- 1College of Biological and Food Engineering, Guangdong University of Petrochemical Technology, Maoming, Guangdong, China
- 2Institute for Quality & Safety and Standards of Agricultural Products Research, Jiangxi Academy of Agricultural Sciences, Nanchang, China
- 3Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, Guangdong, China
Nitrogen (N) is an essential element in the biosynthesis of key cellular components, such as proteins and nucleic acids, in all living organisms. Nitrite, as a form of nitrogen utilization, is the main nutrient for microbial growth. However, nitrite is a potential carcinogen that combines with secondary amines, which are breakdown products of proteins, to produce N-nitroso compounds that are strongly carcinogenic. Nitrite reductase (Nir) produced by microorganisms can reduce nitrite. Binding of GlnR to the promoter of nitrogen metabolism gene can regulate the expression of Nir operon. In this study, nitrite-resistant Lactobacillus plantarum WU14 was isolated from Pickles and its protease Nir was analyzed. GlnR-mediated regulation of L. plantarum WU14 Nir gene was investigated in this study. New GlnR and Nir genes were obtained from L. plantarum WU14. The regulation effect of GlnR on Nir gene was examined by gel block test, yeast two-hybrid system, bacterial single hybrid system and qRT-RCR. Detailed analysis showed that GlnR ound to the Nir promoter region and interacted with Nir at low nitrite concentrations, positively regulating the expression of NIR. However, the transcription levels of GlnR and Nir decreased gradually with increasing nitrite concentration. The results of this study improve our understanding of the function of the Nir operon regulatory system and serve as the ground for further study of the signal transduction pathway in lactic acid bacteria.
Introduction
Nitrite and nitrate are the main components of chemical fertilizers. The increased dependence on chemical fertilizers in agricultural production is the main factor that increases soil nitrogen content (Bryan and Grinsven, 2013). Nitrite combines with secondary amines (secondary amines, tertiary amines, amides, and amino acids), the protein decomposition products in fermented food products (pickled vegetables), to form potent cancerogenic nitrosamines in the gastral cavity that are known to cause digestive system cancers (Lin et al., 2013; Dellavalle et al., 2014). In addition, nitrite treatment of horsefish embryos results in yolk sac edema and developmental abnormalities (Simmons et al., 2012; Keshari et al., 2016). Therefore, vegetables with high nitrate content increase the risk of mutagenesis and carcinogenesis in people consuming such fermented food over a long period of time. Moreover, as people become more aware of food safety, they begin to pay more attention to nitrite contamination of fermented foods, and as a result, there is an urgent need to develop measures to degrade the nitrites in fermented food (Sen et al., 2001).
The genus Lactobacillus comprises 262 species that are extremely diverse at phenotypic, ecological, and genotypic levels. Zheng et al. (2020) proposed a new taxonomic revision of the Lactobacillus genus, reclassifying it into 25 genera which includes host-adapted organisms that have been referred to as the L. delbrueckii group, Paralactobacillus and 23 novel genera. Lactobacillus planturum, Streptococcus lactis, Streptococcus thermophilus, Lactobacillus casei and Lactobacillus brevis contain high NiR (Zhang et al., 2002; Chi and Zhang, 2017). In 1919, Orla-Jensen confirmed that L. plantarum is a facultative hetero-fermentative lactic acid bacterium of the Streptobacterium plantarum (Andersson et al., 2005). L. plantarum strain WU14 was also found to significantly reduce the concentration of nitrites (Ying et al., 2015). Four types of nitrate reductase have been identified, namely cytochrome C nitrite reductase (nrfA), nitrite assimilation reductase (nasB), copper nitrite reduction enzyme (nirK) and cytochrome cd1 nitrite reductase (nirS) (He et al., 2015).
LAB are the dominant species during vegetable fermentation. In recent years, the use of pure starter cultures has been shown to be more effective in reducing nitrite concentration compared with spontaneous fermentation (Oh et al., 2004; Yan et al., 2008; Jagannath et al., 2012). Liu et al. (2014) found that L. casei were effective in degrading nitrites both in the pickle fermentation system and in MRS medium (Liu et al., 2014). Fang et al. (2016) isolated Lactobacillus Coryneformis BBE-H3 from naturally fermented pickles, which showed good nitrite degradation activity (Fang et al., 2016). Therefore, food-grade lactic acid bacteria producing Nir can be a safe and effective strategy to dynamically degrade nitrite produced during pickling and deal with nitrite pollution (Liu et al., 2012; Chen et al., 2017).
Genes involved in nitrogen metabolism respond to nitrogen availability in the culture. GlnR and GlnRII, two regulatory genes, control the expression of several nitrogen metabolism genes at the transcriptional level. Most of the GlnRs, including the GlnR from Saccharopolyspora erythraea, belong to the OmpR family (Amon et al., 2008). However, the GlnR of Bacillus subtilis belongs to the MerR family (Yao et al., 2014). The OmpR-type response regulator GlnR is known to control nitrogen metabolism-related genes. However, it shares no similarity with the MerR-type regulator GlnR in B. subtilis (Brown et al., 2010). Streptomyces coelicolor has GlnR that regulates nine genes to improve nitrogen transport and metabolic factors in a nitrogen-starved state and conserve cellular resources at high nitrogen levels (Wang et al., 2012). For instance, the GlnR of B. subtilis acts as a repressor without binding with RNA polymerase (Ogawa et al., 1995). Typically, it binds to target genes in the promoter and controls the transcription of genes related to nitrogen metabolism (Jiang et al., 2014).
GlnR activates nirB and other genes encoding a putative large subunit of nitrite reductase. In addition, the GlnR coactivator NnaR binds to the nirB promoter (Amin et al., 2012) to promote nirB expression. This study analyzed and verified the regulatory mechanism of Nir operon in lactic acid bacteria and the effect of transcription regulator GlnR on Nir expression and the findings contribute to a better understanding of microbial responses to electron acceptor modifications and optimization of nitrate dependent biodegradation strategies.
Materials and methods
Strains, vectors, and chemicals
L. plantarum strain WU14 was isolated from pickles at the Microorganism Bioengineering Laboratory of the Jiangxi Agricultural University. Escherichia coli strains Trans1-T1 and BL 21 (DE3) were obtained from TransGen (Beijing, China). E. coli strains were cultured in Luria-Bertani medium (LB; 0.5% yeast extract, 1% tryptone, and 1% NaCl) supplemented with 100 μg/ml kana or AMP. Plasmids T3, pEASY-T3 and pET-30a were availed for cloning and expression, and the pGBKT7 and pGADT7 plasmids were applied in yeast two-hybrid assays (Wang et al., 1995; Guo et al., 2007). The pB1H1 and pH3U3 plasmids were used as bait and expression vectors, and the pSUP202 vector was used to construct the gene knockout plasmids. DNA purification kit, LA Taq DNA polymerase, and restriction endonucleases were sourced from TaKaRa (Otsu, Japan), and IPTG was purchased from Sigma (Amresco, United States). All chemicals used in this study were of analytical grade.
Construction, cloning, and expression of GlnR and Nir recombinant plasmids
The whole genomic DNA of L. plantarum WU14 was extracted by CTAB (Arseneau et al., 2017) method. Primers were designed using DANMAN (Table 1). The PCR product was cloned into the T3 vector and transferred into E. coli Trans 1 competent cells for verification and sequencing. Putative transformants were then selected on LB plates containing AMP. The digested T3-GlnR and T3-Nir plasmids were ligated into the pET-30a (Kang et al., 2019) vector using T4DNA ligase, respectively, followed by transformation into E. coli BL21 (DE3) cells and spreading on Kana containing LB plates.
GlnR and Nir protein expression and purification
E. coli BL21 competent (Jones et al., 2021) cells harboring recombinant expression plasmids pET-30a-GlnR and pET-30a-Nir were grown overnight in LB medium containing kana at 37°C in a shaker. The seed culture obtained was inoculated (1%) in LB medium containing AMP and incubated on a shaker at 37°C until OD600 reached 0.6 to 0.8. IPTG (Wandrey et al., 2016) was then added to the medium and cultured in the shaker at 30°C for 6 h to induce protein expression. The liquid culture was centrifuged (12,000 g, 10 min, and 4°C), and the collected cells were suspended in 20 ml of NTA-0 buffer. After disruption of the cells by sonication (ice bath), the crude extract was collected by centrifugation (12,000 g, 10 min, and 4°C). The protein in the crude extract was analyzed by SDS-PAGE, purified using nickel column (Ni Sepharose Fast Flow; China), and analyzed again by SDS-PAGE (Scheller et al., 2021).
Electro mobility shift assay to verify the interaction of GlnR and Nir
Pnir, the Nir promoter, was amplified by PCR with LA-taq and cloned into the T3 vector. E. coli Trans 1 competent cells were transformed using this cloned vector, and the putative transformants were confirmed using colony PCR and sequencing. Positive transformants were selected on LB plates (AMP). The digested T3-Pnir fragment and the GlnR protein were subjected to a binding reaction. Subsequent to the reaction, the GlnR-Pnir complex was analyzed on a 12% polyacrylamide gel (Maizel, 2000).
Yeast two-hybrid assay
The digested empty vectors pGBKT7 and pGADT7 were ligated with GlnR and Nir, respectively, using the Assemble Mix (Beijing Golden Biological Co., Ltd). The recombinant vectors pGBKT7-GlnR and pGADT7-Nir were transformed into Trans1 competent cells and the putative transformants were selected on LB plates containing AMP (Park et al., 2009).
The recombinant plasmid pBD-GlnR was transformed into AH109 yeast competent cells, coated on SD/−Trp/X-α-Gal, SD/−Leu/−Trp/X-α-Gal, and SD/−Leu/−Trp/-His plates and incubated at 30°C for 72 h to detect the toxicity of pBD-GlnR protein to the host cells (Lu et al., 2005). Two recombinant vectors pGBKT7-GlnR and pGADT7-Nir were co-transfected into the AH109 yeast competent cells with the corresponding empty vectors pGBKT7 and pGADT7 as controls and transferred onto SD/−Leu/−Trp/X-α-Gal and SD/−Leu/−Trp/-His+X-α-Gal plates coated with the substrate X-α-Gal for growth analysis (Lu et al., 2005; Song et al., 2018).
Bacterial one-hybrid assay
Construction of pB1H1-GlnR bait vector and pH3U3-Nir recombinant vector
GlnR gene was amplified by PCR, cloned into T3 vector and transformed into Trans 1 competent cells. The transformed cells were screened by colony PCR and sequencing. Positive transformants were selected on LB plates (AMP). The digested T3-GlnR recombinant plasmid and the empty vector PB1H1 were ligated with the T4 DNA ligase, followed by transformation into Trans1 competent cells and spreading on LB plates (Amp) for verification. The pH3U3/Nir recombinant vector was also constructed following the same method (Meng et al., 2005; Xie et al., 2011).
Interaction of GlnR and pNir
The pH3U3-pNir confirmed via restriction digestion were electroporated into the constructed US0/pB1H1-GlnR competent cells. After PCR of colony, the migration of the different DNAs was assayed via gel electrophoresis to verify the results. Finally, US0/pH3U3, US0/pH3U3-pNir and US1/pH3U3-pNir cultures were diluted; 5 μl of each dilution was placed on the positive and negative sieve medium and incubated at 37°C for 36 h to monitor bacterial growth (Hawkins and Guest, 2017).
qRT-PCR analysis of GlnR and Nir under nitrite stress
Activated L. plantarum WU14 was inoculated (1%) in a 50 ml fresh MRS medium containing 0 mg/ml, 5 mg/ml, 10 mg/ml, and 15 mg/ml NaNO2 and the absorbance (OD600) of the culture was measured every hour. Lastly, the growth curve of L. plantarum WU14 under different levels of nitrite stress was outlined. The nitrite concentration suitable for the growth of the strain was selected to quantify the regulatory mechanism of GlnR on Nir under nitrite stress.
Total RNA was extracted (remove DNA midway) from L. plantarum WU14 culture using Trizol reagent and reverse transcribed into cDNA using Vazyme HiScriptII1st Strand cDNA Synthesis Kit. Shorter fluorescent quantitative PCR amplification product could ensure amplification efficiencies close to 100% for each round. When measured with a real-time PCR instrument, the optimum assay conditions were to keep the synthesized product fragment at 200 bp. Two pairs of primers were adopted to amplify GlnR and Nir genes with PCR products of 200 bp and 150 bp. Finally, the fluorescence quantitative PCR analysis was performed using the SYBR Green kit.
Statistical analysis
All data other than those associated with optimization were statistically analyzed using one-way analysis of variance (ANOVA) and a Tukey–Kramer multiple comparison test.
Results
Cloning, expression, and purification of GlnR and Nir
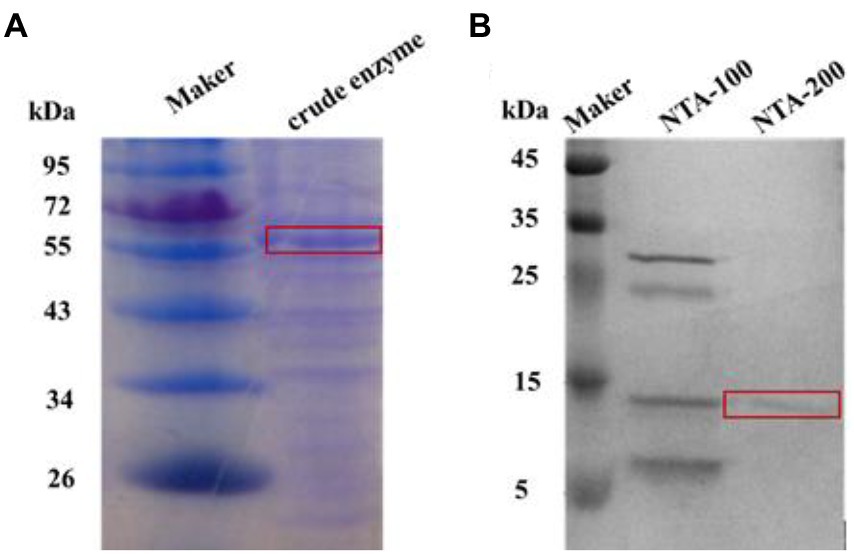
After successful expression of both proteins in E. coli, the crude enzymes were purified by nickel column (Ni Sepharose Fast Flow; China). SDS-PAGE analyses indicated the apparent molecular weights of purified TaBgl1 and IuBgl1 were approximately 62.0 and 14.7 kDa, respectively (Figure 1).
EMSA indicates the GlnR protein-Nir interaction in vitro
EMSA was used to assess the interaction between Nir DNA and GlnR proteins and subsequent regulation of Nir transcription. The results suggest that the migration rate of GlnR protein, which does not bind to the promoter of nitrite reductase (Pnir) on non-denaturing gel, is higher than that of Pnir-GlnR complex and the band is below the active gel, while the band of Pnir-GlnR complex is above the band without binding to DNA due to the low mobility (Figure 2). It was shown that GlnR protein could recognize the specific sequence above the NiR operon, promote the binding of GlnR protein with Pnir, and therefore, regulate the transcription of NiR gene, which confirmed that there are interactions between GlnR regulatory protein and NiR.
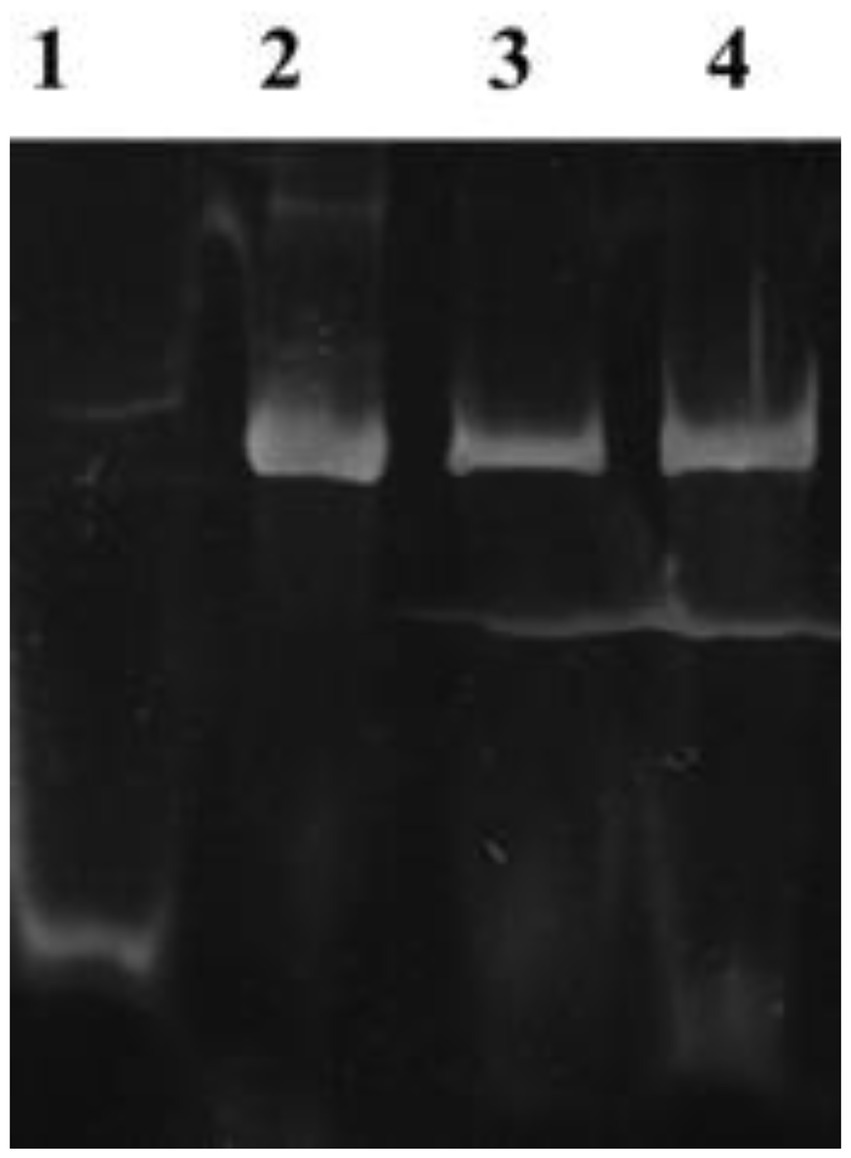
Figure 2. EMSA. Lame 1: GlnR protein without Pnir gene. Lame 2–4: The complex of GlnR protein binding to NiR promoter gene.
Yeast two-hybrid assay to investigate the interaction between GlnR and Nir
Yeast two-hybrid assay was used to study the GlnR and Nir. In this study, the GlnR and Nir genes of L. plantarum WU14 were cloned, and the bait vector and the reporter vector were constructed; Recombinant plasmid pBD-GlnR was transformed into AH109 competent cells and coated with SD/-Trp/X-α-Gal double-deficit SD/-Leu/-Trp/X-α-Gal and SD/-Leu/-Trp/-His triple-deficit plates. When the plates were incubated upside down at 30°C for 72 h, pink colonies were found on the missing plate, as shown in Figures 3–7. The results reveal that recombinant vector pBD-GlnR does not self-activate and the transcription factor GAL4 is not activated, resulting in the expression of downstream galactosidase gene being inhibited and no galactosidase being produced, so the substrate of X-α-Gal on the plate is not degraded and therefore cannot form blue colonies (Figure 3A). Recombinant vectors pGBKT7-GlnR and pGADT7-NiR were co-transfected into AH109 yeast competent cells with the corresponding empty vectors pGBKT7 and pGADT7 as controls. Pink colonies were observed growing on the second plate of the co-transfected empty vectors pGBKT7 and pGADT7 (Figure 3B); however, blue colonies grew on the third plate of the two recombinant plasmids co-transfected (Figure 3C), demonstrating an interaction between Nir and GlnR. This specific interaction may promote the proximity of the two structural domains (DNA binding domain and BD activation domain), and consequently, initiat transcription of the downstream galactosidase gene in the Y2H assay.
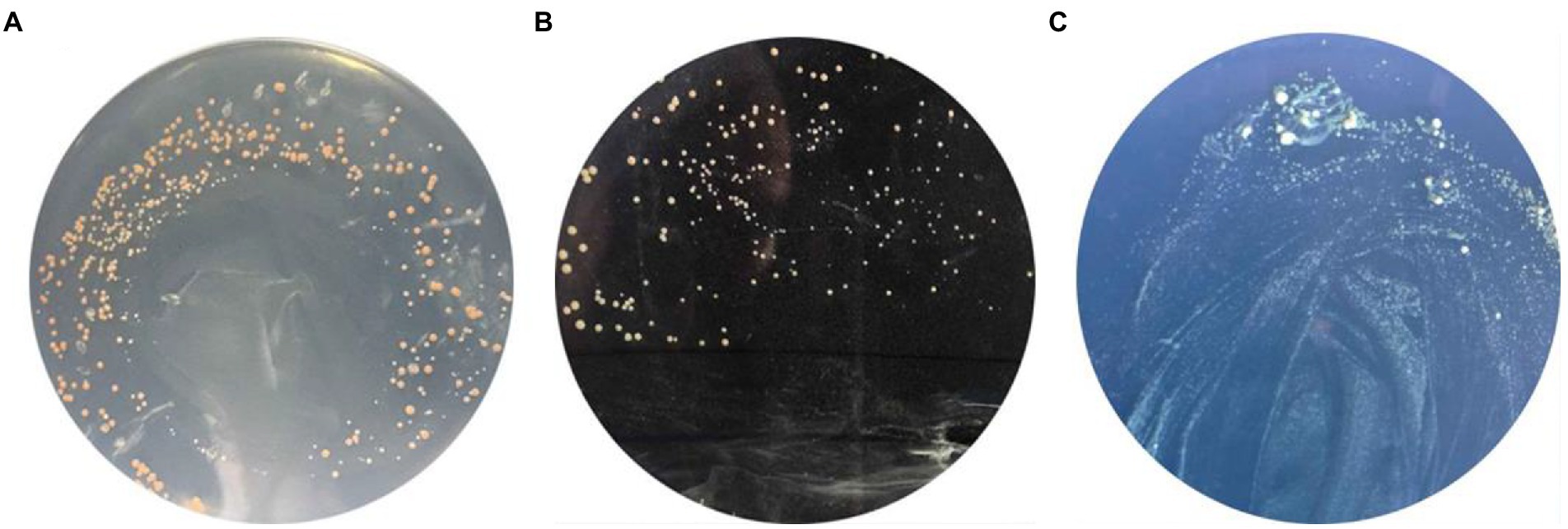
Figure 3. Yeast two-hybrid assay. Self-activation of recombinant vector pBD-GlnR (A). The result of empty gsensor conversion of pGBKT7 and pGADT7 (B). The result of interaction of NiR and GlnR Proteins (C).
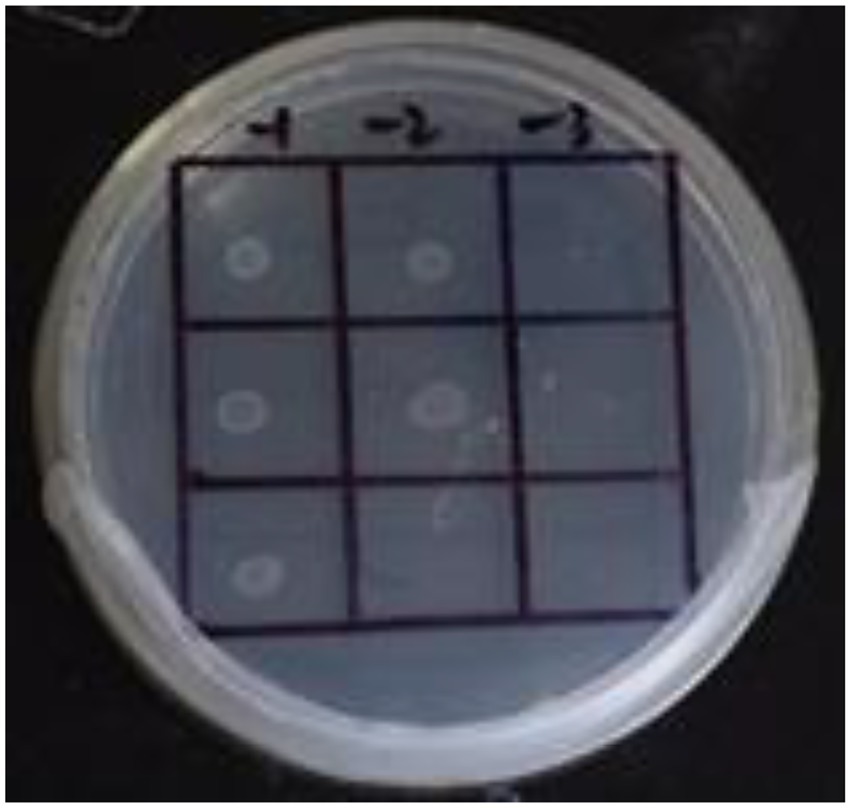
Figure 4. Self-activation of promoter. The first row: US0/pH3U3; the second row: US0/pH3U3-pNir; the second row: US1/pH3U3-pNir. −1, −2 and −3 indicate that the three columns of point samples from left to right are 10−1, 10−2 and 10−3 in the figure of gradually reducing concentrations of the bacterial solution mixed with water.
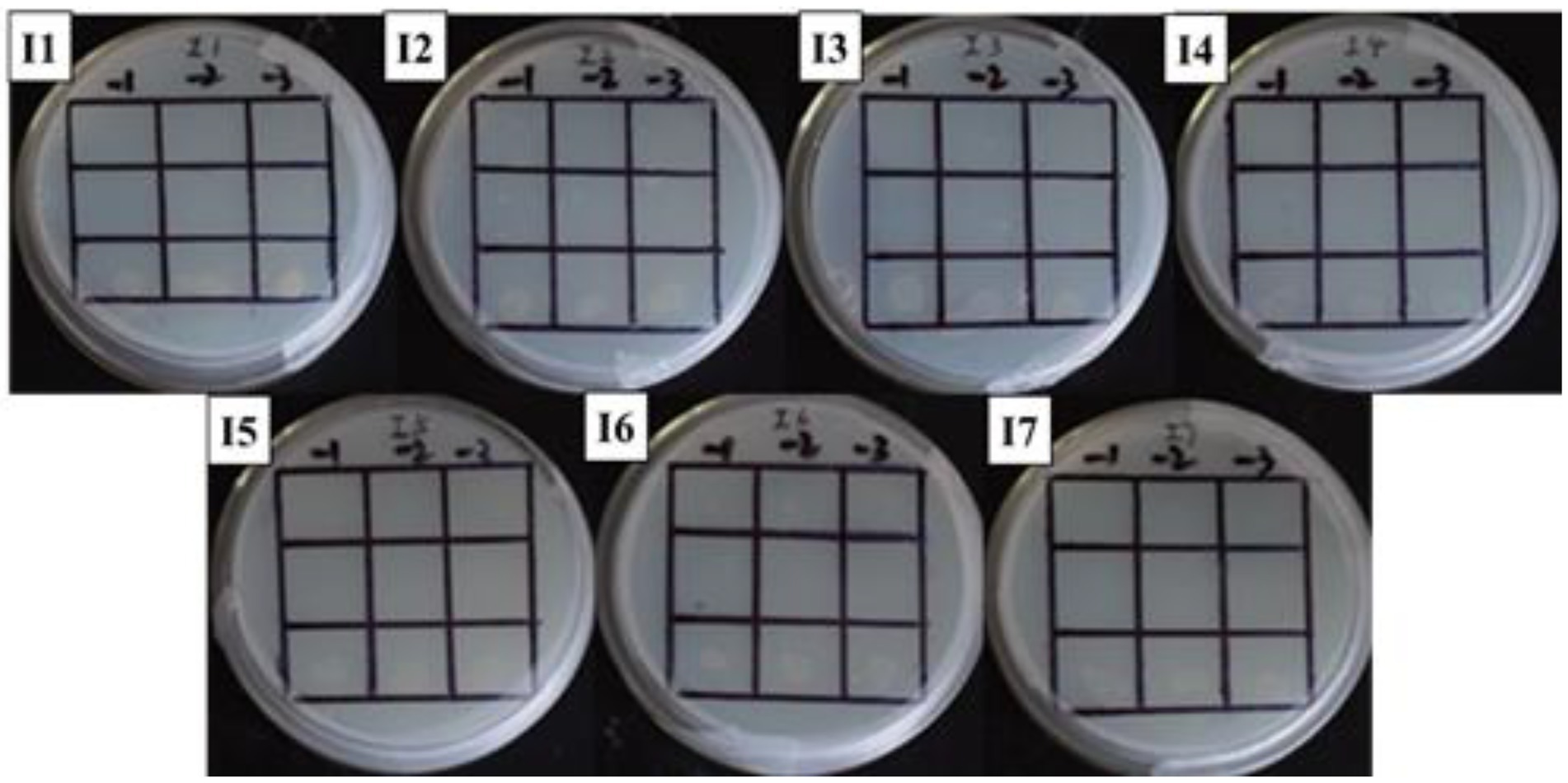
Figure 5. Positive sieve plate with histidine, 0 μg/ml NaNO2 (I1), 20 μg/ml NaNO2 (I2), 40 μg/ml NaNO2 (I3), 60 μg/ml NaNO2 (I4), 80 μg/ml NaNO2 (I5), 100 μg/ml NaNO2 (I6) and 120 μg/ml NaNO2 (I7). The first row: US0/pH3U3; the second row: US0/pH3U3-pNir; the second row: US1/pH3U3-pNir. −1, −2, and −3 indicate that the three columns of point samples from left to right are 10−1, 10−2, and 10−3 in the figure of gradually reducing concentrations of bacterial solution mixed with water.
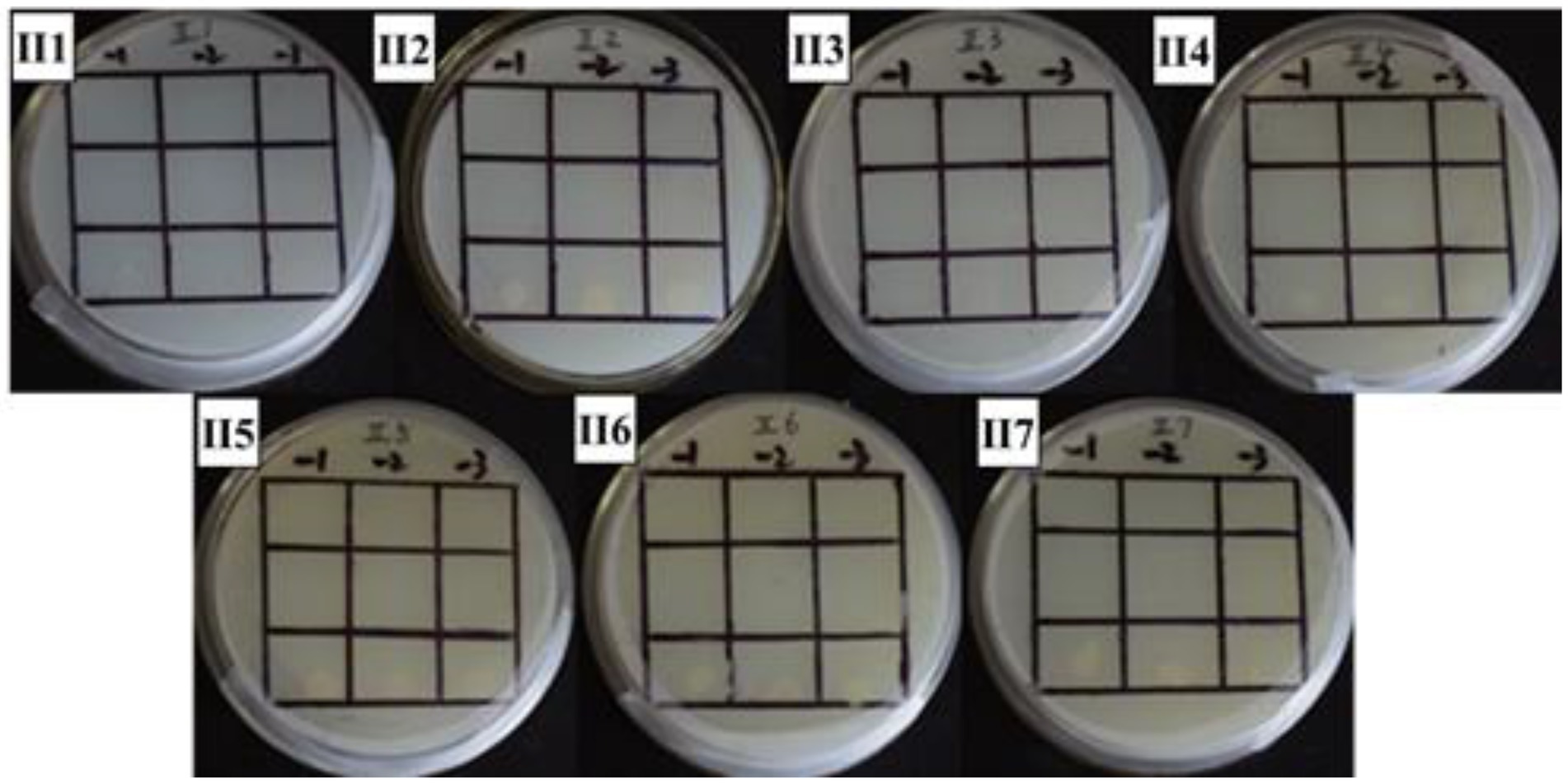
Figure 6. Positive sieve plate with 1 mM 3’AT, 0 μg/ml NaNO2 (II1), 20 μg/ml NaNO2 (II2), 40 μg/ml NaNO2 (II3), and 60 μg/ml NaNO2 (II4)80 μg/ml NaNO2 (II5), 100 μg/ml NaNO2 (II6) and 120 μg/ml NaNO2 (II7). The first row: US0/pH3U3; the second row: US0/pH3U3-pNir; the second row: US1/pH3U3-pNir. −1, −2, and −3 indicate that the three columns of point samples from left to right are 10−1, 10−2 and 10−3 in the figure of gradually reducing concentrations of bacterial solution mixed with water.
Figure 7. Positive sieve plate with 2 mM 3’AT, 0 μg/ml NaNO2 (III1), 20 μg/ml NaNO2 (III2), 40 μg/ml NaNO2 (III3), 60 μg/ml NaNO2 (III4), 80 μg/ml NaNO2 (III5), 100 μg/ml NaNO2 (III6), and 120 μg/ml NaNO2 (III7). The first row: US0/pH3U3; the second row: US0/pH3U3-pNir; the second row: US1/pH3U3-pNir.
Bacterial one-hybrid assay
In this study, the GlnR gene was inserted into the bait vector to construct the pB1H1-GlnR, while the Nir gene was inserted into the reporter vector to construct the pH3U3-pNir. E. coli cells were killed by 5-fluoroorotic acid or failed to survive at low concentrations and grow when ura3 was activated (Figure 4). In the present experiment, the US1/pH3U3-pNir strain (the third row) did not grow on the negative sieve medium, which ruled out inaccurate autoactivation. In contrast, the US0/pH3U3 (the first row) and the US0/pH3U3-pNir (the second row) strains grew as larger colonies, indicating a lack of ura3 expression in US0/pH3U3 and US0/pH3U3-pNir strains. These observations indicate the absence of promoter autoactivation in the negative sieve medium.
The interaction between GlnR protein and pNir was confirmed in the positive screening medium. On the positive screening medium without sodium nitrite, the first row of US0/pH3U3 and the second row of US0/pH3U3-pNir failed to grow, while the third row of strains interacting with target genes generated larger colonies on the positive screening medium (Figure 5, I1). This indicated that the reporter gene his3 was expressed in the positive screening medium for the strain interacting with the protein and the target gene and the strain could grow normally on the His-deficient medium. Similarly, the interaction between GlnR protein and pNir under nitrite stress was observed in the medium supplemented with appropriate concentration of nitrite (Figures 5–7). Such results are identical to those obtained with positive sieve medium without nitrite. Therefore, the bacterial single hybridization experiment reveals that GlnR protein can bind to pNir in the cells of US0 strain, and GlnR protein can also interact with pNir under nitrite stress. In the positive screening medium supplemented with histidine, US0/pH3U3 and US0/pH3U3-pNir did not grow properly. The normal growth of US1/pH3U3-pNir strain might be caused by the insufficient concentration of histidine in the His-deficient medium (Figures 6, 7). Therefore, the bacterial single hybridization experiments described above demonstrate that GlnR protein interacts with Nir promoter fragment.
qRT-PCR analysis of GlnR and Nir under nitrite stress
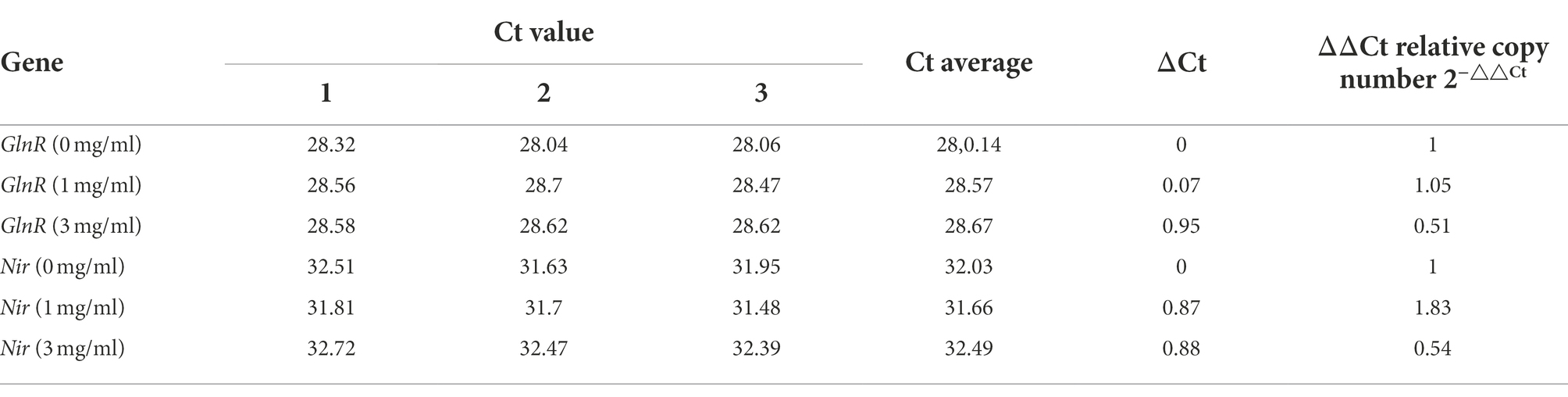
The transcript levels of GlnR and Nir genes significantly increased as the nitrite concentration increased to 1 mg/ml; however, their levels significantly reduced with an increase in the nitrite concentration to 3 mg/ml (Table 2). These observations indicate that under low concentrations of nitrite, GlnR positively regulates the Nir gene, and the transcript level of the Nir gene increase, increasing the amount of nitrite reductase. However, with the gradual increase in nitrite concentration, the transcript levels of GlnR and Nir genes decrease significantly, which indicates that high nitrite concentration is not conducive to bacterial growth. Furthermore, there is a decrease in the production of GlnR protein and a consequent decrease in the production of nitrite reductase.
Discussion
There are eight modes of nitrogen cycling in nature, six of which are attributed to microorganisms. Among the six ways, two are the breakdown of nitrite in the presence of nitrite reductase (Kuypers et al., 2018). Namely, denitrification and nitrite ammoniation, with nitrite reductase (Nir) being the rate-limiting enzyme among enzymes that catalyze denitrification (Chen et al., 2017). Researchers had identified four types of Nir: nrfA, nasB, nirK, nirS. Meanwhile, the reduction of nitrite to ammonium was catalyzed by the cytochrome C nitrite reductase encoded by nrfA and the nitrite assimilation reductase encoded by nasB (Olmo-Mira et al., 2006).
Studies on the biochemical and molecular mechanisms of Nir have been mainly focused on denitrifying nitrogen-fixing bacteria such as Pseudomonas stutzeri and Paracoccus Dentifrice. Theoretically, nitrite degradation is involved in two pathways. In the 1980s, the NO−2 degradation pathway was reported in nitrifying bacteria, but the nitrite degradation pathway of the Lactobacillus genus is still unclear. However, the molecular mechanism of remains unclear (Liu et al., 2014; Paik and Lee, 2014) as there are few studies reporting the structure, function and regulatory mechanism of Nir operon and the transcription regulatory protein of lactic acid bacteria.
GlnR of S. coelicolor is known to activate or inhibit ten target genes in the operon responsible for nitrogen absorption and regulation (amtB-glnK-glnD) and encoding the nitrite and nitrate degradation enzymes (nirB and nasA), urease (ureA), and glutamate and glutamine synthetase (glnA, glnII, and gdhA) (Tiffert et al., 2008; Sola et al., 2013). GlnR is a transcriptional regulator widely distributed in microorganisms and it has been divided into the MerR family and the OmpR family (Ying et al., 2015). It has been found that GlnR regulators are instrumental in the regulation of nitrogen metabolism. Many nitrogen metabolism-related genes, such as gdhA, glnPQ, amtB, nirBD, ureA, glnK and glnD are regulated by GlnR. GlnR is a transcriptional regulator of nitrogen metabolism genes, such as glnA (Arcondéguy et al., 2001; Amon et al., 2008; He et al., 2015).
The GlnR of lactic acid bacterial strain Lactococcus lactis MG1363 also possesses ten target genes, including the direct target glnRA operon, the ammonium transport and sensor operon amtB-glnK, and the glutamine/glutamate ABC transporter encoding gene glnPQ (Larsen et al., 2006). However, based on the protein alignment and the predicted three-dimensional structure, GlnR of L. plantarum WU14 belongs to the MerR family, which is consistent with the GlnR in of B. subtilis (Fisher, 1999). Typically, transcriptional proteins of MerR family adopts a helix - aAngle - helix model, similar to that of GlnR. The most prominent features of MerR proteins are their conserved sequences at the N-terminal and the specific sequences at the C-terminal, which are also reflected in GlnR (Helmann et al., 1990).
In this study, the structure and function of GlnR, a transcriptional regulator of L. plantarum W14 were analyzed, and accordingly, it was found to induce the expression of Nir and GlNR-mediated regulation of nitrogen metabolism in food-grade plant lactic acid bacteria cells. The interaction between GlnR and the NIR operon and promoter in nitrogen metabolism of L. plantarum W14 was demonstrated by bacterial single hybrid experiments, which preliminarily confirmed a major contribution of GlnR in the positive regulation of NIR gene transcription in L. plantarum WU14 at low nitrite concentrations. The transcriptional levels of GlnR and Nir genes at different concentrations of nitrogen sources were analyzed by fluorescence quantitative PCR. The results demonstrate that GlnR is involved not only in ammonium assimilation, but also in ammonium supply. In the absence of GlnR gene, even nitrate-providing bacteria cannot grow and the expression of at least one gene required for nitrate reduction is compromised. GlnR targets in L. plantarum W14 share similarities with some other Gram-positive bacteria, with a number of differences, though. Further research efforts will target on verifying the importance of GlnR in the regulation of nitrogen metabolism in L. plantarum and finding new targets of GlnR in Lactobacillus brevi. In addition, the specific mechanism of Nir gene regulation in L. plantarum WU14 are yet to be investigated.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
HQ: conducted experiments and wrote the papers. XC: analyzed data. YL: provided technical support. FS: proofread the papers. BX: hosted the project and provided financial and technical support. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2019A1515011696 and 2022A1515012380), and the Guangdong Special Project on Key Fields of Colleges and Universities (Rural Revitalization, 2020ZDZX1020). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
L. planturum, Lactobacillus planturum; Amp, Ampicillin; Kana, Kanamycin sulfate; IPTG, Isopropyl-β-D-thiogalactopyranoside; SDS-PAGE, Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis; EMSA, Electrophoretic Mobility Shift Assay.
References
Amin, R., Reuther, J., Bera, A., Wohlleben, W., and Mast, Y. (2012). A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 158, 1172–1182. doi: 10.1099/mic.0.054817-0
Amon, J., Bräu, T., Grimrath, A., Hänssler, E., Hasselt, K., Höller, M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. doi: 10.1128/JB.00855-08
Andersson, U., Molenaar, D., Rådström, P., and de Vos, W. M. (2005). Unity in organisation and regulation of catabolic operons in Lactobacillus plantarum, Lactococcus lactis and Listeria monocytogenes. Syst. Appl. Microbiol. 28, 187–195. doi: 10.1016/j.syapm.2004.11.004
Arcondéguy, T., Jack, R., and Merrick, M. (2001). PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65, 80–105. doi: 10.1128/MMBR.65.1.80-105.2001
Arseneau, J. R., Steeves, R., and Laflamme, M. (2017). Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 17, 686–693. doi: 10.1111/1755-0998.12616
Brown, N. L., Stoyanov, J. V., Kidd, S. P., and Hobman, J. L. (2010). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. doi: 10.1016/S0168-6445(03)00051-2
Bryan, N. S., and Grinsven, H. V. (2013). Chapter three–the role of nitrate in human health. Adv. Agron. 119, 153–182. doi: 10.1016/B978-0-12-407247-3.00003-2
Chen, H., Chen, S. M., Liu, D. M., Huang, J., Chen, S. R., Wu, H., et al. (2017). Isolation and purification of NiR and electron donor from Lactobacillus casei LCR6013 and their synergistic degradation of nitrite. Mod. Food Technol. 33, 125–131.
Chi, X. M., and Zhang, Q. F. (2017). Evaluation of highly efficient degradation of NO 2– fermentation by lactic acid bacteria in food. Food. Ferment. Ind. 43, 78–84.
Dellavalle, C. T., Xiao, Q., Yang, G., Shu, X. O., Aschebrookkilfoy, B., Zheng, W., et al. (2014). Dietary nitrate and nitrite intake and riskof colorectal cancer in the Shanghai Women’s health study. Int. J. Gynecol. Cancer 134, 2917–2926. doi: 10.1002/ijc.28612
Fang, F., Feng, T., Du, G., and Chen, J. (2016). Evaluation of the impact on foodsafety of a Lactobacillus coryniformis strain from pickled vegeta-bles with degradation activity against nitrite and other undesirablecompounds. Food Addit. Contam. 33, 623–630. doi: 10.1080/19440049.2016.1156774
Fisher, S. H. (1999). Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32, 223–232. doi: 10.1046/j.1365-2958.1999.01333.x
Guo, F., Wang, Y., and Zhang, Y. Z. (2007). Construction of two recombination yeast two-hybrid vectors by in vitro recombination. Mol. Biotechnol. 36, 38–43. doi: 10.1007/s12033-007-0018-0
Hawkins, S., and Guest, P. C. (2017). Multiplex analyses using real-time quantitative PCR. Methods Mol. Biol. 1546, 125–133. doi: 10.1007/978-1-4939-6730-8_8
He, T. T., Gong, G. M., and Gao, R. (2015). Study on nitrite reductase related genes in Lactobacillus plantarum. Food Ind. 36, 191–194.
Helmann, J. D., Ballard, B. T., and Walsh, C. T. (1990). The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal- bridged dimer. Science 247, 946–948. doi: 10.1126/science.2305262
Jagannath, A., Raju, P. S., and Bawa, A. S. (2012). A two-step controlled lacticfermentation of cabbage for improved chemical and microbiologi-cal qualities. J Food Q. 35, 13–20. doi: 10.1111/j.1745-4557.2011.00427.x
Jiang, H., Tian, J., Ying, B., Wu, N. F., and Xu, B. (2014). Research progress on GlnR-mediated regulation. Curr. Biotechnol. 4, 90–95.
Jones, C. M., Korosh, T. C., Nielsen, D. R., and Pfleger, B. F. (2021). Optimization of a T7-RNA polymerase system in Synechococcus sp. PCC 7002 mirrors the protein overproduction phenotype from E. coli BL21 (DE3). Appl. Microbiol. Biotechnol. 105, 1147–1158. doi: 10.1007/s00253-020-11085-x
Kang, T. L., Velappan, R. D., Kabir, N., Mohamad, J., Rashid, N. N., and Ismail, S. (2019). The ABA392/pET30a protein of Pasteurella multocida provoked mucosal immunity against HS disease in a rat model. Microb. Pathog. 128, 90–96. doi: 10.1016/j.micpath.2018.12.042
Keshari, V., Adeeb, B., Simmons, A. E., Simmons, T. W., and Diep, C. Q. (2016). Zebrafish as a model to assess the teratogenic potential of nitrite. J. Vis. Exp. 108:53615. doi: 10.3791/53615
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Larsen, R., Kloosterman, T. G., Kok, J., and Kuipers, O. P. (2006). GlnR-mediated regulation of nitrogen metabolism in Lactococcus lactis. J. Bacteriol. 188, 4978–4982. doi: 10.1128/JB.00025-06
Lin, Y., Totsuka, Y., and He, Y. (2013). Epidemiology of esophageal cancer in Japan and China. J. Epidemiol. 23, 233–242. doi: 10.2188/jea.JE20120162
Liu, D. M., Wang, P., Zhang, X. Y., Xu, X. L., Wu, H., and Li, L. (2014). Characterization of nitrite degradation by Lactobacillus casei subsp. Rhamnosus LCR 6013. PLoS One 9:e93308. doi: 10.1371/journal.pone.0093308
Liu, Z. W., Yuan, W. J., Zhang, S. Y., Luo, J., Chen, Z. L., Li, K. T., et al. (2012). Screening and preliminary identification of nitrite-degrading lactic acid bacteria from pickles in Sanjiang town. Food Sci. 33, 166–169.
Lu, Y. Y., Cheng, J., Yang, Y. P., Liu, Y., Wang, L., Li, K., et al. (2005). Cloning and characterization of a novel hepatitis B virus core binding protein C12. World J. Gastroenterol. 11, 5666–5671. doi: 10.3748/wjg.v11.i36.5666
Maizel, J. V. (2000). SDS polyacrylamide gel electrophoresis. Trends Biochem. Sci. 25, 590–592. doi: 10.1016/s0968-0004(00)01693-5
Meng, X., Brodsky, M. H., and Wolfe, S. A. (2005). A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23, 988–994. doi: 10.1038/nbt1120
Ogawa, K., Akagawa, E., Yamane, K., Sun, Z. W., LaCelle, M., Zuber, P., et al. (1995). The nas B operon and nas a gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J. Bacteriol. 177, 1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995
Oh, C. K., Oh, M. C., and Kim, S. H. (2004). The depletion of sodium nitrite bylactic acid bacteria isolated from kimchi. J. Med. Food 7, 38–44. doi: 10.1089/109662004322984680
Olmo-Mira, M. F., Cabello, P., Pino, C., Martínez-Luque, M., Richardson, D. J., Castillo, F., et al. (2006). Expression and characterization of the assimilatory NADH-nitrite reductase from the phototrophic bacterium Rhodobacter capsulatus E1F1. Arch. Microbiol. 186, 339–344. doi: 10.1007/s00203-006-0149-x
Paik, H. D., and Lee, J. Y. (2014). Investigation of reduction and tolerance capability of lactic acid bacteria isolated from kimchi against nitrate and nitrite in fermented sausage condition. Meat Sci. 97, 609–614. doi: 10.1016/j.meatsci.2014.03.013
Park, S. J., Lee, B. H., and Kim, D. J. (2009). Identification of proteins that interact with podocin using the yeast 2-hybrid system. Yonsei Med. J. 50, 273–279. doi: 10.3349/ymj.2009.50.2.273
Scheller, C., Krebs, F., Wiesner, R., Wätzig, H., and Oltmann-Norden, I. (2021). A comparative study of CE-SDS, SDS-PAGE, and simple Western-precision, repeatability, and apparent molecular mass shifts by glycosylation. Electrophoresis 42, 1521–1531. doi: 10.1002/elps.202100068
Sen, N. P., Seaman, S. W., Baddoo, P. A., Burgess, C., and Weber, D. (2001). Formation of N-nitroso-N-methylurea in various samples of smoked/dried fish, fish sauce, seafood, and ethnic fermented/pickled vegetables following incubation with nitrite under acidic conditions. J. Agric. Food Chem. 49, 2096–2103. doi: 10.1021/jf0011384
Simmons, A. E., Karimi, I., Talwar, M., and Simmons, T. W. (2012). Effects ofnitrite on development of embryos and early larval stages of the Zebrafish (Danio rerio). Zebrafish 9, 200–206. doi: 10.1089/zeb.2012.0746
Sola, L. A., Rodríguez, G. A., Amin, R., Wohlleben, W., and Martín, J. F. (2013). Competition between the GlnR and pho P regulators for the gln a and amt B promoters in Streptomyces coelicolor. Nucleic Acids Res. 41, 1767–1782. doi: 10.1093/nar/gks1203
Song, C., Lee, J., Kim, T., Hong, J. C., and Lim, C. O. (2018). VOZ1, a transcriptional repressor of DREB2C, mediates heat stress responses in Arabidopsis. Planta 247, 1439–1448. doi: 10.1007/s00425-018-2879-9
Tiffert, Y., Supra, P., Wurm, R., Wohlleben, W., Wagner, R., and Reuther, J. (2008). The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880. doi: 10.1111/j.1365-2958.2007.06092.x
Wandrey, G., Bier, C., Binder, D., Hoffmann, K., Jaeger, K. E., Pietruszka, J., et al. (2016). Light-induced gene expression with photocaged IPTG for induction profiling in a high-throughput screening system. Microb. Cell Fact. 15:63. doi: 10.1186/s12934-016-0461-3
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amt B in Streptomyces coelicolor inferred a pho P/GlnR competitive binding mechanism for transcriptional regulation of amt B. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Wang, H., Peters, G. A., Zeng, X., Tang, M., Ip, W., and Khan, S. A. (1995). Yeast two-hybrid system demonstrates that estrogen receptor dimerization is ligand-dependent in vivo. J. Biol. Chem. 270, 23322–23329. doi: 10.1074/jbc.270.40.23322
Xie, Z., Hu, S., Qian, J., Blackshaw, S., and Zhu, H. (2011). Systematic characterization of protein-DNA interactions. Cell. Mol. Life Sci. 68, 1657–1668. doi: 10.1007/s00018-010-0617-y
Yan, P. M., Xue, W. T., Tan, S. S., Zhang, H., and Chang, X. H. (2008). Effect of inocu-lating lactic acid bacteria starter cultures on the nitrite concen-tration of fermenting Chinese paocai. Food Control 19, 50–55. doi: 10.1016/j.foodcont.2007.02.008
Yao, L. L., Liao, C. H., Huang, G., Zhou, Y., Rigali, S., Zhang, B., et al. (2014). GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 98, 7935–7948. doi: 10.1007/s00253-014-5878-1
Ying, B., Chang, X. Y., Liu, Z. W., Zhou, T., Chen, Y., Zhong, P. A., et al. (2015). Study on food-level high-efficiency induced expression and enzymatic properties of Lactobacillus plantarum WU14 nitrite reductase under nitrite stress. Sci. Agric. Sin. 48, 1415–1427.
Zhang, Q. F., Chi, N. Y., Zheng, Y., Wang, S. Q., Feng, Y., Meng, X. J., et al. (2002). The study on mechanism of nitrite degradation by lactic acid bacteria. Food Ferment. Ind. 28, 27–31.
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: Lactobacillus plantarum, GlnR, nir, bacterial one-hybrid system, qRT-RCR, two-hybrid system, EMSA
Citation: Qiu H, Chang X, Luo Y, Shen F, Yin A, Miao T, Li Y, Xiao Y, Hai J and Xu B (2022) Regulation of Nir gene in Lactobacillus plantarum WU14 mediated by GlnR. Front. Microbiol. 13:983485. doi: 10.3389/fmicb.2022.983485
Edited by:
Pasquale Russo, University of Foggia, ItalyReviewed by:
Sazzad Hossen Toushik, Gyeongsang National University, South KoreaMd. Ashrafudoulla, Chung-Ang University, South Korea
Pantu Kumar Roy, Gyeongsang National University, South Korea
Copyright © 2022 Qiu, Chang, Luo, Shen, Yin, Miao, Li, Xiao, Hai and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Xu, eHVibzU4M0BzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Hulin Qiu1†
Hulin Qiu1† Bo Xu
Bo Xu