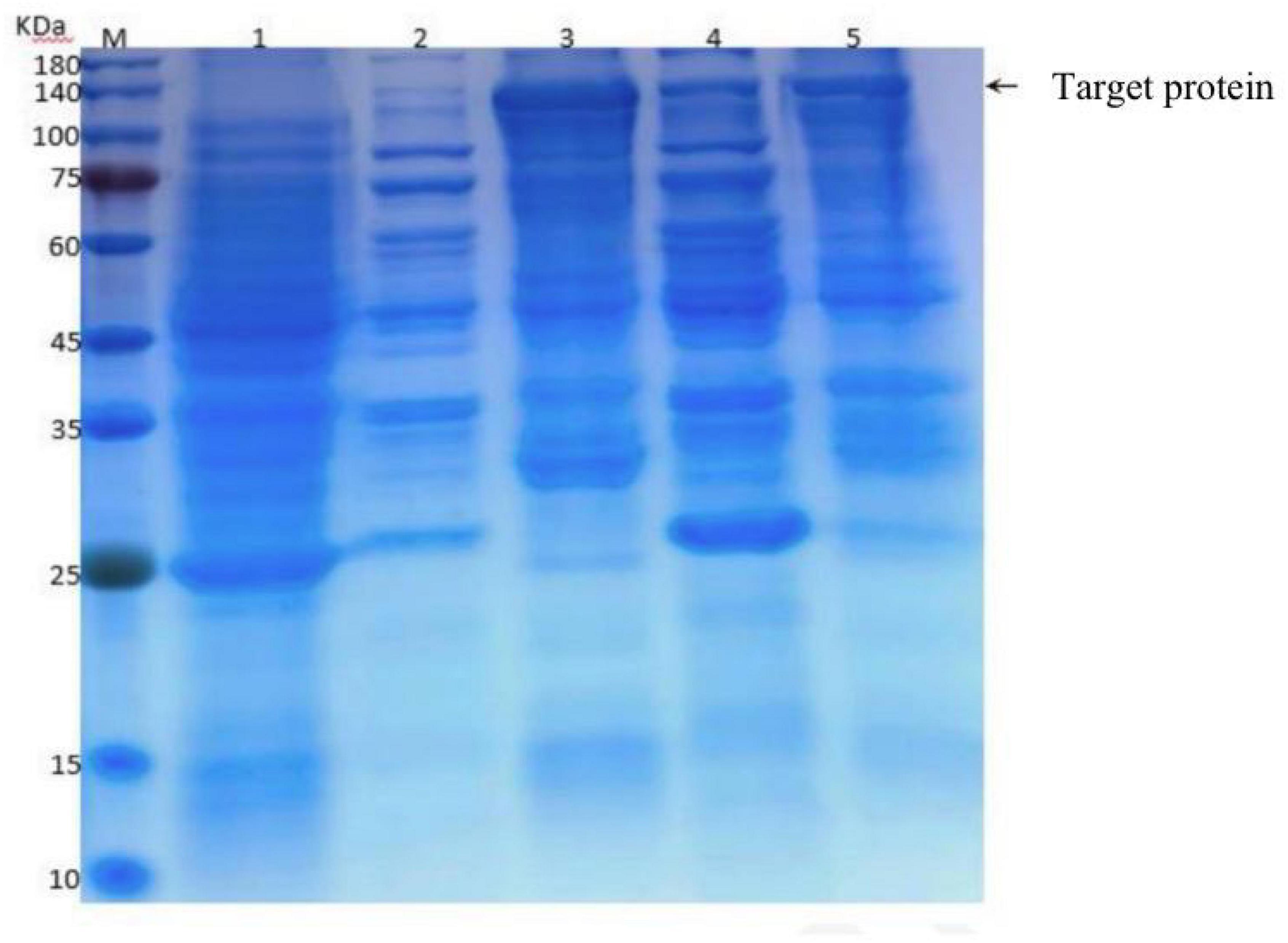
Figure 7. Induced expression of the recombinant plasmid at different temperatures. M: Protein marker; 1: Total protein before induction; 2–5: BL21-pET-DN676-induced supernatants at 20°C, precipitates at 20°C, supernatants at 37°C, and precipitates at 37°C.
Samples were collected from culture medium and growth was assessed at 600 nm via ultraviolet spectrophotometry, while Diquat levels were measured via HPLC (Figure 8). A typical growth curve was observed for the BL21-pET-DN676 strain, whereas the BL21-pET microbes exhibited negligible growth under these conditions. The concentration of Diquat decreased throughout the experimental period such that 12.49% of Diquat (100 mg/L) was removed by the BL21-pET-DN676 strain and 4.01% was removed by the BL21-pET strain after a 7-day incubation period. These results thus confirmed the Diquat-degrading activity of the DN676 gene.
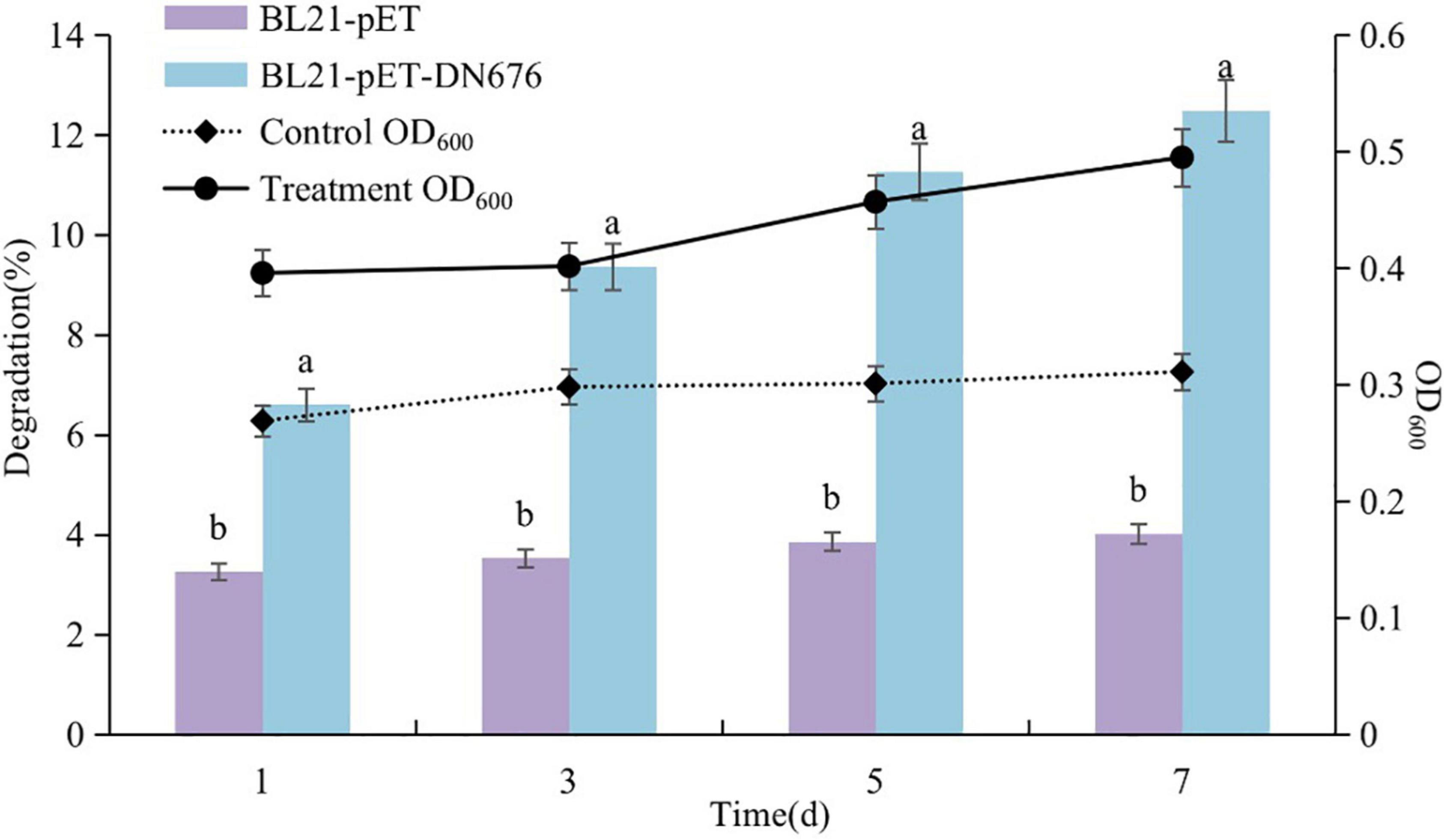
Figure 8. Growth and Diquat degradation of BL21-pET and BL21-pET-DN676 strains. Error bars represent the standard deviation of three replicates. Different letters indicate significant differences between different treatments (P < 0.05).
Funderburk (1969) has previously described a strain of Lipomyces starkeyi capable of degrading Diquat, but their experimental details remain to be published. Current evidence suggests that L. starkeyi can degrade both paraquat and Diquat, utilizing them as a sole nitrogen source to support cell growth. In the present study, a novel yeast strain was isolated from soil exposed to long-term Diquat treatment. In total, 42.51% of Diquat (100 mg/L) was removed by this strain after a 7-day incubation, with this strain thus exhibiting a high degree of Diquat degradation efficiency. Current research on Diquat degradation has focused on the isolation and the characterization of specific microorganisms. In contrast, there have been few studies of the metabolic genes responsible for Diquat degradation. A greater knowledge of the factors that regulate gene expression in specific species may be gained by analysis of microbial RNA-Seq data that reveals the mechanisms of action for particular regulatory genes. As such, RNA-Seq was performed in this study to assess changes in gene expression in this yeast strain over the course of Diquat degradation, revealing 63 and 151 upregulated and downregulated genes, respectively. KEGG pathway enrichment analysis revealed these genes to be most highly enriched in the carbohydrate metabolism pathway. Functional annotation, gene expression analyses, and qRT-PCR assays further revealed DN676 as a candidate Diquat-degrading gene.
RNA-Seq is an efficient means of mining for degraded genes. For example, Zhang et al. (2019) employed an RNA-Seq approach to explore the mechanisms whereby Klebsiella jilinsis 2N3 was able to degrade chlorimuron-ethyl, finding it to do so through a mechanism linked to the sulfur metabolism pathway, with genes encoding carboxylesterases, monooxygenases, glycosyltransferases, and cytochrome P450 being significantly upregulated in treated cells in the context of chlorimuron-ethyl degradation. Through subsequent knockout experiments, they determined that cytochrome P450 enzymes and the Kj-CysJ and Kj-SsuD genes are likely to play central roles in chlorimuron-ethyl degradation (Zhang et al., 2019). Herein, a series of RNA-Seq, qRT-PCR, GO, and KEGG enrichment analyses led to the identification of candidate genes associated with Diquat degradation. These analyses revealed that the carbohydrate metabolism pathway was the most profoundly enriched in the context of Diquat treatment, in line with the similar findings pertaining to sulfur metabolism in the study conducted by Zhang et al. (2019). The DN676 gene was then screened as a candidate associated with Diquat degradation. In contrast to the cytochrome P450 genes identified by Zhang et al. (2019), DN676 is a member of the PDR family, and its function is poorly characterized.
With the exception of Adp1p, PDR members are all full-size ABC proteins. The PDR family is unusual in that it is only found in fungi and plants. Herbicides, phospholipids, peptides, steroids, and anticancer medications are all transported via specific PDR-transporters (Schüller et al., 2003). Yeast cells can quickly counteract toxic environmental challenges through efficient detoxification systems such as the PDR machinery. It’s critical to understand that PDR not only makes cells hypertolerant to numerous unrelated exogenous medications or xenobiotics, but also shields cells against the negative effects of harmful endogenous metabolites (Helmut and Karl, 2006). Several S. cerevisiae PDR genes, such as PDR5 and SNQ2, have been functionally characterized in detail. They confer resistance to a large number of toxic compounds with no, or few common structural or functional properties, such as fungicides, herbicides, pesticides, antibiotics, and detergents. The cytotoxic and mutagenic properties of several of these compounds suggest the involvement of these transporters in cell detoxification and cell resistance (Bauer et al., 1999; Rogers et al., 2001). PDR transporters are also involved in the fungicide resistance of pathogenic fungi such as Candida albicans (Prasad and Kapoor, 2005) or Penicillium digitatum (Nakaune et al., 1998). In the present study, the outcomes demonstrated that the growth of the BL21-pET and BL21-pET-DN676 strains steadily grew over time, and there was a substantial difference between the two (P < 0.05). This indicated that the DN676 gene had increased the resistance to Diquat and had the same result as above. The molecular processes through which members of the ABC protein family members function have become better understood as a result of intensive research efforts in recent years. Many eukaryotic ABC pump proteins’ cellular substrates and physiological roles, however, are yet unknown. Given as PDR transporters remove a large number of potentially harmful metabolites and hundreds of structurally and functionally unrelated cytotoxic chemicals, detoxification is still one of their most plausible physiological activities (Kolaczkowska and Goffeau, 1999). Few studies have explored whether these proteins degrade herbicides or change their structures in the context of detoxification, leading to the selection of DN676 as a candidate Diquat degradation gene for functional verification. In these analyses, when the pET-DN676 vector was expressed in E. coli BL21, this strain was able to remove 12.49% of provided Diquat (100 mg/L) over the course of a 7-day incubation. The DN676 gene increased the degradation of Diquat by BL21, thus confirming that the DN676 gene is related to the degradation of Diquat. However, how the DN676 gene facilitates Diquat degradation warrants further study. Even so, these results suggest that PDR transporters may play an important role in herbicide degradation, potentially opening an entirely new field of research with promising implications for the fields of biotechnology and agriculture.
Zhang et al. (2019) verified the relationship between their genes of interest and chlorimuron-ethyl degradation via a genetic knockout’ approach, we instead verified the role of our target gene of interest by expressing it in E. coli and generated an engineered strain with the ability to degrade Diquat. This method is simple, and allowed for the rapid production strains with specific functions. E. coli strains are very common, and their application in agricultural contexts will not adversely impact local flora. As such, these engineered microbes may represent a valuable tool for future biodegradation and bioremediation efforts. In addition, further RNA-Seq analyses of Diquat degrading strains may provide new insights into the environmental effects of Diquat stress and may facilitate the discovery of new functional genes of interest.
In summary, the Meyerozyma guilliermondii Wyslmt yeast strain was herein isolated from soil that had been exposed to Diquat for an extended period. In total, 42.51% of Diquat (100 mg/L) was removed by this strain after a 7-day incubation. The molecular mechanisms whereby these yeast degrade Diquat were then explored via RNA-Seq. KEGG enrichment analyses indicated that the carbohydrate metabolism pathway exhibited the highest enrichment ratio, and qRT-PCR demonstrated that the treatment group’s levels of mRNA expression of the seven genes during the Diquat degradation process were significantly greater than those of the control group. E. coli gene transfer experiments further demonstrated that when a pET-DN676 vector was expressed in E. coli BL21, 12.49% of Diquat (100 mg/L) was removed by this strain after a 7-day incubation. As such, these results may highlight an effective approach to generating engineered microbial strains with the ability to degrade Diquat.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
FW and BT: conceptualization and methodology. FW: validation, data curation, and writing—original draft preparation. FW, YH, LK, JG, XS, and BT: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Transgenic Major Special Fund of China (2016ZX08004001).
We wish to thank all colleagues who assisted in this research and provided technical advice.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.993721/full#supplementary-material
Arora, P. K., Srivastava, A., Garg, S. K., and Singh, V. P. (2018). Recent advances in degradation of chloronitrophenols. Bioresour. Technol. 250, 902–909. doi: 10.1016/j.biortech.2017.12.007
Bauer, B. E., Wolfger, H., and Kuchler, K. (1999). Inventory and function of yeast ABC proteins: About sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta 1461, 217–236. doi: 10.1016/s0005-2736(99)00160-1
Camachomorales, R. L., Karina, G. N., and Jose, E. S. (2017). Degradation of the herbicide paraquat by macromycetes isolated from southeastern Mexico. Biotech 7, 324–334. doi: 10.1007/s13205-017-0967-3
Chen, S., Chang, C., Deng, Y., An, S., Dong, Y. H., Zhou, J., et al. (2014). Fenpropathrin biodegradation pathway in Bacillus sp. DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agric. Food Chem. 62, 2147–2157. doi: 10.1021/jf404908j
Chen, S., Deng, Y., Chang, C., Lee, J., Cheng, Y., Cui, Z., et al. (2015). Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 5:8784. doi: 10.1038/srep08784
Cheng, Y., Zang, H. L., Wang, H. L., Li, D., and Li, C. (2018). Global transcriptomic analysis of Rhodococcus erythropolis D310-1 in responding to chlorimuron-ethyl. Ecotoxicol. Environ. Safe. 157, 111–120. doi: 10.1016/j.ecoenv.2018.03.074
Cycon, M., and Piotrowska-Seget, Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: A review. Front. Microbiol. 7:1463. doi: 10.3389/fmicb.2016.01463
Ducrot, V., Pery, A. R. R., and Lagadic, L. (2010). Modelling effects of Diquat under realistic exposure patterns in genetically differentiated populations of the gastropod Lymnaea stagnalis. Phil. Trans. R. Soc. B 365, 3485–3494. doi: 10.1098/rstb.2010.0047
Funderburk, H. H. (1969). “Diquat and paraquat,” in Degradation of herbicides, eds P. C. Kearney and D. D. Kaufman (New York: Marcel Dekker), 283.
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261.
Hata, S., Shirata, K., and Takagishi, H. (1986). Degradation of paraquat and Diquat by the yeast Lipomyces starkeyi. J. Gen. Appl. Microbiol. 32, 193–202.
Helmut, J., and Karl, K. (2006). Yeast ABC transporters – A tale of sex, stress, drugs and aging. FEBS Lett. 580, 1131–1138. doi: 10.1016/j.febslet.2005.12.050
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopaedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kolaczkowska, A., and Goffeau, A. (1999). Regulation of pleiotropic drug resistance in yeast. Drug Resist. Updates 2, 403–414.
Li, Y., Ge, X. Z., Wang, X. Y., and Gao, R. (2017). The invention discloses a compound bacterial agent used to degrade paraquat and a preparation method. China. Patent No CN 106520618 A. Beijing: National Intellectual Property Administration.
Liu, J., Chen, S., Ding, J., Xiao, Y., Han, H., and Zhong, G. (2015). Sugarcane bagasse as support for immobilization of Bacillus pumilus HZ-2 and its use in bioremediation of mesotrione-contaminated soils. Appl. Microbiol. Biotechnol. 99, 10839–10851. doi: 10.1007/s00253-015-6935-0
Nakaune, R., Adachi, K., Nawata, O., Tomiyama, M., Akutsu, K., and Hibi, T. (1998). A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitum. Appl. Environ. Microb. 64, 3983–3988. doi: 10.1128/AEM.64.10.3983-3988.1998
Prasad, R., and Kapoor, K. (2005). Multidrug resistance in yeast Candida. Int. Rev. Cytol. 242, 215–248.
Reiner, A., Yekutieli, D., and Benjamini, Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375.
Rogers, B., Decottignies, A., Kolaczkowski, M., Carvajal, E., Balzi, E., and Goffeau, A. (2001). The pleiotropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Biotechnol. 3, 207–214.
Schüller, C., Bauer, B. E., and Kuchler, K. (2003). “Inventory and Evolution of Fungal ABC Protein Genes,” in ABC proteins: From bacteria to man, eds I. B. Holland, S. P. C. Cole, K. Kuchler, and C. F. Higgins (Amsterdam: Academic Press–Elsevier Science), 279–293.
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2011). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wang, X. H., Souders, C. L., Zhao, Y. H., and Martyniuk, C. J. (2017). Mitochondrial bioenergetics and locomotor activity are altered in zebrafifish (Danio rerio) after exposure to the bipyridylium herbicide Diquat. Toxicol. Lett. 283, 13–20. doi: 10.1016/j.toxlet.2017.10.022
Yang, J., Feng, Y., Zhan, H., Liu, J., Yang, F., Zhang, K., et al. (2018). Characterization of a pyrethroid-degrading Pseudomonas fulva strain P31 and biochemical degradation pathway of D-phenothrin. Front. Microbiol. 9:1003. doi: 10.3389/fmicb.2018.01003
Yuan, G., Li, R., Zhao, Q., Kong, X., Wang, Y., Wang, X., et al. (2020). Simultaneous determination of paraquat and Diquat in human plasma by HPLC-DAD: Its application in acute poisoning patients induced by these two herbicides. J. Clin. Lab. Anal. 35:e23669. doi: 10.1002/jcla.23669
Keywords: Diquat, Meyerozyma guilliermondii Wyslmt, biodegradation, RNA-Seq, qRT-PCR, prokaryotic expression
Citation: Wang F, Kong L, Guo J, Song X, Tao B and Han Y (2022) RNA-sequencing analysis of the Diquat-degrading yeast strain Meyerozyma guilliermondii Wyslmt and the discovery of Diquat degrading genes. Front. Microbiol. 13:993721. doi: 10.3389/fmicb.2022.993721
Received: 14 July 2022; Accepted: 15 August 2022;
Published: 02 September 2022.
Edited by:
Hari Prasanna Deka Boruah, North East Institute of Science and Technology (CSIR), IndiaReviewed by:
Xiangchao Cui, Xinyang Normal University, ChinaCopyright © 2022 Wang, Kong, Guo, Song, Tao and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Tao, Ym90YW9sQDE2My5jb20=; Yujun Han, aGFueWo5MjBAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.