- 1Department of Public Health Laboratory Sciences, West China School of Public Health and West China Fourth Hospital, Sichuan University, Chengdu, China
- 2Shen Zhen Biomed Alliance Biotech Group Co., Ltd., Shenzhen, China
Listeria monocytogenes (LM) induces efficient and specific T-cell immune responses in the host. Listeriolysin O (LLO) is the main virulence protein of LM. LLO helps LM escape from the lysosome. However, the pronounced pathogenicity of LM limits its practical application as a live bacterial vector. Listeria ivanovii (LI) also displays intracellular parasitic abilities, cell to cell transfer, and other LM properties, with an elevated biosafety relative to LM. We have confirmed that LI can be used as a viable bacterial vaccine vector. However, we have also observed in vivo that LI vector vaccine candidates survive in the immune organ (spleen) for a shorter time compared with the survival time of LM and elicit weaker immune responses compared with LM. Studies have confirmed that hemolysin correlates with some important biological properties of Listeria, including cell invasion, intracellular proliferation, and the ability to induce immune responses. We speculated that the weaker immunogenicity of LI compared to LM may be related to the function of ivanolysin O (ILO). Here, we established a hemolysin gene deletion strain, LIΔilo, and a modified strain, LIΔilo:hly, whose ilo was replaced by hly. The hemolysin-modified strain was attenuated; however, it led to significantly improved invasive and proliferative activities of antigen-presenting cells, including those of RAW 264.7 macrophages, compared with the effects of LI. Mice immunized twice with LIΔilo:hly showed higher cytokine levels and better challenge protection rates than LI-immunized mice. This is the first description in Listeria carrier vaccine research of the modification of LI hemolysin to obtain a better vaccine carrier than LI. The recombinant strain LIΔilo:hly showed good biosafety and immunogenicity, and thus appears to be a good vector strain for vaccine development.
Introduction
Listeria monocytogenes (LM) is a gram-positive food-borne pathogen that is widely distributed in nature. LM can cause listeriosis, which mainly manifests as mild gastroenteritis, but which can be severe and cause meningitis, gastroenteritis, and sepsis (Farber and Peterkin, 1991; Wallecha et al., 2009). LM can proliferate intracellularly. This process is generally considered to be related to listeriolysin O (LLO). LLO is encoded by hly, which is located on LM pathogenicity island 1 (LPI-1). LLO is a major virulence factor of LM (Vázquez-Boland et al., 2001). LLO degrades lysosomal membranes to help bacteria escape phagocytic vesicles and enter host cytoplasm for growth. LLO also has an immunomodulatory role and is thus a potential adjuvant for cancer immunotherapy. A previous study showed that fusing LLO protein to human papillomavirus E7 protein improved the tumoricidal function of E7-specific CD8+ T cells and increased the number of antigen-specific CD8+ T cells in the tumor (Lamikanra et al., 2001), resulting in an improved antitumor therapeutic effect. The unique intracellular lifecycle of LM allows antigens present in or expressed by LM to elicit antigen-specific cellular immune responses through both MHC class I and MHC class II molecular pathways (Harty et al., 1996; Ruan et al., 2016). Therefore, LM is considered to be a promising vector for constructing vaccines that mainly induce cellular immune responses. However, since LM is pathogenic to humans, it must be attenuated before it can be used as a vaccine vector. Many reports have described preventive and therapeutic vaccines that utilize attenuated LM strains as carriers. For example, Yang et al. (2014) described an attenuated LM strain, deleting the dal and dat genes and carrying the human CD24 gene, led to Hepa1-6-CD24-induced tumor regression and increased tumor-free survival in mice. Tumor therapeutic vaccines based on LM have also shown expected results in clinical trials (Le et al., 2019). Vaccine vectors must have reliable biological safety. However, whether the virulence of LM strains remains attenuated after knocking out virulence or metabolic genes is a major safety issue that remains to be investigated.
Listeria ivanovii (LI) was first isolated from a lamb with congenital listeriosis in Bulgaria in 1955. LI rarely infects humans. Thus it displays better biosafety than LM in humans. LI has properties similar to those of LM, such as intracellular growth and cell-to-cell spread, and can enter antigen-presenting cells and proliferate intracellularly (Frehel et al., 2003). We immunized mice with a recombinant LI strain carrying Mycobacterium tuberculosis antigen in its genome. We found that the recombinant strain induced antigen-specific CD8+ T cell immune responses, confirming that it can be used as a vaccine carrier. However, the immune responses were not very strong (Lin et al., 2015). Ivanolysin O (ILO) differs from LLO in some respects. One study demonstrated that the recombinant strain LIΔilo:hly, constructed by replacing hly with ILO coding gene ilo, proliferated in mouse livers, but not spleens (Zhou et al., 2016). Since LLO is important for intracellular parasitism and induction of cellular immune responses by LM, we speculated that the weak immunogenicity of LI may be related to the function of ILO. We assumed that the immunogenicity of LI can be improved by replacing its hemolysin with LLO. Improved immunogenicity combined with LI’s increased biosafety may render the modified strain a safe and effective vaccine carrier.
In this study, we constructed a hemolysin gene deletion strain LIΔilo by knocking out ilo. From this strain, we constructed a modified strain, LIΔilo:hly whose ilo has been replaced by hly. We studied the in vitro growth rate, hemolytic titer, and changes in biochemical characteristics of the strain after hemolysin replacement. Next, we studied the adhesion, invasion, and proliferative ability of cells in vitro, and the biosafety, immune effect, and immune protection rate in vivo of LIΔilo:hly. Replacement of the hemolysin gene did not significantly affect bacterial growth, and LIΔilo:hly maintained biological properties similar to LI. Compared with LI, the in vitro proliferation ability of LIΔilo:hly in RAW 264.7 macrophages was significantly improved, but in vivo virulence was attenuated; after immunizing animals twice, LIΔilo:hly induced higher cytokine levels and improved the challenge protection rate. The recombinant strain LIΔilo:hly displays good biosafety and immunogenicity, and appears to be a good vector strain for vaccine construction.
Materials and methods
Bacteria, plasmids, cells, and animals
Bacteria strains LM 10403s and LI PAM55 were provided by Dr. Hao Shen (Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States), and plasmid pCW-107 was constructed for this study (Wang et al., 2014; Zhou et al., 2016; Mahdy et al., 2019). RAW 264.7, and Hepa1-6 cells were maintained in DMEM (Gibco, New York, NY, United States). Specific-pathogen-free, female C57BL/6 mice (6–8 weeks old) were purchased from Vital River Laboratory Animal Technology Co., Ltd., (Zhejiang, China) and housed at the Animal Center of the School of Public Health in Sichuan University. Animal experiments were approved by the Animal Care and Use Committee of Sichuan University.
Construction of recombinant strains
Plasmid pCW107 was extracted and digested with Xba I and Not I restriction enzymes to obtain a linear fragment of the targeting plasmid vector. The LI genome was used as a template to amplify the upstream and downstream homologous sequences of ilo. The upstream homologous sequence was inserted into the pCW107 Xba I and Not I sites to generate plasmid pCW618. This plasmid was digested with Spe I and Not I, and the downstream homologous sequence was inserted to obtain pCW619. Plasmid pCW619 was cut with Not I, then inserted into lacZ or hly fragments to obtain pCW620 carrying lacZ or pCW621 carrying hly, respectively. Plasmid pCW620 was electroporated into LI, and strain LIΔilo:lacZ was constructed by homologous recombination (Liu et al., 2020). Plasmids pCW619 and pCW621 were electroporated into LIΔilo:lacZ to construct strains LIΔilo and LIΔilo:hly, respectively. The strategy used for recombinant bacterial strain construction is shown in Supplementary Figure 1. The primers used are listed in Supplementary Table 1.
Basic characteristics
In vitro growth characteristics
Single colonies of LM, LI, LIΔilo, and LIΔilo:hly were inoculated into 5 mL BHI broth and cultured at 37°C for 18∼24 h. A portion of each culture was inoculated into 40 mL of BHI broth. The inoculum volume that was used resulted in an initial absorbance at 600 nm (A600) of each resulting suspension of 0.05. Each culture was incubated in a constant-temperature shaker at 200 rpm and at 37°C. Three milliliters were withdrawn every 1 h for 10 h to measure and record the A600 value. These measurements were used to plot growth curves.
Recombinant strain protein expression
LIΔilo:hly was inoculated into BHI broth medium and cultured to the logarithmic phase of growth. The culture supernatant and bacterial precipitate were respectively collected after centrifugation. The total protein from the culture supernatant was extracted by TCA-acetone precipitation, and the total protein from the bacterial precipitate was extracted by ultrasonic breaking and TCA-acetone precipitation. The protein samples were subjected to SDS-PAGE electrophoresis and then transferred onto a PVDF membrane. The membrane was incubated with rabbit anti-listeriolysin O antibody (1:1000) (Abcam, Cambridge, United Kingdom) as the primary antibody, and horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (H + L) (1:1,000) (Beyotime Biotechnology Co., Ltd., Shanghai, China) as the secondary antibody. The HRP chemiluminescence substrate was applied for color development and the results were analysis after incubation.
Bacterial biochemical identification test
Fresh single colonies of LM, LI, LIΔilo, and LIΔilo:hly were collected and individually added to a tube filled with 4 mL of sterile water. A bacterial suspension was prepared with a McFarland turbidity ranging from 0.4 to 0.6. A bacterial biochemical identification reagent card was placed in the bacterial suspension to sample the bacteria. After removing the bacterial liquid, the reagent card was placed in an automatic microbial identification and drug susceptibility analyzer (Merier Biotechnology Co., Lyon, France) to culture and biochemically identify bacteria.
Determination of hemolytic titer
Five milliliters of each logarithmic phase bacterial culture was centrifuged at 13,000 rpm for 5 min. Supernatants were pipetted after centrifugation into wells of a U-shaped 96-well plate and diluted two-fold to 2–8. PBS was used instead of culture supernatants to prepare the same dilution for use as a negative control. Thirty microliters of 1% sheep erythrocyte suspension was added to both sample and control wells, and then mixed, and incubated at 37°C for 2 h. The hemolytic titer of each strain was recorded.
Intracellular proliferation, adhesion and invasion to the cells
Proliferation in RAW 264.7 cells
RAW264.7 macrophages were seeded in wells of a 24-well plate at a density of 1 × 106 cells/well and cultured to 70–80% confluency. Next, 100 μL of LM, LI, LIΔilo, or LIΔilo:hly bacterial suspensions were added to each well at a multiplicity of infection (MOI) of 20:1 and incubated at 37°C for 1 h. Culture medium was discarded, cells were washed with PBS, 1 mL of DMEM medium containing 200 μg/mL gentamicin and 10% fetal bovine serum was added to each well, and cells were cultured at 37°C for 1 h to kill extracellular bacteria. Culture medium was then discarded, cells were washed with PBS, and 0.1% Triton X-100 was added to 2 wells to lyse the cells. After a 1:10 dilution, 20 μL of the suspension was dropped onto the surface of a BHI plate and cultured at 37°C for 48 h. The colony forming unit (CFU) was defined as the total number of bacteria that invaded the cells. The remaining wells were added to DMEM containing 100 μg/mL gentamicin and 10% fetal bovine serum, and incubation was continued. Cells were lysed 2, 4, and 6 h later, and the number of viable bacteria was counted using the above method. Fold changes = (total number of bacteria at each time point)/(total number of bacteria invading cells at 2 h); Multiplication factor (%) = (total number of bacteria at each time point)/(total number of bacteria initially added).
Laser scanning confocal microscopy observation of bacteria growth in RAW 264.7 cells
The pCW-gfp plasmid expressing green fluorescent protein was electroporated into LM, LI, and LIΔilo:hly to obtain LM-gfp, LI-gfp, and LIΔilo:hly-gfp. RAW264.7 cells were plated on a 24-well plate with slides. After the cells had grown to 70–80% confluency, fresh bacterial suspension was added at an MOI of approximately 10:1. After culturing for 1 h, the culture medium was replaced with DMEM containing 30 μg/mL gentamicin. A 24-well plate was removed after 1 and 7 h, and 200 μL immunostaining fixative was added to each well. Cells were fixed at room temperature for 15 min. Supernatants were discarded, and cells were washed with PBS, followed by addition of 200 μL of PBS containing 0.1% Triton X-100 to each well to lyse cell membranes for 10 min at room temperature. After washing, 200 μL PBS-Tween buffer containing 1% BSA and 22.52 mg/ml glycine was added to each well and cells were blocked at room temperature for 30 min. Supernatants were discarded, and pellets were washed. Phagosome staining (Beuzón et al., 2000) was performed using rabbit anti-lysosomal-associated membrane protein 1 (LAMP-1) antibody (Abcam, Cambridge, United Kingdom) and Alexa Fluor 647-conjugated AffiniPure donkey anti-rabbit secondary antibody (Jackson ImmunoResearch, Pennsylvania, United States). After washing, staining solution containing 10 μg/mL 4’,6-diamidino-2-phenylindol (DAPI; Solarbio, Beijing, China) was added to each well in the dark and incubated at room temperature for 5 min. The staining solution was discarded followed by thorough washing, LSCM observation, and photography.
Adhesion and invasion rate of the strains to Hepa1-6 cells
Hepa1-6 cells were seeded in 24-well plates at a density of 1 × 106 cells/well and cultured to 70–80% confluency. One hundred microliters of LM, LI, LIΔilo, or LIΔilo:hly bacterial suspensions were added to each well at an MOI of approximately 20:1.
To determine the adhesion rate, after culturing for 1 h, the 24-well plate was washed with PBS, and 0.1% Triton X-100 solution was added to lyse the cells. Suspensions were then diluted 1:10. Twenty microliters of the diluted suspension was spread onto the surface of a BHI plate and cultured at 37°C for 48 h. CFU was defined as the number of adherent bacteria. The bacterial adhesion rate (%) was calculated as [(number of adherent bacteria)/(total number of bacteria added to cells)] × 100.
To calculate the invasion rate, after culturing for 1 h, the 24-well plate was removed and washed with PBS. DMEM containing 200 μg/mL gentamicin and 10% fetal bovine serum was added to each well and cultured for 1 h. The subsequent operations were the same as described above. CFU is considered to represent the number of invading bacteria. The bacterial invasion rate (%) was calculated as = [(number of invading bacteria)/(total number of bacteria added to cells)] × 100.
In vivo safety and immunogenicity
LD50 of each strain in C57BL/6 mice
The 50% lethal dose (LD50) of LI was previously determined to be 6.3 × 106 CFU per mouse (Lin et al., 2015). To determine LD50 for each of the other strains, C57BL/6 mice were injected with 100 μL of the bacterial suspension through the tail vein. The dose groups and inoculation volumes for each strain are shown in Supplementary Table 4. Deaths among the mice were recorded for 10 days. LD50 values were determined using an improved Karber method.
Determination of bacterial load in organs
C57BL/6 mice were intravenously inoculated with LM, LI, LIΔilo, or LIΔilo:hly at a dose of 0.1 × LD50 of each strain. The mice were observed daily, and their body weights were recorded. Mice were sacrificed 1, 3, 5, 7, 9, and 14 days post-injection. The liver, spleen, and lungs were collected aseptically and homogenized in PBS containing 0.1% Triton X-100. The homogenate (50 μL) was diluted 1:10, and 20 μL was dropped onto the surface of a BHI plate. Plates were incubated at 37°C for 48 h. Colonies were counted to determine CFU values.
Determination of serum alanine aminotransferase and aspartate aminotransferase levels
At 1, 3, 5, 7, 9, and 14 days after injection, blood was collected from the mice. The serum was separated to detect the levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST).
Tissue sections and pathological observation
Liver, spleen, and lungs were aseptically dissected on days 3 and 4 after injection. Tissues were perfused with 4% paraformaldehyde solution and sent to Sewell Biotechnology Co., Ltd., (Wuhan, China) for section preparation and observation/pathological grading.
Determination of cytokine levels
Mice were randomly divided into four groups. Each mouse was intravenously inoculated twice at an interval of 7 days with LM, LI, or LIΔilo:hly at a dose of 0.1 × LD50 of each strain, or with PBS as a blank control. The mice were sacrificed 9 and 40 days after the second immunization. Splenocyte suspensions were added to 24-well plates. Mixed LLO peptides (LLO318-329 AFDAAVSGKSVS; LLO297-304 AYGRQVYL; LLO253-264 QIYYNVNVNEPT; LLO91-99 GYKDGNEYI; LLO190-201 NEKYAQAYPNVS) were used as stimulants to stimulate LM and LIΔilo:hly groups for 48 h. ILO protein was used for the LI group. Plates were centrifuged at 1,200 rpm for 20 min and supernatants were collected. Levels of tumor necrosis factor-alpha (TNF-α), interleukin (IL)-12, IL-6, IL-4, and interferon-gamma (IFN-γ) were determined according to ELISA kit (Elabscience, China) instructions.
Determination of immune protection rate
The LM, LI, LIΔilo, and LIΔilo:hly groups were immunized with the corresponding strains, as described above. Challenges were conducted 7 days after the second immunization with 5 × LD50 of LM for the LM group and 5 × LD50 of LI for the LI, LIΔilo, LIΔilo:hly, and PBS groups via the tail vein. Mice were observed for 14 days, deaths were recorded, and protection rates were calculated.
Statistical analysis
Data were analyzed using SPSS 21.0 (IBM, United States). The data with a normal distribution are expressed as means ± standard deviation. One-way ANOVA was used for parametric tests, and the LSD test was used for pairwise comparisons between groups. Non-parametric data were analyzed using the Kruskal-Wallis non-parametric test. Count data were analyzed using Fisher’s probability method. P < 0.05 indicated statistical significance.
Results
Targeting plasmid and recombinant strain construction
Four recombinant plasmids (pCW618, pCW619, pCW620, and pCW621) were obtained by amplifying target fragments using PCR, followed by enzyme digestion, ligation, and transformation. The lacZ gene fragment carried by the pCW620 plasmid was integrated into LI by electrotransformation and homologous recombination. LIΔilo:lacZ was obtained, and pCW619 and pCW621 were electro-transformed into LIΔilo:lacZ. After homologous recombination, blue-spot and white-spot screening, antibiotic screening, genome PCR, and sequencing were conducted to verify the successful construction of LIΔilo and LIΔilo:hly (Supplementary Figure 2).
Growth of the strains
The growth of LM, LI, LIΔilo, and LIΔilo:hly reached the logarithmic phase after 2 h of culture. Growth plateaued 9 h after inoculation. After 3 h, the growth rate of LM was higher than that of LI, LIΔilo, and LIΔilo:hly. The growth rate differences between LM and the other three strains were statistically significant. The rate and trend of the growth of LIΔilo:hly and LIΔilo were consistent with those observed for LI at each time point (Figure 1). The growth difference was not statistically significant, indicating that ilo knockout and hly complementation did not affect the in vitro growth of the recombinant strains.
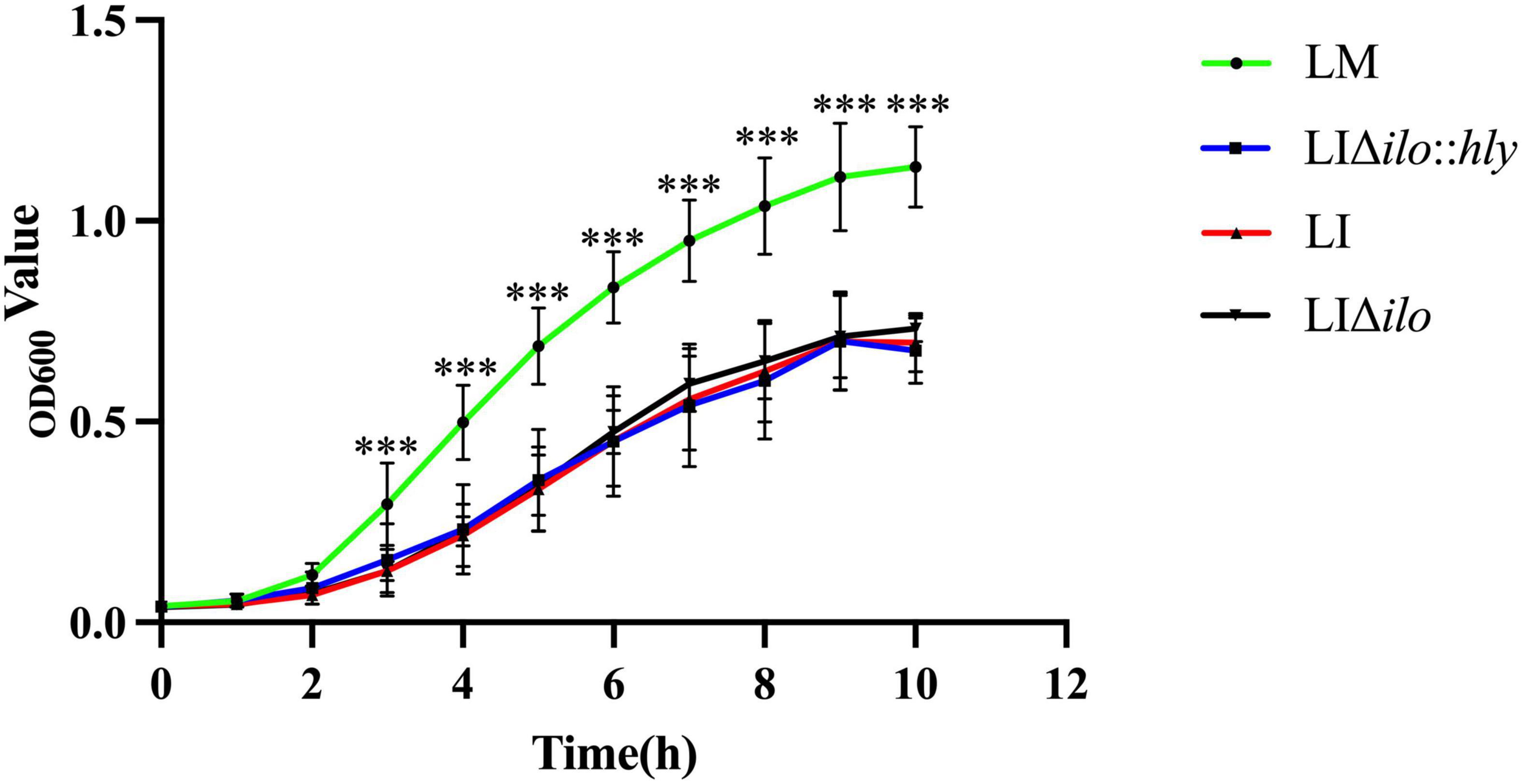
Figure 1. The growth curves of the strains. ***P < 0.001 vs. Listeria monocytogenes (LM). Experiments were carried out at least three times.
Analysis of protein expression of recombinant strain
The culture supernatant and bacterial pellet of LIΔilo:hly were collected, and the total protein was extracted from culture supernatant and bacterial pellet, respectively. The obtained protein samples were denatured and subjected to SDS-PAGE electrophoresis and western blot. The specific-band with expected length (58 KD) was obviously visible in both the medium supernatant and the cell lysate protein samples, indicating that the recombinant bacteria successfully expressed and secreted the LLO protein (Figure 2).
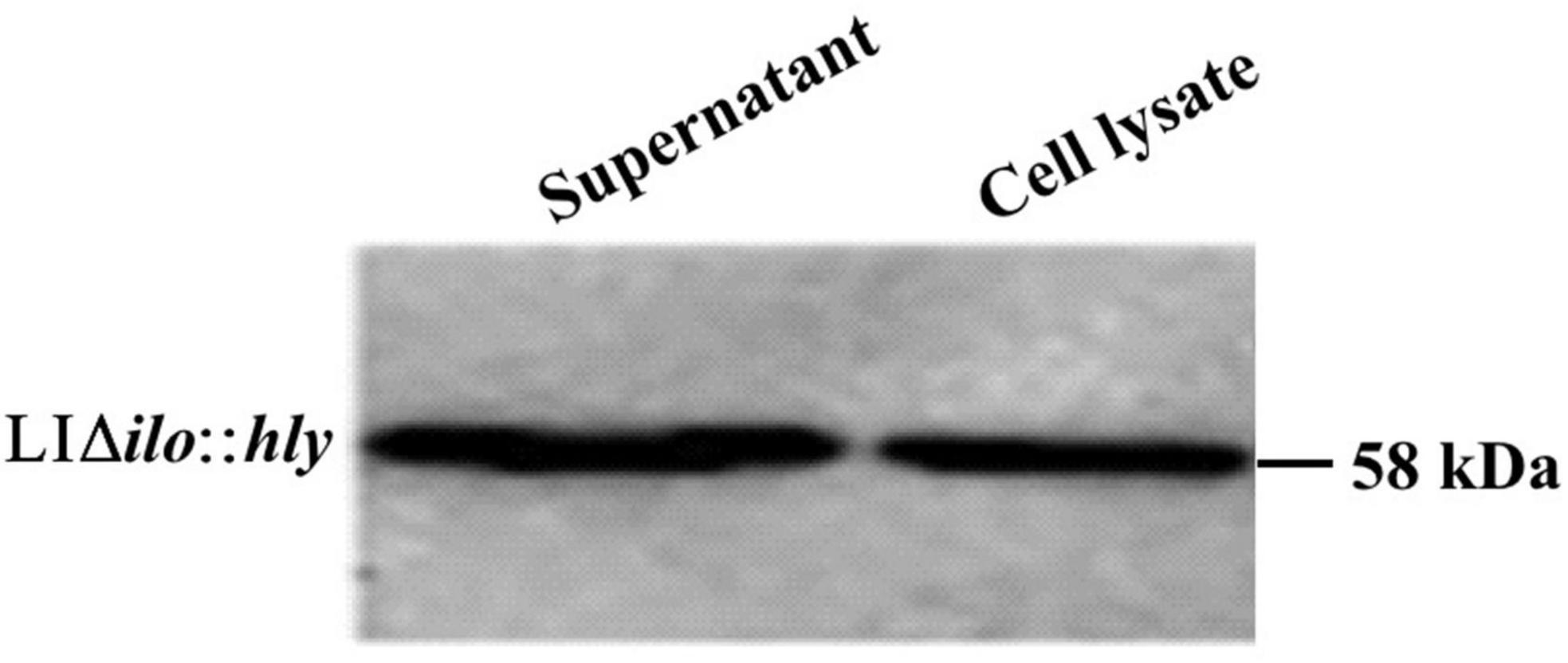
Figure 2. Western blot detection results of hemolysin-modified strain LIΔilo:hly. Supernant: the sample was the protein extracted from 35 mL medium supernatant; Cell lysate: the sample was the protein extracted from the bacterial pellet collected from 6 mL bacteria culture.
Biochemical properties of strains
Most of the biochemical properties of the hemolysin-deletion strain LIΔilo and hemolysin-modified strain LIΔilo:hly were consistent with those of LI (Supplementary Table 1). The hemolytic titer of LIΔilo:hly was higher than that of LM, but lower than that of LI (Supplementary Figure 3).
Intracellular proliferation, cell adhesion, invasion, and lysosome escape
RAW264.7 macrophages were infected with LM, LI, LIΔilo:hly, and LIΔilo, and intracellular bacterial numbers were determined at 2, 4, 6, and 8 h post-infection. All strains except LIΔilo proliferated in macrophages. LIΔilo:hly showed the same proliferation trend as that of LI. Its intracellular proliferation ability was weaker than that of LM but stronger than that of LI, indicating that the intracellular proliferation ability of the modified strain was improved to a certain extent (Figures 3A,B).
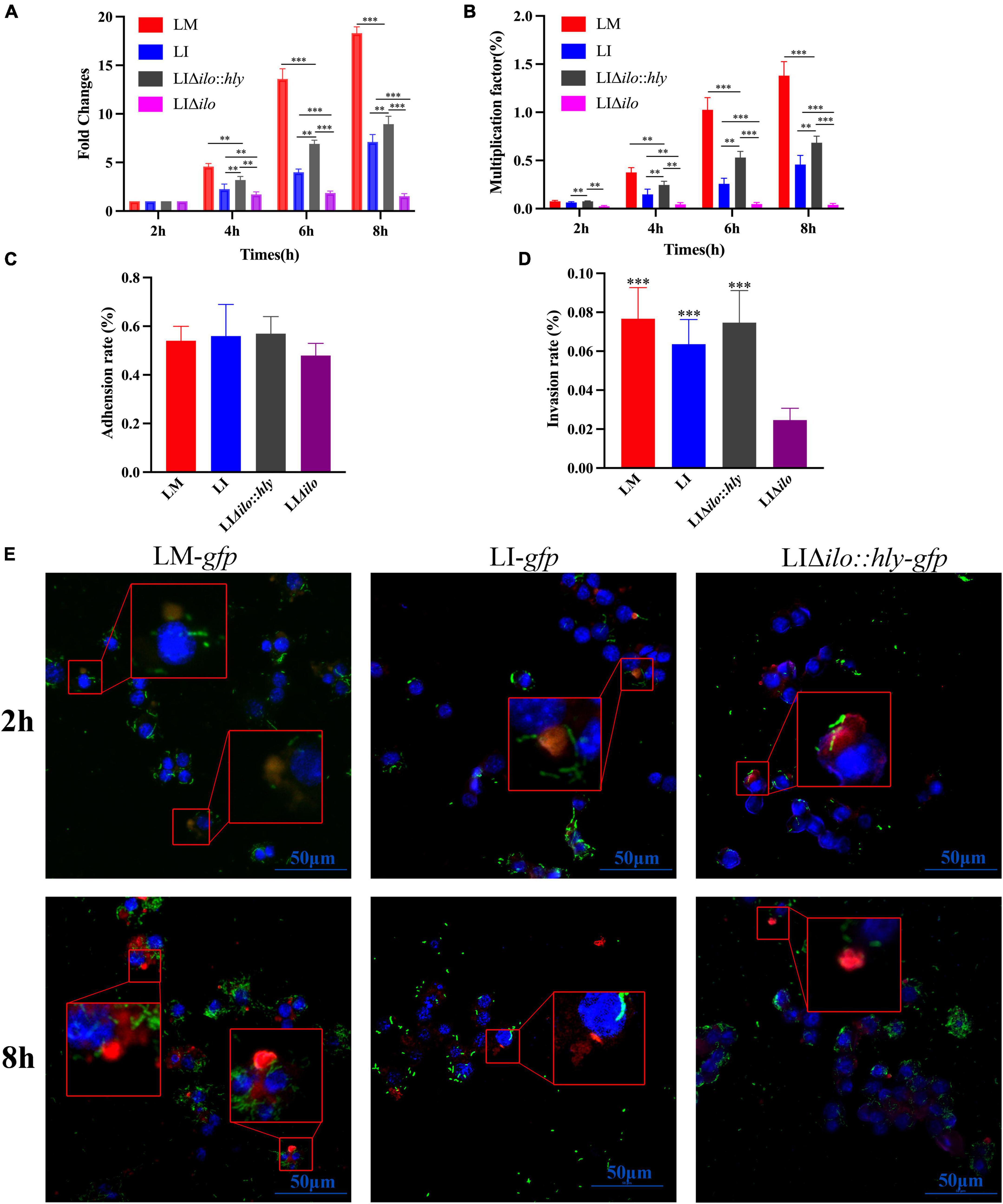
Figure 3. Intracellular proliferation, cell adhesion and invasion and lysosome escape ability. The fold changes (A) and multiplication factor (B) of each strain in mouse macrophage RAW 264.7, **P < 0.01, ***P < 0.001; The adhesion (C) and invasion (D) ability of each strain to Hepa1-6 cells, the experiments were performed with biological triplicates, and results are expressed as means ± SEM per group, ***P < 0.001, vs. LIΔilo. Experiments were carried out at least three times. The proliferation of each strain at 2 and 8 h post infection in RAW 264.7 observed by Laser scanning confocal microscopy (LSCM) (E), blue, green, and red were labeled the nucleus, bacteria and lysosomes respectively. The lysosomes in Figure 3D were orange or pink at 2 h (bacteria have not escaped), but were red at 8 h (bacteria had escaped). The scale bar in Figure 3D is 50 μm.
The adhesion rates of LM, LI, LIΔilo, and LIΔilo:hly to Hepa1-6 cells were 0.54, 0.56, 0.57, and 0.48%, respectively (Figure 3C). Knockout of ilo did not affect the ability to adhere to mouse hepatocellular carcinoma cells. The invasion rates of LM, LI, LIΔilo:hly, and LIΔilo into Hepa1-6 cells were 0.076, 0.062, 0.075, and 0.024%, respectively (Figure 3D). LIΔilo had the lowest invasion rate, indicating that knockout of ilo affected invasiveness, and that replacement with hly restored the strain’s invasiveness.
Laser scanning confocal microscopy (LSCM) of the infected macrophages (Figure 3E) showed that the bacterial numbers at 8 h were significantly higher than those at 2 h. LM-gfp showed the strongest proliferative ability, followed by LIΔilo:hly-gfp and LI-gfp. At 2 h post-infection, there were more green fluorescent bacteria in the lysosome. At 8 h post-infection, there were almost no bacteria in the lysosome and many bacteria appeared in the cytoplasm, indicating that the bacteria had escaped from the lysosomes. Because the total number of bacteria at 8 h was different between the groups, the escape ability of the strains could not be accurately compared.
In vivo biosafety evaluation
Determination of LD50, bacterial load in organs, and levels of alanine aminotransferase and aspartate aminotransferase
To evaluate the virulence of each strain in vivo, the LD50 of LM, LIΔilo:hly and LIΔilo in C57BL/6 mice was determined. LD50 results were 4.3 × 104 CFU/mouse for LM, 1.1 × 107 CFU/mouse for LIΔilo:hly, and 4.3 × 107 CFU/mouse for LIΔilo. The LD50 of LI was 6.3 × 106 CFU/mouse (determined in our laboratory) (Lin et al., 2015). Virulence in vivo was significantly attenuated by ilo knockout. Although virulence was restored after hly compensation, the strain still had reduced virulence compared with LI, confirming the increased biosafety of LIΔilo:hly (Figures 4A–C).
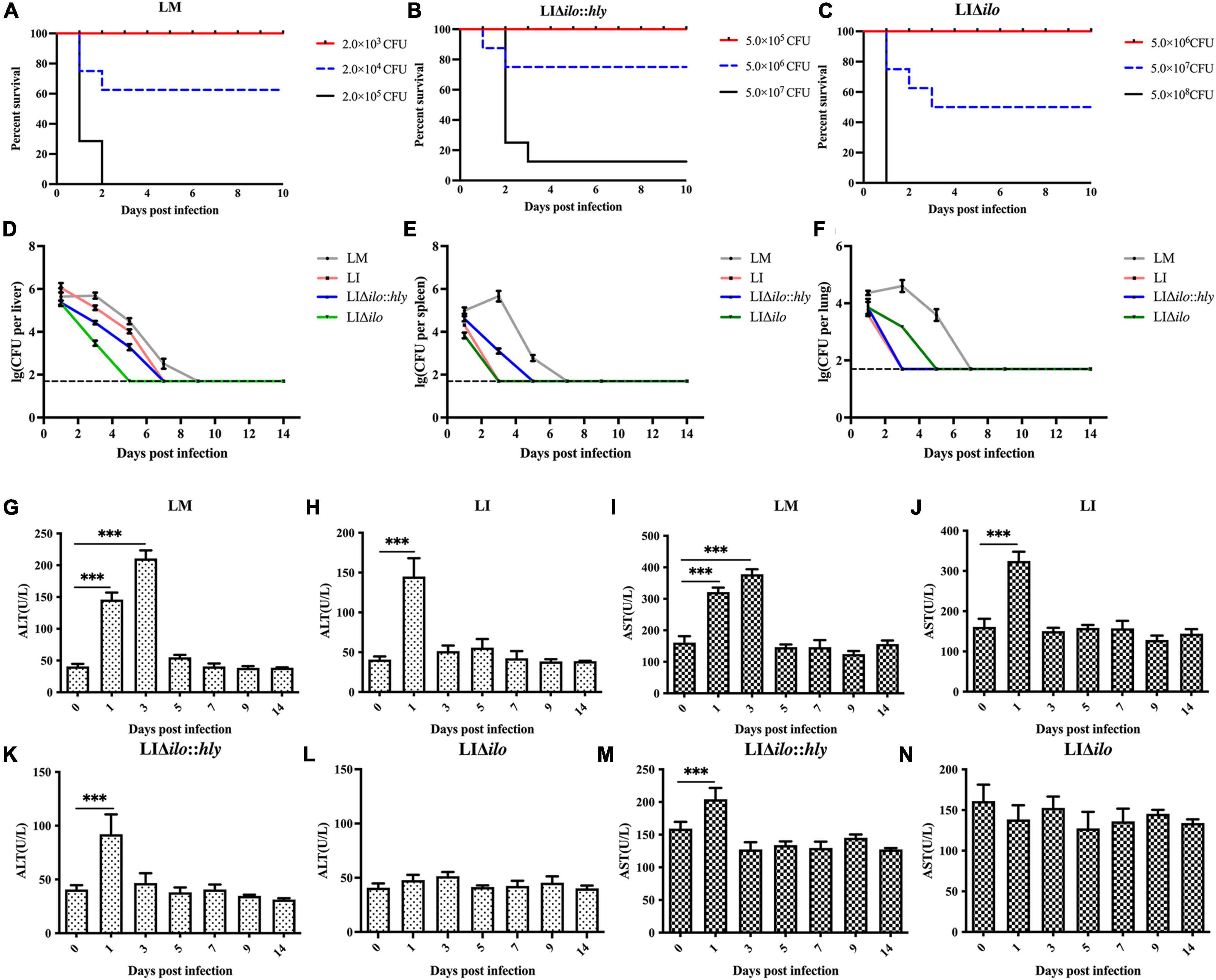
Figure 4. In vivo biosafety evaluation of each strain. Survival curves of C57BL/6 mice intravenously inoculated of Listeria monocytogenes (LM) (A), LIΔilo:hly (B), and LIΔilo (C); Eight mice per group in this experiment. The bacteria load in liver (D), spleen (E), and lung (F) of mice intravenously inoculated of each strain; each point represents the mean ± SEM for a group of three mice from one independent experiment. Mice serum ALT levels after inoculation of LM (G), LI (H), LIΔilo:hly (K), and LIΔilo (L), and mice serum AST levels after inoculation of LI (J), LM (I), LIΔilo:hly (M), and LIΔilo (N). Results are expressed as means ± SEM per group, ***P < 0.005. The experiments were performed with biological triplicates.
C57BL/6 mice were inoculated with 0.1 × LD50 of each strain as the immunizing dose. The growth of the strains in the main organs of the mice is shown in Figures 4D–F. Bacterial numbers in the liver, spleen, and lung of the LM immunized mice group reached a peak on day 3, while peak bacterial numbers in the liver, spleen, and lung of the LI, LIΔilo:hly, and LIΔilo immunized mice groups were observed on day 1. Bacterial numbers gradually decreased thereafter, indicating that the body quickly began to clear bacteria. Notably, the mice required two more days to completely remove LIΔilo:hly from the spleen than to clear LI from the spleen.
Serum ALT and AST levels reflect the status of the liver. As shown in Figures 4G–N, the ALT and AST levels in the serum of mice after LIΔilo inoculation did not change significantly, indicating that LIΔilo inoculation caused no obvious damage to the liver. At day 1 after inoculation, the levels of ALT and AST in the serum of LM-, LI-, and LIΔilo:hly inoculated mice were significantly higher than those before inoculation (P < 0.001). Afterward, they began to decrease and returned to the level of the control group on day 5 in the LM group or on day 3 in the LIΔilo:hly group. This trend is consistent with the above bacterial organ load trend.
Organ pathological examination
After tail vein inoculation, the pathological changes in the liver, spleen, and lung showed a consistent trend in each group. A visible pathological change occurred on day 3, but the pathological status was restored to normal by day 14 (Figures 5A–C). The pathological status of the LIΔilo:hly group at day 3 was better than that of the LM and LI groups. In the liver, a small amount of inflammatory cell focal infiltration in the hepatic lobule (black arrow) was accompanied by inflammatory cell punctate infiltration (red arrow). In the spleen, there was occasional pyknosis and fragmentation accompanied by a small amount of inflammatory cell infiltration. The pathological degree of the spleen was weaker than that of the LM group and the LI group. In the lung, a small amount of alveolar wall around the airway of the lung tissue was slightly thickened (black arrow), the alveolar space was narrow, and inflammatory cell infiltration was scattered. Pathological scores for each organ are shown in Supplementary Table 3. The overall pathological severity of the LIΔilo:hly group was lower than that of the LM and LI groups, indicating that hly supplementation improved the biosafety of the strain.
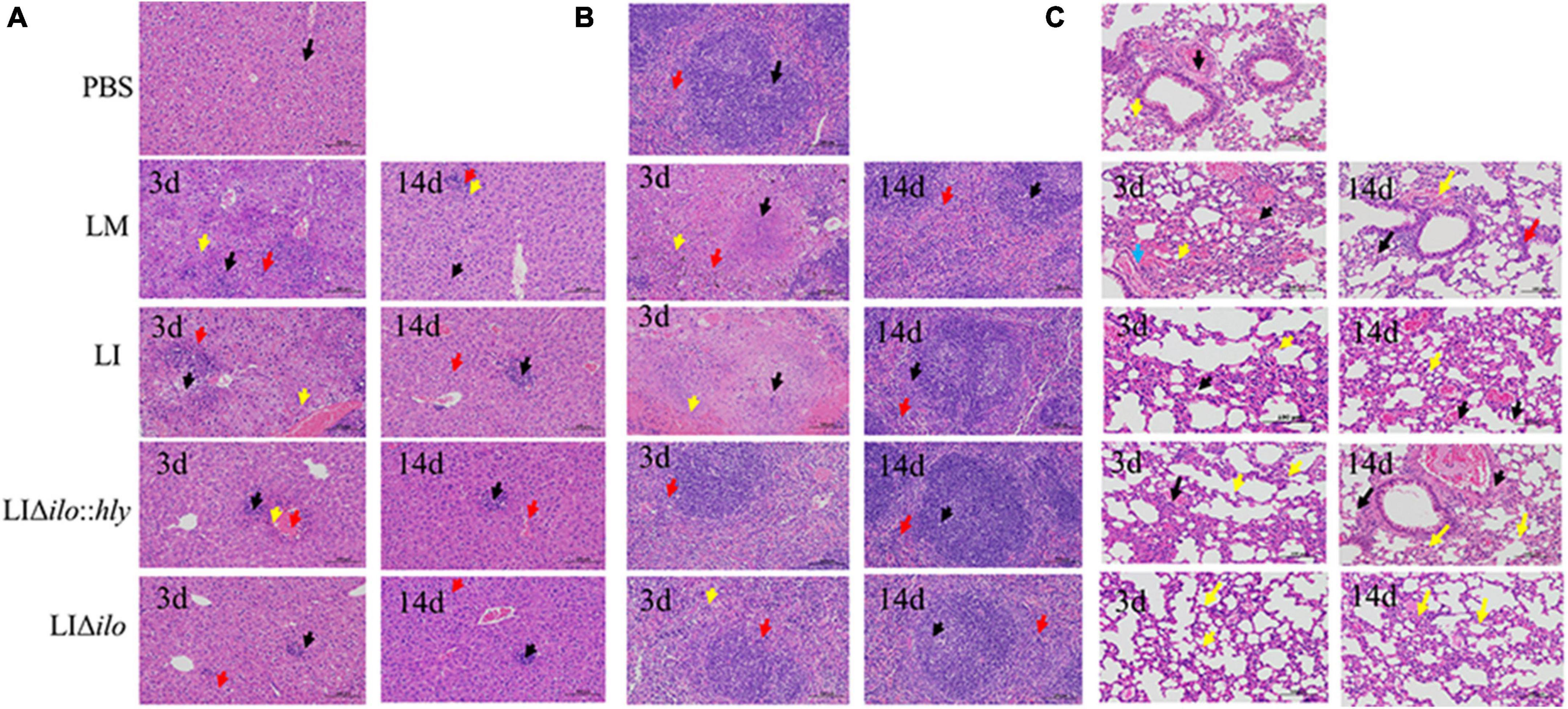
Figure 5. Pathological changes of mouse organs. Pathological changes of Liver (A), spleen (B), and lung (C) after inoculation of each strain (200×). The arrows in panel (A) indicated the lesions as below: ballooning or cytoplasmic vacuolation (black arrows); small foci of necrosis (yellow arrows), and inflammatory cell infiltration (red arrows); The arrows in panel (B) indicated the lesions as below: central expansion of the white pulp or pyknotic fragmentation (black arrows); extensive hemorrhage or multinucleated giant cells (yellow arrows), and inflammatory cell infiltration (red arrows); The arrows in panel (C) indicated the lesions as below: minor hemorrhage in the alveolar space (blue arrow); central dilation of the white pulp or pyknosis and fragmentation (black arrow); small hemorrhage (red arrow), and inflammatory cell infiltration (yellow arrow).
In vivo immunogenicity and immune protection
The mice in each group were immunized twice by tail vein injection. Spleens were collected aseptically at on days 9 and 40 after the second immunization to prepare splenocyte suspensions for cytokine determination. TNF-α, IL-4, IL-12, IL-6, and IFN-γ levels in the LM, LI, and LIΔilo:hly groups were higher than those in the PBS control group. The levels of TNF-α, IL-6, and IFN-γ at day 9 and the levels of IL-6 and IFN-γ at day 40 in the LIΔilo:hly group were higher than those in the LI group (Figures 6A–E). Mice were challenged 1 week after the second immunization. The protection rates of the LM, LI, LIΔilo:hly, LIΔilo, and PBS groups were 60, 30, 40, 0, and 0%, respectively. There was a statistically significant difference in the protection rate between LIΔilo:hly and PBS groups. The protection rate of the LIΔilo:hly group was 10% higher than that of the LI group. However, the difference was not significant (Figure 6F).
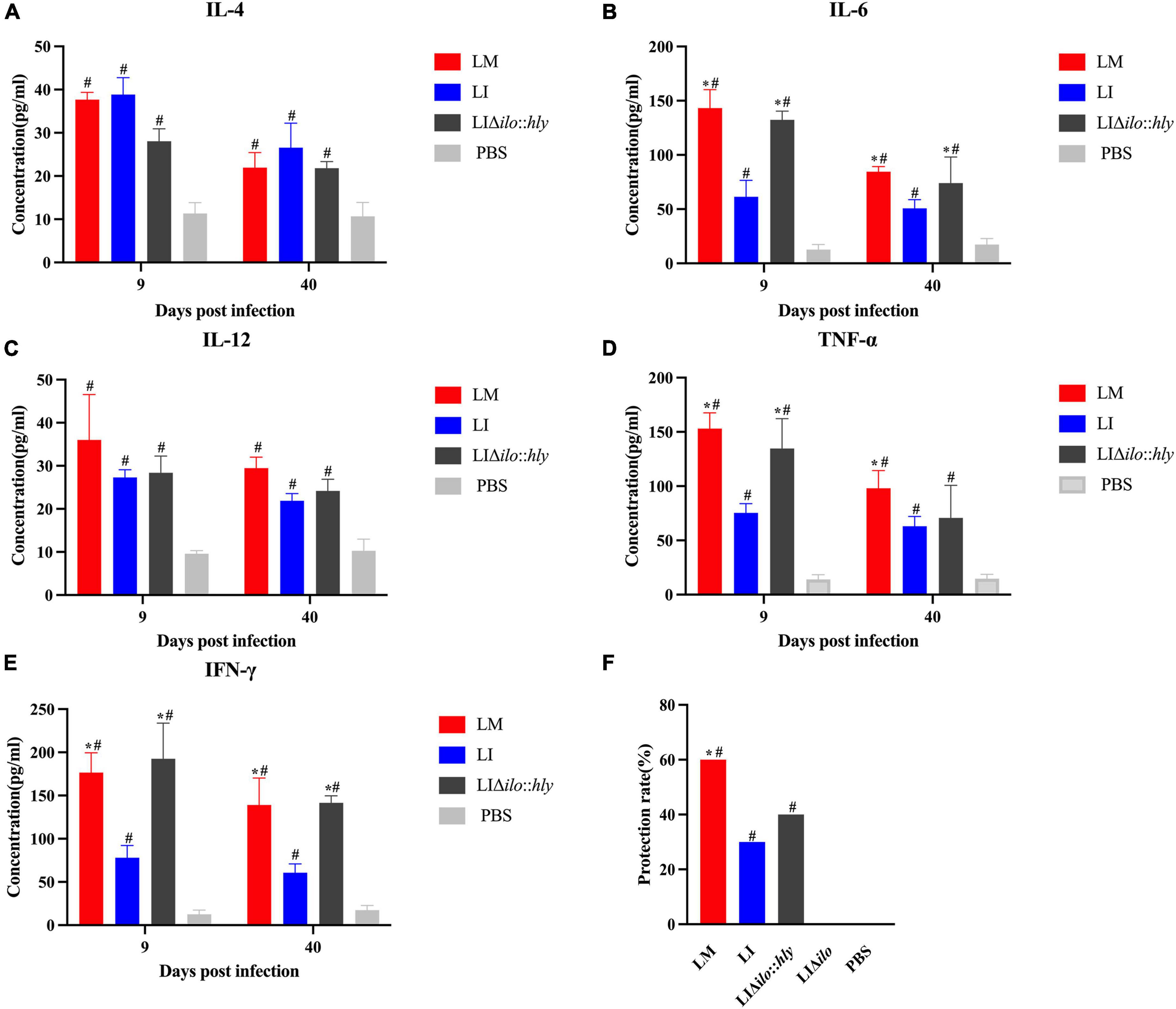
Figure 6. Immunogenicity and immune protection of each strain. Cytokines IL-4 (A), IL-6 (B), IL-12 (C), TNF-α (D), and IFN-γ (E) at the 9th and 40th day after the secondary immunization of each strain, *P < 0.05, vs. Listeria ivanovii (LI); #P < 0.05, vs. PBS, the experiments were performed with biological triplicates. Each bar represents the mean ± SEM for a group of three mice from one independent experiment; Challenge protection rate after twice immunizations (F), *P < 0.05, vs. LI; #P < 0.05, vs. PBS. Ten mice per group in this experiment.
Discussion
A qualified live bacterial vaccine carrier should be safe for the host, and should simultaneously stimulate the host immune system to generate a corresponding immune response. Improving the biosafety of LM as a vaccine carrier is an important goal for researchers. Most attenuation strategies adopted for LM are based on inactivating virulence. Although virulence has been greatly reduced, potential biosafety problems remain. Sometimes, deletion of the key virulence genes of Listeria will affect its ability to grow and proliferate in vitro and in vivo, and also affect its ability to stimulate cellular immune responses and the efficiency of antigen presentation (Maciag et al., 2009; Shahabi et al., 2011; Fares et al., 2019). Insufficient attenuation of LM may lead to potential safety problems. Conversely, excess attenuation of LM may lead to low immunogenicity. Therefore, we used a different approach by selecting LI, which displays good biosafety, as the vaccine carrier. To improve the immunogenicity of LI, we replaced the ilo gene with hly. LLO is an important virulence protein in LM that triggers innate and adaptive immune responses (Mandal and Lee, 2002; Radford et al., 2002). LLO is also utilized as an adjuvant when constructing subunit vaccines to improve the immunogenicity of the antigen. LLO can stimulate the innate immune system and induce the expression of IL-1, IL-12, and IL-18 in macrophages, and the production of IFN-γ by natural killer cells (Kohda, 2002; Carrero et al., 2012). A previous study constructed a recombinant Escherichia coli strain expressing LLO and ovalbumin (OVA) and found that it could kill OVA-expressing melanoma cells (B16-OVA) and effectively inhibit tumor growth in mice. Additionally, LLO can mediate bacterial escape from phagosomes and promote bacterial proliferation during infection. LLO plays a crucial role in the control and specific regulation of immune responses. Therefore, in this study, we modified the hemolysin gene of LI with the expectation of full use of the unique advantages of LLO to obtain an excellent vaccine vector.
In this study, hly was complemented by ilo deletion in the LI strain. The cell invasion and proliferation abilities, biosafety, and immunogenicity of the modified strain LIΔilo:hly were evaluated. The modified strain grew stably in vitro and maintained a growth trend similar to that of wild type LI, and it successfully expressed and secreted LLO protein. It could proliferate in phagocytic cells, and its proliferative capacity was stronger than that of LI, confirming that the intracellular proliferative capacity is closely related to hemolysin, and LLO is more helpful in phagocytic vesicle escape and intra-cytoplasm proliferation. Knockout of ilo significantly reduced cell invasion ability, and replacement with hly restored invasion, indicating that hemolysin is also related to cell invasion. In addition to hemolysin, the internalins of Listeria are also closely related to its adhesion and invasion properties. Internalins A (InlA) and Internalins B (InlB) play their respective roles by binding to specific receptors through leucine rich repeats (LRRs), of which InlA mainly binds Cadherin mediates the passage of bacteria across epithelial cells, and InlB mainly binds to the hepatocyte growth factor receptor (HGFR) and mediates the entry of bacteria into fibroblasts, hepatocytes and epithelial cells (Vázquez-Boland et al., 2001; Domínguez-Bernal et al., 2006; Hamon et al., 2006). The LD50 of LIΔilo:hly was higher than that of LI, and the overall pathological status of the liver, spleen, and lungs was better than that of LI, suggesting that it is less virulent and safer than LI. We speculate that replacement of ilo by hly may affect the expression levels of virulence genes encoded by Listeria pathogenicity islands (LIPI). LIPI is recognized as unstable chromosomal regions, carrying genes related to specific metabolic activities, antibiotic resistance, or pathogenesis, which can be horizontally transferred between bacteria, and contains the essential virulence genes (prfA, hly, plcA, mpl, actA, and plcB) and internalin genes of Listeria (Hacker and Kaper, 2000; Vázquez-Boland et al., 2001). We also speculate that the synergy effect of hly gene with other virulence genes is weakened within the genome environment of LI. After LIΔilo:hly infection, it was maintained in the liver of mice with a higher bacterial number and a longer duration than in other organs, indicating its superiority in colonizing the liver. This is mainly because the internalin inlB of Listeria can bind to specific receptors on the surface of liver cells, resulting in the targeted invasion of the liver by Listeria (Khelef et al., 2006; Pentecost et al., 2010). The clearance time for LIΔilo:hly from the spleen was two more days than that for LI, indicating that LLO has better spleen cell adaptability than ILO. This result also confirmed a previous report that LMΔhly:ilo cannot proliferate in the spleen (Frehel et al., 2003). We speculate that the longer survival time of LIΔilo:hly in the spleen may elicit stronger immune responses. After mice were immunized twice with each strain, we detected levels of IFN-γ, TNF-α, IL-4, IL-6, and IL-12. These cytokines coordinate an effective immune response during Listeria infection, and are closely related to the establishment of protection. Their interactions promote the proliferation and activation of immune cells, resulting in a stronger and longer-lasting cellular immune response. The results showed that on days 9 and 40 after the secondary immunization of mice with LIΔilo:hly, these cytokines were significantly higher than those in the PBS group, and were comparable to those in the LM group. Especially, the levels of IFN-γ, IL-6, and TNF-α (at day 9) were higher than those of LI group. In addition, the challenge results showed that the immune protection rate of LIΔilo:hly was higher than that of LI. These results are consistent with our hypotheses. A longer survival time in the spleen can result in a more durable, specific immune response. LIΔilo:hly has comparable potency to LM in causing a T helper 1 (Th1) cell-mediated immunity response and inducing various cytokines and chemokines.
The recombinant strain LIΔilo:hly was successfully constructed. Through evaluations at both the cellular and animal levels, we confirmed its safety and immunogenicity. Thus, it can be used as a vaccine carrier. In the future, the heterologous antigen genes can be introduced into the recombinant LI strain by genome recombination technique described previously (Wang et al., 2014) or by recombinant plasmid that carrying heterologous antigen genes, thus to acquire the potential vaccine candidates. Subsequently, it is necessary to explore whether the vaccine candidates can stably express and secret foreign genes by western blot, and through animal experiments to determine antibody titers and measure T/B cell immune responses, to evaluate the safety and efficacy of vaccine candidate strains (Xiao et al., 2021; Clint et al., 2022). Thus to prove that LIΔilo:hly is an excellent antigen carrier for vaccine development, contributing to cancer immunotherapy and infectious diseases prevention.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Sichuan University.
Author contributions
CW and ZC conceived and designed the research. QL, RL, SL, YZ, ST, and QO performed the experiments and acquired, interpreted, and analyzed the data. QL, RL, SL, and CW wrote the manuscript. All the authors read and critically reviewed the manuscript and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 31570924) and the Key Program of Sichuan Science and Technology Department (Nos. 2021YFQ0060 and 2021YFS0005).
Conflict of interest
QL, RL, and ZC were employed by Shen Zhen Biomed Alliance Biotech Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.962326/full#supplementary-material
References
Beuzón, C. R., Méresse, S., Unsworth, K. E., Ruíz-Albert, J., Garvis, S., Waterman, S. R., et al. (2000). Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19, 3235–3249.
Carrero, J. A., Vivanco-Cid, H., and Unanue, E. R. (2012). Listeriolysin O Is Strongly Immunogenic Independently of Its Cytotoxic Activity. PLoS One 7:e32310. doi: 10.1371/journal.pone.0032310
Clint, A. H., Rafael, K. C., Sasha, R. A., Tiffany, F. K., Naomi, L. F., et al. (2022). Venezuelan equine encephalitis virus V3526 vaccine RNA-Dependent RNA polymerase mutants increase vaccine safety through restricted tissue tropism in a murine model. Zoonoses 2:2. doi: 10.15212/zoonoses-2021-0016
Domínguez-Bernal, G., Müller-Altrock, S., González-Zorn, B., Scortti, M., Herrmann, P., Monzó, H. J., et al. (2006). A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mole. Microb. 59, 415–432. doi: 10.1111/j.1365-2958.2005.04955.x
Farber, J. M., and Peterkin, P. (1991). Listeria monocytogenes, a food-borne pathogen. Microbiolog. Rev. 55, 476–511.
Fares, E., Mccloskey, C. B., Gutierrez, A., Princiotta, M., and Drevets, D. A. (2019). Vaccine strain Listeria monocytogenes bacteremia occurring 31 months after immunization. Infection 47, 489–492. doi: 10.1007/s15010-018-1249-7
Frehel, C., Lety, M. A., Autret, N., Beretti, J. L., Berche, P., Charbit, A., et al. (2003). Capacity of ivanolysin O to replace listeriolysin in phagosomal escape and in vivo survival of Listeria monocytogenes. Microbiology 149, 611–620. doi: 10.1099/mic.0.25986-0
Hacker, J., and Kaper, J. B. (2000). Pathogenicity islands and the evolution of microbes. Rev. Microbiol. 54, 641–679. doi: 10.1146/annurev.micro.54.1.641
Hamon, M., Bierne, H., and Cossart, P. (2006). Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4, 423–434. doi: 10.1038/nrmicro1413
Harty, J. T., Lenz, L. L., and Bevan, M. J. (1996). Primary and secondary immune responses to Listeria monocytogenes. Curr. Opin. Immunol. 8, 526–530.
Khelef, N., Lecuit, M., Bierne, H., and Cossart, P. (2006). Species specificity of the Listeria monocytogenes InlB protein. Cell Microbiol. 8, 457–470. doi: 10.1111/j.1462-5822.2005.00634.x
Kohda, C. (2002). Dissociated Linkage of Cytokine-Inducing Activity and Cytotoxicity to Different Domains of Listeriolysin O from Listeria monocytogenes. Infect. Immun. 70, 1334–1341. doi: 10.1128/IAI.70.3.1334-1341.2002
Lamikanra, A., Pan, Z.-K., Isaacs, S. N., Wu, T.-C., and Paterson, Y. (2001). Regression of established Human papillomavirus type 16 (HPV-16) immortalized tumors in vivo by vaccinia viruses expressing different forms of HPV-16 E7 correlates with enhanced CD8+ T-cell responses that home to the tumor site. J. Virol. 75, 9654–9664. doi: 10.1128/JVI.75.20.9654-9664.2001
Le, D. T., Picozzi, V. J., Ko, A. H., Wainberg, Z. A., Kindler, H., Wang-Gillam, A., et al. (2019). Results from a phase IIb, randomized, multicenter study of GVAX pancreas and CRS-207 compared with chemotherapy in adults with previously treated metastatic pancreatic adenocarcinoma (ECLIPSE study). Clin. Can. Res. 25, 5493–5502. doi: 10.1158/1078-0432.CCR-18-2992
Lin, Q., Zhou, M., Xu, Z., Khanniche, A., Shen, H., Wang, C., et al. (2015). Construction of two Listeria ivanovii attenuated strains expressing Mycobacterium tuberculosis antigens for TB vaccine purposes. J. Biotechnol. 196, 20–26. doi: 10.1016/j.jbiotec.2015.01.008
Liu, S.-J., Tian, S.-C., Zhang, Y.-W., Tang, T., Zeng, J.-M., Fan, X.-Y., et al. (2020). Heterologous Boosting With Listeria-Based Recombinant Strains in BCG-Primed Mice Improved Protection Against Pulmonary Mycobacterial Infection. Front. Immunol. 11:2036. doi: 10.3389/fimmu.2020.02036
Maciag, P. C., Radulovic, S., and Rothman, J. (2009). The first clinical use of a live-attenuated Listeria monocytogenes vaccine: a Phase I safety study of Lm-LLO-E7 in patients with advanced carcinoma of the cervix. Vaccine 27, 3975–3983. doi: 10.1016/j.vaccine.2009.04.041
Mahdy, S. E., Liu, S., Su, L., Zhang, X., Chen, H., Pei, X., et al. (2019). Expression of the VP1 protein of FMDV integrated chromosomally with mutant Listeria monocytogenes strain induced both humoral and cellular immune responses. Appl. Microbiol. Biotechnol. 103, 1919–1929. doi: 10.1007/s00253-018-09605-x
Mandal, M., and Lee, K. D. (2002). Listeriolysin O-Liposome-Mediated cytosolic delivery of macromolecule antigen in vivo: enhancement of antigen-specific cytotoxic T lymphocyte frequency, activity, and tumor protection. Biochimica et Biophysica Acta 1563, 7–17. doi: 10.1016/s0005-2736(02)00368-1
Pentecost, M., Kumaran, J., Ghosh, P., and Amieva, M. R. (2010). Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 6:e1000900. doi: 10.1371/journal.ppat.1000900
Radford, K. J., Higgins, D. E., Pasquini, S., Cheadle, E. J., Carta, L., Jackson, A. M., et al. (2002). A recombinant E. coli vaccine to promote MHC class I-dependent antigen presentation: application to cancer immunotherapy. Gene Therapy 9, 1455–1463. doi: 10.1038/sj.gt.3301812
Ruan, Y., Rezelj, S., Bedina Zavec, A., Anderluh, G., and Scheuring, S. (2016). Listeriolysin O membrane damaging activity involves arc formation and lineaction–implication for Listeria monocytogenes escape from phagocytic vacuole. PLoS Pathog. 12:e1005597. doi: 10.1371/journal.ppat.1005597
Shahabi, V., Seavey, M. M., Maciag, P. C., Rivera, S., and Wallecha, A. (2011). Development of a live and highly attenuated Listeria monocytogenes-based vaccine for the treatment of Her2/neu-overexpressing cancers in human. Cancer Gene Ther. 18, 43–62. doi: 10.1038/cgt.2010.48
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domıìnguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/CMR.14.3.584-640.2001
Wallecha, A., Carroll, K. D., Maciag, P. C., Rivera, S., Shahabi, V., and Paterson, Y. (2009). Multiple effector mechanisms induced by recombinant Listeria monocytogenes anticancer immunotherapeutics. Adv. Appl. Microbiol. 66, 1–27. doi: 10.1016/S0065-2164(08)00801-0
Wang, C., Zhang, F., Yang, J., Khanniche, A., and Shen, H. (2014). Expression of Porcine Respiratory and Reproductive Syndrome Virus Membrane-Associated Proteins in Listeria ivanovii via a Genome Site-Specific Integration and Expression System. J. Mole. Microbiol. Biotechnol. 24, 191–195. doi: 10.1159/000363450
Xiao, X., Zhou, K., Gu, M., Li, G., Li, Y., and Liao, et al. (2021). The immungenicity and Cross-Neutralizing Activity of Enterovirus 71 Vaccine Candidate Strains. Zoonoses 1:9. doi: 10.15212/ZOONOSES-2021-0008
Yang, Y., Hou, J., Lin, Z., Zhuo, H., Chen, D., Zhang, X., et al. (2014). Attenuated Listeria monocytogenes as a cancer vaccine vector for the delivery of CD24, a biomarker for hepatic cancer stem cells. Cell. Mole. Immunol. 11, 184–196. doi: 10.1038/cmi.2013.64
Keywords: Listeria monocytogenes, Listeria ivanovii, listeriolysin O, ivanolysin O, immune effect
Citation: Liang Q, Li R, Liu S, Zhang Y, Tian S, Ou Q, Chen Z and Wang C (2022) Recombinant Listeria ivanovii strain expressing listeriolysin O in place of ivanolysin O might be a potential antigen carrier for vaccine construction. Front. Microbiol. 13:962326. doi: 10.3389/fmicb.2022.962326
Received: 06 June 2022; Accepted: 30 June 2022;
Published: 22 July 2022.
Edited by:
Wei Wang, Jiangsu Institute of Parasitic Diseases (JIPD), ChinaCopyright © 2022 Liang, Li, Liu, Zhang, Tian, Ou, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaobin Chen, Y2hlbnpiLm1kQHZpcC4xNjMuY29t; Chuan Wang, d2FuZ2NodWFuQHNjdS5lZHUuY24=
†These authors share first authorship
 Qian Liang
Qian Liang Ruidan Li1,2†
Ruidan Li1,2† Yunwen Zhang
Yunwen Zhang Sicheng Tian
Sicheng Tian Chuan Wang
Chuan Wang