
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 September 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.970135
 Cary P. Davies1*
Cary P. Davies1* Thomas Jurkiw2
Thomas Jurkiw2 Julie Haendiges2
Julie Haendiges2 Elizabeth Reed2
Elizabeth Reed2 Nathan Anderson3
Nathan Anderson3 Elizabeth Grasso-Kelley3
Elizabeth Grasso-Kelley3 Maria Hoffmann2
Maria Hoffmann2 Jie Zheng2
Jie Zheng2Low moisture foods (LMFs) have traditionally been recognized as safe for consumption, as most bacteria require higher water content to grow. However, outbreaks due to LMF foods are increasing, and the microbial pathogen Salmonella enterica is frequently implicated. S. enterica can survive in LMFs for years, but few serovars have been studied, and the mechanisms which underlie this longevity are not well understood. Here, we determine that S. enterica serovars S. Tennessee, S. Anatum, and S. Reading but not S. Oranienburg can survive in the ground black pepper for 6 years. S. Reading was not previously associated with any LMF. Using both Illumina and Pacific Biosciences sequencing technologies, we also document changes in the genomes and methylomes of the surviving serovars over this 6-year period. The three serovars acquired a small number of single nucleotide polymorphisms (SNPs) including seven substitutions (four synonymous, two non-synonymous, and one substitution in a non-coding region), and two insertion-deletions. Nine distinct N6-methyladenine (m6A) methylated motifs across the three serovars were identified including five which were previously known, Gm6ATC, CAGm6AG, BATGCm6AT, CRTm6AYN6CTC, and CCm6AN7TGAG, and four novel serovar-specific motifs, GRTm6AN8TTYG, GAm6ACN7GTA, GAA m6ACY, and CAAm6ANCC. Interestingly, the BATGCAT motif was incompletely methylated (35–64% sites across the genome methylated), suggesting a possible role in gene regulation. Furthermore, the number of methylated BATGCm6AT motifs increased after storage in ground black pepper for 6 years from 475 to 657 (S. Tennessee), 366 to 608 (S. Anatum), and 525 to 570 (S. Reading), thus warranting further study as an adaptive mechanism. This is the first long-term assessment of genomic changes in S. enterica in a low moisture environment, and the first study to examine the methylome of any bacteria over a period of years, to our knowledge. These data contribute to our understanding of S. enterica survival in LMFs, and coupled with further studies, will provide the information necessary to design effective interventions which reduce S. enterica in LMFs and maintain a healthy, safe food supply.
Low moisture foods (LMFs) are naturally low in moisture or are produced from higher moisture foods through drying or dehydration processes (water activity aw < 0.85). Cereals, grains, confections (e.g., chocolate), powdered-protein products (e.g., dairy and egg powders), dried fruits and vegetables, honey, spices, seeds, nuts, and nut-based products (e.g., peanut butter) are among LMF products (Podolak et al., 2010; Beuchat et al., 2013; Finn et al., 2013a). In recent years, there has been an increase in the number of foodborne outbreaks associated with microbial contamination of LMFs, and the pathogen Salmonella enterica is often implicated (Vij et al., 2006; Scott et al., 2009; Podolak et al., 2010; Beuchat et al., 2013; Dey et al., 2013; Finn et al., 2013a; Van Doren et al., 2013b). Cereals, grains, and dried protein products are most frequently associated with outbreaks, followed by spices and dried herbs (FAO/WHO, 2014).
This surge in outbreaks is in part due to difficulties in controlling pathogen levels in LMFs. Traditional processing methods which effectively reduce pathogen levels in high moisture foods do not reduce microbial contamination by significant margins (e.g., >5 logs) or to non-detectable levels in LMFs (Beuchat et al., 2013; Finn et al., 2013a). For example, spices and seasonings are often treated with ethylene oxide, propylene oxide, steam treatment, or irradiation to reduce the risk of microbial contamination (Van Doren et al., 2013a), but modifications to these methods are sometimes needed to further reduce levels of S. enterica (Newkirk et al., 2018). Contamination may also occur during the production and processing stages, even after a lethal processing step. Furthermore, Salmonella strains isolated from low aw environments are more likely to acquire tolerance to multiple stresses such as heat, bile salts, and sanitizers than those isolated from high moisture foods (Gruzdev et al., 2011; Smith et al., 2016). Thus, inhibiting the growth of S. enterica in LMFs is a continuing challenge.
Survival of Salmonella in LMFs for periods of multiple years is well documented (Kotzekidou, 1998; Burnett et al., 2000; Hiramatsu et al., 2005; Uesugi et al., 2007; Keller et al., 2013; Haendiges et al., 2021). Moreover, comparative genomics of S. enterica serovar Agona isolates from recurrent multistate outbreaks associated with cereal between 1998 and 2008 demonstrated that Salmonella Agona isolates from both outbreaks only differed by a mean of eight single nucleotide polymorphisms (SNPs), highlighting the persistence of Salmonella over time in food processing facilities (Hoffmann et al., 2020). The mechanisms by which S. enterica responds to the harsh conditions of these environments, however, are not well understood. Populations of S. enterica decrease during the initial desiccation period, after which a subset of cells often enters long-term stationary phase (Finn et al., 2013c). Upon desiccation, Salmonella undergoes multiple physiological processes (Finn et al., 2013a). Solutes such as trehalose accumulate to regulate cell osmolarity (Csonka, 1989; Csonka and Hanson, 1991), filaments may form (Mattick et al., 2000), and changes occur in cell membrane composition and structure (Garmiri et al., 2008; Finn et al., 2013b). Transcriptomic studies yield mechanistic insights into these processes as genes involved in the heat and cold shock response, DNA protection, and gene regulation among others are either up- or downregulated (Deng et al., 2012; Finn et al., 2013a; Crucello et al., 2019).
Less information is available regarding the physiological events and associated mechanisms during long-term survival of S. enterica in low moisture environments. Long-term transcriptional studies have not been performed, and it is unclear whether transcription remains dynamic over a multi-year period after cells enter long-term stationary phase or dormancy. Gene acquisition and loss may affect long-term survival. One study leveraging mutagenesis and sequencing found that genes involved in DNA recombination and repair, osmolarity regulation, lipopolysaccharide biogenesis, stringent response, and stress-induced sigma factors play a role in the survival of S. enterica in pistachio, a low aw food (Jayeola et al., 2020). In addition, increased rates of DNA mutation of up to ∼10–10 mutations per bp per generation can occur as a stress response and some mutations may confer a selective advantage (Bjedov et al., 2003; Foster, 2007). The methyl-directed mismatch repair system or error-prone DNA polymerases may have a role in the generation of this genetic diversity (Goodman, 2002; Foster, 2007). It is therefore possible that mutations which arise and persist in Salmonella populations in LMFs may contribute to desiccation tolerance. However, few studies have examined genome-wide mutations in S. enterica under conditions associated with food production and processing (Knöppel et al., 2018; Baert et al., 2021), and these were not long-term studies. Data spanning extended periods of time are necessary to understand the mechanisms by which S. enterica persists in the food environment and are also critical in the interpretation of data from food safety outbreak traceback investigations such as that of Hoffmann et al. (2020) described above. Such investigations rely on SNP-based phylogenetic analyses to determine isolate relatedness (Allard et al., 2012), and interpretation of these data are contingent on an understanding of genomic changes, particularly over long time periods.
In a previous study, the growth and survival of four S. enterica serovars was studied during storage in ground black pepper for 8 months (Keller et al., 2013). Here, the duration of the storage of ground black pepper and paprika contaminated with the same isolates was extended to a period of 6 years to mimic some of the extreme conditions seen in past recurrent outbreaks with LMF. Genomes and DNA methylation patterns of the isolates were compared before and after the long-term storage study. Desiccation tolerance is achieved through multiple adaptive processes (Finn et al., 2013a; Lebre et al., 2017). These genomic and methylomic data will therefore complement existing physiological and transcriptomic approaches by providing a new perspective on the molecular mechanisms which underpin survival processes of Salmonella in LMFs.
Four S. enterica subsp. enterica serovars were previously obtained from L. Beuchat, University of Georgia, and stored as stock cultures in tryptic soy broth (TSB) (BD Difco, Becton, Dickinson and Company, Sparks, MD, United States) containing 25% glycerol at −80°C in culture collection of the Division of Microbiology, Center for Food Safety and Applied Nutrition (CFSAN), U.S. Food and Drug Administration (U.S. FDA, College Park, MD, United States). All the isolates including S. Tennessee, S. Anatum, S. Reading, and S. Oranienburg were associated with LMFs (Table 1). CFSAN076210 was isolated from a patient in a 2006 peanut associated outbreak in the United States, CFSAN076215 was isolated from raw peanuts in the United States, CFSAN076211 was isolated from raw pecans in the United States, and CFSAN080607 was isolated in the United States (no additional information available). Cultures were plated onto trypticase soy agar with 0.6% yeast (TSAYE, Becton, Dickinson and Company, Sparks, MD, United States) added. Inoculum were prepared as previously described (Keller et al., 2013).
Salmonella inoculated in ground black pepper and paprika samples from a previous study (Keller et al., 2013) were used in this study. In brief, samples were prepared at the Moffett campus, CFSAN, U.S. FDA in 2011. Ground black pepper and paprika were obtained from commercial sources (Keller et al., 2013). Briefly, 25 g quantities of ground black pepper and paprika (not sterilized) were placed into sterile Whirl-Pak filter™ bags (Fisher Scientific, Pittsburgh, PA, United States) and inoculated with 100 μl of undiluted cocktail mix prepared as described earlier to achieve a starting concentration of 1010 CFU/g. Inoculum and ground black pepper/paprika were thoroughly mixed and then stored at 25°C at ambient (low) humidity. The aw of all 25 g samples was 0.5572 ± 0.0432 at the start of the storage period and was measured using an AquaLab dew point water activity meter model 4TE (Meter Foods, Pullman, WA, United States). During storage, Whirlpak bags were left open in a temperature and relative humidity (RH) controlled incubator equipped with a temperature and RH logger and an emergency power source. Reports indicated that no catastrophic events occurred over the duration of the study. After 6 years of storage, these Salmonella inoculated spice samples were shipped to the Division of Microbiology, CFSAN, U.S. FDA for further analysis.
The weight of each sample with the sterile Whirl-Pak bag was recorded. A volume of universal preenrichment broth was added to samples at a 1:9 sample-to-broth ratio and incubated at 35 ± 2°C for 24 ± 2 h. The BAM Salmonella culture method for high microbial load foods was followed thereafter (Wallace et al., 2022). Briefly, aliquots of 1.0 and 0.1 ml from the incubated pre-enrichments were subcultured to 10 ml tetrathionate (TT) broth (Becton, Dickinson and Company, Sparks, MD, United States) and to 10 ml Rappaport-Vassiliadis (RV) medium, respectively. TT broth was incubated for 24 ± 2 h at 43 ± 0.2°C, and RV medium for 24 ± 2 h at 42 ± 0.2°C. Each incubated selective enrichment broth was streaked to bismuth sulfite (BS; Becton, Dickinson and Company, Sparks, MD, United States), Hektoen enteric (Becton, Dickinson and Company, Sparks, MD, United States), and xylose lysine desoxycholate (Becton, Dickinson and Company, Sparks, MD, United States) agar plates, which were incubated for 24 ± 2 h at 35 ± 2°C. Five presumptive positive colonies were transferred to triple sugar iron (TSI; BD, Franklin Lakes, NJ, United States) and lysine iron (LIA; BD, Franklin Lakes, NJ, United States) agar slants and incubated for 24 ± 2 h at 35 ± 2°C. Growth from presumptive-positive TSI slants was confirmed as Salmonella with VITEK MS (bioMérieux, Durham, NC, United States).
Enrichment broth RV and TT from each sample and Salmonella positive colonies were serotyped using the Luminex xMAP® Salmonella Serotyping Assay (Luminex, Madison, WI, United States) to determine composition of surviving serotypes in spice samples. Briefly, DNA was extracted using Bio-Rad InstaGene matrix (Bio-Rad, Hercules, CA, United States) following the manufacturer’s protocol from 1 ml enrichment broth or 1 ml TSB culture from bacterial colony. The standard protocol for the molecular determination of serotype in Salmonella from the Centers for Disease Control and Prevention (CDC) was then followed (Fitzgerald et al., 2007; McQuiston et al., 2011).
Original Salmonella strains were grown overnight in TSB (Becton, Dickinson and Company, Sparks, MD, United States) and strains recovered from ground black pepper samples after long-term storage were plated onto TSA (Becton, Dickinson and Company, Sparks, MD, United States) and incubated overnight at 37°C. (No strains were recovered from paprika therefore these were not sequenced.) The genomic DNA was extracted using the QiaCube Instrument with the QIAamp DNA Mini kit from Qiagen (Qiagen, CA, United States). Concentrations of gDNA were obtained using the Qubit High Sensitivity dsDNA Kit (Thermo Fisher Scientific, Waltham, MA, United States). All samples were analyzed at the exponential stage of growth.
Seven strains, including one representative of each of the three surviving serovars sampled before and after storage in ground black pepper, as well as the S. Oranienburg pre-pepper strain were sequenced using PacBio technology (Table 1). No post-pepper isolate was available for S. Oranienburg, as it did not survive in pepper after 6 years of storage. In brief, DNA was sheared to approximately 10 kb using a Covaris g-TUBE (Covaris, Inc., Woburn, MA, United States). SMRTbell 10 kb template libraries were prepared using DNA Template Prep Kit 2.0 and the Low-Input 10 kb Library Protocol (Pacific Biosciences, Menlo Park, CA, United States). DNA was concentrated, repaired, ligated to hairpin adapters, and purified. Incompletely formed SMRTbell templates were digested with a combination of Exonucleases III and VII. Adapters were annealed, and SMRT sequencing was carried out on the PacBio RS II (Pacific Biosciences, Menlo Park, CA, United States) using standard protocols.
Thirteen isolates were sequenced using Illumina technology, including a single pre-pepper isolate, and three post-pepper isolates per serovar (Table 1). Libraries were constructed using the Nextera DNA Prep Library kit (Illumina Inc., San Diego, CA, United States) following the manufacturer’s protocols (Illumina Inc., San Diego, CA, United States). Paired-end DNA sequencing was performed on the Illumina MiSeq using v2 500-cycle sequencing chemistry (2 × 250 bp) (Illumina, Inc., San Diego, CA, United States). Coverage was at least 50 × for all isolates.
Analysis of PacBio sequence reads was implemented using SMRT Analysis 2.3.0 and the SMRT Portal 2.0 platform (Pacific Biosciences; Menlo Park, CA). De novo assembly was performed using the Hierarchical Genome Assembly Process (HGAP) with default parameters (Bjedov et al., 2003), as described previously (Pirone-Davies et al., 2015). Genomes were annotated using the NCBI (National Center for Biotechnology Information) Prokaryotic Genomes Automatic Annotation Pipeline (Goodman, 2002).1 Adapters were removed from MiSeq reads using bcl2fastq (Illumina, 2017) and reads were trimmed to phred 20 with Trimmomatic v. 0.36 (Bolger et al., 2014), assembled with SPAdes v3.13.0 (Bankevich et al., 2012), and annotated with Prokka (Seemann, 2014).
Single nucleotide polymorphismSNP differences between pre- and post-pepper isolates were identified using the CFSAN-SNP pipeline v. 2.2.0 (Davis et al., 2015) with default parameters. This pipeline maps Illumina reads to a reference genome with Bowtie2, processes the mapping (BAM) files using SAMtools, and identifies variant sites using VarScan. A separate analysis was performed for each serovar, with MiSeq reads as input, and the PacBio pre-pepper assemblies used as the reference genomes (Tennessee, CFSAN076210; Anatum, CFSAN076215; and Reading, CFSAN080607). To verify results of the CFSAN Pipeline, and also to detect single nucleotide insertion-deletions (indels) in post-pepper isolates, the SNP detection workflow of mummer v. 3.23 (Kurtz et al., 2004) was used with default parameters to compare pre- and post-pepper genome assemblies. As with the CFSAN pipeline analyses, the PacBio pre-pepper assemblies were used as the pre-pepper reference genomes, and MiSeq assemblies were used to represent each post-pepper isolate. Last, mummer was also used to compare the PacBio pre- and post-pepper assemblies. However, no new SNPs were identified using this method. Furthermore, indels identified in homopolymeric stretches could not be distinguished from sequencing errors as PacBio sequences are prone to errors in these regions. Thus, we do not report results from these analyses. In addition, a gene presence-absence analysis was conducted on isolates from each serovar using Roary 3.10.2 (Page et al., 2015). Sequences identified as present in some, but not all isolates, were manually checked to ensure sequences were not split between two contigs, and positioned at contig edges. If they were, we assumed these homologs were truncated in the annotation protein fasta and were likely excluded during the filtering step of Roary which removes partial protein sequences. We therefore did not count these as differentially identified among isolates. In other cases, homologs had been classified into distinct Roary clusters, even when sharing high sequence similarity (pident ≥ 95%, qcov ≥ 95%). These sequences were therefore added into the appropriate existing clusters.
Motif Detection and Analysis was also carried out using SMRT Analysis 1.1 and the RS_Modification_and_Motif_Analysis.1 protocol as described at https://www.pacb.com/wp-content/uploads/2015/09/WP_Detecting_DNA_Base_Modifications_Using_SMRT_Sequencing.pdf. Interpulse durations (IPDs) were measured based on the kinetic signals (Flusberg et al., 2010) and processed as described previously (Clark et al., 2012). IPD is the amount of time between emission pulses indicative of base incorporation events during sequencing, and is used to determine the methylation status of a base. At each position in the genome, the observed IPD was compared to the IPD of an in silico control using a two-sample t-test, and a QV score was calculated as QV = −10 log (p-value). Bases were accepted as modified based on a minimum QV threshold value. QV 30 was used as a threshold for preliminary analyses. A plot of QV versus coverage was then constructed using publicly available R scripts found at: https://github.com/topics/motif-finding. The observed bimodal distribution of kinetic data, resulting from modified and unmodified positions, was then used to determine a more stringent QV threshold. Only sites with a minimum of 25× coverage were included. Motifs were identified using the algorithm MotifMaker.2 m6A and m4C motifs can be reliably detected with 25× coverage across all positions in the genome, but m5C requires either significantly higher coverage (∼250×) or Tet-methylation for confident detection. In this study, we consider only m6A and m4C methylations.
Custom python scripts were used to determine whether methylated BATGCAT motifs were present in coding sequence regions (CDSs) or non-coding regions (between CDSs), with a CDS region defined by the biopython packages Bio.SeqIO and Bio.SeqFeature with feature = CDS (Cock et al., 2009; Mckinney, 2010). Sequences of methylations present in coding and non-coding regions of pre- and post-pepper isolates were compared using blastn (Altschul et al., 1990) (pident ≤ 95, qcov ≤ 95). Sequences of methylated coding and non-coding regions were compared across the three serovars using cd-hit (Li and Godzik, 2006), with sequences clustered at 95% identity. Methylated BATGCAT motifs present on plasmids were not considered for these comparisons, as there was a high proportion of sites in the plasmids which did not reach the minimum of 25× coverage required for accurate signal detection.
Salmonella was recovered from all five ground black pepper samples, while none of the paprika samples had a detectable level of Salmonella. Among Salmonella-positive ground black pepper samples, serotypes Reading and Anatum were recovered from all RV and TT samples. Serotype Tennessee was recovered from four of the ground black pepper samples. It was noted that more TT samples were found to have serotype Tennessee than RV samples. None of the samples were observed to contain serotype Oranienburg with both culture method and direct molecular serotyping from enrichment broth.
Few SNP differences between pre- and post-pepper isolates were observed. In S. Tennessee, the CFSAN-SNP pipeline reported the presence of one substitution shared by CFSAN076155 and CFSAN076150, and one additional substitution present in CFSAN076155. In S. Anatum, a single substitution was observed in the three post-pepper isolates, each of which was unique. Of the four S. Reading post-pepper isolates, two isolates, CFSAN076165 and CFSAN076177, each contained a unique SNP substitution (Table 2).
The mummer pipeline identified all SNPs identified in the CFSAN pipeline, as well as several single nucleotide insertion-deletions (indels). All indels were present in homopolymeric stretches of cytosines or guanines. After manual examination of MiSeq reads from all pre- and post-pepper isolates against the PacBio reference pre-pepper genomes, most indels were deemed to be the result of PacBio sequencing errors in homopolymeric stretches, except for two indels in serovars S. Tennessee and S. Anatum.
No differences in gene presence-absence were observed between the pre- and post-pepper isolates in any serovar. Roary identified sequences which were present in only a subset of the isolates examined. However, after blastn analyses, it was determined that homologs of the sequences were present in all genomes (pident ≥ 95%, qcov ≥ 95%).
We observed nine distinct N6-methyladenine (m6A) methylated motifs across the three serovars, with a total of five in S. Tennessee, four in S. Anatum, and six in S. Reading (Figure 1). The motifs Gm6ATC, CAGm6AG, and BATGCm6AT or BATGCm6ATV (B = C,G, or T; V = A,C, or G) were identified in all serovars (Figure 1). In S. Tennessee, GRT m6AN8TTYG, and GAm6ACN7GTA, were also identified. In S. Anatum, CCm6AN8TGAG was observed, and in S. Reading, CRTm6AYN6CTC, GAA m6ACY, and CAA m6ANCC were identified. At least 93% of all 6mA sites within the identified motifs were methylated throughout the genomes, except for the motif BATGCAT, which was partially methylated in all three serovars (35–64%). In S. Anatum, two 4mC motifs, Gm4CSBYVNBG and Gm4CGSNNTBH, were detected in the post-pepper isolate CFSAN076166, and a single 4mC motif, Gm4CGBNDNTB, was detected in the pre-pepper isolate CFSAN076215. However, the mean motif scores for each motif were less than 100, and the percentage of each motif that was methylated throughout the genome was low (6–10%). These motifs were not confidently identified and are thus not included in our final motif list (Figure 1).
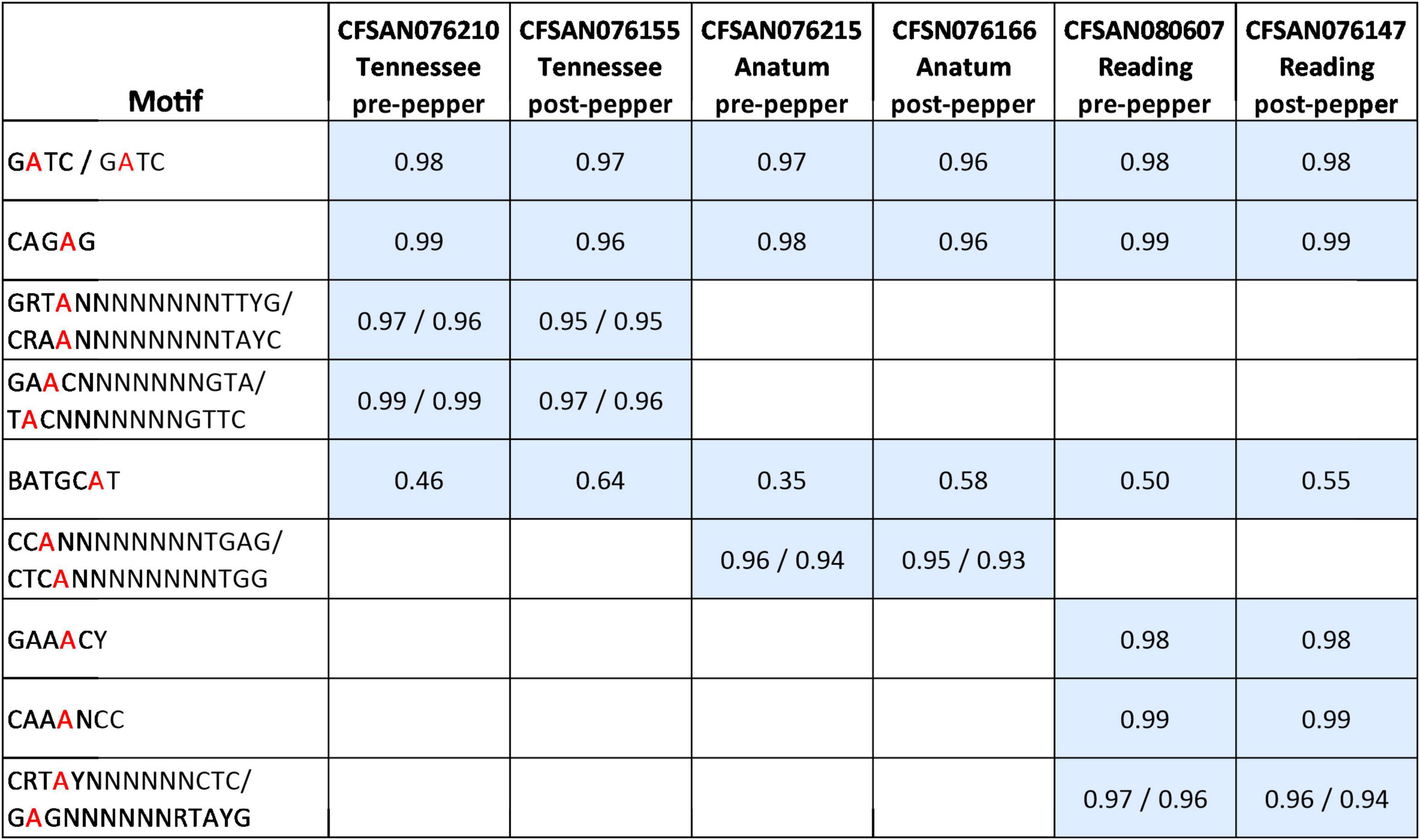
Figure 1. Methylated motifs identified in pre- and post-pepper isolates of three S. enterica serovars, with the percentage of methylation of each motif shown in gray squares (number of methylated motifs/total number of motifs present in genome). BATGCAT is the only motif which is partially methylated (i.e., fewer than 95% of sites are methylated throughout the genome).
In CFSAN076215 (S. Anatum, pre-pepper genome) and CFSAN080607 (S. Reading, pre-pepper genome), the motif BATGCm6ATV (B = C or G or T; V = A or C or G) was identified, while BATGCm6AT was identified in the remaining genomes. In all six genomes, the modifications.gff files from the SMRTPortal pipeline were parsed to determine the percentage of methylated ATGCm6ATT sites. We found that a similar but slightly lower number of ATGCm6ATT sites were methylated in CFSAN076215 and CFSAN080607 (9 and 11%, respectively) compared to the remaining genomes (13–16%), which likely led to the differential reporting of BATGCm6ATV and BATGCm6AT in the MotifMaker output. The percentages of ATGCm6ATT sites methylated across all six genomes do not differ greatly, thus we report on the methylation of BATGCm6AT in all genomes for the remainder of the paper, both for the sake of consistency and to be able to compare the frequency of methylation across pre- and post-pepper isolates. Additional work is needed to determine whether these are true biases in the motifs identified.
The percentage of each motif that was methylated throughout the genome was similar before and after immersion in black pepper, except for that of BATGCm6AT (Figures 1, 2). The total number of methylated BATGCm6AT motifs increased after immersion in pepper in all three serovars from 475 to 657 (S. Tennessee), 366 to 608 (S. Anatum), and 525 to 570 (S. Reading). Approximately equal numbers of methylated BATGCm6AT motifs were present in coding and non-coding regions across all three serovars (Table 3). For example, the pre-pepper Tennessee genome CFSAN076210 contained a total of 475 BATGCm6AT methylations, 231 of which were present within 214 coding regions, and 244 of which occurred in 217 non-coding regions. No methylations of this motif were present in any regions annotated as a non-gene Genbank feature, including different types of RNAs (ncRNA, rRNA, tRNA, and tmRNA), repeat regions, or miscellaneous features. The majority of the methylations in pre-pepper isolates were maintained after immersion in pepper, and additional sites were methylated in each post-pepper genome (Figure 2).
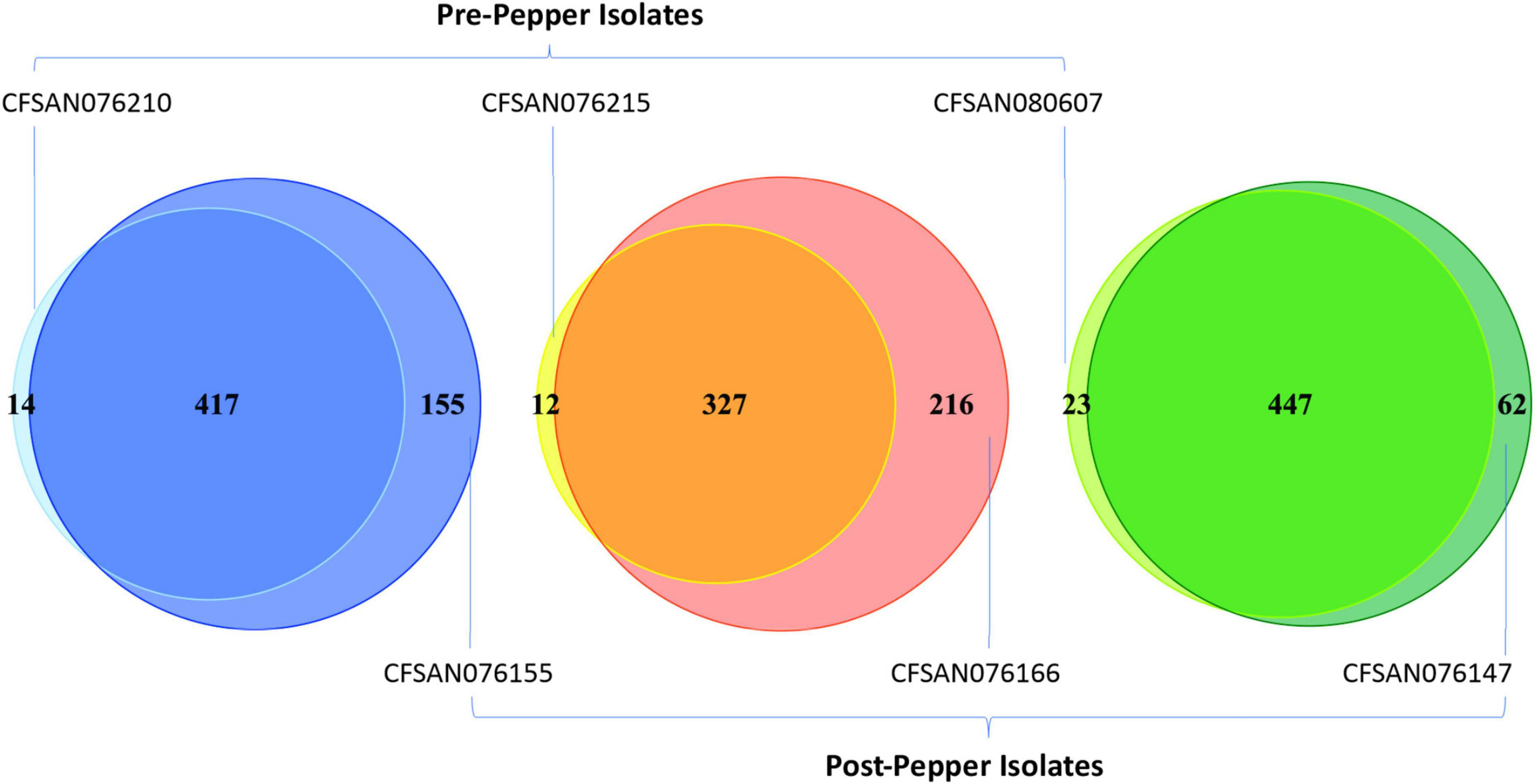
Figure 2. Venn diagrams showing the number of BATGCm6AT motifs before and after immersion in black pepper for the three serovars (from left to right, Tennessee, Anatum, Reading). Most motifs remain methylated throughout storage in black pepper, and some additional motifs are methylated.
We also assessed whether methylated BATGCm6AT motifs occurred in homologous regions across the genomes of the three serovars, with a region defined as the CDS or inter-CDS region in which the methylated adenine was located (see methods for details). Before immersion in pepper, there were 222 methylated regions which were homologous among all three serovars, while after immersion in pepper, this number increased to 377.
Dried ground black pepper is an inhospitable environment for S. enterica. Not only are moisture levels low, but multiple antimicrobial compounds are also present, including 1-phellandrene and caryophyllene (Jirovetz et al., 2002). Still, we recovered serovars S. Tennessee, S. Anatum, and S. Reading from dried ground black pepper after 6 years. S. Reading was not previously associated with LMFs. These results extend our current knowledge on the longevity of S. enterica in low aw foods (Kotzekidou, 1998; Burnett et al., 2000; Hiramatsu et al., 2005; Uesugi et al., 2007; Keller et al., 2013). It is noted that serovar Tennessee was recovered more frequently from TT than RV media.
It is unclear why Salmonella was not recovered from paprika samples. To determine whether Salmonella was present but in a dormant state, we attempted to resuscitate Salmonella in nutrient rich medium, TSB, for 20 h. Still, Salmonella was not recovered. Given the observed serovar-specific survival of Salmonella in black pepper, it is possible that Salmonella survival in paprika may also be serovar-specific. Salmonella serovars Saint Paul, Javiana, and Rubislaw were isolated from paprika and paprika products associated with an outbreak (Lehmacher et al., 1995), and S. Typhimurium in paprika was studied under controlled laboratory conditions (Shankarrao Shirkole et al., 2021). However, the majority of Salmonella serovars have not been studied, and it is likely that a differential response to the low moisture and phytochemicals present in paprika could affect survival.
Transcriptome studies of Salmonella in LMFs provide information on the molecular mechanisms underlying short-term responses to low aw environments. In peanut oil, only 1.36–4.42% of the S. Enteritidis genome was expressed by hours 216 and 528, suggested a significant reduction in overall metabolic activity. Sigma factor RpoE, and cold and heat shock proteins were among the few differentially expressed genes (DEGs) (Deng et al., 2012). In a comparison of S. Typhimurium in black pepper, milk chocolate, powdered milk, and dried pet food, few DEGs were reported to be shared at 24 h except for cold shock genes, but this number increased at 72 h, suggesting a convergence of gene expression across all products at this later timepoints (Crucello et al., 2019). In black pepper, DEGs included ribosomal genes, tRNA genes, and plasmid borne genes involved in horizontal gene transfer (Crucello et al., 2019).
Few studies have examined genomic mutations in Salmonella in LMFs. Baert et al. (2021) reported on SNPs in S. enterica subjected to low aw conditions (Knöppel et al., 2018; Baert et al., 2021). Salmonella was exposed to 10 cycles of heat treatment while in a low aw, high fat matrix. Resulting isolates differed by 0–28 SNPs (Baert et al., 2021), but isolates were cultured in between cycles, thus these results may not be representative of genomic changes which occur outside of the laboratory. In a food safety outbreak traceback investigation, Hoffmann et al. (2020) showed that S. Agona strains can survive and persist in the processing environment for 10 years with only 6 SNPs difference in average (Hoffmann et al., 2020).
Here, over a period of 6 years, we detected seven substitutions and two indels among the three serovars stored in ground black pepper (Table 2), and four of the mutations occur in genes involved in stress-related processes. Four synonymous mutations were identified across the serovars. Synonymous mutations can alter messenger RNA (mRNA) structure and affect translation efficiency, mRNA stability, or protein folding (Lind et al., 2010). Thus, we predicted the effects of the mutations on mRNA secondary structure using the program RNAsnp.3 Of the four mutations, only those found in the enolase (p-value = 0.1716) and a homoserine/homoserine lactone efflux protein (p-value = 0.0752), were predicted to result in structural disruptions, but the p-values were high. We also examined whether the four mutations could affect tRNA usage and ultimately affect translation efficiency (Frumkin et al., 2018; Hanson and Coller, 2018). However, the four mutations are each present in the third codon position and therefore do not likely affect tRNA recognition. It is therefore unlikely that these mutations contribute to Salmonella survival in pepper.
Two non-synonymous mutations were detected, one in S. Anatum, and one in S. Reading. In S. Anatum, a cytosine was replaced with a thymine within a gene encoding a methylated-DNA–protein-cysteine methyltransferase, homologous to ogt, which is involved in the repair of DNA alkylation damage (Daniels and Tainer, 2000; Pegg, 2000). This mutation results in the translation of a lysine instead of a glutamate and leads to a reversal in charge from negative to positive. According to the crystal structure of a homolog from Sulfolobus (Perugino et al., 2015), the mutation occurs in a charged loop region distant from the enzyme active site. DNA damage can be caused in stressed cells due to the accumulation of reactive oxygen species, therefore if this function is enhanced by this mutation, it could confer a fitness benefit in a dry environment. The second non-synonymous mutation occurred in the fepE gene of S. Reading which encodes for a membrane protein involved in the biosynthesis of long chain O-antigens in lipopolysaccharides (Murray et al., 2003; Morona et al., 2009). The mutation occurs at the end of an alpha helix near the protein-protein interaction surface between fepE monomers (Tocilj et al., 2008), and would result in the translation of an arginine in place of a serine. FepE interacts with other Wzz proteins to determine O-antigen length. Efforts to determine the exact mechanism by which this occurs are ongoing, but mutations in multiple sites of fepE and other Wzz genes can alter O-antigen length (Tran and Morona, 2013; Wiseman et al., 2021). Variable O-antigen length can affect the interactions between Salmonella, host, and the environment (Murray Gerald et al., 2006; May and Groisman, 2013), and changes in O-antigen length facilitated by fepE contribute to desiccation tolerance in Cronobacter (Qian et al., 2021). Interestingly, a mutation in the gene gtr which also contributes to O-antigen length arose multiple times when Salmonella was repeatedly exposed to heat in a low aw high fat matrix (Baert et al., 2021). However, the authors conclude that further studies are needed to determine whether the mutation contributes to fitness under stressful conditions.
A single substitution was detected in a non-coding region of the genomes of two S. Tennessee isolates, CFSAN076150 and CFSAN076155. The mutation occurred 75 bases upstream of an ATP-dependent protease, and 277 bases upstream of the IlvGMEDA operon leader peptide. Two single nucleotide indels were observed across all three serovars. In S. Anatum, an insertion occurred in a flotillin family protein in all three isolates and would lead to a frameshift resulting in the translation of two small proteins (52 and 506 amino acids) instead of one longer one (559 amino acids). Flotillins are inner membrane proteins which facilitate the formation of membrane domains and alter membrane fluidity, and thus contribute to many physiological processes (Bach and Bramkamp, 2013; Padilla-Vaca et al., 2019). Flotillin has not been previously reported to play a role in desiccation. However, changes in membrane structure and function including increases in membrane fluidity do occur in Salmonella upon desiccation (Gruzdev et al., 2012; Lebre et al., 2017), therefore the role of flotillin in the response of Salmonella in LMFs should be further investigated.
In S. Tennessee, a deletion occurred in mazG, a nucleoside triphosphate pyrophosphohydrolase which plays a role in adaptation to stress (Zhang and Inouye, 2002; Gross et al., 2006; Lu et al., 2010). This enzyme can hydrolyze non-canonical nucleotides which may otherwise be erroneously incorporated into DNA and RNA (Lu et al., 2010; Lyu et al., 2013), and it also interferes with the stringent stress response and programmed cell death (PCD) induced by the toxin mazF (Gross et al., 2006). The resultant frameshift would lead to a truncation in the mazG protein (266 to 210 amino acids), and a loss of one of the two mazG domains, the Nucleoside Triphosphate Pyrophosphohydrolase (EC 3.6.1.8) C-terminal tandem-domain (cd11528). In Escherichia coli, mazG contains both C- and N-terminal tandem domains, but there are other mazG homologs which contain only a single domain (Robinson et al., 2007; Lu et al., 2010). Thus, it is possible that this mutation does not result in a loss of gene function. In addition, a second open reading frame (orf) is present in the mutated sequence which, when translated, would be 87 amino acids in length and span a full-length C-terminal mazG domain. Either the truncated protein, or the two novel proteins, may remain active, as it would be advantageous to maintain the functioning of this gene in an inhospitable environment such as black pepper.
The documentation of nucleotide mutations in S. enterica over a 6-year period in ground black pepper provides a first look at how the genomes of S. enterica change over long periods of time in LMFs. Although few mutations were observed, four of nine mutations occurred in genes related to stress responses within the cell, and further studies will determine whether any of the mutations confer a selective advantage to S. enterica under stressful conditions. This information will be useful in understanding whether DNA mutations contribute to long-term isolate survival in LMFs, and for the interpretation of data for S. enterica food safety outbreak traceback investigations. Such investigations often determine isolate similarity based on the construction of whole genome sequencing (WGS) phylogenies and pairwise calculation of SNP distances between isolates (Allard et al., 2012). In some cases, similarities between isolates collected over periods of multiple years must be determined (Hoffmann et al., 2020). Mutation rates for S. enterica in specific environments are useful the interpretation of these studies, but these data are not available for S. enterica in LMFs over multi-year periods. Thus, observations regarding the acquisition of mutations among the three serovars over 6 years will aid in these interpretations.
DNA methylation may affect the ability of S. enterica to adapt and survive in the black pepper environment. In prokaryotes, methylation of the N-6 position of adenine (6mA) is the most common DNA modification and is achieved by a methyltransferase enzyme (MTase) (Fujimoto et al., 1965; Casadesus and Low, 2006). MTases, along with a cognate restriction endonuclease, often occur as a restriction-modification system (RM system), which protects cells from foreign DNA (Roberts, 2003; Loenen et al., 2013). However, DNA methylation is also involved in diverse cellular functions including DNA replication initiation and DNA repair, virulence, and gene regulation, among others (Low et al., 2001; Wion and Casadesus, 2006; Shell et al., 2013; Adhikari and Curtis, 2016; Beaulaurier et al., 2019). In S. enterica, the most well-studied MTase is deoxyadenosine methyltransferase (Dam), which methylates the 5′-GATC-3′ motif (Adhikari and Curtis, 2016). Dam regulates the expression of some virulence genes including the plasmid-encoded fimbriae (pef) locus and the std fimbrial operon in S. enterica (Nicolson and Low, 2000; Jakomin et al., 2008). Little is known about the functions of additional methylated motifs in bacteria, and to our knowledge no studies have focused on the function of motifs in Salmonella aside from GATC. However, our prior research showed that the motif ATGCAT is partially methylated (Pirone-Davies et al., 2015), which may indicate the involvement of the motif in gene regulation (Lluch-Senar et al., 2013; Blow et al., 2016). Thus, as a pilot study, we documented 6mA motifs in the genomes of one pre- and one post- pepper isolate from each serovar (Table 1) to determine whether changes occur in methylation after immersion in black pepper, as changes may imply a role in the adaption to the black pepper environment, possibly via differential gene regulation.
We detected a total of nine methylated motifs in the three Salmonella serovars, four of which are not previously described, GRTm6AN8TTYG, GAm6ACN7GTA, GAA m6ACY, and CAA m6ANCC. Gm6ATC, CAGm6AG, and BATGCm6AT, were methylated across all three serovars, while CRTm6AYN6CTC, CCm6AN8TGAG, GRTm6AN8TTYG, GAm6ACN7GTA, GAA m6ACY, and CAA m6ANCC were serovar-specific (Figure 1). These results are similar to those of a previous study, which also observed the conservation of the three motifs across serovars, in addition to serovar-specific motifs (Pirone-Davies et al., 2015). Further comparison of data from this study with that of the previous suggests that within-serovar variation of methylation motifs exists. All motifs in the two S. Anatum genomes examined here were identical to those previously identified in S. Anatum ATCC-BAA1592 (Pirone-Davies et al., 2015), except for a 4mC motif that was identified in S. Anatum ATCC-BAA1592, m4CCWWGG, but not in this study. This motif is methylated by a Type II methyltransferase M.SenAnaIV (deposited in the REBASE database (rebase.neb.com); locus tag SEEA1592_11855). We performed a blastn search of this enzyme against the genomes of S. Anatum CFSAN076215 and CFSAN076166, and did not find any hits, further suggesting that this motif is not likely methylated across all S. Anatum genomes. Previous studies in Listeria monocytogenes (Chen et al., 2017), S. enterica (Pirone-Davies et al., 2015), and Bifidobacterium breve (Bottacini et al., 2018) indicate variations in MTases and methylated motifs occur among species and serovars, but only identical 6mA motifs were identified within different strains of the same serovars (S. Enteritidis and S. Heidelberg) (Pirone-Davies et al., 2015).
A single motif, BATGCm6AT, was incompletely methylated (35–64% of all genome sites methylated), as was found previously (Pirone-Davies et al., 2015). Incomplete methylation may play a role in gene regulation (Lluch-Senar et al., 2013; Blow et al., 2016). Thus, it is possible that methylation of this motif may provide a means for S. enterica to adapt to the stressful environment of black pepper, perhaps by altering gene expression. Thus, we further examined patterns of BATGCm6AT methylation across the three serovars.
Of the methylated BATGCm6AT motifs present in non-coding regions, four occur in promoter regions for genes including the molecular chaperone dnaJ, two distinct MFS transporters, and a hypothetical protein. These four motifs were present in all six genomes examined, except CFSAN076215 only contained this motif in one MFS transporter. In addition, the two S. Tennessee genomes contained a methylated BATGCm6AT in the promoter regions of the genes Ag (+)-translocating P-type ATPase SilP and a multidrug efflux MFS transporter. Transcriptional studies are needed to determine the effect of these and other methylations on gene expression, if any.
Approximately half of all methylated BATGCm6AT motifs (50–57%) in pre-pepper genomes occurred in regions homologous across the serovars, and this percentage increased after immersion in pepper [Tennessee, 51–66%, Anatum, 57–69%, and 50–74% (# methylated BATGCm6AT motifs located in homologous regions/# total methylated BATGCm6AT motifs per genome, before and after immersion in pepper)]. This methylation of BATGCm6AT across conserved regions of the serovars suggests a functional role for the methylations.
To our knowledge, this is the first study to characterize long-term changes in the methylome of Salmonella. Further studies which characterize the role of the methylation of various motifs in S. enterica, particularly of BATGCm6AT, in black pepper and other environments are needed. One limitation of this study is the analysis of the post-pepper methylomes after growth in media, instead of directly after immersion in black pepper as growth in broth may alter the methylome. However, pre-pepper isolates were also immersed in broth in an identical procedure for sequencing before storage in pepper, and consistent changes in the methylation of BATGCm6AT were still observed across all three serovars. This suggests that methylomic signals due to storage in pepper were detectable.
An increasing number of outbreaks are associated with LMFs contaminated with S. enterica. Thus, an enhanced understanding of the mechanisms by which this pathogen adapts to and survives in low moisture environments is needed. In this pilot study, we demonstrate that three S. enterica serovars survive in black pepper for 6 years and acquire a small number of single nucleotide mutations. We also show that among all serovars, the number of BATGCm6AT sites increased after immersion in pepper, and a high proportion of BATGCm6AT sites were present in homologous regions of the genomes from the three serovars. No mutations occurred within methylated motifs. Additional studies which determine the precise mutation rates of S. enterica in LMFs and elucidate the role of nucleotide and methylation changes in S. enterica physiology are needed. Together, these data will provide the basis for the design of effective intervention strategies which eliminate S. enterica from LMFs and safeguard the food supply.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SRX12580390, SRX125803 97, SRX12580393, SRX12580402, SRX12580365, SRX12580364, SRX12580363, SRX12580396, SRX12580391, SRX12580394, SRX12580395, SRX12580410, SRX12580392, CP033338.1, CP0 33339.1, CP033344.1, CP033345.1, CP033346.1, CP068783.1, CP068784.1, CP068785.1, CP068786.1, CP068787.1, and CP068788.1.
CD, JZ, MH, and TJ: data interpretation. CD, JZ, MH, TJ, NA, and EG-K: conceptualization. CD, JZ, TJ, JH, and MH: formal analysis. TJ, JH, JZ, MH, ER, and NA: data generation. JZ, MH, and CD: supervision. CD, JZ, and TJ: writing original draft. JZ, CD, MH, TJ, JH, NA, and EG-K: writing—review and editing. All authors have read and approved the final manuscript.
This research was funded by the U.S. Food and Drug Administration.
We acknowledge the assistance of Susanne “Sue” E. Keller. We appreciate conversations with James Pettengill regarding the CFSAN SNP pipeline and Roary.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adhikari, S., and Curtis, P. D. (2016). DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 40, 575–591.
Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman, D. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Allard, M. W., Luo, Y., Strain, E., Li, C., Keys, C. E., Son, I., et al. (2012). High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32. doi: 10.1186/1471-2164-13-32
Bach, J. N., and Bramkamp, M. (2013). Flotillins functionally organize the bacterial membrane. Mol. Microbiol. 88, 1205–1217.
Baert, L., Gimonet, J., Barretto, C., Fournier, C., and Jagadeesan, B. (2021). Genetic changes are introduced by repeated exposure of Salmonella spiked in low water activity and high fat matrix to heat. Sci. Rep. 11:8144. doi: 10.1038/s41598-021-87330-8
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477.
Beaulaurier, J., Schadt, E. E., and Fang, G. (2019). Deciphering bacterial epigenomes using modern sequencing technologies. Nat. Rev. Genet. 20, 157–172. doi: 10.1038/s41576-018-0081-3
Beuchat, L. R., Komitopoulou, E., Beckers, H., Betts, R. P., Bourdichon, F., Fanning, S., et al. (2013). Low-water activity foods: Increased concern as vehicles of foodborne pathogens. J. Food Prot. 76, 150–172. doi: 10.4315/0362-028X.JFP-12-211
Bjedov, I., Tenaillon, O., Gérard, B., Souza, V., Denamur, E., Radman, M., et al. (2003). Stress-induced mutagenesis in bacteria. Science 300, 1404–1409.
Blow, M. J., Clark, T. A., Daum, C. G., Deutschbauer, A. M., Fomenkov, A., Fries, R., et al. (2016). The epigenomic landscape of prokaryotes. PLoS Genet. 12:e1005854. doi: 10.1371/journal.pgen.1005854
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120.
Bottacini, F., Morrissey, R., Roberts, R. J., James, K., van Breen, J., Egan, M., et al. (2018). Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve. Nucleic Acids Res. 46, 1860–1877. doi: 10.1093/nar/gkx1289
Burnett, S. L., Gehm, E. R., Weissinger, W. R., and Beuchat, L. R. (2000). Survival of Salmonella in peanut butter and peanut butter spread. J. Appl. Microbiol. 89, 472–477.
Casadesus, J., and Low, D. (2006). Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856.
Chen, P., Den Bakker, H. C., Korlach, J., Kong, N., Storey, D. B., Paxinos, E. E., et al. (2017). Comparative genomics reveals the diversity of restriction-modification systems and DNA methylation sites in Listeria monocytogenes. Appl. Environ. Microbiol. 83, e2091–e2016. doi: 10.1128/AEM.02091-16
Clark, T. A., Murray, I. A., Morgan, R. D., Kislyuk, A. O., Spittle, K. E., Boitano, M., et al. (2012). Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40:e29.
Cock, P. J. A., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423.
Crucello, A., Furtado, M. M., Chaves, M. D. R., and Sant’ana, A. S. (2019). Transcriptome sequencing reveals genes and adaptation pathways in Salmonella typhimurium inoculated in four low water activity foods. Food Microbiol. 82, 426–435. doi: 10.1016/j.fm.2019.03.016
Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53, 121–147.
Csonka, L. N., and Hanson, A. D. (1991). Prokaryotic osmoregulation: Genetics and physiology. Annu. Rev. Microbiol. 45, 569–606.
Daniels, D. S., and Tainer, J. A. (2000). Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase. Mutat. Res. 460, 151–163. doi: 10.1016/s0921-8777(00)00024-0
Deng, X., Li, Z., and Zhang, W. (2012). Transcriptome sequencing of Salmonella enterica serovar enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 30, 311–315. doi: 10.1016/j.fm.2011.11.001
Dey, M., Mayo, J. A., Saville, D., Wolyniak, C., and Klontz, K. C. (2013). Recalls of foods due to microbiological contamination classified by the U.S. food and drug administration, fiscal years 2003 through 2011. J. Food Prot. 76, 932–938. doi: 10.4315/0362-028X.JFP-12-464
FAO/WHO (2014). Ranking of low moisture foods in support of microbiological risk management. Rome: FAO.
Finn, S., Condell, O., Mcclure, P., Amezquita, A., and Fanning, S. (2013a). Mechanisms of survival, responses and sources of Salmonella in low-moisture environments. Front. Microbiol. 4:331. doi: 10.3389/fmicb.2013.00331
Finn, S., Hinton, J. C. D., Mcclure, P., Amezquita, A., Martins, M., and Fanning, S. (2013c). Phenotypic characterization of Salmonella isolated from food production environments associated with low–water activity foods. J. Food Prot. 76, 1488–1499. doi: 10.4315/0362-028X.JFP-13-088
Finn, S., Condell, O., Mcclure, P., Amézquita, A., and Fanning, S. (2013b). Mechanisms of survival, responses and sources of Salmonella in low-moisture environments. Front. Microbiol. 4:331.
Fitzgerald, C., Collins, M., van Duyne, S., Mikoleit, M., Brown, T., and Fields, P. (2007). Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45, 3323–3334.
Flusberg, B. A., Webster, D. R., Lee, J. H., Travers, K. J., Olivares, E. C., Clark, T. A., et al. (2010). Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465.
Foster, P. L. (2007). Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397.
Frumkin, I., Lajoie, M. J., Gregg, C. J., Hornung, G., Church, G. M., and Pilpel, Y. (2018). Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc. Natl. Acad. Sci. U.S.A. 115:E4940. doi: 10.1073/pnas.1719375115
Fujimoto, D., Srinivasan, P. R., and Borek, E. (1965). On the nature of the deoxyribonucleic acid methylases. biological evidence for the multiple nature of the enzymes*. Biochemistry 4, 2849–2855. doi: 10.1021/bi00888a041
Garmiri, P., Coles, K. E., Humphrey, T. J., and Cogan, T. A. (2008). Role of outer membrane lipopolysaccharides in the protection of Salmonella enterica serovar typhimurium from desiccation damage. FEMS Microbiol. Lett. 281, 155–159. doi: 10.1111/j.1574-6968.2008.01093.x
Goodman, M. F. (2002). Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71, 17–50.
Gross, M., Marianovsky, I., and Glaser, G. (2006). MazG – a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59, 590–601. doi: 10.1111/j.1365-2958.2005.04956.x
Gruzdev, N., Mcclelland, M., Porwollik, S., Ofaim, S., Pinto, R., and Saldinger-Sela, S. (2012). Global transcriptional analysis of dehydrated Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 78, 7866–7875. doi: 10.1128/AEM.01822-12
Gruzdev, N., Pinto, R., and Sela, S. (2011). Effect of desiccation on tolerance of salmonella enterica to multiple stresses. Appl. Environ. Microbiol. 77, 1667–1673. doi: 10.1128/AEM.02156-10
Haendiges, J., Davidson, G. R., Pettengill, J. B., Reed, E., Blessington, T., Miller, J. D., et al. (2021). Genomic evidence of environmental and resident Salmonella senftenberg and montevideo contamination in the pistachio supply-chain. PLoS One 16:e0259471. doi: 10.1371/journal.pone.0259471
Hanson, G., and Coller, J. (2018). Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30.
Hiramatsu, R., Matsumoto, M., Sakae, K., and Miyazaki, Y. (2005). Ability of shiga toxin-producing Escherichia coli and Salmonella spp. to survive in a desiccation model system and in dry foods. Appl. Environ. Microbiol. 71, 6657–6663. doi: 10.1128/AEM.71.11.6657-6663.2005
Hoffmann, M., Miller, J., Melka, D., Allard, M. W., Brown, E. W., and Pettengill, J. B. (2020). Temporal dynamics of Salmonella enterica subsp. Enterica serovar Agona isolates from a recurrent multistate outbreak. Front. Microbiol. 11:478. doi: 10.3389/fmicb.2020.00478
Jakomin, M., Chessa, D., Baumler, A. J., and Casadesus, J. (2008). Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190, 7406–7413. doi: 10.1128/JB.01136-08
Jayeola, V., Mcclelland, M., Porwollik, S., Chu, W., Farber, J., and Kathariou, S. (2020). Identification of novel genes mediating survival of Salmonella on low-moisture foods via transposon sequencing analysis. Front. Microbiol. 11:726. doi: 10.3389/fmicb.2020.00726
Jirovetz, L., Buchbauer, G., Ngassoum, M. B., and Geissler, M. (2002). Aroma compound analysis of Piper nigrum and Piper guineense essential oils from Cameroon using solid-phase microextraction-gas chromatography, solid-phase microextraction-gas chromatography-mass spectrometry and olfactometry. J. Chromatogr. A 976, 265–275. doi: 10.1016/s0021-9673(02)00376-x
Keller, S. E., Vandoren, J. M., Grasso, E. M., and Halik, L. A. (2013). Growth and survival of Salmonella in ground black pepper (Piper nigrum). Food Microbiol. 34, 182–188.
Knöppel, A., Knopp, M., Albrecht, L. M., Lundin, E., Lustig, U., Näsvall, J., et al. (2018). Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enterica. Front. Microbiol. 9:756. doi: 10.3389/fmicb.2018.00756
Kotzekidou, P. (1998). Microbial stability and fate of Salmonella enteritidis in halva, a low-moisture confection. J. Food Prot. 61, 181–185. doi: 10.4315/0362-028x-61.2.181
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
Lebre, P. H., De Maayer, P., and Cowan, D. A. (2017). Xerotolerant bacteria: Surviving through a dry spell. Nat. Rev. Microbiol. 15, 285–296. doi: 10.1038/nrmicro.2017.16
Lehmacher, A., Bockemühl, J., and Aleksic, S. (1995). Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 115, 501–511. doi: 10.1017/s0950268800058660
Li, W., and Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659.
Lind, P. A., Berg, O. G., and Andersson, D. I. (2010). Mutational robustness of ribosomal protein genes. Science 330:825.
Lluch-Senar, M., Luong, K., Llorens-Rico, V., Delgado, J., Fang, G., Spittle, K., et al. (2013). Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 9:e1003191. doi: 10.1371/journal.pgen.1003191
Loenen, W. A., Dryden, D. T., Raleigh, E. A., and Wilson, G. G. (2013). Type I restriction enzymes and their relatives. Nucleic Acids Res. 42, 20–44.
Low, D. A., Weyand, N. J., and Mahan, M. J. (2001). Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197.
Lu, L. D., Sun, Q., Fan, X. Y., Zhong, Y., Yao, Y. F., and Zhao, G. P. (2010). Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J. Biol. Chem. 285, 28076–28085. doi: 10.1074/jbc.M109.088872
Lyu, L. D., Tang, B. K., Fan, X. Y., Ma, H., and Zhao, G. P. (2013). Mycobacterial MazG safeguards genetic stability via housecleaning of 5-OH-dCTP. PLoS Pathog. 9:e1003814. doi: 10.1371/journal.ppat.1003814
Mattick, K. L., Jørgensen, F., Legan, J. D., Cole, M. B., Porter, J., Lappin-Scott, H. M., et al. (2000). Survival and filamentation of Salmonella enterica serovar enteritidis PT4 and Salmonella enterica serovar typhimurium DT104 at low water activity. Appl. Environ. Microbiol. 66, 1274–1279. doi: 10.1128/AEM.66.4.1274-1279.2000
May, J. F., and Groisman, E. A. (2013). Conflicting roles for a cell surface modification in Salmonella. Mol. Microbiol. 88, 970–983. doi: 10.1111/mmi.12236
Mckinney, W. (2010). “Data structures for statistical computing in python,” in Proceedings of the 9th python in science conference, eds S. Van Der Walt and J. Millan (Austin, TX: SciPy).
McQuiston, J. R., Waters, R. J., Dinsmore, B. A., Mikoleit, M. L., and Fields, P. I. (2011). Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array. J. Clin. Microbiol. 49, 565–73
Morona, R., Purins, L., Tocilj, A., Matte, A., and Cygler, M. (2009). Sequence-structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem. Sci. 34, 78–84. doi: 10.1016/j.tibs.2008.11.001
Murray Gerald, L., Attridge Stephen, R., and Morona, R. (2006). Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006
Murray, G. L., Attridge, S. R., and Morona, R. (2003). Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47, 1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x
Newkirk, J. J., Wu, J., Acuff, J. C., Caver, C. B., Mallikarjunan, K., Wiersema, B. D., et al. (2018). Inactivation of Salmonella enterica and surrogate Enterococcus faecium on whole black peppercorns and cumin seeds using vacuum steam pasteurization. Front. Sustain. Food Syst. 2:48. doi: 10.3389/fsufs.2018.00048
Nicolson, B., and Low, D. (2000). DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol. Microbiol. 35, 728–742. doi: 10.1046/j.1365-2958.2000.01743.x
Padilla-Vaca, F., Vargas-Maya, N. I., Elizarrarás-Vargas, N. U., Rangel-Serrano, Á, Cardoso-Reyes, L. R., Razo-Soria, T., et al. (2019). Flotillin homologue is involved in the swimming behavior of Escherichia coli. Arch. Microbiol. 201, 999–1008. doi: 10.1007/s00203-019-01670-8
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693.
Perugino, G., Miggiano, R., Serpe, M., Vettone, A., Valenti, A., Lahiri, S., et al. (2015). Structure-function relationships governing activity and stability of a DNA alkylation damage repair thermostable protein. Nucleic Acids Res. 43, 8801–8816. doi: 10.1093/nar/gkv774
Pirone-Davies, C., Hoffmann, M., Roberts, R. J., Muruvanda, T., Timme, R. E., Strain, E., et al. (2015). Genome-wide methylation patterns in Salmonella enterica subsp. Enterica serovars. PLoS One 10:e0123639. doi: 10.1371/journal.pone.0123639
Podolak, R., Enache, E., Stone, W., Black, D. G., and Elliott, P. H. (2010). Sources and risk factors for ccontamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 73, 1919–1936. doi: 10.4315/0362-028x-73.10.1919
Qian, C., Huang, M., Du, Y., Song, J., Mu, H., Wei, Y., et al. (2021). Chemotaxis and shorter o-antigen chain length contribute to the strong desiccation tolerance of a food-isolated Cronobacter sakazakii strain. Front. Microbiol. 12:779538. doi: 10.3389/fmicb.2021.779538
Roberts, R. J. (2003). A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31, 1805–1812. doi: 10.1093/nar/gkg274
Robinson, A., Guilfoyle, A. P., Harrop, S. J., Boucher, Y., Stokes, H. W., Curmi, P. M. G., et al. (2007). A putative house-cleaning enzyme encoded within an integron array: 1.8 Å crystal structure defines a new MazG subtype. Mol. Microbiol. 66, 610–621. doi: 10.1111/j.1365-2958.2007.05932.x
Scott, V. N., Chen, Y. U. H., Freier, T. A., Kuehm, J., Moorman, M., Meyer, J., et al. (2009). Control of Salmonella in low-moisture foods I: Minimizing entry of Salmonella into a processing facility. Food Prot. Trends 29, 342–353.
Shankarrao Shirkole, S., Sadashiv Mujumdar, A., Jayabalan, R., and Prakash Sutar, P. (2021). Dry pasteurization of paprika (Capsicum annuum L.) by short time intensive microwave-infrared radiation: Inactivation of Salmonella typhimurium and Aspergillus flavus considering quality degradation kinetics. Food Chem. 338:128012. doi: 10.1016/j.foodchem.2020.128012
Shell, S. S., Prestwich, E. G., Baek, S. H., Shah, R. R., Sassetti, C. M., Dedon, P. C., et al. (2013). DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 9:e1003419. doi: 10.1371/journal.ppat.1003419
Smith, D. F., Hildebrandt, I. M., Casulli, K. E., Dolan, K. D., and Marks, B. P. (2016). Modeling the effect of temperature and water activity on the thermal resistance of Salmonella enteritidis PT 30 in wheat flour. J. Food Prot. 79, 2058–2065. doi: 10.4315/0362-028X.JFP-16-155
Tocilj, A., Munger, C., Proteau, A., Morona, R., Purins, L., Ajamian, E., et al. (2008). Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15, 130–138. doi: 10.1038/nsmb.1374
Tran, E. N. H., and Morona, R. (2013). Residues located inside the Escherichia coli FepE protein oligomer are essential for lipopolysaccharide O-antigen modal chain length regulation. Microbiology (Reading) 159, 701–714. doi: 10.1099/mic.0.065631-0
Uesugi, A. R., Danyluk, M. D., Mandrell, R. E., and Harris, L. J. (2007). Isolation of Salmonella enteritidis phage type 30 from a single almond orchard over a 5-year period. J. Food Prot. 70, 1784–1789. doi: 10.4315/0362-028x-70.8.1784
Van Doren, J. M., Neil, K. P., Parish, M., Gieraltowski, L., Gould, L. H., and Gombas, K. L. (2013b). Foodborne illness outbreaks from microbial contaminants in spices, 1973–2010. Food Microbiol. 36, 456–464. doi: 10.1016/j.fm.2013.04.014
Van Doren, J. M., Kleinmeier, D., Hammack, T. S., and Westerman, A. (2013a). Prevalence, serotype diversity, and antimicrobial resistance of Salmonella in imported shipments of spice offered for entry to the United States, FY2007–FY2009. Food Microbiol. 34, 239–251. doi: 10.1016/j.fm.2012.10.002
Vij, V., Ailes, E., Wolyniak, C., Angulo, F. J., and Klontz, K. C. (2006). Recalls of spices due to bacterial contamination monitored by the U.S. Food and drug administration: The predominance of Salmonellae. J. Food Prot. 69, 233–237. doi: 10.4315/0362-028x-69.1.233
Wallace, H. A., Hua, W., Andrew, J., Beilei, G., Guodong, Z., and Thomas, H. (2022). BAM chapter 5: Salmonella. Available online at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella
Wion, D., and Casadesus, J. (2006). N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192. doi: 10.1038/nrmicro1350
Wiseman, B., Nitharwal, R. G., Widmalm, G., and Högbom, M. (2021). Structure of a full-length bacterial polysaccharide co-polymerase. Nat. Commun. 12, 369–369. doi: 10.1038/s41467-020-20579-1
Keywords: Salmonella, SNP, methylome, low moisture food, food safety
Citation: Davies CP, Jurkiw T, Haendiges J, Reed E, Anderson N, Grasso-Kelley E, Hoffmann M and Zheng J (2022) Changes in the genomes and methylomes of three Salmonella enterica serovars after long-term storage in ground black pepper. Front. Microbiol. 13:970135. doi: 10.3389/fmicb.2022.970135
Received: 15 June 2022; Accepted: 16 August 2022;
Published: 09 September 2022.
Edited by:
Monica Alejandra Delgado, CONICET Higher Institute of Biological Research (INSIBIO), ArgentinaReviewed by:
Leonard Koolman, University College Dublin, IrelandCopyright © 2022 Davies, Jurkiw, Haendiges, Reed, Anderson, Grasso-Kelley, Hoffmann and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cary P. Davies, Y2FyeS5kYXZpZXNAdXNkYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.