
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 November 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.991105
Previous studies have demonstrated that symbiotic gut bacteria in insects are involved in the development of insecticide resistance, but the relationship between the symbiotic gut bacteria and resistance to Lysinibacillus sphaericus C3-41 in Culex pipiens quinquefasciatus remains unclear. In this study, the abundance and diversity of gut symbionts of Cx. quinquefasciatus that were resistant (RLCql) or susceptible (SLCql) to L. sphaericus C3-41 were analyzed by high-throughput 16S rRNA sequencing. The main phyla among the symbiotic gut bacterial communities of Cx. quinquefasciatus were Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes. However, the relative abundance of Firmicutes, Proteobacteria, and unidentified Bacteria in the gut of the resistant strain of Cx. quequinfasciatus (RLCql colony) was higher compared to the susceptible strain (SLCql colony). The NMDS (Non-Metric Multi-Dimensional Scaling) and unweighted unifrac PCoA analyses also revealed significant differences between the symbiotic gut bacterial communities from the resistant and susceptible strains, suggesting that bacterial insecticides can alter bacterial composition. Ultimately, the changes in the bacterial community likely occurred after the development of resistance to L. sphaericus C3-41. These results provide guidance for further research into the mechanisms of gut symbionts involved in resistance against L. sphaericus C3-41 in Cx. quinquefasciatus.
Culex quinquefasciatus Say (Diptera: Culicidae) is an important medical insect that is distributed throughout the world (Samy et al., 2016; Zhang et al., 2019a). Cx. quinquefasciatus breed better in water that is contaminated with organic debris, such as rotten vegetation or human and animal excrement (Shah et al., 2016). Therefore, the larvae of this species are often discovered in urban sewer systems, irrigation ditches, cesspools, septic tanks, used tires, or containers of non-potable water (Shah et al., 2016; Zhang et al., 2019b; Díaz-Nieto et al., 2021). Cx. quinquefasciatus is the primary vector of Japanese encephalitis virus (JEv), St. Louis encephalitis virus (SLEv), West Nile virus (WNv), Western equine encephalitis virus (WEEv), Rift Valley fever virus (RVF), and the filarial nematode (Reid et al., 2018; Zhang et al., 2019a). These pathogens may be transmitted when humans are bitten during the day or night (Guo et al., 2016). These vector-borne diseases pose a major threat to global health and increase the economic burden of individuals and families (Mainali et al., 2017; Redekop et al., 2017). It is widely accepted that these diseases can be controlled or eliminated by targeting vector mosquitoes (Zhang et al., 2019b); thus, insecticides are extensively and intensively used to control Cx. quinquefasciatus worldwide (Yang et al., 2021). However, widespread broad-spectrum insecticide usage has given rise to concerns regarding environmental impact and the evolution of insecticide resistance in mosquitoes. Therefore, the development of bacteria as alternative control agents to insecticides has become highly appealing. Examples may include Bacillus thuringiensis and Lysinibacillus sphaericus Niede (Yu et al., 2017).
Lysinibacillus sphaericus is a globally distributed, aerobic, gram-positive, mesophilic, and spore-forming bacterium (Fu et al., 2017a). L. sphaericus is widely distributed in nature, including soil and aquatic environments, and its spores may even recirculate and survive in unusual environments, such as the bodies of dead mosquito larvae (Fu et al., 2017b). L. sphaericus produces insecticidal binary (Bin) protoxin as crystalline inclusions during bacterial sporulation, which have high larvicidal activity. Therefore, L. sphaericus has been extensively used for controlling the genus Culex in China, Vietnam, Thailand, India, Cuba, Denmark, Belgium, France, the USA, and Brazil (Xiong et al., 2010). Unfortunately, due its action on a single toxin receptor within the larval midgut, resistance to L. sphaericus in Culex has developed quickly in a short period of time (Guo et al., 2013). According to the Arthropod Pesticide Resistance Database (APRD), current field surveys report different levels of resistance to L. sphaericus in Cx. quinquefasciatus and Cx. pipiens populations from China, Thailand, India, France, and Brazil (Yuan et al., 2000; Chenniappan and Ayyadurai, 2012). These studies demonstrated that such high levels of resistance to L. sphaericus are predominantly due to the absence and alteration of the functional receptors in the midgut leading to the failure of Bin toxin binding (Guo et al., 2013).
Lysinibacillus sphaericus C3-41 was isolated in 1987 by the Wuhan institute of Virology, Chinese Academy of Sciences and is highly toxic to Culex larvae (Xiong et al., 2010). A L. sphaericus C3-41 liquid formulation (Jianbao®) has been successfully applied to control urban mosquito larvae in China for more than three decades and has demonstrated good efficacy (Xiong et al., 2010). However, under laboratory conditions, after 13 generations of exposure to high concentrations of L. sphaericus C3-41, a susceptible colony developed high-level resistance against this larvicide (resistance ratio, RR > 140,000-fold) (Pei et al., 2002). Moreover, high-level resistance to this species in a field mosquito colony was also recorded, further hindering the application of this biopesticide in mosquito control (Yuan et al., 2000).
Insects possess diverse symbiotic microbiota in their gut lumen that likely play an essential role in several physiological functions (Wang and Qu, 2017). For example, these symbiotic microorganisms may regulate the growth and development of host insects, as well as their reproduction, aggregation, movement, immune responses, and detoxification mechanisms (Wei et al., 2018). Symbiotic bacteria may also confer chemical insecticide resistance in agricultural pests (Kikuchi et al., 2012; Cheng et al., 2017; Hao et al., 2018; Wang et al., 2021). Conversely, some symbiotic bacteria can invade the body cavity or hemocoel through toxin-induced epithelial lesions and promote the death of medically important insects (Wei et al., 2017). Moreover, shifts in susceptibility to insecticides in insects have been linked with changes in the gut bacterial loads and diversity, as described above.
In this study, we hypothesize that there may be a close relationship between the changes in gut symbionts and L. sphaericus C3-41 resistance in Cx. quinquefasciatus. Using amplicon sequencing, we investigated the gut symbiont composition of the Cx. quinquefasciatus in resistant and susceptible populations, with the aim of elucidating the possible role of symbiotic gut bacteria on the development of L. sphaericus C3-41 resistance in mosquitoes.
The susceptible Cx. quinquefasciatus (SLCql), which was established from a laboratory-reared colony more than 10 years ago, was reared without exposure to any insecticide. The resistant Cx. quinquefasciatus (RLCq1) was derived from the susceptible colony and selection was performed by exposure to L. sphaericus C3-41 under laboratory conditions, with a resistance ratio of more than 140,000-fold. Larvae of the two colonies were reared in enamel pans filled with dechlorinated tap water and fed with a mixture of yeast powder and wheat mill or cat chow. The pupae were removed from the pans every day and placed in screen cages for emergence. The adults were allowed to feed on 10% sucrose solution, and the females were fed with horse serum purchased from Shanghai Yuanye Biological Technology Co., Ltd., (Shanghai, China). All larvae and adults were held at 27 ± 1°C with a photoperiod of 12:12 h (light-dark).
The L. sphaericus C3-41 was grown on an MBS (medium Bacillus sphaericus) medium plate at 30°C for 72–120 h or in MBS (medium B. sphaericus) at 30°C for 36 h. The bacteria were scraped off and rinsed with phosphate buffer before freeze-drying into powder. The MBS medium contained 6.8 g KH2PO4, 0.3 g MgSO4⋅7H2O, 0.2 g CaCl2⋅2H2O, 0.002 g MnSO4⋅H2O, 0.002 g Fe2(SO4)3, 0.002 g ZnSO4⋅6H2O, 10 g tryptone, 2.0 g yeast extract, and 20 g agar powder per liter of medium (pH 7.0).
Bioassays were carried out according to the standard method recommended by the World Health Organization (Zhang et al., 2019a). Twenty third-instar larvae were transferred into 100 mL of distilled water in a 200 mL plastic cup. Three replicates for each dose of insecticide and nine doses in total were performed, and the controls were treated with dechlorinated tap water. All treatments were maintained at a temperature of 27 ± 1°C and 40–50% relative humidity with a 12 h light/12 h dark photoperiod. Mortality was assessed after exposure to insecticide for 48 h. The LC50 values with 95% confidence interval, slopes with standard error (SE), and chi-square value (χ2) with degrees of freedom (df) were calculated using a regression model based on a probit transformation of mortalities and a logarithmic transformation of concentrations tested, i.e., a log-probit model by Polo Plus software.
The third instar larvae of the SLCq1 and RLCq1 strains were used for sample preparation. These larvae were surface-washed in a sterile petri dish containing 75% ethanol for 90 s, and then rinsed four times with sterilized water. The guts and gut contents from 60 insects were collected under a stereomicroscope on a surface-disinfected bench for each independent biological replicate. Then, the guts and gut contents were transferred into a microcentrifuge tube containing 200 μL phosphate-buffered saline (PBS, pH 7.8) and homogenized. The homogenization buffer was frozen at –80°C before DNA extraction.
The samples for 16S rRNA gene Illumina sequencing were dissected from 50 individuals in each generation for 6 generations; thus, each strain had six independent biological replicates (SLCql1, SLCql2, SLCql3, SLCql4, SLCql5, SLCql6, and RLCql1, RLCql2, RLCql3, RLCql4, RLCql5, and RLCql6).
The total genomic DNA of symbiotic bacteria was extracted from the prepared samples using the QIAamp DNA Stool Mini Kit following the manufacturer’s protocol. DNA concentration was detected using a Qubit® 2.0 (Invitrogen). DNA purity was monitored on 1% agarose gels.
16S rRNA genes from distinct regions (16S V4) were amplified used specific primers (e.g., 16S V4: 515F-806R) with barcodes. All PCR reactions were carried out using the Phusion® High-Fidelity PCR Master Mix (New England Biolabs). Equal volumes of 1 × loading buffer (containing SYBR green) were mixed with PCR products and 2% agarose gel electrophoresis was performed for detection. PCR products were mixed in equidensity ratios. Then, mixed PCR products were purified with the GeneJET™ Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using the Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific) following the manufacturer’s recommendations. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific). Finally, the library was sequenced on an Ion S5TM XL platform and 400 bp/600 bp single-end reads were generated.
OTU abundance data were normalized using standard sequence numbers corresponding to the sample with the least sequences. Subsequent analyses of alpha diversity and beta diversity were performed based on the normalized data.
Alpha diversity describes the complexity of species diversity for a sample. Two indices were used: the observed species and Shannon indices, which were both calculated using QIIME (Version 1.7.0) and illustrated using R software (Version 2.15.3).
Beta diversity analysis was performed to evaluate differences across samples in the context of species complexity. Beta diversity analyses on both weighted and unweighted unifrac distances were calculated using QIIME software (Version 1.7.0).
Principal Coordinate Analysis (PCoA) was performed to obtain the principal coordinates and visualize complex, multidimensional data. A previously obtained weighted or unweighted unifrac distance matrix was transformed to a new set of orthogonal axes, with first principal coordinate representing the factor accounting for the greatest variation, the second principal coordinate representing the next most important factor, and so on. PCoA plots were constructed using the WGCNA package, stat package, and ggplot2 package in R software (Version 2.15.3).
Unweighted Pair-group Method with Arithmetic Means (UPGMA) clustering is a hierarchical clustering method for interpreting the distance matrix using average linkage. UPGMA clustering was conducted with QIIME software (Version 1.7.0).
To identify significant differences between the symbiotic bacteria of SLCql and RLCql, a statistical method introduced in Metastats software was used. A p-value of ≤ 0.05 was considered significant.
Bioassay results showed that the L. sphaericus C3-41 freeze-dried powder had a high activity against the susceptible colony (SLCq1), with a 50% lethal concentration (LC50) of 0.0098 mg/L. However, little toxicity was observed in the resistant colony (RLCq1), in which the LC50 was greater than 1,000 mg/L. The RLCq1 has a resistance ratio of > 100, 000-fold to C3-41 (Table 1).
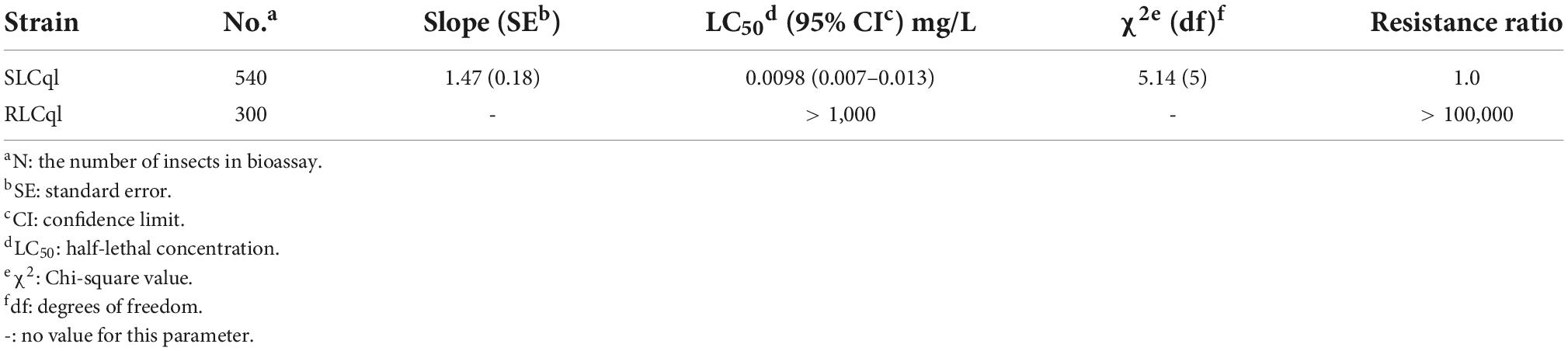
Table 1. Toxicity of Lysinibacillus sphaericus C3-41 (Ls C3-41) to susceptible (SLCql) and resistant (RLCql) Culex quinquefasciatus.
Based on high-throughput sequencing of the V4 hypervariable region of the 16S rRNA gene in gut bacteria, a total of 424, 268 and 503, 677 raw reads were obtained from the guts of Cx. quinquefasciatus from SLCql and RLCql, respectively (Table 2). After removing the adaptors, low-quality sequences, overlapping PE reads, and chimeras, an average of 64, 998 and 77, 314 clean reads with an average length of more than 420 bp were obtained from SLCql and RLCql, respectively (Table 2). The greatest number of clean reads was observed in RLCql6, and the fewest clean reads was found in SLCql5 (Table 2).
Generally speaking, a sequence that clusters into an OTU with more than 97% sequence consistency is considered to be derived from the same species. The most intuitive OTU-based alpha diversity evaluation is the number of OTUs, which can reflect species richness. Analysis of 16S RNA sequencing results showed that 288 and 343 OTUs were attained from the symbiotic gut bacteria of the strains of Cx. Quinquefasciatus that were resistant (RLCql) and susceptible (SLCql) to L. sphaericus C3-41, respectively (Figure 1A). These OTUs corresponded to 176 bacterial species in RLCql and 183 in SLCql (Figures 1B,C). Although a greater number of bacterial species were observed in SLCql than RLCql, this difference was not significant and the abundance distribution of SLCql was also not significantly different to that of RLCql (Figures 1C,D and Table 3). In addition to the number of OTUs, observed species and Shannon indices were used to estimate the total species richness, both of which yielded similar results (Table 3). In addition, the rarefaction curve of the sequencing data tended to be smooth, demonstrating that the sequencing data volume increased at the expected rates. Moreover, the sequencing depth showed good coverage of all species in the sample. Therefore, the sequencing results are a good reflection of the bacteria in the sample, and any further addition of sequencing depth would be unlikely to significantly influence the alpha diversity (Figure 1B). This contrasts many 16S rRNA gene sequencing studies, which ultimately do not obtain smooth rarefaction curves for alpha diversity due to sequencing errors and other reasons.
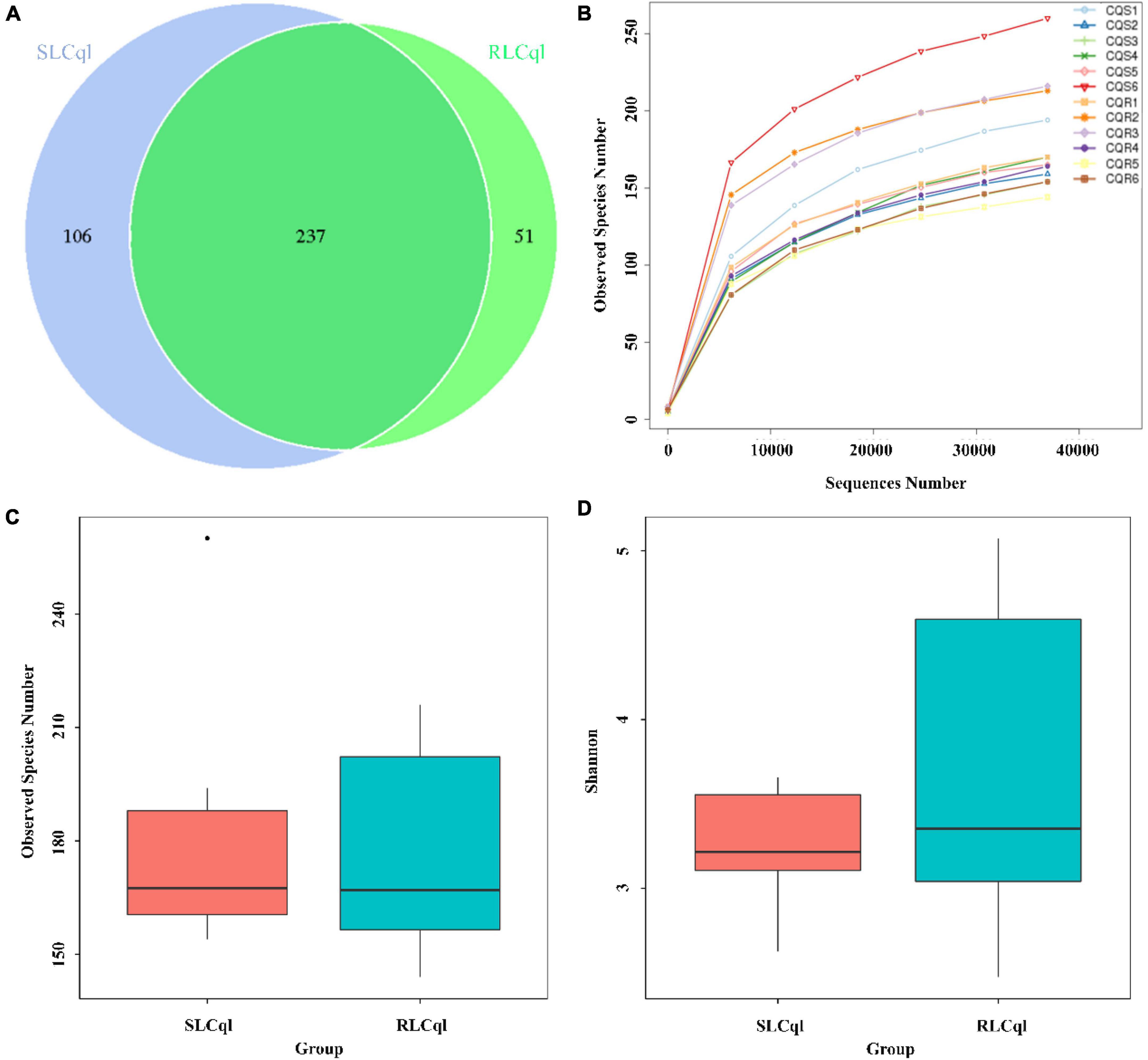
Figure 1. Alpha diversity of symbiotic gut bacteria from strains that are susceptible and resistant to Lysinibacillus sphaericus C3-41 [(A) Venn Graph, (B) Rarefaction curve, (C) Observed species, (D) Shannon index].
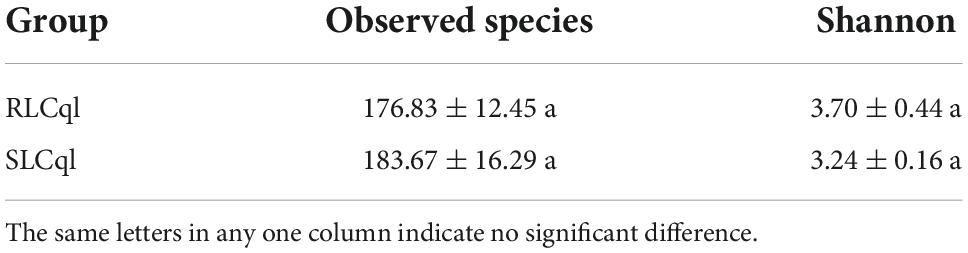
Table 3. Alpha diversity indices of gut bacteria from third-instar Culex quinquefasciatus strains that are susceptible and resistant to Lysinibacillus sphaericus C3-41.
The top 10 phyla with the highest abundance of symbiotic gut bacteria were illustrated in a cumulative histogram of the different bacterial compositions in RLCql and SLCql. The symbiotic gut bacteria of the susceptible colony were mainly comprised of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes (Figures 2A,B), while the resistant colony mainly consisted of Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and unidentified bacteria (Figures 2A,B). The relative abundances of the symbiotic bacterial communities in RLCql were markedly different to those in SLCql, indicating that the gut bacterial flora may have a functional role in the development of insecticide resistance in mosquitoes.
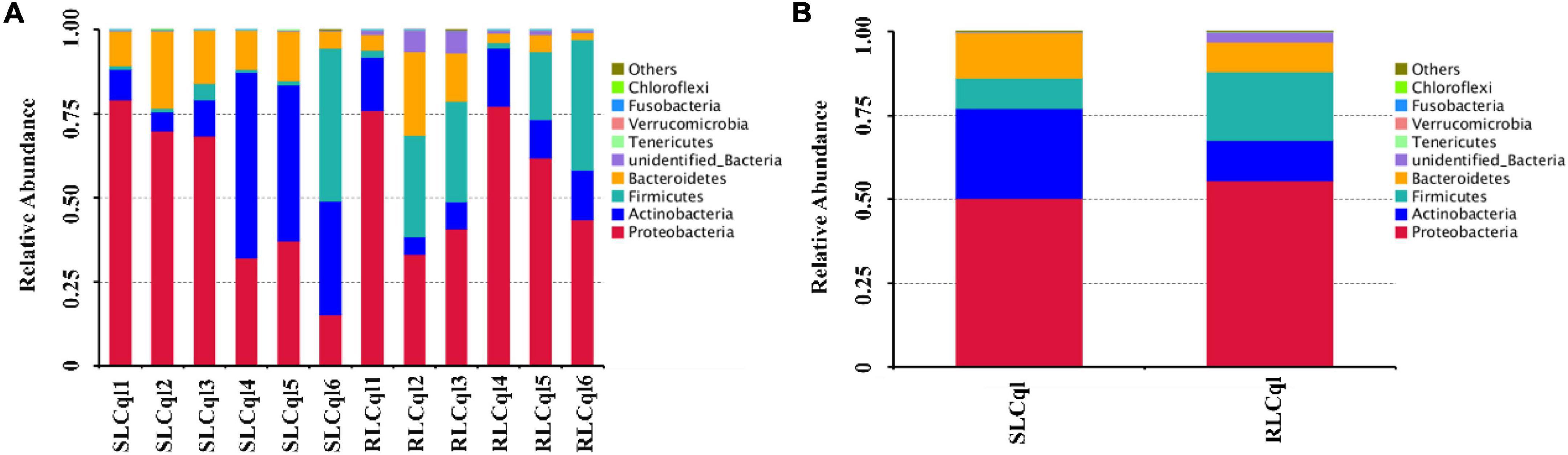
Figure 2. Symbiotic gut bacterial composition at the phylum level in strains that are susceptible and resistant to Lysinibacillus sphaericus C3-41. (A) The taxonomic composition of the dominant phyla per replicate of susceptible and resistant strains of Cx. quinquefasciatus, (B) the average taxonomic composition of the dominant phylum of the susceptible and resistant strains of Cx. quinquefasciatus.
The NMDS (Non-Metric Multi-Dimensional Scaling) and unweighted unifrac PCoA (Principal Coordinate Analysis) were used for beta diversity (β-diversity) analysis of the symbiotic gut bacterial communities of RLCql and SLCql (Figure 3A). The results of the NMDS analyses revealed that the symbiotic gut bacterial communities in the resistant strain differed significantly from those of the susceptible strain (Figure 3A). A similar pattern was observed in the PCoA analysis of unweighted unifrac distances (Figure 3B), which showed that the first and second principal coordinates explained 30.20 and 17.23% of the variation in the microbial β-diversity data, respectively. Therefore, the gut bacterial composition was likely altered in the resistant Cx. quinquefasciatus (Figure 3B).
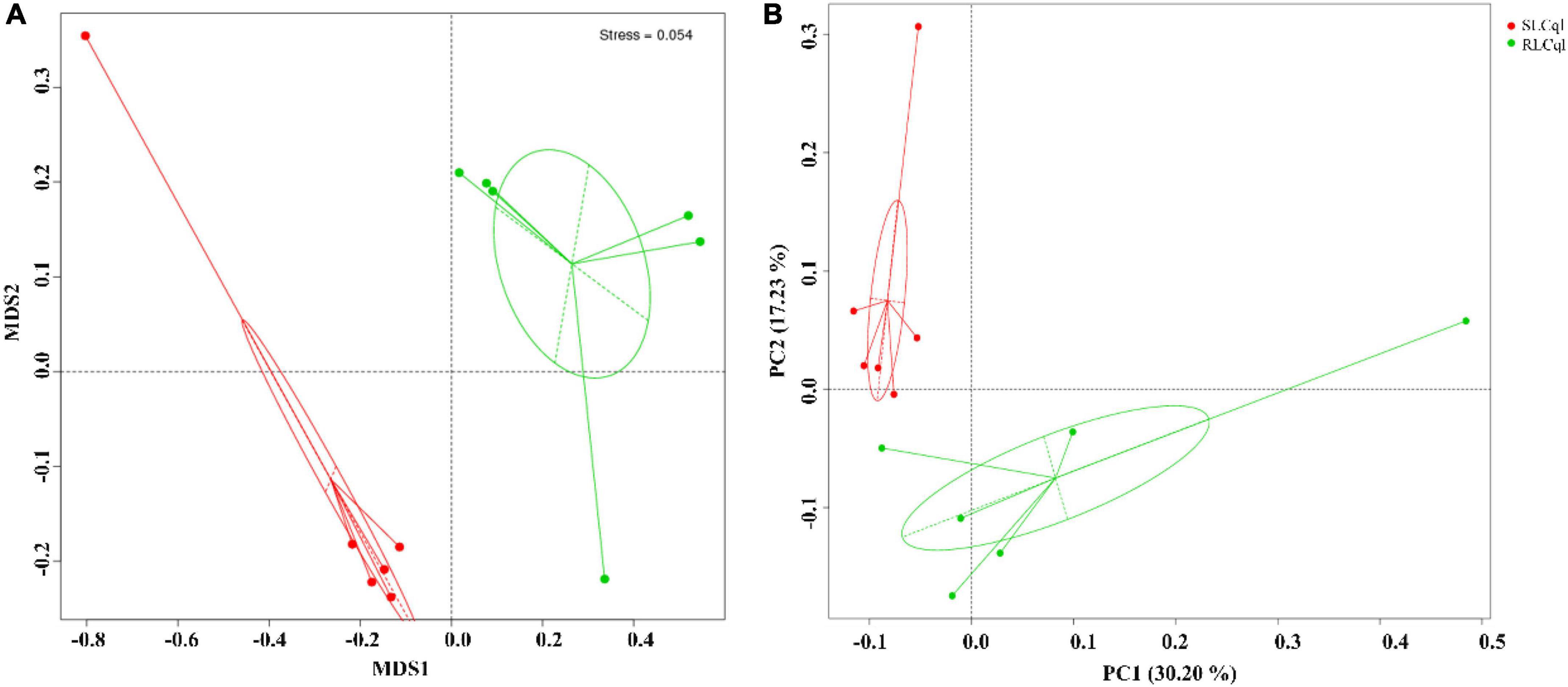
Figure 3. Beta diversity analysis of symbiotic gut bacteria from strains that are susceptible and resistant to Lysinibacillus sphaericus C3-41. (A) Non-metric multidimensional scaling (NMDS), (B) unconstrained principle coordinate analysis (PCoA).
Compared with the susceptible strain, the relative abundance of the genera Rahnella and unidentified Rhodocyclaceae was significantly lower in the resistant strain (Figure 4). In addition, the relative abundance of Rhodocyclaceae bacterium MORI9 in the resistant strain was also significantly lower than that in the susceptible strain (Figure 4). The differences in the relative abundances of the symbiotic bacterial communities between the SLCql and RLCql populations suggest that a critical role for gut symbionts in L. sphaericus C3-41 resistance in Cx. quinquefasciatus. Furthermore, the Rhodocyclaceae bacterium in the six replicates of RLCql demonstrated a lower genus-level contribution compared to SLCql (Figure 5). Taken together, these data suggest that gut symbionts may play an important role in the development of resistance against L. sphaericus C3-41.
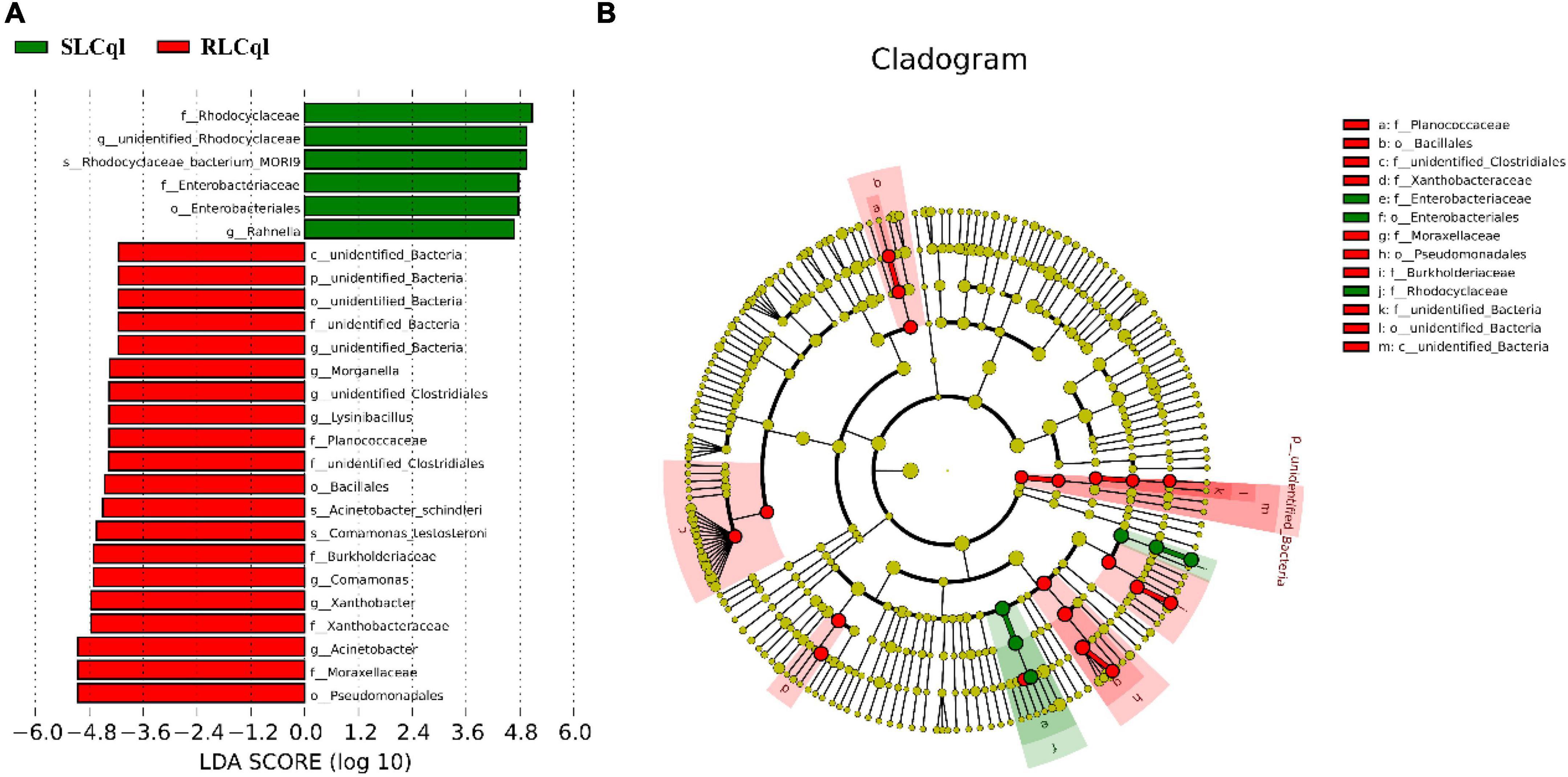
Figure 4. Taxons identified from Line Discriminant analysis (LDA) Effect Size (LEFSe) for strains of Culex quinquefasciatus that are susceptible (SLCql) and resistant (RLCql) to Lysinibacillus sphaericus C3-41. (A) Taxonomic cladogram. Taxa enriched in RLCql or SLCql are colored red or green, respectively. (B) SLCq1-enriched taxa are shown with a positive LDA score (green), and taxa enriched in RLCql are shown with a negative score (red). Only taxa meeting an LDA significance threshold > 4 and p < 0.05 are shown.
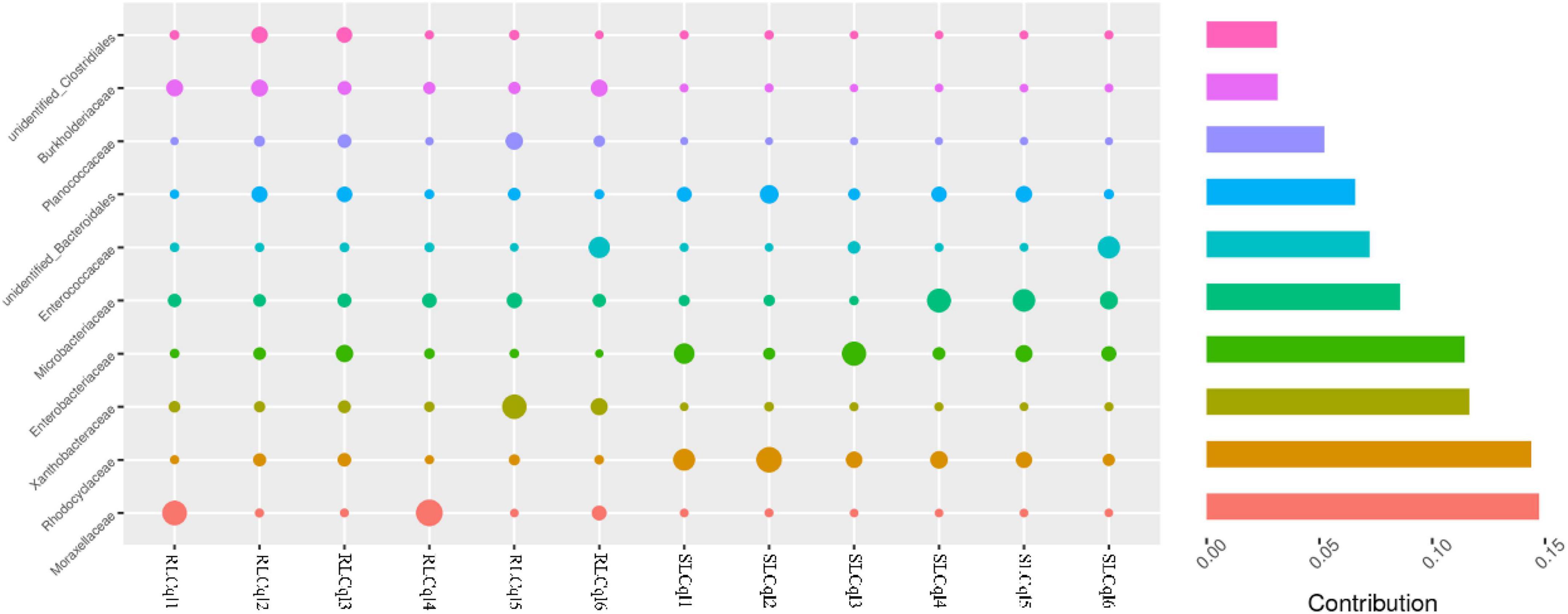
Figure 5. Genus-level contributions in strains of Culex quinquefasciatus that are susceptible and resistant to Lysinibacillus sphaericus C3-41.
Symbiotic gut bacterial communities have been shown to impact several key aspects of host biology, including host nutrition, growth and reproduction, metabolism, immunity, and defense (Engel and Moran, 2013). Therefore, the insect gut microbiome may be considered a virtual organ that plays an integral role in the physiology of the host and is essential for the host’s health (Wang and Qu, 2017). In line with this, this study demonstrated that the symbiotic gut bacterial communities of Cx. quinquefasciatus were mainly distributed in the Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes phyla (Figures 2A,B). Similarly, previous studies have also identified four main phyla, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, in Aedes aegypti and Anopheles gambiae larvae (Coon et al., 2016; Pennington et al., 2016; Guégan et al., 2018). Additionally, Proteobacteria, the most prominent phylum, accounted for approximately 15–79% of the total phyla, indicating that it may play a key role in the gut of Cx. quinquefasciatus. Actinobacteria and Bacteroidetes are also likely to be important, as they can impact mosquito development (Coon et al., 2014).
The structure of the symbiotic gut bacterial communities of the resistant strain was different to that of the susceptible strain. The enrichment of different bacterial groups yields a competitive advantage under insecticide selection (Dada et al., 2018). However, to some extent, the composition of the microbial community is affected by the feeding behavior of the larvae (Strand, 2019). During the selection and maintenance of resistance to L. sphaericus C3-41, the larvae of Cx. quinquefasciatus would ingest a large amount of L. sphaericus C3-41, which may alter the microbial diversity and community structure in the gut. In this study, the relative abundances of Firmicutes, Proteobacteria, and unidentified Bacteria in the gut of susceptible Cx. quequinfasciatus were lower than those in resistant strain (Figures 2A,B), indicating that these phyla may play a critical role in the development of resistance against L. sphaericus C3-41.
Moreover, the observed genus-level variations included significantly attenuated levels of the relative abundance of Rahnella, Rhodocyclaceae bacterium, and unidentified Rhodocyclaceae, and significantly higher levels of the relative abundance of Xanthobacter, Comamonas, Leucobacter, Pseudochrobactrum, Empedobacter, Hydrogenoanaerobacterium, Sphingobacterium, unidentified Bacteria, and Morganella in the resistant strain. These changes are likely related to the role of the gut microbiome as a functioning organ. To the best of our knowledge, however, these genera have not yet been reported in association with insecticide resistance, especially Rhodocyclaceae bacterium. Therefore, further studies are warranted to determine the roles of these bacteria in the mechanisms of insecticide resistance.
In conclusion, the structural differences in the symbiotic gut bacterial communities between resistant and susceptible Cx. quequinfasciatus were identified. Insecticide selection was found to significantly alter the composition of gut symbionts in Cx. Quequinfasciatus, and changes in the bacterial communities may occur after the development of resistance. This study provides a basis for future studies on the mechanisms of resistance to L. sphaericus C3-41 in Cx. quequinfasciatus, as well as a theoretical basis for in-depth study on the interaction between symbiotic gut bacterial communities and their hosts.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
XH and ZY conceived and designed the research. XZ and HM conducted the experiments, analyzed the data, and wrote the manuscript. All authors have read, corrected, and approved the manuscript.
This research was supported by the National Science and Technology Major Project of 13th Five-Year Plan, China (2017ZX10303404).
We are thankful to Quanxin Cai (Wuhan Institute of Virology) for kindly providing the susceptible Culex quequinfasciatus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cheng, D. F., Guo, Z. J., Riegler, M., Xi, Z. Y., Liang, G. W., and Xu, Y. J. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5:13. doi: 10.1186/s40168-017-0236-z
Chenniappan, K., and Ayyadurai, N. (2012). Synergistic activity of Cyt1A from Bacillus thuringiensis subsp. israelensis with Bacillus sphaericus B101 H5a5b against Bacillus sphaericus B101 H5a5b-resistant strains of Anopheles stephensi Liston (Diptera: Culicidae). Parasitol. Res. 110, 381–388. doi: 10.1007/s00436-011-2502-5
Coon, K. L., Brown, M. R., and Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25, 5806–5826. doi: 10.1111/mec.13877
Coon, K. L., Vogel, K. G., Brown, M. R., and Strand, M. R. (2014). Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23, 2727–2739. doi: 10.1111/mec.12771
Dada, N., Sheth, M., Liebman, K., Pinto, J., and Lenhart, A. (2018). Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep. 8:2084. doi: 10.1038/s41598-018-20367-4
Díaz-Nieto, L. M., Gil, M. F., Lazarte, J. N., Perotti, M. A., and Berón, C. M. (2021). Culex quinquefasciatus carrying Wolbachia is less susceptible to entomopathogenic bacteria. Sci. Rep. 11:1094. doi: 10.1038/s41598-020-80034-5
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Fu, P., Xiang, X., Ge, Y., Yuan, Z. M., and Hu, X. M. (2017a). Differential expression of duplicated binary toxin genes binA/binB in Lysinibacillus sphaericus C3-41. Lett. Appl. Micro. 65, 90–97. doi: 10.1111/lam.12752
Fu, P., Ge, Y., Wu, Y. M., Zhao, N., Yuan, Z. M., and Hu, X. M. (2017b). The LspC3-41I restriction-modification system is the major determinant for genetic manipulations of Lysinibacillus sphaericus C3-41. BMC Microbiol. 17:116. doi: 10.1186/s12866-017-1014-6
Guégan, M., Zouache, K., Démichel, C., Minard, G., Van, V. T., Potier, P., et al. (2018). The mosquito holobiont: Fresh insight into mosquito-microbiota interactions. Microbiome 6:49. doi: 10.1186/s40168-018-0435-2
Guo, Q. Y., Cai, Q. X., Yan, J. P., Hu, X. M., Zheng, D. S., and Yuan, Z. M. (2013). Single nucleotide deletion of cqm1 gene results in the development of resistance to Bacillus sphaericus in Culex quinquefasciatus. J. Insect Physiol. 59, 967–973. doi: 10.1016/j.jinsphys.2013.07.002
Guo, X. X., Li, C. X., Deng, Y. Q., Xing, D., Liu, Q. M., Wu, Q., et al. (2016). Culex pipiens quinquefasciatus: A potential vector to transmit Zika virus. Emerg. Microbes Infect. 5:e102. doi: 10.1038/emi.2016.102
Hao, X., Zhang, X., Duan, B., Huo, S., Lin, W., Xia, X., et al. (2018). Screening and genome sequencing of deltamethrin-degrading bacterium ZJ6. Curr. Microbiol. 75, 1468–1476. doi: 10.1007/s00284-018-1546-5
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. PNAS 109, 8618–8622. doi: 10.1073/pnas.1200231109
Mainali, S., Lamichhane, R. S., Clark, K., Beatty, S., Fatouros, M., Neville, P., et al. (2017). “Looking over the backyard fence”: Householders and mosquito control. Int. J. Environ. Res. Public Health 14:246. doi: 10.3390/ijerph14030246
Pei, G. F., Oliveira, C. M. F., Yuan, Z. M., Nielsen-LeRoux, C., Silva-Filha, M. H., Yan, J. P., et al. (2002). A strain of Bacillus sphaericus causes slower development of resistance in Culex quinquefasciatus. Appl. Environ. Microb. 68, 3003–3009. doi: 10.1128/AEM.68.6.3003-3009.2002
Pennington, M. J., Prager, S. M., Walton, W. E., and Trumble, J. T. (2016). Culex quinquefasciatus larval microbiomes vary with instar and exposure to common wastewater contaminants. Sci. Rep. 6:21969. doi: 10.1038/srep21969
Redekop, W. K., Lenk, E. J., Luyendijk, M., Fitzpatrick, C., Niessen, L., Stolk, W. A., et al. (2017). The socioeconomic benefit to individuals of achieving the 2020 targets for five preventive chemotherapy neglected tropical diseases. PLoS Negl. Trop. Dis. 11:e0005289. doi: 10.1371/journal.pntd.0005289
Reid, W. R., Zhang, L., Gong, Y. H., Li, T., and Liu, N. N. (2018). Gene expression profiles of the southern house mosquito Culex quinquefasciatus during exposure to permethrin. Insect Sci. 25, 439–453. doi: 10.1111/1744-7917.12438
Samy, A. M., Elaagip, A. H., Kenawy, M. A., Ayres, C. F. J., Peterson, A. T., and Soliman, D. E. (2016). Climate change influences on the global potential distribution of the mosquito Culex quinquefasciatus, vector of West nile virus and Lymphatic filariasis. PLoS One 11:e0163863. doi: 10.1371/journal.pone.0163863
Shah, R. M., Alam, M., Ahmad, D., Waqas, M., Ali, Q., Binyamin, M., et al. (2016). Toxicity of 25 synthetic insecticides to the field population of Culex quinquefasciatus say. Parasitol. Res. 115, 4345–4351. doi: 10.1007/s00436-016-5218-8
Strand, M. R. (2019). Composition and functional roles of the gut microbiota in mosquitoes. Curr. Opin. Insect Sci. 28, 59–65. doi: 10.1016/j.cois.2018.05.008
Wang, S. B., and Qu, S. (2017). Insect symbionts and their potential application in pest and vector-borne disease control. Bull. Chin. Acad. Sci. 32, 863–872. doi: 10.16418/j.issn.1000-3045.2017.08.008
Wang, Y. T., Shen, R. X., Xing, D., Zhao, C. P., Gao, H. T., Wu, J. H., et al. (2021). Metagenome sequencing reveals the midgut microbiota makeup of Culex pipiens quinquefasciatus and its possible relationship with insecticide resistance. Front. Microbiol. 12:625539. doi: 10.3389/fmicb.2021.625539
Wei, G., Bai, L., Qu, S., and Wang, S. B. (2018). Insect microbiome and their potential application in the insect pest and vector-borne disease control. Acta Microbiol. Sin. 58, 1090–1102. doi: 10.13343/j.cnki.wsxb.20180028
Wei, G., Lai, Y. L., Wang, G. D., Chen, H., Li, F., and Wang, S. B. (2017). Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. PNAS 114, 5994–5999. doi: 10.1073/pnas.1703546114
Xiong, W. H., Hu, X. M., and Yuan, Z. M. (2010). Application of Bacillus sphaericus in mosquito and vector control. Chin. J. Vector Biol. Control. 21, 1–4.
Yang, T., Li, T., Feng, X., Li, M., and Liu, N. (2021). Multiple cytochrome p450 genes: Conferring high levels of permethrin resistance in mosquitoes. Sci. Rep. 11:9041. doi: 10.1038/s41598-021-88121-x
Yu, S., Ji, C., Zhu, X. B., Xue, J. W., Wang, L. H., and Wang, Y. (2017). Impact of Bacillus sphaericus exposure on Anopheles dirus’s fecundity and resistance development. Parasitol. Res. 116, 859–864. doi: 10.1007/s00436-016-5358-x
Yuan, Z. M., Zhang, Y. M., Cai, Q. X., and Liu, E. Y. (2000). High-level field resistance to Bacillus sphaericus C3-41 in Culex quinquefasciatus from Southern China. Biocontrol. Sci. Techn. 10, 41–49.
Zhang, X. L., Karungu, S., Cai, Q. X., Yuan, Z. M., and Hu, X. M. (2019a). Effects of propoxur exposure on insecticidal susceptibility and developmental traits in Culex pipiens quinquefasciatus. Insects 10:288. doi: 10.3390/insects10090288
Keywords: Culex quinquefasciatus, Lysinibacillus sphaericus C3–41, gut symbiotic bacteria, resistance, 16S rRNA
Citation: Zhang X, Meng H, Hu X and Yuan Z (2022) Diversity and functional profile of gut symbiotic bacteria between Lysinibacillus sphaericus C3-41 susceptible and resistant Culex quinquefasciatus Say as revealed by 16S rRNA gene high-throughput sequencing. Front. Microbiol. 13:991105. doi: 10.3389/fmicb.2022.991105
Received: 11 July 2022; Accepted: 18 October 2022;
Published: 03 November 2022.
Edited by:
Jose Luis Ramirez, United States Department of Agriculture, United StatesReviewed by:
Boonhiang Promdonkoy, National Science and Technology Development Agency, ThailandCopyright © 2022 Zhang, Meng, Hu and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiming Yuan, eXptQHdoLmlvdi5jbg==; Xiaomin Hu, aHV4bUB3aC5pb3YuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.