- 1Institute of Nutrition, Mahidol University, Nakhon Pathom, Thailand
- 2National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathum Thani, Thailand
Lactiplantibacillus pentosus 9D3, a prominent gamma-aminobutyric acid (GABA)-producing bacteria isolated from Thai pickled weed was characterized for its safety and probiotic properties via whole-genome analysis and in vitro testing. The whole-genome sequence of L. pentosus 9D3 was determined using a hybrid-sequencing approach, combining PacBio and Illumina technologies. A 3.81-Mbp genome of L. pentosus 9D3 consisting of one 3.65-Mbp chromosome and six plasmids (1.9–71.9 Kbp) was identified with an estimated GC content of 46.09% and 3,456 predicted genes. The strain was confirmed to be Lactiplantibacillus pentosus according to the high average nucleotide identity value of >95% and digital DNA–DNA hybridization scores of >70% to the L. pentosus type strain. Comparative genome analysis with other L. pentosus strains showed that the GABA-producing capability was specific to the strain 9D3. Genes related to GABA biosynthesis and transport were identified on a plasmid, pLPE-70K, indicating the acquired nature of this property. The safety of L. pentosus 9D3 was demonstrated through the lack of genes related to the production of toxins, biogenic amines, and antimicrobial drugs. Although the strain exhibited resistance to ampicillin and chloramphenicol, none of the antimicrobial resistance (AMR) genes were associated with mobile elements, i.e., plasmids and prophages. Therefore, the strain is considered to have low risk of transferring the AMR genes to other, potentially pathogenic bacteria. In addition, L. pentosus 9D3 showed good survivability in the gastrointestinal tract environment and was able to adhere to the intestinal cell in vitro. Therefore, L. pentosus 9D3 is concluded to be safe, with the potential to be used as a probiotic, exerting its health benefit through GABA production in the food system. The GABA-producing capability of the strain in vivo is the subject of further investigation.
Introduction
Lactic acid bacteria (LAB) are biotechnologically important groups of gram-positive bacteria that are non-motile and tolerant to low pH. They usually produce lactic acid as the common metabolic end product of carbohydrate fermentation (Wang et al., 2021). Many of them are considered probiotics due to their potential health benefits related to the gut microbiome, gut function, and immune system (Hajela et al., 2015). Lactiplantibacillus pentosus (previously known as Lactobacillus pentosus) has gained increasing attention due to its immunostimulatory activity, cholesterol-lowering property, and gamma-aminobutyric acid (GABA)-producing capacity (Izumo et al., 2010; Kotani et al., 2010; Bendali et al., 2017; Kittibunchakul et al., 2021; Phuengjayaem et al., 2021). GABA is an inhibitory neurotransmitter that has been reported to be associated with anti-obesity and reverse diabetes (Soltani et al., 2011; Sato et al., 2021). A previous study by Patterson et al. (2019) showed that dietary administration of GABA-producing LAB could improve metabolism and depressive-like behavior in a mouse model (Patterson et al., 2019). Our previous study screened LAB from Thai fermented foods for their ability to grow in brown rice milk and produce GABA (Kittibunchakul et al., 2021). The results showed that L. pentosus 9D3, a GABA-producing strain isolated from pickled weeds, was able to produce high levels of GABA. The 9D3 strain was successfully used to create a plant-based GABA drink using brown rice as a raw material. Additionally, the rice milk fermented with L. pentosus 9D3 contained a high number of live bacteria (8.6 log CFU/ml; Kittibunchakul et al., 2021) prompting the drink to be classified as a probiotic drink, if the strain was characterized as probiotic. Thus, the safety and probiotic properties of the strain 9D3 should be thoroughly investigated.
Previous studies used the advent of whole-genome sequencing technologies and bioinformatics for evaluating the safety and potential benefits of the microorganisms used in fermented food such as fermented pork and fermented olives (Abriouel et al., 2017a; Chokesajjawatee et al., 2020). Therefore, this study aimed to sequence the L. pentosus 9D3 genome and evaluate the safety profile and probiotic properties of the strain. For safety evaluation, we followed the procedural guideline proposed by Chokesajjawatee et al. (2020). The guideline provides details on microbial strain safety assessment via whole-genome sequence (WGS) analysis, including the selection of appropriate sequencing platforms, unambiguous species identification, identification of virulence and undesirable genes, identification of functional and transferrable antimicrobial resistance (AMR) genes, and identification of antimicrobial drug production capability. This guideline was based on a decision tree for microbial safety evaluation proposed by Pariza et al. (2015), in which the rationale for each assessment step was provided in detail. For probiotic properties, the strain’s ability to survive conditions mimicking in vitro digestion and adhesion to the intestinal cell line was analyzed to determine the strain’s probiotic potential (Hill et al., 2014).
Materials and methods
Culture condition for DNA extraction
Lactiplantibacillus pentosus 9D3 was obtained from the culture collection of the Institute of Nutrition, Mahidol University, Thailand. The bacterium was stored at – 80°C in de Man, Rogosa and Sharp (MRS) broth containing 20% glycerol. To obtain cells for DNA extraction, the strain was grown on MRS agar (Difco Laboratories Inc., United States) and incubated at 30°C for 1–2 days. Then, the purified single colony was sub-cultured into 5 ml MRS broth and incubated at 30°C for 12–16 h. The cell pellet obtained from the fresh overnight culture was used for DNA extraction.
Genomic and plasmid DNA extraction
Genomic DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega Corporation, United States) according to the manufacturer’s instructions with some modifications as recommended by Chokesajjawatee et al. (2020). The DNA pellet was dissolved in DNase/RNase-free water. The concentration and purity of the DNA were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, United States). The integrity of the gDNA was assessed using agarose gel electrophoresis. Intact, high purity gDNA with an OD260/OD280 ratio of 1.8–2.0 and an OD260/OD230 ratio of 2.0–2.2 was used for whole-genome sequencing. The plasmid DNA was extracted using a ZymoPURE Plasmid Miniprep kit (Zymo Research Corporation, CA, United States) with some modifications as recommended by Chokesajjawatee et al. (2020). The existence and size of the plasmid, if present, were determined using 0.5% agarose gel electrophoresis.
WGS and de novo assembly process
The gDNA was subjected to two complementary next-generation sequencing platforms, i.e., long-read and short-read sequencing. For long-read sequencing, the gDNA was sequenced using PacBio RSII cell by a certified service provider (DNA Link, Inc., Republic of Korea). For short-read sequencing, the gDNA was sequenced using Illumina HiSeq2000 by a service provided by Macrogen Inc., (Republic of Korea). Several assembly protocols were tested, and the protocol that provided the most accurate (based on manual investigation of the actual reads in problematic areas) and complete (the ability to detect the smallest plasmid presence in the genome) genome was selected. Firstly, the genome was assembled from the long-read sequencing data using the Unicycler pipeline involving the Miniasm assembler and Racon 2 polishing process (Wick et al., 2017). Next, the base accuracy was improved by the short-read sequencing data using the Snippy base-correction protocol (Seemann, 2015). The quality of genome assembly was assessed in terms of the continuity and completeness of orthologous genes using QUAST version 5.0 (Gurevich et al., 2013) and BUSCO version v4 (Simão et al., 2015), respectively. For detection of the small plasmids that may be missed by the initial assembly, the Unicycler hybrid assembly was used with the parameter mode set to “bold.” Candidate plasmids from the Unicycler were subsequently verified by mlplasmids (Arredondo-Alonso et al., 2018) and PlasFlow programs (Krawczyk et al., 2018). The plasmids verified by at least two programs were retrieved.
WGS characterization, gene functional annotation, and comparative genome analysis
Upon the completion of the de novo assembly process, the WGS was characterized through the NCBI prokaryotic genome analysis pipeline for predicting protein-coding genes and genes for noncoding RNAs, e.g., rRNA and tRNA (Tatusova et al., 2016). In addition, the WGS was then characterized using different bioinformatic tools. For AMR gene prediction, the WGS in FASTA format was uploaded and analyzed under default settings (>90% identity and 60% minimum length) in the web server ResFinder 4.11 (accessed on: 6 August 2022) (Florensa et al., 2022). For the detection of prophages, the WGS in FASTA format was uploaded and analyzed under default settings in the online web server PHASTER2 (accessed on: 29 March 2022; Arndt et al., 2016). Moreover, the sequence homology analysis was employed to identify genes potentially associated with AMR by searching all protein-coding genes against a dataset of 2,979 sequences of AMR genes from the comprehensive antibiotic resistance database [CARD;3 (accessed on: 7 September 2021)] (Alcock et al., 2020) using BLASTP (Altschul et al., 1990) with ≥ 80% identity and ≥ 80% coverage cut-off (EFSA, 2021). For gene functional annotation, the protein-coding genes were subsequently analyzed through eggNOG-mapper for clusters of orthologous gene (COG) annotation (Huerta-Cepas et al., 2017, 2019), PANNZER2 for gene ontology (GO) annotation (Törönen et al., 2018), and GhostKOALA for KEGG orthology (KO) annotation (Kanehisa et al., 2016). Additionally, a similarity search was performed using the GenBank database (non-redundant GenBank CDS translations + PDB + SwissProt + PIR + PRF excluding environmental samples from WGS projects, updated date: 31 May 2022, number of sequences: 483,768,206) and BLASTP available at4 (accessed on: 1 June 2022). Graphical genome visualization was generated by CGView (Grant and Stothard, 2008). For comparative genome analysis, all seven publicly available complete genomes of L. pentosus were obtained from the GenBank database (accessed on: 4 August 2022). The seven genomes included L. pentosus SLC13 (GCA_002211885.1), BGM48 (GCA_002850015.1), ZFM222 (GCA_003627295.1), ZFM94 (GCA_003627375.1), DSM 20314 (GCA_003641185.1), KW1 (GCA_023972895.1), and KW2 (GCA_023980805.1). The orthologous gene set was constructed to identify core-and pan-genome using CMG-biotools package version 2.2 (Vesth et al., 2013). Briefly, all predicted protein sequences were compared to each other using BLAST (Binnewies et al., 2005). In which, the two proteins are grouped as the same “gene family” when their BLAST alignment is at least 50% identical and covers 50% of the length of the longest sequence. The core-genome consists of gene families found in all analyzed genomes. The pan-genome is the entire set of gene families from all genomes in the comparison.
Species identification and phylogenetic tree analysis
The 16S rRNA gene was used for initial species identification. Similarities of the 16S rRNA gene to the type/reference strain database were searched using the nucleotide basic local alignment search tool available at NCBI4 (accessed on: 1 June 2022). Phylogenetic tree analysis was performed using the Neighbor-joining method with the bootstrap test of 1000 replicates by MEGA11 (Saitou and Nei, 1987; Stecher et al., 2020; Tamura et al., 2021). The evolutionary distances in the units of the number of base substitutions per site were computed using the Kimura 2-parameter method (Kimura, 1980). The resulting phylogenetic tree was drawn by using Interactive Tree Of Life (iTOL) version 5 (Letunic and Bork, 2021). The species of 9D3 was then confirmed by calculating the average nucleotide identity (ANI) to the type strains of related species using JSpeciesWS (Richter et al., 2016). The strains used for ANI calculation included: L. pentosus DSM 20314T, Lactiplantibacillus argentoratensis DSM 16365T, Lactiplantibacillus plantarum subsp. plantarum ATCC 14917T, Lactiplantibacillus xiangfangensis LMG 26013T, and Lacticaseibacillus fabifermentans DSM 21115T. The ANI value of ≥ 95% was used as a criterion to confirm the species identification (Richter and Rosselló-Móra, 2009). In addition, the WGS similarities expressed as digital DNA–DNA hybridization (dDDH) scores were calculated using the Type (Strain) Genome Server (Meier-Kolthoff and Göker, 2019). For dDDH, a 70% threshold was used as the criterion for species designation (Wayne et al., 1987).
In vitro tests for hemolytic activity, bile-salt deconjugation activity, and antimicrobial susceptibility
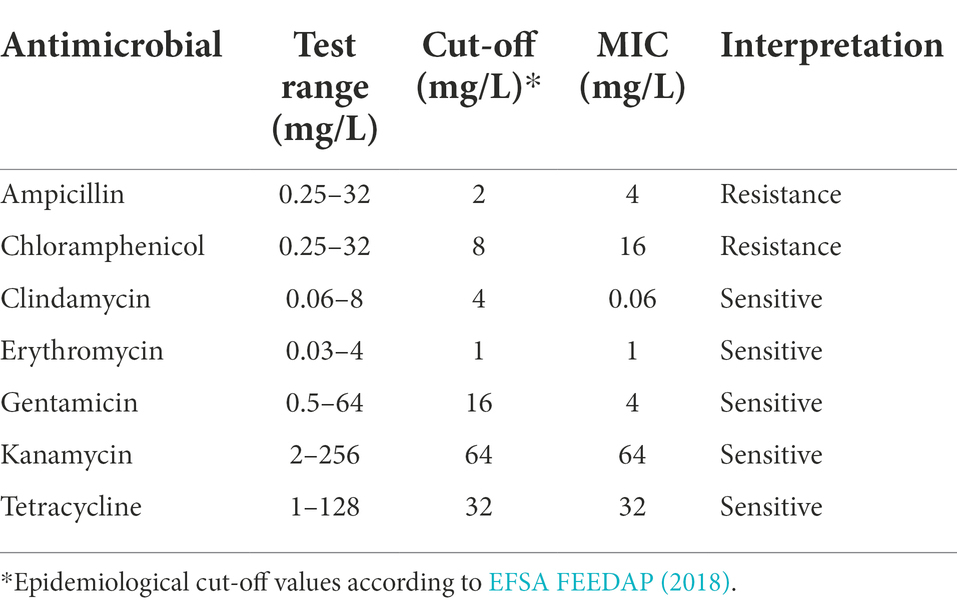
The hemolytic activity was identified by observing the zone around the growth on Columbia blood agar containing 5% sheep blood (Oxoid Ltd., United Kingdom) after being anaerobically incubated at 37°C for 48 h. The bile salt deconjugation activity was determined by observing the precipitation in or around the growth on MRS containing 0.5% taurodeoxycholic acid, or 0.5% glycodeoxycholic acid after 72 h of incubation at 37°C under anaerobic conditions (Dashkevicz and Feighner, 1989). The resistance phenotype of L. pentosus 9D3 was investigated as recommended by the European Food Safety Authority (EFSA FEEDAP, 2018). The strain’s susceptibility to seven antimicrobials, i.e., ampicillin, gentamicin, kanamycin, erythromycin, clindamycin, tetracycline, and chloramphenicol was determined. The minimum inhibitory concentration (MIC) for each antimicrobial was evaluated using the microdilution method following the international standard (International Organization for Standardization (ISO) and International Dairy Federation (IDF), 2010). The MICs were determined in LSM broth [90% ISO-sensitest broth (Oxoid Ltd., United Kingdom): 10% MRS], after incubation of the strain at 37°C under anaerobic conditions (10% H2, 10% CO2, 80% N2) for 48 h.
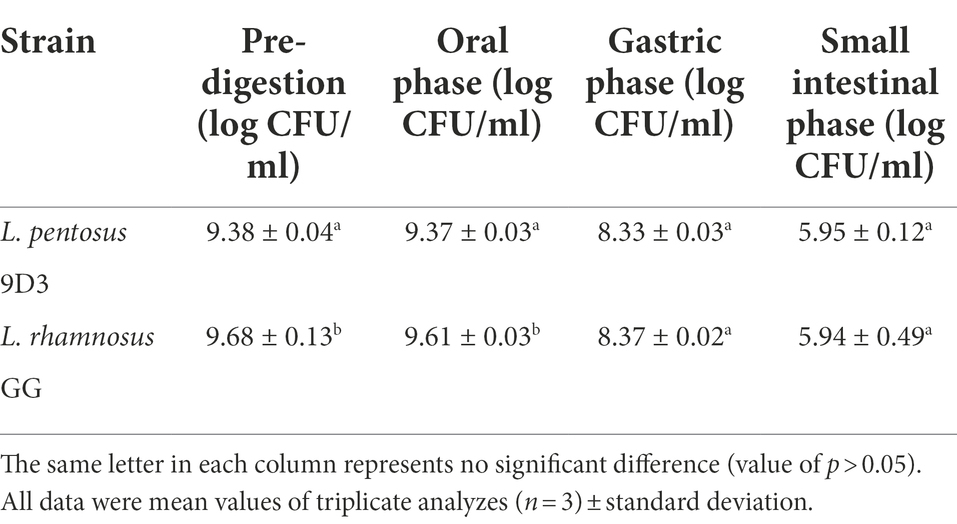
In vitro digestion
In vitro digestion consisted of oral, gastric, and small intestinal phases and was performed following the method previously described (Garrett et al., 1999; Thakkar et al., 2007) with some modifications. For the oral phase, the bacterium was incubated in basal salt solution (120 mM NaCl, 5 mM KCl, 6 mM CaCl2) containing 300 units/ml α-amylase, incubated at 37°C with agitation at 95 rpm for 5 min. For the subsequent gastric phase, the cell suspension was adjusted to pH 2.0 with HCl, and pepsin was added to the final concentration of 2 mg/ml. The suspension was incubated with agitation at 37°C for 1 h. For the subsequent small intestinal phase, the cell suspension was adjusted to pH 6.0 and porcine bile extract, pancreatin, and lipase were added to the final concentrations of 2.4, 0.4, and 0.2 mg/ml, respectively. The solution was incubated at 37°C with agitation at 95 rpm for 2 h. At the end of each digestion phase, the viable bacterial cell counts were determined by plating appropriate 10-fold dilutions onto MRS agar and incubating at 30°C for 48 h. The assay was performed in triplicate. A strain of commercial probiotic, Lacticaseibacillus rhamnosus GG (ATCC 53103) was used for comparison. Student’s t-test was performed to determine statistical differences between 9D3 and L. rhamnosus GG. A p-value of 0.05 was considered a significant difference.
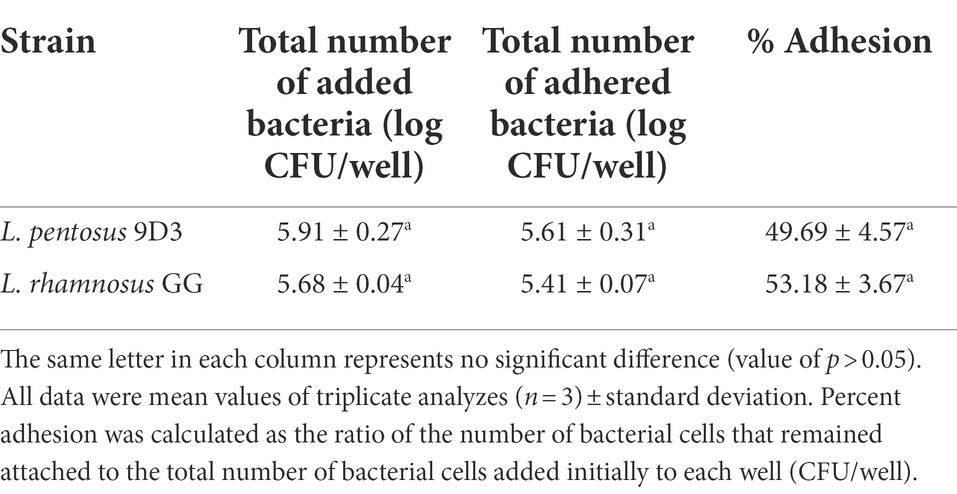
In vitro adhesion
The human colonic carcinoma cell line (Caco-2) was prepared according to the method previously described (Garrett et al., 1999; Chitchumroonchokchai et al., 2004) with some modifications. The Caco-2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 15% heat-inactivated fetal bovine serum (FBS), 2.5 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 25 μg/ml amphotericin B. The cells were maintained under a humidified atmosphere containing 5% CO2 at 37°C. The spent medium was changed with a fresh medium every other day until reaching 80% confluency. For the adhesion assay, 4 × 104 cells were seeded into 6-well culture plates and maintained as previously described. After confluency, the cells were cultured in the complete medium containing 7.5% FBS for 11–14 days. The culture medium was replaced with antibiotic-free DMEM supplemented with 7.5% FBS 1 day before the experiment. For the adhesion reaction, the Caco-2 cells were washed and maintained in the DMEM basal medium (without FBS and antibiotics). After the in vitro digestion, the bacterial cell suspension was prepared in the same basal medium and added to the Caco-2 cells, then incubated at 37°C in an incubator containing 5% CO2 for 1 h. The Caco-2 cells were then washed twice with sterile PBS to remove non-adhering bacteria, and treated with 0.05% TritonX-100 in sterile PBS to release the attached cells. The number of viable bacteria adhering to the Caco-2 cells was determined by plating appropriate 10-fold dilutions onto MRS agar. Percent adhesion was calculated as the ratio of the number of bacterial cells that remained attached to the total number of bacterial cells added initially to each well in the unit of CFU/well (Argyri et al., 2013). The assay was performed in triplicate. L. rhamnosus GG was used for comparison. Student’s t-test was performed to determine statistical differences between 9D3 and L. rhamnosus GG. A p-value of 0.05 was considered a significant difference.
Results and discussion
WGS of Lactiplantibacillus pentosus 9D3
A highly accurate and complete WGS of L. pentosus 9D3 was obtained. The complete genome has a total size of 3.81 Mbp and a GC content of 46.09%. This genome consists of a 3.65-Mbp chromosome and six plasmids in sizes ranging from 1.9 to 71.9 Kbp (Figure 1). A total of 3,456 genes were predicted with an average length of 923.9 bp, and these occupied 83.8% of the genome. Among them, 3,356 genes were predicted as protein-coding genes and the remaining 100 genes were tRNA (80 genes), rRNA (16 genes), and other noncoding RNA (4 genes).
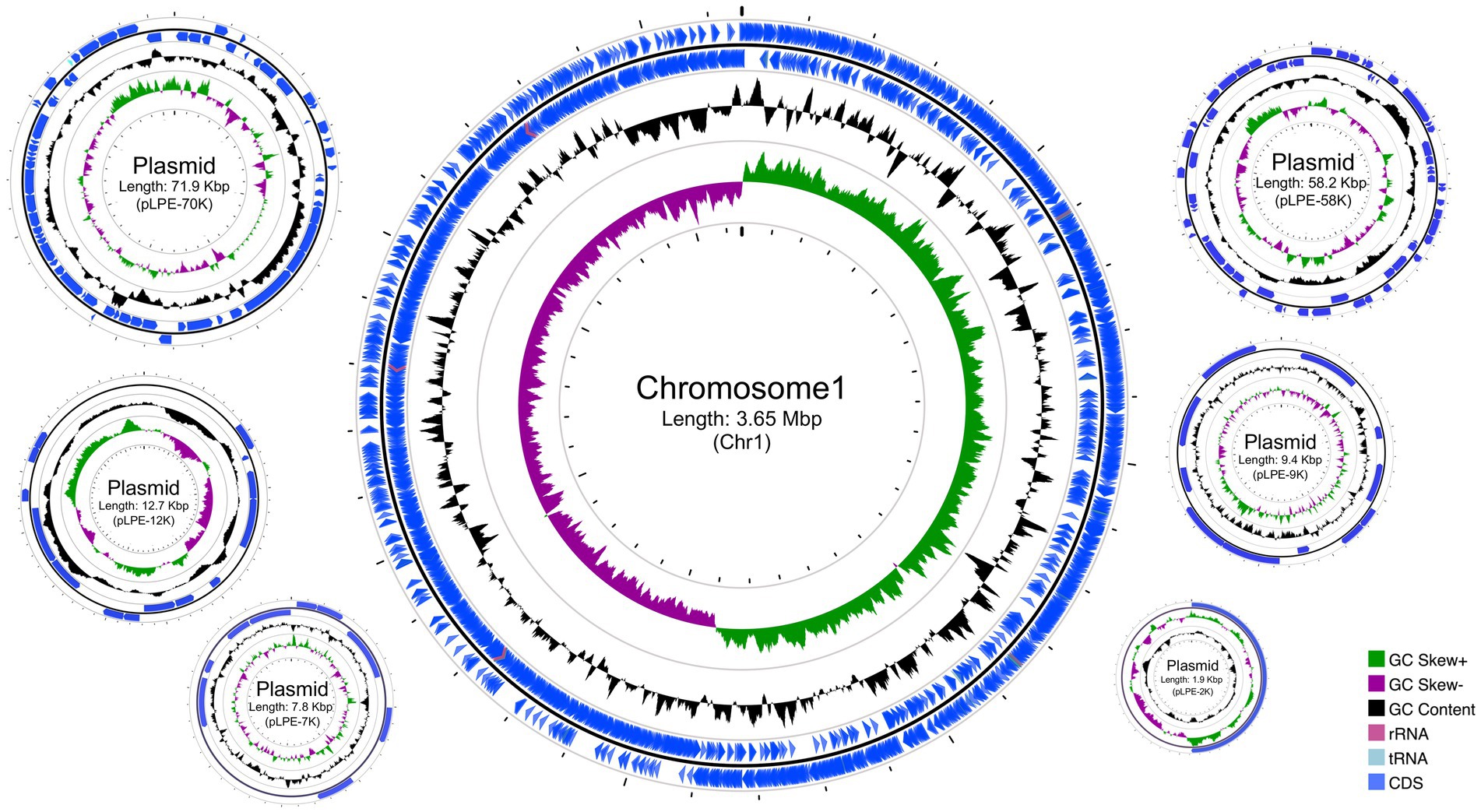
Figure 1. Circular genome maps of Lactiplantibacillus pentosus 9D3. From outside to center: Rings 1 and 2 demonstrate the protein-coding genes (CDS) (blue), tRNA (light blue), and rRNA (light purple) on the forward and reverse strands, respectively; ring 3 represents the GC content plot; ring 4 represents the positive and negative GC skew in green and purple, respectively.
The genome size and gene number of the 9D3 strain were similar to those of L. pentosus strains isolated from fermented olives, such as IG4 (3.81 Mbp, 3,522 genes) and MP-10 (3.70 Mbp, 3,558 genes) with very similar GC contents, varied from 45.97 to 46.32% (Abriouel et al., 2017a; Calero-Delgado et al., 2018). Nevertheless, the 9D3 strain showed a larger genome size and higher gene number than L. pentosus KCA1 isolated from a vaginal source (3.4 Mbp, 2,992 genes) (Anukam et al., 2013). This may be due to the presence of plasmids in L. pentosus 9D3 (6 plasmids) similar to IG4 (6 plasmids) and MP-10 (5 plasmids), while the KCA1 has no plasmid. The variations in the genome size and gene number may reflect different environmental nitches of the strains.
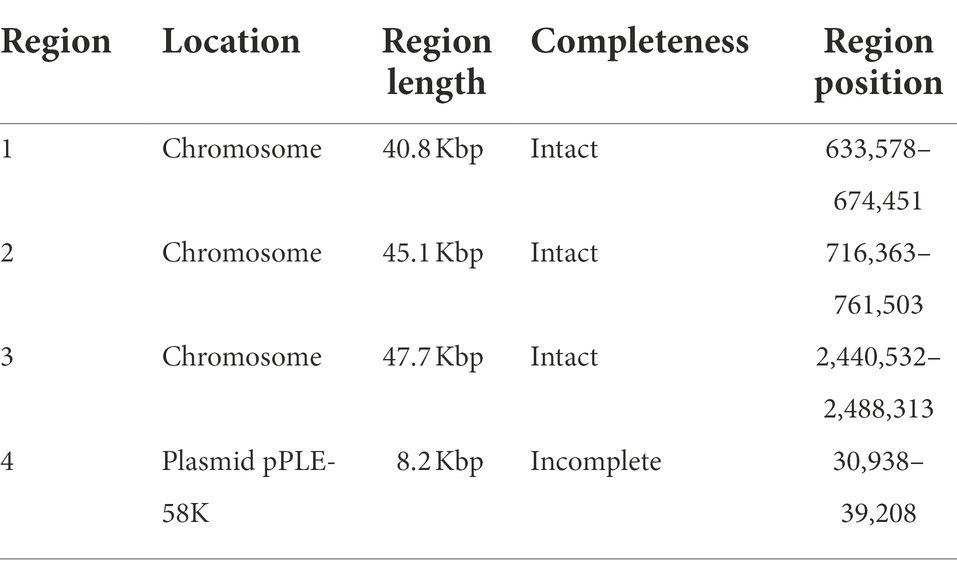
In addition, four potential prophages, including three intact prophages ranging from 40.8 to 47.7 Kbp integrated into the chromosome, and one incomplete prophage located in the plasmid pPLE-58K were found in the 9D3 genome (Table 1). The presence of intact prophages in a range of 1–5 per genome is found to be common in lactobacilli strains (Pei et al., 2021). For L. pentosus, previous studies reported the presence of two and four intact prophages in strains isolated from fermented olives and fermented sausages, respectively (Abriouel et al., 2017a; Stergiou et al., 2021). This is in line with our result in which three intact prophages were found in the genome. The presence of prophages in the bacterial genome may confer fitness to the harsh environment (Pei et al., 2021).
Species identification and phylogenomic profiling of Lactiplantibacillus pentosus 9D3
For initial species identification, five copies of the 16S rRNA gene were identified in the 9D3 genome. These nucleotide sequences were highly similar but not identical, with 98.85–99.81% identity to each other (Supplementary Table S1). The sequences showed the highest sequence similarity to those of L. pentosus and L. plantarum (99.61–99.80%), and L. paraplantarum (99.47–99.67%); (Supplementary Table S2). A phylogenetic tree created from the 16S rRNA gene sequences of the 9D3 strain and the type strains of closely related species is shown in Figure 2. From the high sequence similarities of the 16S rRNA gene among several closely-related species in this group, the species identification of the 9D3 strain is inconclusive. For definitive species identification, the 9D3 strain showed the highest ANI value and dDDH score for the type strain of L. pentosus at 97.24 and 80.6%, respectively. Both values were above the species cut-off values of 95% for ANI and 70% for dDDH. In contrast, the other species indistinguishable by the 16S rRNA gene marker analysis showed clear distances to the strain with much lower ANI and dDDH scores, as detailed in Supplementary Table S3. Therefore, the 9D3 strain was unambiguously identified as L. pentosus.
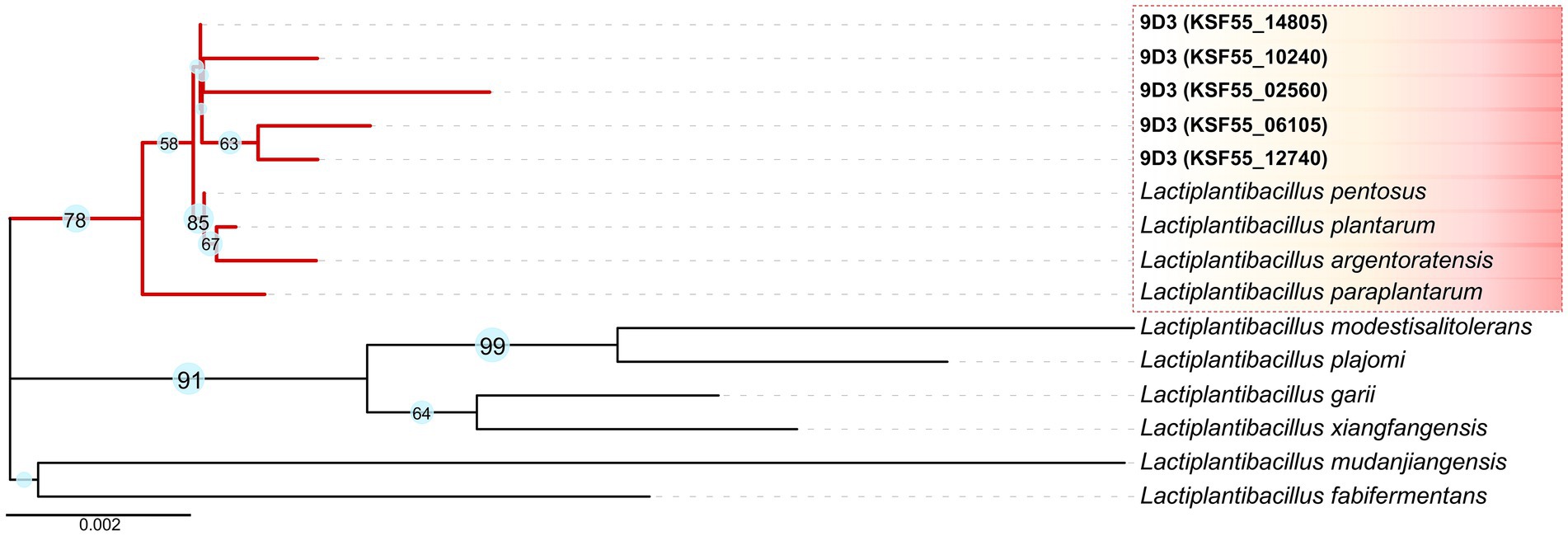
Figure 2. 16S rRNA gene marker analysis of the 9D3 strain and the selected type strains of closely related species. The phylogenetic tree was constructed by the Neighbor-Joining method with 1,000 bootstrap replicates using MEGA11 (Saitou and Nei, 1987; Stecher et al., 2020; Tamura et al., 2021). Evolutionary distances in the units of the number of base substitutions per site were computed using the Kimura 2-parameter method (Kimura, 1980), and the visualization of the phylogenetic tree was drawn by iTOL version 5 (Letunic and Bork, 2021), in which only bootstrap values ≥ 50% are shown next to the branches.
Gene function annotation and comparative analysis of Lactiplantibacillus pentosus 9D3 genome
Among the 3,356 protein-coding genes, 3,031 genes could be assigned with functions regarding the different protein databases, i.e., COG (2,825), GO (2,813), and KEGG (1,883), as shown in Figure 3A and detailed in Supplementary Table S4. Analysis of gene function distribution across the different COG functional categories revealed that metabolism-related genes were the most abundant, accounting for 37.8% of the total COG annotation. Among them, the majority were involved in the metabolism and transport of carbohydrates, as shown in Figure 3B. This finding is in agreement with a previous study, which showed that L. pentosus is a versatile and flexible species that can grow on a wide variety of carbon sources (Abriouel et al., 2017b).
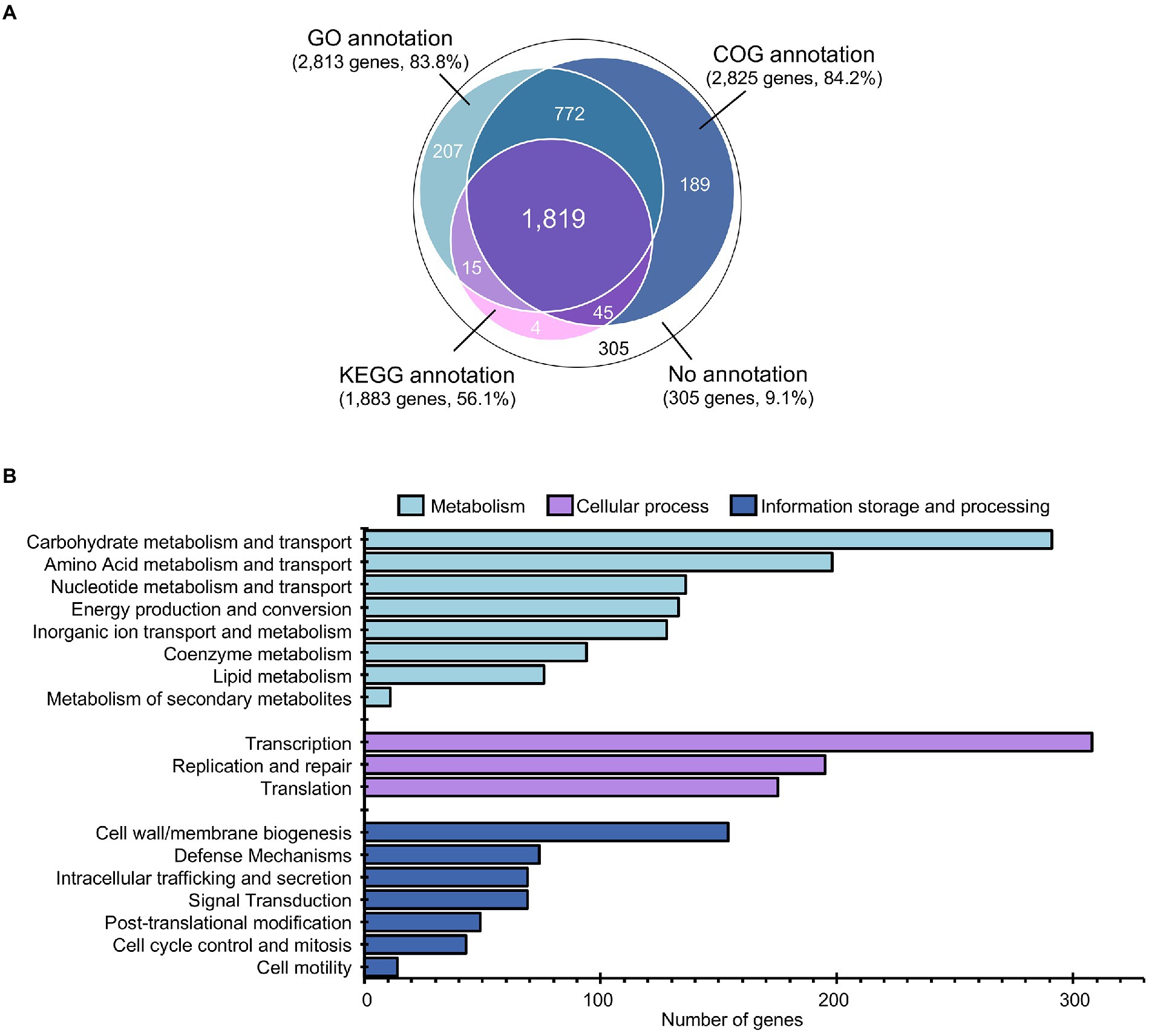
Figure 3. Annotation and gene function analysis of L. pentosus 9D3 genome. (A) Numbers of annotated genes from different databases. (B) Distribution of annotated genes across the different COG functional categories.
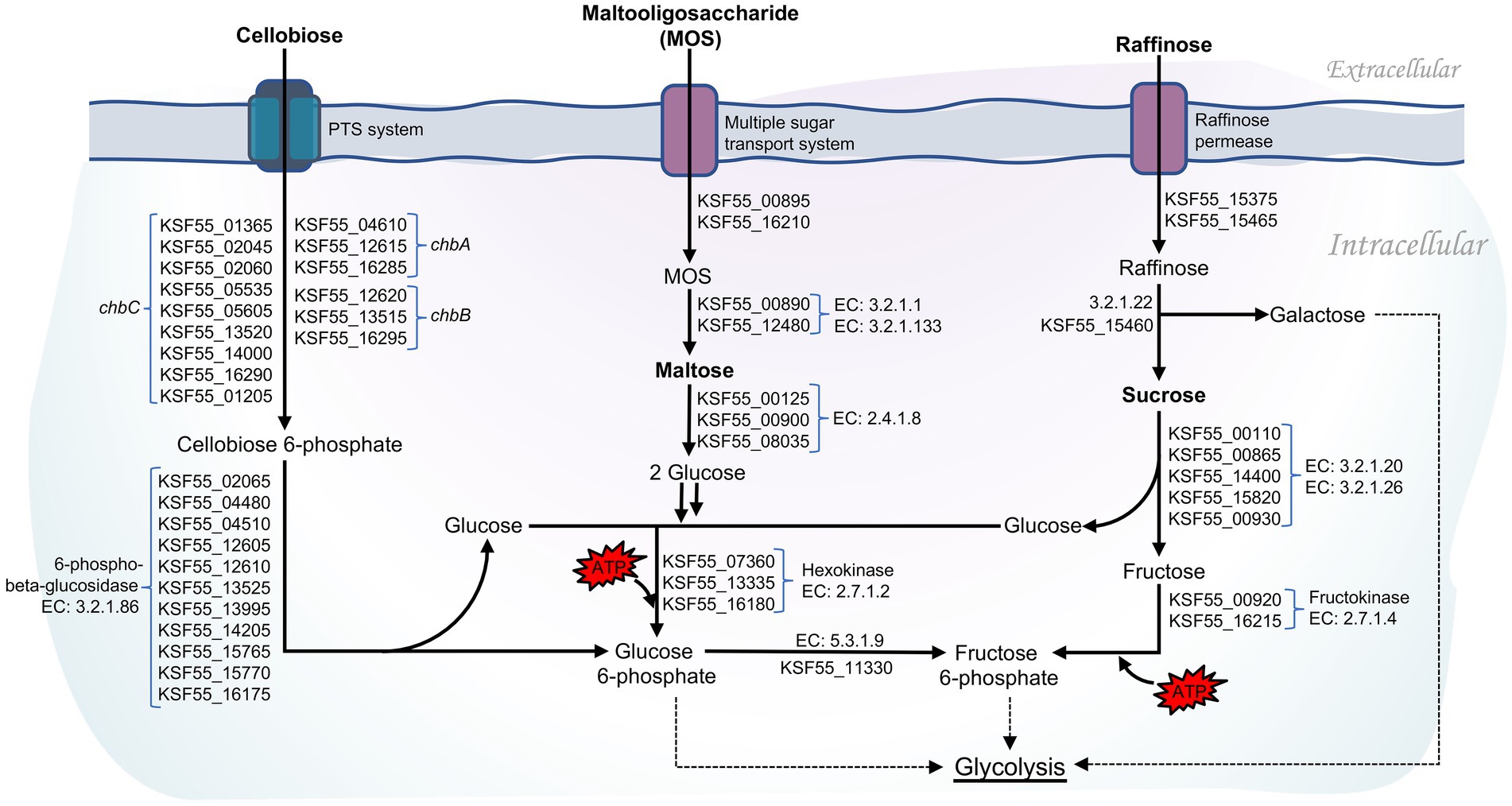
Intriguingly, as many as 58 genes encoded for phosphotransferase systems (PTS) were identified in the 9D3 strain (Supplementary Table S5). The PTS catalyzes the phosphorylation of the incoming carbohydrates concomitant with their translocation across the cell membrane (McCoy et al., 2015). During the translocation process by PTS, the glycolytic conversion of phosphoenolpyruvate to pyruvate is activated, thereby increasing the production of glycolytic and TCA cycle intermediates (Gosset, 2005). The presence of PTS in L. pentosus may increase carbon transport capacity and thus enhance bacterial growth (Abriouel et al., 2017b). Notably, eleven copies of 6-phospho-beta-glucosidase (EC: 3.2.1.86), which hydrolyzes cellobiose 6-phosphate to yield glucose 6-phosphate and glucose, were identified in L. pentosus 9D3 genome (Figure 4) in addition to the large number of genes associated with PTS for cellobiose (15 genes). Compared to other disaccharides such as maltose and sucrose, the 6-phospho-beta-glucosidase pathway required only one ATP to metabolize cellobiose for glycolysis (Figure 4). This pathway is more energy efficient for disaccharide metabolism. Thus, the redundant cellobiose PTS and 6-phospho-beta-glucosidase genes suggest that L. pentosus 9D3 is well-adapted for growth in a cellobiose-rich environment. Besides, the prevalence of genes coding for disaccharides, monosaccharides, and polyols transport proteins supports that 9D3 is capable of utilizing a wide range of carbohydrates. These included sucrose, lactose, maltose, mannose, fructose, arabinose, galactitol, sorbitol, mannitol, and inositol (Supplementary Table S5). In addition, the genes involved in the transport and utilization of prebiotics such as maltooligosaccharide and raffinose were also identified, as shown in Figure 4. These results supported the versatility of the strain to survive and adapt to various environments, including the gastrointestinal (GI) environment, where simple sugars are scarce and non-digestible sugars are the main nutrient source.
In comparison with the other seven L. pentosus strains, 2,357 and 4,837 gene families were identified as core-genome and pan-genome, respectively (Figure 5). A total of 165 genes were unique for L. pentosus 9D3 (Supplementary Table S6). Interestingly, a gene responsible for GABA production, i.e., glutamate decarboxylase (KSF55_17165: gadA) and secretion, i.e., glutamate-GABA antiporter (KSF55_17160: gadC) were uniquely identified on the pLPE-70 K plasmid of 9D3. This glutamate decarboxylase (GAD) system converts glutamate into GABA and helps to maintain cytosolic pH homeostasis. The system was shown to play a significant role in conferring acid resistance and allows LAB to survive in acidic environments (Wu et al., 2017). Moreover, the localization of the GAD genes in the plasmid infers a previous event of horizontal gene transfer of these genes into the genome of L. pentosus 9D3, providing the unique GABA-producing property of the strain when compared to the other analyzed L. pentosus. The genes related to the GABA-producing property of 9D3 exhibited high sequence similarity and synteny to Lactiplantibacillus plantarum KB1253, a GABA-producing reference strain (Nakatani et al., 2019), Figure 6. The result suggested that the GABA biosynthesis genes in 9D3 may be acquired from, or have a common ancestor with those of the GABA-producing L. plantarum. Since the GABA-producing capability of 9D3 is associated with a plasmid, maintenance of the plasmid is crucial for the strain application concerning this property. On the other hand, this finding also opens the possibility to develop new GABA-producing cultures by allocating the GABA-producing plasmid of L. pentosus 9D3 to other desirable strains suitable for the specific application.
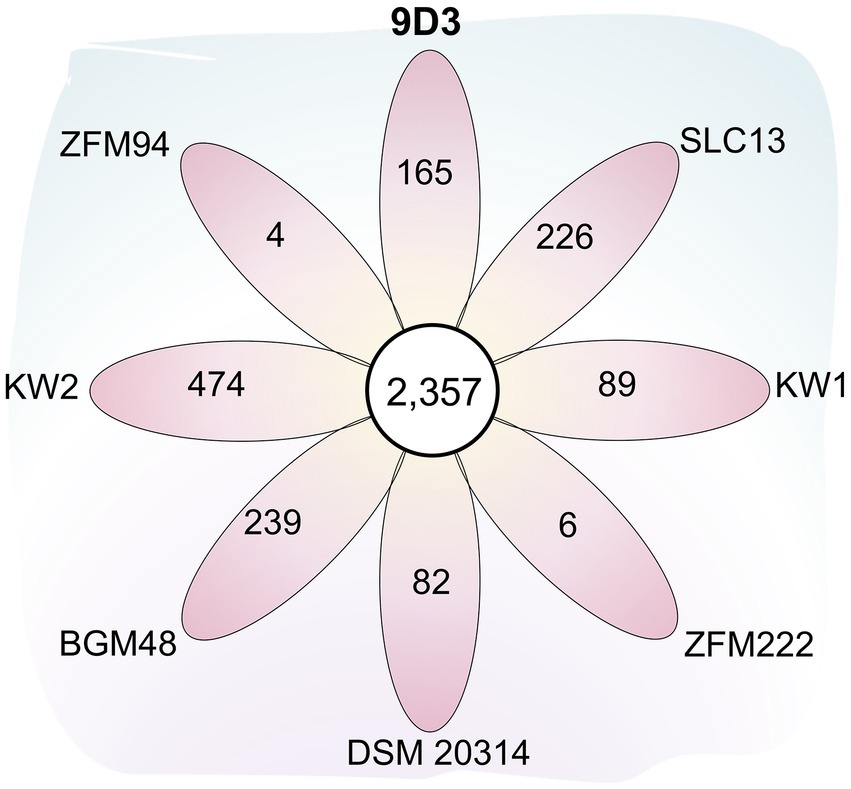
Figure 5. Flower plot diagram showing the numbers of core and unique genes among eight L. pentosus strains. The central circle shows the number of core gene families common to all strains while the petals show the numbers of genes unique to each strain. Abbreviated strain names are given outside each petal.
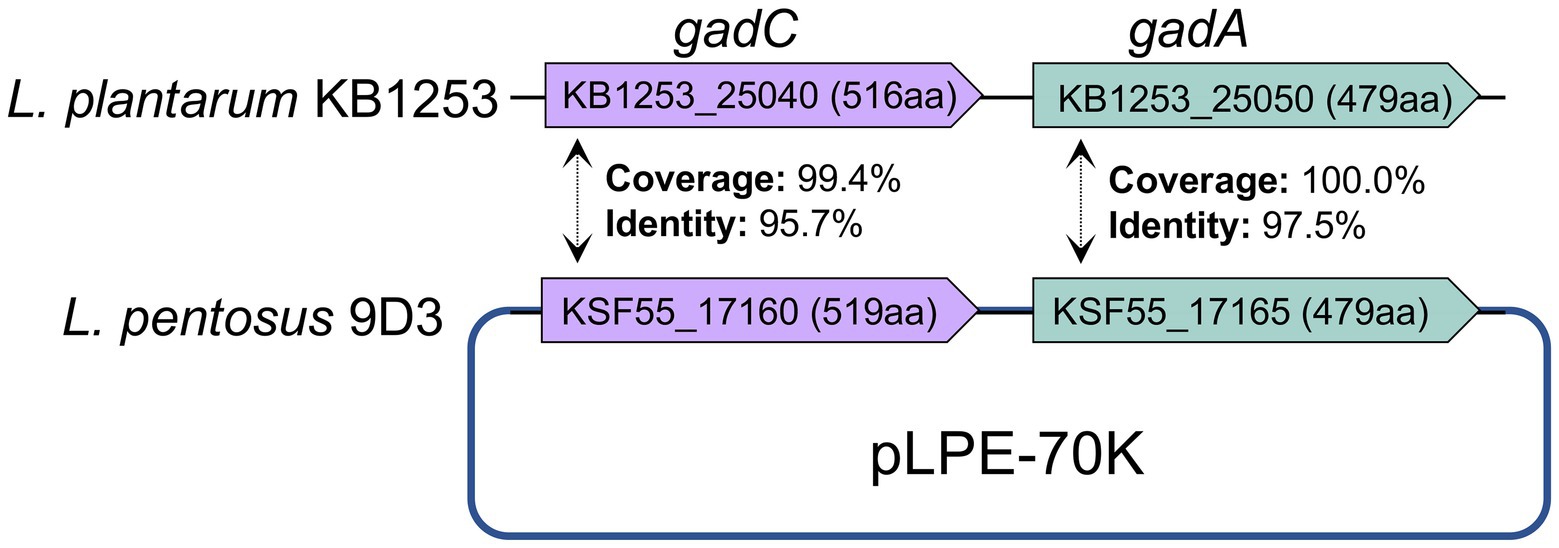
Figure 6. Comparison of genes related to GABA production of L. pentosus 9D3 and L. plantarum KB1253, a GABA-producing reference strain. gadA, glutamate decarboxylase; gadC, glutamate-GABA antiporter.
Identification of genes implicated in the safety of Lactiplantibacillus pentosus 9D3
The safety of L. pentosus 9D3 was evaluated in terms of virulence and undesirable properties, AMR, and antimicrobial drug biosynthesis capability.
Virulence and undesirable properties
Hemolysis
Functional analysis based on KEGG annotation identified one bacterial toxin gene, hlyIII, encoding hemolysin III (KSF55_14680). The gene was found in many lactobacilli probiotic strains and concluded to involve no safety concerns. Hemolysis identified using sheep blood agar showed no visible clear zone around the growth, indicating the strain was nonhemolytic. However, a slight color change similar to the reaction exhibited by a probiotic control strain, L. plantarum 299 V was observed (Supplementary Figure S1). The slight color change from red to yellow around the growth was considered a result of acidity produced by the strain rather than the activity of toxins. Therefore, it was concluded that the L. pentosus 9D3 strain is safe in this aspect.
D-lactate formation
The result of the KEGG pathway analysis showed that the 9D3 genome contained a copy of the gene encoding for lactate racemase (EC: 5.1.2.1; KSF55_00515) and two copies of D-lactate dehydrogenase (EC: 1.1.1.28; KSF55_04355 and KSF55_09370), indicating its capability to convert L-lactate to D-lactate. D-lactate production is a property universally expressed in LAB since it is an essential component in cell wall peptidoglycan and is considered an intrinsic property of LAB. Specific concern on this property was due to historical records of D-lactic acidosis associated with probiotic consumption. However, this condition was very rare and occurred only in specific sub-populations, e.g., patients with carbohydrate malabsorption conditions that failed to control their carbohydrate consumption (Chokesajjawatee et al., 2020). This property was concluded to be of no safety concern for the general population.
Bile salts deconjugation
Identification of the genes involved in bile salts deconjugation activity was performed by KEGG pathway analysis. No gene encoding for the enzyme choloylglycine hydrolase/bile salt hydrolase (BSH) (EC: 3.5.1.24) was found in the 9D3 genome. The absence of bile salts deconjugation activity was confirmed by the absence of the precipitates in or around the growth on MRS containing 0.5% taurodeoxycholic acid or 0.5% glycodeoxycholic acid (Supplementary Figure S2). It should be noted that lacking this ability should not be interpreted as bile intolerance since the two properties are unrelated (Moser and Savage, 2001). The undesirability of BSH activity was controversial. It was mentioned among other properties to be characterized for safety assurance but also listed among the main characteristics tested for probiotic properties (FAO/WHO, 2002). A comprehensive review by Begley et al. (2006) discussed the possible impact of the BSH activity on the host in both aspects. On the beneficial side, oral administration of BSH-positive probiotics has been shown to significantly reduce cholesterol levels in humans and mice. The deconjugated bile salts are less efficiently reabsorbed, hence increasing the demand for cholesterol for de novo synthesis of bile acids. Also, deconjugated bile salts are less efficient in the solubilization of lipids, including cholesterol, thereby reducing the absorption of fat and cholesterol in the gut. However, the reduced emulsification of dietary lipids may compromise normal lipid digestion and impair fatty acids and monoglycerides absorption. Another possible safety concern is related to secondary bile acids resulting from the subsequent modification of unconjugated bile salts. The secondary bile acids may cause DNA damage, promote colon cancer, impair colonic mucosal function, and promote gallstone formation (Begley et al., 2006).
Biogenic amines formation
Genes related to the formation of biogenic amines, i.e., lysine decarboxylase (EC: 4.1.1.18), ornithine/lysine decarboxylase (EC: 4.1.1.116), arginine decarboxylase (EC: 4.1.1.19), agmatinase (EC: 3.5.3.11), spermidine synthase (EC: 2.5.1.16), arginase (EC: 3.5.3.1), ornithine decarboxylase (EC: 4.1.1.17), histidine decarboxylase (EC: 4.1.1.22), tyrosine decarboxylase (EC: 4.1.1.25), and tryptophan decarboxylase (EC: 4.1.1.28) were not found in the 9D3 genome. Therefore, food produced using this strain is safe from biogenic amine accumulation.
AMR property
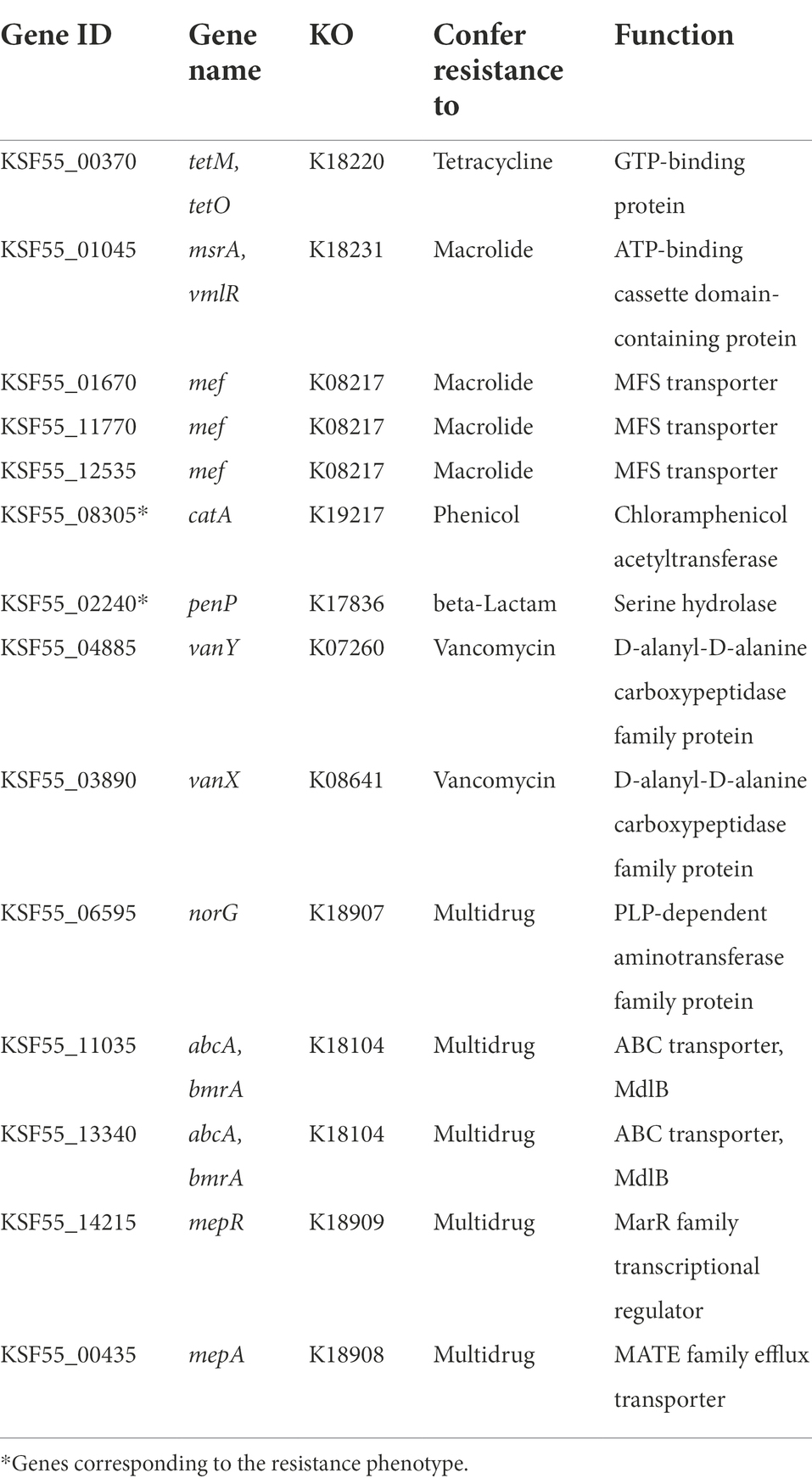
According to the in vitro antimicrobial susceptibility test, the 9D3 strain showed resistance to two antimicrobials, ampicillin and chloramphenicol, indicating acquired resistance to these two drugs, as shown in Table 2. The search for the genetic determinants responsible for the Ampr Chlr phenotype by sequence similarity search using the EFSA (2021) recommended a cut-off value of 80% identity, and 80% coverage yielded no predicted AMR genes from the CARD database (all hits are shown in Supplementary Table S7). Further similarity search using the ResFinder database with the default parameters (90% identity, 60% coverage) also returned no-hit for acquired AMR genes. However, the Hidden-Markov model profile-based KO annotation and KEGG mapping analysis identified 14 possible AMR genes, as listed in Table 3. The beta-lactamase and chloramphenicol acetyltransferase genes were identified as the genetic determinants conferring the resistance phenotype to ampicillin and chloramphenicol, respectively. To determine the transferability of these AMR genes, the locations of all 14 putative AMR genes were compared to those of the mobile elements, plasmids and prophages, in the 9D3 genome. We focused on these two mobile elements since they pose a more immediate risk for horizontal gene transfer through conjugation/transformation and transduction, whereas other mobile elements such as transposons translocate genes within the genome and require additional steps/vehicles before the genes can be transferred to other organisms.
Considering the locations of the AMR genes, it could be concluded that the genes responsible for the Ampr Chlr phenotype of 9D3 have a low risk of transfer since neither gene is associated with plasmids or prophages. Except for one, the rest of the putative AMR genes were not associated with any of the mobile elements of concern. The remaining AMR gene, KSF55_11035 (located at 2,443,645 bp - 2,445,408 bp), was found within an intact prophage region, raising concern about the possibility of transfer. From a similarity search using the GenBank database, the translated amino acids from this gene had 100% identity to an ABC transporter ATP-binding protein/permease, multidrug resistance-like ATP-binding protein (MdlB) of L. pentosus (accession number: WP_152705378). Although the gene was identified as a putative drug transporter, it was not confirmed experimentally. In contrast, a previous study reported that an E. coli strain transformed with a plasmid carrying the mdlB gene exhibited no increase in the resistance phenotype (Nishino and Yamaguchi, 2001). Since the strain showed no resistance to other antimicrobials except for the Ampr Chlr phenotype and the search using other AMR databases did not identify this gene as AMR associated with pathogens, we concluded that this transport gene might have other functions not related to AMR. Therefore, we concluded that the chance of transferring these AMR determinants to other organisms is low even though the strain was resistant to ampicillin and chloramphenicol.
Antimicrobial drug biosynthesis
Microbial culture used in food and feed should not produce any antimicrobial drug of clinical importance. This requirement aims to avoid the creation of a selective environment for the emergence of new AMR strains (Pariza et al., 2015). The biosynthesis pathways for critically important antimicrobial drugs, as identified in the World Health Organization’s complete list of critically important antimicrobials (WHO CIA list; WHO, 2019), were assessed in silico. No complete pathways for any of the CIAs were found in the 9D3 genome. A detail of the pathways analyzed and search results are shown in Supplementary Table S8. Historically, the species L. pentosus has never been noted for its ability to produce antimicrobial drugs of medical importance. Therefore, we concluded that L. pentosus 9D3 poses no safety concern in this regard.
Evaluating the probiotic potential of Lactiplantibacillus pentosus 9D3
To exert its health benefit and function as a probiotic, the bacterium should be able to survive and remain within the host’s target site, i.e., the GI tract in the case of probiotics intentionally add to the food system. The potential of L. pentosus 9D3 to be used as a probiotic for food was assessed by its ability to survive during transit through a GI tract and its ability to adhere to intestinal epithelial cell line Caco-2.
Survival of Lactiplantibacillus pentosus 9D3 in the GI tract conditions
This in vitro study tested the ability of L. pentosus 9D3 to survive conditions mimicking the environment the bacterium is expected to encounter, from the oral phase to the intestinal phase. L. pentosus 9D3 survived well at the oral digestion stage, which contains α-amylase enzymes, and the viable counts remained high at 9.37 ± 0.03 log CFU/ml compared with pre-digestion (9.38 ± 0.04 log CFU/ml; Table 4). The strain also exhibited good survivability in the hostile stomach condition, i.e., pH 2.0 in the presence of digestive enzyme pepsin for 1 h, with approximately 1 log reduction. In the intestinal phase, where the acid-stressed cells encountered several digestive enzymes and bile salts as well as a slightly acidic condition at pH 6.0, the 9D3 strain showed moderate survivability, with approximately 2.4 log reduction. This result was in concert with an earlier study in which a similar log reduction among L. pentosus strains, 0.5–2 log in simulated gastric juice, and 1–3 log in simulated intestinal fluid were reported (Guantario et al., 2018). This response was comparable to that of the popularly used probiotic strain L. rhamnosus GG.
Adhesion ability of Lactiplantibacillus pentosus 9D3
Another important property of a probiotic is the ability to adhere to the intestinal mucosa. This property is important because it prevents the rapid elimination of probiotic cells by peristalsis and thus represents an ecologically competitive advantage in the ecosystem of the GI tract. A simple in vitro intestinal tissue model Caco-2 was used in this study. The cells were grown in culture to form a homogeneous polar monolayer of mature enterocytes resembling the tissue of the small intestine. After going through in vitro digestion, bacterial cells were used for the adhesion experiment to resemble the actual conditions the bacterial cells may encounter in vivo. L. pentosus 9D3 showed good adhesion ability, in which ca. 50% of the bacterial cells were attached to the Caco-2. This adhesion ability is comparable to the positive control L. rhamnosus GG (Table 5). The attachment ability was shown to be strain-specific and can be varied widely. The adhesion ability of L. pentosus strains at comparable levels to that of L. rhamnosus GG was also reported in the study of Argyri et al. (2013).
Overall, L. pentosus 9D3 exhibited the basic requirement properties of a probiotic, i.e., capable to survive the digestive condition and attachment to the intestinal epithelial. However, according to the Guidelines for the Evaluation of Probiotics in Food (FAO/WHO, 2002), the health benefit of the strain should be identified in vivo. Therefore, the benefit to the host after administration of this strain is the subject for further investigation.
Conclusion
In the present study, L. pentosus 9D3, a GABA-producing strain isolated from Thai pickled weed was evaluated for its safety and probiotic properties. Through WGS analysis, L. pentosus 9D3 was shown to be safe and free from functional and transferable AMR genes. The genome contains no genes related to the production of toxins, biogenic amines, and antimicrobial drugs of clinical importance. In addition, the genetic determinants conferring the GABA-producing capability of the strain were identified. A precaution about the acquired nature of this property is noted. The probiotic potential of the strain was shown by its good survivability in the simulated human GI tract conditions and good adhesion ability. The results can be used to support further investigation to identify the strain and the food it produces as probiotic products.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/739022, NCBI-SRA repository under the BioProject accession number: PRJNA739022.
Author contributions
NC and NR conceptualization. NC, PS, WM, and NR methodology. CS and WV resources. NC, WM, and NR data curation. NC, PS, and NR visualization. NC and NR writing original draft preparation. NC, CS, and NR writing review and editing. CS project administration. WV and CS funding acquisition. NR and NC supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by The Agricultural Research Development Agency (ARDA) of Thailand (grant number: 2556NRCT512244); Institute of Nutrition, Mahidol University; the National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Thailand (Project Code P-17-52209).
Acknowledgments
The authors are grateful to Institute of Nutrition, Mahidol University and the National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Thailand for financial support, computing facilities, and resources for the laboratory. The authors would like to thank Miss Kanyawee Whanmek and Miss Kemika Praengam for their technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.969548/full#supplementary-material
Footnotes
References
Abriouel, H., Pérez Montoro, B., Casado Muñoz, M. D. C., Knapp, C. W., Gálvez, A., and Benomar, N. (2017a). In silico genomic insights into aspects of food safety and defense mechanisms of a potentially probiotic Lactobacillus pentosus MP-10 isolated from brines of naturally fermented Aloreña green table olives. PLoS One 12:e0176801. doi: 10.1371/journal.pone.0176801
Abriouel, H., Pérez Montoro, B., Casimiro-Soriguer, C. S., Pérez Pulido, A. J., Knapp, C. W., Caballero Gómez, N., et al. (2017b). Insight into potential probiotic markers predicted in Lactobacillus pentosus MP-10 genome sequence. Front. Microbiol. 8:891. doi: 10.3389/fmicb.2017.00891
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Anukam, K. C., Macklaim, J. M., Gloor, G. B., Reid, G., Boekhorst, J., Renckens, B., et al. (2013). Genome sequence of Lactobacillus pentosus KCA1: vaginal isolate from a healthy premenopausal woman. PLoS One 8:e59239. doi: 10.1371/journal.pone.0059239
Argyri, A. A., Zoumpopoulou, G., Karatzas, K. A., Tsakalidou, E., Nychas, G. J., Panagou, E. Z., et al. (2013). Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 33, 282–291. doi: 10.1016/j.fm.2012.10.005
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Arredondo-Alonso, S., Rogers, M. R. C., Braat, J. C., Verschuuren, T. D., Top, J., Corander, J., et al. (2018). Mlplasmids: a user-friendly tool to predict plasmid-and chromosome-derived sequences for single species. Microb. Genom. 4:e000224. doi: 10.1099/mgen.0.000224
Begley, M., Hill, C., and Gahan, C. G. (2006). Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006
Bendali, F., Kerdouche, K., Hamma-Faradji, S., and Drider, D. (2017). In vitro and in vivo cholesterol lowering ability of Lactobacillus pentosus KF923750. Benef. Microbes 8, 271–280. doi: 10.3920/bm2016.0121
Binnewies, T. T., Hallin, P. F., Stærfeldt, H. H., and Ussery, D. W. (2005). Genome update: proteome comparisons. Microbiology 151, 1–4. doi: 10.1099/mic.0.27760-0
Calero-Delgado, B., Martín-Platero, A. M., Pérez-Pulido, A. J., Benítez-Cabello, A., Casimiro-Soriguer, C. S., Martínez-Bueno, M., et al. (2018). Draft genome sequences of six Lactobacillus pentosus strains isolated from brines of traditionally fermented Spanish-style green table olives. Genome Announc. 6:e00379-18. doi: 10.1128/genomeA.00379-18
Chitchumroonchokchai, C., Schwartz, S. J., and Failla, M. L. (2004). Assessment of lutein bioavailability from meals and a supplement using simulated digestion and Caco-2 human intestinal cells. J. Nutr. 134, 2280–2286. doi: 10.1093/jn/134.9.2280
Chokesajjawatee, N., Santiyanont, P., Chantarasakha, K., Kocharin, K., Thammarongtham, C., Lertampaiporn, S., et al. (2020). Safety assessment of a Nham starter culture Lactobacillus plantarum BCC9546 via whole-genome analysis. Sci. Rep. 10:10241. doi: 10.1038/s41598-020-66857-2
Dashkevicz, M. P., and Feighner, S. D. (1989). Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 55, 11–16. doi: 10.1128/aem.55.1.11-16.1989
EFSA (2021). EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 19:e06506. doi: 10.2903/j.efsa.2021.6506
EFSA FEEDAP (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206. doi: 10.2903/j.efsa.2018.5206
FAO/WHO (2002). “Guidelines for the Evaluation of Probiotics in Food ”. (London, Ontario: Food and Agriculture Organization of the United Nations/World Health Organization).
Florensa, A. F., Kaas, R. S., Clausen, P., Aytan-Aktug, D., and Aarestrup, F. M. (2022). ResFinder - an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 8:000748. doi: 10.1099/mgen.0.000748
Garrett, D. A., Failla, M. L., and Sarama, R. J. (1999). Development of an in vitro digestion method to assess carotenoid bioavailability from meals. J. Agric. Food Chem. 47, 4301–4309. doi: 10.1021/jf9903298
Gosset, G. (2005). Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Factories 4:14. doi: 10.1186/1475-2859-4-14
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Guantario, B., Zinno, P., Schifano, E., Roselli, M., Perozzi, G., Palleschi, C., et al. (2018). In vitro and in vivo selection of potentially probiotic lactobacilli from Nocellara del Belice table olives. Front. Microbiol. 9:595. doi: 10.3389/fmicb.2018.00595
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hajela, N., Ramakrishna, B. S., Nair, G. B., Abraham, P., Gopalan, S., and Ganguly, N. K. (2015). Gut microbiome, gut function, and probiotics: implications for health. Indian J. Gastroenterol. 34, 93–107. doi: 10.1007/s12664-015-0547-6
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by egg NOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
International Organization for Standardization (ISO) and International Dairy Federation (IDF) (2010). Milk and milk products-Determination of the minimal inhibitory concentration (MIC) sof antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB) (ISO 10932|IDF 223). Retrieved from https://www.iso.org/standard/46434.html
Izumo, T., Maekawa, T., Ida, M., Noguchi, A., Kitagawa, Y., Shibata, H., et al. (2010). Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int. Immunopharmacol. 10, 1101–1106. doi: 10.1016/j.intimp.2010.06.012
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Biol. 16, 111–120. doi: 10.1007/bf01731581
Kittibunchakul, S., Yuthaworawit, N., Whanmek, K., Suttisansanee, U., and Santivarangkna, C. (2021). Health beneficial properties of a novel plant-based probiotic drink produced by fermentation of brown rice milk with GABA-producing Lactobacillus pentosus isolated from Thai pickled weed. J. Funct. Foods 86:104710. doi: 10.1016/j.jff.2021.104710
Kotani, Y., Shinkai, S., Okamatsu, H., Toba, M., Ogawa, K., Yoshida, H., et al. (2010). Oral intake of Lactobacillus pentosus strain b240 accelerates salivary immunoglobulin a secretion in the elderly: a randomized, placebo-controlled, double-blind trial. Immun. Ageing 7:11. doi: 10.1186/1742-4933-7-11
Krawczyk, P. S., Lipinski, L., and Dziembowski, A. (2018). PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 46:e35. doi: 10.1093/nar/gkx1321
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
McCoy, J. G., Levin, E. J., and Zhou, M. (2015). Structural insight into the PTS sugar transporter EIIC. Biochim. Biophys. Acta 1850, 577–585. doi: 10.1016/j.bbagen.2014.03.013
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Moser, S. A., and Savage, D. C. (2001). Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67, 3476–3480. doi: 10.1128/AEM.67.8.3476-3480.2001
Nakatani, Y., Fukao, M., and Fukaya, T. (2019). Genome sequence of lactobacillus plantarum KB1253, a gamma-aminobutyric acid (GABA) producer used in GABA-enriched tomato juice production. Microbiol. Resour. Announc. 8:e00158-19. doi: 10.1128/mra.00158-19
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001
Pariza, M. W., Gillies, K. O., Kraak-Ripple, S. F., Leyer, G., and Smith, A. B. (2015). Determining the safety of microbial cultures for consumption by humans and animals. Regul. Toxicol. Pharmacol. 73, 164–171. doi: 10.1016/j.yrtph.2015.07.003
Patterson, E., Ryan, P. M., Wiley, N., Carafa, I., Sherwin, E., Moloney, G., et al. (2019). Gamma-aminobutyric acid-producing lactobacilli positively affect metabolism and depressive-like behaviour in a mouse model of metabolic syndrome. Sci. Rep. 9:16323. doi: 10.1038/s41598-019-51781-x
Pei, Z., Sadiq, F. A., Han, X., Zhao, J., Zhang, H., Ross, R. P., et al. (2021). Comprehensive scanning of prophages in Lactobacillus: distribution, diversity, antibiotic resistance genes, and linkages with CRISPR-Cas systems. mSystems 6:e0121120. doi: 10.1128/mSystems.01211-20
Phuengjayaem, S., Booncharoen, A., and Tanasupawat, S. (2021). Characterization and comparative genomic analysis of gamma-aminobutyric acid (GABA)-producing lactic acid bacteria from Thai fermented foods. Biotechnol. Lett. 43, 1637–1648. doi: 10.1007/s10529-021-03140-y
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sato, K., Komaru, T., Arima, T., Jardson, C., Yanaka, N., and Kumrungsee, T. (2021). Dietary GABA and its combination with vigabatrin mimic calorie restriction and induce antiobesity-like effects in lean mice. J. Funct. Foods 78:104367. doi: 10.1016/j.jff.2021.104367
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling from NGS Reads. Available at: https://github.com/tseemann/snippy (Accessed June 15, 2021).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Soltani, N., Qiu, H., Aleksic, M., Glinka, Y., Zhao, F., Liu, R., et al. (2011). GABA exerts protective and regenerative effects on islet beta cells and reverses diabetes. Proc. Natl. Acad. Sci. U. S. A. 108, 11692–11697. doi: 10.1073/pnas.1102715108
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239. doi: 10.1093/molbev/msz312
Stergiou, O. S., Tegopoulos, K., Kiousi, D. E., Tsifintaris, M., Papageorgiou, A. C., Tassou, C. C., et al. (2021). Whole-genome sequencing, phylogenetic and genomic analysis of Lactiplantibacillus pentosus L33, a potential probiotic strain isolated from fermented sausages. Front. Microbiol. 12:746659. doi: 10.3389/fmicb.2021.746659
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thakkar, S. K., Maziya-Dixon, B., Dixon, A. G. O., and Failla, M. L. (2007). β-Carotene micellarization during in vitro digestion and uptake by Caco-2 cells is directly proportional to β-carotene content in different genotypes of cassava. J. Nutr. 137, 2229–2233. doi: 10.1093/jn/137.10.2229
Törönen, P., Medlar, A., and Holm, L. (2018). PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 46, W84–W88. doi: 10.1093/nar/gky350
Vesth, T., Lagesen, K., Acar, Ö., and Ussery, D. (2013). CMG-biotools, a free workbench for basic comparative microbial genomics. PLoS One 8:e60120. doi: 10.1371/journal.pone.0060120
Wang, Y., Wu, J., Lv, M., Shao, Z., Hungwe, M., Wang, J., et al. (2021). Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 9:612285. doi: 10.3389/fbioe.2021.612285
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464. doi: 10.1099/00207713-37-4-463
WHO (2019). “Critically Important Antimicrobials for Human Medicine, 6th Revision ”. (Geneva: World Health Organization).
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Keywords: whole-genome sequence, Lactiplantibacillus pentosus, GABA-producing strain, safety evaluation, probiotic
Citation: Raethong N, Santivarangkna C, Visessanguan W, Santiyanont P, Mhuantong W and Chokesajjawatee N (2022) Whole-genome sequence analysis for evaluating the safety and probiotic potential of Lactiplantibacillus pentosus 9D3, a gamma-aminobutyric acid (GABA)-producing strain isolated from Thai pickled weed. Front. Microbiol. 13:969548. doi: 10.3389/fmicb.2022.969548
Edited by:
Xiaoyuan Wang, Jiangnan University, ChinaReviewed by:
Gabriela Nicoleta Tenea, Universidad Técnica del Norte, EcuadorAlex Galanis, Democritus University of Thrace, Greece
Hayriye Sebnem Harsa, Izmir Institute of Technology, Turkey
Copyright © 2022 Raethong, Santivarangkna, Visessanguan, Santiyanont, Mhuantong and Chokesajjawatee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nachon Raethong, bmFjaG9uLnJhZUBtYWhpZG9sLmFjLnRo; Nipa Chokesajjawatee, bmlwYS5jaG9AYmlvdGVjLm9yLnRo
 Nachon Raethong
Nachon Raethong Chalat Santivarangkna
Chalat Santivarangkna Wonnop Visessanguan
Wonnop Visessanguan Pannita Santiyanont
Pannita Santiyanont Wuttichai Mhuantong
Wuttichai Mhuantong Nipa Chokesajjawatee
Nipa Chokesajjawatee