- Organismal and Evolutionary Biology Research Program, Faculty of Biological and Environmental Sciences, and Viikki Plant Science Centre, University of Helsinki, Helsinki, Finland
The phyllosphere is a complex habitat for diverse microbial communities. Under natural conditions, multiple interactions occur between host plants and phyllosphere resident microbes, such as bacteria, oomycetes, and fungi. Our understanding of plant associated yeasts and yeast-like fungi lags behind other classes of plant-associated microbes, largely due to a lack of yeasts associated with the model plant Arabidopsis, which could be used in experimental model systems. The yeast-like fungal species Protomyces arabidopsidicola was previously isolated from the phyllosphere of healthy wild-growing Arabidopsis, identified, and characterized. Here we explore the interaction of P. arabidopsidicola with Arabidopsis and found P. arabidopsidicola strain C29 was not pathogenic on Arabidopsis, but was able to survive in its phyllosphere environment both in controlled environment chambers in the lab and under natural field conditions. Most importantly, P. arabidopsidicola exhibited an immune priming effect on Arabidopsis, which showed enhanced disease resistance when subsequently infected with the fungal pathogen Botrytis cinerea. Activation of the mitogen-activated protein kinases (MAPK), camalexin, salicylic acid, and jasmonic acid signaling pathways, but not the auxin-signaling pathway, was associated with this priming effect, as evidenced by MAPK3/MAPK6 activation and defense marker expression. These findings demonstrate Arabidopsis immune defense priming by the naturally occurring phyllosphere resident yeast species, P. arabidopsidicola, and contribute to establishing a new interaction system for probing the genetics of Arabidopsis immunity induced by resident yeast-like fungi.
Introduction
The phyllosphere is the above ground external surface environment of plants and a complex habitat for microbes. The relationships of phyllosphere microbial residents with host plants can be pathogenic, mutualistic, or frequently commensal. This growth space represents a significant reservoir for latent pathogens, the ecological role of which remains poorly defined (Jarvis, 1994; Saikkonen et al., 1998; Slippers and Wingfield, 2007). Plant beneficial effects induced by plant-associated microbes, such as protection against pathogens and growth promotion, have gained attention recently. Priming is the process of initiating enhanced immune defense responses upon encountering pathogenic microbes, non-pathogenic microbes, or a variety of compounds, both natural and synthetic (Balmer et al., 2015; Conrath et al., 2015). Systemic-acquired resistance activated by necrotizing pathogens, as well as induced systemic resistance triggered by yeast-like fungi, filamentous fungi, and rhizobacteria in soil, are believed to be associated with priming of defense responses (Conrath et al., 2006, 2015; Hossain et al., 2007; Buxdorf et al., 2013a; Balmer et al., 2015; Narusaka et al., 2015). Signaling pathways involving hormones, mRNA, protein kinases, transcriptional factors, and epigenetic regulation all have demonstrated roles in plant immune defense (Beckers et al., 2009; Pieterse et al., 2014).
Yeasts and yeast-like fungi modulate plant growth and health (Nassar et al., 2005; Nafady et al., 2019; Mukherjee et al., 2020). Plant growth-promoting traits and induction of defense response by yeasts occur in both cultivated plants and postharvest fruits (Bergauer et al., 2005; Liu et al., 2013; Nutaratat et al., 2014). Several yeasts with so-called pathogen antagonistic activity have the ability to suppress plant diseases caused by various pathogenic fungi (Liu et al., 2013; Mukherjee et al., 2020; Eitzen et al., 2021). Yeasts, such as Candida, Pichia, and Cryptococcus species, inhibit fungal invasion in postharvest biocontrol systems through induction of host defense (Droby et al., 2002; El Ghaouth et al., 2003; Chan et al., 2007; Zheng and Chen, 2009; Macarisin et al., 2010; Xu et al., 2013). Thus, some yeasts are used as bio-agents for promotion of plant growth and immunity in sustainable agriculture (Liu et al., 2013; Mukherjee et al., 2020).
Several plants exhibit multiple responses to various yeast elicitor preparations; such as autoclaved baker’s yeast (Saccharomyces cerevisiae), preparations from S. cerevisiae cell walls, or commercial yeast extract. These responses include, increased immunity against pathogens, induction of reactive oxygen species accumulation, and activation of known immune signaling pathways, in the model plant Arabidopsis thaliana (referred to hereafter as Arabidopsis; Raacke et al., 2006; Khokon et al., 2010; Minami et al., 2011; Ye et al., 2013; Narusaka et al., 2015; Yaguchi et al., 2017) and other plants (Sun et al., 2018a, 2018b). Raacke et al. (2006) used reverse genetics with Arabidopsis to demonstrate that yeast treatments induced salicylic acid (SA) dependent immunity against Pseudomonas syringae; however, induced resistance against the fungal pathogen Botrytis cinerea (hereafter referred to as Botrytis) was independent of the SA, jasmonate, and camalexin pathways. Although immune priming occurs in Arabidopsis treated with various yeast species isolated from other plants (Buxdorf et al., 2013a,b; Ferreira-Saab et al., 2018), a specific interaction system with a naturally occurring phyllosphere resident yeast and the model plant Arabidopsis has not been established.
Protomyces species are yeast-like members of the subphylum Taphrinomycotina (Ascomycotina; Reddy and Kramer, 1975; Kurtzman, 2011). Protomyces species are all dimorphic; they exist in a non-infectious haploid resident yeast phase and infect their hosts as dikaryotic hyphal pathogenic phase after conjugating with another cell of a compatible mating (MAT) type. Nearly all members of this genus are plant pathogens, mostly of wild plant species, but some crops are affected. For instance, P. macrosporus, which causes coriander stem-gall disease in many regions, threatens coriander seed production (Pavgi and Mukhopadhyay, 1972; Malhotra et al., 2016; Khan and Parveen, 2018). Species in the genus Protomyces cause symptoms such as tumors or galls on flowers, leaf veins, petioles, and stems (Reddy and Kramer, 1975; Kurtzman, 2011; Streletskii et al., 2019). Until recently, the definition of the genus Protomyces encompassed only species pathogenic of host plants within the families Asteraceae and Apiaceae (Reddy and Kramer, 1975; Kurtzman, 2011). However, a phyllosphere resident Protomyces species was isolated from healthy wild growing Arabidopsis, characterized, and named Protomyces arabidopsidicola (type strain C29; Wang et al., 2016, 2021). Genomic, morphological, and physiological features of P. arabidopsidicola differentiate it from other described Protomyces species. Its occurrence on Arabidopsis, a member of the Brassicaceae, suggests Protomyces host-range may not be restricted only to the Asteraceae and Apiaceae families (Wang et al., 2021). However, the relationship of P. arabidopsidicola with Arabidopsis remains undetermined.
Our understanding of plant-associated yeasts lags behind that of other classes of microbes. This is especially true for yeasts associated with Arabidopsis, a model plant widely used when studying the molecular basis of plant-microbe interactions (Boutrot and Zipfel, 2017). Plants have a two tiered immune system (Jones and Dangl, 2006), with pattern recognition receptors (PRRs) that perceive pathogen associated molecular patterns (PAMPs), which are also present in non-pathogenic microbes and thus are also often referred to as microbe associated molecular patterns (MAMPs). PAMP perception leads to activation of basal immunity, termed PAMP-triggered immunity (PTI), which limits infections by pathogens not specifically adapted to that plant. Adapted pathogens overcome PTI by deploying small-secreted proteins and toxins, termed effectors. In turn, plants counter effectors with nucleotide-binding, leucine-rich repeat immune receptors, which perceive effector action to activate immune responses. The dynamics of plant immune function in a molecular evolutionary arms race between host and pathogen is summarized in the zigzag model (Jones and Dangl, 2006; Pritchard and Birch, 2014).
Plants and metazoans share a structurally and functionally conserved innate immune system (Nurnberger and Brunner, 2002; Nurnberger et al., 2004). Although the receptors and mechanisms of perception differ, plants and metazoans have independently evolved the ability to perceive many of the same PAMPs using receptors with similar conserved-domain architectures (Nurnberger and Brunner, 2002; Nurnberger et al., 2004). The human immune system includes a complement of PRRs that specifically recognize PAMPs from yeast cell walls (Jouault et al., 2009; Saijo et al., 2010; Patin et al., 2019). Plants have the ability to perceive some molecules present in yeast cell walls, such as chitin, mannopeptides, and β-glucans, which function as PAMPs (Hahn and Albersheim, 1978; Basse et al., 1992; Felix et al., 1993; De Jonge et al., 2010; Mélida et al., 2018; Christita et al., 2021). However, most of the plant receptors responsible for detecting these PAMPs remain unidentified or are not well characterized. Only chitin has a well-described set of PRRs in plants (Faulkner et al., 2013; Xue et al., 2019). Arabidopsis perceives chitin with receptor complexes containing members of the lysin motif (LysM) receptor kinases; LYK1 (also known as CERK1), LYK2, LYK4, and LYK5 (Miya et al., 2007; Wan et al., 2008; Faulkner et al., 2013; Cao et al., 2014; Xue et al., 2019). Additionally, CERK1 is required for perception of linear β-1,3-glucan (Mélida et al., 2018). The LysM domain is responsible for chitin binding and is found in both plant chitin receptors and fungal effector proteins that bind and sequester chitin to suppress immunity (Doehlemann and Hemetsberger, 2013).
Microbe recognition triggers mitogen-activated protein kinase (MAPK) activation. This is a key early event in PAMP-induced immune signaling. MAPK3 and MAPK6 are differentially required for immunity against the broad host-range fungal pathogen Botrytis induced by oligogalacturonides or flagellin (Galletti et al., 2011). In addition, perception of the fungal PAMP chitin results rapid activation of a MAPK cascade in Arabidopsis and rice (Miya et al., 2007; Wan et al., 2008; Yamada et al., 2017). MAPK activation kinetics can vary greatly, lasting for minutes to hours, depending on the MAPK involved and the activating stimulus (Bigeard and Hirt, 2018). The plant immunity network also involves complex and interconnected hormone signaling pathways, including SA, jasmonic acid (JA), ethylene, and others. Additionally, many microbes, including Protomyces are known to produce plant hormones, which play multiple roles including promoting survival in the phyllosphere and manipulating host defense signaling to promote virulence (Vorholt, 2012; Chanclud and Morel, 2016; Patkar and Naqvi, 2017). Our knowledge of plant-pathogen interactions is advancing rapidly. The existence of yeast-like fungi in the phyllosphere of wild growing Arabidopsis is increasingly documented (Agler et al., 2016; Wang et al., 2016; Regalado et al., 2020; Brachi et al., 2021; Almario et al., 2022), but Arabidopsis interactions with this class of fungi are currently not well understood.
We address this utilizing Arabidopsis and the Arabidopsis-associated yeast-like fungal species, P. arabidopsidicola strain C29 (Wang et al., 2021). The objective of this study is to address the influence of this resident yeast on plant immunity. These results introduce a novel experimental system involving a naturally occurring phyllosphere yeast and the host from which it was isolated, the genetic model plant Arabidopsis.
Materials and methods
Microbial strains and culture
A culture of the P. inouyei type strain YB-4354 was provided by the ARS culture collection.1 Protomyces arabidopsidicola strain C29, which was isolated from wild Arabidopsis thaliana (Arabidopsis; Wang et al., 2016) and subsequently described and named (Wang et al., 2021), has been deposited in the following culture collections; HAMBI, HAMBI3697 and DSMZ, DSM 110145. For the current study, all Protomyces species were purified twice from single colonies and cultured on yeast, peptone, dextrose (YPD; 0.5, 0.5, and 2%, respectively) agar (1.5%) at 21°C, unless otherwise indicated. Botrytis cinerea strain BO.510 was obtained from the lab of Tapio Palva, Helsinki, Finland. Botrytis growth and infection was performed as previously described (Cui et al., 2016; Vuorinen et al., 2021). Briefly, Botrytis was cultivated on potato dextrose broth (PDB) with 15 g/l agar in the dark at room temperature. Mycelium with conidia was harvested with forceps, suspended in 0.5× PDB, mixed vigorously, filtered through miracloth, and diluted for infections. Botrytis spore density was determined microscopically using a hemocytometer. A Paenibacillus strain, obtained from Ansa Palojärvi at the University of Helsinki, is known to produce antifungal compounds active against Botrytis and was used as a positive control for direct microbe-microbe interactions. Co-cultivation of Botrytis with P. arabidopsidicola and the Paenibacillus strain was performed on YPD agar plates at 21°C.
Arabidopsis growth, infection, and trypan blue staining
Wild type Columbia-0 (Col-0) accession seeds of Arabidopsis were from the Nottingham Arabidopsis Stock Centre (NASC).2 The local accession Kivikko (Kvk1) was collected at the site where Arabidopsis–associated yeast isolation samples were previously collected (Wang et al., 2016). Standard conditions for Arabidopsis cultivation were as follows. Seeds were sown at high density on wet vermiculite peat mix (1,1, with type B2 peat; Kekkilä; www.kekkila.fi). Seeds were stratified in the dark at 4°C for 72 h, before transfer to a growth chambers (Fitotron SGC120, Weiss Technik)3 with 12/12 h day/night photoperiod with 170 μmol m−2 s−1 illumination, at 23°C/18°C and 65%/75% relative humidity. One-week-old seedlings were transplanted one per pot in 6 cm square pots or five per pot in 10 cm square pots with the same soil mixture. Three-week-old plants were used for infection experiments. Standard infection conditions were done using drop infections by pipetting 5 μl of an OD600 = 1 cell suspension of P. arabidopsidicola in 10 mM MgCl2 onto the adaxial side of the leaves, left from the midvein. Infected plants were evaluated for symptoms every 1–2 days for 2 weeks and photographed at 10 dpi for documentation, as needed. P. arabidopsidicola was monitored on and in infected Arabidopsis leaves by trypan blue staining. Trypan blue stain stock solution (0.05%) in lactophenol (1.1:1, glycerol:85% lactic acid: phenol) was diluted 1:2 in 95% ethanol for staining. Infected leaves were covered in trypan blue staining solution and incubated in a boiling water bath until staining solution started boiling (approx. 3 min). Samples were de-stained for 48 h in 2.5 g/ml chloral hydrate solution in water and placed in 60% glycerol for storage and mounting on slides. Samples were examined under a compound microscope (Leica Model MZ 2500)4 with a 10× eye piece and 20× or 100× objective and photographed with an attached photo documentation system (Leica model DFC490).
Protomyces growth assays
Three-day-old cultures in YPD liquid medium were harvested and washed twice with Milli-Q (MQ) water. Protomyces inouyei (OD600 = 0.14, 5 μl/leaf) and P. arabidopsidicola strain C29 (OD600 = 0.10, 5 μl/leaf) suspensions were drop inoculated on the adaxial side of 24-day-old Arabidopsis leaves, aseptically grown on 0.5× Murashige and Skoog (MS) agar plates. Sterile foil on an agar plate was used as a control for non-specific surface growth. Growth in the phyllosphere was assayed with re-isolation of Protomyces from inoculated leaves by placing leaves in 2 ml Eppendorf tubes with 1 ml 0.025% Silwet-L77 in water and shaking (800 rpm; VWR microplate shaker) for 1 h, after which 5 μl wash solution was serial diluted and plated on YPD agar for colony counting. Pooled data from five biological repeats were analyzed using a linear mixed model with nlme package in R (v3.5.1).
Protomyces arabidopsidicola cell wall preparation and root growth assay
For P. arabidopsidicola cell wall isolation, cells were cultivated in potato dextrose broth (PDB; Carl Roth) in 200 ml flasks on a rotary shaker for 4 days at room temperature. Cells were collected and washed twice in sterile MQ water, re-suspended in a small volume of sterile MQ water, and freeze-dried for 2 days. Dry cells were ground in a tissue homogenizer (Precellys 24)5 using 425–600 μm glass beads (5×15 s at 6,800 rpm, cooling at-20°C between runs). Cell disruption was confirmed using a compound microscope (Leica Model MZ 2500)6 with 40× objective. The insoluble cell walls were separated from the soluble fraction by centrifugation in MQ water at 1,000 g for 20 min in + 4°C. This crude cell wall preparation was then freeze dried as described above, and weighed.
Sterile Arabidopsis Col-0 accession seeds were stratified at + 4°C for 72 h and germinated vertically on 0.5× MS 0.8% agar plates for 4 days in a growth chamber (Fitotron SGC120, Weiss Technik)7 at ~170 μmol m−2 s−1 illumination, at +23/+18°C. Seedlings were transplanted to either 0.5× MS agar control plates or identical plates supplemented with 0.9 g/l P. arabidopsidicola cell wall preparation. Seedlings were grown in the same conditions for 10 more days, photographed, and root length measured using imageJ software.8
Protomyces arabidopsidicola pre-treatment, Botrytis infection, and qPCR
Live and autoclaved P. arabidopsidicola cultures were harvested, washed, and suspended at OD600 = 1 (7 × 106 CFU/ml) in sterile water. Three-week-old soil-grown Arabidopsis were pre-treated by spraying with one of the P. arabidopsidicola cell suspensions (0.2 ml/plant), or water as a control, then infected with a Botrytis a 3 μl drop of a 2 × 106 spore ml−1 spore suspension 3 days after treatments with P. arabidopsidicola cells. Plants were grown in a chamber with 12/12 h (light/dark), 23/18°C, and 65/75% relative humidity. Pre-treated and infected plants were collected at 30 and 60 min for western blot, and at 24 h and 72 h for qPCR. Four or five plants were pooled, frozen in liquid nitrogen, and stored in –80°C. Leaf lesions were photographed at 72 h and diameters measured in ImageJ.9 Lesion size data was statistically analyzed with scripts in R (v3.5.1) using the nlme package. The model contrasts were estimated with multcomp package, and the estimated p-values were subjected to single-step correction. RNA isolation and quantitative reverse transcriptase PCR (qPCR) were performed as previously described (Cui et al., 2016). Three genes TIP41, YLS8 and PP2AA3, which have stable expression levels in a wide variety of conditions (Czechowski et al., 2005), were simultaneously used as reference genes and are listed in (Supplementary Table S1). Stability of multiple reference gene expression and raw cycle threshold values were analyzed with Qbase+2.1 (Hellemans et al., 2007). Expression levels relative to the multiple reference genes using three technical replicates with correction for primer efficiencies and error propagation were first calculated in Qbase+2.1 (Hellemans et al., 2007) and then relative expression was calculated to facilitate comparisons within but not between the two experiments; i.e. in the yeast treatment experiment, treatment values were normalized to the 24 h control and in the yeast pre-treatment with Botrytis infection experiment, treatment values were normalized to the Botrytis-treated 24 h control. Primers used and their primer efficiencies are listed in the supplementary materials (Supplementary Table S1). Significance of differential expression was estimated by glht package in R (3.5.1) using false discovery rate adjustment of p-values.
Protein extraction, SDS-PAGE, and western blotting
Total protein was isolated from 100 mg frozen rosettes in 100 μl Lacus buffer (Brader et al., 2007). Protein concentration determined by Bradford assay10 with BSA as standard. Total protein (100 μg) was loaded in SDS-PAGE, transferred to a membrane (PVDF type; Immobilon–FL), and scanned with LI-COR Odyssey scanner. Equal loading was monitored by amido black staining. Primary anti-TEpY (Phospho-p44/42 MAPK (Erk1/2) Thr202/Tyr204; CST®) rabbit monoclonal antibody specific for the activated form of MAPKs was used at a 1:2,000 dilution and the secondary anti-rabbit IgG antibody (IRDye, 800CW Goat anti-Rabbit) at 1:10,000.
Experimental field infections
Plants for long-term field infections were sown and germinated in September in a covered cage under ambient conditions using seeds of the accessions Col-0 and a local accession Kvk1. Seven-day-old seedlings were transplanted 5 per pot in 10 cm square plastic pots with a 1:1 mix of peat and vermiculite and further grown under the covered cage under ambient conditions. Pots containing two-week-old plants were buried in sand in the Helsinki University Viikki campus experimental field in a triplicate block design at three different sites in the field and spray inoculated with water (mock) or P. arabidopsidicola cell suspensions (OD600 = 0.1 and 1.0, ca. 0.5 ml per pot). The field site was not pre-tested for the presence of the yeast, which is assumed to be wind dispersed and possibly present. Plants remained in the field over the course of the next 7 months (until early May), with snow cover for about 4 months, and were visually examined and photographed at various times. In May, final photos and visual examinations were done and representative mock control and infected plants were sampled for yeast isolations, performed as previously described (Wang et al., 2016). Briefly, the identity of reisolated yeasts were confirmed as P. arabidopsidicola by cleaved restriction fragment length polymorphism analysis of ITS PCR fragments using Taq I.
Results
Protomyces arabidopsidicola strain C29 is not pathogenic on Arabidopsis
The genome of P. arabidopsidicola strain C29 harbors a heterothallic MAT locus of the P-type suggesting that it should not be able to infect Arabidopsis as a single strain (Wang et al., 2019). To confirm that strain C29 was not able to cause disease on Arabidopsis, infected plants were examined using many Arabidopsis-infection protocols, including chamber experiments at low temperatures and long-term field infections (Table 1), which mimicked the environmental conditions, from which P. arabidopsidicola was isolated. The local Arabidopsis accession, Kvk1, which was collected at the site where P. arabidopsidicola was isolated, and the accession Col-0 were used for infections. In field experiments, P. arabidopsidicola was recovered from infected plants, but not control plants that were mock infected with water. In all field and chamber experiments, infected Arabidopsis leaves were examined for formation of typical disease symptoms caused by Protomyces species, such as galls and tumors, as well as visible necrosis, chlorosis, and other more general disease symptoms. As expected, P. arabidopsidicola infection with the single strain C29, did not result in any disease symptoms on Kvk1 (not shown) or Col-0 (Figure 1A), which was representative of results obtained with both accessions. These results (Figure 1) were obtained with plants grown in chambers under standard growth conditions, but are typical for all experiments, as disease symptoms were not found under any of the tested conditions (Table 1). In field experiments, some damaged leaves were present. However, this damage was also found in control plants, suggesting they are not specific to infected plants and due to growth conditions. The location and morphology of P. arabidopsidicola cells was examined in infected Arabidopsis leaves by trypan blue staining (Figure 1B). P. arabidopsidicola was not observed in control uninfected plants, but clearly visible on the leaf surface of inoculated plants in its non-infectious yeast form, especially concentrated in the depressions between epidermal pavement cells (Figure 1B, center panel). Examination under high magnification confirmed the typical Protomyces yeast cell type morphology and no infectious hyphal cell types were found (Figure 1B, right panel). These findings support that P. arabidopsidicola strain C29 was a non-pathogenic resident yeast in the Arabidopsis phyllosphere. In field infections, P. arabidopsidicola survived overwinter in the Arabidopsis phyllosphere and was re-isolated the following spring only from plants that were inoculated in autumn with P. arabidopsidicola.
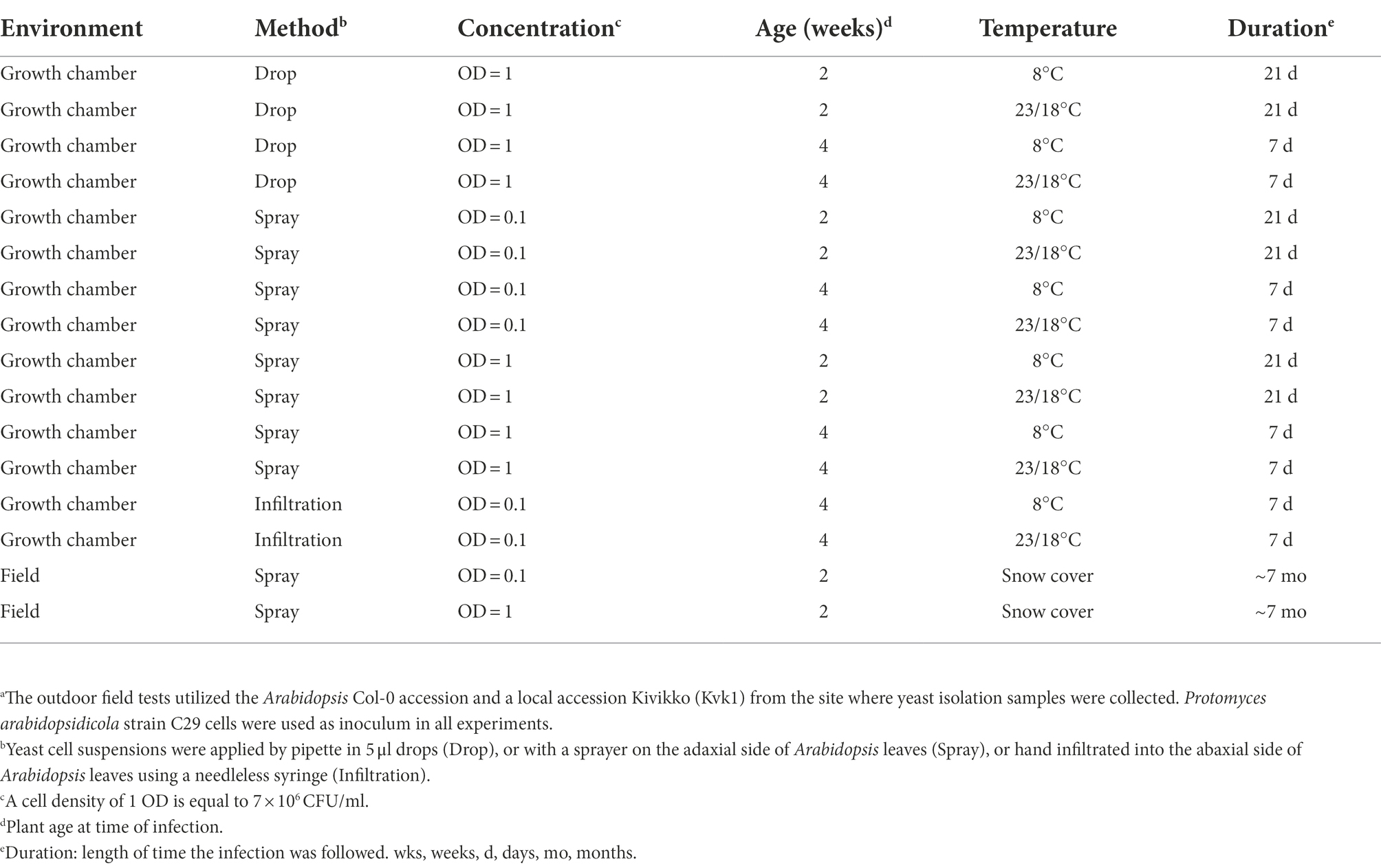
Table 1. Methods used to test infection of Arabidopsis with P. arabidopsidicola strain C29a.
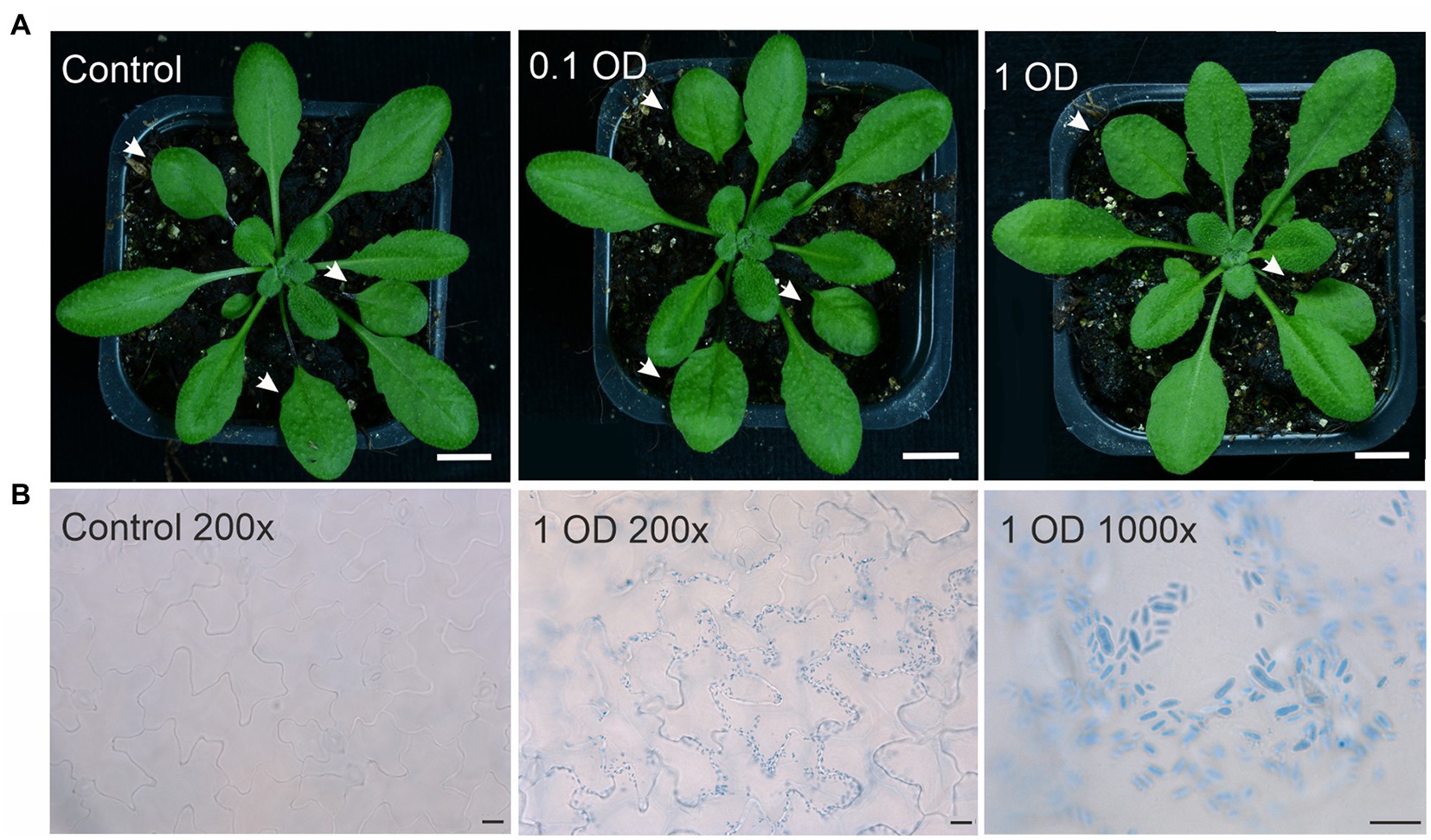
Figure 1. Arabidopsis leaves infected with Protomyces arabidopsidicola. (A) Arabidopsis (Col-0 accession) plants either mock treated with 10 mM MgCl2 (control) or drop infected in the middle of the left hand leaf half with a 0.1 OD or a 1 OD suspension of P. arabidopsidicola cells in 10 mM MgCl2 and photographed at ten days post infection. Treated leaves are indicated with white arrows. White scale bars in all photos are 1.0 cm. (B) Photomicrograph of Arabidopsis leaves either mock treated with 10 mM MgCl2 (control 200×) in the right panel or infected with a 1 OD suspension of P. arabidopsidicola cells (1 OD 200×) in the middle panel. Leaves were stained with trypan blue to visualize P. arabidopsidicola cells. The right hand panel (1 OD 1,000×) is a close up showing the typical Protomyces yeast-type cell structure of the visualized cells. Scale bars: left 20 μm, middle 20 μm, right 10 μm. All experiments used 21-day-old soil-grown Arabidopsis plants. Experiments were repeated 2–3 times with similar results and representative results are shown.
Protomyces arabidopsidicola persists in the Arabidopsis phyllosphere
Since P. arabidopsidicola was able to overwinter in the Arabidopsis phyllosphere under field conditions, P. arabidopsidicola and its most closely related species, P. inouyei, were tested for the ability to persist on the leaf surface of aseptic in vitro grown Arabidopsis. In these experiments, a high level of drop-inoculated P. arabidopsidicola persisted on the Arabidopsis leaf surface, with a stable level of CFUs maintained over a 10-day period (Figure 2). All recovered microbes cultivated from infected plants had typical colony characteristics of P. arabidopsidicola and control plates (not shown) were free of microbes. The Arabidopsis phyllosphere supported a similar but slightly lower level of P. inouyei, which is a close relative P. arabidopsidicola. At the 10 days post infection, the level of P. arabidopsidicola was only about 10-fold higher than that of P. inouyei. This demonstrated that over the time-period studied, the ability to persist in the Arabidopsis phyllosphere was common to both P. arabidopsidicola and P. inouyei. Further, as a negative control, P. arabidopsidicola survival was tested on sterile foil placed inside the same agar medium plates. After 10 days, the level of recovered CFUs dropped to near zero, indicating it was not able to persist on surfaces non-specifically (Figure 2).
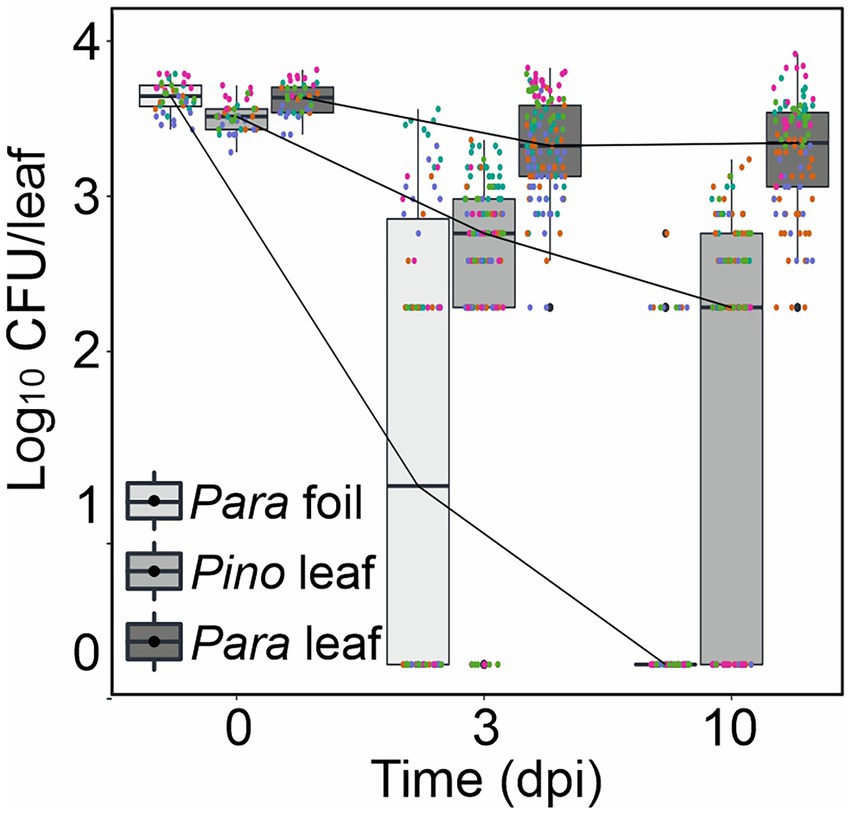
Figure 2. Protomyces arabidopsidicola strain C29 persistence on Arabidopsis. Persistence assay of P. arabidopsidicola (Para) and P. inouyei (Pino) on the surface of Arabidopsis leaves aseptically grown on 0.5 x MS agar. Cells were re-isolated from drop inoculated plants at the indicated times and plated to determine cell numbers, which are presented as the number of colony forming units (Log10 CFU/leaf). Persistence on sterile foil was used as a control for the ability to survive non-specifically on surfaces. Pooled data from five independent biological repeats (n = 25) were analyzed by computing a linear mixed model in R (v3.5.1). The pooled data is displayed in standard boxplots, all data points are plotted with a different color for each independent biological repeat.
Protomyces arabidopsidicola immunity priming on Arabidopsis
The activation of defenses and growth are finely balanced and mutually inhibitory; accordingly, activation of immune defense responses results in a well-known effect where growth is inhibited (Belkhadir et al., 2014; Huot et al., 2014). This effect was used to explore the activation of Arabidopsis immune signaling by P. arabidopsidicola. In in vitro root growth assays, including P. arabidopsidicola cell wall preparations in the growth medium resulted in significant inhibition of root growth (Figure 3). This result supports that P. arabidopsidicola cell walls contain immune stimulating molecular patterns.
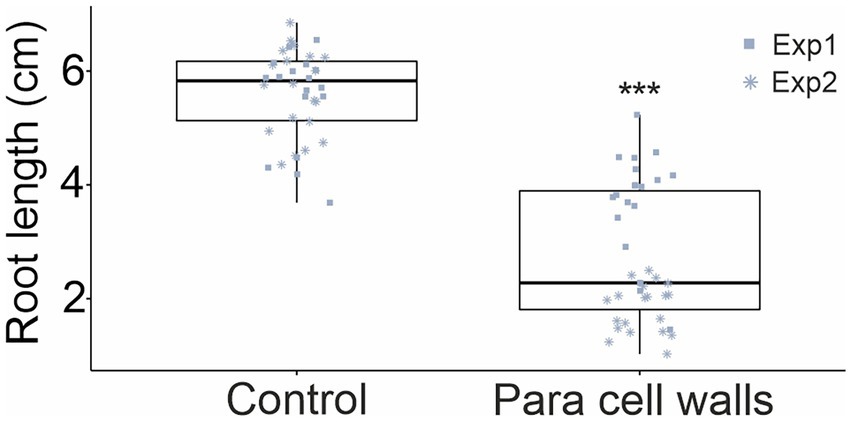
Figure 3. Inhibition of Arabidopsis growth by P. arabidopsidicola cell wall preparations. Arabidopsis growth was quantified in an in vitro root growth assay that measured root length of 14-day-old seedlings that had been grown for 10 days on vertically oriented 0.5× MS agar plates without (Control) or with addition of 0.9 g/l P. arabidopsidicola cell walls (Para cell walls). The experiment was replicated in two independent biological repeats and pooled data is presented (n = 19 in each repeat, total n = 38). Statistical significance was tested using two-way ANOVA in R (v3.5.1), the treatment effect was significant (***p = 2 × 10−16). The pooled data is displayed in standard boxplots; all data points are plotted with a different shape for each independent biological repeat.
To further explore if P. arabidopsidicola can engage Arabidopsis immunity, the ability of strain C29 to alter resistance against a broad-host-range fungal pathogen was tested. Plants were pre-inoculated with water, a suspension of live strain C29 cells, or a suspension of strain C29 cells killed by autoclaving. Plants were then infected with the necrotrophic fungal pathogen Botrytis and disease progression monitored as the size of Botrytis-induced spreading lesions. Significantly smaller lesion diameter was observed in Arabidopsis pre-treated with both live and killed P. arabidopsidicola cell-suspensions (Figures 4A,B), suggesting increased resistance. Additionally, plants pre-treated with live P. arabidopsidicola cells had significantly smaller lesions than those treated with dead cells (Figures 4A,B). This suggests that yeast PAMP molecules liberated from dead P. arabidopsidicola cells could trigger plant immunity, but the full effect required live cells. In order to test for direct microbe-microbe interactions, Botrytis and P. arabidopsidicola were co-cultivated on PDB agar, a medium known to support growth of both of these fungi. This demonstrated that P. arabidopsidicola cultures were not able to inhibit Botrytis growth in vitro (Figure 4C), arguing against the possible direct antagonistic effect by P. arabidopsidicola. These results suggest P. arabidopsidicola on the Arabidopsis leaf surface may inhibit Botrytis disease spread via interaction with the plant to activate immune signaling, rather than via direct microbe-microbe interaction.
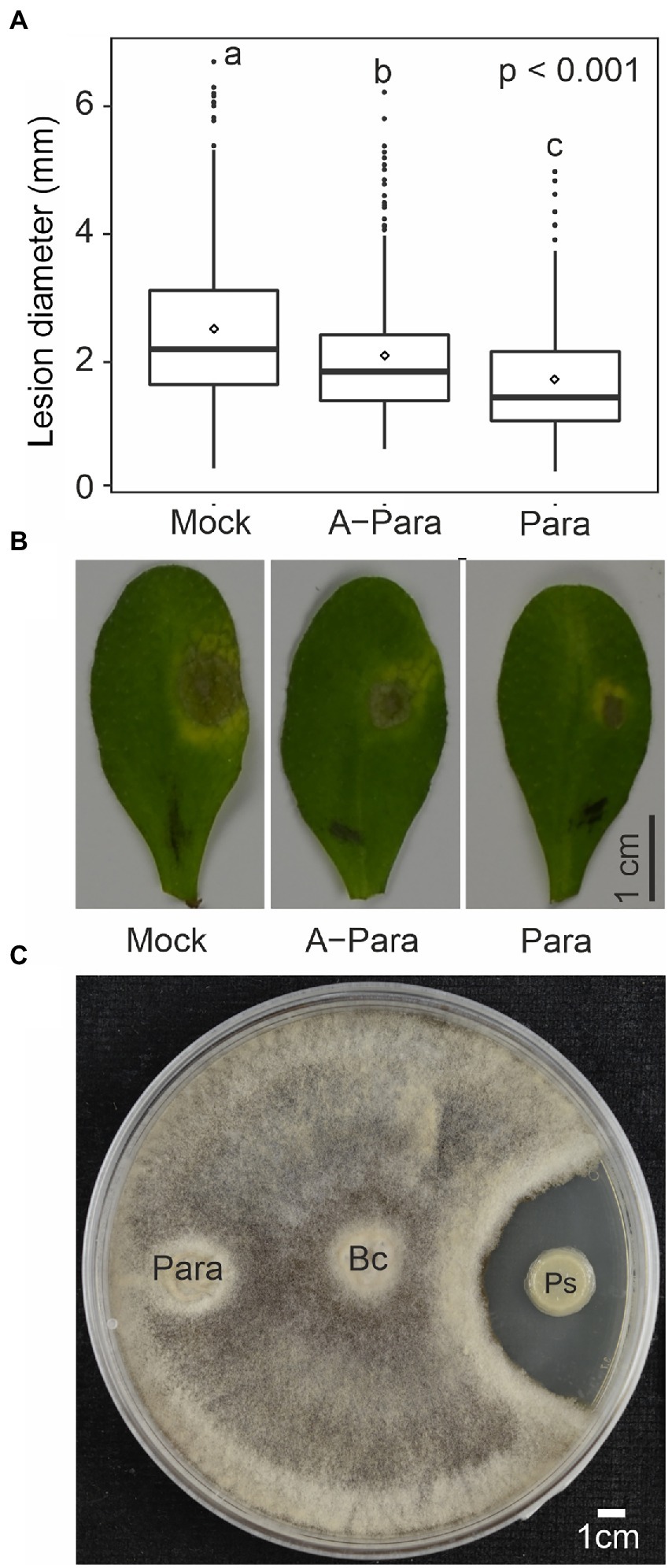
Figure 4. Priming effect by P. arabidopsidicola. (A) Lesion diameters from drop infection of Botrytis spores on leaves of Arabidopsis pre-treated with water (mock), autoclave killed Protomyces arabidopsidicola C29 (A-Para), or live Para. The pooled data from six independent biological repeats (total n = 24) is displayed in standard boxplots (central 50% of the data range in the box, with a line showing the median, bars depict the range of remaining data, and dots display outlier data points). Statistics performed with pooled data from by computing a linear mixed model in R (v3.5.1). (B) Photos of Botrytis-induced necrosis on leaves of Arabidopsis that were pre-treated with water (mock), autoclave killed Para (A-Para), or live Para. (C) No direct growth inhibition by P. arabidopsidicola strain C29 (Para) was observed against Botrytis cinerea (Bc) in co-cultivation assays. As a control, a Paenibacillus sp. (Ps) known to have antifungal activity against Botrytis was used. Yeast extract-peptone-dextrose agar plates were used for cultivation. Photos were taken 7 days post inoculation. Experiment was repeated three time with similar results and one representative result is shown.
Protomyces arabidopsidicola activates MAPK and immune signaling pathways
Mitogen-activated protein kinases (MAPK) activation is among the earliest events in Arabidopsis immune responses. MAPK activation in response to pre-treatment with P. arabidopsidicola cells was assayed, both alone and with subsequent Botrytis infection. Western blots probed with the pTEpY antibody, specific for the active phosphorylation site of MAPKs, revealed slight activation of Arabidopsis MAPK3 and MAPK6 (MAPK3/6) in response to treatment with both live or dead P. arabidopsidicola cells at 30 min and this response increased to a higher level by 60 min (Figure 5A). When plants were infected with Botrytis, at 30 min all samples exhibited MAPK3/6 activation, which had returned to near the control level by 60 min (Figure 5B). Some mock-infection controls showed moderate MAPK-activation, likely due to slight wounding during the mock treatment. Plants pre-treated with live P. arabidopsidicola, dead P. arabidopsidicola, or mock treatment showed a similar degree of MAPK3/6 activation at 30 min after Botrytis infection. We conclude that single treatments with live or dead P. arabidopsidicola, or Botrytis, rapidly activated MAPK3/6 in Arabidopsis.
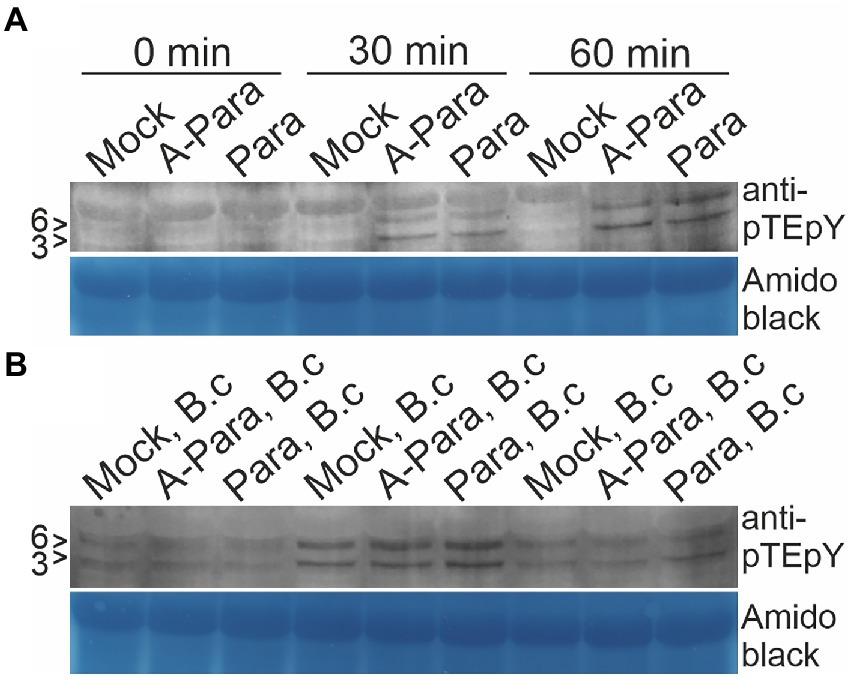
Figure 5. Arabidopsis MAPK activation by P. arabidopsidicola. Arabidopsis mitogen activated protein kinase (MAPK)3 and MAPK6 (MAPK3/6) activation was monitored by western blot with anti-phospho MAPK antibodies following pre-treatment with autoclaved P. arabidopsidicola strain C29 (A-Para) or live P. arabidopsidicola strain C29 (Para). The positions of the 43 and 45 kDa AtMPK3 and AtMPK6 are indicated to the left of the protein blots with the respective markers (3 > and 6 >). MAPK3/6 were activated by both A-Para and Para pre-treatment at 30 and 60 min. (A) Leaf samples were collected at 30 and 60 min after A-Para or Para pre-treatment. (B) Leaf samples were collected at 30 and 60 min after Botrytis infection was applied to plants which has been pre-treated with A-Para or Para for 3 days. Equal loading was confirmed by amido black staining. Three independent experiments were repeated with the same result, of which representative results are shown. B.c: Botrytis cinerea.
To further explore activation of defense signaling, qPCR was employed to monitor expression of defense signaling marker genes (Supplementary Table S1). Strikingly, evidence of enhanced activation of immune signaling was observed. This was seen as enhanced Botrytis-induced transcriptional responses, in samples pre-treated with live or dead P. arabidopsidicola cells, for PDF1.1, a JA marker; CYP71a13 and PAD3, markers of camalexin, which is the primary antimicrobial compound in Arabidopsis; and PR1, a marker of the late SA response (Figure 6A). Live or dead P. arabidopsidicola cells alone had limited effect on defense signaling activation, showing induction of PAD3 at 72 hpi (Figure 6A). Taken together, the all the results presented above indicate P. arabidopsidicola cells had immunity priming effect in Arabidopsis, and activated some of the known canonical defense pathways that involve JA, SA, and camalexin. Markers for other hormone signaling pathways that have also been implicated in immune signaling, such as ethylene, ABA, and auxin, exhibited no significant changes (Figure 6B), suggesting the response to treatment with P. arabidopsidicola is independent of these pathways.
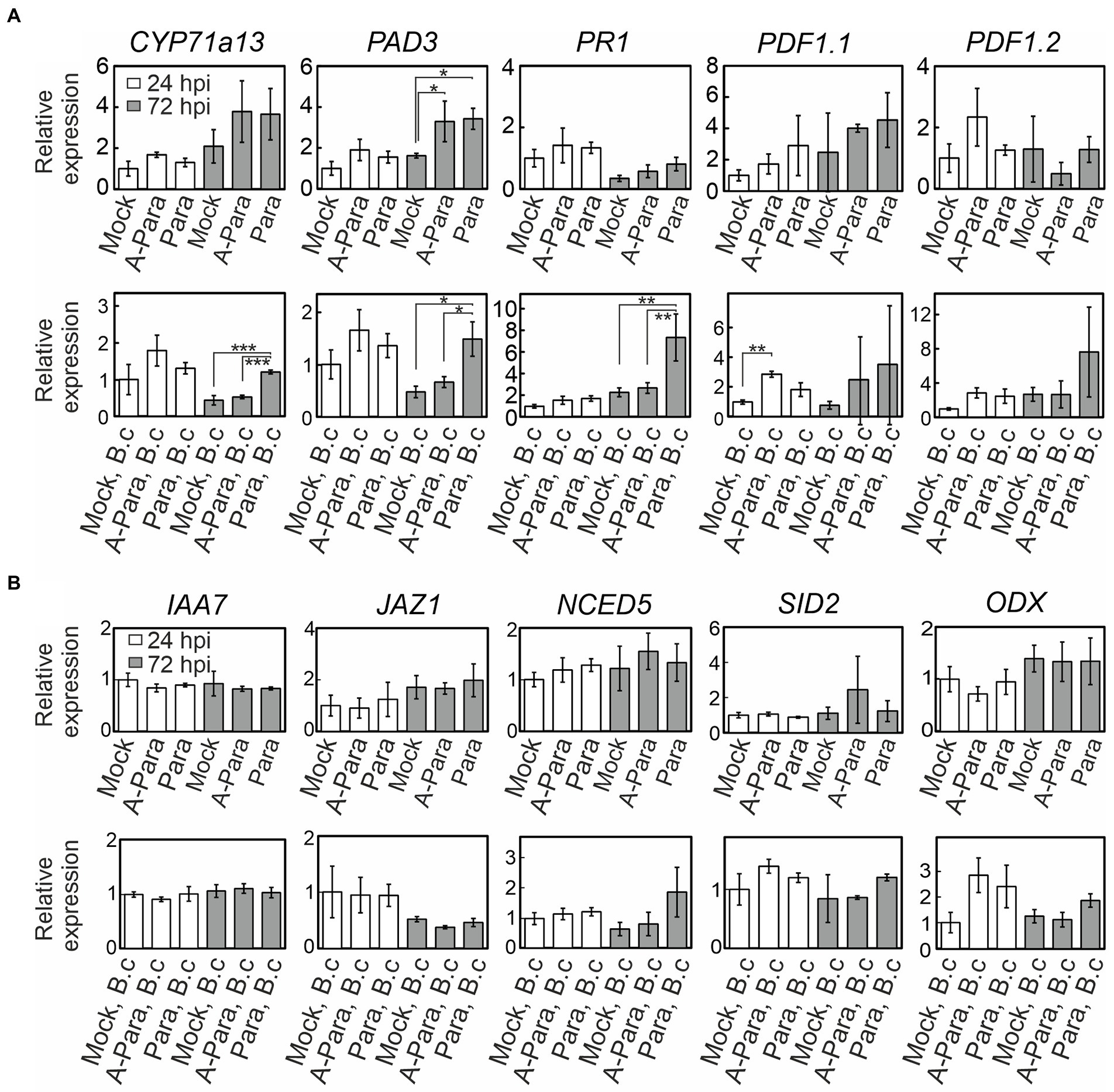
Figure 6. Plant defense and hormone signaling marker expression. Relative gene expression of plant defense and hormone signaling genes at 24 (white bar) and 72 (grey bar) hours post infection (hpi) following P. arabidopsidicola strain C29 treatment only (top row), and with Botrytis infections following strain C29 pre-treatments (bottom row). Relative expression is calculated to facilitate comparisons within but not between the top and bottom rows; i.e. top row is normalized to the 24 h control and the bottom row is normalized to the Botrytis-treated 24 h control. All time points are relative to the last treatment; relative to the strain C29 treatment in the top row and relative to the Botrytis treatment in the bottom row. (A) Expression of the genes: CYP71a13, CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13, a marker for camalexin biosynthesis; PAD3, PHYTOALEXIN DEFICIENT 3, a camalexin marker; PR1, PATHOGENESIS-RELATED GENE 1, a late salicylic acid-response marker; PDF1.1, PLANT DEFENSIN 1.1, a jasmonic acid-response marker; PDF1.2, PLANT DEFENSIN 1.2, a jasmonic acid-response marker. (B) Expression of the genes: IAA7, INDOLE-3-ACETIC ACID 7, an auxin-response marker; JAZ1, JASMONATE-ZIM-DOMAIN PROTEIN1, an early jasmonic acid-response marker; NCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, an ABA-response marker; SID2, SALICYLIC ACID INDUCTION DEFICIENT 2, a salicylic acid-response marker; ODX, 2-OXOACID-DEPENDENT DIOXYGENASE, a salicylic acid-response marker. Data are presented as mean relative expression levels ± SD of three pooled biological repeats. Statistics was performed with glht package in R (v3.5.1) with fdr method for adjusted p values. Para: P. arabidopsidicola strain C29; A-Para: autoclaved P. arabidopsidicola strain C29; B.c: Botrytis cinerea infected. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
In this study, we aimed to establish an experimental system for the study of immune system interaction between the genetic model plant Arabidopsis and a naturally occurring phyllosphere resident yeast-like fungus. Protomyces arabidopsidicola has been found in virtually all studies addressing fungal residents of the Arabidopsis phyllosphere (Agler et al., 2016; Wang et al., 2016, 2019; Regalado et al., 2020; Brachi et al., 2021). The occurrence of P. arabidopsidicola on wild and field grown Arabidopsis, together with its ability to persist in the Arabidopsis phyllosphere, both under growth chamber and experimental field conditions, support that this yeast is naturally associated with Arabidopsis and can utilize its phyllosphere as a living space.
The question of specificity in this yeast-plant interaction remains only partially resolved. Protomyces inouyei, a close relative of P. arabidopsidicola, was also able to persist in Arabidopsis phyllosphere in in vitro experiments, but to a lesser extent. This suggests that the other closely related Protomyces species, P. inouyei, P. lactucaedebilis, and P. pachydermus, which form a distinct clade with P. arabidopsidicola (Wang et al., 2019, 2021), may be able to grow on Arabidopsis, at least on aseptic in vitro grown plants. Further persistence assays and growth assays with a broader selection of Protomyces species will be needed in future studies to test this. However, there are many surveys of Arabidopsis phyllosphere yeasts that have identified P. arabidopsidicola, but none of these studies found any of the other above mentioned Protomyces species (Agler et al., 2016; Wang et al., 2016, 2019; Regalado et al., 2020; Brachi et al., 2021; Lind and Pollard, 2021; Almario et al., 2022). This suggests that P. arabidopsidicola could be better adapted to the Arabidopsis leaf surface in the wild and further implies that factors other than the host, possibly the host microbiome, participate in limiting the ability of yeasts to survive in their host phyllosphere. Accordingly, study of the Arabidopsis phyllosphere microbiome reveals the formation of complex functionally interrelated microbial communities (Agler et al., 2016; Chaudhry et al., 2021; Almario et al., 2022). Protomyces arabidopsidicola was also found in a number of other environmental samples, including several plant species, such as bean and cereal crop-related wild grasses (Prior et al., 2017; Wang et al., 2019; Sun et al., 2020; Wang et al., 2021), supporting that it can also survive in the phyllosphere of plants other than Arabidopsis. Although P. arabidopsidicola may not be specific to only Arabidopsis, the current study still illustrates a broadly distributed yeast living in the phyllosphere of multiple plants with the ability to modulate the health of Arabidopsis and potentially other plants. This suggests that work with P. arabidopsidicola may be relevant beyond just the genetic model plant Arabidopsis.
The nature of this yeast–plant interaction also requires further clarification and confirmation. Nearly all Protomyces species are heterothallic (Reddy and Kramer, 1975; Kurtzman, 2011; Wang et al., 2019), and thus require a strain of the opposite MAT type in order to enter the phytopathogenic hyphal state. Genome sequencing has confirmed MAT loci consistent with heterothallism in six of seven Protomyces species examined (Wang et al., 2019). The presence of a heterothallic MAT locus of the P-type within the genome of P. arabidopsidicola strain C29 (Wang et al., 2019) predicts that it cannot infect Arabidopsis as a single strain, i.e., in the absence of a second strain of the compatible M mating type. Results from Arabidopsis infection experiments presented here confirm this prediction. Further isolations to obtain matM type strains of P. arabidopsidicola and then coinfection tests with two strains of compatible MAT types infecting multiple different accessions will be required to test if P. arabidopsidicola is pathogenic on Arabidopsis. Regardless of the outcome of these tests, the P. arabidopsidicola–Arabidopsis system remains relevant; either for testing interactions with a non-pathogenic phyllosphere resident, or the very interesting prospect of testing the interactions of a host plant with a latent pathogen that resides as a single strain in the phyllosphere while it waits for a compatible mating partner.
The findings presented here illustrate the utility of this new experimental system. Importantly, this work demonstrates the ability of a native phyllosphere resident yeast to activate Arabidopsis defense signaling and induce immunity against Botrytis. This provides evidence supporting the existence of ecologically relevant yeast PAMPs that trigger plant immunity. The human innate immune system deploys a distinct set of PRRs activated by mannose containing linkages that are highly abundant in the outer layers of cell walls of human pathogenic yeasts, such as Candida albicans (Jouault et al., 2009; Saijo et al., 2010). Plant pattern-recognition receptors that perceive yeast PAMPs remain largely unknown, although many studies demonstrate the ability of yeasts to activate plant immune signaling. For example, yeast cell wall extract of the budding yeast Saccharomyces pastorianus activates JA and azelaic acid signaling to induce plant disease resistance (Yaguchi et al., 2017). In Arabidopsis, yeast cell wall extract activates the JA, ethylene, and SA signaling pathways, and SA accumulation and signaling were required for induced resistance (Narusaka et al., 2015). Autoclaved cell suspensions of S. cerevisiae also engaged Arabidopsis immune signaling; specifically, SA signaling, camalexin biosynthesis, and enhanced resistance against other pathogens (Raacke et al., 2006). Accordingly, in the current study, P. arabidopsidicola cell walls caused root growth inhibition and live cell suspensions primed the SA- and camalexin-signaling pathways, enhanced Botrytis resistance, and activated MAPK signaling. MAPK activation induced by some PAMPs is quite rapid and transient; however activation kinetics can be quite varied depending on which MAPK is activated and the activation stimuli (Bigeard and Hirt, 2018). Our study tracked MAPK3 and MAPK6 activation at 30 and 60 min, which is well within known kinetics for these MAPKs.
Protomyces species are well known to produce auxin (Streletskii et al., 2016); including P. arabidopsidicola, where this was demonstrated with multiple lines of evidence. Protomyces arabidopsidicola accumulated indolic compounds in culture filtrates that activated Arabidopsis auxin signaling assessed by expression of the auxin responsive DR5::GUS promoter-reporter system and by root bioassays (Wang et al., 2016, 2019). Analysis of the P. arabidopsidicola genome also revealed a full pathway for IAA biosynthesis via the indole-3-pyruvic acid pathway (Wang et al., 2019). Remarkably, infection of Arabidopsis with P. arabidopsidicola did not result in activation of the auxin responsive marker gene IAA7. Auxin of microbial origin is multifunctional; acting in the microbe to control development, promote phyllosphere survival, in microbe–microbe interactions, and in the host plant, for instance to suppress defense responses (Spaepen et al., 2007). Thus, future studies will be required to further test for in planta IAA production by P. arabidopsidicola and understand its potential functions.
Live P. arabidopsidicola pre-treatment lead to significantly greater immunity against Botrytis than pre-treatment with dead (autoclaved) cells, supporting that live P. arabidopsidicola cells are required for the full priming effect. The ability of dead cells to illicit these responses supports the involvement of PAMPs, which are passive and do not require active metabolism for their activity. However, there are alternative explanations that may account for the higher priming by live cells. The persistence of living yeast cells might provide prolonged stimuli and thus stronger priming effects. Additionally, small secreted proteins/peptides from live P. arabidopsidicola, which contains large number of effector candidates (Wang et al., 2019), might be involved in this stronger priming. However, further studies will be required to resolve this question.
Multiple studies have previously documented enhanced Botrytis resistance in Arabidopsis pre-treated with either autoclaved or live yeast cells. Antagonistic phyllosphere microbes, including yeasts (Liu et al., 2013; Eitzen et al., 2021), can effectively suppress diseases that are caused by pathogenic fungi through a variety of known mechanisms (Legein et al., 2020; Chaudhry et al., 2021), such as direct inhibition effects, and indirect effects such as nutrient and space competition or complex microbiome interactions. In the current study, there was no obvious Botrytis growth inhibition by P. arabidopsidicola in an in vitro co-cultivation experiment. In comparison, the inhibitory effect of a Paenibacillus species known to produce antifungal compounds active against Botrytis was clearly observed. This suggests the enhanced immunity against Botrytis induced by P. arabidopsidicola was due to defense priming, rather than direct antagonistic effects. However, in vitro studies do not reflect the complex environment of the leaf, where nutrient and space competition by live P. arabidopsidicola and Botrytis might occur. Additionally, the leaf microbiome may be modified by the presence of P. arabidopsidicola. This activity could be mediated via activation of the host plant immune system or via indirect microbe–microbe interactions (Chaudhry et al., 2021). Notably, Brachi et al. (2021) reported an Arabidopsis-associated Protomyces strain that acts as a phyllosphere microbiome reorganizing hub species and was among the most abundant fungi in the Arabidopsis phyllosphere. In light of these alternate possibilities, complex indirect effects cannot be fully excluded at this time.
Protomyces and Taphrina are sister genera and share similar lifecycles and virulence strategies (Reddy and Kramer, 1975; Fonseca and Rodrigues, 2011; Kurtzman, 2011; Wang et al., 2021; Christita et al., 2022a,b). Both of these genera invade hosts in their hyphal form, which contains chitin (Valadon et al., 1962; Fonseca and Rodrigues, 2011; Kurtzman, 2011). Indeed, all fungi have chitin in their cell walls, and it is especially abundant and a major PAMP in filamentous (hyphal) fungal pathogens (Latge, 2007; Doehlemann and Hemetsberger, 2013). However, Schizosaccharomyces pombe, Pneumocystis species, and some Taphrina species, which all reside in the Taphrinomycotina together with Protomyces species, have reduced chitin content in many structures (Matsuo et al., 2004; Christita et al., 2021). Accordingly, reduced complement of chitin biosynthesis genes was also seen in a Taphrina strain that is pathogenic on Arabidopsis (Christita et al., 2021). Genomes of pathogenic fungi contain LysM domain effectors, which block chitin-induced immune responses (Kombrink and Thomma, 2013). Remarkably, many human pathogenic yeasts, whose chitin cell wall layers are buried deep beneath layers of beta-glucan and mannoproteins (Perez-Garcia et al., 2011) lack LysM effectors (Kombrink and Thomma, 2013). The sequencing of the genomes of P. arabidopsidicola and six other Protomyces species revealed an absence of the expected candidate effector proteins bearing the LysM domain (Wang et al., 2019). This suggests that they either have reduced chitin, or have their chitin layers sequestered by other structures. Effector candidates bearing the legume (L)-type lectin domain were present in all but one Protomyces genome (Wang et al., 2019). The L-lectin domain is known to mediate the binding of mannose linkages (Itin et al., 1996; Satoh et al., 2006, 2007). The cell wall composition has been addressed in only one Protomyces species. P. inundatus has a cell wall composed of glucan and mannose, similar to other yeasts (Valadon et al., 1962). Taken together, these results support the need for further examination of mannose-linkages, or other yeast MAMPs, in the interactions of Protomyces with their plant hosts. Screens aimed at discovering yeast-perceiving PRRs are underway.
Conclusion
We present evidence that Protomyces arabidopsidicola strain C29 was not pathogenic on Arabidopsis, the premiere genetic model plant and the host from which it was originally isolated. However, it was able to persist in the Arabidopsis phyllosphere and activate immune signaling pathways to induce immunity against subsequent challenge with the generalist necrotrophic fungal pathogen, Botrytis. These finding contribute toward establishing the P. arabidopsidicola–Arabidopsis interaction as a model system, which can facilitate future genetic studies into the biology of Protomyces species, the evolution of fungal virulence, host interactions with phyllosphere yeasts, and plant immunity against yeasts.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
KW and AA performed the experiments. KO provided supervision and project administration. KW and KO wrote the manuscript. KW, AA, and KO were involved in designing experiments, analyzing data, and interpreting data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the following grants: Academy of Finland Fellowship (decisions no. 251397, 256073, and 283254) to KO and the Academy of Finland Center of Excellence in Primary Producers 2014-2019 (decisions #271832 and 307335). Lecturer Betty Väänänen grant from the Kuopio Naturalists’ Society (KLYY) fund and a grant from the August Johannes and Aino Tiura Agricultural Research Foundation to AA. KW was a member of the University of Helsinki Doctoral Program in Plant Science (DPPS) and AA has a PhD fellowship from the Microbiology and Biotechnology Doctoral Program.
Acknowledgments
We thank Tuomas Puukko, Airi Lamminmäki, and Leena Grönholm, for excellent technical support and Katariina Vuorinen for assistance with Botrytis infection assays, Mikeal Broché and Tiina Blomster for advice of qPCR, Julia Krasensky-Wrzaczek for instruction on MAPK work, Timo Sipilä, and Ansa Palojärvi (Joint ‘SUCCESS’ project) for the control Paenibacillus strain with antifungal activity. We gratefully acknowledged the USDA ARS culture collection for providing the Protomyces inouyei strain. KW was a member of the University of Helsinki Doctoral Program in Plant Science (DPPS) and AA has a PhD fellowship from the Microbiology and Biotechnology Doctoral Program. We thank Dominik Begerow, Daniel Croll, and Mikael Brosché for their critical comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.956018/full#supplementary-material
Footnotes
1. ^https://nrrl.ncaur.usda.gov/
4. ^https://www.leica-microsystems.com
5. ^https://www.bertin-instruments.com
6. ^https://www.leica-microsystems.com
References
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S.-T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Almario, J., Mahmudi, M., Kroll, S., Agler, M., Placzek, A., Mari, A., et al. (2022). The leaf microbiota of Arabidopsis displays reproducible dynamics And patterns Throughout The growing season. mBio 13, e02825–e02821. doi: 10.1128/mbio.02825-21
Balmer, A., Pastor, V., Gamir, J., Flors, V., and Mauch-Mani, B. (2015). The ‘prime-ome’: towards a holistic approach to priming. Trend Plant Sci. 20, 443–452. doi: 10.1016/j.tplants.2015.04.002
Basse, C., Bock, K., and Boller, T. (1992). Elicitors and suppressors of the defense response in tomato cells. Purification and characterization of glycopeptide elicitors and glycan suppressors generated by enzymatic cleavage of yeast invertase. J. Biol. Chem. 267, 10258–10265. doi: 10.1016/S0021-9258(19)50012-6
Beckers, G. J., Jaskiewicz, M., Liu, Y., Underwood, W. R., He, S. Y., Zhang, S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. doi: 10.1105/tpc.108.062158
Belkhadir, Y., Yang, L., Hetzel, J., Dangl, J. L., and Chory, J. (2014). The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem. Sci. 39, 447–456. doi: 10.1016/j.tibs.2014.06.006
Bergauer, P., Fonteyne, P.-A., Nolard, N., Schinner, F., and Margesin, R. (2005). Biodegradation of phenol and phenol-related compounds by psychrophilic and cold-tolerant alpine yeasts. Chemosphere 59, 909–918. doi: 10.1016/j.chemosphere.2004.11.011
Bigeard, J., and Hirt, H. (2018). Nuclear signaling of plant MAPKs. Front. Plant Sci. 9:469. doi: 10.3389/fpls.2018.00469
Boutrot, F., and Zipfel, C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Brachi, B., Filiault, D., Whitehurst, H., Darme, P., Le Gars, P., Le Mentec, M., et al. (2021). Plant genetic effects on microbial hubs impact fitness across field trials. bioRxiv [Epub ahead of preprint]. doi: 10.1101/181198
Brader, G., Djamei, A., Teige, M., Palva, E. T., and Hirt, H. (2007). The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol. Plant-Microbe Interact. 20, 589–596. doi: 10.1094/MPMI-20-5-0589
Buxdorf, K., Rahat, I., Gafni, A., and Levy, M. (2013a). The epiphytic fungus Pseudozyma aphidis induces jasmonic acid-and salicylic acid/Nonexpressor of PR1-independent local and systemic resistance. Plant Physiol. 161, 2014–2022. doi: 10.1104/pp.112.212969
Buxdorf, K., Rahat, I., and Levy, M. (2013b). Pseudozyma aphidis induces ethylene-independent resistance in plants. Plant Signal. Behav. 8:e26273. doi: 10.4161/psb.26273
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3:e03766. doi: 10.7554/eLife.03766
Chan, Z., Qin, G., Xu, X., Li, B., and Tian, S. (2007). Proteome approach to characterize proteins induced by antagonist yeast and salicylic acid in peach fruit. J. Proteome Res. 6, 1677–1688. doi: 10.1021/pr060483r
Chanclud, E., and Morel, J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. doi: 10.1111/mpp.12393
Chaudhry, V., Runge, P., Sengupta, P., Doehlemann, G., Parker, J. E., and Kemen, E. (2021). Shaping the leaf microbiota: plant–microbe–microbe interactions. J. Ex. Bot 72, 36–56. doi: 10.1093/jxb/eraa417
Christita, M., Auzane, A., and Overmyer, K. (2022a). “Witches’ broom disease of birch,” in Forest Microbiology Volume 3: Tree Diseases and Pests. eds. F. O. Asiegbu and A. Kovalchuk (Academic Press: United Kingdom).
Christita, M., Sipilä, T. P., Auzane, A., and Overmyer, K. (2022b). Distinct Taphrina strains from the phyllosphere of birch exhibiting a range of witches’ broom disease symptoms. Environ. Microbiol. doi: 10.1111/1462-2920.16037 (in press).
Christita, M., Auzane, A., Wang, K., Sipilä, T., Auvinen, P., Paulin, L., et al. (2021). A Taphrina strain infecting the model plant Arabidopsis thaliana. bioRxiv [Epub ahead of preprint], doi: 10.1101/2021.09.16.460675
Conrath, U., Beckers, G. J., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071. doi: 10.1094/MPMI-19-1062
Conrath, U., Pieterse, C. M., and Mauch-Mani, B. (2015). Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. doi: 10.1016/S1360-1385(02)02244-6
Cui, F., Brosché, M., Lehtonen, M. T., Amiryousefi, A., Xu, E., Punkkinen, M., et al. (2016). Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol. Plant 9, 926–938. doi: 10.1016/j.molp.2016.04.001
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
De Jonge, R., Van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., Bours, R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. doi: 10.1126/science.1190859
Doehlemann, G., and Hemetsberger, C. (2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198, 1001–1016. doi: 10.1111/nph.12277
Droby, S., Vinokur, V., Weiss, B., Cohen, L., Daus, A., Goldschmidt, E., et al. (2002). Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila. Phytopathology 92, 393–399. doi: 10.1094/PHYTO.2002.92.4.393
Eitzen, K., Sengupta, P., Kroll, S., Kemen, E., and Doehlemann, G. (2021). A fungal member of the Arabidopsis thaliana phyllosphere antagonizes Albugo laibachii via a GH25 lysozyme. elife 10:e65306. doi: 10.7554/eLife.65306
El Ghaouth, A., Wilson, C. L., and Wisniewski, M. (2003). Control of postharvest decay of apple fruit with Candida saitoana and induction of defense responses. Phytopathology 93, 344–348. doi: 10.1094/PHYTO.2003.93.3.344
Faulkner, C., Petutschnig, E., Benitez-Alfonso, Y., Beck, M., Robatzek, S., Lipka, V., et al. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. U. S. A. 110, 9166–9170. doi: 10.1073/pnas.1203458110
Felix, G., Regenass, M., and Boller, T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316. doi: 10.1046/j.1365-313X.1993.04020307.x
Ferreira-Saab, M., Formey, D., Torres, M., Aragon, W., Padilla, E. A., Tromas, A., et al. (2018). Compounds released by the biocontrol yeast Hanseniaspora opuntiae protect plants Against Corynespora cassiicola and Botrytis cinerea. Front. Microbiology 9:1596. doi: 10.3389/fmicb.2018.01596
Fonseca, Á., and Rodrigues, M. G. (2011). “Chapter 73—Taphrina fries (1832),” in The Yeasts (Fifth Edition). eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout (London: Elsevier), 823–858.
Galletti, R., Ferrari, S., and De Lorenzo, G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814. doi: 10.1104/pp.111.174003
Hahn, M. G., and Albersheim, P. (1978). Host-pathogen interactions: XIV. Isolation and partial characterization of an elicitor from yeast extract. Plant Physiol. 62, 107–111. doi: 10.1104/pp.62.1.107
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19. doi: 10.1186/gb-2007-8-2-r19
Hossain, M. M., Sultana, F., Kubota, M., Koyama, H., and Hyakumachi, M. (2007). The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol. 48, 1724–1736. doi: 10.1093/pcp/pcm144
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Itin, C., Roche, A. C., Monsigny, M., and Hauri, H. P. (1996). ERGIC-53 is a functional mannose-selective and calcium-dependent human homologue of leguminous lectins. Mol. Biol. Cell 7, 483–493. doi: 10.1091/mbc.7.3.483
Jarvis, W. R. (1994). Latent infections in the Preharvest and postharvest environment. HortScience 29, 749–751. doi: 10.21273/Hortsci.29.7.749
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jouault, T., Sarazin, A., Martinez-Esparza, M., Fradin, C., Sendid, B., and Poulain, D. (2009). Host responses to a versatile commensal: PAMPs and PRRs interplay leading to tolerance or infection by Candida albicans. Cell. Microbiol. 11, 1007–1015. doi: 10.1111/j.1462-5822.2009.01318.x
Khan, M. R., and Parveen, G. (2018). Supplementing biocontrol agents with botanicals improved growth and yield of coriander (Coriandrum sativum L.) infected with Protomyces macrosporus Unger. Curr. Plant Biol. 15, 44–50. doi: 10.1016/j.cpb.2018.10.005
Khokon, M. A., Hossain, M. A., Munemasa, S., Uraji, M., Nakamura, Y., Mori, I. C., et al. (2010). Yeast elicitor-induced stomatal closure and peroxidase-mediated ROS production in Arabidopsis. Plant Cell Physiol. 51, 1915–1921. doi: 10.1093/pcp/pcq145
Kombrink, A., and Thomma, B. P. (2013). LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 9:e1003769. doi: 10.1371/journal.ppat.1003769
Kurtzman, C. P. (2011). “Protomyces Unger (1833),” in The Yeasts: a Taxonomic Study. 5th Edn. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout (Amsterdam: Elsevier), 725–731.
Latge, J. P. (2007). The cell wall: a carbohydrate Armour for the fungal cell. Mol. Microbiol. 66, 279–290. doi: 10.1111/j.1365-2958.2007.05872.x
Legein, M., Smets, W., Vandenheuvel, D., Eilers, T., Muyshondt, B., Prinsen, E., et al. (2020). Modes of action of microbial biocontrol in the phyllosphere. Front. Microbiol. 11:1619. doi: 10.3389/fmicb.2020.01619
Lind, A. L., and Pollard, K. S. (2021). Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome 9, 1–18. doi: 10.1186/s40168-021-01015-y
Liu, J., Sui, Y., Wisniewski, M., Droby, S., and Liu, Y. (2013). Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 167, 153–160. doi: 10.1016/j.ijfoodmicro.2013.09.004
Macarisin, D., Droby, S., Bauchan, G., and Wisniewski, M. (2010). Superoxide anion and hydrogen peroxide in the yeast antagonist–fruit interaction: a new role for reactive oxygen species in postharvest biocontrol? Postharvest Biol. Technol. 58, 194–202. doi: 10.1016/j.postharvbio.2010.07.008
Malhotra, S., Kakani, R., Sharma, Y., Saxena, S., and Vashishtha, B. (2016). Comparative superiority of coriander variety NRCSS ACr-1 for yield and stem gall disease tolerance. Indian J. Hortic. 73, 453–455. doi: 10.5958/0974-0112.2016.00097.9
Matsuo, Y., Tanaka, K., Nakagawa, T., Matsuda, H., and Kawamukai, M. (2004). Genetic analysis of chs1+ and chs2+ encoding chitin synthases from Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68, 1489–1499. doi: 10.1271/bbb.68.1489
Mélida, H., Sopeña-Torres, S., Bacete, L., Garrido-Arandia, M., Jordá, L., López, G., et al. (2018). Non-branched β-1, 3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 93, 34–49. doi: 10.1111/tpj.13755
Minami, T., Tanaka, T., Takasaki, S., Kawamura, K., and Hiratsuka, K. (2011). In vivo bioluminescence monitoring of defense gene expression in response to treatment with yeast cell wall extract. Plant Biotechnol. 28, 481–484. doi: 10.5511/plantbiotechnology.11.1020a
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Mukherjee, A., Verma, J. P., Gaurav, A. K., Chouhan, G. K., Patel, J. S., and Hesham, A. E.-L. (2020). Yeast a potential bio-agent: future for plant growth and postharvest disease management for sustainable agriculture. Appl. Microbiol. Biotech. 104, 1497–1510. doi: 10.1007/s00253-019-10321-3
Nafady, N. A., Hashem, M., Hassan, E. A., Ahmed, H. A., and Alamri, S. A. (2019). The combined effect of arbuscular mycorrhizae and plant-growth-promoting yeast improves sunflower defense against Macrophomina phaseolina diseases. Biol. Control 138:104049. doi: 10.1016/j.biocontrol.2019.104049
Narusaka, M., Minami, T., Iwabuchi, C., Hamasaki, T., Takasaki, S., Kawamura, K., et al. (2015). Yeast cell wall extract induces disease resistance against bacterial and fungal pathogens in Arabidopsis thaliana and brassica crop. PLoS One 10:e0115864. doi: 10.1371/journal.pone.0115864
Nassar, A. H., El-Tarabily, K. A., and Sivasithamparam, K. (2005). Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol. Fertil. Soils 42, 97–108. doi: 10.1007/s00374-005-0008-y
Nurnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. doi: 10.1016/s1369-5266(02)00265-0
Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Nutaratat, P., Srisuk, N., Arunrattiyakorn, P., and Limtong, S. (2014). Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol. 118, 683–694. doi: 10.1016/j.funbio.2014.04.010
Patin, E. C., Thompson, A., and Orr, S. J. (2019). Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 89, 24–33. doi: 10.1016/j.semcdb.2018.03.003
Patkar, R. N., and Naqvi, N. I. (2017). Fungal manipulation of hormone-regulated plant defense. PLoS Pathog. 13:e1006334. doi: 10.1371/journal.ppat.1006334
Pavgi, M., and Mukhopadhyay, A. (1972). Development of coriander fruit infected by Protomyces macrosporus Unger. Cytologia 37, 619–627. doi: 10.1508/cytologia.37.619
Perez-Garcia, L., Diaz-Jimenez, D., Lopez-Esparza, A., and Mora-Montes, H. (2011). Role of cell wall polysaccharides during recognition of Candida albicans by the innate immune system. Glycobiology 1:2. doi: 10.4172/2168-958X.1000102
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopath. 52, 347–375. doi: 10.1146/annurev-phyto-082712–102340
Prior, R., Mittelbach, M., and Begerow, D. (2017). Impact of three different fungicides on fungal epi-and endophytic communities of common bean (Phaseolus vulgaris) and broad bean (Vicia faba). J. Environ. Sci. Health B Pestic. Food Contam. Agric. Wastes 52, 376–386. doi: 10.1080/03601234.2017.1292093
Pritchard, L., and Birch, P. R. J. (2014). The zigzag model of plant-microbe interactions: is it time to move on? Mol. Plant Pathol. 15, 865–870. doi: 10.1111/mpp.12210
Raacke, I. C., von Rad, U., Mueller, M. J., and Berger, S. (2006). Yeast increases resistance in Arabidopsis against pseudomonas syringae and Botrytis cinerea by salicylic acid-dependent as well as-independent mechanisms. Mol. Plant Microbe Interact. 19, 1138–1146. doi: 10.1094/MPMI-19-1138
Regalado, J., Lundberg, D. S., Deusch, O., Kersten, S., Karasov, T., Poersch, K., et al. (2020). Combining whole-genome shotgun sequencing and rRNA gene amplicon analyses to improve detection of microbe–microbe interaction networks in plant leaves. ISME J. 14, 2116–2130. doi: 10.1038/s41396-020-0665-8
Saijo, S., Ikeda, S., Yamabe, K., Kakuta, S., Ishigame, H., Akitsu, A., et al. (2010). Dectin-2 recognition of α-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 32, 681–691. doi: 10.1016/j.immuni.2010.05.001
Saikkonen, K., Faeth, S. H., Helander, M., and Sullivan, T. J. (1998). Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 29, 319–343. doi: 10.1146/annurev.ecolsys.29.1.319
Satoh, T., Cowieson, N. P., Hakamata, W., Ideo, H., Fukushima, K., Kurihara, M., et al. (2007). Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36. J. Biol. Chem. 282, 28246–28255. doi: 10.1074/jbc.M703064200
Satoh, T., Sato, K., Kanoh, A., Yamashita, K., Yamada, Y., Igarashi, N., et al. (2006). Structures of the carbohydrate recognition domain of Ca2+−independent cargo receptors Emp46p and Emp47p. J. Biol. Chem. 281, 10410–10419. doi: 10.1074/jbc.M512258200
Slippers, B., and Wingfield, M. J. (2007). Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol. Rev. 21, 90–106. doi: 10.1016/j.fbr.2007.06.002
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574–6976.2007.00072.x
Streletskii, R. A., Kachalkin, A. V., Glushakova, A. M., Demin, V. V., and Chernov, I. Y. (2016). Quantitative determination of indole-3-acetic acid in yeasts using high performance liquid chromatography-tandem mass spectrometry. Microbiology 85, 727–736. doi: 10.1134/S0026261716060187
Streletskii, R. A., Kachalkin, A. V., Glushakova, A. M., Yurkov, A. M., and Demin, V. V. (2019). Yeasts producing zeatin. PeerJ. 7:e6474. doi: 10.7717/peerj.6474
Sun, C., Fu, D., Jin, L. F., Chen, M. Y., Zheng, X. D., and Yu, T. (2018a). Chitin isolated from yeast cell wall induces the resistance of tomato fruit to Botrytis cinerea. Carbohydr. Polym. 199, 341–352. doi: 10.1016/j.carbpol.2018.07.045
Sun, C., Fu, D., Lu, H. P., Zhang, J. H., Zheng, X. D., and Yu, T. (2018b). Autoclaved yeast enhances the resistance against Penicillium expansum in postharvest pear fruit and its possible mechanisms of action. Biol. Control 119, 51–58. doi: 10.1016/j.biocontrol.2018.01.010
Sun, X., Kosman, E., Sharon, O., Ezrati, S., and Sharon, A. (2020). Significant host-and environment-dependent differentiation among highly sporadic fungal endophyte communities in cereal crops-related wild grasses. Environ. Microbiol. 22, 3357–3374. doi: 10.1111/1462-2920.15107
Valadon, L. R. G., Manners, J. G., and Myers, A. (1962). Studies on the life-history and taxonomic position of Protomyces inundatus Dangeard. Trans. Br. Mycol. Soc. 45, 573–575. doi: 10.1016/S0007-1536(62)80018-7
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Vuorinen, K., Zamora, O., Vaahtera, L., Overmyer, K., and Brosche, M. (2021). Dissecting contrasts in cell death, hormone, and defense signaling in response to Botrytis cinerea and reactive oxygen species. Mol. Plant-Microbe Interact. 34, 75–87. doi: 10.1094/MPMI-07-20-0202-R
Wan, J., Zhang, X.-C., Neece, D., Ramonell, K. M., Clough, S., Kim, S.-y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Wang, K., Sipilä, T. P., and Overmyer, K. (2016). The isolation and characterization of resident yeasts from the phylloplane of Arabidopsis thaliana. Sci. Rep. 6:39403. doi: 10.1038/srep39403
Wang, K., Sipilä, T., and Overmyer, K. (2021). A novel Arabidopsis phyllosphere resident Protomyces species and a re-examination of genus Protomyces based on genome sequence data. IMA Fungus 12, 1–18. doi: 10.1186/s43008-021-00054-2
Wang, K., Sipilä, T., Rajaraman, S., Safronov, O., Laine, P., Auzane, A., et al. (2019). A novel phyllosphere resident Protomyces species that interacts with the Arabidopsis immune system. bioRxiv [Epub ahead of preprint]: doi: 10.1101/594028
Xu, B., Zhang, H., Chen, K., Xu, Q., Yao, Y., and Gao, H. (2013). Biocontrol of postharvest Rhizopus decay of peaches with Pichia caribbica. Curr. Microbiol. 67, 255–261. doi: 10.1007/s00284-013-0359-9
Xue, D.-X., Li, C.-L., Xie, Z.-P., and Staehelin, C. (2019). LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Ex. Bot. 70, 5507–5516. doi: 10.1093/jxb/erz313
Yaguchi, T., Kinami, T., Ishida, T., Yasuhara, T., Takahashi, K., and Matsuura, H. (2017). Induction of plant disease resistance upon treatment with yeast cell wall extract. Biosci. Biotechnol. Biochem. 81, 2071–2078. doi: 10.1080/09168451.2017.1379351
Yamada, K., Yamaguchi, K., Yoshimura, S., Terauchi, A., and Kawasaki, T. (2017). Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 58, 993–1002. doi: 10.1093/pcp/pcx042
Ye, W., Muroyama, D., Munemasa, S., Nakamura, Y., Mori, I. C., and Murata, Y. (2013). Calcium-dependent protein kinase CPK6 positively functions in induction by yeast elicitor of stomatal closure and inhibition by yeast elicitor of light-induced stomatal opening in Arabidopsis. Plant Physiol. 163, 591–599. doi: 10.1104/pp.113.224055
Keywords: beneficial microbes, Botrytis, defenses against pathogens, MAPK, plant–fungal interactions, Protomycetaceae, Taphrinomycetes, yeast-like fungi
Citation: Wang K, Auzane A and Overmyer K (2022) The immunity priming effect of the Arabidopsis phyllosphere resident yeast Protomyces arabidopsidicola strain C29. Front. Microbiol. 13:956018. doi: 10.3389/fmicb.2022.956018
Edited by:
Samantha Chandranath Karunarathna, Qujing Normal University, ChinaReviewed by:
Nimali Indeewari De Silva, Chiang Mai University, ThailandShahzad Munir, Yunnan Agricultural University, China
Milan C. Samarakoon, Chiang Mai University, Thailand
Copyright © 2022 Wang, Auzane and Overmyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kirk Overmyer, a2lyay5vdmVybXllckBoZWxzaW5raS5maQ==; Kai Wang, d2FuZ2thaTEwNzJAMTYzLmNvbQ==; a2FpLndhbmdAaGVsc2lua2kuZmk=
†Present address: Kai Wang,College of Forestry, Fujian Agriculture and Forestry University,Fuzhou, China
 Kai Wang
Kai Wang Agate Auzane
Agate Auzane Kirk Overmyer
Kirk Overmyer