
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 September 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.988871
This article is part of the Research TopicRising Stars in Hydrothermal Vents and Cold Seeps: 2021View all 10 articles
In seafloor sediments, the anaerobic oxidation of methane (AOM) consumes most of the methane formed in anoxic layers, preventing this greenhouse gas from reaching the water column and finally the atmosphere. AOM is performed by syntrophic consortia of specific anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria (SRB). Cultures with diverse AOM partners exist at temperatures between 12°C and 60°C. Here, from hydrothermally heated sediments of the Guaymas Basin, we cultured deep-branching ANME-1c that grow in syntrophic consortia with Thermodesulfobacteria at 70°C. Like all ANME, ANME-1c oxidize methane using the methanogenesis pathway in reverse. As an uncommon feature, ANME-1c encode a nickel-iron hydrogenase. This hydrogenase has low expression during AOM and the partner Thermodesulfobacteria lack hydrogen-consuming hydrogenases. Therefore, it is unlikely that the partners exchange hydrogen during AOM. ANME-1c also does not consume hydrogen for methane formation, disputing a recent hypothesis on facultative methanogenesis. We hypothesize that the ANME-1c hydrogenase might have been present in the common ancestor of ANME-1 but lost its central metabolic function in ANME-1c archaea. For potential direct interspecies electron transfer (DIET), both partners encode and express genes coding for extracellular appendages and multiheme cytochromes. Thermodesulfobacteria encode and express an extracellular pentaheme cytochrome with high similarity to cytochromes of other syntrophic sulfate-reducing partner bacteria. ANME-1c might associate specifically to Thermodesulfobacteria, but their co-occurrence is so far only documented for heated sediments of the Gulf of California. However, in the deep seafloor, sulfate–methane interphases appear at temperatures up to 80°C, suggesting these as potential habitats for the partnership of ANME-1c and Thermodesulfobacteria.
In anoxic deep-sea sediments, the greenhouse gas methane is produced abiotically by thermocatalytic decay of buried organic matter or biotically by methanogens (Whiticar, 1999). Anaerobic oxidation of methane (AOM) mitigates the flux of methane to the water column and eventually to the atmosphere by consuming 90% of the methane produced in the deep sediments (Hinrichs and Boetius, 2002; Reeburgh, 2007; Regnier et al., 2011). In marine sediments, AOM primarily couples to sulfate reduction in a 1:1 stoichiometry:
AOM is mediated by anaerobic methanotrophic archaea (ANME) that oxidize methane to CO2 by reversing the methanogenesis pathway (Hallam et al., 2004; Meyerdierks et al., 2010; Wang et al., 2013). ANME do not encode respiratory pathways, but they pass the reducing equivalents liberated during AOM to sulfate-reducing partner bacteria (SRB), forming characteristic consortia (Boetius et al., 2000; Michaelis et al., 2002; Orphan et al., 2002; McGlynn et al., 2015; Wegener et al., 2015). The nature of this syntrophic association and the mechanisms involved in the transfer of reducing equivalents from ANME toward SRB are not completely resolved at the molecular level. Originally, it was proposed that the archaeal partners produce molecular hydrogen that is consumed by the bacterial partners (Hoehler et al., 1994). However, most ANME do not code for hydrogenases (Chadwick et al., 2022). Previous studies support the hypothesis of direct interspecies electron transfer (DIET) involving multiheme cytochromes and pilus proteins (Meyerdierks et al., 2010; McGlynn et al., 2015; Wegener et al., 2015). The partner SRBs use the AOM-derived electrons for anaerobic respiration with sulfate as final electron acceptor (Boetius et al., 2000; Wegener et al., 2015; Laso-Pérez et al., 2016). The limited energy yield of sulfate-dependent AOM [equation (1), ΔG°′ = −16.67 kJ mol−1 at standard conditions and ΔG = −20 to −40 kJ mol−1 in marine AOM habitats] needs to be shared between ANME and their syntrophic partner SRB (Thauer, 2011).
ANME inhabit a variety of marine habitats including cold seeps (Boetius et al., 2000; Orphan et al., 2001), mud volcanoes (Niemann et al., 2006), gas hydrates (Lanoil et al., 2001; Orcutt et al., 2004), hydrothermal vents (Inagaki et al., 2006; Biddle et al., 2012) and deep subsurface sediments (Roussel et al., 2008). ANME are polyphyletic and fall into three distinct phylogenetic groups (ANME-1, ANME-2, and ANME-3). ANME-3 often dominate AOM at mud volcanoes, where they form consortia with Desulfobulbus-related bacteria (Niemann et al., 2006; Lösekann et al., 2007). Cultivation attempts of ANME-3 have not been successful so far. ANME-2 are globally distributed in a variety of benthic habitats and are typically found associated with Desulfosarcina/Desulfococcus bacteria (DSS, Seep-SRB1 and Seep-SRB2 clades; Knittel et al., 2003, 2005; Boetius and Knittel, 2010). ANME-2 are dominant at cold seeps with high methane fluxes and temperatures below 20°C (Knittel et al., 2005). Cultivation attempts at temperatures ≤ 20°C resulted in the enrichment of ANME-2c (Holler et al., 2009; Wegener et al., 2016).
ANME-1 prevail in most deep sulfate–methane transition zones (SMTZs; Ishii et al., 2004; Niemann et al., 2005; Treude et al., 2005), in hydrothermally heated sediments in the Guaymas Basin (Teske et al., 2002; Schouten et al., 2003; Ruff et al., 2015; Dombrowski et al., 2018), and in the Auka vent field, in the Pescadero Basin (Gulf of California; Speth et al., 2022). Meso- and thermo-philic AOM cultures have been obtained from Guaymas Basin sediments at 37°C, 50°C, and 60°C (Holler et al., 2011; Wegener et al., 2016). These cultures consisted of ANME-1a and HotSeep-1 (Ca. Desulfofervidus) as partner bacteria. Ca. Desulfofervidus sequences are also found in situ at these sites (McKay, 2014; Dowell et al., 2016).
Previous short-term incubations revealed AOM activity at temperatures up to 75°C or 85°C, but the microorganisms performing AOM under these conditions were not assessed (Kallmeyer and Boetius, 2004; Holler et al., 2011; Adams et al., 2013). Here, we obtained an active AOM culture at 70°C (AOM70) from Guaymas Basin hydrothermal sediments consisting of a previously uncultured ANME-1 subgroup (Teske et al., 2002) and an apparently obligate syntrophic Thermodesulfobacterium partner. We describe their function and interaction based on physiological experiments and molecular data.
Sediment push cores from gas-rich hydrothermal vents of the Guaymas Basin (Gulf of California) were collected by the submersible Alvin at 2013 m depth during RV Atlantis cruise AT42-05 (November 2018). Sediments for this AOM enrichment came from cores 4,991–13 and 4,991–14 in the Cathedral Hill area (27°00.6848′ N, 111° 24.2708′ W) collected on 17 November 2018, in an area covered by dense orange-white Beggiatoaceae mats, where temperatures at 50 cm depth reached at least 80°C. On board sediment samples were transferred to glass bottles sealed with butyl rubber stoppers, the headspace was exchanged to argon. Sediments were stored at 4°C until further processing. Sediment slurries were prepared following protocols previously described (Laso-Pérez et al., 2018). Anoxic sediments were mixed with sulfate-reducer medium (Widdel and Bak, 1992) in a 1/10 ratio (v/v) in serum vials sealed with rubber stoppers. The headspace of the serum vials was replaced with 2 atm methane:CO2 (90:10). The dry weight of the original slurries was 60 g L−1. The slurries were incubated at 70°C in the dark. Methane-dependent sulfide production was measured with the copper sulfate assay (Cord-Ruwisch, 1985). Incubations with methane-dependent sulfide productions at 70°C are referred to as AOM70 culture. These cultures were diluted 1/5 with new medium when sulfide levels reached > 10 mM. Cultures were virtually sediment-free after four dilutions.
DNA samples for long-read sequencing were prepared according to previous protocols with few modifications (Zhou et al., 1996; Hahn et al., 2020). In short, 50 ml culture were collected in a Falcon tube and biomass was pelleted by centrifugation at RT (4,000 rpm for 20 min). After removing the supernatant, 800 μl extraction buffer was added (100 mM tris–HCl, 100 mM sodium EDTA, 100 mM sodium phosphate, 1.5 M NaCl, 1% CTAB, pH 8). For physical lysis of cell envelopes, the pellet suspension was frozen twice in liquid nitrogen and thawed in a water bath at 65°C. For enzymatic lysis, 1,000 μl extraction buffer with 60 μl proteinase K (20 mg mL−1) was used at 37°C for 1.5 h with constant shaking. Chemical lysis was done with 300 μl 20% SDS at 65°C for 2 h. Cell debris was pelleted again by centrifugation at RT (13,000 × g for 20 min). The clear supernatant was transferred to a new tube and 2 ml of chloroform-isoamyl alcohol (16:1, v:v) were added. The samples were mixed by inverting the tubes and centrifuged at RT (13,000× g for 20 min). The aqueous phase was transferred to a new tube and mixed with 0.6 volumes isopropanol. DNA was precipitated overnight at −20°C. After precipitation, DNA was re-dissolved at 65°C in a water bath for 5 min and samples were centrifuged at RT (13,000× g for 40 min). Supernatant was removed and the pellet was washed with ice-cold 80% ethanol. Samples were centrifuged at 13,000× g for 10 min and the ethanol was removed. Dried pellets were resuspended in 100 μl PCR-grade water. Long-read (>10 kb) genomic DNA was sequenced on a Sequel IIe (Pacific Biosciences) at the Max Planck Genome Centre in Cologne. Read length distributions and abundances are compiled in Supplementary Table 1.
Triplicates of 30 ml culture were filtered onto 0.2 μl polycarbonate filters under gentle vacuum. Filters were soaked immediately with RNAlater (Invitrogen) preheated at 70°C for 10–15 min. RNAlater was removed by filtration and the filters were stored at −20°C until further processing. For RNA extraction, ¼ of a filter was put into a bead-beating tube (Lysing Matrix E, MPBio) together with 600 μl RNA lysis buffer (Quick-RNA MiniPrep kit, Zymo Research). Tubes were vortexed at maximum speed for 20 min. Biomass was pelleted by centrifugation at RT (10,000× g for 5 min). The supernatant was collected and RNA was extracted with the Quick-RNA MiniPrep kit (Zymo Research) including a DNA digestion step with DNase I. Total RNA libraries were sequenced in an Illumina HiSeq2500 machine at the Max Planck Genome Centre (Cologne, Germany). We obtained 4 Mio 2 × 250 bp paired-end reads.
Metagenomic long-reads were assembled using Flye v. 2.9 (Kolmogorov et al., 2020). Shotgun metatranscriptomic short reads were quality trimmed using BBduk from the BBtools package v. 38.871 with the parameters minlength = 50 mink = 6 hdist = 1 qtrim = r trimq = 20. Metagenomic reads were mapped to the general assembly. Long reads were mapped using minimap2 v. 2.21 (Li, 2018) with default parameters. Open reading frames in metagenomic contigs were predicted with prodigal v. 2.6.3 (Hyatt et al., 2010) and genes were annotated with PFAMs, TIGRFAMs, COGs, KEGGs, and RNAmmer (Kanehisa and Goto, 2000; Haft et al., 2001; Lagesen et al., 2007; Galperin et al., 2015; Mistry et al., 2021). CXXCH motifs in putative multiheme cytochromes were searched with a custom script.2 Predicted hydrogenase sequences were classified into subgroups with the hydrogenase database (HydDB; Søndergaard et al., 2016). Subcellular localization of heme-containing proteins and hydrogenases was predicted with PSORTb 3.0 (Yu et al., 2010). Metagenomic binning based on differential coverage across metagenomic samples was done with maxbin v 2.2.7 (Wu et al., 2016). Bins were manually refined in anvi’o v. 6 (Eren et al., 2015, 2020) by removing contigs with low coverage from high-coverage bins.
Triplicate metatranscriptomes were mapped to curated bins using bowtie 2 (Langmead and Salzberg, 2012). The rRNA and tRNA gene sequences were removed before calculating gene expression levels. Center-log ratio (CLR) values for relative gene expression were calculated according to the formula:
where are the reads mapped to a specific gene and is the length of the gene in kbp. A 0.5 factor was added to read-mapping values to avoid zero values.
To analyze the similarity of cytochrome-like proteins in ANME-1 and sulfate-reducing bacteria, amino acid sequences from sulfate-reducing partner bacteria genomes were downloaded from NCBI (Krukenberg et al., 2016, 2018). BLAST databases were created from the cytochrome sequences in the reference SRB genomes using makeblastdb (BLAST v. 2.10.1; Altschul et al., 1990). Cytochrome-like proteins in AOM70 cultures were queried against the custom database with BLASTp v 2.5.
16S rRNA genes from long-read metagenomic assemblies were extracted with Metaxa2 (Bengtsson-Palme et al., 2015). Full-length 16S rRNA gene sequences were aligned to the SILVA database release 138.1 using the SINA aligner within the ARB software (Ludwig et al., 2004; Pruesse et al., 2012; Quast et al., 2013). Long reads were mapped against 16S rRNA genes using minimap2 (Li, 2018). Shotgun metatranscriptomic reads were aligned to 16S rRNA gene using bowtie2 (Langmead and Salzberg, 2012). Maximum-likelihood 16S rRNA phylogenetic trees with selected ANME or Thermodesulfobacteria sequences were calculated using RAxML with 1,000 bootstraps and a 50% frequency base filter (Stamatakis, 2014).
Archaea and Bacteria genomes were downloaded from public databases (Supplementary Table 3). For ANME-1 phylogenomic analysis, the genomes were annotated with HMMs of 38 conserved archaeal marker genes (Supplementary Table 3; Darling et al., 2014). For Thermodesulfobacteria, the genomes were annotated with HMMs of 71 conserved bacterial marker genes (Rinke et al., 2013). The amino acid sequences of each set were aligned and concatenated using MUSCLE (Edgar, 2004). Maximum likelihood phylogenomic trees were calculated with IQTree using the –test option to estimate the best substitution model for each protein in the partition file and using 100 bootstraps (Chernomor et al., 2016; Kalyaanamoorthy et al., 2017; Minh et al., 2020). Reference hydrogenase sequences (Supplementary Table 5) were downloaded from the hydrogenase database (HydDB; Søndergaard et al., 2016). ANME-1 hydrogenases and reference hydrogenases were aligned with muscle (Edgar, 2004). Maximum likelihood phylogenetic trees of the alignment were calculated with IQTree with 100 bootstraps (Chernomor et al., 2016; Kalyaanamoorthy et al., 2017; Minh et al., 2020). Trees were visualized and edited on the Interactive Tree Of Life (iTOL) online server (Letunic and Bork, 2011).
To prepare catalyzed reporter deposition-fluorescent in situ hybridization (CARD-FISH) samples, 5 ml culture were fixed at 1% formaldehyde concentration over night at 4°C. Fixed samples were sonicated (15 s, 30% power, 20% cycle) to detach cells from sediment particles to detach larger aggregates. Samples were then filtered onto 0.2 μm polycarbonate filters and fixed with 0.2% low-melting agarose before CARD-FISH. Samples were stored at −20°C until further processing. CARD-FISH was performed as described previously (Pernthaler et al., 2002). In short, endogenous peroxidases were inactivated with a solution of 0.15% H2O2 in methanol for 30 min at RT. Cell walls were permeabilized with lysozyme (Sigma Aldrich, 10 mg mL−1 lysozyme in 50 mM EDTA, 100 mM Tris–HCl; 60 min incubation at 37°C), proteinase K (15 μg mL−1 proteinase K in 50 mM EDTA, 100 mM Tris–HCl, 500 mM NaCl; 10 min incubation at RT) and HCl (0.1 M HCl; 1 min incubation at RT). Horseradish peroxidase-labeled probes were diluted in hybridization buffer at the adequate formamide concentration for each probe (Supplementary Table 2). Probes were hybridized at 46°C for 2 h. Signal amplification with fluorescent tyramides was done for 45 min at 46°C. For double hybridizations, peroxidases of the prior hybridization step were inactivated by incubating the filter in 0.30% H2O2 in methanol for 30 min at RT.
For cultures under AOM conditions, hydrogen formation in the headspace was measured by gas chromatography coupled to reducing compound photometry (RCP, Peak Performer 1 RCP, Peak Laboratories). For cultures under methanogenic conditions, methane formation in the headspace was measured via gas chromatography and flame ion detection (GC-FID, Focus GC, Thermo).
A slurry produced from hydrothermally-heated sediments from the Guaymas Basin and sulfate-reducer medium was supplemented with a methane:CO2 headspace and incubated at 70°C. These incubations showed methane-dependent sulfate reduction, as measured by an increase of sulfide in the medium (Figure 1B; Supplementary Figure 1). These incubations produced sulfide 12 to 15 mM sulfide within about 100 days. The slurries were diluted 1/10 (v/v) in fresh SRB medium, and a fresh methane:CO2 headspace was added and the incubation was proceeded. After four additional incubation and dilution steps, the produced AOM70 cultures were virtually sediment-free, contained microbial aggregates visible with the naked eye and produced approximately. 100 μmol sulfide L–1 d–1. To our knowledge, this is the first long-term cultivation of AOM-performing microorganisms above 60°C. The culture showed strongly decreased sulfide production at 60°C. It tolerated a transfer to 75°C, but became inactive at 80°C, confirming prior results on the upper-temperature limit of AOM made in short-term incubations with Guaymas Basin sediments (Holler et al., 2011; McKay et al., 2016).
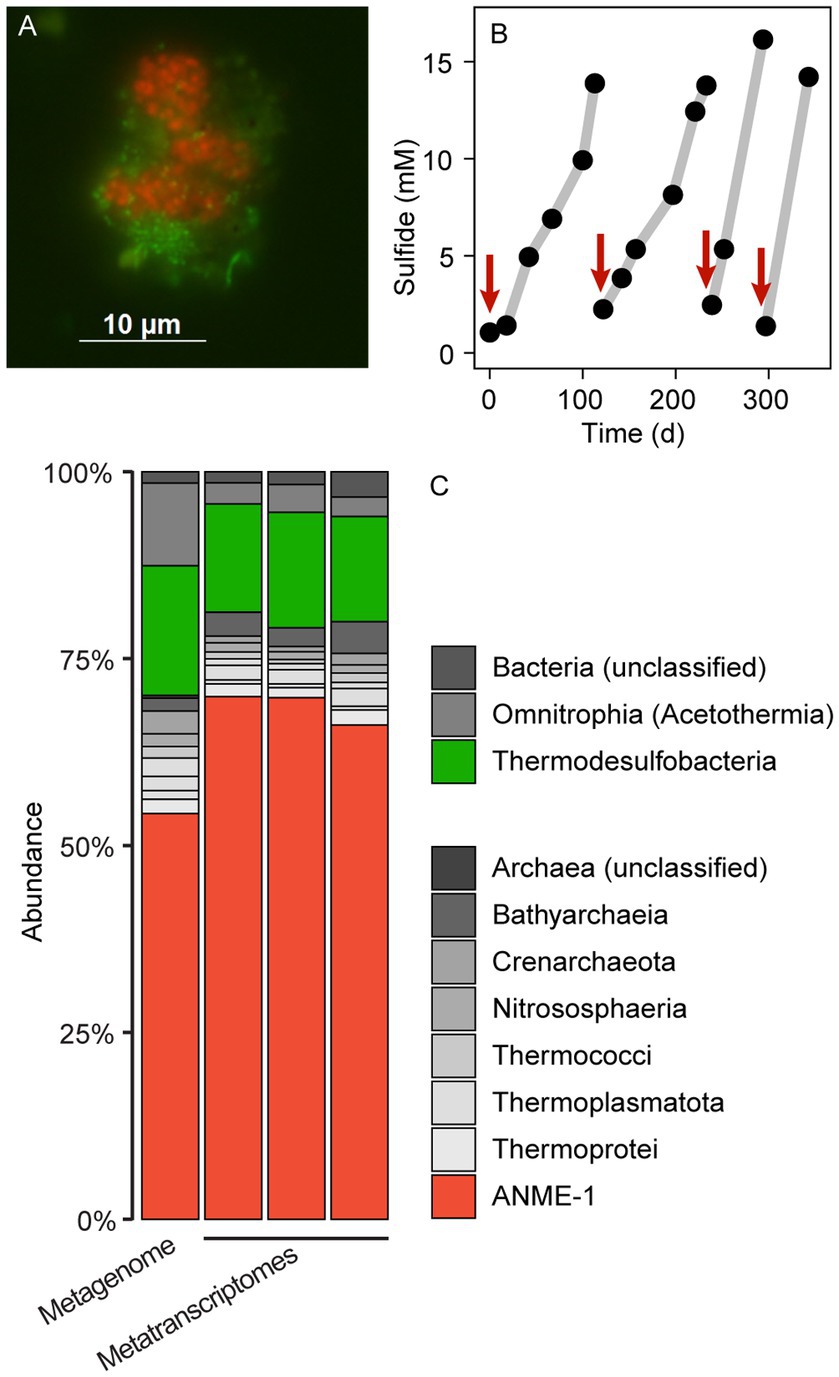
Figure 1. Microbial composition and growth of thermophilic AOM cultures. (A) CARD-FISH on AOM aggregates with the probes EUB388 I-III (green) and ANME-1-389 (red). Bacteria (green) and archaea (red) form shell-type consortia. (B) Methane-dependent sulfide production in AOM cultures. The red arrows indicate when culture medium was replaced. (C) 16S rRNA gene relative abundance in long-read metagenomic reads and shotgun metatranscriptomic reads (triplicate metatranscriptomes). The enrichment is dominated by ANME-1 and sulfate reducers of the class Thermodesulfobacteria. Other bacteria and archaea such as Acetothermia and Bathyarchaeia are side community members from original sediment samples.
To resolve the microbial community composition of the AOM70 culture, we obtained a long-read metagenome and triplicate short-read metatranscriptomes. The community consisted mainly of ANME-1 (~50% relative abundance of mapped reads) and Thermodesulfobacteria (~20% relative abundance) based on 16S rRNA gene fragments recruited from the metagenome (Figure 1C) that were rare in the original sediment samples (Supplementary Figure 2). Metatranscriptomic samples were also dominated by ANME-1 (~70% relative abundance) and Thermodesulfobacteria (~15% relative abundance), forming the active AOM community at 70°C. Both the metagenome and metatranscriptome revealed noticeable populations of Bathyarchaeota and Acetothermia (<10% relative abundance of mapped reads). Yet, we were not able to reconstruct MAGs of these organisms; hence, their potential functions are unknown. Previous studies suggest that these organisms ferment or oxidize biomolecules produced by the AOM community (Kellermann et al., 2012; Dombrowski et al., 2017; Hao et al., 2018; Zhu et al., 2022).
After long-read metagenome assembly and binning, we obtained two high-quality MAGs of the two members of the AOM consortium (Table 1). The Thermodesulfobacterium MAG has a size of 1.7 Mbp and GC content of 29%. The bin is almost complete (98.6%) and has no contamination based on the presence of 104 bacterial single-copy marker genes (CheckM; Parks et al., 2015). The ANME-1 bin has a size of 1.5 Mbp and a GC content of 47.8%. The bin is 90.8% complete and has contamination of 7.9% based on 149 archaeal marker genes (CheckM; Parks et al., 2015).
We attempted to visualize the enriched ANME-1 using a previously established probe targeting the whole ANME-1 clade (ANME-1-350, Supplementary Table 2; Boetius et al., 2000). In situ hybridization with the ANME-1-350 failed, because the probe has two mismatches with the 16S rRNA sequence of the enriched ANME-1. The 16S rRNA gene of this ANME-1 belong to a clade ancestral to all ANME-1a/b, namely ANME-1c (Supplementary Figure 4 and Discussion below; Laso-Pérez et al., 2022). A newly developed ANME-1-389 probe specifically binds to ANME-1c cells. Because the probes available for partner SRB do not target the 16S rRNA sequence of Thermodesulfobacteria, we designed three candidate probes to target this clade (Supplementary Table 2). Unfortunately, none of these probes hybridized the 16S rRNA of this organism after various CARD-FISH attempts (Supplementary Table 2). Thermodesulfobacteria are likely the partner bacteria of ANME-1c during AOM at 70°C based on the abundance of bacterial cells and their gene content (see Discussion below). Furthermore, all genes coding for dissimilatory sulfate reductase (dsr) in the metagenome belong to the Thermodesulfobacterium MAG. Double hybridization with the ANME-1c and the general bacterial probes (Supplementary Table 2) revealed a dominance of “shell-type” aggregates consisting of ANME-1c and partner bacteria (Figure 1A; Supplementary Figure 3). These consortia consist of clumps of ANME-1c cells, surrounded by smaller rod-shaped bacterial cells. These shell-type aggregates differ from the predominantly mixed-type aggregates of moderately thermophilic consortia growing at 50°C–60°C (Holler et al., 2011; Wegener et al., 2015). A shell-type growth morphology is often observed in cold-adapted ANME (Knittel et al., 2005). The reason for the different association types is unknown.
On the basis of whole genome comparison, the ANME-1 population detected in the AOM70 culture falls into the recently named ANME-1c clade (Figure 2A; Laso-Pérez et al., 2022; Speth et al., 2022).The ANME-1c group is basal to its sister groups ANME-1a and ANME-1b within the order ANME-1 (Ca. Methanophagales). The 16S rRNA gene phylogenetic tree supports this phylogenetic placement (Supplementary Figure 4). ANME-1c belong to the class Syntrophoarchaeia with the ANME-1, Ca. Syntrophoarchaeales and Ca. Alkanophagales. Considering an average nucleotide identity (ANI) of <83% for distinct species and >95% for the same species (Jain et al., 2018) the ANME-1c clade consists of two distinct species clusters (Supplementary Figure 5). The ANME-1c MAG from the AOM70 culture belongs to the cluster of Ca. Methanoxibalbensis ujae from Pescadero Basin (Laso-Pérez et al., 2022). ANME-1c 16S rRNA gene sequences have been detected in hydrothermal sediments of the Guaymas Basin and the Juan de Fuca Ridge (Supplementary Figure 4; Teske et al., 2002; Merkel et al., 2013; McKay et al., 2016), and a MAG of the ANME-1c clade (accessions: SAMN09215218, GCA_003661195.1) was derived from Guaymas Basin hydrothermal sediments (Dombrowski et al., 2018). ANME-1c are also present in rock samples from hydrothermal fields in Pescadero Basin (Gulf of California; Speth et al., 2022). The ANME-1c clade was originally named “ANME-1b” by Teske and coworkers to differentiate this lineage from previously described cold-seep ANME-1 (Teske et al., 2002) and later renamed to ANME-1Guaymas because it was predominantly recovered from Guaymas Basin (Biddle et al., 2012; Merkel et al., 2013; Dowell et al., 2016). These sequences originate from sediment cores with sulfate-reducing activity at temperatures between 65°C and 90°C, showing that these archaea are likely all thermophiles (Biddle et al., 2012). Furthermore, the high GC content (>60%) of ANME-1c 16S rRNA genes indicates that these archaea might have temperature optima in the upper range of thermophily above 70°C (Merkel et al., 2013).
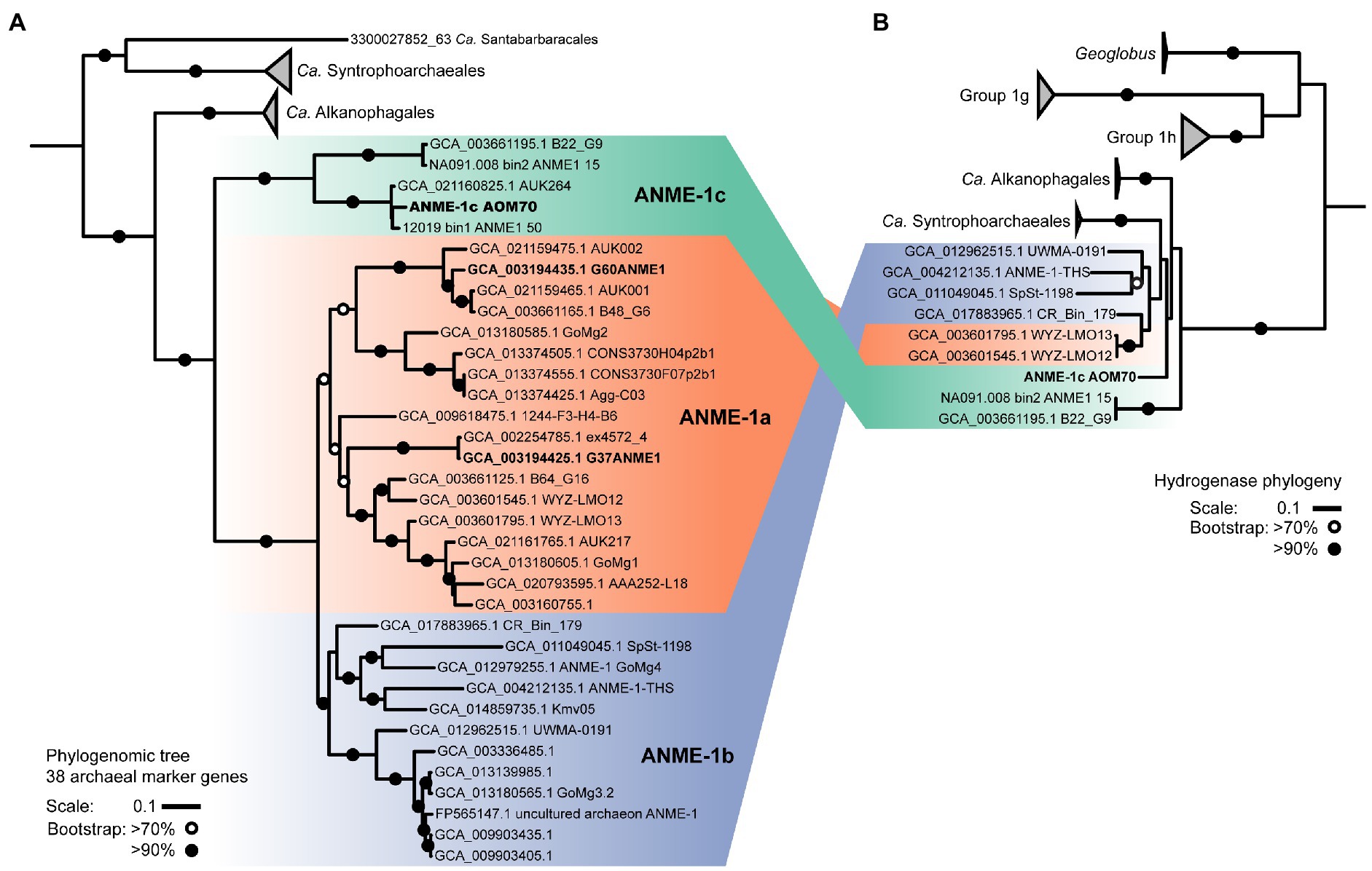
Figure 2. ANME-1 phylogenomic tree and hydrogenase phylogeny. (A) Phylogeny of ANME-1 order (Ca. Methanophagales) with related Ca. Alkanophagales, Ca. Syntrophoarchaeales, and Ca. Santabarbaracales. Maximum likelihood phylogenomic tree based on an alignment of 38 archaeal conserved genes from 55 genomes (Supplementary Table 3). Geoglobus sequences were the outgroup to set the tree root (not shown). (B) Hydrogenase phylogeny. ANME-1, Ca. Alkanophagales and Ca. Syntrophoarchaeales hydrogenases are located at the base of groups 1 g and 1 h of NiFe hydrogenases. A complete hydrogenase tree is shown in Supplementary Figure 11. MAGs from cultured ANME are depicted in bold. Shading in both trees indicates the three subdivisions of the ANME-1: ANME-1c, ANME-1a and ANME-1b. ANME-1 AOM70 is the genome discussed in the main text. Scales indicate nucleotide substitution per site. Bootstrap support is based on 100 iterations above 70% and above 90%.
ANME-1c codes for a complete methanogenesis pathway including a canonical methane-active Mcr (Figure 3). The mcrABC genes in ANME-1c have the highest expression (CLR > 7) among all genes in the dataset. This high expression of mcr confirms previous transcriptomic work in ANME (Haroon et al., 2013; Krukenberg et al., 2018). The activation of methane is the rate-limiting step of AOM, and ANME would promote this reaction by producing large amounts of Mcr (Scheller et al., 2010; Thauer, 2011). Similar to other ANME-1 archaea, ANME-1c does not encode a N5,N10-methylene-H4MPT reductase (mer). This gene might be substituted by a 5,10-methylenetetrahydrofolate reductase (met; Stokke et al., 2012; Krukenberg et al., 2018). The function of this bypass has not been verified yet. All other genes of the methanogenesis pathway show a relatively high expression with CLR values between 0.1 and 3.4 (Supplementary Table 4), supporting a catabolic function of the encoded genes. ANME-1c encodes and expresses the methanogenesis-related membrane-bound complex H+-translocating F420:quinone oxidoreductase (fqo) that catalyzes the transfer of electrons from reduced cofactors to the quinone pool (Pereira et al., 2011). ANME-1c encodes an ATP synthase, which is a common feature in ANME to enable the oxidative phosphorylation of ATP, coupled to the influx of protons. ANME-1c encodes a sulfate adenylyltransferase (cysN; low expression, CLR = −0.02) and an adenylylsulfate kinase (cysC; high expression, CLR = 2.21) that could be used for assimilatory sulfate metabolism, but it lacks the key genes for dissimilatory sulfate reduction. The ANME-1c MAG lacks a complete nitrogenase operon, suggesting it is incapable of nitrogen fixation. The capability for nitrogen fixation has been shown only in ANME-2 archaea but not in ANME-1 (Dekas et al., 2009, 2015; Orphan et al., 2009; Krukenberg et al., 2018). The nitrogenase subunits nifDH detected in ANME-1c and other ANME-1 genomes (Meyerdierks et al., 2010) are likely paralogs of cfbCD because they are located in an operon with genes encoding the biosynthetic pathway of coenzyme F430 (Zheng et al., 2016; Moore et al., 2017). Coenzyme F430 functions as a prosthetic group that binds to the active site of McrA, and is therefore a key molecule for methanogens and methanotrophs (Friedmann et al., 1990; Ermler et al., 1997; Shima et al., 2012).
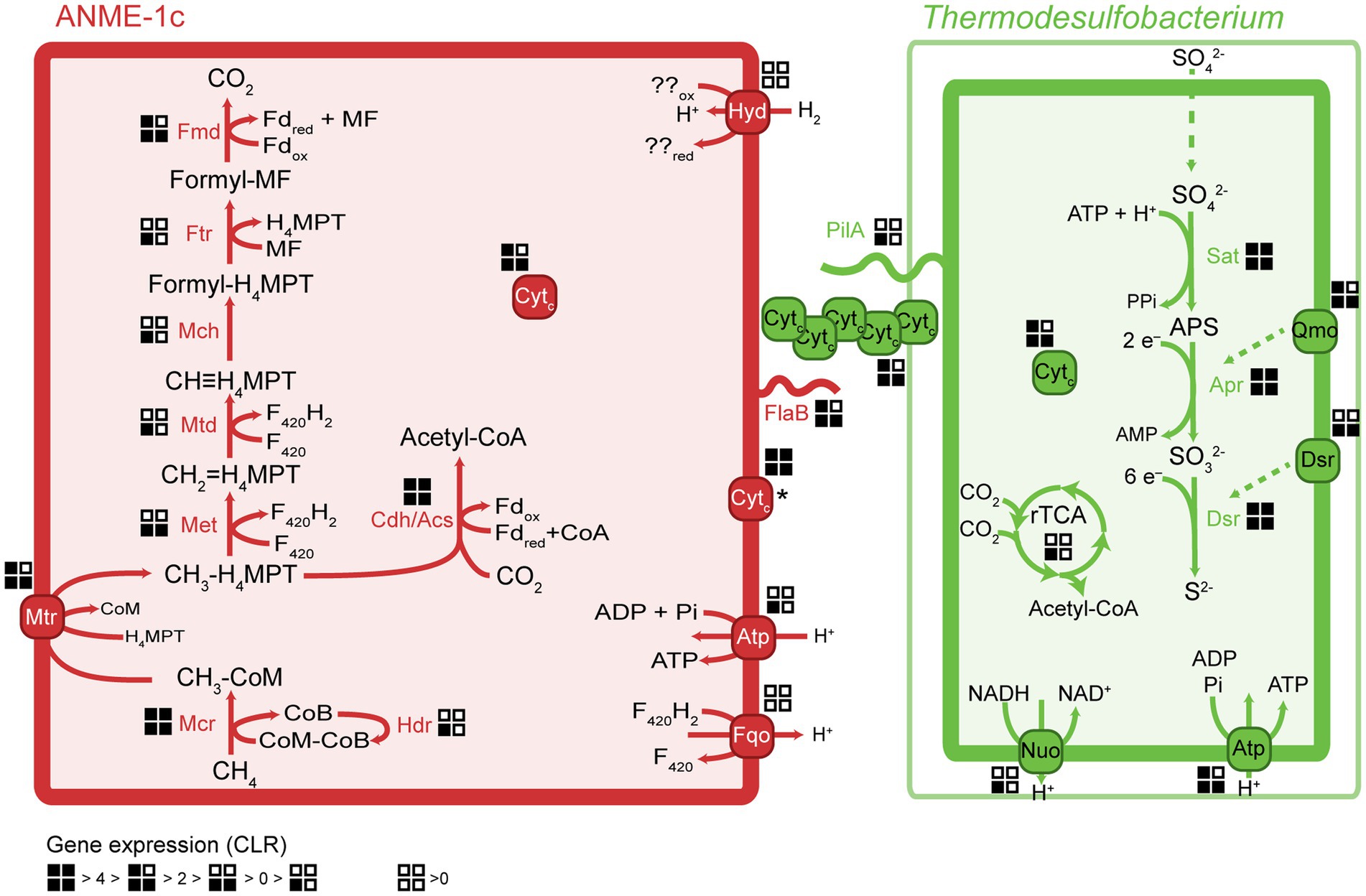
Figure 3. Key metabolic pathways in ANME-1c and Ca. Thermodesulfobacterium torris and metatranscriptomic expression during AOM. Gene expression values were normalized to centered-log ratios (CLR). A CLR value of 0 represents the mean expression of all genes in a genome. The asterisk next to the ANME-1c cytochrome indicates unknown cell localization. H4MPT, tetrahydromethanopterin; MF, methanofuran; Fd, ferredoxin; Mcr, methyl-coenzyme M reductase; Mtr, tetrahydromethanopterin S-methyltransferase; Met, 5,10-methylenetetrahydrofolate reductase; Mtd, methylenetetrahydromethanopterin dehydrogenase; Mch, methenyltetrahydromethanopterin cyclohydrolase; Ftr, formylmethanofuran-tetrahydromethanopterin formyltransferase; Fmd, formylmethanofuran dehydrogenase; Cdh/Acs, CO dehydrogenase/acetyl-coenzyme A synthase complex; rTCA, reductive tricarboxylic acid cycle; Sat, sulfate adenylyltransferase; Apr, adenylylsulfate reductase; Dsr, dissimilatory sulfate reductase; Fqo, ferredoxin: quinone oxidoreductase; Atp, ATP synthase; Hyd, hydrogenase; Qmo, quinone-modifying oxidoreductase; Nuo, NADH:ubiquinone oxidoreductase; Cytc, multiheme cytochrome c-like protein; PilA, bacterial pilus protein; FlaB, archaeal flagellum protein (archaellum).
ANME-1c likely performs autotrophic carbon fixation via the carbon monoxide dehydrogenase/acetyl-CoA synthase complex (Cdh/Acs; Kellermann et al., 2012). All the cdh transcripts are highly abundant (CLR between 0.4 and 2.0, Supplementary Table 4), supporting the use of this pathway for autotrophy. ANME-1c does not encode other complete carbon fixation pathways. The reductive tricarboxylic acid (rTCA) cycle is incomplete, lacking the key enzyme pyruvate carboxylase. The rTCA cycle genes have relatively low expression (CLR −0.7 to 1.8, Supplementary Table 4). Enzymes of this pathway may play a role in the biosynthesis of cell building blocks (Meyerdierks et al., 2010). Like all ANME-1, ANME-1c contains a β-oxidation pathway. The phylogenetically related multi-carbon alkane oxidizers, Ca. Syntrophoarchaeales, Ca. Alkanophagales and Ca. Santabarbaracales harbor several copies of the β-oxidation genes and use the encoded pathway to split alkane-derived acyl-CoA into acetyl-CoA units (Laso-Pérez et al., 2016; Wang et al., 2021). However, the expression of β-oxidation genes in ANME-1c is relatively low, especially the first two reactions (CLR −0.2 to 0.5, Supplementary Table 4). Furthermore, ANME-1c lacks the electron transfer flavoprotein (etfAB) needed to oxidize acyl-CoA to enoyl-CoA. Hence, β-oxidation may not serve a catabolic function in ANME-1c, but play a role in biosynthesis of cell compounds. Wang and colleagues suggested that the ancestor of Syntrophoarchaeia (family including ANME-1, Ca. Syntrophoarchaeales and Ca. Alkanophagales) activated multi-carbon alkanes with their multi-carbon-alkane specific Mcr (Acrs) forming the corresponding alkyl-CoM as intermediate (Laso-Pérez et al., 2016; Wang et al., 2021). It was proposed that ANME-1 acquired a methane-activating Mcr from methylotrophic methanogens, likely from the clade Ca. Methanofastidiosa/Ca. Nuwarchaeia, and later lost the acr genes (Borrel et al., 2019; Wang et al., 2021).
We compared the Thermodesulfobacterium MAG in AOM70 cultures with the Thermodesulfobacteria MAGs from Pescadero Basin and to MAGs retrieved from databases (NCBI and JGI). Our AOM70 Thermodesulfobacterium shares >95% ANI with a MAG of a Thermodesulfobacterium from Pescadero Basin (Laso-Pérez et al., 2022; Speth et al., 2022; Supplementary Figure 7). Based on 16S rRNA phylogeny (Supplementary Figure 6), the Thermodesulfobacterium AOM70 sequences form a cluster with sequences originating from Guaymas Basin and Pescadero Basin hydrothermal seeps (McKay et al., 2016; Lagostina et al., 2021; Pérez Castro et al., 2021; Speth et al., 2022). Several species of Thermodesulfobacterium have been isolated from hot springs (Zeikus et al., 1983; Sonne-Hansen and Ahring, 1999; Hamilton-Brehm et al., 2013), petroleum reservoirs (Rozanova and Khudiakova, 1974) and hydrothermal vents (Jeanthon et al., 2002; Moussard et al., 2004). The 16S rRNA gene sequence of our AOM70 Thermodesulfobacterium is 96% identical to the closest cultured representative, Thermodesulfobacterium geofontis, isolated from Obsidian Pool, Yellowstone National Park (Hamilton-Brehm et al., 2013). Considering an ANI <83% for distinct species and >95% for the same species (Jain et al., 2018) the Thermodesulfobacteria MAG from the AOM70 culture metagenome and the Pescadero MAG are a new candidate species in the genus Thermodesulfobacterium (Supplementary Figure 8). We propose the taxon name Candidatus Thermodesulfobacterium torris (torris “firebrand” referring to the thermophilic lifestyle and the formation of black aggregates in the cultures).
Members of the Thermodesulfobacteria family have not been previously reported as partner bacteria in AOM. All Thermodesulfobacteria isolates are sulfate-reducing (hyper) thermophiles with growth optima between 65°C and 90°C. They differ in the range of electron donors or carbon sources they use, which include molecular hydrogen, formate, lactate, and pyruvate (Zeikus et al., 1983; Sonne-Hansen and Ahring, 1999; Jeanthon et al., 2002; Moussard et al., 2004). Similar to other members, Ca. T. torris encodes a complete dissimilatory sulfate reduction pathway, including sulfate adenylyltransferase (sat), adenylylsulfate reductase (apr), and dissimilatory sulfite reductase (dsr). In Ca. T. torris this pathway is highly expressed during AOM (average CLR values between 4.0 and 6.5, Supplementary Table 4). In addition, Ca. T. torris contains and expresses the Dsr-associated membrane complex (dsrKMOP) which takes up electrons from the periplasmic cytochrome c pool to reduce a disulfide bond in the cytoplasmic DsrC (Pereira et al., 2011; Venceslau et al., 2014). The quinone-modifying oxidoreductase (qmoABC) genes are present in an operon together with the Apr genes. In fact, the Qmo membrane complex interacts with Apr through a third unknown protein and channels electrons from the membrane ubiquinones via electron confurcation (Ramos et al., 2012). Both the dsrKMOP and the qmoABC transcripts have high expression (Supplementary Table 4). Other cytoplasmic enzymes commonly associated with heterodisulfide reductases, such as the methylviologen reducing hydrogenase (Mvh/Hdr), were not found in the dataset. For energy conservation, Ca. T. torris uses a membrane-bound NADH:ubiquinone oxidoreductase (Nuo) and an ATP synthase (Atp). Nuo couples the reduction of NAD+ by reduced ubiquinones in the cytoplasmic membrane to the translocation of protons to the periplasmic space. The proton gradient generated enables oxidative phosphorylation in the ATP synthase.
The reductive acetyl-CoA pathway (Wood-Ljungdahl pathway) for carbon fixation is incomplete in the genome. Ca. T. torris does not encode a formate dehydrogenase (fdh) or a carbon monoxide dehydrogenase/acetyl-CoA complex (cdh/acs), but it encodes the enzymes catalyzing C1-tetrahydrofolate transformations. These reactions are necessary for several cell processes including nucleic acid biosynthesis (Ducker and Rabinowitz, 2017). Instead, Ca. T. torris likely fixes carbon via the rTCA cycle. The genome codes for an almost complete rTCA cycle, lacking a succinyl-CoA synthetase. This enzyme is likely substituted by a putative acetyl-CoA synthetase encoded in the genome and highly expressed (CLR = 3.39, locus MW689_000791). Acetyl-CoA synthetases have sequence homology with succinyl-CoA synthetases and are also active toward succinate with reduced affinity (Sánchez et al., 2000). Similarly, the thermophilic partner bacterium Ca. Desulfofervidus auxilii and other non-symbiotic thermophilic SRB fix carbon via the rTCA cycle (Schauder et al., 1987; Krukenberg et al., 2016). By contrast, meso- and psychrophilic AOM partner bacteria fix carbon using the Wood-Ljungdahl pathway (Skennerton et al., 2017).
We aimed to enrich Ca. T. torris by incubating aliquots of the AOM70 culture with H2, formate, lactate, or pyruvate as electron donors. None of the substrates resulted in immediate sulfide production (Supplementary Figure 9). Pyruvate caused sulfide production after 15 days, which likely indicates the growth of originally rare microorganisms, similar as shown for mesophilic AOM cultures (Zhu et al., 2022). These incubations suggest that Ca. T. torris is an obligate syntrophic bacterium that fully depends on the transfer of reducing equivalents in AOM.
Because ANME have no own respiratory pathways, they need to transfer the reducing equivalents liberated during AOM to their sulfate-reducing partners. Multiple mechanisms have been proposed for syntrophic fermentation, including interspecies hydrogen transfer (Schink, 1997). A canonical syntrophy based on interspecies hydrogen transfer would require membrane-bound hydrogenases in both partners. Notably, the ANME-1c MAGs code for a complete nickel-iron hydrogenase, a feature that is rare in other ANME-1 genomes. The hydrogenase database (HydDB) annotation classifies this hydrogenase within the group 1 g of hydrogenases that are typically found in thermophilic organisms (Brock et al., 1972; Fischer et al., 1983; Huber et al., 2000; Laska et al., 2003). Yet this hydrogenase is only poorly expressed (CLR < −0.3, Supplementary Table 4). In contrast, the Ca. T. torris MAG lacks hydrogenases. The addition of molecular hydrogen to the culture did not stimulate sulfide production in the AOM culture, which confirms that Ca. T. torris cannot grow on hydrogen. Based on these observations we exclude hydrogen as electron carrier from ANME-1c toward Ca. T. torris. Our results confirm thermodynamic models which excluded hydrogen exchange in AOM consortia (Sørensen et al., 2001). Most other AOM partner bacteria such as SeepSRB-1a and SeepSRB2 are also obligate syntrophs and do not encode hydrogenases (Nauhaus et al., 2002; Wegener et al., 2016; Krukenberg et al., 2018). Ca. D. auxilii, performs DIET when growing as partner in AOM or short-chain alkane oxidation at 50°C–60°C (Wegener et al., 2015; Laso-Pérez et al., 2016; Krukenberg et al., 2018; Hahn et al., 2020), but it also shows growth on hydrogen (Krukenberg et al., 2016). It has been shown that DIET allows more efficient growth than interspecies hydrogen transfer (Summers et al., 2010).
In AOM and short-chain alkane-oxidizing consortia, cells are densely packed and the intercellular space contains cytochromes and nanowire-like structures (McGlynn et al., 2015; Wegener et al., 2015; Laso-Pérez et al., 2016; Krukenberg et al., 2018). In these mesophilic and thermophilic consortia, both partners express cytochrome and pilA genes (Laso-Pérez et al., 2016; Krukenberg et al., 2018). The genes pilA (bacterial pilin) in Ca. T. torris and flaB (archaeal flagellin) in ANME-1c show a high expression in the metatranscriptomes (CLR values of 1.8 and 3.2, respectively, Supplementary Table 4). The archaeal flagellum (archaellum) is highly similar to bacterial type IV pili (Albers and Jarrell, 2015) and might also be involved in electron transfer over longer distances. The conductivity of the archaellum from the methanogen Methanospirillum hungatei was demonstrated, yet its possible role in interspecies electron transfer is unclear (Walker et al., 2019). Conductive filaments that enable the transport of electrons across long distances toward extracellular electron acceptors have been widely studied in Geobacter (Reguera et al., 2005; Malvankar et al., 2011; Shrestha et al., 2013; Adhikari et al., 2016). Reguera et al. (2005) showed that pilus-deficient Geobacter mutants could not transfer electrons to extracellular electron acceptors, suggesting an involvement of pilin proteins in this process (Reguera et al., 2005).
The molecular basis of DIET in sulfate-dependent AOM has been intensively discussed in the past years (McGlynn et al., 2015; Wegener et al., 2015; Chadwick et al., 2022; Yu et al., 2022). McGlynn et al. (2015) proposed a model based on direct interspecies electron transfer via multiheme cytochromes for AOM consortia, using evidence from single-cell activities, microscopic observations and genomics (McGlynn et al., 2015). Krukenberg et al. (2018) showed that both partners highly express cytochromes with a low number of heme groups (3–5 heme binding motifs) during thermophilic AOM (Krukenberg et al., 2018). In AOM consortia at 60°C, it was observed that SRB Ca. Desulfofervidus auxilii expressed pili genes and that the intercellular space was filled with nanowire structures similar to syntrophic consortia of Geobacter (Wegener et al., 2015). Indeed it was recently shown that the filaments in Geobacter sulfurreducens are not formed by pilin proteins (PilA), but rather by stacked OmcS hexaheme cytochromes (Wang et al., 2019). Instead, PilA might be involved in secretion of OmcS cytochromes (Gu et al., 2021). Both ANME-1c and Ca. T. torris encode several multiheme cytochromes. ANME-1c codes for several proteins with 2 to 8 heme-binding motifs (Figure 4; Supplementary Table 4). A cytochrome c7 and a protein without annotation, both with 3 heme groups, are among the top expressed genes (CLR values of 5.9 and 2.96, respectively, Supplementary Table 4). However, the predicted subcellular localizations of these putative cytochromes are unknown, as reported by PSORTb. Interestingly, these cytochromes are highly similar to extracellular cytochromes that were highly expressed in ANME-1 during AOM at 60°C (Supplementary Table 4; Krukenberg et al., 2018). Ca. T. torris also contains numerous multiheme cytochromes, with up to 26 heme-binding motifs. A cytochrome-like gene with five heme-binding motifs and predicted extracellular localization shows high expression levels similar to the dsrA (CLR value of 4.0, Figure 4). This putative pentaheme cytochrome shares high sequence identity (<40% identity) with a Ca. Desulfofervidus auxilii OmcS-like protein (Wegener et al., 2015; Krukenberg et al., 2018) (locus tag HS1_000170, Supplementary Figure 10). We hypothesize that the extracellular pentaheme cytochromes from Ca. T. torris are likely involved in receiving electrons derived from methane oxidation. ANME-1c cytochromes with undetermined cell localization might also be involved in interspecies electron transfer.
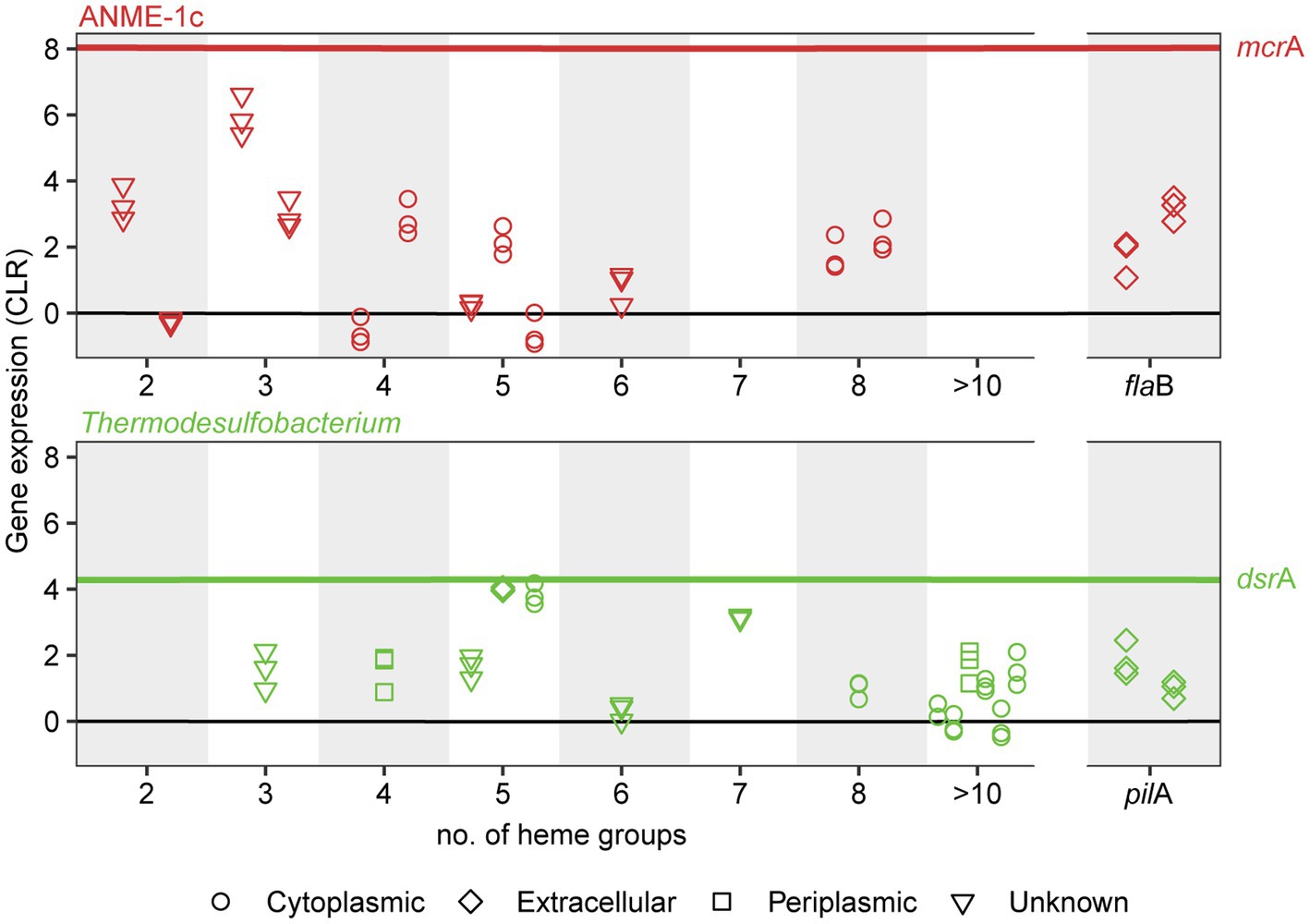
Figure 4. Expression and subcellular localization of multiheme cytochromes and cellular appendages in ANME-1c (top) and Thermodesulfobacterium (bottom) in AOM70 cultures. Gene expression is noted as centered-log ratio values, with 0 as the mean expression of all genes in each genome. mcrA and dsrA mean CLR values are displayed as a reference (red and green lines, respectively). Symbols show the predicted subcellular localization of cytochromes (PSORTb). Three symbols in a vertical line correspond to CLR values of a specific locus in triplicate metatranscriptomes.
Some ANME-1c MAGs code for a NiFe membrane-bound hydrogenase, which is an uncommon feature in most ANME genomes (Stokke et al., 2012; Wegener et al., 2015; Krukenberg et al., 2018). This hydrogenase forms a clade with those of Ca. Syntrophoarchaeum and Ca. Alkanophagales (Figure 2B; Supplementary Figure 11; Laso-Pérez et al., 2016; Wang et al., 2021). In Ca. Syntrophoarchaeum, the hydrogenase is highly expressed during anaerobic propane and butane oxidation, albeit its function is also unknown (Laso-Pérez et al., 2016). In contrast, in ANME-1c the NiFe-hydrogenase has low expression (Figure 3; Supplementary Table 4). A recent study described the ANME-1c as an ancestral clade at the base of ANME-1a/1b and as a sister branch of Ca. Syntrophoarchaeales (Laso-Pérez et al., 2022). ANME-1c were proposed to be facultative methanogens based on the encoded hydrogenase and the unclear association with partner bacteria in environmental samples (Laso-Pérez et al., 2022). The capability of ANME-1 to perform methanogenesis has been repeatedly suggested (Seifert et al., 2006; Treude et al., 2007; Orcutt et al., 2008). ANME-1 16S rRNA and mcrA genes and transcripts were found in methanogenic sediment horizons at White Oak River estuary and in the sulfate–methane transition zone (Lloyd et al., 2011; Kevorkian et al., 2021). The authors weighted this as an argument for a potential role of ANME-1 in methanogenesis. There is no genomic evidence that ANME-1 from these environment contain hydrogenases, which are required to perform methanogenesis from CO2, and this question should be addressed in future metagenomic studies. To test whether ANME-1c are capable of methanogenesis, we transferred AOM70 culture aliquots to sulfate-free medium and exchanged the methane in the headspace with an H2/CO2 atmosphere. Hydrogen addition did not stimulate production of methane in AOM70 cultures in the course of 4-month incubations (Supplementary Figure 12). According to these results, ANME-1c are incapable of hydrogenotrophic methanogenesis. The habitable zones of the hydrothermally-heated sediments in Guaymas Basin are rich in sulfate due to hydrothermal circulation and seawater advection (Ramírez et al., 2021). This provides additional evidence for ANME-1c being obligate methane oxidizers that depend on syntrophic partnerships with sulfate-reducers. The hydrogenase in ANME-1c might be a remnant from the common ancestor of the Ca. Syntrophoarchaeum/Ca. Alkanophagales/ANME-1 clade. These ancestral alkanotrophic archaea might have performed interspecies electron transfer based on hydrogen transfer. In the course of evolution that capability was replaced by an apparently more efficient DIET mechanism via extracellular multiheme cytochromes (Summers et al., 2010).
Here we cultured a thermophilic AOM consortium at 70°C consisting of methane-oxidizing archaea from the ANME-1c clade with Thermodesulfobacteria as sulfate-reducing partner bacteria. Our study bridges the temperature gap between AOM activity previously observed by pore water profiles and tracer experiments, and its in vitro demonstration in cultures. ANME-1c is a basal lineage to the ANME-1a/b clade. Interestingly, ANME-1c MAGs encode a hydrogenase operon that is not present in the ANME-1a/b clades. ANME-1c neither produces nor consumes hydrogen. The hydrogenase genes have low expression and this enzyme is likely a remnant of the ancestor of the Ca. Syntrophoarchaeia, an organism that was likely a multi-carbon alkane oxidizer. The function of this hydrogenase in ANME-1, but also other members of the Syntrophoarchaeia is unresolved. Based on indirect evidence ANME-1 were repeatedly suggested to be facultative methanogens (Seifert et al., 2006; Treude et al., 2007; Orcutt et al., 2008; Lloyd et al., 2011; Kevorkian et al., 2021). Here, we demonstrated that even ANME-1c that encode a hydrogenase are not able to reverse their metabolism toward net methanogenesis. Cultivation-based approaches should be used to test whether the ANME-1a/b that encode hydrogenases are capable of methanogenesis.
Most likely, ANME and their partners interact via DIET. The partner Thermodesulfobacterium encodes an extracellular pentaheme c-type cytochrome with high expression. This cytochrome is highly similar to Ca. Desulfofervidus auxilii cytochromes that have high expression during AOM at 60°C. This evidence suggests a central role of this pentaheme cytochrome of Thermodesulfobacteria in DIET. In addition, multiheme cytochromes and flagella from ANME-1c are highly expressed under AOM conditions. However, the role of these cytochromes with unknown subcellular location in DIET needs further investigation.
Our study provides culture-based evidence for the feasibility of AOM at high-temperature conditions and the versatility and evolutionary diversity of the organisms mediating AOM in marine environments. To our knowledge, this is the first report of syntrophic consortia of ANME and Thermodesulfobacteria. ANME-1c and Ca. T. torris co-occur in environmental samples from the Guaymas Basin and Pescadero Basin (Gulf of California; Laso-Pérez et al., 2022; Speth et al., 2022). This limited geographical distribution is likely a result of undersampling of heated methane-rich environments. Apart from the rather rare methane-rich hydrothermal vents, consortia of these phylotypes might inhabit deep sulfate–methane interfaces. Indeed, these sulfate–methane interfaces occur at depths up to 150 m below the seafloor and at temperatures of up to 80°C (Teske et al., 2021; Beulig et al., 2022). The question of whether these sulfate–methane interfaces are a habitat for AOM needs to be addressed. Future studies should aim to search for the ANME-1c and Thermodesulfobacteria co-occurring in such deep-sea sediments.
The metagenome-assembled genomes (MAGs) and metagenomic contigs are available on NCBI under BioProject PRJNA805391. Assembled metagenomic contigs are available under BioSample ID SAMN30121676. ANME-1c and Ca. Thermodesulfobacterium torris annotated genomes are available under BioSample IDs SAMN27514932 and SAMN27514933, respectively.
DBM and GW designed the study. GW did sampling on board. AT planned and organized the cruise. DBM and HZ did cultivation experiments. DBM performed laboratory experiments and ‘omics analyses and wrote the manuscript with contributions from all coauthors. All authors contributed to the article and approved the submitted version.
The study was funded by the Max Planck Society and the DFG under Germany’s Excellence Initiative/Strategy through the Clusters of Excellence EXC 2077 “The Ocean Floor—Earth’s Uncharted Interface” (project no. 390741601). The Guaymas Basin expedition was supported by the National Science Foundation, Biological Oceanography grant no. 1357238 to AT (Collaborative Research: Microbial Carbon cycling and its interactions with Sulfur and Nitrogen transformations in Guaymas Basin hydrothermal sediments).
The authors thank the captain and crew of R/V Atlantis, and the Alvin group for excellent work during Expedition AT42-05. We thank Susanne Menger for technical support in the laboratory. We also thank Rafael Laso Pérez for providing ANME-1c MAGs from Pescadero Basin.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor SER declared past co-authorships with one of the authors GW.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988871/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Sulfide production in initial sediment slurry and in AOM cultures at 70°C.
SUPPLEMENTARY FIGURE 2 | 16S rRNA community composition of original sediments.
SUPPLEMENTARY FIGURE 3 | CARD-FISH micrographs of AOM the culture at 70°C.
SUPPLEMENTARY FIGURE 4 | 16S rRNA phylogeny of ANME-1, including ANME-1c.
SUPPLEMENTARY FIGURE 5 | Average nucleotide identity of ANME-1c genomes.
SUPPLEMENTARY FIGURE 6 | 16S rRNA phylogeny of Thermodesulfobacteria.
SUPPLEMENTARY FIGURE 7 | Phylogenomic tree of Thermodesulfobacteria.
SUPPLEMENTARY FIGURE 8 | Average nucleotide identity of selected Thermodesulfobacteria genomes.
SUPPLEMENTARY FIGURE 9 | Sulfide production in incubations with substrates for Thermodesulfobacteria.
SUPPLEMENTARY FIGURE 10 | Alignment of Ca. Thermodesulfobacterium torris and partner sulfate-reducing bacteria multiheme cytochromes likely involved in interspecies electron transfer.
SUPPLEMENTARY FIGURE 11 | NiFe hydrogenase tree including ANME-1 hydrogenases.
SUPPLEMENTARY FIGURE 12 | The AOM70 enrichment does not produce methane under methanogenesis conditions.
SUPPLEMENTARY TABLE 1 | Abundance and length distribution of PacBio metagenomic long reads.
SUPPLEMENTARY TABLE 2 | CARD-FISH probes used in this study.
SUPPLEMENTARY TABLE 3 |Genomes and conserved marker genes used for archaeal/bacterial phylogenomic tree calculation.
SUPPLEMENTARY TABLE 4 |Annotation and metatranscriptomic expression of main pathways of ANME-1c and Ca. Thermodesulfobacterium torris discussed in the main text.
SUPPLEMENTARY TABLE 5 |Reference hydrogenase accession numbers used for hydrogenase phylogenetic tree calculation.
1. ^https://sourceforge.net/projects/bbmap/
2. ^https://github.com/dbenitom/Metagenomics_scripts/blob/main/CXXCH_search_anvio_import.sh
Adams, M. M., Hoarfrost, A. L., Bose, A., Joye, S. B., and Girguis, P. R. (2013). Anaerobic oxidation of short-chain alkanes in hydrothermal sediments: potential influences on sulfur cycling and microbial diversity. Front. Microbiol. 4:110. doi: 10.3389/fmicb.2013.00110
Adhikari, R. Y., Malvankar, N. S., Tuominen, M. T., and Lovley, D. R. (2016). Conductivity of individual Geobacter pili. RSC Adv. 6, 8354–8357. doi: 10.1039/C5RA28092C
Albers, S. V., and Jarrell, K. F. (2015). The archaellum: how Archaea swim. Front. Microbiol. 6, 1–12. doi: 10.3389/FMICB.2015.00023
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bengtsson-Palme, J., Hartmann, M., Eriksson, K. M., Pal, C., Thorell, K., Larsson, D. G. J., et al. (2015). Metaxa 2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 15, 1403–1414. doi: 10.1111/1755-0998.12399
Beulig, F., Schubert, F., Adhikari, R. R., Glombitza, C., Heuer, V. B., Hinrichs, K.-U., et al. (2022). Rapid metabolism fosters microbial survival in the deep, hot subseafloor biosphere. Nat. Commun. 131, 1–9. doi: 10.1038/s41467-021-27802-7
Biddle, J. F., Cardman, Z., Mendlovitz, H., Albert, D. B., Lloyd, K. G., Boetius, A., et al. (2012). Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6, 1018–1031. doi: 10.1038/ismej.2011.164
Boetius, A., and Knittel, K. (2010). “Habitats of anaerobic methane oxidizers,” in Handbook of Hydrocarbon and Lipid Microbiology. ed. K. N. Timmis (Berlin, Heidelberg: Springer Berlin Heidelberg), 2193–2202.
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Borrel, G., Adam, P. S., McKay, L. J., Chen, L.-X., Sierra-García, I. N., Sieber, C. M. K., et al. (2019). Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 4, 603–613. doi: 10.1038/s41564-019-0363-3
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. für Mikrobiol. 84, 54–68. doi: 10.1007/BF00408082
Chadwick, G. L., Skennerton, C. T., Laso-Pérez, R., Leu, A. O., Speth, D. R., Yu, H., et al. (2022). Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea. PLoS Biol. 20:e3001508. doi: 10.1371/journal.pbio.3001508
Chernomor, O., Von Haeseler, A., and Minh, B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997–1008. doi: 10.1093/SYSBIO/SYW037
Cord-Ruwisch, R. (1985). A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4, 33–36. doi: 10.1016/0167-7012(85)90005-3
Darling, A. E., Jospin, G., Lowe, E., Matsen, F. A., Bik, H. M., and Eisen, J. A. (2014). Phylo sift: phylogenetic analysis of genomes and metagenomes. Peer J 2:e243. doi: 10.7717/peerj.243
Dekas, A. E., Connon, S. A., Chadwick, G. L., Trembath-Reichert, E., and Orphan, V. J. (2015). Activity and interactions of methane seep microorganisms assessed by parallel transcription and FISH-NanoSIMS analyses. ISME J. 103, 678–692. doi: 10.1038/ismej.2015.145
Dekas, A. E., Poretsky, R. S., and Orphan, V. J. (2009). Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426. doi: 10.1126/science.1178223/
Dombrowski, N., Seitz, K. W., Teske, A. P., and Baker, B. J. (2017). Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5:106. doi: 10.1186/s40168-017-0322-2
Dombrowski, N., Teske, A. P., and Baker, B. J. (2018). Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 9:4999. doi: 10.1038/s41467-018-07418-0
Dowell, F., Cardman, Z., Dasarathy, S., Kellermann, M. Y., Lipp, J. S., Ruff, S. E., et al. (2016). Microbial communities in methane- and short chain alkane-rich hydrothermal sediments of Guaymas Basin. Front. Microbiol. 7:17. doi: 10.3389/fmicb.2016.00017
Ducker, G. S., and Rabinowitz, J. D. (2017). One-carbon metabolism in health and disease. Cell Metab. 25, 27–42. doi: 10.1016/J.CMET.2016.08.009
Edgar, R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 1–19. doi: 10.1186/1471-2105-5-113/FIGURES/16
Eren, A. M., Esen, Ö. C., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi’o: An advanced analysis and visualization platform for ‘omics data. Peer J 3:e1319. doi: 10.7717/peerj.1319
Eren, A. M., Kiefl, E., Shaiber, A., Veseli, I., Miller, S. E., Schechter, M. S., et al. (2020). Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 61, 3–6. doi: 10.1038/s41564-020-00834-3
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., and Thauer, R. K. (1997). Crystal structure of methyl-coenzyme M reductase: The key enzyme of biological methane formation. Science 278, 1457–1462. doi: 10.1126/science.278.5342.1457
Fischer, F., Zillig, W., Stetter, K. O., and Schreiber, G. (1983). Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature 301, 511–513. doi: 10.1038/301511a0
Friedmann, H. C., Klein, A., and Thauer, R. K. (1990). Structure and function of the nickel porphinoid, coenzyme F430 and of its enzyme, methyl coenzyme M reductase. FEMS Microbiol. Rev. 7, 339–348. doi: 10.1111/J.1574-6968.1990.TB04934.X
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/NAR/GKU1223
Gu, Y., Srikanth, V., Salazar-Morales, A. I., Jain, R., O’Brien, J. P., Yi, S. M., et al. (2021). Structure of Geobacter pili reveals secretory rather than nanowire behaviour. Nature 597, 430–434. doi: 10.1038/s41586-021-03857-w
Haft, D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T., et al. (2001). TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 29, 41–43. doi: 10.1093/NAR/29.1.41
Hahn, C. J., Laso-Pérez, R., Vulcano, F., Vaziourakis, K.-M., Stokke, R., Steen, I. H., et al. (2020). “Candidatus Ethanoperedens,” a thermophilic genus of archaea mediating the anaerobic oxidation of ethane. MBio 11. doi: 10.1128/mBio.00600-20
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. M., et al. (2004). Reverse Methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462. doi: 10.1126/science.1100025
Hamilton-Brehm, S. D., Gibson, R. A., Green, S. J., Hopmans, E. C., Schouten, S., van der Meer, M. T. J., et al. (2013). Thermodesulfobacterium geofontis sp. nov., a hyperthermophilic, sulfate-reducing bacterium isolated from obsidian Pool, Yellowstone National Park. Extremophiles 17, 251–263. doi: 10.1007/S00792-013-0512-1
Hao, L., McIlroy, S. J., Kirkegaard, R. H., Karst, S. M., Fernando, W. E. Y., Aslan, H., et al. (2018). Novel prosthecate bacteria from the candidate phylum Acetothermia. ISME J. 12, 2225–2237. doi: 10.1038/s41396-018-0187-9
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Hinrichs, K.-U., and Boetius, A. (2002). The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry. Ocean Margin Syst., 457–477. doi: 10.1007/978-3-662-05127-6_28
Hoehler, T. M., Alperin, M. J., Albert, D. B., and Martens, C. S. (1994). Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cycles 8, 451–463. doi: 10.1029/94GB01800
Holler, T., Wegener, G., Knittel, K., Boetius, A., Brunner, B., Kuypers, M. M. M., et al. (2009). Substantial 13C/12C and D/H fractionation during anaerobic oxidation of methane by marine consortia enriched in vitro. Environ. Microbiol. Rep. 1, 370–376. doi: 10.1111/J.1758-2229.2009.00074.X
Holler, T., Widdel, F., Knittel, K., Amann, R., Kellermann, M. Y., Hinrichs, K.-U., et al. (2011). Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5, 1946–1956. doi: 10.1038/ismej.2011.77
Huber, H., Burggraf, S., Mayer, T., Wyschkony, I., Rachel, R., and Stetter, K. O. (2000). Ignicoccus gen. Nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp nov and Ignicoccus pacificus sp nov. and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 50, 2093–2100. doi: 10.1099/00207713-50-6-2093
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 1–11. doi: 10.1186/1471-2105-11-119
Inagaki, F., Kuypers, M. M. M., Tsunogai, U., Ishibashi, J. I., Nakamura, K. I., Treude, T., et al. (2006). Microbial community in a sediment-hosted CO2 lake of the southern Okinawa trough hydrothermal system. Proc. Natl. Acad. Sci. U. S. A. 103, 14164–14169. doi: 10.1073/PNAS.0606083103
Ishii, K., Mußmann, M., Mac Gregor, B. J., and Amann, R. (2004). An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50, 203–213. doi: 10.1016/J.FEMSEC.2004.06.015
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 91, 1–8. doi: 10.1038/s41467-018-07641-9
Jeanthon, C., L’Haridon, S., Cueff, V., Banta, A., Reysenbach, A.-L., and Prieur, D. (2002). Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int. J. Syst. Evol. Microbiol. 52, 765–772. doi: 10.1099/00207713-52-3-765
Kallmeyer, J., and Boetius, A. (2004). Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70, 1231–1233. doi: 10.1128/AEM.70.2.1231-1233.2004
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). Model finder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/NAR/28.1.27
Kellermann, M. Y., Wegener, G., Elvert, M., Yoshinaga, M. Y., Lin, Y.-S., Holler, T., et al. (2012). Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc. Natl. Acad. Sci. 109, 19321–19326. doi: 10.1073/pnas.1208795109
Kevorkian, R. T., Callahan, S., Winstead, R., and Lloyd, K. G. (2021). ANME-1 archaea may drive methane accumulation and removal in estuarine sediments. Environ. Microbiol. Rep. 13, 185–194. doi: 10.1111/1758-2229.12926
Knittel, K., Boetius, A., Lemke, A., Eilers, H., Lochte, K., Pfannkuche, O., et al. (2003). Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia margin, Oregon). Geomicrobiol J. 20, 269–294. doi: 10.1080/01490450303896
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Kolmogorov, M., Bickhart, D. M., Behsaz, B., Gurevich, A., Rayko, M., Shin, S. B., et al. (2020). Meta Flye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110. doi: 10.1038/s41592-020-00971-x
Krukenberg, V., Harding, K., Richter, M., Glöckner, F. O., Gruber-Vodicka, H. R., Adam, B., et al. (2016). Candidatus Desulfofervidus auxilii, a hydrogenotrophic sulfate-reducing bacterium involved in the thermophilic anaerobic oxidation of methane. Environ. Microbiol. 18, 3073–3091. doi: 10.1111/1462-2920.13283
Krukenberg, V., Riedel, D., Gruber-Vodicka, H. R., Buttigieg, P. L., Tegetmeyer, H. E., Boetius, A., et al. (2018). Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia. Environ. Microbiol. 20, 1651–1666. doi: 10.1111/1462-2920.14077
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/NAR/GKM160
Lagostina, L., Frandsen, S., Mac Gregor, B. J., Glombitza, C., Deng, L., Fiskal, A., et al. (2021). Interactions between temperature and energy supply drive microbial communities in hydrothermal sediment. Commun. Biol. 41, 4, 1–14. doi: 10.1038/s42003-021-02507-1
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 94, 357–359. doi: 10.1038/nmeth.1923
Lanoil, B. D., Sassen, R., La Duc, M. T., Sweet, S. T., and Nealson, K. H. (2001). Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67, 5143–5153. doi: 10.1128/AEM.67.11.5143-5153.2001
Laska, S., Lottspeich, F., and Kletzin, A. (2003). Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149, 2357–2371. doi: 10.1099/MIC.0.26455-0/CITE/REFWORKS
Laso-Pérez, R., Krukenberg, V., Musat, F., and Wegener, G. (2018). Establishing anaerobic hydrocarbon-degrading enrichment cultures of microorganisms under strictly anoxic conditions. Nat. Publ. Gr. 13, 1310–1330. doi: 10.1038/nprot.2018.030
Laso-Pérez, R., Wegener, G., Knittel, K., Widdel, F., Harding, K. J., Krukenberg, V., et al. (2016). Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401. doi: 10.1038/nature20152
Laso-Pérez, R., Wu, F., Crémière, A., Speth, D. R., Magyar, J. S., Krupovic, M., et al. (2022). Evolutionary diversification of methanotrophic Ca. Methanophagales (ANME-1) and their expansive virome. bio Rxiv [preprint].
Letunic, I., and Bork, P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Li, H. (2018). Minimap 2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/BIOINFORMATICS/BTY191
Lloyd, K. G., Alperin, M. J., and Teske, A. (2011). Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ. Microbiol. 13, 2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x
Lösekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, A., et al. (2004). ARB: A software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/NAR/GKH293
Malvankar, N. S., Vargas, M., Nevin, K. P., Franks, A. E., Leang, C., Kim, B. C., et al. (2011). Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 69, 573–579. doi: 10.1038/nnano.2011.119
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
McKay, L. J. (2014). Microbial ecology of a manmade oil spill in the Gulf of Mexico and a natural, hydrothermal oil seep in the Gulf of California. Available at: http://search.proquest.com/docview/1613182914?accountid=14468%5Cnhttp://wx7cf7zp2h.search.serialssolutions.com/?ctx_ver=Z39.88-2004&ctx_enc=info:ofi/enc:UTF-8&rfr_id=info:sid/ProQuest+Dissertations+&+Theses+Global&rft_val_fmt=info:ofi/fmt:kev:mtx:disserta.
McKay, L., Klokman, V. W., Mendlovitz, H. P., Larowe, D. E., Hoer, D. R., Albert, D., et al. (2016). Thermal and geochemical influences on microbial biogeography in the hydrothermal sediments of Guaymas Basin, Gulf of California. Environ. Microbiol. Rep. 8, 150–161. doi: 10.1111/1758-2229.12365
Merkel, A. Y., Huber, J. A., Chernyh, N. A., Bonch-Osmolovskaya, E. A., and Lebedinsky, A. V. (2013). Detection of putatively thermophilic anaerobic methanotrophs in diffuse hydrothermal vent fluids. Appl. Environ. Microbiol. 79, 915–923. doi: 10.1128/AEM.03034-12
Meyerdierks, A., Kube, M., Kostadinov, I., Teeling, H., Glöckner, F. O., Reinhardt, R., et al. (2010). Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12, 422–439. doi: 10.1111/J.1462-2920.2009.02083.X
Michaelis, W., Seifert, R., Nauhaus, K., Treude, T., Thiel, V., Blumenberg, M., et al. (2002). Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297, 1013–1015. doi: 10.1126/SCIENCE.1072502
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/MOLBEV/MSAA015
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/NAR/GKAA913
Moore, S. J., Sowa, S. T., Schuchardt, C., Deery, E., Lawrence, A. D., Ramos, J. V., et al. (2017). Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature 543, 78–82. doi: 10.1038/nature21427
Moussard, H., L’Haridon, S., Tindall, B. J., Banta, A., Schumann, P., Stackebrandt, E., et al. (2004). Thermodesulfatator indicus gen. Nov., sp. nov., a novel thermophilic chemolithoautotrophic sulfate-reducing bacterium isolated from the central Indian ridge. Int. J. Syst. Evol. Microbiol. 54, 227–233. doi: 10.1099/IJS.0.02669-0
Nauhaus, K., Boetius, A., Krüger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4, 296–305. doi: 10.1046/J.1462-2920.2002.00299.X
Niemann, H., Elvert, M., Hovland, M., Orcutt, B., Judd, A., Suck, I., et al. (2005). Methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosciences 2, 335–351. doi: 10.5194/BG-2-335-2005
Niemann, H., Lösekann, T., De Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Orcutt, B. N., Boetius, A., Lugo, S. K., Mac Donald, I. R., Samarkin, V. A., and Joye, S. B. (2004). Life at the edge of methane ice: microbial cycling of carbon and sulfur in Gulf of Mexico gas hydrates. Chem. Geol. 205, 239–251. doi: 10.1016/J.CHEMGEO.2003.12.020
Orcutt, B., Samarkin, V., Boetius, A., and Joye, S. (2008). On the relationship between methane production and oxidation by anaerobic methanotrophic communities from cold seeps of the Gulf of Mexico. Environ. Microbiol. 10, 1108–1117. doi: 10.1111/j.1462-2920.2007.01526.x
Orphan, V. J., Hinrichs, K.-U., Ussler, W., Paull, C. K., Taylor, L. T., Sylva, S. P., et al. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U. S. A. 99, 7663–7668. doi: 10.1073/PNAS.072210299/ASSET/06C54137-68B2-457C-8C76-FEA01D3D60DE/ASSETS/GRAPHIC/PQ1122102003.JPEG
Orphan, V. J., Turk, K. A., Green, A. M., and House, C. H. (2009). Patterns of 15N assimilation and growth of methanotrophic ANME-2 archaea and sulfate-reducing bacteria within structured syntrophic consortia revealed by FISH-SIMS. Environ. Microbiol. 11, 1777–1791. doi: 10.1111/J.1462-2920.2009.01903.X
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pereira, I. A. C., Ramos, A. R., Grein, F., Marques, M. C., da Silva, S. M., and Venceslau, S. S. (2011). A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front. Microbiol. 2, 1–22. doi: 10.3389/FMICB.2011.00069
Pérez Castro, S., Borton, M. A., Regan, K., Hrabe de Angelis, I., Wrighton, K. C., Teske, A. P., et al. (2021). Degradation of biological macromolecules supports uncultured microbial populations in Guaymas Basin hydrothermal sediments. ISME J. 15, 3480–3497. doi: 10.1038/s41396-021-01026-5
Pernthaler, A., Pernthaler, J., and Amann, R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/BIOINFORMATICS/BTS252
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/NAR/GKS1219
Ramírez, G. A., Mara, P., Sehein, T., Wegener, G., Chambers, C. R., Joye, S. B., et al. (2021). Environmental factors shaping bacterial, archaeal and fungal community structure in hydrothermal sediments of Guaymas Basin, Gulf of California. PLoS One 16:e0256321. doi: 10.1371/JOURNAL.PONE.0256321
Ramos, A. R., Keller, K. L., Wall, J. D., and Cardoso Pereira, I. A. (2012). The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5′-phosphosulfate reductase in sulfate reducing bacteria. Front. Microbiol. 3, 1–10. doi: 10.3389/FMICB.2012.00137
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Regnier, P., Dale, A. W., Arndt, S., LaRowe, D. E., Mogollón, J., and Van Cappellen, P. (2011). Quantitative analysis of anaerobic oxidation of methane (AOM) in marine sediments: A modeling perspective. Earth-Science Rev. 106, 105–130. doi: 10.1016/J.EARSCIREV.2011.01.002
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. doi: 10.1038/nature03661
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Roussel, E. G., Bonavita, M. A. C., Querellou, J., Cragg, B. A., Webster, G., Prieur, D., et al. (2008). Extending the sub-sea-floor biosphere. Science 320:1046. doi: 10.1126/SCIENCE.1154545
Rozanova, E., and Khudiakova, A. (1974). Novyĭ bessporovyĭ termofil’nyĭ organizm, vosstanavlivaiushchiĭ sul’faty, Desulfovibrio thermophilus nov. sp [A new non-spore forming thermophilic organism, reducing sulfates, Desulfovibrio thermophilus nov. sp]. Mikrobiologiia 43, 1069–1075.
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. U. S. A. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Sánchez, L. B., Galperin, M. Y., and Müller, M. (2000). Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J. Biol. Chem. 275, 5794–5803. doi: 10.1074/JBC.275.8.5794
Schauder, R., Widdel, F., and Fuchs, G. (1987). Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch. Microbiol. 148, 218–225. doi: 10.1007/BF00414815
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. doi: 10.1038/nature09015
Schink, B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61, 262–280. doi: 10.1128/MMBR.61.2.262-280.1997
Schouten, S., Wakeham, S. G., Hopmans, E. C., and Damsté, J. S. S. (2003). Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl. Environ. Microbiol. 69, 1680–1686. doi: 10.1128/AEM.69.3.1680-1686.2003
Seifert, R., Nauhaus, K., Blumenberg, M., Krüger, M., and Michaelis, W. (2006). Methane dynamics in a microbial community of the Black Sea traced by stable carbon isotopes in vitro. Org. Geochem. 37, 1411–1419. doi: 10.1016/J.ORGGEOCHEM.2006.03.007
Shima, S., Krueger, M., Weinert, T., Demmer, U., Kahnt, J., Thauer, R. K., et al. (2012). Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481, 98–101. doi: 10.1038/nature10663
Shrestha, P. M., Rotaru, A. E., Summers, Z. M., Shrestha, M., Liu, F., and Lovley, D. R. (2013). Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl. Environ. Microbiol. 79, 2397–2404.
Skennerton, C. T., Chourey, K., Iyer, R., Hettich, R. L., Tyson, G. W., and Orphan, V. J. (2017). Methane-fueled Syntrophy through extracellular electron transfer: uncovering the genomic traits conserved within diverse bacterial Partners of Anaerobic Methanotrophic Archaea. MBio 8, 1–14. doi: 10.1128/mBio.00530-17
Søndergaard, D., Pedersen, C. N. S., and Greening, C. (2016). HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 6, 1–8. doi: 10.1038/srep34212
Sonne-Hansen, J., and Ahring, B. K. (1999). Thermodesulfobacterium hveragerdense sp. nov., and Thermodesulfovibrio islandicus sp. nov., two thermophilic sulfate reducing bacteria isolated from a Icelandic hot spring. Syst. Appl. Microbiol. 22, 559–564. doi: 10.1016/S0723-2020(99)80009-5
Sørensen, K. B., Finster, K., and Ramsing, N. B. (2001). Thermodynamic and kinetic requirements in anaerobic methane oxidizing consortia exclude hydrogen, acetate, and methanol as possible electron shuttles. Microb. Ecol. 421, 1–10. doi: 10.1007/S002480000083
Speth, D. R., Yu, F. B., Connon, S. A., Lim, S., Magyar, J. S., Peña-Salinas, M. E., et al. (2022). Microbial communities of Auka hydrothermal sediments shed light on vent biogeography and the evolutionary history of thermophily. ISME J. 16, 1750–1764. doi: 10.1038/s41396-022-01222-x
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/BIOINFORMATICS/BTU033
Stokke, R., Roalkvam, I., Lanzen, A., Haflidason, H., and Steen, I. H. (2012). Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ. Microbiol. 14, 1333–1346. doi: 10.1111/J.1462-2920.2012.02716.X
Summers, Z. M., Fogarty, H. E., Leang, C., Franks, A. E., Malvankar, N. S., and Lovley, D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415. doi: 10.1126/science.1196526
Teske, A., Hinrichs, K. U., Edgcomb, V., De Vera Gomez, A., Kysela, D., Sylva, S. P., et al. (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002
Teske, A., Lizarralde, D., and Höfig, T. W. (2021). Guaymas Basin tectonics and biosphere. Proc. Int. Ocean Discov. Progr. 385. doi: 10.14379/IODP.PROC.385.2021
Thauer, R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299. doi: 10.1016/J.MIB.2011.03.003
Treude, T., Krüger, M., Boetius, A., and Jørgensen, B. B. (2005). Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic). Limnol. Oceanogr. 50, 1771–1786. doi: 10.4319/LO.2005.50.6.1771
Treude, T., Orphan, V., Knittel, K., Gieseke, A., House, C. H., and Boetius, A. (2007). Consumption of methane and CO 2 by Methanotrophic microbial Mats from gas seeps of the anoxic Black Sea. Appl. Environ. Microbiol. 73, 2271–2283. doi: 10.1128/AEM.02685-06
Venceslau, S. S., Stockdreher, Y., Dahl, C., and Pereira, I. A. C. (2014). The “bacterial heterodisulfide,” Dsr C is a key protein in dissimilatory sulfur metabolism. Biochim. Biophys. Acta 1837, 1148–1164. doi: 10.1016/J.BBABIO.2014.03.007
Walker, D. J. F., Martz, E., Holmes, D. E., Zhou, Z., Nonnenmann, S. S., and Lovley, D. R. (2019). The archaellum of Methanospirillum hungatei is electrically conductive. MBio 10, 1–16. doi: 10.1128/MBIO.00579-19
Wang, F., Gu, Y., O’Brien, J. P., Yi, S. M., Yalcin, S. E., Srikanth, V., et al. (2019). Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177, 361–369.e10. doi: 10.1016/j.cell.2019.03.029
Wang, Y., Wegener, G., Williams, T. A., Xie, R., Hou, J., Tian, C., et al. (2021). A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci. Adv. 7, 1–12. doi: 10.1126/sciadv.abj1453
Wang, F. P., Zhang, Y., Chen, Y., He, Y., Qi, J., Hinrichs, K. U., et al. (2013). Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J. 8, 1069–1078. doi: 10.1038/ismej.2013.212
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E., and Boetius, A. (2015). Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590. doi: 10.1038/nature15733
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K. (2016). Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7, 1–16. doi: 10.3389/fmicb.2016.00046
Whiticar, M. J. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314. doi: 10.1016/S0009-2541(99)00092-3
Widdel, F., and Bak, F. (1992). Gram-negative mesophilic sulfate-reducing bacteria. Prokaryotes 183, 3352–3378. doi: 10.1007/978-1-4757-2191-1_21
Wu, Y.-W., Simmons, B. A., and Singer, S. W. (2016). Max bin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Yu, H., Skennerton, C. T., Chadwick, G. L., Leu, A. O., Aoki, M., Tyson, G. W., et al. (2022). Sulfate differentially stimulates but is not respired by diverse anaerobic methanotrophic archaea. ISME J. 16, 168–177. doi: 10.1038/S41396-021-01047-0
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Zeikus, J. G., Dawson, M. A., Thompson, T. E., Ingvorsen, K., and Hatchikian, E. C. (1983). Microbial ecology of volcanic sulphidogenesis: isolation and characterization of Thermodesulfobacterium commune gen. Nov. and sp. nov. Microbiology 129, 1159–1169. doi: 10.1099/00221287-129-4-1159
Zheng, K., Ngo, P. D., Owens, V. L., Yang, X., and Mansoorabadi, S. O. (2016). The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 354, 339–342. doi: 10.1126/science.aag2947
Zhou, J., Bruns, M. A., and Tiedje, J. M. (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322. doi: 10.1128/aem.62.2.316-322.1996
Keywords: anaerobic oxidation of methane, ANME-1, archaea, deep sea, hydrothermal vents
Citation: Benito Merino D, Zehnle H, Teske A and Wegener G (2022) Deep-branching ANME-1c archaea grow at the upper temperature limit of anaerobic oxidation of methane. Front. Microbiol. 13:988871. doi: 10.3389/fmicb.2022.988871
Received: 07 July 2022; Accepted: 15 August 2022;
Published: 23 September 2022.
Edited by:
S. Emil Ruff, Marine Biological Laboratory (MBL), United StatesReviewed by:
Maxim Rubin-Blum, Israel Oceanographic and Limnological Research (IOLR), IsraelCopyright © 2022 Benito Merino, Zehnle, Teske and Wegener. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Benito Merino, ZGJlbml0b0BtcGktYnJlbWVuLmRl; Gunter Wegener, Z3dlZ2VuZXJAbXBpLWJyZW1lbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.