
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 November 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.991703
This article is part of the Research Topic Molecular Diagnostics for Infectious Diseases: Novel approaches, Clinical Applications and Future Challenges View all 15 articles
 Junli Zhang1,2†
Junli Zhang1,2† Zhengan Wang1,2†
Zhengan Wang1,2† Yan Chen1,2
Yan Chen1,2 Zhihui Zhou1,2
Zhihui Zhou1,2 Qing Yang3
Qing Yang3 Ying Fu2,4
Ying Fu2,4 Feng Zhao2,4
Feng Zhao2,4 Xi Li5
Xi Li5 Qiong Chen6
Qiong Chen6 Li Fang1,2
Li Fang1,2 Yan Jiang1,2*
Yan Jiang1,2* Yunsong Yu1,2*
Yunsong Yu1,2*Cryptococcus spp. is a complex species that often causes cryptococcosis, which is one of the most common opportunistic infections in adults living with HIV and has very high morbidity and mortality rates. This study aimed to investigate the antifungal susceptibility profiles and epidemiological characteristics of the Cryptococcus neoformans species complex (CNSC) and the Cryptococcus gattii species complex (CGSC) in Zhejiang Province, China. A total of 177 CNSC and 3 CGSC isolates were collected, and antifungal susceptibility was tested by FUNGUS 3 and verified with an E-test. Moreover, multiple classification methods and genomic analyses were performed. The majority of the isolates (96.11%) were C. neoformans (formerly C. neoformans var. grubii) (ST5-VNI-A-α). Our study highlights that most of the patients with cryptococcosis were non-HIV patients in China, and nearly half of them did not have underlying diseases that led to immune insufficiency. Most of the Cryptococcus spp. isolates in this study were sensitive to common antifungal drugs. Two 5-flucytosine (5-FC)-resistant strains were identified, and FUR1 mutation was detected in the 5-FC-resistant isolates. Typing based on whole-genome sequencing (WGS) showed better discrimination than that achieved with multilocus sequence typing (MLST) and indicated a clear population structure. A phylogenetic analysis based on WGS included more genomic information than traditional classification methods.
Cryptococcosis is primarily caused by infection with the Cryptococcus neoformans species complex (CNSC) or Cryptococcus gattii species complex (CGSC). CNSC has been classified as C. neoformans (serotype A, formerly C. neoformans var. grubii) which includes four genotypes: VNI, VNII, VNBI, and VNBII, C. deneoformans (serotype D, genotype VNIV, formerly C. neoformans var. neoformans) and C. neoformans × C. deneoformans hybrid (serotype AD, genotype VNIII). C. gattii was classified into distinct species, including C. gattii (genotype VGI, formerly C. neoformans var. gattii), C. deuterogattii (VGII), C. bacillisporus (VGIII), C. decagattii (VGIIIc/VGIV hybrid), C. tetragattii (VGIV) and other unnamed species (VGV), which can be classified into serotypes B or C (Hagen et al., 2015; Kwon-Chung et al., 2017; Montoya et al., 2021). Cryptococcus spp. often causes clinical invasive fungal diseases (IFDs), such as cryptococcal meningitis/meningoencephalitis (CM), pulmonary cryptococcosis and cryptococcal sepsis, and among these, CM is the most common and serious type of cryptococcosis. It is estimated that 223,100 cases of cryptococcal meningitis are reported worldwide every year (Rajasingham et al., 2017). Cryptococcosis is known as one of the most common opportunistic infections in adults living with HIV (Rajasingham et al., 2017), but it also occurs in non-HIV populations with underlying diseases, such as diabetes, chronic liver disease, kidney disease, lung disease, malignancies, long-term steroid therapy or solid organ transplants, that may affect the immune status (Xiaobo Feng et al., 2008; Liaw et al., 2010; Zhu et al., 2010; Williamson et al., 2017).
For the clinical treatment of cryptococcosis, particularly CM, the combination of amphotericin B (AMB) and 5-flucytosine (5-FC) followed by fluconazole (FLC) maintenance therapy is recommended (Perfect et al., 2010; Liu et al., 2018). Although research on antifungal drug resistance is important, there is no defined clinical breakpoint (CBP) for antifungal agents except AMB (resistant if MIC >1 mg/L). The epidemiological cut-off values (ECOFFs) indicated in the European Committee on Antimicrobial Susceptibility Testing (2022) guidelines for wild-type (WT) C. neoformans are defined as MIC ≤1 mg/L for AMB and MIC ≤0.5 mg/L for posaconazole and voriconazole, and no ECOFFs for other antifungal agents have been established. For C. gattii, the ECOFFs are 0.5 mg/L and 1 mg/L for AMB and posaconazole, respectively, no ECOFFs have been indicated for other antifungal agents, and no CBPs have been reported for any antifungal agents (European Committee on Antimicrobial Susceptibility Testing, 2022). It is very important to further study the mechanisms of resistance to these drugs because this resistance increases the failure rate of induction treatment (Perfect et al., 2010; May et al., 2016). Further studies are needed to aid the establishment of cryptococcal CBPs in the future.
Several molecular typing systems have been applied for Cryptococcus spp. The commonly used methods include genotype (Meyer et al., 2009), serotype (T G Mitchell JRP., 1995), and mating type (Chaturvedi et al., 2000) analyses. The genotype can be determined with many methods. In this study, we used two band-based methods, M13-based PCR fingerprinting (FP) and URA5-RFLP analysis, and two sequence-based methods, MLST and whole-genome sequencing (WGS). The serotype and mating type can be identified by target sequencing using PCR. MLST (Meyer et al., 2009) provides high discriminatory power, has good repeatability across different laboratories, and is inexpensive. Thus, MLST is a major and important method for strain typing in epidemiological studies of Cryptococcus spp. (Meyer et al., 2009). However, the MLST system (Meyer et al., 2009) is not suitable for the molecular typing of hybrids because it fails to amplify certain alleles (Li et al., 2012; Samarasinghe and Xu, 2018; Cogliati et al., 2020). To overcome this problem, new primers were designed to enable the typing of AD hybrids in 2020 (Cogliati et al., 2020).
Multilocus sequence typing was used to determine the internal 469 ~ 723-bp nucleotide sequence of seven housekeeping genes for further typing (Meyer et al., 2009). ST5/VNI is the major molecular type of CNSC in China. Its appearance suggests low genetic diversity because the current genotyping method covers few genes, but the genomes of Cryptococcus neoformans var. neoformans JEC21 and Cryptococcus gattii WM276 are 18.57 Mb and 18.4 Mb, respectively, and include 14 chromosomes (Loftus et al., 2005; D’Souza et al., 2011). Previous analysis showed that ST5 has different subtypes, which was observed following the detection of single nucleotide polymorphism (SNPs) variants (Zhou et al., 2022), and WGS can further be used to analyse resistance mechanisms and simultaneously acquire other typing features (Zang et al., 2022; Zhou et al., 2022). Considering the complexity of Cryptococcus spp. genomes and the development of genome sequencing technology, typing based on WGS has achieved great success in the discrimination of species (Firacative et al., 2016; Wongsuk et al., 2020) and may further reveal the evolutionary relationship and population structure of Cryptococcus spp. isolates.
In previous studies of Cryptococcus spp. in Zhejiang, 98.1% (51/52) of cryptococcemia isolates were identified as C. neoformans var. grubii ST5-VNI-α. Most patients (64.80–71.4%) with cryptococcosis were not infected with HIV, according to data from a general hospital in Zhejiang Province, which is also a designated HIV hospital (Fang et al., 2020; Zhao et al., 2021). To collect more strains to further and more comprehensively explore the clinical characteristics, drug susceptibility and molecular characteristics of Cryptococcus spp. in Zhejiang Province, we collected all preserved strains of clinically isolated Cryptococcus spp. in 4 hospitals in Zhejiang Province from 2012 to 2018, and antimicrobial susceptibility testing (AST), molecular typing, and genomic analysis were performed.
In total, 180 nonrepetitive isolates were collected from four hospitals (99, 64, 14, and 3 from each hospital) in Zhejiang Province in China, and these included 177 CNSC isolates and three CGSC isolates. All 180 patients were diagnosed as exhibiting positive Cryptococcus spp. culture; among these, 68.33% (123/180), 16.67% (30/180) and 13.89% (25/180) were diagnosed with cryptococcal meningitis, pulmonary cryptococcosis and sepsis, respectively, and 1 patient diagnosed with soft tissue infection and 1 patient diagnosed with abdominal infection were also observed. Correspondingly, 123 strains, which included 3 isolates of CGSC, were isolated from the cerebrospinal fluid, and 30, 25, one and one strains were isolated from lung-related specimens, blood culture, skin-soft tissue, and ascites, respectively.
The patients were classified into three groups based on immune status: HIV-associated (12.78%, 23/180), non-HIV but with underlying diseases that may influence the immune system (45.55%, 82/180), and non-HIV and without underlying diseases (41.67%, 75/180). The number of HIV-related cases of infection was lower than the number of cases observed in the other two groups, as shown in Figure 1. Pearson correlation analysis was performed to assess the increasing trend of each group over time. No statistical significance was observed in each group (p > 0.5).
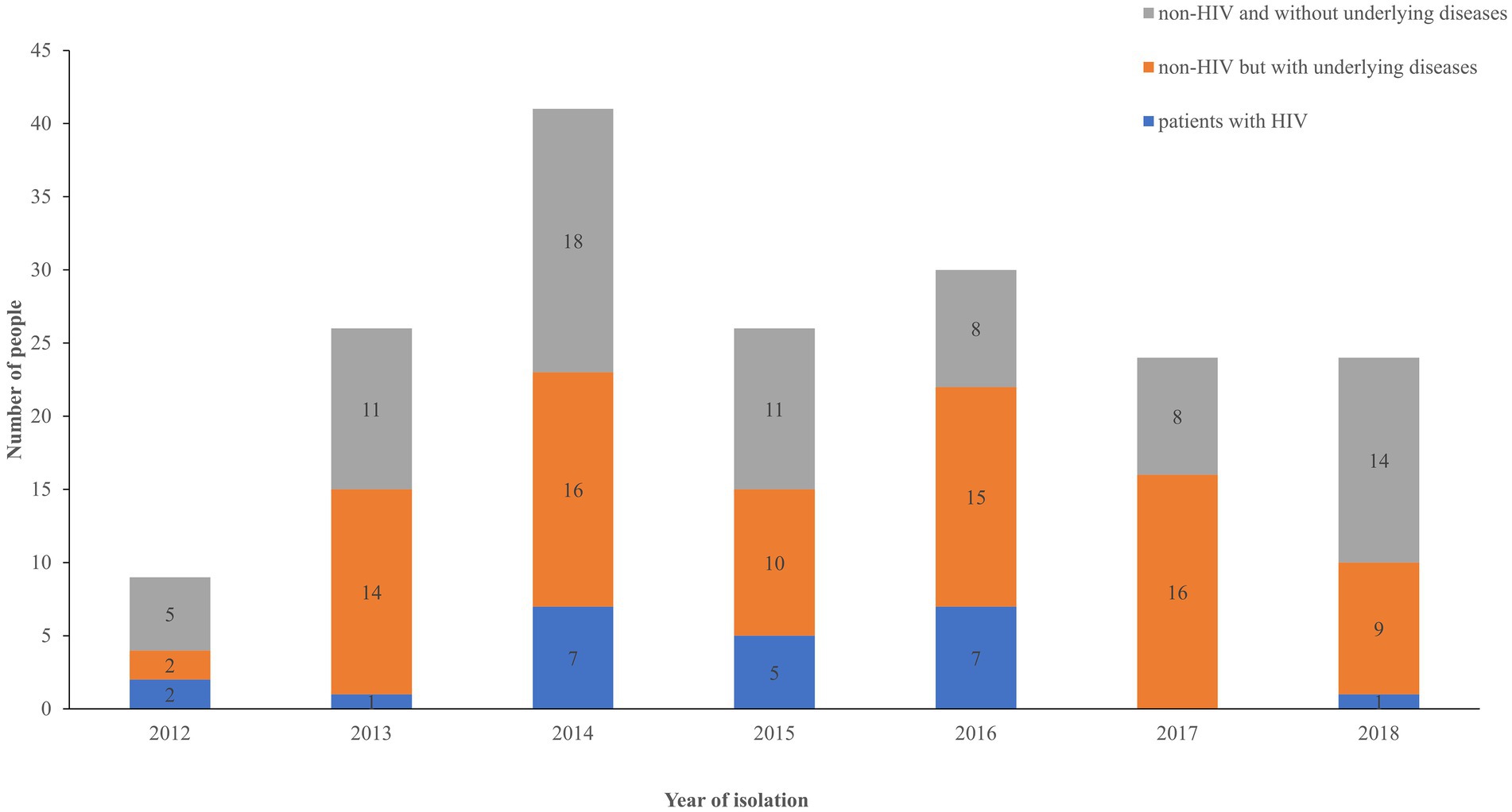
Figure 1. Distribution of patients among the three groups over time. Blue, HIV group; orange, non-HIV group with underlying diseases; gray, non-HIV group without underlying diseases.
Among 177 CNSC isolates, 173 were C. neoformans represented by the genotype VNI, serotype A, and MAT α. Another 4 isolates of CNSC were AD hybrids: RM007 was a hybrid of VNIII-AD, MAT α/a; ZY056 and SR017 were hybrids of VNIII-AD-α; and SR008 was also an AD-MAT α hybrid. Notably, the genotypes of SR008 that were observed using the two genotyping methods were inconsistent; VNIII was genotyped by M13-based PCR fingerprinting (FP), whereas the result of URA5-RFLP was VNIV. The genotyping electrophoresis diagrams of 4 AD hybrids and four reference strains are shown in Supplementary Figures S1A,B. Three isolates of CGSC exhibited the same mating type, MAT α, but different genotypes, including VGI (n = 1) and VGII (n = 2).
Through MLST typing, 173 isolates of C. neoformans were identified into 5 STs: ST5 was the predominant ST (n = 155), followed by ST31 (n = 8), ST79 (n = 5), ST81 (n = 3) and ST359 (n = 2). The CGSC isolates (n = 3) included three STs, namely, ST215 (RM009 VGI, from Taizhou), ST328 (ZY002 VGII, from Shaoxing) and the novel ST577 (SR06 1 VGII, from Taizhou). However, this approach failed to type the 4 AD hybrids even with the newly designed primers (Cogliati et al., 2020). Specifically, 12 ATs, including GPDVNIV 4, PLBVNIV 2, LACVNI 2, IGSVNI 1, IGSVNIV 2, and CAP59VNI 1, could not be amplified.
The phylogenetic tree obtained based on WGS grouped the 180 isolates distinctly, which indicated that the CNSCs and CGSCs belonged to two major clusters (Figure 2). Furthermore, CNSCs were classified into three subclusters: the ST5 cluster, ST31 cluster and hybrid cluster. The ST5 cluster included 165 isolates with sequence types that included ST5, ST79, ST81, and ST359 was the dominant cluster with genotype VNI, serotype A, and MAT 𝛼 and was persistent during the time period in the study region, indicating its wide and continuous epidemic spread. The ST31 cluster contained 8 ST31 isolates. The phylogenetic tree grouped the AD hybrid isolates well, although MLST typing failed. The hybrid isolates were substantially different from those in the other two clusters. Among the hybrid clusters, three strains with the same mating type were closer to each other than to isolate RM007, which had a mating type of a/𝛼 (the other 179 strains in this study were all MAT 𝛼).
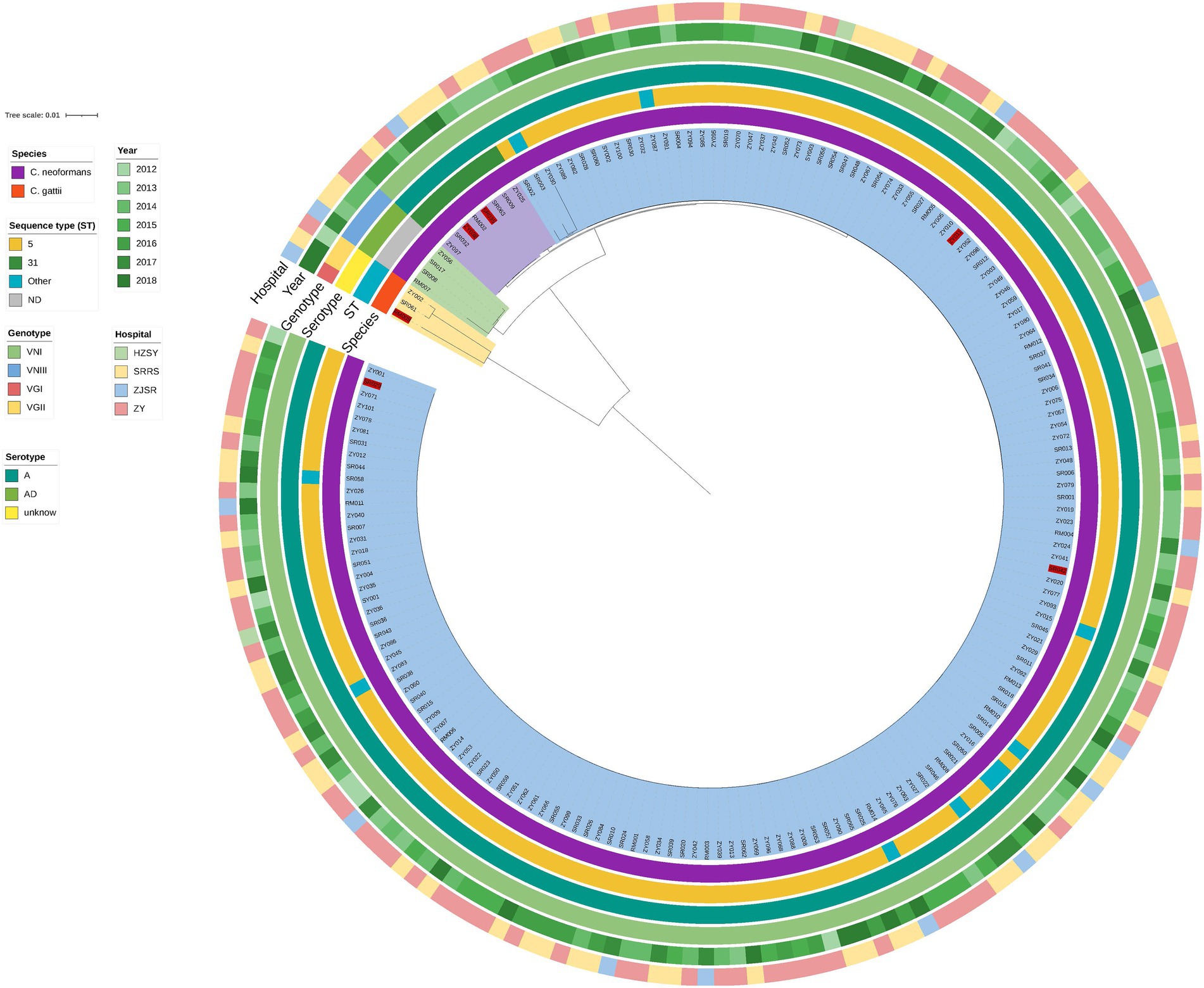
Figure 2. Phylogenetic tree of 180 isolates based on WGS. The innermost circle is the phylogenetic tree based on WGS. All strains were classified into two large clusters. The cluster indicated in yellow (corresponding to the bright orange of the “species” in the first circle) represents the CGSCs, and the other (corresponding to the dark purple of the “species”) represents C. neoformans. The red labelled strains represent strains with high MICs. The second, third, and fourth circles correspond to the sequence type (ST), genotype and serotype, respectively. The cluster of CGSCs contains three strains, RM009-ST215-VGI-α, SR061-ST552-VGII-α, and ZY002-ST328-VGII-α, and these molecular types were significantly different from C. neoformans. In the C. neoformans cluster, the strains were divided into three clusters (green, purple, and blue); the cluster indicated in green had four hybrids, RM007-VNIII-AαDa, ZY056, SR008, and SR017, which were all VNIII-AD-α; the cluster indicated in purple had 8 strains, all of which were ST31-VNI-A-α; the large cluster, which is represented in blue, had 165 strains, including ST5 (n = 155, the dominant ST), ST79 (n = 5), ST81 (n = 3), and ST359 (n = 2), all of which were VNI-A-MAT α. The isolation year (the fifth circle) and hospital (the sixth circle) showed a scattered distribution, and no significant difference in the corresponding strain species was detected.
Only a few strains with high MICs were detected. The MIC range, geometric mean (GM), MIC50 and MIC90 of 5 common antifungal agents are shown in Table 1. Although no AMB-resistant strains were found, 6 isolates with high MICs (Table 2), 2 for 5-FC and 4 for azoles, were identified. Among isolates with high MICs for azoles, 1 isolate had high MICs for all 3 drugs and belonged to cluster 31 (FLC 16 mg/L, ITR 1.5 mg/L, and VRC 0.64 mg/L), 1 isolate had high MICs for FLC (16 mg/L) and ITR (1 mg/L) and belonged to cluster 5, and the other 2 isolates with high MICs (16 mg/L and 24 mg/L) for FLC belonged to clusters 5 and 31 (Figure 2). Two strains had high MICs for 5-FC, with MICs greater than 32 mg/L. RM009 is a type of C. gattii (ST215, VGI, MAT α), and ZY011 is a type of C. neoformans var. grubii (ST5, VNI, MAT α).
The potential drug resistance mechanism was explored. However, no mutation in the ERG11 gene was observed in strains with high MICs for azoles after comparison with the sequence of the reference gene from H99 (ERG11, JQ044790, CNAG_00040). To determine the mechanism underlying 5-FC resistance, mutations in genes belonging to the FCY2-FCY1-FUR1 pathway were detected. Regarding RM009, there were 5 synonymous mutations in the FCY2 and FCY3 genes, 4 synonymous mutations and 2 meaningful mutations (T54C (Ser-Gly) and G460A (Ser-Phe)) in the FCY1 gene, and 3 synonymous mutations in the FUR1 gene. Another strain, ZY011 (C. neoformans-ST5-VNI-A-α), exhibited no mutations in the FCY2-4 and FCY1 genes after comparison with the sequence of the reference gene from strain H99 (VNI, ST2, serotype A), but the sequence alignment of the FUR1 gene was significantly different from the reference sequence, which lacked approximately 200 bp. This missing region started at the 206th amino acid (Figure 3A). The sequence integrity was verified by PCR and Sanger sequencing. The missing region is close to the 5-fluorouracil (5-FU) binding site and may influence the binding of 5-FU and mediate the increase in the MIC for 5-FC (Figure 3B).
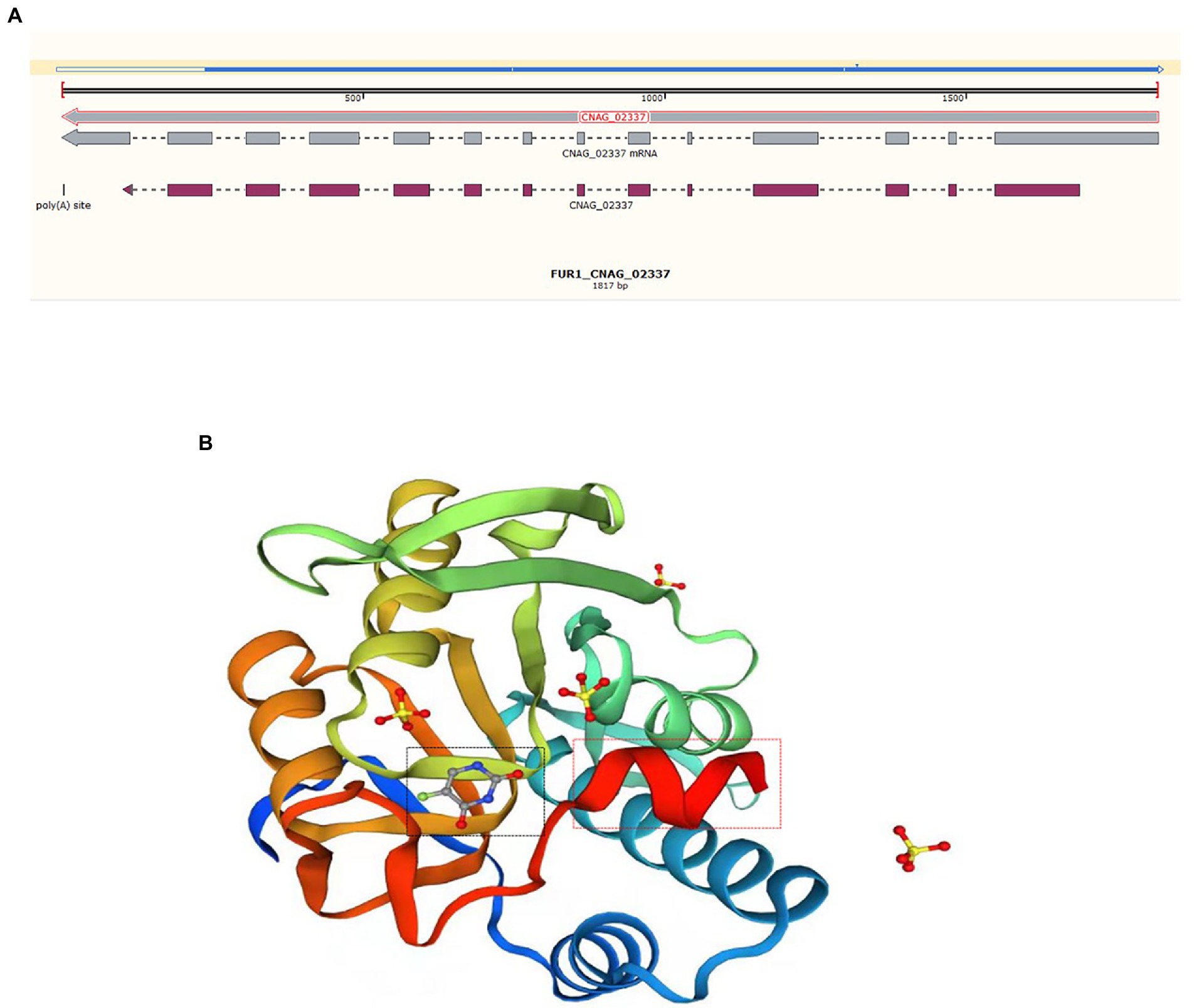
Figure 3. Sequence alignment and protein structure of FUR1. (A) DNA sequence alignment of the FUR1 gene of ZY011 and the reference sequence (CNAG_02337). The blue line above refers to the sequence of ZY011, and the different sequences are illustrated with a hollow line. The gray line is representative of the reference sequence of the gene and mRNA, and the mauve line represents the exons. (B) We revealed the protein structure of FUR1, and the different sequences are highlighted with red rectangles. This region is near the 5-FU-binding site (black rectangle).
As revealed by previous studies (Zhu et al., 2010; Fang et al., 2020; Fu et al., 2020; Zhou et al., 2020; Zhao et al., 2021), there are many more non-HIV patients in China than in other countries. In our study, we observed the same phenomenon: the proportion of patients with HIV was 12.78%. This finding may be due to the lower prevalence of HIV infection and the implementation of primary FLC prophylaxis in patients with CD4 counts <100 cells/μL in China (Organization WH, 2011; Parkes-Ratanshi et al., 2011; Oladele et al., 2017; Williamson et al., 2017). More importantly, this phenomenon indicates the importance of monitoring non-HIV-associated cryptococcosis even in apparently immunocompetent individuals (Williamson et al., 2017). In the United States, 20% of HIV-negative patients have no underlying conditions, such as a history of steroid therapy or malignant tumours (Williamson et al., 2017). Among non-HIV patients in this study, the presence of diseases affecting the immune system did not appear to increase the risk of Cryptococcus spp. infection since the proportions of patients with and without underlying diseases were nearly equal (45.55% vs. 41.67%). CNS-related infections were the main type of disease observed among HIV-negative patients (67.52%, 106/157). We reviewed the previous reports of Chinese researchers (Zhu et al., 2010) reported the clinical characteristics of 154 patients with cryptococcal meningitis over a 10-year period, the major of patients (66.9%) were otherwise apparently healthy and only 33.1% patients had predisposition factors (Zhu et al., 2010), there are also more reports from China that about 60% of cryptococcosis were from immunocompetent individuals, it is speculated that Chinese population might be more susceptible to cryptococcal infections than other ethnic groups (Fang et al., 2015).
In previous Cryptococcus spp. studies, many typing methods, such as RAPD, PCR fingerprinting, AFLP, MLMT, and MLST, have been employed and have mainly focused on genotyping (Hong et al., 2021). Due to their variable discrimination power, each of these methods has expanded knowledge regarding the genomic diversity of Cryptococcus spp. (Meyer et al., 2009; Hagen et al., 2015; Hong et al., 2021). We used band-based methods with the molecular types VNI–VNIV, VNB of CNSC and VGI-VGV of CGSC to assess this type of genomic diversity. With the development of sequencing technology, sequence base typing methods have provided more discriminatory power, repeatability and comparability. Thus, MLST and WGS are becoming increasingly popular. Using the MLST system, we identified lineages using sequence types. However, because MLST only includes the analysis of seven housekeeping genes, MLST may provide unilateral results, unlike WGS typing. Genome analysis in eukaryotes, including assembly, annotation, SNP calling, and phylogenetic tree reconstruction, is extremely difficult and requires expert bioinformatic knowledge and high-performance computers (Desjardins et al., 2017). In our study, we proved that even with roughly processed genomes, the results presented real genomic diversity and were clearly distinguished at the species level and lineage level. Using genome assemblies, Mashtree was used to reconstruct the phylogenetic tree efficiently. Not only can CNSCs and CGSCs be clearly distinguished, but subclusters in the major cluster can also be classified, as reported by Ziyi Zhou et al. in 2022 in a study that showed that ST5 has different subtypes based on the detection of single nucleotide polymorphism (SNPs) variants (Zhou et al., 2022), and the ST5 cluster and ST31 cluster can be roughly considered CC5 and CC31 (Fan et al., 2016; Cogliati et al., 2019; Thanh et al., 2019). Furthermore, the relationship between AD hybrids and other CCs could be clearly distinguished. These results improved the typing methods and revealed the biodiversity of Cryptococcus spp. in China (Zang et al., 2022; Zhou et al., 2022).
The latest research has shown that the nonsusceptibility rate (intermediate and resistant) of Cryptococcus spp. with FLC reached 25.9% (Simwami et al., 2011), which was significantly higher than the value reported 9 years ago (9.5%) (Wang et al., 2012; Xiao et al., 2018). This finding raises concerns about the increased drug resistance rate. Mutation of the ERG11 gene is the most common mechanism underlying azole resistance. It has been reported that the G484S, Y145F and G470R mutations of ERG11 are related to azole resistance (Rodero et al., 2003; Sionov et al., 2012; Gago et al., 2017; Zhang et al., 2019). However, no mutation was found in ERG11 in these high-MIC strains in our study.
In particular, two strains with high MICs of 5-FC (>32 mg/L) were found in this study: 1 strain of C. gattii and 1 strain of C. neoformans. To date, the drug resistance rate of 5-FC with Cryptococcus spp. is 1–25% (Cuenca-Estrella, 2001; Govender et al., 2011; Espinel-Ingroff et al., 2012; Chang et al., 2021). The metabolic pathway of FCY2-FCY1-FUR1 is related to 5-FC biochemical reactions and may cause 5-FC resistance due to its mutations. The heterozygous G/T mutation at position 145 of the FCY2 gene and the nonsense mutation C505T have been reported. In addition, the T26C point mutation of the FCY1 gene leads to a change in the protein (M9T), which can lead to cross-resistance to 5-FC and FLC (Papon et al., 2007; Florent et al., 2009). Considering the mutations identified in the C. gattii isolate RM009, we assumed that the two nonsynonymous mutations in the FCY1 gene may be responsible for 5-FC resistance, but this finding needs further research. There were substantial differences regarding the C. neoformans isolate (ZY011); no exon mutation was found in the FCY2-4 and FCY1 genes, but the sequence of the FUR1 gene was significantly different from the reference sequence. The identical sequence ended in an exon sequence, and because there were no other ways to identify the sequence of mRNA or protein, we could not predict the actual change in this gene. Considering that the changed region is near the 5-FU-binding site, mutations in the FUR1 gene may cause the binding of pyrimidines to be blocked or inefficient, which may be the main factor underlying resistance.
The MICs of the 5 anti-cryptococcal drugs in this study were generally not high, there was no amphotericin B-resistant strain, most of the Cryptococcus spp. strains were sensitive to azoles, the MICs of several NWTs were generally not high, only two 5-FC-resistant strains were identified, these provides a more detailed reference for the empirical therapy of cryptococcosis, and our study supplements the available information regarding cryptococcal infections in Zhejiang Province for the global fungal database. These findings prove that simply processed genome sequences also provide convincing evidence of lineages.
In mainland China, the non-HIV population is the most commonly susceptible to cryptococcosis, and Cryptococcus neoformans (ST5-VNI-A-α) is the predominant pathogen. The isolated hybrids and CGSCs were rare; of particular interest, three strains of C. gattii, which are rare in the clinic, all caused meningitis and occurred in younger patients without underlying diseases. Phylogenetic analysis based on WGS is not band-based, covers more genomic information and therefore shows higher discriminatory power than traditional typing methods. Anti-cryptococcal drug-resistant isolates were rare, but isolates with high MICs were identified. Mutations in FCY2-FCY1-FUR1 were detected in the 5-FC-resistant isolate, and this finding deserves further study.
However, there are also some limitations in this study. First, data may be outdated or incomplete because many data were collected retrospectively through electronic medical records. Second, the proportion of HIV in underlying diseases would be biased in that only one of the four hospitals in the study is a designated hospital for HIV treatment. Third, due to the limitation of diagnosis methods and the lack of understanding of cryptococcosis in early years, not all Cryptococcus spp. strains in four hospitals were collected at the same time, resulting in sample bias. A better epidemiological investigation design is needed in the future.
The study was approved by the ethics review board (found in Supplementary material).
Isolates of CNSC and CGSC from four hospitals were collected between 2012 and 2018. All isolates were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Clinical information, such as age, sex and underlying diseases, was collected by reviewing the patients’ electronic medical records. Based on this information, all cases were classified into three groups: (Kwon-Chung et al., 2017) HIV-associated (Hagen et al., 2015) non-HIV but with underlying diseases that may influence the immune status, and (Montoya et al., 2021) non-HIV and without underlying diseases. The following circumstances were considered underlying diseases: receipt of solid organ transplantation, connective tissue disease, aggressive cancer treatment, use of immunosuppressants or glucocorticoids, malignancies, haematologic malignancies, cirrhosis, diabetes mellitus, and uraemia (Zhu et al., 2010; Williamson et al., 2017; Beardsley et al., 2019). The risk level of HBV reactivation is moderate or high when treat with medium (10–20 mg/day) or high dose (≥20 mg/day) prednisone for ≥4 weeks, medium (10–20 mg/day) or high dose (≥20 mg/day) for ≥4 weeks (Lau et al., 2021), and prednisone therapy ≥7.5 mg/day were associated with a higher herpes zoster risk (Pappas et al., 2015). In the present study, patients receiving continuous glucocorticoids therapy that due to connective tissue diseases were included in the glucocorticoid group (excluding glucocorticoid containing chemotherapy); All patients with diabetes were type 2 diabetes. The “non-HIV but without underlying disease” group included patients who had no disease or some diseases that were well controlled, such as gout, hypertension, chronic hepatitis B, chronic kidney disease, and anaemia.
Further statistical analysis was performed, and the results are expressed as the means ± standard deviations.
Eight reference strains were used as controls: WM148 (VNI, serotype A, MATα), WM626 (VNII, serotype A, MATα), WM628 (VNIII, serotype AD, MATα/a), WM629 (VNIV, serotype D, MATα), WM179 (VGI, serotype B, MATα), WM178 (VGII, serotype B, MATα), WM161 (VGIII, serotype B, MATα) and WM779 (VGIV, serotype C, MATα) (Meyer et al., 2009).
Genomic DNA was extracted from the collected isolates with ZR Fungal/Bacterial DNA Kits (catalogue no. D6005, The Epigenetics Company) according to the manufacturer’s recommendations. The extracted DNA was used for MLST, genotype, serotype and next-generation sequencing analyses.
The genotypes were verified using two methods, M13-based PCR fingerprinting (FP) and URA5-RFLP analysis, according to previously described protocols (Meyer et al., 2009). The serotype and mating type were determined by PCR, and the six related primer pairs of C. neoformans were used to determine the strain serotype and mating type according to previously described protocols (Chaturvedi et al., 2000; Dou et al., 2015; Wu et al., 2015).
Next,-generation sequencing of the strains was performed on the Illumina HiSeq X Ten platform. The raw genomic sequence data passed a quality control assessment, as demonstrated using fastqc (Andrews, 2010) and multiqc (Ewels et al., 2016). Genomes were then assembled using Shovill (Seemann et al., 2019, unpublished)1 with an average depth of 77.1. Utilizing the whole-genome sequence, a mash distance-based phylogenetic analysis was performed with Mashtree (Katz et al., 2019), and the results were visualized using the iTOL web service.2
ST type was determined by screening the genome data from each isolate on Cryptococcus spp. MLST database.3 The alleles that could not be matched were verified using the consensus ISHAM protocol by PCR and by sequencing seven loci: CAP59, IGS1, GPD1, LAC1, PLB1, SOD1, and URA5 (Meyer et al., 2009; Hagen et al., 2015; Cogliati et al., 2020).
The AST of 5 common antifungal agents, including amphotericin B (AMB), 5-flucytosine (5-FC), fluconazole (FLC), itraconazole (ITR), and voriconazole (VRC), was performed using an ATB FUNGUS 3 kit (bioMérieux, France). If the MICs were abnormally high or exhibited strong trailing growth, they were then verified using an E-test (bioMérieux). The preliminary determination of drug sensitivity was based on the Fungus 3 kit instructions and the CBPs of Candida spp. recommended by CLSI/NCCLS and the literature (CLSI, 2008; Tewari et al., 2012; Bongomin et al., 2018; Breakpoint Tables for Interpretation of MICs for Antifungal Agents, 2020). Quality control was performed using Candida parapsilosis (ATCC 22019) and Candida krusei (ATCC 6258) (Torres-Rodriguez and Alvarado-Ramirez, 2007; Performance Standards for Antifungal Susceptibility of Testing Yeasts, 2018).
To explore the possible drug resistance mechanism, the ERG11 sequences of the high-MIC strains were compared with the reference sequences (ERG11, JQ44790, CNAG_00040) using BLAST.4 The changes in the metabolic pathway of FCY2-FCY1-FUR1 were responsible for 5-FC resistance. Among C. neoformans isolates, pathway-related genes were compared with the reference gene of strain H99 (FCY1 CNAG_00613, FCY2 CNAG_01681, FCY3 CNAG_04982, FCY4 CNAG_04276, FUR1 CNAG_02337). For C. gattii isolates, we used E566 (VGI, serotype B) as the reference genome. The missing sequence of the FUR1 gene was verified by PCR and Sanger sequencing with the primers F-TGGATGAGATCATATGCCTG and R-GATTGCTGATTGGAAGGAC.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
YY, YJ, and ZZ: conceptualization. JZ, QY, YF, FZ, XL, and LF: data curation. JZ and ZZ: formal analysis. YY: funding acquisition. JZ, ZW, and LF: investigation. YC, YJ, and YY: methodology. JZ, ZW, and YY: project administration. ZZ, QY, YF, FZ, QC, and LF: resources. ZW, YC, XL, and YJ: software. YJ: supervision. JZ and ZW: validation and visualization. JZ: writing—original draft. ZW, YJ, and YY: writing—review and editing. All authors contributed to the article and approved the submitted version.
This research was funded by the clinical research project of Zhejiang Provincial Department of Health (grant number 2018ZD030) and General Research Plan for Medical and Health of Zhejiang Province (Class A) (grant number 2020KY604).
We thank Wanqing Liao for providing the standard strains of Cryptococcus spp. and Ying Zhang for providing the quality control strains of AST.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.991703/full#supplementary-material
1. ^Seemann, T., Edwards, R., Da Silva, A., and Kiil, K. Shovill. (2019) (unpublished).
Andrews, S. Fast QC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. (2010).
Beardsley, J., Sorrell, T. C., and Chen, S. C. (2019). Central nervous system Cryptococcal infections in non-HIV infected patients. J. Fungi 5: 71. doi: 10.3390/jof5030071
Bongomin, F., Oladele, R. O., Gago, S., Moore, C. B., and Richardson, M. D. (2018). A systematic review of fluconazole resistance in clinical isolates of Cryptococcus species. Mycoses 61, 290–297. doi: 10.1111/myc.12747
Breakpoint Tables for Interpretation of MICs for Antifungal Agents. (2020). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs for antifungal agents, version 10.0. Available online at: http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/.
Chang, Y. C., Lamichhane, A. K., Cai, H., Walter, P. J., Bennett, J. E., and Kwon-Chung, K. J. (2021). Moderate levels of 5-fluorocytosine cause the emergence of high frequency resistance in cryptococci. Nat. Commun. 12:3418. doi: 10.1038/s41467-021-23745-1
Chaturvedi, S., Fan, B. R. J., McClelland, C. M., Wickes, B. L., and Chaturvedi, V. (2000). Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, Ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J. Clin. Microbiol. 38, 2007–2009. doi: 10.1128/JCM.38.5.2007-2009.2000
CLSI. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard-third edition. CLSI document M27-A3. Wayne, PA: Clinical and Laboratory Standards Institute. (2008).
Cogliati, M., Desnos-Ollivier, M., McCormick-Smith, I., Rickerts, V., Ferreira-Paim, K., Meyer, W., et al. (2019). Genotypes and population genetics of Cryptococcus neoformans and Cryptococcus gattii species complexes in Europe and the mediterranean area. Fungal Genet. Biol. 129, 16–29. doi: 10.1016/j.fgb.2019.04.001
Cogliati, M., Roger, F., Meyer, W., Robert, V., and Bertout, S. (2020). New multilocus sequence typing primers to enable genotyping of AD hybrids within the Cryptococcus neoformans species complex. Med. Mycol. 58, 1005–1009. doi: 10.1093/mmy/myaa047
Cuenca-Estrella, M. T. M. D., Mellado, E., and Rodríguez-Tudela, J. L. (2001). Flucytosine primary resistance in Candida species and Cryptococcus neoformans. Eur. J. Clin. Microbiol. Infect. Dis. 20, 276–279. doi: 10.1007/PL00011265
Desjardins, C. A., Giamberardino, C., Sykes, S. M., Yu, C. H., Tenor, J. L., Chen, Y., et al. (2017). Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 27, 1207–1219. doi: 10.1101/gr.218727.116
Dou, H. T. Y. X., Wang, H. Z., and Li, T. S. (2015). Molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii in China between 2007 and 2013 using multilocus sequence typing and the Diversi lab system. Eur. J. Clin. Microbiol. Infect. Dis. 34, 753–762. doi: 10.1007/s10096-014-2289-2
D’Souza, C. A., Kronstad, J. W., Taylor, G., Warren, R., Yuen, M., Hu, G., et al. (2011). Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio 2, e00342–e00310. doi: 10.1128/mBio.00342-10
Espinel-Ingroff, A., Aller, A. I., Canton, E., Castanon-Olivares, L. R., Chowdhary, A., Cordoba, S., et al. (2012). Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 56, 5898–5906. doi: 10.1128/AAC.01115-12
European Committee on Antimicrobial Susceptibility Testing. (2022). Overview of antifungal ECOFFs and clinical breakpoints for yeasts, moulds and dermatophytes using the EUCAST E.Def 7.3, E.Def 9.4 and E.Def 11.0 procedures. Version 3. Available at: http://www.eucast.org.
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. J. B. (2016). Multi QC: summarize analysis results for multiple tools and samples in a single report, J. Bioinform. 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fan, X., Xiao, M., Chen, S., Kong, F., Dou, H. T., Wang, H., et al. (2016). Predominance of Cryptococcus neoformans var. grubii multilocus sequence type 5 and emergence of isolates with non-wild-type minimum inhibitory concentrations to fluconazole: a multi-Centre study in China. Clin. Microbiol. Infect. 22:887.e1-.e9. doi: 10.1016/j.cmi.2016.07.008
Fang, W., Fa, Z., and Liao, W. (2015). Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genet. Biol. 78, 7–15. doi: 10.1016/j.fgb.2014.10.017
Fang, L. F., Zhang, P. P., Wang, J., Yang, Q., and Qu, T. T. (2020). Clinical and microbiological characteristics of cryptococcosis at an university hospital in China from 2013 to 2017. Braz. J. Infect. Dis. 24, 7–12. doi: 10.1016/j.bjid.2019.11.004
Firacative, C., Roe, C. C., Malik, R., Ferreira-Paim, K., Escandon, P., Sykes, J. E., et al. (2016). MLST and whole-genome-based population analysis of Cryptococcus gattii VGIII links clinical, veterinary and environmental strains, and reveals divergent serotype specific sub-populations and distant ancestors. PLoS Negl. Trop. Dis. 10:e0004861. doi: 10.1371/journal.pntd.0004861
Florent, M., Noel, T., Ruprich-Robert, G., Da Silva, B., Fitton-Ouhabi, V., Chastin, C., et al. (2009). Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob. Agents Chemother. 53, 2982–2990. doi: 10.1128/AAC.00880-08
Fu, Y., Xu, M., Zhou, H., Yao, Y., Zhou, J., and Pan, Z. (2020). Microbiological and clinical characteristics of cryptococcemia: a retrospective analysis of 85 cases in a Chinese hospital. Med. Mycol. 58, 478–484. doi: 10.1093/mmy/myz089
Gago, S., Serrano, C., Alastruey-Izquierdo, A., Cuesta, I., Martin-Mazuelos, E., Aller, A. I., et al. (2017). Molecular identification, antifungal resistance and virulence of Cryptococcus neoformans and Cryptococcus deneoformans isolated in Seville, Spain. Mycoses. 60, 40–50. doi: 10.1111/myc.12543
Govender, N. P., Patel, J., van Wyk, M., Chiller, T. M., and Lockhart, S. R. (2011). Group for Enteric R, et al. trends in antifungal drug susceptibility of Cryptococcus neoformans isolates obtained through population-based surveillance in South Africa in 2002–2003 and 2007–2008. Antimicrob. Agents Chemother. 55, 2606–2611. doi: 10.1128/AAC.00048-11
Hagen, F., Khayhan, K., Theelen, B., Kolecka, A., Polacheck, I., Sionov, E., et al. (2015). Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78, 16–48. doi: 10.1016/j.fgb.2015.02.009
Hong, N., Chen, M., and Xu, J. (2021). Molecular markers reveal epidemiological patterns and evolutionary histories of the human pathogenic Cryptococcus. Front. Cell. Infect. Microbiol. 11:683670. doi: 10.3389/fcimb.2021.683670
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., Zhang, S., den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4:1762. doi: 10.21105/joss.01762
Kwon-Chung, K. J., Bennett, J. E., Wickes, B. L., Meyer, W., Cuomo, C. A., Wollenburg, K. R., et al. (2017). The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere. 2: e00357–16. doi: 10.1128/mSphere.00357-16
Lau, G., Yu, M. L., Wong, G., Thompson, A., Ghazinian, H., Hou, J. L., et al. (2021). APASL clinical practice guideline on hepatitis B reactivation related to the use of immunosuppressive therapy. Hepatol. Int. 15, 1031–1048. doi: 10.1007/s12072-021-10239-x
Li, W., Averette, A. F., Desnos-Ollivier, M., Ni, M., Dromer, F., and Heitman, J. (2012). Genetic diversity and genomic plasticity of Cryptococcus neoformans AD hybrid strains. G3 2, 83–97. doi: 10.1534/g3.111.001255
Liaw, S. J., Wu, H. C., and Hsueh, P. R. (2010). Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin. Microbiol. Infect. 16, 696–703. doi: 10.1111/j.1469-0691.2009.02930.x
Liu, Z. Y., Wang, G. Q., Zhu, L. P., Lyu, X. J., Zhang, Q. Q., Yu, Y. S., et al. (2018). Expert consensus on the diagnosis and treatment of cryptococcal meningitis. Zhonghua Nei Ke Za Zhi 57, 317–323. doi: 10.3760/cma.j.issn.0578-1426.2018.05.003
Loftus, B. J., Fung, E., Roncaglia, P., Rowley, D., Amedeo, P., Bruno, D., et al. (2005). The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307, 1321–1324. doi: 10.1126/science.1103773
May, R. C., Stone, N. R., Wiesner, D. L., Bicanic, T., and Nielsen, K. (2016). Cryptococcus: from environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 14, 106–117. doi: 10.1038/nrmicro.2015.6
Meyer, W., Aanensen, D. M., Boekhout, T., Cogliati, M., Diaz, M. R., Esposto, M. C., et al. (2009). Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47, 561–570. doi: 10.1080/13693780902953886
Montoya, M. C., Magwene, P. M., and Perfect, J. R. (2021). Associations between Cryptococcus genotypes, phenotypes, and clinical parameters of human disease: A Review. J. Fungi 7: 260. doi: 10.3390/jof7040260
Oladele, R. O., Bongomin, F., Gago, S., and Denning, D. W. (2017). HIV-associated Cryptococcal disease in resource-limited settings: a case for "prevention is better than cure"? J. Fungi 3: 67. doi: 10.3390/jof3040067
Organization WH. (2011). Rapid advice: Diagnosis, prevention and management of cryptococcal disease in HIV-infected adults, adolescents and children. Geneva: World Health Organization. Available online at: https://appswhoint/iris/handle/10665/44786.
Papon, N., Noel, T., Florent, M., Gibot-Leclerc, S., Jean, D., Chastin, C., et al. (2007). Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob. Agents Chemother. 51, 369–371. doi: 10.1128/AAC.00824-06
Pappas, D. A., Hooper, M. M., Kremer, J. M., Reed, G., Shan, Y., Wenkert, D., et al. (2015). Herpes zoster reactivation in patients with rheumatoid arthritis: analysis of disease characteristics and disease-modifying Antirheumatic drugs. Arthritis Care Res. 67, 1671–1678. doi: 10.1002/acr.22628
Parkes-Ratanshi, R., Wakeham, K., Levin, J., Namusoke, D., Whitworth, J., Coutinho, A., et al. (2011). Primary prophylaxis of cryptococcal disease with fluconazole in HIV-positive Ugandan adults: a double-blind, randomized, placebo-controlled trial. Lancet Infect. Dis. 11, 933–941. doi: 10.1016/S1473-3099(11)70245-6
Perfect, J. R., Dismukes, W. E., Dromer, F., Goldman, D. L., Graybill, J. R., Hamill, R. J., et al. (2010). Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin. Infect. Dis. 50, 291–322. doi: 10.1086/649858
Performance Standards for Antifungal Susceptibility of Testing Yeasts. Performance standards for antifungal susceptibility of testing yeasts. Clinical and Laboratory Standards Institute, Wayne, PA. (2018).
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Rodero, L., Mellado, E., Rodriguez, A. C., Salve, A., Guelfand, L., Cahn, P., et al. (2003). G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47, 3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003
Samarasinghe, H., and Xu, J. (2018). Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes. Infect. Genet. Evol. 66, 245–255. doi: 10.1016/j.meegid.2018.10.011
Simwami, S. P., Khayhan, K., Henk, D. A., Aanensen, D. M., Boekhout, T., Hagen, F., et al. (2011). Low diversity Cryptococcus neoformans variety Grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog. 7:e1001343. doi: 10.1371/journal.ppat.1001343
Sionov, E., Chang, Y. C., Garraffo, H. M., Dolan, M. A., Ghannoum, M. A., and Kwon-Chung, K. J. (2012). Identification of a Cryptococcus neoformans cytochrome P 450 lanosterol 14alpha-demethylase (erg 11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 56, 1162–1169. doi: 10.1128/AAC.05502-11
T G Mitchell, JRP (1995). Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8, 515–548. doi: 10.1128/CMR.8.4.515
Tewari, A., Behera, B., Mathur, P., and Xess, I. (2012). Comparative analysis of the Vitek 2 antifungal susceptibility system and E-test with the CLSI M27-A3 broth microdilution method for susceptibility testing of Indian clinical isolates of Cryptococcus neoformans. Mycopathologia 173, 427–433. doi: 10.1007/s11046-012-9528-9
Thanh, L. T., Phan, T. H., Rattanavong, S., Nguyen, T. M., Van Duong, A., Dacon, C., et al. (2019). Multilocus sequence typing of Cryptococcus neoformans var. Grubii from Laos in a regional and global context. Med. Mycol. 57, 557–565. doi: 10.1093/mmy/myy105
Torres-Rodriguez, J. M., and Alvarado-Ramirez, E. (2007). In vitro susceptibilities to yeasts using the ATB FUNGUS 2 method, compared with Sensititre yeast one and standard CLSI (NCCLS) M27-A2 methods. J. Antimicrob. Chemother. 60, 658–661. doi: 10.1093/jac/dkm247
Wang, H., Xiao, M., Chen, S. C., Kong, F., Sun, Z. Y., Liao, K., et al. (2012). In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance net (CHIF-NET) study. J. Clin. Microbiol. 50, 3952–3959. doi: 10.1128/JCM.01130-12
Williamson, P. R., Jarvis, J. N., Panackal, A. A., Fisher, M. C., Molloy, S. F., Loyse, A., et al. (2017). Cryptococcal meningitis: epidemiology, immunology, diagnosis and therapy. Nat. Rev. Neurol. 13, 13–24. doi: 10.1038/nrneurol.2016.167
Wongsuk, T., Homkaew, A., Faksri, K., and Thongnak, C. (2020). Multi-locus sequence typing and whole genome sequence analysis of Cryptococcus neoformans isolated from clinical specimens in Vajira hospital, Bangkok, Thailand. Mycopathologia 185, 503–514. doi: 10.1007/s11046-020-00456-7
Wu, S. Y., Lei, Y., Kang, M., Xiao, Y. L., and Chen, Z. X. (2015). Molecular characterization of clinical Cryptococcus neoformans and Cryptococcus gattii isolates from Sichuan province. China. Mycoses. 58, 280–287. doi: 10.1111/myc.12312
Xiao, M., Chen, S. C., Kong, F., Fan, X., Cheng, J. W., Hou, X., et al. (2018). Five-year China hospital invasive fungal surveillance net (CHIF-NET) study of invasive fungal infections caused by non-candidal yeasts: species distribution and azole susceptibility. Infect. Drug Resist. 11, 1659–1667. doi: 10.2147/IDR.S173805
Xiaobo Feng, Z. Y., Ren, D., Liao, W., and Jingsong, W. (2008). Genotypeandmating typeanalysis of Cryptococcusneoformans and Cryptococcusgattii isolates from China thatmainlyoriginated fromnon-HIV-infectedpatients. FEMS Yeast Res. 8, 930–938. doi: 10.1111/j.1567-1364.2008.00422.x
Zang, X., Ke, W., Wang, L., Wu, H., Huang, Y., Deng, H., et al. (2022). Molecular epidemiology and microbiological characteristics of Cryptococcus gattii VGII isolates from China. PLoS Negl. Trop. Dis. 16:e0010078. doi: 10.1371/journal.pntd.0010078
Zhang, J., Li, L., Lv, Q., Yan, L., Wang, Y., and Jiang, Y. (2019). The fungal CYP51s: their functions, structures, related drug resistance, and inhibitors. Front. Microbiol. 10:691. doi: 10.3389/fmicb.2019.00691
Zhao, H., Zhou, M., Zheng, Q., Zhu, M., Yang, Z., Hu, C., et al. (2021). Clinical features and outcomes of Cryptococcemia patients with and without HIV infection. Mycoses 64, 656–667. doi: 10.1111/myc.13261
Zhou, L. H., Jiang, Y. K., Li, R. Y., Huang, L. P., Yip, C. W., Denning, D. W., et al. (2020). Risk-based estimate of human fungal disease burden, China. Emerg Infect Dis. 26, 2137–2147. doi: 10.3201/eid2609.200016
Zhou, Z., Zhu, C., Ip, M., Liu, M., Zhu, Z., Liu, R., et al. (2022). Molecular epidemiology and antifungal resistance of Cryptococcus neoformans from human immunodeficiency virus-negative and human immunodeficiency virus-positive patients in eastern China. Front. Microbiol. 13:942940. doi: 10.3389/fmicb.2022.942940
Keywords: Cryptococcus neoformans, Cryptococcus gattii, antifungal susceptibility testing, multilocus sequence typing (MLST), mating type (MAT), genotype, whole-genome sequencing (WGS)
Citation: Zhang J, Wang Z, Chen Y, Zhou Z, Yang Q, Fu Y, Zhao F, Li X, Chen Q, Fang L, Jiang Y and Yu Y (2022) Antifungal susceptibility and molecular characteristics of Cryptococcus spp. based on whole-genome sequencing in Zhejiang Province, China. Front. Microbiol. 13:991703. doi: 10.3389/fmicb.2022.991703
Received: 11 July 2022; Accepted: 31 October 2022;
Published: 17 November 2022.
Edited by:
Kamal El Bissati, The University of Chicago, United StatesReviewed by:
Jianping Xu, McMaster University, CanadaCopyright © 2022 Zhang, Wang, Chen, Zhou, Yang, Fu, Zhao, Li, Chen, Fang, Jiang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunsong Yu, eXZ5czExOUB6anUuZWR1LmNu; Yan Jiang, amlhbmd5QHpqdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.