
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 September 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.964009
Salmonella is a leading cause of foodborne illness worldwide and is a common concern in food safety. Salmonella enterica displaying resistance to extended-spectrum cephalosporins (ESCs) and fluoroquinolone (FQs) has been deemed a high-priority pathogen by the World Health Organization. Co-resistance to ESCs and FQs has been reported in S. enterica serovar Thompson (S. Thompson). However, the genetic context of ESCs and FQs resistance genes in S. Thompson lacks sufficient characterization. In this study, we characterized a multi-drug resistant (MDR) S. Thompson isolate recovered from a retail ready-to-eat (RTE) pork product in China. Short- and long-read sequencing (HiSeq and MinION) of the genome identified the presence of blaCMY−2, qnrS1, and qepA8, along with 11 additional acquired antimicrobial resistance genes, residing on a 152,940 bp IncA/C plasmid. Specifically, the blaCMY−2, qnrS1, and qepA8 genes were located in insertion sequences (ISs) and integron mediated mobile genetic structure, sugE-blc-blaCMY−2-ISEc9, IS26-orf6-qnrS1-orf5-ISKpn19, and intl1-qepA8-orf10-IS91-orf1-dfrA12-orf11-aadA2-qacEΔ1-sul1, respectively. Each gene was identified in various bacteria species, indicating their high transfer ability. The plasmid was found to be transferable to Escherichia coli J53 by conjugation and resulted in the acquiring of multiple resistances in the transconjugants. The plasmid is closely related to plasmids from two human S. Thompson strains isolated in different regions and years in China. Moreover, core-genome Multi Locus Sequence Typing (cgMLST) and phylogenetic analysis based on global 1,868 S. Saintpaul isolates showed that the S. Thompson isolate was highly epidemiologically linked to a human isolate in China. Our findings suggest that Chinese RTE pork products are a possible source of human pathogenic ESCs and FQs co-resistant S. Thompson. Furthermore, the results underline the important role of conjugative plasmids in acquiring and transmission of ESCs and FQs resistance in S. Thompson isolates, which need continuous investigation.
Salmonella is a leading cause of food-borne illness worldwide (Chen et al., 2019). There are over 2,600 serovars of Salmonella. Among them, S. enterica subsp. enterica serovar Thompson (S. Thompson) is one of the most frequent Salmonella serovars in humans and is commonly associated with poultry (Shah et al., 2017; Eun et al., 2019; Qi et al., 2019; Zhou et al., 2019). S. Thompson contaminated meat products have been reported as frequent causes of human salmonellosis (Gaulin et al., 2017; Suijkerbuijk et al., 2017; Marder et al., 2018; Eun et al., 2019). Antimicrobial therapy (e.g., ciprofloxacin in adults and ceftriaxone in children) can be lifesaving in these patients (Crump et al., 2015). There is increasing concern over the emergence and increased incidence of multi-drug resistant (MDR) Salmonella strains, especially those resistant to extended-spectrum cephalosporins (ESCs) and fluoroquinolones (FQs), which have been deemed a high-priority pathogen by the World Health Organization (Collignon et al., 2016).
Cephalosporins resistance is mediated predominantly by extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and carbapenemase (Arlet et al., 2006). Various β-lactamases, including blaCTX−M, blaDHA−1, and blaSHV, have been reported in S. Thompson isolates (Zhou et al., 2019; Elbediwi et al., 2021). Reduced susceptibility to FQs is associated with chromosomal mutations and acquisition of AMR genes, such as efflux pumps encoding genes and plasmid-mediated quinolone resistance (PMQR) genes (qnr, aac(6′)-Ib-cr, oqxAB, and qepA) (Cuypers et al., 2018). Co-occurrence of ESCs and FQs resistance genes has so far only been reported in a S. Thompson isolate from Chicken in China (Zhou et al., 2019). The co-existence and co-transfer of ESCs and FQs resistance genes in Salmonella may seriously compromise treatment options, especially for invasive salmonellosis (Crump et al., 2015). However, the genetic context and transferability of ESCs resistance and PMQR genes have not been thoroughly investigated in S. Thompson.
The aim of this study was to characterize the genetic context of ESCs and FQs resistance genes in a S. Thompson isolate recovered from a ready-to-eat (RTE) pork product in Guangzhou, China, and to analyze its possible origin as well as transferability, in order to gain insight into their public health impact.
During our routine surveillance of foodborne pathogens on various food products, a Salmonella isolates (named GSJ/2017-Sal.-009, hereafter 17Sal009) was recovered from a retail RTE dumpling with pork and cabbage stuffing in Guangzhou, Southern China, in 2017. The isolate was identified by biochemical confirmation using API 20E test identification test strips (bioMérieux, France), as well as amplification of the invA gene by PCR (Bai et al., 2016). The serovar was determined by the slide agglutination test, using Salmonella antisera (SSI Diagnostica, Denmark) according to the Kauffmann–White scheme.
E. coli ATCC 25922 and E. coli J53 (sodium azide resistant) were used as the quality control for antimicrobial susceptibility testing and recipient strain for conjugation, respectively. All the strain was routinely grown for 12–24 h at 37°C on either Luria–Bertani (LB) broth or LB agar (Guangdong Huankai Microbial Sci &Tech, Guangzhou, China).
Susceptibility of S. Thompson 17Sal009 to a panel of antimicrobial drugs (Hangzhou Microbial Reagent Co., Ltd., China), including amikacin (30 μg), ampicillin (10 μg), amoxicillin clavulanic acid (20/10 μg), ampicillin-sulbactam sodium (10/10 μg), azithromycin (15 μg), aztreonam (30 μg), cefazolin (30 μg) (1st generation), cefoxitin (30 μg) (2nd generation), cefuroxime (30 μg) (2nd generation), cefotaxime (30 μg) (3rd generation), ceftazidime (30 μg) (3rd generation), cefepime (30 μg) (4th generation), chloramphenicol (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), ertapenem (10 μg), fosfomycin (200 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), netilmicin (30 μg), piperacillin (100 μg), streptomycin (10 μg), tigecycline (15 μg), tetracycline (30 μg), tobramycin (10 μg), and trimethoprim/sulfamethoxazole (23.75/1.25 μg) (Hangzhou Microbial Reagent Co., Ltd., China), were determined by disk diffusion antimicrobial susceptibility testing (Clinical Laboratory Standards Institute (CLSI)., 2018). Production of ESBL was confirmed by the disk diffusion clavulanate inhibition test using ceftazidime and cefotaxime and an ESBL producing S. typhimurium isolates 17Sal008 was used as a positive control [Li et al., 2022; Clinical Laboratory Standards Institute (CLSI)., 2018]. Minimum inhibitory concentrations (MICs) of 17Sal009, E. coli J53, and transconjugants to ciprofloxacin, nalidixic acid, and cefotaxime (Sigma-Aldrich, St. Louis, MO) were determined by broth microdilution (Clinical Laboratory Standards Institute (CLSI)., 2018). The final concentrations of antibiotics used were 0, 0.0075, 0.015, 0.03, 0.06, 0.12, 0.24, 0.5, 1, 2, and 4 μg/ml for ciprofloxacin, 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 μg/ml for nalidixic acid and 0, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, and 256 μg/ml for cefotaxime. Results were interpreted according to the CLSI breakpoints [Clinical and Laboratory Standards Institute (CLSI), 2018]. For ciprofloxacin, isolates with MICs ≤0.06 μg/ml were considered susceptible, while those with MICs of ≥ 1 μg/ml were considered resistant. For nalidixic acid, isolates with MICs ≤16 μg/ml were considered susceptible, while those with MICs of ≥32 μg/ml were considered resistant. For cefotaxime, Salmonella isolates with MICs of ≤1 μg/ml were considered susceptible, and those with MICs ≥4 μg/ml were categorized as resistant. Reference strain E. coli ATCC 25922 served as quality control. Diameters of disks were presented as mean values from replications with standard errors. All measurements were performed in duplicates and each experiment was repeated three times. MDR referred to non-typhoidal Salmonella isolates that were resistant to ampicillin, chloramphenicol, and trimethoprim/sulfamethoxazole (Gordon et al., 2008).
Genomic DNA of isolate 17Sal009 was extracted using a commercial DNA extraction kit (Magen, Guangzhou, China) following the manufacturer's recommendations. The whole genome of the isolate was sequenced on Illumina Hiseq × 10 with 150 bp paired-end reads (MajorBio Co., Shanghai, China) and MinION (Oxford Nanopore, Oxford, United Kingdom). For the Illumina platform, Initial data quality inspection was performed with FastQC (v0.11.9, https://www.bioinformatics.babraham.ac.uk./projects/fastqc) and then reads were filtered and trimmed using Cutadapt (v1.17) to discard the low-quality reads that contained ambiguous nucleotides or a quality score lower than 20 (Martin, 2011). For the MinION platform, the library was prepared using the ONT 1D ligation sequencing kit (SQK-LSK109) with the native barcoding expansion kit (EXP-NBD104). Fast5 files were basecalled using Guppy (v3.15) and output DNA sequence reads with Q > 7 were saved as fastq files. We assessed read statistics including quality scores and read lengths using NanoStat (v1.1.2, https://github.com/wdecoster/nanostat). The genome was assembled using a combination of short- and long-reads by SPAdes V3.14.0 (Bankevich et al., 2012) and Unicycler hybrid assembler V0.4.8 (Wick et al., 2017), and annotated by Prokka V1.14.6 (Seemann, 2014).
Clonal analysis was assessed by MLST 2.0 (https://cge.food.dtu.dk/services/MLST/). PlasmidFinder V2.1 was used to identify plasmid replicon types (Carattoli and Hasman, 2020). The presence of acquired antimicrobial resistance genes and mutations in the quinolone resistance-determining regions (QRDR) (gyrA, gyrB, parC, and parE) was assessed by ResFinder V4.1 (Bortolaia et al., 2020), and were further confirmed (based on 100% sequence coverage and ≥99% nucleotide identity) by BLASTn against nr database with default parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Virulence factors were predicted by the Virulence Factor Database (VFBD; http://www.mgc.ac.cn/VFs/ (accessed on 30 Jul 2022) (Chen, 2004). The complete plasmid sequence was BLASTn against the nr database with default parameters. The sequences showed 100% coverage and ≥99% nucleotide identity, as well as selected plasmids sharing highly similar backbone, were selected for comparison. The map of plasmid comparison was generated by BRIG 0.95-dev.0004 (Alikhan et al., 2011).
In order to assess the relatedness of 17Sal009 with other S. Thompson strains from different sources and countries, we retrieved all 1,868 genome sequences of S. Thompson that have been released from EnteroBase databases and performed core-genome Multi Locus Sequence Typing (cgMLST) (cgMLST scheme available on EnteroBase, https://enterobase.warwick.ac.uk, accessed on 18 Oct 2021) (Supplementary Table S1). Similar but non-identical strains [strains showing different core genome Sequence Types (cgST)] were identified in EnteroBase by using the hierarchical clustering method (HierCC) that allows for the grouping of strains into hierarchical clusters (HCs) that can differ up to a specified and fixed number of cgMLST alleles. This number is indicated by the suffix following “HC” (e.g., HC5 for 5 cgMLST allelic differences). Isolates belonging to the same HC10 cluster were considered possible epidemiologically linked, and isolates belonging to the same HC5 cluster were considered highly probably epidemiological linked (Bonifait et al., 2021). A minimum-spanning tree was created from cgMLST allelic differences in EnteroBase using GrapeTree with the RapidNJ algorithm (Zhou et al., 2019).
The transferability of the plasmid was assessed by performing the conjugation experiment, using solid mating on a filter (Whatman, Maidstone, UK), in which the sodium azide-resistant E. coil strain J53 was used as a recipient strain (Hammerum et al., 2016). Briefly, recipient and donor strains were inoculated into LB broth and cultured overnight at 37°C. The next day, cells were harvested, washed with saline, mixed together in a ratio of 1:1, and spotted on to 0.45 μm-pore-size filter (Millipore) on LB plates. They were also spotted individually on LB plates as controls. After overnight incubation at 37°C, mating spots were washed and resuspended in saline; and different dilutions were plated on LB media containing 150 μg/ml sodium azide and 4, 8, or 16 μg/ml of cefotaxime to select transconjugants. Control spots were transferred to the same selective media to make sure that no growth was observed.
The conjugation frequency was calculated as the ratio of transconjugants over the number of recipients. The transfer of the plasmid was confirmed by PCR targeting the blaCMY-2 gene with primer CMYF (5′- CTCGACACGGACAGGGTTAG−3′) and CMYR (5′- TATTCCGGGTATGGCCGTTG−3′), as well as the E. coli uidA household gene with the primers UIDF (5′-TGGAATTTCGCCGATTTTGC-3′) and UIDF (5′-ATTGTTTGCCTCCCTGCTGC-3′) (Heijnen and Medema, 2016). The plasmid DNA was extracted from a selected transconjugant by a commercial plasmid extraction kit (Magen, Guangzhou, China) following the manufacturer's recommendations, and further sequenced on the Illumina Hiseq platform (MajorBio Co., Shanghai, China).
The assembly genome sequences of S. Thompson and pSal009 were deposited in GenBank under the accession number: CP050833.1, CP050832. The raw Illumina sequence data were deposited in the Enterobase database under the barcode numbers: SAL_JB2919AA.
The isolate was confirmed as S. enterica serovar Thompson, serotype 6,7:r:1,5 by biochemical confirmation, 16S rRNA gene sequencing, and serotyping.
Disk diffusion antimicrobial susceptibility testing showed the isolate was resistant to ampicillin, amoxicillin-clavulanic acid, chloramphenicol, cefazolin, cefotaxime, ceftazidime, tetracycline, doxycycline, and trimethoprim/sulfamethoxazole, intermediate resistant to ciprofloxacin, aztreonam, cefoxitin, and cefuroxime, and was susceptible to gentamicin, tigecycline, fosfomycin, tobramycin, amikacin, piperacillin, ertapenem, imipenem, meropenem, netilmicin, cefepime, ampicillin-sulbactam sodium and produce ESBL. The isolate exhibited MIC values of ciprofloxacin, nalidixic acid, and cefotaxime for 2, 32, and 128 mg/L, respectively.
The complete genome sequence of S. Thompson 17Sal009 contained a circular 4,897,518 bp chromosome with the G+C content of 52.3%, and a plasmid denoted as pSal009. There were 4,875 predicated CDs in the whole genome sequence, including 107 RNA genes. Multi-locus sequence typing analysis showed that 17Sal009 belongs to sequence type 26 (ST26).
A total of 14 acquired antimicrobial resistance genes were identified by ResFinder, which encoded resistance to eight different antimicrobial classes, including cephalosporins, fluoroquinolones, phenicol, aminoglycoside, sulphonamide, trimethoprim, macrolide, and tetracycline. Mutations were not identified in the quinolone resistance-determining regions (gyrA, gyrB, parC, and parE) in 17Sal009. All 14 genes were located on the plasmid (Table 1 and Figure 1).

Table 1. The antimicrobial resistance profile and drug resistance genes of S. Thompson 17Sal009, the selected transformant (17Sal009T), and the recipient (E.coli J53).
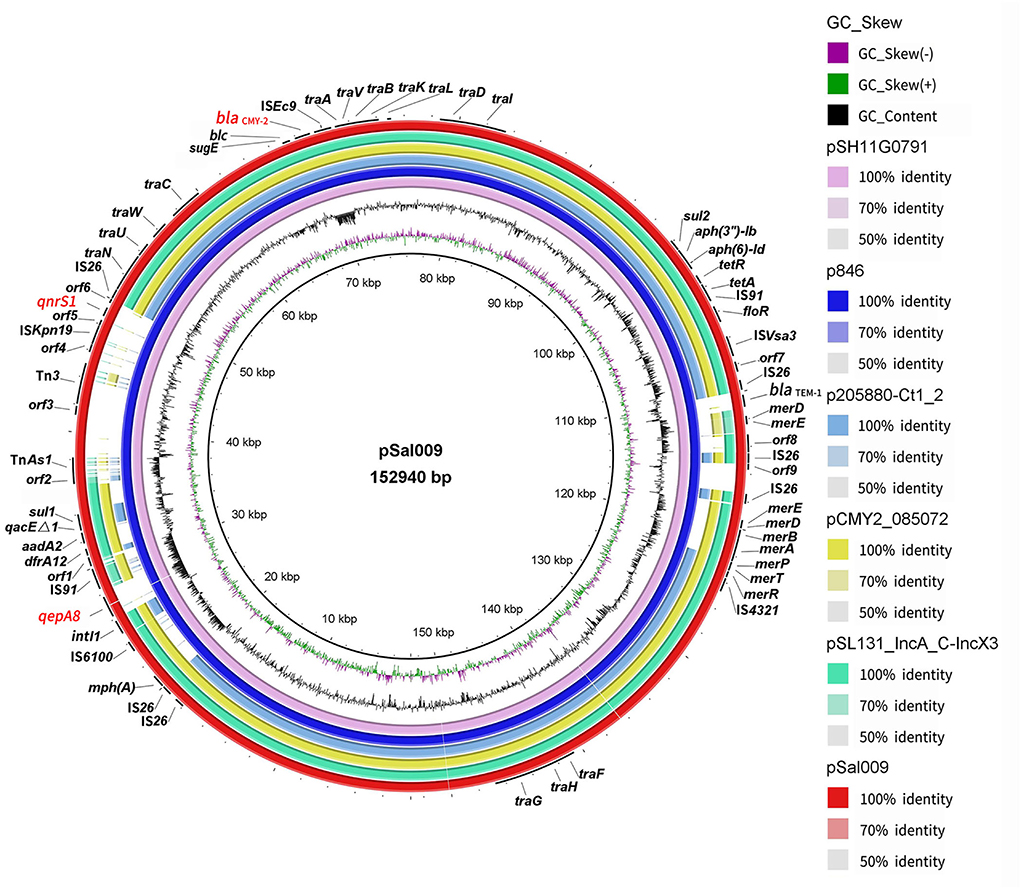
Figure 1. Sequence comparison of plasmid pSal009 identified in S. Thompson 17Sal009 with closely related plasmid pSH11G0791 and p846 as well as similar plasmids (p205880-Ct1/2, pCMY2_085072, and pSL131_IncA/C-IncX3) sharing similar backbone in different bacteria species in BRIG. GC content and GC skew of pSal009 are depicted in the inner rings. The genes located on pSal009 are annotated at the outside black ring. The blaCMY−2, qnrS1, and qepA8 genes are marked red.
The S. Thompson isolate 17Sal009 was predicted to have a total of 74 virulence genes implicated in different mechanisms of virulence and pathogenicity, such as Salmonella pathogenicity island (SPI)-1 (hilA, hilC, hilD, invA, invC, invE, invG, invJ, prgH, sipD, spaO, spaR, and spas), SPI-2 (ssaC, ssaD, ssaL, ssaN, ssaQ, ssaU, ssaV, sseC, and ssrA), which were existed in human isolate (Table 2).
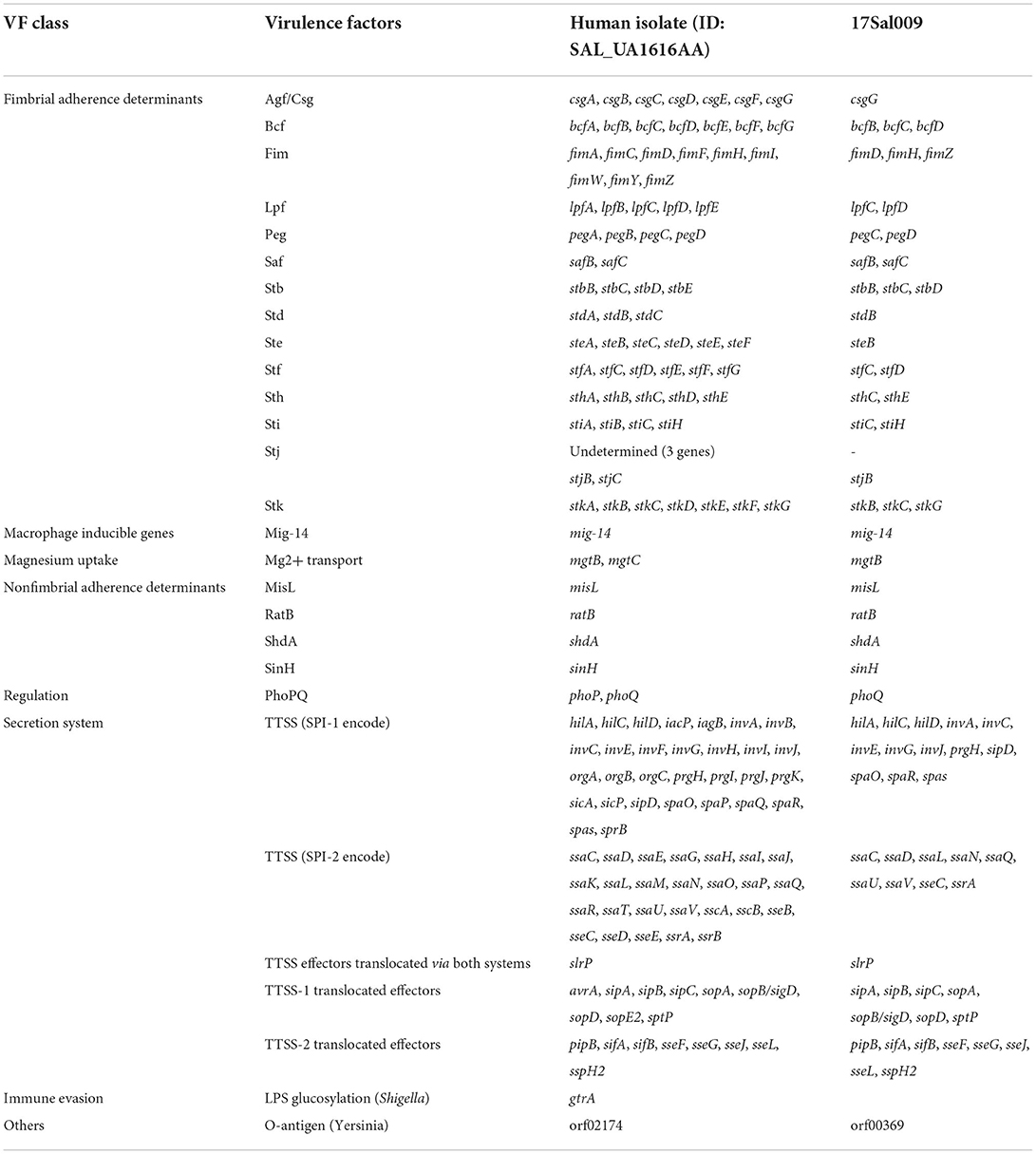
Table 2. Virulence genes predicted to exist in S. Thompson 17Sal009 and closely related human isolate.
pSal009 is a 152,940 bp plasmid, with 194 predicated CDSs and an average GC content of 53.4%. The replicon regions were identified as IncA/C by PlasmidFinder. The plasmid includes the core region (traFHG and traNUWCAVBKLDI) involved in the conjugative transfer, including plasmid replication, horizontal transfer, and stability and maintenance functions, which defines the plasmid backbone (Figure 1) (Call et al., 2010). Various antimicrobial resistance genes were identified on the plasmid, including floR encoding phenicol resistance, tet(A) and tet(R) for tetracycline resistance, qnrS1 and qepA8 for quinolone resistance, dfrA12 for trimethoprim resistance, mph(A) for macrolide resistance, sul1 and sul2 for sulfonamide resistance, aadA2, aph(3”)-Ib, and aph(6)-Id for aminoglycoside resistance, and blaCMY−2 and blaTEM−1 for beta-lactam resistance (Figure 1). The plasmid also harbors quaternary ammonium resistance genes (qacEΔ1 and sugE). A mer gene cluster encoding putative regulatory proteins (MerR, MerD), transporters (MerT, MerP, MerE, and MerF) and the mercuric reductase (MerA, MerB), conferring mercurial resistance were identified on the plasmid. In addition, class 1 integron integrase IntI1 and uncharacterized integrase, and copies of transposases and recombinase family protein were observed.
BLASTn comparison of the entire plasmid sequence to microbial sequences in GenBank indicated that it was closely related to pSH11G0791 (GenBank accession number CP041172) (with 99% nucleotide identity and 100% sequence coverage) from a S. Thompson strain isolated from human feces in Shanghai, China in 2011, and p846 (GenBank accession number CP029249) (with 99% nucleotide identity and 100% sequence coverage) from a S. Thompson strain isolated from children's feces in China in 2014 (Figure 1). Other plasmids with similar backbone include pCMY2_085072 (with 100% nucleotide identity and 86% sequence coverage) (GenBank accession number CP028804) from a clinical K. pneumoniae strain, p205880-Ct1/2 (with nucleotide 99% identity and 75% sequence coverage) (GenBank accession number MF344573) from a clinical K. pneumoniae strain, and pSL131_IncA/C-IncX3 (with 100% nucleotide identity and 85% sequence coverage) (GenBank accession number MH105050) from a clinical S. Lomita strain (Figure 1). The plasmids pCMY2_085072, p205880-Ct1/2 and pSL131_IncA/C-IncX3 carry blaCMY−2 but lacking qnrS1, qepA8, blaTEM−1, mph(A), part of mer genes and several insertion sequences (ISs), such as transposases and recombinase family protein.
blaCMY−2 gene is flanked by ISEc9 and linked with sugE gene and blc encoding lipocalin family protein, sugE-blc-blaCMY−2-ISEc9 (Figure 2A). This structure was found to mainly exist in E.coli (blastn resulted 89 sequences of 100% identity matches), and present in small amounts of S. Typhimurium, S. Anatum, Salmonella serovar 1,4,[5],12:i:-, S. Heidelberg and K. pneumoniae (Supplementary Table S2).
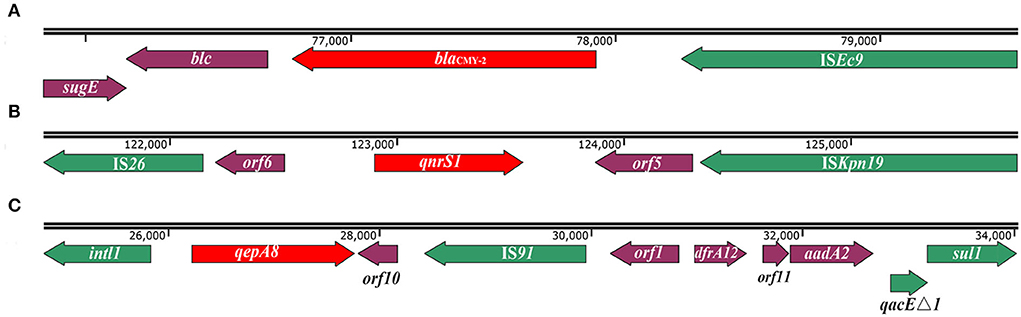
Figure 2. Schematic of genetic context of blaCMY−2, qnrS1, and qepA8 genes in S. Thompson 17Sal009. (A) The genetic context of blaCMY−2 gene. (B) The genetic context of the qnrS1 gene. The orf5 is predicated to encode recombinase family protein, and orf6 is predicated to encode transposase. (C) The genetic context of the qepA8 gene. The orf1 is predicted to encode an uncharacterized site-specific integrase, orf10 is predicted to encode mechanosensitive ion channel, and orf11 is predicated to encode DUF1010 domain-containing protein. The arrows indicate open reading frames. Green colors depict mobile genetic elements. Red colors depict blaCMY−2, qnrS1, and qepA8 genes.
The qnrS1 gene was located in a composite transposon, IS26-orf6-qnrS1-orf5-ISKpn19, in which the orf6 encoding transposase and the orf5 encoding recombinase family protein (Figure 2B). In addition to pSH11G0791 and p846, the similar genetic context has also been found in E.coli, K. pneumoniae, Shigella sonnei, and E. fergusonii (Supplementary Table S3), in which different gene clusters inserted between IS26 and orf6 (data not shown). Interestingly, the gene cluster orf6-qnrS1-orf5-ISKpn19 has been found in various bacterial species, mainly in K. pneumoniae (52 sequences of 100% identity matches), E.coli (23 sequences of 100% identity matches), and sporadically in other Salmonella serotypes (like S. Agona, S. Muenster, Salmonella serovar 1,4,[5], 12:i:-, S. Typhi, S. Thompson, S. typhimurium) or other species including Shigella sonnei, Serratia liquefaciens, Yokenella regensburgei, Leclercia adecarboxylata, and Citrobacter sp. (Supplementary Table S4).
The qepA8 gene was located in a class 1 integron with complete structure, intl1-qepA8-orf10-IS91-orf1-dfrA12-orf11-aadA2-qacEΔ1-sul1, in which the orf10 encoding mechanosensitive ion channel, orf1 encoding an uncharacterized site-specific integrase, and orf11encoding DUF1010 domain-containing protein (Figure 2C). Except for pSH11G0791 and p846, the same integron has been found in a plasmid in E.coli (GenBank accession number CP023959.1) from Urine samples in Canada in 2014 and plasmid in E.coli (GenBank accession number MK291500.1) from retail meat from Pakistan in 2018.
PCR and sequencing results confirmed the successful transfer of the plasmid pSal009 to a plasmid-free recipient E. coli J53. Antimicrobial susceptibility testing revealed the acquisition of the plasmid by E. coli caused at least a 256-fold increase for cefotaxime, 133-fold increase for ciprofloxacin, and 256-fold increase for nalidixic acid, respectively (Table 1). The conjugation rate was 7.8 × 10−6 ± 0.5 transconjugant per recipient cell.
Five major clusters are seen for the 1868 S. Thompson isolates from different countries (Figure 3A). These isolates belong to 19 classical MLST types, with the most frequent being ST26 (94.4%) and ST2125 (2.8%) (Supplementary Figure S1).
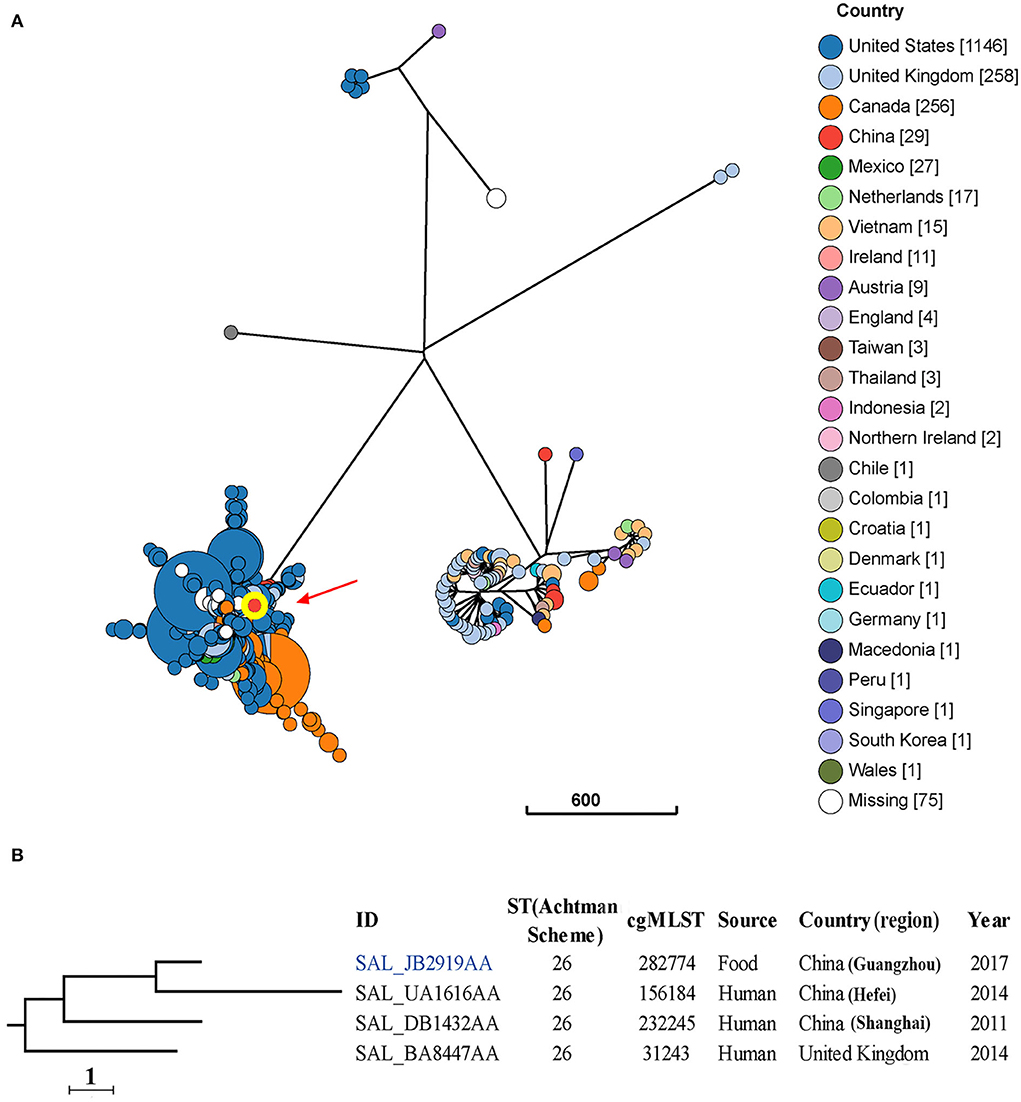
Figure 3. Phylogenetic analysis of 1868 S. Thompson isolates from different sources and countries. (A) A minimum-spanning tree based on cgMLST analysis. The position of isolate 17Sal009 is indicated by red arrow and highlighted circle. Each circle represents a cgMLST group and the size of the circle is proportional to the number of isolates in that group. (B) Detailed information of strains in the branch contained 17Sal009. The isolate 17Sal009 is marked blue.
cgMLST and phylogenetic analysis showed that the isolate 17Sal009 harbored a unique cgST profile (Figure 3B, Supplementary Table S5), and displayed the closest relationship to one human isolate (ID: SAL_UA1616AA) from Hefei, China in 2014 (Figure 3B), and closely related to a human isolate (ID: SAL_BA8447AA) from Shanghai, China in 2011 as well as a human isolate from the United Kingdom in 2014 (ID: SAL_BA8447AA). cgMLST results differentiated the isolate 17Sal009 with the closest related human isolate up to HC5 level (a maximum of 5 cgMLST allelic variations) and with another two close related isolates up to HC10 level (a maximum of 10 cgMLST allelic variations) (Supplementary Table S4). Therefore, it is inferred that isolate 17Sal009 was highly epidemiologically linked with the human isolate (ID: SAL_UA1616AA) from China in 2014 and was possible epidemiological linked with other two human isolates (ID: SAL_BA8447AA, ID: SAL_BA8447AA).
Salmonella resistance to clinically critically important antibiotics ESCs and FQs is a major public health concern (Zhou et al., 2019; Elbediwi et al., 2021). To better understand the origin of the ESCs and FQs resistance in Salmonella and its potential effects on human health, it is of major importance to uncover the resistance mechanism as well as their transmission route through the food chain. In this study, we characterized the genetic context of ESCs and FQs resistance genes in an MDR S. Thompson isolate recovered from RTE pork product in China.
Previously, MDR profiles in S. Thompson have been identified in raw meat (Zhou et al., 2019), RTE meat (Wang et al., 2019), poultry (Xu et al., 2020; Elbediwi et al., 2021), and human (Nair et al., 2016), including ESCs and FQs resistance. However, the genetic determinants of ESCs and FQs resistance in S. Thompson have only been sporadically reported, and their genetic contexts as well as transferability remain obscure. A S. Thompson isolates from poultry has been identified to carry blaCTX−M−15 and qnrB genes in China (Zhou et al., 2019). Several S. Thompson isolates from dead chicks in China were reported to contain blaCTX−M−14, blaDHA−1, blaSHV−12, and qnrB4 genes (Elbediwi et al., 2021), but the co-occurrence patterns of these resistance genes in individual strains were not available. By retrieving the plasmid sequences from the NCBI database, we found two S. Thompson isolates co-harbored the blaCMY−2, qnrS1, and qepA8 genes, and both of them were obtained from human feces. In this study, to the best of our knowledge, we report the first time a S. Thompson isolate from RTE pork product harboring plasmid-mediated blaCMY−2, qnrS1, and qepA8 genes.
The blaCMY−2, qnrS1, and qepA8 genes were found to be located on an IncA/C plasmid, which was also identified in two S. Thompson isolate of human origin in China. The blaCMY−2-carrying plasmids were most frequently IncA/C-, IncHI2- or IncX-type plasmids, which were readily transferable between Salmonella and E. coli from food animals and humans (Shahada et al., 2011). The IncA/C plasmid was observed to be transferable to E. coli in the current study, suggesting its transmission potential, which may lead to the development of the ESCs and FQs resistance among different bacterial species.
blaCMY−2 was found to be located in a transposon-like element consisting of sugE-blc-blaCMY−2-ISEc9, in which ISEc9 was responsible for mobilization of blaCMY−2 from the chromosome and transfer onto a plasmid (Fang et al., 2018). This unit has been identified as a typical carrier of the blaCMY−2 gene (Su et al., 2006; Sidjabat et al., 2014; Yassine et al., 2015) and has been found in many bacterial species, suggesting the highly transferability of blaCMY−2 gene via this unit.
PMQR genes have been regarded as contributive factors to the development of only low-level resistance to fluoroquinolones (ciprofloxacin MICs typically 0.125–2 mg/L) in Salmonella (Robicsek et al., 2006; Jiang et al., 2014). In this study, 17Sal009 exhibited a ciprofloxacin MIC of 2 mg/L, which might be due to the synergistic effects of efflux pumps qepA8 and qnrS1, as indicated by the results of a previous study (Chen et al., 2019). Moreover, both qnrS1 and qepA8 genes are flanked by mobile genetic elements, orf6-qnrS1-orf5-ISKpn19 and intl1-qepA8-orf10-IS91-orf1-dfrA12-orf11-aadA2-qacEΔ1-sul1, which have also been identified in various species, indicating that these mobile structures could readily transfer from one plasmid to another resulting in the progressive development of ciprofloxacin resistance.
In addition, quaternary ammonium resistance genes (qacEΔ1 and sugE) were found to be located on the plasmid, which has been reported to be associated with resistance and enhanced fitness in the intensive-farming environment (Aviv et al., 2014, 2016). Plasmids with similar backbone carrying blaCMY−2 but lacking many resistance genes and mobile genetic elements were identified in various species, suggesting the blaCMY−2-bearing plasmid in S. Thompson was likely recombined by ISs and integron-mediated recombination activities.
Phylogenomic analysis showed that 17Sal009 was highly epidemiologically linked with a human isolate, suggesting that S. Thompson isolates isolated from RTE pork products and humans might come from the same source. Importantly, 17Sal009 contained most of the virulence genes that existed in the closely related human isolate. These genes have been proven to contribute to the strong pathogenicity of S. Thompson isolates (Elbediwi et al., 2021; Zhang et al., 2022), suggesting the potential high pathogenicity of 17Sal009 in humans.
The identification of transferable IncA/C plasmid carrying ESCs and FQs resistance genes in Chinese S. Thompson isolate from RTE pork product represent potential clinical and food safety issues that need to be monitored since they may transmit to human through the food chain and may lead to reduced susceptibility of Salmonella to front-line drugs of choice for treating severe Salmonella infections.
To summarize, this study reports for the first time a S. Thompson isolate derived from RTE pork product in China containing a transferable plasmid harboring blaCMY−2, qnrS1 and qepA8 genes. These genes are localized in mobile structures. The transfer of the plasmid and the mobile structures may contribute to the dissemination of blaCMY−2, qnrS1, and qepA8 genes among different bacterial species and accelerate the development of isolates co-resistant to ESCs and FQs, and this warrants continuous investigations.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
LL performed the experiment and wrote the manuscript. RO revised the manuscript. HM conceptualized and designed the study. ML and JX provided the strains. SP contributed reagents, materials, and analysis tools. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (Grant Numbers 31901789 and 32001796), Natural Science Foundation of Guangdong Province (Grant Numbers 2022A1515011685 and 2020A1515010218), Basic Research Project of Guangzhou (Grant Number 202002030630020049), and Jiangsu Innovative and Enterpreneurial Talent Program (Grant Number JSSCBS20211458).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.964009/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Arlet, G., Barrett, T. J., Butaye, P., Cloeckaert, A., Mulvey, M. R., and White, D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes. Infect. 8, 1945–1954. doi: 10.1016/j.micinf.2005.12.029
Aviv, G., Rahav, G., and Gal-Mor, O. (2016). Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. Mbio 7, e01395–e01417. doi: 10.1128/mBio.01395-16
Aviv, G., Tsyba, K., Steck, N., Salmon-Divon, M., Cornelius, A., Rahav, G., et al. (2014). A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 16, 977–994. doi: 10.1111/1462-2920.12351
Bai, L., Zhao, J., Gan, X., Wang, J., Zhang, X., Cui, S., et al. (2016). Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and foodproducing animals in China. Antimicrob. Agents Chemother. 60, 3365–3371. doi: 10.1128/AAC.02849-15
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bonifait, L., Thépault, A., Baugé, L., Rouxel, S., Le Gall, F., and Chemaly, M. (2021). Occurrence of Salmonella in the cattle production in France. Microorganisms 9, 872. doi: 10.3390/microorganisms9040872
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Call, D. R., Singer, R. S., Meng, D., Broschat, S. L., Orfe, L. H., Anderson, J. M., et al. (2010). blaCMY−2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54, 590–596. doi: 10.1128/AAC.00055-09
Carattoli, A., and Hasman, H. (2020). PlasmidFinder and In Silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Chen, K., Dong, N., Chan, E. W., and Chen, S. (2019). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes. Infect. 8, 857–865. doi: 10.1080/22221751.2019.1626197
Chen, L. (2004). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, 325–328. doi: 10.1093/nar/gki008
Clinical and Laboratory Standards Institute (CLSI). (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Collignon P. J. Conly J. M. Andremont A. McEwen S. A. Aidara-Kane A. World Health Organization Advisory G.roup. (2016). World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. doi: 10.1093/cid/ciw475
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicrobial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Cuypers, W. L., Jacobs, J., Wong, V., Klemm, E. J., Deborggraeve, S., and Van Puyvelde, S. (2018). Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb. Genom. 4, e000195. doi: 10.1099/mgen.0.000195
Elbediwi, M., Tang, Y., Shi, D., Ramadan, H., Xu, Y., Xu, S., et al. (2021). Genomic investigation of antimicrobial-resistant Salmonella enterica isolates from dead chick embryos in China. Front. Microbial. 12, 684400. doi: 10.3389/fmicb.2021.684400
Eun, Y., Jeong, H., Kim, S., Park, W., Ahn, B., Kim, D., et al. (2019). A large outbreak of Salmonella enterica serovar Thompson infections associated with chocolate cake in Busan, Korea. Epidemiol. Health 41, e2019002. doi: 10.4178/epih.e2019002
Fang, L. X., Li, X. P., Li, L., Chen, M. Y., Wu, C. Y., Li, L. L., et al. (2018). ISEcp1-mediated transposition of chromosome-borne blaCMY-2 into an endogenous ColE1-like plasmid in Escherichia coli. Infect. Drug Resist. 11, 995–1005. doi: 10.2147/IDR.S159345
Gaulin, C., Fiset, M., Duchesne, C., Ramsay, D., Savard, N., Urbanek, A., et al. (2017). Salmonella Thompson outbreak associated with consumption of chicken shawarma and the usefulness of genome sequencing in the investigation. Can. Commun. Dis. Rep. 43, 186–192. doi: 10.14745/ccdr.v43i09a05
Gordon, M. A., Graham, S. M., Walsh, A. L., Wilson, L., Phiri, A., Molyneux, E., et al. (2008). Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 46, 963–969. doi: 10.1086/529146
Hammerum, A. M., Hansen, F., Nielsen, H. L., Jakobsen, L., Stegger, M., Andersen, P. S., et al. (2016). Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 71, 3117–3124. doi: 10.1093/jac/dkw289
Heijnen, L., and Medema, G. (2016). Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J. Water Health. 4, 487–498. doi: 10.2166/wh.2006.0032
Jiang, H. X., Song, L., Liu, J., Zhang, X. H., Ren, Y. N., Zhang, W. H., et al. (2014). Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43, 242–247. doi: 10.1016/j.ijantimicag.2013.12.005
Li, L., Wan, X., Olsen, R. H., Xiao, J., Wang, C., Xu, X., et al. (2022). Genomic characterization of mcr-1-carrying foodborne Salmonella enterica serovar Typhimurium and identification of a transferable plasmid carrying mcr-1, blaCTX-M-14, qnrS2, and oqxAB genes from ready-to-eat pork product in China. Front. Microbiol. 13, 903268. doi: 10.3389/fmicb.2022.903268
Marder, E. P., Griffin, P. M., Cieslak, P. R., Dunn, J., Hurd, S., Jervis, R., et al. (2018). Preliminary incidence and trends of infections with pathogens transmitted commonly through food-Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006-2017. MMWR. Morb. Mortal. Wkly. Rep. 67, 324–328. doi: 10.15585/mmwr.mm6711a3
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Nair, S., Ashton, P., Doumith, M., Connell, S., Painset, A., Mwaigwisya, S., et al. (2016). WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J. Antimicrob. Chemother. 71, 3400–3408. doi: 10.1093/jac/dkw318
Qi, X., Li, P., Xu, X., Yuan, Y., Bu, S., and Lin, D. (2019). Epidemiological and molecular investigations on Salmonella responsible for gastrointestinal infections in the southwest of Shanghai from 1998 to 2017. Front. Microbial. 10, 2025. doi: 10.3389/fmicb.2019.02025
Robicsek, A., Jacoby, G. A., and Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640. doi: 10.1016/S1473-3099(06)70599-0
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shah, D. H., Paul, N. C., Sischo, W. C., Crespo, R., and Guard, J. (2017). Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 96, 687–702. doi: 10.3382/ps/pew342
Shahada, F., Sekizuka, T., Kuroda, M., Kusumoto, M., Ohishi, D., Matsumoto, A., et al. (2011). Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 beta-lactamase gene located on a multidrug resistance genomic island. Antimicrob. Agents Chemother. 55, 4114–4121. doi: 10.1128/AAC.00560-11
Sidjabat, H. E., Seah, K. Y., Coleman, L., Sartor, A., Derrington, P., Heney, C., et al. (2014). Expansive spread of IncI1 plasmids carrying blaCMY−2 amongst Escherichia coli. Int. J. Antimicrob. Agents. 44, 203–208. doi: 10.1016/j.ijantimicag.2014.04.016
Su, L. H., Chen, H. L., Chia, J. H., Liu, S. Y., Chu, C., Wu, T. L., et al. (2006). Distribution of a transposon-like element carrying blaCMY−2 among Salmonella and other Enterobacteriaceae. J. Antimicrob. Chemother. 57, 424–429. doi: 10.1093/jac/dki478
Suijkerbuijk, A., Bouwknegt, M., Mangen, M. J., de Wit, G. A., van Pelt, W., Bijkerk, P., et al. (2017). The economic burden of a Salmonella Thompson outbreak caused by smoked salmon in the Netherlands, 2012-2013. Eur. J. Public Health 27, 325–330. doi: 10.1093/eurpub/ckw205
Wang, J., Sheng, H., Xu, W., Huang, J., Meng, L., Cao, C., et al. (2019). Diversity of serotype, genotype, and antibiotic susceptibility of Salmonella prevalent in pickled ready-to-eat meat. Front. Microbial. 10, 2577. doi: 10.3389/fmicb.2019.02577
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Xu, Y., Zhou, X., Jiang, Z., Qi, Y., Ed-Dra, A., and Yue, M. (2020). Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell Infect. Microbiol. 10, 497. doi: 10.3389/fcimb.2020.00497
Yassine, H., Bientz, L., Cros, J., Goret, J., Bebear, C., Quentin, C., et al. (2015). Experimental evidence for IS1294b-mediated transposition of the blaCMY−2 cephalosporinase gene in Enterobacteriaceae. J. Antimicrob. Chemother. 70, 697–700. doi: 10.1093/jac/dku472
Zhang, J., Liu, J., Chen, C., Wang, Y., Chen, X., Li, X., et al. (2022). Resistance and pathogenicity of Salmonella Thompson isolated from incubation end of a poultry farm. Vet. Sci. 9, 349. doi: 10.3390/vetsci9070349
Keywords: Salmonella Thompson, blaCMY−2, qnrS1, qepA8, ready-to-eat pork product
Citation: Li L, Olsen RH, Xiao J, Liang M, Meng H and Peng S (2022) Characterization of extended-spectrum cephalosporins and fluoroquinolone resistance of a Salmonella enterica serovar Thompson isolate from ready-to-eat pork product in China. Front. Microbiol. 13:964009. doi: 10.3389/fmicb.2022.964009
Received: 21 June 2022; Accepted: 24 August 2022;
Published: 15 September 2022.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Yujie Hu, China National Center for Food Safety Risk Assessment, ChinaCopyright © 2022 Li, Olsen, Xiao, Liang, Meng and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hecheng Meng, ZmVtZW5naGNAc2N1dC5lZHUuY24=; Shifu Peng, cGVuZ0Bqc2NkYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.