
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 August 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.983963
This article is part of the Research Topic Global Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and Treatment Volume II View all 11 articles
 Yihai Gu1,2
Yihai Gu1,2 Wei Zhang2
Wei Zhang2 Jine Lei3
Jine Lei3 Lixia Zhang4
Lixia Zhang4 Xuan Hou2
Xuan Hou2 Junqi Tao2
Junqi Tao2 Hui Wang2
Hui Wang2 Minghui Deng2
Minghui Deng2 Mengrong Zhou2
Mengrong Zhou2 Rui Weng2
Rui Weng2 Jiru Xu1*
Jiru Xu1*Bloodstream infection (BSI) caused by Acinetobacter baumannii poses a serious threat to health and is correlated with high mortality in patients with hospital-acquired infections, so the molecular epidemiology and antimicrobial resistance characteristics of this pathogen urgently need to be explored. A. baumannii isolates from BSI patients were collected in three tertiary hospitals in northwest China from 2009 to 2018. Antimicrobial susceptibility testing was used to determine the MICs of the A. baumannii isolates. Whole-genome sequencing based on the Illumina platform was performed for molecular epidemiological analyses and acquired resistance gene screening. The efflux pump phenotype was detected by examining the influence of an efflux pump inhibitor. The expression of efflux pump genes was evaluated by RT-PCR. In total, 47 A. baumannii isolates causing BSI were collected and they presented multidrug resistance, including resistance to carbapenems. Clone complex (CC) 92 was the most prevalent with 30 isolates, among which a cluster was observed in the phylogenetic tree based on the core genome multi-locus sequence type, indicating the dissemination of a dominant clone. BSI-related A. baumannii isolates normally harbour multiple resistance determinants, of which oxacillinase genes are most common. Except for the intrinsic blaOXA-51 family, there are some carbapenem-resistant determinants in these A. baumannii isolates, including blaOXA-23, which is encoded within the Tn2006, Tn2008 or Tn2009 transposon structures and blaOXA-72. The transfer of blaOXA-72 was suggested by XerC/D site-specific recombination. The AdeABC efflux pump system contributed to carbapenem resistance in A. baumannii isolates, as evidenced by the high expression of some of its encoding genes. Both the clone dissemination and carbapenem resistance mediated by oxacillinase or efflux pumps suggest an effective strategy for hospital infection control.
Acinetobacter spp. are opportunistic pathogens observed during clinical infections, among which Acinetobacter baumannii is the most clinically significant (Wong et al., 2017). A. baumannii normally colonises on the surface of the skin, mucosa, throat and respiratory tract, causing severe infections including bloodstream infections (BSIs), respiratory infections, skin and soft tissue infections, urinary tract infections, meningitis (Harding et al., 2018; Ramirez et al., 2020). The high mortality associated with BSI caused by A. baumannii reached up to 45% in a previous study and is a major concern for nosocomial infection control (Zhou et al., 2019; Gu et al., 2021).
Carbapenems have been known as last-resort antibiotics for A. baumannii infections, but unfortunately, carbapenem-resistant A. baumannii (CRAB) has spread worldwide and the positive rate observed during clinical screening has continued to increase in recent decades, from 1% in 2003 to 58% in 2008, resulting in a major threat to human health and clinical settings (Reddy et al., 2010). The average positive rate for CRAB in China was 53.7% in 2020, as determined using the CARSS surveillance data.1
The resistance mechanism of A. baumannii against carbapenems is closely related to the hyperproduction of β-lactamases, including some AmpC β-lactamases, extended-spectrum β-lactamases (ESBLs) and carbapenemases (Patel and Bonomo, 2013; Stewart et al., 2019). In A. baumannii, the most prevalent mechanism responsible for carbapenem resistance is the production of carbapenem-hydrolysing Ambler class D β-lactamases, such as the OXA-23, OXA-24/40, OXA-58, OXA-143 and OXA-235 types (Peleg et al., 2008; Hammoudi and Ayoub, 2020), among which OXA-23-type carbapenemases are most common in CRAB strains spreading worldwide in nosocomial environments (Potron et al., 2015). Carbapenemase-encoding genes are normally located on chromosomes and/or plasmids, and most of them correspond to mobile genetic elements (MGEs), such as insertion sequences (ISs), integrons and transposons. MGEs are responsible for acquiring, transferring or regulating resistance genes within the host. Several studies have described the dissemination of carbapenem resistance genes by MGEs in A. baumannii, resulting in difficulties in treating infectious diseases (Roca et al., 2012; Cornejo-Juarez et al., 2020). Sometimes plasmid-borne carbapenemases are flanked by short DNA sequences providing potential recognition sites for the host XerC and XerD site-specific tyrosine recombinases, contributing to the translocation of these resistance genes (Cameranesi et al., 2018). Furthermore, some multidrug efflux systems, such as the resistance-nodulation-cell division (RND) family efflux pump AdeABC, mediate multidrug resistance, including resistance to carbapenems (Coyne et al., 2011).
The goal of this study was to explore the molecular epidemiology and resistance mechanism of A. baumannii isolated from BSI patients, providing an efficient therapy choice and reducing the mortality due to BSI caused by A. baumannii.
In total, 47 A. baumannii isolates from BSI patients were collected, including four strains collected from Shaanxi Provincial People’s Hospital in 2018, 25 strains from the First Affiliated Hospital of Xi’an Jiaotong University from 2015 to 2018 and 18 strains from the 3201 Hospital from 2009 to 2018. The species of all the isolates were identified by MALDI-TOF (Bruker, Germany) and confirmed by 16S rDNA sequencing and whole-genome sequencing. This study was approved by the Ethics Committees of 3,201 Hospital of Xi’an Jiaotong University School of Medicine (2020005) with a waiver of informed consent because of the retrospective nature of the study.
The microbroth dilution method was employed to determine the MICs of 47 A. baumannii isolates against several antimicrobial agents, including piperacillin/tazobactam, ampicillin/sulbactam, cefepime, ceftazidime, ceftriaxone, cefotaxime, meropenem, imipenem, colistin, gentamicin, amikacin, levofloxacin, ciprofloxacin, tigecycline and cefoperazone/sulbactam. The susceptibility breakpoint was interpreted as recommended by the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2019), except the breakpoint of tigecycline, which was as recommended by the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2018). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality controls.
Total genomic DNA from the 47 A. baumannii isolates was extracted using the QIAamp DNA Minikit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. The whole genomes of all isolates were sequenced on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA, United states) via the 2 × 150 bp paired-end protocol and were subsequently assembled by using CLC genomic workbench version 8.0, and the draft genome contigs were screened for acquired resistance genes by using ResFinder 2.1 on the CGE server.2 The genetic structure surrounding the resistance gene was annotated by BLAST.3 The XerC/D-specific recombination site was recognised by PdifFinder.4
Multi-locus sequence typing (MLST) analysis was performed by screening the assembly contig sequences of each genome using MLST tool version 2.0 on the CGE website.5 The Oxford MLST allele scheme was employed for typing with seven housekeeping genes, including gltA, gyrB, gdhB, recA, cpn60, gpi and rpoD. Ridom SeqSphere + software version 4.1.9 (Ridom GmbH Münster, Germany) was used to illustrate the MLST and core genome (cg) MLST relationship. cgMLST analysis was carried out using Paul G. Higgins’ scheme, which employed 2,390 genes in the A. baumannii genome as core genes. The paired isolates that differed by less than ten core genes were deemed closely related. A phylogenetic tree based on cgMLST was generated by using the minimum spanning tree (AST) algorithm.
Overexpression of the efflux pump phenotype is usually observed when there is a significant increase in carbapenem susceptibility when an isolate is incubated with a carbapenem and the appropriate efflux pump inhibitor (Mmatli et al., 2020). Carbonyl cyanide m-chlorophenylhydrazine (CCCP), phenylalanine-arginine β-naphthylamide (PAβN) and 1-(1-naphthylmethyl)-piperazine (NMP) were used as inhibitors to assess the potential decrease of MIC of carbapenem in BSI-related A. baumannii isolates.
The expression of the genes adeA and adeB, which belong to the multidrug efflux pump AdeABC, was assessed in efflux phenotype-positive isolates by RT-PCR. RNA from the isolates was extracted by using the PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, United States) in the exponential growth period of bacterial cells and was subsequently reverse transcribed to cDNA by the PrimeScript™ RT Reagent Kit (Takara, Kyoto, Japan). The gene expression level was evaluated by using TB Green™ Premix Ex Taq™ (Takara, Kyoto, Japan) in a LightCycler 480 system (Roche, Rotkreuz, Switzerland) with triplicate samples for each isolate, and three replicates were performed independently using the 2–ΔΔCT method with previously reported primers. Genes for which the fold change in expression was greater than 2 were considered to be differentially expressed. The housekeeping gene rpoB was used as the internal reference, and the strain ATCC17978 was used as a reference control.
All 47 A. baumannii isolates causing BSI presented high-level resistance against most antimicrobial agents, including β-lactams/β-lactamase inhibitors, third/fourth generation cephalosporins, quinolones, tetracyclines, aminoglycosides and even carbapenem, the last-resort antibiotic for severe infections caused by Gram-negative bacteria. The resistance rates for piperacillin/tazobactam, ceftriaxone, cefepime, meropenem, imipenem, ciprofloxacin, tigecycline and gentamicin were greater than 70%. Colistin showed the highest susceptibility rate (97.9%) among all the tested antimicrobial agents, followed by ceftazidime (Table 1). During the decade of strain collection, we further selected two periods that possessed relatively more isolates to observe the trend of carbapenem resistance. The early period had 13 isolates and was from 2013 to 2015, and the later period had 33 isolates from 2016 to 2018. We found that the carbapenem resistance rate of BSI-related A. baumannii increased from 61.5% during the early stage to 72.7% during the later stage.
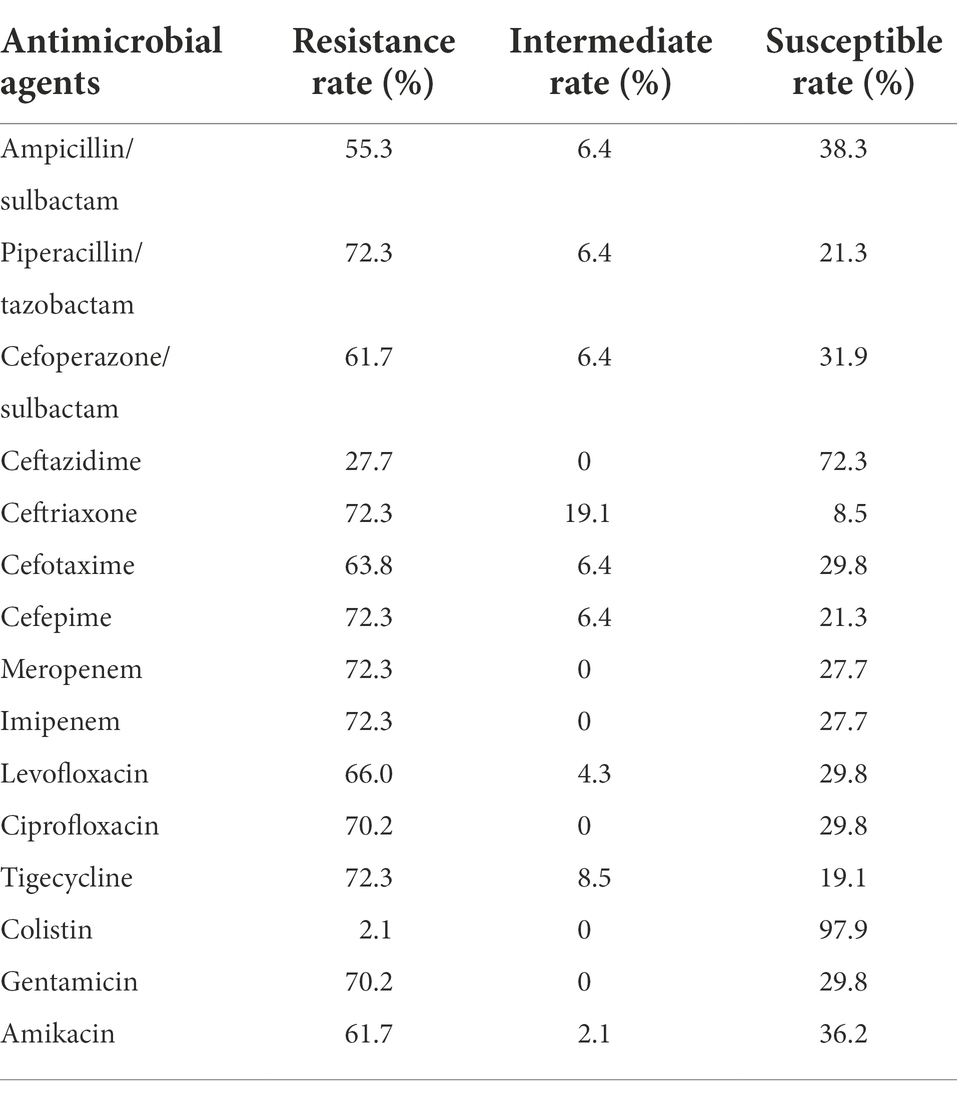
Table 1. Antimicrobial susceptibility testing results of 47 Acinetobacter baumannii isolates causing bloodstream infection (BSI).
All 47 A. baumannii isolates were distributed into 22 STs based on the Oxford MLST scheme, among which ST195 was 0the most dominant, with 17 isolates (36.2%), followed by ST208 with 8 isolates (17.0%). Except for two ST218 and two ST1818 isolates, each of the remaining isolates belonged to a single ST. Clone complex (CC) 92 was the most prevalent with 30 isolates (63.8%) and encompassed six STs, including ST195 and ST208. Strikingly, all of the CC92 isolates (30/30, 100%) exhibited resistance against carbapenems, whereas 4 of 17 (23.5%) of the other ST isolates were carbapenem-resistant. cgMLST analysis was subsequently performed to assess the phylogenetic relationship of these BSI-related A. baumannii isolates with higher resolution based on genome sequences. There was a large relevant cluster (cluster 1 in Figure 1) observed in the phylogenetic tree, the isolates in which originated from all three different hospitals and belonged to ST195, indicating that a dominant clone was disseminated among the hospitals.
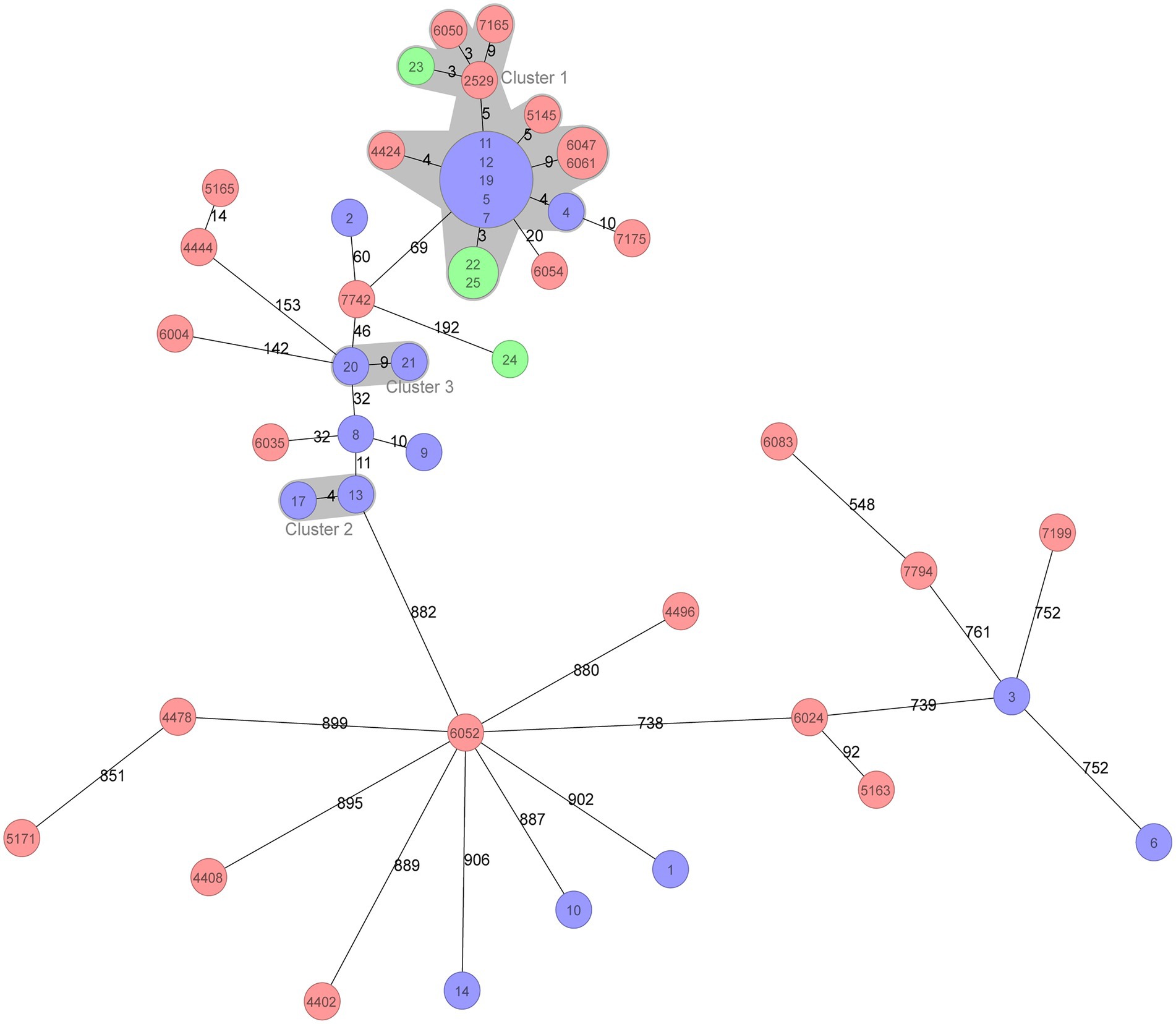
Figure 1. Minimum spanning tree of 47 BSI Acinetobacter baumannii isolates based on cgMLST. cgMLST profiles are represented by circles, and the isolate name is marked on the circle. The size of the circle is proportional to the number of isolates with an identical cgMLST profile. The different colours in the circle represent the three hospitals from which the isolates were collected. The number on the line connecting the cgMLST circles is the number of core genes that differ between the isolates within the circles. The grey zone surrounding a group of circles represents the closely related isolates that differed by less than ten core genes.
Most of these BSI-related A. baumannii isolates (46/47) harboured intrinsic blaOXA-51-like or blaOXA-213-like genes, including blaOXA-51, blaOXA-66, blaOXA-80, blaOXA-88, blaOXA-106, blaOXA-111, blaOXA-120, blaOXA-132, blaOXA-430, blaOXA-273, blaOXA-421, blaOXA-500, blaOXA-526 and blaOXA-533, by which the oxacillinase expression did not mediate carbapenem resistance. Several other OXA-type genes, such as blaOXA-23 and blaOXA-72, were mainly responsible for carbapenem resistance. The blaOXA-23 carbapenemase gene was most common and was present in 32 isolates, one of which co-harboured another metal-β-lactamase, blaNDM-1. There were also 15 blaOXA-72-positive isolates. Moreover, 31/47 isolates harboured the 16S rRNA methylase gene armA, which was responsible for high-level resistance against aminoglycosides. Aminoglycoside-modifying enzymes that commonly mediate low-or medium-level resistance to aminoglycosides, such as aph(3′)-Ia, aph(6)-Id and aadA-1, were detected in 32 A. baumannii isolates. Among other antimicrobial resistance genes, we also screened the macrolide resistance genes mph(E) and msr(E), sulphonamide resistance genes sul1 and sul2, tigecycline resistance gene tet(B), phenicol resistance gene catB8 and fluoroquinolone resistance gene qnrS1 (Figure 2).
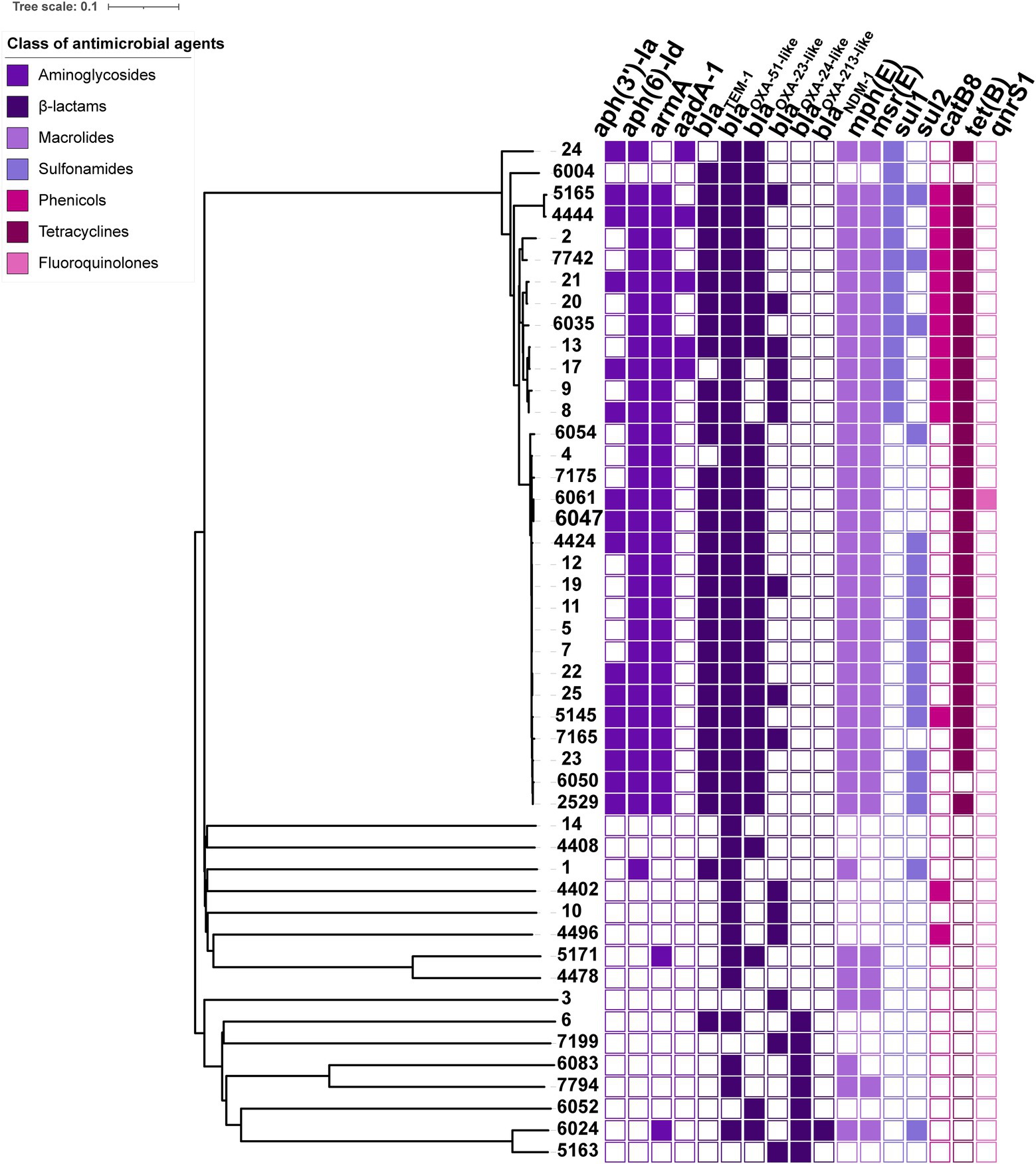
Figure 2. Phylogenetic relationship based on the core genome among 47 BSI A. baumannii isolates and the heatmap of resistance determinants. In the heatmap, colour-filled rectangles indicate the positive resistance determinants, and colour-empty rectangles indicate the negative resistance determinants. Different colours represent the classes of antimicrobial agents to which the isolate confers resistance.
The genetic structure of the acquired carbapenemase genes was further analysed. The spread of the carbapenemase gene blaOXA-23 is normally mediated by several known transposons, such as Tn2006, Tn2007, Tn2008 and Tn2009, among which the distinct genetic structure was found to be distributed on ISAba1/ISAba4 or the dual-copy insertion of ISAba1. The known transposon structures were screened in all blaOXA-23-positive isolates, and the comparison results indicated that 21/32 (65.6%) were connected to Tn2006, 8/32 (25.0%) were Tn2009 and the remaining 3/32 (9.4%) were connected to Tn2008. Conjugation assays showed that the blaOXA-23 gene was located on the plasmid in at least 9/32 blaOXA-23-positive isolates, which successfully acquired the blaOXA-23-harbouring plasmid.
The blaOXA-40 variant blaOXA-72 belonged to the blaOXA-24 cluster and was identified in 15/47 (31.9%) A. baumannii isolates. The blaOXA-72-containing contig sequences were extracted from the genome data, among which six contigs from isolates 8, 9, 13, 17, 20 and 7,199 were greater than 2 kbp in size and were screened for homology against the GenBank database. The results showed that these six blaOXA-72-containing contigs were approximately 6–15 kb in size and 100% identical or partly similar to plasmids pA2503 (MN495626) and pA2485 (MN495625), which are both 15,405 bp in size. Notably, no mobile element, such as a transposon or IS, was found surrounding the blaOXA-72 gene (Figure 3). However, interestingly, the genetic structure comparison illustrated that several insertions, deletions or inversions occurred among these plasmid segments, and on the border of the fragment, we found some pairs of XerC/XerD-like sites, which could provide active pairs for site-specific recombination mediating horizontal gene transfer. For example, a pair of XerC/XerD-like sites were found at the border of a 5 kbp inversion between isolate 8 and isolate 13 (or 17). Similarly, XerC/XerD-like sites also emerged at the border of in/del segments between isolates 20 and 9, isolates 9 and 8, isolates 7,199 and 17, etc. Crucially, the blaOXA-72 gene was observed as a segment flanked closely by XerC/XerD-like sites, suggesting that XerC/XerD-like site-mediated recombination may be responsible for mobilisation of the blaOXA-72 gene in the BSI-related A. baumannii isolates in our study (Figure 3).
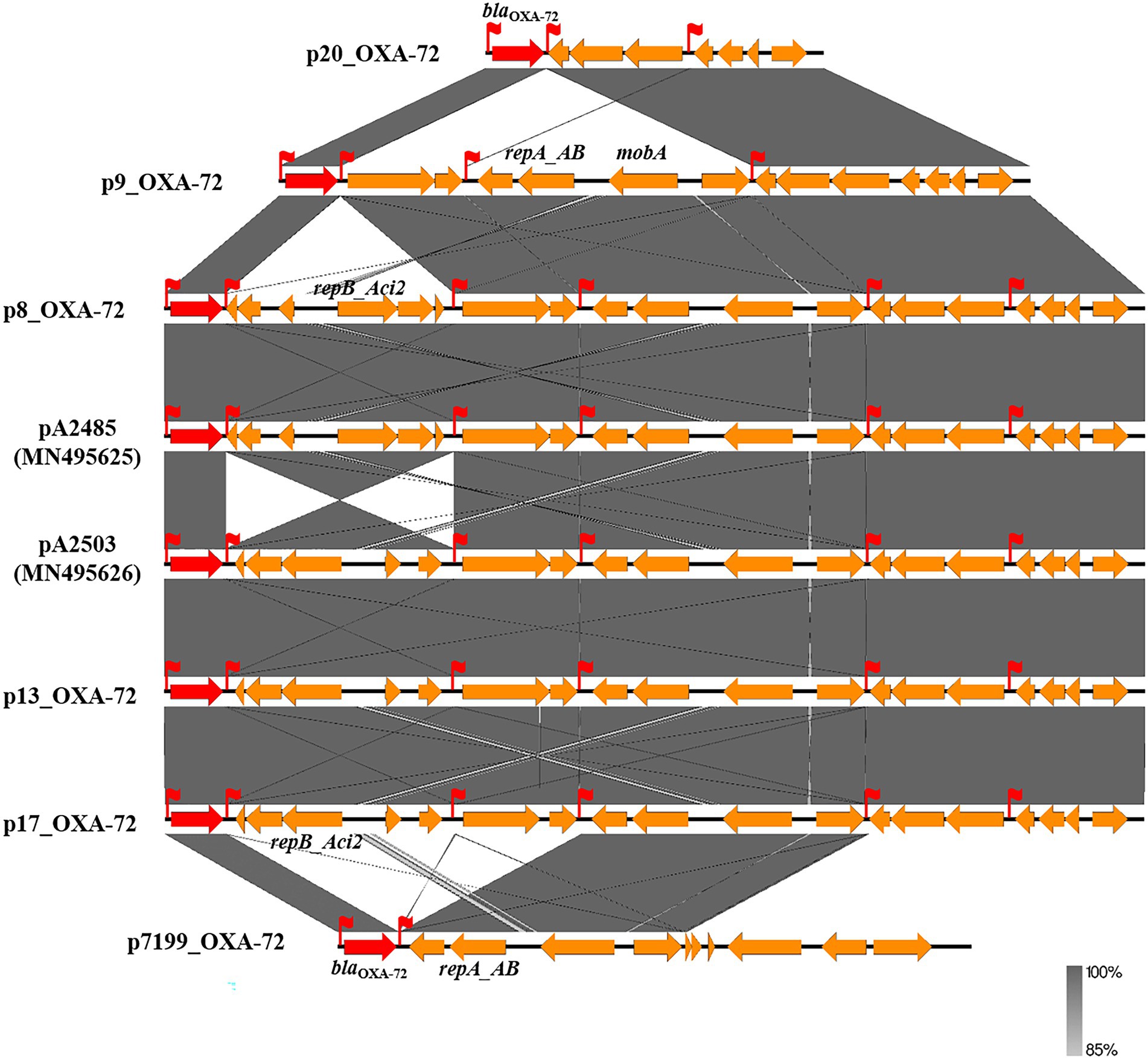
Figure 3. Sketch and comparison of the genetic structure of blaOXA-72-harbouring large contigs. Orange rectangular arrows on the line represent the open reading frames (ORFs) encoded by the contig sequence, and the red arrows represent the blaOXA-gene. The dark grey shadows indicate the 72 identical sequence segment between two contigs. The red flags represent the XerC/XerD-like combination site.
In total, 25/47 (53.2%) isolates presented an efflux pump phenotype that decreased MIC of meropenem or imipenem by 4-fold or more. Most (23/25, 92.0%) of the efflux pump positive isolates were identified under the CCCP inhibitor, and six isolates tested positive under PAβN. The inhibitor NMP did not induce an efflux pump phenotype in any of the isolates. Eighteen isolates that showed a more than 64-fold decrease in the MIC of meropenem were selected for subsequent evaluation of the expression of the genes adeA and adeB in the multidrug efflux pump system AdeABC. The results showed that 17/18 (94.4%) isolates presented higher expression of adeA or adeB than the reference strain ATCC 17978, among them 16 isolates presented a more than 2-fold (significant) increase in expression of at least one pump gene (Figure 4). The last isolate exhibited no expression of either the adeA or adeB gene due to deletion of the AdeABC efflux pump, which was confirmed by PCR on chromosome DNA.
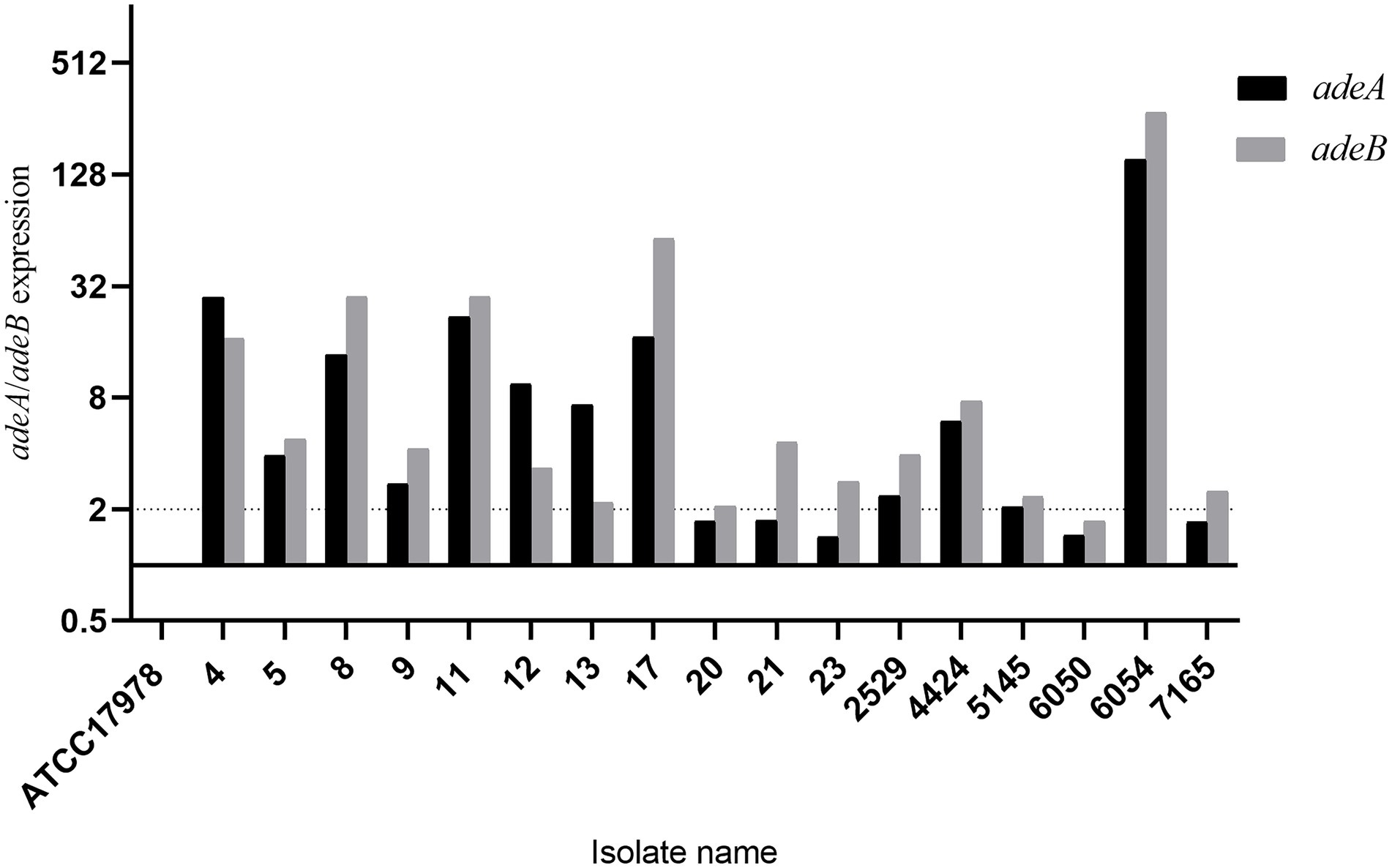
Figure 4. Expression of the AdeABC efflux pump genes adeA and adeB. ATCC17978 was used as a baseline.
BSI caused by A. baumannii is associated with high patient mortality and is one of the most urgent threats to public health. In this study, 47 A. baumannii isolates causing BSI were collected in three tertiary hospitals over a decade. Antimicrobial susceptibility testing and whole-genome sequencing were performed to assess the molecular epidemiology and antimicrobial resistance characteristics of the isolates. Multidrug resistance, especially resistance against carbapenem, was observed in these BSI-related A. baumannii isolates, leaving limited choice for therapy. Colistin, a cationic polypeptide belonging to the polymyxin family, has been introduced into clinical practice as an important therapeutic option for carbapenem-resistant Gram-negative bacterial infections (Falagas et al., 2011). A previous study indicated that colistin monotherapy was associated with a better outcome than colistin-meropenem combination therapy (Dickstein et al., 2019). However, using colistin as a therapy choice still depends on PK/PD and underlying complications due to its potential renal toxicity.
Molecular epidemiology based on genomic data facilitated the investigation of the dissemination and phylogenetic relationship of BSI-related A. baumannii isolates with higher distinguishability. CC92 clone dissemination was observed in this study accounting for more than half of the BSI-related A. baumannii isolates. CC92 is the most common clone complex in A. baumannii from China (Karah et al., 2012). More importantly, all isolates belonging to the CC92 clone were carbapenem-resistant, which was one of the crucial reasons they survived under high antibiotic pressure and spread widely, consequently inducing BSI. cgMLST was used to further discover the diffusion of a dominant clone among three different hospitals in northwest China, as evidenced by the presence of a cluster comprising genetically indistinguishable isolates.
The BSI-related A. baumannii isolates harboured multiple resistance determinants, among which the oxacillinase genes were the most common. The OXA-type enzymes in A. baumannii are normally divided into four clusters based on their genetic similarities, namely, the OXA-51, OXA-23, OXA-24 and OXA-58 clusters (Peleg et al., 2008). The enzymes of the OXA-51 cluster are naturally occurring enzymes in A. baumannii given their chromosomal location and minimal effect on carbapenem susceptibility. More than ten variants of blaOXA-51 or blaOXA-213 were observed in 46 of the BSI-related A. baumannii isolates, indicating the diversity of the intrinsic oxacillinase gene cluster.
In the Acinetobacter genus, the acquired carbapenem hydrolysing oxacillinases contribute to carbapenem resistance. OXA-23 was the first identified carbapenemase, and its role in carbapenem-hydrolysis appeared to be elevated in the presence of the upstream ISAba1 element. In our study, the blaOXA-23-positive isolates were associated with several known transposons, Tn2006, Tn2008 and Tn2009, and all of them encompassed the ISAba1 element that possibly mediated their high-level carbapenem resistance (Chen et al., 2017). Yang’s study reported 58 A. baumannii strains carrying blaOXA-23 gene in China, 47 isolates (47/58, 81.0%) were associated with Tn2009 and 8 isolates (8/58,13.8%) were associated with Tn2006 (Yang et al., 2019). Another study reported by Cerezalesa showed that 51 carbapenem-resistant A. baumannii strains carried the blaOXA-23 gene in transposon Tn2008 (Cerezales et al., 2019). In our study, the transposons that harboured the blaOXA-23 gene in BSI-related A. baumannii in Shaanxi province presented diverse, and the Tn2006 was the most common (65.6%) in our report, suggesting a distinction from the results of previous studies.
The blaOXA-40 variant blaOXA-72 belongs to the blaOXA-24 cluster, and its presence has been reported in several previous studies on CRAB (Kuo et al., 2013; Dortet et al., 2016; Chen et al., 2018). Analysis of the blaOXA-72 genetic environment revealed a potential transfer mechanism corresponding to a recombination site. In A. baumannii and most bacteria, dimers are resolved to monomers by site-specific recombination, which is a process that is performed by two chromosomally encoded tyrosine recombinases (XerC and XerD). Several studies have reported that plasmid-borne blaOXA-containing structures are bordered by short sequences exhibiting homology with the 28-nucleotide dif motif located at the bacterial chromosome replication terminus and are recognised by XerC/D site-specific recombinases, leading to the hypothesis that their mobilisation could be mediated by site-specific recombination (Merino et al., 2010). In our study, all blaOXA-72-containing contigs were found to have a pair of 28-nucleotide XerC/XerD-like sites that closely flanked the blaOXA-72 gene, indicating that recombination through the Xer system likely occurs to mediate transfer of the carbapenem-resistance gene to the nosocomial environment.
Efflux pumps often play a crucial role in multidrug resistance in A. baumannii, including by mediating a possibly significant increase in carbapenem susceptibility (Abdi et al., 2020). In this study, the carbapenem susceptibility in more than half of the BSI-related A. baumannii isolates was influenced by efflux pump inhibitors. The inhibitor CCCP seemed more efficient for the majority of isolates with an efflux pump phenotype, and in contrast, the inhibitor NMP showed no impact. The AdeABC efflux pump is a member of the RND family and can pump out multiple antibiotics, and overexpression of the AdeABC efflux pump may confer high-level resistance to carbapenems (Zhu et al., 2013). The majority of efflux pump phenotype-positive isolates showed higher expression of AdeABC efflux pump genes, except one isolate, which lacked these genes. The presence of other functional efflux pumps in that isolate could potentially explain the efflux pump-positive phenotype.
In conclusion, this study of molecular epidemiology and antimicrobial resistance characteristics revealed that A. baumannii isolates causing BSI presented clone dissemination and multidrug resistance. The multidrug-resistant clone CC92 had spread among distinct hospitals in northwest China over decade-long period of our study. The BSI-related A. baumannii isolates consistently exhibited resistance against carbapenems, which was attributed to the wide distribution of oxacillinases OXA-23 and OXA-72. In addition to the carbapenemases produced, the efflux pump harboured by the A. baumannii isolates also plays an important role, and the efflux pump genes were suggested to exhibit significantly increased expression. BSI caused by A. baumannii isolates poses a serious threat to health and is correlated with high mortality in patients with hospital-acquired infections. Additional strategies for nosocomial infection control urgently needed to prevent these multidrug-resistant A. baumannii clones from becoming endemic.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA844406.
JX conceived and designed this study. YG, JL, and LZ collected strains of bacteria. YG, JL, and JT collected the isolates and clinical data. WZ and HX performed the antimicrobial susceptibility testing. WZ, HW, JT, MD, and MZ carried out whole genome sequencing and analysis. YG and RW structured the variables and performed the statistical analyses. YG wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Key R&D project of Shaanxi Provincial Department of Science and Technology (no. 2019SF-220).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdi, S. N., Ghotaslou, R., Ganbarov, K., Mobed, A., Tanomand, A., Yousefi, M., et al. (2020). Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect Drug Resist 13, 423–434. doi: 10.2147/IDR.S228089
Cameranesi, M. M., Moran-Barrio, J., Limansky, A. S., Repizo, G. D., and Viale, A. M. (2018). Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58-and TnaphA6-resistance module in Acinetobacter baumannii. Front. Microbiol. 9:66. doi: 10.3389/fmicb.2018.00066
Cerezales, M., Xanthopoulou, K., Wille, J., Bustamante, Z., Seifert, H., Gallego, L., et al. (2019). Acinetobacter baumannii analysis by core genome multi-locus sequence typing in two hospitals in Bolivia: endemicity of international clone 7 isolates (CC25). Int. J. Antimicrob. Agents 53, 844–849. doi: 10.1016/j.ijantimicag.2019.03.019
Chen, Y., Gao, J., Zhang, H., and Ying, C. (2017). Spread of the blaOXA-23-containing Tn2008 in Carbapenem-resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front. Microbiol. 8:163. doi: 10.3389/fmicb.2017.00163
Chen, Y., Yang, Y., Liu, L., Qiu, G., Han, X., Tian, S., et al. (2018). High prevalence and clonal dissemination of OXA-72-producing Acinetobacter baumannii in a Chinese hospital: a cross sectional study. BMC Infect. Dis. 18:491. doi: 10.1186/s12879-018-3359-3
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing: 29th Informational Supplement, M100-S29. Wayne, PA: Clinical and Laboratory Standards Institute.
Cornejo-Juarez, P., Cevallos, M. A., Castro-Jaimes, S., Castillo-Ramirez, S., Velazquez-Acosta, C., Martinez-Oliva, D., et al. (2020). High mortality in an outbreak of multidrug resistant Acinetobacter baumannii infection introduced to an oncological hospital by a patient transferred from a general hospital. PLoS One 15:e0234684. doi: 10.1371/journal.pone.0234684
Coyne, S., Courvalin, P., and Perichon, B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953. doi: 10.1128/AAC.01388-10
Dickstein, Y., Lellouche, J., Ben Dalak Amar, M., Schwartz, D., Nutman, A., Daitch, V., et al. (2019). Treatment outcomes of colistin-and carbapenem-resistant Acinetobacter baumannii infections: an exploratory subgroup analysis of a randomized clinical trial. Clin. Infect. Dis. 69, 769–776. doi: 10.1093/cid/ciy988
Dortet, L., Bonnin, R. A., Bernabeu, S., Escaut, L., Vittecoq, D., Girlich, D., et al. (2016). First occurrence of OXA-72-Producing Acinetobacter baumannii in Serbia. Antimicrob. Agents Chemother. 60, 5724–5730. doi: 10.1128/AAC.01016-16
EUCAST (2018). The European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.1. Basel: European Committee on Antimicrobial Susceptibility Testing.
Falagas, M. E., Karageorgopoulos, D. E., and Nordmann, P. (2011). Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 6, 653–666. doi: 10.2217/fmb.11.49
Gu, Y., Jiang, Y., Zhang, W., Yu, Y., He, X., Tao, J., et al. (2021). Risk factors and outcomes of bloodstream infections caused by Acinetobacter baumannii: a case-control study. Diagn. Microbiol. Infect. Dis. 99:115229. doi: 10.1016/j.diagmicrobio.2020.115229
Hammoudi, D., and Ayoub, C. (2020). The current burden of Carbapenemases: review of significant properties and dissemination among Gram-Negative bacteria. Antibiotics (Basel) 9. doi: 10.3390/antibiotics9040186
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, O. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Kuo, S. C., Yang, S. P., Lee, Y. T., Chuang, H. C., Chen, C. P., Chang, C. L., et al. (2013). Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne bla(OXA-72) in Taiwan. BMC Infect. Dis. 13:319. doi: 10.1186/1471-2334-13-319
Merino, M., Acosta, J., Poza, M., Sanz, F., Beceiro, A., Chaves, F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. doi: 10.1128/AAC.01674-09
Mmatli, M., Mbelle, N. M., Maningi, N. E., and Osei Sekyere, J. (2020). Emerging transcriptional and genomic mechanisms mediating carbapenem and polymyxin resistance in Enterobacteriaceae: a systematic review of current reports. mSystems 5:e00783-20 doi: 10.1128/mSystems.00783-20
Patel, G., and Bonomo, R. A. (2013). "Stormy waters ahead": global emergence of carbapenemases. Front. Microbiol. 4:48. doi: 10.3389/fmicb.2013.00048
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Potron, A., Poirel, L., and Nordmann, P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int. J. Antimicrob. Agents 45, 568–585. doi: 10.1016/j.ijantimicag.2015.03.001
Ramirez, M. S., Bonomo, R. A., and Tolmasky, M. E. (2020). Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace. Biomol. Ther. 10:720. doi: 10.3390/biom10050720
Reddy, T., Chopra, T., Marchaim, D., Pogue, J. M., Alangaden, G., Salimnia, H., et al. (2010). Trends in antimicrobial resistance of Acinetobacter baumannii isolates from a metropolitan Detroit health system. Antimicrob. Agents Chemother. 54, 2235–2238. doi: 10.1128/AAC.01665-09
Roca, I., Espinal, P., Vila-Farres, X., and Vila, J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Stewart, N. K., Smith, C. A., Antunes, N. T., Toth, M., and Vakulenko, S. B. (2019). role of the hydrophobic bridge in the carbapenemase activity of class D beta-lactamases. Antimicrob. Agents Chemother. 63:e02191-18. doi: 10.1128/AAC.02191-18
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., and Spellberg, B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058-16
Yang, Y., Xu, Q., Li, T., Fu, Y., Shi, Y., Lan, P., et al. (2019). OXA-23 is a prevalent mechanism contributing to sulbactam resistance in diverse Acinetobacter baumannii clinical strains. Antimicrob. Agents Chemother. 63:e01676-18. doi: 10.1128/AAC.01676-18
Zhou, H., Yao, Y., Zhu, B., Ren, D., Yang, Q., Fu, Y., et al. (2019). Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: a retrospective study from a Chinese hospital. Medicine (Baltimore) 98:e14937. doi: 10.1097/MD.0000000000014937
Keywords: Acinetobacter baumannii, bloodstream infection, cgMLST, oxacillinase, XerC/D
Citation: Gu Y, Zhang W, Lei J, Zhang L, Hou X, Tao J, Wang H, Deng M, Zhou M, Weng R and Xu J (2022) Molecular epidemiology and carbapenem resistance characteristics of Acinetobacter baumannii causing bloodstream infection from 2009 to 2018 in northwest China. Front. Microbiol. 13:983963. doi: 10.3389/fmicb.2022.983963
Received: 01 July 2022; Accepted: 26 July 2022;
Published: 22 August 2022.
Edited by:
Fang He, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Xiaoyan Li, Southern Medical University, ChinaCopyright © 2022 Gu, Zhang, Lei, Zhang, Hou, Tao, Wang, Deng, Zhou, Weng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiru Xu, eHVqaXJ1QG1haWwueGp0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.