
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 August 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.976334
 Peng-xuan Liu1,2
Peng-xuan Liu1,2 Xiao-yun Zhang1
Xiao-yun Zhang1 Quan Wang1
Quan Wang1 Yang-yang Li1,2
Yang-yang Li1,2 Wei-dong Sun2
Wei-dong Sun2 Yu Qi1
Yu Qi1 Kai Zhou3
Kai Zhou3 Xian-gan Han1
Xian-gan Han1 Zhao-guo Chen1
Zhao-guo Chen1 Wei-huan Fang4
Wei-huan Fang4 Wei Jiang1*
Wei Jiang1*Vibrio parahaemolyticus is a marine pathogen thought to be the leading cause of seafood-borne gastroenteritis globally, urgently requiring efficient management methods. V. parahaemolyticus encodes 12 resistance/nodulation/division (RND) efflux systems. However, research on these systems is still in its infancy. In this study, we discovered that the inactivation of VmeL, a membrane fusion protein within the RND efflux systems, led to reduction of the ability of biofilm formation. Further results displayed that the decreased capacity of Congo red binding and the colony of ΔvmeL is more translucent compared with wild type strains, suggested reduced biofilm formation due to decreased production of biofilm exopolysaccharide upon vmeL deletion. In addition, the deletion of vmeL abolished surface swarming and swimming motility of V. parahaemolyticus. Additionally, deletion of vmeL weakened the cytotoxicity of V. parahaemolyticus towards HeLa cells, and impaired its virulence in a murine intraperitoneal infection assay. Finally, through RNA-sequencing, we ascertained that there were 716 upregulated genes and 247 downregulated genes in ΔvmeL strain. KEGG enrichment analysis revealed that quorum sensing, bacterial secretion systems, ATP-binding cassette transporters, and various amino acid metabolism pathways were altered due to the inactivation of vmeL. qRT-PCR further confirmed that genes accountable to the type III secretion system (T3SS1) and lateral flagella were negatively affected by vmeL deletion. Taken together, our results suggest that VmeL plays an important role in pathogenicity, making it a good target for managing infection with V. parahaemolyticus.
Vibrio parahaemolyticus is a gram-negative halophilic bacterium, primarily found in warm marine or estuarine environments (Broberg et al., 2011). V. parahaemolyticus, discovered almost 70 years ago, has been identified as a leading cause of foodborne illness worldwide, and could lead septicemia through wound infection (Su and Liu, 2007; Ghenem et al., 2017). In recent years, the rate of V. parahaemolyticus infection has increased in many countries, including the United States and China (Banu et al., 2018; Lei et al., 2020). V. parahaemolyticus exerts its virulence through diverse factors (Li et al., 2019). Of these, T3SSs are prominent virulence factors, and can inject diverse effectors into eukaryotic cells through a transmembrane apparatus. However, some V. parahaemolyticus isolates can induce acute gastroenteritis without T3SS2 and hemolysin genes (Ottaviani et al., 2012). This indicates alternative mechanisms that contribute to the pathogenicity of V. parahaemolyticus.
As a ubiquitous marine bacterium and human pathogen, V. parahaemolyticus has evolved several motility mechanisms to promote colonization of deep sea vents or the bodies of animals. V. parahaemolyticus exhibits multiple cell types that are appropriate for life under different circumstance (McCarter, 1999). Swimmer cells synthesize a single polar flagellum for adaption to life in liquid environments, and swarmer cells propelled by many proton-powered lateral flagella, for movement through highly viscous environments, colonize surfaces, and form multicellular communities which sometimes display highly periodic architecture (Boles and McCarter, 2002; Wadhwa and Berg, 2021). Motility is essential for adaptability, survival, and infection of the human host (Taw et al., 2015). In addition, V. parahaemolyticus could improve its survival in adverse surroundings through biofilm formation, enhancing antibiotic resistance and leading to the development of intractable infections (Yildiz and Visick, 2009; Harshey and Partridge, 2015; Hall and Mah, 2017; Hotinger et al., 2021).
Various efflux pumps are important contributors to antibiotic resistance in microbes (Uddin et al., 2021). Resistance/nodulation/division (RND) efflux systems are ubiquitous in Gram-negative bacteria, and are linked to antimicrobial resistance (Alvarez-Ortega et al., 2013). RND efflux systems consist of an inner membrane protein, two membrane fusion proteins and an outer membrane pore protein (Alav et al., 2021). The three core constituents operate collectively to efflux several substrates from the cytoplasm and periplasm to the external environment. Interestingly, an increasing number of studies have demonstrated that RND efflux systems have many functions in diverse phenotypes including metabolism, biofilm, and virulence. For instance, the loss of six efflux systems in V. cholerae represses the production of several virulence factors (Bina et al., 2018). The inactivation of abeD, which encodes a membrane transporter, reduced virulence toward a nematode model in Acinetobacter baumannii (Srinivasan et al., 2015). In addition, the inner membrane protein AdeJ could negatively influenced surface motility and biofilm formation in A. nosocomialis (Knight et al., 2018). Thus, previous studies have suggested that RND efflux systems in bacteria were involved not only in antimicrobial resistance, but also in biological and pathophysiological processes.
Twelve RND-type efflux transporter genes in V. parahaemolyticus have been estimated in a previous study, and their finding indicated that resistance nodulation cell division-type efflux transporters contribute not only to intrinsic resistance but also to exerting the virulence of V. parahaemolyticus (Matsuo et al., 2013). However, little is known about the contribution of RND efflux systems to V. parahaemolyticus pathogenicity and host adaptation. The membrane fusion proteins, also known as the periplasmic adaptor proteins, play central role in the primacy of the determinants of substrate specificity and the assemble of the RND efflux systems (Alav et al., 2021). Despite their central importance for the efflux process, the function of membrane fusion proteins remains least-well understood. Therefore, in this study, the biological contributions of membrane fusion protein VmeL in surface motility, biofilm formation, and virulence of V. parahaemolyticus were further examined. Our findings offer insight into the roles of this protein, and can serve as a basis for the control of V. parahaemolyticus contamination and infection.
The wild-type (WT) strain V. parahaemolyticus SH112 (GenBank: JACYGZ000000000.1) stocked in the China General Microbiological Culture Collection Center under the accession number CGMCC 1.90013. V. parahaemolyticus was cultured in MLB (Lysogeny broth with 2% sodium chloride) at 37 °C with constant shaking at 180 rpm. Escherichia coli CC118 λpir was cultivated in LB medium at 37 °C with shaking at 180 rpm; 70 μg/mL kanamycin and 10 μg/mL chloramphenicol were supplemented for specific plasmids. Table 1 displays all WT and derivative strains of V. parahaemolyticus and E. coli.
HeLa cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, United States) with 10% fatal bovine serum (FBS, Gibco) and 100 units/mL of streptomycin and penicillin (Invitrogen, Thermo Fisher Scientific) at 37°C with 5% CO2 humidity.
All primers used for the strain and plasmid construction in this study are shown in Table 2. The deletion mutant was constructed through homologous recombination, as previously described (Lian et al., 2021). First, vmeL-A/B and vmeL-C/D were utilized to amplify the upstream and downstream regions flanking vmeL, respectively. A fragment comprised of flanking sequences was generated using these two fragments as templates, and connected with the suicide plasmid pYAK1, yielding the recombinant plasmid pYAK1-vmeL. This pYAK1-vmeL was transferred into V. parahaemolyticus SH112 through triparental conjugation. After double crossover recombination, the successful mutant was designated as ΔvmeL.
The complemented strain was constructed similarly to the construction of the mutant using pMMB207. Briefly, the total vmeL open reading frame was amplified utilizing the primer pMMB207-vmeL-F/R. The purified PCR product was linked to the pMMB207 plasmid with BamHI and SphI enzymes, resulting in the plasmid pMMB207-vmeL. Then, the plasmid was transformed into ΔvmeL using a method similar to that used for the ΔvmeL mutant. The complemented strain was designated as CΔvmeL.
The overnight cultures were diluted at 1:100 in fresh MLB medium or in DMEM medium containing 10% FBS and incubated at 37°C. Aliquots (200 μL) were transferred to clear 96-well plates every hour, and the optical density of the culture at 600 nm (OD600) was assessed utilizing a spectrophotometer (Thermo Fisher Scientific) at 1 h intervals for 10∼14 h. In the following experiments, the cells were utilized at logarithmic phage (OD600 = 0.2 ± 0.02) or about 1 × 108 cells per mL.
The minimum inhibitory concentrations (MICs) of several antibiotics were examined in Mueller-Hinton (MH) broth, as described previously (Matsuo et al., 2007). Briefly, MICs were processed in MH broth containing antibiotics in a two-fold dilution series. Bacteria were cultured in the test medium at 37°C for 24 h. Each compound’s MIC is defined as the lowest concentration that prevents visible growth.
Crystal violet staining assays were tested as previously described (Zhang et al., 2021a) with some modifications. Overnight cultures were diluted at 1:100 in fresh MLB medium, 200 μL aliquots of the test organism suspension were cultured in 96-well microtiter dishes (Costar, 42592), and then incubated for 48 h at 30 °C without shaking. Then, each well was rinsed with phosphate-buffered saline (PBS), and 250 μL of 0.1% crystal violet solution was added to each well for 15 min at 20–25°C. The wells were rinsed with PBS. 200 μL of 95% (v/v) ethanol was used to dissolve crystal violet, and the opacity of each wells was assessed by measuring OD595.
Both assays were performed as described formerly (Enos-Berlage et al., 2005; Zhang et al., 2021a). For morphology analysis, overnight cultures were diluted at 1:100 in fresh MLB medium, then, the logarithmic bacterial culture was homogenized, and 2 μL aliquots were spotted on heart infusion medium (HI) (ELITE Biotech, Shanghai, China) plates containing 2% NaCl and 2% agar, then incubated at 37°C for at least 48 h. For the CR binding assay, the mixed bacterial culture was spotted on CR plates [HI plates added 80 ug/ml Congo red (Yuanye Bio-Technology, Shanghai, China)], and then incubated at 30°C for 8 d. Images of each strain were recorded every two days.
Motility assays were conducted as described previously (Whitaker et al., 2014), with some modifications. Briefly, overnight cultures were diluted at 1:100 in fresh MLB medium and swimming motility assays were performed using 2 μL of logarithmic bacterial culture spotted onto surface of LB plates containing 1% NaCl and 0.3% agar. Images of each plate were captured after incubating for 4–5 h at 37°C. Swarming motility assays were performed similarly but on HI plates containing 2% NaCl and 1.5% agar. Strains were incubated at 30°C for 15–20 h before images were taken. For both motility assays, the colony diameters were measured and recorded.
Transmission electron microscopy of the lateral flagella was performed as previously described (Gu et al., 2019), with some modifications. All strains were cultured on HI agar plates for 15 h, rinsed lightly with 0.01 M PBS. Subsequently, 5 μL of bacterial suspension was dropped onto the copper grid and left until the mesh was dry enough to cover. Then, samples were observed by TEM (Tecnai G2 Spirit, FEI Company, Hillsboro, OR, United States).
According to a previous report (Tandhavanant et al., 2018), assays of infection of HeLa cells were conducted. HeLa cells were plated in 24-well dishes at a density of 1.5 × 105 per cell, grown for 12 h, and then infected with WT, ΔvmeL, and CΔvmeL V. parahaemolyticus at MOI of 10 for 2 h. After 2 h f co-incubation, the cells were rinsed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. 0.5% Triton-X was used to permeabilize the cells for 10 minat room temperature. Then, the cells were probed with rhodamine-phalloidin (SBS Genetech, Shanghai, China) to stain F-actin and DAPI (Beyotime, Shanghai, China) to highlight HeLa cell DNA. Images were captured using an inverted fluorescence microscope (Axio observer Z1, ZEISS).
HeLa cells were plated in 96-well dishes at 2 × 104 cells per well, cultured for 12 h, and over-night bacterial suspensions were diluted in fresh MLB medium and cultivated to the logarithmic phase. The next day cells were infected with WT, ΔvmeL, and CΔvmeL V. parahaemolyticus in four replicates at a multiplicity of infection (MOI) of 10. LDH release was measured using a CytoTox96 kit (Promega, Madison, WI, United States), and absorbance was measured using a spectrophotometer (Multiskan Go, Thermo Fisher Scientific).
Animal infection experiments were conducted as previously described (Li et al., 2022). Over-night bacterial suspensions were diluted in fresh MLB medium and cultivated to the logarithmic phase. The logarithmic bacterial culture of each strain was washed three times with PBS. Then, 1 107 CFU of each strain was inoculated intraperitoneally into female Institute of Cancer Research mice at three to four weeks of age. After infection, the symptoms of inoculated mice and number of deaths were recorded. The Animal Ethics Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences approved all animal infection studies (no. SYXK<HU > 2020-0027).
When cultured strains reached the exponential phase in MLB medium, total RNA was isolated using Bacteria RNA Extraction Kit (Vazyme, Nanjing, China). Three independent biological samples were conducted, then cDNA libraries were prepared utilizing a customized procedure (Sangon Biotech, Shanghai, China). Low-quality counts were removed utilizing the R package DESeq2 and to filter differential expression in accordance with the criteria of an absolute log2 fold-change value of >1.5, and a p-value of < 0.01. To analyze the differential expression of genes between the WT and ΔvmeL strains, differentially expressed genes (DEGs) were processed through Kyoto Encyclopedia of Genes and Genomes (KEGG) database using the ClusterProfiler package, and the Q-value was regarded as a screening criterion.
Quantitative reverse-transcription PCR was used to examined the transcriptional levels of different genes in the WT and mutant strains. Overnight cultures of WT, ΔvmeL, and CΔvmeL V. parahaemolyticus were adjusted to an OD600 of 0.2, and 2.5 μL aliquots were spread separately onto swarming plates. Cells were collected from swarming plates after 15 h of cultivation, and were suspended in 1 mL of TRIzol (Yu et al., 2019). Planktonic bacteria were cultivated in liquid MLB medium to the logarithmic phase with shaking at 37°C, and the swarming cells were collected. Then, total RNA was isolated from each sample following manufacturer protocol (Bacteria RNA Extraction Kit, Vazyme, Nanjing, China). RNA was reverse transcribed to cDNA. cDNA was utilized as a template for quantitative qRT-PCR. For each gene, reactions were performed for three RNA samples and each reaction was performed in triplicate. The transcriptional level was quantified using the 2–ΔΔCt method using the housekeeping gene gapA (Genbank: BAC61233.1) for normalization (Livak and Schmittgen, 2001). The primers for qRT-PCR are provided in Supplementary Table 1.
Graphpad Prism was used for data analysis. Images were processed using Microsoft Office PowerPoint. Basic Local Alignment Search Tool (BLAST) analysis was conducted using the available tool1. The qRT-PCR results were analyzed using a two-way analysis of variance (ANOVA) and a one-way ANOVA was performed for biofilm formation, motility, and cytotoxicity. The survival percentage was analyzed with Gehan-Breslow-Wilcoxon and log rank tests.
Vibrio parahaemolyticus strain SH112 was used to amplify the nucleotide sequence of membrane fusion protein VmeL, and the sequence of vmeL (GenBank: OL347638) displayed 99.83% identity with that of V. parahaemolyticus RIMD2210633 (GenBank: BA000032.2). BLASTn analyses indicated that the nucleotide sequence similarity between a total of 143 V. parahaemolyticus isolates ranged from 97.84 to 99.91% (Supplementary Figure 1A). Furthermore, a BLASTp search against GenBank suggested that VmeL in V. parahaemolyticus SH112 shared 94.03, 92.47, 92.99, 93.51, and 92.21% similarity with orthologous proteins in V. diabolicus (GenBank: WP_005392685.1), V. harveyi (GenBank: WP_005441462.1), V. alginolyticus (GenBank: EGQ8497134.1), V. chemaguriensis (GenBank: WP_225460886.1), and V. rotiferianus (GenBank: WP_038882841.1), respectively (Supplementary Figure 1B). These findings show that VmeL is an evolutionarily conserved membrane fusion protein found in V. parahaemolyticus and several Vibrio species.
The resulting null mutant strain ΔvmeL and the complemented strain CΔvmeL were examined by PCR and qRT-PCR (Supplementary Figure 2). No significant distinction in growth rate was identified between WT, ΔvmeL, and CΔvmeL strains in MLB medium or in DMEM medium containing 10% FBS (Supplementary Figure 3).
To test whether the deletion of vmeL could influence antimicrobial resistance in V. parahaemolyticus, we performed an antimicrobial susceptibility test. The results showed that loss of vmeL in V. parahaemolyticus had no significant influence on response to any of the tested antimicrobial compounds. The MICs were identified for streptomycin (22.25 μg/mL), cefalotin sodium (3.75 μg/mL), gentamicin (6.25 μg/mL), ampicillin (> 100 μg/mL), florfenicol (0.94 μg/mL), chloramphenicol (0.46 μg/mL), tetracycline (0.36 μg/mL), kanamycin (17.5 μg/mL).
To assay whether VmeL could affect the ability of biofilm formation in V. parahaemolyticus, the crystal violet staining assay was performed. The results showed that biofilm formation by ΔvmeL was significantly reduced compared to that of the WT strain (P < 0.01), while the capacity of biofilm formation was restored in complemented stains (Figure 1A). Since colony morphology is closely related to the synthesis of abundant exopolysaccharides (Enos-Berlage et al., 2005), we next examined whether the loss of vmeL could influence the colony morphology of V. parahaemolyticus. ΔvmeL produced more translucent colonies than those of the WT strain, while CΔvmeL produced opaque colonies similar to those of the WT strain (Figure 1B). In addition to colony morphology, the capacity of CR binding is closely related with the content of biofilm exopolysaccharide in V. parahaemolyticus (Enos-Berlage et al., 2005). At the center of the colony, the ΔvmeL strain displayed a smoother and lighter red color than that of the WT strain under our experimental conditions (Figure 1C). These observations indicated that VmeL positively modulates biofilm formation.
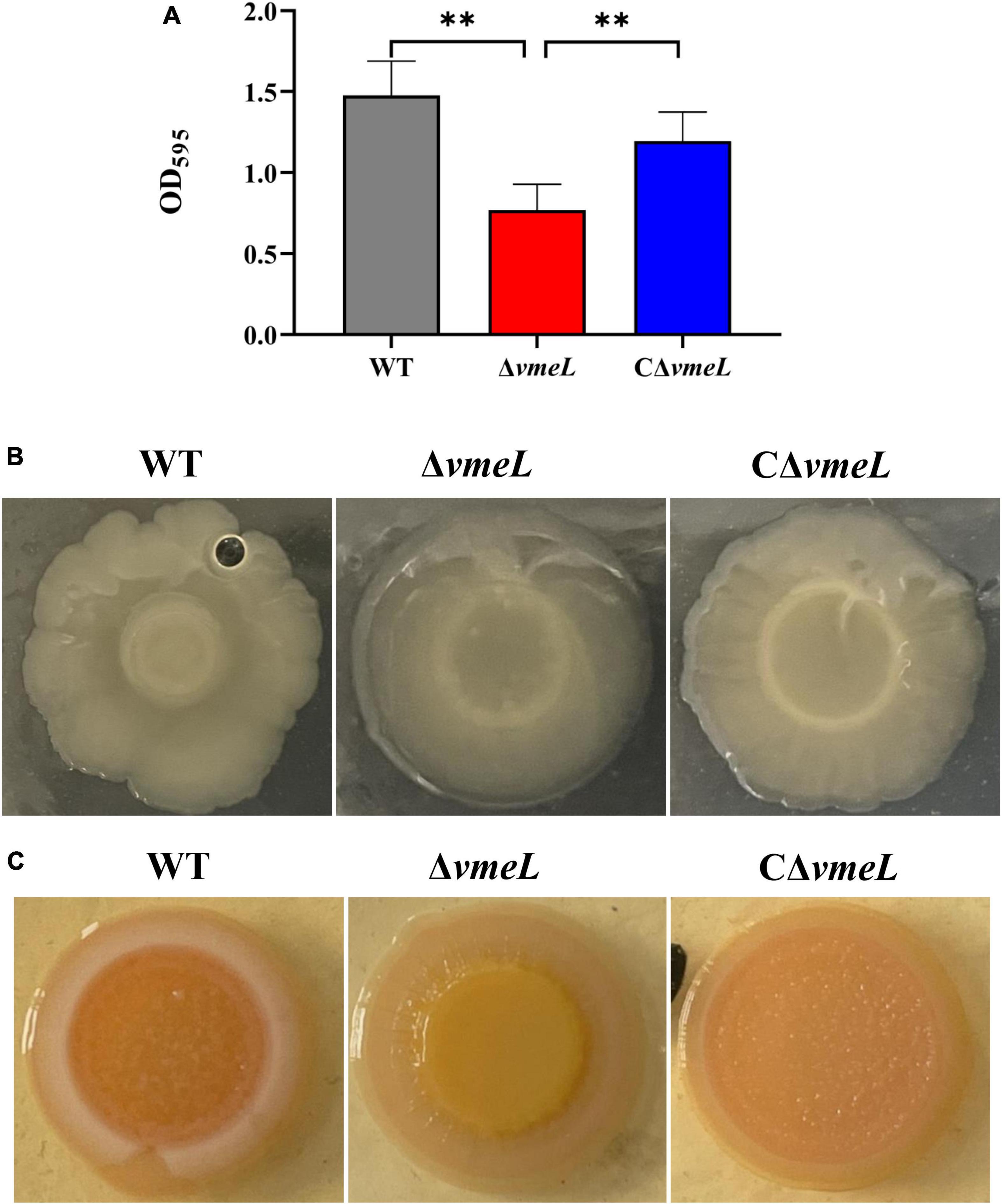
Figure 1. VmeL is required for biofilm formation by Vibrio parahaemolyticus. (A) Optical density at 595 nm (OD595) for crystal violet biofilm assays. Data shown are the mean of six replicates. (B) Colony morphology of strains cultivated on heart infusion plates for at least 48 hours. (C) Strains cultivated on Congo Red plates for eight days and photographed. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain. ** indicates statistical significance at P < 0.01.
To determine whether VmeL affects motility, surface swarming and swimming motilities of the ΔvmeL mutant were compared to those of the WT strain. The ΔvmeL strain did not have the capacity of surface swarming and swimming motility compared to the WT strain (Figures 2A,C). the colony diameter of WT and ΔvmeL on swarming plates were 29.3 ± 0.7 and 7.6 ± 0.6 mm respectively, and the colony of CΔvmeL was 27 ± 2 mm (Figure 2B). In addition, the colony diameters of WT and ΔvmeL on swimming plates were 30 ± 2 and 5.75 ± 0.75 mm, respectively, and the colony of CΔvmeL was 28.25 ± 5.25 mm (Figure 2D). The complemented strains restored the surface motility. Moreover, there is an extra zone of swarming for the complemented strain and the swimming colonies of complemented strains displayed irregular margins. These results indicated that the inactivation of vmeL had negative effects both on swarming motility and swimming motility in V. parahaemolyticus.
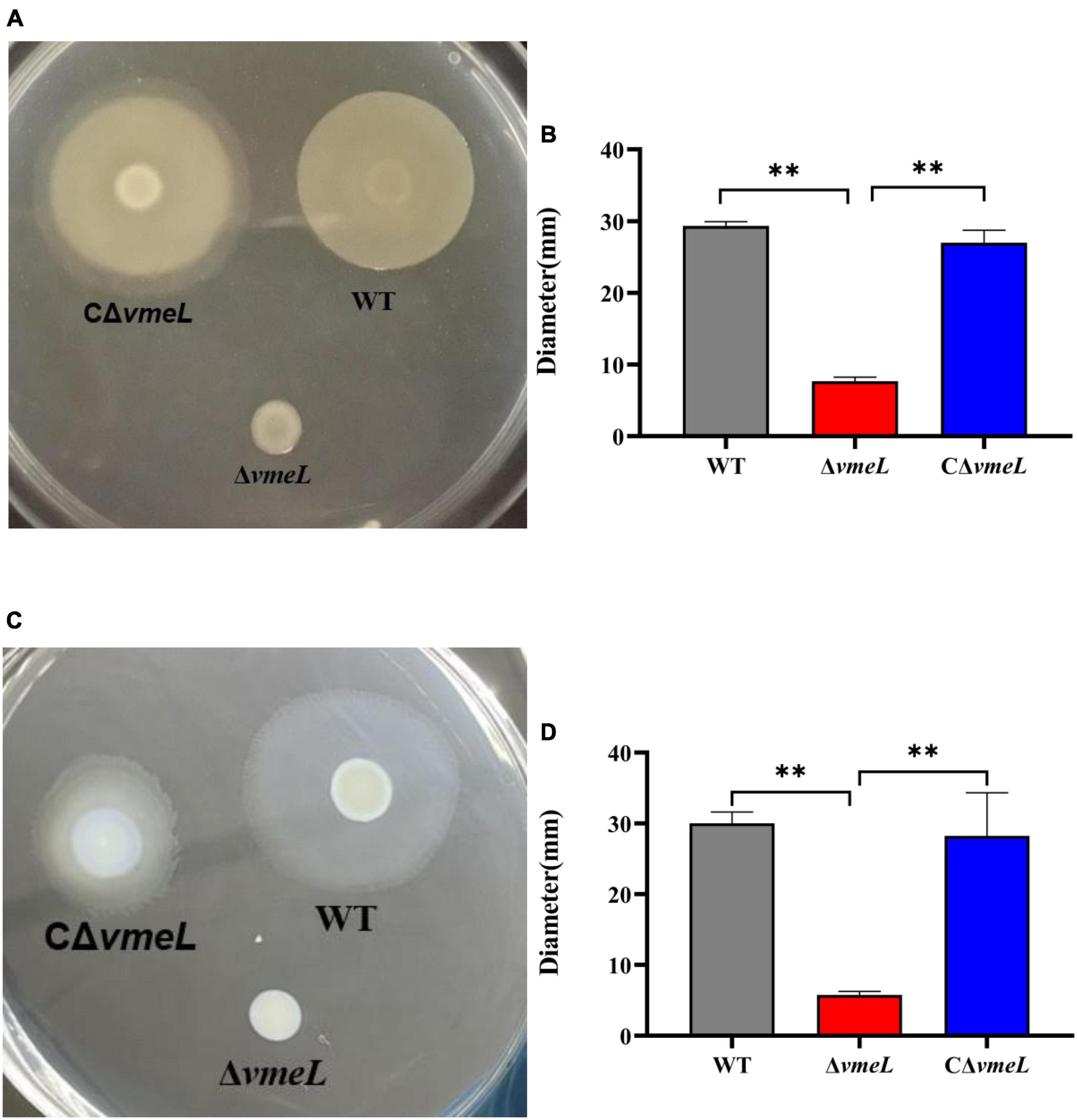
Figure 2. VmeL is required for surface motility of Vibrio parahaemolyticus. (A) Swarming motility was tested on swarming plates (heart infusion medium with 2% NaCl and 1.5% agar) at 30°C, and images were taken after 18 h. (C) Swimming motility was tested on swimming plates (Lysogeny broth supplemented with 1% NaCl and 0.3% agar) at 37°C, and images were taken after 4–5 h. (B,D) Representation of the swarming and swimming separately colony diameters, measured in three independent assays. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain. ** indicates statistical significance at P < 0.01.
To examine lateral flagella morphology of WT, ΔvmeL, and CΔvmeL strains, we collected samples from swarming agar plates. TEM results showed that the lateral flagella of ΔvmeL cells were absent and the polar flagella (red arrow) were still present, whereas each WT and CΔvmeL cell exhibited multiple peripheral flagella (blue arrows) and a polar flagellum (Figure 3). The presence of polar flagella among these cells suggested that swimming inhibition of ΔvmeL was due to flagellar malfunction rather than a default in flagellum biosynthesis.
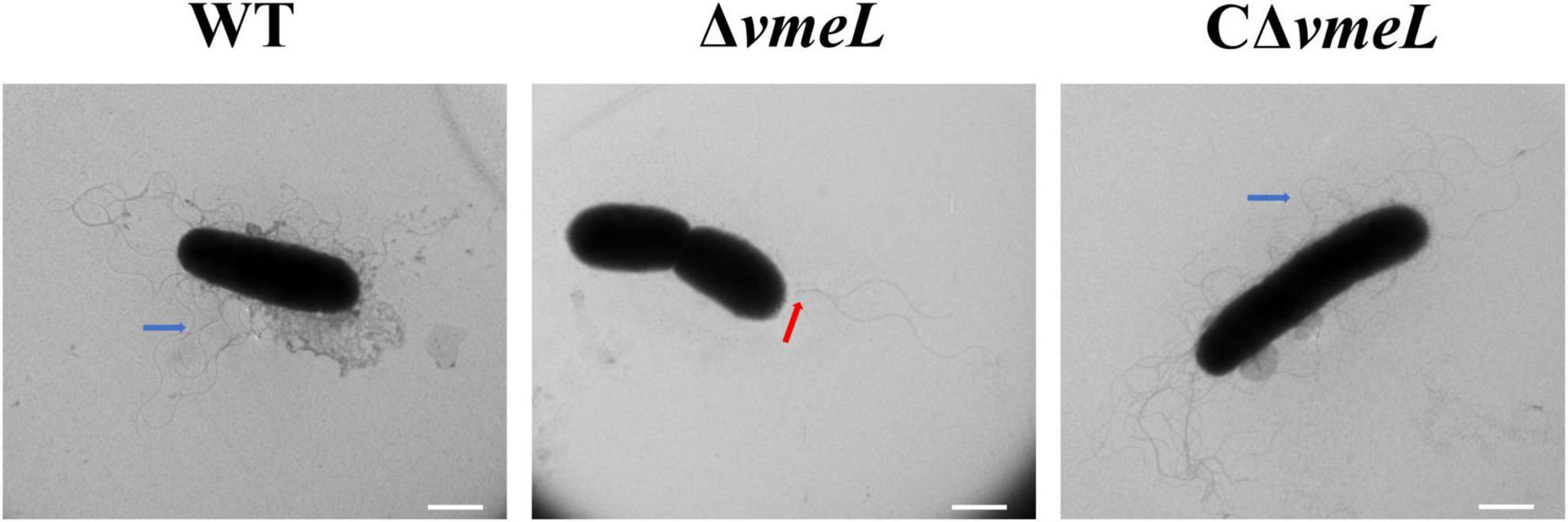
Figure 3. Transmission electron microscope (TEM) images of Vibrio parahaemolyticus flagella. Representative images of flagella in WT, ΔvmeL and CΔvmeL were observed by TEM and shown. Scale bar = 1 μm. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain.
To further investigate the role of VmeL, we next assessed whether the WT and ΔvmeL strains exhibited different levels of cytotoxicity towards HeLa cells. Morphological changes induced in HeLa cells by each strain were tested through staining with DAPI and phalloidin. The results showed that the number of cells on coverslips changed considerably after co-incubation with ΔvmeL, WT and CΔvmeL. Compared with WT and CΔvmeL, more HeLa cells remained on coverslips after incubating with ΔvmeL. As showed in Figure 4A, compared with untreated cells, the infected cells displayed morphological changes including rounding and lysis. Compared with WT, the cells incubated with ΔvmeL displayed lesser morphological changes, and the cells incubated with CΔvmeL displayed a significant cytoplasmic lysis phenomenon. These phenomenon suggested ΔvmeL strains exhibited lower cytotoxicity towards HeLa cells than did other strains. Further LDH assays sustained our conjecture. After co-incubation for 2 h, a significant decrease in LDH release was observed in the HeLa cells co-incubated with ΔvmeL strains compared with those incubated with the WT strain (Figure 4B). Together, these results suggested that VmeL affected the virulence of V. parahaemolyticus at the cellular level.
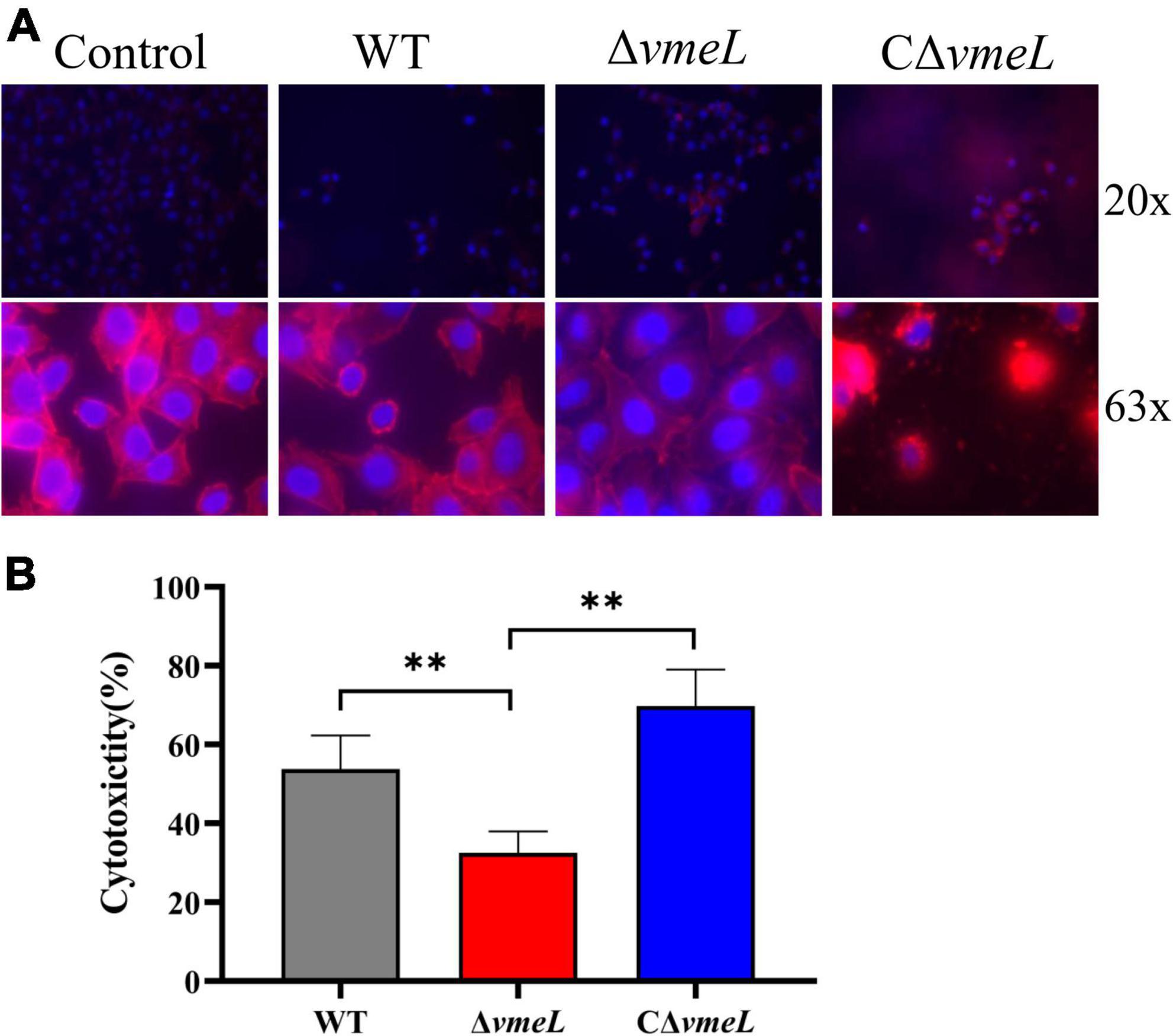
Figure 4. VmeL-mediated virulence toward HeLa cells. (A) Representative images showed HeLa cells infected with WT, ΔvmeL, and CΔvmeL strains, and one uninfected control group. DNA is shown in blue, and actin is in red. The top panels were photographed at 20 x magnification and the bottom panels were photographed at 63 x magnification. (B) Lactate dehydrogenase released by HeLa cells infected with WT, ΔvmeL, and CΔvmeL strains of Vibrio parahaemolyticus were tested at 2 h. Data represent the means of six replicates. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain. ** indicates statistical significance at P < 0.01.
We next aimed to determine whether VmeL of V. parahaemolyticus is involved in virulence. As seen in Figure 5, mice infected with the WT strain exhibited zero survival, while those infected with the ΔvmeL showed 40% survival 48 h after infection. In addition, the mice infected with CΔvmeL exhibited zero survival. Meanwhile, all negative control mice survived. After 24 h of infection, no further lethal symptoms were seen in the mice infected with the vmeL deletion mutant. These results suggested that VmeL in V. parahaemolyticus was involved in pathogenesis towards mouse.
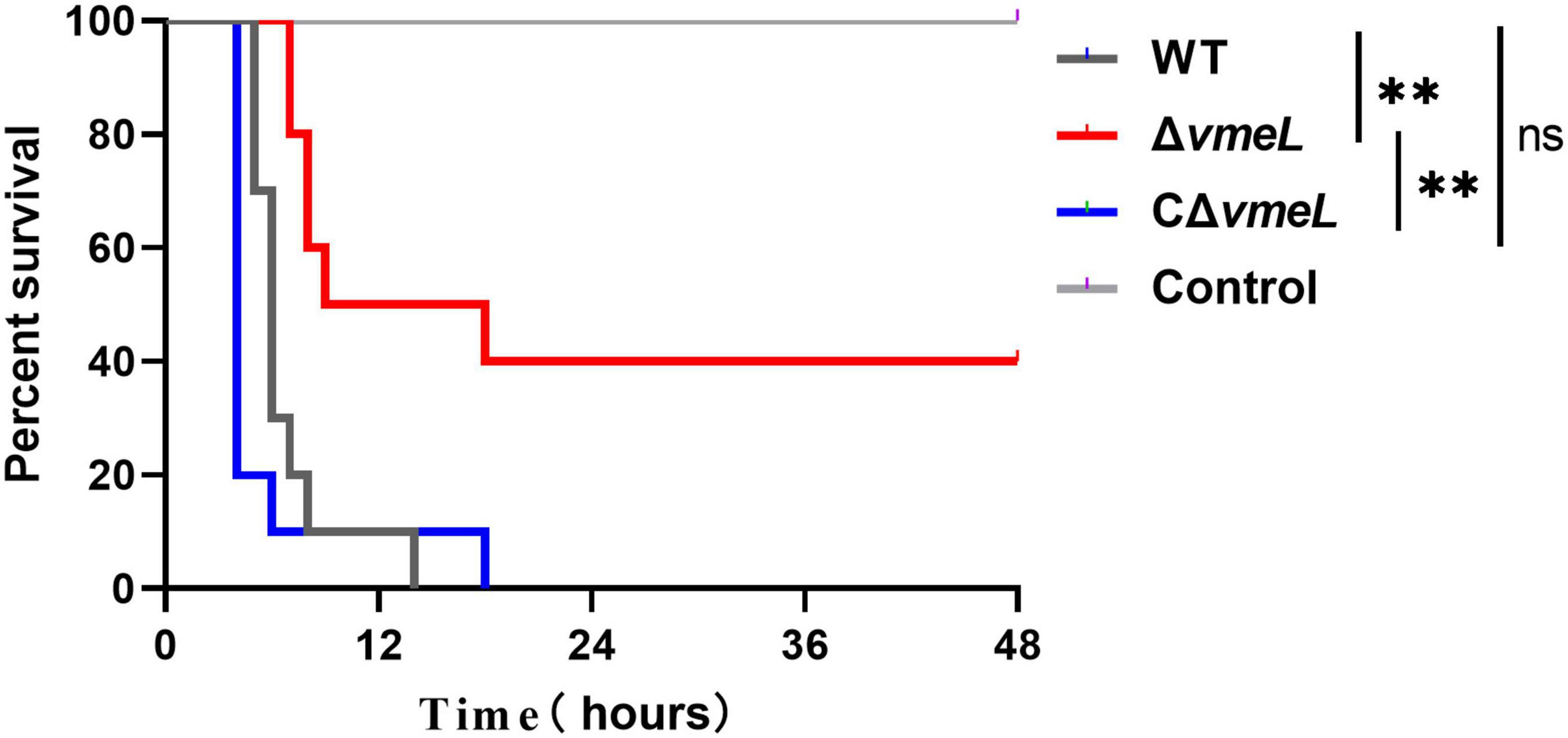
Figure 5. Institute of Cancer Research female mice (n = 10) were infected through intraperitoneal injection with WT, ΔvmeL, or CΔvmeL strains of Vibrio parahaemolyticus. Mouse mortality was recorded at the times indicated. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain. ** indicates statistical significance at P < 0.01.
To identify genes affected by VmeL, we performed transcriptome analysis of ΔvmeL cells using RNA-seq. Total RNA was isolated from ΔvmeL and WT strains cultured in MLB medium. In three biological replicates, the transcriptomes of the WT strain were compared with those of the ΔvmeL strain (P < 0.01, log2| FoldChange| > 1.5). RNAseq analysis showed the transcripts of the other 11 RND-efflux systems, as well as the outer membrane protein vpoc, were not influenced by the loss of vmeL. However, RNA-seq identified 963 genes that were differentially expressed in ΔvmeL (Supplementary Material). Of these DEGs, 716 genes were upregulated in ΔvmeL when compared to WT (Figure 6A and Supplementary Figure 4), indicating that many functions were repressed through mechanisms involving VmeL.
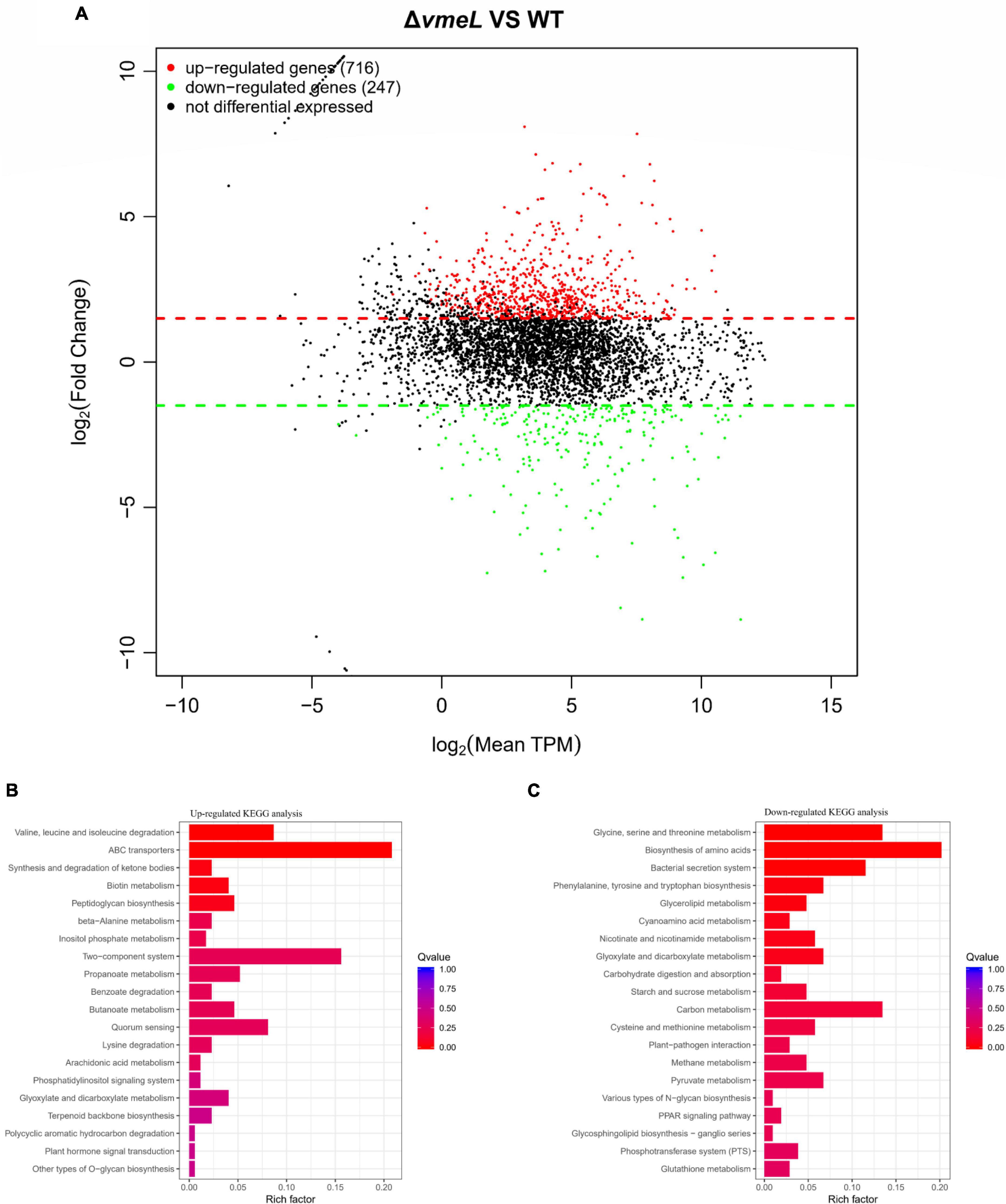
Figure 6. Transcriptomic analysis between ΔvmeL and WT strains of Vibrio parahaemolyticus. (A) MA plot shows the DEGs of ΔvmeL and WT. DEGs with log2FC > 1.5 are denoted in red, and DEGs with log2FC < 1.5 are denoted in green (P < 0.01). TPM indicates transcripts per kilobase of exon model per million mapped reads. (B,C) KEGG pathway enrichment analysis for ΔvmeL and WT strains. The color of the bar corresponds to different ranges of Q values (adjusted P values). WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain.
To further identify the functions of these DEGs, transcriptional profiles were analyzed using the KEGG database (Figures 6B,C). Twelve genes classified as part of the bacterial secretion system (ko03070) pathway were downregulated. Interestingly, all of these genes were related to T3SS1. In addition, a total of 36 ATP-binding cassette (ABC) transporter genes (ko02010) were upregulated in the vmeL mutant. 27 genes involved in two-component systems (ko02020) were upregulated, and 14 quorum sensing genes were upregulated. Furthermore, a large number of DEGs were enriched in the microbial metabolism pathways. For instance, 15 genes involved in valine, leucine, and isoleucine degradation (ko00280) were upregulated. Taken together, the results of transcriptional profiling suggested that VmeL is associated with many metabolic and virulence processes in V. parahaemolyticus.
Transcriptional levels of some differently expressed genes obtained from transcriptome data were further confirmed by qRT-PCR. The results indicated that the inactivation of vmeL decreased the relative expression of fourteen T3SS1-related genes, including eight T3SS1 structural genes and six T3SS1 effector gene (Figures 7A,B). On the other hand, the transcriptional levels were complemented in CΔvmeL indicating T3SS1-related genes in V. parahaemolyticus are affected by VmeL. But qRT-PCR data showed that tdh, which encodes thermostable direct hemolysin, was not altered.
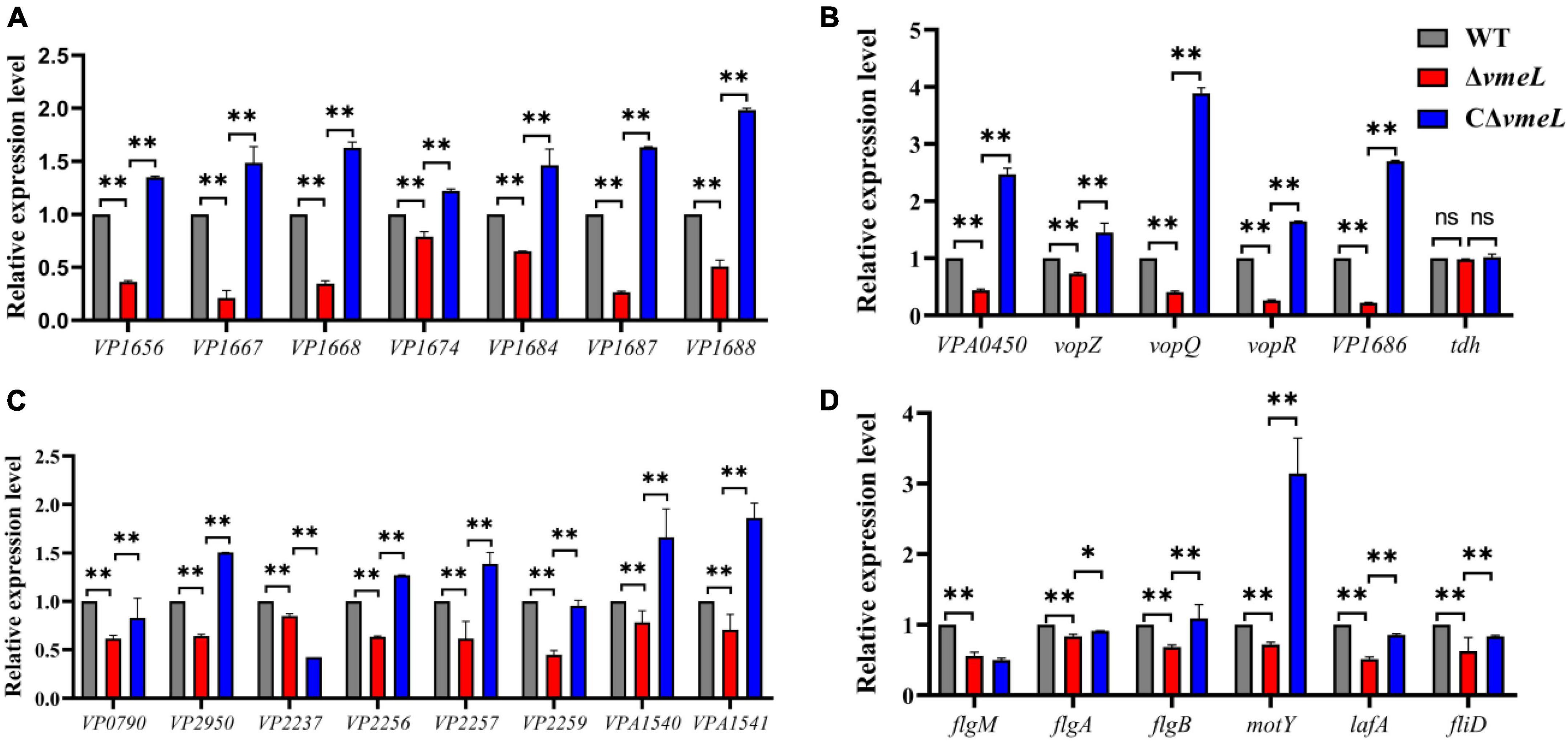
Figure 7. Quantitative reverse transcription-PCR verification of target genes in strains WT, ΔvmeL, CΔvmeL. The transcriptional levels of (A) T3SS1 structural genes, (B) T3SS1 effector genes and (C) flagellar-related genes were tested using the templates that collected from strains incubated in liquid medium. (D) The first gene of every operon was chosen to verify the transcriptional levels of lateral flagella genes in ΔvmeL as compared to those in the wild type (WT), and the templates that collected from strains incubated on swarming plates. WT, wild-type; ΔvmeL, vmeL deletion mutant; CΔvmeL, deletion mutant complemented strain. ** indicates statistical significance at P < 0.01. ns, no significant difference.
Furthermore, qRT-PCR analysis was used to verify the expression of flagellar-related genes that are shown to be altered in RNA-seq. the results demonstrated that the six polar flagellar assembly–related genes (VP0790, VP2950, VP2237, VP2256, VP2257, and VP2259) were downregulated (Figure 7C) and two lateral flagellar genes (VPA1540 and VPA1541), which encode flagellar motor switch proteins, were downregulated. In addition, RNA samples of strains were collected from swarming plates, and some specific genes were chosen as readouts to reflect the transcriptional level of lateral flagellar genes. The results showed that the transcriptional levels of flgM, flgA, flgB, motY, lafA, and fliD in V. parahaemolyticus were significantly downregulated when vmeL was deleted (Figure 7D). The transcriptional levels of these lateral flagellar genes, except for flgM, were all restored after vmeL was complemented.
The purpose of this study was to further investigate the biological contributions of the RND efflux protein VmeL in V. parahaemolyticus. We found that vmeL-deletion did not affect antimicrobial resistance, which is similar to a previous study that they found that single deletion of vmeLM show the same drug susceptibility as the wild type strain V. parahaemolyticus AQ334 (Matsuo et al., 2013). Moreover, our results firstly showed that the membrane fusion protein VmeL modulated biofilm formation, motility, and virulence, and possibly affected multiple metabolic pathways in V. parahaemolyticus.
Biofilms are aggregates of microorganisms embedded in an extracellular polymeric substances matrix and often adhere to a surface, they are highly tolerant to many antimicrobial agents, including antibiotics, heavy metals, and dyes (Yildiz and Visick, 2009; Flemming et al., 2016). In other bacteria, including A. nosocomialis (Knight et al., 2018) and A. baumannii (Srinivasan et al., 2015; Kim et al., 2021), researchers found that biofilm formation can be modulated by efflux pumps. In the present study, we discovered similar results that the loss of vmeL significantly decreased the biofilm formation of V. parahaemolyticus SH112. Similarly, a non-RND membrane fusion protein gene mfpC (VPA1443) mutant exhibited poor biofilm formation and produced smooth colonies on CR plates (Enos-Berlage et al., 2005). Previous studies have shown that Congo red binding and colony morphology is closely related with the synthesis of exopolysaccharides (Bassis and Visick, 2010; Chen et al., 2010; Wang et al., 2013; Reichhardt et al., 2016). Our results confirmed that the loss of vmeL in V. parahaemolyticus produced a more translucent colony on the HI plate and exhibited poorer binding ability with Congo red dye compared with WT strains. The exopolysaccharide plays a key role in the maturing of the biofilm (Yildiz and Visick, 2009). In V. parahaemolyticus, regulation of exopolysaccharide production and biofilm formation is complex, and involves numerous transcriptional regulators and operons, particularly two component signal transduction, quorum sensing regulators and the VPA1403-1412 (cpsA-J) operon (Boles and McCarter, 2002; Yildiz and Visick, 2009; Zhang et al., 2018). RNA-seq results showed the transcriptional level of cpsA is downregulated, while cpsG and cpsJ were upregulated, in the vmeL mutant, compared with wild type. In addition, alterations in gene expressions of quorum sensing and two component systems were found in transcriptomic data. Taken together, we speculated that membrane fusion proteins VmeL could affect biofilm formation in V. parahaemolyticus by means of decreasing the amount of extracellular matrix, especially exopolysaccharides. However, it is worth noting that extracellular DNA and proteins are also essential for biofilm formation of V. parahaemolyticus (Li et al., 2020). Therefore, we need to further clarify the causation of VmeL could influence biofilm formation of V. parahaemolyticus.
Flagellar motility in vibrio spp. is associated with several cellular processes, such as movement, colonization, adhesion, biofilm formation, and virulence (Taw et al., 2015; Khan et al., 2020). Surprisingly, swarming motility was eliminated in V. parahaemolyticus when vmeL was deleted. Swarming is considered a form of flagellum-driven bacterial group motility on a semi-solid surface, that facilitates the acquisition of nutrients and resistance to adverse environmental conditions (Jose and Singh, 2020). Similar results have only been found for the plant pathogen Pseudomonas syringae pv. tabaci 6605, in which inner membrane protein mexF deletion, abrogated swarming motility (Ichinose et al., 2020). Based on the TEM results, we inferred that VmeL influences the swarming motility of V. parahaemolyticus by affecting lateral flagellar assembly. However, in the DEG profile, only two lateral flagellar genes, VPA1540 and VPA1541, were downregulated. A reasonable explanation is that the sample used for RNA sequencing was collected through strains cultivated in MLB medium, but the lateral flagella of V. parahaemolyticus are not activated in a fluid environment (McCarter, 2004). Thus, there was no effect on the DEG profile, and the further qRT-PCR analysis affirmed this suspicion. The deletion of vmeL when incubated on swarming plates led to reduction in the transcriptional level of flgM, flgA, flgB, motY, lafA, fliD, that could be seen as the readouts of every operon of lateral flagellar genes (Stewart and McCarter, 2003). These results suggested that the swarming repression in the ΔvmeL mutant occurred through alterations to lateral flagellar biosynthesis.
Moreover, Swimming motility was abolished when vmeL was deleted in V. parahaemolyticus., and a similar phenomenon was observed in A. nosocomialis (Knight et al., 2018) and A. baumannii (Srinivasan et al., 2015). Although the qRT-PCR results confirmed that some polar flagellar genes were downregulated when vmeL was deleted, the TEM results showed that ΔvmeL cells exhibited polar flagella. These results suggest that RND efflux pumps can affect the swimming motility of bacteria through affecting the function of flagella rather than depressing flagellar assembly. The basal body, the hook and the filament worked together to structure effective bacterial flagellum, which assembled in a hierarchical manner starting at the inner cytoplasmic membrane and ultimately outside the cell (Echazarreta and Klose, 2019). Interestingly, all components of RND efflux systems are also located in the membrane structure, including membrane fusion proteins that exist in the periplasmic space (Venter et al., 2015). Based on these, we infer that RND efflux systems could interactively function with bacterial polar flagella, thereby led the abolishment of swimming motility.
The pathogenicity of V. parahaemolyticus is closely related to a variety of virulence factors, which enable it to cause not only gastroenteritis but also wound infections and septicemia (Daniels et al., 2000; Broberg et al., 2011). Previous studies have proved that the murine intraperitoneal infection model was successfully established and employed to investigate the pathogenicity of V. parahaemolyticus (Hiyoshi et al., 2010; Matsuda et al., 2019; Liu et al., 2021). In our study, the virulence, towards HeLa cells and ICR mouse, of V. parahaemolyticus was found to be lessened when in a vmeL mutant compared with wild type. Bacterial virulence is a sophisticated phenomenon that requires the coordinated expression of various genes types (Lee et al., 2006). Thermostable direct hemolysin (TDH), TDH-related hemolysin (TRH), and type 3 secretion systems (T3SSs) were seen as major pathogenic factors in V. parahaemolyticus (Li et al., 2019). T3SS1 mainly accounts for cytotoxicity, while T3SS2 mainly accounts for enterotoxicity (Hiyoshi et al., 2010). In the present work, the results of qRT-PCR displayed that most T3SS1-related genes were downregulated in ΔvmeL compared with wild type strains. A similar study found that RND null mutants increased leuO transcription in V. cholera (Bina et al., 2018), and the leuO-homologous protein CalR repressed the transcription of T3SS1 in V. parahaemolyticus (Gode-Potratz et al., 2010; Zhang et al., 2021b). Therefore, we inferred that VmeL mediates cytotoxicity by indirectly modulating the transcription of T3SS1 by an unknown mechanism. TDH has been shown to be associated with lethality in the murine intraperitoneal infection model (Matsuda et al., 2019). But inactivation of vmeL has no effect on the transcription of tdh. In addition, several studies demonstrated that the biofilm formation and surface motility of V. parahaemolyticus show a positive correlation with virulence toward mice (Wang et al., 2013; Whitaker et al., 2014; Karan et al., 2021; Li et al., 2022). Consequently, we speculated that VmeL-mediated lethality in the murine intraperitoneal infection on the ground of affecting surface motility, biofilm formation and the transcription of T3SS1-related genes.
RNA-seq results showed that various pathways were altered by vmeL deletion, including those related to ABC transporter systems, bacterial secretion systems, two-component systems, quorum sensing, and amino acid metabolism, suggesting that the global transcription in a vmeL mutant is disrupted compared with wild type. However, it is important to note that VmeL is an intrinsic membrane fusion protein that needs to be coupled with inner membrane protein VmeM to form an effective RND efflux system (Alvarez-Ortega et al., 2013; Matsuo et al., 2013), rather than a transcriptional regulator. Moreover, membrane fusion protein and inner membrane protein were often found in an operon (Martinez et al., 2009; Matsuo et al., 2013). In addition, qRT-PCR showed that the transcriptional level of vmeM dropped about 55% in the vmeL mutant compared with wild type, while vmeM expression was restored in the CΔvmeL strain (Supplementary Figure 2). These results indicated that vmeL positively regulates the transcription of the vmeLM operon. In V. cholerae, the accumulation of vibriobactin leaded to activation of the stress response Cpx two-component system in the absence of vexGH belonging to RND efflux systems (Kunkle et al., 2017). Two-component systems are cell-signaling circuitries that could maintain cellular hemostasis through sensing environmental signals (Papon and Stock, 2019). Similar to our RNA-seq results Bina et al. (2018) found that RND efflux systems had wide-ranging effects on V. cholerae transcriptome, such as several two-component systems. Therefore, we inferred that VmeL may affect multiple aspects of bacterial physiology presumable owing to the accumulation of substrates that could activate the two-component systems, which are normally from the periplasm by the VmeLM RND efflux systems. Notably Matsuo et al. (2013) tested the MICs of all 12 efflux systems deletion strains were decreased significantly, but also found most single deletions of RND efflux systems in V. parahaemolyticus AQ443 had no effect on drug resistance. This suggested the deficiency of one efflux systems gene deletion could be supplemented by other efflux systems. In our study, we found that vmeL-deletion did not affect antimicrobial resistance, and the RNA-seq analysis showed that the transcripts of the other 11 RNA-efflux systems or the outer membrane protein gene vpoC, were not impacted by the loss of vmeL (Supplementary Table 2). Besides, ΔvmeL strains exhibited transcriptional changes in 36 ABC transporter genes, which are similar with RND efflux systems, could efflux various substrates, including antibiotics (Alvarez-Ortega et al., 2013). These results suggested that there is an unknown functional interplay among different RND efflux systems or maybe among different kinds of transporter systems that functions to maintain cellular metabolic homeostasis.
In the present study, we found that VmeL in V. parahaemolyticus SH112 influences various biological functions including biofilm formation, swarming motility, swimming motility, and virulence towards HeLa cells and mice. Understanding the roles of VmeL can be useful for developing effective methods to target RND efflux systems to control both contamination and clinical infection caused by V. parahaemolyticus. Nevertheless, the mechanism by which VmeL affect various phenotypes have not been fully elucidated, as this will requires extensive experimentation. In view of our findings, we suggest that VmeL of V. parahaemolyticus is a potential target for the developments of vaccines to control contamination and infection by V. parahaemolyticus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA797934.
All animal infection experiments were approved by the Animal Ethics Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (no. SYXK2020-0027).
WJ conceived and designed the study. P-XL, X-YZ, YQ, and Y-YL carried out the experiments. P-XL and WJ analyzed the data. P-XL wrote the first draft of the manuscript and all authors contributed to subsequent revisions. All authors read and approved the final manuscript.
This study was funded by the National Natural Science Foundation of China (no. 31702277), the Shanghai Natural Science Foundation of China (nos. 21ZR1477000 and 17ZR1437200), and the Shanghai Science and Technology Commission Research Project (21N31901000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.976334/full#supplementary-material
Alav, I., Kobylka, J., Kuth, M. S., Pos, K. M., Picard, M., Blair, J. M. A., et al. (2021). Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 121, 5479–5596.
Alvarez-Ortega, C., Olivares, J., and Martinez, J. L. (2013). RND multidrug efflux pumps: What are they good for? Front. Microbiol. 4:7.
Banu, S. F., Rubini, D., Murugan, R., Vadivei, V., Gowrishankar, S., Pandian, S. K., et al. (2018). Exploring the antivirulent and sea food preservation efficacy of essential oil combined with DNase on Vibrio parahaemolyticus. Lwt Food Sci. Technol. 95, 107–115.
Bassis, C. M., and Visick, K. L. (2010). The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J. Bacteriol. 192, 1269–1278. doi: 10.1128/JB.01048-09
Bina, X. R., Howard, M. F., Taylor-Mulneix, D. L., Ante, V. M., Kunkle, D. E., and Bina, J. E. (2018). The Vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog 14:e1006804. doi: 10.1371/journal.ppat.1006804
Boles, B. R., and McCarter, L. L. (2002). Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184, 5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001.
Chen, Y., Dai, J., Morris, J. G. Jr., and Johnson, J. A. (2010). Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. doi: 10.1186/1471-2180-10-274
Daniels, N. A., Mackinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M., et al. (2000). Vibrio parahaemolyticus infections in the united states, 1973-1998. J. Infect. Dis. 181, 1661–1666.
Echazarreta, M. A., and Klose, K. E. (2019). Vibrio flagellar synthesis. Front. Cell Infect. Microbiol. 9:131.
Enos-Berlage, J. L., Guvener, Z. T., Keenan, C. E., and Mccarter, L. L. (2005). Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55, 1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575.
Ghenem, L., Elhadi, N., Alzahrani, F., and Nishibuchi, M. (2017). Vibrio parahaemolyticus: A review on distribution, pathogenesis, virulence determinants and epidemiology. Saudi J. Med. Med. Sci. 5, 93–103.
Gode-Potratz, C. J., Chodur, D. M., and Mccarter, L. L. (2010). Calcium and iron regulate swarming and type III secretion in vibrio parahaemolyticus. J. Bacteriol. 192, 6025–6038. doi: 10.1128/JB.00654-10
Gu, D., Meng, H. M., Li, Y., Ge, H. J., and Jiao, X. A. (2019). A GntR family transcription factor (vpa1701) for swarming motility and colonization of Vibrio parahaemolyticus. Pathogens 8:235. doi: 10.3390/pathogens8040235
Hall, C. W., and Mah, T. F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301.
Hiyoshi, H., Kodama, T., Iida, T., and Honda, T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780. doi: 10.1128/IAI.01051-09
Hotinger, J. A., Morris, S. T., and May, A. E. (2021). The case against antibiotics and for anti-virulence therapeutics. Microorganisms 9:2049.
Ichinose, Y., Nishimura, T., Harada, M., Kashiwagi, R., Yamamoto, M., Noutoshi, Y., et al. (2020). Role of two sets of RND-type multidrug efflux pump transporter genes, mexAB-oprM and mexEF-oprN, in virulence of Pseudomonas syringae pv. tabaci 6605. Plant Pathol. J. 36, 148–156. doi: 10.5423/PPJ.OA.11.2019.0273
Jose, R., and Singh, V. (2020). Swarming in bacteria: A tale of plasticity in motility behavior. J. Indian Instit. Sci. 100, 515–524.
Karan, S., Garg, L. C., Choudhury, D., and Dixit, A. (2021). Recombinant FimH, a fimbrial tip adhesin of Vibrio parahaemolyticus, elicits mixed T helper cell response and confers protection against Vibrio parahaemolyticus challenge in murine model. Mol. Immunol. 135, 373–387. doi: 10.1016/j.molimm.2021.05.005
Khan, F., Tabassum, N., Anand, R., and Kim, Y. M. (2020). Motility of vibrio spp.: Regulation and controlling strategies. Appl. Microbiol. Biotechnol. 104, 8187–8208. doi: 10.1007/s00253-020-10794-7
Kim, C. M., Park, G., Ko, Y. J., Kang, S. H., and Jang, S. J. (2021). Relationships between relative expression of RND efflux pump genes, H33342 efflux activity, biofilm-forming activity, and antimicrobial resistance in Acinetobacter baumannii clinical isolates. JPN J. Infect. Dis. 74, 499–506. doi: 10.7883/yoken.JJID.2020.765
Knight, D. B., Rudin, S. D., Bonomo, R. A., and Rather, P. N. (2018). Acinetobacter nosocomialis: defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front. Microbiol. 9:1902. doi: 10.3389/fmicb.2018.01902
Kunkle, D. E., Bina, X. R., and Bina, J. E. (2017). The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8, e00126–17. doi: 10.1128/mBio.00126-17
Lee, D. G., Urbach, J. M., Wu, G., Liberati, N. T., Feinbaum, R. L., Miyata, S., et al. (2006). Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Geno. Biol. 7:R90. doi: 10.1186/gb-2006-7-10-r90
Lei, T., Jiang, F., He, M., Zhang, J., Zeng, H., Chen, M., et al. (2020). Prevalence, virulence, antimicrobial resistance, and molecular characterization of fluoroquinolone resistance of Vibrio parahaemolyticus from different types of food samples in China. Int. J. Food. Microbiol. 317:108461. doi: 10.1016/j.ijfoodmicro.2019.108461
Li, L., Meng, H., Gu, D., Li, Y., and Jia, M. (2019). Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 222, 43–51.
Li, W., Wang, J. J., Qian, H., Tan, L., Zhang, Z., Liu, H., et al. (2020). Insights into the role of extracellular DNA and extracellular proteins in biofilm formation of Vibrio parahaemolyticus. Front. Microbiol. 11:813. doi: 10.3389/fmicb.2020.00813
Li, Y., Sun, W., Wang, Q., Yu, Y., Wan, Y., Zhou, K., et al. (2022). The GntR-like transcriptional regulator HutC involved in motility, biofilm-forming ability, and virulence in Vibrio parahaemolyticus. Microb Pathog 167, 105546. doi: 10.1016/j.micpath.2022.105546
Lian, L., Xue, J., Li, W., Ren, J., Tang, F., Liu, Y., et al. (2021). VscF in T3SS1 helps to translocate VPA0226 in Vibrio parahaemolyticus. Front. Cell Infect. Microbiol. 11:652432. doi: 10.3389/fcimb.2021.652432
Liu, J. F., Qin, K. W., Wu, C. L., Fu, K. F., Yu, X. J., and Zhou, L. J. (2021). De novo sequencing provides insights into the pathogenicity of foodborne Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 11:652957. doi: 10.3389/fcimb.2021.652957
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408.
Martinez, J. L., Sanchez, M. B., Martinez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449.
Matsuda, S., Okada, R., Tandhavanant, S., Hiyoshi, H., Gotoh, K., Iida, T., et al. (2019). Export of a Vibrio parahaemolyticus toxin by the sec and type III secretion machineries in tandem. Nat. Microbiol. 4, 781–788. doi: 10.1038/s41564-019-0368-y
Matsuo, T., Hayashi, K., Morita, Y., Koterasawa, M., Ogawa, W., Mizushima, T., et al. (2007). VmeAB, an RND-type multidrug efflux transporter in Vibrio parahaemolyticus. Microbiology (Reading) 153, 4129–4137. doi: 10.1099/mic.0.2007/009597-0
Matsuo, T., Nakamura, K., Kodama, T., Mikami, T., Hiyoshi, H., Tsuchiya, T., et al. (2013). Characterization of all RND-type multidrug efflux transporters in Vibrio parahaemolyticus. Microbiologyopen 2, 725–742. doi: 10.1002/mbo3.100
McCarter, L. (1999). The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1, 51–57.
McCarter, L. L. (2004). Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7, 18–29. doi: 10.1159/000077866
Morales, V. M., Backman, A., and Bagdasarian, M. (1991). A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47. doi: 10.1016/0378-1119(91)90007-x
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Ottaviani, D., Leoni, F., Serra, R., Serracca, L., Decastelli, L., Rocchegiani, E., et al. (2012). Nontoxigenic Vibrio parahaemolyticus strains causing acute gastroenteritis. J. Clin. Microbiol. 50, 4141–4143. doi: 10.1128/JCM.01993-12
Reichhardt, C., Mccrate, O. A., Zhou, X., Lee, J., Thongsomboon, W., and Cegelski, L. (2016). Influence of the amyloid dye congo red on curli, cellulose, and the extracellular matrix in E. coli during growth and matrix purification. Anal. Bioanal. Chem. 408, 7709–7717. doi: 10.1007/s00216-016-9868-2
Srinivasan, V. B., Venkataramaiah, M., Mondal, A., and Rajamohan, G. (2015). Functional characterization of AbeD, an RND-type membrane transporter in antimicrobial resistance in Acinetobacter baumannii. PLoS One 10:e0141314. doi: 10.1371/journal.pone.0141314
Stewart, B. J., and McCarter, L. L. (2003). Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185, 4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Tandhavanant, S., Matsuda, S., Hiyoshi, H., Iida, T., and Kodama, T. (2018). Vibrio parahaemolyticus senses intracellular K+ to translocate type III secretion system 2 effectors effectively. mBio 9:e01366–18. doi: 10.1128/mBio.01366-18
Taw, M., Lee, H. I., Lee, S. H., and Chang, W. S. (2015). Characterization of MocR, a GntR-like transcriptional regulator, in Bradyrhizobium japonicum: Its impact on motility, biofilm formation, and soybean nodulation. J. Microbiol. 53, 518–525. doi: 10.1007/s12275-015-5313-z
Uddin, T. M., Chakraborty, A. J., Khusro, A., Zidan, B. R. M., Mitra, S., Emran, T. B., et al. (2021). Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health. 14, 1750–1766. doi: 10.1016/j.jiph.2021.10.020
Venter, H., Mowla, R., Ohene-Agyei, T., and Ma, S. (2015). RND-type drug e ffl ux pumps from gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6:377. doi: 10.3389/fmicb.2015.00377
Wadhwa, N., and Berg, H. C. (2021). Bacterial motility: Machinery and mechanisms. Nat. Rev. Microbiol. 20, 161–173.
Wang, L., Ling, Y., Jiang, H., Qiu, Y., Qiu, J., Chen, H., et al. (2013). AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int. J. Food Microbiol. 160, 245–251. doi: 10.1016/j.ijfoodmicro.2012.11.004
Whitaker, W. B., Richards, G. P., and Boyd, E. F. (2014). Loss of sigma factor RpoN increases intestinal colonization of vibrio parahaemolyticus in an adult mouse model. Infect. Immun. 82, 544–556. doi: 10.1128/IAI.01210-13
Yildiz, F. H., and Visick, K. L. (2009). Vibrio biofilms: So much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
Yu, M. J., Zheng, L., Wang, X. B., Wu, M. F., Qi, M., Fu, W. D., et al. (2019). Comparative transcriptomic analysis of surf clams (Paphia undulate) infected with two strains of Vibrio spp. reveals the identity of key immune genes involved in host defense. BMC Geno. 20:988. doi: 10.1186/s12864-019-6351-4
Yu, Y., Yang, H., Li, J., Zhang, P., Wu, B., Zhu, B., et al. (2012). Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 194, 827–835. doi: 10.1007/s00203-012-0816-z
Zhang, L., Weng, Y., Wu, Y., Wang, X., Yin, Z., Yang, H., et al. (2018). H-NS is an activator of exopolysaccharide biosynthesis genes transcription in Vibrio parahaemolyticus. Microb Pathog 116, 164–167. doi: 10.1016/j.micpath.2018.01.025
Zhang, Y., Qiu, Y., Gao, H., Sun, J., Li, X., Zhang, M., et al. (2021a). OpaR controls the metabolism of c-di-GMP in Vibrio parahaemolyticus. Front. Microbiol. 12:676436. doi: 10.3389/fmicb.2021.676436
Keywords: Vibrio parahaemolyticus, VmeL, swarming, biofilm, RNA-sequencing, pathogenicity
Citation: Liu P-x, Zhang X-y, Wang Q, Li Y-y, Sun W-d, Qi Y, Zhou K, Han X-g, Chen Z-g, Fang W-h and Jiang W (2022) Biological and transcriptional studies reveal VmeL is involved in motility, biofilm formation and virulence in Vibrio parahaemolyticus. Front. Microbiol. 13:976334. doi: 10.3389/fmicb.2022.976334
Received: 23 June 2022; Accepted: 19 July 2022;
Published: 09 August 2022.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Nicky O’Boyle, University College Cork, IrelandCopyright © 2022 Liu, Zhang, Wang, Li, Sun, Qi, Zhou, Han, Chen, Fang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Jiang, amlhbmd3ZWlqdzk5QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.