- 1College of Animal Husbandry and Veterinary Medicine, Southwest University for Nationalities, Chengdu, China
- 2Animal Disease Prevention and Control Center of Aba Tibetan and Qiang Autonomous Prefecture, Markang, China
Tick-borne diseases have become a global health concern in recent decades. Spotted fever group (SFG) rickettsiae have been recognized as important pathogens of human tick-borne diseases worldwide. In this study, Dermacentor everestianus (n = 646) and Haemaphysalis qinghaiensis (n = 172) ticks were collected from yaks (Bos grunniens) in Shiqu county, eastern Tibetan Plateau, China. SFG rickettsiae were identified and characterized in these ticks. A total of 49.9% (408/818) ticks were infected by Rickettsia spp. with infection rates of 58.1% (100/172) and 46.7% (308/646) detected in H. qinghaiensis and D. everestianus ticks, respectively. Furthermore, 95% of Rickettsia spp. were Rickettsia raoultii-like bacteria, and 5% were related to Candidatus Rickettsia longicornii. To the best of our knowledge, this is the first time that SFG rickettsiae infections were firstly reported in Shiqu county for these tick species. Our results indicated that H. qinghaiensis and D. everestianus ticks from Shiqu county became highly infected with a R. raoultii-like bacteria during their feeding process. This observation is alarming because of the zoonotic potentiality of these species. Overall, the present study detected a widespread of R. raoultii-like bacteria in ticks that are considered a serious threat to domestic animals and humans in Shiqu county. The prevalence of R. raoultii-like bacteria in human and wildlife hosts should be further investigated in the future.
Introduction
Rickettsioses are important emerging or reemerging vector-borne diseases that are distributed worldwide and have increasingly challenged public health services in recent years. The significance of ticks (Acari: Ixodida) has long been recognized due to their ability to feed on a variety of host species and transmit rickettsial pathogens that can infect various vertebrate hosts, including humans. Members of the genus Rickettsia have been classified into five major groups, including the spotted fever group (SFG), the typhus group (TG), the transitional group (TRG), the Rickettsia bellii group and the Rickettsia canadensis group (Weinert et al., 2009; Merhej et al., 2014). In China, many SFG rickettsiae belong to Rickettsia sibirica group, including: (1) R. sibirica subsp. sibirica, a North Asian tick typhus pathogen, detected in Dermacentor silvarum and Dermacentor sinicus found in northern China (Yu et al., 1993); and (2) R. sibirica subsp. mongolitimonae, a lymphangitis-associated rickettsiosis pathogen, isolated from Hyalomma asiaticum from Inner Mongolia (Yu et al., 1993). Most recently, Zhang et al. (2006) have reported that the Rickettsia identified in Haemaphysalis flava from Cangxi county, Sichuan province, China, was most closely related to a member of the Candidatus Rickettsia gannanii subgroup identified in Haemaphysalis qinghaiensis (Yang et al., 2016). Other Rickettsia species causing SFG rickettsiosis have been discovered in China, including Rickettsia heilongjiangensis, R. sibirica, Rickettsia raoultii, Rickettsia slovaca, Rickettsia felis, Rickettsia aeschlimannii, and Rickettsia massiliae (Wei et al., 2015; Guo et al., 2016).
However, there are very few reports on the occurrence of SFG rickettsiae and their vectors in the eastern Tibetan Plateau especially in Shiqu county (average altitude of 4,200 m above sea level) (Wang et al., 2012). In Shiqu, yaks (Bos grunniens) are the largest population of local livestock (around 600,000 individuals) and are the main source of income for local residents, producing butter, milk and meat. Due to their traditional lifestyle, yaks live in close contact with local residents (especially yak farmers), and severe tick infestation has often been observed. Therefore, in this study, we aim to investigate the prevalence of Rickettsia spp. in ticks collected from yaks in Shiqu county and identify the degree of spread of these pathogens and their impact on livestock in the region, which would provide preliminary data for further human infection risk research.
Materials and methods
Sample collection and molecular identification
Ticks were collected from yaks in Derongma, Changxuganma, Arizha, and Maga villages in Shiqu county (35°58′ 50.77″N, 98°06′ 10.58″E) (Figure 1) from June to August 2018. Ticks were stored in 70% ethanol at 4°C. Approximately 5–6 ticks were collected from each yak. All tick samples were identified by morphological characteristics with standard taxonomic keys (Deng and Jiang, 1991) and further confirmed using PCR amplification targeting the small subunit 16S rRNA gene (Black and Piesman, 1994). Primer sequences are listed in Supplementary Table 1. Detailed information regarding tick collection is listed in Supplementary Table 2. In addition, 120 blood samples were collected from slaughtered yaks in an abattoir in November 2018.
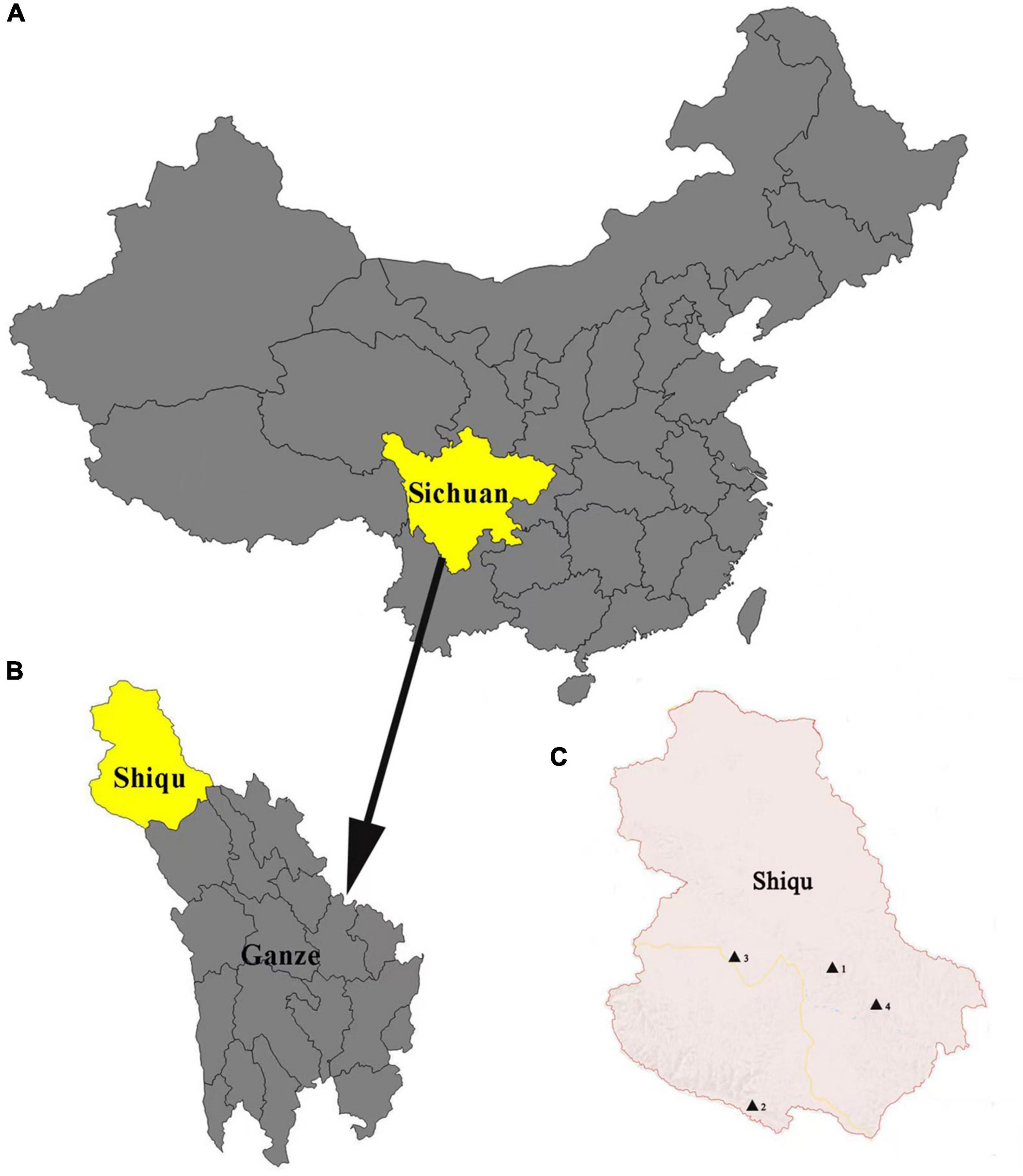
Figure 1. Map of Shiqu county. (A) Map of China where Sichuan province is marked in yellow. (B) Map of Ganze, Tibetan autonomous prefecture, where Shiqu county is marked in yellow. (C) Map of Shiqu country, where the four village locations in this study (1. Ariza, 2. Maga, 3. Derongma, and 4. Changxgma) are marked by black triangles.
DNA extraction and PCR amplification
All ticks were sectioned longitudinally and processed individually for DNA extraction using a TIANamp Genomic DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China, Cat. No. DP304) according to the manufacturer’s protocol. DNA of all blood samples was extracted with a Blood Genomic DNA Kit (Transgen Biotech Co., Ltd., Beijing, China, Cat. No. EE121-11) and stored at −20°C until analysis. PCR amplification was performed targeting the ompA (Oteo et al., 2006) and ompB genes (Fernandez de Mera et al., 2013) according to previously published criteria (Raoult et al., 2005). Primer sequences are listed in Supplementary Table 1. PCR was performed in a total reaction volume of 25 μl, including 2 μl template DNA, 12.5 μl 2 × PCR mix (TransGen Biotech Co., Ltd., Beijing, China, Cat. No. AS111), and 20 pmol of each primer (Sangon Biotech Co., Ltd., Shanghai, China). After an initial denaturation for 3 min at 95°C, 40 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55°C and elongation for 30 s at 72°C were performed, followed by a final extension step at 72°C for 7 min. The final amplification products were sent for sequencing (Sangon Biotech Co., Ltd., Shanghai, China).
Bioinformatics/phylogenetic analysis
Firstly, the obtained sequences were analyzed and manually edited using the BioEdit program (Version 7.0.9). Then sequences were compared to each other using DNASTAR v.7.1.0. Subsequently, in order to assign them identities, each sequence underwent a homology comparison using the previously described BLAST tool in order to compare with sequences deposited in GenBank (Altschul et al., 1990; Benson et al., 2002). Phylogenetic trees were constructed with Neighbor-Joining using MEGA 6 software based on the ompA and ompB genes, respectively. The evolutionary distance was computed using Kimura 2-parameter method with 1,000 bootstrap replicates. The phylogenetic tree was drawn to scale and branch lengths were measured as the number of substitutions per site.
Statistics
A Pearson Chi-square test using SPSS (ver. 19.0, IBM, New York, NY, United States) was applied to compare the prevalence of Rickettsia spp. among the different sampling locations and tick species. Results with P < 0.05 were considered significant.
Results
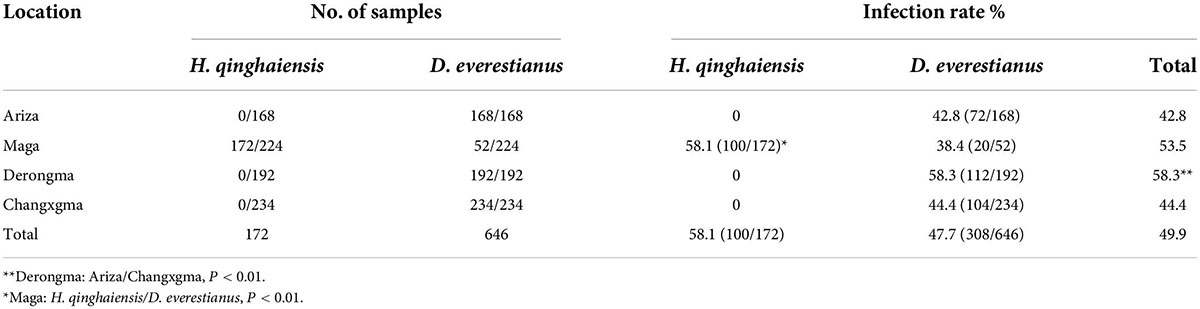
In total, 818 adult ticks (Changxuganma, n = 234; Maga, n = 224; Derongma, n = 192; and Arizha, n = 168) were collected and identified. Molecular identification of ticks based on the mitochondrial subunit 16S rRNA gene confirmed the presence of two different tick species belonging to Dermacentor everestianus (n = 646) and H. qinghaiensis (n = 172). Ticks were first screened using the Rickettsia spp. ompA gene. ompA-positive samples were then screened for the ompB gene. According to the defined criteria for Rickettsia species, 49.9% of the samples (408/818) were identified as SFG rickettsiae (Table 1). However, all yak blood samples were negative.
Based on the analysis of ompA and ompB amplicons, we discovered 7 unique sequences from the 408 positively identified samples and they were submitted to GenBank with accession numbers as follows: MT361017, MT361018, MT361019, and MT361020 for the ompA gene; MT361021, MT361022, and MT361023 for the ompB gene. Sequence analysis of the ompA gene indicated that the MT361017, MT361018, and MT361019 sequences were closely related to R. raoultii (JQ792162 and JQ792137) with a sequence identity of 99.8–100%. Sequence MT361020 was 98.2–100% identical to Candidatus R. longicornii (MG906676) and an uncultured Rickettsia sp. isolated from Korea and Qinghai (MG228270), respectively. For the ompB gene, sequences MT361021 and MT361022 were 99.3–99.5% identical to R. raoultii (MH532272 and DQ365798). Sequence MT361023 showed 100% identity to Rickettsia sp. (KC888953) and Candidatus R. longicornii (MG906675) isolated from Korea. Furthermore, according to defined threshold identity for the SFG rickettsiae species (ompA ≥ 98.8% and ompB ≥ 99.2%) (Raoult et al., 2005), ompA and ompB-based phylogenetic analysis supported the classification and detection of Rickettsia spp. in this study (Figure 2). The sequences are available in the Supplementary Material.
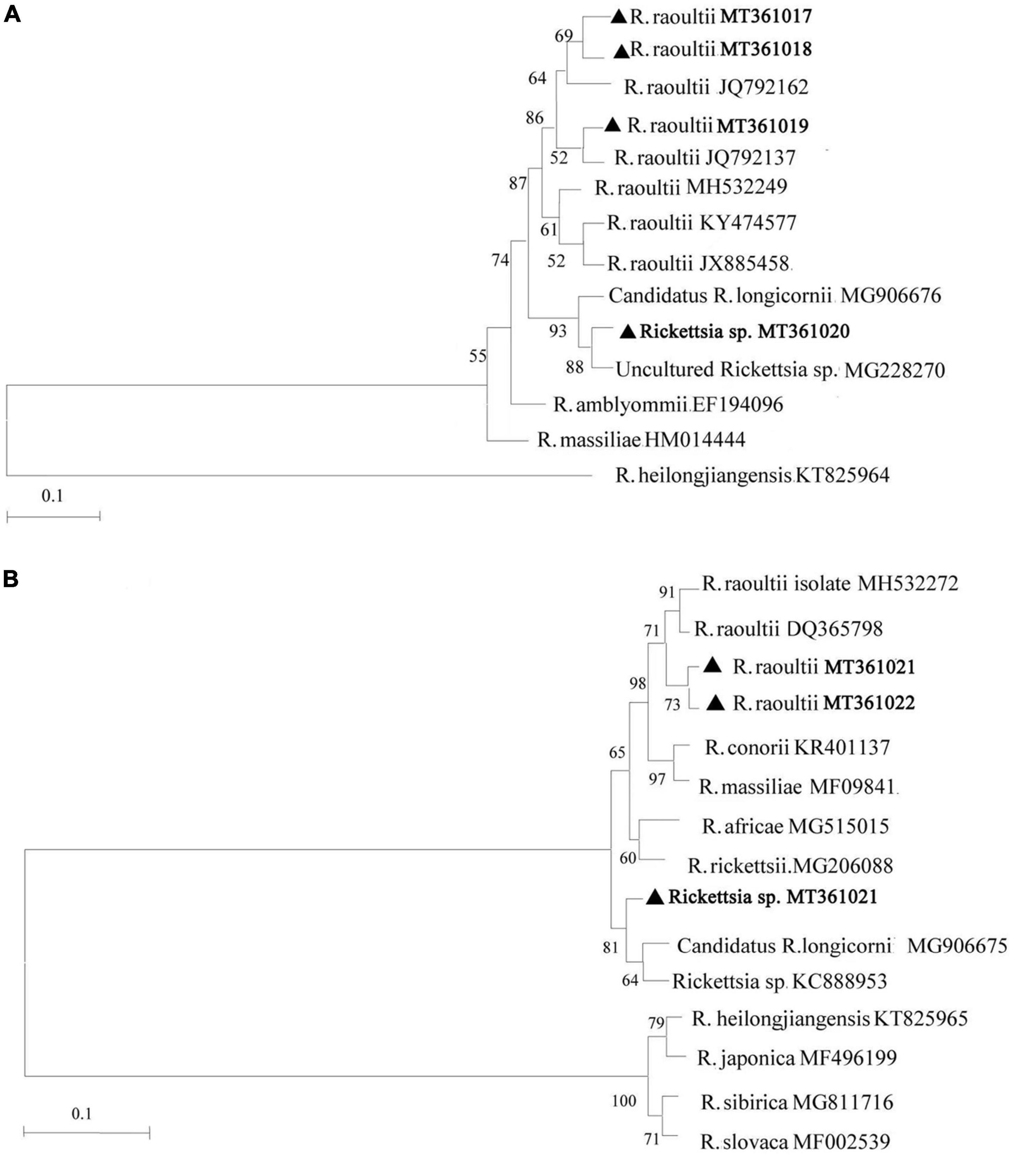
Figure 2. Phylogenetic analysis of Rickettsia spp. identified in Shiqu county. Neighbor-joining (NJ) phylogenetic trees were constructed based on Rickettsia ompA (A) and ompB (B), respectively. The sequences obtained in this study are marked with black triangles.
Rickettsia raoultii-like bacteria were positively identified in 95% of cases, and Rickettsia spp., accounting for 5% of cases was only detected in H. qinghaiensis ticks. The infection rate of R. raoultii-like bacteria in Derongma village was 58.3%, significantly higher than in Ariza and Changxuganma villages (CI = 95%, P < 0.01, Table 1). In Maga village, the infection rate (58.1%) in H. qinghaiensis was significantly higher than D. everestianus (38.4%; CI = 95%, P < 0.01, Table 1). Overall tick infection rates in Shiqu county with Rickettsia spp. were 58.1% in H. qinghaiensis and 46.7% in D. everestianus, respectively.
Discussion
Two tick species (H. qinghaiensis and D. everestianus) infesting yaks from Shiqu county were identified. H. qinghaiensis was only identified in yak from Maga village, whereas D. everestianus has earlier mainly been found in all four villages. Previously D. everestianus was only reported in northwestern China and Nepal (Chen et al., 2014) at an altitude of 2,600–4,700 m (Apanaskevich et al., 2014). Larvae and nymphs from this tick species often infect lagomorphs and rodents, while adult ticks usually infect medium-size domestic and wild mammals such as hare (Lepus oiostolus), sheep, yaks, and horses (Apanaskevich et al., 2014; Chen et al., 2014). However, H. qinghaiensis has only been recorded in China (Gao et al., 2007a,b, 2008, 2009; Li et al., 2007), and is particularly prevalent in Qinghai, Gansu, Sichuan, and Tibet provinces (Weinert et al., 2009). H. qinghaiensis are usually more active at low than high altitudes. In this study, Arizha, Changxuganma, and Derongma villages belong to a sub-frigid zone, with an altitude between 4,300–4,600 m, while Maga village is located in a cold temperate zone, with an altitude of 3,799 m. There are significant differences in altitude between Maga and the other three villages, which is probably the reason H. qinghaiensis was only found in Maga.
Rickettsia raoultii was first isolated from ticks in Russia in 1999 (Rydkina et al., 1999) and has been found mainly in Dermacentor spp. ticks in several countries in Europe (Speck et al., 2012; Foldvari et al., 2013). Human infections of R. raoultii have been reported in recent years. In 2012, R. raoultii DNA was detected in two patients from Mudanjiang, China (Jia et al., 2014). In 2015 and 2016, 26 cases of humans infected of R. raoultii were reported in Inner Mongolia (Jia et al., 2014; Li et al., 2018), and in Henan and Shandong provinces, China (Li et al., 2018). The Khabarovsk R. raoultii strain was isolated from the blood of a patient (Li et al., 2018), which showed high genetic identity with the R. raoultii-like bacteria identified in our study.
Rickettsia raoultii has been found in at least 26 tick species belonging to 7 genera, including Dermacentor (Jia et al., 2014), Haemaphysalis (Li et al., 2018; Zheng et al., 2018), Amblyomma (Parola et al., 2013), Ixodes (Shpynov et al., 2009; Rar et al., 2017), and Hyalomma (Yin et al., 2018), although Dermacentor spp. ticks are the main vector (Mediannikov et al., 2008; Wang et al., 2012). In China, R. raoultii-like bacteria were first detected in Tibet (Wang et al., 2012). In this study, infection rates of R. raoultii-like bacteria in D. everestianus and H. qinghaiensis were 47.6 and 58.1%, respectively, suggesting that these tick species may be possible vectors for R. raoultii in Shiqu county, although Haemaphysalis species including Haemaphysalis longicornis, Haemaphysalis erinacei, and Haemaphysalis verticalis, may also be efficient vectors of R. raoultii.
Our study did not find Rickettsia spp. DNA in yak blood samples (n = 120). Because of local religious customs regarding yaks, it was impossible to obtain blood samples from live animals without the permission of their owners. Instead, all blood samples were collected from slaughtered yaks in an abattoir in November 2018 without Rickettsia spp. DNA detected. In order to obtain more detailed prevalence information of Rickettsia spp. in ticks, tick samples should be collected from other hosts, including sheep, horse, rodents, and plateau pika (Ochotona curzoniae). It would be much more important to collect unfed questing ticks from the vegetation and investigate their infection rates. In Shiqu county, there are about 1.3 billion plateau pikas and other rodents (Eospalax fontanierii, Lasiopodomys fuscus, etc.), which represent the largest mammalian populations in the area. These small mammals are natural hosts of D. everestianus larvae and nymphs and might play important roles in the natural transmission cycle and dispersal of Rickettsia spp. Future studies could also investigate Rickettsia spp. in spleen and/or blood samples from these small mammals. Also, it would be much more important to investigate their reservoir competence for the involved Rickettsia species. Alternatively, seasonal infection risk for human and animals could be estimated by dragging blankets across grasslands to capture and calculate the numbers of unfed ticks in a given area. At present, no reports were found of SFG rickettsiae (identified in H. qinghaiensis and D. everestianus) linked to human cases from the Tibet Plateau. Due to their traditional lifestyle, local people (and particularly herdsmen) do not tend to go to hospital unless they have serious medical issues, resulting in very few reported or recorded clinical cases. However, in the future, we should pay close attention to human tick bite cases in Shiqu.
Conclusion
In summary, we have shown, for the first time high prevalences of Rickettsia spp. in D. everestianus and H. qinghaiensis ticks sampled from yaks in Shiqu county. In this region, key mammalian tick hosts are domesticated yak and wild mammals such as rodents and plateau pika. A more comprehensive study of Rickettsial pathogens to further assess the prevalence of Rickettsia spp. in other livestock and wildlife hosts from Shiqu county should be made in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT361017, MT361018, MT361019, MT361020, MT361021, MT361022, and MT361023.
Author contributions
LH conceptualized and designed the experiments and wrote the manuscript. BL and YT performed the experiments and analyzed the data. BL analyzed the data. All authors have reviewed and agreed with the manuscript.
Funding
This project was supported by Fundamental Research Funds for the Central Universities, Southwest Minzu University (ZYN2022001).
Acknowledgments
We thank colleagues at Center for Animal Disease Control and Prevention in Sichuan Province, Chengdu, China for their help with sample collection. We thank the Dr. Cheng Du (HVRI) for careful editing and proofing the manuscript. We are also grateful to the editors and reviewers for their invaluable comments to improve this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.968793/full#supplementary-material
Supplementary Table 1 | Primer sequences used for tick and Rickettsia spp. identification.
Supplementary Table 2 | Tick sampling information.
Supplementary Text 1 | ompA gene sequence.
Supplementary Text 2 | ompB gene sequence.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignmentsearch tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Apanaskevich, D. A., Duan, W., Apanaskevich, M. A., Filippova, N. A., and Chen, J. (2014). Redescription of Dermacentor everestianus Hirst (Acari: ixodidae), a parasite of mammals in mountains of China and Nepal with synonymization of D. abaensis Teng and D. birulai Olenev. J. Parasitol. 100, 268–278. doi: 10.1645/13-369.1
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A., and Wheeler, D. L. (2002). GenBank. Nucleic Acids Res. 30, 17–20. doi: 10.1093/nar/30.1.17
Black, W. C. T., and Piesman, J. (1994). Phylogeny of hard- and soft-tick taxa (Acari: ixodida) based onmitochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 91, 10034–10038. doi: 10.1073/pnas.91.21.10034
Chen, Z., Li, Y., Ren, Q., Luo, J., Liu, Z., Zhou, X., et al. (2014). Dermacentor everestianus Hirst,1926(Acari: ixodidae): phylogenetic status inferred from molecular characteristics. Parasitol. Res. 113, 3773–3779. doi: 10.1007/s00436-014-4043-1
Deng, G. F., and Jiang, Z. J. (1991). Economic Insect Fauna of China. Ixodes [M]. Beijing: Science Press.
Fernandez de Mera, I. G., Ruiz-Fons, F., de la Fuente, G., Mangold, A. J., Gortazar, C., and de la Fuente, J. (2013). Spotted fever group rickettsiae in questing ticks, central Spain. Emerg. Infect. Dis. 19, 1163–1165. doi: 10.1016/j.ttbdis.2017.10.001
Foldvari, G., Rigo, K., and Lakos, A. (2013). Transmission of Rickettsia slovaca and Rickettsia raoultii by male Dermacentor marginatus and Dermacentor reticulatus ticks to humans. Diagn. Microbiol. Infect. Dis. 76, 387–389. doi: 10.1016/j.diagmicrobio.2013.03.005
Gao, J., Luo, J., Fan, R., Fingerle, V., Guan, G., Liu, Z., et al. (2008). Cloning and characterization of a cDNA clone encoding calreticulin from Haemaphysalis qinghaiensis (Acari: ixodidae). Parasitol. Res. 102, 737–746. doi: 10.1007/s00436-007-0826-y
Gao, J., Luo, J., Fan, R., Guan, G., Ren, Q., Ma, M., et al. (2007a). Molecular characterization of a myosin alkali light chain-like protein, a “concealed” antigen from the hard tick Haemaphysalis qinghaiensis. Vet. Parasitol. 147, 140–149. doi: 10.1016/j.vetpar.2007.03.007
Gao, J., Luo, J., Li, Y., Fan, R., Zhao, H., Guan, G., et al. (2007b). Cloning and characterization of a ribosomal protein L23a from Haemaphysalis qinghaiensis eggs by immuno screening of a cDNA expression library. Exp. Appl. Acarol. 41, 289–303. doi: 10.1007/s10493-007-9065-2
Gao, J., Luo, J., Fan, R., Schulte-Spechtel, U. C., Fingerle, V., Guan, G., et al. (2009). Characterization of a concealed antigen Hq05 from the hard tick Haemaphysalis qinghaiensis and its effect as a vaccine against tick infestation in sheep. Vaccine 27, 483–490. doi: 10.1016/j.vaccine.2008.10.067
Guo, L. P., Jiang, S. H., Liu, D., Wang, S. W., Chen, C. F., and Wang, Y. Z. (2016). Emerging spotted fever group rickettsiae in ticks, northwestern China. Ticks Tick Borne Dis. 7, 1146–1150. doi: 10.1016/j.ttbdis.2016.08.006
Jia, N., Zheng, Y. C., Ma, L., Huo, Q. B., Ni, X. B., Jiang, B. G., et al. (2014). Human infections withRickettsia raoultii China. Emerg. Infect. Dis. 20, 866–868. doi: 10.3201/eid2005.130995
Li, H., Zhang, P. H., Huang, Y., Du, J., Cui, N., Yang, Z. D., et al. (2018). Isolation and Identification of Rickettsia raoultii in Human Cases: a Surveillance Study in 3 Medical Centers in China. Clin. Infect. Dis. 66, 1109–1115. doi: 10.1093/cid/cix917
Li, Y., Luo, J., Liu, Z., Guan, G., Gao, J., Ma, M., et al. (2007). Experimental transmission of Theileria sp. (China 1) infective for small ruminants by Haemaphysalis longicornis and Haemaphysalis qinghaiensis. Parasitol. Res. 101, 533–538. doi: 10.1007/s00436-007-0509-8
Mediannikov, O., Matsumoto, K., Samoylenko, I., Drancourt, M., Roux, V., Rydkina, E., et al. (2008). Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int. J. Syst. Evol. Microbiol. 58, 1635–1639. doi: 10.1099/ijs.0.64952-0
Merhej, V., Angelakis, E., Socolovschi, C., and Raoult, D. (2014). Genotyping, evolution and epidemiological findings of Rickettsia species. Infect. Genet. Evol. 25, 122–137. doi: 10.1016/j.meegid.2014.03.014
Oteo, J. A., Portillo, A., Santibanez, S., Blanco, J. R., Perez-Martinez, L., and Ibarra, V. (2006). Cluster of cases of human Rickettsia felis infection from Southern Europe (Spain) diagnosed by PCR. J. Clin. Microbiol. 44, 2669–2671. doi: 10.1128/JCM.00366-06
Parola, P., Paddock, C. D., Socolovschi, C., Labruna, M. B., Mediannikov, O., Kernif, T., et al. (2013). Update on tick-borne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev. 26, 657–702. doi: 10.1128/CMR.00032-13
Raoult, D., Fournier, P. E., Eremeeva, M., Graves, S., Kelly, P. J., Oteo, J. A., et al. (2005). Naming of Rickettsiae and rickettsial diseases. Ann. N. Y. Acad. Sci. 1063, 1–12. doi: 10.1196/annals.1355.002
Rar, V., Livanova, N., Tkachev, S., Kaverina, G., Tikunov, A., Sabitova, Y., et al. (2017). Detection and genetic characterization of a wide range of infectious agents in Ixodes pavlovskyi ticks in Western Siberia Russia. Parasit Vectors 10:258. doi: 10.1186/s13071-017-2186-5
Rydkina, E., Roux, V., Rudakov, N., Gafarova, M., Tarasevich, I., and Raoult, D. (1999). New Rickettsiae in ticks collected in territories of the former soviet union. Emerg. Infect. Dis. 5, 811–814. doi: 10.3201/eid0506.990612
Shpynov, S., Fournier, P. E., Rudakov, N., Arsen’eva, I., Granitov, M., Tarasevich, I., et al. (2009). Tick-borne rickettsiosis in the Altay region of Russia. Clin. Microbiol. Infect. 15, 313–314. doi: 10.1111/j.1469-0691.2008.02255.x
Speck, S., Derschum, H., Damdindorj, T., Dashdavaa, O., Jiang, J., Kaysser, P., et al. (2012). Rickettsia raoultii, the predominant Rickettsia found in Mongolian Dermacentor nuttalli. Ticks Tick Borne Dis. 3, 227–231. doi: 10.1016/j.ttbdis.2012.04.001
Wang, Y., Liu, Z., Yang, J., Chen, Z., Liu, J., Li, Y., et al. (2012). Rickettsia raoultii-like bacteria in Dermacentor spp. ticks. Tibet, China. Emerg. Infect. Dis. 18, 1532–1534. doi: 10.3201/eid1809.120644
Wei, Q. Q., Guo, L. P., Wang, A. D., Mu, L. M., Zhang, K., Chen, C. F., et al. (2015). The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasit Vectors 8:631. doi: 10.1186/s13071-015-1242-2
Weinert, L. A., Werren, J. H., Aebi, A., Stone, G. N., and Jiggins, F. M. (2009). Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6. doi: 10.1186/1741-7007-7-6
Yang, J., Tian, Z., Liu, Z., Niu, Q., Han, R., Li, Y., et al. (2016). Novel spotted fever group rickettsiae in Haemaphysalis qinghaiensis ticks from Gansu Northwest China. Parasit Vectors 9:146. doi: 10.1186/s13071-016-1423-7
Yin, X., Guo, S., Ding, C., Cao, M., Kawabata, H., Sato, K., et al. (2018). Spotted Fever Group Rickettsiae in Inner Mongolia. China, 2015-2016. Emerg. Infect. Dis. 24, 2105–2107. doi: 10.3201/eid2411.162094
Yu, X., Jin, Y., Fan, M., Xu, G., Liu, Q., and Raoult, D. (1993). Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J. Clin. Microbiol. 31, 83–88. doi: 10.1128/jcm.31.1.83-88.1993
Zhang, L., Jin, J., Fu, X., Raoult, D., and Fournier, P. E. (2006). Genetic differentiation of Chinese isolates of Rickettsia sibirica by partial ompA gene sequencing and multispacer typing. J. Clin. Microbiol. 44, 2465–2467. doi: 10.1128/JCM.02272-05
Keywords: SFG rickettsiae, ticks, Shiqu, high prevalence, yaks (Bos grunniens)
Citation: Lin B, Ta Y and Hao L (2022) High prevalence of spotted fever group rickettsiae in ticks collected from yaks (Bos grunniens) in Shiqu county, eastern Tibetan Plateau, China. Front. Microbiol. 13:968793. doi: 10.3389/fmicb.2022.968793
Received: 14 June 2022; Accepted: 04 July 2022;
Published: 28 July 2022.
Edited by:
Qiaocheng Chang, Shantou University, ChinaReviewed by:
Guo-Hua Liu, Hunan Agricultural University, ChinaNan Hou, Chinese Academy of Medical Sciences & Peking Union Medical College, China
Panagiotis Karanis, University of Nicosia, Cyprus
Copyright © 2022 Lin, Ta and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Hao, bGVlbGVlX2hhb0AxMjYuY29t
 Baoshan Lin
Baoshan Lin Yin Ta
Yin Ta Lili Hao
Lili Hao