- 1Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China
- 2Hainan Institute, Zhejiang University, Hangzhou, China
- 3Institute of Preventive Veterinary Sciences and Department of Veterinary Medicine, Zhejiang University College of Animal Sciences, Hangzhou, China
- 4Zhejiang Provincial Key Laboratory of Preventive Veterinary Medicine, Hangzhou, China
- 5State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Bacillus cereus is a major food-borne bacterial pathogen in the world, which can cause diarrhea and emetic syndrome. This study aimed to reveal the quantitative prevalence of B. cereus in ready-to-eat (RTE) rice products in Eastern China and to gain essential information on the characteristics of B. cereus isolates. A total of 91 out of the 1071 samples were positive for B. cereus. The contamination level of B. cereus in 0.5 % of RTE rice product samples outnumbered 103 CFU/g. The number of B. cereus attained 105−106 CFU/g in one sample. The distribution patterns of virulence genes in B. cereus isolates were identified. 84.6% of the B. cereus isolates had at least one enterotoxin or emetic toxin gene. The predominant pattern was XXV. 9.9% of isolates belonged to it and possessed one enterotoxin gene entFM. The occurrence rate of hblACD and nheABC was 36.3% and 47.3%, respectively. Antimicrobial susceptibility tests revealed a high resistance rate toward penicillin, and 23.1% of the isolates were multi-drug resistant. B. cereus isolates were genotyped by using ERIC-PCR. 89 genotypes were determined. The Hunter Gaston Discriminatory Index (HGDI) attained 0.9995. Relationships analysis revealed that Group A B. cereus isolates tended to carry hblA, hblC, hblD, nheA, nheB, and show resistance to penicillin/trimethoprim/sulfamethoxazole. This study was useful for updating the knowledge of the contamination status of B. cereus in RTE rice products in China.
Introduction
Bacillus cereus is a major causative agent of food poisoning outbreaks worldwide. It causes two types of food-borne illnesses, including diarrheal and emetic syndrome. The diarrheal variant is characterized by abdominal pain and watery diarrhea, and is usually linked to the intake of enterotoxin-producing B. cereus vegetative cells (e.g., Nhe, Hbl, and CytK). The emetic type, characterized by vomiting symptoms, is caused by ingestion of cereulide, a toxic peptide released by B. cereus emetic strains (Senesi and Ghelardi, 2010; Paudyal et al., 2018; Rouzeau-Szynalski et al., 2020; Yue et al., 2020, 2021).
Bacillus cereus can persist in a variety of natural environments, including soil and plants, due to its strong survival ability. The endospore resistance to multiple stresses, the formation of biofilms (Glasset et al., 2021; Li et al., 2022a), and even withstanding most cleaning and decontamination processes at food processing steps also help its survival in the environment (Merzougui et al., 2014; Ramarao et al., 2015). B. cereus is frequently found in processed products/prepared food items, according to data on the prevalence of Bacillus in food and animals in the European Union (European Food Safety Authority, and European Centre for Disease Prevention, and Control., 2017). Rice is a very popular food around the world and is often used as a raw material for the preparation of diet food dishes in many countries. During the cultivation, harvesting, and handling process, rice might be contaminated with vegetative cells and endospores of B. cereus (Vasiee et al., 2016; Kindle et al., 2019; Rodrigo et al., 2021). Although vegetative cells can be killed during some cooking processes, such as heating, however, cereulide and endospores generally survive due to high-stress resistance. Under normal conditions, endospores can germinate and become vegetative cells (Rouzeau-Szynalski et al., 2020; Tsugukuni et al., 2020). The possible safety risk of food-borne pathogens in ready-to-eat (RTE) foods is increasingly gaining public attention, because no additional sterilization steps, during cooking, baking, or pasteurization, are normally conducted before the consumption (Chon et al., 2015; Yu et al., 2019; Martelli et al., 2021). Although the contamination status of B. cereus in some kinds of food including dairy products, infant foods, aquatic products (Zhang et al., 2017, 2020; Gao et al., 2018; Zhao et al., 2020), etc. has been revealed, the data for B. cereus distribution in rice, especially in RTE rice products was still sparse in China. In this study, we investigated the quantitative prevalence of B. cereus in RTE rice products sampled in Zhejiang Province, located in Eastern China. The virulence gene distribution profiles, genotyping and antimicrobial susceptibility of these isolates were also studied.
Materials and methods
Sampling and isolation of Bacillus cereus
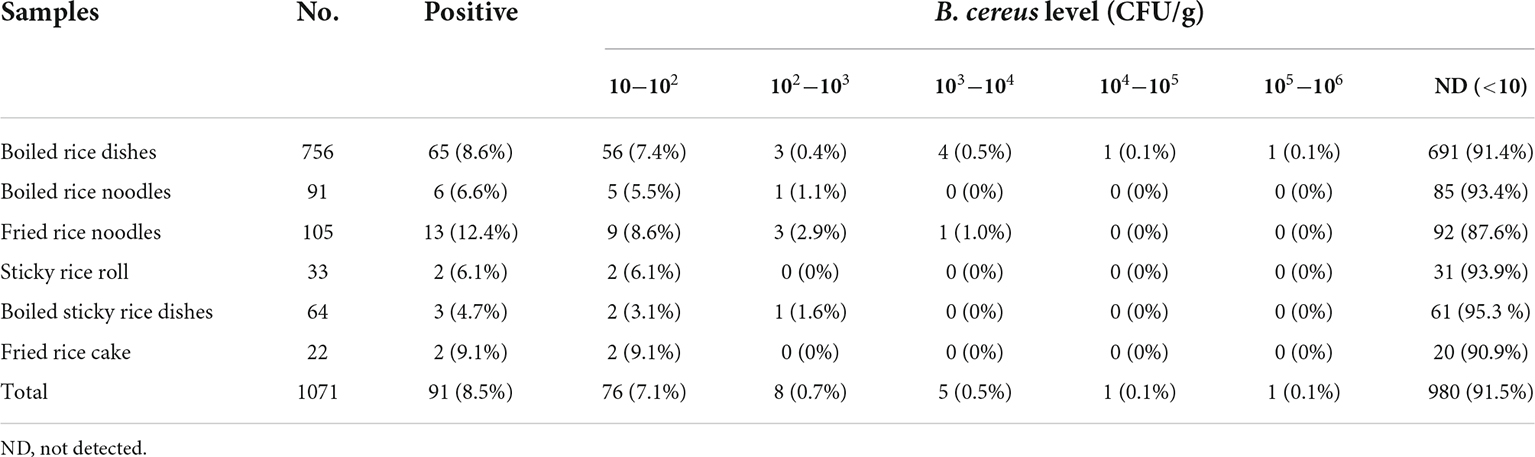
A total of 1071 RTE rice product samples were collected from 11 cities covering the whole Zhejiang Province, Eastern China, during 2017−2019. The samples included 756 boiled rice dishes, 91 boiled rice noodles, 105 fried rice noodles, 33 sticky rice rolls, 64 boiled sticky rice dishes, and 22 fried rice cakes. Quantitative detection of B. cereus in each sample was performed by using the direct plating method (Liu et al., 2021; Yue et al., 2021; Anwar et al., 2022). Briefly, Twenty-five grams of each sample was suspended in 225 mL of PBS and subsequently homogenized for 2 mins. The homogenate was 10-fold serially diluted in sterilized normal saline. The dilutions were spread on Mannitol-Egg-Yolk-Polymyxin (MYP) agars separately in duplicate. Plates were incubated at 30°C for 24 h. Five presumptive colonies with typical morphology on each plate were selected for further identification. Suspected colonies were then identified using Gram staining and the VITEK2 compact system (BioMerieux, France), followed by rhizoid growth and parasporal crystal formation tests to differentiate B. cereus from Bacillus thuringiensis and Bacillus mycoides. GB/T 4789.14-2014 (Ministry of Health of the People’s Republic of China, 2014) was used to calculate B. cereus numbers. One B. cereus isolate from each positive sample was stored for further characterization.
Detection of virulence genes
Genomic DNA was extracted from B. cereus by using a bacterial DNA extraction Kit (Omega, United States), according to manufacturer’s instructions. The primers and PCR protocol for eleven virulence genes were used as previously described ces (Ehling-Schulz et al., 2005), hblA (Zhou et al., 2008), hblC, hblD, nheA, nheB, nheC (Melnick et al., 2012), bceT (in’t Veld et al., 2001), cytK1, cytK2 (Guinebretiere et al., 2006), and entFM (Ngamwongsatit et al., 2008). Individual PCR reaction (25 μL) contain 50 ng of DNA template, 0.5 μL of each primer (10 μM), 0.125 U of Taq polymerase (TaKaRa, Japan), 2.5 μL of 10 × PCR buffer (Mg2+ free), 1.5 μL of MgCl2 (25 mM), and 2 μL of dNTP Mixture (2.5 mM). The amplicon was analyzed with 1% agarose gel. The gels were visualized by a UV Imaging System. A 100 bp DNA ladder (TaKaRa, Japan) was used as a DNA marker.
Antimicrobial susceptibility tests
Antimicrobial susceptibility assay of B. cereus isolates was tested by using the broth micro-dilution minimum inhibitory concentrations (MICs) method according to the standard Clinical and Laboratory Standard Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2015). After 18 h of cultivation on nutrient agar at 37°C, B. cereus was suspended in 0.85 per cent (w/v) NaCl solution to 1.0 MCF, followed by dilution with Mueller-Hinton broth to the final concentration of 1∼2 × 105 CFU/mL. Each 100 μL of the bacterial inoculum was added to 96-well plates containing antibiotics and incubated at 37°C for 20 h. Twelve antimicrobials from different classes were employed, including imipenem (1−64 μg/mL), penicillin (0.06−8 μg/mL), chloramphenicol (2−128 μg/mL), ceftriaxone (4−128 μg/mL), vancomycin (1−128 μg/mL), amikacin (8−128 μg/mL), erythrocin (0.25−32 μg/mL), tetracycline (2−32 μg/mL), ciprofloxacin (0.5−16 μg/mL), clindamycin (0.12−16 μg/mL), trimethoprim/sulfamethoxazole (0.5/9.5−16/304 μg/mL), and rifampin (0.5−8 μg/mL). The MIC results were analyzed based on the breakpoints for Bacillus species as per CLSI guidelines (Clinical and Laboratory Standards Institute, 2015). The breakpoint for ceftriaxone was from CLSI documents M45-A2 (Clinical and Laboratory Standards Institute, 2010). The isolates resistant to three or more types of antimicrobial classified into different antimicrobial categories were defined as multi-drug resistant (Li et al., 2022b). Staphylococcus aureus ATCC 29213 was used as a positive control.
ERIC-PCR analysis
All 91 B. cereus isolates were genotyped by ERIC-PCR using the following primers ERIC-1: 5′-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC-2: 5′-AAGTAAGTGACTGGGGTGAGCG-3′ (Dorneles et al., 2012; Dorneles et al., 2014). The PCR mixture (25 μL) was 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 0.3 mM of each dNTP, 1 U of Taq DNA polymerase (Takara, Dalian, China), 0.4 μM of each primer and 75 ng of DNA template. PCR reaction was carried out as follows: 95°C for 3 min, 35 cycles of 94°C for 30 s, 46°C for 40 s, 72°C for 3 min and a final incubation at 72°C for 10 min. Amplicons size was analyzed by 2.0% agarose gel. The gels were visualized by a UV Imaging System. A 100 bp DNA ladder (TaKaRa, Japan) was used as a marker. A 100% of similarity in bands pattern was defined as an ERIC-PCR genotype according to previous report (Magyar et al., 2019).
Genetic typing analysis
The software BioNumerics 7.6 (Applied Maths, Sint-Martens-Latem, Belgium) was applied to estimate the band size of ERIC-PCR amplicons and analyze the genotypes. Clustering analysis was based on the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA). The Hunter and Gaston Diversity Index (HGDI) was calculated to evaluate the discriminatory capability of ERIC-PCR (Shi et al., 2021). Isolates that share 100% similarity of amplicon bands pattern were grouped into one genotype.
Statistical analysis
Chi-square analysis was performed using the SPSS v 21.0 software package to determine if a significant difference existed in the prevalence distribution of B. cereus in different RTE rice products. The p-Value of <0.05 was used as a significance level. Relationships between genotype groups and virulence gene distribution, and antibiotic resistance profiles were analyzed by carrying out Pearson’s chi-square test and Fisher’s exact test with the Bonferroni correction.
Results and discussion
Quantitative prevalence of Bacillus cereus in ready-to-eat rice product
The prevalence of B. cereus in 1071 RTE rice product samples examined in this study was described in the Table 1. B. cereus was detected in 8.49% (91/1076) of all samples collected, out of which 65/91 (71.4%) were from boiled rice dishes, 6/91 (6.6%) were from boiled rice noodles, 13/91 (14.3%) were from fried rice noodles, 2/91 (2.2%) were from sticky rice roll, 3/91 (3.3%) were from boiled sticky rice dishes and 2/91 (2.2%) were from fried rice cake. According to previously published data, there are significant variances in the detection rate of B. cereus in various types of food samples from different regions of the world (Wang et al., 2019; Xu et al., 2020; Qiu et al., 2021; Shi et al., 2021; Wu et al., 2021). The total occurrence rate of B. cereus in our study was similar to a previous study in which B. cereus was isolated from dairy products, rice and flour products in China (Zhao et al., 2020). The prevalence of B. cerous in our study was lower than in a previous study isolated from artisan cheeses made in Mexico and powdered food products in Switzerland (Heini et al., 2018; Adame-Gomez et al., 2020). There was no statistically significant difference (p > 0.05) in the prevalence of B. cereus across the six types of rice products in our study. B. cereus is an opportunistic pathogen found in food. Ingestion of 105−108 vegetative cells or 8 μg of emetic toxin per kg of body weight may lead to gastroenteritis or/and vomiting syndrome in adults (Paananen et al., 2002; Schoeni and Wong, 2005). According to our findings, the number of B. cereus detected in 1.0 % fried rice noodles and 0.8% boiled rice meal samples varied from 103 to 106 CFU/g. Food poisoning can occur after consuming a specific amount of these highly contaminated meals (Zeng et al., 2021, 2022).
A number of safety criteria for B. cereus in RTE meals have been developed. In Canada and the United Kingdom, an acceptable threshold of 104 CFU/g is recommended. In South Korea, Australia, and New Zealand, a lower permissible threshold (103 CFU/g) is adopted (Nsw Food Authority, 2009; Health Canada, 2010; Chon et al., 2015). In our study, 91.5% of the samples had less than 103 CFU/g of B. cereus. However, 0.5% of the samples of RTE rice products had more than 103 CFU/g of B. cereus, which is more than the acceptable level in some countries. Although B. cereus in 91.5% of the samples was <10 CFU/g in our study, B. cereus in 0.5 % of RTE rice product samples outnumbered 103 CFU/g that could exceed the acceptable level of some countries.
Virulence gene profile of Bacillus cereus isolates
For many years, scientists have been studying the molecular mechanisms of B. cereus virulence. The diarrheal and emetic syndromes have been linked to several virulence factors including, secreted hemolysin BL (Hbl), necrotic enterotoxin (CytK), non-hemolytic enterotoxin (Nhe), enterotoxin FM (EntFM), BceT, and emetic toxin cereulide (Granum and Lund, 1997; Ehling-Schulz et al., 2005; Schoeni and Wong, 2005; Senesi and Ghelardi, 2010). Hbl or Nhe can promote fluid accumulation in ligated rabbit ileal loops due to their hemolytic, dermonecrotic, and vascular permeability activities (Schoeni and Wong, 2005; Griffiths and Schraft, 2017). Both of these enterotoxins comprise the tripartite complex. Three components are required for their maximal biological activity: proteins B, L1 and L2 in Hbl, and proteins A, B, and C in Nhe. Toxin activity has not been detected in any individual components (Arora, 2021; Sornchuer et al., 2022). The genes encoding Hbl and Nhe components are hblA, hblC, hblD, as well as nheA, nheB, and nheC, are located on two different operons (Sastalla et al., 2013). BceT, EntFM, CytK are all single-protein enterotoxins. BceT has cytotoxic, vascular permeability activities and can cause fluid accumulation in ligated mouse ileal loops (Agata et al., 1995). The necrotic enterotoxin CytK, which presents highly cytotoxic, necrotic and hemolytic activities, was initially incriminated in a severe gastroenteritis outbreak causing three patients’ death in France (Lund et al., 2000; Alonzo et al., 2015).
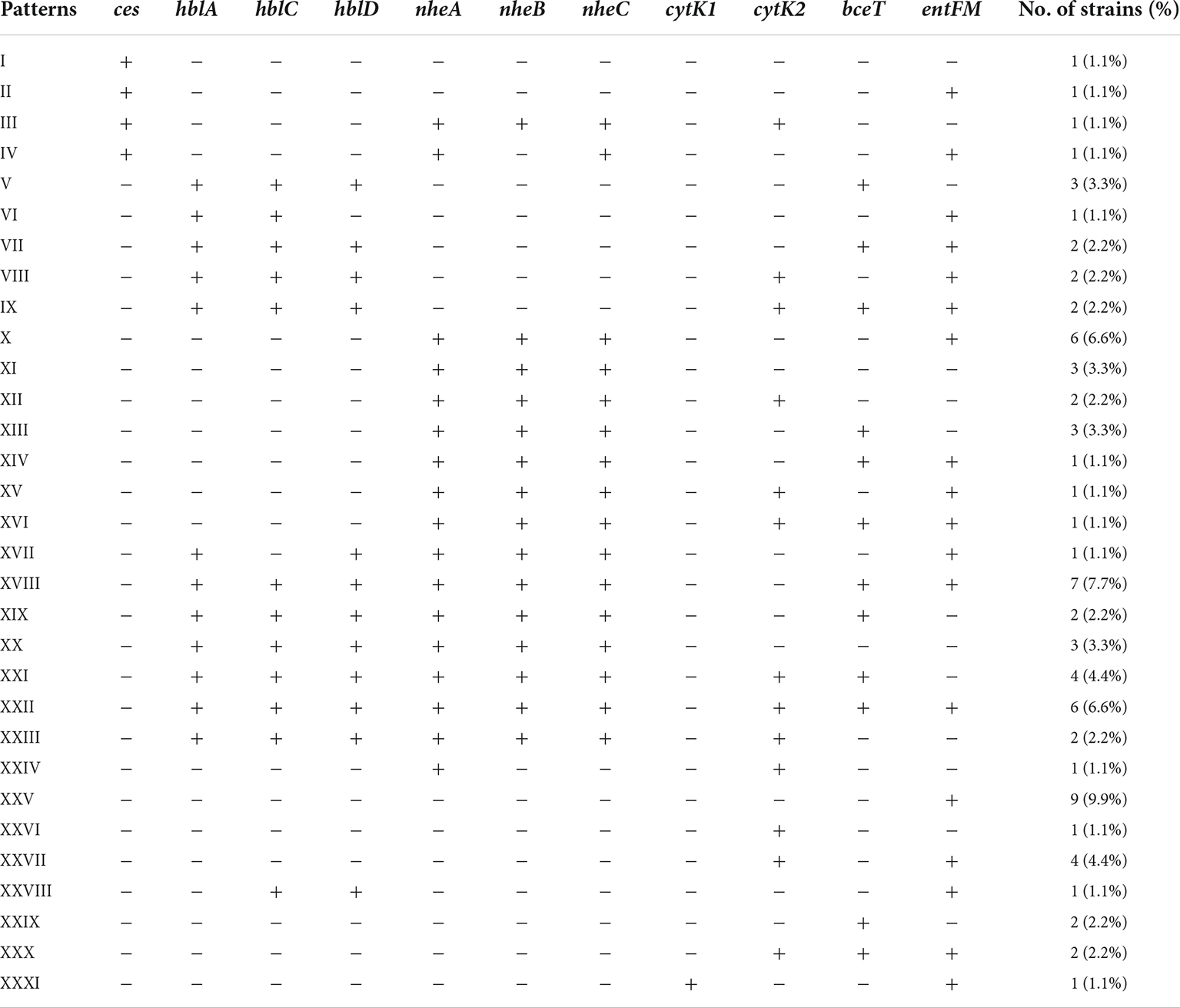
In this study, the distribution of associated encoding genes of the above toxins in B. cereus isolates was investigated. 84.6% of the B. cereus isolates had at least one enterotoxin or emetic toxin gene. A total of 31 distribution patterns of virulence genes were determined in our study (Table 2). The predominant one was XXV, 9.9% of isolates belonged to it and possessed only one enterotoxin gene entFM. The nheABC genes were present in 47.3% of the isolates, this frequency was lower than in B. cereus isolates from various food source samples and clinical isolates associated with foodborne outbreaks in previous studies (Kim et al., 2011; Glasset et al., 2016; Zhang et al., 2017). The occurrence rate of hblACD was 36.3%, which is similar to the previous reports isolated from milk products (Hwang and Park, 2015; Zhang et al., 2017), and it is lower than that of ready-to-eat foods, including vegetables, infant rice flour, rice, and grain-based foods (Chon et al., 2015; Hwang and Park, 2015; Zhang et al., 2017). The coexistence of hblACD and nheABC was found in 24/91 (26.4%) isolates. Six isolates (6.6%) were found to possess all enterotoxin encoding genes detected in this study.
Two distinct variants of CytK have been reported: CytK1 and CytK2. CytK1 is more harmful than CytK2. Although CytK2 proteins are hemolytic and toxic to Vero cells and human intestinal Caco-2 cells, their toxicity was only around 20% CytK1 (Fagerlund et al., 2004). Furthermore, CytK1 has been linked to major B. cereus outbreaks (Fagerlund et al., 2004; Guinebretiere et al., 2006). According to our findings, 33.0% of B. cereus isolates had either cytK1 or cytK2. CytK1 was found in one strain, accounting for 3.3% of all cytK-positive isolates. A previous study also observed this significant variation in cytK1 and cytK2 detection rates in B. cereus isolates from Chinese infant meals (Zhang et al., 2017). Foodborne B. cereus isolates may be slightly mild when producing diarrhea, according to the researchers. Meanwhile, in China, there was a risk of a B. cereus outbreak driven by a cytK1-positive strain. The ces gene for emetic toxin cereulide production was found in 4.4% of B. cereus isolates, consistent with the fact that B. cereus with ces was rarely isolated from food and environmental materials (Arslan et al., 2014; Chon et al., 2015; Zhang et al., 2017).
Prevalence of antimicrobial resistance
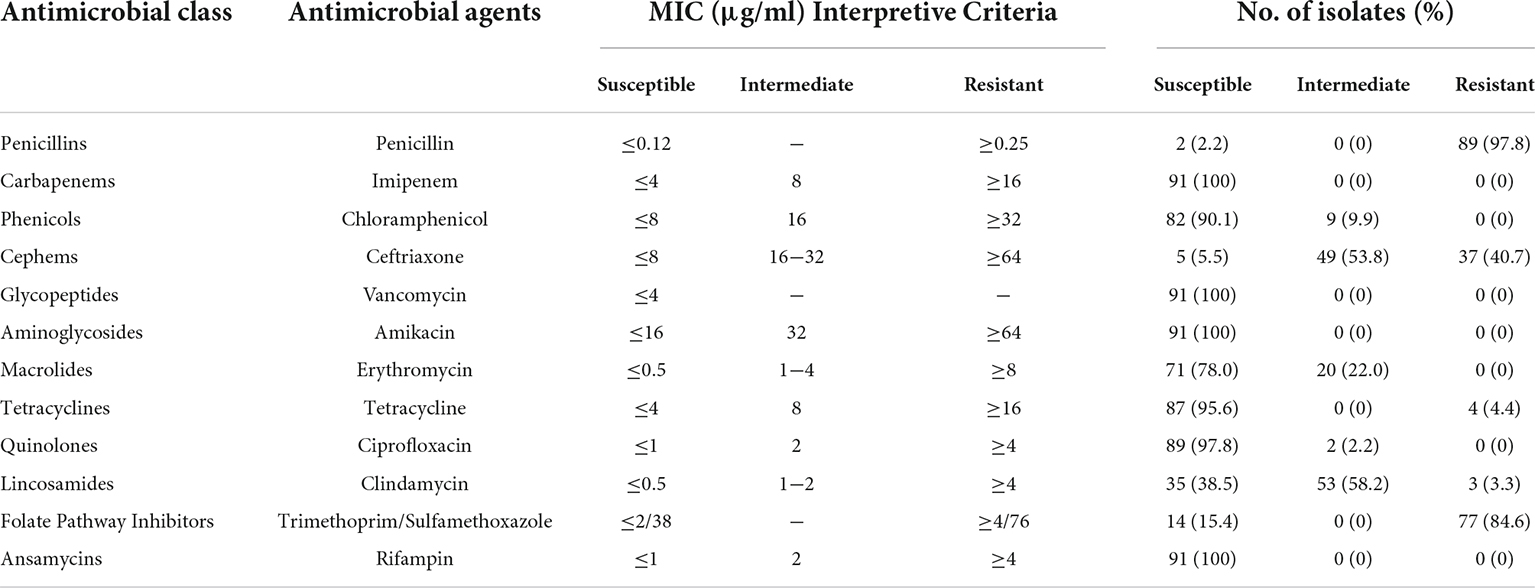
The antimicrobial susceptibility profile of the B. cereus isolates is shown in Table 3. Various susceptibility patterns against 12 types of antibiotics were exhibited. All isolates were susceptible to vancomycin, amikacin, imipenem, and rifampin. 90.1%, 78.0%, 95.6%, and 97.8% of the isolates showed susceptibility to chloramphenicol, erythromycin, tetracycline, and ciprofloxacin, respectively. 97.8% of isolates were resistant to penicillin, consistent with published reports that B. cereus isolates from either clinical or food sources were mostly resistant to penicillins (Park et al., 2009; Merzougui et al., 2014; Zhang et al., 2017). A high rate of antimicrobial resistance (84.6% isolates) was also detected against trimethoprim/sulfamethoxazole. All B. cereus isolates were classified into eight antibiotic resistance patterns (Figure 1). Resistance to penicillin/trimethoprim/sulfamethoxazole was the most common in our study. 21 isolates (23.1%) were multidrug resistance, with 85.7% were resistant to penicillin/ceftriaxone/trimethoprim/sulfamethoxazole.
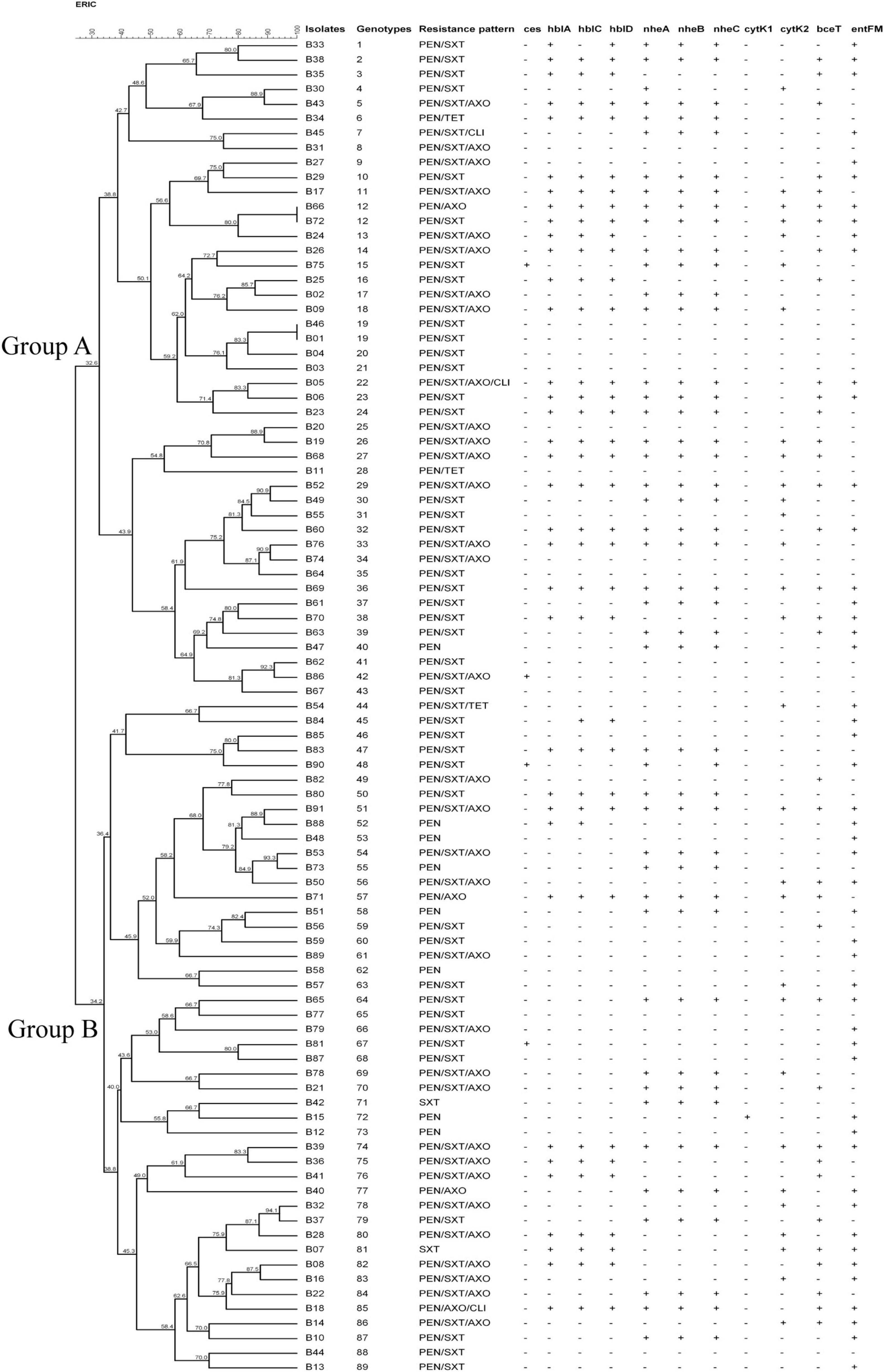
Figure 1. Characteristics of Bacillus cereus isolated from ready-to-eat rice products. The UPGMA tree was constructed by using BioNumerics 7.6. PEN, Penicillin; SXT, Trimethoprim/Sulfamethoxazole; AXO, Ceftriaxone; TET, Tetracycline; CLI, Clindamycin.
ERIC-PCR genotyping
All 91 B. cereus isolates were fingerprinted and assigned genotypes by using ERIC-PCR. The size of amplicons bands ranged approximately from 100 bp to 2000 bp. Each isolate produces 3-12 DNA bands. Considering 100% similarity in band pattern as a cut-off criteria, 89 genotypes were obtained, as a PCR-mediated fingerprinting typing approach. ERIC-PCR is more straightforward and rapid than PFGE and ribotyping (Dorneles et al., 2014; Magyar et al., 2019). It was initially applied to B. cereus for genetic discrimination by PO-REN Hsueh et al. (1999). However, a low number of B. cereus strains and low genetic diversity made it insufficient to evaluate the discriminatory capability of ERIC-PCR for B. cereus. In subsequent reports, ERIC-PCR was utilized to distinguish the strains of different species in the Bacillus genus (Shangkuan et al., 2000). According to our results, the calculated Hunter Gaston Discriminatory Index (HGDI) of ERIC-PCR on B. cereus genotyping attained 0.9995 using the optimal PCR system, demonstrating a high discriminatory capability.
Cluster analysis was performed based on UPGMA (Figure 1). Two major genotype groups (Group A and Group B) were defined in our study. 49.5% (45/91) of the isolates belonged to Group A, and 50.5% of the isolates belonged to Group B. Relationships analysis results between genotype groups and virulence gene distribution, antimicrobial resistance profiles demonstrated that there was no association between genotype groups and nheC (χ2 = 3.167, p = 0.075), bceT (χ2 = 3.167, p = 0.075), cytK1 (χ2 = 0.000, p = 0.987), cytK2 (χ2 = 0.088, p = 0.767), and ces (χ2 = 0.239, p = 1.000). Meanwhile, Group A B. cereus tended to carry hblA (χ2 = 6.018, p = 0.014), hblC (χ2 = 4.09, p = 0.043), hblD (χ2 = 4.967, p = 0.026), nheA (χ2 = 3.963, p = 0.046), and nheB (χ2 = 3.957, p = 0.047), and to be resistant to penicillin/trimethoprim/sulfamethoxazole (χ2 = 4.643, p = 0.031). Considering a limitation of ERIC-PCR as to the repetitive capabilities, genotypic diversity analysis of B. cereus based on the more reproducible methods, i.e., MLST and genomic sequencing (Nguyen and Tallent, 2019; Shen et al., 2021; Zhang et al., 2020) might be more informative in the future study.
Conclusion
Overall, an initial investigation was conducted of the quantitative prevalence and characterization of B. cereus isolated from ready-to-eat rice products in Zhejiang Province, Eastern China. A relatively high level of contamination was detected in ready-to-eat rice products, posing a risk of food poisoning and significant public health concern. Differences in the detection rate between the enterotoxin genes and emetic toxin genes revealed that B. cereus in ready-to-eat rice products was able to cause diarrhea and lead to food poisoning. B. cereus isolates presented high genetic diversity using ERIC-PCR with an HGDI of 0.9995. According to genetic relationships analysis, genotype Group A B. cereus isolates tended to carry hblA, hblC, hblD, nheA, nheB, and show resistance to penicillin/trimethoprim/sulfamethoxazole. This study provided essential data for addressing the microbial safety of ready-to-eat rice products in China (Peng et al., 2022), accordingly, might improve the appropriate safety criteria and policy.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
MY did the conceptualization, wrote, reviewed, and edited the manuscript, and carried out the project administration and funding acquisition. JC, JZ, and LZ investigated the data. HC and ZZ validated the data. JC and JZ carried out the data analysis. JZ wrote the original draft preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Program on Key Research Project of China (2019YFE0103900) as well as the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 861917 – SAFFI, the Natural Science Foundation of Zhejiang Province (LY20H190001), the Medical Scientific Research Foundation of Zhejiang Province (2019KY354), and Zhejiang Provincial Key R&D Program of China (2022C02024, 2021C02008, and 2020C02032).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.964823/full#supplementary-material
Supplementary Table 1 | The complete meta-data information for isolates in this study.
References
Adame-Gomez, R., Munoz-Barrios, S., Castro-Alarcon, N., Leyva-Vazquez, M. A., Toribio-Jimenez, J., and Ramirez-Peralta, A. (2020). Prevalence of the strains of Bacillus cereus group in artisanal Mexican Cheese. Foodborne Pathog Dis. 17, 8–14. doi: 10.1089/fpd.2019.2673
Agata, N., Ohta, M., Arakawa, Y., and Mori, M. (1995). The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology (Reading) 141(Pt 4), 983–988. doi: 10.1099/13500872-141-4-983
Alonzo, D. A., Magarvey, N. A., and Schmeing, T. M. (2015). Characterization of cereulide synthetase, a toxin-producing macromolecular machine. PLoS One 10:e0128569. doi: 10.1371/journal.pone.0128569
Anwar, T. M., Pan, H., Chai, W., -Dra, A., Fang, W., Li, Y., et al. (eds) (2022). Genetic diversity, virulence factors, and antimicrobial resistance of Listeria monocytogenes from food, livestock, and clinical samples between 2002 and 2019 in China. Int. J. Food Microbiol. 366:109572. doi: 10.1016/j.ijfoodmicro.2022.109572
Arora, P. K. (2021). Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. Microbial Products Health Environ. Agriculture 14:4524. doi: 10.2903/j.efsa.2016.4524
Arslan, S., Eyi, A., and Kucuksari, R. (2014). Toxigenic genes, spoilage potential, and antimicrobial resistance of Bacillus cereus group strains from ice cream. Anaerobe 25, 42–46. doi: 10.1016/j.anaerobe.2013.11.006
Chon, J. W., Yim, J. H., Kim, H. S., Kim, D. H., Kim, H., Oh, D. H., et al. (2015). Quantitative prevalence and toxin gene profile of Bacillus cereus from ready-to-eat vegetables in South Korea. Foodborne Pathog Dis. 12, 795–799. doi: 10.1089/fpd.2015.1977
Clinical and Laboratory Standards Institute (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline, 2nd Edn. Wayne: CLSI.
Clinical and Laboratory Standards Institute (2015). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline, 3rd Edn. Wayne: CLSI.
Dorneles, E. M., Santana, J. A., Andrade, G. I., Santos, E. L., Guimaraes, A. S., Mota, R. A., et al. (2012). Molecular characterization of Corynebacterium pseudotuberculosis isolated from goats using ERIC-PCR. Genet. Mol. Res. 11, 2051–2059.
Dorneles, E. M., Santana, J. A., Ribeiro, D., Dorella, F. A., Guimaraes, A. S., Moawad, M. S., et al. (2014). Evaluation of ERIC-PCR as genotyping method for Corynebacterium pseudotuberculosis isolates. PLoS One 9:e98758. doi: 10.1371/journal.pone.0098758
Ehling-Schulz, M., Vukov, N., Schulz, A., Shaheen, R., Andersson, M., Martlbauer, E., et al. (2005). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113. doi: 10.1128/AEM.71.1.105-113.2005
European Food Safety Authority, and European Centre for Disease Prevention, and Control. (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in. EFSA J. 16:e05500.
Fagerlund, A., Ween, O., Lund, T., Hardy, S. P., and Granum, P. E. (2004). Genetic and functional analysis of the cytK family of genes in Bacillus cereus. Microbiology (Reading) 150(Pt 8), 2689–2697. doi: 10.1099/mic.0.26975-0
Gao, T., Ding, Y., Wu, Q., Wang, J., Zhang, J., Yu, S., et al. (2018). Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from pasteurized milk in China. Front. Microbiol. 9:533. doi: 10.3389/fmicb.2018.00533
Glasset, B., Herbin, S., Guillier, L., Cadel-Six, S., Vignaud, M. L., Grout, J., et al. (2016). Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterisation. Euro. Surveill. 21:30413. doi: 10.2807/1560-7917.ES.2016.21.48.30413
Glasset, B., Sperry, M., Dervyn, R., Herbin, S., Brisabois, A., and Ramarao, N. (2021). The cytotoxic potential of Bacillus cereus strains of various origins. Food Microbiol. 98:103759.
Granum, P. E., and Lund, T. (1997). Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157, 223–228.
Griffiths, M. W., and Schraft, H. (2017). Bacillus cereus Food Poisoning. Foodborne Diseases, 3rd Edn. Amsterdam: Elsevier.
Guinebretiere, M. H., Fagerlund, A., Granum, P. E., and Nguyen-The, C. (2006). Rapid discrimination of cytK-1 and cytK-2 genes in Bacillus cereus strains by a novel duplex PCR system. FEMS Microbiol. Lett. 259, 74–80. doi: 10.1111/j.1574-6968.2006.00247.x
Health Canada (2010). Microbial Guidelines for Ready-to-Eat Foods— A Guide for the Conveyance Industry and Environmental Health Officers (EHO). Ottawa, ON: Health Canada.
Heini, N., Stephan, R., Ehling-Schulz, M., and Johler, S. (2018). Characterization of Bacillus cereus group isolates from powdered food products. Int. J. Food Microbiol. 283, 59–64. doi: 10.1016/j.ijfoodmicro.2018.06.019
Hsueh, P. R., Teng, L. J., Yang, P. C., Pan, H. L., Ho, S. W., and Luh, K. T. (1999). Nosocomial pseudoepidemic caused by Bacillus cereus traced to contaminated ethyl alcohol from a liquor factory. J. Clin. Microbiol. 37, 2280–2284. doi: 10.1128/JCM.37.7.2280-2284.1999
Hwang, J. Y., and Park, J. H. (2015). Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J. Dairy Sci. 98, 1652–1660. doi: 10.3168/jds.2014-9042
in’t Veld, P. H., Ritmeester, W. S., Delfgou-van, Asch EH, Dufrenne, J. B., Wernars, K., Smit, E., et al. (2001). Detection of genes encoding for enterotoxins and determination of the production of enterotoxins by HBL blood plates and immunoassays of psychrotrophic strains of Bacillus cereus isolated from pasteurised milk. Int. J. Food Microbiol. 64, 63–70. doi: 10.1016/s0168-1605(00)00443-8
Kim, J. B., Kim, J. M., Cho, S. H., Oh, H. S., Choi, N. J., and Oh, D. H. (2011). Toxin genes profiles and toxin production ability of Bacillus cereus isolated from clinical and food samples. J. Food Sci. 76, T25–T29. doi: 10.1111/j.1750-3841.2010.01958.x
Kindle, P., Etter, D., Stephan, R., and Johler, S. (2019). Population structure and toxin gene profiles of Bacillus cereus sensu lato isolated from flour products. FEMS Microbiol. Lett. 366:fnz240. doi: 10.1093/femsle/fnz240
Li, Y., Ed-Dra, A., Tang, B., Kang, X., Müller, A., Kehrenberg, C., et al. (2022a) Predominant Salmonella serovars circulating in the antibiotic-free feed farms have a higher tolerance to environmental stresses. J. Hazard. Mater. 438:129476. doi: 10.1016/j.jhazmat.2022.129476
Li, Y., Kang, X., Ed-Dra, A., Zhou, X., Jia, C., Müller, A, et al. (2022b). Genome-based assessment of antimicrobial resistance and virulence potential for non-Pullorum/Gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol. Spectr. e0096522. doi: 10.1128/spectrum.00965-22 [Epub ahead of print].
Liu, Y., Jiang, J., -Dra, A., Li, X., Peng, X., Xia, L., et al. (eds) (2021). Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang. China. Food Res. Int. 142:110198. doi: 10.1016/j.foodres.2021.110198
Lund, T., De Buyser, M. L., and Granum, P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38, 254–261. doi: 10.1046/j.1365-2958.2000.02147.x
Magyar, T., Gyuris, É, Ujvári, B., Metzner, M., and Wehmann, E. (2019). Genotyping of Riemerella anatipestifer by ERIC-PCR and correlation with serotypes. Avian. Pathol. 48, 12–16. doi: 10.1080/03079457.2018.1535693
Martelli, F., Marrella, M., Lazzi, C., Neviani, E., and Bernini, V. (2021). Microbiological contamination of ready-to-eat algae and evaluation of Bacillus cereus behavior by microbiological challenge test. J. Food Prot. 84, 1275–1280. doi: 10.4315/JFP-20-407
Melnick, R. L., Testen, A. L., Poleatewich, A. M., Backman, P. A., and Bailey, B. A. (2012). Detection and expression of enterotoxin genes in endophytic strains of Bacillus cereus. Lett. Appl. Microbiol. 54, 468–474. doi: 10.1111/j.1472-765X.2012.03232.x
Merzougui, S., Lkhider, M., Grosset, N., Gautier, M., and Cohen, N. (2014). Prevalence, PFGE typing, and antibiotic resistance of Bacillus cereus group isolated from food in Morocco. Foodborne Pathog Dis. 11, 145–149. doi: 10.1089/fpd.2013.1615
Ministry of Health of the People’s Republic of China (2014). GB 4789.14-2014 National Food Safety Standards, Food Microbiology Test for Bacillus cereus. Beijing: China Standard Press.
Ngamwongsatit, P., Buasri, W., Pianariyanon, P., Pulsrikarn, C., Ohba, M., Assavanig, A., et al. (2008). Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 121, 352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013
Nguyen, A. T., and Tallent, S. M. (2019). Screening food for Bacillus cereus toxins using whole genome sequencing. Food Microbiol. 78, 164–170. doi: 10.1016/j.fm.2018.10.008
Nsw Food Authority (2009). Microbiological Quality Guide for Ready-to-eat foods. Newington, NSW: NSW Food Authority.
Paananen, A., Mikkola, R., Sareneva, T., Matikainen, S., Hess, M., Andersson, M., et al. (2002). Inhibition of human natural killer cell activity by cereulide, an emetic toxin from Bacillus cereus. Clin. Exp. Immunol. 129, 420–428. doi: 10.1046/j.1365-2249.2002.01898.x
Park, Y. B., Kim, J. B., Shin, S. W., Kim, J. C., Cho, S. H., Lee, B. K., et al. (2009). Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J. Food Prot. 72, 612–617. doi: 10.4315/0362-028x-72.3.612
Paudyal, N., Pan, H., Liao, X., Zhang, X., Li, X., Fang, W., et al. (2018). A meta-analysis of major foodborne pathogens in Chinese Food Commodities Between 2006 and 2016. Foodborne Pathog Dis. 15, 187–197. doi: 10.1089/fpd.2017.2417
Peng, X., Ed-Dra, A., and Yue, M. (2022). Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. Online ahead of print.
Qiu, Y. F., Nambiar, R. B., Xu, X. B., Weng, S. T., Pan, H., Zheng, K. C., et al. (2021). Global genomic characterization of Salmonella enterica serovar telelkebir. Front. Microbiol. 12:704152. doi: 10.3389/fmicb.2021.704152
Ramarao, N., Lereclus, D., and Sorokin, A. (2015). The Bacillus cereus group. Mol. Med. Microbiol. 2, 1041–1078.
Rodrigo, D., Rosell, C. M., and Martinez, A. (2021). Risk of Bacillus cereus in relation to rice and derivatives. Foods 10:302.
Rouzeau-Szynalski, K., Stollewerk, K., Messelhausser, U., and Ehling-Schulz, M. (2020). Why be serious about emetic Bacillus cereus: cereulide production and industrial challenges. Food Microbiol. 85:103279. doi: 10.1016/j.fm.2019.103279
Sastalla, I., Fattah, R., Coppage, N., Nandy, P., Crown, D., Pomerantsev, A. P., et al. (2013). The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One 8:e76955. doi: 10.1371/journal.pone.0076955
Schoeni, J. L., and Wong, A. C. (2005). Bacillus cereus food poisoning and its toxins. J. Food Prot. 68, 636–648.
Senesi, S., and Ghelardi, E. (2010). Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2, 1690–1703.
Shangkuan, Y. H., Yang, J. F., Lin, H. C., and Shaio, M. F. (2000). Comparison of PCR-RFLP, ribotyping and ERIC-PCR for typing Bacillus anthracis and Bacillus cereus strains. J. Appl. Microbiol. 89, 452–462. doi: 10.1046/j.1365-2672.2000.01134.x
Shen, L., Zang, X., Sun, K., Chen, H., Che, X., Sun, Y., et al. (2021). Complete genome sequencing of Bacillus sp. TK-2, analysis of its cold evolution adaptability. Sci. Rep. 11:4836. doi: 10.1038/s41598-021-84286-7
Shi, D., Anwar, T. M., Pan, H., Chai, W., Xu, S., and Yue, M. (2021). Genomic determinants of pathogenicity and antimicrobial resistance for 60 global Listeria monocytogenes isolates responsible for invasive infections. Front. Cell Infect. Microbiol. 11:718840. doi: 10.3389/fcimb.2021.718840
Sornchuer, P., Saninjuk, K., Prathaphan, P., Tiengtip, R., and Wattanaphansak, S. (2022). Antimicrobial susceptibility profile and whole-genome analysis of a strong biofilm-forming Bacillus Sp. B87 strain isolated from food. Microorganisms 10:252. doi: 10.3390/microorganisms10020252
Tsugukuni, T., Shigemune, N., Nakayama, M., and Miyamoto, T. (2020). Morphological changes in spores during germination in Bacillus cereus and Bacillus subtilis. Biocontrol. Sci. 25, 203–213.
Vasiee, A., Behbahani, B. A., Yazdi, F. T., and Moradi, S. (2016). Optimization of the production conditions of the lipase produced by Bacillus cereus from rice flour through Plackett-Burman Design (PBD) and response surface methodology (RSM). Microb. Pathog. 101, 36–43. doi: 10.1016/j.micpath.2016.10.020
Wang, X., Biswas, S., Paudyal, N., Pan, H., Li, X., Fang, W., et al. (2019). Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 10:985. doi: 10.3389/fmicb.2019.00985
Wu, B., -Dra, A., Pan, H., Dong, C., Jia, C., and Yue, M. (eds) (2021). Genomic investigation of Salmonella isolates recovered from a pig slaughtering process in hangzhou, China. Front. Microbiol. 12:704636. doi: 10.3389/fmicb.2021.704636
Xu, X., Chen, Y., Pan, H., Pang, Z., Li, F., Peng, X., et al. (eds) (2020). Genomic characterization of Salmonella uzaramo for human invasive infection. Microb Genom. 6:mgen000401. doi: 10.1099/mgen.0.000401
Yu, S., Yu, P., Wang, J., Li, C., Guo, H., Liu, C., et al. (2019). A study on prevalence and characterization of Bacillus cereus in ready-to-eat foods in China. Front. Microbiol. 10:3043. doi: 10.3389/fmicb.2019.03043
Yue, M., Bai, L., Song, H., and Fang, W. (2021). Impacts of microbial food safety in China and beyond. Foodborne Pathog Dis. 18, 508–509.
Yue, M., Song, H., and Bai, L. (2020). Call for special issue papers: food safety in china: current practices and future needs. Foodborne Pathog Dis. 17:295.
Zeng, X., Li, Q., Yang, C., Yu, Y., Fu, Z., Wang, H., et al. (2021). Effects of Clostridium butyricum- and Bacillus spp.-based potential probiotics on the growth performance, intestinal morphology, immune responses, and caecal microbiota in broilers. Antibiotics (Basel) 10:624. doi: 10.3390/antibiotics10060624
Zeng, Z., Zhou, Y., Xu, Y., Wang, S., Wang, B., Zeng, Z., et al. (2022). Bacillus amyloliquefaciens SC06 alleviates the obesity of ob/ob mice and improves their intestinal microbiota and bile acid metabolism. Food Funct. 13, 5381–5395. doi: 10.1039/d1fo03170h
Zhang, Y., Chen, J., Feng, C., Zhan, L., Zhang, J., Li, Y., et al. (2017). Quantitative prevalence, phenotypic and genotypic characteristics of Bacillus cereus isolated from retail infant foods in China. Foodborne Pathog Dis. 14, 564–572. doi: 10.1089/fpd.2017.2287
Zhang, Y., Chen, M., Yu, P., Yu, S., Wang, J., Guo, H., et al. (2020). Prevalence, virulence feature, antibiotic resistance and MLST typing of Bacillus cereus isolated from retail aquatic products in China. Front. Microbiol. 11:1513. doi: 10.3389/fmicb.2020.01513
Zhao, S., Chen, J., Fei, P., Feng, H., Wang, Y., Ali, M. A., et al. (2020). Prevalence, molecular characterization, and antibiotic susceptibility of Bacillus cereus isolated from dairy products in China. J. Dairy Sci. 103, 3994–4001. doi: 10.3168/jds.2019-17541
Keywords: Bacillus cereus, quantitative prevalence, rice products, virulence gene, antimicrobial resistance, ERIC-PCR
Citation: Chen J, Zhang J, Zhan L, Chen H, Zhang Z, Huang C and Yue M (2022) Prevalence and antimicrobial-resistant characterization of Bacillus cereus isolated from ready-to-eat rice products in Eastern China. Front. Microbiol. 13:964823. doi: 10.3389/fmicb.2022.964823
Received: 09 June 2022; Accepted: 28 June 2022;
Published: 15 July 2022.
Edited by:
Zhong Peng, Huazhong Agricultural University, ChinaReviewed by:
Jian Ji, Jiangnan University, ChinaQingli Dong, University of Shanghai for Science and Technology, China
Copyright © 2022 Chen, Zhang, Zhan, Chen, Zhang, Huang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Huang, Y2hodWFuZ0BjZGMuemouY24=; Min Yue, bXl1ZUB6anUuZWR1LmNu
 Jiancai Chen
Jiancai Chen Junyan Zhang1
Junyan Zhang1 Li Zhan
Li Zhan Honghu Chen
Honghu Chen Cheng Huang
Cheng Huang Min Yue
Min Yue