- 1Infectious Diseases Programme, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore
- 2Department of Chemical Immunology, Leiden University Medical Center, Leiden, Netherlands
- 3Pathogen Biology Laboratory, Department of Biotechnology and Bioinformatics, University of Hyderabad, Hyderabad, India
The diverse microbial community that colonizes the gastrointestinal tract has remarkable effects on the host immune system and physiology resulting in homeostasis or disease. In both scenarios, the gut microbiota interacts with their host through ligand-receptor binding whereby the downstream signaling processes determine the outcome of the interaction as disease or the counteractive immune responses of the host. Despite several studies on microbe-host interactions and the mechanisms by which this intricate process happens, a comprehensive and updated inventory of known ligand-receptor interactions and their roles in disease is paramount. The ligands which originate as a result of microbial responses to the host environment contribute to either symbiotic or parasitic relationships. On the other hand, the host receptors counteract the ligand actions by mounting a neutral or an innate response. The varying degrees of polymorphic changes in the host receptors contribute to specificity of interaction with the microbial ligands. Additionally, pathogenic microbes manipulate host receptors with endogenous enzymes belonging to the effector protein family. This review focuses on the diversity and similarity in the gut microbiome-host interactions both in health and disease conditions. It thus establishes an overview that can help identify potential therapeutic targets in response to critically soaring antimicrobial resistance as juxtaposed to tardy antibiotic development research.
Introduction
Facultative anaerobic enteric bacteria form a part of the normal intestinal microbiota of humans and animals (Morales-López et al., 2019; Dougnon et al., 2020; Janda and Abbott, 2021). These bacteria which mostly belong to the Enterobacteriaceae family are responsible for shaping both the metabolic and physiological processes in addition to the development of the immune system in the host body (Martin et al., 2019; Bajinka et al., 2020). A majority of gastrointestinal (GI) microbes form a complex bacterial community and maintain a commensal relationship with the host. Some bacterial species of the genera Escherichia, Shigella, Salmonella, Proteus and Yersinia constantly invade the host GI tract, the blood and vascular system and the genitourinary tract thereby causing and spreading infection (Dekker and Frank, 2015; Hamilton et al., 2018; Gu et al., 2019; Aljahdali et al., 2020; Ramos et al., 2020; Yang et al., 2020). Among various defense mechanisms, the formation of a mucus layer by the host cells in the gut acts as a natural barrier blocking foreign pathogens from invading the intestinal epithelial lining. This mucus membrane acts as a major component of the innate immunity (Okumura and Takeda, 2017; Sicard et al., 2017). A large number of intestinal microbes inhabit the outer mucus layer. The constant degradation and replenishment of the mucin layer helps resist colonization and promote commensalism with the gut microbiota (Josenhans et al., 2020; Fang et al., 2021). Nevertheless, invasive bacterial strains residing in the intestine have evolved strategies to adhere to or swim out of the mucus barrier by the production of adhesins, flagella, and fimbriae, leading to a chronic and sometimes symptomatic colonization in the gut. A compromised mucus barrier can lead to various pathological conditions (Cornick et al., 2015; Paone and Cani, 2020). In response to mucin degradation, the host responds with a myriad of defensive measures against the microbial attack. Among them is the escalation of the host immune response through recognition of pathogen- or damage-associated molecular patterns (PAMPs or DAMPs) by the pattern recognition receptors (PRRs). The PAMPs can be polymers of carbohydrates, proteins or even nucleic acids that are essential for microbial survival and hence their modifications could be detrimental (Patten and Collett, 2013). The PRRs are specialized immune receptors on the host epithelial cells which act upon binding with microbial PAMPs. The most abundant PRRs are the toll-like receptors (TLRs) that are germ line-encoded in the immune system (Akira et al., 2006; Nie et al., 2018). Downstream to PAMP–PRR interaction both on the cell surface or in the intracellular environment, several proinflammatory and anti-microbial responses are triggered by the activation of a wide range of intracellular signaling pathways (Akira et al., 2006). Phagocytic and antigen-presenting cells (APCs) then mediate the innate immune responses. Furthermore, the PRR-induced signaling pathways result in the synthesis of molecules such as chemokines, cytokines, cell adhesion molecules, and immunoreceptors underpinning an early host response to infection (Iwasaki and Medzhitov, 2004; Kogut et al., 2020). To counteract reactions elicited by these receptors, microbes express effector proteins which have the ability to perturb host defense mechanisms. These effector proteins facilitate microbial infection whilst deriving nutrients from the host (Santos, 2015; Popa et al., 2016). In turn, the immune system monitoring the microbial communities acts by inducing inflammation against potential pathogens while retaining tolerance towards commensals in the gut (Tel et al., 2016; Zheng et al., 2020). To this end, many studies have reported on how different microbial ligands modulate microbial sensing of the host. While studies have focused rather exclusively on a few host receptors, a summarized information on the diversity of the host receptors and microbial ligands is limited. This review focuses on consolidating the diversity of enterobacterial interactions with microbial sensors to gain an understanding on how bacteria could possibly modify recognition by host receptors. Furthermore, a brief outlook into the potential therapeutic values of the host-microbiome interactions and the players involved in this process are also addressed.
Human host receptors
The gut mucosa has the ability to readily identify and neutralize microbial infections in addition to distinguishing a microbial attack by pathogens from symbiotic relationship of non-pathogens. Human host receptors of the innate immune system, namely the toll-like receptors (TLRs), nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), C-type lectin receptors (CTLRs) and the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) play a crucial role in host-microbiota communication (Delbridge and O’Riordan, 2007; Kawai and Akira, 2011; Dierking and Pita, 2020). These receptors differ from two other PRRs, i.e., G-protein coupled receptors (GPCRs) and peptidoglycan (PG) recognition proteins (PGRPs) by their specificity to recognize distinct microbial molecules (Dierking and Pita, 2020). The specificity with which PRRs bind with the ligands from pathogenic microbes (PAMPs) are based on different molecular structures and composition of these ligands. Furthermore, different structures of these PAMPs are essential for microbial viability and are specific to microorganisms. Thus, the host PRRs have evolved to distinctly communicate with commensal microbes and pathogens (Chu and Mazmanian, 2013; Li and Wu, 2021).
Toll-like receptors and NOD-like receptors
Among the aforementioned receptors, the TLRs and NLRs distinctly recognize microbe-associated molecular patterns (MAMPs) which are generally expressed by the resident microbes, and the PAMPs usually expressed by the invasive microbes (Gao et al., 2017; Negi et al., 2019). Upon recognition of these molecular patterns, the TLRs trigger signaling from the surface of the cell or endosomes, whereas the NLRs are activated in the cytosol by molecules derived from bacteria, such as flagellins, toxins, RNA and PG (Delbridge and O’Riordan, 2007).
Functionally, the TLRs are categorized into cell membrane TLRs and intracellular TLRs (nucleic acid sensors). For example, the heterodimers of TLR2 in conjunction with TLR1 or TLR6 besides TLR4, TLR5, and TLR10, express on the cell surface and belong to the cell membrane TLRs category (Gay et al., 2014). Whereas TLR3, TLR7, TLR8, and TLR9 which are localized to the intracellular compartments of the cell such as endosomes and lysosomes are the intracellular TLRs (Kawasaki and Kawai, 2014; Sellge and Kufer, 2015). TLRs produce pro-inflammatory cytokines upon binding with PAMPs (Vidya et al., 2018). The TLR is activated by PAMPs, leading to nuclear translocation of a pro-inflammatory transcription factor called activator protein 1 (AP-1), interferon regulatory factor (IRF) 3, and the nuclear factor kappa-B (NF-kB) by the recruitment of adapter proteins. These adapter proteins subsequently recruit interleukin-1R (IL-1R)-associated protein kinases (IRAK) 1, 2, 4, M, transforming growth factor-β-activated kinase (TAK) 1-binding proteins (TAB) 2 and tumor necrosis factor receptor-associated factor 6 (TRAF6; El-Zayat et al., 2019). Specific genes are then transcribed by these transcription factors encoding a different set of proteins such as chemokines [C-X-C motif chemokine ligand (CXCL 8 and CXCL10), type 1 interferons (IFN-α, β)], pro-inflammatory cytokines [tumor necrosis factor alpha (TNF-α), IL-1β and IL-6], and even antimicrobial peptides (El-Zayat et al., 2019).
NLR classification and its activity is divided into four subfamilies based on their domains: an acidic domain—NLRA, a baculovirus inhibitor of apoptosis repeat (BIR) domain—NLRB, a caspase activation and recruitment domain (CARD)—NLRC, and a pyrin domain (PYD)—NLRP. With the exception of NLRX1 comprising a non-identified CARD-related X effector domain (Parlato and Yeretssian, 2014), NLRs are responsible for the formation of inflammasomes and lead to the activation of IRF, mitogen-activated protein kinase (MAPK), and NF-κB pathways initiating a robust immune response during pathogen attack (Zhong et al., 2013).
C-type lectin receptors and RLRs
C-type lectin-like domain proteins (CTLD) are CTLRs which function by binding glycans on the surface of microbial pathogens to regulate both innate and adaptive immune responses. Other CTLRs play a regulatory role in immune homeostasis by binding endogenous ligands (self-antigens; Mayer et al., 2017; Dierking and Pita, 2020). CTLRs contribute to the homing of immune cell-trafficking, leukocytes, pathogen recognition and subsequent T cell activation (Mayer et al., 2017). RLRs in turn are cytoplasmic sensors of viral RNA. RLR signaling leads to activation of players such as NF-κB, MAPK and IRFs which promote induction of type I IFN and some of the pro-inflammatory cytokines (Kawai and Akira, 2009; Lavelle et al., 2010).
G-protein coupled receptors and PGRPs
GPCRs and PGRPs are another group of human host receptors which recognize microbiota-derived signals. The GPCRs recognize the microbiome through a signal peptide or short chain fatty acids (Dierking and Pita, 2020). GPCRs such as FPR1 (formyl peptide receptor 1), FPR2, GPCR41 or FFAR3 (free fatty acid receptor 3) and GPCR43 or FFAR2 can detect bacterial MAMPs (Bloes et al., 2015). While FPRs carry important roles in inflammation and immune cell activation, FFARs are essential nutritional components which function as signaling molecules that regulate cellular and physiological processes (He and Ye, 2017; Kimura et al., 2020). On the other hand, the mammalian PGRPs have a regular antibacterial activity by contributing to the activation of macrophages during immune responses (Dierking and Pita, 2020).
Polymorphisms of microbial sensors
Immuno-polymorphism is a significant aspect of infectious diseases biology that has been explored genetically in population-wide studies shedding light on the association of polymorphisms with epidemiology of several infectious diseases. Polymorphisms in the nucleotide or amino acid sequences of host receptors can influence downstream signaling pathways which in turn promote the resistance or susceptibility to various infections.
Polymorphism in TLRs and NLRs
A number of polymorphisms are directly associated with different phenotypes of infectious diseases of the gut (Table 1). TLR4 polymorphism (D299G and T399I) was reported as the first genetic variant in TLRs to reduce lipopolysaccharides-receptor interactions, which increased susceptibility to sepsis-inducing Gram-negative bacterial infections (Zhang et al., 2021). There are other common residues exhibiting single nucleotide polymorphisms (SNPs) in TLRs, namely, TLR1-I602S, TLR2-R677W, TLR3-P554S, TLR4-D299G, TLR5-L616F, TLR6-P249S, TLR7-Q11L, TLR8-M1V, TLR9-G1174A and TLR10-G1031T (Mukherjee et al., 2019). Polymorphisms in the TLR receptors are known to be associated with disease phenotypes such as ulcerative colitis, pancolitis, decreased or increased incidence and susceptibility to diseases such as inflammatory bowel disease and Crohn’s disease (CD; Mukherjee et al., 2019). CD is a chronic inflammatory bowel disease (IBD) which causes inflammation of the digestive tract mucosa with symptoms such as abdominal pain, fistulas, severe diarrhea, weight loss and malnutrition.
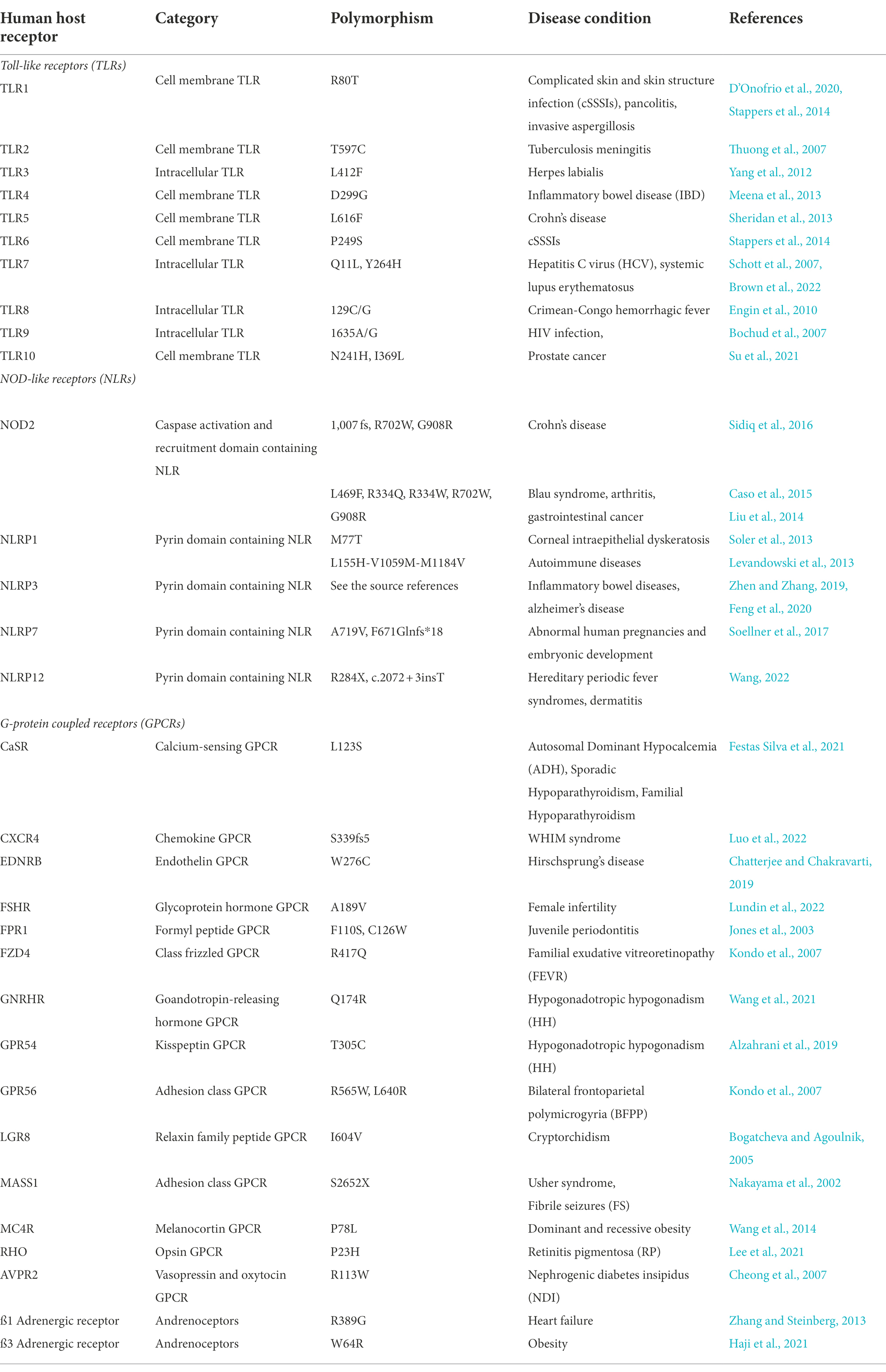
Table 1. Table showing important examples of human host receptors, their polymorphisms and their associated diseases.
NLR mutations on the other hand, are quite often associated with syndromes of autoinflammatory mechanisms, hinting at a convoluted role of cytosolic surveillance in systemic innate immunity (Delbridge and O’Riordan, 2007; Table 1). NOD1 and NOD2 are PG sensors which specifically respond to the PG components found in the rigid cell wall of the bacteria. A genetic association between NOD1 and NOD2 mutations and autoinflammatory diseases shows the importance of NOD proteins in inflammation regulation (Hugot et al., 2001; Girardin et al., 2005; McGovern et al., 2005). However, a direct contribution of NOD proteins to patients with immunodeficiency is not yet established (Delbridge and O’Riordan, 2007). NOD2 with its three coding variants (R702W, G908R, and L1007fs) resulting in decreased muramyl-dipeptide (MDP) responsiveness has been associated with a strong genetic risk factor for development of CD (Parlato and Yeretssian, 2014). On the other hand, genetic studies on NOD1 variants with susceptibility to IBD need further investigation due to conflicting information in literature (McGovern et al., 2005; Tremelling et al., 2006). The implications of NOD1 and NOD2 in bacterial sensing, primarily through the strong association of NOD2 with CD, has been under speculation that an altered detection of commensal microbes might lead to the impairment of homeostasis in the intestines thereby leading to intestinal inflammation (Parlato and Yeretssian, 2014).
Polymorphism in CTLRs and RLRs
Multiple CTLRs and RLRs variants have been reported in literature against non-enteric infectious microbes. In CTLRs, two variants, G54D (rs1800450) and N104S (rs2617170) in MBL2 and KLRC4, respectively, contribute to the susceptibility of Behcet’s disease (Yang et al., 2017). Behcet’s (beh-CHETS) disease is an autoimmune disease that causes systemic blood vessel inflammation. In particular, G1186A mutation in a CTLR known as MRC1 (mannose receptor 1) was linked to an increased risk of pulmonary tuberculosis among the Chinese population (Zhang et al., 2012). Recently, four CLEC4E SNPs (rs10841845, rs10841847, rs10841856, and rs4620776) were identified to be a cause of pulmonary tuberculosis (Bowker et al., 2016). One such finding reports broad spectrum MBL2 polymorphisms associated with human immunodeficiency virus (HIV) and tuberculosis co-infection across diverse populations (Garcia-Laorden et al., 2006). In RLRs, mutations at residues threonine 770, serine 854 and serine 85, were shown to constitutively activate RIG-I resulting in IFN induction (Sun et al., 2011). A non-synonymous polymorphism due to mutation of arginine to cysteine at amino acid position 7 leads to the expression of a functional RIG-I associated with an increased antiviral signaling (Shigemoto et al., 2009; Hu et al., 2010; Loo and Gale Jr, 2011).
Polymorphism in GPCRs and PGRPs
GPCRs, like TLRs and NLRs play an important role in the recognition of MAMPs. The transcription in cells may be affected in a tissue-selective manner which in turn may be affected by the polymorphisms of the GPCR promoter regions genes (Insel et al., 2007; Table 1) while genetic variants in PGLYRP1, PGLYRP2, PGLYRP3 and PGLYRP4 genes were shown to be associated with CD and ulcerative colitis (Zulfiqar et al., 2013; Dierking and Pita, 2020).
Microbial ligands
Microbial metabolites are chemical compounds which have the ability to regulate host immune responses by activating human host receptors (Rooks and Garrett, 2016). On the other hand, microbial ligands which bind to the receptors as mentioned above, also referred to as the PAMPs recognized by PRRs, can be grouped into three main classes, i.e., glycans, nucleic acids, and the proteins (Kawai and Akira, 2009). These microbial ligands do not share structural similarity but they share features which reflect the evolutionary progression of response to innate immunity. First, these are produced by the microbe (commensal/ pathogenic) and not by the host. This is the basis of discrimination of the self and non-self-antigens which in turn enables the host to mount an innate or adaptive immune response against microbial agents. Second, these ligands are conserved among a certain class of pathogens (Rana et al., 2015). Hence, recognition of a conserved portion of the lipopolysaccharide (LPS) lets PRR to identify the presence of any Gram-negative bacteria. Third, as these PAMPs carry out functions which are physiologically essential for microbial survival, any mutation in PRRs would be toxic or detrimental to the host itself (Medzhitov and Janeway Jr, 2000; Mogensen, 2009).
Microbial effector proteins
Microbial effector proteins are known to encompass catalytic domains within their primary sequences and exhibit enzymatic activities (Popa et al., 2016). Effector proteins are classified into four main categories based on the mode of action on the host (Scott and Hartland, 2017): 1. As competitive inhibitors which directly bind with host receptors (NleF—Escherichia, VipD—Legionella), 2. As proteins which functionally mimic the host proteins by post translational modifications (PTMs) such as lipidation (SopE—Shigella, SifA—Salmonella), 3. As mediators of PTM to block host protein functions (AnkX—Legionella, OspF—Shigella), and 4. As proteolytic enzymes which alter the host proteome composition and its spatial organization (NleC—Escherichia, RavZ—Legionella; Paul, 2012). Effector proteins expressed by different bacteria modify host cell proteins aiming to suppress the host defense mechanisms. This in turn allows the pathogens to source essential nutrients from the host and cause infection. Bacterial effector proteins are reported to be distinctively associated with different receptors and activate different downstream signaling pathways—thus establishing the differences between commensals and pathogens (Weigele et al., 2017; Mak and Thurston, 2021).
Among the various functions of the effector proteins, the ability to alter the host protein signaling pathways using PTMs is the most interesting one from a therapeutic perspective (Forrest and Welch, 2020). Pathogenic bacteria, both intracellular and extracellular, secrete effector proteins into the host cell which intervenes in the immuno-modulatory response by mimicking the natural process in the host cell. Among the modifications are phosphorylation, acetylation, methylation, ribosylation, adenylation, lipidation, glycosylation, ubiquitination and the reversal of these modifications (Forrest and Welch, 2020). Irreversible changes like proteolysis, eliminylation and deamidation also perturb the host response pathway. Given the fact that the actual modification occurs due to enzymatic reaction by the effector proteins, the therapeutic interventions in these pathways find a spotlight as potential alternative treatment(s), especially against drug-resistant infections (Stévenin and Neefjes, 2022). Along with the host protein signaling pathways, the host receptors themselves go through a series of PTMs whose role in pathogenesis, if further deciphered would expand our understanding of the molecular and cellular basis of innate immunity (Liu et al., 2016; Patwardhan et al., 2021).
Phosphorylation is a well-known PTM in which a phosphoryl group is transferred to a hydroxyl group of serine, threonine or tyrosine residues and is critical to host cell functions. Infectious bacteria exploit this pathway by secreting their own (de)phosphorylating enzymes to alter the host cell response. For example, in entero-haemorrhagic E. coli (EHEC) O157:H7 (Hua et al., 2020), the effector protein EspF interacts with annexin A6 (ANXA6) of the host protein resulting in phosphorylation of myosin light chain (MLC) and activation of protein kinase C (PKC) leading to the disruption of tight junctions between host cells, thus weakening the intercellular integrity. Lysine acetylation and methylation are other well-known PTMs where an acetyl group or a methyl group is transferred, respectively, to the epsilon-amino group in the side chain of a lysine residue in a reversible manner. In contrast, invasive pathogens such as Salmonella hijack their own process via acetylation to mediate and downregulate the effector proteins (Sang et al., 2016). In the case of Shigella, the effector protein OspZ methylates cysteines in NF-kB activators TAB2 and TAB3 preventing the release of pro-inflammatory cytokine IL-8 (Zheng et al., 2016). In contrast to addition of chemical groups on to the host proteins, ubiquitination (and ubiquitin-like protein conjugation) involves the addition of a small ubiquitin (or ubiquitin-like) protein on to the amino-group of a lysine residue or the N-terminal methionine residue in the host protein. A concerted action of ubiquitinating enzymes facilitate this process while proteases called deubiquitinases (DUBs) reverse the ubiquitination. Several enteric pathogens are known to hijack this process to modulate the immune response. For example, Salmonella uses SseL for deubiquitinating polyubiquitin chains (Rytkönen et al., 2007) while AvrA removes ubiquitin from nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα) and β-catenin resulting in the regulation of the NF-κB and β-catenin signaling pathways (Ye et al., 2007). In some cases, the bacterial effector proteins also mimic the ubiquitinating enzymes of the host protein. Such proteins are called Novel E3 ligases (NELs). For example, Shigella secretes NEL called IpaH9.8 which contributes to the splicing of U2AF35 mRNA resulting in the suppression of host immune response (Rytkönen et al., 2007).
Possible targets for discovery
Given the myriad of host receptor proteins modulated by pathogenic effector proteins in an enzymatic fashion, such processes are considered high profile therapeutic targets for drug interventions (Figure 1). The emergence of antibiotic resistance, compounded by the growth-lag in developing new antibiotics also necessitates the identification of alternative targets for antimicrobial therapy. In this aspect, new avenues are constantly being explored. We have discussed the important interactions in the previous section. The underlying biochemical mechanisms of such interactions and the PTMs associated with pathogenesis could potentially unlock the therapeutic potential of targeting these enzymes. Though in its infancy, the existing state-of-the-art high throughput screening (HTS) facilities and the ever-expanding repertoire of chemical inhibitors and small molecule libraries has pushed the frontiers in the struggle to control enteropathogenic infections with the help of new therapeutic molecules.
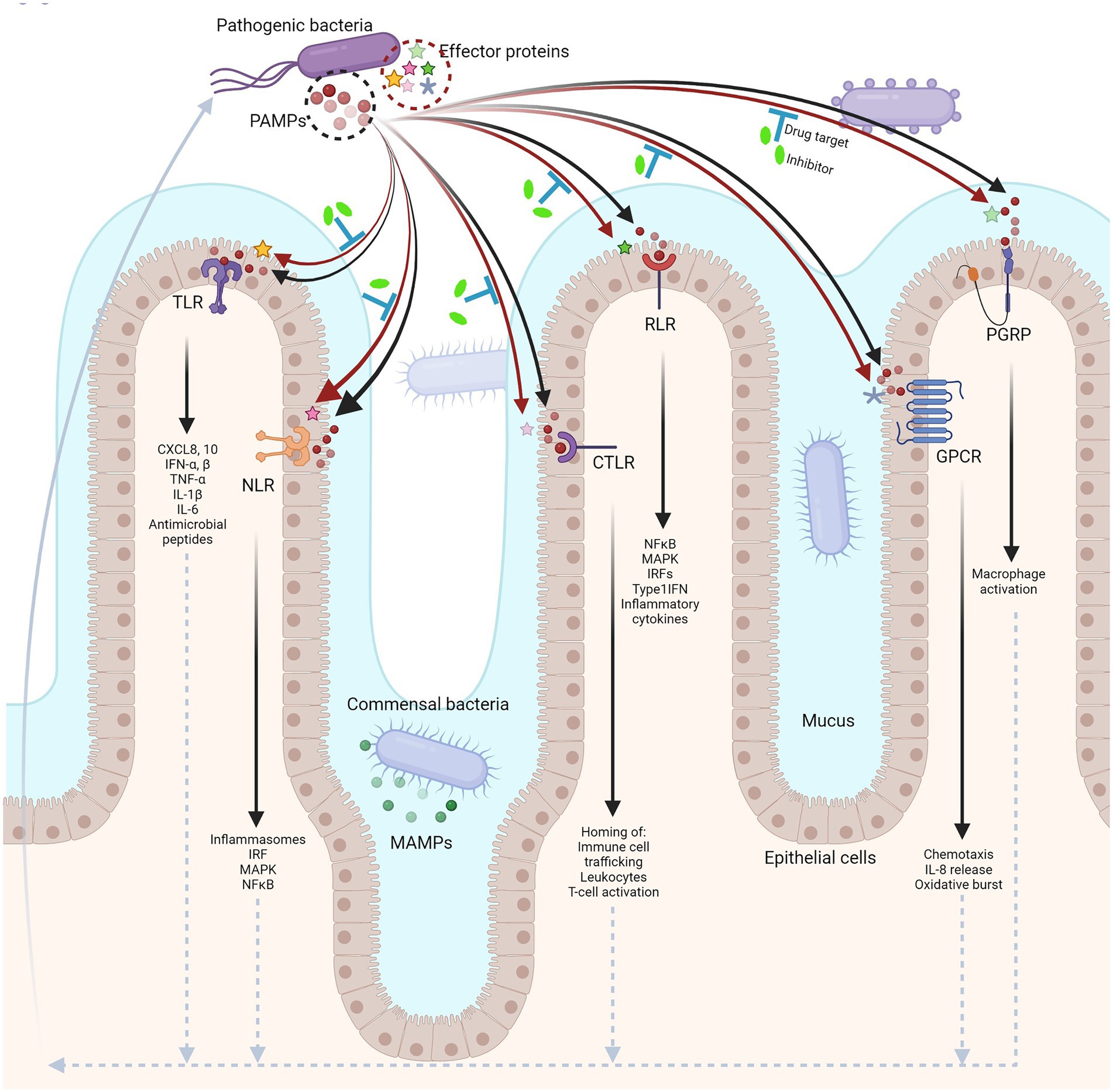
Figure 1. Host–microbe interface in the human gut. Different pattern recognition receptors (PRRs) in the host distinctly recognize PAMPs from pathogenic bacteria (black arrows), eliciting an immune response (upward arrow at far left, dotted arrows). In turn, pathogens secrete effector proteins that can modulate the pathways involved in host-immunity (red arrows). The enzymatic functions of effector proteins can be targeted for drug interventions contributing to alternative therapies against infections (blue ‘T’ bars; created with BioRender.com).
Some of the biochemical pathways involved in infection have been extensively studied to identify small molecule inhibitors that can influence the activity of PTM-promoting effector proteins. For example, Mycobacterium tyrosine phosphatases namely mPTPA and mPTPB were targeted for identifying inhibitors that can potentially lead to therapeutic application (Ruddraraju et al., 2020). Such techniques can be applied to the effector proteins identified in several species of enteric bacteria using similar screening techniques.
There are several mechanisms through which pathogenic bacteria develop drug resistance, including drug inactivation, modification of bacterial target sites, reduced antibiotic uptake and the formation of biofilms. These mechanisms explain how WHO “priority status” listed ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) have developed fitness against a wide-range of compounds such as lipopeptides, fluoroquinolones, tetracyclines, β-lactams and antibiotics that are the last line of defense (De Oliveira et al., 2020). One way to circumvent this problem is to use a multi-drug target instead of a single target thereby overcoming the susceptibility to resistance due to mutations. Single drug-target systems could be prone to drug resistance due to mutations (evolutionarily unavoidable). Therefore, multiple drug-target systems linked by a chemical linker are more effective. For example, in Helicobacter, using a multi-drug system such as the quadruple therapy is effective given that development of multiple phenotypic resistance is often difficult and slow. Similarly, targeting homomers with multiple binding sites (MBS) is better than targeting a single binding site (SBS; Abrusán and Marsh, 2019). Another avenue that is recently being explored is the host-directed therapy (HDT). HDT involves interfering with host factors important for pathogens to invade and cause disease (Ruddraraju et al., 2020). This can be carried out in two steps: directly interfering with host response pathways and enhancing immune response to infection (Kaufmann et al., 2018; Abrusán and Marsh, 2019). Furthermore, advances in the ‘omics’ research of the gut-microbe interactions have opened up possibilities of finding additional targets for drug discovery. Most of these interactions are followed up using mass spectrometric techniques using samples from diseased patients. Such data are consolidated into an interaction map providing a holistic approach for both diagnosis and treatment for various gut-related diseases (Lloyd-Price et al., 2019; Whon et al., 2021).
In addition to this, SNPs are known to be one of the major elements associated with the fate of a microbial infection (Mukherjee et al., 2019). Studies conducted in a German cohort showed correlation of SNPs, G743A and T1805G with cell surface TLR1 expression deficiency resulting in susceptibility to M. tuberculosis infection. Two additional SNPs, S150G and V220M which are associated with the structural organization of the LRR motif of TLR1 further refined the understanding of susceptibility to M. tuberculosis in the context of receptor recognition ability of mycobacteria (Bustamante et al., 2011; Uciechowski et al., 2011). Another interesting observation reported in Asian patients having acute febrile malaria demonstrates the presence of polymorphisms (rs5743551) on G allele of TLR1 leading to reduced parasitemia and thereby a host directed control on severity of the disease (Hahn et al., 2016). From an evolutionary thinking, this points to the possibility that G allele of TLR1 rs5743551 occurs in higher frequencies in non-Caucasian hosts and imparts a fitness advantage in malaria endemic zones. The state-of-the-art CRISPR/Cas9-based genome editing could be employed to target the microbes which cause pathogenesis in the host (Zheng et al., 2022). Polymorphisms in host receptors can further be exploited to understand the virulence potential of pathogenic bacteria that can be targeted for suppression by CRISPR-Cas9 based therapies (Ramachandran and Bikard, 2019; Nath et al., 2022).
Conclusion
Sustained efforts directed at understanding the gut microbiome and the immune interactions have contributed to our understanding of the role of dysbiosis in the immune system concerning a range of infections and disease phenotypes. However, due to a relentless rise in antibiotic resistance, the human gut, which is home to the largest ecosystem of microbes, is at risk of becoming a hot-zone of a wide range of potential infections and pathologies. In future, alternative therapies are critical to replace current infection control regimens by targeting the receptor-ligand interactions and their downstream processes in the gut. In this regard, it is important to understand how commensal and pathogenic bacteria interact with the host in order to maintain symbiosis or trigger an infection. A few interesting aspects need to be addressed in relation to the axis of host-pathogen-environment: colonization of the mucus layer by commensal bacteria while in symbiosis with the host in the presence of specific MAMPs; existence of commensal MAMPs alongside pathogenic variants which can be distinguished from each other based on their environment; and restoring commensality to the pathogenic PAMPs by altering the host environment. Considering the fact that even a single polymorphism might facilitate a commensal-to-pathogen transition, it is important to develop a holistic approach towards an assay platform that profiles pathogenic PAMPs and the commensal MAMPs based on molecular, physiological and host genetic factors. This can be exploited in developing molecular screens targeting the host-pathogen interface in addition to better understanding the relationship between microbial sensors and enteric bacteria and how they affect susceptibility or resistance to an infection.
Author contributions
ST and DSH contributed to writing of the review and editing. NA contributed to narrative building, discussion and editing. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank the Indian Council of Medical Research for a grant awarded to NA (AMR/257/2021/ECD-II). We would like to acknowledge the University of Hyderabad for common facilities and a small grant support (Institution of Eminence–PDF; financial support to UoH-IoE by MHRD [F11/9/2019-U3A]).
Conflict of interest
The authors declare that the article was compiled in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrusán, G., and Marsh, J. A. (2019). Ligands and receptors with broad binding capabilities have common structural characteristics: an antibiotic design perspective. J. Med. Chem. 62, 9357–9374. doi: 10.1021/acs.jmedchem.9b00220
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cells 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Aljahdali, N. H., Sanad, Y. M., Han, J., and Foley, S. L. (2020). Current knowledge and perspectives of potential impacts of salmonella enterica on the profile of the gut microbiota. BMC Microbiol. 20:353. doi: 10.1186/s12866-020-02008-x
Alzahrani, A. J., Ahmad, A., Alhazmi, T., and Ahmad, L. (2019). An isolated Hypogonadotropic Hypogonadism due to a L102P inactivating mutation of KISS1R/GPR54 in a large family. Case Rep. Pediatr. 2019, 1–6. doi: 10.1155/2019/3814525
Bajinka, O., Darboe, A., Tan, Y., Abdelhalim, K. A., and Cham, L. B. (2020). Gut microbiota and the human gut physiological changes. Ann. Microbiol. 70, 1–9. doi: 10.1186/s13213-020-01608-2
Bloes, D. A., Kretschmer, D., and Peschel, A. (2015). Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Microbiol. 13, 95–104. doi: 10.1038/nrmicro3390
Bochud, P.-Y., Hersberger, M., Taffé, P., Bochud, M., Stein, C. M., Rodrigues, S. D., et al. (2007). Polymorphisms in toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS 21, 441–446. doi: 10.1097/QAD.0b013e328012b8ac
Bogatcheva, N. V., and Agoulnik, A. I. (2005). INSL3/LGR8 role in testicular descent and cryptorchidism. Reprod. Biomed. Online 10, 49–54. doi: 10.1016/S1472-6483(10)60803-6
Bowker, N., Salie, M., Schurz, H., van Helden, P. D., Kinnear, C. J., Hoal, E. G., et al. (2016). Polymorphisms in the pattern recognition receptor Mincle gene (CLEC4E) and association with tuberculosis. Lung 194, 763–767. doi: 10.1007/s00408-016-9915-y
Brown, G. J., Cañete, P. F., Wang, H., Medhavy, A., Bones, J., Roco, J. A., et al. (2022). TLR7 gain-of-function genetic variation causes human lupus. Nature 605, 349–356. doi: 10.1038/s41586-022-04642-z
Bustamante, J., Tamayo, E., Flórez, S., Telleria, J. J., Bustamante, E., López, J., et al. (2011). Toll-like receptor 2 R753Q polymorphisms are associated with an increased risk of infective endocarditis. Rev. Esp. Cardiol. 64, 1056–1059. doi: 10.1016/j.recesp.2011.02.024
Carrera, W., Ng, C., Desler, C., Jumper, J. M., and Agarwal, A. (2021). Novel FZD4 and LRP5 mutations in a small cohort of patients with familial exudative vitreoretinopathy (FEVR). Ophthalmic Genet. 42, 200–203. doi: 10.1080/13816810.2020.1855664
Caso, F., Galozzi, P., Costa, L., Sfriso, P., Cantarini, L., and Punzi, L. (2015). Autoinflammatory granulomatous diseases: from Blau syndrome and early-onset sarcoidosis to NOD2-mediated disease and Crohn’s disease. RMD Open 1:e000097. doi: 10.1136/rmdopen-2015-000097
Chatterjee, S., and Chakravarti, A. (2019). A gene regulatory network explains RET-EDNRB epistasis in Hirschsprung disease. Hum. Mol. Genet. 28, 3137–3147. doi: 10.1093/hmg/ddz149
Cheong, H. I., Cho, H. Y., Park, H. W., Ha, I. S., and Choi, Y. (2007). Molecular genetic study of congenital nephrogenic diabetes insipidus and rescue of mutant vasopressin V2 receptor by chemical chaperones. Nephrology 12, 113–117. doi: 10.1111/j.1440-1797.2006.00759.x
Chiang, N.-Y., Hsiao, C.-C., Huang, Y.-S., Chen, H.-Y., Hsieh, I.-J., Chang, G.-W., et al. (2011). Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J. Biol. Chem. 286, 14215–14225. doi: 10.1074/jbc.M110.183830
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Cornick, S., Tawiah, A., and Chadee, K. (2015). Roles and regulation of the mucus barrier in the gut. Tissue Barriers 3:e982426. doi: 10.4161/21688370.2014.982426
D’Onofrio, V., Monnier, A. A., Kremer, C., Stappers, M. H. T., Netea, M. G., and Gyssens, I. C. (2020). Lesion size is associated with genetic polymorphisms in TLR1, TLR6, and TIRAP genes in patients with major abscesses and diabetic foot infections. Eur. J. Clin. Microbiol. Infect. Dis. 39, 353–360. doi: 10.1007/s10096-019-03732-7
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33: e00181–19. doi: 10.1128/CMR.00181-19
Dekker, J. P., and Frank, K. M. (2015). Salmonella, Shigella, and yersinia. Clin. Lab. Med. 35, 225–246. doi: 10.1016/j.cll.2015.02.002
Delbridge, L. M., and O’Riordan, M. X. D. (2007). Innate recognition of intracellular bacteria. Curr. Opin. Immunol. 19, 10–16. doi: 10.1016/j.coi.2006.11.005
Dierking, K., and Pita, L. (2020). Receptors mediating host-microbiota communication in the Metaorganism: the invertebrate perspective. Front. Immunol. 11:1251. doi: 10.3389/fimmu.2020.01251
Dougnon, V., Assogba, P., Anago, E., Déguénon, E., Dapuliga, C., Agbankpè, J., et al. (2020). Enterobacteria responsible for urinary infections: a review about pathogenicity, virulence factors and epidemiology. J. Appl. Biol. Biotechnol. 8, 117–124. doi: 10.7324/JABB.2020.80118
El-Zayat, S. R., Sibaii, H., and Mannaa, F. A. (2019). Toll-like receptors activation, signaling, and targeting: an overview. Bull. Natl. Res. Centre 43, 1–12. doi: 10.1186/s42269-019-0227-2
Engin, A., Arslan, S., Kizildag, S., Oztürk, H., Elaldi, N., Dökmetas, I., et al. (2010). Toll-like receptor 8 and 9 polymorphisms in Crimean-Congo hemorrhagic fever. Microbes Infect. 12, 1071–1078. doi: 10.1016/j.micinf.2010.07.012
Fang, J., Wang, H., Zhou, Y., Zhang, H., Zhou, H., and Zhang, X. (2021). Slimy partners: the mucus barrier and gut microbiome in ulcerative colitis. Exp. Mol. Med. 53, 772–787. doi: 10.1038/s12276-021-00617-8
Feng, Y.-S., Tan, Z.-X., Wu, L.-Y., Dong, F., and Zhang, F. (2020). The involvement of NLRP3 inflammasome in the treatment of Alzheimer’s disease. Ageing Res. Rev. 64:101192. doi: 10.1016/j.arr.2020.101192
Festas Silva, D., De Sousa Lages, A., Caetano, J. S., Cardoso, R., Dinis, I., Gomes, L., et al. (2021). A variant in the CASR gene (c.368T>C) causing hypocalcemia refractory to standard medical therapy. Endocrinol. Diabetes Metab. Case Rep. 2021: 21–0005. doi: 10.1530/EDM-21-0005
Forrest, S., and Welch, M. (2020). Arming the troops: post-translational modification of extracellular bacterial proteins. Sci. Prog. 103:36850420964317. doi: 10.1177/0036850420964317
Gao, W., Xiong, Y., Li, Q., and Yang, H. (2017). Inhibition of toll-like receptor signaling as a promising therapy for inflammatory diseases: a journey from molecular to Nano therapeutics. Front. Physiol. 8:508. doi: 10.3389/fphys.2017.00508
Garcia-Laorden, M. I., Pena, M. J., Caminero, J. A., Garcia-Saavedra, A., Campos-Herrero, M. I., Caballero, A., et al. (2006). Influence of mannose-binding lectin on HIV infection and tuberculosis in a Western-European population. Mol. Immunol. 43, 2143–2150. doi: 10.1016/j.molimm.2006.01.008
Gay, N. J., Symmons, M. F., Gangloff, M., and Bryant, C. E. (2014). Assembly and localization of toll-like receptor signalling complexes. Nat. Rev. Immunol. 14, 546–558. doi: 10.1038/nri3713
Girardin, S. E., Jéhanno, M., Mengin-Lecreulx, D., Sansonetti, P. J., Alzari, P. M., and Philpott, D. J. (2005). Identification of the critical residues involved in peptidoglycan detection by Nod1. J. Biol. Chem. 280, 38648–38656. doi: 10.1074/jbc.M509537200
Gu, W., Tong, P., Liu, C., Wang, W., Lu, C., Han, Y., et al. (2019). The characteristics of gut microbiota and commensal Enterobacteriaceae isolates in tree shrew (Tupaia belangeri). BMC Microbiol. 19:203. doi: 10.1186/s12866-019-1581-9
Hahn, W. O., Harju-Baker, S., Erdman, L. K., Krudsood, S., Kain, K. C., Wurfel, M. M., et al. (2016). A common TLR1 polymorphism is associated with higher parasitaemia in a southeast Asian population with plasmodium falciparum malaria. Malar. J. 15:12. doi: 10.1186/s12936-015-1071-y
Haji, E., Al Mahri, S., Aloraij, Y., Malik, S. S., and Mohammad, S. (2021). Functional characterization of the obesity-linked variant of the β3-adrenergic receptor. Int. J. Mol. Sci. 22: 5721. doi: 10.3390/ijms22115721
Hamilton, A. L., Kamm, M. A., Ng, S. C., and Morrison, M. (2018). Proteus spp. as putative gastrointestinal pathogens. Clin. Microbiol. Rev. 31: e00085–15. doi: 10.1128/CMR.00085-17
He, H.-Q., and Ye, R. D. (2017). The Formyl peptide receptors: diversity of ligands and mechanism for recognition. Molecules 22: 455. doi: 10.3390/molecules22030455
Hu, J., Nistal-Villán, E., Voho, A., Ganee, A., Kumar, M., Ding, Y., et al. (2010). A common polymorphism in the caspase recruitment domain of RIG-I modifies the innate immune response of human dendritic cells. J. Immunol. 185, 424–432. doi: 10.4049/jimmunol.0903291
Hua, Y., Wu, J., Fu, M., Liu, J., Li, X., Zhang, B., et al. (2020). Enterohemorrhagic Escherichia coli effector protein EspF interacts with host protein ANXA6 and triggers myosin light chain kinase (MLCK)-dependent tight junction Dysregulation. Front. Cell Dev. Biol. 8:613061. doi: 10.3389/fcell.2020.613061
Hugot, J. P., Chamaillard, M., Zouali, H., Lesage, S., Cézard, J. P., Belaiche, J., et al. (2001). Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603. doi: 10.1038/35079107
Insel, P. A., Tang, C.-M., Hahntow, I., and Michel, M. C. (2007). Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochim. Biophys. Acta 1768, 994–1005. doi: 10.1016/j.bbamem.2006.09.029
Iwasaki, A., and Medzhitov, R. (2004). Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995. doi: 10.1038/ni1112
Janda, J. M., and Abbott, S. L. (2021). The changing face of the family Enterobacteriaceae (order: “Enterobacterales”): new members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 34(2): e00174–20. doi: 10.1128/CMR.00174-20
Jones, B. E., Miettinen, H. M., Jesaitis, A. J., and Mills, J. S. (2003). Mutations of F110 and C126 of the formyl peptide receptor interfere with G-protein coupling and chemotaxis. J. Periodontol. 74, 475–484. doi: 10.1902/jop.2003.74.4.475
Josenhans, C., Müthing, J., Elling, L., Bartfeld, S., and Schmidt, H. (2020). How bacterial pathogens of the gastrointestinal tract use the mucosal glyco-code to harness mucus and microbiota: new ways to study an ancient bag of tricks. Int. J. Med. Microbiol. 310:151392. doi: 10.1016/j.ijmm.2020.151392
Kaufmann, S. H. E., Dorhoi, A., Hotchkiss, R. S., and Bartenschlager, R. (2018). Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 17, 35–56. doi: 10.1038/nrd.2017.162
Kawai, T., and Akira, S. (2009). The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21, 317–337. doi: 10.1093/intimm/dxp017
Kawai, T., and Akira, S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. doi: 10.1016/j.immuni.2011.05.006
Kawasaki, T., and Kawai, T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5:461. doi: 10.3389/fimmu.2014.00461
Kimura, I., Ichimura, A., Ohue-Kitano, R., and Igarashi, M. (2020). Free fatty acid receptors in health and disease. Physiol. Rev. 100, 171–210. doi: 10.1152/physrev.00041.2018
Kogut, M. H., Lee, A., and Santin, E. (2020). Microbiome and pathogen interaction with the immune system. Poult. Sci. 99, 1906–1913. doi: 10.1016/j.psj.2019.12.011
Kondo, H., Qin, M., Tahira, T., Uchio, E., and Hayashi, K. (2007). Severe form of familial exudative vitreoretinopathy caused by homozygous R417Q mutation in frizzled-4 gene. Ophthalmic Genet. 28, 220–223. doi: 10.1080/13816810701663543
Lavelle, E. C., Murphy, C., O’Neill, L. A. J., and Creagh, E. M. (2010). The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 3, 17–28. doi: 10.1038/mi.2009.124
Lee, E.-J., Chan, P., Chea, L., Kim, K., Kaufman, R. J., and Lin, J. H. (2021). ATF6 is required for efficient rhodopsin clearance and retinal homeostasis in the P23H rho retinitis pigmentosa mouse model. Sci. Rep. 11:16356. doi: 10.1038/s41598-021-95895-7
Levandowski, C. B., Mailloux, C. M., Ferrara, T. M., Gowan, K., Ben, S., Jin, Y., et al. (2013). NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 110, 2952–2956. doi: 10.1073/pnas.1222808110
Li, D., and Wu, M. (2021). Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 6:291. doi: 10.1038/s41392-021-00687-0
Liu, J., He, C., Xu, Q., Xing, C., and Yuan, Y. (2014). NOD2 polymorphisms associated with cancer risk: a meta-analysis. PLoS One 9:e89340. doi: 10.1371/journal.pone.0089340
Liu, J., Qian, C., and Cao, X. (2016). Post-translational modification control of innate immunity. Immunity 45, 15–30. doi: 10.1016/j.immuni.2016.06.020
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Loo, Y.-M., and Gale, M. Jr. (2011). Immune signaling by RIG-I-like receptors. Immunity 34, 680–692. doi: 10.1016/j.immuni.2011.05.003
Lundin, K., Sepponen, K., Väyrynen, P., Liu, X., Yohannes, D. A., Survila, M., et al. (2022). Human pluripotent stem cell-derived cells endogenously expressing follicle-stimulating hormone receptors: modeling the function of an inactivating receptor mutation. Mol. Hum. Reprod. 28: gaac012. doi: 10.1093/molehr/gaac012
Luo, J., De Pascali, F., Richmond, G. W., Khojah, A. M., and Benovic, J. L. (2022). Characterization of a new WHIM syndrome mutant reveals mechanistic differences in regulation of the chemokine receptor CXCR4. J. Biol. Chem. 298:101551. doi: 10.1016/j.jbc.2021.101551
Mak, H., and Thurston, T. L. M. (2021). Interesting biochemistries in the structure and function of bacterial effectors. Front. Cell. Infect. Microbiol. 11:608860. doi: 10.3389/fcimb.2021.608860
Martin, A. M., Sun, E. W., Rogers, G. B., and Keating, D. J. (2019). The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front. Physiol. 10:428. doi: 10.3389/fphys.2019.00428
Mayer, S., Raulf, M.-K., and Lepenies, B. (2017). C-type lectins: their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 147, 223–237. doi: 10.1007/s00418-016-1523-7
McGovern, D. P. B., Hysi, P., Ahmad, T., van Heel, D. A., Moffatt, M. F., Carey, A., et al. (2005). Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum. Mol. Genet. 14, 1245–1250. doi: 10.1093/hmg/ddi135
Medzhitov, R., and Janeway, C. Jr. (2000). Innate immunity. N. Engl. J. Med. 343, 338–344. doi: 10.1056/NEJM200008033430506
Meena, N. K., Verma, R., Verma, N., Ahuja, V., and Paul, J. (2013). TLR4 D299G polymorphism modulates cytokine expression in ulcerative colitis. J. Clin. Gastroenterol. 47, 773–780. doi: 10.1097/MCG.0b013e31828a6e93
Mogensen, T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/CMR.00046-08
Morales-López, S., Yepes, J. A., Prada-Herrera, J. C., and Torres-Jiménez, A. (2019). Enterobacteria in the 21st century: a review focused on taxonomic changes. J. Infect. Dev. Ctries. 13, 265–273. doi: 10.3855/jidc.11216
Mukherjee, S., Huda, S., and Sinha Babu, S. P. (2019). Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand. J. Immunol. 90:e12771. doi: 10.1111/sji.12771
Nakayama, J., Fu, Y.-H., Clark, A. M., Nakahara, S., Hamano, K., Iwasaki, N., et al. (2002). A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann. Neurol. 52, 654–657. doi: 10.1002/ana.10347
Nath, A., Bhattacharjee, R., Nandi, A., Sinha, A., Kar, S., Manoharan, N., et al. (2022). Phage delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiome. Biomed. Pharmacother. 151:113122. doi: 10.1016/j.biopha.2022.113122
Negi, S., Das, D. K., Pahari, S., Nadeem, S., and Agrewala, J. N. (2019). Potential role of gut microbiota in induction and regulation of innate immune memory. Front. Immunol. 10:2441. doi: 10.3389/fimmu.2019.02441
Nie, L., Cai, S.-Y., Shao, J.-Z., and Chen, J. (2018). Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front. Immunol. 9:1523. doi: 10.3389/fimmu.2018.01523
Okumura, R., and Takeda, K. (2017). Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 49:e338. doi: 10.1038/emm.2017.20
Paone, P., and Cani, P. D. (2020). Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 69, 2232–2243. doi: 10.1136/gutjnl-2020-322260
Parlato, M., and Yeretssian, G. (2014). NOD-like receptors in intestinal homeostasis and epithelial tissue repair. Int. J. Mol. Sci. 15, 9594–9627. doi: 10.3390/ijms15069594
Patten, D. A., and Collett, A. (2013). Exploring the immunomodulatory potential of microbial-associated molecular patterns derived from the enteric bacterial microbiota. Microbiology 159, 1535–1544. doi: 10.1099/mic.0.064717-0
Patwardhan, A., Cheng, N., and Trejo, J. (2021). Post-translational modifications of G protein-coupled receptors control cellular signaling dynamics in space and time. Pharmacol. Rev. 73, 120–151. doi: 10.1124/pharmrev.120.000082
Popa, C. M., Tabuchi, M., and Valls, M. (2016). Modification of bacterial effector proteins inside eukaryotic host cells. Front. Cell. Infect. Microbiol. 6:73. doi: 10.3389/fcimb.2016.00073
Ramachandran, G., and Bikard, D. (2019). Editing the microbiome the CRISPR way. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 374:20180103. doi: 10.1098/rstb.2018.0103
Ramos, S., Silva, V., Dapkevicius, M. d. L. E., Caniça, M., Tejedor-Junco, M. T., Igrejas, G., et al. (2020). Escherichia coli as commensal and pathogenic bacteria among food-producing animals: health implications of extended Spectrum β-lactamase (ESBL) production. Animals (Basel) 10: 2239 doi: 10.3390/ani10122239
Rana, A., Ahmed, M., Rub, A., and Akhter, Y. (2015). A tug-of-war between the host and the pathogen generates strategic hotspots for the development of novel therapeutic interventions against infectious diseases. Virulence 6, 566–580. doi: 10.1080/21505594.2015.1062211
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Ruddraraju, K. V., Aggarwal, D., and Zhang, Z.-Y. (2020). Therapeutic targeting of protein tyrosine phosphatases from mycobacterium tuberculosis. Microorganisms 9(11): 14. doi: 10.3390/microorganisms9010014
Rytkönen, A., Poh, J., Garmendia, J., Boyle, C., Thompson, A., Liu, M., et al. (2007). SseL, a salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. U. S. A. 104, 3502–3507. doi: 10.1073/pnas.0610095104
Sang, Y., Ren, J., Ni, J., Tao, J., Lu, J., and Yao, Y.-F. (2016). Protein acetylation is involved in salmonella enterica Serovar Typhimurium virulence. J. Infect. Dis. 213, 1836–1845. doi: 10.1093/infdis/jiw028
Santos, R. L. (2015). “Chapter 72- non-typhoidal salmonella interactions with host cells” in Molecular Medical Microbiology (Second Edition). eds. Y.-W. Tang, M. Sussman, D. Liu, I. Poxton, and J. Schwartzman (Boston, MA: Academic Press), 1307–1317.
Schott, E., Witt, H., Neumann, K., Taube, S., Oh, D.-Y., Schreier, E., et al. (2007). A toll-like receptor 7 single nucleotide polymorphism protects from advanced inflammation and fibrosis in male patients with chronic HCV-infection. J. Hepatol. 47, 203–211. doi: 10.1016/j.jhep.2007.03.021
Scott, N. E., and Hartland, E. L. (2017). Post-translational mechanisms of host subversion by bacterial effectors. Trends Mol. Med. 23, 1088–1102. doi: 10.1016/j.molmed.2017.10.003
Sellge, G., and Kufer, T. A. (2015). PRR-signaling pathways: learning from microbial tactics. Semin. Immunol. 27, 75–84. doi: 10.1016/j.smim.2015.03.009
Sheridan, J., David, R., Mack, D. K., Amre, D. M., Israel, A., Cherkasov, H., et al. (2013). “A Non-Synonymous Coding Variant (L616F) in the TLR5 Gene Is Potentially Associated with Crohn’s Disease and Influences Responses to Bacterial Flagellin.”. PLoS One 8(4): e61326. doi: 10.1371/journal.pone.0061326
Shigemoto, T., Kageyama, M., Hirai, R., Zheng, J., Yoneyama, M., and Fujita, T. (2009). Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J. Biol. Chem. 284, 13348–13354. doi: 10.1074/jbc.M809449200
Sicard, J.-F., Le Bihan, G., Vogeleer, P., Jacques, M., and Harel, J. (2017). Interactions of intestinal bacteria with components of the intestinal mucus. Front. Cell. Infect. Microbiol. 7:387. doi: 10.3389/fcimb.2017.00387
Sidiq, T., Yoshihama, S., Downs, I., and Kobayashi, K. S. (2016). Nod2: a critical regulator of Ileal microbiota and Crohn’s disease. Front. Immunol. 7:367. doi: 10.3389/fimmu.2016.00367
Soellner, L., Begemann, M., Degenhardt, F., Geipel, A., Eggermann, T., and Mangold, E. (2017). Maternal heterozygous NLRP7 variant results in recurrent reproductive failure and imprinting disturbances in the offspring. Eur. J. Hum. Genet. 25, 924–929. doi: 10.1038/ejhg.2017.94
Soler, V. J., Tran-Viet, K. N., Galiacy, S. D., Limviphuvadh, V., Klemm, T. P., St Germain, E., et al. (2013). Whole exome sequencing identifies a mutation for a novel form of corneal intraepithelial dyskeratosis. J. Med. Genet. 50, 246–254. doi: 10.1136/jmedgenet-2012-101325
Stappers, M. H. T., Thys, Y., Oosting, M., Plantinga, T. S., Ioana, M., Reimnitz, P., et al. (2014). TLR1, TLR2, and TLR6 gene polymorphisms are associated with increased susceptibility to complicated skin and skin structure infections. J. Infect. Dis. 210, 311–318. doi: 10.1093/infdis/jiu080
Stévenin, V., and Neefjes, J. (2022). Control of host PTMs by intracellular bacteria: an opportunity toward novel anti-infective agents. Cell Chem Biol 29, 741–756. doi: 10.1016/j.chembiol.2022.04.004
Su, S.-B., Tao, L., Deng, Z.-P., Chen, W., Qin, S.-Y., and Jiang, H.-X. (2021). TLR10: insights, controversies and potential utility as a therapeutic target. Scand. J. Immunol. 93:e12988. doi: 10.1111/sji.12988
Sun, Z., Ren, H., Liu, Y., Teeling, J. L., and Gu, J. (2011). Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 85, 1036–1047. doi: 10.1128/JVI.01734-10
Tel, J., Benitez-Ribas, D., Janssen, E. M., Smits, E. L. J., and Jacobs, J. F. M. (2016). Dendritic cells as vaccines: key regulators of tolerance and immunity. Mediators Inflamm 2016, 1–2. doi: 10.1155/2016/5789725
Thuong, N. T. T., Hawn, T. R., Thwaites, G. E., Chau, T. T. H., Lan, N. T. N., Quy, H. T., et al. (2007). A polymorphism in human TLR2 is associated with increased susceptibility to Tuberculous meningitis. Genes Immun. 8, 422–428. doi: 10.1038/sj.gene.6364405
Tremelling, M., Hancock, L., Bredin, F., Sharpstone, D., Bingham, S. A., and Parkes, M. (2006). Complex insertion/deletion polymorphism in NOD1 (CARD4) is not associated with inflammatory bowel disease susceptibility in East Anglia panel. Inflamm. Bowel Dis. 12, 967–971. doi: 10.1097/01.mib.0000234131.89971.e5
Uciechowski, P., Imhoff, H., Lange, C., Meyer, C. G., Browne, E. N., Kirsten, D. K., et al. (2011). Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J. Leukoc. Biol. 90, 377–388. doi: 10.1189/jlb.0409233
Vidya, M. K., Kumar, V. G., Sejian, V., Bagath, M., Krishnan, G., and Bhatta, R. (2018). Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 37, 20–36. doi: 10.1080/08830185.2017.1380200
Wang, H.-F. (2022). NLRP12-associated systemic autoinflammatory diseases in children. Pediatr. Rheumatol. Online J. 20:9. doi: 10.1186/s12969-022-00669-8
Wang, L., Lin, W., Li, X., Zhang, L., Wang, K., Cui, X., et al. (2021). A case report of congenital idiopathic hypogonadotropic hypogonadism caused by novel mutation of GNRHR gene. Medicine 100:e24007. doi: 10.1097/MD.0000000000024007
Wang, X.-H., Wang, H.-M., Zhao, B.-L., Yu, P., and Fan, Z.-C. (2014). Rescue of defective MC4R cell-surface expression and signaling by a novel pharmacoperone Ipsen 17. J. Mol. Endocrinol. 53, 17–29. doi: 10.1530/JME-14-0005
Weigele, B. A., Orchard, R. C., Jimenez, A., Cox, G. W., and Alto, N. M. (2017). A systematic exploration of the interactions between bacterial effector proteins and host cell membranes. Nat. Commun. 8:532. doi: 10.1038/s41467-017-00700-7
Whon, T. W., Shin, N.-R., Kim, J. Y., and Roh, S. W. (2021). Omics in gut microbiome analysis. J. Microbiol. 59, 292–297. doi: 10.1007/s12275-021-1004-0
Yang, J., Chen, W., Xia, P., and Zhang, W. (2020). Dynamic comparison of gut microbiota of mice infected with Shigella flexneri via two different infective routes. Exp. Ther. Med. 19, 2273–2281. doi: 10.3892/etm.2020.8469
Yang, C.-A., Raftery, M. J., Hamann, L., Guerreiro, M., Grütz, G., Haase, D., et al. (2012). Association of TLR3-hyporesponsiveness and functional TLR3 L412F polymorphism with recurrent herpes labialis. Hum. Immunol. 73, 844–851. doi: 10.1016/j.humimm.2012.04.008
Yang, Y., Tan, H., Deng, B., Yu, H., Su, G., Hu, J., et al. (2017). Genetic polymorphisms of C-type lectin receptors in Behcet’s disease in a Chinese Han population. Sci. Rep. 7:5348. doi: 10.1038/s41598-017-05877-x
Ye, Z., Petrof, E. O., Boone, D., Claud, E. C., and Sun, J. (2007). Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am. J. Pathol. 171, 882–892. doi: 10.2353/ajpath.2007.070220
Zhang, X., Jiang, F., Wei, L., Li, F., Liu, J., Wang, C., et al. (2012). Polymorphic allele of human MRC1 confer protection against tuberculosis in a Chinese population. Int. J. Biol. Sci. 8, 375–382. doi: 10.7150/ijbs.4047
Zhang, Y., Liu, J., Wang, C., Liu, J., and Lu, W. (2021). Toll-like receptors gene polymorphisms in autoimmune disease. Front. Immunol. 12:672346. doi: 10.3389/fimmu.2021.672346
Zhang, F., and Steinberg, S. F. (2013). S49G and R389G polymorphisms of the β₁-adrenergic receptor influence signaling via the cAMP-PKA and ERK pathways. Physiol. Genomics 45, 1186–1192. doi: 10.1152/physiolgenomics.00087.2013
Zhen, Y., and Zhang, H. (2019). NLRP3 Inflammasome and inflammatory bowel disease. Front. Immunol. 10:276. doi: 10.3389/fimmu.2019.00276
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Zheng, L., Tan, Y., Hu, Y., Shen, J., Qu, Z., Chen, X., et al. (2022). CRISPR/Cas-based genome editing for human gut commensal Bacteroides species. ACS Synth. Biol. 11, 464–472. doi: 10.1021/acssynbio.1c00543
Zheng, Z., Wei, C., Guan, K., Yuan, Y., Zhang, Y., Ma, S., et al. (2016). Bacterial E3 ubiquitin ligase IpaH4.5 of Shigella flexneri targets TBK1 to dampen the host antibacterial response. J. Immunol. 196, 1199–1208. doi: 10.4049/jimmunol.1501045
Zhong, Y., Kinio, A., and Saleh, M. (2013). Functions of NOD-like receptors in human diseases. Front. Immunol. 4:333. doi: 10.3389/fimmu.2013.00333
Keywords: bacteria host interaction, receptor modification, microbiota, antimicrobial resistance (AMR), infection and immunity
Citation: Tiruvayipati S, Hameed DS and Ahmed N (2022) Play the plug: How bacteria modify recognition by host receptors? Front. Microbiol. 13:960326. doi: 10.3389/fmicb.2022.960326
Edited by:
Ibrahim M. Sayed, Assiut University, EgyptReviewed by:
Soumita Das, California College San Diego, United StatesSidharth Prasad Mishra, University of South Florida, United States
Copyright © 2022 Tiruvayipati, Hameed and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niyaz Ahmed, bml5YXouYWhtZWRAdW9oeWQuYWMuaW4=, YWhtZWQubml6aUBnbWFpbC5jb20=
 Suma Tiruvayipati
Suma Tiruvayipati Dharjath S. Hameed
Dharjath S. Hameed Niyaz Ahmed
Niyaz Ahmed