- 1Research Institute of the University of Bucharest (ICUB), Bucharest, Romania
- 2Department of Microbiology and Immunology, Faculty of Biology, University of Bucharest, Bucharest, Romania
- 3Department of Microbiology and Immunology, Louisiana State University Health Science Center, Shreveport, LA, United States
When bacteria sense cues from the host environment, stress responses are activated. Two component systems, sigma factors, small RNAs, ppGpp stringent response, and chaperones start coordinate the expression of virulence factors or immunomodulators to allow bacteria to respond. Although, some of these are well studied, such as the two-component systems, the contribution of other regulators, such as sigma factors or ppGpp, is increasingly gaining attention. Pseudomonas aeruginosa is the gold standard pathogen for studying the molecular mechanisms to sense and respond to environmental cues. Bordetella spp., on the other hand, is a microbial model for studying host-pathogen interactions at the molecular level. These two pathogens have the ability to colonize the lungs of patients with chronic diseases, suggesting that they have the potential to share a niche and interact. However, the molecular networks that facilitate adaptation of Bordetella spp. to cues are unclear. Here, we offer a side-by-side comparison of what is known about these diverse molecular mechanisms that bacteria utilize to counteract host immune responses, while highlighting the relatively unexplored interactions between them.
Introduction
Respiratory infections caused by bacterial pathogens lead to changes in the structure of the airways and can eventually limit respiratory function. Though viruses and some bacteria, such as Bordetella pertussis, initiate an acute “bronchitis” in healthy individuals, a vast number of patients with disrupted lung immunity or mucosal clearance suffer from infections that typically do not resolve with antibiotic treatment (Pragman et al., 2016).
Haemophilus influenzae and Pseudomonas aeruginosa are the most common pathogens found in the sputum of non-cystic fibrosis bronchiectasis patients. P. aeruginosa is also the most frequent opportunistic pathogen associated with severe outcome and high mortality in ventilated and cystic fibrosis patients (Demko et al., 1995). Many P. aeruginosa clones that cause severe chronic lung infections are well adapted to their niche and are often unable to grow under standard culture conditions in vitro (Penterman et al., 2014; Pragman et al., 2016). Genotypic and phenotypic diversity can affect the microbiome of the airways and the levels of antimicrobial resistance, making it difficult to eradicate the infection (Feltner et al., 2016; Darch et al., 2017; Sønderholm et al., 2017; Azimi et al., 2020). Chronic lungs exhibit biofilm-type infection with P. aeruginosa which is accompanied by regular release of planktonic bacteria, possibly due to host factors and/or specific environmental signals (Zegans et al., 2002; Furukawa et al., 2006), allowing the biofilm to spread and grow.
In 1993, Muller and Hildebrandt did phylogenic analysis on the 3 classical Bordetella spp. using 23S rRNA analysis. Their results revealed >99% homology between the three classical species of Bordetella. Interestingly, the results also revealed certain degree of homology between these Bordetella species and Pseudomonas aeruginosa (Müller and Hildebrandt, 1993). Both bacteria follow under the Phylum Pseudomonadota, from which then it is divided into Gammaproteobacteria (Pseudomonas spp.) or betaproteobacteria (Bordetella spp.). Bordetella species are most closely related to bacteria classified in the genera Alcaligenes (Varley, 1986; Gerlach et al., 2001) and Achromobacter (Gross et al., 2008). While Pseudomonas spp. are most similar to Acinetobacter spp. or Moraxella spp. Although Bordetella spp. is from the family Alcaligeneaceae (Vandamme et al., 1996; Gerlach et al., 2001), Alcaligenes spp. has also genetic and metabolic similarities with Pseudomonas spp. (Gross et al., 2008; Brickman and Armstrong, 2018). Both have evolved from environmental origins (Mathee et al., 2008), and both have evolved a variety of virulence mechanisms that allows them to be adaptable to different environmental stressors, including host immune responses (Hauser, 2011; Gestal et al., 2019a,b; Qin et al., 2022). B. pertussis is the causative agent of whooping cough, also known as pertussis disease or the 100-day cough (Kilgore et al., 2016). The bacterium is highly transmissible, with an R0 as high as 17 and significant mortality before a vaccine was introduced (Kilgore et al., 2016). Unlike other respiratory pathogens, pertussis may not always show signs of an inflammatory response due to the bacteria’s ability to modulate the host immune responses (Gestal et al., 2018, 2019a,b) or evade them via biofilms (Cattelan et al., 2017). Despite the traditional view of Bordetella infection as a pediatric disease, the number of adolescent and adult patients with underlying conditions who are infected with Bordetella bronchiseptica (Spilker et al., 2008; Bos et al., 2011; Register et al., 2012; Brady et al., 2014; Sukumar et al., 2014; El Khatib et al., 2015; Menetrey et al., 2021) or non-classical Bordetella spp. (Zelazny et al., 2013; Biederman et al., 2015) is increasing, shifting the classical view of these pathogen as exclusive for the infant population (Funke et al., 1996; Coenye et al., 2002; De Schutter et al., 2003; Moissenet et al., 2005; Homola et al., 2012; Karamooz et al., 2018; O’Grady et al., 2019). Importantly, infections with Bordetella spp. have three distinct stages, the catarrhal stage, which is characterized by the mucosal discharge; the paroxysmal stage, which associates with the violent and constant coughing; and finally, the pneumonic stage that in infants can lead to death (Kilgore et al., 2016). Although Bordetella spp. infection might not 100% match the classical description of chronic infection, it is a long term disease that once over leaves the patients with a number of sequelae (Kilgore et al., 2016).
Pseudomonas aeruginosa and Bordetella spp. are both respiratory pathogens that require mucosal responses to be cleared from the lungs of infected patients (Blackwood et al., 2020). Recent and exciting results suggest that P. aeruginosa and Bordetella spp. Interact (Irie et al., 2005), and even share a niche, in the lungs of patients with chronic infections; however, the interface between these two organisms remains unexplored. Both species harbor highly sophisticated mechanisms to manipulate host immune responses and colonize the respiratory tract in similar ways (Blackwood et al., 2020). Therefore, complementary studies using the strengths of each model will bring a better understanding of the complex regulation of bacterial immunomodulatory pathways.
In this review, we contextualize the molecular markers which were recently being related to the infectious process of P. aeruginosa and Bordetella spp. in the respiratory tract. We have chosen these two pathogens because P. aeruginosa is one of the models for the study of quorum sensing (QS) and interkingdom communication, while Bordetella spp. is the gold standard model for the study of host-pathogen interaction. Moreover, Bordetella spp. is increasingly infecting chronic lung patients such as those suffering from Cystic Fibrosis or COPD (Wallet et al., 2002; Spilker et al., 2008; Menetrey et al., 2022; Moore et al., 2022) and naturally cause infection in cystic fibrosis animal models (Rogers et al., 2008; Darrah et al., 2017), suggesting that Bordetella spp. and Pseudomonas spp. can share or compete for the same niche. Understanding Bordetella spp. and Pseudomonas spp. signaling mechanisms, biofilm formation, their pathogenesis in chronic lung disease patients, and their interactions with other microbes is critical as infections produced by these microbes are increasing.
Host interaction and colonization
Pseudomonas aeruginosa and Bordetella spp. follow similar steps in the development of lung infection, utilizing well investigated virulence factors responsible for adherence, invasion, and host immune modulation. However, in recent years the roles of multiple molecules involved in the control of microbial virulence during infection is being elucidated. As studies progress, the molecular connections between bacteria and their host, as well as particularities among different bacteria pathogens during infection are offering a wider perspective regarding the complexity of pathogen-pathogen and host-pathogen interactions (Otero-Asman et al., 2020; Sultan et al., 2021; Trouillon et al., 2021). Here we will briefly describe the main mechanisms to suppress host immune response, but our special focus will be on the regulatory pathways that allow to respond to host stressors helping colonization.
Sensing cues: Two component systems
Once in the host, the first challenge is to sense host-inflammatory cues, and host environment, a successful sensing and adaptation to that new environment will allow pathogens to successfully colonize the host. Bacteria have mechanisms that allow them to sense external cues and respond accordingly to better improve their fitness (Gestal et al., 2019b). To accomplish a successful colonization, bacteria need to express cell-associated virulence factors (i.e., appendages involved in adherence and immune modulation), whose expression occurs after bacteria detect the host environment and inflammatory signals. One of the best studied mechanisms that bacteria utilize to respond to environmental cues are the two component systems (TCS). The simplest TCSs are composed of a protein that senses the external stimuli (e.g., the sensor kinase) that, once phosphorylated, transfers the phosphate group to the effector protein (response regulators), which then regulates gene expression (West and Stock, 2001).
Pseudomonas aeruginosa uses its TCS to detect host molecules and respond, and for that they are critical in host-pathogen signaling (Turkina and Vikström, 2019). Host iron, cytokines, and stress hormones are detected by P. aeruginosa (Holban et al., 2016; Turkina and Vikström, 2019) and Bordetella spp. (Gestal et al., 2019b) via TCS and are used as signaling molecules that regulate bacterial virulence. As a response, bacterial molecules are produced to modulate host phenotypes, involved in inflammatory responses such as chemotaxis, cell migration, phagocytosis, cell differentiation, and apoptosis (Holban et al., 2014a; Turkina and Vikström, 2019). Flagellum, type IV pili, type 3 and type 6 secretion systems (T3SS and T6SS), exopolysaccharides (i.e., alginate), lipopolysaccharides, and secreted proteases (LasA, LasB, AprA, Protease IV) are the traditional virulence factors employed in initial host interaction and colonization by P. aeruginosa (Faure et al., 2018). These virulence factors are involved in tissue colonization, invasion, and evasion of the host immune response. The expression of these virulence factors is constantly regulated by the molecular signals detected by the bacteria via TCSs, which are specialized in sensing, responding, and adapting to external cues (Gestal et al., 2019b). P. aeruginosa uses multi-kinase networks for sensing and integrating multiple signals to increase bacterial fitness and survival. Over 45 conserved response regulators were identified in the core TCSs regulatory network in P. aeruginosa (Trouillon et al., 2021). These TCSs ensure the switch between acute and chronic phases, and modulate phenotypes that are required for infection, including biofilm, motility, and virulence (Preston et al., 1998; Workentine et al., 2009; Ortiz-Martín et al., 2010; Moscoso et al., 2011; Rasamiravaka et al., 2015; Chambonnier et al., 2016; Francis et al., 2017). The main functions targeted by TCS in P. aeruginosa include porin activity, DNA-binding, and transcription factor activity (Trouillon et al., 2021). One of the most important TCS in P. aeruginosa is the GacS-GacA system, which upregulates small RNAs (sRNAs) involved in polysaccharide, rhamnolipid, and lectin production, extracellular DNA release, QS signaling, iron metabolism (Francis et al., 2017), and motility; all highly involved in host colonization (Pérez-Martínez and Haas, 2011). Other TCS and sRNAs (which will be described in more detail later) that control P. aeruginosa virulence include: (1) FleSR, GacSA, CreCB, CarSR, PilSR, FimS-AlgR, ChpA-PilG (which regulates motility and may be involved in the initial host colonization); (2) FleSR, PilSR, RocS1-RocR, RocA1, FimS-AlgR, KinB-AlgB, BfiSR, GacSA, RetS, RcsCB, PvrSR, MifSR, BfmSR, PprAB, BqsSR (which regulates biofilm formation and could impact on the acute/chronic infection switch); and (3) TtsSR, GtrS-GltR, GacS-LadS-RetS, CsrA/RsmA, RsmA, LadS, RocS1-RocR, RocA1, CbrAB, PA2573-PA2572, PhoRB, FimS-AlgR (which controls the expression of soluble enzymes and toxins, which are involved in host invasion) (Sultan et al., 2021).
In Bordetella spp., the primary TCS master regulon (Bordetella virulence genes, Bvg) (Moon et al., 2017) controls virulence (Coutte et al., 2020). A protein-protein blast of BvgA revealed that it shares 98% homology with P. aeruginosa response regulator transcription factors some such as the well-known Lux-family (Venturi, 2006). Some of the genes regulated by Bvg include important virulence factors such as filamentous hemagglutinin (FHA), fimbriae (Fim), and adenylate cyclase toxin (ACT) (Nicholson, 2007; Espinosa-Vinals et al., 2021). But Bvg similar to the Gac system in P. aeruginosa, regulates not only virulence factors but also sigma factors (Nicholson et al., 2009; Moon et al., 2017; Coutte et al., 2020) and sRNAs (Moon et al., 2021), which are also regulators, and the molecular mechanisms that interconnect them, remain not fully understood (Gestal et al., 2019b). The Bvg two component systems is not the only TCS in Bordetella spp. and our knowledge about Bordetella spp. sensory mechanisms is rapidly increasing. Up to date, the other TCS are the RisAS that is important for intracellular survival (Jungnitz et al., 1998; Zimna et al., 2001), and the PlrSR which is critical for colonization of the lower respiratory tract (Bone et al., 2017). Many TCSs in Bordetella spp. are not yet characterized, leaving the question of how they interact with each other and orchestrate the response signals unanswered. Understanding the intracellular signaling mechanisms that dictate the responses of Bordetella spp. is still in early stages (Gestal et al., 2019b), but we can use the knowledge gained on P. aeruginosa as a model to guide us in our investigations as the protein homology between the two component systems of both organisms is remarkable (Table 1).
Adapting to colonize
After bacteria sense inflammatory signals and alter gene expression to respond to external stimuli, the battle with the host immune response begins. Now is when the colonization and grow really start, spreading to new tissues and escaping immune clearance. P. aeruginosa and Bordetella spp. have regulatory systems for controlling the switch between acute and persistent infection. We will discuss three main aspects of the colonization process related with bacterial motility and adhesion, bacterial toxins, and finally we will focus on one of the most powerful bacterial immune-suppressive mechanisms, secretion systems.
Motility
Motility is a very important characteristic of bacterial pathogens. Flagella not only increase attachment that leads to biofilm formation, but flagella are also a very strong activator of the Toll-Receptor 5 (TLR5) (López-Boado et al., 2005) which will lead to a strong activation of the host immune response. Due to their great antigenicity surface molecules in bacteria must be tightly regulated to escape immune recognition. In Pseudomonas spp. an initial step in host interaction is mediated by polar pili that activate the host NF-κB signaling pathway promoting pro-inflammatory responses (Sadikot et al., 2005). Contact between the pili of P. aeruginosa induce the expression of virulence genes (Laventie et al., 2019). Overall, P. aeruginosa utilizes pili to manipulate host immune responses. Similarly, Bordetella spp. pili modulate host immunity and possess antigenic activity (Lee et al., 1986). In Bordetella spp. pili are mostly known as fimbriae (Melvin et al., 2014; Scheller and Cotter, 2015). They are major adhesins that promote biofilm formation and attachment of Bordetella spp. (Cotter et al., 1998; Irie et al., 2004; Mishra et al., 2005; Ulett et al., 2007; Lasaro et al., 2009; Wurpel et al., 2014; Scavone et al., 2016). Fimbriae-null mutants fail to persist in the mouse trachea due to impaired initial colonization (Mattoo et al., 2000). Overall, in both bacteria pili/fimbriae are critical for adhesion and initial colonization.
Another mechanism that facilitates motility is flagella. P. aeruginosa expresses polar flagella, which are necessary for motility and binding to TLR5, resulting in activation of interleukin-8 (IL8) (Sadikot et al., 2005). Flagella motility is common in Pseudomonas aeruginosa but interestingly clinical isolates present highly variable flagella motility and even in contact with sputum from cystic fibrosis patients, P. aeruginosa downregulates flagella expression (Lavoie et al., 2011), possibly to reduce the activation of inflammatory responses in the host. In P. aeruginosa, flagella is critical to trigger neutrophil traps activation (Floyd et al., 2016), a very powerful host pro-inflammatory response (Williams et al., 2020). Similar, in B. bronchiseptica, flagella initiate attachment (Nicholson et al., 2012; Cattelan et al., 2016) and play a critical role in modulating host immune responses (Akerley et al., 1995; Gestal et al., 2019a). Ectopic expression of flagella in Bordetella spp. promote rapid clearance from the respiratory tract (Akerley et al., 1995; Gestal et al., 2019a), possibly due to the activation of TLR5 (Gestal M. et al., 2019). Importantly, one of the hallmarks of Bordetella spp. infection is that during the virulence phase, Bordetella spp. downregulates flagella via Bvg TCS (Stibitz et al., 1988; Irie et al., 2004), as possible mechanism to evade immune response by avoiding TLR5 recognition.
Toxins: Suppressing host immune responses
After initial recognition escape, and while starting the colonization and grow processes, bacteria have to counteract immune defense utilizing several mechanisms, such as secreted toxins or direct delivery of toxins into host cells. Innate immunity plays an important role in both infections settings and neutrophils are in charge of clearance of early infection by both bacteria (Andreasen and Carbonetti, 2009; Rada, 2017). There are many different toxins that bacteria utilize to suppress host immune responses and neutrophil and other innate cells function and here we will focus on the most representative of each bacterium.
Pseudomonas aeruginosa synthesizes pigments that function as toxins to promote infection and colonization (Yabuuchi and Ohyama, 1972). Pigments such as pyocyanin inhibits phagocytosis by macrophages, induces apoptosis in neutrophils (Managò et al., 2015), and causes cellular damage, aiding in lung colonization and persistence, especially in cystic fibrosis patients (Lau et al., 2004). The antibacterial properties of pyocyanin have been thought to aid in eliminating competing bacteria, which allows P. aeruginosa to control many other pathogens and grow unimpeded in patients (Armstrong et al., 1971). In addition, pyocyanin modulates host airway epithelial cells by altering the expression of chemokines IL8 and RANTES and suppressing cilia beating (Denning et al., 1998; Look et al., 2005). Pyocyanin is not only related with neutrophil recruitment via IL8 and leukotriene B4 (Hall et al., 2016) but pyocyanin also suppresses the generation of efficient neutrophil responses by promoting the formation of neutrophils traps (Rada et al., 2013) and neutrophil death via aopotosis (Allen et al., 2005; Managò et al., 2015). A heat-labile hemolytic substance, which is suspected to be phospholipase C (PLC), is produced by P. aeruginosa (Sierra, 1960; Ostroff et al., 1989; McClure and Schiller, 1992). Amongst its functions, is to promote the activation of neutrophils (Jackon et al., 2000). The combination of hemolytic glycolipid and PLC has been hypothesized to produce extensive cytopathology in lung tissue amid P. aeruginosa pulmonary infection and contributes to P. aeruginosa colonization in the lungs (Holm et al., 1991). The extracellular polysaccharide Psl is expressed by non-mucoid P. aeruginosa strains and protects the pathogen by reducing neutrophil phagocytosis and limiting complement-mediated opsonization (Mishra et al., 2012). Another virulence factor identified to functionally inhibit host phagocytosis is the elastolytic metalloproteinase LasB, which cleaves host protease-activated receptors such as PAR2 (Dulon et al., 2005). The alkaline protease (AprA) of P. aeruginosa has been associated with bacterial virulence and is known to interfere with complement-mediated lysis of erythrocytes, while recent studies suggest it blocks complement activation via the classical and lectin pathways (Laarman et al., 2012).
Similar to Pseudomonas, Bordetella spp. also harbor toxins that block and suppress the generation of inflammatory responses. Bordetella harbor many mechanisms that can interfere with phagocytosis (Weingart and Weiss, 2000), but one of the most versatile toxins is the Bordetella spp. pertussis toxin (PT). PT is an AB5 toxin, that interferes with innate immunity by manipulating macrophages (Saukkonen et al., 1991; Carbonetti et al., 2003, 2007), neutrophils (Andreasen and Carbonetti, 2008, 2009), immune signals (Tonon et al., 2002; Wakatsuki et al., 2003; Andreasen et al., 2009), and T-cell responses (Chen et al., 2006; Tonon et al., 2006; Schneider et al., 2007). PT also impairs the generation of antibodies (Kirimanjeswara et al., 2005) and even affects metabolism such as glucose homeostasis by targeting G-protein coupled receptors (GPCRs) (Freyberg and Harvill, 2017). PT not only exerts its role on host immune responses, but it can modulate neuronal responses (Vega et al., 2020), which impacts not only disease pathogenesis (Murphy et al., 2020; Paramonov et al., 2020), but also future investigations and treatment (Zhou et al., 2020) of neurological disorders. Another powerful toxin of Bordetella spp. is the adenylate cyclase toxin (ACT), a pore forming toxin, which is one of the major immunomodulators of Bordetella spp. It is involved in manipulation of phagocytosis (Martín et al., 2015), suppression of antigen presenting cells (Fedele et al., 2017), blockade of neutrophil bactericidal activities (Cerny et al., 2017), modulation of macrophage differentiation (Cheung et al., 2008; Ahmad and Sebo, 2020), adhesion to epithelial cells (Angely et al., 2017), and T-cell differentiation (Paccani et al., 2008; Dunne et al., 2010; Svedova et al., 2016). Bordetella spp. also breaks down erythrocytes, one of the main mechanisms is via adenylate cyclase toxin (ACT) (Bemis and Plotkin, 1982; Vojtová et al., 2006). ACT also is responsible for the lysis of other immune cells including monocytes (Basler et al., 2006), macrophages (Hewlett et al., 2006), and even interferes with neutrophil trap formation (Eby et al., 2014).
Overall, P. aeruginosa and Bordetella spp. have similar strategies that allow them to overcome host responses by utilizing similar toxin-mediated mechanisms to block, suppress, and manipulate host immunity (Figure 1).
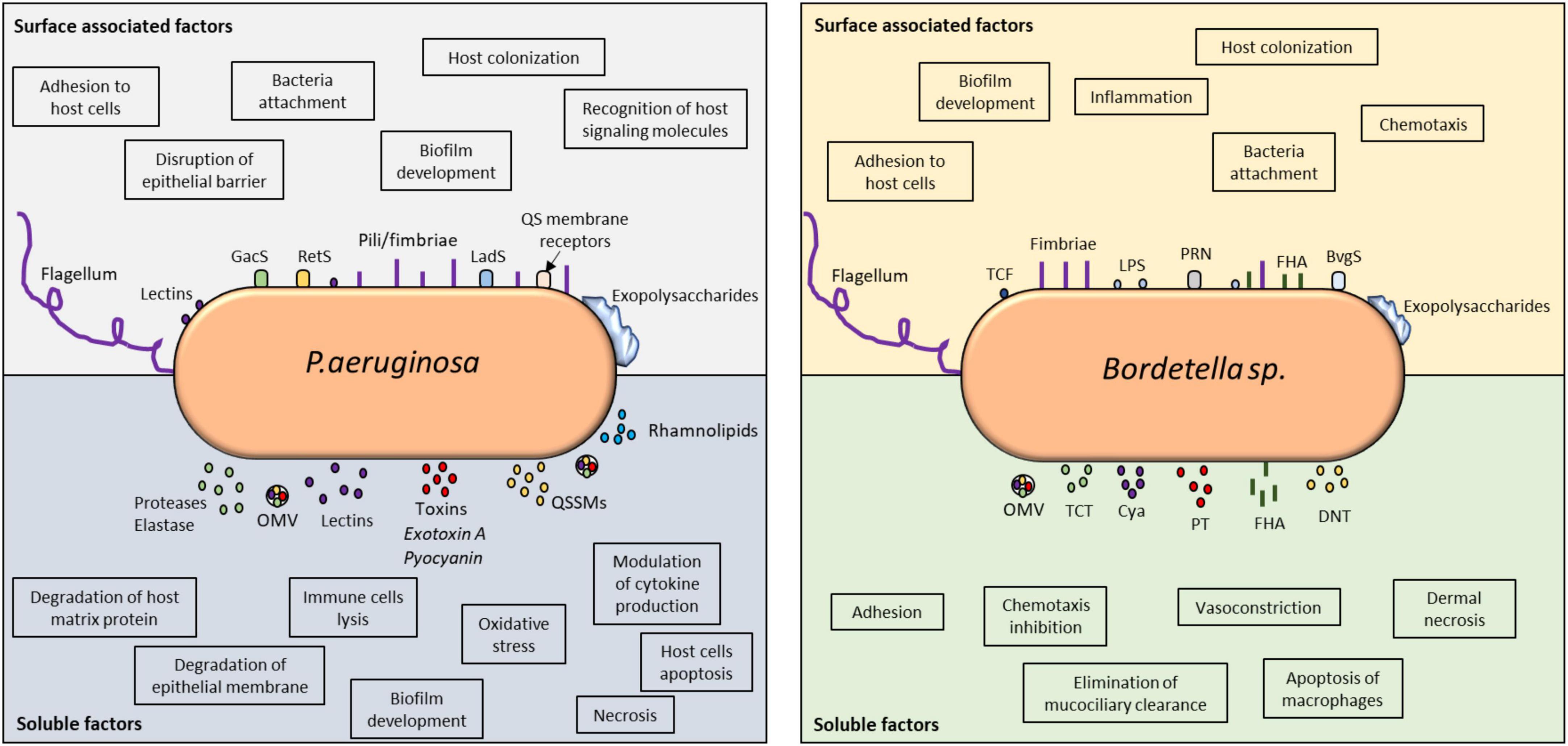
Figure 1. Virulence factors involved in immune detection and evasion by P. aeruginosa and Bordetella spp. Comparative analysis of the surface-associated and soluble virulence factors of these two microorganisms revealed the utilization of classical adhesins for cellular attachment, host colonization, and invasion. The impact of their virulence factors on host immune modulation can be found in the highlighted boxes. Both microorganisms utilize TCSs to detect host signals and modulate their behavior. In P. aeruginosa, QS receptors can also detect host molecules, such as hormones and cytokines. Their soluble factors, especially toxins and lysins, are vital in host invasion and immune evasion because they control inflammation and can kill immune cells. Biofilms produced by these microorganisms are highly controlled at the molecular level and responsible for persistent infection. QS, quorum sensing; OMV, outer membrane vesicle; QSSM, quorum sensing signaling molecule; PT, pertussis toxin; TCF, tracheal colonizing factor; Cya, adenylate cyclase; TCT, tracheal cytotoxin; DNT, dermonecrotic toxin; PRN, pertactin; FHA, filamentous hemagglutinin.
Secretion systems
Secretions systems are one of the most powerful mechanisms to manipulate host immune cells (Coburn et al., 2007). Six secretion systems have been described in P. aeruginosa but their roles in infection are still being investigated. The Type 3 secretion system (T3SS) is associated with acute infection, the type 4 secretion system (T4SS) with biofilm development are involved in chronic infection (Zhu et al., 2016a,b), and the T6SS is important for QS signaling, host-pathogen communication (Gallique et al., 2017), and competition with other microbiota (Weyrich et al., 2014).
T3SS is critical for several aspects of the infection process of Pseudomonas spp. and Bordetella spp., including adherence and invasion of host cells (Ridley, 2006), intracellular survival (Rivera et al., 2019), cytotoxicity against several immune cells (Hauser, 2009; Bayram et al., 2020; Kamanova, 2020), and manipulation of immune cell signaling (Hauser, 2009, 2011). Vance et al. demonstrated that the P. aeruginosa T3SS is vital for bacterial survival when the infection reaches the blood, further indicating that is required to combat immune cells that P. aeruginosa might encounter in the bloodstream (Vance et al., 2005; Furukawa et al., 2006). There is mounting evidence that demonstrate that the T3SS in P. aeruginosa, and related Pseudomonas spp. strains, is regulated via QS by an intricate network of bacterial signals that respond to host inflammatory cues (Pena et al., 2019; O’Malley and Anderson, 2021).
In B. bronchiseptica, the T3SS consists of approximately 30 proteins encoded by the bsc (Kamanova, 2020) locus and it regulation is tighten to the sigma factor brpL (Moon et al., 2017), also known as btrS. Expression of the T3SS in Bordetella spp. also respond to external cues, such as CO2 (Hester et al., 2012), iron (Brickman et al., 2011; Kurushima et al., 2012b), or blood (Gestal et al., 2018; Petráçková et al., 2020; Drzmisek et al., 2021); however, it is still not clear how the molecular network of regulators is connected. It was thought that the T3SS was regulated by the Bvg TCS (Nicholson, 2007; Moon et al., 2017); yet, recent evidence demonstrate that this is not correct. Instead the Bvg regulates btrS, which downstream controls the expression of the T3SS locus (Coutte et al., 2020). Nevertheless, these findings demonstrate that the regulation of the T3SS is more complex than only one regulator (Gutierrez et al., 2022), highlighting the importance of the meticulous regulated network of virulence factors. In Bordetella spp. the T3SS is critical for a successful colonization and persistence in the host as its role during immune suppression is critical. T3SS suppress the migration of antigen presenting cells (Skinner et al., 2005), induces macrophage and neutrophil apoptosis (Stockbauer et al., 2003), differentially activates inflammatory pathways (Yuk et al., 2000), and modifies other immune signals (Pilione and Harvill, 2006), all of which is mostly mediated by the secreted effector proteins, BteA and BopN (Bayram et al., 2020).
Secretion systems in many bacteria are one of the greatest bacterial immunomodulator, it is highly conserved amongst different bacterial species, and in fact, they are required for acute and/or chronic infection for many microbes. Albeit there is a lot of research demonstrating the critical roles of several secretion systems in bacterial interactions with host immune responses, there are still new roles and mechanisms that are still not fully understood. The in-depth understanding of the molecular mechanisms by which T3SS suppresses host immune responses might present a new avenue for therapeutic development.
Orchestration of the bacterial responses
Bacteria can very efficiently sense and respond to host cues some of which will provide a significant advantage during the infection settings. Bacterial ability to hijack host metabolism (Freyberg and Harvill, 2017) to scavenge nutrients (Murdoch and Skaar, 2022) is a very important feature of bacterial pathogens. Iron (Fe) is required by virtually all vertebrates but also bacterial pathogens, therefore, following invasion by pathogenic bacteria, the host limits bacterial access to Fe through a systemic reprogramming of Fe homeostasis by sequestering Fe in macrophages, hepatocytes, and enterocytes, while simultaneously reducing uptake of Fe from the diet. Bacterial pathogens can scavenge Fe through concerted action of secreted siderophores, uptake of host haem or Fe-containing molecules (transferrin and calprotectin) and uptake of ferrous Fe (Murdoch and Skaar, 2022). Pseudomonas sp. secrete haemolysins that integrate into erythrocyte membranes and result in osmotic lysis (Martins et al., 2016). Bacterial haem acquisition permits bacterial survival amidst the presence of host Fe-chelating molecules. In the presence of the Fe-sequestering protein calprotectin, haem availability is essential for the survival of P. aeruginosa, highlighting the importance of haem as an Fe source within the host. The host counteracts bacterial haem scavenging using haptoglobin, which binds haemoglobin and reduces accessibility of haem while promoting clearance by host cells (Murdoch and Skaar, 2022). In Bordetella spp. iron is also a critical nutrient and several studies have focused on how Bordetella sense the iron and how it scavenges it. Via ACT toxin and other hemolysins, Bordetella spp. breaks down erythrocytes to have access to the iron. The iron starvation response is mediated by Fur (Brickman and Armstrong, 1995, 2018), which responds to the presence of cathecolamines (Brickman et al., 2011) or blood and serum (Gestal et al., 2018). But iron stress even promotes binding to the respiratory epithelia (Vidakovics et al., 2007) indicating that is not simply a nutritional exchange but also it is involved in the virulence and attachment.
Zinc (Zn) is essential to host immune function, and even mild zinc insufficiency leads to widespread defects in both innate and adaptive immunity, resulting in impaired clearance of pathogens. P. aeruginosa have developed zinc uptake systems (ZnuABC, HmtA, and ZrmABCD) which are regulated by QS and specific molecules, such as the zinc uptake regulator (Zur) (Wang et al., 2021). Although Zn uptake systems are not yet clearly understood, to ensure a successful infection, P. aeruginosa must adapt to the environment of zinc deficiency, and this can provide new ideas and methods for the development of new drugs targeting P. aeruginosa zinc uptake systems and corresponding treatments for infection (Wang et al., 2021). In Bordetella spp. MerR also responds to nutrients such as Zn (Kidd and Brown, 2003). However, recent research focus on understanding how Bordetella spp. can also modulate virulence in response to other host metabolites (Gonyar et al., 2019) including copper (Rivera-Millot et al., 2021; Roy et al., 2022), manganese (Čapek et al., 2021) or glumatate (Keidel et al., 2018).
In response to nutrients, hormones, and other host cues, bacteria can meticulously regulate the expression of the aforementioned virulence factors which are critical for the successful colonization and infection of the host. Flagella, toxins, and secretions systems allow bacteria to colonize and conquer the host, but how do bacteria regulate gene expression once the cue has been sensed by the TCS? To orchestrate how bacteria sense and respond to environmental cues, a system needs to be in place allowing for bacterial communication, this mechanism is known as quorum sensing (QS). QS is very well studied in various model organisms including, Vibrio spp. or P. aeruginosa and contrary in other pathogens, such as Bordetella spp., the lack of understanding of bacterial communication is noticeable. Importantly, QS is not the only mechanism that bacteria utilize to regulate and finely tune gene expression and amongst the classical bacterial stress response other mechanisms of regulation are included such as sigma factors, small RNAs, alarmones, and chaperones which we will briefly discuss below.
Quorum sensing
Pseudomonas aeruginosa coordinate gene expression in a cell density-dependent manner via an intricate QS network with four intertwined QS systems: las, rhl, pqs, and iqs (Lee and Zhang, 2015; Schütz and Empting, 2018; Meng et al., 2020). Two acyl-homoserine-lactone circuits, LasI-LasR and RhlI-RhlR (Turkina and Vikström, 2019), that are required to activate many genes, including those encoding virulence factors (Kostylev et al., 2019). In P. aeruginosa, the role of QS during host interaction and inter-kingdom communication, has been widely studied and many papers and reviews in depth explore these aspects (Holban et al., 2014a; Turkina and Vikström, 2019). In fact, QS molecules themselves have immunomodulatory activity. N-(3-Oxododecanoyl)-l-Homoserine Lactone (3-Oxo-C12-HSL) reduces the production of proinflammatory cytokines such as IL-12 (Telford et al., 1998), inhibits T cell differentiation (Ritchie et al., 2005, 2007), and promote anti-inflammatory responses in several cells including macrophages (Coquant et al., 2020), indicating that QS molecules not only function in bacterial communication, but also in host-interactions.
Despite the obvious critical role of QS during the infectious process, in Bordetella spp. there is still no evidence of a similar mechanism (Irie et al., 2005). Nevertheless, Bordetella species exhibit key virulence behaviors associated with QS, such as biofilm development or preventing biofouling in membrane bioreactor systems suggesting that a QS-like mechanism in Bordetella spp. might be present (Ergön-Can et al., 2020).
Sigma factors
After the TCS activates the signaling cascade downstream, sigma factors are one of the regulators that will be induced in order to facilitate bacterial adaptability to the stressor. Sigma (σ) factors are critical during bacterial stress response. They are dissociable units of prokaryotic RNA polymerase that direct the holoenzyme to recognize conserved DNA motifs to modulate gene expression (Boor, 2006). During infection, σ factors regulate virulence factors, including secretion systems and secreted proteins, in P. aeruginosa (Otero-Asman et al., 2020) and Bordetella spp. (Ahuja et al., 2016), which are critical during host immune-suppression.
The σECF factor family are important signal-responsive regulatory proteins in P. aeruginosa. Most of the P. aeruginosa sigma factors belong to iron starvation (IS) responses which are essential during host colonization and dissemination of P. aeruginosa during infection (Damron et al., 2016). The second most abundant σECF group in P. aeruginosa is the RpoE-like σECF factors, which are activated in response to cell envelope stress and activate the expression of genes involve in stress mitigation and bacterial cell envelope integrity, ensuring pathogen survival (Potvin et al., 2008; Chevalier et al., 2019). σECF factors are critical for host immunomodulation and successful colonization (Skopelja-Gardner et al., 2019).
Similarly, in Bordetella spp. there are many different sigma factors that respond to stress and that play a role during colonization such as RpoE (Hanawa et al., 2013; Barbier et al., 2017). The Bordetella spp. σECF factor btrS/brpL has been the best studied (Mattoo et al., 2004). brpL/btrS has been known for its role coordinating expression of the type 3 secretion system (T3SS) in conjunction with an anti-sigma factor known as btrA (Ahuja et al., 2016; Nakamura et al., 2019). Moreover, brpL/btrS was shown to suppress host immune responses in a variety of ways to promote long-term infection and even re-infection (Gestal et al., 2019a, 2020, 2022). It is important to highlight protein sequence of RpoE shares up to 76% sequence homology between Pseudomonas spp. and Bordetella spp. Moreover, in P. aeruginosa sigma factors such as rpoE, are regulated by the GacAS system. Similarly, the brpL/btrS,of Bordetella spp., is regulated by the BvgAS two component systems aforementioned (Coutte et al., 2020).
Although there are commonalities on the regulation of sigma factors, the precise mechanism by which TCS modulate sima factors and the connections of sigma factors with other regulators are still not clear. But overall, sigma factors regulate many different genes including those critical for host-immune suppression.
Small RNAs
Another regulatory signaling pathway that bacteria utilize to respond to the external stimuli are small RNAs (sRNAs). sRNAs are non-coding small molecules of RNA that inhibit the expression of the targeted genes via post-translational or chromatin-depending silencing (Zhang, 2009). These play critical roles in the regulation of important phenotypes, including virulence and host immunomodulation (Han and Lory, 2021).
In P. aeruginosa, more than half of the characterized sRNAs are dependent on or mediated by the Hfq chaperone, which we will discuss later, and or two component systems such as GacAS (Pusic et al., 2021). Examples of sRNAs include, RsmY and RsmZ, both of which are involved in the switch from the motile to the sessile mode of life, modulate the expression of the T3SS and type 6 secretion system (T6SS) (Mikkelsen et al., 2011) and, thus, the transition from acute to persistent infection (Li et al., 2017). Another example is the PrrF which is critical to maintain iron homeostasis during murine lung infection (Reinhart et al., 2017) which is a battle that needs to happen prior to bacterial colonization and during infection.
In Bordetella spp. most of the sRNAs identified in B. pertussis in vitro are regulated by Bvg (Hot et al., 2011) which is the master virulence regulon. Differences in their expression are dictated by the phase growth, with some expressed exclusively during the exponential phase (Hot et al., 2011). One example of these sRNAs is RgtA, that regulates expression of proteins related to glutamate transport, which appears to be critical for the adaptation to nutritional stress and the change to persistent infection in the upper respiratory tract (Keidel et al., 2018). It has also been shown that in Bordetella spp. sRNAs play a role during colonization and preliminary data revealed new sRNAs in B. pertussis isolated from the murine trachea 4 days post-infection suggests a different response in vivo (Hiramatsu et al., 2020).
The complexity of sRNAs networks is only starting to being uncovered in Pseudomonas spp. (Zhang et al., 2017) but in Bordetella spp. this field is still at early stages, and although there are groups that are focused on investigating these exciting regulatory mechanisms, there are still multiple opportunities to unravel this fascinated interactions between sRNAs, TCS and virulence factors.
Alarmones
A less studied mechanism part of the bacteria stress response is the nucleotides guanosine tetraphosphate and pentaphosphate ((p)ppGpp). (p)ppGpp are molecular signals known as alarmones, that regulate gene transcription during stress responses such as signals of host immune resposnes (Pita et al., 2018). In other bacteria alarmones have an important role in allowing bacteria to overcome immune responses, such as tolerance to oxidative stress (Fritsch et al., 2020) and stress survival (Léger et al., 2021). In P. aeruginosa, alarmones modulate several aspects of tolerance to host responses (Khakimova et al., 2013), and pathogenesis and virulence, including QS, biofilms, and antibiotic tolerance (Khakimova et al., 2013; Diez et al., 2020) critical during acute infection (Xu et al., 2016). Similarly, in Bordetella spp. they are involved in the regulation of biofilm formation and maturation (Sugisaki et al., 2013; Cattelan et al., 2016), and up-regulation of the T3SS (Hanawa et al., 2016). The role of alarmones during virulence and pathogenesis is still not fully understood, but it is clear that they are an alternative or supplementary stress response crucial for the infectious process.
Chaperones
Chaperones assist in conformational folding or unfolding, and the assembly or disassembly of macromolecular structures (Henderson et al., 2006). There is increasing evidence indicating that chaperones have immunomodulatory functions. In P. aeruginosa, chaperones play a role in pathogenesis, such as biofilms (Vallet et al., 2001), the T3SS (Shen et al., 2008), and mediating antibiotic resistance (Klein et al., 2019). Similarly, in Bordetella spp., chaperones play critical roles in the regulation of multiple virulence factors, such as the T3SS (Kurushima et al., 2012a), or FHA (Baud et al., 2011).
One of the best studied chaperone in bacteria and that is common to P. aeruginosa (Sonnleitner et al., 2006) and Bordetella spp. (Bibova et al., 2013) is Hfq. Although, Hfq is not involved in protein folding, it regulates gene expression, including the expression of sRNA. Hfq has not only being studied in these two model organisms but also in other pathogens where similar results were found. The high degree of conservation of hfq, combined with the overlapping function in multiple pathogens, suggest that the role of this chaperone might be critical during pathogenesis. In fact, hfq might have been critical during evolution (Mura et al., 2013).
Biofilm development and chronic lung disease
Once colonization has occurred and undergo acute phase of disease, it is time to progress toward a chronic establishment of the infection. During the establishment of chronic infection, biofilms play important biological roles in bacterial pathogenesis, increasing survival under arduous conditions (Holban et al., 2014b), such as in the lungs of patients with cystic fibrosis (Vallet et al., 2004) and chronic obstructive pulmonary disease (COPD) (Garcia-Vidal et al., 2009; Versteegh, 2012; Hashemi et al., 2015; Blasi et al., 2020; Wilkinson et al., 2021), and leading to antibiotic resistance (Fux et al., 2005), providing a strong positive selective advantage for biofilm-forming bacteria. Thus, as compared to acute infections, which are characterized by an enhanced host immune response, chronic lung infections are characterized by a silenced immune response, combined with the formation of biofilms. Bacteria within the biofilm are protected from host immune effectors, antimicrobials, antibiotics. However they also have to adjust to an hypoxic, nutrient-deprived and acidic niche that further promotes immune depression (Arciola et al., 2018). Infectious biofilms determine abnormal activation of immune suppressive cells (i.e., regulatory T cells), while exhausting the antigen presenting cells (APCs), invaliding T cell priming, and waning antibody responses by inducing insufficient somatic hypermutations in B cells (Campbell et al., 2020; Heim et al., 2020).
In P. aeruginosa transition toward chronic lung infection consists of the following steps; (1) bacterial flagellar and pilus-dependent motility is reduced limiting the invasion capability, in the host recognition and phagocytosis are dampened; (2) in bacteria there is an increased expression of T3SS and T6SS which modulates interactions with host cells, but also PldA and PldB correlated to bacterial internalization in non-phagocytic cells while repressing the expression of regulatory molecules (RetS/GacS, cyclic-di-GMP); (3) bacteria have impaired production of secreted proteases (i.e., LasA, LasB, AprA, protease (4) leading to decreased host recognition and reduced host tissue destruction; (5) bacteria increase exopolysaccharides production, including alginate, Psl, Pel (Leid et al., 2005; Colvin et al., 2011; Sønderholm et al., 2017), and extracellular adhesins, such as CdrA, which promote bacterial aggregation and biofilm formation (Reichhardt et al., 2020); and finally (6) bacteria undergo lipopolysaccharide (LPS) modification, such as mutations in lipid A or loss of antigen O, leading to immune evasion (Faure et al., 2018).
In whooping cough Bordetella pertussis infections are not associated with a classical chronic infection, however Bordetella can develop biofilms and persist in the trachea and lungs. In Bordetella spp. biofilms, extracellular DNA (eDNA) plays a critical role (Holban et al., 2016), together with virulence factors associated with motility, such as flagella and fimbriae, or filamentous hemagglutinin (Serra et al., 2011), are pivotal to initial attachment and microcolony development (de Kievit, 2009; Holubova et al., 2022). In Bordetella spp., similarly, exopolysaccharides (Sloan et al., 2007) and adhesins are critical for initial attachment and aggregation, whereas eDNA promotes the formation and stability of three-dimensional biofilm structures (Cattelan et al., 2016). Although there is still debate about the clinical relevance of biofilms during Bordetella spp. infection, biofilms have been identified in patients with whooping cough, and clinical B. pertussis strains tend to produce more robust biofilms than lab-adapted strains (Arnal et al., 2015; Cattelan et al., 2017). This suggest that the reduced biofilm formation ability identified in lab strains can be a consequence of adaptation to the laboratory environment and an effect of passaging without a mammalian host.
It is clear that both bacteria produce biofilm however the molecular signaling mechanisms that trigger and regulate biofilm formation in response to external cues is not as well define and understood for Bordetella spp. as it is for P. aeruginosa (Figure 2). We propose that using the knowledge of biofilm gain from Pseudomonas spp. research can guide us on the investigations that can further clarify the role of Bordetella spp. biofilm in disease progression and clinical manifestation in patients with chronic lung disease or neonates, in whom biofilm can be fatal.
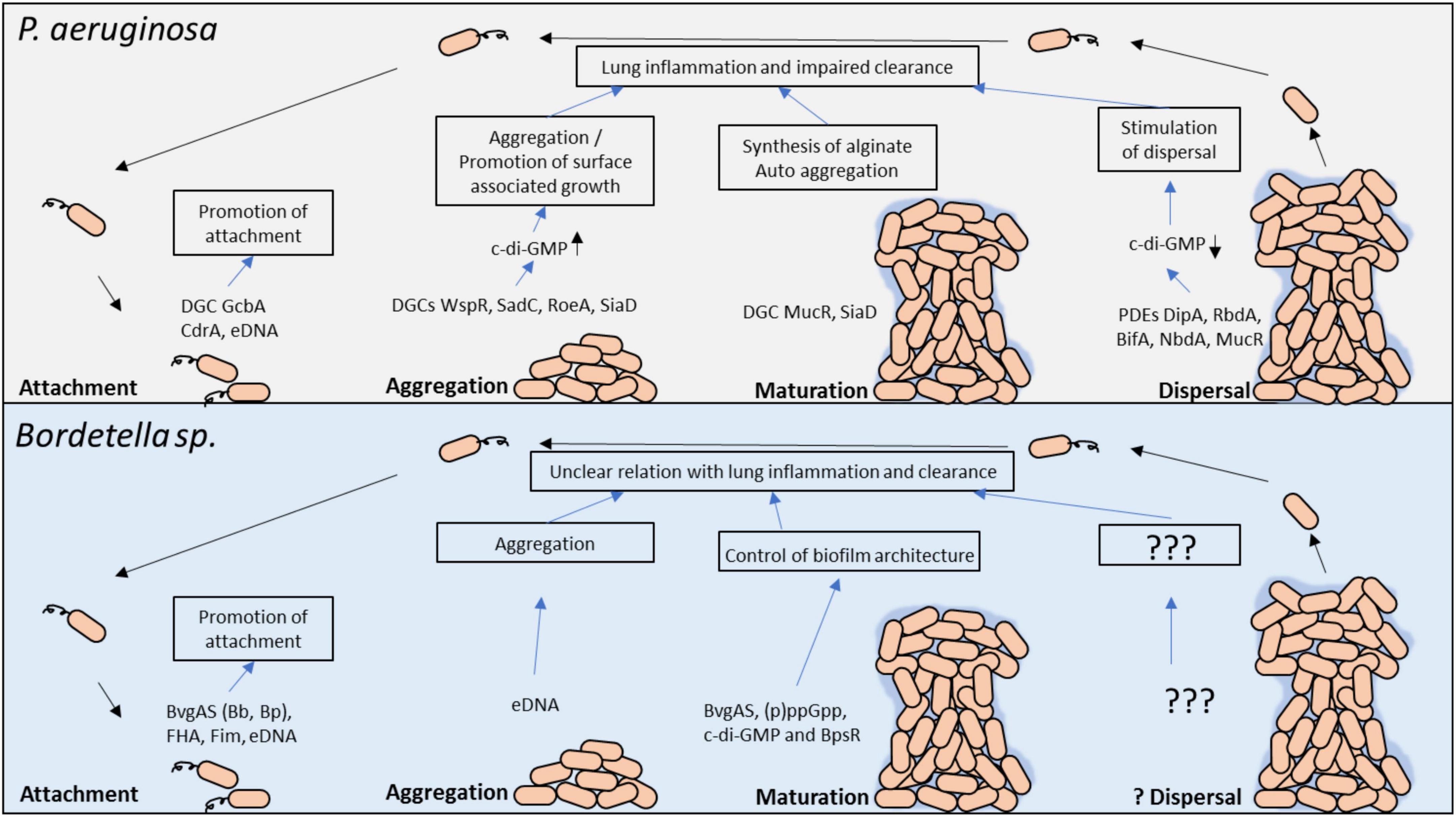
Figure 2. Molecular regulators in P. aeruginosa and Bordetella spp. biofilm development and their potential role in persistent lung infection. Each stage of biofilm development and the main molecular modulators are shown. The gray box above highlights the key factors involved in P. aeruginosa biofilm formation, whereas the light blue box below highlights the known factors in Bordetella spp. biofilm regulation. c-di-GMP, cyclic dimeric guanosine monophosphate; PDE, phosphodiesterase; DGC, diguanylate cyclase; eDNA, extracellular DNA.
Signaling applied to therapeutic development of lung co-infections
Nascent findings on the interactions between P. aeruginosa and B. bronchiseptica demonstrate that rhamnolipids from P. aeruginosa have the ability to successfully disrupt B. bronchiseptica biofilms, without antibacterial activity against planktonic B. bronchiseptica (Irie et al., 2005), suggesting a sensing/signaling mechanism between these two species. Interestingly, there is no molecular signatures that can relate to quorum sensing mechanisms in Bordetella spp. but the fact that biofilms are disrupted while planktonic bacteria are still viable, indicates that there must be a mechanism that allows Bordetella spp. to very tightly coordinate specific responses against these P. aeruginosa molecules (Irie et al., 2005). These findings are further supported by the previous findings that Bordetella spp. are able to respond to host cues such as CO2 (Hester et al., 2012), iron (Fe) (Brickman et al., 2011), blood, or serum (Gestal et al., 2018). Recently, immunomodulatory therapies are becoming cutting-edge research for not only for bacterial treatment but also as anti-biofilm strategies. Some therapies are decreasing lung pathology and reducing the detrimental effects caused by infection (Ernst et al., 2021; Gallop et al., 2021). Some research is focusing on enhancing the performance of innate immune cells such as neutrophils and macrophages. However, these therapies fail to demonstrate anti-biofilm effects, decreasing the enthusiasm. Next generation of infectious immunotherapy may be catalyzed by inducing targeting neutralizing antibodies and long-lasting memory responses against biofilms. However, there are several limitations on the design of adaptive immunity modulatory strategies focusing on biofilm eradication, and some of these limitations including the poorly immunogenic bacterial associated antigens in the biofilm fortress, which would result in deficient antigen presentation and suppressing T cell priming and activation.
Concluding remarks and future perspectives
Bacteria have a very finely tune network of regulators that become critical during the battle with the host. In order to successfully colonize and cause long term infection, bacteria need to very precisely regulate expression of various immunomodulatory/virulence factors that suppress host immune response in a variety of ways. Understanding the molecular mechanisms underlying this fine regulation, can provide novel avenues for vaccine (Gestal et al., 2019b,a; Gestal M. et al., 2019) and therapeutic (Gestal M. et al., 2019) development. However, our current models have limitations and until now most of our investigations have been done in vitro. Combining the molecular knowledge available in P. aeruginosa and animal model of Bordetella spp. we can investigate the molecular mechanisms by which pathogens suppress host immune responses. These interesting and novel areas would shed light on the pathogenesis of bacterial communities within the lungs and provide knowledge for the development of efficacious therapeutic targets.
Author contributions
AH and MG wrote the initial draft, edited and approved the manuscript for submission. CG wrote, edited and approved the manuscript for submission. All authors contributed to the article and approved the submitted version.
Funding
We would like to acknowledge the support of the funding bodies: NIH COBRE award P20GM134974-01A1 and PG20GM134974-02, Louisiana Board of Regents LEQSF(2022-25)-RD-A-33, Center of Excellence for Arthritis and Rheumatology Intramural Award, Intramural Research Council Seed Package Support from LSU Health, Shreveport, and Start-Up Package from LSU Health, Shreveport.
Acknowledgments
We would like to acknowledge Martin Sapp, Joanna Goldberg, Sheyda Azimi, Rajendar Deaora, and Audra Fullen for their ideas and support during the edition process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, J. N., and Sebo, P. (2020). Adenylate Cyclase Toxin Tinkering With Monocyte-Macrophage Differentiation. Front. Immunol. 11:2181. doi: 10.3389/fimmu.2020.02181
Ahuja, U., Shokeen, B., Cheng, N., Cho, Y., Blum, C., Coppola, G., et al. (2016). Differential regulation of type III secretion and virulence genes in Bordetella pertussis and Bordetella bronchiseptica by a secreted anti-σ factor. Proc. Natl. Acad. Sci. U.S.A. 113, 2341–2348. doi: 10.1073/pnas.1600320113
Akerley, B. J., Cotter, P. A., and Miller, J. F. (1995). Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80, 611–620. doi: 10.1016/0092-8674(95)90515-4
Allen, L., Dockrell, D. H., Pattery, T., Lee, D. G., Cornelis, P., Hellewell, P. G., et al. (2005). Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J. Immunol. 174, 3643–3649. doi: 10.4049/jimmunol.174.6.3643
Andreasen, C., and Carbonetti, N. H. (2008). Pertussis toxin inhibits early chemokine production to delay neutrophil recruitment in response to Bordetella pertussis respiratory tract infection in mice. Infect. Immun. 76, 5139–5148. doi: 10.1128/IAI.00895-08
Andreasen, C., and Carbonetti, N. H. (2009). Role of neutrophils in response to Bordetella pertussis infection in mice. Infect. Immun. 77, 1182–1188. doi: 10.1128/IAI.01150-08
Andreasen, C., Powell, D. A., and Carbonetti, N. H. (2009). Pertussis toxin stimulates IL-17 production in response to Bordetella pertussis infection in mice. PLoS One 4:e7079. doi: 10.1371/journal.pone.0007079
Angely, C., Nguyen, N. M., Andre Dias, S., Planus, E., Pelle, G., Louis, B., et al. (2017). Exposure to Bordetella pertussis adenylate cyclase toxin affects integrin-mediated adhesion and mechanics in alveolar epithelial cells. Biol. Cell 109, 293–311. doi: 10.1111/boc.201600082
Arciola, C. R., Campoccia, D., and Montanaro, L. (2018). Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409. doi: 10.1038/s41579-018-0019-y
Armstrong, A. V., Stewart-Tull, D. E., and Roberts, J. S. (1971). Characterisation of the Pseudomonas aeruginosa factor that inhibits mouse-liver mitochondrial respiration. J. Med. Microbiol. 4, 249–262. doi: 10.1099/00222615-4-2-249
Arnal, L., Grunert, T., Cattelan, N., de Gouw, D., Villalba, M. I., Serra, D. O., et al. (2015). Bordetella pertussis Isolates from Argentinean Whooping Cough Patients Display Enhanced Biofilm Formation Capacity Compared to Tohama I Reference Strain. Front. Microbiol. 6:1352. doi: 10.3389/fmicb.2015.01352
Azimi, S., Roberts, A. E. L., Peng, S., Weitz, J. S., McNally, A., Brown, S. P., et al. (2020). Allelic polymorphism shapes community function in evolving Pseudomonas aeruginosa populations. ISME J. 14, 1929–1942. doi: 10.1038/s41396-020-0652-0
Barbier, M., Boehm, D. T., Sen-Kilic, E., Bonnin, C., Pinheiro, T., Hoffman, C., et al. (2017). Modulation of Pertussis and Adenylate Cyclase Toxins by Sigma Factor RpoE in Bordetella pertussis. Infect. Immun. 85:e00565. doi: 10.1128/IAI.00565-16
Basler, M., Masin, J., Osicka, R., and Sebo, P. (2006). Pore-forming and enzymatic activities of Bordetella pertussis adenylate cyclase toxin synergize in promoting lysis of monocytes. Infect. Immun. 74, 2207–2214. doi: 10.1128/IAI.74.4.2207-2214.2006
Baud, C., Gutsche, I., Willery, E., de Paepe, D., Drobecq, H., Gilleron, M., et al. (2011). Membrane-associated DegP in Bordetella chaperones a repeat-rich secretory protein. Mol. Microbiol. 80, 1625–1636. doi: 10.1111/j.1365-2958.2011.07672.x
Bayram, J., Malcova, I., Sinkovec, L., Holubova, J., Streparola, G., Jurnecka, D., et al. (2020). Cytotoxicity of the effector protein BteA was attenuated in Bordetella pertussis by insertion of an alanine residue. PLoS Pathog. 16:e1008512. doi: 10.1371/journal.ppat.1008512
Beaudoin, T., Zhang, L., Hinz, A. J., Parr, C. J., and Mah, T. F. (2012). The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J. Bacteriol. 194, 3128–3136. doi: 10.1128/JB.06178-11
Bemis, D. A., and Plotkin, B. J. (1982). Hemagglutination by Bordetella bronchiseptica. J. Clin. Microbiol. 15, 1120–1127. doi: 10.1128/jcm.15.6.1120-1127.1982
Benkert, B., Quäck, N., Schreiber, K., Jaensch, L., Jahn, D., and Schobert, M. (2008). Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 154, 3053–3060. doi: 10.1099/mic.0.2008/018929-0
Bhagirath, A. Y., Pydi, S. P., Li, Y., Lin, C., Kong, W., Chelikani, P., et al. (2017). Characterization of the Direct Interaction between Hybrid Sensor Kinases PA1611 and RetS That Controls Biofilm Formation and the Type III Secretion System in Pseudomonas aeruginosa. ACS Infect. Dis. 3, 162–175. doi: 10.1021/acsinfecdis.6b00153
Bhuwan, M., Lee, H. J., Peng, H. L., and Chang, H. Y. (2012). Histidine-containing phosphotransfer protein-B (HptB) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J. Biol. Chem. 287, 1903–1914. doi: 10.1074/jbc.M111.256586
Bibova, I., Skopova, K., Masin, J., Cerny, O., Hot, D., Sebo, P., et al. (2013). The RNA chaperone Hfq is required for virulence of Bordetella pertussis. Infect. Immun. 81, 4081–4090. doi: 10.1128/IAI.00345-13
Biederman, L., Rosen, M. R., Bobik, B. S., and Roberts, A. L. (2015). Bordetella petrii recovered from chronic pansinusitis in an adult with cystic fibrosis. IDCases 2, 97–98. doi: 10.1016/j.idcr.2015.09.004
Bielecki, P., Jensen, V., Schulze, W., Gödeke, J., Strehmel, J., Eckweiler, D., et al. (2015). Cross talk between the response regulators PhoB and TctD allows for the integration of diverse environmental signals in Pseudomonas aeruginosa. Nucleic Acids Res. 43, 6413–6425. doi: 10.1093/nar/gkv599
Blackwood, C. B., Sen-Kilic, E., Boehm, D. T., Hall, J. M., Varney, M. E., Wong, T. Y., et al. (2020). Innate and Adaptive Immune Responses against Bordetella pertussis and Pseudomonas aeruginosa in a Murine Model of Mucosal Vaccination against Respiratory Infection. Vaccines 8:647. doi: 10.3390/vaccines8040647
Blasi, F., Bonanni, P., Braido, F., Gabutti, G., Marchetti, F., and Centanni, S. (2020). The unmet need for pertussis prevention in patients with chronic obstructive pulmonary disease in the Italian context. Hum. Vaccin. Immunother. 16, 340–348. doi: 10.1080/21645515.2019.1652517
Bone, M. A., Wilk, A. J., Perault, A. I., Marlatt, S. A., Scheller, E. V., Anthouard, R., et al. (2017). Bordetella PlrSR regulatory system controls BvgAS activity and virulence in the lower respiratory tract. Proc. Natl. Acad. Sci. U.S.A. 114:E1519–E1527. doi: 10.1073/pnas.1609565114
Boor, K. J. (2006). Bacterial stress responses: What doesn’t kill them can make then stronger. PLoS Biol. 4:e23. doi: 10.1371/journal.pbio.0040023
Bos, A. C., Beemsterboer, P., Wolfs, T. F., Versteegh, F. G., and Arets, H. G. (2011). Bordetella species in children with cystic fibrosis: What do we know? The role in acute exacerbations and chronic course. J. Cyst. Fibros. 10, 307–312. doi: 10.1016/j.jcf.2011.06.003
Brady, C., Ackerman, P., Johnson, M., and McNamara, J. (2014). Bordetella bronchiseptica in a pediatric Cystic Fibrosis center. J. Cyst. Fibros. 13, 43–48. doi: 10.1016/j.jcf.2013.08.002
Brickman, T. J., and Armstrong, S. K. (1995). Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J. Bacteriol. 177, 268–270. doi: 10.1128/jb.177.1.268-270.1995
Brickman, T. J., and Armstrong, S. K. (2018). The Bordetella bronchiseptica nic locus encodes a nicotinic acid degradation pathway and the 6-hydroxynicotinate-responsive regulator BpsR. Mol. Microbiol. 108, 397–409. doi: 10.1111/mmi.13943
Brickman, T. J., Cummings, C. A., Liew, S. Y., Relman, D. A., and Armstrong, S. K. (2011). Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J. Bacteriol. 193, 4798–4812. doi: 10.1128/JB.05136-11
Caille, O., Rossier, C., and Perron, K. (2007). A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568. doi: 10.1128/JB.00095-07
Campbell, C., McKenney, P. T., Konstantinovsky, D., Isaeva, O. I., Schizas, M., and Verter, J. (2020). Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479. doi: 10.1038/s41586-020-2193-0
Čapek, J., Procházková, I., Matoušek, T., Hot, D., and Veèerek, B. (2021). A Unique Reverse Adaptation Mechanism Assists Bordetella pertussis in Resistance to Both Scarcity and Toxicity of Manganese. mBio 12:e0190221. doi: 10.1128/mBio.01902-21
Carbonetti, N. H., Artamonova, G. V., Mays, R. M., and Worthington, Z. E. (2003). Pertussis toxin plays an early role in respiratory tract colonization by Bordetella pertussis. Infect. Immun. 71, 6358–6366. doi: 10.1128/IAI.71.11.6358-6366.2003
Carbonetti, N. H., Artamonova, G. V., Van Rooijen, N., and Ayala, V. I. (2007). Pertussis toxin targets airway macrophages to promote Bordetella pertussis infection of the respiratory tract. Infect. Immun. 75, 1713–1720. doi: 10.1128/IAI.01578-06
Cattelan, N., Dubey, P., Arnal, L., Yantorno, O. M., and Deora, R. (2016). Bordetella biofilms: A lifestyle leading to persistent infections. Pathog. Dis. 74:ftv108. doi: 10.1093/femspd/ftv108
Cattelan, N., Jennings-Gee, J., Dubey, P., Yantorno, O. M., and Deora, R. (2017). Hyperbiofilm Formation by Bordetella pertussis Strains Correlates with Enhanced Virulence Traits. Infect. Immun. 85:e373–e317. doi: 10.1128/IAI.00373-17
Cerny, O., Anderson, K. E., Stephens, L. R., Hawkins, P. T., and Sebo, P. (2017). cAMP Signaling of Adenylate Cyclase Toxin Blocks the Oxidative Burst of Neutrophils through Epac-Mediated Inhibition of Phospholipase C Activity. J. Immunol. 198, 1285–1296. doi: 10.4049/jimmunol.1601309
Chambonnier, G., Roux, L., Redelberger, D., Fadel, F., Filloux, A., Sivaneson, M., et al. (2016). The Hybrid Histidine Kinase LadS Forms a Multicomponent Signal Transduction System with the GacS/GacA Two-Component System in Pseudomonas aeruginosa. PLoS Genet. 12:e1006032. doi: 10.1371/journal.pgen.1006032
Chand, N. S., Clatworthy, A. E., and Hung, D. T. (2012). The two-component sensor KinB acts as a phosphatase to regulate Pseudomonas aeruginosa Virulence. J. Bacteriol. 194, 6537–6547. doi: 10.1128/JB.01168-12
Chand, N. S., Lee, J. S., Clatworthy, A. E., Golas, A. J., Smith, R. S., and Hung, D. T. (2011). The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J. Bacteriol. 193, 2989–2999. doi: 10.1128/JB.01546-10
Chen, X., Winkler-Pickett, R. T., Carbonetti, N. H., Ortaldo, J. R., Oppenheim, J. J., and Howard, O. M. (2006). Pertussis toxin as an adjuvant suppresses the number and function of CD4+CD25+ T regulatory cells. Eur. J. Immunol. 36, 671–680. doi: 10.1002/eji.200535353
Cheung, G. Y., Dickinson, P., Sing, G., Craigon, M., Ghazal, P., Parton, R., et al. (2008). Transcriptional responses of murine macrophages to the adenylate cyclase toxin of Bordetella pertussis. Microb. Pathog. 44, 61–70. doi: 10.1016/j.micpath.2007.08.007
Chevalier, S., Bouffartigues, E., Bazire, A., Tahrioui, A., Duchesne, R., Tortuel, D., et al. (2019). Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. Biochim. Biophys. Acta. Gene. Regul. Mech. 1862, 706–721. doi: 10.1016/j.bbagrm.2018.04.008
Coburn, B., Sekirov, I., and Finlay, B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549. doi: 10.1128/CMR.00013-07
Coenye, T., Goris, J., Spilker, T., Vandamme, P., and LiPuma, J. J. (2002). Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40, 2062–2069. doi: 10.1128/JCM.40.6.2062-2069.2002
Colvin, K. M., Gordon, V. D., Murakami, K., Borlee, B. R., Wozniak, D. J., Wong, G. C., et al. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. doi: 10.1371/journal.ppat.1001264
Coquant, G., Grill, J. P., and Seksik, P. (2020). Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 11:1827. doi: 10.3389/fimmu.2020.01827
Cotter, P. A., Yuk, M. H., Mattoo, S., Akerley, B. J., Boschwitz, J., Relman, D. A., et al. (1998). Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66, 5921–5929. doi: 10.1128/IAI.66.12.5921-5929.1998
Coutte, L., Antoine, R., Slupek, S., Solans, L., Derop, J., Bonnefond, A., et al. (2020). Combined RNAseq and ChIPseq Analyses of the BvgA Virulence Regulator of Bordetella pertussis. mSystems 5:e208–e220. doi: 10.1128/mSystems.00208-20
Damron, F. H., Oglesby-Sherrouse, A. G., Wilks, A., and Barbier, M. (2016). Dual-seq transcriptomics reveals the battle for iron during Pseudomonas aeruginosa acute murine pneumonia. Sci. Rep. 6:39172. doi: 10.1038/srep39172
Darch, S. E., Ibberson, C. B., and Whiteley, M. (2017). Evolution of Bacterial “Frenemies”. mBio 8:e675–e617. doi: 10.1128/mBio.00675-17
Darrah, R., Bonfield, T., LiPuma, J. J., Litman, P., Hodges, C. A., Jacono, F., et al. (2017). Cystic Fibrosis Mice Develop Spontaneous Chronic Bordetella Airway Infections. J. Infect. Pulm. Dis. 3:10.16966/2470–3176.128. doi: 10.16966/2470-3176.128
de Bentzmann, S., Giraud, C., Bernard, C. S., Calderon, V., Ewald, F., Plésiat, P., et al. (2012). Unique biofilm signature, drug susceptibility and decreased virulence in Drosophila through the Pseudomonas aeruginosa two-component system PprAB. PLoS Pathog. 8:e1003052. doi: 10.1371/journal.ppat.1003052
de Kievit, T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
De Schutter, I., Malfroot, A., Dab, I., Hoebrekx, N., Muyldermans, G., Piérard, D., et al. (2003). Molecular typing of Bordetella pertussis isolates recovered from Belgian children and their household members. Clin. Infect. Dis. 36, 1391–1396. doi: 10.1086/375071
Dean, C. R., Neshat, S., and Poole, K. (1996). PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178, 5361–5369. doi: 10.1128/jb.178.18.5361-5369.1996
Demko, C. A., Byard, P. J., and Davis, P. B. (1995). Gender differences in cystic fibrosis: Pseudomonas aeruginosa infection. J. Clin. Epidemiol. 48, 1041–1049. doi: 10.1016/0895-4356(94)00230-N
Denning, G. M., Wollenweber, L. A., Railsback, M. A., Cox, C. D., Stoll, L. L., and Britigan, B. E. (1998). Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect. Immun. 66, 5777–5784. doi: 10.1128/IAI.66.12.5777-5784.1998
Dieppois, G., Ducret, V., Caille, O., and Perron, K. (2012). The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One 7:e38148. doi: 10.1371/journal.pone.0038148
Diez, S., Ryu, J., Caban, K., Gonzalez, R. L., and Dworkin, J. (2020). The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence. Proc. Natl. Acad. Sci. U.S.A. 117, 15565–15572. doi: 10.1073/pnas.1920013117
Drzmisek, J., Stipl, D., Petrackova, D., Vecerek, B., and Dienstbier, A. (2021). Omics Analysis of Blood-Responsive Regulon inBordetella pertussis Identifies a Novel Essential T3SS Substrate. Int. J. Mol. Sci. 22:736. doi: 10.3390/ijms22020736
Dulon, S., Leduc, D., Cottrell, G. S., D’Alayer, J., Hansen, K. K., Bunnett, N. W., et al. (2005). Pseudomonas aeruginosa elastase disables proteinase-activated receptor 2 in respiratory epithelial cells. Am. J. Respir. Cell Mol. Biol. 32, 411–419. doi: 10.1165/rcmb.2004-0274OC
Dunne, A., Ross, P. J., Pospisilova, E., Masin, J., Meaney, A., Sutton, C. E., et al. (2010). Inflammasome activation by adenylate cyclase toxin directs Th17 responses and protection against Bordetella pertussis. J. Immunol. 185, 1711–1719. doi: 10.4049/jimmunol.1000105
Eby, J. C., Gray, M. C., and Hewlett, E. L. (2014). Cyclic AMP-mediated suppression of neutrophil extracellular trap formation and apoptosis by the Bordetella pertussis adenylate cyclase toxin. Infect. Immun. 82, 5256–5269. doi: 10.1128/IAI.02487-14
El Khatib, N., Ferroni, A., Le Bourgeois, M., Chedevergne, F., Clairicia, M., Avril, H., et al. (2015). Persistent Bordetella bronchiseptica infection in a child with cystic fibrosis: Relationship to bacterial phenotype. J. Cyst. Fibros. 14:E13–E15. doi: 10.1016/j.jcf.2015.03.014
Ergön-Can, T., Köse-Mutlu, B., Koyuncu, I., and Lee, C.-H. (2020). The Use of the New Quorum Quenching Isolate of Bordetella hinzii S3 to Prevent Biofouling in Membrane Bioreactor Systems. Waste Biomass Valor 11, 3513–3523. doi: 10.1007/s12649-019-00700-2
Ernst, K., Mittler, A. K., Winkelmann, V., Kling, C., Eberhardt, N., Anastasia, A., et al. (2021). Pharmacological targeting of host chaperones protects from pertussis toxin in vitro and in vivo. Sci. Rep. 11:5429. doi: 10.1038/s41598-021-84817-2
Espinosa-Vinals, C. A., Masin, J., Holubova, J., Stanek, O., Jurnecka, D., Osicka, R., et al. (2021). Almost half of the RTX domain is dispensable for complement receptor 3 binding and cell-invasive activity of the Bordetella adenylate cyclase toxin. J. Biol. Chem. 297:100833. doi: 10.1016/j.jbc.2021.100833
Faure, E., Kwong, K., and Nguyen, D. (2018). Pseudomonas aeruginosa in Chronic Lung Infections: How to Adapt Within the Host? Front. Immunol. 9:2416. doi: 10.3389/fimmu.2018.02416
Fedele, G., Schiavoni, I., Adkins, I., Klimova, N., and Sebo, P. (2017). Invasion of Dendritic Cells, Macrophages and Neutrophils by the Bordetella Adenylate Cyclase Toxin: A Subversive Move to Fool Host Immunity. Toxins 9:293. doi: 10.3390/toxins9100293
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa. mBio 7:e1513–e1516. doi: 10.1128/mBio.01513-16
Floyd, M., Winn, M., Cullen, C., Sil, P., Chassaing, B., Yoo, D. G., et al. (2016). Swimming Motility Mediates the Formation of Neutrophil Extracellular Traps Induced by Flagellated Pseudomonas aeruginosa. PLoS Pathog. 12:e1005987. doi: 10.1371/journal.ppat.1005987
Francis, V. I., Stevenson, E. C., and Porter, S. L. (2017). Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 364:fnx104. doi: 10.1093/femsle/fnx104
Freyberg, Z., and Harvill, E. T. (2017). Pathogen manipulation of host metabolism: A common strategy for immune evasion. PLoS Pathog. 13:e1006669. doi: 10.1371/journal.ppat.1006669
Fritsch, V. N., Loi, V. V., Busche, T., Tung, Q. N., Lill, R., Horvatek, P., et al. (2020). The alarmone (p)ppGpp confers tolerance to oxidative stress during the stationary phase by maintenance of redox and iron homeostasis in Staphylococcus aureus. Free Radic. Biol. Med. 161, 351–364. doi: 10.1016/j.freeradbiomed.2020.10.322
Funke, G., Hess, T., von Graevenitz, A., and Vandamme, P. (1996). Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J. Clin. Microbiol. 34, 966–969. doi: 10.1128/jcm.34.4.966-969.1996
Furukawa, S., Kuchma, S. L., and O’Toole, G. A. (2006). Keeping their options open: Acute versus persistent infections. J. Bacteriol. 188, 1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006
Fux, C. A., Costerton, J. W., Stewart, P. S., and Stoodley, P. (2005). Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40. doi: 10.1016/j.tim.2004.11.010
Gallique, M., Bouteiller, M., and Merieau, A. (2017). The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 8:1454. doi: 10.3389/fmicb.2017.01454
Gallop, D., Scanlon, K. M., Ardanuy, J., Sigalov, A. B., Carbonetti, N. H., and Skerry, C. (2021). Triggering Receptor Expressed on Myeloid Cells-1 (TREM-1) Contributes to Bordetella pertussis Inflammatory Pathology. Infect. Immun. 89:e0012621. doi: 10.1128/IAI.00126-21
Garcia-Vidal, C., Almagro, P., Romaní, V., Rodríguez-Carballeira, M., Cuchi, E., Canales, L., et al. (2009). Pseudomonas aeruginosa in patients hospitalised for COPD exacerbation: A prospective study. Eur. Respir. J. 34, 1072–1078. doi: 10.1183/09031936.00003309
Gerlach, G., von Wintzingerode, F., Middendorf, B., and Gross, R. (2001). Evolutionary trends in the genus Bordetella. Microbes Infect. 3, 61–72. doi: 10.1016/S1286-4579(00)01353-8
Gestal, M., Jonhson, H., and Harvill, E. (2019). Immunomodulation as a Novel Strategy for Prevention and Treatment of Bordetella spp. Infections. Front. Immunol. 10:2869. doi: 10.3389/fimmu.2019.02869
Gestal, M. C., Howard, L. K., Dewan, K. K., and Harvill, E. T. (2022). Bbvac: A Live Vaccine Candidate That Provides Long-Lasting Anamnestic and Th17-Mediated Immunity against the Three Classical. mSphere 7:e0089221. doi: 10.1128/msphere.00892-21
Gestal, M. C., Blas-Machado, U., Johnson, H. M., Rubin, L. N., Dewan, K. K., Bryant, C., et al. (2020). Disrupting Bordetella Immunosuppression Reveals a Role for Eosinophils in Coordinating the Adaptive Immune Response in the Respiratory Tract. Microorganisms 8:1808. doi: 10.3390/microorganisms8111808
Gestal, M. C., Howard, L. K., Dewan, K., Johnson, H. M., Barbier, M., Bryant, C., et al. (2019a). Enhancement of immune response against Bordetella spp. by disrupting immunomodulation. Sci. Rep. 9:20261. doi: 10.1038/s41598-019-56652-z
Gestal, M. C., Rivera, I., Howard, L. K., Dewan, K. K., Soumana, I. H., Dedloff, M., et al. (2018). Blood or Serum Exposure Induce Global Transcriptional Changes, Altered Antigenic Profile, and Increased Cytotoxicity by Classical Bordetellae. Front. Microbiol. 9:1969. doi: 10.3389/fmicb.2018.01969
Gestal, M. C., Whitesides, L. T., and Harvill, E. T. (2019b). Integrated Signaling Pathways Mediate Bordetella Immunomodulation, Persistence, and Transmission. Trends Microbiol. 27, 118–130. doi: 10.1016/j.tim.2018.09.010
Gonyar, L. A., Gelbach, P. E., McDuffie, D. G., Koeppel, A. F., Chen, Q., Lee, G., et al. (2019). In vivo Gene Essentiality and Metabolism in Bordetella pertussis. mSphere 4:e00694. doi: 10.1128/mSphere.00694-18
Gross, R., Guzman, C. A., Sebaihia, M., dos Santos, V. A., Pieper, D. H., and Koebnik, R. (2008). The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Geno. 9:449. doi: 10.1186/1471-2164-9-449
Guragain, M., King, M. M., Williamson, K. S., Pérez-Osorio, A. C., Akiyama, T., Khanam, S., et al. (2016). The Pseudomonas aeruginosa PAO1 Two-Component Regulator CarSR Regulates Calcium Homeostasis and Calcium-Induced Virulence Factor Production through Its Regulatory Targets CarO and CarP. J. Bacteriol. 198, 951–963. doi: 10.1128/JB.00963-15
Gutierrez, M. P., Wong, T. Y., Damron, F. H., Fernández, J., and Sisti, F. (2022). Cyclic di-GMP Regulates the Type III Secretion System and Virulence in Bordetella bronchiseptica. Infect. Immun. 90:e0010722. doi: 10.1128/iai.00107-22
Hall, S., McDermott, C., Anoopkumar-Dukie, S., McFarland, A. J., Forbes, A., Perkins, A. V., et al. (2016). Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 8:236. doi: 10.3390/toxins8080236
Han, K., and Lory, S. (2021). Toward a Comprehensive Analysis of Posttranscriptional Regulatory Networks: A New Tool for the Identification of Small RNA Regulators of Specific mRNAs. mBio 12:e3608–e3620. doi: 10.1128/mBio.03608-20
Hanawa, T., Kamachi, K., Yonezawa, H., Fukutomi, T., Kawakami, H., and Kamiya, S. (2016). Glutamate Limitation, BvgAS Activation, and (p)ppGpp Regulate the Expression of the Bordetella pertussis Type 3 Secretion System. J. Bacteriol. 198, 343–351. doi: 10.1128/JB.00596-15
Hanawa, T., Yonezawa, H., Kawakami, H., Kamiya, S., and Armstrong, S. K. (2013). Role of Bordetella pertussis RseA in the cell envelope stress response and adenylate cyclase toxin release. Pathog. Dis. 69, 7–20. doi: 10.1111/2049-632X.12061
Hashemi, S. H., Nadi, E., Hajilooi, M., Seif-Rabiei, M. A., and Samaei, A. (2015). High Seroprevalence of Bordetella pertussis in Patients with Chronic Obstructive Pulmonary Disease: A Case-Control Study. Tanaffos 14, 172–176.
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 7, 654–665. doi: 10.1038/nrmicro2199
Hauser, A. R. (2011). Pseudomonas aeruginosa: So many virulence factors, so little time. Crit. Care Med. 39, 2193–2194. doi: 10.1097/CCM.0b013e318221742d
Heim, C. E., Bosch, M. E., Yamada, K. J., Aldrich, A. L., Chaudhari, S. S., Klinkebiel, D., et al. (2020). Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 5, 1271–1284. doi: 10.1038/s41564-020-0756-3
Henderson, B., Allan, E., and Coates, A. R. (2006). Stress wars: The direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 74, 3693–3706. doi: 10.1128/IAI.01882-05
Hester, S. E., Lui, M., Nicholson, T., Nowacki, D., and Harvill, E. T. (2012). Identification of a CO2 responsive regulon in Bordetella. PLoS One 7:e47635. doi: 10.1371/journal.pone.0047635
Hewlett, E. L., Donato, G. M., and Gray, M. C. (2006). Macrophage cytotoxicity produced by adenylate cyclase toxin from Bordetella pertussis: More than just making cyclic AMP! Mol. Microbiol. 59, 447–459. doi: 10.1111/j.1365-2958.2005.04958.x
Hiramatsu, Y., Suzuki, K., Motooka, D., Nakamura, S., and Horiguchi, Y. (2020). Expression of small RNAs of Bordetella pertussis colonizing murine tracheas. Microbiol. Immunol. 64, 469–475. doi: 10.1111/1348-0421.12791
Holban, A. M., Bleotu, C., Chifiriuc, M. C., Bezirtzoglou, E., and Lazar, V. (2014a). Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence 5, 303–310. doi: 10.4161/viru.27571
Holban, A. M., Gestal, M. C., and Grumezescu, A. M. (2014b). New molecular strategies for reducing implantable medical devices associated infections. Curr. Med. Chem. 21, 3375–3382. doi: 10.2174/0929867321666140304103810
Holban, A. M., Gestal, M. C., and Grumezescu, A. M. (2016). Control of biofilm-associated infections by signaling molecules and nanoparticles. Int. J. Pharm. 510, 409–418. doi: 10.1016/j.ijpharm.2016.02.044
Holm, B. A., Keicher, L., Liu, M. Y., Sokolowski, J., and Enhorning, G. (1991). Inhibition of pulmonary surfactant function by phospholipases. J. Appl. Physiol. 71, 317–321. doi: 10.1152/jappl.1991.71.1.317
Holubova, J., Stanek, O., Juhasz, A., Hamidou Soumana, I., Makovicky, P., and Sebo, P. (2022). The Fim and FhaB adhesins play a crucial role in nasal cavity infection and Bordetella pertussis transmission in a novel mouse catarrhal infection model. PLoS Pathog. 18:e1010402. doi: 10.1371/journal.ppat.1010402
Homola, L., Holčíková, A., Zárošská, E., Krbková, L., Gaillyová, R., Hanslianová, M., et al. (2012). [Pertussis in a 4-month-old infant with unrecognized cystic fibrosis]. Klin Mikrobiol Infekc Lek 18, 196–197.
Hot, D., Slupek, S., Wulbrecht, B., D’Hondt, A., Hubans, C., Antoine, R., et al. (2011). Detection of small RNAs in Bordetella pertussis and identification of a novel repeated genetic element. BMC Geno. 12:207. doi: 10.1186/1471-2164-12-207
Huangyutitham, V., Güvener, Z. T., and Harwood, C. S. (2013). Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4:e242–e213. doi: 10.1128/mBio.00242-13
Hurley, B. P., Goodman, A. L., Mumy, K. L., Murphy, P., Lory, S., and McCormick, B. A. (2010). The two-component sensor response regulator RoxS/RoxR plays a role in Pseudomonas aeruginosa interactions with airway epithelial cells. Microbes Infect 12, 190–198. doi: 10.1016/j.micinf.2009.11.009
Inclan, Y. F., Persat, A., Greninger, A., Von Dollen, J., Johnson, J., Krogan, N., et al. (2016). A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol. Microbiol. 101, 590–605. doi: 10.1111/mmi.13410
Irie, Y., Mattoo, S., and Yuk, M. H. (2004). The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186, 5692–5698. doi: 10.1128/JB.186.17.5692-5698.2004
Irie, Y., O’toole, G. A., and Yuk, M. H. (2005). Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol. Lett. 250, 237–243. doi: 10.1016/j.femsle.2005.07.012
Jackon, J. K., Tudan, C., and Burt, H. M. (2000). The involvement of phospholipase C in crystal induced human neutrophil activation. J. Rheumatol. 27, 2877–2885.
Jungnitz, H., West, N. P., Walker, M. J., Chhatwal, G. S., and Guzmán, C. A. (1998). A second two-component regulatory system of Bordetella bronchiseptica required for bacterial resistance to oxidative stress, production of acid phosphatase, and in vivo persistence. Infect. Immun. 66, 4640–4650. doi: 10.1128/IAI.66.10.4640-4650.1998
Kamanova, J. (2020). Bordetella Type III Secretion Injectosome and Effector Proteins. Front. Cell Infect. Microbiol. 10:466. doi: 10.3389/fcimb.2020.00466
Karamooz, E., Yap, V. L., Barker, A. F., and Metersky, M. L. (2018). Bordetella bronchiseptica in non-cystic fibrosis bronchiectasis. Respir. Med. Case Rep. 25, 187–188. doi: 10.1016/j.rmcr.2018.08.023
Keidel, K., Amman, F., Bibova, I., Drzmisek, J., Benes, V., Hot, D., et al. (2018). Signal transduction-dependent small regulatory RNA is involved in glutamate metabolism of the human pathogen. RNA 24, 1530–1541. doi: 10.1261/rna.067306.118
Khakimova, M., Ahlgren, H. G., Harrison, J. J., English, A. M., and Nguyen, D. (2013). The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J. Bacteriol. 195, 2011–2020. doi: 10.1128/JB.02061-12
Kidd, S. P., and Brown, N. L. (2003). ZccR–a MerR-like regulator from Bordetella pertussis which responds to zinc, cadmium, and cobalt. Biochem. Biophys. Res. Commun. 302, 697–702. doi: 10.1016/S0006-291X(03)00249-3
Kilgore, P. E., Salim, A. M., Zervos, M. J., and Schmitt, H. J. (2016). Pertussis: Microbiology, Disease, Treatment, and Prevention. Clin. Microbiol. Rev. 29, 449–486. doi: 10.1128/CMR.00083-15
Kirimanjeswara, G. S., Agosto, L. M., Kennett, M. J., Bjornstad, O. N., and Harvill, E. T. (2005). Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J. Clin. Invest. 115, 3594–3601. doi: 10.1172/JCI24609
Klein, K., Sonnabend, M. S., Frank, L., Leibiger, K., Franz-Wachtel, M., Macek, B., et al. (2019). Deprivation of the Periplasmic Chaperone SurA Reduces Virulence and Restores Antibiotic Susceptibility of Multidrug-Resistant Pseudomonas aeruginosa. Front. Microbiol. 10:100. doi: 10.3389/fmicb.2019.00100
Kostylev, M., Kim, D. Y., Smalley, N. E., Salukhe, I., Greenberg, E. P., and Dandekar, A. A. (2019). Evolution of the. Proc. Natl. Acad. Sci. U.S.A. 116, 7027–7032. doi: 10.1073/pnas.1819796116
Kuchma, S. L., Connolly, J. P., and O’Toole, G. A. (2005). A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187, 1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005
Kurushima, J., Kuwae, A., and Abe, A. (2012b). Iron starvation regulates the type III secretion system in Bordetella bronchiseptica. Microbiol. Immunol. 56, 356–362. doi: 10.1111/j.1348-0421.2012.00442.x
Kurushima, J., Kuwae, A., and Abe, A. (2012a). Btc22 chaperone is required for secretion and stability of the type III secreted protein Bsp22 in Bordetella bronchiseptica. FEMS Microbiol. Lett. 331, 144–151. doi: 10.1111/j.1574-6968.2012.02561.x
Laarman, A. J., Bardoel, B. W., Ruyken, M., Fernie, J., Milder, F. J., van Strijp, J. A., et al. (2012). Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J. Immunol. 188, 386–393. doi: 10.4049/jimmunol.1102162
Lasaro, M. A., Salinger, N., Zhang, J., Wang, Y., Zhong, Z., Goulian, M., et al. (2009). F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75, 246–251. doi: 10.1128/AEM.01144-08
Lau, C. H., Krahn, T., Gilmour, C., Mullen, E., and Poole, K. (2015). AmgRS-mediated envelope stress-inducible expression of the mexXY multidrug efflux operon of Pseudomonas aeruginosa. Microbiol. Open 4, 121–135. doi: 10.1002/mbo3.226
Lau, G. W., Hassett, D. J., Ran, H., and Kong, F. (2004). The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10, 599–606. doi: 10.1016/j.molmed.2004.10.002
Laventie, B. J., Sangermani, M., Estermann, F., Manfredi, P., Planes, R., Hug, I., et al. (2019). A Surface-Induced Asymmetric Program Promotes Tissue Colonization by Pseudomonas aeruginosa. Cell Host Microbe 25, 140–152.e6. doi: 10.1016/j.chom.2018.11.008
Lavoie, E. G., Wangdi, T., and Kazmierczak, B. I. (2011). Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect. 13, 1133–1145. doi: 10.1016/j.micinf.2011.07.011
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Lee, S. W., Way, A. W., and Osen, E. G. (1986). Purification and subunit heterogeneity of pili of Bordetella bronchiseptica. Infect. Immun. 51, 586–593. doi: 10.1128/iai.51.2.586-593.1986
Léger, L., Byrne, D., Guiraud, P., Germain, E., and Maisonneuve, E. (2021). NirD curtails the stringent response by inhibiting RelA activity in Escherichia coli. eLife 10:e64092. doi: 10.7554/eLife.64092
Leid, J. G., Willson, C. J., Shirtliff, M. E., Hassett, D. J., Parsek, M. R., and Jeffers, A. K. (2005). The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175, 7512–7518. doi: 10.4049/jimmunol.175.11.7512
Li, K., Yang, G., Debru, A. B., Li, P., Zong, L., Xu, T., et al. (2017). SuhB Regulates the Motile-Sessile Switch in Pseudomonas aeruginosa through the Gac/Rsm Pathway and c-di-GMP Signaling. Front. Microbiol. 8:1045.
Li, W., and Lu, C. D. (2007). Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189, 5413–5420. doi: 10.1128/JB.00432-07
Look, D. C., Stoll, L. L., Romig, S. A., Humlicek, A., Britigan, B. E., and Denning, G. M. (2005). Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. J. Immunol. 175, 4017–4023. doi: 10.4049/jimmunol.175.6.4017
López-Boado, Y. S., Cobb, L. M., and Deora, R. (2005). Bordetella bronchiseptica flagellin is a proinflammatory determinant for airway epithelial cells. Infect. Immun. 73, 7525–7534. doi: 10.1128/IAI.73.11.7525-7534.2005
Managò, A., Becker, K. A., Carpinteiro, A., Wilker, B., Soddemann, M., Seitz, A. P., et al. (2015). Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid Redox Signal. 22, 1097–1110. doi: 10.1089/ars.2014.5979
Martín, C., Etxaniz, A., Uribe, K. B., Etxebarria, A., González-Bullón, D., Arlucea, J., et al. (2015). Adenylate Cyclase Toxin promotes bacterial internalisation into non phagocytic cells. Sci. Rep. 5:13774. doi: 10.1038/srep13774
Martins, R., Maier, J., Gorki, A. D., Huber, K. V., Sharif, O., Starkl, P., et al. (2016). Heme drives hemolysis-induced susceptibility to infection via disruption of phagocyte functions. Nat. Immunol. 17, 1361–1372. doi: 10.1038/ni.3590
Mathee, K., Narasimhan, G., Valdes, C., Qiu, X., Matewish, J. M., Koehrsen, M., et al. (2008). Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 3100–3105. doi: 10.1073/pnas.0711982105
Mattoo, S., Miller, J. F., and Cotter, P. A. (2000). Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect. Immun. 68, 2024–2033. doi: 10.1128/IAI.68.4.2024-2033.2000
Mattoo, S., Yuk, M. H., Huang, L. L., and Miller, J. F. (2004). Regulation of type III secretion in Bordetella. Mol. Microbiol. 52, 1201–1214. doi: 10.1111/j.1365-2958.2004.04053.x
McClure, C. D., and Schiller, N. L. (1992). Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J. Leukoc. Biol. 51, 97–102. doi: 10.1002/jlb.51.2.97
McPhee, J. B., Bains, M., Winsor, G., Lewenza, S., Kwasnicka, A., Brazas, M. D., et al. (2006). Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188, 3995–4006. doi: 10.1128/JB.00053-06
Melvin, J. A., Scheller, E. V., Miller, J. F., and Cotter, P. A. (2014). Bordetella pertussis pathogenesis: Current and future challenges. Nat. Rev. Microbiol. 12, 274–288. doi: 10.1038/nrmicro3235
Menetrey, Q., Aujoulat, F., Chiron, R., Jumas-Bilak, E., Marchandin, H., and Dupont, C. (2021). A new perspective on opportunistic pathogens of the genus Bordetella in cystic fibrosis. J. Cyst. Fibros. 21, 344–347.
Menetrey, Q., Aujoulat, F., Chiron, R., Jumas-Bilak, E., Marchandin, H., and Dupont, C. (2022). A new perspective on opportunistic pathogens of the genus Bordetella in cystic fibrosis. J. Cyst. Fibros. 21, 344–347. doi: 10.1016/j.jcf.2021.07.010
Meng, X., Ahator, S. D., and Zhang, L. H. (2020). Molecular Mechanisms of Phosphate Stress Activation of Pseudomonas aeruginosa Quorum Sensing Systems. mSphere 5:e119–e120. doi: 10.1128/mSphere.00119-20
Mikkelsen, H., Hui, K., Barraud, N., and Filloux, A. (2013). The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Mol. Microbiol. 89, 450–463. doi: 10.1111/mmi.12287
Mikkelsen, H., McMullan, R., and Filloux, A. (2011). The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 6:e29113. doi: 10.1371/journal.pone.0029113
Mishra, M., Byrd, M. S., Sergeant, S., Azad, A. K., Parsek, M. R., McPhail, L., et al. (2012). Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 14, 95–106. doi: 10.1111/j.1462-5822.2011.01704.x
Mishra, M., Parise, G., Jackson, K. D., Wozniak, D. J., and Deora, R. (2005). The BvgAS signal transduction system regulates biofilm development in Bordetella. J. Bacteriol. 187, 1474–1484. doi: 10.1128/JB.187.4.1474-1484.2005
Moissenet, D., Bingen, E., Arlet, G., and Vu-Thien, H. (2005). [Use of 16S rRNA gene sequencing for identification of “Pseudomonas-like” isolates from sputum of patients with cystic fibrosis]. Pathol. Biol. 53, 500–502. doi: 10.1016/j.patbio.2005.06.004
Moon, K., Bonocora, R. P., Kim, D. D., Chen, Q., Wade, J. T., Stibitz, S., et al. (2017). The BvgAS Regulon of Bordetella pertussis. MBio 8:e1526–e1517. doi: 10.1128/mBio.01526-17
Moon, K., Sim, M., Tai, C. H., Yoo, K., Merzbacher, C., Yu, S. H., et al. (2021). Identification of BvgA-Dependent and BvgA-Independent Small RNAs (sRNAs) in Bordetella pertussis Using the Prokaryotic sRNA Prediction Toolkit ANNOgesic. Microbiol. Spectr. 9:e0004421. doi: 10.1128/Spectrum.00044-21
Moore, J. E., Rendall, J. C., and Millar, B. C. (2022). A doggy tale: Risk of zoonotic infection with Bordetella bronchiseptica for cystic fibrosis (CF) patients from live licenced bacterial veterinary vaccines for cats and dogs. J. Clin. Pharm. Ther. 47, 139–145. doi: 10.1111/jcpt.13492
Moscoso, J. A., Mikkelsen, H., Heeb, S., Williams, P., and Filloux, A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x
Müller, M., and Hildebrandt, A. (1993). Nucleotide sequences of the 23S rRNA genes from Bordetella pertussis, B.parapertussis, B.bronchiseptica and B.avium, and their implications for phylogenetic analysis. Nucleic Acids Res. 21:3320. doi: 10.1093/nar/21.14.3320
Mund, A., Diggle, S. P., and Harrison, F. (2017). The Fitness of Pseudomonas aeruginosa Quorum Sensing Signal Cheats Is Influenced by the Diffusivity of the Environment. MBio 8:e353–e317. doi: 10.1128/mBio.00816-17
Mura, C., Randolph, P. S., Patterson, J., and Cozen, A. E. (2013). Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 10, 636–651. doi: 10.4161/rna.24538
Murdoch, C. C., and Skaar, E. P. (2022). Nutritional immunity: The battle for nutrient metals at the host-pathogen interface. Nat. Rev. Microbiol. [Epub ahead of print]. doi: 10.1038/s41579-022-00745-6
Murphy, K. L., Fischer, R., Swanson, K. A., Bhatt, I. J., Oakley, L., Smeyne, R., et al. (2020). Synaptic alterations and immune response are sexually dimorphic in a non-pertussis toxin model of experimental autoimmune encephalomyelitis. Exp. Neurol. 323:113061. doi: 10.1016/j.expneurol.2019.113061
Nakamura, K., Shinoda, N., Hiramatsu, Y., Ohnishi, S., Kamitani, S., Ogura, Y., et al. (2019). BspR/BtrA, an Anti-σ Factor, Regulates the Ability of Bordetella bronchiseptica To Cause Cough in Rats. mSphere 4:e93–e19. doi: 10.1128/mSphere.00093-19
Nicholson, T. L. (2007). Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8:220. doi: 10.1186/1471-2164-8-220
Nicholson, T. L., Buboltz, A. M., Harvill, E. T., and Brockmeier, S. L. (2009). Microarray and functional analysis of growth phase-dependent gene regulation in Bordetella bronchiseptica. Infect. Immun. 77, 4221–4231. doi: 10.1128/IAI.00136-09
Nicholson, T. L., Conover, M. S., and Deora, R. (2012). Transcriptome profiling reveals stage-specific production and requirement of flagella during biofilm development in Bordetella bronchiseptica. PLoS One 7:e49166. doi: 10.1371/journal.pone.0049166
O’Grady, K. F., Cripps, A. W., and Grimwood, K. (2019). Paediatric and adult bronchiectasis: Vaccination in prevention and management. Respirology 24, 107–114. doi: 10.1111/resp.13446
Okkotsu, Y., Little, A. S., and Schurr, M. J. (2014). The Pseudomonas aeruginosa AlgZR two-component system coordinates multiple phenotypes. Front. Cell Infect. Microbiol. 4:82. doi: 10.3389/fcimb.2014.00082
O’Malley, M. R., and Anderson, J. C. (2021). Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms 9:1227. doi: 10.3390/microorganisms9061227
Ortiz-Martín, I., Thwaites, R., Mansfield, J. W., and Beuzón, C. R. (2010). Negative regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 23, 682–701. doi: 10.1094/MPMI-23-5-0682
Ostroff, R. M., Wretlind, B., and Vasil, M. L. (1989). Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 57, 1369–1373. doi: 10.1128/iai.57.5.1369-1373.1989
Otero-Asman, J. R., Quesada, J. M., Jim, K. K., Ocampo-Sosa, A., Civantos, C., Bitter, W., et al. (2020). The extracytoplasmic function sigma factor σ. Sci. Rep. 10:3139. doi: 10.1038/s41598-020-60197-x
Overhage, J., Lewenza, S., Marr, A. K., and Hancock, R. E. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189, 2164–2169. doi: 10.1128/JB.01623-06
Paccani, S. R., Dal Molin, F., Benagiano, M., Ladant, D., D’Elios, M. M., Montecucco, C., et al. (2008). Suppression of T-lymphocyte activation and chemotaxis by the adenylate cyclase toxin of Bordetella pertussis. Infect. Immun. 76, 2822–2832. doi: 10.1128/IAI.00200-08
Paramonov, V. M., Sahlgren, C., Rivero-Müller, A., and Pulliainen, A. T. (2020). iGIST-A Kinetic Bioassay for Pertussis Toxin Based on Its Effect on Inhibitory GPCR Signaling. ACS Sens. 5:3438–3448. doi: 10.1021/acssensors.0c01340
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 10:1100. doi: 10.3389/fmicb.2019.01100
Penterman, J., Nguyen, D., Anderson, E., Staudinger, B. J., Greenberg, E. P., Lam, J. S., et al. (2014). Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 6, 293–300. doi: 10.1016/j.celrep.2013.12.019
Pérez-Martínez, I., and Haas, D. (2011). Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 3399–3405. doi: 10.1128/AAC.01801-10
Petráçková, D., Farman, M. R., Amman, F., Linhartová, I., Dienstbier, A., Kumar, D., et al. (2020). Transcriptional profiling of human macrophages during infection with. RNA Biol. 17, 731–742. doi: 10.1080/15476286.2020.1727694
Petrova, O. E., and Sauer, K. (2009). A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668. doi: 10.1371/journal.ppat.1000668
Petrova, O. E., and Sauer, K. (2011). SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 193, 6614–6628. doi: 10.1128/JB.00305-11
Pilione, M. R., and Harvill, E. T. (2006). The Bordetella bronchiseptica type III secretion system inhibits gamma interferon production that is required for efficient antibody-mediated bacterial clearance. Infect. Immun. 74, 1043–1049. doi: 10.1128/IAI.74.2.1043-1049.2006
Pita, T., Feliciano, J. R., and Leitão, J. H. (2018). Small Noncoding Regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia Complex. Int. J. Mol. Sci. 19:3759. doi: 10.3390/ijms19123759
Potvin, E., Sanschagrin, F., and Levesque, R. C. (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55. doi: 10.1111/j.1574-6976.2007.00092.x
Pragman, A. A., Berger, J. P., and Williams, B. J. (2016). Understanding persistent bacterial lung infections: Clinical implications informed by the biology of the microbiota and biofilms. Clin. Pulm. Med. 23, 57–66. doi: 10.1097/CPM.0000000000000108
Preston, G., Deng, W. L., Huang, H. C., and Collmer, A. (1998). Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180, 4532–4537. doi: 10.1128/JB.180.17.4532-4537.1998
Pusic, P., Sonnleitner, E., and Bläsi, U. (2021). Specific and Global RNA Regulators in Pseudomonas aeruginosa. Int. J. Mol. Sci. 22:8632. doi: 10.3390/ijms22168632
Qin, S., Xiao, W., Zhou, C., Pu, Q., Deng, X., Lan, L., et al. (2022). Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal. Transduct. Target. Ther. 7:199. doi: 10.1038/s41392-022-01056-1
Rada, B. (2017). Interactions between Neutrophils and Pseudomonas aeruginosa in Cystic Fibrosis. Pathogens 6:10. doi: 10.3390/pathogens6010010
Rada, B., Jendrysik, M. A., Pang, L., Hayes, C. P., Yoo, D. G., Park, J. J., et al. (2013). Pyocyanin-enhanced neutrophil extracellular trap formation requires the NADPH oxidase. PLoS One 8:e54205. doi: 10.1371/journal.pone.0054205
Rasamiravaka, T., Labtani, Q., Duez, P., and El Jaziri, M. (2015). The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015:759348. doi: 10.1155/2015/759348
Register, K. B., Sukumar, N., Palavecino, E. L., Rubin, B. K., and Deora, R. (2012). Bordetella bronchiseptica in a paediatric cystic fibrosis patient: Possible transmission from a household cat. Zoonoses Public Health 59, 246–250. doi: 10.1111/j.1863-2378.2011.01446.x
Reichhardt, C., Jacobs, H. M., Matwichuk, M., Wong, C., Wozniak, D. J., and Parsek, M. R. (2020). The Versatile Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Promotes Aggregation through Different Extracellular Exopolysaccharide Interactions. J. Bacteriol. 202:e216–e220. doi: 10.1128/JB.00216-20
Reinhart, A. A., Nguyen, A. T., Brewer, L. K., Bevere, J., Jones, J. W., Kane, M. A., et al. (2017). The Pseudomonas aeruginosa PrrF Small RNAs Regulate Iron Homeostasis during Acute Murine Lung Infection. Infect. Immun. 85:e764–e716.
Ridley, A. J. (2006). Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529. doi: 10.1016/j.tcb.2006.08.006
Ritchie, A. J., Jansson, A., Stallberg, J., Nilsson, P., Lysaght, P., and Cooley, M. A. (2005). The Pseudomonas aeruginosa quorum-sensing molecule N-3-(oxododecanoyl)-L-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect. Immun. 73, 1648–1655. doi: 10.1128/IAI.73.3.1648-1655.2005
Ritchie, A. J., Whittall, C., Lazenby, J. J., Chhabra, S. R., Pritchard, D. I., and Cooley, M. A. (2007). The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-L-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol. Cell Biol. 85, 596–602. doi: 10.1038/sj.icb.7100090
Rivera, I., Linz, B., Dewan, K. K., Ma, L., Rice, C. A., Kyle, D. E., et al. (2019). Conservation of Ancient Genetic Pathways for Intracellular Persistence Among Animal Pathogenic Bordetellae. Front. Microbiol. 10:2839. doi: 10.3389/fmicb.2019.02839
Rivera-Millot, A., Slupek, S., Chatagnon, J., Roy, G., Saliou, J. M., Billon, G., et al. (2021). Streamlined copper defenses make Bordetella pertussis reliant on custom-made operon. Commun. Biol. 4:46. doi: 10.1038/s42003-020-01580-2
Rogers, C. S., Abraham, W. M., Brogden, K. A., Engelhardt, J. F., Fisher, J. T., McCray, P. B., et al. (2008).. The porcine lung as a potential model for cystic fibrosis. Am. J. Physiol. Lung. Cell Mol. Physiol. 295:L240–L263. doi: 10.1152/ajplung.90203.2008
Roy, C. R., Miller, J. F., and Falkow, S. (1989). The bvgA gene of Bordetella pertussis encodes a transcriptional activator required for coordinate regulation of several virulence genes. J. Bacteriol. 171, 6338–6344. doi: 10.1128/jb.171.11.6338-6344.1989
Roy, G., Antoine, R., Schwartz, A., Slupek, S., Rivera-Millot, A., Boudvillain, M., et al. (2022). Posttranscriptional Regulation by Copper with a New Upstream Open Reading Frame. mBio e0091222. [Epub ahead of print]. doi: 10.1128/mbio.00912-22
Sadikot, R. T., Blackwell, T. S., Christman, J. W., and Prince, A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. doi: 10.1164/rccm.200408-1044SO
Sarwar, Z., Wang, M. X., Lundgren, B. R., and Nomura, C. T. (2020). MifS, a DctB family histidine kinase, is a specific regulator of α-ketoglutarate response in. Microbiology 166, 867–879. doi: 10.1099/mic.0.000943
Saukkonen, K., Cabellos, C., Burroughs, M., Prasad, S., and Tuomanen, E. (1991). Integrin-mediated localization of Bordetella pertussis within macrophages: Role in pulmonary colonization. J. Exp. Med. 173, 1143–1149. doi: 10.1084/jem.173.5.1143
Scavone, P., Iribarnegaray, V., Caetano, A. L., Schlapp, G., Härtel, S., and Zunino, P. (2016). Fimbriae have distinguishable roles in Proteus mirabilis biofilm formation. Pathog. Dis. 74:ftw033. doi: 10.1093/femspd/ftw033
Scheller, E. V., and Cotter, P. A. (2015). Bordetella filamentous hemagglutinin and fimbriae: Critical adhesins with unrealized vaccine potential. Pathog. Dis. 73:ftv079. doi: 10.1093/femspd/ftv079
Schneider, O. D., Weiss, A. A., and Miller, W. E. (2007). Pertussis toxin utilizes proximal components of the T-cell receptor complex to initiate signal transduction events in T cells. Infect. Immun. 75, 4040–4049. doi: 10.1128/IAI.00414-07
Schütz, C., and Empting, M. (2018). Targeting the Pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. Beilstein J. Org. Chem. 14, 2627–2645. doi: 10.3762/bjoc.14.241
Serra, D. O., Conover, M. S., Arnal, L., Sloan, G. P., Rodriguez, M. E., Yantorno, O. M., et al. (2011). FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 6:e28811. doi: 10.1371/journal.pone.0028811
Shen, D. K., Quenee, L., Bonnet, M., Kuhn, L., Derouazi, M., Lamotte, D., et al. (2008). Orf1/SpcS chaperones ExoS for type three secretion by Pseudomonas aeruginosa. Biomed. Environ. Sci. 21, 103–109. doi: 10.1016/S0895-3988(08)60014-8
Sierra, G. (1960). Hemolytic effect of a glycolipid produced by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 26, 189–192. doi: 10.1007/BF02539004
Sionov, R. V., and Steinberg, D. (2022). Targeting the Holy Triangle of Quorum Sensing, Biofilm Formation, and Antibiotic Resistance in Pathogenic Bacteria. Microorganisms 10:1239. doi: 10.3390/microorganisms10061239
Sivaneson, M., Mikkelsen, H., Ventre, I., Bordi, C., and Filloux, A. (2011). Two-component regulatory systems in Pseudomonas aeruginosa: An intricate network mediating fimbrial and efflux pump gene expression. Mol. Microbiol. 79, 1353–1366. doi: 10.1111/j.1365-2958.2010.07527.x
Skinner, J. A., Pilione, M. R., Shen, H., Harvill, E. T., and Yuk, M. H. (2005). Bordetella type III secretion modulates dendritic cell migration resulting in immunosuppression and bacterial persistence. J. Immunol. 175, 4647–4652. doi: 10.4049/jimmunol.175.7.4647
Skopelja-Gardner, S., Theprungsirikul, J., Lewis, K. A., Hammond, J. H., Carlson, K. M., Hazlett, H. F., et al. (2019). Regulation of Pseudomonas aeruginosa-Mediated Neutrophil Extracellular Traps. Front. Immunol. 10:1670. doi: 10.3389/fimmu.2019.01670
Sloan, G. P., Love, C. F., Sukumar, N., Mishra, M., and Deora, R. (2007). The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 189, 8270–8276. doi: 10.1128/JB.00785-07
Sønderholm, M., Kragh, K. N., Koren, K., Jakobsen, T. H., Darch, S. E., and Alhede, M. (2017). Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics. Appl. Environ. Microbiol. 83:e113–e117. doi: 10.1128/AEM.00113-17
Sonnleitner, E., Schuster, M., Sorger-Domenigg, T., Greenberg, E. P., and Bläsi, U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x
Spilker, T., Liwienski, A. A., and LiPuma, J. J. (2008). Identification of Bordetella spp. in respiratory specimens from individuals with cystic fibrosis. Clin. Microbiol. Infect. 14, 504–506. doi: 10.1111/j.1469-0691.2008.01968.x
Stibitz, S., Aaronson, W., Monack, D., and Falkow, S. (1988). The vir locus and phase-variation in Bordetella pertussis. Tokai J. Exp. Clin. Med. 13, 223–226.
Stockbauer, K. E., Foreman-Wykert, A. K., and Miller, J. F. (2003). Bordetella type III secretion induces caspase 1-independent necrosis. Cell Microbiol. 5, 123–132. doi: 10.1046/j.1462-5822.2003.00260.x
Sugisaki, K., Hanawa, T., Yonezawa, H., Osaki, T., Fukutomi, T., Kawakami, H., et al. (2013). Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiology 159, 1379–1389. doi: 10.1099/mic.0.066597-0
Sukumar, N., Nicholson, T. L., Conover, M. S., Ganguly, T., and Deora, R. (2014). Comparative analyses of a cystic fibrosis isolate of Bordetella bronchiseptica reveal differences in important pathogenic phenotypes. Infect. Immun. 82, 1627–1637. doi: 10.1128/IAI.01453-13
Sultan, M., Arya, R., and Kim, K. K. (2021). Roles of Two-Component Systems in Pseudomonas aeruginosa Virulence. Int. J. Mol. Sci. 22:12152. doi: 10.3390/ijms222212152
Svedova, M., Masin, J., Fiser, R., Cerny, O., Tomala, J., Freudenberg, M., et al. (2016). Pore-formation by adenylate cyclase toxoid activates dendritic cells to prime CD8+ and CD4+ T cells. Immunol. Cell Biol. 94, 322–333. doi: 10.1038/icb.2015.87
Tatke, G., Kumari, H., Silva-Herzog, E., Ramirez, L., and Mathee, K. (2015). Pseudomonas aeruginosa MifS-MifR Two-Component System Is Specific for α-Ketoglutarate Utilization. PLoS One 10:e0129629. doi: 10.1371/journal.pone.0129629
Telford, G., Wheeler, D., Williams, P., Tomkins, P. T., Appleby, P., Sewell, H., et al. (1998). The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect. Immun. 66, 36–42. doi: 10.1128/IAI.66.1.36-42.1998
Tian, Z. X., Yi, X. X., Cho, A., O’Gara, F., and Wang, Y. P. (2016). CpxR Activates MexAB-OprM Efflux Pump Expression and Enhances Antibiotic Resistance in Both Laboratory and Clinical nalB-Type Isolates of Pseudomonas aeruginosa. PLoS Pathog. 12:e1005932. doi: 10.1371/journal.ppat.1005932
Tonon, S., Badran, B., Benghiat, F. S., Goriely, S., Flamand, V., Willard-Gallo, K., et al. (2006). Pertussis toxin activates adult and neonatal naive human CD4+ T lymphocytes. Eur. J. Immunol. 36, 1794–1804. doi: 10.1002/eji.200535697
Tonon, S., Goriely, S., Aksoy, E., Pradier, O., Del Giudice, G., Trannoy, E., et al. (2002). Bordetella pertussis toxin induces the release of inflammatory cytokines and dendritic cell activation in whole blood: Impaired responses in human newborns. Eur. J. Immunol. 32, 3118–3125. doi: 10.1002/1521-4141(200211)32:11<3118::AID-IMMU3118>3.0.CO;2-B
Trouillon, J., Imbert, L., Villard, A. M., Vernet, T., Attrée, I., and Elsen, S. (2021). Determination of the two-component systems regulatory network reveals core and accessory regulations across Pseudomonas aeruginosa lineages. Nucleic Acids Res. 49, 11476–11490. doi: 10.1093/nar/gkab928
Turkina, M. V., and Vikström, E. (2019). Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J. Innate. Immun. 11, 263–279. doi: 10.1159/000494069
Ulett, G. C., Mabbett, A. N., Fung, K. C., Webb, R. I., and Schembri, M. A. (2007). The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153, 2321–2331. doi: 10.1099/mic.0.2006/004648-0
Vallet, I., Diggle, S. P., Stacey, R. E., Cámara, M., Ventre, I., Lory, S., et al. (2004). Biofilm formation in Pseudomonas aeruginosa: Fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186, 2880–2890. doi: 10.1128/JB.186.9.2880-2890.2004
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A., and Filloux, A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 98, 6911–6916. doi: 10.1073/pnas.111551898
Vance, R. E., Rietsch, A., and Mekalanos, J. J. (2005). Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73, 1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005
Vandamme, P., Heyndrickx, M., Vancanneyt, M., Hoste, B., De Vos, P., Falsen, E., et al. (1996). Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Rüger and Tan 1983. Int. J. Syst. Bacteriol. 46, 849–858. doi: 10.1099/00207713-46-4-849
Varley, J. (1986). The characterisation of Bordetella/Alcaligenes-like organisms and their effects on turkey poults and chicks. Avian. Pathol. 15, 1–22. doi: 10.1080/03079458608436262
Vasil, M. L., and Ochsner, U. A. (1999). The response of Pseudomonas aeruginosa to iron: Genetics, biochemistry and virulence. Mol. Microbiol. 34, 399–413. doi: 10.1046/j.1365-2958.1999.01586.x
Vega, S. C., Leiss, V., Piekorz, R., Calaminus, C., Pexa, K., Vuozzo, M., et al. (2020). Selective protection of murine cerebral G. J. Mol. Med. 98, 97–110. doi: 10.1007/s00109-019-01854-1
Venturi, V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30, 274–291. doi: 10.1111/j.1574-6976.2005.00012.x
Versteegh, F. G. (2012). Don’t forget Bordetella. Am. J. Respir. Crit. Care Med. 185, 1027–1028. doi: 10.1164/ajrccm.185.9.1027
Vidakovics, M. L., Lamberti, Y., Serra, D., Berbers, G. A., van der Pol, W. L., and Rodriguez, M. E. (2007). Iron stress increases Bordetella pertussis mucin-binding capacity and attachment to respiratory epithelial cells. FEMS Immunol. Med. Microbiol. 51, 414–421. doi: 10.1111/j.1574-695X.2007.00320.x
Vojtová, J., Kofronová, O., Sebo, P., and Benada, O. (2006). Bordetella adenylate cyclase toxin induces a cascade of morphological changes of sheep erythrocytes and localizes into clusters in erythrocyte membranes. Microsc. Res. Tech. 69, 119–129. doi: 10.1002/jemt.20277
Wakatsuki, A., Borrow, P., Rigley, K., and Beverley, P. C. (2003). Cell-surface bound pertussis toxin induces polyclonal T cell responses with high levels of interferon-gamma in the absence of interleukin-12. Eur. J. Immunol. 33, 1859–1868. doi: 10.1002/eji.200323675
Wallet, F., Perez, T., Armand, S., Wallaert, B., and Courcol, R. J. (2002). Pneumonia due to Bordetella bronchiseptica in a cystic fibrosis patient: 16S rRNA sequencing for diagnosis confirmation. J. Clin. Microbiol. 40, 2300–2301. doi: 10.1128/JCM.40.6.2300-2301.2002
Wang, D., Seeve, C., Pierson, L. S., and Pierson, E. A. (2013). Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genomics 14:618. doi: 10.1186/1471-2164-14-618
Wang, S., Cheng, J., Niu, Y., Li, P., Zhang, X., and Lin, J. (2021). Strategies for Zinc Uptake in Pseudomonas aeruginosa at the Host-Pathogen Interface. Front. Microbiol. 12:741873. doi: 10.3389/fmicb.2021.741873
Weingart, C. L., and Weiss, A. A. (2000). Bordetella pertussis virulence factors affect phagocytosis by human neutrophils. Infect. Immun. 68, 1735–1739. doi: 10.1128/IAI.68.3.1735-1739.2000
West, A. H., and Stock, A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. doi: 10.1016/S0968-0004(01)01852-7
Weyrich, L. S., Feaga, H. A., Park, J., Muse, S. J., Safi, C. Y., Rolin, O. Y., et al. (2014). Resident microbiota affect Bordetella pertussis infectious dose and host specificity. J. Infect. Dis. 209, 913–921. doi: 10.1093/infdis/jit597
Wilkinson, T. M. A., Van den Steen, P., Cheuvart, B., Baudson, N., Dodet, M., Turriani, E., et al. (2021). Seroprevalence of Bordetella pertussis Infection in Patients With Chronic Obstructive Pulmonary Disease in England: Analysis of the AERIS Cohort. COPD 18, 341–348. doi: 10.1080/15412555.2021.1920904
Williams, T. L., Rada, B., Tandon, E., and Gestal, M. C. (2020). NETs and EETs, a Whole Web of Mess. Microorganisms 8:1925. doi: 10.3390/microorganisms8121925
Workentine, M. L., Chang, L., Ceri, H., and Turner, R. J. (2009). The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: A bacterial two-hybrid analysis. FEMS Microbiol. Lett. 292, 50–56. doi: 10.1111/j.1574-6968.2008.01445.x
Wurpel, D. J., Totsika, M., Allsopp, L. P., Hartley-Tassell, L. E., Day, C. J., Peters, K. M., et al. (2014). F9 fimbriae of uropathogenic Escherichia coli are expressed at low temperature and recognise Galβ1-3GlcNAc-containing glycans. PLoS One 9:e93177. doi: 10.1371/journal.pone.0093177
Xu, X., Yu, H., Zhang, D., Xiong, J., Qiu, J., Xin, R., et al. (2016). Role of ppGpp in Pseudomonas aeruginosa acute pulmonary infection and virulence regulation. Microbiol. Res. 192, 84–95. doi: 10.1016/j.micres.2016.06.005
Yabuuchi, E., and Ohyama, A. (1972). Characterization of “Pyomelanin”-Producing Strains of Pseudomonas aeruginosa. Int. J. Syst. Bacteriol. 22 N2:11. doi: 10.1099/00207713-22-2-53
Yeung, A. T., Bains, M., and Hancock, R. E. (2011). The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 193, 918–931. doi: 10.1128/JB.00911-10
Yuk, M. H., Harvill, E. T., Cotter, P. A., and Miller, J. F. (2000). Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 35, 991–1004. doi: 10.1046/j.1365-2958.2000.01785.x
Zamorano, L., Moyà, B., Juan, C., Mulet, X., Blázquez, J., and Oliver, A. (2014). The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob. Agents Chemother. 58, 5084–5095. doi: 10.1128/AAC.02556-14
Zegans, M. E., Becker, H. I., Budzik, J., and O’Toole, G. (2002). The role of bacterial biofilms in ocular infections. DNA Cell Biol. 21, 415–420. doi: 10.1089/10445490260099700
Zelazny, A. M., Ding, L., Goldberg, J. B., Mijares, L. A., Conlan, S., Conville, P. S., et al. (2013). Adaptability and persistence of the emerging pathogen Bordetella petrii. PLoS One 8:e65102. doi: 10.1371/journal.pone.0065102
Zhang, Y. F., Han, K., Chandler, C. E., Tjaden, B., Ernst, R. K., and Lory, S. (2017). Probing the sRNA regulatory landscape of P. aeruginosa: Post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 106, 919–937. doi: 10.1111/mmi.13857
Zhou, F., Liu, R., Han, P., Zhang, X., Li, Z., Zhang, S., et al. (2020). Pertussis Toxin Ameliorates Microglial Activation Associated With Ischemic Stroke. Front. Cell Neurosci. 14:152. doi: 10.3389/fncel.2020.00152
Zhu, M., Zhao, J., Kang, H., Kong, W., and Liang, H. (2016a). Modulation of Type III Secretion System in Pseudomonas aeruginosa: Involvement of the PA4857 Gene Product. Front. Microbiol. 7:7. doi: 10.3389/fmicb.2016.00007
Zhu, M., Zhao, J., Kang, H., Kong, W., Zhao, Y., Wu, M., et al. (2016b). Corrigendum: Modulation of Type III Secretion System in Pseudomonas aeruginosa: Involvement of the PA4857 Gene Product. Front. Microbiol. 7:881. doi: 10.3389/fmicb.2016.00881
Keywords: Bordetella, Pseudomonas, immunosuppression, host-pathogen, stress response, biofilm, virulence
Citation: Holban AM, Gregoire CM and Gestal MC (2022) Conquering the host: Bordetella spp. and Pseudomonas aeruginosa molecular regulators in lung infection. Front. Microbiol. 13:983149. doi: 10.3389/fmicb.2022.983149
Received: 30 June 2022; Accepted: 17 August 2022;
Published: 26 September 2022.
Edited by:
Ibrahim M. Sayed, Assiut University, EgyptReviewed by:
Tridib Ganguly, University of Florida, United StatesOsvaldo Miguel Yantorno, National University of La Plata, Argentina
Copyright © 2022 Holban, Gregoire and Gestal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monica C. Gestal, bW9uaWNhLmNhcnRlbGxlZ2VzdGFsQGxzdWhzLmVkdQ==; bWNhcmdlc0BnbWFpbC5jb20=
 Alina M. Holban
Alina M. Holban Courtney M. Gregoire
Courtney M. Gregoire Monica C. Gestal
Monica C. Gestal