- Grassland Science, School of Animal Technology, Shihezi University, Shihezi, China
Condensed tannins (CT) from sainfoin have a high capacity to inhibit proteolysis. A previous study reported that CT from sainfoin can inhibit lactic acid bacteria activity and decrease ammonium-nitrogen (N) content during sainfoin ensiling; however, no study has focused on the metabolome of ensiled sainfoin. The objective of the present study was to investigate the effects of CT [following supplementation of deactivated CT with polyethylene glycol (PEG)] on protease activity, keystone bacteria, and metabolome during sainfoin ensiling. According to the results, PEG amendment increased non-protein N, amino acid, and soluble protein contents significantly (in the 49.08–59.41, 116.01–64.22, and 23.5–41.94% ranges, respectively, p < 0.05) during ensiling, whereas neutral detergent-insoluble protein and acid detergent-insoluble protein were decreased significantly (in the 55.98–64.71 and 36.58–57.55% ranges, respectively, p < 0.05). PEG supplementation increased aminopeptidase and acid protease activity after 3 days of ensiling (p < 0.05) and increased carboxypeptidase activity during the entire ensiling process (p < 0.05). The keystone bacteria changed following PEG addition (Stenotrophomonas, Pantoea, and Cellulosimicrobium in the control vs. Microbacterium, Enterococcus, and Brevundimonas in the PEG-treated group). In total, 510 metabolites were identified after 60 days of sainfoin ensiling, with 33 metabolites annotated in the Kyoto Encyclopedia of Genes and Genomes database. Among the metabolites, phospholipids were the most abundant (72.7% of 33 metabolites). In addition, 10 upregulated and 23 downregulated metabolites were identified in the PEG-treated group when compared with the control group, after 60 days of ensiling (p < 0.05). Pediococcus (correlated with 20 metabolites, R2 > 0.88, p < 0.05) and Lactobacillus (correlated with 16 metabolites, R2 > 0.88, p < 0.05) were the bacteria most correlated with metabolites. The results suggested antagonistic effects between Lactobacillus and Pediococcus during ensiling. The decreased proteolysis during sainfoin ensiling was mainly attributed to the inhibition of protease activity by CT, particularly carboxypeptidase activity. In addition, proteolysis decreased partly due to CT inhibiting Pediococcus activity during ensiling, with Pediococcus being significantly and positively correlated with dopamine after 60 days of ensiling (R2 = 0.8857, p < 0.05).
Introduction
During ensiling, especially in the case of legumes with high protein contents, proteolysis occurs driven by plant enzymes and microbial activity, which leads to the production of high non-protein nitrogen (NPN) contents (McDonald et al., 1991). During rumen microbial protein synthesis, less NPN from silage is utilized than NPN from fresh forages (Wetherall et al., 1995). Several studies have observed that condensed tannins (CT) reduce protein degradation during ensiling, due to the formation of stable compounds following the binding of CT to protein and inhibition of microbial activity (Peng et al., 2018; He et al., 2020). CT are plant secondary metabolites that are widely distributed in numerous plants, such as sericea lespedeza (Lespedeza cuneata Dum.-Cours.), crown vetch (Coronilla varia L.), hedysarum (Hedysarum alpinum L.), birdsfoot trefoil (Lotus corniculatus L.), and sainfoin (Onobrychis viciifolia). Among the plants above, CT from sainfoin is favored considering it exhibits the highest capacity to bind protein, and inhibits cellulose digestion by rumen bacteria the least so that it delivers a greater amount of protein to the abomasum without adverse effects on rumen microbial activity (McAllister et al., 2005).
Sainfoin is widespread in North America, Europe, and the Middle East, as a perennial forage and fodder legume (Carbonero et al., 2011). CT from sainfoin reportedly reduces protein degradation (Aufrere et al., 2014), reduces methane production (Wang et al., 2021), and has antihelmintic potential against gastrointestinal nematodes (Novobilsky et al., 2013) in the rumen, which improves animal performance.
During alfalfa ensiling, NPN contents decreased with an increase in sainfoin application rate, which was attributed to the CT in sainfoin inhibiting protein degradation (Wang et al., 2021). Similar results have been observed by other researchers when polyethylene glycol (PEG) is used to minimize the effect of CT from sainfoin (Peng et al., 2018; He et al., 2020). However, to the best of our knowledge, few researchers have explored the effects of CT from sainfoin on protease activity and the metabolome during sainfoin ensiling (Theodoridou et al., 2012). A previous study found that CT inhibits Pediococcus activity without affecting Enterobacter activity. Enterobacter can produce amino acid nitrogen during ensiling; however, CT decreases amino nitrogen (AN) contents during the entire ensiling process (Huang et al., 2022). Considering the protein degradation rate is correlated with plant enzyme and microbial activity, CT of sainfoin could have strong effects on protease activity. However, considering the relative abundance of Enterobacter has been reported to be 1.04–17.96% during sainfoin ensiling (Huang et al., 2022), the relationship between microbial activity and protein degradation remains unclear. Consequently, profiling the silage metabolome could enhance our understanding of the biological processes underlying silage fermentation (Guan et al., 2020). Therefore, the present study aimed to investigate the effects of CT on protease activity and the sainfoin metabolome during sainfoin ensiling.
Materials and methods
Forage and silage preparation
Sainfoin (O. viciifolia) was planted in Shihezi City, Xinjiang Province, China. Whole-plant sainfoin was harvested in July 2021 at the early flower stage. After wilting for 12 h to a dry matter (DM) content of approximately 240 g/kg fresh weight, the samples were chopped into 1— to 2-cm pieces. The samples were then sprayed with a solution of 640 g/l PEG (Sigma, molecular weight, 6,000) at a rate of 217 ml/kg DM to achieve a CT:PEG ratio of 1:2 in the samples (Peng et al., 2018). According to Jones and Mangan (1977), PEG inactivates condensed tannins’ biological activity due to its specific form of stable PEG-CT complexes attributed to binds with CT by H-bonds. Three separate piles were prepared and treated separately and bagged randomly for the control and PEG-treated groups. The control samples were sprayed with an equivalent amount of distilled water. After spraying, 1,000 g samples of both the chopped treated and control sainfoin were packed into polyethylene plastic bags (30 × 50 cm), then compacted and sealed using a vacuum sealer. Three replicates were prepared for the control and treatment samples. The bags were stored indoors at 23°C. After ensiling, samples were obtained after different ensilage days (3, 7, 14, 30, and 60) and stored at −20°C for later analyses.
Characteristics of ensiled sainfoin
Silage samples from separate bags for 3 (five fermentation days) × 2 (two treatments) × 3 (replicates) of each silage treatment were collected. Silage samples were dried at 65°C for 48 h and ground using a 1.0-mm in preparation for DM content determination. The NPN, neutral detergent-insoluble protein (NDIP), and acid detergent-insoluble protein (ADIP) concentrations were analyzed according to Licitra et al. (1996). Soluble protein (SOLP) concentrations were analyzed according to Bradford and Marion (1976). The protein fraction dynamics during sainfoin ensiling were analyzed according to the Cornell Net Carbohydrate and Protein System (CN) by Sniffen et al. (1992).
Proteolytic enzyme activity analysis
Samples (10 g) were obtained from each silage treatment, blended in a homogenized (L-1BA, Kuansons Biotechnology Co. Ltd., Shanghai, China) 50 ml of 0.1 M sodium phosphate buffer (pH 6.5), and then centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was stored at −80°C until proteolytic enzyme activity analysis. L-leucine-p-nitroanilide was used as the substrate for aminopeptidases (APs), N-carbobenzoxy-L-phenylalanine-L-alanine was used as the substrate for carboxypeptidase (CP), and azocasein was used as the substrate for acid protease (ACP). Proteolytic enzyme activity analysis was carried out according to Guo et al. (2007).
Metabolite analysis
Sample preparation and metabolite extraction
After 60 days of ensiling, 50-g samples of the ensiled sainfoin treatments were obtained and metabolites extracted using 400-μl methanol:water (4:1, v/v) solution. The mixture was allowed to settle at −20°C and treated using a high throughput tissue crusher (Wonbio-96c Shanghai Wanbo Biotechnology Co., Ltd., Shanghai, China) at 50 Hz for 6 min, followed by vortexing for 30 s and ultrasonication at 40 kHz for 30 min at 5°C. Afterward, the samples were placed at −20°C for 30 min to precipitate proteins. After centrifugation at 13,000 × g at 4°C for 15 min, the supernatant was collected for ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) analysis.
Ultra-high performance liquid chromatography-tandem mass spectrometry analysis
Chromatographic separation of the metabolites was performed on a Thermo UHPLC system equipped with an ACQUITY BEH C18 column (100 mm × 2.1 mm i.d., 1.7 μm; Waters, Milford, MA, United States). The mobile phases consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile:isopropanol (1:1, v/v) solution (solvent B). The solvent gradient was as follows: from 0 to 3 min, 95% (A):5% (B) to 80% (A):20% (B); from 3 to 9 min, 80% (A):20% (B) to 5% (A):95% (B); from 9 to 13 min, 5% (A):95% (B) to 5% (A):95% (B); from 13 to 13.1 min, 5% (A):95% (B) to 95% (A):5% (B), from 13.1 to 16 min, 95% (A):5% (B) to 95% (A):5% (B) for equilibrating the systems. The sample injection volume was 2 μl and the flow rate was 0.4 ml/min. The column temperature was maintained at 40°C. The mass spectrometric data were collected using a Thermo UHPLC-Q Exactive Mass Spectrometer equipped with an electrospray ionization (ESI) source operating in either positive or negative ion mode. The optimal conditions were set as follows: Aus gas heater temperature, 400°C; Sheath gas flow rate 40 psi; Aus gas flow rate 30 psi; ion-spray voltage floating (ISVF), −2,800 V in negative mode and 3,500 V in positive mode, respectively; normalized collision energy, 20–40–60 V rolling for MS/MS. Data acquisition was performed in the Data Dependent Acquisition (DDA) mode. The detection was carried out over a mass range of 70–1,050 m/z.
Metabolite analysis
Multivariate statistical analysis was performed using the ropls package (v1.6.21) in R on the Majorbio Cloud Platform.2 Principal component analysis (PCA) based on an unsupervised method was applied to obtain an overview of the metabolic data. General clustering, trends, or outliers were visualized. All the metabolite variables were scaled to unit-variances before conducting the PCA. Orthogonal partial least squares discriminate analysis (OPLS-DA) was used for statistical analysis to determine global metabolic changes between comparable groups. All the metabolite variables were subjected to Pareto Scaling before the OPLS-DA. The model validity was evaluated based on model parameters R2 and Q2, which provide information on interpretability and predictability, respectively, of the model, and minimize over-fitting risk. Variable importance in projection (VIP) was calculated using the OPLS-DA model. The p-values were estimated using paired Student’s t-test on single-dimensional statistical analysis.
Statistical significance among groups was identified with VIP values >1 and p < 0.05. The Majorbio Cloud Platform (Majorbio Bio-pharm Technology Corporation, Shanghai, China3) was used for further analyses.
Statistical analysis
The sainfoin silage data were subjected to a two-way analysis of variance, 2 × 5 factorial Complete Randomized Design (PEG and control treatments × five ensiling times days). Data were analyzed using IBM SPSS 22 Statistics (IBM Corp., Armonk, NY, United States). Significant differences between treatments were determined using Tukey’s test at p < 0.05. The ammonium nitrogen (NH4+-N), cured protein (CP), and bacteria sequence (SRA number for bacteria: PRJNA796365) data used were obtained from our former study (Huang et al., 2022).
Results
Nitrogen distribution in sainfoin silage
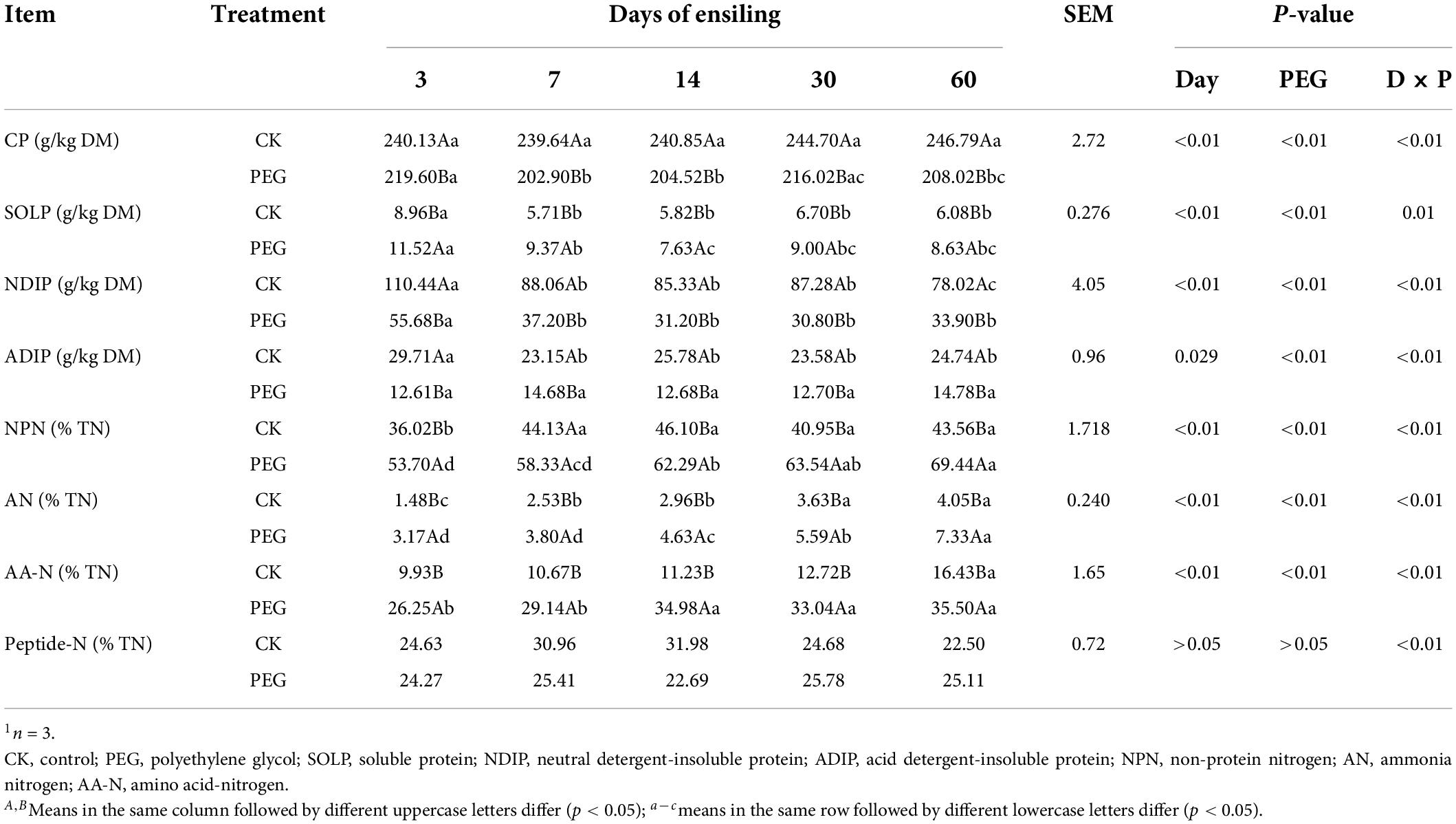
Nitrogen distribution during sainfoin ensiling is listed in Table 1. SOLP content decreased in both the control and treated groups with the prolongation of ensiling; SOLP content in the control decreased significantly after 7 days of ensiling (8.96 g/kg DM vs. 5.71 g/kg DM, p < 0.05), then stabilized with the prolongation of ensiling; SOLP content was the lowest in the PEG-treated group after 14 days of ensiling, which was 7.63 g/kg DM. However, the control had the lowest SOLP content during the entire ensiling process (p < 0.05). Similar results were observed for the effect of ensiling time on NDIP and ADIP contents. PEG addition decreased NDIP and ADIP contents markedly during ensiling by up to 100%, when compared with the contents in the control (p < 0.05). NPN and SOLP content trends in the control were similar. In addition, PEG supplementation increased NPN content significantly during the entire ensiling process (p < 0.05), by up to 59.41%, when compared with the contents in the control after 60 days of ensiling. Ensiling time increased amino acid (AA) contents in both the control and PEG-treated groups (p < 0.05), by 16.43 and 35.50%, respectively, after 60 days of ensiling. Furthermore, the PEG-treated group had the highest AA contents during the entire ensiling process (p < 0.05), which were up to twofold the contents in the control group.
Protein fractions of sainfoin silage
The protein fraction dynamics during sainfoin ensiling determined based on the CNCPS system are shown in Figures 1A–E. PA contents increased rapidly during 7 days of ensiling in both the control and PEG-treated groups. The PEG-treated groups had the highest PA contents during the entire ensiling process (p < 0.05). PB1 content decreased to 49.61% of the initial contents after 7 days of ensiling and then stabilized over time in the control sainfoin silage. However, the PEG-treated groups still had 86.42% of the initial PB1 content after 60 days of ensiling. In addition, PEG-treated groups had higher PB1 contents than the control group during the entire ensiling process (p < 0.05). However, PB2 content was higher in the control group than in the PEG-treated group after 60 days of ensiling, which were 22.26 (% CP) and 10.08 (% CP), respectively. PB3 content decreased rapidly during 14 days of ensiling in both the control and PEG-treated silage. PB3 content was higher in the control group than in the PEG-treated group during the entire ensiling process (p < 0.05). After 60 days of ensiling, PB3 content in the control was 134% higher than that in the PEG-treated group. PC content was higher in the control group than in the PEG-treated group after 60 days of ensiling, which were 10.02 (% CP) and 7.11 (% CP), respectively.
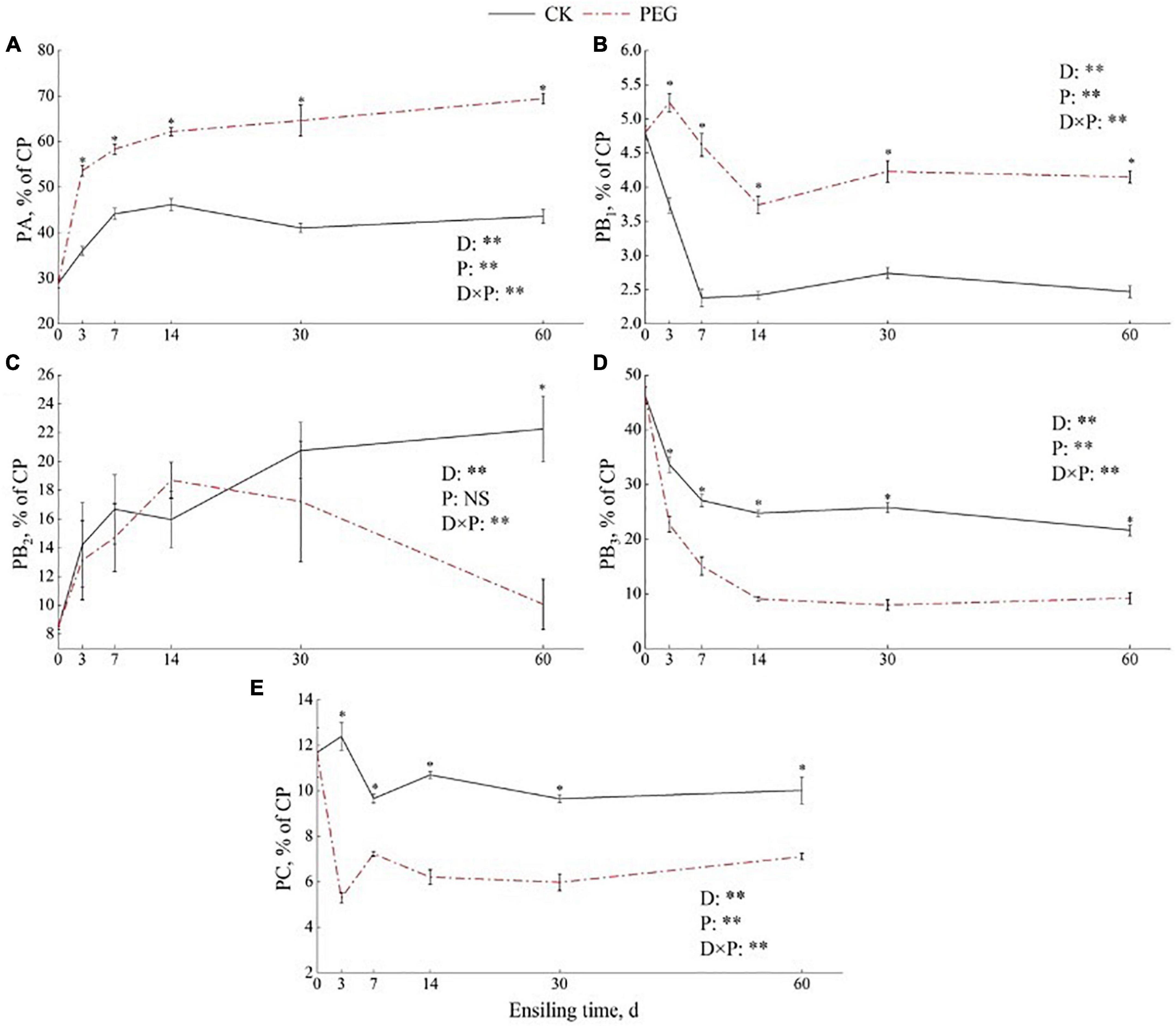
Figure 1. Effect of intrinsic tannins on protein fraction of (A) PA, (B) PB1, (C) PB2, (D) PB3, and (E) PC during sainfoin ensiling. PA (% CP) = NPN (% SOLP) × 0.01 × SOLP (% CP); PB1 = SOLP (% CP)-PA (% CP); PB2 = 100-PA (% CP)-PB1 (% CP)-PB3 (% CP)-PC (% CP); PB3 = NDIN (% CP)-ADIN (% CP); PC = ADIN (% CP). PEG, polyethylene glycol. D = effects of ensiling days; P = effects of addition with PEG. D × P = effects of interaction between ensiling days and PEG treated. Error bars mean SEM. Asterisks (*p < 0.05, **p < 0.01) indicate that there was a significant difference between the control and PEG-treated groups.
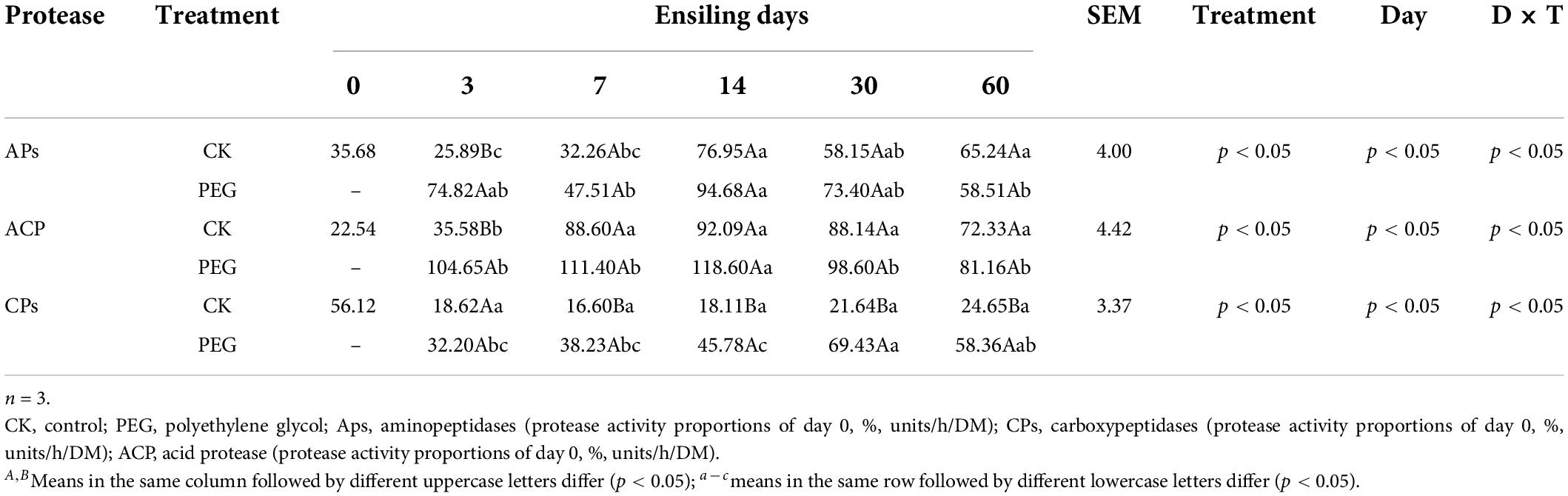
Protease activity in sainfoin silage
Protease activity during sainfoin ensiling is presented in Table 2. PEG addition only increased AP and ACP activities after 3 days of ensiling (p < 0.05), which increased by 188.99% and 194.13%, respectively, when compared with the levels in the control. The PEG-treated group exhibited the highest CP activity at 7–60 days of ensiling (p < 0.05), which was 38.23–69.43% of the protease activity at 0 day. AP and ACP activities both increased with an increase in the ensiling period in the control group (p < 0.05). In the PEG treatment, both AP and ACP activities were the highest after 14 days of ensiling; however, they decreased after 60 days of ensiling (p < 0.05). CP activity did not change with change in the ensiling period in the control group (p > 0.05); however, CP activity increased after 30 days of ensiling in the PEG-treated group (p < 0.05).
Bacterial community structure in sainfoin silage
Bacterial community network analysis results are illustrated in Figures 2A,B, based on betweenness centrality, degree centrality, and closeness centrality values. In the control treatment, the keystone bacteria were Sphingobacterium, Stenotrophomonas, Pantoea, and Cellulosimicrobium. In the PEG treatment group, the keystone bacteria were Microbacterium, Enterococcus, Sphingobacterium, and Brevundimonas.
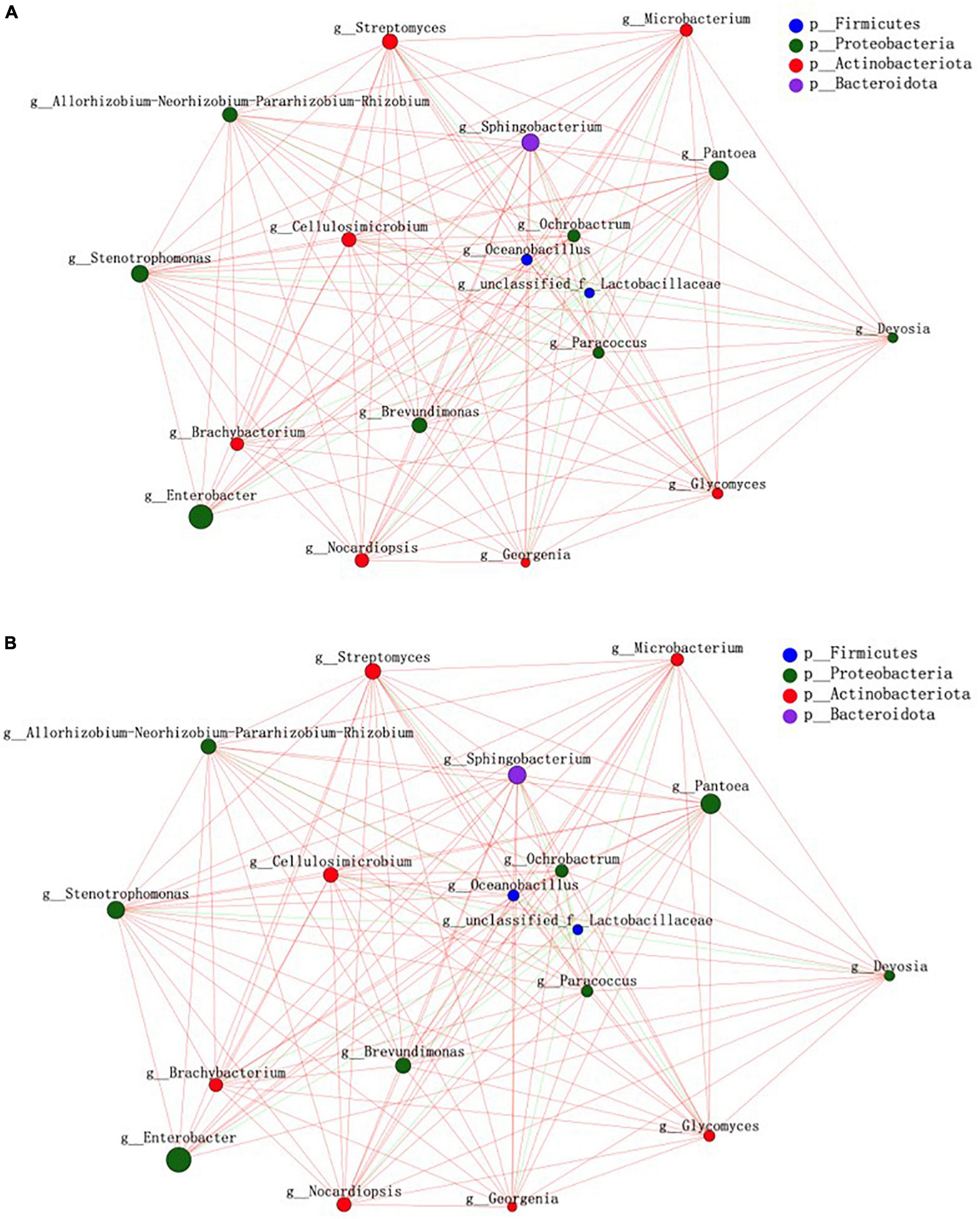
Figure 2. Bacterial community structure in sainfoin silage. (A) Bacterial network analysis of sainfoin silage in the control. (B) Bacterial network analysis of sainfoin silage in the PEG-treated group.
Metabolomic profiles
In total, 510 metabolites (Supplementary Table 1) were identified after 60 days of sainfoin ensiling, among which 33 metabolites were annotated based on the KEGG database, as shown in Figure 3. The metabolites consisted of lipids, nucleic acids, organic acids, peptides, carbohydrates, hormones and transmitters, and steroids. Lipids were the most abundant metabolites (72%) among the 33 metabolites. There were clear distinctions in metabolites between the control and the PEG-treated group based on the PCA and volcano plot analysis results (Figures 4, 5). Based on specific screening conditions (VIP > 1, p < 0.05), 10 upregulated metabolites and 23 downregulated metabolites were identified in the PEG-treated group when compared with the control after 60 days of ensiling, as shown in Figure 6. Among the upregulated metabolites, phospholipids (four metabolites, including cardiolipin, phosphatidylethanolamine, phosphatidylserine, and phosphatidate) were the most abundant, followed by L-methionine, prostaglandin F3a, dopamine, guanosine, taurocholic acid, and triacylglycerol. Similar trends were observed with regard to the upregulated metabolites, with 13 metabolites belonging to phospholipids (such as phosphatidate, phosphatidylcholine, phosphatidylglycerol, and phosphatidylserine) followed by eicosanoic acids (four metabolites, namely, 15-hydroxy-9-oxoprosta, prostaglandin C1, leukotriene E4, and prostaglandin E1), two nucleic acids (cytosine and deoxyadenosine), and four other metabolites (gamabufogenin, citric acid, D-tagatose, and dodecanoic acid). The most affected pathways between the control and PEG-treated groups were “cAMP signaling” (three metabolites were associated with the “cAMP signaling” pathway), followed by “regulation of lipolysis in adipocytes,” “morphine addiction,” “fat digestion and absorption,” “choline metabolism,” “cholesterol metabolism,” and “Alcoholism” with two metabolites were associated with the “Alcoholism” pathway (Figure 7).
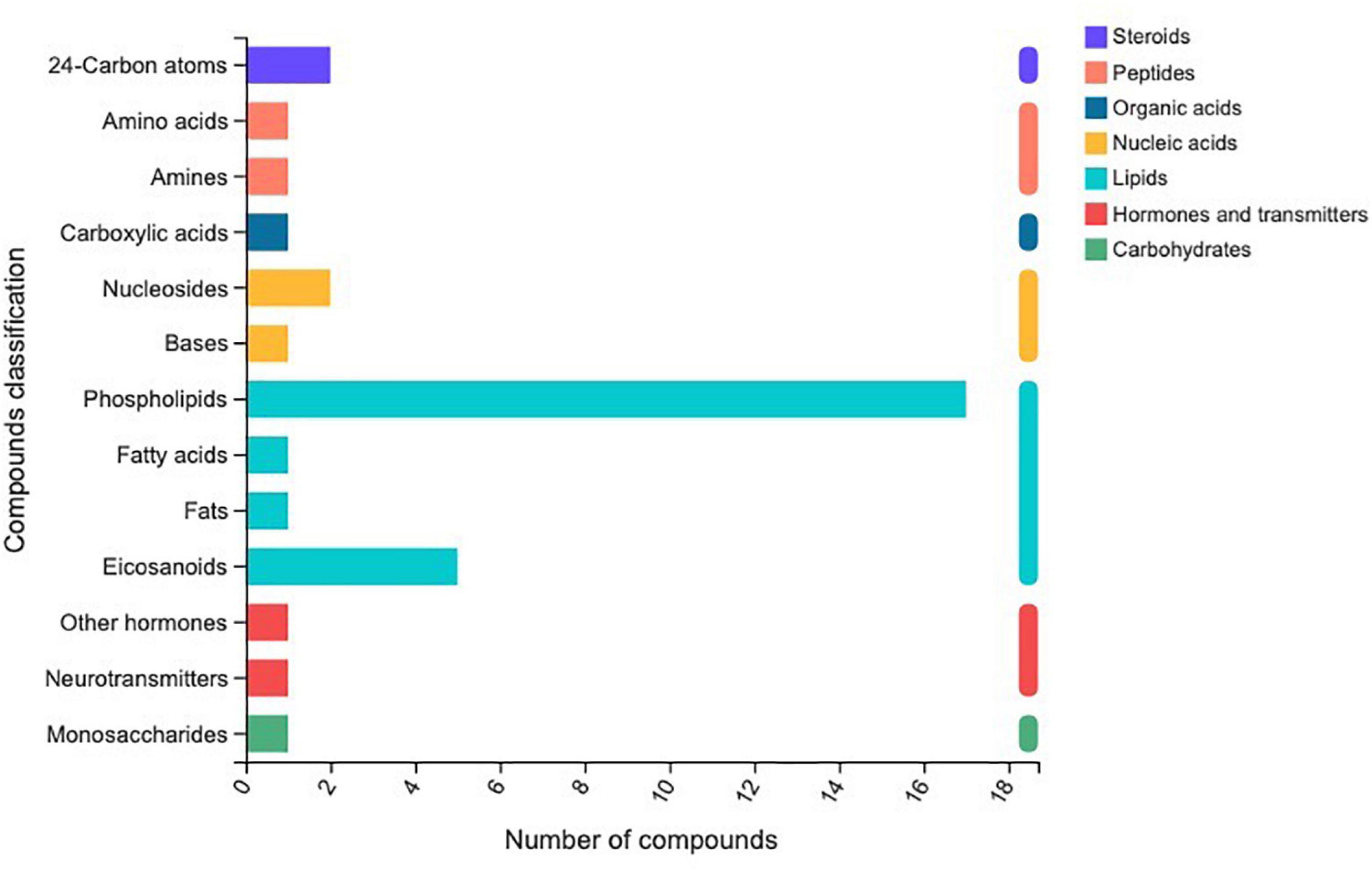
Figure 3. Different metabolites annotated by Kyoto Encyclopedia of Genes and Genomes in sainfoin silage.
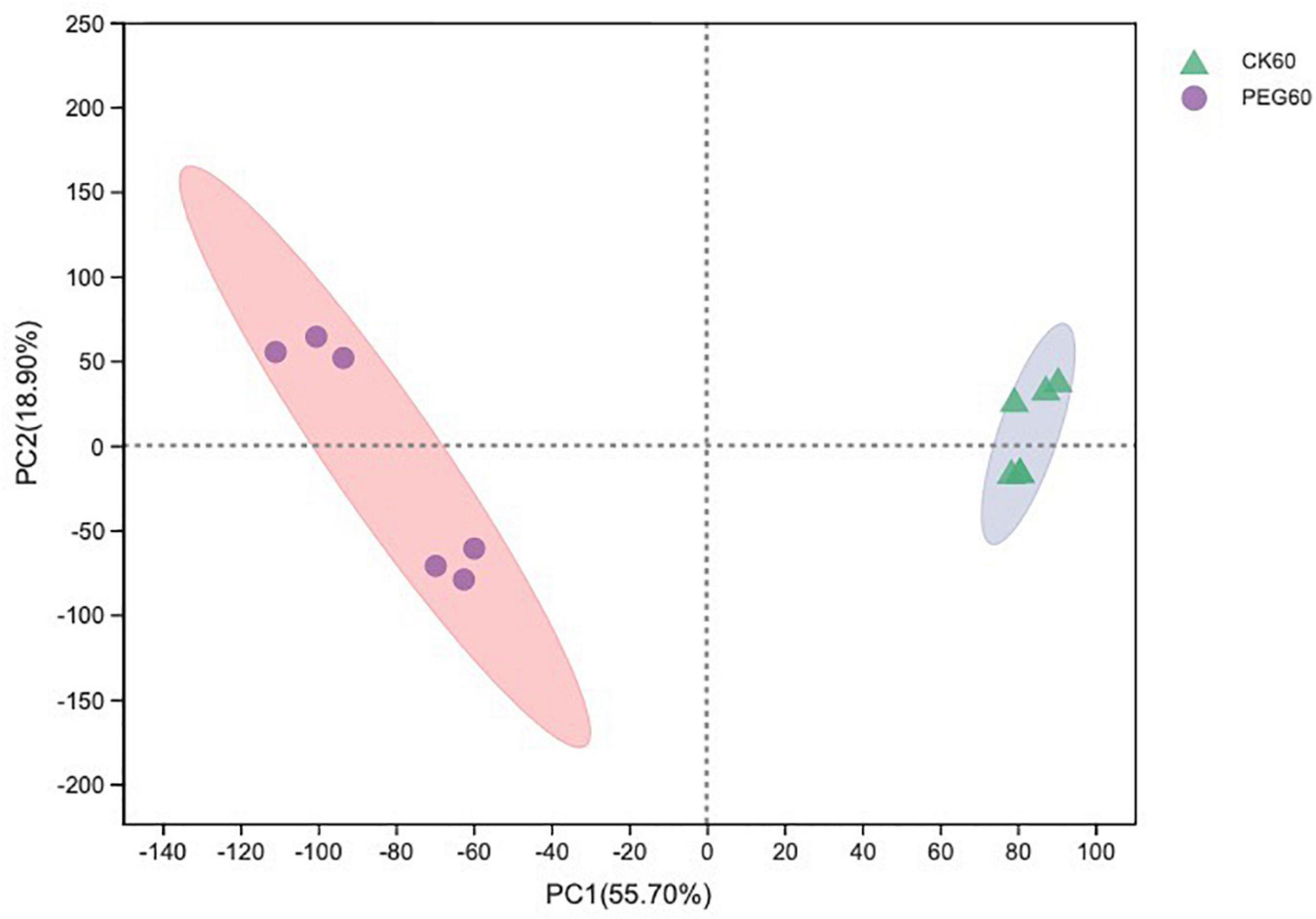
Figure 4. The principal component analysis (PCA) score plot of the metabolites in sainfoin silage (CK 60, control group after 60 days of ensiling; PEG 60, PEG-treated group after 60 days of ensiling).
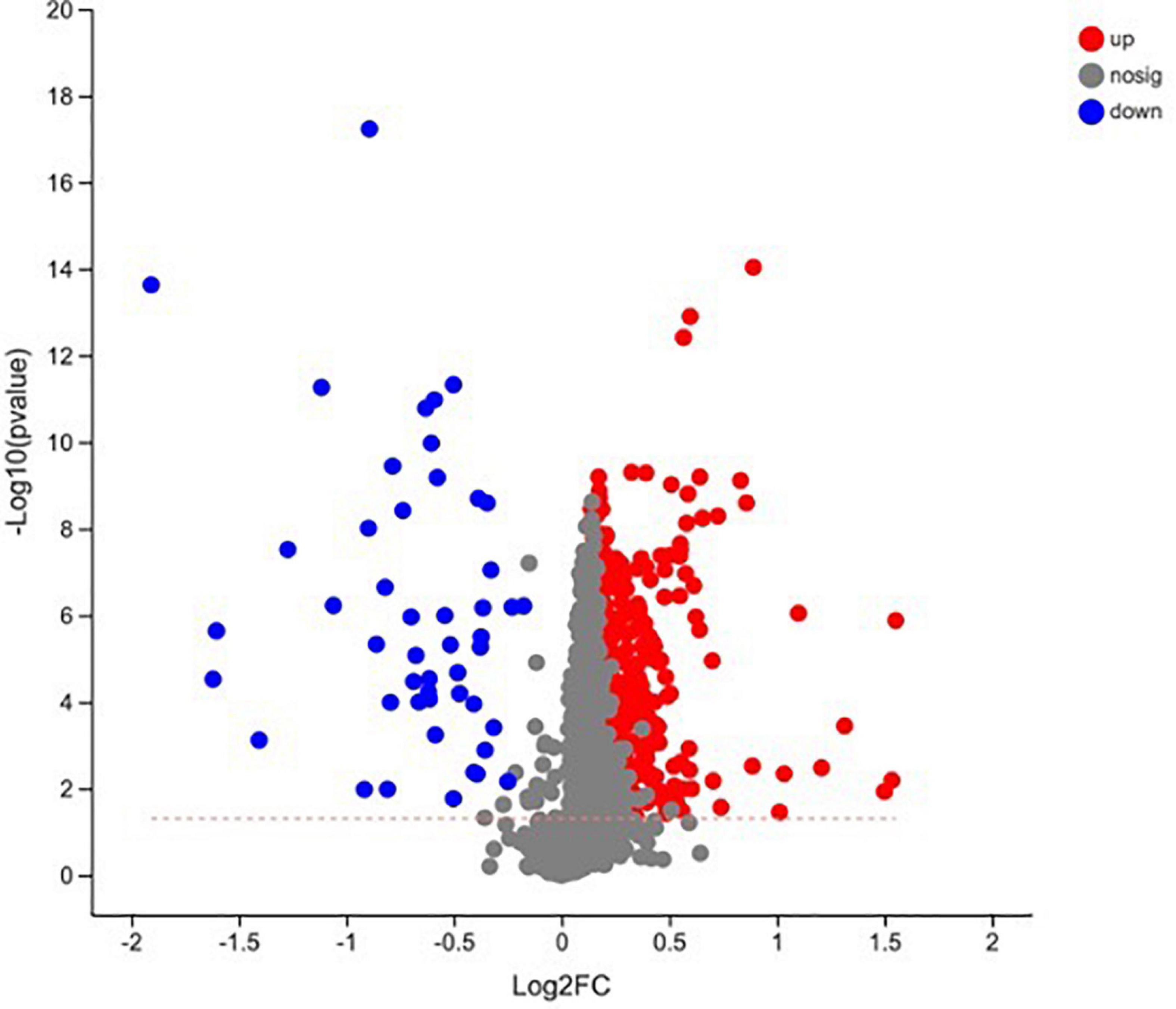
Figure 5. Volcano plot analysis of the metabolites in sainfoin silage (CK 60, control group after 60 days of ensiling; PEG 60, PEG treated group after 60 days of ensiling).

Figure 6. Heatmap of the differentially accumulated metabolites in sainfoin silage (CK 60, control group after 60 days of ensiling; PEG 60, PEG treated group after 60 days of ensiling). *p < 0.05, **0.01 < p < 0.05, ***p < 0.01.
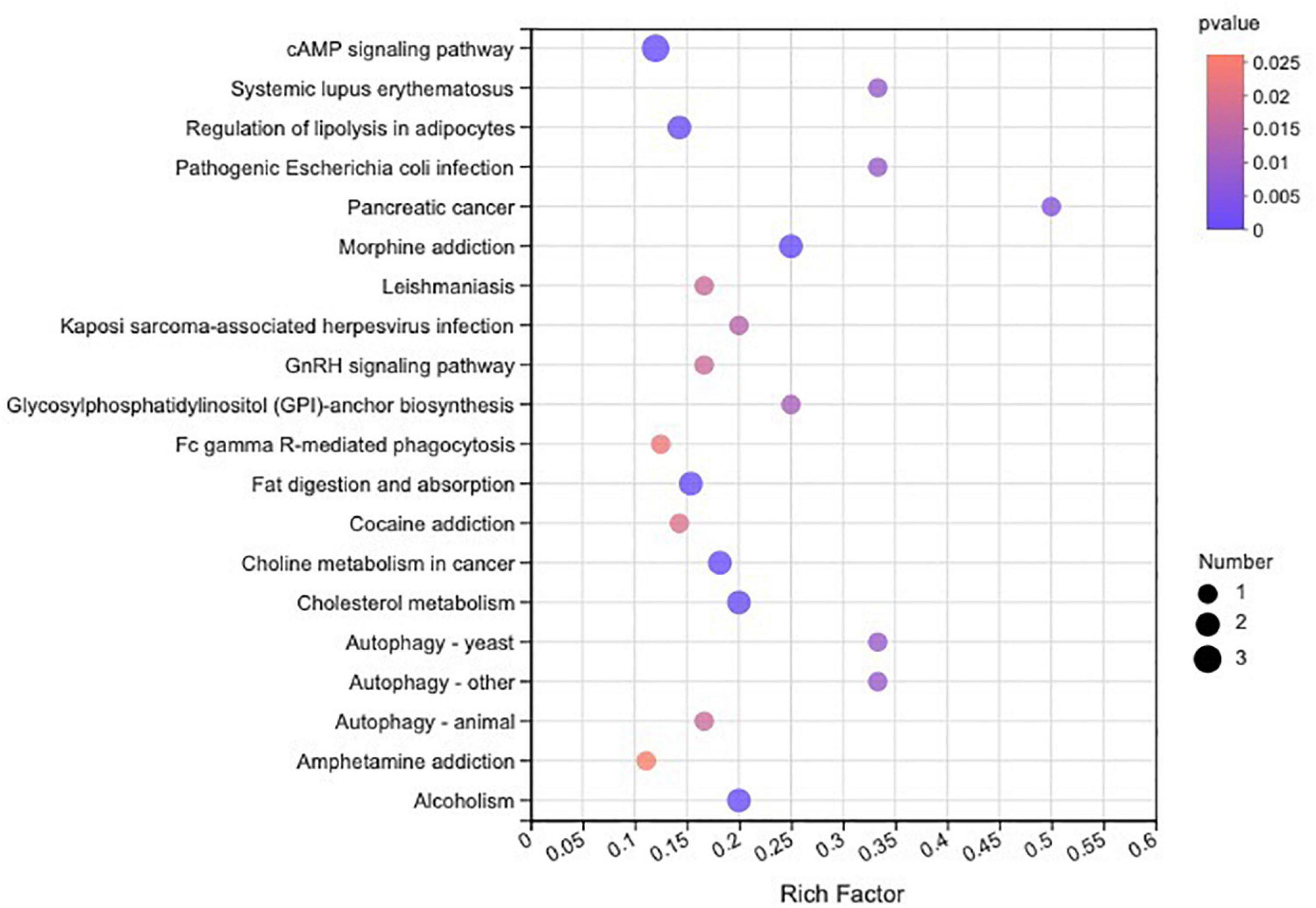
Figure 7. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for differentially accumulated metabolites in sainfoin silage.
Correlation between bacteria relative abundance and metabolites
The results of correlation analysis between bacterial abundance and metabolites in sainfoin silage are shown in Figure 8 and Supplementary Table 3. There was no significant correlation between Enterobacter abundance and metabolites (p > 0.05). Lactobacillus were significantly and highly positively correlated with prostaglandin E1, phosphatidylglycerol (two metabolites), leukotriene E4, D-Tagatose, phosphatidate, prostaglandin C1, deoxyadenosine, dodecanoic acid, and gamabufogenin (R2 = 0.8286, 0.8286, 0.8286, 0.8286, 0.8286, 0.9429, 0.9429, 0.8857, 0.9429, and 0.9429, respectively, p < 0.05), and were significantly negatively correlated with L-methionine, dopamine, cardiolipin, phosphatidylethanolamine, taurocholic acid, and guanosine (R2 = −0.8286, −0.8857, −0.8857, −0.9429, −0.8286, and −0.9429, respectively, p < 0.05). Pediococcus were significantly negatively correlated with phosphatidylserine, phosphatidate (four metabolites), phosphatidylcholine (three metabolites), phosphatidylglycerol (two metabolites), citric acid, leukotriene E4, D-Tagatose, prostaglandin C1, and deoxyadenosine (R2 = −0.8286, −0.8286, −0.8857, −0.9429, −0.9429, −0.8286, −0.8286, −0.8286, −0.8286, −0.9429, −1, −1, −0.9429, −0.9429, and −0.8857, respectively, p < 0.05), and were significantly positively correlated with triacylglycerol, dopamine, cardiolipin, phosphatidylethanolamine, and taurocholic acid (R2 = 0.8857, 0.8857, 0.8857, 0.9429, and 0.8286, respectively, p < 0.05). Weissella was significantly positively correlated with prostaglandin F3a (R2 = 0.8857, p < 0.05).
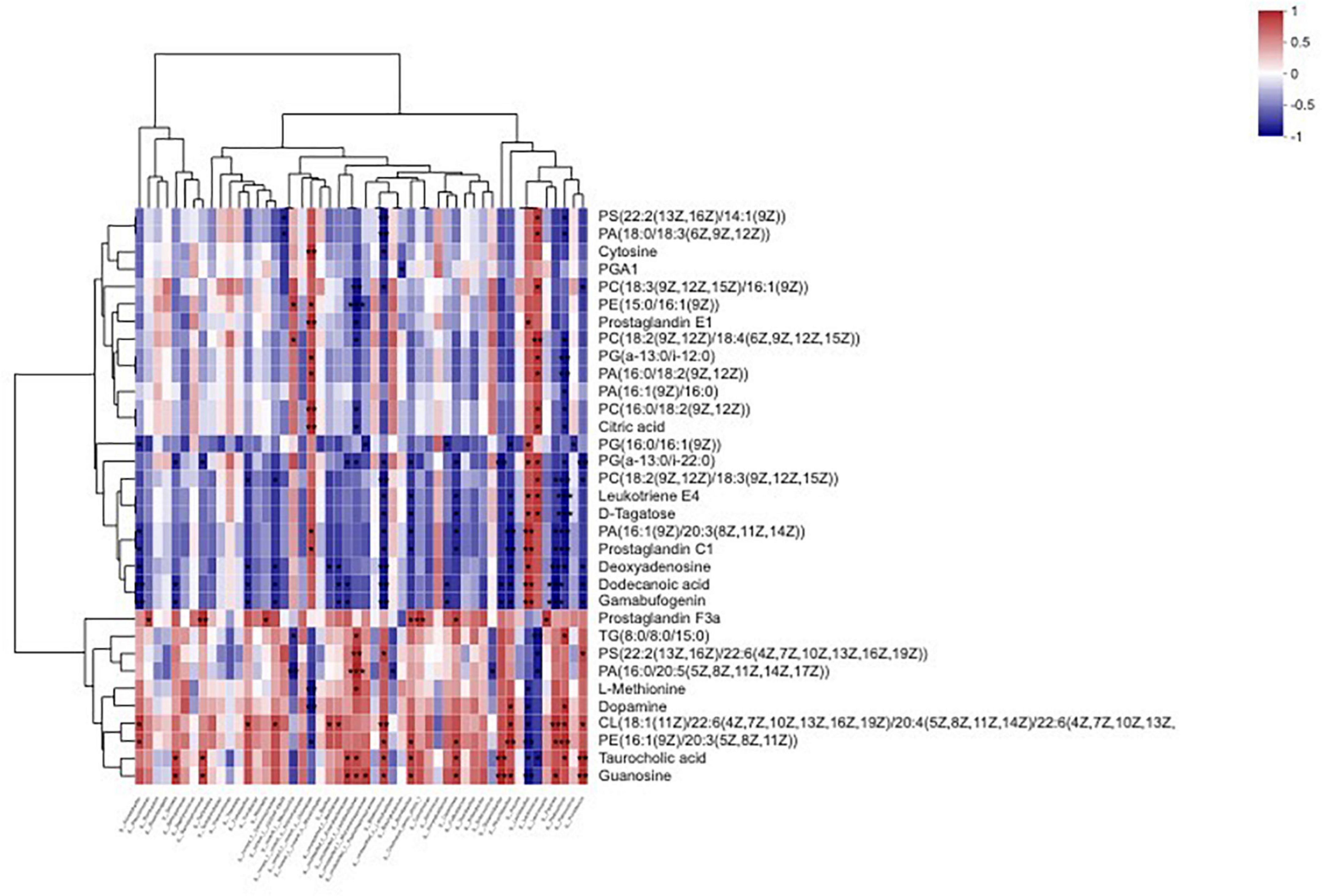
Figure 8. Heatmap of the correlation analysis for bacteria and metabolites in sainfoin silage. *p < 0.05, **0.01 < p < 0.05, ***p < 0.01.
Three keystone bacteria were correlated with metabolites, including Enterococcus, Stenotrophomonas, and Pantoea. Enterococcus was significantly negatively correlated with one metabolite, phosphatidate (one metabolite (R2 = −0.829, p < 0.05). Stenotrophomonas was significantly positively correlated with prostaglandin F3a, taurocholic acid, and guanosine (R2 = 0.9411, 0.8197, and 0.8197, respectively, p < 0.05). Pantoea was significantly negatively correlated with phosphatidylcholine, phosphatidate, prostaglandin C1, deoxyadenosine, dodecanoic acid, and gamabufogenin (R2 = −0.8286, −0.8857, −0.8857, −0.9249, −1, and −1, respectively, p < 0.05), and were significantly positively correlated with cardiolipin, phosphatidylethanolamine, and guanosine (R2 = 0.9249, 0.8857, and 0.8857, respectively, p < 0.05).
Discussion
Nitrogen distribution and protein fractions in sainfoin silage
Proteolysis in silage generally takes place via two steps. First, plant proteases hydrolyze proteins into peptides and free amino acids (FAA); afterward, further degradation by microbial activity occurs, usually mediated by Clostridium and Enterobacter (McDonald et al., 1991). Usually, Clostridium is more active in moist environments with moisture levels >75% (Muck, 2010). However, in a previous study, no Clostridium activity was detected during sainfoin silage with a DM content of 23% (Huang et al., 2022). As a direct indicator of protein hydrolysis during ensiling, NPN consists of peptides, FAA, and NH4+-N (Licitra et al., 1996). According to the results of the present study, NPN content in the control treatment stabilized after 7 days of ensiling; however, in the PEG-treated group, NPN increased with an increase in ensiling time. The results suggest that CT decreased protein degradation during the early silage fermentation stage. AN contents in silage, which are produced by both plant proteases and microbial activities, are generated from protein hydrolysis (Li X. et al., 2018). A previous study observed that PEG addition had no effect on Enterobacter relative abundance during the entire process of sainfoin ensiling (fermentation 60 days); however, AN content was the highest in PEG-treated sainfoin silage during the entire process of sainfoin ensiling (fermentation 60 days). Furthermore, AN content increased in both the control and PEG-treated group with an increase in ensiling time (Huang et al., 2022). The results suggested that Enterobacter relative abundance may not influence AN content in sainfoin silage. However, NH4+-N content in silage formed by deamination from FAA is mainly attributed to bacterial enzyme activity rather than plant enzyme activity (McDonald et al., 1991). Metagenomic analysis revealed that NH4+-N contents were associated with nitrite reductase, with the gene encoding nitrite reductase mainly assigned to Enterobacter (Li R.R. et al., 2021). Considering the present results, AA contents were lower in the control group than in the PEG-treated group during the entire process. The higher AN contents in the PEG-treated silage were likely due to higher AA contents than in the control group. PEG addition increased NH4+-N content significantly during the entire ensiling process (p < 0.05). Other researchers have observed that PEG causes high levels of buffer soluble N in silage, which is attributed to the strong tannin-protein binding effect of CT in sainfoin (Lorenz et al., 2010). Compared to other tannin-rich legume forage, CT from sainfoin may have the lowest NH4+-N levels in silage, which confirms the capacity of CT from sainfoin to strongly inhibit proteolysis during ensiling (Brinkhaus et al., 2017). In addition, according to Huhtanen et al. (2002), high-quality grass silage has AN contents that account for <5% of the total N. In the present study, NH4+-N contents in the control (4.0% of total N) were less than those in the PEG-treated group (7.3% of total N) after 60 d. Therefore, CT from sainfoin could facilitate the maintenance of high-quality sainfoin silage. Among the N distributed in silage, N in peptides may be more useful for rumen microbial N synthesis, as a previous in vitro study observed that certain bacteria from rumen utilize N from peptides more efficiently than N from FAA or from ammonia (Cruz Soto et al., 1994).
According to the results of the present study, there were no significant differences between the control and PEG-treated groups during the entire ensiling process (p > 0.05). However, in the case of Megasphaera elsdenii from the rumen, which is one of the main lactate-fermenting rumen bacterial species, AA N would be more useful than peptide N (Nsereko et al., 1998). PEG-treated silage had higher AA contents than the control silage over the entire ensiling process in the present study. Both the control and PEG-treated groups had the highest AA contents after 60 days of ensiling, which were 16.43 and 35.50%, respectively. The results suggest that PEG addition increased AA contents during sainfoin ensiling, which may promote the growth of certain bacteria in the rumen, with no effects on peptide N. The increased susceptibility of protein to degradation following PEG addition could be attributed to the high capacity of PEG to bind tannins, which could cause either an exchange of tannin-bound protein with PEG or prevent tannin-bound protein formation (Lorenz et al., 2010). In addition, microflora composition during ensiling is influenced by AAs, such as arginine, particularly when pH is not decreased considerably (Winters et al., 2000). In a previous study, PEG addition increased lactic acid bacteria (LAB) activity (Pediococcus relative abundance) during ensiling (Huang et al., 2022). Hence, the inhibition of LAB activity by CT from sainfoin is partly attributable to decreased AA contents.
Remarkably, PEG addition increased the SOLP content, while decreasing NDIP and ADIP contents during the entire ensiling process. Similar results were observed by Peng et al. (2018), although NDIP content remained unchanged in their study. In addition, in the present study, the PB3 fraction decreased with PEG addition, and similar results were observed by Brinkhaus et al. (2017), where the protein from the PB1 and PB2 fraction shifted to the PB3 and PC (insoluble in acid detergent protein) fractions during sainfoin ensiling, suggesting that CT can decrease protein degradation during ensiling.
Protease activity in sainfoin silage
During ensiling, proteolysis can occur in the absence of microbial growth through sterilized herbage, which suggests that plant enzymes are the main agents driving degradation during proteolysis (McKersie and Buchanan-Smith, 1982). ACP, CPs, and APs are three major proteases formed during ensiling; CP and ACP exhibit activities at pH 5.2, ACP exhibits optimal activity under a silage pH of 4.5, whereas APs lose their function (McKersie, 1981). In the present study, sainfoin silage pH was 4.5–4.69 at 3–30 days (pH results can find at Huang et al., 2022); however, the APs still exhibited 25.89 to 94.68% activity when compared to 0 day protease activity. A previous study reported that CP and AP activities stabilized at 14 days but intensified after 30 days of ensiling; however, the pH decreased from 4.1 to 3.9 over a 30-day ensiling period (He et al., 2020). Several studies have suggested that proteolysis still takes place at pH <4.0 (Carpintero et al., 1979; Henderson, 1993). Hence, the silage environment, in which APs maintain activity at pH <4.1 during ensiling, could be much more complex than the herbage incubation environment. Furthermore, PEG had no effect on pH when compared with the control (pH results can find at Huang et al., 2022); however, the PEG group exhibited the highest AP and ACP activity after 3 days of ensiling. Considering there was no difference in pH between the PEG-treated and control groups, the difference in protease activity was mainly attributed to inhibition by CT. The results suggest that CT inhibit AP and ACP activity only in the early fermentation stages. Additionally, CT inhibited CP activity at 7–60 days of ensiling. However, He et al. (2019) observed that CT from Neolamarckia cadamba leaf (NCL) had no effect on CP activity during 30 days of ensiling. The mechanisms via which CT inhibit protease activity, such as tyrosinase, including competition with substrates for active enzyme sites and binding to the enzyme substrate complex, which occur through hydrogen bonding and hydrophobic interactions (Song et al., 2020). Several factors, such as molecular size, mean degree of polymerization (mDP), presence of galloyl groups, and interflavan-3-ol linkages, influence CT affinity for proteins (Zeller, 2019). Previous studies have reported that CT from different materials, such as Rhododendron pulchrum leaves (Chai et al., 2015), sour jujube (Ziziphus jujuba Mill. var. spinosa [Bunge] Hu ex. H. F. Chow) (Song et al., 2020), and Pinus thunbergii needles (Yang et al., 2021) show varied ability to inhibit tyrosinase activity, with IC50 values ranging from 43.7 to 145.35 μg/ml. Therefore, the ability of CT from sainfoin to inhibit protease activity during ensiling might vary from that of NCL.
Cellulosimicrobium is a Gram-positive anaerobic bacteria that secretes several enzymes, such as glycosidases, galactosidases, glucosidases, and esterases; it exploits different types of carbon sources (arabinose, xylose, glucose, etc.) to produce organic acids, in addition to being able to secrete nitrate reductase, pyrazinamidases, and pyrrolidine acyl aromatic amide enzymes (Hamada et al., 2016). Cellulosimicrobium became keystone bacteria in silage result as all of these enzymes could be involved in different metabolic pathways. In the present study, Cellulosimicrobium did not represent keystone bacteria in the PEG-treated silage. The difference could be partly attributed to CT combining with protein and inhibiting enzyme activity.
Bacteria community in sainfoin silage
Keystone bacteria in microecological environments are increasingly attracting the attention of researchers, supported by the proliferation of high-throughput sequencing technologies. Microbial community structure and function regulation by keystone bacteria is not associated with their relative abundance. On the contrary, bacteria with low relative abundance can often be keystone bacteria, and keystone bacteria can selectively regulate microbial community structures (Banerjee et al., 2018). In the present study, PEG addition altered the distribution of keystone bacterial communities in silage significantly. The results suggested that CT had a significant effect on keystone bacterial community composition during sainfoin ensiling. In the present study, Sphingobacterium were the keystone bacteria in both the control and PEG-treated groups. Sphingobacterium were also abundant in fresh corn silage and barley silage, especially in corn silage (Xu et al., 2019; Liu et al., 2020); however, the relative abundance of Sphingobacterium decreased rapidly to below 1% and may participate in ethanol production during ensiling (Ni et al., 2017). Additionally, Sphingobacterium is involved in biotic carbon dioxide (CO2) sequestration as it was negatively correlated with CO2 production during ensiling (Chen et al., 2021). Similar to Sphingobacterium, during anaerobic autotrophic growth, Stenotrophomonas can convert CO2 into organic acids through microbial electrosynthesis (Bian et al., 2020). Both the bacteria could use different WSCs to produce organic acid (Palleroni and Bradbury, 1993). Therefore, the two bacteria could compete with LAB during ensiling. However, a previous study observed that the presence of Sphingobacterium and Stenotrophomonas in silage was associated with increased richness and relative abundance of LAB in a bacterial community (Li M. et al., 2021). The difference may be attributed to the relative abundance of keystone bacteria being the lowest when compared with LAB results as competition for WSC is weakened; however, keystone bacteria regulate microbial activity via different metabolic pathways. Ogunade et al. (2018) first observed that Pantoea was significantly negatively corrected with AN in alfalfa silage. Several studies have reported similar results in silage (Dong et al., 2020; Yang et al., 2020). High Pantoea relative abundance was observed in silage but it decreased by a treated group company, with butyrate acid contents reduced. A study on corn silage indicated that Pantoea was more active under high moisture contents, with a positive correlation with butyrate acid contents (Guan et al., 2018). Such studies indicate that the metabolic pathway of Pantoea is more complex, which was attributed to the capacity of Pantoea not only to convert WSC to organic acid but also to decrease AN content in silage. Therefore, considering the low Pantoea relative abundance when compared with LAB, there could be different mechanisms of reducing pH during ensiling between Pantoea and LAB. A study on sainfoin and alfalfa silage mixture indicated that Pantoea relative abundance increased with an increase in the proportion of sainfoin in the mixture (Wang et al., 2021), which could be related to CT. In the present study, Pantoea was not among the keystone bacteria in the PEG-treated silage. Hence, CT might affect keystone bacteria community structure through its effect on Pantoea during sainfoin ensiling. Microbacterium exists in fresh corn with a relative abundance of 3.5–6.35%, which decreases considerably with a prolongation of ensiling time (Xu et al., 2020b). Microbacterium foliorum could use fructose during ensiling (Wang et al., 2020). To the best of our knowledge, there are few studies on Microbacterium activity during ensiling. Therefore, further studies on the mechanism via which Microbacterium become keystone bacteria during sainfoin ensiling are required.
Enterococcus exhibit greater activity during the early fermentation stages of silage, with Leuconostocs, Pediococci, and Lactococci involved in the initiation of silage fermentation through lactic acid production; however, all the bacteria above are replaced gradually by Lactobacillus with the prolongation of ensiling time (Cai and Benno, 1998). Additionally, Enterococcus could transform FAA to produce AN and bioamines through different metabolic pathways through corresponding deaminases and decarboxylases (Oladosu et al., 2016); for example, lysine decarboxylase forms cadaverine, tyrosine decarboxylase forms tyramine, and arginine decarboxylase forms arginine (Li R.R. et al., 2021). Furthermore, AN synthesis involves different metabolic pathways and enzymes, such as nitrite reductase, nitrogenase, and hydroxylamine reductase (Huang et al., 2020). In the present study, Enterococcus was not among the keystone bacteria in the control attributed to the deactivation of enzymes through that action of CT in combination with protein.
Metabolome profile of sainfoin silage
A total of 510 metabolites (Supplementary Table 1) were identified after 60 days of sainfoin ensiling, with 300 metabolites distinct between the CK and PEG-treated groups. The most distinct metabolites were phospholipids (17 metabolites), followed by eicosanoids (5 metabolites), nucleosides and 24-carbon atoms (5 metabolites), and others (1 metabolite). Several studies have observed that catechins inhibit bacteria by damaging the lipid bilayer membrane (Tsuchiya, 2001; Nakayama et al., 2002; Caturla et al., 2003), via hydrogen bonding between their polyhydroxyl groups and phosphocholine moiety, and interaction with phospholipids (Li Z. et al., 2018). Considering CT contains four types of catechins (Zeller, 2019), the results suggest that CT from sainfoin has strong effects on bacterial phospholipid synthesis. The PCA analysis results revealed distinct metabolites between the control and PEG-treated groups. However, a previous study showed that control and PEG-treated groups had similar microbial community structures at the genus level (Huang et al., 2022). Therefore, the main reason for the variation could be attributed to CT, which had a strong effect on microbial metabolism during ensiling.
One amino acid (L-methionine) was upregulated following PEG addition, whereas citric acid was downregulated following PEG. Furthermore, two nucleosides, including cytosine and deoxyadenosine, were downregulated and one nucleoside, guanosine, was upregulated following PEG addition. Catechin also inhibits bacteria via DNA degradation, and it is more toxic to Gram-positive bacteria than Gram-negative bacteria (Fathima and Rao, 2016). Enterobacter, a Gram-negative bacteria, was not affected by CT during sainfoin ensiling, but Pediococcus, a Gram-positive bacteria, was highly inhibited by CT during sainfoin ensiling (Huang et al., 2022). The results suggest that CT affected Gram-positive bacteria through inhibition of phospholipid activity and gene synthesis during ensiling.
During ensiling, various metabolites are produced through complex metabolic reactions, which are regulated by different metabolic pathways (Xu et al., 2020a). The most affected metabolic pathways were annotated to the “cAMP signaling pathway,” with five phospholipids, deoxyadenosine, and dopamine associated with the pathway, after 60 days of sainfoin ensiling. Furthermore, sainfoin ensiling could alter the metabolome, including altering the dynamics of isoflavone, proanthocyanidins, polyphenol, and so on, through the flavonoid and isoflavonoid biosynthesis pathways. However, inoculation of sainfoin with LAB eliminated the effects, suggesting that LAB inoculation could mainly affect the flavonoid and isoflavonoid biosynthesis pathways (Xu et al., 2020a). In the present study, PEG addition had no effect on the flavonoid biosynthesis pathway (Supplementary Table 2), which indicates that CT biosynthesis is mainly affected by the ensiling process, as CT consists of flavan-3-ol subunits (Zeller, 2019).
In the present study, PEG addition led to the upregulation of 10 metabolites and downregulation of 23 metabolites. Lactobacillus were significantly negatively correlated with six metabolites among the upregulated metabolites and were significantly positively correlated with 10 metabolites among the downregulated metabolites. On the contrary, Pediococcus was significantly positively correlated with five metabolites among the upregulated metabolites, and significantly negatively correlated with 15 metabolites among the downregulated metabolites. Additionally, phospholipids were the most affected metabolites and were correlated with both Lactobacillus and Pediococcus. The results suggested the antagonistic effects between Lactobacillus and Pediococcus during ensiling. Among the metabolites upregulated following PEG addition, dopamine is converted by tyrosine. Pediococcus was significantly positively correlated with dopamine, but Lactobacillus was significantly negatively correlated with dopamine. The cell-free supernatants of some Pediococcus (Pediococcus acidophilus) have stronger stimulation effects on dopamine production by Enterobacter (Escherichia coli) than Lactobacillus (Lactococcus lactis subsp. lactis) (Ozogul et al., 2017). According to the results, PEG upregulated dopamine after 60 days of sainfoin ensiling, which was correlated with CT strongly inhibiting Pediococcus activity during sainfoin ensiling.
Conclusion
Polyethylene glycol addition decreased NDIP and ADIP contents, with NPN contents increased, during sainfoin ensiling. CT from sainfoin decreased proteolysis mainly via the inhibition of protease activity, particularly CP activity. In addition, the decrease in proteolysis was partly attributed to CT inhibiting Pediococcus activity, based on the highly positive correlation between Pediococcus and dopamine. CT also affected bacteria community structure, potentially through shifts in keystone bacteria. The most distinct metabolites between the control and PEG-treated groups were phospholipids, which indicated that CT inhibited bacterial activity through decreased phospholipid synthesis during ensiling.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RH: conceptualization, methodology, and writing – review and editing. FZ: conceptualization, resources, validation, and supervision. XW: resources and investigation. CM: conceptualization, methodology, and visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the China Agriculture Research System of MOF and MARA (CARS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.976118/full#supplementary-material
Footnotes
- ^ http://bioconductor.org/packages/release/bioc/html/ropls.html
- ^ https://cloud.majorbio.com
- ^ https://www.majorbio.com
References
Aufrere, J., Theodoridou, K., Mueller-Harvey, I., Yu, P., and Andueza, D. (2014). Ruminal dry matter and nitrogen degradation in relation to condensed tannin and protein molecular structures in sainfoin (Onobrychis viciifolia) and lucerne (Medicago sativa). J. Agric. Sci. 152, 333–345. doi: 10.1017/S0021859613000452
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of mi-crobiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576.
Bian, B., Bajracharya, S., Xu, J. J., and Pant, D. (2020). Microbial electrosynthesis from CO2: Challenges, opportunities and perspectives in the context of circular bioeconomy. Bio Res. Technol. 302:122863. doi: 10.1016/j.biortech.2020.122863
Bradford Marion, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brinkhaus, A. G., Wyss, U., Arrigo, Y., Girard, M., Bee, G., Zeitza, J. O., et al. (2017). In vitro ru-minal fermentation characteristics and utilisable CP supply of sainfoin and birdsfoot trefoil silages and their mixtures with other legumes. Animal 11, 590–590. doi: 10.1017/S1751731116001816
Cai, Y., and Benno, Y. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/aem.64.8.2982-2987.1998
Carbonero, C. H., Mueller-Harvey, I., and Smith, L. (2011). Sainfoin (Onobrychis viciifolia): A beneficial forage legume. Plant Genet. Res. Charact. Utilizat. 9, 70–85. doi: 10.1017/S1479262110000328
Carpintero, M. C., Henderson, A. R., and McDonald, P. (1979). The effect of some pre-treatments on proteolysis during the ensiling of herbage. Grass Forage Sci. 34, 311–315. doi: 10.1111/j.1365-2494.1979.tb01483.x
Caturla, N., Vera-Samper, E., Villalain, J., Mateo, C. R., and Micol, V. (2003). The relationship be-tween the antioxidant and the antibacterial properties of galloylated catechins and the struc-ture of phospholipid model membranes. Free Rad. Biol. Med. 34, 648–662.
Chai, W.-M., Wang, R., Wei, M.-K., Zou, Z.-R., Deng, R.-G., Liu, W.-S., et al. (2015). Proanthocya-nidins extracted from Rhododendron pulchrum leaves as source of tyrosinase inhibitors: Structure, activity, and mechanism. PLoS One 10:e0145483. doi: 10.1371/journal.pone.0145483
Chen, D. K., Zheng, M. Y., Guo, X., Chen, X. Y., and Zhang, Q. (2021). Altering bacterial commu-nity_ A possible way of lactic acid bacteria inoculants reducing CO2 production and nutrient loss during fermentation. Bio. Technol. 329:124915.
Cruz Soto, R., Muhammed, S. A., Newbild, C. J., Stewart, C. S., and Wallace, R. J. (1994). Influence of peptides, amino acids and urea on microbial activity in the rumen of sheep receiving grass hay and on the growth of rumen bacteria in vitro. Animal Feed Sci. Technol. 49, 151–161.
Dong, L. F., Zhang, H. S., Gao, Y. H., and Diao, Q. Y. (2020). Dynamic profiles of fermentation characteristics and bacterial community composition of Broussonetia papyrifera ensiled with perennial ryegrass. Bio. Technol. 310:123396. doi: 10.1016/j.biortech.2020.123396
Fathima, A., and Rao, J. R. (2016). Selective toxicity of catechin—A natural flavonoid towards bac-teria. Appl. Microbiol. Biotechnol. 100, 6395–6402. doi: 10.1007/s00253-016-7492-x
Guan, H., Shuai, Y., Ran, Q. F., Yan, Y. H., Wang, X., Li, D. D., et al. (2020). The microbiome and metabolome of napier grass silages prepared with screened lactic acid bacteria during ensiling and aerobic exposure. Animal Feed Sci. Technol. 269:114673. doi: 10.1016/j.anifeedsci.2020.114673
Guan, H., Yan, Y. H., Li, X. L., Li, X. M., Shuai, Y., Feng, G. Y., et al. (2018). Microbial communi-ties and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bio. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, X., Zhou, H., Yu, Z., and Zhang, Y. (2007). Changes in the distribution of nitrogen and plant enzymatic activity during ensilage of lucerne treated with different additives. Grass For. Sci. 62, 35–43.
Hamada, M., Shibata, C., Tamura, T., Nurkanto, A., Ratnakomala, S., Lisdiyanti, P., et al. (2016). Cellulosimicrobium marinum sp. nov., an actinobacterium isolated from sea sediment. Arch. Microb. 198, 439–444. doi: 10.1007/s00203-016-1204-x
He, L., Lv, H., Xing, Y., Chen, X., and Zhang, Q. (2020). Intrinsic tannins affect ensiling characteris-tics and proteolysis of Neolamarckia cadamba leaf silage by largely altering bacterial community. Bioresour. Technol. 311:123496. doi: 10.1016/j.biortech.2020.123496
He, L. W., Wang, C., Xing, Y. Q., Zhou, W., Pian, R. Q., Yang, F. Y., et al. (2019). Dynamics of proteolysis, protease activity and bacterial community of Neolamarckia cadamba leaves silage and the effects of formic acid and Lactobacillus farciminis. Bioresour. Technol. 294:122127. doi: 10.1016/j.biortech.2019.122127
Henderson, N. (1993). Silage additives. Animal Feed Sci. Technol. 45, 35–56. doi: 10.1016/0377-8401(93)90070-Z
Huang, R. Z., Zhang, F. F., Wang, T., Zhang, Y. L., Li, X., Chen, Y. C., et al. (2022). Effect of in-trinsic tannins on the fermentation quality and associated with the bacterial and fungal community of sainfoin silage. Microorganisms 10:844. doi: 10.3390/microorganisms10050844
Huang, X., Yang, X. M., Zhu, J., and Yu, J. H. (2020). Microbial interspecific interaction and nitro-gen metabolism pathway for the treatment of municipal wastewater by iron carbon based constructed wetland. Bio. Technol. 315:123841. doi: 10.1016/j.biortech.2020.123814
Huhtanen, P., Khalili, H., Nousianinen, J. I., Rinne, M., Jaakkola, S., Heikkila, T., et al. (2002). Pre-diction of relative intake potential of grass silage bydairy cows. Livest. Prod. Sci. 73, 111–130. doi: 10.1016/S0301-6226(01)00279-2
Jones, W. T., and Mangan, J. L. (1977). Complexes of the condensed tannins of sainfoin (Onobrychis viciifolia scop.) with fraction 1 leaf protein and with Submaxillary mucoprotein, and their reversal by polyethylene glycol and pH. J. Sci. Food Agric. 28, 126–136. doi: 10.1002/jsfa.2740280204
Li, M., Lv, R. L., Zhang, L. D., Zi, X. J., Zhou, H. L., and Tang, J. (2021). Melatonin is a promising silage additive/ evidence from microbiota and metabolites. Front. Microbiol. 12:670764. doi: 10.3389/fmicb.2021.670764
Li, R. R., Zheng, M. L., Zheng, M. H., Cai, R., Cui, X. Y., Wang, Y., et al. (2021). Metagenomic analysis reveals the linkages between bacteria and the functional enzymes responsible for po-tential ammonia and biogenic amine production in alfalfa silage. J. Appl. Microbiol. 132, 2594–2604. doi: 10.1111/jam.15411
Li, X., Tian, J., Zhang, Q., Jiang, Y., Wu, Z., and Yu, Z. (2018). Effects of mixing red clover with alfalfa at different ratios on dynamics of proteolysis and protease activities during ensiling. J. Dairy Sci. 101, 8954–8964. doi: 10.3168/jds.2018-14763
Li, Z., Xie, J., Tian, X., Li, K., Hou, A., and Wang, Y. (2018). Proteomic changes in EHEC O157:H7 under catechin intervention. Microb. Pathog. 123, 9–17. doi: 10.1016/j.micpath.2018.06.034
Licitra, G., Hernandez, T. M., and Van Soest, P. J. (1996). Standardization of procedures for nitrogen fractionation of ruminant feeds. Animal Feed Sci. Technol. 57, 347–358. doi: 10.1016/0377-8401(95)00837-3
Liu, B. Y., Yang, Z. Q., Huan, H. L., Gu, H. R., Xu, N. X., and Ding, C. L. (2020). Impact of molas-ses and microbial inoculants on fermentation quality, aerobic stability, and bacterial and fun-gal microbiomes of barley silage. Sci. Rep. 10:5342. doi: 10.1038/s41598-020-62290-7
Lorenz, M. M., Eriksson, T., and Uden, P. (2010). Effect of wilting, silage additive, PEG treatment and tannin content on the distribution of N between different fractions after ensiling of three different sainfoin (Onobrychis viciifolia). Grass Forage Sci. 65, 175–184. doi: 10.1111/j.1365-2494.2010.00736.x
McAllister, T. A., Martinez, T., Bae, H., Muir, A., Yanke, L., and Jones, G. (2005). Characterization of condensed tannins purified from legume forages: Chromophore production, protein precip-itation, and inhibitory effects on cellulose digestion. J. Chem. Ecol. 31, 2049–2068. doi: 10.1007/s10886-005-6077-4
McDonald, P., Henderson, A. R., and Herson, S. (1991). The Biochemistry Of Silage. Kingston, Kent, UK: Chalcombe Publications.
McKersie, B. D. (1981). Proteinases and peptidases of alfalfa herbage. Can. J. Plant Sci. 61, 53–59. doi: 10.3168/jds.2012-5383
McKersie, B. D., and Buchanan-Smith, B. (1982). Changes in the levels of proteolytic enzymes in en-siled alfalfa forage. Can. J. Plant Sci. 62, 111–116. doi: 10.4141/cjps82-107
Muck, R. E. (2010). Silage microbiology and its control through additives. Rev. Brasil. Zo Otecnia 39, 183–191. doi: 10.1590/S1516-35982010001300021
Nakayama, T., Ichiba, M., Kuwabare, M., Kajiya, K., and Kumazawa, S. (2002). Mechanisms and structural specificity of hydrogen peroxide formation during oxidation of catechins. Food Sci. Technol. Res. 8, 261–267. doi: 10.3136/fstr.8.261
Ni, K. K., Minh, T. T., Tu, T. T. M., Tsuruta, T., Pang, H. L., and Nishino, N. (2017). Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl. Microbiol. Biotechnol. 101, 1385–1394. doi: 10.1007/s00253-016-7900-2
Novobilsky, A., Stringano, E., Carbonero, C. H., Smith, L. M. J., Enemark, H. L., Mueller-Harvey, I., et al. (2013). In vitro effects of extracts and purified tannins of sainfoin (Onobrychis vicii-folia) against two cattle nematodes. Vet. Parasitol. 196, 532–537. doi: 10.1016/j.vetpar.2013.03.024
Nsereko, V. L., Rooke, J. A., Newbold, C. J., and Wallace, R. J. (1998). Influence of protease inhibi-tors on nitrogen distribution in ensiled perennial ryegrass and the utilisation of silage nitrogen for growth by rumen bacteria in vitro. Animal Feed Sci. Technol. 76, 51–63. doi: 10.1016/S0377-8401(98)00211-9
Ogunade, I. M., Jiang, Y., Cervantes, A. A. P., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analysed by illumina miseq sequenc-ing: Effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Oladosu, Y., Rafii, M. Y., Abdullah, N., Magaji, U., Hussin, G., Ramli, A., et al. (2016). Fermenta-tion quality and additives: A case of rice straw silage. Bio. Res. Int. 2016:7985167. doi: 10.1155/2016/7985167
Ozogul, F., Toy, N., Ozogul, Y., and Hamed, I. (2017). Function of cell-free supernatants of leuco-nostoc, lactococcus, streptococcus, pediococcus strains on histamine formation by foodborne pathogens in histidine decarboxylase broth. J. Food Proc. Preservat. 41:e13208.
Palleroni, N. J., and Bradbury, J. F. (1993). Stenotrophomonas, a new bacterial genus for xan-thomonas maltophilia (Hugh 1980) swings, et al. 1983. Int. J. Syst. Bacteriol. 43, 606–609. doi: 10.1099/00207713-43-3-606
Peng, K., Jin, L., Niu, Y. D., Huang, Q., McAllister, T. A., Yang, H. E., et al. (2018). Condensed tannins affect bacterial and fungal microbiomes and mycotoxin production during ensiling and upon aerobic exposure. Appl. Environ. Microbiol. 84:e02274. doi: 10.1128/AEM.02274-17
Sniffen, C. J., O’Connor, J. D., Van Soest, P. J., Fox, D. G., and Russell, J. B. (1992). A net carbo-hydrate and protein system for evaluating cattle diets: II. Carbohydrate and protein availabil-ity. Journal of animal science 70, 3562–3577.
Song, W., Liu, L.-L., Ren, Y.-J., Wei, S.-D., and Yang, H.-B. (2020). Inhibitory effects and molecu-lar mechanism on mushroom tyrosinase by condensed tannins isolation from the fruit of Ziziphus jujuba mill. var. spinosa (bunge) hu ex H. F. chow. Int. J. Biol. Macromol. 165, 1813–1821. doi: 10.1016/j.ijbiomac.2020.09.259
Theodoridou, K., Aufrere, J., Andueza, D., Le Morvan, A., Picard, F., Pourrat, J., et al. (2012). Ef-fects of condensed tannins in wrapped silage bales of sainfoin (Onobrychis viciifolia) on in vivo and in situ digestion in sheep. Animal 6, 245–253. doi: 10.1017/S1751731111001510
Tsuchiya, H. (2001). Stereospecificity in membrane effects of catechins. Chem. Bilog. Int. 134, 41–54.
Wang, B., Gao, R., and Wu, Z. (2020). Functional analysis of sugars in modulating bacterial communities and metabolomics profiles of medicago sativa silage. Front. Microbiol. 11:641. doi: 10.3389/fmicb.2020.00641
Wang, M. S., Chen, M. Y., Bai, J., Zhang, J. Y., Su, R. N., Franco, M., et al. (2021). Ensiling char-acteristics, in vitro rumen fermentation profile, methane emission and archaeal and protozoal community of silage prepared with alfalfa, sainfoin and their mixture. Animal Feed Sci. Technol. 284:115154. doi: 10.1016/j.anifeedsci.2021.115154
Wetherall, J. A., Armstrong, D. G., Finlayson, H. J., and Rooke, J. A. (1995). Reduction of proteoly-sis during ensilage of perennial ryegrass by protease inhibitors. J. Sci. Food Agric. 68, 497–505.
Winters, A. L., Cockburn, J. E., Dhanoa, M. S., and Merry, R. J. (2000). Effects of lactic acid bacte-ria in inoculants on changes in amino acid composition during ensilage of sterile and non-sterile ryegrass. J. Appl. Microbiol. 89, 442–451. doi: 10.1046/j.1365-2672.2000.01133.x
Xu, D. M., Ding, W. R., Ke, W. C., Li, F. H., Zhang, P., and Guo, X. S. (2019). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofer-mentative Lactobacillus plantarum and heterofermentative Lactobacillus buchneri. Front. Microbiol. 9:3299. doi: 10.3389/fmicb.2018.03299
Xu, D. M., Ding, Z. T., Wang, M. S., Bai, J., Ke, W. C., Zhang, Y. X., et al. (2020a). Characteriza-tion of the microbial community, metabolome and biotransformation of phenolic compounds of sainfoin (Onobrychis viciifolia) silage ensiled with or without inoculation of Lactobacillus plantarum. Bioresource Technology 316, 123910. doi: 10.1016/j.biortech.2020.123910
Xu, D. M., Wang, N., Rinne, M., Ke, W. C., Weinberg, Z. G., Da, M., et al. (2020b). The bacterial community and metabolome dynamics and their interactions modulate fermentation pro-cess of whole crop corn silage prepared with or without inoculants. Microbial. Biotechnol. 14, 561–576. doi: 10.1111/1751-7915.13623
Yang, F. Y., Wang, Y. P., Zhao, S. S., and Wang, Y. (2020). Lactobacillus plantarumInoculants de-lay spoilage of high moisture alfalfa silages by regulating bacterial community composition. Front. Mcrobiol. 2020:1989. doi: 10.3389/fmicb.2020.01989
Yang, H., Wang, Z., Song, W., and Zhao, Z. (2021). Isolation of proanthocyanidins from Pinus thunbergii needles and tyrosinase inhibition activity. Proc. Biochem. 100, 245–251. doi: 10.1016/j.procbio.2020.10.003
Keywords: sainfoin, silage, metabolites, proteolysis, protease
Citation: Huang RZ, Wang X, Ma C and Zhang F (2022) Effects of intrinsic tannins on proteolysis dynamics, protease activity, and metabolome during sainfoin ensiling. Front. Microbiol. 13:976118. doi: 10.3389/fmicb.2022.976118
Received: 23 June 2022; Accepted: 25 July 2022;
Published: 18 August 2022.
Edited by:
Jayesh Jagannath Ahire, Unique Biotech Limited, IndiaReviewed by:
Qing Zhang, South China Agricultural University, ChinaPratik Mohan Bagkar, Advanced Enzyme Technologies, India
Copyright © 2022 Huang, Wang, Ma and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunhui Ma, Y2h1bmh1aW1hQDEyNi5jb20=; Fanfan Zhang, emhhbmdmYW5mYW5Ac2h6dS5lZHUuY24=
 Rong Zheng Huang
Rong Zheng Huang Xuzhe Wang
Xuzhe Wang