
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 08 September 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.964019
Phenylacetic acid (PAA) is a central intermediate metabolite involved in bacterial degradation of aromatic components. The bacterial PAA pathway mainly contains 12 enzymes and a transcriptional regulator, which are involved in biofilm formation and antimicrobial activity. They are present in approximately 16% of the sequenced bacterial genome. In this review, we have summarized the PAA distribution in microbes, recent structural and functional study progress of the enzyme families of the bacterial PAA pathway, and their role in bacterial pathogenicity and antibiotic resistance. The enzymes of the bacterial PAA pathway have shown potential as an antimicrobial drug target for biotechnological applications in metabolic engineering.
Aromatic hydrocarbons with diverse chemical structures and resistance to degradation are among the most abundant sources of organic carbon in nature (Bugg et al., 2011; Wang et al., 2017). They include plant-soluble secondary metabolic products and structural polymer lignin, some common environmental pollutants, such as petroleum derivatives BTEX (benzene, toluene, ethylbenzene, and xylene), polycyclic aromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), pentachlorophenol (Cao et al., 2009; Fuchs et al., 2011).
Due to limited reactivity, aromatic compounds are predominantly degraded by microbes, which have evolved enzymatic pathways under aerobic and anaerobic conditions (Metzler, 2003; Carmona et al., 2009; Fuchs et al., 2011). The strategy of converging different peripheral pathways by producing a few central intermediates, like phenylacetic acid (PAA), which are then degraded by shared enzymes in subsequent pathways, enables microbes to utilize various aromatic compounds with high efficiency (Fuchs et al., 2011; Liu et al., 2020). Under anaerobic conditions, the intermediate benzoyl-CoA is formed, and two enzymes are responsible for reduction: a class I reductase driven by ATP hydrolysis (Boll and Fuchs, 1995), and a multisubunit enzyme ATP-independent class II benzoyl-CoA reductase (Kung et al., 2009). There are two aerobic strategies to break down aromatic rings: the introduction of two hydroxyl groups into the aromatic ring by a dioxygenase forming catechol, which cleaves the bond adjacent to the carboxyl in an oxygen-dependent manner (Fuchs, 2008); and the attachment of coenzyme A (CoA) to aromatic compounds, which is then catalyzed by a series of monooxygenases (Ismail et al., 2003; Rather et al., 2010; Teufel et al., 2010).
The PAA degradation pathway is the central aromatic compound metabolic pathway utilizing CoA. This pathway contains both aerobic and anaerobic elements and is present in more than 16% of the currently sequenced bacterial genomes (Teufel et al., 2010), as well as in archaea such as Ferroglobus placidus (Aklujkar et al., 2014) and Thermoprofundales (Liu et al., 2020). To demonstrate remote evolution and propose the possibility of biotechnological application of this pathway from historical findings as well as recent progress, in this review, we focused on the following aspects of the PAA catabolic pathway: distribution and function of PAA, paa cluster components and their structural characteristics, the relationship between PAA metabolic and bacterial pathogenicity, and antimicrobial resistance.
PAA is widely distributed across bacteria, fungi, algae, and terrestrial plants. Pseudomonas species can use aromatic molecules as the sole carbon source for growth (Dagley, 1975). In other genera such as Clostridium (Elsden et al., 1976) and Bacteroides (Mayrand, 1979), PAA is as a metabolic product, but its function remains unknown. PAA is a weak acid that is toxic at certain concentrations and pH values, and its wide spread among bacteria and fungi is responsible for deterring non-specific species and preventing habitat loss (Burkhead et al., 1998; Hwang et al., 2001; Kim et al., 2004a,b; Somers et al., 2005). In Pseudomonas aeruginosa, 1.47 mM PAA disrupts quorum sensing and attenuates biofilm formation (Musthafa et al., 2012). PAA from the soil bacterium Bacillus licheniformis is effective against Staphylococcus aureus and Escherichia coli (Kim et al., 2004a,b). In fungi, PAA is a direct precursor for penicillin G (Moyer and Coghill, 1947), and affects metabolism at the transcriptional and protein levels (Harris et al., 2009; Jami et al., 2018). Based on the application of the above findings, genetic manipulation to decrease PAA degradation achieved penicillin overproduction in P. chyrysogeneum (Rodríguez-Sáiz et al., 2001, 2005). As a ubiquitous plant auxin, PAA promotes plant growth, specifically cell expansion, elongation and differentiation, cubature, callus growth, and lateral root induction, as well as antimicrobial activity (Cook, 2019). The PAA catabolic pathway has not been detected in plants, and animals have only limited capacity to metabolize aromatic compounds (Metzler, 2003).
Microbes including bacteria and archaea are the main organisms in aromatic compound metabolism, suggesting a remote origin of the PAA pathway (Metzler, 2003). In addition to the numerous studies in bacteria, an increasing number of studies in archaea have also demonstrated that they exploit aromatic compounds as energy and carbon sources (Erdoğmuş et al., 2013; Aklujkar et al., 2014). Halophilic archaea use dioxygenases, while hyperthermophilic archaea use the BCoA pathway to mineralize aromatic compounds (Fairley et al., 2006; Schmid et al., 2015). Genomic and biochemical evidence shows that Thermoprofundales can utilize aromatic compounds through the PAA pathway under extreme conditions, such as pH 5–7, and relatively high optimal temperatures (Liu et al., 2020), which could enable application of PAA pathway-related enzymes in biotechnology.
The gene cluster associated with PAA catabolism is the paa operon, which mainly encodes 12 enzymes or enzymatic subunits: PaaZ, PaaA, PaaB, PaaC, PaaE, PaaF, PaaG, PaaH, PaaJ, PaaK, PaaY, and PaaI; transcription regulator PaaX or PaaR in different species; and PaaD protein with unknown function. The PAA pathway can be artificially divided into two parts, the early and late steps (Figure 1), which are similar to that of the benzoate degradation pathway and fatty acid β-oxidation process, respectively. With XI being the precursor for tropone natural products, a recent review has highlighted the structure features of the main enzymes in PAA catabolic pathway to form XI (Grishin and Cygler, 2015; Duan et al., 2020), while we focus on the entire PAA catabolic process. The overview of the catalytic properties of some enzymes are listed in Table 1.
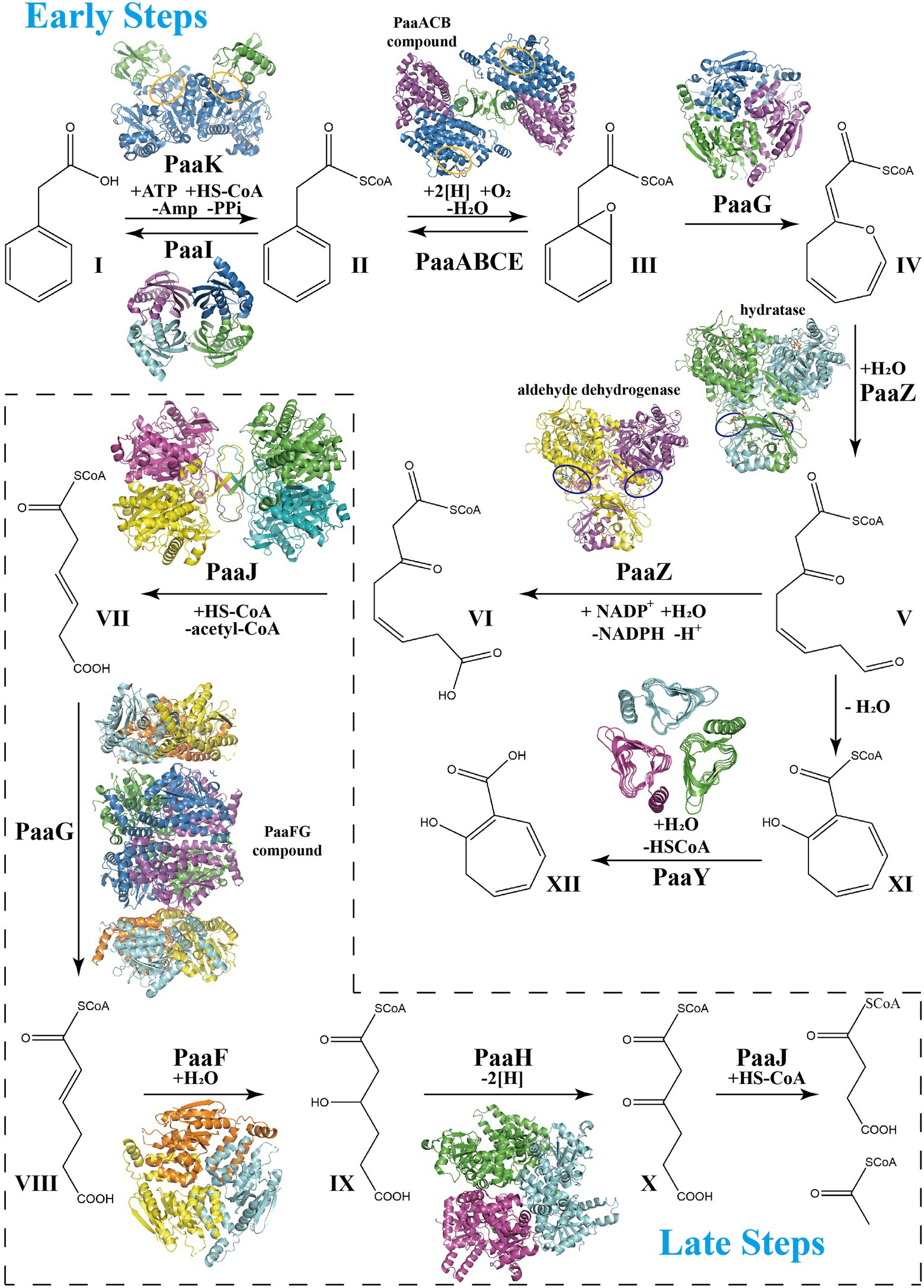
Figure 1. Schema of phenylacetate acid degradation pathway. Early PAA pathway: Step 1, Phenylacetate (I, PA) is converted into phenylacetyl-CoA (II, PA-CoA), catalyzed by a phenylacetate-CoA ligase PaaK. Thioesterase PaaI could lead this step into a reversible direction when toxic accumulation occurs. Step 2, Epoxidation induced by the monooxygenase complex PaaABCDE in ring 1,2-epoxyphenylacetyl-CoA (III, ep-CoA). Step 3, PaaG isomerize ep-CoA into 2-oxepin-2(3H)-ylideneacetyl-CoA (IV, oxepin-CoA), an oxygen-containing heterocycle with three double bonds. Step 4, hydrolysis mediated by the bifunctional enzyme PaaZ induces the oxepin-CoA ring-opening and conversion into 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde (V). Compound V spontaneously rearranges to 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA (XI), which inhibits the enzymatic activity of PaaZ. Thioesterase PaaY could relieve this inhibition by converting XI into 2-hydroxycyclohepta-1,4,6-triene (XII). Step 5, PaaZ also induces the oxidation of the terminal aldehyde group in V, finally producing 3-oxo-5,6-dehydrosuberyl-CoA (VI). Late PAA pathway: Step 6, Thiolase PaaJ induces degradation of long-chain intermediates VI into a C6-intermediate, 3,4-dehydroadipyl-CoA (VII). Step 7, The isomerization of converting VII into 2,3-dehydroadipyl-CoA (VIII) also mediated by isomerase PaaG. Step 8, Hydratase PaaF generates 3-hydroxyadipyl-CoA (IX) from VIII. PaaF could form stable complex with PaaG, which may speed up Step 7 and 8 in vivo. Step 9, Dehydrogenase PaaH oxidizes IX to 3-oxoadipyl-CoA (X). Step 10, The last step of the PAA pathway, degradation of the C6-intermediate into succinyl-CoA and acetyl-CoA, also induced by thiolase PaaJ. The structural representations of the key enzymes in the PAA pathway are also shown in corresponding catalyzing steps. The repressors PaaX and PaaR are not shown.
The phenylacetate-CoA ligase PaaK acts as the initial enzyme in the PAA pathway, controlling the influx of the substrate phenylacetic acid. CoA is attached to phenylacetate by PaaK, which is dependent on Mg2+ and ATP, with high specificity and relatively high heat stability (El-Said Mohamed, 2000; Erb et al., 2008).
PaaK belongs to the adenylate-forming enzyme superfamily and functions as an activator of short-to-long fatty acids, aromatic compounds, and biosynthesis of peptide antibiotics and siderophores (Gulick, 2009). This family member exhibits two α/β domains, with an active site located at the interface between the N-terminal and C-terminal domains (Figures 2A–D; Gulick, 2009). A dimer is mainly formed and maintained by residues from the N-terminal domain, leaving the C-terminal domain free for conformational changes during adenylation catalysis (Law and Boulanger, 2011). Law et al. characterized the structure of two BcePaaK proteins from Burkholderia cenocepacia, BcePaaK1 (PDB:2Y4N) and BcePaaK2 (PDB:2Y4O), providing a comprehensive view of PaaK conformation. Briefly, BcePaaK1 catalyzes the adenylation reaction in which lysine (Lys422) at the active site is essential for substrate binding and nucleophilic attack (Figures 2A,C). Conversely, BcePaaK2 exhibits a different conformation during catalysis. In BcePaaK2, the C-terminal domain rotation removes the equivalent lysine (Lys429) from the active site as in BcePaaK1 and provides an alternative platform for the thioesterification reaction (Figures 2B,D).
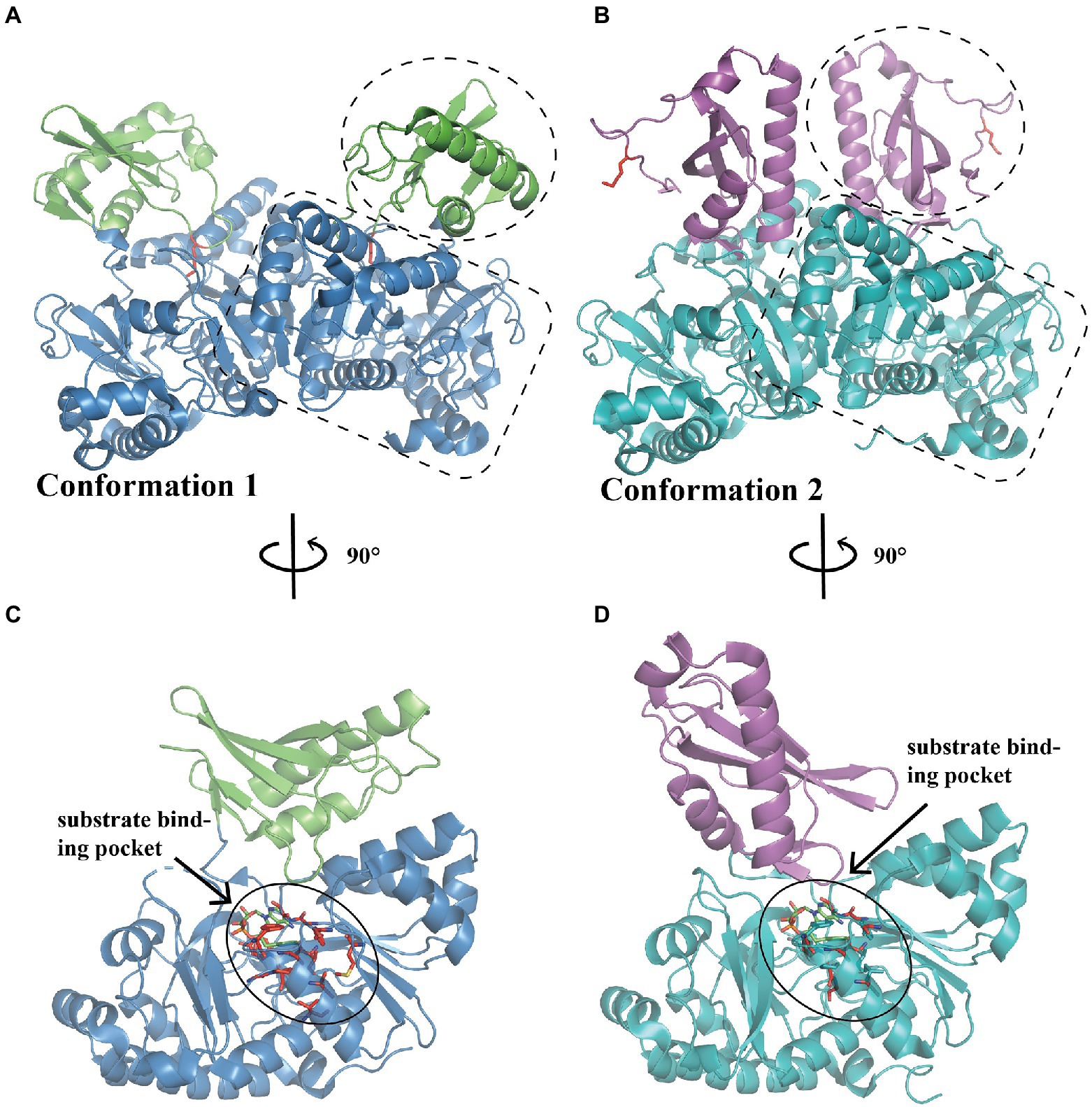
Figure 2. Overall structure of PaaK. (A) The overall structure of dimer BcePaaK1 (PDB: 2Y4N) in conformation 1. The N-terminal domain and C-terminal domain are circled by a dotted line in blue and green, respectively, the Lys422 at the active site is shown as sticks in the side chain in red. (B) The overall structure of dimer BcePaaK2 (PDB: 2Y4O) in conformation 2 with a similar N-terminal domain and a rotated C-terminal domain. The N-terminal domain and C-terminal domain are circled as in (A) in teal and magenta, respectively. The corresponding Lys429 is shown as in (A). The PaaK co-structures with phenylacetyl-adenylate are captured in alternate conformations (C,D). The residues involved in the binding pocket are shown as sticks in red and substrates are shown as sticks in green.
When the native phenylacetic acid substrate was bound, the paralogs displayed a difference in kinetics. BcePaaK1 showed a lower Km than BcePaaK2 (62 μM vs.150 μM, respectively), but a similar Kcat (250 min−1 and 300 min−1 for BcePaaK1 and BcePaaK2, respectively), which may be caused by a deeper aryl binding pocket (Figures 2C,D; Law and Boulanger, 2011). In addition, BcePaaK1 displayed wider substrate specificity than BcePaaK2 for phenylacetic acid derivatives. In Thermus thermophilus, Vmax of TthPaaK was 24 U/mg under the most active reaction conditions at a pH of 7.5–8.0 and temperature of 75°C. The Kcat was 20 s−1 per subunit, and Km was 6, 30, and 50 μM for phenylacetate, CoA, and ATP, respectively. Mg2+ is required for enzymatic reactions and can be substituted by Mn2+ with 90% activity (Erb et al., 2008).
The oxygenase complex catalyzes the most crucial step of the pathway, comprising PaaA, B, C, D, and E, inducing oxygen into the aromatic ring of II (Fernández et al., 2006). Through co-expression and pull-down assay of each component, PaaAC, PaaBC, and PaaABC were confirmed to form stable complexes, while PaaD and PaaE could not be detected (Teufel et al., 2010; Grishin et al., 2011). Further in vitro reconstitution experiments demonstrated that PaaA, B, C, and E subunits are required for the oxidation reaction and formation of epoxide (Grishin et al., 2011). The reductase subunit PaaE, catalytic subunit PaaA, structural subunit PaaC, and bridging subunit PaaB form the overall complex composition PaaA2B3-4C2E1, which contains six iron atoms, two of which belong to the iron–sulfur cluster of PaaE and four to the two molecules of PaaA (Teufel et al., 2012). In their studies, a model for PaaABCE catalysis and catalytic di-iron center-state conversion was proposed. First, the ground-state di-iron core was reduced in an NADPH-dependent manner. The reduced diferric compound could then interact with oxygen, producing a high-valent intermediate or alternatively abstracting the epoxy-oxygen to reverse the reaction. Finally, the high-valent intermediate oxidate II produced III and a ground state di-iron core (Teufel et al., 2012).
In E. coli, the heterotetramer PaaAC (PDB:3PW8) is similar to PaaA and PaaC subunits (Figure 3A; Grishin et al., 2011). Briefly, the core consisted of six long α-helices: B, C, E, F, G, and H, as previously reported (Grishin et al., 2011; Figure 3B). Helices B and C from each subunit form an antiparallel four-helix bundle that is involved in heterodimer oligomerization (Figure 3C). The substrate binds only to the PaaA subunit, located in a tunnel extending from the protein surface toward the center. The structure of the PaaACB complex from Klebsiella pneumonia (PDB:4IIt) reveals a 2:2:2 combination ratio, and the heterohexameric PaaACB is more likely in a PaaAC-(PaaB)2-PaaAC pattern (Grishin et al., 2013). In this model, the PaaB dimer was located in the middle part, forming a plateau for the two PaaAC heterodimer assemblies (Figure 3D; Grishin et al., 2013). The PaaAC subcomplex showed the same conformation as the heterodimer, with the PaaB subunit binding to both PaaA and PaaC subunits in a cleft near the PaaA/PaaC interface (Figure 3D).
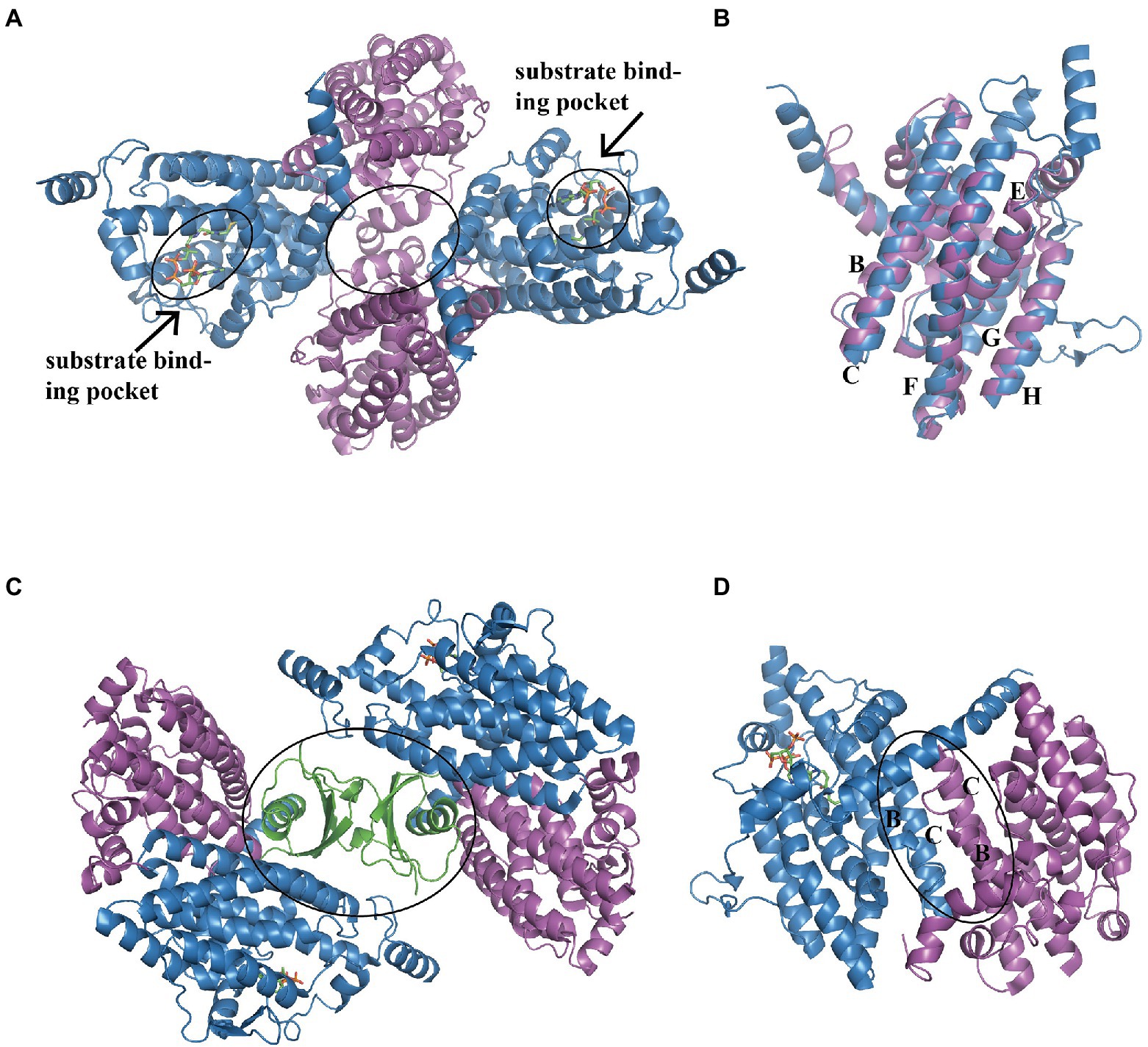
Figure 3. Overall structure of the PaaACB complex. (A) The overall structure of heterotetramer EcoPaaAC (PDB:3PW8), the substrate acetyl-CoA in binding pocket is shown as sticks, the complex is formed by interactions in PaaC, circled with a solid line. (B) Structure superposition of PaaA and PaaC; the core helixes are labeled as reported previously (Grishin et al., 2011). (C) The PaaAC heterodimer formed by interactions between helixes B and C in each subunit. (D) The (PaaACB)2 heterohexamer from Klebsiella pneumoniae (PDB:4IIt) maintained by the dimerization of PaaB, constructing a β-barrel in the center part. PaaA, PaaC, and PaaB are in blue, magenta, and green, respectively.
The end-products measured by LC–MS demonstrated that PaaA, B, C, and E subunits constitute the optimum reaction mixture, while the PaaD subunit showed no effect on the reaction in vitro (Teufel et al., 2010; Grishin et al., 2011). However, previous E. coli knockout mutant studies indicated that PaaD is essential for this reaction in vivo, suggesting that PaaD may induce maturation of the monooxygenase complex, rather than have a direct involvement in catalysis. In addition, PaaB is important for product concentration, which shows a more than 100-fold reduction in its absence (Grishin et al., 2011).
Using 13C-labeled substrates and detecting by 13C-NMR spectroscopy, the Py2PaaABCE complex catalyzed the reaction induced by the compound II, mediating NADPH consumption and depending strictly on oxygen with approximately 1 μmol min−1 mg−1 protein (Teufel et al., 2010). In another study, the features of PaaABCE were well described (Teufel et al., 2012). Under optimum conditions of 30°C and pH 8.0, epoxidase compound activity was approximately 1.0 U mg−1 with NADPH and 0.05 U mg−1 with NADH, and apparent Km were 23 μM, 6 μM, 3 μM, and 17 μM for NADPH, the compound II, O2, and the compound III, respectively. PaaE belongs to the class IA reductases, associated with dioxygenases, with an N-terminal NADPH- and FAD-binding domain and a C-terminal [2Fe-2S] ferredoxin-like domain (Figure 4). The absorption spectrum of MBP-EcoPaaE showed that the spectrum maxima closely resemble those of spinach [2Fe-2S] ferredoxin (Grishin et al., 2011). PaaE is thought to transfer electrons from NADPH through FAD and the iron–sulfur cluster to the iron atoms in the active center of PaaA. The oxidoreductase activity of SpePaaE was demonstrated by the nitroblue tetrazolium (NBT) reduction assay, where NBT serves as an electron acceptor and specifically utilizes NADH to transfer electrons. In addition, flavin obtained from heat-denatured PaaE and confirmed by HPLC provided evidence of the presence of the FAD-binding domain (Niraula et al., 2010). None of the structures of PaaE are available. The phthalate dioxygenase reductase from Burkholderia cepacia (PDB code 2PIA, Correll et al., 1992) is a structural homolog with 22% sequence identity.
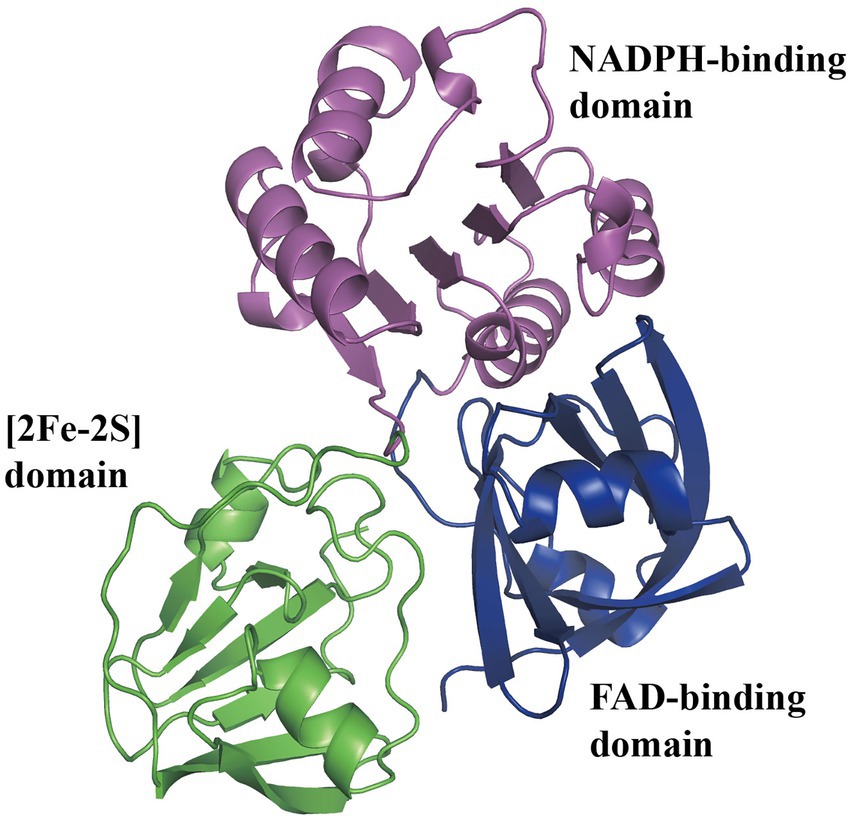
Figure 4. Predicted structure of PaaE. The overall predicted structure of EcoPaaE from AlphaFold. The PaaE is formed by three domains individually, N-terminal FAD-binding domain, NADPH-binding domain, and C-terminal [2Fe-2S] ferredoxin-like domain, shown in blue, magenta, and green, respectively.
After the compound III (Figure 1) is produced during the monooxygenation of the aromatic ring, PaaG isomerizes and finally introduces an α,β-unsaturated CoA-thioester motif. PaaG belongs to the enoyl-CoA hydratase/isomerase family, together with PaaF (late steps). These family members strictly depend on CoA-thioester substrates because of the negatively charged transition state in the form of thioester enolate, stabilized by a dedicated pocket at the active site (Spieker et al., 2019). To evaluate the kinetic parameters of Py2PaaG and TthPaaG, substrates at different catalytic steps were identified using HPLC, spectrophotometry, or UPLC–MS. TthPaaG showed a specific activity of 138 U mg−1 in isomerizing the compound III in the early steps of PAA pathway and 182 U mg−1 for Py2PaaG. As native cis-VII is unstable, the more stable isomer, trans-VII, which could be converted to the same product, was used to evaluate PaaG activity in the late PAA pathway. The specific activity was 7.6 × 10−3 and 116, Km was 564 and 142 μM, Kcat was 3.5 × 10−3 and 60 × 10−3 s−1, and catalytic efficiencies in Py2PaaG and TthPaaG were 6 × 10−6 and 4 × 10−4 s−1 μM−1, respectively (Spieker et al., 2019). Additionally, an aspartate side chain (D136) at the catalytic site of Pseudomonas sp. Y2 PaaG acts as a proton relay amino acid and substantially affects enzyme functionality in mutants (Spieker et al., 2019).
The structure of PaaG reveals the fold features of the enoyl-CoA hydratase/isomerase (crotonase) superfamily. The first report of PaaG structure was based on Thermus thermophilus in 2009 (Kichise et al., 2009). A recent study revealed the isomerase TthPaaG in complex with its native ligands, providing a comprehensive view of PaaG-isomerizing substrates in different steps. The disk-shaped trimer PaaG is related to local three-fold symmetry (Figure 5A; Spieker et al., 2019). During the interaction with different ligands, the adenine moiety in CoA is located at the bottom of an open binding pocket, and the acyl moiety of the ligands is oriented toward a shallow hydrophobic site (Figure 5B).
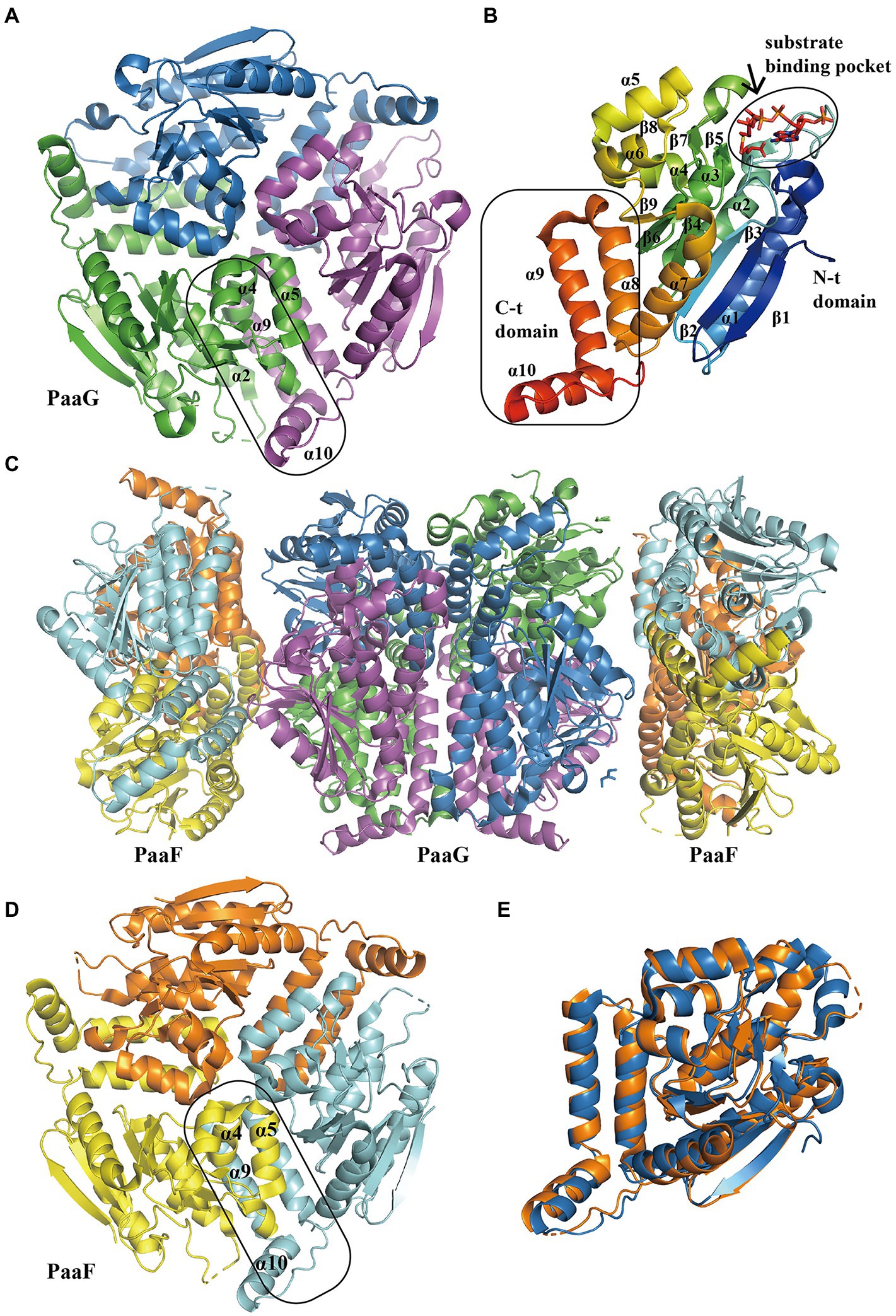
Figure 5. Overall structure of PaaG and PaaF. (A) The overall structure of trimer TthPaaG (PDB:6SLB). The interface is circled with a solid line. The different chains are shown in blue, violet, and green, respectively. (B) The secondary structure elements in PaaG monomer with the substrate trans-3,4-didehydroadipyl-CoA binding in N-spiral domain. The N-terminal and C- terminal domains are circled with a solid line. The monomer is colored as the rainbow and the substrate is shown as sticks. (C) The overall structure of EcoPaaFG complex (PDB:4FZW). The dimer of trimer PaaG is covered with two PaaF trimers. PaaF chains are shown in orange, yellow, and cyan, and PaaG is shown in the same colors as in (A). (D) The overall structure of trimer PaaF, reveals a similar shape and interaction site as PaaG trimer as PaaG. (E) The structure alignment between PaaF and PaaG monomer reveals a similar crotonase fold feature.
In the next two steps, the bifunctional enzyme PaaZ catalyzes the hydrolysis of the compound IV to V (Figure 1), and oxidation of the terminal aldehyde group, finally producing the compound VI. Previous studies have shown that EcoPaaZ is crucial for the removal of toxic intermediates in the early steps of the PAA pathway, with a specific activity of approximately 20 μmol min−1 mg−1 protein in the compound IV ring cleavage (Teufel et al., 2011). A subsequent study determined the rate of PaaZ-catalyzed hydrolysis to be 33 U mg−1 via a photometric test (Teufel et al., 2011). They also reported that the proposed open-chain aldehyde, after hydrolytic ring fission, could rapidly form a seven-membered carbon ring through Knoevenagel-type condensation and finally rearrange into a more stable enol-form compound, inhibiting PaaZ activity (Teufel et al., 2011). In addition, the dehydrogenase activity was approximately 32 U mg−1 by measuring NADPH formation using a photometric assay. The Km was 11 μM and 56 μM for catalyzing the compound IV and NADP+ respectively (Teufel et al., 2011).
The tri-lobed hexamer PaaZ was maintained by the inner core formed by the C-hydratase domain of three dimers. The hydratase domain consists of a mixture of α-helices and β-strands, where the α-helices are involved in dimer oligomerization (Figures 6A,C,D). The dehydrogenase domain was further divided into three regions: co-factor binding, catalytic, and dimerization motifs (Figure 6B). Previous studies have shown that the product of the hydratase domain is a substrate for dehydrogenase (Teufel et al., 2011). A recent study of PaaZ structures with open-ring mimics, octanoyl CoA (Figure 6C) and crotonyl-CoA (Figure 6D), provides a possible model for substrate transfer. The bifunctional enzyme PaaZ presents a positively charged surface at the entrance of the hydratase and dehydrogenase domains, which is complementary to the negatively charged coenzyme A in the substrate. Through this tunnel, by electrostatic pivoting of the CoA part, the key intermediate can transfer from one active site to another internally without being released into the bulk solvent (Sathyanarayanan et al., 2019).
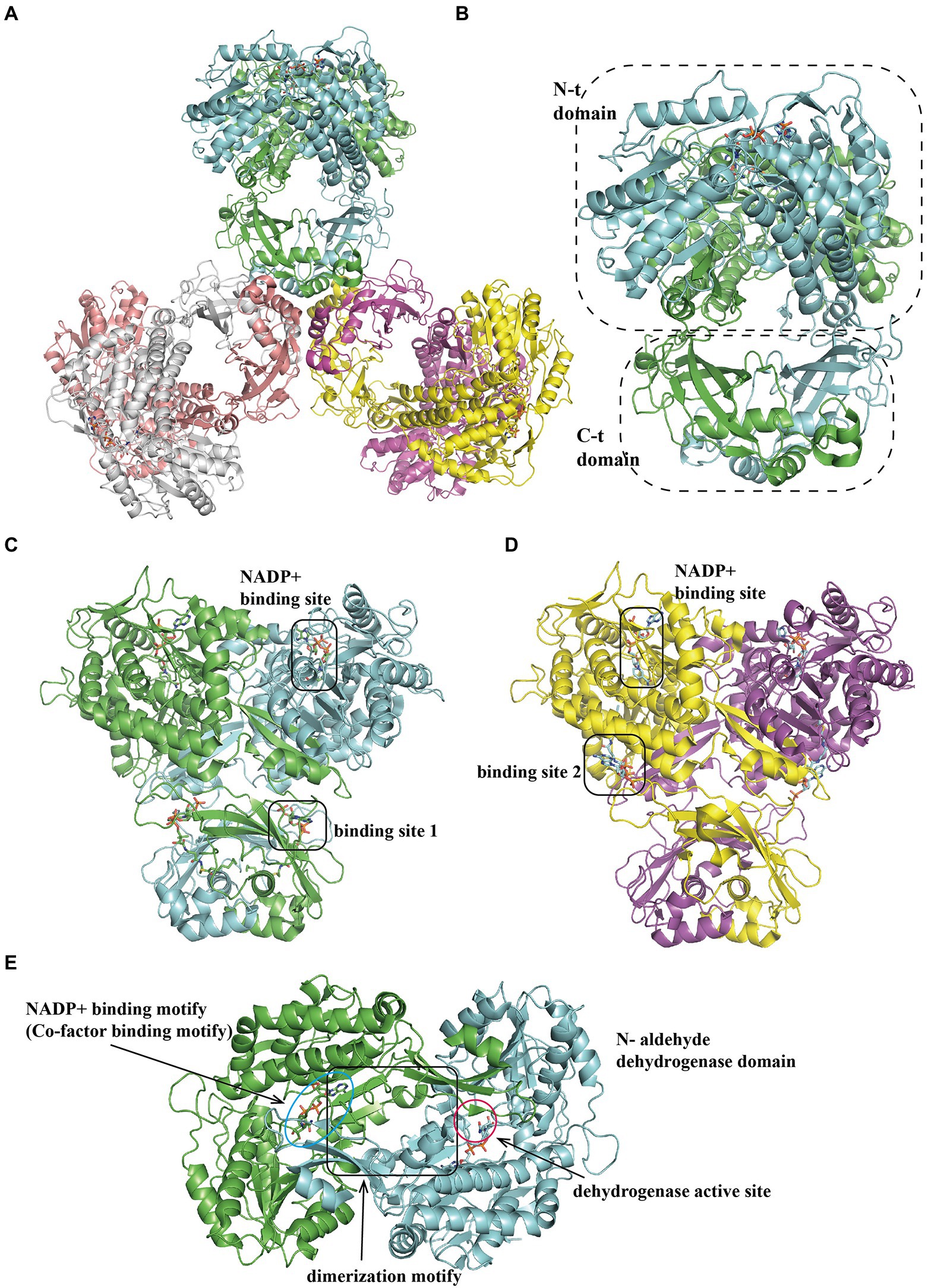
Figure 6. Overall structure of PaaZ. (A) The overall structure of the tri-lobed architecture hexamer EcoPaaZ (PDB:6JQM) is shown in different colors for each chain. The architecture unit PaaZ dimer reveals an N-terminal aldehyde dehydrogenase domain and the C-terminal R-specific hotdog fold hydratase domain, circled with a dotted line. For clarity, in the substrate binding domain, the rotated views from (A) are shown in (B) with OCoA (PDB:6JQN) and with CCoA (PDB:6JQO; C) in different binding site 1 or 2, respectively. The NADP+-binding site is the same in (B) and (C). (D) The N-terminal dehydrogenase domain can be divided into three sub-domains: co-factor binding, catalytic and dimerization motif, shown with lines and annotation, respectively.
Interestingly, PaaZ contains only an aldehyde dehydrogenase domain in several phenylacetate-degrading organisms such as Aromatoleum aromaticum. A hotdog-fold hydratase encoded by a gene outside the paa operon has been identified to perform the compound IV hydrolysis, capable of hydrating crotonyl-CoA with high activity, which could replace the missing hydratase function in PaaZ (Teufel et al., 2011).
Multiple epoxides produced during the Paa catabolic pathway are toxic to cells, and several mechanisms have evolved in bacteria to control these toxic metabolites.
The first epoxides generated in the PAA pathway are mediated by the PaaABCE complex epoxidation of the aromatic ring. The oxygenase complex itself can also perform reverse reaction-deoxygenation to yield the compound II (Figure 1), as discussed above. The PaaABCE complex assists in excess epoxide removal when the inadequate processing by downstream enzymes, PaaG and PaaZ, leads to substrate accumulation (Teufel et al., 2012).
To protect PaaABCE from overloading and avoiding subsequent toxic accumulation, PaaI catalyzes a reverse reaction to the compound II ligase PaaK, removing excess the compound II (Teufel et al., 2012). PaaI exhibited a narrow substrate specificity to the compound II in relevant pathway intermediates, with an activity of 1.6 U mg−1 measured at 22°C (Teufel et al., 2012). The EcoPaaI Km was 9.6 μM and Kcat was 4.1·10−1 s−1 under optimum conditions of 25°C and pH 7.5. And for AevPaaI, the Km and Kcat for II was 9.6 μM and 4.1·10−1 s−1, respectively (Song et al., 2006). In another study, enzyme kinetic analysis revealed that SphPaaI shows high activity against decanoyl for the compound II with Km of 90 μM, Kcat of 6.5 s−1, and specificity constant of 7.2 × 104 m−1 s−1, while a higher activity was observed for medium-chain fatty acyl-CoA substrates (decanoyl/C10-CoA) with corresponding values of 183 μM, 32.8 s−1, and 1.8 × 105 m−1 s−1, respectively (Khandokar et al., 2016).
The representative hotdog-fold structure for PaaI consists of a core α-helix enveloped by an aβ-sheet formed by six strands on one side (Figure 7A). The native molecule works as a homotetramer; the β-sheet from two monomers stack together to form a continuous 12-stranded antiparallel β-sheet (Figure 7B), which associates back-to-back with a second dimer (Figure 7C). The core α-helix also participates in the oligomerization of dimers. The ligand acyl-CoA is located on the dimer surface and binds to the acyl-thioester moieties (Song et al., 2006).
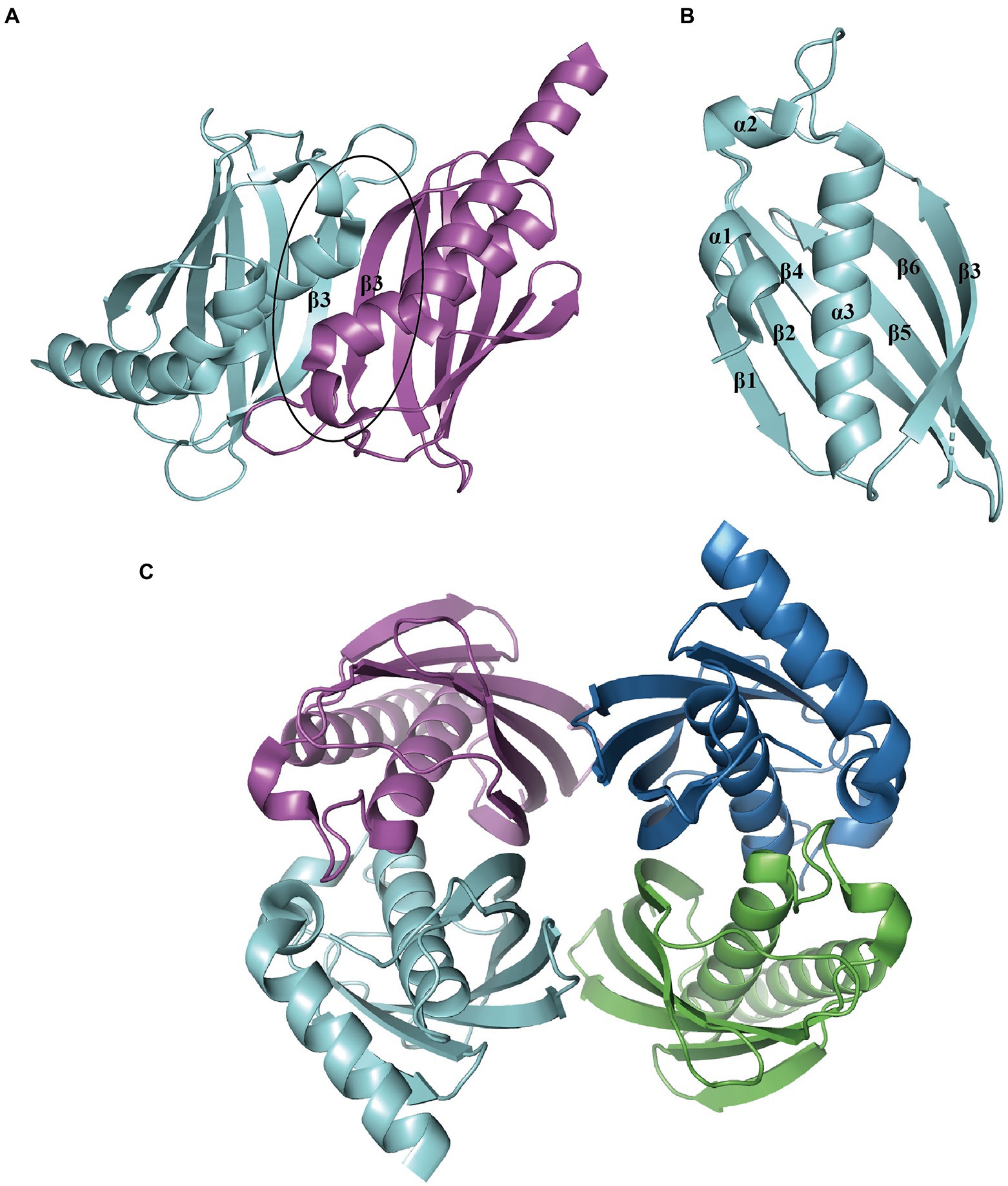
Figure 7. Overall structure of PaaI. (A) The PaaI reveals a hotdog fold shape with a core α-helix (α3) enveloped by a six-strand β-sheet (β1-β6). (B) The architecture unit PaaI dimer is formed by the interaction between β-sheets in each monomer. (C) The overall structure of tetramer TthPaaI (PDB:1J1Y) is shown in different colors for each chain.
Thioesterase PaaY is necessary for the efficient degradation of PAA in E. coli. It serves as a regulatory protein in the PAA pathway, which could specifically hydrolyze the compound XI, an inhibitor to PaaZ when NADP+ deficiency occurs and labile aldehyde oxidation is impaired in the cell, with an activity of 7.6 U mg−1 and Km 35 μM at 22°C (Teufel et al., 2011, 2012) The disruption of EcoPaaY and PpuPaaY showed no effect on PAA catabolism (Ferrández et al., 1998; Olivera et al., 1998), but revealed an obvious lag phase in growth and morphological changes when using PAA as the sole carbon source (Fernández et al., 2014). EcoPaaY associates with trimers containing Ca2+ and Zn2+ ions and shows a wider substrate range of CoA derivatives. The optimal reaction conditions for EcoPaaY were pH 8.0 and 45°C; in addition, thioesterase activity increased in the presence of Co2+ whereas Cu2+, Mn2+, and Ni2+ had an inhibitory effect (Teufel et al., 2010; Fernández et al., 2014). A homolog of PaaY with 33% similarity from Geobacillus kaustophilus was structurally characterized (PDB:3VNP) as a homotrimer (Figure 8A; Paper unpublished). A new function of PaaY and its connection with the regulation of the paa gene cluster need to be investigated.
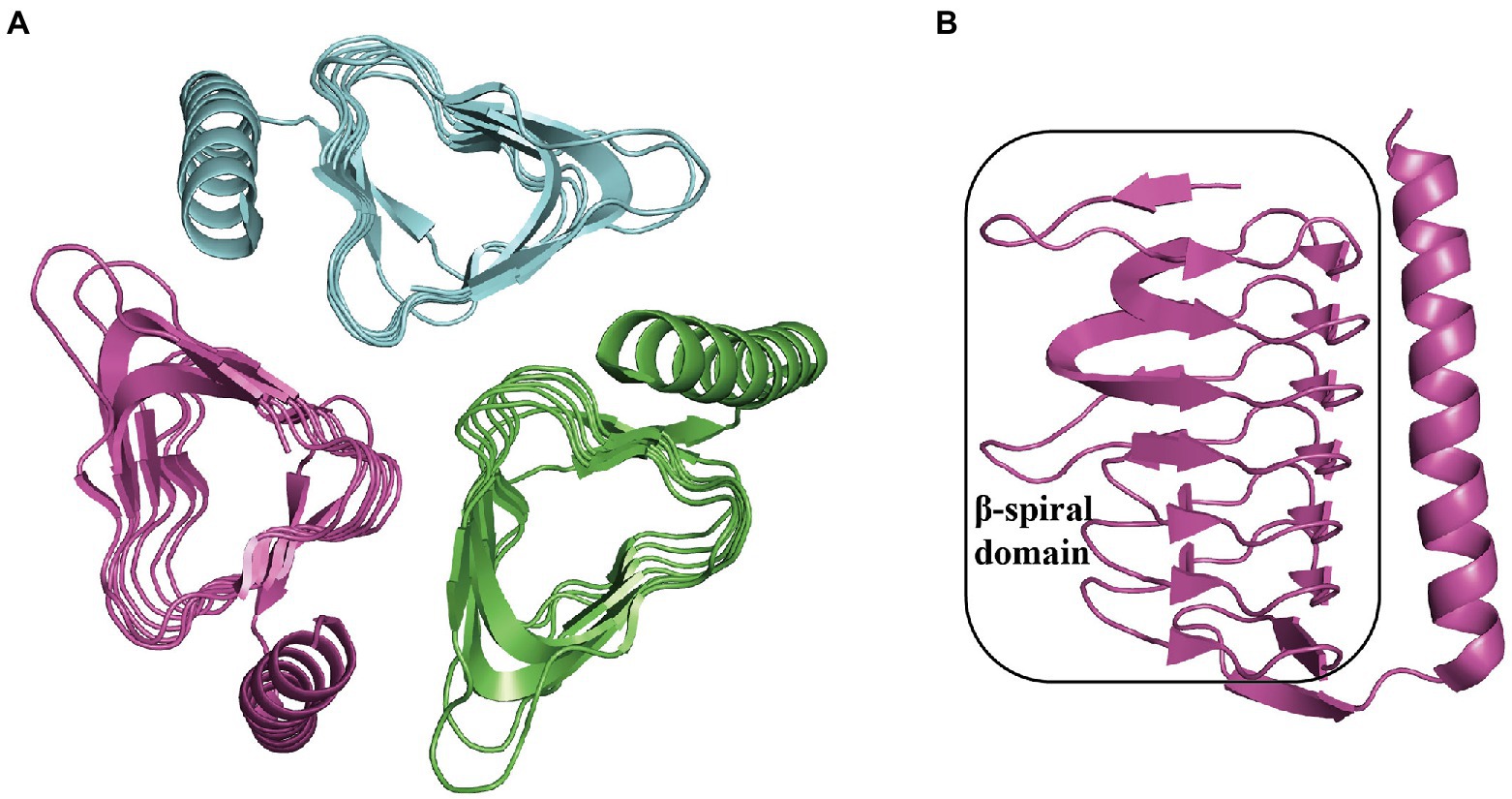
Figure 8. Overall structure in PaaY. (A) The overall structure of trimer GkaPaaY (PDB:3VNP) is shown with a different color for each chain. (B) The PaaY consists of a large β-spiral containing one C-terminal α-helix connected by an extraβ-strand.
The crucial step in the late part of Paa pathway is the catalysis of long-chain intermediate fission, mediated by thiolase PaaJ. PaaJ serves as a β-ketoadipyl-CoA thiolase to clear the C8-intermediate formed by PaaZ, finally forming a C6-intermediate for subsequent processes.
TthPaaJ (PDB:1ULQ) belongs to the thiolase superfamily; the overall structure of TthPaaJ reveals a two-lobe-tetramer linked by a central β-barrel composed of four monomers (Figure 9A). The N- and C-termini in the monomer share a similar “βαβαβαβ” topology, in which the β-strands fold into a five-stranded or four-stranded mixed-sheet sandwich covered by α-helices (Pfündel, 2021; Figure 9B). A loop domain between Nβ4 and Nβ5 extending from the N-terminal β-sheet is mainly composed of α-helices and folds on top of the thiolase core domain, involving CoA in binding and determining substrate specificity (Kiema et al., 2014; Figures 9C,D). In addition, this loop is crucial for the function of human mitochondrial 3-ketoacyl-CoA thiolase, a homolog of PaaJ (PDB:4CJ2; Kiema et al., 2014; Figure 9D).

Figure 9. Overall structure of PaaJ. (A) The overall structure of TthPaaJ (PDB: 1ULQ) reveals a tetramer shown in a different color for each chain with two lobes and a central β-barrel composed of four monomers circled with a solid line. (B) The PaaJ monomer is colored by secondary structure, red, yellow, and green for the α-helix, β-strands, and loop, respectively. The N-terminal and C-terminal region reveals a similar βαβαβαβ topology (the secondary structure elements in the N-terminal are annotated). (C) The rotated view in (B), reveals a sandwich architecture where the N-terminal and C-terminal β-sheet are covered by two-layer α-helices. N, C and M for N-terminal, C-terminal and medial, respectively. (D) The extended loop domain between Nβ4 and Nβ5 in the N-terminal β-sheet is involved in CoA binding. The loop is shown in magenta while the rest is in cyan, and the binding pockets are circled with a solid line.
The S-specific hydratase PaaF, together with PaaG, belong to the crotonase-fold superfamily. In the next two steps, PaaG isomerizes the unsaturated thioester and the PaaF hydrated compound, finally generating the compound IX (Figure 1). The features of the PaaG isomerase are discussed above. Previous studies using mass spectrometry have shown that purified PaaF can catalyze the reversible conversion of substrates between the compound VIII and the compound IX. In addition to PaaABCE, PaaF-PaaG was the only stable complex in the late steps of the PAA pathway, which may be an evolutionary adaptation to speed up subsequent reactions in the pathway (Grishin et al., 2012). The PaaFG complex contained a stack of four homotrimeric discs assembled by two PaaF discs in the center sandwiched between PaaG discs at each end (Figure 5C). The PaaF monomer exhibits a crotonase fold that is highly similar to that of PaaG (Figure 5D), and the active sites are located on the external surfaces of the disc structure, similarly to those of PaaG (Figure 5E; Grishin et al., 2012).
The 3-hydroxyadipyl-CoA dehydrogenase PaaH oxidizes substrates the compound IX to X, depending on NAD+ (Teufel et al., 2010). The EcoPaaH trimer (PDB:3MOG) has been observed (Figure 10A; Paper unpublished). The monomer revealed a sandwich shape with three regions, a medial region located at the interface of the trimer packaged by two similar domains (N-terminal domain and a reduced domain, mainly consisting of a β-sheet surrounded by α-helices) on both sides. The medial region consists of two similar but discontinuous parts, both mainly formed by five α-helices (Figure 10B). Finally, the last step of the pathway, cleaving of X is also catalyzed by PaaJ. Using an HPLC chromatogram measuring end-products in vitro, EcoPaaJ was confirmed to generate acetyl-CoA and succinyl-CoA due to thiolytic fission of β-ketoadipyl-CoA (Nogales et al., 2007).
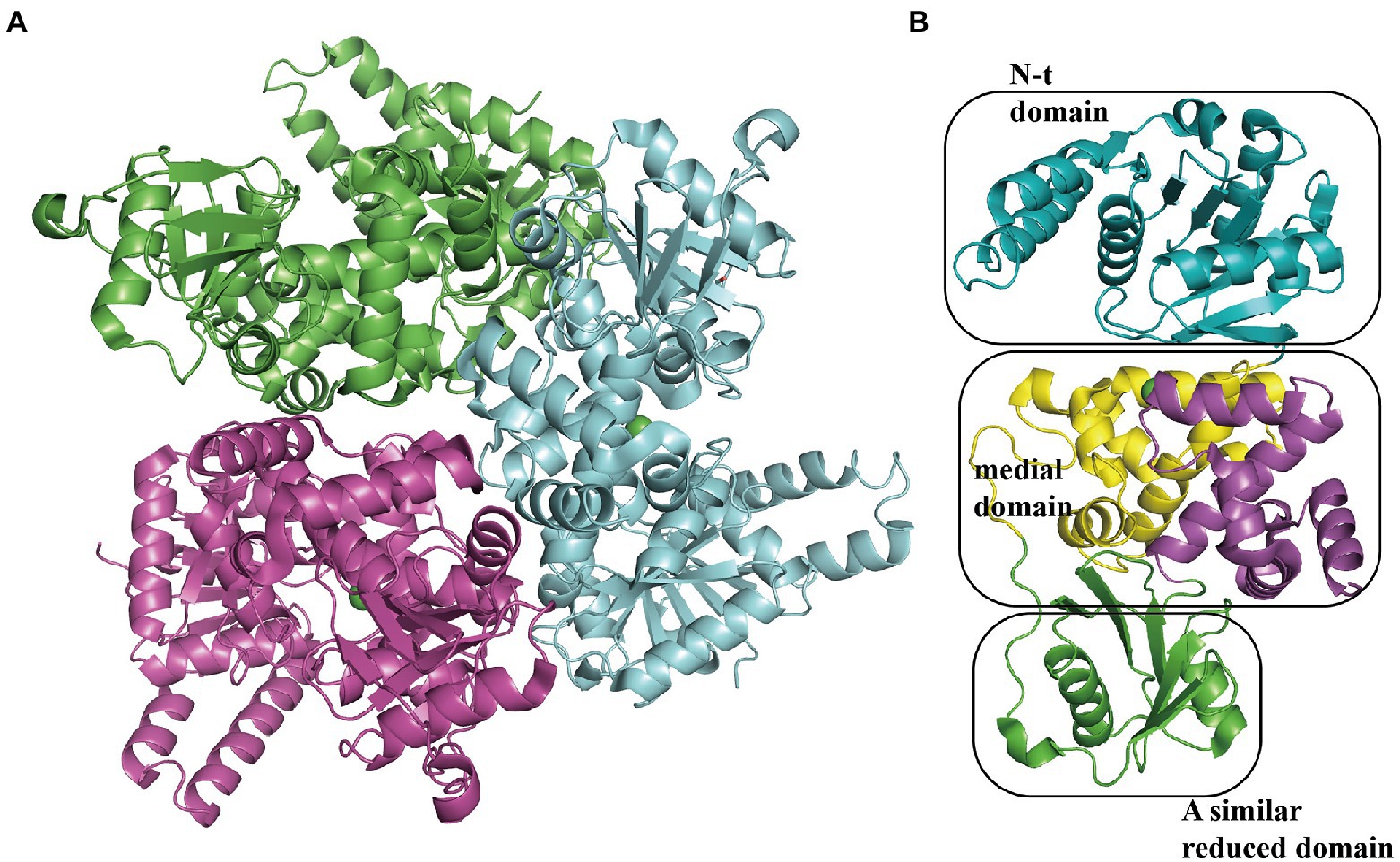
Figure 10. Overall structure of PaaH. (A) The overall structure of the EcoPaaH trimer (PDB:3MOG). The PaaH trimer reveals a central symmetric structure in space, shown in green, magenta, and teal by chains. (B) The PaaH monomer is constructed from three regions, N-terminal domain, and a reduced domain (mainly consisting of a β-sheet surrounded by α-helices) in teal and green, respectively. In addition, a medial region consists of two discontinuous parts sharing a similar five-helixes structure which could be seen as a ‘dimer’ conformation, shown in yellow and magenta in each part, respectively.
GntR-type and TetR-type are two existing systems that regulate PAA catabolism, both in response to II as an inducer. In E. coli, P. putida, and other Pseudomonas species, PaaX is a representative GntR-type regulatory protein (Ferrández et al., 2000; García et al., 2000; del Peso-Santos et al., 2006). Conversely, PaaR is a TetR-type protein present in T. thermophilus, C. glutamicum, and B. cenocepacia (Hamlin et al., 2009; Sakamoto et al., 2011; Chen et al., 2012).
In E. coli, the Pz and Pa promoters control two divergently transcribed operons, paaZ and paaABCDEFGHIJK, respectively, which are negatively regulated by PaaX and are transcribed on an adjacent Px operon (Ferrández et al., 2000). The Px promoter in charge of the paaXY operon expressing the paaX regulatory gene and thioesterase PaaY is repressed by its own product PaaX, based on the steric hindrance of RNAP binding to the Px promoter (Fernández et al., 2014). In addition, II could specifically inhibit the binding of PaaX to the target sequences of Pa or Pz, confirming the first intermediate the compound II in the PAA pathway as the true inducer, but not PAA (Ferrández et al., 1998; Fernández et al., 2014). Jccs1PaaX (PDB 3 L09; Figure 11A) revealed an N-terminal winged helix-turn-helix (wHTH) DNA-binding domain, a dimerization motif, and a C-terminal extended domain (Figure 11B). The binding base sequence of EcoPaaX is TGATTC(N27)GAATCA (Kim et al., 2004a,b), and a similar sequence was found in Py2PaaX (del Peso-Santos et al., 2006). The specific C-terminus binds II to activate the N-terminal domain (Ferrández et al., 2000). PaaX competes with RNA polymerase to bind to the regulatory Px and Pz promoters, but the mechanism of binding to the Pa promoter is different (Fernández et al., 2014). The complex structure of PaaX and its binding operator bases have not yet been elucidated.
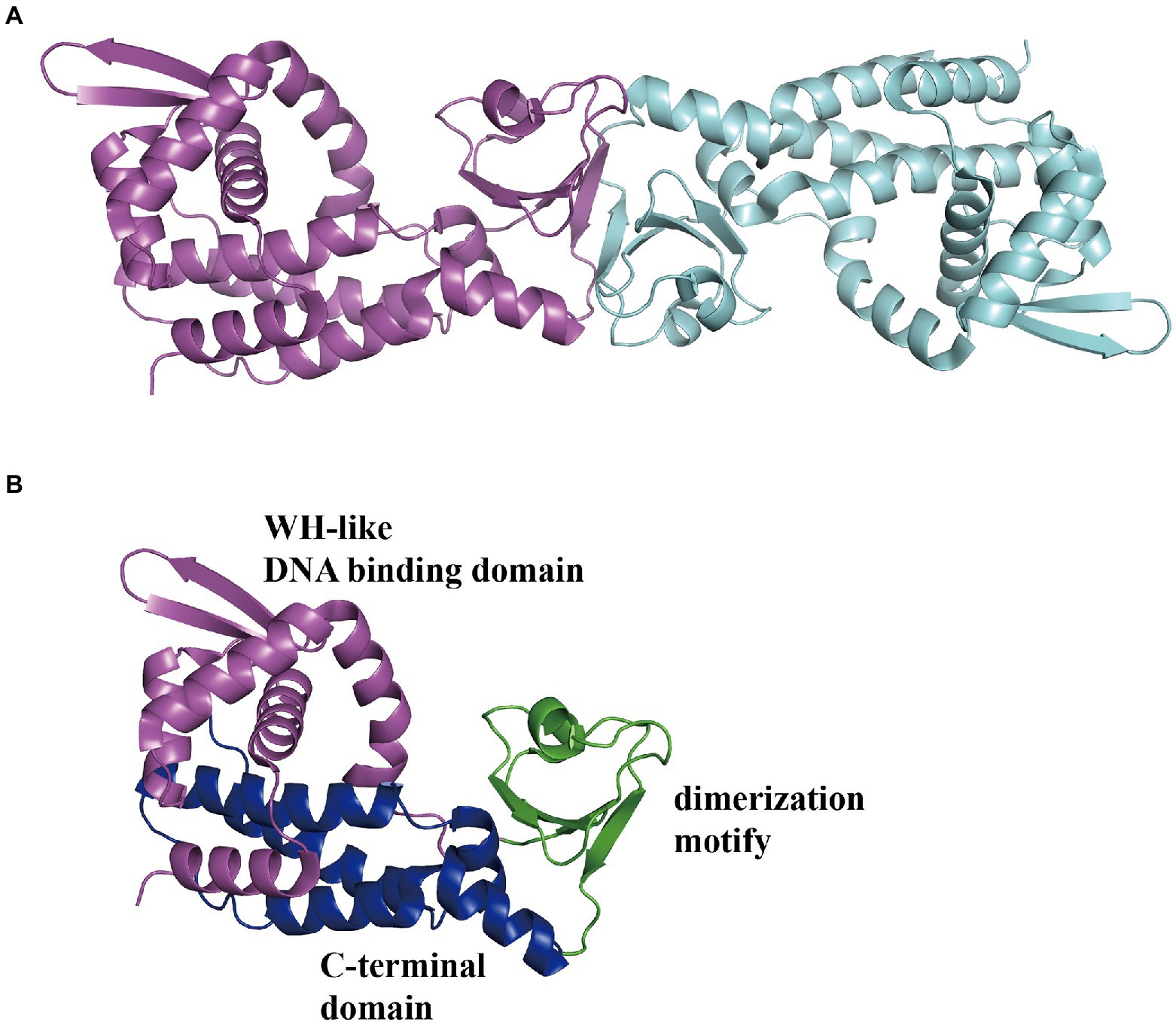
Figure 11. Overall structure of PaaX. (A) The overall structure of dimer Jccs1PaaX (PDB:3 L09) is shown in a different color for each chain. (B) The PaaX could be divided into three regions as an N-terminal Winged helix-like DNA-binding domain, a dimerization motif, and a C-terminal extended domain consisting of five α-helices, shown in magenta, green, and blue, respectively.
In B. cenocepacia, PA-related genes are located in three separate clusters: paaABCDE, paaFZJGIJK1, and paaHK2. A regulatory gene, PaaR, was identified downstream of the paaABCDE gene cluster oriented in the same direction, but in a separate transcriptional unit different from paaABCDE (Hamlin et al., 2009). The N-terminal region of BcePaaR shows high similarity to the TetR-type regulator of the multi-drug efflux pump EcoAcrR (Hamlin et al., 2009). Through insertional mutagenesis, PaaR was confirmed to be a negative regulator of the promoters of PpaaA, PpaaH, and PpaaZ (Hamlin et al., 2009). In addition, a 15 bp inverted repeat (IR) sequence site serving as the operator site in PpaaA, PpaaH, and PpaaZ (ACCGACCGGTCGGTT in PpaaA and PpaaZ, ACCAACCGGTCGGTT in PpaaH) was validated by constructing eGFP translational fusion plasmids and reporter activity measurements (Hamlin et al., 2009). In a subsequent study, electrophoretic mobility shift assay (EMSA) was used to confirm the binding capacity between PaaR and target sequences (Yudistira et al., 2011). In addition, the compound II, but not the compound I, can dissociate PaaR from the target promoter regions (Yudistira et al., 2011).
Another study reported the same conclusion regarding the features of Thermus thermophilus TthPaaR (Sakamoto et al., 2011). Using the BIAcore biosensor assay, TthPaaR was confirmed to bind two DNAs with a consensus sequence of CNAACGNNCGTTNG and similar values of association rate constant, dissociation rate constant, and dissociation constant at 9.3 × 105 M−1 s−1, 1.0 × 10−3 s−1, and 1.1 nM, and 9.8 × 105 M−1 s−1, 0.9 × 10−3 s−1, and 0.9 nM, for TthPaaR binding site 1 and 2, respectively (Sakamoto et al., 2011). The DNA binding ability of PaaR decreased in a concentration-dependent manner in the compound II. In addition, ITC results revealed that the compound II binding with PaaR was Kd at 4.1 × 107 M−1, and the number of binding sites on PaaR was approximately 1.3 (Sakamoto et al., 2011). The putative structure of TthPaaR revealed a typical TetR family member with a predicted N-terminal DNA-binding domain consisting of a helix-turn-helix motif with a positively charged surface (Sakamoto et al., 2011).
The well-studied PAA pathway, as the final degradation mechanism for multiple aromatic hydrocarbons, has been widely applied to specific products generated in vivo via the genetic engineering technology (Chen et al., 2018; Li et al., 2019). Additionally, our knowledge of its native role in bacteria remains limited. Here, we concluded that the PAA pathway plays a role in bacterial pathogenicity and antibiotic resistance.
The PAA pathway is closely linked to antibiotic resistance. The side reaction intermediate products XI of the PAA catabolism is the proposed universal precursor for tropone natural products and their derivatives (Teufel et al., 2011; Brock et al., 2014; Duan et al., 2020, 2021), among which tropodithietic acid (TDA) is a broad-spectrum antimicrobial compound (Geng et al., 2008; Henriksen et al., 2022). TDA also works as a signaling molecule, which influences phenotypic traits like motility and biofilm formation, and gene expression of other bacteria and antibiotic production in the producer (Geng et al., 2008; Beyersmann et al., 2017; Duan et al., 2020).
Penicillin G acylase (PGA), which hydrolyzes penicillin G to 6-aminopenicillanic acid (6-APA) and PAA, is thought to serve as a scavenger of many different natural esters and amides of PAA or its derivatives in EcoPaaX has been shown to specifically bind the Ppga promoter This binding effect could be inhibited by PA-CoA, as discussed above (Galán et al., 2004; Kim et al., 2004a,b). Despite PGA showing no functions in bacterial antibiotic resistance, these findings lay the foundation for the connection between the PAA pathway and antibiotic resistance.
In B. cenocepacia, global gene expression analysis suggested that multiple enzymes in the PAA degradation pathway are upregulated in response to meropenem exposure, indicating a potential connection between PAA and antibiotic resistance (Sass et al., 2011). Coincidentally, the PAA degradation pathway in A. baumannii is significantly upregulated in response to ceftazidime (Alkasir et al., 2018) and lethal concentrations of ciprofloxacin (Kashyap et al., 2021), suggesting that the PAA pathway is a suitable pathogen control target. Furthermore, paa genes were found to be downregulated in a ∆adeIJK mutant of A. baumannii, with AdeIJK efflux as a broad-spectrum pump, especially for amphiphilic compounds (Damier-Piolle et al., 2008; Leus et al., 2020).
A recent study provided more evidence that treatment with antibiotics at a subinhibitory concentration led to an approximately 7-fold increase in the expression of paaA and paaB to impact intracellular PAA levels in A. baumannii (Hooppaw et al., 2022). They also reported that PAA catabolism is important for A. baumannii in multiple antibiotic stress conditions, especially in the presence of cytoplasmic targets such as ciprofloxacin, erythromycin, and tetracycline, where the biofilm formation ability is repressed in the WT strain but not impacted in a deletion mutant strain of paaB (Hooppaw et al., 2022).
High levels of PAA inhibit the pathogenicity of the fungus Rhizoctonia solani; 7.5 mM PAA in the growth medium reduced the biomass to 50% (Bartz et al., 2012). In A. baumannii, a deletion mutant of paaE in a mouse septicemia model showed significantly attenuated virulence (Cerqueira et al., 2014). PA-CoA attenuated CepIR-regulated virulence in B. cenocepacia, suggesting that a metabolic signal can activate virulence in the absence of QS signaling molecules (Lightly et al., 2019).
Biofilms are formed by A. baumannii on abiotic and biotic surfaces to survive in human serum and infection, resistance to desiccation stress, and starvation in the nosocomial environment (Zeidler and Muller 2019). A recent study on the PAA pathway under antibiotic treatment in A. baumannii suggested that PAA could induce biofilm formation depending on the expression of Csu, a pili protein, which is one of the main determinants of biofilm formation (Hooppaw et al., 2022). The exogenous addition of PAA can reverse the inhibition of Csu during antibiotic treatment (Hooppaw et al., 2022).
Previous studies have demonstrated a connection between the PAA pathway and oxidative stress. Expression of the paa operon was downregulated in a deletion mutant A. baumannii strain of MumR, a transcriptional regulator involved in Mn2+ uptake and H2O2 tolerance (Green et al., 2020). A deletion mutant ΔpaaJKXYI in the PAA pathway, rather than WT, was more susceptible to the lethal effects of H2O2, but not restricted to growth, which may occur in high concentrations of H2O2 (Green et al., 2020).
The quorum sensing (QS) system that works on the production and detection of signaling molecules is vital in bacterial intercellular communication; further, this system can bind the transcriptional regulator to activate the expression of virulence factors in several opportunistic pathogens. The relationship between the PAA pathway and bacterial pathogenicity has been well described in the opportunistic pathogen Burkholderia cenocepacia, which establishes persistent infections in humans the genetic diseases, cystic fibrosis and pulmonary cystic fibrosis (Law et al., 2008; Imolorhe and Cardona, 2011; Pribytkova et al., 2014; Lightly et al., 2019).
In insertional mutant B. cenocepacia strains, researchers first confirmed that PaaA and PaaE are important for infection through displaying attenuated pathogenicity in Caenorhabditis elegans without defects in growth and colonization in the host (Law et al., 2008). Subsequent studies have provided compelling evidence connecting the PAA pathway and QS system. Researchers have found that exogenous addition of PAA attenuates the pathogenicity of the ΔpaaABCDE strain, and further studies have demonstrated that the signal molecules in QS are inhibited, which is vital for virulence factor expression. Thus, PAA can participate in the QS-regulated pathogenic responses. Meanwhile, QS (CepIR)-regulated virulence traits, and cepI and cepR promoter activity were downregulated in the ΔpaaABCDE strain (Pribytkova et al., 2014). Taken together, these findings highlight a direct connection between PAA metabolism and QS-regulated pathogenic responses.
However, a recent study reported that the PaaK knockout mutant strain is more virulent, which is in contrast to the less virulent ΔpaaABCDE strain. By constructing deletions of the cepI and cepR genes in the PAA pathway mutant backgrounds, they suggested that there is an alternative signaling pathway to activate virulence in theΔpaaK1 paaK2 ΔcepR mutant, in which PAA-CoA or a derivative, but not PAA, is the central molecule (Lightly et al., 2019). Further studies are needed to uncover the complex internal regulatory mechanisms of the PAA pathway and the QS system.
In A. baumannii, the entire paa operon is controlled by GacSR, a two-component regulatory system sensor kinase, which is also a global virulence regulator responsible for inducing the expression of 674 genes, including biofilm formation and virulence-related genes, responsible for toxicity (Cerqueira et al., 2014). PAA was characterized from the culture supernatants of attenuated cytotoxicity Pseudomonas aeruginosa at high cell density, and PAA can downregulate the expression of virulence-related genes such as T3SS and its related regulatory genes (Wang et al., 2013). In a zebrafish infection model, PAA serves as a neutrophil chemoattractant, and the PAA pathway is crucial in tissue responses to acute infection, whose inhibition contributes to neutrophil response and invader clearance (Cerqueira et al., 2014; Bhuiyan et al., 2016; Kröger et al., 2016). Thus, the PAA pathway is involved in immune evasion and disease progression during the interaction between A. baumannii and its host.
Taken together, the PAA pathway shows the ability to crosstalk among multiple systems involved in bacterial pathogenicity and is a potential target in infection treatment. More work is needed to determine the regulatory relationship of the PAA pathway and measure its potential in restricting microbial infection.
MJ, WH, ZO, QS, and YW wrote the manuscript. WH made all the structural figures under the supervision of YW. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. 31870132, 82072237, and 82102402), Shaanxi Province Natural Science Funding (No. 2021JQ-381), and Institutional Foundation of the First Affiliated Hospital of Xi’an Jiaotong University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
BTEX – benzene, toluene, ethylbenzene, and xylene, CoA – coenzyme A, LC–MS –, NBT – nitroblue tetrazolium, PAA – phenylacetic acid pathway, PAHs – polycyclic aromatic hydrocarbons, PCBs – polychlorinated biphenyls, PGA – penicillin G acylase, QS – quorum sensing, TDT – tropodithietic acid,
Aklujkar, M., Risso, C., Smith, J., Beaulieu, D., Dubay, R., Giloteaux, L., et al. (2014). Anaerobic degradation of aromatic amino acids by the hyperthermophilic archaeon Ferroglobus placidus. Microbiology (Reading) 160, 2694–2709. doi: 10.1099/mic.0.083261-0
Alkasir, R., Ma, Y., Liu, F., Li, J., LVN,, Lv, N., et al. (2018). Characterization and transcriptome analysis of Acinetobacter baumannii persister cells. Microb. Drug Resist. 24, 1466–1474. doi: 10.1089/mdr.2017.0341
Bartz, F. E., Glassbrook, N. J., Danehower, D. A., and Cubeta, M. A. (2012). Elucidating the role of the phenylacetic acid metabolic complex in the pathogenic activity of Rhizoctonia solani anastomosis group 3. Mycologia 104, 793–803. doi: 10.3852/11-084
Beyersmann, P. G., Tomasch, J., Son, K., Stocker, R., Göker, M., Wagner-Döbler, I., et al. (2017). Dual function of tropodithietic acid as antibiotic and signaling molecule in global gene regulation of the probiotic bacterium Phaeobacter inhibens. Sci. Rep. 7:730. doi: 10.1038/s41598-017-00784-7
Bhuiyan, M. S., Ellett, F., Murray, G. L., Kostoulias, X., Cerqueira, G. M., Schulze, K. E., et al. (2016). Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 113, 9599–9604. doi: 10.1073/pnas.1523116113
Boll, M., and Fuchs, G. (1995). Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234, 921–933. doi: 10.1111/j.1432-1033.1995.921_a.x
Brock, N. L., Nikolay, A., and Dickschat, J. S. (2014). Biosynthesis of the antibiotic tropodithietic acid by the marine bacterium Phaeobacter inhibens. Chem. Commun. (Camb.) 50, 5487–5489. doi: 10.1039/c4cc01924e
Bugg, T. D., Ahmad, M., Hardiman, E. M., and Rahmanpour, R. (2011). Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28, 1883–1896. doi: 10.1039/c1np00042j
Burkhead, K. D., Slininger, P. J., and Schisler, D. A. (1998). Biological control bacterium Enterobacter cloacae S11:T:07 (NRRL B-21050) produces the antifungal compound phenylacetic acid in Sabouraud maltose broth culture. Soil Biol. Biochem. 30, 665–667. doi: 10.1016/S0038-0717(97)00170-3
Cao, B., Nagarajan, K., and Loh, K. C. (2009). Biodegradation of aromatic compounds: current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 85, 207–228. doi: 10.1007/s00253-009-2192-4
Carmona, M., Zamarro, M. T., Blázquez, B., Durante-Rodríguez, G., Juárez, J. F., Valderrama, J. A., et al. (2009). Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 73, 71–133. doi: 10.1128/MMBR.00021-08
Cerqueira, G. M., Kostoulias, X., Khoo, C., Aibinu, I., Qu, Y., Traven, A., et al. (2014). A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 210, 46–55. doi: 10.1093/infdis/jiu024
Chen, X., Kohl, T. A., Rückert, C., Rodionov, D. A., Li, L. H., Ding, J. Y., et al. (2012). Phenylacetic acid catabolism and its transcriptional regulation in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78, 5796–5804. doi: 10.1128/AEM.01588-12
Chen, X., Xu, M., Lü, J., Xu, J., Wang, Y., Lin, S., et al. (2018). Biosynthesis of tropolones in Streptomyces spp.: interweaving biosynthesis and degradation of phenylacetic acid and hydroxylations on the tropone ring. Appl. Environ. Microbiol. 84, e00349–e00318. doi: 10.1128/AEM.00349-18
Cook, S. D. (2019). An historical review of phenylacetic acid. Plant Cell Physiol. 60, 243–254. doi: 10.1093/pcp/pcz004
Correll, C. C., Batie, C. J., Ballou, D. P., and Ludwig, M. L. (1992). Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258, 1604–1610. doi: 10.1126/science.1280857
Dagley, S. (1975). A biochemical approach to some problems of environmental pollution. Essays Biochem. 11, 81–138.
Damier-Piolle, L., Magnet, S., Brémont, S., Lambert, T., and Courvalin, P. (2008). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 557–562. doi: 10.1128/AAC.00732-07
del Peso-Santos, T., Bartolomé-Martín, D., Fernández, C., Alonso, S., García, J. L., Díaz, E., et al. (2006). Coregulation by phenylacetyl-coenzyme A-responsive PaaX integrates control of the upper and lower pathways for catabolism of styrene by pseudomonas sp. strain Y2. J. Bacteriol. 188, 4812–4821. doi: 10.1128/JB.00176-06
Duan, Y., Petzold, M., Saleem-Batcha, R., and Teufel, R. (2020). Bacterial tropone natural products and derivatives: overview of their biosynthesis, bioactivities, ecological role and biotechnological potential. Chembiochem 21, 2384–2407. doi: 10.1002/cbic.201900786
Duan, Y., Toplak, M., Hou, A., Brock, N. L., Dickschat, J. S., and Teufel, R. (2021). A flavoprotein dioxygenase steers bacterial tropone biosynthesis via coenzyme A-ester Oxygenolysis and ring epoxidation. J. Am. Chem. Soc. 143, 10413–10421. doi: 10.1021/jacs.1c04996
El-Said Mohamed, M. (2000). Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 182, 286–294. doi: 10.1128/JB.182.2.286-294.2000
Elsden, S. R., Hilton, M. G., and Waller, J. M. (1976). The end products of the metabolism of aromatic amino acids by Clostridia. Arch. Microbiol. 107, 283–288. doi: 10.1007/BF00425340
Erb, T. J., Ismail, W., and Fuchs, G. (2008). Phenylacetate metabolism in thermophiles: characterization of phenylacetate-CoA ligase, the initial enzyme of the hybrid pathway in Thermus thermophilus. Curr. Microbiol. 57, 27–32. doi: 10.1007/s00284-008-9147-3
Erdoğmuş, S. F., Mutlu, B., Korcan, S. E., Güven, K., and Konuk, M. (2013). Aromatic hydrocarbon degradation by halophilic archaea isolated from Camalti Saltern, Turkey. Water Air Soil Pollut. 224, 1449. doi: 10.1007/s11270-013-1449-9
Fairley, D. J., Wang, G., Rensing, C., Pepper, I. L., and Larkin, M. J. (2006). Expression of gentisate 1,2-dioxygenase (gdoA) genes involved in aromatic degradation in two Haloarchaeal genera. Appl. Microbiol. Biotechnol. 73, 691–695. doi: 10.1007/s00253-006-0509-0
Fernández, C., Díaz, E., and García, J. L. (2014). Insights on the regulation of the phenylacetate degradation pathway from Escherichia coli. Environ. Microbiol. Rep. 6, 239–250. doi: 10.1111/1758-2229.12117
Fernández, C., Ferrández, A., Miñambres, B., Díaz, E., and García, J. L. (2006). Genetic characterization of the phenylacetyl-coenzyme A oxygenase from the aerobic phenylacetic acid degradation pathway of Escherichia coli. Appl. Environ. Microbiol. 72, 7422–7426. doi: 10.1128/AEM.01550-06
Ferrández, A., García, J. L., and Díaz, E. (2000). Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J. Biol. Chem. 275, 12214–12222. doi: 10.1074/jbc.275.16.12214
Ferrández, A., Miñambres, B., García, B., Olivera, E. R., Luengo, J. M., García, J. L., et al. (1998). Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273, 25974–25986. doi: 10.1074/jbc.273.40.25974
Fuchs, G. (2008). Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 1125, 82–99. doi: 10.1196/annals.1419.010
Fuchs, G., Boll, M., and Heider, J. (2011). Microbial degradation of aromatic compounds - from one strategy to four. Nat. Rev. Microbiol. 9, 803–816. doi: 10.1038/nrmicro2652
Galán, B., García, J. L., and Prieto, M. A. (2004). The PaaX repressor, a link between penicillin G acylase and the phenylacetyl-coenzyme A catabolon of Escherichia coli W. J. Bacteriol. 186, 2215–2220. doi: 10.1128/JB.186.7.2215-2220.2004
García, B., Olivera, E. R., Miñambres, B., Carnicero, D., Muñiz, C., Naharro, G., et al. (2000). Phenylacetyl-coenzyme A is the true inducer of the phenylacetic acid catabolism pathway in pseudomonas putida U. Appl. Environ. Microbiol. 66, 4575–4578. doi: 10.1128/AEM.66.10.4575-4578.2000
Geng, H., Bruhn, J. B., Nielsen, K. F., Gram, L., and Belas, R. (2008). Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74, 1535–1545. doi: 10.1128/AEM.02339-07
Green, E. R., Juttukonda, L. J., and Skaar, E. P. (2020). The manganese-responsive transcriptional regulator MumR protects Acinetobacter baumannii from oxidative stress. Infect. Immun. 88, e00762–e00719. doi: 10.1128/IAI.00762-19
Grishin, A. M., Ajamian, E., Tao, L., Bostina, M., Zhang, L., Trempe, J. F., et al. (2013). Family of phenylacetyl-CoA monooxygenases differs in subunit organization from other monooxygenases. J. Struct. Biol. 184, 147–154. doi: 10.1016/j.jsb.2013.09.012
Grishin, A. M., Ajamian, E., Tao, L., Zhang, L., Menard, R., and Cygler, M. (2011). Structural and functional studies of the Escherichia coli phenylacetyl-CoA monooxygenase complex. J. Biol. Chem. 286, 10735–10743. doi: 10.1074/jbc.M110.194423
Grishin, A. M., Ajamian, E., Zhang, L., Rouiller, I., Bostina, M., and Cygler, M. (2012). Protein-protein interactions in the β-oxidation part of the phenylacetate utilization pathway: crystal structure of the PaaF-PaaG hydratase-isomerase complex. J. Biol. Chem. 287, 37986–37996. doi: 10.1074/jbc.M112.388231
Grishin, A. M., and Cygler, M. (2015). Structural organization of enzymes of the phenylacetate catabolic hybrid pathway. Biology (Basel). 4, 424–442. doi: 10.3390/biology4020424
Gulick, A. M. (2009). Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827. doi: 10.1021/cb900156h
Hamlin, J. N., Bloodworth, R. A., and Cardona, S. T. (2009). Regulation of phenylacetic acid degradation genes of Burkholderia cenocepacia K56-2. BMC Microbiol. 9, 222. doi: 10.1186/1471-2180-9-222
Harris, D. M., van der Krogt, Z. A., Klaassen, P., Raamsdonk, L. M., Hage, S., van den Berg, M. A., et al. (2009). Exploring and dissecting genome-wide gene expression responses of Penicillium chrysogenum to phenylacetic acid consumption and penicillin G production. BMC Genomics 10, 75. doi: 10.1186/1471-2164-10-75
Henriksen, N. N. S. E., Lindqvist, L. L., Wibowo, M., Sonnenschein, E. C., Bentzon-Tilia, M., and Gram, L. (2022). Role is in the eye of the beholder-the multiple functions of the antibacterial compound tropodithietic acid produced by marine Rhodobacteraceae. FEMS Microbiol. Rev. 46, fuac007. doi: 10.1093/femsre/fuac007
Hooppaw, A. J., McGuffey, J. C., Di Venanzio, G., Ortiz-Marquez, J. C., Weber, B. S., Lightly, T. J., et al. (2022). The phenylacetic acid catabolic pathway regulates antibiotic and oxidative stress responses in Acinetobacter, The Phenylacetic acid catabolic pathway regulates antibiotic and oxidative stress responses in Acinetobacter. mBio 13:e0186321. doi: 10.1128/mbio.01863-21
Hwang, B. K., Lim, S. W., Kim, B. S., Lee, J. Y., and Moon, S. S. (2001). Isolation and in vivo and in vitro antifungal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl. Environ. Microbiol. 67, 3739–3745. doi: 10.1128/AEM.67.8.3739-3745.2001
Imolorhe, I. A., and Cardona, S. T. (2011). 3-hydroxyphenylacetic acid induces the Burkholderia cenocepacia phenylacetic acid degradation pathway - toward understanding the contribution of aromatic catabolism to pathogenesis. Front. Cell. Infect. Microbiol. 1:14. doi: 10.3389/fcimb.2011.00014
Ismail, W., El-Said Mohamed, M., Wanner, B. L., Datsenko, K. A., Eisenreich, W., Rohdich, F., et al. (2003). Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270, 3047–3054. doi: 10.1046/j.1432-1033.2003.03683.x
Jami, M. S., Martín, J. F., Barreiro, C., Domínguez-Santos, R., Vasco-Cárdenas, M. F., Pascual, M., et al. (2018). Catabolism of phenylacetic acid in Penicillium rubens. Proteome-wide analysis in response to the benzylpenicillin side chain precursor. J. Proteome 187, 243–259. doi: 10.1016/j.jprot.2018.08.006
Kashyap, S., Sharma, P., and Capalash, N. (2021). Potential genes associated with survival of Acinetobacter baumannii under ciprofloxacin stress. Microbes Infect. 23:104844. doi: 10.1016/j.micinf.2021.104844
Khandokar, Y. B., Srivastava, P., Sarker, S., Swarbrick, C. M. D., Aragao, D., Cowieson, N., et al. (2016). Structural and functional characterization of the PaaI thioesterase from Streptococcus pneumoniae reveals a dual specificity for phenylacetyl-CoA and medium-chain fatty acyl-CoAs and a novel CoA-induced fit mechanism. J. Biol. Chem. 291, 1866–1876. doi: 10.1074/jbc.M115.677484
Kichise, T., Hisano, T., Takeda, K., and Miki, K. (2009). Crystal structure of phenylacetic acid degradation protein PaaG from Thermus thermophilus HB8. Proteins. 76, 779–786. doi: 10.1002/prot.22455
Kiema, T. R., Harijan, R. K., Strozyk, M., Fukao, T., Alexson, S. E., and Wierenga, R. K. (2014). The crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase (T1): insight into the reaction mechanism of its thiolase and thioesterase activities. Acta Crystallogr. D Biol. Crystallogr. 70, 3212–3225. doi: 10.1107/S1399004714023827
Kim, Y., Cho, J. Y., Kuk, J. H., Moon, J. H., Cho, J. I., Kim, Y. C., et al. (2004b). Identification and antimicrobial activity of phenylacetic acid produced by bacillus licheniformis isolated from fermented soybean. Chungkook-Jang. Curr. Microbiol. 48, 312–317. doi: 10.1007/s00284-003-4193-3
Kim, H. S., Kang, T. S., Hyun, J. S., and Kang, H. S. (2004a). Regulation of penicillin G acylase gene expression in Escherichia coli by repressor PaaX and the cAMP-cAMP receptor protein complex. J. Biol. Chem. 279, 33253–33262. doi: 10.1074/jbc.M404348200
Kröger, C., Kary, S. C., Schauer, K., and Cameron, A. D. (2016). Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8, 12. doi: 10.3390/genes8010012
Kung, J. W., Löffler, C., Dörner, K., Heintz, D., Gallien, S., Van Dorsselaer, A., et al. (2009). Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc. Natl. Acad. Sci. U. S. A. 106, 17687–17692. doi: 10.1073/pnas.0905073106
Law, A., and Boulanger, M. J. (2011). Defining a structural and kinetic rationale for paralogous copies of phenylacetate-CoA ligases from the cystic fibrosis pathogen Burkholderia cenocepacia J2315. J. Biol. Chem. 286, 15577–15585. doi: 10.1074/jbc.M111.219683
Law, R. J., Hamlin, J. N., Sivro, A., McCorrister, S. J., Cardama, G. A., and Cardona, S. T. (2008). A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 190, 7209–7218. doi: 10.1128/JB.00481-08
Leus, I. V., Adamiak, J., Trinh, A. N., Smith, R. D., Smith, L., Richardson, S., et al. (2020). Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol. Microbiol. 114, 1049–1065. doi: 10.1111/mmi.14594
Li, Y., Wang, M., Zhao, Q., Shen, X., Wang, J., Yan, Y., et al. (2019). Shunting phenylacetic acid catabolism for tropone biosynthesis. ACS Synth. Biol. 8, 876–883. doi: 10.1021/acssynbio.9b00013
Lightly, T. J., Frejuk, K. L., Groleau, M. C., Chiarelli, L. R., Ras, C., Buroni, S., et al. (2019). Phenylacetyl coenzyme A, not phenylacetic acid, attenuates CepIR-regulated virulence in Burkholderia cenocepacia. Appl. Environ. Microbiol. 85, e01594–e01519. doi: 10.1128/AEM.01594-19
Liu, W. W., Pan, J., Feng, X., Li, M., Xu, Y., Wang, F., et al. (2020). Evidences of aromatic degradation dominantly via the phenylacetic acid pathway in marine benthic Thermoprofundales. Environ. Microbiol. 22, 329–342. doi: 10.1111/1462-2920.14850
Mayrand, D. (1979). Identification of clinical isolates of selected species of Bacteroides: production of phenylacetic acid. Can. J. Microbiol. 25, 927–928. doi: 10.1139/m79-138
Moyer, A. J., and Coghill, R. D. (1947). Penicillin: X. The effect of phenylacetic acid on penicillin production. J. Bacteriol. 53, 329–341. doi: 10.1128/jb.53.3.329-341.1947
Musthafa, K. S., Sivamaruthi, B. S., Pandian, S. K., and Ravi, A. V. (2012). Quorum sensing inhibition in Pseudomonas aeruginosa PAO1 by antagonistic compound phenylacetic acid. Curr. Microbiol. 65, 475–480. doi: 10.1007/s00284-012-0181-9
Niraula, N. P., Shrestha, P., Oh, T. J., and Sohng, J. K. (2010). Identification and characterization of a NADH oxidoreductase involved in phenylacetic acid degradation pathway from Streptomyces peucetius. Microbiol. Res. 165, 649–656. doi: 10.1016/j.micres.2009.11.011
Nogales, J., Macchi, R., Franchi, F., Barzaghi, D., Fernández, C., García, J. L., et al. (2007). Characterization of the last step of the aerobic phenylacetic acid degradation pathway. Microbiology (Reading) 153, 357–365. doi: 10.1099/mic.0.2006/002444-0
Olivera, E. R., Miñambres, B., García, B., Muñiz, C., Moreno, M. A., Ferrández, A., et al. (1998). Molecular characterization of the phenylacetic acid catabolic pathway in pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U. S. A. 95, 6419–6424. doi: 10.1073/pnas.95.11.6419
Pfündel, E. E. (2021). Simultaneously measuring pulse-amplitude-modulated (PAM) chlorophyll fluorescence of leaves at wavelengths shorter and longer than 700 nm. Photosynth. Res. 147, 345–358. doi: 10.1007/s11120-021-00821-7
Pribytkova, T., Lightly, T. J., Kumar, B., Bernier, S. P., Sorensen, J. L., Surette, M. G., et al. (2014). The attenuated virulence of a Burkholderia cenocepacia paaABCDE mutant is due to inhibition of quorum sensing by release of phenylacetic acid. Mol. Microbiol. 94, 522–536. doi: 10.1111/mmi.12771
Rather, L. J., Knapp, B., Haehnel, W., and Fuchs, G. (2010). Coenzyme A-dependent aerobic metabolism of benzoate via epoxide formation. J. Biol. Chem. 285, 20615–20624. doi: 10.1074/jbc.M110.124156
Rodríguez-Sáiz, M., Barredo, J. L., Moreno, M. A., Fernández-Cañón, J. M., Peñalva, M. A., and Díez, B. (2001). Reduced function of a phenylacetate-oxidizing cytochrome P450 caused strong genetic improvement in early phylogeny of penicillin-producing strains. J. Bacteriol. 183, 5465–5471. doi: 10.1128/JB.183.19.5465-5471.2001
Rodríguez-Sáiz, M., Díez, B., and Barredo, J. L. (2005). Why did the Fleming strain fail in penicillin industry? Fungal genet. Biol. 42, 464–470. doi: 10.1016/j.fgb.2005.01.014
Sakamoto, K., Agari, Y., Kuramitsu, S., and Shinkai, A. (2011). Phenylacetyl coenzyme A is an effector molecule of the TetR family transcriptional repressor PaaR from Thermus thermophilus HB8. J. Bacteriol. 193, 4388–4395. doi: 10.1128/JB.05203-11
Sass, A., Marchbank, A., Tullis, E., LiPuma, J. J., and Mahenthiralingam, E. (2011). Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 12, 373. doi: 10.1186/1471-2164-12-373
Sathyanarayanan, N., Cannone, G., Gakhar, L., Katagihallimath, N., Sowdhamini, R., Ramaswamy, S., et al. (2019). Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway. Nat. Commun. 10, 4127. doi: 10.1038/s41467-019-11931-1
Schmid, G., René, S. B., and Boll, M. (2015). Enzymes of the benzoyl-coenzyme A degradation pathway in the hyperthermophilic archaeon Ferroglobus placidus. Environ. Microbiol. 17, 3289–3300. doi: 10.1111/1462-2920.12785
Somers, E., Ptacek, D., Gysegom, P., Srinivasan, M., and Vanderleyden, J. (2005). Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acetic acid biosynthesis. Appl. Environ. Microbiol. 71, 1803–1810. doi: 10.1128/AEM.71.4.1803-1810.2005
Song, F., Zhuang, Z., Finci, L., Dunaway-Mariano, D., Kniewel, R., Buglino, J. A., et al. (2006). Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J. Biol. Chem. 281, 11028–11038. doi: 10.1074/jbc.M513896200
Spieker, M., Saleem-Batcha, R., and Teufel, R. (2019). Structural and mechanistic basis of an oxepin-CoA forming isomerase in bacterial primary and secondary metabolism. ACS Chem. Biol. 14, 2876–2886. doi: 10.1021/acschembio.9b00742
Teufel, R., Friedrich, T., and Fuchs, G. (2012). An oxygenase that forms and deoxygenates toxic epoxide. Nature 483, 359–362. doi: 10.1038/nature10862
Teufel, R., Gantert, C., Voss, M., Eisenreich, W., Haehnel, W., and Fuchs, G. (2011). Studies on the mechanism of ring hydrolysis in phenylacetate degradation: a metabolic branching point. J. Biol. Chem. 286, 11021–11034. doi: 10.1074/jbc.M110.196667
Teufel, R., Mascaraque, V., Ismail, W., Voss, M., Perera, J., Eisenreich, W., et al. (2010). Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107, 14390–14395. doi: 10.1073/pnas.1005399107
Wang, J., Dong, Y., Zhou, T., Liu, X., Deng, Y., Wang, C., et al. (2013). Pseudomonas aeruginosa cytotoxicity is attenuated at high cell density and associated with the accumulation of phenylacetic acid. PLoS One 8:e60187. doi: 10.1371/journal.pone.0060187
Wang, Y., Zhang, R., He, Z., Van Nostrand, J. D., Zheng, Q., Zhou, J., et al. (2017). Functional gene diversity and metabolic potential of the microbial community in an estuary-shelf environment. Front. Microbiol. 8:1153. doi: 10.3389/fmicb.2017.01153
Yudistira, H., McClarty, L., Bloodworth, R. A., Hammond, S. A., Butcher, H., Mark, B. L., et al. (2011). Phenylalanine induces Burkholderia cenocepacia phenylacetic acid catabolism through degradation to phenylacetyl-CoA in synthetic cystic fibrosis sputum medium. Microb. Pathog. 51, 186–193. doi: 10.1016/j.micpath.2011.04.002
Keywords: phenylacetic acid pathway, aromatic metabolites, enzyme structure, pathogenicity, antibiotic resistance
Citation: Jiao M, He W, Ouyang Z, Shi Q and Wen Y (2022) Progress in structural and functional study of the bacterial phenylacetic acid catabolic pathway, its role in pathogenicity and antibiotic resistance. Front. Microbiol. 13:964019. doi: 10.3389/fmicb.2022.964019
Received: 04 July 2022; Accepted: 08 August 2022;
Published: 08 September 2022.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Robin Teufel, University of Basel, SwitzerlandCopyright © 2022 Jiao, He, Ouyang, Shi and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yurong Wen, WXVyb25nLldlbkB4anR1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.