
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 September 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.992582
This article is part of the Research Topic Worldwide Emergence of Drug-Resistant fungi: from Basic to Clinic, Volume II View all 13 articles
 Penghao Guo1†
Penghao Guo1† Jianlong Chen1†
Jianlong Chen1† Yiwei Tan2
Yiwei Tan2 Li Xia3
Li Xia3 Weizheng Zhang4
Weizheng Zhang4 Xiaojie Li5
Xiaojie Li5 Yujie Jiang6
Yujie Jiang6 Ruiying Li7
Ruiying Li7 Chunmei Chen8
Chunmei Chen8 Kang Liao1*
Kang Liao1* Yaqin Peng1*
Yaqin Peng1*Background: Fusarium species are opportunistic causative agents of superficial and disseminated human infections. Fast and accurate identification and targeted antifungal therapy give help to improve the patients’ prognosis.
Objectives: This study aimed to evaluate the effectiveness of matrix-assisted laser desorption ionisation time of flight mass spectrometry (MALDI-TOF MS) for Fusarium identification, and investigate the epidemiology and antifungal susceptibility profiles of clinical Fusarium isolates in Southern China.
Methods: There were 95 clinical Fusarium isolates identified by DNA sequencing of translation elongation factor 1-alpha (TEF1α) and MALDI-TOF MS, respectively. Antifungal susceptibility testing of isolates was performed by broth microdilution according to the CLSI approved standard M38-A3 document.
Results: Seven species complexes (SC) with 17 Fusarium species were identified. The most prevalent SC was the F. solani SC (70.5%, 67/95), followed by the F. fujikuroi SC (16.8%, 16/95). F. keratoplasticum within the F. solani SC was the most prevalent species (32.6%, 31/95). There were 91.6% (87/95) of isolates identified by MALDI-TOF MS at the SC level. In most of species, amphotericin B and voriconazole showed lower MICs compared to itraconazole and terbinafine. The F. solani SC showed higher MICs to these antifungal agents compared to the other SCs. There were 10.5% (10/95) of strains with high MICs for amphotericin B (≥8 μg/ml), terbinafine (≥32 μg/ml) and itraconazole (≥32 μg/ml) simultaneously, mostly focusing on F. keratoplasticum (9/10).
Conclusion: MALDI-TOF MS exhibited good performance on the identification of Fusarium strains at the SC level. The F. solani SC was the most prevalent clinical SC in Southern China. The MICs varied significantly among different species or SCs to different antifungal agents.
The genus Fusarium is an important phytopathogen; only a few species can cause a broad spectrum of human infections (Al-Hatmi et al., 2016b; Van Diepeningen and de Hoog, 2016). Almost 70 Fusarium species have been reported as opportunistic human pathogens, with the increasing rates of infection over the past years (Tortorano et al., 2014; Triest et al., 2015). The clinical manifestations of Fusarium disease are diverse, depending largely on the immune status of the host and the portal of entry (Tortorano et al., 2014). In immunocompetent patients, Fusarium species mainly lead to superficial infections such as keratitis and onychomycosis, while the invasive or disseminated infections tend to affect critically ill and immunosuppressed patients with a high mortality rate (Zhao et al., 2021).
The clinically relevant Fusarium species are mainly grouped into six species complexes (SC), including the F. solani SC (FSSC), F. oxysporum SC (FOSC), F. fujikuroi SC (FFSC), F. dimerum SC (FDSC), F. incarnatum-equiseti SC (FIESC), and F. chlamydosporum SC (FCSC; Triest et al., 2015). It has been found that antifungal susceptibility may vary among different species within a single species complex (O’Donnell et al., 2008; Al-Hatmi et al., 2015b; Song et al., 2021), which indicates it is necessary to identify the aetiological agent up to the species level for clinical treatment. In the clinical laboratory, these closely related species are often morphologically indistinguishable. Molecular analysis can provide the gold standard for species identification, while it has the disadvantages of being time-consuming and costly. A rapid, simple, cost-effective, and reproducible tool has received increasingly interest for mold identification, i.e., matrix-assisted laser desorption ionisation time of flight mass spectrometry (MALDI-TOF MS; Triest et al., 2015; Al-Hatmi et al., 2015a; Normand et al., 2021). This approach has been found to enhance the correct identification rate of non-Aspergillus filamentous fungi with a 31%–61% increase (Ranque et al., 2014). However, more data are needed for the verification and standardization of Fusarium identification due to the potential impacts of different instrument platforms and reference spectrum databases on its performance.
In clinic, amphotericin B and azole drugs, e.g., voriconazole and itraconazole, are commonly used for Fusarium infection (Nucci and Anaissie, 2007; Tortorano et al., 2014; Oliveira et al., 2020). Amphotericin B or voriconazole is used for the disseminated infections as first-line drugs (Al-Hatmi et al., 2018). Fusarium keratitis is mainly treated with voriconazole and natamycin, and the treatment of onychomycosis should include terbinafine, voriconazole and sometimes itraconazole (Al-Hatmi et al., 2018). However, it has been reported that clinical Fusaria have relatively decreased susceptibility to these commonly used antifungal drugs (Taj-Aldeen et al., 2016; Rosa et al., 2019). Different patterns of in vitro susceptibility have been found in different Fusarium species (Song et al., 2021). Remarkably, since neither clinical breakpoints nor epidemiological cutoff values have been established for Fusarium according to Clinical and Laboratory Standards Institute (CLSI) M59-3ed (CLSI, 2020) and EUCAST database,1 information on the correlation between minimum inhibitory concentration (MIC) and drug efficacy is not clear. Given that a limited number of studies on in vitro susceptibility are available, more data are necessary for the epidemiology and therapy purpose.
Studies on clinical fusaria are limited in Asia, especially in Southern China. In this study, we aim to investigate the prevalence characteristics and antifungal susceptibility profiles of clinical Fusarium strains collected from eight hospitals in Southern China. And the effectiveness of Fusarium identification by MALDI-TOF MS was also investigated.
Ninety-five clinical Fusarium strains were collected from eight hospitals in Southern China between January 2018 and December 2020. These isolates were recovered from corneal scrapings (47.4%, 45/95) and skin secretions (40.0%, 38/95), followed by pus (4.2%, 4/95), blood (4.2%, 4/95), sputum (3.2%, 3/95) and urine (1.0%, 1/95). Duplicated isolates were excluded if they were obtained from the same patient. Given samples were totally collected during routine patient care in this retrospective investigation, the need for informed consent was waived by the institutional review board of the First Affiliated Hospital of Sun Yat-sen University.
The Fusarium strains were cultured for 5 days on potato dextrose agar medium at 28°C. All cultures were handled within a class II biological safety cabinet.
A single colony was picked up in a 1.5-ml Eppendorf (EP) tube containing 1.0 ml PBS, with the turbidity adjusted to 1.0 McFarland. DNA extraction was performed using the Yeast Genomic DNA Rapid Extraction Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions.
The sequence of the translation elongation factor 1-alpha gene (TEF1α) was amplified using the primers EF1 (5′-ATGGGTAAGGARGACAAGAC-3′) and EF2 (5′-GGARGTACCAGTSATCATGTT-3′) as previously described with some modifications (O’Donnell et al., 2008). The PCR amplification was conducted in a 50-μL reaction mixture containing 10 μl 10 × PCR buffer, 5 μl templates, 1 μl forward primer, 1 μl reverse primer, 0.5 μl Taq enzyme, 8 μl dNTP mixture, and 24.5 μl double-distilled water. The PCR amplification condition is as follows: 1 cycle of 95°C 10 min; 40 cycles of 95°C 30 s, 56°C 30 s, 72°C 30 s; 1 cycle of 72°C 10 min. The PCR products were subjected to Sanger sequencing (Sangon Biotech, China). The sequences were identified by BLAST analysis in GenBank2 (Da et al., 2021).
The colonies were picked by sterile swabs in a 1.5-mL EP tube containing 0.9 ml 75% ethanol and 20–30 glass beads, mixed for 2 min. Then the suspension was removed to a new tube for centrifugation at 13,000 rpm for 2 min. The supernatant was removed, and 40 μl freshly prepared 70% formic acid was added to the tube and mixed for 1 min. Then, 40 μl acetonitrile was added to the tube and mixed for 1 min. The tube was centrifuged at 13,000 rpm for 2 min. One μl of supernatant was added on the spot of the target plate, and 1 μl CHCA matrix was added after the 1-μl supernatant dried. After the matrix dried, the target plate was taken to the mass spectrometer’s ionization chamber. The mass spectra of the strains were acquired using a VITEK MS Plus (bioMérieux, France) in IVD mode and analyzed by the IVD knowledge base V3.2 for Fusarium identification.
The dendrogram showing taxonomic relationships was carried out using VITEK MS RUO/SARAMIS (bioMérieux, France) according to the manufacturer’s instructions. Firstly, spectra were manually imported to the SARAMIS™ RUO database version 4.17 using the button “import spectra to spectra database.” Then the dendrogram was generated according to the whole spectra. Consensus spectra were analyzed with a single link agglomerative clustering algorithm, applying the relative taxonomy analysis tool of SARAMIS premium software to show the resulting dendrogram with differences and similarities in relative terms (percent matching masses).
For instrument calibration, the Escherichia coli strain (ATCC 8739) was applied. And the Candida glabrata strain (ATCC MYA-2950) was used as quality control.
Four commonly antifungal agents (Shanghai Aladdin Bio-Chem Technology Co., Ltd., China), i.e., amphotericin B, voriconazole, itraconazole and terbinafine were included and dissolved in dimethyl sulfoxide to 3.2 mg/ml as stock solutions. The work concentrations of these agents ranged from 0.06 to 32 μg/ml. The broth microdilution was performed according to CLSI M38-A3 method (CLSI, 2017). The colonies were picked up and transferred into a 1.5-ml EP tube containing 1.0 ml PBS, with turbidity adjusted to 0.5 McFarland. The suspensions were then diluted in RPMI 1640 to the desired concentration of 0.4 × 104–5 × 104 CFU/ml by counting on a hemocytometer, 100 μl of which were added in the microdilution plates for 48-h incubation at 35°C. The MICs were defined as the lowest concentration with complete growth inhibition compared to the drug-free growth. MIC50 and MIC90 values were defined as the lowest concentrations that inhibited the growth of 50% or 90% of the strains. WHONET software version 5.6 was used for determining MIC50, MIC90, geometric mean (GM) and MIC range.
The strains of Candida parapsilosis (ATCC 22019) and Candida krusei (ATCC 6258) were used as quality controls.
All sequences identified in this study were deposited in GenBank (ON959267–ON959361).
The 95 isolates were identified by DNA sequencing of TEF1α as members of 7 species complexes (SC) with 17 Fusarium species (Table 1): FSSC (70.5%, 67/95), FFSC (16.8%, 16/95), FOSC (7.4%, 7/95), FDSC (2.1%, 2/95), one isolate of FIESC, FCSC and F. nisikadoi SC (FNSC), respectively. The FSSC was the most prevalent SC, including F. keratoplasticum (32.6%, 31/95), F. falciforme (20.0%, 19/95), F. solani sensu stricto (6.3%, 6/95), F. ambrosium (5.3%, 5/95), F. petroliphilum (4.2%, 4/95) and F. lichenicola (2.1%, 2/95). The FFSC included F. proliferatum (7.4%, 7/95), F. sacchari (3.2%, 3/95), F. concentricum (3.2%, 3/95), F. verticillioides (2.1%, 2/95) and F. napiforme (1.1%, 1/95). The FOSC included F. oxysporum (6.3%, 6/95) and one isolate of F. acutatum.

Table 1. Comparison of identification results of 95 clinical Fusarium strains using DNA sequencing of TEF1α and MALDI-ToF MS methods.
For the 45 isolates obtained from cornea scrapings, the detection rates of FSSC, FFSC and FOSC were 73.3% (33/45), 20.0% (9/45) and 4.4% (2/45), respectively. Both F. keratoplasticum (28.9%, 13/45) and F. falciforme (28.9%, 13/45) within FSSC were the most common species from cornea scrapings (Figure 1). And 63.2% (24/38) of isolates originating from skin secretions belonged to FSSC, followed by FFSC (13.2%, 5/38) and FOSC (13.2%, 5/38). The most prevalent species from skin secretions was F. keratoplasticum (34.2%, 13/38; Figure 1).
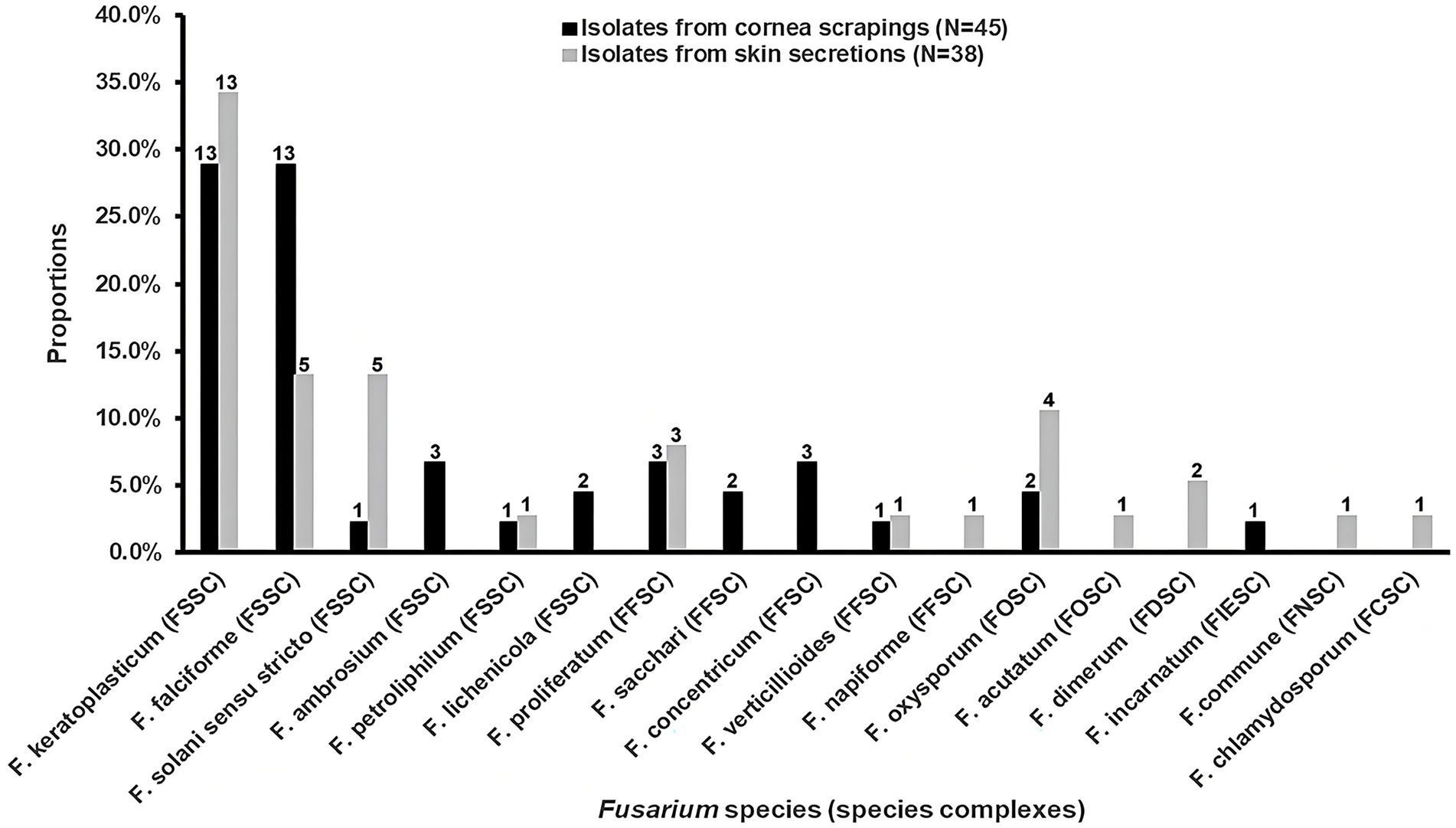
Figure 1. The distributions and proportions of Fusarium strains among isolates from cornea scrapings and skin secretions, respectively. FSSC, F. solani species complex (SC); FFSC, F. fujikuroi SC; FOSC, F. oxysporum SC; FDSC, F. dimerum SC; FIESC, F. incarnatum-equiseti SC; FNSC, F. nisikadoi SC; FCSC, F. chlamydosporum SC.
Comparison of data with DNA sequencing and MALDI-TOF MS is listed in Table 1. The results showed that 91.6% (87/95) of isolates were identified at the SC level by MALDI-TOF MS. For FSSC (n = 67) and FDSC (n = 2), all the isolates were correctly recognized. Most of isolates were also identified by MALDI-TOF MS for FFSC (75.0%, 12/16) and FOSC (85.7%, 6/7). However, MALDI-TOF MS correctly identified 11.6% (11/95) of the isolates down to the species level, including all isolates of F. proliferatum (n = 7), F. verticillioides (n = 2) and F. dimerum (n = 2). One isolate of F. concentricum and two isolates of F. sacchari were misidentified as F. proliferatum but were correct at the SC level. Further, we analyzed the MALDI-TOF MS profiles of Fusarium species corresponding to the morphological characteristics of cultures. Although it was hard to differentiate them by morphology, the discrepancies of MS profile characteristics were observed significantly among these species (Figure 2).
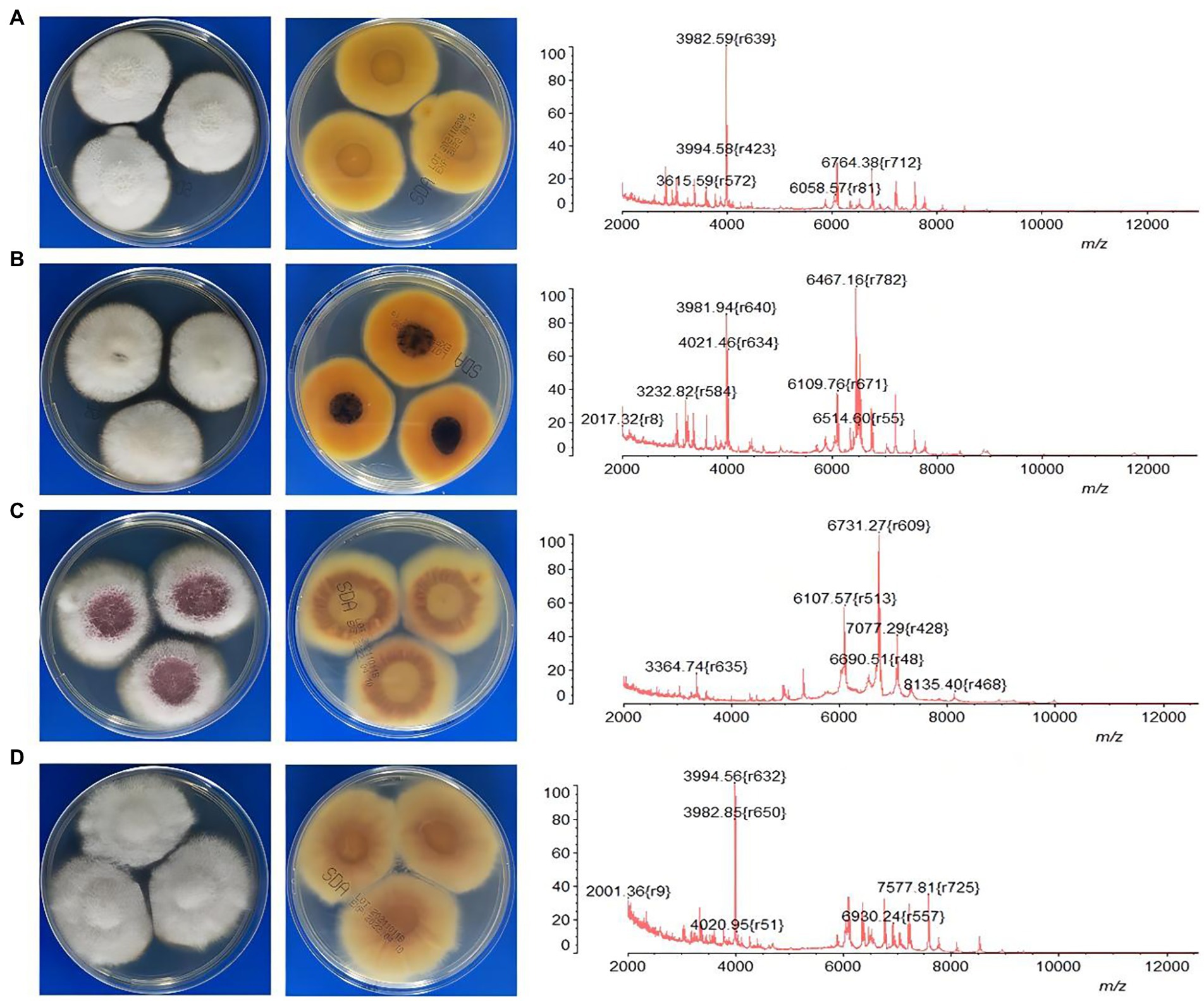
Figure 2. The characteristics of MALDI-TOF MS profiles corresponding to the morphologies of four common Fusarium species. (A) F. keratoplasticum; (B) F. falciforme; (C) F. proliferatum; (D) F. oxysporum.
In the MALDI-TOF dendrogram, almost all of members were found to cluster together in the FSSC except F. lichenicola (Figure 3). However, members of FFSC and FOSC were randomly interspersed with those of other species complexes. The strains of the F. keratoplasticum within FSSC were found to cluster together in the dendrogram. Differences between F. proliferatum and other strains were also unambiguous.
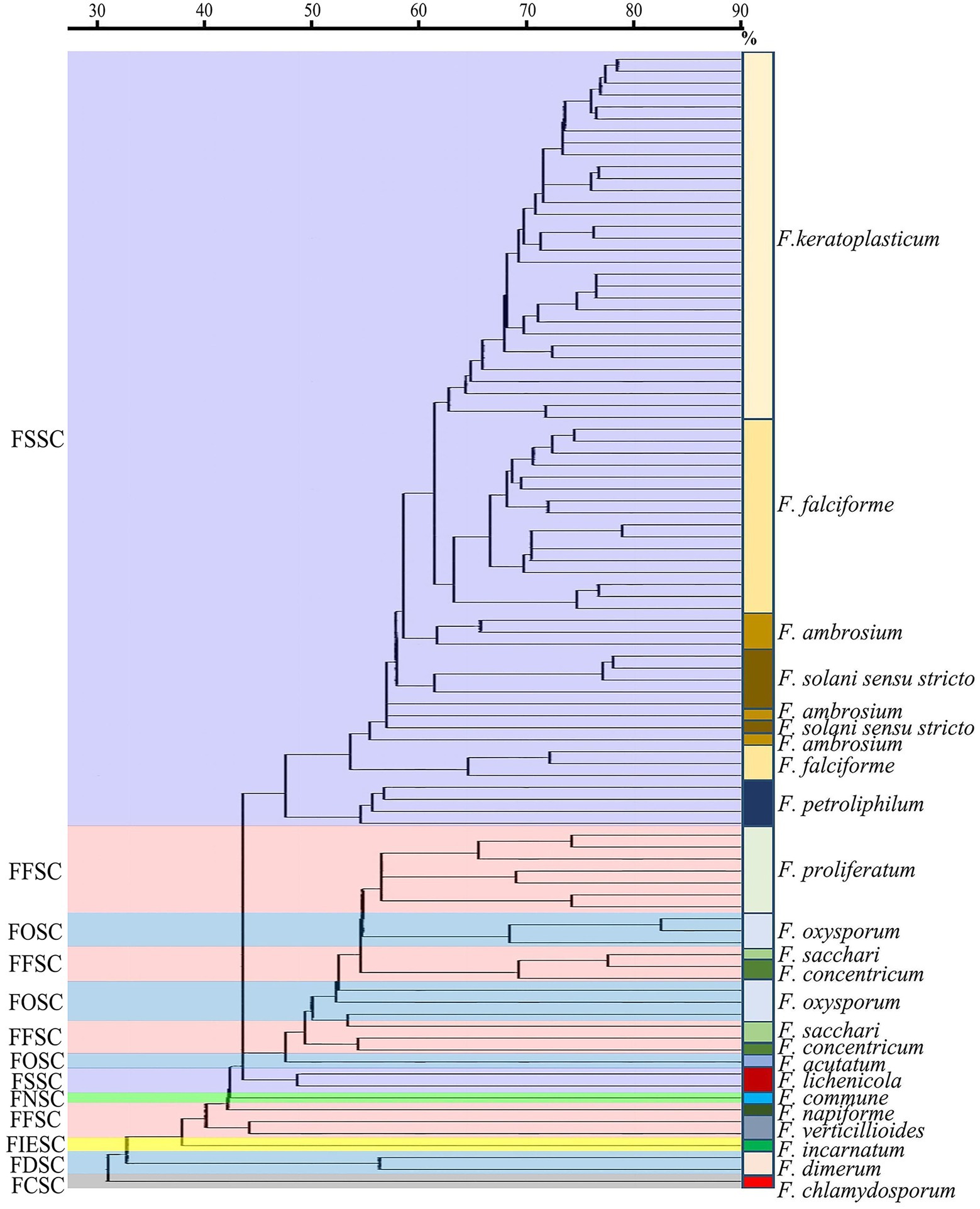
Figure 3. The MALDI-ToF dendrogram of 95 clinical Fusarium strains. FSSC, F. solani species complex (SC); FFSC, F. fujikuroi SC; FOSC, F. oxysporum SC; FNSC, F. nisikadoi SC; FIESC, F. incarnatum-equiseti SC; FDSC, F. dimerum SC; FCSC, F. chlamydosporum SC.
The MICs varied among different species complexes to these antifungal agents (Table 2). Compared to itraconazole and terbinafine, voriconazole and amphotericin B showed lower MICs to most of species. Fusarium isolates showed variable MICs to voriconazole ranging between 0.5 and 16 μg/ml. Amphotericin B had good activity against most of species, with 1–16 μg/ml in FSSC, 1–4 μg/ml in FFSC and 1–2 μg/ml in FOSC, respectively. Interestingly, 10.5% (10/95) of strains for amphotericin B had high MICs (≥8 μg/ml), totally belonging to the FSSC. For itraconazole, 93.7% (89/95) of strains showed high MICs (≥32 μg/ml). There were 76.8% (73/95) of strains with high MICs (≥8 μg/ml) for terbinafine. And terbinafine showed low MICs in FFSC (GM = 2.3 μg/ml) and FCSC (1 μg/ml). Compared to the other species complexes, FSSC presented relatively higher MICs to these antifungal agents.
We further analyzed antifungal activities of species within FSSC (Table 3). The MICs of Fusarium isolates to voriconazole ranged from 1 to 16 μg/ml. All strains within FSSC showed high MICs (≥32 μg/ml) for itraconazole. For terbinafine, there were 65.3% (62/95) of strains with highest MICs (≥32 μg/ml). Among the 10 strains with high MICs (≥8 μg/ml) for amphotericin B, nine strains belonged to F. keratoplasticum and only one were in F. falciforme. Remarkably, high MICs (≥ 32 μg/ml) both for terbinafine and itraconazole were observed among these 10 strains.
Along with the rising numbers of severely immunocompromised patients in recent decades, invasive or disseminated Fusarium infections with high mortality have been found to increase remarkably (Muhammed et al., 2013; Al-Hatmi et al., 2016a). Considering the relatively low susceptibility of Fusarium species to most of commonly used antifungal drugs, the prevalence and resistance profile of clinical Fusarium species can contribute to enhance the management of the infection (O’Donnell et al., 2008; Guarro, 2013). As a major challenge, it is lack of an accurate, quick and easy to operate approach for the identification of clinical Fusarium strains so far. In most of clinical laboratories, Fusarium identification mainly depends on different morphological characteristics of size and shape of macro- and microconidia and presence or absence of chlamydospores as well as colony appearance (Najafzadeh et al., 2020; Da et al., 2021). However, a series of factors can affect the morphological characteristics of cultures such as the temperature, the culture medium and maybe the thickness of the medium (Da et al., 2021). Fusarium at the SC level are usually hard to be distinguished by this conventional and time-consuming approach if not for experienced experts.
We observed that MALDI-TOF MS had excellent performance of Fusarium identification at the SC level with the correct rate up to 91.6% (87/95), taking DNA sequencing of TEF1α as the gold standard (Herkert et al., 2019; Oliveira et al., 2020; Da et al., 2021). Similar results were achieved by Paziani et al. (94.4%) and Song et al. (95.2%; Paziani et al., 2019; Song et al., 2021). To a large extent, it attributed to a success ratio of 100% correct identifications for the most prevalent SC (FSSC; Table 1). High correct rates were also observed for FDSC (100%, 2/2) and FOSC (85.7%, 6/7). For FFSC (n = 16), there were four strains unable to be identified by MALDI-TOF MS which were F. sacchari (n = 1), F. concentricum (n = 2) and F. napiforme (n = 1), respectively. Some studies showed good performance of Fusarium identification by MALDI-TOF MS down to the species level (Triest et al., 2015; Song et al., 2021). Regrettably, only 11.6% (11/95) of isolates could be correctly identified to the species level in this study. It might be limited by small species and strain representations in commercial libraries (Sleiman et al., 2016). Triest’s study presented a correct rate of the identifications (91.0%) to the species level by constructing an in-house reference spectrum database combined with a standardized MALDI-TOF MS assay (Triest et al., 2015). Song et al. found MALDI-TOF MS recognized 89.04% of Fusarium species though a combination of the Bruker library and an expanded version in the BMU database (Song et al., 2021). Further studies will be needed to improve species identification in our laboratory. In the dendrogram, we found all strains except one clustered together in the FSSC, which was similar as Triest’s finding (Triest et al., 2015). However, most of members of the other species complexes were randomly distributed. Normand et al. also demonstrated about 30% of the strains clustered correctly in the dendrograms (Herkert et al., 2019). Given the identification probably depends on recognition of a limited number of conserved proteins regardless of intraspecific variability, phylogenetic interpretation of MALDI-TOF data is not recommended.
The discrepancy of Fusarium distribution has been thought to be associated with several factors such as geographical regions, clinical patient populations and infection sites. When being judged from numerous literature data, members of fusaria encountered in human infections are mostly found in three species complexes: FSSC, FFSC, and FOSC. FSSC is considered as the most frequently detected SC worldwide, mainly causing superficial infections such as keratitis and onychomycosis under tropical and subtropical climatic conditions, especially in Asia and Latin America (Castro López et al., 2009; Salah et al., 2015; Sun et al., 2015; Guevara-Suarez et al., 2016; Muraosa et al., 2017; Rosa et al., 2017; Tupaki-Sreepurna et al., 2017; Dallé da Rosa et al., 2018; Najafzadeh et al., 2020). Several studies showed FFSC to be the prevalent SC in some areas such as Iran and Turkey, whereas FOSC was more common in Europe (Dalyan Cilo et al., 2015; Abastabar et al., 2018; Oliveira et al., 2019; Najafzadeh et al., 2020; Walther et al., 2021). Our results demonstrated FSSC (70.5%, 67/95) was the most prevalent group mainly originating from corneal scrapings (33/45), followed by FFSC (16.8%, 16/95) and FOSC (7.4%, 7/95). The prevalence of Fusarium SC here showed similar as Song’s finding in Northern China and Sun’s finding in central China (Sun et al., 2015; Song et al., 2021).
There were 40.0% (38/95) of isolates in this study that were obtained from skin secretions, a proportion of which were collected from inpatients with burns or diabetes mellitus (data not shown). Severe burns and poorly controlled diabetes are thought to be high risk factors for invasive mold infections (Nucci and Anaissie, 2007; Enoch et al., 2017). However, little is known about the epidemiology of Fusarium strains causing locally invasive skin infection in patients with burns or diabetes mellitus, limited by sporadic case reports (Nucci and Anaissie, 2002; Taj-Aldeen et al., 2006; Pai et al., 2010; Atty et al., 2014; Rosanova et al., 2016; Karadag et al., 2020; Tram et al., 2020; Liza et al., 2021). We observed 63.2% (24/38) of isolates from skin secretions belonged to FSSC. Limited by incomplete clinical data here, further studies will be needed to investigate the association of Fusarium strains and locally invasive skin infection among these patients. Remarkably, we found one isolate of F. commune obtained from skin secretion. F. commune within FNSC has been reported as a plant pathogen (Mezzalama et al., 2021; Wang et al., 2022). To the best of our knowledge, this is the first to report this species in clinical specimens.
In Nucci’s review, F. solani sensu stricto was regarded as the most common species, followed by F. oxysporum and F. verticillioides (Nucci and Anaissie, 2007). However, the three most common species were F. falciforme and F. keratoplasticum, followed by F. oxysporum in Al-Hatmi’s review (Al-Hatmi et al., 2016a). Song et al. demonstrated the most prevalent species was F. solani sensu stricto (93.8%, 135/144) within the FSSC, and F. verticillioides (60.6%, 40/66) within the FFSC (Song et al., 2021). Walther et al. presented F. petroliphilum within the FSSC was the most prevalent species (Walther et al., 2021). We here found that 46.3% (31/67) of isolates belonged to F. keratoplasticum within the FSSC, followed by F. falciforme (28.4%, 19/67) and F. solani sensu stricto (9.0%, 6/67). For FFSC, F. proliferatum (43.8%, 7/16) was the most common species. Given species-specific differences in antifungal susceptibility, the discrepancy of species distribution should be considered on the treatment options.
Currently, most of Fusarium infection still based on empirical antifungal therapy. A limited number of studies on in vitro susceptibility were available, showing variable results. In this study, antifungal susceptibility profiles of 95 strains were analyzed for four commonly used agents, i.e., amphotericin B, voriconazole, itraconazole and terbinafine. Our results showed high MICs for itraconazole (93.7%, MIC ≥ 32 μg/ml) and terbinafine (76.8%, MIC ≥ 8 μg/ml) in most of species. Rosa et al. presented higher MICs (≥64 μg/ml) for itraconazole and terbinafine in general (Rosa et al., 2017), while more than 50% of Fusarium strains were sensitive to these agents in Sun’s study (Sun et al., 2015). Here, terbinafine showed low MICs in FFSC (GM = 2.3 μg/ml), showing similar results as Song’s study (Song et al., 2021). However, Song et al. presented good activities for terbinafine against FSSC (GM = 2.4 μg/ml) and FOSC (GM = 2.5 μg/ml), which were significantly different from our results (Table 3). For voriconazole, it is thought to be clinically effective against Fusarium spp., despite variable in vitro activity (Walther et al., 2021). Similarly, the MICs for voriconazole here ranged from 0.5 to 16 μg/ml. Castro López et al. showed F. solani sensu stricto had the highest MIC for voriconazole (Castro López et al., 2009). Interestingly, here the MIC of all the F. solani sensu stricto strains was 4 μg/ml for voriconazole. In line with our results, several studies showed low MICs for amphotericin B to the majority of isolates (Al-Hatmi et al., 2015b; Rosa et al., 2017; Oliveira et al., 2019, 2020). Remarkably, we observed 10.5% (10/95) of strains with high MICs for amphotericin B (≥8 μg/ml), terbinafine (≥32 μg/ml) and itraconazole (≥32 μg/ml) simultaneously, which were totally belonged to the FSSC. More attentions should be paid on these multi-resistance strains within the FSSC. It is worth noting that information on the relationships between low MIC and clinical response to therapy is still unavailable due to lack of species-specific clinical breakpoints.
Our study has some limitations. Clinical data was not fully collected, preventing us to decipher whether these clinical isolates were related to proven fusariosis or could be associated with contamination of organs. In summary, our results demonstrated that MALDI-TOF MS exhibited good performance on the identification of Fusarium strains at the SC level. In most of species, amphotericin B and voriconazole showed lower MICs compared to itraconazole and terbinafine. F. keratoplasticum within the FSSC was the most prevalent species in southern China, showing relatively high MICs for these antifungal agents. Further studies will be needed for investigating the correlations of low and high MICs with the prognosis of patients as well as the resistance mechanisms of Fusarium strains.
The data presented in the study are deposited in the GenBank repository, accession number ON959267–ON959361.
KL and YP participated in research design and data analysis. PG participated in the writing of the manuscript and data analysis. JC performed the experiments. YT, LX, WZ, XL, YJ, RL, and CC participated in the collection of Fusarium strains. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abastabar, M., Al-Hatmi, A., Vafaei, M. M., de Hoog, G. S., Haghani, I., Aghili, S. R., et al. (2018). Potent activities of luliconazole, lanoconazole, and eight comparators against molecularly characterized Fusarium species. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.00009-18
Al-Hatmi, A., Bonifaz, A., Ranque, S., Sybren, D. H. G., Verweij, P. E., and Meis, J. F. (2018). Current antifungal treatment of fusariosis. Int. J. Antimicrob. Agents 51, 326–332. doi: 10.1016/j.ijantimicag.2017.06.017
Al-Hatmi, A. M., Hagen, F., Menken, S. B., Meis, J. F., and de Hoog, G. S. (2016a). Global molecular epidemiology and genetic diversity of Fusarium, a significant emerging group of human opportunists from 1958 to 2015. Emerg. Microbes Infect. 5:e124. doi: 10.1038/emi.2016.126
Al-Hatmi, A. M., Meis, J. F., and de Hoog, G. S. (2016b). Fusarium: molecular diversity and intrinsic drug resistance. PLoS Pathog. 12:e1005464. doi: 10.1371/journal.ppat.1005464
Al-Hatmi, A. M., Normand, A. C., van Diepeningen, A. D., Hendrickx, M., de Hoog, G. S., and Piarroux, R. (2015a). Rapid identification of clinical members of Fusarium fujikuroi complex using MALDI-TOF MS. Future Microbiol. 10, 1939–1952. doi: 10.2217/fmb.15.108
Al-Hatmi, A. M., van Diepeningen, A. D., Curfs-Breuker, I., de Hoog, G. S., and Meis, J. F. (2015b). Specific antifungal susceptibility profiles of opportunists in the Fusarium fujikuroi complex. J. Antimicrob. Chemother. 70, 1068–1071. doi: 10.1093/jac/dku505
Atty, C., Alagiozian-Angelova, V. M., and Kowal-Vern, A. (2014). Black plaques and white nodules in a burn patient. Fusarium and Mucormycosis. JAMA Dermatol 150, 1355–1356. doi: 10.1001/jamadermatol.2014.2463
Castro López, N., Casas, C., Sopo, L., Rojas, A., del Portillo, P., Cepero de García, M. C., et al. (2009). Fusarium species detected in onychomycosis in Colombia. Mycoses 52, 350–356. doi: 10.1111/j.1439-0507.2008.01619.x
CLSI (2017). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi. CLSI Standard M38, 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (Ed.) (2020). “Epidemiological cutoff values for antifungal susceptibility testing,” in CLSI supplement M59. 3rd Edn. (Wayne, PA: Clinical and Laboratory Standards Institute)
Da, R. P., Aquino, V., Fuentefria, A. M., and Goldani, L. Z. (2021). Diversity of Fusarium species causing invasive and disseminated infections. J. Mycol. Med. 31:101137. doi: 10.1016/j.mycmed.2021.101137
Dallé da Rosa, P., Nunes, A., Borges, R., Batista, B., Meneghello Fuentefria, A., and Goldani, L. Z. (2018). In vitro susceptibility and multilocus sequence typing of Fusarium isolates causing keratitis. J. Mycol. Med. 28, 482–485. doi: 10.1016/j.mycmed.2018.05.001
Dalyan Cilo, B., al-Hatmi, A. M. S., Seyedmousavi, S., Rijs, A. J., Verweij, P. E., Ener, B., et al. (2015). Emergence of fusarioses in a university hospital in Turkey during a 20-year period. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1683–1691. doi: 10.1007/s10096-015-2405-y
Enoch, D. A., Yang, H., Aliyu, S. H., and Micallef, C. (2017). The changing epidemiology of invasive fungal infections. Methods Mol. Biol. 1508, 17–65. doi: 10.1007/978-1-4939-6515-1_2
Guarro, J. (2013). Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1491–1500. doi: 10.1007/s10096-013-1924-7
Guevara-Suarez, M., Cano-Lira, J. F., Cepero de García, M. C., Sopo, L., de Bedout, C., Cano, L. E., et al. (2016). Genotyping of Fusarium isolates from onychomycoses in Colombia: detection of two new species within the Fusarium solani species complex and in vitro antifungal susceptibility testing. Mycopathologia 181, 165–174. doi: 10.1007/s11046-016-9983-9
Herkert, P. F., al-Hatmi, A. M. S., de Oliveira Salvador, G. L., Muro, M. D., Pinheiro, R. L., Nucci, M., et al. (2019). Molecular characterization and antifungal susceptibility of clinical Fusarium species from Brazil. Front. Microbiol. 10:737. doi: 10.3389/fmicb.2019.00737
Karadag, A. S., Cebeci, F., Aslan Kayıran, M., Özakkaş, F., Çobanoğlu, B., Kuru, B. C., et al. (2020). Fusarium solani infection in a diabetic patient treated with itraconazole and debridement. Dermatol. Ther. 33:e14203. doi: 10.1111/dth.14203
Liza, D., Divya, D., Kirti, G., Mahesh, P., Bhanu, M., et al. (2021). Eumycetoma of the foot due to Fusarium solani in a person with diabetes mellitus: report of a case and review of literature. Mycopathologia 186, 277–288. doi: 10.1007/s11046-020-00524-y
Mezzalama, M., Guarnaccia, V., Martino, I., Tabome, G., and Gullino, M. L. (2021). First report of Fusarium commune causing root and crown rot on maize in Italy. Plant Dis. 105:4156. doi: 10.1094/PDIS-01-21-0075-PDN
Muhammed, M., Anagnostou, T., Desalermos, A., Kourkoumpetis, T. K., Carneiro, H. A., Glavis-Bloom, J., et al. (2013). Fusarium infection: report of 26 cases and review of 97 cases from the literature. Medicine (Baltimore) 92, 305–316. doi: 10.1097/MD.0000000000000008
Muraosa, Y., Oguchi, M., Yahiro, M., Watanabe, A., Yaguchi, T., and Kamei, K. (2017). Epidemiological study of Fusarium species causing invasive and superficial fusariosis in Japan. Med. Mycol. J. 58, E5–E13. doi: 10.3314/mmj.16-00024
Najafzadeh, M. J., Dolatabadi, S., de Hoog, S., Esfahani, M. K., Haghani, I., Aghili, S. R., et al. (2020). Phylogenetic analysis of clinically relevant Fusarium species in Iran. Mycopathologia 185, 515–525. doi: 10.1007/s11046-020-00460-x
Normand, A. C., Imbert, S., Brun, S., al-Hatmi, A. M. S., Chryssanthou, E., Cassaing, S., et al. (2021). Clinical origin and species distribution of Fusarium spp. isolates identified by molecular sequencing and mass spectrometry: a European multicenter hospital prospective study. J. Fungi. 7:246. doi: 10.3390/jof7040246
Nucci, M., and Anaissie, E. (2002). Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: implications for diagnosis and management. Clin. Infect. Dis. 35, 909–920. doi: 10.1086/342328
Nucci, M., and Anaissie, E. (2007). Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704. doi: 10.1128/CMR.00014-07
O’Donnell, K., Sutton, D. A., Fothergill, A., McCarthy, D., Rinaldi, M. G., Brandt, M. E., et al. (2008). Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46, 2477–2490. doi: 10.1128/JCM.02371-07
Oliveira, D. S. C., Kolwijck, E., van der Lee, H. A., Tehupeiory-Kooreman, M. C., al-Hatmi, A. M. S., Matayan, E., et al. (2019). In vitro activity of Chlorhexidine compared with seven antifungal agents against 98 Fusarium isolates recovered from fungal keratitis patients. Antimicrob. Agents Chemother. 63:e02669-18. doi: 10.1128/AAC.02669-18
Oliveira, D. S. C., Kolwijck, E., van Rooij, J., Stoutenbeek, R., Visser, N., Cheng, Y. Y., et al. (2020). Epidemiology and clinical management of Fusarium keratitis in the Netherlands, 2005-2016. Front. Cell. Infect. Microbiol. 10:133. doi: 10.3389/fcimb.2020.00133
Pai, R., Boloor, R., Shreevidya, K., and Shenoy, D. (2010). Fusarium solani: an emerging fungus in chronic diabetic ulcer. J. Lab. Physicians 2, 037–039. doi: 10.4103/0974-2727.66710
Paziani, M. H., Tonani Carvalho, L., Melhem, M. S. C., Almeida, M. T. G., Nadaletto Bonifácio da Silva, M. E., Martinez, R., et al. (2019). First comprehensive report of clinical Fusarium strains isolated in the state of Sao Paulo (Brazil) and identified by MALDI-TOF MS and molecular biology. Microorganisms 8:66. doi: 10.3390/microorganisms8010066
Ranque, S., Normand, A. C., Cassagne, C., Murat, J. B., Bourgeois, N., Dalle, F., et al. (2014). MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses 57, 135–140. doi: 10.1111/myc.12115
Rosa, P. D., Heidrich, D., Correa, C., Scroferneker, M. L., Vettorato, G., Fuentefria, A. M., et al. (2017). Genetic diversity and antifungal susceptibility of Fusarium isolates in onychomycosis. Mycoses 60, 616–622. doi: 10.1111/myc.12638
Rosa, P., Ramirez-Castrillon, M., Borges, R., Aquino, V., Meneghello, F. A., Zubaran, G. L., et al. (2019). Epidemiological aspects and characterization of the resistance profile of Fusarium spp. In patients with invasive fusariosis. J. Med. Microbiol. 68, 1489–1496. doi: 10.1099/jmm.0.001059
Rosanova, M. T., Brizuela, M., Villasboas, M., Guarracino, F., Alvarez, V., Santos, P., et al. (2016). Fusarium spp. infections in a pediatric burn unit: nine years of experience. Braz. J. Infect. Dis. 20, 389–392. doi: 10.1016/j.bjid.2016.04.004
Salah, H., Al-Hatmi, A. M., Theelen, B., Abukamar, M., Hashim, S., van Diepeningen, A. D., et al. (2015). Phylogenetic diversity of human pathogenic Fusarium and emergence of uncommon virulent species. J. Infect. 71, 658–666. doi: 10.1016/j.jinf.2015.08.011
Sleiman, S., Halliday, C. L., Chapman, B., Brown, M., Nitschke, J., Lau, A. F., et al. (2016). Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Aspergillus, Scedosporium, and Fusarium spp. in the Australian clinical setting. J. Clin. Microbiol. 54, 2182–2186. doi: 10.1128/JCM.00906-16
Song, Y., Liu, X., Yang, Z., Meng, X., Xue, R., Yu, J., et al. (2021). Molecular and MALDI-TOF MS differentiation and antifungal susceptibility of prevalent clinical Fusarium species in China. Mycoses 64, 1261–1271. doi: 10.1111/myc.13345
Sun, S., Lyu, Q., Han, L., Ma, Q., Hu, H., He, S., et al. (2015). Molecular identification and in vitro susceptibility of Fusarium from fungal keratitis in Central China. Zhonghua Yan Ke Za Zhi 51, 660–667. doi: 10.3760/cma.j.issn.0412-4081.2015.09.005
Taj-Aldeen, S. J., Gene, J., Al, B. I., Buzina, W., Cano, J. F., and Guarro, J. (2006). Gangrenous necrosis of the diabetic foot caused by Fusarium acutatum. Med. Mycol. 44, 547–552. doi: 10.1080/13693780500543246
Taj-Aldeen, S. J., Salah, H., Al-Hatmi, A. M., Hamed, M., Theelen, B., van Diepeningen, A. D., et al. (2016). In vitro resistance of clinical Fusarium species to amphotericin B and voriconazole using the EUCAST antifungal susceptibility method. Diagn. Microbiol. Infect. Dis. 85, 438–443. doi: 10.1016/j.diagmicrobio.2016.05.006
Tortorano, A. M., Richardson, M., Roilides, E., van Diepeningen, A., Caira, M., Munoz, P., et al. (2014). ESCMID and ECMM joint guidelines on diagnosis and management of hyalohyphomycosis: Fusarium spp., Scedosporium spp. and others. Clin. Microbiol. Infect. 20, 27–46. doi: 10.1111/1469-0691.12465
Tram, Q. A., Minh, N. T. N., Anh, D. N., Lam, N. N., Dung, T. N., Thi Minh Chau, N., et al. (2020). A rare case of fungal burn wound infection caused by Fusarium solani in Vietnam. J. Investig. Med. High Impact Case Rep. 8:232470962091212. doi: 10.1177/2324709620912122
Triest, D., Stubbe, D., De Cremer, K., Pierard, D., Normand, A. C., Piarroux, R., et al. (2015). Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of molds of the Fusarium genus. J. Clin. Microbiol. 53, 465–476. doi: 10.1128/JCM.02213-14
Tupaki-Sreepurna, A., Al-Hatmi, A. M., Kindo, A. J., Sundaram, M., and de Hoog, G. S. (2017). Multidrug-resistant Fusarium in keratitis: a clinico-mycological study of keratitis infections in Chennai, India. Mycoses 60, 230–233. doi: 10.1111/myc.12578
Van Diepeningen, A. D., and de Hoog, G. S. (2016). Challenges in Fusarium, a trans-kingdom pathogen. Mycopathologia 181, 161–163. doi: 10.1007/s11046-016-9993-7
Walther, G., Zimmermann, A., Theuersbacher, J., Kaerger, K., von Lilienfeld-Toal, M., Roth, M., et al. (2021). Eye infections caused by filamentous fungi: spectrum and antifungal susceptibility of the prevailing agents in Germany. J. Fungi 7:511. doi: 10.3390/jof7070511
Wang, H., Hou, X., Huang, X., Gao, M., Chen, T., Gao, Q., et al. (2022). First report of Fusarium commune causing leaf spot disease on Bletilla striata in China. Plant Dis. 106:1070. doi: 10.1094/PDIS-07-21-1486-PDN
Keywords: Fusarium, humans, sequence analysis, mass spectrometry, microbial sensitivity tests
Citation: Guo P, Chen J, Tan Y, Xia L, Zhang W, Li X, Jiang Y, Li R, Chen C, Liao K and Peng Y (2022) Comparison of molecular and MALDI-TOF MS identification and antifungal susceptibility of clinical Fusarium isolates in Southern China. Front. Microbiol. 13:992582. doi: 10.3389/fmicb.2022.992582
Received: 12 July 2022; Accepted: 22 August 2022;
Published: 10 October 2022.
Edited by:
Weihua Pan, Shanghai Changzheng Hospital, ChinaReviewed by:
Mahdi Abastabar, Mazandaran University of Medical Sciences, IranCopyright © 2022 Guo, Chen, Tan, Xia, Zhang, Li, Jiang, Li, Chen, Liao and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kang Liao, bGlhb2thbmcxOTcxQDE2My5jb20=; Yaqin Peng, cHlxZHJlYW1AMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.