- 1Department of Biotechnology and Food Science, Durban University of Technology, Durban, South Africa
- 2Water and Environmental Management Research Group, Engineering and Agriculture, University of Venda, Thohoyandou, South Africa
Enteric viruses are common waterborne pathogens found in environmental water bodies contaminated with either raw or partially treated sewage discharge. Examples of these viruses include adenovirus, rotavirus, noroviruses, and other caliciviruses and enteroviruses like coxsackievirus and polioviruses. They have been linked with gastroenteritis, while some enteric viruses have also been implicated in more severe infections such as encephalitis, meningitis, hepatitis (hepatitis A and E viruses), cancer (polyomavirus), and myocarditis (enteroviruses). Therefore, this review presents information on the occurrence of enteric viruses of public health importance, diseases associated with human exposure to enteric viruses, assessment of their presence in contaminated water, and their removal in water and wastewater sources. In order to prevent illnesses associated with human exposure to viral contaminated water, we suggest the regular viral monitoring of treated wastewater before discharging it into the environment. Furthermore, we highlight the need for more research to focus on the development of more holistic disinfection methods that will inactivate waterborne viruses in municipal wastewater discharges, as this is highly needed to curtail the public health effects of human exposure to contaminated water. Moreover, such a method must be devoid of disinfection by-products that have mutagenic and carcinogenic potential.
Introduction
The world is increasingly faced with daunting challenges in meeting the expanding requirements for clean water as the existing supplies of freshwater are in short supply owing to (i) prolonged droughts; (ii) population growth; (iii) strict health-based guidelines; and (iv) contending demands from diverse users (Mancosu et al., 2015). Due to these facts and many others, water protection against possible biological and chemical contaminants is becoming critical in water resources management. Enteric viruses are one of the emerging biological contaminants responsible for the sporadic outbreak of waterborne illnesses worldwide (La Rosa et al., 2012; Upfold et al., 2021). Compared with bacteria and protozoa, viruses are smaller, measuring between 20 and 350 nm in diameter (Health Canada, 2019). Viruses are obligate intracellular parasites containing bundles of gene strands of either RNA or DNA as a core nucleic acid surrounded by a protective coat called protein capsid (Clemente et al., 2012). Sometimes, the capsid protein is enclosed in an additional spikey envelope. In an enveloped virus, the nucleocapsid is bounded by a lipid bilayer which results from the modified host cell membrane with an outer layer of virus envelope glycoproteins (Zwart et al., 2009; Lázaro et al., 2018). Unlike bacterial cells, which are free-living entities, viruses employ the host cell environment to multiply. Studies have shown that viruses can latch onto host cells, as seen in several outbreaks including the current coronavirus (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Shang et al., 2020).
According to Clemente et al. (2012), patients suffering from gastroenteritis may defecate about 105–1011 virus particles per gram of stool which in turn gain entrance into environmental media through the discharge of contaminated wastewater (Wu et al., 2020; Yeo et al., 2020). Though enteric viruses are the smallest enteric pathogens with a low infective dose, they are robust and difficult to detect and inactivate (Gonzales-Gustavson et al., 2019; Farkas et al., 2020a) hence, they serve as a route of potential health risk at a low exposure dose (Zwart et al., 2009; Zhu et al., 2018). Thus, the transmission of human enteric viruses via the water route is becoming more widely recognized as a potential cause of human diseases such as gastroenteritis, encephalitis, meningitis, and hepatitis, among others (World Health Organization, 2014; Tang et al., 2020).
There are several cases where viruses are detected in water deemed compliant with bacterial indicators, and the consumption of such water has been reported to result in waterborne diseases (Umesha et al., 2008; Chen et al., 2021). Even when subjected to conventional wastewater treatments, only about 20–80% of the enteric viruses get inactivated due to their resistance to chemical disinfectants (Jiang et al., 2001; La Rosa et al., 2012), thus leading to the discharge of viruses into the aquatic environment. They can survive unfavorable conditions such as temperature and pH among others for an extended period in environmental waters owing to their small size (Bertrand et al., 2012; Meixell et al., 2013), inertness, and the presence of viral capsid.
Several studies have detected enteric viruses in seawater, surface and groundwater, municipal wastewater influent, and inadequately treated wastewater effluent (Saxena et al., 2015; Shih et al., 2017; Hendriksen et al., 2019; Pang et al., 2019; Verani et al., 2019; Shaheen et al., 2020; Tang et al., 2020; Farkas et al., 2020a). However, their occurrence is not limited to those water matrices, but they have also been found in drinking water systems and recreational water (Umesha et al., 2008; Upfold et al., 2021). Another route of enteric virus transmission is food products contaminated with irrigation water (Benko et al., 2002; Bouseettine et al., 2020). They could be transmitted via food, such as shellfish grown in contaminated water, wastewater irrigated farm produce, open defecation, untreated contaminated surface water, and exposure to inadequately treated wastewater due to poor infrastructure (Umesha et al., 2008; Haramoto et al., 2018). Therefore, this review focuses on the occurrence of enteric viruses in water and wastewater sources, their public health implications, and associated diseases due to human exposure to waterborne enteric viruses. Finally, we examined how the identified public health concerns can be minimized.
Enteric viruses of public health importance
Worldwide, more than 150 enteric viruses are associated with waterborne diseases. The prevalence of Hepatitis A Virus (HAV), Adenovirus (AdV), Rotaviruses (RV) and Enteroviruses (EVs), Astroviruses (AstVs), Noroviruses (NoVs), and bacteriophages in surface waters (Radin, 2014), dams and treated drinking water (Fernandez-Cassi et al., 2018; Opere, 2019), treated effluent from wastewater treatment plants (WWTPs; Gonzales-Gustavson et al., 2019) and the detection in post chlorinated water is not exempted (La Rosa et al., 2012). Figure 1 shows both enveloped (e.g., coronaviruses) and non-enveloped viruses found in treated and untreated wastewater. In the recent SARS-CoV-2 outbreak, scientists discovered that the virus responsible for the pandemic; SARS-CoV-2 is shed in feces, detected in the sewerage system, and treated effluent (Ahmed et al., 2020; Haramoto et al., 2020; Randazzo et al., 2020). Furthermore, NoV, AdV, AstV, EV, HAV, RV, and hepatitis E virus (HEV) are transmitted via water (Gibson, 2014; Bouseettine et al., 2020).
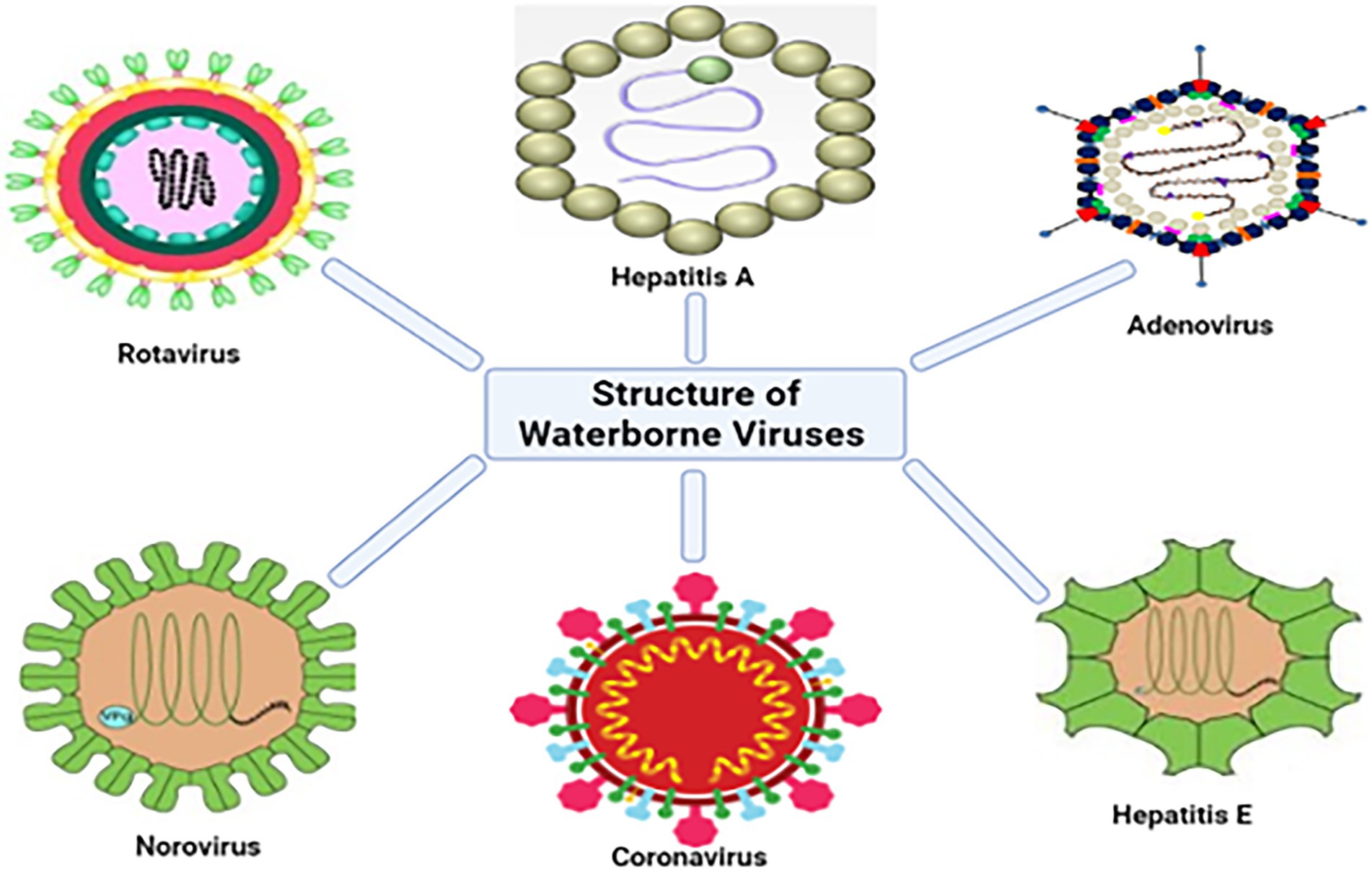
Figure 1. Structures of some of the viruses that have been detected in water sources (adapted from Altintas et al., 2015).
Adenovirus
Human adenoviruses (HAdV) are icosahedral capsid structures, non-segmented, non-enveloped, and double-stranded DNA viruses with a diameter between 90 and 100 nm (Opere, 2019). The family Adenoviridae is divided into five genera: Genus Mastadenovirus, which infects mammals; Aviadenovirus, which infects birds; Siadenovirus, which infects birds and frogs and Atadenovirus, which infects a wide variety of hosts such as aves, reptiles, and marsupial hosts (Arnold and MacMahon, 2017) and lastly, the newly proposed genera, Ichtadenovirus (Benko et al., 2002). Human adenovirus (HAdV) is a nonlinear envelope single-stranded DNA in the Adenoviridae family and genus Mastadenoviruses (Crenshaw et al., 2019). Within the Mastadenovirus genus, there are presently 103 HAdV serotypes, categorized into seven species (A–G) (Kosulin, 2019; Mennechet et al., 2019).
Many adenoviruses are shed in high amounts with more than 1011 particles per gram of faeces, urine, or respiratory excretions (Hewitt et al., 2013). Across the world, these viruses have been found in wastewater, both influent and effluent (Mena and Gerba, 2008; Iaconelli et al., 2017a). Specifically, many HAdV species have been detected in surface water (Sibanda and Okoh, 2012; Opere, 2019) and wastewater (Osuolale and Okoh, 2015; Iaconelli et al., 2017b). Generally, they can survive as extra circular chromosomes or through integration into the DNA of the host (Fu et al., 2019).
In general, HAdVs are prevalent etiological agents of intestinal, respiratory, and ophthalmic diseases. Several clinical features are caused in the respiratory tract, eyes, gastrointestinal tract, and other organs due to the initial propagation, which may occur in the mucosa of the pharynx, conjunctiva, or intestinal mucosa (Howley and Lowy, 2007). Symptoms of infection include gastroenteritis, upper and lower respiratory tract infection, conjunctivitis, pneumonia, myocarditis, and encephalitis (Russell, 2009; Akello et al., 2020). They are generally mild and self-limiting, but in immunocompromised individuals, they can evolve into severe infections with unique manifestations (Khanal et al., 2018). In both developed and developing nations, the occurrence of enteric HAdVs in childhood diarrhea range from 1 to 8% and 2 to 31%, respectively (Meqdam and Thwiny, 2007; Hassou et al., 2020). Between 2007 and 2019, HAdV was found in about 10.8% of all gastroenteritis incidents among children below five in Sub-Saharan Africa (Oppong et al., 2020).
Rotavirus
Rotavirus (RV), belonging to the Reoviridae family, is a relevant and significant waterborne pathogen (Atabakhsh et al., 2021). The genus comprises five species (A–E), two tentative species (F and G) as well as an unassigned species (ADRV-N) which is referred to as RVH (Desselberger, 2014). The rotavirus genome has 11 segments of dsRNA with a size that ranges from 0.6 to 33 kilobase pairs (Bouseettine et al., 2020). Segmentation in the RV genome allows the rearrangement mechanism and production of new strains with different combinations of genome segments resulting in genetic diversity, boosting the evolution, and emerging new strains of RVs each year (Esona et al., 2010; Menezes et al., 2020).
In humans, serogroups A–C cause gastroenteritis, while group A causes severe diarrhea in young children (Bouseettine et al., 2020). Group A is the most common cause of gastrointestinal disease in children below the age of 5, with severe outcomes such as hospitalization and death (Prez et al., 2015; Magana-Arachchi and Wanigatunge, 2020). The principal agents of infectious dehydrating diarrhea in infants are the rotavirus group A [RVA], which is linked to most human RV infections. RVA genotypes G1 P[8], G2 P[4], G3 P[8], and G9 P[8] are associated with most human gastroenteritis and easily undergo genetic recombination (Leite et al., 2008; Tate et al., 2010). Wa-like (G1-P[8]-I1-R1-C1- M1-A1-N1- T1-E1-H1) and DS-1-like (G2-P[4]-I2-R2-C2-M2-A2- N2-T2-E2-H2) are the two main genogroups identified from the analyses of the complete genome sequences of human rotavirus (HRV) strains to contain the majority of wild-type HRV strains (Tate et al., 2010). The G1P [8] strain is the most predominant in Europe, with about 69.4% of infections in America and some Asian countries (Fongaro et al., 2015). However, there has been a wide level of reduction in the cases of severe childhood diarrhea in countries where routine childhood vaccination against rotavirus has been entrenched. In the same vein, there seems to be a reduction in the number of unvaccinated children due to the protection already offered to vaccinated children, thereby suggesting herd protection because of the vaccination (Fongaro et al., 2015).
Rotavirus infection has been linked to about 258 million cases of diarrhea in children under 5 years worldwide (Troeger et al., 2018). Nevertheless, RV continues to be the leading cause of annual childhood deaths associated with diarrhea worldwide despite the effective introduction of vaccines in more than 106 countries (Kim et al., 2021). The virus has been estimated to be responsible for 122,000–215,000 diarrheic child deaths between 2013 and 2017 annually (Tate et al., 2016; Troeger et al., 2018). In a study conducted to investigate the link between the contaminated water source and rotaviral infection among 184 children under 5 years with acute watery diarrhea, the highest rotaviral infection was observed in children that consume contaminated pond water (94.44%). It was followed by children that were not breastfed exclusively (83.87%) and 64.00% in children living in areas with lower socio-economic conditions (Habib et al., 2021).
Enterovirus
Enterovirus (EV) is a spherical non-enveloped virus containing a 7,500-nucleotide positive-sense single-stranded RNA genome. They are 7.3–7.4 kb in length and are protected by an icosahedral capsid (Opere, 2019). Enteroviruses belong to the family Picornaviridae and are extremely small, with diameters ranging from 22 to 30 nm (Magana-Arachchi and Wanigatunge, 2020). The genus enterovirus has more than 300 known serotypes and 15 species (Chen et al., 2020). The different species include polioviruses, coxsackieviruses A and B, enteroviruses, and echoviruses and they have been linked with human infections (Table 1; Bouseettine et al., 2020).
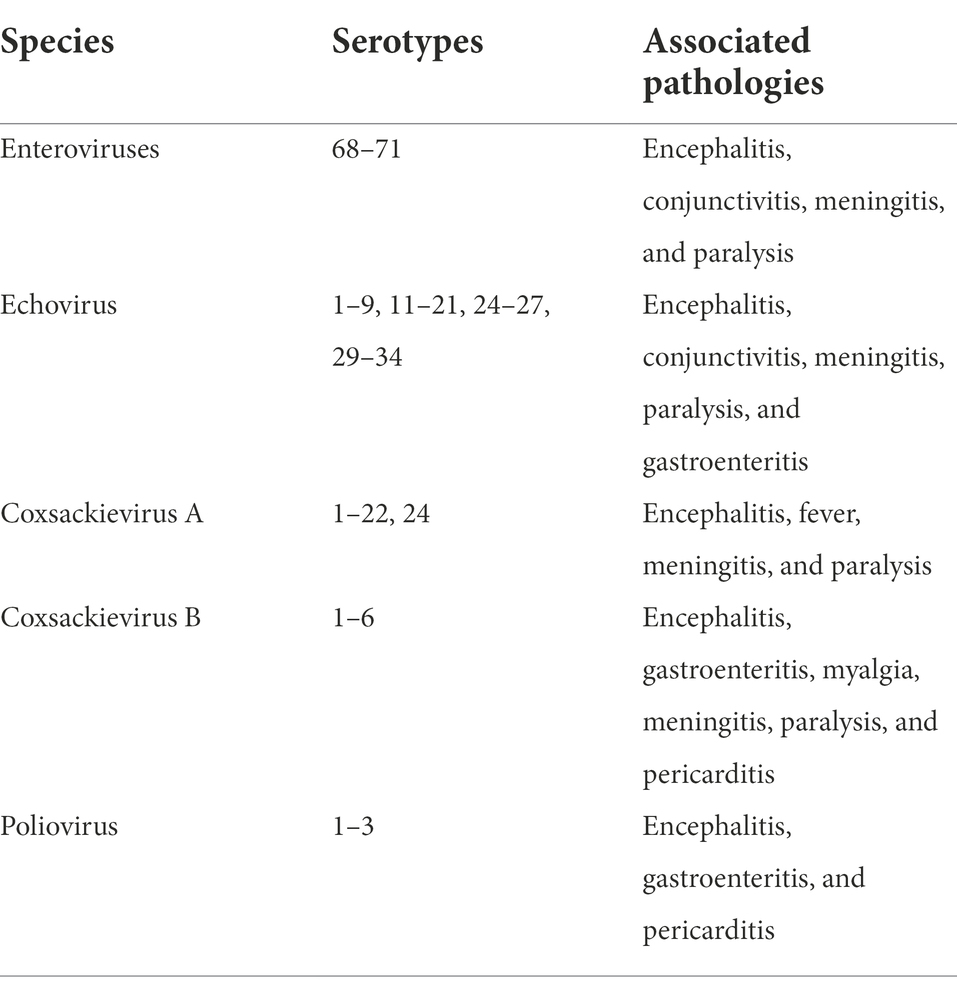
Table 1. Species of human enteroviruses and associated pathologies (adapted from Bouseettine et al., 2020).
Millions of people worldwide have been infected with enteroviruses, which have caused the irregular outbreak of illnesses in both advanced and evolving nations (Lugo and Krogstad, 2016; Cassidy et al., 2018; Smuts et al., 2018; Puenpa et al., 2019). Of the seven species to which human EVs belong, only four species (A–D) infect the gastrointestinal tract (Baggen et al., 2018). Enterovirus 71 (EV-A71) is a human enteric virus linked with hand-foot-and-mouth disease (HFMD), while EV-D68 is associated with acute flaccid paralysis (Lee and Chi, 2014; Baggen et al., 2018). A total of 2,967 cases of EV and Parechovirus infections were documented in the United States of America between 2014 and 2016. The most frequently reported enterovirus is the EV-D68 which is responsible for 68% of identified types in 2014 and accounted for 56% of all documented types between 2014 and 2016. Other enteroviruses that were also frequently reported include echovirus 30 (13.1%), coxsackievirus A6 (12.5%), echovirus 18 (9.5%), and coxsackievirus B3 (9%) (Abedi et al., 2018).
The mode of transmission of enterovirus is the faecal-oral route through different sources, such as contaminated water, food, or person-to-person contact. The faecal-oral route becomes problematic when there is exposure to aerosols that are transmitted from any surface or groundwater sources (Gao et al., 2012). Enteroviruses are robust organisms as they can survive critical changes in both pH and temperature (Okoh et al., 2010). Therefore, they have been proposed as a criterion for assessing viral contamination of environmental waters because they are common and shed for an extended period in the environment (La Rosa et al., 2010a). Studies that focused on their removal at WWTPs revealed that they are more resistant to treatment when compared to some other enteric viruses such as adenoviruses and noroviruses (La Rosa et al., 2010b). Several studies have identified them in both raw and treated sewage (La Rosa et al., 2010b; Simmons et al., 2011).
Norovirus
Noroviruses, which were initially referred to as Norwalk or Norwalk-like viruses, are small non-enveloped single-stranded RNA viruses of 27–38 nm in diameter (Rani et al., 2021). They are members of the family Caliciviridae (Upfold et al., 2021). Norwalk virus is composed of a single strand of RNA and bounded by many copies of a single protein organized into a protective capsid (Wang et al., 2018b). According to Thorne and Goodfellow (2014), it has been frequently revealed that Norovirus is widespread and that infections occur more in the human population. Noroviruses are classified into 10 different genogroups (GI–GX; Laconi et al., 2020). Generally, NoVs GI, GII, and GIV infect man, while GI is less found in patients with acute gastroenteritis across the globe than in GII strains (Koopmans et al., 2002). The most common genogroup in man is the three distinct clusters in GII to which the porcine NoVs belong. This has raised public health concerns regarding the potential NoVs recombinant strains in causing zoonotic transmission (Shen et al., 2012; Okada et al., 2019; Laconi et al., 2020). Generally, gastroenteritis linked with NoVs is self-limiting, although young children, the elderly, and immunocompromised individuals could experience long-term symptoms (Trivedi et al., 2013; Petrignani et al., 2018). Norovirus GI and GII are majorly responsible for human infections; however, for about 10 years now, the genogroup GII.4 has been linked with most human NoVs diseases (Siebenga et al., 2009; Vinjé, 2015; Mans et al., 2016). A novel strain of genotype GII.17 (GII.P17-GII.17) was reported in Asia to be responsible for gastroenteritis linked with NoV in the year 2014 and has subsequently been documented worldwide (Chan et al., 2015, 2017; Lu et al., 2015; Hoa-Tran et al., 2017; Zhou et al., 2019).
The transmission of noroviruses is primarily via the faecal-oral route. It transpires through the intake of contaminated water or by direct contact with persons already infected and on environmental surfaces (Zhou et al., 2016; Boonchan et al., 2017; Kittigul et al., 2019). Drinking water and recreational water polluted from sewage are the sources of several outbreaks of NoVs (Kittigul et al., 2019; Prado et al., 2019; Tryfinopoulou et al., 2019). According to de Graaf et al. (2016), GI variants are more linked with transmission through the water route than variant GII.4, which relates to person–person contact. The GI strains of Norovirus are believed to be stable in water compared to the GII strains because they are more likely to be transmitted through water than other transmission routes. Across the globe, NoVs have been detected in different water bodies, including sewages, rivers, recreational waters, municipal water, and groundwater (La Rosa et al., 2007, 2008, 2010a; Miura et al., 2019).
Astrovirus
Human astroviruses (HAstVs) are the third most frequent cause of gastroenteritis, and they were first detected in 1975 (Vu et al., 2017). They are small with sizes ranging from 28 to 30 nm and are non-enveloped viruses having single-stranded RNA (+ssRNA) genomes with about 6,400–7,900 nucleotides. Genera Mamastrovirus (MAstV) and Avastrovirus (AAstV) make up the family Astroviridae (Appleton, 1975; De Benedictis et al., 2011; Donato and Vijaykrishna, 2017). Human astroviruses (HAstVs) are a vital cause of gastroenteritis in infants and young children (De Benedictis et al., 2011). They are found in surface and groundwaters meant for drinking purposes, marine waters, and wastewater effluents (Magana-Arachchi and Wanigatunge, 2020). A study on the molecular detection of gastroenteritis viruses reported that HAstV had a lesser occurrence than EV, RV, NoV, and AdV (Chitambar et al., 2012). The aged and individuals with weak immune system are susceptible to gastroenteritis linked with HAstV (Vu et al., 2017; Wohlgemuth et al., 2019). This could be credited to the pathogenic role of the virus being performed in immunocompromised individuals. In addition, HAstVs have been linked to central nervous system (CNS) infections including encephalitis and acute flaccid paralysis in people with compromised immunity (Cordey et al., 2016; Vu et al., 2016). It could also be due to the inability of the individuals to mount an active inflammatory response to the virus hence a reflection of their highly immunocompromised state.
Hepatitis A virus
Hepatitis A virus (HAV), which is a member of the Hepatovirus genus belonging to the family Picornaviridae, is non-enveloped with an icosahedral structure of an average of 30 nm in diameter and a naked RNA genome (Smith and Simmonds, 2018; Bouseettine et al., 2020). The capsid contains a densely packed icosahedral configuration of 60 protomers, each of which includes three polypeptides VP1, VP2, and VP3 (McKnight and Lemon, 2018). Studies have reported that about 1.5 million people are infected with HAV annually, which is underestimated due to the asymptomatic presentation of the virus and limited epidemiologic information on the virus (World Health Organization, 2017b; Lemon et al., 2018).
As the major causative agent of non-parenteral hepatitis in developing nations as well as endemic infections in developing countries, the primary mechanism of transmission of the HAV is the faecal-oral route and direct contact with an infected person (Mehta and Reddivari, 2021). The outbreaks of HAV infection are mostly associated with water supplies (Jothikumar et al., 2005). The consumption of raw or improperly cooked oysters and clams from sewage-contaminated water has led to many outbreaks of HAV infection (Elbashir et al., 2018). Fever, anorexia, malaise, abdominal discomfort, nausea, and jaundice are some of the numerous symptoms that equally result from HAV infection and could to liver damage (La Rosa et al., 2012).
Hepatitis A virus is found in the feces and urine of diseased individuals through which it contaminates the soil and water. The virus is detected in water and wastewater globally; however, an area’s sanitary conditions determine the virus’s prevalence (World Health Organization, 2012c). People living in areas with poor sanitation infrastructure are more prone to contracting the virus, especially among children (World Health Organization, 2012b). According to Rodriguez-Lazaro et al. (2012), HAV can subsist for about 60 days in tap water, while it can stay alive for more than 6 weeks in river water, can stay above 8 weeks in groundwater, and for about 30 weeks in seawater. The virus has been found in different water environments such as raw wastewaters (Osuolale and Okoh, 2015), treated effluents (Van Zyl et al., 2019), surface waters (Marie and Lin, 2017; Shaheen et al., 2019), and drinking waters (Moreno et al., 2009).
Hepatitis E virus
Hepatitis E virus (HEV) is a member of the family Hepeviridae and belongs to the Orthohepevirus genus and the Orthohepevirus A species. HEV is a non-enveloped, positive sense, ssRNA virus (Denner, 2019) that causes significant acute hepatitis worldwide thereby resulting in sporadic infections and epidemics (Raji et al., 2021). The infection is usually asymptomatic in the general population (Khuroo and Kamili, 2003). The virus is regarded as zoonotic with swine and other animals such as rabbits serving as the reservoir for human infections (Ruggeri et al., 2013; Syed et al., 2018). Hepatitis E virus is another waterborne pathogen threatening global health in developing and developed countries (Raji et al., 2021). Currently, eight HEV genotypes have been identified (Upfold et al., 2021), and genotypes 1–4 and 7 are known to be the main threats to humans. They are transmitted through the ingestion of contaminated water, causing acute hepatitis (Purdy et al., 2017).
Ruggeri et al. (2013), posited that HEV strains that belong to genotypes 1 and 2 are the cause of the most sporadic outbreak of hepatitis in developing countries of Africa, Asia, and Mexico (Smith and Simmonds, 2018). Genotypes 3, 4, and 7 are basically associated with zoonotic transmission (Marek et al., 2010) and are generally connected with irregular and grouped infections in advanced countries (Nimgaonkar et al., 2018). It is transmitted via the faecal-oral pathway and spreads readily through water bodies polluted with human faeces. Globally, the virus has been detected in different water bodies including rivers (Iaconelli et al., 2015), and raw wastewater (Iaconelli et al., 2017a; Wang et al., 2018a), among others.
Aichivirus
Aichivirus (AiV) belongs to the Kobuvirus genus and the family Picornaviridae (Rivadulla and Romalde, 2020). It is one of the significant causative organisms of gastroenteritis in humans and is transmitted via the faecal-oral route from contaminated food or water (Yamashita et al., 2000; Adams et al., 2013; Kebe et al., 2021). Aichivirus consists of Human AiV (HAiV) 1, Murine Kobuvirus (MuKV) 1, and Canine Kobuvirus (CaKV) 1. Human AiV is composed of three genotypes (A–C; Yamashita et al., 2000; Shaheen et al., 2020; Kebe et al., 2021), and the infection rate of AiV in gastroenteritis cases worldwide is low between 0.4 and 6.5% (Rivadulla and Romalde, 2020; Taghinejad et al., 2020; Upfold et al., 2021). However, specific antibodies produced against the virus are found in about 80–90% of adults (Kitajima and Gerba, 2015; Rivadulla and Romalde, 2020), which is a pointer to the asymptomatic condition of most infections (Bergallo et al., 2017). Furthermore, this virus is often detected in connection with other enteric pathogens (Oh et al., 2006; Ambert-Balay et al., 2008; Kaikkonen et al., 2010; Japhet et al., 2019). Diarrhea, abdominal pain, vomiting, and fever are some clinical signs and symptoms of human AiV virus infection (Yamashita et al., 2000). Aichivirus has been reported in Africa, Asia, South America, and Europe (Oh et al., 2006; Lodder et al., 2013). The virus is normally passed out in the faeces of humans directly or after discharge of treated or untreated sewage (Lodder et al., 2013; Kebe et al., 2021), and can be adopted as a potential indicator of wastewater reclamation system (Kitajima et al., 2014; Farkas et al., 2020b).
Coronaviruses
Coronaviruses (CoVs) are positive-sense single-stranded RNA viruses (Qu et al., 2020; Yeo et al., 2020) with more than 30 different species and have the biggest genome of RNA viruses of 30 kb (Amirian, 2020; Amoah et al., 2020). Coronaviruses are classified into four types: Alphacoronavirus (Alpha-CoV), Betacoronavirus (Beta-CoV), Gammacoronavirus (Gamma-CoV), and Deltacoronavirus (Delta-CoV; Delta-CoV; Qu et al., 2020). The coronavirus virion is typically spherical, with a diameter of 60–140 nm, and is enclosed by an outer viral envelope covered by projections (9–12 nm; Zhu et al., 2020). CoVs were thought to be insignificant human pathogens until the beginning of this century. The latest outbreak in Wuhan, China toward the end of 2019 was the emergence of a novel coronavirus (Lu et al., 2020). It was initially referred to as HCoV-19 but later officially named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2); because the novel virus is most closely linked to the SARS-CoV virus that was implicated in the 2003 SARS outbreak (Lu et al., 2020; World Health Organization, 2020).
The SARS-CoV-2 is responsible for a variety of common cold-like and acute respiratory diseases (Qu et al., 2020; Yeo et al., 2020). COVID-19 is the respiratory disease induced by SARS-CoV-2 (World Health Organization, 2020), with clinical symptoms including diarrhea, nausea, fever, cough, rhinorrhea, dyspnea, or severe pneumonia and myalgia (Guan et al., 2020; Liu et al., 2020; Yeo et al., 2020). Nevertheless, a considerable proportion of individuals remain symptomless despite testing positive for SARS-CoV-2 (Bai et al., 2020; Lai et al., 2020; Liu et al., 2020; Rothe et al., 2020). Evidence from the present COVID-19 outbreak proposes that about 2–35% of patients develop gastrointestinal (GI) symptoms such as diarrhea, abdominal discomfort, and vomiting; however, this is less common than respiratory symptoms (Yeo et al., 2020; Wang et al., 2020a), and this has resulted in the presence of the virus in feces and sewage (Amirian, 2020; Pan et al., 2020). Therefore, this virus has been detected in untreated wastewater and rivers (La Rosa et al., 2020; Medema et al., 2020; Rimoldi et al., 2020; Sherchan et al., 2020; Pillay et al., 2021), as well as in treated wastewaters (Haramoto et al., 2020; Randazzo et al., 2020).
Prevalence of enteric viruses in freshwater and wastewater sources
Critical tools for detection and prevention of the further spread of diseases and outbreaks include clinical surveillance and monitoring of waterborne pathogens. However, disease prevalence is under-reported through clinical testing because it is typically restricted to those who are ill to the extent of seeking treatment and testing (Cacciò and Chalmers, 2016). Hence, there is a lag indicator for predicting an outbreak in a community. Therefore, there is a desperate need for cost-effective and improved monitoring methods to detect numerous waterborne diseases in a community, preferably in real-time. To this end, wastewater-based epidemiology (WBE) has recently garnered much attention as an early warning technique for a range of waterborne infectious diseases (Zahedi et al., 2021).
Wastewater-based epidemiology (WBE) has been employed before now to monitor drug abuse within a community (Castiglioni et al., 2006) and other chemical pollutants (Choi et al., 2018), as well as the prevalence of poliovirus (Pöyry et al., 1988; Berchenko et al., 2017). Broadly, it entails the detection of nucleic acids or other biomarkers excreted in feces and urine in wastewater to offer detailed health information about a community (Mao et al., 2020). The approaches encompass the detection of pathogens released in saliva, sputum, mucus, vomitus, and phlegm that are frequently trapped in wastewater (Zahedi et al., 2021). Accordingly, WBE is thus equal to the collection and analysis of a large community-based combined sample of faeces, saliva, vomitus, sputum, urine, shed skin, and other substances released during personal cleansing, washing, bathing, and excretion. Hence, it provides a sensitive technique for tracking temporal alterations and variety in pathogen concentrations within a community (Xagoraraki and O’Brien, 2020). Another benefit of the direct analysis of wastewater samples is the presence of higher populations of pathogens in wastewater compared to environments where inadequately treated wastewater is discharged (Zahedi et al., 2021).
Wastewater passes through a series of treatment processes, including oxidation ponds, coagulation, activated sludge, chlorination, and ozonation until good effluent quality is achieved. However, about 50–90% of the waterborne viruses can only be removed via different treatment processes, thereby releasing a significantly high viral load into the environment that humans become exposed to with accompanying public health diseases (Gersberg et al., 1988; Zhu et al., 2005; Charles et al., 2008; Okoh et al., 2010; La Rosa et al., 2012; Sano et al., 2016). Supplementary Table 1 shows the prevalence of selected waterborne enteric viruses across different countries in various water environments, which portends the potential risk of the transmission of the virus. The employment of bacterial indicator species to assess the quantity of the virus load in wastewater effluents is one of such limitations. This has been faulted as an inefficient technique for monitoring the quality of wastewater (Gersberg et al., 1988; Hunter, 1997; Zhu et al., 2005; Bofill-Mas et al., 2006; Charles et al., 2008; Karmakar et al., 2008; Okoh et al., 2010; Farkas et al., 2020b).
The prevalence of HAdV in both 60 stool samples from children with acute gastroenteritis and 96 sewage samples collected from the Zenin wastewater treatment plant was investigated within a community in Egypt from January to December 2017 (Elmahdy et al., 2019). The virus was detected in 17 (28.3%) of stool, 27 (84.4%) of raw sewage, 16 (50%) of treated sewage, and 25 (78%) of sludge samples throughout a whole year of sample collection. According to Elmahdy et al. (2019), the occurrence of HAdV in the treated effluent of the WWTP portends a serious public health problem. Similarly, the prevalence of AdVs in treated wastewater in Brazil was reported by Quintão et al. (2021). The study reported the occurrence of AdVs in 27.2% (61/224) of the investigated samples. It was observed that the occurrence of the virus was higher in downstream samples than that in upstream samples. In another study, F species HAdVs serotype 41 (79.2%) and C species PAdVs serotype 5 (18.1%) were higher than other serotypes in the water samples collected from Puzi River, Taiwan. On the other hand, the prevalence of NoV GII was more than GI in the same river. Specifically, GII.4 (21.2%) and GII.17 (18.2%) were reported as the predominant genotypes. The occurrence of both AdVs and NoVs was higher in the winter compared to the spring, summer, and autumn seasons.
Furthermore, Iaconelli et al. (2017a), reported the prevalence of EV, AdV, HAV, HEV, and NoV in raw and treated wastewater. Adenovirus was the most frequently detected virus in raw wastewater (81%) compared to 33% in treated effluents, followed by EV, with 13 samples detected in raw sewage and 3 samples in treated effluents. Norovirus GI or GII were detected in 10 raw wastewater and 4 treated effluents. Likewise, HEV was detected in one sewage sample, while HAV was 33% in raw sewage and 19% in treated effluents (Iaconelli et al., 2017a). Conversely, Hepatitis A and E virus outbreaks were linked to contaminated drinking water in 72% (109/151) and 49 (38%) of the 128 outbreaks (Kumar et al., 2015). Also, Miao et al. (2018), assessed the prevalence of EV, RV, AstV, NoV GII, and AdV in Jinhe River, China. The detection frequency differs as follows; 91.7% for AdV, 81.3% for NoV GII, 79.2% for EV and AstV, and 70.8% for RV. In addition, the authors reported a seasonal pattern concerning the prevalence of the detected viruses in which there was an abundance of EVs in summer whereas RVs, AstVs, NoV GII, and AdVs showed opposite seasonal trends.
The prevalence of different astrovirus strains in different water matrices such as wastewater in the US, groundwater, and river in Nepal has also been reported (Hata et al., 2018). The identified strains include types 6 and 7 classical human astroviruses, emerging type 5 VA-astroviruses, and putative recombinants. The prevalence of classical and VA-astroviruses was reported during the cooler months while it was during the warmer months that MLB-astroviruses were detected (Hata et al., 2018). Aichivirus was found in Nepal in different water sources such as sewage pipes, rivers, groundwater, and a house that received its water from a tanker (Haramoto and Kitajima, 2017). Variations in AiV detection with a significant prevalence of AiV B were reported in the study. When compared to shallow tube wells, where AiV was detected in 1 out of 15 samples (7%), the frequency of AiV detection was substantially higher in shallow dug wells, where it was discovered in 10 out of 22 samples (45%). This variation could be attributed to the fragile structure of dug wells often composed of stone or brick compared to that of tube wells. Conversely, AiV was detected in 50% of river water samples examined in a study conducted in Iran (Azhdar et al., 2019). According to a survey carried out by Moreira and Bondelind (2017), Cryptosporidium, norovirus, Giardia, Campylobacter, and rotavirus were the waterborne pathogens responsible for drinking waterborne outbreaks during 2000–2014. Authors further reported that contamination of surface water sources affected most consumers which led to gastrointestinal diseases, while the distribution network was responsible for most individual incidents (Moreira and Bondelind, 2017).
Also, rotaviruses in surface water and wastewater have been reported in different parts of the world, especially in developing countries (Taylor et al., 2001; Verheyen et al., 2009; Rezaeinejad et al., 2014). Nevertheless, they are less often detected in environmental samples compared to AdVs (Spilki et al., 2013). The distribution of RV infection is seasonal, with the incidence reaching the peak during winter (Suzuki et al., 2005). However, it has been reported all year round worldwide (Osuolale and Okoh, 2016). The RV strains are linked with diseases like acute diarrhea in humans and animals (Almeida et al., 2018; Menezes et al., 2020). The transmission of rotavirus is majorly via the ingestion of food and water that has been contaminated with human wastes. In most cases, newborns or young children with gastroenteritis are infected with subclinical illness from an older sibling or mother. The shedding of the virus from the intestinal tract occurs before diarrhea sets in or even after it has been reported (Mukhopadhya et al., 2013).
During the global COVID-19 outbreak in Japan, Hata et al. (2021) investigated the presence of SARS-CoV-2 RNA in wastewater samples and the number of confirmed COVID-19 cases in the study area was compared. During the study period, a total of 45 influent samples were collected from five wastewater treatment plants in Ishikawa and Toyama regions in Japan. The virus was detected in 21 out of the 45 influent samples. There was an increase in the frequency of detection as the total number of confirmed cases in 100,000 people surpassed 10 in each region; however, SARS-CoV-2 RNA could still be detected even when there was a reduction in the number of confirmed cases. This could be attributed to the continuous shedding of the virus from discharged asymptomatic individuals. Hata et al. (2021), opined that the viral monitoring of wastewater could be adopted as an early warning signal of COVID-19 outbreaks in Japan.
Considering the occurrence of these viruses in water bodies, there is a need to carefully monitor treated wastewater before discharge; however, the employed methods have some drawbacks. The number of viruses contained in environmental media can vary significantly; however, it is a function of the type of sample being examined. High viral concentration could be detected and quantified in wastewater or sludge from treatment plants using a very small sample volume (Haramoto et al., 2018). Nevertheless, a significantly small volume which could be less than 1 ml that will be concentrated from the wastewater sample is required for the downstream detection experiments. Ultrafiltration, adsorption/elution, flocculation, and ultracentrifugation, among other techniques, have been utilized to concentrate viruses from wastewater (Prata et al., 2012; Haramoto et al., 2018). A dependable and appropriate method of concentration should have a capacity for high viral recovery, repeatable results, be suitable for detecting a wide array of viruses, create a minimal volume of viral concentrate, be rapid and cost-effective to operate (Bosch, 1998).
Unfortunately, all these criteria have not been found in any single technique for the concentration of viruses in water. This has made the concentration of viruses in water considerably more difficult. Hence, rapid action is necessary to track viruses in fresh and wastewater sources. Even though no strict standards have been set in addressing the concentration of viruses in treated wastewater effluents before disposal. Their elimination before discharge into environmental media is critical to minimize potential outbreaks and accompanying diseases due to exposure to humans (Haramoto et al., 2018). It is crucial to optimize and evaluate the available concentration techniques to improve virus recovery and build a highly effective process that will go a long way in combating virus outbreaks in the future hence, research should be geared toward achieving this goal (Ibrahim et al., 2021).
Public health implications of waterborne enteric viral diseases
The genomic content and capsid proteins of enteric viruses differ, nevertheless, they share some similar attributes making them more of public health threat regarding the risk of drinking contaminated water. The non-enveloped virus can remain active in water bodies for extended periods (Reynolds et al., 2008). Considering those attributes, drinking inadequately disinfected water contaminated with faeces could lead to waterborne disease outbreaks (Gall et al., 2015a; Sano et al., 2016; Adelodun et al., 2021). It is also noteworthy that viruses could be transmitted through three major routes; ingestion, inhalation, and direct contact via interaction with skin and eyes (swimming) leading to respiratory and ocular infections (Gall et al., 2015a).
Usually, waterborne enteric viruses are associated with gastrointestinal diseases, epidemics, and acute hepatitis. Adenovirus, AstVs, HAV, HEV, RVs, NoVs, and other caliciviruses, and enteroviruses, including coxsackieviruses and polioviruses are classified by WHO as waterborne viral pathogens with modest to high health importance (WHO, 2017c). These viruses are linked with gastroenteritis and diarrhea coupled with the severity of other symptoms such as fever and abdominal cramps among others. Table 2 shows the different waterborne enteric viruses and related diseases. Gastroenteritis is the most frequent pathology connected with enteric virus’ infections (Bányai et al., 2018). Various gastroenteritis outbreaks have been associated with drinking sewage-contaminated water containing some of these enteric viruses which include AdVs, EVs, NoVs, RVs, AstVs, AiVs among others (Maunula et al., 2009; Räsänen et al., 2010; Kauppinen et al., 2019). For instance, a minimum of 33 outbreaks were connected to drinking contaminated water between 2009 and 2014 in the United States of America (Control and Prevention, 2013). The most important cause of gastrointestinal illness across the world is human noroviruses. Due to increased environmental robustness, human noroviruses genogroup I is usually involved in waterborne cases (Matthews et al., 2012), while genogroup II with the large majority of cases is transmitted from person to person and presumed to be connected with food (Ahmed et al., 2014).
Likewise, diarrhea has also emerged as the world’s second-biggest cause of death in children under 5 years (Murray and Newby, 2012). It is one of the major recurrent waterborne infections, with 1.7 billion cases recorded each year (WHO, 2017a), and 525,000 children’s deaths (Pooi and Ng, 2018). According to the global burden of disease investigated in 2015, 1.2 million deaths and 71.7 million disability-adjusted life years (DALYs) coupled with 1.1 million deaths and 61.1 million DALYs from diarrheal diseases all resulted from contaminated water sources (Forouzanfar et al., 2016). The primary cause of diarrhea contracted by drinking contaminated water is bacteria; however, little or no attention is being paid to viral pathogens in water sources as well as their impact on public health. Gibney et al. (2017), conducted a study in 2010 on the disease burden of cryptosporidiosis, campylobacteriosis, giardiasis, nontyphoidal salmonellosis, and norovirus ascribed to the waterborne transmission of selected enteric pathogens in Australia. Most waterborne disease cases were linked to norovirus, a waterborne enteric virus (479,632; 95% confidence interval [UI]: 0–1,111,874), followed by giardiasis and campylobacteriosis. Based on a global study of groundwater-related enteric illness outbreaks survey, an alarming rise in groundwater-related acute gastrointestinal infections (AGI) was reported between 1948 and 2015, with 649 incidents identified (Murphy et al., 2017).
Recently, Carol et al. (2021) investigated the causative agent responsible for an outbreak of acute gastroenteritis (AGE) among 174 pupils that were involved in a school trip between 30 January and 3 February 2017 at a holiday camp in Catalonia. The authors discovered about 41 episodes of AGE with symptoms ranging from abdominal pain (73.8%), nausea (64.3%), vomiting (54.8%), diarrhea (45.2%), and headache (42.9%). The outbreak also was linked to the consumption of NoV GII contaminated water samples (crude RR: 1.72, 95% CI: 1.01–2.92; adjusted RR: 1.88, 95% CI 1.03–3.56). According to the World Health Organization/United Nations Children Emergency Fund, (2015) reports, untreated water is used by 663 million people worldwide. Although, individuals living in poverty-stricken or rural areas and developing regions are affected by a lack of access to safe drinking water. Notwithstanding, even people living in developed countries with advanced water and wastewater treatment facilities are not spared from waterborne diseases (Adelodun et al., 2021). Furthermore, it has been estimated that by the year 2030, approximately 1.6 billion people (19% of the global population) will lack clean water, leading to more waterborne outbreaks and illnesses (World Health Organization/United Nations Children Emergency Fund, 2021).
The Integrated Disease Surveillance Programme (IDSP) reported 804,782 hepatitis cases and 291 outbreaks in India between 2011 and 2013. Hepatitis A testing revealed 44,663 (7.4%) positive cases out of 599,605 total cases, and HEV testing revealed 19,508 (10.4%) positive cases out of 187,040 total cases. Hepatitis E virus accounted for 78 (48%) out of 163 (56%) outbreaks with known etiologies as hepatitis A accounted for 54 (33%), while hepatitis A and E accounted for 19 (12%), (Kumar et al., 2015). In a hepatitis A and E surveillance carried out between 2015 and 2017 in India, 23 disease outbreaks were reported of which 4 outbreaks occurred in 2015, 12 in 2016, and 7 in 2017 (Kadri et al., 2018). Twelve of the total outbreaks were related to hepatitis A infection, 10 to hepatitis E infection, while 1–8 cases of jaundice with no hepatitis A or hepatitis E virus were identified. During the study period, a total of 393 cases of hepatitis A or E were detected. Of the 50 water samples that were examined, 38 were unsuitable for human consumption hence the authors opined that both HAV and HEV outbreaks were due to human exposure to contaminated water (Kadri et al., 2018).
In addition, there was an increase in diarrhea cases reported in Mgcawu District, Northern Cape province, and eThekwini Metropolitan Municipality, KwaZulu-Natal province, South Africa (Shonhiwa et al., 2020). The reported diarrhea outbreaks that affected children under age 5 coincided with the yearly South Africa rotavirus season in 2013 (Shonhiwa et al., 2020). In the same vein, a gastroenteritis outbreak linked to swimming in the lagoon in KwaZulu-Natal Coast, South Africa, resulted in more than 600 cases within 3 weeks in December 2016/January 2017 (Sekwadi et al., 2018). Also, RVs, AstVs, NoVs GI.6, GII.3, and GII.6, were the waterborne viruses identified in the lagoon water samples. The prevalence of the viruses is due to the discharge from WWTPs effluent into the river that feeds the lagoon (Sekwadi et al., 2018). Similarly, in 2017, there was a rotavirus outbreak in India, with a 22.8% attack rate due to drinking water from contaminated wells (Joshi et al., 2019). The Hepatitis E virus was also linked with the largest viral waterborne outbreak in India in which about 80,000 people were affected (Naik et al., 1992; Farkas et al., 2020b).
With above 6 million deaths worldwide as of March 2022, COVID-19, the highly contagious infectious disease caused by SARS-CoV-2, had a devastating impact on the world’s demographics and is emerging as the most significant global health crisis since the period of the influenza pandemic of 1918 (Cascella et al., 2022). The COVID-19 pandemic has resulted in the emergence of significant health challenges owing to its contagious nature and the absence of efficient medical treatment (Coccia, 2021). Respiratory symptoms are the most common in COVID-19 individuals. However, research suggests that gastrointestinal (GI) symptoms such as diarrhea, nausea/vomiting, and abdominal discomfort are common in COVID-19 patients, with a frequency of up to 31.9% (Cholankeril et al., 2020; Remes-Troche et al., 2020). The possibility of the waterborne transmission of SARS-CoV-2 began to draw increasing attention due to the detection of its genetic marker in different water matrices as highlighted earlier. However, there is currently no scientific evidence that the viral RNA is infectious in water and wastewater and can be contacted in water. This gives room for more future studies to look comprehensively into this.
Apart from the loss of lives associated with the unavailability of safe drinking water and poor sanitation, the World Bank estimated an annual economic loss of US$260 billion globally (World Health Organization, 2012a). The infections associated with contaminated water and enteric viral outbreaks are considered under-reported irrespective of the country’s socio-economic condition because the symptoms are generally mild, and people rarely seek medical treatment for self-limiting illnesses (Control and Prevention, 2013). Generally, in healthy individuals, viral infections are self-limiting; however, in children under the age of five, the aged, pregnant women, and immunocompromised patients, greater morbidity could result. Hence, it makes the surveillance of diseases associated with enteric viruses challenging (Cortez et al., 2017; Li et al., 2017). Unfortunately, few broad-spectrum antiviral drugs exist for the treatment of those diseases (Gall et al., 2015a).
Strategies for the removal of waterborne viruses
Detecting and quantifying the various types of viruses in wastewater are critical for preventing diseases and creating strategic responses to outbreaks. Nevertheless, removing enteric viruses from wastewater is equally essential to avert their spread through food, water, or other pathways. However, the transmission of waterborne viruses is common in developing countries due to their poor sanitation. For instance, there has been a threat to global polio eradication. This is due to the possible movement of indigenous wild poliovirus (WPV) into polio-free countries. Also, it could be through the importation of WPV from polio-endemic countries as well as the release of WPV into the environment from laboratory stocks (Sharma et al., 2015). Hence, every country must detect WPV circulation and other waterborne viruses via a sensitive surveillance system.
Although, physical removal of pathogens via conventional methods, ultraviolet light or chemical oxidants like chlorine, chloramines, and ozone have been employed, unfortunately, the capacity to resist disinfection due to viral particle size (Ibrahim et al., 2021), and the presence of viral capsid make virus removal a challenging task for most available technologies as well as wastewater epidemiologist. Till date, chlorine disinfectant is being used for water treatment to enhance the deactivation of pathogens and maintain a residual concentration in the distribution line (Crittenden et al., 2012). The availability of chlorine made it a common disinfectant in water purification (Rosario-Ortiz et al., 2016); however, it raises the risk of creating possibly mutagenic and carcinogenic disinfection by-products like bromate and chlorite (Sharma et al., 2014; Gall et al., 2015b) that could pose major health risks to humans (Agency, 2006; Richardson and Postigo, 2011).
Generally, the side effects and limitations associated with the use of conventional techniques for the disinfection of water made it inappropriate in the long run. As highlighted, harmful disinfection by-products are formed when chlorine reacts with natural organic matter found in water sources (Hrudey and Charrois, 2012). The by-products released from the use of those conventional oxidants as disinfectants have been reported to be harmful to human health. Another danger for humans that is associated with the use of conventional disinfectants is their non-specificity as higher doses are needed for disinfection (Kumar et al., 2020). Specifically, studies have shown that the reoviruses may be more sensitive to chlorine disinfection than enteroviruses (Betancourt and Gerba, 2016). Unfortunately, attaining complete sterilization and disinfection has not been confirmed to be achieved by any conventional wastewater treatment procedures (Sano et al., 2016). Hence, finding solutions to reduce this problem has become an issue of keen interest. Therefore, research should be geared toward the development of an effective method of disinfection for the removal of waterborne enteric viruses. In addition, such methods must be devoid of the formation of disinfection by-products which characterizes the chemical methods of disinfection owing to the reaction of residual chlorine with organic matter to form possible mutagenic and carcinogenic by-products.
Conclusion
This article reviews the public health implications of human exposure to viral contaminated water. The presence of enteric viruses in water comes with accompanying public health implications that cannot be ignored. Although the presence of other microbes in wastewater has been the focus of research before now, the risk of diseases associated with the presence of viruses in water is by far greater than that of other microbes as just an insignificant quantity of viruses is adequate to cause diseases when compared to other microorganisms.
The detection and quantification of the various human viruses in environmental water are critical for public health concerning the prevention of diseases as well as response to outbreaks. Hence, conducting regular viral monitoring of treated wastewater discharged into the environment is important for the prevention of diseases associated with exposure to viral contaminated water. Meanwhile, the presence of viral particles in water and wastewater does not mean that the particles are infectious but are pointers to potential infections and health risk burdens that could emanate from the matrices. Hence, there is an urgent need for further studies to highlight infectious fractions of viral particles disseminated via wastewater and water resources for effective risk assessment and epidemiologic purposes. Nevertheless, removing viruses from wastewater using other alternative treatment methods in a bid to stop their transmission via the ingestion of contaminated water should be considered. Therefore, a search for a more holistic and cost-effective disinfection method that will inactivate waterborne viruses in water is highly needed to curtail the public health effects of human exposure to contaminated water.
Author contributions
AL, AE-F, and FS conceived the review idea. AL wrote the first draft. AL, AE-F, SS, and JE reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Water Research Commission (WRC) of South Africa (Project no. K5/C2020-2021-00181). We were also supported by our institution, the Durban University of Technology, South Africa.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.981896/full#supplementary-material
References
Abedi, G. R., Watson, J. T., Nix, W. A., Oberste, M. S., and Gerber, S. I. (2018). Enterovirus and parechovirus surveillance—United States, 2014–2016. Morb. Mortal. Wkly Rep. 67, 515–518. doi: 10.15585/mmwr.mm6718a2
Adams, M., King, A., and Carstens, E. (2013). Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2013). Arch. Virol. 158, 2023–2030. doi: 10.1007/s00705-013-1688-5
Adelodun, B., Ajibade, F. O., Ighalo, J. O., Odey, G., Ibrahim, R. G., Kareem, K. Y., et al. (2021). Assessment of socioeconomic inequality based on virus-contaminated water usage in developing countries: a review. Environ. Res. 192:110309. doi: 10.1016/j.envres.2020.110309
Agency, U. E. P. (2006). National primary drinking water regulations: stage 2 disinfectants and disinfection byproducts rule. Fed. Regist. 71, 387–493.
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J. W., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728:138764. doi: 10.1016/j.scitotenv.2020.138764
Ahmed, S. M., Hall, A. J., Robinson, A. E., Verhoef, L., Premkumar, P., Parashar, U. D., et al. (2014). Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 14, 725–730. doi: 10.1016/S1473-3099(14)70767-4
Akello, J. O., Kamgang, R., Barbani, M. T., Suter-Riniker, F., Leib, S. L., and Ramette, A. (2020). Epidemiology of human adenoviruses: a 20-year retrospective observational study in hospitalized patients in Bern, Switzerland. Clin. Epidemiol. 12, 353–366. doi: 10.2147/CLEP.S246352
Almeida, P. R., Lorenzetti, E., Cruz, R. S., Watanabe, T. T., Zlotowski, P., Alfieri, A. A., et al. (2018). Diarrhea caused by rotavirus A, B, and C in suckling piglets from southern Brazil: molecular detection and histologic and immunohistochemical characterization. J. Vet. Diagn. Investig. 30, 370–376. doi: 10.1177/1040638718756050
Altintas, Z., Gittens, M., Pocock, J., and Tothill, I. E. (2015). Biosensors for waterborne viruses: detection and removal. Biochimie 115, 144–154. doi: 10.1016/j.biochi.2015.05.010
Ambert-Balay, K., Lorrot, M., Bon, F., Giraudon, H., Kaplon, J., Wolfer, M., et al. (2008). Prevalence and genetic diversity of Aichi virus strain in stool samples from community and hospitalized patients. J. Clin. Microbiol. 46, 1252–1258. doi: 10.1128/JCM.02140-07
Amirian, E. S. (2020). Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 95, 363–370. doi: 10.1016/j.ijid.2020.04.057
Amoah, I. D., Kumari, S., and Bux, F. (2020). Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 143:105962. doi: 10.1016/j.envint.2020.105962
Appleton, H. (1975). Viruses and gastroenteritis in infants. Lancet 305:1297. doi: 10.1016/S0140-6736(75)92581-7
Arnold, A., and MacMahon, E. (2017). Adenovirus infections. Medicine 45, 777–780. doi: 10.1016/j.mpmed.2017.09.016
Assis, A. S. F., Fumian, T. M., Miagostovich, M. P., Drumond, B. P., and da Rosa e Silva, M. L. (2018). Adenovirus and rotavirus recovery from a treated effluent through an optimized skimmed-milk flocculation method. Environ. Sci. Pollut. Res. 25, 17025–17032. doi: 10.1007/s11356-018-1873-x
Atabakhsh, P., Kargar, M., and Doosti, A. (2021). Detection and evaluation of rotavirus surveillance methods as viral indicator in the aquatic environments. Braz. J. Microbiol. 52, 811–820. doi: 10.1007/s42770-020-00417-8
Azhdar, Z., Ghaderi, M., and Mousavi-Nasab, S. D. (2019). Optimization of RT-qPCR for detection of Aichi virus in sewage and river water samples in Karaj, Iran. Arch. Iran. Med. 22, 242–246.
Baggen, J., Thibaut, H. J., Strating, J. R., and van Kuppeveld, F. J. (2018). The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 16, 368–381. doi: 10.1038/s41579-018-0005-4
Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D. Y., Chen, L., et al. (2020). Presumed asymptomatic carrier transmission of COVID-19. JAMA 323, 1406–1407. doi: 10.1001/jama.2020.2565
Bányai, K., Estes, M. K., Martella, V., and Parashar, U. D. (2018). Viral gastroenteritis. Lancet 392, 175–186. doi: 10.1016/S0140-6736(18)31128-0
Benko, M., Élo, P., Ursu, K., Ahne, W., LaPatra, S. E., Thomson, D., et al. (2002). First molecular evidence for the existence of distinct fish and snake adenoviruses. J. Virol. 76, 10056–10059. doi: 10.1128/JVI.76.19.10056-10059.2002
Berchenko, Y., Manor, Y., Freedman, L. S., Kaliner, E., Grotto, I., Mendelson, E., et al. (2017). Estimation of polio infection prevalence from environmental surveillance data. Sci. Transl. Med. 9, 1–8. doi: 10.1126/scitranslmed.aaf6786
Bergallo, M., Galliano, I., Montanari, P., Rassu, M., and Daprà, V. (2017). Aichivirus in children with diarrhea in northern Italy. Intervirology 60, 196–200. doi: 10.1159/000487051
Bertrand, I., Schijven, J., Sánchez, G., Wyn-Jones, P., Ottoson, J., Morin, T., et al. (2012). The impact of temperature on the inactivation of enteric viruses in food and water: a review. J. Appl. Microbiol. 112, 1059–1074. doi: 10.1111/j.1365-2672.2012.05267.x
Betancourt, W. Q., and Gerba, C. P. (2016). Rethinking the significance of reovirus in water and wastewater. Food Environ. Virol. 8, 161–173. doi: 10.1007/s12560-016-9250-8
Beyer, S., Szewzyk, R., Gnirss, R., Johne, R., and Selinka, H. C. (2020). Detection and characterization of hepatitis E virus genotype 3 in wastewater and urban surface waters in Germany. Food Environ. Virol. 12, 137–147. doi: 10.1007/s12560-020-09424-2
Bofill-Mas, S., Albinana-Gimenez, N., Clemente-Casares, P., Hundesa, A., Rodriguez-Manzano, J., Allard, A., et al. (2006). Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72, 7894–7896. doi: 10.1128/AEM.00965-06
Boonchan, M., Motomura, K., Inoue, K., Ode, H., Chu, P. Y., Lin, M., et al. (2017). Distribution of norovirus genotypes and subtypes in river water by ultra-deep sequencing-based analysis. Lett. Appl. Microbiol. 65, 98–104. doi: 10.1111/lam.12750
Bortagaray, V., Girardi, V., Pou, S., Lizasoain, A., Tort, L. F. L., Spilki, F. R., et al. (2020). Detection, quantification, and microbial risk assessment of group A rotavirus in rivers from Uruguay. Food Environ. Virol. 12, 89–98. doi: 10.1007/s12560-019-09416-x
Bosch, A. (1998). Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1, 191–196.
Bouseettine, R., Hassou, N., Bessi, H., and Ennaji, M. M. (2020). “Waterborne transmission of enteric viruses and their impact on public health” in Emerging and Reemerging Viral Pathogens. ed. M. M. Ennaji (London: Elsevier), 907–932.
Cacciò, S., and Chalmers, R. (2016). Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 22, 471–480. doi: 10.1016/j.cmi.2016.04.021
Carol, M., Guadalupe-Fernández, V., Rius, C., Soldevila, N., Razquin, E., Guix, S., et al. (2021). A waterborne gastroenteritis outbreak caused by a GII Norovirus in a holiday camp in Catalonia (Spain), 2017. Viruses 13:1792. doi: 10.3390/v13091792
Cascella, M., Rajnik, M., Aleem, A., Dulebohn, S. C., and Di Napoli, R. (2022). Features, Evaluation, and Treatment of Coronavirus (COVID-19), United States: Statpearls Publishing.
Cassidy, H., Poelman, R., Knoester, M., Leer-Buter, V., Coretta, C., and Niesters, H. G. (2018). Enterovirus D68–the new polio? Front. Microbiol. 9:2677. doi: 10.3389/fmicb.2018.02677
Castiglioni, S., Zuccato, E., Crisci, E., Chiabrando, C., Fanelli, R., and Bagnati, R. (2006). Identification and measurement of illicit drugs and their metabolites in urban wastewater by liquid chromatography-tandem mass spectrometry. Anal. Chem. 78, 8421–8429. doi: 10.1021/ac061095b
Chan, M. C., Hu, Y., Chen, H., Podkolzin, A. T., Zaytseva, E. V., Komano, J., et al. (2017). Global spread of norovirus GII. 17 Kawasaki 308, 2014–2016. Emerg. Infect. Dis. 23, 1359–1364. doi: 10.3201/eid2308.161138
Chan, M. C., Lee, N., Hung, T. N., Kwok, K., Cheung, K., Tin, E. K., et al. (2015). Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII. 17 variant in late 2014. Nature. Communications 6, 1–9. doi: 10.1038/ncomms10061
Charles, K. J., Souter, F. C., Baker, D. L., Davies, C. M., Schijven, J. F., Roser, D. J., et al. (2008). Fate and transport of viruses during sewage treatment in a mound system. Water Res. 42, 3047–3056. doi: 10.1016/j.watres.2008.02.032
Chen, L., Deng, Y., Dong, S., Wang, H., Li, P., Zhang, H., et al. (2021). The occurrence and control of waterborne viruses in drinking water treatment: a review. Chemosphere 281:130728. doi: 10.1016/j.chemosphere.2021.130728
Chen, B. S., Lee, H. C., Lee, K. M., Gong, Y. N., and Shih, S. R. (2020). Enterovirus and encephalitis. Front. Microbiol. 11:261. doi: 10.3389/fmicb.2020.00261
Chitambar, S., Gopalkrishna, V., Chhabra, P., Patil, P., Verma, H., Lahon, A., et al. (2012). Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western India. Int. J. Environ. Res. Public Health 9, 895–915. doi: 10.3390/ijerph9030895
Choi, P. M., Tscharke, B. J., Donner, E., O'Brien, J. W., Grant, S. C., Kaserzon, S. L., et al. (2018). Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 105, 453–469. doi: 10.1016/j.trac.2018.06.004
Cholankeril, G., Podboy, A., Aivaliotis, V. I., Tarlow, B., Pham, E. A., Spencer, S. P., et al. (2020). High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology 159, 775–777. doi: 10.1053/j.gastro.2020.04.008
Cioffi, B., Ianiro, G., Iaccarino, D., D'Apice, F., Ferraro, A., Race, M., et al. (2021). A potential risk assessment tool to monitor pathogens circulation in coastal waters. Environ. Res. 200:111748. doi: 10.1016/j.envres.2021.111748
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
Coccia, M. (2021). Pandemic prevention: lessons from COVID-19. Encyclopedia 1, 433–444. doi: 10.3390/encyclopedia1020036
Control and Prevention (2013). Prevention and control of seasonal influenza with vaccines. Recommendations of the advisory committee on immunization practices--United States, 2013-2014. MMWR 62, 1–43.
Cooksey, E. M., Singh, G., Scott, L. C., and Aw, T. G. (2019). Detection of coliphages and human adenoviruses in a subtropical estuarine lake. Sci. Total Environ. 649, 1514–1521. doi: 10.1016/j.scitotenv.2018.08.322
Cordey, S., Brito, F., Vu, D.-L., Turin, L., Kilowoko, M., Kyungu, E., et al. (2016). Astrovirus VA1 identified by next-generation sequencing in a nasopharyngeal specimen of a febrile Tanzanian child with acute respiratory disease of unknown etiology. Emerg. Microb. Infect. 5, 1–3. doi: 10.1038/emi.2016.67
Cortez, V., Meliopoulos, V. A., Karlsson, E. A., Hargest, V., Johnson, C., and Schultz-Cherry, S. (2017). Astrovirus biology and pathogenesis. Ann. Rev. Virol. 4, 327–348. doi: 10.1146/annurev-virology-101416-041742
Crenshaw, B. J., Jones, L. B., Bell, C. R., Kumar, S., and Matthews, Q. L. (2019). Perspective on adenoviruses: epidemiology, pathogenicity, and gene therapy. Biomedicine 7:61. doi: 10.3390/biomedicines7030061
Crittenden, J. C., Trussell, R. R., Hand, D. W., Howe, K. J., and Tchobanoglous, G. (2012). MWH's Water Treatment: Principles and Design. United States: John Wiley & Sons.
De Benedictis, P., Schultz-Cherry, S., Burnham, A., and Cattoli, G. (2011). Astrovirus infections in humans and animals–molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 11, 1529–1544. doi: 10.1016/j.meegid.2011.07.024
de Graaf, M., van Beek, J., and Koopmans, M. P. (2016). Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 14, 421–433. doi: 10.1038/nrmicro.2016.48
Dias, J., Pinto, R. N., Vieira, C. B., and de Abreu Corrêa, A. (2018). Detection and quantification of human adenovirus (HAdV), JC polyomavirus (JCPyV) and hepatitis A virus (HAV) in recreational waters of Niterói, Rio de Janeiro, Brazil. Mar. Pollut. Bull. 133, 240–245. doi: 10.1016/j.marpolbul.2018.05.031
Donato, C., and Vijaykrishna, D. (2017). The broad host range and genetic diversity of mammalian and avian astroviruses. Viruses 9:102. doi: 10.3390/v9050102
Dongdem, J. T., Soyiri, I., and Ocloo, A. (2009). Public health significance of viral contamination of drinking water. Afr. J. Microbiol. Res. 3, 856–861. doi: 10.5897/AJMR.9000610
Elbashir, S., Parveen, S., Schwarz, J., Rippen, T., Jahncke, M., and DePaola, A. (2018). Seafood pathogens and information on antimicrobial resistance: a review. Food Microbiol. 70, 85–93. doi: 10.1016/j.fm.2017.09.011
Elmahdy, E. M., Ahmed, N. I., Shaheen, M. N., Mohamed, E. C. B., and Loutfy, S. A. (2019). Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health 17, 287–294. doi: 10.2166/wh.2019.303
Elmahdy, E. M., Shaheen, M. N., Rizk, N. M., and Saad-Hussein, A. (2020). Quantitative detection of human adenovirus and human rotavirus group A in wastewater and el-Rahawy drainage canal influencing River Nile in the north of Giza, Egypt. Food Environ. Virol. 12, 218–225. doi: 10.1007/s12560-020-09429-x
Esona, M. D., Mijatovic-Rustempasic, S., Conrardy, C., Tong, S., Kuzmin, I. V., Agwanda, B., et al. (2010). Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg. Infect. Dis. 16, 1844–1852. doi: 10.3201/eid1612.101089
Farkas, K., Cooper, D. M., McDonald, J. E., Malham, S. K., de Rougemont, A., and Jones, D. L. (2018). Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 634, 1174–1183. doi: 10.1016/j.scitotenv.2018.04.038
Farkas, K., Hillary, L. S., Malham, S. K., McDonald, J. E., and Jones, D. L. (2020a). Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr. Opinion Environ. Sci. Health 17, 14–20. doi: 10.1016/j.coesh.2020.06.001
Farkas, K., Walker, D. I., Adriaenssens, E. M., McDonald, J. E., Hillary, L. S., Malham, S. K., et al. (2020b). Viral indicators for tracking domestic wastewater contamination in the aquatic environment. Water Res. 181:115926. doi: 10.1016/j.watres.2020.115926
Fernandez-Cassi, X., Timoneda, N., Martínez-Puchol, S., Rusinol, M., Rodriguez-Manzano, J., Figuerola, N., et al. (2018). Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 618, 870–880. doi: 10.1016/j.scitotenv.2017.08.249
Fongaro, G., Silva, H. D. D., Ibrahim, E. M. E., Schissi, C. D., Moreira, M., Lanna, M. C. D. S., et al. (2015). Enteric viruses as contaminants and bioindicators in environmental samples. Virus Rev. Res. 20, 1–20. doi: 10.17525/vrrjournal.v20i2.255
Forouzanfar, M. H., Afshin, A., Alexander, L. T., Anderson, H. R., Bhutta, Z. A., Biryukov, S., et al. (2016). Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet 388, 1659–1724. doi: 10.1016/S0140-6736(16)31679-8
Fu, Y., Tang, Z., Ye, Z., Mo, S., Tian, X., Ni, K., et al. (2019). Human adenovirus type 7 infection causes a more severe disease than type 3. BMC Infect. Dis. 19, 36–11. doi: 10.1186/s12879-018-3651-2
Gall, A. M., Mariñas, B. J., Lu, Y., and Shisler, J. L. (2015a). Waterborne viruses: a barrier to safe drinking water. PLoS Pathog. 11:e1004867. doi: 10.1371/journal.ppat.1004867
Gall, A. M., Shisler, J. L., and Mariñas, B. J. (2015b). Analysis of the viral replication cycle of adenovirus serotype 2 after inactivation by free chlorine. Environ. Sci. Technol. 49, 4584–4590. doi: 10.1021/acs.est.5b00301
Gao, F., Wang, Y. P., Mao, Q. Y., Yao, X., Liu, S., Li, F. X., et al. (2012). Enterovirus 71 viral capsid protein linear epitopes: identification and characterization. Virol. J. 9, 1–7. doi: 10.1186/1743-422X-9-26
Gersberg, R., Lyon, S., Brenner, R., and Elkins, B. (1988). Performance of a clay-alum flocculation (CCBA) process for virus removal from municipal wastewater. Water Res. 22, 1449–1454. doi: 10.1016/0043-1354(88)90103-0
Gibney, K. B., O'Toole, J., Sinclair, M., and Leder, K. (2017). Burden of disease attributed to waterborne transmission of selected enteric pathogens, Australia, 2010. Am. J. Trop. Med. Hygiene 96, 1400–1403. doi: 10.4269/ajtmh.16-0907
Gibson, K. E. (2014). Viral pathogens in water: occurrence, public health impact, and available control strategies. Curr. Opin. Virol. 4, 50–57. doi: 10.1016/j.coviro.2013.12.005
Girardi, V., Demoliner, M., Rigotto, C., Schneider, V. E., Paesi, S., and Spilki, F. R. (2018). Assessment of diversity of adenovirus DNA polymerase gene in recreational waters facilitated by ultracentrifugal concentration. J. Water Health 16, 102–111. doi: 10.2166/wh.2017.144
Goh, S. G., Saeidi, N., Gu, X., Vergara, G. G. R., Liang, L., Fang, H., et al. (2019). Occurrence of microbial indicators, pathogenic bacteria and viruses in tropical surface waters subject to contrasting land use. Water Res. 150, 200–215. doi: 10.1016/j.watres.2018.11.058
Gonzales-Gustavson, E., Rusiñol, M., Medema, G., Calvo, M., and Girones, R. (2019). Quantitative risk assessment of norovirus and adenovirus for the use of reclaimed water to irrigate lettuce in Catalonia. Water Res. 153, 91–99. doi: 10.1016/j.watres.2018.12.070
Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720. doi: 10.1056/NEJMoa2002032
Habib, F. B., Rahman, M. M., Choudhury, R., Ahmed, N., Chowdhury, S., Tanni, N. N., et al. (2021). Relation between water source and Rotaviral infection: An observational study. Archiv. Microbiol. Immunol. 5, 353–361. doi: 10.26502/ami.93650068
Haramoto, E., and Kitajima, M. (2017). Quantification and genotyping of Aichi virus 1 in water samples in the Kathmandu Valley, Nepal. Food Environ. Virol. 9, 350–353. doi: 10.1007/s12560-017-9283-7
Haramoto, E., Kitajima, M., Hata, A., Torrey, J. R., Masago, Y., Sano, D., et al. (2018). A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 135, 168–186. doi: 10.1016/j.watres.2018.02.004
Haramoto, E., Malla, B., Thakali, O., and Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 737:140405. doi: 10.1016/j.scitotenv.2020.140405
Hassou, N., Bouseettine, R., Abouchoaib, N., and Ennaji, M. M. (2020). “Enteric adenoviruses: emerging of a public health threat” in Emerging and Reemerging Viral Pathogens. ed. M. M. Ennaji (London: Elsevier), 879–905.
Hata, A., Hara-Yamamura, H., Meuchi, Y., Imai, S., and Honda, R. (2021). Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 758:143578. doi: 10.1016/j.scitotenv.2020.143578
Hata, A., Kitajima, M., Haramoto, E., Lee, S., Ihara, M., Gerba, C. P., et al. (2018). Next-generation amplicon sequencing identifies genetically diverse human astroviruses, including recombinant strains, in environmental waters. Sci. Rep. 8, 11837–11839. doi: 10.1038/s41598-018-30217-y
Health Canada (2019). Guidelines for Canadian Drinking Water Quality: Guideline Technical Document—Enteric Viruses. Water and Air Quality Bureau, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario.
Hendriksen, R. S., Lukjancenko, O., Munk, P., Hjelmsø, M. H., Verani, J. R., Ng’eno, E., et al. (2019). Pathogen surveillance in the informal settlement, Kibera, Kenya, using a metagenomics approach. PLoS One 14:e0222531. doi: 10.1371/journal.pone.0222531
Hewitt, J., Greening, G. E., Leonard, M., and Lewis, G. D. (2013). Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res. 47, 6750–6761. doi: 10.1016/j.watres.2013.09.001
Hoa-Tran, T., Nakagomi, O., Dao, A., Nguyen, A., Agbemabiese, C., Vu, H., et al. (2017). Molecular epidemiology of noroviruses detected in Vietnamese children with acute gastroenteritis from 2012 to 2015. J. Med. Microbiol. 66, 34–45. doi: 10.1099/jmm.0.000417
Howley, P. M., and Lowy, D. (2007). “Papillomaviridae” in Fields Virology. eds. D. M. Knipe and P. M. Howley (Philadelphia: Lippincott Williams and Wilkins).
Hrudey, S. E., and Charrois, J. W. (2012). Disinfection By-Products and Human Health. United Kingdom: IWA publishing.
Iaconelli, M., Muscillo, M., Della Libera, S., Fratini, M., Meucci, L., De Ceglia, M., et al. (2017a). One-year surveillance of human enteric viruses in raw and treated wastewaters, downstream river waters, and drinking waters. Food Environ. Virol. 9, 79–88. doi: 10.1007/s12560-016-9263-3
Iaconelli, M., Purpari, G., Della Libera, S., Petricca, S., Guercio, A., Ciccaglione, A., et al. (2015). Hepatitis A and E viruses in wastewaters, in river waters, and in bivalve molluscs in Italy. Food Environ. Virol. 7, 316–324. doi: 10.1007/s12560-015-9207-3
Iaconelli, M., Valdazo-González, B., Equestre, M., Ciccaglione, A., Marcantonio, C., Della Libera, S., et al. (2017b). Molecular characterization of human adenoviruses in urban wastewaters using next generation and sanger sequencing. Water Res. 121, 240–247. doi: 10.1016/j.watres.2017.05.039
Ibrahim, C., Hammami, S., Mejri, S., Mehri, I., Pothier, P., and Hassen, A. (2017a). Detection of Aichi virus genotype B in two lines of wastewater treatment processes. Microb. Pathog. 109, 305–312. doi: 10.1016/j.micpath.2017.06.001
Ibrahim, C., Mehri, I., Hammami, S., Mejri, S., Hassen, A., and Pothier, P. (2017b). Removal of human astroviruses from hospital wastewater by two biological treatment methods: natural oxidizing lagoons and rotating biodisks. Desalin. Water Treat. 89, 287–296. doi: 10.5004/dwt.2017.21356
Ibrahim, Y., Ouda, M., Kadadou, D., Banat, F., Naddeo, V., Alsafar, H., et al. (2021). Detection and removal of waterborne enteric viruses from wastewater: a comprehensive review. J. Environ. Chem. Eng. 9:105613. doi: 10.1016/j.jece.2021.105613
Jahne, M. A., Brinkman, N. E., Keely, S. P., Zimmerman, B. D., Wheaton, E. A., and Garland, J. L. (2020). Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: implications for onsite reuse. Water Res. 169:115213. doi: 10.1016/j.watres.2019.115213
Janahi, E. M., Mustafa, S., Parkar, S. F., Naser, H. A., and Eisa, Z. M. (2020). Detection of enteric viruses and bacterial indicators in a sewage treatment center and shallow Water Bay. Int. J. Environ. Res. Public Health 17:6483. doi: 10.3390/ijerph17186483
Japhet, M., Famurewa, O., Adesina, O., Opaleye, O., Wang, B., Höhne, M., et al. (2019). Viral gastroenteritis among children of 0-5 years in Nigeria: characterization of the first Nigerian aichivirus, recombinant noroviruses and detection of a zoonotic astrovirus. J. Clin. Virol. 111, 4–11. doi: 10.1016/j.jcv.2018.12.004
Jiang, S., Noble, R., and Chu, W. (2001). Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67, 179–184. doi: 10.1128/AEM.67.1.179-184.2001
Joshi, M. S., Lole, K. S., Barve, U. S., Salve, D. S., Ganorkar, N. N., Chavan, N. A., et al. (2019). Investigation of a large waterborne acute gastroenteritis outbreak caused by group B rotavirus in Maharashtra state, India. J. Med. Virol. 91, 1877–1881. doi: 10.1002/jmv.25523
Jothikumar, N., Cromeans, T., Sobsey, M., and Robertson, B. (2005). Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 71, 3359–3363. doi: 10.1128/AEM.71.6.3359-3363.2005
Kadri, S. M., Rehana, K., Benetou, D. R., Ahmad, D. F., Abdullah, A., and Chattu, V. K. (2018). Hepatitis A and E outbreak surveillance during 2015–2017 in Kashmir, India: is the water to blame? J. Epidemiol. Global Health 8, 203–207. doi: 10.2991/j.jegh.2018.04.101
Kaikkonen, S., Räsänen, S., Rämet, M., and Vesikari, T. (2010). Aichi virus infection in children with acute gastroenteritis in Finland. Epidemiol. Infect. 138, 1166–1171. doi: 10.1017/S0950268809991300
Karmakar, S., Rathore, A. S., Kadri, S. M., Dutt, S., Khare, S., and Lal, S. (2008). Post-earthquake outbreak of rotavirus gastroenteritis in Kashmir (India): An epidemiological analysis. Public Health 122, 981–989. doi: 10.1016/j.puhe.2008.01.006
Kauppinen, A., Pitkänen, T., Al-Hello, H., Maunula, L., Hokajärvi, A. M., Rimhanen-Finne, R., et al. (2019). Two drinking water outbreaks caused by wastewater intrusion including Sapovirus in Finland. Int. J. Environ. Res. Public Health 16:4376. doi: 10.3390/ijerph16224376
Kebe, O., Fernandez-Garcia, M. D., Fall, A., Dia, H., Bidalot, M., Ambert-Balay, K., et al. (2021). Prevalence and genetic diversity of Aichi virus 1 from urban wastewater in Senegal. Intervirology 64, 96–101. doi: 10.1159/000512130
Keller, R., Pratte-Santos, R., Scarpati, K., Martins, S. A., Loss, S. M., Fumian, T. M., et al. (2019). Surveillance of enteric viruses and thermotolerant coliforms in surface water and bivalves from a mangrove estuary in southeastern Brazil. Food Environ. Virol. 11, 288–296. doi: 10.1007/s12560-019-09391-3
Khanal, S., Ghimire, P., and Dhamoon, A. S. (2018). The repertoire of adenovirus in human disease: the innocuous to the deadly. Biomedicine 6:30. doi: 10.3390/biomedicines6010030
Khuroo, M., and Kamili, S. (2003). Aetiology, clinical course and outcome of sporadic acute viral hepatitis in pregnancy. J. Viral Hepat. 10, 61–69. doi: 10.1046/j.1365-2893.2003.00398.x
Kim, A. H., Hogarty, M. P., Harris, V. C., and Baldridge, M. T. (2021). The complex interactions between rotavirus and the gut microbiota. Front. Cell. Infect. Microbiol. 10:586751. doi: 10.3389/fcimb.2020.586751
Kitajima, M., and Gerba, C. P. (2015). Aichi virus 1: environmental occurrence and behavior. Pathogens 4, 256–268. doi: 10.3390/pathogens4020256
Kitajima, M., Iker, B. C., Pepper, I. L., and Gerba, C. P. (2014). Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci. Total Environ. 488-489, 290–296. doi: 10.1016/j.scitotenv.2014.04.087
Kittigul, L., Rupprom, K., Che-arsae, M., Pombubpa, K., Thongprachum, A., Hayakawa, S., et al. (2019). Occurrence of noroviruses in recycled water and sewage sludge: emergence of recombinant norovirus strains. J. Appl. Microbiol. 126, 1290–1301. doi: 10.1111/jam.14201
Kiulia, N. M., Gonzalez, R., Thompson, H., Aw, T. G., and Rose, J. B. (2021). Quantification and trends of rotavirus and Enterovirus in untreated sewage using reverse transcription droplet digital PCR. Food Environ. Virol. 13, 154–169. doi: 10.1007/s12560-020-09455-9
Koopmans, M., von Bonsdorff, C. H., Vinjé, J., de Medici, D., and Monroe, S. (2002). Foodborne viruses. FEMS Microbiol. Rev. 26, 187–205. doi: 10.1111/j.1574-6976.2002.tb00610.x
Kosulin, K. (2019). Intestinal HAdV infection: tissue specificity, persistence, and implications for antiviral therapy. Viruses 11:804. doi: 10.3390/v11090804
Kumar, S., Ahlawat, W., Bhanjana, G., Heydarifard, S., Nazhad, M. M., and Dilbaghi, N. (2014). Nanotechnology-based water treatment strategies. J. Nanosci. Nanotechnol. 14, 1838–1858. doi: 10.1166/jnn.2014.9050
Kumar, S., Gupta, A. K., Maurya, A., and Singh, M. K. (2020). “Chemical treatment for removal of waterborne pathogens,” in Waterborne pathogens. eds. M. N. Prasad and A. Grobelak (United Kingdom: Elsevier), 205–218.
Kumar, T., Shrivastava, A., Kumar, A., Laserson, K. F., Narain, J. P., Venkatesh, S., et al. (2015). Viral hepatitis surveillance—India, 2011–2013. Morb. Mortal. Wkly Rep. 64, 758–762. doi: 10.15585/mmwr.mm6428a3
La Rosa, G., Fontana, S., Di Grazia, A., Iaconelli, M., Pourshaban, M., and Muscillo, M. (2007). Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 73, 4152–4161. doi: 10.1128/AEM.00222-07
La Rosa, G., Fratini, M., Della Libera, S., Iaconelli, M., and Muscillo, M. (2012). Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanita 48, 397–406. doi: 10.4415/ANN_12_04_07
La Rosa, G., Iaconelli, M., Mancini, P., Ferraro, G. B., Veneri, C., Bonadonna, L., et al. (2020). First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 736:139652. doi: 10.1016/j.scitotenv.2020.139652
La Rosa, G., Iaconelli, M., Pourshaban, M., and Muscillo, M. (2010a). Detection and molecular characterization of noroviruses from five sewage treatment plants in Central Italy. Water Res. 44, 1777–1784. doi: 10.1016/j.watres.2009.11.055
La Rosa, G., Pourshaban, M., Iaconelli, M., and Muscillo, M. (2008). Recreational and drinking waters as a source of norovirus gastroenteritis outbreaks: a review and update. Environ. Biotechnol. 1, 15–22. doi: 10.1007/s12560-008-9002-5
La Rosa, G., Pourshaban, M., Iaconelli, M., and Muscillo, M. (2010b). Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Ann. Ist. Super. Sanita 46, 266–273. doi: 10.4415/ANN_10_03_07
La Rosa, G., Proroga, Y., De Medici, D., Capuano, F., Iaconelli, M., Della Libera, S., et al. (2018). First detection of hepatitis E virus in shellfish and in seawater from production areas in southern Italy. Food Environ. Virol. 10, 127–131. doi: 10.1007/s12560-017-9319-z
Laconi, A., Cavicchio, L., Tassoni, L., Cunial, G., Milani, A., Ustulin, M., et al. (2020). Identification of two divergent swine Noroviruses detected at the slaughterhouse in north East Italy. Porcine Health Manag. 6, 9–6. doi: 10.1186/s40813-020-00147-1
Lai, C. C., Liu, Y. H., Wang, C. Y., Wang, Y. H., Hsueh, S. C., Yen, M. Y., et al. (2020). Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 53, 404–412. doi: 10.1016/j.jmii.2020.02.012
Lázaro, G. R., Mukhopadhyay, S., and Hagan, M. F. (2018). Why enveloped viruses need cores—the contribution of a nucleocapsid core to viral budding. Biophys. J. 114, 619–630. doi: 10.1016/j.bpj.2017.11.3782
Lee, H. F., and Chi, C. S. (2014). Enterovirus 71 infection–associated acute flaccid paralysis: a case series of long-term neurologic follow-up. J. Child Neurol. 29, 1283–1290. doi: 10.1177/0883073813516193
Leite, J. P. G., Carvalho-Costa, F. A., and Linhares, A. C. (2008). Group A rotavirus genotypes and the ongoing Brazilian experience: a review. Mem. Inst. Oswaldo Cruz 103, 745–753. doi: 10.1590/S0074-02762008000800001
Lemon, S. M., Ott, J. J., Van Damme, P., and Shouval, D. (2018). Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 68, 167–184. doi: 10.1016/j.jhep.2017.08.034
Li, L. L., Liu, N., Yu, J. M., Ao, Y. Y., Li, S., Stine, O. C., et al. (2017). Analysis of Aichi virus and Saffold virus association with pediatric acute gastroenteritis. J. Clin. Virol. 87, 37–42. doi: 10.1016/j.jcv.2016.12.003
Liu, R., Ma, Q., Han, H., Su, H., Liu, F., Wu, K., et al. (2020). The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clin. Chem. Lab. Med. 58, 1121–1124. doi: 10.1515/cclm-2020-0220
Lizasoain, A., Burlandy, F. M., Victoria, M., Tort, L. F. L., da Silva, E. E., and Colina, R. (2018). An environmental surveillance in Uruguay reveals the presence of highly divergent types of human enterovirus species C and a high frequency of species a and B types. Food Environ. Virol. 10, 343–352. doi: 10.1007/s12560-018-9351-7
Lodder, W. J., Rutjes, S. A., Takumi, K., and de Roda Husman, A. M. (2013). Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 19, 1222–1230. doi: 10.3201/eid1908.130312
Lu, J., Sun, L., Fang, L., Yang, F., Mo, Y., Lao, J., et al. (2015). Gastroenteritis outbreaks caused by norovirus GII. 17, Guangdong Province, China, 2014–2015. Emerg. Infect. Dis. 21, 1240–1242. doi: 10.3201/eid2107.150226
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Lugo, D., and Krogstad, P. (2016). Enteroviruses in the early 21st century: new manifestations and challenges. Curr. Opin. Pediatr. 28, 107–113. doi: 10.1097/MOP.0000000000000303
Magana-Arachchi, D., and Wanigatunge, R. (2020). “Ubiquitous waterborne pathogens” in Waterborne Pathogens. eds. M. N. Prasad and A. Grobelak (United Kingdom: Elsevier), 15–42.
Malla, B., Ghaju Shrestha, R., Tandukar, S., Bhandari, D., Thakali, O., Sherchand, J. B., et al. (2019). Detection of pathogenic viruses, pathogen indicators, and fecal-source markers within tanker water and their sources in the Kathmandu Valley, Nepal. Pathogens 8:81. doi: 10.3390/pathogens8020081
Mancosu, N., Snyder, R. L., Kyriakakis, G., and Spano, D. (2015). Water scarcity and future challenges for food production. WaterSA 7, 975–992. doi: 10.3390/w7030975
Mans, J., Armah, G. E., Steele, A. D., and Taylor, M. B. (2016). Norovirus epidemiology in Africa: a review. PLoS One 11:e0146280. doi: 10.1371/journal.pone.0146280
Mao, K., Zhang, K., Du, W., Ali, W., Feng, X., and Zhang, H. (2020). The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opinion Environ. Sci. Health 17, 1–7. doi: 10.1016/j.coesh.2020.04.006
Marek, A., Bilic, I., Prokofieva, I., and Hess, M. (2010). Phylogenetic analysis of avian hepatitis E virus samples from European and Australian chicken flocks supports the existence of a different genus within the Hepeviridae comprising at least three different genotypes. Vet. Microbiol. 145, 54–61. doi: 10.1016/j.vetmic.2010.03.014
Marie, V., and Lin, J. (2017). Viruses in the environment–presence and diversity of bacteriophage and enteric virus populations in the Umhlangane River, Durban, South Africa. J. Water Health 15, 966–981. doi: 10.2166/wh.2017.066
Masachessi, G., Pisano, M. B., Prez, V. E., Martínez, L., Michelena, J., Martínez-Wassaf, M., et al. (2018). Enteric viruses in surface waters from Argentina: molecular and viable-virus detection. Appl. Environ. Microbiol. 84, e02327–e02317. doi: 10.1128/AEM.02327-17
Matthews, J., Dickey, B., Miller, R., Felzer, J., Dawson, B., Lee, A., et al. (2012). The epidemiology of published norovirus outbreaks: a review of risk factors associated with attack rate and genogroup. Epidemiol. Infect. 140, 1161–1172. doi: 10.1017/S0950268812000234
Maunula, L., Klemola, P., Kauppinen, A., Söderberg, K., Nguyen, T., Pitkänen, T., et al. (2009). Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ. Virol. 1, 31–36. doi: 10.1007/s12560-008-9004-3
McCall, C., Wu, H., Miyani, B., and Xagoraraki, I. (2020). Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 184:116160. doi: 10.1016/j.watres.2020.116160
McKnight, K. L., and Lemon, S. M. (2018). Hepatitis A virus genome organization and replication strategy. Cold Spring Harb. Perspect. Med. 8:a033480. doi: 10.1101/cshperspect.a033480
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., and Brouwer, A. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 7, 511–516. doi: 10.1021/acs.estlett.0c00357
Meixell, B. W., Borchardt, M. A., and Spencer, S. K. (2013). Accumulation and inactivation of avian inluenza virus by filter-feeding invertebrate Daphnia magna. Appl. Environ. Microbiol. 79, 7249–7255. doi: 10.1128/AEM.02439-13
Mena, K. D., and Gerba, C. P. (2008). Waterborne adenovirus. Rev. Environ. Contam. Toxicol. 133-167. doi: 10.1007/978-0-387-09647-6_4
Menezes, P. Q., Silva, T. T., Simas, F. B., Brauner, R. K., Bandarra, P., Demoliner, M., et al. (2020). Molecular detection of human adenovirus and rotavirus in Faeces of white-eared opossums. EcoHealth 17, 326–332. doi: 10.1007/s10393-020-01497-6
Mennechet, F. J., Paris, O., Ouoba, A. R., Salazar Arenas, S., Sirima, S. B., Takoudjou Dzomo, G. R., et al. (2019). A review of 65 years of human adenovirus seroprevalence. Expert Rev. Vaccines 18, 597–613. doi: 10.1080/14760584.2019.1588113
Meqdam, M. M., and Thwiny, I. R. (2007). Prevalence of group a rotavirus, enteric adenovirus, norovirus and astrovirus infections among children with acute gastroenteritis in Al-Qassim, Saudi Arabia. Pakistan J. Med. Sci. 23:551.
Miao, J., Guo, X., Liu, W., Yang, D., Shen, Z., Qiu, Z., et al. (2018). Total coliforms as an indicator of human enterovirus presence in surface water across Tianjin city. BMC Infect. Dis. 18, 1–9. doi: 10.1186/s12879-018-3438-5
Miura, T., Gima, A., and Akiba, M. (2019). Detection of norovirus and rotavirus present in suspended and dissolved forms in drinking water sources. Food Environ. Virol. 11, 9–19. doi: 10.1007/s12560-018-9361-5
Moazeni, M., Nikaeen, M., Hadi, M., Moghim, S., Mouhebat, L., Hatamzadeh, M., et al. (2017). Estimation of health risks caused by exposure to enteroviruses from agricultural application of wastewater effluents. Water Res. 125, 104–113. doi: 10.1016/j.watres.2017.08.028
Moreira, N. A., and Bondelind, M. (2017). Safe drinking water and waterborne outbreaks. J. Water Health 15, 83–96. doi: 10.2166/wh.2016.103
Moreno, S., Alvarado, M. V., Bermúdez, A., and Gutiérrez, M. F. (2009). DNA sequence analysis indicates human origin of rotavirus and hepatitis A virus strains from western Colombia. Biomedica 29, 209–217. doi: 10.7705/biomedica.v29i2.23
Mukhopadhya, I., Sarkar, R., Menon, V. K., Babji, S., Paul, A., Rajendran, P., et al. (2013). Rotavirus shedding in symptomatic and asymptomatic children using reverse transcription-quantitative PCR. J. Med. Virol. 85, 1661–1668. doi: 10.1002/jmv.23641
Murphy, H. M., Prioleau, M. D., Borchardt, M. A., and Hynds, P. D. (2017). Review: epidemiological evidence of groundwater contribution to global enteric disease, 1948–2015. Hydrogeol. J. 25, 981–1001. doi: 10.1007/s10040-017-1543-y
Murray, C., and Newby, H. (2012). Data resource profile: United Nations children’s fund (UNICEF). Int. J. Epidemiol. 41, 1595–1601. doi: 10.1093/ije/dys185
Naik, S., Aggarwal, R., Salunke, P., and Mehrotra, N. (1992). A large waterborne viral hepatitis E epidemic in Kanpur, India. Bull. World Health Organ. 70, 597–604.
Nasiri, M., Ghalejoogh, Z. Y., Ataei-Pirkooh, A., Bokharaei-Salim, F., Monavari, S. H., Tavakoli, A., et al. (2021). Detection and phylogenetic analysis of hepatitis A virus in the wastewater treatment Plant of Ekbatan Town in Tehran, Iran. Hepat. Mon. 21:121270. doi: 10.5812/hepatmon.121270
Nimgaonkar, I., Ding, Q., Schwartz, R. E., and Ploss, A. (2018). Hepatitis E virus: advances and challenges. Nat. Rev. Gastroenterol. Hepatol. 15, 96–110. doi: 10.1038/nrgastro.2017.150
Nour, I., Hanif, A., Zakri, A. M., Al-Ashkar, I., Alhetheel, A., and Eifan, S. (2021). Human adenovirus molecular characterization in various water environments and seasonal impacts in Riyadh, Saudi Arabia. Int. J. Environ. Res. Public Health 18:4773. doi: 10.3390/ijerph18094773
Oh, D. Y., Silva, P., Hauroeder, B., Diedrich, S., Cardoso, D., and Schreier, E. (2006). Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 151, 1199–1206. doi: 10.1007/s00705-005-0706-7
Okada, A., Kobayashi, S., and Inoshima, Y. (2019). Detection frequency of porcine noroviruses in healthy pigs in Japan. Jpn. Agric. Res. Q. 53, 305–310. doi: 10.6090/jarq.53.305
Okoh, A. I., Sibanda, T., and Gusha, S. S. (2010). Inadequately treated wastewater as a source of human enteric viruses in the environment. Int. J. Environ. Res. Public Health 7, 2620–2637. doi: 10.3390/ijerph7062620
Onosi, O., Upfold, N. S., Jukes, M. D., Luke, G. A., and Knox, C. (2019). The first molecular detection of Aichi virus 1 in raw sewage and mussels collected in South Africa. Food Environ. Virol. 11, 96–100. doi: 10.1007/s12560-018-9362-4
Opere, W. M. (2019). Detection of Pathogenic Human Adenoviruses in water Samples Collected from Lake Victoria along Homa Bay Town, Homa Bay Country: Kenya Kenyatta University
Opere, W. M., John, M., and Ombori, O. (2021). Molecular detection of human enteric adenoviruses in water samples collected from Lake Victoria waters Along Homa Bay town, Homa Bay County, Kenya. Food Environ. Virol. 13, 32–43. doi: 10.1007/s12560-020-09444-y
Oppong, T., Yang, H., Amponsem-Boateng, C., Kyere, E. D., Abdulai, T., Duan, G., et al. (2020). Enteric pathogens associated with gastroenteritis among children under 5 years in sub-Saharan Africa: a systematic review and meta-analysis. Epidemiol. Infect. 148:e64. doi: 10.1017/S0950268820000618
Osuolale, O., and Okoh, A. (2015). Incidence of human adenoviruses and hepatitis A virus in the final effluent of selected wastewater treatment plants in eastern Cape Province, South Africa. Virol. J. 12, 1–8. doi: 10.1186/s12985-015-0327-z
Osuolale, O., and Okoh, A. (2016). Human enteric bacteria and viruses in five wastewater treatment plants in the eastern cape, South Africa. J. Infect. Public Health 10, 541–547. doi: 10.1016/j.jiph.2016.11.012
Pan, Y., Zhang, D., Yang, P., Poon, L. L., and Wang, Q. (2020). Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 20, 411–412. doi: 10.1016/S1473-3099(20)30113-4
Pang, X., Qiu, Y., Gao, T., Zurawell, R., Neumann, N. F., Craik, S., et al. (2019). Prevalence, levels and seasonal variations of human enteric viruses in six major rivers in Alberta, Canada. Water Res. 153, 349–356. doi: 10.1016/j.watres.2019.01.034
Petrignani, M., Verhoef, L., de Graaf, M., Richardus, J. H., and Koopmans, M. (2018). Chronic sequelae and severe complications of norovirus infection: a systematic review of literature. J. Clin. Virol. 105, 1–10. doi: 10.1016/j.jcv.2018.05.004
Pillay, L., Amoah, I. D., Deepnarain, N., Pillay, K., Awolusi, O. O., Kumari, S., et al. (2021). Monitoring changes in COVID-19 infection using wastewater-based epidemiology: a south African perspective. Sci. Total Environ. 786:147273. doi: 10.1016/j.scitotenv.2021.147273
Pooi, C. K., and Ng, H. Y. (2018). Review of low-cost point-of-use water treatment systems for developing communities. NPJ Clean Water 1:11. doi: 10.1038/s41545-018-0011-0
Pöyry, T., Stenvik, M., and Hovi, T. (1988). Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54, 371–374. doi: 10.1128/aem.54.2.371-374.1988
Prado, T., de Castro Bruni, A., Barbosa, M. R. F., Garcia, S. C., Moreno, L. Z., and Sato, M. I. Z. (2019). Noroviruses in raw sewage, secondary effluents and reclaimed water produced by sand-anthracite filters and membrane bioreactor/reverse osmosis system. Sci. Total Environ. 646, 427–437. doi: 10.1016/j.scitotenv.2018.07.301
Prata, C., Ribeiro, A., Cunha, Â., Gomes, N. C., and Almeida, A. (2012). Ultracentrifugation as a direct method to concentrate viruses in environmental waters: virus-like particle enumeration as a new approach to determine -the efficiency of recovery. J. Environ. Monit. 14, 64–70. doi: 10.1039/C1EM10603A
Prez, V. E., Gil, P. I., Temprana, C. F., Cuadrado, P. R., Martinez, L. C., Giordano, M. O., et al. (2015). Quantification of human infection risk caused by rotavirus in surface waters from Cordoba, Argentina. Sci. Total Environ. 538, 220–229. doi: 10.1016/j.scitotenv.2015.08.041
Prez, V. E., Poma, H. R., Giordano, G. G., Victoria, M., Nates, S. V., Rajal, V. B., et al. (2020). Rotavirus contamination of surface waters from the northwest of Argentina. J. Water Health 18, 409–415. doi: 10.2166/wh.2020.005
Puenpa, J., Wanlapakorn, N., Vongpunsawad, S., and Poovorawan, Y. (2019). The history of enterovirus A71 outbreaks and molecular epidemiology in the Asia-Pacific region. J. Biomed. Sci. 26, 75–11. doi: 10.1186/s12929-019-0573-2
Purdy, M. A., Harrison, T. J., Jameel, S., Meng, X. J., Okamoto, H., Van der Poel, W., et al. (2017). ICTV virus taxonomy profile: Hepeviridae. J. Gen. Virol. 98, 2645–2646. doi: 10.1099/jgv.0.000940
Purpari, G., Macaluso, G., Di Bella, S., Gucciardi, F., Mira, F., Di Marco, P., et al. (2019). Molecular characterization of human enteric viruses in food, water samples, and surface swabs in Sicily. Int. J. Infect. Dis. 80, 66–72. doi: 10.1016/j.ijid.2018.12.011
Qu, G., Li, X., Hu, L., and Jiang, G. (2020). An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19). Environ. Sci. Technol. 54, 3730–3732. doi: 10.1021/acs.est.0c01102
Quintão, T. S. C., Silva, F. G., Pereira, A. L., Araújo, W. N., Oliveira, P. M., Souza, M. B. L. D., et al. (2021). Detection and molecular characterization of enteric adenovirus in treated wastewater in the Brazilian Federal District. SN Appl. Sci. 3, 1–11. doi: 10.1007/s42452-021-04678-2
Radin, D. (2014). New trends in food-and waterborne viral outbreaks. Arch. Biol. Sci. 66, 1–9. doi: 10.2298/ABS1401001R
Raji, Y. E., Toung, O. P., Taib, N. M., and Sekawi, Z. B. (2021). Hepatitis E virus: An emerging enigmatic and underestimated pathogen. Saudi J. Biol. Sci. 29, 499–512. doi: 10.1016/j.sjbs.2021.09.003
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., and Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181:115942. doi: 10.1016/j.watres.2020.115942
Rani, M., Rajyalakshmi, S., Pakalapaty, S., and Kammilli, N. (2021). “Norovirus structure and classification” in Norovirus, vol. 31
Räsänen, S., Lappalainen, S., Kaikkonen, S., Hämäläinen, M., Salminen, M., and Vesikari, T. (2010). Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 138, 1227–1234. doi: 10.1017/S0950268809991671
Remes-Troche, J. M., Ramos-de-la-Medina, A., Manríquez-Reyes, M., Martínez-Pérez-Maldonado, L., Lara, E. L., and Solís-González, M. A. (2020). Initial gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2 infection in 112 patients from Veracruz in southeastern Mexico. Gastroenterology 159, 1179–1181. doi: 10.1053/j.gastro.2020.05.055
Reynolds, K. A., Mena, K. D., and Gerba, C. P. (2008). Risk of waterborne illness via drinking water in the United States. Rev. Environ. Contam. Toxicol. 192, 117–158. doi: 10.1007/978-0-387-71724-1_4
Rezaeinejad, S., Vergara, G., Woo, C., Lim, T., Sobsey, M., and Gin, K. (2014). Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 58, 122–131. doi: 10.1016/j.watres.2014.03.051
Richardson, S. D., and Postigo, C. (2011). “Drinking water disinfection by-products” in Emerging Organic Contaminants and Human Health. ed. D. Barcelo (Berlin, Heidelberg: Springer), 93–137.
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., et al. (2020). Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 744:140911. doi: 10.1016/j.scitotenv.2020.140911
Rivadulla, E., and Romalde, J. L. (2020). A comprehensive review on human Aichi virus. Virol. Sin. 35, 501–516. doi: 10.1007/s12250-020-00222-5
Rodriguez-Lazaro, D., Cook, N., Ruggeri, F. M., Sellwood, J., Nasser, A., Nascimento, M. S. J., et al. (2012). Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 36, 786–814. doi: 10.1111/j.1574-6976.2011.00306.x
Rosario-Ortiz, F., Rose, J., Speight, V., Von Gunten, U., and Schnoor, J. (2016). How do you like your tap water? Science 351, 912–914. doi: 10.1126/science.aaf0953
Rosiles-González, G., Ávila-Torres, G., Moreno-Valenzuela, O. A., Cháidez-Quiroz, C., Hernández-Flores, C. I., Acosta-González, G., et al. (2019). Norovirus and human adenovirus occurrence and diversity in recreational water in a karst aquifer in the Yucatan peninsula, Mexico. J. Appl. Microbiol. 127, 1255–1269. doi: 10.1111/jam.14385
Rothe, C., Schunk, M., Sothmann, P., Bretzel, G., Froeschl, G., Wallrauch, C., et al. (2020). Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971. doi: 10.1056/NEJMc2001468
Ruggeri, F. M., Di Bartolo, I., Ponterio, E., Angeloni, G., Trevisani, M., and Ostanello, F. (2013). Zoonotic transmission of hepatitis E virus in industrialized countries. New Microbiol. 36, 331–344.
Russell, W. (2009). Adenoviruses: update on structure and function. J. Gen. Virol. 90, 1–20. doi: 10.1099/vir.0.003087-0
Salvador, D., Caeiro, M. F., Serejo, F., Nogueira, P., Carneiro, R. N., and Neto, C. (2020). Monitoring waterborne pathogens in surface and drinking waters. Are water treatment plants (WTPs) simultaneously efficient in the elimination of enteric viruses and fecal indicator bacteria (FIB)? WaterSA 12:2824. doi: 10.3390/w12102824
Sano, D., Amarasiri, M., Hata, A., Watanabe, T., and Katayama, H. (2016). Risk management of viral infectious diseases in wastewater reclamation and reuse. Environ. Int. 91, 220–229. doi: 10.1016/j.envint.2016.03.001
Santiso-Bellón, C., Randazzo, W., Pérez-Cataluña, A., Vila-Vicent, S., Gozalbo-Rovira, R., Muñoz, C., et al. (2020). Epidemiological surveillance of norovirus and rotavirus in sewage (2016–2017) in Valencia (Spain). Microorganisms 8:458. doi: 10.3390/microorganisms8030458
Saxena, G., Bharagava, R. N., Kaithwas, G., and Raj, A. (2015). Microbial indicators, pathogens and methods for their monitoring in water environment. J. Water Health 13, 319–339. doi: 10.2166/wh.2014.275
Sedji, M. I., Varbanov, M., Meo, M., Colin, M., Mathieu, L., and Bertrand, I. (2018). Quantification of human adenovirus and norovirus in river water in the north-east of France. Environ. Sci. Pollut. Res. 25, 30497–30507. doi: 10.1007/s11356-018-3045-4
Sekwadi, P., Ravhuhali, K., Mosam, A., Essel, V., Ntshoe, G., Shonhiwa, A., et al. (2018). Waterborne outbreak of gastroenteritis on the KwaZulu-natal coast, South Africa, december 2016/january 2017. Epidemiol. Infect. 146, 1318–1325. doi: 10.1017/S095026881800122X
Shaheen, M. N., El-Daim, A., Sahar, E., Ahmed, N. I., and Elmahdy, E. M. (2020). Environmental monitoring of Aichi virus and human bocavirus in samples from wastewater treatment plant, drain, and River Nile in Egypt. J. Water Health 18, 30–37. doi: 10.2166/wh.2019.075
Shaheen, M. N., Elmahdy, E. M., and Chawla-Sarkar, M. (2019). Quantitative PCR-based identification of enteric viruses contaminating fresh produce and surface water used for irrigation in Egypt. Environ. Sci. Pollut. Res. 26, 21619–21628. doi: 10.1007/s11356-019-05435-0
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., et al. (2020). Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 117, 11727–11734. doi: 10.1073/pnas.2003138117
Sharma, D. K., Nalavade, U. P., and Deshpande, J. M. (2015). Real-time reverse transcription-polymerase chain reaction assays for identification of wild poliovirus 1 & 3. Indian J. Med. Res. 142, 471–478. doi: 10.4103/0971-5916.169216
Sharma, V. K., Zboril, R., and McDonald, T. J. (2014). Formation and toxicity of brominated disinfection byproducts during chlorination and chloramination of water: a review. J. Environ. Sci. Health B 49, 212–228. doi: 10.1080/03601234.2014.858576
Shen, Q., Zhang, W., Yang, S., Yang, Z., Chen, Y., Cui, L., et al. (2012). Recombinant porcine norovirus identified from piglet with diarrhea. BMC Vet. Res. 8:155. doi: 10.1186/1746-6148-8-155
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., et al. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 743:140621. doi: 10.1016/j.scitotenv.2020.140621
Shih, Y. J., Tao, C. W., Tsai, H. C., Huang, W. C., Huang, T. Y., Chen, J. S., et al. (2017). First detection of enteric adenoviruses genotype 41 in recreation spring areas of Taiwan. Environ. Sci. Pollut. Res. 24, 18392–18399. doi: 10.1007/s11356-017-9513-4
Shonhiwa, A. M., Ntshoe, G., Crisp, N., Olowolagba, A. J., Mbuthu, V., Taylor, M. B., et al. (2020). Investigation of two suspected diarrhoeal-illness outbreaks in northern cape and KwaZulu-Natal provinces, South Africa, April–July 2013: The role of rotavirus. South. Afr. J. Infect. Dis. 35, 159. doi: 10.4102/sajid.v35i1.159
Sibanda, T., and Okoh, A. I. (2012). Assessment of the incidence of enteric adenovirus species and serotypes in surface waters in the eastern cape province of South Africa: Tyume River as a case study. Sci. World J. 2012:949216. doi: 10.1100/2012/949216
Siebenga, J. J., Vennema, H., Zheng, D. P., Vinjé, J., Lee, B. E., Pang, X. L., et al. (2009). Norovirus illness is a global problem: emergence and spread of norovirus GII. 4 variants, 2001–2007. J. Infect. Dis. 200, 802–812. doi: 10.1086/605127
Simmons, F. J., Kuo, D. H. W., and Xagoraraki, I. (2011). Removal of human enteric viruses by a full-scale membrane bioreactor during municipal wastewater processing. Water Res. 45, 2739–2750. doi: 10.1016/j.watres.2011.02.001
Smith, D. B., and Simmonds, P. (2018). Classification and genomic diversity of enterically transmitted hepatitis viruses. Cold Spring Harb. Perspect. Med. 8:a031880. doi: 10.1101/cshperspect.a031880
Smuts, H., Cronje, S., Thomas, J., Brink, D., Korsman, S., and Hardie, D. (2018). Molecular characterization of an outbreak of enterovirus-associated meningitis in Mossel Bay, South Africa, December 2015–January 2016. BMC Infect. Dis. 18, 709–711. doi: 10.1186/s12879-018-3641-4
Spilki, F. R., Luz, R. B. D., Fabres, R. B., Soliman, M. C., Kluge, M., Fleck, J. D., et al. (2013). Detection of human adenovirus, rotavirus and enterovirus in water samples collected on dairy farms from Tenente Portela, northwest of Rio Grande do Sul, Brazil. Braz. J. Microbiol. 44, 953–957. doi: 10.1590/S1517-83822013000300046
Staggemeier, R., Heck, T. M., Demoliner, M., Ritzel, R. G., Röhnelt, N. M., Girardi, V., et al. (2017). Enteric viruses and adenovirus diversity in waters from 2016 Olympic venues. Sci. Total Environ. 586, 304–312. doi: 10.1016/j.scitotenv.2017.01.223
Suzuki, H., Sakai, T., Tanabe, N., and Okabe, N. (2005). Peak rotavirus activity shifted from winter to early spring in Japan. Pediatr. Infect. Dis. J. 24, 257–260. doi: 10.1097/01.inf.0000154327.00232.4d
Syed, S. F., Zhao, Q., Umer, M., Alagawany, M., Ujjan, I. A., Soomro, F., et al. (2018). Past, present and future of hepatitis E virus infection: zoonotic perspectives. Microb. Pathog. 119, 103–108. doi: 10.1016/j.micpath.2018.03.051
Sylvestre, É., Prévost, M., Burnet, J.-B., Pang, X., Qiu, Y., Smeets, P., et al. (2021). Demonstrating the reduction of enteric viruses by drinking water treatment during snowmelt episodes in urban areas. Water Res. X 11:100091. doi: 10.1016/j.wroa.2021.100091
Taghinejad, M., Ghaderi, M., and Mousavi-Nasab, S. D. (2020). Aichivirus with acute gastroenteritis in Iran. Pediatr. Infect. Dis. J. 39, 576–579. doi: 10.1097/INF.0000000000002638
Tang, A., Bi, X., Li, X., Li, F., Liao, X., Zou, J., et al. (2020). The inactivation of bacteriophage MS2 by sodium hypochlorite in the presence of particles. Chemosphere 266:129191. doi: 10.1016/j.chemosphere.2020.129191
Tate, J. E., Burton, A. H., Boschi-Pinto, C., Parashar, U. D., World Health Organization–Coordinated Global Rotavirus Surveillance Network, Agocs, M., et al. (2016). Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin. Infect. Dis. 62, S96–S105. doi: 10.1093/cid/civ1013
Tate, J. E., Patel, M. M., Steele, A. D., Gentsch, J. R., Payne, D. C., Cortese, M. M., et al. (2010). Global impact of rotavirus vaccines. Expert Rev. Vaccines 9, 395–407. doi: 10.1586/erv.10.17
Taylor, M., Cox, N., Vrey, M., and Grabow, W. (2001). The occurrence of hepatitis A and astroviruses in selected river and dam waters in South Africa. Water Res. 35, 2653–2660. doi: 10.1016/S0043-1354(00)00551-0
Thorne, L. G., and Goodfellow, I. G. (2014). Norovirus gene expression and replication. J. Gen. Virol. 95, 278–291. doi: 10.1099/vir.0.059634-0
Trivedi, T. K., Desai, R., Hall, A. J., Patel, M., Parashar, U. D., and Lopman, B. A. (2013). Clinical characteristics of norovirus-associated deaths: a systematic literature review. Am. J. Infect. Control 41, 654–657. doi: 10.1016/j.ajic.2012.08.002
Troeger, C., Khalil, I. A., Rao, P. C., Cao, S., Blacker, B. F., Ahmed, T., et al. (2018). Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 172, 958–965. doi: 10.1001/jamapediatrics.2018.1960
Tryfinopoulou, K., Kyritsi, M., Mellou, K., Kolokythopoulou, F., Mouchtouri, V. A., Potamiti-Komi, M., et al. (2019). Norovirus waterborne outbreak in Chalkidiki, Greece, 2015: detection of GI.P2_GI.2 and GII.P16_GII.13 unusual strains. Epidemiol. Infect. 147, e227. doi: 10.1017/S0950268819000852
Umesha, K. R., Bhavani, N. C., Venugopal, M. N., Karunasagar, I., Krohne, G., and Karunasagar, I. (2008). Prevalence of human pathogenic enteric viruses in bivalve molluscan shellfish and cultured shrimp in south west coast of India. Int. J. Food Microbiol. 122, 279–286. doi: 10.1016/j.ijfoodmicro.2007.12.024
Upfold, N. S., Luke, G. A., and Knox, C. (2021). Occurrence of human enteric viruses in water sources and shellfish: A focus on Africa. Food Environ. Virol. 13, 1–31. doi: 10.1007/s12560-020-09456-8
Van Zyl, W. B., Zhou, N. A., Wolfaardt, M., Matsapola, P. N., Ngwana, F. B., Symonds, E. M., et al. (2019). Detection of potentially pathogenic enteric viruses in environmental samples from Kenya using the bag-mediated filtration system. Water Supply 19, 1668–1676. doi: 10.2166/ws.2019.046
Verani, M., Federigi, I., Donzelli, G., Cioni, L., and Carducci, A. (2019). Human adenoviruses as waterborne index pathogens and their use for quantitative microbial risk assessment. Sci. Total Environ. 651, 1469–1475. doi: 10.1016/j.scitotenv.2018.09.295
Verheyen, J., Timmen-Wego, M., Laudien, R., Boussaad, I., Sen, S., Koc, A., et al. (2009). Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl. Environ. Microbiol. 75, 2798–2801. doi: 10.1128/AEM.01807-08
Vinjé, J. (2015). Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 53, 373–381. doi: 10.1128/JCM.01535-14
Vu, D. L., Bosch, A., Pintó, R. M., and Guix, S. (2017). Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9:33. doi: 10.3390/v9020033
Vu, D. L., Cordey, S., Brito, F., and Kaiser, L. (2016). Novel human astroviruses: novel human diseases? J. Clin. Virol. 82, 56–63. doi: 10.1016/j.jcv.2016.07.004
Wang, Y., Hao, L., Pan, L., Xue, C., Liu, Q., Zhao, X., et al. (2018b). Age, primary symptoms, and genotype characteristics of norovirus outbreaks in Shanghai schools in 2017. Sci. Rep. 8:15238. doi: 10.1038/s41598-018-33724-0
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020a). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323, 1061–1069. doi: 10.1001/jama.2020.1585
Wang, H., Kjellberg, I., Sikora, P., Rydberg, H., Lindh, M., Bergstedt, O., et al. (2020b). Hepatitis E virus genotype 3 strains and a plethora of other viruses detected in raw and still in tap water. Water Res. 168:115141. doi: 10.1016/j.watres.2019.115141
Wang, H., Sikora, P., Rutgersson, C., Lindh, M., Brodin, T., Björlenius, B., et al. (2018a). Differential removal of human pathogenic viruses from sewage by conventional and ozone treatments. Int. J. Hyg. Environ. Health 221, 479–488. doi: 10.1016/j.ijheh.2018.01.012
WHO (2017a). Diarrhoeal disease. Fact Sheets.1–4. Available at: https://www.who.int/en/news-room/fact-sheets/detail/diarrhoeal-disease (December 7, 2020).
WHO (2017c). Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First Addendum. Geneva, Switzerland: WHO
Wohlgemuth, N., Honce, R., and Schultz-Cherry, S. (2019). Astrovirus evolution and emergence. Infect. Genet. Evol. 69, 30–37. doi: 10.1016/j.meegid.2019.01.009
World Health Organization (2012a). Global Costs and Benefits of Drinking-water Supply and Sanitation Interventions to reach the MDG Target and universal Coverage. Geneva: WHO
World Health Organization (2012b). Position paper on hepatitis A vaccines. Weekly Epidemiol. Rec. 87, 261–276.
World Health Organization (2012c). WHO position paper on hepatitis A vaccines–June 2012. Weekly Epidemiol. Rec. 87, 261–276.
World Health Organization (2014). Waterborne Outbreaks of Hepatitis E: Recognition, Investigation and Control. World Health Organization.
World Health Organization (2020). Naming the coronavirus disease (COVID-19) and the virus that causes it. Braz. J. Implantol. Health Sci. 2, 1–4.
World Health Organization/United Nations Children Emergency Fund (2015). Progress on sanitation and drinking water: 2015 update and MDG assessment.
World Health Organization/United Nations Children Emergency Fund (2021). Progress on Household Drinking water, Sanitation, and Hygiene 2000–2020: Five Years into the SDGs. Geneva, Switzerland.
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., et al. (2020). Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435. doi: 10.1016/S2468-1253(20)30083-2
Xagoraraki, I., and O’Brien, E. (2020). “Wastewater-based epidemiology for early detection of viral outbreaks” in Women in Water Quality. ed. D. O’Bannon (Switzerland: Springer), 75–97.
Yamashita, T., Sugiyama, M., Tsuzuki, H., Sakae, K., Suzuki, Y., and Miyazaki, Y. (2000). Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 38, 2955–2961. doi: 10.1128/JCM.38.8.2955-2961.2000
Yang, Q., Rivailler, P., Zhu, S., Yan, D., Xie, N., Tang, H., et al. (2021). Detection of multiple viruses potentially infecting humans in sewage water from Xinjiang Uygur autonomous region, China. Sci. Total Environ. 754:142322. doi: 10.1016/j.scitotenv.2020.142322
Yeo, C., Kaushal, S., and Yeo, D. (2020). Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 5, 335–337. doi: 10.1016/S2468-1253(20)30048-0
Zahedi, A., Monis, P., Deere, D., and Ryan, U. (2021). Wastewater-based epidemiology—surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, cryptosporidium and giardia. Parasitol. Res. 120, 4167–4188. doi: 10.1007/s00436-020-07023-5
Zhou, X., Kong, D. G., Li, J., Pang, B. B., Zhao, Y., Zhou, J. B., et al. (2019). An outbreak of gastroenteritis associated with GII. 17 Norovirus-contaminated secondary water supply system in Wuhan, China, 2017. Food Environ. Virol. 11, 126–137. doi: 10.1007/s12560-019-09371-7
Zhou, N., Lin, X., Wang, S., Tao, Z., Xiong, P., Wang, H., et al. (2016). Molecular epidemiology of GI and GII noroviruses in sewage: 1-year surveillance in eastern China. J. Appl. Microbiol. 121, 1172–1179. doi: 10.1111/jam.13218
Zhu, B., Clifford, D. A., and Chellam, S. (2005). Virus removal by iron coagulation–microfiltration. Water Res. 39, 5153–5161. doi: 10.1016/j.watres.2005.09.035
Zhu, H., Yuan, F., Yuan, Z., Liu, R., Xie, F., Huang, L., et al. (2018). Monitoring of Poyang lake water for sewage contamination using human enteric viruses as an indicator. Virol. J. 15, 3–9. doi: 10.1186/s12985-017-0916-0
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Keywords: enteric viruses, gastroenteritis, outbreak, wastewater, wastewater-based epidemiology
Citation: Lanrewaju AA, Enitan-Folami AM, Sabiu S, Edokpayi JN and Swalaha FM (2022) Global public health implications of human exposure to viral contaminated water. Front. Microbiol. 13:981896. doi: 10.3389/fmicb.2022.981896
Edited by:
Anthony Ayodeji Adegoke, University of Uyo, NigeriaReviewed by:
Timothy Sibanda, University of Namibia, NamibiaAbdennaceur Hassen, Centre de Recherches et des Technologies des Eaux, Tunisia
Copyright © 2022 Lanrewaju, Enitan-Folami, Sabiu, Edokpayi and Swalaha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abimbola Motunrayo Enitan-Folami, ZW5pdGFuYWJpbWJvbGFAZ21haWwuY29t
 Adedayo Ayodeji Lanrewaju
Adedayo Ayodeji Lanrewaju Abimbola Motunrayo Enitan-Folami
Abimbola Motunrayo Enitan-Folami Saheed Sabiu
Saheed Sabiu Joshua Nosa Edokpayi
Joshua Nosa Edokpayi Feroz Mahomed Swalaha
Feroz Mahomed Swalaha