- 1Department of Microbiology, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand
- 2Department of Veterinary Biosciences and Veterinary Public Health, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 3Department of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 4Department of Veterinary and Animal Science, Northern Faculty, Scotland’s Rural College, Inverness, United Kingdom
- 5Department of Bacteriology, Animal and Plant Health Agency, Weybridge, United Kingdom
- 6Department of Chemical Engineering, Loughborough University, Loughborough, United Kingdom
- 7Department of Genetics and Genome Biology, University of Leicester, Leicester, United Kingdom
- 8Research Center of Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
- 9Center of Multidisciplinary Technology for Advanced Medicine, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand
Acute non-typhoidal salmonellosis (NTS) caused by a Gram-negative bacterium Salmonella enterica serovar Typhimurium (S. Tm) is one of the most common bacterial foodborne diseases worldwide. Bacteriophages (phages) can specifically target and lyse their host bacteria, including the multidrug-resistant strains, without collateral damage to other bacteria in the community. However, the therapeutic use of Salmonella phages in vivo is still poorly investigated. Salmonella phages ST-W77 and SE-W109 have previously been shown by our group to be useful for biocontrol properties. Here, we tested whether phages ST-W77 and SE-W109 can reduce Salmonella invasion into cultured human cells and confer a therapeutic benefit for acute NTS in a mammalian host. Human colonocytes, T84 cells, were treated with phages ST-W77, SE-W109, and its combination for 5 min before S. Tm infection. Gentamicin protection assays demonstrated that ST-W77 and SE-W109 significantly reduced S. Tm invasion and inflammatory response in human colonocytes. Next, streptomycin-pretreated mice were orally infected with S. Tm (108 CFU/mouse) and treated with a single or a combination of ST-W77 and SE-W109 (1010 PFU/mouse for 4 days) by oral feeding. Our data showed that phage-treated mice had lower S. Tm numbers and tissue inflammation compared to the untreated mice. Our study also revealed that ST-W77 and SE-W109 persist in the mouse gut lumen, but not in systemic sites. Together, these data suggested that Salmonella phages ST-W77 and SE-W109 could be further developed as an alternative approach for treating an acute NTS in mammalian hosts.
Introduction
Salmonella enterica serovar Typhimurium (S. Tm) is a Gram-negative bacterium in the family Enterobacteriaceae. Human infection caused by S. Tm through the fecal-oral route transmission is called “acute non-typhoidal salmonellosis (NTS).” Common symptoms of an acute NTS are high-grade fever, abdominal pain, and diarrhea, which usually develop on days two or three after ingesting S. Tm contaminated food or drink. Although most acute NTS patients with proper supportive treatments would recover within a few days, some immunocompromised patients could develop an invasive NTS (iNTS), a life-threatening form of NTS (Morpeth et al., 2009; Mather et al., 2018; Appiah et al., 2021). Appropriate antibiotics are essential to decrease the morbidity and mortality rate in iNTS patients.
Intravenous antibiotics are essential to treat an iNTS patient. However, the rise of multidrug resistance (MDR) S. Tm worldwide, including in Southeast Asia, increased the challenge (Mather et al., 2018; Whistler et al., 2018). Clinical strains of S. Tm from Thailand and worldwide exhibit resistance to several groups of the commonly used antibacterial agents such as fluoroquinolones and cephalosporins (Patchanee et al., 2015, 2017; Hengkrawit and Tangjade, 2022). These concerns raise the urgent need to find alternative approaches to an antibiotic for acute NTS, such as probiotics or bacteriophages (Deriu et al., 2013; Nikkhahi et al., 2017).
Bacteriophages (phages) are a group of particular viruses that infect and kill their bacterial hosts. Phages are usually found in common environmental reservoirs where their hosts are found (Clokie et al., 2011). Most tailed phages belong to the family Myoviridae, Podoviridae, and Siphoviridae (Weinbauer, 2004). For several decades, phages have been considered an alternative treatment to MDR bacteria in human and animal infectious diseases (Salmond and Fineran, 2015; Harada et al., 2018; Jamal et al., 2019; Au et al., 2021). For example, a local application of a phage cocktail at a very low concentration gradually decreased P. aeruginosa burden in a human burn wound clinical trial (Jault et al., 2019). Recently, phage therapy improved the symptoms of disseminated cutaneous Mycobacterium chelonae infection in immunocompromised patient (Little et al., 2022). Moreover, the anti-Salmonella therapeutic effect of bacteriophages has been already demonstrated in agriculture, livestock, farm animals, and their related food products (Fiorentin et al., 2005; Svircev et al., 2018; Au et al., 2021; Thanki et al., 2022).
Our previous study in Galleria mellonella, a larvae model for several bacterial infections, demonstrated that the prophylactic regimen with a phage cocktail significantly increased the survival rate of Salmonella-infected larvae (Nale et al., 2020). Compared with co-infection and remedial regimens, the preventive phage regimen showed better protection and control against Salmonella infection in the haemolymph of the Galleria. We also found that Salmonella phages ST-W77 and SE-W109 isolated from sewages in Thailand conferred the highest broad host range activity to several serovars of Salmonella enterica (Phothaworn et al., 2020). ST-W77 belongs to Myovirus within the Viunalikevirus genus, and SE-W109 belongs to Siphovirus within the Jerseylikevirus genus. Both ST-W77 and SE-W109 phages conferred an obligate lytic lifestyle and lack of lysogeny-related genes in their genomes (Phothaworn et al., 2020). ST-W77 and SE-W109 can reduce Salmonella numbers in food products (milk and chicken meat) and hence have therapeutic potential for further development as a biocontrol agent in food (Phothaworn et al., 2020).
According to our previous studies (Nale et al., 2020; Phothaworn et al., 2020), the application of phages ST-W77 and SE-W109 as potential candidates for alternative treatment of MDR Salmonella infection in mammalian hosts should be explored. It had been reported that therapeutic administration of Salmonella phage increased the survival rate up to 60% in mice (Tang et al., 2019). Preadministration of a lytic phage reduced gut dysbiosis and intestinal inflammation in S. Tm-infected mice (Bao et al., 2022). However, the therapeutic effect of phages ST-W77 and SE-W109 in a mammalian host infected with S. Tm is still unknown.
In this study, we performed a gentamicin protection assay on human colonocytes, T84 cells, challenge with S. Tm and phages to investigate the efficacy of phages ST-W77 and SE-W109 in the prevention of S. Tm invasion. Then, we further investigated bacterial proliferation, gut and systemic tissue inflammation in S. Tm-infected mice treated with a single oral administration of phages ST-W77, SE-W109, and its combination. A viable bacterial count, qualitative polymerase chain reaction (qPCR), and histopathological study were used to determine the degree of S. Tm proliferation and tissue inflammation in mice.
Materials and methods
Ethical statement
The animal experiment protocol was reviewed and approved by the Animal Care and Use Committee, Chiang Mai University, Thailand, in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines (Approval no.2563/MC-0002). This work was approved by the Institutional Biosafety Committee, Faculty of Medicine, Chiang Mai University (Approval no. CMUIBC 02033/2562).
Bacterial strain and cell culture
Salmonella enterica serovar Typhimurium strain IR715 (ATCC 14028 derivative with nalidixic acid resistance; S. Tm) was used (Stojiljkovic et al., 1995). S. Tm was grown at 37°C aerobically with shaking in Luria-Bertani (LB) broth (10 g/l tryptone, 5 g/l yeast extract, and 10 g/l NaCl; Difco, United States) for 16–18 h. A nalidixic acid (0.05 mg/ml; AppliChem, Germany) was used as a selective antibiotic for S. Tm in liquid and solid media. The list of bacterial and phage strains used in this study shown in Supplementary Table S1.
The human colonic epithelial cell T84 (ATCC CCL-248) purchased from the American Type Culture Collection (ATCC) was used for the invasion assay in this study. A completed media for the T84 was prepared by adding 5% fetal bovine serum (FBS; Hyclone, Singapore) and 1% Penicillin/Streptomycin (Gibco, United States) into Dulbecco’s Modified Eagle Medium (DMEM): F12 (Gibco, United States) contained 2.5 mM L-glutamine and 15 mM HEPES. Cells were grown at 37°C with 5% CO2 in a humidified condition.
Phage preparation by spot assay
Salmonella phage ST-W77 and SE-W109 isolated from sewage samples in Thailand were prepared by a double-layer agar assay as previously described (Phothaworn et al., 2020). S. Tm lawn was formed by adding 100 μl of mid-log (exponential) phase liquid culture of S. Tm into 4 ml of 0.35% melted top LB agar supplemented with 10 mM CaCl2. Then, 10 μl of ten-fold serially diluted phage solution was spotted on the lawn, and a clear zone was observed at 18 h after the incubation at 37°C. Plaque-forming unit (PFU)/mL of phage solution was calculated by a ten-fold serially diluted phage solution.
Invasion assay (gentamicin protection assay)
The invasion assay of S. Tm into human colonocytes was adapted from the previously described work (Winter et al., 2009). Briefly, 0.5 ml of T84 cells resuspended in the complete medium were seeded into 24-well plates at a density of ~105 cells/well and incubated at 37°C with 5% CO2 for 24 h. The complete growth medium was replaced by the DMEM: F12 without FBS or antibiotics for an additional 24 h to synchronize the cells. Then, T84 cells were treated with 10 μl of phages 5 × 1010 PFU/mL in SM buffer into each well for 5 min before S. Tm IR715 adding at the multiplicity of infection (MOI) of 25 at 37°C for 1 h. The cells were subsequently washed with Dulbecco’s phosphate-buffered saline (DPBS; Hyclone, Singapore) and treated with 500 μl/well of 100 μg/ml gentamycin sulfate (AppliChem, Germany), and incubated at 37°C for 90 min. T84 cells were then washed with DPBS before adding 0.5 ml of 1% Triton-X-100 (Thermo Fisher Scientific, United States) in PBS to lyse the cells. The lysate was ten-fold serially diluted and enumerated on LB agar to determine the numbers of intracellular bacteria.
Gene expressions in T84 cells by a qPCR
A qPCR from T84 cell culture was performed as previously described (Winter et al., 2009). In brief, 2 ml of T84 cells resuspended in the complete medium were seeded into 6-well plates at a density of ~106 cells/well and incubated at 37°C with 5% CO2 for 24 h. The complete medium was replaced at 24 h prior to the assay by the DMEM: F12 without FBS and antibiotics. Then, T84 cells were treated by adding 10 μl of 1 × 1011 PFU/mL phages and S. Tm IR715 simultaneously at the MOI of 25 at 37°C for 3 h. Cells were harvested by adding 1 ml of Trizol (Ambion, United States) into each well. RNA from T84 cells was extracted by using TRIzol reagent (Thermo Fisher Scientific, United States) according to the manufacturer’s protocol. Then, complementary DNA (cDNA) was generated by using TaqMan reverse transcription reagents (Applied Biosystems, United States). A qPCR was performed using SYBR-Green based real-time PCR (Bioline, Tennessee, United States) in ViiA7 Real-Time PCR system (Applied Biosystems, United States) with primer pairs shown in Supplementary Table S2. Relative fold change of mRNA expressions was calculated by the comparative Ct method (2−ΔΔCT). The expression levels of target mRNA genes (IL-8, MIP-3A, and IL-1B) were normalized with the housekeeping gene GAPDH as previously described (Winter et al., 2009).
Mouse experiment
Female C57BL/6NJcl mice (Mus musculus) aged between 6 to 8-week-old were purchased from Nomura Siam International Company (Bangkok, Thailand). All mice were housed at the Laboratory Animal Center at Chiang Mai University in a temperature-controlled room (25°C) under a 12/12-h light/dark cycle with free access to food and water (ad libitum) for at least 1 week for acclimatization. Mice were divided into 4 groups (7 mice per group; Appiah et al., 2021) Saline-magnesium sulfate (SM) buffer-treated (Morpeth et al., 2009) ST-W77-treated, (Mather et al., 2018) SE-W109, and (Whistler et al., 2018) ST-W77 combined with SE-W109 (Supplementary Figure S1). All mice were orally fed with 100 μl of 200 mg/ml streptomycin sulfate (Lot number 5 K013479, AppliChem, Germany) 1 day prior to S. Tm infection. A streptomycin pre-treatment transiently impairs mouse gut colonization resistance. This treatment allows S. Tm to cause a gut inflammation similar to the pathology found in human acute NTS (Santos et al., 2001).
Streptomycin-pretreated mice were orally infected with 100 μl of 109 CFU/ml S. Tm with an oral feeding. Then, all S. Tm-infected mice were orally fed with 100 μl of 1011 PFU/mL phage solution in SM buffer (1010 PFU/mouse) or SM buffer started at 1 h after S. Tm infection. Then, phage solutions were orally given to mice once daily for three consecutive days. The SM buffer was used as a control in the untreated group. Mouse feces were collected every day to enumerate the phage and S. Tm shedding. On day 4 post-infection (p.i.), all mice were euthanized, and mouse tissues were collected as previously described (Sarichai et al., 2020).
Enumerations of Salmonella and phages from mouse fecal pellet and tissues
Mouse fecal pellets (2–3 pellets/mouse) were collected on day 1, 2, 3, and 4 p.i. Pellets were immediately weighed and homogenized using 1.0 mm diameter zirconium/silica beads (Biospec Products, United States) with a bead-beating machine. Then, ST-W77 and SE-W109 numbers (PFU/gm fecal pellet or tissue) were calculated by a double-layer agar assay.
Mouse tissues (colon content, cecal content, colon, cecum, ileum, liver, and spleen) were collected on day four p.i. when the most prominent gut inflammation occurred (Tsolis et al., 2011). Tissues were immediately weighed and homogenized using 1.0 mm diameter zirconium/silica beads (Biospec Products, Bartesville, United States) with a bead-beating machine. S. Tm colony-forming unit (CFU)/gm tissues were determined by a serial diluting technique on the LB agar with nalidixic acid (0.05 mg/ml).
Gene expressions in mouse tissues by a qPCR
Mouse tissues (colon and spleen) were harvested at day 4 p.i. and immediately kept in RNA preservation (RNAstore Reagent, TIANGEN, China), then stored at -20°C until use according to the manufacturer’s instruction. Mouse colonic and splenic mRNA were isolated as previously described (Thiennimitr et al., 2011). A qPCR was performed using SYBR-Green-based real-time PCR (Bioline, Tennessee, United States) in ViiA7 Real-Time PCR system (Applied Biosystems, United States) with primer pairs shown in Supplementary Table S2. Relative fold change of mouse mRNA expressions (Kc, IL-17, Zo-1, Ifn-g and Nos-2) were calculated by the comparative Ct method using a mouse Gapdh as a housekeeping gene.
Cecal histopathological study
Segments of mouse ceca were fixed in 10% buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E; Sarichai et al., 2020). The slides were blindly scored by the veterinary pathologist using the criteria shown in Supplementary Table S3.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism (version 7) programs (GraphPad Software). Data were analyzed using the Student’s t-test to compare two groups and using one-way analysis of variance (ANOVA) when comparing three or more groups. The multiple comparison test by the GraphPad Prism 7.0 program assigned a p-value of <0.05, indicating a statistically significant difference. *, ** and *** indicate p-values <0.05, 0.01, and 0.001, respectively. Bars represent a geometric mean with the standard errors of the mean (SEM).
Results
Phage ST-W77 and SE-W109 reduced Salmonella invasiveness and inflammatory response in the human gut epithelial cells
Lytic activity of phage ST-W77 and SE-W109 on different serovars of non-typhoidal Salmonella strains was previously shown (Phothaworn et al., 2019). However, the lytic activity of ST-W77 and SE-W109 on S. Tm strain IR715 (the nalidixic acid derivatives of ATCC 14028) has never been reported. This study investigated whether ST-W77 and SE-W109 can lyse S. Tm IR715 in vitro. The spotting of 10 μl of each phage solution on the lawn of S. Tm IR715 resulted in clear zones for both phages compared to the SM buffer negative control (Figure 1).
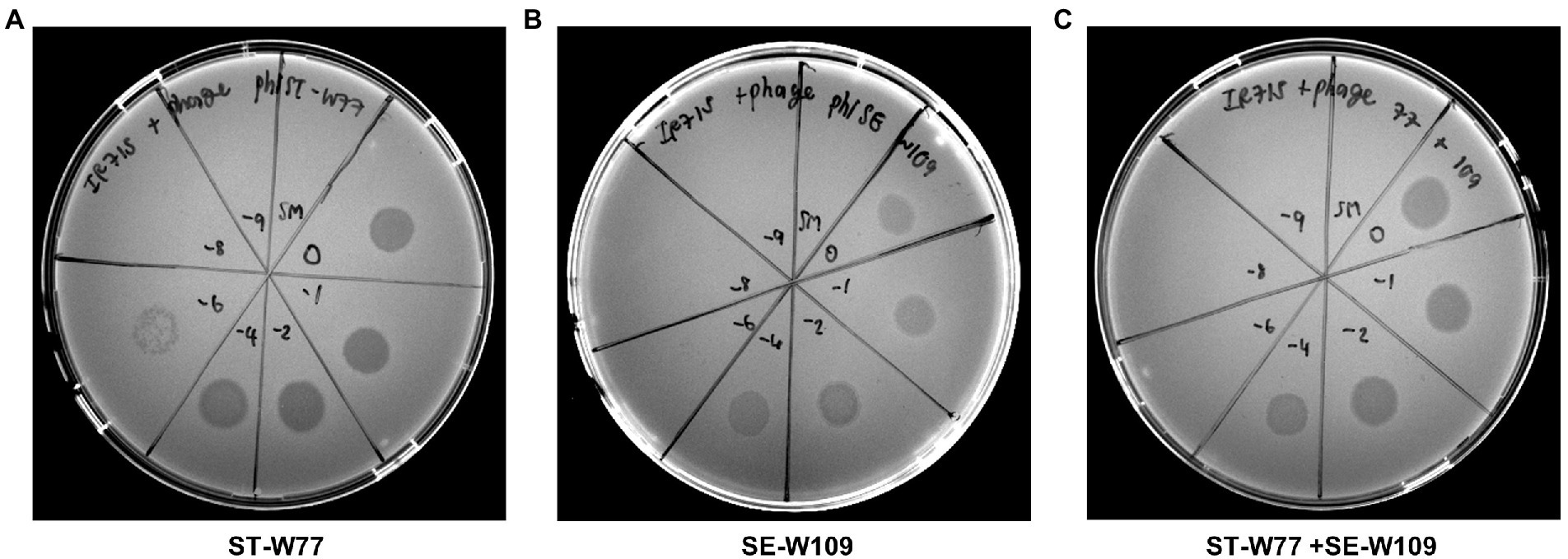
Figure 1. Susceptibility of S. Tm IR715 against phages ST-W77 and SE-W109. 10 μl of different phage dilutions in SM buffer (undiluted (10−0), 10−2, 10−4, 10−6, 10−8, and 10−9) were spotted on the S. Tm lawn, and clear zones were observed at 18 h after the incubation at 37°C. ST-W77 (A), SE-W109 (B), and a 1:1 combination of ST-W77 and SE-W109 (C). SM buffer was used as a negative control.
S. Tm is an invasive enteropathogen that uses several virulence factors to invade the host gut epithelium. Here, we investigated whether phages ST-W77 and SE-W109 can affect S. Tm invasiveness. The T84 cell, a human colonic epithelium, was used to assess S. Tm invasion using a gentamicin protection assay. Our data showed that both phages added to the cells concurrently with S. Tm significantly reduced the numbers (about 1.5 logs, p-value <0.001) of the invading S. Tm after 90 min of incubation (Figure 2A).
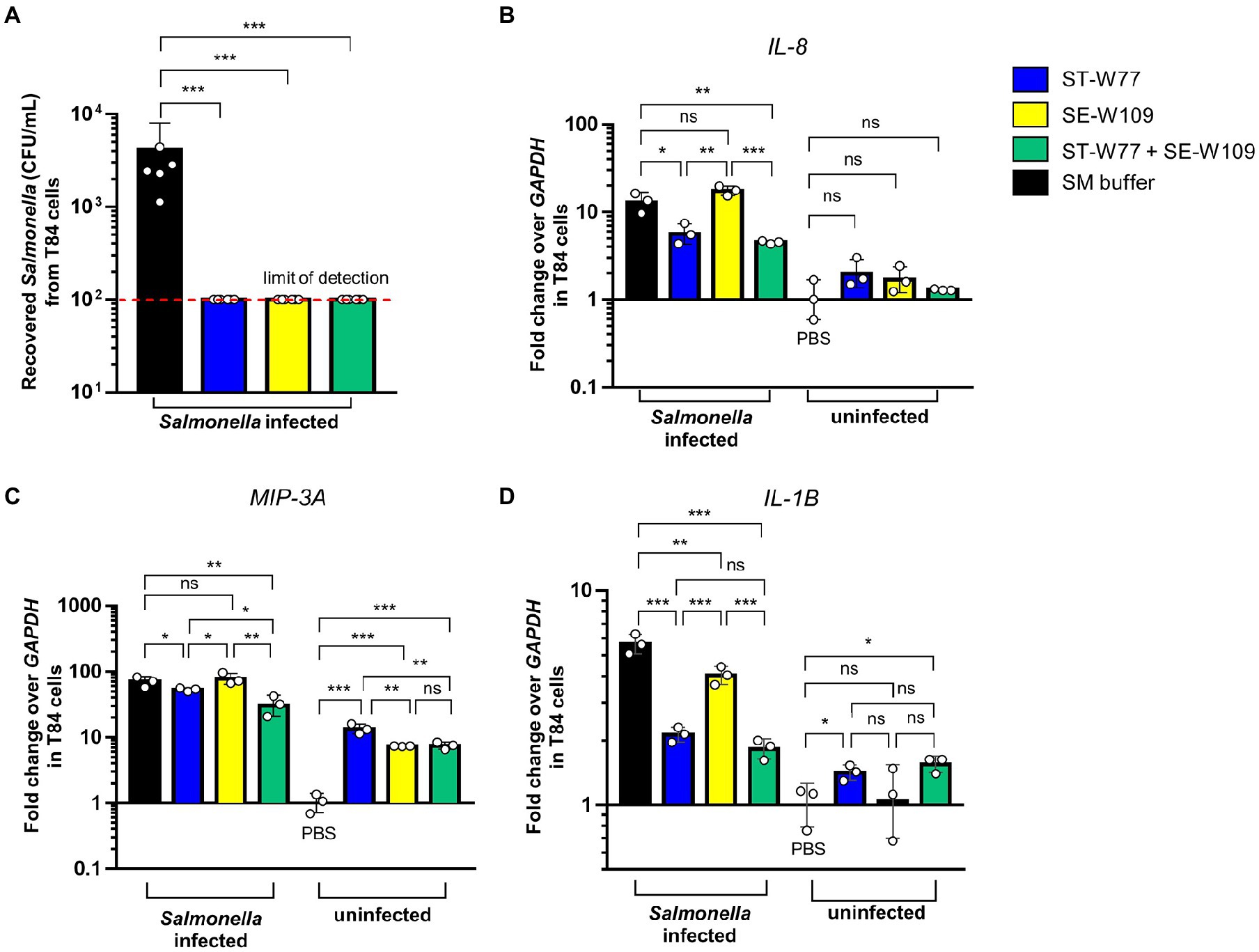
Figure 2. Efficiency of phages ST-W77 and SE-W109 in the protection against S. Tm invasion of human colonic epithelium. Human colonic epithelium (T84 cells) were seeded in a 24-well plate at a density of 105 cells/well. Each well was added 10 μl of 5 × 1010 PFU/mL phages or SM buffer for 5 min before S. Typhimurium infection (MOI = 25). After the gentamicin protection for 90 min, cells were lysed and recovered bacterial numbers (CFU/mL) were counted (A). The invasion assay was repeated in a 6-well-plate with the same MOI to observe proinflammatory gene expressions. Fold change of gene expressions of human IL-8 (B), MIP-3A (C), and IL-1B (D) were determined by a qPCR using GAPDH as a housekeeping gene. Bars represent the geometric mean; error bars indicate geometric standard deviation. *, **, *** indicated p < 0.05, 0.01 and 0.001, respectively. ns, a non-statistically significant difference.
Next, we performed a qPCR to determine the relative fold change in mRNA expression of critical proinflammatory genes in T84 cells. We found that phage ST-W77 significantly decreased (p-value <0.05) the expression of two significant chemotactic cytokine genes (IL-8 and MIP-3A; Figures 2B,C). Moreover, both phages reduced the fever-producing (pyrogenic) cytokine IL-1β gene (IL-1B) expression (Figure 2D). IL-8, encoding for interleukin (IL)-8 or chemokine (C-X-C motif) ligand 8 (CXCL8), is the important neutrophil chemotactic factor (Modi et al., 1990). MIP-3A, encoding for macrophage inflammatory protein-3 alpha (MIP-3A), is a macrophage proinflammatory chemokine in human (Rossi et al., 1997). IL-1β is a member of the IL-1 family that play a critical role in inflammation (Smirnova et al., 2002). However, phage SE-W109 reduced only IL-1B significantly (p-value <0.01; Figure 2D). The combination of ST-W77 and SE-W109 showed similar results to the single ST-W77 treatment (decreased IL-8, MIP-3A, and IL-1B expression).
Moreover, we tested whether each phage alone would activate an innate immune response in T84 cells without the S. Tm infection. We added 10 μl of 1 × 1011 PFU/mL phages in SM buffer into T84 cells resuspended in 6-well plates (106 cells/well) without S. Tm challenge. Our data demonstrated that neither of the phages induce the expression of IL-8 and IL-1B (Figures 2B,D). Interestingly, both phages increased the expression of MIP-3A by about ten folds in T84 cells without S. Tm infection (Figure 2C). Taken together, these data suggested that ST-W77 reduced proinflammatory response better than that of SE-W109 in human colonic epithelium infected with S. Tm. Moreover, expression of MIP-3A but not the IL-8 and IL-1B, can increase in T84 cells after phage challenges without S. Tm infection. These suggested that phages ST-W77 and SE-W109 might some how activate expression of some human proinflammatory cytokines. However, the contamination of other bacterial components, especially a lipopolysaccharide (LPS or endotoxin), in a crude phage lysate might activate the host immune system (Boratynski and Szermer-Olearnik, 2017). Therefore, the removal of bacterial component contaminations are essential before phage applications.
Phage ST-W77 and SE-W109 persist in mouse gut but not a systemic site
To investigate the therapeutic effect of ST-W77 and SE-W109 in vivo, a streptomycin pre-treated mouse colitis model was used. The mice were orally infected with 108 CFU of S. Tm IR715 and either treated with phages or with SM buffer in a control group. Mouse feces were collected on days 1, 2, 3, and 4 p.i. to determine the amount of shedding Salmonella (CFU/gm). Both phages significantly reduced Salmonella numbers in mouse feces compared to untreated mice on days 1 and 4 but not on days 2 and 3 p.i. (Figure 3). On day 4 p.i., all mice were euthanized, and recovered phages from mouse tissues were enumerated by plating (Figure 4). Our data suggest that both phages predominantly persisted in the mouse colon content (between 106 to 1011 PFU/gm) and with the lower PFU (between 105 to 109 PFU/gm) in the mouse colon tissue. Phage numbers in the mouse tissues were similar for each of the two phages. No phage was detected from the mouse spleens in any of the groups. These data suggested that phages persist in the gut lumen and in colon tissue but not in systemic sites.
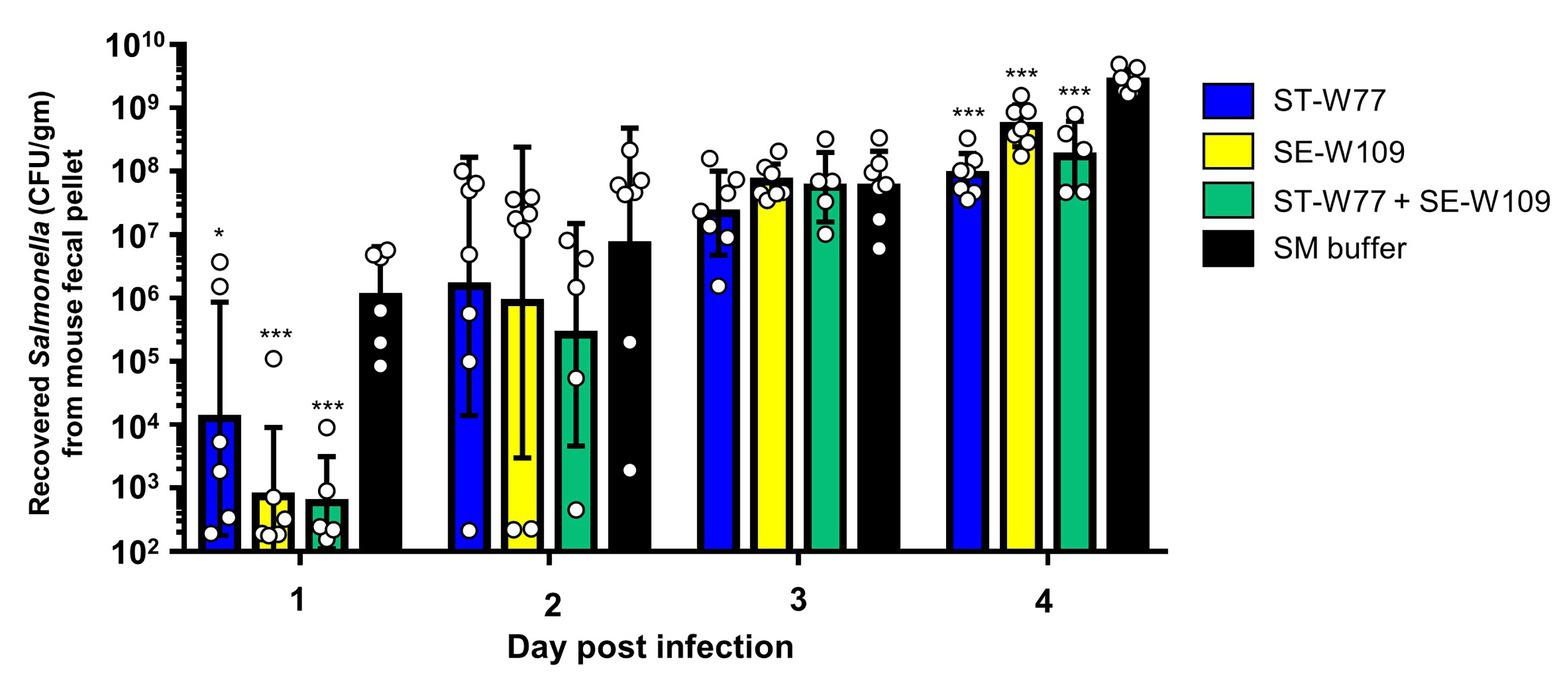
Figure 3. Oral feeding of phages (ST-W77 and SE-W109) reduced Salmonella shedding in mouse feces. The streptomycin-pretreated mouse colitis model of acute non-typhoidal salmonellosis was used. Orally treatment with phages (1010 PFU/mouse) was started 1 h after S. Tm infection and once daily for three consecutive days. On days 1, 2, 3, and 4 p.i., mouse fecal pellets from all groups, including ST-W77, SE-W109, and combination (ST-W77 + SE-W109), were collected for S. Tm CFU/gm. Bars represent the geometric mean; error bars indicate geometric standard deviation.* and *** indicated p < 0.01 and 0.001, respectively, compared to the SM buffer-treated group (black bar).
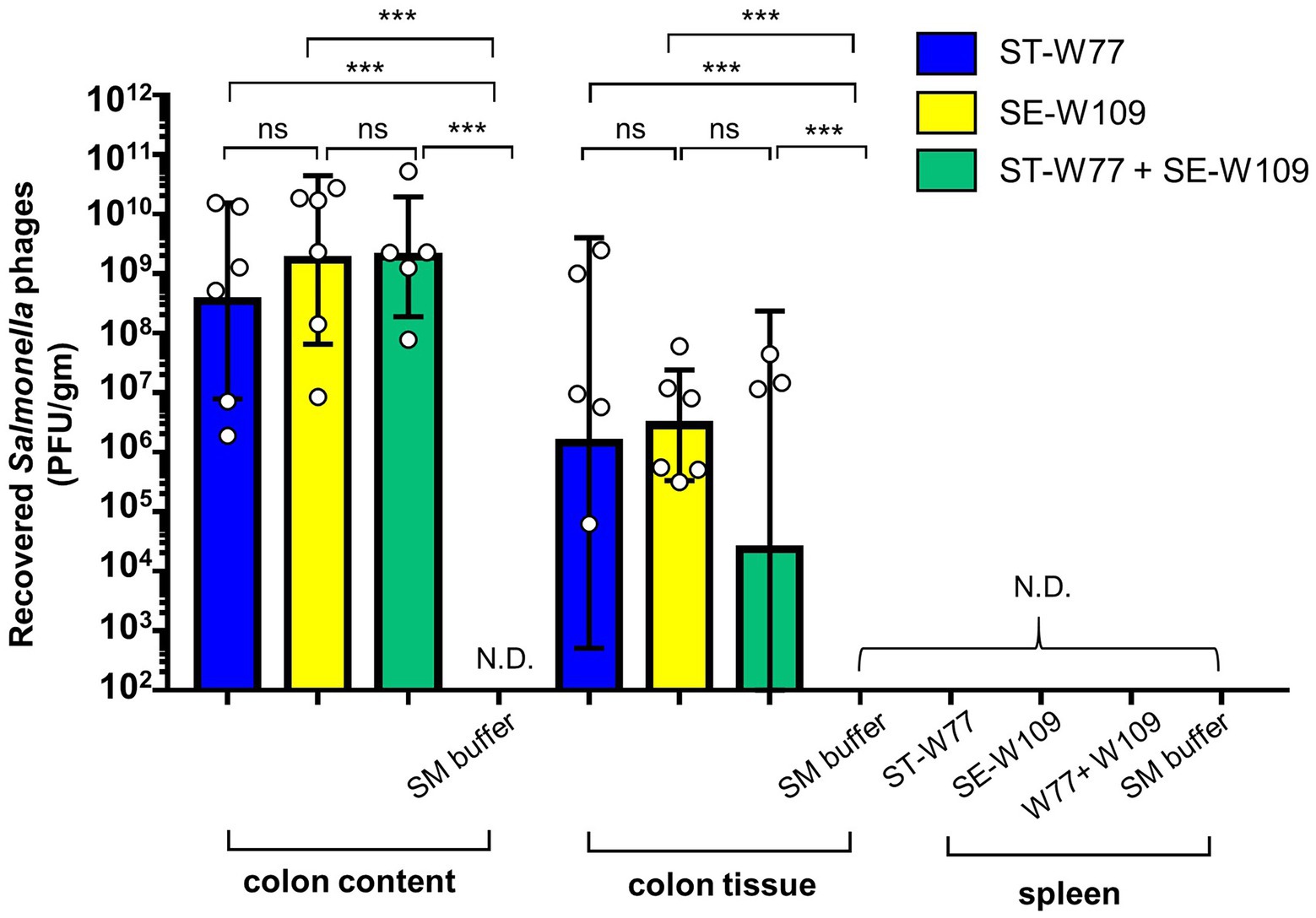
Figure 4. Phages ST-W77 and SE-W109 were recovered from mouse colon content, colon tissue but not spleen of Salmonella-infected mouse. All mice were orally fed with streptomycin sulfate 1 day prior to S. Tm infection (108 CFU/mouse). Orally treatment with phages (1010 PFU/mouse) was started 1 h after S. Tm infection and once daily for three consecutive days. On day 4 p.i., mouse tissues were harvested for enumerated phage in each tissue (PFU/gm) by a double-layer plaque assay. Bars represent the geometric mean; error bars indicate geometric standard deviation. *** indicated p < 0.001 and ns, a non-statistically significant difference; N.D., not detectable.
Oral treatment with phage ST-W77 and SE-W109 significantly reduced Salmonella proliferation and inflammation in the mouse
After 4 days of oral treatment of S. Tm infected mice with phages (1010 PFU/mouse once daily), mice were euthanized, and tissues were collected and homogenized; the presence of bacteria was then assessed by plating (Figures 5A–G). Our data clearly demonstrate that infected mice treated with either of the phages had significantly fewer S. Tm in their tissues compared to the control SM buffer-treated mice. However, the treatment with ST-W77 caused more significant reduction in S. Tm numbers than treatment with SE-W109 alone or ST-W77 combined with SE-W109, in the cecum but not the colon. Treatment with either of the phages markedly reduced S. Tm numbers in the ileum (about 3–4 logs; p-value <0.001; Figure 5E). The systemic disseminating population of S. Tm in the liver and spleen was also significantly reduced in the phage-treated mice (Figures 5F,G). Interestingly, treatment with SE-W109 caused a greater reduction of S. Tm in the liver than treatment with ST-W77.
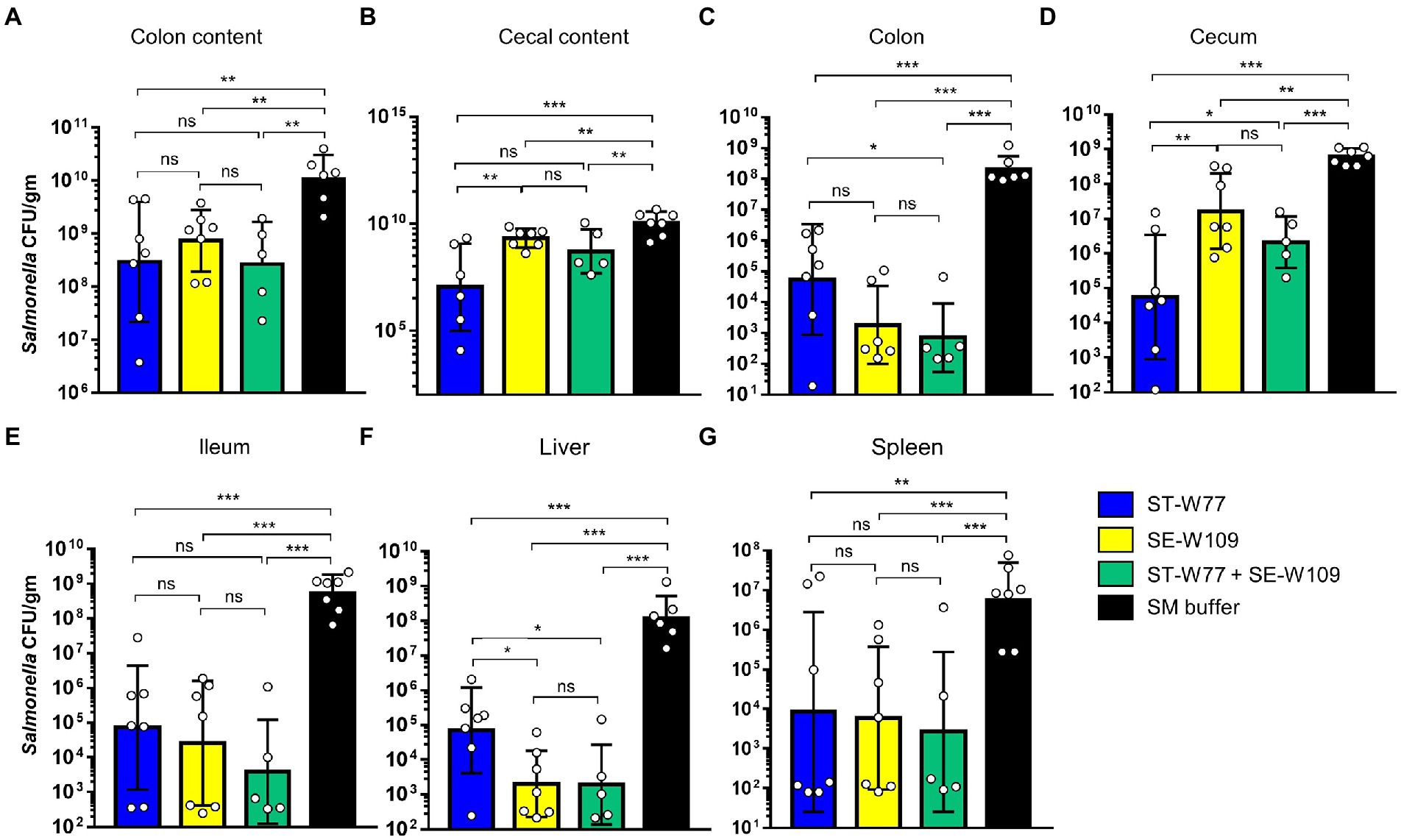
Figure 5. Both phages ST-W77 and SE-W109 significantly decreased S. Tm numbers in mouse gastrointestinal tract and systemic sites. All mice were orally fed with streptomycin sulfate 1 day prior to S. Tm infection (108 CFU/mouse) and treated with phages (1010 PFU/mouse) for three consecutive days. On day 4 p.i., all mice were euthanized tissues were collected to detect Salmonella numbers in each tissue (CFU/gm) by a plating method. Numbers of S. Tm in the gut lumen (A,B), local gut tissues (C-E), and systemic tissues (F,G) were investigated. Bars represent the geometric mean; error bars indicate geometric standard deviation.*, **, *** indicated p < 0.05, 0.01 and 0.001, respectively. ns, a non-statistically significant difference.
Next, we determined therapeutic effect of phages on the inflammatory response caused by S. Tm infection in mice using qPCR assay and cecal histopathological study. ST-W77, SE-W109, and their combination conferred anti-inflammatory effects in both colon (Figures 6A–C) and spleen (Figures 6D–F). Our qPCR results indicated that the phages attenuated mouse gut inflammatory response. The lower expression of the colonic proinflammatory cytokine, keratinocyte chemoattractant (Kc), and IL-17 encoded by Kc and IL-17 genes, respectively, were shown in phage-treated groups (Figures 6A,B) compared to the control group. In addition, increased colonic tight junction protein-1 or Zonula occludens-1 (Zo-1) expression encoded by the Zo-1 gene (Figure 6C) was observed in both phage-treated groups.
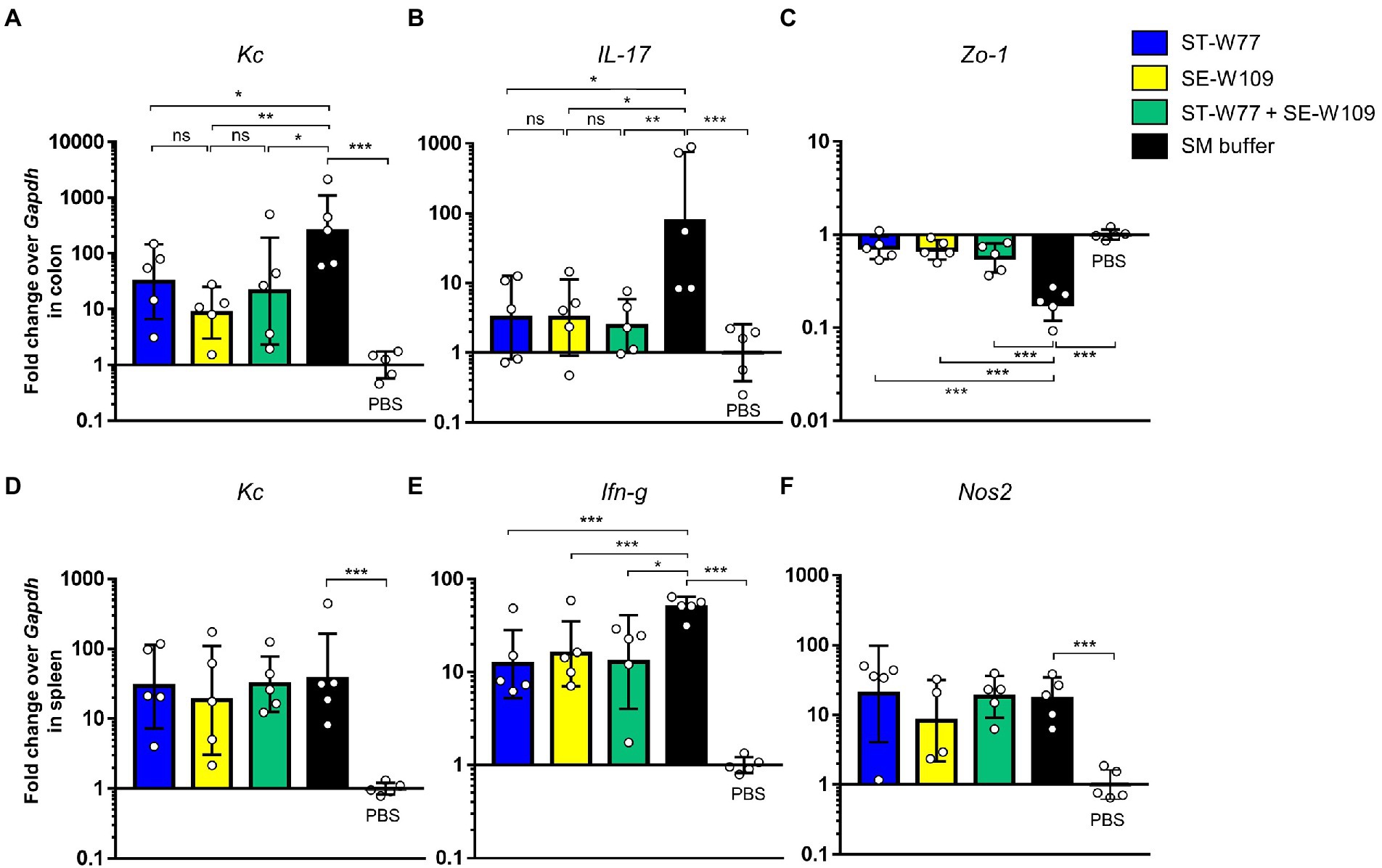
Figure 6. A reduction of proinflammatory makers in S. Tm infected mice orally treated with phages ST-W77 and SE-W109. Mouse tissues (colon and spleen) were collected for mRNA extraction and qPCR analysis. Colonic proinflammatory genes Kc (A), IL-17 (B), and tight junction Zo-1 (C) gene expressions were investigated. Expression of splenic proinflammatory genes Kc (D), Ifn-g (E), and Nos2 (F) was shown. Bars represent the geometric mean; error bars indicate geometric standard deviation. *, **, *** indicated p < 0.05, 0.01 and 0.001, respectively. ns, non-statistically significant difference; PBS, phosphate-buffered saline.
To evaluate the effect of phages in mouse systemic inflammation, expression of three proinflammatory genes (Kc, Ifn-g and Nos2) in the spleen were determined. In both phage-treated groups reduced expression only Ifn-g but not Kc and Nos2 (Figures 6D-F) was observed. Interferon (IFN)-γ, encoding by Ifn-g, is an important cytokine in a systemic phase of Salmonella infection (Santos, 2014). Expressions of Kc and Nos2 (encoding for an inducible nitric oxide synthase or iNOS) were not different in the spleens of mice in all groups. We also evaluated the change in histopathology of mouse ceca by staining with haematoxylin and eosin (H & E). Our data showed that all phage-treated mice had significantly lower cecal histopathological scores compared to untreated mice (Figure 7). These data indicated that phage ST-W77 and SE-W109 attenuated inflammatory response caused by S. Tm infection in mice.
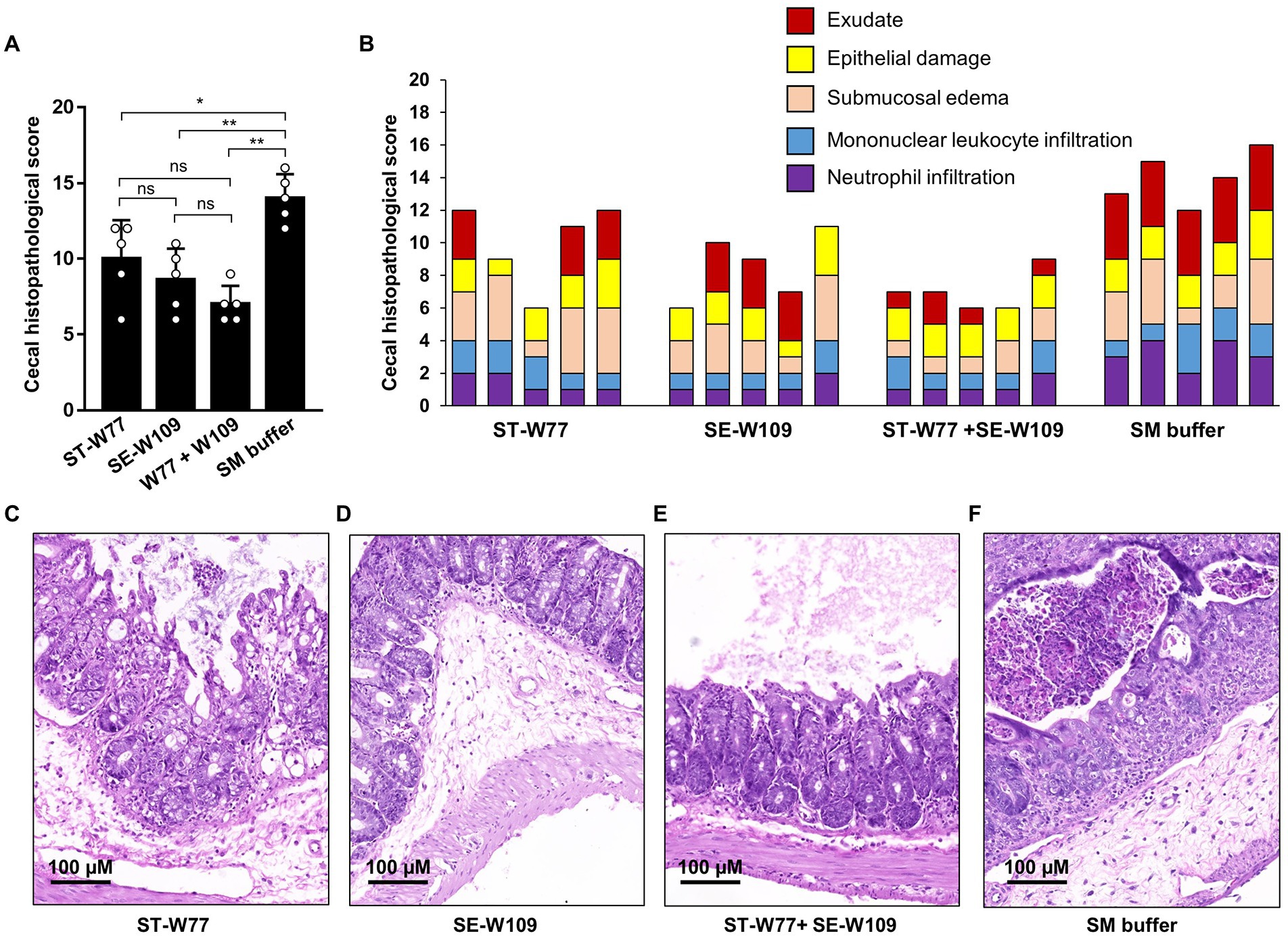
Figure 7. A significantly lower cecal histopathological score of S. Tm-infected mice orally treated with phages ST-W77 and SE-W109. Tips of mouse ceca were collected in 10% formalin and then hematoxylin–eosin (H&E) stained and double-blind scored by a veterinary pathologist using a histopathological score in Supplementary Table S3. The combined histopathological score (A) and the individual score with each criterion (B). The representative picture of mouse cecum treated with ST-W77 (C), SE-W109 (D), combined ST-W77 and SE-W109 (E), and SM buffer (F). Bars represent the mean; error bars indicate standard deviation. * and **indicated p < 0.05 and 0.01, respectively. ns, a non-statistically significant difference.
Discussion
S. Tm is one of the major Salmonella enterica serovars that cause an acute gastroenteritis or acute NTS in human and farm animals such as swine, cattle and poultry (Santos et al., 2003, 2009). Considerable health problems and economic losses can occur after S. Tm contamination in the food supply chain. In addition, the increasing rates of MDR S. Tm raised concerns about the treatment of iNTS. The anti-Salmonella effect of two recently isolated obligate lytic Salmonella phages: ST-W77 and SE-W109, has been demonstrated in the previous studies in vitro (Nale et al., 2020; Phothaworn et al., 2020). Furthermore, a phage cocktail composed of ST-W77, SE-W109, and SPFM17 reduced the mortality rate of Salmonella infection in a Galleria model (Nale et al., 2020). To provide further justification and evidence-based support for the therapeutic usage of the two phages, we investigated the anti-Salmonella effect of ST-W77 and SE-W109 in a streptomycin-pretreated mouse colitis model of an acute NTS.
Phages ST-W77 and SE-W109 were able to lyse S. Tm IR715 strain and reduced its invasiveness to human colonic epithelium cells (Figures 1, 2, respectively). Reduced numbers of invading S. Tm has a potential significant advantage in the disease management as diminishing the bacterial invasion rate in the human gut, which is critical for the treatment of S. Tm infection. Our data indicated that the treatment with ST-W77 significantly reduced expression of three important proinflammatory cytokine genes (IL-8, MIP-3A, and IL-1B) better than SE-W109. This observation may be linked to the originally-isolated strain differences with ST-W77 specifically targeting the Typhimurium serovar., while SE-W109, the serovar Enteritidis. We observed that ST-W77 and SE-W109 did not induce the expression of IL-8 and IL-1B but induced MIP-3A gene expression in the uninfected T84 cells (Figure 2C). Nonetheless, the observed activation of MIP-3A by ST-W77 and SE-W109 (about 10-fold increase) is still significantly lower than that in S. Tm-infected cells (about 100-fold increase). Another possibility of this observation might be explained by the contamination of bacterial components (e.g., DNA, protein or LPS) in the crude phage lysate (Boratynski and Szermer-Olearnik, 2017). In the absence of a Salmonella host, it is expected that phages should not activate a mammalian immune system. Nonetheless, other contributing factors such as microbial homeostasis, mucosal immunity, and immunological tolerance might play a role in phage-eukaryote interaction should be considered (Chatterjee and Duerkop, 2018). Taken together, our findings suggested that the purification of crude phage lysate before its application, especially for the phage therapy, to eliminate the contaminated bacterial components is essential. In addition, some phages may confer an immunomodulatory effect on mammalian cells even without their bacterial host and concurs with other reports (Gorski et al., 2012; Khatami et al., 2021; Safari et al., 2022).
Mammalian host immune response to both bacteria and phages could also determine the outcome of phage therapy (Hodyra-Stefaniak et al., 2015). In this study, both phages were not detected in the mouse spleen, indicating the gastrointestinal tract localized anti-Salmonella effect with possible low immunogenicity to mouse systemic immune response. To our knowledge, the effect of Salmonella lytic phage therapy on gut inflammation in mice is still poorly understood. Oral treatment with ST-W77 and SE-W109 reduced inflammation caused by S. Tm infection by reducing proinflammatory response in the colon and spleen. This anti-inflammatory effect of both phages could result from the fewer numbers of invading S. Tm compared to the untreated mice.
Phages ST-W77 and SE-W109 reduced the shedding amount of S. Tm in mouse feces on days 1 and 4 but not on days 2 and 3 p.i (Figure 3). On day 1 p.i., we found that both phages significantly reduce the proliferation of S. Tm. Later, S. Tm numbers were increased on days 2, 3, and 4 p.i. probably due to the change in gut micro-environment from the induced inflammation that enhances S. Tm growth (Lupp et al., 2007). In the mouse colitis model of NTS, the most prominent inflammation occurs on day 4 p.i. and we found that both phages were significantly reduced S. Tm at this time point. Both phages can persist well in the gut lumen and tissue, including the mucus layer but not in the spleen (Figure 4). A previous study showed that phage adherence to the mucus layer increases the protection against pathogenic bacteria by providing phages a medium for interacting with invading bacteria (Almeida et al., 2019). Nonetheless, in this study, we did not quantify mucus-specific phage numbers.
Recently, there are growing pieces of evidence that support the benefit of using a mouse model to evaluate the efficacy and safety of Salmonella phage in vivo (Lamy-Besnier et al., 2021; Kumar et al., 2022). Here, we also showed that a mouse colitis model for acute NTS could be used to determine the efficacy of phages in reducing Salmonella numbers and Salmonella-induced inflammation.
Conclusion
Therapeutic administration of Salmonella phages ST-W77 and SE-W109 can reduce S. Tm numbers and S. Tm-induced inflammation in both in vitro (human gut epithelium) and in vivo (mouse colitis model). ST-W77 and SE-W109 reduced proliferating and invading S. Tm subpopulations in a mammalian inflamed gut, resulting in decreased intestinal inflammation. A mouse model for acute human NTS should be a valuable tool to investigate the efficacy and safety of phage therapy in a pre-clinical setting.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee, Chiang Mai University, Thailand (Approval number 2563/MC-0002).
Author contributions
PT and SK generated the research and critically reviewed the final version of manuscript. PT, SK, and CS designed and conceptualized the research. CS, SB, AT, and PT performed the research. TK performed the histopathological study. CS, SB, AT, SK, and PT analyzed the data. CS and PT drafted the manuscript. PP, JN, AL-G, MA, MFA, DM, EG, and MC supervised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Biotechnology and Biological Sciences Research Council (BBSRC), grant number RM38G0140 awarded to MC, and the National Science and Technology Development Agency (NSTDA), grant number P-18-50454 awarded to SK, the Faculty of Medicine Research Fund (MIC-2561-05411), Chiang Mai University, Chiang Mai, Thailand and the Thailand Research Fund (RGNS 63-066) awarded to PT. CS was awarded by the Faculty of Medicine Chiang Mai University graduate student scholarship. AT was awarded by the Chiang Mai University (CMU) Presidential scholarship.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.955136/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | The schematic of phage treatment in S. Typhimurium-infected mouse model established in this study. After one-week acclimatization, 6–8 week-old female C57BL/6NJcl mice were orally fed with 100 μl of 200 mg/ml streptomycin sulfate 1 day before S. Typhimurium oral infection with 100 μl of 109 CFU/ml (108 CFU/mouse). Infected mice were divided into 4 groups (7 mice per group) and orally fed 100 μl of 1011 PFU/mL (1010 PFU/mouse) of ST-W77, SE-W109, ST-W77 combined with SE-W109, and SM buffer as the indicated time point. All mice were euthanized on day 4 post-infection.
SUPPLEMENTARY TABLE S1 | The list of bacterial and phage strains used in this study.
SUPPLEMENTARY TABLE S2 | The list of primers used in this study.
SUPPLEMENTARY TABLE S3 | The criteria for mouse cecal histopathological scoring used in this study (Buddhasiri et al., 2021).
References
Almeida, G. M. F., Laanto, E., Ashrafi, R., and Sundberg, L. R. (2019). Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. MBio 10, 1–12. doi: 10.1128/mBio.01984-19
Appiah, G. D., Mpimbaza, A., Lamorde, M., Freeman, M., Kajumbula, H., Salah, Z., et al. (2021). Salmonella bloodstream infections in hospitalized children with acute febrile illness-Uganda, 2016–2019. Am. J. Trop. Med. Hyg. 105, 37–46. doi: 10.4269/ajtmh.20-1453
Au, A., Lee, H., Ye, T., Dave, U., and Rahman, A. (2021). Bacteriophages: combating antimicrobial resistance in food-borne bacteria prevalent in agriculture. Microorganisms 10, 1–18. doi: 10.3390/microorganisms10010046
Bao, H., Zhang, H., Zhou, Y., Zhu, S., Pang, M., Zhang, X., et al. (2022). Dysbiosis and intestinal inflammation caused by Salmonella Typhimurium in mice can be alleviated by preadministration of a lytic phage. Microbiol. Res. 260:127020. doi: 10.1016/j.micres.2022.127020
Boratynski, J., and Szermer-Olearnik, B. (2017). Endotoxin removal from Escherichia coli bacterial lysate using a biphasic liquid system. Methods Mol. Biol. 1600, 107–112. doi: 10.1007/978-1-4939-6958-6_10
Buddhasiri, S., Sukjoi, C., Kaewsakhorn, T., Nambunmee, K., Nakphaichit, M., Nitisinprasert, S., et al. (2021). Anti-inflammatory effect of probiotic Limosilactobacillus reuteri KUB-AC5 against Salmonella infection in a mouse colitis model. Front. Microbiol. 12:716761. doi: 10.3389/fmicb.2021.716761
Chatterjee, A., and Duerkop, B. A. (2018). Beyond bacteria: bacteriophage-eukaryotic host interactions reveal emerging paradigms of health and disease. Front. Microbiol. 9:1394. doi: 10.3389/fmicb.2018.01394
Clokie, M. R., Millard, A. D., Letarov, A. V., and Heaphy, S. (2011). Phages in nature. Bacteriophage 1, 31–45. doi: 10.4161/bact.1.1.14942
Deriu, E., Liu, J. Z., Pezeshki, M., Edwards, R. A., Ochoa, R. J., Contreras, H., et al. (2013). Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37. doi: 10.1016/j.chom.2013.06.007
Fiorentin, L., Vieira, N. D., and Barioni, W. Jr. (2005). Oral treatment with bacteriophages reduces the concentration of Salmonella enteritidis PT4 in caecal contents of broilers. Avian Pathol. 34, 258–263. doi: 10.1080/01445340500112157
Gorski, A., Miedzybrodzki, R., Borysowski, J., Dabrowska, K., Wierzbicki, P., Ohams, M., et al. (2012). Phage as a modulator of immune responses: practical implications for phage therapy. Adv. Virus Res. 83, 41–71. doi: 10.1016/B978-0-12-394438-2.00002-5
Harada, L. K., Silva, E. C., Campos, W. F., Del Fiol, F. S., Vila, M., Dabrowska, K., et al. (2018). Biotechnological applications of bacteriophages: state of the art. Microbiol. Res. 212-213, 38–58. doi: 10.1016/j.micres.2018.04.007
Hengkrawit, K., and Tangjade, C. (2022). Prevalence and trends in antimicrobial susceptibility patterns of multi-drug-resistance non-typhoidal Salmonella in Central Thailand, 2012–2019. Infect. Drug Resist. 15, 1305–1315. doi: 10.2147/IDR.S355213
Hodyra-Stefaniak, K., Miernikiewicz, P., Drapala, J., Drab, M., Jonczyk-Matysiak, E., Lecion, D., et al. (2015). Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep. 5:14802. doi: 10.1038/srep14802
Inagaki-Ohara, K., Sawaguchi, A., Suganuma, T., Matsuzaki, G., and Nawa, Y. (2005). Intraepithelial lymphocytes express junctional molecules in murine small intestine. Biochem. Biophys. Res. Commun. 331, 977–983. doi: 10.1016/j.bbrc.2005.04.025
Jamal, M., Bukhari, S., Andleeb, S., Ali, M., Raza, S., Nawaz, M. A., et al. (2019). Bacteriophages: an overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 59, 123–133. doi: 10.1002/jobm.201800412
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y. A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Johanesen, P. A., and Dwinell, M. B. (2006). Flagellin-independent regulation of chemokine host defense in Campylobacter jejuni-infected intestinal epithelium. Infect. Immun. 74, 3437–3447. doi: 10.1128/IAI.01740-05
Jung, H. C., Eckmann, L., Yang, S. K., Panja, A., Fierer, J., Morzycka-Wroblewska, E., et al. (1995). A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J. Clin. Invest. 95, 55–65. doi: 10.1172/JCI117676
Khatami, A., Lin, R. C. Y., Petrovic-Fabijan, A., Alkalay-Oren, S., Almuzam, S., Britton, P. N., et al. (2021). Bacterial lysis, autophagy and innate immune responses during adjunctive phage therapy in a child. EMBO Mol. Med. 13:e13936. doi: 10.15252/emmm.202113936
Kumar, R. N., Surekha, M. V., Ramalingam, B., Kumar, P. U., Polasa, K., Hemalatha, R., et al. (2022). Oral toxicity study for Salmonella killing lytic bacteriophage NINP13076 in BALB/c mice and its effect on probiotic microbiota. Curr. Microbiol. 79:89. doi: 10.1007/s00284-021-02754-9
Lamy-Besnier, Q., Chaffringeon, L., Lourenco, M., Payne, R. B., Trinh, J. T., Schwartz, J. A., et al. (2021). Prophylactic administration of a bacteriophage cocktail is safe and effective in reducing Salmonella enterica serovar Typhimurium burden in vivo. Microbiol. Spectr. 9:e0049721. doi: 10.1128/Spectrum.00497-21
Little, J. S., Dedrick, R. M., Freeman, K. G., Cristinziano, M., Smith, B. E., Benson, C. A., et al. (2022). Bacteriophage treatment of disseminated cutaneous mycobacterium chelonae infection. Nat. Commun. 13:2313. doi: 10.1038/s41467-022-29689-4
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., et al. (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129. doi: 10.1016/j.chom.2007.06.010
Mather, A. E., Phuong, T. L. T., Gao, Y., Clare, S., Mukhopadhyay, S., Goulding, D. A., et al. (2018). New variant of multidrug-resistant Salmonella enterica serovar Typhimurium associated with invasive disease in immunocompromised patients in Vietnam. MBio 9, 1–11. doi: 10.1128/mBio.01056-18
Modi, W. S., Dean, M., Seuanez, H. N., Mukaida, N., Matsushima, K., and O'Brien, S. J. (1990). Monocyte-derived neutrophil chemotactic factor (MDNCF/IL-8) resides in a gene cluster along with several other members of the platelet factor 4 gene superfamily. Hum. Genet. 84, 185–187.
Morpeth, S. C., Ramadhani, H. O., and Crump, J. A. (2009). Invasive non-Typhi Salmonella disease in Africa. Clin. Infect. Dis. 49, 606–611. doi: 10.1086/603553
Nale, J. Y., Vinner, G. K., Lopez, V. C., Thanki, A. M., Phothaworn, P., Thiennimitr, P., et al. (2020). An optimized bacteriophage cocktail can effectively control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 11:609955. doi: 10.3389/fmicb.2020.609955
Nikkhahi, F., Soltan Dallal, M. M., Alimohammadi, M., Rahimi Foroushani, A., Rajabi, Z., Fardsanei, F., et al. (2017). Phage therapy: assessment of the efficacy of a bacteriophage isolated in the treatment of salmonellosis induced by Salmonella enteritidis in mice. Gastroenterol. Hepatol. Bed. Bench. 10, 131–136.
Patchanee, P., Boonkhot, P., Kittiwan, N., Tadee, P., and Chotinun, S. (2015). Dissemination of Salmonella enterica sequence types among asean economic community countries. Southeast Asian J. Trop. Med. Public Health 46, 707–719.
Patchanee, P., Eiamsam-Ang, T., Vanaseang, J., Boonkhot, P., and Tadee, P. (2017). Determination of regional relationships among Salmonella spp. isolated from retail pork circulating in the Chiang Mai municipality area using a WGS data approach. Int. J. Food Microbiol. 254, 18–24. doi: 10.1016/j.ijfoodmicro.2017.05.006
Phothaworn, P., Dunne, M., Supokaivanich, R., Ong, C., Lim, J., Taharnklaew, R., et al. (2019). Characterization of flagellotropic, chi-like Salmonella phages isolated from Thai poultry farms. Viruses 11, 1–18. doi: 10.3390/v11060520
Phothaworn, P., Supokaivanich, R., Lim, J., Klumpp, J., Imam, M., Kutter, E., et al. (2020). Development of a broad-spectrum Salmonella phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand. Food Microbiol. 92:103586. doi: 10.1016/j.fm.2020.103586
Raffatellu, M., Chessa, D., Wilson, R. P., Dusold, R., Rubino, S., and Baumler, A. J. (2005). The vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 73, 3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005
Rossi, D. L., Vicari, A. P., Franz-Bacon, K., McClanahan, T. K., and Zlotnik, A. (1997). Identification through bioinformatics of two new macrophage proinflammatory human chemokines: MIP-3alpha and MIP-3beta. J. Immunol. 158, 1033–1036.
Safari, Z., Sadeghizadeh, M., Asgaritarghi, G., Bardania, H., Sadeghizadeh, D., and Soudi, S. (2022). M13 phage coated surface elicits an anti-inflammatory response in BALB/c and C57BL/6 peritoneal macrophages. Int. Immunopharmacol. 107:108654. doi: 10.1016/j.intimp.2022.108654
Salmond, G. P., and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786. doi: 10.1038/nrmicro3564
Santos, R. L. (2014). Pathobiology of Salmonella, intestinal microbiota, and the host innate immune response. Front. Immunol. 5:252. doi: 10.3389/fimmu.2014.00252
Santos, R. L., Raffatellu, M., Bevins, C. L., Adams, L. G., Tukel, C., Tsolis, R. M., et al. (2009). Life in the inflamed intestine, Salmonella style. Trends Microbiol. 17, 498–506. doi: 10.1016/j.tim.2009.08.008
Santos, R. L., Tsolis, R. M., Baumler, A. J., and Adams, L. G. (2003). Pathogenesis of Salmonella-induced enteritis. Braz. J. Med. Biol. Res. 36, 03–12. doi: 10.1590/S0100-879X2003000100002
Santos, R. L., Zhang, S., Tsolis, R. M., Kingsley, R. A., Adams, L. G., and Baumler, A. J. (2001). Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3, 1335–1344. doi: 10.1016/S1286-4579(01)01495-2
Sarichai, P., Buddhasiri, S., Walters, G. E., Khantawa, B., Kaewsakhorn, T., Chantarasakha, K., et al. (2020). Pathogenicity of clinical Salmonella enterica serovar Typhimurium isolates from Thailand in a mouse colitis model. Microbiol. Immunol. 64, 679–693. doi: 10.1111/1348-0421.12837
Smirnova, M. G., Kiselev, S. L., Gnuchev, N. V., Birchall, J. P., and Pearson, J. P. (2002). Role of the pro-inflammatory cytokines tumor necrosis factor-alpha, interleukin-1 beta, interleukin-6 and interleukin-8 in the pathogenesis of the otitis media with effusion. Eur. Cytokine Netw. 13, 161–172.
Stojiljkovic, I., Baumler, A. J., and Heffron, F. (1995). Ethanolamine utilization in Salmonella Typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177, 1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995
Stylianou, E., Yndestad, A., Sikkeland, L. I., Bjerkeli, V., Damas, J. K., Haug, T., et al. (2002). Effects of interferon-alpha on gene expression of chemokines and members of the tumour necrosis factor superfamily in HIV-infected patients. Clin. Exp. Immunol. 130, 279–285. doi: 10.1046/j.1365-2249.2002.01980.x
Svircev, A., Roach, D., and Castle, A. (2018). Framing the future with bacteriophages in agriculture. Viruses 10, 1–13. doi: 10.3390/v10050218
Tang, F., Zhang, P., Zhang, Q., Xue, F., Ren, J., Sun, J., et al. (2019). Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb. Pathog. 126, 193–198. doi: 10.1016/j.micpath.2018.10.042
Thanki, A. M., Clavijo, V., Healy, K., Wilkinson, R. C., Sicheritz-Ponten, T., Millard, A. D., et al. (2022). Development of a phage cocktail to target Salmonella strains associated with swine. Pharmaceuticals (Basel) 15, 1–19. doi: 10.3390/ph15010058
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Tsolis, R. M., Xavier, M. N., Santos, R. L., and Baumler, A. J. (2011). How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79, 1806–1814. doi: 10.1128/IAI.01369-10
Weinbauer, M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181. doi: 10.1016/j.femsre.2003.08.001
Whistler, T., Sapchookul, P., McCormick, D. W., Sangwichian, O., Jorakate, P., Makprasert, S., et al. (2018). Epidemiology and antimicrobial resistance of invasive non-typhoidal salmonellosis in rural Thailand from 2006-2014. PLoS Negl. Trop. Dis. 12:e0006718. doi: 10.1371/journal.pntd.0006718
Winter, S. E., Thiennimitr, P., Nuccio, S. P., Haneda, T., Winter, M. G., Wilson, R. P., et al. (2009). Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infect. Immun. 77, 1904–1916. doi: 10.1128/IAI.01341-08
Keywords: bacteriophage therapy, Salmonella enterica Typhimurium, foodborne pathogen, acute non-typhoidal salmonellosis, mouse colitis model, inflammatory response
Citation: Sukjoi C, Buddhasiri S, Tantibhadrasapa A, Kaewsakhorn T, Phothaworn P, Nale JY, Lopez-Garcia AV, AbuOun M, Anjum MF, Malik DJ, Galyov EE, Clokie MRJ, Korbsrisate S and Thiennimitr P (2022) Therapeutic effects of oral administration of lytic Salmonella phages in a mouse model of non-typhoidal salmonellosis. Front. Microbiol. 13:955136. doi: 10.3389/fmicb.2022.955136
Edited by:
Sílvio B. Santos, University of Minho, PortugalReviewed by:
Shabarinath Srikumar, United Arab Emirates University, United Arab EmiratesYi Li, Yangzhou University, China
Copyright © 2022 Sukjoi, Buddhasiri, Tantibhadrasapa, Kaewsakhorn, Phothaworn, Nale, Lopez-Garcia, AbuOun, Anjum, Malik, Galyov, Clokie, Korbsrisate and Thiennimitr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunee Korbsrisate, c3VuZWUua29yQG1haGlkb2wuZWR1; Parameth Thiennimitr, cGFyYW1ldGgudEBjbXUuYWMudGg=, cGFyYW1ldGhAZ21haWwuY29t
†ORCID: Parameth Thiennimitr, https://orcid.org/0000-0002-6339-8744
 Chutikarn Sukjoi
Chutikarn Sukjoi Songphon Buddhasiri
Songphon Buddhasiri Arishabhas Tantibhadrasapa
Arishabhas Tantibhadrasapa Thattawan Kaewsakhorn
Thattawan Kaewsakhorn Preeda Phothaworn
Preeda Phothaworn Janet Y. Nale
Janet Y. Nale Angela V. Lopez-Garcia5
Angela V. Lopez-Garcia5 Manal AbuOun
Manal AbuOun Muna F. Anjum
Muna F. Anjum Danish J. Malik
Danish J. Malik Edouard E. Galyov
Edouard E. Galyov Martha R. J. Clokie
Martha R. J. Clokie Sunee Korbsrisate
Sunee Korbsrisate Parameth Thiennimitr
Parameth Thiennimitr