- 1College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot, China
- 2Key Laboratory of Animal Clinical Treatment Technology, Ministry of Agriculture, Hohhot, China
- 3Xuzhou Vocational College of Bioengineering, Xuzhou, Jiangsu, China
Introduction: The unscientific and irrational use of antimicrobial drugs in dairy farms has led to the emergence of more serious drug resistance in Escherichia coli.
Methods: In this study, cases of calf diarrhea in cattle farms around the Hohhot area were studied, and Escherichia coli were identified by PCR and biochemical methods, while the distribution of virulence and drug resistance genes of the isolates was analyzed.
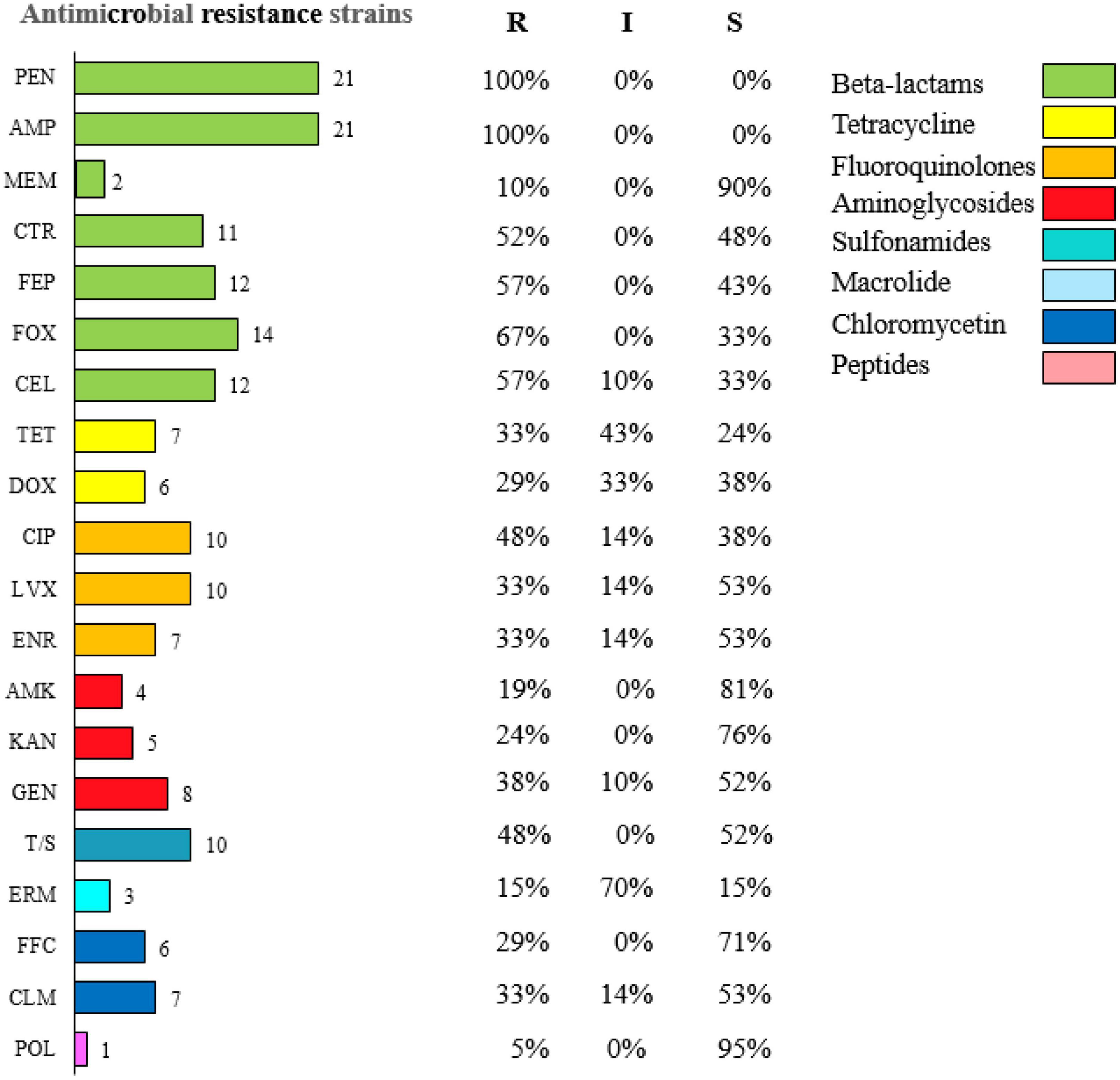
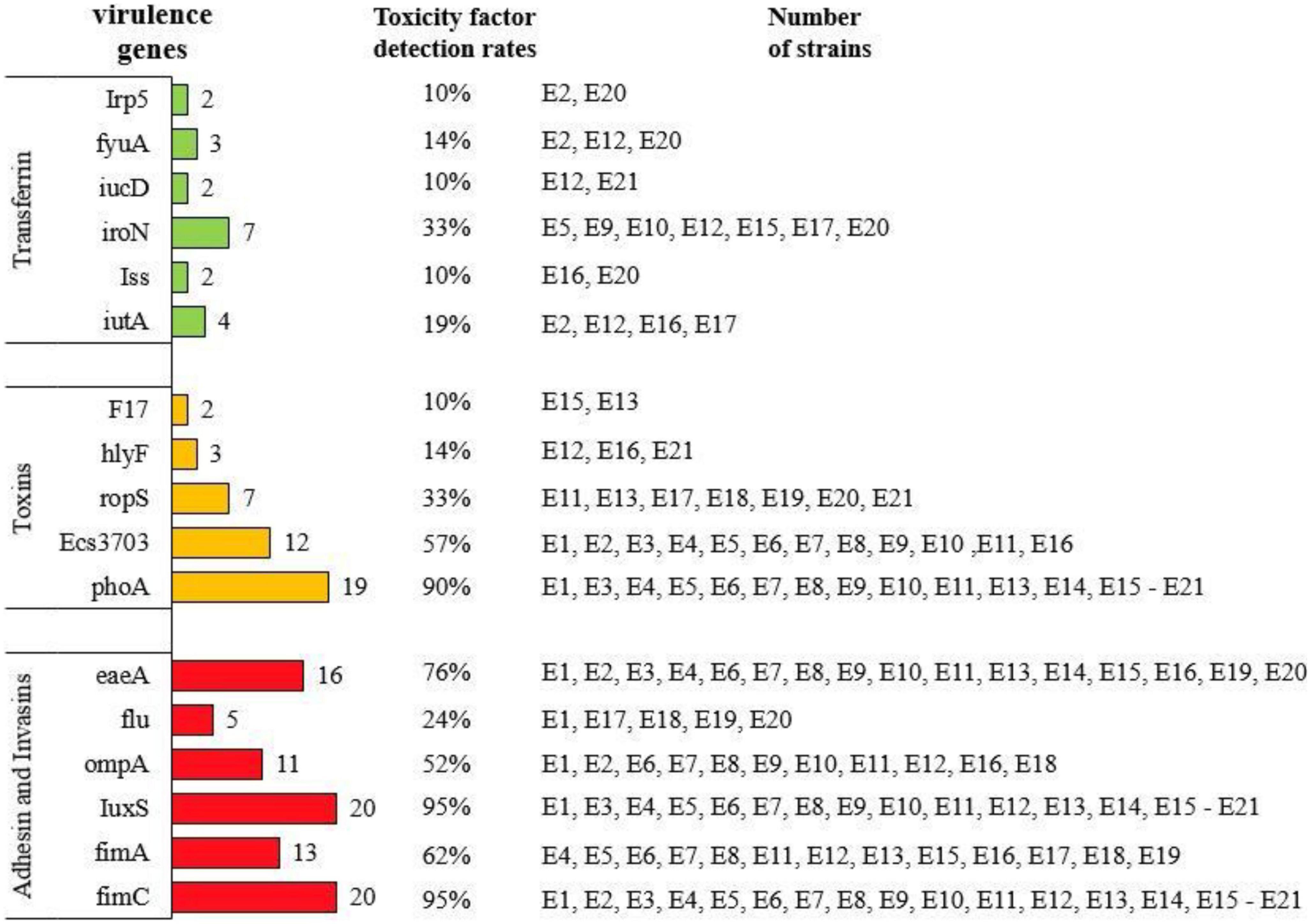
Results: The results showed that 21 strains of Escherichia coli were isolated from the diseased materials, and the isolation rate was 60%. The isolated strains belong to 15 ST types. The drug resistance levels of the isolated strains to 20 kinds of antimicrobial agent viz., penicillin, ampicillin, cefotaxime, cefepime, cefoxitin, and ceftriaxone were more than 50%. The resistance rate to meropenem was 10%. The resistance rates to tetracycline and doxycycline were 33% and 29%, to ciprofloxacin, levofloxacin and enrofloxacin were 48%, 33%, and 33%, to amikacin, kanamycin and gentamicin were 19%, 24% and 38%, to cotrimoxazole and erythromycin were 48% and 15%, to florfenicol, chloramphenicol and polymyxin B were 29%, 33%, and 5%. Nine strains of pathogenic calf diarrhea Escherichia coli were isolated by mouse pathogenicity test. The detection rates of virulence genes for the adhesion class were fimC (95%), IuxS (95%), eaeA (76%), fimA (62%), ompA (52%), and flu (24%). The detection rates for iron transporter protein like virulence genes were iroN (33%), iutA (19%), fyuA (14%), irp5 (9.5%), Iss (9.5%), and iucD (9.5%). The detection rates for toxin-like virulence genes were phoA (90%), Ecs3703 (57%), ropS (33%), hlyF (14%), and F17 (9.5%). The detection rates of tetracycline resistance genes in isolated strains were tetB (29%), tetA (19%) and tetD (14%). The detection rates for fluoroquinolone resistance genes were parC (Y305H, P333S, R355G) (9.5%), gyrA (S83L, D87N) (28%), qnrD (43%), and qnrS (9.5%). The detection rates for β-lactam resistance genes were blaCTX–M (29%), blaTEM (29%), and blaSHV (9.5%). The detection rates for aminoglycoside resistance genes were strA-B (57%), aacC (33%), aac(3′)-IIa (29%), and aadAI (24%). The detection rates of chloramphenicol resistance genes floR and sulfa resistance genes sul2 were 24 and 33%.
Conclusion: Pathogenic Escherichia coli causing diarrhea in calves contain abundant virulence genes and antibiotic resistance genes.
Introduction
Calf diarrhea can cause diarrhea in newborn calves and affect normal calf development. This disease is seasonal, with high morbidity and mortality (Hur et al., 2013). A variety of bacteria can cause diarrhea in calves, among which Escherichia coli (E. coli) is more common. The clinical manifestations are often light brown foul-smelling stools, and later gray watery stools containing bubbles and blood, accompanied by abdominal pain, and occasionally acute toxic cases, which can lead to death or growth retardation (Foster and Smith, 2009).
It is now customary to classify pathogenic E. coli as Enterohemorrhagic E. coli (EHEC)/Shiga-like toxigenic E. coli (STEC), which can secrete characteristic genes such as Shiga toxin (stx) and Hemolysin (Melton-Celsa et al., 2012). As well as Enteropathogenic E. coli (EPEC) causes adhesion and effacing (A/E) lesions in pathological small intestinal tissues via the outer membrane protein adhesion and effacing factor (eaeA), which is responsible for encoding a protein located in the enterocyte effacing locus. The EPEC attaching factor (EAF) plasmid encodes the type IV bundle forming pili (BFP) gene, which causes pathogenesis in the organism. The atypical EPEC (aEPEC) strain contains only eaeA genes. Also, the characteristic genes eaeA and bfp can be identified to determine whether E. coli belongs to EPEC (Katsowich et al., 2017). Enterotoxigenic E. coli (ETEC) producing enterotoxin can cause disease by producing adhesinogenic hairs K88, K99, 987P, F17, F18, F41, etc., and heat-stable enterotoxin (ST) and heat-unstable enterotoxin (LT) genes acting on intestinal epithelial cells. Whether E. coli belongs to ETEC was determined by identifying STa, STb, LTI, and LTII subtype genes among ST and LT signature genes, respectively. Enteric Aggregate E. coli (EAEC), Enteroinvasive E. coli (EIEC), and Diffusely adherent E. coli (DAEC) were identified by aggregative adhesion genes (aggR), invasive plasmid antigen H gene (ipaH), and Afa/Dr adhesin gene (Robins et al., 2016).
In veterinary practice, antibiotics are often used to treat bacterial calf diarrhea, however, with the continuous and irrational use of antibiotics, the spectrum of E. coli resistance is gradually expanding and resistance is becoming increasingly serious. The main mechanisms of drug resistance in E. coli include: inactivating or passivating enzymes produced by bacteria that can disrupt the structure of a drug or inhibit its antimicrobial activity (Deckert et al., 2010; Ghotaslou et al., 2014). The mode of bacterial drug resistance gene transmission is mainly plasmid-mediated (Paterson and Bonomo, 2005). The structure of the target of antimicrobial drug action is altered, resulting in the inability of the antimicrobial drug to bind to it (Andrade et al., 2014; Misawa et al., 2018). Bacteria pump antibacterial drugs out of the body by efflux pump (EP) (Fernandez and Hancock, 2013). E. coli reduces the accumulation of drugs in cells by altering the permeability of the extracellular membrane (Davies and Davies, 2010). Bacterial biofilm (BF) is regulated by population-sensing (QS)-related genes luxS, mqsR, regulation of type I hairs (fimB), stress regulators (rpoS), etc., to make bacteria resistant to drugs (Sharma et al., 2016).
In this study, we analyzed the pathogenicity, drug resistance, strain grouping, and strain sequence type of 21 E. coli strains from Hohhot, Inner Mongolia, to provide a test basis for the control of calf diarrhea caused by E. coli in the region. The virulence genes and drug resistance genes of the isolated strains were tested to facilitate the understanding of the distribution and prevalence of virulence genes in the sampled herds and the mechanisms of drug resistance of the bacteria in different cattle farms.
Materials and methods
Source of strains
Laboratory personnel used disposable samplers to collect anal swabs 35 calves comprising of 26 female and 9 male Holstein calves within 1 month of age with diarrhea symptoms from cattle farms of different sizes around Hohhot, and none of the sampled calves had been given antibiotic treatment. A total of 21 strains of E. coli, numbered E1–E21, were isolated from the 35 samples, and E. coli ATCC 25922 was kept in our laboratory as a reference strain for subsequent experiments.
Isolation culture
The 35 samples were first inoculated on normal medium to complete proliferation. Then they were purified and identified by E. coli selective medium Eosin-methylene blue medium (EMB) plates and MacConkey Agar (MAC) plates (All the above media were purchased from Qingdao Hi-tech Industrial Park Haibo Biotechnology Co., Qingdao City, China) (Kim et al., 2010).
Identification
The biochemical reactions were performed on the bacterial solution separately according to the biochemical identification tube instructions. The biochemical characteristics of E. coli were positive for maltose, glucose (GLU), mannitol and lactose (MAL), indole, and methyl red (M-R), and negative for acetyl methyl methanol (V-P), Citrate, urea, and hydrogen sulfide (H2S) were all negative. The E. coli-specific gene 16SrRNA was used as a primer to amplify the isolates. E. coli ATCC 25922 was used as the standard strain for all tests in this paper (Kim et al., 2010).
Multilocus sequence typing of E. coli
The housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA of isolates strains were amplified according to the E. coli multilocus sequence typing (MLST) typing scheme provided by the E. coli MLST database1 (Wirth et al., 2006).
Drug sensitivity testing
The drug susceptibility testing of isolates E1–E21 was performed by referring to the Kirby-Bauer disk agar diffusion method (K-B) paper diffusion method recommended by the Clinical and Laboratory Standards Institute [CLSI; Weinstein and Lewis (2020)]. The concentration of E. coli solution to be tested was adjusted to 1.5 × 108 CFU/ml by turbidimetry, and three replicate groups were made for each strain. The resistance pattern of ATCC 25922 to antibiotics is resistant to ampicillin. ATCC 25922 was used as the control group for the E. coli drug susceptibility test, and the criteria for the drug susceptibility test are shown in Supplementary Table 1; Kong et al., 2016.
Pathogenicity test in mice
The clean-grade mice were randomly divided into 21 test groups and 1 control group, with 4 mice in each group. The 21 test groups of mice were injected intraperitoneally with 0.4 ml of E1–E21 at a concentration of 1.5 × 108 CFU/ml, and the control mice were injected intraperitoneally with an equal amount of saline. The mice were observed continuously for 3 days. The time of onset and death was recorded, and the dead mice were dissected for clinical observation and microscopic examination. The experiments were performed using clean-grade Kunming mice, male and female, weighing about 20–22 g. The mice were purchased from the Animal Experiment Center of Inner Mongolia Medical University, Animal Certificate of Conformity No.: SCXK (Meng) 2020-0001.
Detection of virulence and resistance genes
Polymerase chain reaction was used to detect the virulence and resistance gene carriage of the isolates, and specific primer sequences were designed for the virulence and resistance gene-specific conserved regions of E. coli with reference to the literature. The primer sequences and amplification conditions of E. coli virulence and antibiotic resistance genes are shown in Supplementary Tables 2, 3; Paton and Paton, 1998; Kong et al., 1999; Park et al., 2006; Chase et al., 2008; Dallenne et al., 2010; Gündoğdu et al., 2011; Chen et al., 2012; Liu et al., 2016; Belaynehe et al., 2018; Gundran et al., 2019.
Results
Results of the identification of E. coli
E1–E21 were all consistent with the biochemical characteristics of E. coli. The positive control ATCC 25922 and the test samples E1–E21 both amplified specific bands close to the size of the target fragment 16SrRNA (1438 bp). the sequencing results of E1–E21 were compared by BLAST in NCBI and showed more than 99% similarity with the existing sequences in GenBank.
Results of drug sensitivity tests
The resistance test results of E1–E21 could be seen in Figure 1, the highest resistance rate to penicillin and ampicillin was 100%, and the resistance rate to cephalosporins was higher than 50%, but the resistance rate of E1–E21 to meropenem was 10%; The resistance rates to tetracycline and doxycycline were 33 and 29%, to ciprofloxacin, levofloxacin, and enrofloxacin were 48, 33, and 33%, to amikacin, kanamycin, and gentamicin were 19, 24, and 38%, to cotrimoxazole and erythromycin were 48 and 15%, to florfenicol, chloramphenicol, and polymyxin B were 29, 33, and 5%.
Results of multilocus sequence typing
As shown in Figure 2, E1 to E21 were classified into 15 ST types. E4, E16, E17, and E19 were ST10 types, which were the dominant types in this study. E9 and E10 were ST6594 types, E6 and E8 were ST362 types, and the remaining 13 strains of E. coli belonged to 13 sequence types, respectively. No new alleles or new sequence types were found.
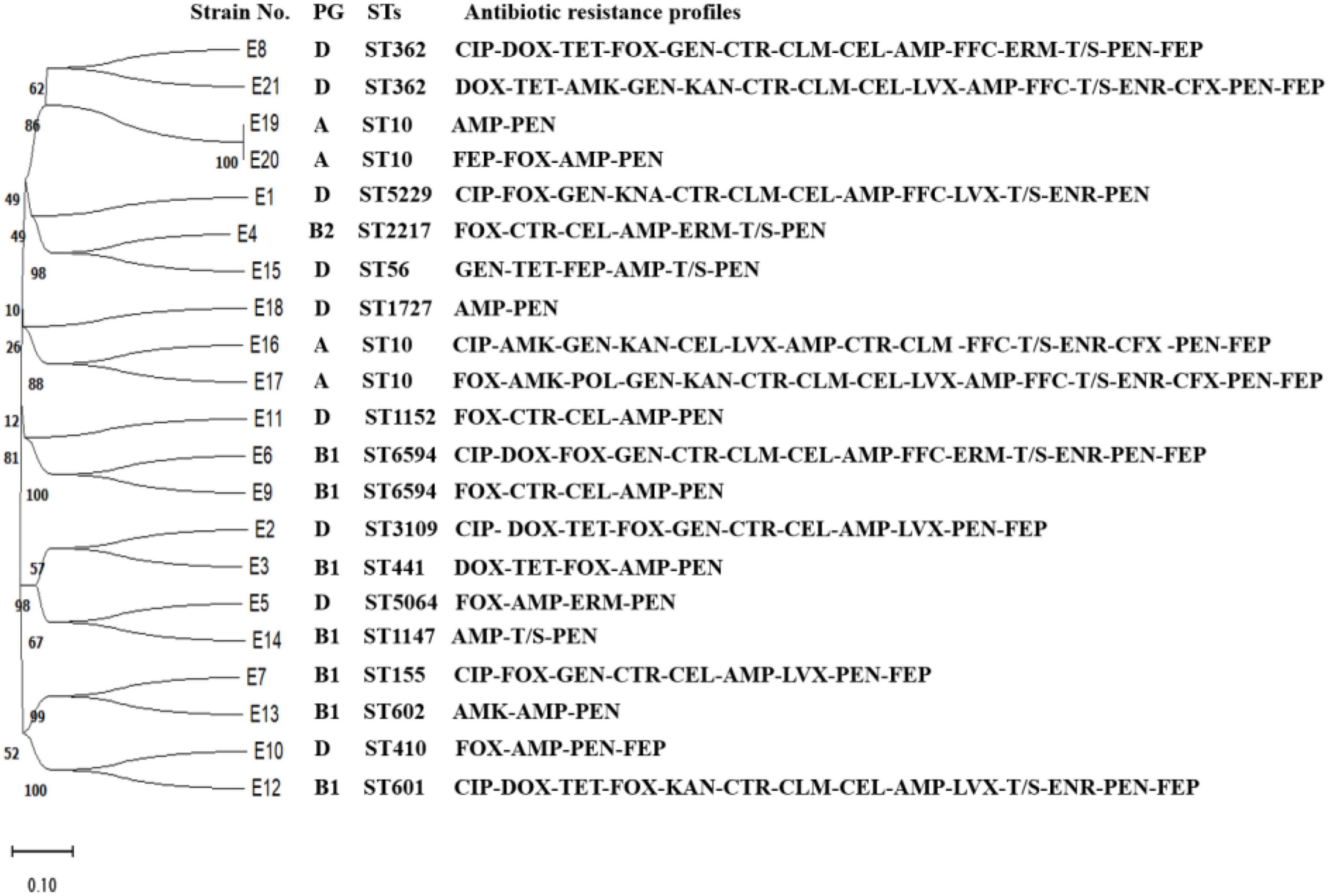
Figure 2. Evolutionary tree of ST types and drug resistance profile in diarrheagenic calves source Escherichia coli.
Results of pathogenicity tests in mice
The pathogenicity of E1–E21 was identified by a mouse pathogenicity test, and the results showed that E12, E20, E1, E2, E3, E6, E7, E8, and E16 isolates were 100, 100, 50, 75, 25, 25, 25, 75, and 25% lethal to mice, respectively.
The detection rate of virulence genes of E. coli in calves with diarrhea
As shown in Figure 3, the detection rates of eaeA, fimC, fimA, IuxS, ompA, and flu in the adhesion class of virulence factors from E1 to E21 were 76 (16/21), 95 (20/21), 62 (13/21), 95 (20/21), 52 (11/21), and 24% (5/21), respectively. The detection rates of fyuA, iroN, iutA, irp5, Iss, and iucD among iron transport protein-like virulence factors were 14 (3/21), 33 (7/21), 19 (4/21), 9.5 (2/21), 9.5 (2/21), and 9.5% (2/21), respectively. The detection rates of toxin-like virulence factors F17, hlyF, ropS, phoA, and Ecs3703 were 9.5 (2/21), 14 (3/21), 33 (7/21), 90 (19/21), and 57% (12/21), respectively.
Association between drug-resistant strains with mutations in the gryA and parC genes and drug resistance
E1, E2, E6, E12, and E21 were all highly resistant to enrofloxacin, levofloxacin, and ciprofloxacin, and only E8 was sensitive to enrofloxacin. All of the above six strains had S83L and D87N double-site mutations in gyrA, with E1 and E12 having three point mutations in parC, R355G, P333S, and Y305H.
The detection rate of resistance genes of E. coli in calves with diarrhea
As shown in Figure 4, the detection rates of tetracycline resistance genes tetA, tetB, and tetD in E1–E21 were 19% (4/21), 29% (6/21), and 14% (3/21), respectively. The detection rates of quinolone resistance genes gyrA, gyrB, parC, parE, qnrD, and qnrS were 76% (16/21), 95% (20/21), 95% (20/21), 100% (21/21), 43% (9/21), and 9.5% (2/21), respectively. The detection rate of mutants of gyrA (S83L and D87N) was 28%, and the detection rate of mutants of parC (Y305H, P333S, and R355G) was 9.5%. β-lactam resistance genes blaCTX–M, blaTEM, and blaSHV were detected at 29% (6/21), 29% (6/21), and 9.5% (2/21), respectively. The detection rate of chloramphenicol resistance gene floR was 24% (5/21). The detection rate of sul2, a sulfonamide resistance gene, was 33% (7/21). The aminoglycoside resistance genes aac(3′)-IIa, aacC, aadAI, and strA-B were detected at 29% (6/21), 33% (7/21), 24% (5/21), and 57% (12/21), respectively.
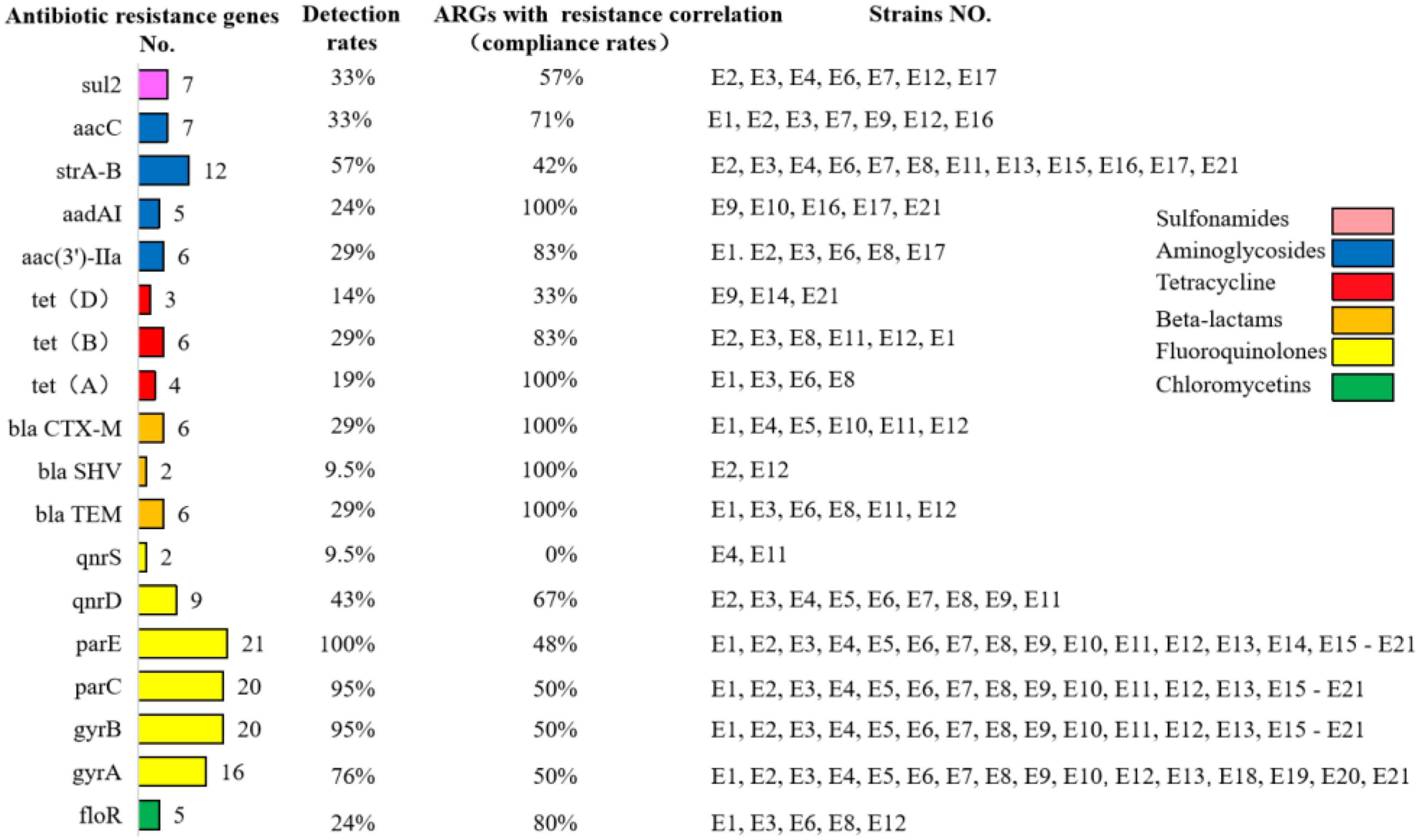
Figure 4. Detection rates of antibiotic resistance genes in diarrheagenic calves source Escherichia coli and compliance with correlation with antibiotic resistance correlation.
Distribution of virulence and drug resistance genes in E. coli with calf diarrhea
From Figure 5, eaeA, ompA, phoA, iutA, fimA/C, and IuxS are present in groups A, B1, B2, and D. Ecs3703 and iroN are distributed in groups B1, B2, and D in that order, and relatively less in group A. Iss is distributed in group A. hlyF is distributed in groups A and B1. aadAI, aac(3′)-IIa, strA-B, sul2, gyrA, gyrB, parC, and parE were found in all four groups. floR and qnrD were distributed in groups B1 and D. The distribution trends of virulence and resistance genes were generally consistent between E1–E21.

Figure 5. Comparison of virulence and resistance genes in diarrheagenic calves source Escherichia coli.
Correlation of antibiotic resistance profiles and resistance genes in calves diarrhea E. coli
The compliance rate of resistance among genotype-positive strains in E1–E21 was also positive. The phenotypically resistant antibiotics in the β-lactam class were penicillin, ampicillin, ceftriaxone, cefepime, cefoxitin, and ceftiofur, while their corresponding resistance genes blaCTX–M, blaTEM, and blaSHV were all 100% consistent with the resistance correlation. The phenotypically resistant antibiotics in the tetracycline class were tetracycline and doxycycline, and the corresponding prevalent resistance genes were tetA, tetB, and tetD with resistance correlations of 100, 83, and 33%, respectively. The phenotypically resistant antibiotics in the chloramphenicol class were florfenicol and chloramphenicol, and the corresponding prevalent resistance genes were floR with a resistance correlation rate of 80%. The phenotypically resistant antibiotics in the sulfonamides class were cotrimoxazole, corresponding to the prevalent resistance gene of sul2 and the correlation rate of resistance was 57%. The phenotypically resistant antibiotics among the aminoglycosides were streptomycin, kanamycin, and gentamicin, corresponding to the popular resistance genes strA-B, aac(3′)-IIa, aacC, and aadAI with resistance correlation rates of 42, 83, 71, and 100%, respectively. The phenotypically resistant antibiotics in the fluoroquinolones were ciprofloxacin, enrofloxacin, and levofloxacin, corresponding to the prevalent resistance genes gyrA/B, parC/E, and qnrD/S with resistance correlations of 50, 50, 50, 48, 67, and 0%, respectively. The prevalence of resistance and resistance genotypes in the tested strains corresponded to each other; however, due to the limited number of antimicrobials and resistance genes selected in this study, although some resistance genes and resistance phenotypes were more prevalent, it was not possible to determine whether they corresponded to their corresponding resistance phenotypes and resistance genotypes, respectively.
Discussion
In this study, the results of in vitro drug susceptibility tests of 20 antimicrobial drugs from E1–E21 were analyzed for resistance profiles, and it was found that the resistance profiles of the strains varied greatly, with strains that were more than 10 heavy resistance accounted for 38% of the total isolated strains. It can be seen that the current E. coli in Hohhot Holstein cattle farm is highly resistant to drugs. The resistance rates of E1–E21 to penicillin and ampicillin were 100%, and the resistance rates to cephalosporins were higher than 50%, however, the resistance rate of E1–E21 to meropenem was 10% similar to the results of domestic and international studies on calf-derived E. coli resistance to β-lactam antibiotics (Fazel et al., 2019). E1–E21 were 19, 24, and 38% resistant to amikacin, kanamycin, and gentamicin, respectively, while Algammal found 93.7% resistant to amikacin in calves with E. coli in Egypt (Algammal et al., 2020). The resistance rates of E1–E21 to polymyxin B and cotrimoxazole were 5 and 48%, respectively. This finding is consistent with the results of resistance studies on E. coli in calves with diarrhea in other countries (Khachatryan et al., 2004). This may be due to the fact that E. coli is more susceptible to resistance to sulfonamides. Although multidrug-resistant E. coli was shown to be sensitive to polymyxin by Poirel et al. (2017). The resistance rate of E1–E21 to tetracycline was 33%, while Carballo found 66.7% resistance to tetracycline in beef cattle E. coli (Carballo et al., 2013). It indicates that the distribution of resistance of various types of antimicrobial drugs to E. coli in calf diarrhea at home and abroad showed differences. The resistance to antimicrobial drugs varies among isolates. The epidemiology of drug-resistant E. coli in calves may be multifactorial, so veterinarians should use antimicrobial drugs appropriately according to the level of resistance of E. coli in different pastures.
In this study, the 21 E. coli strains were classified into 15 ST types, indicating that E. coli of Hohhot calf origin are characterized by genetic diversity and distant affinity among strains. Because of the limited number of tested strains in this study, it was not possible to accurately describe the association between each ST type category and the germline taxa. Genetic target genes of the corresponding pathogenicity species of IPEC were identified for E1–E21, eaeA, and bfp genes for EPEC; STa, STb, and LTI and LTII genes for ETEC; aggR gene for EAEC; ipaH gene for EIEC; Afa/Dr gene for DAEC; and stx1 and stx2 genes for EHEC/STEC. It was found that eaeA genes could be detected in 16 strains from E1 to E21, while the representative genes of other pathogenic species were not detected. This indicates that the main prevalent pathogenic type of E. coli in calves with diarrhea around the Hohhot area is atypical enteropathogenic E. coli (aEPEC).
Only sul2 (33%) and no sul1 and sul3 were detected among the sulfonamide resistance genes in E1–E21, indicating that the sul2 gene was predominantly prevalent in E. coli of Hohhot calf origin, a result consistent with the results of Blahna et al. (2006) who detected sul2 as the most prevalent gene in E. coli of European and Canadian cattle origin. In this study, mobile colistin resistance-1 (MCR-1) was not detected which is consistent with the results of Brennan et al. (2016) on the detection rate of MCR-1 in feces and milk of cattle in France and Germany and Bai et al. (2018) and Zheng et al. (2019) on the carriage rate of MCR-1 by E. coli in dairy farms in several regions of China. The MCR-1 of dairy-derived E. coli showed low levels of prevalence.
In this study, we examined multiple resistance genes contained in ultra broad-spectrum beta-lactamases (ESBLs) and AmpC enzymes. blaCTX–M, blaTEM, and blaSHV were detected at 29, 29, and 9.5%, respectively, and blaPSE, NDM-1, blaOXA, and AmpC were not detected. It has been shown that ESBLs and AmpC enzymes are widely prevalent in E. coli of animal origin with the increased frequency of β-lactamases antimicrobial drugs, with blaCTX–M and blaTEM types being the more widely distributed genotypes at present (Tangden et al., 2010; Gautam et al., 2019).
E1 to E21 carried tetA (19%), tetB (29%), and tetD (14%), and tetC and tetE were not detected. tetA (35%) and tetB (63%) were found to be the main prevalent resistance genes for tetracycline in different animal organisms of E. coli by Shapir et al. (2004). This may be due to the different prevalence of resistance genes in cows versus beef cattle. gyrA (76%), gyrB (95%), parC (95%), parE (100%), qnrD (43%), and qnrS (9.5%) were carried in E1–E21, and qnrA/B/C, oqxA/B, and qepA were not detected. Detection of these genes does not necessarily imply that they will exhibit resistance, as these genes may remain unexpressed in bacteria (Olonitola et al., 2015). No NDM-related genes were detected in ST410 E. coli in this study, but it carried drug resistance genes such as gyrA, gyrB, parC, parE, CTX-M, and aadAI (Gamal et al., 2016). In this study, amino acid substitutions in S83L and D87N were found to occur in the Quinolone-Resistant Determining Region (QRDR) region encoded by the gyrA gene in six strains, and amino acid substitutions in R355G, P333S, and Y305H were found for the first time in the parC subunit. It has been shown earlier that double mutation sites (S83L and D87N, D87G, or D87Y) exist in gyrA, while S80I, S80R, or E84K single point mutations exist in parC and S459A or N463D single point mutations are found in parE (Babus et al., 1998; Piddock, 1999). The amino acid substitution site of gyrA found in this study was consistent with related reports from home and abroad, while the type of amino acid substitution on the parC subunit was found for the first time.
The resistance and resistance gene carriage of E1–E21 showed roughly positive correlation, such as kanamycin, gentamicin, and amikacin with resistance genotypes aac(3′)-IIa, aacC, and aadAI conformed to 83, 71, and 100%, respectively. Streptomycin had 42% concordance with resistance genotype strA-B; tetracycline and doxycycline had 100, 83, and 33% concordance with resistance genotype tetA/B/D, respectively; florfenicol and chloramphenicol had 80% concordance with resistance genotype floR; cotrimoxazole had 57% concordance with resistance genotype sul2; penicillin, ampicillin, and cephalosporins had 100% concordance with resistance blaCTX–M, blaSHV, and blaTEM were all 100%; ciprofloxacin, levofloxacin, and enrofloxacin were 50, 50, 50, 48, 67, and 0%, respectively with the resistant genotypes gyrA/B, parC/E, and qnrD/S. The strains with amino acid substitutions in the parC and gyrA subunits in this test showed severe resistance to enrofloxacin, levofloxacin, and ciprofloxacin. The amino acid substitutions on the parC and gyrA subunits of strains with the same resistance phenotype varied greatly. It has been suggested that carrying the corresponding resistance genes may lead to the development of corresponding drug resistance (Han et al., 2013). The lower detection rate of genes encoding β-lactam resistance than phenotypic resistance in this study may be attributed to the fact that other β-lactamase genes were not studied or other resistance mechanisms were not addressed.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors. The data presented in the study are deposited in the GenBank Overview (nih.gov) repository, accession number 2654460.
Ethics statement
All animal experiments were performed according to regulations of the Administration of Affairs Concerning Experimental Animals in China. The experimental protocol was approved by the Animal Welfare and Research Ethics Committee of the Inner Mongolia Agricultural University (Approval ID: NND202103).
Author contributions
WM, BL, SZ, XX, and JC: preparation, creation, presentation of the published work, and specifically writing the initial draft (including substantive translation). YJ: methodology, data analysis, and original draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Research and development of key technology for prevention and control of bovine diarrhea disease and demonstration and promotion of standardized operation protocols (2020ZD0006).
Acknowledgments
We are grateful for the support of the research and development of key technologies for the prevention and control of bovine diarrhea and the demonstration and promotion project of standardized operating procedures. As well as the supported by program for young talents of science and technology in universities of Inner Mongolia autonomous region (NJYT22041).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.992111/full#supplementary-material
Footnotes
References
Algammal, A. M., El-Kholy, A. W., Riad, E. M., Mohamed, H. E., Elhaig, M. M., Yousef, S. A. A., et al. (2020). Genes encoding the virulence and the antimicrobial resistance in enterotoxigenic and shiga-toxigenic E. coli isolated from Diarrheic Calves. Toxins 12, 383–396. doi: 10.3390/toxins12060383
Andrade, J. C., Morais Braga, M. F., Guedes, G. M., Tintino, S. R., Freitas, M. A., Menezes, I. R., et al. (2014). Enhancement of the antibiotic activity of aminoglycosides by alph-tocopherol and other cholesterol derivates. Biomed. Pharmacother. 68, 1065–1069. doi: 10.1016/j.biopha.2014.10.011
Babus, S. D., Bebear, C. M., Charron, A., Bébéar, C., and de Barbeyrac, B. (1998). Squencing of grase and topoi somerase IV quinolone resistance determining regions of Chamydia trachomatis and characterization of quinolone resistant nlatants obtained in vitro. Antimicrob. Agents Chemother. 42, 2474–2481. doi: 10.1128/AAC.42.10.2474
Bai, F., Li, X., Niu, B., Zhang, Z., Malakar, P. K., Liu, H., et al. (2018). A mcr-1-carrying conjugative IncX4 plasmid in colistin-resistant Escherichia coli ST278 strain isolated from dairy cow feces in Shanghai, China. Front. Microbiol. 9:2833. doi: 10.3389/fmicb.2018.02833
Belaynehe, K. M., Shin, S. W., and Yoo, H. S. (2018). Interrelationship between tetracycline resistance determinants, phylogenetic group affiliation and carriage of class 1 integrons in commensal Escherichia coli isolates from cattle farms. BMC Vet. Res. 14:340. doi: 10.1186/s12917-018-1661-3
Blahna, M. T., Zalewski, C. A., Reuer, J., Kahlmeter, G., Foxman, B., and Marrs, C. F. (2006). The role of horizontal gene transfer in the spread of trimethoprim sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57, 666–672. doi: 10.1093/jac/dkl020
Brennan, E., Martins, M., McCusker, M. P., Wang, J., Alves, B. M., Hurley, D., et al. (2016). Multidrug resistant Escherichia coli in Bovine animals, Europe. Emerg. Infect. Dis. 22, 1650–1652. doi: 10.3201/eid2209.160140
Carballo, M., Esperon, F., Sacriston, C., González, M., Vázquez, B., Aguayo, S., et al. (2013). Occurrence of tetracycline residues and antimicrobial resistance in gram negative bacteria isolates from cattle farms in Spain. Adv. Biosci. Biotechnol. 4, 295–303. doi: 10.4236/abb.2013.42A040
Chase, E. G., Thenesa, J. O., Chistopher, M. W., Barletta, F., and Cleary, T. G. (2008). Detection of Diarrheagenic Escherichia coli by use of melting curve analysis and real time multiplex PCR. J. Clin. Microbiol. 46, 1752–1757. doi: 10.1128/JCM.02341-07
Chen, X., Zhang, W., Pan, W., Yin, J., Pan, Z., Gao, S., et al. (2012). Prevalence of qnr, aac (6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 56, 3423–3427. doi: 10.1128/AAC.06191-11
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Davies, J., and Davies, D. (2010). Origins and evolution of antimicrobial resistance. Microbiol. Mol. Biol. Rev. 74, 417–433.
Deckert, A., Gow, S., Rosengren, L., Léger, D., Avery, B., Daignault, D., et al. (2010). Canadian integrated program for antimicrobial resistance surveillance (CIPARS) farm program:Results from finisher pig surveillance. Zoonoses Public Health 57, 71–84.
Fazel, F., Jamshidi, A., and Babak, K. (2019). Phenotypic and genotypic study on antimicrobial resistance patterns of E. coli isolates from bovine mastitis. Microbial. Pathog. 132, 355–361. doi: 10.1016/j.micpath.2019.05.018
Fernandez, L., and Hancock, R. E. W. (2013). Adaptive and mutational resistance: Role of forms and efflux pumps in drug resistance. Clin. Microbiol. Rev. 26, 163–163. doi: 10.1186/1471-2180-14-185
Foster, D. M., and Smith, G. W. (2009). Pathophysiology of diarrhea in Calves. Vet. Clin. North Am. Food Anim. Pract. 25, 13–36. doi: 10.1016/j.cvfa.2008.10.013
Gamal, D., Fernandez, M., Defrawy, I., Ocampo-Sosa, A. A., and Martínez-Martínez, L. (2016). First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg. Microbes Infect. 5, 30–32. doi: 10.1038/emi.2016.24
Gautam, V., Thakur, A., Sharma, M., Singh, A., Bansal, S., Sharma, A., et al. (2019). Molecular characterization of extended spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: A multicentric study from tertiary care hospitals in India. Indian J. Med. Res. 149, 208–215. doi: 10.4103/ijmr.IJMR_172_18
Ghotaslou, R., Aghazadeh, M., Ahangarzadeh Rezaee, M., Moshafi, M. H., Forootanfar, H., Hojabri, Z., et al. (2014). The prevalence of aminoglycoside-modifying enzymes among coagulase-negative staphylococci in Iranian pediatric patients. J. Infect. Chemother. 20, 569–573. doi: 10.1016/j.jiac.2014.05.004
Gündoğdu, A., Long, Y. B., Vollmerhausen, T. L., and Katouli, M. (2011). Antimicrobial resistance and distribution of sul genes and integron associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J. Med. Microbiol. 60, 1633–1642. doi: 10.1099/jmm.0.034140-0
Gundran, R. S., Cardenio, P. A., Villanueva, M. A., Sison, F. B., Benigno, C. C., Kreausukon, K., et al. (2019). Prevalence and distribution of blaCTX–M, blaSHV, blaTEM genes in extended spectrum β-lactamase producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 15:227. doi: 10.1186/s12917-019-1975-9
Han, J. H., Nachamkin, I., Tolomeo, P., Mao, X., Bilker, W. B., and Lautenbach, E. (2013). Temporal changes in resistance mechanisms in colonizing Escherichia coli isolates with reduced susceptibility to fluoroquinolones. Diagn. Microbiol. Infect. Dis. 76, 491–496. doi: 10.1016/j.diagmicrobio.2013.04.018
Hur, T. Y., Jung, Y. H., Choe, C. Y., Cho, Y. I., Kang, S. J., Lee, H. J., et al. (2013). The dairy calf mortality: The causes of calf death during ten years at a large dairy farm in Korea. Korean J. Vet. Res. 53, 103–108. doi: 10.14405/kjvr.2013.53.2.103
Katsowich, N., Elbaz, N., Pal, R. R., Mills, E., Kobi, S., Kahan, T., et al. (2017). Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355, 735–739.
Khachatryan, A. R., Hancock, D. D., Besser, T. E., and Call, D. R. (2004). Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in Dairy Calves. Appl. Environ. Microbiol. 70, 752–757. doi: 10.1128/AEM.70.2.752-757.2004
Kim, T. W., Kim, Y. H., Kim, S. E., Lee, J. H., Park, C. S., and Kim, H. Y. (2010). Identification and distribution of Bacillus species in Doenjang by whole cell protein patterns and 16SrRNA Gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214. doi: 10.4014/jmb.1002.02008
Kong, C., Chen, J., Zou, X., Zhang, Y., and Zhu, L. (2016). Study on drug sensitivity against comamonas testosteroni by kirby-bauer disk diffusion method. J. Cent. South Univ. 41, 856–859. doi: 10.11817/j.issn.1672-7347
Kong, R. Y. C., So, C. L., Law, W. F., and Wu, R. S. S. (1999). A sensitive and versatile multiplex PCR system for the rapid detection of enterotoxigenic (ETEC), enterohaemorrhagic (EHEC) and enteropathogenic (EPEC) strains of Escherichia coli. Mar. Pollut. Bull. 38, 1207–1215. doi: 10.1016/S0025-326X(99)00164-2
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Melton-Celsa, A., Mohawk, K., Teel, L., and O’Brien, A. (2012). Pathogenesis of Shiga toxin producing Escherichia coli. Curr. Top. Microbiol. Immunol. 357, 67–103.
Misawa, K., Tarumoto, N., Tamura, S., Osa, M., Hamamoto, T., Yuki, A., et al. (2018). Single nucleotide polymorphisms in genes encoding penicillin-binding proteins in β-lactamase negative ampicillin-resistant Haemophilus influenzae in Japan. BMC Res. Notes 11:53. doi: 10.1186/s13104-018-3169-0
Olonitola, O. S., Fahrenfeld, N., and Pruden, A. (2015). Antibiotic resistance profiles among mesophilic aerobic bacteria in Nigerian chicken litter and associated antibiotic resistance genes. Poult. Sci. 94, 867–874. doi: 10.3382/ps/pev069
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., and Hooper, D. C. (2006). Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Paterson, D. L., and Bonomo, R. A. (2005). Extended spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 18, 657–686.
Paton, A. W., and Paton, J. C. (1998). Detection and characterization of shiga Toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J. Clin. Microbiol. 36, 598–602. doi: 10.1128/JCM.36.2.598-602
Piddock, L. J. (1999). Mechanisms of fIuoroquinolone resistance: An update 1994-1998. Drugs 58, 11–18. doi: 10.2165/00003495-199958002-00003
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Robins, R. M., Holt, K. E., Ingle, D. J., Hocking, D. M., Yang, J., and Tauschek, M. (2016). Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing. Front. Cell. Infect. Microbiol. 6:141. doi: 10.3389/fcimb.2016.00141
Shapir, N., Bryan, A., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/AEM.70.4
Sharma, G., Sharma, S., Sharma, P., Chandola, D., Dang, S., Gupta, S., et al. (2016). Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 121, 309–319. doi: 10.1111/jam.13078
Tangden, T., Cars, O., and Melhus, A. (2010). Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M type extendedspectrum beta-lactamases: A prospective study with Swedish volunteers. Antimicrob. Agents Chemother. 54, 3564–3568. doi: 10.1128/AAC.00220-10
Weinstein, M. P., and Lewis, J. S. (2020). Commentary: The clinical and laboratory standards institute (CLSI) subcommittee on antimicrobial susceptibility testing: Background, organization, functions, and processes. J. Clin. Microbiol. 58:e01864-19. doi: 10.1128/JCM.01864-19
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958
Keywords: Escherichia coli, calf diarrhea, drug resistance genes, virulence genes, antimicrobial resistance genes
Citation: Jia Y, Mao W, Liu B, Zhang S, Cao J and Xu X (2022) Study on the drug resistance and pathogenicity of Escherichia coli isolated from calf diarrhea and the distribution of virulence genes and antimicrobial resistance genes. Front. Microbiol. 13:992111. doi: 10.3389/fmicb.2022.992111
Received: 12 July 2022; Accepted: 01 December 2022;
Published: 22 December 2022.
Edited by:
Yue Xie, Sichuan Agricultural University, ChinaReviewed by:
Dharmendra Sinha, Indian Veterinary Research Institute (IVRI), IndiaJoshua Mbanga, National University of Science and Technology, Zimbabwe
Copyright © 2022 Jia, Mao, Liu, Zhang, Cao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojing Xu, ✉ eHV4aWFvamluZzcyQHNpbmEuY29t; Jinshan Cao, ✉ amluc2hhbmNhb0BpbWF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Yan Jia
Yan Jia Wei Mao1,2†
Wei Mao1,2† Bo Liu
Bo Liu Jinshan Cao
Jinshan Cao