- 1College of Chemistry and Environmental Engineering, Shenzhen University, Shenzhen, China
- 2Department of Gastroenterology and Hepatology, Shenzhen University General Hospital, Shenzhen, China
Microbial bromate reduction plays an important role in remediating bromate-contaminated waters as well as biogeochemical cycling of bromine. However, little is known about the molecular mechanism of microbial bromate reduction so far. Since the model strain Shewanella oneidensis MR-1 is capable of reducing a variety of oxyanions such as iodate, which has a high similarity to bromate, we hypothesize that S. oneidensis MR-1 can reduce bromate. Here, we conducted an experiment to investigate whether S. oneidensis MR-1 can reduce bromate, and report bromate reduction mediated by a dimethylsulfoxide reductase encoded with dmsA. S. oneidensis MR-1 is not a bromate-respiring bacterium but can reduce bromate to bromide under microaerobic conditions. When exposed to 0.15, 0.2, 0.25, 0.5, and 1 mM bromate, S. oneidensis MR-1 reduced bromate by around 100, 75, 64, 48, and 23%, respectively, within 12 h. In vivo evidence from gene deletion mutants and complemented strains of S. oneidensis MR-1 indicates that MtrB, MtrC, CymA, GspD, and DmsA are involved in bromate reduction, but not NapA, FccA, or SYE4. Based on our results as well as previous findings, a proposed molecular mechanism for bromate reduction is presented in this study. Moreover, a genomic survey indicates that 9 of the other 56 reported Shewanella species encode proteins highly homologous to CymA, GspD, and DmsA of S. oneidensis MR-1 by sequence alignment. The results of this study contribute to understanding a pathway for microbial bromate reduction.
Introduction
Bromate (), an oxyanion of bromine, has been classified by the World Health Organization as a possible human carcinogen (WHO, 2011). Bromate contamination has been detected in various environments (e.g., drinking water, wastewater, surface water, and groundwater) posing human health risks (Butler et al., 2005; Jahan et al., 2021). Microbial bromate reduction is a promising method for bioremediating bromate-contaminated waters, and has attracted extensive attention (Zhong et al., 2018; Lv et al., 2019; Liu et al., 2020; Jahan et al., 2021).
To date, there are only a few reports on bromate reduction by isolates (e.g., Rhodococcus sp. Br-6, Dechloromonas sp. PC1, Klebsiella variicola Glu3, and Shewanella decolorationis Ni1-3) (Davidson et al., 2011; Tamai et al., 2016; Jahan et al., 2021; Wang D. et al., 2022; Wang Y. et al., 2022). Rhodococcus sp. Br-6 reduced bromate to bromide under transition conditions (from aerobic to anaerobic conditions), and that reaction was significantly dependent on both ferric iron and a redox mediator, 2,6-dichloroindophenol (Tamai et al., 2016). In addition, terminal reductases purified from bacteria, such as (per)chlorate reductase (PcrA) (Kengen et al., 1999), chlorate reductase (ClrA) (Thorell et al., 2003), nitrate reductase (NarG) (Morpeth and Boxer, 1985; Maria Martinez-Espinosa et al., 2015), selenate reductase (SerA) (Ridley et al., 2006), and trimethylamine-N-oxide reductase (TorA) (Shimokawa and Ishimoto, 1979), have only shown bromate-reducing activity in vitro, but whether they can mediate bromate reduction in vivo remains unclear. So far, little is known about microbial bromate reduction because of limited number of available isolates and paucity of information for key genes involved in that reaction (Jahan et al., 2021). Moreover, a variety of isolates can reduce oxidative oxyanions [i.e., nitrate and Cr(VI)] in the presence of oxygen (Pradhan et al., 2017; Huang et al., 2020; Yang et al., 2020; Karimi-Maleh et al., 2021; Zhang et al., 2021), but the (micro)aerobic reduction of bromate by pure cultures is poorly understood at present.
Shewanella species are facultative anaerobic bacteria well-known for their remarkable respiratory diversity (Hau and Gralnick, 2007; Fredrickson et al., 2008; Lemaire et al., 2020). The Shewanella genus currently includes around 70 species that are widely distributed in aquatic environments such as freshwater and marine sediments around the world (Lemaire et al., 2020). Knowledge of the respiratory diversity of Shewanella species is mainly derived from the model strain Shewanella oneidensis MR-1, which can reduce diverse oxyanions including iodate, sulfite, nitrate, U(VI), Cr(VI), and selenite (Shirodkar et al., 2011; Li et al., 2014; Beblawy et al., 2018; Lemaire et al., 2020; Vettese et al., 2020; Guo et al., 2022; Shin et al., 2022). Bromate is a halogen oxyanion with a molecular structure and chemical properties similar to iodate. Additionally, a recent study shows that S. decolorationis Ni1-3 can perform bromate reduction, and that its genome shares an average nucleotide identity (ANI) of 85% with S. oneidensis MR-1 (Wang Y. et al., 2022). Taken together, we anticipate that the model strain S. oneidensis MR-1 is able to reduce bromate. Based on this hypothesis, we intend to address what enzymes mediate bromate reduction by S. oneidensis MR-1.
Previous studies suggest that nitrate reductase might be responsible for bromate reduction, and FccA (periplasmic fumarate reductase) was shown to mediate selenite reduction by S. oneidensis MR-1 (Hijnen et al., 1995, 1999; Li et al., 2014). Hence, we hypothesize that NapA (periplasmic nitrate reductase) or FccA may contribute to bromate reduction by S. oneidensis MR-1. As a powerful oxidant, bromate can induce oxidative stress in cells (Ahmad et al., 2015). SYE4 sequences of S. oneidensis MR-1 and the NemA [Cr(VI) reductase of Escherichia coli] share an identity of 42%, both belonging to the old yellow enzyme (a NAPDH oxidoreductase) family, and SYE4 has been reported to be induced under oxidative stress (Brigé et al., 2006; Thatoi et al., 2014). Moreover, recent evidence suggests that sye4 of S. decolorationis Ni1-3 was highly induced in response to bromate (Wang Y. et al., 2022). Therefore, we also hypothesize that SYE4 may contribute to bromate reduction by S. oneidensis MR-1.
This study aims (1) to test the hypothesis that S. oneidensis MR-1 can reduce bromate, (2) to explore the bromate reductase of S. oneidensis MR-1, and (3) to identify whether all Shewanella species possess key proteins related to bromate reduction. The experimental strategy consists of the following steps: (1) batch cultivation under both anaerobic and microaerobic conditions to test the bromate-reducing capacity of S. oneidensis MR-1, (2) construction of in-frame deletion mutants and complemented strains and subsequent measurement of bromate-reducing capacity, and (3) identification of a homologous protein required for bromate reduction among the other 56 Shewanella species.
Materials and methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. S. oneidensis and E. coli strains were routinely cultured aerobically at 30 and 37°C in lysogeny broth (LB) (10 g/L NaCl, 5 g/L yeast extract, and 10 g/L tryptone). When required, the LB medium was supplemented with chemicals at the following concentrations: 2,6-diaminopimelicacid 50 μg/ml, gentamycin 15 μg/ml, and kanamycin 50 μg/ml. Bromate reduction by S. oneidensis strains was performed under both anaerobic and microaerobic conditions in a bromate reduction (BR) medium (pH = 7.2), which contained 2.24 g sodium lactate (20 mM), 1.2 g Na2HPO4, 0.8 g KH2PO4, 1 g (NH4)2HPO4, 0.1 g yeast extract, 0.1 g tryptone, 10 ml vitamin solution (Wolin et al., 1963), and 1 ml trace element solution per liter (Supplementary Table 1).
Bromate reduction
The bromate reduction by S. oneidensis strains was conducted at 30°C. S. oneidensis MR-1 was incubated aerobically in the LB medium for ∼12 h, and cell pellets were collected and washed with phosphate buffered solution (PBS) and transferred into a serum bottle that contained 100 ml BR medium, and the bromate was added to the culture before aeration. Oxygen was purged with high-purity nitrogen gas for 10 min, and the culture was incubated anaerobically and shaken at 150 rpm. For microaerobic bromate reduction, S. oneidensis strains were incubated aerobically in the LB medium for ∼12 h, and cell pellets were collected and washed once with PBS and transferred into a 250-ml Erlenmeyer flask containing 100 ml BR medium at an initial OD600 (optical density at 600 nm) of 0.12. The culture was added with bromate and incubated and shaken at 150 rpm.
Mutagenesis and complementation
In-frame markerless deletion strains were constructed by seamless cloning and SacB-based counterselection as described by Jin et al. (2013). Briefly, two fragments (500–1,000 bp in length) flanking the target gene and linearized pHGM01 were recombined using a Hi-Fusion Cloning Mix V2 kit (Monad, China) according to the manufacturer’s instructions. The resulting plasmids were maintained in Escherichia coli WM3064 and subsequently transferred into S. oneidensis strains by conjugation. Verified transconjugants were grown in LB medium without NaCl and subsequently plated on LB agar plates supplemented with 10% sucrose. Sucrose-resistant and gentamicin-sensitive colonies were screened by PCR for the intended deletion. Deletion mutants were then verified by Sanger sequencing. For complementation of genes, a fragment containing the gene of S. oneidensis MR-1 wild type was generated by PCR and cloned into pHGE. After verification by Sanger sequencing, the resultant plasmids were transferred into relevant strains by conjugation. The primers used for mutagenesis and complementation are listed in Supplementary Table 2.
Determination of biomass and chemical assays
The OD600 of the cultures was detected using a Synergy HTX multi-mode plate reader (BioTek, United States). For total protein quantification, cell pellets were collected by centrifugation and resuspended in 0.85% (w/v) NaCl solution, and cells were disrupted by sonication at 200 W for 5 min. Protein concentrations were then determined using the method described by Bradford (1976). Bromate and bromide concentrations were determined by the 883 Basic IC plus ion chromatograph (Metrohm, Switzerland) using a Metrosep A Supp 7-250/4.0 column and an eluent consisting of 3.6 mmol/L Na2CO3 with 2% (v/v) acetonitrile at 0.7 ml/min. The dissolved oxygen (DO) concentration was measured using a portable JPB-607A DO meter (REX, China). The pH value of the cultures was determined using a LAQUAtwin pH-11 meter (Horiba, Japan).
Sequence alignment and phylogenetic analysis
Reference genomes of 57 species of Shewanella genus (S. oneidensis MR-1 included) were downloaded from NCBI database (Supplementary Table 3). In this study, a database was constructed using the CymA, GspD, and DmsA protein sequences of S. oneidensis MR-1. Based on BLAST (Camacho et al., 2009) alignment, we identified whether the other 56 Shewanella species contain homologous protein sequences of CymA, GspD, and DmsA. Positive alignments were improved based on the criteria (identity > 70%, query coverage > 80%, and e-value < 10–5) described by Assis et al. (2017). The MEGA X software (Kumar et al., 2018) was used to align the identified homolog sequences to DmsA with those of other DMSO reductases and construct a phylogenetic tree based on the neighbor-joining method (Naruya and Masatoshi, 1987). The phylogenetic tree was visualized using the iTOL software (Letunic and Bork, 2021).
Other analysis
A Mann–Whitney U test was conducted using the GraphPad Prism software for pairwise comparisons of groups.
Results
Bromate reduction by Shewanella oneidensis MR-1
To test whether S. oneidensis MR-1 could be a bromate-respiring bacterium, a set of batch cultivation was performed with bromate as the sole electron acceptor under anaerobic conditions. It was found that only about 7% (24 h) of bromate was reduced with the dosage of 1 mM bromate, and that almost no bromate reduction occurred within 24 h when the dosage of 2 mM bromate was used (Supplementary Figure 1A). Moreover, the biomass of S. oneidensis MR-1 did not increase in the presence of bromate under anaerobic conditions (Supplementary Figure 1B).
Another set of batch cultivation was carried out to test whether S. oneidensis MR-1 could reduce bromate under microaerobic conditions. Obvious bromate reduction was observed within 12 h under microaerobic conditions, and with increase in bromate concentration, the bromate-reducing efficiency of S. oneidensis MR-1 was decreased (Figure 1A). Bromate was completely reduced by S. oneidensis MR-1 with the dosage of 0.15 mM, and when the dosage of bromate was 0.2, 0.25, 0.5, and 1 mM, the bromate-reducing efficiencies (12 h) reached around 75, 64, 48, and 23%, respectively. Under the microaerobic conditions of this study, the DO concentration (0–12 h) in the culture was maintained at around 3.5 mg/L (Supplementary Figure 2). Besides, measurable growth of S. oneidensis MR-1 was observed under microaerobic conditions, and bromate at these concentrations (0.15–1 mM) appeared to neither promote nor inhibit the growth of S. oneidensis MR-1 (Supplementary Figure 3). The bromide concentration increased as the bromate concentration was decreased, indicating that bromate was eventually reduced to bromide (Figure 1B).
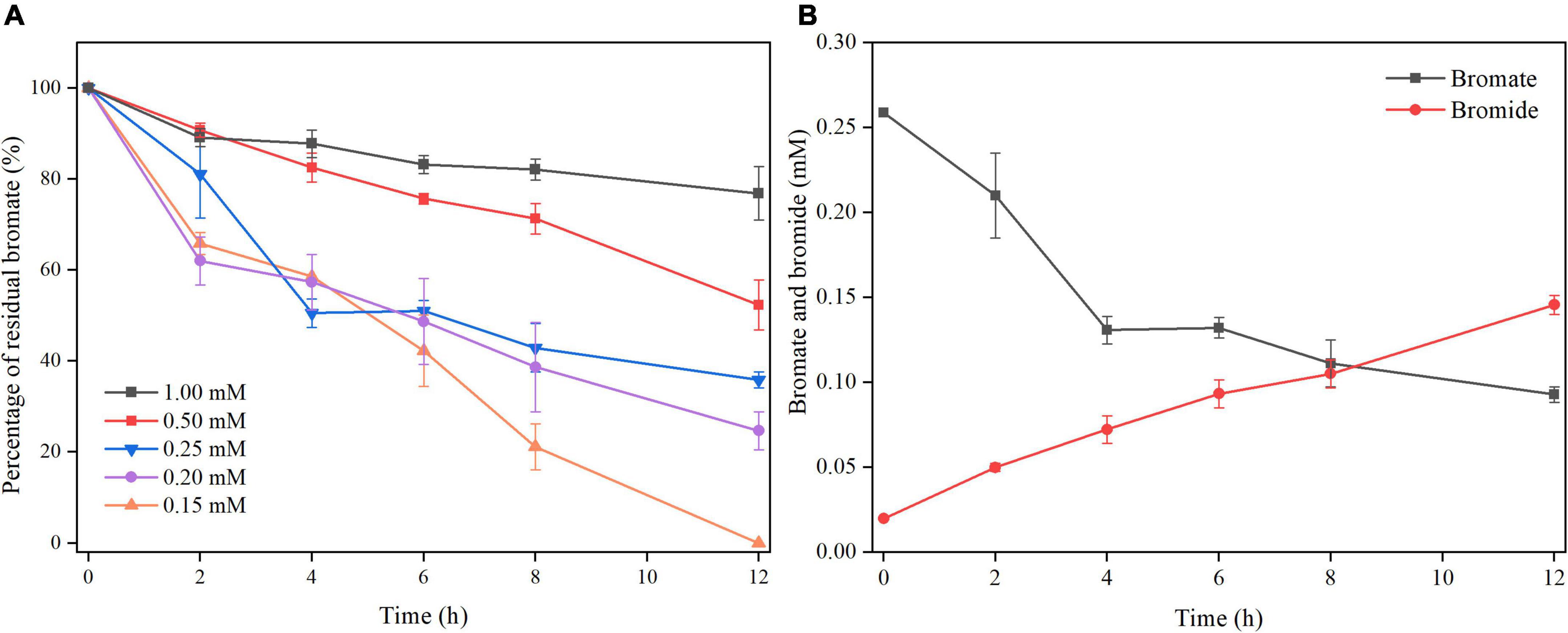
Figure 1. Microaerobic bromate reduction by S. oneidensis MR-1. (A) The strain was cultured with the dosage of bromate at 0.15, 0.2, 0.25, 0.5, and 1 mM. (B) The strain was cultured with the dosage of bromated at 0.25 mM. Error bars represent standard deviations of triplicate samples.
To demonstrate whether microaerobic bromate reduction is dependent on biological process, two control experiments were performed with the dosage of of 0.25 mM bromate. Only about 6% of bromate was reduced using heat-killed cells (Supplementary Figure 4), but no bromide was detected. In the absence of lactate, almost no bromate reduction occurred (Supplementary Figure 4). The above results suggest that bromate reduction is dependent on the metabolism of S. oneidensis MR-1.
Bromate-reducing capacities of ΔnapA, ΔfccA, and Δsye4 mutants
To identify the key reductase involved in microaerobic bromate reduction, a number of in-frame deletion mutants derived from the S. oneidensis MR-1 wild type (WT) were constructed, and their bromate-reducing capacities were evaluated at an identical bacterial concentration (OD600 = 0.12). The putative bromate reductase-encoding genes (napA, fccA, and sye4) described in the introduction were first knocked out. However, compared to WT, ΔnapA, ΔfccA, and Δsye4 all showed no significant (p ≥ 0.2) difference in bromate-reducing rate and efficiency (Figures 2A,B). The results suggest that NapA, FccA, and SYE4 are not required in microaerobic bromate reduction by S. oneidensis MR-1.
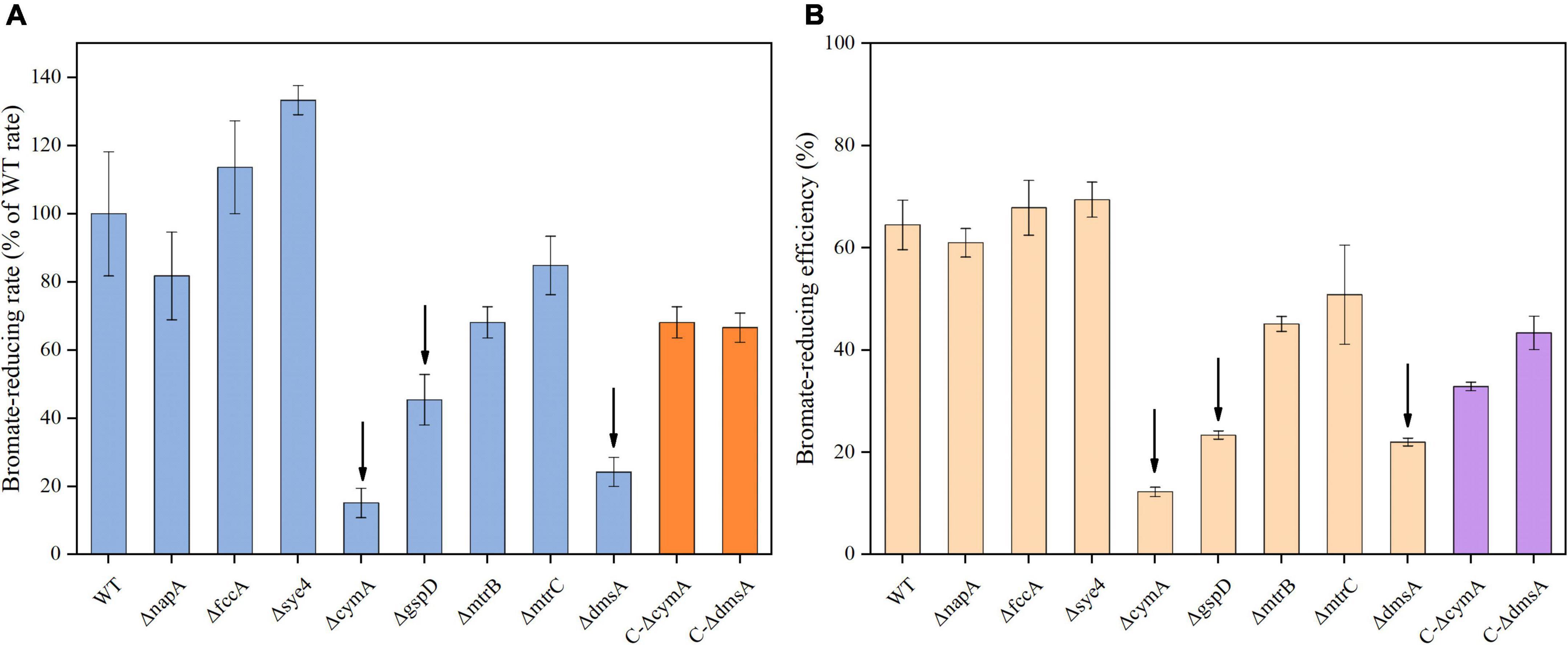
Figure 2. Microaerobic reduction of bromate at 0.25 mM by the S. oneidensis MR-1 wild type (WT), in-frame deletion mutants (ΔnapA, ΔfccA, Δsye4, ΔcymA, ΔgspD, ΔmtrB, ΔmtrC, and ΔdmsA), and complemented strains (C-ΔcymA and C-ΔdmsA, bromate reduction was performed in the presence of 0.2 mM IPTG). (A) Bromate-reducing rate (μMh– 1 mg protein– 1) was calculated from the first 2 h of incubation and normalized to total protein concentration. (B) Bromate-reducing efficiency represents the proportion of reduced bromate (at 12 h) in the initial bromate. Error bars represent standard deviations of triplicate samples.
Involvement of the terminal reductase located in the outer membrane
To determine whether cytoplasmic, periplasmic, or extracellular reductase mediates microaerobic bromate reduction, ΔcymA and ΔgspD were constructed. Interestingly, the deletion of cymA or gspD severely impaired the bromate-reducing capacity of S. oneidensis MR-1. Compared to the WT, the bromate-reducing rate and efficiency of ΔcymA were decreased by 85 and 52%, respectively; correspondingly those of ΔgspD were decreased by 55 and 41%, respectively (Figures 2A,B). The results indicate that CymA, GspD, and outer membrane proteins are responsible for bromate reduction. Furthermore, we constructed three in-frame deletion mutants (i.e., ΔmtrB, ΔmtrC, and ΔdmsA) to identify which outer membrane protein is required for bromate reduction. By deletion of mtrB or mtrC, S. oneidensis MR-1 exhibited a slight defect in bromate-reducing capacity (Figures 2A,B). With dmsA deleted, the bromate-reducing ability of S. oneidensis MR-1 was impaired to a degree close to that of ΔcymA and greater than that of ΔgspD. Moreover, the bromate-reducing capacities of complemented strains C-ΔcymA and C-ΔdmsA were recovered relative to the gene deletion mutant strains (i.e., ΔcymA and ΔdmsA).
Identification of homologous proteins and phylogenetic relationship
To explore whether all the Shewanella species possess key proteins related to bromate reduction, protein sequence alignment was performed. According to the alignment standard of the present study, it was found that 9 of the 56 other Shewanella species with a whole-genome sequence possess homologs to DmsA, GspD, and CymA of S. oneidensis MR-1 (Figure 3). In order to explore the phylogenetic relationship between the DmsA sequences of Shewanella species (the DmsA sequence of Shewanella glacialipiscicola was eliminated because of incompleteness) and 28 other DMSO reductase sequences, a phylogenetic tree was constructed. As shown in Figure 4, the DmsA of Shewanella species and E. coli belongs to the same major clade, which is different from the other DMSO reductases, including PcrA, ClrA, NarG, and TorA (these reductases have shown bromate-reducing activity in vitro), and the phylogenetic relationship of this clade and the TorA clade is the closest.
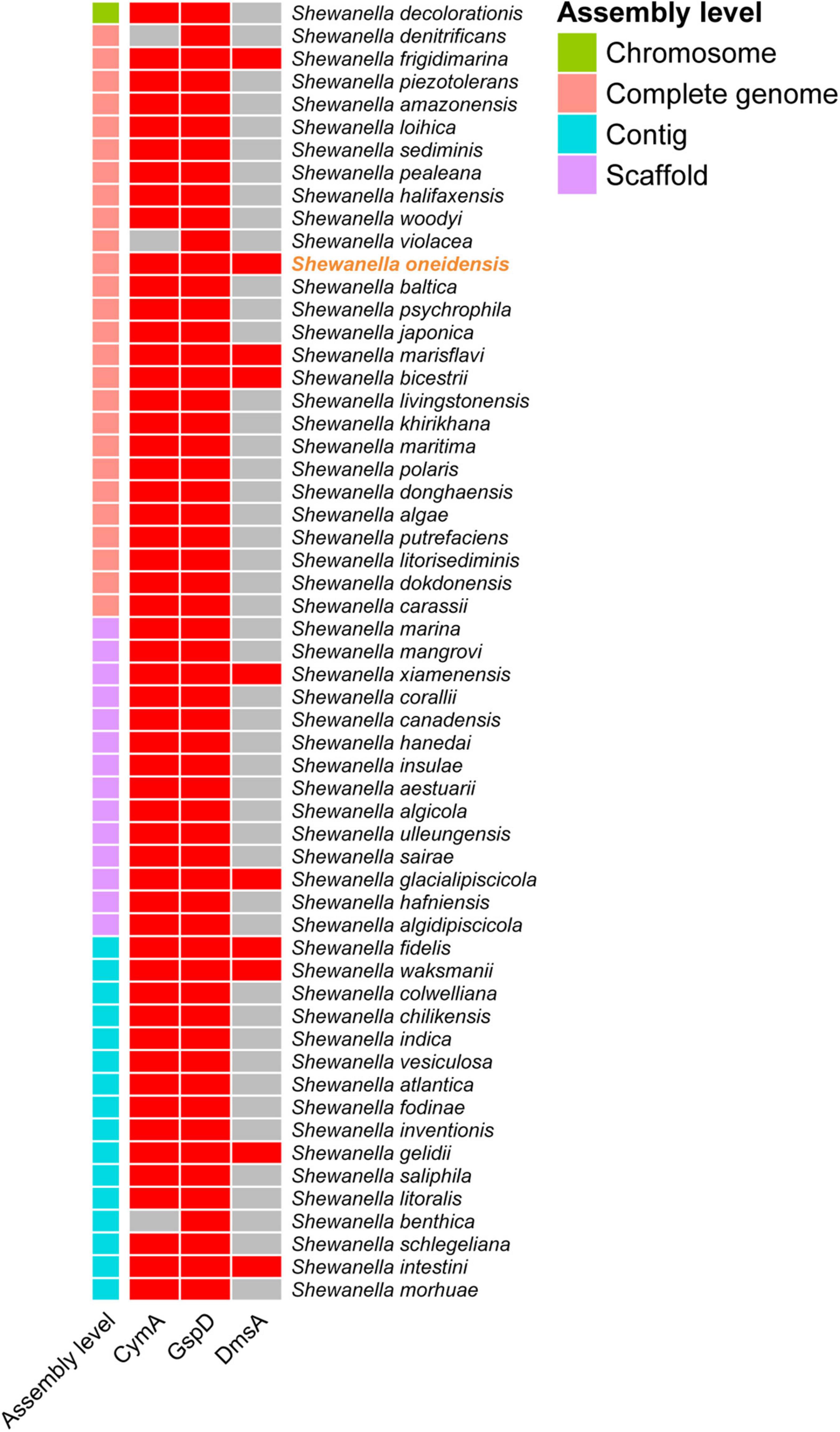
Figure 3. Homologous protein identification of Shewanella species. Cells colored in red or gray indicate if the specific protein is identified or not, respectively.
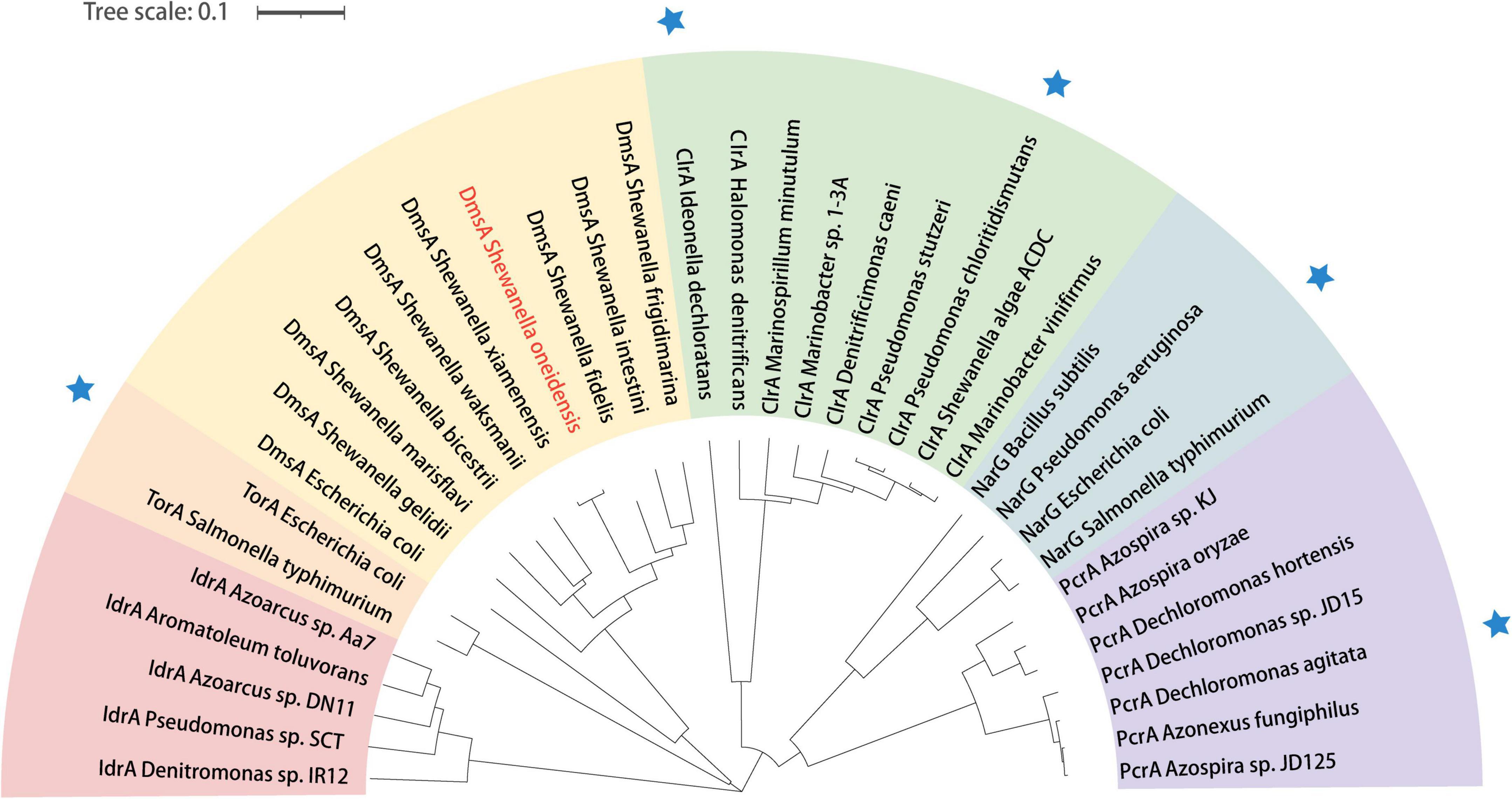
Figure 4. Phylogenetic analysis of DmsA of Shewanella species in relation to other reductases in the DMSO reductase family. The phylogenetic tree is constructed using the neighbor-joining method, and the bootstrap values of all nodes in the tree are not less than 50%. Blue star indicates that the protein has exhibited bromate-reducing activity in vitro. IdrA, iodate reductase; TorA, trimethylamine-oxide reductase; DmsA, DMSO reductase; ClrA, chlorate reductase; PcrA, perchlorate reductase.
Discussion
Shewanella oneidensis MR-1 did not exhibit the characteristic of bromate-respiring under anaerobic conditions (Supplementary Figures 1A,B), indicating that S. oneidensis MR-1 may not respire a high concentration of bromate under the anaerobic conditions of this study. However, according to previous findings (Toporek et al., 2019; Guo et al., 2022; Shin et al., 2022), when S. oneidensis MR-1 has sufficient biomass, it is still possible to perform anaerobic reduction of bromate at low concentrations. As expected, it was found that S. oneidensis MR-1 and the bromate-reducing bacterium S. decolorationis Ni1-3 have a similar bromate-reducing ability, and that both of them can reduce bromate to bromide with high efficiency (Wang Y. et al., 2022; Figures 1A,B). S. oneidensis MR-1 can grow at relatively high bromate concentrations under microaerobic conditions, which is similar to a previous study on perchlorate-reducing bacteria that some halophilic bacteria can grow in the presence of perchlorate at as high as 0.4 M under aerobic conditions (Oren et al., 2014). Although there are several reports on reduction of bromate under (micro)aerobic conditions in the biologically active carbon (BAC) filter, no available isolate capable of (micro)aerobic bromate reduction has been isolated (Kirisits and Snoeyink, 1999; Kirisits et al., 2001, 2002; Liu et al., 2012). Recently, a transcriptome analysis has provided insights into the tolerance and aerobic reduction of S. decolorationis Ni1-3 to bromate, but no bromate reductase has been identified (Wang Y. et al., 2022).
Previous studies have shown that dissimilatory nitrate reductase (NarG) can reduce bromate in vitro (Morpeth and Boxer, 1985; Maria Martinez-Espinosa et al., 2015). S. oneidensis MR-1 has only one dissimilatory nitrate reductase, NapA, which is homologous to NarG. However, the results suggest that NapA is not required for microaerobic bromate reduction by S. oneidensis MR-1. This finding is similar to a previous study that NapA is not involved in iodate reduction by S. oneidensis MR-1 (Mok et al., 2018). The in vivo evidence from the mutants (ΔfccA and Δsye4) also disproves our hypothesis that FccA and SYE4 are involved in microaerobic bromate reduction by S. oneidensis MR-1. Membrane-anchored CymA is a key component of the electron transport chain in the extracellular and periplasmic spaces (McMillan et al., 2012). GspD is an important protein in the type II secretion system, which transports extracellular terminal reductases (e.g., MtrC, OmcA, and DmsA) to the outer membrane surface (Rondelet and Condemine, 2013). In the present study, the bromate-reducing ability of ΔcymA and ΔgspD was severely impaired, suggesting that CymA, GspD, and outer membrane proteins are involved in bromate reduction. It should be noted that the bromate-reducing ability of ΔcymA, ΔgspD, and ΔdmsA is not completely lost, and whether TorA located in the periplasm is responsible for small partial bromate reduction needs to be further determined.
MtrC and OmcA, complexed together in a ratio of 1:2, are typical extracellular terminal reductases of S. oneidensis MR-1, which can reduce U(VI), Cr(VI), V(V), and Tc(VII) (Beblawy et al., 2018). MtrAB is responsible for transferring electron to MtrC (Beblawy et al., 2018). The results of this study rule out the possibility that MtrC is the major terminal bromate reductase, but that MtrCAB can contribute to microaerobic reduction of bromate. The purified reductases of bacteria (i.e., PcrA, ClrA, NarG, SerA, and TorA) with bromate-reducing activity all belong to the DMSO reductase family (Miralles-Robledillo et al., 2019). DmsEFAB, the complex protein of S. oneidensis MR-1, has been proved to mediate the dissimilatory reduction of DMSO and the extracellular reduction of iodate (Gralnick et al., 2006; Guo et al., 2022; Shin et al., 2022). DmsA is located in the outer membrane and is the catalytic subunit; it also belongs to the DMSO reductase family (Gralnick et al., 2006). The in vivo evidence from the present study indicates that the terminal reductase DmsA mediates microaerobic bromate reduction by S. oneidensis MR-1, and that both CymA and GspD are also required in that process. Previous studies have shown that the DMSO reductase of MR-1 belongs to the anaerobic respiration system, but it can be expressed under aerobic conditions, although its expression level is less than that under anaerobic conditions (Gralnick et al., 2006). In this study, the cultures were not sparging with air or oxygen, shaking was not violent, and microaerobic or anoxic zones were easily formed in the cultures. Thus, the DMSO reductase could be expressed and perform a limited function. In addition, when the cells of S. oneidensis MR-1 get into the stationary phase, oxygen is not the preferred electron acceptor; other electron acceptors such as nitrite can be respired (Dong et al., 2012). Similarly, S. oneidensis MR-1 quickly got into the stationary phase (Supplementary Figure 3), so that the DMSO reductase might be available for extracellular reduction of bromate during that time.
It is well-known that respiratory reductases belonging to the DMSO reductase family use molybdenum as a cofactor and catalyze two-electron-transferring reactions, such as perchlorate → chlorate, chlorate → chlorite, selenate → selenite, nitrate → nitrite, and DMSO → dimethyl sulfide (DMS) (McEwan et al., 2002; Sparacino-Watkins et al., 2014; Miralles-Robledillo et al., 2019), whereas the final product of microaerobic bromate reduction by S. oneidensis MR-1 is bromide, and the valence of bromine is from positive hexavalent to negative monovalent, requiring six electrons. We therefore assume that there must be an intermediate, bromite, or hypobromous acid, in microaerobic bromate reduction by S. oneidensis MR-1. A recent study has shown that the DmsEFAB of S. oneidensis MR-1 is responsible for the reduction of iodate to hypoiodous acid while producing hydrogen peroxide, and that MtrCAB is involved in scavenging hydrogen peroxide, which then facilitates iodate reduction by S. oneidensis MR-1 (Guo et al., 2022). In addition, when mtrCAB was knocked out, there were still other reactive oxygen species scavengers (ROSSs) (i.e., catalases and peroxidases) that can replace MtrCAB to complete the reduction of hydrogen peroxide in S. oneidensis MR-1 (Guo et al., 2022). As mentioned in the introduction, bromate and iodate are quite similar, and considering that the results of this study are also consistent with those of iodate reduction by S. oneidensis MR-1, hypobromous acid is very likely to be the intermediate of bromate reduction. The final product of iodate reduction by S. oneidensis MR-1 is iodide, but how the intermediate hypoiodous acid is decomposed remains unclear (Guo et al., 2022). The intermediate of (per)chlorate (halogen oxyanions) reduction, chlorite, is decomposed into chloride and oxygen by chlorite dismutase (Cld); thus (per)chlorate-respiring bacteria can utilize high concentrations of (per)chlorate as the sole electron acceptor to gain energy for growth (Youngblut et al., 2016). Besides, the iodate-respiring bacterium Pseudomonas sp. SCT also has a Cld-like protein, which may reduce the intermediate of iodate reduction, hypoiodous acid, to iodide and oxygen (Yamazaki et al., 2020). S. oneidensis MR-1 has no protein homologous to Cld, supporting the idea that S. oneidensis MR-1 is not a bromate-respiring bacterium, and that the possible intermediate hypobromous acid may be scavenged by abiotic reaction. Hypobromous acid and hypochlorous acid share a high similarity, and hypochlorous acid can react with the antioxidant reduced glutathione (GSH) to form chloride (Winterbourn and Brennan, 1997; Fang and Dehaen, 2021); thus, hypobromous acid may also be reduced to bromide by GSH. GSH is ubiquitous in proteobacteria, and S. oneidensis MR-1 is found to possess two genes, gshA and gshB, necessary for synthesis of GSH (Masip et al., 2006). Based on these findings, a molecular mechanism was proposed for microaerobic bromate reduction by S. oneidensis MR-1 (Figure 5). It is assumed that bromate is reduced to hypobromous acid and hydrogen peroxide by DmsEFAB. Subsequently, hypobromous acid is reduced to bromide by GSH, and hydrogen peroxide is reduced to H2O by MtrCAB or other ROSSs; these two processes may limit the bromate-reducing ability of S. oneidensis MR-1. Future research should focus on determining whether hypobromous acid and hydrogen peroxide are the intermediates and the role of GSH in bromate reduction.
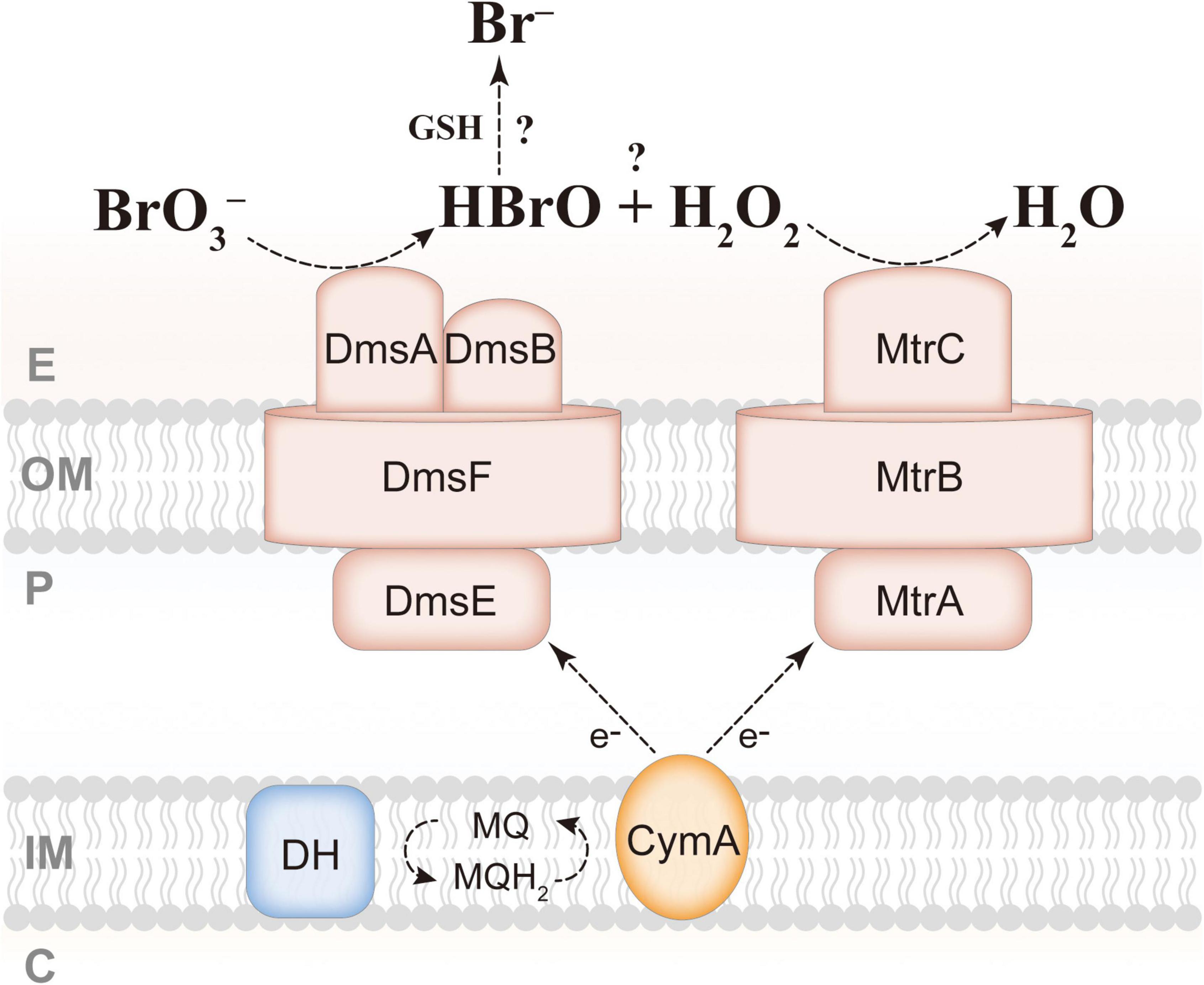
Figure 5. Proposed mechanism for extracellular reduction of bromate by S. oneidensis MR-1. GSH, reduced glutathione; E, extracellular space; OM, outer membrane; P, periplasm; IM, inner membrane; C, cytoplasm; DH, dehydrogenase; MQ, menaquinone.
The result of the sequence alignment indicates that 9 Shewanella species (i.e., S. frigidimarina, S. marisflavi, S. bicestrii, S. xiamenensis, S. glacialipiscicola, S. fidelis, S. waksmanii, S. gelidii, and S. intestini) most likely possess a bromate-reducing capacity and may play important roles in biogeochemical cycling of bromine. The result of the phylogenetic analysis shows that the DmsA of Shewanella species and other DMSO reductases belong to different major clades. Interestingly, the DMSO reductases of the four different major clades exhibit a bromate-reducing activity. In the future, it would be interesting to explore whether the catalytically active centers of the DMSO reductases are quite similar, and how many of the remaining DMSO reductases have a bromate-reducing activity.
In summary, we demonstrated that S. oneidensis MR-1 can effectively reduce bromate under microaerobic conditions, and this process is mediated by the extracellular terminal reductase DmsA. The microbial reduction process of bromate also requires membrane-anchored CymA and the type II protein secretion system. Moreover, by protein sequence alignment, it was found that a total of 9 Shewanella species possess homologs to DmsA, GspD, and CymA of S. oneidensis MR-1. The results of this study provide new insights into the molecular mechanism of microbial bromate reduction and indicate that Shewanella strains may play roles in biogeochemical cycling of bromine.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YW, JF, YS, FY, ZF, and QY performed the experiments. YW wrote the manuscript. JF, DW, XC, and YM reviewed and revised the manuscript. YM supervised the whole study. All authors read and approved the final version of the manuscript.
Funding
This study was funded by the Natural Science Foundation of China (41907214), Natural Science Foundation of Guangdong Province (2022A1515011961), Stable Support Program of Colleges and Universities in Shenzhen (20200813153536001), Special Fund for Guangdong Climbing Plan (pdjh2022b0448), the Mentoring Program at Jutu College of Shenzhen University, and Natural Science Foundation of Shenzhen University (860-000002110245).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.955249/full#supplementary-material
References
Ahmad, M. K., Khan, A. A., Ali, S. N., and Mahmood, R. (2015). Chemoprotective effect of taurine on potassium bromate-induced DNA damage, DNA-protein cross-linking and oxidative stress in rat intestine. PLoS One 10:e0119137. doi: 10.1371/journal.pone.0119137
Assis, F. L., Franco-Luiz, A. P. M., Santos, R. N. D., Campos, F. S., Dornas, F. P., Borato, P. V. M., et al. (2017). Genome characterization of the first mimiviruses of lineage C isolated in Brazil. Front. Microbiol. 8:2562. doi: 10.3389/fmicb.2017.02562
Beblawy, S., Bursac, T., Paquete, C., Louro, R., Clarke, T. A., and Gescher, J. (2018). Extracellular reduction of solid electron acceptors by Shewanella oneidensis. Mol. Microbiol. 109, 571–583. doi: 10.1111/mmi.14067
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brigé, A., Van Den Hemel, D., Carpentier, W., De Smet, L., and Van Beeumen, J. J. (2006). Comparative characterization and expression analysis of the four Old Yellow Enzyme homologues from Shewanella oneidensis indicate differences in physiological function. Biochem. J. 394, 335–344. doi: 10.1042/BJ20050979
Butler, R., Godley, A., Lytton, L., and Cartmell, E. (2005). Bromate environmental contamination: review of impact and possible treatment. Crit. Rev. Environ. Sci. Technol. 35, 193–217. doi: 10.1080/10643380590917888
Camacho, C., Coulouris, G., Avagyan, V., Ning, M., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Davidson, A. N., Chee-Sanford, J., Lai, H. Y., Ho, C.-H., Klenzendorf, J. B., and Kirisits, M. J. (2011). Characterization of bromate-reducing bacterial isolates and their potential for drinking water treatment. Water Res. 45, 6051–6062. doi: 10.1016/j.watres.2011.09.001
Dong, Y., Wang, J., Fu, H., Zhou, G., Shi, M., and Gao, H. (2012). A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS One 7:e51643. doi: 10.1371/journal.pone.0051643
Fang, Y., and Dehaen, W. (2021). Fluorescent probes for selective recognition of hypobromous acid: achievements and future perspectives. Molecules 26:363. doi: 10.3390/molecules26020363
Fredrickson, J. K., Romine, M. F., Beliaev, A. S., Auchtung, J. M., Driscoll, M. E., Gardner, T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603. doi: 10.1038/nrmicro1947
Gralnick, J. A., Vali, H., Lies, D. P., and Newman, D. K. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U.S.A. 103, 4669–4674. doi: 10.1073/pnas.0505959103
Guo, J. Z., Jiang, Y. G., Hu, Y. D., Jiang, Z., Dong, Y. R., and Shi, L. (2022). The roles of DmsEFAB and MtrCAB in extracellular reduction of iodate by Shewanella oneidensis MR-1 with lactate as the sole electron donor. Environ. Microbiol. Online ahead of print doi: 10.1111/1462-2920.16130
Hau, H. H., and Gralnick, J. A. (2007). Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61, 237–258. doi: 10.1146/annurev.micro.61.080706.093257
Hijnen, W. A. M., Jong, R., and Van der Kooij, D. (1999). Bromate removal in a denitrifying bioreactor used in water treatment. Water Res. 33, 1049–1053. doi: 10.1016/s0043-1354(98)00306-6
Hijnen, W., Voogt, R., Veenendaal, H. R., van der Jagt, H., and van der Kooij, D. (1995). Bromate reduction by denitrifying bacteria. Appl. Environ. Microbiol. 61, 239–244. doi: 10.1128/aem.61.1.239-244.1995
Huang, X., Weisener, C. G., Ni, J., He, B., Xie, D., and Li, Z. (2020). Nitrate assimilation, dissimilatory nitrate reduction to ammonium, and denitrification coexist in Pseudomonas putida Y-9 under aerobic conditions. Bioresour. Technol. 312:123597. doi: 10.1016/j.biortech.2020.123597
Jahan, B. N., Li, L., and Pagilla, K. R. (2021). Fate and reduction of bromate formed in advanced water treatment ozonation systems: a critical review. Chemosphere 266:128964. doi: 10.1016/j.chemosphere.2020.128964
Jin, M., Jiang, Y. M., Sun, L. L., Yin, J. H., Fu, H. H., Wu, G. F., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. doi: 10.1371/journal.pone.0075610
Karimi-Maleh, H., Ayati, A., Ghanbari, S., Orooji, Y., Tanhaei, B., Karimi, F., et al. (2021). Recent advances in removal techniques of Cr(VI) toxic ion from aqueous solution: a comprehensive review. J. Mol. Liq. 329:115062. doi: 10.1016/j.molliq.2020.115062
Kengen, S. W. M., Rikken, G. B., Hagen, W. R., van Ginkel, C. G., and Stams, A. J. M. (1999). Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181, 6706–6711. doi: 10.1128/jb.181.21.6706-6711.1999
Kirisits, M. J., and Snoeyink, V. L. (1999). Reduction of bromate in a BAC filter. J. Am. Water Works Ass. 91, 74–84. doi: 10.1002/j.1551-8833.1999.tb08684.x
Kirisits, M. J., Snoeyink, V. L., Chee-Sanford, J. C., Daugherty, B. J., Brown, J. C., and Raskin, L. (2002). Effect of operating conditions on bromate removal efficiency in BAC filters. J. Am. Water Works Ass. 94, 182–193. doi: 10.2307/41298509
Kirisits, M. J., Snoeyink, V. L., Inan, H., Chee-Sanford, J. C., Raskin, L., and Brown, J. C. (2001). Water quality factors affecting bromate reduction in biologically active carbon filters. Water Res. 35, 891–900. doi: 10.1016/s0043-1354(00)00334-1
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lemaire, O. N., Mejean, V., and Iobbi-Nivol, C. (2020). The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 44, 155–170. doi: 10.1093/femsre/fuz031
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D.-B., Cheng, Y.-Y., Wu, C., Li, W.-W., Li, N., Yang, Z.-C., et al. (2014). Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 4:3735. doi: 10.1038/srep03735
Liu, C., Li, W., Liu, L., Yu, H., Liu, F., and Lee, D.-J. (2020). Autotrophic induced heterotrophic bioreduction of bromate in use of elemental sulfur or zerovalent iron as electron donor. Bioresour. Technol. 317:124015. doi: 10.1016/j.biortech.2020.124015
Liu, J., Yu, J., Li, D., Zhang, Y., and Yang, M. (2012). Reduction of bromate in a biological activated carbon filter under high bulk dissolved oxygen conditions and characterization of bromate-reducing isolates. Biochem. Eng. J. 65, 44–50. doi: 10.1016/j.bej.2012.04.004
Lv, X., Wang, D., Iqbal, W., Yang, B., and Mao, Y. (2019). Microbial reduction of bromate: current status and prospects. Biodegradation 30, 365–374. doi: 10.1007/s10532-019-09882-x
Maria Martinez-Espinosa, R., Richardson, D. J., and Jose Bonete, M. (2015). Characterisation of chlorate reduction in the haloarchaeon Haloferax mediterranei. Biochim. Biophys. Acta 1850, 587–594. doi: 10.1016/j.bbagen.2014.12.011
Masip, L., Veeravalli, K., and Georgioui, G. (2006). The many faces of glutathione in bacteria. Antioxid. Redox Sign. 8, 753–762. doi: 10.1089/ars.2006.8.753
McEwan, A. G., Ridge, J. P., McDevitt, C. A., and Hugenholtz, P. (2002). The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19, 3–21. doi: 10.1080/014904502317246138
McMillan, D. G., Marritt, S. J., Butt, J. N., and Jeuken, L. J. (2012). Menaquinone-7 is specific cofactor in tetraheme quinol dehydrogenase CymA. J. Biol. Chem. 287, 14215–14225. doi: 10.1074/jbc.M112.348813
Miralles-Robledillo, J. M., Torregrosa-Crespo, J., Martínez-Espinosa, R. M., and Pire, C. (2019). DMSO reductase family: phylogenetics and applications of extremophiles. Int. J. Mol. Sci. 20:3349. doi: 10.3390/ijms20133349
Mok, J. K., Toporek, Y. J., Shin, H.-D., Lee, B. D., Lee, M. H., and DiChristina, T. J. (2018). Iodate reduction by Shewanella oneidensis does not involve nitrate reductase. Geomicrobiol. J. 35, 570–579. doi: 10.1080/01490451.2018.1430189
Morpeth, F. F., and Boxer, D. H. (1985). Kinetic analysis of respiratory nitrate reductase from Escherichia coli K12. Biochemistry 24, 40–46. doi: 10.1021/bi00322a007
Naruya, S., and Masatoshi, N. (1987). The neighbor-joining method : a new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4:406. doi: 10.1093/oxfordjournals.molbev.a040454
Oren, A., Bardavid, R. E., and Mana, L. (2014). Perchlorate and halophilic prokaryotes: implications for possible halophilic life on Mars. Extremophiles 18, 75–80. doi: 10.1007/s00792-013-0594-9
Pradhan, D., Sukla, L. B., Sawyer, M., and Rahman, P. K. S. M. (2017). Recent bioreduction of hexavalent chromium in wastewater treatment: a review. J. Ind. Eng. Chem. 55, 1–20. doi: 10.1016/j.jiec.2017.06.040
Ridley, H., Watts, C. A., Richardson, D. J., and Butler, C. S. (2006). Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl. Environ. Microbiol. 72, 5173–5180. doi: 10.1128/aem.00568-06
Rondelet, A., and Condemine, G. (2013). Type II secretion: the substrates that won’t go away. Res. Microbiol. 164, 556–561. doi: 10.1016/j.resmic.2013.03.005
Shi, M. M., Wu, L., Xia, Y., Chen, H. J., Luo, Q. X., Sun, L. L., et al. (2013). Exoprotein production correlates with morphotype changes of nonmotile Shewanella oneidensis mutants. J. Bacteriol. 195, 1463–1474. doi: 10.1128/jb.02187-12
Shimokawa, O., and Ishimoto, M. (1979). Purification and some properties of inducible tertiary amine N-oxide reductase from Escherichia coli. J. Biochem. 86, 1709–1717. doi: 10.1093/oxfordjournals.jbchem.a132691
Shin, H.-D., Toporek, Y., Mok, J. K., Maekawa, R., Lee, B. D., Howard, M. H., et al. (2022). Iodate reduction by Shewanella oneidensis requires genes encoding an extracellular dimethylsulfoxide reductase. Front. Microbiol. 13:852942. doi: 10.3389/fmicb.2022.852942
Shirodkar, S., Reed, S., Romine, M., and Saffarini, D. (2011). The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environ. Microbiol. 13, 108–115. doi: 10.1111/j.1462-2920.2010.02313.x
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706. doi: 10.1039/c3cs60249d
Tamai, N., Ishii, T., Sato, Y., Fujiya, H., Muramatsu, Y., Okabe, N., et al. (2016). Bromate reduction by Rhodococcus sp. Br-6 in the presence of multiple redox mediators. Environ. Sci. Technol. 50, 10527–10534. doi: 10.1021/acs.est.6b02261
Thatoi, H., Das, S., Mishra, J., Rath, B. P., and Das, N. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J. Environ. Manage. 146, 383–399. doi: 10.1016/j.jenvman.2014.07.014
Thorell, H. D., Stenklo, K., Karlsson, J., and Nilsson, T. (2003). A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl. Environ. Microbiol. 69, 5585–5592. doi: 10.1128/AEM.69.9.5585-5592.2003
Toporek, Y. J., Mok, J. K., Shin, H. D., Lee, B. D., Lee, M. H., and DiChristina, T. J. (2019). Metal reduction and protein secretion genes required for iodate reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 85:e02115-18. doi: 10.1128/AEM.02115-18
Vettese, G. F., Morris, K., Natrajan, L. S., Shaw, S., Vitova, T., Galanzew, J., et al. (2020). Multiple lines of evidence identify U(V) as a key Intermediate during U(VI) reduction by Shewanella oneidensis MR1. Environ. Sci. Technol. 54, 2268–2276. doi: 10.1021/acs.est.9b05285
Wang, D., Wang, Y., Lv, X., Cai, X., Iqbal, W., Yang, B., et al. (2022). Isolation of anaerobic bromate-reducing bacteria using different carbon sources and transcriptomic insights from Klebsiella variicola Glu3. Front. Microbiol. 13:851844. doi: 10.3389/fmicb.2022.851844
Wang, Y., Cai, X., Fan, J., Wang, D., and Mao, Y. (2022). Transcriptome analysis provides new insights into the tolerance and aerobic reduction of Shewanella decolorationis Ni1-3 to bromate. Appl. Microbiol. Biotechnol. 106, 4749–4761. doi: 10.1007/s00253-022-12006-w
Winterbourn, C. C., and Brennan, S. O. (1997). Characterization of the oxidation products of the reaction between reduced glutathione and hypochlorous acid. Biochem. J. 326, 87–92. doi: 10.1042/bj3260087
Wolin, E. A., Wolin, M. J., and Wolfe, R. S. (1963). Formation of methane by bacterial extracts. J. Biol. Chem. 238, 2882–2886. doi: 10.1016/S0021-9258(18)67912-8
Yamazaki, C., Kashiwa, S., Horiuchi, A., Kasahara, Y., Yamamura, S., and Amachi, S. (2020). A novel dimethylsulfoxide reductase family of molybdenum enzyme, Idr, is involved in iodate respiration by Pseudomonas sp. SCT. Environ. Microbiol. 22, 2196–2212. doi: 10.1111/1462-2920.14988
Yang, J., Feng, L., Pi, S., Cui, D., Ma, F., Zhao, H.-P., et al. (2020). A critical review of aerobic denitrification: insights into the intracellular electron transfer. Sci. Total Environ. 731:139080. doi: 10.1016/j.scitotenv.2020.139080
Youngblut, M. D., Wang, O. W., Barnum, T. P., and Coates, J. D. (2016). (Per)chlorate in biology on earth and beyond. Annu. Rev. Microbiol. 70, 435–457. doi: 10.1146/annurev-micro-102215-095406
Zhang, H., Ma, B., Huang, T., and Shi, Y. (2021). Nitrate reduction by the aerobic denitrifying actinomycete Streptomyces sp. XD-11-6-2: performance, metabolic activity, and micro-polluted water treatment. Bioresour. Technol. 326:124779. doi: 10.1016/j.biortech.2021.124779
Keywords: Shewanella oneidensis, bromate reduction, bromate, bromide, molecular mechanism, DMSO reductase, genomic survey
Citation: Wang Y, Fan J, Shen Y, Ye F, Feng Z, Yang Q, Wang D, Cai X and Mao Y (2022) Bromate reduction by Shewanella oneidensis MR-1 is mediated by dimethylsulfoxide reductase. Front. Microbiol. 13:955249. doi: 10.3389/fmicb.2022.955249
Received: 28 May 2022; Accepted: 09 August 2022;
Published: 30 August 2022.
Edited by:
George F. Wells, Northwestern University, United StatesReviewed by:
Alice C. Dohnalkova, Pacific Northwest National Laboratory (DOE), United StatesBenjamin K. Keitz, University of Texas at Austin, United States
Hyun-Dong Shin, Bereum Co., Ltd., South Korea
Copyright © 2022 Wang, Fan, Shen, Ye, Feng, Yang, Wang, Cai and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanping Mao, bWFveUBzenUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yicheng Wang
Yicheng Wang Jiale Fan
Jiale Fan Yonglin Shen
Yonglin Shen Fan Ye
Fan Ye Zhiying Feng
Zhiying Feng Qianning Yang
Qianning Yang Dan Wang
Dan Wang Xunchao Cai
Xunchao Cai Yanping Mao
Yanping Mao